ABSTRACT
Increasing occurrence of multidrug-resistant (MDR) and hypervirulent (hv) Klebsiella pneumoniae (MDR-hvKp) convergent clones is being observed. Those strains have the potential of causing difficult-to-treat infections in healthy adults with an increased capacity for mortality. It is therefore crucial to track their dissemination to prevent their further spread. The aim of our study was to investigate the occurrence of carbapenemase-producing hvKp isolates in Switzerland and to determine their genetic profile. A total of 279 MDR carbapenemase-producing K. pneumoniae from patients hospitalized all over Switzerland was investigated, and a rate of 9.0% K. pneumoniae presenting a virulence genotype was identified. Those isolates produced either KPC, NDM, or OXA-48 and had been either recovered from rectal swabs, urine, and blood. A series of previously reported K. pneumoniae clones such as ST23-K1, ST395-K2, and ST147-K20 or ST147-K64 were identified. All the isolates defined as MDR-hvKp (4.7%) possessed the aerobactin and the yersiniabactin clusters. The ST23-K1s were the only isolates presenting the colibactin cluster and achieved higher virulence scores. This study highlights the occurrence and circulation of worrisome MDR-hvKp and MDR nonhypervirulent K. pneumoniae (MDR-nhv-Kp) isolates in Switzerland. Our findings raise an alert regarding the need for active surveillance networks to track and monitor the spread of such successful hybrid clones representing a public health threat worldwide.
KEYWORDS: Klebsiella pneumoniae, hypervirulence, carbapenemase, multidrug resistance, convergent clones
INTRODUCTION
Since its first clinical report, back in Taiwan 1986 (1), hypervirulent Klebsiella pneumoniae (hvKp) causing invasive infections have been progressively increasingly reported worldwide (2–4). These isolates are usually susceptible to almost all antibiotics (4, 5). The hvKp clinical pathotype is mainly associated with community-acquired infections, affecting healthy patients and causing diverse severe diseases, such as liver abscesses, endophthalmitis, and meningitis (6).
Intrinsically, K. pneumoniae exhibits a so-called opportunistic pathogenicity, causing invasive hospital-acquired infections among high-risk patients, corresponding to the “classical” K. pneumoniae (cKp) (7). Urinary tract infections, pneumonia, and bacteremia are some of the infections caused by cKp that may be multidrug-resistant (MDR), including carbapenemase producers (CpKp), which are nowadays widely disseminated around the globe (8, 9).
Although hvKp and CpKp populations are recognized for nonoverlapping clonal groups and sequence types (STs), respectively, circulating mainly in the community and hospitals (10), some of their virulence- and resistance-associated genes can be genetically transmitted and therefore exchanged between each other (11, 12). Hence, this worrisome evolution may lead to convergent clones of MDR hypervirulent K. pneumoniae (MDR-hvKp). These MDR-hvKps have the potential of causing very-difficult-to-treat infections in previously healthy adults (13, 14). These isolates have been detected in Argentina, China, Egypt, Japan, Switzerland, Taiwan, the United Kingdom, and the United States (14–22).
Although the MDR-hvKp population has been diversely distributed worldwide, the most prevalent STs are the KPC-producing K. pneumoniae ST11 and the dominant hvKp lineage ST23 (13, 23). The recent emergence of the latter in Europe led to a public alarm raised by the European Centre for Disease Prevention and Control (ECDC) (13).
It is therefore of utmost importance to track the spread of such clones to better trace their dissemination and reinforce measures to prevent their further spread. The goal of this study was to investigate the occurrence of carbapenemase-producing hvKp isolates in Switzerland and to determine their genetic profile.
RESULTS
Among the studied 279 carbapenemase-producing K. pneumoniae isolates, 58.4% were isolated from rectal swab, 21.5% from urine, 6.1% from respiratory tract, 3.9% from wounds, 3.6% from blood culture, and 6.5% from other biological sites. A total of 9.0% (25 of 279) MDR K. pneumoniae carrying important virulence genotype isolates were identified, distributed among 10 different Swiss cantons. The prevalence of such isolates along the years was 0%, 14%, 9.4%, and 6.1% for years 2017, 2018, 2019, and 2020, respectively. These isolates were recovered from rectal swabs (56.0%), urine (28.0%), blood culture (8.0%), tracheal fluids (4.0%), and punctures (4.0%). Antimicrobial susceptibility testing showed that 48%, 80%, and 100% of those isolates presented reduced susceptibilities to imipenem, meropenem, and ertapenem, respectively. A large proportion (84%) of the isolates was resistant to the fluoroquinolones ciprofloxacin and norfloxacin, as well as to aminoglycosides (52%, 84%, and 88% to amikacin, gentamicin, and tobramycin, respectively). Furthermore, 88% of them were resistant to sulfamethoxazole-trimethoprim. All phenotypic resistance profiles are shown in Table S1. The most prevalent carbapenemase identified was NDM-1 (44%), followed by KPC (KPC-3, 28%; KPC-41, 8%) and OXA-48 (20%). The extended-spectrum β-lactamase (ESBL) gene blaCTX-M-15 was detected in 64% of the isolates. Hence, co-occurrence of blaCTX-M-15 with different carbapenemase genes was frequently identified, being, respectively, 12%, 20%, and 32% for blaKPC-3, blaOXA-48, and blaNDM-1 genes. Complete data showing the corresponding resistomes are given in Table S1.
Among all isolates studied, five main groups could be identified with respect to their capsular types. Group 1 and 2 isolates possess the typical hypervirulent capsular types K1 and K2, respectively, and were thus named MDR-hvKp (4.7%); The remaining groups were considered MDR nonhypervirulent K. pneumoniae (MDR-nhv-Kp) strains (4.3%), since they are either common MDR clones that acquired a virulence plasmid or clones presenting with nontypically hypervirulent capsular types, as detailed below. Summarized and complete characteristics of all selected isolates are shown, respectively, in Table 1 and Table S1.
TABLE 1.
Characteristics of carbapenemase-producing K. pneumoniae isolates from our collectiona
|
Strain |
ST | K-type | Virulence score | Definition | rmpABC | rmpA2 | iucABCD_iutA | ybt | clb | kfu | No. virulence genes | Virulence plasmids | Carbapenemase | Canton | Source | Date |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N330 | 23 | K1 | 5 | hvKp | − | +; trunc | + | + | + | + | 35 | pST23_OXA-48 | OXA-48 | VD | Rectal swab | 2018 |
| N488 | 23 | K1 | 5 | hvKp | − | +; trunc | + | + | + | + | 35 | pST23_OXA-48 | OXA-48 | VD | Rectal swab | 2019 |
| N593 | 23 | K1 | 5 | hvKp | − | − | + | + | + | + | 34 | pST23_OXA-48 | OXA-48 | VD | Rectal swab | 2019 |
| N911 | 14 | K2 | 4 | hvKp | − | +; trunc | + | + | − | + | 20 | pKpCH_aerob | OXA-48 | GE | Rectal swab | 2019 |
| N321 | 395 | K2 | 4 | hvKp | − | +; trunc | + | + | − | − | 17 | pKpCH_aerob | KPC-3 | VS | Urine | 2018 |
| N346 | 395 | K2 | 4 | hvKp | − | − | + | + | − | − | 16 | pKpCH_aerob | KPC-41 | VS | Rectal swab | 2018 |
| N447 | 395 | K2 | 4 | hvKp | − | +; trunc | + | + | − | − | 17 | pKpCH_aerob | KPC-3 | VS | Blood culture | 2018 |
| N1052 | 395 | K2 | 4 | hvKp | − | − | + | + | − | − | 16 | pKpvST147B-like | NDM-1 | TI | Urine | 2019 |
| N570 | 395 | K2 | 4 | hvKp | − | +; trunc | + | + | − | − | 17 | pKpCH_aerob | KPC-3 | VD | Tracheal fluid | 2018 |
| N732 | 395 | K2 | 4 | hvKp | − | − | + | + | − | − | 16 | pKpCH_aerob | KPC-3 | VS | Rectal swab | 2019 |
| N793 | 395 | K2 | 4 | hvKp | − | +; trunc | + | + | − | − | 17 | pKpCH_aerob | KPC-3 | BL | Urine | 2019 |
| N435 | 395 | K2 | 4 | hvKp | − | − | + | + | − | − | 16 | pKpCH_aerob | KPC-41 | VD | Rectal swab | 2018 |
| N649 | 395 | K2 | 4 | hvKp | − | +; trunc | + | + | − | − | 17 | pKpCH_aerob | OXA-48 | BE | Rectal swab | 2019 |
| N621 | 147 | K20 | 3 | nhv-Kp | + | +; trunc | + | − | − | − | 9 | pKpvST147B-like | NDM-1 | VD | Rectal swab | 2019 |
| N969 | 147 | K20 | 3 | nhv-Kp | + | +; trunc | + | − | − | − | 9 | pKpvST147B-like | NDM-1 | LU | Urine | 2019 |
| N829 | 147 | K20 | 3 | nhv-Kp | +; A_trunc | − | + | − | − | − | 8 | pKpvST147B-like | NDM-1 | LU | Urine | 2019 |
| N1259 | 147 | K20 | 3 | nhv-Kp | + | +; trunc | + | − | − | − | 9 | pKpvST147B-like | NDM-1 | LU | Urine | 2020 |
| N1498 | 147 | K20 | 3 | nhv-Kp | + | +; trunc | + | − | − | − | 9 | pKpvST147B-like | NDM-1 | SG | Urine | 2020 |
| N1712 | 147 | K64 | 4 | nhv-Kp | − | − | + | + | − | − | 16 | pKpvST147B-like | NDM-1 | ZH | Puncture | 2020 |
| N1071 | 147 | K64 | 4 | nhv-Kp | + | − | + | + | − | − | 19 | pKpvST147B-like | NDM-1 | GE | Rectal swab | 2020 |
| N1448 | 147 | K64 | 4 | nhv-Kp | + | +; trunc | + | + | − | − | 20 | pKpvST147B-like | NDM-1 | ZH | Rectal swab | 2020 |
| N1692 | 147 | K64 | 4 | nhv-Kp | − | − | + | + | − | − | 16 | pKpvST147B-like | NDM-1 | ZH | Blood culture | 2020 |
| N896 | 101 | K17 | 4 | nhv-Kp | − | +; trunc | + | + | − | + | 20 | pKpCH_aerob | KPC-3 | GE | Rectal swab | 2019 |
| N1325 | 101 | K17 | 4 | nhv-Kp | − | − | + | + | − | + | 19 | pKpCH_aerob | KPC-3 | GR | Rectal swab | 2020 |
| N608 | 16 | K51 | 4 | nhv-Kp | + | +; trunc | + | + | − | − | 20 | pKpvST147B-like | NDM-1 | VS | Rectal swab | 2019 |
rmpABC and rmpA2 are mucoid phenotype regulators. iucABCD_iutA is an aerobactin cluster, ybt is a yersiniabactin cluster; and clb is a colibactin cluster. kfu shows ferric iron uptake. The virulence plasmids used have the following accession numbers: pST23_OXA-48, OP690161; pKpCH_aerob, OP690162; and pKpvST147, CP040726.1. −, absence; +, presence; BE, Bern; BL, Basel-Landschaft; GE, Geneva; GR, Graubünden; LU, Lucerne; SG, St-Gallen; TI, Ticino; VD, Vaud; VS, Valais; ZH, Zürich; hvKp, hypervirulent K. pneumoniae; nhv-Kp, nonhypervirulent K. pneumoniae; trunc, truncated.
Group 1.
Three MDR-hvKp were identified as belonging to the worldwide spread ST23, possessing the conserved capsular type K1 (ST23-K1) (24). They had a virulence score of 5 (presence of aerobactin, colibatin, and yersiniabactin) and carried the aerobactin cluster (iucABCD_iutA) along with the yersiniabactin cluster (ybtAEPQSTUX, irp1, irp2, and fyuA), the colibactin cluster (clbABCDEFGHILMNOPQ), and the ferric uptake system cluster (kfuABC). Two of those isolates additionally carried a truncated rmpA2 mucoid regulator. The magA gene encoding a component of K1 capsule formation was detected in all ST23-K1 isolates. It is noteworthy that all those ST23-K1 hvKp produced the OXA-48 carbapenemase and had been recovered from rectal swabs (Table 1). All of them presented a plasmid 98% similar to pVir-CR-HvKP4 (MF437313.1) (15), which was detected here and which we named pKpST23_OXA-48 (221,352 bp; OP690161). A schematic representation of plasmid pKpST23_OXA-48 is shown in Fig. S1.
Group 2.
Ten MDR-hvKps possessed the K2 capsular type, and all had a virulence score of 4 (presence of aerobactin and yersiniabactin, but no colibactin). Nine of them were ST395 (ST395-K2), and one was ST14 (ST14-K2). We detected the aerobactin cluster and yersiniabactin genes in all these isolates. Six strains possessed a truncated rmpA2 gene, and the remaining four strains lacked this gene. Of note, a single ST14-K2 isolate possessed the ferric uptake cluster. The ST395-K2 isolates produced different carbapenemases, namely, KPC-3 (55.6%, 5 of 9), KPC-41 (22.2%, 2 of 9), NDM-1 (11.1%, 1 of 9), or OXA-48 (11.1%, 1 of 9). The single ST14-K2 produced the OXA-48 carbapenemase. Isolates N911 and N1052 (respectively, ST14 and ST395) were the only isolates giving positive results to the string test. Acquisition and dissemination of the hypervirulence trait among K2-type producers was mainly due to a given plasmid identified here and named pKpCH_aerob (297,473bp; OP690162) (Fig. S2) that has 86.7% of similarity to plasmid pBA813_1 (MK649825.1) (25). The exception was only one ST395 isolate that had a pKpvST147B-like (CP040726.1) (26). Altogether, those MDR-hvKp possessing the K2 capsular type were detected from diverse biological sources and originated from six different Swiss cantons (Fig. 1).
FIG 1.
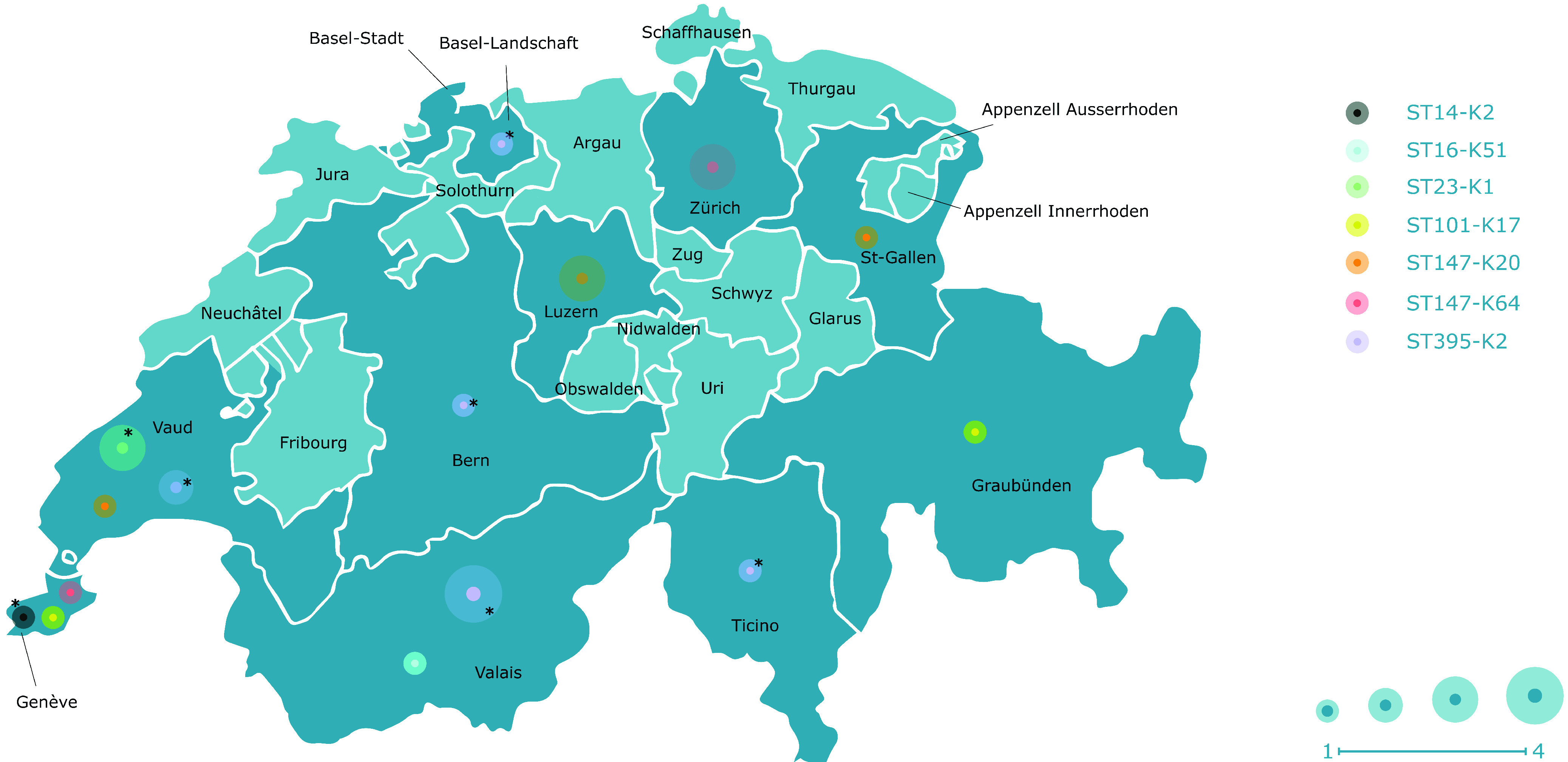
Cantons of Switzerland where the MDR-hvKp and MDR-nhv-Kp isolates were detected from January 2017 to December 2020. Cantons from which an MDR-hvKp or MDR-nhv-Kp isolate was detected are colored dark blue, and the ones where no isolate was detected are colored light blue. *, MDR-hvKp.
Group 3.
Five isolates presented a K20 capsular type, all belonging to ST147 (ST147-K20). All of them had a virulence score of 3 (presence of aerobactin, and absence of yersiniabactin- or colibactin-encoding genes). They carried the aerobactin cluster, and additionally, all but one had a complete rmpACD cluster, the remaining isolate having a truncation in the rmpA gene and an intact rmpCD. The former four isolates also presented the other mucoid phenotype regulator, namely, rmpA2, but in all cases, the gene was truncated. They all produced the NDM-1 carbapenemase and were recovered from urine (four isolates) and rectal swab (one isolate) from patients hospitalized in three Swiss cantons (Fig. 1). The occurrence of virulence genotype in those MDR-nhv-Kp isolates was related to the dissemination of a pKpvST147B plasmid (CP040726.1) (26).
Group 4.
Four MDR-nhv-Kp ST147 exhibited a K64 capsular type. These four isolates had a virulence score of 4 and were positive for aerobactin and yersiniabactin clusters. Two of them possessed an additional rmpACD cluster, and one of these two had also a truncated rmpA2 gene. They were all NDM-1 producers and had been recovered from blood culture, puncture, and rectal swabs in two cantons (Fig. 1). Likewise, they carried the aforementioned pKpvST147B plasmid (26).
Group 5.
Three MDR-nhv-Kp strains belonged to ST16-K51 (n = 1), and ST101-K17 (n = 2). These isolates had a virulence score of 4. All of them possessed the aerobactin and yersiniabactin clusters. Additionally, the ST101 isolates had the ferric uptake cluster, and one of them also had a truncated rmpA2 gene. They produced the KPC-3 carbapenemase and originated from rectal swabs collected in two cantons. The ST16 isolate originating from a rectal swab produced NDM-1 and also carried the whole mucoid phenotype regulator cluster rmpACD and a truncated rmpA2 gene. The pKpvST147B plasmid was identified in the ST16 isolate and both ST101 strains possessed the pKpCH_aerob plasmid, first identified in this study.
DISCUSSION
MDR-hvKp isolates have been increasingly reported during the last decade, even if it is still difficult to evaluate whether this really corresponds to an emerging phenomenon due to the paucity of epidemiological data in relation with that subject. Although these isolates are now commonly identified in Asia (15, 16, 21), it is difficult to evaluate to what extend they have disseminated around the globe (14, 17, 20, 22). In Switzerland, a study reported the first two cases of severe multifocal infections caused by hypervirulent K. pneumoniae in 2017 (27), those two isolates being of wild-type phenotype in terms of antibiotic susceptibility.
Following the evolution timeline and confirming the silent spread of convergent MDR-hvKp isolates in the country, Blanc et al. (19) recently reported the occurrence of an ST23-K1 OXA-48-producing K. pneumoniae in Switzerland. This isolate was obtained from the rectal microbiota of a young male patient hospitalized at the University Hospital of Lausanne (Vaud Canton) in 2018. The present study confirmed that K. pneumoniae possessing an hypervirulence genotype are quite commonly either infecting or colonizing patients in Switzerland, being identified all over the country, since 2018. Hence, we detected one MDR-hvKp ST23-K1 isolate from 2018 and two from 2019. Noticeably, the frequency of detection identified during the years in our collection argues for a significant increasing trend, gathering different clonal backgrounds. Among those, there were isolates producing either the K1 or the K2 capsular type.
The capsular type K1 is the main serogroup causing hvKp infections in Taiwan (28, 29). K1-type strains often belong to ST23, being for that reason considered so-called relatively monophyletic (24). Noticeably, the ST23-K1 isolates identified in this study were all very similar to the ST23 isolate previously reported in Switzerland (19), being also OXA-48 producers, and carrying the colibactin, aerobactin, and yersiniabactin clusters, as well as the rmpA2 gene (except in isolate N593) (19). However, this gene was found to be truncated in two other isolates in our collection. They were all positive for the magA chromosomal gene, involved in capsule biosynthesis of K1 isolates (24, 29). The three ST23-K1 identified in our study had the highest virulence score (virulence score of 5) among all the other 22 isolates. This finding is in line with data from the ECDC report establishing that the majority (82.4%) of the isolates from clade ST23-K1 had a virulence score of 5 (13). Additionally, the kfuABC cluster was also identified in the ST23 strains of our collection. This cluster has been associated with increased virulence by enabling lineages to use iron from diverse human and environmental sources (30).
Those ST23-K1 isolates carried a plasmid detected in our study that we named pKpST23_OXA-48. This genetic mobile element carried the aerobactin cluster, a truncated rmpA2 gene, and a series of resistance genes, including blaCTX-M-15, sul2, blaTEM-1, and qnrB1. Moreover, this plasmid has two replicons, namely, IncFIB and IncHI1B. Interestingly, this plasmid had a lower similarity with commonly ST23- or K1-associated plasmids such as plasmids pSGH10 (86%; CP025081.1) (31) and pK2044 (87%; AP006726.1) (32) compared to the 98% of similarity with plasmid pVir-CR-HvKP4 (MF437313.1) (15). The latter is a pLVPK-like plasmid that had been identified in a ST11 carbapenem-resistant hvKp responsible for causing a fatal outbreak in a Chinese Hospital (15). Noteworthy, as opposed to the aforementioned plasmid detected in China, the ST23-K1 detected in our study (all from rectal swabs) was negative for the salmochelin-encoding iroE gene. Considering the salmochelin cluster has been proven to be associated with invasive K. pneumoniae infections (8, 33) and enhancement of the bacterial growth (34), it might be speculated that those isolates lacking this gene would be less virulent.
As opposed to K1, K2-type MDR-hvKp isolates are not strongly linked to a particular sequence type (ST). Nevertheless, ST65 and ST86 strains are often associated with the K2 serotype (35), but none of them were identified in our study. Two different sequence types were detected within the K2 isolates, confirming the molecular heterogeneous characteristics of this group. One of those isolates corresponded to an OXA-48-producing ST14-K2 isolate. Although ST14 is not often related to K2 in K. pneumoniae isolates, Fursova et al. (36) reported cases of severe systemic infections associated with these organisms that evolved to fatal outcomes. According to our findings, the ferric iron uptake system was also detected in the isolates from the mentioned study, as well as in other studies reporting ST14-K2 isolates (37, 38). Interestingly, the ST14-K2 isolate identified in our study was the only one carrying the aerobactin cluster compared to those identified in Russia, Japan, and Algeria (36–38).
ST395-K2 strains were the most frequent MDR-hvKp strains identified in our study. All the nine isolates presented the yersiniabactin and the aerobactin clusters. This combination of features enables bacteria to increase iron acquisition from host transport proteins, enhances the ability to replicate and survive in different biological sites, and finally causes invasive infections (39, 40). Of note, one of the ST395-K2 isolates identified in our study was recovered from a blood culture and produced the KPC-3 carbapenemase. It exhibited a similar virulence profile compared to hvKp isolates with same ST and capsular serotype identified in Russia (41). In that latter study, results from a mouse lethality assay showed that the MDR hvKp-ST395-K2 isolates harboring rmpA2, aerobactin, yersiniabactin, and additional virulence genes displayed a higher lethality in the model compared with the ones lacking rmpA2 and aerobactin but harboring yersiniabactin genes (41). In contrast, four ST395-K2 isolates of our study were negative for the rmpA2 mucoid phenotype regulator, and the remaining five had truncations in the gene, making them unable to upregulate capsule production (42). This finding might explain the relatively high percentage of strains belonging to this group originating from biological surveillance sources (e.g., 44.4%, 4 of 9 from rectal swabs) rather than from infections.
A novel pKpCH_aerob plasmid was detected in most of the K2-type isolates from our collection. This virulence plasmid first described here possesses two replication sites: IncFIB and IncHI1B. It carries the aerobactin cluster and a truncated rmpA2 gene, as well as some resistance genes such as sul1, catA1, and ant(2′′)Ia. This plasmid is 86.7% similar to the pBA813_1 (MK649825.1) detected in a ST2096-K64 K. pneumoniae in India (25). As opposed to the plasmid detected in the Indian isolate, the ST14-K2 and ST395-K2 isolates identified in our study lacked the salmochelin cluster. The pBA813_1 plasmid also carries the sul1 resistance gene, but in contrast to pKpCH_aerob, it harbors other acquired resistance genes such as blaCTX-M-15, blaOXA-232, and blaTEM-54, among others (25). Surprisingly, one ST395-K2 isolate had a plasmid similar to the pKpvST147 virulence plasmid, which is mainly associated with ST147 K. pneumoniae isolates (26).
The well known MDR lineage ST147 was detected in our study presenting two different serotypes: the hypervirulent-associated K20 and the MDR-related K64. Few studies had detected MDR-hvKp ST147-K20 isolates so far. A study performed in Russia identified NDM-1-producing MDR-hvKp ST147-K20 isolates presenting rmpA, rmpA2, and the aerobactin cluster but lacking the yersiniabactin genes (41). The data shown in the Russian study are similar to the virulome profile of our ST147-K20 isolates, all of them being negative to the ybt cluster. Unfortunately, the limitation of our study is that no in vivo virulence assay (e.g., Galleria mellonella as an infection model) has been performed; thus, the ST147-K20 isolates identified in our study have been defined as MDR-nhv-Kp. Another study performed in the United Kingdom reported an NDM-producing ST147 K. pneumoniae carrying the rmpA, rmpA2, aerobactin, and yersiniabactin genes; however, this isolate had a K35 capsular type (43).
The ST147 isolates of K64 are commonly recognized as classical K. pneumoniae (cKp), but they have also recently been described as hybrid MDR-hvKp due to virulence plasmids acquisition (26). Here, we described four NDM-1-producing MDR-Kp belonging to ST147-K64 that all carried the aerobactin and the ybt clusters, with two of them possessing the two mucoid phenotype regulators often associated with hypervirulence, namely, rmpACD and rmpA2. Capsular type K64 isolates have been recently shown to possess an hypervirulence phenotype in the G. mellonella model (44); thus, we might also consider that the ST147-K64 isolates from our study could be regarded as hypervirulent. Of note, isolates belonging to the ST147-K64 lineage that were responsible for a large regional outbreak in Italy carried a similar virulence profile (ybt, iuc, rmpACD, and rmpA2) compared to our isolates (44). That latter study showed that the virulence potential of that lineage was variable after infection using G. mellonella as an experimental model (44). A multicenter analysis conducted in China showed that ST11-K64 carbapenem-resistant K. pneumoniae was the most prevalent hypervirulent profile, being confirmed by an in vivo infection model (45). Furthermore, the same capsular type K64 and ST11 profile was reported in a fatal bacteremia in Brazil (46), reinforcing that the K64 isolates could be hypervirulent and transmissible.
The ST147 isolates detected in our study had a virulence plasmid that shared ca. 98.5% of nucleotide identity with plasmid pKpvST147B (CP040726.1) described by Turton et al. (26), which was first detected from a rectal swab of a patient in the United Kingdom (26). Here, we identified such pKpvST147B plasmid among different NDM-producing MDR-nhv-Kp ST147 strains disseminated across the country. We found evidence that this plasmid was commonly associated with virulence factors in our strains, such as (i) the aerobactin cluster, which was carried by this plasmid in all ST147 isolates, and (ii) the mucoid phenotype regulators, being identified in 77.8% (7 of 9) of our isolates. Di Pilato et al. (44) recently demonstrated that hybrid pKpvST147B-like plasmids are mobilizable by conjugation, highlighting their ability to spread among successful MDR clones of K. pneumoniae. Indeed, the emergence of these plasmids has been noticed not only in K. pneumoniae ST147, but also ST307, ST11, ST15, ST16 (present study), ST395 (present study), and ST383 since 2017 in different countries, suggesting that the distribution of these elements carrying virulence and resistance genes may be wider than expected (22, 26, 47–49).
K. pneumoniae ST101-K17 isolates have been reported in Italy and Brazil (50–52). Virulence genotype of the different ST101-K17 isolates published in these studies includes the yersiniabactin cluster and ferric uptake system, but in contrast with our findings, the aerobactin cluster was lacking. Aerobactins have been associated with a significant increase in survival in serum, human ascites, and systemic and pulmonary infection in mouse models (40). These data support the hypothesis that aerobactin is an important virulence determinant among hvKp, enabling such isolates to cause systemic infection. As mentioned above, we did not perform an in vivo virulence test to further characterize our isolates; thus, ST101-K17 isolates identified in our study have been defined as MDR-nhv-Kp.
Isolates producing the K51 capsular type are rarely described. ST16-K51 nonhypervirulent K. pneumoniae isolates that were described in Thailand originated from biological sources from three different hospitals (53). In our study, a single NDM-1-producing MDR-nhv-Kp ST16-K51 was identified. The isolates from Thailand had only the yersiniabactin cluster, while the one described in our study possesses the aerobactin, yersiniabactin, and mucoid phenotype regulatory clusters. This isolate harbored a plasmid sharing 99% of nucleotide identity with the pKpvST147B virulence plasmid, known to be associated with the ST147 lineage (26). This finding therefore highlights that this given virulence plasmid might be disseminating within different strain backgrounds.
Phenotypic detection of hypermucoviscosity through the string test has been used to putatively define hypervirulence (54). However, the accuracy of using such a test to extrapolate any correlation with the hvKp phenotype infections remains highly questionable. Reports in the literature addressing this issue indicated that such association might range from 51% to 98% (54, 55). Another recent study reported a sensitivity of 28.7% of the string test compared with the presence of the iucA gene by PCR (56).
On the other hand, positive string test results have also been reported among “classical” K. pneumoniae phenotypes (57). Recently, Dey et al. (58) reported a K. pneumoniae isolate recovered from a patient’s solid cystic lesion that presented a hypermucoviscous phenotype; however, this isolate lacked key virulence determinants such as aerobactin, yersiniabactin, and salmochelin biosynthesis clusters, as well as the rmpACD and rmpA2 genes, which are associated with enhanced capsule synthesis and hypermucoviscosity. Among our collection of 13 MDR-hvKp isolates, only a single ST14-K2 and a single ST395-K2 turned positive for the string test, although both were negative for the rmpACD cluster, and one (ST14) had a truncated rmpA2 gene. These data also suggest a more critical analysis when dealing with hypervirulence definition.
Interestingly, all our isolates that carried the rmpA2 mucoid phenotype regulator (60%, 15 of 25) had truncations after, in average, 50.3% length of the intact amino acid sequence, generating a probable loss of protein function. This plasmid-borne mucoid regulator seems to be independent of rmpA and produces a RmpA2 protein that can help the K2 capsule serotype biosynthesis interacting with its promoters (59). As mentioned above, 60% (6 of 10) of the K2 isolates had rmpA2, but it was always truncated. In fact, truncations in the mucoid phenotype regulators seems to be common, and a significant proportion of the recorded rmpA or rmpA2 genes carries frameshift mutations arising from insertion or deletion of nucleotides at a poly(G) sequence, causing loss of function (60).
Conclusion.
This study including isolates recovered during a 4-year period in Switzerland confirms the occurrence and dissemination of worrisome convergent carbapenemase-producing and hypervirulent K. pneumoniae isolates. Our findings highlight the importance of an active surveillance network to track and monitor the evolution of those successful hybrid clones. These strains represent a substantial public health threat, and strict monitoring seems indeed crucial to prevent large-scale dissemination and potential outbreaks at the source of very-difficult-to-treat infections and increased mortality/morbidity. We might now consider that screening of carbapenemase genes could be associated with screening of virulence genes when dealing with MDR K. pneumoniae isolates. In fact, a new paradigm could be to consider the identification of hvKp strains (and therefore of the responsible virulence markers) as quite systematic when dealing with such MDR strains, with corresponding information being eventually reported to clinicians. Along with the accurate identification of resistance patterns, such systematic screening of virulence traits might be considered useful and help the clinician when a patient develops uncommon clinical and infectious signs.
MATERIALS AND METHODS
Strain collection.
A total of 413 carbapenem-resistant K. pneumoniae (CrKp) were received at the Swiss National Reference Center for Emerging Antibiotic Resistance (NARA) between 2017 and 2020. From these isolates, 279 (67.6%) were identified as carbapenemase-producing K. pneumoniae (CpKp) and were initially included in the study. They had been recovered from different sources, being mostly urine, blood cultures, rectal swabs, and respiratory tract specimens.
Antimicrobial susceptibility testing.
Antibiotic susceptibility testing was performed by disk diffusion and interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (61). Multidrug resistance (MDR) was defined as proposed by Magiorakos et al. (62).
Carbapenemase detection.
Screening of the isolates for carbapenemase genes was performed by PCR for the blaKPC, blaNDM, blaIMP, blaVIM, and blaOXA-48 genes (63–66), and the corresponding amplicons were then sequenced (Microsynth AG, Switzerland). Sequence analysis was performed using MEGA software 11.0.11 (67) and the nucleotide Basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Hypermucoviscosity detection.
The string test was performed as described by Fang et al. (54). Briefly, after an overnight incubation at 37°C on a 5% sheep blood agar, the colonies were stretched using a standard bacteriologic loop. Positive strains were defined as those forming a viscous string >5 mm in length.
Virulence features.
A set of virulence features was selected to screen the carbapenemase-producing collection for hvKp by using a PCR approach. The focus was made on the mucoid phenotype regulatory genes rmpA and rmpA2 (68, 69), the aerobactin siderophore cluster genes iucABCD_iutA (69–71); the yersiniabactin siderophore cluster was detected using the ybtA, ybtS, and ybtU genes (72, 73); and the genotoxin colibactin cluster was detected using the clbABNQ genes (74), the ferric iron uptake gene kfu (30), and the capsular types K1, K2, K5, K20, K54, and K57 (28, 54, 75). The gene magA was detected to double check the presence of K1 (69).
Hypervirulence definition.
Preselection of hvKp candidates among CpKp was performed using the virulence score based on the presence of the genes encoding yersiniabactin (ybt), colibactin (clb), and aerobactin (iuc) proposed by Lam et al. (76). Using this scheme, virulence scores vary from 0 to 5. Isolates with a score equal to 0 to 2 were not further considered in this study, and those with scores of 4 to 5 were included in the study. Additionally, isolates with virulence score of 3 simultaneously presenting the molecular biomarker rmpACD and rmpA2 genes (mucoid phenotype regulators) were also included (77). Finally, those strains presenting the typical hypervirulent capsular type K1 and K2 were named MDR-hvKp. The remaining isolates were considered MDR nonhypervirulent K. pneumoniae (MDR-nhv-Kp) strains; they were either common MDR clones that acquired a virulence plasmid or present nontypically hypervirulent capsular types.
Whole-genome sequencing (WGS).
Genomic DNA paired-end libraries were generated using the Nextera sample preparation kit (Illumina, Inc., USA). The libraries were sequenced using the Illumina MiSeq platform (Illumina, https://www.illumina.com) with 2 × 150 bp paired-end reads. Raw sequence data were previously submitted to the National Center for Biotechnology Information’s Sequence Read Archive (BioProject no. PRJNA890192). Reads were assembled into contigs using a SPAdes (78) tool-based pipeline called Shovill (https://github.com/tseemann/shovill). Sequence types, the presence of acquired resistance genes, species, and plasmid replicons were detected using, respectively, MLST version 2.0, ResFinder version 4.1 (79), KmerFinder version 3.2 (80), and PlasmidFinder 2.1, on the Center for Genomic Epidemiology platform (https://www.genomicepidemiology.org/); the presence of virulence-associated features, including capsular type, and the virulence scores were confirmed using Kleborate tool (https://github.com/katholt/Kleborate); and contigs were annotated using Prokka (81).
Two isolates representing the well known hypervirulent associated K1 (ST23;_isolate N330) and K2 (ST395;_isolate N447) capsular types were selected for long-read sequencing as described elsewhere (82). Briefly, genomic DNA was extracted using QIAamp DNA minikit (Qiagen, Valencia, CA, USA) and sequenced using the MinION Mk1C (Oxford Nanopore Technologies, Oxford, UK). The libraries were prepared using a one-dimensional chemistry ligation sequencing kit (SQK-LSK109, Oxford Nanopore Technologies). Sequencing was performed in a R9.4.1 Flongle flow cell (FLO-FLG001, Oxford Nanopore Technologies). The sequences were first assembled using a Canu tool (83). Finally, trimmed long-read sequences and short-read sequences were used to construct hybrid assembles with UniCycler tool (84).
Worldwide-disseminated virulence plasmid sequences were downloaded from GenBank to generate a plasmid reference database for mapping analyses. CLC Genomics Workbench 20.0.4 Qiagen Aarhus A/S (Qiagen, Redwood City, CA, USA) was used to confirm the presence of the plasmids by mapping sequences against the reference database. Then the Mauve Multiple Genome Alignment tool (85) was used to reorder and analyze plasmids sequence matches and manually mitigate against false-positive sequences. Finally, we used >95% coverage and identity to define significant matches.
ACKNOWLEDGMENTS
This work was financed by the University of Fribourg (Fribourg, Switzerland) and by the NARA.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Liu YC, Cheng DL, Lin CL. 1986. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med 146:1913–1916. 10.1001/archinte.1986.00360220057011. [DOI] [PubMed] [Google Scholar]
- 2.Siu LK, Yeh K-M, Lin J-C, Fung C-P, Chang F-Y. 2012. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis 12:881–887. 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 3.Shon AS, Bajwa RPS, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Virulence 4:107–118. 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, Andersen PS, Driebe EM, Keim P, Krogfelt KA. 2015. Mapping the evolution of hypervirulent Klebsiella pneumoniae. mBio 6:e00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo Y, Wang Y, Ye L, Yang J. 2014. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect 20:O818–O824. 10.1111/1469-0691.12664. [DOI] [PubMed] [Google Scholar]
- 6.Prokesch BC, TeKippe M, Kim J, Raj P, TeKippe EM, Greenberg DE. 2016. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect Dis 16:e190–e195. 10.1016/S1473-3099(16)30021-4. [DOI] [PubMed] [Google Scholar]
- 7.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen K, Van Nguyen TV, Dao TT, Mensink M, Le Minh V, Nhu NTK, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 112:E3574–E3581. 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei T, Zou C, Qin J, Tao J, Yan L, Wang J, Du H, Shen F, Zhao Y, Wang H. 2022. Emergence of hypervirulent ST11-K64 Klebsiella pneumoniae poses a serious clinical threat in older patients. Front Public Heal 10:765624. 10.3389/fpubh.2022.765624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard A-S, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine M-H, Decré D, Brisse S. 2014. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 20:1812–1820. 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Queenan AM, Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 20:440–458. 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, Brisse S, Holt KE. 2018. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 4:e000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emergence of hypervirulent Klebsiella pneumoniae ST23 carrying carbapenemase genes in EU/EEA countries. 2021. European Centre for Disease Prevention and Control, Stockholm, Sweden. https://www.ecdc.europa.eu/en/publications-data/risk-assessment-emergence-hypervirulent-klebsiella-pneumoniae-eu-eea. Accessed 13 June 2022. [Google Scholar]
- 14.Karlsson M, Stanton RA, Ansari U, McAllister G, Chan MY, Sula E, Grass JE, Duffy N, Anacker ML, Witwer ML, Rasheed JK, Elkins CA, Halpin AL. 2019. Identification of a carbapenemase-producing hypervirulent Klebsiella pneumoniae isolate in the United States. Antimicrob Agents Chemother 63:e00519-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW-C, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 16.Zhang R, Lin D, Chan EW, Gu D, Chen G-X, Chen S. 2016. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother 60:709–711. 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cejas D, Fernández Canigia L, Rincón Cruz G, Elena AX, Maldonado I, Gutkind GO, Radice MA. 2014. First isolate of KPC-2-producing Klebsiella pneumoniae sequence type 23 from the Americas. J Clin Microbiol 52:3483–3485. 10.1128/JCM.00726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yonekawa S, Mizuno T, Nakano R, Nakano A, Suzuki Y, Asada T, Ishii A, Kakuta N, Tsubaki K, Mizuno S, Ogawa M, Yano H, Kasahara K, Mikasa K. 2020. Molecular and epidemiological characteristics of carbapenemase-producing Klebsiella pneumoniae clinical isolates in Japan. mSphere 5:e00490-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanc DS, Poirel L, Van Singer M, Greub G, Nordmann P. 2021. Hypervirulent Klebsiella pneumoniae ST23 producing OXA-48 in Switzerland. Int J Antimicrob Agents 58:106457. 10.1016/j.ijantimicag.2021.106457. [DOI] [PubMed] [Google Scholar]
- 20.Roulston KJ, Bharucha T, Turton JF, Hopkins KL, Mack DJF. 2018. A case of NDM-carbapenemase-producing hypervirulent Klebsiella pneumoniae sequence type 23 from the UK. JMM Case Rep 5:e005130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y-H, Chou S-H, Liang S-W, Ni C-E, Lin Y-T, Huang Y-W, Yang T-C. 2018. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother 73:2039–2046. 10.1093/jac/dky164. [DOI] [PubMed] [Google Scholar]
- 22.Ahmed M-GE-S, Yang Y, Yang Y, Yan B, Chen G, Hassan RM, Zhong L-L, Chen Y, Roberts AP, Wu Y, He R, Liang X, Qin M, Dai M, Zhang L, Li H, Yang F, Xu L, Tian G-B. 2021. Emergence of hypervirulent carbapenem-resistant Klebsiella pneumoniae coharboring a blaNDM-1-carrying virulent plasmid and a blaKPC-2-carrying plasmid in an Egyptian hospital. mSphere 6:e00088-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang M, Kong X, Hao J, Liu J. 2020. Epidemiological characteristics and formation mechanisms of multidrug-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol 11:581543. 10.3389/fmicb.2020.581543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harada S, Doi Y. 2018. Hypervirulent Klebsiella pneumoniae: a call for consensus definition and international collaboration. J Clin Microbiol 56:e00959-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyres KL, Nguyen TNT, Lam MMC, Judd LM, van Vinh Chau N, Dance DAB, Ip M, Karkey A, Ling CL, Miliya T, Newton PN, Lan NPH, Sengduangphachanh A, Turner P, Veeraraghavan B, Vinh PV, Vongsouvath M, Thomson NR, Baker S, Holt KE. 2020. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med 12:11. 10.1186/s13073-019-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turton J, Davies F, Turton J, Perry C, Payne Z, Pike R. 2019. Hybrid resistance and virulence plasmids in “high-risk” clones of Klebsiella pneumoniae, including those carrying blaNDM-5. Microorganisms 7:326. 10.3390/microorganisms7090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Babouee Flury B, Donà V, Buetti N, Furrer H, Endimiani A. 2017. First two cases of severe multifocal infections caused by Klebsiella pneumoniae in Switzerland: characterization of an atypical non-K1/K2-serotype strain causing liver abscess and endocarditis. J Glob Antimicrob Resist 10:165–170. 10.1016/j.jgar.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Fang C-T, Lai S-Y, Yi W-C, Hsueh P-R, Liu K-L, Chang S-C. 2007. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 45:284–293. 10.1086/519262. [DOI] [PubMed] [Google Scholar]
- 29.Chuang Y, Fang C, Lai S, Chang S, Wang J. 2006. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 193:645–654. 10.1086/499968. [DOI] [PubMed] [Google Scholar]
- 30.Ma L, Fang C, Lee C, Shun C, Wang J. 2005. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J Infect Dis 192:117–128. 10.1086/430619. [DOI] [PubMed] [Google Scholar]
- 31.Lam MMC, Wyres KL, Duchêne S, Wick RR, Judd LM, Gan YH, Hoh CH, Archuleta S, Molton JS, Kalimuddin S, Koh TH, Passet V, Brisse S, Holt KE. 2018. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 9:2703. 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu K-M, Li L-H, Yan J-J, Tsao N, Liao T-L, Tsai H-C, Fung C-P, Chen H-J, Liu Y-M, Wang J-T, Fang C-T, Chang S-C, Shu H-Y, Liu T-T, Chen Y-T, Shiau Y-R, Lauderdale T-L, Su I-J, Kirby R, Tsai S-F. 2009. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 191:4492–4501. 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jun J-B. 2018. Klebsiella pneumoniae liver abscess. Infect Chemother 50:210–218. 10.3947/ic.2018.50.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miethke M, Marahiel MA. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71:413–451. 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin J-C, Koh TH, Lee N, Fung C-P, Chang F-Y, Tsai Y-K, Ip M, Siu LK. 2014. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog 6:21. 10.1186/1757-4749-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fursova NK, Astashkin EI, Ershova ON, Aleksandrova IA, Savin IA, Novikova TS, Fedyukina GN, Kislichkina AA, Fursov MV, Kuzina ES, Biketov SF, Dyatlov IA. 2021. Multidrug-resistant Klebsiella pneumoniae causing severe infections in the neuro-ICU. Antibiotics 10:979. 10.3390/antibiotics10080979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harada S, Ishii Y, Saga T, Aoki K, Tateda K. 2018. Molecular epidemiology of Klebsiella pneumoniae K1 and K2 isolates in Japan. Diagn Microbiol Infect Dis 91:354–359. 10.1016/j.diagmicrobio.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Zemmour A, Dali-Yahia R, Maatallah M, Saidi-Ouahrani N, Rahmani B, Benhamouche N, Al-Farsi HM, Giske CG. 2021. High-risk clones of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolated from the University hospital establishment of Oran, Algeria (2011–2012). PLoS One 16:e0254805. 10.1371/journal.pone.0254805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holden VI, Bachman MA. 2015. Diverging roles of bacterial siderophores during infection. Metallomics 7:986–995. 10.1039/c4mt00333k. [DOI] [PubMed] [Google Scholar]
- 40.Russo TA, Olson R, MacDonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. 2014. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 82:2356–2367. 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazareva I, Ageevets V, Sopova J, Lebedeva M, Starkova P, Likholetova D, Lebedeva M, Gostev V, Moiseenko V, Egorenkov V, Navatskaya A, Mitroshina G, Myasnikova E, Tsvetkova I, Lobzin Y, Sidorenko S. 2020. The emergence of hypervirulent blaNDM-1-positive Klebsiella pneumoniae sequence type 395 in an oncology hospital. Infect Genet Evol 85:104527. 10.1016/j.meegid.2020.104527. [DOI] [PubMed] [Google Scholar]
- 42.Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. 2010. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 192:3144–3158. 10.1128/JB.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turton JF, Payne Z, Coward A, Hopkins KL, Turton JA, Doumith M, Woodford N. 2018. Virulence genes in isolates of Klebsiella pneumoniae from the UK during 2016, including among carbapenemase gene-positive hypervirulent K1-st23 and ‘non-hypervirulent’ types ST147, ST15 and ST383. J Med Microbiol 67:118–128. 10.1099/jmm.0.000653. [DOI] [PubMed] [Google Scholar]
- 44.Di Pilato V, Henrici De Angelis L, Aiezza N, Baccani I, Niccolai C, Parisio EM, Giordano C, Camarlinghi G, Barnini S, Forni S, Righi L, Mechi MT, Giani T, Antonelli A, Rossolini GM. 2022. Resistome and virulome accretion in an NDM-1-producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: a genotypic and phenotypic characterisation. Lancet Microbe 3:e224–e234. 10.1016/S2666-5247(21)00268-8. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Jin L, Ouyang P, Wang Q, Wang R, Wang J, Gao H, Wang X, Wang H, Kang H, Gu B, Wang C, Cao B, Yang C, Jin L, Liao K, Zhang X, Ma X, Xie L, Zheng R, Zou H, Wang S, Pei F, Man S, Li W, Zhang Y, Cui Q, Jia X, Guo D, Fu Q, Zhang Z, Guo Z, Li Z, Xu Y, Ma X, Li Y, Jin Y, Liu Z, Zeng J, Li X, Zou C, Ji P, Jin C, Huang J, Tian J, Wu W, Xu X, Wen H, Yuan J. 2020. Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother 75:327–336. 10.1093/jac/dkz446. [DOI] [PubMed] [Google Scholar]
- 46.de Campos TA, Gonçalves LF, Magalhães KG, de Paulo Martins V, Pappas Júnior GJ, Peirano G, Pitout JDD, Gonçalves GB, Furlan JPR, Stehling EG, Pitondo-Silva A. 2018. A fatal bacteremia caused by hypermucousviscous KPC-2 producing extensively drug-resistant K64-ST11 Klebsiella pneumoniae in Brazil. Front Med 5:265. 10.3389/fmed.2018.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heiden SE, Hübner N-O, Bohnert JA, Heidecke C-D, Kramer A, Balau V, Gierer W, Schaefer S, Eckmanns T, Gatermann S, Eger E, Guenther S, Becker K, Schaufler K. 2020. A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med 12:113. 10.1186/s13073-020-00814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chudejova K, Kraftova L, Mattioni Marchetti V, Hrabak J, Papagiannitsis CC, Bitar I. 2021. Genetic plurality of OXA/NDM-encoding features characterized from Enterobacterales recovered from Czech hospitals. Front Microbiol 12:641415. 10.3389/fmicb.2021.641415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Starkova P, Lazareva I, Avdeeva A, Sulian O, Likholetova D, Ageevets V, Lebedeva M, Gostev V, Sopova J, Sidorenko S. 2021. Emergence of hybrid resistance and virulence plasmids harboring New Delhi metallo-β-lactamase in Klebsiella pneumoniae in Russia. Antibiotics 10:691. 10.3390/antibiotics10060691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roe CC, Vazquez AJ, Esposito EP, Zarrilli R, Sahl JW. 2019. Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front Microbiol 10:542. 10.3389/fmicb.2019.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gona F, Bongiorno D, Aprile A, Corazza E, Pasqua B, Scuderi MG, Chiacchiaretta M, Cirillo DM, Stefani S, Mezzatesta ML. 2019. Emergence of two novel sequence types (3366 and 3367) NDM-1- and OXA-48-co-producing K. pneumoniae in Italy. Eur J Clin Microbiol Infect Dis 38:1687–1691. 10.1007/s10096-019-03597-w. [DOI] [PubMed] [Google Scholar]
- 52.Andrade LN, Novais Â, Stegani LMM, Ferreira JC, Rodrigues C, Darini ALC, Peixe L. 2018. Virulence genes, capsular and plasmid types of multidrug-resistant CTX-M(-2, -8, -15) and KPC-2-producing Klebsiella pneumoniae isolates from four major hospitals in Brazil. Diagn Microbiol Infect Dis 91:164–168. 10.1016/j.diagmicrobio.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Loraine J, Heinz E, De Sousa Almeida J, Milevskyy O, Voravuthikunchai SP, Srimanote P, Kiratisin P, Thomson NR, Taylor PW. 2018. Complement susceptibility in relation to genome sequence of recent Klebsiella pneumoniae isolates from Thai hospitals. mSphere 3:e00537-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang C-T, Chuang Y-P, Shun C-T, Chang S-C, Wang J-T. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Y-C, Lu M-C, Tang H-L, Liu H-C, Chen C-H, Liu K-S, Lin C, Chiou C-S, Chiang M-K, Chen C-M, Lai Y-C. 2011. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol 11:50. 10.1186/1471-2180-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raj S, Sharma T, Pradhan D, Tyagi S, Gautam H, Singh H, Sood S, Dhawan B, Das BK, Kapil A, Chaudhry R, Mohapatra S. 2022. Comparative analysis of clinical and genomic characteristics of hypervirulent Klebsiella pneumoniae from hospital and community settings: experience from a tertiary healthcare center in India. Microbiol Spectr 10:e0037622. 10.1128/spectrum.00376-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee C-H, Liu J-W, Su L-H, Chien C-C, Li C-C, Yang K-D. 2010. Hypermucoviscosity associated with Klebsiella pneumoniae-mediated invasive syndrome: a prospective cross-sectional study in Taiwan. Int J Infect Dis 14:e688–e692. 10.1016/j.ijid.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 58.Dey T, Chakrabortty A, Kapoor A, Warrier A, Nag VL, Sivashanmugam K, Shankar M. 2022. Unusual hypermucoviscous clinical isolate of Klebsiella pneumoniae with no known determinants of hypermucoviscosity. Microbiol Spectr 10:e0039322. 10.1128/spectrum.00393-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lai Y-C, Peng H-L, Chang H-Y. 2003. RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol 185:788–800. 10.1128/JB.185.3.788-800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu W-L, Lee M-F, Tang H-J, Chang M-C, Chuang Y-C. 2015. Low prevalence of rmpA and high tendency of rmpA mutation correspond to low virulence of extended spectrum β-lactamase-producing Klebsiella pneumoniae isolates. Virulence 6:162–172. 10.1080/21505594.2015.1016703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2022. Breakpoint tables for interpretation of MICs and zone diameters, version 12, 1–110.
- 62.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 63.Kaase M, Nordmann P, Wichelhaus TA, Gatermann SG, Bonnin RA, Poirel L. 2011. NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J Antimicrob Chemother 66:1260–1262. 10.1093/jac/dkr135. [DOI] [PubMed] [Google Scholar]
- 64.Pillai DR, Melano R, Rawte P, Lo S, Tijet N, Fuksa M, Roda N, Farrell DJ, Krajden S. 2009. Klebsiella pneumoniae carbapenemase, Canada. Emerg Infect Dis 15:827–829. 10.3201/eid1505.081536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. 2011. Characterization of OXA-181, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4896–4899. 10.1128/AAC.00481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 67.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nadasy KA, Domiati-Saad R, Tribble MA. 2007. Invasive Klebsiella pneumoniae syndrome in North America. Clin Infect Dis 45:e25–e28. 10.1086/519424. [DOI] [PubMed] [Google Scholar]
- 69.Xu M, Fu Y, Fang Y, Xu H, Kong H, Liu Y, Chen Y, Li L. 2019. High prevalence of KPC-2-producing hypervirulent Klebsiella pneumoniae causing meningitis in Eastern China. Infect Drug Resist 12:641–653. 10.2147/IDR.S191892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ling J, Pan H, Gao Q, Xiong L, Zhou Y, Zhang D, Gao S, Liu X. 2013. Aerobactin synthesis genes iucA and iucC contribute to the pathogenicity of avian pathogenic Escherichia coli O2 strain E058. PLoS One 8:e57794. 10.1371/journal.pone.0057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. 1995. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol 12:85–90. 10.1111/j.1574-695X.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 72.Hsieh P, Lin T, Lee C, Tsai S, Wang J. 2008. Serum‐induced iron‐acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis 197:1717–1727. 10.1086/588383. [DOI] [PubMed] [Google Scholar]
- 73.Compain F, Babosan A, Brisse S, Genel N, Audo J, Ailloud F, Kassis-Chikhani N, Arlet G, Decré D. 2014. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol 52:4377–4380. 10.1128/JCM.02316-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lan Y, Zhou M, Jian Z, Yan Q, Wang S, Liu W. 2019. Prevalence of pks gene cluster and characteristics of Klebsiella pneumoniae‐induced bloodstream infections. J Clin Lab Anal 33:e22838. 10.1002/jcla.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turton JF, Baklan H, Siu LK, Kaufmann ME, Pitt TL. 2008. Evaluation of a multiplex PCR for detection of serotypes K1, K2 and K5 in Klebsiella sp. and comparison of isolates within these serotypes. FEMS Microbiol Lett 284:247–252. 10.1111/j.1574-6968.2008.01208.x. [DOI] [PubMed] [Google Scholar]
- 76.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russo TA, Olson R, Fang C-T, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR, Backer M, Bajwa R, Catanzaro AT, Crook D, de Almeda K, Fierer J, Greenberg DE, Klevay M, Patel P, Ratner A, Wang J-T, Zola J. 2018. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 56:e00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Larsen MV, Cosentino S, Lukjancenko O, Saputra D, Rasmussen S, Hasman H, Sicheritz-Pontén T, Aarestrup FM, Ussery DW, Lund O. 2014. Benchmarking of methods for genomic taxonomy. J Clin Microbiol 52:1529–1539. 10.1128/JCM.02981-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 82.Findlay J, Perreten V, Poirel L, Nordmann P. 2022. Molecular analysis of OXA-48-producing Escherichia coli in Switzerland from 2019 to 2020. Eur J Clin Microbiol Infect Dis 41:1355–1360. 10.1007/s10096-022-04493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aac.01424-22-s0001.xlsx, XLSX file, 0.02 MB (24.3KB, xlsx)
Supplemental material. Download aac.01424-22-s0002.pdf, PDF file, 0.8 MB (804.2KB, pdf)


