Abstract
Cancer-associated fibroblasts (CAFs) can exert their immunosuppressive effects by secreting various effectors that are involved in the regulation of tumor-infiltrating immune cells as well as other immune components in the tumor immune microenvironment (TIME), thereby promoting tumorigenesis, progression, metastasis, and drug resistance. Although a large number of studies suggest that CAFs play a key regulatory role in the development of head and neck squamous cell carcinoma (HNSCC), there are limited studies on the relevance of CAFs to the prognosis of HNSCC. In this study, we identified a prognostic signature containing eight CAF-related genes for HNSCC by univariate Cox analysis, lasso regression, stepwise regression, and multivariate Cox analysis. Our validation in primary cultures of CAFs from human HNSCC and four human HNSCC cell lines confirmed that these eight genes are indeed characteristic markers of CAFs. Immune cell infiltration differences analysis between high-risk and low-risk groups according to the eight CAF-related genes signature hinted at CAFs regulatory roles in the TIME, further revealing its potential role on prognosis. The signature of the eight CAF-related genes was validated in different independent validation cohorts and all showed that it was a valid marker for prognosis. The significantly higher overall survival (OS) in the low-risk group compared to the high-risk group was confirmed by Kaplan-Meier (K-M) analysis, suggesting that the signature of CAF-related genes can be used as a non-invasive predictive tool for HNSCC prognosis. The low-risk group had significantly higher levels of tumor-killing immune cell infiltration, as confirmed by CIBERSORT analysis, such as CD8+ T cells, follicular helper T cells, and Dendritic cells (DCs) in the low-risk group. In contrast, the level of infiltration of pro-tumor cells such as M0 macrophages and activated Mast cells (MCs) was lower. It is crucial to delve into the complex mechanisms between CAFs and immune cells to find potential regulatory targets and may provide new evidence for subsequently targeted immunotherapy. These results suggest that the signature of the eight CAF-related genes is a powerful indicator for the assessment of the TIME of HNSCC. It may provide a new and reliable potential indicator for clinicians to predict the prognosis of HNSCC, which may be used to guide treatment and clinical decision-making in HNSCC patients. Meanwhile, CAF-related genes are expected to become tumor biomarkers and effective targets for HNSCC.
Keywords: Cancer-associated fibroblasts, Prognostic signature, Immune cell infiltration, Head and neck squamous cell carcinoma
Abbreviations: CAFs, Cancer-associated fibroblasts; CSCs, cancer stem cells; DCs, Dendritic cells; EMT, epithelial mesenchymal transition; GEO, Gene Expression Omnibus; GEPIA, Gene Expression Profiling Interactive Analysis; GO, Gene Ontology; GSEA, Gene Set Enrichment Analysis; HNSCC, head and neck squamous cell carcinoma; HR, Hazard Ratio; KEGG, Kyoto Encyclopedia of Genes and Genomes; K-M, Kaplan-Meier; MCs, Mast cells; NFs, normal fibroblasts; OS, overall survival; OSCC, oral squamous cell carcinomas; TME, tumor microenvironment; TIME, tumor immune microenvironment; TAMs, tumor-associated macrophages; TCGA, The Cancer Genome Atlas; ROC, receiver operating characteristic
Summary
We identified a prognostic signature of head and neck squamous cell carcinoma (HNSCC) containing eight Cancer-associated fibroblasts (CAFs) related genes by bioinformatic analysis. Our validation in primary cultures of CAFs from human HNSCC and four HNSCC cell lines confirmed that these eight genes are indeed characteristic markers of CAFs. Immune cell infiltration differences analysis according to the eight CAF-related genes signature hinted CAFs regulatory roles in the tumor immune microenvironment (TIME), further revealing its potential role on prognosis. The eight CAF-related genes may be a new potential indicator for HNSCC prognosis, which used to guide treatment and clinical decision-making in HNSCC patients.
1. Introduction
Head and neck squamous cell carcinoma (HNSCC), the seventh most frequent type of cancer and responsible for many deaths worldwide, is a major health problem [1]. Cancerous cells, adjacent epithelial, stromal, and immune cells, and their surrounding matrix together constitute the tumor microenvironment (TME) [2]. Among them, cancer-associated fibroblasts (CAFs) and phenotypically altered fibroblasts are the dominant cells of TME, which can facilitate tumor proliferation, angiogenesis, invasion, immune escape, drug resistance, and metastasis [3,4].
CAFs interact with nearby cells through various mechanisms, including autocrine, paracrine, and direct actions. Especially, through the secretion of exosomes, CAFs can form complex networks and perform their biological functions [5]. CAFs interact with tumor-infiltrating immune cells and other immune components within the TME through the secretion of various cytokines, growth factors, chemokines, exosomes, and other effector molecules, thus resulting in an immunosuppressive TME that allows cancer cells to evade the surveillance of the immune system. In-depth studies of CAFs and immune microenvironment interactions, particularly the complicated mechanisms connecting CAFs with immune cells, might provide novel strategies for subsequent targeted immunotherapies [6].
For many cancers, the presence of CAFs means a boost in malignancy features as they interact with cancer cells and other components of the TME to shape a tumor-supportive environment [[7], [8], [9]]. CAFs can be used as biomarkers of immunosuppressive TME [10] and be associated with the poor prognosis of many different types of cancer [3]. In a recent study, correlational analysis of clinical oral squamous cell carcinomas (OSCC) datasets with TGFβ axis differentially expressed genes revealed that the summed expression of FN1, TGFβ2, TGFBR2, and TGFBI, dubbed as the CAF index, is a powerful predictor in dichotomized survival analysis for patients of diverse cancer subtypes [11]. This study confirmed the significant advantages of the CAF index compared to the epithelial mesenchymal transition (EMT) score. However, it also had its limitations, such as being validated only in OSCC and the absence of studies on the mechanisms of immune cell regulation. To assess the prognosis of HNSCC patients more comprehensively and accurately, and to provide a reliable basis for further exploration of the immunomodulatory mechanisms of CAFs on HNSCC, we constructed a CAF-related prognosis signature and validated it in multiple HNSCC validation cohorts. In our current study, we integrated bioinformatics analyses to construct and validate a prognosis signature of eight CAF-related genes for HNSCC through univariate regression, lasso regression, stepwise regression, and multivariate regression analysis in training and testing cohorts. The immune cell infiltration analysis suggested that CAF-related genes might regulate tumorigenesis and progression through their immunomodulatory effects. These results might provide potential therapeutic targets for HNSCC patients.
2. Materials and methods
2.1. Acquisition and processing of data sets
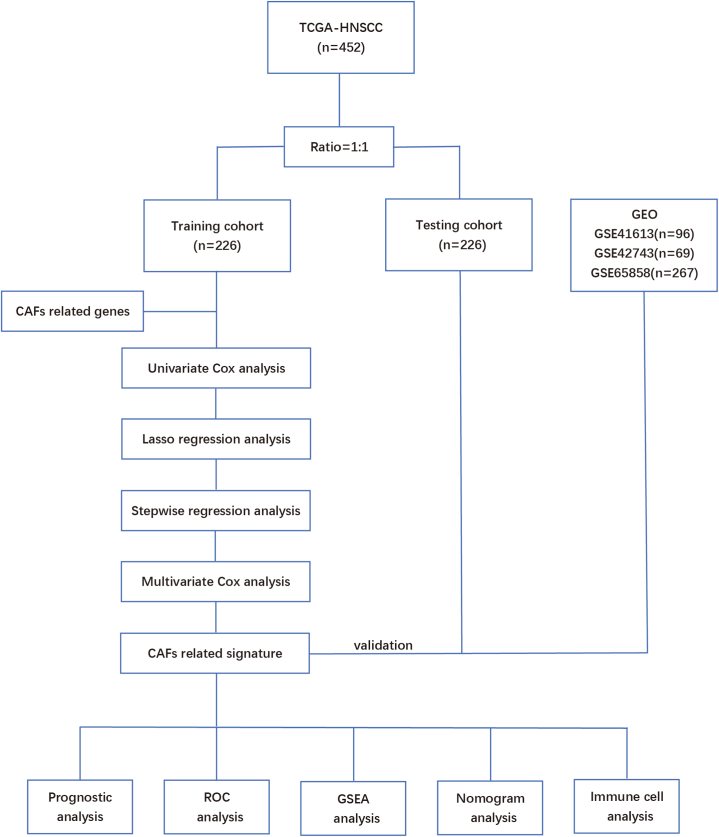
The RNA-seq and clinical data of patients with HNSCC were obtained from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/), and TCGA-HNSCC were downloaded from UCSC Xena (http://xena.ucsc.edu/) database. GSE41613, GSE42743, and GSE65858 datasets were obtained from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database. To ensure the accuracy of the follow-up analysis, we re-annotated the expression matrix and homogenized the gene symbol of the four queue matrices. The FPKM values were converted to transcripts per kilobase million (TPM) values and normalized and standardized by transforming the expression matrix into log2 (x + 1). This study included patients with complete follow-up data, a survival time of more than 30 days, and complete clinicopathological data. The TCGA cohort was divided into a training cohort and an internal verification cohort at the ratio of 1:1, and the three GEO data sets were used as external independent verification cohorts. The GSE41613 dataset consisting of 96 HNSCC patients was set as verification cohort 1. The GSE42743 dataset consisting of 69 HNSCC patients was set as verification cohort 2. The GSE65858 dataset consisting of 267 HNSCC patients was set as verification cohort 3 (Fig. 1).
Fig. 1.
Flowchart of the study.
2.2. The relationship between CAF score and HNSCC prognosis
We calculated the CAF score of TCGA-HNSCC patients through TIDE online website (http://tide.dfci.harvard.edu/) [12], Based on tumor expression profiles, TIDE can score multiple transcriptomic biomarkers of CAFs. TCGA-HNSCC patients were divided into high and low CAF score groups [[13], [14], [15]], based on the best intercept value, and the prognosis of the high and low CAF score groups was analyzed by the Survminer package in R software [16].
2.3. Acquisition of CAF-related genes
Two subpopulations of CAFs were identified in the study of Sidharth V. Puram et al. through re-evaluating the bulk RNA-seq data of 6000 cells from 18 patients [17]. In the study of Phillip M. Galbo Jr et al., six CAF subtypes were also identified according to their differentially expressed marker genes through FindMarkers function with a Wilcoxon rank-sum test [18]. All the related genes of those CAF subsets were sorted out and combined, which were called CAF-related genes. Afterwards, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of CAF-related genes were performed using clusterProfiler package in the R software.
2.4. Screening of CAF-related genes signature
To determine the prognosis related genes, we performed univariate Cox analysis of the CAF-related genes. In univariate Cox analysis, genes with P < 0.05 were included in Lasso regression. To further improve the prediction efficiency of signatures, genes with significant values were reconfirmed using stepwise regression and finally incorporated into subsequent multivariate Cox analysis to establish a prognostic signature.
2.5. Construction and evaluation of the prognostic signature based on eight CAF-related genes
The multivariate Cox regression model was used to establish an independent prognostic signature. The risk score of each HNSCC patient was calculated as the expression value of each gene multiplied by the sum of their weights in the multivariable Cox model. According to the optimal intercept value, HNSCC patients were divided into high- and low-risk groups. Kaplan-Meier (K-M) survival analysis was used to compare the survival differences between high- and low-risk groups using the Survminer R package, and the “timeROC” R package was used to evaluate the predictive value of the model. To further verify the effectiveness of our prognostic markers, we verified them in the internal verification cohort and three external verification cohorts.
2.6. Clinical independent prognostic factors analysis
Univariate and multivariate Cox regression analyses were used to analyze the relationship between clinical data, such as age, sex, TNM stage, risk score, and survival prognosis, to determine whether the risk score was an independent prognostic indicator. We constructed a prediction signature with the help of a nomogram and used the calibration curves of 3 years and 5 years to test the accuracy of the prediction model using the RMS and survival packages in the R software [16].
2.7. GSEA enrichment analysis and immune infiltration level analysis
Evaluation of gene enrichment function in the high- and low-risk groups was performed using Gene Set Enrichment Analysis (GSEA) by the clusterProfiler package in the R software. We constructed protein-protein interaction networks and screened the hub-genes in prognostic signature using the String database (https://cn.string-db.org/). Moreover, the relationships between immune cells and prognostic hub-genes were explored through the TIMER database (https://cistrome.shinyapps.io/timer/). To quantify the proportion of immune cells in HNSCC patients, we used CiberSort (https://cibersort.stanford.edu/) to compare the level of immune cell infiltration in the high- and low-risk groups.
2.8. Cell culture
Human HNSCC cancer tissue (patient samples were ethically approved by Yantai Yuhuangding Hospital and informed consent was obtained from patients) was cut into 1 mm3 pieces, suspended in DMEM/F12 medium (Biological Industries, Israel) with 0.1% type I collagenase (Coolaber, China), incubated for 2 h at 37 °C in a shaker, filtered through a 70 μm filter, and centrifuged to pellet cells (1500r, 5 min). Cell precipitates were resuspended in DMEM/F12 medium containing 10% fetal bovine serum (Biological Industries, Israel) and 1% penicillin + streptomycin (Sparkjade, China) and incubated at 37 °C in a 5% CO2 incubator.
Human hypopharyngeal carcinoma cells (FaDu), human laryngeal carcinoma cells (AMC–HN–8, TU212), and human nasopharyngeal carcinoma cells (5–8F) were purchased from ATCC (Virginia, USA), and were cultured in DMEM medium (Biological Industries, Israel) containing 10% fetal bovine serum (Biological Industries, Israel). All Cells were incubated at 37 °C in a 5% CO2 incubator.
2.9. RNA isolation and quantitative PCR
Total RNA in CAFs, FaDu cells, AMC–HN–8 cells, TU212 cells, and 5–8F cells was extracted using TRIzol reagent (SparkJade, China) according to the manufacturer's instructions. cDNA was synthesized using an Evo M-MLV Reverse transcription Kit (Applied Biological Materials, Canada). Quantitative PCR reactions were performed using a SYBR Green qPCR Mix kit (SparkJade, China) on a FTC-3000A fluorescence quantitative PCR instrument (Funglyn Biotech, Canada). Gene expression levels were quantified using the 2-ΔΔCT method based on CT values and normalized to a reference gene, GAPDH. All samples were prepared in triplicates, and the mean value was used for comparative analyses. The primers used for amplification of mRNA and genes were synthesized by Sango Biotech, China, and are listed in Supplementary Table 1.
3. Results
3.1. CAF score was associated with poor prognosis of HNSCC, and acquisition and function analysis of CAF-related genes
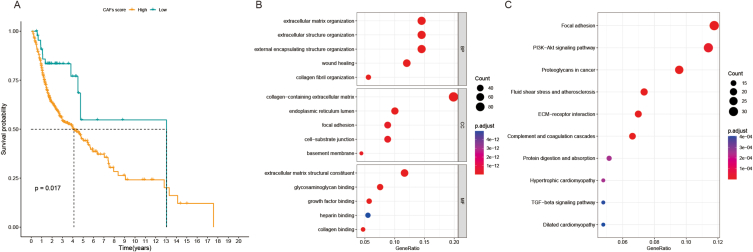
The CAF score of TCGA-HNSCC patients is shown in Supplementary Table 2. Through the survminer package in the R software, the best intercept value (cut-off = −0.14) was calculated. Based on the cut-off value, the 452 patients were divided into the high CAF score group (314 patients) and the low CAF score group (138 patients). The K-M curve showed that the prognosis of patients with high CAF score was poorer than that in patients with low CAF score (p = 0.017) (Fig. 2A).
Fig. 2.
A. K-M survival analysis was used to analyze the CAF score of OS curve in HNSCC patients. B. GO analysis of CAF-related genes. C. KEGG analysis of CAF-related genes.
Based on the study by Sidharth V. Puram et al. [17] and Phillip M. Galbo Jr et al. [18], a total of 661 CAF-related genes were identified (Supplementary Table 3). As illustrated in Fig. 2B and C, GO enrichment analysis indicated that the 661 CAF-related genes were mainly enriched in extracellular matrix organization, collagen fibril organization, collagen-containing extracellular matrix, and extracellular matrix structural constituent(Supplementary Table 4), while KEGG analysis showed that these 661 CAF-related genes were mainly enriched in focal adhesion, ECM-receptor interaction, complement, and coagulation cascades, proteoglycans in cancer, and PI3K-Akt signaling pathway (Supplementary Table 5).
3.2. Construction of a prognostic signature based on CAF-related genes
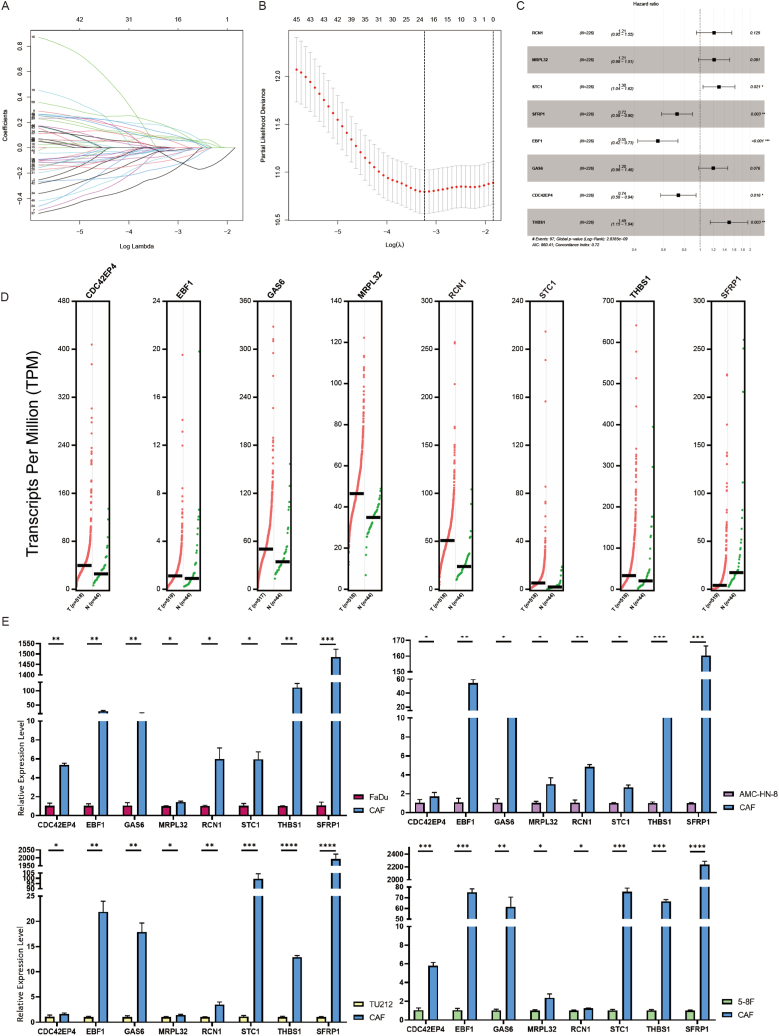
Compared with the TCGA expression matrix of the training cohort, 472 CAF-related genes were included in the follow-up analysis. 50 genes related to prognosis were screened from the CAF-related mRNAs by univariate Cox analysis (Supplementary Table 6). 21 genes were obtained by Lasso regression (Fig. 3A and B). To further screen the meaningful influencing factors and eliminate the unrelated factors, we conducted a stepwise regression analysis and obtained eight genes with the highest prognostic values, including RCN1, MRPL32, STC1, SFRP1, EBF1, GAS6, CDC42EP4, and THBS1, which were considered to have significant characteristic values.
Fig. 3.
The acquisition of CAF-related genes signature. (A) Lasso coefficient distribution in the training cohort. (B) The coefficient profile is generated according to the logarithmic λ sequence. Selection of optimal parameter λ in the lasso model. (C) Forest plot of eight CAF-related genes signature. (D) GEPIA2 database analysis of the expression of eight CAF-related genes in HNSCC. (E) qPCR validation of the expression of eight CAF-related mRNAs in primary cultured HNSCC CAFs versus human hypopharyngeal carcinoma cells (FaDu), human laryngeal carcinoma cells (AMC–HN–8, TU212) and human nasopharyngeal carcinoma cells (5–8F). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Multivariate Cox analysis was used to construct prognostic biomarkers using the eight mRNA (Fig. 3C). Five risk factors, namely RCN1, MRPL32, STC1, GAS6, and THBS1, were identified for the prognosis of patients with HNSCC using the cutoff value of Hazard Ratio (HR) > 1. Meanwhile, three protective factors (SFRP1, EBF1, and CDC42EP4) were identified for the prognosis of patients with HNSCC with the cut-off value of HR < 1. Expression status of the eight CAF-related genes in HNSCC derived from the Gene Expression Profiling Interactive Analysis (GEPIA2) database (http://gepia2.cancer-pku.cn/#index (Fig. 3D) and their expression status in the CAFs of HNSCC as verified by our experiments were shown (Fig. 3E). The overall risk score for prognosis was calculated as Risk score = where Expi is the expression value of each mRNA, and Coef is the regression coefficient of the multivariate Cox analysis for the target mRNA.
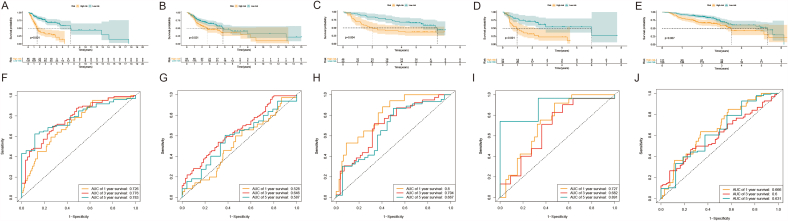
3.3. Verification of the prognostic signature based on the CAF-related genes
According to the optimal intercept value (cut-off = 1.217744), the HNSCC samples were divided into high- and low-risk groups. The K-M curve showed that the overall survival (OS) rate of the high-risk group was poorer than that of the low-risk group (Fig. 4A), indicating that the prognostic signature of the risk score was effective. For the 1-, 3- and 5-year survival rates, the AUC values analyzed by receiver operating characteristic (ROC) were 0.726, 0.776, and 0.783 (Fig. 4F), respectively. To further verify the reliability of these prognostic markers, we carried out verification in the external independent verification cohorts. The results showed that in different independent verification cohorts, according to the risk stratification of the best intercept value, the K-M curve showed that the OS of the high-risk group was poorer than that of the low-risk group (Fig. 4B–E), indicating that the prognostic signature of the risk score was effective. For 1-, 3-, and 5-year survival rates, the AUC values of ROC analysis were in the range of 0.6–0.8 (Fig. 4G–J).
Fig. 4.
Verification of CAF-related genes prognostic signature. The risk score level of the model-based classifier, K-M survival analysis was used to analyze the risk of death in the (A) training cohort, (B) testing cohort, (C) GSE41613 data sets, (D) GSE42743 data sets, and (E) GSE65858 data sets. ROC analysis of the sensitivity and specificity of the survival for the eight CAF-related genes signature risk score in (F) training cohorts, (G) testing cohorts, (H) GSE41613 data sets, (I) GSE42743 data sets, and (J) GSE65858 data sets.
3.4. The eight CAF-related genes signature as an independent predictive factor
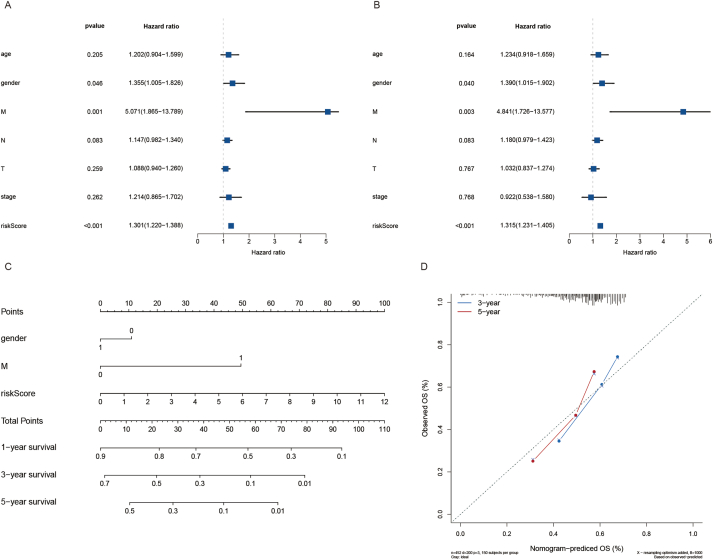
Univariate and multivariate Cox regression analyses were used to explore whether the above eight CAF-related genes were prognostic factors independent of age, sex, pathological stage, and other clinicopathological factors. The HR of risk scores in univariate Cox regression analysis and multivariate Cox regression were 1.301 and 1.220–1.388 (P < 0.001) (Fig. 5A), 1.315 and 1.231–1.405 (P < 0.001) (Fig. 5B), respectively, suggesting that the eight genes were independent prognostic factors in patients with HNSCC. In addition, to improve the accuracy of prediction, a nomogram was integrated and constructed based on the statistically significant eigenvalues of the above multivariate Cox analysis results, including gender, M, and risk score (Fig. 5C). The calibration curves of 3-year and 5-year survival probabilities predicted by nomogram were in good agreement with the actual observation results (Fig. 5D).
Fig. 5.
Risk score analysis and nomogram construction to evaluate OS in HNSCC patients. (A) The independent prognostic value of the risk score was evaluated by Cox regression analysis. Univariate Cox regression analysis of models in TCGA cohort. (B) The independent prognostic value of the risk score was evaluated by Cox regression analysis. Multivariate Cox regression analysis of models in TCGA cohort. (C) Nomogram for predicting the OS rate of HNSCC. (D) Nomogram calibration chart during 3-year and 5-year follow-up.
3.5. Gene enrichment analysis
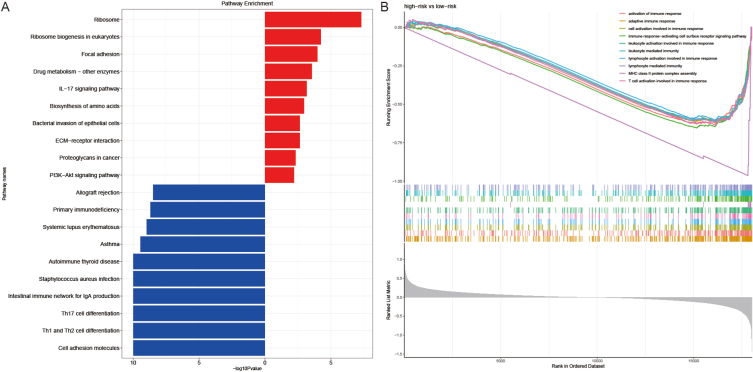
To explore the potential mechanism of prognostic signals, GSEA was used to analyze the functional enrichment of different subgroups. The results showed that the high-risk group was significantly enriched in cell adhesion molecules, Th1 and Th2 cell differentiation, and Th17 cell differentiation, while the focal adhesion, IL-17 signaling pathway, biosynthesis of amino acids, and ECM-receptor interaction of the low-risk group were more abundant (Fig. 6A). Moreover, high-risk patients were significantly less enriched than the low-risk patients in adaptive immune response, activation of the immune response, cell activation involved in the immune response, lymphocyte activation involved in immune response, and T cell activation involved in the immune response. These findings suggested that there were differences in these related genes and signal pathways between the high-risk and low-risk groups, which may partly explain the significant differences in prognosis between subgroups (Fig. 6B).
Fig. 6.
GSEA analysis. Results of functional enrichment in different subgroups.
3.6. Immune infiltration analysis
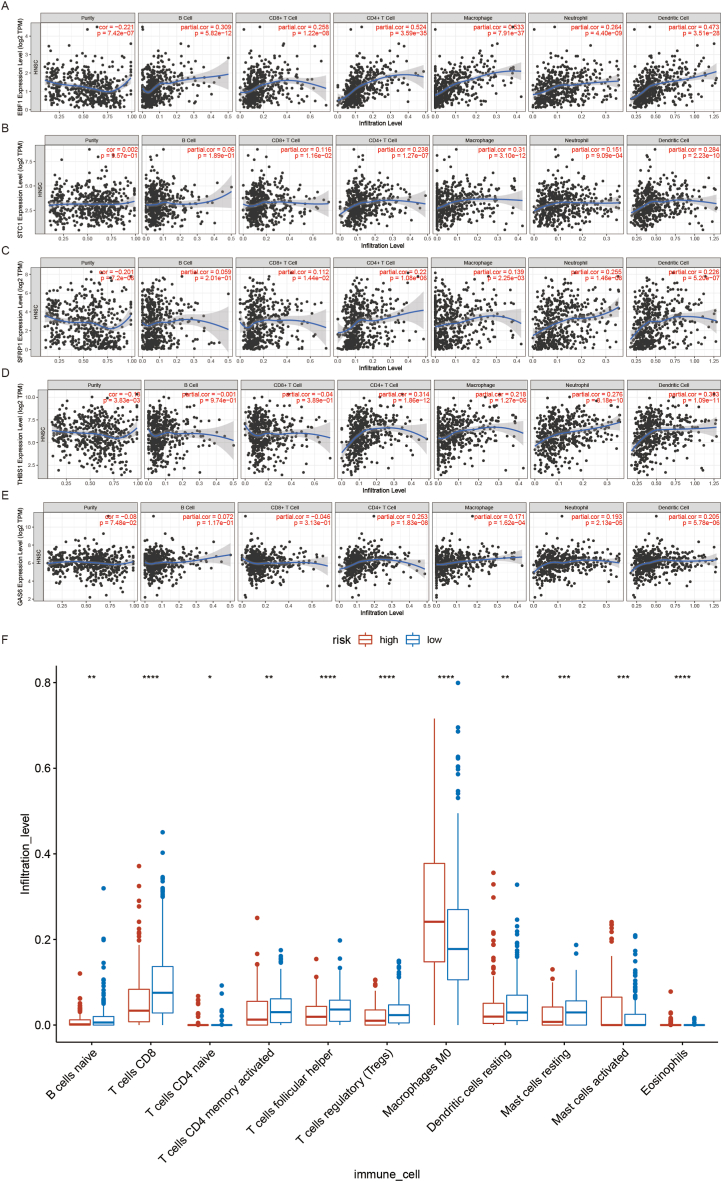
Five hub-genes from the signature were screened by reviewing a large amount of literature and combining correlation analysis with the String database. To better understand the characteristics of immune cells and their relations with CAF-related signature, the TIMER database was used to analyze the correlation between the abundance of immune cells and the five prognostic genes (EBF1, STC1, SFRP1, THBS1, and GAS6). The results showed that the five prognostic genes were positively associated with CD4+ T cell. Moreover, EBF1 and STC1 were positively associated with Macrophages and Dendritic cells (DCs). SFRP1 was positively associated with neutrophils. THBS1 and GAS6 were positively associated with DCs (Fig. 7A–E). To study the role of a prognostic signature in TME, CiberSort result showed that there were significant increases in naive B cells, CD8+ T cells, memory activated CD4 T cells, follicular helper T cells, T regulatory cells (Tregs), and resting Mast cells (MCs) in the low-risk group. Activated MCs and M0 macrophages were more common in the high-risk group than in the low-risk group. These results suggested that low-risk patients had a better immune microenvironment than high-risk patients (Fig. 7F).
Fig. 7.
Correlation between genes and HNSCC immune microenvironment. (A) Correlation analysis of EBF1 and immune cell infiltration in HNSCC. (B) Correlation analysis of STC1 and immune cell infiltration in HNSCC. (C) Correlation analysis of SFRP1 and immune cell infiltration in HNSCC. (D) Correlation analysis of THBS1 and immune cell infiltration in HNSCC. (E) Correlation analysis of GAS6 and immune cell infiltration in HNSCC. (F) The difference in the expression of infiltrating immune cells between the high-risk group and the low-risk group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
4. Discussion
CAFs have immunomodulatory effects by interacting with T lymphocytes, B lymphocytes, macrophages, DCs, natural killer cells, and MCs in the TME, and play roles in immune escape and resistance to immunotherapy in solid cancers, thereby regulating tumorigenesis and progression [19]. In recent years, accumulating studies have confirmed the importance of CAF-related genes expression on the prognosis and immune cell infiltration of non-small cell lung cancer [20], bladder cancer [21], glioma [22], and OSCC [11] cancer, suggesting that CAF-related genes signature might help optimize risk stratification and provide new insight into individual treatment for cancers. However, whether CAF-related genes play important roles in HNSCC prognosis remains not deep enough. In our study, we identified and validated a CAF-related genes prognostic signature that could contribute to a more personalized risk assessment for the treatment of HNSCC through its modulatory effects on immune cells. The prognostic reliability of the CAF signature was confirmed and validated by K-M analysis, ROC analysis, and multivariate Cox regression analysis. Eight genes closely associated with prognosis were screened from a relevant database of CAF-related genes, which were considered as the most possible candidate genes for prognosis. The eight CAF-related genes signature was constructed and verified to be a prognostic factor for HNSCC, independent of the clinicopathological factors. And immune infiltration analysis was performed to further explore potential immune regulatory mechanisms of the eight CAF-related genes signature. However, the clinical decision-making effect of the prognostic signature for cancer patients is still unclear and needs further studies.
The eight CAF-related genes were RCN1, MRPL32, STC1, SFRP1, EBF1, GAS6, CDC42EP4, and THBS1, and our results confirmed that RCN1, MRPL32, STC1, GAS6, and THBS1 were upregulated in HNSCC when compared with control, while SFRP1, EBF1, and CDC42EP4 were down-regulated. Except for MRPL32 and CDC42EP4, which are rarely reported, aberrant expression of six other genes correlates with the prognosis of a variety of tumors. In general, overexpression of RCN1, STC1, and GAS6 in various tumors always predict a poor prognosis, indicating a role in tumorigenesis and invasion [[23], [24], [25]]. In contrast, due to the loss of its expression in many human cancers, THBS1, EBF1 and SFPR1 have been classified as tumor suppressor genes [[26], [27], [28]]. However, expression was inconsistent in breast cancer, gastric cancer, and urothelial cancer [[29], [30], [31], [32]]. These genes play critical regulatory roles in the development of multiple types of cancers, including ovarian cancer, pancreatic cancer, liver cancer, lung cancer, thyroid cancer, esophageal cancer, colorectal cancer, breast cancer, prostate cancer, gastric cancer, and HNSCC [[33], [34], [35], [36], [37], [38]].
Numerous studies have demonstrated that these genes are intricately cross-linked to CAFs in TME. Overexpression of THBS1 in gastric cancer was significantly associated with CAFs and positively correlated with immunosuppressive genes, causing an unfavorable prognosis [39]. CAFs derived STC1 affects metastasis in lymph nodes and distant organs and enhances the ability of cancer cells to intravasate, which in turn drives metastasis in colorectal cancer [40]. Additionally, STC1 upregulated several markers of EMT and could mediate the transformation of normal fibroblasts (NFs) to CAFs in the TME, suggesting that STC1 plays a crucial role in the tumor environment on the metastasis of ovarian cancer [41]. SFRP1 strongly decreased the number of CAFs and induced an EMT phenotype in colorectal cancer cells [42]. CAF derived GAS6 is a key factor in enhancing the metastasis and survival of gastric cancer cells by stimulating many signal transduction pathways, including AKT, STAT3, and ERK signaling [43]. In recent years, corresponding studies on the mechanisms of these eight genes in HNSCC have been reported. THBS1 is the major tumor-specific extracellular matrix protein that is specifically induced in the TME. THBS1 produced by OSCC cells can stimulate cancer cell migration and expression of matrix metalloproteinases with TGFβ1, thereby promoting OSCC invasion [44]. OSCC cells release exosomal THBS1 to polarize macrophages into M1-like tumor-associated macrophages (TAMs), promoting tumor cell migration in OSCC [45]. And a positive feedback loop was formed between M1-like TAMs and OSCC cells in the regulation of EMT and cancer stem cells (CSCs) [46]. Multiple studies have shown that THBS1 expression is negatively associated with inferior disease-free survival and cancer progression and recurrence in patients with HNSCC [47]. It has also been shown that methylation and silencing of the SFRP1 in OSCC contribute to the activation of the Wnt signaling pathway, which leads to cell proliferation during oral carcinogenesis [48]. RCN1 dysregulation is present in nasopharyngeal and OSCC [49,50], and RCN1 knockdown inhibits the migration and invasive ability of laryngeal cancer cells [51]. Our findings are consistent with the regulatory roles of these genes as mentioned above.
Gene function and pathway enrichment analyses showed that these eight genes were enriched in Th1 and Th2 cell differentiation, Th17 cell differentiation, IL-17 signaling pathways, ECM-receptor interactions, cell adhesion molecules, focal adhesion, and other pathways that are closely associated with CAFs and play important roles in cancers [[52], [53], [54], [55], [56], [57], [58], [59]]. Researches have shown that CAFs facilitate Th17 cell differentiation in vivo and promote disease development by producing TGFβ1 [52,53]. Cytokines and chemokines produced by CAFs in the absence of IL17A favor the recruitment of Th1 and cytotoxic CD8+ T cells and restrict the recruitment of suppressive myeloid cells and Tregs [54]. Altogether, CAFs modulate the transformation of most Th cells into immunoinhibitory subpopulations in tumors to create an immunosuppressive and cancer-adaptive TME and then exert a pro-invasive effect on cancer cells [6]. The interaction between CAFs and ECM also plays a very important role in tumor progression [55]. A positive feedback loop between CAFs and the ECM has been identified [56]. CAFs are capable of upregulating the expression of several cytoskeletal regulators such as anillin and diaphanous-related formin-3 through activated YAP, to contribute to ECM stiffening. When the matrix becomes stiffer in the ECM, isometric tension within CAFs significantly increases and further facilitates YAP activation by stimulating Src family kinases, consequently maintaining the CAF phenotype and its cancer-promoting properties [56]. It has been shown that CAFs exert physical forces on cancer cells that enable their collective invasion [57]. Force transmission is mediated by a heterophilic adhesion involving N-cadherin on the CAFs membrane and E-cadherin on the cancer cell membrane [58]. Impairment of E-cadherin/N-cadherin adhesion abrogates the ability of CAFs to guide collective cell migration and blocks cancer cell invasion [58]. In addition, studies have shown that focal adhesion kinase deficiency reduces the ability of CAFs to promote tumor cell migration by impairing CAFs derived exosomes, which significantly inhibits breast cancer metastasis [59]. These suggest that differentially expressed genes between the high- and low-risk subgroup patients may regulate mostly immune cell responses, ECM, and focal transmission to mediate prognosis for HNSCC.
HNSCC patients were divided into the high-risk group and the low-risk group according to the eight CAF-related genes prognostic signature, and significant differences in immune cell infiltration in the two groups were found. For example, anti-tumor T cells such as CD8+ T cells, CD4 memory-activated T cells, follicular helper T cells, Treg cells, B cells, and DCs were significantly more infiltrated in the low-risk group compared to the high-risk group. Tumor-promoting cells such as macrophages and activated MCs were detected in patients in the high-risk group compared to the low-risk group. CAFs facilitate the cancer-promoting phenotype transition of naive T cells, enhancing immune inhibitory T lymphocyte function, and suppressing the activity of effector T lymphocytes, thereby resulting in immune suppression in the TME [5,6,60,61]. Adaptive immune CD8+ and CD4+ T cells are considered to be the main driver of anti-tumor immunity [62], and they could reshape the tumor immune microenvironment (TIME) and promote tumor clearance [63,64]. CAFs induce the expression of immune checkpoint proteins on CD4+ and CD8+ T cells, which contributes to a diminished immune function [65]. Increased infiltration of CD8+ T cells and activated CD4 T cells favors the prognosis of HNSCC [66], which is consistent with our results as much more CD8+ and CD4+ T cell infiltration was observed in the low-risk HNSCC patients. In addition, CAFs have been found to stimulate the migration of Treg cells and induce Treg cells by releasing growth factor VEGF-A, resulting in a significant increase in their frequency of appearance at tumor sites [[67], [68], [69]]. CAFs also promote phase transformation to ultimately induce immunosuppression [70]. Innate immune cells such as macrophages, MCs, and DCs could be regulated by CAFs. CAFs regulate macrophages recruitment, polarization, and functions [71]. Additionally, CAFs preferentially induce the transformation of M0 macrophages into a tumor-promoting (M2) phenotype in vitro [72]. CAFs can potentiate MCs proliferation, migration, and inflammatory cytokine secretion in the TME and thus increases intra-tumor microvessel density, promotes tumor growth and tumor invasion, and contributes to an overall poor clinical outcome [73,74]. Meanwhile, MCs are also capable of promoting the activity of CAFs, such as enhancing the proliferation and secretion of CAFs through the TGFβ signaling pathway, thereby increasing the protumor effects of CAFs [75]. In recent years, several investigations have illustrated that CAFs can drive immune evasion of tumor cells by blocking DCs maturation, antigen presentation, and their associated adaptive immune responses [6]. The results of our study are consistent with these reports. It is thus suggested that the CAF signature has important implications in the prognosis of HNSCC patients.
Unlike NFs, CAFs suppress the immune response in the TME [76]. High levels of CAFs in tumors are associated with poorer treatment outcomes and prognosis [[77], [78], [79], [80]]. This theory was also confirmed in our study, where the TIME was more conducive to tumor killing in the low-risk group. In our study, we combined bioinformatics analysis with knowledge of the TIME to identify and validate CAF signature for HNSCC prognosis. We believe that the CAF signature is a good indicator of the TIME, allowing new strategies for the stratification of patients with HNSCC.
5. Limitations of the study
This study proposes a non-invasive predictive tool, an eight CAF-related genes signature, which provides clinicians with a valuable tool for predicting the prognosis of HNSCC. However, the clinical decision-making role of the CAF prognostic signature for HNSCC patients has not been clinically validated, and the regulatory mechanisms of CAF-related genes on TME in HNSCC are unclear, and require further clinical studies and mechanistic exploration. Future studies should attempt to clarify the mechanisms by which CAF-related genes regulate TIME in HNSCC, so as to further improve the discriminatory efficacy of the signature and provide new targets for HNSCC immunotherapy in the future.
Author contribution statement
Lei Dong: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.Qi Sun: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.Fei Song: Performed the experiments; Analyzed and interpreted the data.Xiaoyu Song; Congxian Lu: Performed the experiments.Xi-cheng Song; Yumei Li: Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data associated with this study has been deposited at The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/); Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/); TIDE (http://tide.dfci.harvard.edu/); String (https://cn.string-db.org/); TIMER (https://cistrome.shinyapps.io/timer/); CiberSort (https://cibersort.stanford.edu/); UCSC Xena (http://xena.ucsc.edu/); Gene Ontology (GO, http://geneontology.org/); Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/); GSEA (http://www.gsea-msigdb.org/); Gene Expression Profiling Interactive Analysis (GEPIA2, http://gepia2.cancer-pku.cn/#index).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e14003.
Contributor Information
Lei Dong, Email: dl830408@163.com.
Qi Sun, Email: sunqi_1125@126.com.
Fei Song, Email: genierui@163.com.
Xiaoyu Song, Email: 18254577511@163.com.
Congxian Lu, Email: lu_olivia@126.com.
Yumei Li, Email: myheart1263@163.com.
Xicheng Song, Email: drxchsong@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Alsahafi E., et al. Clinical update on head and neck cancer: molecular biology and ongoing challenges. Cell Death Dis. 2019;10(8):540. doi: 10.1038/s41419-019-1769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curry J.M., et al. Tumor microenvironment in head and neck squamous cell carcinoma. Semin. Oncol. 2014;41(2):217–234. doi: 10.1053/j.seminoncol.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Cirri P., Chiarugi P. Cancer associated fibroblasts: the dark side of the coin. Am J Cancer Res. 2011;1(4):482–497. [PMC free article] [PubMed] [Google Scholar]
- 4.Alcolea S., et al. Interaction between head and neck squamous cell carcinoma cells and fibroblasts in the biosynthesis of PGE2. J. Lipid Res. 2012;53(4):630–642. doi: 10.1194/jlr.M019695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An Y., et al. Crosstalk between cancer-associated fibroblasts and immune cells in cancer. J. Cell Mol. Med. 2020;24(1):13–24. doi: 10.1111/jcmm.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao X., et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol. Cancer. 2021;20(1):131. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchsbaum R.J., Oh S.Y. Breast cancer-associated fibroblasts: where we are and where we need to go. Cancers. 2016;8(2) doi: 10.3390/cancers8020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun D.Y., et al. Cancer-associated fibroblast regulate proliferation and migration of prostate cancer cells through TGF-β signaling pathway. Life Sci. 2019;235 doi: 10.1016/j.lfs.2019.116791. [DOI] [PubMed] [Google Scholar]
- 9.Ham I.H., Lee D., Hur H. Role of cancer-associated fibroblast in gastric cancer progression and resistance to treatments. JAMA Oncol. 2019;2019 doi: 10.1155/2019/6270784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato T., et al. Cancer-associated fibroblasts affect intratumoral CD8(+) and FoxP3(+) T cells via IL6 in the tumor microenvironment. Clin. Cancer Res. 2018;24(19):4820–4833. doi: 10.1158/1078-0432.CCR-18-0205. [DOI] [PubMed] [Google Scholar]
- 11.Ko Y.C., et al. Index of cancer-associated fibroblasts is superior to the epithelial-mesenchymal transition score in prognosis prediction. Cancers. 2020;12(7) doi: 10.3390/cancers12071718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang P., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heagerty P.J., Lumley T., Pepe M.S. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu X., et al. PRRC2A promotes hepatocellular carcinoma progression and associates with immune infiltration. J. Hepatocell. Carcinoma. 2021;8:1495–1511. doi: 10.2147/JHC.S337111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue S., et al. Corrigendum: USP5 promotes metastasis in non-small cell lung cancer by inducing epithelial-mesenchymal transition via wnt/β-catenin pathway. Front. Pharmacol. 2020;11:948. doi: 10.3389/fphar.2020.00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Q., et al. Identification and validation of 17-lncRNA related to regulatory T cell heterogeneity as a prognostic signature for head and neck squamous cell carcinoma. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.782216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puram S.V., et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171(7):1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galbo P.M., Jr., Zang X., Zheng D. Molecular features of cancer-associated fibroblast subtypes and their implication on cancer pathogenesis, prognosis, and immunotherapy resistance. Clin. Cancer Res. 2021;27(9):2636–2647. doi: 10.1158/1078-0432.CCR-20-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa A., et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33(3):463–479.e10. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Navab R., et al. Prognostic gene-expression signature of carcinoma-associated fibroblasts in non-small cell lung cancer. Proc. Natl. Acad. Sci. U. S. A. 2011;108(17):7160–7165. doi: 10.1073/pnas.1014506108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Y., et al. The cancer-associated fibroblasts related gene CALD1 is a prognostic biomarker and correlated with immune infiltration in bladder cancer. Cancer Cell Int. 2021;21(1):283. doi: 10.1186/s12935-021-01896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z., et al. Prognosis and immunotherapy significances of a cancer-associated fibroblasts-related gene signature in gliomas. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.721897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J.W., et al. RCN1 induces sorafenib resistance and malignancy in hepatocellular carcinoma by activating c-MYC signaling via the IRE1α-XBP1s pathway. Cell Death Dis. 2021;7(1):298. doi: 10.1038/s41420-021-00696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao F., et al. Expression, function and clinical application of stanniocalcin-1 in cancer. J. Cell Mol. Med. 2020;24(14):7686–7696. doi: 10.1111/jcmm.15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu G., et al. Molecular insights of Gas6/TAM in cancer development and therapy. Cell Death Dis. 2017;8(3):e2700. doi: 10.1038/cddis.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaur S., et al. Functions of thrombospondin-1 in the tumor microenvironment. Int. J. Mol. Sci. 2021;22(9) doi: 10.3390/ijms22094570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao D. Emerging roles of the EBF family of transcription factors in tumor suppression. Mol. Cancer Res. 2009;7(12):1893–1901. doi: 10.1158/1541-7786.MCR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baharudin R., et al. Epigenetics of SFRP1: the dual roles in human cancers. Cancers. 2020;12(2) doi: 10.3390/cancers12020445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice A.J., Steward M.A., Quinn C.M. Thrombospondin 1 protein expression relates to good prognostic indices in ductal carcinoma in situ of the breast. J. Clin. Pathol. 2002;55(12):921–925. doi: 10.1136/jcp.55.12.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyata Y., Sakai H. Thrombospondin-1 in urological cancer: pathological role, clinical significance, and therapeutic prospects. Int. J. Mol. Sci. 2013;14(6):12249–12272. doi: 10.3390/ijms140612249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernemann C., et al. Influence of secreted frizzled receptor protein 1 (SFRP1) on neoadjuvant chemotherapy in triple negative breast cancer does not rely on WNT signaling. Mol. Cancer. 2014;13:174. doi: 10.1186/1476-4598-13-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu Y., et al. High levels of secreted frizzled-related protein 1 correlate with poor prognosis and promote tumourigenesis in gastric cancer. Eur. J. Cancer. 2013;49(17):3718–3728. doi: 10.1016/j.ejca.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 33.Poland J., et al. Study of therapy resistance in cancer cells with functional proteome analysis. Clin. Chem. Lab. Med. 2002;40(3):221–234. doi: 10.1515/CCLM.2002.037. [DOI] [PubMed] [Google Scholar]
- 34.Liu G., et al. Stanniocalcin 1 and ovarian tumorigenesis. J. Natl. Cancer Inst. 2010;102(11):812–827. doi: 10.1093/jnci/djq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meric F., et al. Expression profile of tyrosine kinases in breast cancer. Clin. Cancer Res. 2002;8(2):361–367. [PubMed] [Google Scholar]
- 36.Isenberg J.S., et al. Regulation of nitric oxide signalling by thrombospondin 1: implications for anti-angiogenic therapies. Nat. Rev. Cancer. 2009;9(3):182–194. doi: 10.1038/nrc2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson J.W., et al. Expression of Id helix-loop-helix proteins in colorectal adenocarcinoma correlates with p53 expression and mitotic index. Cancer Res. 2001;61(24):8803–8810. [PubMed] [Google Scholar]
- 38.Atschekzei F., et al. SFRP1 CpG island methylation locus is associated with renal cell cancer susceptibility and disease recurrence. Epigenetics. 2012;7(5):447–457. doi: 10.4161/epi.19614. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X., et al. Upregulation of THBS1 is related to immunity and chemotherapy resistance in gastric cancer. Int. J. Gen. Med. 2021;14:4945–4957. doi: 10.2147/IJGM.S329208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peña C., et al. STC1 expression by cancer-associated fibroblasts drives metastasis of colorectal cancer. Cancer Res. 2013;73(4):1287–1297. doi: 10.1158/0008-5472.CAN-12-1875. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y., et al. Stanniocalcin 1 in tumor microenvironment promotes metastasis of ovarian cancer. OncoTargets Ther. 2019;12:2789–2798. doi: 10.2147/OTT.S196150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosa M.H., et al. A wnt-induced phenotypic switch in cancer-associated fibroblasts inhibits EMT in colorectal cancer. Cancer Res. 2020;80(24):5569–5582. doi: 10.1158/0008-5472.CAN-20-0263. [DOI] [PubMed] [Google Scholar]
- 43.Bae C.A., et al. Inhibiting the GAS6/AXL axis suppresses tumor progression by blocking the interaction between cancer-associated fibroblasts and cancer cells in gastric carcinoma. Gastric Cancer. 2020;23(5):824–836. doi: 10.1007/s10120-020-01066-4. [DOI] [PubMed] [Google Scholar]
- 44.Pal S.K., et al. THBS1 is induced by TGFB1 in the cancer stroma and promotes invasion of oral squamous cell carcinoma. J. Oral Pathol. Med. 2016;45(10):730–739. doi: 10.1111/jop.12430. [DOI] [PubMed] [Google Scholar]
- 45.Xiao M., et al. M1-like tumor-associated macrophages activated by exosome-transferred THBS1 promote malignant migration in oral squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2018;37(1):143. doi: 10.1186/s13046-018-0815-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You Y., et al. M1-like tumor-associated macrophages cascade a mesenchymal/stem-like phenotype of oral squamous cell carcinoma via the IL6/Stat3/THBS1 feedback loop. J. Exp. Clin. Cancer Res. 2022;41(1):10. doi: 10.1186/s13046-021-02222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sepiashvili L., et al. Potentially novel candidate biomarkers for head and neck squamous cell carcinoma identified using an integrated cell line-based discovery strategy. Mol. Cell. Proteomics. 2012;11(11):1404–1415. doi: 10.1074/mcp.M112.020933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sogabe Y., et al. Epigenetic inactivation of SFRP genes in oral squamous cell carcinoma. Int. J. Oncol. 2008;32(6):1253–1261. doi: 10.3892/ijo_32_6_1253. [DOI] [PubMed] [Google Scholar]
- 49.Huang Z.H., et al. Reticulocalbin-1 knockdown increases the sensitivity of cells to Adriamycin in nasopharyngeal carcinoma and promotes endoplasmic reticulum stress-induced cell apoptosis. Cell Cycle. 2020;19(13):1576–1589. doi: 10.1080/15384101.2020.1733750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ueda S., et al. Salivary NUS1 and RCN1 levels as biomarkers for oral squamous cell carcinoma diagnosis. Vivo. 2020;34(5):2353–2361. doi: 10.21873/invivo.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang H., et al. Development and validation of an immune-related signature for the prediction of recurrence risk of patients with laryngeal cancer. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.683915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Monte L., et al. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J. Exp. Med. 2011;208(3):469–478. doi: 10.1084/jem.20101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gutcher I., et al. Autocrine transforming growth factor-β1 promotes in vivo Th17 cell differentiation. Immunity. 2011;34(3):396–408. doi: 10.1016/j.immuni.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mucciolo G., et al. IL17A critically shapes the transcriptional program of fibroblasts in pancreatic cancer and switches on their protumorigenic functions. Proc. Natl. Acad. Sci. U. S. A. 2021;118(6) doi: 10.1073/pnas.2020395118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Najafi M., Farhood B., Mortezaee K. Extracellular matrix (ECM) stiffness and degradation as cancer drivers. J. Cell. Biochem. 2019;120(3):2782–2790. doi: 10.1002/jcb.27681. [DOI] [PubMed] [Google Scholar]
- 56.Calvo F., et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 2013;15(6):637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaggioli C., et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007;9(12):1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 58.Labernadie A., et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat. Cell Biol. 2017;19(3):224–237. doi: 10.1038/ncb3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu H.J., et al. FAK signaling in cancer-associated fibroblasts promotes breast cancer cell migration and metastasis by exosomal miRNAs-mediated intercellular communication. Oncogene. 2020;39(12):2539–2549. doi: 10.1038/s41388-020-1162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrett R., Puré E. Cancer-associated fibroblasts: key determinants of tumor immunity and immunotherapy. Curr. Opin. Immunol. 2020;64:80–87. doi: 10.1016/j.coi.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barrett R.L., Puré E. Cancer-associated fibroblasts and their influence on tumor immunity and immunotherapy. Elife. 2020;9 doi: 10.7554/eLife.57243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Leun A.M., Thommen D.S., Schumacher T.N. CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer. 2020;20(4):218–232. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borst J., et al. CD4(+) T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018;18(10):635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- 64.Mandal R., Chan T.A. Personalized oncology meets immunology: the path toward precision immunotherapy. Cancer Discov. 2016;6(7):703–713. doi: 10.1158/2159-8290.CD-16-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorchs L., et al. Human pancreatic carcinoma-associated fibroblasts promote expression of Co-inhibitory markers on CD4(+) and CD8(+) T-cells. Front. Immunol. 2019;10:847. doi: 10.3389/fimmu.2019.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carstens J.L., et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat. Commun. 2017;8 doi: 10.1038/ncomms15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacobs J., et al. Unveiling a CD70-positive subset of cancer-associated fibroblasts marked by pro-migratory activity and thriving regulatory T cell accumulation. OncoImmunology. 2018;7(7):e1440167. doi: 10.1080/2162402X.2018.1440167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bourhis M., et al. Direct and indirect modulation of T cells by VEGF-A counteracted by anti-angiogenic treatment. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.616837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wada J., et al. The contribution of vascular endothelial growth factor to the induction of regulatory T-cells in malignant effusions. Anticancer Res. 2009;29(3):881–888. [PubMed] [Google Scholar]
- 70.Chen W., et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003;198(12):1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mills C.D., et al. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000;164(12):6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 72.Zhang A., et al. Cancer-associated fibroblasts promote M2 polarization of macrophages in pancreatic ductal adenocarcinoma. Cancer Med. 2017;6(2):463–470. doi: 10.1002/cam4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watermann C., et al. Recurrent HNSCC harbor an immunosuppressive tumor immune microenvironment suggesting successful tumor immune evasion. Clin. Cancer Res. 2021;27(2):632–644. doi: 10.1158/1078-0432.CCR-20-0197. [DOI] [PubMed] [Google Scholar]
- 74.Ellem S.J., et al. A pro-tumourigenic loop at the human prostate tumour interface orchestrated by oestrogen, CXCL12 and mast cell recruitment. J. Pathol. 2014;234(1):86–98. doi: 10.1002/path.4386. [DOI] [PubMed] [Google Scholar]
- 75.Yang F.C., et al. Nf1+/- mast cells induce neurofibroma like phenotypes through secreted TGF-beta signaling. Hum. Mol. Genet. 2006;15(16):2421–2437. doi: 10.1093/hmg/ddl165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kraman M., et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 2010;330(6005):827–830. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- 77.Herrera M., et al. Cancer-associated fibroblast-derived gene signatures determine prognosis in colon cancer patients. Mol. Cancer. 2021;20(1):73. doi: 10.1186/s12943-021-01367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ligorio M., et al. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell. 2019;178(1):160–175.e27. doi: 10.1016/j.cell.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song M., et al. Cancer-associated fibroblast-mediated cellular crosstalk supports hepatocellular carcinoma progression. Hepatology. 2021;73(5):1717–1735. doi: 10.1002/hep.31792. [DOI] [PubMed] [Google Scholar]
- 80.Qin X., et al. Cancer-associated fibroblast-derived IL-6 promotes head and neck cancer progression via the osteopontin-NF-kappa B signaling pathway. Theranostics. 2018;8(4):921–940. doi: 10.7150/thno.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/); Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/); TIDE (http://tide.dfci.harvard.edu/); String (https://cn.string-db.org/); TIMER (https://cistrome.shinyapps.io/timer/); CiberSort (https://cibersort.stanford.edu/); UCSC Xena (http://xena.ucsc.edu/); Gene Ontology (GO, http://geneontology.org/); Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/); GSEA (http://www.gsea-msigdb.org/); Gene Expression Profiling Interactive Analysis (GEPIA2, http://gepia2.cancer-pku.cn/#index).