Abstract
Genetic studies in rodent and chicken mutant cell lines have suggested that Rad51 paralogs (XRCC2, XRCC3, Rad51B/Rad51L1, Rad51C/Rad51L2 and Rad51D/Rad51L3) play important roles in homologous recombinational repair of DNA double-strand breaks and in maintaining chromosome stability. Previous studies using yeast two- and three-hybrid systems have shown interactions among these proteins, but it is not clear whether these interactions occur simultaneously or sequentially in vivo. By utilizing immunoprecipitation with extracts of human cells expressing epitope-tagged Rad51 paralogs, we demonstrate that XRCC2 and Rad51D, while stably interacting with each other, co-precipitate with Rad51C but not with XRCC3. In contrast, Rad51C is pulled down with XRCC3, whereas XRCC2 and Rad51D are not. In addition, Rad51B could be pulled down with Rad51C and Rad51D, but not with XRCC3. These results suggest that Rad51C is involved in two distinct in vivo complexes: Rad51B–Rad51C–Rad51D–XRCC2 and Rad51C–XRCC3. In addition, we demonstrate that Rad51 co-precipitates with XRCC3 but not with XRCC2 or Rad51D, suggesting that Rad51 can be present in an XRCC3–Rad51C–Rad51 complex. These complexes may act as functional units and serve accessory roles for Rad51 in the presynapsis stage of homologous recombinational repair.
INTRODUCTION
Homologous recombination is an important mechanism for maintaining genomic stability by accurately repairing double-stranded DNA breaks arising during DNA replication or induced by DNA-damaging agents. Saccharomyces cerevisiae and human Rad51 proteins, the homologs of RecA in Escherichia coli (1,2), play a central role in homologous recombinational repair (HRR) by mediating DNA strand pairing and strand exchange (3–5). Like RecA, Rad51 polymerizes on single-stranded DNA and forms nucleoprotein filaments that promote homologous pairing. Several other proteins in S.cerevisiae Rad52 epistasis group (namely Rad52, Rad54, Rad55, Rad57 and RPA) promote the strand-exchange activity of Rad51 in vitro (6–8). Since Rad55 and Rad57 share distant sequence similarity with Rad51, they are referred to as paralogs of Rad51, meaning that these proteins probably share a common ancestral gene but have gained new functions during evolution. Genetic and biochemical studies suggest that the Rad55–Rad57 complex functions as an accessory factor to facilitate the strand transfer activity of Rad51 (6,9).
Five vertebrate Rad51 paralogs have been identified, namely XRCC2, XRCC3, Rad51B/Rad51L1, Rad51C/Rad51L2 and Rad51D/Rad51L3 (recently reviewed in 10). These proteins show a low level of sequence identity (20–30%) with Rad51 and with each other. Accumulating evidence suggests that they may have a function in HRR analogous to that of Rad55 and Rad57. The XRCC2 and XRCC3 genes were isolated by functional complementation of mitomycin C (MMC) sensitivity in hamster mutants irs1 (11,12) and irs1SF (13), respectively, which are cross-sensitive to ionizing radiation. Rad51B–D were identified based on their similarity to Rad51 (14–18). The hamster XRCC2 and XRCC3 mutants have remarkably similar phenotypes, i.e. mild sensitivity to ionizing radiation and UV, and extreme sensitivity to cross-linking agents (19,20). These mutants also have elevated spontaneous and induced chromosomal aberrations (11,21,22). Each of the vertebrate Rad51 paralogs has been knocked out in chicken DT40 cells, and all of the mutants show hypersensitivity to DNA damaging agents and chromosome instability, suggesting that the role of each of the Rad51 paralogs in DNA repair is very similar (23,24). Moreover, targeted disruption of XRCC2, Rad51B and Rad51D in mice causes early to mid-gestation embryonic lethality (25–27), indicating that the paralogs, like Rad51 (28,29), are essential for mouse embryonic development.
Protein interactions among Rad51 paralogs and with Rad51 have been identified by yeast two- or three-hybrid studies and by co-expression and co-immunoprecipitation in insect cells (30). Each of the Rad51 paralogs was found to interact with one or more of the others, whereas no self-interactions were observed. The two-hybrid interaction of XRCC3 with Rad51 was confirmed by immunoprecipitation using human cell extracts (11). Similar experiments demonstrated in vivo and in vitro interactions between XRCC2 and Rad51D (31). Recently, an XRCC3–Rad51C complex was purified from bacteria and baculovirus-infected insect cells co-expressing these proteins (32,33). However, it is not known which of these interactions occur in human cells and whether these proteins form a single multiprotein complex.
In this study, we characterized in vivo protein interactions among Rad51 paralogs and showed that these proteins form at least two distinct multiprotein complexes, each of which contains Rad51C. XRCC2, Rad51B and Rad51D are found to interact with Rad51C in one complex, whereas only XRCC3 is bound to Rad51C in another complex.
MATERIALS AND METHODS
Cell lines and γ-irradiation
Hamster and HeLa cell lines were grown in monolayer or in suspension culture in α-minimum essential media (MEM) supplemented with 10% fetal bovine serum and antibiotics as described (11). The hamster cell lines used were irs1 (xrcc2 mutant) and V79 (wild type). Human cell lines HD16, HD19, HD21 and HD25 were derived from HeLaS3 cells (a suspension derivative of HeLa) (34), after transfection with HA-tagged Rad51D expression vector pDS179 and selection for G418 resistance (G418R). HD19 and HD25 express HA-Rad51D, while HD16 and HD21 show little or no expression. HeLa158 is a transformant cell line of HeLaS3 that expresses HA-XRCC3 (11). Cells in suspension (HeLa cells and transformants) or in monolayer (hamster cell lines) were exposed to 8 Gy of 137Cs γ-rays at 1.83 Gy/min and incubated at 37°C for 2 h before harvesting.
Plasmid transfection and cell extract preparation
Plasmid pDS179 contains the human Rad51D cDNA fused with a C-terminal HA-tag (a nine amino acid epitope, YPYDVPDYA). Plasmid pDS200 contains human XRCC2 cDNA fused with an N-terminal Flag-tag (an eight amino acid epitope, DYKDDDDK). Both pDS179 and pDS200 were constructed in pcDNA3 vector in which expression is driven by the cytomegalovirus (CMV) promotor (Invitrogen, Carlsbad, CA). Plasmid pGFP-XRCC2 was made in pEGFP-N1 vector (Clontech, Palo Alto, CA) and contains human XRCC2 cDNA fused with a C-terminal green fluorescent protein (GFP). The plasmids were transfected into cells by electroporation or using lipofectomine 2000 (Life Technologies, Rockville, MD) by the manufacturer’s instructions. Stable transformants were obtained after selection for G418R (0.8 mg/ml for HeLa and 1.7 mg/ml for hamster cell lines). To prepare cell extracts, exponentially growing cells were suspended in extraction buffer (50 mM Tris pH 7.5, 100 mM NaCl, 5 mM EDTA and 0.5% NP-40), supplemented with 50 mM NaF, 1 mM PMSF, 20 µg/ml aprotinin and 10 µg/ml leupeptin, and incubated on ice for 30 min. The cell extracts were centrifuged (Backman, microfuge R) at 15 000 r.p.m. for 10 min at 4°C and the clear supernatant was stored at –80°C. In some experiments, cell pellets were lysed in 2× SDS buffer followed by boiling for 5 min.
Antibodies
Anti-XRCC2 (α-XRCC2) polyclonal antibodies were produced in rabbit (by HTI, Ramona, CA) or in mouse by immunizing the animals with recombinant human XRCC2, and are referred to as α-XRCC2r and α-XRCC2m, respectively. XRCC2 protein (with an N-terminal pelB leading sequence and N- and C-terminal His6 tags) was expressed in bacterial strain B834 bearing the plasmid pET29/XRCC2 and purified on a Ni2+ column. The resulting XRCC2 antiserum or mouse ascites were purified on an affinity column coupled with the antigen. Rabbit anti-human Rad51B and Rad51C antibodies (α-Rad51B and α-Rad51C) and rabbit anti-human Rad51 (α-HsRad51) antiserum were kindly provided by Dr Patrick Sung (University of Texas, San Antonio, TX). The α-HsRad51 antiserum was purified on an HsRad51 affinity column kindly provided by Dr Patrick Sung. The rabbit antibody raised against mouse Rad51 (α-MmRad51) was kindly provided by Dr Randy Legerski (M. D. Anderson Cancer Center, Houston, TX). The polyclonal antibodies against peptides of XRCC2, XRCC3, Rad51B and Rad51D were obtained from Novus Biologicals (Littleton, CO). Affinity matrices and epitope-specific monoclonal antibodies were purchased from Babco (Berkeley, CA).
Immunoprecipitation and immunoblotting
Immunoprecipitation was done with 2 mg cell extracts in a final volume of 200 µl of the extract buffer (described above) by gently agitating the extracts with epitope-affinity matrix or antibodies bound to protein A beads for 3 h at 4°C. The beads were washed with 300 µl extract buffer four times and the bound proteins were eluted in 40 µl of elution buffer (50 mM Tris pH 6.8, 0.1 M dithiothreitol, 10% glycerol, 2% SDS) by incubation at 37°C for 10 min. The eluates (20 µl) were separated on 10% SDS–PAGE gel and transferred at room temperature onto PMDF membranes in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol, pH 8.3) at 15 V overnight. Western blotting was performed at room temperature as described previously (11). The membranes were blocked with 5% blocking reagent (Amersham) in PBS supplemented with 0.05% Tween-20. The primary or HRP-conjugated secondary antibodies (Amersham) were diluted in PBS containing 2.5% blocking agents and 0.025% Tween-20 and incubated with the membranes for 1 h with gentle agitation. Each incubation was followed by four washes with PBS (with 0.05% Tween-20). The blots were incubated with the substrates in an ECL Plus kit (Amersham).
RESULTS
Expression of HA-Rad51D in HeLa cells and co-precipitation of HA-Rad51D with XRCC2
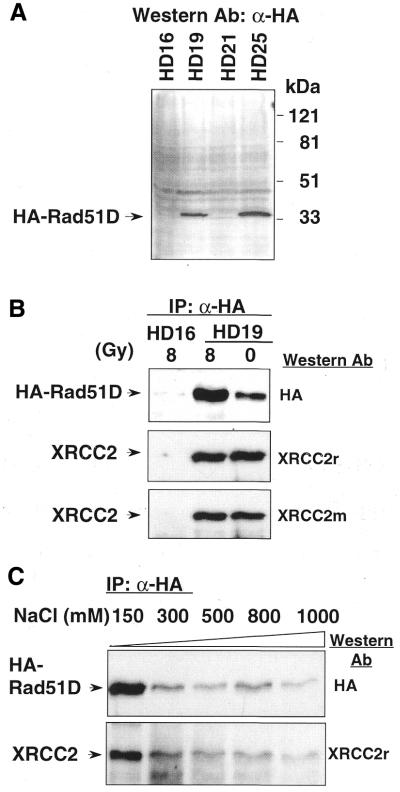
To improve the sensitivity and specificity for detecting Rad51D, HeLaS3 cells were transfected with plasmid pDS179, which expresses the human Rad51D cDNA tagged at the N-terminus with an epitope of influenza hemagglutinin (HA). Several stable G418R clones were isolated and the expression of HA-Rad51D was analyzed by immunoblotting. About one-third of the G418R clones analyzed over-expressed HA-Rad51D. As shown in Figure 1A, a single band at ∼36 kDa of HA-Rad51D was specifically detected in two of the four G418R clones, namely HD19 and HD25, while clones HD16 and HD21 showed little or no expression of HA-Rad51D. HD19 and HD25 cell extracts were then subjected to immunoprecipitation with α-HA. HA-Rad51D was specifically pulled down by α-HA in HD19 (Fig. 1B, top) and in HD25 (data not shown), but not in the HD16 extract (Fig. 1B, top).
Figure 1.
Expression of HA-Rad51D in human cells and interaction of XRCC2 with HA-Rad51D. (A) Western blots with α-HA antibody of cell extracts from HeLaS3 subclones transfected with HA-Rad51D expression vector. Clones HD19 and HD25 expressed HA-Rad51D at a molecular weight at ∼36 kDa while clones HD16 and HD21 show little or no expression. (B) Immunoprecipitation of HA-Rad51D with HA affinity matrix in HD16 and HD19 extracts with or without irradiation. HA-Rad51D was pulled down in HD19 and detected on western blot with HA antibody. Native XRCC2 was specifically detected in the HA-Rad51D precipitates using either rabbit XRCC2 (α-XRCC2r) or mouse XRCC2 (α-XRCC2m) antibody. (C) HA-Rad51D was immunoprecipitated in HD19 extracts using HA affinity matrix at increasing concentrations of NaCl. XRCC2 was co-precipitated under all the conditions tested.
The in vivo interaction between XRCC2 and HA-Rad51D before and after ionizing radiation was examined. In HD19 extract prepared after γ-irradiation with 0 or 8 Gy, XRCC2 was detected at ∼31 kDa in HA-Rad51D precipitates using either rabbit anti-human XRCC2 (α-XRCC2r) or mouse anti-human XRCC2 (α-XRCC2m) antibodies (Fig. 1B). No obvious modification in terms of phosphorylation was observed 2 h after irradiation since the mobility of HA-Rad51D and XRCC2 remained unchanged. The lower level of HA-Rad51D in the unirradiated extract was caused by sample loading error because this difference was not seen in other experiments, e.g. Figure 3 below. To examine the stability of the XRCC2–Rad51D complex, the protein-bound beads were washed with increasing NaCl concentrations. Figure 1C shows that XRCC2 could be pulled down with HA-Rad51D even at 1 M NaCl, indicating a relatively stable interaction between the two proteins. At 300 mM NaCl, the amount of HA-Rad51D bound to α-HA matrix started to decrease sharply compared with that at 150 mM, presumably caused by disrupting the association between HA antibody and the HA-tag.
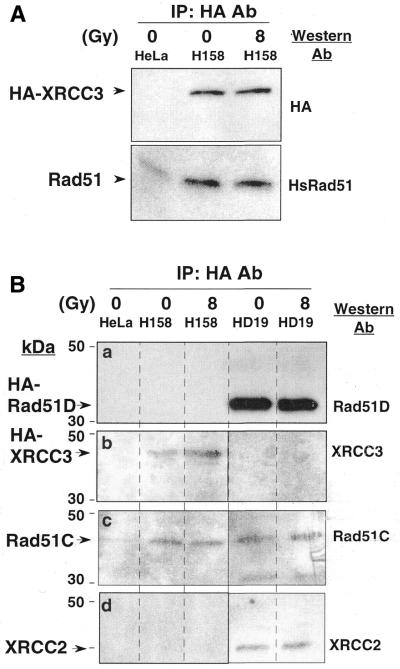
Figure 3.
Interaction of XRCC3 with Rad51C but not with Rad51D and XRCC2. The cells were incubated for 2 h after 8 Gy γ-irradiation. (A) Immunoprecipitation of control HeLa and H158 (expressing HA-XRCC3) cell extracts with HA affinity matrix. The western blots were incubated with α-HA (top) or α-HsRad51 (bottom). HA-XRCC3 is detected in H158 cells but not in the untransfected control, and Rad51 co-precipitates with HA-XRCC3. (B) Immunoprecipitation analysis in H158 and HD19 (expressing HA-Rad51D) was done using HA affinity matrix. The western blots were incubated with: a, α-Rad51D; b, α-XRCC3; c, α-Rad51C; and d, α-XRCC2r.
Co-immunoprecipitation of Rad51C with Rad51D and XRCC2
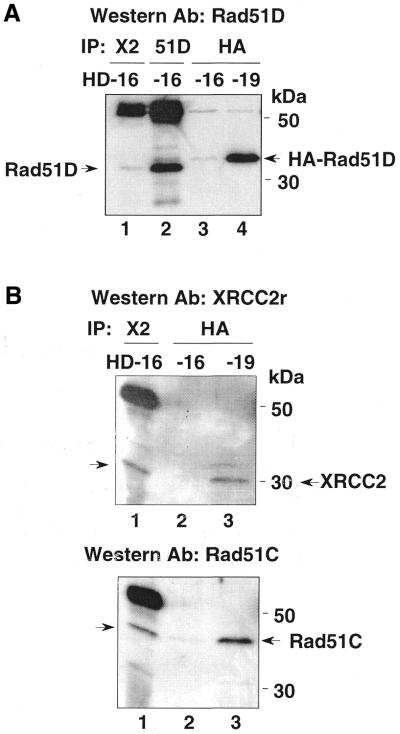
The studies using the yeast two-hybrid and baculovirus overexpression systems showed that Rad51C directly interacts with Rad51D, but Rad51C has no direct interaction with XRCC2 (30). To test for an interaction of Rad51C with Rad51D in human cell extracts, and the possible indirect association of Rad51C with XRCC2 through Rad51D, we performed immunoprecipitation using α-XRCC2 (Novus) and α-Rad51D (Novus) in HeLa transformants HD16 and HD19. HA antibody was also used to pull down HA-Rad51D in HD19 extracts. Figure 2A shows that these antibodies specifically precipitated Rad51D in HD16 or HA-Rad51D in HD19 extracts. α-XRCC2 (lane 1) or α-Rad51D (lane 2) precipitated non-tagged Rad51D in HD16 cell extracts, while α-HA precipitated HA-Rad51D in HD19 extract but very little in the HD16 extract in which HA-Rad51D expression is very low (lanes 4 and 3, respectively). As shown in Figure 2B, XRCC2 was detected at ∼31 kDa in the α-XRCC2 precipitate using a different XRCC2 antibody from the one used for immunoprecipitation (α-XRCC2r) (top, lane 1), as well as in the HA-Rad51D precipitate in HD19 extract (top, lane 3). When the blot shown in the upper panel of Figure 2B was re-probed with α-Rad51C (bottom), Rad51C at ∼42 kDa was detected in the precipitates using α-XRCC2 antibody with HD16 extract (lane 1) and using α-HA with HD19 extract (lane 3), but very little was detected with HD16 extract (lane 2). These data show that Rad51C is specifically associated with Rad51D and XRCC2. Since XRCC2 and Rad51C are pulled down with either HA-Rad51D (from HD19) or native Rad51D (from HD 16), we conclude that the HA tag does not alter these interactions. Based on the previous findings (30), the interaction of XRCC2 with Rad51C may be indirect and bridged by the direct interaction between Rad51C and Rad51D.
Figure 2.
Co-immunoprecipitation of Rad51D, Rad51C and XRCC2. (A) Immunoprecipitation of HD16 and HD19 extracts with α-XRCC2 (Novus, lane 1), α-Rad51D (Novus, lane 2) and α-HA (lanes 3 and 4). The western blot was probed with α-Rad51D. The native Rad51D and HA-Rad51D are indicated. The dark band at ∼55 kDa (lanes 1 and 2) is the heavy chain of rabbit IgG [the same in (B)]. (B) Immunoprecipitation of HD16 and HD19 extracts with α-XRCC2 (Novus, lane 1) and α-HA (lanes 2 and 3). The XRCC2 (top) and Rad51C (bottom) were detected using α-XRCC2r and α-Rad51C antibody, respectively. Rad51C co-precipitates with XRCC2 (lane 1) or HA-Rad51D (lane 3).
Interaction of XRCC3 with Rad51C but not with Rad51D and XRCC2
To examine whether XRCC3 is present in the complex of Rad51C, Rad51D and XRCC2, we performed immunoprecipitation in HeLa transformant H158 cell extracts (expressing HA-XRCC3) and in HD19 cell extracts (expressing HA-Rad51D). Previous work showed that HA-XRCC3 transfection corrected the hypersensitivity of mutant irs1SF to MMC as well as did the untagged XRCC3 cDNA, confirming the functionality of the tagged protein (11). To test for the possible influence of ionizing radiation on complex formation, the cells were irradiated with 8 Gy γ-rays and incubated at 37°C for 2 h before extraction. In the H158 extract, HA-XRCC3 at ∼40 kDa was pulled down with α-HA (Fig. 3A, top). The HA-XRCC3 band was not seen in the control HeLa extract, indicating a specific pull down of HA-XRCC3 with the α-HA antibody. The level of HA-XRCC3 was unaltered by irradiation (Fig. 3A, top). In the XRCC3 precipitates, Rad51 was detected at a level above background, i.e. untransfected HeLa cells (Fig. 3A, bottom), and the amount of precipitated Rad51 was similar in cell extracts with or without irradiation. These results confirm that Rad51 is associated with XRCC3, in agreement with our previous finding (11). The HA-XRCC3 that precipitated with HA-antibody was also specifically detected with an α-XRCC3 antibody in H158 extract but not in the control HeLa extract (Fig. 3B, b).
Using α-Rad51C, Rad51C was detected in HA-XRCC3 precipitates as well as in HA-Rad51D precipitates (Fig. 3B, c), indicating that Rad51C is bound to both XRCC3 and Rad51D. However, neither Rad51D nor XRCC2 was co-precipitated with XRCC3 in H158 extracts (Fig. 3B, a and d). Similarly, when HA-Rad51D was pulled down with α-HA in HD19 extracts (Fig. 3B, a), XRCC3 was not detected in the precipitates (Fig. 3B, b). Both Rad51C and XRCC2 were co-precipitated with HA-Rad51D (Fig. 3B, c and d). To test for a weak indirect interaction between XRCC3 and Rad51D, we reduced the binding and washing stringency by lowing the NP-40 concentration in the cell extracts from 0.5 to 0.05%. The results showed that HA-Rad51D did not co-precipitate with XRCC3. Conversely, XRCC3 did not co-precipitate with HA-Rad51D either (data not shown). Taken together, these results clearly show that XRCC3 interacts with Rad51C but does not interact with Rad51D or XRCC2. Interestingly, Rad51C can interact with either XRCC3 or with Rad51B–Rad51D–XRCC2 and, thus, is likely to be partitioned between these two complexes in cells.
Co-precipitation of Rad51B with Rad51C and Rad51D but not with XRCC3
Rad51B was shown to bind to Rad51C in a yeast two-hybrid system (30). Since Rad51C interacts with both XRCC3 and Rad51D–XRCC2, it is of interest to test whether Rad51B is associated with one or both of the complexes. We did immunoprecipitation in HD16 extracts and tested for co-precipitation of Rad51B with Rad51C, Rad51D or XRCC3. As shown in Figure 4A, Rad51C was pulled down with Rad51B antibodies from two suppliers although in lane 2 the band is barely detectable. Conversely, Rad51B, which migrates at ∼45 kDa, was found in Rad51C precipitates (Fig. 4B, lane 1), confirming the interaction suggested in Figure 4A. Furthermore, Rad51D was detected in Rad51B precipitates (Fig. 4C, lane 1) and vice versa (Fig. 4B, lane 3). Interestingly, Rad51B is not found in XRCC3 precipitates with α-XRCC3 (Fig. 4B, lane 2). These results suggest that Rad51B is associated with Rad51C–Rad51D–XRCC2, but not with XRCC3–Rad51C.
Figure 4.
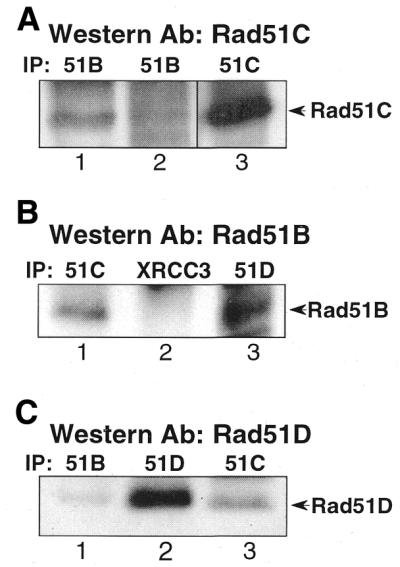
Association of Rad51B with Rad51D and Rad51C. Western blot was performed with the antibody indicated at the top of each panel. (A) Rad51B was co-precipitated with Rad51C (lanes 1 and 2). Rad51B antibodies from Novus (lane 1) or from Dr Patrick Sung (lane 2) were used for immunoprecipitation. Lane 3 is a control of Rad51C that was pulled down by α-Rad51C. (B) Rad51B was not co-precipitated with XRCC3 (lane 2), while it was co-precipitated with Rad51C (lane 1) and Rad51D (lane 3). (C) Lane 1, Rad51D was co-precipitated with Rad51B (Novus antibody). Lanes 2 and 3 are controls showing the position of Rad51D, which is pulled down with α-Rad51D or α-Rad51C.
Absence of Rad51B–Rad51C–Rad51D–XRCC2 complex interaction with Rad51
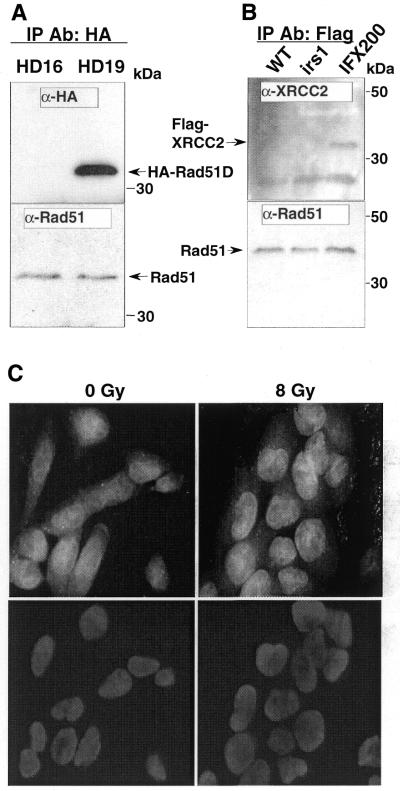
The possible association of the Rad51B–C–D–XRCC2 complex with Rad51 was examined by immunoprecipitation since a weak interaction between Rad51C and Rad51 was suggested by yeast two-hybrid studies (30). We used α-HA to pull down HA-Rad51D, and the co-precipitation of Rad51 with HA-Rad51D was tested in HD16 and HD19 cells. HA-Rad51D was specifically pulled down in HD19 cells (Fig. 5A), and a low level of Rad51 was detected in both HD16 and HD19 (Fig. 5A). Since the Rad51 level was not significantly higher in HD19 precipitate, the presence of Rad51 in the precipitates presumably resulted from non-specific binding of Rad51 to the matrix. This result indicates that Rad51 does not specifically co-precipitate with HA-Rad51D.
Figure 5.
Lack of association of XRCC2 with Rad51. (A) Immunoprecipitation with HA affinity matrix in HD16 and HD19 cell extracts. The blots were incubated with α-HA (top) or α-HsRad51 (bottom). (B) Immunoprecipitation with Flag affinity matrix in wild type, irs1 and Flag-XRCC2 transformants of irs1 (IFX200). The blots were incubated with α-XRCC2r (top) or α-MmRad51 (bottom). (C) Immunofluorescence detection of GFP-XRCC2 in stable transformants of wild-type V79 cells 2 h after 0 or 8 Gy γ-irradiation (top). The same cells stained with DAPI (bottom).
Although the yeast two-hybrid study suggested that XRCC2 does not physically interact with Rad51 (11), we tested whether XRCC2 is associated with Rad51 through an indirect interaction. A Flag-XRCC2 expression vector was constructed and tested for its functionality in the XRCC2 mutant irs1 cells. Transfection of irs1 cells with Flag-XRCC2 vector rescued the mutant from killing by MMC at 15 nM, a concentration that kills the mutant (data not shown). A pool of stable transformants of irs1 (IFX200) that was resistant to 200 nM MMC was used for immunoprecipitation. From extracts of these cells, Flag-XRCC2 was pulled down using Flag affinity matrix and analyzed on western blot with XRCC2 antibody. The 32 kDa Flag-XRCC2 was detected specifically in IFX200 cells, and not in untransfected V79 and irs1 cells (Fig. 5B, top). Again, Rad51 protein was detected in all of the precipitates, and the Rad51 level was not significantly higher in the Flag-XRCC2 precipitate (Fig. 5B, bottom). These results indicate that Rad51 does not associate with either XRCC2 or Rad51D. Therefore, Rad51 may not stably interact with the Rad51B–C–D–XRCC2 complex.
By expressing GFP–XRCC2 in hamster cells, we tested whether XRCC2 forms nuclear foci that might co-localize with Rad51 in cells irradiated with 8 Gy γ-rays. GFP–XRCC2 fusion protein was stably expressed in hamster cells, and the protein was visualized using fluorescence microscopy. The functionality of the fusion protein was tested by growing GFP–XRCC2 transfected irs1 cells in medium containing 30 nM MMC, which is highly toxic to irs1 cells. The results showed that GFP–XRCC2 expression conferred MMC resistance to irs1 cells (data not shown). The results in Figure 5C show that the fluorescent fusion protein was localized primarily to nuclei but did not form foci before or 2 h after γ-irradiation, at which time Rad51 foci are prominently displayed in nuclei (35).
DISCUSSION
We have identified two discrete protein complexes containing Rad51 paralogs in human cell extracts. One contains XRCC2, Rad51B, Rad51C and Rad51D, and possibly other proteins; the other contains Rad51C and XRCC3 at the least. Rad51C is involved in both complexes, whereas XRCC3 is not associated with Rad51B, Rad51D or XRCC2. Since XRCC2 was previously found to interact in a yeast two-hybrid system with Rad51D but no other Rad51 paralogs (30), we suggest that Rad51C is bound only to Rad51D so that XRCC2 interacts with Rad51C indirectly through Rad51D. Although our protein interaction data are primarily qualitative in nature, they provide a basis for a model and further studies. Another study has recently obtained results similar to our data (36). Therefore, a complex containing Rad51B–C–D–XRCC2, and possibly other proteins, may exist under physiological conditions.
The interactions of XRCC2 with Rad51D (31) and of XRCC3 with Rad51C (33) in human cell extracts were recently reported. Our data not only confirm the presence of these interactions in human cells, but also demonstrate that a molecule of Rad51C binds Rad51D and XRCC3 separately rather than simultaneously. This conclusion is also consistent with the findings from the yeast three-hybrid system, in which no evidence could be found that Rad51C can interact simultaneously with XRCC3 and Rad51D (30). Therefore, the Rad51 paralogs form at least two distinct complexes rather than a speculated multiprotein complex as suggested by results obtained using yeast two- and three-hybrid analyses and proteins overexpressed in insect cells (30). Our data do not exclude the presence of an XRCC2–Rad51D dimer in cell extracts (31), but argue that at least a portion of XRCC2 and Rad51D exist in a complex that includes Rad51C. The same situation might also exist for Rad51B, e.g. a Rad51B–Rad51C dimer as well as an Rad51B–C–D–XRCC2 complex. In this regard, it is notable that Rad51D binds only weakly to Rad51C in a yeast two-hybrid assay (30). XRCC2, and possibly other proteins including Rad51B, may help stabilize the Rad51C–Rad51D interaction.
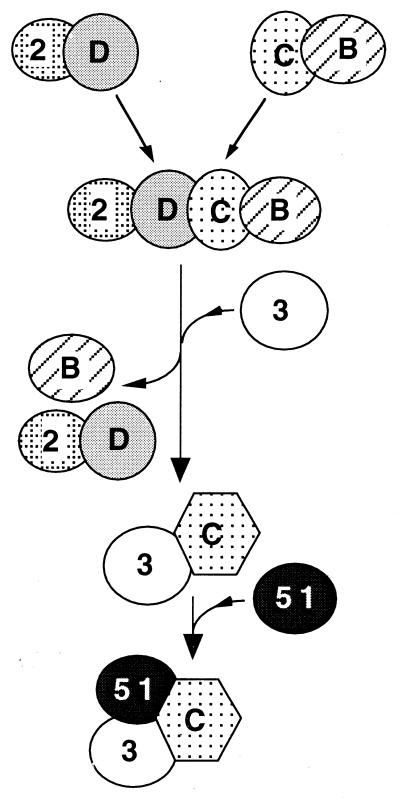
Based on our results and findings in the yeast three-hybrid system (30), we suggest the dynamic interactions among the Rad51 paralogs depicted in Figure 6. We suggest that the interaction of XRCC3 with Rad51C releases XRCC2–Rad51D in a step that is possibly associated with a conformational change in Rad51C. Rad51B may help promote this step because it was found in three-hybrid analysis to enhance the interaction of Rad51C and XRCC3 (30). Thus, it will be important to determine whether the same or different regions of Rad51C bind Rad51D and XRCC3. There may be additional stable complexes besides those shown in Figure 6.
Figure 6.
Speculative model of complexes and dynamic interactions among Rad51 paralogs and HsRad51. The diagram takes into account the results by Braybrooke and co-workers showing an XRCC2–Rad51D dimer in cell extracts (31) and the data from yeast two-hybrid analyses showing that Rad51B and Rad51C can form a dimer (30). XRCC2–Rad51D and Rad51B–Rad51C dimers combine to form the Rad51B–C–D–XRCC2 complex. XRCC3 may promote the dissociation of Rad51C from this complex to produce the XRCC3–Rad51C dimer that binds to Rad51. A possible conformational change as depicted for Rad51C could contribute to the specificity of interactions.
Although the precise biochemical functions of the Rad51 paralogs in HRR are unknown, they may act as cofactors in mediating Rad51 filament formation on single-stranded DNA. It was reported that both Rad51D (31) and the Rad51C–XRCC3 dimer (32,33) preferentially bind to single-stranded, rather than double-stranded, DNA. The specificity of binding to single-stranded DNA would be consistent with a role of the Rad51 paralogs in facilitating the formation of Rad51 nucleoprotein filaments that initiate homologous pairing (37). One report suggested that the XRCC3–Rad51C dimer also has homologous pairing activity as determined by D-loop formation between single-stranded and double-stranded oligonucleotides (32). Interestingly, the XRCC3–Rad51C complex forms protein–DNA networks in vitro (33). Our results also suggest that Rad51 is associated with the XRCC3–Rad51C complex (Fig. 4), but not with the Rad51B–C–B–XRCC2 complex.
In the context of ionizing radiation-induced damage, many proteins in the HRR pathway, including Rad51, undergo phosphorylation (reviewed by Thompson and Schild; 10), but this is not seen for the Rad51 paralogs by electrophoretic shift (Figs 1B and 4). However, minor phosphorylations in a protein may not result in detectable molecular weight increases, so metabolic labeling experiments are needed for further assessment. It is also curious that XRCC2, XRCC3 and Rad51C are all expressed in brain tissue (11,16,32) where there is no opportunity for recombinational repair to occur between sister chromatids. Recombination between homologous chromosomes is unlikely to occur at a significant rate in mammalian cells (38). These findings suggest that the Rad51 paralogs might have other functions besides their association with Rad51.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Patrick Sung and Randy Legerski for kindly supplying the Rad51B–C, human and mouse Rad51 antibodies. Thanks also go to Mona Hwang for producing XRCC2 antibody in mouse and to Marilyn J. Ramsey for assisting with the fluorescence microscopy. This work was supported partly by grant CA84407-01 from the National Cancer Institute (to N.L.), and was performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under contract No. W-7405-ENG-48. D.S. was supported by GM30990 administered under DOE Contract No. DE-AC03-76SF00098.
REFERENCES
- 1.Shinohara A., Ogawa,H. and Ogawa,T. (1992) Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell, 69, 457–470. [DOI] [PubMed] [Google Scholar]
- 2.Shinohara A., Ogawa,H., Matsuda,Y., Ushio,N., Ikeo,K. and Ogawa,T. (1993) Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nature Genet., 4, 239–243. [DOI] [PubMed] [Google Scholar]
- 3.Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science, 265, 1241–1243. [DOI] [PubMed] [Google Scholar]
- 4.Baumann P., Benson,F.E. and West,S.C. (1996) Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell, 87, 757–766. [DOI] [PubMed] [Google Scholar]
- 5.Gupta R.C., Bazemore,L.R., Golub,E.I. and Radding,C.M. (1997) Activities of human recombination protein Rad51. Proc. Natl Acad. Sci. USA, 94, 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung P. (1997) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. [DOI] [PubMed] [Google Scholar]
- 7.New J.H., Sugiyama,T., Zaitseva,E. and Kowalczykowski,S.C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature, 391, 407–410. [DOI] [PubMed] [Google Scholar]
- 8.Solinger J.A., Lutz,G., Sugiyama,T., Kowalczykowski,S.C. and Heyer,W.D. (2001) Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol., 307, 1207–1221. [DOI] [PubMed] [Google Scholar]
- 9.Johnson R.D. and Symington,L.S. (1995) Functional differences and interactions among the putative RecA homologs Rad51, Rad55 and Rad57. Mol. Cell. Biol., 15, 4843–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson L.H. and Schild,D. (2001) Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res., 477, 131–153. [DOI] [PubMed] [Google Scholar]
- 11.Liu N., Lamerdin,J.E., Tebbs,R.S., Schild,D., Tucker,J.D., Shen,M.R., Brookman,K.W., Siciliano,M.J., Walter,C.A., Fan,W. et al. (1998) XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol. Cell, 1, 783–793. [DOI] [PubMed] [Google Scholar]
- 12.Cartwright R., Tambini,C.E., Simpson,P.J. and Thacker,J. (1998) The XRCC2 DNA repair gene from human and mouse encodes a novel member of the recA/RAD51 family. Nucleic Acids Res., 26, 3084–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tebbs R.S., Zhao,Y., Tucker,J.D., Scheerer,J.B., Siciliano,M.J., Hwang,M., Liu,N., Legerski,R.J. and Thompson,L.H. (1995) Correction of chromosomal instability and sensitivity to diverse mutagens by a cloned cDNA of the XRCC3 DNA repair gene. Proc. Natl Acad. Sci. USA, 92, 6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albala J.S., Thelen,M.P., Prange,C., Fan,W., Christensen,M., Thompson,L.H. and Lennon,G.G. (1997) Identification of a novel human RAD51 homolog, RAD51B. Genomics, 46, 476–479. [DOI] [PubMed] [Google Scholar]
- 15.Cartwright R., Dunn,A.M., Simpson,P.J., Tambini,C.E. and Thacker,J. (1998) Isolation of novel human and mouse genes of the recA/RAD51 recombination-repair gene family. Nucleic Acids Res., 26, 1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dosanjh M.K., Collins,D.W., Fan,W., Lennon,G.G., Albala,J.S., Shen,Z. and Schild,D. (1998) Isolation and characterization of RAD51C, a new human member of the RAD51 family of related genes. Nucleic Acids Res., 26, 1179–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pittman D.L., Weinberg,L.R. and Schimenti,J.C. (1998) Identification, characterization and genetic mapping of Rad51d, a new mouse and human RAD51/RecA-related gene. Genomics, 49, 103–111. [DOI] [PubMed] [Google Scholar]
- 18.Rice M.C., Smith,S.T., Bullrich,F., Havre,P. and Kmiec,E.B. (1997) Isolation of human and mouse genes based on homology to REC2, a recombinational repair gene from the fungus Ustilago maydis. Proc. Natl Acad. Sci. USA, 94, 7417–7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones N.J., Cox,R. and Thacker,J. (1987) Isolation and cross-sensitivity of X-ray-sensitive mutants of V79-4 hamster cells. Mutat. Res., 183, 279–286. [DOI] [PubMed] [Google Scholar]
- 20.Fuller L.F. and Painter,R.B. (1988) A Chinese hamster ovary cell line hypersensitive to ionizing radiation and deficient in repair replication. Mutat. Res., 193, 109–121. [DOI] [PubMed] [Google Scholar]
- 21.Tucker J.D., Jones,N.J., Allen,N.A., Minkler,J.L., Thompson,L.H. and Carrano,A.V. (1991) Cytogenetic characterization of the ionizing radiation-sensitive Chinese hamster mutant irs1. Mutat. Res., 254, 143–152. [DOI] [PubMed] [Google Scholar]
- 22.Cui X., Brenneman,M., Meyne,J., Oshimura,M., Goodwin,E.H. and Chen,D.J. (1999) The XRCC2 and XRCC3 repair genes are required for chromosome stability in mammalian cells. Mutat. Res., 434, 75–88. [DOI] [PubMed] [Google Scholar]
- 23.Takata M., Sasaki,M.S., Sonoda,E., Fukushima,T., Morrison,C., Albala,J.S., Swagemakers,S.M., Kanaar,R., Thompson,L.H. and Takeda,S. (2000) The Rad51 paralog Rad51B promotes homologous recombinational repair. Mol. Cell. Biol., 20, 6476–6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takata M., Sasaki,M.S., Tachiiri,S., Fukushima,T., Sonoda,E., Schild,D., Thompson,L.H. and Takeda,S. (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol., 21, 2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shu Z., Smith,S., Wang,L., Rice,M.C. and Kmiec,E.B. (1999) Disruption of muREC2/RAD51L1 in mice results in early embryonic lethality which can be partially rescued in a p53(–/–) background. Mol. Cell. Biol., 19, 8686–8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deans B., Griffin,C.S., Maconochie,M. and Thacker,J. (2000) Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J., 19, 6675–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pittman D.L. and Schimenti,J.C. (2000) Midgestation lethality in mice deficient for the RecA-related gene, Rad51d/Rad51l3. Genesis, 26, 167–173. [DOI] [PubMed] [Google Scholar]
- 28.Lim D.S. and Hasty,P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol., 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuzuki T., Fujii,Y., Sakumi,K., Tominaga,Y., Nakao,K., Sekiguchi,M., Matsushiro,A., Yoshimura,Y. and Morita,T. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA, 93, 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schild D., Lio,Y.c., Collins,D.W., Tsomondo,T. and Chen,D.J. (2000) Evidence for simultaneous protein interactions between human Rad51 paralogs. J. Biol. Chem., 275, 16443–16449. [DOI] [PubMed] [Google Scholar]
- 31.Braybrooke J.P., Spink,K.G., Thacker,J. and Hickson,I.D. (2000) The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J. Biol. Chem., 275, 29100–29106. [DOI] [PubMed] [Google Scholar]
- 32.Kurumizaka H., Ikawa,S., Nakada,M., Eda,K., Kagawa,W., Takata,M., Takeda,S., Yokoyama,S. and Shibata,T. (2001) Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C. Proc. Natl Acad. Sci. USA, 98, 5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masson J.Y., Stasiak,A.Z., Stasiak,A., Benson,F.E. and West,S.C. (2001) Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc. Natl Acad. Sci. USA, 98, 8440–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen T.R. (1988) Re-evaluation of HeLa, HeLa S3 and HEp-2 karyotypes. Cytogenet. Cell Genet., 48, 19–24. [DOI] [PubMed] [Google Scholar]
- 35.Haaf T., Golub,E.I., Reddy,G., Radding,C.M. and Ward,D.C. (1995) Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl Acad. Sci. USA, 92, 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiese C., Collins,D.W., Albala,J., Thompson,L.H., Kronenberg,A. and Schild,D. (2002) Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 30, 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazin A.V., Zaitseva,E., Sung,P. and Kowalczykowski,S.C. (2000) Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J., 19, 1148–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson R.D. and Jasin,M. (2001) Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans, 29, 196–201 [DOI] [PubMed] [Google Scholar]