Abstract
Estimated glomerular filtration rate (eGFR) reflects kidney function. Progressive eGFR-decline can lead to kidney failure, necessitating dialysis or transplantation. Hundreds of loci from genome-wide association studies (GWAS) for eGFR help explain population cross section variability. Since the contribution of these or other loci to eGFR-decline remains largely unknown, we derived GWAS for annual eGFR-decline and meta-analyzed 62 longitudinal studies with eGFR assessed twice over time in all 343,339 individuals and in high-risk groups. We also explored different covariate adjustment. Twelve genome-wide significant independent variants for eGFR-decline unadjusted or adjusted for eGFR-baseline (11 novel, one known for this phenotype), including nine variants robustly associated across models were identified. All loci for eGFR-decline were known for cross-sectional eGFR and thus distinguished a subgroup of eGFR loci. Seven of the nine variants showed variant-by-age interaction on eGFR cross section (further about 350,000 individuals), which linked genetic associations for eGFR-decline with age-dependency of genetic cross-section associations. Clinically important were two to four-fold greater genetic effects on eGFR-decline in high-risk subgroups. Five variants associated also with chronic kidney disease progression mapped to genes with functional in-silico evidence (UMOD, SPATA7, GALNTL5, TPPP). An unfavorable versus favorable nine-variant genetic profile showed increased risk odds ratios of 1.35 for kidney failure (95% confidence intervals 1.03–1.77) and 1.27 for acute kidney injury (95% confidence intervals 1.08–1.50) in over 2000 cases each, with matched controls). Thus, we provide a large data resource, genetic loci, and prioritized genes for kidney function decline, which help inform drug development pipelines revealing important insights into the age-dependency of kidney function genetics.
Keywords: acute kidney injury, diabetes, chronic kidney disease, gene expression
INTRODUCTION
Glomerular filtration rate (GFR) is accepted as best overall index of kidney function1. A GFR<60 mL/min/1.73m2 defines chronic kidney disease (CKD)2, which affects about 10% of adults3. A decline in GFR over time is characteristic for CKD-progression, which can lead to kidney failure4 requiring dialysis or kidney transplantation with a high risk of premature mortality5. In population studies on kidney function, estimated GFR (eGFR) is usually derived from serum creatinine6 and annual eGFR-decline as the difference between two such assessments divided by the years between these assessments. Decline in eGFR is age-related, with a physiological loss of ~1 mL/min/1.73m2 per year2 generally and 3 mL/min/1.73m2 per year in the presence of diabetes mellitus (DM), a major risk factor for CKD-progression7,8. Therapeutic options to decelerate kidney function decline are limited. In addition to pharmacological inhibitors of the RAAS-system9, the recent introduction SGLT2 inhibitors show promising reno-protective effects10,11. An understanding of the mechanisms of kidney function decline and the developing of new therapeutic options is thus of high clinical and public health relevance7,12.
Genes underneath genome-wide association study (GWAS) loci for diseases and biomarkers help identify new therapies13. Open access GWAS summary statistics from large sample sizes are a highly queried resource, also for causal inference studies14. Hundreds of loci and genes are identified by cross-sectional GWAS for eGFR, i.e. GWAS for eGFR based on a single serum creatinine measurement15–18, which help explain population variability. However, the mechanisms underlying a genetic variant association with lower but stable eGFR over time might not always be disease-relevant. GWAS on parameters more directly linked to disease progression are thought to better inform drug development19.
Current evidence from GWAS on annual eGFR-decline is limited, owed to substantial logistics in conducting longitudinal studies and thus small sample sizes. Only one variant, in the UMOD-PDILT locus, has been identified at genome-wide significance20 (n~60,000). With an estimated heritability of 38% for annual eGFR-decline20, comparable to 33%−39% estimated for cross-sectional eGFR in general populations21,15, much more can be expected in larger sample sizes. Further three loci were genome-wide significant in an extreme phenotype approach, comparing individuals with large eGFR-decline or steep drop into CKD with respective controls22. While these are important binary clinical endpoints, methodological literature supports the use of regression methods on undichotomized variables23.
The limited availability of longitudinal GWAS is not only an issue for kidney function decline, but also generally: e.g. change in lung function (n=27,24924), glucose (n=13,80725), or blood pressure (n=33,72026); consequently, locus findings on biomarker change are few and often unstable14. A challenge beyond power is limited experience in longitudinal GWAS with regard to covariate adjustment: clinical trials for disease-related biomarker change require control for differences in baseline levels between therapy groups27. However, covariate adjustment in GWAS requires a careful choice28: it can reveal important mediator effects (e.g. DM adjusted for BMI29), alter the phenotype (e.g. waist-to-hip ratio “unexpected” by body-mass-index28,30), yield artefacts from heritable covariates (collider bias28) or non-sense association (e.g. sex adjusted for height31). The impact of covariate adjustment on longitudinal GWAS on eGFR-decline, and biomarker change generally, is not well explored.
We thus aimed to identify genetic loci associated with annual eGFR-decline and CKD-progression (defined as eGFR-decline among individuals with CKD at baseline) and to prioritize genes that may inform drug development for slowing down eGFR-decline and CKD-progression. We also aimed to fill the gap of large-data genome-wide SNP summary statistics for annual eGFR-decline and CKD-progression, to help future meta-analyses and Mendelian randomization studies. Finally, we wanted to understand the impact of different covariate adjustment and whether a SNP associated with eGFR-decline showed an age-dependent association on eGFR cross-sectionally (i.e. SNP-by-age interaction on eGFR cross-sectionally). By this, we aimed to contribute to a better understanding of the interpretation of genetic findings for eGFR-decline and other progression traits.
To achieve these aims, we (i) increased sample size for GWAS on annual eGFR-decline to >340,000 individuals based on the CKDGen consortium32 and UK Biobank33, (ii) applied a suite of covariate adjustment models, (iii) analyzed SNP-by-age interaction on eGFR cross-sectionally in >350,000 individuals independent of the GWAS on decline, and (v) conducted genetic risk score (GRS) analyses for acute kidney injury (AKI) and end-stagekidney disease (ESKD).
METHODS
We conducted GWAS meta-analysis based on study-specific summary statistics. Each study utilized data on two measurements of serum creatinine over time and genome-wide SNP-information imputed to 1000 Genomes34 phase 1 or phase 3, the Haplotype Reference Consortium35 v1.1 or similar (Table S1&S2). Serum creatinine measured at baseline and follow-up were used to estimate eGFR at baseline and follow-up, respectively, according to the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation6. Annual eGFR-decline was defined as “-(eGFR at follow-up - eGFR at baseline) / number of years of follow-up”. GWAS analyses were conducted separately by ancestry (if applicable), where ancestry was defined by genetic principal components or participants’ self-report. GWAS were based on linear regression with different covariate adjustment conducted overall and focused on individuals with DM or CKD at baseline.
Study-specific genome-wide summary statistics and detailed phenotype information were transferred to the meta-analysis center. For each SNP, summary statistics were pooled and genomic control corrected. Significant genetic variants were identified and respective locus regions selected.
Additionally, we investigated identified SNPs for SNP-by-age interaction on cross-sectional eGFR (based on creatinine or cystatin C, eGFRcrea, eGFRcys) using UK Biobank data that was independent of the SNP identification step (excluding the individuals in the decline GWAS). We computed the GRS and its association on eGFR-decline in the HUNT study via linear regression and provided odds ratios (OR) for GRS association in case-control studies on AKI and ESKD via logistic regression.
Detailed methods are provided in the Supplementary Methods.
RESULTS
Overview across studies and models for GWAS
This GWAS meta-analysis included 343,339 individuals from 62 studies (Supplementary Table S1&S3, Supplementary Figure S1, Methods) and 12,403,901 analyzable SNPs. Most studies were population-based (76%) and of European ancestry (74%). Study-specific median annual eGFR-decline was independent of sample size and follow-up length (Supplementary Figure S2A&S2B) and the median across studies was 1.32 mL/min/1.73m2 per year; follow-up length was 1–21 years (median [25th, 75th] = 5 years [4,7]); median age ranged from 33 to 77 years (Supplementary Figure S2C).
All analyses were adjusted for age-, sex, and study-specific covariates, which is not mentioned further from here on (stable across different modes of age-adjustment, Supplementary Figure S3). We had five GWAS results for eGFR-decline (Methods): (i) “unadjusted”, (ii) “DM-adjusted”, (iii) “adjusted for eGFR-baseline”, (iv) restricted to individuals with DM at baseline (unadjusted), and (v) restricted to individuals with CKD at baseline (unadjusted).
Similarities and differences across different model adjustments
There is, to date, no standard conduct for GWAS on eGFR-decline with regard to covariate adjustment. We explored the impact of two potentially important covariates additional to age and sex: (i) DM, as an important risk factors for eGFR-decline and potential mediator, and (ii) eGFR at baseline, as adjustment for baseline levels in analyses of change over time has noted pros (larger effects, better detectability) and cons (biased effects)36,37.
With regard to DM-adjustment, this model was computed in all studies (n=343,339; 62 studies) and compared to unadjusted results for a subset of studies of varying scope (n=103,970). DM-adjusted SNP-associations on eGFR-decline were precisely the same as unadjusted, in terms of beta-estimates and standard errors (Supplementary Figure S4A, Supplementary Note S1). We therefore did not distinguish these two models further.
In contrast, adjustment for eGFR-baseline altered SNP-associations on eGFR-decline (Supplementary Figure S4B). Therefore, results from both eGFR-decline unadjusted and adjusted for eGFR-baseline were evaluated in the following. GWAS summary statistics for eGFR-decline adjusted for eGFR-baseline were formula-derived from GWAS summary statistics for unadjusted eGFR-decline and for eGFR-baseline together with study-specific phenotypic information (Supplementary Note S2). In a subset of studies (n=103,970), we validated that the formula-approach worked very well in our setting (Supplementary Note S3, Supplementary Figure S4C&D). Meta-analysis yielded GWAS results for eGFR-decline adjusted for eGFR-baseline for 320,737 individuals (50 studies, Supplementary Figure S1).
Twelve variants identified for eGFR-decline unadjusted or adjusted for eGFR-baseline
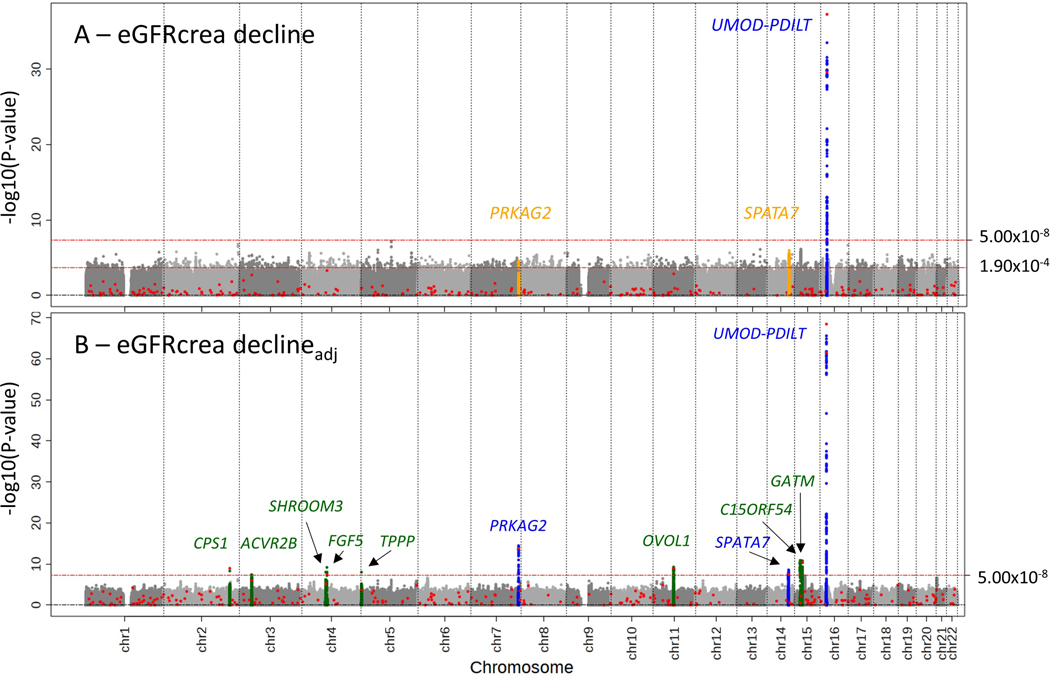
First, our genome-wide screen for eGFR-decline unadjusted for eGFR-baseline (n=343,339) identified two genome-wide significant independent variants near UMOD-PDILT (; Figure 1A, Table 1A): rs34882080, highly correlated with rs12917707 identified previously for this phenotype (r2=1.00)20, and rs77924615, known for altering UMOD expression and urine uromodulin15 and genome-wide significant for eGFR-decline for the first time.
Figure 1: Eleven loci identified by GWAS for eGFR-decline unadjusted and/or adjusted for eGFR-baseline.
We conducted GWAS for eGFR-decline baseline-unadjusted and baseline-adjusted (n up to 343,339 or 320,737, respectively). Shown are association P-values versus genomic position, identified loci annotated by nearest gene: (A) association for eGFR-decline baseline-unadjusted identified one genome-wide significant locus for decline (P<5×108) and two Bonferroni-corrected significant loci among the 263 lead variants for cross-sectional eGFR15 outside of UMOD-PDILT (red dots, P<0.05/263=1.90×10−4; known locus for decline marked in blue; novel loci for this phenotype in orange); (B) association for eGFR-decline baseline-adjusted identified 8 additional loci (novel loci marked in green; known loci or loci already identified in (A) marked in blue). Altogether, 11 loci were identified with genome-wide significance for eGFR-decline unadjusted and/or adjusted for eGFR-baseline.
Table 1: Twelve independent variants in 11 loci identified for association with eGFR-decline unadjusted and adjusted for eGFR-baseline.
We conducted GWAS for eGFR-decline baseline-unadjusted and baseline-adjusted (“decline”, n up to 343,339; declineadj, n up to 320,737). This identified (A) 2 variants with genome-wide significance for eGFR-decline baseline-unadjusted (UMOD-PDILT, Pdecline<5×10−8) and 2 further variants in a candidate search of the 263 variants known for cross-sectional eGFR15 outside UMOD-PDILT, judged at Bonferroni-corrected significance (Pdecline<0.05/263=1.90×10−4; PRKAG2, SPATA7), (B) 5 variants with genome-wide significance for eGFR-decline baseline-adjusted AND Bonferroni-corrected significant baseline-unadjusted (Pdecline-adj-BL<5×10−8, Pdecline <0.05/12=4.17×10−3), (C) 3 variants with genome-wide significance for eGFR-decline baseline-adjusted but not significantly associated baseline-unadjusted (Pdecline-adj-BL<5×10−8, Pdecline≥4.17×10−3). For each identified variant, we show results for decline (baseline-unadjusted), for decline baseline-adjusted, and for cross-sectional eGFR15. Beta-estimates are in mL/min/1.732 per year and per faster-decline allele; significant P-values are stated in bold.
| decline | declineadj | cross-sectional | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPID | Locus Name | Chr | Pos | EA/OA | EAF | Beta | P | Beta | P | Beta | P |
| A from GWAS/candidate search for decline (baseline-unadjusted) | |||||||||||
|
| |||||||||||
| rs34882080 | UMOD-PDILT | 16 | 20,361,441 | a/g | 0.815 | 0.065 | 2.45×10 −30 | 0.092 | 3.31×10 −62 | −0.009 | 2.86×10 −95 |
| rs77924615 | UMOD-PDILT | 16 | 20,392,332 | g/a | 0.798 | 0.074 | 5.30×10 −38 | 0.099 | 3.75×10 −69 | −0.010 | 1.45×10 −138 |
| rs10254101 | PRKAG2 * | 7 | 151,415,536 | t/c | 0.276 | 0.020 | 4.10×10 −05 | 0.037 | 1.78×10 −14 | −0.007 | 1.85×10 −67 |
| rs1028455 | SPATA7 * | 14 | 88,829,975 | t/a | 0.657 | 0.021 | 5.90×10 −06 | 0.024 | 3.43×10 −08 | −0.002 | 4.78×10 −10 |
|
| |||||||||||
| B from GWAS for declineadj, with association for decline (baseline-unadjusted) | |||||||||||
|
| |||||||||||
| rs1458038 | FGF5 | 4 | 81,164,723 | c/t | 0.690 | 0.019 | 3.87×10 −05 | 0.028 | 6.85×10 −10 | −0.003 | 7.49×10 −24 |
| rs4930319 | OVOL1 | 11 | 65,555,458 | c/g | 0.333 | 0.015 | 9.93×10 −04 | 0.028 | 5.27×10 −10 | −0.003 | 2.21×10 −24 |
| rs434215 | TPPP § | 5 | 699,046 | a/g | 0.277 | 0.020 | 3.70×10 −04 | 0.032 | 7.19×10 −09 | −0.003 | 7.63×10−06 |
| rs28857283 | C15ORF54 † | 15 | 39,224,711 | g/a | 0.656 | 0.021 | 1.47×10 −06 | 0.030 | 1.31×10 −11 | −0.002 | 6.20×10 −09 |
| rs13095391 | ACVR2B | 3 | 38,447,232 | a/c | 0.502 | 0.017 | 1.77×10 −04 | 0.025 | 4.03×10 −08 | −0.003 | 6.57×10 −15 |
|
| |||||||||||
| C from GWAS for declineadj, without association for decline (baseline-unadjusted) | |||||||||||
|
| |||||||||||
| rs9998485 | SHROOM3 | 4 | 77,362,445 | a/g | 0.466 | 0.007 | 0.156 | 0.027 | 9.84×10 −09 | −0.005 | 1.22×10 −41 |
| rs1047891 | CPS1 | 2 | 211,540,507 | a/c | 0.293 | 0.004 | 0.441 | 0.029 | 1.15×10 −09 | −0.007 | 1.18×10 −75 |
| rs2453533 | GATM | 15 | 45,641,225 | a/c | 0.422 | 0.002 | 0.710 | 0.029 | 1.72×10 −11 | −0.009 | 4.57×10 −141 |
SNPID=Variant identifier on GRCh37, Locus name=Nearest Gene, Chr and Position=Chromosome and Position on GRCh37, EA/OA=Effect allele / other allele, EAF=effect allele frequency, beta and P=genetic effect coefficient of association and association P-value.
In PRKAG2 and SPATA7 loci, variants with smallest Pdecline (rs73158188 and rs7160717, respectively) were highly correlated with these candidate-based variants (r2=1.00 and 0.93, respectively).
Since the TPPP locus lead variant had imputation quality <0.6 in 45% of the studies (median 0.64), we analyzed this locus omitting the imputation quality filter (with filter: declineadj beta=0.033, P=1.00×10−8; decline beta=0.015, P=0.039; median imputation quality=0.74).
In the C15ORF54 locus, the identified lead variant for decline was highly correlated with a 2nd signal lead variant for cross-sectional eGFR (rs28833881, r2=0.90), but not with the 1st signal lead variant (rs12913015, r2=0.04).
Second, we evaluated the 263 additional lead variants known for cross-sectional eGFR GWAS15 for association with baseline-unadjusted eGFR-decline (candidate approach); we had a prior hypothesis that cross-sectionally known variants might also show association with eGFR-decline. We identified two additional variants for eGFR-decline near PRKAG2 and SPATA7, both new loci for this phenotype, at Bonferroni-corrected significance (; Table 1A).
Third, our genome-wide screen for eGFR-decline adjusted for eGFR-baseline (n=320,737) identified 12 independent variants across 11 loci (, Figure 1B), including the four variants already identified by the baseline-unadjusted analyses (directly or via high correlation, r2≥0.9). The 8 variants additionally identified pointed to novel loci for this phenotype. Of these, 5 variants also showed directionally consistent, significant association for eGFR-decline unadjusted for eGFR-baseline (Bonferroni-corrected, ; near FGF5, OVOL1, TPPP, C15ORF54, and ACVR2B; Table 1B), but 3 variants did not ( from 0.156 to 0.710; near GATM, CPS1, SHROOM3, Table 1C).
Overall, we found 12 variants across 11 loci with genome-wide significant association for eGFR-decline unadjusted and/or adjusted for eGFR-baseline ( or ). All but one variant/locus were novel for this phenotype. All resided in loci known for eGFR cross-sectional GWAS15, but none was associated with DM-status (Supplementary Table S4).
The 12 variants’ associations showed no between-ancestry heterogeneity, stable statistics in various sensitivity analyses, and no impact by DM-adjustment (Supplementary Table S5&S6). Meta-analysis restricted to African American (n=9,038) did not identify associations for published APOL1 risk variants38, but two other suggestive variants (Supplementary Table S7).
The 12 variants included 9 variants with non-zero effects on eGFR-decline unadjusted for eGFR-baseline (i.e. Bonferroni-corrected significant, i.e. ).
SNP-effects for eGFR-decline were larger when baseline-adjusted than baseline-unadjusted
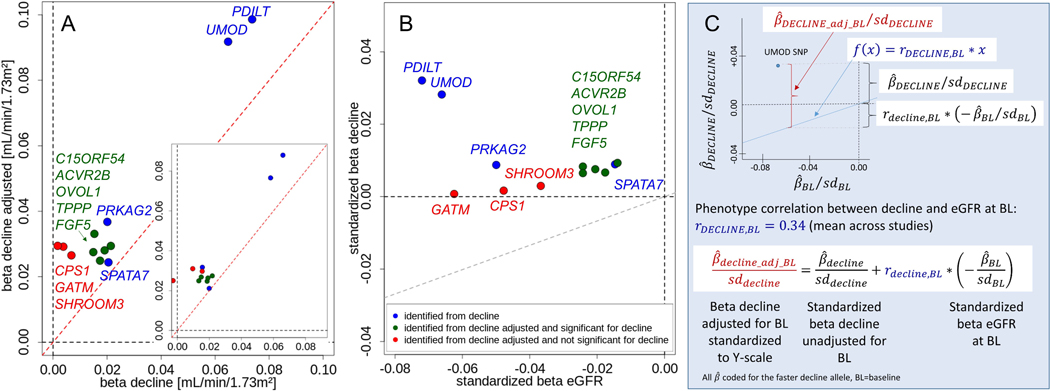
Several interesting aspects emerged when comparing genetic effect sizes of the 12 identified variants across models. First, we observed consistently larger effects for eGFR-decline baseline-adjusted than baseline-unadjusted (Figure 2A), also when restricting to studies where the baseline-adjusted model was directly computed (inserted small panel, Figure 2A). This, together with the smaller standard errors (Supplementary Figure S4B), explained the larger yield of genome-wide significant loci in the baseline-adjusted GWAS.
Figure 2: Relationship of SNP-effects on eGFR-decline baseline-unadjusted with baseline-adjusted effects for the 12 identified variants.
Shown are: (A) SNP-effects per year and allele for eGFR-decline baseline-unadjusted (“decline”) versus eGFR-decline baseline-adjusted in all studies (ndecline=343,339; ndecline-adj=320,737) and restricted to studies where baseline-adjusted results were computed rather than formula-derived (inserted panel, n=103,970); red line indicates identify line); (B) standardized SNP-effects per year and allele for eGFR-decline baseline-unadjusted (, n=343,339) and per allele for cross-sectional eGFR on ln-scale (, n=765,348 15); grey line indicates phenotype correlation line y=0.34*x (0.34=mean phenotype correlation across studies). For A&B: coding allele is the faster-decline allele (=cross-sectional eGFR-lowering allele). Color codes whether SNP was identified for decline baseline-unadjusted and/or baseline-adjusted. (C) Illustration of the SNP-effect for eGFR-decline baseline-adjusted (standardized to Y-scale) as a sum of the SNP-effect baseline-unadjusted (standardized) and the correlation-weighted SNP-effect on eGFR at baseline (standardized).
Second, we contrasted effect sizes for eGFR-decline unadjusted for eGFR-baseline with those for cross-sectional eGFR15 (Figure 2B). Three variants showed relatively extreme cross-sectional effects and no effect on decline (near GATM, SHROOM3, CPS1). For the other 9 variants, the faster-decline allele was always the cross-sectional eGFR-lowering allele (Spearman correlation coefficient=−0.32). A similar more schematic presentation (Figure 2C) illustrates the mathematical relationship between baseline-adjusted and baseline-unadjusted effect sizes (Supplementary Note S4). This yields a corollary on the directionality of baseline-adjusted effect sizes: when the faster-decline allele (i.e. ) coincides with the baseline eGFR-lowering allele (i.e. ), then the baseline-adjusted eGFR-decline effect size is larger than baseline-unadjusted (i.e. ) – in theory. Our data confirmed this empirically (Figure 2A). The larger genetic effect sizes for eGFR-decline adjusted for eGFR-baseline are thus a direct consequence of the phenotypic and genetic correlation between eGFR-decline and eGFR-baseline. The genetic effect for eGFR-decline unadjusted for eGFR-baseline provides the relevant effect size for further use and to distinguish between a “genuine association with eGFR-decline” (9 variants) and a pure “collider bias” effect (3 variants).
Four genes with compelling biological in-silico evidence mapped to novel eGFR-decline loci
All 11 identified loci for eGFR-decline coincided with loci detected for cross-sectional eGFR: among the 12 identified variants, 11 variants were genome-wide significant for cross-sectional eGFR15 and the variant near TPPP showed P=7.63×10−6 cross-sectionally with genome-wide significant variants nearby (Supplementary Figure S5A-C, Supplementary Note S5).
The 8 loci with genuine association for eGFR-decline included the well-known UMOD-PDILT locus. Biological evidence at the other seven loci was summarized using the Gene PrioritiSation tool18 generated from GWAS data on cross-sectional eGFR including evidence for SNP-modulated gene expression (eQTL, false-discovery-rate < 0.05): four lead variants or highly correlated proxies were eQTLs in tubule-interstitial kidney tissue with upregulating effects for SPATA7 and GALNTL5 (in PRKAG2 locus, kidney-tissue specific), a downregulating effect for FGF5 (kidney-tissue specific), and an upregulating effect for TPPP using NEPTUNE39. This supported these four genes in novel loci for eGFR-decline as kidney-tissue relevant and potentially causal genes for the association signals.
SNPs for eGFR-decline showed SNP-by-age interaction on cross-sectional eGFR
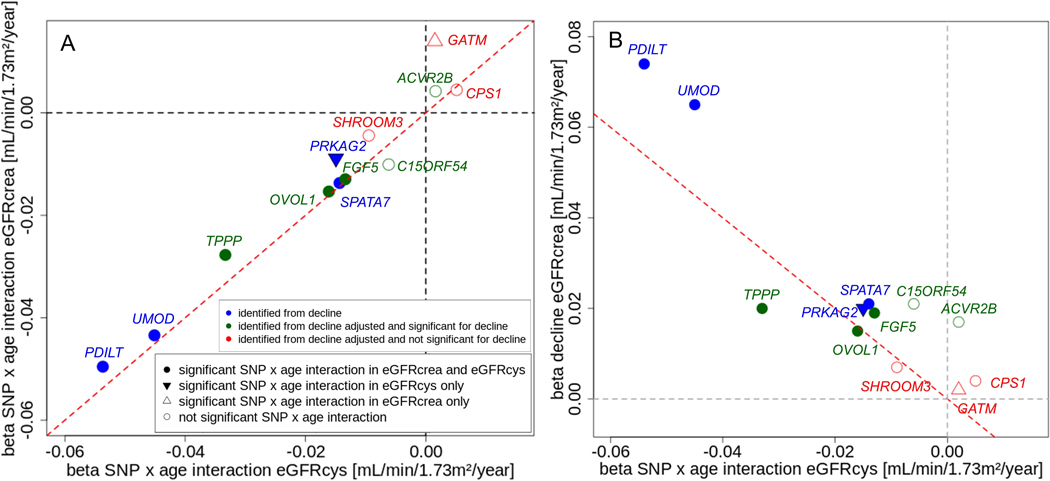
In the absence of birth cohort effects, we hypothesized that a SNP associated with eGFR-decline might also show an age-dependent association on cross-sectional eGFR, which is SNP-by-age interaction on cross-sectional eGFR. Of note, the age-effect on eGFR should reflect the age-effect on filtration rate, not on creatinine metabolism, within limits of uncertainty of the CKD-EPI formula6. To empirically assess this hypothesis, we tested the identified 12 SNPs for SNP-by-age interaction on cross-sectional eGFRcrea or eGFRcys in UK Biobank data, which was independent from and similarly-sized as the decline GWAS (n=351,462 or 351,601 for eGFRcrea or eGFRcys, respectively; Methods). For 8 of the 12 SNPs, we found SNP-by-age interaction for eGFRcrea and/or eGFRcys at Bonferroni-corrected significance (, Table 2). Interaction effect sizes were similar between eGFRcrea and eGFRcys (Figure 3A), except for the SNP near GATM.
Table 2: SNP-by-age interaction for cross-sectional eGFR for the 12 identified variants.
For the 12 identified variants, we conducted SNP-by-age interaction analysis for cross-sectional eGFRcrea and eGFRcys in UK Biobank (excluding individuals from decline GWAS; n=351,462 for eGFRcrea, n=351,601 for eGFRcys; main age effect modelled non-linearly, main SNP effect linearly, age centered at 50 years). The interaction term (age effect and SNP effect modelled linearly) was judged at Bonferroni-corrected significance level (P<0.05/12=4.17×10−3). Beta-estimates are in mL/min/1.732 per year and per cross-sectional eGFR-lowering allele (which was equivalent to faster-decline allele for each SNP); significant P-values are stated in bold.
| SNPID | Locus Name | EA/OA | SNP x age interaction eGFRcrea |
SNP x age interaction eGFRcys |
||
|---|---|---|---|---|---|---|
| Beta | P | Beta | P | |||
| A from GWAS/candidate search for decline (baseline-unadjusted) | ||||||
|
| ||||||
| rs34882080 | UMOD-PDILT | a/g | −0.043 | 5.53×10 −22 | −0.045 | 2.37×10 −17 |
| rs77924615 | UMOD-PDILT | g/a | −0.050 | 2.55×10 −29 | −0.054 | 6.59×10 −25 |
| rs10254101 | PRKAG2 | t/c | −0.009 | 0.0263 | −0.015 | 9.84×10 −04 |
| rs1028455 | SPATA7 | t/a | −0.014 | 2.19×10 −04 | −0.014 | 1.06×10 −03 |
|
| ||||||
| B from GWAS for declineadj, with association for decline (baseline-unadjusted) | ||||||
|
| ||||||
| rs1458038 | FGF5 | c/t | −0.013 | 7.11×10 −04 | −0.013 | 3.12×10 −03 |
| rs4930319 | OVOL1 | c/g | −0.015 | 2.55×10−05 | −0.016 | 1.84×10−04 |
| rs434215 | TPPP | a/g | −0.028 | 1.02×10 −10 | −0.033 | 5.02×10−11 |
| rs28857283 | C15ORF54 | g/a | −0.010 | 5.09×10−03 | −0.006 | 0.148 |
| rs13095391 | ACVR2B | a/c | 0.004 | 0.227 | 0.002 | 0.695 |
|
| ||||||
| C from GWAS for declineadj, without association for decline (baseline-unadjusted) | ||||||
|
| ||||||
| rs9998485 | SHROOM3 | a/g | −0.004 | 0.206 | −0.009 | 0.022 |
| rs1047891 | CPS1 | a/c | 0.004 | 0.228 | 0.005 | 0.244 |
| rs2453533 | GATM | a/c | 0.014 | 9.71×10 −05 | 0.002 | 0.722 |
SNPID=Variant identifier on GRCh37, Locus name=Nearest Gene, EA/OA=Effect allele / other allele, Beta and P=genetic effect and association P-value. The TPPP variant rs434215 is well-imputed in the UK Biobank (imputation quality=0.82).
Figure 3: Relationship of SNP-by-age interaction effects for eGFRcys with those of eGFRcrea and with SNP-effects for eGFR-decline for the 12 identified variants.
Shown are SNP-by-age interaction effect sizes per year and allele for cross-sectional eGFRcys (UK Biobank individuals independent from GWAS, nSNPxage=351,601; main age effect modelled non-linearly, main SNP-effect linearly, age effect and SNP effect in interaction term linearly, age centered at 50 years) versus: (A) SNP-by-age interaction effects on cross-sectional eGFRcrea (nSNPxage=351,462), (B) SNP-effects on eGFR-decline baseline-unadjusted per year and allele (ndecline=343,339). Coding allele is the faster-decline allele (=cross-sectional eGFR-lowering allele); color code as in Figure 2; red line indicates identity line; symbol types code significance of interaction term (P< 0.05/12). Among the 9 SNPs with genuine eGFR-decline association, 7 SNPs showed interaction for eGFRcrea or eGFRcys (all negative), and all 3 SNPs without genuine eGFR-decline association showed no interaction for eGFRcys (one with positive significant interaction for eGFRcrea).
The age-dependency of all SNP-effects and main age-effects were approximately linear (Supplementary Figure S6, Supplementary Note S6). The SNP-by-age interaction effect size can also be interpreted as the genetically modified age-effect on eGFR. This effect was large: e.g., 5 unfavorable alleles decreased eGFRcys by −0.136 mL/min/1.73m2 per year, which was ~10% of the overall age-effect on eGFRcys (−1.024 mL/min/1.73m2per year, Supplementary Note S6). SNP-by-age interaction effects on eGFRcys were highly correlated with SNP-effects on eGFR-decline (both in units of mL/min/1.73m2 per allele and year: “per year of age-difference between individuals” and “per year of person’s aging”, respectively; Figure 3B).
There was a noteworthy pattern with regard to presence and direction of SNP-by-age interaction: (i) among the 9 variants with genuine association for eGFR-decline, 7 variants showed significant SNP-by-age interaction on cross-sectional eGFRcys (Table 2A&B). All interaction effects were negative, i.e. the cross-sectional SNP-effect became larger (in absolute value) with older age. (ii) Among the three SNPs without genuine association for eGFR-decline, two showed no SNP-by-age interaction; the third (near GATM) showed SNPby-age interaction, but only for eGFRcrea and with positive direction (, ). Thus, the GATM SNP-effect on cross-sectional eGFRcrea gets smaller (in absolute value) by higher age. This might be explained by GATM being the rate-limiting enzyme in creatine synthesis in muscle, age-related loss of muscle mass, and thus decreased creatinine production with increasing age - in line with the lack of interaction with eGFRcys, which is unrelated to muscle mass.
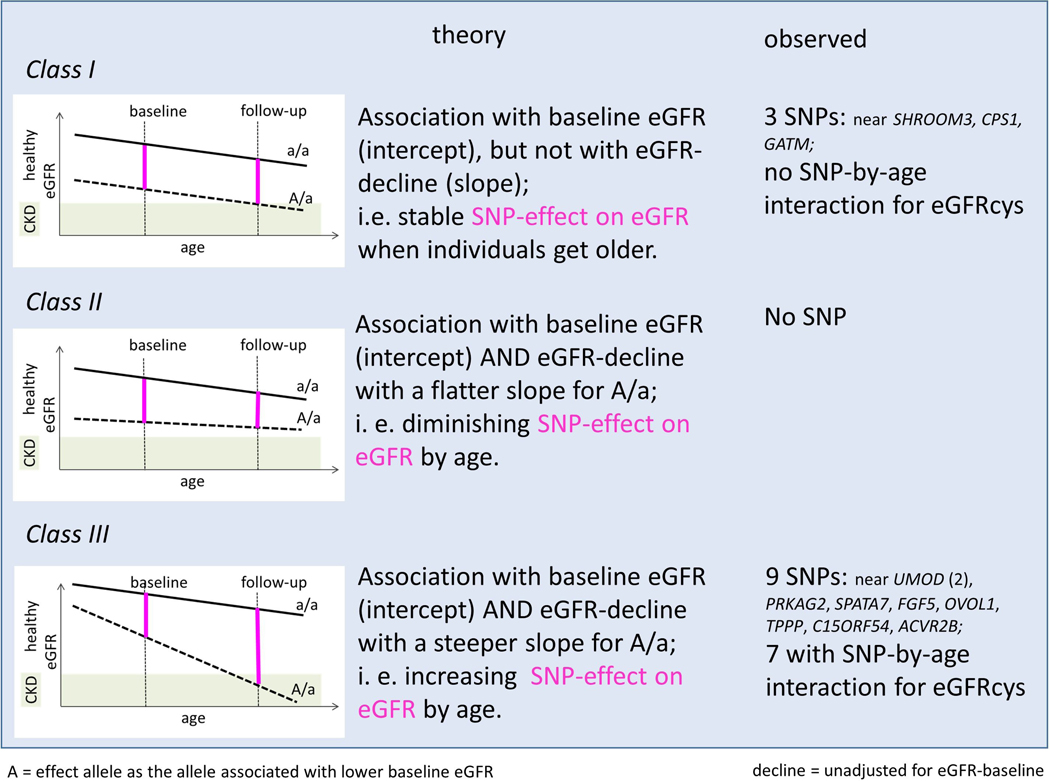
A concept of three classes of SNPs for cross-sectional eGFR distinguished by their eGFR-decline association
Our results suggested that SNPs for eGFR-decline were found among SNPs associated with eGFR cross-sectionally. This motivated the idea of, in theory, three classes of SNP-associations on cross-sectional eGFR (intercept) distinguished their eGFR-decline association unadjusted for eGFR-baseline (slope; Figure 4): no association with slope (class I), association of the eGFR-baseline lowering allele with flatter slope (class II), or association of the eGFR-baseline lowering allele with steeper slope (class III).
Figure 4: A concept for three classes of SNP-associations on cross-sectional eGFR distinguished by the presence and direction of the SNP-association with eGFR-decline.
Let A/a be the genotype group of individuals with, on average, lower cross-sectional eGFR compared to a/a (A=effect allele). Let’s further assume that eGFR-declines monotonously by age (approximated as linear decline) and that there is no “cross-over” between genotype groups. Shown are (left) a graphical scheme, (middle) the theoretical association, (right) the observed SNPs in line with the respective class. In the three graphical schemes, black lines illustrate mean eGFR-decline by genotype group; SNP-effects on eGFR for these individuals captured cross-sectionally at different ages are magenta. When a cross-sectional study captures individuals of relevant ages, the SNP-effects on eGFR should show an interaction by age for class II and class III SNPs (positive and negative, respectively). The 9 variants with genuine eGFR-decline association were class III, while the other 3 variants were class I.
In our data, we found (i) three of the 12 SNPs as class I, in line with the lack of SNP-by-age interaction on eGFR cross-sectionally (judged for eGFRcys). (ii) No variant was class II, consistent with the lack of positive SNP-by-age interaction on eGFRcys. (iii) The 9 variants with genuine eGFR-decline association were class III, and 7 of these showed negative SNP-by-age interaction on eGFR. Thus, our data supported two classes of genetic effects on eGFR: no association with slope or steeper slope for the eGFR-lowering allele.
Larger SNP-effects for eGFR-decline were observed in high-risk subgroups
Individuals with DM and/or CKD (defined as eGFR<60 mL/min/1.73m2) are at higher risk for CKD-progression and kidney failure, prompting us to quantify SNP-effects on eGFR-decline in these high-risk subgroups (meta-analysis for eGFR-decline unadjusted for eGFR-baseline restricted to DM or CKD at baseline, n= 37,375 or 26,653 respectively, Methods). For the 9 variants with genuine eGFR-decline association, we found almost all effects to be two- to four-fold larger in DM or in CKD compared to the overall analysis (Table 3, average effect size [mL/min/1.73m2/year and allele]: 0.061 in DM, 0.079 in CKD, compared to 0.030 overall).
Table 3: The 9 variants’ effects on eGFR-decline unadjusted for eGFR-baseline in high-risk subgroups.
Shown are the 9 variants with genuine association for eGFR-decline for their association with eGFR-decline restricted to individuals with baseline diabetes mellitus (DM, n up to 38,206) or baseline CKD (i.e. eGFR<60 mL/min/1.73m2, n up to 26,653). Beta-estimates and 95% confidence intervals (CI) are in mL/min/1.73m2 per year and per faster-decline allele.
| SNPID | Locus Name | Decline among DM at baseline |
Decline among CKD at baseline |
Decline among all |
|||
|---|---|---|---|---|---|---|---|
| Beta | 95% CI | Beta | 95% CI | Beta 95% CI | |||
| A from GWAS/candidate search for decline (baseline-unadjusted) | |||||||
|
| |||||||
| rs34882080 | UMOD-PDILT | 0.159* | 0.108, 0.211 | 0.138* | 0.074, 0.203 | 0.065 | 0.054, 0.076 |
| rs77924615 | UMOD-PDILT | 0.136* | 0.084, 0.189 | 0.167* | 0.099, 0.235 | 0.074 | 0.063, 0.085 |
| rs10254101 | PRKAG2 | 0.065 | 0.020, 0.110 | 0.095* | 0.042, 0.148 | 0.020 | 0.010, 0.030 |
| rs1028455 | SPATA7 | 0.030 | −0.011, 0.071 | 0.085* | 0.034, 0.135 | 0.021 | 0.012, 0.029 |
|
| |||||||
| B from GWAS for declineadj, with association for decline (baseline-unadjusted) | |||||||
|
| |||||||
| rs1458038 | FGF5 | 0.030 | −0.013, 0.072 | 0.040 | −0.013, 0.092 | 0.019 | 0.010, 0.028 |
| rs4930319 | OVOL1 | 0.021 | −0.021, 0.062 | 0.031 | −0.019, 0.080 | 0.015 | 0.006, 0.024 |
| rs434215 | TPPP § | 0.031 | −0.024, 0.086 | 0.112* | 0.043, 0.180 | 0.020 | 0.006, 0.035 |
| rs28857283 | C15ORF54 | 0.046 | 0.005, 0.086 | 0.042 | −0.007, 0.091 | 0.021 | 0.013, 0.030 |
| rs13095391 | ACVR2B | 0.029 | −0.021, 0.080 | 0.006 | −0.054, 0.066 | 0.017 | 0.008, 0.026 |
|
| |||||||
| Average | 0.061 | 0.079 | 0.030 | ||||
SNPID=Variant identifier on GRCh37, Locus name=Nearest Gene, Beta=genetic effect of genetic association where the effect alleles is the same as in Table 1 and Table 2, 95% CI = 95% confidence interval of Beta (Beta±1.96*standard error of the association).
Statistically significant different from zero (P< 0.05/9=5.56×10−3).
Since the lead variant had imputation quality <0.6 in 45% of the studies (median 0.64), we analyzed this variant omitting the imputation quality filter (with filter: decline among DM at baseline beta=−0.093, P=0.338, n=927; decline among eGFR <60 mL/min/1.73m2 beta=0.022, P=0.618, n=2924; median imputation quality=0.74).
To get an idea of the magnitude, we scaled the effects to “per 5 unfavorable average alleles” resulting in a decline of 0.305 in DM, 0.395 in CKD, compared to 0.150 mL/min/1.73m2/year overall. This compared well to the 9-variant weighted GRS effect on eGFR-decline per 5 unfavorable average alleles in the HUNT study (n=2,235 with DM, n=502 with CKD, n=46,328 overall; Methods): 0.219 in DM, 0.262 in CKD, and 0.102 mL/min/1.73m2/year overall (one-sided P=1.57×10−5, P=0.0193, and P=1.06×10−34, respectively).
The genetic effect sizes were also larger in the two subgroups when viewed relative to the phenotype variance (on the example of HUNT, Methods): rs77924615 variant (UMOD-PDILT locus) explained 0.38% of the eGFR-decline variance in DM, 0.47% in CKD, and 0.22% overall; the 9-variants jointly explained 1.14%, 1.48%, and 0.51%, respectively. Of note, the explained variance of eGFR-decline overall was comparable to the explained variance of cross-sectional eGFR (rs77924615: 0.21%; 9 variants: 0.62%), but narrow-sense heritability was smaller (Supplementary Note S7).
GALNTL5, SPATA7, and TPPP were identified as candidates for CKD-progression
Variants associated with CKD-progression and mapped genes might help identify drug targets against disease progression19. We queried the 9 SNPs with genuine association for eGFR-decline for significant association with CKD-progression, i.e. whether they still showed significant association with eGFR-decline when focusing on individuals with CKD at baseline (judged at P<0.05/9=5.56×10−3, n up to 26,547). We found five such SNPs: (i) two in the UMOD-PDILT locus, which confirmed UMOD for a role in CKD-progression, (ii) three SNPs in novel loci for eGFR-decline, which mapped to three genes with eQTL in kidney tissue (GALNTL5 in PRKAG2 locus, kidney-tissue specific; SPATA7, and TPPP), making these compelling candidates as CKD-progression genes.
Unfavorable GRS increased the risk for ESKD and AKI
Finally, we wanted to understand the cumulative impact of the 9 genuine eGFR-decline variants for severe clinical endpoints. We thus evaluated the 9-variant weighted GRS in cases-control studies for ESKD and AKI via logistic regression (ncases=2,068 and 3,878, ncontrols=4,640 and 11,634, respectively; Methods). The GRS effect per 5 unfavorable average alleles showed a significant OR=1.12 for ESKD (95%CI=0.99–1.23; one-sided P=0.033) and OR=1.18 for AKI (95% CI=1.09–1.27; one-sided P<0.0001 Table 4). When comparing the individuals with GRS ≥90th versus ≤10th percentile (i.e. ≥14.6 unfavorable alleles versus ≤8.3 in UK Biobank), we found a significant OR=1.35 for ESKD (95%CI=1.03–1.77, one-sided P=0.0157) and OR=1.27 (95%CI=1.08–1.50, one-sided P=0.002, Table 4).
Table 4: Genetic risk score (GRS) analyses for end-stage kidney disease (ESKD) and Acute Kidney Injury (AKI).
In 3 case-control studies for ESKD and one for AKI, we computed the weighted GRS across the 9 eGFR-decline variants (counting the faster-decline alleles, weighted by effect size for eGFR-decline unadjusted for eGFR-baseline; divided by sum of weights and multiplied by 9, i.e. scaled as 0 to 18). Shown are odds ratios (OR), 95% confidence intervals (CI) and P-values (one-sided) for the quantitative GRS association (per 5 “average” unfavorable alleles) and for a high versus low GRS association (≥95th versus ≤5th, ≥90th versus ≤10th GRS percentiles derived in UK Biobank) with (A) ESKD and (B) AKI. Associations are derived by logistic regression adjusted for matching variables age-group and sex (AKI additionally for principal components).
| Per 5 unfavorable average alleles |
High versus low GRS group |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5% versus 95% |
10% versus 90% |
||||||||||
| Study | Number of Cases | Number of Controls | OR | 95% CI | P (1-sided) | OR | 95% CI | P (1-sided) | OR | 95% CI | P (1-sided) |
| (A) ESKD (cases: ICD10 code N18.0 or N18.5; controls: no ICD10 code N18, eGFR>60 mL/min/1.73m2, frequency-matched by age-group and sex) | |||||||||||
| 4D_KORA-F3 | 1,100 | 1,601 | 1.122 | 0.925,1.362 | 0.121 | 1.26 | 0.669,2.377 | 0.237 | 1.526 | 0.978,2.379 | 0.0313 |
| GENDIAN_KORA-F4 | 470 | 1,545 | 1.146 | 0.923,1.423 | 0.108 | 0.954 | 0.468,1.946 | 0.449 | 1.036 | 0.625,1.719 | 0.445 |
| UKBBCa_co | 498 | 1,494 | 1.085 | 0.885,1.330 | 0.216 | 1.220 | 0.639,2.329 | 0.273 | 1.479 | 0.921,2.373 | 0.0525 |
|
| |||||||||||
| Meta-analysis | 2,068 | 4,640 | 1.117 | 0.993,1.256 | 0.0329 | 1.150 | 0.785,1.686 | 0.236 | 1.349 | 1.027,1.773 | 0.0157 |
|
| |||||||||||
|
| |||||||||||
| (B) AKI (cases: ICD 10 code N17; controls: no ICD10 code N17, eGFR>60 mL/min/1.73m2, frequency-matched by age-group and sex) | |||||||||||
|
| |||||||||||
| UKBBCaCo | 3,878 | 11,634 | 1.179 | 1.095,1.270 | 6.47×10−06 | 1.524 | 1.204,1.931 | 4.70×10−04 | 1.272 | 1.080,1.499 | 1.97×10−03 |
Study=Study name, OR=Odds Ratio of the GRS-association, 95% CI=95% confidence interval of the association, P (1-sided)=1-sided association P-value, ESKD=End-stage Kidney Disease, Individuals analyzed here are distinct from the eGFR-decline GWAS except for the KORA-F3 and KORA-F4 controls. AKI=Acute Kidney Injury, UKBBCaCo=cases and controls from UK Biobank distinct from UK Biobank study participants used in the GWAS for eGFR decline.
DISCUSSION
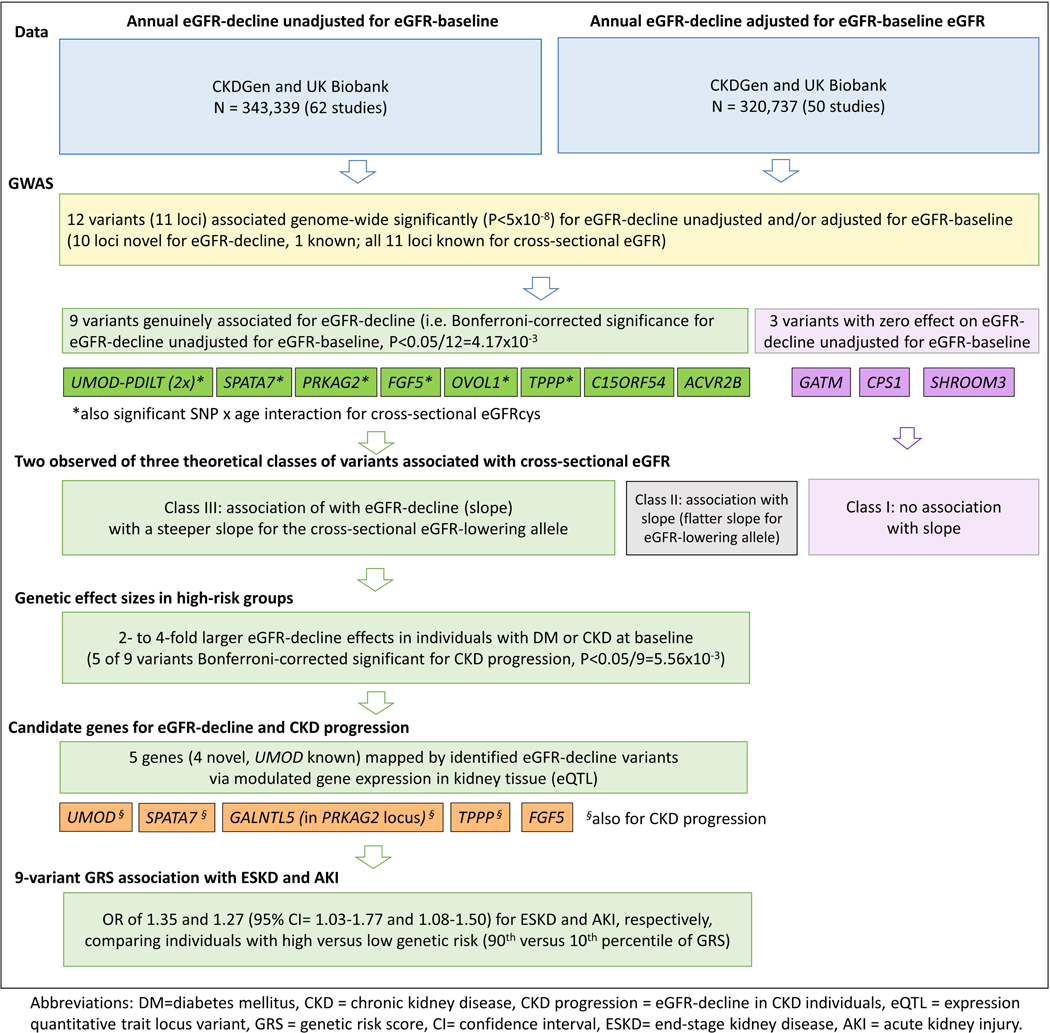
Here, we provide data and results on a large longitudinal GWAS on annual eGFR-decline with >340.000 individuals from mostly population-based studies – to our knowledge the largest GWAS on annual eGFR-decline so far and probably one of the largest longitudinal GWAS of any trait. We identified 12 variants across 11 loci as genome-wide significant for annual eGFR-decline unadjusted and/or adjusted for eGFR-baseline (Figure 5). These included 9 variants across 8 loci with non-zero association unadjusted for eGFR-baseline, which we termed “genuinely” associated with eGFR-decline. Seven of these 9 variants also showed SNP-by-age interaction on cross-sectional eGFR in independent data of >350,000 individuals, while the three variants without genuine association did not. We generated and provide genome-wide summary statistics for eGFR-decline, CKD-progression, and eGFR-decline in DM. This data resource is informative for future meta-analyses, causal inference studies via Mendelian Randomization40, and drug development pipelines.
Figure 5: Data, analyses, and results in a nutshell.
Clinically very important is our finding of the two-to four-fold larger genetic effects of almost all identified variants when focusing on individuals with DM or CKD at baseline, since these individuals are already at higher risk of kidney failure. This observation is in line with a “horse-racing effect”41 (“a faster horse is more likely observed up front”): individuals with an accumulation of faster eGFR-decline alleles are more likely observed with low eGFR at a given point in time, implying that these genetic effects might partly explain lower eGFR at baseline. A part of the larger eGFR-decline effect among CKD individuals might reflect collider bias. However, DM-status does not fulfill the characteristics of a collider for the SNP-associations with eGFR-decline (no impact by adjusting for DM-status, no SNP-association with DM-status), rendering the higher eGFR-decline effects in DM genuine.
The clinical relevance is further underscored by the 9-variant GRS being associated with increased risk of AKI and ESKD. This observation requires further analyses in future larger data. If substantiated, this may indicate a genetic risk of incomplete kidney function recovery after AKI and a genetic predisposition for ESKD.
The 9 identified variants across 8 loci included the UMOD-PDILT locus associated with eGFR-decline and CKD-progression, which is largely confirmatory but serves as proof-of-concept. A variant near MIR378C previously identified for CKD-progression42 (n~3000) was not confirmed here. Our other 7 loci are novel for eGFR-decline (near/in PRKAG2-GALNTL5, SPATA7, FGF5, OVOL1, TPPP, C15ORF54, and ACVR2B). These included at least three loci associated with CKD-progression (defined as eGFR-decline in individuals with CKD at baseline), mapping to the genes GALNTL5, SPATA7 and TPPP by SNP-modulated expression in tubolo-interstitium15,18. These associations and genes for CKD-progression are in strong demand as genetic information on a disease progression phenotype, in order to help identify treatment19. Our data particularly flags TPPP by its locus’ large effect on eGFR-decline and CKD-progression, making it second only after UMOD. This also documents the value of longitudinal GWAS in revealing relevance of genes like TPPP: the TPPP locus was one of hundreds of small effect loci cross-sectionally, but among the few loci longitudinally.
Our results highlight some overlap of quantitative eGFR-decline genetics with binary extreme decline genetics22, but also distinction. All loci identified here were directionally consistent, nominally significant with “rapid3” and/or “CKDi25” (one-sided P<0.05) and two were genome-wide significant for rapid3 or CKDi25 (UMOD-PDILT, PRKAG2-GALNTL5). Particularly the loci identified here for CKD-progression, which is among individuals with CKD at baseline, complement the previously reported associations with CKDi25, which is among individuals without CKD at baseline. Methodologically, regression applied to a quantitative rather than dichotomized outcome has larger power and statistical advantages.
While all variants identified for eGFR-decline captured loci known from cross-sectional eGFR15, these associations are important on various accounts. First, the mere fact that eGFR-decline genetics is a subgroup of cross-sectional eGFR genetics is informative for future searches. Second, the finding that the full genetic signals were the same enabled the use of fine-mapping results from cross-sectional GWAS in >1 million individuals18 to prioritize genes also for longitudinal eGFR-decline. Third, all faster-decline alleles were the cross-sectional eGFR-lowering alleles. Together, this supported two classes of genetic variants for cross-sectional eGFR, distinguished by lack or presence of a slope effect, with steeper slope for the cross-sectional eGFR-lowering allele. The data rendered the third theoretical option, i.e. presence of a slope effect with flatter slope for the cross-sectional eGFR-lowering allele, void.
Some limitations warrant mentioning. Although this GWAS is currently the largest GWAS on eGFR-decline so far, more loci for eGFR-decline and CKD-progression might be detectable upon further increased sample size. The yield of eGFR-decline loci in >340,000 individuals was comparably low considering older GWAS for cross-sectional eGFR having already detected >50 loci in 170,000 individuals43. We used the CKD-EPI formula containing an ancestry term (Levey et al., Ann Intern Med), accounted for by ancestry-specific GWAS; future work should utilize the new ancestry-term-free CKD-EPI formula 2021 (Inker et al., NEJM). Evaluating the potential existence of sex-specific genetic effects on eGFR-decline is of interest, but was not addressed in this project. The target population is primarily population-based, including kidney diseases proportional to respective prevalence, and primarily European ancestry. Larger all-ancestry meta-analyses on eGFR-decline will open up opportunities to also utilize differential linkage disequilibrium between ancestries to help narrow down causal variants and genes. The interpretability of the SNP-by-age interaction on cross-sectional eGFR is limited to the age spectrum in the data (40–70 years) and by the power given the sample size; still, the sample size used was large and the age range typical also for most eGFR-decline GWAS studies. Two aspects need mentioning regarding the phenotype definition: uncertainty in eGFR-decline may be larger for studies with shorter follow-up, which decreases power, but measurement error in the outcome does not induce bias in linear regression44. By defining annual eGFR-decline from two eGFR assessments over time, our SNP associations capture only the linear component of decline. Serial eGFR assessments are better to characterize eGFR-trajectories, but at the cost of limiting sample size, since such studies are few and typically small. Furthermore, generalized additive mixed models for nonlinear eGFR-trajectories are complex and require particularly large sample sizes. The linear modelling of eGFR-decline is a reasonable approximation of monotonous decline, maintaining large sample sizes and limiting model complexity to be applicable for GWAS. Overall, the choice of the adjustment, target population, and phenotype definition are important to consider when interpreting results. While some modelling aspects are addressed here, other covariate adjustment or relative decline as phenotype might reveal further or other genetic loci. Future work is warranted to quantify effects in different target populations and the genetically determined shape of the decline, which requires more – and larger – longitudinal studies, ideally with more than two eGFR assessments over time.
Methodologically unique is our contrasting of GWAS SNP-associations on eGFR-decline for different covariate adjustment, which fills an important gap and helps design future studies. This is highly relevant, since covariate adjustment can alter GWAS findings and interpretation28–31,45. Adjusting for baseline DM-status had no impact, but genetic effects for eGFR-decline were larger when restricting to DM-individuals; this suggests DM-status as modulator for the SNP-association with eGFR-decline rather than mediator (i.e. in the causal pathway from SNP to eGFR-decline) or collider (i.e. generating biased association). Adjustment for eGFR-baseline yielded larger eGFR-decline effects and more genome-wide significant variants. Glymour et al. highlight that adjustment for baseline levels in analyses of change may help detect effects, but can induce spurious associations when the rate of change observed after baseline reflects a rate of change experienced in the past36. This might reflect the situation here rendering the larger genetic effects adjusted for eGFR-baseline - and the larger genetic effects when restricting to individuals with CKD at baseline – reflective of collider bias. Glymour et al. recommend the documentation of change effects without baseline adjustment36. In line with this, we considered a variant’s association with eGFR-decline genuine, when the variant reached genome-wide significance baseline-unadjusted or baseline-adjusted and Bonferroni-corrected significance baseline-unadjusted. The baseline-unadjusted model provides the relevant genetic effect sizes for eGFR-decline.
Interestingly, two of the three associations without genuine eGFR-decline association may relate to biomarker generation rather than kidney function: GATM and CPS1, known for a role in creatine biosynthesis41 and urea cycle42, respectively, reside in loci without supporting association with cross-sectional cystatin-based eGFR18. Conversely, the SHROOM3 locus was associated with cystatin-based eGFR18,15 and experimental studies support a role of SHROOM3 in kidney pathology46–48; thus, SHROOM3 appears to have an effect on cross-sectional kidney function, but not on kidney function decline within the limits of detectability by sample size.
A further unique aspect of our work is the empirical evidence for a link between SNP-effects on eGFR-decline with SNP-by-age interaction effects on cross-sectional eGFR. By this, we provide important insights into the age-dependency of kidney function genetics as well as into the genetic dependency of aging eGFR in adult general populations, where “aging” includes onset of age-related diseases as they develop in populations. Considering the much broader availability of cross-sectional than longitudinal data, the further parallel exploitation of SNP-by-age interaction might be a promising route to help improve our understanding of the mechanisms of kidney function decline over time.
In summary, we provide GWAS summary statistics, identified genetic loci, and prioritized genes for kidney function decline and CKD-progression. While UMOD has drawn attention already, GALNTL5, SPATA7, and TPPP may now receive more focus as therapeutic targets for disease progression. Our exploration of different covariate adjustment and the comparison to age-dependency of SNP-effect on eGFR cross-sectional provides important insights into the interpretation of these effects. With the emerging large biobank data linking medical records, longitudinal GWAS will become very important in the future. Our methodological framework is informative and applicable also generally for longitudinal phenotypes.
Availability of data and materials
To support future work, we provide genome-wide summary statistics on eGFR-decline unadjusted for eGFR-baseline (adjusted for age, sex and DM-status) overall and restricted to individuals with DM or CKD at baseline (all adjusted for age and sex) (https://www.uniregensburg.de/decline and http://ckdgen.imbi.uni-freiburg.de). The summary statistics on eGFR-decline in individuals with CKD at baseline can be considered genetic effects on CKD-progression. We also provide genome-wide summary statistics on eGFR-decline adjusted for eGFR-baseline (additionally to adjustment for age and sex), but these summary statistics should be used with great care and an understanding that beta-estimates are subject to collider bias. For quantification of the genetic effect on eGFR-decline, the results unadjusted for eGFR-baseline should be utilized.
Supplementary Material
Supplementary Methods
Note S1. Equivalence of DM-adjusted versus not DM-adjusted GWAS on eGFR-decline in the validation meta-analysis
Note S2. Formula-based covariate adjustment using GWAS summary statistics
Note S3. Validation of the formula-derived association for eGFR-decline adjusted for eGFR-baseline
Note S4. Graphical illustration of the relationship between SNP-effects on eGFR-decline unadjusted and adjusted for eGFR-baseline
Note S5. Comparison of the signals for eGFR-decline unadjusted and adjusted for eGFR-baseline and cross-sectional eGFR for the 11 identified loci
Note S6. Age-dependency of SNP-effects and main age effect on eGFR
Note S7. Narrow-sense heritability
Figure S1. Meta-analysis workflow
Figure S2. Study-specific median annual eGFR-decline versus sample size, follow-up time and median age
Figure S3. Influence of alternative adjustments for age on eGFR-decline in UK Biobank
Figure S4A. No influence from adjusting SNP-associations for eGFR-decline for diabetes mellitus (DM)
Figure S4B. Differences between SNP-association for eGFR-decline unadjusted versus adjusted for eGFR-baseline
Figure S4C. Validation of formula-derived adjustment for eGFR-baseline in eGFR-decline associations (part 1).
Figure S4D. Validation of formula-derived adjustment for eGFR-baseline in eGFR-decline associations (part 2)
Figure S5. Region plots of loci identified for eGFR-decline unadjusted and adjusted for eGFR-baseline
Figure S6. Age-dependency of eGFR and age-dependency of the variant effects on eGFR in UK Biobank
Table S1. Description of participating studies: study design
Table S2. Description of participating studies: genotyping and imputation
Table S3. Description of participating studies: phenotype distribution
Table S4. The 12 identified variants for eGFR-decline were associated with other kidney phenotypes, but not with DM-status
Table S5. The 12 identified variants for eGFR-decline do not show heterogeneity between ancestries and FHS is not an influential study
Table S6. No influence by DM-adjustment versus no DM-adjustment or by model-based versus formula-based adjusting for baseline eGFR (BL) on the 12 variants’ association with eGFR-decline
Table S7. Association of APOL1 risk variants in African American and European CKDGen studies
Extended acknowledgements, study funding information and author contributions
Supplementary References
Author contributions
ACKNOWLEDGEMENTS
The Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) supported the meta-analysis – Project-ID 387509280 – SFB1350 (Subproject C6 to I.M.H.). We conducted this research using the UK Biobank resource under the application number 20272. Extended acknowledgements and funding information are provided in the Supplementary Online Material.
DISCLOSURE STATEMENT
JÄ reports personal fees from AstraZeneca, Boehringer Ingelheim and Novartis, outside the submitted work. Sanofi Genzyme currently employs KeH. WKo reports modest consultation fees for advisory board meetings from Amgen, DalCor, Kowa, Novartis, Pfizer and Sanofi; modest personal fees for lectures from Amgen, AstraZeneca, Novartis, Pfizer and Sanofi, outside the scope of this work. CL received Grants/ Research Support from Bayer Ag/ Novo Nordisk, Husband works for Vertex. KBS, LMY, DMW and MAL are full-time employees of GlaxoSmithKline. MLO received grant support from GlaxoSmithKline, MSD, Eisai, AstraZeneca, MedCo and Janssen. BMP serves on steering committee of the Yale Open Data Access Project funded by Johnson & Johnson. PR received fees to his institution for research support from AstraZeneca and Novo Nordisk; for steering group participation from AstraZeneca, Gilead, Novo Nordisk, and Bayer; for lectures from Bayer, Eli Lilly and Novo Nordisk; and for advisory boards from Sanofi and Boehringer Ingelheim outside of this work. LWal received institutional grants from GlaxoSmithKline, AstraZeneca, BMS, Boehringer-Ingelheim, Pfizer, MSD and Roche Diagnostics. HW received grants and non-financial support from GlaxoSmithKline, during the conduct of the study, grants from Sanofi-Aventis, Eli Lilly, the National Institute of Health, Omthera Pharmaceuticals, Pfizer New Zealand, Elsai Inc. and Dalcor Pharma UK; honoraria and non-financial support from AstraZeneca; and is on advisory boards for Sirtex/ Acetilion and received personal fees from CSL Behring and American Regent outside the scope of this work. GS, DG, HH, IO, KStef, PS and UT are employees of deCODE/Amgen Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All other authors declared no competing interests.
REFERENCES
- 1.Levey AS, Coresh J, Tighiouart H, Greene T, Inker LA. Measured and estimated glomerular filtration rate: current status and future directions. Nat Rev Nephrol. 2020;16(1):51–64. [DOI] [PubMed] [Google Scholar]
- 2.Andrassy KM. Comments on “KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease.” Kidney Int. 2013;84(3):622–623. [DOI] [PubMed] [Google Scholar]
- 3.Eckardt K-U, Coresh J, Devuyst O, et al. Evolving importance of kidney disease: from subspecialty to global health burden. Lancet (London, England). 2013;382(9887):158–169. [DOI] [PubMed] [Google Scholar]
- 4.Neuen BL, Weldegiorgis M, Herrington WG, Ohkuma T, Smith M, Woodward M. Changes in GFR and Albuminuria in Routine Clinical Practice and the Risk of Kidney Disease Progression. Am J Kidney Dis. 2021;78(3):350–360.e1. [DOI] [PubMed] [Google Scholar]
- 5.Meguid El Nahas A, Bello AK. Chronic kidney disease: the global challenge. Lancet (London, England). 2005;365(9456):331–340. [DOI] [PubMed] [Google Scholar]
- 6.Levey AS, Stevens LA, Schmid CH, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150(9):604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin A, Stevens PE, Bilous RW, et al. Notice. Kidney Int Suppl. 2013;3(1):1. [Google Scholar]
- 8.Hemmelgarn BR, Zhang J, Manns BJ, et al. Progression of kidney dysfunction in the community-dwelling elderly. Kidney Int. 2006;69(12):2155–2161. [DOI] [PubMed] [Google Scholar]
- 9.Garcia Sanchez JJ, Thompson J, Scott DA, et al. Treatments for Chronic Kidney Disease: A Systematic Literature Review of Randomized Controlled Trials. Adv Ther. 2022;39(1):193–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heerspink HJL, Stefánsson BV., Correa-Rotter R, et al. Dapagliflozin in Patients with Chronic Kidney Disease. N Engl J Med. 2020;383(15):1436–1446. [DOI] [PubMed] [Google Scholar]
- 11.Borges-Júnior FA, Silva dos Santos D, Benetti A, et al. Empagliflozin Inhibits Proximal Tubule NHE3 Activity, Preserves GFR, and Restores Euvolemia in Nondiabetic Rats with Induced Heart Failure. J Am Soc Nephrol. 2021;32(7):1616–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbafati C, Abbas KM, Abbasi-Kangevari M, et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2020;396(10258):1204–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King EA, Davis JW, Degner JF. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 2019;15(12):e1008489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buniello A, Macarthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wuttke M, Li Y, Li M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet. 2019;51(6):957–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellwege JN, Velez Edwards DR, Giri A, et al. Mapping eGFR loci to the renal transcriptome and phenome in the VA Million Veteran Program. Nat Commun. 2019;10(1):3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers JC, Zhang W, Lord GM, et al. Genetic loci influencing kidney function and chronic kidney disease. Nat Genet. 2010;42(5):373–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanzick KJ, Li Y, Schlosser P, et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nat Commun. 2021;12(1):4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paternoster L, Tilling K, Davey Smith G. Genetic epidemiology and Mendelian randomization for informing disease therapeutics: Conceptual and methodological challenges. PLOS Genet. 2017;13(10):e1006944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorski M, Tin A, Garnaas M, et al. Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int. 2015;87(5):1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox CS, Yang Q, Cupples LA, et al. Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: the Framingham Heart Study. J Am Soc Nephrol. 2004;15(9):2457–2461. [DOI] [PubMed] [Google Scholar]
- 22.Gorski M, Jung B, Li Y, et al. Meta-analysis uncovers genome-wide significant variants for rapid kidney function decline. Kidney Int. 2021;99(4):926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacCallum RC, Zhang S, Preacher KJ, Rucker DD. On the practice of dichotomization of quantitative variables. Psychol Methods. 2002;7(1). [DOI] [PubMed] [Google Scholar]
- 24.Tang W, Kowgier M, Loth DW, et al. Large-scale genome-wide association studies and meta-analyses of longitudinal change in adult lung function. PLoS One. 2014;9(7):e100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C-T, Merino J, Rybin D, et al. Genome-wide Association Study of Change in Fasting Glucose over time in 13,807 non-diabetic European Ancestry Individuals. Sci Rep. 2019;9(1):9439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouveia MH, Bentley AR, Leonard H, et al. Trans-ethnic meta-analysis identifies new loci associated with longitudinal blood pressure traits. Sci Rep. 2021;11(1):4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vickers AJ, Altman DG. Statistics notes: Analysing controlled trials with baseline and follow up measurements. BMJ. 2001;323(7321):1123–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aschard H, Vilhjálmsson BJ, Joshi AD, Price AL, Kraft. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am J Hum Genet. 2015;96(2):329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winkler TW, Günther F, Höllerer S, et al. A joint view on genetic variants for adiposity differentiates subtypes with distinct metabolic implications. Nat Commun. 2018;9(1):1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day FR, Loh P-R, Scott RA, Ong KK, Perry JRB. A Robust Example of Collider Bias in a Genetic Association Study. Am J Hum Genet. 2016;98(2):392–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köttgen A, Pattaro C. The CKDGen Consortium: ten years of insights into the genetic basis of kidney function. Kidney Int. 2020;97(2):236–242. [DOI] [PubMed] [Google Scholar]
- 33.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The 1000 Genomes Project Consortium. An integrated map of genetic variation Supplementary Material. Nature. 2012;135:1–113. [Google Scholar]
- 35.McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glymour MM, Weuve J, Berkman LF, Kawachi I, Robins JM. When is baseline adjustment useful in analyses of change? An example with education and cognitive change. Am J Epidemiol. 2005;162(3):267–278. [DOI] [PubMed] [Google Scholar]
- 37.Yanez ND, Kronmal RA, Shemanski LR. The effects of measurement error in response variables and tests of association of explanatory variables in change models. Stat Med. 1998;17(22):2597–2606. [DOI] [PubMed] [Google Scholar]
- 38.Parsa A, Kao WHL, Xie D, et al. APOL1 Risk Variants, Race, and Progression of Chronic Kidney Disease. N Engl J Med. 2013;369(23):2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gillies CE, Putler R, Menon R, et al. An eQTL Landscape of Kidney Tissue in Human Nephrotic Syndrome. Am J Hum Genet. 2018;103(2):232–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davey Smith G, Paternoster L, Relton C. When Will Mendelian Randomization Become Relevant for Clinical Practice and Public Health? JAMA. 2017;317(6):589–591. [DOI] [PubMed] [Google Scholar]
- 41.Peto R. The horse-racing effect. Lancet (London, England). 1981;2(8244):467–468. [DOI] [PubMed] [Google Scholar]
- 42.Parsa A, Kanetsky PA, Xiao R, et al. Genome-wide association of CKD progression: The chronic renal insufficiency cohort study. J Am Soc Nephrol. 2017;28(3):923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pattaro C, Köttgen A, Teumer A, et al. Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet. 2012;8(3):e1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carroll RJ, Ruppert D, Stefanski LA, Crainiceanu CM. Measurement Error in Nonlinear Models: A Modern Perspective. Vol 2. 2nd ed.; 2006. [Google Scholar]
- 45.Vansteelandt S, Goetgeluk S, Lutz S, et al. On the adjustment for covariates in genetic association analysis: A novel, simple principle to infer direct causal effects. Genet Epidemiol. 2009;33(5):394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeo NC, O’Meara CC, Bonomo JA, et al. Shroom3 contributes to the maintenance of the glomerular filtration barrier integrity. Genome Res. 2015;25(1):57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khalili H, Sull A, Sarin S, et al. Developmental Origins for Kidney Disease Due to Shroom3 Deficiency. J Am Soc Nephrol. 2016;27(10):2965–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuura R, Hiraishi A, Holzman LB, et al. SHROOM3, the gene associated with chronic kidney disease, affects the podocyte structure. Sci Rep. 2020;10(1):21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Note S1. Equivalence of DM-adjusted versus not DM-adjusted GWAS on eGFR-decline in the validation meta-analysis
Note S2. Formula-based covariate adjustment using GWAS summary statistics
Note S3. Validation of the formula-derived association for eGFR-decline adjusted for eGFR-baseline
Note S4. Graphical illustration of the relationship between SNP-effects on eGFR-decline unadjusted and adjusted for eGFR-baseline
Note S5. Comparison of the signals for eGFR-decline unadjusted and adjusted for eGFR-baseline and cross-sectional eGFR for the 11 identified loci
Note S6. Age-dependency of SNP-effects and main age effect on eGFR
Note S7. Narrow-sense heritability
Figure S1. Meta-analysis workflow
Figure S2. Study-specific median annual eGFR-decline versus sample size, follow-up time and median age
Figure S3. Influence of alternative adjustments for age on eGFR-decline in UK Biobank
Figure S4A. No influence from adjusting SNP-associations for eGFR-decline for diabetes mellitus (DM)
Figure S4B. Differences between SNP-association for eGFR-decline unadjusted versus adjusted for eGFR-baseline
Figure S4C. Validation of formula-derived adjustment for eGFR-baseline in eGFR-decline associations (part 1).
Figure S4D. Validation of formula-derived adjustment for eGFR-baseline in eGFR-decline associations (part 2)
Figure S5. Region plots of loci identified for eGFR-decline unadjusted and adjusted for eGFR-baseline
Figure S6. Age-dependency of eGFR and age-dependency of the variant effects on eGFR in UK Biobank
Table S1. Description of participating studies: study design
Table S2. Description of participating studies: genotyping and imputation
Table S3. Description of participating studies: phenotype distribution
Table S4. The 12 identified variants for eGFR-decline were associated with other kidney phenotypes, but not with DM-status
Table S5. The 12 identified variants for eGFR-decline do not show heterogeneity between ancestries and FHS is not an influential study
Table S6. No influence by DM-adjustment versus no DM-adjustment or by model-based versus formula-based adjusting for baseline eGFR (BL) on the 12 variants’ association with eGFR-decline
Table S7. Association of APOL1 risk variants in African American and European CKDGen studies
Extended acknowledgements, study funding information and author contributions
Supplementary References
Author contributions
Data Availability Statement
To support future work, we provide genome-wide summary statistics on eGFR-decline unadjusted for eGFR-baseline (adjusted for age, sex and DM-status) overall and restricted to individuals with DM or CKD at baseline (all adjusted for age and sex) (https://www.uniregensburg.de/decline and http://ckdgen.imbi.uni-freiburg.de). The summary statistics on eGFR-decline in individuals with CKD at baseline can be considered genetic effects on CKD-progression. We also provide genome-wide summary statistics on eGFR-decline adjusted for eGFR-baseline (additionally to adjustment for age and sex), but these summary statistics should be used with great care and an understanding that beta-estimates are subject to collider bias. For quantification of the genetic effect on eGFR-decline, the results unadjusted for eGFR-baseline should be utilized.