Abstract
Background.
Currently, the precise mechanisms that underline male infertility are still unclear. Accumulating data implicate non-coding RNA (ncRNA) cargo of seminal plasma extracellular vesicles (spEVs) due to their association with poor semen quality and higher expression level relative to vesicle-free seminal plasma.
Method.
We assessed sperm-free spEV ncRNA profiles from 91 semen samples collected from male participants of couples seeking infertility treatment. Men were classified into two groups (poor, n=32; normal, n=59) based on World Health Organization semen cutoffs. Small RNA-seq reads were mapped to standard biotype-specific transcriptomes in the order miRNA>tRNA>piRNA>rRNA> “other” RNA>circRNA>lncRNA using STAR. Differential expression of normalized ncRNA read counts between the two groups was conducted by EdgeR (Fold change ≥1.5 & FDR<0.05).
Result.
Small RNA-seq identified a wide variety of spEV ncRNA biotypes including miRNA, rRNAs, piRNAs, tRNA, lncRNAs as well as circRNAs and fragments associated with pseudogenes, and nonsense mediated decay. The expression levels of 57 spEV ncRNAs (miRNA:6, piRNA:4, rRNA:6, circRNA:34, lncRNA:7) were altered in men with poor semen quality relative to normal semen parameters, many (60%) of which were circRNA species. Ontology analysis of differentially expressed miRNAs and circRNAs showed enrichment in functional terms related to cellular communication and early development.
Conclusion.
This is the first study to generate comprehensive spEV ncRNA profiles in a clinical setting and to determine their differences between men with normal and abnormal semen parameters. Thus, our study suggests that spEV ncRNAs may represent novel biomarkers of male reproductive phenotypes.
Keywords: Seminal plasma, extracellular vesicles, small non-coding RNA, infertility, IVF
INTRODUCTION
Infertility, defined as the inability to achieve pregnancy within 12 months of unprotected sexual intercourse, remains a global problem affecting 15% of heterosexual couples with the desire to conceive. Infertility affects both males and females equally, such that up to 50% of all cases have a male factor contribution (1). While male infertility is a multi-factorial medical condition involving endo- and exogenous factors that may impair spermatogenesis (2), semen parameter analysis remains the most prevalent diagnostic tool for determining clinical male infertility diagnosis and includes sperm concentration, morphology and motility (3). However, these measures of semen quality have an unclear underlying biology and connection to sperm function. Biomarkers that directly measure characteristics of semen may provide insights into male reproductive health beyond conventional methods. Thus, recent research has focused on other potential markers of male infertility such as the role of extracellular vesicles (EV) that are present in the male reproductive tract and other secondary sex organs. (4).
EVs are nano-sized lipid bilayer particles secreted from cells into the extracellular matrix and are important for cell-to-cell communication. EVs are found in all body fluids and their abundance in seminal plasma from healthy male has been documented with average concentrations ranging from 2.89×109 particle/mL (5) to 1.37 × 1013 particle/mL (6) of semen. Despite the small size range of EVs in seminal plasma (50 – 302 nm) (5, 6), they contain cargo of biologically active and regulatory molecules including proteins (7), coding and non-coding RNAs (ncRNAs) (6, 8). Interestingly, emerging data show seminal plasma EVs (spEVs) cargo can be shuttled into sperm during post-testicular maturation and ejaculation (9–11), resulting in modification of sperm RNA profiles. This suggest a cross-talk between sperm and spEVs derived from somatic cells (11, 12) and represents the last opportunity for modification of post-testicular sperm prior to fertilization (13).
Numerous ncRNA biotypes have been reported in spEVs from healthy semen donors including micro RNAs (miRNAs), transfer RNAs (tRNAs), and piwi-interacting RNAs (piRNAs) (6). Thus, spEV ncRNAs have been linked to poor semen quality and reproductive outcomes. For example, decreased expression levels of piRNA (5) and lncRNA fragments (14) in spEVs have been reported in men with low sperm motility compared to normal controls. Similarly, miRNAs (15) and a panel of lncRNA fragments (16) in spEVs were observed to predict spermatogenic impairment and have been proposed as important molecular markers for the absence of testicular spermatozoa (17). However, these studies had several limitations including use of a single semen parameter for diagnosis of male infertility, limited sample size and lack of comprehensive profiling of spEV ncRNAs, thereby limiting analyses of other ncRNA constituents of spEVs such as circular RNAs (circRNAs). Thus, we performed small RNA-seq to determine the profiles of spEV ncRNAs among a clinical cohort of men undergoing IVF treatment and then examined whether distinct ncRNA expression profiles were present in men with poor semen quality compared to normal controls.
MATERIALS AND METHODS
Study population
The Sperm Environmental Epigenetics and Developments Study (SEEDS) is a prospective observational cohort study designed to investigate the associations of male preconception endocrine disrupting chemical exposures with sperm epigenetics and subsequent early-life development. The current study includes a subset of enrolled men of couples (n = 91) undergoing IVF treatment at Baystate Medical Center, Springfield, Massachusetts. The inclusion criteria were male partners between 18 and 55 years old without vasectomy and fresh ejaculate sperm used for IVF treatment. Relevant demographics (race, age, height, and weight), lifestyle factors (current and past alcohol and cigarette use), and medical history (diagnoses of infertility) data were collected by clinic personnel during the IVF cycle for both partners. Male fertility diagnoses were based on semen quality parameters according to World Health Organization (WHO) reference values (18) and Kruger strict morphology criteria (19). Written informed consent was obtained from eligible males who showed voluntary interest in participating in the study by the attending physicians. The study is approved by the institutional review boards at Baystate Medical Center and at the University of Massachusetts Amherst (IRB number: BH-12–190).
Isolation of EVs from seminal plasma and RNA extraction
Fresh ejaculate semen samples (n = 96) were collected in a sterile plastic cup following a recommended 2–3-day abstinence period as per standard IVF protocol. Semen samples were processed using a two-step (40% and 80%) gradient fractionation method and the resulting seminal plasma was stored in −80°C until subsequent analysis. Frozen seminal plasma samples were thawed for 20 min at room temperature and EV extraction was performed using Qiagen exoRNAeasy Midi Kit (Cat. #77144) according to manufacturer’s protocol (20). Equal volumes of PBS and seminal plasma were mixed and centrifuged at 12,000 g for 45 min at 4⁰C to pellet any remaining sperm and cell debris. To exclude non-EV particles, supernatants were filtered through a 0.45 μm membrane (Whatman PURADISC 25, Cat. #6780–2505). Next, the filtered seminal plasma was thoroughly mixed with an equal volume of DNA binding buffer (XBP) prior to centrifugation at 500 g for 3 min on an exoEasy spin column. To purify the column-bound EVs, 3.5mL of buffer XWP was added to the column and the suspension was centrifuged at 5000 g for 5 min. The flow-through was carefully discarded and the column was transferred to a new collection tube followed by elution in buffer XE. Nanoparticle tracking analysis was performed on a subset of the total samples (n = 5) to evaluate the concentration and size distribution of the eluted EVs using NanoSight NS300 (Malvern Instruments, UK) and were imaged using transmission electron microscopy (TEM).
The purified EVs were lysed with QIAzol, followed by phase-separation method with chloroform to separate the total RNA. The aqueous phase containing RNA was mixed with 2 volumes of 100% ethanol and transferred onto RNeasy MinElute spin column. RNA purification was performed using buffers RWT and RPE, followed by final washing in 80% ethanol and elution in nuclease free water. Total RNA quantification was performed using nanodrop and fluorometer while fragment analyzer was used for quality assessment.
Small RNA library preparation and sequencing
Libraries construction and sequencing were conducted at the UMass Medical Genomics Core Facility. First, end repair of the isolated total RNA using T4 Polynucleotide kinase (NEB, Cat #M0201) according to manufacturer’s instructions. Polyadenylated RNA library preparation, reverse transcription cDNA synthesis, and 6 cycles of PCR amplification steps followed modified Clontech SMARTer smRNA-Seq protocol. To avoid small RNA sequencing bias, SPRI beads were used for library cleanup. Size selection and quality assessment were performed using AMPure X beads and fragment analyzer respectively. Samples were multiplexed prior to loading onto the flow cell using Illumina barcodes (1 – 24) and 50-bp single-end reads were sequenced on an Illumina HiSeq 4000.
Small RNA sequencing analysis
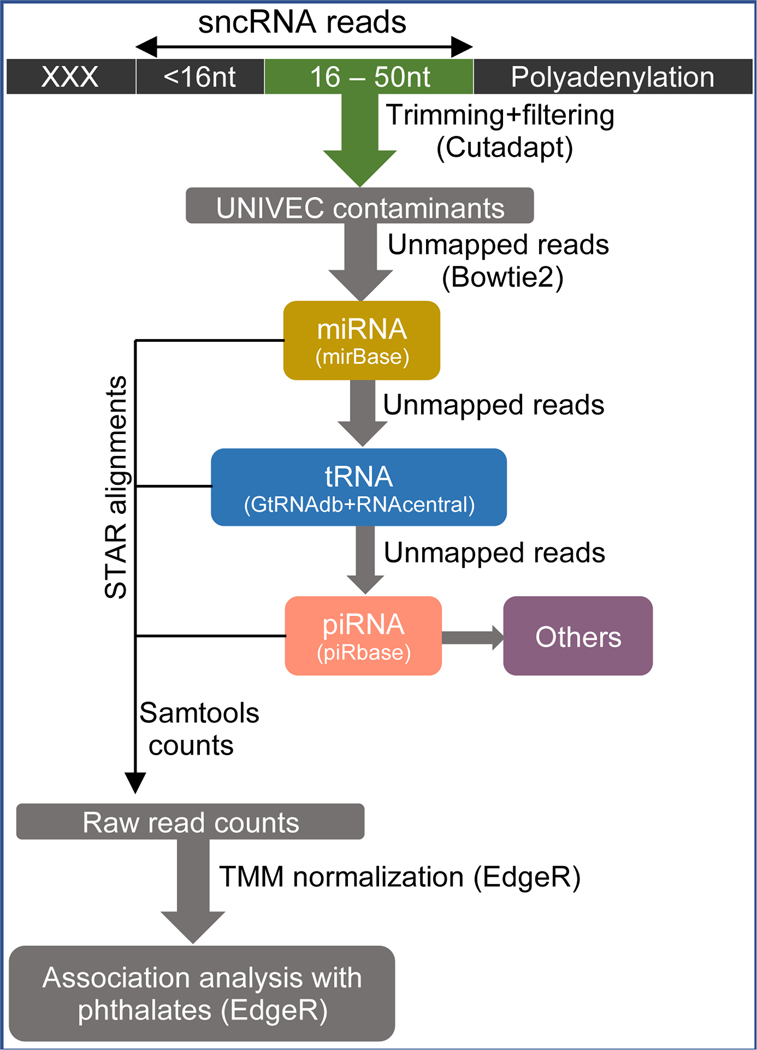
Figure 1 presents the flowchart of our bioinformatics pipeline. Raw sequencing read quality was assessed using FastQC (21). As recommended by smRNA-seq protocol, the first three nucleotides, 3’ adaptor and polyA sequences were trimmed using cutadapt (22). Low quality reads (PHRED score < 20) and reads shorter than 16 nt were also filtered, resulting in average read length ranging from 16 – 47 nt with a GC of 40%. Reads were aligned to Homo sapiens transcriptomes following the Extracellular RNA Communication Consortium (ERCC) pipeline (23) with slight modifications. The adaptor-free reads were first mapped to the UniVec database to remove sequencing contaminants using Bowtie2 (24).
Figure 1: Flow chart of bioinformatic analysis.
XXX: Extra bases from SMART template switching activity.
To identify known selected ncRNAs, the resulting unmapped reads were then sequentially aligned to standard, human annotated reference transcriptome databases in the following order: miRbase 22.1 release (http://www.mirbase.org/) for miRNA (precursor and mature), GtRNAdb (http://gtrnadb.ucsc.edu/) and RNAcentral v18 (https://rnacentral.org/) for mature tRNA with 3’-CCA tail and no introns, and piRBase v.1.7.5 (http://www.regulatoryrna.org/database/piRNA/) for piRNA. The reference genomes for miRNA and tRNA databases were modified with a U->T conversion to get standard DNA fasta files prior to mapping. Unaligned reads to piRBase were subsequently mapped to transcriptomes of ribosomal RNA (rRNA), “other” RNAs (small nucleolar, snoRNA; small nuclear, snRNA; pseudogenes; Y-RNA; mitochondrial, Mt-RNA; antisense; guide; miscellaneous, misc; non-stop and nonsense-mediated decay (NSD & NMD, respectively); precursor; ribozyme; RNase; small cajal body-specific, scaRNA, vault and Y), circular RNA (circRNA) from circBase (http://www.circbase.org) and lncRNA. rRNA, “other” RNAs and lncRNA transcriptomes were derived from biotype-specific sequences annotated in Ensembl (http://useast.ensembl.org/), Gencode (https://www.gencodegenes.org/) and RNAcentral. Merging of RNA-biotype specific transcript sequences and duplicate removal were performed using “cat” and seqkit (v. 0.14.0)(25) functions in Linux (cat | seqkit rmdup -s).
All transcriptome alignments were performed using STAR (v.2.7.6a) (26) with the following parameters: --outFilterMismatchNoverLmax 0.05 --outFilterMatchNmin 16 --outFilterScoreMinOverLread 0 --outFilterMatchNminOverLread 0 --alignIntronMax 1 --outMultimapperOrder Random --runRNGseed 2021 --outSAMmultNmax 1 --seedSearchLmax 28 --seedSplitMin 9 --outSAMtype BAM Unsorted --outReadsUnmapped Fastx --outSAMattributes Standard.
Multi-mapping reads were assigned to the alignment with the best mapping quality and in cases with matching quality, reads were randomly assigned. We allowed incremental mismatch: no mismatches in the reads ≤ 25 bases and 1 mismatch in 26 to 50 bases. Samtools (v. 1.9) (27) was used for SAM/BAM data manipulation and counting of mapped reads was performed using samtools idxstats. Biotype-specific raw read counts from all samples were then imported into R and joined to produce raw data tables. All raw sequences were deposited into the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE203494.
Data analysis
Visualization of spEV ncRNA raw read count across a total of 96 samples was performed using ggplot2 in R. Biotype-specific library size counts were normalized using weighted trimmed mean of M-values TMM (28) and species with normalized count ≥ 1 RPM in all samples were utilized for downstream analyses. We excluded five samples in which sperm morphology or concentration was missing. Male participants (n=91) were categorized as having poor semen characteristics if they were below WHO reference cutoffs (1) in at least one of the following parameters: sperm concentration, motility or morphology (poor, n = 32; normal, n = 59) (Table 1 & Figure S1). Differential expression analysis between groups was performed using EdgeR (29). ncRNA species with a fold change (FC) ≥ 1.5 and FDR<0.05 were considered statistically significant. Student’s t-test or the nonparametric Mann–Whitney U test was used to compare participants’ demographic and clinical characteristics between groups (p<0.05).
Table 1:
Demographic and clinical features of all SEEDS participants (n = 91) and stratification by semen quality status.
| Features | All participants (n = 91) Mean ± SD/count (%) |
Normal Semen Quality (n = 59) Mean ± SD/count (%) |
Poor Semen Quality (n=32) Mean ± SD/count (%) |
Pvalue† |
|---|---|---|---|---|
| Age (years) | 36.2 ± 5.9 | 36.1 ± 6.2 | 36.4 ± 5.5 | 0.84 |
| BMI (kg/m2) a | 29.9 ± 5.5 | 29.6 ± 5.3 | 30.4 ± 5.9 | 0.55 |
| Race b | ||||
| Hispanic white | 73 (80.2%) | 50 (90.1%) | 23 (82.1%) | 0.42 |
| Others | 10 (11.0%) | 5 (9.1%) | 5 (17.9%) | |
|
| ||||
| Semen parameters | 5th centile WHO references (n, %<WHO) | Median (Range) | Median (Range) | Pvalue |
|
| ||||
| Concentration (106/mL) | 15 (8, 8.8) | 78.8 (23.0 – 667.5) | 30.9 (0.8 – 170.6) | 2.40×10−5 |
| Total motility (%) | 40 (13, 14.3) | 67 (40.0 – 91.0) | 46.5 (5.0 – 87.0) | 2.50×10−6 |
| Normal morphology (%) c | 4 (20, 22.0) | 9.2 (4.0 – 21.0) | 3.2 (0.5 – 10.5) | 2.60×10−9 |
| Count (106/ejaculate) | 39 (9, 9.8) | 185.4 (54.0 – 1,117.0) | 99.8 (4.2 – 563.0) | 9.80×10−4 |
| Semen volume (mL) | 1.5 (16, 17.6) | 2.9 (0.5 – 7.5) | 3.2 (0.7 – 6.5) | 0.12 |
Missing data:
BMI (n = 3);
race (n = 8);
normal morphology (n = 4);
Represents statistical comparison (Student’s t-test/Mann–Whitney U test) of parameters between normal (n = 59) semen quality vs poor (n = 32) semen quality groups. Stratification was based on poor semen quality in one of sperm concentration, morphology or motility.
Gene target determination and functional annotations
We utilized suitable algorithms for the differentially expressed ncRNA biotypes. Given miRNAs has been well studied, we combined two experimentally validated databases (Mirtarbase, Tarbase), executed through multiMiR package in R (v.1.14.0) (30). To identify potential gene targets of differentially expressed piRNAs, rRNAs and lncRNAs, we annotated to non-repeat genomic region obtained from piRBase and total human genome (https://www.gencodegenes.org/human/release_38lift37.html) using BEDTools (31). Lastly, for the differentially expressed circRNAs, functional analysis was explored based on its role in competing endogenous RNA networks (32). Given interaction between circRNA and miRNAs targets has been mostly validated in carcinogenesis (33, 34) and could potentially result in biased functional analyses, predicted hits were utilized for gene targets of differentially expressed circRNAs. Thus, we first identified the predicted circRNA-miRNA pairs using miRNA species (top 5%) curated either in miranda or targetscan database. To keep the number of gene targets of the identified circRNA-miRNA pairs within manageable level, predicted miRNA-gene pairs were restricted to conserved genes that overlap in both miranda and targetscan. Ontology and pathway analyses of target genes were performed using metascape (https://metascape.org/gp/index.html#/main/step1) (35).
RESULTS
Demographic and clinical characteristics
This study investigated the expression profiles of spEV small non-coding RNAs (sncRNAs) between men with poor and normal semen quality as determined by WHO semen parameter guidelines and Kruger’s morphological strict criteria. In the poor semen quality group (n = 32), 24 (75%) had either poor concentration, motility or morphology only; 7 (22%) had a combination of two poor parameters, while only 1 (3%) participant had poor semen quality for all three parameters (Figure S1). Table 1 presents the overall demographic and clinical characteristics of men (n = 91) who were enrolled as part of the SEEDS study. Participants’ average age (years) and BMI (kg/m2) were 36.1±5.9 and 29.9±5.4, respectively and were mostly non-Hispanic white (71.9%) with BMI ranging from 20 – 48 kg/m2. Participants did not differ in socio-demographic characteristics such as age, BMI and race (p>0.05 for all) between the two groups. As expected, semen characteristics (Median values for normal vs poor: sperm concentration, 78.8 vs 30.9 (106/mL); motility, 67.0 vs 46.5 %; morphology, 9.2 vs 3.2%; and total count, 185.4 vs 99.8 (106/ejaculates)) were significantly lower among the men with poor semen quality (p < 0.001 for all) (Table 1).
spEV characterization and ncRNA profiles
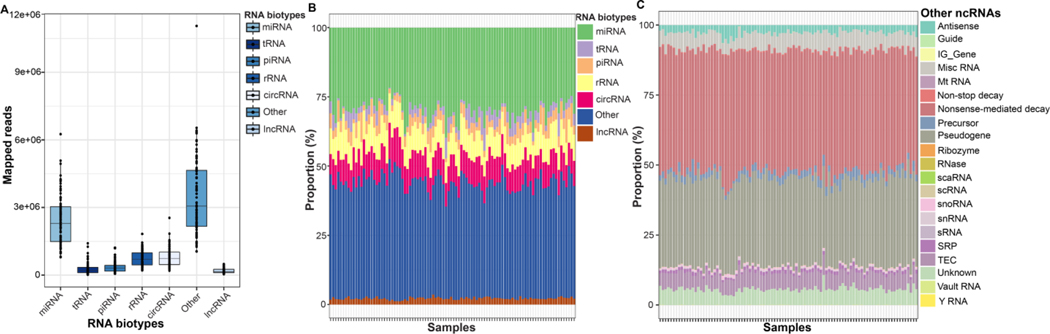
The characterization of particles of EVs in seminal plasma from a subset of the total samples is presented in Figure S2. The average size distribution of the EVs were 120±1.1nm (Figure S2A). TEM imaging and fragment analyzer electropherogram confirmed the presence of EVs (Figure S2B) and total RNA composition after extraction (Figure 2C), respectively. The distribution of read counts across spEV ncRNA biotypes is presented in Figure 2A. Of the total raw reads (814,508,936) obtained from our samples, 96 % (780,817,829) were successfully aligned to biotype-specific ncRNA transcriptomes. “Other” RNA had the highest (median (range): 3,058,422 (1,47,957 – 11,057,341), followed by miRNA (2,286,544 (792,604 – 6,257,318)) while lncRNA had the lowest (150,559 (42,462– 489,906)) mapped reads.
Figure 2: Profiles seminal plasma EV ncRNA.
(A) Total number of reads mapped to RNA biotypes (B) Sample specific relative abundance of all seminal plasma extracellular vesicles nRNA biotypes and (C) Other ncRNAs.
Next, we determined the percent distribution of the biotype-specific mapped reads relative to other species within each sample (Figure 2B). The relative compositions (mean±SD) across samples and for each biotype were: miRNA (29.8±3.5%), tRNA (3.0±2.0%), piRNA (4.3±2.3%), rRNA (9.2±1.4%), circRNA (9.4±1.2%), “other” RNAs (42.1±3.3%), and lncRNA fragments (2.2±0.5%). Given the highest proportion was observed among the “other” RNA category, its relative composition was further identified with transcripts corresponding to NMD and pseudogene representing the most dominant (74.5%) species (Figure 2C). The remaining biotypes with relatively low enrichment in the “other” RNA category included, but not limited to, misc-RNA (6.0%), antisense (2.8%), precursor (2.5%) and SRP (1.1%) (Figure 2C).
Identification and functional annotation of differentially expressed ncRNAs in men with poor semen quality compared to healthy controls
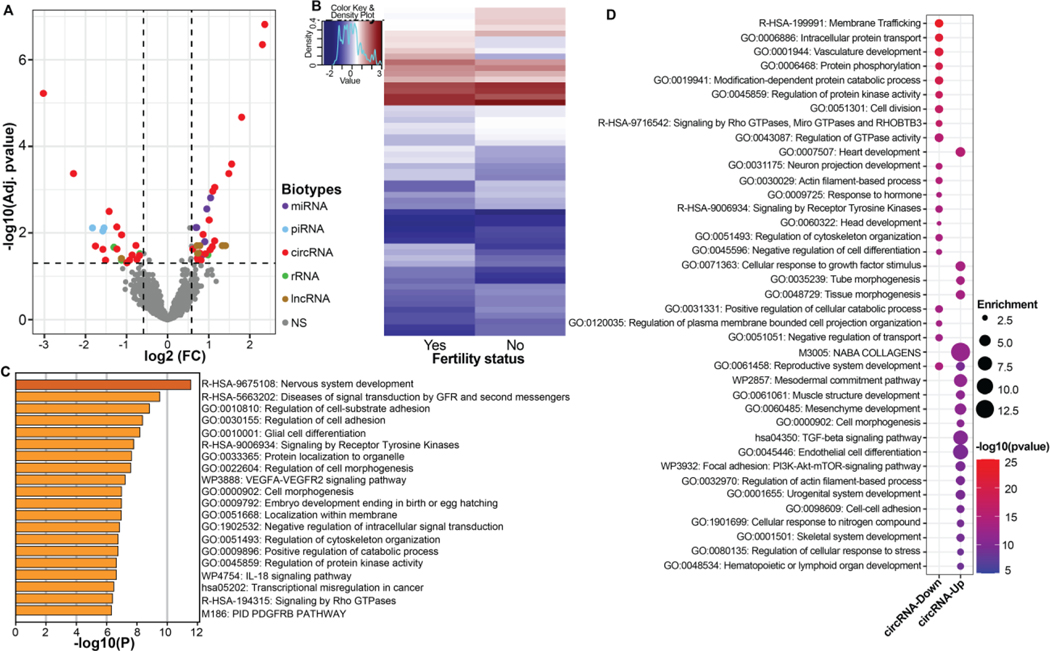
Using a cutoff of fold change ≥ 1.5 and FDR<0.05, our analysis identified a total of 57 differentially expressed ncRNA biotypes among men with poor semen quality, 35 (61%) of which were upregulated. Of these, most (n = 34) were circRNA species while others were 6 miRNAs, 4 piRNAs, 6 rRNA and 7 lncRNAs (Figure 3A & Table S1). A heatmap of all the differentially expressed ncRNAs is shown in Figure 3B with profiles showing distinct expression levels between the two groups. Next, we identified potential gene targets of our differentially expressed ncRNAs and performed functional analyses. All differentially expressed miRNAs were upregulated in the poor semen quality group and targeted genes (n = 824) that were enriched in pathways related to development including cell morphogenesis, VEGFA-VEGFR2 signaling pathway and embryo development ending in birth or egg hatching (Figure 3C).
Figure 3: ncRNA expression levels by semen quality status (Normal/poor, referent: normal).
(A) Volcano plot of differential expression of all identified non-coding RNAs biotypes. Dashed lines represent cutoff points (horizontal line = FDR of 0.05; vertical lines = absolute value of log2 fold change (FC) ≥ 0.585). Colored and grayed datapoints are differentially expressed and non-differentially expressed ncRNAs, respectively. NS represents non-significant (B) Heatmaps of all differentially expressed ncRNAs (C) Barchart of ontology analysis of differentially expressed miRNA gene targets. Bar colors and heights represent enrichments and pvalues respectively. (D) Dotplot of ontology analysis of differentially expressed circRNA gene targets. *GFR: growth factor receptors.
Of the 34 differentially expressed spEV circRNAs, 19 were up-regulated in men with poor semen quality and were associated with 78 miRNAs and subsequently 706 conserved predicted genes. Whereas the 15 down-regulated differentially expressed circRNAs were associated with 153 unique miRNAs and subsequently 1,384 conserved predicted genes. The top enriched ontology terms using the conserved predicted genes of downregulated circRNAs included membrane trafficking, intracellular protein transport, vasculature development, and cell division; whereas top enriched terms for upregulated circRNA targets were heart development, cellular response to growth factor stimulus, tube and tissue morphogenesis, endothelial cell differentiation and Focal adhesion: PI3k-Akt-mTOR-signaling pathway (Figure 3A & 3D).
For the four differentially expressed piRNAs, three were downregulated in men with poor semen quality and overlapped with genomic features such as protein coding genes (DLG2, PID1), lncRNA (CTD-2126E3.4) and pseudogenes (RP11–1315F8.2). Of the seven differentially expressed lncRNAs, six were upregulated in men poor semen quality and were annotated to lncRNA (LINC02775), as well as protein coding gene (FARS2) and pseudogenes (RP11–313J2.1, RPSAP68). Lastly, four of the six differentially expressed rRNAs were annotated to small ribosomal subunit regions (Figure 3A & Table S1).
DISCUSSION
In the present study, we conducted comprehensive ncRNA profiling of spEVs obtained from ejaculates of male partners of couples seeking infertility treatment. Sequenced reads were sequentially mapped to combined biotype-specific small and long ncRNA transcriptomes curated in standard databases. spEVs were found to be enriched in various classes of ncRNAs with 58% of the total reads mapping to different biotypes including miRNA (29.8%), piRNA (4.3%), rRNA (9.2), tRNA (3.0%), circRNA (9.4%) and lncRNA fragments (2.2%). The remaining reads (42%) were mapped to “other” ncRNAs, which were predominantly fragments of pseudogenes and NMD. Next, we identified a total of 57 differentially expressed spEV ncRNA biotypes in men with poor semen quality (n = 32) compared to normal controls (n = 59). The majority (n = 34, 59.6%) were circRNA species while other biotypes were 6 miRNAs, 4 piRNAs, 6 rRNAs and 7 lncRNAs. Ontology analysis of gene targets of differentially expressed circRNAs and miRNAs revealed enrichments in functional terms related cell communications and development.
Inter-cellular communication has been observed between EVs secreted from various epithelial cells in male reproductive system and sperm during maturation (36, 37) and ejaculation (38). In doing so, EVs deliver their protein and RNA cargo to modify the molecular composition of sperm (12, 37). In the present study, we observed that spEVs were composed of different ncRNAs biotypes with little variability in relative proportions across participants, suggesting a consistent loading of spEV RNA cargo. These observations were consistent with previous findings (6) that showed the presence of distinctive ncRNA repertoire in spEVs obtained from a cohort of healthy donors.
Importantly, we report for the first time that spEV ncRNAs mapped to known circRNA transcripts. circRNAs are a novel class of competing endogenous noncoding RNAs with covalently closed loop structures without 5′ caps and 3′ poly tails. They have recently attracted scientific attention due to their stability, and gene regulatory function by miRNA sponging (32, 39, 40). In mouse, the expression of circRNAs across testicular spermatogenic cell types has been documented (41). Also, in humans, circRNAs have not only been reported in ejaculated spermatozoa, they were differentially localization in both head and tail relative to intact sperm (42). This indicates the potential role of circRNAs across multiple functional processes during spermatogenesis. Interestingly, testes-derived circRNAs were also observed in seminal plasma (43). While testes-derived circRNA stability in seminal plasma has been largely attributed to its circular morphology and protein binding (43), our observation of circRNAs in spEVs suggests additional mechanism for the protection of circRNAs with potential transfer to spermatozoa during epididymal maturation, storage and ejaculation. Overall, our data indicate that spEVs are more enriched with non-coding RNA biotypes, including less-commonly reported species like circRNAs, pseudogenes, and NMD fragments than previously reported (6).
Further understanding of the contribution on spEV is expected to provide new avenues of biomarker development for reproductive phenotypes such as male infertility (4, 44). Previous studies have shown sperm uptake of spEVs carrying regulatory ncRNAs results in acquisition of sperm motility and fertilization capabilities (38) with the potential to affect downstream embryo development and offspring phenotypes (12, 45, 46). For example, a panel of spEV lncRNA fragments (16) has been proposed to predict the absence of mature spermatozoa in testicular tissues. Similarly, based on sperm motility criterion, other studies have reported differential expression levels of spEV piRNA (5) and lncRNA (14) between normal semen quality and poor semen quality patients.
In our study, we compared spEV ncRNA profiles of men with poor semen quality compared to normal controls and found a total of 57 differentially expressed ncRNAs, including 6 miRNAs, 4 piRNAs, 34 circRNAs, 6 rRNAs, and 7 lncRNAs. Ontology analysis of differentially expressed miRNAs showed enrichment in functional terms related to development including cell morphogenesis and embryo development ending in birth or hatching, suggesting that the identified miRNA species likely play a functional role beyond fertilization. It must be noted that the identified differentially expressed miRNAs, piRNAs and lncRNAs did not overlap with previous studies that evaluated spEV ncRNA biomarkers of infertility (5, 14, 15, 47), which may be due to technical variabilities such as study design, library preparations, and sequence mapping. For example, Abu-Halima et al (47) reported 36 miRNAs to be differentially expressed among oligoasthenozoospermia men compared to men with normal semen parameters by microarray. However, only nine miRNAs were present in our study and had between 0.23 to 57 RPM across 91 participants, none of which passed our filtering method of ≥ 1 RPM in all samples. This highlights issues of comparability between sequencing and array-based methods, the latter which are more prone to false positives (48) especially when combined with limited sample sizes, suggesting the need for standardization of methods.
Intriguingly, the majority (60%) of the identified differentially expressed ncRNAs in our study were circRNAs, which were found to be both up- and down-regulated in men with poor semen quality. Similar findings were reported by Manfrevola, Chioccarelli, Cobellis, Fasano, Ferraro, Sellitto, Marella, Pierantoni and Chianese (49) who utilized micro-array technique and identified >1,400 differentially expressed circRNAs in poor quality sperm relative to motile spermatozoa fraction with expression levels proceeding in both directions. However, these circRNAs (49) were not publicly available for comparison with our findings. While the role of EV circRNAs (33) in carcinogenesis has been well documented, the function of spEV-derived circRNAs in reproductive phenotypes is limited. Here, our differentially expressed circRNAs targeted genes that were enriched in functional terms related to cellular communication and development. Consistent with this study, Chioccarelli, Manfrevola, Ferraro, Sellitto, Cobellis, Migliaccio, Fasano, Pierantoni and Chianese (42) also observed enrichment of up-and down-regulated EV-mediated circRNA gene targets in mTOR, focal adhesion and PI3k-Akt-pathways in sperm with poor quality selected based on motility and morphology. PI3k-Akt-mTOR signaling is not only important in regulation of spermatogenesis (50), it has been demonstrated to play critical developmental roles ranging from fertilization to birth (51). Given previous observation showing physical interaction between both human and mice sperm circRNAs and pre-fertilized oocyte miRNAs (40), our data indicate that altered expression levels of spEV circRNAs in men with poor semen quality likely target cellular processes which are relevant in reproductive outcomes.
Our study has several notable strengths. To our knowledge, this is the first study to utilize small RNA sequencing method to uncover the comprehensive ncRNA biotypes present in spEVs from a large cohort (n = 91) of men seeking IVF treatment. Also, we found several biotypes in spEVs that were not identified previously including circRNAs, pseudogenes and NMD, which may be important sncRNA markers for male fertility and reproductive outcomes. Furthermore, our prospective clinical cohort design will allow us to further characterize the relevance of these differentially expressed sncRNAs on fertilization, embryo development and live birth outcomes.
We note, however, that this study has some limitations. Since our focus was to compare the spEV ncRNA profiles of men with abnormal semen parameters to those with normal parameters in IVF population, we only recruited men that were part of a couple seeking infertility treatment. As such, our study may limit the generalizability of findings to other populations, such as those in the general population with proven fertility status. It must be noted that spEVs prior to RNA extraction were not treated with RNAse, thereby we cannot completely rule out the introduction of some non-EV RNA in our extractions. However, RNAses found in seminal plasma are highly active even on double-stranded RNA (52), which will minimize the presence of non-EV RNA. Furthermore, exoRNeasy workflow incorporates washing steps to remove non-EV RNA contamination (20) and previous studies have shown that RNAse pretreatment of plasma EVs did not appreciable change total RNA yield of sample processed with exoRNeasy as compared to ultracentrifugation isolation (53) and did not significantly alter detection of RNA targets via PCR. Also, we recognize that EV subtypes including microvesicles, exosomes and apoptotic bodies have different particle sizes, biogenesis, and cargo (54). Thus, we used the term “EV” throughout this paper as the sncRNA payloads of specific EV subtypes were not evaluated. However, our objective was to determine the total spEV ncRNA payload rather than ncRNA profiles of different EV subtypes. We classified participating males into groups based on single diagnostic semen analysis. This may be a limitation in our study due to temporal variability in semen quality (55) and biomarker measurements. Future studies should evaluate intra-individual variation of spEV snRNAs from consecutive semen samples to determine their stability over time.
In summary, this study revealed the presence of a diverse repertoire of spEV RNA cargo including small and long ncRNA biotypes such as miRNA, piRNA, tRNA, rRNA circRNA and lncRNA fragments. Differential expression of ncRNAs, the majority of which were circRNAs, were found in men with poor semen parameters compared to normal controls and were enriched in pathways related to early development and cell communication. Thus, our findings expand the ncRNA payloads of spEVs and suggest that these ncRNAs may be important molecular markers of male infertility status. Future studies should investigate the role of spEV ncRNAs in sperm function as it relates to couple-level characteristics and reproductive outcomes.
Supplementary Material
Funding:
This work was funded in part by grants from the National Institute of Environmental Health Sciences, National Institutes of Health (R01 ES030942; PI: J.R.P. and P30 ES020957).
Footnotes
Competing interest: The authors have no conflict of interest to disclose.
References
- 1.WHO. Mother or nothing: the agony of infertility. Bull World Health Organ. 2010;88(12):881–2. doi: doi: 10.2471/BLT.10.011210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahmasbpour E, Balasubramanian D, Agarwal A. A multi-faceted approach to understanding male infertility: gene mutations, molecular defects and assisted reproductive techniques (ART). Journal of Assisted Reproduction and Genetics. 2014;31(9):1115–37. doi: 10.1007/s10815-014-0280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell MJ, Lotti F, Baldi E, Schlatt S, Festin MPR, Björndahl L, Toskin I, Barratt CLR. Distribution of semen examination results 2020 – A follow up of data collated for the WHO semen analysis manual 2010. Andrology. 2021;9(3):817–22. doi: 10.1111/andr.12983. [DOI] [PubMed] [Google Scholar]
- 4.Ayaz A, Houle E, Pilsner JR. Extracellular vesicle cargo of the male reproductive tract and the paternal preconception environment. Syst Biol Reprod Med. 2021;67(2):103–11. Epub 2021/02/26. doi: 10.1080/19396368.2020.1867665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong Y, Wu Y, Zhang J, Yu C, Shen L, Chen H, Chen L, Zhou X, Gao F. Decreased piRNAs in Infertile Semen Are Related to Downregulation of Sperm MitoPLD Expression. Front Endocrinol (Lausanne). 2021;12:696121. Epub 2021/07/31. doi: 10.3389/fendo.2021.696121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, Hladik F. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42(11):7290–304. Epub 2014/05/20. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y, Liang A, He Y, Li Z, Li Z, Wang G, Sun F. Proteomic analysis of seminal extracellular vesicle proteins involved in asthenozoospermia by iTRAQ. Molecular Reproduction and Development. 2019;86(9):1094–105. doi: 10.1002/mrd.23224. [DOI] [PubMed] [Google Scholar]
- 8.Yang C, Guo WB, Zhang WS, Bian J, Yang JK, Zhou QZ, Chen MK, Peng W, Qi T, Wang CY, Liu CD. Comprehensive proteomics analysis of exosomes derived from human seminal plasma. Andrology. 2017;5(5):1007–15. doi: 10.1111/andr.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, Rando OJ. Small RNAs Are Trafficked from the Epididymis to Developing Mammalian Sperm. Developmental Cell. 2018;46(4):481–94.e6. doi: 10.1016/j.devcel.2018.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natt D, Ost A. Male reproductive health and intergenerational metabolic responses from a small RNA perspective. J Intern Med. 2020;288(3):305–20. Epub 2020/05/18. doi: 10.1111/joim.13096. [DOI] [PubMed] [Google Scholar]
- 11.Reilly JN, McLaughlin EA, Stanger SJ, Anderson AL, Hutcheon K, Church K, Mihalas BP, Tyagi S, Holt JE, Eamens AL, Nixon B. Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Scientific Reports. 2016;6(1):31794. doi: 10.1038/srep31794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, Rando OJ. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–6. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, Hauser R, Krawetz SA, Pilsner JR. Environmental Susceptibility of the Sperm Epigenome During Windows of Male Germ Cell Development. Current Environmental Health Reports. 2015;2(4):356–66. doi: 10.1007/s40572-015-0067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu H, Xu D, Wang P, Sun W, Xue X, Hu Y, Xie C, Ma Y. RNA-sequencing and bioinformatics analysis of long noncoding RNAs and mRNAs in the asthenozoospermia. Bioscience Reports. 2020;40(7). doi: 10.1042/bsr20194041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barceló M, Mata A, Bassas L, Larriba S. Exosomal microRNAs in seminal plasma are markers of the origin of azoospermia and can predict the presence of sperm in testicular tissue. Human Reproduction. 2018;33(6):1087–98. doi: 10.1093/humrep/dey072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Y, Yao J, Zhang X, Chen J, Gao Y, Zhang C, Chen H, Wang Z, Zhao Z, Chen W, Lv L, Li Y, Gao F, Xie M, Zhang J, Zhao L, Wang Z, Liang X, Sun X, Zou X, Deng C, Liu G. A panel of extracellular vesicle long noncoding RNAs in seminal plasma for predicting testicular spermatozoa in nonobstructive azoospermia patients. Human Reproduction. 2020;35(11):2413–27. doi: 10.1093/humrep/deaa184. [DOI] [PubMed] [Google Scholar]
- 17.Guzick DS, Overstreet JW, Factor-Litvak P, Brazil CK, Nakajima ST, Coutifaris C, Carson SA, Cisneros P, Steinkampf MP, Hill JA, Xu D, Vogel DL. Sperm Morphology, Motility, and Concentration in Fertile and Infertile Men. New England Journal of Medicine. 2001;345(19):1388–93. doi: 10.1056/nejmoa003005. [DOI] [PubMed] [Google Scholar]
- 18.Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, Behre HM, Haugen TB, Kruger T, Wang C, Mbizvo MT, Vogelsong KM. World Health Organization reference values for human semen characteristics. Human reproduction update. 2010;16(3):231–45. Epub 2009/11/26. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 19.Kruger TF, Acosta AA, Simmons KF, Swanson RJ, Matta JF, Oehninger S. Predictive value of abnormal sperm morphology in in vitro fertilization. Fertility and Sterility. 1988;49(1):112–7. doi: 10.1016/s0015-0282(16)59660-5. [DOI] [PubMed] [Google Scholar]
- 20.Enderle D, Spiel A, Coticchia CM, Berghoff E, Mueller R, Schlumpberger M, Sprenger-Haussels M, Shaffer JM, Lader E, Skog J, Noerholm M. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLOS ONE. 2015;10(8):e0136133. doi: 10.1371/journal.pone.0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews S. FastQC: a quality control tool for high throughput sequence data.2010.
- 22.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17(1):10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 23.Rozowsky J, Kitchen RR, Park JJ, Galeev TR, Diao J, Warrell J, Thistlethwaite W, Subramanian SL, Milosavljevic A, Gerstein M. exceRpt: A Comprehensive Analytic Platform for Extracellular RNA Profiling. Cell Systems. 2019;8(4):352–7.e3. doi: 10.1016/j.cels.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen W, Le S, Li Y, Hu F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLOS ONE. 2016;11(10):e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biology. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler R, Mahaffey S, Rossi S, Calin GA, Bemis L, Theodorescu D. The multiMiR R package and database: integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Research. 2014;42(17):e133-e. doi: 10.1093/nar/gku631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou W-Y, Cai Z-R, Liu J, Wang D-S, Ju H-Q, Xu R-H. Circular RNA: metabolism, functions and interactions with proteins. Molecular Cancer. 2020;19(1). doi: 10.1186/s12943-020-01286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng X, Lin X, Zhang Y, Li Q, Guo Y, Fang C, Wang H. Exosomal circular RNA sorting mechanisms and their function in promoting or inhibiting cancer (Review). Oncology Letters. 2020. doi: 10.3892/ol.2020.11449. [DOI] [PMC free article] [PubMed]
- 34.Wang S, Dong Y, Gong A, Kong H, Gao J, Hao X, Liu Y, Wang Z, Fan Y, Liu C, Xu W. Exosomal circRNAs as novel cancer biomarkers: Challenges and opportunities. International Journal of Biological Sciences. 2021;17(2):562–73. doi: 10.7150/ijbs.48782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications. 2019;10(1). doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rimmer MP, Gregory CD, Mitchell RT. The transformative impact of extracellular vesicles on developing sperm. Reproduction and Fertility. 2021;2(3):R51–R66. doi: 10.1530/raf-20-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, Rando OJ. Small RNAs Are Trafficked from the Epididymis to Developing Mammalian Sperm. Dev Cell. 2018;46(4):481–94 e6. Epub 2018/07/31. doi: 10.1016/j.devcel.2018.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murdica V, Giacomini E, Alteri A, Bartolacci A, Cermisoni GC, Zarovni N, Papaleo E, Montorsi F, Salonia A, Vigano P, Vago R. Seminal plasma of men with severe asthenozoospermia contain exosomes that affect spermatozoa motility and capacitation. Fertil Steril. 2019;111(5):897–908 e2. Epub 2019/04/29. doi: 10.1016/j.fertnstert.2019.01.030. [DOI] [PubMed] [Google Scholar]
- 39.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–13. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragusa M, Barbagallo D, Chioccarelli T, Manfrevola F, Cobellis G, Di Pietro C, Brex D, Battaglia R, Fasano S, Ferraro B, Sellitto C, Ambrosino C, Roberto L, Purrello M, Pierantoni R, Chianese R. CircNAPEPLD is expressed in human and murine spermatozoa and physically interacts with oocyte miRNAs. RNA Biology. 2019;16(9):1237–48. doi: 10.1080/15476286.2019.1624469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X, Han M, Cheng L, Chen J, Zhang Z, Shen T, Wang M, Wen B, Ni T, Han C. Expression dynamics, relationships, and transcriptional regulations of diverse transcripts in mouse spermatogenic cells. RNA Biology. 2016;13(10):1011–24. doi: 10.1080/15476286.2016.1218588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chioccarelli T, Manfrevola F, Ferraro B, Sellitto C, Cobellis G, Migliaccio M, Fasano S, Pierantoni R, Chianese R. Expression Patterns of Circular RNAs in High Quality and Poor Quality Human Spermatozoa. Front Endocrinol (Lausanne). 2019;10:435. Epub 2019/07/25. doi: 10.3389/fendo.2019.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong W-W, Li H-M, Qing X-R, Huang D-H, Li H-G. Identification and characterization of human testis derived circular RNAs and their existence in seminal plasma. Scientific Reports. 2016;6(1):39080. doi: 10.1038/srep39080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vickram AS, Srikumar PS, Srinivasan S, Jeyanthi P, Anbarasu K, Thanigaivel S, Nibedita D, Jenila Rani D, Rohini K. Seminal exosomes - An important biological marker for various disorders and syndrome in human reproduction. Saudi J Biol Sci. 2021;28(6):3607–15. Epub 2021/06/15. doi: 10.1016/j.sjbs.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan JC, Morgan CP, Adrian Leu N, Shetty A, Cisse YM, Nugent BM, Morrison KE, Jašarević E, Huang W, Kanyuch N, Rodgers AB, Bhanu NV, Berger DS, Garcia BA, Ament S, Kane M, Neill Epperson C, Bale TL. Reproductive tract extracellular vesicles are sufficient to transmit intergenerational stress and program neurodevelopment. Nature Communications. 2020;11(1). doi: 10.1038/s41467-020-15305-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng G, Peng H, Zhang X, Zhang Y, Qian J, Duan E, Zhai Q, Zhou Q. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. SCIENCE. 2016;351(6271):397–400. doi: DOI: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 47.Abu-Halima M, Ludwig N, Hart M, Leidinger P, Backes C, Keller A, Hammadeh M, Meese E. Altered micro-ribonucleic acid expression profiles of extracellular microvesicles in the seminal plasma of patients with oligoasthenozoospermia. Fertility and Sterility. 2016;106(5):1061–9.e3. doi: 10.1016/j.fertnstert.2016.06.030. [DOI] [PubMed] [Google Scholar]
- 48.Nersisyan S, Shkurnikov M, Poloznikov A, Turchinovich A, Burwinkel B, Anisimov N, Tonevitsky A. A Post-Processing Algorithm for miRNA Microarray Data. International Journal of Molecular Sciences. 2020;21(4):1228. doi: 10.3390/ijms21041228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manfrevola F, Chioccarelli T, Cobellis G, Fasano S, Ferraro B, Sellitto C, Marella G, Pierantoni R, Chianese R. CircRNA Role and circRNA-Dependent Network (ceRNET) in Asthenozoospermia. Front Endocrinol (Lausanne). 2020;11:395. Epub 2020/08/06. doi: 10.3389/fendo.2020.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Busada JT, Chappell VA, Niedenberger BA, Kaye EP, Keiper BD, Hogarth CA, Geyer CB. Retinoic acid regulates Kit translation during spermatogonial differentiation in the mouse. Dev Biol. 2015;397(1):140–9. Epub 2014/12/03. doi: 10.1016/j.ydbio.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Land SC, Scott CL, Walker D. mTOR signalling, embryogenesis and the control of lung development. Seminars in Cell & Developmental Biology. 2014;36:68–78. doi: 10.1016/j.semcdb.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 52.Roccode DP, Salvatore S, Enzo L, Massimo L. A ribonuclease from human seminal plasma active on double -stranded RNA. Biochim Biophys Acta. 1984;14;788(3):356–63. doi: 10.1016/0167-4838(84)90049-9. [DOI] [PubMed] [Google Scholar]
- 53.Gutierrez Garcia G, Galicia Garcia G, Zalapa Soto J, Izquierdo Medina A, Rotzinger-Rodriguez M, Casas Aguilar GA, Lopez Pacheco CP, Aguayo A, Aguilar-Hernandez MM. Analysis of RNA yield in extracellular vesicles isolated by membrane affinity column and differential ultracentrifugation. PLoS One. 2020;15(11):e0238545. Epub 2020/11/07. doi: 10.1371/journal.pone.0238545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doyle L, Wang M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells. 2019;8(7):727. doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiu Y-H, Edifor R, Rosner BA, Nassan FL, Gaskins AJ, Mínguez-Alarcón L, Williams PL, Tanrikut C, Hauser R, Chavarro JE. What Does a Single Semen Sample Tell You? Implications for Male Factor Infertility Research. American Journal of Epidemiology. 2017;186(8):918–26. doi: 10.1093/aje/kwx169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.