Abstract
Translation is a key determinant of gene expression and an important biotechnological engineering target. In bacteria, 5′-untranslated region (5′-UTR) and coding sequence (CDS) are well-known mRNA parts controlling translation and thus cellular protein levels. However, the complex interaction of 5′-UTR and CDS has so far only been studied for few sequences leading to non-generalisable and partly contradictory conclusions. Herein, we systematically assess the dynamic translation from over 1.2 million 5′-UTR-CDS pairs in Escherichia coli to investigate their collective effect using a new method for ultradeep sequence-function mapping. This allows us to disentangle and precisely quantify effects of various sequence determinants of translation. We find that 5′-UTR and CDS individually account for 53% and 20% of variance in translation, respectively, and show conclusively that, contrary to a common hypothesis, tRNA abundance does not explain expression changes between CDSs with different synonymous codons. Moreover, the obtained large-scale data provide clear experimental evidence for a base-pairing interaction between initiator tRNA and mRNA beyond the anticodon-codon interaction, an effect that is often masked for individual sequences and therefore inaccessible to low-throughput approaches. Our study highlights the indispensability of ultradeep sequence-function mapping to accurately determine the contribution of parts and phenomena involved in gene regulation.
INTRODUCTION
Translation is a key step of gene expression and an important engineering target in synthetic biology. To this end, genetic parts that influence translation are modified to alter absolute and relative expression levels to engineer biosystems through control of individual genes, pathways, and even entire metabolic networks (1–3). In prokaryotes, initiation of translation is the rate-limiting step in the translational process, during which ribosomes assemble on the mRNA to start the templated elongation of the nascent polypeptide (4–8). At the onset of this step, the 30S ribosomal subunit attaches to the ribosome binding site (RBS) in the 5′-untranslated region (5′-UTR) upstream of the coding sequence (CDS). The 3′-end of the 16S rRNA hybridises with the Shine-Dalgarno (SD) motif, a conserved five to eight nucleotide (nt) sequence located upstream of the start codon, which facilitates translation (9–13). However, since Shine and Dalgarno's discovery in 1973 (9), various additional influencing factors and sequence determinants affecting translation initiation were identified. For example, the distance between SD motif and start codon, the type of start codon, and interactions between distant 5′-UTR parts and the ribosome play important roles (14–22). Remarkably, in some cases SD-like motifs are not required for translation, an observation hinting at the existence of other mechanisms besides ‘canonical’ translation initiation (23–27). Further, the influence of mRNA secondary structures was studied under the hypothesis that the required unfolding of such structures during translation initiation might decrease expression (15,21,28–41). For example, stable secondary structures around the start codon were found to hinder translation, while structures further up- or downstream had less pronounced effects (36).
Moreover, codon usage was found to influence translation. Genome-wide analyses of Escherichia coli and other organisms revealed an overrepresentation of rare codons in the first five to ten triplets of the CDS in native genes, and their occurrence in this region was found to coincide with high expression (30,42–45). These observations led to two different hypotheses that differ fundamentally in terms of the underlying causality. The first hypothesis is related to the fact that cellular tRNA concentrations correlate with the occurrence frequency of their cognate codons (46–48). It was postulated that rare codons (with low-abundant cognate tRNAs) may have been evolutionary selected for within the N-terminal CDS to slow down early translation elongation and reduce premature termination due to clashing ribosomes (39,49–59). These ‘translational ramps’ were postulated to be causally responsible for elevated expression of genes rich in rare codons at the CDS’s 5′-end. As an alternative explanation independent of tRNA abundance, a second hypothesis has been proposed based on the fact that many rare codons are (or happen to be) AT-rich (30,54). Their occurrence is therefore associated with a lower tendency to form stable mRNA secondary structures (30,34,44), which are known to hinder translation initiation.
In the context of these two hypotheses, several studies have been conducted to investigate the impact of codon usage on expression focussing either on the N-terminal codons alone (30,34,38,44,45) or the entire CDS (30,60) while applying different metrics of codon usage such as the codon adaptation index (CAI) (42), the frequency of ‘optimal’ codons pairing with the most abundant tRNAs (46), and the tRNA adaptation index (tAI) (61), as discussed in detail elsewhere (45,62–64). Remarkably, while there is clear evidence for a high degree of interactivity between 5′-UTR and CDS (15,21,34,36–38), these two mRNA parts were handled separately in these studies: commonly only one of the two parts (either 5′-UTR or CDS) was diversified at a time, and systematic testing of larger numbers of 5′-UTR-CDS combinations to assess their interaction was not performed (16,21,34). Thus, due to the strong interdependence, the measured effects could not be clearly assigned to individual sequence parameters, and thus their contribution to overall expression likely not accurately quantified (15,21,34,36–38). Moreover, many early studies relied on experimental testing of few sequences (usually <100 variants) due to limitations in experimental throughput or library generation (note that the CDS cannot be freely mutated, since amino acid substitution may result in change or loss of reporter protein activity). Although these valuable previous studies critically contributed to our understanding of translation initiation, the underlying empiric efforts have so far not allowed for the establishment of generalisable rules and means to quantitatively measure the effects of sequence parameters, which in some cases even led to contradictory conclusions (38,39,53,54,56,65–69).
For example, the question of whether tRNA abundance has a significant impact on translation initiation or whether the observed effect is caused by mRNA secondary structures alone remains inconclusively answered. Enabled by advances in DNA synthesis and sequencing, some recent works assessed larger numbers of 5′-UTRs or CDSs, again only diversifying one of the two sequence parts at a time (21,34,37,39,70,71). In a recent study, Arkin et al. combined full-factorial in silico design with DNA synthesis on arrays to evaluate the principles of sequence design for translation in a systematic manner (38). They tested synthetic sequences combining a single bicistronic 5′-UTR (16,72) with 244 000 CDSs using fluorescence-activated cell sorting combined with next-generation sequencing (NGS). Several relevant sequence parameters such as AT-content, codon usage, and mRNA folding were varied and combined in a statistically full-factorial manner. This was achieved using a sophisticated modular design approach based on a priori hypotheses, which, however, bears the risk of introducing ‘user-borne’ bias.
Herein, we describe our efforts to further deepen our knowledge about the impact of different mRNA parts and sequence parameters on translation initiation with the goal to assess and accurately quantify their effect. We combine randomly generated 5′-UTRs and CDSs following different assembly strategies to obtain libraries of random, combinatorial and full-factorial 5′-UTR-CDS combinations. Using a recently developed method for ultradeep sequence-function mapping (71), we dynamically assess translation of more than 1.2 million 5′-UTR-CDS pairs in more than 9.5 million sequence-function data points and different genetic backgrounds. The extremely high throughput and the modular assembly strategy applied herein allow us to systematically disentangle and assess individual and combined effects of 5′-UTR and CDS, and to quantify the contribution of various sequence parameters including individual bases and positions, predicted mRNA folding energies, 16S-rRNA hybridisation, and codon usage.
MATERIALS AND METHODS
Reagents
All chemicals were obtained from Sigma Aldrich (Buchs, Switzerland). Restriction enzymes were obtained from New England Biolabs (Ipswich, USA). PCR was performed using Q5 DNA polymerase from New England Biolabs (Ipswich, USA). Oligonucleotides (Supplementary Table S1) were obtained from Microsynth AG (Balgach, Switzerland). All primers containing degenerate bases were ordered PAGE-purified. Custom duplex DNA adapters and gene fragments were obtained from Integrated DNA Technologies (Leuven, Belgium). Plasmid DNA for cloning was extracted with the ZR Plasmid Miniprep kit from Zymo research (Irvine, USA). Plasmid DNA from cultures used for subsequent sample preparation for NGS was extracted with the QIAprep Spin Miniprep kit from Qiagen (Hilden, Germany). Gel extraction of DNA was performed using Zymoclean Gel DNA Recovery Kits from Zymo research (Irvine, USA).
Strains, cultivation conditions and growth analysis
Escherichia coli TOP10 ΔrhaA (l-rhamnose isomerase) was used throughout the study. The generation of this rhamnose utilisation-deficient strain is described elsewhere (71). For experiments with plasmid-borne variants of tRNAfMet, the strain E. coli TOP10 ΔrhaA ΔmetZWV was generated by additional replacement of the chromosomal metZWV locus with a spectinomycin resistance cassette using the method described by Datsenko and Wanner (73). The spectinomycin resistance cassette was PCR-amplified from a commercial gene fragment (Suppl. Note 1) using primers p1 and p2 (Supplementary Table S1) to generate the linear fragment for transformation complementary to 41 bp both up- and downstream of the chromosomal metZWV locus. Transformants were verified for successful integration by colony PCR using primers p3 and p4 and subsequent Sanger sequencing. The exact genotypes of both E. coli strains are provided in Supplementary Table 2. E. coli cells were generally cultivated in lysogeny broth (LB) supplemented with 50 mg l−1 kanamycin, 50 mg l−1 streptomycin, and 10 g l−1d-glucose for repression of the rhamnose-inducible promoter where appropriate. 15 g l−1 agar were added for plate cultures. Cells were grown at 37°C in an incubator (plates) or shaking incubator at 200 rpm (shake flasks cultivations). Doubling times of strains with different tRNAfMet variants were determined in biological triplicate cultures as follows. E. coli TOP10 ΔrhaA (‘WT’) and E. coli TOP10 ΔrhaA ΔmetZWV (‘ΔmetZWV’) were transformed with pSEVA361 (empty vector (74)), ptRNAfMet-A37, ptRNAfMet-A37G or ptRNAfMet-A37U, respectively (Supplementary Table S3). Transformants of each strain bearing sequence-verified plasmids were used to inoculate an overnight pre-culture in LB (34 mg l−1 chloramphenicol, 12.5 mg l−1 spectinomycin for ΔmetZWV). After, 120 ml main cultures in baffled shake flasks (1 l) were inoculated to a starting OD600 of 0.01 and incubated shaking (37°C, 200 rpm). The optical density at 600 nm (OD600) was measured in intervals of 15–30 min and doubling times were determined by dividing ln(2) by the specific growth rate during exponential growth.
Plasmid and library construction
A list of plasmids used in this study is provided in Supplementary Table 3. Plasmids were constructed by conventional restriction-ligation cloning. To enable facile library cloning, plasmid pASPIre4 (Supplementary Figure S1) was generated as a derivative of the previously published pASPIre3 (71). pASPIre4 additionally contains a SpeI restriction site within the CDS of bxb1 to enable diversification of the 5′-UTR and codons 2–16 of bxb1.
Library inserts were generated by PCR with degenerate primers to diversify the respective regions and inserted into the pASPIre4 backbone thereafter. The fully randomised 5′-UTR-CDS library was generated via PCR using pASPIre4 as template and primers p5 and p6. After, the PCR product and pASPIre4 were digested with SpeI and PstI (37°C, 3 h), gel purified and ligated (16°C, T4 ligase, overnight). The ligation mixture was purified and used to electroporate freshly prepared E. coli TOP10 ΔrhaA cells (75). After 60 min recovery at 37°C in LB with 10 g l−1d-glucose, transformants were plated in different dilutions for colony counting on LB agar plates. After overnight incubation (37°C), 10 ml LB were added to the plates and approximately 400 000 colonies were scraped off with a spatula. Glycerol was added to the cell suspension to a final concentration of 150 g l−1 and OD600 of the glycerol stock was adjusted to 5.0 before freezing of aliquots in liquid nitrogen and storage at −80°C. This pool of clones was designated Librandom, and the corresponding plasmid architecture was termed pASPIre4lib (Supplementary Figure S2). For the uASPIre with mutated tRNAfMet variants, a glycerol stock of Librandom was plated on LB agar and plasmid DNA of approximately 50 000 clones was extracted and subsequently used to transform E. coli bearing the respective plasmids for the expression of tRNAfMet (see below).
Combinatorial and full factorial libraries combining different 5′-UTRs and CDSs were generated in a stepwise procedure as illustrated in Supplementary Figure S3. First, 5′-UTR and CDS half-libraries (Supplementary Figures S4 and S5) were cloned separately as described above. The 5′-UTR half-library was generated by PCR with primers p5 and p7 on pASPIre4 as template and subsequently inserted into the pASPIre4 backbone using PstI and NotI. Primer p7 introduces degeneracy in the 5′-UTR and a BbsI site between the randomised 5′-UTR and the NotI site (Supplementary Figure S3). The CDS half-library was generated by PCR with primers p8 and p9 on pASPIre4 as template and inserted into the pASPIre4 backbone using PstI and NotI. Primer p8 introduces degeneracy in the CDS and a BbsI site between the CDS and the PstI site (Supplementary Figure S3). Transformants of both half-libraries were plated separately in various dilutions. Depending on the libraries to be created afterwards, a desired number of colonies was scraped off with a spatula and plasmid DNA was extracted: for Libcomb1, approximately 1000 colonies of the 5′-UTR half-library and approximately 1000 colonies of the CDS half-library; for Libcomb2, approximately 100 colonies of the 5′-UTR half-library and approximately 10 000 colonies of the CDS half-library. For Libfact, 10 plates of approximately 100 colonies each of the 5′-UTR half-library and ten plates of approximately 100 colonies each of the CDS half-library were scraped off. In a second step, 5′-UTR and CDS half-libraries were combined to generate libraries Libcomb1, Libcomb2 and Libfact. To achieve this, plasmid DNA from the different 5′-UTR half-libraries was PCR-amplified with primers p9 and p10 and the PCR product was digested with BbsI and PvuI. Subsequently, these half-libraries were ligated into plasmid backbones isolated from the individual CDS half-libraries via digestion with PvuI and BbsI. Note that the BbsI type IIS restriction site enables scarless joining of 5′-UTR and CDS half-libraries using ATGC (start codon ATG + first downstream base) as sticky ends for ligation. Libcomb1 (approx. 1000 5′-UTRs combined with approx. 1000 CDSs) and Libcomb2 (approx. 100 5′-UTRs combined with approx. 10 000 CDSs) were used to transform E. coli TOP10 ΔrhaA yielding approximately 1.5 million and 2.3 million colonies, respectively. Libfact was transformed in ten separate batches (10 times 100 5′-UTRs combined with 100 CDSs) yielding ten full-factorial sub-libraries. Each of these should contain a maximum of approximately 10 000 different 5′-UTR-CDS combinations, amongst which theoretically all 5′-UTRs are combined with all CDSs and vice versa. Colonies of these ten sub-libraries were scraped off plates and pooled to equivalent cell densities according to their OD600.
All plasmids for overexpression of tRNAfMet variants are derivatives of pSEVA361. We selected the chromosomal metY locus including promoters and terminators of E. coli TOP10 as a scaffold since it is monocistronic and therefore simpler to mutate compared to the metZWV locus. In this scaffold we introduced an A-to-G point mutation at position 47 of the tRNAfMet to match the sequence of metZWV (note that the metY-derived tRNA differs by this one base from metZWV tRNAs, which are three identical tRNAfMet copies). The resulting monocistronic design was obtained as commercial gene fragment in four versions containing the wild-type base (A) as well as three mutants (C, G and T) at position 37 of tRNAfMet, respectively. The gene fragments were cloned into pSEVA361 (p15A replicon, chloramphenicol resistance) via KpnI and SpeI sites using standard procedures and sequence verified. The resulting plasmids were designated ptRNAfMet-A37, ptRNAfMet-A37C, ptRNAfMet-A37G and ptRNAfMet-A37U (Supplementary Figure S6, Supplementary Table S3) and used to transform E. coli TOP10 ΔrhaA and E. coli TOP10 ΔrhaA ΔmetZWV. Note that transformants of ptRNAfMet-A37C failed to grow and could thus not be included in further experiments. To assess the effect of tRNAfMet mutations, E. coli TOP10 ΔrhaA and E. coli TOP10 ΔrhaA ΔmetZWV bearing the plasmids for tRNA overexpression were each co-transformed with the pool of 50 000 variants of Librandom (see above).
Library cultivation, sample preparation and NGS
The different libraries were separately grown in independent shake flask cultivations. Libfact was cultivated in two biological replicates. Cultivations were conducted in 600 ml LB with 50 mg l−1 kanamycin and, in case of tRNAfMet overexpression, 34 mg l−1 chloramphenicol in 5 l baffled shake flasks. Pre-warmed (37°C) LB was inoculated from glycerol stocks of the respective libraries to an initial OD600 of 0.05. Cultures were grown at 37°C in a shaking incubator at 200 rpm. At an OD600 of approximately 0.5, expression of bxb1 was induced by addition of 2 g l−1l-rhamnose. Samples were drawn at 0, 95, 225, 290, 360 and 480 min after induction and immediately diluted in an excess of ice-cold PBS. Cell suspensions were centrifuged (4000 × g, 10 min, 4°C) and pellets were snap frozen on dry ice. Afterwards, plasmid DNA was extracted and digested with SpeI and NcoI (4 h, 37°C). Target fragments containing the 5′-UTR-CDS region and the Bxb1 recombination substrate were purified via gel electrophoresis (2.5% agarose). Afterwards, duplex DNA adapters for Illumina NGS with sample-specific indices (Supplementary Table S4) were ligated to the target fragments and full-length ligation products were purified via gel electrophoresis (2% MetaPhor agarose, Lonza, Basel, Switzerland). Purity and concentration of extracted fragments were determined using capillary electrophoresis (Fragment Analyser, Agilent) and samples were pooled in equimolar ratios. The pool was spiked with 15% PhiX DNA to increase sample diversity and afterwards sequenced on an Illumina NovaSeq6000 platform (SP flow cell, paired-end reading with at least 30 cycles forward and 100 cycles reverse read). Primary sequencing data were processed with Illumina RTA version V3.4.4 and bcl2fastq to obtain *.fastq files for further processing (see below).
NGS data processing
NGS raw data analysis was performed using a combination of bash and R scripts (R version 4.2.1) running on a Red Hat Enterprise Linux Server (release 7.9).
In brief, forward and reverse reads from *.fastq files were paired. From the forward reads, the identity of the sample-specific index (six options) and the state of the Bxb1 substrate (either unflipped or flipped), were extracted through alignment against all possible twelve combinations allowing a maximum of three mismatches between read and reference to avoid data loss due to sequencing errors. Afterwards, a similar procedure was applied to the reverse reads to identify the second sample-specific index (six options). Next, the sample-specific combination of forward and reverse indices was used to split the data and assign reads to the different libraries and sampling time points (Supplementary Table S5).
Next, NGS reads with a frameshift within the CDS (e.g. due to sequencing errors or undesired mutations) were removed by filtering for the correct positioning of the constant first five nucleotides (ATGCG) of the bxb1 CDS. Then, all 40 randomised nucleotides of 5′-UTR (25 nt) and CDS (each third nucleotide in codons 2–16; in total 15 nt) were extracted for each read, serving as unique identifier for each variant (i.e. 5′-UTR-CDS combination). To rescue reads with sequencing errors in the variable regions (<5% of total reads), a clustering procedure was applied to Libcomb1, Libcomb2 and Libfact to map them to actual (i.e. physically present) variants. This clustering can be applied since the extremely large theoretical sequence space of these variable regions (40 nt randomised; >1023 possible permutations) renders the occurrence of highly similar sequences virtually impossible. First, variants were sorted based on their total read number across all time points. Then, starting with the most frequent variant, all other variants with a Hamming distance of 1 (i.e. maximum of one substitution) were mapped back to this variant. This procedure was continued with the next most abundant variant until all remaining variants were further than one substitution apart from all others. 5′-UTRs and CDSs were treated separately to keep the computational complexity manageable. For Librandom, clustering was omitted since all 5′-UTRs and CDSs in this library are unique rendering the mapping process computationally infeasible. Afterwards, the number of reads with unflipped and flipped Bxb1 substrates was counted for the remaining variants and for each time sample to obtain time-resolved flipping profiles.
Lastly, an additional filtering step was performed to ensure high data quality, which excludes variants with less than 10 reads in at least one of the six time points. Moreover, variants containing an unintended non-synonymous codon mutation in the CDS were removed (227 variants).
This data processing procedure resulted in 1 214 438 high-quality variants split across the four libraries with an average of 464.3 reads per variant or 77.4 reads per variant and time point. For the uASPIre of tRNAfMet mutants, this procedure resulted in 44 289 high-quality variants. In total, this amounts to 9 589 692 sequence-function pairs obtained from three NGS runs. The relative trapezoidal area under the flipping curve (termed ‘integral of the flipping profile’, IFP) was calculated for each variant. For Libfact, the average IFP of the two biological replicates was used.
Correlation of Bxb1 recombination with cellular Bxb1-sfGFP levels
To convert Bxb1-catalysed flipping into relative cellular Bxb1 concentrations, we used the same approach as described previously, which relies on translational fusion of Bxb1 to the superfolder green fluorescent protein (sfGFP) and the use of internal standard RBSs (71). In brief, we first recorded the sfGFP fluorescence of 31 manually constructed RBSs controlling translation of the Bxb1-sfGFP fusion. These RBSs span a wide range of RBS strengths (from low to high) as previously shown in triplicate shake flask cultivations (71). A pool of these 31 standard RBSs was cultivated in a separate shake flask in parallel to the cultivations of Librandom, Libcomb1 and Libcomb2 and processed alongside the different libraries as described above. From the resulting NGS data, we obtained the IFP for the standard RBSs and constructed a calibration curve between IFP and the aforementioned sfGFP fluorescence measurements (71). A LOESS fit (locally estimated scatterplot smoothing) was used to correlate the IFP with the slope of the cell-specific sfGFP signal between 0 and 290 minutes after induction (slope GFP0–290 min) using the function loess from the R package stats. Relying on the LOESS function, the IFP values of all library members were converted into the corresponding slope GFP0-290min. The resulting values were normalised to the maximum slope GFP0–290 min in the entire data and the normalised slope GFP0–290 min was designated relative translation rate (rTR) and used for all further analyses.
Splitting of full-factorial sub-libraries
Since Libfact consists of ten full-factorial sub-libraries that were sequenced in bulk, the resulting data had to be computationally split into the sub-libraries for further analysis. Therefore, we sequenced at least three clones (reference variants) from each sub-library by Sanger sequencing covering both the randomised 5′-UTR and CDS regions. From the resulting reference sequences, we reconstructed and split the ten individual sub-libraries as follows: all variants that shared either the 5′-UTR or CDS with one of the reference sequences were assigned to the corresponding sub-library. To obtain full-factorial sub-libraries (i.e. libraries in which the majority of 5′-UTRs is combined with each CDS and vice versa), we further removed all variants with a 5′-UTR that occurred in combination with less than 50 CDSs as well as all variants with a CDS that occurred in combination with less than 50 5′-UTRs.
Label-free proteomics
E. coli TOP10 ΔmetZWV cells bearing ptRNAfMet plasmids were cultivated in four biological replicates in shake flasks (100 ml LB, 34 mg l−1 chloramphenicol, 37°C, 200 rpm). Cultures were inoculated from glycerol stocks to an initial OD600 of 0.05 and grown to an OD600 of approximately 0.4. After, 50 ml of culture were centrifuged (4000 × g, 10 min, 4°C) and the pellet was frozen in liquid nitrogen. The subsequent steps were carried out by the Functional Genomics Center Zurich (University of Zurich, Switzerland). Briefly, cells were lysed and protein was extracted followed by LC–MS/MS and differential expression analysis against an empty vector strain as described previously (76).
For data analysis, a set of functions implemented in the R package prolfqua was used (77). Data were filtered keeping only peptides/proteins detectable in all four biological replicates of all four strains (empty vector and three tRNA-overexpressing strains). Further, we obtained the genomic sequence of parent strain E. coli TOP10 ΔrhaA through commercial sequencing (Novogene, Cambridge, United Kingdom) and annotation using Geneious Prime 2022.2.2 with E. coli DH10B NC_010473.1 as reference genome (78). Based on this genomic sequence, we extracted the nucleotide (A, C, U or G) directly upstream of the AUG start codon for all genes whose corresponding proteins appeared in the filtered proteomics data set. For this, only identical gene/protein names and only single-copy genes were considered, which resulted in a remaining total of 1098 genes/proteins.
Data analysis
Data analysis was conducted in R (version 4.2.1) (79) and figures were produced using the package ggplot2. For ANOVA of positional effects, variants from Librandom were split according to their respective base in each of the 40 randomised positions within 5′-UTR and CDS (i.e. 40 splits for 40 position). After, type II ANOVA was performed using the R function Anova (package car) treating each positional group as covariate to determine the contribution of each covariate/position to the variance of the rTR in the entire library assuming additive behaviour. For the assessment of effects of single bases, we calculated the average rTR of all variants in Librandom with a given base at a given position and divided the resulting value by the average rTR of all variants with any other base at this position. For example, the effect of U at 5′-UTR position −1 was calculated by dividing the average rTR of all variants with U at 5′-UTR position −1 (0.185) by the average rTR of all other variants (0.150). The resulting value (example: 1.233) represents the average relative in- or decrease in rTR for a given base and position. In the example above this means that the rTR of variants with U at 5′-UTR position −1 is on average 23.3% increased over the rest of the library. To assess the enrichment of bases amongst strong variants, variants in Librandom were first split into two groups with rTR ≥ 0.5 (strong) and rTR < 0.5 (weak). After, the relative occurrence of each base at each position was calculated within each group. The ratio between the occurrences in the two groups represents the relative enrichment/depletion of a given base in a given position amongst strong variants over weak variants.
For calculations related to mRNA folding, bash scripts were used. Minimum free energy (mfe), ensemble free energy (efe) and mRNA accessibility (acc) were each predicted using two models for base pairing, the Turner energy model (T) and the CONTRAfold model (C) (80,81), resulting in six different metrics (mfeT, mfeC, efeT, efeC, accT, accC). For mfeT and efeT, RNAfold (ViennaRNA package, version 2.4.18) and default parameters were used (82). For mfeC and efeC, default parameters were applied. For accT and accC, the Raccess program was used (83). Next, Spearman's correlation was calculated between each metric and the rTR. Note that Spearman's correlation was used since rTR values do not follow a normal distribution (P-value of 1.1 × 10−79 according to Shapiro-Wilk normality test). Squared Spearman's coefficient (ρ2) is reported as a measure of correlation between the respective folding metric and the rank of the rTR. Accordingly, the higher ρ2 of a metric, the more it explains the observed variance in the rTR. To identify the optimal mRNA sequence window that leads to the highest correlation between folding and rTR, mfeT and efeT were calculated for all possible sequence windows of lengths between 10 and 200 nucleotides within the first 200 positions of the mRNA. For computational reasons, this analysis was performed only on the 10 000 variants of Librandom with the highest number of NGS reads. The best correlation between folding energy and rTR was achieved using the first 80 positions of the mRNA (i.e. between positions −27 and + 53) (Supplementary Figure S7). This ‘optimal’ sequence window was then used to calculate mfeT, mfeC, efeT, efeC, accT and accC for all variants in all libraries. For accT and accC, the access length was set to 80 nucleotides in Raccess. For accessibility scanning, the correlation between the accessibility of each position and the rTR was determined applying an access length of 10 nucleotides in Raccess (accT10nt and accC10nt).
To calculate 16S rRNA hybridisation energies, RNAduplex from the ViennaRNA package (82) was used, which only allows intermolecular base pairing. Allowing intramolecular base pairing would favour 5′-UTR-internal folds and thus disregard interactions with the 16S rRNA. Specifically, hybridisation energy was calculated between 5′-UTR (positional window: −18 to −4) and the 16S rRNA 3′-end (5′-ACCUCCUUA-3′). As an alternative, we also calculated a positional hybridisation energy between 16S rRNA 3′-end and a 9-nt sliding window along the entire mRNA.
The minimum edit distance was determined using the stringdist function of the R package stringdist and corresponds to the Levenshtein distance between the 7-bp long canonical SD motif AGGAGGU and a sliding 7-nt window within 5′-UTR positions −18 and −4. Levenshtein distance is the minimum number of operations (substitutions, deletions, and insertions) to transform one string into another.
The random forest model was built using h2o.randomForest from the R package h2o (https://github.com/h2oai/h2o-3). Variants of Librandom were split into a randomly selected training set (90%) and a test set (10%), which was strictly held out during training. Sequences were encoded using one-hot encoding, a position-wise accessibility score accC1nt (compare above), GC-content, minimum edit distance to the SD motif AGGAGGU, 16S rRNA hybridisation energy, the position of 16S rRNA hybridisation on the mRNA, as well as the folding metrics mfeT, mfeC, efeT, efeC, accT and accC (see above). Using tenfold cross-validation, the model was then trained with default parameters using 50 trees, and its performance was validated on the strictly held-out test set.
To quantify the contributions of UTR and CDS, we first grouped variants from Libcomb1, Libcomb2 and Libfact by their 5′-UTR and then calculated the average rTR of all CDSs in each group. Similarly, we also grouped variants by their CDS and calculated the average rTR of all 5′-UTRs in each group.
Codon adaptation index (CAI) and tRNA adaptation index (tAI) were calculated using the cai function from the R package seqinr. Codon weights and frequencies (Supplementary Table S6) were used as presented in Sharp et al. (42) and dos Reis et al., respectively (61).
All sequence variants and their calculated parameters were combined into a single dataset and further analysed.
RESULTS
High-throughput characterisation of 5′-UTR-CDS combinations
It is challenging to investigate the impact of different mRNA parts on translation initiation due to the vast sequence space of possible variants. For instance, even for a comparably short 5′-UTR of twelve nucleotides, >16 million (412) sequences are possible. The sequence space becomes even larger if different parts are diversified simultaneously, which is required to analyse interactions and combined effects. Such combinatorial complexity cannot be addressed appropriately by measuring the expression of a few handpicked sequences. Instead, it requires high-throughput methodology capable of linking sequences to corresponding expression levels at large scale. To achieve this for 5′-UTR-CDS combinations, we capitalise herein on a recently developed technology for ultradeep Acquisition of Sequence-Phenotype Interrelations (uASPIre) (71). Briefly, uASPIre uses the phage recombinase Bxb1 to record functional information in DNA. This DNA-recorder enables, for instance, to determine both sequence and corresponding gene expression of gene regulatory elements via NGS at extremely high throughputs, which we have recently demonstrated in a proof-of-concept study (71).
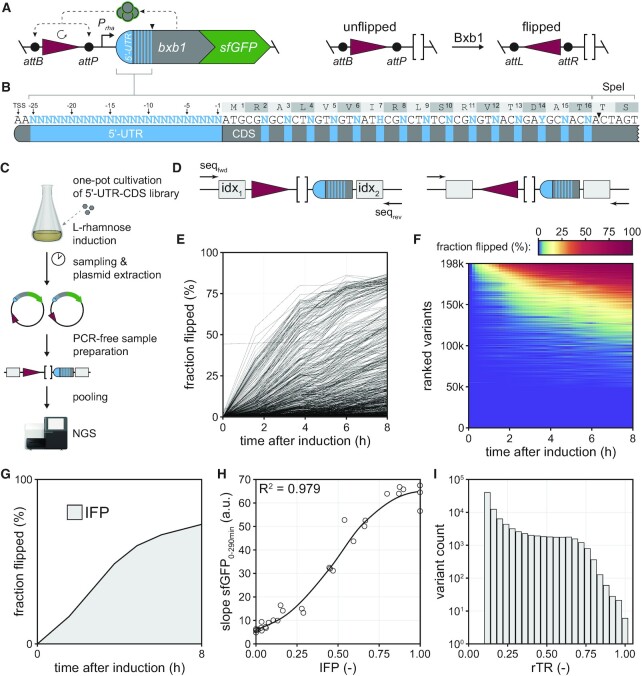
To make uASPIre amenable for the characterisation of 5′-UTR-CDS combinations, we created the plasmid architecture shown in Figure 1A, which contains a gene encoding a Bxb1-sfGFP fusion (71) controlled by an l-rhamnose-inducible promoter (Prha) and a 150-bp stretch of silent DNA flanked by Bxb1’s cognate attachment sites attB and attP in opposite orientation (74). Furthermore, a SpeI site is introduced in codons 17 and 18 of the bxb1 CDS via silent mutation (Figure 1A, B), which enables facile exchange of the 5′-UTR and the first 16 codons of the bxb1 CDS as well as NGS sample preparation (Materials and Methods). Once expressed, Bxb1-sfGFP converts its attB-/P-flanked DNA substrate from its initial (‘unflipped’ hereafter) to an inverted (‘flipped’ hereafter) state (Figure 1A). Thus, Bxb1-sfGFP expression can be read out by determining the state of the substrate DNA by sequencing. Importantly, the flipping rate directly correlates with the cellular Bxb1-sfGFP concentration, and sequencing of many copies of this architecture via NGS can be used to determine the fraction of flipped DNA substrates (‘fraction flipped’ hereafter) amongst all copies of a given variant. This ‘oversampling’ facilitates a precise, quantitative readout for Bxb1-sfGFP expression, whose resolution solely depends on the sequencing depth (i.e. number of reads obtained per variant) as we have previously shown (71).
Figure 1.
Ultradeep characterisation of 5′-UTR-CDS combinations. (A) Plasmid architecture for the uASPIre of 5′-UTR-CDS pairs. A bxb1-sfGFP gene (translational fusion) controlled by Prha is placed on the same DNA molecule as the substrate modifiable by Bxb1-sfGFP, which is flanked by Bxb1 attachment sites (attB/P). A SpeI site in codons 17 and 18 of bxb1-sfGFP allows for seamless exchange of 5′-UTR and N-terminal CDS. Once expressed, Bxb1-sfGFP irreversibly inverts its substrate from an unflipped into a flipped state creating recombined attachment sites (attL/R). (B) Design of Librandom. The 25 nucleotides preceding the start codon are fully randomised. Additionally, the third positions of codons 2–16 are mutated allowing only synonymous codon replacements. Sequences follow the IUPAC nucleotide code (N: A/C/G/T, H: A/C/T, Y: C/T). TSS: transcriptional start site of Prha. (C) Experimental workflow for the uASPIre of 5′-UTR-CDS pairs. Pooled transformants of Librandom are grown in LB and bxb1-sfGFP expression is induced by l-rhamnose addition. After, samples are taken at different time points followed by plasmid extraction and preparation of NGS fragments followed by pooling of samples and NGS (Methods). NGS fragments are flanked by duplex adapters with sample-specific index combinations (grey boxes). (D) Close-up view of target fragments for paired-end NGS using forward (seqfwd) and reverse (seqrev) sequencing primers. Forward reads are used to identify the first index (idx1) and the state of the recombinase substrate. Reverse reads are used to obtain the second index (idx2) and the sequence of 5′-UTR and CDS. (E) Representative flipping profiles of 5′-UTR-CDS variants from Librandom. For clarity, only the 1000 most abundant variants are displayed. (F) Flipping profiles of all 198174 Librandom members above high-quality read-count threshold (Methods). Horizontal lines are time series of individual variants coloured according to the fraction flipped and ranked by the average fraction flipped across all time points from high (top) to low (bottom). (G) Illustration of the IFP (grey area), i.e. the normalised integral of the flipping profile. (H) Correlation between IFP and slope sfGFP0-290min as shown for 31 standard RBSs (Methods). A LOESS function (black line) can be used to interconvert IFP and slope sfGFP0-290min with high confidence. (I) Histogram of the rTR of all variants from Librandom.
Next, we generated a first library through simultaneous diversification of the 5′-UTR and CDS of bxb1-sfGFP with the goal to characterise the impact on bacterial translation initiation in a highly parallelised fashion relying on uASPIre (Figure 1B, Materials and Methods). We mutated the 25 nucleotides directly upstream of the start codon applying full randomisation (i.e. N25-mer, N: equimolar mixture of A, C, G and T). This corresponds to the entire 5′-UTR in our setup except for two consecutive A’s at the 5′-end of the mRNA, which were fixed to match the native transcriptional start of Prha and thus avoid changes in transcription rates (84). Further, we mutated the third positions of codons 2–16 downstream of the start codon (ATG itself was kept constant) to additionally diversify the CDS. We selected this region since the first 30–50 nucleotides of CDSs reportedly affect translation initiation whereas sequence changes further downstream show only negligible effects on expression (30,34). Importantly, in this region we only allowed synonymous (‘silent’) codon replacements to maintain the same Bxb1 amino acid sequence and hence specific recombination activity for all library members, which is crucial to study only translational effects. This library is designated Librandom hereafter pointing to the full randomisation of 5′-UTR and N-terminal CDS.
Librandom was used to transform E. coli yielding approximately 400 000 individual transformants. Specifically, we used the rhamnose-utilisation deficient strain TOP10 ΔrhaA to ensure temporally stable induction due to the lack of inducer consumption (71). Afterwards, transformants were pooled and cultivated in a single shake flask (Figure 1C). In parallel, we cultivated 31 5′-UTR variants (‘standard RBSs’ hereafter) controlling the same bxb1-sfGFP fusion, which were constructed and characterised in a previous study (71). These standard RBSs span a wide range of expression levels and serve as internal standard sequences to compare different experiments. Further, they are used to convert the fraction flipped time series into practically more relevant metrics for protein expression relying on calibration curves generated from individual sfGFP fluorescence measurements (see below, Materials and Methods) (71). After induction by addition of l-rhamnose, six samples each were drawn over the course of eight hours from both cultures (Librandom and standard RBSs), and plasmid DNA was extracted followed by NGS sample preparation (Materials and Methods). Note that sample preparation was carried out without PCR amplification, which avoids non-linear PCR bias (71). The final target DNA fragments are flanked by NGS adapters with sample-specific indices and contain the DNA substrate modifiable by Bxb1 and the randomised 5′-UTR-CDS region. NGS adapters, substrate and 5′-UTR-CDS region were sequenced in an Illumina platform yielding approximately 108 paired-end reads for Librandom (Figure 1D).
Next, we processed the NGS data to obtain time series of Bxb1-mediated flipping (‘flipping profiles’) using a previously developed computational pipeline adapted to the new plasmid architecture (Materials and Methods) (71). This procedure yielded flipping profiles for 198 174 5′-UTR-CDS pairs above an applied minimal threshold of ten reads per time point and variant (i.e. high-quality data, average of 433.7 reads per variant). The base composition in Librandom was homogeneously distributed across all diversified positions (Supplementary Figure S8). Library members showed a diverse range of translational activities from low to high and a skew towards weaker variants as to be expected for full randomisation of the 5′-UTR (Figure 1E, F) (85). Notably, the behaviour of the standard RBSs correlated strongly with results from our previous study even though the experiments were carried out approximately two years apart from each other (Supplementary Figure S9) (71). This confirms the validity of the recorded data and indicates a high reproducibility and robustness of the uASPIre method in general. Next, we calculated the trapezoid integral of the flipping profiles (IFP, Figure 1G), which constitutes a robust metric correlating well with rates of cellular Bxb1-sfGFP accumulation as previously shown (71). Indeed, the IFP of the 31 standard RBSs as determined in this study correlated well with the linear slope of the cell-specific Bxb1-sfGFP fluorescence between 0 and 290 min after induction (slope sfGFP0–290 min, Figure 1H, Materials and Methods). Therefore, IFP values can be converted into the slope sfGFP0–290 min relying on a fit applied between the two metrics for the standard RBSs. Specifically, we performed locally estimated scatterplot smoothing (LOESS) (Figure 1H) and used the resulting fit function to convert the IFPs of Librandom members into the corresponding slope sfGFP0–290 min normalised to the strongest variant found in this study (Figure 1I, Materials and Methods). This normalised parameter was designated relative translation rate (rTR) and used for all further analyses, because it represents a practically more relevant metric for translation initiation directly corresponding to cell-specific protein accumulation.
Analysis of positional and base-specific effects on translation initiation
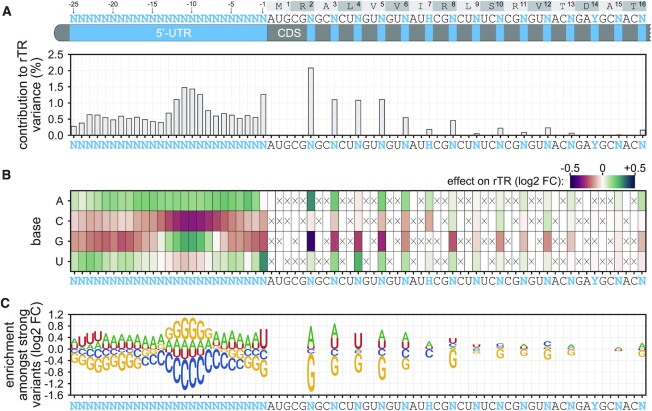
Relying on the data generated for Librandom, we investigated the impact of different positions, nucleotides, and sequence motifs on expression. To assess positional effects, we performed analysis of variance (ANOVA) treating each variable position in the 5′-UTR (−25 to −1) and CDS (third positions of codons 2–16) as a covariate and calculated the contribution to the observed variance in rTR (Figure 2A, Materials and Methods). Individual positions in the 5′-UTR explain between 0.3 and 1.5% of the variance. The most pronounced effect was observable for positions −13 to −8, which corresponds to an anticipated SD region, and, more unexpectedly, position −1. Within the CDS, the impact of codons decreases with increasing distance from the start codon with codon 2 showing the highest contribution (2.1%). Codons 2 to 8 show a marked effect, which strongly decreases to a negligible degree thereafter. Notably, the cumulative contribution of all 40 randomised positions only amounts to about 25.0% of which about 17.5% and 7.4% are attributed to 5′-UTR and CDS, respectively (Supplementary Figure S10). The remaining high fraction of unexplained variance (about 75%) points towards a strong interaction between positions leading to non-additive behaviour. Next, we calculated the effect of specific bases at the variable positions by dividing the average rTR of variants with a given base at a position by the average rTR of all other variants (Figure 2B). Generally, C and G tend to have a negative, and A and U a positive effect on translation initiation, which is stronger in the 5′-UTR and weaker in the CDS decreasing with increasing distance to the start codon. A striking exception to that end are positions −14 to −7 (SD region), for which the effect of G is highly positive. The strongest negative effect is observable for CGG as the second codon (Arg) with corresponding variants being on average 26.3% weaker than those with CGA, CGC or CGU in this codon. The strongest positive impact is associated with U at 5′-UTR position −1 amounting to a mean rTR increase of 23.3%. Finally, to identify characteristic sequence determinants of strong variants, we split the data from Librandom into two sets of strong variants (i.e. rTR ≥ 0.5; 11 212 sequences) and weaker variants (i.e. rTR < 0.5; 186 962 sequences) and calculated the relative enrichment or depletion of each base at each position in the strong over the weaker subset (Figure 2C, Materials and Methods). This analysis confirmed the finding above that both 5′-UTR and CDS of strong variants are generally enriched for A and U, and depleted for G and C except for a G-favouring region at positions −14 to −7. The latter shows a strong resemblance to archetypal AG-rich SD motifs, which commonly follow a consensus of AGGAGA/G in E. coli. In the CDS, we again observed a consistent decrease in positional importance with increasing codon number and a sharp drop of effect size after codon 8. Moreover, the aforementioned significance of U (but not A!) at 5′-UTR position −1 and the strong negative impact of CGG in codon 2 are confirmed by this analysis of strong sequences. The high and base-specific impact of these two positions prompted us to perform further analyses and experiments towards the causality of these effects (see below).
Figure 2.
Positional and base-specific effects on translation initiation. (A) Contribution of variable mRNA positions to the observed rTR variance. The relative sum of squares calculated by ANOVA with each position as covariate is displayed. (B) Base-specific effects of the randomised positions. Displayed effects are log2-transformed fold changes (log2 FC) of the mean rTR of variants with a given base at the respective position over the mean rTR of variants with any other base permitted at that position. Positive and negative values correspond to translation-increasing or -decreasing effects, respectively. Crossed boxes indicate unvaried bases. (C) Enrichment of bases amongst strong variants. The log2 FC of a base's relative occurrence amongst strong variants (rTR ≥ 0.5) over its relative occurrence amongst weak variants (rTR < 0.5) is displayed.
Quantification of sequence parameters and their effect on translation initiation
Since <30% of the variance in translation could be explained by global analysis of individual positions, we sought to examine the impact of different sequence parameters on the level of individual variants. Specifically, we computed several parameters known or hypothesised to influence rTR for all members of Librandom and calculated their correlation with rTR. This analysis included parameters related to GC-content, hybridisation between mRNA and 16S rRNA, predicted mRNA folding and other features. Since rTR values follow a non-normal distribution (P-value = 1.1 × 10−79, Shapiro–Wilk normality test) and some sequence parameters are likely to non-linearly correlate with rTR, we also report Spearman's correlation (coefficient ρ) as a metric of rank correlation between parameters and rTR.
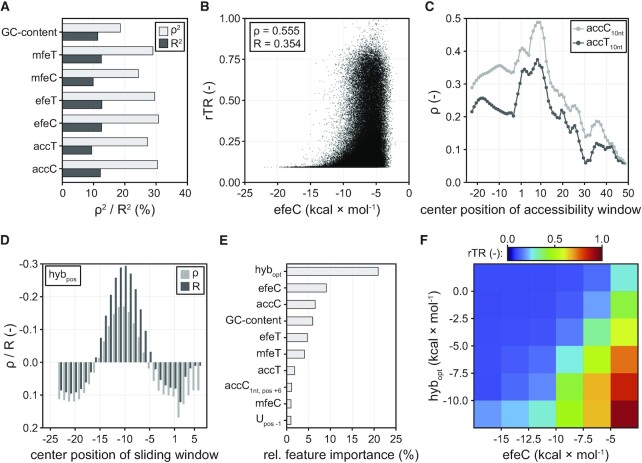
Overall GC-content shows significant correlation with the rTR (ρ2 = 18.6%, R2 = 11.3%) and its impact is higher in the 5′-UTR than the CDS (Figure 3A, Supplementary Figure S11). In particular high GC-content is strongly associated with low rTRs (Supplementary Figure S11), likely due to a tendency of GC-rich sequences to form stable secondary structures, which are known to counteract translation initiation (28). Further, we predicted the minimum free energy (mfe), ensemble free energy (efe) and mRNA accessibility (acc) using two models for base pairing, the Turner energy model (T) and the CONTRAfold (C) model (80,81), resulting in six metrics which all relate to mRNA folding: mfeT, mfeC, efeT, efeC, accT and accC (Figure 3A, Materials and Methods). In brief, mfe and efe are energies required for the unfolding of the most likely and the ensemble of possible mRNA secondary structure(s), respectively, whereas acc is a predicted accessibility score for a defined sequence window along the mRNA corresponding to the probability of this window being embedded within a secondary structure (19). Folding of mRNA showed a clear impact on rTR across all tested metrics (Figure 3A). The latter show a positive correlation with the rTR, which is stronger than for GC-content and highest for efeC (ρ2 = 30.8%, R2 = 12.6%) and accC (ρ2 = 30.4%, R2 = 12.2%) (Figure 3A, B). In particular very strong folding (e.g. efeC < −15 kcal × mol−1) completely abolishes efficient translation initiation (Figure 3B). We investigated further the impact of the positioning of predicted secondary structures by calculating mRNA accessibility within a window of ten nucleotides. Correlation of the resulting scores (accT/C10nt) with rTR is highest around the first few codons followed by the SD region, and sharply decreases further downstream in the CDS (Figure 3C).
Figure 3.
Effect of different sequence parameters on translation initiation in Librandom. (A) Correlation of GC-content and different mRNA folding metrics with rTR. Spearman's ρ2 and Pearson's R2 are displayed. (B) Scatterplot between rTR and the best-correlating mRNA folding parameter efeC. (C) Correlation of rTR with local mRNA accessibility. Parameters accT10nt and accC10nt correspond to the mRNA accessibility of a 10-nt window centred around the mRNA position specified on the horizontal axis. Endings C and T denote base pairing calculated by two different energy models (Methods). (D) Correlation of hybridisation energy between 16S rRNA and different mRNA positions with rTR. Positional hybridisation energy (hybpos) is displayed for 9-bp windows centred around the indicated mRNA position (horizontal axis). (E) Relative feature importance of a random forest model trained on Librandom. The ten most important of 248 features are displayed. hybopt: best-correlating hybridisation parameter (see main text). accC1nt, pos+6: accC score for position +6 of the mRNA. Upos –1: one-hot encoded U at position −1 of the mRNA. (F) Mean rTR of variants in Librandom as grouped by the two most predictive features of the random forest, hybopt and efeC. Tick labels mark the boundaries of the respective bins (boxes).
Next, we investigated the impact of interactions between mRNA and 16S rRNA. As expected, the hybridisation energy hybSD between E. coli’s 16S rRNA (sequence: 5′-ACCUCCUUA-3′) and the approximate SD region in the 5′-UTR (window between positions −18 and −4) shows a clear correlation with the rTR (Supplementary Figure S12, Materials and Methods) (82). This observation is further supported by the fact that similarity with the canonical SD motif AGGAGGU in this window is strongly associated with high rTRs (Supplementary Figure S13). Since the position of hybridisation is known to be critical for efficient translation initiation, we further calculated positional hybridisation energies hybpos sliding the 9-nt 16S rRNA sequence along the mRNA (Figure 3D, Materials and Methods). We found that hybpos is negatively correlated with rTR between 5′-UTR positions −15 and −6 indicating that stronger hybridisation (i.e. lower hybpos) has a translation-favouring effect in this region. Outside of this window, a negative effect on rTR is observable. The 9-nt hybridisation window with the strongest correlation to rTR is centred on position −10 corresponding to a binding of the 16S rRNA 3′-end to the 5′-UTR between positions −14 and −6. A more systematic analysis of hybridisation windows and positions (Supplementary Table S7) revealed the mean of hybridisation energies at positions −11 and −10 (hybopt) as the parameter with the highest correlation with rTR (ρ2 = 2.9%, R2 = 8.9%).
Based on those findings, we sought to quantify the utility of different sequence parameters for predictive modelling. To this end, we used the data from Librandom to train a random forest regressor with the goal to predict the rTR from different features including primary sequence information as well as the above-mentioned secondary parameters (Materials and Methods). The model was trained using tenfold cross-validation (Supplementary Figure S14) and its performance was evaluated on a test set strictly held out during training (randomly selected 10% of data). The resulting model predicts rTR values with good confidence (R2 = 58.0%, Supplementary Figure S15). More importantly, we extracted the relative importance of features of the random forest (Figure 3E). Remarkably, while the 16S rRNA hybridisation parameter hybopt had shown only moderate correlation coefficients ρ and R, it was by far the most important model feature (20.9%) followed by the folding parameters efeC (9.1%) and accC (6.5%). The over-proportional importance of hybopt could imply that successful hybridisation with the 16S rRNA must be fulfilled to obtain strong translation initiation rendering hypopt a critical, early decision criterion for the model. Furthermore, U at 5′-UTR position −1 ranked 10th (1.0%) amongst the total of 248 encodings constituting the most important single-nucleotide feature. Most features (227) exhibited a relative importance below 0.5% pointing towards the multifactorial, interactive nature of the translation initiation process and likely to a high degree of redundancy between the tested encodings.
Lastly, we binned the variants from Librandom according to the two most important features of the random forest, hybopt and efeC, and calculated the average rTR of each bin (Figure 3F). Interestingly, we found that the appearance of very high rTRs (i.e. >0.5) is co-dependent on strong 16S rRNA hybridisation and weak mRNA folding. Variants with strong secondary structures (efeC < −15.0 kcal × mol−1) only exhibit significant translation initiation if they hybridise well with the 16S rRNA. By contrast, variants with weak mRNA folding can exhibit intermediate-to-strong translation initiation even in the absence of SD motifs.
Codon usage and interaction between 5′-UTR and CDS
A long-standing question is how strong the impact of the CDS on translation initiation is, both in absolute terms and relative to the 5′-UTR. Changes in the CDS affect critical determinants of translation initiation such as codon usage and mRNA folding. Importantly, testing many different CDSs in combination with a single 5′-UTR (as amply done in previous studies) is insufficient to unambiguously assign observed effects to different sequence parameters and to quantify their contribution in a precise fashion, since some parameters also depend on and change with the 5′-UTR in place. Thus, it remains unclear if and how strong any observed effect is causally related to a sequence parameter change in a generalisable fashion, or whether it is merely a context-specific artefact only occurring for the selected 5′-UTR. Similarly, full randomization (as in Librandom in this work) only delivers unique pairs of 5′-UTRs and CDSs, which again prohibits unambiguous attribution of effects to either of the two mRNA parts (5′-UTR or CDS). This problem can only be circumvented by testing large numbers of 5′-UTR-CDS combinations in a combinatorial manner with sufficient overlap allowing to average out case-specific artefacts.
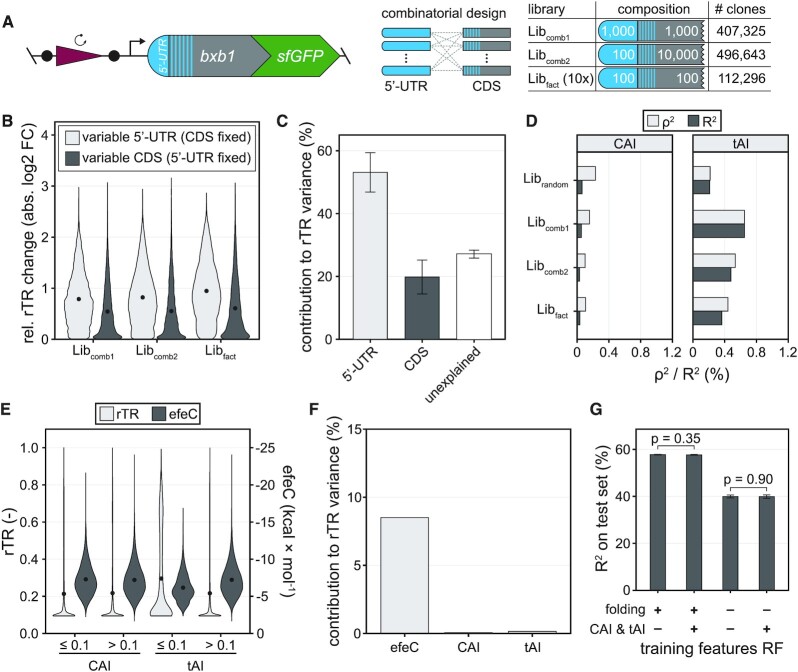
Therefore, to investigate the individual impact of 5′-UTR and CDS independently, we generated three additional libraries of combinatorial (Libcomb1, Libcomb2) and full-factorial (Libfact) 5′-UTR-CDS pairs, which were constructed through combination of defined half-libraries (Figure 4A, Materials and Methods): Libcomb1 combines about 1000 5′-UTRs with about 1000 CDSs, Libcomb2 is a combination of approximately 100 5′-UTRs with approximately 10 000 CDSs, and Libfact features ten independently cloned batches of about 100 5′-UTRs combined with about 100 CDSs each. Note that Libfact was designed such that in each batch every 5′-UTR is combined with every CDS and vice versa (i.e. full-factorial design). Next, we recorded the activity of variants from the three libraries applying the same uASPIre workflow as described for Librandom above. Processing of NGS data yielded time series for 407 325, 496 643, 112 296 unique variants above high-quality read count threshold for Libcomb1, Libcomb2 and Libfact, respectively. For Libfact, two independent biological replicates were tested which showed a high degree of reproducibility (Pearson's R2 = 0.989, Supplementary Figure S16). We then grouped variants according to the 5′-UTR (or CDS) in place and analysed the diversity of the rTR amongst all CDSs (or 5′-UTRs) appearing with the respective fixed 5′-UTR (or CDS). Exchanging either 5′-UTR or CDS (while maintaining the other) can lead to strong up- and downshifts in expression (Figure 4B). Shifts are on average much stronger for an exchange of the 5′-UTR than of the CDS, and in many cases cover a large fraction of the rTR range (Figure 4B, Supplementary Figure S17). We further quantified the individual impact of 5′-UTR and CDS performing an ANOVA with the mean rTRs of all 5′-UTRs and CDSs (Figure 4C, Materials and Methods). This analysis was performed exclusively on Libfact, since full-factorial design is required to exclude case-specific artefacts and achieve a precise quantification of each part's individual contribution (see above). The ANOVA revealed that the 5′-UTR explains on average 53.1 ± 6.3% and the CDS 19.8 ± 5.4% of rTR variance, and thus a significantly higher impact of the 5′-UTR compared to the CDS. 27.0 ± 1.3% of variance remain unexplained in the additive model and must therefore be caused by non-linear interactions between 5′-UTR and CDS, which clearly demonstrates a high degree of interdependence between the two parts. Crucially, this quantification of the mean individual contribution of 5′-UTR and CDS is only possible due to the applied full-factorial library design and the high throughput of uASPIre.
Figure 4.
Overall impact of 5′-UTR, CDS and codon usage on translation initiation. (A) Three additional libraries of combinatorial (Libcomb1, Libcomb2) and full-factorial (Libfact) design were assessed via uASPIre. Libcomb1: combinatorial combination of about 1000 5′-UTRs and 1000 CDSs. Libcomb2: combinatorial combination of about 100 5′-UTRs and 10 000 CDSs. Libfact: ten independent batches, each a full factorial combination of approx. 100 5′-UTRs and 100 CDSs. Libfact was tested in two independent biological replicates. The number of analysed clones is indicated for each library. (B) Impact of the exchange of 5′-UTRs or CDSs on translation initiation. The rTR change (absolute value) of a given 5′-UTR upon exchanging its CDS versus the mean rTR of all variants with that same 5′-UTR is displayed (and vice versa). Black circles within violins are mean relative rTR changes. (C) ANOVA with the mean rTRs of all 5′-UTRs and CDSs in Libfact. Error bars: standard deviation between ten independent batches of Libfact. (D) Correlation of codon usage indices CAI and tAI with rTR. (E) Comparison of rTRs and predicted folding energies (efeC) of variants with low (≤0.1) and high (>0.1) CAI/tAI in all libraries. Black circles within violins are mean rTR/efeC values. (F) Contribution of efeC, CAI and tAI to the rTR variance in all libraries according to an ANOVA with only the three parameters as covariates. (G) Impact of folding and codon usage metrics on the performance of random forest (RF) models trained on Librandom. Sequence parameters for mRNA folding (mfeT, mfeC, efeT, efeC, accT and accC) and codon usage (CAI and tAI) were added or omitted during training. Error bars: Standard deviation of five training repeats with 10-fold cross-validation each. P-values were calculated with Welch two sample t-tests.
A controversially discussed sequence feature of the CDS is codon usage, which is well known to influence translation initiation. To this end, the appearance of rare codons within the first few triplets of the CDS was found to coincide with high expression (30,42–45). Thus, we first analysed the impact of two commonly used metrics for codon usage, CAI and tAI (Supplementary Table S6) (42,61), on rTR, which indicated a weak (R2 and ρ2 consistently below 0.7%) yet significant correlation in all libraries (Figure 4D). However, it remains unclear whether this is caused by differential abundance of the corresponding tRNAs in the cell or by changes in mRNA folding. Since folding is also co-dependent on the 5′-UTR in place, combinatorial testing of 5′-UTR-CDS pairs is also essential in this case to unambiguously test if and to which extent the two aforementioned hypotheses are correct. Accordingly, we first compared the rTR of Libfact variants rich in rare codons (i.e. CAI/tAI ≤ 0.1) with the other variants (i.e. CAI/tAI > 0.1). Variants with low CAI exhibit a mean rTR of 0.213, which is virtually indifferent from high-CAI variants (0.217) (Figure 4E). This finding is further corroborated by the fact that the mean rTRs of CDSs and CAI do not correlate significantly (P-value = 0.256, one sample t-test) in Libfact (Supplementary Figure S18). Low-tAI variants, by contrast, exhibit on average a higher rTR than the control group (Figure 4E, Supplementary Figure S18). At the same time, however, predicted mRNA folding is significantly weaker (P-value < 10−300, one-sided Welch two sample t-test) in low- versus high-tAI variants, which is not the case for the corresponding CAI groups (P-value = 1.0, Figure 4E). We repeated this analysis with applying different CAI/tAI cut-offs, which showed the same trend (Supplementary Figure S19). Moreover, the codon frequency of E. coli showed only very small and inconsistent effects on the rTR for the randomised codons (Supplementary Figure S20). Collectively, these findings clearly speak against tRNA abundance being causally responsible for differences in expression levels between CDSs with different synonymous codons. Therefore, we further analysed to which extent mRNA folding can explain the rTR’s dependence on codon usage. An ANOVA with only efeC, CAI and tAI as covariates indicated that the overwhelming majority of variance in rTR explainable by these parameters is attributed to efeC (8.5%), whereas the contribution CAI and tAI was about 155- and 53-fold lower, respectively (Figure 4F). To test if the rather strong effect of secondary structures masks any potential small effect of codon usage and tRNA abundance, we further performed a similar ANOVA restricted to variants with weak secondary structure potential (approximately 60% of all variants), which again indicated that the overwhelming majority of variance in rTR is attributed to efeC whereas the effects of CAI and tAI remained incrementally small (Supplementary Figure S21).
Furthermore, we re-trained the former random forest model (see above) with different sets of sequence parameters including CAI and tAI (Figure 4G). Remarkably, while removal of mRNA folding parameters led to a substantial decrease in model performance, addition of CAI and tAI did neither increase accuracy of the initial random forest nor was it able to compensate for the performance loss in the absence of folding parameters. Accordingly, the relative feature importance of CAI and tAI was very low (Supplementary Figure S22). Collectively, these findings strongly suggest that any influence of codon usage on rTR can be virtually completely explained by mRNA folding. On the contrary, a causal connection to cellular tRNA abundance or the previously postulated translational ramps could not be established and is either insignificant or negligible amongst the over 1.2 million sequences tested in this study.
Assessment of translational anomalies of arginine codon 2 and 5′-UTR position −1
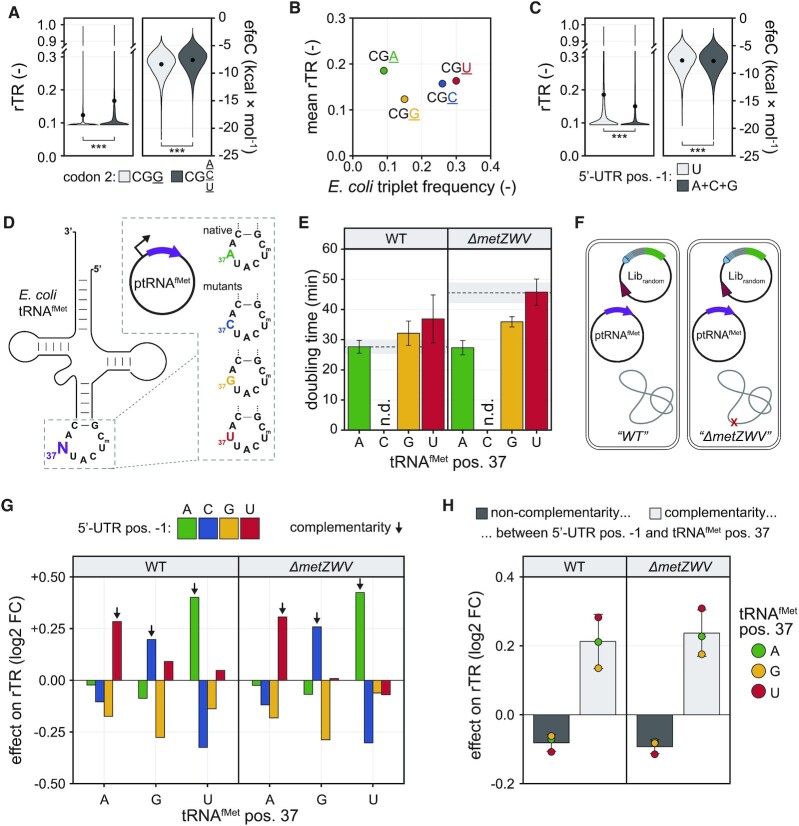
Lastly, we sought to decipher the reasons for the behaviour of the two nucleotide positions in the mRNA exhibiting the strongest positive and negative effect observable in our data, respectively (see above). To this end, the presence of G in the third position of codon 2 (arginine) and U in position −1 of the 5′-UTR exhibit a profound impact on the rTR, which is negative in the former and positive in the latter case (compare Figure 2). Variants with CGG as the second codon show an average decrease in rTR of 26.3% compared to variants carrying A, C or U in the third position (Figure 5A). This different behaviour is likely not caused by codon frequencies or tRNA availability, since both arginine codons with higher (CGC, CGU) and lower (CGA) frequency show significantly higher mean rTRs (Figure 5B). By contrast, the average predicted folding energy of CGG-bearing variants is significantly lower (ΔefeC = −0.9 kcal × mol−1) than for the other codons (Figure 5A), pointing again to mRNA folding (and not tRNA availability) as the mechanistic reason for the differential expression of synonymous codons.
Figure 5.
Assessment of translational anomalies of arginine codon 2 and 5′-UTR position −1 in Librandom. (A) Effect of different synonymous codons in the second triplet of the CDS on rTR and predicted mRNA folding energy (efeC). Black circles within violins are mean rTR/efeC values. *** denote P-values < 10−16 in a Welch two sample t-test. (B) Relationship between relative triplet frequency in E. coli and rTR for the four synonymous triplets in arginine codon 2. (C) Effect of different bases in 5′-UTR position −1 on rTR and efeC. Black circles within violins are mean rTR/efeC values. *** denote P-values < 10−16 in a Welch two sample t-test. (D) Plasmids for the overexpression of native initiator tRNAfMet and mutants thereof (Supplementary Figure S6, Methods). Position 37 (3′-adjacent to the CAU anticodon) of tRNAfMet is mutated from A to C, G or T/U. (E) Growth of E. coli strains carrying plasmids for tRNAfMet overexpression in shake flask cultivations (LB, 37°C). Bars are mean doubling times of independent biological triplicate cultivations with standard deviation as error bars. Dashed lines are the mean doubling time of the respective strain without tRNA overexpression (i.e. empty vector control) with standard deviation as grey shaded areas. For tRNAfMet-A37C, doubling times were not determined (n.d.) due to severe growth inhibition (see main text). (F) Approximately 50 000 variants of Librandom were tested in the presence of overexpressed tRNAfMet variants in E. coli strains containing (WT) and lacking (ΔmetZWV) the chromosomal metZWV locus. (G) Impact of tRNAfMet mutations on the rTR of variants from Librandom. Displayed effects are log2-transformed fold-changes (log2 FC) of the average rTR of variants with a given base at 5′-UTR position −1 over the average rTR of variants with any other base at this position. Black arrows indicate complementarity between 5′-UTR position −1 and position 37 of the tRNAfMet variant. (H) Impact of complementarity between 5′-UTR position −1 and tRNAfMet position 37. Circles are log2-transformed fold-changes (log2 FC) of the average rTR of variants with complementarity or non-complementarity between mRNA and tRNA over the mean rTR of all variants in the same group (i.e. same tRNAfMet variant and strain). Bars are the mean log2 FCs of the three tRNAfMet variants for each case and strain with standard deviation as error bars.
For variants with U at position −1 in the 5′-UTR, the mean rTR is 23.3% higher than for those with any other base in this position (Figure 5C). In this case, the average folding energy is only marginally increased for U (ΔefeC = +0.1 kcal × mol−1) likely excluding mRNA folding as the reason (Figure 5C). Therefore, we sought to investigate further reasons for the unexpectedly large effect of U at 5′-UTR position −1. As an alternative explanation, we suspected that an interaction of this U with the initiator tRNA (tRNAfMet) could be responsible for the observed effect. In E. coli, initiator tRNAs are encoded by one monocistronic (metY) and one tricistronic (metZWV) transcriptional unit, and their sequences are identical except for position 46 (G in metY, A in metZWV). Importantly, methionine elongator tRNAs (metT, metU) do not initiate translation (86), and all tRNAfMet copies carry an A in position 37 directly 3′ to the CAU anticodon, which could preferentially hybridise with mRNAs carrying a U directly 5′ to the start codon.
Several previous studies have postulated or shown that the presence of U in this position favours formation of the prokaryotic ribosomal initiation complex and/or translation of the corresponding genes in vitro and in vivo (37,71,87–95). These effects were attributed to a proposed interaction of A37 in tRNAfMet and U in 5′-UTR position −1, for which further evidence was later provided in algal chloroplasts through compensatory mutation of tRNAfMet position 37 (96). Furthermore, structural analyses have shown that A37 is released from internal base pairing upon reaching the ribosomal P-site (97), which would render this position available for Watson-Crick base pairing with nucleotide(s) upstream of the start codon. Collectively, these prior works highlight the importance of bases directly upstream of the start codon and point to a potential interaction of mRNA and tRNAfMet beyond the codon-anticodon hybridisation. A causal link between any observed impact on translation and an interaction with the 5′-UTR position −1 was, however, so far not conclusively established. A potential reason for this could be that only few mRNA sequence variants were tested prohibiting generalisable statements due to the high context dependence of translation initiation.
We therefore investigated whether the proposed interaction between mRNA and tRNAfMet could be substantiated relying on systematic high-throughput sequence-function mapping. We first constructed plasmids for the overexpression of tRNAfMet with the native A37 as well as the mutants A37C, A37G and A37U (Figure 5D, Materials and Methods). To reduce the background from the chromosomal tRNAfMet copies, we further deleted the metZWV locus of E. coli TOP10 ΔrhaA (‘WT’) yielding strain TOP10 ΔrhaA ΔmetZWV (‘ΔmetZWV’) and transformed both strains with the tRNA plasmids. Note that simultaneous knockout of metZWV and metY failed in our hands despite complementation via plasmid-borne tRNAfMet. Remarkably, transformants of ptRNAfMet-A37C showed severe growth inhibition (colonies visible only few days after transformation), whereas the native tRNAfMet-A37 and the other two mutants (tRNAfMet-A37G, tRNAfMet-A37U) were tolerated with minor effects on growth in both strains (Figure 5E). While in the case of the WT strain a small increase of doubling times was observable, ΔmetZWV showed an improvement of growth upon overexpression of all tRNAfMet variants, likely due to compensation of the reduced level of chromosomally derived tRNAfMet copies in this strain. The apparent toxicity of tRNAfMet-A37C could stem from global dysregulation of translation initiation, and due to its prohibitively slow growth we excluded this variant from further experiments. Next, we tested approximately 50 000 variants from Librandom in both strains (WT, ΔmetZWV) in presence of the remaining tRNAfMet plasmids via uASPIre (Figure 5F, Supplementary Figure S23, Materials and Methods). We analysed the resulting NGS data comparing 44 289 common 5′-UTR-CDS variants above high-quality read count threshold that appeared in all six conditions (i.e. two strains with three plasmids). Specifically, we determined for each condition the effects of 5′-UTR position −1 by dividing the mean rTR of variants with a given base at this position by the mean rTR of all other variants (Figure 5G). This analysis confirmed the above-mentioned (Figure 2) strong, base-specific impact of this position, and, beyond that, revealed a significant dependence of the effect on the base present in position 37 of tRNAfMet. To this end, we observed a strong increase in the rTR for variants whose base upstream of the start codon is complementary to position 37 of the overexpressed tRNAfMet in both the WT and ΔmetZWV strain. Non-complementarity consistently leads to a lower expression compared to the complementarity case across both strains and all tRNAfMet variants (Figure 5H). Similarly, a small yet significant positive impact on rTR is observable for the major wobble base pair G-U/U-G, which appears consistently for both directions of interaction (G in position 37 of tRNAfMet with U in 5′-UTR position −1 and vice versa) and both strains (Supplementary Figure S24). Interestingly, a U at 5′-UTR position −1 leads to a small rTR-boosting effect also in presence of the non-complementary initiators tRNAfMet-A37G and tRNAfMet-A37U only in the WT strain (Figure 5G). This can be explained by the presence of chromosomally encoded, endogenous tRNAfMet-A37 copies, since this positive effect is neutralised or slightly inverted in the ΔmetZWV strain. The effects at all other randomised positions in the mRNA were highly similar to the ones obtained for Librandom without overexpression of tRNAfMet variants (Supplementary Figure S25 compare Figure 2B).
Based on these findings and prompted by the observed growth impairment in our attempts to overexpress tRNAfMet-A37C (Figure 5E, Supplementary Figure S23), we lastly investigated how expression of endogenous genes is affected upon modification of position 37 of the initiator tRNA. To this end, we subjected E. coli ΔmetZWV overexpressing tRNAfMet-A37, tRNAfMet-A37G or tRNAfMet-A37U to label-free proteomics (Materials and Methods). Corroborating the results for the synthetic reporter, we observed that endogenous genes with 5′-UTR position −1 complementary to position 37 of the overexpressed initiator show on average a statistically significant increase in expression (Supplementary Figure S26). Notably, this effect, while statistically significant, is not consistent across all genes, which may be attributed to consequential effects of global dysregulation of genes and/or growth. For instance, altering the expression of regulators such as transcription factors will inevitably lead to secondary and tertiary effects on many other genes, which are hard to predict.
In summary, our findings strongly suggest a direct base-pairing interaction of 5′-UTR position −1 with the nucleotide following the anticodon in tRNAfMet (position 37), which leads to a significant positive effect on translation initiation upon successful hybridisation. Thus, our analyses confirm previous hypotheses and empirical observations to that end in a statistically solid manner based on more than 132 000 mRNA-tRNAfMet combinations, which were kinetically assessed in two different genetic backgrounds.
DISCUSSION
In this study, we systematically investigated and quantified the impact of 5′-UTR and N-terminal CDS on translation initiation through mapping of more than 1.2 million mRNA sequence variants to their corresponding kinetically recorded expression levels in E. coli constituting, to the best of our knowledge, the by far deepest experimental assessment of translational sequence determinants performed to date. This ultradeep characterisation effort provided strong experimental evidence for a base-pairing interaction between initiator tRNA and mRNA outside the codon-anticodon interaction and led to the rejection of a standing hypothesis on the impact of tRNA abundance on translation initiation. Furthermore, in combination with random and combinatorial library design, our ultrahigh-throughput approach allowed us to critically assess various sequence parameters known or supposed to influence translation initiation enabling precise quantification of effect sizes and correction for sequence-specific artefacts via solid statistical analyses.
To this end, we assessed mean effects of individual bases and positions in 5′-UTR and CDS along with various higher-order sequence parameters of the mRNA. We found that 25% of variance in our data can be explained by individual nucleotides and that GC-content, hybridisation with the 16S-rRNA and mRNA folding are the most significant determinants of translation initiation in line with previous studies (9–13,21,28–31,33–38,40,41,98).
Using a simplistic machine learning approach, we compared the predictive potential of 248 parameters, which ranked 16S-rRNA hybridisation highest (20.9%) followed by various predicted mRNA folding features (between 1.0% and 9.1%) and GC-content (6.0%) and pointed to a high degree of interaction and redundancy amongst parameters (Figure 3E). Furthermore, we discovered an unexpectedly large, base-specific contribution of two individual nucleotides, the negative impact of G in the third position of codon 2 (arginine) and the positive effect of U in position −1 of the 5′-UTR (Figure 2). Follow-up analyses revealed that the former is not causally related to tRNA availability in the cell but can likely be attributed to a stronger tendency of variants with CGG as second codon to form mRNA secondary structures (Figure 5A, B). Notably, mRNA accessibility at this position ranked amongst the most important features (accC1nt, pos. +6) of a predictive random forest model (Figure 3E), which confirms the relation of the observed effect to mRNA folding. Similar effects were also observable for codons further downstream and independent of the encoded amino acid, with effect sizes decreasing with the distance to the start codon. Thus our study clearly supports the common notion that the impact of mRNA folding is highest near the start codon. In this context, it should be mentioned that there can be cases in which translation is limited by mRNA regions further downstream, for instance for very stable secondary structures or due to ribosome stalling (36,99). Furthermore, it was found that specific arrays of triplets in both pro- and eukaryotes can have a pronounced impact on elongation rates unrelated to codon frequency or mRNA secondary structures (39,55,56,69). While arguably important, such case-specific phenomena were not subject of this study and should be avoided in synthetic constructs by re-designing the CDS wherever possible.
The positive effect of U directly upstream of the start codon, by contrast, was not linked to folding or any other mRNA parameter (Figure 5C), which prompted further experiments to that end. Specifically, we assessed whether a base-pairing interaction of 5′-UTR position −1 with the base in 3′ to the anticodon in initiator tRNAfMet (position A37) could be responsible for the effect. This hypothesis could be confirmed through compensatory mutation of tRNAfMet position 37, which led to a translation-favouring effect in all cases of complementarity between tRNA and 5′-UTR (Figure 5G, H). This could also be confirmed for endogenous genes/proteins as tested by label-free proteomics corroborating the observations for the synthetic reporter (Supplementary Figure S26). Several previous studies had shown that a U upstream of the start codon favours ribosome assembly and/or translation initiation in vitro and in vivo (37,71,87–95). A link of these effects to the aforementioned base-pairing interaction was postulated but not experimentally confirmed in these studies. Esposito et al. (96) provided further evidence in favour of this hypothesis in algal chloroplasts by substitution of position A37 in tRNAfMet. Notably, the variant tRNAfMet-A37C was not generated in their study, which showed severe growth inhibition and was thus excluded also in our work. For the tested reporter gene (petA), substitution of A37 indeed led to a translation-favouring effect only in cases of complementarity between tRNAfMet and the base upstream of the start codon. However, this observation was made on the basis of only three 5′-UTR position −1 variants of petA carrying a non-native weak UAA start codon and could not be confirmed for several other analysed genes. Whether the impact for petA is specific to this gene (context) or the weak start codon, or indeed related to the interaction between tRNAfMet and the base upstream of the start codon therefore remained unclear. Herein, we assessed 45 258 mRNA sequences tested with three tRNAfMet variants and in two strains of normal and reduced endogenous expression of native tRNAfMet-A37. This did not only substantiate the previously proposed quadruplet interaction in a statistically firm fashion but also allowed us to quantify comparably subtle phenomena such as wobble base pairing (Supplementary Figure S24), which are frequently masked for individual sequences and thus inaccessible to low-throughput approaches. Interestingly, this effect seems to be stronger for wobbling between G in 5′-UTR position −1 and U at tRNAfMet position 37 than for the inverted case (Supplementary Figure S24), the reasons of which remain unclear. We hypothesize that this could be related to base-specific structural changes in the tRNA upon modification since its 3D conformation is known to be critical for functionality (100).
Lastly, we constructed and assessed more than a million combinatorial and full-factorial 5′-UTR-CDS combinations, which, in view of the high degree of interactivity, is indispensable to correctly assign observed effects to different mRNA parts and sequence parameters, and to precisely measure their contribution. This allowed us to quantify the mean individual contribution of the 5′-UTR and CDS to translational variance in a manner that would not be possible otherwise (e.g. using fully random libraries), which amount to approximately 53% and 20%, respectively. Moreover, we capitalised on the combinatorial libraries and the large data basis to revise different hypotheses on the causal relationship between relative translation rate and codon usage. Similar to previous studies (e.g. (29,30,34,38,54,101)), our data confirmed a strong dependence of the rTR on the N-terminal CDS and a decreasing impact of codons with increasing distance to the start codon (Figures 2, 4B, C). While this dependence unquestionably exists, the underlying mechanistic reasons remain less clear and were linked to both differences in mRNA folding and cellular tRNA abundance in the past. The codon ramp hypothesis was developed on the basis of ribosome profiling data and suggests that ribosomes decelerate at rare codons directly downstream of the start codon preventing ribosome collisions thereby promoting efficient translation (53). Later, it was shown that technical and chemical reasons (cycloheximide treatment) could explain these findings, and the hypothesis could also not be confirmed in metagenomic analyses (68,102). Nonetheless, the codon ramp hypothesis was picked up in several later studies including recent works (30,34,38,39,53–59,64–67,99,103–116), which promoted us to further investigate the in our view still unresolved topic. In our data, we found a small (R2/ρ2 < 0.7%) yet significant correlation of the rTR with codon usage metrics (Figure 4D). However, the corresponding variance of the rTR can be explained by mRNA folding to an overwhelming degree while the contribution of codon usage metrics is extremely low (Figure 4F). This low impact is further corroborated by the fact that none of the codon usage metrics was capable to increase the prediction accuracy of a random forest model, whereas mRNA folding had a very large impact (Figure 4G). In summary, amongst the 1.2 million unique 5′-UTR-CDS combinations tested in this study the influence of different codons is virtually fully explainable by mRNA folding, whereas a causal connection to cellular tRNA abundance was either insignificant or negligibly small. The small apparent correlation between codon usage indices and rTR thus likely stems from differences in GC-content between rare and frequent codons, which leads to different tendencies to form secondary mRNA structures.
Consequently, our study highlights the importance of ultradeep sequence-function mapping for the accurate determination of the contribution of parts and phenomena involved in gene regulation. It should be mentioned that several other factors are known to influence translation initiation, which have not been addressed in this study. These include the use of different start codons, long-range interactions between ribosome and 5′-UTR, and limitations of translation elongation (e.g. related to protein folding). Nonetheless, the presented methodology can be applied to scrutinise these additional factors, which, together with the results from this study, could serve as a basis to improve on inaccuracies of currently available models for the prediction and forward design of prokaryotic protein expression.
DATA AVAILABILITY
Plasmid pASPIre4 and plasmids for tRNA overexpression are available via Addgene under IDs 196656, 196657, 196658 and 196659, respectively.
The raw data for all NGS experiments are provided in the NCBI SRA under accession codes SAMN27867644 (NGS run 1), SAMN27867645 (NGS run 2), SAMN27867646 (NGS run 3). An assignment of NGS runs to different libraries and samples is provided in Supplementary Table 5.
Time series data including IFP and cellular Bxb1-sfGFP values (rTR) for each variant including annotated scripts for data processing, statistical analyses and plotting are available under https://doi.org/10.5281/zenodo.7536586 or https://github.com/JeschekLab/uASPIre_UTR_CDS.
Supplementary Material
ACKNOWLEDGEMENTS
We kindly thank Dr Christian Beisel (ETH Zurich) and all members of the Genomics Facility Basel for support with NGS experiments and the Functional Genomics Centre Zurich for support with proteomics analyses. Further, we thank Dr Nico Claassens, Dr Thijs Nieuwkoop and Prof. Sven Panke for critical reading of the manuscript.
Author contributions: M.J. and S.H. conceived the study and planned experiments. S.H. performed experiments and computational works. S.H. and M.J. analysed data. M.J. coordinated the study. S.H. and M.J. wrote the manuscript.
Contributor Information
Simon Höllerer, Department of Biosystems Science and Engineering, Swiss Federal Institute of Technology – ETH Zurich, Basel CH-4058, Switzerland.
Markus Jeschek, Department of Biosystems Science and Engineering, Swiss Federal Institute of Technology – ETH Zurich, Basel CH-4058, Switzerland; Institute of Microbiology, Synthetic Microbiology Group, University of Regensburg, Regensburg D-93053, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Commission [MADONNA, 766975]; Swiss National Science Foundation under the NCCR ‘Molecular Systems Engineering’.
Conflict of interest statement. None declared.
This paper is linked to: doi:10.1093/nar/gkad035.
REFERENCES
- 1. Wang H.H., Isaacs F.J., Carr P.A., Sun Z.Z., Xu G., Forest C.R., Church G.M.. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009; 460:894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pullmann P., Ulpinnis C., Marillonnet S., Gruetzner R., Neumann S., Weissenborn M.J.. Golden mutagenesis: an efficient multi-site-saturation mutagenesis approach by Golden Gate cloning with automated primer design. Sci. Rep. 2019; 9:10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xu W., Klumbys E., Ang E.L., Zhao H.. Emerging molecular biology tools and strategies for engineering natural product biosynthesis. Metab Eng. Commun. 2020; 10:e00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vellanoweth R.L., Rabinowitz J.C.. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 1992; 6:1105–1114. [DOI] [PubMed] [Google Scholar]
- 5. Laursen B.S., Sorensen H.P., Mortensen K.K., Sperling-Petersen H.U.. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005; 69:101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lovmar M., Ehrenberg M.. Rate, accuracy and cost of ribosomes in bacterial cells. Biochimie. 2006; 88:951–961. [DOI] [PubMed] [Google Scholar]
- 7. Hersch S.J., Elgamal S., Katz A., Ibba M., Navarre W.W.. Translation initiation rate determines the impact of ribosome stalling on bacterial protein synthesis. J. Biol. Chem. 2014; 289:28160–28171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tietze L., Lale R.. Importance of the 5′ regulatory region to bacterial synthetic biology applications. Microb. Biotechnol. 2021; 14:2291–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shine J., Dalgarno L.. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steitz J.A., Jakes K.. How ribosomes select initiator regions in mRNA: base pair formation between the 3′ terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:4734–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jacob W.F., Santer M., Dahlberg A.E.. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:4757–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dalboge H., Carlsen S., Jensen E.B., Christensen T., Dahl H.H.. Expression of recombinant growth hormone in Escherichia coli: effect of the region between the Shine-Dalgarno sequence and the ATG initiation codon. DNA. 1988; 7:399–405. [DOI] [PubMed] [Google Scholar]
- 13. Li G.W., Oh E., Weissman J.S.. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012; 484:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen H., Bjerknes M., Kumar R., Jay E.. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994; 22:4953–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salis H.M., Mirsky E.A., Voigt C.A.. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009; 27:946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mutalik V.K., Guimaraes J.C., Cambray G., Lam C., Christoffersen M.J., Mai Q.A., Tran A.B., Paull M., Keasling J.D., Arkin A.P.et al.. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013; 10:354–360. [DOI] [PubMed] [Google Scholar]
- 17. Osterman I.A., Evfratov S.A., Sergiev P.V., Dontsova O.A.. Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res. 2013; 41:474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Espah Borujeni A., Channarasappa A.S., Salis H.M.. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 2014; 42:2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonde M.T., Pedersen M., Klausen M.S., Jensen S.I., Wulff T., Harrison S., Nielsen A.T., Herrgard M.J., Sommer M.O.. Predictable tuning of protein expression in bacteria. Nat. Methods. 2016; 13:233–236. [DOI] [PubMed] [Google Scholar]
- 20. Hecht A., Glasgow J., Jaschke P.R., Bawazer L.A., Munson M.S., Cochran J.R., Endy D., Salit M.. Measurements of translation initiation from all 64 codons in E. Coli. Nucleic Acids Res. 2017; 45:3615–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuo S.T., Jahn R.L., Cheng Y.J., Chen Y.L., Lee Y.J., Hollfelder F., Wen J.D., Chou H.D.. Global fitness landscapes of the Shine-Dalgarno sequence. Genome Res. 2020; 30:711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Komarova E.S., Chervontseva Z.S., Osterman I.A., Evfratov S.A., Rubtsova M.P., Zatsepin T.S., Semashko T.A., Kostryukova E.S., Bogdanov A.A., Gelfand M.S.et al.. Influence of the spacer region between the Shine-Dalgarno box and the start codon for fine-tuning of the translation efficiency in Escherichia coli. Microb. Biotechnol. 2020; 13:1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fargo D.C., Zhang M., Gillham N.W., Boynton J.E.. Shine-dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplasts or in Escherichia coli. Mol. Gen. Genet. 1998; 257:271–282. [DOI] [PubMed] [Google Scholar]
- 24. Zheng X., Hu G.Q., She Z.S., Zhu H.. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics. 2011; 12:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beck H.J., Fleming I.M., Janssen G.R.. 5′-Terminal augs in Escherichia coli mRNAs with shine-dalgarno sequences: identification and analysis of their roles in non-canonical translation initiation. PLoS One. 2016; 11:e0160144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakagawa S., Niimura Y., Gojobori T.. Comparative genomic analysis of translation initiation mechanisms for genes lacking the Shine-Dalgarno sequence in prokaryotes. Nucleic Acids Res. 2017; 45:3922–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saito K., Green R., Buskirk A.R.. Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing. Elife. 2020; 9:e55002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Smit M.H., van Duin J.. Control of translation by mRNA secondary structure in Escherichia coli. A quantitative analysis of literature data. J. Mol. Biol. 1994; 244:144–150. [DOI] [PubMed] [Google Scholar]
- 29. Voges D., Watzele M., Nemetz C., Wizemann S., Buchberger B.. Analyzing and enhancing mRNA translational efficiency in an Escherichia coli in vitro expression system. Biochem. Biophys. Res. Commun. 2004; 318:601–614. [DOI] [PubMed] [Google Scholar]
- 30. Kudla G., Murray A.W., Tollervey D., Plotkin J.B.. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009; 324:255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Simonetti A., Marzi S., Jenner L., Myasnikov A., Romby P., Yusupova G., Klaholz B.P., Yusupov M.. A structural view of translation initiation in bacteria. Cell. Mol. Life Sci. 2009; 66:423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Na D., Lee S., Lee D.. Mathematical modeling of translation initiation for the estimation of its efficiency to computationally design mRNA sequences with desired expression levels in prokaryotes. BMC Syst. Biol. 2010; 4:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seo S.W., Yang J.S., Kim I., Yang J., Min B.E., Kim S., Jung G.Y.. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab. Eng. 2013; 15:67–74. [DOI] [PubMed] [Google Scholar]
- 34. Goodman D.B., Church G.M., Kosuri S.. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013; 342:475–479. [DOI] [PubMed] [Google Scholar]
- 35. Reeve B., Hargest T., Gilbert C., Ellis T.. Predicting translation initiation rates for designing synthetic biology. Front. Bioeng. Biotechnol. 2014; 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Espah Borujeni A., Cetnar D., Farasat I., Smith A., Lundgren N., Salis H.M.. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Res. 2017; 45:5437–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yus E., Yang J.S., Sogues A., Serrano L.. A reporter system coupled with high-throughput sequencing unveils key bacterial transcription and translation determinants. Nat. Commun. 2017; 8:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cambray G., Guimaraes J.C., Arkin A.P.. Evaluation of 244,000 synthetic sequences reveals design principles to optimize translation in Escherichia coli. Nat. Biotechnol. 2018; 36:1005–1015. [DOI] [PubMed] [Google Scholar]
- 39. Verma M., Choi J., Cottrell K.A., Lavagnino Z., Thomas E.N., Pavlovic-Djuranovic S., Szczesny P., Piston D.W., Zaher H.S., Puglisi J.D.et al.. A short translational ramp determines the efficiency of protein synthesis. Nat. Commun. 2019; 10:5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Terai G., Asai K.. Improving the prediction accuracy of protein abundance in Escherichia coli using mRNA accessibility. Nucleic Acids Res. 2020; 48:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cetnar D.P., Salis H.M.. Systematic quantification of sequence and structural determinants controlling mRNA stability in bacterial operons. ACS Synth. Biol. 2021; 10:318–332. [DOI] [PubMed] [Google Scholar]
- 42. Sharp P.M., Li W.H.. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987; 15:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eyre-Walker A., Bulmer M.. Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res. 1993; 21:4599–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bentele K., Saffert P., Rauscher R., Ignatova Z., Bluthgen N.. Efficient translation initiation dictates codon usage at gene start. Mol. Syst. Biol. 2013; 9:675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hanson G., Coller J.. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018; 19:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ikemura T. Correlation between the abundance of Escherichia coli transfer rnas and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981; 146:1–21. [DOI] [PubMed] [Google Scholar]
- 47. Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985; 2:13–34. [DOI] [PubMed] [Google Scholar]
- 48. Dong H., Nilsson L., Kurland C.G.. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996; 260:649–663. [DOI] [PubMed] [Google Scholar]
- 49. Gustafsson C., Govindarajan S., Minshull J.. Codon bias and heterologous protein expression. Trends Biotechnol. 2004; 22:346–353. [DOI] [PubMed] [Google Scholar]
- 50. Mitarai N., Sneppen K., Pedersen S.. Ribosome collisions and translation efficiency: optimization by codon usage and mRNA destabilization. J. Mol. Biol. 2008; 382:236–245. [DOI] [PubMed] [Google Scholar]
- 51. Zhang G., Ignatova Z.. Generic algorithm to predict the speed of translational elongation: implications for protein biogenesis. PLoS One. 2009; 4:e5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dobrzynski M., Bruggeman F.J.. Elongation dynamics shape bursty transcription and translation. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:2583–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., Pan T., Dahan O., Furman I., Pilpel Y.. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010; 141:344–354. [DOI] [PubMed] [Google Scholar]
- 54. Tuller T., Waldman Y.Y., Kupiec M., Ruppin E.. Translation efficiency is determined by both codon bias and folding energy. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:3645–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arthur L., Pavlovic-Djuranovic S., Smith-Koutmou K., Green R., Szczesny P., Djuranovic S.. Translational control by lysine-encoding A-rich sequences. Sci. Adv. 2015; 1:e150015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chu D., Kazana E., Bellanger N., Singh T., Tuite M.F., von der Haar T.. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 2014; 33:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boel G., Letso R., Neely H., Price W.N., Wong K.H., Su M., Luff J., Valecha M., Everett J.K., Acton T.B.et al.. Codon influence on protein expression in E. Coli correlates with mRNA levels. Nature. 2016; 529:358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Osterman I.A., Chervontseva Z.S., Evfratov S.A., Sorokina A.V., Rodin V.A., Rubtsova M.P., Komarova E.S., Zatsepin T.S., Kabilov M.R., Bogdanov A.A.et al.. Translation at first sight: the influence of leading codons. Nucleic Acids Res. 2020; 48:6931–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zahurancik W.J., Szkoda B.E., Lai L.B., Gopalan V.. Ramping recombinant protein expression in bacteria. Biochemistry. 2020; 59:2122–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nieuwkoop T., Terlouw B.R., Stevens K.G., Scheltema A.R., de Ridder D., van der Oost J., Claassens N.J.. Revealing determinants of translation efficiency via whole-gene codon randomisation and machine learning. Nucleic Acids Res. 2023; 10.1093/nar/gkad035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. dos Reis M., Wernisch L., Savva R.. Unexpected correlations between gene expression and codon usage bias from microarray data for the whole Escherichia coli K-12 genome. Nucleic Acids Res. 2003; 31:6976–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Plotkin J.B., Kudla G.. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 2011; 12:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Quax T.E., Claassens N.J., Soll D., van der Oost J.. Codon bias as a means to fine-tune gene expression. Mol. Cell. 2015; 59:149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nieuwkoop T., Finger-Bou M., van der Oost J., Claassens N.J.. The ongoing quest to crack the genetic code for protein production. Mol. Cell. 2020; 80:193–209. [DOI] [PubMed] [Google Scholar]
- 65. Etchegaray J.P., Inouye M.. Translational enhancement by an element downstream of the initiation codon in Escherichia coli. J. Biol. Chem. 1999; 274:10079–10085. [DOI] [PubMed] [Google Scholar]
- 66. Martin-Farmer J., Janssen G.R.. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol. Microbiol. 1999; 31:1025–1038. [DOI] [PubMed] [Google Scholar]
- 67. Sato T., Terabe M., Watanabe H., Gojobori T., Hori-Takemoto C., Miura K.. Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J. Biochem. 2001; 129:851–860. [DOI] [PubMed] [Google Scholar]
- 68. Gerashchenko M.V., Gladyshev V.N.. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014; 42:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu C.H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M.S., Liu Y.. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell. 2015; 59:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kosuri S., Goodman D.B., Cambray G., Mutalik V.K., Gao Y., Arkin A.P., Endy D., Church G.M.. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:14024–14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hollerer S., Papaxanthos L., Gumpinger A.C., Fischer K., Beisel C., Borgwardt K., Benenson Y., Jeschek M.. Large-scale DNA-based phenotypic recording and deep learning enable highly accurate sequence-function mapping. Nat. Commun. 2020; 11:3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Claassens N.J., Finger-Bou M., Scholten B., Muis F., de Groot J.J., de Gier J.W., de Vos W.M., van der Oost J.. Bicistronic design-based continuous and high-level membrane protein production in Escherichia coli. ACS Synth. Biol. 2019; 8:1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Silva-Rocha R., Martinez-Garcia E., Calles B., Chavarria M., Arce-Rodriguez A., de Las Heras A., Paez-Espino A.D., Durante-Rodriguez G., Kim J., Nikel P.I.et al.. The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2013; 41:D666–D675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sambrook J.F., Russel D.W.. Molecular Cloning: A Laboratory Manual. 2001; Cold Spring Harbor Laboratory Press. [Google Scholar]
- 76. Schneider T.D., Roschitzki B., Grossmann J., Kraemer T., Steuer A.E.. Determination of the time since deposition of blood traces utilizing a liquid chromatography-mass spectrometry-based proteomics approach. Anal. Chem. 2022; 94:10695–10704. [DOI] [PubMed] [Google Scholar]
- 77. Wolski W.E., Nanni P., Grossmann J., d’Errico M., Schlapbach R., Panse C.. prolfqua: a comprehensive R-package for Proteomics Differential Expression Analysis. 2022; bioRxiv doi:09 June 2022, preprint: not peer reviewed 10.1101/2022.06.07.494524. [DOI] [PMC free article] [PubMed]
- 78. Durfee T., Nelson R., Baldwin S., Plunkett G., Burland V., Mau B., Petrosino J.F., Qin X., Muzny D.M., Ayele M.et al.. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 2008; 190:2597–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. R Core Team 4.0.3 ed. R Foundation for Statistical Computing. 2017; Vienna, Austria. [Google Scholar]
- 80. Do C.B., Woods D.A., Batzoglou S.. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics. 2006; 22:e90–e98. [DOI] [PubMed] [Google Scholar]
- 81. Mathews D.H., Sabina J., Zuker M., Turner D.H.. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999; 288:911–940. [DOI] [PubMed] [Google Scholar]
- 82. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kiryu H., Terai G., Imamura O., Yoneyama H., Suzuki K., Asai K.. A detailed investigation of accessibilities around target sites of siRNAs and miRNAs. Bioinformatics. 2011; 27:1788–1797. [DOI] [PubMed] [Google Scholar]
- 84. Egan S.M., Schleif R.F.. A regulatory cascade in the induction of rhaBAD. J. Mol. Biol. 1993; 234:87–98. [DOI] [PubMed] [Google Scholar]
- 85. Jeschek M., Gerngross D., Panke S.. Rationally reduced libraries for combinatorial pathway optimization minimizing experimental effort. Nat. Commun. 2016; 7:11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Seong B.L., RajBhandary U.L.. Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:8859–8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ganoza M.C., Fraser A.R., Neilson T.. Nucleotides contiguous to AUG affect translational initiation. Biochemistry. 1978; 17:2769–2775. [DOI] [PubMed] [Google Scholar]
- 88. Eckhardt H., Luhrmann R.. Recognition by initiator transfer ribonucleic acid of a uridine 5′ adjacent to the AUG codon: different conformational states of formylatable methionine-accepting transfer ribonucleic acid at the ribosomal peptidyl site. Biochemistry. 1981; 20:2075–2080. [DOI] [PubMed] [Google Scholar]
- 89. Ganoza M.C., Sullivan P., Cunningham C., Hader P., Kofoid E.C., Neilson T.. Effect of bases contiguous to AUG on translation initiation. J. Biol. Chem. 1982; 257:8228–8232. [PubMed] [Google Scholar]
- 90. Hui A., Hayflick J., Dinkelspiel K., de Boer H.A. Mutagenesis of the three bases preceding the start codon of the beta-galactosidase mRNA and its effect on translation in Escherichia coli. EMBO J. 1984; 3:623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ganoza M.C., Marliere P., Kofoid E.C., Louis B.G.. Initiator tRNA may recognize more than the initiation codon in mRNA: a model for translational initiation. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:4587–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gross G., Mielke C., Hollatz I., Blocker H., Frank R.. RNA primary sequence or secondary structure in the translational initiation region controls expression of two variant interferon-beta genes in Escherichia coli. J. Biol. Chem. 1990; 265:17627–17636. [PubMed] [Google Scholar]
- 93. Esposito D., Hicks A.J., Stern D.B.. A role for initiation codon context in chloroplast translation. Plant Cell. 2001; 13:2373–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Krishnan K.M., Van Etten W.J. 3rd, Janssen G.R.. Proximity of the start codon to a leaderless mRNA’s 5′ terminus is a strong positive determinant of ribosome binding and expression in Escherichia coli. J. Bacteriol. 2010; 192:6482–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Krishnan K.M. Ribosome-mRNA interactions that contribute to recognition and binding of a 5’-terminal AUG start codon. 2010; Dissertation Miami University; ETH: 5378https://europepmc.org/article/ETH/5378. [Google Scholar]
- 96. Esposito D., Fey J.P., Eberhard S., Hicks A.J., Stern D.B.. In vivo evidence for the prokaryotic model of extended codon-anticodon interaction in translation initiation. EMBO J. 2003; 22:651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Barraud P., Schmitt E., Mechulam Y., Dardel F., Tisne C.. A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis. Nucleic Acids Res. 2008; 36:4894–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Na D., Lee D. RBSDesigner: software for designing synthetic ribosome binding sites that yields a desired level of protein expression. Bioinformatics. 2010; 26:2633–2634. [DOI] [PubMed] [Google Scholar]
- 99. Samatova E., Daberger J., Liutkute M., Rodnina M.V.. Translational control by ribosome pausing in bacteria: how a non-uniform pace of translation affects protein production and folding. Front Microbiol. 2020; 11:619430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. de Crecy-Lagard V., Jaroch M. Functions of bacterial tRNA modifications: from ubiquity to diversity. Trends Microbiol. 2021; 29:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Presnyak V., Alhusaini N., Chen Y.H., Martin S., Morris N., Kline N., Olson S., Weinberg D., Baker K.E., Graveley B.R.et al.. Codon optimality is a major determinant of mRNA stability. Cell. 2015; 160:1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Mohammad F., Green R., Buskirk A.R.. A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. Elife. 2019; 8:e42591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Fredrick K., Ibba M. How the sequence of a gene can tune its translation. Cell. 2010; 141:227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cannarozzi G., Schraudolph N.N., Faty M., von Rohr P., Friberg M.T., Roth A.C., Gonnet P., Gonnet G., Barral Y.. A role for codon order in translation dynamics. Cell. 2010; 141:355–367. [DOI] [PubMed] [Google Scholar]
- 105. Gamble C.E., Brule C.E., Dean K.M., Fields S., Grayhack E.J.. Adjacent codons act in concert to modulate translation efficiency in yeast. Cell. 2016; 166:679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Navon S.P., Kornberg G., Chen J., Schwartzman T., Tsai A., Puglisi E.V., Puglisi J.D., Adir N.. Amino acid sequence repertoire of the bacterial proteome and the occurrence of untranslatable sequences. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:7166–7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Burkhardt D.H., Rouskin S., Zhang Y., Li G.W., Weissman J.S., Gross C.A.. Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. Elife. 2017; 6:e22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Szavits-Nossan J., Ciandrini L.. Inferring efficiency of translation initiation and elongation from ribosome profiling. Nucleic Acids Res. 2020; 48:9478–9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Xu Y., Liu K., Han Y., Xing Y., Zhang Y., Yang Q., Zhou M.. Codon usage bias regulates gene expression and protein conformation in yeast expression system P. Pastoris. Microb. Cell Fact. 2021; 20:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hia F., Takeuchi O.. The effects of codon bias and optimality on mRNA and protein regulation. Cell. Mol. Life Sci. 2021; 78:1909–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Xu K., Tong Y., Li Y., Tao J., Li J., Zhou J., Liu S.. Rational design of the N-terminal coding sequence for regulating enzyme expression in Bacillus subtilis. ACS Synth Biol. 2021; 10:265–276. [DOI] [PubMed] [Google Scholar]
- 112. Ferreira M., Ventorim R., Almeida E., Silveira S., Silveira W.. Protein abundance prediction through machine learning methods. J. Mol. Biol. 2021; 433:167267. [DOI] [PubMed] [Google Scholar]
- 113. Diez M., Medina-Munoz S.G., Castellano L.A., da Silva Pescador G., Wu Q., Bazzini A.A.. iCodon customizes gene expression based on the codon composition. Sci. Rep. 2022; 12:12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kim D.J., Kim J., Lee D.H., Lee J., Woo H.M.. DeepTESR: a deep learning framework to predict the degree of translational elongation short ramp for gene expression control. ACS Synth. Biol. 2022; 11:1719–1726. [DOI] [PubMed] [Google Scholar]
- 115. Miller J.B., Meurs T.E., Hodgman M.W., Song B., Miller K.N., Ebbert M.T.W., Kauwe J.S.K., Ridge P.G.. The Ramp Atlas: facilitating tissue and cell-specific ramp sequence analyses through an intuitive web interface. NAR Genom. Bioinform. 2022; 4:lqac039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wang C., Zhang W., Tian R., Zhang J., Zhang L., Deng Z., Lv X., Li J., Liu L., Du G.et al.. Model-driven design of synthetic N-terminal coding sequences for regulating gene expression in yeast and bacteria. Biotechnol. J. 2022; 17:e2100655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmid pASPIre4 and plasmids for tRNA overexpression are available via Addgene under IDs 196656, 196657, 196658 and 196659, respectively.
The raw data for all NGS experiments are provided in the NCBI SRA under accession codes SAMN27867644 (NGS run 1), SAMN27867645 (NGS run 2), SAMN27867646 (NGS run 3). An assignment of NGS runs to different libraries and samples is provided in Supplementary Table 5.
Time series data including IFP and cellular Bxb1-sfGFP values (rTR) for each variant including annotated scripts for data processing, statistical analyses and plotting are available under https://doi.org/10.5281/zenodo.7536586 or https://github.com/JeschekLab/uASPIre_UTR_CDS.