Abstract
Introduction:
Rare genetic diseases affect millions of people worldwide. Most of them are caused by defective genes that impair quality of life and can lead to premature death. As genetic therapies aim to fix or replace defective genes, they are considered the most promising treatment for rare genetic diseases. Yet, as these therapies are still under development, it is still unclear whether they will be successful in treating these diseases. This study aims to address this gap by assessing researchers’ opinions on the future of genetic therapies for the treatment of rare genetic diseases.
Methods:
We conducted a global cross-sectional web-based survey of researchers who recently authored peer-reviewed articles related to rare genetic diseases.
Results:
We assessed the opinions of 1430 researchers with high and good knowledge about genetic therapies for the treatment of rare genetic diseases. Overall, the respondents believed that genetic therapies would be the standard of care for rare genetic diseases before 2036, leading to cures after this period. CRISPR-Cas9 was considered the most likely approach to fixing or replacing defective genes in the next 15 years. The respondents with good knowledge believed that genetic therapies would only have long-lasting effects after 2036, while those with high knowledge were divided on this issue. The respondents with good knowledge on the subject believed that non-viral vectors are more likely to be successful in fixing or replacing defective genes in the next 15 years, while most of the respondents with high knowledge believed viral vectors would be more successful.
Conclusion:
Overall, the researchers who participated in this study expect that in the future genetic therapies will greatly benefit the treatment of patients with rare genetic diseases.
Keywords: expert opinion, future, genetic therapies, rare genetic diseases, survey
Plain Language Summary
A global survey of researchers on the future of genetic therapies for rare genetic diseases
Rare genetic diseases are caused by defective genes that result from one or more mutations in the genome. Today, the therapeutic options for these diseases are limited, and there are approved treatments for about 5% of them. In the future, genetic therapies (a group of techniques developed to correct defective genes) are expected to revolutionize the treatment of rare genetic diseases. Although promising, most of these therapies are currently under development and have a long way to go before their efficacy and safety can be proved. The uncertainty surrounding this topic therefore means the success of genetic therapies in treating or curing rare genetic diseases is not yet assured. To address this knowledge gap, we surveyed 1430 researchers working in rare genetic diseases about the future of genetic therapies for the treatment of these diseases over the next 15 years. Most of them expected gene therapies to be the standard of care for rare genetic diseases before 2036 and to be able to cure them after this date. CRISPR-Cas9 was felt to be the gene editing approach that was most likely to succeed in fixing or replacing defective genes in the next 15 years. The respondents with high knowledge about gene therapies for the treatment of rare diseases believed gene therapies would have long-lasting effects before 2036, while those with good knowledge expected this to be the case only after 2036. The former believed in viral vectors and the latter in non-viral vectors to fix or replace defective genes in the next 15 years.
Introduction
Rare diseases are estimated to affect 3.5–5.9% of the world’s population. 1 The classification of a rare disease varies. In the European Union (EU), a disease is considered rare if it affects one person in 2000 (European Commission: ec.europa.eu/info/research-and-innovation/research-area/health-research-and-innovation/rare-diseases_en), whereas in the United States, it must affect fewer than 200,000 people to be rare (US Food & Drug Administration: fda.gov/patients/rare-diseases-fda). Six to seven thousand rare diseases are currently reported in the medical literature, and the causes of many are still unknown. Some are infections, cancers, and autoimmune diseases, but most are genetic in origin (Orphanet: orpha.net/consor/cgi-bin/Education_AboutRareDiseases.php?lng=EN).
Rare genetic diseases are caused by defective genes that result from one or more mutations in the genome. 2 The malfunction of one or more genes impairs quality of life and can lead to premature death.1,3 Today, the therapeutic options for these diseases are limited, 3 and there are approved treatments for only about 5% of them. 4 In the future, however, genetic therapies are expected to revolutionize the treatment of rare genetic diseases.5–7 The hope is that these therapies will reduce the symptoms of the diseases or even cure them.8–10
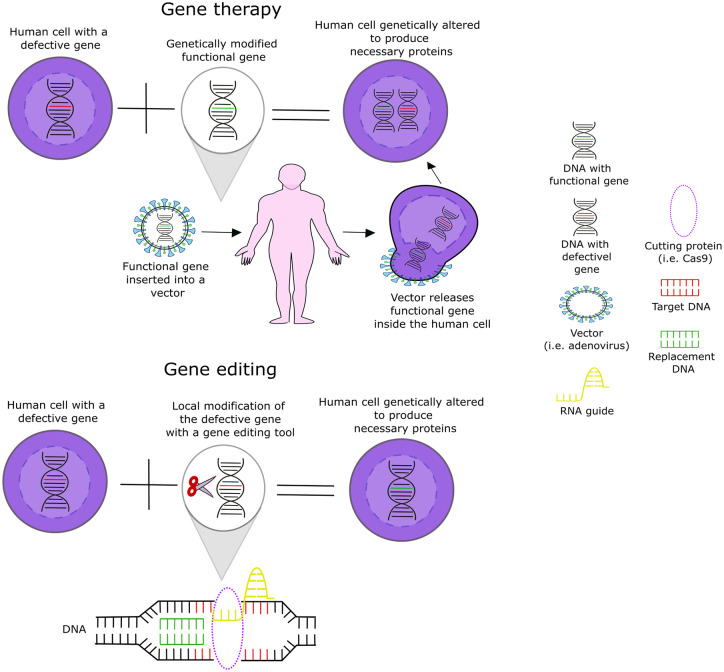
Genetic therapies are a group of techniques developed to correct defective genes. 2 Two of the techniques used are gene therapy and gene editing7,11 (Figure 1). Gene therapy adds a functional version of the defective gene inside the cells,2,6,12 whereas gene editing corrects or edits the defective gene by making a local modification in the genome to restore its function. Whether by adding a new functional gene or correcting a defective one, the target is to eliminate the cause of the disease 2 and thus achieve a cure.8,13–16
Figure 1.
Gene therapy and gene editing approaches.
The ideal genetic therapy should fix defective genes without activating oncogenes or causing off-target effects or immune or inflammatory reactions.17–19 Proto-oncogenes are genes responsible for cell growth and proliferation. When there is a change in the chromosome, they can suffer a mutation and become an oncogene. 20 When activated, oncogenes can promote uncontrolled cell growth, possibly leading to cancer. 21 The development of acute T cell lymphoblastic leukemia has been reported as an adverse effect in clinical trials of genetic therapies. 22 Off-target effects are caused by unintended modifications in the DNA that occur when the gene editing unintentionally modifies a non-target area. 23 Studies using animal models have shown that off-target effects resulting from CRISPR-Cas9 therapies can delete non-target genes such as vital or tumor suppressor genes. 21 These effects can be harmful to patients, reducing the safety of these therapies, and must be addressed before they progress through clinical trial phases. 18 Immune reactions to viral vectors are the most common adverse effects of genetic therapies. 24 An immune reaction occurs when the body recognizes an exogenous substance as harmful 25 and responds by creating an inflammatory reaction to fight it. 26 Modifying viral vectors to avoid immune reactions is still a challenge in genetic therapy research.26,27
In the United States, the Food and Drug Administration (FDA) can grant orphan drug status to rare disease drugs under development. Similarly, in the EU, the European Medicines Agency (EMA) can grant these developing drugs the status of orphan medicine. 24 Genetic therapies for the treatment of rare diseases can be granted these statuses 28 and then receive regulatory (e.g. differentiated clinical trial rules and fast-track approval), financial (reduced fees and taxes), and commercial (7 years of market exclusivity in the United States and 10 years in the EU) advantages. These advantages are expected to foster the transition from research to the market.24,29 Today, there are only two genetic therapies for rare diseases approved by the FDA: Zolgensma, for spinal muscular atrophy, and Luxturna, for leber congenital amaurosis, 30 both designated as orphan drugs. In addition to Luxturna and Zolgensma, the EMA has also approved and given orphan medicine status to Strimvelis and Zynteglo, the former for the treatment of severe combined immunodeficiency (ADA-SCID) and the latter for beta-thalassemia. 31
To have an idea of the current patent and clinical trial landscape related to genetic therapies for rare diseases, we searched for data in the Derwent Innovations Index and ClinicalTrials.gov (the queries we used are available in Supplemental Material I). We retrieved 1565 records of patents (with priority years between 1986 and 2022) and 30 ongoing clinical trials. According to the patent titles, the most frequent diseases addressed are cystic fibrosis (230), hemophilia (222), muscular dystrophy (114), and retinitis pigmentosa (58). In turn, the conditions with the highest number of ongoing clinical trials are Fabry disease (5), thalassemia (4), X-linked chronic granulomatous disease (3), and limb-girdle muscular dystrophy (3). Most of these clinical trials are related to gene therapy. One of them is for the biological compound ST-920, which is a gene therapy that uses a recombinant adeno-associated virus (AAV) vector to treat Fabry disease. An example of a clinical trial related to gene editing is the biological compound CTX001. It uses CRISPR-Cas9 technology and is being tested to treat thalassemia and sickle cell disease.
Once genetic therapies reach the market, they can be an alternative to currently available treatments for rare genetic diseases, such as enzyme replacements, autologous transplants, and antibody therapies.14,22,24,32 Yet, the expected high cost of genetic therapies combined with the high costs associated with their use – which involves the creation of new capabilities (such as new professional skills) and the adaptation of existing infrastructure (hospitals, medical centers, etc.), regulations, and health systems – may be a major barrier to their diffusion in the future. 33
Although promising, most genetic therapies are still under development and have a long way to go before their efficacy and safety can be proved.8,11,34,35 The uncertainty surrounding this topic therefore means the success of genetic therapies in treating or curing rare genetic diseases is not yet assured. This study aims to address this knowledge gap by anticipating the future of genetic therapies for the treatment of rare genetic diseases, considering the next 15 years. To do so, we performed a global cross-sectional web-based survey of researchers of rare genetic diseases who recently authored peer-reviewed articles indexed in the Web of Science Core Collection (WoS).
Some studies have already tried to anticipate the potential of genetic therapies for rare genetic diseases. Most are literature review articles reporting on genetic therapy research for a specific rare genetic disease, such as hemophilia, 36 cystic fibrosis,17,37 spinal muscular atrophy, 38 retinitis pigmentosa, 39 achromatopsia, 40 Bardet–Biedl syndrome, 11 and Leigh syndrome. 41 Others have investigated their potential in other areas, such as cancer,42–44 Parkinson’s disease, 45 Alzheimer’s disease, 45 and immunodeficiencies.21,43 Therefore, our study contributes to the current knowledge by providing a more comprehensive and long-term perspective on the future drawn from the opinions of more than 1400 researchers of rare genetic diseases from around the world.
Methods
Literature review and questionnaire
To conduct this global cross-sectional web-based survey of researchers of rare genetic diseases, we first performed a literature review of recent review articles on genetic therapies for the treatment of rare diseases indexed in the WoS. This literature review allowed us to identify the relevant topics related to the future of the study subject and then develop the survey questionnaire. Table 1 shows the strategies used to search the WoS in May 2021. We restricted the search to review articles published recently (2016 to May 2021) in journals of science using the Science Citation Index Expanded (SCI-EXPANDED). Strategies 2 and 4 searched the titles, abstracts, and keywords of review articles and used descriptors related to rare diseases (Strategy 2) and genetic therapies (Strategy 4) from the Medical Subject Headings (MeSH), US National Library of Medicine (MeSH: ncbi.nlm.nih.gov/mesh). Strategy 1 searched only the titles of review articles and used descriptors related to rare genetic diseases collected from Orphanet (Orphanet: orphadata.org/cgi-bin/index.php). Orphanet’s database Orphadata provides XML schemas with its own classification of rare diseases. This classification is based on scientific publications periodically reviewed by experts. 1 We collected the descriptors for search Strategy 1 from the XML schema ‘Rare Genetic Diseases’ (Orphanet: orphadata.org/cgi-bin/rare_free.html). In total, 6514 terms related to rare genetic diseases were retrieved: 1320 disease groups, 4429 diseases, and 765 disease subtypes (Supplemental Material I).
Table 1.
Search strategies for identifying review articles.
| Set | Search strategies | Results |
|---|---|---|
| 5 | 4 AND 3 | 486 |
| 4 | TS = (‘Gene* Therap*’ OR ‘DNA Therap*’ OR ‘RNAi Therap*’ OR
‘RNAi-Based Therap*’ OR ‘Gene* Repair*’ OR ‘Mutation
Repair*’ OR ‘Gene* Correction*’ OR ‘Gene* edit*’) AND
DOCUMENT TYPES: (Review) Indexes = SCI-EXPANDED, Timespan = 2016–2021 |
5399 |
| 3 | 2 OR 1 | 8518 |
| 2 | TS = (‘rare genetic disease*’ OR ‘rare disease*’ OR ‘orphan
genetic disease*’ OR ‘orphan disease*’) AND DOCUMENT TYPES:
(Review) Indexes = SCI-EXPANDED, Timespan = 2016–2021 |
1957 |
| 1 | TI = (#) AND DOCUMENT TYPES:
(Review) Indexes = SCI-EXPANDED, Timespan = 2016–2020 |
6829 |
#Supplemental Material I.
The 486 records of review articles identified in search Strategy 5 were imported into the data/text mining software VantagePoint 11.0, where, after reading their titles and abstracts, 214 records were pre-selected. Next, we generated a list of their Digital Object Identifiers (DOIs) and imported it into the Citavi 6.8 reference management software, where we conducted the literature review and managed the references. Then, we downloaded the full articles and, after reading them, selected the 66 references used in the literature review that led to the survey questionnaire.2,5–19,22,25,32,34–37,39–41,46–85
We structured the questionnaire in six parts (see questionnaire in Supplemental Material II). The first introduced the survey and informed the respondent about data collection, voluntary participation, informed consent, and anonymization of results. The second qualified/disqualified respondents for the survey according to their self-assessed level of knowledge about genetic therapies for the treatment of rare genetic diseases. High-knowledge respondents (HKRs), good-knowledge respondents (GKRs), and some-knowledge respondents (SKRs) were qualified for the survey, while no-knowledge respondents (NKRs) were disqualified and did not answer the questionnaire. The third part asked respondents to report which of the 33 groups of the Orphanet classification of rare genetic diseases (Orphanet: orpha.net/consor/cgi-bin/Disease_Classif.php?lng=EN&data_id=156&PatId=13071&search=Disease_Classif_Simple&new=1) was most closely related to their area of expertise.
The fourth part consisted of five questions about the future of genetic therapies in the treatment of rare genetic diseases. Three of these were statements about the future, for which the respondents were asked to indicate the period within which they believed they would likely occur. The response options were as follows: likely before 15 years, likely after 15 years, unlikely, and unknown. The statements referred to the likelihood of genetic therapies (1) becoming the standard of care for rare genetic diseases, (2) having long-lasting effects, not requiring repeated interventions, and (3) leading to a cure for these diseases. The two other questions asked respondents (1) to report which of the two types of vectors (viral and non-viral) would be most likely to succeed in fixing or replacing defective genes, and (2) to rank (on a scale of 1 to 4, where 1 = most likely and 4 = least likely) four main approaches to fixing or replacing defective genes: transcription activator-like effector nucleases (TALENs), zinc finger nucleases (ZFNs), meganucleases (MNs), and clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9 (CRISPR-Cas9). The ranking was based on the weighted average of the respondents’ individual classifications, assigning Weight 4 for the approach ranked first, Weight 3 for second place, Weight 2 for third place, and Weight 1 for fourth place.
The fifth and sixth parts were optional and were not considered for calculating the number of completed questionnaires, and so did not affect the survey’s confidence level and margin of error. The fifth part allowed respondents to share comments, criticisms, suggestions, and so on. The last part consisted of six demographic questions: (1) rare disease expertise, (2) education level, (3) experience in the field, (4) place of work, (5) professional occupation, and (6) region of residence.
Identifying and collecting respondents in scientific publications
The survey respondents were authors of review articles related to genetic therapies for the treatment of rare diseases indexed in WoS. They were identified following the search strategies depicted in Table 2. Strategies 1 and 2 are the same as in Table 1 except for the addition of articles and a shorter period (2018 to June 2021). The search was conducted in June 2021 and retrieved 43,017 records of articles and review articles (Strategy 3), which were then imported into VantagePoint 11.0, where we identified 48,796 unique author emails. We then imported a CSV file containing the emails, author names, and title of publications into our Python-based author name and email-linking software, which linked 89.13% of the emails to their owners. This procedure allowed us to forward personalized emails to most respondents.
Table 2.
Search strategies for identifying respondents.
| Set | Search strategies | Results |
|---|---|---|
| 3 | 2 OR 1 | 43,017 |
| 2 | TS = (‘rare genetic disease*’ OR ‘rare disease*’ OR ‘orphan
genetic disease*’ OR ‘orphan disease*’) AND DOCUMENT TYPES:
(Article OR Review) Indexes = SCI-EXPANDED, Timespan = 2018–2021 |
5718 |
| 1 | TI = (#) AND DOCUMENT TYPES: (Article OR
Review) Indexes = SCI-EXPANDED, Timespan = 2018–2021 |
38,312 |
#Supplemental Material I.
Survey procedure, ethics, and limitations
The list of respondents containing all 48,796 emails was imported into the professional online survey platform SurveyMonkey, where we prepared the questionnaire and conducted the survey. After importing, the number of valid emails was reduced to 46,921 because of 1215 bounced and 660 opt-out emails.
We validated the questionnaire through a pilot study with a random sample of 2363 emails (about 5% of the total). After the invitation email was sent, the questionnaire was available for completion for 8 days. During this period, up to three reminder emails were sent to those who did not answer the questionnaire. Before answering the questionnaire, respondents were informed the following: (1) the study was for research purposes, (2) sensitive data would not be collected, (3) data collected would be anonymized in the results, (4) participation would be voluntary, and (5) informed consent would be given by responding to the online questionnaire. The 90 researchers who participated in the pilot study did not report problems with the questions or the survey design or leave any comments that prompted changes to the questions. As the survey proper used the same questionnaire and administration procedure as the pilot, data collected in the pilot were included in the survey results. The pilot and the survey proper were conducted in July 2021. This study follows Clauses I and V of Article 1 of Brazilian Resolution number 510, dated 7 April 2016, which exempts this type of study from registration or evaluation by a research ethics committee (Official Federal Gazette: in.gov.br/materia/-/asset_publisher/Kujrw0TZC2Mb/content/id/22917581).
The methods employed followed previous future-oriented web-based surveys.86–92 As such, it had the same limitations. One limitation relates to the identification of respondents from scientific articles. As a consequence, the sample was predominantly formed of professors and researchers affiliated to universities and research organizations, limiting respondent variability. In addition, because they were experts invested in the technologies they studied and/or helped develop, they may be optimistic about the future of these technologies, including how successful these technologies may be and how long it will take for success to be achieved. Accordingly, the results of this survey may be subject to a higher degree of optimism bias than surveys with greater respondent variability. Experts are nevertheless the most qualified to anticipate future developments of new and emerging technologies, and are widely regarded as a reliable source of information for research surveys. Another limitation is related to the respondents’ self-attributed level of knowledge because it was not possible to evaluate a posteriori whether their level of knowledge actually corresponded to the level they reported in the questionnaire. Yet, as they were all authors of recently published peer-reviewed scientific articles on the subject, the chance of the survey gathering the opinions of non-knowledgeable respondents was low.
Statistical analysis of the sample
In this study, the population was the number of emails sent to researchers working with rare genetic diseases (43,921). Significance was set at 95% [so the margin of error (e) was 5% (observed power = 0.95)]. Applying Slovin’s equation, the minimum required sample size was 396.38. Although some SKR qualified for the survey and answered the questionnaire, we chose to analyze only the answers of the respondents with the highest levels of knowledge (HKRs and GKRs). As we will see below, their responses totaled 1430, yielding a sample size that was more than large enough to generate consistent results.
We applied two nonparametric tests to analyze the sample distribution: the Shapiro–Wilk test and the Kolmogorov–Smirnov test with Lilliefors correction. We used the Shapiro–Wilk test because it requires no prior knowledge of the data distribution and the Kolmogorov–Smirnov test because it is one of the most widely used to compare two samples. The Lilliefors correction compares the data with the reference mean and variance of a normal distribution, allowing the data to be compared with a normally distributed sample. As such, the Lilliefors correction serves to make the nonparametric Kolmogorov–Smirnov test a normal distribution test. 93
The results of the Shapiro–Wilk and Kolmogorov–Smirnov tests showed that the sample does not follow a normal distribution (Supplemental Material I). Thus, we used three nonparametric tests to analyze the data collected in the survey: the binomial test, the Wilcoxon signed-rank test, and the Mann–Whitney U test. The binomial test is used when there are two distinct dichotomous groups within the sample. It was used to analyze whether the sample follows a 50% distribution for each group of respondents (HKRs and GKRs). This test shows whether one group overlaps the other and whether a predominant group influences the results. The Wilcoxon signed-rank test was used to assess the standard response of the respondents. This test ranks responses in ascending order (i.e. 1 for the observation with the fewest responses, 2 for the observation with the second fewest responses, etc.). Based on this classification, we analyzed the median of the sample to assess whether there was a statistically significant pattern of responses. As there were two distinct groups of respondents, we used the Mann–Whitney U test to assess whether their level of knowledge interfered with the results: that is, whether the responses followed a statistically homogeneous pattern. All statistical analyses were performed using IBM-SPSS Statistics 26, and the complete results are available in Supplemental Material I.
Results
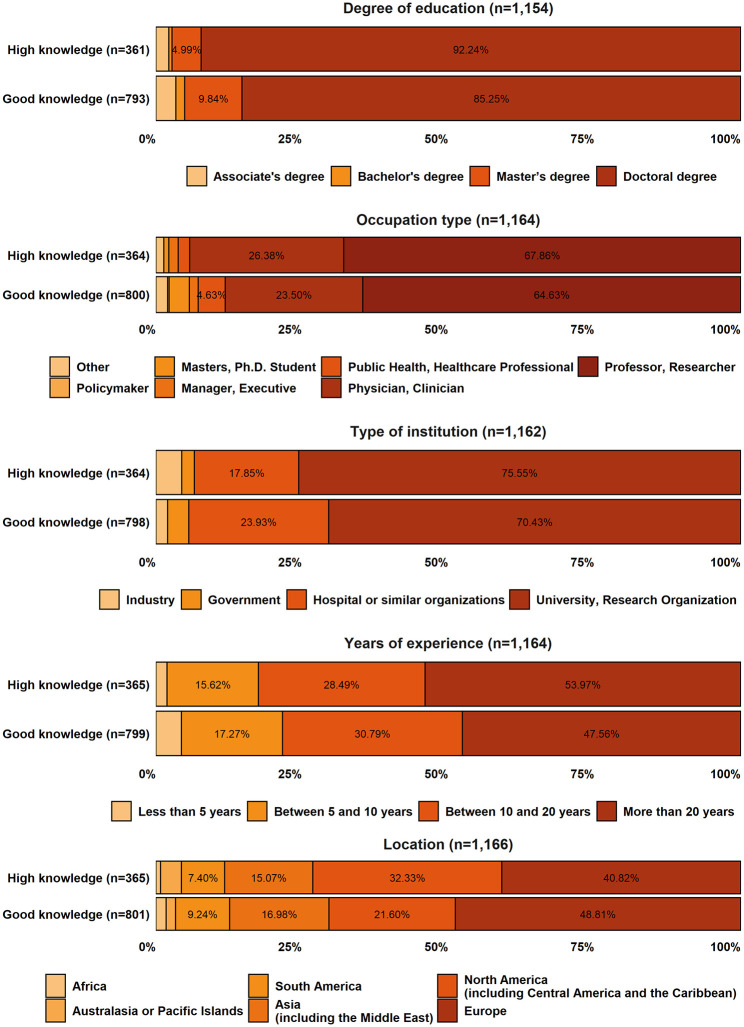
Of the 46,921 researchers invited, 2977 agreed to participate in the study, meaning a response rate of 6.34%. The breakdown of the respondents by knowledge group was as follows: 14.75% HKRs, 33.29% GKRs, 44.98% SKRs, and 6.99% NKRs. As mentioned in the section ‘Methods’, SKRs were not included in the results reported here (but their responses are available in Supplemental Material I), and the NKRs were disqualified from the survey. The HKRs’ and GKRs’ responses totaled 1430, 72.94% of which were fully completed. This gives us a representative sample with a 95% confidence level and a margin error of 2.6%. Figure 2 shows the demographics of the HKRs and GKRs (percentages less than 4.5% are available in Supplemental Material I).
Figure 2.
Demographics of the respondents.
Descriptive analysis
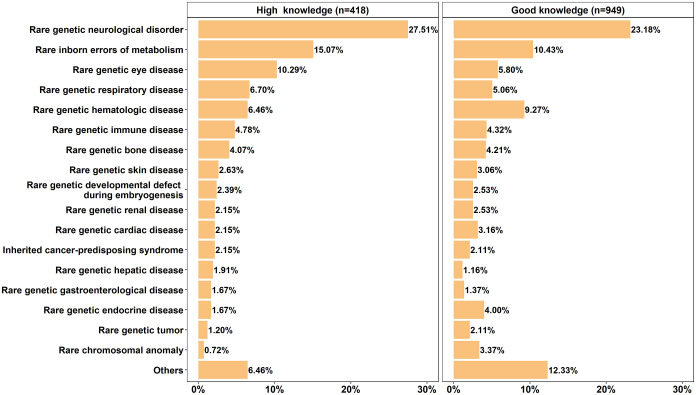
The Orphanet groups of rare genetic diseases representing the respondents’ expertise are depicted in Figure 3 (only groups with a frequency of at least 1.5% in at least one of the two knowledge levels). From the 33 Orphanet groups, only sea blue histiocytosis was not selected by any respondent. The group ‘rare genetic neurological disorder’ had the highest frequency among HKRs and GKRs (27.51% and 23.18%, respectively). The group ‘rare inborn errors of metabolism’ was the second highest (15.07% and 10.43% for the HKRs and GKRs, respectively). ‘Rare genetic eye diseases’ ranked third among the HKRs (10.29%), while for the GKRs (9.27%), the third most representative group was ‘rare genetic hematologic disease’.
Figure 3.
Distribution of respondents among Orphanet groups of rare genetic diseases, according to their research expertise.
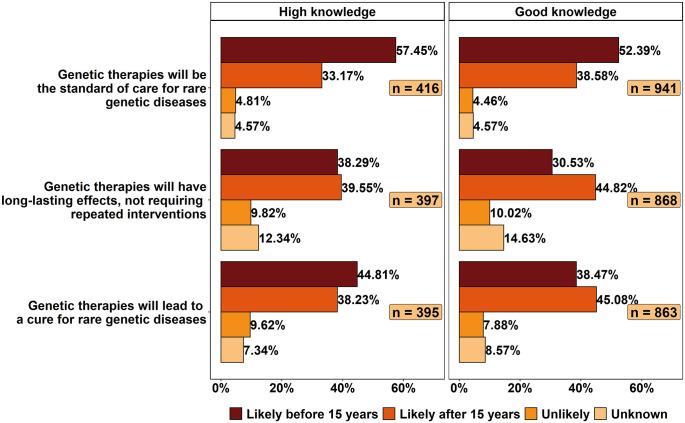
Most of the respondents (57.45% of HKRs and 52.39% of GKRs) expected genetic therapies to be the standard of care for rare genetic diseases in less than 15 years (Figure 4). Slightly more HKRs expected genetic therapies to have long-lasting effects after 2036 (39.55%) than before it (38.29%). The GKRs’ opinions on this topic were more divided, with 44.82% expecting long-lasting effects to be achieved only after 2036 and 30.53% expecting this to happen before 2036. The HKRs appeared to be slightly more optimistic than the GKRs about the likely time frame in which genetic therapies would be able to cure rare genetic diseases, as 44.81% of them expect this to happen within 15 years (versus 38.47% for the GKRs). Nonetheless, both groups were largely in agreement (more than 80%) that this was likely to occur sometime in the future.
Figure 4.
Likelihood of genetic therapies becoming standard of care, having long-lasting effects, and curing rare genetic diseases.
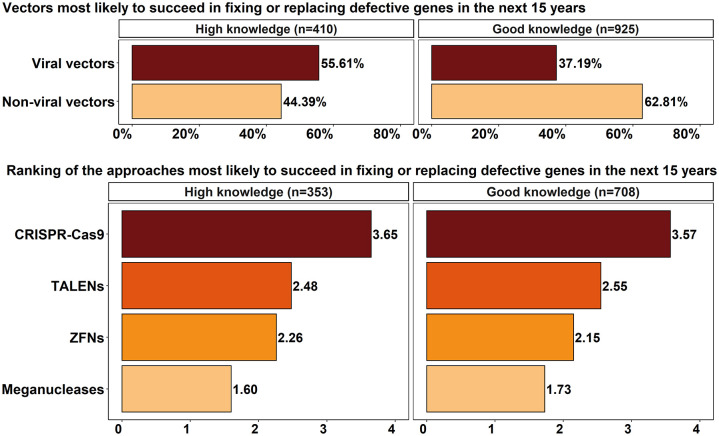
The majority of the HKRs (55.61%) believed viral vectors were the most likely to succeed in fixing or replacing defective genes in the next 15 years, while most of the GKRs (62.82%) chose non-viral vectors (Figure 5). In turn, the opinions of the HKRs and GKRs were similar regarding the likelihood of CRISPR-Cas9, TALENs, ZFNs, and meganucleases succeeding in fixing or replacing defective genes in the next 15 years. Although there were some differences in the values shown in the ranking, the order of preference of the HKRs and GKRs was the same, with CRISPR-Cas9 being the most likely to succeed and meganucleases the least likely.
Figure 5.
Likelihood of success in fixing or replacing defective genes in the next 15 years.
Statistical analysis
The binomial test showed that the sample was not homogeneously distributed, with a predominance of GKRs. For the questions that the Mann–Whitney U test showed that the level of knowledge influenced the results, the frequency distribution of responses suggests the following: (1) the GKRs believed that genetic therapies were likely to have long-lasting effects after 2036 (44.8%), while HKRs were divided as to how long it might take (38.3% before and 39.5% after 15 years); (2) the HKRs were more positive about the likely success of viral vectors (55.61%), while the GKRs’ preference was for non-viral vectors (62.81%); and (3) the HKRs expect CRISPR-Cas 9 to be the most likely to fix or replace defective genes in the next 15 years (81.8%) and did not expect meganucleases to achieve this in the same time frame (58.6%). Finally, the Wilcoxon signed-rank test showed that genetic therapies (1) will likely be the standard of care for rare genetic diseases before 15 years; (2) are likely to have long-lasting effects after 2036, not requiring repeated interventions; and (3) will likely lead to a cure for rare genetic diseases after 15 years. This test also shows that viral vectors are more likely to successfully fix or replace defective genes than non-viral vectors.
Discussion
There are no approved treatments for approximately 95% of rare diseases. 94 About 30% of patients with rare diseases die before their fifth birthday, and therapeutic options are limited to symptom control and comfort care. 4 For the 5% of rare diseases with approved treatments, the therapeutic options include small-molecule drugs, protein or enzyme replacement, antibody therapies, autologous transplants, and, more recently, genetic therapies.24,29,94
Genetic therapies began to be developed in the last quarter of the 20th century and since then have been a source of hope for patients with rare genetic diseases. 69 Their target is to either replace or restore the function of a gene or to suppress the action of a pathogenic gene. 24 By replacing, removing, or silencing the genetic code causing the disease, they are expected to be a solution for rare monogenic genetic diseases. 28
Globally, about four genetic therapies for rare genetic diseases have been approved and marketed. 28 Yet, most of our survey respondents believed genetic therapies would be the standard of care for rare genetic diseases before 15 years. Glybera was the first genetic therapy product to enter the market. It was approved for use in Europe in 2012 to treat lipoprotein lipase deficiency, a rare genetic disease. It was, however, withdrawn from the market in 2017, before obtaining FDA approval for use in the United States. 48 The withdrawal was not related to safety issues; it occurred because it was an extremely expensive treatment that did not offer long-lasting benefits to patients. 95 Also in 2017, Luxturna became the first genetic therapy approved by the FDA to treat leber congenital amaurosis, a rare genetic disease. 96 In 2019, another genetic therapy was approved by the FDA: Zolgensma for spinal muscular atrophy. 96
Luxturna and Zolgensma are single-dose treatments 96 designed to induce long-lasting curative effects. 30 Yet, research with animal models has indicated that long-lasting therapies without repeated interventions may not be possible for all monogenic diseases. 97 These studies suggest that the effects of genetic therapy last when a chromosomal integration between the functional gene and the patient’s cells is established. 97 While most of the GKRs to our survey expected genetic therapies to have long-lasting effects and not require repeated interventions only after 2036, the HKRs were divided about how long it would take. Long-lasting genetic therapies could increase the quality and expectancy of patients’ lives71,74 and amortize the costs. 83
Gene therapy and gene editing promise to go beyond controlling symptoms to actually fix or replace the source of the problem, the defective gene.2,6 Thus, genetic therapy is expected to lead to a cure for most monogenic disorders.9,10,13,14,16,32,54 According to most of our respondents, a cure for rare genetic diseases via genetic therapies is not likely to occur in less than 15 years. This expectation seems reasonable given the challenge of curing these diseases and the current stage of development of genetic therapies. Today, for example, researchers are still trying to identify the best vectors and approaches to fix or replace defective genes.22,53,77
The effectiveness of gene therapy – which uses a vector to deliver a functional gene into the cell – is directly related to the vectors’ ability to transfer the gene without activating oncogenes or generating immune or inflammatory reactions.15,17 Activation of oncogenes has been reported in clinical trials of gene therapies in which patients developed leukemia related to the viral vector used in the treatment, in this case retrovirus.2,53 For its part, immune and inflammatory reactions may reduce the effectiveness of the treatment and even make new attempts impossible because the patient’s immune system may start recognizing and rejecting the vector.17,32
While viral vectors such as AAV, lentivirus, and retrovirus may cause immune or inflammatory reactions, they are widely used in gene therapy because they are very effective in delivering the functional gene into the cell.35,46 For example, Luxturna and Zolgensma, the FDA-approved gene therapies for genetic diseases, are both AAV-based. 48 Compared with viral vectors, the therapy with non-viral vectors such as liposomes, electroporation, plasmids, peptides, and mRNA are less likely to cause immune and inflammatory reactions, activate oncogenes, or generate off-target effects, and they are also low cost. They are, however, less effective than viral vectors in gene transfer.7,19
For gene editing, the type of vector is also relevant as this is what is responsible for delivering the nuclease safely inside the cell. 98 The first CRISPR-based therapy to enter clinical trials uses an AAV5 to deliver a Cas9 enzyme, which mutates the CEP290 gene, the cause of leber congenital amaurosis, a rare genetic disease. 99 However, AAV vectors have a limited packaging capacity and may not be appropriate for carrying larger nucleases like some CRISPR-Cas systems. 23 Lentivirus vectors are an alternative as they offer a higher packaging capacity, yet they have a higher propensity to generate off-target effects. 23 Non-viral vectors have a higher packaging capacity and are more tolerable by immune systems than viral vectors, but despite these advantages they have been used less than viral vectors in gene editing research. 100 Overall, the survey’s respondents had different expectations about which vectors would be better for fixing or replacing defective genes in the next 15 years. The HKRs believed viral vectors were the most likely and the GKRs thought non-viral vectors were more promising.
Gene editing techniques are still at the clinical trial stage, but their potential to replace, remove, or add fragments to DNA to correct a defective gene has already been demonstrated in animal models. 47 Two examples are SB-FIX and CTX001. The former is a candidate therapy that uses ZFNs and is being tested for hemophilia, 13 and the latter is a candidate therapy that uses CRISPR-Cas9 and is being tested for thalassemia and sickle cell disease. 49 CRISPR-Cas9 is considered the most promising gene editing tool,22,101 which is in line with the respondents’ expectations. Compared with ZFNs, TALENs, and meganucleases, CRISPR-Cas9 is less complex, more accurate, and costs less.47,63 Yet, the other approaches are less likely to generate off-target effects. 76
Conclusion
This article presented the opinions of 1430 researchers working with rare diseases from around the world on the future of genetic therapies to treat rare genetic diseases. Overall, they expect genetic therapies to be the standard of care for rare genetic diseases before 2036 and to cure them after this period. They also believe CRISPR-Cas9 is the most likely gene editing approach to succeed in fixing or replacing defective genes in the next 15 years. In turn, while the respondents with a good level of knowledge believe genetic therapies will have long-lasting effects after 2036, those with high knowledge are divided on this. Respondents with good and high levels of knowledge also have different opinions on the most likely delivery method for fixing or replacing defective genes in the next 15 years. The former believes in non-viral vectors and the latter in viral vectors.
Supplemental Material
Supplemental material, sj-docx-1-trd-10.1177_26330040221100840 for Future of genetic therapies for rare genetic diseases: what to expect for the next 15 years? by Luiza Amara Maciel Braga, Carlos Gilbert Conte Filho and Fabio Batista Mota in Therapeutic Advances in Rare Disease
Supplemental material, sj-pdf-2-trd-10.1177_26330040221100840 for Future of genetic therapies for rare genetic diseases: what to expect for the next 15 years? by Luiza Amara Maciel Braga, Carlos Gilbert Conte Filho and Fabio Batista Mota in Therapeutic Advances in Rare Disease
Acknowledgments
The authors thank the respondents who participated in this study, the anonymous reviewers, and associate editor Dr Suvra Moulick for their comments and suggestions, which greatly contributed to the improvement of the manuscript. The authors also thank Dr Luiz Anastácio (Laboratory of Cellular Communication, Oswaldo Cruz Institute, Fiocruz) for reviewing and contributing to the final version of the manuscript.
Footnotes
Author contribution(s): Luiza Amara Maciel Braga: Conceptualization; Formal analysis; Investigation; Methodology; Writing – original draft.
Carlos Gilbert Conte Filho: Conceptualization; Data curation; Formal analysis; Methodology; Software; Validation; Writing – original draft.
Fabio Batista Mota: Conceptualization; Formal analysis; Investigation; Project administration; Supervision; Writing – review & editing.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics statement: This study follows the Brazilian Resolution number 510, dated 7 April 2016, which exempts this type of study from registration or evaluation by a research ethics committee. All participants in this study gave us their informed consent by responding to the online questionnaire.
ORCID iD: Luiza Amara Maciel Braga  https://orcid.org/0000-0002-1726-2643
https://orcid.org/0000-0002-1726-2643
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Luiza Amara Maciel Braga, Faculty of Economics, Fluminense Federal University, Niteroi, Brazil.
Carlos Gilbert Conte Filho, Department of Economics, Federal University of Santa Maria, Santa Maria, Brazil.
Fabio Batista Mota, Laboratory of Cellular Communication, Oswaldo Cruz Institute, Oswaldo Cruz Foundation, Av. Brasil, 4.365, Pavilhão 108, Manguinhos, Rio de Janeiro RJ 21040-360, Brazil.
References
- 1.Nguengang Wakap S, Lambert DM, Olry A, et al. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur J Hum Genet 2020; 28: 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delhove J, Osenk I, Prichard I, et al. Public acceptability of gene therapy and gene editing for human use: a systematic review. Hum Gene Ther 2020; 31: 20–46. [DOI] [PubMed] [Google Scholar]
- 3.Kole A, Hedley V.Recommendations from the rare 2030 foresight study: the future of rare diseases starts today, EURORDIS, Europe 2021, https://download2.eurordis.org/rare2030/Rare2030_recommendations.pdf
- 4.Roessler HI, Knoers NVAM, van Haelst MM, et al. Drug repurposing for rare diseases. Trends Pharmacol Sci 2021; 42: 255–267. [DOI] [PubMed] [Google Scholar]
- 5.Matsoukas IG.Prime editing: genome editing for rare genetic diseases without double-strand breaks or donor DNA. Front Genet 2020; 11: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guggino WB, Cebotaru L.Adeno-associated virus (AAV) gene therapy for cystic fibrosis: current barriers and recent developments. Expert Opin Biol Ther 2017; 17: 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pranke I, Golec A, Hinzpeter A, et al. Emerging therapeutic approaches for cystic fibrosis. From gene editing to personalized medicine. Front Pharmacol 2019; 10: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soni S.Gene therapies for transfusion dependent β-thalassemia: current status and critical criteria for success. Am J Hematol 2020; 95: 1099–1112. [DOI] [PubMed] [Google Scholar]
- 9.Schneider-Futschik EK.Beyond cystic fibrosis transmembrane conductance regulator therapy: a perspective on gene therapy and small molecule treatment for cystic fibrosis. Gene Ther 2019; 26: 354–362. [DOI] [PubMed] [Google Scholar]
- 10.Verhoeyen E, Roman-Rodriguez FJ, Cosset FL, et al. Gene therapy in Fanconi anemia: a matter of time, safety and gene transfer tool efficiency. Curr Gene Ther 2017; 16: 297–308. [DOI] [PubMed] [Google Scholar]
- 11.Forsythe E, Kenny J, Bacchelli C, et al. Managing Bardet-Biedl syndrome – now and in the future. Front Pediatr 2018; 6: 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Fernandez De La Camara C, Nanda A, Salvetti AP, et al. Gene therapy for the treatment of X-linked retinitis pigmentosa. Expert Opin Orph Drug 2018; 6: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reiss UM, Zhang L, Ohmori T.Hemophilia gene therapy – new country initiatives. Haemophilia 2021; 27(Suppl. 3): 132–141. [DOI] [PubMed] [Google Scholar]
- 14.Hodges CA, Conlon RA.Delivering on the promise of gene editing for cystic fibrosis. Genes Dis 2019; 6: 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlon MS, Vidović D, Birket S.Roadmap for an early gene therapy for cystic fibrosis airway disease. Prenat Diagn 2017; 37: 1181–1190. [DOI] [PubMed] [Google Scholar]
- 16.Hanenberg H, Roellecke K, Wiek C.Stem cell genetic therapy for Fanconi anemia – a new hope. Curr Gene Ther 2017; 16: 309–320. [DOI] [PubMed] [Google Scholar]
- 17.Parsons D, Donnelley M.Will airway gene therapy for cystic fibrosis improve lung function? New imaging technologies can help us find out. Hum Gene Ther 2020; 31: 973–984. [DOI] [PubMed] [Google Scholar]
- 18.Asher DR, Thapa K, Dharia SD, et al. Clinical development on the frontier: gene therapy for Duchenne muscular dystrophy. Expert Opin Biol Ther 2020; 20: 263–274. [DOI] [PubMed] [Google Scholar]
- 19.Vera LNP, Baldo G. The potential of gene therapy for mucopolysaccharidosis type I. Expert Opinion on Orphan Drugs 2020; 8: 33–41. [Google Scholar]
- 20.Bilezikian JP, Cusano NE, Khan AA, et al. Primary hyperparathyroidism. Nat Rev Dis Primers 2016; 2: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B.CRISPR/Cas gene therapy. J Cell Physiol 2021; 236: 2459–2481. [DOI] [PubMed] [Google Scholar]
- 22.Zhang ZY, Thrasher AJ, Zhang F.Gene therapy and genome editing for primary immunodeficiency diseases. Genes Dis 2020; 7: 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, Li L, Jiang J, et al. Applications and challenges of CRISPR-Cas gene-editing to disease treatment in clinics. Precis Clin Med 2021; 4: 179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tambuyzer E, Vandendriessche B, Austin CP, et al. Therapies for rare diseases: therapeutic modalities, progress and challenges ahead. Nat Rev Drug Discov 2020; 19: 93–111. [DOI] [PubMed] [Google Scholar]
- 25.Brimble MA, Reiss UM, Nathwani AC, et al. New and improved AAVenues: current status of hemophilia B gene therapy. Expert Opin Biol Ther 2016; 16: 79–92. [DOI] [PubMed] [Google Scholar]
- 26.Bessis N, GarciaCozar FJ, Boissier MC.Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther 2004; 11(Suppl. 1): S10–S17. [DOI] [PubMed] [Google Scholar]
- 27.Chan YK, Wang SK, Chu CJ, et al. Engineering adeno-associated viral vectors to evade innate immune and inflammatory responses. Sci Transl Med 2021; 13: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huml RA.Rare disease drug development. Cham: Springer International Publishing, 2021. [Google Scholar]
- 29.Sun W, Zheng W, Simeonov A.Drug discovery and development for rare genetic disorders. Am J Med Genet A 2017; 173: 2307–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkarni JA, Witzigmann D, Thomson SB, et al. The current landscape of nucleic acid therapeutics. Nat Nanotechnol 2021; 16: 630–643. [DOI] [PubMed] [Google Scholar]
- 31.Maldonado R, Jalil S, Wartiovaara K.Curative gene therapies for rare diseases. J Community Genet 2021; 12: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perrin GQ, Herzog RW, Markusic DM.Update on clinical gene therapy for hemophilia. Blood 2019; 133: 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carvalho M, Sepodes B, Martins AP.Patient access to gene therapy medicinal products: a comprehensive review. BMJ Innov 2021; 7: 123–134. [Google Scholar]
- 34.Keith AR, Twaroski K, Ebens CL, et al. Leading edge: emerging drug, cell, and gene therapies for junctional epidermolysis bullosa. Expert Opin Biol Ther 2020; 20: 911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Géléoc GGS, El-Amraoui A. Disease mechanisms and gene therapy for Usher syndrome. Hear Res 2020; 394: 1–12. [DOI] [PubMed] [Google Scholar]
- 36.Doshi BS, Arruda VR.Gene therapy for hemophilia: what does the future hold. Ther Adv Hematol 2018; 9: 273–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan Z, McCray PB, Engelhardt JF.Advances in gene therapy for cystic fibrosis lung disease. Hum Mol Genet 2019; 28: R88–R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen TL, Gøtzsche CR, Woldbye DPD. Current and future prospects for gene therapy for rare genetic diseases affecting the brain and spinal cord. Front Mol Neurosci 2021; 14: 695937–695927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fahim A.Retinitis pigmentosa: recent advances and future directions in diagnosis and management. Curr Opin Pediatr 2018; 30: 725–733. [DOI] [PubMed] [Google Scholar]
- 40.Pascual-Camps I, Barranco-Gonzalez H, Aviñó-Martínez J, et al. Diagnosis and treatment options for achromatopsia: a review of the literature. J Pediatr Ophthalmol Strabismus 2018; 55: 85–92. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Cui Y, Jiang D, et al. Management of Leigh syndrome: current status and new insights. Clin Genet 2018; 93: 1131–1140. [DOI] [PubMed] [Google Scholar]
- 42.Morgan MA, Galla M, Grez M, et al. Retroviral gene therapy in Germany with a view on previous experience and future perspectives. Gene Ther 2021; 28: 494–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bulaklak K, Gersbach CA.The once and future gene therapy. Nat Commun 2020; 11: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diallo J-S.Viral vectors for treatment of human disease: therapeutic and manufacturing challenges. In: Filice M, Ruiz-Cabello J. (eds) Nucleic Acid Nanotheranostics. Amsterdam: Elsevier, 2019, pp. 213–246. [Google Scholar]
- 45.Zhu X, Zhang Y, Yang X, et al. Gene therapy for neurodegenerative disease: clinical potential and directions. Front Mol Neurosci 2021; 14: 618171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robles-Rodríguez OA, Pérez-Trujillo JJ, Villanueva-Olivo A, et al. Advances in gene therapy for hemophilia. J Biosci 2020; 45: 1–8. [PubMed] [Google Scholar]
- 47.Pradhan A, Kalin TV, Kalinichenko VV.Genome editing for rare diseases. Curr Stem Cell Rep 2020; 6: 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li C, Samulski RJ.Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet 2020; 21: 255–272. [DOI] [PubMed] [Google Scholar]
- 49.Zeng J, Wu Y, Ren C, et al. Therapeutic base editing of human hematopoietic stem cells. Nat Med 2020; 26: 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goncalves-Maia M, Magnaldo T.Genetic therapy of Xeroderma Pigmentosum: analysis of strategies and translation. Exp Opin Orph Drug 2017; 5: 5–17. [Google Scholar]
- 51.Pipe SW.Gene therapy for hemophilia. Pediatr Blood Cancer 2018; 65: 1–8. [DOI] [PubMed] [Google Scholar]
- 52.Crawford KJ, Downey DG.Theratyping in cystic fibrosis. Curr Opin Pulm Med 2018; 24: 612–617. [DOI] [PubMed] [Google Scholar]
- 53.Ahmed B, Zafar M, Qadir MI.Review: oncogenic insertional mutagenesis as a consequence of retroviral gene therapy for X-linked severe combined immunodeficiency disease. Crit Rev Eukaryot Gene Expr 2019; 29: 511–520. [DOI] [PubMed] [Google Scholar]
- 54.Ajufo E, Cuchel M.Recent developments in gene therapy for homozygous familial hypercholesterolemia. Curr Atheroscler Rep 2016; 18: 22–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arruda VR, Doshi BS.Gene therapy for hemophilia: facts and quandaries in the 21st century. Mediterr J Hematol Infect Dis 2020; 12: e2020069–e2020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arruda VR, Samelson-Jones BJ.Gene therapy for immune tolerance induction in hemophilia with inhibitors. J Thromb Haemost 2016; 14: 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aubourg P.Gene therapy for leukodystrophy: progress, challenges and opportunities. Expert Opinion on Orphan Drugs 2016; 4: 359–367. [Google Scholar]
- 58.Batty P, Lillicrap D.Advances and challenges for hemophilia gene therapy. Hum Mol Genet 2019; 28: R95–R101. [DOI] [PubMed] [Google Scholar]
- 59.Booth C, Romano R, Roncarolo MG, et al. Gene therapy for primary immunodeficiency. Hum Mol Genet 2019; 28: R15–R23. [DOI] [PubMed] [Google Scholar]
- 60.Cai Y, Shi Q.Platelet-targeted FVIII gene therapy restores hemostasis and induces immune tolerance for hemophilia A. Front Immunol 2020; 11: 964–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cavazzana M, Six E, Lagresle-Peyrou C, et al. Gene therapy for X-linked severe combined immunodeficiency: where do we stand. Hum Gene Ther 2016; 27: 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guggino WB, Cebotaru L.Gene therapy for cystic fibrosis paved the way for the use of adeno-associated virus in gene therapy. Hum Gene Ther 2020; 31: 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hagemeijer MC, Siegwart DJ, Strug LJ, et al. Translational research to enable personalized treatment of cystic fibrosis. J Cyst Fibros 2018; 17: S46–S51. [DOI] [PubMed] [Google Scholar]
- 64.Hampson DR, Hooper AWM, Niibori Y.The application of adeno-associated viral vector gene therapy to the treatment of fragile X syndrome. Brain Sci 2019; 9: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han HA, Pang JKS, Soh BS.Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J Mol Med (Berl) 2020; 98: 615–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iskrov G, Vasilev G, Stefanov R.What could gene therapies learn from orphan drugs’ post-regulatory approval access in the EU? Expert Opinion on Orphan Drugs 2019; 7: 407–414. [Google Scholar]
- 67.Julkowska D, Austin CP, Cutillo CM, et al. The importance of international collaboration for rare diseases research: a European perspective. Gene Ther 2017; 24: 562–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karaarslan C.Leber’s hereditary optic neuropathy as a promising disease for gene therapy development. Adv Ther 2019; 36: 3299–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khamsi R.Gene therapy could offer an inclusive cure for cystic fibrosis. Nature 2020; 583: S12–S14. [Google Scholar]
- 70.Martin F, Gutierrez-Guerrero A, Sánchez S, et al. Genome editing: an alternative to retroviral vectors for Wiskott-Aldrich Syndrome (WAS) gene therapy? Expert Opinion on Orphan Drugs 2016; 4: 281–289. [Google Scholar]
- 71.Maule G, Arosio D, Cereseto A.Gene therapy for cystic fibrosis: progress and challenges of genome editing. Int J Mol Sci 2020; 21: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miah KM, Hyde SC, Gill DR.Emerging gene therapies for cystic fibrosis. Expert Rev Respir Med 2019; 13: 709–725. [DOI] [PubMed] [Google Scholar]
- 73.Panchal N, Booth C, Cannons JL, et al. X-linked lymphoproliferative disease type 1: a clinical and molecular perspective. Front Immunol 2018; 9: 666–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paul-Smith MC, Bell RV, Alton WE, et al. Gene therapy for cystic fibrosis: recent progress and current aims. Expert Opinion on Orphan Drugs 2016; 4: 649–658. [Google Scholar]
- 75.Piekarowicz K, Machowska M, Dzianisava V, et al. Hutchinson-Gilford progeria syndrome-current status and prospects for gene therapy treatment. Cells 2019; 8: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Navarro S, Giorgetti A, Raya A, et al. Induced pluripotency and gene editing in fanconi anemia. Curr Gene Ther 2017; 16: 321–328. [DOI] [PubMed] [Google Scholar]
- 77.Verghese SC, Kurre P.Anchored lentiviral vector episomes for stem cell gene therapy in fanconi anemia. Curr Gene Ther 2017; 16: 329–337. [DOI] [PubMed] [Google Scholar]
- 78.Adair JE, Sevilla J, Heredia CD, et al. Lessons learned from two decades of clinical trial experience in gene therapy for fanconi anemia. Curr Gene Ther 2017; 16: 338–348. [DOI] [PubMed] [Google Scholar]
- 79.Samelson-Jones BJ, Arruda VR.Protein-engineered coagulation factors for hemophilia gene therapy. Mol Ther Methods Clin Dev 2019; 12: 184–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samelson-Jones BJ, Arruda VR.Translational potential of immune tolerance induction by AAV liver-directed factor VIII gene therapy for hemophilia A. Front Immunol 2020; 11: 618–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sherman A, Biswas M, Herzog RW.Innovative approaches for immune tolerance to factor VIII in the treatment of hemophilia A. Front Immunol 2017; 8: 1604–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shukla V, Seoane-Vazquez E, Fawaz S, et al. The landscape of cellular and gene therapy products: authorization, discontinuations, and cost. Hum Gene Ther Clin Dev 2019; 30: 102–113. [DOI] [PubMed] [Google Scholar]
- 83.Yamaguti-Hayakawa GG, Ozelo MC.Gene therapy: paving new roads in the treatment of hemophilia. Semin Thromb Hemost 2019; 45: 743–750. [DOI] [PubMed] [Google Scholar]
- 84.Yue Y, Binalsheikh IM, Leach SB, et al. Prospect of gene therapy for cardiomyopathy in hereditary muscular dystrophy. Expert Opin Orphan Drugs 2016; 4: 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Massaro G, Geard AF, Liu W, et al. Gene therapy for lysosomal storage disorders: ongoing studies and clinical development. Biomolecules 2021; 11: 1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pereira Cabral B, Bonventre JV, Wieringa F, et al. Probing expert opinions on the future of kidney replacement therapies. Artif Organs 2021; 45: 79–87. [DOI] [PubMed] [Google Scholar]
- 87.Mota FB, Braga LAM, Cabral BP, et al. What is the future of lab-on-a-chip diagnostic devices? Assessing changes in experts’ expectations over time. FS 2021; 23: 640–654. [Google Scholar]
- 88.Cabral BP, Braga LAM, Mota FB.Expert opinions on the most promising treatments and vaccine candidates for COVID-19: global cross-sectional survey of virus researchers in the early months of the pandemic. JMIR Public Health Surveill 2021; 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mota F, Braga L, Rocha L, et al. 3D and 4D bioprinted human model patenting and the future of drug development. Nat Biotechnol 2020; 38: 689–694. [DOI] [PubMed] [Google Scholar]
- 90.Rocha LFM, Braga LAM, Mota FB.Gene editing for treatment and prevention of human diseases: a global survey of gene editing-related researchers. Hum Gene Ther 2020; 31: 852–862. [DOI] [PubMed] [Google Scholar]
- 91.Pereira Cabral B, da Graça Derengowski Fonseca M, Mota FB. Long term prevention and vector control of arboviral diseases: what does the future hold. Int J Infect Dis 2019; 89: 169–174. [DOI] [PubMed] [Google Scholar]
- 92.Cabral BP, Da Graça Derengowski Fonseca M, Mota FB.What is the future of cancer care? A technology foresight assessment of experts’ expectations. Economics of Innovation and New Technology 2019; 28: 635–652. [Google Scholar]
- 93.Miot HA.Avaliação da normalidade dos dados em estudos clínicos e experimentais. J Vasc Bras 2017; 16: 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kaufmann P, Pariser AR, Austin C.From scientific discovery to treatments for rare diseases – the view from the National Center for Advancing Translational Sciences – Office of rare diseases research. Orphanet J Rare Dis 2018; 13: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Senior M.After Glybera’s withdrawal, what’s next for gene therapy? Nat Biotechnol 2017; 35: 491–492. [DOI] [PubMed] [Google Scholar]
- 96.Prado DA, Acosta-Acero M, Maldonado RS.Gene therapy beyond luxturna: a new horizon of the treatment for inherited retinal disease. Curr Opin Ophthalmol 2020; 31: 147–154. [DOI] [PubMed] [Google Scholar]
- 97.Brommel CM, Cooney AL, Sinn PL.Adeno-associated virus-based gene therapy for lifelong correction of genetic disease. Hum Gene Ther 2020; 31: 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shim G, Kim D, Park GT, et al. Therapeutic gene editing: delivery and regulatory perspectives. Acta Pharmacol Sin 2017; 38: 738–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.First CRISPR therapy dosed. Nat Biotechnol 2020; 38: 381–382. [DOI] [PubMed] [Google Scholar]
- 100.Sainz-Ramos M, Gallego I, Villate-Beitia I, et al. How far are non-viral vectors to come of age and reach clinical translation in gene therapy? Int J Mol Sci 2021; 22: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rautela I, Uniyal P, Thapliyal P, et al. An extensive review to facilitate understanding of CRISPR technology as a gene editing possibility for enhanced therapeutic applications. Gene 2021; 785: 1–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-trd-10.1177_26330040221100840 for Future of genetic therapies for rare genetic diseases: what to expect for the next 15 years? by Luiza Amara Maciel Braga, Carlos Gilbert Conte Filho and Fabio Batista Mota in Therapeutic Advances in Rare Disease
Supplemental material, sj-pdf-2-trd-10.1177_26330040221100840 for Future of genetic therapies for rare genetic diseases: what to expect for the next 15 years? by Luiza Amara Maciel Braga, Carlos Gilbert Conte Filho and Fabio Batista Mota in Therapeutic Advances in Rare Disease