Abstract
Precise spatiotemporal organization and regulation of signal transduction networks is essential for cellular response to internal and external cues. To understand how this biochemical activity architecture impacts cellular function, many genetically encodable tools which regulate kinase activity at a subcellular level have been developed. In this review, we highlight various types of genetically encodable molecular tools, including tools to regulate endogenous kinase activity and biorthogonal techniques to perturb kinase activity. Finally, we emphasize the use of these tools alongside biosensors for kinase activity to measure and perturb kinase activity in real time for a better understanding of the cellular biochemical activity architecture.
Introduction
Spatial organization of biochemical processes, including signal transduction networks, is essential for proper cellular function. Compartmentation of signaling machinery by membranes, organelles, and other discrete microdomains allows cells to encode specificity within networks and ensure appropriate cellular responses [1,2]. Of particular importance to these networks are protein kinases, which phosphorylate much of the proteome to regulate cellular behavior [3]. Through subcellular compartmentation, cells can locally direct kinase activity towards specific targets to achieve precise spatial and temporal control of signaling and accurate signal decoding. Our understanding of compartmentalized kinase activity has been expanded by genetically encoded fluorescent protein (FP)-based biosensors, which provide a real-time view of kinase activity dynamics in live cells [4]. Most kinase biosensors utilize a kinase-specific substrate and phosphoamino acid-binding domain (PAABD) as the sensing unit, which are linked to one or more FPs as the reporting unit. Binding of the PAABD to the phosphorylated substrate induces a conformational change that alters the fluorescence of the associated FP(s), such as by modulating Forster Resonance Energy Transfer (FRET) efficiency, increasing intensity, or shifting the excitation/emission wavelength. Alternatively, kinase translocation reporters (KTR) couple a substrate peptide to nuclear localization (NLS) and export (NES) sequences and use nuclear-to-cytoplasmic shuttling of an FP as a readout for kinase activity (see [4-6] for more detailed discussion of biosensors). These versatile molecular tools are an adaptable platform for observing the activities of a variety of kinases [5]. Notably, integration of subcellular targeting motifs that function as “molecular zip codes” enables biosensors to selectively monitor local kinase activities at discrete microdomains, including near organelle membranes and within spatial gradients [1,7]. This approach has greatly informed our understanding of the precise coordination of cellular events by biochemical activity architectures [7].
In addition to the detailed spatiotemporal readout of kinase activity offered by biosensors, molecular tools that enable similarly precise manipulation of kinase activities promise to further transform investigations into how compartmentalized signaling events regulate the behavior and function in living cells while maintaining the native biochemical activity architecture. For example, correlating the specific activation or inhibition of kinase activity at one organelle with downstream cellular events would help illuminate the distinct impact of local pools of kinase activity on cell function. Towards this end, great strides have been made in designing and engineering various molecular tools to enable spatiotemporally selective modulation of kinase activity. These include fully genetically encoded modulators such as inhibitory peptides, intrabodies, and direct optogenetic regulation of kinases, as well as genetically encodable chemical biology (i.e., chemogenetic) approaches. In this review, we highlight recent advances and strategies for modulating kinase activity in real time in single cells and emphasize the joint applicability of these tools with biosensors to concurrently measure and perturb kinase activities within live cells and elucidate the mechanisms underlying local kinase signaling.
Modulating endogenous kinases via genetically encoded binders
Genetically encoded binders are protein-based molecular tools that modulate the target kinase via direct binding to achieve inhibition. Here, we describe several techniques and designs which have been successfully deployed to regulate endogenous kinase targets.
Genetically encoded inhibitory peptides
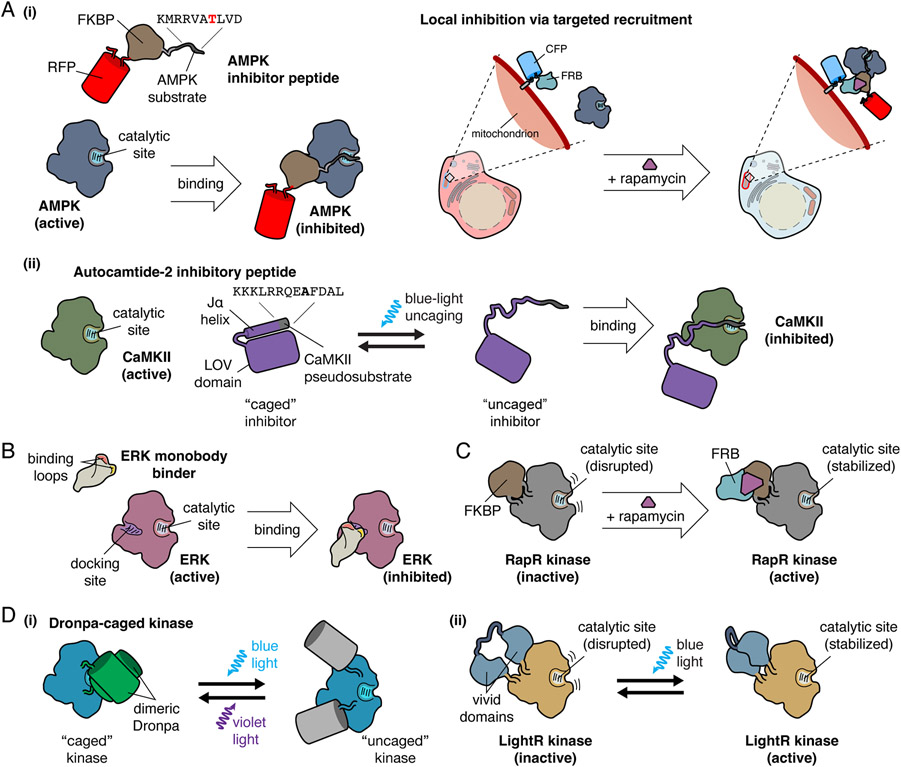
Within cells, kinase activity is often gated by substrate-like sequences found within intrinsic regulatory domains or accessory proteins. These pseudosubstrates bind the catalytic site and block phosphorylation of downstream substrates until upstream signals trigger a conformational change to promote activation [8-11]. Endogenous inhibitory proteins such as the protein kinase A (PKA) inhibitor (PKI) similarly utilize pseudosubtrates to regulate signaling via competitive inhibition [12]. The high specificity and selectivity of these peptides has inspired the development of genetically encoded inhibitory peptides, often based on pseudosubstrates or actual substrate sequences, as a general strategy for modulating local kinase signaling in living cells via targeted competitive inhibition. As with biosensors, spatial selectivity is achieved using localization sequences to target distinct subcellular compartments. This approach has been successfully utilized for over a decade to dissect spatial signaling via localized inhibition of several kinases, including PKA [13-15], AMP-activated protein kinase (AMPK) [16,17], calcium-calmodulin kinase II (CaMKII) [18,19], myosin light chain kinase [15], and Akt [20]. Modifying inhibitory peptide designs to incorporate chemically inducible dimerization (CID) or optogenetic switches can further allow for temporally precise manipulation of kinase activity. For example, we previously combined an AMPK-inhibitory peptide with CID to dynamically block mitochondrial AMPK activity [16] (Fig 1a, i), while Murakoshi et al more recently deployed an optically caged CaMKII inhibitory peptide to probe the timing of CaMKII activity during synaptic plasticity [19] (Fig 1a, ii).
Figure 1. Design of Genetically Encodable Molecular Tools.
A, Examples of competitive kinase inhibitory peptides. (i) AMPK inhibitory peptide is a substrate that binds to the active site of AMPK, functioning as a competitive inhibitor. Local inhibition of AMPK is accomplished through fusion of AMPK inhibitory peptide to FKBP and a fluorescent protein for visualization. CFP-FRB targeted to the mitochondria recruits the FKBP-fused inhibitor to the mitochondria upon the addition of rapamycin, resulting in targeted inhibition of mitochondrial AMPK activity. (ii) A CaMKII pseudosubstrate, termed autocamtide-2 inhibitory peptide, binds to the catalytic site of CaMKII, and functions as a competitive inhibitor. For temporal inhibition of CaMKII, autocamtide-2 inhibitory peptide was fused to the Jα helix of the LOV domain, which is caged in the absence of blue light. Upon blue light irradiation, the autocamtide-2 inhibitory peptide is uncaged, and competitively inhibits CaMKII.
B, ERK monobody binder is a monobody engineered from the fibronectin type III domain to specifically bind the docking site on ERK, preventing ERK interaction with downstream effectors, inhibiting ERK activity.
C, The RapR system achieves temporal regulation of kinase activity via chemically inducible dimerization through the insertion of FKBP, which disrupts the kinase catalytic site. Addition of rapamycin recruits FRB, which binds to FKBP and relieves active site inhibition, enabling kinase activity.
D, Optogenetic regulation of kinase activity by insertion of light-regulated domains into kinases. (i) Kinases can be caged by the inclusion of a dimeric Dronpa within the catalytic domain, where dimerization of Dronpa blocks kinase activity by sterically hindering the active site. Illumination with blue light triggers Dronpa photodissociation and uncaging of the kinase active site, which can be reversed by illumination with violet light. (ii) In LightR, regulation of kinases by light is accomplished through insertion of tandem vivid photoreceptors into the kinase domain backbone. In the dark state, the catalytic site is disrupted, but illumination with blue light induces vivid homodimerization and restoration of the catalytic site.
Intrabodies for regulating kinase activity
Tools to perturb kinase activity must have high specificity to achieve efficient target modulation. Antibodies, typically produced by B cells for defense against extracellular pathogens, are highly specific for their target antigens. Recently, antibodies engineered for intracellular use (intracellular antibodies, or intrabodies) have been derived from single-chain antibodies in camelids or cartilaginous fish (nanobodies) or from fibronectin type III domains (monobodies) [21,22]. Rather than being secreted, intrabodies are retained inside cells and can be engineered or evolved to recognize a wide range of intracellular targets for a variety of applications [23]. Their versatility and specificity make intrabodies an attractive option for genetically encoded perturbation of kinase signaling. Intrabodies have been developed that bind substrate docking sites, interaction interfaces necessary for activity, and regulatory sites [24-29]. Intrabodies can globally modulate kinase activity but subcellular targeting using localization sequences enables spatially selective regulation of kinase activity. For example, by locally inhibiting plasma membrane ERK using a targeted ERK monobody binder [29], Keyes et al were able to demonstrate that plasma membrane ERK signaling regulates Rac1 GTPase activity to control cell morphology and membrane protrusions (Fig 1b) [30].
Direct modulation of kinase activity
In contrast to inhibitory peptides and intrabodies, which modulate kinase activity by binding to the kinase of interest, several methods have been developed that directly manipulate the target kinase, either by regulating kinase localization or through allosteric modulation of kinase activity. These approaches allow for activation and/or inhibition of kinase activity for more precise tuning with minimal off-target effects. Here, we highlight techniques for directly regulating kinase activity using chemo- and optogenetic approaches.
Regulation of kinase activity using chemically inducible dimerization
CID systems, such as the rapamycin-induced interaction between FK506 binding protein (FKBP) and the FKBP-rapamycin binding (FRB) domain of mechanistic target of rapamycin complex 1 (mTORC1) [31], have long been popular tools for modulating signaling networks via the recruitment of regulatory or effector proteins [16,20,32,33]. More recently, Karginov et al developed the RapR kinase system, wherein kinase activity is allosterically regulated by rapamycin [34] (Fig 1c). In RapR, a small fragment of FKBP is inserted into a flexible loop in the kinase catalytic domain near the active site, effectively disrupting the active site conformation and preventing activity. Addition of rapamycin induces FKBP dimerization with FRB, which stabilizes the kinase active site, turning the kinase on. Initially applied to focal adhesion kinase, RapR has been adapted to regulate multiple kinases, including Src [35,36]. An alternative CID approach deployed by Diaz et al involves FKBP-FRB-driven fragment complementation of a target kinase. Specifically, Abl kinase was split into two fragments, with one half fused to FRB and the other to FKBP, such that rapamycin induced FRB-FKBP dimerization reconstitutes full-length, active Abl [37]. This system was used to characterize downstream targets of Abl. Deletion of an internal NLS in Abl sequestered the kinase to the cytoplasm and allowed for interrogation of cytoplasmic-specific Abl targets. As kinase domain folds are highly conserved [38], the authors were also able to apply this technique to other kinases, yielding split Src and split Akt [37]. Importantly, CID provides temporal control over kinase activity, as the kinase can only turn on in the presence of rapamycin, providing a “window” of activity. While the FKBP-FRB system is largely irreversible, alternative CID systems have been developed that could enable reversible manipulation of kinase activity [39,40]. Although not utilized in these examples, subcellular targeting of these tools via localization sequences can permit greater spatial control, enabling allosteric, spatiotemporal regulation of kinase activity.
Optogenetic regulation of kinase activity
Optogenetic approaches to reversibly regulate kinase activity have great appeal due to the high spatial and temporal precision that can be achieved. Kinase activity can be spatially regulated by selectively illuminating specific cellular regions, while localization sequences can also be added to optogenetic tools to achieve even greater selectivity. Thus, many optogenetic approaches have been developed to directly regulate kinases and their signaling cascades [41-43]. For example, the blue light-inducible interaction of Arabidopsis cryptochrome 2 photosensor (CRY2PHR) with itself or its binding partner cryptochrome-interacting basic helix-loop-helix (CIB1) [44] has been used to engineer light-controlled fibroblast growth factor receptor (FGFR) and Src activation [45,46]. Optogenetic switches can also be incorporated directly into a kinase backbone (Fig 1d). An approach developed by Zhou et al uses the light-induced dissociation of an engineered Dronpa FP pair (photodissociable dimeric Dronpa; pdDronpa) to reversibly cage and uncage kinase activity [47]. pdDronpa monomers are inserted into flexible loops N- and C-terminal to the kinase domain, such that dark-state pdDronpa dimerization sterically hinders the catalytic site. pdDronpa is photo-dissociated by blue light to expose the catalytic site and activate the kinase, which is reversed by violet light. pdDronpa was successfully used to regulate the MAPK signaling cascade in single cells and in vivo. Inspired by the RapR CID system, Shaaya et al engineered their LightR system based on the insertion of a tandem pair of vivid photoreceptor domains (VVD) from Neurospora crassa, which homodimerize upon blue-light illumination, into a flexible region of a kinase catalytic domain [43]. In the dark, LightR is monomeric, which disrupts the catalytic domain conformation, whereas homodimerization of LightR VVD under blue light restores the catalytic domain to activate the kinase. LightR has been applied to control several kinases, including Src, Abl, and BRAF. Liaunardy-Jopeace et al utilized an alternative approach, deploying non-natural amino acids to enable optical control over the Src-family kinase LCK [41]. A lysine critical to LCK enzymatic activity was replaced with a photocaged lysine, granting selective stimulation of LCK activity. While incorporation of non-natural amino acids is experimentally challenging, they potentially offer a more universal avenue for optically controlling kinase activity that relies less heavily on protein engineering.
Measuring and perturbing kinase activity in real time
Genetically encodable kinase modulators can be used with a variety of assays to study kinase signaling and downstream impacts on cellular function. One exciting application for these tools is combined monitoring of kinase activity dynamics using biosensors. This approach enables concurrent observation and modulation of kinase activity in the same cell. Such studies provide a unique and unparalleled view of compartmentalized signaling. Furthermore, using modulators in conjunction with biosensors enables assessment of single-cell kinase activity dynamics that could be missed using ensemble measurements. Here, we highlight applications combining genetically encodable kinase modulators with biosensors, emphasizing how pairing these tools enabled mechanistic investigation of spatiotemporal kinase signaling.
Inhibitory peptides unravel the regulation of nuclear mTORC1 activity
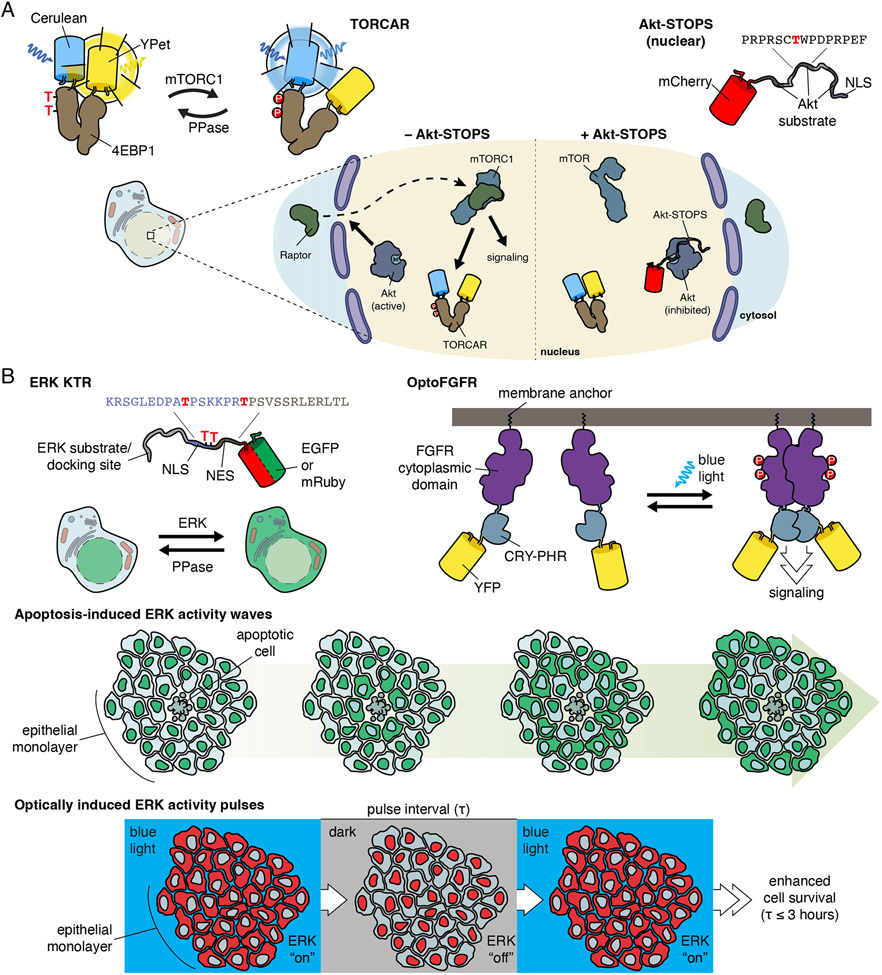
mTORC1 is a key metabolic regulator which senses and responds to a variety of intra- and extracellular cues, including growth factors, amino acids, and cellular energy status [48]. While canonical mTORC1 signaling is associated with the lysosome, mTORC1 has been detected at many subcellular locations. Using targeted versions of a genetically encoded mTORC1 activity reporter, TORCAR, which consists of the mTORC1 substrate eIF4E binding protein 1 flanked by a cyan and yellow FP FRET pair [49], we were previously able to measure growth factor-stimulated mTORC1 activity at various subcellular locations, including the nucleus. However, the mechanism(s) controlling this nuclear pool of mTORC1 were unclear. Nuclear mTORC1 activity could be due to either nuclear trafficking of active cytosolic mTORC1 or direct activation of nuclear-resident mTORC1. To probe these possibilities, we focused on Akt, which acts at multiple steps to promote growth factor-stimulated mTORC1 activity [50]. We hypothesized that a nuclear pool of mTORC1 would depend on nuclear Akt for activation and thus sought to selectively perturb nuclear Akt activity. We developed Akt Substrate-based Tandem Occupancy Peptide Sponge (Akt-STOPS) [20], a genetically encoded tool for location-specific Akt blockade, by fusing three tandem Akt substrate sequences derived from FOXO1 to mCherry. Targeting Akt-STOPS to the nucleus completely inhibited the growth factor-induced response measured using nuclear TORCAR, suggesting in situ regulation of nuclear mTORC1 activity by nuclear Akt. Indeed, further mechanistic dissection revealed that nuclear Akt activity controls the nuclear recruitment of an mTORC1 subunit to enable mTORC1 activity (Fig 2a). Combined subcellular application of both TORCAR and Akt-STOPS thus allowed us to precisely interrogate mechanisms of local mTORC1 regulation.
Figure 2. Examples of measuring and perturbing kinase activity in real time.
A, Using the FRET-based mTORC1 activity reporter TORCAR with local inhibition of Akt using the genetically encoded Akt inhibitor, Akt-STOPS, it was found that nuclear Akt signaling regulates nuclear mTORC1 activation through the recruitment of the mTORC1 subunit Raptor.
B, The ERK reporter, ERK KTR, was used to reveal that apoptotic cells induce ERK activity waves in epithelial monolayers. Through optogenetic regulation of ERK activity using OptoFGFR, it was found that these pulses of ERK activity enhance cell survival.
Optogenetic ERK regulation unveils a role for ERK in regulating apoptosis
As ERK signaling regulates cellular growth and proliferation, how short- and long-term ERK activity regulate cell behavior is an area of intense study. Previous studies using an ERK biosensor to investigate long-term ERK signaling in starved epithelial monolayers demonstrated that EGF treatment stimulated pulses of ERK activity over the course of days to regulate cell proliferation [51]. To better understand how these ERK activity pulses impact cell behavior, Gagliardi et al used ERK-KTR to examine ERK activity [52]. ERK-KTR contains the FP-tagged Elk1 ERK docking site fused to an NES and NLS whose effectiveness is regulated by phosphorylation [53]. Using ERK-KTR in monolayers of starved epithelial cells stimulated with EGF, Gagliardi et al observed waves of ERK activity that propagated outward from apoptotic cells. To identify the signaling events that induce these ERK activity waves, as well as their functional impact on cell health, Gagliardi et al used two optogenetic regulators, optoFGFR and optoRAF, to achieve growth factor-independent control of ERK activity. optoFGFR consists of FGFR fused to CRY2, where blue-light-induced CRY2 homodimerization induces FGFR dimerization, mimicking growth factor stimulation [45]. optoRAF uses the blue-light-induced interaction of CRY2 and plasma membrane-localized CIBN to recruit CRY2-tagged RAF to the plasma membrane and initiate ERK signaling independent of receptors [54]. OptoFGFR and optoRAF induced ERK activity pulses like those triggered by apoptotic cells, as visualized with ERK-KTR (Fig 2b). When ERK activity was stimulated with optoFGFR or optoRAF frequently over the course of hours, starvation-induced apoptosis was reduced, revealing a role for pulsatile ERK activity in promoting cell survival. By combining optogenetic modulators for different nodes of the ERK cascade with an ERK biosensor, these studies were able to demonstrate that ERK activity, independent of growth factor signaling, is sufficient to protect against apoptosis, illuminating the importance of ERK activity waves for cell growth and survival.
Future Perspectives
Genetically encodable modulators of kinase activity have allowed for investigation of mechanisms and functional roles of protein kinases in situ. Key to the success of these tools will be expanded application beyond a limited set of kinases. In this regard, further development of inhibitory peptides based on substrate or pseudosubstrate designs, aided by dedicated efforts to identify and characterize kinase substrate motifs [55], presents a straightforward opportunity to engineer genetically encodable modulators targeting more of the kinome [16,20]. Meanwhile, ongoing tool development efforts continue to yield new strategies for regulating cellular behavior. For example, Yu et al engineered the light-induced dimerization of a split intrabody, which they used to modulate the activity of gelsolin and β2-adrenergic receptor [56]. Other tools like peptide disrupters of kinase localization allow for the determination of the impacts of spatial kinase activity by relocalizing kinases [57-59]. We hope to see new approaches to modulate kinase activity using genetically encodable tools that take inspiration from these emerging designs, allowing for new strategies to regulate kinase activity.
Here, we differentiated between perturbation of endogenous kinases, including the use of binding peptides and intrabodies, and direct modulation of kinases, including the use of chemo- and optogenetic approaches. Regulation of endogenous kinases allows for kinase activity to be perturbed while still maintaining the biochemical activity architecture. Alternatively, using an engineered kinase where activity is allosterically regulated by CID or light allows for a biorthogonal approach to directly modulate and tune kinase activity independent of typical signaling cues. Use of these tools rely on overexpression systems, but as CRISPR/Cas9 gene editing becomes more accessible, knock-in of these tools will allow for regulation of cellular signaling without overexpression and maintain the integrity of the cellular biochemical activity architecture.
In this review, we highlighted efforts to combine application of genetically encodable modulators with genetically encoded biosensors to both observe and perturb kinase activity inside the same cell. This multiplexed approach provides a unique and powerful perspective on spatiotemporal regulation of cellular signaling and can be used to uncover mechanisms responsible for precisely tuning kinase signaling that might otherwise be missed. While many existing modulators, including inhibitory peptides, intrabodies, and CID systems are well-suited for use with biosensors, optogenetic systems pose a greater challenge given their wide reliance on blue light, which overlaps with the wavelengths used to visualize many popular biosensor classes [60]. Nevertheless, optogenetic tools are highly attractive for their high spatiotemporal precision. Better compatibility with biosensors could be achieved through incorporation of red-shifted FPs into biosensors by taking inspiration from red-shifted biosensors for small molecules [61,62], or by using recently developed red-shifted optogenetic systems [65]. KTRs, whose readout involves only FP translocation, are easily modified [52]. Ultimately, these efforts will enable a deeper understanding of the mechanisms governing cellular signaling.
Acknowledgements
This work was supported by the National Institutes of Health (NIH/NIGMS K12 GM068524 to D.L.S.; R35 CA197622, R01 H162302 and R01 DE030497 to J.Z.) and the Air Force Office of Scientific Research (FA9500-18-1-0051 to J.Z.).
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Zhang J-F, Mehta S, Zhang J: Signaling Microdomains in the Spotlight: Visualizing Compartmentalized Signaling Using Genetically Encoded Fluorescent Biosensors. Annu Rev Pharmacol 2021, 61:587–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang JZ, Mehta S, Zhang J: Liquid–liquid phase separation: a principal organizer of the cell’s biochemical activity architecture. Trends Pharmacol Sci 2021, 42:845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma K, D’Souza RCJ, Tyanova S, Schaab C, Wiśniewski JR, Cox J, Mann M: Ultradeep Human Phosphoproteome Reveals a Distinct Regulatory Nature of Tyr and Ser/Thr-Based Signaling. Cell Reports 2014, 8:1583–1594. [DOI] [PubMed] [Google Scholar]
- 4.Schmitt DL, Mehta S, Zhang J: Illuminating the kinome: Visualizing real-time kinase activity in biological systems using genetically encoded fluorescent protein-based biosensors. Curr Opin Chem Biol 2020, 54:63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenwald EC, Mehta S, Zhang J: Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem Rev 2018, 118:11707–11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X, Mehta S, Zhang J: Genetically Encodable Fluorescent and Bioluminescent Biosensors Light Up Signaling Networks. Trends Biochem Sci 2020, 45:889–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta S, Zhang J: Biochemical Activity Architectures Visualized–Using Genetically Encoded Fluorescent Biosensors to Map the Spatial Boundaries of Signaling Compartments. Accounts Chem Res 2021, 54:2409–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott JW, Ross FA, Liu JD, Hardie DG: Regulation of AMP-activated protein kinase by a pseudosubstrate sequence on the γ subunit. Embo J 2007, 26:806–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stratton MM, Chao LH, Schulman H, Kuriyan J: Structural studies on the regulation of Ca2+/calmodulin dependent protein kinase II. Curr Opin Struc Biol 2013, 23:292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor SS, Wu J, Bruystens JGH, Rio JCD, Lu T-W, Kornev AP, Eyck LFT: From structure to the dynamic regulation of a molecular switch: A journey over 3 decades. J Biological Chem 2021, 296:100746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baffi TR, Newton AC: Protein kinase C: release from quarantine by mTORC2. Trends Biochem Sci 2022, 47:518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knighton DR, Zheng J, Eyck LFT, Xuong N, Taylor SS, Sowadski JM: Structure of a Peptide Inhibitor Bound to the Catalytic Subunit of Cyclic Adenosine Monophosphate-Dependent Protein Kinase. Science 1991, 253:414–420. [DOI] [PubMed] [Google Scholar]
- 13.Sroubek J, McDonald TV: Protein Kinase A Activity at the Endoplasmic Reticulum Surface Is Responsible for Augmentation of Human ether-a-go-go-related Gene Product (HERG)*. J Biol Chem 2011, 286:21927–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tkachenko E, Sabouri-Ghomi M, Pertz O, Kim C, Gutierrez E, Machacek M, Groisman A, Danuser G, Ginsberg MH: Protein Kinase A Governs a RhoA-RhoGDI Protrusion-Retraction Pacemaker in Migrating Cells. Nat Cell Biol 2011, 13:660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi JJ, Wang H, Vilela M, Danuser G, Hahn KM: Manipulation of Endogenous Kinase Activity in Living Cells Using Photoswitchable Inhibitory Peptides. Acs Synth Biol 2014, 3:788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamoto T, Rho E, Sample V, Akano H, Magari M, Ueno T, Gorshkov K, Chen M, Tokumitsu H, Zhang J, et al. : Compartmentalized AMPK Signaling Illuminated by Genetically Encoded Molecular Sensors and Actuators. Cell Rep 2015, 11:657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunniff B, McKenzie AJ, Heintz NH, Howe AK: AMPK activity regulates trafficking of mitochondria to the leading edge during cell migration and matrix invasion. Molecular Biology of the Cell 2016, 27:2662–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bok J, Wang Q, Huang J, Green SH: CaMKII and CaMKIV mediate distinct prosurvival signaling pathways in response to depolarization in neurons. Mol Cell Neurosci 2007, 36:13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *19. Murakoshi H, Shin ME, Parra-Bueno P, Szatmari EM, Shibata ACE, Yasuda R: Kinetics of Endogenous CaMKII Required for Synaptic Plasticity Revealed by Optogenetic Kinase Inhibitor. Neuron 2017, 94:37–47.e5. Development of a light-gated CaMKII inhibitory peptide, allowing for the temporal role of CaMKII in long-term potentiation and memory formation.
- **20. Zhou X, Zhong Y, Molinar-Inglis O, Kunkel MT, Chen M, Sun T, Zhang J, Shyy JY-J, Trejo J, Newton AC, et al. : Location-specific inhibition of Akt reveals regulation of mTORC1 activity in the nucleus. Nat Commun 2020, 11:163–14. Here, the authors developed an inhibitory peptide for Akt, which when subcellularly targeted and paired with targeted biosensors for kinase activity, enabled the interrogation of an Akt-dependent mechanism for nuclear mTORC1 activity.
- 21.Marschall AL, Dübel S, Böldicke T: Specific in vivo knockdown of protein function by intrabodies. Mabs 2015, 7:1010–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sha F, Salzman G, Gupta A, Koide S: Monobodies and other synthetic binding proteins for expanding protein science. Protein Sci Publ Protein Soc 2017, 26:910–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher D, Helma J, Schneider AFL, Leonhardt H, Hackenberger CPR: Nanobodies: Chemical Functionalization Strategies and Intracellular Applications. Angewandte Chemie Int Ed Engl 2018, 57:2314–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wojcik J, Hantschel O, Grebien F, Kaupe I, Bennett KL, Barkinge J, Jones RB, Koide A, Superti-Furga G, Koide S: A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat Struct Mol Biol 2010, 17:519–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sha F, Gencer EB, Georgeon S, Koide A, Yasui N, Koide S, Hantschel O: Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains. Proc National Acad Sci 2013, 110:14924–14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann JK, Wood JF, Stephan AF, Tzanakakis ES, Ferkey DM, Park S: Epitope-Guided Engineering of Monobody Binders for in Vivo Inhibition of Erk-2 Signaling. ACS Chem Biol 2013, 8:608–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, Santana D, Gupta A, Jacobs M, Herrero-Garcia E, et al. : Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol 2017, 13:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zorba A, Nguyen V, Koide A, Hoemberger M, Zheng Y, Kutter S, Kim C, Koide S, Kern D: Allosteric modulation of a human protein kinase with monobodies. Proceedings of the National Academy of Sciences 2019, 116:13937–13942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teng KW, Tsai ST, Hattori T, Fedele C, Koide A, Yang C, Hou X, Zhang Y, Neel BG, O’Bryan JP, et al. : Selective and noncovalent targeting of RAS mutants for inhibition and degradation. Nat Commun 2021, 12:2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **30. Keyes J, Ganesan A, Molinar-Inglis O, Hamidzadeh A, Zhang J, Ling M, Trejo J, Levchenko A, Zhang J: Signaling diversity enabled by Rap1-regulated plasma membrane ERK with distinct temporal dynamics. eLife 2020, 9:e57410. Here, the authors used a localized inhibitory monobody against ERK in conjunction with biosensors to interrogate the role of plasma membrane ERK activity in cellular dynamics.
- 31.DeRose R, Miyamoto T, Inoue T: Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflügers Archiv - European J Physiology 2013, 465:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T: An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nature Methods 2005, 2:415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu HD, Kikuchi M, Dagliyan O, Aragaki AK, Nakamura H, Dokholyan NV, Umehara T, Inoue T: Rational design and implementation of a chemically inducible heterotrimerization system. Nat Methods 2020, 17:928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karginov AV, Ding F, Kota P, Dokholyan NV, Hahn KM: Engineered allosteric activation of kinases in living cells. Nat Biotechnol 2010, 28:743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karginov AV, Tsygankov D, Berginski M, Chu P-H, Trudeau ED, Yi JJ, Gomez S, Elston TC, Hahn KM: Dissecting motility signaling through activation of specific Src-effector complexes. Nat Chem Biol 2014, 10:286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *36. Klomp JE, Shaaya M, Matsche J, Rebiai R, Aaron JS, Collins KB, Huyot V, Gonzalez AM, Muller WA, Chew T-L, et al. : Time-Variant SRC Kinase Activation Determines Endothelial Permeability Response. Cell Chem Biol 2019, 26:1081–1094.e6. Here, the authors used chemically induced dimerization gated Src for temporal control over Src activity to identify a role for Src in endothelial barrier integrity.
- *37. Diaz JE, Morgan CW, Minogue CE, Hebert AS, Coon JJ, Wells JA: A Split-Abl Kinase for Direct Activation in Cells. Cell Chem Biol 2017, 24:1250–1258.e4. Development of a chemically induced dimerization split Abl kinase, which was used to identify subcellular targets of Abl.
- 38.Taylor SS, Kornev AP: Protein kinases: evolution of dynamic regulatory proteins. Trends in Biochemical Sciences 2011, 36:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu P, Calderon A, Konstantinidis G, Hou J, Voss S, Chen X, Li F, Banerjee S, Hoffmann J, Theiss C, et al. : A Bioorthogonal Small-Molecule-Switch System for Controlling Protein Function in Live Cells. Angewandte Chemie Int Ed 2014, 53:10049–10055. [DOI] [PubMed] [Google Scholar]
- 40.Feng S, Laketa V, Stein F, Rutkowska A, MacNamara A, Depner S, Klingmüller U, Saez-Rodriguez J, Schultz C: A Rapidly Reversible Chemical Dimerizer System to Study Lipid Signaling in Living Cells. Angewandte Chemie Int Ed 2014, 126:6838–6841. [DOI] [PubMed] [Google Scholar]
- 41.Liaunardy-Jopeace A, Murton BL, Mahesh M, Chin JW, James JR: Encoding optical control in LCK kinase to quantitatively investigate its activity in live cells. Nat Struct Mol Biol 2017, 24:1155–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leopold AV, Chernov KG, Verkhusha VV: Optogenetically controlled protein kinases for regulation of cellular signaling. Chem Soc Rev 2018, 47:2454–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaaya M, Fauser J, Zhurikhina A, Conage-Pough JE, Huyot V, Brennan M, Flower CT, Matsche J, Khan S, Natarajan V, et al. : Light-regulated allosteric switch enables temporal and subcellular control of enzyme activity. Elife 2020, 9:e60647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL: Rapid blue light induction of protein interactions in living cells. Nat Methods 2010, 7:973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim N, Kim JM, Lee M, Kim CY, Chang K-Y, Heo WD: Spatiotemporal Control of Fibroblast Growth Factor Receptor Signals by Blue Light. Chem Biol 2014, 21:903–912. [DOI] [PubMed] [Google Scholar]
- *46. Kerjouan A, Boyault C, Oddou C, Hiriart-Bryant E, Grichine A, Kraut A, Pezet M, Balland M, Faurobert E, Bonnet I, et al. : Control of SRC molecular dynamics encodes distinct cytoskeletal responses by specifying signaling pathway usage. J Cell Sci 2021, 134:jcs254599. Development of light-gated Src, which was used to decode spatiotemporal Src signaling networks regulating cellular adhesive site dynamics.
- **47. Zhou XX, Fan LZ, Li P, Shen K, Lin MZ: Optical control of cell signaling by single-chain photoswitchable kinases. Science 2017, 355:836–842. Here, the authors developed photoswitchable kinases, enabling temporal control over kinase activity across scales.
- 48.Kim J, Guan K-L: mTOR as a central hub of nutrient signalling and cell growth. Nat Cell Biol 2019, 21:63–71. [DOI] [PubMed] [Google Scholar]
- 49.Zhou X, Clister TL, Lowry PR, Seldin MM, Wong GW, Zhang J: Dynamic Visualization of mTORC1 Activity in Living Cells. Cell Rep 2015, 10:1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manning BD, Toker A: AKT/PKB Signaling: Navigating the Network. Cell 2017, 169:381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albeck JG, Mills GB, Brugge JS: Frequency-Modulated Pulses of ERK Activity Transmit Quantitative Proliferation Signals. Mol Cell 2013, 49:249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **52. Gagliardi PA, Dobrzyński M, Jacques M-A, Dessauges C, Ender P, Blum Y, Hughes RM, Cohen AR, Pertz O: Collective ERK/Akt activity waves orchestrate epithelial homeostasis by driving apoptosis-induced survival. Dev Cell 2021, 56:1712–1726.e6. Here, the authors used optogenetically-regulated FGFR and RAF alongside a biosensor for ERK activity to determine the functional role of ERK activity waves in epithelial monolayers.
- 53.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW: High-Sensitivity Measurements of Multiple Kinase Activities in Live Single Cells. Cell 2014, 157:1724–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aoki K, Kondo Y, Naoki H, Hiratsuka T, Itoh RE, Matsuda M: Propagating Wave of ERK Activation Orients Collective Cell Migration. Dev Cell 2017, 43:305–317.e5. [DOI] [PubMed] [Google Scholar]
- 55.Johnson JL, Yaron TM, Huntsman EM, Kerelsky A, Song J, Regev A, Lin T-Y, Liberatore K, Cizin DM, Cohen BM, et al. : A global atlas of substrate specificities for the human serine/threonine kinome. BioRxiv 2022, doi: 10.1101/2022.05.22.492882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *56. Yu D, Lee H, Hong J, Jung H, Jo Y, Oh B-H, Park BO, Heo WD: Optogenetic activation of intracellular antibodies for direct modulation of endogenous proteins. Nat Methods 2019, 16:1095–1100. Here, the authors developed a light-gated split-intrabody approach to regulate cellular activity with high spatiotemporal resolution.
- 57.Burns-Hamuro LL, Ma Y, Kammerer S, Reineke U, Self C, Cook C, Olson GL, Cantor CR, Braun A, Taylor SS: Designing isoform-specific peptide disruptors of protein kinase A localization. Proc National Acad Sci 2003, 100:4072–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lim CJ, Kain KH, Tkachenko E, Goldfinger LE, Gutierrez E, Allen MD, Groisman A, Zhang J, Ginsberg MH: Integrin-mediated Protein Kinase A Activation at the Leading Edge of Migrating Cells. Mol Biol Cell 2008, 19:4930–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Depry C, Allen MD, Zhang J: Visualization of PKA activity in plasma membrane microdomains. Mol Biosyst 2010, 7:52–58. [DOI] [PubMed] [Google Scholar]
- 60.Lee HN, Mehta S, Zhang J: Recent advances in the use of genetically encodable optical tools to elicit and monitor signaling events. Curr Opin Cell Biol 2020, 63:114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akerboom J, Calderón NC, Tian L, Wabnig S, Prigge M, Tolö J, Gordus A, Orger MB, Severi KE, Macklin JJ, et al. : Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci 2013, 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qian Y, Piatkevich KD, Larney BM, Abdelfattah AS, Mehta S, Murdock MH, Gottschalk S, Molina RS, Zhang W, Chen Y, et al. : A genetically encoded near-infrared fluorescent calcium ion indicator. Nat Methods 2019, 16:171–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mehta S, Zhang Y, Roth RH, Zhang J, Mo A, Tenner B, Huganir RL, Zhang J: Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nature Cell Biol 2018, 20:1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, Liu B, Hong I, Mo A, Roth RH, Tenner B, Lin W, Zhang JZ, Molina RS, Drobizhev M, et al. : An ultrasensitive biosensor for high-resolution kinase activity imaging in awake mice. Nat Chem Biol 2020, 118:11707–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaberniuk AA, Shemetov AA, Verkhusha VV: An optogenetic system based on bacterial phytochrome controllable with near-infrared light. Nat Methods 2016, 13:591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]