Abstract
Protein footprints of the enhancer-dependent σ54 protein, upon binding the Escherichia coli RNA polymerase core enzyme or upon forming closed promoter complexes, identified surface-exposed residues in σ54 of potential functional importance at the interface between σ54 and core RNA polymerases (RNAP) or DNA. We have now characterised alanine and glycine substitution mutants at several of these positions. Properties of the mutant σ54s correlate protein footprints to activity. Some mutants show elevated DNA binding suggesting that promoter binding by holoenzyme may be limited to enable normal functioning. One such mutant (F318A) within the DNA binding domain of σ54 shows a changed interaction with the promoter regulatory region implicated in transcription silencing and fails to silence transcription in vitro. It appears specifically defective in preferentially binding to a repressive DNA structure believed to restrict RNA polymerase isomerisation and is largely intact for activator responsiveness. Two mutants, one in the regulatory region I and the other within core interacting sequences of σ54, failed to stably bind the activator in the presence of ADP-aluminium fluoride, an analogue of ATP in the transition state for hydrolysis. Overall, the data presented describe a collection σ54 mutants that have escaped previous analysis and display an array of properties which allows the role of surface-exposed residues in the regulation of open complex formation and promoter DNA binding to be better understood. Their properties support the view that the interface between σ54 and core RNAP is functionally specialised.
INTRODUCTION
Bacterial DNA-dependent RNA polymerases (RNAP) are multisubunit enzymes (α2, β, β′, ω, σ; holoenzyme) that catalyse the transcription of DNA into RNA. In Escherichia coli, seven σ subunits function as specificity factors and direct the RNAP holoenzyme to promoter sites on DNA for the transcription of different sets of genes. The E.coli σ factors are grouped into two classes: the σ70 class and the σ54 class. The σ54 containing RNAP holoenzyme is distinctive in its ability to remain transcriptionally silent prior to activation (1). Productive transcription initiation by the σ54 RNAP holoenzyme only occurs in response to an interaction with an enhancer-bound activator protein in a nucleotide hydrolysis-dependent manner (2,3), whereas for the σ70 RNAP holoenzyme, activators often function by simply recruiting the RNAP to promoter sites. Determinants within σ54 required for activator-dependent interactions and conformational changes within the holoenzyme that lead to transcription are localised in the N-terminal region I and within the minimal DNA binding domain (residues 329–477 in Klebsiella pneumoniae σ54) (4,5). In the σ54 RNAP, which is regulated at the DNA melting step, these activation determinants localise over the start site proximal promoter element where DNA melting originates. This protein–DNA arrangement is termed the regulatory centre of the σ54 RNAP closed complex (6). To catalyse DNA melting, oligomers of activator protein, competent for hydrolysing ATP, must interact with the regulatory centre to induce conformational re-arrangements in the nucleoprotein complex formed between the σ54 RNAP holoenzyme and promoter DNA to allow the progression of the closed promoter complex to a transcription initiation competent open complex (7–11).
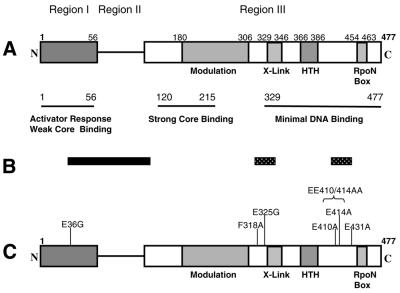
The domain organisation and functions of σ54 have been explored previously using mutagenesis and biochemical methods (Fig. 1A). Previous protein footprinting studies have identified surface-exposed regions of σ54 that likely contribute to locally interacting surfaces involved in binding core RNAP, forming the closed complex and used during the isomerisation of the closed complex to open complex upon activation (11,12) (Fig. 1B). Two distinct regions in σ54 were protected from protease attack upon interaction with core RNAP: (i) a patch including region I consisting of residues 36–100 and (ii) a sequence in region III spanning residues 315–334. The core RNAP-protected regions lie outside the minimal core RNAP binding domain (residues 120–215) but within regions involved in respose to activation (residues 1–50 and 315–346) and near or within the DNA binding domain (residues 329–477) (1). In the closed complex, residues E410, E414 and E431 that lie within the minimal DNA binding domain were protected from protease attack, possibly by their proximity to the promoter DNA in the closed complex (12).
Figure 1.
Domain organisation of the K.pneumoniae σ54. (A) The N-terminal 56 residues (region I) have roles in activator responsiveness. The major DNA binding determinants of σ54 are localised in region III which contains a patch of 18 amino acids (residues 329–346) that UV-crosslink to DNA (1), a putative helix–turn–helix (HTH) motif (residues 366–386) that has been implicated in interacting with the start site proximal promoter element (the –12 region) (16,42) and a σ54 characteristic motif of 10 amino acids (residues 454–463) known as the RpoN box that has been implicated in interacting with start site distal promoter element (the –24 region) (4,43). Residues 120–215 in region III of σ54 contain the major core RNAP binding determinant (23). (B) Summary of protease-sensitive sites (12). The black bar indicates core RNAP-specific protection and dotted bars show closed complex-specific protection. (C) Sites where single mutations were introduced at surface-exposed positions.
In the present work, surface-exposed residues in region III (F318, E410, E414 and E431) that were protected from protease attack by core RNAP (residue F318) or promoter DNA (residues E410, E414 and E431) in the closed complex were subsituted with alanine or glycine (Fig. 1C). A residue in region I (E36) that became hypersensitive to protease attack in open promoter complexes was substituted with glycine in an attempt to increase the conformational flexibility of region I around residue 36 in the anticipation that this mutation could have an altered response to activation. Previously, residue 36 was shown to localise close to the active site of the core RNAP and to the –12 promoter region within the holoenzyme and closed complex, respectively (6,13). Similarly, residue E325 that was protected from protease attack by core RNAP was substituted with glycine. Although many residues in σ54 can be altered without obtaining a clear phenotype (4,14,15), the σ54 mutants described here all show a clear phenotype and, thus, the results presented below indicate a positive correlation between protein footprints and the functional importance of the targeted residues. Interestingly, a residue (F318) was identified from footprints of core RNAP upon σ54 which functions in DNA binding. We show that F318 has a regulatory role in keeping the σ54 RNAP holoenzyme silent for transcription prior to activation, supporting the view that the interface of σ54 and core RNAP is important for controlling the activity of σ54.
MATERIALS AND METHODS
Site-directed mutagenesis
Region I (E36G) and region III (F318A, E325G, E410A, E414A, EE410/414A and E431A) of K.pneumoniae σ54 point mutations were constructed as N-terminal His6-tagged proteins using the Quickchange mutagenesis kit (Stratagene) as described previously (16). Briefly, the low copy number plasmid pMM83 (17) carrying the K.pneumoniae rpoN gene was used as the template with a large molar excess of complementary mutagenic primers. Following mutagenesis, DNA was transformed into E.coli strain XL2B and mutant clones were indentified by sequencing. The E36G mutant was also constructed as an N-terminal His6-tagged protein using pET28::rpoN (pMTHσN) template DNA (18) for overexpression (see later) using the Quickchange mutagenesis kit.
In vivo activity assays (β-galactosidase assay)
Mutant plasmids were transformed into K.pneumoniae strain UNF2792 (hisD2, ΔrpoN71::kan, recA56, sbl300::Tn10) (17) containing K.pneumoniae nifH::lacZ translational fusion reporter plasmid pMB1 (19). Three to four independent colonies were grown in nutrient broth overnight. A sample (100 µl) of this culture was used to inoculate nitrogen-free Davis and Mingoli (NFDM) media (20) and grown overnight at 30°C under anaerobic nitrogen limiting conditions. Chromosomal nifA expression was activated by supplementing NFDM with 100 µg/ml of aspartic acid. Subsequently, β-galactosidase activity was measured.
Immunoblotting
Cells (1 ml) of K.pneumoniae strain UNF2792, grown under conditions described above (see in vivo activity assays), were collected by centrifugation and resuspended in 100 µl of sterile H2O. Aliquots of 20 µl of concentrated cells were lysed with 20 µl of 2× SDS sample buffer, heated at 95°C and 10 µl used for loading. Proteins were separated on denaturing 7.5% SDS–PAGE mini-gels and blotted onto PVDF membranes (0.2 µm pore size for western blotting; Millipore). Anti-σ54 (21) and alkaline phosphatase-conjugated anti-rabbit IgG (Promega) antibodies were used for detection.
Protein expression and purification
For overexpression of σ54 mutants, BamHI–HindIII fragments carrying the region III point mutants F318A, E325G, E410A, E414A, EE410/414AA and E431A were cloned into a pET28b+ based pMT1/306 (18) to form clones carrying full-length rpoN. Overexpression plasmids pSRW-E36G, pSRW-F318A, pSRW-E325G, pSRW-E410A, pSRW-E414A, pSRW-EE410/414AA and pSRW-E431A were transformed ino E.coli strain BL21 (pLysS) (Novagen). Mutant σ54 proteins were expressed and purified by Ni-affinity chromatography using FPLC and eluted with an imidazole gradient as decribed previously (16). The DNA binding mutant form of the activator E.coli phage shock protein F (PspFΔHTH) was overexpressed and purified as a His6-tagged fusion protein from pMJ15 (22). For the activator assays, heart muscle kinase (HMK)-tagged PspFΔHTH was overexpressed from pSRW-HMKPSPFΔHTH (10) and purified as a His6-tagged fusion protein essentially as described above. The purfied mutant σ54s were stored in TGED buffer [10 mM Tris–Cl pH 8.0, 50 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol (DTT) and 50% (v/v) glycerol] at –70°C (long term) or –20°C (short term). PspFΔHTH was aliquoted and stored in TGED buffer at –70°C. Escherichia coli core RNAP was purchased from Epicentre Technologies.
Core RNAP binding assays
These were performed essentially as described previously as 10 µl reactions in Tris–NaCl buffer [40 mM Tris–HCl pH 8.0, 10% (v/v) glycerol, 0.1 mM EDTA, 1 mM DTT and 100 mM NaCl] (18). Briefly, E.coli core RNAP (250 nM) and different amounts of mutant σ54 proteins were mixed together and incubated at 30°C for 10 min, followed by the addition of glycerol–bromophenol blue loading dye. The reactions were loaded onto Bio-Rad native 4.5% polyacrylamide Mini-Protean II gels and run at 50 V for 2 h at room temperature in Tris–glycine buffer (25 mM Tris and 200 mM glycine). Complexes were visualised by Coomassie blue staining of the gels.
Gel mobility shift assays
32P-End-labelled 88 bp homoduplex or heteroduplex fragments mismatched at positions –12 to –11 (early melted probe) and –10 to –1 (late melted probe) consisting of the –60 to +28 Sinorhizobium meliloti nifH promoter sequence were formed as described previously (23) and used as probes. A typical σ54 or σ54 holoenzyme (formed with σ54 at a 4-fold molar excess over core RNAP) binding assay contained 16 nM DNA and σ54 or σ54 holoenzyme (concentrations as indicated in figures or corresponding legends) in STA buffer [25 mM Tris–acetate pH 8.0, 8 mM magnesium acetate, 10 mM KCl, 1 mM DTT and 3.5% (w/v) PEG 6000] and incubated for 10 min at 30°C. For activation, 4 µM PspFΔHTH activator protein and 4 mM dGTP were used. Briefly, core RNAP, σ54 and DNA were pre-incubated at 30°C for 10 min and then nucleotide and activator were added for 10 min and, if required, heparin (final concentration 100 µg/ml) for a further 5 min prior to gel loading. Samples were then loaded onto native 4.5% polyacrylamide gels and run at 60 V for 80 min at room temperature in Tris–glycine buffer. DNA–protein complexes were detected and quantified by phosphorimager analysis.
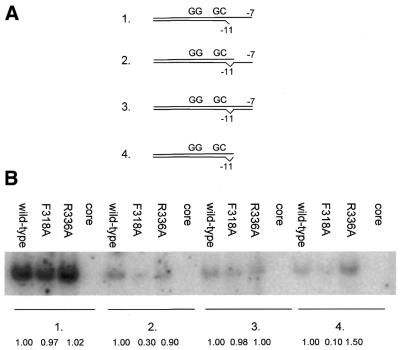
Fork junction shift assays were done as described previously (4). Briefly, probes were prepared by annealing two complementary DNA strands. The lengths of the template and non-template strands and the mismatch created are specified in Figure 10A. The annealing mixtures contained 4 pmol of labelled template strand and 6 pmol of non-template strand in 10 mM HEPES pH 7.9 and 80mM NaCl. Binding reactions (10 µl) contained 1 nM DNA and 7.5 nM σ54 (holoenzyme) Eσ54 in 1× HEPES buffer [50 mM HEPES–HCl pH 7.9, 100 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.05 µg/ml BSA, 2.8% (w/v) PEG 8000 and 5 ng/µl poly(dI–dC)]. Following incubation at 37°C for 10 min, the samples were separated on a 4.5% native polyacrylamide gel. Gels were run at 4°C and analysed as described above.
Figure 10.
Fork junction DNA binding by F318A holoenzyme. (A) The fork junction probes used for the binding assays based on E.coli glnHp2 promotor. (B) Autoradiograph of binding of holoenzymes to the fork junction probes 1,2,3 and 4. The fraction of complexes formed with respect to wild-type σ54 is shown for each fork junction probe.
In vitro transcription assays
The template for the transcription assays was the supercoiled plasmid pMKC28 carrying the S.meliloti nifH promoter in pTE103 (5,24). The transcription assays were performed in STA buffer as outlined previously (6). Briefly, 50 nM Eσ54 (50 nM core RNAP: 200 nM σ54) and 10 nM DNA was used. For activation, 4 µM PspFΔHTH was added with 4 mM ATP. The reactions were incubated for 10 min to allow open complexes to form. The remaining rNTPs (100 nM), 3 µCi [α-32P]UTP and heparin (100 µg/ml) were added and incubated for a further 10 min. The S.meliloti nifH start sequence is GGG (+1 to +3). To allow activator-independent initiated complex formation, 4 mM GTP was incubated with the Eσ54 prior to challenge with heparin and the addition of remaining nucleotide. The reactions were stopped with 4 µl of formamide loading buffer and 7 µl aliquots were loaded on 6% denaturing sequencing gels. The dried gel was analysed on a phosphorimager.
Activator interaction assays
These were conducted essentially in STA buffer as described by Chaney et al. (10). Briefly, mutant σ54 (10 µM) or holoenzyme (300 nM) complexes thereof were incubated with 32P-labelled PspFΔHTH (400 nM), ADP (0.2 mM) and NaF (5 mM) for 5 min at 37°C. After addition of AlCl3 (0.2 mM) the reactions were incubated for a further 10 min and directly loaded onto a 4.5% native gel. For the closed complex-activator binding assays, 32P-labelled early melted probe (heteroduplex at –11 and –12) was incubated with 300 nM holoenzyme and reactions conducted essentially as described above.
RESULTS
In vivo activity of σ54 mutants
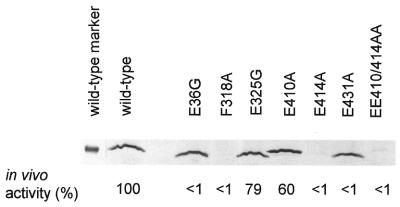
β-Galactosidase assays were used to assess the in vivo activity of the σ54 mutants listed in Figure 1C. For assays, K.pneumoniae strain UNF2792, which lacks a functional σ54, was transformed with a mutant rpoN in a low copy number vector and pMB1 which contains the β-galactosidase gene under the control of σ54-dependent K.pneumoniae nifH promoter. Activator nitrogen fixation protein A (NifA) was chromosomally expressed under the nitrogen limiting growth conditions. Figure 2 (also see Table 1) shows the in vivo β-galactosidase activity measured for each mutant. With the exception of the E325G and E410A mutants, all others are apparently inactive (<5% of wild-type activity) for in vivo promoter activation. The in vivo promoter activation experiments were performed at least three times to enhance reliability. The standard error range for the data shown in Figure 2 was ±6%. Promoter activation assays using pMB210.1, which harbours the β-galactosidase gene under the control of the S.meliloti nifH promoter (25), which represents a higher affinity binding site for σ54, did not reveal any activity for E36G, F318A, E414A and EE410/414AA mutants (data not shown).
Figure 2.
In vivo activity and immunoblot of cell extracts prepared from K.pneumoniae UNF2792 cells containing mutant plasmids.
Table 1. Summary of the in vivo and in vitro properties of the σ54 mutants described in this work.
| Assay Mutant | In vivo activity (% wild-type) | Homoduplex probe binding (σ54) (% wild-type) | Early melted probe binding (σ54) (% wild-type) | Isomerisation (%) | Homoduplex probe binding (Eσ54) (% wild-type) | Eσ54 heparin stability on early melted probe (% wild-type) | Late melted probe binding (Eσ54) (% wild-type) |
|---|---|---|---|---|---|---|---|
| Wild-type | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| E36G | <1 | <5 | 19 | 57 | 83 | 19 | 80 |
| F318A | <1 | 210 | 83 | 62 | 75 | <4 | 120 |
| E325G | 79 | <5 | 8 | 86 | 80 | <4 | 35 |
| E410A | 60 | 190 | 93 | 88 | 57 | 111 | 100 |
| E414A | <1 | <5 | 80 | 88 | 52 | 73 | 120 |
| E431A | <1 | <5 | 45 | 150 | 52 | 96 | 96 |
| EE410/414AA | <1 | 345 | 116 | 88 | 117 | 119 | 106 |
| Assay Mutant | Eσ54 heparin stability on late melted probe (activated) (% wild-type) | σ54–activator complexes (% wild-type) | Eσ54–activator complexes (% wild-type) | Eσ54–activator complexes (% wild-type) on late melted probe | In vitro transcription at 37°C (% wild-type) | In vitro transcription at 15°C (% wild-type) | In vitro ‘bypass’ transcription |
|---|---|---|---|---|---|---|---|
| Wild-type | 100 | 100 | 100 | 100 | 100 | 100 | No |
| E36G | 75 | <5 | 10 | 86 | 78 | 88 | Yes |
| F318A | 124 | 78 | 80 | 49 | 70 | 98 | Yes |
| E325G | 26 | <5 | <5 | 32 | 23 | 25 | No |
| E410A | 134 | 56 | 96 | 88 | 76 | 120 | No |
| E414A | 124 | 54 | 82 | 91 | 68 | 118 | No |
| E431A | 122 | 109 | 101 | 96 | 82 | 122 | No |
| EE410/414AA | 124 | 45 | 80 | 92 | 59 | 124 | No |
One explanation for the lack of in vivo activity of some of the σ54 mutants could be protein instability. Thus, the levels of mutant σ54 expressed in K.pneumoniae UNF2792 cells under the same conditions as used in the activation assays were measured by immunoblotting. Strikingly, the mutants F318A, E414A and EE410/414AA, which were inactive for promoter activation in vivo, were not stably expressed (Fig. 2). To further confirm the instability of the F318A and E414A mutants in vivo, the pET28b+ based overexpression plasmids harbouring the F318A and E414A were transformed into E.coli TH1 cells (ΔrpoN mutant strain). Immunoblots of cell extract prepared from E.coli TH1 cells under in vivo promoter activation conditions consistently showed that F318A and E414A were not stably expressed, but the wild-type σ54 was detected (data not shown and 26). However, the E36G and E431A mutants were found to be expressed at near wild-type levels (Fig. 2). Thus, for the E36G and E431A mutants protein instability appears not to be the cause for the loss of in vivo activity.
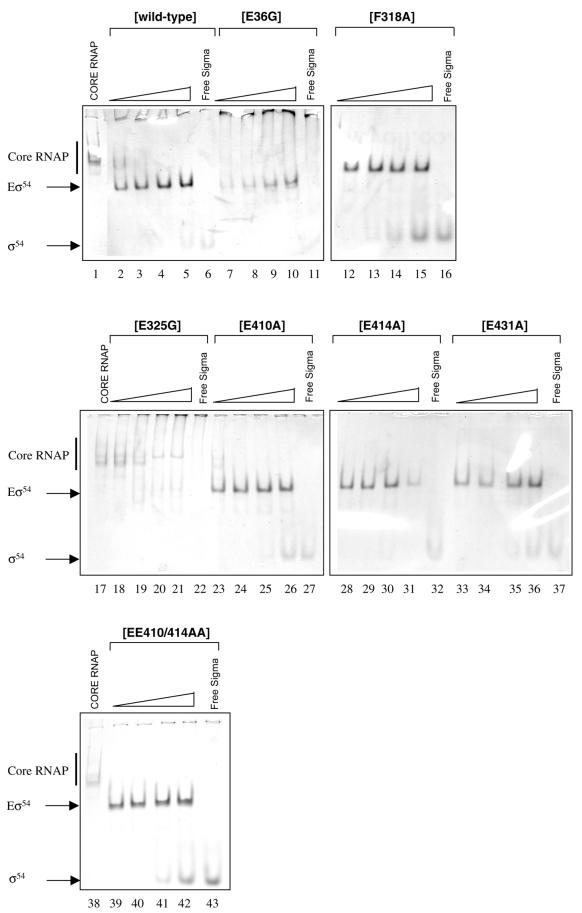
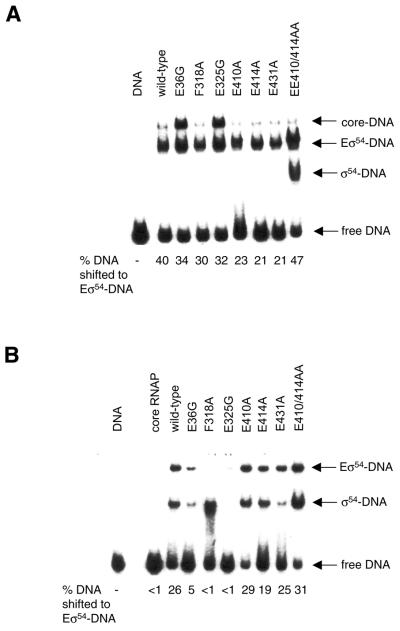
Core RNAP binding properties of purified σ54 mutants
To characterise holoenzymes formed with the mutant σ54 proteins, purified proteins were initially assayed for their ability to bind to E.coli core RNAP using a native gel assembly assay. As shown in Figure 3, F318A, E410A, E414A, E431A and the double mutant EE410/414AA bound the core RNAP well, as judged by the depletion of the bands corresponding to the core RNAP and the appearance of the faster migrating band corresponding to the holoenzyme (Eσ54). The mutant E325G, and to a lesser extent mutant E36G, showed clear defects in holoenzyme formation. Mutants E325G and E36G did not migrate cleanly into the native gel, and no free fast-running σ54 protein was detected in the lower part of the gel, contrasting with the behaviour of the wild-type protein (Fig. 3, compare lane 6 with lanes 11 and 22) and possibly suggesting aggregation of the purified proteins. The E325G and E36G mutants were needed at a ratio of 8:1 (σ54 to core RNAP) to saturate the core RNAP. This may reflect a reduced availability of E325G and E36G due to aggregation (Fig. 3, compare lanes 2–5 with lanes 7–10 and 17–21). Alternatively, as residues E36 and E325 lie outside the high affinity core RNAP binding determinant (residues 120–215) in σ54 but within sequences that are proximal to the core RNAP in the holoenzyme (13,18), changing E36 and E325 to glycine may have introduced conformational changes that result in significant defects in core RNAP binding.
Figure 3.
Escherichia coli core RNAP binding by purified σ54 mutants. Holoenzymes (Eσ54) were formed at 1:1 (lanes 2, 7, 12, 18, 23, 28, 33 and 39), 1:2 (lanes 3, 8, 13, 19, 24, 29, 34 and 40), 1:4 (lanes 4, 9, 14, 20, 25, 30, 35 and 41) and 1:8 (lanes 5, 10, 15, 21, 26, 31, 36 and 42) ratios of core RNAP (250 nM) to σ54. Free core RNAP (lanes 1, 17 and 38) and free σ54:wild-type (lane 6), E36G (lane 11), F318A (lane 16), E325G (lane 22), E410A (lane 27), E414A (lane 32), E431A (lane 37) and EE410/414AA (lane 43) are also shown.
Interaction of mutant σ54 with promoter DNA
For the DNA binding assays, in addition to a double-stranded homoduplex S.meliloti nifH DNA probe, we used a heteroduplex promoter DNA probe that represents a DNA structure in which the nucleation of DNA melting has occurred in the closed complex (early melted DNA probe) (Fig. 4A). This ‘early melted’ DNA probe is believed to represent the locally distorted structure that forms downstream of the consensus GC element in σ54 holoenzyme closed complexes (7). The wild-type σ54 shows a 6-fold higher binding activity for the early melted DNA probe than for the homoduplex probe (7), presumably because binding is energetically favoured.
Figure 4.
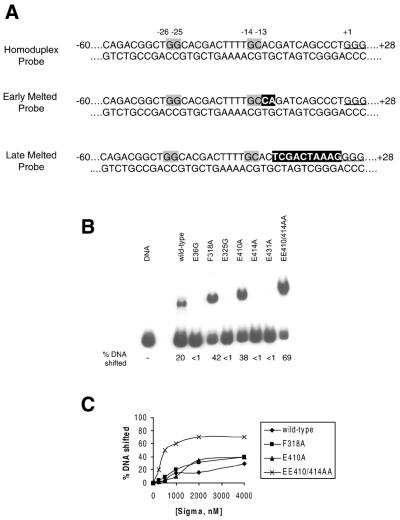
Homoduplex DNA binding. (A) Sinorhizobium meliloti nifH promoter probes used for the DNA binding assays (see text for details). The consensus GG and GC of the σ54 binding sites are highlighted and the mismatched regions are boxed in black. (B) Binding of wild-type and mutant σ54 proteins (at 1 µM) to homoduplex promoter DNA probe. (C) Titration of wild-type, F318A, E410A and EE414/410AA on the homoduplex promoter probe.
Binding to homoduplex probe. Results in Figure 4B (also see Table 1) show that E36G, E325G, E414A and E431A σ54 proteins did not detectably bind the homoduplex DNA probe. The E36G mutant is striking as alanine scans of region I have failed to identify amino acids with such a strong DNA binding defect (27). Residues E325, E414 and E431 are within the minimal DNA binding domain in σ54 (residues 329–477) and E414 and E431 are located between the putative helix–turn–helix motif (residues 366–386) and the RpoN box (residues 454–463) (1,12 and Fig. 1A). Residue 36-tethered FeBABE footprinting of promoter DNA showed that residue 36 is close to the consensus GC promoter element (6). Therefore, given that residues E414 and E431 are also protected in closed complexes by DNA from protease attack (12), it is possible that glutamate residues at positions 36, 325, 414 and 431 make or contribute to sequence-specific contacts with promoter DNA. Strikingly, mutating E414 to A in the context of the E410 mutation apparently rescues homoduplex DNA binding (Fig. 4B), suggesting a dominant role for residue E410 in DNA binding. Further, the binding assays reveal that the double mutant EE414/410AA has a 4-fold increased binding activity compared with the wild-type σ54 protein for the homoduplex probe. Strikingly, such an increased binding activity is also evident with the F318A and E410A mutant proteins. σ54 titration assays show that EE410/414AA, and to a lesser extent F318A and E410A, saturate homoduplex promoter DNA binding at a much earlier point in the binding curve when compared with the wild-type σ54 protein (Fig. 4C). Therefore, it appears that mutating surface-exposed residues F318, E410A and E414 (in combination with E410A) increases σ54 DNA binding activity.
Binding to early melted DNA probe. Binding assays using the early melted promoter probe revealed two classes of mutants. First, the F318A, E410A and EE410/414AA mutant σ54 proteins bind the early melted DNA probe with near wild-type activity, contrasting their improved binding to the homoduplex DNA probe. Secondly, the σ54 mutants (E36G, E325G and E431A) that were very defective for homoduplex DNA binding show moderate improvement in their binding to the early melted probe (Fig. 5 and Table 1). Strikingly, we note that E414A, which did not detectably bind the homoduplex probe, has a near wild-type level of binding activity to the early melted DNA probe. Overall, our early melted probe binding assays with the σ54 mutants show that the region I residue E36 and region III residue E325 in σ54 are involved in binding to both homoduplex and early melted DNA probes and residues F318, E410 and E410/E414 contribute to distinguishing between early melted DNA and homoduplex DNA. Residue E414 appears important for binding to homoduplex DNA (Fig. 4B), less so for early melted DNA binding.
Figure 5.
Binding (lanes 2, 4, 6, 8, 10, 12, 14 and 16) and isomerisation (lanes 3, 5, 7, 9, 11, 13, 15 and 17) of wild-type and mutant σ54 in the presence of PspFΔHTH and dGTP on the early melted promoter probe. The amount of DNA bound (σ54–DNA) and isomerised (ssσ54–DNA) complexes formed are indicated as % DNA shifted and % isomerised, respectively.
σ54 isomerisation on early melted DNA probe. σ54 bound to the early melted DNA probe responds to the activator in a nucleotide hydrolysis-dependent manner to form a new slower migrating ‘supershifted’ DNA complex in which σ54 has isomerised (7). We attempted to form the isomerised σ54–DNA (ssσ–DNA) species using the activator protein PspFΔHTH and hydrolysable nucleotide dGTP (Fig. 5) to test the activator responsiveness of the σ54 mutants. For each mutant, a comparison of the fraction of initially bound DNA converted (in an activator-dependent and nucleotide-requiring reaction) to the isomerised (ssσ–DNA) species was made (shown as percent isomerisation in Fig. 5 and Table 1). For all mutants, the efficiency of conversion to the isomerised state was 34–90%, compared with 60% for the wild-type, suggesting that the mutant σ54 proteins are largely functional for isomerisation, and that some are more active than wild-type σ54. Hence, residues E36 in region I and region III residues F318, E325, E410, E414 and E431 seem dispensable for σ54 productive interaction with activator PspFΔHTH, at least within the scope of the isomerisation assay. The EE410/414AA mutation apparently has increased the efficiency of isomerisation (Fig. 5, lane 15).
Interaction of mutant holoenzymes with promoter DNA
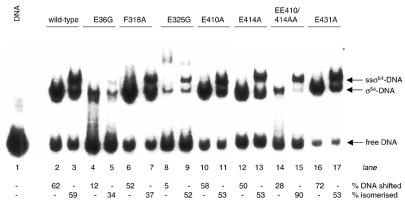
Holoenzyme–homoduplex DNA interactions. We compared the DNA binding activities of holoenzymes (formed with 1:4 ratio of core RNAP to σ54) for the homoduplex promoter probe. As shown in Figure 6A, it seems that core RNAP can recover some of the lost DNA binding activity of the E36G, E325G, E414A and E431A mutants seen in the σ54-DNA binding assays (compare Figs 4A and 6A). The reduced affinity of E36G and E325G for core RNAP is evident as judged by the free core RNAP–DNA complexes present in reactions containing the E36G and E325G holoenzymes (compare Figs 3 and 6A). We note that the increased DNA binding activity of the F318A, E410A and E431A mutants is not readily evident in their holoenzyme–DNA binding patterns or in an increase in heparin stability (data not shown).
Figure 6.
σ54 Holoenzyme (Eσ54)-promoter–DNA interactions. (A) Binding of 100 nM mutant holoenzyme [formed with 1:4 core RNAP (E) to σ54 ratio] to the homoduplex promoter DNA. (B) Stability of mutant holoenzyme on the early melted DNA probe following a 5 min heparin challenge. The percentage of Eσ54–DNA complexes formed on both promoter probes are indicated in (A) and (B), respectively.
Holoenzyme–early melted DNA interactions. All mutant holoenzymes, with the exception of the E325G holoenzyme, bound the early melted probe with a similar pattern to that of homoduplex binding (data not shown). The wild-type holoenzyme binds to the early melted DNA probe to form a heparin resistant complex (27). Figure 6B and Table 1 show that holoenzymes formed with the mutant σ54 proteins E36G, E410A, E414, E431A and EE410/414AA form heparin stable complexes on the early melted DNA probe suggesting that determinants required for the acquisition of heparin stability on the early melted probe are largely intact in the mutant holoenzymes. Holoenzyme containing the E325G mutant σ54 is defective for binding to the early melted DNA probe in a heparin stable manner, a defect probably associated with the promoter DNA (Figs 4A and 6A) and/or core RNAP binding of that mutant (Fig. 3). Strikingly, even though the F318A mutant holoenzyme binds the early melted DNA probe with wild-type levels of activity (data not shown), it fails to form heparin stable complexes on the early melted DNA probe (Fig. 6B). It is possible that the inability of F318A to distinguish homoduplex and early melted DNA (see above) may correlate with its failure to form a stable complex with the core on the early melted DNA. F318A may fail to engage with the early melted DNA structure in a way that then confers heparin resistance upon the complex with core RNAP.
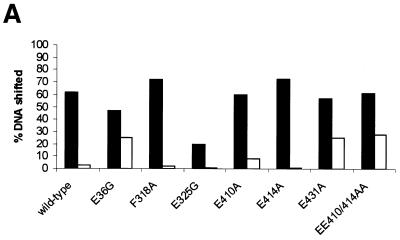
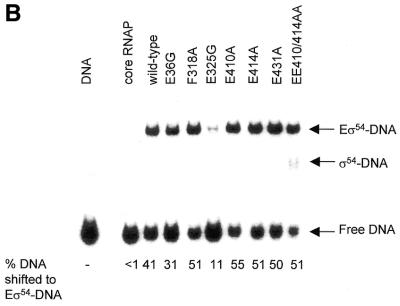
Activator-independent holoenzyme–late melted DNA interactions. The late melted DNA probe is believed to represent the conformation of promoter DNA in the open complex where sequences from –10 to –1 are heteroduplex (23; Fig. 4A). Results shown in Figure 7A and Table 1 suggest that all except the E325G mutant holoenzyme bind the late melted DNA probe with a similar activity as the wild-type holoenzyme. However, following a heparin challenge, only the mutant holoenzymes harbouring E36G, E431A and EE410/414AA σ54 formed heparin resistant complexes with the late melted DNA probe. Acquisition of heparin resistance independent of activation is normally a property of mutant σ54 holoenzymes that are deregulated for transcription. Such mutants bypass the requirement for activation and initiate transcription via an unstable open complex. Consistent with this it has been reported that σ54 harbouring a cysteine at position 36 (E36C) is partially deregulated (13). However, it also appears that alanine substitution at residues E431 and EE410/414AA confer upon the holoenzyme an ability to interact with the late melted DNA in an activator-independent manner to give heparin resistant complexes. Thus, it is possible that mutant holoenzymes (E431A and EE410/E414AA) and the wild-type σ54 holoenzymes represent two distinct functional states. Interestingly, we note that mutant holoenzymes harbouring the E410A or E414A mutations alone are not able to form heparin stable complexes on the late melted DNA probe independent of activation. It appears that both glutamic acid residues have to be replaced by alanine to confer heparin resistance to the σ54 holoenzyme. Further, the activator-independent heparin resistant complex formation by E431A and EE410/414AA holoenzymes predicts activator bypass transcription by these holoenzymes (4,5).
Figure 7.
σ54 Holoenzyme stability on the late melted promoter probe. (A) Binding (black bars) and stability (white bars) of wild-type and mutant σ54 holoenzyme complexes formed on the late melted promoter probe following a 5 min heparin challenge. (B) Stability of activated wild-type and mutant closed complexes formed on the late melted probe following a 5 min heparin challenge.
Activator-dependent holoenzyme–late melted DNA interactions. Efficient formation of heparin stable complexes by the wild-type σ54 holoenzyme on the late melted DNA probe requires activation, indicating that a conformational change in the holoenzyme must occur to allow heparin stable interactions with late melted DNA (2,23). We observed a clear activator-dependent increase in the number of heparin stable holoenzyme complexes formed on the late melted DNA probe, suggesting that the σ54 mutant holoenzymes are functional in responding to the activator protein and can engage with the late melted DNA in order to confer the holoenzyme heparin stability (Fig. 7B and Table 1). We note that the mutant E431A and EE410/414AA holoenzymes respond to the activator as evidenced by the increased number of heparin stable complexes formed (Fig. 7A). The holoenzyme with the E325G mutant forms the fewest activator-dependent heparin stable complexes on the late melted probe, probably owing to its core RNAP binding defects (see above) and activator interaction defects (see later).
Activator–σ54 interactions
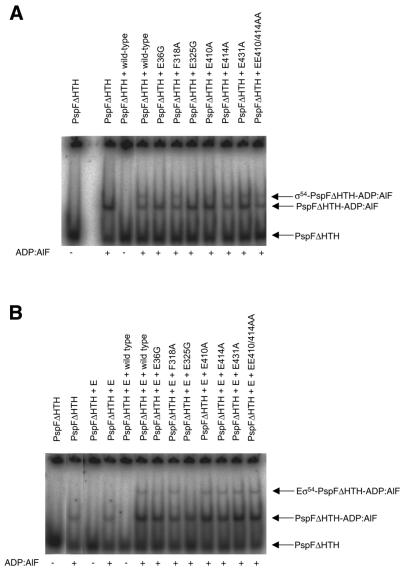
σ54 constitutes a major target within its holoenzyme for activator interaction (10). To seek a correlation between the activator-dependent interactions of mutant σ54 proteins and their holoenzymes with DNA probes and to assess the ability of the mutant σ54 proteins to stably interact with activator, we conducted direct σ54–activator interaction assays. The basic assay consisted of incubating PspFΔHTH and σ54 or the σ54 holoenzyme complex with nucleotide combinations and appropriate inorganic ions to mimic the pentavalent state of the γ-phosphate of ATP at the point of hydrolysis (ADP-AlF), then resolving the mixture on a native polyacrylamide gel (10). Results in Figure 8A and Table 1 show that the E36G and E325G mutant σ54 do not detectably form a stable nucleotide-dependent complex with PspFΔHTH. Residue 36 is part of the regulatory centre (6) and emerging data imply that it constitutes an important part of the target with which activator interacts (10). Given that the E36G mutant σ54 and the holoenzyme thereof are responsive to activator in DNA binding (see above) and transcription assays (see below), it is probable that increased conformational flexibility around region I due to the presence of glycine at position 36 prevents the E36G mutant from making a stable interaction with PspFΔHTH. The failure of E325G to form a nucleotide-dependent complex with PspFΔHTH argues that other determinants outside region I of σ54 also contribute to the interface σ54 makes with activator proteins.
Figure 8.
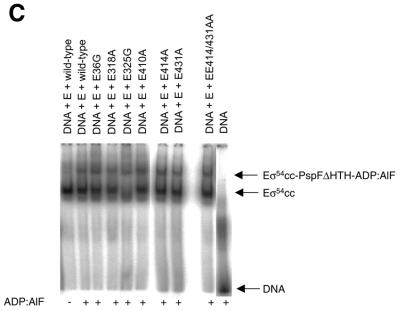
Nucleotide-dependent σ54–activator complex formation. (A) Nucleotide-dependent binding of wild-type and mutant σ54 proteins and (B) the holoenzymes thereof to 32P-labelled PspFΔHTH. (C) Nucleotide-dependent binding of wild-type and mutant closed complexes formed on the late melted DNA probe to PspFΔHTH.
In the context of the holoenzyme, some σ54 mutants (notably E414A and EE410/414AA) show an improved interaction with PspFΔHTH (Fig. 8B and Table 1). However, the E36G and E325G mutant σ54 proteins still failed to stably interact with PspFΔHTH. We note that E36G and E325G mutants also show defects in core RNAP binding and display strong aggregation (Fig. 3). Thus, the lack of any clear interaction of the E36G and E325G mutants with PspFΔHTH when present in the holoenzyme could also be attributed to their defects in core RNAP binding and/or migration difficulties into native gels.
Holoenzymes formed with E36G and E325G mutants formed a significant number of activation-dependent heparin stable complexes on the late melted DNA probe (Fig. 7B), implying that determinants within the E36G and E325G holoenzymes are nevertheless functional for activator responsiveness. Thus, we considered the possibility that the DNA-bound holoenzyme complex could be a preferred target for the PspFΔHTH. We investigated this possibility by forming complexes with the E36G and E325G holoenzymes on the late melted DNA probe and assaying whether PspFΔHTH could form a nucleotide-dependent stable complex with the late melted promoter-bound holoenzyme. Results shown in Figure 8C and Table 1 argue that PspFΔHTH is able to stably interact with the complexes formed with the E36G and E325G holoenzymes and the late melted DNA. Overall, the activator interaction assays confirm that region I of σ54, notably residue 36, contributes to the integrity of the interface of σ54 with PspFΔHTH. Further, region III residue E325 is either directly or indirectly involved in activator interaction, consistent with the idea that region I and region III of σ54 interact to form the regulatory centre (6,11).
In vitro transcription assays
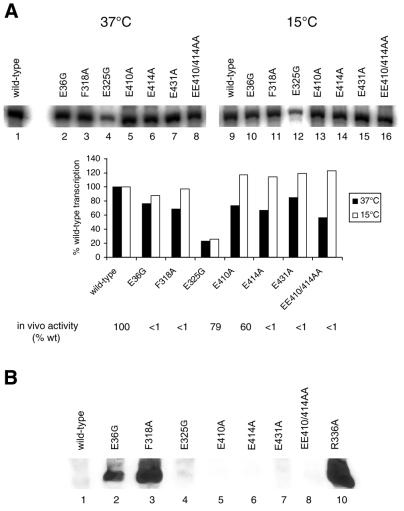
Activator-dependent transcription at 37°C. To correlate the in vitro DNA binding data, in vitro transcription assays were conducted from a supercoiled S.meliloti nifH promoter template (pMKC28) (5). As shown in Figure 9A and Table 1, all mutant holoenzymes were active for in vitro transcription (40–70 ± 4% wild-type holoenzyme activity). The reduced activity (19 ± 4% of wild-type holoenzyme activity) of the holoenzyme containing the E325G mutant σ54 correlates well with the amount of activation-dependent heparin stable complexes formed by this holoenzyme on the late melted DNA probe (Fig. 7B and Table 1). Holoenzymes containing the F318A, E410A, E414A, E431A and EE410/414AA mutant σ54 proteins formed more activator-dependent heparin stable complexes on the late melted promoter probe compared with the wild-type holoenzyme (Fig. 7B). Interestingly, this pattern of activity is not reflected in the in vitro transcription assays. Possibly, the holoenzymes containing the F318A, E410A, E414A, E431A and EE410/414AA mutant σ54 proteins are defective in some later stages in the transcription initiation process. In order to correlate the in vivo and in vitro activities of the mutant σ54s, we conducted in vitro transcription assays using NifA as the activator and pNH8 (which harbours the K.pneumoniae nifH promoter; 28) at 30°C. We did not detect any differences in the transcription pattern when using NifA/pNH8 or PspFΔHTH/pMKC28 (data not shown).
Figure 9.
In vitro transcription from S.meliloti nifH promoter. (A) Activated transcription by wild-type and mutant holoenzymes at 37 and 15°C. The in vivo activity of the mutants are shown for direct comparison (see also Fig. 2). (B) Activator-independent ‘bypass’ transcription.
Activator-dependent transcription at 15°C. Based on the expectation that open complex formation and DNA melting would be cold sensitive, the single round activator-dependent transcription assay was repeated at 15°C to see whether any phenotypes of the mutants were more distinguishable. Under these conditions, the E410A, E414A, E431A and EE410/414AA mutant holoenzymes produce ∼10–15 ± 3% more transcripts compared with the wild-type (Fig. 9A and Table 1). The improved DNA binding (E410 and EE410/414) and increased amount of activation-dependent heparin stable complex formation on the late melted probe (E410A, E414A, E431A and EE410/414AA) seem to correlate well with the transcription pattern of the mutants at 15°C. We note that the F318A mutant, albeit having increased DNA binding activity and activation-dependent heparin stable complex formation on late melted probe (like the E410 and EE410/414AA mutants), fails to give elevated levels of transcripts at 15°C (unlike the E410A and EE410/414AA mutants; Fig. 9A).
Activator-independent transcription assays. Mutations in region I of σ54 lead to activator-independent in vitro transcription (11,29). Recently, two charged residues (R336 and K388) in the region III DNA binding domain of σ54 have also been implicated in silencing activator-independent transcription (4,5,30). To determine whether any of our mutants bypassed the requirement of activator for transcript production, we tested all mutant holoenzymes in an in vitro activator-independent transcription assay. In this assay, the wild-type, E325G, E410A, E414A, E431A and EE410/414AA holoenzymes showed very low (<5%) levels of activator-independent transcript production. Consistent with previous observations, holoenzyme harbouring the E36G mutant σ54 was active for low levels of activator-independent transcription (11,13). Strikingly, the F318A holoenzyme gave very significant levels of activator-independent transcription (Fig. 9B). Thus, the F318A mutant appears to belong to a small group of region III residues that silences transcription.
σ54 F318A mutant is defective for DNA melting and template strand binding but not for creating a fork junction structure
Besides region III residues R336 and K388, residue F318 in σ54 appears important for silencing transcription initiation in the absence of activation (4,5; Fig. 9B). However, the F318A mutant differs significantly from R336A and K388A in that it is able to bind well to the early melted DNA probe, contrasting the binding properties of R336A and K388A (4,6). A fork junction-related structure is created when σ54 holoenzyme, but not when certain deregulated holoenzymes, binds promoters (31) as detected with the conformationally sensitive DNA footprinting reagent ortho-copper phenanthroline (o-Cup). Despite being deregulated for transcription, holoenzymes formed with F318A σ54 gave a clear o-Cup signal next to the promoter GC sequence (data not shown). Together with the early melted probe binding results (Fig. 5), the o-Cup data demonstrate that F318A mutant is not defective in creating an initial fork junction structure in the promoter DNA. Thus, in order to elucidate the basis for deregulation, we characterised the DNA binding properties of the F318A and its holoenzyme during the early stages of promoter opening by potassium permanganate footprinting and gel mobility shift assays using fork junction promoter DNA probes.
KMnO4 footprints of F318A–early melted DNA complexes. As the binding and isomerisation on the early melted DNA probe measures an early step during regulated (activator-dependent) open complex formation, we conducted KMnO4 footprinting on the F318A–early melted DNA complex and the isomerised complex thereof. Isomerisation of the wild-type σ54 on the early melted DNA is associated with the spreading of DNA melting away from the stably nucleated DNA structure towards the transcription start site (7,32). When KMnO4 was used to probe the template strand in F318A–DNA and isomerised F318A–DNA complexes formed on the early melted E.coli glnHp2 promoter, we failed to see any extensive deregulated (activator-independent) melting of DNA in the F318A–σ54 complex (data not shown). This suggests melting is transient for activator-independent transcription, occurring preferentially on supercoiled DNA. Interestingly, contrasting the extended KMnO4 footprint of isomerised (KMnO4 reactivity at thymines at positions –11, –9 and –7) wild-type σ54 complex, in the isomerised F318A–DNA complex, we detected limited KMnO4 reactivity at position –7 suggesting that either transient or no DNA melting has occurred beyond position –9 in isomerised complexes formed with the mutant F318A (data not shown). The apparent downstream DNA melting defects of the F318A mutant could explain the inability of its holoenzyme to form heparin stable complexes on the late melted DNA probe and could be linked to reduced transcription at lower temperatures. The significance of this melting defect for the bypass properties of F318A may have a basis in interactions with early melted DNA that help stop activator-independent transcription (see Discussion).
Fork junction DNA binding by F318A. σ54 needs to retain the ability to recognise promoter templates with single-stranded DNA near the start site to transcribe efficiently. Efficient binding and recognition of single-stranded DNA sequences is of regulatory importance in σ54-dependent transcription initiation (33,34). To assess this property and to extend results with heteroduplex DNA, we carried out binding assays for F318A, R336A and wild-type holoenzyme with fork junction probes based on the E.coli glnHp2 promoter. Four types of promoter probes were employed in our binding assays (Fig. 10A) that contained a frayed (A:A mismatch) at position –11, thus creating an optimal binding fork junction for σ54 binding (4). All three holoenzymes bound probe 1, which extends on the top strand to –7, equally well (Fig. 10B). In contrast to the R336A and wild-type holoenzyme, the F318A holoenzyme bound probe 2, which extends to –7 on the bottom strand, poorly, suggesting a weak interaction with bottom single-stranded DNA and a requirement for non-template strand sequences for stable binding. Consistent with this, all three holoenzymes bound similarly to the double-stranded probe (probe 3). However, only the wild-type and the R336A holoenzyme bound to the short double-stranded probe 4 which stops at –11, contrasting the F318A holoenzymes and demonstrating that the latter holoenzyme requires downstream non-template strand interactions for binding. Thus, it appears that the F318A holoenzyme is defective for template strand single-stranded DNA binding, a characteristic property of deregulated σ54 mutants (4,33,34).
DISCUSSION
The σ54 holoenzyme is distinctive in its ability to from a transcriptionally silent closed complex that only isomerises to a transcription competent open complex in response to an interaction with an enhancer-bound activator protein in a reaction requiring the hydrolysis of nucleoside triphosphates. To further understand regulated transcription initiation by the σ54 holoenzyme, we have targeted certain amino acids and determined that they are associated with particular functionalities by making substitutions guided by protein footprinting results. Surface-exposed residues in region III in σ54 that were protected from protease attack by the core RNAP in the holoenzyme or by promoter DNA in the closed complex were substituted with alanine (F318, E410, E414 and E431) or glycine (E325) to seek a correlation between the protein footprints and the functional importance of the amino acid. The in vivo and in vitro activity of the mutant σ54 proteins reported here strongly suggest that the protease-protected residues have a functionally significant role in σ54-dependent transcription initiation (summarised in Table 1).
Residue E36
Results from residue 36-tethered FeBABE footprinting experiments led to the conclusion that E36 is part of the regulatory nucleoprotein complex in closed complexes formed with the σ54 holoezyme (6,13) and constitutes a target for the activator protein to act upon. A glycine substitution at this position probably confers increased conformational flexibility of region I around position 36. Consistent with properties of other substitutions at E36, activator-independent transcription initiation by the holoenzyme harbouring the E36G mutant σ54 was evident (13). The importance of residue 36 in σ54 function is also highlighted by protein footprinting studies on the σ54–PspFΔHTH complex in which residue 36 is protected by PspFΔHTH from protease attack (10). Overall, the phenotype of the E36G mutant described here contributes to the significance of this residue in region I function (6,10,13,29).
Residue F318
Even though this residue lies outside the minimal σ54–DNA binding domain (residues 329–477), an alanine substitution at F318 (F318A) has significant consequences for the DNA binding and regulated transcription properties of the σ54 holoenzyme. The F318A mutant σ54 shows an increased binding activity to promoter DNA compared with the wild-type protein. It appears that mutating F318 has ‘unmasked’ extra DNA binding in σ54, perhaps by removing an energetically unfavourable interaction, one in which F318 contributes to early DNA melting. The observation that holoenzyme containing the F318A mutant σ54 gave increased activator-independent transcription initiation is likely linked to its altered DNA interactions. It is possible that the F318 protein contributes directly to single-stranded DNA binding with the early melted DNA required for regulated (activated) transcription initiation. Accordingly, DNA binding assays with F318A using –12 mismatched promoter probes containing different nucleotides at position –12 of the non-template strand reveal that F318A has an altered binding pattern compared with the wild-type (S.R.Wigneshweraraj and M.Buck, unpublished observations). Binding assays with fork junction probes suggest that the F318A holoenzyme is defective for the interaction with the template strand and requires the presence of the non-template strand for stable binding. The failure to see downstream DNA melting (towards the transcription start site) within the isomerised F318A σ54–early melted promoter complexes suggests a role for residue F318 in DNA melting. The role of phenylalanine residues in σ54 (35) and other proteins (36) in single-stranded DNA binding and DNA mismatch recognition has been reported.
Contrasting the wild-type holoenzyme, the F318A holoenzyme is unable to form heparin stable complexes on the early melted probe. Given its altered DNA binding property and proximity to core RNAP (12), it is plausible that residue F318 also functions to communicate promoter structure to core RNAP. As F318A binds the core RNAP well (Fig. 3) it is likely that F318A is defective in recognising promoter structures that lead to heparin stability of the holoenzyme. Consistently, the F318A holoenzyme is defective in binding single-stranded template DNA. This binding defect is probably associated with the heparin instability of the F318A holoenzyme on the late melted DNA probe.
The bypass properties of the F318A holoenzyme suggests that residue F318 is part of the regulatory centre in σ54 that includes the –12 promoter region, the DNA-crosslinking patch of σ54 and region I (6). It seems F318A is different to the previously described mutants [notably R336A (5) and K338A (4)] in σ54 region III that transcribe without activator. The latter fail to generate o-Cup-sensitive sites in footprinting reactions and bind fork junction probes weakly (4,6,30). The ability of F318A to transcribe without activator in vitro may be related to a defect closely related to changes in interactions that follow fork junction binding. In contrast to the previously described activator bypass σ54 mutants (in region I, R336A and K388A), the F318A σ54 binds well to the early melted probe. Dissociation assays showed that F318A binds the early melted probe with a similar affinity as the wild-type protein (S.R.Wigneshweraraj and M.Buck, unpublished observations). Nevertheless and in common with other bypass mutants, relative to the homoduplex DNA, binding is not increased with early melted DNA. A key property for limiting bypass transcription seems to be preferred binding to the early melted DNA. Unlike other bypass mutants (e.g. R336A), the F318A holoenzyme does not efficiently and stably engage with late melted DNA independent of activation, implying it has not deregulated in the same way. F318A falls within the interface of σ54 and core RNAP as deduced by protein footprinting (12). The F318A mutant may have a changed interaction with core RNAP that may also help explain its ability to transcribe in vitro without activator.
Residue E325
The mutant σ54 protein harbouring the E325G mutation is very defective for holoenzyme formation and DNA binding. This phenotype is reflected in the reduced in vitro transcription activity of the holoenzyme harbouring the E325G mutant σ54. An additional role for residue E325 in activator binding cannot be excluded, consistent with it being protected from protease attack when complexed with activator (10). The overall phenotype of the E325G mutant argues that E325 is involved in holoenzyme formation and activator interaction. The E325G mutant and to a lesser extent the E36G mutant displayed aggregation and did not readily enter into the native gel (Fig. 3). When interpreting DNA and activator binding results for these two mutants, this non-ideal behaviour needs consideration.
Residues E410, E414 and E431
These residues were protected from protease attack in the closed complex. The closed complex-specific protection can be interpreted as conformational changes that occur in σ54 upon DNA binding, but because the protected residues lie within the DNA binding domain, it is also possible that protection is due to their proximity to promoter DNA (12). The phenotypes of mutant σ54 proteins with alanine substitutions at E410, E414 and E431 support a role for these residues in promoter DNA interactions. Reduced DNA binding activity of the E431A mutant σ54 and its holoenzyme could be a reason for the lack of in vivo activity. Strikingly, like the F318A mutant, the E410A and EE410/414AA mutants showed increased DNA binding activity compared with the wild-type σ54. The region I deleted σ54 protein has an increased DNA binding activity (6,23). Mutating surface-exposed glutamate (and phenylalanine) residues in region III of σ54 may result indirectly in conformational changes in region I such that extra DNA binding activity is unmasked. Indeed region I and region III interact (6). The increased promoter DNA binding activity of the mutants (E410A, EE410/414A and F318A) compared with the wild-type protein is not reflected as an increased in vitro transcription activity. This implies that the mutant holoenzymes are defective in one or several steps after closed complex formation. Also, the increased DNA binding activity seen with the E410A EE410/414AA and F318A mutant σ54 proteins may impair efficient promoter clearance by the mutant holoenzymes once transcription is initiated. Interestingly, in an analysis of amino acid–DNA interactions in crystal structures, it appears that glutamate residues have a distinct preference for cytosine residues (37). Given that the σ54 binding site consensus is GC-rich, 5′-CGTGGCG-N5-TTGC-3′ (38), glutamate residues that are protected from protease attack in the closed complex (E410, E414 and E431) as well as the core RNAP-protected E325 may make specific contacts with promoter cytosine residues. Results suggest that certain glutamate residues in σ54 function to limit DNA binding. It is known that the promoter DNA sequence that is the strongest binding site for the σ54 holoenzyme is not the best promoter (39), and so limiting promoter binding may be important to allow, for example, changes in σ-promoter–DNA contacts as occurs when the open complex is formed from the closed complex, and when polymerase has to escape the promoter.
Residues E410 and E414 appear to have different roles in σ54-dependent transcription, probably contributing to the different conformational states adopted by σ54 during transcription initiation. However, the precise nature of this contribution remains unclear. The E431A and EE410/414AA holoenzymes form heparin stable complexes on the late melted DNA probe independent of activation, normally a property of deregulated holoenzymes. The failure to see activator-independent transcription with the E431A and EE410/414AA holoenzymes can probably be attributed to other defects in σ54-promoter–DNA interactions during open complex formation.
Summary
We have described the phenotypes of σ54 proteins carrying substitutions at surface-exposed residues that were protected from protease attack by either the core RNAP or by their proximity to the promoter DNA in the closed complex. The results presented here leave little doubt about the positive correlation of the protease-protected residues and their functional importance. For instance, residue E325, which was protected from protease attack by the core RNAP, when mutated to glycine, resulted in a mutant form of σ54 that was defective for holoenzyme formation, DNA binding and activator interaction. Similarly, residue F318, which is proximal to core RNAP, when substituted with alanine, resulted in a σ54 mutant with altered DNA binding and regulatory properties. The F318A mutant, as well as other mutants with alanine substitutions at residues E410, EE410/414AA that were protected by promoter DNA from protease cleavage, showed enhanced DNA binding activity to certain promoter conformations (homoduplex) but reduced binding activity on others (early melted promoter). The double mutant EE410/414AA had properties that were not simply the additive properties of the single mutants (E410A and E414A). Possibly the properties of the single mutants are partially masked by the functionality contributed by the second mutation. Several mutants displayed properties in vitro that were not easily reconciled with their in vivo properties (for example, E36G and E325G). Such differences have been observed before (14,15,40,41) and may relate to non-ideal behaviour of the mutant proteins in in vitro assays and issues of protein stability in vivo, although their precise bases remain unknown. Overall, our study of surface-exposed residues in σ54 by site-directed mutagenesis guided by previous protease footprinting studies has yielded a number of mutant σ54 proteins with a rich array of phenotypes and has identified residues and regions in σ54 of functional and regulatory significance that otherwise would have escaped analysis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Wendy Cannon for oligonucleotides and o-Cup phenanthroline footprinting and Matthew Chaney for helpful discussion and suggestions. This work was supported by a project grant from the BBSRC to M.B.
REFERENCES
- 1.Buck M., Gallegos,M.T., Studholme,D.J., Guo,Y. and Gralla,J.D. (2000) The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol., 182, 4129–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wedel A. and Kustu,S. (1995) The bacterial enhancer-binding protein NtrC is a molecular machine: ATP hydrolysis is coupled to transcriptional activation. Genes Dev., 9, 2042–2052. [DOI] [PubMed] [Google Scholar]
- 3.Popham D.L., Szeto,D., Keener,J. and Kustu,S. (1989) Purification of the alternative sigma factor, sigma 54, from Salmonella typhimurium and characterisation of sigma 54-holoenzyme. Science, 243, 629–635. [PubMed] [Google Scholar]
- 4.Wang L. and Gralla,J.D. (2001) Roles for the C-terminal region of sigma 54 in transcriptional silencing and DNA-binding. J. Biol. Chem., 276, 8979–8986. [DOI] [PubMed] [Google Scholar]
- 5.Chaney M. and Buck,M. (1999) The sigma 54 DNA binding domain includes a determinant of enhancer responsiveness. Mol. Microbiol., 33, 1200–1209. [DOI] [PubMed] [Google Scholar]
- 6.Wigneshweraraj S.R., Chaney,M.K., Ishihama,A. and Buck,M (2001) Regulatory sequences in sigma 54 localise near the start of DNA melting. J. Mol. Biol., 306, 681–701. [DOI] [PubMed] [Google Scholar]
- 7.Cannon W.V., Gallegos,M.T. and Buck,M. (2000) Isomerisation of a binary sigma-promoter DNA complex by enhancer binding transcription activators. Nature Struct. Biol., 7, 594–601. [DOI] [PubMed] [Google Scholar]
- 8.Rombel I., North,A., Hwang,I., Wyman,C. and Kustu,S. (1998) The bacterial enhancer-binding protein NtrC as a molecular machine. In Inglis,J. (ed), Cold Spring Harbor Symposia on Quantitative Biology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 58, 157–166. [DOI] [PubMed]
- 9.Wyman C., Rombel,I., North,A., Bustamante,C. and Kustu,S. (1997) Unusual oligomerisation required for activity of NtrC, a bacterial enhancer-binding protein. Science, 275, 1658–1661. [DOI] [PubMed] [Google Scholar]
- 10.Chaney M., Grande,R., Wigneshweraraj,S.R., Cannon,W., Casaz,P., Gallegos,M.T., Schumacher,J., Jones,S., Elderkin,S., Dago,A.E, Morett,E. and Buck,M. (2001) Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action. Genes Dev., 15, 2282–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casaz P. and Buck,M. (1999) Region I modifies DNA binding domain conformation of sigma 54 within the holoenzyme. J. Mol. Biol., 285, 507–514. [DOI] [PubMed] [Google Scholar]
- 12.Casaz P. and Buck,M. (1997) Probing the assembly of transcription initiation complexes through changes in sigma N protease sensitivity. Proc. Natl Acad. Sci. USA, 94, 12145–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wigneshweraraj S.R., Fujita,N., Ishihama,A. and Buck,M. (2000) Conservation of sigma-core RNA polymerase proximity relationships between the enhancer independent and enhancer dependent sigma classes. EMBO J., 19, 3038–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syed A. and Gralla,J.D. (1998) Identification of an N-terminal region of sigma 54 required for enhancer responsiveness. J. Bacteriol., 180, 5619–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y. and Gralla,J.D. (1997) DNA binding determinants of σ54 as deduced from libraries of mutations. J. Bacteriol., 179, 1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wigneshweraraj S.R., Ishihama,A. and Buck,M. (2001) In vitro roles of invariant helix-turn-helix motif residue R383 in σ54 (σN). Nucleic Acids Res., 29, 1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coppard J.R. and Merrick,M.J. (1991) Cassette mutagenesis implicates a helix-turn-helix motif in promoter recognition by the novel RNA polymerase sigma factor sigma 54. Mol. Microbiol., 5, 1309–1317. [DOI] [PubMed] [Google Scholar]
- 18.Gallegos M.T. and Buck,M. (1999) Sequences in sigma N determining holoenzyme formation and properties. J. Mol. Biol., 288, 539–553. [DOI] [PubMed] [Google Scholar]
- 19.Buck M., Kahn,R. and Dixon,R. (1985) Site-directed mutagenesis of the Klebsiella pneumoniae nifL and nifH promoters and in vivo analysis of promoter activity. Nucleic Acids Res., 13, 7621–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixon R., Eady,R.R., Espin,G., Hill,S., Iaccarino,M., Kahn,D. and Merrick,M.J. (1980) Analysis of regulation of Klebsiella pneumoniae nitrogen fixation (nif) gene cluster with gene fusions. Nature, 286, 128–132. [DOI] [PubMed] [Google Scholar]
- 21.Jishage M. and Ishihama,A. (1996) Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol., 178, 5447–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jovanovic G., Rajonjac,J. and Model,P. (1999) In vivo and in vitro activities of the Escherichia coli sigma 54 transcription activator PspF and its DNA binding mutant PspFΔHTH. J. Mol. Biol., 285, 469–456. [DOI] [PubMed] [Google Scholar]
- 23.Cannon W.V., Gallogos,M.T., Casaz,P. and Buck,M. (1999). Amino-terminal sequences of sigma N (sigma 54) inhibit RNA polymerase isomerisation. Genes Dev., 13, 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elliot S. and Geiduschek,E.P. (1994) Defining a bacteriophage T4 late promoter: absence of a –35 region. Cell, 36, 211–219. [DOI] [PubMed] [Google Scholar]
- 25.Buck M. and Cannon,W.V. (1992) Specific binding of the transcription factor sigma 54 to promoter DNA. Nature, 358, 422–428. [DOI] [PubMed] [Google Scholar]
- 26.Casaz P., Gallegos,M.T. and Buck,M. (1999) Systemic analysis of σ54 N-terminal sequences identifies regions involved in positive and negative regulation of transcription. J. Mol. Biol., 292, 229–239. [DOI] [PubMed] [Google Scholar]
- 27.Gallegos M.T. and Buck,M. (2000) Sequences in σ54 region I required for binding to early melted DNA and their involvement in sigma-DNA interactions. J. Mol. Biol., 297, 849–859. [DOI] [PubMed] [Google Scholar]
- 28.Wassem R., de Sousa,E.M., Yates,M.G., de Oliviera Pedrosa,F. and Buck,M. (2000) Two roles for integration host factor at an enhancer-dependent nifA promoter. Mol. Microbiol., 35, 756–764. [DOI] [PubMed] [Google Scholar]
- 29.Wang J.T., Syed,A., Hsieh,M. and Gralla,J.D. (1995) Converting Escherichia coli RNA polymerase into an enhancer-responsive enzyme: role of an NH2-terminal leucine patch in sigma 54. Science, 270, 992–994. [DOI] [PubMed] [Google Scholar]
- 30.Chaney M.K., Pitt,M. and Buck,M. (2000) Sequences within the DNA-crosslinking patch of sigma 54 involved in promoter recognition, σ isomerisation and open complex formation. J. Biol. Chem., 275, 22104–22113. [DOI] [PubMed] [Google Scholar]
- 31.Morris L., Cannon,W.V., Cleverie-Martin,F., Austin,S. and Buck,M. (1994) DNA distortion and nucleation of local DNA unwinding within σ54 (σN) holoenzyme closed promoter complexes. J. Biol. Chem., 269, 11563–11571. [PubMed] [Google Scholar]
- 32.Cannon W.V., Gallegos,M.T. and Buck,M. (2001) DNA melting within a binary sigma(54)-promoter DNA complex. J. Biol. Chem., 276, 386–394. [DOI] [PubMed] [Google Scholar]
- 33.Guo Y., Wang,L. and Gralla,J.D. (1999) A fork junction DNA-binding switch that controls promoter melting by the bacterial enhancer-dependent sigma factor. EMBO J., 18, 3736–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y. and Gralla,J.D. (2000) Promoter opening by σ54 and σ70 RNA polymerase: σ factor-directed alterations in the mechanism and tightness control. Genes Dev., 14, 2242–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oguiza J.A. and Buck,M. (1997) DNA-binding domain mutants of sigma N (sigma 54) defective between closed and stable open promoter complex formation. Mol. Microbiol., 26, 141–155. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto A., Schofield,M.J., Biswas,I. and Hsieh,P. (2000) Requirement for Phe36 for DNA binding and mismatch repair by Escherichia coli MutS protein. Nucleic Acids Res., 28, 3564–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandel-Gutfreund Y., Schueler,O. and Maraglit,H. (1995) Comprehensive analysis of hydrogen bonds in regulatory protein DNA-complexes: in search of common principles. J. Mol. Biol., 253, 370–382. [DOI] [PubMed] [Google Scholar]
- 38.Barrios H., Valderamam,B. and Morrett,E. (1999) Compilation and analysis of sigma 54 dependent promoter sequences. Nucleic Acids Res., 15, 4305–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L. and Gralla,J.D. (1998) Multiple in vivo roles for the –12 region elements of sigma 54 promoters. J. Bacteriol., 180, 5626–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Syed A. and Gralla,J.D. (1997) Isolation and properties of enhancer-bypass mutants of sigma 54. Mol. Microbiol., 23, 1239–1245. [DOI] [PubMed] [Google Scholar]
- 41.Hsieh M. and Gralla,J.D. (1994) Analysis of the N-terminal leucine heptad and hexad repeats of sigma 54. J. Mol. Biol., 239, 15–24. [DOI] [PubMed] [Google Scholar]
- 42.Merrick M.J. and Chambers,S. (1992) The helix-turn-helix motif of sigma 54 is involved in recognition of the –13 promoter region. J. Bacteriol., 174, 7221–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor M., Butler,R., Chambers,S., Casimiro,M., Badii,F. and Merrick,M.J. (1996) The RpoN box motif of the RNA polymerase sigma factor sigma N plays a role in promoter recognition. Mol. Microbiol., 22, 1045–1054. [DOI] [PubMed] [Google Scholar]