Abstract
The Saccharomyces cerevisiae RSP5 gene encodes an essential HECT E3 ubiquitin-protein ligase. Rsp5p contains an N-terminal C2 domain, three WW domains in the central portion of the molecule, and a C-terminal catalytic HECT domain. A diverse group of substrates of Rsp5p and vertebrate C2 WW-domain-containing HECT E3s have been identified, including both nuclear and membrane-associated proteins. We determined the intracellular localization of Rsp5p and the determinants necessary for localization, in order to better understand how Rsp5p activities are coordinated. Using both green fluorescent protein fusions to Rsp5p and immunogold electron microscopy, we found that Rsp5p was distributed in a punctate pattern at the plasma membrane, corresponding to membrane invaginations that are likely sites of endosome formation, as well as at perivacuolar sites. The latter appeared to correspond to endocytic intermediates, as these structures were not seen in a sla2/end4-1 mutant, and double-immunogold labeling demonstrated colocalization of Rsp5p with the endosomal markers Pep12p and Vps32p. The C2 domain was an important determinant of localization; however, mutations that disrupted HECT domain function also caused mislocalization of Rsp5p, indicating that enzymatic activity is linked to localization. Deletion of the C2 domain partially stabilized Fur4p, a protein previously shown to undergo Rsp5p- and ubiquitin-mediated endocytosis; however, Fur4p was still ubiquitinated at the plasma membrane when the C2 domain was deleted from the protein. Together, these results indicate that Rsp5p is located at multiple sites within the endocytic pathway and suggest that Rsp5p may function at multiple steps in the ubiquitin-mediated endocytosis pathway.
Eucaryotic cells internalize extracellular materials and portions of the cell surface through the process of endocytosis. This provides for the selective uptake of nutrients as well as for down-regulation of membrane receptors, permeases, and channel proteins (41, 51). In higher eucaryotic cells, membrane proteins to be internalized are surrounded by an area of plasma membrane that buds off inside the cell in a process that is mediated by clathrin and many accessory factors (58). The protein contents of endocytic vesicles are subsequently sorted and trafficked to the vacuole or lysosome for degradation, or, alternatively, recycled to the plasma membrane (54, 75). An interesting link has emerged between the lysosomal-vacuolar degradation pathway and the other major cellular pathway for protein degradation, the ubiquitin proteolysis system. The ubiquitin proteolysis system, in its most common form, consists of two general steps: the covalent conjugation of ubiquitin to substrate proteins and the recognition and degradation of the ubiquitinated proteins by the 26S proteasome (25). It was therefore surprising to find that the rapid degradation of several plasma membrane proteins was dependent on ubiquitin conjugation and vacuolar proteases, rather than on the 26S proteasome. This has led to the model in which ubiquitination can serve as a degradation signal for membrane proteins by serving as a signal for endocytosis (21, 22). Ubiquitin-mediated endocytosis is emerging as a common pathway for regulated clearance of proteins from the plasma membrane in both yeast and mammalian cells; however, many aspects of this pathway remain poorly understood.
An early indication that ubiquitination could serve as a signal for vacuolar degradation came from studies with the yeast Ste6p a-factor transporter (34). It was shown that Ste6p accumulates in a ubiquitinated form in endocytosis mutants and that a ubc4,5 mutant exhibits delayed degradation of Ste6p and accumulation of Ste6p at the cell surface. Studies with the Ste2p α-factor receptor definitively showed that ubiquitination was required for its ligand-stimulated endocytosis (23), leading to the model that ubiquitination of membrane proteins can serve as a signal for endocytosis and degradation in the vacuole. Other proteins undergoing ubiquitin-mediated endocytosis in Saccharomyces cerevisiae include the Ste3p a-factor receptor (55, 56), the Fur4p uracil permease (14, 15), the Gap1p general amino acid permease (62), the Gal2p galactose permease (27), the Mal61p maltose permease (43), the multidrug resistance-like transporter Pdr5p/Sts1p (9), and the Tat2p tryptophan permease (5). In addition, examples of ubiquitinated cell surface proteins targeted for lysosomal degradation in mammalian cells include the amiloride-sensitive epithelial sodium channel, colony-stimulating factor 1 receptor, the epidermal growth factor receptor, and the growth hormone receptor (reference 21 and references therein). In some cases, phosphorylation of the membrane proteins appears to be a prerequisite or a signal for ubiquitination (24, 40).
Protein ubiquitination cascades begin with the E1 ubiquitin-activating enzyme, which activates ubiquitin in an ATP-dependent reaction by forming a thioester bond with the C terminus of ubiquitin. E1 then transfers ubiquitin to the active-site cysteine of one of a number of E2 ubiquitin-conjugating enzymes, maintaining a thioester linkage. E2 enzymes cooperate with a diverse set of E3 ubiquitin-protein ligases to catalyze substrate ubiquitination, with E3 enzymes providing the direct link to the substrate (20). In the case of the HECT family of E3 proteins, the E2 transfers ubiquitin to an active-site cysteine within the HECT domain, again in the form of a thioester, and the E3 then directly catalyzes substrate ubiquitination (57). In the case of other types of E3 proteins (SCF E3s, APC-like E3s, VHL, and Cb1/RING finger E3s), the E3 appears to act more as a docking protein for both the E2 and substrate, with ubiquitination being catalyzed directly by the E2 (36, 37, 42, 46, 74). E3s or E2-E3 complexes generally link multiple ubiquitin molecules to the substrate in the form of polyubiquitin chains, with lysine side chains of ubiquitin serving as acceptor sites for additional ubiquitin molecules. Polyubiquitin chains linked through Lys48 are required for recognition by the proteasome (47); however, ubiquitin-mediated endocytosis can in some cases be triggered by monoubiquitination or by short ubiquitin chains linked through Lys63 of ubiquitin (14, 60, 64, 68).
In yeast, the essential Rsp5p HECT E3 has been shown to be important for ubiquitin-mediated endocytosis of several proteins, including Gap1p (63), Fur4p (15, 18, 19), Mal61p (43), the hexose transporter (Hxt6/7) (35), and Tat2p (5). Deletion of the C2 domain of Rsp5p was shown to result in ubiquitination without subsequent endocytosis of Gap1p (63). This suggested that Rsp5p may play a broader role in endocytosis, which is an idea that is further supported by the localization results presented here. Rsp5p also has non-membrane-associated substrates, including at least two nuclear proteins, the largest subunit of RNA polymerase II (Rpb1p) (31) and Rfa1p, a subunit of replication protein A (10). Ubiquitination of Rpb1p is stimulated by UV irradiation, suggesting that this may be part of the response to DNA damage (3). Rsp5p also plays important roles in the mitochondrion-cytoplasm distribution of Mod5p (a tRNA-modifying enzyme) and in the inheritance of mitochondria to daughter cells (12, 79), although the direct substrate(s) of Rsp5p related to these events is not known. Recent work has shown that the essential function of Rsp5p is the ubiquitination of the Spt23p and Mga2p transcription factors (26), which control transcription of the OLE1 gene, which encodes an enzyme required for synthesis of oleic and palmitoleic acid. This appears to be an example of ubiquitination serving as a signal for proteolytic processing of the substrate, which releases Spt23p from its endoplasmic reticulum tether so that it is free to enter the nucleus and activate OLE1 transcription. This link of Rsp5p to unsaturated fatty acid synthesis may also explain the effect of Rsp5p on mitochondrial distribution, since ole1 mutants are also defective in this process (65).
Rsp5p is one of the smallest HECT E3s described (92 kDa), with the largest HECT E3s being over 500 kDa. The HECT E3s have only a conserved C-terminal 40-kDa catalytic domain in common (29). The large and divergent N-terminal regions of these proteins have been proposed to contain determinants for substrate recognition, based largely on work with yeast Rsp5p and human E6AP (30, 73). Rsp5p contains a C2 domain at its extreme N terminus and three WW domains between the C2 domain and the HECT domain. C2 domains mediate interactions with membranes by binding to phospholipids, inositol polyphosphates, and/or proteins, often in a Ca2+-dependent manner (48, 52). WW domains are approximately 30-amino-acid protein-protein interaction modules that recognize polyproline ligands (7), which are similar to but distinct from SH3 ligands (45). Structure-function studies demonstrated that the HECT domain and the region spanning the second and third WW domains are sufficient for complementation of rsp5-1 temperature sensitivity (73), while Hoppe et al. showed that only the third WW domain and the HECT domain are required for the essential function of Rsp5p at standard growth temperatures (26).
The diverse set of functions and substrates of Rsp5p led us to explore the intracellular localization of Rsp5p and the determinants responsible for its localization. We show here that Rsp5p is found primarily at sites consistent with its role in ubiquitin-mediated endocytosis. Both the C2 domain and an intact catalytic HECT domain are critical for proper localization as well as for normal turnover of Fur4, a membrane-associated substrate.
MATERIALS AND METHODS
Yeast strains and plasmids.
Yeast genetic manipulations and media were according to standard methods (17). Generation of haploid strain GW003 was described previously (73). Other S. cerevisiae strains used in these studies are listed in Table 1. The centromere-containing plasmid pFL38gF (URA3 GAL-Fur4) (59) carries the Fur4 gene (33) under the control of the GAL10 promoter and was used in experiments that examined Fur4p protein levels and activities. For these experiments, cells were grown at 30°C in synthetic medium supplemented with 4% galactose and 0.05% glucose plus appropriate nutrients, including Casamino Acids (0.2%) without tryptophan.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Plasmid |
|---|---|---|
| GW003 | MATarsp5Δ::LEU2 ade2-1 his3-11 ura3-1 trp1-1 can1-100 | pRS416-RSP5 URA3 |
| GW047 | MATarsp5Δ::LEU2 ade2-1 his3-11 ura3-1 trp1-1 can1-100 | pRS414-HA-RSP5 TRP1 |
| GW048 | MATarsp5Δ::LEU2 ade2-1 his3-11 ura3-1 trp1-1 can1-100 | pRS414-rsp5-1(ts) TRP1 |
| GW050 | MATarsp5Δ::LEU2 ade2-1 his3-11 ura3-1 trp1-1 can1-100 | pRS414-HA-RSP5ΔC2 TRP1 |
| GW072 | MATarsp5Δ::LEU2 ade2-1 his3-11 ura3-1 trp1-1 can1-100 | pRS414gal-GFP-HA-RSP5 TRP1 |
| GW073 | MATarsp5Δ::LEU2 ade2-1 his3-11 ura3-1 trp1-1 can1-100 | pRS414gal-GFP-HA-RSP5ΔC2 TRP1 |
| GW082 | MATarsp5Δ::LEU2 ade2-1 his3-11 ura3-1 trp1-1 can1-100 | pRS414gal-GFP-rsp5-1(ts) TRP1 |
| GW100 | MATasla2/end4-1 | pRS414-gal-GFP-HA-RSP5 TRP1 |
Wild-type green fluorescent protein (GFP) under control of the GAL1 promoter was cloned into the pRS414 vector (containing a centromere and TRP1-selectable marker), generating pRS414galGFP. The pRS414gal-GFP-RSP5, -rsp5-1, and -RSP5ΔC2 plasmids were generated by subcloning the EcoRI/NotI fragments of pYES2-RSP5, rsp5-1, and RSP5ΔC2 plasmids (73) into pRS414galGFP. These were transformed into yeast strain GW003, followed by counterselection with 5-fluoroorotic acid, generating the GW072, GW082, and GW073 strains. These were maintained in rich galactose-containing medium. For observation with fluorescence microscopy, cells were grown in trp- and leu-dropout media with 2% galactose supplemented with adenine (80 μg/ml, final concentration).
The plasmid YEp96 (2μm TRP1 UB) contains a synthetic yeast ubiquitin gene under the control of the copper-inducible CUP1 promoter (8). The multicopy plasmid YEp96-fF (2μm URA3 CUP1-Ub FUR4) was constructed as follows. The CUP1-UB gene (including CYC1 terminator) was amplified by PCR using YEp96 as a template and the oligonucleotides L1 (CGGGATCCCATTACCGACATTTC) and L2 (AAGCTTGCAAATTAAAGCCTTCGAGCGCG). The resulting PCR fragment was digested with BamHI and cloned at the unique BamHI site of the plasmid YEp352fF (2μm URA3 FUR4) (15).
Immunoblotting.
Yeast protein extracts for immunoblotting (see Fig. 1) were made as described previously (73). Primary antibodies were anti-GFP rabbit polyclonal antibody (Santa Cruz Biotechnology) and anti-Rsp5p mouse monoclonal antibody (31). Fur4p was detected using an antiserum to the last 10 residues of uracil permease.
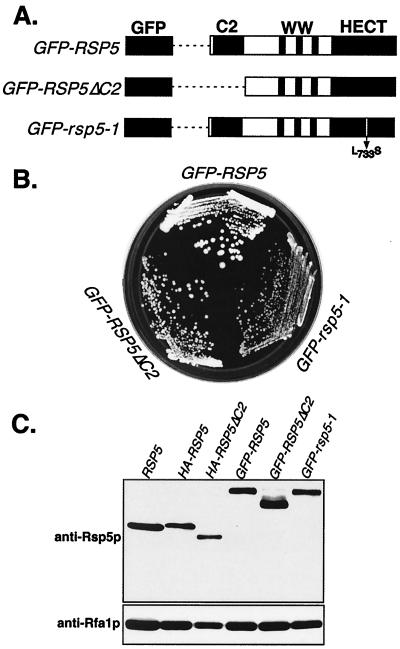
FIG. 1.
GFP-Rsp5p fusion proteins are functional in yeast. (A) Schematic representation of GFP fused to wild-type Rsp5p, Rsp5ΔC2p, and the rsp5-1p temperature-sensitive mutant allele. (B) Growth of haploid rsp5Δ cells expressing the indicated plasmid-borne GFP fusion proteins. Cells expressing GFP alone were inviable. (C) Western analysis to confirm the expression of full-length GFP fusion proteins, by using anti-Rsp5p antibody. The first lane shows Rsp5p expression in the undisrupted W303 strain; all other lanes are in the rsp5Δ background expressing the indicated plasmid-borne HA- or GFP-tagged Rsp5p proteins. The lower panel shows levels of a control protein, Rfa1p.
Uracil uptake.
Uracil uptake was measured in exponentially growing cells as previously described (72). One milliliter of yeast culture was incubated with 5 μM [14C]uracil (Amersham) for 20 s at 30°C and then quickly filtered through Whatman GF/C filters which were washed twice with ice-cold water. Uptake of [14C]uracil was quantitated by scintillation counting.
Fluorescence microscopy and EM.
GFP fluorescence was observed in living yeast using an Olympus IX-70 microscope at either ×600 or ×1,000 magnification. The immunogold electron microscopy (EM) on yeast cells was performed essentially as described previously (50). Exponentially growing cells at 30°C were pelleted at 300 × g for 5 min, resuspended in 4% formaldehyde in 1× phosphate-buffered saline (PBS; pH 7.4), and allowed to fix for 1 h at room temperature followed by 18 to 24 h at 4°C. The cells were then washed briefly in PBS and resuspended in 1% low-melting-temperature agarose. After cooling, the agarose blocks were trimmed into 1-mm3 pieces; cryoprotected by infiltration with a mixture of 2.3 M sucrose–20% polyvinylpyrrolidone (10K) (pH 7.4) for 2 h; and mounted onto cryopins and rapidly frozen in liquid nitrogen. Ultrathin cryosections were cut on a Leica UCT ultramicrotome equipped with an FC-S cryoattachment and collected onto formvar-carbon-coated nickel grids. The nickel grids were washed through several drops of 1× PBS containing 5% fetal calf serum (FCS), 10 mM glycine (pH 7.4); blocked in 10% FCS for 30 min; and incubated overnight in 20 μg of monoclonal antihemagglutinin (anti-HA) antibody (Babco) per ml, anti-GFP monoclonal antibody (Santa Cruz Biotechnology), anti-Vps32p rabbit polyclonal antibody (provided by Markus Babst and Scott Emr), or anti-Pep12p rabbit polyclonal antibody (provided by Chris Burd and Scott Emr). After washing, the grids were incubated for 2 h in 5-nm-diameter Au–donkey anti-mouse antibody conjugate or 10-nm-diameter Au–anti-rabbit antibody conjugate (Jackson Immunoresearch Labs). The grids were then washed through several drops of PBS, followed by several drops of double-distilled H2O, and subsequently embedded in an aqueous solution containing 3.2% polyvinyl alcohol (10K), 0.2% methyl cellulose (400 centiposes), and 0.1% uranyl acetate. The grids were observed and photographed on a Philips 420 transmission electron microscope at 80 kV.
Fractionation by differential centrifugation.
GW047 and GW050 cells expressing the HA-Rsp5p and HA-Rsp5ΔC2p proteins, respectively, were grown to mid-log phase, spheroplasted, and homogenized in lysis buffer (0.2 M sorbitol, 50 mM potassium acetate, 20 mM HEPES, 2 mM EDTA [pH 6.8], with protease inhibitors) with 10 strokes in a Thomas Dounce homogenizer (76). The lysate was subjected to differential centrifugations of 300 × g for 5 min, 13,000 × g for 10 min, and 100,000 × g for 1 h. For detergent solubilizations, the cells were lysed in the above buffer with 1% Triton X-100 and held for 30 min on ice prior to centrifugation. The supernatant and pellet fractions were precipitated on ice with 10% trichloroacetic acid and washed twice with ice-cold acetone, and protein pellets were solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Proteins were separated by SDS–10% PAGE and transferred to nitrocellulose for Western blotting with antibodies against the HA epitope to detect HA-Rsp5p, Pep12p, and glucose-6-phosphate dehydrogenase (G6PDH) (Sigma, St. Louis, Mo.). Each fraction analyzed represented protein from an equivalent number of cells.
Equilibrium density centrifugation.
Cell organelles were fractionated on equilibrium density gradients essentially as described previously (34, 53). Exponentially growing cultures (A600, 40) were arrested by the addition of 10 mM sodium azide, washed once in 10 mM sodium azide, and broken by vigorous agitation after the addition of 0.2 ml of glass beads and 0.2 ml of STET (10% [wt/wt] sucrose, 10 mM Tris-HCl [pH 7.6], 10 mM EDTA plus protease inhibitors [Complete cocktail; Boehringer]). After low-speed centrifugation (1,000 × g, 3 min), 0.4 ml of the cleared extracts was layered on top of a 5-ml 20-to-60% linear sucrose gradient made up in 10 mM Tris-HCl (pH 7.6), 10 mM EDTA. Samples were centrifuged for 18 h at 100,000 × g in a SW50.1 rotor (Beckman). Fractions were collected from the top of the gradient, and proteins were precipitated with 10% trichloroacetic acid. After 30 min of incubation on ice, proteins were pelleted by centrifugation and resuspended in 40 μl of 1 M Tris base plus 80 μl of 2× sample buffer (100 mM Tris-HCl [pH 6.8], 4 mM EDTA, 4% SDS, 20% glycerol, 0.002% bromophenol blue) containing 2% 2-mercaptoethanol and heated at 95°C for 4 min. Proteins in each gradient fraction were analyzed by Western blotting. Monoclonal antibodies against Pep12p, an integral membrane protein marker of late endosome, and Vat2p, a vacuolar membrane protein, were obtained from Molecular Probes (Eugene, Oreg.). Polyclonal antibodies against the plasma membrane [H+] ATPase were a gift from C. Slayman, and polyclonal antibodies against Sss1p, an integral membrane protein of the endoplasmic reticulum (11), were a gift from F. Kepes.
In vivo Fur4p ubiquitination.
Induction of the CUP1 promoter was for 1 h in the presence of 0.1 mM CuSO4. Yeast cells (A600, 40) in the exponential growth phase were harvested by centrifugation in the presence of 10 mM sodium azide, washed once in distilled water plus 10 mM sodium azide, and used to prepare membrane-enriched fractions (13,000 × g pellet; P13) as previously described (15), except that lysis buffer (0.1 M Tris-HCl [pH 7.5], 0.15 M NaCl, 5 mM EDTA, plus a mixture of protease inhibitors [Complete; Roche]) containing freshly prepared N-ethylmaleimide (25 mM) in order to prevent deubiquitination.
RESULTS
Expression of GFP-Rsp5p proteins in yeast.
Plasmid constructs were generated for expression of GFP-Rsp5p fusion proteins as shown in Fig. 1A. The GFP open reading frame (ORF) was fused in frame to the 5′ end of the full-length RSP5 ORF, the RSP5-ΔC2 ORF (with amino acids 2 to 138 deleted), and the rsp5-1 ORF, corresponding to a temperature-sensitive allele. The fusion proteins were expressed from a TRP1-based centromere-containing vector under control of the GAL1 promoter. Each plasmid was transformed into the rsp5Δ haploid strain (GW003), which harbored a complete deletion of the chromosomal RSP5 gene (rsp5Δ) and a URA3-based plasmid copy of wild-type RSP5, whose expression was driven by the natural RSP5 promoter. Following transformation, loss of the URA3-based plasmid was selected on galactose- and 5-fluoroorotic acid-containing plates. Viable cells were obtained from all three transformants but not from a transformant expressing only GFP, indicating that all three GFP-Rsp5p proteins were capable of supporting the essential in vivo function of Rsp5p (Fig. 1B). The GFP–rsp5-1 strain still displayed temperature-sensitive growth like that of the original rsp5-1 mutant. Immunoblotting with anti-Rsp5p antibody confirmed that each transformant expressed the predicted full-length GFP-Rsp5p fusion protein (Fig. 1C) and that this was the sole source of Rsp5p in the cells. Thus, fusion of GFP to the N terminus of Rsp5p does not interfere with the essential function of Rsp5p and, as shown below (see Fig. 8), GFP-Rsp5p also fully supports normal turnover of a membrane-associated substrate, Fur4p. The amounts of GFP-Rsp5p fusion proteins expressed in the transformants were very similar, if not slightly lower, than the amount of Rsp5p expressed in the parental haploid strain expressing Rsp5p from its natural chromosomal position. The only exception was GFP-Rsp5ΔC2, which was expressed two- to threefold higher than Rsp5p. Similar expression levels were also seen with GAL1 promoter-driven HA-tagged Rsp5p proteins, as shown in Fig. 1C and as reported previously (73).
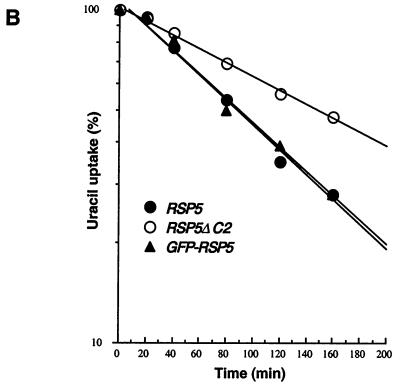
FIG. 8.
Normal turnover of Fur4p, but not ubiquitination, is dependent on C2 domain. Strains GW047 (RSP5), GW050 (RSP5-ΔC2), and GW072 (GFP-RSP5) were transformed with plasmid p38gF (GAL10-FUR4) and grown as described in Materials and Methods. (A) Cycloheximide (CHX; 100 μg/ml) was added to log-phase cultures, and extracts were prepared at the indicated times after CHX addition. Fur4p protein levels were determined by immunoblotting. A control protein remained stable during the experiment. (B) Uracil uptake was measured at the times indicated after addition of CHX. Results are expressed as a percentage of the activity at time zero. ●, GW047 (RSP5), ○, GW050 (RSP5-ΔC2), ▴, GW072 (GFP-RSP5). (C) Deletion of C2 domain does not impair Fur4p ubiquitination. GWO47 (RSP5) and GW050 (RSP5ΔC2) cells transformed with YEp96fF (2μm URA3 FUR4 CUP1-UB) were grown in yeast nitrogen base medium plus glucose as carbon source, and ubiquitin overexpression was induced for 1 h in the presence of CuSO4 (0.1 mM). Cells were collected during the exponential growth phase (A600, 0.8) and used to prepare membrane-enriched fractions (P13). Aliquots of the P13 pellets were analyzed by Western immunoblotting for uracil permease. Longer exposure of the blot revealed in both types of cells a small amount of additional Ub-Fur4p species of higher molecular weight.
The C2 domain is a determinant of Rsp5p localization.
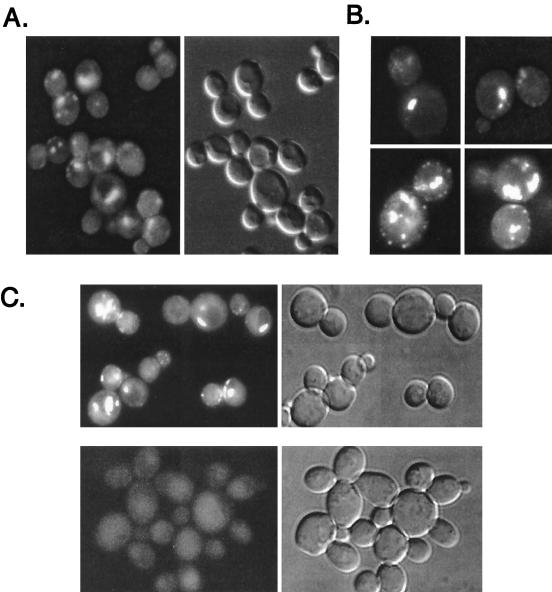
Cells expressing the GFP-Rsp5p fusion protein were examined by using fluorescence and differential interference contrast (DIC) microscopy. The GFP-RSP5 wild-type (wt) strain exhibited punctate fluorescence on the plasma membrane and at structures within the cytoplasm and adjacent to the vacuole (Fig. 2A and B). DAPI (4′,6′-diamidino-2-phenylindole) staining showed that GFP-Rsp5p was not detectably present in the nucleus, and 4-nitroquinoline oxide or UV irradiation did not induce visible nuclear localization (data not shown), a possibility suggested by the fact that UV irradiation stimulates ubiquitination of Rpb1p (3). While the C2 domain of Rsp5p is not required for cell viability (63, 73), deletion of the C2 domain dramatically affected distribution of a GFP fusion protein, as shown in Fig. 2C. GFP-Rsp5ΔC2p appeared to be distributed diffusely throughout the cell, with loss of specific localization at the plasma membrane and at internal structures.
FIG. 2.
Localization of GFP-Rsp5p and GFP-Rsp5p-ΔC2. GW072 cells expressing GFP-Rsp5p were examined by fluorescence (A and B) and DIC (A, right panel) microscopy. (C) Top panels show GW072 cells (GFP-Rsp5p) and bottom panels show GW073 cells (GFP- Rsp5p-ΔC2) as seen with fluorescence (left panels) or DIC (right panels) microscopy.
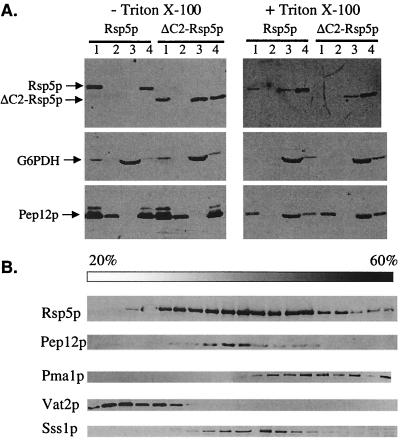
Cell fractionation experiments were done to further explore the effect of C2 deletion on Rsp5p localization. Cell lysates from strains GW047 and GW050, expressing HA-tagged Rsp5p and Rsp5ΔC2p, respectively, were subjected to differential centrifugation and fractions were analyzed for Rsp5p by immunoblotting (Fig. 3A), using G6PDH and Pep12p as markers for soluble cytoplasmic and endosomal proteins, respectively. Wild-type Rsp5p was found in both low-speed (300 × g) and high-speed (100,000 × g) pellet fractions (P3 and P100 fractions) and was not detected in the 100,000 × g supernatant (S100), indicating that nearly all of the Rsp5p is associated with larger structures. Triton X-100 (1%) solubilized a large fraction of the Rsp5p in the low-speed pellet, suggesting that the Rsp5p in this fraction was peripherally associated with membranes. Pep12p was split evenly between low- and high-speed pellet fractions, and detergent solubilized the material in both of these fractions, consistent with previous reports for Pep12p fractionation (4, 16). The fractionation of Rsp5p with the high-speed pellet was therefore consistent with possible endosome localization; however, the Rsp5p present in this fraction was not solubilized by 1% Triton X-100. This suggests that Rsp5p is linked to the P100 fraction by protein-protein interactions and therefore that Rsp5p in this fraction probably does not reflect endosome association. Rsp5p is a multifunctional protein, having both nuclear and endoplasmic reticulum-associated substrates in addition to plasma membrane substrates, and it is possible that Rsp5p present in the P100 fraction reflects its role in another process.
FIG. 3.
Fractionation of Rsp5p and Rsp5p-ΔC2. (A) GW047 and GW050 cells expressing HA-Rsp5p and HA-Rsp5ΔC2p were spheroplasted and lysed, and lysates were incubated without (left panel) or with (right panel) 1% Triton X-100 prior to sequential differential centrifugation. Immunoblotting of fractions was performed with antibodies against Rsp5p, G6PDH, and Pep12p. G6PDH and Pep12p served as controls for soluble and vacuolar proteins, respectively. Lanes 1 to 4 represent the 300 × g pellet (lane 1), the 13,000 × g pellet (lane 2), the 100,000 × g supernatant (lane 3), and the 100,000 × g pellet (lane 4). Each lane represents protein from an equivalent number of cells. (B) Cell lysates were prepared from exponentially growing wild-type RSP5 cells (W303) in yeast nitrogen base medium with glucose as a carbon source. Lysate was fractionated on a 20-to-60% sucrose density gradient. Aliquots of the various fractions were analyzed by immunoblotting for Rsp5p, Pma1p (plasma membrane marker), Pep12p (late endosome marker), Vat2p (vacuolar marker), and Sss1p (endoplasmic reticulum marker).
In contrast to the wild-type protein, a significant fraction of Rsp5ΔC2p was found in the high-speed supernatant in the absence of detergent treatment. The protein remaining in the low-speed pellet was, like the wild-type protein, solubilized by 1% Triton X-100, while the high-speed pellet material was again resistant. These results indicate that Rsp5p is associated with membrane-containing fractions that are dependent in part on the C2 domain. Sedimentation of Rsp5p was also examined by sucrose gradient fractionation (Fig. 3B). These results also indicated that Rsp5p is associated with large structures. It was distributed in fractions containing endosomes (Pep12p-positive fractions), although Golgi- and endoplasmic reticulum-containing fractions (Sss1p positive) were not clearly resolved from endosomes by this technique. In addition, the broad distribution of Rsp5p overlapped with plasma membrane-containing fractions (Pma1p positive). Deletion of the C2 domain had essentially no effect on sucrose gradient sedimentation (results not shown).
Rsp5p catalytic activity is a determinant of localization.
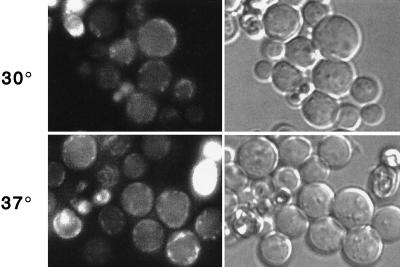
To test whether Rsp5p catalytic activity was linked to its localization, we observed cells expressing the GFP–rsp5-1 temperature-sensitive allele. This allele contains a Leu733Ser substitution within the HECT domain that impairs ubiquitin-thioester formation in vitro (73). Based on analogy to the E6AP HECT domain crystal structure (28), this alteration is likely to destabilize the folded state of the hydrophobic core of the C lobe of the HECT domain. At the permissive temperature (30°), GFP-rsp5-1p was localized primarily at the plasma membrane; however, it was in a less punctate distribution than that of GSP-Rsp5p (wt) and in larger patches on the membrane (Fig. 4). In addition, the strongly fluorescing structures adjacent to the vacuole were less prominent. After shifting to the nonpermissive temperature (37°) for 4 h, the signal was observed almost exclusively at the plasma membrane. The same temperature shift did not detectably affect localization of wild-type GFP-Rsp5p. Therefore, the catalytic activity of Rsp5p is a determinant of its localization. We also observed RSP5 cells expressing GFP fusions to the active-site cys-to-ala mutant of Rsp5p or to a mutant lacking the last six amino acids. Both of these proteins are catalytically inactive and noncomplementing (73); however, their overall structure, based on the X-ray crystal structure of the HECT domain of E6AP (28), is unlikely to be affected by the mutations. Both of these proteins were localized similarly to the rsp5-1p protein (data not shown), further indicating that the catalytic activity of Rsp5p is linked to its localization.
FIG. 4.
Localization of temperature-sensitive GFP–rsp5-1p. Fluorescence and phase-contrast microscopy of cells at 30 and 37°C (top and bottom, respectively), expressing GFP-Rsp5p (GW072; left) or GFP–Rsp5-1p (GW082; right).
Localization of Rsp5p is affected in a sla2/end4-1 mutant.
The sla2/end4-1 mutant is defective in the internalization step of endocytosis, and normal turnover of many plasma membrane proteins is blocked in sla2/end4 mutants (49). GFP-Rsp5p (wt) was expressed in the sla2/end4-1 temperature-sensitive mutant to determine if the cytoplasmic sites of GFP-Rsp5p localization might correspond to endocytic compartments downstream of SLA2/END4 function. While GFP-Rsp5p was still localized at the plasma membrane in the sla2/end4-1 mutant, virtually all localization to internal structures was lost at both 30° and 37°C (Fig. 5). This suggests that the cytoplasmic Rsp5p-containing perivacuolar structures may be intermediate or late endocytic compartments, while the peripheral compartments do not require SLA2/END4 function for Rsp5p localization.
FIG. 5.
Localization of GFP-Rsp5p in the sla2/end4-1 mutant. Plasmid pRS414gal-GFP-RSP5 was introduced into sla2/end4-1 cells, and cells were grown in dextrose-containing medium. Cells were switched into galactose-containing medium for 4 h at 30 and 37°C, respectively, and then examined by fluorescence and phase-contrast microscopy.
Localization of Rsp5p by immunogold EM.
Immunogold EM was performed to independently confirm the localization results based on GFP fusion proteins. Strain GW047, expressing HA-tagged wild-type Rsp5p from the RSP5 promoter as the sole source of Rsp5p, grows identically to an isogenic RSP5 strain. Immunogold EM using anti-HA antibody showed that HA-Rsp5p was localized to plasma membrane invaginations that are likely to correspond to sites of endosome formation (Fig. 6C and D) as well as at membranous structures immediately adjacent to the vacuole (Fig. 6A and B). As with the GFP fusion proteins, no specific signal was detected at either the plasma membrane or adjacent to the vacuole for the HA-tagged C2 deletion mutant (data not shown).
FIG. 6.
Localization of Rsp5p by immunogold EM. GW047 cells harboring the plasmid expressing HA-Rsp5p were prepared for electron microscopy and probed with anti-HA antibody. Labeling was in the cytoplasm and adjacent to the vacuole (A and B) and at sites of invaginations at the plasma membrane (C and D). V, vacuole; pm, plasma membrane. Bar = 90 nm.
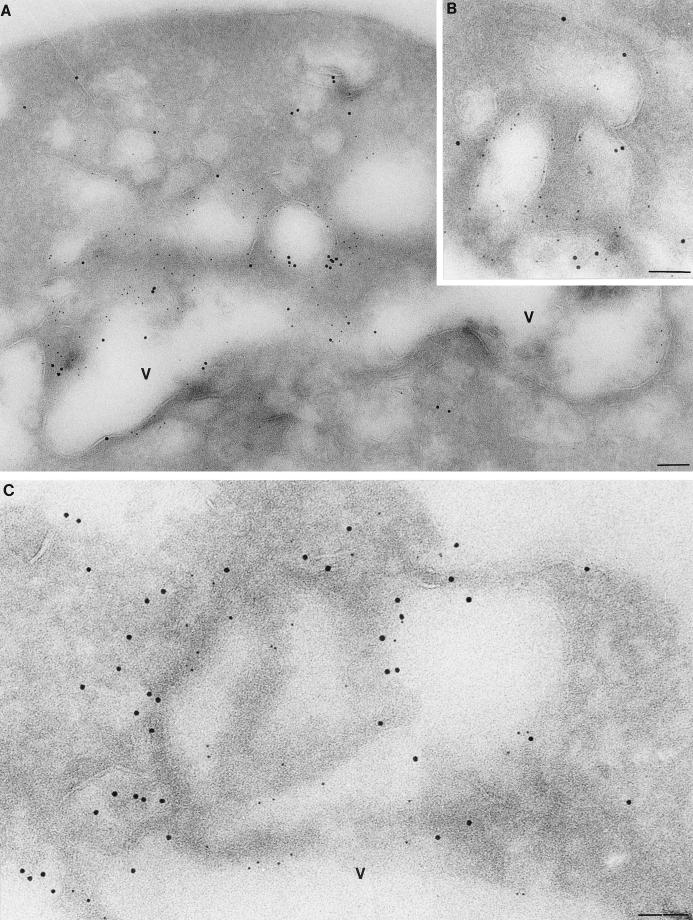
Double-immunogold labeling was also done on the GFP-RSP5p-expressing strain (GW0072) with antibodies against GFP and either Pep12p, a t-SNARE component of endosomes, or Vps32/Snf7p, a class E late endosomal marker (2). As shown in Fig. 7, GFP-Rsp5p (5-nm-diameter gold particles) was closely associated with both Pep12p and Vps32p (10-nm gold particles) at perivacuolar regions. The peripheral plasma membrane structures containing Rsp5p were not double-labeled with these marker proteins (data not shown). Together, the EM results further support the idea that Rsp5p is localized at sites of early endosome formation at the plasma membrane as well at later endocytic intermediates adjacent to the vacuole.
FIG. 7.
Double-label immunogold EM. GW072 (GFP-HA-Rsp5p) cells were prepared for EM and probed with anti-GFP antibody to detect Rsp5p, using a 5-nm-diameter Au bead-conjugated secondary antibody (small gold particles), and either anti-Pep12p (A and B) or anti-Vps32p (C) with a 10-nm-diameter Au bead-conjugated secondary antibody. V, vacuole. Scale bar = 85 nm.
Normal turnover of Fur4p is dependent on C2 domain of Rsp5p.
Since the C2 domain is the primary determinant of Rsp5p localization at the plasma membrane in vivo (Fig. 2C), deletion of the C2 domain might be predicted to affect the turnover of Fur4p, a protein subject to Rsp5p-dependent ubiquitin-mediated endocytosis. The decay rate of Fur4p was compared in RSP5 and RSP5ΔC2 strains by cycloheximide treatment of log-phase cells, followed by immunoblotting with anti-Fur4p antibody (Fig. 8A). The apparent half-life of Fur4p was increased at least twofold in the RSP5ΔC2 strain, a difference equal to that shown previously for the rsp5/npi1 mutant, which expresses a much-reduced level of Rsp5p (15, 19). Uracil permease activity, a measure of cell surface permease, was also tested after cycloheximide addition (Fig. 8B). Permease activity decreased more rapidly in the RSP5 strain than in the RSP5ΔC2 strain, indicating that deletion of the C2 domain affected permease internalization. In addition, both Fur4p protein and activity decay were similar in the RSP5 and GFP-RSP5 strains, indicating that GFP-Rsp5p is fully functional in ubiquitin-mediated endocytosis of Fur4p.
The decrease in Fur4p turnover and internalization after deletion of the C2 domain of Rsp5p prompted us to check whether this decrease resulted from reduced ubiquitination of Fur4p, which is required for its internalization. Fur4p-ubiquitin conjugates were examined in membrane-enriched fractions prepared from cells overexpressing ubiquitin. It has been shown that ubiquitin overproduction in wild-type cells does not modify the rate of Fur4p internalization or degradation or the overall pattern of Fur4p ubiquitination (14, 15) and that Fur4p is ubiquitinated on two target lysines in the N-terminal region of the protein, each of which accepts one or two ubiquitin molecules (14, 39). Figure 8C shows that both RSP5 and RSP5ΔC2 cells contained, in addition to nonubiquitinated Fur4p, at least three additional higher-molecular-weight species that correspond to Fur4p-ubiquitin conjugates (14, 15, 44). These results indicate that deletion of the C2 domain does not impair Fur4p ubiquitination at the plasma membrane or affect the number of ubiquitin residues added per target lysine. This result is consistent with a previous report that Gap1p was ubiquitinated by a form of Rsp5p with C2 deleted (63). Similar to our results described above for Fur4p, Gap1p internalization was also impaired in rsp5ΔC2p cells. Together, these results suggest that Rsp5p may have two separable functions at the plasma membrane: direct ubiquitination of substrates (independent of the C2 domain) and a C2-dependent function in the internalization process.
DISCUSSION
It has been previously demonstrated that Rsp5p is involved in down-regulation of several membrane-associated permeases, including Fur4p, Gap1p, Mal61p, and Tat2p (5, 15, 43, 62). Rsp5p-dependent ubiquitination of these proteins has been proposed to serve as a signal for endocytosis, resulting in transport to the vacuole, where they are degraded. The localization results presented here are consistent with the role of Rsp5p in ubiquitin-mediated endocytosis of these proteins, and they further suggest that Rsp5p may play a role at multiple steps in this pathway.
Rsp5p was localized by immunogold EM at peripheral compartments that appear to correspond to plasma membrane invaginations, as well as at perivacuolar structures where it colocalized with the t-SNARE protein Pep12p and the class E endocytic protein Vps32/Snf7p. Perivacuolar localization of GFP-Rsp5p was lost in both rsp5-1 and sla2/end4-1 mutant cells. Sla2p/End4p is required for the internalization step of endocytosis, as well as in organization of the actin network (49, 77). GFP-Rsp5p was still localized at the plasma membrane in the sla2/end4-1 cells, consistent with the perivacuolar structures being postinternalization endocytic compartments. Experiments with GFP–rsp5-1p (temperature sensitive) indicate that loss of Rsp5p catalytic activity alters localization at both the peripheral and internal structures. The GFP–rsp5-1p mutant protein was localized to the plasma membrane at the nonpermissive temperature; however, it was present in larger patches on the surface, while localization at internal structures was lost nearly completely. Together, these observations suggest that Rsp5p functions not only in directly ubiquitinating membrane-associated substrates but also in promoting both the internalization process and perhaps events further downstream in the endocytic pathway. We imagine that this could be due to Rsp5p-catalyzed ubiquitination of one or more protein components of the endocytic pathway, although whether such a ubiquitination event leads to protein degradation or serves an alternative function is unknown. Consistent with a role for Rsp5p in the internalization step, it has been noted that Rsp5p is also required for internalization of proteins carrying non-ubiquitin-dependent signals (21).
C2 domains are found in many proteins and mediate interactions with membrane phospholipids and/or membrane proteins (48). Rsp5p was not found to be distributed evenly over the entire surface of the plasma membrane, suggesting that C2-mediated localization is specific for structures or proteins at the plasma membrane. Based on immunogold EM, the punctate Rsp5-containing structures at the membrane appear to mark sites of membrane invagination and early endosome formation. Cell fractionation experiments showed nearly all Rsp5p to be associated with large structures (both low-speed and high-speed pellet fractions), and detergent solubilized only a portion of the Rsp5p present in the low-speed pellet without affecting Rsp5p in the high-speed pellet fraction. Deletion of the C2 domain released about half of the low-speed pellet material into the soluble fraction, and detergent solubilized the remaining material in this fraction but again did not affect the high-speed pellet material. Together, these findings suggest that there may be at least three distinct pools of Rsp5p: (i) low-speed pellet, membrane associated; (ii) low-speed pellet, non-membrane-associated; and (iii) high-speed pellet, non-membrane-associated. The fact that the C2 domain can be deleted while membrane association is maintained is consistent with the finding that Fur4p and Gap1p can be ubiquitinated at the membrane (but not internalized) by the protein with C2 deleted. The non-detergent-soluble Rsp5p may be linked to multiprotein complexes in both the low- and high-speed pellet fractions through protein-protein interactions, perhaps mediated by the WW domain or the HECT domain. While it is difficult to directly correlate the fractionation results with the localization results, this may be a reflection of the multiple substrates of Rsp5p that are located in diverse locales. In addition, at least some HECT E3s have been shown to be associated with the proteasome (78), and preliminary evidence indicates that WW HECT E3s, including Rsp5p, are proteasome associated (C. Salvat and J. M. Huibregtse, unpublished data). The fractionation results may therefore be further complicated by sedimentation of Rsp5p with the proteasome as well as fractionation of the proteasome with other complexes in the cell.
An additional indication that Rsp5p may function at multiple steps in endocytosis comes from its genetic interaction with PAN1 (76), which in turn has been linked genetically and biochemically to a complex that includes clathrin, End3p, Ent1p, and Sla1p (66, 67, 76). This complex of proteins has been found to play essential roles in both the organization of the actin cytoskeleton and endocytosis. Pan1p is similar to the mammalian EH domain-containing protein Eps15, a component of clathrin-coated pits and vesicles that plays a critical role in endocytosis at the plasma membrane and endosomes (61, 70). Furthermore, it has been suggested that, like Eps15, Pan1p protein complexes participate in multiple steps in the endocytic pathway in concert with Ent1p, acting at both the internalization and endosomal sorting steps (76). Interestingly, Eps15 becomes monoubiquitinated in response to stimulation with transforming growth factor α (71). Whether Pan1p or other components of the Pan1p protein complex are ubiquitinated by Rsp5p is currently being investigated.
A finding that lends strong support to the idea that ubiquitin metabolism plays an important role in the later stages of endocytosis is that mutations in genes encoding six different class E vacuolar protein-sorting factors, including Vps32p, were isolated as suppressors of the doa4-1 mutation (1). Doa4p is a deubiquitinating enzyme that plays an important role in ubiquitin recycling, and Doa4p itself was found at late endocytic structures (1). This finding, together with the findings that Rsp5p was localized by EM to Pep12p- and Vps32p-containing compartments and that catalytic activity is linked to Rsp5p localization, suggests that a dynamic ubiquitination-deubiquitination process may be linked to endosome trafficking, either at the level of ubiquitination of the endocytosed cargo or the endocytic machinery itself. Ubiquitination of components of the machinery is suggested by Eps15 ubiquitination, as noted above, and by the finding that the Drosophila Liquid facets protein, an epsin, is a target of the Fat facets deubiquitinating enzyme in photoreceptor organization (6).
An unresolved problem is how Rsp5p recognizes its integral membrane protein substrates. Rsp5p WW domains may interact directly with integral membrane protein substrates; however, this has not been demonstrated in vitro, and Fur4p or Gap1p does not have an obvious consensus WW domain binding site. In some cases, WW domains can also recognize phosphorylated ligands (38), and phosphorylation of Fur4p at PEST-like sequences has been shown to be a prerequisite for Rsp5p-dependent ubiquitination (40). Therefore, it remains possible that recognition of Fur4p by Rsp5p is direct and specific for the phosphorylated form of the substrate. Alternatively, the recognition of membrane-associated substrates might be indirect and possibly mediated by additional cellular factors. Interestingly, ubiquitination of all Rsp5p substrates identified so far—Rpb1p (RNA polymerase II large subunit), Spt23p, and the plasma membrane substrates—requires WW domain function but not the C2 domain (26, 73). The only function so far assigned to the C2 domain with respect to this set of substrates is the internalization of the ubiquitinated integral membrane proteins.
Rsp5p has been shown to interact with and ubiquitinate Rpb1p in response to UV irradiation (3). Rfa1p, a subunit of replication protein A, also has been reported to be a nuclear substrate of Rsp5p (10). Although we cannot rule out that a small percentage of Rsp5p is nuclear, it was not detected in the nucleus by either GFP fluorescence or immunogold EM. An intriguing possibility is that nuclear substrates might be exported to the cytoplasm prior to their ubiquitination and degradation. This idea has precedent in the case of ubiquitination of the mammalian p53 and p27Kip1 proteins (13, 69). We also did not see evidence of Rsp5p at the endoplasmic reticulum, which is the site of localization of the Spt23p substrate (26), although it is again possible that the amount of Rsp5p at this site is below the limits of detection or that Rsp5p is never stably associated with the endoplasmic reticulum.
The results presented here indicate that the determinants for appropriate localization of Rsp5p are important for its function in the endocytic pathway, and they further suggest that Rsp5p may have multiple functions at both early and late stages of endocytosis. A large amount of data now indicate that the ubiquitination pathways are involved in endocytosis, although ubiquitination has also been implicated in other trafficking pathways that lead to the vacuole. For instance, ubiquitination appears to be involved in delivery of mutant yeast α-factor receptor from an intracellular site to the vacuole (32). Ubiquitin-dependent endocytosis of the plasma membrane-associated Tat2p (tryptophan permease) appears to be similar to that of other permeases; however, Tat2p can also be diverted from the secretory pathway to the vacuole (5). It will be important to determine if Rsp5p plays a role in multiple trafficking pathways that converge at late stages of the endocytic pathway.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health to J.M.H. (CA72943) and B.W. (GM60979), a Burroughs Wellcome Fund New Investigator Award to B.W., and a grant from the Association pour la Recherche sur le Cancer (ARC grant 9773) to R.H.-T.
REFERENCES
- 1.Americk A Y, Nowak J, Swaminathan S, Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst M, Wendland B, Estepa E J, Emr S D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosomal function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaudenon S L, Huacani M R, Wang G, McDonnell D P, Huibregtse J M. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becherer K A, Rieder S E, Emr S D, Jones E W. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck T, Schmidt A, Hall M N. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–1238. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadavid A L, Ginzel A, Fischer J A. The function of the Drosophila Fat facets deubiquitinating enzyme in limiting photoreceptor cell number is intimately associated with endocytosis. Development. 2000;127:1727–1736. doi: 10.1242/dev.127.8.1727. [DOI] [PubMed] [Google Scholar]
- 7.Chen H I, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ecker D J, Ishaq Khan M, Marsh J, Butt T R, Crooke S T. Chemical synthesis and expression of a cassette adapted ubiquitin gene. J Biol Chem. 1987;262:3524–3527. [PubMed] [Google Scholar]
- 9.Egner R, Kuchler K. The yeast multidrug transporter Pdr5 of the plasma membrane is ubiquitinated prior to endocytosis and degradation in the vacuole. FEBS Lett. 1996;378:177–181. doi: 10.1016/0014-5793(95)01450-0. [DOI] [PubMed] [Google Scholar]
- 10.Erdeniz N, Rothstein R. Rsp5, a ubiquitin-protein ligase, is involved in degradation of the single-stranded DNA binding protein Rfa1 in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:224–232. doi: 10.1128/mcb.20.1.224-232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esnault Y, Feldheim D, Blondel M O, Schekman R, Kepes F. The yeast SSS1 gene is essential for secretory protein translocation and encodes a conserved protein of the endoplasmic reticulum. EMBO J. 1993;12:4083–4093. doi: 10.1002/j.1460-2075.1993.tb06092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisk H A, Yaffe M P. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1199–1208. doi: 10.1083/jcb.145.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galan J, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galan J M, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 16.Gerrard S R, Bryant N J, Stevens T H. VPS21 controls entry of endocytosed and biosynthetic proteins into the yeast prevacuolar compartment. Mol Biol Cell. 2000;11:613–626. doi: 10.1091/mbc.11.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guthrie C, Fink R G, editors. Guide to yeast genetics and molecular biology. Vol. 194. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 18.Hein C, Andre B. A C-terminal di-leucine motif and nearby sequences are required for NH4(+)-induced inactivation and degradation of the general amino acid permease, Gap1p, of Saccharomyces cerevisiae. Mol Microbiol. 1997;24:607–616. doi: 10.1046/j.1365-2958.1997.3771735.x. [DOI] [PubMed] [Google Scholar]
- 19.Hein C, Springael J Y, Volland C, Haguenauer-Tsapis R, Andre B. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 20.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 21.Hicke L. Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- 22.Hicke L. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 1997;11:1215–1226. doi: 10.1096/fasebj.11.14.9409540. [DOI] [PubMed] [Google Scholar]
- 23.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 24.Hicke L, Zanolari B, Riezman H. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J Cell Biol. 1998;141:349–358. doi: 10.1083/jcb.141.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 26.Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich H D, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 27.Horak J, Wolf D H. Catabolite inactivation of the galactose transporter in the yeast Saccharomyces cerevisiae: ubiquitination, endocytosis, and degradation in the vacuole. J Bacteriol. 1997;179:1541–1549. doi: 10.1128/jb.179.5.1541-1549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley P M, Huibregtse J M, Pavletich N P. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 29.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huibregtse J M, Scheffner M, Howley P M. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huibregtse J M, Yang J C, Beaudenon S L. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenness D D, Li Y, Tipper C, Spatrick P. Elimination of defective α-factor pheromone receptors. Mol Cell Biol. 1997;17:6236–6245. doi: 10.1128/mcb.17.11.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jund R, Weber E, Chevallier M R. Primary structure of the uracil transport protein of Saccharomyces cerevisiae. Eur J Biochem. 1988;171:417–424. doi: 10.1111/j.1432-1033.1988.tb13806.x. [DOI] [PubMed] [Google Scholar]
- 34.Kolling R, Hollenberg C P. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krampe S, Stamm O, Hollenberg C P, Boles E. Catabolite inactivation of the high-affinity hexose transporters Hxt6 and Hxt7 of Saccharomyces cerevisiae occurs in the vacuole after internalization by endocytosis. FEBS Lett. 1998;441:343–347. doi: 10.1016/s0014-5793(98)01583-x. [DOI] [PubMed] [Google Scholar]
- 36.Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu P-J, Zhou X Z, Shen M, Lu K P. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 39.Marchal C, Haguenauer-Tsapis R, Urban-Grimal D. Casein kinase I-dependent phosphorylation within a PEST sequence and ubiquitination at nearby lysines signal endocytosis of yeast uracil permease. J Biol Chem. 2000;275:23608–23614. doi: 10.1074/jbc.M001735200. [DOI] [PubMed] [Google Scholar]
- 40.Marchal C, Haguenauer-Tsapis R, Urban-Grimal D. A PEST-like sequence mediates phosphorylation and efficient ubiquitination of yeast uracil permease. Mol Cell Biol. 1998;18:314–321. doi: 10.1128/mcb.18.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsh M, McMahon H T. The structural era of endocytosis. Science. 1999;285:215–220. doi: 10.1126/science.285.5425.215. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Noel G, Niedenthal R, Tamura T, Harbers K. A family of structurally related RING finger proteins interacts specifically with the ubiquitin-conjugating enzyme UbcM4. FEBS Lett. 1999;454:257–261. doi: 10.1016/s0014-5793(99)00823-6. [DOI] [PubMed] [Google Scholar]
- 43.Medintz I, Jiang H, Michels C A. The role of ubiquitin conjugation in glucose-induced proteolysis of Saccharomyces maltose permease. J Biol Chem. 1998;273:34454–34462. doi: 10.1074/jbc.273.51.34454. [DOI] [PubMed] [Google Scholar]
- 44.Moreau V, Galan J-M, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen J T, Turck C W, Cohen F E, Zuckermann R N, Lim W A. Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors. Science. 1998;282:2088–2092. doi: 10.1126/science.282.5396.2088. [DOI] [PubMed] [Google Scholar]
- 46.Peters J M. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 47.Pickart C M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 48.Plant P J, Yeger H, Staub O, Howard P, Rotin D. The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+-dependent plasma membrane localization. J Biol Chem. 1997;272:32329–32336. doi: 10.1074/jbc.272.51.32329. [DOI] [PubMed] [Google Scholar]
- 49.Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rieder S E, Banta L M, Kohrer K, McCaffery J M, Emr S D. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riezman H. The ins and outs of protein translocation. Science. 1997;278:1728–1729. doi: 10.1126/science.278.5344.1728. [DOI] [PubMed] [Google Scholar]
- 52.Rizo J, Sudhof T C. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 53.Roberg K J, Rowley N, Kaiser C A. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J Cell Biol. 1997;137:1469–1482. doi: 10.1083/jcb.137.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson M S, Watts C, Zerial M. Membrane dynamics in endocytosis. Cell. 1996;84:13–21. doi: 10.1016/s0092-8674(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 55.Roth A F, Davis N G. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth A F, Sullivan D M, Davis N G. A large PEST-like sequence directs the ubiquitination, endocytosis, and vacuolar degradation of the yeast a-factor receptor. J Cell Biol. 1998;142:949–961. doi: 10.1083/jcb.142.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheffner M, Nuber U, Huibregtse J M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 58.Schmid S L. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- 59.Seron K, Blondel M O, Haguenauer-Tsapis R, Volland C. Uracil-induced down-regulation of the yeast uracil permease. J Bacteriol. 1999;181:1793–1800. doi: 10.1128/jb.181.6.1793-1800.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shih S C, Sloper-Mould K E, Hicke L. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sorkina T, Bild A, Tebar F, Sorkin A. Clathrin, adaptors and eps15 in endosomes containing activated epidermal growth factor receptors. J Cell Sci. 1999;112:317–327. doi: 10.1242/jcs.112.3.317. [DOI] [PubMed] [Google Scholar]
- 62.Springael J Y, Andre B. Nitrogen-regulated ubiquitination of the gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1253–1263. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Springael J Y, De Craene J O, Andre B. The yeast Npi1/Rsp5 ubiquitin ligase lacking its N-terminal C2 domain is competent for ubiquitination but not for subsequent endocytosis of the gap1 permease. Biochem Biophys Res Commun. 1999;257:561–566. doi: 10.1006/bbrc.1999.0505. [DOI] [PubMed] [Google Scholar]
- 64.Springael J Y, Galan J M, Haguenauer-Tsapis R, Andre B. NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J Cell Sci. 1999;112:1375–1383. doi: 10.1242/jcs.112.9.1375. [DOI] [PubMed] [Google Scholar]
- 65.Stewart L C, Yaffe M P. A role for unsaturated fatty acids in mitochondrial movement and inheritance. J Cell Biol. 1991;115:1249–1257. doi: 10.1083/jcb.115.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang H Y, Munn A, Cai M. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang H Y, Xu J, Cai M. Pan1p, End3p, and Sla1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 69.Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent kinase inhibitor p27kip1 is instigated by Jab1. Nature. 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 70.Torrisi M R, Lotti L V, Belleudi F, Gradini R, Salcini A E, Confalonieri S, Pelicci P G, Di Fiore P P. Eps15 is recruited to the plasma membrane upon epidermal growth factor receptor activation and localizes to components of the endocytic pathway during receptor internalization. Mol Biol Cell. 1999;10:417–434. doi: 10.1091/mbc.10.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Delft S, Govers R, Strous G J, Verkleij A J, van Bergen en Henegouwen P M. Epidermal growth factor induces ubiquitination of Eps15. J Biol Chem. 1997;272:14013–14016. doi: 10.1074/jbc.272.22.14013. [DOI] [PubMed] [Google Scholar]
- 72.Volland C, Garnier C, Haguenauer-Tsapis R. In vivo phosphorylation of the yeast uracil permease. J Biol Chem. 1992;267:23767–23771. [PubMed] [Google Scholar]
- 73.Wang G, Yang J, Huibregtse J M. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waterman H, Levkowitz G, Alroy I, Yarden Y. The RING finger of c-Cb1 mediates desensitization of the epidermal growth factor receptor. J Biol Chem. 1999;274:22151–22154. doi: 10.1074/jbc.274.32.22151. [DOI] [PubMed] [Google Scholar]
- 75.Wendland B, Emr S D, Riezman H. Protein traffic in the yeast endocytic and vacuolar protein sorting pathways. Curr Opin Cell Biol. 1998;10:513–522. doi: 10.1016/s0955-0674(98)80067-7. [DOI] [PubMed] [Google Scholar]
- 76.Wendland B, Steece K E, Emr S D. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wesp A, Hicke L, Palecek J, Lombardi R, Aust T, Munn A L, Riezman H. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2291–2306. doi: 10.1091/mbc.8.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie Y, Varshavsky A. Physical association of ubiquitin ligases and the 26S proteasome. Proc Natl Acad Sci USA. 2000;14:2497–2502. doi: 10.1073/pnas.060025497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zoladek T, Tobiasz A, Vaduva G, Boguta M, Martin N C, Hopper A K. MDP1, a Saccharomyces cerevisiae gene involved in mitochondrial/cytoplasmic protein distribution, is identical to the ubiquitin-protein ligase gene RSP5. Genetics. 1997;145:595–603. doi: 10.1093/genetics/145.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]