Summary
Background
Neurological damage caused by coronavirus disease 2019 (COVID-19) has attracted increasing attention. Recently, through autopsies of patients with COVID-19, the direct identification of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in their central nervous system (CNS) has been reported, indicating that SARS-CoV-2 might directly attack the CNS. The need to prevent COVID-19-induced severe injuries and potential sequelae is urgent, requiring the elucidation of large-scale molecular mechanisms in vivo.
Methods
In this study, we performed liquid chromatography-mass spectrometry-based proteomic and phosphoproteomic analyses of the cortex, hippocampus, thalamus, lungs, and kidneys of SARS-CoV-2-infected K18-hACE2 female mice. We then performed comprehensive bioinformatic analyses, including differential analyses, functional enrichment, and kinase prediction, to identify key molecules involved in COVID-19.
Findings
We found that the cortex had higher viral loads than did the lungs, and the kidneys did not have SARS-COV-2. After SARS-CoV-2 infection, RIG-I-associated virus recognition, antigen processing and presentation, and complement and coagulation cascades were activated to different degrees in all five organs, especially the lungs. The infected cortex exhibited disorders of multiple organelles and biological processes, including dysregulated spliceosome, ribosome, peroxisome, proteasome, endosome, and mitochondrial oxidative respiratory chain. The hippocampus and thalamus had fewer disorders than did the cortex; however, hyperphosphorylation of Mapt/Tau, which may contribute to neurodegenerative diseases, such as Alzheimer's disease, was found in all three brain regions. Moreover, SARS-CoV-2-induced elevation of human angiotensin-converting enzyme 2 (hACE2) was observed in the lungs and kidneys, but not in the three brain regions. Although the virus was not detected, the kidneys expressed high levels of hACE2 and exhibited obvious functional dysregulation after infection. This indicates that SARS-CoV-2 can cause tissue infections or damage via complicated routes. Thus, the treatment of COVID-19 requires a multipronged approach.
Interpretation
This study provides observations and in vivo datasets for COVID-19-associated proteomic and phosphoproteomic alterations in multiple organs, especially cerebral tissues, of K18-hACE2 mice. In mature drug databases, the differentially expressed proteins and predicted kinases in this study can be used as baits to identify candidate therapeutic drugs for COVID-19. This study can serve as a solid resource for the scientific community. The data in this manuscript will serve as a starting point for future research on COVID-19-associated encephalopathy.
Funding
This study was supported by grants from the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences, the National Natural Science Foundation of China, and the Natural Science Foundation of Beijing.
Keywords: SARS-CoV-2, K18-hACE2 mouse, Brain, Multiple organs, Label-free proteomics and phosphoproteomics
Research in context.
Evidence before this study
COVID-19-induced clinical symptoms, especially cerebral manifestations, continue to be of concern. Perhaps owing to technical limitations, early autopsy studies usually reported the absence of or a very low SARS-CoV-2 load in the brains of patients with COVID-19. However, the direct identification of SARS-CoV-2 in the brains of patients with COVID-19 during autopsies has recently been reported. SARS-CoV-2 may persist in neurons for several months after the onset of symptoms. To date, COVID-19-associated brain research has mainly focused on whether and how SARS-CoV-2 infects the brain. Considering that COVID-19-induced cerebral symptoms have been confirmed, we believe that proteomics- and phosphoproteomics-derived molecular atlases of cerebral regions after SARS-CoV-2 infection are necessary. The lung is the main target of SARS-CoV-2, and a low estimated glomerular filtration rate (eGFR) without acute kidney injury has been observed in people who recovered from COVID-19. Therefore, we performed proteomic and phosphoproteomic analyses of the cortex, hippocampus, thalamus, lung, and kidney of K18-hACE2 mice, which have not been reported to date.
Added value of this study
This study demonstrated the commonness and individuality of functional alterations in the cortex, hippocampus, thalamus, lung, and kidney of SARS-CoV-2-infected K18-hACE2 mice. After SARS-CoV-2 infection, RIG-I-associated virus recognition, antigen processing and presentation, and complement and coagulation cascades were activated to different degrees in all five organs, especially the lungs. Notably, SARS-CoV-2 was related to the hyperphosphorylation of multiple phosphosites in Mapt/Tau, which may contribute to neurodegenerative diseases such as Alzheimer's disease. This study demonstrated the complexity of SARS-CoV-2 tissue infection and injury using comprehensive molecular maps. This study provided high-quality proteomic and phosphoproteomic datasets for SARS-CoV-2-infected cerebral regions.
Implications of all the available evidence
This study provides high-quality proteomic and phosphoproteomic datasets for further SARS-CoV-2-associated studies. The differentially expressed proteins and predicted kinases in this study can be used as baits to identify candidate therapeutic drugs for COVID-19 in mature drug databases. The data in this manuscript will serve as a starting point for future research on COVID-19-associated encephalopathy.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndromes coronavirus 2 (SARS-CoV-2) has been ongoing for over 3 years since December 2019. Multiple organ injuries have been observed in patients with COVID-19 and those in long-term recovery,1, 2, 3 and potential neurological damage has not been overlooked.4,5 Anosmia, the loss of smell, is a common symptom among patients with COVID-19.6 Other neuropathogeneses and neurologic manifestations, including ageusia, headaches, strokes, impairment of consciousness, seizures, and encephalopathy, have also been reported.4 SARS-CoV-2 RNA and the neuronal injury markers neurofilament light and Tau have been detected in the cerebrospinal fluid (CSF) of some patients with COVID-19.7 Zhang et al.8 have summarised COVID-19-induced brain histopathological damages, such as neurophagocytosis, microglia nodules, satellite phenomena, extensive oedemas, focal haemorrhages, infarctions, and mononuclear cell infiltration. Some studies do not consider neuronal viral load to be the direct cause of COVID-19-associated neurological symptoms.9, 10, 11, 12, 13 However, evidence of the direct identification of SARS-CoV-2 in the central nervous system (CNS) has been increasing. Song et al.14 detected SARS-CoV-2 in cortical neurons in autopsied patients who died of COVID-19. Through autopsies of patients with COVID-19, Stein et al.15 showed not only that SARS-CoV-2 was distributed and had replicated in the brain at the early stage of infection, but also that it could persist there for several months following symptom onset. SARS-CoV-2 directly targets neurons in rhesus monkeys, the neuro-derived SK-N-SH cell line, the glial-derived U251 cell line,16 and Ramani's three-dimensional human brain organoids.17 SARS-CoV-2 has been detected in multiple brain regions in K18-hACE2 mice.18 Using hamsters19,20 and human angiotensin-converting enzyme 2 (hACE2) transgenic mice,18,21,22 studies have confirmed that damage to the nervous system was present during or even after SARS-CoV-2 infection. Thus, COVID-19-induced neurological injury and potential sequelae must be prevented.
The lack of large-scale molecular mechanisms makes it difficult to prevent COVID-19-associated acute or chronic injuries or potential sequelae, including encephalopathy. Proteomic analyses help to simultaneously evaluate alterations in thousands of proteins or post-translational modification (PTM) sites and offer an opportunity for global molecular profiling during physiological or pathophysiological processes. Phosphorylation is a noteworthy PTM because it is a quick and flexible way for organisms to respond to physiological or pathological changes.23 COVID-19-associated proteomics studies have used autopsies of end-stage patients,24, 25, 26, 27 human sera,28, 29, 30, 31 CSF,7,32 tissues from model animals,33 or intact SARS-CoV-2 viruses.34 However, proteomic and phosphoproteomic data from SARS-CoV-2-infected cerebral tissues in vivo have not yet been reported.
In this study, we performed proteomic and phosphoproteomic analyses of the cortex, hippocampus, thalamus, lung, and kidney of COVID-19-infected K18-hACE2 mice, providing cerebral proteomic datasets related to COVID-19 in vivo. This study was designed to record COVID-19-associated molecular alterations in multiple organs, especially the cerebral regions. We chose the lung and kidney for proteomic detection because the lung is the main target of SARS-CoV-2, and a low estimated glomerular filtration rate (eGFR) without acute kidney injury has been observed during COVID-19 recovery, which has attracted attention.1,35,36
The results revealed that K18-hACE2 mice had higher viral loads in the cortex than in the lungs. Consistent with the results of previous reports,37 we observed that SARS-CoV-2 induced minimal abnormalities in the renal tubular lumen; however, we found no viral RNA or protein in the kidney. After SARS-CoV-2 infection, the cortex presented with extensive protein expression disorders, including dysregulated spliceosome, ribosome, peroxisome, proteasome, endosome, and mitochondrial oxidative respiratory chain. The numbers of protein disorders in the hippocampus and thalamus were lower than those in the cortex. However, hyperphosphorylation of Mapt/Tau was observed in all three cerebral regions after infection. The Retinoic acid-inducible gene I (RIG-I) signalling pathway, antigen processing and presentation, and complement and coagulation cascades were activated to different degrees in each organ, and were most obvious in the lung. The kidney showed strong hACE2 expression and antigen presentation; however, the virus was not detected. Activated lysosomes, dysregulated metabolism, and extracellular matrix accumulation were more conspicuous in the kidney than in the other organs. In summary, SARS-CoV-2-induced protein changes differed in each organ. We observed different proteomic reactions in the cortex, hippocampus, thalamus, lung, and kidney of SARS-CoV-2-infected K18-hACE2 mice. This study provides both in vivo observations and datasets for COVID-19 mechanisms and therapeutic research.
Methods
Ethics and biosafety statement
Murine studies were performed in an animal biosafety level 3 (ABSL3) facility using high-efficiency particulate air-filtered isolators. All animal procedures were reviewed and approved by the Institutional Animal Care and Use Committee of the Institute of Laboratory Animal Science, Peking Union Medical College (LJN22005 and LJN22009, ILAS, PUMC).
Animal experimental procedures
Female specific-pathogen-free (SPF) transgenic (K18-hACE2) mice provided by GemPharmatech Co., Ltd. were used in this study. SARS-CoV-2 (SARS-CoV-2/WH-09/human/2020/CHN; GenBank: MT093631.2) was provided by ILAS (PUMC, China). Eight 10-week-old K18-hACE2 mice were equally divided into virus-infected (n = 4) and control (n = 4) groups. Mice in the virus-infected group were challenged with SARS-CoV-2 at a dose of 1 × 105 TCID50 by intranasal inoculation, and mice in the control group were inoculated with an equal volume of phosphate-buffered saline (PBS). Mice were dissected at 5 dpi to collect the cortex, hippocampus, thalamus, lung, and kidney to screen for virus loads, histopathological changes, and proteomic and phosphoproteomic profiling.
In wild-type mice, ACE2 is located on the X chromosome; therefore, human ACE2 was inserted into the X chromosome of hACE2 mice. Male hACE2 mice were mated with female wild-type mice to rapidly produce hACE2 mice. Only female mice harbour human ACE2 on the X chromosome; therefore, we infected female hACE2 mice.
Pathological examination and immunohistochemistry staining
The paraffin sections (3–4 μm in thickness) of organs were stained with H&E and immunohistochemistry (IHC), and observed under light microscopy as described previously. Antibody (GTX635654, rabbit monoclonal antibody, GeneTex) for SARS-CoV-2 Spike S1 protein was used.38,39
RNA extraction and quantitative PCR with reverse transcription (RT–qPCR)
The total RNA was extracted and reverse transcription was performed as described previously.40 RT–qPCR reactions were performed using the PowerUp SYBG Green Master Mix Kit (Applied Biosystems, A25741). Samples were processed using the following cycling protocol and primers: 50 °C for 2 min, 95 °C for 2 min, then followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s, and final incubations at 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 45 s. The primer sequence of SARS-CoV-2 based on the envelope (E) gene was as follows: forward primer, 5′-TCGTTTCGGAAGAGACAGGT-3′; reverse primer, 5′-GCGCAGTAAGGATGGCTAGT-3′.
Sample preparation and LC-MS/MS analyses
Sample preparation and LC-MS/MS analyses were performed as previously reported.41 Briefly, the dissected tissues were added with lysis buffer (1% SDS, 1% protease inhibitor, and 1% phosphatase inhibitor), homogenized, and heated at >56 °C for 30 min in the ABL3 facility. The biosafety compliant samples were stored at −80 °C until use. Approximately 150 mg of tissue per sample was used for the protein extraction. After removing the debris by centrifugation at 12,000 × g at 4 °C for 10 min, the extracted protein supernatant was collected for protein concentration detection and trypsin (Promega, SRT200601) digestion (1:50 trypsin-to-protein mass ratio for the first digestion overnight and 1:100 trypsin-to-protein mass ratio for a second 4 h-digestion). The collected peptides were desalted and prepared for subsequent high-performance liquid chromatography (HPLC) fractionation.
An Agilent 300 Extend C18 column (5 μm particles, 4.6 mm ID, and 250 mm length) was used for high pH reverse-phase HPLC to fractionate peptides. Peptides for proteomic analysis were separated into 10 fractions/sample and dried by vacuum centrifugation. Peptides for phosphoproteomic analysis were incubated with the Fe-IMAC microsphere suspension to enrich phosphopeptides. After removing the non-specifically absorbed peptides, the Fe-IMAC microspheres were washed with elution buffer. The eluted supernatant containing enriched phosphopeptides was collected, separated into fractions (two fractions/sample for the cortex, hippocampus, lung, and kidney; one fraction/sample for the thalamus), and lyophilized.
An Orbitrap Exploris™ 480 mass spectrometer (Thermo Fisher Scientific) equipped with a FAIMS device and an EASY-nLC 1200 UPLC system was used for LC-MS analysis. Tandem mass spectrometry and data-dependent analysis (DDA) strategies were used to identify and quantify the peptides, proteins, and phosphosites.
Statistics
A database search was conducted as previously reported.41 Briefly, mass spectrometry (MS) raw files were uploaded to MaxQuant42 version 1.6.15.0, and the data were compared with those of the SwissProt mouse FASTA database downloaded on 14/12/2020. Basic data rearrangements, including normalisation and imputation, were processed using Perseus43 version 1.6.5.0 as previously reported.41,44 For quality control of the LC-MS data, we constructed homemade pipelines. For proteomics, we assessed the peptide-spectrum match (PSM), peptide length, spectral count per peptide, Andromeda score, and mass error. For phosphoproteomics, we assessed the PSM, peptide length, spectral count per phosphorylated peptide, Andromeda score, mass error, count of phosphosite per phosphorylated peptide, and localization probability of phosphosite.44 Using our homemade quality control pipelines, all samples passed quality control and were retained for subsequent bioinformatics analyses.
Differential expression analysis was performed using Student's t test, and the cut-off was set to P < 0.05. The fold change (FC) of each molecule was calculated as (mean value of the virus-infected group)/(mean value of the control group) for each organ. Upregulated molecules were defined as t test P value < 0.05 and FC > 1.5. Downregulated molecules were defined as t test P value < 0.05 and FC < (1/1.5). Upregulated and downregulated molecules were combined as differentially expressed molecules in each organ.41
The KEGG database45 was used for functional annotation and pathway enrichment analysis (Fisher's exact test; Benjamini and Hochberg method for FDR correction). Pathways with P value < 0.05 (Fisher's exact test) were defined as enriched. The proteins in the enriched pathways were submitted to the STRING database (https://string-db.org/) for protein–protein interaction (PPI) analysis. PPI networks were presented using Cytoscape software.46
Kinase prediction was based on kinase-substrate relationships recorded by iGPS1.0.47 The iGPS software utilized the theory that short linear motifs around phosphosites are specifically recognized by specific kinases. The threshold parameter was set to ‘medium’. The interactions parameter was set to ‘Exp./String’ to filter potentially false-positive hits. Kinase activity prediction was based on gene set enrichment analysis (GSEA) algorithm48 because the intensity of phosphosites could reflect the activity of corresponding kinases. The .rnk file was set to the log-transformed FC of phosphosites, and the .gmt file was set to the corresponding kinase-substrate relationships. The generated normalized enrichment score (NES) of the enrichment results was regarded as the kinase activity score. The cut-off of the predicted activated kinases was set to NES >0, and the cut-off of the inhibited kinases was set to NES <0. Kinases with significantly altered activity were set to have a cut-off of GSEA P value < 0.05 and NES >1/<−1. The kinome tree was modified from Cell Signaling Technology Inc. (www.cellsignal.com) and annotated using the Kinome Render.49
Viral-host protein interactions were predicted as previously reported41 using the SARS-CoV-2 protein interaction map provided by Gordon et al.50
Principal component analysis (PCA), fuzzy C-means clustering,51 heat maps, line charts, bar plots, circos plots, volcano plots, and Venn diagrams are generated by R.
Role of funders
The funders were not involved in study design, data collection, data analyses, interpretation, or writing of this study.
Results
Global proteomic and phosphoproteomic profiling of the cortex, hippocampus, thalamus, lung, and kidney in control and SARS-CoV-2 infected K18-hACE2 mice
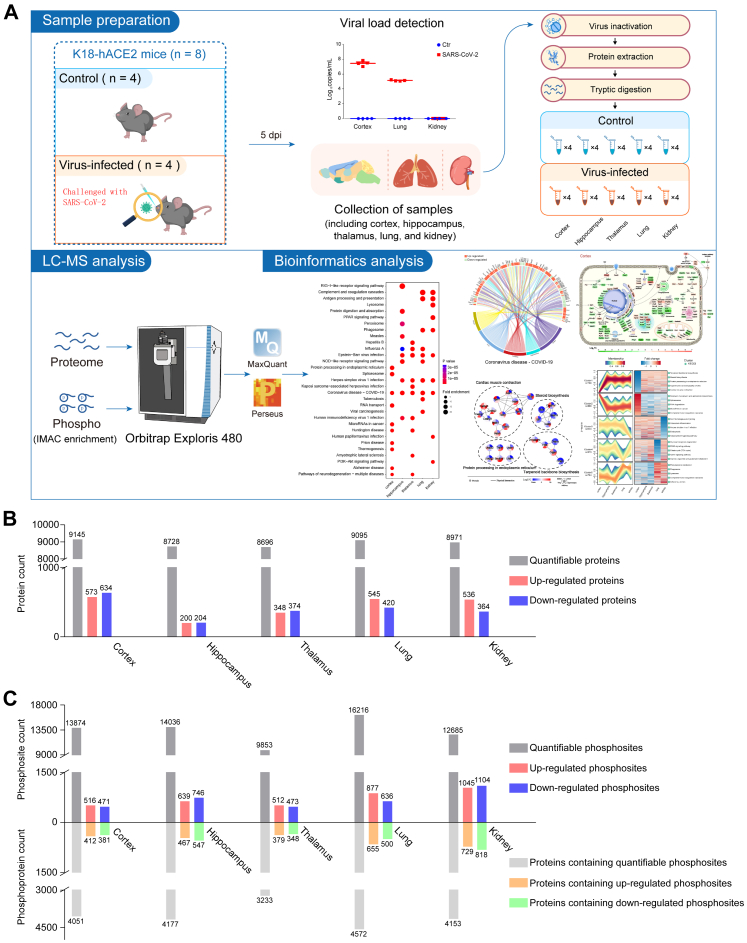
Eight 10-week-old female K18-hACE2 mice were equally divided into control (n = 4) and virus-infected (n = 4) groups. Mice in the virus-infected group were challenged with 1 × 105 median tissue culture infective dose (TCID50) of SARS-CoV-2 by intranasal inoculation, and mice in the control group were treated with equal volumes of PBS. Five days post infection (dpi), all mice were euthanized, and their cortex, hippocampus, thalamus, lung, and kidney were harvested for viral load detection, morphological analysis, and LC-MS-based proteome and phosphoproteome analysis (Fig. 1A, Supplementary Table S1).
Fig. 1.
Workflow and global proteomic and phosphoproteomic profiling of the cortex, hippocampus, thalamus, lung, and kidney in SARS-CoV-2 infected K18-hACE2 mice. A. Brief workflow of the study. Eight 10-week-old female K18-hACE2 mice were divided equally into control and virus-infected groups. Mice in the virus-infected group were infected with 1 × 105 TCID50 SARS-CoV-2 by intranasal inoculation and the control mice were treated with equal volumes of PBS. On the fifth day post infection (dpi), all the eight mice were euthanized. The cortex, hippocampus, thalamus, lung, and kidney were extracted for viral load detection, morphological analyses, protein extraction, and LC-MS/MS detection. After MaxQuant-based database search and Perseus-based data rearrangement, proteomic and phosphoproteomic data were processed into bioinformatics analyses and data mining. B and C. Each organ in the virus-infected group was compared with the corresponding organ in the control group. Quantifiable and differentially expressed proteins in each organ are presented in figure B. Quantifiable and differentially expressed phosphosites and their corresponding proteins are presented in figure C.
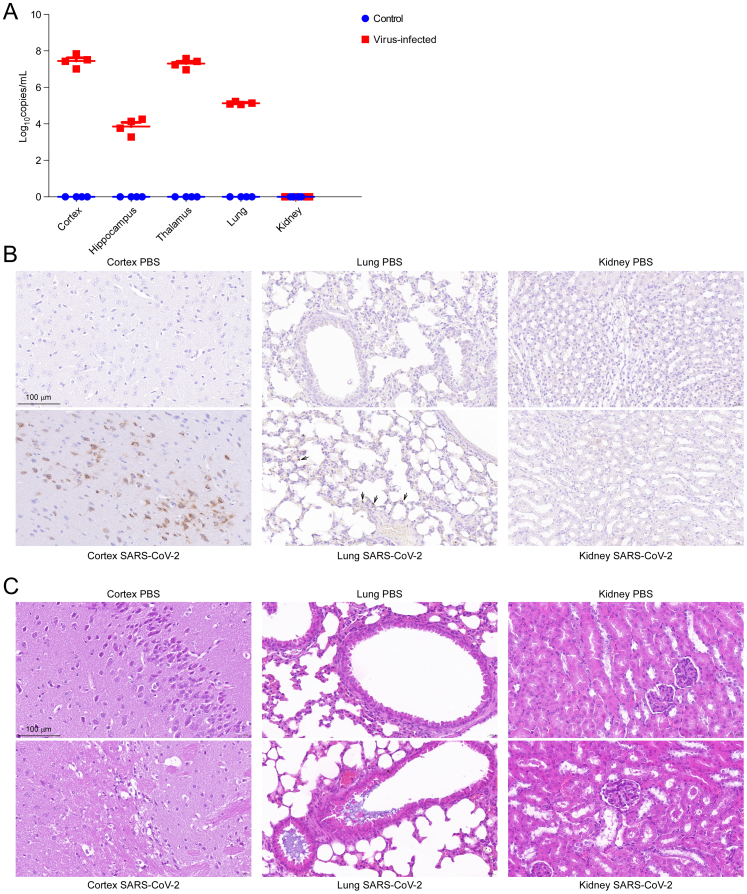
Viral nucleic acid load detection (Fig. S1A) and IHC staining of SARS-CoV-2 Spike S1 protein (Fig. S1B) showed that the cortex had higher SARS-CoV-2 viral loads than the lung in the infected mice. After SARS-Cov-2 infection, the K18-hACE2 mice showed marked widening of the alveolar septa, degeneration of the alveolar epithelial cells, mild exudation, interstitial congestion and hemorrhage, and basophilic exudation in the bronchi. Beneath the cortical areas, there are scattered vacuolated degenerating neurons with swollen cytoplasm and cerebral vasculitis. In the cortex layer, we observed vasculitis, infiltration of inflammatory cell, and denatured and necrotic cells (Fig. S1C). Although SARS-CoV-2 viral proteins were not detected in kidneys (Fig. S1A and B), the infected kidney showed pathological manifestations; for example, the eosinophilic protein-like tubular structure in the renal tubular lumen was increased (Fig. S1C).
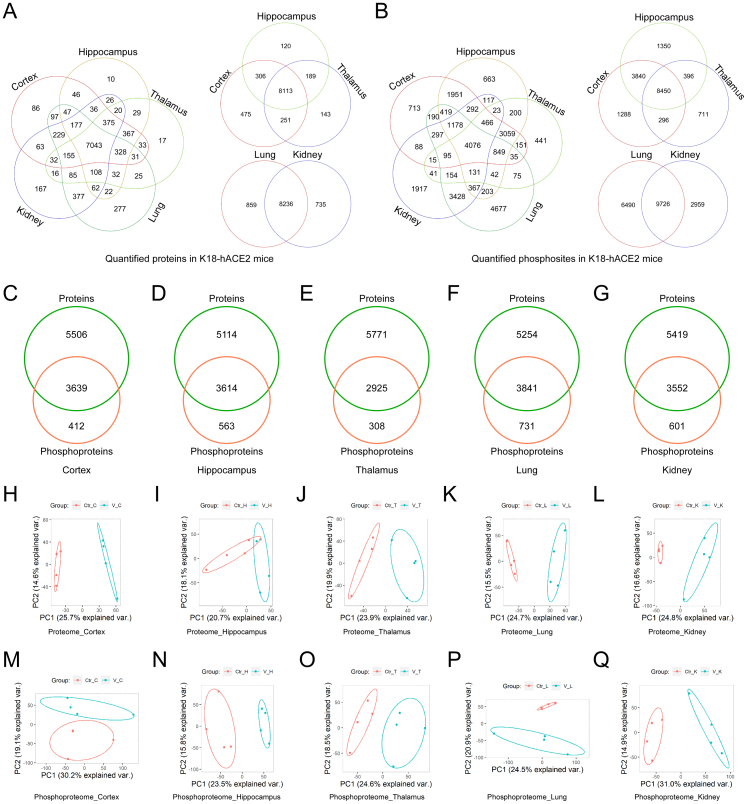
For basic bioinformatic analysis, each organ of the virus-infected group was compared to the corresponding organ in the control group. Student's t test was used to perform differential analysis. The number of quantifiable proteins in the proteome of the cortex, hippocampus, thalamus, lung, and kidney, was 9145 (573 up- and 634 downregulated), 8728 (200 up- and 204 downregulated), 8696 (348 up- and 374 downregulated), 9095 (545 up- and 420 downregulated), and 8971 (536 up- and 364 downregulated), respectively (Fig. 1B and Supplementary Tables S2 and S4); the number of quantifiable phosphosites and phosphoproteins in the phosphoproteome was 13,874 (516 up- and 471 downregulated) sites in 4051 proteins, 14,036 (639 up- and 746 downregulated) sites in 4177 proteins, 9853 (512 up- and 473 downregulated) sites in 3233 proteins, 16,216 (877 up- and 636 downregulated) sites in 4572 proteins, and 12,685 (1045 up- and 1104 downregulated) sites in 4153 proteins (Fig. 1C and Supplementary Tables S3 and S4).
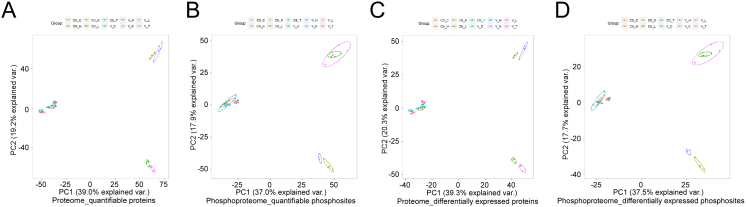
Venn diagram briefly presents commonness and individuality of elements among several datasets. In this study, Venn diagrams showed that 7043 proteins and 4076 phosphosites were shared by the cortex, hippocampus, thalamus, lungs, and kidneys. However, each organ had its unique proteins and phosphosites, which means that SARS-CoV-2-induced biological responses are likely to be different in different organs (Fig. S2A and B). The proportion of post-translationally modified peptides in all peptides was too low to be detected by conventional proteomics method. Fe-IMAC microspheres were used to obtain highly concentrated phosphorylated peptides for phosphosites identification. The enrichment of phosphopeptides by Fe-IMAC led to the discovery of some proteins that were not detected in the proteomic datasets (Fig. S2C–G). Principal component analysis (PCA) is an unsupervised clustering method that evaluates the overall similarity among samples. PCA of quantifiable proteins (Fig. S2H–L) and phosphosites (Fig. S2M–Q) confirmed that SARS-CoV-2 infection induced holistic molecular alterations in the cortex, hippocampus, thalamus, lungs, and kidneys of K18-hACE2 mice.
Commonness and individuality of functional alterations in the cortex, hippocampus, thalamus, lung, and kidney of K18-hACE2 mice after SARS-CoV-2 infection
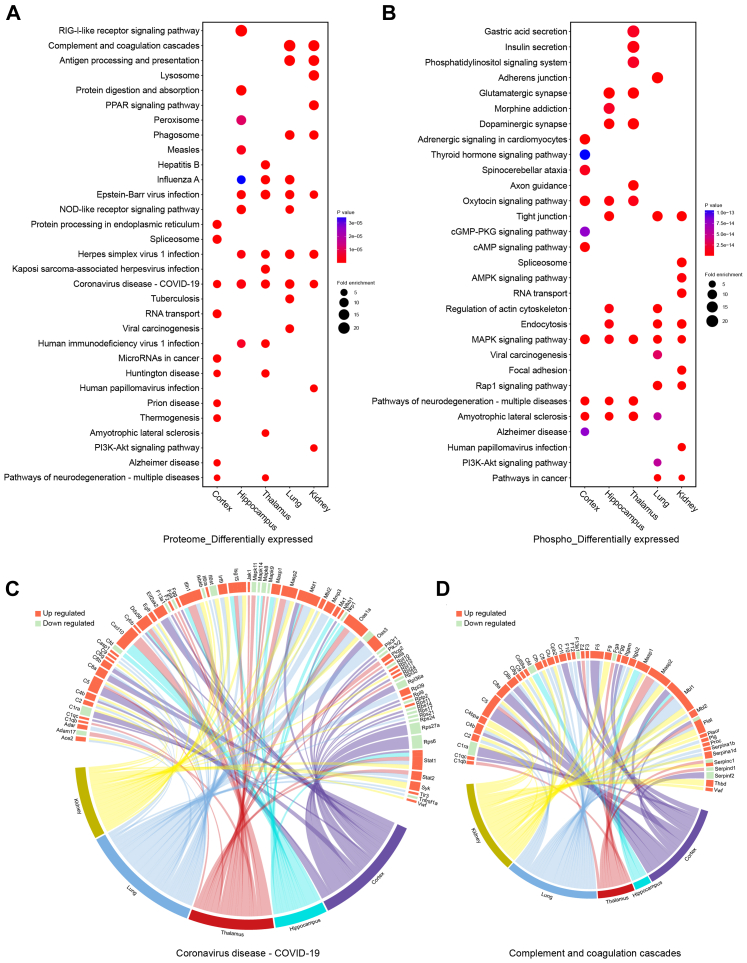
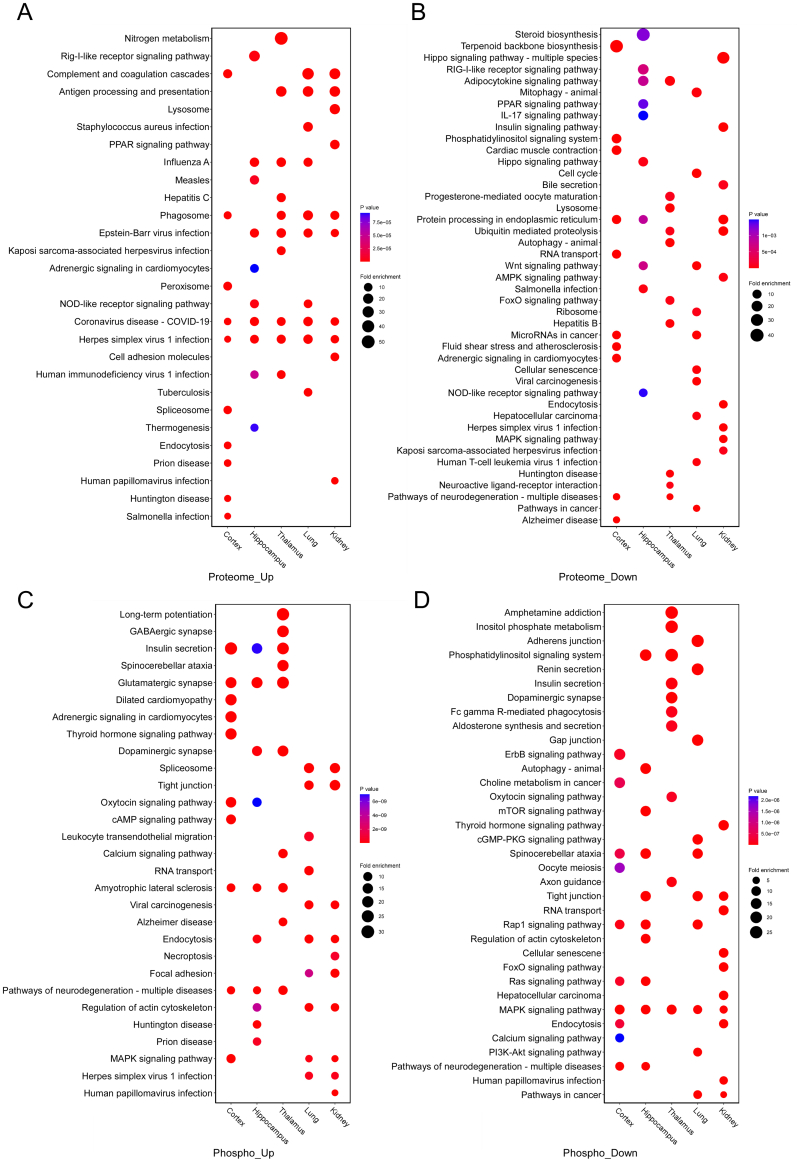
Differential analysis in the previous step defined the protein or phosphosite with a Student's t test P value <0.05, and FC >1.5 or <1/1.5 in one organ as differentially expressed after infection. Differentially expressed, upregulated, or downregulated proteins, and proteins containing differentially expressed, upregulated, or downregulated phosphosites in each organ were separately enriched for analysis of KEGG pathways (Fig. 2A and B, Fig. S3A–D, Supplementary Tables S5 and S6). The 10 pathways with the highest enrichment levels (the lowest P value of Fisher's exact test) in each organ were selected and integrated into bubble plots (Fig. 2A and B and Fig. S3A–D). We then observed commonness and individuality pathways in the cortex, hippocampus, thalamus, lungs, and kidneys of K18-hACE2 mice after SARS-CoV-2 infection.
Fig. 2.
Functional analyses for differentially expressed proteins in the proteome and proteins containing differentially expressed phosphosites in the phosphoproteome. KEGG pathways enriched for the cortex, hippocampus, thalamus, lung, and kidney are integrated into bubble plots. A. Functional enrichment analyses for differentially expressed proteins in the five organs. Fold change (FC) and P value of Student's t test were separately calculated in each organ. That is, FCprotein = (mean value of four infected samples)/(mean value of corresponding four control samples) and each organ had differentially expressed proteins independent of other organs. Differentially expressed proteins contained both up- and downregulated proteins. Upregulated proteins were defined as FCprotein >1.5 and P < 0.05 (Student's t test, n = 4). Downregulated proteins were defined as FCprotein <1/1.5 and P < 0.05 (Student's t test, n = 4). The top 10 KEGG pathways enriched in each organ were integrated and presented in the bubble plots. B. Functional enrichment analyses for proteins containing differentially expressed phosphosites in the five organs. FC and P value of Student's t test were separately calculated in each organ. FCphosphosite = (mean value of four infected samples)/(mean value of corresponding four control samples) and each organ had differentially expressed phosphosites independent of other organs. Differentially expressed phosphosites contained both up- and downregulated phosphosites. Upregulated phosphosites were defined as FCphosphosite >1.5 and P < 0.05 (Student's t test, n = 4). Downregulated phosphosites were defined as FCphosphosite <1/1.5 and P < 0.05 (Student's t test, n = 4). Proteins containing differentially expressed, upregulated, or downregulated phosphosites were used for functional enrichment analysis. The top 10 KEGG pathways enriched in each organ are integrated and presented in the bubble plots. C. Circos plot showing the correspondence between organs and differentially expressed proteins in a KEGG pathway. Differentially expressed proteins belonging to the COVID-19 pathway in each organ were presented in the plot. Red segments show upregulated proteins and green segments show downregulated proteins. The length of each segment corresponds to the sum of |log2 FC| of each protein. D. Circos plot showing the correspondence between organs and differentially expressed proteins in a KEGG pathway. Differentially expressed proteins belonging to complement and coagulation cascades in each organ are presented in the plot. Red segments show upregulated proteins and green segments show downregulated proteins. The length of each segment corresponds to the sum of |log2 FC| of each protein.
From the proteomic data, we found that differentially expressed proteins in all five organs were enriched in the KEGG-annotated COVID-19 pathway (Fig. 2A). Compared with the three brain regions, the peripheral lung and kidney showed relatively apparent antigen processing and presentation, complement and coagulation cascades, and phagosome processes (Fig. 2A). Upregulated proteins in the five organs were enriched in multiple pathways of virus infection (Fig. S3A). However, the downregulated proteins were enriched in different pathways in each organ (Fig. S3B).
From the phosphoproteomic data, proteins containing differentially expressed phosphosites in all five organs were enriched in the MAPK signalling pathway. The oxytocin signalling pathway and pathways of neurodegeneration were consistently enriched for proteins containing differentially expressed phosphosites in the cortex, hippocampus, and thalamus, but not in the lungs and kidneys (Fig. 2B). The spliceosome, tight junction, viral carcinogenesis, and focal adhesion showed consistent enrichment of proteins containing upregulated phosphosites in the lung and kidney, but not in the three brain regions (Fig. S3C). Except for the consistency of the MAPK signalling pathway, proteins containing downregulated phosphosites were enriched in different pathways (Fig. S3D).
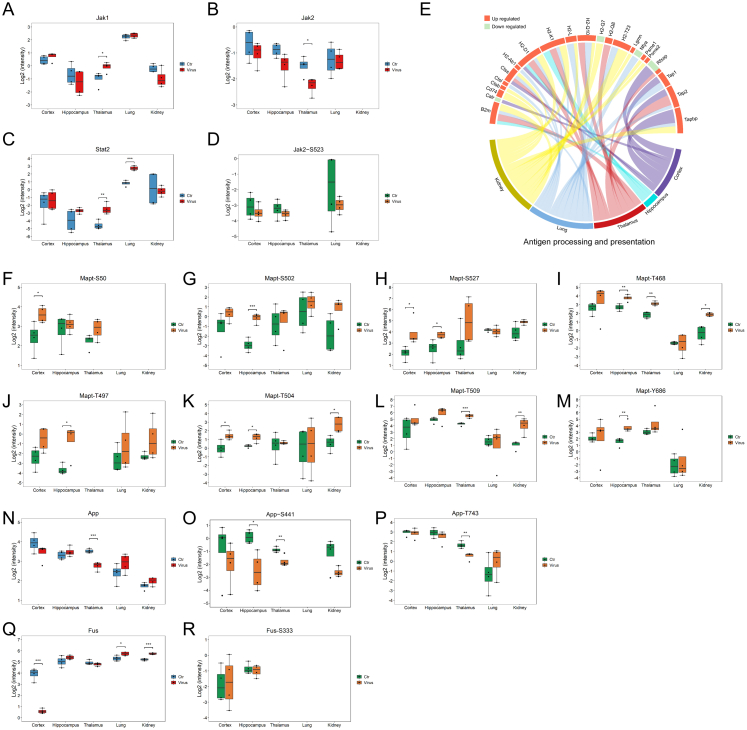
To obtain concrete and detailed functional alterations, we extracted molecules from specific enriched pathways. For example, the COVID-19 pathway, which was highly enriched in the proteome, showed that proteins in the complement and coagulation cascades (such as C2, C4b, Vwf, etc.), Mapk signalling pathway (such as Mapk8, Mapk9, Mapk11, and Mapk14), Jak-Stat signalling pathway (such as Jak1 and Stat2), and ribosome (such as Rpl10, Rps11, etc.) were differentially regulated in the five organs (Fig. 2C). Ddx58 in the RIG-I signalling pathway was upregulated in all five organs; however, Eif2ak2, Ifih1, Isg15, Nfkb1, and some other proteins in the RIG-I pathway had different expression patterns in each organ (Fig. 2C). Specific observations of complement and coagulation cascades revealed that the lungs and kidneys had a greater number of upregulated complements (upregulated C2, C4b, C4bp, C8g, C9, Cfh, Mbl2, etc.) and coagulation-related proteins (upregulated F3, F10, F12, Plg, Vwf, etc.) than the three brain regions (downregulated Cfd, F13a1, Fga, Plat, Serpinf2, etc.; Fig. 2D). This indicated that complement and coagulation cascades were strongly activated in the lungs and kidneys, but were weakly activated or dysregulated in the three brain regions after infection, which confirmed that SARS-CoV-2-induced proteomic responses were different in different organs.
FC and fuzzy C-means-based observation of the functional heterogeneity in the five organs in K18-hACE2 mice after SARS-CoV-2 infection
The above analyses revealed that each organ had distinct molecular and functional alterations in the infected K18-hACE2 mice. We then extracted intersecting quantifiable or differentially expressed proteins/phosphosites and performed PCA to identify the heterogeneity of SARS-CoV-2-induced differences among the organs (Fig. S4A–D). PCA results showed that the three brain regions gathered closely in both principal component 1 (PC1) and principal component 2 (PC2). The lung and kidney were highly consistent in PC1 and dispersed in PC2 (Fig. S4A–D).
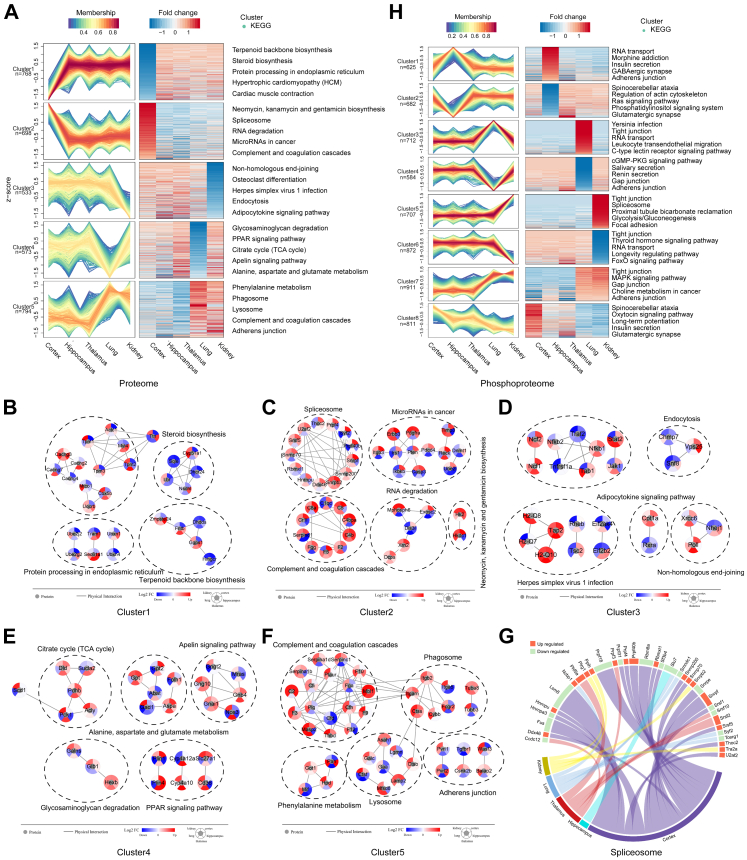
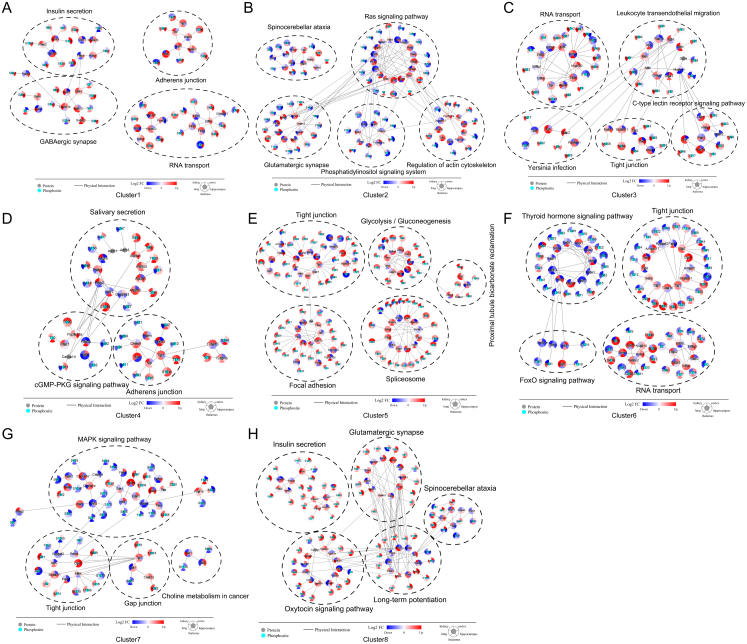
To determine organ specificity after SARS-CoV-2 infection, we collected differentially expressed proteins from each organ and converted them into a union set. Fuzzy C-means clustering algorithm is one of the typical clustering algorithms in data mining.51 We then used the fuzzy C-means algorithm and separated the FC in the five organs into five clusters (Fig. 3A and Supplementary Table S7). STRING database-based protein–protein interaction (PPI) analysis for the top 5 pathways showed proteins with specific expression patterns in each cluster (Fig. 3B–F).
Fig. 3.
FC of differentially expressed proteins or phosphosites were separated by fuzzy C-means and each resulting cluster showed specific characteristics of organs. A. Differentially expressed proteins in each organ were integrated into a union set. Based on protein FC in each organ, fuzzy C-means separated differentially expressed proteins into five clusters. KEGG functional enrichment analysis was performed for proteins in each cluster. B. PPI network of proteins in the enriched top5 KEGG pathways in cluster1 of A. The red to blue colour gradient is the log2 FC of corresponding proteins in each organ. C. PPI network of proteins in the enriched top5 KEGG pathways in cluster2 of A. The red to blue colour gradient is the log2 FC of corresponding proteins in each organ. D. Circos plot that presents the correspondence between organs and differentially expressed proteins in a KEGG pathway. Differentially expressed proteins belonging to the spliceosome in each organ are presented in the plot. Red segments show upregulated proteins and green segments show downregulated proteins. The length of each segment corresponds to the sum of |log2 FC| of each protein. E. PPI network of proteins in the enriched top5 KEGG pathways in cluster3 of A. The red to blue coluor gradient is the log2 FC of corresponding proteins in each organ. F. PPI network of proteins in the enriched top 5 KEGG pathways in cluster4 of A. The red to blue colour gradient is the log2 FC of corresponding proteins in each organ. G. PPI network of proteins in the enriched top5 KEGG pathways in cluster5 of A. The red to blue colour gradient is the log2 FC of corresponding proteins in each organ. H. Differentially expressed phosphosites in each organ were integrated into a union set. Based on phosphosite FC in each organ, fuzzy C-means separated differentially expressed phosphosites into 8 clusters. KEGG functional enrichment analysis was performed for proteins containing phosphosites in each cluster.
Cluster1 and cluster2 showed that the cortex had distinct proteomic characteristics, including small molecular metabolism, protein processing in the endoplasmic reticulum, spliceosome, and RNA degradation (Fig. 3A). In the cortex, proteins associated with steroid biosynthesis (Cyp51a1, Dhcr24, Lbr, Nsdhl, and Sqle), terpenoid backbone biosynthesis (Dhdds, Fntb, Ggps1, Hmgcr, and Zmpste24), calcium transport (Cacng2, Cacng3, Cacng4, and Cacng7), cytoskeleton (Tpm1, Tpm2, Tpm3, Myl4, etc.), and protein processing and proteases (Sec61a1, Tram1, Ube2g2, Ube2j2, Ubxn1, and Ubxn4) were downregulated after SARS-CoV-2 infection in the five organs (Fig. 3B). RNA-associated proteins, such as those related to the spliceosome (Srsf5, Snrpf, Snrnp70, etc.) and RNA degradation (Dis3l, Dcps, Exosc2, Mphosph6, and Xrn2), were upregulated in the cortex (Fig. 3C). Considering the obviously dysregulated spliceosome identified in the cortex (Fig. 2A), we drew a circus plot focusing on differentially expressed proteins belonging to the spliceosome and found that the spliceosome in the cortex exhibited the highest number of disorders of the five organs after infection (Fig. 3G). Although there were a limited number of differentially expressed proteins in the complement and coagulation cascades, the cortex had some highly expressed complements (C4b, C4bpa, C5, C8a, etc.) and coagulation factors (F2, F5, Fgg, etc.; Fig. 3C). Cluster3 revealed kidney-specific molecular characteristics such as non-homologous end-joining, endocytosis, and adipocytokine signalling pathways (Fig. 3A). In the infected kidney, we found downregulation of Nfkb2 in the NF-κB signalling pathway, Jak1 and Stat2 in the Jak-Stat signalling pathway, Tsc2 and Rheb in the mTor signalling pathway, and Nhej1 in the non-homologous end-joining process (Fig. 3D). Cluster4 revealed lung-specific metabolic characteristics, such as glycosaminoglycan degradation, PPAR signalling pathway, and citrate cycle (Fig. 3A). Multiple metabolic proteins were downregulated in the infected lungs (Fig. 3E). Cluster5 showed differences between the three brain regions and the two peripheral organs, such as phagosomes, lysosomes, complement and coagulation cascades, and adherens junction (Fig. 3A). Of the proteins, Ctsb and Ctss were highly expressed in the infected lung and kidney; these proteins which are reported to assist SARS-CoV-2 with entry into cells and to promote inflammatory injury (Fig. 3F).24
The FC of differentially expressed phosphosites in the five organs was separated into eight clusters (Fig. 3H and Supplementary Table S8). We observed hippocampus-specific (cluster1 and cluster2), lung-specific (cluster3 and cluster4), and kidney-specific (cluster5 and cluster6) molecular and functional characteristics. In the infected hippocampus, we found hyperphosphorylated proteins associated with adherens junctions, GABAergic synapse, and RNA transport (Fig. S5A), and hypophosphorylated proteins associated with glutamatergic synapse, phosphatidylinositol signalling, Ras signalling, and actin cytoskeleton regulation (Fig. S5B). The infected lung showed hyperphosphorylated proteins associated with RNA transport, leukocyte transendothelial migration, tight junctions, and C-type lectin receptor signalling pathway (Fig. S5C), and hypophosphorylated proteins associated with secretion and cGMP-PKG signalling pathway (Fig. S5D). The infected kidney showed hyperphosphorylated proteins associated with focal adhesion, glycolysis/gluconeogenesis, the spliceosome, and proximal tubule bicarbonate reclamation (Fig. S5E), and hypophosphorylated proteins associated with FoxO signalling pathway and RNA transport (Fig. S5F). Cluster7 distinguished the three brain regions and two peripheral organs. The MAPK signalling pathway, tight junctions, and choline metabolism-associated proteins tended to be hyperphosphorylated in the lung and kidney but hypophosphorylated in the three brain regions after SARS-CoV-2 infection (Fig. S5G). Cluster8 showed a phosphorylation tendency that was different in each organ, involving proteins in glutamatergic synapses, the oxytocin signalling pathway, and long-term potentiation (Fig. 3H and Fig. S5H). The frequently enriched tight junction and RNA transport processes in the phosphoproteome suggest that phosphorylation is regulated during SARS-CoV-2 infection. Thus, we performed kinase prediction to identify the regular pattern of SARS-CoV-2 virus-associated phosphorylation.
Kinase prediction collated and extended phosphorylation patterns of each organ in K18-hACE2 mice
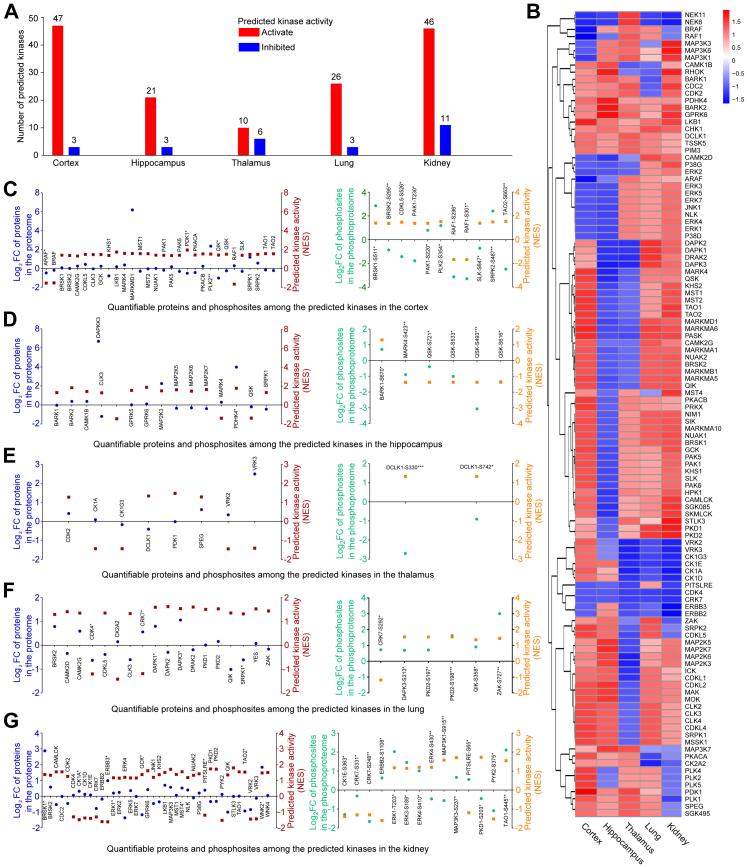
At the molecular level, phosphosites show extremely complicated expression patterns. To extend our knowledge of SARS-CoV-2-induced molecular alterations in vivo and identify a regular pattern, we applied kinase prediction to phosphoproteome data.
Quantifiable phosphosites were submitted to iGPS47 to obtain phosphosite-kinase relationships. Kinase activity was predicted using the GSEA algorithm.48 In total, 254 kinases were predicted in at least one organ. Kinases filtered by GSEA P < 0.05 and NES >1 (activated) or <−1 (inhibited) were defined as having significantly altered activity. We found that the number of activated kinases (cortex, 47; hippocampus, 21; thalamus, 10; lung, 26; kidney, 46) was more than that of inhibited kinases (cortex, 3; hippocampus, 3; thalamus, 6; lung, 3; kidney, 11) in all five organs after SARS-CoV-2 infection (Fig. 4A and Supplementary Table S9). Kinases predicted to have significantly altered activity in each organ were clustered into a union of 120 kinases. After excluding missing values, the NES values of 113 kinases were plotted using a heat map to compare the kinase activity patterns in the cortex, hippocampus, thalamus, lung, and kidney. We found that no kinases showed significantly altered activity in all five organs, revealing that the kinase activity pattern was unique in each organ after infection (Fig. 4B). For preliminary verification, we matched the predicted kinases with quantifiable proteins and phosphosites. Of the union of the 120 kinases, 87 kinases (cortex, 30; hippocampus, 15; thalamus, 8; lung, 18; kidney, 40) in the proteome and 85 kinases containing 405 phosphosites (cortex, 126 sites in 28 proteins; hippocampus, 33 sites in 11 proteins; thalamus, 15 sites in 3 proteins; lung, 82 sites in 17 proteins; kidney, 86 sites in 26 proteins) in the phosphoprotoeme were quantifiable in at least one of the five organs. The FC and NES of all quantifiable proteins and differentially expressed phosphosites in the group of 120 predicted kinases are presented by dot plots (Fig. 4C–G). We found that the trends of NES and FC were volatile, which means that the predicted activated kinases might have upregulated or downregulated proteins/phosphosites and vice versa. Then, we determined kinases whose NES and FC had the same trends, especially kinases corresponding to proteins/phosphosites with Student's t test P value < 0.05, in each organ. In the cortex, PLK2, BRSK1-S511, PAK1-S220, PLK2-S354, and SRPK2-S487 were upregulated, whereas ARAF, RAF1-S296, and RAF1-S301 were downregulated (Fig. 4C). The hippocampus showed upregulated PDHK4 and BARK1-S670 and downregulated MARK4-S423, QSK-S271, QSK-S533, QSK-S493, and QSK-S616 (Fig. 4D). Very few kinases were predicted to be present in the thalamus (Fig. 4E). Upregulated DAPK1, DAPK3, DAPK3-S313, PKD2-S197, PKD2-S198, QIK-S358, and ZAK-S727, and downregulated CDK4 were found in the lungs (Fig. 4F). Kinases found in the kidney included upregulated BRSK1, ERK1, MST4, WNK2, ERK1-T203, ERK3-S189, ERK4-S410, MAP3K3-S237, and TAO1-S445, and downregulated CK1E-S363, CRK7-S331, CRK7-S248, and PYK2-S375 (Fig. 4G). The functions of these kinases associated with synaptic plasticity and memory regulation in the central nervous system, and inflammation in peripheral organs should be further discussed.
Fig. 4.
Kinase prediction and kinase activity prediction based on the phosphoproteome. A. Number of predicted kinases in each organ after SARS-CoV-2 infection. Based on quantification of phosphosites, kinase prediction was performed using the iGPS algorithm and kinase activity prediction was performed using the GSEA algorithm. NES values derived from the GSEA algorithm reflect predicted kinase activity. The threshold of predicted activated kinases is set at NES >0. The threshold of predicated inhibited kinases is set at NES <0. B. Heat map of predicted kinases that may have had significantly altered activity after SARS-CoV-2 infection. Kinase activity is represented by NES values. C–G. Comparison of the predicted activity of kinases and actual measured FC of proteins and phosphosites in the cortex (C), hippocampus (D), thalamus (E), lung (F), and kidney (G) after SARS-CoV-2 infection (n = 4, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (Student's t test)). The predicted kinases were matched to quantifiable proteins or phosphosites in each organ. The left panel shows quantification of alterations of proteins in the proteome and the corresponding kinase activity. The right panel shows quantification of alterations of phosphosites in the phosphoproteome and the corresponding kinase activity. Red and orange squares represent log2 FC of protein or phosphosites, respectively. Blue and green points represent the NES of the kinases.
SARS-CoV-2 viral-host protein interactions in the cortex, hippocampus, thalamus, lung, and kidney of K18-hACE2 mice
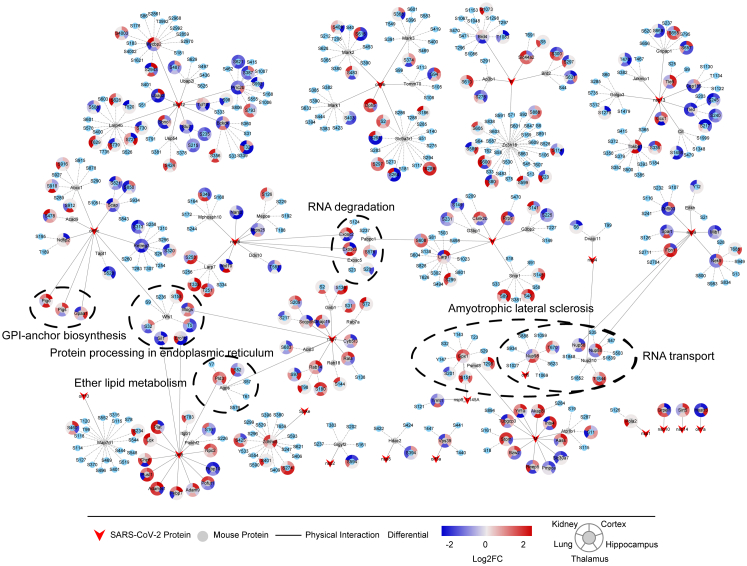
As previously reported,41 we predicted viral-host protein interactions in K18-hACE2 mice based on the SARS-CoV-2 protein interaction map by Gordon et al.50 We found that 23 SARS-CoV-2 proteins potentially interacted with 117 host proteins containing 402 phosphosites (Fig. 5 and Supplementary Table S10). To identify the potential functions and mechanisms of the proteins that interacted with SARS-CoV-2 proteins, we matched the 117 proteins to KEGG pathway maps. Of the annotated 22 KEGG pathways containing 34 proteins, six pathways containing 19 proteins were significantly enriched (Fig. 5). Pigo, Pigs, and Gpaa1 interacting with viral orf9c were associated with GPI-anchor biosynthesis. Sil1 and Ero1b interacting with viral orf8, Mogs interacting with viral nsp7, and Wfs1 (containing phosphosites T5, S32, and S158) interacting with orf9c were associated with protein processing in endoplasmic reticulum. Pld3 interacting with viral orf8 and Agps (containing phosphosite S52) interacting with viral nsp7 were associated with ether lipid metabolism. Exosc2, Exosc3, and Exosc5 (containing phosphosite S20) interacting with viral nsp9 and Pabpc1 (containing phosphosite S576) interacting with the viral N protein were enriched in the RNA degradation pathway. Nup98 (containing phosphosite T670) interacting with viral orf6, Nup210 (containing phosphosite T1844) interacting with viral nsp4, and Nup58 and Nup88 interacting with viral nsp9 were enriched in the RNA transport pathway and were associated with amyotrophic lateral sclerosis, a chronic progressive neurodegenerative disease. These viral-host protein interactions may serve as therapeutic targets for COVID-19.
Fig. 5.
Predicted interactions between SARS-CoV-2 proteins and host mouse proteins in the five organs of K18-hACE2 mice. Based on the host-SARS-CoV-2 protein interaction data by Gordon et al., we constructed predicted interactions between SARS-CoV-2 and host mouse proteins in the five organs of K18-hACE2 mice. A total of 23 SARS-CoV-2 proteins potentially interact with 117 host mouse proteins containing 402 phosphosites. The 117 host proteins were submitted to the KEGG database for functional prediction, and 19 proteins were enriched in 6 KEGG pathways that are shown in figure. Red polygons represent SARS-CoV-2 viral proteins. Circles with gray centres represent host proteins, and circles with blue centres represent host phosphosites. The red to blue colour gradient around circles represents the quantification of alterations of proteins or phosphosites in each organ after infection.
Integrated proteomic alteration maps in the cortex, hippocampus, thalamus, lung, and kidney of K18-hACE2 mice after SARS-CoV-2 infection
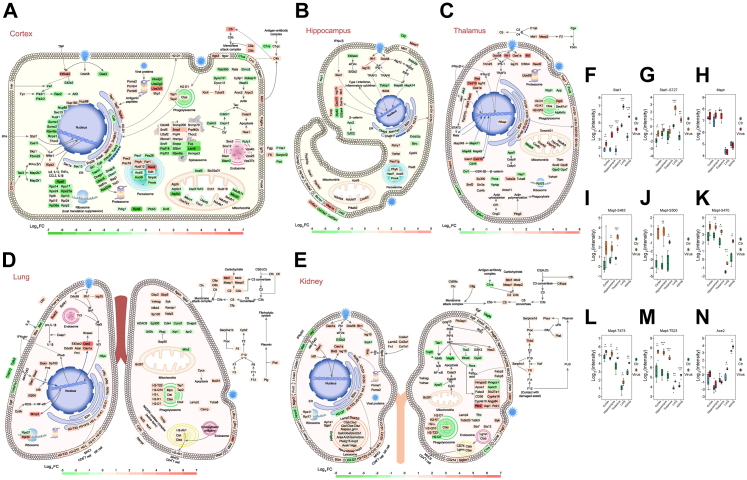
To further obtain comprehensive and intuitive functional alterations after SARS-CoV-2 infection, we matched differentially expressed molecules in significantly enriched pathways in cellular and subcellular diagrams (Fig. 6A–E). We found that the RIG-I signalling pathway, antigen processing and presentation, and complement and coagulation cascades were activated to different degrees in the five organs. In all five organs, the recognition of viral RNA by Ddx58, interferon production, secondary activation of the Jak-Stat signalling pathway, and subsequent antiviral responses were observed. We observed increased Stat1 and Stat1-pS727 levels (Fig. 6F and G and Fig. S6A–D) in the quantifiable proteins in the Jak-Stat signaling pathway. Major histocompatibility class I (MHC I) molecules were upregulated in all five organs (H2-D1 in the cortex, H2-D1 and H2-K1 in the hippocampus, and H2-D1, H2-K1, H2-Q8, and H2-Q10 in the thalamus), especially in the lung and kidney (H2-D1, H2-K1, H2-L, H2-Q10, and H2-T23). Tap1, Tap2, Tapbp, and B2m, proteins responsible for transferring MHC I molecules, were also upregulated after infection (Fig. 6A–E and Fig. S6E). However, the MHC II molecule, H2-Ab1, was upregulated only in the lung. The lung seemed to have stronger antigen presentation than the other four organs. Again, we observed that the lung and kidney seemed to exhibit a stronger complement attack and a higher coagulation risk than the three brain regions. Complement and coagulation reactions in the hippocampus were especially mild (Fig. 6A–E).
Fig. 6.
Maps of COVID-19-induced altered signalling pathway in each organ of K18-hACE2 mice and quantification alteration of specific molecules. A–E. Map of proteins in significantly altered signalling pathways in the cortex (A), hippocampus (B), thalamus (C), lung (D), and kidney (E) after SARS-CoV-2 infection. The pink to green colour gradient represents the log2 FC of the proteins measured in each organ. F–G. Quantification of Stat1 (F) and Stat1-S727 (G) in five organs of K18-hACE2 mice. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (Student's t test, n = 4). H–M. Quantification of Mapt/Tau (H), Mapt-S483 (I), Mapt-S500 (J), Mapt-S470 (K), Mapt-T473 (L), and Mapt-T523 (M) in five organs of K18-hACE2 mice. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (Student's t test, n = 4). N. Quantification of hACE2 in five organs of K18-hACE2 mice. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (Student's t test, n = 4).
Next, we observed each organ individually, which helped to determine the organic specificity after infection. The cortex had the highest viral load of the five organs. Correspondingly, the quantities of proteins and pathways were altered in the cortex after infection. Multiple proteins in the spliceosome (upregulated Ddx46, Hnrnpu, Prpf4, Prpf40b, Rbmxl1, Snrnp70, Snrnp200, Snrpf, Srsf5, Thoc2, and U2af2; downregulated Fus, Hnrnpa3, Lsm8, Prpf18, Prpf31, Rbm8a, Sf3b4, Snrpe, and Srsf1), peroxisome (upregulated Far1, Hao2, Nudt12, Pex3, Pex19, Phyh, and Xdh; downregulated Acot8, Acsl5, Gnpat, Pex1, Pex26, and Pmvk), proteasome (upregulated Psma2, Psmb4, Psmb6, Skp1, and Ube2d1; downregulated Ube2g2 and Ube2j2), ribosome (upregulated Rpl9, Rpl15, and Rpl24; downregulated Rps6, Rps14, Rps17, Rps24, Rps27a, Rpl10, Rpl36a, and Rplp2), cytoskeleton (upregulated Dync1i1, Klc4, Tubal3, Cyfip1, Arpc2, and Arpc3; downregulated Dync1li1, Nckap1l, and Wasf3), nuclear pore complex (upregulated Nup88, Nup160, and Thoc2; downregulated Nup85, Nupl1, Rbm8a, Rnps1, Sec13, Sumo1, and Sumo2), protein processing in endoplasmic reticulum (upregulated Dnajb11, Erp29, Man1b1, Pdia6, and Sar1a; downregulated Calr, Sil1, Prkcsh, Tusc3, etc.), and mitochondrial oxidative respiratory chain (upregulated Ndufa1, Ndufv1, Ndufv2, Cyc1, Cox7a1, and Atp5i; downregulated Mtnd3, Ndufb11, Uqcrb, Mtco1, Atp5d, and Atp5c11) were affected; however, it was difficult to determine a pattern (Fig. 6A). Thus, we inferred that cortical function was severely impaired after SARS-CoV-2 infection. The levels of differentially expressed proteins in the hippocampus were lower than those in the cortex. Protein processing in the endoplasmic reticulum (upregulated Dnajb11; downregulated Ckap4 and Man1a2), peroxisomes (upregulated Pex6, Pex11g, Hacl1, and Phyh; downregulated Acsl5 and Pmvk), mitochondrial oxidative respiratory chain (upregulated Ndufa4, Ndufaf1, and Xox4i2), membrane transporters (upregulated Cacng3, Atp1a4, and Slc8a2; downregulated Slc1a1), and extracellular matrix (upregulated Col5a1; downregulated Col4a1 and Col25a1) was dysregulated in the infected hippocampus; however, the alterations were mild (Fig. 6B). The thalamus did not show the same severe protein expression disorder as that in the cortex. We found several alterations in the processes of the infected thalamus, including the mitochondrial oxidative respiratory chain (upregulated Ndufb3, Uqcrfs1, Uqcr10, Cox4i2, and Cox6c; downregulated Mtatp8) and cytoskeleton (upregulated Gng3, Cfl1, Plcg2, Tuba3a, and Tuba8; Fig. 6C). In the nervous system, we focused on the factors that may cause long-term neurodegeneration. Thus, Mapt/Tau, whose pathological phosphorylation and aggregation may induce Alzheimer's disease,52, 53, 54 was specifically assessed; the protein expression levels did not change significantly after SARS-CoV-2 infection (Fig. 6H); however, Mapt/Tau phosphorylation showed an obvious upward trend. Mapt-pS483 (Fig. 6I) and Mapt-pS500 (Fig. 6J) were upregulated in the three brain regions, and Mapt-pS470 (Fig. 6K), Mapt-pT473 (Fig. 6L), and Mapt-pT523 (Fig. 6M) were upregulated in all five organs. Moreover, unstable upregulation of pS50, pS502, pS507, pT468, pT497, pT504, pT509, and pY686 in Mapt/Tau should also be further investigated (Fig. S6F–M). We also found downregulated App, App-pS441, and App-pT743 in the thalamus, downregulated App-pS441 in the hippocampus (Fig. S6N–P), and downregulated Fus in the cortex (Fig. S6Q and R). However, App and Fus did not seem to be major contributors to potential neurological dysfunctions.
Antigen processing and presentation, antiviral processes, and complement and coagulation cascades were evident in the lungs. In addition, we found an increase in the FC of the active form of the NADPH oxidase complex (Ncf1, Ncf2, Cybb, and Nampt), indicating a high oxidation state in the infected lung (Fig. 6D). The kidney had the highest number of activated lysosomes (upregulated Ctsb, Ctsd, Ctsf, Ctss, Ctsz, Napsa, Lgmn, Galc, Gba, Gla, Glb1, Arsa, Arsb, Galns, Gns, Pla2g15, Acp2, Asah1, Aga, Lamp2, Scarb2, Npc2, Abca2, and Acp2) and exhibited the most apparent accumulation of extracellular matrix (upregulated Col1a1, Col2a1, Col4a1, Fn1, and Lamb2) of the five organs (Fig. 6E). Multiple membrane proteins (upregulated Cadm1, CD274/PD-L1, Egfr, L1cam, Pvrl1, Siglec1, Esam, Vcam1, etc.; downregulated Il6st, Jag1, etc.) and fatty acid metabolism-associated proteins (upregulated Hmgcs2, Apoa1, Fabp3, Slc27a1, CD36, Cyp4a14, Cyp4a10, Angptl14, Plin2, Ucp1, and Pck1; downregulated Apoc3 and Hmgcs1) were also dysregulated in the kidney. Although there were no viral nucleic acid and proteins (Fig. S1A and B) detected, the kidneys of the infected K18-hACE2 mice contained differentially expressed proteins, including proteins involved in antigen processing and presentation, Ddx58-based viral identification, and subsequent antiviral responses (Fig. 6E). Furthermore, high expression of hACE2 was observed in the kidneys, and the expression level of hACE2 was even higher in the kidneys and lungs after infection (Fig. 6N). This suggests that the high expression of hACE2 is not necessarily associated with SARS-CoV-2 infection in tissues. Large-scale molecular alterations suggest that kidney damage may occur after SARS-CoV-2 infection. However, the mechanism by which kidney damage is induced by systemic SARS-CoV-2 infection is not clear.
Discussion
In this study, we performed proteomic and phosphoproteomic analyses of the cortex, hippocampus, thalamus, lung, and kidney of control and SARS-CoV-2-infected K18-hACE2 mice. The RIG-I signalling pathway, antigen processing and presentation, and complement and coagulation cascades were identified in all five organs and were particularly robust in the lungs. Among the five organs, the cortex had the highest viral load and showed severe protein expression disorders in multiple organelles and biological processes. After infection, Mapt/Tau phosphorylation was significantly increased in all three brain regions. Although the virus was not detected, SARS-CoV-2-associated antiviral responses, metabolic alterations, and elevated extracellular matrix accumulation were observed in the kidneys. This study describes the observed SARS-CoV-2-induced multiorgan responses in K18-hACE2 mice and provides a cerebral proteomic dataset related to COVID-19 in vivo.
Although brain injury has been confirmed in patients with COVID-19, the mechanism by which SARS-CoV-2 infects the CNS remains unclear. Some studies do not consider the neuronal viral load to be the direct reason for COVID-19-associated neurological symptoms. For example, anosmia is likely to be attributed to SARS-CoV-2 infection of the respiratory and olfactory mucosae instead of olfactory sensory neurons or the olfactory bulb in patients with COVID-19.9, 10, 11, 12, 13 The findings of Pellegrini et al. and Wang et al. support that SARS-CoV-2 has a tropism for choroid plexus epithelial cells or pericyte-like cells instead of neurons or glial cells in human stem cell-derived brain organoids.55, 56, 57 Zhang et al.8 stated that COVID-19-associated brain damage might be non-specific owing to ischaemia, haemorrhage, inflammation, and aging. However, direct identification of SARS-CoV-2 in the CNS has been increasing. Song et al.14 used three independent approaches, namely human brain organoids, mice overexpressing human ACE2, and autopsies of patients who died of COVID-19, to probe the capacity of SARS-CoV-2 to infect the brain. Stein et al.15 implemented short post-mortem intervals, a comprehensive standardized approach to tissue collection, dissection of the brain before fixation, preservation of tissue in RNAlater, and flash freezing of fresh tissue. Stein et al.15 identified SARS-CoV-2 in brain autopsies of patients with COVID-19, indicating that the CNS might be directly attacked by SARS-CoV-2. Stein et al. fundamentally improved the understanding of SARS-CoV-2 cellular distribution and persistence in the human body and brain. Considering that the outbreak of the COVID-19 pandemic was unexpected, technological limitations may have prevented the detection of SARS-CoV-2 in the human brain in early studies.
Clinical samples are preferred in basic medical research; however, they are rare, and samples are usually embedded in paraffin. Thus, conducting intervention experiments is more feasible in animals than in humans. Under many conditions, the SARS-CoV-2-infected animal model samples in a controlled setting are more stable and high-quality than the clinic samples. Based on existing research, COVID-19-associated encephalopathy research models have mainly been conducted in vitro. In human brain organoids, robust infections of pericyte-like cells, choroid plexus epithelial cells, and neurons and disruptions of the blood-CSF barrier have been observed.17,55,57 K18-hACE2 mice are viable for in vivo studies of SARS-CoV-2-associated encephalopathy. Consistent with the results of a previous report,21,58 high SARS-CoV-2 viral loads were detected in the cortex of the infected K18-hACE2 mice. Post infection, the SARS-CoV-2-infected K18-hACE2 mice experienced significant weight loss from 3 to 6 dpi. The infected mice began to die from day 3, and the surviving mice showed signs of diseases, such as bradykinesia and laboured breathing. Neurological symptoms included ruffled fur, arched back, tremors, ataxic gait, and decreased responses to external stimuli. As an animal model, K18-hACE2 mice maintained susceptibility to SARS-CoV-2 in the CNS; thus, the model combined the advantages of in vitro and in vivo COVID-19 neuronal infection patterns to some extent. K18-hACE2 mice showed amplified clinical neurological manifestations and helped us perform basic medical studies. K18-hACE2 mice are a useful model for studying the pathological basis of mild and lethal COVID-19 and for assessing therapeutic interventions.18
The cortex, hippocampus, and thalamus responded differently to SARS-CoV-2 infection. The cortex contained the highest number of quantifiable and differentially expressed proteins among the five organs. However, only a few proteins were differentially expressed in the hippocampus. Compared with the lung and kidney, the hippocampus and thalamus showed reduced RIG-I signalling pathway activity, antigen processing and presentation, and complement and coagulation cascades. In both the proteome and phosphoproteome, differentially expressed proteins (or proteins containing differentially expressed phosphosites) were enriched in the neurodegeneration pathways in three brain regions. Phosphorylation is a quick and flexible method for organisms to respond to physiological or pathological changes. A change in kinase activity can be predicted according to the intensity of phosphosites, providing substantially increased information. In this study, some predicted kinases potentially regulated synaptic plasticity, axon guidance, and neurodegeneration in the brain. For example, in the cortex, BRSK1 (containing upregulated phosphosite S511), also known as SadB, may regulate the polarisation of cortical and hippocampal neurons by regulating mitochondrial dynamics and Tau phosphorylation.59,60 PAK1 (containing upregulated phosphosite S220 in the infected cortex) plays a crucial role in the formation of dendritic spines, synaptic stability, and synaptic transmission by regulating cytoskeleton dynamics, cell motility, and morphology.61, 62, 63 MARK4, a member of the microtubule affinity-regulating kinase family,64 contains downregulated phosphosite S423 in the infected hippocampus. Predicted activated SRPK2 (containing the upregulated phosphosite S487 in the infected cortex) may regulate splicing and promote neuronal apoptosis.65, 66, 67, 68
We also specifically focused on a contributor to Alzheimer's disease (AD), Mapt/Tau.52 Although Mapt/Tau did not significantly change at the protein level; 39 phosphosites in Mapt/Tau were found in this study. Post-translational modifications of Mapt/Tau have been reported to occur in an ordered manner and may lead to Tau aggregation,69 and hyperphosphorylation tends to induce structural reorganization to form insoluble Tau-paired helical filaments and accelerate AD.53,54 In this study, phosphosites S500, S527, T473, T504, and T523 in mouse Mapt match AD-associated S208, S235, T181, T212, and T231 in human Tau40, respectively.69 This study identified multiple phosphosites in Mapt/Tau and revealed that SARS-CoV-2 infection could pose a high risk for potential neurodegenerative diseases.
We investigated SARS-CoV-2-infected lungs and kidneys because the lungs are the main target of SARS-CoV-2, and COVID-19 recoverers were observed to have low eGFR at follow-up.1,35,36 Comprehensive proteomic analyses of the central nervous system and peripheral organs have revealed some notable phenomena.
For example, although no virus was detected, antigen processing and presentation and the RIG-I-mediated virus recognition system were activated in the infected kidney. Moreover, dysregulated intracellular metabolism, lysosomal processes, and extracellular matrix were observed in the kidneys of SARS-CoV-2-infected K18-hACE2 mice. This revealed that COVID-19-associated virus recognition, anti-virus responses, and tissue/cellular damage could not only be directly induced by SARS-CoV-2, but also indirectly induced by SARS-CoV-2 caused other systematic alterations. Therefore, therapeutic approaches to COVID-19 should be multi-pronged.
A recent report found that macrophages and some CD8+ T cells were predominant in the perivascular infiltration area in brain autopsy; however, only rare CD4+ T cells were present.70 In this study, we found that MHC I molecules were widely upregulated in multiple infected organs. However, MHC II molecules, such as H2-Ab1, were only upregulated in the infected lungs. MHC I and MHC II molecules present antigenic peptides to cytotoxic CD8+ and CD4+ T cells, respectively.71, 72, 73 Based on these proteomic results, we inferred that cytotoxic T cells were widely activated in multiple organs after SARS-CoV-2 infection. Nevertheless, SARS-CoV-2 can activate both cytotoxic and CD4+ T cells in the lungs. Moreover, of the predicted kinases, the infected lung and kidney seem to experience complicated regulation of apoptosis and inflammation. Death-associated protein kinases, DAPK1 and DAPK3, mediate apoptosis74, 75, 76, 77 and were upregulated in the infected lung. ZAK (containing the upregulated phosphosite, S727, in the infected lung) can induce apoptosis and cell cycle arrest,78, 79, 80 and we identified downregulated CDK4 concurrent. In the kidneys, MAPK family kinases undergo complex regulation. Potentially activated MST4-ERK and TAO1-MAPK signalling pathways regulate cell growth, apoptosis, and autophagy.81, 82, 83, 84 Meanwhile, MAP3K3, ERK, p38 MAPK, and PI3K play vital roles in inflammation by regulating NF-κB and other inflammatory modulators.85
As for the limitations of this study, artificial transgenic construction might lead to different responses in K18-hACE2 mice than in humans. Comparisons of mice and humans should be homologously matched when applying these datasets to new research. In addition, cortex, hippocampus, and thalamus were selected in this study because they have strong correlation with clinical symptoms and occupy most of the brain. In the pre-experiment, we separated olfactory bulb for evaluation. However, we abandoned olfactory bulb in formal experiment because its sample size in one mouse was too low to ensure the quality of the experiment. It is surely a pity that olfactory bulb and other parts of the brain were not involved in this study. We will consider to increase investment, change experimental design, and improve technical methods to investigate other parts of the brain in future studies. Moreover, the SARS-CoV-2 viral-host protein interactions that we considered are largely based on overexpression assays.50 However, many viral-host interactions have not been verified in the context of infectious viruses. For example, orf9c has not yet been identified in infected cells. In the context of infectious viruses, further studies are required to confirm the interactions between the virus and host proteins. At last, the lack of distinction between cell types during sample preparation is another limitation of this study. The heat inactivation of the virus ensures biosafety, but also induces some interference in the samples. A single-cell transcriptomic landscape of the lungs in patients with COVID-19 has been reported.86 In the future, more single-cell studies, including single-cell microproteomics, in ABSL3 or ABSL4 may help in the discovery of other mechanisms of COVID-19.
In conclusion, these datasets provide comprehensive proteomic and phosphoproteomic profiles of the cortex, hippocampus, thalamus, lung, and kidney after SARS-CoV-2 infection in vivo. In mature drug databases, differentially expressed proteins and predicted kinases of this study can be used as baits to pick out candidate therapeutic drugs for COVID-19. This study will be a solid resource for the community. The data in the manuscript will serve as a starting point for future investigations.
Contributors
L.D., L.J.N., and Y.J. conceived of and designed the experiments. L.J.N. and P.W. generated the SARS-CoV-2-infected K18-hACE2 mouse models and performed viral load detection and histopathological analyses. L.J.F., Y.J., W.Y., W.S., and Y.Y. performed proteomic and phosphoproteomic analyses. L.J.N., P.W., L.J.F., Y.J., and W.S. verified the underlying data. L.J.F., P.W., W.Y., W.S., and Y.J. wrote the manuscript with inputs from all co-authors. All authors have read and approved the final version of the manuscript.
Data sharing statement
All proteomic and phosphoproteomic raw data have been deposited in the ProteomeXchange Consortium through iProx87 with the identifier PXD035474.
Declaration of interests
The authors declare no conflict of interest.
Acknowledgements
This study was supported by grants from the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences, China (CIFMS2022-I2M-1-011, CIFMS2019-I2M-1-004, CIFMS2021-I2M-1-016), the National Natural Science Foundation of China (Grant No. 32070543 and 92169210), and the Natural Science Foundation of Beijing (Grant No. Z210014).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104518.
Contributor Information
De-Pei Liu, Email: liudp@pumc.edu.cn.
Jiang-Ning Liu, Email: liujn@cnilas.org.
Jun-Tao Yang, Email: yangjt@pumc.edu.cn.
Appendix A. Supplementary data
Figure S1.
Figure S2.
Figure S3.
Figure S4.
Figure S5.
Figure S6.
References
- 1.Huang C., Huang L., Wang Y., et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397(10270):220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang L., Yao Q., Gu X., et al. 1-Year outcomes in hospital survivors with COVID-19: a longitudinal cohort study. Lancet. 2021;398(10302):747–758. doi: 10.1016/S0140-6736(21)01755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zubair A.S., McAlpine L.S., Gardin T., et al. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: a review. JAMA Neurol. 2020;77(8):1018–1027. doi: 10.1001/jamaneurol.2020.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballering A.V., van Zon S.K.R., Olde Hartman T.C., et al. Persistence of somatic symptoms after COVID-19 in the Netherlands: an observational cohort study. Lancet. 2022;400(10350):452–461. doi: 10.1016/S0140-6736(22)01214-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xydakis M.S., Albers M.W., Holbrook E.H., et al. Post-viral effects of COVID-19 in the olfactory system and their implications. Lancet Neurol. 2021;20(9):753–761. doi: 10.1016/S1474-4422(21)00182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virhammar J., Kumlien E., Fallmar D., et al. Acute necrotizing encephalopathy with SARS-CoV-2 RNA confirmed in cerebrospinal fluid. Neurology. 2020;95(10):445–449. doi: 10.1212/WNL.0000000000010250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang P.P., He Z.C., Yao X.H., et al. COVID-19-associated monocytic encephalitis (CAME): histological and proteomic evidence from autopsy. Signal Transduct Target Ther. 2023;8(1):24. doi: 10.1038/s41392-022-01291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosentino G., Todisco M., Hota N., et al. Neuropathological findings from COVID-19 patients with neurological symptoms argue against a direct brain invasion of SARS-CoV-2: a critical systematic review. Eur J Neurol. 2021;28(11):3856–3865. doi: 10.1111/ene.15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang A.C., Kern F., Losada P.M., et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature. 2021;595(7868):565–571. doi: 10.1038/s41586-021-03710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brann D.H., Tsukahara T., Weinreb C., et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci Adv. 2020;6(31) doi: 10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan M., Yoo S.J., Clijsters M., et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell. 2021;184(24):5932–5949.e15. doi: 10.1016/j.cell.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zazhytska M., Kodra A., Hoagland D.A., et al. Non-cell-autonomous disruption of nuclear architecture as a potential cause of COVID-19-induced anosmia. Cell. 2022;185(6):1052–1064.e12. doi: 10.1016/j.cell.2022.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song E., Zhang C., Israelow B., et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med. 2021;218(3) doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein S.R., Ramelli S.C., Grazioli A., et al. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature. 2022;612(7941):758–763. doi: 10.1038/s41586-022-05542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao L., Yang Y., Yu W., et al. The olfactory route is a potential way for SARS-CoV-2 to invade the central nervous system of rhesus monkeys. Signal Transduct Target Ther. 2021;6(1):169. doi: 10.1038/s41392-021-00591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramani A., Muller L., Ostermann P.N., et al. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020;39(20):e106230. doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng J., Wong L.R., Li K., et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589(7843):603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferren M., Favede V., Decimo D., et al. Hamster organotypic modeling of SARS-CoV-2 lung and brainstem infection. Nat Commun. 2021;12(1):5809. doi: 10.1038/s41467-021-26096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Melo G.D., Lazarini F., Levallois S., et al. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci Transl Med. 2021;13(596) doi: 10.1126/scitranslmed.abf8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oladunni F.S., Park J.G., Pino P.A., et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun. 2020;11(1):6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang R.D., Liu M.Q., Chen Y., et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182(1):50–58.e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robles M.S., Humphrey S.J., Mann M. Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. 2017;25(1):118–127. doi: 10.1016/j.cmet.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Nie X., Qian L., Sun R., et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell. 2021;184(3):775–791.e14. doi: 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leng L., Cao R., Ma J., et al. Pathological features of COVID-19-associated lung injury: a preliminary proteomics report based on clinical samples. Signal Transduct Target Ther. 2020;5(1):240. doi: 10.1038/s41392-020-00355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leng L., Cao R., Ma J., et al. Pathological features of COVID-19-associated liver injury-a preliminary proteomics report based on clinical samples. Signal Transduct Target Ther. 2021;6(1):9. doi: 10.1038/s41392-020-00406-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu M., Chen Y., Xia H., et al. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc Natl Acad Sci U S A. 2020;117(45):28336–28343. doi: 10.1073/pnas.2018030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen B., Yi X., Sun Y., et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villar M., Urra J.M., Rodriguez-Del-Rio F.J., et al. Characterization by quantitative serum proteomics of immune-related prognostic biomarkers for COVID-19 symptomatology. Front Immunol. 2021;12:730710. doi: 10.3389/fimmu.2021.730710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Alessandro A., Thomas T., Dzieciatkowska M., et al. Serum proteomics in COVID-19 patients: altered coagulation and complement status as a function of IL-6 level. J Proteome Res. 2020;19(11):4417–4427. doi: 10.1021/acs.jproteome.0c00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leng L., Li M., Li W., et al. Sera proteomic features of active and recovered COVID-19 patients: potential diagnostic and prognostic biomarkers. Signal Transduct Target Ther. 2021;6(1):216. doi: 10.1038/s41392-021-00612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., Zhang Z., Zhou J., et al. Next-generation sequencing and proteomics of cerebrospinal fluid from COVID-19 patients with neurological manifestations. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.782731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suresh V., Mohanty V., Avula K., et al. Quantitative proteomics of hamster lung tissues infected with SARS-CoV-2 reveal host factors having implication in the disease pathogenesis and severity. FASEB J. 2021;35(7) doi: 10.1096/fj.202100431R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorne L.G., Bouhaddou M., Reuschl A.K., et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature. 2021;602(7897):487–495. doi: 10.1038/s41586-021-04352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cortinovis M., Perico N., Remuzzi G. Long-term follow-up of recovered patients with COVID-19. Lancet. 2021;397(10270):173–175. doi: 10.1016/S0140-6736(21)00039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang L., Gu X., Wang Y., et al. Long-term effects of COVID-19 on kidney function–authors' reply. Lancet. 2021;397(10287):1807–1808. doi: 10.1016/S0140-6736(21)00875-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudose S., Batal I., Santoriello D., et al. Kidney biopsy findings in patients with COVID-19. J Am Soc Nephrol. 2020;31(9):1959–1968. doi: 10.1681/ASN.2020060802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng W., Lv Q., Li F., et al. Sequential immunizations confer cross-protection against variants of SARS-CoV-2, including Omicron in Rhesus macaques. Signal Transduct Target Ther. 2022;7(1):124. doi: 10.1038/s41392-022-00979-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song Z., Bao L., Deng W., et al. Integrated histopathological, lipidomic, and metabolomic profiles reveal mink is a useful animal model to mimic the pathogenicity of severe COVID-19 patients. Signal Transduct Target Ther. 2022;7(1):29. doi: 10.1038/s41392-022-00891-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao L., Deng W., Huang B., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 41.Liu J.F., Zhou Y.N., Lu S.Y., et al. Proteomic and phosphoproteomic profiling of COVID-19-associated lung and liver injury: a report based on rhesus macaques. Signal Transduct Target Ther. 2022;7(1):27. doi: 10.1038/s41392-022-00882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 43.Tyanova S., Temu T., Sinitcyn P., et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 44.Liu J.F., Wu Y., Yang Y.H., et al. Phosphoproteome profiling of mouse liver during normal aging. Proteome Sci. 2022;20(1):12. doi: 10.1186/s12953-022-00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M., Araki M., Goto S., et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song C., Ye M., Liu Z., et al. Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol Cell Proteomics. 2012;11(10):1070–1083. doi: 10.1074/mcp.M111.012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chartier M., Chenard T., Barker J., et al. Kinome Render: a stand-alone and web-accessible tool to annotate the human protein kinome tree. PeerJ. 2013;1:e126. doi: 10.7717/peerj.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gordon D.E., Jang G.M., Bouhaddou M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Han J. Differential privacy fuzzy C-means clustering algorithm based on Gaussian kernel function. PLoS One. 2021;16(3):e0248737. doi: 10.1371/journal.pone.0248737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Congdon E.E., Sigurdsson E.M. Tau-targeting therapies for Alzheimer disease. Nat Rev Neurol. 2018;14(7):399–415. doi: 10.1038/s41582-018-0013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wegmann S., Biernat J., Mandelkow E. A current view on Tau protein phosphorylation in Alzheimer's disease. Curr Opin Neurobiol. 2021;69:131–138. doi: 10.1016/j.conb.2021.03.003. [DOI] [PubMed] [Google Scholar]
- 54.Rani L., Mallajosyula S.S. Phosphorylation-induced structural reorganization in tau-paired helical filaments. ACS Chem Neurosci. 2021;12(9):1621–1631. doi: 10.1021/acschemneuro.1c00084. [DOI] [PubMed] [Google Scholar]
- 55.Pellegrini L., Albecka A., Mallery D.L., et al. SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell. 2020;27(6):951–961.e5. doi: 10.1016/j.stem.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L., Sievert D., Clark A.E., et al. A human three-dimensional neural-perivascular 'assembloid' promotes astrocytic development and enables modeling of SARS-CoV-2 neuropathology. Nat Med. 2021;27(9):1600–1606. doi: 10.1038/s41591-021-01443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacob F., Pather S.R., Huang W.K., et al. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism predominates in choroid plexus epithelium. Cell Stem Cell. 2020;27(6):937–950.e9. doi: 10.1016/j.stem.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bi Z., Hong W., Yang J., et al. Animal models for SARS-CoV-2 infection and pathology. MedComm (2020) 2021;2(4):548–568. doi: 10.1002/mco2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller M., Lutter D., Puschel A.W. Persistence of the cell-cycle checkpoint kinase Wee1 in SadA- and SadB-deficient neurons disrupts neuronal polarity. J Cell Sci. 2010;123(Pt 2):286–294. doi: 10.1242/jcs.058230. [DOI] [PubMed] [Google Scholar]
- 60.Di Meo D., Ravindran P., Sadhanasatish T., et al. The balance of mitochondrial fission and fusion in cortical axons depends on the kinases SadA and SadB. Cell Rep. 2021;37(12):110141. doi: 10.1016/j.celrep.2021.110141. [DOI] [PubMed] [Google Scholar]
- 61.Cernohorska M., Sulimenko V., Hajkova Z., et al. GIT1/betaPIX signaling proteins and PAK1 kinase regulate microtubule nucleation. Biochim Biophys Acta. 2016;1863(6 Pt A):1282–1297. doi: 10.1016/j.bbamcr.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 62.Harms F.L., Kloth K., Bley A., et al. Activating mutations in PAK1, encoding p21-activated kinase 1, cause a neurodevelopmental disorder. Am J Hum Genet. 2018;103(4):579–591. doi: 10.1016/j.ajhg.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manabe R., Kovalenko M., Webb D.J., et al. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J Cell Sci. 2002;115(Pt 7):1497–1510. doi: 10.1242/jcs.115.7.1497. [DOI] [PubMed] [Google Scholar]
- 64.Trinczek B., Brajenovic M., Ebneth A., et al. MARK4 is a novel microtubule-associated proteins/microtubule affinity-regulating kinase that binds to the cellular microtubule network and to centrosomes. J Biol Chem. 2004;279(7):5915–5923. doi: 10.1074/jbc.M304528200. [DOI] [PubMed] [Google Scholar]
- 65.Jang S.W., Liu X., Fu H., et al. Interaction of Akt-phosphorylated SRPK2 with 14-3-3 mediates cell cycle and cell death in neurons. J Biol Chem. 2009;284(36):24512–24525. doi: 10.1074/jbc.M109.026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong Y., Jang S.W., Ye K. The N-terminal fragment from caspase-cleaved serine/arginine protein-specific kinase2 (SRPK2) translocates into the nucleus and promotes apoptosis. J Biol Chem. 2011;286(1):777–786. doi: 10.1074/jbc.M110.193441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H.Y., Lin W., Dyck J.A., et al. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140(4):737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathew R., Hartmuth K., Mohlmann S., et al. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat Struct Mol Biol. 2008;15(5):435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 69.Wesseling H., Mair W., Kumar M., et al. Tau PTM profiles identify patient heterogeneity and stages of Alzheimer's disease. Cell. 2020;183(6):1699–1713.e13. doi: 10.1016/j.cell.2020.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee M.H., Perl D.P., Steiner J., et al. Neurovascular injury with complement activation and inflammation in COVID-19. Brain. 2022;145(7):2555–2568. doi: 10.1093/brain/awac151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pishesha N., Harmand T.J., Ploegh H.L. A guide to antigen processing and presentation. Nat Rev Immunol. 2022;22(12):751–764. doi: 10.1038/s41577-022-00707-2. [DOI] [PubMed] [Google Scholar]
- 72.Xiong Y., Bosselut R. CD4-CD8 differentiation in the thymus: connecting circuits and building memories. Curr Opin Immunol. 2012;24(2):139–145. doi: 10.1016/j.coi.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Singer A., Adoro S., Park J.H. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8(10):788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tian X., Shi Y., Liu N., et al. Upregulation of DAPK contributes to homocysteine-induced endothelial apoptosis via the modulation of Bcl2/Bax and activation of caspase 3. Mol Med Rep. 2016;14(5):4173–4179. doi: 10.3892/mmr.2016.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tu W., Xu X., Peng L., et al. DAPK1 interaction with NMDA receptor NR2B subunits mediates brain damage in stroke. Cell. 2010;140(2):222–234. doi: 10.1016/j.cell.2009.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y., Yang Y., Chen L., et al. Death-associated protein kinase 1 mediates ventilator-induced lung injury in mice by promoting alveolar epithelial cell apoptosis. Anesthesiology. 2020;133(4):905–918. doi: 10.1097/ALN.0000000000003464. [DOI] [PubMed] [Google Scholar]
- 77.Wang Q., Lin Y., Zhong W., et al. Regulatory non-coding RNAs for death associated protein kinase family. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.649100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu T.C., Huang C.J., Chu Y.C., et al. Cloning and expression of ZAK, a mixed lineage kinase-like protein containing a leucine-zipper and a sterile-alpha motif. Biochem Biophys Res Commun. 2000;274(3):811–816. doi: 10.1006/bbrc.2000.3236. [DOI] [PubMed] [Google Scholar]
- 79.Gross E.A., Callow M.G., Waldbaum L., et al. MRK, a mixed lineage kinase-related molecule that plays a role in gamma-radiation-induced cell cycle arrest. J Biol Chem. 2002;277(16):13873–13882. doi: 10.1074/jbc.M111994200. [DOI] [PubMed] [Google Scholar]
- 80.Tosti E., Waldbaum L., Warshaw G., et al. The stress kinase MRK contributes to regulation of DNA damage checkpoints through a p38gamma-independent pathway. J Biol Chem. 2004;279(46):47652–47660. doi: 10.1074/jbc.M409961200. [DOI] [PubMed] [Google Scholar]
- 81.Lin J.L., Chen H.C., Fang H.I., et al. MST4, a new Ste20-related kinase that mediates cell growth and transformation via modulating ERK pathway. Oncogene. 2001;20(45):6559–6569. doi: 10.1038/sj.onc.1204818. [DOI] [PubMed] [Google Scholar]
- 82.Huang T., Kim C.K., Alvarez A.A., et al. MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer Cell. 2017;32(6):840–855.e8. doi: 10.1016/j.ccell.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma X., Zhao H., Shan J., et al. PDCD10 interacts with Ste20-related kinase MST4 to promote cell growth and transformation via modulation of the ERK pathway. Mol Biol Cell. 2007;18(6):1965–1978. doi: 10.1091/mbc.e06-07-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J., Liu Z., Wang L., et al. Thousand and one kinase 1 protects MCAO-induced cerebral ischemic stroke in rats by decreasing apoptosis and pro-inflammatory factors. Biosci Rep. 2019;39(10) doi: 10.1042/BSR20190749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yeung Y.T., Aziz F., Guerrero-Castilla A., et al. Signaling pathways in inflammation and anti-inflammatory therapies. Curr Pharm Des. 2018;24(14):1449–1484. doi: 10.2174/1381612824666180327165604. [DOI] [PubMed] [Google Scholar]
- 86.Wang S., Yao X., Ma S., et al. A single-cell transcriptomic landscape of the lungs of patients with COVID-19. Nat Cell Biol. 2021;23(12):1314–1328. doi: 10.1038/s41556-021-00796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma J., Chen T., Wu S., et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47(D1):D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.