Abstract
One of the problems that hamper the use of antisense DNAs as effective drugs is the non-specific binding of chemically-modified oligonucleotides to cellular proteins. We previously showed that the affinity of a model ssDNA-binding protein, the Ff gene 5 protein (g5p), was >300-fold higher for phosphorothioate-modified DNA (S-DNA) than for unmodified dA36, consistent with the propensity of S-DNA to bind indiscriminately to proteins. The current work shows that g5p binding is also sensitive to sugar and pyrimidine modifications used in antisense oligomers. Binding affinities of g5p for 10 36mer oligomers were quantitated using solution circular dichroism measurements. The oligomers contained C-5-propyne (prC), 2′-O-methyl (2′-O-Me) or 2′-OH (RNA) groups, alone or combined with the phosphorothioate modification. In agreement with reported increases in antisense activity, the addition of prC or 2′-O-Me modifications substantially reduced the affinity of oligomers for g5p by ∼2-fold compared with the same DNA oligomer sequences containing only phosphorothioate linkages. That is, such modifications moderated the propensity of the phosphorothioate group to bind tightly to the g5p. The Ff g5p could be a useful model protein for assessing non-specific binding effects of antisense oligomer modifications.
INTRODUCTION
The potential control of gene expression by antisense drugs has relied on the development of suitably modified oligonucleotides and their analogs. The effectiveness of an antisense oligonucleotide (ASO) in inhibiting gene expression depends on many factors, of which three that have been typically evaluated are: (i) the nuclease resistance of the ASO, (ii) the thermal stability of the ASO·RNA hybrid formed with the ASO, and (iii) the ability of RNase H to degrade the RNA of an ASO·RNA hybrid (1–5). As one means of optimizing the ASO composition and antisense activity, chimeric oligonucleotides containing several modifications have been synthesized and tested for gene-specific inhibition of the targeted mRNA (6,7).
The importance of non-specific binding of ASOs to cellular proteins has been recognized as an additional factor that influences antisense activity. It is generally accepted that the sulfur-substituted backbone of a phosphorothioate ASO (S-ASO) causes it to have a greater affinity than normal DNA for various cellular proteins, and this effect may contribute to the non-specific antisense effects of S-ASOs (8–13). ASOs that contain propyne or 2′-O-methyl (2′-O-Me) modifications form ASO·RNA hybrids with increased stability (5) and can have increased antisense effects in comparison with phosphorothioate oligonucleotides of the same sequences (11). An important question is how the antisense effects of such promising DNA modifications are moderated by non-specific binding to cellular proteins.
In order to evaluate the non-specific protein binding of important ASO modifications, we compared the binding affinities of DNA 36mer oligonucleotides containing C-5 propynyl-modified cytosines (prC), and RNA 36mer oligomers containing 2′-O-Me sugars or normal ribose sugars, with or without phosphorothioate backbones, to the Ff gene 5 protein (g5p). Binding was monitored by circular dichroism (CD) measurements. The g5p encoded by filamentous Ff phages is a well characterized single-stranded nucleic acid binding protein that shares binding characteristics with other ssDNA-binding proteins, such as Pf3 g5p, T4 g32p, Escherichia coli SSB, and human replication protein A (RPA) (14–17). The homodimeric structure of g5p has been determined by X-ray crystallography and NMR (18,19). The structures of g5p·ssDNA complexes have been extensively studied by electron microscopy, solution scattering and modeling techniques (20–23). The g5p is usually considered to be a non-specific binding protein since it binds cooperatively to newly formed phage genomic ssDNA molecules in the course of phage morphogenesis. A dominant mode of binding is with 4 nt bound per g5p monomer (24,25), but the stoichiometric mode depends on the template sequence and solution conditions. The binding affinity (Kω) of g5p is the product of the intrinsic binding constant, K, and a cooperativity factor, ω. Kω values for binding of g5p to different ssDNA sequences can vary by over two orders of magnitude, and the binding affinities to ssDNA depend inversely on base–base stacking interactions in unmodified ssDNA sequences (17,25–27). In previous work, we showed that the protein has >300-fold greater affinity for phosphorothioate S-dA36 than for unmodified dA36, and the increased binding affinity is nearly proportional to the number of phosphorothioate linkages in the sequence (28).
Results in this paper compare the binding affinities of g5p for unmodified or phosphorothioate-modified DNA and RNA oligomers, with or without prC or 2′-O-Me modifications. Binding affinities for 10 36mer oligomers, extrapolated to 0.2 M NaCl, were in the decreasing order of S-d(AAC)12 > S-d(AAprC)12 > S-dA36 > S-2′-O-MeA36 >> d(AAC)12 > d(AAprC)12 > S-rA36 > 2′-O-MeA36 > rA36 > dA36. The effect of the phosphorothioate linkage on the binding affinities of d(AAC)12 or dA36 was greatly reduced when accompanied by prC or 2′-O-Me modifications, respectively, indicating the potential usefulness of combined modifications in reducing non-specific protein binding effects of S-ASOs.
MATERIALS AND METHODS
Oligonucleotides and protein purification
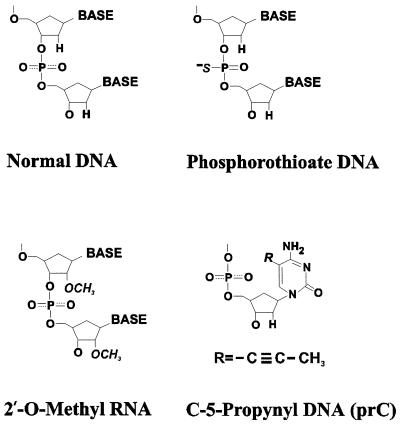
The 10 oligomers used in this study were all 36 nt long. The chemical modifications are shown in Figure 1. The dA36 and S-dA36 were purchased from Oligos, Etc (Wilsonville, OR). The d(AAC)12, S-d(AAC)12, rA36, S-rA36, 2′-O-MeA36, S-2′-O-MeA36, d(AAprC)12 and S-d(AAprC)12 were purchased from Midland Certified Reagent Co. (Midland, TX). The dA36 and S-dA36 were gel purified, and electrophoresis on 12% polyacrylamide sizing gels (Oligos, Etc) showed no fragmentation (at a detection limit of <2% for a given size fragment) due to depurination. Other oligonucleotides were purified by HPLC, and mass spectroscopy (Midland) showed no detectable fragmentation. The oligomers were all provided as lyophilized ammonium salts and were dissolved in 2 mM Na+ (phosphate buffer, pH 7.0) for the experiments.
Figure 1.
Structures of normal and modified nucleotides. The phosphorothioate modification was also used in combination with the 2′-O-Me and prC modifications.
Concentrations of oligomers were determined from UV absorption spectra, using nearest-neighbor calculated ɛ260 values at 20°C of 12 050 M–1 cm–1 for all the A36 oligomers and 10 610 M–1 cm–1 for d(AAC)12 and S-d(AAC)12 (29). It was assumed that extinction coefficients were the same for the unmodified and phosphorothioate-modified oligomers. An ɛ260 value at 90°C of 10 020 M–1 cm–1 was used for the prC-modified sequences, d(AAprC)12 and S-d(AAprC)12; this extinction coefficient was the sum of monomer values calculated using an ɛ260 value for the prC monomer of 5100 M–1 cm–1, which was provided by the manufacturer (Glen Research Corporation, Sterling, VA).
Wild-type Ff g5p was isolated and characterized as described previously (28,30,31). The isolated protein was finally dialyzed into 2 mM Na+ (phosphate buffer, pH 7.0).
Spectroscopy
UV absorption spectra were collected using an Olis-modified Cary 14 spectrophotometer (On-Line Instrument Co., Bogart, GA), and CD spectra were collected using a Jasco Model J715 spectropolarimeter (Jasco, Easton, MD). Samples were measured in 1.0 or 2.0 cm cuvettes at a temperature of 20 ± 0.5°C, except as noted. CD spectra were acquired as described by Mou et al. (26). Smoothed CD spectra are plotted at 1 nm intervals as ɛL – ɛR in units of M–1 cm–1, per mole of nucleotide.
Titrations
CD titrations were performed as described by Mou et al. (28). After the addition of aliquots of concentrated g5p (∼1 × 10–4 M) to the oligomers in 2 mM Na+ (phosphate buffer, pH 7.0), the samples were gently mixed and allowed to reach equilibrium for ≥20 min. Final nucleotide concentrations were 3.5–5.5 × 10–5 M in the titrated samples. Light scattering was monitored as the relative absorption at 320 nm to the absorption at 260 nm, which was at most 8% at the highest [P]/[N] ratios. (The [P]/[N] ratio is the molar ratio of protein monomer to nucleotide.)
Salt dissociation and binding affinities
The binding affinities of Ff g5p to individual oligomers were obtained by monitoring the CD spectra of complexes as they were dissociated by increasing the NaCl concentration with aliquots of 4 M NaCl as described by Kansy et al. (25). Complexes were first formed in 2 mM Na+ (phosphate buffer, pH 7.0) by adding g5p to a given oligonucleotide until the [P]/[N] ratio needed for saturation was reached. The [P]/[N] ratio needed for saturation was 0.25 for all the oligomers except for 2′-O-MeA36, for which the saturation [P]/[N] ratio was 0.3. The final g5p monomer concentrations in these samples of complexes were in the range 2–10 × 10–5 M. The sample solutions were weighed after each addition of NaCl, and the density of the added NaCl solution was taken into account in calculating the dilution of the sample. The percent dissociation was monitored by CD measurements at 270 nm [for rA36, S-rA36, 2′-O-MeA36, S-2′-O-MeA36, d(AAC)36 and S-d(AAC)36] or 300 nm [for d(AAprC)12 and S-d(AAprC)12], and plotted as a function of [NaCl]. The salt concentration required for 50% dissociation of a complex was determined from the dissociation curves as described by Mou et al. (26) with the assumption that the complexes were dissociated in an all-or-none manner (26,28). Thus, the apparent value of the binding constant product (Kωapp) per dimer for a given finite lattice of 36 nt was derived by setting Kωapp = 1/(2L), where L was the free protein dimer concentration at 50% dissociation. The factor of two corrected for the 2-fold rotational symmetry of the g5p dimer, yielding the Kωapp values for the binding of the g5p dimer in one orientation to two DNA strands (24,31,32).
Hyperchromicity
Optical density (OD) changes for the free oligonucleotides were monitored at 260 nm. The percent hyperchromicity (%H) upon heating from 20 to 95°C was calculated as %H = 100 × [OD (95°C) – OD (20°C)]/[OD (20°C)]. Temperatures were controlled to ±1°C with an accuracy of ±0.5°C.
RESULTS AND DISCUSSION
Effects of modifications on the CD and absorption properties of the single-stranded oligomers
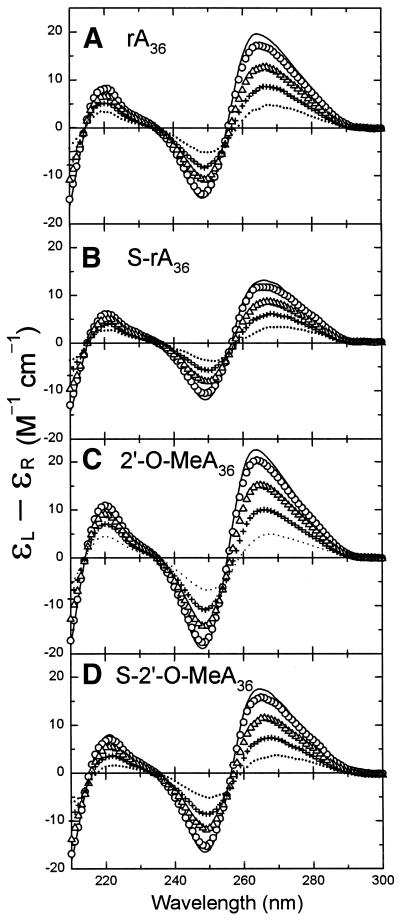
We previously showed that the CD spectrum of dA36 does not change significantly when phosphodiester linkages are substituted with different proportions of phosphorothioate linkages (28). The CD spectrum of free S-rA36 was also qualitatively similar to that of rA36, as shown in Figure 2A and B. Both spectra had positive CD bands at 264 and 220 nm and a negative CD band at 248 nm, CD features that were the same as for poly[r(A)] (33). However, the magnitude of the positive CD band at 264 nm in the spectrum of S-rA36 at 20°C was only ∼65% of that for rA36. The CD spectra of 2′-O-MeA36 and S-2′-O-MeA36 (Fig. 2C and D) were also quite similar in shape to the spectra of rA36 and S-rA36, except that the 2′-O-Me modification resulted in increases in the magnitudes of the long-wavelength CD bands with respect to spectra of rA36 and S-rA36, so that the combination of the phosphorothioate and 2′-O-Me modifications gave a spectrum with band magnitudes within 10% of those of the unmodified rA36. Thus, constraints on the base–base interaction in S-rA36 caused by the phosphorothioate modification were mostly compensated by the 2′-O-Me modification in S-2′-O-MeA36. The similarity of the CD spectra for these oligomers, and the fact that they all had long-wavelength CD bands much larger than observed for deoxyadenosine-containing oligomers (28) (Fig. S1), was persuasive evidence that oligomers having ribose sugars with either -O-H or -O-Me at the 2′ position were in similar RNA A conformations. Other workers have found that 2′-O-Me-modified oligomers can adopt the C3′-endo sugar pucker conformation that is typical of RNA (34–36).
Figure 2.
Representative CD spectra of normal and modified rA36 oligomers during heating. CD changes of (A) rA36, (B) S-rA36, (C) 2′-O-MeA36 and (D) S-2′-O-MeA36 at temperatures of 10°C (line), 20°C (circles), 38°C (triangles), 57°C (plus) and 86°C (dotted). CD data above and in Figures 3 and 4 are plotted as ɛL–ɛR in units of M–1 cm–1, per mole of nucleotide. Molar nucleotide concentrations of the oligomers were in the range of 5.7–8.0 × 10–5 M.
The prC modification caused the CD spectra of d(AAprC)12 and S-d(AAprC)12 to have small positive CD bands above 270 nm that were decreased ∼50% and red-shifted in comparison with the long-wavelength positive bands of d(AAC)12 and S-d(AAC)12, respectively (Fig. S1). The shift of CD bands to longer wavelengths for the prC-modified DNAs was in agreement with UV absorption spectra, which showed long-wavelength CD bands that extended above 300 nm (data not shown). CD bands above 300 nm have also been reported for propynyl-modified d(CCUCCUU) (5). Phosphorothioate modifications of d(AAC)12 and d(AAprC)12 did not affect their CD spectra dramatically, except for reductions of ∼10% in the magnitudes of the respective long-wavelength CD bands (Fig. S1).
Figure 2 shows representative temperature-dependent CD spectra for rA36, S-rA36, 2′-O-MeA36, and S-2′-O-MeA36 in 2 mM Na+ (phosphate buffer, pH 7.0). The CD magnitudes of these four oligomers decreased in a monotonic fashion as the temperature increased from 10 to 86°C, which is typical of temperature-dependent CD changes for single-stranded oligomers. For these RNA-like oligomers, the temperature-dependent CD changes were similar and exhibited the same three isodichroic points near 215, 235 and 255 nm. There was no cooperative transition in the UV absorbance melting profiles of these oligomers over the range of 4–90°C (not shown).
The %H values at 260 nm were measured for all 10 oligomers upon heating from 20 to 90°C and are shown in Table 1. The %H values are an indication of base stacking within the free oligomers. The %H values of the RNA-like oligomers rA36, S-rA36, 2′-O-MeA36 and S-2′-O-MeA36 were similar and were in the range of 23.6–26.5%. Phosphorothioate and 2′-O-Me modifications apparently had little effect on the response to heating, with the %H value being reduced by at most 2.5% (from 26.1 to 23.6) by the phosphorothioate modification of the RNA-like oligomers. The phosphorothioate effect on the hyperchromicities of two of the DNAs, those that did not carry prC modifications [S-dA36 and S-d(AAC)36] was greater, with reductions in the %H value of ∼4% relative to the unmodified oligomers. This effect of phosphorothioate modification on the DNAs seems significant, given that the total %H for the DNAs was less than that of the RNAs. That is, the phosphorothioate modification slightly reduced the base stacking (or base stacking response to heating) for these two DNAs.
Table 1. Hyperchromicities of chemically-modified oligonucleotides.
| Percent hyperchromicity (%H)b,c | |||||
|---|---|---|---|---|---|
| Backbone modificationa | rA36 | 2′-O-MeA36 | dA36 | d(AAC)12 | d(AAprC)12 |
| P-linkages | 26.1 ± 0.5 | 26.5 ± 0.5 | 22.1 ± 0.4 | 14.2 ± 0.1 | 20.9 ± 0.2 |
| S-linkages | 23.6 ± 0.4 | 25.4 ± 1.6 | 18.1 ± 1.0 | 9.9 ± 1.0 | 18.7 ± 0.5 |
aP- and S-linkages denote oligonucleotides with normal phosphodiester linkages and phosphorothioate-modified linkages, respectively.
bThe nucleotide concentrations of the oligomers were 5–7 × 10–5 M.
cThe %H upon heating from 20 to 90°C was monitored at 260 nm and was calculated as 100 × [OD (90°C) – OD (20°C)]/[OD (20°C)]. %H values are the averages and the errors are the ranges of values from two or three experiments.
In contrast, the prC modification caused an increase in %H of ∼7–9%, from 14% [for d(AAC)12] to 21% [for d(AAprC)12] and from 10% [for S-d(AAC)12] to 19% [for S-d(AAprC)12], consistent with an increased stacking of the prC bases with the neighboring adenine bases in both the unmodified DNA and the phosphorothioate-modified DNA. The phosphorothioate and prC modifications appeared to have opposing effects on DNA hyperchromicity, with the prC modification being the dominant modification.
Since the CD and absorbance spectral effects of the different modifications could be explained as simply the alterations of base–base interactions and/or the addition of a long-wavelength absorbing propyne group, our overall conclusion was that the chemical modifications used in this work caused no dramatic structural changes in the oligomers and that they all remained single stranded.
Binding stoichiometries and changes in CD spectra during g5p binding
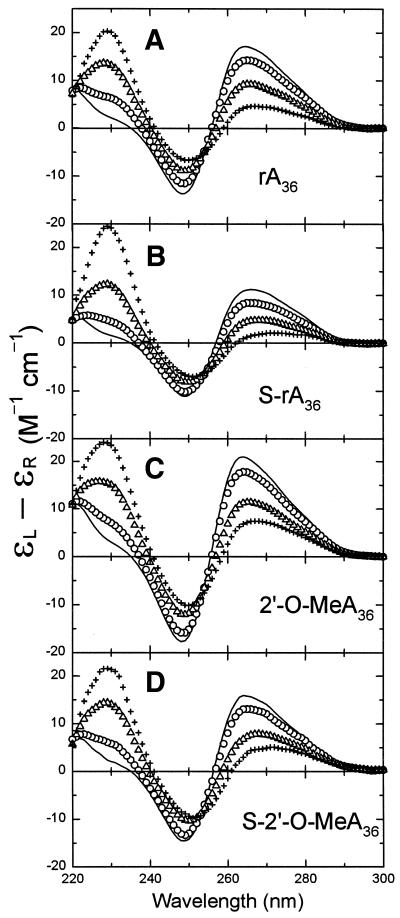
The binding stoichiometries of Ff g5p for individual oligomers were determined from titration experiments in which g5p was added to solutions of the oligomers in a buffer of 2 mM Na+ (phosphate buffer, pH 7.0) at 20°C. Figure 3 shows representative spectra from CD titrations of rA36, S-rA36, 2′-O-MeA36, and S-2′-O-MeA36 with g5p. The increase in the positive CD band at 229 nm during protein titrations was due to the large tyrosyl 229 nm band of the added g5p (30). At wavelengths above 250 nm the CD contribution of g5p is negligible on this scale (30), and CD changes were due to base unstacking and alterations in the environment of the nucleic acid as the oligomer became saturated with protein to form a left-handed superhelical complex (20,23,37). During binding of g5p, spectral changes above 250 nm for all four RNA-like oligomers were comparable and gave the titration endpoints of [P]/[N] ≈ 0.25 (or 0.30 for 2′-O-MeA36), indicating that the protein bound these oligomers with the binding stoichiometries of 4 or 3 nt/protein monomer (Figs 3 and 4B and C). The CD changes above 250 nm upon saturation of rA36, S-rA36, 2′-O-MeA36 and S-2′-O-MeA36 with the g5p were similar to the CD changes upon heating the respective oligomers, as may be seen by comparing Figures 2 and 3. This suggested that the adenine bases were significantly unstacked in the g5p-bound forms of all four oligomers. The phosphorothioate-modified RNAs differed slightly in their CD response to g5p binding and heating in that binding of g5p caused a noticeable red-shift in the long-wavelength crossover (Fig. 3). The persistent positive long-wavelength CD bands of all four g5p-bound RNA oligomers suggested that a fraction of bases remained stacked in a right-handed conformation within an oligomer that must follow an overall left-handed helical path when complexed with g5p. This inference was previously made from a CD study of g5p·poly[r(A)] complexes (27).
Figure 3.
Representative CD spectra for titrations of the normal and modified rA36 oligomers with g5p at 20°C. (A) Titration of rA36 with g5p at [P]/[N] ratios of 0.0 (line), 0.05 (circles), 0.20 (triangles) and 0.25 (plus). (B) Titration of S-rA36 with g5p at [P]/[N] ratios of 0.0 (line), 0.05 (circles), 0.19 (triangles) and 0.24 (plus). (C) Titration of 2′-O-MeA36 with g5p at [P]/[N] ratios of 0.0 (line), 0.06 (circles), 0.18 (triangles) and 0.30 (plus). (D) Titration of S-2′-O-MeA36 with g5p at [P]/[N] ratios of 0.0 (line), 0.04 (circles), 0.21 (triangles) and 0.26 (plus).
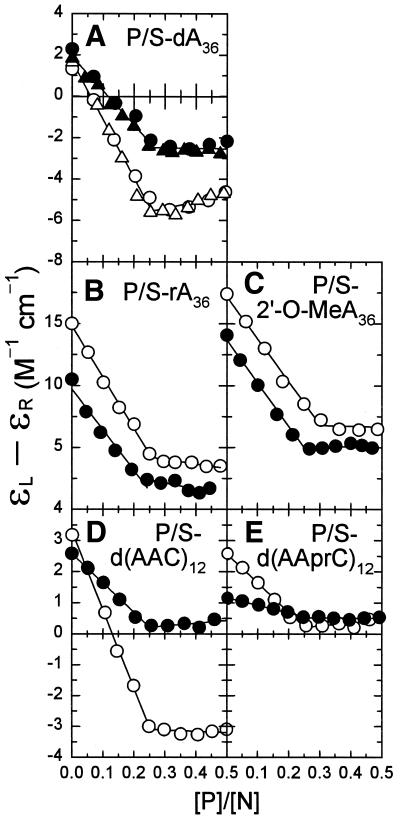
Figure 4.
Plots showing the change in nucleic acid CD as a function of [P]/[N] ratio during titrations with g5p. Titrations shown as open and closed symbols are for oligomers with normal phosphodiester (P) and phosphorothioate-modified (S-) linkages, respectively. (A) dA36 and S-dA36. (B) rA36 and S-rA36. (C) 2′-O-MeA36 and S-2′-O-MeA36. (D) d(AAC)12 and S-d(AAC)12. (E) d(AAprC)12 and S-d(AAprC)12. The molar CD is plotted at 270 nm for all of the oligomers except for d(AAprC)12 and S-d(AAprC)12 in (E), where the CD is plotted at 300 nm.
The nucleic acid CD changes at 270 or 300 nm are plotted as a function of [P]/[N] molar ratio in Figure 4 for titrations of all 10 oligomers. The binding of g5p to S-dA36 has been shown to cause smaller long wavelength CD changes than found for binding to dA36 (Fig. 4A) (28). In a consistent fashion, a reduced long-wavelength CD perturbation was observed in the titrations of the two other S-DNAs, S-d(AAC)12 and S-d(AAprC)12, compared with d(AAC)12 (Fig. 4D) and d(AAprC)12 (Fig. 4E), respectively. The CD effects of binding of g5p to the modified as well as to the unmodified nucleic acids were proportional to the amount of added g5p until titration endpoints were reached at [P]/[N] ratios of ∼0.25 or ∼0.30, indicating that g5p bound to all of the oligomers in n = 4 or 3 stoichiometric binding modes (i.e. with 4 or 3 nt bound per g5p monomer; see footnote a to Table 2).
Table 2. Dependence of binding affinities on NaCl concentration and extrapolated binding affinities per mole of g5p dimer from titrations of 36mer oligonucleotides with Ff g5pa.
| Slopeb | Kωapp (× 10–5) (M–1) at 0.2 M NaClc | ΔΔG° (20°C)d at 0.2 M NaCl | |
|---|---|---|---|
| (–∂log(Kωapp)/∂log[NaCl]) | |||
| dA36 | 3.68 ± 0.29 | 0.71 ± 0.05 | |
| S-dA36 | 2.70 ± 0.28 | 246.0 ± 21 | –0.42 ± 0.06e |
| rA36 | 4.23 ± 0.41 | 1.3 ± 0.13 | |
| S-rA36 | 3.51 ± 0.37 | 8.3 ± 0.85 | –0.13 ± 0.05e |
| 2′-O-MeA36 | 3.78 ± 0.38 | 3.8 ± 0.40 | –0.08 ± 0.07f |
| S-2′-O-MeA36 | 3.76 ± 0.51 | 125 ± 17 | –0.25 ± 0.07e |
| d(AAC)12 | 3.05 ± 0.07 | 19.3 ± 0.01 | |
| S-d(AAC)12 | Not determined | >1000 | –0.29e (minimum) |
| d(AAprC)12 | 4.07 ± 0.62 | 13.9 ± 2.1 | +2.03 ± 0.09g |
| S-d(AAprC)12 | 2.68 ± 0.38 | 553 ± 65 | –0.27 ± 0.09e |
aTitration endpoints were at [P]/[N] = 0.30 for 2′-O-MeA36 and at [P]/[N] = 0.24–0.26 for all the other oligomers. SDs of the titration endpoints, determined by fitting CD titration data at 270 nm [or 300 nm for d(AAprC)12 and S-d(AAprC)12] with two linear regressions, were generally ±0.01; endpoint data for S-d(AAprC)12 were less precise with an error of ±0.11.
bSlope and SDs from linear regressions of the data in Figures 5 and 6.
cExperimental data were extrapolated to 0.2 M NaCl.
dΔΔG° values (kcal/mol, 20°C, 0.2 M NaCl, per mole of nucleotide) were calculated as ΔG°(modified oligomer) –ΔG°(unmodified oligomer) = RT ln[(Kω)modified/(Kω)unmodified]/n, where R is the gas constant, T is the absolute temperature, and n is the number of modified nucleotides bound per g5p dimer; n = 8 when all of the nucleotides were modified and n = 2.7 when one-third of the nucleotides were modified.
eChange in ΔG° (kcal/mol per phosphorothioate linkage) of binding with respect to the same oligomer without phosphorothioate modification, for example to S-dA36 with respect to dA36. (The minus sign means that the binding is more favorable for the phosphorothioate nucleic acid. The value for S-d(AAC)12 is a minimum negative value.)
fChange in ΔG° per nucleotide with respect to rA36, i.e. the effect of changing 2′-OH to 2′-O-Me.
gChange in ΔG° per propyne group with respect to d(AAC)12, i.e. the effect of changing C to prC.
In summary, the oligomers with different modifications displayed comparable stoichiometries of binding, and within the sets of six DNA and four RNA-like oligomers qualitatively similar CD changes were displayed upon binding of g5p. Therefore, it appeared that the normal and modified oligomers were bound in the same g5p sites.
Effect on g5p binding affinities of the phosphorothioate modification
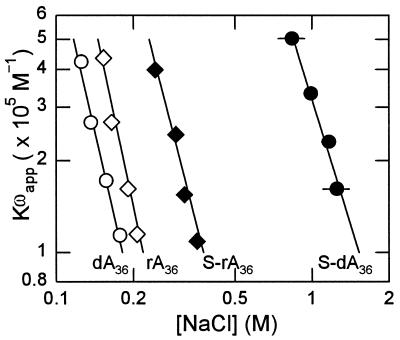
First, we consider the binding of g5p to the dA36, rA36, S-dA36 and S-rA36 oligomers in order to compare the results for the RNA with those previously reported for the DNA (28). As shown in Figure 5, the logarithm of the binding affinity, log[Kωapp], was linearly related to the logarithm of the salt concentration, log[NaCl], for all four oligomers. The slope of a log[Kωapp] versus log[NaCl] plot gives the number of ions released per protein dimer when forming a complex. The values of the slopes of the four plots in Figure 5, listed in Table 2, showed that there were about four ions released per g5p dimer during binding to unmodified dA36 and rA36, about one ion more than during binding to the phosphorothioate-modified S-dA36 and S-rA36. The slope of 4.23 ± 0.41 for binding to rA36 was comparable with the slope of 4.0 for binding to unmodified poly[r(A)] (17).
Figure 5.
Comparison of the dependence of log[Kωapp] on log[NaCl] for phosphorothioate and unmodified DNA and RNA oligonucleotides. Complexes of g5p with dA36 (open circles), S-dA36 (closed circles), rA36 (open diamonds) and S-rA36 (closed diamonds). Complexes of oligomers with g5p were formed at [P]/[N] = 0.25 in 2 mM Na+ (phosphate buffer, pH 7.0). Total protein concentrations in these complexes were in the range 5.0–20.0 × 10–6 M. Kωapp values here and in Figure 6 were determined as the reciprocals of the free concentrations and are plotted versus the NaCl concentrations that were required for 50% dissociation of the respective complexes (Materials and Methods). Error bars were generally within the size of the symbols shown.
The binding affinities of g5p for unmodified dA36 and rA36 extrapolated to 0.2 M NaCl were relatively weak and were only slightly higher for the RNA than for the DNA. The binding affinities for dA36 and rA36 were comparable with those reported for the polymers poly[d(A)] and poly[r(A)] [Kωapp = ∼1 × 105 M–1 at 0.23 M NaCl, 37°C (17)].
The phosphorothioate-substituted backbone in the S-dA36 and S-rA36 oligomers increased the binding affinities of these oligomers over those of the unmodified DNA and RNA oligomers. The extrapolated value of Kωapp at 0.2 M NaCl was increased ∼6-fold for S-rA36 over that for rA36 and was substantially increased ∼300-fold for S-dA36 over that for dA36 (Table 2). That is, the Ff g5p bound to S-dA36 with a 30-fold higher binding affinity than to S-rA36, although the relative binding affinities for the unmodified DNA and RNA were in the reverse order. Another group reported analogous results in that various plasma proteins showed a lower or negligible binding to a 19mer phosphorothioate RNA compared with the binding of these proteins to a phosphorothioate DNA (38).
Differences in binding affinity to g5p are expressed as increases in binding free energy per phosphorothioate linkage in the last column of Table 2. The effects of phosphorothioate modification were to increase the binding free energies of S-dA36 and S-rA36 by –0.42 and –0.13 kcal/mol, respectively. In the three other cases for which data were obtained [S-2′-O-MeA36, S-d(AAC)12 and S-d(AAprC)12], the phosphorothioate modification increased the binding free energy by –0.25 to at least –0.29 kcal/mol. The increased binding affinities to g5p caused by phosphorothioate modification were not explained by a polyelectrolyte effect of the phosphorothioate backbone, since there was either about one fewer ion released [binding to S-dA36, S-rA36 or S-d(AAprC)12] or there was no change in the number of ions released (S-2′-O-MeA36) during g5p binding, compared with binding to the respective unmodified oligomers.
Phosphorothioate linkages can increase binding affinity by an indirect effect on the DNA or RNA conformation. For example, Smith and Nikonowicz (39) found that a phosphorothioate substitution in an RNA hairpin can substantially increase the binding affinity of MS2 coat protein by 20-fold by reducing the amount of structural rearrangement within the hairpin necessary to form the protein–RNA complex. A specific possibility to explain the increased binding affinity of g5p for phosphorothioate-modified oligomers is that base–base interactions are decreased due to increased steric hindrance or electrostatic effects of the phosphorothioate linkages, which then results in an increased binding affinity because of a reduction in the free energy cost of unstacking the bases during binding (28,40). As shown by the generally reduced %H values at 260 nm for the phosphorothioate-modified oligomers compared with the respective oligomers having normal phosphodiester backbones, phosphorothioate modification appears to cause a slight base unstacking or a reduction in the unstacking effect of heating (Table 1). Moreover, a larger reduction in the %H value for S-dA36 than for S-rA36 would be consistent with an enhanced affinity of S-dA36 for g5p. However, such a correlation with %H is not supported in general, because the effect of phosphorothioate modification on %H was barely detectable for S-2′-O-MeA36 and S-d(AAprC)12 relative to 2′-O-MeA36 and d(AAprC)12, respectively, although phosphorothioate modification resulted in significant increases in binding affinities (Tables 1 and 2). A reduction in magnitude of the long-wavelength CD bands of oligomers due to the phosphorothioate modification (Figs 2, 4 and S1), while indicative of a perturbed and possibly less stacked structure, also could not be relied on as a predictor of increased g5p binding affinity, because the 2′-O-Me modification increased the same band magnitudes while still causing an increased binding affinity to g5p. Finally, we note that phosphorothioate modification had a significant effect on the binding of S-d(AAC)12 versus d(AAC)12, although d(AAC)12 is already a relatively unstacked sequence (28).
Therefore, although the spectral properties of the phosphorothioate-modified sequences indicated slight alterations in the free oligomer conformations that may play a role in increasing the binding affinity to g5p, we conclude, in support of previous work (28), that reduced base stacking in the nucleic acid structure probably did not dominate the binding enhancement caused by phosphorothiaote modification. Direct effects of the phosphorothioate, including electrostatic, van der Waals and hydrophobic effects, more likely contributed to the increased binding affinity.
Effect on g5p binding affinities of the 2′-O-Me modification
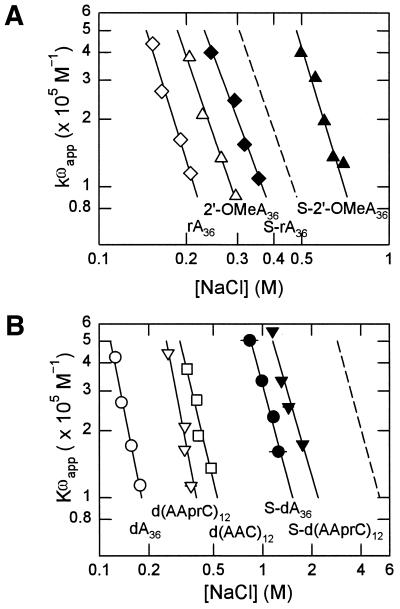
Figure 6A shows the log[Kωapp] versus log[NaCl] dependence for binding to 2′-O-MeA36 and S-2′-O-MeA36. The binding dependences for rA36 and S-rA36 are shown for comparison. The slopes of 3.78 ± 0.38 for 2′-O-MeA36 and 3.76 ± 0.51 for S-2′-O-MeA36 were the same, while the effect of phosphorothioate modification was to reduce the number of ions released per g5p dimer in three other cases [S-dA36, S-rA36 and S-d(AAprC)12] (Table 2). Therefore, this effect of the sulfur substitution did not play a role in altering the binding affinity in the presence of the 2′-O-Me group.
Figure 6.
Dependence of log[Kωapp] on log[NaCl] for all the modified and unmodified oligonucleotides. (A) Comparison of the binding affinities for RNA oligomers; complexes of g5p with rA36 (open diamonds), S-rA36 (closed diamonds), 2′-O-MeA36 (open triangles), and S-2′-O-MeA36 (closed triangles). The dashed line represents the predicted sum of effects on g5p binding from phosphorothioate and 2′-O-Me modifications of rA36 calculated as: log[Kωapp]S-2′-O-MeA = log[Kωapp]2′-O-MeA + log[Kωapp]S-rA – log[Kωapp]rA. (B) Comparison of the binding affinities for DNA oligomers; complexes of g5p with d(AAC)12 (open squares), d(AAprC)12 (open inverted triangles) and S-d(AAprC)12 (closed inverted triangles). Data for dA36 (open circles) and S-dA36 (closed circles) are shown for reference. The dashed line represents the predicted binding affinity for S-d(AAC)12 calculated as: log[Kωapp]S-dAAC = log[Kωapp]dAAC + log[Kωapp]S-dA – log[Kωapp]dA. Complexes of oligomers with g5p were formed at [P]/[N] = 0.25 in 2 mM Na+ (phosphate buffer, pH 7.0), except that complexes with the 2′-O-MeA36 oligomer were formed at [P]/[N] = 0.30. Total protein concentrations in these complexes were in the range 4.8–24.0 × 10–6 M.
The binding affinities, extrapolated to 0.2 M NaCl, of g5p for the 2′-O-Me-modified and unmodified oligomers are compared in Table 2. Methylation of ribose at the 2′ position to give 2′-O-MeA36 increased the binding affinity of g5p by ∼3-fold over that for unmethylated rA36 (Table 2). Furthermore, the binding affinity of g5p was substantially increased by 30-fold when the phosphorothioate modification was added to that of the 2′-O-Me modification. As shown in Figure 6A, the experimental Kωapp for binding to S-2′-O-MeA36 was greater than the Kωapp predicted for the combined, independent effects of 2′-O-Me and phosphorothioate modifications on rA36 (dashed line). That is, when considering the RNA structure, the addition of the 2′-O-Me modification may be of no advantage in decreasing the non-specific binding to a protein such as the g5p. On the other hand, the Kωapp for binding S-2′-O-MeA36 was about one-half of that for binding S-dA36, so that, compared with typical S-DNA oligomers, the addition of the 2′-O-Me modification should be advantageous in reducing non-specific protein binding.
The binding affinities of g5p for the adenylic acid 36mers increased in the order of dA36 < rA36 < 2′-O-MeA36 < S-rA36 << S-2′-O-MeA36 < S–dA36 at the extrapolated salt concentration of 0.2 M NaCl. This order was in agreement with the order of S-RNA << S-2′-O-MeRNA < S-DNA found by another group for the non-specific binding of plasma proteins, such as human serum albumin, γ-globulin and fibrinogen for these oligomer modifications (38).
Oligonucleotides that are methylated at the ribose sugar 2′ position adopt an RNA-like conformation and form a 2′-O-Me-RNA·RNA hybrid that has an A conformation (34,41). A 2′-O-Me-RNA·RNA hybrid has a greater thermal stability than does an S-DNA·RNA hybrid (42). Several studies indicate that antisense activity can be increased by the 2′-O-Me substitution in the ASO. In a cell-free translation assay, a 2′-O-Me RNA demonstrated a better antisense efficiency in repressing the translation of rabbit α-globin mRNA than did the analogous sequence of S-DNA (43). Agrawal et al. (7) showed that the 2′-O-Me modification can increase the antisense effect of phosphorothioate DNA in proportion to the number of modified sugars in the sequences. The inhibitory concentration that is required to suppress HIV-1 replication (IC90) by a 25mer S-DNA can be 20-fold less when 7–11 of the nucleotides are substituted with 2′-O-Me sugars (7). It should be noted that a 2′-O-methylated ASO has to be uniformly phosphorothioate modified in order to be fully nuclease resistant (44).
The higher affinity of g5p for S-2′-O-MeA36 than for 2′-O-MeA36 was consistent with other observations that non-specific protein binding is associated with the phosphorothioate backbone in a 2′-O-methylated ASO. Pitts and Corey (45) found that oligomers with 2′-O-Me and S-2′-O-Me modifications have a comparable efficacy in inhibiting human telomerase. However, the phosphorothioate-modified oligonucleotide had multiple binding sites at the catalytic site of the telomerase protein as well as at the potential telomerase RNA target position (10). In still another experiment, 2′-O-methylated and S-2′-O-methylated ASOs resulted in enhanced antisense repression of human thymidylate synthase protein expression, compared with the expression by unmodified DNA (12,46). In agreement with our work, the S-2′-O-Me modified oligonucleotide had significantly increased non-specific interactions with unrelated mRNAs and/or cellular proteins (12).
Effect on g5p binding affinities of the prC modification
Figure 6B shows the Kωapp values for the d(AAprC)12 and S-d(AAprC)12 oligomers compared with the values for dA36, S-dA36, d(AAC)12 and calculated values for S-d(AAC)12 (dashed line) on a log[Kωapp] versus log[NaCl] graph. Values of Kωapp for S-d(AAC)12 were calculated as described previously by Mou et al. (28). Experimental values for S-d(AAC)12 could not be accurately determined but were at least as great as calculated for a given [NaCl] (28). The Kωapp values for these six unmodified and modified DNA oligomers were extrapolated to 0.2 M NaCl and are listed in Table 2.
Interestingly, the binding affinity of g5p for the prC-modified d(AAprC)12 was significantly reduced relative to that for the unmodified oligomer. The ΔΔG° value {ΔG°[d(AAprC)12] – ΔG°[d(AAC)12]} that represents the effect on binding free energy was more positive by ∼2.03 kcal/mol per prC modification (Table 2). The addition of the phosphorothioate modification to give S-d(AAprC)12 resulted in an increased binding free energy ΔΔG° = –0.27 kcal/mol, comparable with values measured for S-2′-O-MeA36 and calculated for S-d(AAC)12, with respect to the respective unmodified oligomers. This indicated that the effects of the propyne and phosphorothioate modifications were largely independent. Since the effect of the propyne base modification was likely to modify base stacking (see below), the independence of the two effects was consistent with our conclusion above that the increased binding affinity due to phosphorothioate modification was not mainly the result of decreased base stacking.
There was a substantial increase in %H for the single-stranded prC-modified d(AAprC)12 and S-d(AAprC)12 compared with the unpropynylated d(AAC)12 and S-d(AAC)12, respectively (Table 1). This signified that there was an increase in base–base stacking of the propynyl-modified bases. An increase in base stacking could be responsible for the lowered g5p binding affinities of oligomers containing the propyne modification compared with the unpropynylated d(AAC)12 and S-d(AAC)12 as seen in Table 2 (i.e. 13.9 × 105 M–1 compared with 19.3 × 105 M–1 and 553 × 105 M–1 compared with >1000 × 105 M–1). Therefore, the effect of the propyne modification on protein binding might be enthalpic in nature, whereas the increased DNA·RNA hybridization stability due to this base modification has been shown to reside in a more favorable entropic effect (5).
Phosphorothioate ASOs containing the prC modification have been suggested to be potent and effective ASOs (2,6,47,48). ASOs containing propynyl-modified sequences or phosphorothioate backbones can serve as a substrate of RNase H (2), while ASOs with 2′-O-Me modification do not (1). Enhanced antisense activity could be related to more selective hybridization to the complementary RNA, due to the ability of prC substitutions to substantially increase the stability of a DNA·RNA hybrid (2). Wagner et al. (2) used a microinjection assay to demonstrate that prC ASOs were several orders of magnitude more efficient than unmodified phosphorothioate ASOs in antisense inhibition of a reporter gene. Those and other workers further showed that the prC and phosphorothioate modifications can be effective antisense inhibitors regardless of whether the mRNA target locations are in the 5′- or 3′-untranslated regions, in the coding region, or in sequences containing AUG codon (6,47,48).
Whereas others have attributed the antisense effectiveness of the propyne modification to the greater thermal stability of hybrids formed with propynyl-modified sequences, our work shows that a reduced non-specific protein binding of propynyl-modified sequences could also be a factor in the effectiveness of this ASO modification.
CONCLUSION
In this work, we show that the binding of Ff g5p to DNA or RNA is influenced by nucleic acid modifications that have been used in studies of antisense inhibition of translation. DNA oligomers with phosphorothioate backbones, S-dA36 and S-d(AAC)12, had significantly increased g5p affinities compared with dA36 and d(AAC)12, but this increase in binding affinity was moderated when the phosphorothioate was combined with 2′-OH or 2′-O-Me sugars or with the prC modification. A disadvantage of phosphorothioate-modified oligomers is their ability to bind non-specifically to cellular proteins (8–13). However, the antisense effectiveness of phosphorothioate-modified ASOs can be increased when 2′-O-Me or prC modifications are added (2,6,7,12,42–48). Thus, the available data in the literature support the notion that increased specific antisense activities of oligomers with these modifications may be due, in part, to their decreased non-specific binding to cellular proteins.
It would be valuable to have an assay for the non-specific binding to cellular proteins of various oligomer modifications. Recently, Brukner and Tremblay (49) presented an assay to detect the influence of proteins on the ability of a phosphorothioate ASO to form a hybrid with RNA. They evaluated the ability of a crude mouse liver extract to inhibit the formation of an S-dT18·rA18 hybrid by testing whether a hybrid was formed that could be attacked by RNase H. Moderating the usefulness of their assay is the fact that degradative enzymes (including RNase H) can be present in the cellular lysate. Moreover, some ASO modifications, for example, the 2′-O-Me modification, do not activate RNase H. The binding affinities of ASOs for specific eukaryotic proteins, such as basic fibroblast growth factor, might also be relevant in assessing differences in non-specific binding of various nucleic acid modifications (8,9).
The determination of binding affinities to the Ff g5p could be a particularly useful option for evaluating the undesirable, non-specific binding effects of modifications used in constructing ASOs, since the g5p has well defined ssDNA-binding properties. Although the g5p is not a eukaryotic protein, it is a relevant model protein because its DNA-binding site and binding properties are similar to those of the eukaryotic RPA, and photocrosslinking experiments with ssDNA oligomers in the presence of cellular extracts have shown that RPA is one of the major crosslinked protein products (50). The phosphorothioate modification increases the binding affinity of oligomers for g5p as it does for many eukaryotic proteins. The oligomers that we have studied are longer (36mer) than the usual lengths (∼20mer) of ASOs. However, the use of a longer oligomer serves to minimize end effects. The high cooperativity factor of the g5p ensures that oligomers are saturated, and the small size of the g5p binding site (3–4 nt) means that the effects of various nucleotide modifications can be assessed for ssDNAs that are constrained to bind in a given protein binding-site environment.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online. Figure S1 shows representative CD spectra obtained during titrations of the four oligomers d(AAC)12, S-d(AAC)12, d(AAprC)12 and S-d(AAprC)12 with g5p.
Acknowledgments
ACKNOWLEDGEMENTS
This work was performed by T.-C.M. in partial fulfillment of the requirements for the PhD degree in the Department of Molecular and Cell Biology, The University of Texas at Dallas. Support was provided by grants from the Robert A. Welch Foundation (AT-503), the Texas Advanced Technology Program (009741-0021-1999) and a grant from eXegenics, Inc. (Dallas, TX).
REFERENCES
- 1.Inoue H., Hayase,Y., Iwai,S. and Ohtsuka,E. (1987) Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. Nucleic Acids Symp. Ser., 18, 221–224. [PubMed] [Google Scholar]
- 2.Wagner R.W., Matteucci,M.D., Lewis,J.G., Gutierrez,A.J., Moulds,C. and Froehler,B.C. (1993) Antisense gene inhibition by oligonucleotides containing C-5 propyne pyrimidines. Science, 260, 1510–1513. [DOI] [PubMed] [Google Scholar]
- 3.Mercola D. and Cohen,J.S. (1995) Antisense approaches to cancer gene therapy. Cancer Gene Ther., 2, 47–59. [PubMed] [Google Scholar]
- 4.Eckstein F. (2000) Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev., 10, 117–121. [DOI] [PubMed] [Google Scholar]
- 5.Barnes T. and Turner,D. (2001) Long-range cooperativity in molecular recognition of RNA by oligodeoxynucleotides, with multiple C5-(1-propynyl) pyrimidines. J. Am. Chem. Sci., 123, 4107–4118. [DOI] [PubMed] [Google Scholar]
- 6.Moulds C., Lewis,J.G., Froehler,B.C., Grant,D., Huang,T., Milligan,J.F., Matteucci,M.D. and Wagner,R.W. (1995) Site and mechanism of antisense inhibition by C-5 propyne oligonucleotides. Biochemistry, 34, 5044–5053. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal S., Jiang,Z., Zhao,Q., Shaw,D., Cai,Q., Roskey,A., Channavajjala,L., Saxinger,C. and Zhang,R. (1997) Mixed-backbone oligonucleotides as second generation antisense oligonucleotides: in vitro and in vivo studies. Proc. Natl Acad. Sci. USA, 94, 2620–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guvakova M.A., Yakubov,L.A., Vlodavsky,I., Tonkinson,J.L. and Stein,C.A. (1995) Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors and remove it from low affinity binding sites on extracellular matrix. J. Biol. Chem., 270, 2620–2627. [DOI] [PubMed] [Google Scholar]
- 9.Fennewald S.M. and Rando,R.F. (1995) Inhibition of high affinity basic fibroblast growth factor binding by oligonucleotides. J. Biol. Chem., 270, 21718–21721. [DOI] [PubMed] [Google Scholar]
- 10.Cheng X., DeLong,R.K., Wickstrom,E., Kligshteyn,M., Demirdji,S.H., Caruthers,M.H. and Juliano,R.L. (1997) Interactions between single-stranded DNA binding protein and oligonucleotide analogs with different backbone chemistries. J. Mol. Recognit., 10, 101–107. [DOI] [PubMed] [Google Scholar]
- 11.Giles R.V., Spiller,D.G., Grzybowski,J., Clark,R.E., Nicklin,P. and Tidd,D.M. (1998) Selecting optimal oligonucleotide composition for maximal antisense effect following streptolysin O-mediated delivery into human leukaemia cells. Nucleic Acids Res., 26, 1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz J.C., Agrawal,S. and Chu,E. (1998) Repression of human thymidylate synthase mRNA translation by antisense 2′-O-methyl oligoribonucleotides. Antisense Nucleic Acid Drug Dev., 8, 371–378. [DOI] [PubMed] [Google Scholar]
- 13.Matthes E. and Lehmann,C. (1999) Telomerase protein rather than its RNA is the target of phosphorothioate-modified oligonucleotides. Nucleic Acids Res., 27, 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell M.D. and Gray,D.M. (1993) Characterization of the Pf3 single-strand DNA binding protein by circular dichroism spectroscopy. Biochemistry, 32, 12538–12547. [DOI] [PubMed] [Google Scholar]
- 15.Lohman T.M. and Bujalowski,W. (1991) Thermodynamic methods for model-independent determination of equilibrium binding isotherms for protein-DNA interactions: spectroscopic approaches to monitor binding. Methods Enzymol., 208, 258–290. [DOI] [PubMed] [Google Scholar]
- 16.Kim C., Snyder,R.O. and Wold,M.S. (1992) Binding properties of replication protein A from human and yeast cells. Mol. Cell Biol., 12, 3050–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulsink H., Harmsen,B.J. and Hilbers,C.W. (1985) Specificity of the binding of bacteriophage M13 encoded gene-5 protein to DNA and RNA studied by means of fluorescence titrations. J. Biomol. Struct. Dyn., 3, 227–247. [DOI] [PubMed] [Google Scholar]
- 18.Folkers P.J., Nilges,M., Folmer,R.H., Konings,R.N. and Hilbers,C.W. (1994) The solution structure of the Tyr41→His mutant of the single-stranded DNA binding protein encoded by gene V of the filamentous bacteriophage M13. J. Mol. Biol., 236, 229–246. [DOI] [PubMed] [Google Scholar]
- 19.Skinner M.M., Zhang,H., Leschnitzer,D.H., Guan,Y., Bellamy,H., Sweet,R.M., Gray,C.W., Konings,R.N., Wang,A.H. and Terwilliger,T.C. (1994) Structure of the gene V protein of bacteriophage f1 determined by multiwavelength X-ray diffraction on the selenomethionyl protein. Proc. Natl Acad. Sci. USA, 91, 2071–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray C.W. (1989) Three-dimensional structure of complexes of single-stranded DNA-binding proteins with DNA. IKe and fd gene 5 proteins form left-handed helices with single-stranded DNA. J. Mol. Biol., 208, 57–64. [DOI] [PubMed] [Google Scholar]
- 21.Folmer R.H., Nilges,M., Folkers,P.J., Konings,R.N. and Hilbers,C.W. (1994) A model of the complex between single-stranded DNA and the single-stranded DNA binding protein encoded by gene V of filamentous bacteriophage M13. J. Mol. Biol., 240, 341–357. [DOI] [PubMed] [Google Scholar]
- 22.Guan Y., Zhang,H. and Wang,A.H. (1995) Electrostatic potential distribution of the gene V protein from Ff phage facilitates cooperative DNA binding: a model of the GVP-ssDNA complex. Protein Sci., 4, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olah G.A., Gray,D.M., Gray,C.W., Kergil,D.L., Sosnick,T.R., Mark,B.L., Vaughan,M.R. and Trewhella,J. (1995) Structures of fd gene 5 protein.nucleic acid complexes: a combined solution scattering and electron microscopy study. J. Mol. Biol., 249, 576–594. [DOI] [PubMed] [Google Scholar]
- 24.Terwilliger T.C. (1996) Gene V protein dimerization and cooperativity of binding of poly(dA). Biochemistry, 35, 16652–16664. [DOI] [PubMed] [Google Scholar]
- 25.Kansy J.W., Clack,B.A. and Gray,D.M. (1986) The binding of fd gene 5 protein to polydeoxynucleotides: evidence from CD measurements for two binding modes. J. Biomol. Struct. Dyn., 3, 1079–1110. [DOI] [PubMed] [Google Scholar]
- 26.Mou T.C., Gray,C.W. and Gray,D.M. (1999) The binding affinity of Ff gene 5 protein depends on the nearest-neighbor composition of the ssDNA substrate. Biophys. J., 76, 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sang B.C. and Gray,D.M. (1989) Specificity of the binding of fd gene 5 protein to polydeoxyribonucleotides. J. Biomol. Struct. Dyn., 7, 693–706. [DOI] [PubMed] [Google Scholar]
- 28.Mou T.C., Gray,C.W., Terwilliger,T.C. and Gray,D.M. (2001) Ff gene 5 protein has a high binding affinity for single-stranded phosphorothioate DNA. Biochemistry, 40, 2267–2275. [DOI] [PubMed] [Google Scholar]
- 29.Gray D.M., Hung,S.H. and Johnson,K.H. (1995) Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol., 246, 19–34. [DOI] [PubMed] [Google Scholar]
- 30.Mark B.L., Terwilliger,T.C., Vaughan,M.R. and Gray,D.M. (1995) Circular dichroism spectroscopy of three tyrosine-to-phenylalanine substitutions of fd gene 5 protein. Biochemistry, 34, 12854–12865. [DOI] [PubMed] [Google Scholar]
- 31.Thompson T.M., Mark,B.L., Gray,C.W., Terwilliger,T.C., Sreerama,N., Woody,R.W. and Gray,D.M. (1998) Circular dichroism and electron microscopy of a core Y61F mutant of the F1 gene 5 single-stranded DNA-binding protein and theoretical analysis of CD spectra of four Tyr→Phe substitutions. Biochemistry, 37, 7463–7477. [DOI] [PubMed] [Google Scholar]
- 32.McGhee J.D. and von Hippel,P.H. (1974) Theoretical aspects of DNA–protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol., 86, 469–489. [DOI] [PubMed] [Google Scholar]
- 33.Antao V.P. and Gray,D.M. (1993) CD spectral comparisons of the acid-induced structures of poly[d(A)], poly[r(A)], poly[d(C)] and poly[r(C)]. J. Biomol. Struct. Dyn., 10, 819–839. [DOI] [PubMed] [Google Scholar]
- 34.Popenda M., Biala,E., Milecki,J. and Adamiak,R.W. (1997) Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2′-O-Me(CGCGCG)2 under low salt conditions. Nucleic Acids Res., 25, 4589–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishizaki T., Iwai,S., Ohtsuka,E. and Nakamura,H. (1997) Solution structure of an RNA·2′-O-methylated RNA hybrid duplex containing an RNA.DNA hybrid segment at the center. Biochemistry, 36, 2577–2585. [DOI] [PubMed] [Google Scholar]
- 36.Venkateswarlu D., Lind,K.E., Mohan,V., Manoharan,M. and Ferguson,D.M. (1999) Structural properties of DNA:RNA duplexes containing 2′-O-methyl and 2′-S-methyl substitutions: a molecular dynamics investigation. Nucleic Acids Res., 27, 2189–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray D.M. (1996) Circular dichroism of protein-nucleic acid interactions. In Fasman,G.D. (ed.), Circular Dichrosim and the Conformational Analysis of Molecules. Plenum Press, New York, pp. 469–500.
- 38.Kandimalla E.R., Shaw,D.R. and Agrawal,S. (1998) Effects of phosphorothioate oligodeoxyribonucleotide and oligoribonucleotides on human complement and coagulation. Bioorg. Med. Chem. Lett., 8, 2103–2108. [DOI] [PubMed] [Google Scholar]
- 39.Smith J.S. and Nikonowicz,E.P. (2000) Phosphorothioate substitution can substantially alter RNA conformation. Biochemistry, 39, 5642–5652. [DOI] [PubMed] [Google Scholar]
- 40.Ferrari M.E. and Lohman,T.M. (1994) Apparent heat capacity change accompanying a nonspecific one-dimensional protein-DNA interaction. Escherichia coli SSB tetramer binding to oligodeoxyadenylates. Biochemistry, 33, 12896–12910. [DOI] [PubMed] [Google Scholar]
- 41.Adamiak D.A., Milecki,J., Popenda,M., Adamiak,R.W., Dauter,Z. and Rypniewski,W.R. (1997) Crystal structure of 2′-O-Me(CGCGCG)2, an RNA duplex at 1.30 Å resolution. Hydration pattern of 2′-O-methylated RNA. Nucleic Acids Res., 25, 4599–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cummins L.L., Owens,S.R., Risen,L.M., Lesnik,E.A., Freier,S.M., McGee,D., Guinosso,C.J. and Cook,P.D. (1995) Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res., 23, 2019–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein D., Foster,E., Huang,S.B., Weller,D. and Summerton,J. (1997) A specificity comparison of four antisense types: morpholino, 2′-O-methyl RNA, DNA and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev., 7, 151–157. [DOI] [PubMed] [Google Scholar]
- 44.McKay R.A., Miraglia,L.J., Cummins,L.L., Owens,S.R., Sasmor,H. and Dean,N.M. (1999) Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-α expression. J. Biol. Chem., 274, 1715–1722. [DOI] [PubMed] [Google Scholar]
- 45.Pitts A.E. and Corey,D.R. (1998) Inhibition of human telomerase by 2′-O-methyl-RNA. Proc. Natl Acad. Sci. USA, 95, 11549–11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitz J.C., Yu,D., Agrawal,S. and Chu,E. (2001) Effect of 2′-O-methyl antisense ORNs on expression of thymidylate synthase in human colon cancer RKO cells. Nucleic Acids Res., 29, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raviprakash K., Liu,K., Matteucci,M., Wagner,R., Riffenburgh,R. and Carl,M. (1995) Inhibition of dengue virus by novel, modified antisense oligonucleotides. J. Virol., 69, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamel Y., Lacoste,J., Frayssinet,C., Sarasin,A., Garestier,T., Francois,J.C. and Helene,C. (1999) Inhibition of gene expression by anti-sense C-5 propyne oligonucleotides detected by a reporter enzyme. Biochem. J., 339, 547–553. [PMC free article] [PubMed] [Google Scholar]
- 49.Brukner I. and Tremblay,G.A. (2000) Cellular proteins prevent antisense phosphorothioate oligonucleotide (SdT18) to target sense RNA (rA18): development of a new in vitro assay. Biochemistry, 39, 11463–11466. [DOI] [PubMed] [Google Scholar]
- 50.Revers F., Cario,M., Cao,T.L. and Cazenave,C. (1999) Detection of proteins binding to short RNA·DNA hybrids of short antisense oligonucleotides in Xenopus laevis oocytes and human macrophage cell extracts by photoaffinity radiolabeling. Antisense Nucleic Acid Drug Dev., 9, 317–331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.