Abstract
Necroptosis is a form of regulated cell death associated with degenerative disorders, autoimmune and inflammatory diseases, and cancer. To better understand the biochemical mechanisms regulating necroptosis, we constructed a detailed computational model of tumor necrosis factor-induced necroptosis based on known molecular interactions from the literature. Intracellular protein levels, used as model inputs, were quantified using label-free mass spectrometry, and the model was calibrated using Bayesian parameter inference to experimental protein time course data from a well-established necroptosis-executing cell line. The calibrated model reproduced the dynamics of phosphorylated mixed lineage kinase domain-like protein, an established necroptosis reporter. A subsequent dynamical systems analysis identified four distinct modes of necroptosis signal execution, distinguished by rate constant values and the roles of the RIP1 deubiquitinating enzymes A20 and CYLD. In one case, A20 and CYLD both contribute to RIP1 deubiquitination, in another RIP1 deubiquitination is driven exclusively by CYLD, and in two modes either A20 or CYLD acts as the driver with the other enzyme, counterintuitively, inhibiting necroptosis. We also performed sensitivity analyses of initial protein concentrations and rate constants to identify potential targets for modulating necroptosis sensitivity within each mode. We conclude by associating numerous contrasting and, in some cases, counterintuitive experimental results reported in the literature with one or more of the model-predicted modes of necroptosis execution. In all, we demonstrate that a consensus pathway model of tumor necrosis factor-induced necroptosis can provide insights into unresolved controversies regarding the molecular mechanisms driving necroptosis execution in numerous cell types under different experimental conditions.
Significance
Necroptosis is an alternative form of programmed cell death, in which the cell membrane is ruptured, leading to immune response activation. Although many of the molecular species involved in necroptosis have been identified, including receptor-interacting protein kinase-1 (RIP1), RIP3, and mixed lineage kinase domain-like protein, efforts to target necroptosis dysregulation, or leverage it therapeutically, are hindered by the lack of a detailed, mechanistic understanding of the biochemical pathways driving necroptosis execution. In this work, we present a detailed, mechanistic model of necroptosis execution that, by varying initial protein concentrations and rate constants, can resolve seeming contradictions and counterintuitive behaviors reported in the literature. The model thus significantly advances our understanding of the basic biochemical mechanisms underlying necroptosis.
Introduction
Apoptosis is widely recognized as the primary form of programmed cell death, characterized by a concerted dismantling of the cell into apoptotic bodies that can be easily processed by the immune system (1). Conversely, necroptosis is an alternative form of programmed cell death in which the cell membrane is ruptured, leading to immune response activation (2,3). Various human diseases, including neurodegenerative disorders and cancer, have been associated with necroptosis (4). Induction of necroptosis is also currently being explored as an alternative anticancer therapy, since apoptosis resistance is a hallmark of cancer (5,6,7). Although many of the primary molecular species involved in necroptosis have been identified, including receptor-interacting protein kinase-1 (RIP1), RIP3, and mixed lineage kinase domain-like protein (MLKL), efforts to target necroptosis dysregulation, or leverage it therapeutically, are hindered by the lack of a detailed, mechanistic understanding of the biochemical pathways driving necroptosis execution (4).
Previous studies of necroptosis (8,9,10,11,12,13,14,15) identified multiple mechanisms of ubiquitination regulation, including K63, K48, and M1 ubiquitin chains, which lead to phosphorylation of RIP1 and RIP3, phosphorylation and activation of cell death marker MLKL (8), and plasma membrane permeabilization resulting in cell death (16). The K63-specific deubiquitinase CYLD (cylindromatosis lysine 63 deubiquitinase) (17) and the ubiquitin-editing enzyme A20 (tumor necrosis factor [TNF], alpha-induced protein 3) (14) are both known to mediate deubiquitination of RIP1, which precedes RIP1 phosphorylation, by cleaving K63 ubiquitin chains and facilitating the formation of complex II (9,10,11,12,13,14,15). Therefore, both enzymes are generally considered drivers of necroptosis (18). However, CYLD- and A20-driven deubiquitination of RIP1 have been variously reported as pro- and antinecroptotic in different cell types: some studies have shown that CYLD drives RIP1 deubiquitination (11,17,19,20), while others have implicated A20 (13,21,22) or reported equal contributions from both enzymes (23,24,25). These varying reports have led to unresolved controversies within the field regarding the specific molecular mechanisms of complex II formation and subsequent necroptotic cell death (4). For example, Vanlangenakker et al. (25) showed that repression of CYLD in L929 cells, a murine fibrosarcoma cell line, protects from TNF-induced necroptosis but, unexpectedly, A20 repression increases sensitivity to necroptosis. A recent time-resolved analysis of necroptosis rates and network components revealed an incoherent feedforward loop through which NF-κB and A20 counteract pronecroptotic signaling in L929 cells (26), providing one possible explanation for this unexpected behavior. However, it remains unclear how general or cell context-dependent this regulatory control of necroptosis is and whether alternative explanations are possible.
Here, we present a detailed biochemical model of TNF-induced necroptosis that incorporates known biology obtained from decades’ worth of published experimental studies (Table 1). We calibrate the model to experimental phosphorylated MLKL (pMLKL) time course data from TNF-treated mouse fibrosarcoma cells at multiple TNF doses. We then perform a dynamical systems analysis that identifies four modes of necroptosis signal execution. In one case, A20 and CYLD contribute approximately equally to RIP1 deubiquitination, such that both must be knocked out to delay necroptosis induction (knocking out one has no effect, since the signal can be rerouted through the other). In another, RIP1 deubiquitination is driven exclusively by CYLD, with A20 being effectively inactive. In the other two modes, either A20 or CYLD acts as the driver of RIP1 deubiquitination, with the other enzyme, counterintuitively, acting to inhibit necroptosis (consistent with the observation by Vanlangenakker et al. (25) mentioned above). We also perform sensitivity analyses to identify proteins and kinetic parameters that can be targeted within each mode to modulate pMLKL dynamics and time-to-death (TTD) by necroptosis. We find that, for two modes, proteins and rate constants centered around RIP1 ubiquitination regulation in complex I have the most significant effect on necroptosis signal execution. For the other two, potential targets include factors involved in the balance between complex II degradation and necrosome formation. Overall, our results show that a consensus pathway model of TNF-induced necroptosis can explain numerous experimentally observed behaviors, including conflicting and counterintuitive results from multiple studies involving different cell types. Following a detailed description of our proposed model, we present results of the parameter calibration, dynamical systems analysis, in silico knockout (KO) experiments, and sensitivity analyses. We conclude with a discussion of the broader implications of our results, including important insights into the molecular mechanisms of necroptosis execution and the potential for extending and using the model to identify novel pro- and antinecroptosis therapeutic targets.
Table 1.
Key proteins involved in necroptosis
| Protein | Role in necroptosis | Initial amount | UniProt ID | Refs. |
|---|---|---|---|---|
| A20 | Ubiquitin-editing enzyme responsible for deubiquitinating RIP1 in complex I | 0.225 μM (8868 molecules) | F8K9X7 | (13,21) |
| Caspase-8 | Heterodimerizes with cFLIPL (long isoform), leading to cleavage and inactivation of RIP1 and RIP3 in complex II | 0.096 μM (3799 molecules) | Q14790 | (42,107) |
| cFLIPL | Heterodimerizes with caspase-8, leading to cleavage and inactivation of RIP1 and RIP3 in complex II | 0.081 μM (3193 molecules) | O15519 | (43,108) |
| cIAP1/2 | Catalyzes, via its RING domains, the activating K63-linked polyubiquitination of RIP1 | 0.225 μM (8868 molecules) | Q13075, Q13490 | (25,81) |
| CYLD | Deubiquitinates RIP1 in either complex I or within the RIP1-RIP3 necrosome | 0.225 μM (8868 molecules) | Q9NQC7 | (11,17) |
| FADD | TNFR1-interacting scaffold protein in complex II | 0.079 μM (3109 molecules) | Q13158 | (18,109,110) |
| LUBAC | TNFR1-interacting protein complex (composed of HOIL-1 and HOIP) recruited by cIAP1/2 in complex I that promotes RIP1 ubiquitination | 0.180 μM (7095 molecules) | Q9BYM8, Q96EP0 | (25,82) |
| MLKL | Recruited to the necrosome by RIP1, where it is phosphorylated, leading to cell death by membrane rupture | 0.141 μM (5544 molecules) | Q9BYM8, Q96EP0 | (48,111,112) |
| RIP1 | A multifunctional adaptor protein in the necrosome that recruits and activates RIP3 and MLKL | 0.500 μM (19,707 molecules) | Q8NB16 | (25,113,114) |
| RIP3 | Recruited to the necrosome by binding to and cross-phosphorylating RIP1 | 0.270 μM (10,654 molecules) | Q9Y572 | (111,115,116) |
| TNF | Pleiotropic proinflammatory cytokine that activates necroptosis in the absence of caspase activity | 0.1–100 ng/mL (2–2413 molecules) | Q13546 | (117) |
| TNFR1 | TNF receptor superfamily member death receptor that recruits RIP1 to complex 1 | 0.125 μM (4927 molecules) | Q9Y572 | (33,118) |
| TRADD | TNFR1-interacting protein in complexes I and II that serves as a docking adaptor for the binding of RIP1 to TRAF2 | 0.119 μM (4696 molecules) | P01375 | (35,118) |
| TRAF2 | TNFR1-interacting protein that recruits cIAP1/2 to complex I, promoting K63-linked RIP1 ubiquitination | 0.299 μM (11,776 molecules) | P19438 | (109,119) |
Initial amounts are based on either mass spectrometry measurements (this work) or estimates from the literature (see materials and methods). All UniProt IDs (www.uniprot.org) are for human proteins.
Results
A biochemical model of TNF-induced necroptosis describes the formation of key signaling complexes along the path to cell death
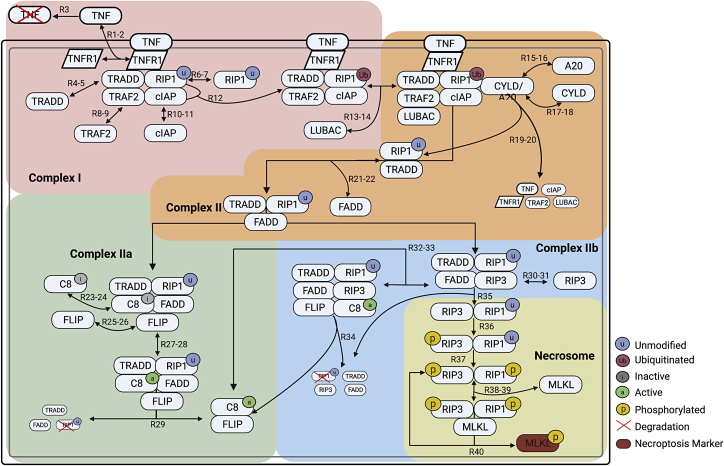
The death receptor ligand TNF (27), an extensively studied inducer of necroptosis and well-known master regulator of inflammation, has been at the forefront of numerous fundamental discoveries concerning the interplay between cell death and survival pathways (25). Here, we propose a detailed, mechanistic model of TNF-induced necroptosis based on an extensive review of the literature (Table 1). The model includes 14 proteins interacting via 40 reactions (all mass action; Fig. 1; Table 2) to produce 37 biochemical species, comprising complex I, complex II, and the necrosome, three key macromolecular complexes along the path from cell death cue to necroptosis execution. Below, we describe in detail how we model the steps involved in the formation of each complex, beginning with TNF binding to TNF receptor 1 (TNFR1) and ending at phosphorylation of the necroptosis cell death reporter MLKL. A model schematic is provided as a visual aid (Fig. 1), with reactions, including association, dissociation, phosphorylation, ubiquitination, deubiquitination, and degradation, denoted as “RN,” where N is the reaction index. Note that protein synthesis is assumed negligible and omitted from the model. Also, while relatively complex, the proposed model is still a highly simplified representation of the true biochemical events involved in necroptosis execution (28). Many proteins in the pathway (e.g., RIP1, TRADD, FADD, caspase-8, and FLIP) are also involved in related pathways, such as apoptosis, which we do not consider in this work (29,30,31). Abstractions and simplifications such as these are standard practice in computational modeling and necessary for gaining critical insights into complex biological phenomena (32).
Figure 1.
Schematic of the necroptosis execution model. The diagram is color coded to highlight the processes involved in formation of complex I, complex II, complex IIa, complex IIb, and the necrosome. Arrows are labeled with “RN” or “RN-M,”, where N and M correspond to reaction indices in the model. In many cases (but not all; see text), “RN-M” denotes a set of reversible reactions, with N the index of the forward reaction and M the index of the reverse. Created with BioRender.com.
Table 2.
Rate constants for all reactions in the necroptosis model
| Parameter | Reaction | Medians | Units |
|---|---|---|---|
| P1 | R1: association of TNF to TNFR | 10 [1.07, 0.39, 0.91, 0.49] | μM−1 min−1 |
| P2 | R2: dissociation of TNF:TNFR | 10 [−2.00, −2.22, −3.40, −2.20] | min−1 |
| P3 | R3: degradation of TNF | 10 [−1.52, −1.70, −1.62, −1.62] | min−1 |
| P4 | R4: association of TRADD to complex I | 10 [0.11, 0.45, 0.71, 1.19] | μM−1 min−1 |
| P5 | R5: dissociation of TRADD from complex I | 10 [1.22, 1.51, 1.84, 1.30] | min−1 |
| P6 | R6: association of RIP1-u to complex I | 10 [6.30, 5.91, 6.19, 5.94] | μM−1 min−1 |
| P7 | R7: dissociation of RIP1-u from complex I | 10 [−1.70, −1.82, −1.70, −1.70] | min−1 |
| P8 | R8: association of TRAF2 to complex I | 10 [5.34, 5.51, 5.22, 5.16] | μM−1 min−1 |
| P9 | R9: dissociation of TRAF2 from complex I | 10 [−2.26, −1.82, −2.40, −1.70] | min−1 |
| P10 | R10: association of cIAP to complex I | 10 [4.70, 4.58, 4.63, 4.57] | μM−1 min−1 |
| P11 | R11: dissociation of cIAP from complex I | 10 [−0.01, −0.33, −0.05, −0.24] | min−1 |
| P12 | R12: ubiquitination of RIP1-u by cIAP in complex I | 10 [−1.52, −2.00, −2.00, −1.70] | min−1 |
| P13 | R13: association of LUBAC to RIP1-Ub in complex I | 10 [2.41, 2.01, 2.12, 2.42] | μM−1 min−1 |
| P14 | R14: dissociation of LUBAC from RIP1-Ub in complex I | 10 [−3.80, −3.74, −4.44, −3.74] | min−1 |
| P15 | R15: association of A20 to RIP1-Ub in complex I | 10 [4.88, 4.05, 5.18, −0.43] | μM−1 min−1 |
| P16 | R16: dissociation of A20 from complex I | 10 [−1.40, −2.00, −2.00, −4.05] | min−1 |
| P17 | R17: association of CYLD to RIP1-Ub in complex I | 10 [7.62, 1.84, 7.43, 1.71] | μM−1 min−1 |
| P18 | R18: dissociation of CYLD from complex I | 10 [0.28, 0.11, 0.59, 0.71] | min−1 |
| P19 | R19: deubiquitination of RIP1-Ub by A20 in complex I | 10 [−1.52, −1.00, −1.10, −1.52] | min−1 |
| P20 | R20: deubiquitination of RIP1-Ub by CYLD in complex I | 10 [−1.22, 2.67, 2.84, 2.59] | min−1 |
| P21 | R21: association of FADD to RIP1-u:TRADD in complex II | 10 [7.47, 7.73, 7.53, 7.37] | μM−1 min−1 |
| P22 | R22: dissociation of FADD from RIP1-u:TRADD in complex II | 10 [−0.17, −0.15, −0.31, −0.10] | min−1 |
| P23 | R23: association of C8i to FADD in complex IIa | 10 [1.49, 1.48, 1.49, 1.15] | μM−1 min−1 |
| P24 | R24: dissociation of C8i from complex IIa | 10 [−2.10, −2.10, −2.00, −1.72] | min−1 |
| P25 | R25: association of FLIP to C8i in complex IIa | 10 [3.73, 3.85, 3.93, 3.72] | μM−1 min−1 |
| P26 | R26: dissociation of FLIP from C8i in complex IIa | 10 [−4.10, −4.70, −5.40, −3.96] | min−1 |
| P27 | R27: activation of C8i in complex IIa | 10 [−2.05, −2.30, −2.30, 1.66] | min−1 |
| P28 | R28: inactivation of C8a in complex IIa | 10 [1.41, 1.53, 1.36, −2.41] | min−1 |
| P29 | R29: degradation of RIP1-u by C8a:FLIP in complex IIa | 10 [−0.12, −0.38, −0.21, −0.10] | min−1 |
| P30 | R30: association RIP3 to RIP1-u in complex IIb | 10 [6.22, 6.10, 6.09, 6.26] | μM−1 min−1 |
| P31 | R31: dissociation of RIP3 from RIP1-u in complex IIb | 10 [−0.79, −0.81, −0.53, −0.97] | min−1 |
| P32 | R32: association of C8a:FLIP to complex IIb | 10 [3.28, 3.83, 3.18, 3.19] | μM−1 min−1 |
| P33 | R33: dissociation of C8a:FLIP from complex IIb | 10 [−3.00, −3.00, −2.21, −2.26] | min−1 |
| P34 | R34: degradation of RIP1-u by C8a:FLIP in complex IIb | 10 [−2.60, −2.40, −3.85, −1.75] | min−1 |
| P35 | R35: dissociation of RIP1-u:RIP3 from complex IIb | 10 [−3.30, −2.44, −2.42, −2.41] | min−1 |
| P36 | R36: phosphorylation of RIP1-p by RIP3 in the necrosome | 10 [−2.28, −2.14, −2.32, −2.31] | min−1 |
| P37 | R37: phosphorylation of RIP3 by RIP1-p in the necrosome | 10 [−2.62, −2.55, −2.61, −3.01] | min−1 |
| P38 | R38: association of MLKL to the necrosome | 10 [5.03, 4.16, 3.98, 4.11] | μM−1 min−1 |
| P39 | R39: dissociation of MLKL from the necrosome | 10 [0.29, 0.47, 0.02, 0.16] | min−1 |
| P40 | R40: phosphorylation of MLKL | 10 [2.10, 2.63, 2.46, 2.53] | min−1 |
C8i, C8a: inactive and active caspase-8; RIP1-u, RIP1-Ub, RIP1-p: unmodified, ubiquitinated, and phosphorylated RIP1; RIP3-p: phosphorylated RIP3. Median values are ordered as 10 [M1,M2,M3,M4].
In the model (Fig. 1), signaling through the necroptosis pathway is initiated when the cytokine TNF binds to the extracellular domain of TNFR1 (R1-2), which protects TNF from degradation (R3) and activates the receptor by causing a conformational change in its intracellular domain (25,33,34). The adaptor protein TRADD (TNFR1-associated death domain) is then recruited to the intracellular domain of TNFR1 (R4-5) to facilitate binding of RIP1 (unmodified; R6-7) and TRAF2 (TNFR-associated factor 2) (R8-9) (35,36,37). TRAF2 recruits and binds cIAP1/2 (cellular inhibitor of apoptosis proteins 1 and 2) (R10-11), which add nondegradative polyubiquitin chains to RIP1 (R12) (8). Ubiquitinated RIP1 recruits other necessary components to the complex, including LUBAC (linear ubiquitin chain assembly complex) (R13-14). We refer to the supramolecular structure that is anchored to the cell membrane and composed of TNF, TNFR1, TRADD, ubiquitinated RIP1, TRAF2, cIAP1/2, and LUBAC as complex I (38,39) (Fig. 1, pink). Biologically, complex I is known to drive multiple pathways in addition to necroptosis, including apoptosis and the inflammatory NF-κB pathway (40).
Formation of complex I is followed by deubiquitination of RIP1 by the enzymes A20 (14,21) and CYLD (11,17,19,20), which competitively bind to RIP1 in its ubiquitinated state (R15-18), causing cleavage, deubiquitination, and release in association with TRADD and the dissolution of complex I (R19-20). The deubiquitinated RIP1:TRADD heterodimer then recruits FADD (Fas-associated protein with death domain) (R21-22), initiating the formation of complex II, also known as the cytosolic death-inducing signaling complex (Fig. 1, orange). Complex II can be modified via two competing paths, one antinecroptotic and one pronecroptotic. We model the antinecroptotic path beginning with FADD, via its death effector domain, mediating the recruitment of inactive caspase-8 (C8i) (R23-24) (41), which subsequently binds long-form FLIP (cellular FADD-like IL-1β-converting enzyme-inhibitory protein) (R25-26), resulting in the complex commonly referred to as complex IIa (25,41) (Fig. 1, green). FLIP then oligomerizes with C8i to produce the active form of caspase-8 in necroptosis (C8a) (R27-28) (42,43), which proceeds to cleave RIP1 for truncation (i.e., degradation), resulting in dissolution of the complex and release of the active C8a:FLIP heterodimer (44,45) (R29) that directly inhibits necroptosis (R32-34).
The pronecroptotic path is modeled as involving formation of complex IIb (Fig. 1, blue), which occurs when deubiquitinated RIP1 in complex II recruits RIP3 (R30-31), blocking C8i recruitment (R23-24). The C8a:FLIP heterodimer can then be recruited to complex IIb (R32-33), which cleaves RIP1 for truncation, leading to dissolution of the complex (R34). Alternatively, RIP3 and deubiquitinated RIP1 can dissociate from complex IIb as a heterodimer (R35) (25). Cross-phosphorylation of RIP3 (R36) and then RIP1 (R37), followed by recruitment of MLKL (R38-39) (46,47), results in the necroptosis signaling complex known as the necrosome (Fig. 1, yellow) (25). Phosphorylation of MLKL (48) in the necrosome by phosphorylated RIP1 and RIP3 is followed by release of pMLKL from the phosphorylated RIP1:RIP3 heterodimer (R40), which is again free to bind MLKL. We assume that dephosphorylation and degradation of the phosphorylated RIP1:RIP3 heterodimer is negligible, consistent with experimental reports (49). Biologically, translocation of pMLKL to the cell membrane (50) then causes rapid plasma membrane rupture and inflammatory response due to the release of damage-associated molecular patterns and cytokines (51), ultimately resulting in cell death. These latter steps are not included in the present model.
Western blots and mass spectrometry enable Bayesian parameter estimation of the necroptosis model
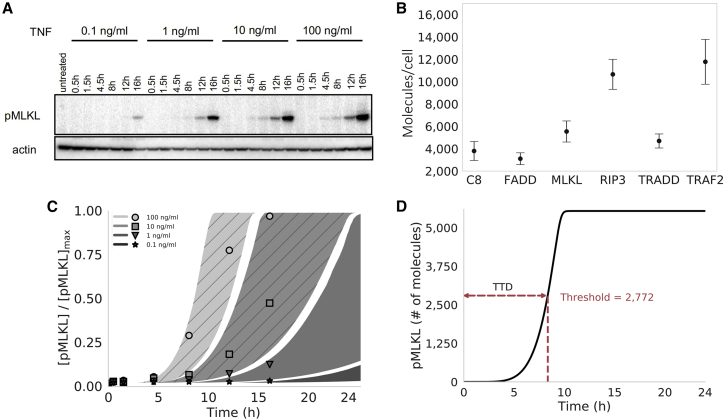
To explore the dynamics of our computational necroptosis model, we first calibrated it to experimental protein time course data using a Bayesian parameter estimation approach (52). In brief, we used L929 cells, a murine fibrosarcoma cell line that is a well-established model system for studying necroptosis (25). Cells were treated with 100, 10, 1, and 0.1 ng/mL of TNF over 16 h and pMLKL levels were estimated at multiple time points via western blot using densitometry (Fig. 2 A). To quantify initial protein abundances, used as inputs to the model, we used label-free mass spectrometry in untreated L929 cells for the proteins C8, FADD, MLKL, RIP3, TRADD, and TRAF2 (Fig. 2 B). All other initial protein levels (other than TNF, which depends on applied dose) were set to values based on biologically plausible assumptions (Table 1). Parameter estimation was then performed using PyDREAM (52) (Fig. 2 C), a multichain Monte Carlo sampling tool, with a multiobjective cost function that included data from the two highest TNF doses (100 and 10 ng/mL; Fig. S1). In all, an ensemble of 10,628 parameter sets was obtained (Fig. S2), all of which reproduce the experimental data reasonably well (53) (see materials and methods for additional details). Model simulations at the two lowest TNF doses (1 and 0.1 ng/mL; Fig. 2 C) showed good correspondence to experimental data, providing a simple validation of the model fits.
Figure 2.
Proteomics, parameter calibration, and time-to-death. (A) Western blots for phosphorylated MLKL (pMLKL) at multiple time points in L929 (murine fibrosarcoma) cells under 0.1–100 ng/mL TNF stimulation. Actin, used as a loading control, is also shown for comparison. (B) Mass spectrometry data from untreated L929 cells for multiple proteins involved in necroptosis execution. Points represent the median of three replicates (used as input to the computational model); error bars span the interquartile range. (C) Simulated pMLKL time courses (plotted as 95% probability envelopes) for 0.1–100 ng/mL TNF stimulation (same concentrations as in (A)) based on 10,628 parameter sets obtained using Bayesian parameter estimation. The model was calibrated to the 100 and 10 ng/mL TNF data only (shaded regions with diagonal lines); time courses for the lowest TNF concentrations (shaded regions with no diagonal lines) amount to a simple model validation. Points correspond to the western blot data in (A), quantified via densitometry. Points and shaded regions are colored the same, based on TNF dose. (D) Illustration of the time-to-death (TTD) metric used to quantify cell death in silico. A hard threshold of 2772 molecules (half the median MLKL level in (B)) was chosen to signify cell death (see materials and methods). MLKL, mixed lineage kinase domain-like protein; TNF, tumor necrosis factor.
A dynamical systems analysis identifies four distinct necroptosis execution modes differing by mechanism of RIP1 ubiquitination regulation
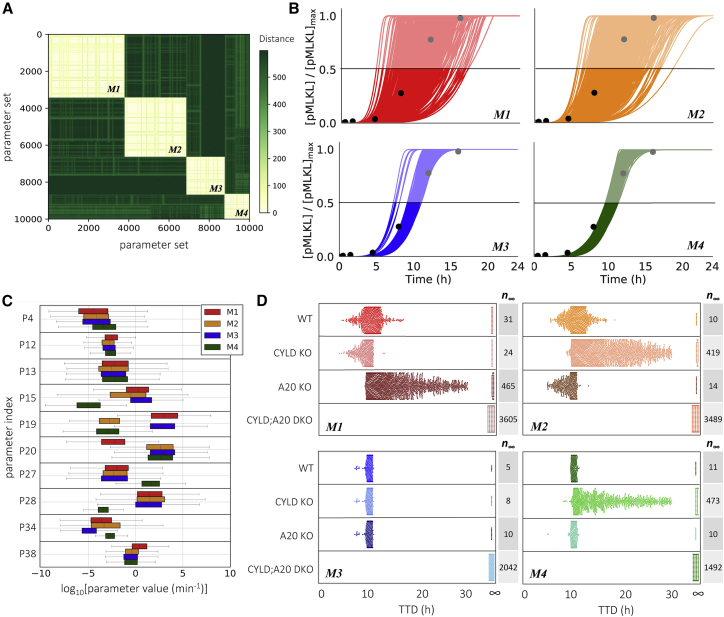
We performed a dynamical systems analysis to explore the possibility that distinct “modes of necroptosis execution” exist within the parameter set ensemble obtained from Bayesian parameter estimation. The rationale is that, while different parameterizations of the model achieve cell death at approximately equal times, they may arrive there via significantly different sequences of molecular events. We utilized a computational tool (54) that identifies subnetworks of reactions that dominate the production or consumption of a target species, pMLKL in this case, at user-specified times along a time course. Each subnetwork is given an integer label and each time point is associated with a subnetwork. Thus, a continuous concentration time course is “digitized” into a sequence of integers, which we refer to as a “dynamical signature.” This transformation enables simple comparisons between time courses obtained with different parameter sets using standard dissimilarity metrics, such as the longest common subsequence (55). Applying this approach to all 10,628 parameter sets obtained from Bayesian parameter estimation of our necroptosis model and clustering the resulting dynamical signatures using a spectral clustering algorithm (56), we obtained four distinct clusters, or modes of necroptosis execution (Figs. 3 A and S3; see materials and methods for additional details).
Figure 3.
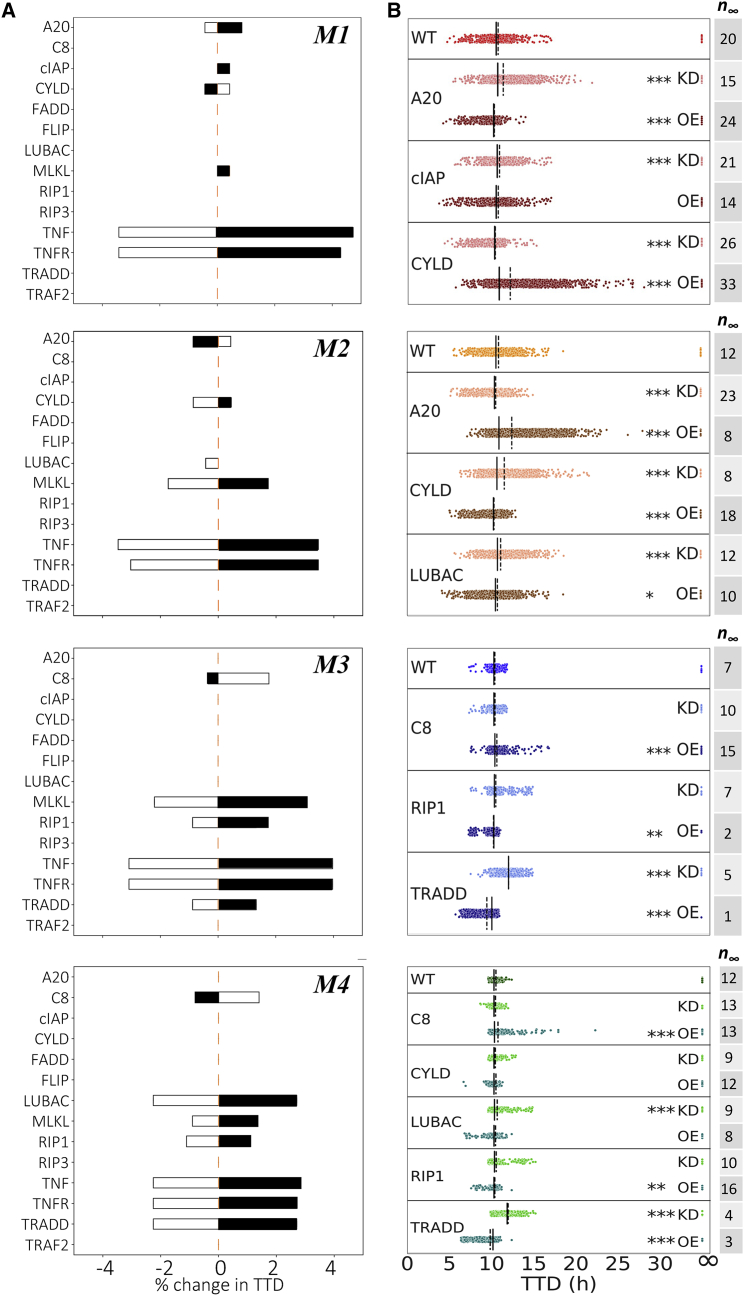
Four modes of necroptosis execution exhibit variability in temporal dynamics and differ in rate constant values and responses to CYLD and A20 knockouts. (A) Clustering analysis of simulated time courses (100 ng/mL TNF) from 10,628 parameter sets reveals four distinct modes of execution (M1, …, M4). Dissimilarity (“distance”) between dynamical signatures (digitized time courses) was quantified using the longest common subsequence (see materials and methods). (B) Simulated time courses (100 ng/mL TNF) of the necroptosis marker, pMLKL, show significantly more variability in TTD (defined as the time at which pMLKL reaches its half-maximal value) in modes 1 and 2 than in modes 3 and 4. Time courses for all parameter sets associated with each mode are shown. Experimental western blot data (black circles; quantified from Fig. 2A) are included to illustrate the model fit for each mode. (C) Variations in the values of 10 rate constants distinguish the four modes of execution. Parameter indices (PN) match reaction indices (RN) in Fig. 1 and Table 2. Note that second-order rate constants (P4, P13, P15, and P38; see Table 2) were converted from units of μM−1 min−1 to min−1 assuming a spherical cell of diameter 5 μm (see materials and methods). (D) Knockouts of CYLD and A20 (100 ng/mL TNF) differentially affect TTD, relative to wild-type (WT), across the four modes of execution (each dot corresponds to a parameter set). Note that CYLD;A20 double knockout inhibits cell death in all cases (TTD = ∞). The number of parameter sets that do not result in cell death (n∞) are included for all modes under all conditions. KO, knockout; DKO, double knockout.
Interestingly, two of the execution modes identified (modes 1 and 2) exhibit significantly more variability in pMLKL temporal dynamics and TTD (defined in Fig. 2 D; see materials and methods) across their associated parameter sets than the other two (Fig. 3 B). This suggests that the modes harbor fundamental differences in rate constant values that lead to differential robustness to parameter variations. To explore this further, we compared the distributions of rate constants across modes and identified eight (out of 40) with significant differences (>7.5-fold) between the largest and smallest mean (Fig. 3 C; additional distributions are shown in Fig. S4). We also consider distributions for two rate constants (P12 and P13; see Fig. 1 and Table 2) with much smaller differences across means (∼threefold in both cases) but for which the model exhibits high sensitivity (discussed in the next subsection). In all, these 10 rate constants correspond to reactions spanning the model topology, starting with the association of TRADD to complex I (P4), which has a somewhat increased rate in mode 4. Further downstream, the rate constant for ubiquitination of RIP1 by cIAP (P12) is slightly larger in mode 1 than in the other modes. Small differences are also seen for the binding rate of LUBAC to complex I (P13). The rate constant for binding of A20 to ubiquitinated RIP1 (P15) is significantly smaller in mode 4 than in the other modes and somewhat smaller in mode 2 relative to modes 1 and 3. Deubiquitination of RIP1 by A20 (P19) is significantly reduced in modes 2 and 4, while, interestingly, the rate constant for RIP1 deubiquitination by CYLD (P20) in mode 1 is reduced by almost the same amount relative to the other modes. For activation/deactivation of C8 in complex IIa, which is a critical step in the pathway for determining whether the cell will progress to necroptosis, mode 4 has both a significantly larger activation (P27) and significantly smaller deactivation (P28) rate constant. The rate constant for subsequent RIP1 degradation by the active C8a:FLIP heterodimer to complex IIb (P34), which inhibits necroptosis, is somewhat smaller in mode 3 and larger in mode 4 relative to the other modes. Finally, the binding rate constant for MLKL to the phosphorylated RIP1:RIP3 heterodimer (P38), the final step in the formation of the necrosome, is somewhat increased in mode 1. These results clearly illustrate that significant differences exist in the values of rate constants across the modes of execution, despite similarities in pMLKL temporal dynamics.
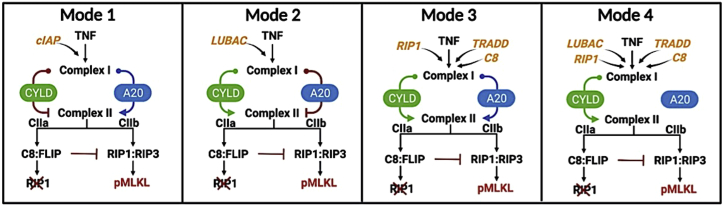
CYLD and A20 are known regulators of RIP1 deubiquitination (9,10,11,12,13,14,15) but have been reported as both drivers and inhibitors of necroptosis in different cell types (11,17,19,20,23,24,25). To investigate the roles of CYLD and A20 in our necroptosis model, we performed in silico CYLD and A20 KO experiments and compared TTD distributions with the unperturbed, i.e., “wild-type”, case (Fig. 3 D). Unsurprisingly, in all cases CYLD;A20 double KO (DKO) prevents cell death (TTD = ∞). However, for single CYLD and A20 KOs, we see highly variable responses across the four modes of execution. For mode 1, we see that knocking out A20 leads to a general increase in TTD (i.e., decrease in necroptosis sensitivity) across the parameter sets, consistent with A20 acting as a regulator of RIP1 ubiquitination and driver of necroptosis (14,21). Conversely, CYLD KO results in a general reduction in TTD (i.e., increase in sensitivity), indicating that CYLD in mode 1 counterintuitively operates as an inhibitor of necroptosis. We see the opposite trends in mode 2: A20 KO reduces TTD, while CYLD KO leads to a general increase in TTD across the parameter sets. Interestingly, this result is consistent with observations by Vanlangenakker et al. (25) that A20 depletion can sensitize cells to death by necroptosis. In mode 3, we see that single KOs of A20 and CYLD have no effect on TTD. Since DKO prevents cell death in all cases, this reveals that A20 and CYLD both drive RIP1 deubiquitination and, hence, when one enzyme is knocked out signal flow diverts through the other. Finally, in mode 4, CYLD KO leads to a general increase in TTD, like mode 2; however, A20 KO has no effect, as in mode 3. In all, the results of in silico KO experiments reveal distinct differences in the roles of A20 and CYLD in RIP1 ubiquitination regulation among the four model-predicted modes of necroptosis execution (summarized in Fig. 4).
Figure 4.
Summary depictions of the four modes of necroptosis execution. Each mode is distinguished by differences in sensitivity to initial protein levels (orange text; see Fig. 5), rate constants (not depicted; see Fig. 6), and mechanism of RIP1 ubiquitination regulation by CYLD and A20. Promotion of complex II formation (and hence necroptosis) by CYLD or A20 is represented by arrows (green or blue); inhibition is represented by a red line with a bar.
Ubiquitination of RIP1 by cIAP in complex I and binding of LUBAC to complex I are global modulators of necroptosis sensitivity across execution modes
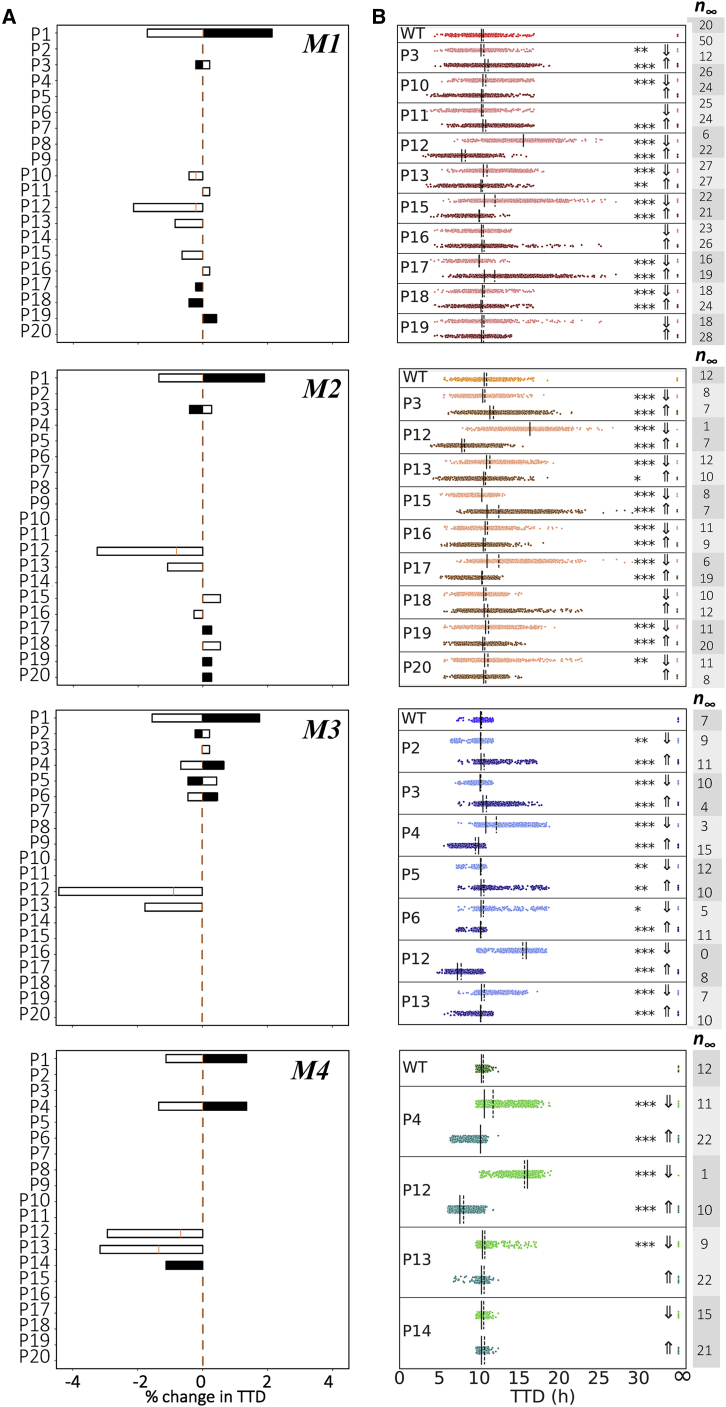
Targeting necroptosis by small-molecule modulators has emerged as a promising approach for both cancer therapy and treatment of inflammatory diseases (57). It is of interest, therefore, to determine if modulating factors exist that are common across all model-predicted modes of execution, which could represent novel therapeutic targets. Toward this end, we performed sensitivity analyses based on “representative” parameter sets for each mode (automatically generated by our dynamical systems analysis tool (54); see materials and methods for details) over the 14 non-zero initial protein concentrations (Fig. 5 A) and 40 rate constants (Figs. 6 A and S5). Changes in necroptosis sensitivity, quantified by changes in TTD (Fig. 2 D), were calculated over ranges ±20% around a reference set of initial protein concentrations (Table 1) and values of the rate constants from the representative parameter set for each mode. We then validated the results of these analyses (i.e., to confirm they are not specific to the representative parameter set) by performing, for all parameter sets associated with each mode, in silico knockdowns (KDs) by 70% and 10-fold overexpressions for the initial concentrations (58,59) (Fig. 5 B) and by varying the rate constant values ±10-fold (Fig. 6 B).
Figure 5.
Sensitivity analyses and model-predicted protein targets for each mode of execution. (A) Changes in TTD for “representative” parameter sets of each mode (see materials and methods). Black shaded regions signify decreases in initial protein concentrations; white shaded regions signify increases. (B) Knockdown (KD) (70%) and overexpression (OE) (10-fold) of potential targets identified in (A) for all parameter sets for each mode. The number of parameter sets that do not result in cell death (n∞) are included to the right. Solid black lines, medians; dashed black lines, means; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Mood’s median test).
Figure 6.
Sensitivity analyses and model-predicted rate constant targets for each mode of execution. (A) Changes in TTD for “representative” parameter sets of each mode (see materials and methods). Black shaded regions signify decreases in rate constant values; white shaded regions signify increases. (B) Decreases (⇓; 10-fold) and increases (⇑; 10-fold) of potential targets identified in (A) for all parameter sets for each mode. The number of parameter sets that do not result in cell death (n∞) are included to the right. Parameter indices (PN) match reaction indices (RN) in Fig. 1 and Table 2. Solid black lines, medians; dashed black lines, means; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Mood’s median test).
Across the four modes of execution, we see three common protein modulators of necroptosis sensitivity: TNF, TNFR, and MLKL (Fig. 5). These are expected, since these proteins are well-known master regulators of TNF-induced necroptosis (28,60). In addition, for the rate constants we see three common modulators across the four modes (Fig. 6), corresponding to the association of TNF to TNFR (P1), ubiquitination of RIP1 by cIAP in complex I (P12), and association of LUBAC (P13) to complex I (see Fig. 1, pink). The former is expected since TNF is the death-inducing stimulus driving necroptosis. The latter two are interesting in that they are consistent with published reports demonstrating that cIAP inhibition by SMAC mimetics (45) and LUBAC inhibition by genetic deletion (61) alter necroptosis sensitivity. For all four modes, we see that increasing the values of these two rate constants (P12 and P13) leads to a significant decrease in TTD (i.e., increased necroptosis sensitivity), and vice versa. Note that the analyses based on the representative parameter set (Fig. 6 A) show only that TTD decreases when these two rate constant values are increased. However, by repeating the analyses over all parameter sets associated with each mode (Fig. 6 B), we confirm that TTD also increases when the rate constant values are decreased.
Sensitivities to initial protein levels and rate constant values reveal execution mode-dependent targets for modulating TTD
We have shown that the four modes of necroptosis execution (Fig. 3 A) exhibit differences in variability in TTD (Fig. 3 B), rate parameter values (Fig. 3 C), and responses to A20 and CYLD KOs (Fig. 3 D). This suggests that, in addition to the global modulators identified above (TNF, TNFR, MLKL, P1, P12, and P13; Figs. 5 and 6), each mode must also have a unique set of factors that drive response. For mode 1, these include proteins (A20, cIAP, and CYLD; Fig. 5, top row) and rate constants (P10, P11, and P15–P19; Fig. 6, top row) associated with RIP1 ubiquitination regulation in complex I (see Fig. 1, orange). The sensitivities to A20 and CYLD are consistent with the results from in silico KO experiments (Fig. 3 D). Intuitively, we can understand these sensitivities as due to competitive binding between A20 and CYLD to complex I, coupled with differences in the rate constants for RIP1 deubiquitination by A20 (P19) and CYLD (P20; see Fig. 3 C). In other words, increasing the amount of A20 leads to increased amounts of A20-bound complex I (and vice versa). Since the rate constant for RIP1 deubiquitination in mode 1 by A20 is much larger than for CYLD (Fig. 3 C), this results in a significant decrease in TTD, i.e., A20 promotes necroptosis, as expected. Conversely, increasing the amount of CYLD leads to more CYLD-bound complex I (and vice versa). Since CYLD is less efficient at deubiquitinating RIP1, this results in a much lower overall rate of RIP1 deubiquitination and a significant increase in TTD, i.e., CYLD, counterintuitively, inhibits necroptosis in this mode. Sensitivities to rate constants associated with these processes (P10, P11, and P15–P19) can be understood similarly.
As in mode 1, potential targets in mode 2 include proteins (A20, CYLD, and LUBAC; Fig. 5 A, second row) and rate constants (P15–P20; Fig. 6, second row) associated with RIP1 ubiquitination regulation. However, the sensitivities to A20 and CYLD are reversed in their effects on TTD as compared with mode 1, i.e., A20 counterintuitively inhibits necroptosis, while CYLD promotes necroptosis, as expected. Again, these results are consistent with in silico KO experiments (Fig. 3 D) and can be understood in terms of competitive binding between A20 and CYLD to complex I and differences in rate constants for RIP1 deubiquitination by A20 and CYLD (Fig. 3 C). Also note that TTD in both modes 1 and 2 are sensitive to the rate constant for TNF degradation (P3; Fig. 6, top and second rows), which is as expected since TNF is the stimulus driving necroptosis.
For mode 3, potential targets are associated with formation of the necrosome from complex IIb, which immediately precedes necroptosis execution (see Fig. 1, blue). Specifically, we see sensitivities to proteins C8, RIP1, and TRADD (Fig. 5, third row), the latter two of which are key components of complex II, and rate constants (P2–P6; Fig. 6, third row) for reactions upstream of complex II that include the association of RIP1 and TRADD to complex I. Intuitively, the comparatively small value of the rate constant in mode 3 for degradation of the C8a:FLIP-bound complex IIb (P34; see Fig. 3 C) is what ultimately drives these sensitivities. Modifying rates of reactions that contribute to complex II formation and/or the rate of binding of C8i to complex II alters the balance between the rates of necrosome formation and the degradation of complex IIb that prevents necroptosis, thus affecting TTD. Also note, in contrast to modes 1 and 2, the lack of sensitivity in mode 3 to variations in the initial concentrations of A20 and CYLD. This is because, in this mode, A20 and CYLD are effectively indistinguishable enzymes, i.e., rate constants for binding and unbinding from complex I (P15–P18) and RIP1 deubiquitination (P19 and P20) are virtually identical for both (Figs. 3 C and S4). Thus, varying the concentration of one is effectively equivalent to varying the concentration of the other by the same amount.
In mode 4, we see the same sensitivities as in mode 3 to varying concentrations of C8, RIP1, and TRADD (Fig. 5, bottom row) and the rate constant for association of TRADD to complex I (P4; Fig. 6, bottom row). These sensitivities can be understood in the same way as in mode 3 in terms of the balance between necrosome formation and complex IIb degradation. However, we see an additional sensitivity in mode 4 to the initial concentration of LUBAC (Fig. 5, bottom row). Interestingly, for the representative parameter set, this is evident for both increases and decreases in LUBAC concentration (Fig. 5 A, bottom row), but when all parameter sets are considered is only statistically significant for the KD experiments (Fig. 6 A, bottom row). Note also that the representative parameter set shows a sensitivity to the dissociation rate of LUBAC from complex I (P14; Fig. 6 A, bottom row) but the effect is not statistically significant when all parameter sets are considered (Fig. 6 B, bottom row). Furthermore, despite the results of in silico KO experiments that show RIP1 deubiquitination in mode 4 is driven exclusively by CYLD (Fig. 3 D), we do not see a sensitivity in TTD to variations in CYLD concentration, even for a 70% KD (Fig. 5, bottom row). We can explain both this result and the one-way sensitivity to variations in LUBAC as due to a severely dysfunctional A20 in this mode, evident in exceedingly small rate constants for A20 binding to complex I (P15) and subsequent RIP1 deubiquitination (P19), coupled with a comparatively large rate constant for C8 activation (P27) and small rate constant for C8 inactivation (P28; Fig. 3 C). Essentially, A20 does not compete with CYLD for binding to complex I (P15 ≪ P16), and since CYLD is in great excess relative to complex I (Fig. S6 A), varying CYLD concentration has little to no effect on TTD except for very large reductions, such as a KO (Figs. 3 D and S6 B). Moreover, the exceedingly fast rate of C8 activation (P28 ÷ P27 ≪ 1) leads to a rapid accumulation of active C8a:FLIP heterodimer, which inhibits necroptosis by binding and degrading complex IIb. This essentially sets a “speed limit” on the rate of pMLKL production, i.e., any increase in complex I concentration due to an increase in the concentration of LUBAC, which would be expected to decrease TTD because of the large excess of CYLD, is counteracted by the increased concentration of C8a:FLIP. However, decreasing complex I concentration by knocking down LUBAC would still be expected to increase TTD, as confirmed by our results.
Discussion
A recent review of TNF-induced necroptosis (28) described signaling along the RIP1-RIP3-MLKL axis in terms of at least three major compartmentalization events: TNFR internalization in complex I, multiprotein assembly of complexes IIa and IIb, and necrosome formation leading to translocation of pMLKL to the plasma membrane. Importantly, the authors emphasized that cues and regulation mechanisms underlying these compartmentalization events are poorly understood and proposed that a network of modulators surrounds the necroptotic signaling core (62,63,64), tuned in a context-, cell-type-, and species-dependent manner. The results presented in this work are consistent with this view: a detailed kinetic model comprising core and complementary necroptotic signaling proteins and associated rate constants (Fig. 1; Tables 1 and 2), calibrated to experimental western blot data (Fig. 2, A–C), can produce cell death dynamics via distinct execution modes (Fig. 3, A and B), distinguished by variations in rate constants (Fig. 3 C) and the roles of A20 and CYLD in RIP1 ubiquitination regulation (Fig. 3 D). Moreover, model sensitivity analyses based on changes in TTD (Fig. 2 D) revealed global and mode-specific modulators of necroptosis sensitivity for each mode (Figs. 5 and 6; summarized in Fig. 4). Global modulators include known effectors, such as TNF, TNFR, MLKL, and rate constants associated with these proteins, as well as two additional modulators, the rate constant for RIP1 ubiquitination by cIAP in complex I (P12) and the binding rate constant for LUBAC to complex I (P13). Mode-specific modulators include, for modes 1 and 2, proteins and rate constants involved in RIP1 ubiquitination regulation (A20, cIAP, CYLD, LUBAC, P10, P11, and P15–P20) and, for modes 3 and 4, factors regulating the balance between complex IIb degradation and necrosome formation (C8, LUBAC, RIP1, TRADD, P2–P6, P14, P27, and P28).
Importantly, although model parameters were inferred from experimental data for only a single mouse fibrosarcoma cell line (L929), because the proposed model is mechanistic, based on decades’ worth of experimental studies involving numerous cell types and experimental conditions, as long as the same proteins and interactions are assumed to exist, it can be applied to other cell types and contexts as well. Differences in behaviors can then be attributed to intrinsic differences among the cell types and/or experimental conditions, which can be mathematically accounted for by varying the rate constant values and initial protein concentrations. With this in mind, numerous published experimental studies in mammalian cells (human and mouse) have shown that RIP1 deubiquitination in complex I is driven by A20, CYLD, or both, depending on cell type. For example, Wertz et al. (13) showed that A20 can deubiquitinate RIP1 in human embryonic kidney cells and mouse embryonic fibroblasts (MEFs). In contrast, Feoktistova et al. (65) reported that deletion of A20 in human T lymphocyte cells has no effect on necroptosis sensitivity. Moreover, Moquin et al. (11) reported that RIP1 deubiquitination in MEFs is mediated by CYLD, but proposed that it occurs in the necrosome rather than complex I, since KD of CYLD had no effect on RIP1 deubiquitination. Vanlangenakker et al. (25) showed in mouse fibrosarcoma cells that RIP1 can be deubiquitinated by both A20 and CYLD but, while inhibition of CYLD protects cells from necroptosis, inhibiting A20, counterintuitively, increases sensitivity to necroptosis. They also observed no effect on necroptosis after KD of TRADD. Hitomi et al. (9) showed that increased CYLD expression reduces necroptosis in human T lymphocyte cells. Similarly, Liu et al. (66) showed in hippocampal neurons that KD of CYLD blocks necroptosis and Wright et al. (20) showed that CYLD deubiquitinates RIP1 in human cervical adenocarcinoma (HCAC) cells.
To reconcile these contrasting reports, we have associated with each of the above experimental studies one or more modes of necroptosis execution identified from our model analysis (Table 3). Specifically, the report by Wertz et al. (13) that A20 deubiquitinates RIP1 in human embryonic kidney cells and MEFs implies that knocking down A20 leads to an increase in TTD (i.e., a decrease in necroptosis sensitivity), which is only consistent with mode 1 (Fig. 3 D). Conversely, the reports by Hitomi et al. (9), Liu et al. (66), and Wright et al. (20) all suggest that knocking down CYLD would increase TTD, which could be explained by either modes 2 or 4 (Fig. 3 D). The observation by Feokstitova et al. (65) that deletion of A20 has no effect on necroptosis sensitivity in HCAC cells is intriguing because it is consistent with both modes 3 and 4 (Fig. 3 D) and they used the same cell line (HeLa) as Wright et al. (20), whose observations are consistent with modes 2 and 4 (as mentioned above). This could indicate that HCAC cells (or HeLa cells, specifically) operate via mode 4, since both studies are consistent with this mode, or that the cells in these experiments are operating via different modes of necroptosis execution because of differences in context, i.e., genetic or epigenetic variations between samples or differences in experimental conditions between laboratories. The report by Moquin et al. (11) is interesting because their observation that CYLD binds to complex I but RIP1 ubiquitination is not affected in CYLD-deficient MEFs led them to conclude that RIP1 ubiquitination is regulated by CYLD in the necrosome, rather than complex I. However, our analysis shows these observations are consistent with mode 4, in which TTD increases for CYLD KO (Fig. 3 D) but there is no effect on TTD for CYLD KD < 90% (Fig. 5 B, bottom row, and Fig. S6B), providing an alternative possible explanation.
Table 3.
Multiple experimental studies of necroptosis can be associated with different model-predicted modes of execution
| Refs. | Cell type | Quote(s) from article | Interpretation | Possible execution mode(s) |
|---|---|---|---|---|
| Feoktistova et al. (65) | HCAC (HeLa) | "[T]he deletion of A20 in HeLa or HaCaT cells had no effect on the TNF-mediated cell death sensitivity" | A20 ⇓ TTD ⇔ | M3, M4 |
| Hitomi et al. (9) | HTL (Jurkat) | "[I]nhibition of CYLD expression in Jurkat cells also attenuated necroptosis" | CYLD ⇓ TTD ⇑ | M2, M4 |
| Liu et al. (66) | HCN (HT-22) | "RIP1 and its deubiquitinase CYLD are required for TNF-induced necrosis of HT-22 cells" | CYLD ⇓ TTD ⇑ | M2, M4 |
| Moquin et al. (11) | MEF | "CYLD regulates RIP1 ubiquitination in the TNF-α-induced necrosome, but not in the TNFR-1 signaling complex"" Although CYLD was recruited to TNFR-1 in a ligand-dependent manner, RIP1 ubiquitination was not affected in CYLD−/− MEFs" |
CYLD ⇓ TTD ⇑ | M4 (see text) |
| Vanlangenakker et al. (25) | MFS (L929) | "[W]e and others previously showed that CYLD repression protects L929 cells from TNF-induced necroptosis"" [W]e were surprised to find that A20 depletion had an opposite effect and greatly sensitized the cells to death"" [W]e found that TRADD depletion in L929 cells did not affect TNF-induced necroptosis" |
CYLD ⇓ TTD ⇑ A20 ⇓ TTD ⇓ TRADD ⇓ TTD ⇔ |
M2 |
| Wertz et al. (13) | HEK (HEK293T) | "Co-transfection of wild-type A20 deubiquitinates RIP in HEK293T cells" | A20 ⇓ TTD ⇑ | M1 |
| Wertz et al. (13) | MEF | "However, in the absence of A20, RIP1 will neither be deubiquitinated nor targeted for proteasomal degradation. Indeed, RIP recruited to activated TNFR1 remained hyperubiquitinated and was stabilized in A20−/− MEFs" | A20 ⇓ TTD ⇑ | M1 |
| Wright et al. (20) | HCAC (HeLa) | "RIP1 ubiquitination [was] inhibited by wild-type (Wt) CYLD but not a catalytically inactive CYLD mutant (Mut)" | CYLD ⇓ TTD ⇑ | M2, M4 |
In the second column, the specific cell line used (if applicable) is included in parentheses. HCAC, human cervical adenocarcinoma; HCN, hippocampal neuron; HEK, human embryonic kidney; HTL, human T lymphocyte; MEF, mouse embryonic fibroblast; MFS, mouse fibrosarcoma. ⇓, decrease; ⇑, increase; ⇔, no change.
Of particular interest is the report by Vanlangenakker et al. (25) that, like Hitomi et al. (9), Liu et al. (66), and Wright et al. (20), suggests that knocking down CYLD increases TTD. However, their study also suggests that knocking down A20, counterintuitively, decreases TTD and knocking down TRADD has no effect. Of the four modes of execution identified in this work (Fig. 4), only mode 2 (Figs. 3 D and 5, second and bottom rows), in which RIP1 deubiquitination is significantly faster by CYLD than by A20 (Fig. 3 C), can explain all these observations. As explained above, binding of A20 to complex I in this mode has the effect of slowing down RIP1 deubiquitination and, hence, inhibiting necroptosis. Interestingly, this explanation for the inhibitory effect of A20 reported by Vanlangenakker et al. (25) differs from that offered in Oliver Metzig et al. (26), where we proposed that TNF-induced necroptosis is driven by an incoherent feedforward loop within a dynamic NF-κB-A20-RIPK3 circuit. Specifically, the feedforward loop acts on A20 through the NF-κB pathway, which is not included in the model proposed here. An open question, therefore, is whether these two mechanisms are possible alternative explanations of the same phenomenon or if both exist and operate simultaneously within cells. If the former, it is of interest to determine which (if either) is the “true” mechanism driving this behavior. If the latter, an important question is whether the two mechanisms operate independently or if there is cross talk. Future work will aim to answer these questions by combining additional experimental data with advanced model selection techniques (67), such as nested sampling (68), to quantify likelihoods of alternative model structures.
Evading apoptosis is a hallmark of cancer (5,6,7) and, thus, inducing necroptosis is currently being explored as a potential anticancer treatment (40,57,69). Moreover, inhibiting necroptosis is crucial for treating a variety of inflammatory diseases, including cardiovascular, liver, and neurodegenerative diseases (4,12). Multiple pivot points are also known to exist, allowing cells to switch between prosurvival signaling and various programmed cell death pathways, including apoptosis and necroptosis (29,70,71). Improving our understanding of the molecular pathways that drive necroptosis and other cell death and survival pathways is thus critical for developing improved therapies against these deadly diseases. We have shown that a consensus model of TNF-induced necroptosis can explain a variety of incommensurate and counterintuitive experimental observations reported in the literature and can predict novel molecular targets for modulating necroptosis sensitivity. The proposed model thus establishes a solid foundation on which future model iterations can be built that include cross talk between signaling pathways that drives survival/death decisions in individual cells. To achieve this, the proposed necroptosis model will need to be integrated with models of, e.g., downstream events involved in MLKL-mediated permeabilization of the plasma membrane (72,73) (Golgi-, microtubule-, and actin-dependent mechanisms), prosurvival (1,26) (e.g., NF-κB) and other programmed cell death pathways (e.g., apoptosis, ferroptosis) (29,74), cell-cycle progression (75), and the immune system (40) (e.g., antigen-induced proliferation of T cells). The model can also be expanded to include additional proteins and small molecules known to play a role in necroptosis (76,77) (e.g., ADAM17, CHIP, TAK1, and necrostatins), additional necroptosis-associated receptors (8) (e.g., TNFR2, CD95, and Toll-like receptors) and ligands (78,79,80) (e.g., LPS, FasL, and TRAIL), both forms of cIAP (81) (i.e., cIAP1 and cIAP2), assembly of the LUBAC trimer complex (82), different RIP1 ubiquitin chains (28) (i.e., M1, K48, and K63), and additional biochemical events involved in the activation of caspase-8 (83) (e.g., binding of pro-caspase-8 to FADD, followed by oligomerization and cleavage) and formation of the necrosome (84) (e.g., RIP3 phosphorylation by CK1 family kinases). Eventually, a comprehensive, mechanistic model of cell fate decisions could aid in efforts to develop new drugs for treating inflammatory diseases and cancer by identifying novel molecular targets that shift the balance of cell fates toward either evasion or promotion of cell death.
Materials and methods
Cell culture and reagents
L929 cells (natural cytotoxic T cell clone 929, derivative of strain L) were purchased from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle medium (Corning, NY) supplemented with 10% fetal bovine serum (Omega Scientific, CA), 1% L-glutamine, and 1% penicillin/streptomycin (Thermo Fisher Scientific, MA) at 5% CO2 and 37°C. Mouse recombinant TNF was purchased from R&D Systems (MN) (cat. no. 410-MT-10).
Immunoblotting
L929 cells (2–3 × 106) were grown in 10-cm dishes for 24 h followed by treatment with TNF (0.1, 1, 10, or 100 ng/mL) for 16 h. Dead cells were removed by washing with ice-cold phosphate-buffered saline. Remaining adherent cells were lysed using radioimmunoprecipitation assay buffer with 1% Triton X-100, protease, and phosphatase inhibitors. Samples were normalized for total protein concentration (Bradford assay; Bio-Rad, CA) and to the number of alive (i.e., adherent) cells, denaturated in 3× sodium dodecyl sulfate sample buffer (5 min at 95°C), and subjected to gel electrophoresis (4–15% Criterion TGX Precast Midi Protein Gel; Bio-Rad) and immunoblotting (PVDF Transfer Membrane; Thermo Fisher Scientific). Membranes were blocked in 5% bovine serum albumin/Tris-buffered saline with Tween 20 (TBS-T) and incubated with the following antibodies: pMLKL (1:1000; Abcam, UK, cat. no. ab196436), actin (1:3000; Santa Cruz Biotechnology (TX), cat. no. sc-1615), anti-rabbit (1:5000; Santa Cruz, cat. no. sc-2004), and anti-goat (1:3000; Santa Cruz, cat. no. sc-2354). Signal was developed using chemiluminescent substrate (SuperSignal West Pico Plus; Thermo Fisher Scientific) and visualized with a ChemiDoc MP imaging system (Bio-Rad). At all time points, pMLKL levels were normalized to actin loading control and then ratios calculated with respect to the maximum measured pMLKL level (i.e., at 16 h for 100 ng/mL TNF; Fig. 2 A). Based on previous work (26), where we showed L929 cell viability is <10% at ∼20 h in 10 ng/mL TNF, we can assume this is the maximum pMLKL level that an individual cell can tolerate.
Initial protein concentrations
Initial concentrations and associated molecule counts for all proteins in the model are provided in Table 1. Expression levels for six proteins (caspase-8, FADD, unmodified MLKL, RIP3, TRADD, and TRAF2) were measured directly in untreated L929 cells using absolute protein quantitation mass spectrometry. As a negative control, cells were collected in three replicate six-well plates and cell lysates were gathered, prepped for protein precipitation, pellet, and digestion in the Vanderbilt Mass Spectrometry Research Center (MSRC) Proteomics Core Laboratory. For the other seven proteins (A20, cIAP, CYLD, FLIP, LUBAC, RIP1, and TNFR), initial concentrations were based on typical values reported in the literature (85,86,87). Molecule counts from mass spectrometry measurements were converted to concentrations, and concentrations from the literature converted to molecule counts, assuming an L929 cell of diameter 5 μm (88). For TNF, molecule counts (for doses of 0.1–100 ng/mL) were calculated assuming an enclosing sphere around the cell of diameter 16 μm and a molecular weight of 51.9 kg/mol for the TNF homotrimer (89).
Model simulations and calculating TTD
All model simulations were run in PySB (90) by numerically integrating the governing ordinary differential equations (ODEs) (Tables S1 and S2) using LSODA (91), as implemented in the Python package SciPy (92). Simulations were run for 30 h of simulated time, with outputs every 0.03 h. Following previous work with a model of extrinsically induced apoptosis (86,93), we estimated TTD as the time point at which the amount of pMLKL exceeded a set threshold of 2772 molecules (Fig. 2 D), which is half the amount measured by mass spectrometry (Fig. 2 B). In previous work (26), we showed there is a direct correlation between percent survival and pMLKL level in L929 cells. We used a hard threshold rather than, e.g., the half-maximal amount of pMLKL, so that the conditions for cell death would not change when we varied the initial MLKL amount in the sensitivity analysis (Fig. 5). Furthermore, note that, although we are assuming in the model that the maximum pMLKL level is 100% the initial MLKL level (5544 molecules; Table 1), this choice has little effect on the TTD calculations because of the steep (i.e., “snap-action” (86)) pMLKL response curve (Fig. 2, C and D).
Bayesian parameter calibration
We estimated parameter values using PyDREAM (52), a Python implementation of the differential evolution adaptive metropolis (DREAM) algorithm (94). We utilized pMLKL western blot data at the two highest TNF doses (100 and 10 ng/mL) and defined the multiobjective cost function
| (1) |
where Θ is the parameter set, xm(t,d) and xe(t,d) are model-predicted and experimentally measured pMLKL concentrations, respectively, at time t and TNF dose d, and σ(t,d) = xe(t,d)/10 (following previous studies (53,93,95)). Parameter sampling was performed using 5 Monte Carlo chains, each run for 50,000 iterations. The first 25,000 samples were considered burn-in and discarded, leaving 125,000 parameter sets for further analysis. Out of these, we extracted an ensemble of 10,628 unique parameter sets. Convergence was achieved for all chains (Fig. S1), assessed using the Gelman-Rubin test (96,97). Starting positions for all PyDREAM chains were determined using particle swarm optimization (PSO) (98), i.e., we performed 100 PSO runs of 500 iterations each, saved the parameter sets from the last iteration of each run, and selected the 5 with the lowest cost function values (Eq. 1). Furthermore, for all parameters, we set previous distributions as log-normal distributions, , where pi is the value of the parameter from the i-th PSO run. Starting rate constant values for the PSO runs were set to physically plausible values (99,100): association = 10−6 min−1, dissociation = 10−3 min−1, ubiquitination/phosphorylation = 1 min−1, and degradation = 1 min−1 (see Table 2).
Identifying modes of signal execution in a parameter set ensemble
Modes of signal execution were identified using PyDyNo, a Python-based software package for dynamical systems analysis of biochemical models with uncertain parameters (54). PyDyNo takes as input a model object (PySB (90) or SBML (101,102) formats), an input file with parameter sets, and a target species (pMLKL, in our case). ODE simulations are run (91,92) for all parameter sets and “digitized” into a sequence of integers, termed a “dynamical signature,” based on “dominant” subnetworks of reactions identified at each time point. Basically, the algorithm identifies, at every time point, the subnetwork of reactions that contribute most to either the production or consumption (depending on user preference; production, in our case) of the target species and assigns to each identified subnetwork an integer index. Each time point is thus associated with an integer index and the entire simulated time course with a sequence of integers, i.e., the dynamical signature. We refer the reader to the original work (54) for further details on how PyDyNo identifies dominant subnetworks from ODE simulations of biochemical models. We repeated this procedure for all 10,628 unique parameter sets obtained from PyDREAM, with all simulations run at the highest TNF dose (100 ng/mL) for 16 h simulated time, in line with experimental data (Fig. 2 A). Dynamical signatures were then clustered using a spectral clustering algorithm (103) with the longest common subsequence (55) as the distance metric. The optimal number of clusters, i.e., modes of execution, was determined using a silhouette score (104) for cluster sizes between 2 and 20 (Fig. S4). For each mode, a “representative” dynamical signature was defined as the one with the minimal sum of distances to all other signatures (105) (i.e., the medoid).
Sensitivity analyses for initial protein concentrations and rate constants
We used a sensitivity analysis tool (106) available in PySB to quantify changes in TTD due to changes in both initial protein concentrations and rate constants. In brief, the sensitivity analysis tool varies pairs of protein concentrations or rate constants over a range of values relative to a reference set (in this case [−20%, …, −2%, 0%, 2%, …, 20%]) and calculates the resulting changes in TTD. For each protein or rate constant, a “single-parameter sensitivity multiset” (106) is then obtained, which summarizes the range of changes in TTD due to changes in protein or rate constant values and can be visualized as a boxplot (Figs. 5 A and 6 A). Reference rate constants are those associated with the representative dynamical signatures obtained for each mode from PyDyNo (see previous subsection). For protein concentration sensitivities, reference concentrations are those obtained from mass spectrometry (Fig. 2 B) or the literature (85,86,87) (Table 1) and all simulations were run using the reference rate constant values. Results of the sensitivity analyses using reference rate constant values were validated by performing, over the full set of rate constant values for each mode, in silico KD (70%) and overexpression (10-fold) experiments for protein concentrations and ±10-fold variations for rate constants (Figs. 5 B and 6 B). This was critical for identifying and discarding from our analyses results that were specific only to the reference parameter set.
Data and materials availability
All western blot data, mass spectrometry data, and Python code used in this study, including the PySB encoding of the Necroptosis Execution Reaction Model (NERMv1.0), are available at github.com/LoLab-MSM/NERM.
Author contributions
Conceptualization, A.H., C.F.L., and G.V.I.; methodology, C.F.L., G.V.I., and L.A.H.; investigation, A.H., C.F.L., G.V.I., L.A.H., and M.O.M.; visualization, C.F.L., G.V.I., and L.A.H.; supervision, A.H., C.F.L., and L.A.H.; writing – original draft, C.F.L., G.V.I., and L.A.H.; writing – review & editing, A.H., C.F.L., G.V.I., L.A.H., and M.O.M.
Acknowledgments
This work is dedicated to the memory of our friend and colleague Melaine N. Sebastian. We thank Sam Beik, Sarah Maddox Groves, Corey Hayford, Michael Irvin, Alex Lubbock, Tolu Omokehinde, Oscar Ortega, James Pino, and Vito Quaranta for useful discussions regarding this work. We further thank Hayes McDonald of the Vanderbilt MSRC Proteomics Core Laboratory for help in obtaining the mass spectrometry data. G.V.I. thanks Beth Bowman, Don Brunson, Roger Chalkley, Christina Keeton, Linda Sealy, and Patricia Mueller of Vanderbilt University, and Michael Aldarando-Jeffries, Arlene Olivierre, and Natalia Toro of the University of Central Florida for continued support. This work was supported by the NSF Graduate Research Fellowship Program under grant nos. 1445197 and 1937963 (to G.V.I.), NIH grants R01-AI132731 (to A.H.), R01-AI127867 (to A.H.), K22-CA237857 (to L.A.H.), U54-CA217450 (to C.F.L.), U01-CA215845 (to C.F.L.), and NSF CAREER award MCB 1942255 (to C.F.L.).
Declaration of interests
The authors declare no competing interests.
Editor: Jianhua Xing.
Footnotes
Leonard A. Harris and Carlos F. Lopez contributed equally to this work.
Geena V. Ildefonso’s present address is Department of Biomedical Engineering, University of Southern California, Los Angeles, California
Marie Oliver Metzig’s present address is Institute of Pathology, University Medical Center Mainz, Mainz, Germany
Carlos F. Lopez’s present address is Altos Laboratories, Redwood City, California
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.01.035.
Contributor Information
Leonard A. Harris, Email: harrisl@uark.edu.
Carlos F. Lopez, Email: c.lopez@vanderbilt.edu.
Supporting material
References
- 1.Vanden Berghe T., Kaiser W.J., et al. Vandenabeele P. Molecular crosstalk between apoptosis, necroptosis, and survival signaling. Mol. Cell. Oncol. 2015;2:e975093. doi: 10.4161/23723556.2014.975093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Degterev A., Huang Z., et al. Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 3.Aldridge B.B., Saez-Rodriguez J., et al. Lauffenburger D.A. Fuzzy logic analysis of kinase pathway crosstalk in TNF/EGF/insulin-induced signaling. PLoS Comput. Biol. 2009;5:e1000340. doi: 10.1371/journal.pcbi.1000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanlangenakker N., Vanden Berghe T., Vandenabeele P. Many stimuli pull the necrotic trigger, an overview. Cell Death Differ. 2012;19:75–86. doi: 10.1038/cdd.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 8.Zhou W., Yuan J. Necroptosis in health and diseases. Semin. Cell Dev. Biol. 2014;35:14–23. doi: 10.1016/j.semcdb.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Hitomi J., Christofferson D.E., et al. Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanlangenakker N., Vanden Berghe T., et al. Bertrand M.J.M. cIAP1 and TAK1 protect cells from TNF-induced necrosis by preventing RIP1/RIP3-dependent reactive oxygen species production. Cell Death Differ. 2011;18:656–665. doi: 10.1038/cdd.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moquin D.M., McQuade T., Chan F.K.M. CYLD deubiquitinates RIP1 in the TNFα-induced necrosome to facilitate kinase activation and programmed necrosis. PLoS One. 2013;8:e76841. doi: 10.1371/journal.pone.0076841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi M.E., Price D.R., et al. Choi A.M.K. Necroptosis: a crucial pathogenic mediator of human disease. JCI Insight. 2019;4:e128834. doi: 10.1172/jci.insight.128834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wertz I.E., O’Rourke K.M., et al. Dixit V.M. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 14.Sun S.C. A20 restricts inflammation via ubiquitin binding. Nat. Immunol. 2020;21:362–364. doi: 10.1038/s41590-020-0632-6. [DOI] [PubMed] [Google Scholar]
- 15.Lork M., Verhelst K., Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: so similar, yet so different. Cell Death Differ. 2017;24:1172–1183. doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X., Li W., et al. Han J. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014;24:105–121. doi: 10.1038/cr.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simonson S.J.S., Wu Z.H., Miyamoto S. CYLD: a DUB with many talents. Dev. Cell. 2007;13:601–603. doi: 10.1016/j.devcel.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Vandenabeele P., Galluzzi L., et al. Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 19.Kovalenko A., Chable-Bessia C., et al. Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 20.Wright A., Reiley W.W., et al. Sun S.C. Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev. Cell. 2007;13:705–716. doi: 10.1016/j.devcel.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Gurung P., Man S.M., Kanneganti T.-D. A20 is a regulator of necroptosis. Nat. Immunol. 2015;16:596–597. doi: 10.1038/ni.3174. [DOI] [PubMed] [Google Scholar]
- 22.Lu T.T., Onizawa M., et al. Ma A. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity. 2013;38:896–905. doi: 10.1016/j.immuni.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dondelinger Y., Darding M., et al. Walczak H. Poly-ubiquitination in TNFR1-mediated necroptosis. Cell. Mol. Life Sci. 2016;73:2165–2176. doi: 10.1007/s00018-016-2191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Draber P., Kupka S., et al. Walczak H. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 2015;13:2258–2272. doi: 10.1016/j.celrep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanlangenakker N., Bertrand M.J.M., et al. Vanden Berghe T. TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell Death Dis. 2011;2:e230. doi: 10.1038/cddis.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver Metzig M., Tang Y., et al. Hoffmann A. An incoherent feedforward loop interprets NFκB/RelA dynamics to determine TNF-induced necroptosis decisions. Mol. Syst. Biol. 2020;16:e9677. doi: 10.15252/msb.20209677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M., Beg A.A. Induction of necrotic-like cell death by tumor necrosis factor alpha and caspase inhibitors: novel mechanism for killing virus-infected cells. J. Virol. 2000;74:7470–7477. doi: 10.1128/jvi.74.16.7470-7477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samson A.L., Garnish S.E., et al. Murphy J.M. Location, location, location: a compartmentalized view of TNF-induced necroptotic signaling. Sci. Signal. 2021;14:6178. doi: 10.1126/scisignal.abc6178. [DOI] [PubMed] [Google Scholar]
- 29.Li X., Zhong C.-Q., et al. Han J. RIP1-dependent linear and nonlinear recruitments of caspase-8 and RIP3 respectively to necrosome specify distinct cell death outcomes. Protein Cell. 2021;12:858–876. doi: 10.1007/s13238-020-00810-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickens L.S., Powley I.R., et al. MacFarlane M. The ‘complexities’ of life and death: death receptor signalling platforms. Exp. Cell Res. 2012;318:1269–1277. doi: 10.1016/j.yexcr.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Hughes M.A., Powley I.R., et al. MacFarlane M. Co-operative and hierarchical binding of c-FLIP and caspase-8: a unified model defines how c-FLIP isoforms differentially control cell fate. Mol. Cell. 2016;61:834–849. doi: 10.1016/j.molcel.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shou W., Bergstrom C.T., et al. Skinner F.K. Theory, models and biology. Elife. 2015;4:e07158. doi: 10.7554/eLife.07158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wajant H., Siegmund D. TNFR1 and TNFR2 in the control of the life and death balance of macrophages. Front. Cell Dev. Biol. 2019;7:91. doi: 10.3389/fcell.2019.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Antwerp D.J., Martin S.J., et al. Verma I.M. Suppression of TNF-α-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 35.Pobezinskaya Y.L., Liu Z. The role of TRADD in death receptor signaling. Cell Cycle. 2012;11:871–876. doi: 10.4161/cc.11.5.19300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng L., Bidere N., et al. Lenardo M. Competitive control of independent programs of tumor necrosis factor receptor-induced cell death by TRADD and RIP1. Mol. Cell Biol. 2006;26:3505–3513. doi: 10.1128/MCB.26.9.3505-3513.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X., Shi F., et al. Cao Y. Post-translational modifications as key regulators of TNF-induced necroptosis. Cell Death Dis. 2016;7:e2293. doi: 10.1038/cddis.2016.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Etemadi N., Chopin M., et al. Silke J. TRAF2 regulates TNF and NF-κB signalling to suppress apoptosis and skin inflammation independently of sphingosine kinase. Elife. 2015;4:e10592. doi: 10.7554/eLife.10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallach D. Preparations of lymphotoxin induce resistance to their own cytotoxic effect. J. Immunol. 1984;132:2464–2469. [PubMed] [Google Scholar]
- 40.Gong Y., Fan Z., et al. Liu C. The role of necroptosis in cancer biology and therapy. Mol. Cancer. 2019;18:100. doi: 10.1186/s12943-019-1029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Micheau O., Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 42.Micheau O., Thome M., et al. Grütter M.G. The long form of FLIP Is an activator of caspase-8 at the Fas death-inducing signaling complex. J. Biol. Chem. 2002;277:45162–45171. doi: 10.1074/jbc.M206882200. [DOI] [PubMed] [Google Scholar]
- 43.Tsuchiya Y., Nakabayashi O., Nakano H. FLIP the switch: regulation of apoptosis and necroptosis by cFLIP. Int. J. Mol. Sci. 2015;16:30321–30341. doi: 10.3390/ijms161226232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McIlwain D.R., Berger T., Mak T.W. Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feoktistova M., Geserick P., et al. Leverkus M. cIAPs block ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moriwaki K., Chan F.K.-M. RIP3: a molecular switch for necrosis and inflammation. Genes Dev. 2013;27:1640–1649. doi: 10.1101/gad.223321.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Declercq W., Vanden Berghe T., Vandenabeele P. RIP kinases at the crossroads of cell death and survival. Cell. 2009;138:229–232. doi: 10.1016/j.cell.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Sun L., Wang H., et al. Wang X. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 49.Cho Y.S., Challa S., et al. Chan F.K.M. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ronan T., Qi Z., Naegle K.M. Avoiding common pitfalls when clustering biological data. Sci. Signal. 2016;9:re6. doi: 10.1126/scisignal.aad1932. [DOI] [PubMed] [Google Scholar]
- 51.Pasparakis M., Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517:311–320. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- 52.Shockley E.M., Vrugt J.A., Lopez C.F. PyDREAM: high-dimensional parameter inference for biological models in Python. Bioinformatics. 2018;34:695–697. doi: 10.1093/bioinformatics/btx626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eydgahi H., Chen W.W., et al. Sorger P.K. Properties of cell death models calibrated and compared using Bayesian approaches. Mol. Syst. Biol. 2013;9:644. doi: 10.1038/msb.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ortega O.O., Ozen M., et al. Lopez C.F. Probability-based mechanisms in biological networks with parameter uncertainty. bioRxiv. 2022 doi: 10.1101/2021.01.26.428266. Preprint at. [DOI] [Google Scholar]
- 55.Studer M., Ritschard G. What matters in differences between life trajectories: a comparative review of sequence dissimilarity measures. J. Roy. Stat. Soc. A. 2016;179:481–511. [Google Scholar]
- 56.Rokach L., Maimon O. In: Data Mining and Knowledge Discovery Handbook. Maimon O., Rokach L., editors. Springer-Verlag; 2005. Clustering methods; pp. 321–349. [Google Scholar]
- 57.Wu Y., Dong G., Sheng C. Targeting necroptosis in anticancer therapy: mechanisms and modulators. Acta Pharm. Sin. B. 2020;10:1601–1618. doi: 10.1016/j.apsb.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taxman D.J., Livingstone L.R., et al. Reed W. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 2006;6:7. doi: 10.1186/1472-6750-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moriya H. Quantitative nature of overexpression experiments. Mol. Biol. Cell. 2015;26:3932–3939. doi: 10.1091/mbc.E15-07-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vercammen D., Vandenabeele P., et al. Fiers W. Tumour necrosis factor-induced necrosis versus anti-Fas-induced apoptosis in L929 cells. Cytokine. 1997;9:801–808. doi: 10.1006/cyto.1997.0252. [DOI] [PubMed] [Google Scholar]
- 61.Taraborrelli L., Peltzer N., et al. Walczak H. LUBAC prevents lethal dermatitis by inhibiting cell death induced by TNF, TRAIL and CD95L. Nat. Commun. 2018;9:3910. doi: 10.1038/s41467-018-06155-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murphy J.M., Czabotar P.E., et al. Alexander W.S. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 63.Najafov A., Mookhtiar A.K., et al. Yuan J. TAM kinases promote necroptosis by regulating oligomerization of MLKL. Mol. Cell. 2019;75:457–468.e4. doi: 10.1016/j.molcel.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 64.Sai K., Parsons C., et al. Ninomiya-Tsuji J. Necroptosis mediators RIPK3 and MLKL suppress intracellular Listeria replication independently of host cell killing. J. Cell Biol. 2019;218:1994–2005. doi: 10.1083/jcb.201810014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feoktistova M., Makarov R., et al. Panayotova-Dimitrova D. A20 promotes ripoptosome formation and TNF-induced apoptosis via cIAPs regulation and NIK stabilization in keratinocytes. Cells. 2020;9:351. doi: 10.3390/cells9020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu S., Wang X., et al. He S. Necroptosis mediates TNF-induced toxicity of hippocampal neurons. BioMed Res. Int. 2014;2014:290182. doi: 10.1155/2014/290182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirk P., Thorne T., Stumpf M.P.H. Model selection in systems and synthetic biology. Curr. Opin. Biotechnol. 2013;24:767–774. doi: 10.1016/j.copbio.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 68.Beik S.P., Harris L.A., et al. Lopez C.F. bioRxiv; 2022. Unified Tumor Growth Mechanisms from Multimodel Inference and Dataset Integration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meng M.-B., Wang H.-H., et al. Wang P. Necroptosis in tumorigenesis, activation of anti-tumor immunity, and cancer therapy. Oncotarget. 2016;7:57391–57413. doi: 10.18632/oncotarget.10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dondelinger Y., Jouan-Lanhouet S., et al. Bertrand M.J.M. NF-κB-independent role of IKKα/IKKβ in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol. Cell. 2015;60:63–76. doi: 10.1016/j.molcel.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 71.Verzella D., Pescatore A., et al. Zazzeroni F. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis. 2020;113:210. doi: 10.1038/s41419-020-2399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang H., Sun L., et al. Wang X. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell. 2014;54:133–146. doi: 10.1016/j.molcel.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 73.Samson A.L., Zhang Y., et al. Murphy J.M. MLKL trafficking and accumulation at the plasma membrane control the kinetics and threshold for necroptosis. Nat. Commun. 2020;11:3151. doi: 10.1038/s41467-020-16887-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ouyang L., Shi Z., et al. Bao J.-K. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Csikász-Nagy A. Computational systems biology of the cell cycle. Briefings Bioinf. 2009;10:424–434. doi: 10.1093/bib/bbp005. [DOI] [PubMed] [Google Scholar]
- 76.Chen J., Kos R., et al. Redegeld F. Molecular insights into the mechanism of necroptosis: the necrosome as a potential therapeutic target. Cells. 2019;8:1486. doi: 10.3390/cells8121486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bolik J., Krause F., et al. Schmidt-Arras D. Inhibition of ADAM17 impairs endothelial cell necroptosis and blocks metastasis. J. Exp. Med. 2022;219:e20201039. doi: 10.1084/jem.20201039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Strasser A., Jost P.J., Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–192. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kearney C.J., Cullen S.P., et al. Martin S.J. Necroptosis suppresses inflammation via termination of TNF-or LPS-induced cytokine and chemokine production. Cell Death Differ. 2015;22:1313–1327. doi: 10.1038/cdd.2014.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Füllsack S., Rosenthal A., et al. Siegmund D. Redundant and receptor-specific activities of TRADD, RIPK1 and FADD in death receptor signaling. Cell Death Dis. 2019;10:122. doi: 10.1038/s41419-019-1396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McComb S., Cheung H.H., et al. Sad S. cIAP1 and cIAP2 limit macrophage necroptosis by inhibiting Rip1 and Rip3 activation. Cell Death Differ. 2012;19:1791–1801. doi: 10.1038/cdd.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haas T.L., Emmerich C.H., et al. Walczak H. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 83.Donepudi M., Mac Sweeney A., et al. Grütter M.G. Insights into the regulatory mechanism for caspase-8 activation. Mol. Cell. 2003;11:543–549. doi: 10.1016/s1097-2765(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 84.Hanna-Addams S., Liu S., et al. Wang Z. CK1α, CK1δ, and CK1ε are necrosome components which phosphorylate serine 227 of human RIPK3 to activate necroptosis. Proc. Natl. Acad. Sci. USA. 2020;117:1962–1970. doi: 10.1073/pnas.1917112117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hua F., Cornejo M.G., et al. Lauffenburger D.A. Effects of Bcl-2 levels on Fas signaling-induced caspase-3 activation: molecular genetic tests of computational model predictions. J. Immunol. 2005;175:985–995. doi: 10.4049/jimmunol.175.2.985. [DOI] [PubMed] [Google Scholar]
- 86.Albeck J.G., Burke J.M., et al. Sorger P.K. Modeling a snap-action, variable-delay switch controlling extrinsic cell death. PLoS Biol. 2008;6:2831–2852. doi: 10.1371/journal.pbio.0060299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu J.Q., Pollard T.D. Cell biology: counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 88.Higuchi A., Tsukamoto Y. Cell separation of hepatocytes and fibroblasts through surface-modified polyurethane membranes. J. Biomed. Mater. Res. 2004;71:470–479. doi: 10.1002/jbm.a.30169. [DOI] [PubMed] [Google Scholar]
- 89.Arakawa T., Yphantis D.A. Molecular weight of recombinant human tumor necrosis factor-alpha. J. Biol. Chem. 1987;262:7484–7485. [PubMed] [Google Scholar]
- 90.Lopez C.F., Muhlich J.L., et al. Sorger P.K. Programming biological models in Python using PySB. Mol. Syst. Biol. 2013;9:646. doi: 10.1038/msb.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Petzold L. Automatic selection of methods for solving stiff and nonstiff systems of ordinary differential equations. SIAM J. Sci. Stat. Comput. 1983;4:136–148. [Google Scholar]
- 92.Virtanen P., Gommers R., et al. S. 1. Contributors SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spencer S.L., Gaudet S., et al. Sorger P.K. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vrugt J.A., Ter Braak C.J.F. DREAM(D): an adaptive Markov chain Monte Carlo simulation algorithm to solve discrete, noncontinuous, and combinatorial posterior parameter estimation problems. Hydrol. Earth Syst. Sci. 2011;15:3701–3713. [Google Scholar]
- 95.Kochen M.A., Lopez C.F. A probabilistic approach to explore signal execution mechanisms with limited experimental data. Front. Genet. 2020;11:686. doi: 10.3389/fgene.2020.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vrugt J.A. Markov chain Monte Carlo simulation using the DREAM software package: theory, concepts, and MATLAB implementation. Environ. Model. Software. 2016;75:273–316. [Google Scholar]
- 97.Csete M.E., Doyle J.C. Reverse engineering of biological complexity. Science. 2002;295:1664–1669. doi: 10.1126/science.1069981. [DOI] [PubMed] [Google Scholar]
- 98.Marini F., Walczak B. Particle swarm optimization (PSO). A tutorial. Chemometr. Intell. Lab. Syst. 2015;149:153–165. [Google Scholar]
- 99.Aldridge B.B., Burke J.M., et al. Sorger P.K. Physicochemical modelling of cell signalling pathways. Nat. Cell Biol. 2006;8:1195–1203. doi: 10.1038/ncb1497. [DOI] [PubMed] [Google Scholar]
- 100.Lawson M.J., Petzold L., Hellander A. Accuracy of the Michaelis–Menten approximation when analysing effects of molecular noise. J. R. Soc. Interface. 2015;12:20150054. doi: 10.1098/rsif.2015.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hucka M., Finney A., et al. Wang J., SBML Forum The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 102.Keating S.M., Waltemath D., et al. SBML Level 3 Community members SBML Level 3: an extensible format for the exchange and reuse of biological models. Mol. Syst. Biol. 2020;16:e9110. doi: 10.15252/msb.20199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Von Luxburg U. A tutorial on spectral clustering. Stat. Comput. 2007;17:395–416. [Google Scholar]
- 104.Rousseeuw P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. [Google Scholar]
- 105.Gabadinho A., Ritschard G., et al. Studer M. Analyzing and visualizing state sequences in R with TraMineR. J. Stat. Software. 2011;40:1–37. [Google Scholar]
- 106.Harris L.A., Nobile M.S., et al. Lopez C.F. GPU-powered model analysis with PySB/cupSODA. Bioinformatics. 2017;33:3492–3494. doi: 10.1093/bioinformatics/btx420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Feltham R., Vince J.E., Lawlor K.E. Caspase-8: not so silently deadly. Clin. Transl. Immunology. 2017;6:e124. doi: 10.1038/cti.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oberst A., Dillon C.P., et al. Green D.R. Catalytic activity of the caspase-8–FLIPL complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hsu H., Shu H.-B., et al. Goeddel D.V. TRADD–TRAF2 and TRADD–FADD interactions define two distinct TNF Receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 110.Lawrence C.P., Chow S.C. FADD deficiency sensitises Jurkat T cells to TNF-α-dependent necrosis during activation-induced cell death. FEBS Lett. 2005;579:6465–6472. doi: 10.1016/j.febslet.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 111.Wegner K.W., Saleh D., Degterev A. Complex pathologic roles of RIPK1 and RIPK3: moving beyond necroptosis. Trends Pharmacol. Sci. 2017;38:202–225. doi: 10.1016/j.tips.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grootjans S., Vanden Berghe T., Vandenabeele P. Initiation and execution mechanisms of necroptosis: an overview. Cell Death Differ. 2017;24:1184–1195. doi: 10.1038/cdd.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.O’Donnell M.A., Ting A.T. RIP1 comes back to life as a cell death regulator in TNFR1 signaling. FEBS J. 2011;278:877–887. doi: 10.1111/j.1742-4658.2011.08016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vandenabeele P., Declercq W., et al. Vanden Berghe T. The role of the kinases RIP1 and RIP3 in TNF-induced necrosis. Sci. Signal. 2010;3:re4. doi: 10.1126/scisignal.3115re4. [DOI] [PubMed] [Google Scholar]
- 115.Feng S., Yang Y., et al. Wu M. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell. Signal. 2007;19:2056–2067. doi: 10.1016/j.cellsig.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 116.He S., Wang L., et al. Wang X. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 117.Carswell E.A., Old L.J., et al. Williamson B. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl. Acad. Sci. USA. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hsu H., Huang J., et al. Goeddel D.V. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 119.Wang C.Y., Mayo M.W., et al. Baldwin A.S. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.