Abstract
Minimized trans-acting HDV ribozyme systems consisting of three (Rz-3) and two (Rz-2) RNA strands were prepared and their folding conformations were analyzed by NMR spectroscopy. The guanosine residues in one of the enzyme components of Rz-3 were labeled with 13C and 15N. Imino proton signals were assigned by analysis of NOESY and HSQC spectra. The results are consistent with the nested double pseudoknot model, which contains novel base pairs (P1.1), as observed in the crystal structure of a genomic HDV ribozyme. The NOE connectivities suggest an additional G:G pair at the bottom of P1.1 and at the top of P4. The effects of temperature and Mg2+ ions on base pairs for Rz-3 were examined. The temperature variation experiment on Rz-3 showed that P3 is the most stable and that P1.1 is as stable as P1 and P2. The imino proton signals of the G:U pair at the bottom of P1 and the top of P1.1, which are close to the cleavage site, showed the largest changes upon Mg2+ titration of Rz-3. The results suggest that the catalytic Mg2+ ion binds to the pocket formed by P1 and L3.
INTRODUCTION
Human hepatitis delta virus (HDV) is a satellite virus that specifically infects cells already infected with hepatitis B virus. The HDV genome consists of a circular, single-stranded RNA of ∼1700 nt in length (1). The genome is replicated through the rolling circle mechanism and self-cleavage reactions of polymeric forms of antigenomic and genomic RNAs are involved in the process. The cleavage reactions occur in the presence of divalent cations, with 2′,3′-cyclic phosphate and 5′-hydroxy groups being produced through transesterification (2,3). A domain of ∼85 nt is necessary for the self-cleavage activity of both the genomic and antigenomic RNAs (4,5).
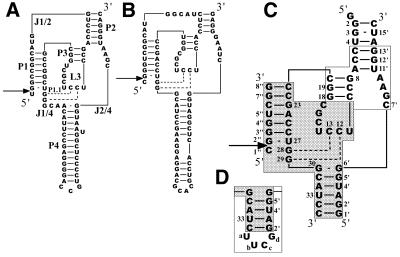
The active domains are assumed to take on a ‘pseudoknot’ structure that includes four double-stranded stems (P1–P4), a hairpin loop (L3) and three internal loops (J1/2, J2/4 and J1/4) (see Fig. 1A and B) (6,7). An X-ray crystallographic study of a modified genomic ribozyme suggested that a part of L3 forms an extra double helical region (P1.1) with a part of J1/4 (Fig. 1A) (8). It turns out that the newly found base pairs are important for the cleavage activity of both the genomic and antigenomic ribozymes (9,10). The crystal structure was obtained for a complex between the ribozyme, which is actually a 3′ cleavage product, and an RNA-binding protein, U1A. Therefore it can be assumed that the solution structure with a proper substrate is somewhat different from the crystal structure. Moreover, no Mg2+ ion, which is required for the cleavage activity, is found around the cleavage site in the crystal structure (8).
Figure 1.
Nucleotide sequences and possible secondary structures of HDV ribozymes. (A) Genomic HDV ribozyme. The stem, loop and junction regions are labeled according to the notations of Been (7). (B) Antigenomic HDV ribozyme. (C) Sequence and numbering for Rz-3, which consists of three RNA oligomer strands. The ribozyme contains a hybrid sequence of the genomic sequence (shown in a white box) and antigenomic sequence (shown in a shaded box). (D) Partial sequence (P4 region) of Rz-2, which consists of two RNA oligomer strands, and its numbering. Arrows indicate the cleavage site.
The solution structure can be determined by NMR spectroscopy. However, an NMR study on an HDV ribozyme system is difficult since the active domain is very large, the molecular size being comparable to that of tRNA. Only a few papers on NMR studies of HDV ribozyme systems have been published (11–13). There have been reports of NMR studies of a catalytic domain of an inactive mutant ribozyme (11) and solution structures of model hairpin RNAs which contain a P3–L3 sequence (12) or mimic P2–P3–L3 segments (13).
We have designed an HDV ribozyme system (Rz-3) which consists of three RNA strands (substrate 8mer and enzyme 16mer plus 35mer) and contains chimeric sequences of genomic and antigenomic ribozymes for a structural study involving NMR (Fig. 1C). The chimeric sequences are employed as a compromised combination of a shorter L3 (7 bases) and shorter J2/4 (5 bases), which are preferable for structural analysis, and a longer P2, which is preferable for ribozyme activity. Rz-3 is derived from an HDV ribozyme system (Rz-2) that was originally designed by Been et al. (14) and consists of two RNA strands (Fig. 1D). Rz-3 contains only essential components of the active domain, the total number of nucleotides being reduced to 59, but retains relatively high activity (15). The three component system enables us to incorporate 13C and 15N labels into each part separately for signal assignment.
In this paper we report a structural analysis of Rz-3 and Rz-2 systems containing a non-cleavable substrate analog by NMR. Analysis of the imino proton NMR spectra showed that the ribozymes indeed take on a ‘nested double pseudoknot’ (9) structure in solution. A titration experiment involving MgCl2 suggests the existence of a Mg2+-binding site near the cleavage site, which is not observed in the crystal structure (8).
MATERIALS AND METHODS
All solutions except for Tris–HCl buffer were treated with diethyl pyrocarbonate and autoclaved to inactivate any traces of RNases. T7 RNA polymerase was prepared by overexpression of its gene in Escherichia coli strain BL21 carrying plasmid pAR1219 (16). Polynucleotide kinase was purchased from Takara Shuzo (Kyoto, Japan). RNase T1, RNase U2, RNase phyM and Bacillus cereus RNase were obtained from Amersham Pharmacia Biotech. 13C,15N-labeled GTP was purchased from Nippon Sanso (Tokyo, Japan). RNA secondary structure prediction based on free energy minimization was carried out using the program RNA Structure v.3.5, downloaded from http://rna.chem.rochester.edu/RNAstructure.html (17).
Preparation of HDV ribozymes
The non-cleavable substrate RNA 8mer (mS8) was synthesized using a DNA/RNA synthesizer (Model 392; Applied Biosystems). For synthesis of the non-cleavable substrate, N4-benzoyl-2′-O-methylcytidine was prepared and incorporated as described previously (18,19). mS8 was purified by denaturing (7 M urea) PAGE. The gel containing the 8mer was crushed and soaked overnight in 0.3 M sodium acetate, pH 7, with shaking at room temperature. The extracted RNA was isolated by two rounds of ethanol precipitation at –25°C for 2 h. The enzyme components, i.e. the 16mer (Eb16) and 35mer (Ea35) of Rz-3 and 53mer (E53) of Rz-2, were prepared by in vitro transcription with T7 RNA polymerase using a fully double-stranded DNA promoter–template. The DNA oligomers, i.e. the 33mer, 52mer and 70mer, for the promoter–template DNAs were synthesized with the DNA/RNA synthesizer and purified in the same manner as described for the substrate RNA oligomer. The transcription reaction was carried out at 37°C for 3 h according to the procedure of Milligan et al. (20), with some modifications. The reaction mixture comprised 2 µM promoter–template DNA, 0.1 mg/ml T7 RNA polymerase, 7.5 mM each NTP, 35 mM MgCl2, 160 mM Tris–HCl, pH 8.1, 5 mM DTT, 2 mM spermidine, 0.01% (v/v) Triton X-100, 10 mg/ml PEG 8000 and 3 U/ml inorganic pyrophosphatase to decompose the pyrophosphate produced during polymerization (21). GMP (20 mM) was also added to the mixture for preparation of non-labeled RNAs (22). The yields of the oligomers on the 200 µl scale were 8.3 (turnover number 21), 11.5 (turnover number 29) and 16 nmol (turnover number 40) for Eb16, Ea35 and E53, respectively. For preparation of the 13C- and 15N-G-labeled oligomer, 13C,15N-labeled GTP was used. The RNA oligomers were identified by sequence determination. The oligomers were labeled at the 5′-ends by means of polynucleotide kinase and [γ-32P]ATP when necessary. The labeled RNA was subjected to partial alkaline hydrolysis (23) and partial digestion with RNase T1, RNase U2, RNase phyM and B.cereus RNase. The degradation products were separated by denaturing 20% PAGE and the sequences were determined by comparison of the product bands. Rz-2 showed the same RNA cleavage activity as that reported by Been et al. (14). The yields and concentrations of RNA oligomers were calculated from the absorbance at 260 nm using the molar absorption coefficients of the component nucleotides.
NMR analysis
All NMR spectra were recorded with a Bruker DRX-600 spectrometer equipped with a quadruple resonance probe with x, y and z gradients of 5–60°C. Each enzyme component (Eb16 plus Ea35 for Rz-3 and E53 for Rz-2) was dissolved in 10 mM sodium phosphate buffer, pH 6.5, 50 mM NaCl, 1 mM NaN3 containing 5% D2O. For measurement of the complete HDV ribozyme system, an equimolar amount of non-cleavable substrate (mS8) was added to the enzyme sample. A Mg2+-containing sample was prepared by adding a concentrated solution of MgCl2. The concentration of the 13C- and 15N-labeled RNA for HSQC spectra was 0.3 mM and those of non-labeled samples were 1 mM (for the NOESY and temperature variation experiments) or 0.3 mM (for the Mg2+ titration experiment). The 1H chemical shifts were measured relative to internal DSS. The 15N chemical shifts were referenced relative to external liquid NH3 by the indirect referencing method (24). NOESY spectra were recorded with 512 data points in the t1 domain using the time-proportional phase incrementation method and the free induction decays (FIDs) (256 scans each) of 2048 data points in the t2 domain were collected with a mixing time of 200 ms. One-dimensional spectra were obtained at a temperature between 5 and 60°C and two-dimensional spectra were obtained at 25°C. By 60°-shifted sine-bell function apodization in both the t1 and t2 domains followed by Fourier transformation, spectra of 2048 × 512 data points were obtained. For NOESY experiments, the jump-and-return pulse (25) was used for suppression of the water resonance. 15N-1H HSQC spectra were recorded with 128 data points in the t1 domain using the echo/anti-echo gradient selection method. The FIDs (128 scans each) of 2048 data points in the t2 domain were collected. By 72°-shifted sine-bell function apodization in both the t1 and t2 domains followed by Fourier transformation, spectra of 2048 × 128 data points were obtained.
RESULTS AND DISCUSSION
Design and preparation of RNA oligomers
We prepared two types of HDV ribozyme systems, Rz-3 and Rz-2, which consist of three and two RNA strands, respectively (Fig. 1C and D). Rz-2, which was originally designed by Been et al. (14), consists of a substrate component (8mer, S8) and an enzyme component (53mer, E53). It was designed to have a secondary structure in which the long P4 stem is shortened and closed with a tetranuclotide loop, UUCG, and J1/2 is removed from the structure of the wild-type ribozymes. Moreover, Rz-2 has a chimeric sequence composed of genomic (P3 and a part of P2) and antigenomic (P1, P4, L3 and J1/4) sequences. The antigenomic L3 and J1/4 sequences are shorter than those of the genomic ribozyme. Rz-3 consists of a substrate component (S8) and two enzyme components (35mer and 16mer, Ea35 and Eb16). The external loop attached to P4 is removed and a base pair is added to the P4 stem when compared with Rz-2. This three component system is advantageous for NMR studies since each RNA strand can be labeled separately.
The non-cleavable substrate 8mer (mS8), which contains a 2′-O-methyl group on the 5′-terminal cytidine residue, was chemically synthesized. The other longer RNA oligomers were synthesized with T7 RNA polymerase using appropriate promoter–template DNAs. Labeled Ea35, in which every guanosine residue was labeled with 13C and 15N, was prepared by polymerization in the presence of 13C,15N-labeled GTP.
Structure of the 35mer fragment (Ea35) of the enzyme component for Rz-3
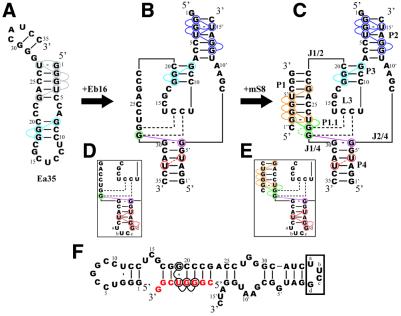
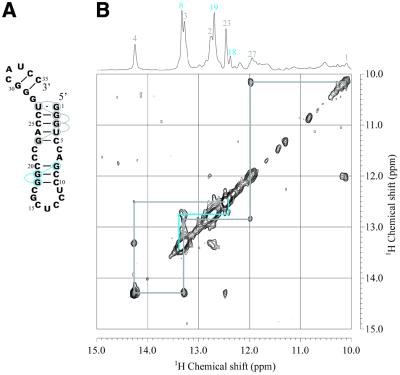
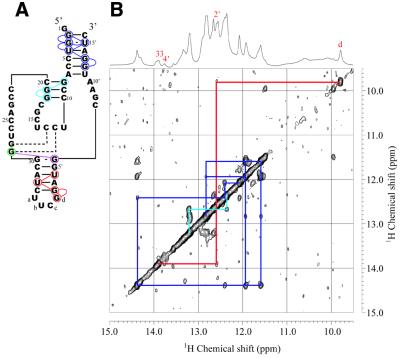
A NOESY spectrum of the longer enzyme component of Rz-3 (Ea35, Fig. 2A) was measured in 50 mM NaCl, 10 mM sodium phosphate buffer, pH 6.5, at 25°C (Fig. 3). An HSQC spectrum of labeled Ea35, in which every guanosine residue was labeled with 13C and 15N, was also measured. In this spectrum, only signals for the labeled guanine 15N-1H are observed. Sequential assignment of the imino proton resonances was performed, as shown in Figures 2A and 3. The resonances for the G1:U27 pair can be uniquely assigned based on their characteristic chemical shifts and the strong NOE between the two imino protons. Therefore sequential NOE connectivity can be safely traced for G1:U27 through C5:G23. The direction of connectivity for the other stem, G8:C20 through C10:G18, is not definite. The G8 and G18 imino proton resonances (13.35 and 12.41 p.p.m., respectively) were assigned in accordance with the chemical shifts (13.3 and 12.6 p.p.m.) reported for a RNA hairpin, which has the same partial sequence corresponding to the P3 and L3 parts of the antigenomic HDV ribozyme (13). The results are consistent with the secondary conformation (Fig. 2A) obtained with the free energy minimization algorithm for RNA secondary structure prediction developed by Mathews et al. (17). This folded conformation contains P3 and L3 of the pseudoknot model of the ribozyme. The same patterns of NOE connectivity as those for the imino protons of G8–G19–G18 were observed for the Ea35:Eb16 complex, E53, the complete Rz-3 complex and the complete Rz-2 complex.
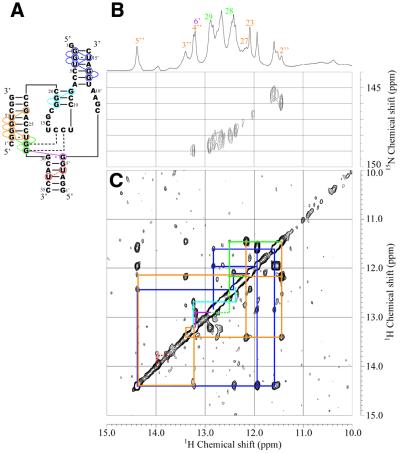
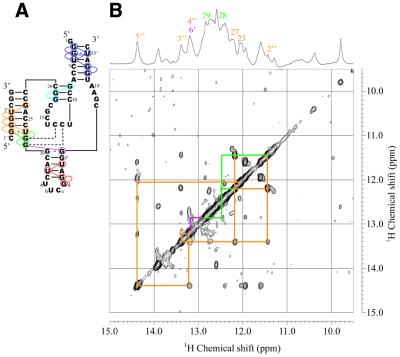
Figure 2.
Secondary structures deduced from NOE connectivities between imino proton resonances for the components and complete systems of Rz-3 and Rz-2. Colored circles indicate the residues for which the imino proton resonances were assigned. Colored lines indicate NOE connectivities. The dotted lines for Rz-3 show that the resonances were assigned, although a NOE cross-peak between them was not observed, by comparison with the corresponding resonances of Rz-2, for which not only a NOE cross-peak between them but also connectivities with the neighboring residues were observed. The residues and NOE connectivity lines are colored differently according to the stem. (A) Ea35 alone. A hairpin structure obtained by calculation is shown. (B) Ea35 + Eb16. (C) Rz-3. (D) E53. (E) Rz-2. In (D) and (E) only partial sequences and NOE connectivities which are different from those for Rz-3 are shown. (F) An alternative, minor secondary structure formed by mS8 and E53 obtained by free energy calculation.
Figure 3.
Secondary structure (A) and NOESY spectrum (B) of Ea35 alone in the imino proton–imino proton region measured in 10 mM sodium phosphate, pH 6.5, 50 mM NaCl, 1 mM NaN3 containing 5% D2O at 25°C. The light blue lines indicate NOEs for the P3 stem and the gray lines for a stem formed through intra-strand association. The one-dimensional 1H spectrum and signal assignments are shown at the top.
Structure of the enzyme part (Ea35:Eb16 complex) of Rz-3 and E53 of Rz-2
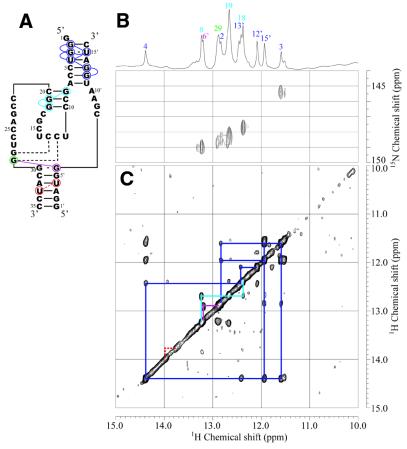
An equimolar amount of Eb16 was added to the Ea35 samples and then NOESY and HSQC spectra were measured (Fig. 4). The imino proton resonances for guanosine residues in the Ea35 component were identified by analysis of the HSQC spectrum of the labeled complex. Resonances for G3:U15′ can be assigned based on the characteristic 15N chemical shift (G3-N1, 145.4 p.p.m.) and strong intra-base pair NOE. Then NOE connectivity can be traced from G2:C16′ to C6:G12′, covering most of the base pairs in P2 (Figs 2B and 4). The same NOE connectivity, G8:C20 through C10:G18, as that observed for P3 of Ea35 alone was observed for Ea35:Eb16.
Figure 4.
Secondary structure (A), 15N-1H HSQC (B) and NOESY (C) spectra of the enzyme part (Ea35:Eb15) for Rz-3 measured in 10 mM sodium phosphate buffer, pH 6.5, 50 mM NaCl containing 5% D2O at 25°C. (B) The sample contained a mixture of 13C,15N-G-labeled Ea35 and non-labeled Eb16 (0.3 mM each). (C) The sample contained a mixture of non-labeled Ea35 and Eb16 (1 mM each). Blue, light blue and purple lines indicate NOE connectivities for P2 and P3 and the NOE between G29 and G6′, respectively. Dotted red lines indicate the putative NOE between U33 and U4′ in P4, which is expected from the assignment for Rz-2. The one-dimensional 1H spectrum and signal assignments are shown at the top.
Analysis of the NOESY spectrum of E53 showed that the NOE connectivities for P2 and P3 were almost identical to those of the Ea35:Eb16 complex (Fig. 5). The one-dimensional spectrum of E53 shows a relatively sharp signal at 9.78 p.p.m., which was not observed for the Ea35:Eb16 complex. This signal can be assigned to an imino proton of the G residue (Gd) in the tetranucleotide loop, UUCG, since similar characteristic signals for the G residue of a UUCG loop are observed for hairpin RNA oligomers with a UUCG loop (26–28). This signal shows a small NOE cross-peak with a resonance at 12.59 p.p.m., which is assumed to be that of G2′:C34. Thus the NOE connectivity can be traced from Gd to U4′:A32, covering most of the base pairs in P4 (Fig. 2D). This is not observed for the Ea35:Eb16 complex, probably due to the lower stability of the P4 stem. In the case of E53, P4 may be greatly stabilized by the presence of the tetranucleotide loop.
Figure 5.
Secondary structure (A) and NOESY spectrum (B) of the enzyme part of Rz-2 (E53, 1 mM) measured in 10 mM sodium phosphate buffer, pH 6.5, 50 mM NaCl containing 5% D2O at 25°C. Blue, light blue and red lines indicate NOE connectivities for the P2, P3 and P4 stems, respectively. The one-dimensional 1H spectrum and signal assignments are shown at the top.
Structure of the complete Rz-3 system (mS8:Ea35:Eb16 complex) and the Rz-2 system (mS8:E53 complex)
An equimolar amount of mS8 was added to the Ea35:Eb16 samples and then NOESY and HSQC spectra were measured (Fig. 6). Upon addition of the substrate analog some new signals appeared in the one-dimensional and NOESY spectra. Among them, two clearly resolved signals at 11.45 and 13.39 p.p.m. were readily recognizable in the one-dimensional spectrum. The former signal (11.45 p.p.m.) shows a strong NOE with a signal at 12.16 p.p.m., which is characteristic of a G:U base pair, suggesting that the NOE cross-peak is between the imino protons of G2″:U27. The latter signal (13.39 p.p.m.) shows NOE cross-peaks with both the signals assumed for G2″:U27. Thus the NOE connectivity can be traced from G2″:U27 to C6″:G23, covering the 5 bp in P1 (Figs 2C and 6). These results suggest that the substrate analog indeed binds to the enzyme part to form a complete system.
Figure 6.
Secondary structure (A), 15N-1H HSQC (B) and NOESY (C) spectra of the complete Rz-3 system (mS8:Ea35:Eb16) measured in 10 mM sodium phosphate buffer, pH 6.5, 50 mM NaCl containing 5% D2O at 25°C. (B) The sample contained a mixture of 13C,15N-G-labeled Ea35, non-labeled mS8 and Eb16 (0.3 mM each). (C) The sample contained non-labeled mS8, Ea35 and Eb16 (1 mM each). Blue, light blue and orange lines indicate NOE connectivities for the P2, P3 and P1 stems, respectively. Dotted red lines indicate the putative NOE between imino protons of U4′ and U33 in the P4 stem, which is expected from the assignment for Rz-2. Green lines indicate NOE connectivities for the P1.1 stem and the dotted part indicates the putative NOE between G28 and G29, which is expected from the assignment for Rz-2. Purple lines indicate a NOE which can be assigned as that between G29 and G6′ of the G30:G6′ pair. The one-dimensional 1H spectrum and signal assignments for P1, P1.1, G6′ and P4 are shown at the top.
The NOESY spectrum obtained with an equimolar mixture of mS8 and E53 for the Rz-2 system was very similar to that obtained for the Rz-3 system (Fig. 7). Similar NOE connectivity for G2″:U27 through C6″:G23 can also be traced, suggesting the formation of a new stem (P1), as expected (Figs 2E and 7). We found further connectivity in the opposite direction. Both the imino proton signals for G2″:U27 of the Rz-2 system show NOE cross-peaks with a signal at 12.46 p.p.m. and this signal in turn shows a NOE cross-peak with a signal at 12.86 p.p.m. These signals are assumed to be those of G28:C13 and G29:C12, which are the 2 bp of P1.1 newly found in the crystal structure (8). It should be noted that even when C1″ was removed from mS8, these NOEs were still observed for S7:E53, suggesting that these NOEs do not include that between G2″:U27 and C1″:G28 (Y.Tanaka, unpublished data).
Figure 7.
Secondary structure (A) and NOESY spectrum (B) of the complete Rz-2 system (mS8:E53) measured in 10 mM sodium phosphate buffer, pH 6.5, 50 mM NaCl containing 5% D2O at 25°C. The sample contained a mixture of mS8 and E53 (1 mM each). Orange and green lines indicate NOE connectivities for the P1 and P1.1 stems, respectively. Purple lines indicate a NOE which can be assigned for that between G29 and G6′ of the G30:G6′ pair. The one-dimensional 1H spectrum and signal assignments for P1, P1.1 and G6′ are shown at the top.
The imino proton signals for G2″:U27 of the Rz-3 system also show NOE cross-peaks with a signal at 12.51 p.p.m., which can be assigned to G28:C13. Although a further NOE was not observed for this signal in this case, the base pair G29:C12 may exist, since a sharp peak is observed at 12.89 p.p.m. in the one-dimensional spectrum and the HSQC spectrum shows a signal for G:C NH at that 1H position. The putative G29 imino proton resonance for both systems shows a further NOE cross-peak with a signal at ∼13.2 p.p.m. We tentatively assigned this resonance to the imino proton of G6′ hydrogen bonded to the G30 base forming a G:G mismatch base pair just above the P4 stem.
Mutation experiments by Been and Perrotta revealed that the formation of a G:G homopurine mismatch pair at the corresponding region in the antigenomic HDV ribozyme is required for optimal self-cleavage activity (29). The crystal structure, which was determined for a genomic ribozyme, contains an A:G homopurine mismatch pair just above the P4 stem (8). Our Rz-3 and Rz-2 systems contain antigenomic sequences in the same region. It is highly probable that the 13.2 p.p.m. resonance which shows a NOE with G29 of G29:C12 is due to the G6′ imino proton of the G30:G6′ pair. This resonance and NOE have already been observed for the Ea35:Eb16 complex (Fig. 4) and the HSQC spectrum shows that the resonance is not due to G residues in Ea35. They have also been observed for E53 alone (Fig. 5). The absence of an imino proton signal for G30 suggests that the imino proton is not involved in hydrogen bonding. A similar chemical shift (13.24 p.p.m.) is observed for a G:G base pair in an AMP–RNA aptamer complex (30), where the N1H (13.24 p.p.m.) and 2-NH2 of a G residue form hydrogen bonds with N7 and O6 of the partner G residue, respectively.
For P2 and P3 of both systems the same connectivities were found as those observed for the enzyme part. The chemical shifts of the imino proton signals of these stems are almost identical with those of the enzyme part alone. For P4 of the Rz-2 system the same connectivity as that observed for E53 was found. A stronger NOE cross-peak between Gd and G2′:C34 was observed for the complete system. Substrate binding may stabilize the P4 stem. On the other hand, for P4 of Rz-3 no NOE connectivity was observed.
Although most of the imino proton resonances were assigned as described above, four resonances at ∼11.3–11.4 (α), 12.2 (β), 12.8 (γ) and 13.4 (δ) p.p.m. remained unassigned. These resonances show small NOEs and a connectivity α–β–γ–δ, suggesting that they are of residues in a stem. Since they were not observed for the Ea35:Eb16 complex but observed for both Rz-2 and Rz-3, the substrate analog may be involved in the stem, which is a part of a minor conformation formed by the substrate and the enzyme. Taking these results and the HSQC spectrum into account, we tentatively assigned these resonances to the imino protons of G19, U5″, G4″ and G3″ in an alternative conformation formed by the substrate and the enzyme, as shown in Figure 2F.
Temperature dependence of the imino proton spectra
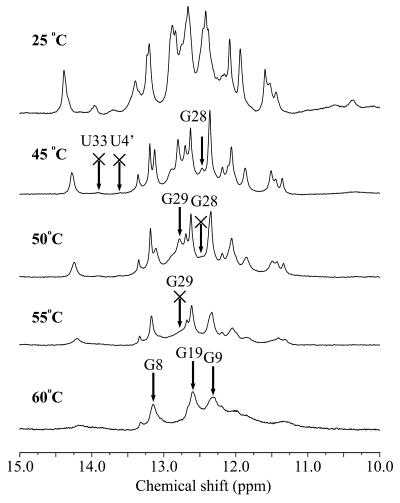
We examined the temperature dependence of the imino proton spectra of Rz-3, which shows sharper signals than does Rz-2, to evaluate the thermal stability of each base pair. With increasing temperature the rate of proton exchange between the hydrogen bonded imino proton and the solvent water protons increases and the signal becomes broader and finally disappears above a certain temperature, usually well below the melting temperature (Tm), depending on the thermal stability of the base pair. One-dimensional spectra of the complete Rz-3 system were measured while increasing the temperature stepwise by 5°C from 25 to 60°C (Fig. 8). At ∼45°C the imino proton signals for U33 and U4′ of P4 disappear. At ∼50–55°C many more signals for P1 and P2 disappear, but even at 60°C all signals for P3 can still be observed. These results suggest that the base pairs in the P3 stem are the most stable ones among the four stems. The base pairs in P3 may be buried inside the ribozyme complex and protected from proton exchange with solvent water molecules. In the crystal structure P3 has tertiary interactions with adenine bases in J4/2 and this may further stabilize the P3 stem.
Figure 8.
Temperature dependence of 1H spectra in the imino proton region for Rz-3 measured in 10 mM sodium phosphate buffer, pH 6.5, 50 mM NaCl containing 5% D2O at the temperatures indicated. The resonances which disappear from the spectrum at the temperature indicated are marked by arrows with crosses.
The P4 stem shows lower stability than the other stems. The wild-type ribozymes have a long P4 stem that contains more than 7 consecutive base pairs and a hairpin loop. This long P4 stem might be necessary for stabilization of the entire active domain of the ribozymes to attain optimal activity.
As for the imino proton signals of the two G:C pairs for the P1.1 stem, the signals of G28 and G29 disappear at ∼50 and 55°C, respectively. Although P1.1 contains only 2 bp, it turns out that the base pairs are as stable as those of the P1 and P2 stems. This suggests that stacking of P1.1 with both P1 and P4, which is extended by 1 bp with the G:G mismatch base pair, may stabilize P1.1. It should also be noted that, of the 2 bp in P1.1, the signal of G28 disappears at a lower temperature than that of G29, suggesting that the former base pair has lower thermal stability.
It has been shown that these 2 bp have a profound effect on the RNA cleavage activity (9,10). Nishikawa and Nishikawa (10) reported that the combination of purine28–purine29 (J1/4) and pyrimidine13–pyrimidine12 (L3) sequences gives higher activity than other combinations when two ribozymes which have the same number of A:U and G:C base pairs in P1.1 are compared. They also reported that when P1.1 contains purine28–purine29:pyrimidine13–pyrimidine12 and one of the G:C pairs is replaced by an A:U pair, kobs for a mutant ribozyme where the upper base pair (G28:C13) is replaced is reduced to 1/100 of that for the wild-type ribozyme, while that of the mutant ribozyme where the lower base pair (G29:C12) is replaced is reduced to only 1/10. This phenomenon can be explained by the lower stability of G28:C13 with respect to that of G29:C12. Replacement of the G28:C13 base pair, which has intrinsically lower stability, by a weaker A:U base pair may further destabilize the essential P1.1 stem to maintain the active ribozyme conformation. On the other hand, replacement of the intrinsically more stable G29:C12 pair may have a smaller influence on the active ribozyme conformation.
Chemical shift changes of imino proton resonances upon Mg2+ addition
A Rz-3 solution (0.3 mM) was titrated with MgCl2 up to a Mg2+ concentration of 10 mM to obtain imino proton NMR spectra (Fig. 9A). The cleavage activity of the wild-type ribozymes, which contain larger catalytic domains, reaches a plateau at low Mg2+ concentration (1–10 mM) (31,32). On the other hand, the activity of Rz-3 increases beyond 10 mM Mg2+ and is still increasing even at 100 mM (15), suggesting a lower affinity for Mg2+ ions. Analysis of the Rz-3 data by means of curve fitting suggested that the Mg2+ concentration dependence of the Rz-3 system is consistent with a three Mg2+ ion binding model, where Kd1, Kd2 and Kd3 are ∼0.8, 6 and 160 mM, respectively, and the second and third Mg2+ ions mainly contribute to RNA cleavage activity (15). In the present titration experiment 10 mM Mg2+ may not have been high enough, but further addition of MgCl2 was not performed since MgCl2 addition caused considerable broadening of the imino proton resonances. The binding of Mg2+ ions to a ribozyme should cause chemical shift changes at imino protons that are close to or in the binding sites and if it causes changes in the secondary structure, disappearance and/or appearance of imino proton signals may be observed.
Figure 9.
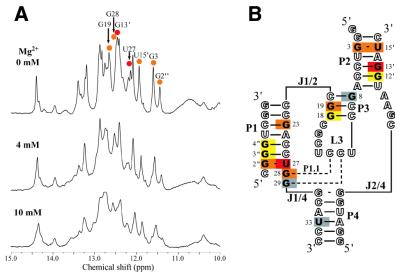
Changes in 1H spectra in the imino proton region upon titration of Rz-3 with MgCl2. (A) 1H spectra in the presence of 0, 4 and 10 mM MgCl2 at 25°C. (B) The chemical shift changes upon addition of 10 mM MgCl2 are mapped on the secondary structure of Rz-3. Gray, yellow, orange and red colors indicate the chemical shift changes at <0.02, 0.03–0.05, 0.06–0.09 and >0.10 p.p.m., respectively. The residues in open letters are those which do not have imino protons or for which the changes are not measurable due to signal overlap.
The addition of MgCl2 causes neither the appearance nor disappearance of imino proton signals, suggesting no big change in the secondary structure (Fig. 9A). Some signals show considerable chemical shift changes, whereas no information on the chemical shift change has been obtained for the resonances (those of G2, U4, G23, U4′, U5″, Gα and Gδ) which are weak and heavily overlapped by other signals. The residues that show significant changes were mapped on the secondary structure model of the ribozyme (Fig. 9B). Upon addition of 10 mM MgCl2 the largest changes (∼0.1 p.p.m.) are observed for the resonances of U27 and G13′ (Table 1) while, in contrast, some resonances, such as G29 and U33, show almost no change. These results suggest that there are Mg2+-binding sites with relatively high affinity around the regions close to the imino protons of U27 and G13′. U27, which shows the largest change, forms a G:U pair with G2″ at the bottom of P1 and, therefore, is close to the cleavage site, C1′′p-.
Table 1. Chemical shifts (p.p.m.) of imino proton signals for each residue of Rz-2 and Rz-3.
| Residue | Chemical shifta | ||||
|---|---|---|---|---|---|
| Rz-2 (1 mM), 0 mM MgCl2 | Rz-3 (1 mM), 0 mM MgCl2 | Rz-3 (0.3 mM), 0 mM MgCl2 | Rz-3 (0.3 mM), 4 mM MgCl2 | Rz-3 (0.3 mM), 10 mM MgCl2 | |
| G2b | 12.84 | 12.84 | 12.83 | 12.88 (0.05) | |
| G3 | 11.60 | 11.60 | 11.60 | 11.56 (0.04) | 11.53 (0.07) |
| U4 | 14.38 | 14.38 | 14.39 | 14.37c | 14.34c |
| G8 | 13.21 | 13.24 | 13.22 | 13.24 (0.02) | 13.24 (0.02) |
| G18 | 12.35 | 12.38 | 12.42 | 12.41 (0.01) | 12.39 (0.03) |
| G19 | 12.67 | 12.68 | 12.66 | 12.71 (0.05) | 12.73 (0.07) |
| G23 | 12.03 | 12.13 | |||
| U27 | 12.18 | 12.16 | 12.15 | 12.08 (0.07) | 12.04 (0.11) |
| G28 | 12.46 | 12.51 | 12.48 | 12.53 (0.05) | 12.56 (0.08) |
| G29 | 12.86 | 12.89 | 12.88 | 12.88 (0.00) | 12.87 (0.01) |
| U33 | 13.92 | 13.96 | 13.96 | 13.95 (0.01) | 13.96 (0.00) |
| Gd | 9.78 | ||||
| G2′ | 12.60 | ||||
| U4′d | 13.73 | 13.77 | 13.69 | ||
| G6′e | 13.18 | 13.19 | |||
| G12′ | 12.08 | 12.09 | 12.09 | 12.08 (0.01) | 12.04 (0.05) |
| G13′ | 12.40 | 12.41 | 12.46 | 12.53 (0.07) | 12.56 (0.10) |
| U15′ | 11.94 | 11.95 | 11.94 | 11.89 (0.05) | 11.87 (0.07) |
| G2″ | 11.43 | 11.45 | 11.44 | 11.40 (0.04) | 11.38 (0.06) |
| G3″ | 13.40 | 13.39 | 13.40 | 13.38 (0.02) | 13.36 (0.04) |
| G4″ | 13.21 | 13.21 | 13.20 | 13.24 (0.04) | 13.25 (0.05) |
| U5″ | 14.38 | 14.38 | 14.39 | 14.37c | 14.34c |
| Gαe | 11.30 | 11.46 | 11.44 | ||
| Uβ | 12.24 | 12.19 | 12.19 | 12.20 (0.01) | 12.23 (0.04) |
| Gγ | 12.75 | 12.75 | 12.75 | 12.74 (0.01) | 12.78 (0.03) |
| Gδe | 13.39 | 13.39 | |||
aNumbers in parentheses represent the chemical shift changes upon MgCl2 addition.
bThe chemical shift for Rz-3 at 10 mM MgCl2 was not determined due to signal broadening.
cThe chemical shift changes are not accurate due to complete overlap of the two resonances for U4 and U5″.
dThe signal for U4′ was too broad in the presence of MgCl2 to trace the chemical shift change.
eThe chemical shift changes for G6′, Gα and Gδ were not determined due to weak signal intensity and signal overlap or broadening.
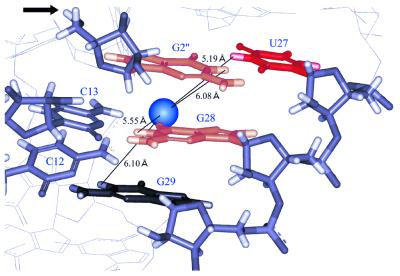
In the crystal structure P1 forms a continuous double helix with P1.1 and P4. In this region the ribozyme contains the same sequences as those of our ribozymes. Ferré-D’Amaré and Doudna reported that the ribozyme does not contain well-ordered bound metal ions in the active site cavity (33). The crystal structure deposited by them in the Protein Data Bank (PDB accession no. 1CX0) contains several magnesium ions in the major groove region. One of them (Mg6) is located close to O6 of G28 (3.2 Å distant) and the distances between the magnesium and imino protons of U27, G28, G29 and G2″ are all ∼5–6 Å (Fig. 10). The chemical shift changes in this region upon addition of 10 mM MgCl2 are 0.04, 0.06, 0.11, 0.08 and 0.01 p.p.m. for G3″, G2″, U27, G28 and G29, respectively (Table 1). It should be noted that remarkable broadening was also observed for G2″ and G3″ (Fig. 9A). These results suggest that the relatively high affinity Mg2+-binding site in solution may be rather closer to the G2″:U27 base pair than the position of Mg6 in the crystal structure. This means that the bound Mg2+ ion could be the putative catalytic Mg2+ ion, which is assumed to be involved in abstraction of the hydrogen of 2′-OH of C1″ at the cleavage site (34).
Figure 10.
The Mg2+-binding site which is closest to the cleavage site in the crystal structure. The blue sphere is a Mg2+ ion which is observed in the crystal structure (33). The distances between the Mg2+ ion and nearby base imino protons are shown. An arrow indicates the 5′-OH group of G2″, the leaving group of the cleavage reaction. The bases are colored according to the extent of the chemical shift change upon addition of 10 mM MgCl2. Black, orange and red indicate <0.02, 0.06–0.09 and >0.10 p.p.m. chemical shift changes, respectively.
A large chemical shift change similar to that for U27 is also observed for G13′ in the P2 stem. G3 and G15′ in P2 also show relatively large chemical shift changes (0.07 p.p.m. each). In the crystal structure two Mg2+ ions are observed in P2 (at the top and center), one of which forms at least two inner sphere coordinations with O6 and N7 of a guanine residue (G82, which corresponds to G70 in the PDB data and to A14′ of Rz-3) (33). It has been reported that a trans-acting antigenomic HDV ribozyme is specifically cleaved at the position corresponding to 11′, at the bottom of P2, in the presence of 10 mM Mg2+ and at pH 9.5 (35), and therefore the presence of a Mg2+ coordination site at around the cleavage site is assumed. Our results also suggest that there is at least one Mg2+-binding site with relatively high affinity in the P2 stem. The relatively large chemical shift change (0.07 p.p.m.) for G19 may suggest another binding site in the P3 stem, whereas no Mg2+ ion is observed in the major groove region in the crystal structure. No evidence was obtained for Mg2+ binding to the P4 stem, where no Mg2+ ion is observed in the crystal structure (33).
The results of the Mg2+ titration experiments suggest that Mg2+ ions bind to a few different sites similar to those found in the crystal structure. In the crystal structure the strongest Mg2+-binding site seems to be in P2. In solution our results suggest that another binding site with comparable affinity is close to U27 and closer to the cleavage site (C1′′p-) than the weak binding site at around G28 in the crystal structure. It has been reported that specific cleavage of the RNA chains of genomic HDV ribozymes occurs in the J2/4 and L3 regions (most intensively at C7′p-) on treatment with Pb2+ ions at pH 7 or other divalent metal ions, including Mg2+, at pH 9 (36,37), suggesting strongly that there is a high affinity metal-binding site spatially close to the cleavage site. The base residue of C7′ is very close to the cleavage site (5′-OH of G2″), the phosphate group of C7′p- is opposite the major groove side of G2″:U27 and C13:G28 and the 2′-O of C7′ is approximately in line with respect to the P and 5′-O of G8′ in the crystal structure. This region coincides with the Mg2+-binding site suggested from our results. A magnesium ion at this site may play a catalytic role in the ribozyme reaction.
Rz-3 shows higher activity in the presence of Mn2+ than that for Mg2+ (15). The Mn2+ ion is known to cause paramagnetic line broadening of proton signals in a highly distance-dependent manner (38). Preliminary Mn2+ titration (up to 7 µM MnCl2) experiments with Rz-3 (0.2 mM) showed that the G2, U27, G12′, G13′ and U15′ imino proton signals were considerably broadened (Supplementary Material, Fig. S1). These results also suggest that there are high affinity divalent cation binding sites in the P2 and P1 regions.
CONCLUSIONS
We measured 1H NMR and 15N-1H HSQC spectra of two small HDV ribozymes, Rz-3 and Rz-2 (Fig. 1C and D), and assigned most of the imino proton resonances. The results are consistent with the ‘nested double pseudoknot’ structure (9) which was actually found in the crystal structure by Ferré-D’Amaré et al. (8,33). Our results suggest that the P1.1 helix, which was found in the crystal structure, also exists in solution and is extended by 1 bp with a mismatch base pair G30:G6′ at the bottom. This extended P1.1 may stabilize coaxial stacking of the P1 and P4 stems on it, making a continuous helix of P1–P4. A mismatch G:A base pair is also found on top of the P4 stem stacked with P1.1 in the crystal structure. G:G pair formation has been suggested by mutation experiments for both genomic and antigenomic HDV ribozymes (29).
Comparison of the temperature at which the imino proton signals of Rz-3 disappear indicates that the P3 stem has the highest stability while the P4 stem has the lowest stability. These results suggest that the P3 stem is buried inside the complex and the nested double pseudoknot conformation as a whole is maintained at the expense of P4 stem stability. The results of the temperature dependence experiments also show that two base pairs of P1.1 have different thermal stabilities, which is assumed to be relevant to the difference in the activities of mutant ribozymes where one of the G:C pairs is replaced by an A:U pair (10).
We examined the chemical shift changes of imino proton resonances for Rz-3 upon titration with MgCl2. The observed large changes for the imino proton resonances of U27, G28 and G2″ suggest that a Mg2+ ion with relatively high affinity lies in the vicinity of G2″:U27 of P1 and C13:G28 of P1.1, while a Mg2+ ion with low affinity is observed in the vicinity of the two base pairs of P1.1 in the crystal structure (34). Although it is not clear how the Mg2+ ion participates in the cleavage reaction, it is highly probable that this Mg2+ ion plays an important role in catalysis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Satoshi Nishikawa for helpful advice and discussions. This work was supported in part by Grants-in-Aid for Scientific Research to S.U. (nos 09278210 and 10174211) and to M.K. (nos 10179102 and 12470487) from the Ministry of Education, Science and Culture of Japan and by the ‘Research for the Future’ Program (JSPS-RFTF97L00503) of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Lai M.M.C. (1995) The molecular biology of hepatitis delta virus. Annu. Rev. Biochem., 64, 259–286. [DOI] [PubMed] [Google Scholar]
- 2.Kuo M.Y.P., Sharmeen,L., Dinter-Gottlieb,G. and Taylor,J. (1988) Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J. Virol., 62, 4439–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharmeen L., Kuo,M.Y.P., Dinter-Gottlieb,G. and Taylor,J. (1988) Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J. Virol., 62, 2674–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu H.-N., Wang,Y.-J., Hung,C.-F., Lee,H.-J. and Lai,M.M.C. (1992) Sequence and structure of the catalytic RNA of hepatitis delta virus genomic RNA. J. Mol. Biol., 223, 233–245. [DOI] [PubMed] [Google Scholar]
- 5.Perrotta A.T. and Been,M.D. (1990) The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant. Nucleic Acids Res., 18, 6821–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrotta A.T. and Been,M.D. (1991) A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature, 350, 434–436. [DOI] [PubMed] [Google Scholar]
- 7.Been M.D. (1994) Cis- and trans-acting ribozymes from a human pathogen, hepatitis delta virus. Trends Biochem. Sci., 19, 251–256. [DOI] [PubMed] [Google Scholar]
- 8.Ferré-D’Amaré A.R., Zhou,K. and Doudna,J.A. (1998) Crystal structure of a hepatitis delta virus ribozyme. Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- 9.Wadkins T.S., Perrotta,A.T., Ferré-D’Amaré,A.R., Doudna,J.A. and Been,M.D. (1999) A nested double pseudoknot is required for self-cleavage activity of both the genomic and antigenomic hepatitis delta virus ribozymes. RNA, 5, 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishikawa F. and Nishikawa,S. (2000) Requirement for canonical base pairing in the short pseudoknot structure of genomic hepatitis delta virus ribozyme. Nucleic Acids Res., 28, 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee B.-S., Wu,H.-N. and Huang,T.-H. (1993) The catalytic domain of human hepatitis delta virus RNA. A proton nuclear magnetic resonance study. FEBS Lett., 324, 296–300. [DOI] [PubMed] [Google Scholar]
- 12.Kolk M.H., Heus,H.A. and Hilbers,C.W. (1997) The structure of the isolated, central hairpin of the HDV antigenomic ribozyme: novel structural features and similarity of the loop in the ribozyme and free in solution. EMBO J., 16, 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch S.R. and Tinoco,I.,Jr (1998) The structure of the L3 loop from the hepatitis delta virus ribozyme: a syn cytidine. Nucleic Acids Res., 26, 980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Been M.D., Perrotta,A.T. and Rosenstein,S.P. (1992) Secondary structure of the self-cleaving RNA of hepatitis delta virus: applications to catalytic RNA design. Biochemistry, 31, 11843–11852. [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto T., Tanaka,Y., Kuwabara,T., Kim,M.H., Kurihara,Y., Katahira,M. and Uesugi,S. (1997) Properties of hepatitis delta virus ribozyme, which consists of three RNA oligomer strands. J. Biochem. (Tokyo), 121, 1123–1128. [DOI] [PubMed] [Google Scholar]
- 16.Davanloo P., Rosenberg,A.H., Dunn,J.J. and Studier,F.W. (1984) Cloning and expression of the gene for bacteriophage T7. Proc. Natl Acad. Sci. USA, 81, 2035–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 18.Inoue T., Hayase,Y., Imura,A., Iwai,S., Miura,K. and Ohtsuka,E. (1987) Synthesis and hybridization studies on two complementary nona(2′-O-methyl)ribonucleotides. Nucleic Acids Res., 15, 6131–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odai O., Kodama,H., Sakata,T., Tanaka,T. and Uesugi,S. (1990) Synthesis and NMR study of ribooligonucleotides forming a hammerhead-type RNA enzyme system. Nucleic Acids Res., 18, 5955–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunningham P.R. and Ofengand,J. (1990) Use of inorganic pyrophosphatase to improve the yield of in vitro transcription reactions catalyzed by T7 RNA polymerase. Biotechniques, 9, 713–714. [PubMed] [Google Scholar]
- 22.Martin C.T. and Coleman,E. (1989) T7 RNA polymerase does not interact with the 5′-phosphate of the initiating nucleotide. Biochemistry, 28, 2760–2762. [DOI] [PubMed] [Google Scholar]
- 23.Kuchino Y. and Nishimura,S. (1989) Enzymatic RNA sequencing, Methods Enzymol., 80, 154–163. [DOI] [PubMed] [Google Scholar]
- 24.Markley J.L., Bax,A., Arata,Y., Hilbers,C.W., Kaptein,R., Sykes,B.D., Wright,P.E. and Wuthrich,K. (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. Eur. J. Biochem., 256, 1–15. [DOI] [PubMed] [Google Scholar]
- 25.Plateau P. and Gueron,M. (1982) Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J. Am. Chem. Soc., 104, 7310–7311. [Google Scholar]
- 26.Sakata T., Hiroaki,H., Oda,Y., Tanaka,T., Ikehara,M. and Uesugi,S. (1990) Studies on the structure and stabilizing factor of the CUUCGG hairpin RNA using chemically synthesized oligonucleotides. Nucleic Acids Res., 18, 3831–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varani G., Cheong,C. and Tinoco,I.,Jr (1991) Structure of an unusually stable RNA hairpin. Biochemistry, 30, 3280–3289. [DOI] [PubMed] [Google Scholar]
- 28.Allain F.H.-T. and Varani,G. (1995) Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol., 250, 333–353. [DOI] [PubMed] [Google Scholar]
- 29.Been M.D. and Perrotta,A.T. (1995) Optimal self-cleavage activity of the hepatitis delta virus RNA is dependent on a homopurine base pair in the ribozyme core. RNA, 1, 1061–1070. [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang F., Fiala,R., Live,D., Kumar,R.A. and Patel,D.J. (1996) RNA folding topology and intermolecular contacts in the AMP-RNA aptamer complex. Biochemistry, 35, 13250–13266. [DOI] [PubMed] [Google Scholar]
- 31.Suh Y.-A., Kumar,P.K.R., Taira,K. and Nishikawa,S. (1993) Self-cleavage activity of the genomic HDV ribozyme in the presence of various divalent metal ions. Nucleic Acids Res., 21, 3277–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano S., Chadalavada,D.M. and Bevilacqua,P.C. (2000) General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science, 287, 1493–1497. [DOI] [PubMed] [Google Scholar]
- 33.Ferré-D’Amaré A.R. and Doudna,J.A. (2000) Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol., 295, 541–556. [DOI] [PubMed] [Google Scholar]
- 34.Takagi Y., Warashina,M., Stec,W.J., Yoshinari,K. and Taira,K. (2001) Recent advances in the elucidation of the mechanism of action of ribozymes. Nucleic Acids Res., 29, 1815–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lafontaine D.A., Ananvoranich,S. and Perreault,J.-P. (1999) Presence of a coordinated metal ion in a trans-acting antigenomic deltaribozyme. Nucleic Acids Res., 27, 3236–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar P.K.R., Jeoung,Y.-H. and Nishikawa,S. (2000) Probing regions of important phosphates of HDV ribozyme. In Krupp,G and Gaur,R.K. (eds), Ribozyme Biochemistry and Biotechnology. Eaton Publishing, Natick, MA, pp. 257–275.
- 37.Matysiak M., Wrzesinski,J. and Ciesiolka,J. (1999) Sequential folding of the genomic ribozyme of the hepatitis delta virus: structural analysis of RNA transcription intermediates. J. Mol. Biol., 291, 283–294. [DOI] [PubMed] [Google Scholar]
- 38.Ott G., Arnold,S. and Limmer,S. (1993) Proton NMR of manganese ion binding to tRNA-derived acceptor arm duplexes. Nucleic Acids Res., 21, 5859–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.