Abstract
Resting-state functional connectivity (RSFC) is a powerful tool for characterizing brain changes, but it has yet to reliably predict higher-order cognition. This may be attributed to small effect sizes of such brain-behavior relationships, which can lead to underpowered, variable results when utilizing typical sample sizes (N∼25). Inspired by techniques in genomics, we implement the polyneuro risk score (PNRS) framework - the application of multivariate techniques to RSFC data and validation in an independent sample. Utilizing the Adolescent Brain Cognitive Development® cohort split into two datasets, we explore the framework’s ability to reliably capture brain-behavior relationships across 3 cognitive scores – general ability, executive function, learning & memory. The weight and significance of each connection is assessed in the first dataset, and a PNRS is calculated for each participant in the second. Results support the PNRS framework as a suitable methodology to inspect the distribution of connections contributing towards behavior, with explained variance ranging from 1.0 % to 21.4 %. For the outcomes assessed, the framework reveals globally distributed, rather than localized, patterns of predictive connections. Larger samples are likely necessary to systematically identify the specific connections contributing towards complex outcomes. The PNRS framework could be applied translationally to identify neurologically distinct subtypes of neurodevelopmental disorders.
Keywords: Neuroimaging, MRI, Reproducibility, Big data, BWAS, PNRS
Highlights
-
•
PNRS framework offers approach for BWAS studies.
-
•
Aggregation of small, globally distributed effects is most predictive of cognition.
-
•
Large, high-powered datasets are necessary for reproducible BWAS analyses.
1. Introduction
An influential discovery in functional neuroimaging was finding that low-frequency blood-oxygen level dependent (BOLD) signal fluctuations in functionally related, yet topographically distinct, grey matter regions are strongly correlated at rest (Biswal et al., 1995). While this work lay dormant for nearly a decade, work by Grecius et al. (2003) and Fox et al. (2005) led to a massive increase in the utilization of this technique to further characterize network organization based on functional connections, i.e., the co-activation patterns of brain areas (Cohen et al., 2008, Dosenbach et al., 2007, Fair et al., 2009, Power et al., 2011, Stevens et al., 2014). Since, resting state functional connectivity (RSFC) has proven to be a useful tool for mapping functional-anatomic networks as well as characterizing brain changes and differences in clinical populations (Cohen et al., 2008, Dosenbach et al., 2007, Fair et al., 2009, Lynch et al., 2021, Power et al., 2011, Stevens et al., 2014, Yeo et al., 2011).
Despite this success, it’s been difficult to postulate or reliably predict higher-order brain-behavior relationships with RSFC (Boekel et al., 2015, Dinga et al., 2019, Marek et al., 2022, Masouleh et al., 2019, Poldrack et al., 2017). For example, the functional architecture representing executive function has been debated for decades (Alvarez and Emory, 2006, McKenna et al., 2017). Several researchers have looked to RSFC as a potential method for defining the construct based on its mechanistic underpinnings. The implicated networks have varied widely between studies (Alvarez and Emory, 2006, Dosenbach et al., 2007, McKenna et al., 2017, Niendam et al., 2012). While there are likely several factors contributing to these inconsistencies, the true effect size of the brain-behavior relationship is likely to play a substantive role (Marek and Tervo-Clemmens et al., 2022).
A recent report by Marek and Tervo-Clemmens et al. (2022) highlights how underpowered studies produce inconsistencies regarding the localization of complex behaviors in brain-wide association studies (BWAS) in today’s academic environment (Fair et al., 2021). In short, effect sizes in population-based studies examining complex behaviors tend to be small, and as such, can lead to underpowered and highly variable results across studies. When typical BWAS sample sizes (N∼25) are used to identify network interactions, the results may produce highly inflated effect sizes and spurious correlations due to sampling variability and publication bias (Dick et al., 2021, Marek et al., 2022).
Historically, similar findings have been well documented in genome-wide association studies (GWAS) (Holland et al., 2016, Korte and Farlow, 2013, Wang et al., 2005, Zhang et al., 2019). To overcome such limitations in genetics, multivariate analytical methods have become a mainstay and have resulted in higher predictive power than univariate models (Lambert et al., 2019). For example, the polygenic risk score (PRS) model combines the cumulative small effects (or beta-weights) across the genome to better predict the risk for disease than a single locus (International Schizophrenia Consortium et al., 2009). This highlights a potential path for similar analyses in BWAS of complex behaviors with magnetic resonance imaging (MRI). Indeed, the integration of these GWAS-inspired multivariate approaches has produced novel techniques for studying brain-behavior relationships with improved predictive power (Marek et al., 2022, Palmer et al., 2021, Zhao et al., 2021).
Using task-based neuroimaging data, Zhao and colleagues (Zhao et al., 2021) introduced a multivariate prediction model that aggregates the small effects across the cortex and produces a summary score of behavioral prediction, similar to a PRS. Here, we apply this approach and other similarly inspired multivariate techniques, partial least squares regression (PLSR) (Miranda-Dominguez et al., 2022, Ragothaman et al., 2022, Ragothaman et al., 2022, Rosipal and Krämer, 2006, Rudolph et al., 2018), to RSFC data from the Adolescent Brain Cognitive Development (ABCD) Study® to test their predictive power on higher-order cognitive functions, while also examining the reproducibility of the prediction using a split-half sample (Marek et al., 2022, Feczko et al., 2021). Specifically, for a given outcome of interest, we calculate a “polyneuro risk score” (PNRS) for each participant based on the most salient RSFC connections, identified in an independent sample. As outcomes of interest, we utilize three higher-order cognitive domains, previously identified using principal component analysis, that explain large amounts of variance in the administered neurocognitive battery (Thompson et al., 2019). We explore the effectiveness of the PNRS framework to reliably capture brain-behavior relationships across our 3 outcomes of interest – general ability, executive function, and learning and memory. We evaluate whether the connectivity patterns for each outcome of interest trend locally or globally across the brain. Lastly, we examine the stability of the observed relationships through replication in a demographically matched, independent sample.
2. Materials and methods
2.1. Description of ABCD dataset
The Adolescent Brain Cognitive Development (ABCD) study is the largest US-based study assessing brain development from adolescence through young adulthood. 11,877 youth, aged 9–10, were primarily recruited through elementary schools across 21 nationally distributed study sites (Casey et al., 2018). Recruitment efforts attempted to ensure representation of the wide socio-demographic diversity of the larger US population. Inclusion and exclusion criteria were intentionally broad with the only exclusionary factors being a lack of English language proficiency, contraindications to MRI scanning, and the presence of severe sensory, intellectual, medical, or neurological issues (Feczko et al., 2021, Garavan et al., 2018). In the ongoing longitudinal study, participants attend annual visits, undergoing a comprehensive protocol including physical, cognitive, social, emotional, environmental, behavioral, and academic assessments. Biannually, visits also include neuroimaging and biospecimen collection (Casey et al., 2018). The baseline data collection for the sample has been completed, and neuroimaging, cognitive, biospecimen, behavioral, youth self-report metrics, parent self-report metrics, and environmental measures are now available for analysis and utilized in the present study (Feczko et al., 2021, Karcher and Barch, 2021). This analysis leveraged the ABCD Reproducible Matched Samples (ARMS) (Feczko et al., 2021), which are defined in detail in Section 2.7.1.
2.2. Calculation of PC scores
For our outcomes of interest, we leveraged principal component scores that characterize 3 general domains of cognition in the ABCD sample. Thompson et al. (Thompson et al., 2019) demonstrated that a Bayesian Probabilistic Principal Components Analysis (BPPCA) model (with random effects for site and family to account for nested covariance structure) could extract three components from the ABCD neurocognitive battery. This battery consists of eleven measures and has been extensively detailed within the scientific literature (Luciana et al., 2018). Nine of the measures were of interest to the Thompson group when calculating the principal component scores. Briefly, 7 cognition measures from the NIH Toolbox were included: Picture Vocabulary measures language skills and verbal intellect, Oral Reading Recognition measures reading and pronunciation abilities, Pattern Comparison Processing Speed measures rapid visual processing, List Sorting Working Memory measures working memory via categorical and perceptual characteristics, Picture Sequence Memory measures memory via sequencing tasks, Flanker measures response inhibition and conflict monitoring, and Dimensional Change Card Sort measures cognitive flexibility (Bleck et al., 2013, Gershon et al., 2013, Hodes et al., 2013). In addition to the NIH Toolbox measures, 2 other tasks from the battery were included in this analysis. The Rey Auditory Verbal Learning Test (RAVLT) measures auditory learning, memory, and recognition (Daniel et al., n.d.). The Little Man Task measures visual-spatial processing, specifically mental rotation (Acker and Acker, 1982). The BPPCA model revealed three principal component scores: general ability (PC1), which is most strongly contributed to by the oral reading, picture vocabulary, and list sort working memory tasks; executive function (PC2), which is most strongly contributed to by flanker, dimensional change card sort, and pattern comparison processing speed tasks; learning & memory (PC3), which is most strongly contributed to by the picture sequence memory and list sort working memory tasks (Thompson et al., 2019). To maintain the integrity of the independent samples, the traits used in this analysis were extracted from ARMS-1 and ARMS-2 independently (Feczko et al., 2021), the two partitions used in this study.
2.3. MRI data acquisition
Scanning sessions were consistent across sites and comprised of a localizer, acquisition of 3D T1-weighted images (TR = 2500 ms, TE = 2.88 ms, 1.0 mm iso-voxels, 176 slices, FOV = 256 ×256 mm), 2 runs of resting-state fMRI (TR = 800 ms, TE = 30 ms, FA =52°, 2.4 mm iso-voxels, 60 slices, FOV = 216 ×216 mm), diffusion-weighted images (TR = 4100 ms, TE = 88 ms, 1.7 mm iso-voxels, 81 slices, FOV = 240 × 240 mm), 3D T2-weighted images (TR = 3200 ms, TE = 565 ms, 1.0 mm iso-voxels, 176 slices, FOV = 256 ×256 mm), 1–2 more runs of resting-state fMRI, and 3 task-based fMRI runs (Casey et al., 2018, Chaarani et al., 2021). This fixed order of scans was followed for all participants, with the exception of the 3 fMRI tasks, which were randomized across subjects. Head motion was monitored using FIRMM (Framewise Integrated Real-time Motion Monitoring) (Dosenbach et al., 2017), a system implemented to detect motion during resting-state fMRI at the Siemens sites. This allows the scan operators to provide real-time feedback to the participants and increase the amount of usable data collected. Overall, participants underwent a scanning protocol of approximately 100–120 min.
2.4. Data processing
All data for this project are from the ABCD BIDS Community Collection (Feczko et al., 2021). Imaging data is stored and organized according to the Brain Imaging Data Structure (BIDS) format (Gorgolewski et al., 2016). Native anatomical data (i.e., T1 or both T1 and T2) serve as the primary input to PreFreesurfer, which normalizes and rigidly aligns the data to the MNI template, preserving the native structure. The MNI-warped anatomical data are input to FreeSurfer, which produces and refines native cortical surface meshes (Fischl, 2012). The processed anatomical data are converted to the CIFTI format in PostFreeSurfer. fMRI data are brought into this CIFTI space by normalizing the volumetric data in fMRIVolume and projecting the normalized data onto the CIFTI format in fMRISurface. DCAN BOLD preprocessing (DBP), or “Preproc”, uses the DCAN (Developmental Cognition and Neuroimaging) Labs connectivity preprocessing program (Developmental Cognition and Neuroimaging Labs, n.d.) on the fMRI CIFTI data, resulting in both dense (dtseries) and parcellated (ptseries) CIFTI datasets (Feczko et al., 2021).
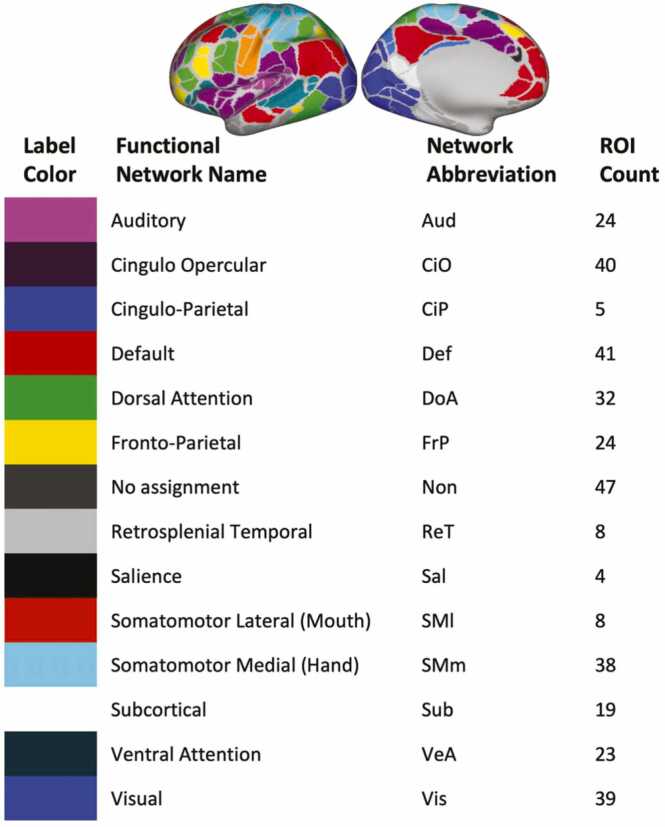
Timecourses were obtained by calculating the average of greyordinates within each ROI as defined by the Gordon parcellation (Gordon et al., 2016). This parcellation schema includes 333 cortical ROIs. To include subcortical structures, we added 19 subcortical ROIs from freesurfer (Fischl et al., 2002, Fischl et al., 2004) for a total of 352 ROIs assigned to 14 functional networks (Fig. 1).
Fig. 1.
An overview of the Gordon Parcellation. Timecourses were calculated using ROIs defined by the Gordon parcellation schema, which includes 333 cortical ROIs. They are color coded on the cerebral hemispheres with the corresponding network name, network abbreviation, and ROI count listed. Networks are ranked alphabetically.
2.5. Motion censoring
The quality of neuroimaging data is heavily influenced by the presence of motion artifacts, and is a major confound in studies of brain development and individual differences. To ameliorate this, a stringent framewise displacement (FD) threshold was used according to methods outlined by Power and colleagues (Power et al., 2014). Subjects were included in this analysis if they had > 8 min of high-quality data (FD < 0.2 mm). We had a total of 6575 survive our motion censoring (ARMS-1 = 3339, ARMS-2 = 3236). The analysis was conducted using a variety of conditions to determine these optimal parameters (Fig. S3).
2.6. Calculation of connectivity matrices using BICEPS
BICEPS is a graphical user interface (GUI) developed in Matlab aimed to calculate connectivity matrices using BIDS derivative parcellated data (i.e., Gordon or HCP parcellation schemas) processed following the surface-based registration pipelines implemented in the DCAN labs where the data are saved as CIFTIs in BIDS format folders (Miranda-Dominguez et al., 2023). BICEPS applies motion censoring and outlier detection and calculates connectivity matrices keeping constant the number of frames across participants (8 min of data in this analysis, selected randomly from surviving frames).
2.7. Statistical analysis
2.7.1. Description of ARMS
In order to conduct cross-validation, we leveraged the ABCD Reproducible Matched Samples (ARMS) (Feczko et al., 2021). ARMS split the ABCD sample into discovery and replication sets (N = 5786) based on nine sociodemographic factors thought to be important for developmental outcomes (site, age, sex, ethnicity, grade, highest level of parental education, handedness, combined family income, and exposure to anesthesia), and accounted for family structure (all X2 p > 0.9). Anesthesia exposure was selected to account for the possible effects on behavioral and neurodevelopmental outcomes (Schneuer et al., 2018). To maximize the relative independence of the two datasets, family members were kept together in the same ARMS and the groups were matched to have equivalent numbers of sibling and twin pairs and triplets (Feczko et al., 2021). All neuroimaging data collected from participants was reviewed for quality control and data that did not meet metrics outlined above were excluded from the final analyses. Abbreviated demographics are reported in Table 1 for participants included in this analysis. Extended demographics are covered in Table S2. Statistical tests, including t-tests, Kolmogorov–Smirnov tests, and Chi-Squared tests, were performed between the matched groups to verify the samples remained matched after participant exclusion.
Table 1.
Cohort Demographic Variables with statistical comparison between ARMS-1 and ARMS-2. “M” indicates Mean.
| Variable |
ARMS-1 (N = 3339) |
ARMS-2 (N = 3236) |
ARMS-1 v. ARMS-2 | P-Value |
|---|---|---|---|---|
| % Male | 48.20 % | 50.60 % | Chi-squared test of homogeneity | 0.047 |
| % Latinx | 18.60 % | 17.86 % | Chi-squared test of homogeneity | 0.522 |
| Represented Races | 8 | 8 | Chi-squared test of homogeneity | 0.575 |
| Represented Sites | 21 | 21 | Chi-squared test of homogeneity | 0.153 |
| M (sd)Highest Parent Education | 17.362 (2.417) |
17.301 (2.503) |
Chi-squared test of homogeneity | 0.937 |
| M (sd)Interview Age (Months) | 119.529 (7.537) | 119.547 (7.465) | Kolmogorov–Smirnov test | 0.748 |
| M (sd)General Ability (PC1) | 0.0798 (0.731) |
0.0783 (0.747) |
T-Test | 0.934 |
| M (sd)Executive Function (PC2) | 0.0612 (0.746) |
0.0599 (0.734) |
T-Test | 0.946 |
| M (sd)Learning & Memory (PC3) | 0.0663 (0.683) |
0.0583 (0.699) |
T-Test | 0.636 |
2.7.2. Development of the PNRS framework
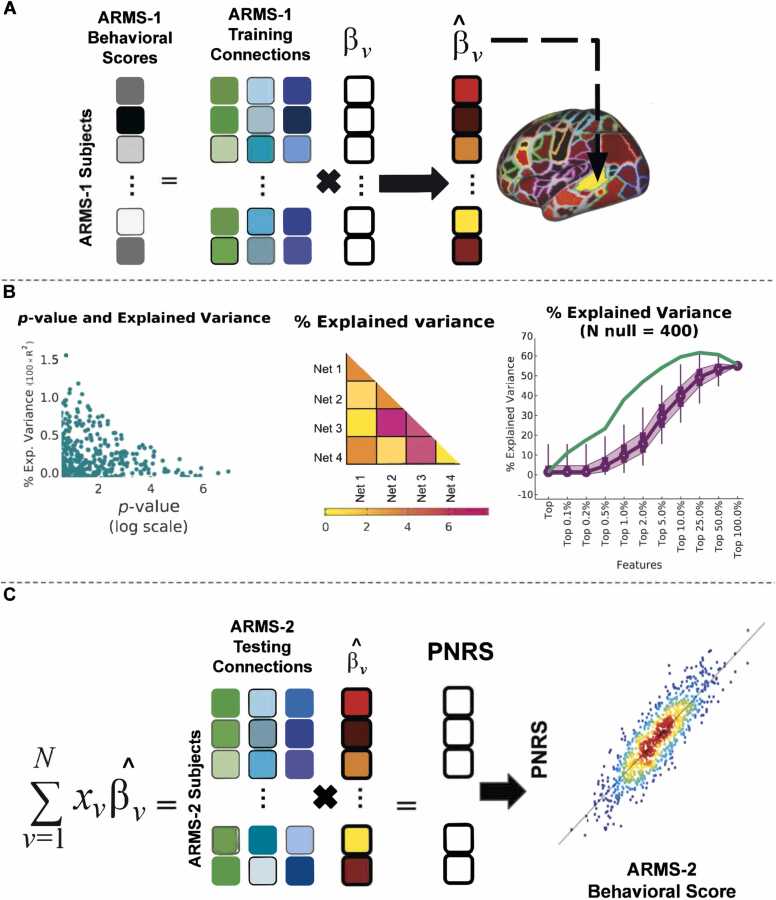
Using a brain-wide association study (BWAS) approach, and following the method proposed by Zhao and colleagues (Zhao et al., 2021), we assume that the relationship between each PC score and functional connectivity is captured by linear models at each connection and that a score for each behavioral outcome can be calculated by the sum of the individual predictions from connections with the most predictive power (Important, while two use-cases are presented here, this framework can support a plethora of multivariate prediction methods. An example of the PNRS framework using partial least squares regression (PLSR) is described below and also presented in supplementary material).
In more detail, and using general ability (GA) as the behavioral outcome of interest, for each connection ν we assume that an estimation of GA () is modeled by:
| (1) |
Where is the connectivity value at the brain feature; is the beta-weight for the least squares solution mapping to y; Covariate i is a variable that could confound that association (site, gender, race, ethnicity, parental education, and age); and is the corresponding weight needed to control for that effect. These estimated beta-weights, were calculated for each connection within each participant within ARMS-1 (Fig. 2A).
Fig. 2.
Polyneuro risk score framework methodology. An in-depth overview of each step within the polyneuro risk score framework. (A) Behavioral measures and functional connectivity from ARMS-1 subjects are used to calculate the weighted contribution, beta-weights, of each connection. (B) Beta-weights can be evaluated by explained variance achieved at three distinct levels: single connection, connections within a network pair, and a percentage of the top connections, when ranked by their p-values. The method by which beta-weights are evaluated dictates which connections are used to generate the PNRS. (C) Estimated beta-weights from (A) are then applied to the functional connectivity data from ARMS-2 subjects to calculate a PNRS. The PNRS can then be compared to the independent behavioral scores for ARMS-2 subjects.
The estimated beta-weights from ARMS-1 highlight the most salient connections in the brain with respect to the outcome of interest, general ability. The evaluation of these beta-weights can be performed assessing its corresponding p-value or by its contribution to the overall explained variance. The resulting p-values were sorted, and connections are selected by their relative position in the ranked list. From this list, we selected the top connection, the top 0.01 % of connections, and so on. Alternatively, features can be selected by their functional network assignment (Fig. 2B).
Finally, a polyneuro risk score (PNRS) of GA is obtained for each ARMS-2 participant by applying the estimated beta-weights and summing them for N connections. This would be the group of connections, either by network pair or imposed threshold, that explained the most variance when previously evaluated:
| (2) |
The resulting output, , is then compared to the independently calculated scores of general ability, PC1, for each of the ARMS-2 participants (Fig. 2C).
To test the specificity of the features selected by p-value, we generated null data by re-sorting features randomly and repeating the analysis (i.e., recalculating the explained variance). This random permutation was repeated 400 times for each one of the different criteria we used to add effects, i.e., by threshold and by networks, which allows us to evaluate whether the unique combination of connections identified by the model explains significantly more variance than what could happen simply by chance.
The entire analytic framework was developed in Matlab. Source code and a containerized version is publicly available via ReadTheDocs.
2.8. The PNRS framework can also incorporate other multivariate techniques
To test the robustness of the framework, we also calculated the polyneuro risk scores using another multivariate technique known as partial least squares regression (PLSR). PLSR models relate predicted and predictor variables after transforming them into a new set of latent variables to maximize the covariance between dependent and independent variables (Rosipal and Krämer, 2006). PLSR consists of a regularized summation of connections and corresponding beta-weights, so that connections with the lowest p-values are weighted more in the calculated PNRS. This contrasts the standard summation in Eq. 2, where all included connections are weighted the same, regardless of p-value.
3. Results
3.1. Evaluation of beta-weights for general ability suggests combining globally distributed features explains the most variance
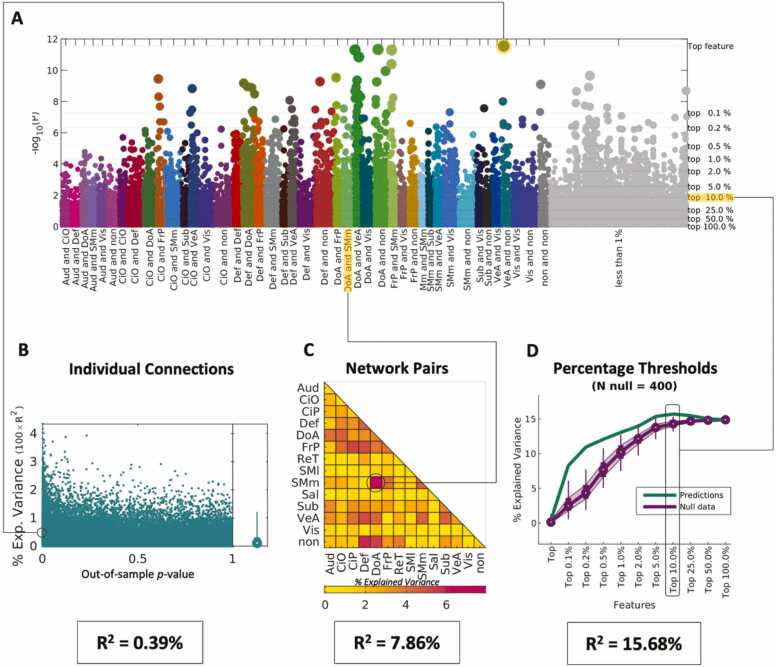
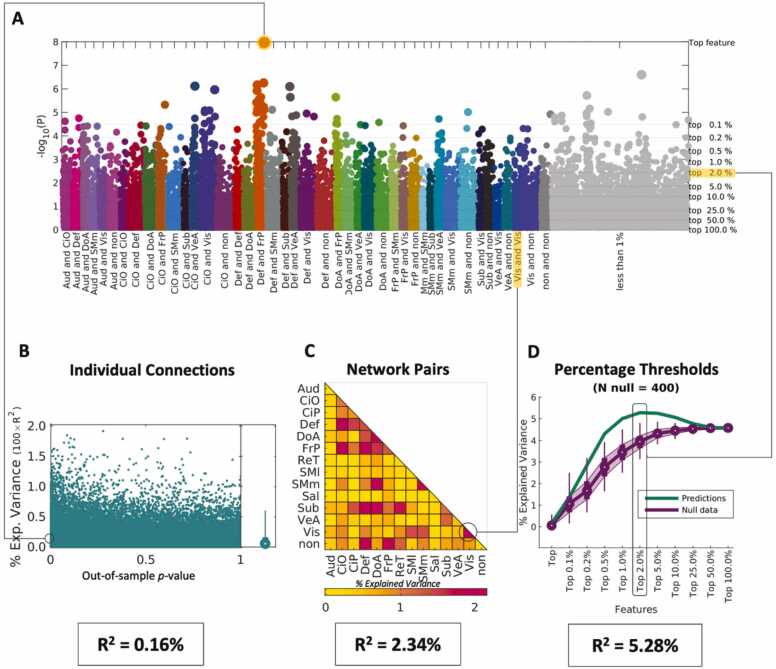
Beta-weights were calculated in the ARMS-1 (N = 3339) sample, with general ability (GA) as the outcome of interest, after controlling for covariates standardly used in ABCD analyses (site, gender, race, ethnicity, parental education, and age). The beta-weights, displayed in the Manhattan plot in Fig. 3A, are ranked by p-value. The top connection, selected from the training sample (ARMS-1), can explain 0.39 % of GA in the independent sample, as shown in Fig. 3, panel B. However, each individual connection can predict between 0 % and 4.31 % of GA in the independent sample (ARMS-2), suggesting that the random selection of connections can lead to overfitting.
Fig. 3.
Evaluation of beta-weights for general ability (GA). Beta-weights were calculated in the ARMS-1 sample with GA as the outcome of interest. (A) Manhattan plot, in logarithmic scale, showing the p-values of each connection. Connections are color coded according to the Gordon parcellation schema. Connections are color-coded by network assignment and ranked by the p-value of their beta-weight calculated in ARMS-1 for GA. The right-hand axis and thin gray horizontal lines denote the imposed thresholds evaluated to predict scores in the independent sample (ARMS-2). (B) Scatterplot showing the explained variance as a function of p-value per individual connection in the independent sample. (C) Explained variance calculated per network pair by summing all the connections within a given network pair. (D) Green line shows the explained variance per imposed threshold for the connections identified in the Manhattan plot (gray lines). Null data testing the specificity of the selected connections is shown in purple. At each threshold the PNRS approach was repeated 400 times after re-sorting connections randomly. Resulting predictions are shown as box plots where mean values are indicated as circles, the interquartile range is indicated in shaded purple and thin lines show the 2.5 % and 97.5 % of the distribution.
Alternatively, we can add the effects from all connections within and between networks (i.e., Auditory-Auditory, Auditory-Cingulo Opercular, …) to calculate the PNRS in the independent sample (Fig. 3C). For GA, connections emerging from brain areas belonging to the Dorsal Attention (DoA) and Somatomotor Medial (SMm) networks appear to be most predictive, explaining 7.86 % of the variance when only connections from this network pair are used in the model.
Lastly, to evaluate the spatial distribution of effects, we can use imposed thresholds that select a top percentage of connections, regardless of network assignment and predict scores in the independent sample. This is where we see our highest explained variance across these three methodologies. The top 10 % of connections, which are widely distributed across the grey matter, explain 15.68 % of the variance. This peak falls outside of the distribution of predicted scores obtained from repeating the approach 400 times after permuting imaging and GA scores (Fig. 3D), indicating that this combination of connections can explain more variance than what is possible when we randomly pick the same number of connections to use in the PNRS calculation.
3.2. Reproducibility across samples requires thousands of participants
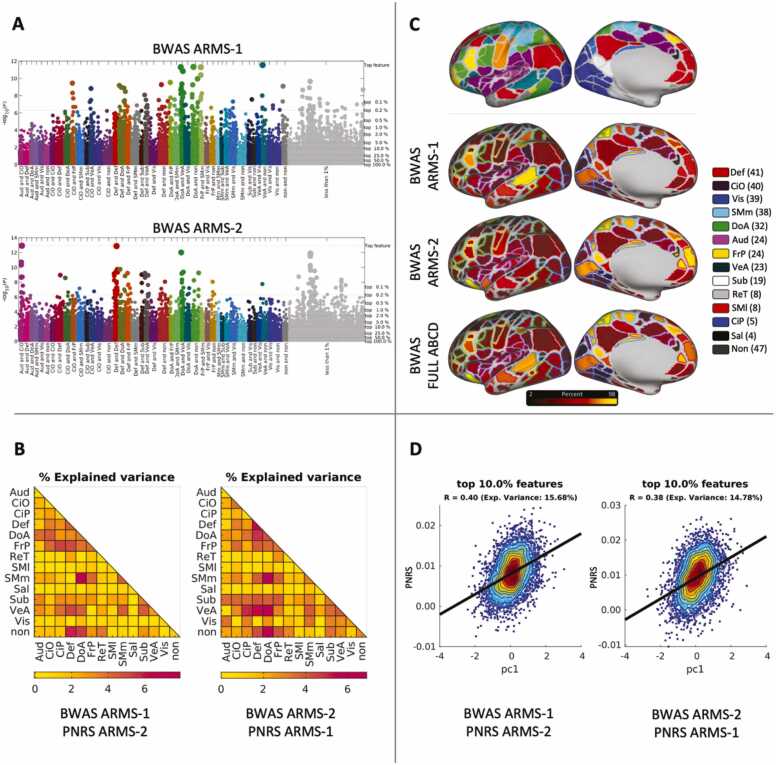
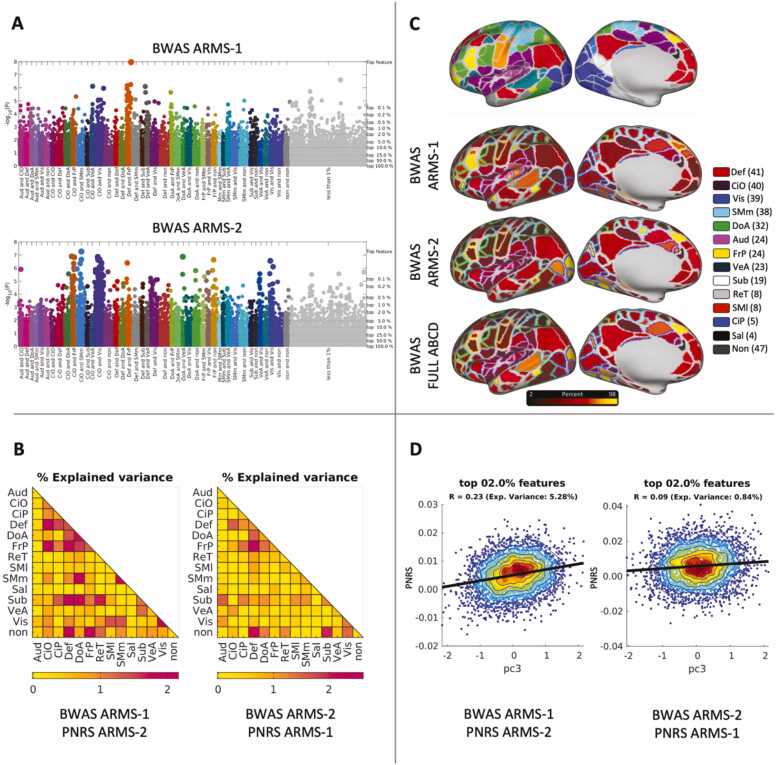
Because we utilized the ABCD Study® Reproducible Matched Samples (ARMS), we can evaluate whether the sample size of each ARMS is enough for reproducible results. We re-calculated the beta-weights using data from ARMS-2 (N = 3236) and then predicted PNRS within ARMS-1 participants. This revealed inconsistencies regarding the exact individual connections implicated in GA (Fig. 4A). While we still see several connections remain salient (for example, several within the Dorsal Attention network (green)) across samples, we also see a more prominent contribution of individual connections from the Cingulo Opercular and Auditory network pair when using ARMS-2 as the modeling sample. We also observe inconsistencies when we inspect the predictive power of adding effects by networks (Fig. 4B). For example, several more network pairs, such as Dorsal Attention-Ventral Attention and Cingulo Parietal-Ventral Attention, appear to explain a higher portion of the variance when training with ARMS-2 rather than ARMS-1.
Fig. 4.
Beta-weights and explained variance across ARMS-1 and ARMS-2 for general ability (GA). To test the reproducibility of our results, beta-weights were calculated within each of the ARMS to allow for evaluation of the stability of the identified connections and resulting explained variance. (A) Manhattan plots across ARMS-1 and ARMS-2 for GA reveal differences in the ranking of individual connections. (B) Explained variance calculated per network pair by summing all the connections within a given network pair. (C) The absolute values of all the beta-weights (100 % threshold) are summed and mapped by their topographical location using the Gordon parcellation schema (Gordon et al., 2016). Differences are noted across the ARMS-1, ARMS-2, and full ABCD beta-weights, but the brain-wide instantiation of the behavior is observed across all 3 models. (D) The calculated polyneuro risk scores are compared to the independently calculated PC1 scores for both ARMS-2 (left) and ARMS-1 (right) participants.
Importantly, despite these connection-specific or network inconsistencies, we observe features that are distributed across much of the cortex for both ARMS, as opposed to being isolated to a given network (Fig. 4C). It is likely that individual feature instability is due to reduced power and the need for even larger sample sizes (Marek and Tervo-Clemmens et al., 2022). With that said, we found that adding effects globally distributed led to the most effective way to predict scores in the independent sample and that the amount of explained variance in the independent samples were high and similar (15.68 % and 14.78 % for each case).
Fig. 4C shows the beta-weights of the combined ARMS (N = 6574).
These beta-weights are more powered and probably closer to the ground truth; however, without an independent dataset to confirm, caution should be taken. Indeed, effect sizes appear to continue to decrease from ABCD sample sizes N∼10,000 to UK Biobank (UKB) sample sizes N∼36,000 (Marek et al., 2019). Nonetheless, the beta-weights continue to be distributed across the whole cortex as opposed to being specified to a given circuit or network. The calculated PNRS also show a strong correlation with the independently calculated scores of GA for both samples (Fig. 4D).
3.3. Changing the behavior of interest yields strikingly different results
We repeated the same analysis for executive function (EF) and learning & memory (LM) as detailed above. Executive function is detailed in the supplemental material (Fig. S1 and S2) and learning & memory results are below.
When the top connection, identified in the training sample, ARMS-1, (Fig. 5A), is used to calculate the PNRS in ARMS-2, we can only explain 0.16 % of the variance (Fig. 5B). When the most predictive network pair is utilized (Visual - Visual), our explained variance is 2.24 % (Fig. 5C). As with GA, we reach our maximum explained variance, 5.28 %, when using a combination of connections distributed across the cortex to calculate the PNRS (Fig. 5D).
Fig. 5.
Evaluation of Beta-weights for Learning & Memory (LM). Caption as in Fig. 2.
However, the results proved to be less stable when the train and test datasets are switched. Specifically, we observe large differences in the individual ranked connections associated with LM across the two ARMS (Fig. 6A). When ARMS-1 is used as the training set, several network pairs can explain a similar proportion of variance, but when ARMS-2 is used as the training set, only a small subset of network pairs reach relatively high levels of explained variance (Fig. 6B). While a brain-wide instantiation of the behavior is similar across ARMS (Fig. 6C), there is instability in the degree to which various networks and ROIs relate to LM. The beta-weights calculated from the full ABCD dataset are likely closer to the ground truth; however, they have not been validated with an independent dataset. Lastly, when comparing ARMS-1 and ARMS-2 PNRS to LM scores per participant, the correlations follow different trends (Fig. 6D).
Fig. 6.
Beta-weights and explained variance across ARMS-1 and ARMS-2 for Learning & Memory (LM). Caption as in Fig. 3.
3.4. Adding scores via regularization leads to similar results
We repeated the PNRS approach but added effects using regularization via PLSR (Table 2). Interestingly, we found similar trends: adding global effects led to the largest predictive power, as compared to adding effects by networks and the spatial distribution of connections was similar, as indicated by the similar count of connections needed, independent of methodology, to achieve the largest predictive value. This was observed for GA, EF, and LM (Table 2 and S1). Additionally, we see a similar result indicating issues of reproducibility across ARMS used for the training and testing of the model.
Table 2.
Results of PLSR PNRS compared to the original multivariate PNRS for general ability and learning & memory.
| Outcome of Interest |
General Ability (GA) |
|||
|---|---|---|---|---|
| Train-Test |
ARMS-1 - ARMS-2 |
ARMS-2 - ARMS-1 |
||
| Feature | Summation |
Regularized Summation (viaPLSR) |
Summation |
Regularized Summation (viaPLSR) |
| Top feature | 2.713 | 2.713 | 0.923 | 0.923 |
| top 00.1 % features | 8.553 | 10.685 | 9.859 | 11.712 |
| top 00.2 % features | 10.933 | 12.866 | 11.025 | 13.300 |
| top 00.5 % features | 12.372 | 13.850 | 13.132 | 16.192 |
| top 01.0 % features | 12.926 | 11.779 | 14.380 | 14.617 |
| top 02.0 % features | 13.958 | 17.848 | 14.976 | 17.764 |
| top 05.0 % features | 15.323 | 14.112 | 15.049 | 17.031 |
| top 10.0 % features | 15.705 | 17.878 | 14.629 | 18.234 |
| top 25.0 % features | 15.430 | 21.425 | 13.561 | 16.638 |
| top 50.0 % features | 15.016 | 13.028 | ||
| top 100.0 % features | 14.893 | 12.909 | ||
| Outcome of Interest |
Learning & Memory (LM) |
|||
|---|---|---|---|---|
| Train-Test |
ARMS-1 - ARMS-2 |
ARMS-2 - ARMS-1 |
||
| Feature | Summation |
Regularized Summation (viaPLSR) |
Summation |
Regularized Summation (viaPLSR |
| Top feature | 0.159 | 0.159 | 0.068 | 0.068 |
| top 00.1 % features | 1.520 | 2.999 | 0.702 | 0.802 |
| top 00.2 % features | 2.887 | 4.290 | 0.955 | 2.806 |
| top 00.5 % features | 4.407 | 5.537 | 0.879 | 1.107 |
| top 01.0 % features | 5.057 | 3.635 | 0.880 | 3.531 |
| top 02.0 % features | 5.265 | 3.747 | 0.896 | 2.650 |
| top 05.0 % features | 5.227 | 4.677 | 0.950 | 3.699 |
| top 10.0 % features | 5.017 | 4.213 | 0.998 | 3.870 |
| top 25.0 % features | 4.750 | 4.111 | 0.921 | 4.831 |
| top 50.0 % features | 4.550 | 0.900 | ||
| top 100.0 % features | 4.544 | 0.874 | ||
4. Discussion
4.1. The PNRS framework shows reproducible brain-wide instantiation for general ability
We show that the PNRS framework is a promising methodology to determine the nature of the distribution of brain areas toward GA. We quantified the amount of variance explained per individual connection, per network, and when combining connections across the grey matter. We found that adding globally distributed effects led to the largest amount of predictive power in an independent dataset. Importantly, the addition of effects seems to reach an optimal threshold and once reached, adding more effects, leads to a decrease in predictive power. Additionally, when combining random, null connections, the maximum achievable explained variance is lower, adding supporting evidence of the relationship between GA and the connections that are identified using the PNRS framework.
Importantly, similar results were achieved when the test and train samples (ARMS-1 and ARMS-2) were switched, supporting the stability of the results. Specifically, the globally distributed nature of the connections was preserved across samples, although the ranked order of such connections fluctuated based on which ARMS was utilized as the training sample. This finding lends itself to the interpretation that numerous small effects distributed across brain networks contribute to specific cognitive functions. Moreover, such findings regarding the wide distribution of connections across the brain contributing to cognitive functions will aid in furthering the field that has thus far focused on specific brain regions or singular networks perhaps resulting in less reproducible results.
While we observe variability in stability and predictive power when using different behavioral outcomes, our findings suggest that the brain features supporting those behaviors are globally distributed. We found that GA is the behavioral outcome that was most stable and easiest to identify. In contrast, our findings were less stable for EF and LM, although the underlying connectivity patterns of these behaviors appear to trend globally rather than locally. For both EF and LM, there are notable differences observed in top connections, network pairs, and combined connections identified when switching the training and testing samples. The explained variance achieved for EF and LM is considerably lower than achieved for GA. This could be a function of the outcomes leveraged in this analysis. Scores produced from a Bayesian Probabilistic Principal Components Analysis (BPPCA) sequentially decrease in their achieved explained variance, therefore we would expect our model’s predictive power to follow the same trend.
4.2. Large datasets facilitate the estimation and reproducibility of brain-behavior associations
The sample size of the ARMS (N = 3339, N = 3236) may still be too small to consistently identify which connections contribute most to higher-order cognitive functions. Because of the complex nature of the cognitive functions assessed, the sample size used was likely inadequate in identifying the distribution of brain connections. Additionally, the nature of the cognitive functions is also likely why using PLSR as the multivariate approach within the framework demonstrates better performance, as it is less susceptible to false-positive correlations and inflated effect estimates. But still, a larger sample size will be needed to consistently identify which connections are the most pertinent to predicting complex behaviors.
The full ABCD sample may be large enough to identify specific features, particularly for general ability, however, we do not have an independent sample to validate the connections identified by the PNRS framework. In addition, as the longitudinal data for ABCD come online the repeated measures may provide significantly more power and thus provides yet another opportunity to characterize the stability of individual connections shown in the full sample.
In this study we only used data that passed stringent quality check criteria. For that reason, we lost approximately 45 % of the ABCD sample due to the motion censoring criteria imposed for this analysis. Image acquisition is a known challenge and head motion is a large limitation faced by many studies, regardless of sample size. It is also likely that the ABCD sample loses demographic diversity due to motion artifacts and data quality. Recent investigations determined that ABCD participants with low-noise data are predominantly older, less diverse, and have higher scores on neurocognitive assessments as well as fewer neurodevelopmental problems (Cosgrove et al., 2022). As a result, our findings may be less generalizable to the population. Methodological changes are needed to properly represent marginalized populations in large consortia studies.
4.3. PNRS is an effective framework to compare different ways to add effects to predict behavioral outcomes
For the complex behavioral outcomes that we assessed, the PNRS framework identified globally distributed patterns of RSFC. These patterns encompass numerous small effects spanning the brain that explain the most variance in the investigated outcomes, particularly GA. However, because the PNRS inspects the explainable variance achieved at the levels of single connections, connections within a network pair, as well as thresholded amounts of the most salient connections, the methodology is equipped to estimate the predictive power of contributing brain connections, regardless of if such connections are locally or globally distributed. The framework is agnostic to the way the behavior is measured or summarized, so it can take composite scores, as shown in this analysis, but it can also accept individual scores as input. While the cognitive scores used in this study point toward a global distribution of associated functional brain circuitry, localized regions could also be identified using this methodology. Moreover, a brain-behavior relationship with a larger effect size may have more predictive power and accuracy using the PNRS framework, even with the sample sizes of publicly available datasets, like ABCD (Jollans et al., 2019). This framework is equipped to handle several different neuroimaging modalities (task fMRI, resting state fMRI, DTI, and anatomical features such as curvature and cortical thickness) to identify brain-behavior associations, as has been demonstrated using similar multivariate models (Palmer et al., 2020, Zhao et al., 2021).
Our BWAS/PNRS framework offers a simple and effective way to derive brain-behavior associations. The additive nature of the approach makes easy to quantify and relate the contribution of brain areas to observed behavior, something that might be problematic when using non-linear approaches. To demonstrate the flexibility of the PNRS framework we also included a unique multivariate method as an example - regularization via PLSR. PLSR is only one of a family of methods that can be used to estimate beta-weights, including Tikhonov regularization, truncated singular value decomposition, lasso regression, canonical correlation analysis or support vector regression to name a few. PLSR in particular decomposes the data such that the covariance between predicted and predictor variables is maximized. Other regularization methods implement different cost functions and might predict out-of-sample data with a different degree of accuracy. Far from being an exhaustive comparison of methods, this report shows that the addition of small effects is a promising approach to characterize brain function. The versatility of the PNRS framework could allow for robust investigation of brain-behavior relationships using a common methodology, increasing the utility of the scores for clinical purposes by allowing for the generation of models or associations on a large training set, then testing on a smaller independent test set (as is done with PRS). The current paper focused on ABCD data only, however, future work should consider a 3rd validation set for robustness as well.
Previous work has indicated a benefit in accounting for spatial correlation across the brain when using this type of methodology (Park and Fiecas, 2022, Vilhjálmsson et al., 2015, Zhao et al., 2021). However, recent findings have suggested that the use of parcellated connectivity data, as used in this analysis, may already account for this spatial correlation, removing the need for an additional adjustment (Mooney et al., 2021). Further exploration is needed to determine best practices for spatial correlation adjustments. This should be considered when utilizing the PNRS framework.
4.4. PNRS has potential translational applications, similar to polygenic risk scores
Polygenic risk scores (PRS) have been developed for and implemented in studies assessing bipolar disorder (Coombes et al., 2020), coronary heart disease (Natarajan et al., 2017), schizophrenia (Agerbo et al., 2015, Frank et al., 2015), and breast cancer (Mavaddat et al., 2019), among other diseases and psychopathologies (Waszczuk et al., 2020). The PRS model combines the cumulative small effects across the genome to better predict the risk for disease than a single locus (International Schizophrenia Consortium et al., 2009). PRS have shown utility in identifying disease/disorder subgroups and disentangling clinical heterogeneity at a genomic level. However, PRS are more often used as a predictive measure rather than as a diagnostic tool given the relatively small percentages of explained variance that the genome typically accounts for regarding a disease state (Frank et al., 2015, Lambert et al., 2019). Relatedly, even though true effect sizes discovered with high-powered consortia level BWAS approaches are small, they show notable potential to be larger than effect sizes typically achieved by high-powered GWAS (Gratton et al., 2022).
Future work could reveal the PNRS framework possesses similar utility to PRS models in terms of risk evaluation for neurodevelopmental disorders. The extent of this application will be dependent upon the effect sizes of the various behaviors of interest. Additionally, the framework has potential to address the heterogeneity problem of several psychiatric disorders (Feczko et al., 2019). Recent work reviewing the current state of brain-based subtyping in psychiatric research has highlighted the overwhelming absence of external validation of the resulting subgroups (Brucar et al., 2022). The PNRS framework has that crucial step built in, offering a solution to this current gap. Furthermore, the individually calculated PNRS could serve as the input for clustering algorithms to examine subgroups within cohorts enriched for a specific disorder of interest.
There is also the potential to extend beyond the current scope of PRS applications. PNRS are not immutable, as the measurement inputs to generate the scores are highly dynamic; the plasticity of the brain allows for changes to occur over time. Because of this, the PNRS may possess a large potential for clinical utility, as they could be used to evaluate the efficacy of therapies and interventions, such as psychiatric medications, on a quantitative basis rather than through trial-and-error observations. Tangentially, smaller, more focused BWAS studies that employ measures to increase the signal-to-noise ratio (SNR) and utilize within-subject designs could also implement the PNRS framework to inform personalized, targeted interventions (Gratton et al., 2022).
5. Conclusion
Though RSFC has been a valuable tool for identifying functional networks and characterizing brain changes as they relate to behavior, advancements in analyses of these data are still needed in order to utilize this technique to characterize certain brain-behavior relationships. Additionally, high-powered large consortia data are required to reliably capture the likely small effect sizes of higher-order cognition. To address these issues, we have implemented the PNRS framework which utilizes multivariate and large, independent sample validation techniques to capture brain-behavior relationships. This methodology supports various multivariate algorithms, and the produced polyneuro risk scores show promising clinical utility given their potential to capture the underlying functional connectivity pattern of both typical and atypical neurodevelopment. Our results indicate that the PNRS framework is a promising way to postulate the distribution of contributing connections toward general ability and may also show utility in characterizing the connectivity of other brain-behavior relationships.
Funding
This work was supported by the National Institutes of Health [U01DA041148, R01MH096773, R37MH125829, R37MH059105].
CRediT authorship contribution statement
Nora Byington: methodology, software, formal analysis, writing – original draft Gracie Grimsrud: methodology, software, formal analysis, writing – original draft Michael A. Mooney: conceptualization, methodology, software, data curation Michaela Cordova: data curation Olivia Doyle: data curation Robert J.M. Hermosillo: data curation, software Eric Earl: data curation Audrey Houghton: data curation Gregory Conan: data curation Timothy J. Hendrickson: data curation Anjanibhargavi Ragothaman: software Cristian Morales Carrasco: software Amanda Rueter: data curation, project administration Anders Perrone: data curation Lucille A. Moore: data curation Alice Graham: resources Joel T. Nigg: conceptualization, resources Wesley K. Thompson: resources Steven M. Nelson: resources, writing – review & editing Eric Feczko: methodology, software, data curation, formal analysis, writing – review & editing Damien A. Fair: conceptualization, methodology, data curation, formal analysis, writing – review & editing, supervision Oscar Miranda-Dominguez: conceptualization, methodology, software, data curation, formal analysis writing – original draft, supervision.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Damien Fair reports a relationship with Nous Imaging, Inc that includes: board membership and equity or stocks. Damien A. Fair has patent REAL TIME MONITORING AND PREDICTION OF MOTION IN MRI issued to Nous Imaging.
Acknowledgments
The ABCD Study® is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, U24DA041147. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/consortium_members/.
This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH, its affiliated Institutes, Centers or offices, or ABCD consortium investigators.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dcn.2023.101231.
Appendix A. Supplementary material
Supplementary material
.
References
- Acker, Acker (1982). Bexley Maudsley automated processing screening and Bexley Maudsley category sorting test manual. Windsor, England: NFER-Nelson.
- Agerbo E., Sullivan P.F., Vilhjalmsson B.J., Pedersen C.B., Mors O., Børglum A.D., Hougaard D.M., Hollegaard M.V., Meier S., Mattheisen M., Others Polygenic risk score, parental socioeconomic status, family history of psychiatric disorders, and the risk for schizophrenia: a Danish population-based study and meta-analysis. JAMA Psychiatry. 2015;72(7):635–641. doi: 10.1001/jamapsychiatry.2015.0346. 〈https://jamanetwork.com/journals/jamapsychiatry/article-abstract/2211892〉 [DOI] [PubMed] [Google Scholar]
- Alvarez J.A., Emory E. Executive function and the frontal lobes: a meta-analytic review. Neuropsychol. Rev. 2006;16(1):17–42. doi: 10.1007/s11065-006-9002-x. [DOI] [PubMed] [Google Scholar]
- Biswal B., Zerrin Yetkin F., Haughton V.M., Hyde J.S. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn. Reson. Med. 1995;Vol. 34(Issue 4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Bleck T.P., Nowinski C.J., Gershon R., Koroshetz W.J. What is the NIH Toolbox, and what will it mean to neurology? Neurology. 2013;Vol. 80(Issue 10):874–875. doi: 10.1212/wnl.0b013e3182872ea0. [DOI] [PubMed] [Google Scholar]
- Boekel W., Wagenmakers E.-J., Belay L., Verhagen J., Brown S., Forstmann B.U. A purely confirmatory replication study of structural brain-behavior correlations. Cortex; J. Devoted Study Nerv. Syst. Behav. 2015;66:115–133. doi: 10.1016/j.cortex.2014.11.019. [DOI] [PubMed] [Google Scholar]
- Brucar L.R., Feczko E., Fair D.A., Zilverstand A. Current approaches in computational psychiatry for the data-driven identification of brain-based subtypes. Biol. Psychiatry. 2022 doi: 10.1016/j.biopsych.2022.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey B.J., Cannonier T., Conley M.I., Cohen A.O., Barch D.M., Heitzeg M.M., Soules M.E., Teslovich T., Dellarco D.V., Garavan H., Orr C.A., Wager T.D., Banich M.T., Speer N.K., Sutherland M.T., Riedel M.C., Dick A.S., Bjork J.M., Thomas K.M., Dale A.M. The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 2018;Vol. 32:43–54. doi: 10.1016/j.dcn.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaarani B., Hahn S., Allgaier N., Adise S., Owens M.M., Juliano A.C., Yuan D.K., Loso H., Ivanciu A., Albaugh M.D., Dumas J., Mackey S., Laurent J., Ivanova M., Hagler D.J., Cornejo M.D., Hatton S., Agrawal A., Aguinaldo L., ABCD Consortium. Baseline brain function in the preadolescents of the ABCD Study. Nat. Neurosci. 2021;24(8):1176–1186. doi: 10.1038/s41593-021-00867-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.L., Fair D.A., Dosenbach N.U.F., Miezin F.M., Dierker D., Van Essen D.C., Schlaggar B.L., Petersen S.E. Defining functional areas in individual human brains using resting functional connectivity MRI. NeuroImage. 2008;41(1):45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes B.J., Markota M., Mann J.J., Colby C., Stahl E., Talati A., Pathak J., Weissman M.M., McElroy S.L., Frye M.A., Biernacka J.M. Dissecting clinical heterogeneity of bipolar disorder using multiple polygenic risk scores. Transl. Psychiatry. 2020;10(1):314. doi: 10.1038/s41398-020-00996-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove K.T., McDermott T.J., White E.J., Mosconi M.W., Thompson W.K., Paulus M.P., Cardenas-Iniguez C., Aupperle R.L. Limits to the generalizability of resting-state functional magnetic resonance imaging studies of youth: An examination of ABCD Study® baseline data. Brain Imaging Behav. 2022;Vol. 16(Issue 4):1919–1925. doi: 10.1007/s11682-022-00665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, Wahlstrom, Zhang (n.d.). Equivalence of Q-interactive and paper administrations of cognitive tasks: WISC-V. Q-Interactive Technical Report. http://www.helloq.com.au/userfiles/830351450329565.pdf.
- Developmental Cognition and Neuroimaging Labs. (n.d.). Github. Retrieved November 21, 2022, from https://github.com/DCAN-Labs.
- Dick A.S., Lopez D.A., Watts A.L., Heeringa S., Reuter C., Bartsch H., Fan C.C., Kennedy D.N., Palmer C., Marshall A., Haist F., Hawes S., Nichols T.E., Barch D.M., Jernigan T.L., Garavan H., Grant S., Pariyadath V., Hoffman E., Thompson W.K. Meaningful associations in the adolescent brain cognitive development study. NeuroImage. 2021;239 doi: 10.1016/j.neuroimage.2021.118262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinga R., Schmaal L., Penninx B.W.J.H., van Tol M.J., Veltman D.J., van Velzen L., Mennes M., van der Wee N.J.A., Marquand A.F. Evaluating the evidence for biotypes of depression: Methodological replication and extension of. NeuroImage. Clin. 2019;22 doi: 10.1016/j.nicl.2019.101796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach N.U.F., Fair D.A., Miezin F.M., Cohen A.L., Wenger K.K., Dosenbach R.A.T., Fox M.D., Snyder A.Z., Vincent J.L., Raichle M.E., Schlaggar B.L., Petersen S.E. Distinct brain networks for adaptive and stable task control in humans. Proc. Natl. Acad. Sci. USA. 2007;104(26):11073–11078. doi: 10.1073/pnas.0704320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach N.U.F., Koller J.M., Earl E.A., Miranda-Dominguez O., Klein R.L., Van A.N., Snyder A.Z., Nagel B.J., Nigg J.T., Nguyen A.L., Wesevich V., Greene D.J., Fair D.A. Real-time motion analytics during brain MRI improve data quality and reduce costs. NeuroImage. 2017;Vol. 161:80–93. doi: 10.1016/j.neuroimage.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair, Dosenbach, Moore . Annual Review of. Antitrust Law Developments. 2021. Developmental cognitive neuroscience in the era of networks and big data: Strengths, weaknesses, opportunities, and threats.〈https://dosenbachlab.wustl.edu/media/papers/annurev-devpsych-121318-085124.pdf〉 [Google Scholar]
- Fair D.A., Cohen A.L., Power J.D. Functional brain networks develop from a “local to distributed” organization. Plos. Comput. Biol. 2009 doi: 10.1371/journal.pcbi.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feczko E., Miranda-Dominguez O., Marr M., Graham A.M., Nigg J.T., Fair D.A. The heterogeneity problem: approaches to identify psychiatric subtypes. Trends Cogn. Sci. 2019;23(7):584–601. doi: 10.1016/j.tics.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feczko, E., Conan, G., Marek, S., Tervo-Clemmens, B., Cordova, M., Doyle, O., Earl, E., Perrone, A., Sturgeon, D., Klein, R., Harman, G., Kilamovich, D., Hermosillo, R., Miranda-Dominguez, O., Adebimpe, A., Bertolero, M., Cieslak, M., Covitz, S., Hendrickson, T., … Fair, D.A. (2021). Adolescent Brain Cognitive Development (ABCD) Community MRI Collection and Utilities. In bioRxiv (p. 2021.07.09.451638). https://doi.org/10.1101/2021.07.09.451638.
- Fischl B. FreeSurfer. NeuroImage. 2012;Vol. 62(Issue 2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Salat D.H., Busa E., Albert M., Dieterich M., Haselgrove C., van der Kouwe A., Killiany R., Kennedy D., Klaveness S., Montillo A., Makris N., Rosen B., Dale A.M. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B., van der Kouwe A., Destrieux C., Halgren E., Ségonne F., Salat D.H., Busa E., Seidman L.J., Goldstein J., Kennedy D., Caviness V., Makris N., Rosen B., Dale A.M. Automatically parcellating the human cerebral cortex. Cereb. Cortex. 2004;14(1):11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Fox M.D., Snyder A.Z., Vincent J.L., Corbetta M., Van Essen D.C., Raichle M.E. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc. Natl. Acad. Sci. USA. 2005;102(27):9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Lang M., Witt S.H., Strohmaier J., Rujescu D., Cichon S., Degenhardt F., Nöthen M.M., Collier D.A., Ripke S., Naber D., Rietschel M. Erratum: Identification of increased genetic risk scores for schizophrenia in treatment-resistant patients. Mol. Psychiatry. 2015;Vol. 20(Issue 7) doi: 10.1038/mp.2015.52. 913–913. [DOI] [PubMed] [Google Scholar]
- Garavan H., Bartsch H., Conway K., Decastro A., Goldstein R.Z., Heeringa S., Jernigan T., Potter A., Thompson W., Zahs D. Recruiting the ABCD sample: design considerations and procedures. Dev. Cogn. Neurosci. 2018;Vol. 32:16–22. doi: 10.1016/j.dcn.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon, R.C., Wagster, M.V., Hendrie, H.C., Fox, N.A., Cook, K.F., & Nowinski, C.J. (2013). NIH toolbox for assessment of neurological and behavioral function. Neurology (Vol. 80, Issues 11, Supplement 3, pp. S2–S6). https://doi.org/10.1212/wnl.0b013e3182872e5f. [DOI] [PMC free article] [PubMed]
- Gordon E.M., Laumann T.O., Adeyemo B., Huckins J.F., Kelley W.M., Petersen S.E. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex. 2016;26(1):288–303. doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski, K.J., Auer, T., Calhoun, V.D., Cameron Craddock, R., Das, S., Duff, E.P., Flandin, G., Ghosh, S.S., Glatard, T., Halchenko, Y.O., Handwerker, D.A., Hanke, M., Keator, D., Li, X., Michael, Z., Maumet, C., Nolan Nichols, B., Nichols, T.E., Pellman, J., … Poldrack, R.A. (2016). The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. In Scientific Data (Vol. 3, Issue 1). https://doi.org/10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed]
- Gratton C., Nelson S.M., Gordon E.M. Brain-behavior correlations: two paths toward reliability [Review of Brain-behavior correlations: Two paths toward reliability] Neuron. 2022;110(9):1446–1449. doi: 10.1016/j.neuron.2022.04.018. [DOI] [PubMed] [Google Scholar]
- Greicius, M.D., Krasnow, B., Reiss, A.L., Menon, V. (2003). Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc. Natl. Acad. Sci. USA, 100(1), 253–258. https://doi.org/10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed]
- Hodes, R.J., Insel, T.R., Landis, S.C., On behalf of the NIH Blueprint for Neuroscience Research. (2013). The NIH Toolbox: Setting a standard for biomedical research. In Neurology (Vol. 80, Issues 11, Supplement 3, pp. S1–S1). https://doi.org/10.1212/wnl.0b013e3182872e90. [DOI] [PMC free article] [PubMed]
- Holland D., Wang Y., Thompson W.K., Schork A., Chen C.-H., Lo M.-T., Witoelar A., Schizophrenia Working Group of the Psychiatric Genomics Consortium, Enhancing Neuro Imaging Genetics through Meta Analysis Consortium, Werge T., O’Donovan M., Andreassen O.A., Dale A.M. Estimating effect sizes and expected replication probabilities from GWAS summary statistics. Front. Genet. 2016;7:15. doi: 10.3389/fgene.2016.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium, Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O’Donovan M.C., Sullivan P.F., Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jollans L., Boyle R., Artiges E., Banaschewski T., Desrivières S., Grigis A., Martinot J.-L., Paus T., Smolka M.N., Walter H., Schumann G., Garavan H., Whelan R. Quantifying performance of machine learning methods for neuroimaging data. NeuroImage. 2019;Vol. 199:351–365. doi: 10.1016/j.neuroimage.2019.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher N.R., Barch D.M. The ABCD study: understanding the development of risk for mental and physical health outcomes. Neuropsychopharmacology. 2021;Vol. 46(Issue 1):131–142. doi: 10.1038/s41386-020-0736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A., Farlow A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods. 2013;9(1):1–9. doi: 10.1186/1746-4811-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S.A., Abraham G., Inouye M. Towards clinical utility of polygenic risk scores. Hum. Mol. Genet. 2019;28(R2):R133–R142. doi: 10.1093/hmg/ddz187. [DOI] [PubMed] [Google Scholar]
- Luciana M., Bjork J.M., Nagel B.J., Barch D.M., Gonzalez R., Nixon S.J., Banich M.T. Adolescent neurocognitive development and impacts of substance use: overview of the adolescent brain cognitive development (ABCD) baseline neurocognition battery. Dev. Cogn. Neurosci. 2018;Vol. 32:67–79. doi: 10.1016/j.dcn.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C.J., Elbau I., Liston C. Improving precision functional mapping routines with multi-echo fMRI. Curr. Opin. Behav. Sci. 2021;40:113–119. doi: 10.1016/j.cobeha.2021.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S., Tervo-Clemmens B., Nielsen A.N., Wheelock M.D., Miller R.L., Laumann T.O., Earl E., Foran W.W., Cordova M., Doyle O., Perrone A., Miranda-Dominguez O., Feczko E., Sturgeon D., Graham A., Hermosillo R., Snider K., Galassi A., Nagel B.J., Dosenbach N.U.F. Identifying reproducible individual differences in childhood functional brain networks: An ABCD study. Dev. Cogn. Neurosci. 2019;Vol. 40 doi: 10.1016/j.dcn.2019.100706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S., Tervo-Clemmens B., Calabro F.J., Montez D.F., Kay B.P., Hatoum A.S., Donohue M.R., Foran W., Miller R.L., Hendrickson T.J., Malone S.M., Kandala S., Feczko E., Miranda-Dominguez O., Graham A.M., Earl E.A., Perrone A.J., Cordova M., Doyle O., Dosenbach N.U.F. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022 doi: 10.1038/s41586-022-04492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masouleh S.K., Eickhoff S.B., Hoffstaedter F., Genon S., Alzheimer’s Disease Neuroimaging Initiative Empirical examination of the replicability of associations between brain structure and psychological variables. eLife. 2019;Vol. 8 doi: 10.7554/elife.43464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavaddat N., Michailidou K., Dennis J., Lush M., Fachal L., Lee A., Tyrer J.P., Chen T.-H., Wang Q., Bolla M.K., Yang X., Adank M.A., Ahearn T., Aittomäki K., Allen J., Andrulis I.L., Anton-Culver H., Antonenkova N.N., Arndt V., Easton D.F. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet. 2019;104(1):21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna R., Rushe T., Woodcock K.A. Informing the structure of executive function in children: a meta-analysis of functional neuroimaging data. Front. Hum. Neurosci. 2017;11:154. doi: 10.3389/fnhum.2017.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O., Ramirez J.S.B., Mitchell A.J., Perrone A., Earl E., Carpenter S., Feczko E., Graham A., Jeon S., Cohen N.J., Renner L., Neuringer M., Kuchan M.J., Erdman J.W., Fair D. Carotenoids improve the development of cerebral cortical networks in formula-fed infant macaques. Sci. Rep. 2022;Vol. 12(Issue 1) doi: 10.1038/s41598-022-19279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O., Reiners P., Lee E., Morales Carrasco C., Conan G. DCAN-Labs/biceps: BICEPS First release (beta) Zenodo. 2023 doi: 10.5281/zenodo.7697404. [DOI] [Google Scholar]
- Mooney, M.A., Hermosillo, R.J.M., Feczko, E., Miranda-Dominguez, O., Moore, L.A., Perrone, A., Byington, N., Grimsrud, G., Rueter, A., Nousen, E., Antovich, D., Feldstein Ewing, S.W., Nagel, B.J., Nigg, J.T., Fair, D.A. (2021). Cumulative effects of resting-state connectivity across all brain networks significantly correlate with ADHD symptoms. In bioRxiv. https://doi.org/10.1101/2021.11.16.21266121.
- Natarajan P., Young R., Stitziel N.O., Padmanabhan S., Baber U., Mehran R., Sartori S., Fuster V., Reilly D.F., Butterworth A., Rader D.J., Ford I., Sattar N., Kathiresan S. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135(22):2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niendam T.A., Laird A.R., Ray K.L., Dean Y.M., Glahn D.C., Carter C.S. Meta-analytic evidence for a superordinate cognitive control network subserving diverse executive functions. Cogn. Affect. Behav. Neurosci. 2012;12(2):241–268. doi: 10.3758/s13415-011-0083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, C.E., Zhao, W., Loughnan, R., Zou, J., Fan, C.C., Thompson, W.K., Jernigan, T.L., Dale, A.M. (2020). Determining the association between regionalisation of cortical morphology and cognition in 10,145 children. In bioRxiv (p. 816025). https://doi.org/10.1101/816025.
- Palmer C.E., Zhao W., Loughnan R., Zou J., Fan C.C., Thompson W.K., Dale A.M., Jernigan T.L. Distinct regionalization patterns of cortical morphology are associated with cognitive performance across different domains. Cereb. Cortex. 2021;31(8):3856–3871. doi: 10.1093/cercor/bhab054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.Y., Fiecas M. CLEAN: leveraging spatial autocorrelation in neuroimaging data in clusterwise inference. NeuroImage. 2022;255 doi: 10.1016/j.neuroimage.2022.119192. [DOI] [PubMed] [Google Scholar]
- Poldrack R.A., Baker C.I., Durnez J., Gorgolewski K.J., Matthews P.M., Munafò M.R., Nichols T.E., Poline J.-B., Vul E., Yarkoni T. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci. 2017;18(2):115–126. doi: 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Cohen A.L., Nelson S.M., Wig G.S., Barnes K.A., Church J.A., Vogel A.C., Laumann T.O., Miezin F.M., Schlaggar B.L., Petersen S.E. Functional network organization of the human brain. Neuron. 2011;72(4):665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Mitra A., Laumann T.O., Snyder A.Z., Schlaggar B.L., Petersen S.E. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage. 2014;Vol. 84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragothaman A., Mancini M., Nutt J.G., Fair D.A., Miranda-Dominguez O., Horak F.B. Resting state functional networks predict different aspects of postural control in Parkinson’s disease. Gait Posture. 2022;Vol. 97:122–129. doi: 10.1016/j.gaitpost.2022.07.003. [DOI] [PubMed] [Google Scholar]
- Ragothaman A., Miranda-Dominguez O., Brumbach B.H., Giritharan A., Fair D.A., Nutt J.G., Mancini M., Horak F.B. Relationship Between Brain Volumes and Objective Balance and Gait Measures in Parkinson’s Disease. J. Parkinson’s Dis. 2022;Vol. 12(Issue 1):283–294. doi: 10.3233/jpd-202403. [DOI] [PubMed] [Google Scholar]
- Rosipal R., Krämer N. Overview and recent advances in partial least squares. Subspace, Latent Structure and Feature Selection. 2006:34–51. doi: 10.1007/11752790_2. [DOI] [Google Scholar]
- Rudolph M.D., Graham A.M., Feczko E., Miranda-Dominguez O., Rasmussen J.M., Nardos R., Entringer S., Wadhwa P.D., Buss C., Fair D.A. Maternal IL-6 during pregnancy can be estimated from newborn brain connectivity and predicts future working memory in offspring. Nat. Neurosci. 2018;Vol. 21(Issue 5):765–772. doi: 10.1038/s41593-018-0128-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneuer F.J., Bentley J.P., Davidson A.J., Holland A.J.A., Badawi N., Martin A.J., Skowno J., Lain S.J., Nassar N. The impact of general anesthesia on child development and school performance: a population-based study. Pediatr. Anesth. 2018;Vol. 28(Issue 6):528–536. doi: 10.1111/pan.13390. [DOI] [PubMed] [Google Scholar]
- Stevens W.D., Dale Stevens W., Nathan Spreng R. Resting-state functional connectivity MRI reveals active processes central to cognition. Wiley Interdiscip. Rev.: Cogn. Sci. 2014;Vol. 5(Issue 2):233–245. doi: 10.1002/wcs.1275. [DOI] [PubMed] [Google Scholar]
- Thompson W.K., Barch D.M., Bjork J.M., Gonzalez R., Nagel B.J., Nixon S.J., Luciana M. The structure of cognition in 9 and 10 year-old children and associations with problem behaviors: findings from the ABCD study’s baseline neurocognitive battery. In. Dev. Cogn. Neurosci. 2019;Vol. 36 doi: 10.1016/j.dcn.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhjálmsson B.J., Yang J., Finucane H.K., Gusev A., Lindström S., Ripke S., Genovese G., Loh P.-R., Bhatia G., Do R., Hayeck T., Won H.-H., Schizophrenia Working Group of the Psychiatric Genomics Consortium, Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) study, Kathiresan S., Pato M., Pato C., Tamimi R., Stahl E., Zaitlen N., Price A.L. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 2015;97(4):576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.Y.S., Barratt B.J., Clayton D.G., Todd J.A. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 2005;6(2):109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- Waszczuk M.A., Eaton N.R., Krueger R.F., Shackman A.J., Waldman I.D., Zald D.H., Lahey B.B., Patrick C.J., Conway C.C., Ormel J., Hyman S.E., Fried E.I., Forbes M.K., Docherty A.R., Althoff R.R., Bach B., Chmielewski M., DeYoung C.G., Forbush K.T., Kotov R. Redefining phenotypes to advance psychiatric genetics: implications from hierarchical taxonomy of psychopathology. J. Abnorm. Psychol. 2020;Vol. 129(Issue 2):143–161. doi: 10.1037/abn0000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo B.T.T., Krienen F.M., Sepulcre J., Sabuncu M.R., Lashkari D., Hollinshead M., Roffman J.L., Smoller J.W., Zöllei L., Polimeni J.R., Fischl B., Liu H., Buckner R.L. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 2011;106(3):1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-M., Jia Z., Dunwell J.M. Editorial: the applications of new multi-locus GWAS methodologies in the genetic dissection of complex traits. Front. Plant Sci. 2019;Vol. 10 doi: 10.3389/fpls.2019.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Palmer C.E., Thompson W.K., Chaarani B., Garavan H.P., Casey B.J., Jernigan T.L., Dale A.M., Fan C.C. Individual differences in cognitive performance are better predicted by global rather than localized BOLD activity patterns across the cortex. Cereb. Cortex. 2021;31(3):1478–1488. doi: 10.1093/cercor/bhaa290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material