Abstract
Fission yeast checkpoint protein Rad17 is required for the DNA integrity checkpoint responses. A fraction of Rad17 is chromatin bound independent of the other checkpoint proteins throughout the cell cycle. Here we show that in response to DNA damage induced by either methyl methanesulfonate treatment or ionizing radiation, increased levels of Rad17 bind to chromatin. Following S-phase stall induced by hydroxyurea or a cdc22 mutation, the chromatin-bound Rad17 progressively dissociates from the chromatin. After S-phase arrest by hydroxyurea in cds1Δ or rad3Δ cells or by replication mutants, Rad17 remains chromatin bound. Rad17 is able to complex in vivo with an Rfc small subunit, Rfc2, but not with Rfc1. Furthermore, cells with rfc1Δ are checkpoint proficient, suggesting that Rfc1 does not have a role in checkpoint function. A checkpoint-defective mutant protein, Rad17(K118E), which has similar nuclear localization to that of the wild type, is unable to bind ATP and has reduced ability in chromatin binding. Mutant Rad17(K118E) protein also has reduced ability to complex with Rfc2, suggesting that Lys118 of Rad17 plays a role in Rad17-Rfc small-subunit complex formation and chromatin association. However, in the rad17.K118E mutant cells, Cds1 can be activated by hydroxyurea. Together, these results suggest that Rad17 binds to chromatin in response to an aberrant genomic structure generated from DNA damage, replication mutant arrest, or hydroxyurea arrest in the absence of Cds1. Rad17 is not required to bind chromatin when genomic structures are protected by hydroxyurea-activated Cds1. The possible checkpoint events induced by chromatin-bound Rad17 are discussed.
For maintaining genome integrity following replication perturbation or DNA damage, eukaryotic cells employ checkpoint mechanisms to delay or arrest the cell cycle, allowing cells to recover from the perturbation or to repair the damage (10, 19, 20). In fission yeast, a group of gene products named Rad1, Rad3, Rad9, Rad17, Rad26, Hus1, and Cut5 (Rad4) are required for the checkpoint (1, 33, 39, 41, 42). Five of the checkpoint gene products, Rad1, Rad3, Rad9, Rad17, and Hus1, are evolutionarily conserved from yeast to humans (8, 33). These checkpoint proteins are thought to function as sensors and transducers in signaling both replication perturbation and DNA damage by activating two downstream kinases, Cds1 and Chk1. These kinases ultimately have an effect on the cell cycle machinery to delay or arrest the cell cycle to prevent inappropriate mitotic entry in maintaining genome integrity (5, 12, 13, 22, 29, 32, 39, 40, 60).
Recent studies have shown that the checkpoint proteins form complexes. In fission yeast and human cells, Hus1 and Rad1 form a stable complex in a Rad9-dependent manner (21, 55). Further studies have shown that Hus1 exists in several forms. The main form, Hus1B, participates in the complex with Rad1 and Rad9 (8). Rad3 has been shown to complex with Rad26 and phosphorylates Rad26 in response to γ radiation in G2 cells independent of other checkpoint proteins (9), suggesting that this type of damage can activate Rad3 kinase directly (27).
Rad17 and its budding yeast homologue RAD24 each contain five domains with sequence homology to replication factor C (Rfc) (17). Rfc is an evolutionarily conserved five-subunit protein complex. During DNA replication, the Rfc complex recognizes the primer-template junction of the initiation DNA structure synthesized by polymerase α and loads the processivity clamp, PCNA, onto DNA to allow polymerase δ to synthesize the main bulk of the DNA (56, 61). Genetic evidence has shown that several Rfc subunits not only function as the PCNA clamp loader in replication, but they are also required for the cell cycle checkpoint (24, 30, 31, 38, 43, 45, 46). Furthermore, budding yeast Rad24p and fission yeast Rad17 have been shown to coimmunoprecipitate with Rfc5p and Rfc3, respectively (30, 43, 44). Coprecipitation of budding yeast Rad24p with all four of the small subunits of Rfc proteins (Rfc2p, Rfc3p, Rfc4p, and Rfc5p) but not with Rfc1p has also been reported (15). We and others have found that Rad17 exists as a large protein complex independent of the other checkpoint Rad proteins (8, 16). Although Rad17 does not associate with the Hus1-Rad1-Rad9 complex, Rad17 is required for the nuclear localization of Hus1 and Rad9 and interacts with Rad1 in two-hybrid reactions (8).
Studies of the budding yeast checkpoint proteins have suggested that following DNA damage, aberrant DNA structure may be first processed by checkpoint proteins Rad17p, Rad24p, and Mec3p (homologues of Schizosaccharomyces pombe Rad1, Rad17, and Hus1, respectively) into a structure that activates Mec1p (the S. pombe Rad3 homologue). Mec1p then activates Rad53p and Chk1p (the S. pombe Cds1 and Chk1 counterparts) for cell cycle arrest (14, 25, 49). The human homologue of fission yeast Rad1 is thought to encode an exonuclease, and human Rad9 has been reported as having 3′-to-5′ exonuclease activity in vitro (3, 35). Finding that fission yeast Rad3-Rad26 responds to damage independently of other checkpoint proteins (9, 27) suggests that Rad17 and the Rad1-Rad9-Hus1B complex may have a role in processing the aberrant DNA structure.
To begin to investigate this hypothesis, we focused on Rad17. We have recently reported that a fraction of the cellular Rad17 protein is chromatin bound throughout the cell cycle. Importantly, chromatin binding of Rad17 is independent of other checkpoint proteins and the Cds1 and Chk1 kinases (16). This finding (16) and the findings by others (15, 30, 31, 43) describing the association of Rad17 with Rfc proteins raise the following questions. (i) What are the chromatin binding statuses of Rad17 in response to DNA damage and replication perturbation? (ii) Does a checkpoint-defective mutant Rad17 protein bind to chromatin? (iii) Is a fraction of the Rfc small subunits always in complex with Rad17 for checkpoint function? (iv) Is Rfc1 not involved in the checkpoint process but only in replication? Here we describe the chromatin association of wild-type and a checkpoint-defective Rad17 protein in response to DNA damage and various S-phase perturbations. Similar to reports by others (15, 31), we showed that Rad17 coprecipitates with Rfc2 but not with Rfc1. We have further demonstrated that rfc1+ is not involved in the checkpoint function and that residue Lys118 of Rad17 is required for ATP binding of Rad17, complex formation with Rfc2, and chromatin association. Together our data suggest that Rad17, in a complex with the small subunits of Rfc, associates with chromatin in response to an aberrant genomic structure. This complex is not required to associate with the chromatin when the genomic structure is protected by the hydroxyurea-activated Cds1. We discuss the possible events induced by the chromatin-bound Rad17-Rfc small-subunit complex in functioning as a part of the checkpoint process.
MATERIALS AND METHODS
Yeast strains, plasmids, media, genetics, and molecular techniques.
All yeast strains were constructed by standard fission yeast genetic techniques as described previously (18), and resulting mutants were isolated by tetrad analysis. Where necessary, strains were also confirmed for the presence of either the hemagglutinin (HA) or myc epitope tags by Western blotting with the respective monoclonal antibody. Fission yeast cells were grown in rich media (YE5S) or Edinburgh minimal media with appropriate supplements as described earlier (28). Molecular biological techniques were performed according to prior methods (26). Myc-tagged rad17 and rad17.K118E strains (17) and pREP41-myc-rad17+ and pREP41-rad17.K118E plasmids were generous gifts from A. M. Carr.
Chromatin fractionation assay.
Chromatin fractionation of myc-tagged Rad17 or myc-tagged Rad17(K118E) was performed as described previously (16) with the following modifications. Logarithmically growing cells (5 × 108 cells) were harvested in 1 mM sodium azide by centrifugation and were washed sequentially with 25 ml of STOP buffer (0.9% NaCl, 1 mM NaN3, 50 mM NaF, 10 mM EDTA), 25 ml of double-distilled water, and 10 ml of 1.2 M sorbitol. Cells were resuspended in 1.125 ml of CB1 (50 mM sodium citrate, 40 mM EDTA, 1.2 M sorbitol) to which 125 μl of CB1 containing 10 mg of lysis enzymes (L2265; Sigma), 10 μg of Zymolyase-20T (ICN), and 2.5 μl of β-mercaptoethanol were added. Digestion of cells was monitored by removing 2 μl of cell sample and adding an equal volume of 10% (wt/vol) sodium dodecyl sulfate (SDS). When cell lysis reached approximately 90% after zymolyase treatment, the digestion was terminated by adding an equal volume of ice-cold 1.2 M sorbitol. Spheroplasts were harvested by centrifugation at 290 × g for 4 min and were washed twice with 1.2 ml of 1.2 M sorbitol. Finally, the spheroplasts were resuspended in 425 μl of 1.2 M sorbitol, frozen in liquid nitrogen, and stored at −80°C. Samples were subsequently thawed on ice and lysed by the addition of 50 μl of 10× lysis buffer (500 mM potassium acetate, 20 mM MgCl2, 200 mM HEPES [pH 7.9]). To the lysates, a Complete Protease Inhibitor EDTA-Free Tablet (Roche Molecular Biochemical) and 20 μl of 25% Triton X-100 (TX-100) were added. Samples were incubated on ice for 10 min, and 50 μl was removed and boiled in an equal volume of 2× SDS sample loading buffer. This was analyzed as the total protein fraction (designated Total). Extracts were subsequently fractionated into soluble and pellet fractions by centrifugation at 12,000 × g for 15 min in a bench-top microcentrifuge. Supernatant was removed and boiled in an equal volume of 2× SDS sample buffer (designated Sup). An insoluble chromatin-enriched pellet fraction was washed once with the lysis buffer without TX-100 and digested with DNase I (100 U; Stratagene) in the presence of 5 mM MgSO4 and protease inhibitors on ice for 30 min. The DNase I-digested chromatin-enriched fraction was centrifuged for 5 min at 14,000 × g. Supernatant was designated the chromatin fraction (designated Chr), while the pellet was designated the cellular scaffold and debris. The chromatin binding of myc-Rad17 and myc-Rad17(K118E) was detected by an anti-myc monoclonal antibody (9E10).
Preparation of cell extract.
Logarithmically growing cells were washed in 20 ml of HB buffer (25 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 15 mM EGTA, 1 mM dithiothreitol [DTT], and proteinase inhibitors as described above) with the addition of 300 mM NaCl and 1% TX-100. Cells were disrupted with glass beads (425 to 600 μm; Sigma) in the above buffer and centrifuged at 14,000 × g for 15 min at 4°C. Supernatant was diluted to 150 mM NaCl–0.5% TX-100 by HB buffer for immunoprecipitation. For preparation of cell extracts from cells expressing an epitope-tagged protein from the nmt1 promoter of pREP41, cells were first grown in medium containing thiamine to repress overexpression. Cells were then harvested by centrifugation, washed with distilled water to remove thiamine, reinoculated into thiamine lacking minimal medium with appropriate nutritional selection, and grown for 18 h.
Immunofluorescence analysis.
For nuclear localization analysis, anti-myc antibody (9E10) was used at 1:1,000 dilution and the secondary antibody (goat anti-mouse immunoglobulin Alexa 488; Molecular Probes) for immunofluorescence was used at 1:250 dilution. The immunofluorescence protocol was as described previously (36), with modification of in-cell-wall digestion with 0.2 mg of Zymolyase 20T (ICN) per ml in buffer for 8 to 10 min at room temperature. DNA was stained with propidium iodide. For photography, cells were dried on polylysine-coated coverslips and mounted in 50% glycerol and phosphate-buffered saline (PBS).
Construction of epitope-tagged rfc1+ and rfc2+ strains.
Three-HA or 13 myc epitope tags were independently constructed at the C terminus of rfc1+, and the three-HA tag was constructed at the C terminus of rfc2+ by a PCR method followed by G418 selection exactly as described previously (2).
Construction of the rfc1Δ strain and analysis of the phenotype of the rfc1Δ germinating spores.
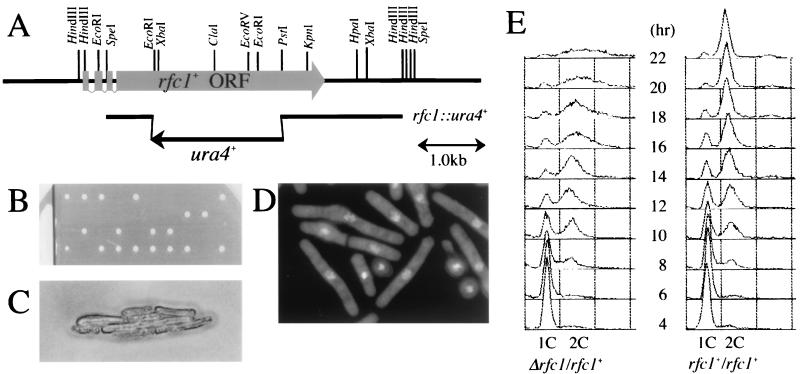
A heterozygous diploid (rfc1+/rfcΔ) was constructed by one-step gene replacement by replacing the 1.7-kb EcoRI-PstI fragment of rfc1+ with a 1.8-kb ura4+ gene cassette as illustrated in Fig. 6A. The 3.8-kb SpeI-HindIII fragment containing the disrupted rfc1 gene was transformed into a diploid strain (h+/h− leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M210/ade6-M216). Stable transformants were selected and verified as heterozygous diploids by Southern blot analysis. The heterozygous diploid cells were sporulated on MEA medium for 7 days. Spores derived from the rfc1+/rfc1Δ heterozygous diploid were washed with double-distilled water and then incubated at 30°C for 18 h in 40 ml of water containing 0.1 ml of helicase (IBF) followed by three further washes with water. The spores were cultured in minimal medium containing adenine and leucine at 30°C to induce preferential germination of ura4+ and rfc1Δ spores. Germinating spores were collected every 2 h, fixed with 70% ethanol, stained with 4′,6′-diamidino-2-phenylindole (DAPI), and analyzed.
FIG. 6.
Cells with deletion of rfc1 are checkpoint proficient. (A) Restriction map and open reading frame (ORF) of rfc1+. A 1.8-kb ura4+ gene cassette was used to replace 1.7 kb of the EcoRI-PstI coding region as described in Materials and Methods. (B) rfc1+ is an essential gene. Spores derived from the heterozygous diploid rfc1+/rfc1Δ were analyzed by tetrad dissection after incubation on yeast extract agar plates at 30°C for 4 days. The 2:2 segregation of the germinating spores indicates that rfc1+ is essential for cell viability. (C) Germinating rfc1Δ spores have elongated cell morphology. Shown are cells derived from a single germinating spore. (D) Cells with rfc1Δ have intact checkpoint processes. DAPI staining of germinating rfc1Δ spores having elongated cell morphology and normal nuclear morphology is shown. (E) FACS profiles. Heterozygous diploid rfc1+/rfc1Δ and control ura4+/ura4D-18 diploid cells were germinated at 30°C in minimal media lacking uracil. Germinating spores were collected every 2 h, fixed with 70% ethanol, and analyzed by FACS as described in Materials and Methods. The positions of 1C and 2C DNA peaks are indicated.
Purification of his-myc-tagged Rad17 and myc-tagged Rad17(K118E) from fission yeast cells.
Rad17 was purified as a side fraction of a large-scale fission yeast DNA polymerase α-primase purification protocol, details of which will be published elsewhere (R. E. Davis and T. S. Wang, unpublished). Briefly, 225 g of fission yeast cells containing his-myc-rad17+ at their endogenous genomic loci (17) were disrupted with glass beads (425 to 600 μm; Sigma) in 2× lysis buffer which contains 300 mM HEPES (pH 7.9), 1 M KCl, 20% glycerol, 1 mM EDTA, 2 mM DTT, and proteinase inhibitors (6 μM leupeptin, 2 μM pepstatin A, 2 mM benzamidine, and 1 mM phenylmethylsulfonyl fluoride). Crude cell lysates were centrifuged at 8,000 × g for 15 min at 4°C. The supernatant was collected, recentrifuged at 90,000 × g for 90 min, diluted to 500 mM KCl, and then applied to a fast-flow Q-Sepharose column pre-equilibrated in 1× lysis buffer to remove nucleic acids. The flow-through fraction was collected and diluted with dilution buffer (1 mM EDTA, 1 mM DTT, 10% glycerol, 100 mM KPO4). Phosphocellulose resin equilibrated in the same buffer was added to the flow-through fraction and rotated end to end at 4°C. The phosphocellulose resin was washed three times with wash buffer (1 mM EDTA, 1 mM DTT, 10% glycerol, protease inhibitors as described above) plus 100 mM KPO4, pH 7.5. Proteins were eluted with wash buffer plus 350 mM KPO4, pH 7.5. After dialysis against a buffer (20 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol, and protease inhibitors), the eluates were loaded onto a high-resolution Q-Sepharose column. Proteins were eluted from the high-resolution Q-Sepharose column with a gradient of 50 to 1,000 mM NaCl. The myc-his-tagged Rad17 protein eluted from the column was monitored by Western blot analysis and was eluted at approximately 800 mM NaCl. Fractions containing the myc-his-Rad17 protein were pooled, dialyzed against MCAC buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, 10% glycerol, protease inhibitors), and loaded onto a Ni+-agarose column (1 ml) pre-equilibrated in the MCAC buffer. The column was washed with five column volumes of MCAC buffer, and myc-his-Rad17 protein was eluted by an MCAC buffer containing 100 mM imidazole. The myc-his-tagged Rad17 protein was monitored by Western blotting with an anti-myc monoclonal antibody (9E10). Purified myc-Rad17 protein was dialyzed against the dialysis buffer and was used for ATP binding studies. Mutant Rad17(K118E) protein was purified from the rad17Δ strain harboring a pREP41-rad17.K118E plasmid by using the same protocol.
ATP binding assay.
A 20-μl reaction mixture containing 0.2 μg of purified Rad17 protein or Rad17(K118E) protein in a solution containing 25 mM Tris-HCl (pH 7.5), 3 mM MgCl2, 1 mM DTT, 50 μg of bovine serum albumin per ml, 0.2 μg of 96-mer oligonucleotide or M13 DNA, and 10 μM [γ-32P]ATP (0.33 μCi/nmol) was incubated at room temperature for 20 min. The reaction mixtures were irradiated with short-wave UV light in a Stratalinker (Stratagene) for 20 min on ice, and the cross-linked products were loaded onto SDS–8% polyacrylamide gels. The gels were dried and exposed to PhosphorImager screens or X-ray films.
Immunoprecipitation and Western blotting.
One milligram of total cell extract was diluted to 300 μl with HB buffer plus 150 mM NaCl and 0.5% TX-100. The diluted cell extract was then incubated with protein G agarose (Calbiochem) which had been prebound with anti-HA monoclonal antibody (3F10 high affinity; Boehringer Mannheim). Immunocomplexes were collected by centrifugation and washed four times with 1 ml of HB buffer, boiled in SDS sample buffer, and fractionated on SDS–8% polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked with Blotto (PBS, 1% fat-free milk powder, 0.05% Tween 20) and incubated in Blotto plus anti-HA monoclonal antibody (12CA5) (1:1,000 dilution) or anti-myc antibody (9E10) at a dilution of 1:1,000. The membranes were then washed in Blotto and incubated with horseradish peroxidase-conjugated secondary antibody (1:2,000 dilution). Chemiluminescent detection of horseradish peroxidase-conjugated secondary antibodies was carried out (NEM).
Cds1 kinase assay.
Cds1 kinase assays were performed as previously described (22) with the following modifications. Cell lysates were prepared from the myc-tagged Cds1 strain by glass bead disruption in HB buffer plus 150 mM NaCl as described above. Cds1 protein was immunoprecipitated from 1 mg of soluble protein in a 300-μl volume using protein G plus A agarose (Calbiochem) which had been prebound with anti-myc monoclonal antibody (9E10). Immunocomplexes were collected by centrifugation and washed four times with 1 ml of HB buffer plus 150 mM NaCl and further washed once with kinase buffer (10 mM HEPES [pH 7.5], 75 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT). Twenty microliters (50% slurry) of the immunocomplex-containing beads were incubated with 10 μl of 2× kinase buffer, 5 μCi of [γ-32P]ATP, 1 μl of 2 mM ATP, and 5 μl of myelin basic protein (1 mg/ml stock) at 30°C for 15 min. One third of the sample was removed and analyzed by Western blotting as a protein-loading control. Reactions were terminated by being boiled in an equal volume of 2× SDS sample buffer, and samples were fractionated on 15% SDS gels. Gels were dried and exposed on PhosphorImager screens or X-ray films.
Flow cytometry analysis.
Cells were harvested, washed in water, and fixed in 70% ethanol prior to staining with DAPI, as described earlier. DNA contents were measured using a FACScan system and CellFIT cell cycle analysis and LYSISII software (Becton Dickinson).
Cytology analysis.
Cells were fixed in 70% ethanol and stained by DAPI as described previously (53).
RESULTS
DNA damage enhances Rad17 chromatin binding.
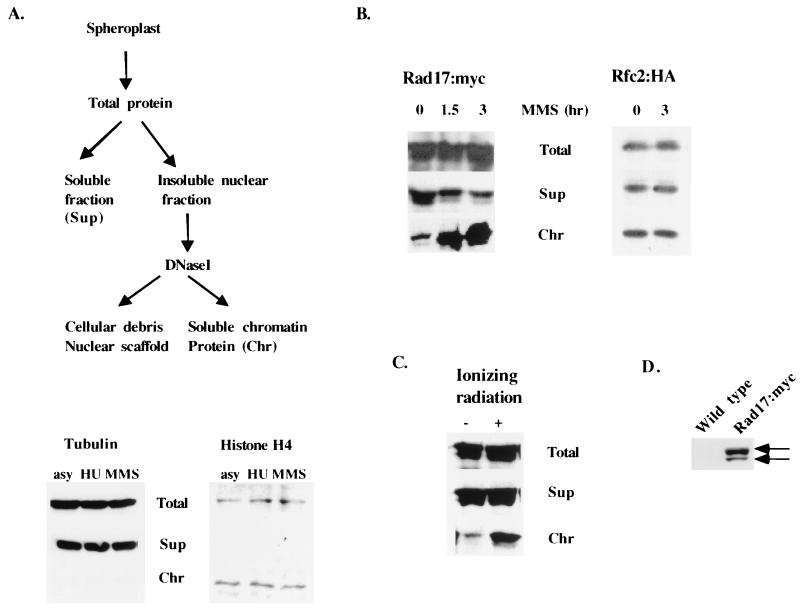
We have previously shown that Rad17 binds to chromatin independent of other checkpoint proteins throughout the cell cycle (16). To investigate the significance of this finding, we first tested Rad17 chromatin binding in response to DNA damage. Since others have reported that Rad17 exists in a complex with the small subunits of the Rfc protein (15, 30, 31, 43), we also used Rfc2 as the representative Rfc small subunit and tested the Rfc2 chromatin binding status. We constructed strains containing HA epitope-tagged rfc1+ or rfc2+ at their respective chromosomal loci (see Materials and Methods). We first analyzed the chromatin associations of Rad17 and Rfc2 in response to DNA damage induced by methyl methanesulfonate (MMS) treatment from the myc-tagged rad17+ strain (17) or the HA-tagged rfc2+ strain. Spheroplasts prepared from cells grown in MMS were made by limited enzymatic digestion followed by lysis with nonionic detergent. Lysates were then fractionated into soluble (Sup) and insoluble nuclear pellet fractions. The insoluble nuclear pellet fraction was treated with DNase I to release the chromatin-bound proteins as soluble chromatin-bound protein fractions (Fig. 1A, Chr), leaving an insoluble pellet of cellular debris and nuclear scaffold-bound proteins. Histone H4 was used as a control for the chromatin-bound protein, and tubulin was used as a control for the non-chromatin-bound protein. We also tested whether MMS and hydroxyurea could affect the chromatin binding of cellular proteins. As shown in the bottom panel of Fig. 1A, tubulin was not chromatin bound and histone H4 was chromatin bound under either hydroxyurea or MMS treatment, indicating that the chromatin binding assay used in our study is a valid analysis. We also tested the viability of cells in MMS. Following MMS treatment for 1.5 h, cells with untagged rad17+ and strains with myc-tagged rad17+ and HA-tagged rfc2+ were 100% viable. After 3 h of MMS treatment, similar to the wild-type cells, 30 to 35% of the myc-tagged rad17+ and HA-tagged rfc2+ cells were viable, whereas less than 1% of the rad17Δ cells were viable after 3 h of MMS treatment.
FIG. 1.
Chromatin binding of Rad17 is enhanced following DNA damage. (A) Schematic outline of the chromatin fractionation assay. Details of the assay are described in Materials and Methods. Histone H4 and α-tubulin were used as controls for chromatin-bound and non-chromatin-bound proteins in either hydroxyurea- or MMS-treated or -untreated cells with anti-acetyl-histone H4 (Lys 16) polyclonal antibody (Upstate Biotechnology) or antitubulin monoclonal antibody, respectively. HU, hydroxyurea. (B) Chromatin association of Rad17 and Rfc2 following MMS treatment. Cells were grown in 0.05% MMS. At the indicated times, cells were fractionated as outlined for panel A and as described in Materials and Methods. Spheroplasts were prepared from cells containing rad17+:myc (left panel) or rfc2+:HA (right panel). Twenty micrograms of total protein, an equivalent volume of the supernatant (Sup), and five volume equivalents of chromatin-bound proteins (Chr) were fractionated on an 8% SDS gel and analyzed by Western blotting with anti-myc monoclonal antibody (9E10). (C) Chromatin association of Rad17 after ionizing radiation. Cells containing rad17:myc were irradiated with 250 Gy and recovered by incubation at 25°C for 20 min and fractionated as described for panel A. (D) Total cell extracts were prepared under denaturing conditions from wild-type (untagged rad17+) and myc-tagged rad17+ cells and were probed with anti-myc antibody. The arrows mark the two forms of Rad17:myc.
We then analyzed chromatin association of Rad17:myc and Rfc2:HA in MMS-treated cells. After incubation in MMS for 1.5 and 3 h, a progressive increase of chromatin-bound Rad17 protein was observed (Fig. 1B, left panel). However, there was no discernable difference in the chromatin-bound Rfc2 protein levels between cells treated or not treated with MMS (Fig. 1B, middle). To test whether the enhancement of chromatin association of Rad17 is specific to the MMS-induced damage, we analyzed the effect of ionizing radiation on Rad17 chromatin association. Cells with myc-tagged rad17+ were irradiated with 250 Gy. At this dosage, more than 70% of the rad17+ cells survived irradiation (17). Similar to the DNA damage induced by MMS treatment, ionizing radiation induced an increase of Rad17 binding to chromatin (Fig. 1C). These results indicate that DNA damage induced by either MMS or ionizing irradiation enhances the Rad17-chromatin association.
We often observed that the myc-tagged Rad17 protein appears as a doublet with a major protein of higher molecular mass and a minor protein with lower molecular mass (Fig. 1D). Both Rad17 protein bands are chromatin bound. Phosphatase treatment did not reduce the two Rad17 proteins to a single species, indicating that the slower mobility form is not a phosphorylated Rad17 protein. Since the extracts were prepared under denaturing conditions, the observed lower-molecular-mass species was not likely to be a degradation product.
Rad17 responds to S-phase arrest induced by nucleotide pool depletion and the replication mutant by binding to chromatin in different ways.
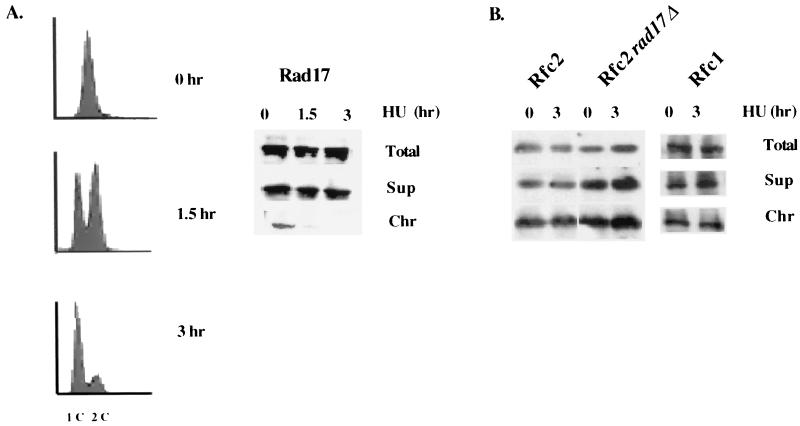
We then investigated the chromatin association of Rad17, Rfc1, and Rfc2 after 1.5 and 3 h of incubation of the cells in 12 mM hydroxyurea (Fig. 2A and B). Since asynchronous fission yeast cells are predominantly in G2 phase, cells had a 2C DNA content before hydroxyurea treatment (Fig. 2A, 0 h FACS profile). After 1.5 h, cells began to progress into S phase. After 3 h in hydroxyurea, a majority population of the cells was arrested in early S phase, with a 1C FACS profile. Interestingly, after 1.5 h in hydroxyurea a large fraction of the chromatin-bound Rad17 dissociated from the chromatin, and after 3 h nearly all of the chromatin-bound Rad17 prior to incubation in hydroxyurea dissociated from the chromatin (Fig. 2A). The dissociation of Rad17 from the chromatin was not due to cell death, since 100% of the cells are viable after 3 h in hydroxyurea (data not shown). These results indicate that Rad17 responds to hydroxyurea-induced S-phase block by dissociating from the chromatin.
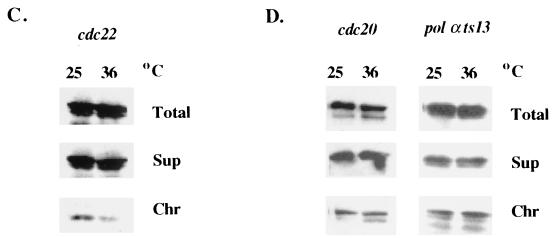
FIG. 2.
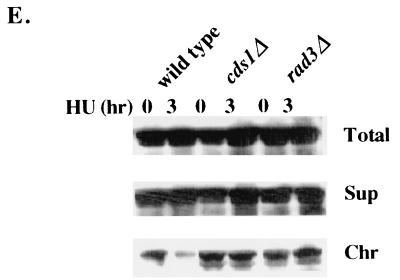
The chromatin binding of Rad17 in response to S-phase perturbation. (A) The mid-log-phase rad17+:myc strain was incubated in medium containing 12 mM hydroxyurea. At the indicated times, cell samples were removed for FACS analysis and chromatin fractionation assay by probing with anti-myc monoclonal antibody. (B) Rfc1 and Rfc2 were chromatin bound in cells grown in hydroxyurea. Mid-log-phase cells expressed Rfc1:HA in a rad17+ background, or Rfc2:HA in a rad17+ or rad17Δ background was incubated in medium with 12 mM hydroxyurea for 3 h. Chromatin fractionation assay was performed and analyzed by Western blotting and was probed with anti-HA monoclonal antibody. (C) Mid-log-phase cells of rad17+:myc in a cdc22-M45 background grown at 25°C were shifted to 36°C for 3 h. Cell samples grown at 25 and 36°C were removed for chromatin fractionation assay. (D) Mid-log-phase cells that expressed Rad17:myc from genomic loci in cdc20 and polαts13 backgrounds grown at 25°C were shifted to 36°C for 2 h. Chromatin fractionation was performed, analyzed by Western blotting, and probed with anti-myc monoclonal antibody. (E) Mid-log-phase cells that expressed Rad17:myc from genomic loci in cds1Δ and rad3Δ backgrounds were incubated in media containing 12 mM hydroxyurea. Chromatin fractionation assay was performed and analyzed by Western blotting and was probed with anti-myc monoclonal antibody.
We then analyzed the chromatin binding statuses of Rfc1 and Rfc2 in the hydroxyurea-arrested cells. After 3 h of incubation in hydroxyurea, Rfc1 in the rad17+ background and Rfc2 in either the rad17+ or rad17Δ background remained chromatin bound (Fig. 2B). Since the Rfc1-5 complex is required for replication, it is not surprising that in spite of Rad17 dissociating from the chromatin or in cells with rad17Δ, there are chromatin-bound Rfc1 and Rfc2 proteins for replication purposes.
To ascertain that Rad17 dissociates from the chromatin in response to nucleotide pool depletion caused by hydroxyurea, we tested the chromatin binding status of Rad17 in response to S-phase stall induced by cdc22-M45 mutant arrest. cdc22 encodes the large subunit of ribonucleotide reductase, which physiologically mimics the effects of hydroxyurea. We constructed a strain with myc-tagged rad17+ in a cdc22-M45 background and compared the chromatin binding status of Rad17 after incubation for 3 h at 25 and 36°C. Similar to that found in hydroxyurea block, the Rad17 that previously bound to chromatin at 25°C dissociated from the chromatin after 3 h at 36°C (Fig. 2C). Together, these results indicate that Rad17 dissociates from the chromatin in response to S-phase stall induced by nucleotide pool depletion.
Finding that Rad17 dissociates from chromatin in response to S-phase stall induced by hydroxyurea and cdc22-M45 mutant arrest led us to investigate Rad17 chromatin binding status in response to S-phase arrest by replication mutants. We constructed strains with the myc-tagged rad17+ at their genomic loci in thermosensitive replication mutant backgrounds and tested the chromatin binding of Rad17 in each strain after incubation for 2 h at the permissive or nonpermissive temperature (Fig. 2D). Rad17 was chromatin bound in cells arrested at G1/S phase by cdc20 (polɛ mutant), consistent with previous findings (16). Rad17 was also chromatin bound in cells arrested in early S phase by polαts13. Rad17 also remained chromatin bound in cells arrested in late S phase by mutants of Polδ subunits, cdc27 and cdc1 (data not shown). Thus, Rad17 binds to chromatin in response to S-phase arrest induced by nucleotide pool depletion and by replication mutants by binding to chromatin in different ways.
Rad17 remains chromatin bound in hydroxyurea-arrested cds1Δ cells.
Cds1 is thought to have a role in maintaining genomic integrity. Upon hydroxyurea block, Cds1 kinase is highly activated and Chk1 becomes phosphorylated in hydroxyurea only when Cds1 is absent (22). Phosphorylation of Chk1 has been correlated to cell cycle arrest (59), while cell cycle arrest by a replication mutant results in moderate activation of Cds1 (4, 47) and phosphorylation of Chk1 (4, 11).
Findings that Rad17 dissociates in hydroxyurea-arrested cells (Fig. 2A) but remains chromatin bound in replication mutant-arrested cells (Fig. 2D) have led us to analyze the chromatin binding status of Rad17 in hydroxyurea-treated cds1Δ and rad3Δ cells. After 3 h in hydroxyurea, Rad17 remained on chromatin in cds1Δ and rad3Δ cells (Fig. 2E). In contrast, in cds1+ cells Rad17 dissociated from chromatin in a manner similar to that observed above for Fig. 2A. Rad17 remained chromatin bound in rad3Δ cells, since Rad3 is required for Cds1 activation by hydroxyurea (22). These results suggest that in response to S-phase arrest by hydroxyurea, Rad17 does not bind to chromatin when Cds1 is fully activated, whereas Rad17 remains chromatin bound when Chk1 is activated by hydroxyurea in the absence of Cds1 or in the replication mutants.
Checkpoint-defective mutant Rad17(K118E) protein localizes in the nucleus but has reduced ability to bind chromatin.
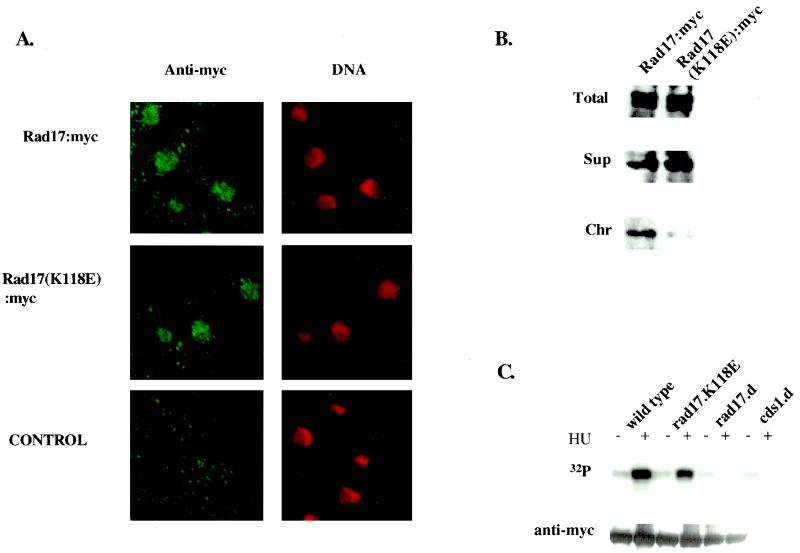
To further investigate the relevance of Rad17-chromatin association in the checkpoint process, we analyzed a checkpoint-defective mutant protein, Rad17(K118E). Mutant rad17.K118E has a checkpoint-defective phenotype of severity approaching that of rad17Δ (17). We first analyzed whether the checkpoint defect of rad17.K118E is due to the mutant Rad17(K118E) protein's inability to enter the nucleus. Both wild-type myc-tagged Rad17 and mutant myc-tagged Rad17(K118E) ectopically expressed from the pREP41 vector in rad17Δ cells were found localized in the nucleus by immunofluorescence microscopy analysis (Fig. 3A).
FIG. 3.
The checkpoint-defective Rad17(k118E) mutant protein localizes in the nucleus but has reduced ability to bind chromatin. (A) Logarithmically growing rad17Δ cells harboring pREP41-myc-rad17+ or pREP41-myc-rad17.K118E and wild-type untagged rad17+ cells were harvested and processed for immunofluorescence analysis as described in Materials and Methods. (B) Chromatin fractionation assay was performed with logarithmically growing rad17Δ cells harboring pREP41-myc-rad17+ or pREP41-myc-rad17.K118E as described in Materials and Methods. (C) Cds1 kinase activities of the rad17+ wild type and mutant rad17.K118E in the presence of 12 mM hydroxyurea. The indicated strains were grown at 25°C in either the absence or presence of 12 mM hydroxyurea for 3 h. Cds1 was immunoprecipitated from 1 mg of soluble protein and assayed for kinase activity as described in Materials and Methods (upper panel, 32P). The relative amounts of Cds1 used in the kinase assay were estimated by Western blotting (lower panel, anti-myc).
We then compared the chromatin association status of wild-type Rad17 and mutant Rad17(K118E). Equal levels of wild-type myc-Rad17 protein and myc-Rad17(K118E) mutant proteins in total spheroplast lysates (Fig. 3B, Total panel) were used for comparison. A significant fraction of wild-type Rad17 protein expressed from the pREP41 vector was chromatin bound (Fig. 3B, Chr, left panel). In contrast, only a nominal amount of mutant Rad17(K118E) protein was chromatin bound (Fig. 3B, Chr). Most of the Rad17(K118E) proteins remained in the soluble cytoplasmic fraction (Fig. 3B, Sup). Thus, there is a striking difference in chromatin binding ability between the Rad17 protein from a checkpoint-proficient strain and that from a checkpoint-deficient mutant strain. Furthermore, the decreased ability of Rad17(K118E) chromatin binding is not due to a failure of nuclear import.
Cds1 kinase is activated by hydroxyurea in the checkpoint-defective rad17.K118E mutant.
Mutant rad17.K118E is highly sensitive to γ irradiation but exhibits a 50% cell viability after 3 h in hydroxyurea, while rad17Δ cells under the same condition have less than 1% cell survival (17). The ability of cells to maintain viability in hydroxyurea has been attributed to the checkpoint Rad-dependent activation of the Cds1 protein kinase to prevent accumulation of aberrant DNA structures and to enable recovery from the stalled replication structure caused by hydroxyurea arrest (22). We tested whether the Cds1 kinase activity could be activated in the checkpoint-defective strain of the rad17.K118E mutant. Cds1 kinase activity in wild-type cells was highly induced after incubation in hydroxyurea, as expected. Cds1 kinase was also substantially activated in the rad17.K118E mutant, whereas Cds1 was not activated by hydroxyurea in rad17Δ cells (Fig. 3C).
Lys118 of Rad17 is involved in ATP binding and complex formation with Rfc2.
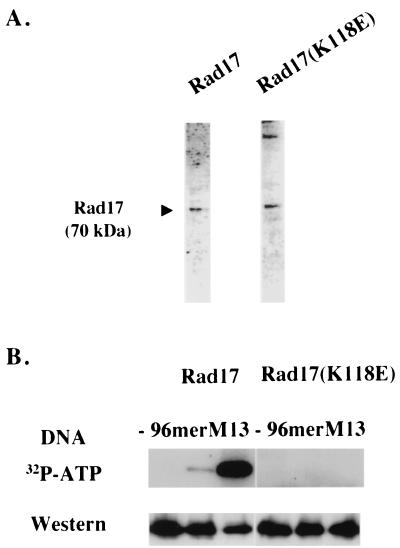
We further compared the biochemical properties of wild-type Rad17 and mutant Rad17(K118E) proteins in vitro. The Lys118 in Rad17 is located within the proposed nucleotide binding site of the Walker type A domain (17). Molecular modeling of the Rad17 protein has suggested that Rad17 protein binds ATP via this residue (54). We therefore tested the abilities of wild-type Rad17 and mutant Rad17(K118E) proteins to bind ATP in vitro. Wild-type myc-tagged Rad17 and mutant myc-tagged Rad17(K118E) proteins were purified from fission yeast cells (Fig. 4A). Neither the purified wild-type Rad17 nor the mutant Rad17(K118E) proteins had detectable ATPase activity (data not shown). However, with identical amounts of wild-type Rad17 or mutant Rad17(K118E) protein (Fig. 4B, lower panel), only wild-type Rad17 was able to bind ATP when ATP was added simultaneously with either oligonucleotides of 96 bases (96 mer) or single-stranded M13 DNA followed by cross-linking with UV irradiation (Fig. 4B, 32P-ATP panel). These results indicate that mutation of Lys118 to Glu abolishes the ATP binding capacity of the Rad17 protein. Thus, the Lys118 of Rad17 protein is involved in nucleotide binding.
FIG. 4.
Purified Rad17 protein, but not the checkpoint-defective Rad17(K118E) mutant protein, binds ATP in vitro. (A) Silver-stained polyacrylamide gel of 0.2 μg of purified wild-type Rad17 and mutant Rad17(K118E) proteins. (B) Wild-type Rad17, but not mutant Rad17(K118E), binds ATP. [γ-32P]ATP was incubated with purified Rad17 (left panel) or purified mutant Rad17(K118E) protein (right panel) in the presence or absence of 96-mer oligonucleotide or M13 DNA and was processed as described in Materials and Methods. The bottom panel shows the Western blot analysis of the purified Rad17 and Rad17(K118E) proteins used for the ATP binding assay.
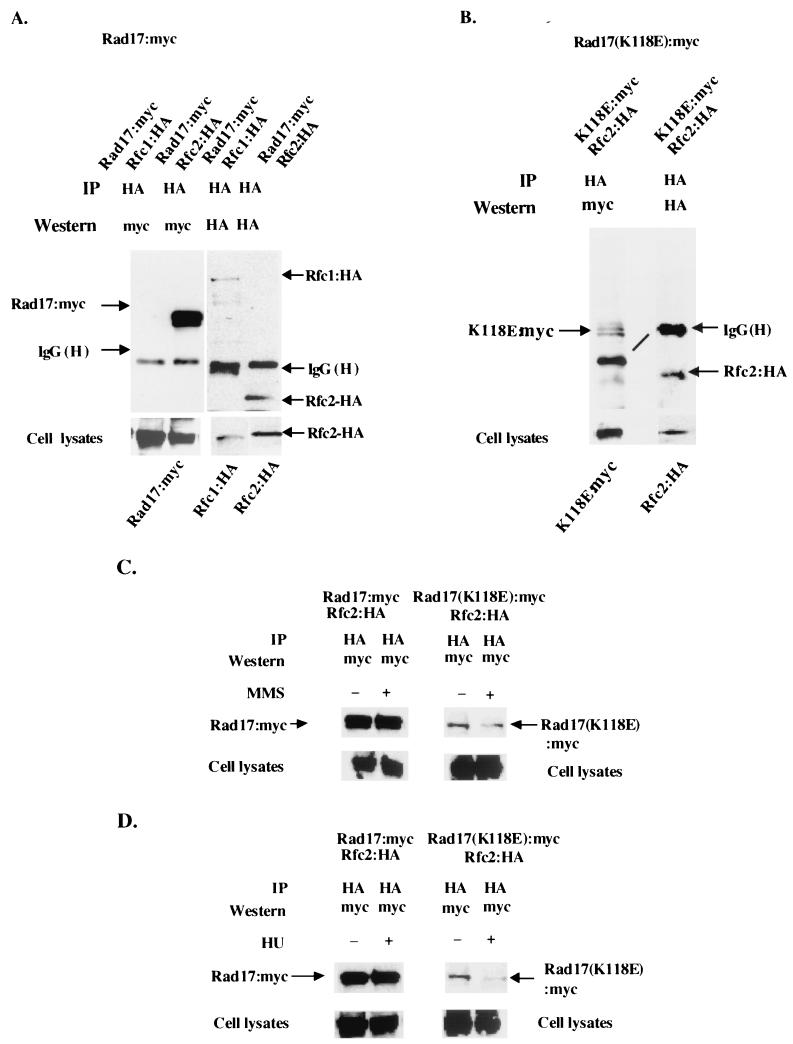
Budding yeast Rad24p coimmunoprecipitates with the four small subunits of Rfc but not with the large subunit, Rfc1p (15), and the interaction between Rad24p and Rfc5p is required for checkpoint function (30, 44). We therefore tested whether the Rad17(K118E) mutant protein could form a complex with the Rfc small subunits in vivo. We transformed either pREP41-myc-rad17+ or pREP41-myc-rad17(K118E) into a rad17Δ strain containing either HA-tagged rfc1+ or HA-tagged rfc2+ at their respective chromosomal loci (see Materials and Methods). Strains of rad17Δ that expressed epitope-tagged Rad17:myc from pREP41 and Rfc1:HA or Rfc2:HA from their genomic loci had identical growth rates and sensitivities to hydroxyurea, UV, and MMS as the wild-type strain (data not shown), indicating that those strains do not have any significant biological perturbations. Using these strains, we tested the coimmunoprecipitation of HA-tagged Rfc1 and HA-tagged Rfc2 with either myc-tagged Rad17(K118E) or myc-tagged wild-type Rad17. Anti-myc antibody (9E10) was unable to immunoprecipitate the 2×-myc-tagged Rad17 under the conditions used. However, the 2×-myc-tagged Rad17 is readily detectable by the anti-myc monoclonal antibody in Western blotting of the crude cell extracts. We therefore used an anti-HA antibody (3F10) to immunoprecipitate HA-Rfc1 or HA-Rfc2 followed by testing the immunoprecipitates for myc-Rad17 or myc-Rad17(K118E) by probing with anti-myc antibody.
Lysates from rad17Δ strains that expressed either Rfc1:HA or Rfc2:HA had comparable levels of Rad17:myc protein (Fig. 5A, cell lysates panel). In immunoprecipitates of anti-HA antibody (3F10), no detectable myc-Rad17 coprecipitated with HA-Rfc1 from cell lysates of the rad17Δ strain expressing Rfc1:HA and Rad17:myc (Fig. 5A, first lane), while the Rfc1:HA protein was present in the immunoprecipitate (Fig. 5A, third lane). In contrast, from lysates of the rad17Δ strain expressing Rfc2:HA and Rad17:myc, coimmunoprecipitation of Rad17:myc protein and Rfc2:HA was readily detected (Fig. 5A, second lane). These results confirm the findings by others (15, 30, 31, 43) that fission yeast Rad17, similar to budding yeast Rad24p, exists in complex with Rfc small subunits in vivo but not with Rfc1 protein.
FIG. 5.
Rad17 from a checkpoint-defective mutant, rad17.K118E, has reduced ability to associate with Rfc2. (A) Wild-type Rad17 can complex with Rfc2 but not with Rfc1. Crude cell lysates were prepared from 5 × 108 rad17Δ cells expressing either the HA-tagged rfc1+ or HA-tagged rfc2+ at their endogenous chromosomal loci and myc-Rad17 from pREP41-myc-rad17+. Cell lysates (300 μl) containing 1 mg of protein were used for immunoprecipitation (IP) by anti-HA antibody. The immunoprecipitates were Western blotted with either anti-HA (right panel) or anti-myc monoclonal antibody (left panel). (B) Mutant Rad17(K118E) from a checkpoint-defective strain has significantly reduced ability to associate with Rfc2. Lysates from 5 × 108 rad17Δ cells containing HA-tagged rfc2+ at its endogenous chromosomal locus and pREP41-myc-rad(K118E) were prepared and immunoprecipitated with anti-HA monoclonal antibody, and the immunoprecipitates were analyzed as for panel A. The cell lysate panels show that all strains have adequate expression of the epitope-tagged proteins. (C) Association of wild-type Rad17 and mutant Rad17(K118E) proteins with Rfc2 following MMS treatment. Strains described for panels A and B were grown in 0.05% MMS for 3 h. Cells (5 × 108) were removed for immunoprecipitation with anti-HA monoclonal antibody and were probed with anti-myc monoclonal antibody. (D) Association of wild-type Rad17 and mutant Rad17(K118E) proteins with Rfc2:HA following hydroxyurea (HU) treatment. Cells (5 × 108) from strains described for panels A and B were grown in media containing 12 mM hydroxyurea for 3 h. Cell lysates were prepared and immunoprecipitated with anti-HA monoclonal antibody and Western blotted with anti-myc monoclonal antibody.
We next tested whether the Rad17(K118E) mutant protein was able to coimmunoprecipitate with HA-tagged Rfc2 (Fig. 5B). With comparable levels of Rfc2:HA being immunoprecipitated, the amount of Rad17(K118E):myc mutant protein coprecipitated with Rfc2 was much smaller than the amount of wild-type Rad17:myc protein (compare Fig. 5B, left panel, with the second lane in panel A). Thus, the Rad17(K118E) mutant protein from the checkpoint-defective strain not only has significantly reduced ability to associate with chromatin (Fig. 3B) and an inability to bind ATP in vitro (Fig. 4B) but also has a substantially reduced ability to complex with Rfc2 protein (Fig. 5B). These results indicate that Lys118 of Rad17 plays an important role in ATP binding, in complex formation with Rfc2, and in chromatin association.
We then investigated whether Rad17-Rfc2 complex formation could be different in the wild-type and a checkpoint-defective mutant Rad17 strain following DNA damage and S-phase perturbation. We compared the ability of wild-type Rad17 and mutant Rad17(K118E) proteins to complex with Rfc2 after MMS treatment. After incubation of the rad17Δ strains that expressed Rfc2:HA from genomic loci and Rad17:myc or Rad17(K118E):myc from pREP41 in MMS for 1.5 h, levels of total Rad17:myc protein expressed and levels of the Rad17:myc protein coimmunoprecipitated with Rfc2:HA were comparable in the MMS-treated and untreated cell lysates (Fig. 5C, left panel). Again, a significantly reduced amount of mutant Rad17(K118E):myc protein was coimmunoprecipitated with Rfc2:HA (Fig. 5C, right panel). After 1.5 h of incubation in MMS, the level of mutant Rad17(K118E) protein in complex with Rfc2 seemed slightly reduced (Fig. 5C, right panel, compare + MMS and − MMS lanes). Thus, DNA damage by MMS treatment does not significantly affect complex formation between wild-type Rad17 and Rfc2 but does reduce the ability of the checkpoint-defective mutant Rad17(K118E) protein to complex with Rfc2.
We next tested whether hydroxyurea could affect the abilities of wild-type Rad17 and mutant Rad17(K118E) proteins to associate with Rfc2. rad17Δ strains that expressed Rfc2:HA from genomic loci and Rad17:myc or Rad17(K118E):myc from pREP41 were grown in hydroxyurea for 3 h. With comparable levels of Rad17:myc and Rad17(K118E):myc expressed in lysates of each strain, wild-type Rad17:myc coimmunoprecipitated with Rfc2:HA with or without hydroxyurea (Fig. 5D, left panel), whereas significantly reduced amounts of the Rad17(K118E):myc protein coimmunoprecipitated with Rfc2:HA protein (Fig. 5D, right panel). Thus, S-phase arrest by hydroxyurea does not affect the ability of Rad17 to complex with Rfc2 but further reduces the ability of Rad17(K118E) to associate with Rfc2.
These experiments suggest that Rad17 exists in a complex with the small subunits of Rfc protein and differentially associates with or dissociates from the chromatin in response to DNA damage or S-phase stall induced by hydroxyurea.
Cells with rfc1Δ are checkpoint proficient.
Cells with deletions or mutations of RFC2, RFC3, and RFC5 from budding yeast or fission yeast have been shown to display checkpoint-deficient phenotypes (31, 38, 43–46). Our finding that Rad17 coimmunoprecipitates with Rfc2 but not with Rfc1 (Fig. 5A) and the findings by others (15, 31, 38, 43–46) strongly suggest that the small subunits of Rfc, Rfc2-5, are involved in checkpoint function as well as in replication. This raises the question, are there two distinct types of Rfc complexes in cells, one essential for DNA replication and having Rfc1 in complex with Rfc2-5 and another involved in the checkpoint process and having Rad17 in complex with Rfc2-5? To answer this question, we analyzed the phenotype of cells with a deletion of rfc1+. A heterozygous rfc1+/rfc1::ura4+ diploid was constructed as described in Materials and Methods. Tetrad analysis of the diploid yielded two viable ura4− spores, indicating that rfc1+ is essential for cell viability (Fig. 6B). Analysis of the phenotype of the germinating spores derived from this heterozygous diploid in selective media for germination of rfc1Δ spores showed that germinating spores with rfc1Δ displayed elongated cell morphology (Fig. 6C). Analysis of the FACS profiles of the germinating rfc1+ wild-type spores showed that the rfc1+ germinating spores completed S phase after 16 h, displaying a 2C DNA profile. The rfc1Δ germinating spores displayed a >2C DNA profile after 14 h which reflected the elongated cdc phenotype (Fig. 6D and E). This result indicates that cells in the absence of rfc1+ are proficient in cell cycle checkpoint response. Thus, Rfc1, in contrast to Rfc2 and Rfc3, does not have a role in checkpoint function.
DISCUSSION
In this study we investigated the significance of Rad17 chromatin binding. We showed the following results. (i) Following DNA damage, increased levels of Rad17 protein bind to chromatin. (ii) In response to S-phase perturbation induced by nucleotide pool depletion, Rad17 dissociates from the chromatin, whereas Rad17 remains chromatin bound during replication mutant arrest. (iii) In contrast to Rad17 protein in cds1+ cells, Rad17 remains chromatin bound in hydroxyurea-treated cds1Δ cells. (iv) Rad17 protein from a checkpoint-defective mutant, rad17.K118E, is unable to bind ATP in vitro and has significantly reduced ability to associate with chromatin and to complex with Rfc2 in vivo; however, Cds1 kinase can be activated by hydroxyurea in these mutant cells. (v) Rad17 exists in a complex with Rfc2 but not with Rfc1. The Rad17 protein associates with Rfc2 even under DNA damage by MMS or S-phase stall by hydroxyurea, while Rfc1 is not involved in checkpoint function. These results suggest that a fraction of Rad17 in a complex with the small subunits of Rfc proteins binds to chromatin in response to the aberrant genomic structures and may function as a part of the checkpoint process. We discuss this hypothesis below and propose a model of how the Rad17-Rfc small-subunit complex responds to different replication and damage structures by differentially binding to chromatin (Fig. 7).
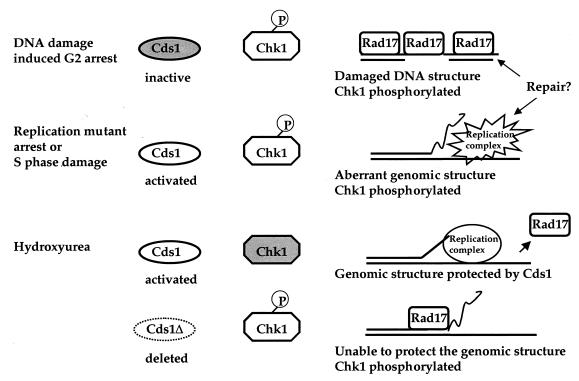
FIG. 7.
A proposed model illustrating how Rad17 binds to chromatin in response to aberrant genomic structures induced by damage, replication mutant arrest, and hydroxyurea block in the presence or absence of an activated Cds1 kinase. P, phosphorylation.
Rad17 binds to chromatin in response to aberrant genomic structures.
We showed in this study that Rad17 binds to chromatin (i) following DNA damage (Fig. 1), (ii) following replication mutant arrest (Fig. 2D), and (iii) in hydroxyurea-arrested cds1Δ cells (Fig. 2E). DNA damage induced by MMS treatment or ionizing radiation causes aberrant genomic structures and Chk1 phosphorylation (59). S-phase arrest by replication mutants compromises the replication complex or delays the progression of replication forking, which could result in a mutagenic genomic structure, such as a long stretch of single-strand template and double-strand breaks (23, 50). These could induce a moderate activation of Cds1 and phosphoryation of Chk1 (4, 47). Hydroxyurea treatment highly activates Cds1 kinase activity, which is thought to protect the genome from deterioration (22). In cds1Δ cells, in the absence of Cds1 the aberrant replication complex or structure might further deteriorate, which induces phosphorylation of Chk1 (22). Since in hydroxyurea arrested cells damage-induced phosphoryation of Chk1 is prevented, hydroxyurea-activated Cds1 is thought to suppress a repair process and Chk1 phosphorylation (6). Together, results of these Cds1 studies suggest that hydroxyurea-activated Cds1 is involved in a process that prevents aberrant genomic structure formation and induction of Chk1 phosphorylation. We showed here that DNA damage enhances Rad17 chromatin association (Fig. 1), and Rad17 remains chromatin bound following a replication mutant arrest and in hydroxyurea-arrested cds1Δ cells (Fig. 2). Our results thus suggest that Rad17 binds to chromatin or remains on the chromatin in response to the aberrant genomic structures that may lead to phosphorylation of Chk1 to prevent cell cycle progression (Fig. 7).
How might Rad17 dissociate from the chromatin in hydroxyurea-treated cds1+ cells? In hydroxyurea-treated cds1+ cells, Cds1 kinase is highly activated. Thus, the integrity of the replication complex or replication fork is being protected from deterioration into aberrant genomic structures by the activated Cds1 (22). If Cds1 is activated by hydroxyurea, there would be no aberrant genomic structure or phosphorylation of Chk1. Thus, Rad17 is not required to bind to chromatin, resulting in dissociation from chromatin (Fig. 2A and 7).
Consistent with these notions, rad17Δ cells in which Cds1 cannot be activated by hydroxyurea (22) are highly sensitive to hydroxyurea, with less than 1% cell survival after 3 h in hydroxyurea, whereas mutant rad17.K118E cells with an activated Cds1 (Fig. 3C) have 50% cell survival. Moreover, in the rad17.K118E mutant, Chk1 is not phosphorylated in MMS-induced DNA damage (data not shown), and mutant Rad17(K118E) protein has a substantially reduced ability to bind chromatin (Fig. 5B). Furthermore, rad17Δ cells with an overexpression of Rad17(K118E) from pREP41 had 80% cell survival in hydroxyurea; however, the overexpression of Rad17(K118E) was unable to suppress the MMS sensitivity of rad17Δ cells (data not shown). Together, these results suggest that a fraction of Rad17 binds to chromatin in response to the presence of aberrant genomic structures, either participating in maintaining the genomic integrity or in signalling downstream checkpoint and repair processes.
These data led us to propose a model where Rad17 is not required to associate with chromatin when the genomic structures are being protected by hydroxyurea-activated Cds1. Rad17 binds to chromatin when genomic structures are aberrant without protection by a hydroxyurea-activated Cds1, and the aberrant genomic structures induce phosphorylation of Chk1 (Fig. 7).
What event(s) might be induced by Rad17 chromatin binding?
It has been suggested that double-strand-break formation is intrinsic to S-phase progression (37). One possible event is that Rad17 remains on the chromatin throughout the cell cycle, and following recognition of an aberrant genomic structure by Rad3, additional Rad17 proteins are induced to bind chromatin to initiate damage processing. Human Rad1 protein has been reported to encode a 3′-to-5′ exonuclease (35), and human Rad9 has been shown in vitro as being a 3′-to-5′ exonuclease (3). Rad1 and Rad9 form a protein complex with Hus1B (8), and molecular modeling has predicted that this complex has structural similarity to the PCNA sliding clamp (8, 48, 54). In two-hybrid reactions, Rad17 interacts with Rad1 and nuclear localization of Hus1B and Rad9 requires Rad17 (8). Two studies of human checkpoint proteins have also shown that in response to diverse DNA damage agents and hydroxyurea, human Rad9 is converted into an extraction-resistant nuclear form (7). In vitro, human Rad17 interacts with human Rad1, Rad9, and Hus1 in a checkpoint clamp-loading complex similar to that of a replication clamp loader (54). It is possible that chromatin binding of the Rad17-Rfc small-subunit complex may function as a signal in loading Rad1-Rad9-Hus1 onto the aberrant DNA structures for processing the lesion.
How could mutation of Rad17 at Lys118 to Glu cause a checkpoint defect?
Of the six site-directed mutations introduced into Rad17, the rad17.K118E mutant is the only mutant that displays a checkpoint-deficient phenotype of severity approaching that of rad17Δ (17). Lys118 is in a domain that has sequence similarity to the Walker type A domain in a proposed nucleotide phosphate binding P-loop (51, 58). Rad17 protein purified from the checkpoint-defective rad17.K118E mutant failed to bind ATP (Fig. 4B). This indicates that the basic Lys118 residue is indeed a residue critical for ATP binding and supports the structure-based prediction that Lys118 in the P-loop directly contacts the phosphate moiety of ATP (54).
The replication complex of Rfc1-5 is the eukaryotic homologue of the prokaryotic replication clamp loader, the γ-complex of Escherichia coli Pol III (34, 52). In E. coli Pol III, the γ-complex harnesses energy from ATP binding, which induces a conformational change of the γ-complex, allowing it to load the β clamp onto DNA for processive DNA synthesis (52). The predicted molecular model of Rad17 has also suggested that ATP binding of Rad17 might induce changes in orientation between the N terminus and linker domains of Rad17 and that mutation of Lys118 to Glu would result in an unfavorable electrostatic interaction with ATP (54). Similar to the prokaryotic γ-complex clamp loader, ATP binding of Rad17 may induce a conformational change which may be required for the proper assembly of the Rad17-Rfc small-subunit complex. The inability of the mutant Rad17(K118E) protein to bind ATP may fail to induce a conformational change of the mutant protein, thus precluding proper assembly of the Rad17-Rfc small-subunit complex and chromatin association. Compared to wild-type Rad17, Rad17(K118E) has a substantially reduced ability to complex with Rfc2, whereas wild-type Rad17 is able to complex with Rfc2 following replication perturbation or DNA damage (Fig. 5B, C, and D). Moreover, the reduced ability of Rad17(K118E) to bind chromatin is not due to its lack of nuclear import ability (Fig. 3A). Thus, a properly assembled Rad17–Rfc2-5 complex associating with chromatin may be a prerequisite for a Rad17-mediated checkpoint process, and mutation of Lys118 in rad17.K118E would have a checkpoint-deficient phenotype.
There are two distinct types of Rfc complexes in cells.
Rfc is a five-subunit protein complex that is an essential factor for DNA replication (57). Genetic studies have shown that Rfc small subunits, in addition to functioning in replication, are also involved in the mitotic checkpoint (31, 38, 43, 46). In support of this notion, we have demonstrated in this study that cells with a deletion of rfc1 are checkpoint proficient (Fig. 6). Thus, there are two distinct types of Rfc complexes in cells. Rfc1-5 complex functions in replication and constitutively binds to chromatin as a replication-processive clamp loader. Rfc2-5–Rad17 complex functions in the checkpoint process and binds to chromatin in response to the presence of aberrant genomic structures.
ACKNOWLEDGMENTS
We thank A. M. Carr for providing us with the myc-tagged rad17 strain, mutant rad17(K118E) strain, and plasmids pREP-myc-rad17 and pREP-myc-rad17(K118E). We also thank Hiroto Okayama for allowing Hiroyuki Tanaka to perform the rfc1 gene disruption experiment in his laboratory. We especially thank members of our laboratory for helpful discussion during the course of this work and R. E. Davis for his help in large-scale Rad17 protein purification.
This study was supported by grant CA54415 from the National Cancer Institute of the National Institutes of Health.
REFERENCES
- 1.Al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J, Lehmann A R, Carr A M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahler J, Wu J Q, Longtine M S, Shah N G, McKenzie A, Steever A B, Wach A, Philippsen P, Pringle J R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 3.Bessho T, Sancar A. Human DNA damage checkpoint protein hRAD9 is a 3′ to 5′ exonuclease. J Biol Chem. 2000;275:7451–7454. doi: 10.1074/jbc.275.11.7451. [DOI] [PubMed] [Google Scholar]
- 4.Bhaumik D, Wang T S-F. Mutational effect of fission yeast Polα on cell cycle events. Mol Biol Cell. 1998;9:2107–2123. doi: 10.1091/mbc.9.8.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinase Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 6.Brondello J-M, Broddy M N, Furnari B, Russell P. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol Cell Biol. 1999;19:4262–4269. doi: 10.1128/mcb.19.6.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burtelow M A, Kaufmann S H, Karnitz L M. Retention of the hRad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J Biol Chem. 2000;275:26343–26348. doi: 10.1074/jbc.M001244200. [DOI] [PubMed] [Google Scholar]
- 8.Caspari T, Dahlen M, Kanter-Smoler G, Lindsay H D, Hoffmann K, Papadimitriou K, Sunnerhagen P, Carr A M. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;74:1254–1262. doi: 10.1128/mcb.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards P J, Bentley N J, Carr A M. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 10.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 11.Francesconi S, Grenon M, Bouvier D, Baldacci G. p56chk1 protein kinase is required for the DNA replication checkpoint at 37°C in fission yeast. EMBO J. 1997;16:1332–1341. doi: 10.1093/emboj/16.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furnari B, Blasina A, Boddy M N, McGowan C H, Russell P. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol Biol Cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 14.Gardner R, Putnam C W, Weinert T. RAD53, DUN1 and PDS1 defines two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 1999;18:3173–3185. doi: 10.1093/emboj/18.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green C M, Erdjument-Bromage H, Tempst P, Lowneds N F. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 1999;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths D, Uchiyama M, Nurse P, Wang T S-F. A novel allele of the chromatin-bound fission yeast checkpoint protein Rad17 separates the DNA structure checkpoints. J Cell Sci. 2000;113:1075–1088. doi: 10.1242/jcs.113.6.1075. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths D J F, Barbet N C, McCready S, Lehmann A R, Carr A M. Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King R C, editor. Handbook of genetics 1. I. New York, N.Y: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- 19.Hartwell L H, Kastan M B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 20.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 21.Kostrub C F, Knudsen K, Subramani S, Enoch T. Hus1p, a conserved fission yeast checkpoint protein, interacts with Rad1p and is phosphorylated in response to DNA damage. EMBO J. 1998;17:2055–2066. doi: 10.1093/emboj/17.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsay H D, Griffiths D J F, Edwards R, Murray J M, Christensen P U, Walworth N, Carr A M. S-phase specific activation of Cds1 kinase defines a subpathway of the checkpoint response in S. pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu V F, Bhaumik D, Wang T S-F. Mutator phenotype induced by aberrant replication. Mol Cell Biol. 1999;19:1126–1135. doi: 10.1128/mcb.19.2.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- 25.Lydall D, Weinert T A. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 27.Michelson R, Weinert T. Sensor-less checkpoint activation? Nat Cell Biol. 1999;1:177–178. doi: 10.1038/15614. [DOI] [PubMed] [Google Scholar]
- 28.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 29.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 30.Naiki T, Shimomura T, Kondo T, Matsumoto K, Sugimoto K. Rfc5, in coorperation with Rad24, conrols DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5888–5896. doi: 10.1128/mcb.20.16.5888-5896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noskov V N, Araki H, Sugino A. The RFC2 gene, encoding the third-largest subunit of the replication factor C complex, is required for an S-phase checkpoint in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:4914–4923. doi: 10.1128/mcb.18.8.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Connell M J, Raleigh J M, Verkade H M, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Connell M J, Walworth N C, Carr A M. The G2-phase DNA damage checkpoint. Trends Cell Biol. 2000;10:296–303. doi: 10.1016/s0962-8924(00)01773-6. [DOI] [PubMed] [Google Scholar]
- 34.O'Donnell M, Onrust R, Dean F B, Chen M, Hurwitz J. Homology in accessory proteins of replicative polymerases—E. coli to humans. Nucleic Acids Res. 1993;21:1–3. doi: 10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker A E, Weyer I V D, Laus M C, Oostveen I, Yon J, Verhasselt P, Luyten W H M L. A human homolog of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J Biol Chem. 1998;273:18332–18339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- 36.Pasion S G, Forsburg S L. Nuclear localization of Schizosaccharomyces pombe Mcm2/Cdc19p requires MCM complex assembly. Mol Biol Cell. 1999;10:4043–4057. doi: 10.1091/mbc.10.12.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrini J H J. The Mre11 complex and ATM: collaborating to navigate S phase. Curr Opin Cell Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 38.Reynolds N, Fantes P A, MacNeill S A. A key role of replication factor C in DNA replication checkpoint function in fission yeast. Nucleic Acids Res. 1999;27:462–469. doi: 10.1093/nar/27.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhind N, Russell P. Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways. J Cell Sci. 2000;113:3889–3896. doi: 10.1242/jcs.113.22.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhind N, Russell P. The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics. 1998;149:1729–1737. doi: 10.1093/genetics/149.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saka Y, Fantes P, Yanagida M. Coupling of DNA replication and mitosis by fission yeast rad4/cut5. J Cell Sci. 1994;18(Suppl.):57–61. doi: 10.1242/jcs.1994.supplement_18.8. [DOI] [PubMed] [Google Scholar]
- 43.Shimada M, Okuzaki D, Tanaka S, Tougan T, Tamai K K, Shimoda C, Nojima H. Rfc3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, is required for both replication and damage checkpoints. Mol Biol Cell. 1999;10:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugimoto K, Ando S, Shimomura T, Matsumoto K. Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol Cell Biol. 1997;17:5905–5914. doi: 10.1128/mcb.17.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugimoto K, Shimomura T, Hashimoto K, Araki H, Sugino A, Matsumoto K. Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc Natl Acad Sci USA. 1996;93:7048–7052. doi: 10.1073/pnas.93.14.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan S, Wang T S-F. Analysis of fission yeast primase defines the checkpoint responses to aberrant S phase initiation. Mol Cell Biol. 2000;20:7853–7866. doi: 10.1128/mcb.20.21.7853-7866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thelen M P, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 49.Torre-Ruiz M A D L, Green C M, Lowndes N F. Rad9 and Rad24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 1998;17:2687–2698. doi: 10.1093/emboj/17.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran H T, Degtyareva N P, Koloteva N N, Sugino A, Msumoto H, Gordenin D A, Resnick M A. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and RAD50 and RAD52 genes. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Traut W. The function and consensus motifs of nine types of peptide segments that form different types of nucleotide binding sites. Eur J Biochem. 1994;222:9–19. doi: 10.1111/j.1432-1033.1994.tb18835.x. [DOI] [PubMed] [Google Scholar]
- 52.Turner J, Hingorani M M, Kelman Z, O'Donnell M. The internal working of a DNA polymerase clamp-loading machine. EMBO J. 1999;18:771–783. doi: 10.1093/emboj/18.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uchiyama M, Galli I, Griffiths D J F, Wang T S-F. A novel mutant allele of Schizosaccharomyces pombe rad26 defective in monitoring S phase progression to prevent premature mitosis. Mol Cell Biol. 1997;17:3103–3115. doi: 10.1128/mcb.17.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Venclovas C, Thelen M P. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volkmer E, Karnitz L M. Human homologs of Schizosaccharomyces pombe Rad1, Hus1, and Rad9 form a DNA damage-responsive protein complex. J Biol Chem. 1999;274:567–570. doi: 10.1074/jbc.274.2.567. [DOI] [PubMed] [Google Scholar]
- 56.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 57.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 58.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinase and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walworth N C, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 60.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]
- 61.Zhang G, Gibbs E, Kelman Z, O'Donnell M, Hurwitz J. Studies on the interactions between human replication factor C and human proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 1999;96:1869–1874. doi: 10.1073/pnas.96.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]