ABSTRACT
Many modern farming practices negatively impact ecosystems on the local and global scales. Here, we assessed the taxonomic structures of 48 soil microbial communities along an agricultural transect using 16S rRNA and internal transcribed spacer (ITS) amplicon sequencing. We further characterized the functional structures of a subsample of 12 microbiomes using whole-genome sequencing.
ANNOUNCEMENT
Microbial communities in agricultural soil play important ecological and economic roles (1, 2). Many modern agricultural practices negatively impact soils and soil microbiomes, degrading nutrients and reducing diversity (3). The use of antimicrobials in agriculture further alters microbial communities in soil and surrounding environments (4). Given increasing pressures on the global food system, understanding how soil microbiomes respond to agricultural practices is critical. Here, we present metagenomic sequences from soil samples collected from an agricultural transect encompassing different proximities to human activity.
Samples were collected from four sites along a transect of previously farmed soil, perpendicular to Esopus Creek, New York (Fig. 1A), in a gradient from most affected by human activity to least, as follows: F8-W, a forested strip between the riverbank and a dirt road; F8-0 (3.05 m), F8-300 (91.44 m), and F8-600 (182.88 m), fallow farm sites. At each site, we collected three 10-g topsoil samples on 6 June, 19 June, and 3 July 2019 using sterile techniques. Samples were transported to the laboratory on ice in a dark cooler and frozen for at least 24 h.
FIG 1.
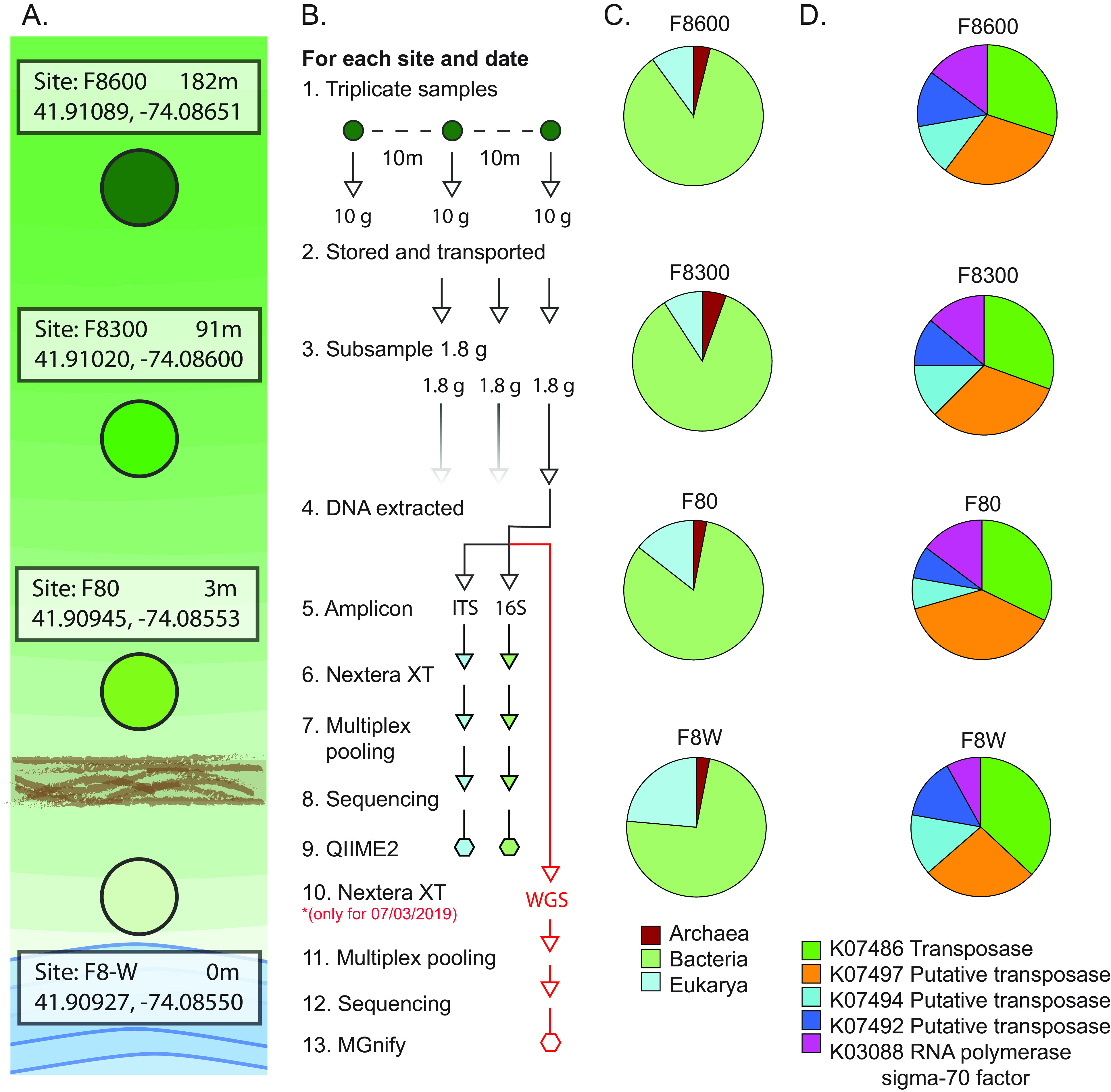
(A) Schematic diagram showing the distance from the water line (white circle) of each site along the F8 transect from which triplicate samples were taken in 2019. (B) Schematic diagram of the sampling, library construction, and sequencing of each sample. (C) Pie charts showing the relative proportions of eukaryotes, prokaryotes, and archaea. (D) Percent abundances of the top five most abundant KO orthologs.
For each sample (Fig. 1B), we extracted DNA from 1.8 g of soil using the Quick-DNA fecal/soil microbe miniprep kit (Zymo), and this DNA was used for 16S rRNA and internal transcribed spacer (ITS) amplicon sequencing; for samples collected on 3 July, these extractions were also used for whole-genome sequencing (WGS). All 16S rRNA amplicon sequencing amplified the V4 region using the 515F and 806R primers (5). ITS amplicon sequencing used the ITS1f and ITS2 primers (5). PCR products for the 16S rRNA and ITS amplicons were pooled separately and purified on a 2% agarose gel using a Qiagen gel extraction kit. The excised regions were ~385 bp for the 16S rRNA amplicons and 200 to 600 bp for the ITS amplicons. WGS library preparation was performed using the Nextera XT DNA library preparation kit (Illumina). All purified libraries were quality checked using an Agilent 2100 BioAnalyzer and a high-sensitivity DNA kit (Agilent) and stored at −20°C until sequencing. Amplicon libraries were sequenced at Wright Labs (Huntingdon, PA) using Illumina MiSeq v2 paired-end sequencing (2 × 250-bp reads) with 20% PhiX spike-in. WGS libraries were sequenced using an Illumina NextSeq 2000 system (2 × 150-bp paired-end reads) with default parameters.
For 16S rRNA and ITS analyses, the QIIME2 pipeline was used with default parameters except for DADA2 (16S rRNA, denoise-paired, –p-trim-left-f 0 –p-trim-left-r 0 –p-trunc-len-f 250 –p-trunc-len-r 250; ITS, denoised-single, –p-trunc-len 150). For WGS, raw-read processing, assembly, and analysis were performed using the MGnify v5.0 pipeline (6) with default parameters for adapter trimming, quality filtering, and subsequent analysis. The results for each sample are shown in Table 1. We found that the majority of small subunit (SSU)- and large subunit (LSU)-containing contigs identified by MGnify belonged to bacteria (82%), eukarya (14%), and archaea (4%) (Fig. 1C). From 149,688 contigs, MGnify identified 208,556 coding sequences associated with 6,039 genome properties (7). Transposases and putative transposases (0.8% of KEGG Orthology [KO] orthologs) accounted for the majority of high-abundance KO orthologs in each sample (11.5% of all KO orthologs assigned) (Fig. 1D).
TABLE 1.
Summary of data
| Samplea | SRA or MGnify accession no. | Mean read length (bp) | Total no. of read pairs submitted | Total no. of read pairs | Total no. of contigs | N50 (bp) |
|---|---|---|---|---|---|---|
| WGS | ||||||
| F8W_T25 | ERR9752702 | 151 | 13,493,283 | |||
| F8W_T26 | ERR9752724 | 151 | 11,497,693 | |||
| F8W_T27 | ERR9752734 | 151 | 9,693,650 | |||
| F80_T28 | ERR9752742 | 151 | 9,510,055 | |||
| F80_T29 | ERR9752750 | 151 | 11,153,342 | |||
| F80_T30 | ERR9752757 | 151 | 10,188,435 | |||
| F8300_T31 | ERR9752762 | 151 | 13,358,533 | |||
| F8300_T32 | ERR9752768 | 151 | 12,361,583 | |||
| F8300_T33 | ERR9752775 | 151 | 11,262,599 | |||
| F8600_T34 | ERR9752780 | 151 | 11,369,775 | |||
| F8600_T35 | ERR9752794 | 151 | 14,442,272 | |||
| F8600_T36 | ERR9760450 | 151 | 16,556,960 | |||
| ITS | ||||||
| F80_1_0607 | ERR10168568 | 151 | 1,444,523 | |||
| F80_1_0619 | ERR10168570 | 151 | 1,444,589 | |||
| F80_1_0703 | ERR10168632 | 151 | 2,185,005 | |||
| F80_2_0607 | ERR10168636 | 151 | 2,328,428 | |||
| F80_2_0619 | ERR10168647 | 151 | 1,453,311 | |||
| F80_2_0703 | ERR10168649 | 151 | 1,011,053 | |||
| F80_3_0607 | ERR10168652 | 151 | 948,188 | |||
| F80_3_0619 | ERR10168654 | 151 | 1,571,348 | |||
| F80_3_0703 | ERR10168656 | 151 | 975,492 | |||
| F8300_1_0607 | ERR10168659 | 151 | 1,095,855 | |||
| F8300_1_0619 | ERR10168660 | 151 | 787,551 | |||
| F8300_1_0703 | ERR10168662 | 151 | 1,072,100 | |||
| F8300_2_0607 | ERR10168666 | 151 | 1,319,891 | |||
| F8300_2_0619 | ERR10168672 | 151 | 1,446,988 | |||
| F8300_2_0703 | ERR10168675 | 151 | 1,152,882 | |||
| F8300_3_0607 | ERR10168677 | 151 | 1,414,719 | |||
| F8300_3_0619 | ERR10168680 | 151 | 23 | |||
| F8300_3_0703 | ERR10168682 | 151 | 1,489,768 | |||
| F8600_1_0607 | ERR10168684 | 151 | 1,456,243 | |||
| F8600_1_0619 | ERR10168688 | 151 | 3,790,963 | |||
| F8600_1_0703 | ERR10168745 | 151 | 1,336,475 | |||
| F8600_2_0607 | ERR10168750 | 151 | 1,154,444 | |||
| F8600_2_0619 | ERR10168753 | 151 | 506,739 | |||
| F8600_2_0703 | ERR10168760 | 151 | 1,315,871 | |||
| F8600_3_0607 | ERR10168776 | 151 | 1,582,067 | |||
| F8600_3_0619 | ERR10168779 | 151 | 1,398,289 | |||
| F8600_3_0703 | ERR10639905 | 151 | 47 | |||
| F8W_1_0607 | ERR10213569 | 151 | 2,199,554 | |||
| F8W_1_0619 | ERR10213571 | 151 | 873,471 | |||
| F8W_1_0703 | ERR10213561 | 151 | 1,256,782 | |||
| F8W_2_0607 | ERR10213574 | 151 | 656,150 | |||
| F8W_2_0619 | ERR10213577 | 151 | 1,178,439 | |||
| F8W_2_0703 | ERR10213582 | 151 | 759,588 | |||
| F8W_3_0607 | ERR10213585 | 151 | 1,327,567 | |||
| F8W_3_0619 | ERR10213587 | 151 | 1,281,001 | |||
| F8W_3_0703 | ERR10213589 | 151 | 926,657 | |||
| 16S rRNA | ||||||
| F80_1_2019_06_07_16S | ERR10169571 | 250.7 | 30,535 | |||
| F80_1_2019_06_19_16S | ERR10169574 | 250.6 | 36,286 | |||
| F80_1_2019_07_03_16S | ERR10169578 | 250.6 | 21,911 | |||
| F80_2_2019_06_07_16S | ERR10169580 | 250.7 | 7,794 | |||
| F80_2_2019_06_19_16S | ERR10169582 | 250.6 | 24,368 | |||
| F80_2_2019_07_03_16S | ERR10169585 | 250.6 | 18,269 | |||
| F80_3_2019_06_07_16S | ERR10169587 | 250.6 | 30,048 | |||
| F80_3_2019_06_19_16S | ERR10169589 | 250.6 | 34,117 | |||
| F80_3_2019_07_03_16S | ERR10169592 | 250.6 | 20,410 | |||
| F8300_1_2019_06_07_16S | ERR10169803 | 250.6 | 33,208 | |||
| F8300_1_2019_06_19_16S | ERR10169804 | 250.6 | 44,023 | |||
| F8300_1_2019_07_03_16S | ERR10169815 | 250.6 | 23,870 | |||
| F8300_2_2019_06_07_16S | ERR10169821 | 250.6 | 28,959 | |||
| F8300_2_2019_06_19_16S | ERR10169825 | 250.5 | 36,641 | |||
| F8300_2_2019_07_03_16S | ERR10169841 | 250.7 | 17,673 | |||
| F8300_3_2019_06_07_16S | ERR10169845 | 250.6 | 27,000 | |||
| F8300_3_2019_06_19_16S | ERR10169847 | 250.6 | 38,461 | |||
| F8300_3_2019_07_03_16S | ERR10169849 | 250.6 | 30,309 | |||
| F8600_1_2019_06_07_16S | ERR10169850 | 250.6 | 28,737 | |||
| F8600_1_2019_06_19_16S | ERR10169852 | 250.6 | 21,212 | |||
| F8600_1_2019_07_03_16S | ERR10169854 | 250.6 | 42,005 | |||
| F8600_2_2019_06_07_16S | ERR10169855 | 250.7 | 29,635 | |||
| F8600_2_2019_06_19_16S | ERR10169856 | 250.6 | 34,151 | |||
| F8600_2_2019_07_03_16S | ERR10169859 | 250.6 | 40,674 | |||
| F8600_3_2019_06_07_16S | ERR10169861 | 250.6 | 50,432 | |||
| F8600_3_2019_06_19_16S | ERR10169885 | 250.6 | 37,360 | |||
| F8600_3_2019_07_03_16S | ERR10169864 | 250.7 | 33,704 | |||
| F8W_1_0607_16S | ERR10213607 | 250.6 | 46,515 | |||
| F8W_1_0619_16S | ERR10213609 | 250.6 | 48,041 | |||
| F8W_1_0703_16S | ERR10213610 | 250.6 | 16,155 | |||
| F8W_2_0607_16S | ERR10213612 | 250.4 | 288 | |||
| F8W_2_0619_16S | ERR10213613 | 250.6 | 44,701 | |||
| F8W_2_0703_16S | ERR10213614 | 250.7 | 15,809 | |||
| F8W_3_0607_16S | ERR10213616 | 250.6 | 34,981 | |||
| F8W_3_0619_16S | ERR10213618 | 250.6 | 54,720 | |||
| F8W_3_0703_16S | ERR10213620 | 250.6 | 39,692 | |||
| MGnify | ||||||
| F80_T28 | MGYA00607100 | 9,016,418 | 9,977 | 562 | ||
| F80_T29 | MGYA00607095 | 10,558,938 | 833 | 560 | ||
| F80_T30 | MGYA00607099 | 9,646,580 | 7,393 | 554 | ||
| F8300_T31 | MGYA00607093 | 12,653,552 | 17,047 | 566 | ||
| F8300_T32 | MGYA00606633 | 11,703,364 | 14,294 | 555 | ||
| F8300_T33 | MGYA00607091 | 10,659,098 | 10,304 | 555 | ||
| F8600_T34 | MGYA00607098 | 10,767,204 | 11,398 | 558 | ||
| F8600_T35 | MGYA00607096 | 13,663,903 | 13,799 | 557 | ||
| F8600_T36 | MGYA00607092 | 15,683,527 | 20,013 | 564 | ||
| F8W_T25 | MGYA00607094 | 12,781,962 | 22,814 | 572 | ||
| F8W_T26 | MGYA00607097 | 10,887,794 | 7,033 | 551 | ||
| F8W_T27 | MGYA00607090 | 9,183,848 | 7,283 | 556 |
Sequencing run statistics are available for WGS, ITS amplicon sequencing, and 16S rRNA V4 amplicon sequencing, including the sample name (for WGS, site_id; for ITS, site_replicate_date; for 16S, site_replicate_date_16S), read accession number, mean read length, and total reads or read pairs submitted. WGS raw reads were used to perform metagenomic analyses using MGnify, which are accessible using the MGnify accession numbers.
Data availability.
All data are available through EMBL/EBI BioProject accession number PRJEB52998 and the MGnify identification number MGYS00006044. Individual accession numbers for each sample are available on Table 1.
ACKNOWLEDGMENTS
This work was funded by the Applied Farmscape Ecology Research Collaborative of the Hudson Valley FarmHub (Hurley, NY, USA) and the Bard Summer Research Institute of Bard College.
We thank Conrad Vispo, Anne Bloomfield, Tejaswee Neupane, Hannah Herrick, Maram Al Zayyad, Benjamin Bryant, Christopher Benincasa, and Maureen Schulz-O’Callaghan for their assistance in the field and in the laboratory.
Contributor Information
Gabriel G. Perron, Email: gperron@bard.edu.
Leighton Pritchard, University of Strathclyde.
REFERENCES
- 1.Kim N, Zabaloy MC, Guan K, Villamil MB. 2020. Do cover crops benefit soil microbiome? A meta-analysis of current research. Soil Biol Biochem 142:107701. doi: 10.1016/j.soilbio.2019.107701. [DOI] [Google Scholar]
- 2.Singh BK, Trivedi P, Egidi E, Macdonald CA, Delgado-Baquerizo M. 2020. Crop microbiome and sustainable agriculture. Nat Rev Microbiol 18:601–602. doi: 10.1038/s41579-020-00446-y. [DOI] [PubMed] [Google Scholar]
- 3.Santos LF, Olivares FL. 2021. Plant microbiome structure and benefits for sustainable agriculture. Curr Plant Biol 26:100198. doi: 10.1016/j.cpb.2021.100198. [DOI] [Google Scholar]
- 4.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. P T 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 5.Bokulich NA, Mills DA. 2013. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl Environ Microbiol 79:2519–2526. doi: 10.1128/AEM.03870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell AL, Almeida A, Beracochea M, Boland M, Burgin J, Cochrane G, Crusoe MR, Kale V, Potter SC, Richardson LJ, Sakharova E, Scheremetjew M, Korobeynikov A, Shlemov A, Kunyavskaya O, Lapidus A, Finn RD. 2020. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res 48:D570–D578. doi: 10.1093/nar/gkz1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson LJ, Rawlings ND, Salazar GA, Almeida A, Haft DR, Ducq G, Sutton GG, Finn RD. 2019. Genome properties in 2019: a new companion database to InterPro for the inference of complete functional attributes. Nucleic Acids Res 47:D564–D572. doi: 10.1093/nar/gky1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available through EMBL/EBI BioProject accession number PRJEB52998 and the MGnify identification number MGYS00006044. Individual accession numbers for each sample are available on Table 1.


