Abstract
Astragalus membranaceus var. mongholicus (AMM), a member of the Leguminosae, is one of the most important medicinal plants worldwide. The dried roots of AMM have a wide range of pharmacological effects and are a traditional Chinese medicine. Here, we report the first chromosome-level reference genome of AMM, comprising nine pseudochromosomes with a total size of 1.47 Gb and 27 868 protein-encoding genes. Comparative genomic analysis reveals that AMM has not experienced an independent whole-genome duplication (WGD) event after the WGD event shared by the Papilionoideae species. Analysis of long terminal repeat retrotransposons suggests a recent burst of these elements at approximately 0.13 million years ago, which may explain the large size of the AMM genome. Multiple gene families involved in the biosynthesis of triterpenoids and flavonoids were expanded, and our data indicate that tandem duplication has been the main driver for expansion of these families. Among the expanded families, the phenylalanine ammonia-lyase gene family was primarily expressed in the roots of AMM, suggesting their roles in the biosynthesis of phenylpropanoid compounds. The functional versatility of 2,3-oxidosqualene cyclase genes in cluster III may play a critical role in the diversification of triterpenoids in AMM. Our findings provide novel insights into triterpenoid and flavonoid biosynthesis and can facilitate future research on the genetics and medical applications of AMM.
Key words: Astragalus membranaceus var. mongholicus, genome sequences, LTR-RTs, triterpenoid, flavonoid
This study reports a chromosome-level reference genome of Astragalus mongholicus (AMM). The PAL family genes are primarily expressed in the roots of AMM, suggesting their roles in the biosynthesis of phenylpropanoid compounds. Furthermore, the functional versatility of OSC genes in cluster III may play a critical role in the diversification of triterpenoids in AMM.
Introduction
Astragalus membranaceus var. mongholicus (AMM) (2n = 18), a perennial herb from the Astragalus genus of the Leguminosae family, is one of the most widely used medicinal plants worldwide (Fu et al., 2014). The dried roots of AMM, also known as “Huang Qi,” have been used as a traditional herbal medicine for more than 2000 years, with tonic, hepatoprotective, diuretic, and expectorant properties (Tang et al., 2010). The medicinal value of Huang Qi was recorded for the first time in Divine Farmer’s Materia Medica (Shennong Bencao Jing), the earliest extant pharmaceutical monograph in China. Modern pharmacological studies have shown that Huang Qi improves immune function, has antioxidant, anti-aging, and anti-fatigue properties, and has protective effects on the cardiovascular and cerebrovascular systems (Hikino et al., 1976; Qin et al., 2013; Shahrajabian et al., 2019; Zheng et al., 2020). Huang Qi is a major component of HSBD (Huashibaidu granules), which were clinically proven to be effective in the treatment of severe COVID-19 patients (Huang et al., 2021). In addition, Huang Qi has been widely used as an ingredient in dietary supplements such as teas, beverages, soups, and trail mix (Song et al., 2008; Zhang et al., 2011).
The main biologically active compounds isolated from AMM are flavonoids and triterpenoids (Ma et al., 2002; Kim et al., 2003; Chu et al., 2010), of which calycosin-7-O-b-D-glucoside and astragaloside IV are used as indexes to evaluate the quality of Huang Qi in China (Ma et al., 2003; Wu et al., 2020). Calycosin-7-O-b-D-glucoside has anti-inflammatory, anti-cancer, and anti-bacterial properties (Choi et al., 2005; Lee et al., 2005), and astragaloside IV has anti-fatigue and anti-viral properties (Wang et al., 2009). Identifying the genes encoding key enzymes in the biosynthesis of active compounds in AMM would be helpful for elucidating their biosynthetic mechanisms at the molecular level and would lay a theoretical foundation for their artificial synthesis in the future. However, few studies have examined the flavonoid and triterpenoid biosynthesis pathways in AMM, and only a few candidate genes have been identified through transcriptome analysis (Li et al., 2017a; Liang et al., 2020).
Despite the commercial importance and increasing demand for AMM, the absence of genome-wide information has hampered comprehensive and systematic research on this plant. In this study, we assembled a chromosome-scale reference genome of AMM (Figure 1A) using wild samples from the Taihang Mountains (the traditional Chinese production region for Huang Qi) with a combination of Illumina sequencing, PacBio sequencing, and high-throughput chromatin conformation capture (Hi-C) technology. We performed phylogenetic and comparative genomic analysis to determine the phylogenetic position, differentiation time, and whole-genome duplication (WGD) events of AMM. Using the assembled genome as a reference, we identified candidate genes for triterpenoid and flavonoid biosynthesis. Comparative genomic analysis of the AMM genome with those of other species show that multiple gene families have expanded in AMM, which may explain the abundance and diversity of its triterpenoids and flavonoids. Overall, our study provides important insights to facilitate molecular-assisted breeding, genome editing, and further exploration of the molecular mechanisms underlying the chemical diversity of active compounds in AMM.
Figure 1.
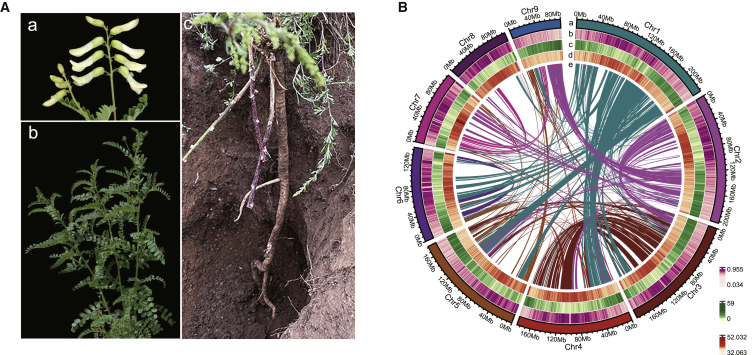
Morphological and genomic characteristics of AMM.
(A) Overview of the botanical characteristics of AMM. (A) Flower; (B) stem and leaf; (C) roots.
(B) Genomic landscape of AMM. (a) chromosome ideogram; (b) TE density; (c) gene density; (d) GC content; and (e) intra-genome collinear blocks connected by curved lines.
Results
Sequencing and assembly of the AMM genome
We performed whole-genome sequencing of AMM using Illumina sequencing, PacBio single-molecule real-time (SMRT) sequencing, and Hi-C technologies. A total of 91.60 Gb of high-quality paired-end reads were generated on the Illumina platform for k-mer (k = 21) analysis to estimate the genome size of AMM (Supplemental Figure 1; Supplemental Tables 1 and 2). The size of the AMM genome is approximately 1.23 Gb, with a heterozygous rate of 1.34% (Supplemental Table 2), indicating that the genome is highly heterozygous, compared with a 0.07% heterozygous rate of Amphicarpaea edgeworthii in Leguminosae (Liu et al., 2021a; 2021b).
Figure 2.
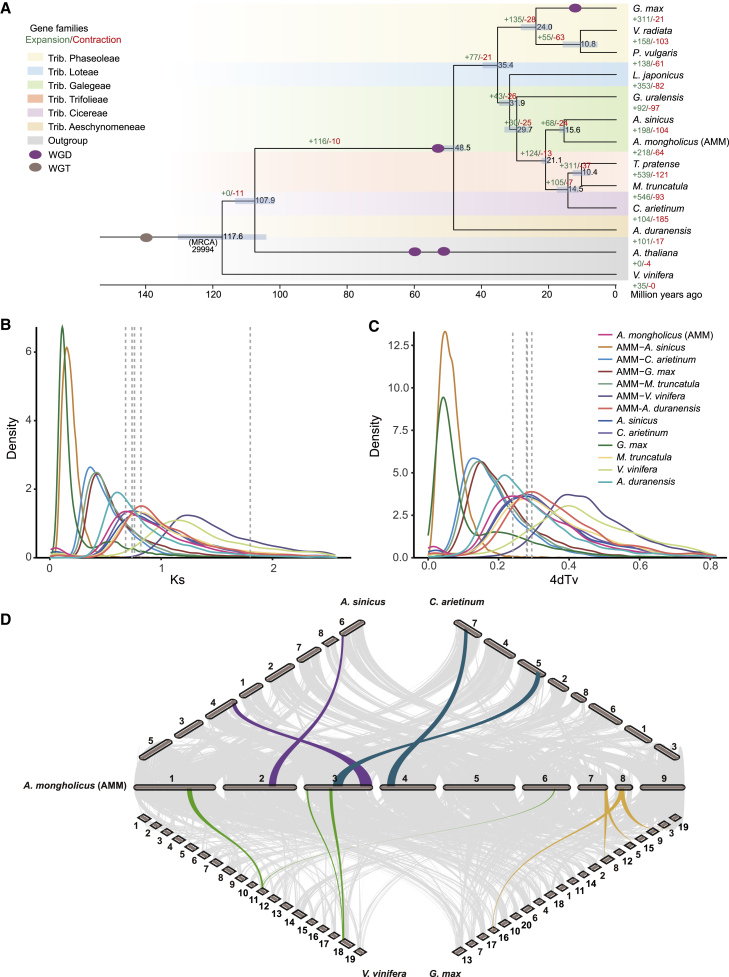
Comparative genomic and evolutionary analysis of AMM.
(A) Phylogenetic tree of 13 plant species and the expansion/contraction of gene families. The numbers on the nodes indicate estimated divergence times (million years ago [Mya]), and the blue bars show the error range. The numbers in green and red indicate the expanded and contracted gene families in the lineage, respectively. The timings of WGD and WGT events are superimposed on the tree. The background color represents the tribe to which the species belongs. All the nodes have 100% bootstrap support. MRCA, most recent common ancestor; WGD, whole genome duplication; WGT, whole genome triplication.
(B and C) Ks and 4dTv distribution between AMM, A. sinicus, C. arietinum, M. truncatula, G. max, and Vitis vinifera. Astragalus membranaceus var. mongholicus is abbreviated as A. mongholicus. The peak location is indicated by dotted lines. Ks, synonymous substitution; 4dTv, four-fold synonymous third-codon transversion position.
(D) Synteny between A. sinicus (8 chromosomes), C. arietinum (8 chromosomes), V. vinifera (19 chromosomes), G. max (20 chromosomes), and AMM (9 chromosomes).
Deep long-read sequencing of AMM produced 58.16 Gb of PacBio Circular Consensus Sequence (CCS) data (approximately 39.54× coverage of the estimated genome) with an average read length of 17 989 bp and an N50 read length of 17 907 bp (Supplemental Table 3). De novo assembly of the CCS reads generated an initial genome of 1.47 Gb (∼1.20-fold of the estimated genome size) consisting of 521 contigs (Table 1). We used 149.20 Gb of clean Hi-C sequencing data, representing ∼120.90× of the total data comprising the AMM genome, to evaluate the assembly and perform scaffolding (Supplemental Table 4). This analysis uncovered 348 855 915 read pairs that uniquely matched to the genome, 207 179 826 (59.39%) of which were valid Hi-C data. After Hi-C error correction and assembly, 1.47 Gb of data (99.75% of sequences) were anchored on nine pseudochromosomes, with a contig N50 of 9.79 Mb and a scaffold N50 of 181.02 Mb (Table 1; Supplemental Tables 5 and 6).
Table 1.
Assembly and annotation statistics of the AMM genome.
| Feature | Value |
|---|---|
| Assembled genome size (Gb) | 1.47 |
| Chromosome number | 9 |
| Contig number | 521 |
| Scaffold number | 304 |
| Chromosome-anchored sequence length (Gb) | 1.47 |
| Chromosome-anchored contigs | 494 |
| Contig N50 (Mb) | 9.79 |
| Scaffold N50 (Mb) | 181.02 |
| GC content (%) | 38.82 |
| Number of protein-coding genes | 27 868 |
| Average number of exons per gene | 5.64 |
| Number of pseudogenes | 266 |
| Number of rRNAs | 17 534 |
| Number of tRNAs | 1859 |
| Number of miRNAs | 143 |
| Number of snRNAs | 833 |
| Number of snoRNAs | 599 |
Figure 6.
Characterization of the triterpenoid biosynthesis gene clusters in the AMM genome.
(A) Gene-level synteny of cluster I genes between AMM and A. sinicus. CYP450 genes are colored in dark red; bAS genes are colored in dark green. Other genes unrelated to triterpenoid biosynthesis are in gray. The arrows indicate the relative positions and directions of genes in the cluster. Double slashes indicate regions that are not displayed.
(B) Gene-level synteny of cluster III genes between AMM and A. sinicus. LUS genes are colored dark brown, LAS genes are colored dark orange, and UGT genes are colored dark pink.
(C) Functional identification of Am07G013660 in yeast.
(D) Functional identification of Am07G013690 in yeast.
(E) MS spectra of target product in the pESC-URA-Am07G013660 strain and the authentic cycloartenol standard.
(F) MS spectra of product in the pESC-URA-Am07G013690 strain and the authentic lupeol standard.
We evaluated the assembly results using three strategies: short-read sequence alignment, CEGMA v2.5 (Core Eukaryotic Genes Mapping Approach), and BUSCO v5 (Benchmarking Universal Single-Copy Orthologs) (Supplemental Table 7). The overall mapping rate of short reads was 99.71%. CEGMA analysis identified 450 (98.25%) of the 458 core eukaryotic genes in the assembled AMM genome. BUSCO analysis showed that 1580 (97.89%) of the 1614 core conserved genes were present in the AMM genome, and the sequences of 1563 (96.84%) were complete (Supplemental Table 7). A heatmap of the genome-wide Hi-C interaction matrix (Supplemental Figure 2) revealed that the intensity of interactions of adjacent sequences was higher than that of non-adjacent sequences, demonstrating the quality of the Hi-C assembly. The high quality and integrity of the assembled AMM genome compare favorably with those of recently sequenced leguminous species (Cui et al., 2021; Pootakham et al., 2021).
Genome annotation
Transposable elements (TEs) make up a significant proportion of many eukaryotic genomes (Amyotte et al., 2012; Zhao and Ma, 2013). In the assembled AMM genome, approximately 1.02 Gb of sequences are TEs, accounting for 69.66% of the total genome length (Figure 1B). Among these TE sequences, 92.04% (943.22 Mb) are retroelements (class I), and 7.96% (81.53 Mb) are DNA transposons (class II) (Supplemental Table 8). In addition, we annotated 107.13 Mb (7.28% of the total genome length) of tandem repeat sequences, 11.66 Mb (0.79%) of microsatellites, 56.09 Mb (3.81%) of minisatellites, and 39.39 Mb (2.68%) of satellites (Supplemental Table 8).
We predicted protein-coding genes using homology, ab initio, and transcriptome prediction (Supplemental Figure 3). In total, 27 868 genes were predicted in the AMM genome, with an average gene length of 4674.34 bp and an average of 5.64 exons per gene (Table 1; Supplemental Figure 4; Supplemental Tables 9 and 10). The number of genes in AMM is larger than that in Cicer arietinum (23 386), smaller than that in Glycine max (56 044), Medicago truncatula (50 444), and Glycyrrhiza uralensis (33 968), and similar to that in Arabidopsis thaliana (27 336). We identified 20 968 non-coding RNAs in the AMM genome, including 17 534 rRNAs, 1859 tRNAs, 143 microRNAs (miRNAs), 833 small nuclear RNAs (snRNAs), and 599 small nucleolar RNAs (snoRNAs) (Table 1). We also predicted 266 pseudogenes in the AMM genome. The 27 868 predicted protein-coding genes were compared with genes in public databases, and our data show that 99.31% were in public databases, including 23 497 genes in the eggNOG database, 23 070 in the GO database, and 20 772 in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Supplemental Table 11), suggesting the high confidence of the gene annotations.
BUSCO analysis revealed the presence of 97.77% of the core conserved genes among the predicted genes, indicating the high quality of the gene prediction (Supplemental Table 12). In addition, 90.87% of the RNA sequencing (RNA-seq) data that mapped to the genome were located in predicted exons, demonstrating the high accuracy of gene prediction (Supplemental Figure 5).
Genome evolution in AMM
We compared the genes in the assembled AMM genome with those in genomes of 10 leguminous species (G. max, G. uralensis, C. arietinum, M. truncatula, Astragalus sinicus, Lotus japonicus, Arachis duranensis, Phaseolus vulgaris, Trifolium pratense, and Vigna radiata) and 2 outgroup species (Vitis vinifera and A. thaliana) (Supplemental Table 13). A total of 46 776 gene families were identified, including 3434 that are common to the 13 species and 169 that are specific to the AMM genome (Supplemental Figure 6A; Supplemental Table 14). Single-copy genes accounted for a large proportion of genes in AMM and in seven other leguminous species, whereas G. max had a higher proportion of gene families with two copies, probably due to the recent WGD event that is unique to G. max (Supplemental Figure 7). We next compared gene families among 5 leguminous species (AMM, A. sinicus, C. arietinum, T. pratense, and M. truncatula). As shown in Supplemental Figure 6B, 11 853 gene families were shared among the 5 leguminous species, and 606 were specific to AMM. KEGG analysis revealed that the AMM-specific genes were enriched in several pathways, including biosynthesis of amino acids, ubiquitin-mediated proteolysis, and diterpenoid biosynthesis (Supplemental Figure 8).
We constructed a high-confidence phylogenetic tree based on 177 single-copy syntenic genes from the 13 species and used the Bayesian relaxation molecular clock method to estimate divergence times (Figure 2A). The species most closely related to AMM among these 12 species was A. sinicus, with a divergence time roughly 15.6 million years ago (Mya). AMM and the lineage of C. arietinum, T. pretense, and M. truncatula diverged from a common ancestor approximately 21.1 Mya. Among the 11 leguminous species, A. duranensis is the most diverged species, with an estimated divergence time approximately 48.5 Mya from the other 10 leguminous species. The three Phaseoleae species (G. max, V. radiata, and P. vulgaris) diverged from the other leguminous species at ∼35.4 Mya.
The expansion and contraction of gene families play an important role in promoting the formation of specific traits and phenotypic diversity in plants (Renny-Byfield and Wendel, 2014). Expanded gene families may acquire new functions and pathways that make plants more adaptable to the environment. Analysis of gene family expansion and contraction showed that 218 gene families were expanded and 64 gene families were contracted in the AMM genome compared with the other 12 genomes (Figure 2A; Supplemental Table 15). Gene ontology (GO) and KEGG enrichment analyses were performed to gain more insights into the expanded and contracted gene families (Supplemental Figure 9). GO analysis revealed that the contracted gene families were enriched in the pathways “rejection of self-pollen,” “defense response,” and “negative regulation of molecular function,” whereas the expanded gene families were enriched in the pathways “DNA integration,” “translation,” and “DNA repair.” KEGG enrichment analysis revealed that most of the expanded gene families in the AMM genome were enriched in the pathways “oxidative phosphorylation,” “protein processing in endoplasmic reticulum,” “photosynthesis,” and “amino sugar and nucleotide sugar metabolism,” pointing to the possible expansion of secondary metabolism in AMM. Notably, some of these expanded gene families are involved in the biosynthesis of triterpenoids and flavonoids (Supplemental Table 16), such as genes encoding cytochrome P450 (CYP450) and uridine diphosphate glycosyltransferase (UGT). The expansion of these gene families has important implications for the biosynthesis and diversity of secondary metabolites, such as flavonoids and triterpenoids, in AMM. Given that the expansion of gene families is often caused by gene duplication within the family (Shang et al., 2020), we analyzed the duplication status of the expanded gene families. Our data show that 93 (42.7%) of the 218 expanded gene families have experienced tandem duplications, suggesting that tandem duplication played an important role in the expansion of gene families in AMM (Supplemental Table 15).
Positive selection of genes is important for the generation of new functions in species (Zhang et al., 2014). We identified 118 genes in AMM that have been subjected to significant positive selection (Supplemental Table 17). KEGG enrichment analysis showed that these genes were mainly involved in, but not limited to, “mRNA surveillance pathway,” “glutathione metabolism,” and “N-glycan biosynthesis,” suggesting that genes involved in perennial growth tended to undergo positive selection (Supplemental Figure 10).
WGDs shared by legume species
WGD, or polyploidy, has been an important contributor of genetic novelty throughout the evolutionary history of eukaryotes (Otto and Whitton, 2000; Adams and Wendel, 2005; Soltis et al., 2015). Polyploidy is particularly widespread among flowering plants, many of which have undergone several rounds of WGD (Blanc and Wolfe, 2004; Jiao et al., 2011; Soltis and Soltis, 2016; Zhao et al., 2017). To understand the evolutionary history of WGD events in AMM, we examined the four-fold synonymous third-codon transversion position (4dTv) and pairwise synonymous substitution (Ks) values of paralogs within the AMM genome. The Ks distribution for paralogous genes in the AMM genome shows two prominent peaks at ∼0.68 and ∼1.79 (Figure 2B), indicating that AMM has experienced two rounds of WGD. The most recent WGD event in AMM is similar to that in A. sinicus, C. arietinum, M. truncatula, and G. max, indicating that this WGD event is shared by these species. We estimated the timing of the most recent WGD event in AMM at ∼53.29 Mya, consistent with an ancestral Papilionoideae WGD event (Kim et al., 2013; Li et al., 2013; Quilbe et al., 2021; Chang et al., 2022). Another WGD event in AMM corresponds to the whole-genome triplication (WGT) event shared by core eudicot species. Our analysis suggested that, after the Papilionoideae WGD event, AMM successively diverged from G. max, M. truncatula, C. arietinum, and A. sinicus, consistent with the results of phylogenetic analysis. Among these five species, only G. max experienced a very recent WGD event roughly 13 Mya, which may explain the larger number of protein-coding genes in the G. max genome compared with the AMM genome (Supplemental Table 18). Overall, the results of 4dTv analysis were consistent with those of Ks analysis (Figure 2C).
To better understand the evolutionary history of AMM, we performed genomic collinearity analysis of AMM, A. sinicus, V. vinifera, C. arietinum, and G. max (Figure 2D). We detected one syntenic region in V. vinifera corresponding to two paralogous segments in AMM (Supplemental Figure 11). By contrast, the ratios of syntenic regions of both A. sinicus and C. arietinum to AMM are about 1:1 (Supplemental Figure 12), suggesting that A. sinicus, C. arietinum, and AMM experienced a WGD event after the divergence from V. vinifera, i.e., the WGD event shared by the Papilionoideae species. G. max shows a 2:1 collinearity relationship with AMM (Supplemental Figure 13), indicating that G. max has experienced another unique WGD event in addition to the WGD event shared among Papilionoideae species (Schmutz et al., 2010). This result echoes the gene family analysis and may partially explain why the G. max genome has the highest proportion of double-copy genes (Supplemental Figure 7).
Proliferation of LTR-RTs led to the large genome size of AMM
Polyploidization and proliferation of TEs have been recognized as the two major contributors to increased genome sizes of many plant genomes (Kreplak et al., 2019). The number of protein-encoding genes in the genome of AMM (27 868) is similar to that in C. arietinum (24 640), P. vulgaris (27 433), V. radiata (26 893), and G. uralensis (33 968). However, the assembled genome size of AMM (1471.09 Mb) is at least 2.7 times higher than the genome sizes of the other four species (378.86–549.6 Mb) (Figure 3A; Supplemental Table 18). Our data demonstrate that no unique WGD event occurred after the divergence of AMM from the other leguminous species (Figure 2B and 2C), suggesting that the larger genome size of AMM was not caused by WGD.
Figure 3.
Analysis of transposable elements in AMM.
(A) Relationship between genome sizes and gene numbers in the investigated legumes. The area of the fan indicates the size of the genome, and the color indicates the number of coding genes.
(B) Correlation between genome sizes and transposon sizes in 19 legume species (TEs, transposable elements). The detailed data are listed in Supplemental Table 19.
(C) Phylogenetic analysis of Ty3-gypsy retrotransposons. The red branch represents AMM, and the green branch denotes A. sinicus.
(D) Phylogenetic analysis of Ty1-copia retrotransposons.
(E) Insertion time and abundance of Ty3-gypsy retrotransposons of different lineages in AMM.
(F) Insertion time and abundance of Ty1-copia retrotransposons of different lineages in AMM.
To determine the causes of the increased size of the AMM genome, we analyzed the repeat sequences in genomes of 19 leguminous species (Supplemental Table 19). Our data show that the proportion of TEs varies greatly among these 19 species, ranging from 29.19% in Aeschynomene evenia to 69.66% in AMM, with a strong correlation between genome size and total TE size (R2 = 0.98, Figure 3B). Long terminal repeat retrotransposons (LTR-RTs) are the major type of TE in plants. Statistical analysis showed that the total lengths of all LTR-RTs, Ty3-gypsy, and Ty1-copia are positively correlated with genome size (R2 = 0.95, R2 = 0.85, and R2 = 0.78, respectively). Next, we examined the insertion times of intact LTR-RTs in AMM and related species, and we found that LTR-RTs have accumulated rapidly within the last 0.13 million years in the AMM genome (Supplemental Figure 14). Further estimates of insertion times of the Ty1-copia and Ty3-gypsy superfamilies show that these two superfamilies have proliferated rapidly within the last 0.1 million years in the AMM genome (Supplemental Figure 15). Our data suggest that the proliferation of LTR-RTs in the AMM genome may have caused its genome expansion.
To shed light on the evolution of LTR-RTs in the AMM genome, we compared LTR-RTs in AMM and the closely related A. sinicus. We de novo annotated Ty3-gypsy and Ty1-copia elements in the assembled genome of A. sinicus using the same pipeline used for AMM (Supplemental Table 20). We then constructed phylogenetic trees using the conserved domain of the reverse transcriptase (RT) of Ty3-gypsy and Ty1-copia elements. As shown in Figure 3C and 3D, Ty3-gypsy and Ty1-copia elements were grouped into five and eight lineages. Among these 13 lineages, 12 and 11 lineages are present in AMM and A. sinicus. Except for Athila, Ikeros, and Angela, all the lineages have consistently higher copy numbers in AMM than in A. sinicus, which may partially explain the three-fold larger genome of AMM. Specifically, the Tekay lineage accounts for only 1.22% of the Ty3-gypsy superfamily in A. sinicus but makes up 38.91% of the total number of Ty3-gypsy elements in AMM, suggesting that the LTR-RT elements in this lineage have been largely deleted from the A. sinicus genome or markedly amplified in the AMM genome.
We next analyzed the insertion times of different LTR-RT lineages in the AMM genome. In the Ty3-gypsy superfamily, rapid expansion of Tekay and Tat began 0.5 Mya, and they became the dominant families of the gypsy superfamily (Figure 3E). Tekay is the youngest lineage with the most intact elements amplified within the last 0.1 million years. In the copia superfamily, Ale and Ivana initially expanded rapidly after 0.2 Mya, but their amplification has decreased since 0.1 Mya (Figure 3F). The most active lineage in the copia superfamily is SIRE. Together, our data suggest the tremendous dynamics of transposon proliferation in the AMM genome.
Next, we classified these LTR-RTs into intact LTR-RTs, incomplete LTR-RTs (truncated LTRs and nested LTRs), and solo LTRs (Ou and Jiang, 2018). The insertion, translocation, and deletion of intact LTR-RTs have occurred continuously beginning 2 Mya, resulting in higher copy numbers of incomplete LTR-RTs than intact and solo LTRs (Supplemental Figure 16).
Identification and evolution of genes involved in flavonoid biosynthesis
Flavonoids are widely found in legumes, including AMM, and play a variety of roles in nodulation development and secondary metabolism (Subramanian et al., 2007). Previous studies identified 101 flavonoid-related genes in AMM and elucidated the partial flavonoid biosynthetic process in AMM (Figure 4A) (Yu et al., 2000; Wu et al., 2020). Based on protein domain and phylogenetic analyses, we identified 51 high-confidence genes encoding seven enzymes in the flavonoid biosynthetic pathway and compared the expression levels of these genes in different tissues, including roots at different developmental stages (Figure 4B; Supplemental Figures 17 and 18; Supplemental Tables 21 and 22). These genes include, but are not limited to, phenylalanine ammonia-lyase (PAL, 10), 4-coumarate:CoA ligase (4CL, 6), chalcone synthase (CHS, 14), and chalcone isomerase (CHI, 3).
Figure 4.
Evolution of genes involved in flavonoid biosynthesis.
(A) A simplified representation of the flavonoid biosynthesis pathway. The enzymes participating in each catalytic step in the pathway are shown in blue boxes.
(B) Expression patterns of key gene family members involved in the flavonoid biosynthesis pathway in different tissues at different developmental stages. The normalized (z-score) FPKM (fragments per kilobase of transcript per million fragments mapped) values of each gene are colored based on expression.
(C) Tandem duplication and phylogenetic analysis of PAL genes. Black lines point to tandemly duplicated genes in AMM.
(D) Syntenic analysis of PAL genes in AMM, M. truncatula, and A. sinicus. The red lines indicate the collinear relationships of PAL genes between AMM, M. truncatula, and A. sinicus. The arrows indicate the directions of the genes. Two slashes indicate that the area is not displayed.
(E) Expression of PAL genes in different tissues of AMM. The gene expression values were determined by qRT–PCR. Values are means ± SD from three independent experiments. Student’s t-test: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001; n.s., not statistically significant.
Other key enzymes in flavonoid biosynthesis include: cinnamate-4-hydroxylase (C4H, CYP73A) (Zhang et al., 2020a), flavanone 6-hydroxylase (F6H, CYP71D9) (Latunde-Dada et al., 2001), isoflavone synthase (IFS, CYP93C) (Sawada et al., 2002), and isoflavone 2′-hydroxylase and isoflavone 3′-hydroxylase (I2′H and I3′H, CYP81E) from the CYP450 superfamily (Liu et al., 2003). Given that genes in the CYP450 superfamily participate in almost half of the enzymatic reactions in flavonoid biosynthesis, we analyzed the CYP450 superfamily and identified 243 CYP450 genes in the AMM genome. Phylogenetic analysis of the CYP450 genes in A. thaliana and AMM revealed that AmCYP450s were found in eight of the nine family clusters endemic to dicotyledons, and 129 AmCYP450s (53.01%) were found in the 71 clan cluster, which is the only A-type CYP450 (Supplemental Figure 19). In addition, the 243 AmCYP450s were grouped into 43 families (Supplemental Table 23). We found that 242 (99.6%) of the 243 AmCYP450s are located on the nine assembled chromosomes (Supplemental Figure 20A), with the greatest enrichment (46, 19%) on chromosome 5. Among these 242 AmCYP450s, 109 (45%) form 39 clusters on all nine chromosomes except for chromosome 8. We next identified 3 C4H, 1 IFS, 12 F6H, 2 I2′H, and 3 I3′H candidate genes based on superfamily classification and phylogenetic analysis with related species (Supplemental Table 21; Supplemental Figures 21–23). MEME analysis of I2′H and I3′H genes in AMM and its related species showed that all I2′H and I3′H homologs contain 10 conserved motifs, except for the two I2′H- and I3′H-like genes Am05G032310 and Am05G030430 in AMM, which lack motif 6. Interestingly, these two genes show almost no expression in roots at any developmental stage, suggesting that motif 6 might be pivotal for maintenance of I2′H and I3′H function in catalyzing the hydroxylation of isoflavones (Supplemental Table 22).
The final step in flavonoid biosynthesis is UGT-catalyzed glycosylation. This step promotes the solubility, stability, and bioactivity of secondary metabolites to enable AMM plants to adapt to environmental changes (Hirotani et al., 2000; Vogt and Jones, 2000; Paquette et al., 2003; Bowles et al., 2006). Isoflavone 7-O-glucosyltransferase (IF7GT) and calycosin 7-O-glucosyltransferase (UCGT) are the main UGTs involved in flavonoid biosynthesis in AMM. In this study, 154 UGTs were identified in the AMM genome, more than those in A. sinicus (115) (Supplemental Table 24). Phylogenetic analysis of all the available UGT protein sequences from AMM, A. thaliana, and Zea mays revealed that the 154 AmUGTs were grouped into all 14 previously identified groups except for groups C and K, which are absent in AMM (Supplemental Figure 24). Most AmUGTs (81%) were clustered into groups A (19), D (42), E (25), G (19), and L (19). These results are consistent with the previous finding that groups A, D, E, G, and L are the most rapidly evolving groups in flowering plants (Caputi et al., 2012). The UGTs were found on all nine chromosomes, including 3 UGTs on chromosome 8 and 26 UGTs on chromosome 9 (Supplemental Figure 20A). Of the 154 UGTs, 97 (63%) were distributed on chromosomes as gene clusters. Based on UGT superfamily classification and phylogenetic analysis with related species, three IF7GT and three UCGT candidate genes were identified (Supplemental Table 21; Supplemental Figures 25 and 26).
The analysis of gene family expansion and contraction shows that CYP450s and UGTs are expanded in AMM (Supplemental Table 16). To elucidate the mechanism of AmUGT and AmCYP450 expansion and evolution, we analyzed potential duplication events in the AMM genome. Based on amino acid sequence homology, we identified 25 paralogous gene pairs and 78 tandemly duplicated genes among the AmCYP450s and 12 paralogous gene pairs and 87 tandemly duplicated genes among the AmUGTs (Supplemental Figure 20B; Supplemental Table 25). Our results suggest that tandem duplication is the main driver contributing to the expansion of these two gene families. We next calculated Ka (nonsynonymous substitutions), Ks, and the Ka/Ks ratio between tandemly duplicated genes. The Ka/Ks ratios of almost all these genes were less than 1.0 (Supplemental Table 26), suggesting that AmCYP450s and AmUGTs have undergone purifying selection.
PAL (EC 4.3.1.24) is the first and key enzyme in the phenylalanine metabolism pathway and was the first “defense gene” identified in plants (Zhang and Liu, 2015). Comparative genomics analysis identified 10 PAL-like genes in AMM and 6 PAL-like genes in A. sinicus and M. truncatula (Supplemental Figure 27). These 10 PAL genes were split into four groups by phylogenetic analysis (Figure 4C). Seven PALs in group I are on chromosome 9 and are tandemly duplicated genes, suggesting that tandem duplication might be the cause of PAL family expansion in AMM. By contrast, these tandemly duplicated PAL genes have only one copy in the A. sinicus and M. truncatula genomes (Figure 4D), suggesting that tandem duplication of PAL genes in AMM occurred after its divergence from A. sinicus and M. truncatula. Ka/Ks analysis of the orthologous gene pairs between AMM and these two species showed that the Ka/Ks ratios were all less than 1.0, indicating that PAL has undergone purifying selection (Supplemental Table 27) (Navarro and Barton, 2003). Transcriptome and quantitative real-time PCR analyses show that PALs in group I were highly expressed in roots, PALs in group II were highly expressed in flowers, the PAL in group III was not expressed in any tissue, and PALs in group IV were highly expressed in stems, highlighting the different expression patterns of PALs in different groups (Figure 4E; Supplemental Table 22). The group I tandemly duplicated PAL genes were highly expressed in roots, which to some extent explains the high stress tolerance and abundant secondary metabolites of AMM.
Genes involved in triterpenoid biosynthesis pathways
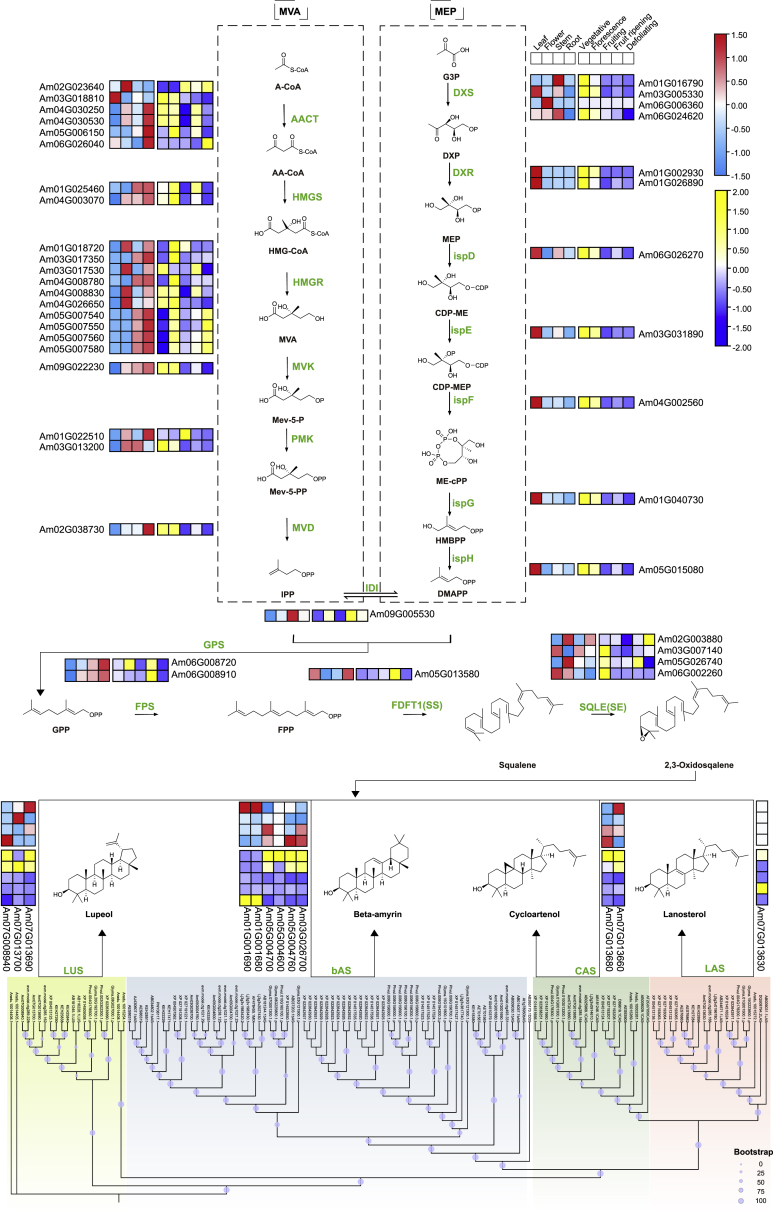
Astragalosides are one of the most abundant secondary metabolites in AMM; they have anti-inflammatory, anti-fibrosis, antioxidant, and anti-tumor properties and function in regulation of the immune system and metabolism (Li et al., 2017b). Astragalosides are a type of triterpenoid. As shown in Figure 4A, the mevalonic acid (MVA) and methylerythritol 4-phosphate (MEP) pathways are upstream pathways of triterpenoid biosynthesis (Sawai and Saito, 2011). Through comparative analysis of homologous genes in A. thaliana, we identified genes encoding 18 enzymes in the triterpenoid biosynthetic pathway, including but not limited to acetoacetyl-CoA thiolase (AACT, 6), hydroxymethylglutaryl-CoA synthase (HMGS, 2), and hydroxymethylglutaryl-CoA reductase (HMGR, 10) (Figure 5; Supplemental Table 28). A previous metabolomic and transcriptomic analysis showed that astragalosides are enriched in the roots of AMM (Wu et al., 2020). Transcriptome analysis of these genes in different tissues, including roots at different developmental stages, showed that genes in the MVA pathway were highly expressed in roots, whereas genes in the MEP pathway were highly expressed in leaves. In general, most triterpenoid biosynthesis genes showed greater expression in roots than in other tissues and were highly expressed in roots at the vegetative stage. (Supplemental Table 29).
Figure 5.
Identification and expression profiles of candidate genes involved in the biosynthesis of triterpenoids in AMM.
The dotted boxes indicate the MVA and MEP pathways of triterpenoid biosynthesis (Vranová et al., 2013). Green fonts represent the enzymes participating in the catalytic steps. Candidate genes and their expression patterns in different tissues at different developmental stages are shown on both sides of the corresponding enzymes. The bottom phylogenetic tree was constructed from OSC genes identified in different species using the maximum likelihood method; tree nodes are indicated by purple dots on branches (1000 bootstrap replicates). The normalized FPKM values of each gene are colored based on expression. A-CoA, acetyl-CoA; AA-CoA, acetoacetyl-CoA; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; MVA, mevalonate; Mev-5-P, mevalonate-5-phosphate; Mev-5-PP, mevalonate-5-diphosphate; IPP, isopentenyl diphosphate; G3P, D-glyceraldehyde 3-phosphate; DXP, 1-deoxy-D-xylulose 5-phosphate; MEP, 2-C-methyl-D-erythritol 4-phosphate; CDP-ME, 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol; CDP-MEP, 2-phospho-4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol; ME-cPP, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate; HMBPP, 4-hydroxy-3-methylbut-2-enyl-diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl diphosphate; FPP, farnesyl diphosphate.
Oxidosqualene cyclase (OSC) catalyzes the conversion of 2,3-oxidated squalene to produce different sterols and triterpenoids and is a key step in the creation of triterpenoid diversity (Haralampidis et al., 2002). OSCs in plants, including cycloartenol synthases (CAS), β-amyrin synthases (bAS), lanosterol synthases (LAS), and lupeol synthases (LUS), catalyze the formation of precursor substances for triterpenoid biosynthesis (Xue et al., 2012). We systematically identified OSCs in the genomes of 10 leguminous species including AMM (Figure 5; Supplemental Table 30). Nine species contained bAS genes, and bAS genes were highly expressed in the roots of AMM (Supplemental Table 29). Because the first step in astragaloside biosynthesis is the formation of cycloartenol from 2,3-oxidosqualene (OS) in the presence of CAS (Chen et al., 2015), we examined the expression of the two CAS genes. Both CAS genes were expressed at higher levels in roots at the vegetative stage than at the other four stages, and their activity might provide sufficient raw skeleton materials for astragaloside biosynthesis.
Functional characterization of OSCs in triterpenoid cluster III
PlantiSMASH analysis revealed that the AMM genome contains 24 potential gene clusters related to secondary metabolism (Supplemental Table 31), including that of 2 alkaloids, 10 saccharides, 1 lignan, 2 terpenes, 1 terpene-polyketide, 2 saccharide-terpenes, and 1 saccharide-alkaloid. Three gene clusters appeared to be involved in triterpenoid biosynthesis because cluster I includes three bAS genes, cluster II includes one LUS gene, and cluster III contains one LAS, two CAS, and two LUS genes. Based on collinearity analysis, we identified syntenic regions associated with clusters I–III between AMM and A. sinicus. Cluster I of A. sinicus was not as complete as that of AMM, and only 36% of the genes in the cluster I region of AMM are collinear with those in A. sinicus (Figure 6A). The three bAS genes in cluster I show a good collinear relationship between the two genomes, indicating that the bAS genes are relatively conserved. We did not find clusters similar to cluster III of AMM in the A. sinicus genome. Nonetheless, there are three OSC-like genes on chromosome 8 of A. sinicus (Figure 6B). Interestingly, these OSCs in AMM are located adjacent to CYP450s and UGTs on chromosome 7, whereas the OSCs in A. sinicus do not have CYP450s and UGTs nearby, suggesting that genes involved in AMM triterpenoid biosynthesis are preferentially present in clusters to promote efficient function of the pathway.
To better understand the function of OSCs in triterpenoid biosynthesis in AMM, we cloned three OSC genes (Am07G013660, Am07G013680, and Am07G013690) in cluster III. We did not clone Am07G013630 and Am07G013700, as they were not expressed in any AMM tissue. The three OSCs were transformed into the yeast strain WAT11v, which synthesizes 2,3-oxidosqualene. The target cycloatenol was detected in strain pESC-URA-Am07G013660, and the target lupeol was detected in strain pESC-URA-Am07G013690 (Figure 6C–6F). By contrast, the target cycloatenol was not detected in strain pESC-URA-Am07G013680, consistent with the low expression level of Am07G013680 in all tissues of AMM (Supplemental Figure 28) and suggesting that this gene may not play a role in cycloatenol synthesis of AMM. Together, these data suggest that that Am07G013660 and Am07G013690 are two potentially functional genes in the synthesis of cycloatenol and lupeol in AMM.
Discussion
The chromosome-level AMM genome provides a benchmark for genetic and functional genomic research
Astragalus membranaceus var. mongholicus is an important medicinal plant in traditional Chinese medicine. It is enriched with flavonoids, triterpenoids, polysaccharides, amino acids, alkaloids, and other recognized biologically active substances (Auyeung et al., 2016). A complete genome sequence provides the basis for studying gene function, phylogeny, and gene evolution in AMM and perhaps in many other medicinal plants. The AMM genome is characterized by high heterozygosity (1.34%) and a large amount of repetitive sequence (70.72%), making it challenging to assemble (Pu et al., 2020; Liu et al., 2021a, 2021b). In this study, we combined Illumina and PacBio sequencing with high-throughput Hi-C technology and generated a high-quality AMM genome with a scaffold N50 of 181.02 Mb and a contig N50 of 9.73 Mb. CEGMA analysis revealed that the assembled genome completely covered 98.25% of the core eukaryotic genes, and BUSCO analysis showed that 96.84% of the core conserved genes were captured in the assembled genome. These results highlight the greater integrity and higher quality of the AMM genome compared with recently sequenced medicinal plant genomes (Zhao et al., 2019; Cheng et al., 2021b; Jiang et al., 2021; Xiong et al., 2021). The AMM reference genome will provide a new perspective for understanding gene structure, composition, and function, gene regulation, and species evolution at the molecular level. It also has great value for improving the agronomic and medicinal characters of AMM through molecular breeding.
LTR-RT expansion led to the large genome size of AMM
Ks and 4DTv analyses showed that AMM has not experienced an Astragalus-specific WGD event after differentiation, but its genome size is generally larger than those of closely related species. In addition to polyploidy, retrotransposon insertion is the main cause of genome expansion (Devos et al., 2002; Kreplak et al., 2019; Xie et al., 2019; Liu et al., 2021a, 2021b). Comparative analysis of LTR-RT insertion times between AMM and related species indicated that an explosive insertion of LTR-RTs occurred in the AMM genome 0.13 Mya, perhaps explaining its large genome size. During the Quaternary glaciation (0.01–2 Mya), the burst of LTR-RT insertions may have helped the population survive in the face of adversity (Yang et al., 2021). The proliferation of different LTR-RT lineages is an important factor that causes differences in plant genome size. For example, the Tat family and the Ogre family are the major contributors to genome amplification of Camellia sinensis var. sinensis (Zhang et al., 2020b) and Pisum sativum (Kreplak et al., 2019), respectively. Although the amplification rate of the Tat family recently declined rapidly in AMM, the Tat family is still the largest LTR-RT lineage after a long period of accumulation and has made a significant contribution to the size of the AMM genome. Analyses of retrotransposon sequences in the genomes of the two Astragalus species revealed that the copy number of most lineages was consistently higher in AMM than in A. sinicus, resulting in the huge difference in genome size between the two species. Based on the historical changes in each LTR-RT lineage in AMM, it appears that changing lineage sizes lead to a constantly changing genome size. Moreover, because of insertions, translocations, and deletions of LTR-RTs during this process, incomplete autonomous LTRs now occupy a considerable proportion of the genome. Although LTR-RTs contribute greatly to genome size diversity, the origin, expression, insertion specificity, evolutionary fate, and potential effects of LTR-RTs on genetic and epigenetic gene regulation still remain largely unexplored (Zhao and Ma, 2013).
Gene expansion driven by tandem duplication promotes the accumulation and diversification of triterpenoids and flavonoids
Triterpenoids and flavonoids are two major active substances in AMM. Triterpenoids have anti-inflammatory, hypotensive, and anti-aging properties, as well as beneficial effects on myocardial ischemia injury and chronic kidney disease (Zhang et al., 2021). Comparative genome analysis confirmed the significant expansion of gene families related to the biosynthesis of substances such as flavonoids and triterpenoids in AMM. Secondary metabolites are the products of adaptation to the environment during the long-term evolution of plants (Deavours and Dixon, 2005; Wang et al., 2011; Zandalinas et al., 2018). AMM is often subjected to drought, strong light, ultraviolet radiation, and nutrient deficiency stress, suggesting that AMM has evolved towards the biosynthesis of secondary metabolites through natural selection (Guo et al., 2020). In this study, 53 and 72 candidate genes in the triterpenoid and flavonoid biosynthetic pathways were identified in AMM, respectively, as revealed by homology searches and functional annotation. Further analysis revealed that tandem duplication has played a key role in the expansion of genes in these pathways, such as CYP450 and UGT genes, which are the key nodes controlling metabolic flow (Seki et al., 2015). The increased copy numbers of these genes may promote the synthesis and diversity of active substances in AMM.
The phenylpropane metabolic pathway is necessary for the growth and development of terrestrial plants and is the result of long-term adaptation of plants to the natural environment (Huang et al., 2010). PAL catalyzes the deamination of phenylalanine to cinnamate and is the key and rate-limiting enzyme in phenylpropane metabolism (Rohde et al., 2004). We identified 13 gene families involved in flavonoid biosynthesis in 9 leguminous species and found that the PAL family was greatly expanded in AMM. After the ancestral polyploidy event 58 Mya, genes encoding flavonoid biosynthetic enzymes were duplicated and retained in Papilionoideae, which may have facilitated flavonoid synthesis and genetic diversity in Papilionoideae species (Li et al., 2013). PAL of AMM experienced tandem duplication events after its divergence from A. sinicus and M. truncatula, leading to further expansion of PAL genes in AMM. Expression levels of all tandemly duplicated PAL genes were higher in roots than in other tissues, suggesting that they are important for the synthesis of phenylpropanoid compounds in roots.
Triterpenoid biosynthetic genes are found in clusters
Triterpenoids are a series of natural compounds with various structures formed by the condensation of triterpenoid saponins with one or more sugar and/or other chemical groups, among which tetracyclic triterpenoids and pentacyclic triterpenoids are common (Thimmappa et al., 2014). We found that OSC, CYP450, and UGT genes are closely linked and clustered in the AMM genome, indicating that there may be regulatory mechanisms for their collaborative expression. Indeed, gene clusters for different secondary metabolite biosynthesis pathways are widely present in dicots and monocots, such as the gene cluster for paclitaxel biosynthesis in Taxus (Nutzmann et al., 2016; Xiong et al., 2021). This phenomenon is conducive to the efficient biosynthesis of secondary metabolites to a certain extent. By comparing AMM with A. sinicus, we found that gene clusters such as cluster I, which includes bAS genes, have similar structures in the two species, suggesting that the biosynthesis pathway of oleanane-type triterpenoids may have a common evolutionary origin. By contrast, there was no gene cluster similar to cluster III in A. sinicus, suggesting that this gene cluster is specific to AMM. This may partially explain why AMM can specifically synthesize tetracyclic triterpenes such as astragaloside IV in large quantities.
OSC is the first rate-limiting enzyme that functions downstream of triterpenoid biosynthesis, guiding OS to complete the cyclization process and generate a triterpenoid skeleton (Haralampidis et al., 2002). OSCs, together with genes encoding post-modification enzymes such as CYP450s and UGTs, affect the diversity of triterpene structures (Sandeep et al., 2019). Identification of OSCs in the unique gene cluster III of AMM suggests that OSCs have contributed to the diversification of triterpenoids in AMM, a finding that provides insight into the biosynthesis of diverse triterpenoids. Future investigation of these gene clusters will be important for understanding the regulatory mechanism underlying triterpenoid biosynthesis and will facilitate the use of triterpenoid products.
In summary, we have assembled a chromosome-level AMM genome, which is an important resource for comprehensive and in-depth study of AMM and many other medicinal plants. Identification of candidate genes involved in flavonoid and triterpenoid biosynthesis lays a foundation for future genetic improvement of AMM, and genome evolution analysis provides new insights into the evolutionary history of leguminous species.
Methods
Plant materials
AMM was collected at Mianshan Forest Farm at Jiexiu City, Shanxi Province (36°53′16.35″ N, 112°1′1.44″ E; 2015.6 m altitude). Healthy young leaves were collected for genome sequencing. Leaves, roots, stems, seeds, and seedlings were collected for RNA sequencing. All collected tissues were immediately quick-frozen with liquid nitrogen and then stored at −80°C. High-quality genomic DNA was extracted from leaves using the improved cetyl trimethyl ammonium bromide method (Murray and Thompson, 1980).
Short-read Illumina sequencing and genome size evaluation
Quality-checked genomic DNA was broken into ∼350-bp fragments by physical fragmentation (ultrasonic shock), and a small-fragment sequencing library was constructed by terminal repair, addition of A, addition of adaptors, target fragment selection, and PCR amplification. Qseq400 and Qubit instruments were used for library fragment size detection and quantification. The library was fixed to the sequencing chip by bridge PCR and sequenced on the Illumina NovaSeq 6000 platform to obtain paired-end 150-bp reads.
The short reads were filtered according to the following protocol: (1) removal of both polyG tails, (2) removal of paired reads shorter than 100 bp, (3) removal of read pairs in which more than 10% of the bases were the same as the next base, (4) removal of reads in which more than 50% of the nucleotides had quality scores less than 10, and (5) removal of read pairs if the end of either read had an average quality score lower than 20. The remaining 91.60 Gb of clean reads were used for subsequent analysis. The genome size, repeat ratio, and heterozygosity were assessed using Jellyfish v2.1.4 (-h 100000) (Marcais and Kingsford, 2011) and GenomeScope software v2.0 (-k 21 -p 2 -m 100000) (Ranallo-Benavidez et al., 2020).
SMRT long-read sequencing
The SMRTbell library (15–20 kb) was constructed according to the standard protocol provided by PacBio (Rao et al., 2014). Total genomic DNA was sheared using a g-TUBE (Covaris), and the fragments were screened with the BluePippin system (Sage Science). High-quality CCS reads were generated using the PacBio HiFi platform.
Genome assembly
High-accuracy CCS data were assembled using hifiasm v0.14 (Cheng et al., 2021a) to obtain the genome sequence. BWA v0.7.10 (Li and Durbin, 2009) was used to map the short reads obtained by Illumina sequencing to the draft genome. The completeness of the assembled genome was evaluated with CEGMA v2.5 (Parra et al., 2007) and BUSCO v5 (Simao et al., 2015).
Hi-C analysis and pseudo-chromosome construction
We constructed Hi-C fragment libraries with a 300–700 bp insert size (Rao et al., 2014) and sequenced them on the Illumina platform. In brief, adaptor sequences of raw reads were trimmed, and low-quality reads were removed. Only uniquely mapped paired-end reads with a mapping quality greater than 20 were retained for further analysis. Invalid read pairs, including dangling-end and self-cycle, re-ligation, and dumped products, were removed using HiC-Pro v2.10.0 (Servant et al., 2015). Before chromosome assembly, we first performed a preassembly for error correction of scaffolds, which required the splitting of scaffolds into segments of 50 kb on average. The Hi-C data were mapped to these segments using BWA. The uniquely mapped data were retained and used to perform assembly with LACHESIS software (parameters: CLUSTER_MIN_RE_SITES = 66; CLUSTER_MAX_LINK_DENSITY = 2; ORDER_MIN_N_RES_IN_TRUNK = 138; ORDER_MIN_N_RES_IN_SHREDS = 156) (Burton et al., 2013). Any two segments that showed inconsistent connection with information from the raw scaffold were checked manually. These corrected scaffolds were then assembled with LACHESIS.
RNA-seq
Total RNA was extracted from roots, stems, leaves, seeds, and seedlings. After multi-tissue mixing, mRNA was enriched by magnetic beads with oligo(dT) and fragmented randomly with fragmentation buffer. cDNA synthesis, purification, terminal repair, and other steps were performed, and the resulting libraries were sequenced on the NovaSeq 6000 platform.
The total mRNA was extracted from leaves, stems, flowers, and root samples, and cDNA libraries were constructed for RNA-seq. We also downloaded additional RNA-seq data from root tissues at different developmental stages (Liang et al., 2020). Quality control of raw reads was first performed using Trimmomatic (Bolger et al., 2014), and the clean reads were then aligned to the genome using HISAT2 (Kim et al., 2015). Gene expression levels were estimated as fragments per kilobase of transcript per million fragments mapped using StringTie 2. TBtools was used for data normalization with the z-score method.
TE and gene annotation
TEs and tandem repeats were annotated with the following workflows. TEs were identified by a combination of de novo and homology-based approaches. We first customized a de novo repeat library for the genome using RepeatModeler v2.0.1 (Flynn et al., 2020), which mainly depended on two de novo repeat-finding programs, RECON v1.08 (Bao and Eddy, 2002) and RepeatScout v1.0.6 (Price et al., 2005). Full-length LTR-RTs (fl-LTR-RTs) were then identified using LTRharvest v1.5.9 (Ellinghaus et al., 2008) and LTR_FINDER v1.1 (Xu and Wang, 2007). The high-quality intact fl-LTR-RTs and non-redundant LTR library were then obtained using LTR_retriever v2.9.0 (Ou and Jiang, 2018). A non-redundant species-specific TE library was constructed by combining the de novo TE sequences identified above with the publicly available TE sequences in the Repbase v19.06 (Jurka et al., 2005), REXdb v3.0 (Neumann et al., 2019), and Dfam v3.2 (Wheeler et al., 2013) databases. This non-redundant TE library was used to search against the entire genome sequence with RepeatMasker v4.10 to identify fragmented TEs in the AMM genome (Tarailo-Graovac and Chen, 2009).
We integrated de novo, homology-based, and transcript-based predictions to annotate protein-coding genes in the genome. The de novo gene models were predicted using two ab initio gene-prediction software tools, Augustus v2.4 (Stanke et al., 2008) and SNAP (2006-07-28) (Korf, 2004). For the homology-based approach, GeMoMa v1.7 (Keilwagen et al., 2016) was performed using reference gene models from A. thaliana, C. arietinum, G. max, G. uralensis, and M. truncatula. For transcript-based prediction, RNA-seq data were mapped to the reference genome using HISAT v2.0.4 and assembled with StringTie v1.2.3 (Pertea et al., 2015). GeneMarkS-T v5.1 (Tang et al., 2015) was used to predict genes from the assembled transcripts. PASA v2.0.2 (Haas et al., 2003) was used to predict genes based on the unigenes (and full-length transcripts from PacBio [ONT] sequencing) assembled by Trinity v2.11 (Grabherr et al., 2011). Gene models from these different approaches were combined using EVM v1.1.1 (Haas et al., 2008) and updated with PASA. The final gene models were annotated by searching the GenBank Non-Redundant (20200921), TrEMBL (202005), Pfam v33.1 (Finn et al., 2006), SwissProt (202005), eukaryotic orthologous groups (KOG, 20110125), Gene Ontology (GO, 20200615), and KEGG (20191220) (Kanehisa et al., 2016) databases.
The genBlastA algorithm v1.0.4 (She et al., 2009) was used to scan whole genomes after masking the predicted functional genes. Putative candidates were then analyzed by searching for non-mature mutations and frame-shift mutations using GeneWise v2.4.1 (Birney et al., 2004). tRNAscan-SE v1.3.1 (Lowe and Eddy, 1997) was used with eukaryote parameters to predict tRNAs. Identification of rRNA genes was performed using Barrnap v0.9.0. The miRNA genes were identified using the miRBase database (Griffiths-Jones et al., 2006). The snoRNA and snRNA genes were predicted using Infernal 1.1 (Nawrocki and Eddy, 2013) against the Rfam database v12.0 (Griffiths-Jones et al., 2005). Motif annotation was performed using InterProScan (5.34–73.0) (Jones et al., 2014).
Gene families and phylogenetic analysis
OrthoFinder v2.4 software (Emms and Kelly, 2019) was used to classify the protein sequences of 13 species into families (DIAMOND alignment, e ≤ 1e−3). These gene families were further annotated according to the PANTHER v15 database (Mi et al., 2019). The clusterProfiler package v3.14.0 (Yu et al., 2012) was used for GO and KEGG enrichment analysis of endemic gene families. A phylogenetic tree of 177 single-copy gene sequences from 13 species was constructed using IQ-Tree v1.6.11 software (Lam-Tung et al., 2015). First, MAFFT v7.205 was used to align the sequences of each single-copy gene family (parameters: --localpair --maxiterate 1000) followed by PAL2NAL v14 (Suyama et al., 2006), which was used to convert protein alignments into nucleotide sequence alignments. Next, Gblocks v0.91 (parameter: -b5 = H) (Talavera and Castresana, 2007) was used to remove regions with poor sequence alignments or with large differences, and all well-aligned gene families of each species were connected from end to end. Finally, ModelFinder (Kalyaanamoorthy et al., 2017) was used to construct the phylogenetic tree using the maximum likelihood method with 1000 bootstrap replicates.
The MCMCTree program in PAML v4.9i software (Yang, 1997) was used to calculate divergence times. Six calibration points were obtained from the TimeTree website (http://www.timetree.org/): 107–135 Mya for V. vinifera and AMM, 15.2–23.7 Mya for A. sinicus and AMM, 4.7–15.6 Mya for P. vulgaris and V. radiata, 36–48 Mya for G. uralensis and AMM, 46–109 Mya for G. max and C. arietinum, and 98–117 Mya for V. radiata and A. thaliana. Another four calibration points were also selected: 20–30 Mya for C. arietinum and L. japonicus, 10–20 Mya for C. arietinum and M. truncatula (Varshney et al., 2013), 16.8–21.6 Mya for A. sinicus and T. pratense, and 7.6–13.0 Mya for T. pratense and M. truncatula (Chang et al., 2022). Then, the parameter gradient and Hessian required by the MCMCTree module in PAML (Puttick, 2019) were estimated. Finally, the maximum likelihood method was used to estimate the divergence times using the correlated molecular clock and the JC69 model, and repeated calculations were performed twice to observe the consistency (the correlation between two iterations in this test was 1). The iteration times of the Markov chain were set to burnin 5 000 000, sampfreq 30, and nsample 10 000 000. CAFE v4.2 (Han et al., 2013) was used to predict the contraction and expansion of gene families relative to their ancestors based on the results of evolutionary trees with divergence times and gene family clustering. The birth and death models were used to estimate the numbers of ancestral gene family members in each branch. The criteria for defining whether significant expansion or contraction had occurred were family-wide P values and Viterbi P values, both of which were less than 0.05. Expanded and contracted gene families in the studied species were extracted, and PANTHER annotation was performed. Finally, clusterProfiler was used for GO and KEGG enrichment analysis of the expanded and contracted gene families in AMM.
Chromosome synteny and WGD analyses
DIAMOND v0.9.29.130 (Buchfink et al., 2015) was used to compare the genomic sequences of two species and identify homologous gene pairs based on sequence similarity (e < 1e−5, C > 0.5). Next, the homologous gene pairs were used to identify collinear blocks with MCScanX (-m 15) (Wang et al., 2012), and the results were visualized with JCVI v0.9.13. Based on the distribution of Ks values of paralogous genes, we calculated the WGD events using wgd v1.1.1 (Zwaenepoel and Van de Peer, 2019). Publicly available scripts (https://github.com/JinfengChen/Scripts) were used to calculate the proportion of conversion mutations to 4dTv bases in each homologous gene pair, and the HKY substitution model was used for correction. According to T = K/2r (r = 6.38 × 10−9), the timing of WGD events was estimated (Xu et al., 2020).
Genome mining for gene families and gene clusters involved in flavonoid and triterpenoid biosynthesis
The protein sequences of the reference gene families involved in flavonoid and triterpenoid biosynthesis in A. thaliana or P. ginseng were downloaded from UniProt. These sequences were used as a query for a BLAST search against the protein sequences of AMM with an e value cutoff of 1e−40 (Cui et al., 2022). Next, all the homologous sequences were submitted to the Pfam database for domain analysis (Mistry et al., 2021). PlantiSMASH software (Kautsar et al., 2017) was used to search for gene clusters potentially involved in plant specialized metabolism.
Identification of members of the OSC, cytochrome P450, and UGT superfamilies
Reference sequences of OSCs are shown in Supplemental Table 32, and reference sequences of CYP450s and UGTs were downloaded from The Arabidopsis Information Resource database (www.arabidopsis.org). After BlastP and hmmsearch searches (HMMER profiles PF13243, PF13249, PF00067, and PF00201 downloaded from the PFAM library), CDD (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) was used to identify the conserved domains of the predicted OSC, CYP450, and UGT protein sequences. Phylogenetic trees were constructed by the maximum likelihood method in MEGA X (Kumar et al., 2018) with 1000 bootstrap replicates.
The protein sequences of AmCYP450 genes were searched against gene sequences in the Arabidopsis Information Resource database and NCBI using BlastP. The predicted P450 genes were classified into families according to the international nomenclature standard for P450 genes (Nelson et al., 1996). Two P450 genes with an amino acid sequence identity of 40%–55% were classified into the same family, and those with an identity >55% were classified into the same subfamily. A linkage map was created using TBtools (Chen et al., 2020). Clusters of CYP450 genes were identified using the following two criteria: (1) the distance between two adjacent P450 genes was ≤200 kb; and (2) the number of non-P450 genes located between the two P450 genes was ≤8 (Huang et al., 2012).
Data validation by quantitative real-time PCR
qRT–PCR analysis of nine PAL genes was performed in leaf, flower, stem, and root tissues with the primers listed in Supplemental Table 33.
Functional identification of candidate OSC genes in yeast
Three candidate OSC genes in gene cluster III were cloned from cDNA of AMM and inserted into the XhoI and KpnI digestion enzyme sites of the pESC-URA vector to generate plasmids for heterologous expression in Saccharomyces cerevisiae. The generated vectors were transformed into competent cells of WAT11v. As controls, yeast with the empty vector (pESC-URA) was also constructed. The transformants were selected on solid synthetic defined (SD) medium without uracil (−URA) to obtain new strains. The positive colonies were inoculated into 10 ml liquid SD-URA medium with 20 g/l glucose, precultured at 30°C for 24 h at 220 rpm, and then transferred to 50 ml of SD-URA for growth. The yeast cells were collected by centrifugation, washed twice with sterile water, resuspended in 50 ml SD-URA medium containing 2% galactose and 1% raffinose for induction, and incubated at 30°C for 5 days at 220 rpm. The cells were harvested by centrifugation, resuspended in 10 ml of 20% KOH (dissolved in 50% ethanol), and lysed by heating at 95°C for 15 min. The resulting lysate was extracted twice with n-hexane, the combined organic phase was evaporated under reduced pressure, and the residue was re-dissolved with 50 μl anhydrous pyridine and 50 μl BSTFA–TMCS (99:1) and silanized at 85°C for 1 h before GC–MS analysis. GC–MS analysis was performed on an Agilent gas chromatograph (split, 20:1; injector temperature, 250°C) with an HP-5MS (30 × 0.25 × 0.25 mm) column. The GC conditions were as follows: 3 μl of the concentrated organic phase was injected under a He flow rate of 1 ml/min. The column heating procedure was an initial temperature of 85°C held for 1 min, followed by a gradient from 40 to 280°C at 20°C/min, a hold for 18 min, then an increase to 320°C at 20°C/min, and a hold for 1 min. The solution delay time was 3 min. The MS parameters were ion trap temperature, 250°C; electron energy, 70 eV; and mass range, 45–800 m/z.
Funding
This work was supported by grants from the City-University Cooperation Project of China (201904710111639).
Author contributions
X.D. and L.S. designed the study. Y.C., P.W., L.W., W.S., and Q.H. prepared the sequencing samples. Y.C., S.D., and R.M. conducted experiments. Y.C., T.F., H.S., and S.D. analyzed the data. Y.C. wrote the manuscript. M.Z., L.S., and X.D. revised the manuscript.
Acknowledgments
We thank Prof. Weidong Cao and Dr. Danna Chang of the Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences, for technical support on analysis of Astragalus sinicus. We thank Xiaoyuan Wu at the Institute of Botany, Chinese Academy of Sciences (IB-CAS) for valuable advice on the comparative genomics analysis. We also thank Prof. Chao Sun of the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, for providing the vector (pESC-URA) and yeast strain WAT11v. No conflict of interest is declared.
Published: October 28, 2022
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Lianjun Sun, Email: sunlj@cau.edu.cn.
Xuehui Dong, Email: xuehuidong@cau.edu.cn.
Accession numbers
The genome sequences and raw sequencing data have been deposited in the Global Pharmacopoeia Genome Database (GPGD) (Liao et al., 2021) at the following URL: http://www.gpgenome.com/species/109.
Supplemental information
References
- Adams K.L., Wendel J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Amyotte S.G., Tan X., Pennerman K., Jimenez-Gasco M.d.M., Klosterman S.J., Ma L.J., Dobinson K.F., Veronese P. Transposable elements in phytopathogenic Verticillium spp.: insights into genome evolution and inter- and intra-specific diversification. BMC Genom. 2012;13:314. doi: 10.1186/1471-2164-13-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auyeung K.K., Han Q.B., Ko J.K. Astragalus membranaceus: A review of its protection against inflammation and gastrointestinal cancers. Am. J. Chin. Med. 2016;44:1–22. doi: 10.1142/s0192415x16500014. [DOI] [PubMed] [Google Scholar]
- Bao Z., Eddy S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R. Genewise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D., Lim E.K., Poppenberger B., Vaistij F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006;57:567–597. doi: 10.1146/annurev.arplant.57.032905.105429. [DOI] [PubMed] [Google Scholar]
- Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O., Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi L., Malnoy M., Goremykin V., Nikiforova S., Martens S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012;69:1030–1042. doi: 10.1111/j.1365-313X.2011.04853.x. [DOI] [PubMed] [Google Scholar]
- Chang D., Gao S., Zhou G., Deng S., Jia J., Wang E., Cao W. The chromosome-level genome assembly of Astragalus sinicus and comparative genomic analyses provide new resources and insights for understanding legume-rhizobial interactions. Plant Commun. 2022;3:100263. doi: 10.1016/j.xplc.2021.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Cheng H., Concepcion G.T., Feng X., Zhang H., Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods. 2021;18:170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Wang X., Liu X., Zhu X., Li Z., Chu H., Wang Q., Lou Q., Cai B., Yang Y., et al. Chromosome-level genome of Himalayan yew provides insights into the origin and evolution of the paclitaxel biosynthetic pathway. Mol. Plant. 2021;14:1199–1209. doi: 10.1016/j.molp.2021.04.015. [DOI] [PubMed] [Google Scholar]
- Chen J., Wu X., Xu Y., et al. Global transcriptome analysis profiles metabolic pathways in traditional herb Astragalus membranaceus Bge. var. mongolicus (Bge.) Hsiao. BMC Genom. 2015;16:S15. doi: 10.1186/1471-2164-16-s7-s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Park S.R., Heo T.R. Inhibitory effect of Astragali Radix on matrix degradation in human articular cartilage. J. Microbiol. Biotechnol. 2005;15:1258–1266. [Google Scholar]
- Chu C., Qi L.W., Liu E.H., Li B., Gao W., Li P. Radix Astragali (Astragalus): latest advancements and trends in chemistry, analysis, pharmacology and pharmacokinetics. Curr. Org. Chem. 2010;14:1792–1807. doi: 10.2174/138527210792927663. [DOI] [Google Scholar]
- Cui J., Lu Z., Wang T., Chen G., Mostafa S., Ren H., Liu S., Fu C., Wang L., Zhu Y., et al. The genome of Medicago polymorpha provides insights into its edibility and nutritional value as a vegetable and forage legume. Hortic. Res. 2021;8:47. doi: 10.1038/s41438-021-00483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Meng F., Pan X., Qiu X., Zhang S., Li C., Lu S. Chromosome-level genome assembly of Aristolochia contorta provides insights into the biosynthesis of benzylisoquinoline alkaloids and aristolochic acids. Hortic. Res. 2022;9:uhac005. doi: 10.1093/hr/uhac005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deavours B.E., Dixon R.A. Metabolic engineering of isoflavonoid biosynthesis in alfalfa. Plant Physiol. 2005;138:2245–2259. doi: 10.1104/pp.105.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M., Brown J.K.M., Bennetzen J.L. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Kurtz S., Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinf. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Mistry J., Schuster-Böckler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J.M., Hubley R., Goubert C., Rosen J., Clark A.G., Feschotte C., Smit A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Wang Z., Huang L., Zheng S., Wang D., Chen S., Zhang H., Yang S. Review of the botanical characteristics, phytochemistry, and pharmacology of Astragalus membranaceus (Huangqi) Phytother Res. 2014;28:1275–1283. doi: 10.1002/ptr.5188. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. MiRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L.P., Zhou L.Y., Kang C.Z., Wang H.Y., Zhang W.J., Wang S., Wang R.S., Wang X., Han B.X., Zhou T., et al. Strategies for medicinal plants adapting environmental stress and “simulative habitat cultivation” of Dao-di herbs. China J. Chin. Mater. Med. 2020;45:1969–1974. doi: 10.19540/j.cnki.cjcmm.20200302.101. [DOI] [PubMed] [Google Scholar]
- Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., White O., Buell C.R., Wortman J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K., Hannick L.I., Maiti R., Ronning C.M., Rusch D.B., Town C.D., et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.V., Thomas G.W.C., Lugo-Martinez J., Hahn M.W. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- Haralampidis K., Trojanowska M., Osbourn A.E. Biosynthesis of triterpenoid saponins in plants. Adv. Biochem. Eng. Biotechnol. 2002;75:31–49. doi: 10.1007/3-540-44604-4_2. [DOI] [PubMed] [Google Scholar]
- Hikino H., Funayama S., Endo K. Hypotensive principle of Astragalus and Hedysarum roots. Planta Med. 1976;30:297–302. doi: 10.1055/s-0028-1097733. [DOI] [PubMed] [Google Scholar]
- Hirotani M., Kuroda R., Suzuki H., Yoshikawa T. Cloning and expression of UDP-glucose: flavonoid 7-O-glucosyltransferase from hairy root cultures of Scutellaria baicalensis. Planta. 2000;210:1006–1013. doi: 10.1007/pl00008158. [DOI] [PubMed] [Google Scholar]
- Huang J., Gu M., Lai Z., Fan B., Shi K., Zhou Y.H., Yu J.Q., Chen Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153:1526–1538. doi: 10.1104/pp.110.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Zhang P., Zhang Z., Youn J.Y., Wang C., Zhang H., Cai H. Traditional Chinese Medicine (TCM) in the treatment of COVID-19 and other viral infections: efficacies and mechanisms. Pharmacol. Ther. 2021;225:107843. doi: 10.1016/j.pharmthera.2021.107843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Gao Y., Liu J., Peng X., Niu X., Fei Z., Cao S., Liu Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012;287:495–513. doi: 10.1007/s00438-012-0696-6. [DOI] [PubMed] [Google Scholar]
- Jiang Z., Tu L., Yang W., Zhang Y., Hu T., Ma B., Lu Y., Cui X., Gao J., Wu X., et al. The chromosome-level reference genome assembly for Panax notoginseng and insights into ginsenoside biosynthesis. Plant Commun. 2021;2:100113. doi: 10.1016/j.xplc.2020.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wickett N.J., Ayyampalayam S., Chanderbali A.S., Landherr L., Ralph P.E., Tomsho L.P., Hu Y., Liang H., Soltis P.S., et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautsar S.A., Suarez Duran H.G., Blin K., Osbourn A., Medema M.H. plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 2017;45:W55–W63. doi: 10.1093/nar/gkx305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J., Wenk M., Erickson J.L., Schattat M.H., Grau J., Hartung F. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016;44:e89. doi: 10.1093/nar/gkw092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Ha H., Kim J.S., Kim Y.T., Kwon S.C., Park S.W. Induction of growth hormone by the roots of Astragalus membranaceus in pituitary cell culture. Arch Pharm. Res. (Seoul) 2003;26:34–39. doi: 10.1007/bf03179928. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Parupalli S., Azam S., Lee S.H., Varshney R.K. Comparative sequence analysis of nitrogen fixation-related genes in six legumes. Front. Plant Sci. 2013;4:300. doi: 10.3389/fpls.2013.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinf. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreplak J., Madoui M.A., Cápal P., Novák P., Labadie K., Aubert G., Bayer P.E., Gali K.K., Syme R.A., Main D., et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019;51:1411–1422. doi: 10.1038/s41588-019-0480-1. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam-Tung N., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latunde-Dada A.O., Cabello-Hurtado F., Czittrich N., Didierjean L., Schopfer C., Hertkorn N., Werck-Reichhart D., Ebel J. Flavonoid 6-hydroxylase from soybean (Glycine max L.), a novel plant P-450 monooxygenase. J. Biol. Chem. 2001;276:1688–1695. doi: 10.1074/jbc.M006277200. [DOI] [PubMed] [Google Scholar]
- Lee Y.M., Choi S.I., Lee J.W., Jung S.M., Park S.M., Heo T.R. Isolation of hyaluronidase inhibitory component from the roots of Astraglus membranaceus Bunge (Astragali radix) Food Sci. Biotechnol. 2005;14:263–267. [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Harata-Lee Y., Denton M.D., Feng Q., Rathjen J.R., Qu Z., Adelson D.L. Long read reference genome-free reconstruction of a full-length transcriptome from Astragalus membranaceus reveals transcript variants involved in bioactive compound biosynthesis. Cell Discov. 2017;3:17031. doi: 10.1038/celldisc.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Hou X., Xu R., Liu C., Tu M. Research review on the pharmacological effects of astragaloside IV. Fundam. Clin. Pharmacol. 2017;31:17–36. doi: 10.1111/fcp.12232. [DOI] [PubMed] [Google Scholar]
- Li Q.G., Zhang L., Li C., Dunwell J.M., Zhang Y.M. Comparative genomics suggests that an ancestral polyploidy event leads to enhanced root nodule symbiosis in the Papilionoideae. Mol. Biol. Evol. 2013;30:2602–2611. doi: 10.1093/molbev/mst152. [DOI] [PubMed] [Google Scholar]
- Liang J., Li W., Jia X., Zhang Y., Zhao J. Transcriptome sequencing and characterization of Astragalus membranaceus var. mongholicus root reveals key genes involved in flavonoids biosynthesis. Genes Genomics. 2020;42:901–914. doi: 10.1007/s13258-020-00953-5. [DOI] [PubMed] [Google Scholar]
- Liao B., Hu H., Xiao S., Zhou G., Sun W., Chu Y., Meng X., Wei J., Zhang H., Xu J., et al. Global Pharmacopoeia Genome Database is an integrated and mineable genomic database for traditional medicines derived from eight international pharmacopoeias. Sci. China Life Sci. 2021;65:809–817. doi: 10.1007/s11427-021-1968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.J., Huhman D., Sumner L.W., Dixon R.A. Regiospecific hydroxylation of isoflavones by cytochrome P450 81E enzymes from Medicago truncatula. Plant J. 2003;36:471–484. doi: 10.1046/j.1365-313X.2003.01893.x. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang X., Han K., Li R., Xu G., Han Y., Cui F., Fan S., Seim I., Fan G., et al. Insights into amphicarpy from the compact genome of the legume Amphicarpaea edgeworthii. Plant Biotechnol. J. 2021;19:952–965. doi: 10.1111/pbi.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang B., Shu S., Li Z., Song C., Liu D., Niu Y., Liu J., Zhang J., Liu H., et al. Analysis of the Coptis chinensis genome reveals the diversification of protoberberine-type alkaloids. Nat. Commun. 2021;12:3276. doi: 10.1038/s41467-021-23611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Tu P., Chen Y., Zhang T., Wei Y., Ito Y. Preparative isolation and purification of two isoflavones from Astragalus membranaceus Bge. var. mongholicus (Bge.) Hsiao by high-speed counter-current chromatography. J. Chromatogr. A. 2003;992:193–197. doi: 10.1016/s0021-9673(03)00315-7. [DOI] [PubMed] [Google Scholar]
- Ma X.Q., Shi Q., Duan J.A., Dong T.T.X., Tsim K.W.K. Chemical analysis of Radix Astragali (Huangqi) in China: a comparison with its adulterants and seasonal variations. J. Agric. Food Chem. 2002;50:4861–4866. doi: 10.1021/jf0202279. [DOI] [PubMed] [Google Scholar]
- Marçais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Barton N.H. Chromosomal speciation and molecular divergence - accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.R., Koymans L., Kamataki T., Stegeman J.J., Feyereisen R., Waxman D.J., Waterman M.R., Gotoh O., Coon M.J., Estabrook R.W., et al. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- Neumann P., Novák P., Hoštáková N., Macas J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mobile DNA. 2019;10:1. doi: 10.1186/s13100-018-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann H.W., Huang A., Osbourn A. Plant metabolic clusters - from genetics to genomics. New Phytol. 2016;211:771–789. doi: 10.1111/nph.13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S.P., Whitton J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Ou S., Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176:1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette S., Møller B.L., Bak S. On the origin of family 1 plant glycosyltransferases. Phytochemistry. 2003;62:399–413. doi: 10.1016/s0031-9422(02)00558-7. [DOI] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genornes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pootakham W., Nawae W., Naktang C., Sonthirod C., Yoocha T., Kongkachana W., Sangsrakru D., Jomchai N., U-thoomporn S., Somta P., et al. A chromosome-scale assembly of the black gram (Vigna mungo) genome. Mol. Ecol. Resour. 2021;21:238–250. doi: 10.1111/1755-0998.13243. [DOI] [PubMed] [Google Scholar]
- Price A.L., Jones N.C., Pevzner P.A. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:I351–I358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Pu X., Li Z., Tian Y., Gao R., Hao L., Hu Y., He C., Sun W., Xu M., Peters R.J., et al. The honeysuckle genome provides insight into the molecular mechanism of carotenoid metabolism underlying dynamic flower coloration. New Phytol. 2020;227:930–943. doi: 10.1111/nph.16552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttick M.N. MCMCtreeR: functions to prepare MCMCtree analyses and visualize posterior ages on trees. Bioinformatics. 2019;35:5321–5322. doi: 10.1093/bioinformatics/btz554. [DOI] [PubMed] [Google Scholar]
- Qin X.M., Li Z.Y., Sun H.F., Zhang L.Z., Zhou R., Feng Q.J., Li A.P. Status and analysis of astragali radix resource in China. China J. Chin. Mater. Med. 2013;38:3234–3238. [PubMed] [Google Scholar]
- Quilbé J., Lamy L., Brottier L., Leleux P., Fardoux J., Rivallan R., Benichou T., Guyonnet R., Becana M., Villar I., et al. Genetics of nodulation in Aeschynomene evenia uncovers mechanisms of the rhizobium-legume symbiosis. Nat. Commun. 2021;12:829. doi: 10.1038/s41467-021-21094-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez T.R., Jaron K.S., Schatz M.C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 2020;11:1432. doi: 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renny-Byfield S., Wendel J.F. Doubling down on genomes: polyploidy and crop plants. Am. J. Bot. 2014;101:1711–1725. doi: 10.3732/ajb.1400119. [DOI] [PubMed] [Google Scholar]
- Rohde A., Morreel K., Ralph J., Goeminne G., Hostyn V., De Rycke R., Kushnir S., Van Doorsselaere J., Joseleau J.P., Vuylsteke M., et al. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino Acid, and carbohydrate metabolism. Plant Cell. 2004;16:2749–2771. doi: 10.1105/tpc.104.023705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandeep, Misra R.C., Chanotiya C.S., Mukhopadhyay P., Ghosh S. Oxidosqualene cyclase and CYP716 enzymes contribute to triterpene structural diversity in the medicinal tree banaba. New Phytol. 2019;222:408–424. doi: 10.1111/nph.15606. [DOI] [PubMed] [Google Scholar]
- Sawada Y., Kinoshita K., Akashi T., Aoki T., Ayabe S.I. Key amino acid residues required for aryl migration catalysed by the cytochrome P450 2-hydroxyisoflavanone synthase. Plant J. 2002;31:555–564. doi: 10.1046/j.1365-313X.2002.01378.x. [DOI] [PubMed] [Google Scholar]
- Sawai S., Saito K. Triterpenoid biosynthesis and engineering in plants. Front. Plant Sci. 2011;2:25. doi: 10.3389/fpls.2011.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Cannon S.B., Schlueter J., Ma J., Mitros T., Nelson W., Hyten D.L., Song Q., Thelen J.J., Cheng J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Seki H., Tamura K., Muranaka T. P450s and UGTs: key players in the structural diversity of triterpenoid saponins. Plant Cell Physiol. 2015;56:1463–1471. doi: 10.1093/pcp/pcv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrajabian M.H., Sun W., Cheng Q. A review of Astragalus species as foodstuffs, dietary supplements, a traditional Chinese medicine and a part of modern pharmaceutical science. Appl. Ecol. Environ. Res. 2019;17:13371–13382. doi: 10.15666/aeer/1706_1337113382. [DOI] [Google Scholar]
- She R., Chu J.S.C., Wang K., Pei J., Chen N. GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 2009;19:143–149. doi: 10.1101/gr.082081.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Tian J., Cheng H., et al. The chromosome-level wintersweet (Chimonanthus praecox) genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biol. 2020;21:200. doi: 10.1186/s13059-020-02088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Soltis P.S., Soltis D.E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 2016;30:159–165. doi: 10.1016/j.pbi.2016.03.015. [DOI] [PubMed] [Google Scholar]
- Soltis P.S., Marchant D.B., Van de Peer Y., Soltis D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015;35:119–125. doi: 10.1016/j.gde.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Song J.Z., Yiu H.H.W., Qiao C.F., Han Q.B., Xu H.X. Chemical comparison and classification of Radix Astragali by determination of isoflavonoids and astragalosides. J. Pharm. Biomed. Anal. 2008;47:399–406. doi: 10.1016/j.jpba.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Stanke M., Diekhans M., Baertsch R., Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Stacey G., Yu O. Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci. 2007;12:282–285. doi: 10.1016/j.tplants.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tang L., Liu Y., Wang Y., Long C. Phytochemical analysis of an antiviral fraction of Radix astragali using HPLC-DAD-ESI-MS/MS. J. Nat. Med. 2010;64:182–186. doi: 10.1007/s11418-009-0381-1. [DOI] [PubMed] [Google Scholar]
- Tang S., Lomsadze A., Borodovsky M. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res. 2015;43:e78. doi: 10.1093/nar/gkv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M., Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2009;25:4. doi: 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- Thimmappa R., Geisler K., Louveau T., O'Maille P., Osbourn A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 2014;65:225–257. doi: 10.1146/annurev-arplant-050312-120229. [DOI] [PubMed] [Google Scholar]
- Varshney R.K., Song C., Saxena R.K., Azam S., Yu S., Sharpe A.G., Cannon S., Baek J., Rosen B.D., Tar'an B., et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 2013;31:240–246. doi: 10.1038/nbt.2491. [DOI] [PubMed] [Google Scholar]
- Vogt T., Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000;5:380–386. doi: 10.1016/s1360-1385(00)01720-9. [DOI] [PubMed] [Google Scholar]
- Vranová E., Coman D., Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013;64:665–700. doi: 10.1146/annurev-arplant-050312-120116. [DOI] [PubMed] [Google Scholar]
- Wang C., Lu J., Zhang S., Wang P., Hou J., Qian J. Effects of Pb stress on nutrient uptake and secondary metabolism in submerged macrophyte Vallisneria natans. Ecotoxicol. Environ. Saf. 2011;74:1297–1303. doi: 10.1016/j.ecoenv.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Wang S., Li J., Huang H., Gao W., Zhuang C., Li B., Zhou P., Kong D. Anti-hepatitis b virus activities of astragaloside IV isolated from Radix Astragali. Biol. Pharm. Bull. 2009;32:132–135. doi: 10.1248/bpb.32.132. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.h., Jin H., Marler B., Guo H., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler T.J., Clements J., Eddy S.R., Hubley R., Jones T.A., Jurka J., Smit A.F.A., Finn R.D. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 2013;41:D70–D82. doi: 10.1093/nar/gks1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li X., Wang W., Shan Y., Wang C., Zhu M., La Q., Zhong Y., Xu Y., Nan P., et al. Integrated metabolomics and transcriptomics study of traditional herb Astragalus membranaceus Bge. var. mongolicus (Bge.) Hsiao reveals global metabolic profile and novel phytochemical ingredients. BMC Genom. 2020;21:697. doi: 10.1186/s12864-020-07005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D., Xu Y., Wang J., Liu W., Zhou Q., Luo S., Huang W., He X., Li Q., Peng Q., et al. The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype. Nat. Commun. 2019;10:5158. doi: 10.1038/s41467-019-13185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Gou J., Liao Q., Li Y., Zhou Q., Bi G., Li C., Du R., Wang X., Sun T., et al. The Taxus genome provides insights into paclitaxel biosynthesis. Native Plants. 2021;7:1026–1036. doi: 10.1038/s41477-021-00963-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Zhang Q., Yuan W., Xu F., Muhammad Aslam M., Miao R., Li Y., Wang Q., Li X., Zhang X., et al. The genome evolution and low-phosphorus adaptation in white lupin. Nat. Commun. 2020;11:1069. doi: 10.1038/s41467-020-14891-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Duan L., Liu D., Guo J., Ge S., Dicks J., ÓMáille P., Osbourn A., Qi X. Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 2012;193:1022–1038. doi: 10.1111/j.1469-8137.2011.03997.x. [DOI] [PubMed] [Google Scholar]
- Yang Y., Bocs S., Fan H., Armero A., Baudouin L., Xu P., Xu J., This D., Hamelin C., Iqbal A., et al. Coconut genome assembly enables evolutionary analysis of palms and highlights signaling pathways involved in salt tolerance. Commun. Biol. 2021;4:105. doi: 10.1038/s42003-020-01593-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu O., Jung W., Shi J., Croes R.A., Fader G.M., McGonigle B., Odell J.T. Production of the isoflavones genistein and daidzein in non-legume dicot and monocot tissues. Plant Physiol. 2000;124:781–794. doi: 10.1104/pp.124.2.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandalinas S.I., Mittler R., Balfagón D., Arbona V., Gómez-Cadenas A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plantarum. 2018;162:2–12. doi: 10.1111/ppl.12540. [DOI] [PubMed] [Google Scholar]
- Zhang B., Lewis K.M., Abril A., Davydov D.R., Vermerris W., Sattler S.E., Kang C. Structure and function of the cytochrome P450 monooxygenase cinnamate 4-hydroxylase from Sorghum bicolor. Plant Physiol. 2020;183:957–973. doi: 10.1104/pp.20.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.H., Yang X., Wei J.R., Chen N.M.H., Xu J.P., Bi Y.Q., Yang M., Gong X., Li Z.Y., Ren K., et al. Ethnopharmacology, phytochemistry, pharmacology, toxicology and clinical applications of Radix Astragali. Chin. J. Integr. Med. 2021;27:229–240. doi: 10.1007/s11655-019-3032-8. [DOI] [PubMed] [Google Scholar]
- Zhang D., Qi J., Yue J., Huang J., Sun T., Li S., Wen J.F., Hettenhausen C., Wu J., Wang L., et al. Root parasitic plant Orobanche aegyptiaca and shoot parasitic plant Cuscuta australis obtained Brassicaceae-specific strictosidine synthase-like genes by horizontal gene transfer. BMC Plant Biol. 2014;14:19. doi: 10.1186/1471-2229-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.J., Liu H.K., Hsiao P.C., Kuo L.M.Y., Lee I.J., Wu T.S., Chiou W.F., Kuo Y.H. New isoflavonoid glycosides and related constituents from Astragali Radix (Astragalus membranaceus) and their inhibitory activity on nitric oxide production. J. Agric. Food Chem. 2011;59:1131–1137. doi: 10.1021/jf103610j. [DOI] [PubMed] [Google Scholar]
- Zhang Q.J., Li W., Li K., Nan H., Shi C., Zhang Y., Dai Z.Y., Lin Y.L., Yang X.L., Tong Y., et al. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons in driving genome size evolution. Mol. Plant. 2020;13:935–938. doi: 10.1016/j.molp.2020.04.009. [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu C.J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol. Plant. 2015;8:17–27. doi: 10.1016/j.molp.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Zhao M., Ma J. Co-evolution of plant LTR-retrotransposons and their host genomes. Protein Cell. 2013;4:493–501. doi: 10.1007/s13238-013-3037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Zhang B., Lisch D., Ma J. Patterns and consequences of subgenome differentiation provide insights into the nature of paleopolyploidy in plants. Plant Cell. 2017;29:2974–2994. doi: 10.1105/tpc.17.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Yang J., Cui M.Y., Liu J., Fang Y., Yan M., Qiu W., Shang H., Xu Z., Yidiresi R., et al. The reference genome sequence of Scutellaria baicalensis provides insights into the evolution of wogonin biosynthesis. Mol. Plant. 2019;12:935–950. doi: 10.1016/j.molp.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Ren W., Zhang L., Zhang Y., Liu D., Liu Y. A review of the pharmacological action of Astragalus polysaccharide. Front. Pharmacol. 2020;11:349. doi: 10.3389/fphar.2020.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel A., Van de Peer Y. Wgd-simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics. 2019;35:2153–2155. doi: 10.1093/bioinformatics/bty915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.