Abstract
Crystal structures of Type II restriction endonucleases demonstrate a conserved common core and active site residues but diverse structural elements involved in DNA sequence discrimination. Comparative structural analysis of restriction enzymes recognizing the same nucleotide sequence might therefore contribute to our understanding of the structural diversity of specificity determinants within restriction enzymes. We have solved the crystal structure of the Bacillus stearothermophilus restriction endonuclease Bse634I by the multiple isomorphous replacement technique to 2.17 Å resolution. Bse634I is an isoschisomer of the Cfr10I restriction enzyme whose crystal structure has been reported previously. Comparative structural analysis of the first pair of isoschisomeric enzymes revealed conserved structural determinants of sequence recognition and catalysis. However, conformations of the N-terminal subdomains differed between Bse634I/Cfr10I, suggesting a rigid body movement that might couple DNA recognition and catalysis. Structural similarities extend to the quaternary structure level: crystal contacts suggest that Bse634I similarly to Cfr10I is arranged as a tetramer. Kinetic analysis reveals that Bse634I is able to interact simultaneously with two recognition sites supporting the tetrameric architecture of the protein. Thus, restriction enzymes Bse634I, Cfr10I and NgoMIV, recognizing overlapping nucleotide sequences, exhibit a conserved tetrameric architecture that is of functional importance.
INTRODUCTION
In order to understand possible structural and mechanistical diversity of Type II restriction endonucleases we have focused on the structural analysis of enzymes recognizing closely related nucleotide sequences. Comparison of the crystal structure of MunI restriction enzyme (recognition sequence C/AATTG) with the previously solved structure of EcoRI (recognition sequence G/AATTC) provided a first case study (1). Analysis of the structural elements employed by MunI and EcoRI for sequence recognition revealed that both enzymes use a conserved mechanism for the interaction with a common AATT target but differ in the recognition of external nucleotides, suggesting a possible modular organization of the specificity determinants.
The conservation of target recognition elements observed for MunI and EcoRI seems not to be a general rule. Structural analysis of the specificity determinants of BglII (recognition sequence A/GATCT) and BamHI (recognition sequence G/GATCC) restriction enzymes indicated that both proteins display different protein–DNA contacts even at the common GATC target (2). Thus, in contrast to MunI–EcoRI, a single base pair difference in the recognition site leads to large differences in the DNA recognition elements of BglII and BamHI, demonstrating that both proteins use independent mechanisms of target recognition.
Comparative structural analysis of restriction enzymes recognizing the same nucleotide sequence might further contribute to our understanding of the structural diversity of specificity determinants within restriction enzymes. In this paper we report the crystal structure of Bse634I restriction enzyme at 2.17 Å resolution. The Bse634I restriction enzyme (3) from Bacillus stearothermophilus recognizes the nucleotide sequence Pu/CCGGPy (cleavage point indicated by /) and is an isoschisomer of the Cfr10I restriction enzyme from Citrobacter freundii (4). The crystal structure of Bse634I complements our previous crystallographic studies of the Cfr10I restriction enzyme (5) and allows, for the first time, a direct structural comparison of two restriction enzymes recognizing the same DNA sequence. Structural comparison reveals a high degree of structural homology between Bse634I and Cfr10I, and suggests that in both enzymes DNA recognition and catalysis are possibly coupled through the rearrangement of the flexible N-terminal subdomains.
MATERIALS AND METHODS
Expression of Bse634I restriction endonuclease
Cloning of the restriction–modification genes of Bse634I was performed using the methyltransferase selection technique (6). pET3c expression vector (Novagen) was used for the initial cloning and expression of Bse634I restriction endonuclease. Primers containing sites of NdeI and BamHI restriction enzymes were used to amplify the gene of Bse634I from the 11.8-kb plasmid pBse634IRM (derived from pACYC177-E). The amplified fragment containing the gene encoding Bse634I was cloned into the NdeI and BamHI sites of pET3c, yielding the 6.6-kb ampicillin resistance (Apr) plasmid pBse634IR6.6. The cloning hosts were obtained by transformation of Escherichia coli ER2267 [recA1 (McrA–) lacIq lacZΔM15 zzf::mini-Tn10 [kanamycin resistance (Kanr)] Δ(mcrC-mrr)] with the 6.2-kb plasmid pHpaIIM (tetracyclin resistance, chloramphenicol resistance) containing the gene encoding HpaII DNA methyltransferase (MTase) (7). The HpaII MTase modifies the internal cytosine within 5′-CCGG-3′, yielding C5-methylcytosine (8). The DNA modified by HpaII MTase becomes resistant to Bse634I endonuclease cleavage. Unfortunately, we were unable to propagate pBse634IR6.6 in strains HMS174(DE3) or BL21(DE3) expressing T7 RNA polymerase. Therefore, the coding sequence of bse634IR was amplified from pBse634IR6.6 using standard primers (Novagen) recognizing the promoter and terminator regions of pET vectors. The obtained PCR fragment was digested with XbaI, blunt ended, and once more digested with BamHI. The resulting DNA fragment was finally cloned through the BamHI and blunt ended Ecl136II sites of pUC18 yielding the 3.7-kb Apr plasmid pBse634IR3.7. In this plasmid, bse634IR was placed under the control of the standard isopropyl-1-thio-β-d-galactopyranoside (IPTG)-inducible lacZ promoter of pUC18. The expression of Bse634I ENase was performed in E.coli ER2267 [recA1 lacIq lacZΔM15 zzf::mini-Tn10(Kanr) Δ(mcrC-mrr)] carrying pHpaIIM. The integrity of expressed protein was monitored by SDS–PAGE and confirmed by determination of the N-terminal sequence.
Protein purification
The bacterial cells of E.coli ER2267 strain carrying compatible plasmids pBse634IR3.7 and pHpaIIM were grown to late logarithmic phase in Luria broth medium containing 50 mg/l ampicillin and 30 mg/l chloramphenicol. After induction with IPTG (0.5 mM, 4 h) the cells were harvested by centrifugation and resuspended in chromatography buffer (10 mM K3PO4, pH 7.4; 100 mM NaCl; 7 mM 2-mercaptoethanol; 1 mM EDTA). Crude cell extract was obtained by sonication, and cell debris was separated by centrifugation. The resulting supernatant was applied to a phosphocellulose (Whatman) column and eluted using a NaCl gradient. The purification of Bse634I endonuclease was monitored by following λDNA cleavage activity (see below) in the fractions. The fractions containing active endonuclease were pooled and dialyzed against the chromatography buffer (see above). Further protein purification was achieved by subsequent chromatography on heparin–Sepharose and blue-Sepharose (Pharmacia) columns. Final fractions containing purified enzyme were pooled and dialyzed against storage buffer (10 mM K3PO4, pH 7.4; 100 mM KCl; 2 mM dithiothreitol; 0.1 mM EDTA; 50% glycerol) and stored at –20°C. The protein was homogeneous according to SDS–PAGE analysis. Protein concentrations were determined spectrophotometrically at 280 nm using an extinction coefficient of 34 400 M–1 cm–1 for a monomer. The concentration of Bse634I endonuclease is given in terms of tetramer.
λ DNA cleavage assay
The cleavage of λ DNA by Bse634I was monitored as described by Skirgaila et al. (9) except that the reaction buffer contained 10 mM Tris–HCl (pH 8.5 at 37°C), 10 mM MgCl2, 100 mM KCl and 1 µg λ DNA.
Crystallization
The Bse634I restriction endonuclease has been crystallized using the sitting drop vapor diffusion technique. The 0.10 mM tetramer protein solution in 20 mM Tris–HCl, 50 mM NaCl and 1 mM EDTA has been mixed with the reservoir solution containing 100 mM Na acetate buffer at pH 5.5, 12% PEG 8000 and 100 mM CaCl2 in a 1:2 ratio (2 µl protein solution and 4 µl reservoir solution) in the depletions of Cryschem“ plates. Drops were equilibrated against 500 µl of the reservoir solution. Crystals grew in 4–7 days.
Data collection and processing
The data for the final refinement have been collected on the BW6 beamline at DESY, Hamburg. Bse634I crystals have been soaked in a cryoprotecting buffer containing 25% (v/v) glycerol, 14% PEG 8000 (other components same as in crystallization buffer) for 1.5 h and frozen in a cold nitrogen stream (90 K) immediately before measurement. Data from the heavy atom derivatives have been collected on a Rigaku RU-200 rotating anode generator equipped with a MAR Research Image plate detector. Oscillation images have been processed using the DENZO program package (10) and scaled with Scalepack; the data collection statistics are shown in Table 1. Difference Patterson maps for the heavy atom derivatives have been calculated using the CCP4 (11) program suite. The Harker sections of the maps have been extracted and searched for possible heavy atom positions using the hara program (S. Grazulis, unpublished). Single isomorphous replacement phases from each derivative were used to search/verify positions in other derivatives. The found heavy atom positions were brought to a common origin and hand using difference Fourier syntheses, then the positions have been refined and the multiple isomorphous replacement phases have been calculated with the help of the mlphare program from the CCP4 suite.
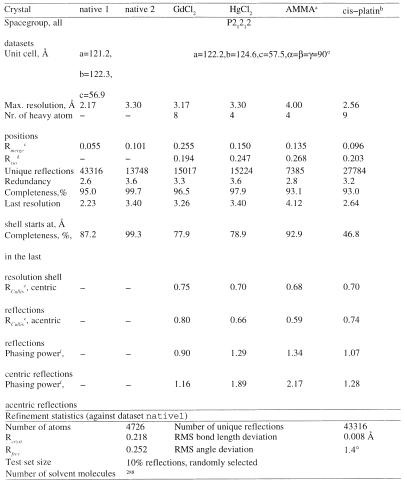
Table 1. Data collection, phasing and refinement statistics.
a4,6-bis-(acetoxymercury)-2-methyl-aniline.
bcis-[PtCl2(NH3)2].
cRmerge=σhσi=1nh(<Ih>–Ihi)2) / Σi Ih2 where Ih is the intensity value of i-th measurement of reflection h, and <Ih> is the average measured intensity of reflection h. nh is the number of measurements of reflection h.
dRiso=Σh|Fp–Fph|/Σh|Fp|, where Fp and Fph are native and derivative structure factors, respectively.
eRCullis=Σh|Fh(obs)–Fh(calc)|/Σh|Fh(obs)|.
fPhasing power=<|Fh(obs)|>/ RMSD(ɛ), where ɛ is lack of closure.
Model building and refinement
The phases from four derivatives gave an interpretable map after solvent flattening, into which three-quarters of the model could be built with the O molecular modeling program (12). The model was then transferred to the native 1 data set (Table 1) using CNS (13). In the successive cycles of building and refinement the rest of the model became visible in the σA weighted 2Fo – Fc electron density maps. The new parts of the model have been built into the model only when the density of these parts was well defined in the maps phased with the truncated model. Finally, ions and water molecules have been added. Coordinates have been deposited in the PDB with accession code 1KNV.
Accessible surface calculations
Accessible surface areas have been calculated using the naccess program (http://wolf.bms.umist.ac.uk/naccess). The surface area buried between any subunits X and Y was calculated as (X + Y) – XY, where X and Y are accessible surface areas of individual chains X and Y, and XY is the accessible surface area of the complex. Calculations of buried surface area for ribonuclease (PDB entry 8rsa) yielded a value of 1812 Å2, which differs <1% from the published reference value 1795 Å2 (14).
Structural comparisons
Structural comparison of Bse634I with other restriction enzymes has been performed by superimposing the active site of Bse634I as a rigid body with active sites of other restriction endonucleases using Kabsch’s method (15). For convenient access the superposition algorithm has been coded in the Perl programming language (16). All molecule figures were prepared with the Molscript (17), Bobscript (18) and Raster3d (19) packages.
Supercoiled plasmid cleavage assay
Supercoiled plasmids pUC19 (20) and pUCAC2 (see below), containing one and two Bse634I recognition sites (5′-ACCGGC), respectively, were used in cleavage experiments. A 28-bp cognate oligonucleotide duplex obtained by annealing two complementary oligonucleotides 5′-CGC GAG ACC CAC GCT CAC CGG CTC CAG A and 5′-TCT GGA GCC GGT GAG CGT GGG TCT CGC G (recognition sequences of Bse634I are shown in bold) was ligated to pUC19 pre-cleaved with SmaI to yield pUCAC2. The sequences flanking the engineered 5′-ACCGGC site in pUCAC2 are identical to those in pUC19. Both plasmids were purified by CsCl centrifugation (21), >90% as supercoiled monomers. Cleavage experiments were performed at 25°C in an assay buffer containing 30 mM Tris–acetate (pH 8.5, 25°C), 70 mM CH3COOK and 0.1 mg/ml bovine serum albumin (BSA). Varying concentrations of Bse634I (0.25–50 nM tetramer) were mixed with 2.3 nM pUC19 or pUCAC2 in the assay buffer, and the reaction was initiated by adding (CH3COO)2Mg to give a final concentration of 10 mM. The effect of the oligonucleotides on the Bse634I cleavage rate of the pUC19 was studied by adding to the reaction mixture 50–400 nM of cognate 28 bp duplex (see above) or non-cognate oligonucleotide duplex obtained by annealing complementary 30-nt oligonucleotides 5′-AGC GTA GCA CTG GGC TGC TGA ACT GTG CTG and 5′-CAG CAC AGT TCA GCA GCC CAG TGC TAC GCT. Aliquots were removed after fixed time intervals (the shortest accessible reaction time was 4 s) and mixed with loading dye solution containing EDTA. The DNA samples were separated in agarose gel, and the amounts of supercoiled (SC), open circular (OC), linear with one double-strand break (L1), and linear with two double-strand breaks (L2, observed only in the case of pUCAC2 cleavage) forms of plasmid DNA were determined by densitometric analysis of ethidium bromide-stained gels (22). Cleavage experiments were performed at 25°C to make the reaction rates slow enough to collect samples manually and avoid melting of the oligonucleotide duplexes. Exponential function was fitted to the supercoiled plasmid depletion curves obtained under excess of enzyme and apparent first-order reaction rate constants (k1) were determined.
RESULTS AND DISCUSSION
General features
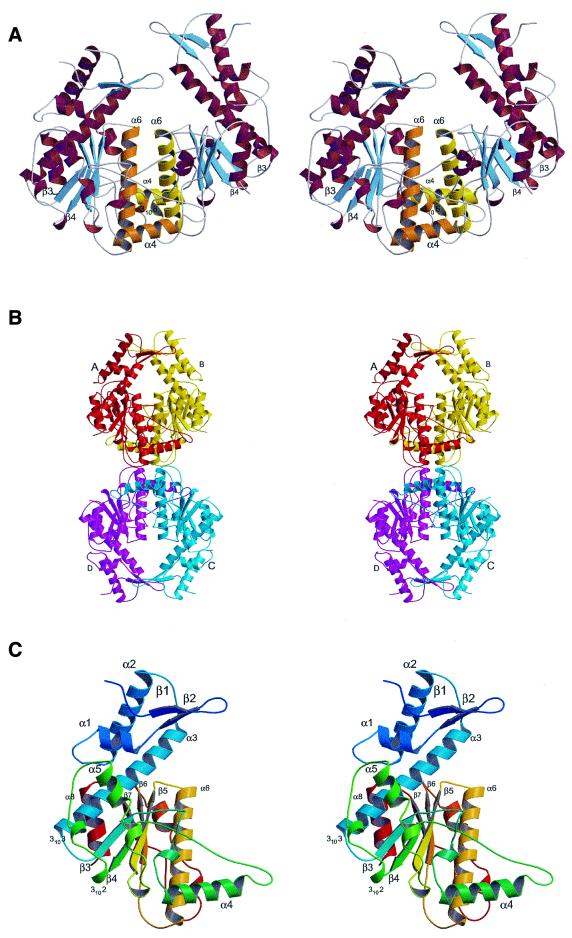
There are two non-crystallographic symmetry (NCS)-related protein chains A and B in the asymmetric unit. Chains A and B build up a U-shaped dimer (Fig. 1A) with a 30 Å wide cleft which is large enough to accommodate a B-DNA molecule. Helices α6 and 3106 from the two monomers related by NCS dominate at the dimer interface. Of note is that structurally equivalent helices are located at the dimer interface of other restriction enzymes that cleave hexanucleotide sequences giving 4-bp 5́-overhangs (1,23,24). Additional intersubunit contacts in the Bse634I dimer come from protein chain segment (α4 helix followed by a loop) located between β3 and β4 strands and extending out of the core of each monomer. Hydrophobic interactions supported by a few hydrogen bonds dominate across the dimer interface. Calculation of the accessible surface area indicates that a total surface of 3100 Å2 is buried at the interface between the two Bse634I monomers. This value is consistent with the values reported by Janin (14) for specific protein–protein contacts of the oligomeric proteins.
Figure 1.
General view of Bse634I restriction endonuclease. (A) Ribbon representation of a Bse634I dimer. Structural elements involved in dimerization are shown in orange (subunit A) and yellow (subunit B); β-sheets are shown in blue. (B) Arrangement of the protein chains in the crystal suggesting possible structure of the Bse634I tetramer. Chain A is in red, chain B is in yellow, chain C is in blue and chain D in magenta. A crystallographic 2-fold axis relating A to C and B to D is perpendicular to the figure plane and passes through the center of the picture. (C) Ribbon representation of the Bse634I monomer in stereo. The color changes from blue to red following residues from N-terminus to C-terminus.
Two dimers in a unit cell related by a 2-fold crystallographic axis (A to B: –X + 1, –Y + 1, Z) are arranged ‘back-to-back’ with their putative DNA-binding clefts in the opposite directions (Fig. 1B). A total surface area of 3400 Å2 (or 1700 Å2 per chain) becomes buried between two dimers AB and CD. This value is characteristic for the specific protein–protein contacts (14) and suggests that the Bse634I tetramer also exists in solution. Indeed, the sedimentation equilibrium analysis experiment of Bse634I (initial concentration 7.2 µM) yields a molecular mass of 123 kDa which is very close to that calculated for a Bse634I tetramer (data not shown). The tetramerization interface is formed by the amino acid residues located at the C-terminal ends of the helices α6 and short loops beyond the 3106 helices (residues 260–264). Hydrophobic interactions dominate at the interface between two dimers. Of note is that helices α6 contribute both to the dimer and tetramer interface. Other contact surface areas between neighboring protein molecules related by crystal symmetry are less than half of the tetramer contact.
Monomer architecture
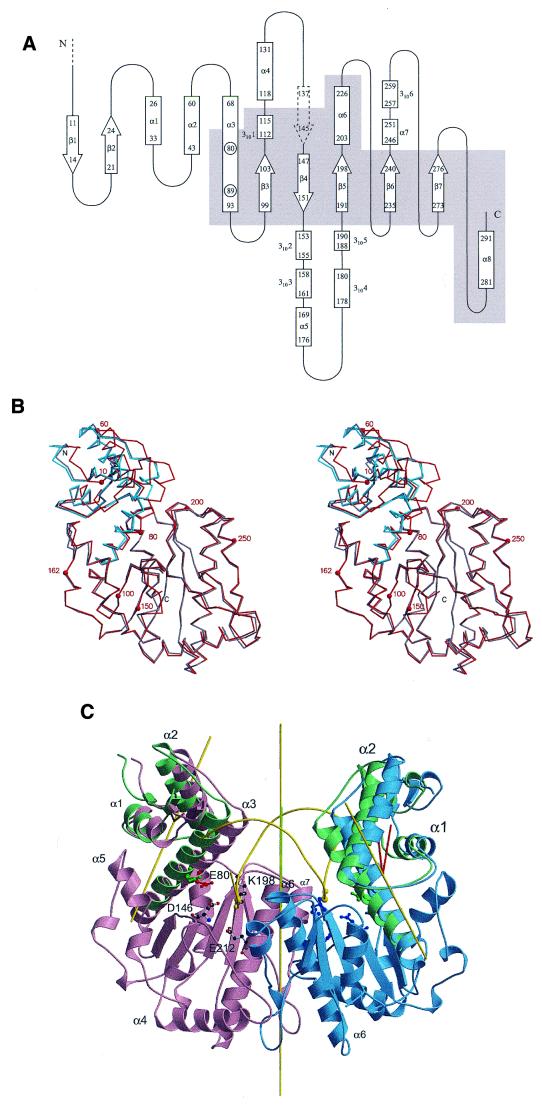
The single Bse634I chain is folded into a compact α/β structure (Fig. 1C) with approximate dimensions of 66 × 57 × 48 Å. The five-stranded β-sheet (strands β3–β7) makes up the core of the protein globule. The central β-sheet is flanked on one side by helix α8 and the C-terminus of helix α3 on the opposite side by the short helix 3101 and helix α6. The general topology (Fig. 2A) of the Bse634I restriction enzyme is similar to that of other restriction enzymes (25) that cleave hexanucleotide sequences giving 4-bp 5′-overhangs.
Figure 2.
(Opposite) Structural comparison of subunits A and B of Bse634I restriction endonuclease. (A) Topology diagram of Bse634I restriction endonuclease. The central core region is shown on the gray background. The first three N-terminal amino acids invisible in the density are denoted as a dashed line. Dashed block arrow represents a part of the chain in extended conformation which does not however belong to the central β-sheet. (B) Superposition of the Cα traces of Bse634I subunits A (red) and B (gray) in stereo. C-terminal subdomains of A and B were superimposed. In blue, the N-terminal subdomain of A subunit is shown after additional rotation that superimposes it with the N-subdomain of the subunit B. (C) Rigid body motion of the N-terminal domains in Bse634I. Two conformational states of the N-terminal subdomain are depicted in green and purple for subunit A and in green and blue for subunit B, respectively. The backbone of the DNA modeled into the putative DNA-binding cleft is shown in yellow. The scissile bond phosphate is shown as a large sphere, and a scissile bond oxygen is shown as small sphere on the DNA backbone. The yellow lines show the 2-fold non-crystallographic axis of the protein dimer and the rotation axes for the subdomains. Red spikes at the subdomain rotation axes depict the subdomain rotation angle (10°). The green line shows the dyad axis of the DNA, which has been brought into superposition with the protein axis.
Comparative analysis of the NCS-related subunits A and B in the Bse634I dimer reveals local conformational differences (Fig. 2B). Indeed, individual chains of subunits A and B of Bse634I could be superimposed only with the RMS deviations of 1.7 Å (for all atoms)/1.5 Å (for Cα atoms). Detailed differences in local conformations of subunits A and B were analyzed by superimposing Cα atoms of protein subfragments. This analysis revealed that the N-terminal parts (N-domain) (residues 1–89, helices α1–α3 and strands β1 and β2; depicted in green in Fig. 2C) and C-terminal parts (C-domain) (residues 90–293, helices 3101–α8 and strands β3–β7) can be superimposed with much lower RMSD than complete chains. Indeed, the best RMS deviations are 1.1 Å (all atoms)/0.61 Å (Cα) for C-domains and 1.0 Å (all atoms)/0.61 Å (Cα) for N-domains. However, extension of the length of the N-terminal subdomain beyond residue 90 sharply increased the RMSD to the value of 1.4–1.5 Å, close to the value for the complete chain. A similar trend is seen for the C-terminal subdomain. This suggests that the Bse634I monomer is comprised of separate N- and C-subdomains connected by a hinge located between residues 70 and 90 in helix α3. Helix α3 dominates the interface between N- and C-terminal subdomains. It contacts the C-terminal subdomain at strands β3 and β4 and helix α8. The C-terminus of the α3 helix becomes sandwiched between the loop protruding between β3 and β4 strands and helix α8. A cluster of hydrophobic residues Ile83, Ala84, Ile85, Trp88, Tyr90 and Val92 exposed on one side of the α3-helix contributes to the interface between α3 and C-terminal subdomain. Hydrophobic residues Phe71 and Trp87 positioned on the opposite side of the helix α3 and a possible salt bridge between Asp48 and Arg75 residues make an interface with an N-terminal part of the protein.
The N-terminal domains of individual subunits A and B (Fig. 2C) appear to be rotated ∼10° around axes that pass through the Cα atom of residue Asn89 in the helix α3, in good agreement with the hinge position estimated from the RMSD analysis. These rotation axes make a 30° angle with the dimer dyad axis and ∼50° angle with helix α3. We suppose that two rigid domains are connected by a relatively flexible joint located at residue Asn89. The movement of the N-terminal domains of the Bse634I protein is most likely induced by crystal packing forces. Since weak lattice interactions appear to be sufficient to displace the N-subdomains of the protein, one might speculate that sequence-specific interactions of Bse634I with DNA might induce even larger N-terminal subdomain movements.
At least two other restriction endonucleases, PvuII and EcoRV, exhibit similar conformational changes as described above for Bse634I. The PvuII restriction endonuclease undergoes transition from an ‘open’ conformation observed in the apo-enzyme (26) to a ‘closed’ DNA-bound form (27).
The relative movement of subdomains has been also analyzed in several EcoRV structures of wild-type and T93A mutant proteins in different crystal environments both in DNA-bound and free states (28). In different lattice environments, DNA-binding subdomains of EcoRV are reported to rotate 6–11°, a value very close to that found in Bse634I structure. Upon DNA binding, the subdomains of EcoRV rotate 22–28° (28), similarly to the subdomains of PvuII. These studies indicate that restriction enzymes Bse634I, EcoRV and PvuII undergo conformational changes that might be described as rigid body movements of the separate subdomains in respect to each other. Such structural rearrangements in the case of PvuII and EcoRV are enhanced upon binding of cognate DNA and play an important role in the sequence recognition and catalysis. We propose a similar mechanism for the Bse634I restriction enzyme.
The conformational changes of Bse634I, PvuII and EcoRV structures differ from structural rearrangements reported for the BamHI restriction enzyme. Comparison of apo–BamHI and BamHI–DNA complex structures revealed an unfolding of the C-terminal helix and 19° rotation of the entire protein subunits in respect to each other upon DNA binding (24). One can speculate that such subunit rotation in BamHI plays the same role as the N-domain motion in Bse634I, in both cases narrowing the DNA-binding cleft and enabling specific DNA–protein contacts that otherwise could not be formed.
The central core of the C-terminal subdomain in the Bse634I is structurally conserved between all known structures of restriction endonucleases. In contrast, conformations of the N-domain of Bse634I and N-terminal parts of other restriction enzymes differ significantly. While most of the contacts to DNA come from the structural elements surrounding the conserved central core of restriction enzymes, in a few cases N-terminal parts provide additional specificity (26,29–32).
Structural comparison of Bse634I with Cfr10I
The Bse634I protein shares 30% sequence identity and 50% similarity with the isoschisomeric Cfr10I protein, which suggests similar folds (33). Indeed, the crystal structures of both proteins are very similar. However, a 9° rotation of the Cfr10I N-terminal subdomain is necessary to superimpose it with subunit A of Bse634I and 13° with subunit B; the rotation axis in both cases passes through the middle of the respective helices α3. On the basis of structural comparisons we propose that Cfr10I has the same subdomain organization and probably undergoes similar conformational changes as Bse634I. These were not observed in Cfr10I previously since there is only one subunit of Cfr10I in the asymmetric unit of the crystal (5). A structural comparison with Bse634I suggests that the N-terminal subdomain of Cfr10I should extend from residue 1 to Glu80 that is a structural counterpart of the Asn89 residue of Bse634I. Each subdomain of Cfr10I can be superimposed onto the corresponding Bse634I domain with RMS deviations of 1.1 Å for the N-terminal domain and 1.3 Å for the C-terminal domain (only the identical residues from the sequence alignment have been used for the superposition of both proteins). However, if the same residues are used for superposition of the entire proteins, the RMS deviation is increased to 2.0 Å, indicating similar subdomain organization in Cfr10I and Bse634I.
Catalytic/metal-binding site
All currently known restriction enzymes except BfiI (34) need magnesium ions for catalysis. In the final Bse634I electron density map there is no density that could be interpreted as a metal ion, but the residues of the catalytic/metal-binding site can be predicted from the structural comparisons with the other restriction endonucleases.
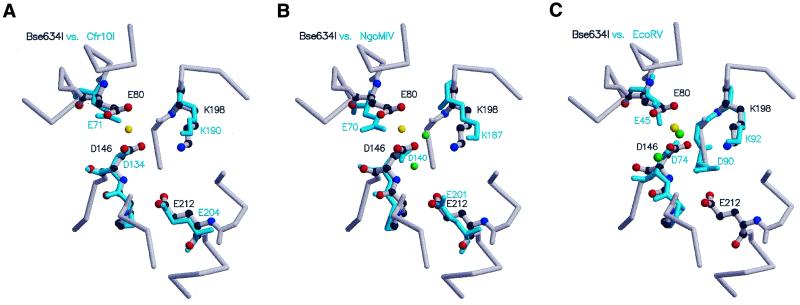
Superposition of the central β-sheets between Bse634I and Cfr10 restriction enzymes revealed that amino acid residues Asp146, Lys198 and Glu212 of Bse634I spatially overlap with Asp134, Lys190 and Glu204 residues of Cfr10I (Fig. 3A) which constitute the catalytic/metal-binding site (5,9). Gly196 residue in Bse634I appeared to be a structural equivalent of the Ser188 residue of Cfr10I. Thus, we suggest that residues Asp146, Lys198 and Glu212 contribute to the catalytic/metal-binding site of Bse634I.
Figure 3.
Catalytic/metal-binding site of the Bse634I restriction endonuclease. (A) Comparison of the catalytic/metal-binding sites of Bse634I and Cfr10I restriction enzymes. Bse634I is shown in gray and Cfr10I is shown in cyan. The yellow sphere shows the position of the Gd3+ ion in the Bse634I heavy atom derivative. (B) Comparison of the catalytic/metal-binding sites of Bse634I and NgoMIV restriction enzymes. Bse634I is shown in gray and NgoMIV restriction enzyme is shown in cyan. Green spheres show the positions of Mg2+ ions in the NgoMIV–DNA complex. The yellow sphere shows the position of the Gd3+ ion in the Bse634I heavy atom derivative. (C) Comparison of the catalytic/metal-binding sites of Bse634I and EcoRV restriction enzymes. Bse634I is shown in gray and EcoRV is shown in cyan. Green spheres show the positions of Mg2+ ions in the EcoRV structure. The yellow sphere shows the position of the Gd3+ ion in the Bse634I heavy atom derivative.
In most restriction enzymes, two acidic residues and a lysine from the conserved sequence motif PDX10–30(D/E)XK are located at the ends of β-strands and comprise the first catalytic metal/binding site (25,35). The Asp146 of Bse634I spatially coincides with the aspartate residue that is invariant in all active sites of restriction enzymes (Table 2). Lys198 of Bse634I superimposes with conserved lysine residues (except for BamHI and BglII) from the active signature motif PDX10–30(D/E)XK. Similarly, the Pro145 of Bse634I was structurally equivalent to the proline present at the active sites of a number of restriction enzymes (Table 2). The best RMSD for the Pro145, Asp146, and Lys198 residues of Bse634I with their structural equivalents in other restriction enzymes ranges from a maximum 1.5 Å for EcoRV to a minimum 0.5 Å for MunI. Of note is that the Gly188 residue of Bse634I was located at the spatial position occupied by a second acidic residue (aspartate or glutamate) at the active sites of other restriction enzymes except for the Cfr10I and NgoMIV (Table 2).
Table 2. Structural correspondence of the catalytic/Mg2+-binding residues of Type II restriction endonucleases.
| Bse634I | Cfr10I | NgoMIV | MunI | EcoRI | EcoRV | BamHI | PvuII | BglI | BglII |
|---|---|---|---|---|---|---|---|---|---|
| E80 | E71 | E70 | - | D59a | E45 | K61a | E55a | E87 | N54a |
| P145 | P133 | P139 | P82 | P90 | P73 | I93 | N57a | P115 | I83 |
| D146 | D134 | D140 | D83 | D91 | D74 | D94 | D58 | D116 | D84 |
| G196 | S188b | S185 | E98 | E111 | D90 | E111 | E68 | D142 | E93 |
| K198 | K190 | K187 | K100 | K113 | K92 | E113 | K70 | K144 | Q95 |
| E212 | E204c | E201 | L125 | N149a | – | K156a | – | Q161a | R108a |
The central β-sheet of Bse634I was superimposed with structurally equivalent β-sheets of other restriction enzymes and residues spatially overlapping with putative catalytic/Mg2+-binding residues of Bse634I were selected.
aResidues that overlap spatially but come from the non-equivalent secondary structure elements or have different functional groups; their correspondence might be casual.
bS188 is not crucial for catalysis of Cfr10I according to Skirgaila et al. (9).
cE204 is catalytically important for Cfr10I and is a structural and functional analog of E98 in MunI and E111 in EcoRI (9).
Glu212 of Bse634I overlaps with Glu204 in Cfr10I and Glu201 in NgoMIV (Fig. 3A and B). In Cfr10I, the Glu204 has been shown to be the structural counterpart of the Asp90 in EcoRV and Glu111 in EcoRI, although it comes from a different part of the sequence (9). Mutational experiments revealed that a ‘swap’ mutant of the Cfr10I S188E/E204S that rebuilds the canonical sequence motif PD...(E/D)XK in Cfr10I retains significant catalytic activity suggesting that spatial rather than sequence conservation plays the dominant role in the formation of the restriction enzymes active sites (9). We infer from the structural similarity that the Glu212 of Bse634I is involved in coordinating the metal ion at the active site similarly to Glu204 in Cfr10I. Mutation of Glu212 to alanine completely abolished DNA cleavage ability of Bse634I (A.Skirgailiene and V.Siksnys, unpublished data) supporting its key role in catalysis/metal-ion binding. Thus, a sequence motif 133PDX51KX13E specifies the first catalytic/metal-binding site of Bse634I and is similar to the conserved active site motifs PDX46–53KX13E in Cfr10I and NgoMIV but differs from the canonical PDX9–18(E/D)XK motif characteristic for most restriction enzymes.
Upon superposition of Asp146 and Lys198 residues of Bse634I with their structural counterparts at the active sites of other restriction enzymes, residue Glu80 of Bse634I overlapped spatially with Glu71 in Cfr10I, Glu70 in NgoMIV and Glu45 in EcoRV (Fig. 3A–C and Table 2), although it has not been included in the calculation of the superposition operator. It has been suggested that Glu45 of EcoRV forms a part of the second metal-ion binding site and is important for catalysis (36,37). Mutational analysis also revealed that the Glu71 of Cfr10I is important for catalysis and suggested its possible role in metal-ion binding (9). A recent crystal structure analysis of NgoMIV in complex with product DNA indicates that the Glu70 residue of NgoMIV is involved in the coordination of the second metal ion at the active site (32). Thus, structural comparisons suggest that Glu80 of Bse634I might form a second metal-binding site similar to the EcoRV, NgoMIV and Cfr10I restriction enzymes. Interestingly, in the gadolinium heavy atom derivative of Bse634I, the Gd3+ ion is complexed by the side chains of Glu80 and Asp146. The position of the ion is spatially equivalent to the position of one of the magnesium ions in the structures of EcoRV (atom MG2 in the PDB entry 1rvc), NgoMIV and BamHI (32,38).
The Glu80 of Bse634I is located on helix α3 of the N-subdomain whereas the rest of the active site residues are positioned at the C-terminal subdomain (Fig. 2C). Analysis of the subdomain motions in the Bse634I protein (see above) indicates that the Cα atom of Glu80 moves 2.3 Å and the Cδ atom moves ∼3 Å (Fig. 2C, red and green positions of Glu80) towards the active site residues located at the C-terminal subdomain. We propose that a similar ‘cantilever’ α3 helix-mediated movement of the N-terminal subdomain in Bse634I (and probably Cfr10I) restriction enzyme during specific DNA binding might build up the optimal geometry for the coordination of Mg2+ ions at the active site and couple catalysis and sequence recognition.
Model of DNA binding
The structure of Bse634I has been solved in the absence of DNA. However, the position of DNA bound to the Bse634I protein can be predicted by structural comparison with available structures of restriction endonuclease–DNA complexes. Recently, the crystal structure of the NgoMIV restriction enzyme specific for the G/CCGGC sequence that overlaps with one of the possible recognition sequences of Bse634I, has been solved in complex with product DNA (32). The superposition of the active sites of NgoMIV and Bse634I positions NgoMIV DNA into the U-shaped cleft of Bse634I (Fig. 2C). The DNA molecule fits remarkably well into the putative DNA-binding cleft of Bse634I, with just a few steric clashes easily avoided by a moderate rotation (<10°) of DNA around the protein dimer axis.
In the Bse634I–DNA model (Fig. 2C) residues E80, D146, K198 and E212 become positioned close to the cleaved phosphate, in accordance with their predicted active site function. The α3 helix bearing the E80 residue fits well into the minor groove of the DNA, thus supporting the hypothesis that E80 acts as a recognition and catalysis coupler in the ‘cantilever-helix’ mechanism.
N-termini of two symmetry-related α6 helices of Bse634I protrude into the major groove of DNA (Fig. 2C). Crystal structure analysis of the NgoMIV–DNA complex (32) revealed that R191, D193 and R194 residues located just upstream of the α7 helices (which are equivalent to the α6 helices of Bse634I) make sequence-specific contacts with a central CCGG tetranucleotide of the recognition site. Structural comparison reveals that Bse634I residues Arg202, Asp204 and Arg205 overlap well with the Arg191, Asp193 and Arg194 residues of NgoMIV (data not shown), suggesting that both enzymes use the same mechanism for the recognition of their common CCGG tetranucleotide.
Structural mechanisms for the discrimination of the outer base pair by NgoMIV and Bse634I seem to be different. Amino acid residues Asp34 and R227 located, respectively, at the N-terminal domain and α8 helix of NgoMIV, specify the outer Gua:Cyt pair (32). However, the N-terminal subdomain of Bse634I appears to be rotated ∼90° in respect to the N-terminal subdomain of the NgoMIV and becomes positioned closer to the outer Gua:Cyt base pair at the opposite recognition half-site than in the case of NgoMIV. In the Bse634I–DNA model, the loop between β1 and β2 strands is located close to the outer Gua base in the major grove; however, amino acid residues involved in the specific interactions with the outer base pair cannot be unequivocally predicted from the current model.
Cleavage of supercoiled plasmid DNA by Bse634I
Similarly to bona fide tetramers Cfr10I (39) and NgoMIV (32), the Bse634I in principle could interact with two recognition sites. Processing at these sites may be independent or cooperative. Cleavage patterns of plasmids containing one or two recognition sites provide a general test whether the restriction enzyme acts at the two copies of the recognition sequence independently or concertedly (40,41). It was demonstrated that tetrameric restriction endonucleases SfiI (40), Cfr10I (39) and NgoMIV (32) cleave supercoiled plasmid substrates containing single recognition site much slower than plasmids containing two sites. However, cleavage of plasmid DNA with a single site was significantly enhanced by the addition of cognate oligonucleotide, indicating that tetrameric restriction enzymes require two recognition sites supplied in cis or in trans for effective catalysis.
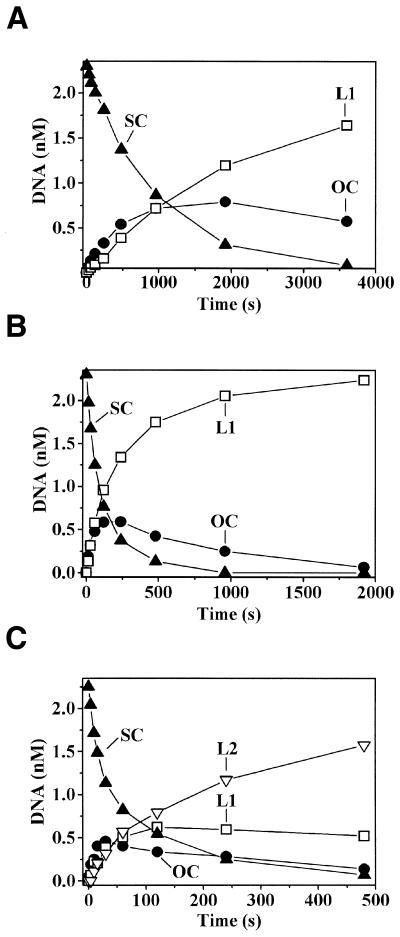
Therefore, we studied the Bse634I cleavage of supercoiled plasmids pUC19 and pUCAC2, containing a single or two copies of the Bse634I recognition sequence 5′-ACCGGC, respectively. In order to eliminate possible effects of substrate binding or product release on the hydrolysis rates, cleavage of pUC19 was studied under single turnover reaction conditions at a saturating Bse634I concentration (2.3 nM substrate, 20 nM enzyme). Under those conditions, the cleavage of pUC19 by Bse634I followed a sequential reaction pathway: supercoiled plasmid DNA was converted into the linear product via an OC DNA intermediate (Fig. 4A). An apparent rate constant k1 value of 0.001 s–1 for the cleavage of the supercoiled form of pUC19 was obtained by fitting an exponential function to the experimental data. Addition of 200 nM of cognate oligonucleotide (Fig. 4B) led to the 10-fold increase of the supercoiled pUC19 cleavage rate (k1 = 0.01 s–1). In contrast, non-cognate oligonucleotide had no effect on pUC19 cleavage (data not shown).
Figure 4.
Cleavage of plasmids containing either one or two recognition sites by Bse634I. The reaction mixtures contained 2.3 nM plasmid DNA, 20 nM Bse634I, 30 mM Tris–acetate (pH 8.5, 25°C), 70 mM CH3COOK, 0.1 mg/ml BSA and 10 mM (CH3COO)2Mg at 25°C. The amounts of SC (closed triangles), OC (closed circles) and linear DNAs with one (L1, open squares) or two (L2, open triangles) double-stranded breaks are shown. (A) Cleavage of supercoiled plasmid pUC19 containing a single recognition site of Bse634I. (B) Cleavage of supercoiled plasmid pUC19 containing a single recognition site of Bse634I in the presence of cognate oligonucleotide. The reaction mixture was supplemented with 200 nM of cognate oligonucleotide duplex. (C) Cleavage of supercoiled plasmid pUCAC2 containing two Bse634I recognition sites.
The requirement of two recognition sites for effective DNA hydrolysis by Bse634I was further tested by the cleavage of plasmid pUCAC2, which contains two recognition sites located in cis. The cleavage profile of pUCAC2 differs significantly from that of pUC19 (Fig. 4C). The supercoiled form of pUCAC2 is cleaved rapidly (k1 = 0.02 s–1) and most of the supercoiled pUCAC2 is converted into the final reaction product—linear DNA with two double-strand breaks. Significant differences in pUC19 and pUCAC2 cleavage by Bse634I were also observed under multiple-turnover reaction conditions (2.3 nM plasmid substrate, 0.25 nM enzyme). The major reaction product with pUC19 was OC DNA, while the major pUCAC2 cleavage products were linear DNAs with one or two double-strand breaks (data not shown).
Similar cleavage patterns for plasmids containing one or two recognition sites were reported previously for the tetrameric restriction enzymes Cfr10I (39) and NgoMIV (32). Thus, differences in the Bse634I cleavage patterns of plasmids containing one or two recognition sites are consistent with tetrameric architecture of the protein and indicate that Bse634I, similarly to SfiI, Cfr10I and NgoMIV, is functional as a tetramer. Thus, the family of restriction endonucleases Bse634I, Cfr10I and NgoMIV recognizing overlapping nucleotide sequences exhibits a conserved tetrameric architecture that is of functional importance.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr G. Burenkov and Dr H. Bartunik for invaluable help with data collection at synchrotron DESY, DORIS/BW6 beamline. We are grateful to Dr I. Gutsche and E. Manakova for careful reading of the manuscript and numerous creative ideas and suggestions. We also acknowledge J. Elhai for sharing data before publication and J. Elhai and S. Halford for their comments on the initial versions of the manuscript. This work was supported in part by the Lithuania Science Foundation grant 387/1999, NATO Linkage grant 960928 and HHMI International Research Scholarship grant 55000336.
DDBJ/EMBL/GenBank accession nos AY046876 and AY046877
REFERENCES
- 1.Deibert M., Grazulis,S., Janulaitis,A., Siksnys,V. and Huber,R. (1999) Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 Å resolution. EMBO J., 18, 5805–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lukacs C.M., Kucera,R., Schildkraut,I. and Aggarwal,A. (2000) Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 Å resolution. Nat. Struct. Biol., 7, 134–140. [DOI] [PubMed] [Google Scholar]
- 3.Repin V.E., Lebedev,L.R., Puchkova,L., Serov,G.D., Tereschenko,T., Chizikov,V.E. and Andreeva,I. (1995) New restriction endonucleases from thermophilic soil bacteria. Gene, 157, 321–322. [DOI] [PubMed] [Google Scholar]
- 4.Janulaitis A., Stakenas,P. and Berlin,Y.A. (1983) A new site-specific endodeoxyribonuclease from Citrobacter freundii. FEBS Lett., 161, 210–212. [DOI] [PubMed] [Google Scholar]
- 5.Bozic D., Grazulis,S., Siksnys,V. and Huber,R. (1996) Crystal structure of Citrobacter freundii restriction endonuclease Cfr10I at 2.15 Å resolution. J. Mol. Biol., 255, 176–186. [DOI] [PubMed] [Google Scholar]
- 6.Szomolanyi I., Kiss,A. and Venetianer,P. (1980) Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene, 10, 219–225. [DOI] [PubMed] [Google Scholar]
- 7.Kulakauskas S., Barsomian,J., Lubys,A., Roberts,R. and Wilson,G. (1994) Organization and sequence of the HpaII restriction-modification system and adjacent genes. Gene, 142, 9–15. [DOI] [PubMed] [Google Scholar]
- 8.Butkus V., Petrauskiene,L., Maneliene,Z., Klimasauskas,S. and Laucys,V. (1987) Cleavage of methylated CCCGGG sequences containing either N4-methylcytosine or 5-methylcytosine with MspI, HpaII, SmaI, XmaI and Cfr9I restriction endonucleases. Nucleic Acids Res., 15, 7091–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skirgaila R., Grazulis,S., Bozic,D., Huber,R. and Siksnys,V. (1998) Structure-based redesign of the catalytic/metal binding site of Cfr10I restriction endonuclease reveals importance of spatial rather than sequence conservation of active centre residues. J. Mol. Biol., 279, 473–481. [DOI] [PubMed] [Google Scholar]
- 10.Otwinowski Z. and Minor,W. (1996) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 11. Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr., D50, 760–763. [DOI] [PubMed] [Google Scholar]
- 12.Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr., A47, 110–119. [DOI] [PubMed] [Google Scholar]
- 13.Brünger A.T., Adams,P.D., Clore,G.M., Delano,W.L., Gros,P., Grosse Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,M., Pannu,N.S. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., D54, 905–921. [DOI] [PubMed] [Google Scholar]
- 14.Janin J. (1997) Specific versus non-specific contacts in protein crystals. Nature Struct. Biol., 4, 973–974. [DOI] [PubMed] [Google Scholar]
- 15.Kabsch W. (1976) A solution for the best rotation to relate two sets of vectors. Acta Crystallogr., A32, 922–923. [Google Scholar]
- 16.Wall L. (1996) Programming Perl, 2nd edn. O’Reilly & Associates, Cambridge, MA.
- 17.Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- 18.Esnouf R.M. (1999) Further additions to Molscript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr., D55, 938–940. [DOI] [PubMed] [Google Scholar]
- 19.Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 20.Yanisch-Perron C., Vieira,J. and Messing,J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and puc19 vectors. Gene, 33, 103–119. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Sasnauskas G., Jeltsch,A., Pingoud,A. and Siksnys,V. (1999) Plasmid DNA cleavage by MunI restriction enzyme: single-turnover and steady-state kinetic analysis. Biochemistry, 38, 4028–4036. [DOI] [PubMed] [Google Scholar]
- 23.Kim Y.C., Grable,J.C., Love,R., Greene,P.J. and Rosenberg,J.M. (1990) Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science, 249, 1307–1309. [DOI] [PubMed] [Google Scholar]
- 24.Newman M., Strzelecka,T., Dorner,L.F., Schildkraut,I. and Aggarwal,A. (1995) Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science, 269, 656–663. [DOI] [PubMed] [Google Scholar]
- 25.Pingoud A. and Jeltsch,A. (2001) Structure and function of type II restriction endonucleases. Nucleic Acids Res., 29, 3705–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Athanasiadis A., Vlassi,M., Kotsifaki,D., Tucker,P.A., Wilson,K.S. and Kokkinidis,M. (1994) Crystal structure of PvuII endonuclease reveals extensive structural homologies to EcoRV. Nature Struct. Biol., 1, 469–475. [DOI] [PubMed] [Google Scholar]
- 27.Horton J.R., Nastri,H.G., Riggs,P.D. and Cheng,X. (1998) Asp34 of PvuII endonuclease is directly involved in DNA minor groove recognition and indirectly involved in catalysis. J. Mol. Biol., 284, 1491–1504. [DOI] [PubMed] [Google Scholar]
- 28.Perona J.J. and Martin,A.M. (1997) Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis. J. Mol. Biol., 273, 207–225. [DOI] [PubMed] [Google Scholar]
- 29.Winkler F.K., Banner,D.W., Oefner,C., Tsernoglou,D., Brown,R.S., Heathman,S.P., Bryan,R.K., Martin,P.D., Petratos,K. and Wilson,K. (1993) The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J., 12, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horton N.C. and Perona,J.J. (1998) Role of protein-induced bending in the specificity of DNA recognition: crystal structure of EcoRV endonuclease complexed with d(AAAGAT) + s(ATCTT). J. Mol. Biol., 277, 779–787. [DOI] [PubMed] [Google Scholar]
- 31.Cheng X., Balendiran,K., Schildkraut,I. and Anderson,J. (1994) Structure of the PvuII endonuclease with cognate DNA. EMBO J., 13, 3927–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deibert M., Grazulis,S., Siksnys,V. and Huber,R. (2000) Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nature Struct. Biol., 7, 792–799. [DOI] [PubMed] [Google Scholar]
- 33.Abagyan R.A. and Batalov,S. (1997) Do aligned sequences share the same fold? J. Mol. Biol., 273, 355–368. [DOI] [PubMed] [Google Scholar]
- 34.Sapranauskas R., Sasnauskas,G., Lagunavicius,A., Vilkaitis,G., Lubys,A. and Siksnys,V. (2000) Novel subtype of type IIs restriction enzymes. J. Biol. Chem., 275, 30878–30885. [DOI] [PubMed] [Google Scholar]
- 35.Aggarwal A.K. (1995) Structure and function of restriction endonucleases. Curr. Opin. Struct. Biol., 5, 11–19. [DOI] [PubMed] [Google Scholar]
- 36.Selent U., Ruter,T., Kohler,E., Liedtke,M., Thielking,V., Alves,J., Oelgeschlager,T., Wolfes,H., Peters,F. and Pingoud,A. (1992) A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry, 31, 4808–4815. [DOI] [PubMed] [Google Scholar]
- 37.Kostrewa D. and Winkler,F.K. (1995) Mg2+ binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 Å resolution. Biochemistry, 34, 683–696. [DOI] [PubMed] [Google Scholar]
- 38.Viadiu H. and Aggarwal,A.K. (1998) The role of metals in catalysis by the restriction endonuclease BamHI. Nature Struct. Biol., 5, 910–916. [DOI] [PubMed] [Google Scholar]
- 39.Siksnys V., Skirgaila,R., Sasnauskas,G., Urbanke,C., Cherny,D., Grazulis,S. and Huber,R. (1999) The Cfr10I restriction enzyme is functional as a tetramer. J. Mol. Biol., 291, 1105–1118. [DOI] [PubMed] [Google Scholar]
- 40.Wentzell L.M., Nobbs,T.J. and Halford,S.E. (1995) The SfiI restriction endonuclease makes a four-strand DNA break at two copies of its recognition sequence. J. Mol. Biol., 248, 581–595. [DOI] [PubMed] [Google Scholar]
- 41.Bilcock D.T. and Halford,S.E. (1999) DNA restriction dependent on two recognition sites: activities of the SfiI restriction-modification system in Escherichia coli. Mol. Microbiol., 31, 1243–1254. [DOI] [PubMed] [Google Scholar]