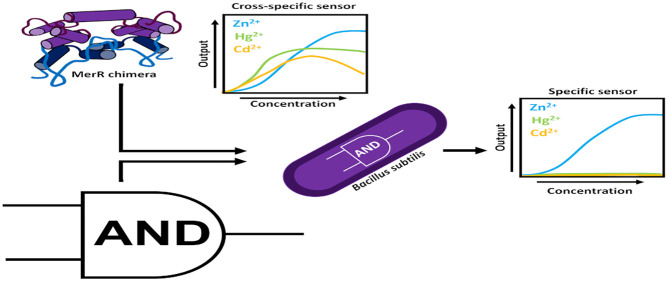
Abstract
Whole-cell biosensors are emerging as promising tools for monitoring environmental pollutants such as heavy metals. These sensors constitute a genetic circuit comprising a sensing module and an output module, such that a detectable signal is produced in the presence of the desired analyte. The MerR family of metal-responsive regulators offers great potential for the construction of metal sensing circuits, due to their high sensitivity, tight transcription control, and large diversity in metal-specificity. However, the sensing diversity is broadest in Gram-negative systems, while chassis organisms are often selected from Gram-positive species, particularly sporulating bacilli. This can be problematic, because Gram-negative biological parts, such as promoters, are frequently observed to be nonfunctional in Gram-positive hosts. Herein, we combined construction of synthetic genetic circuits and chimeric MerR regulators, supported by structure-guided design, to generate metal-sensitive biosensor modules that are functional in the biotechnological work-horse species Bacillus subtilis. These chimeras consist of a constant Gram-positive derived DNA-binding domain fused to variable metal binding domains of Gram-negative origins. To improve the specificity of the whole-cell biosensor, we developed a modular “AND gate” logic system based on the B. subtilis two-subunit σ-factor, SigO-RsoA, designed to maximize future use for synthetic biology applications in B. subtilis. This work provides insights into the use of modular regulators, such as the MerR family, in the design of synthetic circuits for the detection of heavy metals, with potentially wider applicability of the approach to other systems and genetic backgrounds.
Keywords: AND gate, biosensor, synthetic biology, genetic engineering
Introduction
Heavy metal pollution, caused by anthropogenic activities such as metallurgic processes associated with increased industrialization and the overuse of pesticides and fertilizers, poses a risk to the environment and human health.1,2 These metals cannot be broken down and subsequently accumulate within the environment. Furthermore, the presence of heavy metals has been linked to the co-selection of antibiotic resistance genes, as resistance determinants for heavy metals and antibiotics frequently co-occur on mobile genetic elements.3−5 As a result, the persistence of such contaminants in waterways is likely to encourage the dissemination of antibiotic resistance genes in the environment.6,7 It is therefore important to monitor environmental levels of metal contaminants to identify and manage risks, as well as to implement and assess remediation strategies. Traditional analytical techniques such as Atomic Absorption Spectroscopy and Inductively Coupled Plasma Mass Spectrometry (ICP-MS) offer high sensitivity in detection of toxic metals in contaminated environments, but are hampered by cost, lack of in situ monitoring, and do not specifically report the biologically available fractions of polluting metals, which present the most direct risk to human or environmental health. Advances in synthetic biology in combination with decreasing costs of DNA synthesis have made whole-cell biosensors, based on a microbial chassis into which a genetic circuit is built for the detection of an analyte of interest, a potential future option to circumvent these limitations. Indeed, the construction of synthetic circuits in bacteria to develop whole cell biosensors for the monitoring of heavy metals has gained considerable interest.8−11
Metalloregulatory systems offer a source of biological parts for the construction of whole-cell biosensors sensitive to heavy metals.12−14 The MerR protein family is a well described example of metal-responsive regulators.15,16 The corresponding target promoters are characterized by an unusually long spacer region (19–20 bp) between the −10 and −35 elements of a σ70/σA dependent promoter, which places these elements on opposite faces of the DNA. As a result, the promoters are a poor substrate for RNA polymerase binding and transcription initiation.17 The regulator, MerR, binds between the −10 and −35 elements of the promoter and upon binding of an inducer, such as Hg2+ or Cu+ ions, undergoes a conformational change to under-twist the promoter and realign the −10 and −35 elements. This facilitates recognition by RNA polymerase and triggers transcription initiation.17,18 The tightly controlled mechanism of transcription and the high sensitivity and selectivity of MerR regulators for specific metal ions make them ideal for the design of metal sensing circuits.8,9,19 Moreover, as MerR proteins are located cytoplasmically, toxic metals must pass into the cell to evoke a transcriptional response. MerR-based biosensors thus give an indication of the bioavailability of a given contaminant.
MerR regulators have a modular architecture consisting of two discrete domains, an N-terminal DNA binding domain (DBD) responsible for promoter recognition20−22 and a C-terminal domain with a metal binding loop for the coordination of metal ions, referred to here as the metal binding domain (MBD).23−25 The specificity of metal recognition in the MBD is determined by metal coordinating amino acids that allow the coordination of some metals but exclude others. Diversity within these domains facilitates the detection of different metals, providing potential candidates for biosensors with different specificity. These MerR proteins can be used as the sensory modules in biosensors, with their corresponding target promoter fused to a detectable output, e.g., fluorescent or luminescent reporter genes. However, harnessing the sensing diversity of the MerR family requires incorporating a new protein every time the specificity needs to be changed, each of which includes a new DBD that recognizes a different promoter. This necessitates the redesign of the output module to ensure the promoter is recognized, and a signal can be detected.
Furthermore, the largest diversity of metal-specificity in MerR family regulators is found in Gram-negative bacteria—including ZntR (for Zn2+) and CueR (for Cu+).26 Heterologous use of regulators in chassis systems from unrelated species can be problematic due to competition for host transcription and translation machinery resources,27,28 interference from the host genetic background,29 and species-specific differences in the recognition of circuit parts, such as promoters, ribosome binding sites, and other regulatory features.30−32 Synthetic biology approaches can circumvent these problems, for example, through rewiring biological circuits with synthetic promoters to solve transcriptional incompatibilities. It was also shown that a chimeric two-component response regulator produced by domain-swapping could restore functionality, such as seen with Escherichia coli derived NarX-NarL for use in Bacillus subtilis.33 The modular nature of MerR-family regulators may make these proteins well-suited to such approaches.
The Gram-positive, spore-forming bacterium Bacillus subtilis represents an ideal candidate as a chassis for whole cell biosensors given its biotechnological relevance, genetic tractability, and availability of extensive genetic resources.34−36 Numerous examples exist of synthetic biosensor circuits that have been implemented in this chassis organism with application in the detection of pathological biomarkers, antibiotics, antifungal polyenes, and parasites.37−40 Based on these features, we here aimed to use B. subtilis as the host organism to explore the possibility of using domain swapping to engineer MerR-based biosensor circuits with a range of specificities. We hypothesized that combining variable MBDs with a constant DBD that is functional in B. subtilis would allow us to harness the metal binding diversity of proteins from Gram-negative, while ensuring compatibility with the hosts’ transcriptional machinery. Biosensor design would further be simplified using a single, luciferase-based output module.
An additional challenge in developing application-relevant biosensors is the potentially broad substrate specificity of some MerR regulators, which respond to multiple heavy metals and thus do not facilitate differentiation between specific contaminants. A solution for this may be found in logic gates, such as AND gates, which can be introduced into synthetic circuits to improve specificity.9 AND gates require multiple separate inputs to produce an output. In this way, combinations of nonspecific sensing modules can be assembled to produce specific sensors. The use of recombinase-based AND logic circuits has been demonstrated, but these suffer from slow response times, which limits application.37 While many examples of AND gates in the Gram-negative bacterium E. coli exist,29,41,42 there are comparatively fewer examples in B. subtilis. Based on the extensive genetic resources available for B. subtilis, we here sought to design a logic gate to circumvent slow response times as well as improve the specificity of our designed sensors.
The present study describes the design, optimization, and characterization of heavy metal biosensors in B. subtilis based on chimeric MerR transcription factors. We first demonstrate the functionality of a heterologous MerR circuit derived from Staphylococcus aureus TW2043 in B. subtilis for the detection of Hg2+ ions. Subsequently, domain-swapping with two representative Gram-negative MerR-family regulators, ZntR (E. coli) and CueR (E. coli), is used to engineer novel specificity of metal detection in B. subtilis. We demonstrate that rational engineering, guided by protein structure predictions, can be used to improve the functionality of such hybrids. To overcome problems with cross-specificity we engineer a standardized and modular AND gate logic system based on the B. subtilis two-subunit σ-factor system, SigO-RsoA—demonstrating its use in generating an ultraspecific heavy metal detection circuit. Our results establish the basic design rules for functional hybrid MerR-based metal sensors, which should easily be adaptable to broaden the range of detectable metals in the B. subtilis chassis and may enable construction of functional hybrid regulators in other genetic backgrounds.
Results and Discussion
Functional Reconstitution of a Heterologous Hg2+-Sensitive Circuit in B. subtilis W168
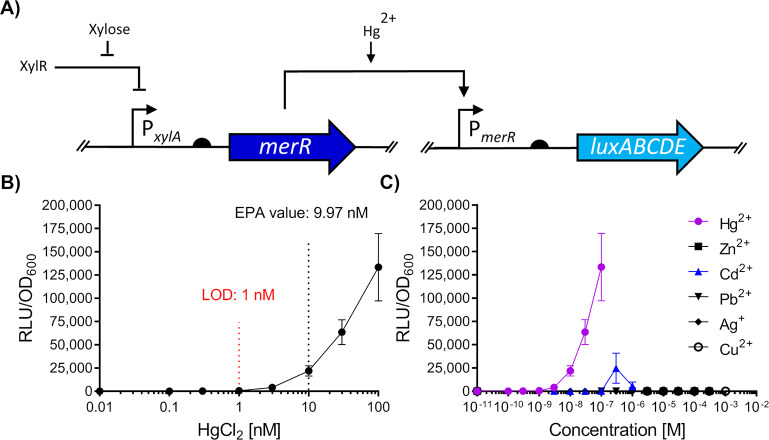
Bacillus subtilis lacks a native metal-sensitive MerR regulatory system. We thus first sought to construct a synthetic metal-sensitive circuit using the regulatory components of the MerR mercury resistance determinant from S. aureus TW20.43 This included the regulator MerR and its cognate promoter PmerR20, which we combined into the whole-cell biosensor genetic circuit shown in Figure 1A. In this circuit, in the absence of any inducing Hg2+ ions, MerR should bind and repress the PmerR20 promoter which possesses an elongated spacer region between the −10 and −35 elements, rendering it a poor substrate for RNAP recognition. As PmerR20 was fused to the bacterial luciferase operon luxABCDE, no luminescence should be detected under these conditions. Binding of Hg2+ (input) promotes conformational changes within MerR44 (whose production is driven by the xylose-inducible promoter, PxylA), facilitating under-twisting of the promoter (PmerR20) and realigning the −10 and −35 element, allowing for RNAP to initiate expression of luxABCDE (output). Thus, luciferase activity of the cells should be correlated with the concentration of exogenous Hg2+ ions.
Figure 1.
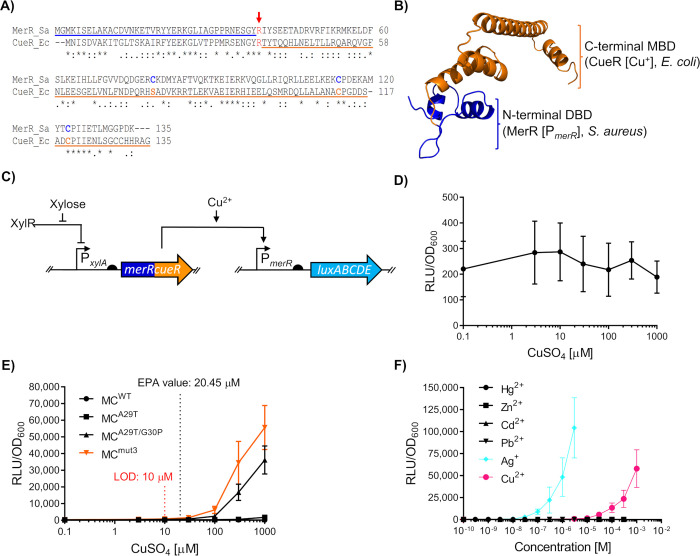
Design and evaluation of a heterologous Hg2+-sensing synthetic circuit in B. subtilis. (A) Schematic representation of the MerR synthetic sensing module in B. subtilis W168 derived from S. aureus TW20. In the circuit, induction with Hg2+ allows MerR to activate luciferase expression as a function of Hg2+ concentration. Bent arrows indicate promoters, flat-head arrows indicate inhibition, black semicircles indicate ribosome binding sites and genes for both merR and luxABCDE coding sequences are indicated by dark and light blue arrows, respectively. (B) Dose response of the Hg2+ circuit. The Environmental Protection Agency (EPA) guideline value is indicated, and the estimated limit of detection (LOD) for the regulatory circuit is shown (red). (C) Metal specificity of the circuit. For panels (B–C), cells were grown to OD600 = ∼0.03 and induced with the concentrations of metals as indicated with luciferase activity output (relative luminescence units [RLU]) normalized to optical density (OD600) values (RLU/OD600) for three time points (35, 40, and 45 min) postinduction. Values are presented as mean and ± standard deviation of either two or three independent replicates.
To test the functionality of this circuit, B. subtilis cells harboring both the PxylA-merR and the PmerR20-luxABCDE constructs (SGB1005) were challenged with subinhibitory concentrations of Hg2+, and promoter activity was monitored. The resulting dose–response behavior (Figure 1B) showed that promoter activity (RLU/OD600) was proportional to the Hg2+ concentration with a limit of detection (LOD) of 1 nM, below the guideline values set by the Environmental Protection Agency (9.97 nM). The sensitivity of this first, simple biosensor was already comparable to previous Hg2+-inducible systems, which had required RBS tuning and amplification circuits to achieve such sensitivity.8 Therefore, we have demonstrated that a heterologous MerR resistance determinant from the Gram-positive S. aureus was functional as part of a synthetic circuit in B. subtilis. In addition, the operational range of this circuit was relevant to application and within the range of values (0.99–17.95 nM) found in environmental samples according to global surveys of total Hg2+ in water over the last 50 years.45
To determine the metal specificity of the circuit we first determined the MIC values of a range of heavy metals (MICs: Cd2+ [10 μM], Ag+ [10 μM], Zn2+ [1 mM], Cu+ [3 mM], Pb2+ [100 μM], and Hg2+ [1000 nM]) and subsequently tested our circuit using sublethal concentrations of these metals (Figure 1C). Strong induction (35–45 min after metal addition) was seen for Hg2+, whereas Zn2+, Cu2+, and Pb2+ did not produce significant signals. The addition of cadmium (Cd2+) at 0.3 μM led to a detectable signal and at higher levels of Cd2+ (1 μM), a drop in RLU/OD600 activity was observed (Figure 1C). Heavy metals such as Cd2+ are known to disrupt protein folding, which includes proteins such as luciferases.46 Therefore, these results could indicate inhibitory effects of Cd2+ on reporter output at this concentration, despite being 10-fold below MIC. As Cd2+ concentrations in the environment over the last 20 years cover a range from ∼0.02–5 μM,47 these may realistically be detected by our sensory circuit. We therefore considered the response profile for this sensing module to be specific for Hg2+ with some cross-reactivity to Cd2+.
Guided Design of Hybrid Regulators to Alter Metal-Specificity of the Sensing Module
Having established a functional sensing module, we then wanted to further explore the use of MerR homologues to construct sensors in B. subtilis with specificities for other metals. Phylogenetic analysis shows that in Gram-positive bacteria, the specificity of MerR regulators appears to be restricted to Hg2+, whereas the diversity of metal specificity appears to be much broader in MerR regulators from Gram-negative species, including metals such as Zn2+, Cu+, Pb2+, and Cd2+.26 Upon initial construction of a metal detection circuit based on Gram-negative-derived biological parts in B. subtilis, we found the promoter to be nonfunctional in the B. subtilis host (Supplementary Figure S1). This is consistent with several other studies that tested promoters from Gram-negatives in B. subtilis, including Ptac,48 PlacUV5,49 the strong synthetic Anderson promoter J23101,31 and the NarX-NarL two-component system target promoter PdcuS77.33 This is likely due to differences in the transcription machinery, for example, in σ-factor stringency between Gram-negative and Gram-positive species.50
To circumvent the issue of host/biological part incompatibility, we investigated the possibility of exploiting the modularity of MerR regulators by using a domain-swap strategy to engineer the metal specificity of the sensing module. We had already demonstrated the compatibility of the S. aureus MerR protein and its target promoter PmerR20 with the B. subtilis host, where the MerR DNA-binding domain (MerRDBD) determined the promoter specificity (see above). We therefore speculated that replacement of the MerR C-terminal metal binding region (conferring metal-specificity) with the corresponding region derived from a MerR homologue of Gram-negative origin may enable us to change the specificity of the sensing module. Indeed, such approaches have been successful using other small one-component regulators such as TetR, LacI, and GalR, for a variety of synthetic circuit applications,51,52 as well as resolving part incompatibility in B. subtilis using chimeric two-component response regulators.33
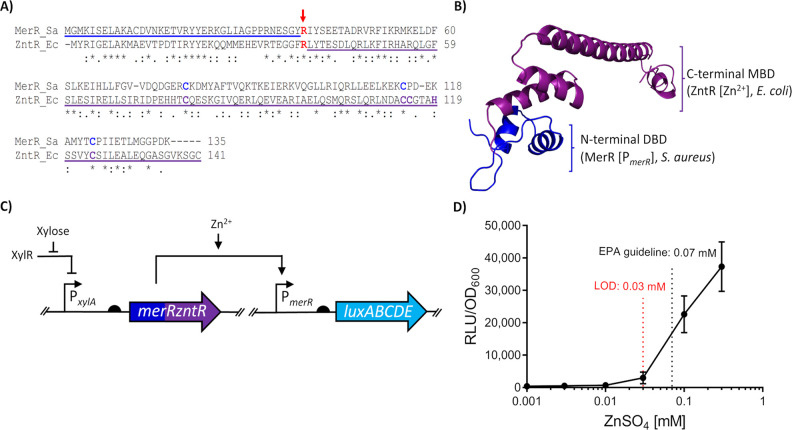
To test this approach, we designed a chimeric MerR regulator, MerRZntR, with the DNA-binding domain derived from the Hg2+-responsive MerR of S. aureus TW20 used above (MerRDBD; residues Met1-Tyr38) and the C-terminal metal binding domain derived from the Zn2+-responsive MerR homologue ZntR from E. coli (ZntRMBD; residues Arg38-Cys141) (Figure 2A). The junction point of Arg38 (ZntR) between the MerRDBD and ZntRDBD was selected based on previous work on ZntR, which showed this region was susceptible to cleavage by trypsin and could separate ZntR into two domains.23 The resulting chimeric amino acid sequence was used to generate a predicted homology model for the structure of MerRZntR, the monomer of which is shown for simplicity (Figure 2B). The analysis of the homology model indicated an overall topology for MerRZntR similar to other MerR regulators, suggesting that the fusion of the two domains would be unlikely to disrupt the overall protein architecture.
Figure 2.
Design and assessment of the chimera, MerRZntR. (A) Sequence alignment of MerR homologues. Both regulators MerR (S. aureus, “Sa”; accession code: CBI50741.1) and ZntR (E. coli, “Ec”; accession code: AAC76317.1) were aligned using the ClustalOmega tool.53 The MerR derived DNA-Binding Domain (residues 1–38) is underlined blue, while the ZntR derived Metal-Binding Domain (residues 38–141) is underlined purple. The fusion point (Arg38, ZntR) is indicated in red with an arrow. Residues involved in Hg2+ coordination by MerR are indicated in dark blue, those involved in Zn2+ coordination by ZntR are indicated in purple. Asterisk (*) indicates fully conserved residues, colon (:) indicates conserved residues with similar properties, and period (.) indicates residues of weakly similar properties. (B) The resulting homology model of MerRZntR using the aforementioned sequence in panel (A) generated using I-TASSER54 is shown; the top-ranking structural analogue was CueR from E. coli (PDB: 1Q05, C-score = 0.71, TM-score = 0.87). The origin of each domain, and their ligands are indicated using the same color scheme as in A. (C) Circuit schematic comprising the designed chimera MerRZntR. Bent arrows indicate promoters, flat-head arrows indicate inhibition, black semi-circles indicate ribosome binding sites, and genes for both merRzntR and luxABCDE coding sequences are indicated by the dark blue/purple and light blue arrows, respectively. (D) Dose response of the MerRZntR chimera sensory circuit. Cells were grown to OD600 = ∼0.03 and induced with the concentrations of Zn2+ indicated, with luciferase activity output (relative luminescence units [RLU]) normalized to optical density (OD600) values (RLU/OD600) recorded and averaged for three time points (35, 40, and 45 min) post-induction. Values are presented as mean ± standard deviation of two or three independent replicates.
To test its functionality, the DNA sequence encoding the chimeric MerRZntR regulator was incorporated into the sensing module developed above, again under control of PxylA, and integrated into the B. subtilis chromosome. Activity of the chimeric protein was again monitored by its ability to control luxABCDE expression from PmerR20 (Figure 2C). The resulting strain (SGB1011) was then tested by measuring promoter activities of cells in exponential growth phase challenged with sublethal concentrations of Zn2+ (Figure 2D). The results showed a Zn2+-concentration dependent response of the promoter, with an LOD of 0.03 mM and a response of 90-fold over unchallenged cells at 0.3 mM (Figure 2D). This clearly indicated that a functional chimera was produced, with the domain-swap leading to a change in metal specificity so that the module could now sense Zn2+. The sensitivity of the module was relevant to environmental levels of contamination, with the EPA guideline value of 0.07 mM falling within the sensitivity range. The data thus demonstrate the feasibility of engineering novel heterologous metal-sensitive biological parts using domain swaps to introduce novel metal specificity into MerR type regulators and overcome problems such as promoter incompatibility in the host organism.
Structure-Guided Mutagenesis Allows Optimization of MerRZntR Activity
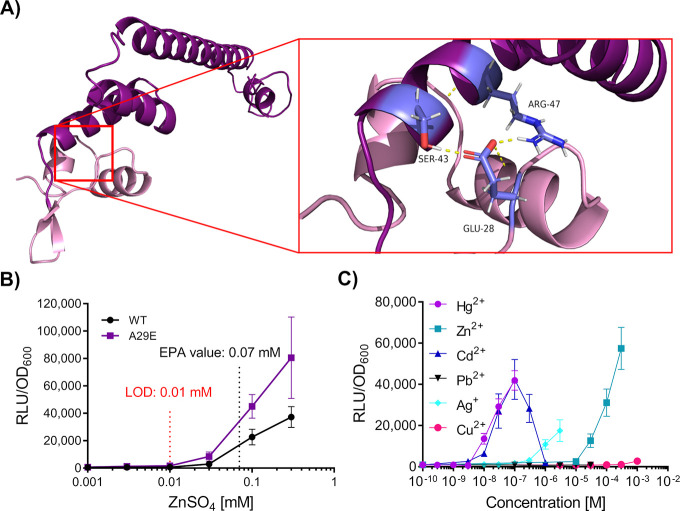
Interdomain amino acid communication within metal-sensitive transcription factors plays a key role in coordinating the binding of a metal with changes in DNA-binding to either activate or repress transcription.55 Examples of such communication can be seen for MerR as well as MerR homologues CueR and SoxR (E. coli), where formation of a hydrogen bonding network upon ligand binding mediates communication between the MBD and DBD to activate transcription by remodelling local promoter topology.17,56 To assess whether this type of interdomain interaction was likely to have been affected by the construction of the MerRZntR hybrid, a homology model of native ZntR was generated (Figure 3A). This was necessary, because the available partial structure of the protein (PDB: 1Q08) lacked both the ZntRDBD and interdomain interactions with its MBD. Hydrogen bonding was observed in the homology model between residues Ser43MBD, Arg47MBD, and Glu28DBD (Figure 3A). However, in the chimera MerRZntR, Ala29DBD is present at the position equivalent to the negatively charged Glu28 in ZntR (Supplementary Figure S2A,B). Seeing as the interaction between Ser43MBD and Glu28DBD may potentially contribute to the signaling mechanism between the two domains, we created an Ala29Glu variant of the MerRZntR hybrid to determine whether we could improve the Zn2+ response. The variant MerRZntRA29E (in strain SGB1035) indeed exhibited greater increases in luminescence across all tested concentrations when compared to MerRZntR, with an improved LOD of 0.01 mM Zn2+ compared to 0.03 mM and a maximum response of 128-fold at 0.3 mM (Figure 3B) compared to 90-fold in MerRZntR. Therefore, restoring the hydrogen bonding between the two protein domains could indeed improve the protein’s activity. Interestingly, upon introduction of additional substitutions in the chimera MerRZntR to re-establish structural interactions, the variants MerRZntRA29E/G30H and MerRZntRA29E/G30H/P32V showed no discernible difference in RLU/OD600 when compared to the single variant A29E (Supplementary Figure S3).
Figure 3.
Structural analysis of ZntR to guide design of the chimera MerRZntRA29E. (A) Homology model of ZntR. I-TASSER54 was used to generate a full-length homology model of ZntR. E. coli, accession code: AAC76317.1, C-score = 0.71, TM score = 0.91). The DNA-Binding Domain is indicated in mauve, while the C-terminal Domain comprising the metal binding loop is indicated in dark purple. Residues involved in interdomain communication (Glu-28, DNA-Binding Domain; Ser-43 and Arg-47, Metal-Binding Domain) are shown in lavender blue with hydrogen bonds shown in yellow. (B) Dose response behavior of the mutant MerRZntRA29E following targeted mutagenesis of MerRZntR. Amino acid substitutions were introduced into the α-helix 2 and α-helix 3 loop region. The Environmental Protection Agency (EPA) guideline value is indicated, and the estimated limit of detection (LOD) for the regulatory circuit is shown (red). The most potent activator, MerRZntRA29E is indicated in purple. (C) Metal-specificity of the MerRZntRA29E based circuit. Inducers are colored using the key shown. For panels (B–C), cells were grown to OD600 = ∼0.03 and induced with the concentrations of metals as indicated, with luciferase activity output (relative luminescence units [RLU]) normalized to optical density (OD600) values (RLU/OD600) for three time points (35, 40, and 45 min) postinduction. Values are presented as mean and ± standard deviation of either two or three independent replicates.
After selecting MerRZntRA29E as the improved Zn2+ sensor, we sought to determine its substrate specificity profile. Strain SGB1035 was subjected to subinhibitory concentrations of various heavy metals (Figure 3C). Cross-specificity was observed for MerRZntRA29E to various heavy metals, including Hg2+, Cd2+, and to a lesser degree Ag+. This cross-specificity could be problematic in environments where these metals are likely to be co-contaminants, as it would make it impossible to distinguish between them and to estimate relevant levels of pollution. Taken together, our data indicate that structure-guided design is a promising approach for optimization of newly designed chimeric biological parts—an approach we have utilized to improve both the overall RLU/OD600 output and LOD of a functional heterologous Zn2+ inducible circuit. However, the cross-specificity of the hybrid MerRZntRA29E has downstream implications for specific monitoring of target heavy metals in potential field applications, which is further addressed below.
Design and Optimization of a Copper Responsive Hybrid MerRCueR through Structure-Guided Mutagenesis
Having demonstrated the feasibility in principle of the domain-swap strategy, we investigated whether this approach could be applied to additional MerR homologues. To test this, we selected the well-characterized copper-responsive MerR homologue CueR of E. coli.17 A chimeric MerRCueR regulator was designed using amino acids 1–38 of MerR (S. aureus), as above, and residues 37–135 of CueR (E. coli), with Arg37 of CueR used as a fusion point between the two domains (Figure 4A). The resulting chimeric amino acid sequence was used to generate a homology model of MerRCueR as before, with a C-score of 0.71 and TM-score of 0.81 (Figure 4B), indicating a close match to the structure of CueR (PDB: 1Q05). The circuit was then assembled in B. subtilis as above, using the PmerR20-luxABCDE reporter (in strain SGB1027) to test functionality (Figure 4C).
Figure 4.
Design, assessment and optimization of the chimera MerRCueR. (A) Sequence alignment of MerR homologues. Both regulators MerR (S. aureus, “Sa”; accession code: CBI50741.1) and CueR (E. coli, “Ec”; accession code: CAD6020341) were aligned using the ClustalOmega tool.53 The MerR derived DNA-Binding Domain (residues 1–38) is underlined blue, while the CueR derived C-terminal Domain (residues 37–135) is underlined orange. The fusion point (Arg37, CueR) is indicated in red with an arrow. Residues involved in Hg2+ coordination by MerR are indicated in dark blue, those involved in Cu+ coordination by CueR are indicated in orange. Note, Ser77 in CueR excludes divalent cations and is not involved in direct metal coordination with Cu+. Asterisk (*) indicates fully conserved residues, colon (:) indicates conserved residues with similar properties, and period (.) indicates residues of weakly similar properties. (B) Homology model of the chimera MerRCueR. I-TASSER54 was used to generated a homology of MerRCueR using the sequence from panel (A). The top-ranking structural analogue was CueR from E. coli (PDB: 1Q05; C-score = 0.71, TM-score = 0.904). The origin of each domain and their ligands are indicated using the same color scheme. (C) Circuit schematic comprising the designed chimera, MerRCueR. Bent arrows indicate promoters, flat-head arrows indicate inhibition, black semicircles indicate ribosome binding sites, and genes for both merRcueR and luxABCDE coding sequences are indicated by the dark blue/orange and light blue arrows, respectively. (D) Dose response of the chimera MerRCueRWT following induction with a range of Cu2+ concentrations. (E) Dose response behavior of MerRCueR mutants following targeted mutagenesis. Amino acid substitutions within the loop region between α-helix 2 and α-helix were introduced into the parent hybrid gene merRcueR generating MerRCueR (MC) variants MCA29T, MCA29T/G30P, and MCmut3 (which carries the substitutions A29T/G30P/P32M). The dose response to CuSO4 is shown for all variants, with the limit of detection (LOD) shown only for MCmut3 (red) and EPA limit indicated. (F) Substrate specificity of the MerRCueR circuit with variant MCmut3. Colors are indicated for the inducing metals only. For panels (D–F), cells were grown to OD600 = ∼0.03 and induced with the concentrations of metal salts indicated, with luciferase activity (relative luminescence units [RLU]) normalized to optical density (OD600) values (RLU/OD600) measured and averaged for three time points (35, 40, and 45 min) postinduction. Values are presented as mean and ± standard deviation of two or three independent replicates.
Exposure of this new strain to Cu2+ at a sublethal concentration of 1 mM failed to induce luciferase expression (Figure 4D). As we had shown earlier that the structural interactions that couple the occupancy of the MBD to movement in the DBD were important for correct functioning of the MerRZntR chimera, we again inspected those residues in the homology model for MerRCueR. This revealed that several interactions between residues were absent in the MerRCueR hybrid17 (Supplementary Figure S4A,B). This included interactions between the side-chains of Glu46MBD and Thr27DBD; the side chain of His43MBD with the Pro28DBD backbone;17 and the backbones of Thr38MBD and Met30DBD. The corresponding residues at these positions in the S. aureus derived MerRDBD are Ala29, Gly30, and Pro32, which would cause some disruption in the interdomain communication network (Supplementary Figure S4B). As residues Thr27DBD, Pro28DBD, and, to a lesser extent, Met30DBD (a preference for a hydrophobic residue at this position) are highly conserved in CueR homologues from several genetic backgrounds (Supplementary Figure S4C), we speculated that, as with the chimera MerRZntR, restoration of the hydrogen bonding network in MerRCueR to match that found natively in CueR would generate a functional copper responsive circuit.
To test this, we sequentially introduced amino acid substitutions A29T, A29T/G30P and A29T/G30P/P32M (strains SGB1028, SGB1029, and SGB1030, respectively). We observed gradual improvements in copper-responsive changes in activity from 1.1-fold with MerRCueRWT to 64.3-fold with MerRCueRA29T/G30P/P32M (from here on termed MerRCueRmut3 for simplicity) upon challenge with 1 mM Cu2+ (Figure 4E). MerRCueRmut3 provided the highest RLU/OD600 output at each concentration and an LOD (10 μM) below the EPA guideline value of 20.45 μM (Figure 4E). Moreover, the operational range was within the range of Cu2+ concentrations found in polluted environments,57−59 validating the potential use of this sensor as a relevant Cu2+ monitoring tool. These results confirmed that structure-guided mutagenesis indeed can be used to restore protein functionality following construction of MerR hybrid proteins. More generally, our results support the importance of residues relaying occupancy of the metal-binding site to movement in the DBD in metalloregulators.17,55
To assess the substrate-specificity, MerRCueRmut3 (SGB1030) was assayed in the presence of Cu2+, Hg2+, Cd2+, Pb2+, and Zn2+ as well as Ag+—a monovalent ion to which E. coli CueR is known to cross-react.60,61 Consistent with previous reports, MerRCueRmut3, did not show any response to divalent cations Hg2+, Cd2+, Pb2+, and Zn2+ (Figure 4F). Cross-specificity was observed for Ag+, which revealed an LOD (0.03 μM) lower than values set by the EPA (0.46 μM) (Supplementary Figure S5) and an operational range of the biosensor strain covering reported values for silver in polluted environments.62 This responsiveness to a monovalent metal ion is consistent with previous work,61 which showed that purified CueR protein in fact responds to monovalent Cu+ ions. In our experiments with living bacterial cells, copper was supplied as CuSO4 and thus Cu2+ ions. But uptake into the reducing conditions of the cytoplasm subsequently leads to conversion to Cu+, where this ion is detected by the CueR MBD, explaining the specificity profile of the biosensor of Cu2+ and Ag+.
Taken together, our results thus far highlighted that the modularity of the MerR regulators can be exploited to generate novel sensing modules, and a chimera-based strategy can be used to overcome species-specific design constraints such as promoter incompatibility. We envisage that a chimeric approach may be applicable to other protein families for import into a heterologous host such as B. subtilis, where using a closely related protein homologue from the desired host, or a related species, for example, S. aureus, could be a suitable donor of DNA-binding domains.
MerR Hybrids Display Preferences in Promoter Spacing Distance
Past research has given detailed insights into how MerR regulators bind and differentiate between metal ions,63−67 which allowed us to develop the functional hybrid regulators described above. However, to fully engineer these proteins for synthetic biology applications, detailed knowledge is also required of the factors that enable MerR regulators to correctly under-twist their respective cognate promoters. MerR target promoters generally possess either a 19- or 20-bp spacer between the −10 and −35 elements. In the following text, a MerR homologue will be denoted with a subscript of spacing in its target promoter, for example, CueR19 acts upon the PcopA19 promoter, which possesses a spacer region of 19 bp. Perturbation of this spacer region in MerR family promoters is known to disrupt correct transcriptional regulation of the promoter.68 Interestingly, previous work on ZntR20 has suggested that the MBD, rather than the DBD, determines the degree of promoter distortion.23 This would imply that regulators CueR19 and ZntR20, which act on promoters with 19- and 20-bp spacer regions, respectively, would distort their respective promoters to different degrees, based on their MBDs, as previously suggested by Brown et al.15 This may have implications for chimeric MerR transcription factors that could prefer a target promoter whose spacer region length is determined by the origin of the MBD, even if the same DBD was used in all chimeras. For example, in the MerRCueRmut3 regulator we described above, we had so far used the 20 bp-spaced promoter PmerR20 to drive expression of the luciferase reporter. This promoter is natively recognized by MerR20 of S. aureus. However, the donor protein of the MBD, E. coli CueR19, natively controls a 19 bp-spaced promoter, PcopA19. If it is indeed the MBD that determines promoter distortion, we might hypothesize that the MerRCueRmut3 chimera should perform better when provided with a promoter with a 19-bp spacer region.
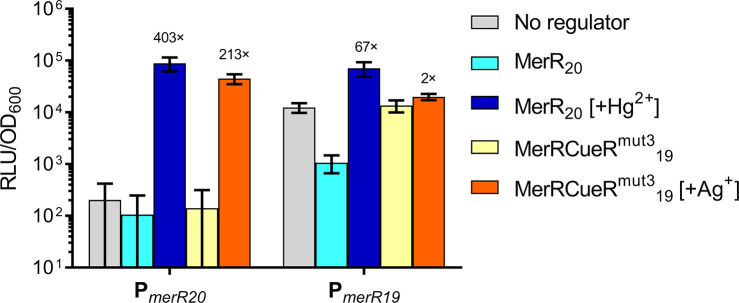
To test the effects of the C-terminal MBD region of our hybrid proteins on promoter regulation, we investigated the ability of MerR and MerRCueRmut3 to regulate the activity of either a 19- or 20-bp spaced promoter. PmerR20 is the S. aureus promoter that we have used so far, and PmerR19 is its derivative in which 1bp was deleted (Supplementary. Figure S6). When MerR20 was used to regulate the activity of the PmerR19-lux reporter, the dynamic range of induction was severely perturbed, with higher basal activity when compared to PmerR20 (Figure 5). This was consistent with previous studies of the mer operon of Tn501 in Gram-negative systems, where increased basal expression was seen when the 19-bp spacer was shortened.68 Thus, MerR of S. aureus worked better when provided with its native promoter in the heterologous B. subtilis system, as expected.
Figure 5.
Regulation of promoters PmerR20 and PmerR19 by wild-type MerR and the chimeric MerRCueRmut3. The activity of either promoters PmerR20 and PmerR19 fused to the luciferase reporter was tested in either the absence of any regulator (gray), or in the presence of the regulator MerR (blue) or MerRCueRmut319 (yellow/orange). The experiments were carried out in the absence (lighter colors) or presence of an inducing metal (MerR, dark blue [100 nM Hg2+], MerRCueRmut319, orange [3 μM Ag+]). Cells were grown to OD600 = ∼0.03 and then induced with the metals as indicated, with luciferase activity output (relative luminescence units [RLU]) normalized to optical density (OD600) values (RLU/OD600) measured and averaged for three time points (35, 40, and 45 min) postinduction. Fold values represent the induction ratio between induced against uninduced. Values are presented as mean ± standard deviation of two or three independent replicates.
Next, we compared the ability of MerRCueRmut3 to regulate the activity of both PmerR20 and PmerR19 transcriptional reporters. Surprisingly, in the system containing the PmerR19 promoter, the presence of the MerRCueRmut3 regulator did not lead to any change in promoter activity compared to cells lacking a regulator. This was not changed whether the inducer (Ag+) was present or not, suggesting the chimeric protein was unable to interact with the shorter spaced target promoter. In contrast, the chimeric regulator was able to elicit a 213-fold increase in the promoter activity of PmerR20 when challenged with Ag+ (Figure 5). This strongly suggests that in the MerRCueRmut3 hybrid, the MBD did not determine optimal promoter spacing, and thus behaved differently from earlier reports on ZntR.23 It is currently not clear whether this is because here the DBD and promoter were derived from a Gram-positive system, where the DBD may play a more important role in determining promoter spacing, or whether there simply is no generalizable rule on which promoter spacing is optimal for hybrid MerR regulators. Different combinations of MBD and DBDs from a variety of systems and donor species would need to be tested using different output modules to answer this. However, we can conclude that for the B. subtilis system used here, we appear to be able to construct and use chimeric regulators with diverse metal specificity without the need of adjusting promoter spacing in the output module.
The Two-Subunit Sigma Factor SigO-RsoA Enables the Design of Modular AND Logic Circuits
Having demonstrated the utility of and some design rules for chimeric regulators as novel metal-responsive circuits, we wanted to test whether we could overcome possible problems with cross-specificity between metals by designing a modular AND logic gate. This type of genetic gate requires the presence of two inputs in order to produce an output.41 To illustrate, none of our metal biosensors responded to only a single metal. But it may be possible to use AND logic to combine two different metal sensors whose substrate specificity overlaps only for one metal. In such a case, this metal would be the only substrate to trigger both regulators, and therefore the output signal would be produced in response only to this single metal, creating a highly specific biosensor. A similar approach has already proved effective in generating an ultraspecific metal sensor circuit in E. coli.9 However, a standardized, easy to use two-input AND gate system, offering modular assembly and fast response times, is currently not available in the suite of genetic toolboxes for B. subtilis.
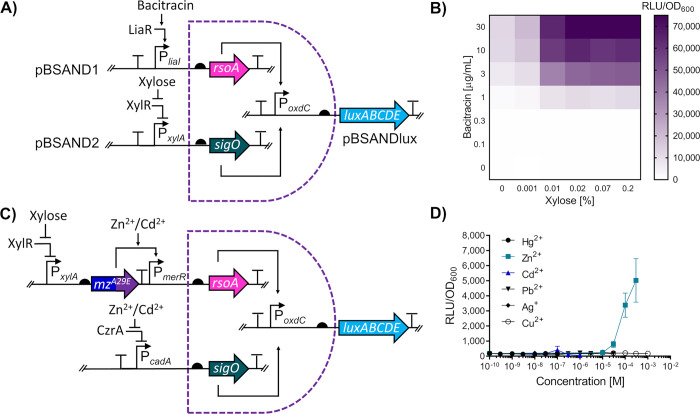
Therefore, to generate such a system, we exploited the B. subtilis two-subunit sigma factor system SigO-RsoA. Various studies have demonstrated that both SigO and RsoA, which constitute domains σ4 and σ2 of the sigma factor, respectively, must cooperate to initiate transcription from the promoter PoxdC.69,70 This system thus effectively acts as a natural biological AND gate, which should be amenable for engineering such regulatory logic in synthetic circuits. To facilitate fast assembly of the AND gate, we generated a series of new plasmids termed SANDBOX (Subtilis AND BOX) based on the Golden Gate assembly format using the type II restriction enzyme BsaI. This toolbox includes vectors pBSAND1 (carrying rsoA), pBSAND2 (carrying sigO), pBSANDlux (carrying the PoxdC-luxABCDE reporter), and a CRISPR based deletion plasmid (pBSANDdel) used to delete the native SigO-RsoA divergent regulon.71 The architecture of all the vectors and the Golden Gate cloning site sequences can be found in Supplementary Figure S7.
For the design and validation of our initial two-input AND gate, we used two well-characterized B. subtilis promoters, PxylA and PliaI,31 to control transcription of SigO and RsoA, respectively. The output from the gate (luciferase activity) was driven by the cognate promoter of this two-subunit σ-factor system, PoxdC. The overall architecture of the circuit is shown in Figure 6A. To assess the functionality and determine optimum induction of the AND gate, we assayed different combinations of bacitracin (PliaI-inducer) and xylose (PxylA-inducer) concentrations, revealing the system behaved correctly in an AND gate manner,41 i.e., reporter induction was only observed if both inducers were added together (Figure 6B). We did observe some leakiness from PxylA, giving weak outputs when bacitracin alone was used, which was consistent with a previous study that characterized the activity of this promoter.31 Overall, our data demonstrated that the new standardized logic system was functional in B. subtilis and should allow for flexible and rapid assembly of two-input AND gate circuits for a variety of applications.
Figure 6.
Assessing the functionality and modularity of the two-subunit SigO-RsoA σ-factor two-input AND gate for B. subtilis. (A) Circuit schematic of the two-input AND gate. To test the functionality of the SigO-RsoA based AND gate, two promoters PliaI and PxylA, inducible by bacitracin and xylose, respectively, were used to induce the expression of both SigO and RsoA, allowing for transcription from the PoxdC promoter to drive luciferase (luxABCDE) expression. The sigO and rsoA genes are indicated via the teal and pink arrows, respectively. (B) Functionality of the SigO-RsoA AND gate using bacitracin and xylose. Cells with the integrated SANDBOX vectors incorporating a bacitracin and xylose inducible promoter were induced with various combinations of xylose and bacitracin with the heatmap used to show luciferase output across the tested concentrations. (C) Circuit schematic for an ultraspecific Zn2+ biosensor used the SigO-RsoA system. SigO and RsoA genes are indicated as previously described. The circuit, which utilizes the chimera MerRZntRA29E, is denoted via the split dark blue and purple arrow and simplified to “mzA29E”. For simplicity, the native B. subtilis metalloregulator CzrA present at a different genomic locus on the chromosome has not been indicated. (D) Dose response and specificity of the SigO-RsoA based Zn2+ detection circuit. For panels (B) and (D), cells were grown to OD600 = ∼0.03 and induced with the concentrations of either xylose and bacitracin (B) or metals (D) as indicated. Luciferase activity output (relative luminescence units [RLU]) was normalized to optical density (OD600) values (RLU/OD600) measured and averaged for three time points (35, 40, and 45 min) postinduction. Values are presented as mean ± standard deviation of two or three independent replicates.
Having confirmed the functionality of our SANDBOX system, we sought to combine it with our chimeric biological parts for metal detection to develop an ultraspecific zinc biosensor. As mentioned above, this required two different metal-inducible regulators with overlapping substrate specificity for one metal. Moreover, the AND gate requires the use of two target promoters without cross-recognition by the two regulators. Given our MerR-based systems all converge on the same output promoter, we had to source the second component of our AND gate system from an unrelated metal sensor. Therefore, we decided to use the optimized chimera MerRZntRA29E, together with CzrA—a native metalloregulator in B. subtilis belonging to the ArsR protein family.72 While we had already determined the specificity profile for MerRZntRA29E (Figure 3C), we needed to determine this for CzrA. For this, we generated a transcriptional luciferase reporter of the CzrA target promoter PcadA and exposed cells harboring this reporter to the same panel of metals used before. This revealed induction of PcadA-luxABCDE in the presence of Zn2+ and, to a minor degree, Cd2+ (Supplementary Figure S8). Thus, both MerRZntRA29E and CzrA responded strongly to Zn2+ but shared no other substrates. They should therefore be a suitable pair for construction of the AND gate biosensor. To note, while both regulators responded to Cd2+, transcriptional output from PcadA was negligible compared to the output produced from MerRZntRA29E (Figure 4C and Supplementary Figure S8 and thus unlikely to create interference.
Based on this information, we proceeded to construct the AND gate-based Zn2+-specific circuit, shown schematically in Figure 6C. Expression of SigO was placed under control of PcadA (CzrA), and RsoA expression was placed under control of PmerR (MerRZntRA29E). To generate a detectable output, luciferase expression was again controlled from PoxdC (SigO-RsoA). When the strain harboring this genetic circuit was tested against the same panel of metals as above, a strong response was only obtained in the presence of Zn2+, showing a clear dose–response behavior (Figure 6D). As anticipated, the signal elicited by the presence of Cd2+ was very low and barely detectable above background. This showed that the use of a simple logic AND gate significantly reduced the noise generated from nontarget metals, such as Cd2+, to generate a highly specific Zn2+ biosensor from individual regulatory parts that each respond to multiple different metals. Furthermore, the resulting data confirmed that the B. subtilis SigO-RsoA system can be exploited for the design of robust AND gates, with the modularity of the system demonstrated through the adaptation of this system in the design of an ultraspecific biosensor circuit.
Conclusions
Monitoring of environmental levels of heavy metal contamination is integral in the management of the risk associated with polluted areas and to assess the efficacy of remediation efforts at these sites. Whole cell biosensors offer an attractive alternative to conventional monitoring methods, which can be expensive and resource heavy. In this work, we demonstrated that novel hybrid transcription factors can be assembled using the modular MerR proteins from different bacteria, and that their functionality as biosensors in B. subtilis can be optimized using structure-guided mutagenesis. This approach will allow researchers to tap into the great diversity of substrate specificity found in the MerR proteins from Gram-negative bacteria for use in Gram-positive chassis organisms, by utilizing their metal-binding domains in a hybrid protein that has a constant DNA-binding domain from a Gram-positive donor. This is important in overcoming problems commonly faced when sourcing genetic components from different species for use in established chassis systems such as B. subtilis, including issues with promoter recognition and compatibility with the host’s transcription machinery.
We initially demonstrated the functionality of a heterologous MerR based regulatory circuit from S. aureus for the detection of Hg2+ ions in B. subtilis and showed that the modular architecture of MerR can be exploited to generate novel chimeric regulators by replacing the metal-binding domain of one regulator with one from another protein with different specificity. Maintaining the same DNA-binding domain in all proteins addresses problems of promoter recognition. We further demonstrated that the functionality and sensitivity of these circuits can be improved through structure-guided design, allowing monitoring of metal contamination in environmentally relevant ranges. While some chimeras with a MBD from Gram-negative-derived proteins may not immediately be functional, we have shown here how engineering of residues in the communication interface between MBD and DBD can be used to restore function.
Finally, we showed that problems with cross-specificity can be resolved by incorporating our novel orthologous regulators into AND gate-based logic circuits that include native B. subtilis metalloregulators. For this, we utilized the B. subtilis two-subunit σ-factor system SigO-RsoA and demonstrated that this can drastically reduce the signal from nontarget contaminants. Initial construction and validation of the circuit was done using well-characterized B. subtilis promoters, with subsequent demonstration of the modularity of this system in the design of an ultraspecific Zn2+ sensor based on overlapping specificities of two metalloregulators.
Apart from the design and testing of novel biosensors, this work has led to the development of a new toolbox of Golden Gate-based vectors, which enables easy construction of two-input AND gates in B. subtilis. We anticipate that based on the modularity of this system, it will not only be useful for the design of a variety of biomonitoring tools, but also can be adopted for a range of applications such as biomedical diagnostics or metabolic engineering. Taken together, this work provides insights into how modular regulators, such as the MerR family, can be exploited in the design of synthetic circuits for the detection of heavy metal contaminants. We have shown how structure-guided design can produce functional sensors even when protein domains are sourced from distinct species, which may also inform work on other proteins with a similar modular architecture.
Materials and Methods
Bacterial Strains, Growth Conditions, and Reagents
All strains used in this study are listed in Table S1 and were routinely grown in Lysogeny Broth (LB; 10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl) at 37 °C with aeration (agitation at 180 rpm). Solid media contained 1.5% (wt/vol) agar. Selective media for B. subtilis contained chloramphenicol (5 μg/mL) or spectinomycin (100 μg/mL), while selective media for E. coli contained ampicillin (100 μg/mL). For luciferase assays, B. subtilis strains were grown in a modified M9 minimal media (MM9). The composition of MM9 was as follows: 1 mM MgSO4, 0.3% fructose, 1% casamino acids, 0.05 mM FeCl3/0.1 mM citric acid solution, deionized water and 1× M9 salts (31.7 mM Na2HPO4, 17.22 mM K2HPO4, 17.11 mM NaCl, 9.34 mM NH4Cl). For transformations of E. coli DH5α (see below), SOC medium was used with the following composition: 2% tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose. For transformations of B. subtilis, MNGE medium was used based on the composition described by Radeck et al.31 Further details about the transformation procedure can be found below. Metal salts Ag(NO3), ZnSO4·7H2O, Pb(NO3), and CdCl2 were obtained from Fisher Scientific, CuSO4·7H2O and HgCl2 were obtained from Sigma-Aldrich.
DNA Manipulation, Plasmid Construction, and Bacterial Transformation
Detailed information regarding strains, plasmids, primers, and genetic sequences for biological parts used in this used in this study are listed in Supporting Information, as Tables S1, S2, S3, and S4, respectively. All cloning steps, including restriction endonuclease digestion, ligation, and PCRs, used enzymes and buffers from New England Biolabs (NEB; Ipswich, MA, USA) according to the relevant NEB protocols. All PCR cleanup kits (Monarch PCR cleanup kit), plasmid (Monarch PCR mini-prep kit), and gel extraction kits (Monarch DNA gel extraction kit) were also obtained from NEB and used according to the manufacturer’s protocols. For ligation of inserts into plasmid vectors used, except for the Golden Gate procedure described below, the NEB T4 DNA ligase protocol M0202 was used. All PCR amplifications were performed using Q5 DNA polymerase (NEB protocol M0491), whereas for colony PCR to confirm ligation of inserts into desired vectors, OneTaq polymerase was used (NEB M0480). Chemically competent DH5α cells were transformed with isolated plasmids or ligation reactions using a heat-shock procedure in which cells were mixed with DNA for 10 min on ice, heat-shocked at 42 °C for 90 s, placed back on ice for 5 min, after which SOC medium was added, and cells were incubated at 37 °C in a shaking incubator (200 rpm) for 1 h before plating onto selective media (see above). Transformations of B. subtilis were carried out as described by Harwood and Cutting73 with integration of plasmids derived from pAH328 at the sacA locus confirmed using colony PCR with sacA up- and down-primers SG0528/SG0529 and SG0530/SG0531, respectively, and integration of pXT-derived plasmids at the thrC locus confirmed via threonine auxotrophy, as described by Radeck et al.31
Plasmid Construction
To amplify PmerR and MerR, S. aureus TW20 genomic DNA (gDNA) was isolated using a GeneJet genomic DNA extraction kit (Thermo Fisher). PmerR20 and MerR were amplified using primers SG0985/SG0986 and SG0987/SG0988, respectively, with 20 ng of S. aureus gDNA as a template. The amplified promoter and regulator were digested with EcoRI-/SpeI-HF and BamHI-/EcoRI-HF, respectively, and ligated into vectors pAH328 and pXT, respectively to generate plasmids pJGlux01 and pJGXT01, respectively. To generate a 1-bp deletion of the PmerR20 promoter (PmerR19), we used site-directed mutagenesis with primers designed as described by Liu and Naismith,74 and amplification performed using the Q5 DNA polymerase protocol as described above using 20 ng of pJGlux01 as a template and 2.5 nM of primers SG1124/SG1125 (50 μL reaction). For this, 12 amplifications cycles, an extension time of 1 min per kb, and an annealing temperature of 60–66 °C were used. Following amplification, DpnI was added directly to the reaction to a final concentration of 400 U/mL, with amplification confirmed using agarose gel electrophoresis. This generated plasmid pJGlux02. To generate the Pveg-luciferase fusion, the promoter was excised from pSB1C3–Pveg31 using EcoRI- /SpeI-HF and ligated into EcoRI-/SpeI-HF digested pAH328 to generate plasmid pJGlux03. To generate the promoter fusion for Gram-negative-derived MerR (PcadA19), primers SG1164/SG1171 were used to amplify the PcadA19 promoter from Pseudomonas aeruginosa PAO1 gDNA (20 ng). The amplified promoter was digested with EcoRI/SpeI-HF and ligated into pAH328 to generate pJGlux04.
Metal Sensitive Chimeras
To construct chimeric regulator MerRZntR, the MerRZntR amino acid sequence was designed on a previous chimeric regulator as described by Brocklehurst et al.23 using residues Met1–Tyr38 of the MerRDBD region and residues Arg38 (used as a junction point between both proteins) to Cys141 of ZntR of E. coli MG1655. The resulting chimeric DNA sequence was flanked a 3′-BamHI and 5′-EcoRI sites and commercially synthesized (GenScript, Rijswijk, Netherlands) into vector pUC19 (pJGUC01). The insert was excised with BamHI-/EcoRI-HF (1 μg) and subcloned into BamHI-/EcoRI-HF digested pXT to generate pJGXT02. To generate variants MerRZntRA29E, MerRZntRA29E/G30H, and MerRZntRA29E/G30H/P32V, site-directed mutagenesis approach was used as described above using mutagenic primer pairs SG1172/SG1173, SG1174/SG1175, and SG1200/SG1201, respectively, which generated plasmids pXTJG15, pXTJG16, and pXTJG23, respectively.
The chimeric regulator MerRCueR was constructed in a manner analogous to that of MerRZntR using amino acid residues Met1–Tyr38 of the MerR DBD (S. aureus) and residues Arg37 (used as a junction point) to Gly135 of CueR of E. coli MG1655. The resulting chimeric DNA sequence was flanked with the previously mentioned restriction sites as for MerRZntR and synthesized into pUC19 (pJGUC02). MerRCueR was subsequently subcloned into pXT as described above for the generation of pJGXT02 (see above), with the resulting plasmid designated pJGXT03. For the construction of MerRCueRA29T and MerRCueRA29T/G30P, site-directed mutagenesis was performed as described above using plasmid pJGXT03 as template with mutagenic primers SG1142/SG1143 and SG1154/SG1155 to generate plasmids pJGXT07 and pJGXT08, respectively. For MerRCueRA29T/G30P/P32M (MerRCueRmut3), plasmid pJGXT08 was used as a template with primers SG1167/SG1168 to generate plasmid pXTJG11.
The Bacillus SANDBOX Plasmids
pBSAND1
The Bacillus BioBrick vector pBS4S31 was used as a parent plasmid for the construction of pBSAND1. The BsaI site in the bla (ampr) gene of pBS4S was removed using primer pairs SG1242/SG1243 to generate plasmid pJGBS4S01. The rfp cassette was amplified from pBS4S using primers SG1272/SG1273 to incorporate a 5′-BsaI site and 5′-terminator sequence and 3′-BsaI and 3′-SfiI site. rsoA was amplified from B. subtilis W168 gDNA (20 ng) using primers SG1275/SG1250 to incorporate a 5′-SfiI site and a 3′- terminator and 3′-PstI site. Amplified rfp and rsoA were digested with EcoRI-/SfiI-HF and SfiI-/PstI-HF, respectively, and ligated into EcoRI-/PstI-HF digested pJGBS4S01 in a single reaction. The removal of BsaI from bla, and the insertion of the new BsaI sites flanking RFP were confirmed by restriction digestion with BsaI, and the ligation of both rfp and rsoA into pJGBS4S01 confirmed using PCR with primers SG601/SG602 and Sanger sequencing (Eurofins Genomics, Germany). The resulting plasmid allowing for expression of RsoA from a promoter of choice was designated pBSAND1 and can be linearized for transformation of B. subtilis using the enzyme ScaI. The terminator sequence used is an in silico designed terminator called “Term 1”,75 while to ensure strong expression of RsoA, the RBS sequence R1 described by Guiziou et al.76 was used. The resulting plasmid was designated pBSAND1 and can be linearized using ScaI to allow for integration in B. subtilis at thrC.
For plasmid pBSAND1-PliaI, PliaI was amplified from B. subtilis gDNA using primers SG1388/SG1389 and assembled into pBSAND1 using Golden Gate cloning (see below) to allow for bacitracin inducible expression of RsoA. To construct pBSAND1-PxylA-MerRZntRA29E-Terminator-PmerR20, PxylA was amplified from pSB1A3-PxylA31 using primers SG1297/SG1410; MerRZntRA29E (including its native RBS) was amplified from pJGXT15 using primers SG1382/SG1383; and PmerR20 (to include a 5′ terminator, “Term 1”) was amplified from pJGlux01 using primers SG1384/1385. The three fragments were assembled into pBSAND1 using Golden Gate to allow for metal-inducible expression of RsoA.
pBSAND2
The Bacillus BioBrick vector pBS2E31 was used as a parent plasmid for the construction of pBSAND2. The BsaI site in the bla gene of pBS2E was removed using SG1242/SG1243 generating pJGBS2E03, after which an NgoMIV site was inserted in the bla gene using primers SG1245/SG1246, generating plasmid pJGBS2E04. The same rfp cassette was used as for pBSAND1, while sigO was amplified from B. subtilis gDNA using primers SG1274/SG1248 to incorporate a 5′-SfiI site, a 3′- terminator and a 3′-PstI site. Both amplified rfp and sigO were digested with EcoRI-/SfiI-HF and SfiI-/PstI-HF, respectively and ligated into EcoRI-/PstI-HF digested pJGBS2E04. Removal of BsaI from bla, the insertion of NgoMIV into bla, and the insertion of new BsaI sites flanking RFP were confirmed by restriction digestion with BsaI and NgoMIV. Ligation of rfp and sigO into pJGBS2E04 was confirmed by PCR using primers SG0245/SG0246 and sequencing (Eurofins Genomics, Germany). The resulting plasmid was designated pBSAND2 and can be linearized using NgoMIV to allow for integration in B. subtilis at lacA.
For plasmid pBSAND2-PxylA, PxylA was amplified from pSB1A3-PxylA31 using primers SG1297/SG1298 and assembled into pBSAND2 using Golden Gate to allow for xylose inducible expression of SigO. For pBSAND2-PcadA, PcadA was amplified from B. subtilis gDNA using primers SG1403/SG1387 and assembled into pBSAND2 using Golden Gate (see below) to allow for metal-inducible expression of SigO.
pBSANDlux
The Bacillus BioBrick vector pBS3Slux was used as a parent plasmid for the construction of pBSANDlux (PoxdC-luxABCDE reporter). The BsaI sites in the luxC and bla genes were removed via site-directed mutagenesis as described above using primer pairs SG1239/SG1240 and SG1242/SG1243, respectively, to generate plasmids pJGBS3Clux02 and pJGBS3Clux03, respectively. The rfp cassette was amplified from pBS4S using primers SG1272/SG1324, to incorporate a 5′ terminator as well as flanking 5′- and 3′-BsaI sites which was subsequently digested using EcoRI/PstI and ligated into EcoRI-/PstI-HF digested pJGBS3Clux03. To confirm the presence of the rfp cassette with a 5′-terminator, colony PCR was performed primers SG1303/SG1325 and constructs sequenced using SG0991. Removal of BsaI sites in bla and luxC, as well as the incorporation of BsaI sites flanking RFP were confirmed using restriction digestion using BsaI. The resulting plasmid pBSGGlux can be linearized using ScaI for integration in B. subtilis at the sacA locus.
Finally for plasmid pBSANDlux, required as part of the Bacillus SANDBOX system to generate 2-input biosensors, PoxdC was amplified using primers SG1299/SG1300 from B. subtilis W168 gDNA (20 ng) and assembled into pBSGGlux using Golden Gate (see below). When utilized with plasmids pBSAND1-PliaI and pBSAND2-PxylA, pBSANDlux allows for bacitracin and xylose inducible luciferase output. When utilized with pBSAND1-PxylA-MerRZntRA29E-Terminator-PmerR, and pBSAND2-PcadA, pBSANDlux allows for metal-inducible luciferase output.
pBSANDdel
The CRISPR-Cas9 deletion plasmid pJOE8999 was used as the parent plasmid for the construction of pBSANDdel, with construction done as described by Altenbuchner.77 To amplify left and right homology regions surrounding the sigO-rsoA operon, primers SG1326/1327 and SG1328/SG1329 were used. These fragments were digested with SfiI-HF and ligated into SfiI-HF digested pJOE8999. The insertion of the flanking homology regions was confirmed using colony PCR with primers SG1347/SG1348. To insert the gRNA to direct the Cas9 machinery, 5 μL of oligonucleotides SG1349/SG1350 (100 μM) were mixed with 90 μL of 30 mM HEPES (pH 7.8), heated to 95 °C for 5 min and then cooled to 4 °C at a rate of 0.1 °C/sec. The annealed oligos (2 μL) were ligated into pJOE8999 containing the left/right homology regions using Golden Gate and the incorporation of the gRNA confirmed using blue-white screening on LB agar supplemented with X-Gal (50 μg/mL). The resulting vector pBSANDdel cuts upstream of the gene rsoI and allows for removal of the entire sigO-rsoA operon in B. subtilis W168.
Golden Gate Assembly
For the cloning of inserts using Golden Gate, the reaction (10 μL) comprised final concentrations of 10 000 U/mL T4 DNA ligase, 1000 U/mL BsaI-HFv2, 1× T4 DNA ligase buffer, 1× CutSmart Buffer, 100 ng of plasmid and equimolar amounts of inserts. Note for the assembly of pBSAND1-PxylA-MerRZntRA29E-Terminator-PmerR, a 3:1 insert to vector ratio was used. The reaction conditions were as follows, 37 °C for 5 min, 5–10 cycles of 37 °C for 5 min and 16 °C for 10 min, followed by 16 °C for 30 min, 50 °C for 5 min, and 80 °C for 10 min. For assembling three or more inserts into a vector, we found increasing the numbers of cycles to 30 or more beneficial.
Homology Modeling and Structural Analysis
Homology models for both MerRZntR and MerRCueR were generated using the online I-TASSER server with default parameters, with the respective homology model with the highest C-score selected for further analysis.54 As only partially resolved structures for ZntR are available in the Protein Data Bank (all of which lack residues 1–68, PDB: 1Q09), a homology model of a full-length ZntR monomer was also constructed using the amino acid sequence of ZntR from E. coli MG1655. For all generated homology models, the closest structural analogue was selected and used as a structural reference for quality control. For models of ZntR, MerRZntR and MerRCueR, the C-scores were 0.71 for all the generated models, all of which shared the same closest structural analogue (CueR, PDB: 1Q05) as determined by I-TASSER. The TM-structural alignment program within I-TASSER compared closest structural analogue, PDB 1Q05, to all the generated structures of ZntR, MerRZntR, and MerRCueR with TM-scores of 0.71, 0.71, 0.81 respectively, where a score of >0.5 indicates a similar fold. For visualization of all structures, the resulting PDB structures generated by I-TASSER were imported into PyMol Version 2.0 (Schrödinger, LLC) for visualization.78
Luciferase Assays
For luciferase reporter assays, overnight cultures of each strain to be tested were inoculated 1:1000 into the modified M9 minimal medium described above with added xylose (0.2% final concentration) and distributed into 96-well microplates (Corning; black, clear, flat bottom), with 95 μL culture volume per well. Wells around the plate edge were filled with water to minimize evaporation. A Tecan Spark microplate reader (Tecan Trading AG, Switzerland) was used to monitor luciferase activity and OD600 values for each strain. Cells were grown with continuous shaking incubation (37 °C; 180 rpm; orbital motion; amplitude, 3 mm) until OD600 = ∼0.03 (corresponding to OD600 = ∼0.3 when measured in cuvettes of 1 cm light path length), after which cells were induced with 5 μL of metal stock solutions to reach varying final concentrations (100 μL final volume). For metal induction experiments, the following metal salts were used: HgCl2, ZnSO4, CuSO4, CdCl2, Pb(NO)3, and Ag(NO)3. Measurements of OD600 and luminescence (relative luminescence units [RLU]) were measured every 5 min for 120 min. RLU and OD600 values were blank-corrected using the average of triplicate measurements of RLU and OD600 for MM9 medium alone. Luminescence activity was normalized to cell density for each time data point and reported as RLU/OD600. For all dose–response and metal-specificity studies, final RLU/OD600 values were the average of three time points (35, 40, and 45 min) after challenge with heavy metal salts. All experiments were performed in biological triplicates. All data were processed using Microsoft Excel and subsequently analyzed in GraphPad Prism 7. To determine the Limit of Detection (LOD) for our sensors, we followed the methods as described by Armbruster and Pry79 and Wan et al.8
Acknowledgments
We acknowledge the Engineering and Physical Sciences Research Council (EPSRC; EP/PO2081X/1) and industrial collaborators/partners for funding the Resilient Materials for Life (RM4L) project. J.G. was supported by a University of Bath Leveraged Studentship Award. We thank the technical staff in the Life Sciences Department for key support.
Data Availability Statement
All data used to generate the figures has been made available in the Supporting Information File S2. Bacterial strains have been deposited with the Bacillus Genetic Stock Center (BGSC) and include plasmids (in Escherichia coli DH5α) pBSANDlux, pBSAND1, pBSAND2, pBSANDdel, and pBSGGlux.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.2c00545.
File S1: Figures and Tables—Table S1: Bacterial strains used in this study; Table S2: Plasmids used in this study; Table S3: Primers and oligonucleotides used in this study; Table S4: Relevant DNA sequences for plasmid construction; Figure S1: Comparison of Gram positive and Gram negative MerR promoter activity in B. subtilis; Figure S2: Comparison of ZntR sequences and structural analysis of the chimera MerRZntR; Figure S3: Activity of double and triple MerRZntR mutants against the wild-type and single mutant MerRZntRA29E; Figure S4: Comparison of interdomain communication between CueR and the chimera MerRCueR; Figure S5: Dose response of PmerR regulated by MerRCueRmut3 in response to Ag+ induction; Figure S6: Wild-type (PmerR20) and mutant (PmerR19) promoters; Figure S7: Maps of the B. subtilis SANDBOX plasmids; Figure S8: Bacillus subtilis metal-sensory circuit controlled by the native CzrA regulator (PDF)
File S2: Numerical data that led to the production of the graphs and plots in Figures 1–6, and Supplementary Figures S1, S3, S5, and S8 (XLSX)
Author Contributions
B.J.R., J.S.G., and S.G. conceived the study; J.G. conducted all experimental work and data analysis; S.G. acquired funding, B.J.R. and S.G. coordinated the work; J.G., B.J.R., and S.G. wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Tchounwou P. B.; Yedjou C. G.; Patlolla A. K.; Sutton D. J. Heavy Metals Toxicity and the Environment. Mol. Clin. Environ. Toxicol. 2012, 101, 133–164. 10.1007/978-3-7643-8340-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah S.; Li Z.; Hassan S.; Ahmad S.; Guo X.; Wanghe K.; Nabi G. Heavy metals bioaccumulation and subsequent multiple biomarkers based appraisal of toxicity in the critically endangered Tor putitora. Ecotoxicol. Environ. Saf. 2021, 228, 113032. 10.1016/j.ecoenv.2021.113032. [DOI] [PubMed] [Google Scholar]
- Baker-Austin C.; Wright M. S.; Stepanauskas R.; McArthur J. V. Co-selection of antibiotic and metal resistance. Trends Microbiol 2006, 14, 176–182. 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Mazhar S. H.; Li X.; Rashid A.; Su J. M.; Xu J.; Brejnrod A. D.; Su J. Q.; Wu Y.; Zhu Y. G.; Zhou S. G.; Feng R.; Rensing C. Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Sci. Total Environ. 2021, 755, 142702. 10.1016/j.scitotenv.2020.142702. [DOI] [PubMed] [Google Scholar]
- Foster T. J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol. Rev. 1983, 47, 361–409. 10.1128/mr.47.3.361-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Gu A. Z.; Cen T.; Li X.; He M.; Li D.; Chen J. Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ. Pollut. 2018, 237, 74–82. 10.1016/j.envpol.2018.01.032. [DOI] [PubMed] [Google Scholar]
- Chen J.; Li J.; Zhang H.; Shi W.; Liu Y. Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in northern China. Front. Microbiol. 2019, 10, 1–12. 10.3389/fmicb.2019.01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X.; Volpetti F.; Petrova E.; French C.; Maerkl S. J.; Wang B. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nat. Chem. Biol. 2019, 15, 540–548. 10.1038/s41589-019-0244-3. [DOI] [PubMed] [Google Scholar]
- Wang B.; Barahona M.; Buck M. A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals. Biosens. Bioelectron. 2013, 40, 368–376. 10.1016/j.bios.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P. T. K.; Berg M.; Viet P. H.; Van Mui N.; Van Der Meer J. R. Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environ. Sci. Technol. 2005, 39, 7625–7630. 10.1021/es050992e. [DOI] [PubMed] [Google Scholar]
- Bereza-Malcolm L. T.; Mann G.; Franks A. E. Environmental Sensing of Heavy Metals Through Whole Cell Microbial Biosensors: A Synthetic Biology Approach. ACS Synth. Biol. 2015, 4, 535–546. 10.1021/sb500286r. [DOI] [PubMed] [Google Scholar]
- Jung J.; Lee S. J. Biochemical and biodiversity insights into heavy metal ion-responsive transcription regulators for synthetic biological heavy metal sensors. J. Microbiol. Biotechnol. 2019, 29, 1522–1542. 10.4014/jmb.1908.08002. [DOI] [PubMed] [Google Scholar]
- Baksh K. A.; Zamble D. B. Allosteric control of metal-responsive transcriptional regulators in bacteria. J. Biol. Chem. 2020, 295, 1673–1684. 10.1074/jbc.REV119.011444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontidean I.; Lloyd J. R.; Hobman J. L.; Wilson J. R.; Csöregi E.; Mattiasson B.; Brown N. L. Bacterial metal-resistance proteins and their use in biosensors for the detection of bioavailable heavy metals. J. Inorg. Biochem. 2000, 79, 225–229. 10.1016/S0162-0134(99)00234-2. [DOI] [PubMed] [Google Scholar]
- Brown N. L.; Stoyanov J. V.; Kidd S. P.; Hobman J. L. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003, 27, 145–163. 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- Hobman J. L.; Wilkie J.; Brown N. L. A design for life: Prokaryotic metal-binding MerR family regulators. BioMetals 2005, 18, 429–436. 10.1007/s10534-005-3717-7. [DOI] [PubMed] [Google Scholar]
- Philips S. J.; Canalizo-Hernandez M.; Yildirim I.; Schatz G. C.; Mondragón A.; O’Halloran T. V. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science (80-.) 2015, 349, 877–881. 10.1126/science.aaa9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A. Z.; Chael M. L.; O’Halloran T. V. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature 1992, 355, 87–89. 10.1038/355087a0. [DOI] [PubMed] [Google Scholar]
- Wu C. H.; Le D.; Mulchandani A.; Chen W. Optimization of a whole-cell cadmium sensor with a toggle gene circuit. Biotechnol. Prog. 2009, 25, 898–903. 10.1002/btpr.203. [DOI] [PubMed] [Google Scholar]
- Humbert M. V.; Rasia R. M.; Checa S. K.; Soncini F. C. Protein signatures that promote operator selectivity among paralog MerR monovalent metal ion regulators. J. Biol. Chem. 2013, 288, 20510–20519. 10.1074/jbc.M113.452797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Audero M. E.; Podoroska B. M.; Ibáñez M. M.; Cauerhff A.; Checa S. K.; Soncini F. C. Target transcription binding sites differentiate two groups of MerR-monovalent metal ion sensors. Mol. Microbiol. 2010, 78, 853–865. 10.1111/j.1365-2958.2010.07370.x. [DOI] [PubMed] [Google Scholar]
- Korostelev Y. D.; Zharov I. A.; Mironov A. A.; Rakhmaininova A. B.; Gelfand M. S. Identification of position-specific correlations between DNA-Binding domains and their binding sites. application to the merr family of transcription factors. PLoS One 2016, 11, e0162681. 10.1371/journal.pone.0162681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocklehurst K. R.; Hobman J. L.; Lawley B.; Blank L.; Marshall S. J.; Brown N. L.; Morby A. P. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 1999, 31, 893–902. 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- Ibáñez M. M.; Cerminati S.; Checa S. K.; Soncini F. C. Dissecting the metal selectivity of MerR monovalent metal ion sensors in Salmonella. J. Bacteriol. 2013, 195, 3084–3092. 10.1128/JB.00153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D.; Ballard B.; Walsh C. T. The MerR metalloregulatory protein binds mercuric ion as a tricoordinate, metal-bridged dimer. Science (80-.) 1990, 247, 946–948. 10.1126/science.2305262. [DOI] [PubMed] [Google Scholar]
- Permina E. A.; Kazakov A. E.; Kalinina O. V.; Gelfand M. S. Comparative genomics of regulation of heavy metal resistance in Eubacteria. BMC Microbiol. 2006, 6, 1–11. 10.1186/1471-2180-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale S.; Arkin A. P. Contextualizing context for synthetic biology - identifying causes of failure of synthetic biological systems. Biotechnol. J. 2012, 7, 856–866. 10.1002/biot.201200085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy J. A. N.; Voigt C. A. Principles of genetic circuit design. Nat. Methods 2014, 11, 508–520. 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Kitney R. I.; Joly N.; Buck M. Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology. Nat. Commun. 2011, 2, 508–509. 10.1038/ncomms1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I.; Svetlov V.; Anthony L.; Burgess R. R.; Landick R. RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J. Bacteriol. 2000, 182, 6027–6035. 10.1128/JB.182.21.6027-6035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeck J.; Kraft K.; Bartels J.; Cikovic T.; Dürr F.; Emenegger J.; Kelterborn S.; Sauer C.; Fritz G.; Gebhard S.; Mascher T. The Bacillus BioBrick Box: Generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J. Biol. Eng. 2013, 10.1186/1754-1611-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns N. I.; Gomes A. L. C.; Yim S. S.; Yang A.; Blazejewski T.; Smillie C. S.; Smith M. B.; Alm E. J.; Kosuri S.; Wang H. H. Metagenomic mining of regulatory elements enables programmable species-selective gene expression. Nat. Methods 2018, 15, 323–329. 10.1038/nmeth.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl S. R.; Ekness F.; Sofjan K.; Daeffler K. N. M.; Brink K. R.; Landry B. P.; Gerhardt K. P.; Dyulgyarov N.; Sheth R. U.; Tabor J. J. Rewiring bacterial two-component systems by modular DNA-binding domain swapping. Nat. Chem. Biol. 2019, 15, 690–698. 10.1038/s41589-019-0286-6. [DOI] [PubMed] [Google Scholar]
- Popp P. F.; Dotzler M.; Radeck J.; Bartels J.; Mascher T. The Bacillus BioBrick Box 2.0: Expanding the genetic toolbox for the standardized work with Bacillus subtilis. Sci. Rep. 2017, 7, 1–13. 10.1038/s41598-017-15107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Liu L.; Li J.; Du G.; Chen J. Synthetic Biology Toolbox and Chassis Development in Bacillus subtilis. Trends Biotechnol 2019, 37, 548–562. 10.1016/j.tibtech.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Michna R. H.; Zhu B.; Mäder U.; Stülke J. SubtiWiki 2.0 - An integrated database for the model organism Bacillus subtilis. Nucleic Acids Res. 2016, 44, D654–D662. 10.1093/nar/gkv1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbet A.; Endy D.; Renard E.; Molina F.; Bonnet J. Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci. Transl. Med. 2015, 10.1126/scitranslmed.aaa3601. [DOI] [PubMed] [Google Scholar]
- Revilla-Guarinos A.; Dürr F.; Popp P. F.; Döring M.; Mascher T. Amphotericin B Specifically Induces the Two-Component System LnrJK: Development of a Novel Whole-Cell Biosensor for the Detection of Amphotericin-Like Polyenes. Front. Microbiol. 2020, 11, 1–16. 10.3389/fmicb.2020.02022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb A. J.; Kelwick R.; Doenhoff M. J.; Kylilis N.; MacDonald J. T.; Wen K. Y.; McKeown C.; Baldwin G.; Ellis T.; Jensen K.; Freemont P. S. A protease-based biosensor for the detection of schistosome cercariae. Sci. Rep. 2016, 6, 1–14. 10.1038/srep24725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobras C. M.; Mascher T.; Gebhard S. Application of a Bacillus subtilis whole-cell biosensor (PliaI-lux) for the identification of cell wall active antibacterial compounds. Metholds Mol. Biol. 2017, 121. 10.1007/978-1-4939-6634-9_7. [DOI] [PubMed] [Google Scholar]
- Anderson J. C.; Voigt C. A.; Arkin A. P. Environmental signal integration by a modular and gate. Mol. Syst. Biol. 2007, 3, 133. 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon T. S.; Lou C.; Tamsir A.; Stanton B. C.; Voigt C. A. Genetic programs constructed from layered logic gates in single cells. Nature 2012, 491, 249–253. 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M. T. G.; Lindsay J. A.; Corton C.; Quail M. A.; Cockfield J. D.; Pathak S.; Batra R.; Parkhill J.; Bentley S. D.; Edgeworth J. D. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW). J. Bacteriol. 2010, 192, 888–892. 10.1128/JB.01255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C.; Lin L. Y.; Zou X. W.; Huang C. C.; Chan N. L. Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR. Nucleic Acids Res. 2015, 43, 7612–7623. 10.1093/nar/gkv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gworek B.; Bemowska-Kałabun O.; Kijeńska M.; Wrzosek-Jakubowska J. Mercury in Marine and Oceanic Waters—a Review. Water. Air. Soil Pollut. 2016, 10.1007/s11270-016-3060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. K.; Goloubinoff P.; Christen P. Heavy metal ions are potent inhibitors of protein folding. Biochem. Biophys. Res. Commun. 2008, 372, 341–345. 10.1016/j.bbrc.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Kubier A.; Wilkin R. T.; Pichler T. Cadmium in soils and groundwater: A review. Appl. Geochem. 2019, 108, 104388. 10.1016/j.apgeochem.2019.104388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P.; Lang N.; LeGrice S. F. J.; Lee G.; Stephens M.; Sonenshein A. L.; Pero J.; Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. MGG Mol. Gen. Genet. 1982, 186, 339–346. 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Henkin T. M.; Sonenshein A. L. Mutations of the Escherichia coli lacUV5 promoter resulting in increased expression in Bacillus subtilis. MGG Mol. Gen. Genet. 1987, 209, 467–474. 10.1007/BF00331151. [DOI] [PubMed] [Google Scholar]
- Forrest D.; Warman E. A.; Erkelens A. M.; Dame R. T.; Grainger D. C. Xenogeneic silencing strategies in bacteria are dictated by RNA polymerase promiscuity. Nat. Commun. 2022, 13, 1–13. 10.1038/s41467-022-28747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimas R. P.; Jordan B. R.; Jiang X. L.; Martini C.; Glavy J. S.; Patterson D. P.; Morcos F.; Chan C. T. Y. Engineering DNA recognition and allosteric response properties of TetR family proteins by using a module-swapping strategy. Nucleic Acids Res. 2019, 47, 8913–8925. 10.1093/nar/gkz666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt S.; Manley M. W.; Becker N. A.; Hessman J. A.; Maher L. J.; Swint-Kruse L. Novel insights from hybrid LacI/GalR proteins: Family-wide functional attributes and biologically significant variation in transcription repression. Nucleic Acids Res. 2012, 40, 11139–11154. 10.1093/nar/gks806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F.; Wilm A.; Dineen D.; Gibson T. J.; Karplus K.; Li W.; Lopez R.; McWilliam H.; Remmert M.; Söding J.; Thompson J. D.; Higgins D. G. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A.; Kucukural A.; Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra A. J.; Giedroc D. P. Metal site occupancy and allosteric switching in bacterial metal sensor proteins. J. Biol. Chem. 2012, 519, 210. 10.1016/j.abb.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A. Z.; Bradner J. E.; O’Halloran T. V. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 1995, 374, 370–375. 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- Arya S.; Williams A.; Reina S. V.; Knapp C. W.; Kreft J. U.; Hobman J. L.; Stekel D. J. Towards a general model for predicting minimal metal concentrations co-selecting for antibiotic resistance plasmids. Environ. Pollut. 2021, 275, 116602. 10.1016/j.envpol.2021.116602. [DOI] [PubMed] [Google Scholar]
- Ashelford M.; Gore D. B. Elemental and mineralogical constraints on environmental contamination from slag at Gulf Creek copper mine. Miner. Eng. 2020, 154, 106407. 10.1016/j.mineng.2020.106407. [DOI] [Google Scholar]
- Luo Y.; Rao J.; Jia Q. Heavy metal pollution and environmental risks in the water of Rongna River caused by natural AMD around Tiegelongnan copper deposit, Northern Tibet, China. PLoS One 2022, 17, e0266700 10.1371/journal.pone.0266700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyanov J. V.; Hobman J. L.; Brown N. L. CueR (Ybbl) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol. Microbiol. 2001, 39, 502–512. 10.1046/j.1365-2958.2001.02264.x. [DOI] [PubMed] [Google Scholar]
- Changela A.; Chen K.; Xue Y.; Holschen J.; Outten C. E.; O’Halloran T. V.; Mondragón A. Molecular Basis of Metal-Ion Selectivity and Zeptomolar Sensitivity by CueR. Science 2003, 301, 1383–1387. 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- Grigoletto J. C.; Segura-Munoz S. I.; Barbosa-Junior F.; Sanches S. M.; Takayanagui A. M. M. Silver discharged in effluents from image-processing services: A risk to human and environmental health. Biol. Trace Elem. Res. 2011, 144, 316–326. 10.1007/s12011-011-9096-8. [DOI] [PubMed] [Google Scholar]
- Ibáñez M. M.; Checa S. K.; Soncini F. C. A single serine residue determines selectivity to monovalent metal ions in metalloregulators of the MerR family. J. Bacteriol. 2015, 197, 1606–1613. 10.1128/JB.02565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D.; Ballard B.; Walsh C. T. The MerR metalloregulatory protein binds mercuric ion as a tricoordinate, metal-bridged dimer. Science (80-.) 1990, 247, 946–948. 10.1126/science.2305262. [DOI] [PubMed] [Google Scholar]
- Liu X.; Hu Q.; Yang J.; Huang S.; Wei T.; Chen W.; He Y.; Wang D.; Liu Z.; Wang K.; Gan J.; Chen H. Selective cadmium regulation mediated by a cooperative binding mechanism in CadR. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 20398–20403. 10.1073/pnas.1908610116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobman J. L.; Julian D. J.; Brown N. L. Cysteine coordination of Pb(II) is involved in the PbrR-dependent activation of the lead-resistance promoter, PpbrA, from Cupriavidus metallidurans CH34. BMC Microbiol. 2012, 12, 109. 10.1186/1471-2180-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.; Brocklehurst K. R.; Jones G. W.; Morby A. P. The functional analysis of directed amino-acid alterations in ZntR from Escherichia coli. Biochem. Biophys. Res. Commun. 2002, 299, 438–445. 10.1016/S0006-291X(02)02660-8. [DOI] [PubMed] [Google Scholar]
- Parkhill J.; Brown N. L. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by. MerR 1990, 18, 5157–5162. 10.1093/nar/18.17.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X.; Davis M. C.; Steeves T.; Bishop A.; Breen J.; MacEacheron A.; Kesthely C. A.; Hsu F. S.; MacLellan S. R. Characterization of a protein-protein interaction within the SigO-RsoA two-subunit σ factor: The σ70 region 2.3-like segment of RsoA mediates interaction with SigO. Microbiol. (United Kingdom) 2016, 162, 1857–1869. 10.1099/mic.0.000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLellan S. R.; Guariglia-Oropeza V.; Gaballa A.; Helmann J. D. A two-subunit bacterial σ-factor activates transcription in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 21323–21328. 10.1073/pnas.0910006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLellan S. R.; Helmann J. D.; Antelmann H. The YvrI alternative σ factor is essential for acid stress induction of oxalate decarboxylase in Bacillus subtilis. J. Bacteriol. 2009, 191, 931–939. 10.1128/JB.01435-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. M.; Gaballa A.; Hui M.; Ye R. W.; Helmann J. D. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol. Microbiol. 2005, 57, 27–40. 10.1111/j.1365-2958.2005.04642.x. [DOI] [PubMed] [Google Scholar]
- Harwood C. R.; Cutting S. M.. Molecular Biological Methods for Bacillus; John Wiley & Sons: Chichester, 1990. [Google Scholar]
- Liu H.; Naismith J. H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol 2008, 8, 91. 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W.; Lin Q.; Hu R.; Han L.; Cheng Z.; Zhang L.; Zhou Z. Data-Driven and in Silico-Assisted Design of Broad Host-Range Minimal Intrinsic Terminators Adapted for Bacteria. ACS Synth. Biol. 2021, 10, 1438–1450. 10.1021/acssynbio.1c00050. [DOI] [PubMed] [Google Scholar]
- Guiziou S.; Sauveplane V.; Chang H. J.; Clerté C.; Declerck N.; Jules M.; Bonnet J. A part toolbox to tune genetic expression in Bacillus subtilis. Nucleic Acids Res. 2016, 44, 7495–7508. 10.1093/nar/gkw624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbuchner J. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2016, 82, 5421–5427. 10.1128/AEM.01453-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, Version 2.0; Schrödinger, LLC, 2015.
- Armbruster D. A.; Pry T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29 (Suppl 1), S49–S52. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used to generate the figures has been made available in the Supporting Information File S2. Bacterial strains have been deposited with the Bacillus Genetic Stock Center (BGSC) and include plasmids (in Escherichia coli DH5α) pBSANDlux, pBSAND1, pBSAND2, pBSANDdel, and pBSGGlux.