Summary
Myeloid cells, comprised of macrophages, dendritic cells, monocytes, and granulocytes, represent a major component of the tumor microenvironment (TME) and are critically involved in the regulation of tumor progression and metastasis. In recent years, single-cell omics technologies identified multiple phenotypically distinct subpopulations. In this review, we discuss recent data and concepts suggesting that the biology of myeloid cells is largely defined by a very limited number of functional states that transcend the narrowly defined cell populations. These functional states are primarily centered around classical and pathological states of activation, with the latter state being commonly defined as myeloid-derived suppressor cells. We discuss a concept that lipid peroxidation of myeloid cells represents a major mechanism that governs their pathological state of activation in the TME. Lipid peroxidation is associated with ferroptosis mediating suppressive activity of these cells and thus could be considered as attractive target for therapeutic intervention.
Introduction
Myeloid cells have evolved as a major component of innate and adaptive immunity fueled by their remarkable function diversity, migratory potential, and abundance in tissues. Their critical role in cancer is now well-established 1. In the vast universe of various myeloid cell types three terminally differentiated cell populations occupy the central place: dendritic cells (DC), macrophages, and granulocytes (primarily neutrophils). Monocytes are bone marrow (BM)-derived precursors that migrate to the tissue and differentiate there to macrophages (BM-derived macrophages, BMDM) and some DCs. In tissues, they are joined by the tissue-resident macrophages (TRMA), that are differentiated in early embryogenesis and are able to proliferate in situ2 (Fig. 1). The functional specialization of TRMA and BMDM is not clearly defined. The details of the origin, function of major populations of myeloid cells, and their contribution to tumor progression were discussed in multiple reviews. We refer readers to several most recent ones 3-8.
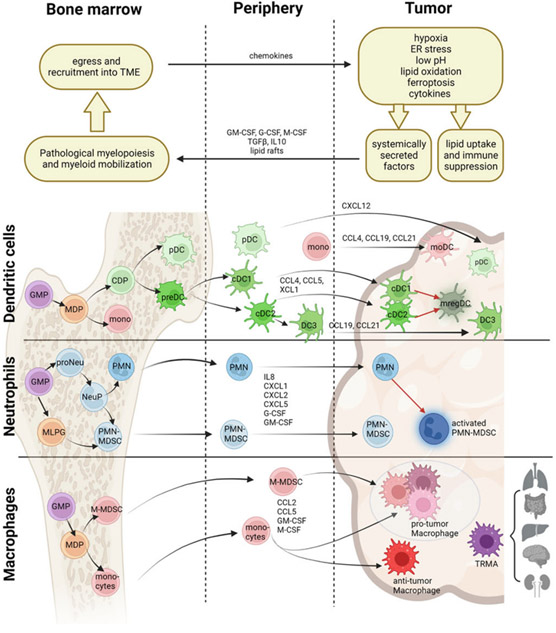
Figure 1. Impact of TME-derived pathological factors on lineage development, differentiation, and recruitment of myeloid cells.
Schematic illustration of lineage development and differentiation of myeloid cells and their identification in the bone marrow, circulation, and tumors. Pathological factors in the TME promote lipid uptake and immune suppression by myeloid cells, while also leading to secretion of systemic factors that impact myelopoiesis and mobilization of myeloid cells from the bone marrow. Figure illustrates how pathological factors impact the differentiation of dendritic cells, neutrophils, and macrophages into functional subsets, and subsequently affect their recruitment and/or differentiation into the tumor microenvironment. Black lines indicate lineage development while red lines indicate activated sub-states. GMP = granulocyte-monocyte percursor; MDP =monocyte-dendritic precursor; M-MDSC = monocytic myeloid-derived suppressor cell; CDP = DC precursor; preDC = pre-dendritic cells, committed precursor of DCs, pDC= plasmacytoid DC; cDC = conventional DC; mono = monocyte; moDC = monocyte-derived DC; mregDC = mature regulatory DC; MLPG = monocyte-like precursor of granulocytes; proNeu = pro-neutrophil; NeuP = neutrophil progenitor; PMN = polymorphonuclear cell; IM = intermediate monocyte; TRMA = tissue-resident macrophage. Created with Biorender.com
Classical/traditional role of myeloid cells is usually ascribed to several major functions: protection of organisms from various pathogens; tissue remodeling after injury or recovery from severe infection; stimulation of adaptive immunity; maintenance of homeostasis in selective tissues to limit potential damage (for example suppression of immune responses in BM, lung, placenta, newborns). The specific mechanisms of classical state of myeloid cell activation are well-characterized 9-11. During the last 20 years, understanding of the alternative state of myeloid cell-activation has emerged, referred to as pathological state of activation. This state is the result of continuous stimulation of myeloid compartment in various pathologic conditions including cancer. Neutrophils and monocytes in this state of activation are termed myeloid-derived suppressor cells (MDSC): PMN-MDSC – to define polymorphonuclear cells that belong to granulocytic lineage and M-MDSC – to define monocytic cells. In recent years, similar term was also applied to macrophages with strong immune suppressive activity. Pathologically activated myeloid cells have several major features: 1) potent suppressive activity of various functions of T cells, natural killer (NK) cells, and B cells; 2) rather distinct transcriptional and proteomic profile, biochemical characteristics, and some phenotypic markers that allow their separation from classically activated neutrophils and monocytes; 3) support metastasis via production of cytokines that promote tissue remodeling and angiogenesis; 4) the functional separation from classicaly activated neutrophils and monocytes is observed on the stage of the precursors in bone marrow but it becomes much more prominent in tissues; 5) although most of MDSC are relatively immature cells, this is not their defining characteristic since mature cells may have the full potential of MDSC. The details of the biology of these cells are described elsewhere 4,9,12 and will not be discussed in this review. Here, we will focus on emergeing issues of functional diversity of myeloid cells in cancer and on the concept that functional specialization of various myeloid cells in cancer can be governed by the state of lipid oxidation.
Transcriptomic clustering and the functional states of myeloid cells
Heterogeneity of myeloid cells and their capacity for plasticity has been a longstanding challenge in dissecting the role of myeloid cells in cancer. The advent of single-cell technologies has transformed the field and enabled tracking the ontogeny, phenotypic characteristics, and potential relevance of various subpopulations of cells. Multiple clusters of myeloid cells were described in many publications 3,4,13,14. However, in the absence of functional characterization, the biological relevance of various myeloid single-cell clusters remained unclear. The question has arisen, do these clusters truly reflect the functional state of myeloid cells? It appears that the answer depends on the type of cells.
Dendritic cells.
Perhaps the best classified myeloid population is DC, for which distinct subpopulations were described prior to single-cell technologies 6. We will focus on the common DC clusters observed in the tumor microenvironment (TME). Single-cell clustering has largely confirmed separation of DCs into the major subsets: classical cDC1, cDC2, plasmacytoid pDC, and the newly identified DC3. These populations have distinct functional characteristics, which helps to place the functionality of these cells in the context of antitumor immune response. For instance, cross-presenting activity of cDC1 is directly implicated in generation and support of antitumor immune responses 15. It is not surprising that these populations are reproducibly identified in different tumor types. Cheng et al. applied single-cell RNA-sequencing (scRNA-seq) to pan-myeloid cells isolated from 210 patients spanning 15 solid tumor types, and identified 4 phenotypic clusters of DCs observed across all 15 tumor types: pDCs, CLEC9A+ cDC1, CD1c+ cDC2, and LAMP3+/CCR7+ cluster they aligned with an activated and migratory phenotype 16. A similar pan-cancer approach spanning 20 patients from 3 distinct indications described the same clusters 17, although they also identified a distinct 5th cluster of DCs that occurred largely in lung cancer samples and was closely related to cDC2 cells. Zillionis et al. dissected DCs into the same 4 clusters in a single-cell analysis of 7 non-small cell lung cancer (NSCLC) patient samples, and furthermore found a clear conservation of these 4 DC subsets in mouse tumors obtained from a syngeneic KP1.9 lung adenocarcinoma model 18. Remarkably all four DC subsets hierarchically clustered together independent of type of tumor, treatment status, or stage 16,18, suggesting a developmental determination rather than environmental polarization defining these populations. While the function of cDC1, cDC2 and pDC is indisputable, the LAMP3+/ CCR7+ cluster could track back to several phenotypes, as these markers have been used to describe DC3 19, mature regulatory DC (mregDC) 20, and inflammatory DCs 21. DC3, characterized as CD163+, CD1c+, are uniquely related to cDC2 22, yet pan-cancer analysis found that the LAMP3+/CCR7+ population emerges from both cDC1 and cDC2 subtypes 16,17, and lacks the characteristic marker phenotype of DC3. Instead, this population better resembles regulatory mregDC, an activated form of both cDC1 and cDC2 20. Deeper interrogation of the cDC2 population across 8 indications identified two minor subclusters within cDC2 with phenotypic characteristic of DC3 (CD5−/low, CD163+). And elsewhere, a significant DC3 cluster was found alongside cDC1, cDC2, and mregDC from a set of 35 NSCLC samples, that was not only the most prevalent DC cluster in these samples, but also considerably enriched in tumor samples over matched normal 23. Taken together, the patterns show consistency in how single-cell technologies are able to resolve clear phenotypic and functional subsets of DCs, in line with known characterization of these subtypes. However, the different picture has emerged in respect of macrophages and neutrophils.
Monocytes and macrophase.
Despite evidence that historically defined “M1” macrophages associate with good clinical outcome while “M2” macrophages have the opposite effect 24,25, the convention of breaking down macrophages into two highly polarized states in TME did not find strong experimental support and now is replaced with the concept of a continuous gradient of monocyte and macrophage functional states. Single-cell technologies supposed to shed light on the plasticity of macrophages. Diverse macrophage clusters were indeed identified, but most subsets were restricted to the specific tissues or patients 16,17, demonstrating the susceptibility of macrophage phenotype to the microenvironment. In one instance, where 12 distinct macrophage clusters were identified, only 5 could be reproducibly identified in >30% of samples tested 16. A lack of phenotypic overlap between mouse and human lung cancer macrophages further supports this notion. Some common features were identified within macrophage subsets spanning diverse tumor indications. One example is the identification of SPP1+ macrophages. SPP1 is an integrin binding sialoprotein that is frequently upregulated in cancer and associated with pro-tumoral traits of macrophages across many independent reports 26-28. However, the largest pan-cancer analysis found distinct SPP1+ macrophages only in about half of the indications examined 16. However, upon closer examination they discovered that the indications lacking SPP1+ macrophages instead had the populations that shared the same angiogenesis signature as the SPP1+ cluster, yet come up with different phenotypic markers (INHBA, VCAN, or FN1). This demonstrates the fluidity of markers across populations. C1QC+ macrophages are another cluster identified in many single-cell studies 16,29 and is generally separated from the aforementioned SPP1+ cluster by functional signatures (phagocytosis for the former vs. angiogenesis for the latter). However, follow-up studies found instances of clear overlap between these two markers 30,31, yet functional significance of this double positive subset termed ‘lipid-associated macrophages” in breast cancer, tracks back to the phagocytosis signatures of the C1QC+ cluster 31 rather than the angiogenesis signatures of the SPP1+ cluster. Thus, separating single-cell clusters based on a subset of genes that show the highest upregulation may steer incorrectly into a perception of phenotypic diversity that may not exist.
While diversity based on transcriptomics may be hard to capture for macrophages, functional diversity appears more conserved. Regardless of cluster annotation, functional signatures attributed to clusters frequently return functional macrophage properties of angiogenesis, tissue remodeling phagocytosis, and inflammatory activity16,30,32,33. These functional features often cannot be ascribed to an individual cluster, and, in fact, have been shown to occur across multiple macrophage clusters simultaneously. For example, in breast cancer, some degree of angiogenesis activity was ascribed to both VCAN+ and SPP1+ macrophages, while phagocytosis activity in lung cancer stemmed from both C1QC+ and PPARG+ macrophages 16. And in several instances, the same cluster returned equal degrees of both angiogenic and phagocytic activity, as seen with ISG15+ macrophages in thymic cancer and IL1B+ and GPNMB+ macrophages in renal cancer16. Plasticity of macrophage function in cancer mirrors the native role for myeloid cells, which can acutely function in a pro-inflammatory capacity in host defense, but switch during resolution phase. By turning on angiogenesis, phagocytosis, and immune suppression pathways, myeloid cells mediate wound healing responses and dampen the pro-inflammatory cascade to protect the host from autoimmunity 34,35. Thus, it is not surprising that this functional conservation trumps their phenotypic characterization in single-cell dissections. An ambitious meta-analysis across healthy and diseases tissues found conserved monocyte-to-macrophage programming across cancer and several inflammatory diseases33, further highlighting the importance of conservation of these functional traits.
The path for monocyte programming towards functional endpoints is likely influenced by the localization. A consistent phenotypic and functional distinction between intratumoral and stromal CD14+ cells was found across 20 melanoma metastases obtained from different anatomical sites. This distinction was not observed for intratumoral vs. stromal T-cells36. Furthermore, stromal CD14+ cells were enriched for a CD14/CD2/LY75 signature that might suggest antigen presentation functionality.This signature correlated with better clinical outcome in several types of cancer (melanoma, DLBCL, sarcoma, adrenal carcinoma)36. Similar findings were reported in the PyMT breast cancer model where tumor-associated macrophages (TAMs) developed different phenotypic and functional profiles depending on whether they localized to the stroma or ductal niches37.
Neutrophils and PMN-MDSC.
In the context of cancer, neutrophils have taken on a binary reputation, with tumor-associated neutrophils largely classified as pro-tumor, yet recognition given that neutrophils can also have anti-tumor functionality. The ontogeny of this distinction has been characterized in a framework of a pathologically activated PMN-MDSC 9. However, single-cell resolution of circulating and tumor-infiltrated neutrophils has resolved up to 5 unique clusters in cancer patients. We interrogated the subclustering of peripheral and tumor neutrophils from LL2 tumor-bearing mice, and identified 3 distinct neutrophil populations in the tumors: PMN1, PMN2, and PMN3, while the peripheral neutrophils from either tumor-bearing or non-tumor bearing mice only clustered into PMN1 and PMN2 38. Based on transcriptional signatures and ex vivo functional analysis, PMN1 appeared to fit characteristics of classical neutrophils, while PMN2 and PMN3 were pro-tumor, immunosuppressive PMN-MDSC. However, PMN3 cells were largely present in tumors and not in spleen of mice, while PMN2 are mostly present in spleens. Trajectory analysis and functional experiments suggested that those populations derived not from one another but from PMN1 and skewed by environmental cues. Sagiv et al. postulated from the studies in 4T1-tumor bearing mice that the low density fraction of neutrophils consisted of PMN-MDSC and classical neutrophils that had degranulated in response to tumor cues into the low density fraction 39, thus mirroring the findings with PMN2 and PMN3. Irrespective of origin, neutrophils in the low density fraction had the same T-cell suppressive function.
Zillionis et al. identified 5 distinct clusters of tumor infiltrated neutrophils in human NSCLC with conservation of these five clusters, along with identification of a mouse-specific 6th, in mouse KP1.9 lung tumors. In mice, mN1 and mN2 enriched in healthy lung tissue, while the remaining four clusters were specifically enriched in tumors, of which mN4-mN6 corresponded to elevated Siglec-F expression previously found to align with pro-tumor functions 18. mN1, mN3 on the other hand tracked with characteristics of classical neutrophils, whereby N2 transcriptionally diverted with a signature of type-I interferon responsiveness. A similar delineation occurred in human tissues, where hN1 and hN3 were transcriptionally similar, tracked closest with classical neutrophils markers, and associated with better prognosis in NSCLC patients than the other subsets. hN5 on the other hand tracked with worst overall prognosis, was found to be tumor specific, and presented phenotypic signatures known to be associated with pro-tumor function. Blood samples lacked a compatible hN5 subset, therefore suggesting that N5 was likely similar to the tumor-derived PMN3 population defined in our study. Like mN2, hN2 was a distinct subset from the remaining clusters that presented with a distinct type-I interferon signature and, like N5, associated with poorer outcome. Perplexingly, this N2 subset was enriched in healthy mouse lung over tumor tissue, suggesting that it did not emerge from pathological activation. Furthermore, the association of type-I interferon response with anti-tumor neutrophils 40,41, postulates that this N2 subset could be aligned with anti-tumor neutrophil function. Ultimately, while 5 distinct human neutrophil clusters were identified in this work, further interrogation coalesced them down to 3 functional states, however unlike the two pro-tumor and one anti-tumor functional divisions identified in other studies, the subsets described here likely break down to two potential anti-tumor fractions. Further analysis would be needed to understand whether the ISG15+ N2 population originates alternatively from classical neutrophils or merely represents an activated state.
The concept of tumor-associated PMN-MDSC was further strengthened by two recent reports. Analysis of 124 liver cancer samples revealed the emergence of distinct neutrophil clusters in the tumors that did not overlap with phenotypes found in peripheral blood or normal adjacent liver42. Moreover, the neutrophil clusters emerging in hepatocellular carcinoma (HCC) patients differed in phenotype and number from those in intrahepatic cholangiocarcinoma (ICC)42, supporting the hypothesis that pathological activation of neutrophils is strongly influenced by TME cues. Mouse models of HCC and ICC recapitulated the emergence of tumor-specific neutrophils that histologically clustered back to their human counterparts42. A retrospective analysis of NSCLC samples across 29 original datasets identified 4 tumor-associated neutrophil (TAN) clusters that differed from the 3 normal-adjacent tissue neutrophils (NAN), with upregulation of genes closely associated with previously identified PMN-MDSC signature: OLR1, VEGFA, CXCR4, and CD83, across all 4 TAN subsets43. NANs broke down into two similar clusters with classical neutrophil traits and a third NAN-3 cluster enriched for ISG genes, perhaps similar to the distinct ISG+ N2 cluster found previously in NSCLC18, with trajectory analysis revealing the NAN-3 cluster to be a starting point for other two NAN clusters 43. Trajectory analysis demonstrated a close link between TAN clusters supporting the importance of pathological TME in shaping the tumor-specific neutrophil phenotype.
An unbiased approach to resolve M-MDSC and PMN-MDSC clusters also identified both subsets separately from classical monocytes and neutrophils, and found common genes related to immunosuppression (eg. Arg2, Il1b, Cd84) in the signatures identifying these subsets 44. This further supports the necessity to analyze single-cell clusters based on functional features rather than markers, as the existence of a universal state of pathological activation may be similar across various myeloid cells, and may not become apparent otherwise. Interestingly, in contrast to macrophages, neutrophils and monocytes, functional state associated with potent immune suppression was not convincingly demonstrated for DCs. It is tempting to speculate that lack of such functional dichotomy in these cells could be a foundation of rather unique stiability of DC clustering in TME.
Conditioning of pathological state of myeloid-cell activation in the TME
Stepping back to the concept of classical vs. pathological activation of myeloid cells helps to further understand the origins of the diversity described above. MDSC emerge from alterations in BM development programs. In tumor-bearing mice, the gradient of functional activity is evident in different tissue compartments: from poorly suppressive/activated MDSC in BM to more potent MDSC in spleen, even more potent cells in non-tumor tissues with maximum activity of MDSC observed in tumors 45. In cancer patients, similar gradient exist between peripheral blood and tumor MDSC. These data suggested that the functional fate of MDSC and, by extension, macrophages is shaped by tissue microenvironment. Following-up on the above described PMN2 and PMN3 populations 38, PMN3 functionally resembled PMN2, and based on enrichement of pro-inflammatory and immune suppressive factors and can be characterized as activated PMN-MDSC. These cells have much stronger suppressive activity than PMN2 cells, thus likely representing a pathologically activated fraction of PMN-MDSSC in tumors. Several factors were implicated in conversion of PMN and monocytes to PMN-MDSC and M-MDSC in tumor tissues. Among them are high concentrations of GM-CSF, ER stress, acidosis, high level of IL-6 and other cytokines 38,40,46. A cluster of tumor infiltrated neutrophils in human NSCLC that was not present in blood was identified 18, suggesting that this cluster also arose from pathological activation in the tissue. By tracing similar occurrence of myeloid clusters that emerge only in the tumor, one can identify similar sets of pathologically activated monocytes or macrophages. A tumor-specific macrophage cluster, designated as MMP9+, emerged from a single-cell survey of lung, colorectal, and ovarian cancer 17. This functional type develops along a monocyte-to-macrophage pathway, of which pseudotime precursors were also detected in paired normal tissue. However the TME apparently induced further evolution of the cells into this tumor-specific macrophage cluster. Interesting observations from myeloid phenotypes in brain metastasis vs. gliomas further highlights the importance of pathological cues impacting myeloid diversity. It was reported that while TAMs in gliomas were largely linked to tissue-resident microglia, TAMs in brain metastatic lesions of melanomas and carcinomas had a more significant proportion of monocyte-derived macrophages 47. Thus differences in pathological cues clearly dictated the ontogeny of TAMs. Perhaps not surprisingly, clusters of myeloid cells uniquely present in tumors are generally associated with functional attributes that support the tumor progression.
However, conditioning of myeloid cells in the TME may not necessarily reflect the true nature of MDSC development. Using an in vitro experimental system where macropages were differentiated from monocytes or M-MDSC from the same cancer patient, we demonstrated that the transcriptional profile of macrophages closely correlated with the type of monocytes these cells were differentiated from 48. Moreover, macrophages generated from M-MDSC were more imunosuppressive than macrophages generated from moncoytes. These results were reproduced in mouse experimental models in vivo and after adoptive transfer of monocytes or M-MDSC to tumor-free recipients. The effect was dependent on increased expression of S100A8/S100A9 proteins in immue suppressive macrophage. A strong link of S100A9 expressing macrophages with negative clinical outcome in patients with head and neck cancer was found 48. Another suggestion that conditioning of myeloid cells prior to migration to the TME is important for the development of potent MDSC, comes from experiments where neutrophils from healthy donors were converted to immune suppressive PMN-MDSC. Activation of neutrophils with various cytokines and LPS failed to generate immune suppressive PMN-MDSC 49. Strong activation of cells with ER stress inducer was required. Up-regulation of the genes associated with ER stress, was one of the very few features of BM neutrophils in mice with early stage tumors that distinguish them from BM neutrophils in tumor-free mice 50. It is possible that conditioning of neutrophils in the BM facilitates their transition to fully functional PMN-MDSC once these cells migrate to the tissues.
One important caveat that needs to be considered is the fact that many transplantable mouse models have a strong inflammatory component with a large number of MDSC, a topic we discussed recently45. It can be illustrated further in recent study where autochthonous tumors arising in the spontaneous PyMT tumor model demonstraed development of distinct stromal vs. ductal TAM programs, whereas orthotopic transplantation of the PyMT tumors into the mammary fat pad of recipient mice yielded only one main TAM differentiation path 37. Markedly different results in monocyte compartment were found between the MMTV-PyMT breast carcinoma model and human breast cancer patients51. Whereas mouse breast cancer was found to influence down-regulation of interferon (IFN) signaling in the monocyte compartmentmonocytes actually demonstrated an upregulation of IFN signaling in cancer patients samples51.These results reiterate a need to use different types of tumor models as well as samples from cancer patients to increase translatability of the results.
Immune suppressive state of activation of myeloid cells is associated with multiple mechanisms including various reactive oxygen (ROS) and nitrogen (RNS) species (superoxide radical, hydroxyl radical, peroxynitrite, nitric oxide), ER stress, immune suppressive cytokines, arginase I, immune suppressive ligands, etc. 9. These mechanisms are usually distinct between PMN-MDSC, macrophages, M-MDSC 52. These and other mechanisms of pathological myeloid cell activation were explored therapeutically 45,53. In recent years, evidence emerged that a of common mechanism exists that transcends various types of immune suppressive cells. These mechanisms involve oxidized lipids and may play a dominant role in regulation of the function of these cells.
Lipid peroxidation in the TME
Growing cancer cells create a microenvironment with a metabolic landscape that supports their proliferation, migration and simultaneously impairing antitumor immunity54. One of the main signaling molecules used for these purposes are lipids. Lipid molecules contain a hydrophilic polar head and a hydrophobic nonpolar tail (Fig. 2A). Lipids spontaneously aggregate in the aqueous environments to yield micelles, bilayers, or vesicles through entropy-driven hydrophobic interactions 55. There are several dozen types of lipids of different classes and an average animal cell may contain close to 109 lipid molecules in its membranes. The most abundant lipid species in cell membranes are phospholipids (PL) 56. Their polar heads are exposed on the membrane surface and linked, via a glycerol backbone, to two fatty acid (FA) residues which constitute the membrane hydrophobic core. FA may be saturated or contain one or more – usually up-to six, cis-double bonds. The latter are called polyunsaturated fatty acids (PUFA) (Fig. 2A). FA may contain from 14 to 26 carbons and their double-bonds are localized in methylene interrupted arrangements starting from the 3rd or 6th carbon thus representing two co-3 and co-6 major families. PUFA-PL are of particular importance due to their ability to undergo free radical-mediated peroxidation (Fig. 2B, C).
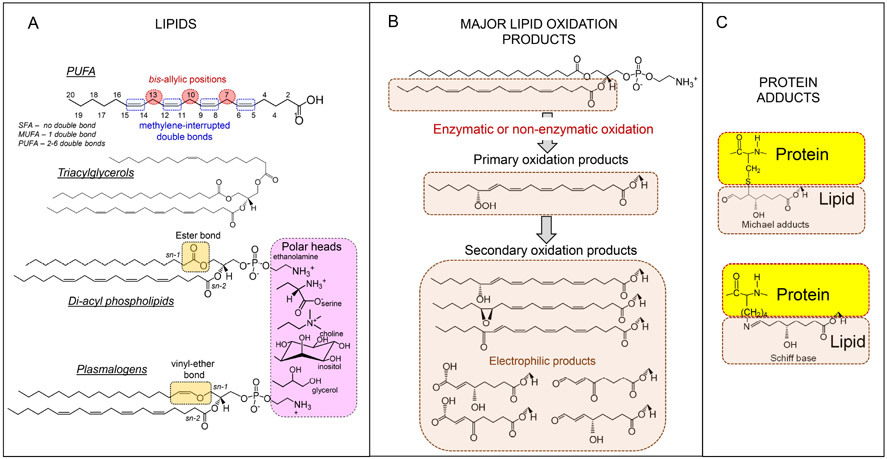
Fig. 2. Polyunsaturated fatty acids (PUFA) lipids and their peroxidation products formed in cells and tissues.
A: structure of different classes of lipids. There are two stereo-specific positions in phospholipid (PL) molecules, sn-1 and sn-2, at which FA can be covalently linked to glycerol. Typically, PLs contain PUFA at the sn-2 position whereas saturated and mono-unsaturated FAs are localized at the sn-1 position and polar head-groups attached at the sn-3 position. In plasmalogens, fatty acids in sn-2 position are connected to glycerol backbone via a vinyl-ether bond. Abbreviations: SFA = satuated fatty acids; MUFA = monounsaturated fatty acids B: Peroxidation can occur as a random chemical reaction or as a enzymatically-catalyzed process in which only a selective group of PUFA-PLs are modified 109. While the former random process is characteristic mostly of disbalanced redox conditions in stressed myeloid cells, the latter represents a physiological genetically controlled reaction. In myeloid cells, peroxidation produces the primary hydroperoxy-PUFA, which can undergo additional redox reactions to yield secondary oxidation products containing keto-, epoxy and hydroxy- functionalities. Alternatively, the hydroperoxy-PUFA-PLs can undergo beta-scission to form oxidatively truncated electrophilic products with shortened hydrocarbon chains.
C: The electrophilic products with shortened hydrocarbon chains can form adducts with several nucleophilic sites in proteins including histidines, cysteines and lysines to form Michael adducts and Schiff bases.
In most solid tumors, the TME is enriched for lipids, which facilitates tumor cell growth and immune escape 57. Profound metabolic reprogramming of tumor cells via enhanced aerobic glycolysis, gene mutations in the tricarboxylic acid (TCA) cycle enzymes, and upregulation of de novo lipid synthesis and glutaminolysis, are pivotal to the development and maintenance of the malignant phenotype of tumor cells 58 and there is a fierce competition between immune cells and tumor cells for nutrients to provide for adaptive survival strategies at the expense of compromised immune functions59.
De novo FA synthesis, uptake of exogenous FAs from the TME and enhanced fatty acid oxidation (FAO) are the features related to the immunosuppressive activity of immune cells. FAO is a catabolic process during which FA are converted into acetyl-CoA, a TCA cycle substrate, where it gets oxidized and, through the interactions with the mitochondrial respiratory chain and oxygen consumption, produces ATP. Recent studies indicate that tumor cells rely on FAO for proliferation, survival, sternness, drug resistance, and metastatic progression. Moreover, FAO is also re-wired in TME immune cells leading to immune suppression 58. In fact, it appears that transition from acute to chronic hypoxia, typical of tumor growth, shifts the cellular respiration from being predominantly pyruvate-based, to FA-oriented oxidation and electron flow (in parallel with enhanced glutamine oxidation) 60.
Both mitochondria and peroxisomes can degrade FA chains. The mitochondrial β-oxidation system feeds the oxidative phosphorylation pathway for ATP synthesis. Very-long-chain FA and long-chain dicarboxylic acids, are exclusively processed by the peroxisomal β-oxidation, whereas other common long-chain FA are oxidized by mitochondria61. If activated mitochondrial FAO “burns” predominantly saturated and monounsaturated FAs with the chain lengths of 14-18 carbons, it is logical to expect that longer chain (C20-C26) polyunsaturated FAs metabolized predominantly by peroxisomes would accumulate in the TME and its cells. This is important because these PUFA-phospholipids may be utilized for the production of signals catalyzed by lypoxygenase (LOX)- and cyclooxygenase (COX)-driven oxygenation reactions leading to the formation of eicosanoids (C20:4), docosapentanoids (C22:5) and docosahexanoids (C22:6) (Fig. 3). Among these oxygenated PUFA are – important immune-suppressive signals (eg, prostaglandin E2, PGE2) that may be generated by different myeloid cells in TME 62. The high activation of mitochondrial FAO vs. much slower peroxisomal oxidation of long-chain and very long-chain PUFA creates conditions for their peroxidation, which along with the avid uptake of PUFA, peroxidized PUFA and peroxidized PUFA-PL, lead to their accumulation in the myeloid cells.
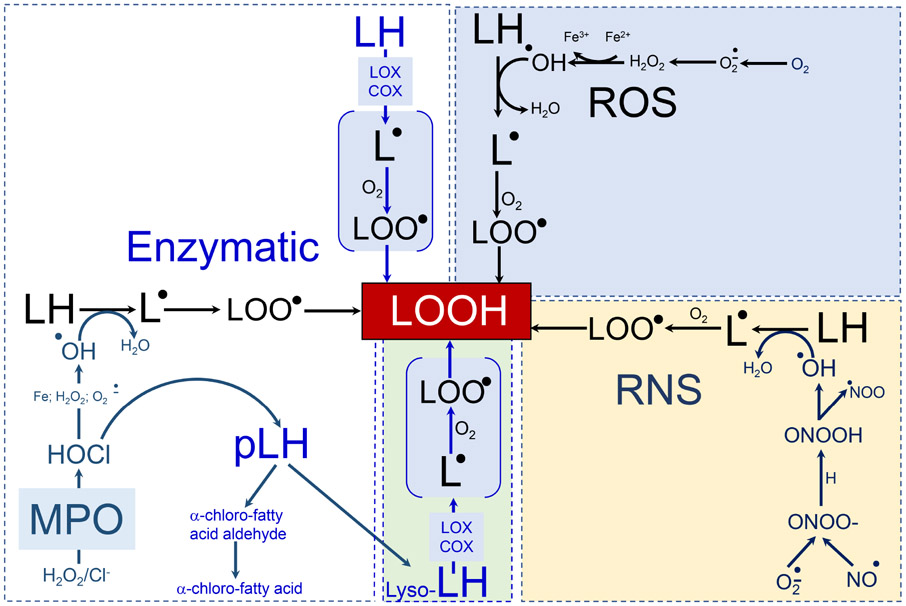
Figure 3. Enzymatic and non-enzymatic (phospho)lipid peroxidation.
White box: the most common enzymatic processes generating lipid hydroperoxides in myeloid cells followed by the formation of secondary peroxidation products that take place in myeloid cells and affect their ability to regulate function of other cells. 15LOX can directly oxidize PUFA-containing (phospho)lipids to form the respective lipid hydroperoxides (including pro-ferroptotic signals). MPO-catalyzed production of hypochlorite can react with superoxide, H2O2 or iron to yield hydroxyl radicals and initiate lipid peroxidation. MPO/HClO can hydrolyze the plasmalogen vinyl bond to yield 2-sn-PUFA-lysophospholipids which can be further oxidized by 15LOX to generate lyso-phospholipid hydroperoxide (green box). Blue box: the free radical reactions of lipid peroxidation. Superoxide formed by NADPH oxidases, xanthine oxidase, and the mitochondrial electron-transport chain can dismutate to generate hydrogen peroxide. Hydroxyl radicals produced in the Fenton reaction abstract hydrogen from the bis-allylic positions of PUFA-lipids and create carbon-centered radical that reacts with molecular oxygen to form a lipid-peroxyl radical. The latter can abstract another hydrogen to generate lipid hydroperoxide. Nitric oxide readily reacts with superoxide to generate peroxynitrite homolytic decomposition of which yields hydroxyl radicals and also initiates lipid peroxidation observed in myeloid cells (yellow box).
Abbreviations: ROS = reactive oxygen species; RNS = reactive nitrogen species; 15LOX =15-lipoxygenase; COX = cyclooxygenase; MPO = myeloperoxidase; H2O2 = hydrogen peroxyde; HOCl = hypochlorous acid; NO = nitric oxide; Fe = iron; Lyso = phospholipids with one of two fatty acids removed; LH= non-oxidized lipid from which a bis-allylic hydrogen can be abstracted; pLH = plasmalogen form of lipid; L = lipid radical; LOO = lipid peroxyl radical; LOOH = lipid peroxide; HO = hydroxyl radical.
Physiologically, the majority of oxygenated free PUFA acting as lipid mediators are generated by COXs and LOXs. However, PUFA-PL are poor substrates for COX, although some of the LOXs – 15LOX, 12LOX – can also utilize PUFA-PL as their substrates 63. Transition metals and their redox-active low molecular weight complexes may be invoveld in the catalysis of lipid peroxidation. However, non-enzymatic lipid peroxidation cannot selectively generate stereospecific oxygenation products which are usually required for the signaling function.
Lipid peroxidation and cell death
Perhaps one of the most important signaling mechanisms by oxygenated PL is associated with different types of regulated cell death. It has been shown that peroxidation of a unique mitochondrial phospholipid, cardiolipin, is a required stage in intrinsic apoptosis. In pyroptosis, peroxidation of cardiolipin participates in the formation of NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome patform. Necroptosis is associated with peroxidation of phosphatidylcholine 64-67. Selective and stereospecific oxygenation of PUFA-PL play central role during ferroptosis 68. Execution of ferroptosis is triggered in cells with impaired redox regulation whereby excessive availability/activity of redox-active iron, induces lipid peroxidation that cannot be adequately controlled by the thiol regulation, particularly by Glutathione peroxidase 4/glutatione GPX4/GSH system. Specific for ferroptosis are oxidation products generated from PUFA-phosphatidylethanolamine (PUFA-PE), particularly those emerging from oxidation of arachidonoyl (C20:4)-PE and adrenoyl (C22:4)-PE 64 (Fig. 4). It has been demonstrated that 15LOX, is a redox catalyst of PUFA-PE peroxidation. The formation of pro-ferroptotic signals requires the co-participation of yet another protein, PE-binding protein 1 (PEBP1) exerting allosteric control over selectivity and specificity of the 15LOX-catalyzed reaction and leading to the accumulation of sn-2-15-hydroperoxy-arachdidonoyl-PE 69.
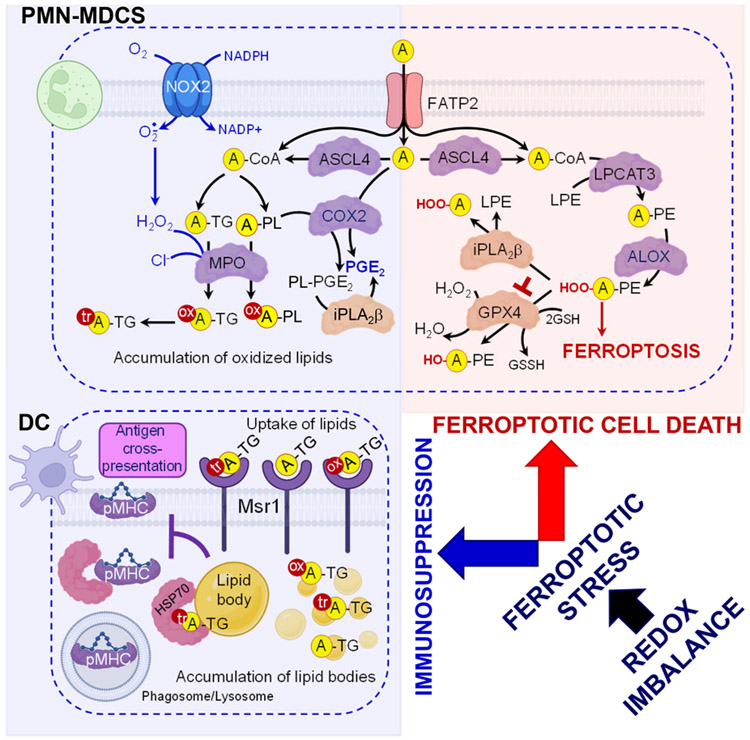
Figure 4. Redox imbalance causes ferroptotic stress which can culminate in ferroptotic death in PMN-MDCSs.
The initiation of the ferroptotic program in redox-imbalanced cells results in the accumulation of polyunsaturated fatty acids- phospholipid (PUFA-PL) peroxidation products. FATP2 transports exogenous arachidonic acid from the TME to PMN-MDCS, and together with ACSL4 and FATP2, contributes to synthesis of PUFA-PL and the production of immunosuppressive signals in PMN-MDSC. Oxidized products specific for ferroptosis can be eliminated via an enzymatic action of either iPLA2β that hydrolyzes sn-2 hydroperoxyl (HOO)-PUFA-residue from hydroperoxy-PE species or by the reduction of HOO-PUFA-PE by GPX4. AA and its COX-catalyzed metabolite, PGE2, are mainly responsible for the immunosuppression. PGE2 can be presented in two forms – as a free oxygenated AA and also as PGE2-esterified into PE. PLA2-catalyzed hydrolysis of PGE2-PE may be required to release immunosuppressive free PGE2. Oxidized and oxidatively-truncated triacylglycerols are produced by PMN-MDSC in a MPO-catalyzed reaction and can be taken-up by dendritic cells. Accumulated oxidatively truncated forms of triacylglycerols prevent trafficking of pMHC from the phagosome/lysosome to the cell surface. These DCs are not able to effectively stimulate antigen-specific T cells due to suppressed antigen cross-presentation. Abbreviations: PMN-MDSC = polymorphonucler myeloid-derived suppressor cells; DC = dendritic cell; FATP2 = fatty acid transport protein 2; ACSL4 = Long chain acyl-CoA synthetase isoform 4; ALOX = arachidonate 15-lipoxygenase; GPX4 = Glutathione Peroxidase 4; GSSG = oxidized glutathione; MPO = myeloperoxidase; NOX2,= NADPH oxidase 2; COX2 = cyclooxygenase-2; iPLA2β = calcium-independent phospholipase A2β; PL = phospholipid; A = arachidonic acid; TG = triacylglycerol; PE = phosphatidylethanolamine; HO = hydroxide; H2O2, = hydrogen peroxide; LPE = lyso-phosphatidylethanolamine; PGE2 = prostaglandin E2; PE-PGE2 = esterified prostaglandin E2; tr = truncated; ox = oxidized; HSP70 = heat shock protein 70; pMHC = MHC class I molecule. Created with Biorender.com
Tumor cells, especially of pro-metastatic mesenchymal type, have increased sensitivity to ferroptosis 70-72. Ferroptosis has been discovered during search of new anti-cancer modalities alternative to pro-apoptotic agents 73. Therefore, ferroptosis was utilized as a potential therapeutic target in cancer therapy using various chemical inhibitors of GPX4 or antiporter system xc-.Strong antitumor effect was observed in vitro and in xenograft systems in vivo 74-81. However, these studies did not take into account contribution of the TME. Wang et al. has demonstrated that genetic manipulation of tumor cells in vitro to decrease their sensitivity to ferroptosis made them less sensitive to IFN-γ and cytotoxic T cells . In contrast, treatment of mice with cysteinase, a ferroptosiss inducer, sensitizes tumor cells to IFN-γ and synergizes effectively with immune checkpoint blockade (ICB) 82. In other words, the suggested mechanism of ferroptosis induced by CD8+ T cells has been primarily ascribed to insufficiency of the thiol (GSH) control of lipid peroxidation. However, other data suggested a more complex role of ferroprosis in cancer. Recent data showed that ferroptosis in tumor infiltrating CD8+ T cells had tumor promoting effects due to decreased T-cell survival and impaired T-cell function 83-84. Increased expression of CD36 in tumor-infiltrating CD8+ T cells in the TME stimulates uptake of FA followed by lipid peroxidation and ferroptosis associated with tumor progression and poor survival in human and murine tumor cells 83. It was further established that elevated levels of oxidized lipids in the TME enhanced expression of CD36 causing uptake and accumulation of oxidized lipids (oxidized LDL) and enhanced lipid peroxidation associated with inhibition of CD8+ T cell effector function preventable by over-expression of GPX4 84.
Oxidized lipids and ferroptosis of myeloid cells in cancer
A recent study clarified the role of ferroptosis in the TME by focusing on myeloid cells, specifically PMN-MDSC. It revealed that ferroptosis does not commonly occur in BM or spleen neutrophils from tumor-free or tumor-bearing mice. However, the active ROS/RNS-generating machinery of PMN-MDSC shatters redox balance and stimulates a predominantly ferroptotic death of the majority of these cells (Fig. 4). This process was controlled by hypoxia-inducible down-regulation of GPX4 85. The death of these immune-suppressive cells may be viewed as a potentially anti-cancer mechanism eliminating the important components of the immune-suppressive TME cell populations. However, this simplistic view was incorrect. Ferroptotic PMN-MDSC demonstrated potent immune supprsssive activity. Moreover, induction of ferroptosis was sufficient to convert neutrophils to immune suppressive PMN-MDSC. In addition, ferroptotic PMN-MDSC released lipid peroxidation products that directly suppress T-cell function. Those products could cause ferroptosis of macrophages that in turn became immune suppressive, thereby spread the wave of immune suppression from one cell to another 85. This concept is in line with the observation demonstrating that oxidized lipids can be transferred from PMN-MDSC to DCs and blunt their ability to cross-present antigens 86. Treatment of immune competent tumor-bearing mice with ferroptosis inhibitor, liproxstatin-1, had anti-tumor activity and the effect of immunotherapy was substantially enhanced 85. In contrast, treatment of mice with a ferroptoss inducer, IKE, promoted tumor growth indicating that in immune competent hosts, ferroptosis in the TME is largely a tumor-promoting force. In neutrophils, immune suppression associated with ferroptosis induction and actual cell death were separated in time. The former occurred much earlier (withn hours after induction of ferroptosis process) whereas the latter take place at least 24 hours later. Whether ferroptosis can be reversed at the stage of immune suppression remains unclear.
Ferroptosis of PMN-MDSC in TME is supported by the accumulation of readily oxidizable PUFA-PL, which can be peroxidized by intracellular pro-oxidant machinery to generate ferrpototic signals as well as additional peroxidized (phospho)lipid species with immunosuppressive propensities. Two enzymes, acyl-CoA synthetase long chain family member 4 (ACSL4) and fatty acid transport protein 2 (FATP2), can deliver, redistribute and activate PUFA to acyl-CoA PUFA 87,88. Both of them are involved in the regulation of ferroptosis and production of immunosuppressive signals in PMN-MDSC 85,89. FATP2 and ACSL4 are highly expressed in ferroptosis sensitive PMN-MDSC and genetic deletion of the genes and their products results in the enhanced tolerance to ferroptosis and immunosuppressive phenotype 85 (Fig. 4). Redox lipidomics analysis discovered that uptake and accumulation of arachidonic acid (AA) and its COX-catalyzed metabolite, PGE2, were mainly responsible for the immunosuppressive activity of FATP2 89.
While PGE2 is a well-characterized immune suppressive factor, it is possible that other individual molecular species of peroxidized PE and other PLs may act as effective regulators of immune responses of PMN-MDSC in TME thus indicating to an area of future exploratory work. The main mechanism of FATP2-mediated suppressive activity involved the uptake of AA and the synthesis of PGE2. The selective pharmacological inhibition of FATP2 abrogated the activity of PMN-MDSC and substantially delayed tumor progression. In combination with ICB, pharmacological inhibition of FATP2 abrogated the immunosuppressive activity of PMN-MDSC and substantially delayed tumor progression 89 thus indicating a potentially promising therapeutic target.
The TME is enriched for macrophages with some features of M2 type polarization. These cells are highly susceptible to hydroperoxy-PUFA-PL peroxidation and ferroptosis. It has been demonstrated that iNOS represents the major ferroptosis regulating mechanism in macrophages. High level of iNOS makes macrophages resistant to PL peroxidation and ferroptosis 90. This is significant because the very low iNOS levels make the macrophage susceptible to ferroptotic cell death 90. Detailed studies revealed that iNOS/NO•, the major antiferroptotic mechanism in macrophages, acts independently of GPX4 and other anti-ferroptotic regulators (eg, FSP1). The tolerance of macrophages to pro-ferroptotic stimulation suggests that there may be another, yet to be identified anti-ferroptotic cascade, specific for TME conditions. Interestingly, another lipid-dependent mechanism of TAMs reported a correlation of their functional diversity with prognostic significance in colorectal liver metastasis. It has been shown that small (S-TAM) and large (L-TAM) macrophages correlated with 5-yr disease-free survival rates of 27.8% and 0.2%, respectively (P < 0.0001)91. RNA sequencing of morphologically distinct macrophages established that up-regulation of genes involved in suppressive metabolism was the most enriched pathway in L-TAMs.
DC in TME are characterized by the robust accumulation of neutral lipids – tri- and di-acylglycerols, cholesterol esters and free fatty acids forming large lipid bodies (LB) 92,93. This has been associated with the suppressed cross-presentation of exogenous, incuding tumor-specific, antigens without inhibition of the endogenous peptides presentation of 94,95. This concept was confirmed and expanded by different groups 96-99. Accumulation of oxidized lipids in tumor associated DC could be also the result of ER stress response.
Impaired cross-presentation by lipids was due to the defects in trafficking of peptide-MHC class I (pMHC) complexes 100. Specific biochemical mechanisms of this association emerged with the understanding of the role of a small fraction of these neutral lipids, oxidizable PUFA species as well as their peroxidation products. Notably, oxidatively truncated forms of tri-acyl glycerols were the prevailing oxidized lipid species in LBs 94,100. While some of these oxidatively-truncated triacylglycerols may be formed in DC101, hypoxic TME conditions and related low expression of GPX4 may create pro-ferroptotic stressory conditions culminating in the accumulation of PUFA LB lipids. However, it is likely that the majority of them are produced by PMN-MDSC in a myeloperoxidase (MPO)-catalyzed reaction as MPO-deficient PMN-MDSC did not affect cross-presentation by DCs 86. Given that MPO is readily released from activated PMN-MDSC, it is also possible that the origin of peroxidized lipids in LBs includes pools of oxidized lipids in TME extracellular compartments taken-up the DC. Experimental analysis and computational modeling demonstrated that these more polar peroxidized lipid species transgressed from the hydrophobic core to the LB surface 102. Oxidatively-truncated lipids form adducts with a major stress-induced peptide chaperone, heat shock protein 70 100. This prevents trafficking of pMHC from the phagosome/lysosome to the cell surface. As a result, DCs are not able to effectively stimulate antigen-specific T cells (Fig. 4). Interestingly, a recent study presented a potential link between ferroptosis-associated lipid peroxidation in TME, particularly in cancer cells, and their effects on DC antigen cross-presentation 103. It was shown that “early” ferroptotic cancer cells decreased maturation of DC and dampened their antigen cross-presentation. DC loaded with ferroptotic, but not necroptotic cancer cells failed to protect against tumor progression. However, other studies reported low-immunogenic potential of “late” but not “early” ferroptotic cells 103,104. These seemingly contradictory results may find explanations in detailed analysis of differences in the composition and identity of lipid peroxidation products accumulating in “early” vs. “late” ferroptotic cancer cells, ie., the concept that ferroptotic stress and ferrototic death are separated in time, which we discssed above.
Oxidised lipid signals causing awakening of dormant cancer cells
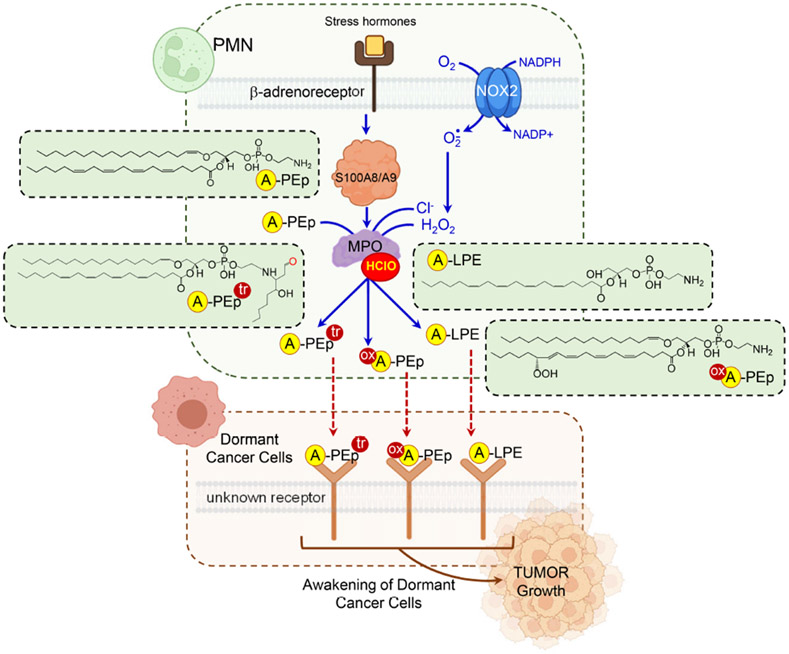
Senescence-like state of dormancy induced by oncogenes, chemotherapy, or radiation therapy is one of the major mechanisms by which tumor cells can persist in tissues long after seemingly successful therapy 105. Tumor recurrence after reactivation of these dormant cells represent a majot clinical problem. In reactivation of dormant cancer cells (DCC) inflammation and myeloid cells seem to play important roles 106. Recently, we described a mechanism enacted by stress hormone stimulation of β2-adreno-receptors on neutrophils. This results in the release of S100A8/A9 proteins and activation of MPO followed by changes in PLs, including their chlorination, hydrolysis and oxidation 107. Using a mouse model of disseminated DCC (lung and ovarian cancer), we demonstrated that MPO activated by S100A8/A9 triggered dramatic changes in a special type of PLs, plasmalogens (Fig. 5). In plasmalogens, attachment of the sn-1-tail occurs via an ether (alkenyl)-bond, in contrast to diacyl-PLs where both FA residues are linked to the glycerol moiety by ester-bonds 108. Plasmalogenic forms are present in all major classes of PLs but most commonly in phosphatidylcholines (PC) and PE - the two most abundant classes of PLs. In S100A8/9-stimulated PMN, plasmalogen alkenyl-acyl species of PE (PE-p) and PC (PC-p) were more predominant than di-acylated PE (PE-d) and PC (PC-d) species. Notably, the alkenyl bond of plasmalogens can be readily hydrolyzed by MPO generated hypochlorite (HOCl). The resultant products represent an unusual type of lyso-PLs, sn1-O-sn2-PUFA-PLs 107. Importantly, PE-p were mostly represented by the molecular species with highly oxidizable arachidonic acid in the sn-2 position. In contrast, PC-p species had saturated and monoenoic acids in the sn-2 position. Not surprisingly, sn1-lyso-sn2-PUFA-PE - were found in S100A8/9-stimulated PMN. These lyso-PE were further chlorinated and oxidized by MPO/HOCl causing the accumulation of aldehydes, particularly 4-hydroxy-nonenal (4-HNE). Reactive 4-HNE can covalently modify amino-groups of proteins and amino-PLs, such as PE. Indeed, increased contents of Michael PE-4HNE adducts and lyso-PE (LPE) were found in mouse and human neutrophils incubated with S100A8/A9 107. The similar profiles of lyso-PUFA-PE were found in neutrophils isolated from mice that underwent experimental stress. Importantly, these mice had much stronger ability to reactivate DCC. PLs extracted from PMN treated with S100A8/A9, but not from control PMN, stimulated proliferation of DCC. Similarly, lipids extracted from PMN isolated from stressed mice also reactivated DCC. However, PL extracts from S100A8/A9-treated MPO deficient PMN failed in induce proliferation of DCC 107. Biochemical characterization of “awakening” by active lipids revealed that the major products generated in MPO driven reaction were PE (18:0p/20:4)-4HNE and lysoPE containing C20:4 in sn-2 position (OH/20:4). These lipids activated tumor cell proliferation via up-regulation of fibroblast growth factor pathway. Given a complex nature of the MPO-driven reaction products generated from p-PUFA-PE, hydrolyzed, peroxidized, chlorinated, oxidatively-truncated metabolites and protein adducts, significant further work will be necessary to narrow-down the list of active compunds and the possible activating receptors. As discussed above, the oxidized derivatives of these PE species are also formed during ferroptotic stress and may be accountable for the ferroptotic cell death. This may be significant in the context of the design and development of effective pro-ferroptotic anti-cancer agents which should not disturb the quiescence of disseminated DCC. Because of high membrane diffusability of PE-derived metabolites, this also raises the question on the role of ferroptotic death of PMN-MDSC in metastatic spreading of the primary tumor.
Figure 5. Lipid signals are involved in awakening of dormant cancer cells by activated PMN.
Stress hormones stimulate β2-adreno-receptors on PMN and cause S100A8/A9-dependent activation of MPO. MPO and its product, hypochlorite, induce changes in PE plasmalogens with highly oxidizable AA in sn-2 position, including their chlorination, hydrolysis and oxidation. MPO driven reaction generates lyso-AA-PE which can be further oxidized by COXs and LOXs to yield hydroperoxy-AA-PE in sn-2 position. These unusual oxygenated lipids can be released by PMN-MDCS and induce “awakening” of dormant tumor cells. Abbreviations: PMN = polymorphonuclear neutrophils; MPO = myeloperoxidase; NOX2 = NADPH oxidase 2; PEp = phosphatidylethanolamine plasmalogen; LPE = lyso-phosphatidylethanolamine. HClO = hypochlorite; H2O2, = hydrogen peroxide; ox = oxidized; tr = truncated. Created with Biorender.com
Conclusions
In this review, we discussed several concepts that may be important for better understanding of the role of myeloid cells in cancer and their potential therapeutic targeting.
First, functional state of actvation of myeoid cells along classical vs. pathological states rather than sub-cluster classification of these cells is a dominant force defining functionality of various myeloid cells. Although many issues require further clarification and validation, it appears that functional signatures transcend specific subsets of these cells and may be useful for the designing effective therapeutic strategies. Interesting exception is DC. It appears that, at least for now, pathological state of activation of these cells is not clearly identified and this is reflected in the fact that phenotypic sub-population of these cells are remarkably conserved and functionally stable. Although data on functionality of mregDC are rather limited, it seems that these cells cannot be fully characterized as pathologically activated cells as it appears to be a functional state of DCs under homeostatic conditions and contribute to regulation of effector T cells indirectly and not utilize mechanisms commonly attributed to TAM and MDSC. The reason for this phenomenon is not clear for us but may be related to the unique differentiation and maturation/activation pathway of these cells.
Second, although critical role of microenvironment in shaping up functional state of myeloid cells in TME seems well-defined, it does not fully explain the changes observed in the TME. We suggest that condiontioning of neutrophils and monocytes in the BM via ER stress and various tumor-derived cytokines play an important role in their conversion to MDSC in the TME. Thus, targeting these cells at very early stages of differentiation may be critical.
Third, high metabolic demands of rapidly proliferating cancer cells create PUFA-rich TME in which hypoxic conditions discourage oxidation resistant phenotypes of immune cells. These cells cannot adequately respond other than by the developing pro-ferroptotic stress and generation of lipid peroxidation products, defining their enhanced vulnerability to ferroptotic death. This process is most likely predominantly driven by PMN-MSC with their powerful oxidants-generating machinery for the production of NO•/O2-• (eg, peroxynitrite, ONOO-) and ClO-. These PMN-MDSC offer the “Trojan horse” gift to other types of immune cells such as DCs, macrophages, T cells, etc. As a result, the entire immune cell community suffers the losses of unpreparedness to aggressive pro-ferroptotic factors triggering “inappropriate” ferroptotic death.
Fourth, ferroptotic response and ferroptotic cell death are not identical events. They are separated in time (in experimental conditions in vitro by 24-48 hours). Immune suppressive effect exerted by myeloid cells undergoing ferroptotic response have profound negative effect on adaptive immune cells in the TME and limited tumor control by these cells. Myeloid cells that eventually die from ferroptosis are rapidly replaced by new cells arriving from the BM. This concept explains the less than satisfactory results of numerous attempts to use pro-ferroptotic agents to overcome the limitations of pro-apoptotic anti-cancer strategies and suggest a paradigm of differential pro-/anti-ferroptotic approaches whereby stimulation of ferroptosis in cancer cells can be combined with anti-ferroptotic protection selectively delivered to immune cells.
van Vlerken-Ysla et al. discuss that functional states of myeloid cells transcend single-cell omics-based cell clusters and outline factors involved in diversification of these cells in the bone marrow, periphery and tumor microenvironment. They propose that lipid peroxidation represents a major mechanism governing their pathological state of activation, potentially providing a therapeutic vulnerability.
van Vlerken-Ysla et al. discuss how functional states of myeloid cells transcend single-cell omics-based cell clusters and outline the factors that contribute to their formation and (functional) diversification in the bone marrow, periphery and tumor microenvironment (TME). They propose that lipid peroxidation represents a major mechanism governing their pathological state of activation in the TME, thereby providing a potential therapeutic vulnerability.
van Vlerken-Ysla et al. discuss how functional states of myeloid cells transcend single-cell omics-based cell clusters and outline the factors that contribute to the formation of myeloid cells in the bone marrow, periphery and tumor microenvironment (TME). They propose that lipid peroxidation represents a major mechanism governing their pathological state of activation in the TME, thereby providing a potential therapeutic vulnerability.
Overview the signals that TME-derived pathological factors on lineage development, differentiation, and recruitment of myeloid cells
In recent years, evidence emerged that a of common mechanism exists that transcends various types of immune suppressive cells. These mechanisms involve oxidized lipids and may play a dominant role in regulation of the function of these cells.
functional signatures transcend specific subsets of these cells and may be useful for the designing effective therapeutic strategies
We discuss a concept that lipid peroxidation of myeloid cells represents a major mechanism that governs their pathological state of activation in the TME. Lipid peroxidation is associated with ferroptosis mediating suppressive activity of these cells and thus could be considered as attractive target for therapeutic intervention.
Acknowledgements
This study was supported by NIH grants AI156924 (VEK), CA243142 (VEK), CA165065 (VEK), CA266342 (VEK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
LVY and DG are employee and shareholders of AstraZeneca. All other authors declare no competing interests.
References
- 1.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, et al. (2018). Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 24, 541–550. 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casanova-Acebes M, Dalla E, Leader AM, LeBerichel J, Nikolic J, Morales BM, Brown M, Chang C, Troncoso L, Chen ST, et al. (2021). Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature 595, 578–584. 10.1038/s41586-021-03651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Locati M, Curtale G, and Mantovani A (2020). Diversity, Mechanisms, and Significance of Macrophage Plasticity. Annu Rev Pathol 15, 123–147. 10.1146/annurev-pathmechdis-012418-012718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veglia F, Sanseviero E, and Gabrilovich DI (2021). Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol 21, 485–498. 10.1038/s41577-020-00490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegde S, Leader AM, and Merad M (2021). MDSC: Markers, development, states, and unaddressed complexity. Immunity 54, 875–884. 10.1016/j.immuni.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kvedaraite E, and Ginhoux F (2022). Human dendritic cells in cancer. Sci Immunol 7, eabm9409. 10.1126/sciimmunol.abm9409. [DOI] [PubMed] [Google Scholar]
- 7.Kalafati L, Hatzioannou A, Hajishengallis G, and Chavakis T (2022). The role of neutrophils in trained immunity. Immunol Rev. 10.1111/imr.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quail DF, Amulic B, Aziz M, Barnes BJ, Eruslanov E, Fridlender ZG, Goodridge HS, Granot Z, Hidalgo A, Huttenlocher A, et al. (2022). Neutrophil phenotypes and functions in cancer: A consensus statement. J Exp Med 219. 10.1084/jem.20220011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veglia F, Perego M, and Gabrilovich D (2018). Myeloid-derived suppressor cells coming of age. Nat Immunol 19, 108–119. 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaillon S, Ponzetta A, Di Mitri D, Santoni A, Bonecchi R, and Mantovani A (2020). Neutrophil diversity and plasticity in tumour progression and therapy. Nat Rev Cancer 20, 485–503. 10.1038/s41568-020-0281-y. [DOI] [PubMed] [Google Scholar]
- 11.Cox N, Pokrovskii M, Vicario R, and Geissmann F (2021). Origins, Biology, and Diseases of Tissue Macrophages. Annu Rev Immunol 39, 313–344. 10.1146/annurev-immunol-093019-111748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Nefedova Y, Lei A, and Gabrilovich D (2018). Neutrophils and PMN-MDSC: Their biological role and interaction with stromal cells. Semin Immunol 35, 19–28. 10.1016/j.smim.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siwicki M, and Pittet MJ (2021). Versatile neutrophil functions in cancer. Semin. Immunol 57, 101538. 10.1016/j.smim.2021.101538. [DOI] [PubMed] [Google Scholar]
- 14.Pittet MJ, Michielin O, and Migliorini D (2022). Clinical relevance of tumour-associated macrophages. Nature reviews. Clinical oncology 19, 402–421. 10.1038/s41571-022-00620-6. [DOI] [PubMed] [Google Scholar]
- 15.Audsley KM, McDonnell AM, and Waithman J (2020). Cross-Presenting XCR1(+) Dendritic Cells as Targets for Cancer Immunotherapy. Cells 9, 565. 10.3390/cells9030565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, Qin S, Zhang L, Ouyang H, Du P, et al. (2021). A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792–809.e723. 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Qian J, Olbrecht S, Boeckx B, Vos H, Laoui D, Etlioglu E, Wauters E, Pomella V, Verbandt S, Busschaert P, et al. (2020). A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell research 30, 745–762. 10.1038/s41422-020-0355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, Krishnan I, Maroni G, Meyerovitz CV, Kerwin CM, et al. (2019). Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity 50, 1317–1334 e1310. 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourdely P, Anselmi G, Vaivode K, Ramos RN, Missolo-Koussou Y, Hidalgo S, Tosselo J, Nuñez N, Richer W, Vincent-Salomon A, et al. (2020). Transcriptional and Functional Analysis of CD1c(+) Human Dendritic Cells Identifies a CD163(+) Subset Priming CD8(+)CD103(+) T Cells. Immunity 53,335–352.e338. 10.1016/j.immuni.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maier B, Leader AM, Chen ST, Tung N, Chang C, LeBerichel J, Chudnovskiy A, Maskey S, Walker L, Finnigan JP, et al. (2020). A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 580, 257–262. 10.1038/s41586-020-2134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coutant F, Pin JJ, and Miossec P (2021). Extensive Phenotype of Human Inflammatory Monocyte-Derived Dendritic Cells. Cells 10. 10.3390/cells10071663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, et al. (2017). Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573. 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leader AM, Grout JA, Maier BB, Nabet BY, Park MD, Tabachnikova A, Chang C, Walker L, Lansky A, Le Berichel J, et al. (2021). Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell 39, 1594–1609.e1512. 10.1016/j.ccell.2021.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becht E, Giraldo NA, Germain C, de Reynies A, Laurent-Puig P, Zucman-Rossi J, Dieu-Nosjean MC, Sautés-Fridman C, and Fridman WH (2016). Immune Contexture, Immunoscore, and Malignant Cell Molecular Subgroups for Prognostic and Theranostic Classifications of Cancers. Adv Immunol 130, 95–190. 10.1016/bs.ai.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Bruni D, Angell HK, and Galon J (2020). The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer 20, 662–680. 10.1038/s41568-020-0285-7. [DOI] [PubMed] [Google Scholar]
- 26.Matsubara E, Komohara Y, Esumi S, Shinchi Y, Ishizuka S, Mito R, Pan C, Yano H, Kobayashi D, Fujiwara Y, et al. (2022). SPP1 Derived from Macrophages Is Associated with a Worse Clinical Course and Chemo-Resistance in Lung Adenocarcinoma. Cancers (Basel) 14, 4374. 10.3390/cancers14184374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng S, Yuan W, Sun Z, Guo X, Ling J, Chang A, Zhao H, and Zhuo X (2022). SPP1 as a key gene in the lymph node metastasis and a potential predictor of poor prognosis in head and neck carcinoma. Journal of oral pathology & medicine : official publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology 51, 620–629. 10.1111/jop.13333. [DOI] [PubMed] [Google Scholar]
- 28.Qi J, Sun H, Zhang Y, Wang Z, Xun Z, Li Z, Ding X, Bao R, Hong L, Jia W, et al. (2022). Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nature communications 13, 1742. 10.1038/s41467-022-29366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong F, Meng Q, Zhang W, Zheng R, Li X, Cheng T, Hu D, and Gao X (2021). Single-Cell Analysis of the Pan-Cancer Immune Microenvironment and scTIME Portal. Cancer Immunol. Res 9, 939–951. 10.1158/2326-6066.Cir-20-1026. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O'Brien SA, He Y, Wang L, Zhang Q, Kim A, et al. (2020). Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 181,442–459.e429. 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Gao Z, Li B, Li J, Ou Y, Yu X, Zhang Z, Liu S, Fu X, Jin H, et al. (2022). Lipid-associated macrophages in the tumor-adipose microenvironment facilitate breast cancer progression. Oncoimmunolog 11, 2085432. 10.1080/2162402x.2022.2085432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan JM, Quintanal-Villalonga Á, Gao VR, Xie Y, Allaj V, Chaudhary O, Masilionis I, Egger J, Chow A, Walle T, et al. (2021). Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell 39, 1479–1496.e1418. 10.1016/j.ccell.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulder K, Patel AA, Kong WT, Piot C, Halitzki E, Dunsmore G, Khalilnezhad S, Irac SE, Dubuisson A, Chevrier M, et al. (2021). Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity 54, 1883–1900.e1885. 10.1016/j.immuni.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Funes SC, Rios M, Escobar-Vera J, and Kalergis AM (2018). Implications of macrophage polarization in autoimmunity. Immunology 154, 186–195. 10.1111/imm.12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavin Y, and Merad M (2013). Macrophages: gatekeepers of tissue integrity. Cancer Immunol. Res 1, 201–209. 10.1158/2326-6066.Cir-13-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinek J, Lin J, Kim KI, Wang VG, Wu TC, Chiorazzi M, Boruchov H, Gulati A, Seeniraj S, Sun L, et al. (2022). Transcriptional profiling of macrophages in situ in metastatic melanoma reveals localization-dependent phenotypes and function. Cell Rep Med 3, 100621. 10.1016/j.xcrm.2022.100621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laviron M, Petit M, Weber-Delacroix E, Combes AJ, Arkal AR, Barthelemy S, Courau T, Hume DA, Combadiere C, Krummel MF, and Boissonnas A (2022). Tumor-associated macrophage heterogeneity is driven by tissue territories in breast cancer. Cell reports 39, 110865. 10.1016/j.celrep.2022.110865. [DOI] [PubMed] [Google Scholar]
- 38.Veglia F, Hashimoto A, Dweep H, Sanseviero E, De Leo A, Tcyganov E, Kossenkov A, Mulligan C, Nam B, Masters G, et al. (2021). Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. J Exp Med 218, e20201803. 10.1084/jem.20201803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sagiv JY, Michaeli J, Assi S, Mishalian I, Kisos H, Levy L, Damti P, Lumbroso D, Polyansky L, Sionov RV, et al. (2015). Phenotypic diversity and plasticity in circulating neutrophil subpopulations in cancer. Cell.Rep. 10, 562–573. 10.1016/j.celrep.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 40.Alicea-Torres K, Sanseviero E, Gui J, Chen J, Veglia F, Yu Q, Donthireddy L, Kossenkov A, Lin C, Fu S, et al. (2021). Immune suppressive activity of myeloid-derived suppressor cells in cancer requires inactivation of the type I interferon pathway. Nat. Commun 12, 1717. 10.1038/s41467-021-22033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalafati L, Kourtzelis I, Schulte-Schrepping J, Li X, Hatzioannou A, Grinenko T, Hagag E, Sinha A, Has C, Dietz S, et al. (2020). Innate Immune Training of Granulopoiesis Promotes Anti-tumor Activity. Cell 183, 771–785.e712. 10.1016/j.cell.2020.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue R, Zhang Q, Cao Q, Kong R, Xiang X, Liu H, Feng M, Wang F, Cheng J, Li Z, et al. (2022). Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature 612, 141–147. 10.1038/s41586-022-05400-x. [DOI] [PubMed] [Google Scholar]
- 43.Salcher S, Sturm G, Horvath L, Untergasser G, Kuempers C, Fotakis G, Panizzolo E, Martowicz A, Trebo M, Pall G, et al. (2022). High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell 40, 1503–1520 e1508. 10.1016/j.ccell.2022.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alshetaiwi H, Pervolarakis N, McIntyre LL, Ma D, Nguyen Q, Rath JA, Nee K, Hernandez G, Evans K, Torosian L, et al. (2020). Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci Immunol 5, eaay6017. 10.1126/sciimmunol.aay6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grover A, Sanseviero E, Timosenko E, and Gabrilovich DI (2021). Myeloid-Derived Suppressor Cells: A Propitious Road to Clinic. Cancer Discov 11, 2693–2706. 10.1158/2159-8290.CD-21-0764. [DOI] [PubMed] [Google Scholar]
- 46.Tcyganov EN, Hanabuchi S, Hashimoto A, Campbell D, Kar G, Slidel TW, Cayatte C, Landry A, Pilataxi F, Hayes S, et al. (2021). Distinct mechanisms govern populations of myeloid-derived suppressor cells in chronic viral infection and cancer. J Clin Invest 131, e145971. 10.1172/JCI145971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friebel E, Kapolou K, Unger S, Núñez NG, Utz S, Rushing EJ, Regli L, Weller M, Greter M, Tugues S, et al. (2020). Single-Cell Mapping of Human Brain Cancer Reveals Tumor-Specific Instruction of Tissue-Invading Leukocytes. Cell 181, 1626–1642.e1620. 10.1016/j.cell.2020.04.055. [DOI] [PubMed] [Google Scholar]
- 48.Kwak T, Wang F, Deng H, Condamine T, Kumar V, Perego M, Kossenkov A, Montaner LJ, Xu X, Xu W, et al. (2020). Distinct Populations of Immune-Suppressive Macrophages Differentiate from Monocytic Myeloid-Derived Suppressor Cells in Cancer. Cell. Rep 33, 108571. 10.1016/j.celrep.2020.108571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Condamine T, Dominguez GA, Youn JI, Kossenkov AV, Mony S, Alicea-Torres K, Tcyganov E, Hashimoto A, Nefedova Y, Lin C, et al. (2016). Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci Immunol 1, aaf8943. 10.1126/sciimmunol.aaf8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel S, Fu S, Mastio J, Dominguez GA, Purohit A, Kossenkov A, Lin C, Alicea-Torres K, Sehgal M, Nefedova Y, et al. (2018). Unique pattern of neutrophil migration and function during tumor progression. Nat Immunol 19, 1236–1247. 10.1038/s41590-018-0229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson A, Burgess M, Webb S, Louwe PA, Ouyang Z, Skola D, Han CZ, Batada NN, Gonzalez-Huici V, Cassetta L, et al. (2022). Systemic Influences of Mammary Cancer on Monocytes in Mice. Cancers (Basel) 14. 10.3390/cancers14030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabrilovich DI (2017). Myeloid-Derived Suppressor Cells. Cancer Immunol Res 5, 3–8. 10.1158/2326-6066.CIR-16-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barry ST, Gabrilovich DI, Sansom OJ, Campbell AD, and Morton JP (2023). Therapeutic targeting of tumour myeloid cells Nat. Rev. Cancer in press. [DOI] [PubMed] [Google Scholar]
- 54.Mohamed E, Al-Khami AA, and Rodriguez PC (2018). The cellular metabolic landscape in the tumor milieu regulates the activity of myeloid infiltrates. Cell Mol Immunol 15, 421–427. 10.1038/s41423-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anishkin A, Loukin SH, Teng J, and Kung C (2014). Feeling the hidden mechanical forces in lipid bilayer is an original sense. Proc Natl Acad Sci U S A 111, 7898–7905. 10.1073/pnas.1313364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, and Peter Walter P (2002). Molecular Biology of the Cell. (Garland Science). [Google Scholar]
- 57.Ma K, and Zhang L (2021). Overview: Lipid Metabolism in the Tumor Microenvironment. Adv Exp Med Biol 1316, 41–47. 10.1007/978-981-33-6785-2_3. [DOI] [PubMed] [Google Scholar]
- 58.Ma Y, Temkin SM, Hawkridge AM, Guo C, Wang W, Wang XY, and Fang X (2018). Fatty acid oxidation: An emerging facet of metabolic transformation in cancer. Cancer Lett 435, 92–100. 10.1016/j.canlet.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng W, Yin X, Jiang Y, Jin L, and Liang W (2021). PPARalpha at the crossroad of metabolic-immune regulation in cancer. The FEBS journal. 10.1111/febs.16181. [DOI] [PubMed] [Google Scholar]
- 60.Fuhrmann DC, Olesch C, Kurrle N, Schnutgen F, Zukunft S, Fleming I, and Brune B (2019). Chronic Hypoxia Enhances beta-Oxidation-Dependent Electron Transport via Electron Transferring Flavoproteins. Cells 8. 10.3390/cells8020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tahri-Joutey M, Andreoletti P, Surapureddi S, Nasser B, Cherkaoui-Malki M, and Latruffe N (2021). Mechanisms Mediating the Regulation of Peroxisomal Fatty Acid Beta-Oxidation by PPARalpha. International journal of molecular sciences 22. 10.3390/ijms22168969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korbecki J, Bobinski R, and Dutka M (2019). Self-regulation of the inflammatory response by peroxisome proliferator-activated receptors. Inflamm Res 68, 443–458. 10.1007/s00011-019-01231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuhn H, Barnett J, Grunberger D, Baecker P, Chow J, Nguyen B, Bursztyn-Pettegrew H, Chan H, and Sigal E (1993). Overexpression, purification and characterization of human recombinant 15-lipoxygenase. Biochim Biophys Acta 1169, 80–89. 10.1016/0005-2760(93)90085-n. [DOI] [PubMed] [Google Scholar]
- 64.Kagan VE, Mao G, Qu F, Angeli JP, Doll S, Croix CS, Dar HH, Liu B, Tyurin VA, Ritov VB, et al. (2017). Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nature chemical biology 13, 81–90. 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaschler MM, Andia AA, Liu H, Csuka JM, Hurlocker B, Vaiana CA, Heindel DW, Zuckerman DS, Bos PH, Reznik E, et al. (2018). FINO2 initiates ferroptosis through GPX4 inactivation and iron oxidation. Nat. Chem. Biol 14, 507–515. 10.1038/s41589-018-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wiernicki B, Dubois H, Tyurina YY, Hassannia B, Bayir H, Kagan VE, Vandenabeele P, Wullaert A, and Vanden Berghe T (2020). Excessive phospholipid peroxidation distinguishes ferroptosis from other cell death modes including pyroptosis. Cell death & disease 11, 922. 10.1038/s41419-020-03118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kagan VE, Tyurina YY, Sun WY, Vlasova II, Dar H, Tyurin VA, Amoscato A, Mallampalli R, van der Wel PCA, He RR, et al. (2020). Redox phospholipidomics of enzymatically generated oxygenated phospholipids as specific signals of programmed cell death. Free Radic Biol Med 147, 231–241. 10.1016/j.freeradbiomed.2019.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascon S, Hatzios SK, Kagan VE, et al. (2017). Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 171, 273–285. 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wenzel SE, Tyurina YY, Zhao J, St Croix CM, Dar HH, Mao G, Tyurin VA, Anthonymuthu TS, Kapralov AA, Amoscato AA, et al. (2017). PEBP1 Wardens Ferroptosis by Enabling Lipoxygenase Generation of Lipid Death Signals. Cell 171, 628–641 e626. 10.1016/j.cell.2017.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow A, Kaffenberger SD, Eaton JK, Shimada K, Aguirre AJ, et al. (2017). Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457. 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hangauer MJ, Viswanathan VS, Ryan MJ, Bole D, Eaton JK, Matov A, Galeas J, Dhruv HD, Berens ME, Schreiber SL, et al. (2017). Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250. 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsoi J, Robert L, Paraiso K, Galvan C, Sheu KM, Lay J, Wong DJL, Atefi M, Shirazi R, Wang X, et al. (2018). Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell 33, 890–904 e895. 10.1016/j.ccell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. (2012). Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072. 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Tan H, Daniels JD, Zandkarimi F, Liu H, Brown LM, Uchida K, O'Connor OA, and Stockwell BR (2019). Imidazole Ketone Erastin Induces Ferroptosis and Slows Tumor Growth in a Mouse Lymphoma Model. Cell Chem Biol 26, 623–633 e629. 10.1016/j.chembiol.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Badgley MA, Kremer DM, Maurer HC, DelGiorno KE, Lee HJ, Purohit V, Sagalovskiy IR, Ma A, Kapilian J, Firl CEM, et al. (2020). Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 368, 85–89. 10.1126/science.aaw9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yi J, Zhu J, Wu J, Thompson CB, and Jiang X (2020). Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc Natl Acad Sci U S A 117, 31189–31197. 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shiromizu S, Yamauchi T, Kusunose N, Matsunaga N, Koyanagi S, and Ohdo S (2019). Dosing Time-Dependent Changes in the Anti-tumor Effect of xCT Inhibitor Erastin in Human Breast Cancer Xenograft Mice. Biol Pharm Bull 42, 1921–1925. 10.1248/bpb.b19-00546. [DOI] [PubMed] [Google Scholar]
- 78.Wu X, Liu C, Li Z, Gai C, Ding D, Chen W, Hao F, and Li W (2020). Regulation of GSK3beta/Nrf2 signaling pathway modulated erastin-induced ferroptosis in breast cancer. Mol Cell Biochem 473, 217–228. 10.1007/s11010-020-03821-8. [DOI] [PubMed] [Google Scholar]
- 79.Shibata Y, Yasui H, Higashikawa K, Miyamoto N, and Kuge Y (2019). Erastin, a ferroptosis-inducing agent, sensitized cancer cells to X-ray irradiation via glutathione starvation in vitro and in vivo. PloS one 14, e0225931. 10.1371/journal.pone.0225931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huo H, Zhou Z, Qin J, Liu W, Wang B, and Gu Y (2016). Erastin Disrupts Mitochondrial Permeability Transition Pore (mPTP) and Induces Apoptotic Death of Colorectal Cancer Cells. PloS one 11, e0154605. 10.1371/journal.pone.0154605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang Y, Luo M, Zhang K, Zhang J, Gao T, Connell DO, Yao F, Mu C, Cai B, Shang Y, and Chen W (2020). Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun 11, 433. 10.1038/s41467-020-14324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang W, Green M, Choi JE, Gijón M, Kennedy PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al. (2019). CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 569, 270–274. 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma X, Xiao L, Liu L, Ye L, Su P, Bi E, Wang Q, Yang M, Qian J, and Yi Q (2021). CD36-mediated ferroptosis dampens intratumoral CD8(+) T cell effector function and impairs their antitumor ability. Cell metabolism 33, 1001–1012 e1005. 10.1016/j.cmet.2021.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu S, Chaudhary O, Rodriguez-Morales P, Sun X, Chen D, Zappasodi R, Xu Z, Pinto AFM, Williams A, Schulze I, et al. (2021). Uptake of oxidized lipids by the scavenger receptor CD36 promotes lipid peroxidation and dysfunction in CD8(+) T cells in tumors. Immunity 54, 1561–1577 e1567. 10.1016/j.immuni.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim R, Hashimoto A, Markosyan N, . , Tyurin VA, Tyurina YY, Kar G, Fu S, Sehgal M, Garcia-Gerique L, Kossenkov A, et al. (2022). Ferroptosis of tumor neutrophils causes immune suppression in cancer. Nature 612, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ugolini A, Tyurin VA, Tyurina YY, Tcyganov EN, Donthireddy L, Kagan VE, Gabrilovich DI, and Veglia F (2020). Polymorphonuclear myeloid-derived suppressor cells limit antigen cross-presentation by dendritic cells in cancer. JCI insight 5, e138581. 10.1172/jci.insight.138581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qiu P, Wang H, Zhang M, Zhang M, Peng R, Zhao Q, and Liu J (2020). FATP2-targeted therapies - A role beyond fatty liver disease. Pharmacol Res 161, 105228. 10.1016/j.phrs.2020.105228. [DOI] [PubMed] [Google Scholar]
- 88.Kuwata H, and Hara S (2019). Role of acyl-CoA synthetase ACSL4 in arachidonic acid metabolism. Prostaglandins Other Lipid Mediat 144, 106363. 10.1016/j.prostaglandins.2019.106363. [DOI] [PubMed] [Google Scholar]
- 89.Veglia F, Tyurin VA, Blasi M, De Leo A, Kossenkov AV, Donthireddy L, To TKJ, Schug Z, Basu S, Wang F, et al. (2019). Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature 569, 73–78. 10.1038/s41586-019-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kapralov AA, Yang Q, Dar HH, Tyurina YY, Anthonymuthu TS, Kim R, St Croix CM, Mikulska-Ruminska K, Liu B, Shrivastava IH, et al. (2020). Redox lipid reprogramming commands susceptibility of macrophages and microglia to ferroptotic death. Nature chemical biology 16, 278–290. 10.1038/s41589-019-0462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Donadon M, Torzilli G, Cortese N, Soldani C, Di Tommaso L, Franceschini B, Carriero R, Barbagallo M, Rigamonti A, Anselmo A, et al. (2020). Macrophage morphology correlates with single-cell diversity and prognosis in colorectal liver metastasis. J Exp Med 217. 10.1084/jem.20191847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dierge E, Debock E, Guilbaud C, Corbet C, Mignolet E, Mignard L, Bastien E, Dessy C, Larondelle Y, and Feron O (2021). Peroxidation of n-3 and n-6 polyunsaturated fatty acids in the acidic tumor environment leads to ferroptosis-mediated anticancer effects. Cell metabolism 33, 1701–1715 e1705. 10.1016/j.cmet.2021.05.016. [DOI] [PubMed] [Google Scholar]
- 93.den Brok MH, Raaijmakers TK, Collado-Camps E, and Adema GJ (2018). Lipid Droplets as Immune Modulators in Myeloid Cells. Trends Immunol 39, 380–392. 10.1016/j.it.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 94.Ramakrishnan R, Tyurin VA, Veglia F, Condamine T, Amoscato A, Mohammadyani D, Johnson JJ, Zhang LM, Klein-Seetharaman J, Celis E, et al. (2014). Oxidized lipids block antigen cross-presentation by dendritic cells in cancer. J Immunol 192, 2920–2931. 10.4049/jimmunol.1302801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Herber DL, Cao W, Nefedova Y, Novitskiy SV, Nagaraj S, Tyurin VA, Corzo A, Cho HI, Celis E, Lennox B, et al. (2010). Lipid accumulation and dendritic cell dysfunction in cancer. Nat Med 16, 880–886. 10.1038/nm.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gao F, Liu C, Guo J, Sun W, Xian L, Bai D, Liu H, Cheng Y, Li B, Cui J, et al. (2015). Radiation-driven lipid accumulation and dendritic cell dysfunction in cancer. Sci. Rep 5, 9613. 10.1038/srep09613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gardner JK, Mamotte CD, Patel P, Yeoh TL, Jackaman C, and Nelson DJ (2015). Mesothelioma tumor cells modulate dendritic cell lipid content, phenotype and function. PloS one 10, e0123563. 10.1371/journal.pone.0123563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walter P, and Ron D (2011). The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. [DOI] [PubMed] [Google Scholar]
- 99.Cubillos-Ruiz JR, Silberman PC, Rutkowski MR, Chopra S, Perales-Puchalt A, Song M, Zhang S, Bettigole SE, Gupta D, Holcomb K, et al. (2015). ER Stress Sensor XBP1 Controls Anti-tumor Immunity by Disrupting Dendritic Cell Homeostasis. Cell 161, 1527–1538. 10.1016/j.cell.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Veglia F, Tyurin VA, Mohammadyani D, Blasi M, Duperret EK, Donthireddy L, Hashimoto A, Kapralov A, Amoscato A, Angelini R, et al. (2017). Lipid bodies containing oxidatively truncated lipids block antigen cross-presentation by dendritic cells in cancer. Nature communications 8, 2122. 10.1038/s41467-017-02186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Demuynck R, Efimova I, Naessens F, and Krysko DV (2021). Immunogenic ferroptosis and where to find it? J Immunother Cancer 9. 10.1136/jitc-2021-003430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mohammadyani D, Tyurin VA, O'Brien M, Sadovsky Y, Gabrilovich DI, Klein-Seetharaman J, and Kagan VE (2014). Molecular speciation and dynamics of oxidized triacylglycerols in lipid droplets: Mass spectrometry and coarse-grained simulations. Free Radic Biol Med 76, 53–60. 10.1016/j.freeradbiomed.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wiernicki B, Maschalidi S, Pinney J, Adjemian S, Vanden Berghe T, Ravichandran KS, and Vandenabeele P (2022). Cancer cells dying from ferroptosis impede dendritic cell-mediated anti-tumor immunity. Nature communications 13, 3676. 10.1038/s41467-022-31218-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alzeibak R, Mishchenko TA, Shilyagina NY, Balalaeva IV, Vedunova MV, and Krysko DV (2021). Targeting immunogenic cancer cell death by photodynamic therapy: past, present and future. J Immunother Cancer 9, e001926. 10.1136/jitc-2020-001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Talukdar S, Bhoopathi P, Emdad L, Das S, Sarkar D, and Fisher PB (2019). Dormancy and cancer stem cells: An enigma for cancer therapeutic targeting. Adv Cancer Res 141, 43–84. 10.1016/bs.acr.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 106.Park SY, and Nam JS (2020). The force awakens: metastatic dormant cancer cells. Exp Mol Med 52, 569–581. 10.1038/s12276-020-0423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perego M, Tyurin VA, Tyurina YY, Yellets J, Nacarelli T, Lin C, Nefedova Y, Kossenkov A, Liu Q, Sreedhar S, et al. (2020). Reactivation of dormant tumor cells by modified lipids derived from stress-activated neutrophils. Sci. Transl. Med 12, eabb5817. 10.1126/scitranslmed.abb5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jimenez-Rojo N, and Riezman H (2019). On the road to unraveling the molecular functions of ether lipids. FEBS Lett 593, 2378–2389. 10.1002/1873-3468.13465. [DOI] [PubMed] [Google Scholar]
- 109.Wagner BA, Buettner GR, and Burns CP (1994). Free radical-mediated lipid peroxidation in cells: oxidizability is a function of cell lipid bis-allylic hydrogen content. Biochemistry 33, 4449–4453. 10.1021/bi00181a003. [DOI] [PubMed] [Google Scholar]