Abstract
Salmonella is one of the most extensively characterized bacterial pathogens and is a leading cause of bacterial gastroenteritis. Despite this, we are only just beginning to understand at a molecular level how Salmonella interacts with its mammalian hosts to cause disease. Studies during the past decade on the genetic basis of virulence of Salmonella have significantly advanced our understanding of the molecular basis of the host-pathogen interaction, yet many questions remain. In this review, we focus on the interaction of enterocolitis-causing salmonellae with the intestinal mucosa, since this is the initiating step for most infections caused by Salmonella. Animal and in vitro cell culture models for the interaction of these bacteria with the intestinal epithelium are reviewed, along with the bacterial genes that are thought to affect this interaction. Lastly, recent studies on the response of epithelial cells to Salmonella infection and how this might promote diarrhea are discussed.
Salmonella is one of the most extensively studied bacterial species in terms of its physiology, genetics, cell structure, and development. It is also one of the most extensively characterized bacterial pathogens and is a leading cause of bacterial gastroenteritis. Despite this, we are only just beginning to understand at a molecular level how Salmonella interacts with its mammalian hosts to cause disease. Studies during the past decade on the genetic basis of virulence of Salmonella have significantly advanced our understanding of the molecular basis of the host-pathogen interaction, yet many questions remain. In this review, we focus on the interaction of salmonellae that cause enterocolitis with the intestinal mucosa, because this is the initiating step for most infections caused by these organisms.
General Background on Salmonellosis
Salmonellae are motile, gram-negative, rod-shaped bacteria belonging to the Enterobacteriaceae family; the species is a close relative of Escherichia coli. Salmonella was first described in 1880 by Eberth and cultured in 1884 by Gaffky (40). Strains were differentiated based on their reaction to sera, and for many decades each new serotype was given a new species designation (e.g., S. typhimurium, S. enteritidis, S. pullorum, and S. dublin). It is generally accepted now that there is only a single species of Salmonella (S. enterica) rather than the over 2,000 named serovars (174), but most investigators continue to write, e.g., “S. typhimurium” rather than “S. enterica serovar Typhimurium” out of convenience and for continuity with the previous literature. Thus, in this review, we refer to the Salmonella serotypes by their “species” names. While some serotypes of Salmonella such as S. typhi and S. pullorum have a restricted host range, most serotypes infect a broad range of warm-blooded animals and are capable of causing disease in humans. The majority of serovars that cause disease in humans belong to subgroup 1 (129, 234).
Salmonella is capable of causing a variety of disease syndromes: enteric fever, bacteremia, enterocolitis, and focal infections. Enterocolitis is by far the most common manifestation of disease caused by Salmonella, but bacteremia and focal infections can accompany or follow enterocolitis. Enteric fever (typhoid fever) is caused primarily by S. typhi and S. paratyphi and occasionally by other serotypes. While approximately 2,000 serotypes of Salmonella have been associated with enterocolitis, at a given time it is a smaller set of about 10 serotypes that accounts for the majority of infections; these typically include S. typhimurium, S. enteritidis, and S. heidelberg (46a, 232). The incubation period is typically 6 to 48 h and is followed by headache, abdominal pain, diarrhea, and vomiting. The diarrhea can contain blood, lymphocytes, and mucus. Fever, malaise, and muscle aches are quite common. Symptoms usually resolve within a week but Salmonella can be shed in the feces for up to 20 weeks by children <5 years of age and for 8 weeks by adults (104). Children, especially those <1 year of age, and those over 60 are most susceptible to disease and tend to have more severe infections (reviewed in references ;0104, 129, 234, and 239;0).
There are approximately 40,000 reported cases of salmonellosis in the USA each year (46b, 16, 18, 56, 151, 234); only 1 to 5% of infections with Salmonella are reported (47), thus there are actually 2 to 4 million cases in the US each year with an estimated annual cost of over $2 billion (234). The incidence of salmonellosis in the United States has steadily increased since World War II (46). The reasons for this are complex and include an increase in the proportion of the population older than 60 years (38, 39), changing agricultural and food distribution methods (33, 122, 214), increased consumption of raw or slightly cooked foods (219), an increase in the number of immunocompromised or chronically ill people, and deterioration of the public health infrastructure (12, 30, 44, 45, 133, 232–234, 253). Transmission of Salmonella to humans is usually by consumption of contaminated food, but human-to-human transmission and direct animal-to-human transmission can occur (205, 234, 254). The most common sources of Salmonella are beef, poultry, and eggs (231). The past two decades have seen an increase in the importance of eggs and egg products in transmission (4, 79, 105, 185, 195, 221, 236). Eggs can be contaminated through cracks in the shell or transovarally from an infected ovary or oviduct to the yolk prior to deposition of the shell (221, 223); internally infected eggs left at room temperature can rapidly achieve Salmonella concentrations of 1011 cells per yolk (261). This mode of transmission can be especially difficult to control because the egg-laying hens are usually asymptomatic (223). S. enteritidis is a common cause of internally contaminated eggs, and as the incidence of S. enteritidis infection in chickens has increased, so has the incidence of S. enteritidis infection in humans (185, 209, 223). S. enteritidis now frequently rivals S. typhimurium as the most common cause of salmonellosis in the United States, and there has been a similar increase in many other parts of the world as well (12, 185, 209).
General Description of Pathogenesis
The first step in the disease process is transmission to a susceptible host. As mentioned above, for Salmonella, this is usually by consumption of contaminated food. It is estimated from volunteer studies that 105 to 1010 bacteria are required to initiate an infection (103), but the exact amount needed varies with the strain (170–172), what is consumed with the bacteria (17), and the physiological state of the host. It is generally believed that a large inoculum is required to overcome the stomach acidity and to compete with the normal flora of the intestinal tract (reviewed in references 234 and 239). There are several lines of evidence to support this. The infectious dose decreases when Salmonella is consumed with food that traverses the stomach rapidly (i.e., liquids) or with food that neutralizes the stomach acidity (i.e., cheese, milk) (234). Individuals with high gastric pH, such as the elderly, are more susceptible to infection (15, 32, 95, 208, 222, 241, 242). It has also been shown that pretreatment of mice with streptomycin, which reduces the amount of the normal flora, decreases the dose of Salmonella required to infect 50% of the mice (35); similar effects of antibiotic treatment have been observed in humans (214).
In an article published in 1966, Kent et al. (142) wrote, “There is very little information on the distribution of the lesions in typical acute human Salmonella gastroenteritis, although textbooks frequently state that salmonellosis is a small intestine disease.” These words are as true 33 years later as when they were written. There is very little information about the pathological lesions found in humans during an infection with nontyphoid Salmonella serotypes. The few reports that do exist suggest that the colon rather than the small intestine is the primary site of involvement (37, 68, 119, 161, 173). What was reported in these cases is that there is a wide range of severity ranging from slight to severe edema with infiltration of polymorphonuclear leukocytes (PMNs) and monocytes, focal inflammation of the lamina propria, degeneration of the mucosa, and extravasation of erythrocytes and PMNs. The small intestines appeared normal. It should be noted that all these cases are from patients admitted to the hospital and that in several instances the infections were fatal (37, 119). It is difficult to ascertain how representative these cases are, because most people infected with Salmonella are never hospitalized, nor do they necessarily consult a physician.
To get information about the progression of infection relevant to the human situation, Kent et al. (142) infected rhesus monkeys and examined the colonization and histopathology of various organs at several time points (1, 2, 4, and 7 days) postinfection. The symptoms that developed in these monkeys were similar to what is seen in humans (142, 213). During the first 4 days, the ileum, cecum, distal colon, and mesenteric lymph nodes (MLN) were colonized in every animal; there was sporadic colonization of the jejunum, liver, and spleen. The MLN was still colonized in all animals on day 7. In the colon (days 1 and 2), infiltration of PMNs into the mucosa, sloughing of the epithelial cells, and a purulent exudate in the lumen were observed. Occasionally, scattered crypt abscesses and microabscesses in the lymphoid patches were observed. By day 4, bacteria were seen in colonic epithelial cells and within macrophages of the lamina propria. By day 7, involvement of the colonic mucosa was more diffuse (less focal) and the lamina propria contained more chronic inflammatory cells. Progression in the ileum was slower than in the colon. On days 1 and 2, only mild lesions were observed, and these were observed only in one of four animals. However, on days 4 and 7, severe mucosal lesions were observed in 5 of 6 monkeys. There was an infiltration of PMNs and macrophages into the lamina propria, elongation of the crypts along with scattered crypt abscesses, and cuboidal changes in the surface and crypt epithelium. These results suggested that a more typical infection with Salmonella involves both the ileum and colon and that the bacteria invade these tissues and colonize (at least transiently) distal sites such as the MLN, liver, and spleen.
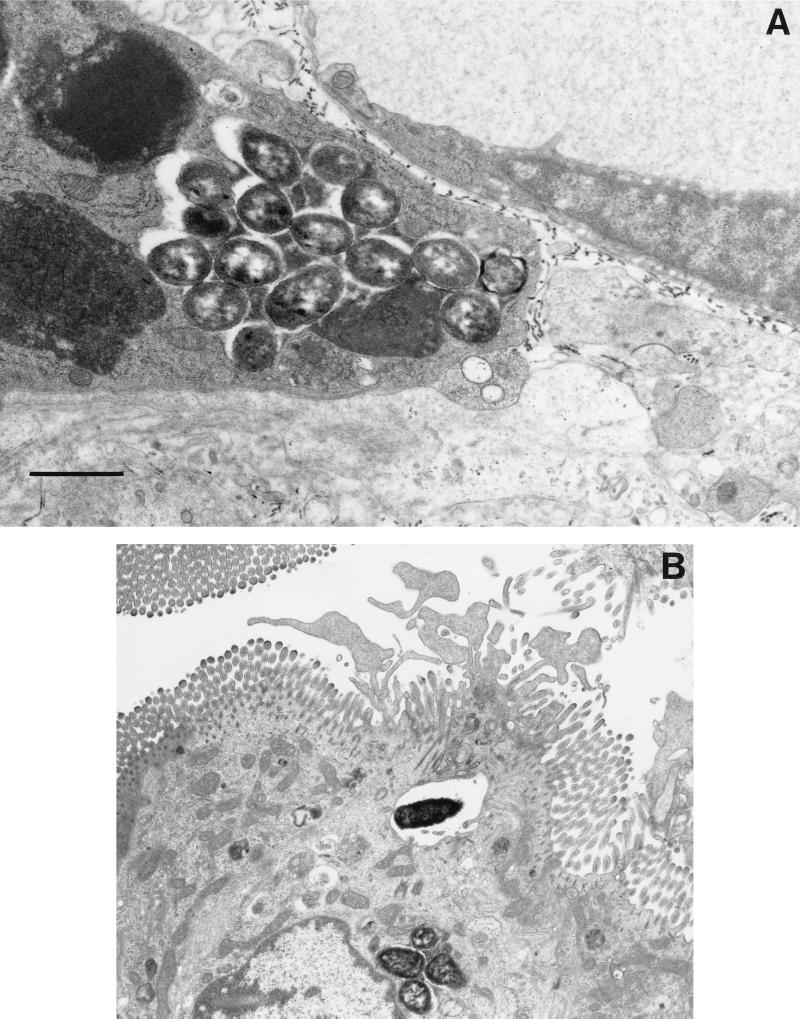
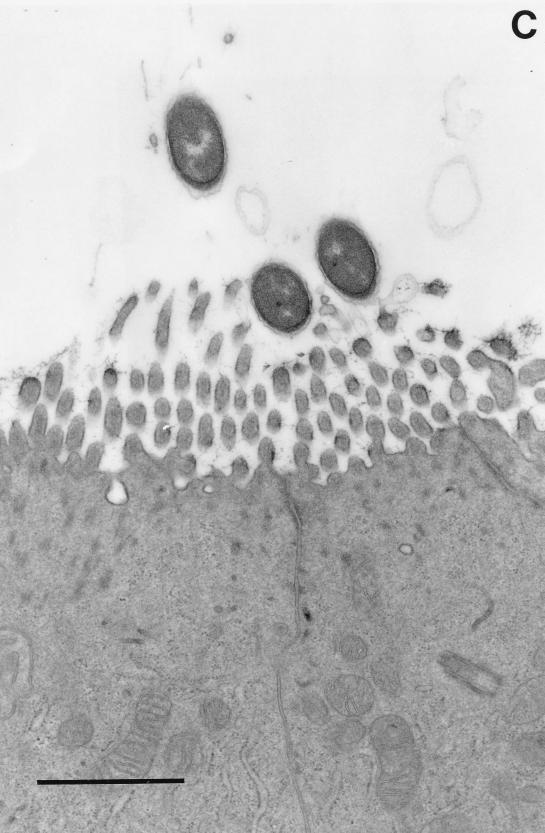
The most detailed ultrastructural study of an infection with Salmonella was done by Takeuchi (229) and Takeuchi and Sprinz (230) using a guinea pig model; these studies represent one of the earliest and best demonstrations of Salmonella traversing the epithelium through the epithelial cells. In this model, the guinea pigs are starved for 4 days prior to oral administration of the bacteria along with opium (to slow intestinal movement); only the ileum was examined in these studies. By 12 h, bacteria could be found within epithelial cells and within PMNs, and some of these PMNs were between epithelial cells or in the lumen. Bacteria that were in very close contact with the brush border seemed to cause degeneration of the microvilli and protruding cytoplasm much like the “ruffles” described in later experiments with mice and cultured cells (137, 146, 259) (Fig. 1B shows an example of ruffles). The initial phagocytic compartments containing bacteria were usually quite large, but they contracted as the vesicles moved to the subnuclear region. Villus rather than crypt epithelium was the primary site of involvement. By 24 h, an intense inflammatory exudate was observed and the brush border was reduced in height. At this time, some bacteria were found within desquamated epithelial cells and within PMNs in the lumen. However, many bacteria were in phagocytic cells within the lamina propria and Peyer’s patches (PP). At 48 h, the inflammation was more extensive and extended to the submucosa where bacteria could be found within phagocytes (Fig. 1A shows an example of bacteria within a phagocyte). Extrusion of epithelial cells, emptying of goblet cells, and elongation of crypt cells were also observed.
FIG. 1.
Interaction of Salmonella with intestinal mucosa. Transmission electron micrographs of non-follicle-associated epithelia from calves infected with either wild-type S. typhimurium ST4/74 (A and B) or with an isogenic invH mutant (C) are shown. (A) Macrophage from the laminae propriae of the absorptive villi with several internalized bacteria 3 h postinfection. (B) Enterocyte with an internalized bacterium. Note the membrane ruffling. (C) Enterocyte with attached bacteria (invH mutant). No ruffling was observed. Bars, 1 μm. Reprinted from reference 248 with permission of the publisher.
Similar interactions of Salmonella with the mucosa were observed in a rabbit ileal-loop model (96). In this model, the bacteria were seen adhering to the brush border as early as 3 h postinfection and the bacteria could be seen on and in epithelial cells and within the lamina propria by 7 h. Epithelium overlying lymphoid tissue was the most heavily involved and showed acute inflammation. By 12 to 18 h postinfection, as with the guinea pig model, PMNs were observed within the epithelium, in the lumen, and in the lamina propria. By this time, the villi were blunted and swollen and the crypts were hyperplastic. Similar results were found in two other studies with the rabbit ileal-loop model (245, 257). This model was also used in conjunction with a HeLa cell assay for invasion to test a variety of different S. typhimurium strains (94); there was a strong correlation between the ability to invade HeLa cells and the ability to invade the rabbit intestinal mucosa. More recently, the infection of calves with Salmonella has been examined as a model because calves exhibit an enteritis similar to that of humans (220). The pathology observed in this model is very similar to that described above (220, 244, 245, 248) (Figure 1). The fact that similar interactions of Salmonella with the intestinal mucosa occur in at least four different mammalian models (monkeys, guinea pigs, calves, and rabbits) suggests that similar interactions occur during human infections with Salmonella.
Mice are used as a primary animal model of infection today because of their ease of use, low cost, and susceptibility to infection with Salmonella. Oral infection of mice leads to colonization initially of the distal ileum and cecum and then of the draining lymph nodes by 48 h (43). In another study, it was found that although the PP represent only a small fraction of the small intestine, nearly 25% of the Salmonella cells within the small intestine were in the PP (127). It was also found that most of the bacteria found in the cecum could be easily washed off the tissue but that most of the bacteria in the small intestine and PP remained associated with the tissue after extensive washing (127).
Ultrastructural analysis of mouse ligated intestinal loops infected with S. typhimurium revealed that the bacteria initially invaded M cells almost exclusively (137); similar results were found in mouse ligated loops infected with S. typhi (146). In both these studies, the M cells frequently were associated with numerous bacteria and were ultimately destroyed by the invading bacteria. For these studies, high concentrations of bacteria were used (108 to 109 cells) in a relatively small ileal segment (4 to 5 cm); thus, it is possible that the destruction of the M cells observed is at least in part a function of the high bacterial load. As with the studies in rabbits, calves, and guinea pigs, host cells in close contact with Salmonella exhibited membrane ruffling during the invasion process (137, 146). It is not clear why exclusive invasion of M cells and destruction of the M cells was observed in the mouse ligated loop model but not in the other models. This difference could be due to the animal model chosen, the method of inoculation, the time point for observation, or differences in the test strains of Salmonella. However, it is clear that regardless of the model chosen, an early manifestation of the host-pathogen interaction is attachment to and invasion of the intestinal epithelium by the bacteria and subsequent inflammation of the lamina propria and lymph nodes. Thus, much of the research on Salmonella virulence has focused on identifying and studying the bacterial products involved in triggering these responses by the host.
ATTACHMENT
Prior to invasion of any cell type, bacteria must encounter and attach to one or more cell types found in intestinal tissue. Such tropism by S. typhimurium may involve several types of fimbriae or pili, four of which have been genetically defined.These include type 1 fimbriae (Fim), plasmid-encoded (PE) fimbriae, long polar (LP) fimbriae, and thin aggregative fimbriae (curli). Some studies suggest that these fimbriae each have a specific tropism for a certain cell type or for cells from particular animal species (25, 26) (Table 1). S. typhimurium induces transepithelial migration of neutrophils in an in vitro assay (168); therefore, it has been hypothesized that fimbriae might play a role in the recruitment of neutrophils to a site of infection (26). Fimbriae might not be directly involved in neutrophil recruitment, but it is possible that they help the bacteria attain close contact with host cells and therefore allow for the interaction of factors that do stimulate transepithelial migration of neutrophils.
TABLE 1.
Description of genetically characterized S. typhimurium fimbrial loci
| Locus | Map positiona | Tissue specificityb | Fold attenuationc |
|---|---|---|---|
| fim | 13 | NDd | 0.3 |
| lpf | 80 | PP | 4.8 |
| pef | pSLT | Villous intestine | 2.4 |
| agf/csg | 26 | ND | 3.3 |
Map positions are indicated in centisomes (23, 63). pSLT is the S. typhimurium virulence plasmid (85).
LD50s were determined for intragastrically infected mice; fold attenuation refers to the ratio of the LD50 of the respective mutant strain to the LD50 of wild-type S. typhimurium. Experiments were done previously (24, 158, 240).
ND, not determined.
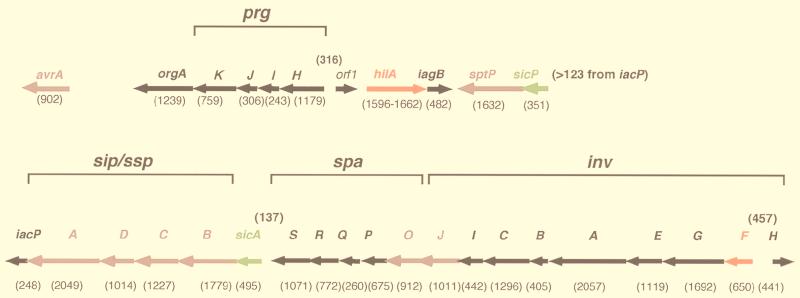
The organization of genes encoding the putative fimbriae of S. typhimurium is similar to that of other characterized fimbrial loci of related members of the family Enterobacteriaceae. The functions assigned to many of the Salmonella fimbrial proteins have been assigned based on information from the best-characterized system, that of type 1 fimbriae (55). It is notable that not all fimbriae (type 1 or otherwise) are exclusively associated with virulence, since nonpathogenic strains may contain one or more of the known fimbrial operons. Following is a description of four genetically defined loci in S. typhimurium and their contributions to pathogenesis (Table 1 and Fig. 2 give summaries of these loci).
FIG. 2.
Organization of genes within the four fimbrial operons of S. typhimurium. Gene lengths (in base pairs) are indicated below each open reading frame. Genes encoding the major fimbrial subunits are indicated in yellow, genes encoding chaperones are indicated in green, genes encoding ushers are indicated in blue, genes encoding minor subunits are indicated in black, and genes encoding regulatory proteins are indicated in red.
Type 1 Fimbriae
S. typhimurium type 1 fimbriae (Fim) are encoded by the fimAICDHF operon at centisome 15 (63, 215) and are morphologically similar to but antigenically distinct from that of the E. coli type 1 fimbriae (147). In E. coli and Salmonella, type 1 fimbriae are defined as peritrichous, 7 nm wide, and 0.2 to 2.0 μm (145, 147). Type 1 fimbriae specifically bind α-d-mannose receptors on various eucaryotic cell types; therefore, binding of bacteria with type 1 fimbriae to eucaryotic cells can be inhibited by the addition of α-d-mannose in vitro (“mannose-sensitive fimbriae”) (145). As in other members of the Enterobacteriaceae, type 1 fimbriae of S. typhimurium consist of a major subunit, FimA (21 kDa), and several other associated proteins including the FimH adhesin (55). Although the function of FimH as an adhesin appears the same in S. typhimurium and E. coli, the respective proteins are not highly similar (147) and do not show identical binding specificities (82). Perhaps these differences dictate to which tissues or surfaces these bacteria can bind.
E. coli and S. typhimurium undergo phase variation between a fimbriated and a nonfimbriated state (72). Although growth conditions that result in reduced or increased expression of fimA have been studied, it is not clear how environmental signals manifest a change in fimbriae expression (54). Moreover, the mechanism of phase variation of type 1 fimbriae in E. coli and S. typhimurium appears to be unrelated. E. coli fimA expression is regulated by a 314-bp promoter region that can undergo an inversion resulting in an on or off state (3). Unlike E. coli, S. typhimurium fimA has not been shown to be regulated by an invertible DNA sequence, since both fimbriated and nonfimbriated strains are observed to contain promoters in an on orientation (54). Perhaps not surprisingly, S. typhimurium does not appear to contain fimB or fimE, which encode putative recombinases involved in the inversion of the E. coli fimA promoter (144). On the other hand, the E. coli fim locus does not contain fimY or fimW, which may play a role in regulation of S. typhimurium fimA (258). In S. typhimurium, FimZ directly binds the fimA promoter and is required for expression of fimA, whereas no ancillary fim gene plays a direct role in the regulation of E. coli fimA. It should be noted, however, that some E. coli strains appear to have a FimZ homologue (71% identity) in the 5′ region of argU (fimU in S. typhimurium) based on sequence comparison analysis (258). However, it is not known if this homologue plays any role in the fimbriation of E. coli. Expression of a S. typhimurium fimA-lacZ reporter in E. coli (turned off) differs from that of the native E. coli fimA, further suggesting that these loci are regulated by different mechanisms (228).
The S. typhimurium type 1 fimbriae mediate adhesion to HeLa cells in vitro; however, they do not appear to play any role in adhesion to various other tissue culture cell types such as the human cell lines HEp-2, T84, and Int-407 or the canine cell line MDCK (25). At least two independent studies have shown that a fim deletion mutant appears to be slightly more virulent than a fim+ strain (158, 240). It has been suggested that type 1 fimbriated S. typhimurium strains as well as E. coli strains are removed by the liver more efficiently in the mouse model (155); therefore, it is not clear what role, if any, type 1 fimbriae play in adhesion or virulence of S. typhimurium in the mouse or other animal hosts.
Long Polar Fimbriae
The lpfABCDE (for “long polar fimbriae”) locus was identified in a search for S. typhimurium chromosomal loci not present in related members of the Enterobacteriaceae (23). This operon was mapped to centisome 80 of the S. typhimurium LT2 chromosome and is flanked by sequences homologous to those in E. coli K-12, suggesting that lpf may have been acquired by horizontal transfer during the evolution of S. typhimurium. Homologues of lpf have not been found in various other pathogenic Enterobacteriaceae, including enterotoxigenic, enteroinvasive, and enteropathogenic E. coli and Shigella. Moreover, at least two Salmonella strains, S. typhi and S. arizoneae, appear to have lost or never acquired lpf (22).
Expression of the lpf operon in a nonfimbriated E. coli strain results in the appearance of polar filaments in a subpopulation of transformants (23), hence the LP designation. Although the fimbriae encoded by lpf suggest a polar location on the bacterial cell surface, it has not been determined conclusively that LP fimbriae are polar on Salmonella. There is no direct evidence that lpf actually encodes the LP fimbriae; it is possible that lpf induces the expression of cryptic fimbrial genes on the E. coli chromosome. However, in addition to evidence described below, the organization of the lpf genes is similar to that of fim and the deduced Lpf protein sequences are homologous to proteins encoded by fim, strongly suggesting that lpf does encode components of fimbriae (23, 26).
LP fimbriae mediate adhesion to the cells of the Peyer’s patches of the small intestine in a mouse model of infection (27). In an in vitro, mouse small intestine model of infection, a mutation in lpfC (which encodes a putative outer membrane usher for fimbrial assembly) resulted in reduced colonization of PP but not of villous enterocytes. Complementation of the lpfC mutation restored the ability of S. typhimurium to associate with the PP and also enhanced its ability to colonize the villous intestine. An lpfC mutation has only a modest effect on virulence in a mouse model (a threefold increase in oral 50% lethal dose [LD50]). A mutation inactivating InvA, a component of the type III secretion system required for invasion, had a moderate effect on virulence in a mouse model (16- to 50-fold increase in oral LD50). However, the lpfC invA double mutant was significantly less virulent than the wild-type strain or either single mutant (150-fold increase in oral LD50) (28). Neither mutation alone or in combination with each other affected virulence by intraperitoneal infection, supporting the hypothesis that lpfC and invA are required for interaction of S. typhimurium with the intestinal mucosa. Perhaps for salmonellae to efficiently invade host tissues by using the type III secretion system, the bacteria must first be associated intimately with target cells. This requirement could be fulfilled by use of LP fimbriae which appear to be specific for cells in the PP, the primary site of infection by salmonellae in the mouse model (137).
A lpf-lacZYA transcriptional fusion reporter strain was tested to characterize the expression of lpf in vitro and in vivo (189). In vitro, phase variation from a Lac+ (on) to a Lac− (off) colony phenotype and from a Lac− to a Lac+ colony phenotype occurred at a frequency of 6.8 × 10−3 and 2.4 × 10−4, respectively. However, mice infected with the lpf-lacZYA reporter strain showed an increased selection for bacteria in an LP-on phase of expression. This selection was observed for bacteria isolated from PP but not for bacteria isolated from the MLN or spleen. Mice that were immunized with a glutathione S-transferase–LpfA (major fimbrial subunit) fusion protein prior to infection showed a strong selection against the LP-on expression state in the PP. These results together suggest that LP fimbriae are important at an early step of disease by an oral route of infection. The mechanism of regulation of expression of LP fimbriae has not yet been characterized, but it is thought that inverted repeats flanking lpfA might mediate phase variation of LP fimbriae by causing inversion of the lpfA gene (23).
Plasmid-Encoded Fimbriae
Several serotypes of Salmonella contain virulence plasmids ranging in size from 50 to 90 kb (108). S. typhimurium contains a 90-kb virulence plasmid (pSLT) which is required for full virulence of the organism after oral infection (110). The role of plasmid-encoded virulence determinants, anything from invasion to serum resistance to survival, is not clear due to conflicting reports on their function (111); however, it has been generally accepted that the virulence plasmid of S. typhimurium is important for causing a systemic infection after oral inoculation of experimental animals (109). Several virulence loci have been cloned and identified from the virulence plasmid, including the spv (for “Salmonella plasmid virulence”) (109) and rck (for “resistance to complement killing”) (113) genes. The spv region is sufficient for complementation of splenic infectivity of a plasmid-cured strain (109). The rck gene (see below) can confer in vitro serum resistance to serum-sensitive strains of E. coli and plasmid-cured Salmonella (53, 113, 120); however, its specific role in S. typhimurium virulence in vivo has not been determined. Upstream of rck, a 13.9-kb region was sequenced and determined to contain another putative fimbrial operon called pefBACD (for “plasmid-encoded fimbriae”) (85). Only four serotypes, S. typhimurium, S. enteritidis, S. choleraesuis, and S. paratyphi C, contain pef sequences, as determined by DNA hybridization to a pefA probe (26). The significance of this finding has yet to be established.
Transmission electron micrographs of nonfimbriated E. coli with pef sequences carried on a cloning vector reveal numerous peritrichous fimbriae (24, 85). E. coli producing PE fimbriae can also adhere to histological sections of murine small intestine more effectively than can nonfimbriated E. coli (24). A mutation in the putative outer membrane usher gene, pefC, does not affect adhesion to HeLa or T84 human cell lines of either S. typhimurium or E. coli carrying the pef locus (25). This is consistent with experiments demonstrating that plasmid-cured strains of Salmonella could still adhere to and invade cultured CHO (Chinese hamster ovary) cells as efficiently as the plasmid-containing strain could (158). The effects of a mutation in pefC were also tested in an intestinal-organ culture (IOC) model. The IOC model in this study involved infection of BALB/c mouse small intestines which had been removed from the mice just prior to infection and maintained under tissue culture conditions. Under these perhaps more native conditions, the S. typhimurium pefC mutant did not attach to the small intestine as well as the wild-type bacteria did when the two strains were used to coinfect the same tissue sample. This result was mirrored by coinfection with nonfimbriated E. coli carrying the pef operon on a plasmid and a plasmidless strain in the IOC model. The authors concluded that pef enhanced attachment to villous small intestine; however, it was not clear in this experiment if “villous intestine” included the M cells. Experiments to distinguish attachment to PP from villous small intestine have been performed with LP-fimbriated strains (27); therefore, it should be possible to test the PE-fimbriated strains in the same way.
The pefC mutant was also tested in an infant suckling-mouse model of infection (24). According to these studies, PE fimbriae were necessary for attachment to the small intestine and for fluid accumulation in infant mice. As a control, mutations in the downstream rck locus and another fimbrial locus (fim) were also tested in this model and shown to have no effect on fluid accumulation. When E. coli carrying the pef genes on a plasmid was tested in this system and compared to a plasmidless parent strain, no effect on fluid accumulation was seen, demonstrating that PE fimbriae were necessary but not sufficient for fluid accumulation in this model.
Thin Aggregative Fimbriae
Thin aggregative fimbriae (3 to 4 nm wide) (curli) were identified and purified from S. enteritidis (62) and found to be antigenically similar to E. coli curli (210). The locus encoding S. typhimurium curli, agfBAC, was cloned (226) and found to be organized identically to the E. coli csgBAC genes (114). S. typhimurium agfBAC maps to centisome 26 on the LT2 chromosome (63). (Due to the location of agf near a breakpoint of an 815-kb inversion that distinguishes S. typhimurium from S. enteritidis, it should be noted that the S. enteritidis agf genes map between centisomes 40 and 43.3 [63].) As in E. coli, agfC does not appear to be part of the agfBA transcript based on Northern analysis with an agfC-specific probe (60, 210). A divergent operon, agfDEFG, homologous to csgDEFG of E. coli was also found (210, 226). Although agf and csg encode similar proteins, they have low identity at the nucleotide level. However, due to the high level of identity between the csg- and agf-encoded components, csg mutants of E. coli can be complemented by corresponding S. typhimurium agf genes (210).
Expression of agf affects colony morphology, since curli-producing bacteria form a rigid multicellular network within the colony, a phenotype referred to as rdar (211). The rdar phenotype can be observed when bacteria are grown at ambient temperatures (i.e., below 37°C), in rich media and low osmolarity, or at 37°C under iron starvation (210). This inducible phenotype has provided a simple assay for studying the regulation of expression of curli genes. The rdar phenotype is dependent on rpoS, which encodes an alternate sigma factor required for transcription of genes in response to stress and starvation conditions (165), and ompR, a transcriptional activator of the ompC and ompF porin genes (210, 218, 235). Both rpoS and ompR are required for expression of agfD/csgD, which encodes a putative transcriptional activator (211). Agf/CsgD is believed to activate the transcription of agf/csgBA, which encodes the surface-exposed nucleator and major fimbrial subunit, respectively, but it is not known if Agf/CsgD directly interacts with the agf/csgD promoter (210, 211). It is also unknown if RpoS and OmpR directly interact with the agf/csgD promoter.
Curli-producing bacteria tend to autoaggregate, a phenomenon which has been suggested to enhance the survival of salmonellae facing hostile barriers such as stomach acid or other biocides they may encounter (61). Another hypothesis suggests that curli are involved in attachment of bacteria to the host cell epithelium. This idea is supported by experiments showing that curli but not SEF 21 (for “Salmonella enteritidis fimbriae 2”) (type 1) or SEF 14 (not found in S. typhimurium) mediate the binding of S. enteritidis to fibronectin, a component of the eucaryotic extracellular matrix (61, 193). An in vitro system has been developed which shows that curli play a significant role in attachment to differentiated murine small intestinal cells (226). In vivo, a preferred site of attachment by curli-producing salmonellae has yet to be determined. The LD50 of an agfB mutant of S. typhimurium was increased only 3.3-fold compared to wild-type bacteria when infection occurred intragastrically (240). Despite this modest effect on LD50, it is clear that agf significantly contributes to pathogenesis. Mouse experiments have revealed that a mutation in agfB, the putative surface-exposed nucleator of AgfA fimbrin subunits (115), dramatically increases the LD50 of a Δfim/pefC/lpfC mutant (AJB12) from 1.7 × 105 to 1.5 × 107 cells (an 88-fold increase in oral LD50). This was especially notable since AJB12 is slightly more virulent than wild-type bacteria, most probably due to a dominant Δfim phenotype.
Rck
Rck was originally identified as a “cryptic plasmid”-encoded protein that conferred high levels of serum resistance to both E. coli and plasmid-cured, serum-sensitive Salmonella strains (113). Rck is a member of a family of outer membrane proteins (OMPs) including PagC (for “phoP-activated gene”) of S. typhimurium (112) and Ail (for “attachment and invasion locus”) of Yersinia enterocolitica (180, 181). Unlike PagC, Rck is more similar to Ail in that both proteins confer invasiveness and serum resistance to noninvasive, serum-sensitive E. coli strains (34, 53, 113, 120, 121, 198). Both proteins are predicted to have multiple transmembrane domains and exposed loops based on amino acid sequence analysis (53, 180). Site-directed mutagenesis and “domain-swapping” experiments have been done with Rck and show that loop 3 is required for serum resistance and invasion in E. coli (53). Unfortunately, most probably due to the presence of at least one other invasion system and rck-independent serum resistance mechanisms (227), a specific role for Rck in S. typhimurium pathogenesis has not been reported. Moreover, virulence plasmid-cured strains carrying only the spv genes can cause disease in a mouse model almost as well as wild-type strains can (109), suggesting that rck is not important in the penetration of the intestinal epithelium. However, it is possible that Rck plays a role in the infection of nonmurine hosts of S. typhimurium.
Functional Redundancy?
The presence of at least four fimbrial systems and Rck suggests that attachment to cellular or noncellular surfaces may be a critical step in the survival of S. typhimurium in the environment. Due to the apparent redundancy of fimbrial operons in Salmonella, it has been difficult to assess the role of each system in an in vitro or in vivo assay, because one system may compensate for another. van der Velden et al. have shown by transmission electron microscopy that even an attenuated quadruple fimbrial mutant can still produce what appears to be a fifth fimbrial structure (240). It will be interesting to see how mutations in five fimbrial loci will affect attachment and virulence in vivo.
MUCOSAL INVASION
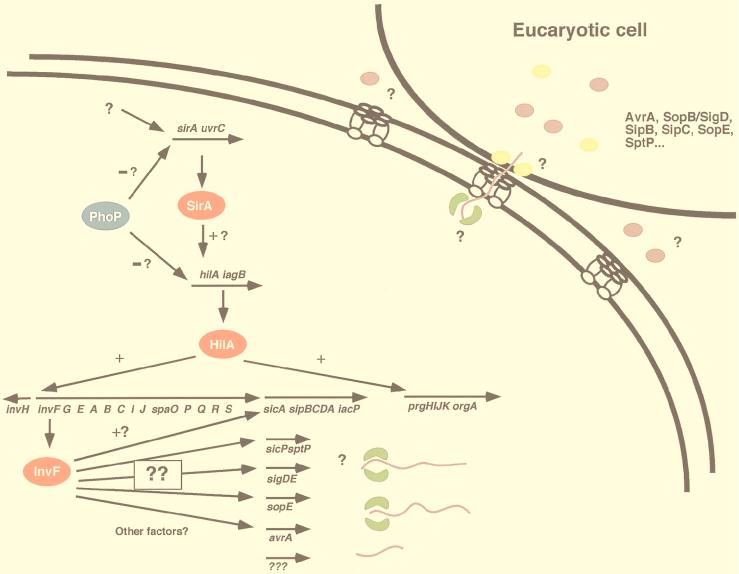
Several studies have shown that salmonellae preferentially attach to and invade cells in the PP of the small intestine in orally infected mice (127, 137, 197). However, bacteria can also be found in nonphagocytic enterocytes (229). The mechanisms of Salmonella invasion, that is, the stimulation of nonphagocytic cells to internalize bacteria, are clearly complex. Putative invasins that stimulate the uptake of bacteria by epithelial cells have been identified in organisms such as Yersinia by introducing genomic libraries of invasive strains into E. coli “laboratory” strains (134, 181). Little success has been achieved in the search for invasion genes by this technique in S. typhimurium. In S. typhi, one locus, invABCD*, was identified by this technique (77); however, nothing is known about how this locus functions in invasion (the asterisk indicates that a nonhomologous locus with the same name was identified in S. typhimurium). Interestingly, S. typhimurium also has the invABCD* region (182) in addition to the genetically linked invasion system encoded at centisome 63 (updated from the old map position of centisome 59 [215]). This locus, known as Salmonella pathogenicity island 1 (SPI1), is believed to have been acquired by horizontal transfer from another pathogenic bacterial species during its evolution (107) (see Fig. 3 for a map of SPI1).
FIG. 3.
Organization of genes in SPI1 at centisome 63. Gene lengths (in base pairs) are indicated below each open reading frame. Gaps longer than 30 bp between genes are indicated above intergenic spaces. Genes encoding regulatory proteins are indicated in red, genes encoding secreted proteins are indicated in mauve, genes encoding chaperones are indicated in green, and genes encoding apparatus proteins or proteins of uncharacterized function are indicated in black.
The first cloned SPI1 genes, invABC(D), were identified by complementing an attenuated, invasion-defective strain of S. typhimurium with a cosmid library of S. typhimurium genomic DNA (89). It was determined subsequently that invD was not actually a SPI1 gene but represented a noncontiguous fragment which had been cloned into the complementing cosmid (182). Since the discovery of invABC, many more genes have been identified in SPI1. The 40-kb SPI1 region encodes at least 33 proteins which include components of a type III secretion apparatus (13, 29, 76, 89, 90, 102, 107, 136, 138, 196), regulatory proteins (19, 135, 138), and secreted effector proteins and their chaperones (59, 87, 131, 139–141, 156). In addition to the SPI1 system, S. typhimurium has two other known type III secretion systems: the SPI2 (Salmonella pathogenicity island 2) system for macrophage survival (centisome 30.5) (123, 124, 191) and the flagellar assembly system for motility (centisomes 27 and 42 to 43) (215) (for a recent review of type III secretion, see reference 130). The hallmark of type III systems is that none of the secreted proteins has a conserved or recognizable signal sequence. Although several type III secretion systems have been identified in many different organisms, the mechanisms by which proteins are secreted has yet to be elucidated.
At least three proteins encoded within SPI1 are known to participate in a supramolecular structure which reaches from the cytoplasmic membrane to the outer membrane. Electron microscopy has visualized this structure, which resembles a syringe (149). This syringe is believed to secrete effector proteins from the bacterium, and these proteins stimulate dramatic cytoskeletal rearrangements in eucaryotic host cells (81, 84, 229). These membrane ruffles facilitate the engulfment of the bacteria by eucaryotic cells. Mutations in genes which encode the apparatus proteins, regulators, and certain secreted proteins eliminate or greatly diminish invasion and the appearance of membrane ruffles (90, 101). In addition to invasion, the SPI1 system appears to function in programmed cell death (apoptosis) of infected cultured macrophages (48, 187) and the recruitment of PMNs to the small intestines in a bovine model of infection (92).
Limited LD50 analysis has been done with nonpolar SPI1 mutants. Transposon insertions in invA (28, 89), invF, invG, hilA, sipC, sipD, spaR, and orgA (197) result in a 16- to 100-fold attenuation of S. typhimurium by oral inoculation. Most of these mutations result in the loss of a functional type III secretion and/or translocation system. Mutations in individual putative effector genes result in subtle in vivo phenotypes, suggesting that no single protein is sufficient for pathogenesis in the mouse model.
Apparatus Genes
The genes encoding the secretion machinery fall into two clusters: inv-spa and prg-org (Fig. 3). The two are separated by several other genes including those which encode secreted proteins (Sip/Ssps, SptP) and an essential regulator (HilA). Mutations in almost any of the apparatus genes are predicted or have been shown to result in the absence of several proteins from culture supernatants.
invH.
InvH (16.5 kDa) was originally identified as both an attachment and an invasion factor for S. typhimurium in vitro (13). Stone et al. also identified invH in a screen for invasion mutants of S. enteritidis; however, a mutation in invH did not affect attachment in vitro (224). Recently, Daefler and Russel elegantly showed that InvH is actually an outer membrane protein required for the proper localization of InvG, a PulD-like outer membrane component of the SPI1 type III secretion system (67). Although InvH localizes to the outer membrane, it does not appear to be an integral membrane protein; it is likely to be anchored to the outer membrane by a lipid moiety. InvH was determined to have a consensus lipoprotein sequence which could be labeled by [3H]palmitic acid. Deletion of the consensus sequence abolished palmitic acid labeling. Secretion of one of the SPI1 type III apparatus substrates, SipC (described below), was eliminated in an invH deletion mutant, confirming InvH participation in the secretion apparatus. Secretion could be restored by invH in trans, but an invH clone missing its lipoprotein signal sequence could not complement the invH mutation (67). In light of the above evidence, it seems unlikely that InvH normally acts as an attachment factor; instead, it is likely to be an essential structural component of the type III secretion apparatus.
Several groups have found mutations in invH which affect invasion in vitro and in vivo (159, 248). Lodge et al. tested the ability of TnphoA mutants defective for invasion in HEp-2 cells in vitro to invade ileal epithelial cells in the rabbit model (159). InvH mutants were recovered in similar numbers to the wild-type parent strain; therefore, the role of InvH in pathogenesis in the rabbit model was not clear. In another study, an invH mutant was shown to be recovered in fewer numbers in bovine ileal loops, demonstrating a role for invH in invasion in cows (248). This result may suggest that InvH (and the SPI1-encoded machinery) is required for invasion of certain host tissues but not others.
invG.
Amino acid sequence analysis of the SPI1 type III syringes demonstrated that InvG is a major component of these structures (149). InvG belongs to a family of proteins known as secretins (206) and is required for the secretion of proteins by the SPI1 type III apparatus (67, 117, 118, 138, 141). InvG is similar to MxiD of Shigella (8), YscC of Yersinia (148, 178), and PulD of the Klebsiella type II pullulanase secretion system (71). Based on these similarities, InvG was assumed to be targeted to the outer membrane (138). Because the PulD secretin requires a lipoprotein for proper localization, it was believed that a lipoprotein may also be involved in the localization of InvG to the outer membrane. Three putative lipoproteins had been identified in SPI1, i.e., InvH (13, 67), PrgH (29, 196), and PrgK (29, 196). InvH, but not PrgH or PrgK, was required for InvG localization (67). In wild-type S. typhimurium, InvG was found in density gradient fractions containing outer membrane proteins; in an invH mutant, InvG was found mostly in the inner membrane fractions. This was confirmed in another study which showed that InvG could form ring-like structures in the outer membranes of E. coli (66). Similar to the previous study, targeting of these structures to the outer membrane required InvH (66). Unlike PulD, which requires the PulS lipoprotein for stabilization of the PulD polypeptide in addition to proper localization, InvG does not require InvH for stabilization (67). Although InvH was shown to be required for the proper localization of InvG in the outer membrane, it is likely that another putative lipoprotein, PrgK (see below), also interacts with InvG, because it was purified from the secretion syringes (149).
invE.
invE was identified upon sequence analysis of the region where invABC was found (102). invE encodes a 43-kDa polypeptide required for invasion in vitro. In addition, membrane ruffling was not observed upon analysis of electron micrographs of cultured cells infected with an invE mutant.
InvE is homologous to YopN, a hypothesized “gatekeeper” for the Yersinia pseudotuberculosis and Y. enterocolitica type III secretion systems (36, 83). Yersinia spp. produce a type III secretion system which secretes Yops (for “Yersinia outer proteins”), proteins required for cytotoxicity against macrophages (for a review, see reference 65). It is believed that YopN prevents the secretion of other accumulated Yops prior to their contact with an appropriate target cell, i.e., a macrophage (36). yopN mutants secrete Yops in an unpolarized manner; that is, Yops are secreted in larger than normal quantities into the extracellular milieu. Moreover, Yops are not efficiently translocated into eucaryotic target cells from a yopN mutant. Most probably as a result of this inefficient translocation of Yops, yopN mutants are not cytotoxic.
YopN localizes to the bacterial cell surface (83), but it is also found in culture supernatants (36, 83). Unlike several of the other Yops, YopN is not internalized by macrophages (36). It is not known if Salmonella InvE is surface localized, secreted, or translocated into eucaryotic cells.
Similar to Yersinia, S. typhimurium appears to undergo an accumulation of at least one secreted protein, InvJ, prior to contact with tissue culture cells or exposure to high (10%) concentrations of bovine calf serum (BCS) (260). InvJ is released into culture medium upon cell contact or exposure to 10% BCS, but it is not known if it is translocated into host cells. An invE mutant is still able to secrete InvJ (260). Perhaps InvE, like YopN, functions in the retention of secreted substrates within the bacterium rather than the translocation of substrates through the bacterial and/or eucaryotic membranes. In light of the evidence that an invE mutant can still secrete at least one protein, it would be interesting to see if the other SPI1 effector proteins are still secreted, perhaps hypersecreted, by this mutant. Although one might initially assume that increased secretion of invasion factors would increase invasiveness (or cytotoxicity), the yopN model demonstrates that secretion of proteins in the proper context may be important for function.
Another phenotype observed with an invE mutant was its ability to influence the intracellular calcium concentrations, [Ca2+]i, of eucaryotic cells. Intracellular Ca2+ is known to be an important regulator of various cellular functions, including phagocytosis (162). Wild-type Salmonella was shown to induce an 18-fold increase in [Ca2+]i over an 1-h time course; in contrast, cultured cells infected with the invE mutant did not stimulate this influx (102). Because InvE may be a part of the secretion apparatus rather than a secreted effector, it is possible that InvE is indirectly involved in the [Ca2+]i levels of host cells. It is more likely that a substrate(s) secreted by the SPI1 secretion apparatus is directly involved in affecting [Ca2+]i. This would imply that mutations in any of the essential apparatus genes would result in the same [Ca2+]i phenotype as an invE mutant. However, this experiment has not been repeated with any of the other SPI1 mutants.
invA.
InvA encodes a putative inner membrane protein of 71 kDa homologous to type III secretion proteins from various animal- and plant-pathogenic bacteria (130). In addition, InvA has homologues in systems involved in the biosynthesis of flagella in E. coli, Salmonella, and other bacterial species. InvA and MxiD of Shigella are so similar that mxiD could complement an invA mutant almost as well as invA itself (100). Yersinia LcrD is also similar to InvA. However, unlike mxiD, lcrD of Y. pseudotuberculosis could not complement an invA mutation (100). TnphoA mutagenesis and amino acid sequence analysis predict that LcrD (in Y. pestis) is an integral membrane protein with eight transmembrane domains and a C-terminal cytoplasmic domain (199, 200). Based on this model, in addition to TnphoA mutagenesis of invA, it is believed that InvA assumes a similar structure in the inner membrane of Salmonella (89, 90). However, the N-terminal sequences of InvA and LcrD were found to be more similar to each other than were the C-terminal sequences. Ginocchio and Galán (100) hypothesized that the C-terminal domains were required for the recognition and secretion of species-specific proteins and that the conserved N-terminal sequences were not as important for this specificity. To test this, chimeric proteins between InvA and LcrD were constructed and tested for their ability to complement an invA mutant. When the N-terminal region of LcrD was fused to the C-terminal domain of InvA, the chimeric protein complemented the invA mutant for invasion nearly as well as invA did. However, a chimeric protein consisting of the N-terminal half of InvA and the C-terminal domain of LcrD could not complement an invA mutant. These results supported the idea that the C-terminal cytoplasmic domains of these proteins were essential for the recognition of species-specific substrates intended for secretion or for species-specific interactions with other apparatus components.
In Yersinia, mutations in lcrD not only affected the secretion of Yops required for virulence but also reduced expression of the genes which encode these proteins (200). LcrD is not believed to be a membrane-bound regulator. Instead, it is believed that a negative-feedback inhibition of yop expression occurs in the absence of a functional secretion apparatus (see “spaS” below [225]). This negative-feedback regulation effect on the SPI1 genes which encode secreted proteins (131, 139, 140) has not been reported for any of the apparatus mutants of Salmonella.
It is notable that InvA is also homologous to the SPI2 type III secretion protein SsaV (124). This poses an interesting problem for the bacterium: how do secreted substrates distinguish one secretion system from another? It is also notable that the SPI1 and SPI2 systems are less similar to each other than the SPI1 or SPI2 system is to other systems such as the Yop secretion (Ysc) system of Yersinia. The ultimate functions (i.e., invasion versus cytotoxicity) of all of these secretion systems, despite their similarities in structure, are quite different. It will be interesting to see how secreted substrates are properly targeted to their respective secretion systems. There is also the intriguing possibility that secreted substrates from one system are capable of using alternate systems for export out of the cell (124).
invB.
invB is located 24 bp downstream of invA and encodes a protein with a predicted molecular mass of 15 kDa (76). Like InvA, InvB is homologous to proteins of other type III secretion systems, but its function is unknown. A kanamycin resistance cassette in invB did not affect invasion in vitro; therefore, its role in the secretion apparatus may not be essential under these conditions. Similarly, in Shigella, a mutation in the invB homologue, spa15, does not affect invasion in vitro (217). InvB has not been analyzed further, probably due to the apparent lack of phenotype associated with invB mutants.
invC.
The start codon of invC overlaps the stop codon of invB, suggesting that the two genes are cotranscribed (76). Unlike invB, a mutation in invC was shown to reduce invasion considerably in vitro. Invasion of cultured epithelial cells by the invC mutant was partially restored by providing invC in trans; however, the mutation in invC (a kanamycin resistance cassette disruption) probably had a polar effect on expression of the downstream genes.
InvC (47 kDa) is a member of a family of proteins similar to the catalytic subunits of F0F1 ATPases (76). Potential nucleotide binding domains, Walker boxes A and B, in InvC prompted Eichelberg et al. (76) to mutate a predicted critical residue in Walker box A of a plasmid encoded invC copy. Although a full-length InvC mutant protein, K165EInvC, was produced, it failed to complement the invC chromosomal mutation. InvC was also tested for its ability to hydrolyze ATP. Wild-type InvC was shown to hydrolyze ATP whereas K165EInvC could not.
The requirement of ATP for the secretion of proteins has been shown for the Sec-dependent secretory pathway (157) and is likely to be important for type III secretion as well. Eichelberg et al. hypothesized that InvC might interact with other components of the type III secretion apparatus to facilitate the translocation of proteins out of the cell (76). The subcellular localization of InvC has not yet been reported, but it is not believed to be an integral membrane protein based on its predicted amino acid sequence (76). Elucidating the precise interaction of InvC with other components of the secretion machinery as well as the secreted substrates might provide insight into how proteins with uncharacterized secretion signals are exported out of the cell.
invI (spaM).
In a continuing effort to sequence SPI1, Collazo et al. identified invI and determined that it was needed for invasion in vitro (59). Groisman and Ochman also identified this and other genes while analyzing sequences found in Salmonella but not E. coli K-12 (107). Unlike many of the type III secretion proteins, InvI/SpaM does not have significant homology to proteins of other organisms. Proteins with low identity to InvI/SpaM have been found in Shigella (SpaM) and Salmonella (FliJ), but their similarity is so low that it is difficult to make any meaningful comparisons between them. It is also not known where these related proteins localize in the cell or how they function.
spaP, spaQ, spaR (surface presentation of antigen).
As mentioned above, Groisman and Ochman identified several genes which were not found in E. coli K-12 but were homologous to genes found in Shigella (107). spaP was identified downstream of spaO (see below) and predicted to encode a protein of 24 kDa. A spaP mutant has a dramatically reduced capability for invasion in vitro (58, 107). The Shigella homologue of SpaP, Spa24, was able to fully complement the spaP mutant for invasion in vitro (107). SpaP is believed to be localized to the inner membrane; however, evidence showing this has not been reported.
spaQ is predicted to encode a small, hydrophobic protein of 9 kDa (107), which is highly similar to SpaQ of Shigella (217) and FliQ of the gram-positive bacterium Bacillus subtilus (31) in addition to proteins from several other organisms (130). Although SpaQ is predicted to be localized to the inner membrane (130), evidence supporting this has not been demonstrated. SpaR is homologous to SpaR/29 of Shigella (107, 217) as well as to proteins from other systems. SpaP, SpaQ, and SpaR are required for invasion and for the secretion of SipB, SipC, and InvJ (58), but the precise function of these proteins in the secretion machinery is unknown. Complementation of spaP, spaQ, and spaR mutants with their respective wild-type alleles restored both invasion and secretion of proteins to wild-type levels (58). In this study, the expression of genes encoding secreted proteins did not appear to be affected because whole-cell lysates of the spaP, spaQ, or spaR mutants contained “comparable” amounts of the withheld proteins to those in wild-type lysates. Quantitative analysis with transcriptional reporters to the pertinent genes would complement and extend these observations.
spaS.
The transcriptional organization of the inv-spa-sip genes has not been confirmed, but it appears that spaS might be the last gene of the inv-spa cluster. This is similar to the organization of genes encoding SpaS homologues in other systems including yscU of pYV (Yersinia virulence plasmid) and ssaU of SPI2 in Salmonella (11, 124). SpaS, like InvA, is a highly conserved protein of about 40 kDa and is homologous to proteins from several other organisms including Yersinia, Shigella, and Pseudomonas (see reference 130 for a complete list). The best-characterized homologue is Y. enterocolitica YscU, which is believed to be an inner membrane protein (11). YscU is required for secretion of Yops; therefore, it has been assumed that SpaS is also required for secretion of proteins from S. typhimurium.
Mutations in yscU affect not only the secretion of Yops but also the transcription of several yop genes (11); yop expression was repressed in a yscU mutant. When expression of yop genes was artificially induced by the addition of the yop transcriptional activator virF in multicopy, Yops could be produced but not secreted by the yscU mutant. This phenotype was not found to be universal to all of the apparatus proteins; another putative membrane protein, YscJ (homologous to PrgK of S. typhimurium), when mutated, did not affect the expression of yop genes (11).
In Yersinia, yop expression is under a negative-feedback control mechanism (for a review, see reference 225); that is, mutations affecting several of the type III secretion proteins also affect the expression of other genes. This is also a well-characterized phenomenon observed for the biosynthesis of flagella in Salmonella. The production of flagella requires the expression of genes organized into three classes, I, II, and III, which are expressed in a sequential manner (150). Expression of flagellar genes requires, in addition to other regulators, the alternate sigma factor FliA (192). However, FliA cannot activate transcription in the presence of the antisigma factor FlgM (99). Upon assembly of the flagellar basal body, FlgM is secreted and FliA can subsequently activate transcription of class III genes encoding components of flagella. In this system, the bacteria prevent the accumulation of proteins secreted by the basal body until the basal body is formed.
Despite the genetic and physical similarities between the SPI1 and the flagellar type III secretion systems, SPI1 gene expression (or Yersinia Ysc expression) has not yet been shown to parallel the expression of the flagellar regulon. Repression of genes encoding secreted substrates prior to the assembly of the SPI1 secretion machinery is a feature which remains to be determined. Although this type of feedback control appears to be functioning in Yersinia with yscU, the mechanism by which it occurs is unknown.
prgH (phoP-repressed gene).
prgH was identified in a screen for genes repressed in a pho-24 mutant (29). A pho-24 mutation [phoQ(Con)] results in a constitutively active form of PhoP, the response regulator of the phoP/phoQ two-component regulatory system (80, 179). Unlike many of the type III secretion proteins, PrgH has not been found to be homologous to any other protein. PrgH is predicted to be a 45-kDa lipoprotein and a major component of the SPI1-associated syringe structures (29, 149, 196). Upon amino acid sequence analysis of the syringe structure components, PrgH did not appear to be processed. Syringe structures containing functional PrgH tagged with an epitope were labeled with monoclonal antibodies specific for the epitope tag; the antibodies appeared to preferentially label the base of the syringe structures (149).
prgI and prgJ.
prgI and prgJ are predicted to encode proteins of 8.8 and 10.9 kDa, respectively (196). Neither protein contains a recognizable signal sequence, and both are highly similar to secretion proteins from Shigella (MxiH and MxiI for PrgI and PrgJ, respectively) (9) and Yersinia (YscF and YscI for PrgI and PrgJ, respectively) (178). Little is known about the function of prgI and prgJ because mutations in these genes have not been constructed and tested for invasion in vitro. Moreover, it is not known where these proteins or their homologues in other systems localize in the bacterium.
prgK.
PrgK (28 kDa), like PrgH, is believed to be a membrane-associated lipoprotein (196) and is a major component of the type III secretion syringe structure (149). Unlike PrgH, PrgK is similar to proteins from other systems including MxiJ of Shigella and YscJ of Yersinia (10, 130, 178). Like PrgI and PrgJ, it is not conclusively known where PrgK localizes or if a mutation in prgK would abolish invasion in vitro. However, due to its participation in the formation of syringe structures, it is predicted that PrgK would be membrane associated and that a prgK mutant would be noninvasive (149).
orgA (oxygen-regulated gene).
orgA was identified in a screen for genes which were expressed under oxygen-limiting conditions (136). OrgA mutants do not secrete invasion-associated proteins into culture supernatants; therefore, OrgA is presumed to be a part of the type III secretion apparatus (197). Little is known about the function of OrgA (48 kDa), but it is required for invasion into cultured epithelial cells and for cytotoxicity to macrophages (187). In addition, a transposon insertion in orgA results in attenuation of S. typhimurium in intragastrically infected mice (197).
Secreted Proteins and Their Chaperones
Ten proteins secreted by the SPI1 secretion apparatus have been genetically characterized, and eight of them are encoded within SPI1 (Table 2). Mutations in the apparatus genes result in the absence of these proteins from culture supernatants; however, it is possible that these proteins are secreted via other type III systems under different conditions (124, 128). Three putative chaperones which themselves are not likely to be secreted have also been identified.
TABLE 2.
Proteins secreted by the SPI1 type III secretion system
| Protein | Host cell localizationa | Functionb | Homologuesc |
|---|---|---|---|
| SipA | Surface | NDd | IpaA (Shigella) |
| SipB | Intracellular | Translocation? Apoptosis? | IpaB (Shigella), EspD (EPECe), EspB (EHECe), Pep/PopB (Pseudomonas aeruginosa) |
| SipC | Intracellular | Translocation? | IpaC (Shigella) |
| SipD | Extracellular | Translocation? | IpaD (Shigella) |
| InvJ/SpaN | ND | Secretion | SpaN (Shigella) |
| SpaO | ND | Secretion | SpaO (Shigella); YscQ (Yersinia); FliNf |
| SptP | Intracellular | Cytoskeletal disruption; splenic colonization | YopE, ExoS (N-term.); YopH (C-term.) |
| SopE | Intracellular | Cytoskeletal reorganization; invasion | None identified |
| SigD/SopB | Intracellular | Invasion; transepithelial signaling of PMNs | IpgD (Shigella) |
| AvrA | Intracellular | ND | YopJ/YopP (Yersinia); AvrAxc (Xanthamonas) |
Localization of the secreted protein with respect to the eucaryotic host cell.
? indicates that the location of these proteins with respect to the host cell has not been determined.
Homologues listed have significant sequence and/or functional similarity to the respective Salmonella protein.
ND, not determined.
EHEC, enterohemorrhagic E. coli; EPEC, enteropathogenic E. coli.
FliN, “flagellar motor switch protein” from E. coli, S. typhimurium, Caulobacter crescentus, and several other gram-negative bacteria.
SPI1-encoded secreted proteins.
(i) invJ (spaN) and spaO.
InvJ is encoded within the inv-spa cluster of genes, which predominantly encodes apparatus proteins (59), and it does not have significant similarity to any other protein. Secretion of InvJ (36 kDa) is stimulated by the presence of 10% BCS or cultured epithelial cells (260). Interestingly, fixed tissue culture cells could not stimulate the secretion of InvJ, the significance of which has not been determined. Immediately downstream of invJ, spaO was identified with the putative spa apparatus genes. Like InvJ, SpaO is a secreted protein dependent upon the SPI1 secretion machinery (107, 156).
Collazo and Galán reported that InvJ and SpaO were required for the secretion of several other proteins including InvJ, SpaO, SipB, and SipC (58). Therefore, a mutation in either invJ or spaO resulted in an invasion-defective phenotype (58, 59). Expression of the genes encoding these proteins appeared unaffected; however, these observations were based on analysis of immunoblots and not transcriptional reporters to these genes. It is unclear how InvJ and SpaO function in the secretion of the other proteins.
(ii) sicA (Salmonella invasion chaperone).
sicA encodes a putative chaperone for one or more of the secreted proteins encoded in the sip operon (140). Chaperones have been identified in other type III secretion systems and are required for the secretion, stabilization, and/or translocation of effector proteins (50, 87, 154, 176, 204, 249, 251, 252). Nothing is known about the interaction between SicA and the secreted polypeptide(s), but it is required for invasion in vitro. Several secreted proteins have been identified in Yersinia, some with specific chaperones (249, 251, 252). It is intriguing that the four Sip proteins may share one chaperone; however, it is not known which proteins actually require SicA. SicA is homologous to IpgC of Shigella, a chaperone required for the stabilization and secretion of the Shigella invasins IpaB and IpaC (for “invasion plasmid antigen”) (176). Perhaps SicA functions similarly and interacts with the Salmonella IpaB and IpaC homologues, SipB and SipC, respectively.
(iii) sipB/sspB (Salmonella invasion protein or Salmonella secreted protein).
Several groups independently identified sipB (the locus containing sipB has been designated sip and ssp, but for simplification, we refer to it as sip) in a cluster of genes encoding secreted proteins required for invasion of S. typhimurium and S. typhi into tissue culture cells (125, 131, 140). sipB is located immediately downstream of the putative chaperone gene sicA and upstream of four additional open reading frames. SipB (63 kDa) is highly similar to Shigella IpaB, a protein required for invasion and cytotoxicity of shigellae in vivo and in vitro (21, 41, 164, 262). Both SipB and IpaB localize within eucaryotic cells (57, 237). For SipB to be translocated into eucaryotic cells, SipC and SipD must also be present (57). Moreover, SipB, SipC, and SipD are required for the translocation of other proteins, including the protein tyrosine phosphatase SptP (88, 141) and SopE (256). It has been proposed that SipB, SipC, and SipD are involved in the translocation of secreted effector proteins into the eucaryotic cytoplasm; however, it is not known how these proteins, which are themselves translocated (except SipD), accomplish this. Unlike InvJ and SpaO, the Sips are not believed to be involved in the secretion of proteins from the bacterium (57).
In addition to a potential role in translocation of effector proteins, SipB may have effector functions required for pathogenesis. As mentioned above, SipB is most closely homologous to IpaB of Shigella. A mutation in ipaB, ipaC, or ipaD results in the inability of Shigella to invade epithelial cells or escape the phagocytic compartments of epithelial cells or macrophages (126, 177, 216). Moreover, ipaB, ipaC, and ipaD mutants are unable to induce apoptosis in infected macrophages (263), but only ipaB is essential for this process (262). Purified IpaB can induce apoptosis if it is microinjected into macrophages, suggesting that it is sufficient for cytotoxicity (49). S. typhimurium also induces apoptosis of cultured macrophages; however, it is not known which effector protein(s) is required for this process (48, 187). Salmonella-induced apoptosis requires the SPI1 type III machinery (48, 187). Due to the homology between SipB and IpaB, SipB is a potential candidate as an effector for induction of apoptosis. Although sipB of S. typhi was previously shown to partially complement an ipaB mutant for invasion and for escape from the phagolysosome (125), it is not known if SipB expressed in a Shigella ipaB mutant can induce apoptosis of macrophages.
It is important to note that the intracellular life-styles of Shigella and Salmonella must somehow dictate how the secreted effector proteins reach the eucaryotic cytoplasm. For shigellae to induce apoptosis, the bacteria must enter cells and escape from phagolysosomal compartments (262). Once in the host cell cytoplasm, Ipa proteins are secreted without barriers. Unlike Shigella, salmonellae do not escape the phagolysosomal compartment (42) and do not need to be internalized to translocate SipB into eucaryotic cells (57). It is not clear how or why SipB reaches the eucaryotic cytoplasm without the bacterium entering the eucaryotic cytoplasm. There is also an apparent discrepancy with respect to Shigella Ipa function. The ipa genes were previously shown to be required for invasion of epithelial cells (126, 177, 216). Therefore, it is not clear how the Ipa proteins, particularly IpaB, function in invasion, phagolysosomal escape, and apoptosis.
A sipB fliM double mutant of S. dublin appears to “hypersecrete” several “non-Sip” proteins into culture supernatants, forming “filaments” (256). It is not known if the larger amount of proteins secreted from this mutant is due to the increased secretion of available protein within the cell or from increased transcription of genes encoding these proteins. A mutation in ipaD, which encodes a secreted protein in Shigella, actually increases the transcription of at least two other genes encoding secreted proteins (70). It is possible that a similar regulatory phenomenon occurs in Salmonella because mutations in sipB and sipD result in the increased secretion of several proteins.
(iv) sipC/sspC.
SipC is a 42-kDa protein homologous to Shigella IpaC and is encoded downstream of sipB in S. typhimurium (131, 140). Like sipB, sipC is required for invasion of Salmonella and for translocation of SipB and SptP into eucaryotic cells (88). SipC is itself translocated into cultured epithelial cells, but its function within the eucaryotic cell is unknown (57). As observed with SipB, salmonellae do not need to be internalized in order to translocate SipC into eucaryotic cells; treatment of the eucaryotic cells with cytochalasin D (which inhibits invasion) prior to infection does not inhibit the appearance of intracellular SipC.
SipC is homologous to the IpaC invasin of Shigella (21, 131, 140). As in Salmonella, IpaC is required for invasion of cells in vitro and in vivo (177). Interestingly, purified IpaC was sufficient to stimulate changes in the phosphoprotein content of tissue culture cells (163). These changes were believed to resemble those induced by invading shigellae. Purified IpaC could also increase the uptake of both wild-type and plasmid-cured (Δipa) shigellae in a dose-dependent manner (163). This suggested that IpaC might be sufficient to stimulate invasion of Shigella into cultured cells; it is notable, however, that invasion was much less efficient in the experiments with the plasmid-cured strain. In another study, inert particles coated with IpaB-IpaC complexes could be internalized by tissue culture cells; however, this study did not distinguish the contribution of each Ipa to the process of internalization (175). Due to the homology between IpaC and SipC, it is possible that purified SipC functions in a similar manner.
Unlike IpaB, IpaC is not required for inducing apoptosis in macrophages (262). Although dependent upon each other for translocation and cytotoxicity, IpaB and IpaC appear to have different but not necessarily exclusive functions: IpaB for cytotoxicity and IpaC for penetration of eucaryotic host cells. It would not be surprising if SipB and SipC function similarly for the pathogenesis of Salmonella. It is important to note, however, that neither Sip protein can behave in an exclusive manner. A sipB mutant cannot translocate SipC and a sipC mutant cannot translocate SipB into eucaryotic cells (57). Perhaps both proteins have dual functions, one for translocation and another for invasion and/or cytotoxicity.
(v) sipD/sspD.
Mutations in sipD result in the reduced invasion of cultured cells (131, 139). Unlike SipB and SipC, SipD (38 kDa) does not appear to enter eucaryotic cells (57), but it is required for the translocation of SipB, SipC, and other putative effector proteins (117). SipD does not appear to function alone, because SipB and SipC are also required for this process.
A sipD mutant was shown to hypersecrete several proteins into culture supernatants, but the mechanism of this increase is unknown (139). In Yersinia, a mutation in yopN, the putative gatekeeper for Yop secretion (see “invE” above), also resulted in the hypersecretion of proteins. Perhaps SipD also functions as a gatekeeper for Sip secretion. As mentioned above, a mutation in ipaD, the sipD homologue in Shigella, results in the increased transcription of at least two genes encoding secreted proteins (70, 177). Therefore, it is possible that SipD does not function to prevent secretion as hypothesized in the YopN model but, rather, functions to repress expression (most probably indirectly) of the other sip genes.
(vi) sipA/sspA.
sipA encodes one of the largest known Salmonella secreted proteins (87 kDa) and is homologous to ipaA of Shigella (131, 139). Neither ipaA nor sipA were thought to be essential for invasion in vitro (131, 139, 177). Recently, however, IpaA has been reported to actually have an effect on invasion by associating with vinculin, a protein involved in the association of actin with the plasma membrane (238). The discrepancy between the two results has not been clarified. SipA, unlike SipB and SipC, is not translocated into eucaryotic cells but localizes to the surface of these cells (57); therefore, it is not clear if SipA, like IpaA, could interact with intracellular proteins like vinculin. Despite the lack of evidence for a role of SipA in Salmonella pathogenesis, the conservation of this protein in at least two pathogenic species suggests its importance at some stage of the life cycle of the bacterium.
(vii) iacP (invasion-associated acyl carrier protein).
iacP is predicted to encode a 9-kDa protein which is homologous to several acyl carrier proteins involved in the biosynthesis of essential fatty acids (139, 160). Like the sip genes, iacP is conserved in the Shigella virulence plasmid; however, the function of iacP in invasion or virulence has not been established by mutational analysis.
(viii) sicP.
SicP is a specific chaperone for the secreted tyrosine phosphatase SptP (see below) (87, 88, 141). Typical of specific chaperones for secreted proteins, sicP is located immediately adjacent to (in this case, upstream of) sptP. SicP shares several features of known chaperones, including small size (13 kDa), acidic isoelectric point (pI = 3.9), and predicted α-helical structure (for a review, see reference 252). SicP is homologous to the Shigella protein IpgA, whose function has not been established (7). A mutation in sicP results in decreased amounts of SptP both in culture supernatants and in intracellular pools (87). Transcription of sptP was not significantly affected in a sicP mutant, suggesting that SicP acts posttranslationally. This was confirmed by pulse-chase analysis, which showed that the absence of SicP resulted in the rapid degradation of the SptP polypeptide. Epitope-tagged SicP also binds SptP in vitro, and amino acids 15 to 100 of SptP are important for SicP binding.
(ix) sptP (secreted protein tyrosine phosphatase).
SptP is a secreted protein tyrosine phosphatase with two putative effector domains (88, 141). The N-terminal part of SptP is homologous to YopE of Yersinia spp. and ExoS of Pseudomonas aeruginosa, which have cytotoxic activity (194, 212). YopE depolymerizes the actin microfilament network of target cells (212). ExoS contains an ADP-ribosylating activity (132) in its C-terminal domain; however, this domain is not in SptP. The C-terminal domain of SptP is similar to the catalytic domain of the tyrosine phosphatase YopH. The tyrosine phosphatase activity of YopH prevents phagocytosis of yersiniae by J774 macrophages (14).
Like YopE, YopH, and ExoS, SptP is translocated into eucaryotic cells. It is secreted by the SPI1 type III secretion system and requires SipB, SipC, and SipD for translocation into the host cell cytoplasm (88). Unlike Yersinia, Salmonella does not attempt to prevent its uptake by macrophages; therefore, it does not seem likely that SptP and YopH have a common function. Purified SptP induces actin depolymerization in cultured epithelial cells; however, unlike YopE and YopH, purified SptP is not cytotoxic for epithelial cells or macrophages. Moreover, cultured epithelial cells infected with an sptP mutant are indistinguishable from cells infected with wild-type salmonellae (88). A mutation in sptP results in a subtle in vivo phenotype characterized by reduced competition for the colonization of spleens by a sptP mutant when coinfected with wild-type bacteria. It is likely that infection of mice with only a sptP strain would not result in a dramatic difference in infectivity.
(x) avrA (avirulence).
AvrA is encoded within SPI1 and is homologous to other type III secreted proteins including AvrRxv of the plant pathogen Xanthamonas campestris and YopJ/YopP of Yersinia pseudotuberculosis/Y. enterocolitica (91, 117, 183). Under typical in vitro culture conditions, AvrA is not abundantly produced and can be visualized only by expressing avrA from a multicopy plasmid in wild-type bacteria or by performing immunoblot analysis (117). AvrA requires the SPI1 secretion apparatus for secretion and is not secreted by strains with mutations in invG, invJ, or spaO. AvrA has also been shown to be translocated into eucaryotic cells and requires SipD for the translocation (117). Despite this, there is no evidence that AvrA plays a role in invasion in vitro or pathogenesis in vivo.
The significance of AvrA homology to an avirulence protein from a plant pathogen is unclear. Avirulence proteins are players in a complex pathogen-host interaction resulting in the localized cell death of infected plant tissue (for a review, see reference 5;). AvrA is actually much more similar to YopJ/YopP, a protein secreted by the Ysc system of Yersinia spp. (183, 186). Y. pseudotuberculosis YopJ was originally reported not to have an effect on virulence in orally infected mice (91). After this original study and the identification of AvrA, YopJ/YopP was shown to be required for the induction of apoptosis of cultured macrophages (183, 186). It is unknown whether AvrA is required for the induction of apoptosis by Salmonella spp.
Non-SPI1-encoded proteins dependent on SPI1 for secretion.
(i) sigDE/sopB (Salmonella invasion gene/Salmonella outer protein).
SigD/SopB (62 kDa) was identified independently by different approaches. In S. dublin, sopB was cloned by sequencing one of five proteins which were hypersecreted into culture supernatants from a sipB/fliM mutant (92). In S. typhimurium, sigDE was identified in a screen for genes required for invasion in vitro (128). In S. dublin, SopB was translocated into eucaryotic cells in an invasion-independent manner; i.e., pretreatment of cells with cytochalasin D did not prevent translocation (92). SopB was required for the inflammation and fluid accumulation in bovine ileal loops (92) and is believed to stimulate the recruitment of PMNs to the site of a Salmonella infection. In S. typhimurium, a sigD or sigE mutant (sigE encodes a putative specific chaperone for SigD) showed reduced invasion into cultured epithelial cells. Interestingly, an invasion defect in a sopB mutant of S. dublin was not observed; therefore, it is not clear if the function of SopB/SigD is for inflammation or the invasion of the intestinal epithelium or both.
The Shigella homologue of SigD/SopB is IpgD, a protein which also appears to be a secreted protein (6). ipgD is part of the ipgDEF operon, which is located between the mxi and spa virulence plasmid-borne loci required for the secretion of virulence factors. ipgE was not characterized, and ipgF was shown to be membrane associated (6). S. typhimurium sigDE is located at centisome 25.5 (linked to putA) of the LT2 chromosome, far from the SPI1 locus containing the Salmonella mxi and spa homologues (128). sigE appears to encode a specific chaperone for SigD, because a transposon mutation in sigE results in an invasion defect identical to that of a sigD mutant (128). Moreover, a sigE mutant does not secrete SigD into culture supernatants (128). The Salmonella sig/sop locus does not include an ipgF homologue; however, there appears to be an ipgF homologue in SPI1 (see “iagB” below) (19). Unlike sigDE, ipgDEF has not been shown to be required for invasion or for virulence when tested in the guinea pig model of infection (6).
Recently, the region around sopB pipC (pipC [pathogenicity island-encoded protein] is the sigE homologue) in S. dublin was sequenced, confirmed to be linked to putA, and named SPI5 (255). Mutations in several of the SPI5 genes resulted in a phenotype similar to that of a sopB mutant. These genes, pipA, pipB, and pipD, are not homologous to genes encoding proteins found in Shigella or Yersinia. PipA shows no sequence similarity to other proteins from other bacterial species; PipB appears to be similar to the HglK protein from Anabaena and Synechocystis; and PipD is homologous to dipeptidases from Lactobacillus spp. (255). It is not known how mutations in these apparently unrelated genes result in similar phenotypes in vivo.
One of the interesting aspects of sigDE in S. typhimurium is its dependence on SPI1 for both secretion and regulation of gene expression. An invA mutant does not secrete SigD, suggesting that SigD requires the SPI1 secretion system (128). In addition, expression of sigDE requires the SPI1-borne activator, hilA (originally it was reported that sigDE expression was dependent on SirA but not HilA; it has since been determined that HilA is also required for sigDE expression) (128). Both sirA and hilA are required for the expression of several SPI1 genes tested to date, but it is not known which promoters they directly activate. Like several of the SPI1 genes (see “Regulators” below), sigDE expression is induced during late-log-phase growth under microaerophilic conditions in S. typhimurium (128).
(ii) sopE.
sopE was originally cloned from S. dublin by reverse genetics (similar to how sopB was cloned) (256) and later found to be located on a cryptic prophage in S. typhimurium at centisome 61 (118). Interestingly, not all S. typhimurium strains contain sopE sequences (118). A mutation in sopE has a modest effect on invasion in vitro (about twofold reduced compared to wild-type bacteria) (118, 256). Like several of the other known effector proteins, SopE is translocated into eucaryotic cells (118, 256) and cannot be secreted from invG, invJ, or spaO mutants (118). A mutation in sipB of S. dublin prevents the translocation of SopE into cultured epithelial cells (256).
Cultured epithelial cells infected with a sopE mutant do not exhibit the same morphological changes in the eucaryotic cytoskeleton as are observed with wild-type bacteria (118). Microinjection of purified SopE or transient expression of sopE within eucaryotic cells stimulated signaling pathways within the target cells (116). SopE appears to activate GDP-GTP nucleotide exchange in several Rho GTPases, resulting in cytoskeletal rearrangements (membrane ruffling) similar to those induced by wild-type Salmonella (116). Membrane ruffling was reversible in vitro, and SopE did not appear to be cytotoxic to cultured cells. Despite these significant observations, a sopE mutant is still fairly invasive and completely virulent in a mouse model of infection (118). Therefore, it is not clear how and when SopE contributes to virulence. It is, however, possible that the effects of SopE on invasion and virulence would be more substantial in a different cell type or host. Moreover, it is possible that a mutation in sopE, in combination with mutations in other putative effector proteins such as sigD, avrA, and sptP, would have a more dramatic effect on invasion.
Regulators
SPI1 genes are maximally expressed at 37°C and under oxygen-limiting conditions (78, 152). In addition, expression is optimal at neutral pH, at high osmolarity, and during late-log-phase growth (20). Invasion gene expression also requires the central regulator HilA, which is encoded in SPI1 (19, 153). Expression of hilA is activated by SirA (see below), but it is not known how environmental signals stimulate its expression or activity (135). Although several regulators have been implicated in the expression of invasion genes, none has been conclusively determined to interact with any promoter. Clearly, much has yet to be done to determine the precise molecular interactions of these proteins and the DNA sequences they bind.
invF.
The first gene in the inv-spa cluster encodes a protein homologous to members of the AraC family of transcriptional regulators (138). A mutation in invF was shown to dramatically reduce invasion into tissue culture cells. Transcriptional reporters to two other inv genes, invE and invA, were assayed for expression in an invF mutant, but their expression was unaffected. Because invF, invE, and invA are likely to be in the same transcriptional unit, it is not surprising that a difference in expression was not observed in the invF mutant. The expression of invH, a gene that is not cotranscribed with invF, was also unaffected by a mutation in invF (138).
Current models have suggested that InvF regulates the expression of the sip genes (135) (Fig. 3); however, no data supporting this model has been published. invF expression is dependent on two transcriptional activators, SirA (135) and HilA (19, 153). The mechanism by which these activators regulate invF expression is not known; however, preliminary evidence suggests that HilA directly binds the invF promoter. A transcriptional reporter plasmid containing the sequences upstream of invF fused to lacZYA can be activated in E. coli DH5α when hilA is provided in trans (19).
AraC-like regulators have been found in other virulence-associated systems, including VirF of the Yersinia Ysc system (64, 250). In Yersinia, VirF is required for the expression of several promoters, including those of effector genes. A consensus sequence was identified for VirF binding; however, the location and orientation of this site varied from promoter to promoter (250). In Salmonella, promoter analysis has not been performed for InvF; therefore, it is not known which promoters it directly affects.
hilA-iagB.
HilA (hyperinvasive locus) is an SPI1-encoded protein required for the expression of the type III secretion apparatus (19, 20, 153). Invasion into epithelial cells and induction of apoptosis of macrophages is absolutely HilA dependent (19, 187). It is not known precisely which promoters HilA activates; however, it does appear that HilA directly binds the invF and prgH promoters. Transcriptional fusions of invF and prgH to lacZ can be activated in E. coli strains expressing hilA (19). However, this result is not conclusive, because it is possible that HilA activated the expression of another regulator which in turn activated invF or prgH expression.
HilA is related to the OmpR/ToxR family of transcriptional regulators. The only conserved sequence in HilA with respect to this family of proteins is the putative DNA binding region. Expression of hilA is activated under environmental signals which stimulate invasion of S. typhimurium (20). Transcription of hilA does not appear to be autoregulated but is activated by another protein, SirA (135). It is not known if SirA directly binds the hilA promoter. hilA expression also is affected by phoPQ (which encodes a two-component regulatory system [see below]), but this is measurable only in the context of a pho-24 mutant (which has constitutive PhoP activity) (20). Whether PhoP acts directly on hilA expression or acts indirectly (perhaps through SirA) is not known. It is not known under what conditions PhoP normally represses hilA expression. It was once commonly believed that after salmonellae were internalized, they no longer required expression of the SPI1 genes. However, because it is now believed that SPI1 may not encode functions exclusively for invasion (48, 187), it is not clear when the SPI1 genes would be repressed.
Downstream of hilA is another gene, called iagB. iagB was identified in S. typhi along with iagA, its hilA homologue, as a gene required for invasion of S. typhi (184). However, this report did not characterize the function of these genes. The function of iagB in S. typhimurium pathogenesis is unknown, since mutational analysis did not reveal any obvious phenotype (19). Moreover, the iagB homologue in Shigella, ipgF, does not appear to have any function in invasion or virulence of Shigella flexneri (6).
sirA (Salmonella invasion regulator).
SirA is a highly conserved protein with homologs in other gram-negative bacteria such as E. coli (UvrY) (188) and Pseudomonas (GacA) (135, 207). Unlike hilA and invF, sirA maps to centisome 42, not to SPI1. In E. coli, uvrY was identified as an open reading frame upstream of uvrC, a gene encoding a protein required for repair of DNA damage (188), but the function of uvrY is not known.
Expression of hilA requires SirA; therefore, a mutation in sirA reduces the invasiveness of S. typhimurium (135). Interestingly, a sirA mutant could be complemented by two unlinked, previously uncharacterized loci, sirB (between centisomes 37 and 40) and the SPI1-linked locus sirC (135). It is not yet known how these loci complement the sirA mutation.
phoPQ.
phoP and phoQ encode a highly conserved two-component regulatory system required for virulence of S. typhimurium in vivo (179). Expression of invasion genes is repressed in vitro only in the context of a pho-24 mutant as previously described (20, 196). It is not known when wild-type PhoP, the response regulator, represses invasion gene expression, nor is it known which invasion gene promoters are directly affected by PhoP repression. Because HilA appears to be the central activator of expression of the SPI1-associated genes, it would be logical for PhoP to repress the expression (directly or indirectly) of hilA under the appropriate conditions.
Operon structure and regulation of SPI1-encoded genes.
Little has been confirmed with respect to the operon structure of the SPI1 genes. invH is likely to be regulated from its own promoter because it is expressed divergently from invF (13); however, the regulation of invH expression has not yet been examined. The remaining inv genes, along with the spa genes, may form a single transcriptional unit, but this has not been confirmed. Downstream of the inv-spa genes is sicA. It is not known if sicA has its own promoter or a promoter shared with the upstream inv-spa genes. sicA is believed to be transcribed in a single mRNA with the downstream sip genes (140), but again, this has yet to be demonstrated. Like the inv-spa genes, expression of sip genes is dependent on sirA (135) and hilA (20). Similarly, the expression of sigDE is also dependent upon sirA and hilA (128). A putative model for invasion gene expression suggests that SirA activates the expression of hilA, which in turn activates the expression of invF, a putative AraC-like regulator (19, 131, 138) (Fig. 4). InvF is proposed to activate the expression of the sic-sip operon; however, no evidence supporting this hypothesis has been published. sip expression is repressed in a pho-24 mutant; therefore, it appears that these genes may be repressed under conditions when PhoP is active (20).
FIG. 4.
Model of expression of virulence genes and localization of their gene products. Putative activators are indicated in red, chaperones are indicated in green, and secreted proteins are indicated in mauve (effectors) and yellow (translocators).
Similar to the sip genes, the prg operon is repressed in a pho-24 mutant (196). In addition, the prg operon is regulated positively by HilA (20). A reporter fusion of the prgH promoter region to lacZYA was activated in E. coli containing hilA in multicopy, suggesting that prgH is directly regulated by HilA (19). However, the precise interactions between PhoP and HilA and the prg regulatory sequences are not known. orgA, whose start codon is within the coding sequence of prgK, also requires the SPI1 transcriptional activator HilA for expression (136). Although the org and prg gene sequences overlap, Northern analysis has shown that the genes are not transcribed together (196). Moreover, prg transcripts are undetectable in a pho-24 mutant whereas orgA transcripts from a pho-24 mutant were present in amounts comparable to orgA transcripts from wild-type bacteria. These data suggest that orgA is not PhoP regulated and that the transcriptional start site of orgA (or at least one start site) is within the coding region of prgK.
Mutations in at least two of the putative apparatus proteins of the SPI2 type III secretion system (for macrophage survival) affect the secretion of at least one SPI1 protein (SipC) and transcription of hilA (69, 124). This finding suggests a possible cross talk between type III secretion systems (69). However, it is notable that complementation of SPI2 mutations affecting SPI1 gene expression has not been accomplished, suggesting a dominant phenotype (69). In addition to regulatory cross talk, it is tempting to consider the possibility that effectors secreted by one type III system are secreted by another system. Perhaps under conditions not yet established in vitro, SPI1-dependent proteins can be secreted by the SPI2 or even the flagellar secretion systems.
DIARRHEA
How do salmonellae cause diarrhea (fluid secretion)? It was clear from a study by Giannella et al. (96), testing a variety of S. typhimurium strains in the rabbit ileal-loop model, that invasion of the mucosa is necessary but not sufficient for fluid accumulation. Similarly, it was found that in a calf ileal-loop model, an isogenic invH mutant of S. typhimurium did not invade the epithelial cells and did not cause fluid accumulation (247, 248). This indicated that the type III secretion apparatus encoded by SPI1 is necessary for enteritis. The role of SPI1 in enteritis does not appear to be simply at the level of mucosal invasion, because a sopB mutant (SopB requires SPI1 for secretion) causes reduced fluid accumulation in calves but still invades the mucosa (92).
A number of studies have described cytotoxic and enterotoxic activities in salmonellae (for a review, see reference 190). However, in most cases, these factors have not been purified and their role in diarrhea or other aspects of Salmonella infections has not been elucidated. The most thoroughly studied of these toxins is Stn of S. typhimurium. The gene for Stn (stn) was cloned and shown to encode a 29-kDa protein with limited sequence similarity to the A subunit of cholera toxin of Vibrio cholerae and heat labile toxin of E. coli (51, 52, 203). Lysates from E. coli expressing stn cause fluid accumulation in rabbit ileal loops, CHO cell elongation, and cyclic AMP accumulation in CHO cells (51, 202, 203). These activities can be neutralized by antibody to cholera toxin or by ganglioside GM1 (202). Sequences homologous to stn have been found in all Salmonella serovars tested except S. bongori (201). It is attractive to think that Stn plays a role in inducing diarrhea during an infection with Salmonella. However, stn mutants of S. typhimurium and S. dublin were tested recently in calf ileal loops, and no difference in fluid secretion compared to that in loops infected by wild-type strains was observed (247).
While it is possible that an enterotoxin is responsible for or contributes to diarrhea associated with Salmonella infections, an alternative model put forth by Giannella and colleagues (97) is that fluid secretion is a result of the effect of prostaglandins released from the PMNs infiltrating Salmonella-infected intestinal tissue. In this model, invasion of the epithelium results in inflammation and infiltration of PMNs. Release of prostaglandins causes an increase in the activity of adenylate cyclase (AC) in the intestinal cells (143), causing an inhibition of Na+ absorption and an increase in Cl− secretion. Several lines of evidence support this model. Increases in AC activity and changes in Na+ and Cl− levels were observed after Salmonella infection (86, 97, 213). It was observed that the severity of diarrhea in monkeys and in rabbit ileal loops correlated with the degree of inflammation and PMN infiltration (86, 97, 213). In later experiments with a calf ligated-loop model, Wallis et al. (245) showed that the initiation of fluid secretion followed a massive influx of PMNs but occurred in the absence of mucosal damage. Subsequently, this PMN influx was quantitated, and it was found that the amount of infiltration was dependent on the dose of the bacterial inoculum. In addition, it was shown that fluid secretion never occurred in the absence of PMN infiltration (243). It was also found that an invH mutant of S. typhimurium has reduced invasion of the epithelium, PMN infiltration, and fluid accumulation (244, 247, 248).
The effect of PMN depletion on fluid secretion was examined by using nitrogen mustard pretreatment to deplete the PMNs (93, 246). It was found that in the rabbit ileal-loop model, this treatment depleted the PMNs effectively and inhibited fluid secretion following infection with Salmonella without causing observable changes in the ileal mucosal morphology (93). A more recent study in the calf ileal-loop model also found that nitrogen mustard effectively depletes PMNs and inhibits fluid secretion following infection (246). However, in this study it was observed that the nitrogen mustard treatment alone caused significant alterations in the ileal mucosal morphology.
Indomethacin has been used as an inhibitor of prostaglandin synthesis and was tested in the rabbit ileal-loop model for its effect on fluid secretion associated with Salmonella infection (97, 106). In these studies, infection of the loops with a strain of Salmonella that is invasive and causes fluid secretion (strain TML) resulted in an increase in AC activity that was not observed when the loops were infected with a strain that is invasive but does not cause fluid secretion. Pretreatment of the rabbits with indomethacin prior to infection with strain TML prevented AC activation and abolished fluid secretion; indomethacin did not affect invasion of the epithelium or dissemination of the bacteria (106). Similar effects of indomethacin on fluid secretion in rhesus monkeys and calf ileal loops infected with Salmonella have been reported (98, 246). While these studies with indomethacin and nitrogen mustard suggest that PMN infiltration and prostaglandin synthesis play a role in fluid secretion induced by Salmonella infection, inhibitor studies should always be interpreted with caution because they can have effects other than the desired ones. In addition, in the calf ileal-loop model, fluid secretion did not always occur after infiltration of PMNs, suggesting that other factors (either host or bacterial) are required (243).
Recent studies with cell culture models have tried to dissect in more detail the signalling events required for PMN influx and the role of prostaglandins in diarrhea. These studies also have begun to analyze which bacterial components are required for these events. Two studies published in 1993 demonstrated that colonic epithelial cells upregulated expression and secretion of the chemokine interleukin-8 (IL-8) in response to infection with Salmonella (73, 166); this required protein synthesis on the part of both the T84 cells and the bacteria (166). IL-8 produced by these T84 cells was preferentially released on the basolateral side of the polarized monolayer (73, 166). IL-8 is a potent chemoattractant for PMNs. However, McCormick et al. (166) demonstrated in an in vitro model for PMN transmigration that IL-8 released by T84 cells in response to Salmonella infection was not necessary for transepithelial migration of PMNs from the basolateral side to the apical surface. In subsequent studies, McCormick et al. (167) demonstrated that infection of polarized T84 cells causes the release of IL-8 on the basolateral side, which then binds to the matrix, setting up a gradient that promotes PMN migration through the matrix. In addition, infection of the polarized T84 cells causes the release of a new PMN chemoattractant, pathogen-elicited epithelial chemoattractant (PEEC), on the apical surface, which stimulates transmigration of the PMNs through the epithelium (169). Thus, the current model based on these data is that infection of the epithelium by Salmonella causes basolateral release of IL-8, resulting in migration of PMNs into the subepithelial space and apical release of PEEC, in turn resulting in intraepithelial migration of the PMNs with subsequent release into the lumen.
In the course of these studies, it was found that IL-8 production increased with increasing numbers of intracellular bacteria and that infection with an invA mutant (which has a 100-fold reduction in invasion) did not stimulate IL-8 production (73, 166). Similarly treatment of the monolayer with cytochalasin D reduced both the number of intracellular bacteria and the amount of IL-8 produced (73). It has also been shown that PMN transepithelial migration correlates well with the ability of a Salmonella strain to cause enteritis in humans; for example, S. typhimurium is PMN migration positive but S. typhi and S. paratyphi are not (168). S. typhi and S. paratyphi were able to invade the epithelium; therefore, it appears that invasion is not sufficient to induce signalling (168). In the PMN transepithelial-migration studies, it was found that S. typhimurium phoP(Con), ΔhilA-prgH, or invA mutants were unable to stimulate migration (168). From these studies, it was concluded that bacterial invasion is necessary to stimulate IL-8 secretion and PMN infiltration. However, these conclusions must be interpreted more cautiously in light of what is now known about the SPI1-encoded type III secretion system and its secreted proteins. All of the above mutations would affect the assembly and activity of the secretion apparatus, resulting in the loss of most if not all of the secreted effector proteins. Not all of the effectors directly affect invasion. Therefore, it will be necessary to test mutants more specifically affected in individual effector proteins to dissect out which Salmonella products mediate which effects on the host cells.
The original model proposed by Giannella and colleagues suggested that prostaglandin (PG) synthesis on the part of the PMNs that are recruited to the infected site is responsible for the fluid secretion observed after infection with Salmonella (93, 96, 97). A recent study by Eckmann et al. (75) supports a role for PG synthesis but suggests that PG synthesis by epithelial cells also plays a role. HT-29 cells (a colonic cell line) produced prostaglandins PGE2 and PGF2α in a dose-dependent manner in response to infection with Salmonella. This production could be completely blocked with indomethacin, which is specific for PGH synthase 2 (PGHS-2), a key enzyme in the synthesis of PGs. An increase in the level of PGHS-2 was also observed after infection of a human intestinal xenograph model in SCID mice (75). Supernatants of HT-29 cells infected with Salmonella, when added to polarized T84 cells, caused an increase in Cl− secretion. The Cl− secretion was due to the presence of PGE2 in the supernatants (75), supporting a role for PGE2 secretion by epithelial cells in fluid accumulation during Salmonella infection. However, Cl− secretion may not be the result of the action of PGE2 alone. Infection of T84 cells with Salmonella also causes an increase in the d-myo-inositol-1,4,5,6-tetrakisphosphate [Ins(1,4,5,6)P4] level (74). This increase in the Ins(1,4,5,6)P4 level results in an inhibition of the epidermal growth factor-induced inhibition of calcium-mediated Cl− secretion from the T84 cells. The net result would be an increase in Cl− secretion. Infection of the T84 cells by an invA mutant of Salmonella did not result in an increase in the Ins(1,4,5,6)P4 level (74), suggesting that the SPI1-encoded type III secretion pathway is necessary for Salmonella to stimulate this event. Interestingly, while many of these events (IL-8 and PGE2 production) are stimulated by many different invasive bacteria (Y. enterocolitica, Shigella, and Listeria monocytogenes) (73, 75), the increase in the Ins(1,4,5,6)P4 level was observed only after infection with Salmonella (74).
A model by which Salmonella infection triggers diarrhea is emerging from these in vivo and in vitro studies. Taken together, these results suggest that the interaction of Salmonella with the epithelium results not only in invasion of the epithelial cells but also in the production of a variety of signalling molecules on the part of the epithelial cells. Production of IL-8 and PEEC by the epithelial cells stimulates inflammation and PMN transmigration. Production of PG by the PMNs and/or the epithelial cells, along with synthesis of Ins(1,4,5,6)P4, causes an increase in Cl− secretion, which triggers fluid secretion that is manifested as diarrhea in the host. It is clear from these studies that these events require a functional type III secretion system on the part of Salmonella, but the effectors that trigger individual events remain to be determined.
CONCLUSIONS
Significant advances in our understanding of how Salmonella interacts with the intestinal mucosa to cause disease have been made during the past decade. This is due in part to the use and development of both in vitro and in vivo models of infection and to the identification of key bacterial genes required for these interactions. The tools are now in hand to further dissect these processes at a molecular level, but this will require collaboration between immunologists and cell biologists studying the epithelial cell responses and microbiologists studying the bacterial responses. Questions remaining to be answered that derive from the above studies include the following. (i) What are the receptors for the individual adhesins that Salmonella produces? (ii) What is the operon structure of the SPI1-borne type III secretion genes and genes encoding secreted proteins? How does the regulatory cascade function to coordinate the synthesis of these products? (iii) Which type III apparatus proteins are required for export of effectors out of the bacterial cells, and which are required for translocation into the host cell? (iv) Which effectors are needed for invasion versus stimulation of IL-8, PEEC, PGE2, or Ins(1,4,5,6)P4 production? (v) How can this knowledge be used for better treatment or prevention of salmonellosis?
ACKNOWLEDGMENTS
We thank Bill Goldman for comments on the manuscript and Tim Wallis for contributing Fig. 1.
REFERENCES
- 1.Reference deleted.
- 2.Reference deleted.
- 3.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. An invertible element of DNA controls phase variation of type 1 fimbriae in Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ager E A, Nelson K E, Galton M M, Boring III J, Jernigan J. Two outbreaks of egg-borne salmonellosis and implications for their prevention. JAMA. 1967;199:372–378. [PubMed] [Google Scholar]
- 5.Alfano J R, Collmer A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr protein, and death. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allaoui A, Ménard R, Sansonetti P J, Parsot C. Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect Immun. 1993;61:1707–1714. doi: 10.1128/iai.61.5.1707-1714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allaoui A, Mounier J, Prevost M C, Sansonetti P J, Parsot C. icsB: a Shigella flexneri virulence gene necessary for the lysis of protrusions during intracellular spread. Mol Microbiol. 1992;6:1605–1616. doi: 10.1111/j.1365-2958.1992.tb00885.x. [DOI] [PubMed] [Google Scholar]
- 8.Allaoui A, Sansonetti P J, Parsot C. MxiD, an outer membrane protein necessary for the secretion of Shigella flexneri invasion protein antigens. Mol Microbiol. 1993;7:759–768. doi: 10.1111/j.1365-2958.1993.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 9.Allaoui A, Sansonetti P J, Parsot C. MxiJ, a lipoprotein involved in the secretion of Shigella Ipa invasins, is homologous to YscJ, a secretion factor of the Yersinia Yop proteins. J Bacteriol. 1992;174:7661–7669. doi: 10.1128/jb.174.23.7661-7669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allaoui A, Schulte R, Cornelis G R. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- 11.Allaoui A, Woestyn S, Sluiters C, Cornelis G R. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol. 1994;176:4534–4542. doi: 10.1128/jb.176.15.4534-4542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altekruse S F, Cohen M L, Swerdlow D L. Emerging foodborne diseases. Emerging Infect Dis. 1997;3:285–293. doi: 10.3201/eid0303.970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altmeyer R M, McNern J K, Bossio J C, Rosenshine I, Finlay B B, Galán J E. Cloning and molecular characterization of a gene involved in Salmonella adherence and invasion of cultured epithelial cells. Mol Microbiol. 1993;7:89–98. doi: 10.1111/j.1365-2958.1993.tb01100.x. [DOI] [PubMed] [Google Scholar]
- 14.Andersson K, Carballeira N, Magnusson K-E, Persson C, Stendahl O, Wold-Watz H, Fällman M. YopH of Yersinia pseudotuberculosis interrupts early phosphotyrosine signalling associated with phagocytosis. Mol Microbiol. 1996;20:1057–1069. doi: 10.1111/j.1365-2958.1996.tb02546.x. [DOI] [PubMed] [Google Scholar]
- 15.Anonymous. Recurrent Salmonella virchow infection in a hospital group. Br Med J. 1977;1:59. [Google Scholar]
- 16.Archer D L, Kvenberg J E. Incidence and cost of foodborne diarrheal disease in the United States. J Food Prot. 1985;48:887–894. doi: 10.4315/0362-028X-48.10.887. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong R W, Fodor T, Curlin G T, Cohen A B, Morris G K, Martin W T, Feldman J. Epidemic salmonella gastroenteritis due to contaminated imitation ice cream. Am J Epidemiol. 1970;91:300–307. doi: 10.1093/oxfordjournals.aje.a121140. [DOI] [PubMed] [Google Scholar]
- 18.Baird-Parker A C. Foodborne salmonellosis. Lancet. 1990;336:1231–1235. doi: 10.1016/0140-6736(90)92844-8. [DOI] [PubMed] [Google Scholar]
- 19.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 20.Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 21.Baudry B, Kaczorek M, Sansonetti P J. Nucleotide sequence of the invasion plasmid antigen B and C genes (ipaB and ipaC) of Shigella flexneri. Microb Pathog. 1988;4:345–357. doi: 10.1016/0882-4010(88)90062-9. [DOI] [PubMed] [Google Scholar]
- 22.Bäumler A J. The record of horizontal gene transfer in Salmonella. Trends Microbiol. 1997;5:318–322. doi: 10.1016/S0966-842X(97)01082-2. [DOI] [PubMed] [Google Scholar]
- 23.Bäumler A J, Heffron F. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J Bacteriol. 1995;177:2087–2097. doi: 10.1128/jb.177.8.2087-2097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bäumler A J, Tsolis R M, Bowe F A, Kusters J G, Hoffmann S, Heffron F. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect Immun. 1996;64:61–68. doi: 10.1128/iai.64.1.61-68.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bäumler A J, Tsolis R M, Heffron F. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun. 1996;64:1862–1865. doi: 10.1128/iai.64.5.1862-1865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bäumler A J, Tsolis R M, Heffron F. Fimbrial adhesins of Salmonella typhimurium. Adv Exp Med Biol. 1997;412:149–158. [PubMed] [Google Scholar]
- 27.Bäumler A J, Tsolis R M, Heffron F. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer’s patches. Proc Natl Acad Sci USA. 1996;93:279–283. doi: 10.1073/pnas.93.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bäumler A J, Tsolis R M, Valentine P J, Ficht T A, Heffron F. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect Immun. 1997;65:2254–2259. doi: 10.1128/iai.65.6.2254-2259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behlau I, Miller S I. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berkelman R L, Bryan R T, Osterholm M T, LeDuc J W, Hughes J M. Infectious disease surveillance: a crumbling foundation. Science. 1994;264:368–370. doi: 10.1126/science.8153621. [DOI] [PubMed] [Google Scholar]
- 31.Bischoff D S, Weinrich M D, Ordal G W. Nucleotide sequences of Bacillus subtilus flagellar biosynthetic genes fliP and fliQ and identification of a novel flagellar gene, fliZ. J Bacteriol. 1992;174:4017–4025. doi: 10.1128/jb.174.12.4017-4025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Black P H, Kunz L J, Swartz M N. Salmonellosis—a review of some unusual aspects. N Engl J Med. 1960;262:811–816. doi: 10.1056/NEJM196004212621606. , 846–870, 921–927. [DOI] [PubMed] [Google Scholar]
- 33.Blaser M J. How safe is our food: lessons from an outbreak of salmonellosis. N Engl J Med. 1996;334:1324–1325. doi: 10.1056/NEJM199605163342009. [DOI] [PubMed] [Google Scholar]
- 34.Bliska J B, Falkow S. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc Natl Acad Sci USA. 1992;89:3561–3565. doi: 10.1073/pnas.89.8.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohnhoff M, Miller C P, Martin W R. Resistance of the mouse’s intestinal tract to experimental Salmonella infection. II. Factors responsible for its loss following streptomycin treatment. J Exp Med. 1964;120:817–828. doi: 10.1084/jem.120.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boland A, Sory M-P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 37.Boyd J F. Pathology of the alimentary tract in Salmonella typhimurium food poisoning. Gut. 1985;26:935–944. doi: 10.1136/gut.26.9.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bureau of the Census. 1990 Census of population: general population characteristics. U.S. Washington, D.C: Government Printing Office; 1990. [Google Scholar]
- 39.Bureau of the Census. Population projections of the United States, by age, sex, race, and Hispanic origin: 1993–2050. U.S. Washington, D.C: Government Printing Office; 1993. [Google Scholar]
- 40.Burrows W. Textbook of microbiology. 7th ed. Philadelphia, Pa: The W. B. Saunders Co.; 1959. [Google Scholar]
- 41.Buysse J M, Stover C K, Oaks E V, Venkatesan M, Kopecko D J. Molecular cloning of invasion plasmid antigen (ipa) genes from Shigella flexneri: analysis of ipa gene products and genetic mapping. J Bacteriol. 1987;169:2561–2569. doi: 10.1128/jb.169.6.2561-2569.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrol M E W, Jackett P S, Aber V R, Lowrie D B. Phagolysosome formation, cyclic adenosine 3′:5′-monophosphate and the fate of Salmonella typhimurium within mouse peritoneal macrophages. J Gen Microbiol. 1979;110:421–429. doi: 10.1099/00221287-110-2-421. [DOI] [PubMed] [Google Scholar]
- 43.Carter P B, Collins F M. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Celum C L, Chaisson R E, Rutherford G W, Barnhart J L, Echenberg D F. Incidence of salmonellosis in patients with AIDS. J Infect Dis. 1987;156:998–1002. doi: 10.1093/infdis/156.6.998. [DOI] [PubMed] [Google Scholar]
- 45.Centers for Disease Control and Prevention. 1993 Revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. Morbid Mortal Weekly Rep. 1992;41:1–19. [PubMed] [Google Scholar]
- 46.Centers for Disease Control and Prevention. Multistate outbreak of Salmonella poona infections: United States and Canada. Morbid Mortal Weekly Rep. 1991;40:549. [PubMed] [Google Scholar]
- 46a.Centers for Disease Control and Prevention. Salmonella surveillance—annual summary—1992. Atlanta, Ga: Centers for Disease Control and Prevention; 1994. [Google Scholar]
- 46b.Centers for Disease Control and Prevention. Salmonella surveillance report—annual summary—1990. Atlanta, Ga: Centers for Disease Control and Prevention; 1991. [Google Scholar]
- 47.Chalker R B, Blaser M J. A review of human salmonellosis. III. Magnitude of Salmonella infection in the United States. Rev Infect Dis. 1988;9:111–124. doi: 10.1093/clinids/10.1.111. [DOI] [PubMed] [Google Scholar]
- 48.Chen L M, Kaniga K, Galán J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y, Smith M R, Thirumalai K, Zychlinsky A. A bacterial invasin induces macrophage apoptosis by binding directly to ICE. EMBO J. 1996;15:3853–3860. [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng L W, Anderson D M, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 51.Chopra A K, Houston C W, Peterson J W, Prasad R, Mekalanos J J. Cloning and expression of the Salmonella enterotoxin gene. J Bacteriol. 1987;169:5095–5100. doi: 10.1128/jb.169.11.5095-5100.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chopra A K, Peterson J W, Chary P, Prasad R. Molecular characterization of an enterotoxin from Salmonella typhimurium. Microb Pathog. 1994;16:85–98. doi: 10.1006/mpat.1994.1010. [DOI] [PubMed] [Google Scholar]
- 53.Cirillo D M, Heffernan E J, Wu L, Harwood J, Fierer J, Guiney D G. Identification of a domain of Rck, a product of the Salmonella typhimurium virulence plasmid, required for both serum resistance and cell invasion. Infect Immun. 1996;64:2019–2023. doi: 10.1128/iai.64.6.2019-2023.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clegg S, Hancox L S, Yeh K-S. Salmonella typhimurium fimbrial phase variation and FimA expression. J Bacteriol. 1996;178:542–545. doi: 10.1128/jb.178.2.542-545.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clegg S, Swenson D L. Salmonella fimbriae. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 105–113. [Google Scholar]
- 56.Cohen M L, Tauxe R V. Drug-resistant Salmonella in the United States: an epidemiologic perspective. Science. 1986;234:964–969. doi: 10.1126/science.3535069. [DOI] [PubMed] [Google Scholar]
- 57.Collazo C M, Galán J E. The invasion-associated type III system of Salmonella typhimurium directs the translocation of the Sip proteins into the host cell. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 58.Collazo C M, Galán J E. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Collazo C M, Zierler M K, Galán J E. Functional analysis of the Salmonella typhimurium invasion genes invI and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol Microbiol. 1995;15:25–38. doi: 10.1111/j.1365-2958.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 60.Collinson S K, Clouthier S C, Doran J L, Banser P A. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Collinson S K, Doig P C, Doran J L, Clouthier S, Trust T J, Kay W W. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol. 1993;175:12–18. doi: 10.1128/jb.175.1.12-18.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Collinson S K, Emödy L, Müller K-H, Trust T J, Kay W W. Purification of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collinson S K, Liu S-L, Clouthier S C, Banser P A, Doran J L, Sanderson K E, Kay W W. The location of four fimbrin-encoding genes, agfA, fimA, sefA and sefD, on the Salmonella enteritidis and/or S. typhimurium XbaI-BlnI genomic restriction maps. Gene. 1996;169:75–80. doi: 10.1016/0378-1119(95)00763-6. [DOI] [PubMed] [Google Scholar]
- 64.Cornelis G R, Sluitiers C, Lambert de Rouvroit C, Michiels T. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol. 1989;171:254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 66.Crago A M, Koronakis V. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol Microbiol. 1998;30:47–56. doi: 10.1046/j.1365-2958.1998.01036.x. [DOI] [PubMed] [Google Scholar]
- 67.Daefler S, Russel M. The Salmonella typhimurium InvH protein is an outer membrane lipoprotein required for the proper localization of InvG. Mol Microbiol. 1998;28:1367–1380. doi: 10.1046/j.1365-2958.1998.00908.x. [DOI] [PubMed] [Google Scholar]
- 68.Day D W, Mandal B K, Morson B C. The rectal biopsy appearances in Salmonella colitis. Histopathology. 1978;2:117–131. doi: 10.1111/j.1365-2559.1978.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 69.Deiwick J, Nokolaus T, Shea J E, Gleeson C, Holden D W, Hensel M. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J Bacteriol. 1998;180:4775–4780. doi: 10.1128/jb.180.18.4775-4780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Demers B, Sansonetti P J, Parsot C. Induction of type III secretion in Shigella flexneri is associated with differential control of transcription of genes encoding secreted proteins. EMBO J. 1998;17:2894–2903. doi: 10.1093/emboj/17.10.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.d’Enfert C, Reyss I, Wandersman C, Pugsley A P. Protein secretion by gram-negative bacteria. Characterization of two membrane proteins required for pullulanase secretion by E. coli K-12. J Biol Chem. 1989;264:17462–17468. [PubMed] [Google Scholar]
- 72.Duguid J P, Anderson E S, Campbell I. Fimbriae and adhesive properties in salmonellae. J Pathol Bacteriol. 1966;92:107–138. doi: 10.1002/path.1700920113. [DOI] [PubMed] [Google Scholar]
- 73.Eckmann L, Kagnoff M F, Fierer J. Epithelial cells secrete the chemokine interleukin-8 in response to bacterial entry. Infect Immun. 1993;61:4569–4574. doi: 10.1128/iai.61.11.4569-4574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eckmann L, Rudolf M T, Ptasnik A, Schultz C, Jiang T, Wolfson N, Tsien R, Fierer J, Shears S B, Kagnoff M F, Traynor-Kaplan A E. d-myo-Inositol 1,4,5,6-tetrakisphosphate produced in human intestinal epithelial cells in response to Salmonella invasion inhibits phosphoinositide 3-kinase signaling pathways. Proc Natl Acad Sci USA. 1997;94:14456–14460. doi: 10.1073/pnas.94.26.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eckmann L, Stenson W F, Savidge T C, Lowe D C, Barrett K E, Fierer J, Smith J R, Kagnoff M F. Role of intestinal epithelial cells in the host secretory response to infection by invasive bacteria: Bacterial entry induces epithelial prostaglandin H synthase-2 expression and prostaglandin E2 and F2a production. J Clin Investig. 1997;100:296–309. doi: 10.1172/JCI119535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eichelberg K, Ginocchio C C, Galán J E. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J Bacteriol. 1994;176:4501–4510. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elsinghorst E A, Baron L S, Kopecko D J. Penetration of human intestinal epithelial cells by Salmonella: molecular cloning and expression of Salmonella typhi invasion determinants in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:5173–5177. doi: 10.1073/pnas.86.13.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ernst R K, Dombroski D M, Merrick J M. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Faddoul G P, Fellows G W. A five-year survey of the incidence of salmonella in avian species. Avian Dis. 1966;10:296–304. [Google Scholar]
- 80.Fields P I, Groisman E A, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 81.Finlay B B, Ruschkowski S, Dedhar S. Cytoskeletal rearrangements accompanying Salmonella entry into epithelial cells. J Cell Sci. 1991;99:283–296. doi: 10.1242/jcs.99.2.283. [DOI] [PubMed] [Google Scholar]
- 82.Firon N, Ofek I, Sharon N. Carbohydrate-binding sites of the mannose-specific fimbrial lectins of enterobacteria. Infect Immun. 1984;43:1088–1090. doi: 10.1128/iai.43.3.1088-1090.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Forsberg Å, Viitanen A-M, Skurnik M, Wolf-Watz H. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol Microbiol. 1991;5:977–986. doi: 10.1111/j.1365-2958.1991.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 84.Francis C L, Ryan T A, Jones B D, Smith S J, Falkow S. Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature. 1993;364:639–642. doi: 10.1038/364639a0. [DOI] [PubMed] [Google Scholar]
- 85.Friedrich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 86.Fromm D, Giannella R A, Formal S B, Quijano R, Collins H. Ion transport across isolated ileal mucosa invaded by Salmonella. Gastroenterology. 1974;66:215–225. [PubMed] [Google Scholar]
- 87.Fu Y, Galán J E. Identification of a specific chaperone for SptP, a substrate of the centisome 63 type III secretion system of Salmonella typhimurium. J Bacteriol. 1998;180:3393–3399. doi: 10.1128/jb.180.13.3393-3399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fu Y, Galán J E. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 89.Galán J E, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Galán J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Galyov E E, Håkansson S, Wolf-Watz H. Characterization of the operon encoding the YpkA Ser/Thr protein kinase and the YopJ protein of Yersinia pseudotuberculosis. J Bacteriol. 1994;176:4543–4548. doi: 10.1128/jb.176.15.4543-4548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Galyov E E, Wood M W, Rosqvust R, Mullan P B, Watson P R, Hedges S, Wallis T S. A secreted effector protein of Salmonella dublin is translocated into eucaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 93.Giannella R A. Importance of the intestinal inflammatory reaction in Salmonella-mediated intestinal secretion. Infect Immun. 1979;23:140–145. doi: 10.1128/iai.23.1.140-145.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Giannella R A. Invasion of HeLa cells by Salmonella typhimurium: a model for study of invasiveness of Salmonella. J Infect Dis. 1973;128:69–75. doi: 10.1093/infdis/128.1.69. [DOI] [PubMed] [Google Scholar]
- 95.Giannella R A, Broitman S A, Zamcheck N. Salmonella enteritis. 1. Role of reduced gastric secretion in pathogenesis. Am J Dig Dis. 1971;16:1000–1006. doi: 10.1007/BF02235012. [DOI] [PubMed] [Google Scholar]
- 96.Giannella R A, Formal S B, Dammin G J, Collins H. Pathogenesis of salmonellosis. Studies of fluid secretion, mucosal invasion, and morphologic reaction in the rabbit ileum. J Clin Investig. 1973;52:441–453. doi: 10.1172/JCI107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Giannella R A, Gots R E, Charney A N, Greenough W B, Formal S B. Pathogenesis of Salmonella-mediated intestinal fluid secretion: activation of adenylate cyclase and inhibition by indomethacin. Gastroenterology. 1975;69:1238–1245. [PubMed] [Google Scholar]
- 98.Giannella R A, Rout W R, Formal S B. Effect of indomethacin on intestinal water transport in Salmonella-infected Rhesus monkeys. Infect Immun. 1977;17:136–139. doi: 10.1128/iai.17.1.136-139.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gillen K L, Hughes K T. Molecular characterization of flgM, a gene encoding a negative regulator of flagellin synthesis in Salmonella typhimurium. J Bacteriol. 1991;173:6453–6459. doi: 10.1128/jb.173.20.6453-6459.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gincocchio C C, Galán J E. Functional conservation among members of the Salmonella typhimurium InvA family of proteins. Infect Immun. 1995;63:729–732. doi: 10.1128/iai.63.2.729-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ginocchio C, Olmsted S B, Wells C L, Galán J E. Contact with epithelial cells induces the formation of surface appendages on Salmonella typhimurium. Cell. 1994;76:717–724. doi: 10.1016/0092-8674(94)90510-x. [DOI] [PubMed] [Google Scholar]
- 102.Ginocchio C, Pace J, Galán J E. Identification and molecular characterization of a Salmonella typhimurium gene involved in triggering the internalization of salmonellae into cultured epithelial cells. Proc Natl Acad Sci USA. 1992;89:5976–5980. doi: 10.1073/pnas.89.13.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Glaser M J, Newman L S. A review of human salmonellosis. I. Infective dose. Rev Infect Dis. 1982;34:1096–1106. doi: 10.1093/clinids/4.6.1096. [DOI] [PubMed] [Google Scholar]
- 104.Gomez H F, Cleary G G. Salmonella. 4th ed. Vol. 1. Philadelphia, Pa: The W. B. Saunders Co.; 1998. [Google Scholar]
- 105.Gordon R F, Tucker J F. The epizoology of Salmonella menston infection of fowls and the effect of feeding poultry food artificially infected with salmonella. Br Poultry Sci. 1965;6:251–264. doi: 10.1080/00071666508415581. [DOI] [PubMed] [Google Scholar]
- 106.Gots R E, Formal S B, Giannella R A. Indomethacin inhibition of Salmonella typhimurium, Shigella flexneri, and cholera-mediated rabbit ileal secretion. J Infect Dis. 1974;130:280–284. doi: 10.1093/infdis/130.3.280. [DOI] [PubMed] [Google Scholar]
- 107.Groisman E A, Ochman H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993;12:3779–3787. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gulig P A. Virulence plasmids of Salmonella typhimurium and other salmonellae. Microb Pathog. 1990;8:3–11. doi: 10.1016/0882-4010(90)90003-9. [DOI] [PubMed] [Google Scholar]
- 109.Gulig P A, Caldwell A L, Chiodo V A. Identification, genetic analysis and DNA sequence of a 7.8-kb virulence region of the Salmonella typhimurium virulence plasmid. Mol Microbiol. 1992;6:1395–1411. doi: 10.1111/j.1365-2958.1992.tb00860.x. [DOI] [PubMed] [Google Scholar]
- 110.Gulig P A, Curtiss R., III Plasmid-associated virulence of Salmonella typhimurium. Infect Immun. 1987;55:2891–2901. doi: 10.1128/iai.55.12.2891-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gulig P A, Danbara H, Guiney D G, Lax A J, Norel F, Rhen M. Molecular analysis of spv virulence genes of the salmonella virulence plasmids. Mol Microbiol. 1993;7:825–830. doi: 10.1111/j.1365-2958.1993.tb01172.x. [DOI] [PubMed] [Google Scholar]
- 112.Gunn J S, Alpuche-Arande C M, Loomis W P, Belden W J, Miller S I. Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J Bacteriol. 1995;177:5040–5047. doi: 10.1128/jb.177.17.5040-5047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hackett J, Wyk P, Reeves P, Mathan V. Mediation of serum resistance in Salmonella typhimurium by an 11-kilodalton polypeptide encoded by the cryptic plasmid. J Infect Dis. 1987;155:540–549. doi: 10.1093/infdis/155.3.540. [DOI] [PubMed] [Google Scholar]
- 114.Hammar M, Bian Z, Normark S. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 115.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hardt W-D, Chen L-M, Schuebel K E, Bustelo X R, Galán J E. S. typhimurium encodes an activator of rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 117.Hardt W-D, Galán J E. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc Natl Acad Sci USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hardt W-D, Urlaub H, Galán J E. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic prophage. Proc Natl Acad Sci USA. 1998;95:2574–2579. doi: 10.1073/pnas.95.5.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hartz P H, Pot A W, Felsenfeld O. Fatal fulminant colitis due to Salmonella saint paul. Am J Clin Pathol. 1950;20:184–187. doi: 10.1093/ajcp/20.2.184. [DOI] [PubMed] [Google Scholar]
- 120.Heffernan E J, Harwood J, Fierer J, Guiney D. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes, including pagC and ail. J Bacteriol. 1992;174:84–91. doi: 10.1128/jb.174.1.84-91.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Heffernan E J, Wu L, Louie J, Okamoto S, Fierer J, Guiney D. Specificity of the complement resistance and cell association phenotypes encoded by the outer membrane protein genes rck from Salmonella typhimurium and ail from Yersinia enterocolitica. Infect Immun. 1994;62:5183–5186. doi: 10.1128/iai.62.11.5183-5186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hennessy T W, Hedberg C W, Slutsker L, et al. A national outbreak of Salmonella enteritidis infections from ice cream. N Engl J Med. 1996;334:1281–1286. doi: 10.1056/NEJM199605163342001. [DOI] [PubMed] [Google Scholar]
- 123.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 124.Hensel M, Shea J E, Raupach B, Monack D, Falkow S, Gleeson C, Kubo T, Holden D W. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol Microbiol. 1997;24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 125.Hermant D, Ménard R, Arricau N, Parsot C, Popoff M Y. Functional conservation of the Salmonella and Shigella effectors of entry into epithelial cells. Mol Microbiol. 1995;17:781–789. doi: 10.1111/j.1365-2958.1995.mmi_17040781.x. [DOI] [PubMed] [Google Scholar]
- 126.High N, Mounier J, Prevost M C, Sansonetti P J. IpaB of Shigella flexneri causes entry into epithelial cells and escape from the phagocytic vesicle. EMBO J. 1992;11:1991–1999. doi: 10.1002/j.1460-2075.1992.tb05253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hohmann A W, Schmidt G, Rowley D. Intestinal colonization and virulence of Salmonella in mice. Infect Immun. 1978;22:763–770. doi: 10.1128/iai.22.3.763-770.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hong K H, Miller V L. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J Bacteriol. 1998;180:1793–1802. doi: 10.1128/jb.180.7.1793-1802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hook E W. Salmonella species (including typhoid fever) 3rd ed. New York, N.Y: Churchill Livingstone, Inc.; 1990. [Google Scholar]
- 130.Hueck C J. Type III protein secretion in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hueck C J, Hantman M J, Bajaj V, Johnston C, Lee C A, Miller S I. Salmonella typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol Microbiol. 1995;18:479–490. doi: 10.1111/j.1365-2958.1995.mmi_18030479.x. [DOI] [PubMed] [Google Scholar]
- 132.Iglewski B H, Sadoff J, Bjorn M J, Maxwell M S. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc Natl Acad Sci USA. 1978;75:3211–3215. doi: 10.1073/pnas.75.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Institute of Medicine. Emerging infections: microbial threats to health in the United States. Washington, D.C: National Academy Press; 1992. [PubMed] [Google Scholar]
- 134.Isberg R R, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 1985;317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 135.Johnston C, Pegues D A, Hueck C J, Lee C A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 136.Jones B D, Falkow S. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the M cells of the Peyer’s patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaniga K, Bossio J C, Galán J E. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 139.Kaniga K, Trollinger D, Galán J E. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kaniga K, Tucker S, Trollinger D, Galán J E. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kaniga K, Uralil J, Bliska J B, Galán J E. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol Microbiol. 1996;21:633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 142.Kent T H, Formal S B, Labrec E H. Salmonella gastroenteritis in Rhesus monkeys. Arch Pathol. 1966;82:272–279. [PubMed] [Google Scholar]
- 143.Kimberg D V, Field M, Johnson J, Henderson A, Gershon E. Stimulation of intestinal mucosal adenylate cyclase by cholera enterotoxin and prostaglandins. J Clin Investig. 1971;50:1218–1230. doi: 10.1172/JCI106599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Klemm P. Two regulatory fim genes, fimB and fimE, control phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Klemm P, Krogfelt K A. Type 1 fimbriae of Escherichia coli. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 9–26. [Google Scholar]
- 146.Kohbata S, Yokoyama H, Yabuuchi E. Cytopathogenic effect of Salmonella typhi GIFU 10007 on M cells of murine ileal Peyer’s patches in ligated ileal loops: an ultrastructural study. Microbiol Immunol. 1986;30:1225–1237. doi: 10.1111/j.1348-0421.1986.tb03055.x. [DOI] [PubMed] [Google Scholar]
- 147.Korhonen T K, Lounatmaa K, Ranta H, Kuusi N. Characterization of type I pili of Salmonella typhimurium LT2. J Bacteriol. 1980;144:800–805. doi: 10.1128/jb.144.2.800-805.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Koster M, Bitter W, de Cock H, Allaoui A, Cornelis G R, Tommassen J. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 149.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J E, Aizawa S-I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 150.Kutsukake K, Ohya Y, Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kvenberg J E, Archer S L. Economic impact of colonization control on foodborne disease. Food Technol. 1987;41:77–88. [Google Scholar]
- 152.Lee C A, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lee C A, Jones B D, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lee V T, Anderson D M, Schneewind O. Targeting of Yersinia Yop proteins into the cytosol of HeLa cells: one-step translocation of YopE across bacterial and eucaryotic membranes is dependent on SycE chaperone. Mol Microbiol. 1998;28:593–601. doi: 10.1046/j.1365-2958.1998.00822.x. [DOI] [PubMed] [Google Scholar]
- 155.Leunk R D, Moon R J. Association of type 1 pili with the ability of livers to clear Salmonella typhimurium. Infect Immun. 1982;36:1168–1174. doi: 10.1128/iai.36.3.1168-1174.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li J, Ochman H, Groisman E A, Boyd E F, Solomon F, Nelson K, Selander R K. Relationship between evolutionary rate and cellular location among the Inv/Spa invasion proteins of Salmonella enterica. Proc Natl Acad Sci USA. 1995;92:7252–7256. doi: 10.1073/pnas.92.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lill R, Cunningham K, Brundage L A, Ito K, Oliver D, Wickner W. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 1989;8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lockman H A, Curtiss R., III Virulence of non-type 1-fimbriated and nonfimbriated nonflagellated Salmonella typhimurium mutants in murine typhoid fever. Infect Immun. 1992;60:491–496. doi: 10.1128/iai.60.2.491-496.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lodge J, Douce G R, Amin I I, Bolton A J, Martin G D, Chatfield S, Dougan G, Brown N L, Stephen J. Biological and genetic characterization of TnphoA mutants of Salmonella typhimurium TML in the context of gastroenteritis. Infect Immun. 1995;63:762–769. doi: 10.1128/iai.63.3.762-769.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Magnuson K, Jackowski S, Rock C O, Cronan J. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mandal B K, Mani V. Colonic involvement in salmonellosis. Lancet. 1976;i:887–888. doi: 10.1016/s0140-6736(76)92100-0. [DOI] [PubMed] [Google Scholar]
- 162.Marks P W, Maxfield F R. Local and global changes in cytosolic free calcium in neutrophils during chemotaxis and phagocytosis. Cell Calcium. 1990;11:181–190. doi: 10.1016/0143-4160(90)90069-7. [DOI] [PubMed] [Google Scholar]
- 163.Marquart M E, Picking W L, Picking W D. Soluble invasion plasmid antigen C (IpaC) from Shigella flexneri elicits epithelial cell responses related to pathogen invasion. Infect Immun. 1996;64:4182–4187. doi: 10.1128/iai.64.10.4182-4187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Maurelli A T, Baudry B, d’Hauteville H, Hale T L, Sansonnetti P J. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect Immun. 1985;49:164–171. doi: 10.1128/iai.49.1.164-171.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McCann M P, Kidwell J P, Matin A. The putative ς factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McCormick B A, Colgan S P, Delp-Archer C, Miller S I, Madara J L. Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils. J Cell Biol. 1993;123:895–907. doi: 10.1083/jcb.123.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.McCormick B A, Hofman P M, Kim J, Carnes D K, Miller S I, Madara J L. Surface attachment of Salmonella typhimurium to intestinal epithelia imprints the subepithelial matrix with gradients chemotactic for neutrophils. J Cell Biol. 1995;131:1599–1608. doi: 10.1083/jcb.131.6.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McCormick B A, Miller S I, Carnes D, Madara J L. Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infect Immun. 1995;63:2302–2309. doi: 10.1128/iai.63.6.2302-2309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.McCormick B A, Parkos C A, Colgan S P, Carnes D K, Madara J L. Apical secretion of a pathogen-elicited epithelial chemoattractant activity in response to surface colonization of intestinal epithelia by Salmonella typhimurium. J Immunol. 1998;160:455–466. [PubMed] [Google Scholar]
- 170.McCullough N B, Eisele C W. Experimental human salmonellosis. 1. Pathogenicity of strains of Salmonella meleagridis and Salmonella anatum obtained from spray-dried whole egg. J Infect Dis. 1951;88:278–289. doi: 10.1093/infdis/88.3.278. [DOI] [PubMed] [Google Scholar]
- 171.McCullough N B, Eisele C W. Experimental human salmonellosis. 3. Pathogenicity of strains of Salmonella newport, Salmonella derby, and Salmonella bareilly obtained from spray-dried whole egg. J Infect Dis. 1951;89:209–213. doi: 10.1093/infdis/89.3.209. [DOI] [PubMed] [Google Scholar]
- 172.McCullough N B, Eisele C W. Experimental human salmonellosis. 4. Pathogenicity of strains of Salmonella pullorum obtained from spray-dried whole egg. J Infect Dis. 1951;89:259–265. doi: 10.1093/infdis/89.3.259. [DOI] [PubMed] [Google Scholar]
- 173.McGovern V J, Slavutin L J. Pathology of salmonella colitis. Am J Surg Pathol. 1979;3:483–490. doi: 10.1097/00000478-197912000-00001. [DOI] [PubMed] [Google Scholar]
- 174.McWhorter-Murlin A C, Hickman-Brenner F W. Identification and serotyping of Salmonella and an update of the Kauffmann-White scheme. Atlanta, Ga: Centers for Disease Control and Prevention; 1994. [Google Scholar]
- 175.Ménard R, Prévost M-C, Gounon P, Sansonetti P J, Dehio C. The secreted Ipa complex of Shigella flexneri promotes entry into mammalian cells. Proc Natl Acad Sci USA. 1996;93:1254–1258. doi: 10.1073/pnas.93.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ménard R, Sansonetti P, Parsot C, Vasselon T. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell. 1994;79:515–525. doi: 10.1016/0092-8674(94)90260-7. [DOI] [PubMed] [Google Scholar]
- 177.Ménard R, Sansonetti P J, Parsot C. Non-polar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Michiels T, Vanooteghem J-C, Lambert de Rouvroit C, China B, Gustin A, Boudry P, Cornelis G. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J Bacteriol. 1991;173:4994–5009. doi: 10.1128/jb.173.16.4994-5009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Miller S I, Kukral A M, Mekalanos J J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Miller V L, Bliska J B, Falkow S. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J Bacteriol. 1990;172:1062–1069. doi: 10.1128/jb.172.2.1062-1069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Miller V L, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mills D M, Bajaj V, Lee C A. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 183.Mills S D, Boland A, Sory M-P, Van der Smissen P, Kerbourch C, Finlay B B, Cornelis G R. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Miras I, Hermant D, Arricau N, Popoff M Y. Nucleotide sequence of iagA and iagB genes involved in invasion of HeLa cells by Salmonella enterica subsp. enterica ser. Typhi. Res Microbiol. 1995;146:17–20. doi: 10.1016/0923-2508(96)80267-1. [DOI] [PubMed] [Google Scholar]
- 185.Mishu B, Koehler J, Lee L A, Rodrigue D, Hickman-Brenner F, Blake P, Tauxe R V. Outbreaks of Salmonella enteritidis infections in the United States, 1985–1991. J Infect Dis. 1994;169:547–552. doi: 10.1093/infdis/169.3.547. [DOI] [PubMed] [Google Scholar]
- 186.Monack D M, Mecsas J, Ghori N, Falkow S. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc Natl Acad Sci USA. 1997;94:10385–10390. doi: 10.1073/pnas.94.19.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Moolenar G F, van Sluis C A, Backendorf C, van de Putte P. Regulation of the Escherichia coli excision repair gene uvrC. Overlap between the uvrC structural gene and the region coding for a 24 kD protein. Nucleic Acids Res. 1987;15:4273–4289. doi: 10.1093/nar/15.10.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Norris T L, Kingsley R A, Bäumler A J. In vivo expression and transcriptional control of the Salmonella typhimurium lpf fimbrial operon by phase variation. Mol Microbiol. 1998;29:311–320. doi: 10.1046/j.1365-2958.1998.00934.x. [DOI] [PubMed] [Google Scholar]
- 190.O’Brien A D, Holmes R K. Protein toxins of Escherichia coli and Salmonella. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2788–2802. [Google Scholar]
- 191.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ohnishi K, Kutsukake K, Suzuki H, Iino T. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol Gen Genet. 1990;221:139–147. doi: 10.1007/BF00261713. [DOI] [PubMed] [Google Scholar]
- 193.Olsén A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 194.Olson J C, McGuffie E M, Frank D W. Effects of differential expression of the 49-kilodalton exoenzyme S by Pseudomonas aeruginosa on cultured eucaryotic cells. Infect Immun. 1997;65:248–256. doi: 10.1128/iai.65.1.248-256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Peel B. Occurrence of Salmonella in raw and pasteurized liquid whole egg. Queensl J Agric Anim Sci. 1976;33:13–21. [Google Scholar]
- 196.Pegues D A, Hantman M J, Behlau I, Miller S I. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 197.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer’s patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 198.Pierson D E, Falkow S. The ail gene of Yersinia enterocolitica has a role in the ability of the organism to survive serum killing. Infect Immun. 1993;61:1846–1852. doi: 10.1128/iai.61.5.1846-1852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Plano G V, Straley S C. LcrD, a membrane-bound regulator of the Yersinia pestis low-calcium response. J Bacteriol. 1991;173:7293–7303. doi: 10.1128/jb.173.22.7293-7303.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Plano G V, Straley S C. Multiple effects of lcrD mutations in Yersinia pestis. J Bacteriol. 1993;175:3536–3545. doi: 10.1128/jb.175.11.3536-3545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Prager R, Fruth A, Tschäpe H. Salmonella enterotoxin (stn) gene is prevalent among strains of Salmonella enterica, but not among Salmonella bongori and other Enterobacteriaceae. FEMS Immunol Med Microbiol. 1995;12:47–50. doi: 10.1111/j.1574-695X.1995.tb00173.x. [DOI] [PubMed] [Google Scholar]
- 202.Prasad R, Chopra A K, Chary P, Peterson J W. Expression and characterization of the cloned Salmonella typhimurium enterotoxin. Microb Pathog. 1992;13:109–121. doi: 10.1016/0882-4010(92)90071-u. [DOI] [PubMed] [Google Scholar]
- 203.Prasad R, Chopra A K, Peterson J W, Pericas R, Houston C W. Biological and immunological characterization of a cloned cholera toxin-like enterotoxin from Salmonella typhimurium. Microb Pathog. 1990;9:315–329. doi: 10.1016/0882-4010(90)90066-y. [DOI] [PubMed] [Google Scholar]
- 204.Price S B, Straley S C. lcrH, a gene necessary for virulence of Yersinia pestis and for the normal response of Y. pestis to ATP and calcium. Infect Immun. 1989;57:1491–1498. doi: 10.1128/iai.57.5.1491-1498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Prost E, Riemann H. Food-borne salmonellosis. Annu Rev Microbiol. 1967;23:495–528. doi: 10.1146/annurev.mi.21.100167.002431. [DOI] [PubMed] [Google Scholar]
- 206.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Riley L W, Cohen M L, Seals J E, Blaser J J, Birkness K A, Hargrett N T, Martin S M, Feldman R A. Importance of host factors in human salmonellosis caused by multiresistant strains of Salmonella. J Infect Dis. 1984;149:878–883. doi: 10.1093/infdis/149.6.878. [DOI] [PubMed] [Google Scholar]
- 209.Rodrigue D C, Tauxe R V, Rowe B. International increase in Salmonella enteritidis: a new pandemic? Epidemiol Infect. 1990;105:21–27. doi: 10.1017/s0950268800047609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Römling U, Bian Z, Hammar M, Sierralta W D, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Römling U, Sierralta W D, Eriksson K, Normark S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol. 1998;28:249–264. doi: 10.1046/j.1365-2958.1998.00791.x. [DOI] [PubMed] [Google Scholar]
- 212.Rosqvist R, Forsberg A, Wolf-Watz H. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect Immun. 1991;59:4562–4569. doi: 10.1128/iai.59.12.4562-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rout W R, Formal S B, Dammin G J, Giannela R A. Pathophysiology of salmonella diarrhea in the Rhesus monkey: intestinal transport, morphological and bacteriological studies. Gastroenterology. 1974;67:59–70. [PubMed] [Google Scholar]
- 214.Ryan C A, Nickels M K, Hargrett-Bean N T, Potter M E, Endo T, Mayer L, Langkop C W, Gibson C, McDonald R C, Kenney R T, Puhr N D, McDonnell P J, Martin R J, Cohen M L, Blake P A. Massive outbreak of antimicrobial-resistant salmonellosis traced to pasteurized milk. JAMA. 1987;258:3269–3274. [PubMed] [Google Scholar]
- 215.Sanderson K E, Hessel A, Rudd K E. Genetic map of Salmonella typhimurium, edition VIII. Microbiol Rev. 1995;59:241–303. doi: 10.1128/mr.59.2.241-303.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sasakawa C, Kamata K, Sakai T, Makino S, Yamada M, Okada N, Yoshikawa M. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J Bacteriol. 1988;170:2480–2484. doi: 10.1128/jb.170.6.2480-2484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sasakawa C, Komatsu K, Tobe T, Suzuki T, Yoshikawa M. Eight genes in region 5 that form an operon essential for invasion of epithelial cells by Shigella flexneri 2a. J Bacteriol. 1993;175:2334–2346. doi: 10.1128/jb.175.8.2334-2346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Slauch J M, Silhavy T J. The porin regulon: a paradigm for the two-component regulatory systems. In: Lin E C C, Lynch A S, editors. Regulation of gene expression. R. G. Austin, Tex: Landes Co.; 1996. pp. 383–404. [Google Scholar]
- 219.Slutsker L, Altekruse S F, Swerdlow D L. Foodborne diseases: emerging pathogens and trends. Infect Dis Clin North Am. 1998;12:199–216. doi: 10.1016/s0891-5520(05)70418-9. [DOI] [PubMed] [Google Scholar]
- 220.Smith H W, Jones J E T. Observations on experimental oral infection with Salmonella dublin in calves and Salmonella choleraesuis in pigs. J Pathol Bacteriol. 1967;93:141–156. doi: 10.1002/path.1700930114. [DOI] [PubMed] [Google Scholar]
- 221.Snoeyenbos G H, Smyser C F, Van Roekel H. Salmonella infections of the ovary and peritoneum of chickens. Avian Dis. 1969;13:668–670. [PubMed] [Google Scholar]
- 222.Steere A C, Hall III W J, Wells J G, Craven P J, Leotsakis N, Farmer III J J I, Gangarosa E J. Person-to-person spread of Salmonella typhimurium after a hospital common source outbreak. Lancet. 1975;i:319–321. doi: 10.1016/s0140-6736(75)91221-0. [DOI] [PubMed] [Google Scholar]
- 223.St. Louis M E, Morse D L, Potter M E, DeMelfi T M, Guzewich J J, Tauxe R V, Blake P A. The emergence of grade A shell eggs as a major source of Salmonella enteritidis infections: new implications for the control of salmonellosis. JAMA. 1988;259:2103–2107. [PubMed] [Google Scholar]
- 224.Stone B J, Garcia C M, Badger J L, Hassett T, Smith R I F, Miller V L. Identification of novel loci affecting entry of Salmonella enteritidis into eukaryotic cells. J Bacteriol. 1992;174:3945–3952. doi: 10.1128/jb.174.12.3945-3952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Straley S C, Plano G V, Skrzypek E, Haddix P L, Fields K A. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol Microbiol. 1993;8:1005–1010. doi: 10.1111/j.1365-2958.1993.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 226.Sukupolvi S, Lorenz R G, Gordon J I, Bian Z, Pfeifer J D, Normark S J, Rhen M. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect Immun. 1997;65:5320–5325. doi: 10.1128/iai.65.12.5320-5325.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Sukupolvi S, Riikonen P, Taira S, Saarilahti H, Rhen M. Plasmid-mediated serum resistance in Salmonella enterica. Microb Pathog. 1992;12:219–225. doi: 10.1016/0882-4010(92)90056-t. [DOI] [PubMed] [Google Scholar]
- 228.Swenson D L, Clegg S. Identification of ancillary fim genes affecting fimA expression in Salmonella typhimurium. J Bacteriol. 1992;174:7697–7704. doi: 10.1128/jb.174.23.7697-7704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Takeuchi A. Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am J Pathol. 1967;50:109–119. [PMC free article] [PubMed] [Google Scholar]
- 230.Takeuchi A, Sprinz H. Electron-microscope studies of experimental Salmonella infection in the preconditioned guinea pig. II. Response of the intestinal mucosa to the invasion by Salmonella typhimurium. Am J Pathol. 1967;50:137–161. [PMC free article] [PubMed] [Google Scholar]
- 231.Tauxe R V. Transmission of human bacterial pathogens through poultry (banquet address). New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 232.Tauxe R V. An update on Salmonella. Health Environ Dig. 1996;10:1–4. [Google Scholar]
- 233.Tauxe R V, Hughes J M. International investigation of outbreaks of foodborne disease. Br Med J. 1996;313:1093–1094. doi: 10.1136/bmj.313.7065.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tauxe R V, Pavia A T. Salmonellosis: nontyphoidal. In: Evans A S, Brachman P S, editors. Bacterial infections of humans: epidemiology and control. 3rd ed. New York, N.Y: Plenum Medical Book Co.; 1998. pp. 613–630. [Google Scholar]
- 235.Taylor R K, Hall M N, Enquist L, Silhavy T J. Identification of OmpR: a positive regulatory protein controlling the expression of the major outer membrane matrix porin proteins of Escherichia coli K-12. J Bacteriol. 1981;147:255–258. doi: 10.1128/jb.147.1.255-258.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Thatcher F S, Montford J. Egg products as a source of Salmonella in processed foods. Can J Public Health. 1962;53:61–69. [PubMed] [Google Scholar]
- 237.Thirumalai K, Kim K-S, Zychlinsky A. IpaB, a Shigella flexneri invasin, colocalizes with interleukin-1β-converting enzyme in the cytoplasm of macrophages. Infect Immun. 1997;65:787–793. doi: 10.1128/iai.65.2.787-793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Tran Van Nhieu G, Ben-Ze’ev A, Sansonetti P J. Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. EMBO J. 1997;16:2717. doi: 10.1093/emboj/16.10.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Turnbull P C B. Food poisoning with special reference to Salmonella—its epidemiology, pathogenesis and control. Clin Gastroenterol. 1979;8:663–714. [PubMed] [Google Scholar]
- 240.van der Velden A W M, Bäumler A J, Tsolis R M, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Voino-Yasenetsky M V. Problems of the pathogenesis of salmonella infection. Budapest, Hungary: Akadémiai Kiadó; 1977. [Google Scholar]
- 242.Waddell S R, Kunz L J. Association of Salmonella enteritidis with operations on the stomach. N Engl J Med. 1956;255:555–559. doi: 10.1056/NEJM195609202551203. [DOI] [PubMed] [Google Scholar]
- 243.Wallis T S, Hawker R J H, Candy D C A, Ql G-M, Clarke G J, Worton K J, Osborne M P, Stephen J. Quantification of the leucocyte influx into rabbit ileal loops induced by strains of Salmonella typhimurium of different virulence. J Med Microbiol. 1989;30:149–156. doi: 10.1099/00222615-30-2-149. [DOI] [PubMed] [Google Scholar]
- 244.Wallis T S, Paulin S M, Plested J S, Watson P R, Jones P W. The Salmonella dublin virulence plasmid mediates systemic but not enteric phases of salmonellosis in cattle. Infect Immun. 1995;63:2755–2761. doi: 10.1128/iai.63.7.2755-2761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wallis T S, Starkey W G, Stephen J, Haddon S J, Osborne M P, Candy D C A. The nature and role of mucosal damage in relation to Salmonella typhimurium-induced fluid secretion in the rabbit ileum. J Med Microbiol. 1986;22:39–49. doi: 10.1099/00222615-22-1-39. [DOI] [PubMed] [Google Scholar]
- 246.Wallis T S, Vaughan T M, Clarke G J, Ql G-M, Worton K J, Candy D C A, Osborne M P, Stephen J. The role of leucocytes in the induction of fluid secretion by Salmonella typhimurium. J Med Microbiol. 1990;31:27–35. doi: 10.1099/00222615-31-1-27. [DOI] [PubMed] [Google Scholar]
- 247.Watson P R, Galyov E E, Paulin S M, Jones P W, Wallis T S. Mutation of invH, but not stn, reduces Salmonella-induced enteritis in cattle. Infect Immun. 1998;66:1432–1438. doi: 10.1128/iai.66.4.1432-1438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Watson P R, Paulin S M, Bland P, Jones P W, Wallis T S. Characterization of intestinal invasion by Salmonella typhimurium and Salmonella dublin and effect of a mutation in the invH gene. Infect Immun. 1995;63:2743–2754. doi: 10.1128/iai.63.7.2743-2754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wattiau P, Bernier B, Deslée P, Michiels T, Cornelis G R. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wattiau P, Cornelis G R. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J Bacteriol. 1994;176:3878–3884. doi: 10.1128/jb.176.13.3878-3884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wattiau P, Cornelis G R. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol Microbiol. 1993;8:123–131. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 252.Wattiau P, Woestyn S, Cornelis G R. Customized secretion chaperones in pathogenic bacteria. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 253.Whimbey E, Gold J W M, Polsky B, Dryjanski J, Hawkins C, Blevins A, Brannon P, Kiehn T E, Brown A E, Armstrong D. Bacteremia and fungemia in patients with the acquired immunodeficiency syndrome. Ann Intern Med. 1986;104:511–514. doi: 10.7326/0003-4819-104-4-511. [DOI] [PubMed] [Google Scholar]
- 254.Wilson R, Feldman R A, Davis J, LaVenture M. Salmonellosis in infants: the importance of intrafamilial transmission. Pediatrics. 1982;69:436–438. [PubMed] [Google Scholar]
- 255.Wood M W, Jones M A, Watson P R, Hedges S, Wallis T S, Galyov E E. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol Microbiol. 1998;29:883–891. doi: 10.1046/j.1365-2958.1998.00984.x. [DOI] [PubMed] [Google Scholar]
- 256.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 257.Worton K J, Candy D C A, Wallis T S, Clarke G J, Osborne M P, Haddon S J, Stephen J. Studies on early association of Salmonella typhimurium with intestinal mucosa in vivo and in vitro: relationship to virulence. J Med Microbiol. 1989;29:283–294. doi: 10.1099/00222615-29-4-283. [DOI] [PubMed] [Google Scholar]
- 258.Yeh K-S, Hancox L S, Clegg S. Construction and characterization of a fimZ mutant of Salmonella typhimurium. J Bacteriol. 1995;177:6861–6865. doi: 10.1128/jb.177.23.6861-6865.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yokoyama H, Ikedo M, Kohbata S, Exaki T, Yabuuchi E. An ultrastructural study of HeLa cell invasion with Salmonella typhi GIFU 10007. Microbiol Immunol. 1987;31:1–11. doi: 10.1111/j.1348-0421.1987.tb03063.x. [DOI] [PubMed] [Google Scholar]
- 260.Zierler M K, Galán J E. Contact with cultured epithelial cells stimulates secretion of Salmonella typhimurium invasion protein InvJ. Infect Immun. 1995;63:4024–4028. doi: 10.1128/iai.63.10.4024-4028.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ziprin R L. Salmonella. Vol. 1. New York, N.Y: Marcel Dekker, Inc.; 1994. [Google Scholar]
- 262.Zychlinsky A, Kenny B, Ménard R, Prevost M C, Holland I B, Sansonetti P J. IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol Microbiol. 1994;11:619–627. doi: 10.1111/j.1365-2958.1994.tb00341.x. [DOI] [PubMed] [Google Scholar]
- 263.Zychlinsky A M C, Prevost M C, Sansonetti P J. Shigella flexneri induces apoptosis in infected macrophages. Nature. 1992;358:167–168. doi: 10.1038/358167a0. [DOI] [PubMed] [Google Scholar]