We read with interest the article entitled, ‘H3.3K27M mutation is not a suitable target for immunotherapy in HLA-A2+ patients with diffuse midline glioma’ (DMG).1 In this report, the investigators isolated an H3.3K27M-specific T cell receptor (TCR) from transgenic mice expressing a diverse human TCR repertoire and human HLA-A*02:01. They observed a high functional avidity of human T cells transduced with the TCR. They also tested another TCR (1H5) we reported.2 However, unlike our published study,2 the T cells transduced with either H3.3K27M-reactive TCR failed to recognize HLA-A*02:01+tumor cells that endogenously express H3.3K27M. The investigators used CRISPR/Cas9 editing to mitigate the mispairing of endogenous versus transgene TCR α- and β-chains. The investigators conclude that the H3.3K27M mutation is not a suitable target for H3.3K27M+ DMG, most likely due to insufficient epitope processing and presentation in HLA-A*02:01.
The investigators had different observations than those in our publication.2 They included HLA-A*02:01+U87 glioma cells transduced with full-length H3.3K27M cDNA. Although a different vector and transduction method may have been used, our study also used U87 cells expressing transgene-derived full-length H3.3K27M cDNA. While their liquid chromatography-tandem mass spectrometry (LC-MS/MS) failed to detect endogenously processed mutant H3.3-derived peptides in U87MG cells overexpressing full-length H3.3K27M cDNA, we reported the detection of such by LC-MS/MS in the HLA class I immunopeptidome of our U87 cells expressing the mutant cDNA.2
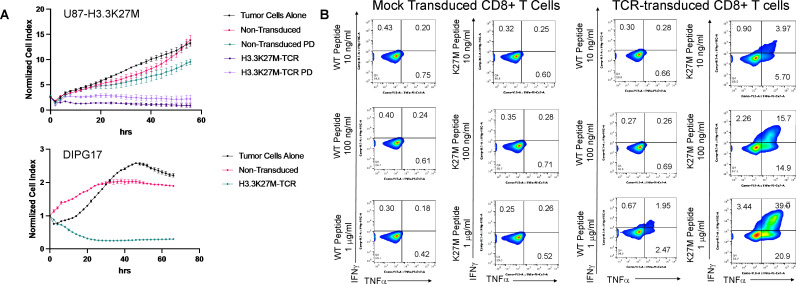
Accordingly, we demonstrated that human T cells transduced with a retroviral vector, which incorporates the TCR (clone 1H5) α-chains and β-chains as well as siRNA targeting constant regions of the endogenous TCR α- and β-chains, mediated an antigen-specific cytotoxic activity against the U87 cells expressing the full-length H3.3K27M cDNA.2 In addition to the lactate dehydrogenase assay data that we reported,2 we recently implemented a live cell imaging system (the xCELLigence system) and reproduced the same data (figure 1). The different observations with the U87 cell line may have been due to the different levels of the mutant H3.3 expression and the use of the retroviral TCR vector that also encoded the siRNA against endogenous TCR. In our hands, the siRNA-based disruption of endogenous TCR has been more effective than the CRISPR/Cas9-based system. Furthermore, these T cells could mediate antigen-specific killing of patient-derived HLA-A*02:01+H3.3K27M+DMG cells (figure 1). Nevertheless, we agree with the investigators that the endogenous presentation of the H3.3K27M epitope in HLA-A*02:01 may not be robust. We are currently evaluating whether standard-of-care treatment and resulting stress on histone and DNA (eg, ionizing radiation and chemotherapy) can enhance the presentation of the epitope.
Figure 1.
Functions of H3.3K27M-TCR-transduced T cells. (A) Cytotoxicity assays using a live cell imaging system. U87-H3.3K27M (upper panel) or HSJD-DIPG-0172 (lower panel) were seeded on laminin-coated plates and were rested for 24 hours to attain optimum cell index. Effector CD8+T cells were added at an E:T ratio of 5:1. U87-H3.3K27M cells (upper panel), were in the assay with control, non-transduced but similarly activated CD8+T cells were included. ‘PD’ refers to ‘process development’, because these cells were derived from GMP-grade manufacturing test cultures of the cells using Miltenyi’s Prodigy system. Cytotoxicity against glioma cells was monitored for 56 (upper panel) or 66 (lower panel) hours using the xCELLigence platform (Agilent). The experiment was performed at least twice with similar results and graphs represent means of triplicate cultures±SD. (B) Intracellular cytokine expression assays. T2 cells were plated at a density of 50,000 cells/well in 100 µL with the indicated concentrations of WT or H3.3K27M peptide. Aliquots of 50,000 control or TCR-transduced CD8+ cells were added to each well in 100 µL. Cells were harvested after 5 hours and stained for intracellular markers. TCR, T cell receptor; WT, wild-type.
There are additional indirect but supporting observations regarding the clinical relevance of the H3.3K27M (26-35) epitope. Around the same time as our report,2 Dr Michael Platten’s group at University of Heidelberg also described H3.3K27M (26-35) as a cytotoxic T lymphocyte (CTL) epitope.3 Importantly, in their studies, H3.3K27M-specific CTLs were exclusively detected in peripheral blood mononuclear cell samples derived from three of three patients with H3.3K27M-mutant gliomas after ex vivo expansion, indicating spontaneous H3.3K27M (26-35)-specific T-cell responses. Our published report independently described the same observations2 in our H3.3K27M-positive HLA-A*02:01 patient cohort. The spontaneous induction of the H3.3K27M (26-35)-specific CD8+T cell responses, observed in these two independent studies,2 3 strongly suggests that the epitope can be processed and presented by the immune system of HLA-A*02:01+H3.3K27M+DMG patients. Of course, non-tumor cells, such as DMG-infiltrating myeloid cells, may theoretically cross-present the DMG cell-derived H3.3K27M (26-35) epitope. However, the tumor microenvironment of DMG is known to have few inflammatory myeloid cells.4 Furthermore, although dural lymphatics may drain the antigens from the cerebral spinal fluid space to cervical lymph nodes, the draining route for protein antigens in the brain parenchyma has not been fully established. These considerations and observations still indicate that DMG cells can directly present the epitope.
The investigators mentioned that our H3.3K27M epitope vaccination study (PNOC007) did not improve the overall outcome in H3.3K27M+patients with DMG.5 However, in this study, the development of H3.3K27M-specific CD8+T cell response was associated with prolonged overall survival and progression-free survival of patients. We were keen to discern whether the observed H3.3K27M-reactive CD8+T cell responses merely reflected general immune competency, which is often compromised in patients with progressive tumors. Our multivariate analysis on bulk CD8+T cells strongly suggested this was not the case. Another concern is the possibility that patients with intrinsically slow-growing tumors may have received more vaccines than those with fast-growing tumors, thereby mounting higher vaccine-reactive responses. Based on our analyses, this does not appear to be the case, as the median number of vaccines preceding immunological responses among responders was the same as the median number of total vaccines among nonresponders. Taken together, our data support that the H3.3K27M-specific CD8+T cell responses may have provided clinical benefits.
Finally, the investigators report that T cells expressing the TCR derived from the 1H5 clone reacted with both mutant and wild-type (WT) peptides. In sharp contrast, our T cells transduced with the retroviral vector encoding the 1H5 TCR and siRNA against endogenous TCR common chains reproducibly produced significantly higher response levels to H3.3K27M than the WT peptide. We demonstrated multiple lines of data to support this. First of all, as the investigators noted, there is a striking difference in the binding affinity of the mutant vs WT peptide to HLA-A*02:01. We included a data set from in vitro binding assays2 in addition to the predicted affinity based on algorithms. Using a competitive binding-inhibition assay, we determined that the half-maximal inhibitory concentration (IC50) values for the mutant and WT peptides are 151 nM and >38 687 nM, respectively. Even if the 1H5 TCR recognizes the common amino acid sequence between the mutant and WT peptides, the marked difference in the HLA-binding capability between the two peptides should lead to remarkably different levels of reactivity. Indeed, although we did report detectable levels of IFN-γ response on T2 cells loaded with extremely high concentrations (~10 µg/mL) of WT peptide, the response was below background levels with lower concentrations of (1–100 ng/mL) WT peptide. In addition to the published data,2 we have now included a data set from our intracellular cytokine assays (figure 1). Our report also includes a dataset from Alanine scanning assays.2 We cocultured J.RT3-T3.5 cells transduced with the 1H5 TCR and evaluated them for IL-2 production in response to T2 cells loaded with a series of synthetic peptides, including the H3.3 mutant and WT peptide (10 µg/mL). Again, we showed a robust level of IL-2 production with the mutant peptide but merely background levels with the WT peptide.
In summary, while we agree with the investigators that the H3.3K27M (26-35) epitope may not be robustly presented by all HLA-A*02:01+DMG cells, our data and observations from both laboratory and clinical trials still support a role of this epitope in immunotherapy for HLA-A*02:01+patients with DMG. Further investigations are warranted to develop strategies to enhance the role of the epitope as a target.
Footnotes
Contributors: All authors equally contributed to writing this letter. BH generated the data set presented in figure 1.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: No, there are no competing interests.
Provenance and peer review: Not commissioned; internally peer reviewed.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1. Immisch L, Papafotiou G, Popp O, et al. H3.3K27M mutation is not a suitable target for immunotherapy in HLA-A2+ patients with diffuse midline glioma. J Immunother Cancer 2022;10:e005535. 10.1136/jitc-2022-005535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chheda ZS, Kohanbash G, Okada K, et al. Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy. J Exp Med 2018;215:141–57. 10.1084/jem.20171046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ochs K, Ott M, Bunse T, et al. K27M-mutant histone-3 as a novel target for glioma immunotherapy. Oncoimmunology 2017;6:e1328340. 10.1080/2162402X.2017.1328340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin GL, Nagaraja S, Filbin MG, et al. Non-Inflammatory tumor microenvironment of diffuse intrinsic pontine glioma. Acta Neuropathol Commun 2018;6:51. 10.1186/s40478-018-0553-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mueller S, Taitt JM, Villanueva-Meyer JE, et al. Mass cytometry detects H3.3K27M-specific vaccine responses in diffuse midline glioma. J Clin Invest 2020;130:6325–37. 10.1172/JCI140378 [DOI] [PMC free article] [PubMed] [Google Scholar]