Abstract
Background
We hypothesized that the high-dose opioid requirement in patients carrying the rs4680-GG variant in the COMT gene encoding catechol-O-methyltransferase would be greater for patients taking morphine than for those taking oxycodone, thus providing a much-needed biomarker to inform opioid selection for cancer pain.
Methods
A randomized, multicenter, open-label trial was conducted at a Japanese hospital’s palliative care service. Patients with cancer pain treated with regular doses of nonsteroidal anti-inflammatory drugs or acetaminophen were enrolled and randomized (1:1) into morphine (group M) and oxycodone (group O) groups. The minimum standard dose of immediate-release (IR) oral opioids was repeatedly administered by palliative care physicians to achieve pain-reduction goals (Pain reduction ≥ 33% from baseline and up to ≤ 3 on a numerical rating scale). The primary endpoint was the proportion of subjects requiring high-dose opioids on day 0 with the GG genotype.
Results
Of 140 participants who developed cancer-related pain among 378 subjects registered and pre-screened for the genotype, 139 were evaluated in the current study. Among patients carrying a COMT rs4680-GG genotype, 48.3% required high-dose opioids in group M, compared with the 20.0% in group O (95% CI, 3.7%-50.8%; P = .029). Of those with the non-GG genotype, 41.5% treated with morphine and 23.1% with oxycodone required high-dose opioids (95% CI, 3.3%-38.3%; P = 0.098).
Conclusion
Using the COMT rs4680 genotype alone is not recommended for selecting between morphine and oxycodone for pain relief.
Keywords: humans, opioid analgesics, cancer pain, morphine, biomarker, genotype
This report evaluates the potential for the COMT rs4680 genotype to serve as a biomarker for opioid choice.
Lessons Learned.
The proportion of patients requiring high-dose opioids was significantly higher in the morphine group when compared with the oxycodone group in those with the COMT-rs4680 GG genotype. However, a similar trend was observed in noncarriers as well suggesting the difference may not have been attributable to the genotype.
No significant interaction between the COMT rs4680 genotypes and dose of analgesia required to alleviate the pain was indicated in the current study. Although a significant difference was seen in dose between immediate-release morphine and oxycodone among the GG genotype, the same trend of more morphine requirement than oxycodone was seen in subjects with a non-GG type although not reaching statistical significance.
Although the recent evidence-based review has shown that if patients are COMT G/G genotype carriers, clinicians need to consider initiating morphine at a higher dose and/or more aggressive dose titration (recommendation grade B), using the COMT rs4680 genotype alone is not recommended for selecting between morphine and oxycodone for pain relief.
Discussion
We conducted a multicenter, randomized, open-label trial to evaluate the potential for the COMT rs4680 genotype to serve as a biomarker for opioid choice, and we have shown that the proportion of patients requiring high-dose opioids was significantly higher in the morphine group when compared with the oxycodone group in those with the COMT-rs4680 GG genotype; a similar trend was observed in noncarriers as well (Table 1, Fisher’s exact test).
Table 1.
Proportion of participants requiring high-dose morphine and oxycodone on day 0 (n = 139).
| Variable | High-dose | Low-dose | P-value |
|---|---|---|---|
| Morphine (GG) no. (%) | 14 (48.3) | 15 (51.7) | P = .029 |
| Oxycodone (GG) no. (%) | 6 (20.0) | 24 (80.0) | |
| Morphine (Non-GG) no. (%) | 17 (41.5) | 24 (58.5) | P = .098 |
| Oxycodone (Non-GG) no. (%) | 9 (23.1) | 30 (76.9) |
This study was based on the observation that interpatient variation in morphine requirement to relieve pain is not purely due to pain intensity. The COMT gene is a leading candidate for driving some of this interpatient variation, but there has been disagreement in the literature regarding how much impact the identified genetic variants could have on the function of this gene. A recent evidence-based review has shown that if patients carry the COMT G/G genotype, clinicians need to consider initiating morphine at a higher dose and/or using a more aggressive dose titration (recommendation grade B).1 In contrast, in a review in 2017 and in guidelines based on a systematic review in 2021, there is limited evidence for an association between COMT rs4680 genotype and analgesia or opioid dosage.2,3 However, COMT is the only gene under consideration for a genetic difference in opioid requirement. The rs4680 (Val158Met) polymorphism is the most studied SNP in the COMT gene because the valine (Val) to methionine (Met) substitution leads to a three-to fourfold reduced activity of the COMT enzyme, hence the Val/Val, Val/Met and Met/Met genotypes predict a high, intermediate, and low COMT enzyme activity, respectively.4
As the COMT enzyme metabolizes catecholamines, a high COMT enzyme activity could result in reduced activation of dopaminergic neurotransmission. It is shown in animal models that the neuronal content of enkephalin peptides is enhanced by the chronic deactivation of dopaminergic neurotransmission. Pain sensitivity is affected by the neuronal content of enkephalin, and enhancement in the enkephalin content is shown to be followed by a downregulation of mu-opioid receptors.4 Taken together, this can explain the influence of variation in the COMT gene on the effect of opioids in pain treatment. As several factors other than genetics, such as etiology and environmental factors, could also affect the opioid response, the impact of genetic differences may vary with respect to their relationship with these factors. Although our current understanding of genetic factors is inadequate to provide individualized pain treatment, further research, such as testing for the interaction of genotype (GG versus non-GG) by treatment (morphine versus oxycodone) is needed to validate the study findings on individualized pain treatment.
| Trial Information | |
|---|---|
| Disease | Cancer pain |
| Stage of disease/treatment | Stage IV |
| Prior therapy | NSAIDs/acetaminophen |
| Type of study | Randomized controlled trial |
| Primary endpoint | The proportion of subjects (morphine compared with oxycodone) requiring high-dose opioids on day 0 with the GG genotype (Table 1). |
| Secondary endpoints | 1. The proportion of subjects (morphine compared with oxycodone) requiring high-dose opioids on day 0 with the Non-GG genotype (Table 1). 2. Hospital anxiety and depression scale (Table 8) 3. Quality of life score (Table 9) 4. The pain catastrophizing scale (Table 7) 4. Adverse events (Tables 4 and 10). |
| Additional details of endpoints or study design | Table 2 details inclusion and exclusion criteria. |
| Investigator’s analysis | Correlative endpoints met but not powered to assess activity |
| Additional analysis | Factors influencing pain numerical rating scale on day 8 (Table 5) Factors influencing high-dose opioid cases for titration response (Table 6) |
| Drug Information: Intervention Arms | Arm 1 | Arm 2 |
|---|---|---|
| Generic/working name | OXINORM | OPSO |
| Company name | Shionogi Pharma | Sumitomo Pharma |
| Drug type | Powder | Liquid |
| Drug class | Class A | Class A |
| Dose | 2.5 | 5 |
| Unit | Mg | mg |
| Route | Oral | Oral |
| Schedule of administration | The minimum standard dose of immediate-release 2.5 mg/dose was repeatedly administered to achieve pain-reduction goals. |
The minimum standard dose of immediate-release 5 mg/dose was repeatedly administered to achieve pain-reduction goals. |
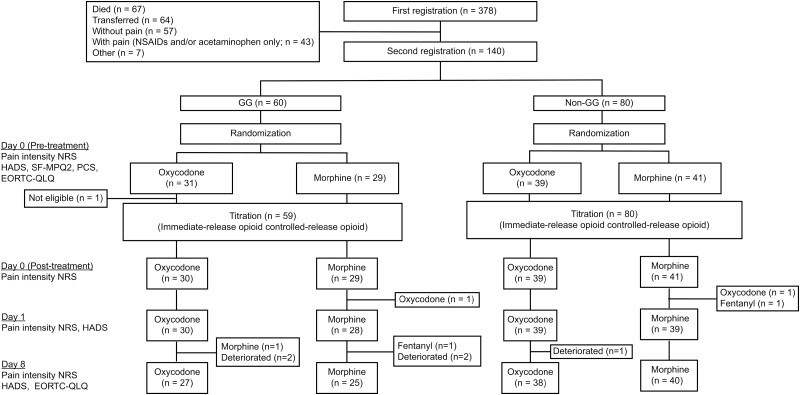
Figure 1.
CONSORT diagram of the study. After genetic classification into the rs4680-GG and non-GG genotype groups, the participants were randomized (1:1 allocation ratio) into the morphine (M) or oxycodone (O) treatment group. Evaluations were performed at baseline (pre-titration) and on days 0 (post-titration), 1, and 8.
| Primary Assessment Method | |
|---|---|
| Title | The proportion of subjects requiring high-dose opioids on day 0 with the GG genotype. |
| Number of patients screened | 378 |
| Number of patients enrolled | 140 |
| Number of patients evaluable for toxicity | 139 |
| Number of patients evaluated for efficacy | 139 |
| Evaluation method | Other: The definition of high-dose opioids was ≥ 10.0 mg morphine or ≥ 7.5 mg oxycodone. |
| Secondary assessment method | |
| Title | The proportion of subjects requiring high-dose opioids on day 0 with the non-GG genotype. |
| Number of patients screened | 378 |
| Number of patients enrolled | 140 |
| Number of patients evaluable for toxicity | 139 |
| Number of patients evaluated for efficacy | 139 |
| Evaluation method | The definition of high-dose opioids was ≥10.0 mg morphine or ≥7.5 mg oxycodone. |
Assessment, Analysis, and Discussion
| Completion | Study completed |
|---|---|
| Investigator’s assessment | Correlative endpoints met but not powered to assess activity |
Opioids are commonly used for cancer pain relief; however, the dose required for quick and potent pain relief remains debatable5 because the opioid dose and associated adverse effects (AEs) vary among patients.6 Specific biomarkers have been sought to monitor therapeutic efficacy and AEs.7-12 Currently, there are no biomarkers for selecting morphine or oxycodone for cancer pain relief.
We previously demonstrated that among various biomarkers including drug metabolites and plasma concentrations, circulating markers, and genetic markers, patients with the single nucleotide polymorphism (SNP) rs4680-GG in catechol-O-methyltransferase (COMT) require a significantly higher dose of morphine than non-GG patients. Further, the plasma morphine concentration 1 day after morphine administration is significantly higher in rs4680-GG patients than in non-GG patients.12,13
Based on these previous findings, we hypothesized that the high-dose opioid requirement rates in patients with the COMT rs4680-GG genotype are higher with morphine than with oxycodone. Thus, we conducted a multicenter, randomized, open-label trial to evaluate the potential of rs4680 genotype as a biomarker,14 and we have shown the proportion of patients requiring high-dose opioids was significantly higher in the morphine group when compared with the oxycodone group in those with the COMT-rs4680 GG genotype. However, a similar trend was observed in noncarriers as well. Results in the patients with COMT-rs4680 GG genotype thus have to be interpreted with caution because: (1) oxycodone was better than morphine for pain relief in both genotype groups, although the difference was not significant in the non-GG group, indicating that individualizing morphine and oxycodone based on the COMT genotype is not a realistic strategy for rapid titration on day 0; and (2) morphine was associated with a higher numerical rating scale on day 8, suggesting that oxycodone is more efficacious than morphine regardless of the genotype.
This trial has several strengths. First, we employed a prospective, multicenter, randomized controlled design using genetic biomarkers. Moreover, changes in concurrent adjuvant analgesics were not allowed during the study period, thus reducing the risk of evaluation bias, which is a bottleneck for evaluating subjective endpoints, such as pain in palliative medicine. Second, selection bias was minimized using a properly stratified population. Third, the dropout rate was extremely low (just 1/140 participants for the primary endpoint). In clinical trials in palliative medicine, a considerably high dropout rate often reduces statistical power and sometimes leads to trial discontinuation. Based on our study eligibility criteria, we selected participants for whom the protocol was feasible, and the treatment was safely and successfully completed. Further, to our knowledge, this is the first randomized controlled trial to explore a potential biomarker for selecting opioids for cancer pain. We made many relevant clinical observations and investigated morphine versus oxycodone as a first-line opioid for cancer pain.
Our study has some limitations. First, we used an unblinded procedure to administer opioids. The physicians and patients knew which opioid was administered, but they were blinded to genotype information. Second, we might have included heterogeneous cases of cancer pain (eg, mixed patients with nociceptive and neuropathic pain), and the results might vary depending on the presence or absence of neuropathic pain. Here, oxycodone might be more effective than morphine, as there were more elements of intermittent and neuropathic pain in group O.15 Third, our definition of high-dose opioid cases is arbitrary in both groups; therefore, the results could be affected by the cut-off values. Fourth, the non-GG group had higher functional status values, which could have biased the results toward no change. Fifth, opioid switching was permitted and performed for four patients in the short 8-day interval of the study but will not affect the primary endpoint on day 0. Finally, only one polymorphism was examined; therefore, the study findings remain inconclusive.
According to a recent review in 20176 and guidelines based on a systematic review in 2021,16 there is limited evidence for an association between COMT rs4680 genotype and analgesia or opioid dosage, however, another recent evidence-based review has shown that if patients are found to have a COMT GG genotype, clinicians need to consider initiating morphine at a higher dose and/or providing for more aggressive dose titration (recommendation grade B).17 As several factors such as etiology, genetics, environmental factors, determine the opioid response, the impact of genetic differences may vary with respect to their relationship with these factors. Although our current understanding of genetic factors is inadequate to provide individualized pain treatment, further research, such as testing for the interaction of genotype (GG versus non-GG) by treatment (morphine versus oxycodone) is needed to validate the study findings on individualized pain treatment.
Table 2.
Inclusion and exclusion criteria at the second registration into the study based on genotype
| Inclusion criteria |
|---|
| • Cancer pain targeted for daily treatment with opioids, although stable regular oral nonsteroidal anti-inflammatory drugs (NSAIDs) or acetaminophen were also administered daily (both inpatients and outpatients) |
| • A numerical rating scale (NRS) of ≥324-26 (average over 24 h) |
| • Opioid treatment-naive within 30 h |
| • No chemotherapy, radiotherapy, or bisphosphonate treatment in the past 2 weeks |
| • No change in any adjuvant analgesic (if applicable) within 72 h before commencement of the study |
| • Provided written informed consent |
| Exclusion criteria |
| • Patients with chronic renal failure (glomerular filtration rate, 30 mL/min) |
| • Patients with severe hepatic (AST (aspartate aminotransferase) > 90 U/L, ALT (alanine aminotransferase) > 126 U/L (male), ALT > 69 U/L (female), total bilirubin > 2.25 mg/dL) |
| • Patients with respiratory failure (respiratory system dysfunction with arterial blood partial pressure of oxygen < 60 Torr during ambient air inhalation) |
| • Patients with planned surgical interventions or recent surgical interventions |
| • Patients deemed ineligible for the study by the study coordinator or a collaborative investigator (eg, neuropathic pain and predominant spontaneous pain only) |
Table 3.
Baseline characteristics.
| Variable | rs4680-GG group (n = 59) | rs4680-Non-GG group (n = 80) | ||||
|---|---|---|---|---|---|---|
| Opioid group | Group M (n = 29) |
Group O (n = 30) |
P value | Group M (n = 41) |
Group O (n = 39) |
P value |
| Age (years), mean (SD) | 66.8 (13.07) | 67.5 (9.23) | .81 | 69.2 (1.2) | 70.5 (1.4) | .51 |
| Sex, no. (%) | .42 | 1.00 | ||||
| Male | 20 (69.0) | 17 (56.7) | 22 (53.7) | 21 (53.8) | ||
| Female | 9 (31.0) | 13 (43.3) | 19 (46.3) | 18 (46.2) | ||
| Performance status, no. (%) | .83 | .25 | ||||
| 0 | 2 (6.9) | 2 (6.7) | 1 (2.4) | 5 (12.8) | ||
| 1 | 15 (51.7) | 16 (53.3) | 26 (63.4) | 22 (56.4) | ||
| 2 | 11 (37.9) | 9 (30.0) | 7 (17.1) | 9 (23.1) | ||
| 3 | 1 (3.4) | 1 (3.3) | 5 (12.2) | 3 (7.6) | ||
| 4 | 0 (0.0) | 2 (6.7) | 2 (4.9) | 0 (0.0) | ||
| Cancer type, no. (%) | .94 | .88 | ||||
| Lung | 12 (41.4) | 7 (23.3) | 15 (36.6) | 12 (30.8) | ||
| Breast | 2 (6.9) | 6 (20.0) | 4 (9.8) | 9 (23.1) | ||
| Colon | 3 (10.3) | 3 (10.0) | 2 (4.9) | 3 (7.7) | ||
| Head and neck | 3 (10.3) | 3 (10.0) | 5 (12.2) | 1 (2.6) | ||
| Esophageal | 2 (6.9) | 0 (0.0) | 5 (12.2) | 3 (7.7) | ||
| Gastric | 2 (6.9) | 3 (10.0) | 1 (2.4) | 1 (2.6) | ||
| Primary unknown | 2 (6.9) | 1 (3.3) | 1 (2.4) | 4 (10.3) | ||
| Pancreas | 0 (0.0) | 2 (6.7) | 3 (7.3) | 1 (2.6) | ||
| Mesothelioma | 0 (0.0) | 2 (6.7) | 1 (2.4) | 2 (5.1) | ||
| Gallbladder/bile duct | 0 (0.0) | 1 (3.3) | 1 (2.4) | 2 (5.1) | ||
| Others | 3 (10.3) | 2 (6.7) | 3 (7.3) | 1 (2.6) | ||
| Most painful areas, No. (%) | .70 | .58 | ||||
| Frontal pain (chest and abdomen) | 12 (41.4) | 11 (36.7) | 15 (36.6) | 12 (30.8) | ||
| Back pain (upper and lower) | 11 (37.9) | 11 (36.7) | 14 (34.1) | 10 (25.6) | ||
| Head and neck | 2 (6.9) | 4 (13.3) | 4 (9.8) | 4 (10.3) | ||
| Extremities | 2 (6.9) | 4 (13.3) | 6 (14.6) | 7 (17.9) | ||
| Others | 2 (6.9) | 0 (0.0) | 2 (4.9) | 6 (15.4) | ||
| BPI item 5, mean (SD) | 5.34 (2.09) | 5.47 (1.96) | .82 | 5.27 (2.10) | 5.46 (2.05) | .67 |
| BPI item 5 ≥ 8, No. (%) | 5 (17.2) | 6 (20.0) | 1.00 | 6 (14.6) | 8 (20.5) | .56 |
| SF-MPQ-2 | ||||||
| Continuous pain | 17.14 (10.59) | 21.57 (13.02) | .16 | 16.98 (9.54) | 20.74 (12.89) | .14 |
| Intermittent pain | 8.17 (12.28) | 18.47 (16.48) | .01 | 10.83 (10.58) | 13.87 (14.80) | .29 |
| Neuropathic pain | 5.66 (7.58) | 12.03 (10.93) | .01 | 7.24 (7.19) | 8.69 (9.36) | .44 |
| Affective descriptors | 7.86 (9.36) | 10.17 (8.98) | .34 | 7.61 (8.27) | 10.05 (10.37) | .25 |
| Total score | 38.83 (34.01) | 62.23 (42.05) | .02 | 42.66 (26.27) | 53.36 (38.42) | .15 |
| HADS ≥ 11, No. (%) | 22 (75.9) | 18 (60.0) | .27 | 33 (80.5) | 29 (74.4) | .60 |
| HADS ≥ 20, No. (%) | 9 (31.0) | 7 (23.3) | .57 | 12 (29.3) | 11 (28.2) | 1.00 |
| EORTC-QLQ-C15-PAL | ||||||
| Overall quality of life | 30.09 (22.16) | 38.77 (26.37) | .18 | 41.05 (22.08) | 35.90 (24.94) | .33 |
| Physical functioning | 58.84 (28.16) | 57.99 (30.67) | .91 | 62.10 (29.18) | 55.38 (26.57) | .29 |
| Emotional functioning | 59.78 (29.88) | 61.79 (31.53) | .80 | 61.12 (33.51) | 57.74 (23.96) | .61 |
| Fatigue | 58.99 (25.06) | 62.83 (24.38) | .56 | 64.77 (27.56) | 64.38 (27.25) | .95 |
| Nausea/vomiting | 8.06 (13.82) | 10.56 (25.33) | .64 | 10.17 (18.96) | 12.40 (24.10) | .65 |
| Pain | 70.69 (25.85) | 76.43 (24.20) | .39 | 73.57 (24.72) | 77.77 (24.58) | .46 |
| Dyspnea | 35.62 (32.04) | 28.88 (25.87) | .38 | 24.38 (26.90) | 33.33 (39.52) | .25 |
| Insomnia | 55.17 (37.04) | 58.89 (38.84) | .71 | 52.84 (36.50) | 52.99 (33.10) | .99 |
| Appetite loss | 36.77 (28.66) | 51.11 (41.74) | .13 | 52.85 (35.74) | 52.13 (38.85) | .93 |
| Constipation | 34.48 (32.72) | 35.63 (39.78) | .91 | 33.33 (30.74) | 29.90 (29.42) | .61 |
| Pain catastrophizing scale (SD) | 34.2 (9.5) | 33.0 (11.7) | .665 | 31.9 (11.3) | 32.1 (13.0) | .921 |
Abbreviations: BPI, Brief Pain Inventory; HADS, Hospital Anxiety and Depression Scale; SD, standard deviation; group M, morphine-treated group; group O, oxycodone-treated group; SF-MPQ-2, Short-Form McGill Pain Questionnaire 2; EORTC, European Organisation for Research and Treatment of Cancer.
Table 4.
Number of participants reporting adverse events by rs4680 genotype group
| Variable | GG | Non-GG | ||||
|---|---|---|---|---|---|---|
| Adverse event | Group M (n = 29) no. (%) |
Group O (n = 30) no. (%) |
P value | Group M (n = 41) no. (%) |
Group O (n = 39) no. (%) |
P value |
| Somnolence | 12 (41.4) | 11 (36.7) | .79 | 18 (43.9) | 13 (33.3) | .49 |
| Nausea | 8 (27.6) | 8 (26.7) | 1.00 | 12 (29.3) | 10 (25.6) | .81 |
| Vomiting | 5 (17.2) | 6 (20.0) | 1.00 | 8 (19.5) | 5 (12.8) | .55 |
| Constipation | 13 (44.8) | 10 (33.3) | .43 | 20 (48.8) | 19 (48.7) | 1.00 |
| Delirium | 1 (3.4) | 0 (0.0) | .49 | 2 (4.9) | 3 (7.7) | .67 |
| Others | 3 (10.3) | 0 (0.0) | .11 | 1 (2.4) | 0 (0.0) | 1.00 |
Abbreviations: group M, morphine-treated group; group O, oxycodone-treated group.
Table 5.
Factors influencing pain numerical rating scale (NRS) on day 8 (multiple linear regression).
| Variables | Simple regression model | Multiple regression model | |||
|---|---|---|---|---|---|
| Partial regression coefficient (95% CI) | P value | Partial regression coefficient (95% CI) | P value | VIF | |
| Age | 0.006 (−0.028 to 0.040) | .715 | |||
| Sex | 0.586 (−0.059 to 1.230) | .075 | 0.510 (−0.101 to 1.120) | .101 | 1.005 |
| Performance status | 0.045 (−0.337 to 0.426) | .817 | |||
| Opioid regimen | 0.565 (−0.076 to 1.206) | .083 | 0.671 (0.038 to 1.304) | .038 | 1.095 |
| OMME | 0.023 (−0.002 to 0.048) | .073 | −0.002 (−0.028 to 0.025) | .902 | 1.258 |
| Neuropathicpain | −0.015 (−0.051 to 0.022) | .423 | |||
| Genotype | 0.452 (−0.207 to 1.110) | .177 | |||
| PretreatmentNRS | 0.290 (0.135 to 0.444) | <.001 | 0.304 (0.138 to 0.471) | <.001 | 1.207 |
| Pretreatment HADS | −0.001 (−0.045 to 0.044) | .969 | |||
Abbreviations: OMME, oral morphine milligram equivalent; NRS, numerical rating scale; HADS, Hospital Anxiety and Depression Scale; VIF, variance inflation factor.
Table 6.
Factors influencing high-dose opioid cases for titration response (multiple logistic regression)
| Variable | Univariate model | Multivariate model | ||
|---|---|---|---|---|
| Odds Ratio (95% CI) | P value | Odds Ratio (95% CI) | P value | |
| Age | 1.056 (1.012–1.102) | .012 | 1.053 (1.003–1.105) | .036 |
| Sex | 1.856 (0.887–3.885) | .101 | ||
| Performance status | 1.038 (0.682–1.578) | .863 | ||
| Opioid regimen | 2.862 (−0.076 to 1.206) | .005 | 3.818 (1.664–8.763) | .002 |
| Neuropathic pain | 1.015 (0.976–1.054) | .461 | ||
| rs4680 genotype | 0.939 (0.460–1.917) | .863 | ||
| Pretreatment NRS | 1.423 (1.179–1.717) | <.001 | 1.445 (1.175–1.777) | <.001 |
| Pretreatment HADS | 1.018 (0.970–1.069) | .471 | ||
Abbreviations: HADS, Hospital Anxiety and Depression Scale; NRS, numerical rating scale.
Table 7.
Pain catastrophizing scale (PCS) scores on day 0 between the treatment groups in each rs4680 genotype group.
| Variable | rs4680-GG | rs4680-Non-GG | ||||
|---|---|---|---|---|---|---|
| Group M mean (SD) |
Group O mean (SD) |
P value | Group M mean (SD) |
Group O mean (SD) |
P value | |
| Overall QOL | 47.33 (20.23) | 40.74 (26.68) | .323 | 47.81 (23.31) | 46.29 (20.36) | .767 |
| Physical functioning | 53.07 (27.93) | 50.24 (34.45) | .742 | 61.04 (27.43) | 56.48 (21.61) | .424 |
| Emotional functioning | 60.68 (24.23) | 65.44 (26.01) | .498 | 71.50 (23.69) | 67.34 (22.61) | .436 |
| Fatigue | 55.10 (21.41) | 57.60 (28.27) | .720 | 55.84 (29.30) | 48.53 (22.60) | .228 |
| Nausea/vomiting | 19.22 (25.25) | 21.41 (24.36) | .746 | 19.25 (27.18) | 9.221 (18.47) | .063 |
| Pain | 55.34 (29.93) | 56.41 (27.11) | .894 | 53.94 (27.24) | 51.30 (24.32) | .802 |
| Dyspnea | 30.76 (37.62) | 22.60 (24.09) | .344 | 24.78 (29.34) | 25.43 (27.33) | .921 |
| Insomnia | 28.20 (30.83) | 30.86 (33.24) | .764 | 40.16 (29.80) | 33.32 (26.85) | .294 |
| Appetite loss | 39.74 (32.70) | 44.04 (34.02) | .638 | 51.28 (35.75) | 43.85 (32.98) | .347 |
| Constipation | 46.66 (33.34) | 54.32 (39.39) | .454 | 39.32 (36.58) | 35.08 (29.97) | .580 |
Abbreviations: QOL, quality of life; group M, morphine-treated group; group O, oxycodone-treated group; SD, standard deviation.
Table 8.
Hospital anxiety and depression scale (HADS) score on day 8 between the treatment groups for each rs4680 genotype.
| Variable | rs4680-GG | ||
|---|---|---|---|
| HADS | Group M (n = 29); mean (SD) | Group O (n = 30); mean (SD) | P value |
| HADS-Anxiety | 6.38 (4.20) | 7.19 (4.39) | .50 |
| HADS-Depression | 9.19 (3.86) | 8.41 (5.71) | .56 |
| HADS-Total | 15.58 (7.69) | 15.59 (9.40) | 1.00 |
| rs4680-Non-GG | |||
| Group M (n = 41); mean (SD) | Group O (n = 39); mean (SD) | P value | |
| HADS-Anxiety | 6.38 (4.24) | 6.21 (3.54) | .85 |
| HADS-Depression | 7.67 (4.30) | 8.37 (4.13) | .47 |
| HADS-Total | 14.05 (7.95) | 14.58 (6.80) | .77 |
Abbreviations: group M, morphine-treated group; group O, oxycodone-treated group, SD, standard deviation; HADS, Hospital Anxiety and Depression Scale.
Table 9.
European Organization for Research and Treatment of Cancer (EORTC)-QLQ-C15-PAL scores on day 8 between the treatment groups for each rs4680 genotype.
| Variables | GG | Non-GG | ||||
|---|---|---|---|---|---|---|
| Group M mean (SD) |
Group O mean (SD) |
P value | Group M mean (SD) |
Group O mean (SD) |
P value | |
| Overall QOL | 47.33 (20.23) | 40.74 (26.68) | .323 | 47.81 (23.31) | 46.29 (20.36) | .767 |
| Physical functioning | 53.07 (27.93) | 50.24 (34.45) | .742 | 61.04 (27.43) | 56.48 (21.61) | .424 |
| Emotional functioning | 60.68 (24.23) | 65.44 (26.01) | .498 | 71.50 (23.69) | 67.34 (22.61) | .436 |
| Fatigue | 55.10 (21.41) | 57.60 (28.27) | .720 | 55.84 (29.30) | 48.53 (22.60) | .228 |
| Nausea/vomiting | 19.22 (25.25) | 21.41 (24.36) | .746 | 19.25 (27.18) | 9.221 (18.47) | .063 |
| Pain | 55.34 (29.93) | 56.41 (27.11) | .894 | 53.16 (20.70) | 51.33 (23.40) | .366 |
| Dyspnea | 30.76 (37.62) | 22.60 (24.09) | .344 | 24.78 (29.34) | 25.43 (27.33) | .921 |
| Insomnia | 28.20 (30.83) | 30.86 (33.24) | .764 | 40.16 (29.80) | 33.32 (26.85) | .294 |
| Appetite loss | 39.74 (32.70) | 44.04 (34.02) | .638 | 51.28 (35.75) | 43.85 (32.98) | .347 |
| Constipation | 46.66 (33.34) | 54.32 (39.39) | .454 | 39.32 (36.58) | 35.08 (29.97) | .580 |
Abbreviations: QOL, quality of life; group M, morphine-treated group; group O, oxycodone-treated group; SD, standard deviation.
Table 10.
Number of participants reporting adverse events in each rs4680 genotype group.
| Item | GG | |||||||
|---|---|---|---|---|---|---|---|---|
| Opioids | Group M (n = 29) | Group O (n = 30) | ||||||
| Adverse event | Grade 1; n (%) | Grade 2; n (%) | Grade 3; n (%) | Grade 4; n (%) | Grade 1; n (%) | Grade 2; n (%) | Grade 3; n (%) | Grade 4; n (%) |
| Somnolence | 7 (24.1) | 5 (17.2) | 0 (0.0) | 0 (0.0) | 11 (36.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Nausea | 6 (20.7) | 2 (6.9) | 0 (0.0) | 0 (0.0) | 5 (16.7) | 3 (10.0) | 0 (0.0) | 0 (0.0) |
| Vomiting | 3 (10.3) | 2 (6.9) | 0 (0.0) | 0 (0.0) | 5 (16.7) | 1 (3.3) | 0 (0.0) | 0 (0.0) |
| Constipation | 7 (24.1) | 6 (20.7) | 0 (0.0) | 0 (0.0) | 5 (16.7) | 5 (16.7) | 0 (0.0) | 0 (0.0) |
| Delirium | 1 (3.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Others | 3 (10.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Item | Non-GG | |||||||
| Opioids | Group M (n = 41) | Group O (n = 39) | ||||||
| Adverse event | Grade 1; n (%) | Grade 2; n (%) | Grade 3; n (%) | Grade 4; n (%) | Grade 1; n (%) | Grade 2; n (%) | Grade 3; n (%) | Grade 4; n (%) |
| Somnolence | 17 (41.5) | 1 (2.4) | 0 (0.0) | 0 (0.0) | 12 (30.8) | 1 (2.6) | 0 (0.0) | 0 (0.0) |
| Nausea | 8 (19.5) | 4 (9.8) | 0 (0.0) | 0 (0.0) | 9 (23.1) | 1 (2.6) | 0 (0.0) | 0 (0.0) |
| Vomiting | 5 (12.2) | 3 (7.3) | 0 (0.0) | 0 (0.0) | 5 (12.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Constipation | 9 (22.0) | 11 (26.8) | 0 (0.0) | 0 (0.0) | 13 (33.3) | 6 (15.4) | 0 (0.0) | 0 (0.0) |
| Delirium | 2 (4.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (5.1) | 1 (2.6) | 0 (0.0) | 0 (0.0) |
| Others | 1 (2.4) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
Abbreviations: group M, morphine-treated group; group O, oxycodone-treated group.
Acknowledgments
This trial was registered at the UMIN Clinical Trials Registry under registration no. UMIN000015579. This study was supported by 2014 Health Labor Sciences Research Grant (H26-Innovative Cancer-General-056) and 2015–2017 Japan Agency for Medical Research and Development (Innovative Clinical Cancer Research: 16ck0106060h0003).
Contributor Information
Hiromichi Matsuoka, Department of Psychosomatic Medicine, Kindai University Faculty of Medicine, Osaka, Japan; Palliative Care Center, Kindai Hospital, Osaka, Japan; Palliative Care Team, National Cancer Center, Tokyo, Japan.
Junji Tsurutani, Advanced Cancer Translational Research Institute, Showa University, Tokyo, Japan; Department of Medical Oncology, Kindai University Faculty of Medicine, Osaka, Japan.
Yasutaka Chiba, Department of Biostatics, Kindai University Faculty of Medicine, Osaka, Japan.
Yoshihiko Fujita, Department of Genome Biology, Kindai University Faculty of Medicine, Osaka, Japan.
Kiyohiro Sakai, Department of Psychosomatic Medicine, Kindai University Faculty of Medicine, Osaka, Japan; Palliative Care Center, Kindai Hospital, Osaka, Japan.
Takeshi Yoshida, Palliative Care Center, Kindai Hospital, Osaka, Japan; Department of Medical Oncology, Kindai University Faculty of Medicine, Osaka, Japan.
Miki Nakura, Department of Psychosomatic Medicine, Kindai University Faculty of Medicine, Osaka, Japan.
Ryo Sakamoto, Department of Psychosomatic Medicine, Kindai University Faculty of Medicine, Osaka, Japan.
Chihiro Makimura, Department of Psychosomatic Medicine, Kindai University Faculty of Medicine, Osaka, Japan.
Yoichi Ohtake, Department of Psychosomatic Medicine, Kindai University Faculty of Medicine, Osaka, Japan; Department of Internal Medicine, Sakai City Medical Center, Osaka, Japan.
Kaoru Tanaka, Department of Medical Oncology, Kindai University Faculty of Medicine, Osaka, Japan.
Hidetoshi Hayashi, Department of Medical Oncology, Kindai University Faculty of Medicine, Osaka, Japan.
Masayuki Takeda, Department of Medical Oncology, Kindai University Faculty of Medicine, Osaka, Japan.
Tatsuya Okuno, Department of Medical Oncology, Kindai University Faculty of Medicine, Osaka, Japan.
Naoki Takegawa, Department of Medical Oncology, Kindai University Faculty of Medicine, Osaka, Japan.
Koji Haratani, Department of Medical Oncology, Kindai University Faculty of Medicine, Osaka, Japan.
Atsuko Koyama, Department of Psychosomatic Medicine, Kindai University Faculty of Medicine, Osaka, Japan; Palliative Care Center, Kindai Hospital, Osaka, Japan.
Kazuto Nishio, Department of Genome Biology, Kindai University Faculty of Medicine, Osaka, Japan.
Kazuhiko Nakagawa, Department of Medical Oncology, Kindai University Faculty of Medicine, Osaka, Japan.
Conflict of Interest
Hiromichi Matsuoka reported personal fees from Daiichi Sankyo, Eisai, and Mundi, outside the submitted work. Junji Tsurutani reported personal fees and other from Daiichi Sankyo, Chugai,Nihonkayaku, Eli Lilly, Pfizer, grants, personal fees, and other from Eisai, and personal fees from Taiho, Novartis, and AstraZeneca, outside the submitted work. Yasutaka Chiba reported personal fees from Chugai Pharmaceutical Co., Ltd., outside the submitted work. Kaoru Tanaka reported personal fees from AstraZeneca, Merck Biopharma, Eisai, Bristol Myers Squibb, Ono Pharmaceutical, MSD, and Kyowa Kirin, outside the submitted work. Hidetoshi Hayashi reported personal fees from AstraZeneca K.K., Boehringer Ingelheim Japan Inc., Bristol Myers Squibb Co., Ltd., Chugai Pharmaceutical Co., Ltd., Eli Lilly Japan K.K., Kyorin Pharmaceutical Co., Ltd., Merck Biopharma Co., Ltd., MSD K.K., Novartis Pharmaceuticals K.K., Ono Pharmaceutical Co., Ltd., Pfizer Japan Inc., Shanghai Haihe Biopharm, Taiho Pharmaceutical Co., Ltd., and Takeda Pharmaceutical Co., Ltd., and grants from AstraZeneca K.K., Boehringer Ingelheim Japan Inc., Chugai Pharmaceutical Co., Ltd., and Ono Pharmaceutical Co., Ltd., outside the submitted work. Masayuki Takeda reported grants, personal fees, and honoraria from Ono Pharmaceutical Co., Boehringer Ingelheim Japan Inc., and Novartis Pharma K.K., outside the submitted work. Koji Haratani reported grants and personal fees from AS ONE Corporation and MSD K.K., and personal fees from AstraZeneca K.K., Bristol Myers Squibb Co., Ltd., Chugai Pharmaceutical Co., Ltd., and Ono Pharmaceutical Co., Ltd., outside the submitted work. Kazuto Nishio reported grants and personal fees from Eli Lilly Japan K.K., during the conduct of the study, grants and personal fees from Otsuka Pharmaceutical Co., Ltd., Nippon Boehringer Ingelheim Co., Ltd., Eli Lilly Japan K.K., personal fees from Life Technologies Japan Ltd., Eisai Co., Ltd., Pfizer Inc., Novartis Pharma K.K., MSD K.K., Ono Pharmaceutical Co., Ltd., Bristol Myers Squibb Co., SymBio Pharmaceuticals Ltd., Solasia Pharma K.K., Yakult Honsha Co., Ltd., Roche Diagnostics K.K., AstraZeneca K.K., Sanofi K.K., Guardant Health Inc, Chugai Pharmaceutical Co., Ltd., and grants from Ignyta Inc., Astellas Pharma Inc., Thoracic Oncology Research Group, and North East Japan Study Group, outside the submitted work. Kazuhiko Nakagawa reported personal fees from Clinical Trial Co., Ltd., Medicus Shuppan Publishers Co., Ltd., Care Net, Inc., Reno Medical K.K., Medical Review Co., Ltd., Roche Diagnostics K.K., Bayer Yakuhin, Ltd, Medical Mobile Communications Co., Ltd., 3H Clinical Trial Inc., Nichi-Iko Pharmaceutical Co., Ltd., Nanzando Co., Ltd., Yodosha Co., Ltd., Nikkei Business Publications, Inc., Thermo Fisher Scientific K.K., Yomiuri Telecasting Co., Nippon Kayaku Co., Ltd., personal fees and other from Kyorin Pharmaceutical Co., Ltd., grants, personal fees, and other from Takeda Pharmaceutical Co., Ltd. And Ono Pharmaceutical Co., grants and personal fees from Taiho Pharmaceutical Co., Ltd., SymBio Pharmaceuticals Ltd., AbbVie Inc., AstraZeneca K.K., Astellas Pharma Inc., MSD K.K., grants from inVentiv Health Japan, ICON Japan K.K., Gritstone Oncology Inc., Parexel International Co., Kissei Pharmaceutical Co., Ltd., EPS Co., Syneos Health, Pfizer R&D Japan G.K., A2 Healthcare Co., Quintiles Inc./IQVIA Services JAPAN K.K., EP-CRSU Co., Ltd., Linical Co., Ltd., Eisai Co., Ltd., CMIC Shift Zero K.K., Kyowa Hakko Kirin Co., Ltd., Bayer Yakuhin, Ltd., EPS International Co., Ltd., Otsuka Pharmaceutical Co., Ltd.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Vieira CMP, Fragoso RM, Pereira D, Medeiros R.. Pain polymorphisms and opioids: an evidence-based review. Mol Med Rep. 2019;19(3):1423-1434. 10.3892/mmr.2018.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Owusu Obeng A, Hamadeh I, Smith M.. Review of opioid pharmacogenetics and considerations for pain management. Pharmacotherapy. 2017;37(9):1105-1121. 10.1002/phar.1986. [DOI] [PubMed] [Google Scholar]
- 3. Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy. Clin Pharmacol Ther. 2021;110(4):888-896. 36. 10.1002/cpt.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zubieta JK, Heitzeg MM, Smith YR, et al. COMT Val158Met genotype affects mu-opioid neurotransmitter responses to a pain stressor. Science. 2003;299(5610):1240-1243. doi: 10.1126/science.1078546. [DOI] [PubMed] [Google Scholar]
- 5. Caraceni A, Hanks G, Kaasa S, et al. ; European Palliative Care Research Collaborative (EPCRC). Use of opioid analgesics in the treatment of cancer pain: evidence-based recommendations from the EAPC. Lancet Oncol. 2012;13(2):e58-e68. doi: 10.1016/S1470-2045(12)70040-2. [DOI] [PubMed] [Google Scholar]
- 6. Owusu Obeng A, Hamadeh I, Smith M.. Review of opioid pharmacogenetics and considerations for pain management. Pharmacotherapy. 2017;37(9):1105-1121. doi: 10.1002/phar.1986. [DOI] [PubMed] [Google Scholar]
- 7. Gilron I, Bailey JM, Tu D, et al. Morphine, gabapentin, or their combination for neuropathic pain. N Engl J Med. 2005;352(13):1324-1334. doi: 10.1056/NEJMoa042580. [DOI] [PubMed] [Google Scholar]
- 8. Dean M. Opioids in renal failure and dialysis patients. J Pain Symptom Manage. 2004;28(5):497-504. doi: 10.1016/j.jpainsymman.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 9. Kotlinska-Lemieszek A, Klepstad P, Haugen DF.. Clinically significant drug–drug interactions involving opioid analgesics used for pain treatment in patients with cancer: a systematic review. Drug Des Devel Ther. 2015;9:5255-5267. doi: 10.2147/DDDT.S86983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuoka H, Yoshiuchi K, Koyama A, et al. Expectation of a decrease in pain affects the prognosis of pain in cancer patients: a prospective cohort study of response to morphine. Int J Behav Med. 2017;24(4):535-541. doi: 10.1007/s12529-017-9644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Campa D, Gioia A, Tomei A, Poli P, Barale R.. Association of ABCB1/MDR1 and OPRM1 gene polymorphisms with morphine pain relief. Clin Pharmacol Ther. 2008;83(4):559-566. doi: 10.1038/sj.clpt.6100385. [DOI] [PubMed] [Google Scholar]
- 12. Matsuoka H, Arao T, Makimura C, et al. Expression changes in arrestin β 1 and genetic variation in catechol-O-methyltransferase are biomarkers for the response to morphine treatment in cancer patients. Oncol Rep. 2012;27(5):1393-1399. doi: 10.3892/or.2012.1660. [DOI] [PubMed] [Google Scholar]
- 13. Matsuoka H, Makimura C, Koyama A, et al. Prospective replication study implicates the catechol-O-methyltransferase Val158Met polymorphism as a biomarker for the response to morphine in patients with cancer. Biomed Rep. 2017;7(4):380-384. doi: 10.3892/br.2017.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsuoka H, Tsurutani J, Chiba Y, et al. Selection of opioids for cancer-related pain using a biomarker: a randomized, multi-institutional, open-label trial (RELIEF study). BMC Cancer. 2017;17(1):674. doi: 10.1186/s12885-017-3664-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watson CPN, Moulin D, Watt-Watson J, Gordon A, Eisenhoffer J.. Controlled-release oxycodone relieves neuropathic pain: a randomized controlled trial in painful diabetic neuropathy. Pain. 2003;105(1-2):71-78. doi: 10.1016/s0304-3959(03)00160-x. [DOI] [PubMed] [Google Scholar]
- 16. Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid t1herapy. Clin Pharmacol Ther. 2021;110(4):888-896. 36. doi: 10.1002/cpt.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vieira CMP, Fragoso RM, Pereira D, Medeiros R.. Pain polymorphisms and opioids: an evidence-based review. Mol Med Rep. 2019;19(3):1423-1434. doi: 10.3892/mmr.2018.9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.