Abstract
The fight against rare cancers faces myriad challenges, including missed or wrong diagnoses, lack of information and diagnostic tools, too few samples and too little funding. Yet many advances in cancer biology, such as the realization that there are tumour suppressor genes, have come from studying well-defined, albeit rare, cancers. Fibrolamellar hepatocellular carcinoma (FLC), a typically lethal liver cancer, mainly affects adolescents and young adults. FLC is both rare, 1 in 5 million, and problematic to diagnose. From the paucity of data, it was not known whether FLC was one cancer or a collection with similar phenotypes, or whether it was genetically inherited or the result of a somatic mutation. A personal journey through a decade of work reveals answers to these questions and a road map of steps and missteps in our fight against a rare cancer.
Subject terms: Liver cancer, Scientific community, Paediatric cancer
Fibrolamellar hepatocellular carcinoma is a rare cancer type and, as such, research into this disease comes with many challenges. In this Perspective, Sanford Simon tells of his personal journey and experiences in the fight against this rare cancer type.
Introduction
The moment I learned my 12-year-old daughter Elana had a rare, lethal liver cancer is, not surprisingly, seared into my memory. It had taken us years to get the diagnosis. We had already been told her incessant abdominal pains might be due to lactose intolerance, or Crohn’s disease, or tween-related stress. None was true. When the pain became acute, we were told to rush to the hospital for a possible appendectomy. A pre-operative scan revealed a 15 cm mass in her liver, ruled a bacterial abscess. However, when a tube was inserted to drain the abscess, little fluid came out. Then, we were told it was a liver cancer, “the fibrolamellar variant of hepatocellular carcinoma”. They said there was nothing that could be done.
Her prospects seemed grim. It was not known whether fibrolamellar hepatocellular carcinoma (FLC) was the result of a somatic mutation or a germline mutation, or both. Nor was it known whether FLC was a single disease or a catch-all term used to describe a collection of cancers with vaguely similar phenotypes. There was no accepted systemic therapy. There were published case reports of liver transplants and partial hepatectomy1,2, which is the path we chose. Scans revealed an additional mass, the size of a golf ball, above her heart, a potential metastatic lesion. During Elana’s surgery at Memorial Sloan Kettering Cancer Center (MSKCC), in April 2008, Michael LaQuaglia, her surgeon, let us know that the ‘golf ball’ was just an inflamed lymph node. This was congruent with LaQuaglia’s report of a large infiltration of immune cells in the primary tumour. The partial hepatectomy was a success. Two days later, as she lay recovering in her hospital room, she opened her laptop and searched for ‘fibrolamellar’. The first paper she found showed a 5-year survival rate of close to zero, with no patients younger than 23 years old surviving3. Recurrence had been reported to be 86% (ref. 4). I recalled the trauma of my younger brother Billy, with a medical ailment when I was 9 years old. At that time, I knew little and could do less. Now, a scientist with a laboratory, still scarred by memories of Billy’s loss, I vowed this time would be different.
As a professor at Rockefeller University focused on cell biology, I had the means to at least try to understand this cancer. Fibrolamellar is rare, estimated at 1 in 5 million5. Fighting a rare disease produces myriad challenges. My wife and I soon learned that our experience of dismissed symptoms and missed diagnosis is a common one for patients with rare diseases. Additionally, there is a paucity of information, tissue samples, funding and good examples of scientific success stories as models. We needed all of them if we were to have a chance of saving Elana.
Over the past decade, I and multiple collaborators who joined my fight successfully took on each of these challenges of information, samples and funding. In the process, we identified the driver mutation of FLC and validated it as a therapeutic target. We also found ways to repurpose drugs that both give insight into the biology and accelerated bringing therapeutics to the patients. We have answered many of the previous unknowns of fibrolamellar; even though, along the way, we got lost down more than a few unproductive rabbit holes. I hope our experiences will not only be relevant to the fight against fibrolamellar but provide a model for others to use in the pursuit of tackling other cancers.
Taking on the challenges
I was trained in cell biology and biophysics and, as I was often reminded by study section reviewers, I am “not a cancer biologist”. My closest collaborator, the cell biologist Gunter Blobel6, with whom I had worked for 25 years, encouraged me to leap off in this new direction, building upon my cell biological skills to fight this cancer. I forged ahead, well aware that 2010 was an auspicious time to enter cancer biology. Decades invested in the human genome project, genetic engineering, chemistry, computational algorithms, biochemistry, microscopy and optics had created a perfect storm in which countless new technologies emerged at the interfaces. This enabled addressing questions that some still thought were in the realm of science fiction.
Funding is tough for rare cancers
Both journals and study sections repeatedly judged that FLC is too rare to justify studying, ignoring the evidence that a well-defined cancer, even if rare, can provide critical insights to reveal general principles. For example, the study of glioblastoma led to the discovery of mutations in isocitrate dehydrogenases, which have since been implicated in many human diseases, including other cancer types7. Also, the study of the rare cancer retinoblastoma led to the discovery of tumour suppressor genes8,9. One reason to study rare cancers is that they tend to be more precisely defined. Often, the term ‘liver cancer’ is used to cover conglomerates of different pathologies that happen to be in the same organ or, in some cases, such as cholangiocarcinoma, tumours in the same cell type. If a cancer can be defined precisely enough, with all tumours being the result of a common molecular dysregulation, this facilitates elucidation of the mechanism as well as enabling the testing of therapeutics. Although rare cancers are considered to account for 20% of all new cancers10, as we improve our ability to characterize the pathogenesis of tumours I believe we will find that many of the more common cancers are actually collections of different rare cancers.
I was fortunate that Gunter Blobel, who had been my postdoctoral adviser, lent not only his personal support but also his financial support. The Development Office at my university is attuned to the essential role of private philanthropy in initiating new projects that are often considered ‘too speculative’ or ‘too preliminary’ to warrant US National Institutes of Health (NIH) support. At the same time, Elana met another patient with FLC who had formed the Fibrolamellar Cancer Foundation, which his parents continued after his passing. In those early years, our work was enabled by private individuals and non-profit foundations. Even now, private individuals and foundations, such as The Sohn Conference, The Rally Foundation, Truth365, Richard Lounsbery Foundation, B+ Foundation, The Murray G. and Beatrice H. Sherman Charitable Trust, Neucrue Cancer Fight and Bear Necessities, provide critical seed money to jump-start projects that otherwise would not exist.
It was not clear where to start our studies. I went to the literature, to seminars and to my colleagues. My son Joel, who was in high school at the time, was puzzled that Elana’s tumour was filled with ‘inflammatory cells’, yet they were not killing the cancer. The immune cells were finding the cancer and, therefore, must be recognizing the tumour cells. Upstairs, one flight from my office, a friend, the immunologist Ralph Steinman, was eager to assist with immunological initiatives. After he unfortunately succumbed to pancreatic cancer, my colleagues Jeffrey Ravetch, who specialized in immunoglobins, and Michel Nussenzweig, a former trainee of Ralph’s who focused on antigen presentation, helped advise my son as he characterized the entire repertoire of amplified B cells and T cells in the patients11 as well as the autoantibodies that were bound in the tumour tissue12.
Elana, now 3 years since her surgery and also in high school, was reading about cancer genomics during an internship studying pancreatic cancer at Mount Sinai medical school. She decided she should sequence her own tumour. She proposed coming to my laboratory to try to characterize the genome (using whole genome sequencing), the coding and non-coding transcriptome (using RNA sequencing) and the proteome (using western blots, immunofluorescence and mass spectrometry) (Box 1).
Elana and I were encouraged by others to try alternative approaches to characterize the genome, either single-nucleotide polymorphism arrays or whole exome sequencing. Although these are the standard first steps, Elana decided to skip them. She reasoned that if there was something obvious, such as a previously identified mutation that one could screen with a single-nucleotide polymorphism array or a mutation in the exome, someone would have found it already. Indeed, as we prepared our submission to the Institutional Review Board for ‘Human Subjects Research’, someone told us “You’re wasting your time, Vogelstein has solved it, he has sequenced the whole thing already”. That was a reference to the renowned cancer biologist Bert Vogelstein. That evening, we wrote him a detailed letter explaining what we were planning on doing and why we were taking that approach. There was no reason to compete — especially as he knew how to do this much better than we did. We asked whether there were things he had not done, or was not planning on doing, that we could pursue. Early the next morning, there was a gracious response in my inbox. In fact, Vogelstein, together with pathologist Michael Torbenson who had taken an interest in FLC, had done whole exome sequencing of ten tumours from patients with FLC and, as we suspected, found nothing remarkable. He encouraged our plans for whole genome sequencing and transcriptome analysis to look for altered splicing. A few days later, we received all of his exome sequencing data. I still have not met him, but he remains one of my heroes. This interaction had the further benefit of introducing me to Torbenson who, a decade later, continues to be one of my close collaborators. Science is a community.
Box 1 Choice of modalities to profile a tumour.
Each approach for characterizing tumours has advantages and limitations.
Single-nucleotide polymorphism array
There are many variations in the human genome. Some are considered ‘normal’, and others have been associated with specific diseases51. A single-nucleotide polymorphism array probes for these identified variants in the DNA. It is the least expensive of the sequencing approaches but can only catch previously identified variants.
Whole genome sequencing
Whole genome sequencing determines most of the DNA sequence of an individual — 3 × 109 bp. It covers both the coding and the non-coding genome. However, as most of the genome is sequenced, it is more expensive. This can either restrict the number of individuals who are sequenced or necessitate a lower number of ‘reads’ or ‘coverage’. This reduces the accuracy, especially when there is some ambiguity in the reads, making it harder to find single nucleotide changes.
Whole exome sequencing
Whole exome sequencing sequences all of the protein-coding regions of genes. As exons account for ~1% of the genome, it is cheaper than sequencing the whole genome. This allows one to either sequence more samples or read them with higher ‘coverage’, which means greater accuracy. It is good for finding structural changes such as copy number variants, inversions, deletions or mutations in the coding genes. Thus, whole exome sequencing is good at finding rare mutations. It does not report on variations in expression of any of the coding or non-coding transcripts.
Bisulfite sequencing
One modification of DNA that leaves ‘epigenetic’ marks, which can repress the expression of a gene, is the addition of a methyl group. Bisulfite sequencing can map the location of these methyl marks on the DNA.
RNA sequencing
RNA sequencing quantifies the RNA transcripts from each gene. Thus, unlike whole exome sequencing and whole genome sequencing, it can report on whether expression of a particular gene is increased or decreased. It can also help to catch changes in splicing of transcripts. However, RNA sequencing is more expensive than whole exome sequencing. If you want to catch mutations, the cost of doing a coverage that is deep enough may be prohibitive.
Mass spectrometry
Mass spectrometry can assess the levels of each protein, as well as post-translational modifications such as phosphorylation, glycosylation or lipidation. Levels of mRNA may not change, but mass spectrometry can catch changes in the rates of protein synthesis or degradation, which would be missed by the other techniques. Of particular interest to many cancers is the ability to quantify changes in the addition of phosphates onto proteins.
Getting patient samples
Obtaining patient samples is yet another challenge for rare cancers. LaQuaglia at MSKCC, my daughter’s surgeon, helped us to obtain our first samples, as well as arrange for some of his surgical fellows to work in my laboratory. Elana, together with another patient with FLC, encouraged other patients, using phone calls, Facebook and a YouTube video titled ‘Fibrolamellar Liver Cancer Research’, to donate their resected tissue. As we were preparing to ship our samples off to a commercial venture in late 2012, some institutions in New York joined resources to form the New York Genome Center. This fortuitous timing was a reflection of the growing power of genomic technologies.
Elana’s first sequencing results examining the transcriptome were encouraging. The coding and non-coding transcriptome revealed thousands of significant changes in coding and non-coding transcripts between the tumour and the adjacent non-transformed tissue, changes that were also found in the proteome13,14. I had not done a transcriptomic analysis before. However, numerous members of the scientific community stepped in to help. Brad Rosenberg, an independent fellow at Rockefeller University (now at Mount Sinai), taught Elana how to quantify and analyse the RNA reads. The authors of DESeq15 and other analysis programs in the R package kindly, patiently and comprehensively responded to Elana’s e-mail queries. Hani Goodarzi, who was a postdoctoral researcher at the time in Sohail Tavazoie’s laboratory at the Rockefeller University (now an independent principal investigator at the University of California, San Francisco), showed Elana how to search for patterns in the promoters of genes. More proof that science is a community.
Alas, the whole genome sequencing, at first, was disappointing. Elana, together with another patient and two of LaQuaglia’s fellows who were working in my laboratory, found there were no recurrent mutations, no inversions and no amplifications16. All the known oncogenes, kinases and genes in liver development were normal. What, then, could account for the transcriptome and phenotypic changes in the tumour?
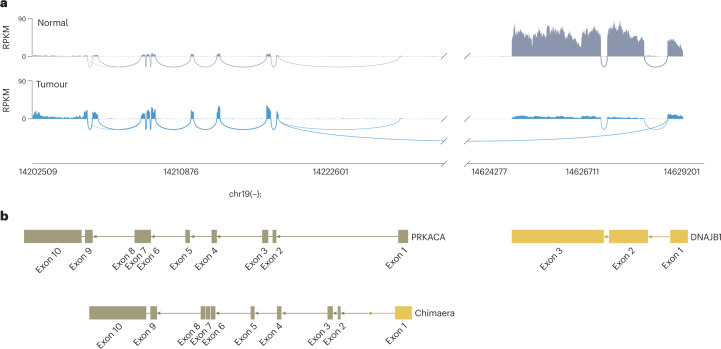
By chance, Elana and I read over a paper together describing how changes in the splicing of RNA can drive breast cancer17. The genes for many proteins of the cell are stored in the genome as a series of discontinuous units, called exons. After they are transcribed from the DNA to make mRNA, the spacers between the exons, called introns, are spliced out. A change in the splicing pattern would alter the sequence and, thus, alter the protein made from the mRNA. Elana examined the transcripts for all the known oncogenes and kinases, and any genes in liver development. The reads from the RNA were mapped on the DNA to quantify the use of different exons, identify the splice junctions and probe for alternative splicing. The splicing of the transcripts for the oncogenes and kinases were unaltered between the FLC tumour and the adjacent normal tissue. There was one exception: the transcript encoding the catalytic subunit of protein kinase A (PKA), PRKACA18. PRKACA is a member of a family of enzymatic proteins that can modify other molecules in the cell by adding a small molecular group called a phosphate. These kinases regulate the activity of other molecules by the addition of this phosphate. This was of particular interest as alterations of kinases have been found in many cancers19. PRKACA mRNA was increased in the tumour, but only exons 2–10 were increased, as were the reads that bridged the exons (Fig. 1). Exon 1 was, if anything, decreased. Also, the reads that should span the junction between exons 1 and 2 were decreased. Instead, there were reads that mapped from the start of exon 2 and reached more than 400,000 bp away to the end of exon 1 of DNAJB1, which encodes a heat shock protein cofactor. It appeared as if there were still normal copies of DNAJB1 and normal copies of PRKACA, but suggestions of an aberrant first exon of DNAJB1 that was joined to the second to the tenth exons of PRKACA. This was found in the first ten patients tested18. We were convinced it was a glitch in the analysis or a mismatch of the reads. After finding it in every patient sample, we extracted RNA from the tumour tissue, reverse transcribed it to DNA, performed Sanger sequencing and found a fusion transcript of DNAJB1–PRKACA in every tumour.
Fig. 1. Mapping reads from the transcriptome onto the genome reveals the fusion transcript.
a, The short nucleotide fragments of RNA sequenced from human fibrolamellar hepatocellular carcinoma (FLC) tumours and adjacent normal tissue mapped onto the known sequence of DNA in the genome. Marks that raise up above the baseline represent RNA fragments that map within a particular exon on the DNA. Loops below the baseline are the RNA fragments that bridge two different exons. DNA has two strands, each of which contain information. b, The genes encoding PRKACA (beige) and DNAJB1 (yellow) are on the negative strand of the DNA and, by convention, are drawn from right to left. The two genes are normally separated by ~400,000 bp. In normal tissue (in grey at the top), all the RNA reads for DNAJB1 are only mapped to the DNA for DNAJB1, and all the RNA reads for PRKACA only map to PRKACA. However, in fibrolamellar tumours, there are RNA reads that bridge between the first exon of DNAJB1 and the second exon of PRKACA. This is not seen in even the adjacent non-transformed tissue from the same patient. Thus, FLC is a somatic mutation — it is not in the germline of the DNA and is therefore not inherited. RPKM, reads per kilobase of transcript per million reads mapped. Modified from ref. 18. © The Authors, some rights reserved; exclusive licensee, AAAS.
I have read about ‘eureka’ moments in science; one thinks of Archimedes in the bathtub. Our observations were anything but. We still harboured concerns that it was a bioinformatic glitch. What led to this fusion transcript at the RNA level? DNA sequencing showed that one chromosome was intact, producing the normal DNAJB1, PRKACA and all the transcripts in between. But there was a deletion, of approximately 400 kb, on the second chromosome copy. That second chromosome led to the DNAJB1–PRKACA fusion transcript.
We still harboured concerns. The presence of the novel transcript did not mean that there was a fusion protein. In previous unrelated studies, we had tried to engineer numerous fusion transcripts (often to GFP) only to find that the fusion protein was not made or was rapidly degraded. However, in all 15 FLC tumour samples tested we found a protein that moved slower on a gel (suggesting that it is larger) compared with either DNAJB1 or PRKACA alone, consistent with the existence of a fusion protein. When we used an antibody to immunoprecipitate DNAJB1 from the tumour tissue, and ran it on a gel, an antibody against PRKACA recognized a single band, the slower moving protein on the gel. This slower moving band was a true fusion protein18. But this only changed the kinds of doubts in our minds. This result still did not mean that the fusion protein was functional. After expressing the fusion protein in cells, we confirmed that the rate at which it added phosphates to proteins was the same as for the normal PRKACA kinase18. Then, the bioinformaticians working with us at the New York Genome Center reported that, computationally, they detected the same novel fusion gene in every sample. These were experts in this kind of analysis. Only then did we write an article in 2014 concluding that “While the role of the DNAJB1–PRKACA chimaera in the pathogenesis of FL-HCC has yet to be addressed, our observations raise the possibility that it contributes to the pathogenesis of the tumour and may represent a therapeutic target”18. Doubts still clawed at us. This is yet an additional challenge to studying a rare disease: we found an alteration in a protein not previously implicated in cancer. In science, it is usually more reassuring to find a solution previously observed by others. Something totally different could be the result of an unforeseen artefact.
Our observations were quickly validated by other groups 1 year later20,21. But this did not mean that the DNAJB1–PRKACA fusion protein was sufficient to cause FLC. Fortunately, across the street at MSKCC, Scott Lowe was an expert at expressing oncogenes in mice. When we used CRISPR–Cas9 to create the same deletion in mice, the mice produced the DNAJB1–PRKACA chimaeric gene and the chimaera protein, and developed tumours in their livers that looked similar to fibrolamellar22 (encouragingly, similar results were observed by another group in a different mouse line)23. However, our results did not resolve the question of whether the tumour was a result of forming this new fusion protein or the loss of the genes in between. Fortunately, the Lowe laboratory was also great at delivering genes via a transposon to mice. Just expression of the fusion gene, without deleting anything in the genome, was sufficient for developing the tumour22. Even then, we had only demonstrated that DNAJB1–PRKACA was sufficient to initiate the tumour. This did not mean that it was a valid therapeutic target.
There never was a ‘eureka’ moment, but rather a series of findings that progressively resolved doubts and provided clarity. By then, we had not only grown close to many other patients but had recruited some to work at the bench. They were invaluable in many ways. As they were the same age as many of the students in the laboratory, the presence of patients brought immediacy to the work. The patients participated in our discussions for guiding our research focus (Box 2). Given that resources are limited, how much effort should be expended on what triggers fibrolamellar, the subsequent pathogenesis, diagnostics or therapeutics? With respect to therapeutics, they helped to balance the discussion between whether to pursue agents that are less than perfect, but could be brought into the clinic sooner, or new precision medicines that would need years of development before they could be in the clinic. They debated pursuing agents that might be extremely efficacious but, with current technology, only available or affordable for a few people, in contrast to those that might be slightly less efficacious but, potentially, cheaper and more accessible. Alas, as work went on, most patients got too ill to work and unfortunately passed away.
One patient with FLC, Jackson Clark, a graduate student in chemistry at Cornell, spent a few years working in our laboratory purifying the oncogenic protein that drives fibrolamellar. His project was to understand how it contributed to pathogenesis and also how to inhibit or degrade this protein. He saw the encouraging progress, signs that in the near future we could manage the disease and rationally design a path (if we could be so bold) to a cure. He was also realistic about his chances. Jackson was fighting to help others survive, even if he did not make it. I am blessed to have worked with these patients and hold memories of them as some of the most painful parts of life and also the most life-affirming. Jackson’s mother, Barbara Lyons, is a chemist at New Mexico State University who, similar to me, has shifted the focus of her laboratory and is a most valuable collaborator. Another patient to work with us in setting up the genomics was an author on our first paper18 but got too weak to continue. Yet another patient, Willow Pickard, worked in the laboratory briefly, before she passed away. Her mother Julie works with us on the patient and family-run Fibrolamellar Registry (discussed below), and her father Douglas is an information technology senior systems administrator and has helped to set up workstations in our laboratory for our molecular dynamics simulations of the DNAJB1–PRKACA fusion protein24,25. In retrospect, even if we had realized that the driver for fibrolamellar was nailed down, it would not have felt like a celebration. At that point, it was all too clear that we needed to focus on better ways of detecting fibrolamellar and developing therapeutic strategies.
Box 2 Steps and missteps.
Science articles are not usually written as a linear historical narrative. Instead, the various data that are judged to be robust and reproducible are arranged in a form to make a compelling set of observations or a test of a hypothesis. This convention can sometimes give the impression that ‘everything worked’. In practice, at least for me, this is far from the case. In most cases, one does not know in advance the proper course, and many experiments do not end up in manuscripts. Sometimes, this is because we discover we have disproved something, and the end result is trivial and, for many, uninteresting. In other cases, we found that an experiment did not work: a technique did not have the resolution or sensitivity desired. Sometimes, we refer to these as dead ends. However, sometimes an approach is dropped before we know whether it will provide useful information. In some cases, it has taken a lot of resources and time, and there is not yet a sign of success. Thus, a decision is made to cut our losses. Alas, one often does not know whether it would have been resolved by one more experiment or whether it would still be unresolved years later. It is perhaps wrong to call these missteps as one needs to continuously re-evaluate projects.
For example, we had tried to form a cell line for the fibrolamellar tumour. With repeat efforts, the only conditions that allowed the fibrolamellar hepatocellular carcinoma (FLC) cells to grow also resulted in a change of their transcriptome and, potentially, their drug response. That is not to say it cannot work.
We studied all the transcripts that were altered in expression when DNAJB1–PRKACA was introduced into cells and tried to identify common transcription factors or motifs in their promoters without success. That is not to say it cannot be done or that it is not worth doing. But, at some point, one has to choose priorities and decide when to hold back. Our priorities were developing better diagnostics and therapeutics. One day, we may return to some of these rabbit holes we left and chase them all the way through.
Finding a therapy
There were many different strategies to pursue. One potential approach was immunological, which includes immune checkpoint inhibitors (ICIs), peptide vaccines or T cell-based therapy. The ICIs are most successful on tumour types with a high mutational burden26. However, FLC has an extremely low level of mutations (see the section ‘Patient engagement’). Thus, we shelved this approach until someone more experienced in immunology develops the methodology for applying these agents to tumours with few mutations.
A second approach was to block oncogenic pathways that were increased in FLC. Thanks to Elana’s participation at The White House Science Fair, where she presented her research to President Obama, she was subsequently invited to introduce President Obama as he launched the nascent Precision Medicine Initiative (Box 3). The idea was to precisely define the disease, not by its organ or histopathology but by its molecular aetiology. There, we met experts who advised us that to find therapeutics we should identify which oncogenes are increased in the transcriptome. We had already characterized the coding and non-coding transcriptome13,14, as well as the proteome, which revealed increases in numerous transcripts known to be oncogenic in other systems, such as Aurora kinase A, signalling molecules of the epidermal growth factor receptor (EGFR) pathway, including ERBB2, and those of the WNT pathway13. These would be a good starting point for identifying targets for therapy.
The challenge was to develop an appropriate system to test for functional relevance. There are numerous models that could be used. Some groups develop a cell line from their tumour that can grow in vitro. However, we could not find conditions that allowed the FLC cells to grow in vitro, without significant shifts in the transcriptome, proteome and drug response of the cells. Thus, the response of these cells may not reflect the response of the patients. An independent approach is to develop organoids. We were fortunate to have the collaboration of Hans Clevers, a pioneer of organoid development. Together, we made and validated organoids both of fibrolamellar and hepatoblastoma27,28. Yet another approach is to implant cells from human FLC tumours into mice with compromised immune systems to form what are called patient-derived xenografts (PDXs). With the help of colleagues Sohail Tavazoie, Ype de Jong, Charlie Rice and their laboratory members, we learned different ways of implanting and assessing the tumours. We could demonstrate that their histology looked similar to fibrolamellar, they expressed both the fusion transcript and the fusion protein, and the transcriptome of these tumours recapitulated that of the patients29. However, when we resected the tumours, dissociated the cells and tested them with drugs that are known to block these recognized oncogenes, such as Aurora kinase A or ERBB2, there was no effect. This was repeated on multiple independent PDXs, all without effect29.
Despite the successes, my attempts to secure grant funding for the work met obstacles. Grant review panels repeated their assertion that I was “not a cancer biologist”. Our failure to see an effect of blocking the oncogenes could appropriately reflect their concerns. Was it a problem in the development of our PDXs or a problem in our testing? Each test on the PDXs showed they were recapitulating the behaviour of the human tumours. We reasoned that perhaps the properties of the tumours changed when we resected them from the mice. So, we tested the drugs on the tumours while they were still growing in the mice. They still had no effect.
We had another potential worry. The initial tumours took 6–9 months to grow after we implanted them. Maybe only a small subset of cells were growing in the mice, a subset that may not represent the bulk of the tumour. We felt we needed to test the entire tumour immediately upon resection from patients, without allowing time for select populations to survive. We were already collecting some tumours directly from the operating room, owing to the help of some caring surgeons such as Erik Schadde (Rush University Medical Center), Tomoaki Kato (Columbia University Irving Medical Center), William Jarnagin (MSKCC), Mark Trudy (Mayo Clinic) and LaQuaglia. Owing to members of my laboratory who were willing to fly on short notice, these live-tissue samples were rushed from various operating rooms to my laboratory, where they were tested against a panel of drugs. The results reflected all the cells from the resection, not just those cells that survived growth in a mouse. Within 3 days, we had the results: the tumour samples taken directly from patients were indistinguishable from those tumours grown in mice. Within 72 h of resection, we had a personalized drug response profile for each patient’s tumour29,30. This validated our PDX models for testing drugs. It confirmed that blocking these overexpressed oncogenes had no effect. The lack of response showed that just because the expression of an oncogene is increased, it does not mean it is causally related to tumorigenesis. These results also left us without a direction.
At this point, I decided that the study sections were correct: I am limited in what I know about cancer, or at least my ability (and perhaps the field’s ability) to predict what should work against a tumour. Instead, we chose to do an agnostic functional screen to determine the vulnerabilities of the tumour, an unbiased acknowledgement of our ignorance. It would still be ‘precision medicine’ but based on function, not transcriptomics. We screened cells dissociated from human FLC tumours with a collection of every drug we could get our hands on. We took advantage of a library of small compounds — some clinically relevant, some ‘exploratory tool compounds’ — and added to it every compound that had been tested in the clinic or implicated to block PRKACA. In total, we examined more than 5,000 compounds. The results yielded numerous agents that we would never have predicted29. One was napabucasin. Although napabucasin is reported to block the signal transducer and activator of transcription (STAT) pathway, this did not appear to be its mechanism of action for fibrolamellar: there were no changes in the STAT pathway, and there was no effect of other compounds that hit this pathway. While we were eliminating other possible mechanisms of action, the company that made the drug was bought, and the new owners decided to discontinue its use. After many unsuccessful attempts to reach the new owners, we dropped this option. Agents that interfere with histone deacetylases were also at the top of our list for efficacy against FLC. These are still being actively pursued. I was fortunate to have started an active dialogue with numerous paediatric oncologists, including Michael Ortiz and Julia Glade-Bender (MSKCC) and Allison O’Neill (Dana-Farber), who contributed their perspective on the use of various agents. Their experience with the histone deacetylase inhibitors with children was still limited, and associated toxicities led them to question whether this was the right avenue to pursue. Another one of the top hits was irinotecan29,30, which inhibits topoisomerase I (a nuclear enzyme, required to relax DNA supercoiling generated by transcription, replication and chromatin remodelling). Irinotecan was effective against every sample from patients. As it blocks topoisomerase, irinotecan is supposed to work against rapidly growing tumours. The paediatric oncologists had the benefit of decades of work with irinotecan, and they presented compelling arguments for its use. Science is a community.
I was perplexed by the efficacy of irinotecan. Fibrolamellar is notoriously slow growing, so why did irinotecan have any effect? It had been observed that patients with Gilbert’s syndrome, who cannot process bilirubin, are very sensitive to irinotecan. These patients have reduced activity from a specific enzyme family, UDP-glucuronosyltransferases, that add a sugar to various molecules to facilitate exporting them from the liver31,32. SN38 is the active metabolite of irinotecan and is one of the molecules whose inactivation depends on these enzymes33. When we examined this family of proteins, they were reduced by more than 40-fold in FLC30. FLC was more sensitive because a protein was decreased, not because a protein was increased. This highlighted the value of functional precision medicine. Finally, some disparate pieces were starting to fit together. The tools that have been developed over the past decades (by a multitude of different scientists) have enabled elucidation of cellular pathways at a subcellular level that allow us to understand drug sensitivities at the patient level.
Yet another top hit was navitoclax, which blocks two different anti-apoptotic molecules: BCL-2 and BCL-XL. Other compounds that selectively inhibit BCL-2, such as venetoclax, were without effect. Compounds, albeit not yet used in the clinic, selective against BCL-XL were very effective. The combination of irinotecan with navitoclax was particularly effective. Alas, navitoclax had a reputation for causing platelet toxicity, which concerned both the paediatric oncologists and members of the NIH Cancer Therapy Evaluation Program (CTEP), in particular Malcolm Smith, who generously offered their time. We were fortunate that, at the time, a new class of therapeutics was being synthesized called proteolysis-targeting chimaeras (PROTACs)34. These are bifunctional agents: at one end, they bind to a target of interest; at the other end, a small molecule engages an E3 ligase that, in turn, has the potential to ubiquitinate your target of interest. If the topological orientation is permissive, if there are available lysines to ubiquitinate and if your target of interest is a good substrate for the proteosome, then there is the potential to selectively degrade your target. A group led by Daohong Zhou (at the time at the University of Florida, now at UT Health San Antonio) had developed just such a PROTAC targeting BCL-XL (ref. 35). They wisely targeted an E3 ligase that is not very active in platelets. Working together with them, we combined irinotecan with their PROTAC against BCL-XL. The results showed not just a reduction of growth but complete remission of the most typical PDXs growing in mice30. Similar results were observed when we tested the combination on cells fresh from a patient surgical resection. Even the most extreme drug-resistant tumour was reduced to stable disease, potentially buying time for patients to have other interventional treatments or until new treatments are developed30. For all of this work, I was fortunate that a collaborator from the University of California, San Francisco, Philip Coffino, who had studied the proteosome for 40 years and, previously, the activity of PRKACA in cells, moved to New York to join us in developing therapeutics. He has now expanded his involvement to all of our FLC projects.
An alternative therapeutic approach is to directly hit DNAJB1–PRKACA. However, this first meant determining whether it was a valid therapeutic target. As mentioned above, perhaps it only triggers FLC but is then no longer needed? If so, therapeutics targeting DNAJB1–PRKACA would have no effect. It was also possible that the use of a therapeutic to eliminate the DNAJB1–PRKACA fusion would result in the cells becoming ‘normal’ hepatocytes rather than simply dying. In this case, continued administration of the therapeutic would be required, opening up the possibility of a mutational escape. We tested these possibilities by introducing a virus into the FLC cells that can induce expression of a short hairpin RNA (shRNA), an antisense to the junction between DNAJB1 and PRKACA. Once we could measure the FLC tumour growing in the mice, we induced production of the shRNA. The DNAJB1–PRKACA fusion disappeared from the cells, and the tumours stopped growing and then shrank36. The same virus and shRNA had no effect on other tumour types36. This showed that DNAJB1–PRKACA was needed for the FLC tumour to continue to grow. Equally importantly, when the oncogenic transcript was eliminated with the shRNA, the cells died, indicating an addiction37,38. This validates DNAJB1–PRKACA as a therapeutic target and is the basis for numerous therapeutics currently under development (Box 4).
Box 3 Advocacy versus privacy.
There are many advantages to recruiting patients into the laboratory. Patient involvement provides a face for the research, it establishes a clear motivation. The patients provide a valuable perspective on the priorities for research and directions for therapeutics. They are also highly motivated workers at the bench and are powerful public advocates. Upon publication of our first paper, there was a flurry of media activity. People loved the concept of a patient turned researcher and, after the first few interviews, Elana realized that her position made her an effective advocate for the childhood and rare disease communities. In the next few months, she spoke at the opening plenary session of the American Association for Cancer Research general meeting, presented her work at The White House Science Fair and appeared on television, from Sunrise Australia to Canada AM and Al-Jazeera. She spoke at fundraisers for childhood cancers for the Sohn Conference Foundation and Solving Kids’ Cancer. The media attention also created opportunities for political engagement. She participated in the launch of the Precision Medicine Initiative with President Obama at the White House and the Cancer Moonshot initiative with President Biden and Pope Francis at the Vatican, as well as walked the halls of Congress advocating for long-term support for basic and translational research. After using her position to help lobby for the Cures Act, she was invited to be on the stage with Obama and Biden for the signing of the legislation. The emotionally compelling aspect of the patient-researcher story is what enabled all these opportunities and put Elana in a position to effectively raise funds for paediatric and rare disease research, while also giving hope to many other patients who still reach out even to this day.
Although this attention can be used to effect important change, it can impose a burden on the patients and have negative side effects. Although Elana received positive feedback, there were also unfortunate comments and criticism from strangers on the Internet. Publishing any research, or speaking out, can always lead to backlash. However, it becomes more complicated when it is not just an academic story but a personal one too. Negativity and bullying on social media have become all too prevalent, and many forums on social media harbour misogyny52,53.
Public involvement in research and advocacy not only places someone in a position to receive public scrutiny but can also reveal a patients’ disease status. Previously, Elana had tried to keep her cancer history private, usually making up stories (such as a shark attack) to explain to the other kids why she had surgical scars. Some patients who have subsequently worked in the laboratory have requested that their names be kept off manuscripts to maintain privacy and avoid the chance that future employers could discriminate based on their condition. Although patient-researchers have the potential opportunity to develop a platform to advocate for important causes, patient involvement is a personal decision, and patients should consider both the pros and cons before getting involved in research and advocacy.
Box 4 Serendipity in science.
Although I was trained to believe that success in science is the result of careful planning, hard work and creative thinking, I have learned that serendipity can sometimes play a critical role. In 2013, when we had our first data suggestive of a fusion gene, my daughter asked me “Why don’t we develop an antisense RNA as a therapy?” In my endless ignorance, I explained that RNA is not very stable in the circulation and it could not succeed.
Four years later, Elana had recruited Willow Pickard, another friend with fibrolamellar hepatocellular carcinoma (FLC) who already was not in the best of health, to the laboratory. She was a big fan of jam bands such as the Grateful Dead and Phish, but she needed to be within a short distance of a bathroom. Phish were coming to New York City. Alas, the only way she could see them was if she got a ticket in one of the corporate suites, which has its own facilities. Through various friends in music, I put out a call looking for two seats in a corporate suite, for a patient who had the same cancer as my daughter and her father. I got an e-mail back from someone saying they knew someone with one of these suites and could they show him my note? The next day, I got an e-mail from Dinakar Singh, the owner of the suite. Coincidently, Dinakar was on the board of my university, and he had seen my daughter and I talking about our work in an interview called ‘Cancer survivor’s brilliant YouTube idea’ on the Today Show (a television show in the United States). He invited us as his guests to the concert. As we spoke, I learned his daughter was born with spinal muscular atrophy. Dinakar described his 16-year journey to help find therapies, which led him to support work on antisense oligonucleotides such as Spinraza, developed by Ravindra Singh (University of Massachusetts), Adrian Krainer (Cold Spring Harbour Laboratory) and C. Frank Bennett (Ionis Pharmaceuticals) and by Ionis and Biogen54. Once again, I should have listened to my daughter sooner. But Dinakar’s story inspired me to pursue antisense strategies. The community of science extends beyond the scientists.
Patient engagement
Lack of information is another major barrier to fighting rare diseases. Each hospital has very few of these patients, and the hospitals were not sharing their data. We realized one workaround was to have the patients share their own data. My wife Rachael Migler, Elana and I, along with other patients and their families, launched a patient-hosted registry called the Fibrolamellar Registry. We wrote an extensive questionnaire with input from a diverse group of stakeholders and experts, including patients, family members, surgeons, oncologists, pathologists, hepatologists, epidemiologists and paediatricians. This questionnaire could be laborious for patients to answer but the rewards for everyone could be great. Patients’ trust was critical to us, and we made the decision that we would not sell the patients’ data to a company. This contributed, we believe, to a large ‘buy-in’ from the patients. We already have data from more than 200 patients from more than 20 countries. As we were run by patients and family members, we could keep in touch with patients. Thus, our records have more detailed outcome information as patients frequently switched doctors and institutions, even between different countries, in the often-futile search for a cure. Some patients were followed between institutions until a molecular analysis revealed that they had been incorrectly diagnosed as having fibrolamellar.
There were numerous observations that surprised us in the patient dataset. Although most liver cancers are more prevalent in men39, FLC is slightly more prevalent in women40. We were surprised to find that women survived longer than men41. It was possible that women were diagnosed earlier, perhaps they are more sensitive to changes in their bodies. But, on average, women were diagnosed at the same age and same stage as men. Furthermore, the increased survival was not related to the use of hormonal supplements. Because of these observations, we are now studying variations in expression of genes on the X chromosome: either immune-related genes that could be protective or tumour suppressor genes, which men would be more sensitive to the loss of. Women can lose one copy of a tumour suppressor and still have a healthy copy, but men have only one copy. Another surprising observation was that survival was not related to the size of the tumour at diagnosis41. A patient’s chance of survival was no different if their tumour was 0.5 cm or 20 cm. What was important was how many metastases were present. But the number of metastases was not dependent on the size of the tumour. I had, previously, naively thought that as the tumour grows, there is a continuous chance that some cells will separate off and metastasize. Instead, the results suggest that some FLC tumours are highly metastatic. Even at their first detection, they are likely to have already spread. Others are more indolent. They can grow to tens of centimetres without metastases (Elana’s tumour was 15 cm). This was surprising as we could not detect any genomic differences between those that were highly metastatic and those that were not. There could be epigenetic modifications that we have missed, or, perhaps, there is something in the genomic background of the patient that is more permissive or restrictive for metastases. Similarly, we find patients fall into discrete groups in their sensitivity to therapeutics29. These results have now led us to start a genome-wide association study to identify background variants that associate either with patient outcomes or drug sensitivity. A similar effect of background genomics has been seen in melanoma where a patient’s germline variant of apolipoprotein E affects the metastatic proclivity of the primary tumour42.
Our relationship with the patients allows us to go back and request additional data. A research team led by Mark Yarchoan at Johns Hopkins University wanted to do a retrospective study of ICIs in FLC. ICIs might not be expected to work on FLC, given the few mutations in the tumour26. However, some oncologists have tried them on patients with FLC, but the results are too sparse in any one institution to make a clear statement. The registry reached out to patients. Those who agreed to participate shared their medical records and scans. After Rachael spent months manually de-identifying all the scans and medical records, they were analysed by radiologists and oncologists. Through data in the registry, it was possible to collect sufficient numbers to determine that ICIs have only modest clinical activity in FLC43. The liver is responsible for removing 70% of the ammonia generated in the body44. In most liver cancers, ammonia builds up due to liver failure, and this can adversely affect the brain producing hyperammonemic encephalopathy. This is a frequent problem in patients with FLC45. However, in FLC, much of the liver is still functional. Data from the Fibrolamellar Registry are helping to elucidate some of the alternative metabolic pathways responsible for hyperammonemic encephalopathy. This progress, as evidenced by the three recent publications involving the Fibrolamellar Registry30,41,43, demonstrates the power of the patient community to advance work, especially on the study of rare diseases.
Seven years ago, at the launch of the Precision Medicine Initiative, I discussed the potential of these patient groups with Francis Collins, the long-term director of the NIH. He emphasized that the success of patient engagement should not be judged by the ability of scientists and clinicians to give the patients a seat at the table. Instead, he said we should bring them into the kitchen and involve them in the planning and preparation of the meal. We have been fortunate to have his guidance. We still need to move forward to make it easier for patients to access their records, to own their records and to share their records together. Patients should own their tissues and be allowed to target them to research. We should remember, science is a community.
Prognosis
It has been less than a decade since we identified the oncogenic driver for FLC (Box 5). In the coming year, we hope to have drugs that are already in the clinic ‘repurposed’ for FLC. In the near future, there will be therapies targeted directly against the driver36 or towards recruitment of the immune system43,46. Still, it does not feel like a moment to celebrate. It is painful: only an instantaneous solution would be fast enough. Most patients with FLC are in their teens or twenties. Many have not only helped to recruit tissue but have worked at the bench side by side with my graduate students and postdoctoral scientists who are their age. They were involved in evaluating data and planning research directions. Most of these patients have succumbed to fibrolamellar.
Despite these losses, we should not lose sight of how fortunate we are, in many ways. For decades, scientists of all disciplines have been telling the public that if we invest in basic research, it will comeback to help all humanity. The motto of my university is Scientia pro bono humani generis, which translates to science for the benefit of humanity. All the advances in my laboratory are the result of decades of work by others in developing bioinformatic software, technologies for engineering genomics, high-resolution microscopy, chemistry, machine learning and even the Internet, which allowed us to reach the patients. Our experiences are shared by many other groups. I cannot think of a more exciting time to enter science.
A public recognition of these advances can be seen in the United States in the Rare Cancer Research Initiative of The Department of Defense and in President Biden’s Cancer Moonshot. The Blue-Ribbon panel that set the priorities for this Moonshot initiative recognized the potential of technological advances, the importance of addressing rare and childhood cancers, and the necessity of making all results ‘open access’, that is, accessible to the public. Owing to the support of private philanthropy and private foundations, our work is now supported with a specific Cancer Moonshot grant for consortiums that study childhood cancers driven by fusion proteins, such as FLC, Ewing sarcoma, rhabdomyosarcoma and many others. A panel of all the principal investigators meet together every 2 months, with at least one longer format meeting each year. The different consortia share their experiences, both positive and negative, with different approaches. This openness, in a protected environment, has helped to stimulate many new collaborations.
The promises issued by agencies such as the NIH and the National Science Foundation (NSF) in the United States, and many others around the world, are more than fulfilling themselves. Just look at the advances in the past 2 years, hepatitis C is en route to being cured47 and, less than a year after SARS-CoV-2 was detected, a new generation of vaccines were being tested that showed astounding success48. Both of these built upon decades of progress. Although some aspects of immunotherapy for cancer are in the early stages, some cancer types such as melanoma are being successfully treated with ICIs49 and some blood tumours with engineered T cells50. For almost any project in modern biology, there is no longer distinct translational research or basic research. There is just a continuum where almost any study on the bench or the computer can impact what happens at the bedside, and information from patients, whether normal physiology or pathology, impacts what happens in the laboratory. Questions that were not addressable a decade ago are now accessible, techniques that seemed to be science fiction are now routine. Alas, there are not improvements everywhere. Social media provided the initial means for patients with rare diseases to find each other. But now discourse is often dominated by a few who are very vocal but not necessarily the best informed. Whereas a decade ago I revelled in the opportunities offered by social media, I now think it is more dangerous to get your medical advice from social media than to get your news from social media.
Just as many scientists joined together to combat SARS-CoV-2, many of my colleagues have joined our FLC venture. Within my laboratory, everyone joins in when there is a fresh tissue resection, splitting up the tasks and backing each other up when experiments pile up. I am intensely grateful to all and regret that I could not include all their contributions nor name the individuals who are in the laboratory. They bring and meld a diversity of invaluable perspectives, including a hepatologist, biochemist, paediatric surgeon, biophysicist, paediatric oncologist, family physician, immunologist, geneticist and computational biologist. They are from Israel, Egypt, Germany, Iran, China, Peru, Colombia, Jordan, Ecuador, Turkey, India, Pakistan, Sweden, the United Kingdom, France and the United States. They are Muslim, Jewish, Christian and agnostic, but they are a unified congregation that believes Scientia pro bono humani generis. Humanity is still far from curing all disease, eliminating all pain and suffering. Yet our abilities as humans to work together to explore, elucidate and engineer provide grounds for hope.
Box 5 Is fibrolamellar hepatocellular carcinoma a single disease?
Fibrolamellar hepatocellular carcinoma (FLC) was first described in 1956 by Hugh A. Edmondson55 based on its very distinctive morphology: large eosinophilic, hepatocyte-like polygonal cells, with prominent nucleoli and pale inclusion bodies. Coursing through the tumour are fibrotic bands or ‘lamellae’, from which the tumour derives its name56. Unfortunately, even with a biopsy, there is often disagreement over the diagnosis. In one test of the precision of histological typing, a set of slides from 25 primary liver tumours, including FLC, were distributed for analysis by a dozen pathologists; what resulted was poor reproducibility among these experts in the pathological diagnosis57. As a result, various histological immune stains have been used to assist with diagnosis, including cytokeratin 7 (CK7) and CD68 (refs. 58,59). Despite these modifications to the diagnostic procedure, it is an ongoing concern that many studies of ‘FLC’ include misdiagnosed cases41. Likewise, some cases of FLC have been mistakenly diagnosed as other tumour types13,57,60.
The expression of DNAJB1–PRKACA is sufficient to produce the pathology of FLC, and the DNAJB1–PRKACA fusion gene is found in the vast majority of patients with FLC. However, there are exceptions. Together with Mike Torbenson, we have found three patients whose tumours looked similar to FLC but they did not have the DNAJB1–PRKACA fusion gene. Instead, their tumours were missing one of the regulatory subunits that inhibits the catalytic activity of PRKACA61. Indeed, they were in families that carried the genetically inherited Carney syndrome that results from mutations in one of the regulatory subunits of protein kinase A (PKA)62. The Zucman-Rossi laboratory found tumours that had previously been characterized as mixed FLC/hepatocellular carcinoma that had an inactivation of BRCA1-associated protein 1 (BAP1), which, amongst other things, increased activation of PKA63. In a few anecdotal cases in other cancers, different genomic rearrangements have been found with aberrations of the catalytic subunit of PKA, including fusions of the first exon of ATP1B1 or DNAJB1 to either PRKACA or PRKACB. These were observed in intraductal oncocytic papillary neoplasms of the pancreas and bile duct64,65.
All these molecular alterations share disruption of PKA signalling. However, some outstanding questions include whether expression of the other fusion genes (that is, ATP1B1–PRKACA) or inactivation of BAP1 is sufficient to produce the tumour, or whether there are other background mutations that are critical, or even more important. To what extent do these develop into overt tumours? We had previously found that overexpression of PRKACA was sufficient to produce many cellular alterations in hepatocytes, but we did not get overt tumours. Addressing these questions is important for understanding the pathogenesis of FLC. To what extent is the increased activity of PKA in tumour cells alone responsible for the pathology of FLC? Knowing the answer is also important for guiding therapeutics for patients. Agents that specifically target DNAJB1–PRKACA at the RNA level will not work for a ATP1B1–PRKACA fusion, but a therapeutic that targets at the protein level might. Yet neither approach would work for inactivation of BAP1.
Thus, for treating patients it is critical to precisely define the molecular alterations in disease. Whether the term ‘FLC’ is used to describe only the cancer type driven by the DNAJB1–PRKACA fusion or any tumour with fibrous bands is a question that I leave to the clinicians. Unfortunately, FLC is still officially characterized as a subvariant of hepatocellular carcinoma in the International Classification of Diseases for Oncology database66, even though it has a different molecular basis and drug response. As a result, many patients with FLC are treated with therapeutics that show no efficacy29,40,67. We have received numerous tumour samples from patients who were diagnosed as having fibrolamellar, but we did not find the DNAJB1–PRKACA fusion gene, nor any of the other fusions, nor an alteration in BAP1. Additionally, the transcriptome, proteome and phosphoproteome of these tumours did not segregate with any other FLC tumours. In each case, when we sent these samples (unlabelled) to Michael Torbenson, who is a pathologist, he said that they were not FLC. Alas, in the interim, the clinicians treated these patients as if they had FLC. These patients, albeit well intentioned, would then go online to private chat groups and share the outcomes of their therapies. This has led to a morass of misinformation amongst patients. Much of this is then shared with their clinicians. Everyone would benefit from fulfilling the promise of precision medicine.
Endnote
It is 14 years since most of Elana’s liver was resected. She entered a PhD programme in biomedical informatics in autumn 2022.
Acknowledgements
The author thanks the fibrolamellar community for their commitment to defeat this beast. The author also thanks all members of the laboratory who have been working together as an incredible team, backing each other up on every step. Additionally, the author thanks colleagues around the world who have so generously given their time and thoughts.
Author contributions
The author handled all aspects of the article.
Peer review
Peer review information
Nature Reviews Cancer thanks the anonymous reviewers for their contribution to the peer review of this work.
Competing interests
The author declares no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
B+ Foundation: https://www.bepositive.org/
Bear Necessities: https://bearnecessities.org/about-bn/
Cancer survivor’s brilliant YouTube idea: https://www.today.com/video/cancer-survivors-brilliant-youtube-idea-228752963974
Fibrolamellar Cancer Foundation: https://fibrofoundation.org/about-fibro/
Fibrolamellar Liver Cancer Research: https://youtu.be/Y5lkp_uK9Ww
Fibrolamellar Registry: http://fibroregistry.org
Neucrue Cancer Fight: https://www.neucruecancerfight.org/
Precision Medicine Initiative: https://www.youtube.com/watch?v=qHD-_NYOcVA
Richard Lounsbery Foundation: https://www.rlounsbery.org/
Solving Kids’ Cancer: https://solvingkidscancer.org/
The Rally Foundation: https://rallyfoundation.org/
The Sohn Conference: https://www.sohnconference.org/
The White House Science Fair: https://youtu.be/q0TYR-MxC3I
Truth365: https://www.thetruth365.org/
Change history
4/20/2023
A Correction to this paper has been published: 10.1038/s41568-023-00575-5
References
- 1.Pinna AD, et al. Treatment of fibrolamellar hepatoma with subtotal hepatectomy or transplantation. Hepatology. 1997;26:877–883. doi: 10.1002/hep.510260412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starzl TE, et al. Treatment of fibrolamellar hepatoma with partial or total hepatectomy and transplantation of the liver. Surg. Gynecol. Obstet. 1986;162:145–148. [PMC free article] [PubMed] [Google Scholar]
- 3.Moreno-Luna LE, et al. Clinical and pathologic factors associated with survival in young adult patients with fibrolamellar hepatocarcinoma. BMC Cancer. 2005;5:142. doi: 10.1186/1471-2407-5-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamashita S, et al. Prognosis of fibrolamellar carcinoma compared to non-cirrhotic conventional hepatocellular carcinoma. J. Gastrointest. Surg. 2016;20:1725–1731. doi: 10.1007/s11605-016-3216-x. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag HB, Davila JA. Is fibrolamellar carcinoma different from hepatocellular carcinoma? A US population-based study. Hepatology. 2004;39:798–803. doi: 10.1002/hep.20096. [DOI] [PubMed] [Google Scholar]
- 6.Simon SM. Gunter Blobel (1936–2018) Nature. 2018;556:32. doi: 10.1038/d41586-018-03849-3. [DOI] [PubMed] [Google Scholar]
- 7.Tommasini-Ghelfi S, et al. Cancer-associated mutation and beyond: the emerging biology of isocitrate dehydrogenases in human disease. Sci. Adv. 2019;5:eaaw4543. doi: 10.1126/sciadv.aaw4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl Acad. Sci. USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friend SH, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 10.Gatta G, et al. Rare cancers are not so rare: the rare cancer burden in Europe. Eur. J. Cancer. 2011;47:2493–2511. doi: 10.1016/j.ejca.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Simon JS, Botero S, Simon SM. Sequencing the peripheral blood B and T cell repertoire — quantifying robustness and limitations. J. Immunol. Methods. 2018;463:137–147. doi: 10.1016/j.jim.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rich BS, et al. Endogenous antibodies for tumor detection. Sci. Rep. 2014;4:5088. doi: 10.1038/srep05088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon EP, et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proc. Natl Acad. Sci. USA. 2015;112:E5916–E5925. doi: 10.1073/pnas.1424894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farber BA, et al. Non coding RNA analysis in fibrolamellar hepatocellular carcinoma. Oncotarget. 2018;9:10211–10227. doi: 10.18632/oncotarget.23325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darcy DG, et al. The genomic landscape of fibrolamellar hepatocellular carcinoma: whole genome sequencing of ten patients. Oncotarget. 2015;6:755–770. doi: 10.18632/oncotarget.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bemmo A, et al. Exon-level transcriptome profiling in murine breast cancer reveals splicing changes specific to tumors with different metastatic abilities. PLoS ONE. 2010;5:e11981. doi: 10.1371/journal.pone.0011981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honeyman JN, et al. Detection of a recurrent DNAJB1–PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–1014. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen P, Cross D, Janne PA. Kinase drug discovery 20 years after imatinib: progress and future directions. Nat. Rev. Drug Discov. 2021;20:551–569. doi: 10.1038/s41573-021-00195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham RP, et al. DNAJB1–PRKACA is specific for fibrolamellar carcinoma. Mod. Pathol. 2015;28:822–829. doi: 10.1038/modpathol.2015.4. [DOI] [PubMed] [Google Scholar]
- 21.Xu L, et al. Genomic analysis of fibrolamellar hepatocellular carcinoma. Hum. Mol. Genet. 2015;24:50–63. doi: 10.1093/hmg/ddu418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kastenhuber ER, et al. DNAJB1–PRKACA fusion kinase interacts with β-catenin and the liver regenerative response to drive fibrolamellar hepatocellular carcinoma. Proc. Natl Acad. Sci. USA. 2017;114:13076–13084. doi: 10.1073/pnas.1716483114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelholm LH, et al. CRISPR/Cas9 engineering of adult mouse liver demonstrates that the Dnajb1–Prkaca gene fusion is sufficient to induce tumors resembling fibrolamellar hepatocellular carcinoma. Gastroenterology. 2017;153:1662–1673. doi: 10.1053/j.gastro.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomasini MD, et al. Conformational landscape of the PRKACA–DNAJB1 chimeric kinase, the driver for fibrolamellar hepatocellular carcinoma. Sci. Rep. 2018;8:720. doi: 10.1038/s41598-017-18956-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao B, et al. Structures of the PKA RIα holoenzyme with the FLHCC driver J-PKAcα or wild-type PKAcα. Structure. 2019;27:816–828.e4. doi: 10.1016/j.str.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayan NJC, et al. Human liver organoids for disease modeling of fibrolamellar carcinoma. Stem Cell Rep. 2022;17:1874–1888. doi: 10.1016/j.stemcr.2022.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saltsman JA, et al. A human organoid model of aggressive hepatoblastoma for disease modeling and drug testing. Cancers. 2020;12:2668. doi: 10.3390/cancers12092668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lalazar G, et al. Identification of novel therapeutic targets for fibrolamellar carcinoma using patient-derived xenografts and direct-from-patient screening. Cancer Discov. 2021;11:2544–2563. doi: 10.1158/2159-8290.CD-20-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shebl B, et al. Targeting BCL-XL in fibrolamellar hepatocellular carcinoma. JCI Insight. 2022;7:e161820. doi: 10.1172/jci.insight.161820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ando Y, et al. UGT1A1 genotypes and glucuronidation of SN-38, the active metabolite of irinotecan. Ann. Oncol. 1998;9:845–847. doi: 10.1023/A:1008438109725. [DOI] [PubMed] [Google Scholar]
- 32.Iyer L, et al. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J. Clin. Invest. 1998;101:847–854. doi: 10.1172/JCI915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oguri T, et al. UGT1A10 is responsible for SN-38 glucuronidation and its expression in human lung cancers. Anticancer Res. 2004;24:2893–2896. [PubMed] [Google Scholar]
- 34.Sakamoto KM, et al. Protacs: chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc. Natl Acad. Sci. USA. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan S, et al. A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat. Med. 2019;25:1938–1947. doi: 10.1038/s41591-019-0668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neumayer C, et al. Oncogenic addiction of fibrolamellar hepatocellular carcinoma to the fusion kinase DNAJB1–PRKACA. Clin. Cancer Res. 2022;29:271–278. doi: 10.1158/1078-0432.CCR-22-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felsher DW. Oncogene addiction versus oncogene amnesia: perhaps more than just a bad habit? Cancer Res. 2008;68:3081–3086. doi: 10.1158/0008-5472.CAN-07-5832. [DOI] [PubMed] [Google Scholar]
- 38.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 39.Ananthakrishnan A, Gogineni V, Saeian K. Epidemiology of primary and secondary liver cancers. Semin. Interv. Radiol. 2006;23:47–63. doi: 10.1055/s-2006-939841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lalazar G, Simon SM. Fibrolamellar carcinoma: recent advances and unresolved questions on the molecular mechanisms. Semin. Liver Dis. 2018;38:51–59. doi: 10.1055/s-0037-1621710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berkovitz A, et al. Clinical and demographic predictors of survival for fibrolamellar carcinoma patients—a patient community registry-based study. Hepatol. Commun. 2022;6:3539–3549. doi: 10.1002/hep4.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ostendorf BN, et al. Common germline variants of the human APOE gene modulate melanoma progression and survival. Nat. Med. 2020;26:1048–1053. doi: 10.1038/s41591-020-0879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen KY, et al. Clinical outcomes in fibrolamellar hepatocellular carcinoma treated with immune checkpoint inhibitors. Cancers. 2022;14:5347. doi: 10.3390/cancers14215347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakvoort TB, et al. Pivotal role of glutamine synthetase in ammonia detoxification. Hepatology. 2017;65:281–293. doi: 10.1002/hep.28852. [DOI] [PubMed] [Google Scholar]
- 45.Surjan RC, Dos Santos ES, Basseres T, Makdissi FF, Machado MA. A proposed physiopathological pathway to hyperammonemic encephalopathy in a non-cirrhotic patient with fibrolamellar hepatocellular carcinoma without ornithine transcarbamylase (OTC) mutation. Am. J. Case Rep. 2017;18:234–241. doi: 10.12659/AJCR.901682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bauer J, et al. The oncogenic fusion protein DNAJB1–PRKACA can be specifically targeted by peptide-based immunotherapy in fibrolamellar hepatocellular carcinoma. Nat. Commun. 2022;13:6401. doi: 10.1038/s41467-022-33746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callaway E, Ledford H. Virologists who discovered hepatitis C win medicine Nobel. Nature. 2020;586:348. doi: 10.1038/d41586-020-02763-x. [DOI] [PubMed] [Google Scholar]
- 48.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 49.Wolchok J. Putting the immunologic brakes on cancer. Cell. 2018;175:1452–1454. doi: 10.1016/j.cell.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 50.Melenhorst JJ, et al. Decade-long leukaemia remissions with persistence of CD4+ CAR T cells. Nature. 2022;602:503–509. doi: 10.1038/s41586-021-04390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.1000 Genomes Project Consortium. et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barker K, Jarasz O. Online misogyny: a challenge for digital feminism. J. Int. Aff. 2019;72:95–114. [Google Scholar]
- 53.Lorenz, T. & Browning, K. Dozens of women in gaming speak out about sexism and harassment. New York Timeshttps://www.nytimes.com/2020/06/23/style/women-gaming-streaming-harassment-sexism-twitch.html (2020).
- 54.Bennett CF. Therapeutic antisense oligonucleotides are coming of age. Annu. Rev. Med. 2019;70:307–321. doi: 10.1146/annurev-med-041217-010829. [DOI] [PubMed] [Google Scholar]
- 55.Edmondson HA. Differential diagnosis of tumors and tumor-like lesions of liver in infancy and childhood. AMA J. Dis. Child. 1956;91:168–186. doi: 10.1001/archpedi.1956.02060020170015. [DOI] [PubMed] [Google Scholar]
- 56.Craig JR, Peters RL, Edmondson HA, Omata M. Fibrolamellar carcinoma of the liver: a tumor of adolescents and young adults with distinctive clinico-pathologic features. Cancer. 1980;46:372–379. doi: 10.1002/1097-0142(19800715)46:2<372::AID-CNCR2820460227>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 57.Malouf G, et al. Is histological diagnosis of primary liver carcinomas with fibrous stroma reproducible among experts? J. Clin. Pathol. 2009;62:519–524. doi: 10.1136/jcp.2008.062620. [DOI] [PubMed] [Google Scholar]
- 58.Abdul-Al HM, Wang G, Makhlouf HR, Goodman ZD. Fibrolamellar hepatocellular carcinoma: an immunohistochemical comparison with conventional hepatocellular carcinoma. Int. J. Surg. Pathol. 2010;18:313–318. doi: 10.1177/1066896910364229. [DOI] [PubMed] [Google Scholar]
- 59.Ross HM, et al. Fibrolamellar carcinomas are positive for CD68. Mod. Pathol. 2011;24:390–395. doi: 10.1038/modpathol.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torbenson M. Fibrolamellar carcinoma: 2012 update. Scientifica. 2012;2012:743790. doi: 10.6064/2012/743790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Graham RP, et al. Fibrolamellar carcinoma in the Carney complex: PRKAR1A loss instead of the classic DNAJB1–PRKACA fusion. Hepatology. 2018;68:1441–1447. doi: 10.1002/hep.29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirschner LS, et al. Mutations of the gene encoding the protein kinase A type Iα regulatory subunit in patients with the Carney complex. Nat. Genet. 2000;26:89–92. doi: 10.1038/79238. [DOI] [PubMed] [Google Scholar]
- 63.Hirsch TZ, et al. BAP1 mutations define a homogeneous subgroup of hepatocellular carcinoma with fibrolamellar-like features and activated PKA. J. Hepatol. 2020;72:924–936. doi: 10.1016/j.jhep.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 64.Singhi AD, et al. Recurrent rearrangements in PRKACA and PRKACB in intraductal oncocytic papillary neoplasms of the pancreas and bile duct. Gastroenterology. 2020;158:573–582.e2. doi: 10.1053/j.gastro.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vyas M, et al. DNAJB1–PRKACA fusions occur in oncocytic pancreatic and biliary neoplasms and are not specific for fibrolamellar hepatocellular carcinoma. Mod. Pathol. 2020;33:648–656. doi: 10.1038/s41379-019-0398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fritz, A.G. et al. (eds) International Classification of Diseases for Oncology: ICD-O (World Health Organization, 2013).
- 67.O’Neill AF, et al. Fibrolamellar carcinoma: an entity all its own. Curr. Probl. Cancer. 2021;45:100770. doi: 10.1016/j.currproblcancer.2021.100770. [DOI] [PubMed] [Google Scholar]