Summary
The elicitation of cross-variant neutralizing antibodies against SARS-CoV-2 represents a major goal for current COVID-19 vaccine strategies. Additionally, natural infection may also contribute to broaden neutralizing responses. To assess the contribution of vaccines and natural infection, we cross-sectionally analyzed plasma neutralization titers of six groups of individuals, organized according to the number of vaccines they received and their SARS-CoV-2 infection history. Two doses of vaccine had a limited capacity to generate cross-neutralizing antibodies against Omicron variants of concern (VOCs) in uninfected individuals, but efficiently synergized with previous natural immunization in convalescent individuals. In contrast, booster dose had a critical impact on broadening the cross-neutralizing response in uninfected individuals, to level similar to hybrid immunity, while still improving cross-neutralizing responses in convalescent individuals. Omicron breakthrough infection improved cross-neutralization of Omicron subvariants in non-previously infected vaccinated individuals. Therefore, ancestral Spike-based immunization, via infection or vaccination, contributes to broaden SARS-CoV-2 humoral immunity.
Subject areas: Biochemistry, Immune response, Microbiology
Graphical abstract
Highlights
-
•
Initial mRNA vaccine schedule achieved limited cross-neutralization of Omicron VOCs
-
•
Previous infection increased Omicron neutralization in vaccinated individuals
-
•
Booster vaccine had a critical impact on broadening cross-neutralizing responses
-
•
Omicron breakthrough infection further improved Omicron cross-neutralization
Biochemistry; Immune response; Microbiology
Introduction
One of the main features of the coronavirus disease 2019 (COVID-19) pandemic is the continuous evolution of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) fueled by the high rate of viral transmission (as illustrated by the > 759 million confirmed cases worldwide to date, World Health Organization (WHO) COVID-19 Dashboard; https://covid19.who.int/ accessed on March 9th 2023). Over the course of the pandemic, the WHO has considered five variants of SARS-CoV-2 (Alpha, Beta, Gamma, Delta, and Omicron) as variants of concern (VOCs), but currently four of them have been de-escalated. At the end of 2021, the world saw the rise of the Omicron (B.1.1.529) family of SARS-CoV-2 VOCs.1 First identified in Botswana and South Africa,2 Omicron BA.1 (B.1.1.529.1) and its derivative BA.1.1 achieved global prevalence, like the Delta (B.1.617.2) VOC before it. BA.1 shows low fusogenicity, shifted tropism, reduced pathogenicity3,4,5 and roughly 3-fold increased transmissibility compared to Delta.6 In addition, Omicron was shown to evade the action of previously acquired or therapeutic neutralizing antibodies7,8,9,10,11,12,13,14,15 by adding mutations in the coding region of the Spike glycoprotein, particularly in the receptor binding domain (RBD), and the N-terminal domain (NTD). In March 2022, a new—incrementally more transmissible16—Omicron VOC, BA.2 (B.1.1.529.2) took the lead and became highly prevalent worldwide.17 While considered to be part of the Omicron lineage and showing similar resistance to neutralization,12 BA.2 carries a different NTD mutation landscape and several additional mutations in the RBD.18 Finally, since June 2022 the Omicron BA.4 (B.1.1.529.4) and mainly the Omicron BA.5 (B.1.1.529.5) replaced BA.2 as the most prevalent VOCs and—at the time of this writing—are associated with a global increase in infection cases.17 BA.4 and BA.5 share an identical Spike glycoprotein that is close to BA.2 although it contains the 69–70 deletion described for BA.1, and the RBD mutation L452R previously detected in the Delta VOC.
Remarkably, COVID-19 vaccination campaigns started less than a year after the beginning of the pandemic. While the precise impact of vaccines on the population worldwide is still evaluated,19 their effectiveness in preventing infection and reducing disease severity at the individual level was clearly demonstrated.20,21 However, while the effect on fatality rate seemed longer lasting,22 protection against symptomatic disease started to wane progressively after 3 months23 due to the combination of both a progressive decrease in the titer of circulating neutralizing antibodies, and the emergence of neutralization escape variants. Not coincidently, the Omicron waves fueled an unprecedented surge of infection worldwide with a large number of mostly mild breakthrough infections.24 Interestingly, the administration of a booster vaccine based on the ancestral Spike, which was initiated concomitantly with rising Omicron cases, showed a remarkable effect in restoring protection against Omicron, although not to optimal levels and probably only for a limited time. This was visible in a large efficacy study25 but also through the measurement of neutralizing antibody titers,10,26,27 which are considered a strong surrogate marker of protection against symptomatic disease.28,29,30,31,32 Indeed, booster vaccine doses based on the ancestral variant have shown a highly positive impact on cross-neutralization of new variants.
Immunity against SARS-CoV-2 is also acquired from natural infection. As a considerable percentage of the global population has already been infected by SARS-CoV-2 at least once especially since the emergence of Omicron variants24 immunity acquired through infection with previously circulating variants becomes an important parameter to understand and predict the level of protection against currently circulating ones. Indeed, immune responses induced by natural infection present some advantages over mRNA vaccines; natural infection was shown to trigger a more sustained B cell evolution leading over time to more potent and more broadly neutralizing antibodies.24 Better yet, previous infection can combine with vaccination, in so-called hybrid immunity, to improve responses in vaccinated convalescent individuals.33,34,35,36,37 Hybrid immunity was also observed following breakthrough infection which improved vaccine induced cross-neutralizing responses.38 Finally, Omicron breakthrough infection was shown to induce poor cross-neutralizing responses in non-vaccinated individuals but this was improved with previous vaccination.39
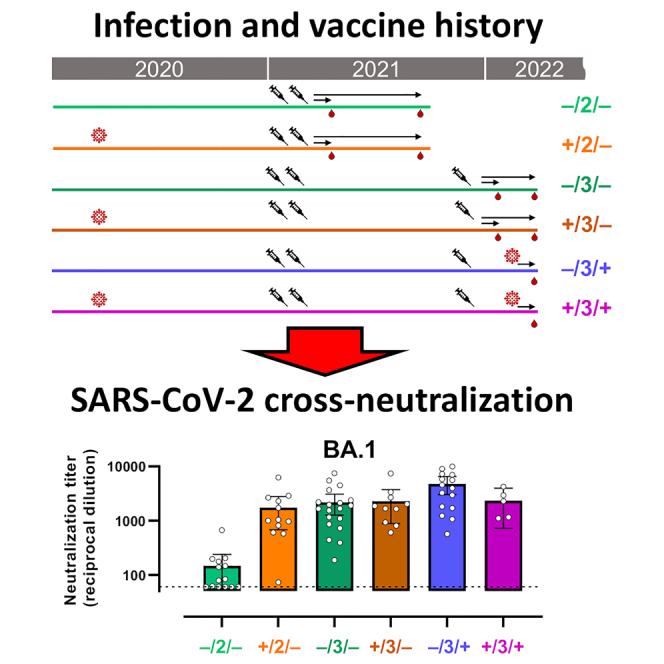
To measure the impact of booster vaccines and hybrid immunity on the cross-neutralization of VOCs, we followed a cohort of vaccinated individuals (mRNA vaccine BNT162b2 from Pfizer-BioNTech or mRNA-1273 Moderna) who received or not an mRNA booster dose and who experienced or not mild infection before vaccination during the first year of the pandemic. In addition, we analyzed individuals who all received a booster dose and subsequently experienced breakthrough infection during the Omicron VOCs onset. We evaluated their neutralizing antibody responses against the ancestral SARS-CoV-2, Delta, and Omicron BA.1, BA.2, and BA.4/5 variants. Our data suggest that the number of vaccine doses and/or infection exposures similarly contribute to cross-neutralization activity.
Results
A total of 76 participants were included in this study and organized in 6 groups (Table 1) according to 3 parameters: status of SARS-CoV-2 infection before all mRNA-based vaccine, status of vaccination (full schedule only or additional booster) and infection following the booster vaccine. All participants were vaccinated with mRNA vaccines (BNT162b2 [Pfizer-BioNTech] or mRNA-1273 [Moderna]) and at the time of sampling, had received either a full vaccine schedule (2 doses with 3–4 weeks interval between first and second dose) or also an additional booster dose (six months interval from full schedule). Notably, 5 participants, under 55 years of age, who experience SARS-CoV-2 infection before vaccination only received 1 mRNA vaccine dose that was considered a full vaccine schedule by the Spanish health authority. However, for simplicity, throughout the manuscript, full vaccine schedule is referred to “2 doses vaccination” while vaccination that included booster are referred to “3 doses vaccination”. Booster vaccine was in large majority mRNA-1273 (Moderna) including for most participants who had received BNT162b2 (Pfizer-BioNTech). The rest of the booster was BNT162b2.
Table 1.
Characteristics of individuals included in the analysis
| Infection/Vaccine status |
||||||
|---|---|---|---|---|---|---|
| −/2/− |
+/2/− |
−/3/− |
+/3/− |
−/3/+ |
+/3/+ |
|
| n = 14 | n = 12 | n = 19 | n = 10 | n = 16 | n = 5 | |
| Age (years), median [IQR] | 45 [28–52] | 43 [35–51] | 42 [26–48] | 37 [24–49] | 49 [35–51] | 43 [38–49] |
| Sex (% female) | 78.6 | 66.7 | 73.7 | 70.0 | 75.0 | 100.0 |
| Time between the first plasma sample collection and the indicated event (median days [IQR]): | ||||||
| Last vaccine dose | 34 [32–38] | 34 [14–46] | 34 [31–37] | 33 [31–41] | 90 [60–92] | 35 [32–70] |
| Pre-vaccine infection | NA | 336 [153–360] | NA | 588 [459–662] | NA | 621 [424–677] |
| Post-vaccine infection | NA | NA | NA | NA | 35 [18–43] | 25 [13–38] |
| Time between the second plasma sample collection and the indicated event (median days [IQR]): | ||||||
| Last vaccine dose | 183 [177–197] | 200 [176–223] | 92 [91–99] | 92 [91–104] | NA | NA |
| Pre-vaccine infection | NA | 488 [333–529] | NA | 697 [502–725] | NA | NA |
Groups are named according to their SARS-CoV-2 infection status prior to infection (+ or –), the number of vaccines received (2 or 3) and the occurrence of breakthrough infection (+ or –).
Infections before vaccination occurred a median of 277 (IQR = 87–296) days before vaccination (Table 1), during the first year of the pandemic, between March 2020 and February 2021, which, in Catalonia Spain, involved mainly ancestral Wuhan-Hu-1 (WH1), B.1, B.1.177, and Alpha (B.1.1.7) variants (Figure S1A, http://covidtag.paseq.org/). Infections after the third vaccine dose happened, from December 2021 to March 2022, when, in Catalonia Spain, Omicron (BA.1 and BA.2) displaced Delta and became largely prevalent (Figure S1B). Breakthrough Infections occurred a median of 39 (IQR = 24–54) days after last vaccine dose (Table 1). All samples from boosted individuals were collected around 4 weeks after the latest event (either vaccination or infection, Table 1). The uninfected status of participants was verified by testing their plasma sample for the absence of anti-nucleocapsid protein antibodies by ELISA (Figure S1C). No significant differences among groups were observed in age or sex (Table 1).
Hybrid immunity is associated with increased and more stable cross-variant neutralization responses
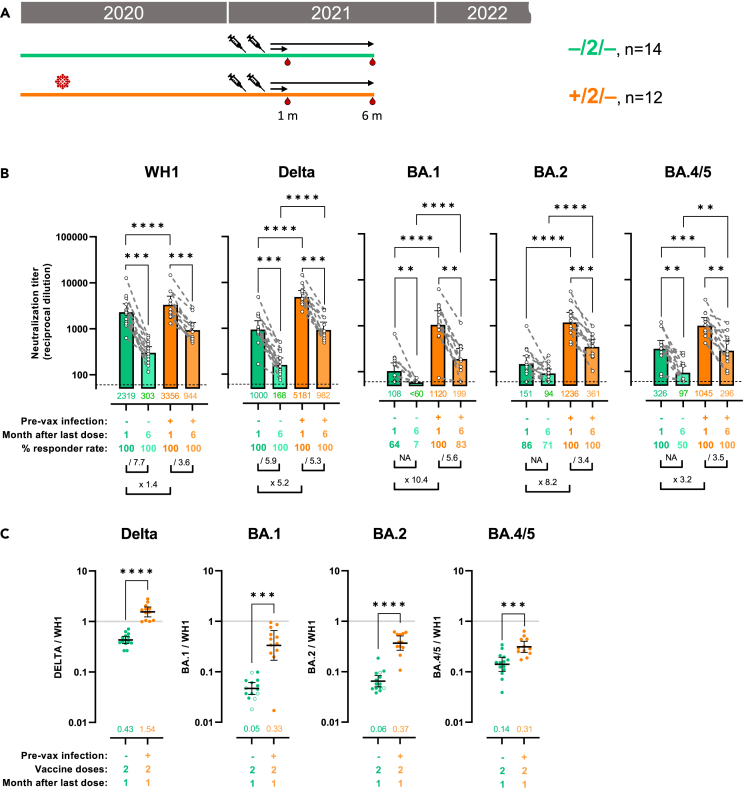
First, we tested the effect of previous infection in combination with 2 doses of mRNA vaccines on the neutralization of WH1, Delta, Omicron BA.1, BA.2, and BA.4/5 variants. In plasma samples collected from uninfected participants 1 month after second dose of mRNA vaccine, neutralization titers (50% inhibitory dilution, ID50) were the highest against WH1 (Figures 1B and S2A: geometric mean titer [GMT] = 2319), decreased, although not significantly, against Delta (GMT = 1000, p = 0.943) and were highly reduced against Omicron subvariants BA.1 (GMT = 108, p < 0.0001), BA.2 (GMT = 151, p < 0.0001), and BA.4/5 (GMT = 326, p = 0.0053).
Figure 1.
Hybrid immunity is associated with increased and more stable cross-variant neutralization responses
(A) Schematic representation of plasma samples collection from individuals previously infected or not who received 2 doses of mRNA vaccine were tested for neutralization activity against pseudo viruses expressing the WH1, B.1.217.2 (Delta), BA.1, BA.2, and BA.4/5 SARS-CoV-2 Spike proteins. Plasma samples were collected at about 1 or 6 months after last vaccination.
(B) Raw ID50 (reciprocal dilutions) titers against the indicated variant Spikes. Horizontal bars and numbers indicate ID50 geometric means (GMT) for each group and error bars indicate 95% confidence intervals. At the bottom are fold changes for the indicated comparisons. For the comparison between 1- and 6-month plasma samples (intragroup), we applied a Wilcoxon paired test or a Prentice Z test when undetectable titers were observed. For intergroup comparison we applied a Mann-Whitney test or a Peto-Peto test when undetectable titers were observed. Significant p values are indicated and were corrected for multiple comparison (∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Dashed lines indicate the assay lower limit of detection (60 reciprocal dilutions). Non-responder rate was defined by undetectable neutralization titers for a specific variant at an initial serum dilution of 1:60 and are indicated.
(C) Ratio of mVOC ID50 over the indicated WH1 ID50 calculated for the groups with plasma samples at 1 month (shown in 1B) after the second vaccine dose. Horizontal bars indicate ratio geometric means for each group and error bars indicate 95% confidence intervals. Bottom numbers indicate the geometric mean of the ratio for each group. Significant p values are indicated for comparisons between uninfected and previously infected groups (Mann-Whitney test, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Non-responder samples are indicated by an open circle. See also Figure S2.
By comparison, in vaccinated participants who were previously infected (and therefore experienced 3 immunogenic impacts), neutralization titers, 1 month after second vaccine dose, were similar against WH1 (Figure 1B: x 1.4, GMT = 3356, p = 0.1759) but significantly increased against Delta (x 5.2, GMT = 5181, p < 0.0001), BA.1 (x 10.4, GMT = 1120, p < 0.0001), BA.2 (x 8.2, GMT = 1236, p < 0.0001), and BA.4/5 (x 3.2, GMT = 1045, p = 0.0003). While Delta neutralization of previously infected individuals was above but not significantly different from the ancestral Spike (Figure S2B: Delta = 5181 vs WH1 = 3356, p > 0.9999), the improved neutralization titers against Omicron subvariants were still about 3-fold reduced compared to WH1 (BA.1 = 1120, p = 0.0368; BA.2 = 1236, p = 0.0241; BA.4/5 = 1045, p = 0.0008).
VOCs cross-neutralization was also assessed by calculating ratios between each VOC and the corresponding WH1 titers from the same individuals (Figure 1C). This approach gives a measurement of the cross-neutralization potential using WH1 response as a reference and provides an alternative analysis with reduced inter-individual variability, which can bias group comparisons when analyzing only raw titers. An increased ratio value indicates increased cross-neutralization. A ratio superior to 1 indicates a better neutralization in comparison to WH1, while a ratio inferior to 1 indicates a worsen neutralization compared to WH1. As suggested by titer comparisons, the cross-neutralization potential against Delta that was inferior in vaccinated-only participant (ratio = 0.43) was increased to an apparent optimal level in previously infected individuals (ratio = 1.54, p < 0.0001). In the case of Omicron BA.1, BA.2, and BA.4/5, cross-neutralization potential was highly reduced in vaccinated-only participants (fold reduction for BA.1 = 0.05, BA.2 = 0.06, BA.4/5 = 0.14, Figure S2C) and sensibly improved in the previously infected group (fold reduction for BA.1 = 0.33, p = 0.0004; BA.2 = 0.37, p = 0.0004; BA.4/5 = 0.31, p = 0.001) albeit still lagging behind the response against Delta (Figure S2D). Importantly, in the non-previously infected group, some individuals showed undetectable neutralization titers against BA.1 (Figure 1B; 64% responders) and BA.2 (86% responders). Therefore, we also analyzed cross-neutralization ratios after removal of undetectable samples, confirming the initial results with slightly weaken statistics (Figures S2E and S2F).
To evaluate the stability of these responses at mid-term, the same participants were tested 5 months later (6 months after second dose). Non-infected participants showed a major decrease in their neutralizing titers against all variants tested (Figure 1B): while titers against WH1 and Delta were reduced 7.7 times (p < 0.0001) and 6 times (p = 0.0016) respectively, many participants reached non-detectable levels (ID50 < 60) against Omicron BA.1, BA.2 and BA.4/5 in our assay. In contrast, previously infected participants showed a more robust stability of their neutralizing response against all variants (WH1 =/3.5; Delta =/5.3; BA.1 =/5.6; BA.2 =/3.4; BA.4/5 =/3.5). Temporal changes in ratios between VOC and WH1 neutralization could not be calculated after six months due to high number of undetectable measurements. We considered that a level superior to 25% undetectable samples could bias the results.
Overall, these results indicate that hybrid immunity greatly improves quantitatively and qualitatively the neutralization capacities against Delta and Omicron. However, neutralization against Omicron variants was still severely reduced after previous infection and 2 vaccine doses.
Booster shot of mRNA vaccine in non-infected individuals generates hybrid immunity-like benefits 1 month after last immunization
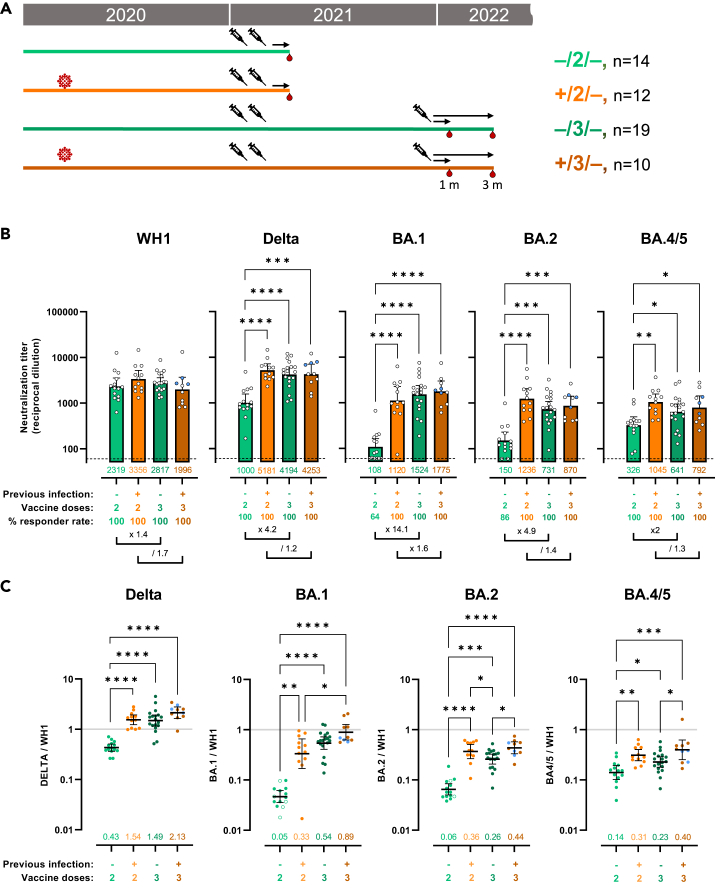
Next, we evaluated the effect of a third vaccine dose, which was administered with a median of 9.2 (IQR = 8–10.3) months after the second dose, comparing participants previously infected or not (Figure 2A). In this analysis, we focused on plasma samples collected with a median of 34 (IQR = 16–40) days after last vaccine dose and, for comparison, we included data on participants who only received 2 vaccine doses, already shown in Figure 1.
Figure 2.
Booster shot of mRNA vaccine in non-infected individuals generates hybrid immunity-like benefits 1 month after last immunization
(A) Schematic representation of plasma samples collection from individuals previously infected or not, who received 2 or 3 doses of mRNA vaccine were tested for neutralization activity against pseudo viruses expressing the WH1, B.1.217.2 (Delta), BA.1, BA.2, and BA.4/5 SARS-CoV-2 Spike proteins. Plasma samples were collected at about 1 month after last vaccination.
(B) Raw ID50 (reciprocal dilutions) titers against the indicated variant Spikes. Horizontal bars and numbers indicate ID50 geometric means (GMT) for each group and error bars indicate 95% confidence intervals. At the bottom are fold changes for the indicated comparisons. Significant p values are indicated for comparisons between all groups (Kruskal-Wallis test with two-stage step-up multiple comparison, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Dashed lines indicate the assay lower limit of detection (60 reciprocal dilutions). Non-responder rate was defined by undetectable neutralization titers for a specific variant at an initial serum dilution of 1:60 and are indicated. Participants (3 from the +/3/− group) who only received 1 dose as part of the full vaccine schedule are indicated in blue.
(C) Ratio of WH1 ID50 over the indicated VOC ID50 calculated for the groups with plasma samples at 1 month (shown in 2A) after last vaccine dose. Horizontal bars indicate ratio geometric means for each group and error bars indicate 95% confidence intervals. Bottom numbers indicate fold reduction compared to WH1. Significant p values are indicated for comparisons between all groups (Kruskal-Wallis test with two-stage step-up multiple comparison, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Non-responder samples are indicated by an open circle. Participants (3 from the +/3/− group) who only received 1 dose as part of the full vaccine schedule are indicated in blue. See also Figures S3 and S4.
When comparing neutralization titers at similar time (1 month) after last vaccination, the effect of the third dose on WH1 neutralization was not statistically different from the 2 doses groups respectively, whether the participants had been previously infected or not (Figure 2B). The third vaccine dose (booster) did, in fact, increase WH1 neutralization titers when compared with decayed neutralization response at 6 months post second dose (2817 vs 303 in Figures 1B and 2B), but it only brought back WH1 neutralization to an apparent maximal activity already reached after 2 doses. In sharp contrast, third dose vaccine increased protection against Delta (x 4.2, p < 0.0001), BA.1 (x 14.1, p < 0.0001), BA.2 (x 4.9, p = 0.0002) and BA.4/5 (x 2, p = 0.0187) to levels not previously reached in vaccinated-only individuals (Figures 2B and S3A).
Of note, we observed a slightly decreased neutralization against WH1 in previously infected individuals compared to vaccinate only after booster; however, the difference was not significative and may result from the limited size of this group. This reduction was not observed against other variants.
When considering raw neutralization titers, previously infected participants who received a third vaccine dose did not show any significant increase of neutralization activity against any of the variant tested when compared to previously infected individuals who received only 2 doses of vaccine (Figures 2B and S3B). However, when considering cross-neutralization ratios (Figures 2C, S3C, and S3D), a trend for a better response against all VOCs was visible for the previously infected group who received the third vaccine dose compared to previously infected group who received 2 doses, but did not reach significance except for BA.1 (+/2/− = 0.33 and +/3/− = 0.89, p = 0.04711). Finally, the benefit of previous infection in the context of boosted vaccination (3 doses) was not visible when considering raw neutralization titers (Figure 2B), although there was still a trend for a better response quality against Omicron variants when considering cross-neutralization ratios (Figure 2C), which only reached significance against BA.2 (p = 0.0314) and BA.4/5 (p = 0.0352).
Of note, the group of individuals, who were previously infected and received booster vaccine, contained 3 individuals who had only received a single vaccine dose as part of the full vaccine schedule. To verify the impact of these individuals on our results, we re-analyzed both neutralization titers and ratios excluding them (Figures S3C, S3F, S4A, and S4B). No clear difference was observed although some statistical significance was lost as a result of group reduction and lower statistical power. We also verified the impact of BA.1 and BA.2 non-responders on the analysis of the cross-neutralization responses (Figure S3G). Results were almost identical.
Finally, we performed a follow up on these individuals at around 3 months after booster vaccine (Figure S4C). Neutralization titers showed an encouraging limited decay for all variants. Additionally, we could observe a trend for an improved stability in previously infected individuals; however, it did not reach significance.
All together these results indicate that hybrid immunity conferred by previous infection provides clear benefits increasing protection against Delta and Omicron variants, but third vaccine dose could provide non-infected participants with a level of protection similar to previously infected individuals.
Omicron breakthrough infection specifically increases neutralization activity against Omicron.
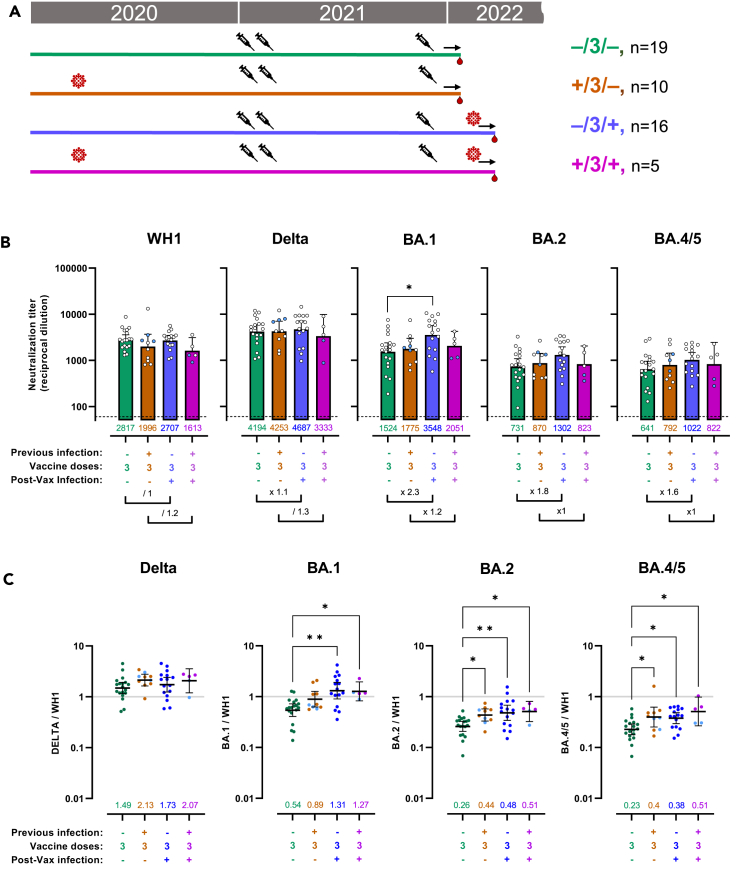
Finally, we tested the effect of breakthrough infection in boosted individuals previously infected or not (Figure 3A). All breakthrough infections happened between December 2021 and March 2022 and probably include mostly Omicron BA.1 and BA.2 infections (Figure S1B). Plasma samples were collected with a median of 35 days (IQR = 18–43) after last infection in the case of participants only infected after the third dose (Table 1) and 25 days (IQR = 13–38) in the case of individuals infected before and after full vaccination. We included for comparison the groups of participants, who received 3 vaccine doses but did not undergo breakthrough infection, already shown Figure 2.
Figure 3.
Omicron breakthrough infection specifically increases neutralization activity against Omicron
(A) Schematic representation of plasma samples collected from individuals previously infected or not, who received 3 doses of mRNA vaccine and were affected or not by Omicron breakthrough infection were tested for neutralization activity against pseudo viruses expressing the WH1, B.1.217.2 (Delta), BA.1, BA.2, and BA.4/5 SARS-CoV-2 Spike proteins. Plasma samples were collected at about 1 month after last vaccination or 1 month after breakthrough infection if the latter happened.
(B) Raw ID50 (reciprocal dilutions) titers against the indicated variant Spikes. Horizontal bars and numbers indicate ID50 geometric means (GMT) for each group and error bars indicate 95% confidence intervals. At the bottom are fold changes for the indicated comparisons. Significant p values are indicated for comparisons between all groups (Kruskal-Wallis test with two-stage step-up multiple comparison, ∗p < 0.05). Dashed lines indicate the assay lower limit of detection (60 reciprocal dilutions). Responder rates were all 100% (ID50 > 60). Participants (3 of the +/3/− group and 2 of the +/3/+ group) who only received 1 dose as part of the full vaccine schedule are indicated in blue.
(C) Ratio of WH1 ID50 over the indicated VOC ID50 calculated for the groups with plasma samples at 1 month (shown in 3A) after last event (infection or vaccination). Horizontal bars indicate ratio geometric means for each group and error bars indicate 95% confidence intervals. Bottom numbers indicate fold reduction compared to WH1. Significant p values are indicated for comparisons between all groups (Kruskal-Wallis test with two-stage step-up multiple comparison, ∗p < 0.05, ∗∗p < 0.01). Participants (3 of the +/3/− group and 2 of the +/3/+ group) who only received 1 dose as part of the full vaccine schedule are indicated in blue. See also Figures S5 and S6.
Omicron breakthrough infection had no effect on neither WH1 nor Delta neutralization titers, whether participants were or not previously infected (Figures 3B, S5A, and S5B). Delta cross-neutralization ratio did not change, either, compared to other 3 vaccine dose groups (Figures 3C, S5D, and S5E). The absence of increased neutralization against WH1 and Delta suggests that recent breakthrough infection had limited cross-variant immunity toward these variants because of a more pronounced antigenic disparity between pre-Omicron and Omicron variants. Alternatively, as the participants had received a booster vaccine soon before breakthrough infection (median of 45 days in between), it is possible that neutralization responses against WH1 and Delta were still at a peak after vaccine. BA.1, BA.2, and BA4/5 raw neutralization titers all showed trends of improvement following breakthrough infection in non-previously infected participants (Figure 3B) but statistical significance was only met for BA.1 (x 2.3, p = 0.0095). When looking at cross-neutralization ratios (Figure 3C), BA.1, BA.2, and BA.4/5 all showed significant improvement when comparing the breakthrough infection groups (−/3/+ and +/3/+) to the 3 doses uninfected group (−/3/−) but not the 3 doses previously infected group (+/3/−).
Of note, the group of individuals who experienced 2 infections (+/3/+) included 2 individuals who received a single dose as part of the full vaccine schedule. To verify their impact on the overall results, we re-analyzed both titers and ratios excluding them (Figures S5C, S5F, S6A, and S6B). Titers and ratios were both increased as a result but statistical analyses were hampered by the group reduction.
Taken together these results show that BA.1/BA.2 breakthrough infection contributed to improve cross-neutralization against all Omicron variants but not Delta. This effect was more evident when analyzing participants who did not experience previous infection with pre-Omicron variants.
Discussion
We face a seemingly relentless race between COVID-19 vaccinations and viral evolution, giving rise to more transmissible and immunity-escaping variants, leading to successive COVID-19 waves. As more and more vaccinated people experience SARS-CoV-2 breakthrough infection, especially since the onset of the Delta and Omicron variants,24,40 individual and global levels of protection get redefined. In line with previous reports,33,37 we show that hybrid immunity, based on previous infection with non-Omicron variants, increased protection against SARS-CoV-2 VOCs following 2 doses vaccine. Importantly, these benefits were observed against all Omicron variants although, the levels of protection obtained were still reduced about 3-folds compared to the ancestral variant. Interestingly, after booster vaccine, hybrid immunity still showed a trend for improving cross-neutralization: This was visible against all the VOCs tested including significative difference against BA.2 and BA.4/5. The waning neutralizing responses following mRNA vaccine is a major concern.41,42,43 We observed an improved stability of neutralization titers at mid-term associated with previous infection status following 2 doses vaccination. Additionally, following booster vaccine, titers showed an encouraging limited decay.
We evaluated the effect of Omicron BA.1 and BA.2 breakthrough infection on neutralization titers. While a modest but significant increase of BA.1 neutralizing titer was detected, most of the qualitative benefits observed after Omicron breakthrough infection on cross-neutralization ratios in uninfected individuals were comparable to the values observed in individuals previously infected with non-Omicron variants. In addition to the strong antigenic differences between Omicron and ancestral variants (particularly in the RBD), this apparent low impact of Omicron breakthrough infections could be explained by intrinsic viral factors of this VOC (tropism, fusogenicity)3,4 or by the time interval and order of antigenic exposures. Certainly, pre-vaccination infections were separated by a median of 277 days from vaccination, while only a median of 39 days separated booster vaccination and breakthrough infection. Timing between infection and vaccination seems to be a major determinant on the extent of boosting of humoral responses with shorter time intervals resulting in lower suboptimal boost.44
Finally, in line with other studies,10,26,33,45,46 we saw a remarkable effect of booster vaccines in improving neutralization titers against Omicron VOCs to similar levels as the ones obtained with hybrid immunity. These and others’ results suggest that the number of antigenic exposures is a critical parameter for the improvement of cross-neutralizing responses. Indeed, it has been shown that booster mRNA vaccines can mobilize additional memory B cells, including Omicron specific B cells.47,48 In addition, recent studies now propose that the generation of more broadly neutralizing antibodies, following immunization with ancestral Spike, requires multiple and sequential antigenic exposures to achieve the activation, differentiation, and expansion of subdominant memory B cells.49,50
Our data indicate that the benefits of Omicron over non-Omicron-based hybrid immunity on the neutralization Omicron VOCs was marginal. The scientific community and the pharmaceutical industry have developed second-generation COVID-19 vaccines that can boost acquired immunity more specifically against new variants and reduce their transmission.51,52,53,54,55 A recent study not yet peer-reviewed predicts a limited added benefits of new Omicron specific COVID-19 mRNA vaccines.56 These data suggest that for the time being ancestral Spike-based immunization remains a valuable strategy to improve and maintain protection against SARS-CoV-2 and limit transmission globally since the number of immunological exposures to the Spike, rather than the specific sequence, seems to be a major determinant of VOC cross-neutralization of humoral immune response.
Limitations of the study
The present work is limited mainly by the number of individuals included in each group, and the lack of parallel data on CD4+ and CD8+ T cells. In addition, there are many variables that have been detailed but not analyzed due to the complexity and limited power of the analyses, including the time elapsed between previous infections and vaccination and between the third dose and breakthrough infection, that could influence on the neutralizing response in the short, mid, and/or long term.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Peroxidase AffiniPure F(ab')₂Fragment Goat Anti-Human IgG, Fcγ Fragment Specific | Jackson ImmunoResearch | Cat# 109-036-098; RRID: AB_2337596 |
| Anti-6x-His clone HIS.H8 | Thermo Fisher Scientific | Cat#MA1-21315; RRID: AB_557403 |
| Bacterial and Virus Strains | ||
| pNL4-3.Luc.R-.E- | NIH ARP | Cat#3418 |
| SARS-CoV-2.SctΔ19 | This paper | N/A |
| pcDNA3.1(+) | GeneArt/Thermo Fisher Scientific | Cat#810330DE |
| pVSV-G | Clontech | Sánchez-Palomino et al.57 |
| Biological Samples | ||
| Pre-pandemic Plasma Samples | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fetal Bovine Serum | Thermo Fisher Scientific | Cat#10270106 |
| Dulbecco’s Modified Eagle Medium | Thermo Fisher Scientific | Cat#41966052 |
| Expi293 Expression Medium | Thermo Fisher Scientific | Cat#A1435102 |
| Opti-MEM I Reduced Serum Medium | Thermo Fisher Scientific | Cat#31985070 |
| ExpiFectamine 293 Transfection Kit | Thermo Fisher Scientific | Cat#A14524 |
| Versene | Thermo Fisher Scientific | Cat#15040033 |
| Puromycin | Thermo Fisher Scientific | Cat#A1113803 |
| DEAE-Dextran | Sigma-Aldrich | Cat#D9885-100G |
| BriteLite Plus Luciferase | PerkinElmer | Cat#6066769 |
| SARS-CoV-2 (2019-nCoV) Nucleocapsid-His Recombinant Protein | Sino Biological Inc | Cat# 40588-V08B |
| MACS BSA Solution | Miltenyi Biotec | Cat# 130-091-376 |
| Phosphate Buffered Saline | Thermo Fisher Scientific | Cat# 10010015 |
| Phosphate-Citrate Buffer with Sodium Perborate | Merck Life Science SLU | Cat# P4922-50CAP |
| o-Phenylenediamine Dihydrochloride | Merck Life Science SLU | Cat# P8787-100TAB |
| H2SO4 | Sigma-Aldrich | Cat# 258105-1L-PC |
| Deposited Data | ||
| Group Geo Means and Raw Neutralization Titers | https://data.mendeley.com/ | https://doi.org/10.17632/3572bvtgsb.1 |
| Experimental Models: Cell Lines | ||
| Expi293F GnTI- cells | Thermo Fisher Scientific | Cat#A39240 |
| HEK293T/hACE2 cells | Integral Molecular | Cat#C-HA101 |
| Software and Algorithms | ||
| GraphPad Prism v9.3.1 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| R v3.6.3 | R Foundation for Statistical Computing | https://www.r-project.org/ |
| Other | ||
| GeneArt Gene Synthesis | Thermo Fisher Scientific | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Benjamin Trinité (btrinite@irsicaixa.es).
Material availability
The plasmids pcDNA3.1 SARS-CoV-2.SctΔ19 of each variant described in this work are available upon request to the lead contact.
Experimental model and subject details
Study overview and subjects
This work includes plasma samples from participants of 2 different cohorts: the KING cohort extension and the KING VAX study. Both studies were approved by the Hospital Ethics Committee Board from Hospital Universitari Germans Trias i Pujol (HUGTiP, PI-20-217 and PI-21-351). This is an observational cohort; no blinding or randomization was applied. All participants provided written informed consent. Both uninfected and infected individuals were included in both studies. Infected ones had a documented positive RT-qPCR result from nasopharyngeal swab, positive antigen test and/or a positive serological diagnostic test. Plasma samples from Uninfected individuals were negative for SARS-CoV-2 specific Nucleocapside antibodies (Figure S1C). At the time of sampling, all participants had received either complete vaccine schedule (2 doses or 1 dose in case of previously infected individuals < 55 years) or booster dose of mRNA vaccines BNT162b2 (Pfizer-BioNTech) and or mRNA-1273 (Moderna). Samples in this study were collected between February 2021 and March 2022, about one month after complete vaccine schedule or booster vaccines or about one month after breakthrough infection. Participants were recruited irrespective of age and all the infected groups included only mild disease cases. Information on sex and age is available in Table 1.
Cell lines
HEK293T cells (presumably of female origin) overexpressing wild type (WT) human ACE-2 (Integral Molecular, USA) were used as target in pseudovirus-based neutralization assay. Cells were maintained in T75 flasks with Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum and 1 μg/ml of Puromycin (Thermo Fisher Scientific, USA).
Expi293F cells (presumably of female origin, Thermo Fisher Scientific) are a HEK293 cell adapted for suspension culture that were used for SARS-CoV-2 pseudovirus production. Cells were maintained in Expi293 expression medium (Thermo Fisher Scientific) under continuous shaking at 125 rpm in Erlenmeyer flasks following manufacturer’s guidelines.
Method details
Pseudovirus generation and neutralization assay
HIV reporter pseudo viruses expressing SARS-CoV-2 S protein and Luciferase were generated. pNL4-3.Luc.R-.E- was obtained from the National Institut of Health (NIH) acquired immune deficiency syndrome (AIDS) Reagent Program.58 SARS-CoV-2.SctΔ19 was generated (GeneArt) from the full protein sequence of the original WH1 SARS-CoV-2 Spike (Genbank: MN908947.3) with a deletion of the last 19 amino acids in C-terminal58, human-codon optimized and inserted into pcDNA3.1(+).58 A similar procedure was followed to generate expression plasmids for the Delta, BA.1, BA.2 and BA.4/5 variants of SARS-CoV-2 S protein according to consensus data (www.outbreak.info/) (see Figure S1A for a detail of the mutations). Expi293F cells were transfected with pNL4-3.Luc.R-.E- and the different SARS-CoV-2.SctΔ19 Spike plasmids using ExpiFectamine 293 Reagent (Thermo Fisher Scientific). Control pseudo viruses were obtained by replacing the S protein expression plasmid with a VSV-G protein expression plasmid as reported.57 Supernatants were harvested 48 hours after transfection, filtered at 0.45 μm, frozen, and titrated on HEK293T cells overexpressing WT human ACE-2 (Integral Molecular, USA).
Neutralization assays were performed in duplicate as previously described.59 Briefly, in Nunc 96-well cell culture plates (Thermo Fisher Scientific), 200 TCID50 of pseudovirus were preincubated with three-fold serial dilutions (1/60–1/14,580) of heat-inactivated (incubated at 56°C for 30 minutes) plasma samples for 1 hour at 37°C. Then, 1x104 HEK293T/hACE2 cells treated with DEAE-Dextran (Sigma-Aldrich) were added. Results were read after 48 hours using the EnSight Multimode Plate Reader and BriteLite Plus Luciferase reagent (PerkinElmer, USA). The values were normalized, and the ID50 (reciprocal dilution inhibiting 50% of the infection) was calculated by plotting and fitting all duplicate neutralization values and the log of plasma samples dilution to a 4-parameters equation in Prism 9.0.2 (GraphPad Software, USA). This assay has been previously validated with a replicative viral inhibition assay.60
ELISA
Confirmation of SARS-CoV-2 serostatus was performed by evaluating anti-N protein IgG responses.61 Nunc MaxiSorp plates were coated with 50 μL of anti-6x-His antibody clone HIS.H8 (2 μg/mL, Thermo Fisher Scientific) in PBS overnight at 4°C. After washing, plates were blocked with 1% BSA in PBS (Miltenyi Biotec, Germany) for two hours at room temperature. Whole nucleocapsid protein (NP, Sino Biological, Germany) were added at 1 μg/mL concentration (50 μL/well) and incubated overnight at 4°C. Plasma samples were heat-inactivated before use (56°C for 30 minutes) and analyzed in duplicate in antigen-coated and antigen-free wells in the same plate. A positive plasma sample and a pool of pre-pandemic plasmas from healthy controls was used as positive and negative controls, respectively. Standards, negative control, and plasma samples were diluted (1/100) in blocking buffer and were incubated (50 μL/well) for one hour at room temperature. The HRP-conjugated (Fab)2 goat anti-human IgG (Fc specific, Jackson ImmunoResearch, UK) was then incubated for 30 minutes at room temperature. Plates were revealed with o-Phenylenediamine dihydrochloride (Sigma-Aldrich, USA) and reaction was stopped using 4N of H2SO4 (Sigma-Aldrich). Optical density (OD) at 492 nm with noise correction at 620 nm were used to calculate specific signal for each antigen after subtracting the antigen-free well signal for each sample.
Quantification and statistical analysis
For cohort analyses (Table 1), continuous variables were described using medians and the interquartile range (IQR, defined by the 25th and 75th percentiles), whereas categorical factors were reported as percentages over available data. Values of n are indicated in Table 1.
Neutralization titers and ratios were shown as geometric means. The Friedman test with Dunn’s multiple comparison was used to compare neutralization of different pseudo viruses (paired comparison). A Wilcoxon paired test or a Prentice Z test (when undetectable titers were observed) were used for longitudinal comparisons. The Mann-Whitney or Kruskal-Wallis test with Dunn’s multiple comparison were used to compare neutralization titers and ratio when comparing 2 groups or multiple groups, respectively. A Peto-Peto test was used when undetectable titers were observed. Comparison involving multiple tests were corrected by false discovery rate. In all analyses, statistical significance was indicated on graphs as follows: ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, and ∗∗∗∗ p < 0.0001. Analyses were performed using R-3.6.3 (R Foundation for Statistical Computing) and Prism 9.4.1 (GraphPad Software).
Acknowledgments
This work was partially funded by Grifols, the Departament de Salut, Generalitat de Catalunya (grants SLD016 to J.B. and SLD015 to J.C.), the Spanish Health Institute Carlos III and the European Regional Development Fund (grant PI17/01518 and PI20/00093 to J.B. and PI18/01332 to J.C.), CERCA Program/Generalitat de Catalunya 2017 SGR 252, and the crowd funding initiatives #joemcorono, BonPreu/Esclat, and Correos. The funders had no role in study design, data collection, and analysis, the decision to publish, or the preparation of the manuscript. E.P. was supported by a doctoral grant from the National Agency for Research and Development of Chile (ANID: 72180406). M.M. was granted with RYC2020-028934-I/AEI/10.13039/501100011033 from Spanish Ministry of Science and Innovation and State Research Agency and the European Social Fund “investing in your future”. A.P-G. was supported by a predoctoral grant from Generalitat de Catalunya and Fons Social Europeu (2022 FI_B 00698). We are grateful to all participants and the Clinical Research Organization from FLS for support in ethical aspects and database construction, the clinical trial unit from Fundació Lluita contra les Infeccions for study coordination and sample collection, and technical staff of IrsiCaixa for sample processing.
Author contributions
J.B., B.T., and B.C. designed and coordinated the study. E.P., S.M., M.T., T.P., A.P.G., R.O., C.R, F.T.-F., C.A-G. performed and analyzed experimental assays. V.U. and E.P. performed statistical analyses. R.T., A.C., M.N.-J., L.M., I.B., E.G., M.M., and J.C. selected patients and coordinated data. J.B., B.T., and E.P. drafted the manuscript, and all authors have made substantial contributions to the revision of the subsequent versions. All authors approved the submitted version of the manuscript and agreed both to be personally accountable for their own contributions, and to ensure the accuracy or integrity of any part of the work.
Declaration of interests
Unrelated to the submitted work, J.B. and J.C. are founders and shareholders of AlbaJuna Therapeutics, SL. B.C. is founder and shareholder of AlbaJuna Therapeutics, SL, and AELIX Therapeutics, SL. The other authors declare no competing interests.
Published: March 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106457.
Contributor Information
Benjamin Trinité, Email: btrinite@irsicaixa.es.
Julià Blanco, Email: jblanco@irsicaixa.es.
Supplemental information
Data and code availability
-
•
De-identified human neutralization data have been deposited at Mendeley Data and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.World Health Organization Tracking SARS-CoV-2 variants. https://www.who.int/activities/tracking-SARS-CoV-2-variants
- 2.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki R., Yamasoba D., Kimura I., Wang L., Kishimoto M., Ito J., Morioka Y., Nao N., Nasser H., Uriu K., et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603:700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shuai H., Chan J.F.W., Hu B., Chai Y., Yuen T.T.T., Yin F., Huang X., Yoon C., Hu J.C., Liu H., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 5.Tarrés-Freixas F., Trinité B., Pons-Grífols A., Romero-Durana M., Riveira-Muñoz E., Ávila-Nieto C., Pérez M., Garcia-Vidal E., Perez-Zsolt D., Muñoz-Basagoiti J., et al. Heterogeneous infectivity and pathogenesis of SARS-CoV-2 variants Beta, Delta and omicron in transgenic K18-hACE2 and wildtype mice. Front. Microbiol. 2022;13:840757. doi: 10.3389/fmicb.2022.840757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long B., Carius B.M., Chavez S., Liang S.Y., Brady W.J., Koyfman A., Gottlieb M. Clinical update on COVID-19 for the emergency clinician: presentation and evaluation. Am. J. Emerg. Med. 2022;54:46–57. doi: 10.1016/J.AJEM.2022.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang J., Novak T., Hecker J., Grubbs G., Zahra F.T., Bellusci L., Pourhashemi S., Chou J., Moffitt K., Halasa N.B., et al. Cross-reactive immunity against the SARS-CoV-2 Omicron variant is low in pediatric patients with prior COVID-19 or MIS-C. Nat. Commun. 2022;13:2979–3010. doi: 10.1038/s41467-022-30649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muik A., Lui B.G., Wallisch A.K., Bacher M., Mühl J., Reinholz J., Ozhelvaci O., Beckmann N., Güimil Garcia R.d.l.C., Poran A., et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science. 2022;375:678–680. doi: 10.1126/SCIENCE.ABN7591/SUPPL_FILE/SCIENCE.ABN7591_MDAR_REPRODUCIBILITY_CHECKLIST.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rössler A., Riepler L., Bante D., von Laer D., Kimpel J. SARS-CoV-2 omicron variant neutralization in serum from vaccinated and convalescent persons. N. Engl. N. Engl. J. Med. 2022;386:698–700. doi: 10.1056/nejmc2119236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pajon R., Doria-Rose N.A., Shen X., Schmidt S.D., O’Dell S., McDanal C., Feng W., Tong J., Eaton A., Maglinao M., et al. SARS-CoV-2 omicron variant neutralization after mRNA-1273 booster vaccination. N. Engl. N. Engl. J. Med. 2022;386:1088–1091. doi: 10.1056/NEJMC2119912/SUPPL_FILE/NEJMC2119912_DISCLOSURES.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J., Collier A.R.Y., Rowe M., Mardas F., Ventura J.D., Wan H., Miller J., Powers O., Chung B., Siamatu M., et al. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. N. Engl. N. Engl. J. Med. 2022;386:1579–1580. doi: 10.1056/NEJMC2201849/SUPPL_FILE/NEJMC2201849_DISCLOSURES.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/J.CELL.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dejnirattisai W., Zhou D., Ginn H.M., Duyvesteyn H.M.E., Supasa P., Case J.B., Zhao Y., Walter T.S., Mentzer A.J., Liu C., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200.e22. doi: 10.1016/J.CELL.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/J.CELL.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyngse F.P., Kirkeby C.T., Denwood M., Christiansen L.E., Mølbak K., Møller C.H., Skov R.L., Krause T.G., Rasmussen M., Sieber R.N., et al. Transmission of SARS-CoV-2 omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. medRxiv. 2022 doi: 10.1101/2022.01.28.22270044. Preprint at. [DOI] [Google Scholar]
- 17.World Health Organization. Weekly epidemiological update on COVID-19 - 27 July 2022. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---27-july-2022
- 18.Mykytyn A.Z., Rissmann M., Kok A., Rosu M.E., Schipper D., Breugem T.I., van den Doel P.B., Chandler F., Bestebroer T., de Wit M., et al. Antigenic cartography of SARS-CoV-2 reveals that Omicron BA.1 and BA.2 are antigenically distinct. Sci. Immunol. 2022;7:eabq4450. doi: 10.1126/SCIIMMUNOL.ABQ4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Huang H., Ju J., Sun R., Zhang J. Impact of vaccination on the COVID-19 pandemic in U.S. states. Sci. Rep. 2022;12:1554–1610. doi: 10.1038/s41598-022-05498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Self W.H., Tenforde M.W., Rhoads J.P., Gaglani M., Ginde A.A., Douin D.J., Olson S.M., Talbot H.K., Casey J.D., Mohr N.M., et al. Comparative effectiveness of Moderna, pfizer-BioNTech, and janssen (johnson & johnson) vaccines in preventing COVID-19 hospitalizations among adults without immunocompromising conditions — United States, march–august 2021. MMWR. MMWR Morb. Mortal. Wkly. Rep. 2021;70:1337–1343. doi: 10.15585/MMWR.MM7038E1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMOA2034577/SUPPL_FILE/NEJMOA2034577_PROTOCOL.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews N., Tessier E., Stowe J., Gower C., Kirsebom F., Simmons R., Gallagher E., Thelwall S., Groves N., Dabrera G., et al. Duration of protection against mild and severe disease by covid-19 vaccines. N. Engl. N. Engl. J. Med. 2022;386:340–350. doi: 10.1056/NEJMOA2115481/SUPPL_FILE/NEJMOA2115481_DISCLOSURES.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pouwels K.B., Pritchard E., Matthews P.C., Stoesser N., Eyre D.W., Vihta K.D., House T., Hay J., Bell J.I., Newton J.N., et al. Effect of Delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat. Med. 2021;27:2127–2135. doi: 10.1038/s41591-021-01548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke K.E.N., Jones J.M., Deng Y., Nycz E., Lee A., Iachan R., Gundlapalli A.V., Hall A.J., MacNeil A. Seroprevalence of infection-induced SARS-CoV-2 antibodies — United States, september 2021–february 2022. MMWR. MMWR Morb. Mortal. Wkly. Rep. 2022;71:606–608. doi: 10.15585/MMWR.MM7117E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O’Connell A.-M., et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. N. Engl. J. Med. 2022;386:1532–1546. doi: 10.1056/NEJMOA2119451/SUPPL_FILE/NEJMOA2119451_DISCLOSURES.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022;28:477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/J.CELL.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cromer D., Steain M., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Kent S.J., Triccas J.A., Khoury D.S., Davenport M.P. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. Lancet. Microbe. 2022;3:e52–e61. doi: 10.1016/S2666-5247(21)00267-6/ATTACHMENT/528CF602-EB89-4974-8E7C-2AE0D29E5454/MMC1.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 31.Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J. Clin. Microbiol. 2020;58:e02107-20–e02111. doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrillo J., Izquierdo-Useros N., Ávila-Nieto C., Pradenas E., Clotet B., Blanco J. Humoral immune responses and neutralizing antibodies against SARS-CoV-2; implications in pathogenesis and protective immunity. Biochem. Biophys. Res. Commun. 2021;538:187–191. doi: 10.1016/j.bbrc.2020.10.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altarawneh H.N., Chemaitelly H., Ayoub H.H., Tang P., Hasan M.R., Yassine H.M., Al-Khatib H.A., Smatti M.K., Coyle P., Al-Kanaani Z., et al. Effects of previous infection and vaccination on symptomatic omicron infections. N. Engl. J. Med. 2022;387:21–34. doi: 10.1056/NEJMOA2203965/SUPPL_FILE/NEJMOA2203965_DISCLOSURES.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds C.J., Pade C., Gibbons J.M., Otter A.D., Lin K.-M., Muñoz Sandoval D., Pieper F.P., Butler D.K., Liu S., Joy G., et al. Immune boosting by B.1.1.529 ( Omicron) depends on previous SARS-CoV-2 exposure. Science. 2022;377:eabq1841. doi: 10.1126/SCIENCE.ABQ1841/SUPPL_FILE/SCIENCE.ABQ1841_MDAR_REPRODUCIBILITY_CHECKLIST.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trinité B., Pradenas E., Marfil S., Rovirosa C., Urrea V., Tarrés-Freixas F., Ortiz R., Rodon J., Vergara-Alert J., Segalés J., et al. Previous sars-cov-2 infection increases b.1.1.7 cross-neutralization by vaccinated individuals. Viruses. 2021;13:e061135. doi: 10.3390/v13061135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldberg Y., Mandel M., Bar-On Y.M., Bodenheimer O., Freedman L.S., Ash N., Alroy-Preis S., Huppert A., Milo R. Protection and waning of natural and hybrid immunity to SARS-CoV-2. N. Engl. J. Med. 2022;386:2201–2212. doi: 10.1056/NEJMOA2118946/SUPPL_FILE/NEJMOA2118946_DISCLOSURES.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altarawneh H.N., Chemaitelly H., Ayoub H.H., Tang P., Hasan M.R., Yassine H.M., Al-Khatib H.A., Smatti M.K., Coyle P., Al-Kanaani Z., et al. Effect of prior infection, vaccination, and hybrid immunity against symptomatic BA.1 and BA.2 Omicron infections and severe COVID-19 in Qatar. medRxiv. 2022 doi: 10.1101/2022.03.22.22272745. Preprint at. [DOI] [Google Scholar]
- 38.Quandt J., Muik A., Salisch N., Lui B.G., Lutz S., Krüger K., Wallisch A.-K., Adams-Quack P., Bacher M., Finlayson A., et al. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci. Immunol. 2022;7:eabq2427. doi: 10.1126/SCIIMMUNOL.ABQ2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suryawanshi R.K., Chen I.P., Ma T., Syed A.M., Brazer N., Saldhi P., Simoneau C.R., Ciling A., Khalid M.M., Sreekumar B., et al. Limited cross-variant immunity from SARS-CoV-2 Omicron without vaccination. Nature. 2022;607:351–355. doi: 10.1038/s41586-022-04865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erikstrup C., Laksafoss A.D., Gladov J., Kaspersen K.A., Mikkelsen S., Hindhede L., Boldsen J.K., Jørgensen S.W., Ethelberg S., Holm D.K., et al. Seroprevalence and infection fatality rate of the SARS-CoV-2 Omicron variant in Denmark: a nationwide serosurveillance study. Lancet Reg. Heal. The Lancet Regional Health - Europe. 2022;21 doi: 10.1016/J.LANEPE.2022.100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., et al. Waning immune humoral response to BNT162b2 covid-19 vaccine over 6 Months. N. Engl. N. Engl. J. Med. 2021;385:e84. doi: 10.1056/NEJMOA2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas S.J., Moreira E.D., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Polack F.P., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine through 6 Months. N. Engl. N. Engl. J. Med. 2021;385:1761–1773. doi: 10.1056/NEJMOA2110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldberg Y., Mandel M., Bar-On Y.M., Bodenheimer O., Freedman L., Haas E.J., Milo R., Alroy-Preis S., Ash N., Huppert A. Waning immunity after the BNT162b2 vaccine in Israel. N. Engl. N. Engl. J. Med. 2021;385:e85. doi: 10.1056/NEJMOA2114228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buckner C.M., Kardava L., El Merhebi O., Narpala S.R., Serebryannyy L., Lin B.C., Wang W., Zhang X., Lopes de Assis F., Kelly S.E.M., et al. Interval between prior SARS-CoV-2 infection and booster vaccination impacts magnitude and quality of antibody and B cell responses. Cell. 2022;185:4333–4346.e14. doi: 10.1016/j.cell.2022.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowen J.E., Addetia A., Dang H.V., Stewart C., Brown J.T., Sharkey W.K., Sprouse K.R., Walls A.C., Mazzitelli I.G., Logue J.K., et al. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science. 2022;377:890–894. doi: 10.1126/SCIENCE.ABQ0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hachmann N.P., Miller J., Collier A.R.Y., Barouch D.H., Yu J., Rowe M., Bondzie E.A., Powers O., Surve N., Hall K., et al. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022;387:1904–1906. doi: 10.1056/NEJMC2206576/SUPPL_FILE/NEJMC2206576_DISCLOSURES.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muecksch F., Wang Z., Cho A., Gaebler C., Ben Tanfous T., DaSilva J., Bednarski E., Ramos V., Zong S., Johnson B., et al. Increased memory B cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature. 2022;607:128–134. doi: 10.1038/s41586-022-04778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goel R.R., Painter M.M., Lundgreen K.A., Apostolidis S.A., Baxter A.E., Giles J.R., Mathew D., Pattekar A., Reynaldi A., Khoury D.S., et al. Efficient recall of Omicron-reactive B cell memory after a third dose of SARS-CoV-2 mRNA vaccine. Cell. 2022;185:1875–1887.e8. doi: 10.1016/J.CELL.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tas J.M.J., Koo J.-H., Lin Y.-C., Xie Z., Steichen J.M., Jackson A.M., Hauser B.M., Wang X., Cottrell C.A., Torres J.L., et al. Antibodies from primary humoral responses modulate the recruitment of naive B cells during secondary responses. Immunity. 2022;55:1856–1871.e6. doi: 10.1016/J.IMMUNI.2022.07.020/ATTACHMENT/E224FB40-4FAA-45DB-99B4-7F8E00C98FA3/MMC1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L., Van Beek M., Wang Z., Muecksch F., Canis M., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C., Chakraborty A.K. Antigen presentation dynamics shape the response to emergent variants like SARS-CoV-2 Omicron strain after multiple vaccinations with wild type strain. bioRxiv. 2022 doi: 10.1101/2022.08.24.505127. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chalkias S., Harper C., Vrbicky K., Walsh S.R., Essink B., Brosz A., McGhee N., Tomassini J.E., Chen X., Chang Y., et al. A bivalent omicron-containing booster vaccine against covid-19. N. Engl. J. Med. 2022;387:1279–1291. doi: 10.1056/NEJMOA2208343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corominas J., Garriga C., Prenafeta A., Moros A., Cañete M., Barreiro A., González-González L., Madrenas L., Güell I., Clotet B., et al. Safety and immunogenicity of the protein-based PHH-1V compared to BNT162b2 as a heterologous SARS-CoV-2 booster vaccine in adults vaccinated against COVID-19: a multicentre, randomised, double-blind, non-inferiority phase IIb trial. medRxiv. 2022 doi: 10.1101/2022.07.05.22277210. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leal L., Pich J., Ferrer L., Nava J., Martí-Lluch R., Esteban I., Pradenas E., Raïch-Regué D., Prenafeta A., Escobar K., et al. Safety and Immunogenicity of a Recombinant Protein RBD Fusion Heterodimer Vaccine against SARS-CoV-2: preliminary results of a phase 1-2a dose-escalating, randomized, double-blind clinical trial. medRxiv. 2022 doi: 10.1101/2022.08.09.22278560. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muik A., Lui B.G., Bacher M., Wallisch A.-K., Toker A., Couto C.I.C., Güler A., Mampilli V., Schmitt G.J., Mottl J., et al. Exposure to BA.4/BA.5 Spike glycoprotein drives pan-Omicron neutralization in vaccine-experienced humans and mice. bioRxiv. 2022 doi: 10.1101/2022.09.21.508818. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheaffer S.M., Lee D., Whitener B., Ying B., Wu K., Liang C.Y., Jani H., Martin P., Amato N.J., Avena L.E., et al. Bivalent SARS-CoV-2 mRNA vaccines increase breadth of neutralization and protect against the BA.5 Omicron variant in mice. Nat. Med. 2023;29:247–257. doi: 10.1038/S41591-022-02092-8. [DOI] [PubMed] [Google Scholar]
- 56.Khoury D.S., Docken S.S., Subbarao K., Kent S.J., Davenport M.P., Cromer D. Predicting the efficacy of variant-modified COVID-19 vaccine boosters. medRxiv. 2022 doi: 10.1101/2022.08.25.22279237. Preprint at. [DOI] [PubMed] [Google Scholar]
- 57.Sánchez-Palomino S., Massanella M., Carrillo J., García A., García F., González N., Merino A., Alcamí J., Bofill M., Yuste E., et al. A cell-to-cell HIV transfer assay identifies humoral responses with broad neutralization activity. Vaccine. 2011;29:5250–5259. doi: 10.1016/J.VACCINE.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 58.Connor R.I., Chen B.K., Choe S., Landau N.R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/VIRO.1995.1016. [DOI] [PubMed] [Google Scholar]
- 59.Pradenas E., Trinité B., Urrea V., Marfil S., Tarrés-Freixas F., Ortiz R., Rovirosa C., Rodon J., Vergara-Alert J., Segalés J., et al. Clinical course impacts early kinetics,magnitude, and amplitude of SARS-CoV-2 neutralizing antibodies beyond 1 year after infection. Cell Rep. Med. 2022;3:100523. doi: 10.1016/j.xcrm.2022.100523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trinité B., Tarrés-Freixas F., Rodon J., Pradenas E., Urrea V., Marfil S., Rodríguez de la Concepción M.L., Ávila-Nieto C., Aguilar-Gurrieri C., Barajas A., et al. SARS-CoV-2 infection elicits a rapid neutralizing antibody response that correlates with disease severity. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-81862-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trigueros M., Pradenas E., Palacín D., Muñoz-López F., Ávila-Nieto C., Trinité B., Bonet-Simó J.M., Isnard M., Moreno N., Marfil S., et al. Reduced humoral response 3 months following BNT162b2 vaccination in SARS-CoV-2 uninfected residents of long-term care facilities. Age Ageing. 2022;51 doi: 10.1093/AGEING/AFAC101. afac101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
De-identified human neutralization data have been deposited at Mendeley Data and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.