SUMMARY
Talaromycosis is an invasive mycosis endemic in tropical and subtropical Asia and is caused by the pathogenic fungus Talaromyces marneffei. Approximately 17,300 cases of T. marneffei infection are diagnosed annually, and the reported mortality rate is extremely high (~1/3). Despite the devastating impact of talaromycosis on immunocompromised individuals, particularly HIV-positive persons, and the increase in reported occurrences in HIV-uninfected persons, diagnostic and therapeutic approaches for talaromycosis have received far too little attention worldwide. In 2021, scientists living in countries where talaromycosis is endemic raised a global demand for it to be recognized as a neglected tropical disease. Therefore, T. marneffei and the infectious disease induced by this fungus must be treated with concern. T. marneffei is a thermally dimorphic saprophytic fungus with a complicated mycological growth process that may produce various cell types in its life cycle, including conidia, hyphae, and yeast, all of which are associated with its pathogenicity. However, understanding of the pathogenic mechanism of T. marneffei has been limited until recently. To achieve a holistic view of T. marneffei and talaromycosis, the current knowledge about talaromycosis and research breakthroughs regarding T. marneffei growth biology are discussed in this review, along with the interaction of the fungus with environmental stimuli and the host immune response to fungal infection. Importantly, the future research directions required for understanding this serious infection and its causative pathogenic fungus are also emphasized to identify solutions that will alleviate the suffering of susceptible individuals worldwide.
KEYWORDS: Talaromyces marneffei, talaromycosis, growth biology, host immune response, infectious disease, pathogenic mechanism
INTRODUCTION
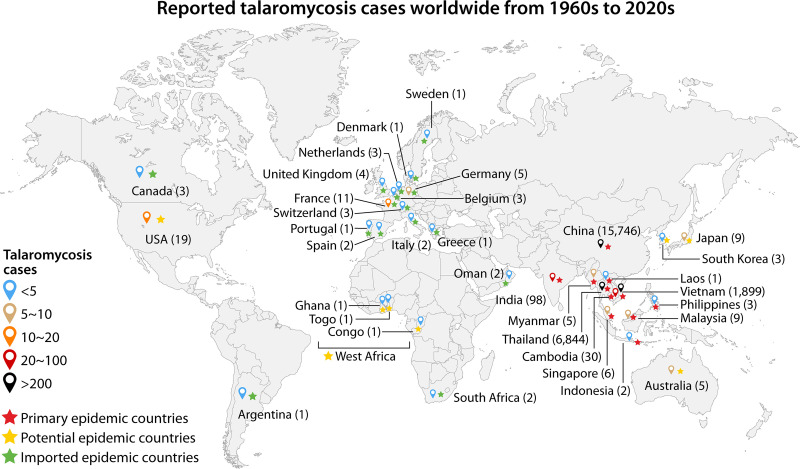
Talaromyces marneffei is a saprophytic fungus with complex mycological growth; it can form multiple cell types, including conidia, hyphae, and yeast, throughout its life cycle. T. marneffei infection causes talaromycosis, an endemic invasive mycosis found primarily in tropical and subtropical Southeast Asia. Travelers who have visited areas of endemicity are also potentially vulnerable (1–6) (Fig. 1). T. marneffei infection was first observed in bamboo rats in 1956 (7, 8), and the first reported case of natural human infection occurred in 1973 (9); in the following 15 years, cases remained sporadic in Thailand, Hong Kong, and southern China (10). However, the number of reported infections subsequently increased with the emergence of the HIV pandemic. Since the 1990s, talaromycosis has become one of the most common illnesses among persons living with HIV in areas of endemicity (11–13). By mid-2022, over 288,000 talaromycosis cases had been reported in 34 countries (Fig. 1), accounting for a pooled prevalence of 3.6% among people living with HIV (6, 14, 15). Along with advancements in cancer therapy and organ transplantation (10), an increased number of T. marneffei infections have also emerged in HIV-uninfected patients, putting people without known immune disorders at risk for talaromycosis. Notably, although T. marneffei was ranked second among the world’s 10 most feared fungi in 2018 (16), talaromycosis diagnostic and treatment modalities have received insufficient worldwide attention. To promote research and development for talaromycosis, scientists from nations around the world where it is endemic issued a plea in 2021 (17) to classify talaromycosis as a neglected tropical disease.
FIG 1.
Talaromycosis cases reported in 34 countries worldwide by the 2020s. The color of the teardrop shape represents the range of T. marneffei infection case numbers; blue, brown, coral, red, and black indicate <5, 5 to 10, 10 to 20, 20 to 100, and >100 infections, respectively. Red, orange, and green stars represent the primary countries where talaromycosis is epidemic, the countries where talaromycosis has the potential to become epidemic, and the countries with epidemic talaromycosis has been imported, respectively. The numbers in parentheses indicate the diagnosed talaromycosis cases in each country until mid-2022. Adapted using the world map from BioRender.com.
AIDS has been linked to numerous fungal diseases, including talaromycosis, candidiasis, cryptococcosis, pneumocystosis, histoplasmosis, and aspergillosis (18). High rates of comorbidities associated with COVID-19 among persons living with HIV have been reported (19), and individuals with severe viral respiratory tract infections, such as COVID-19, were shown to have invasive fungal coinfections, putting a larger population at risk for fungal infections (20, 21), especially aspergillosis and mucormycosis (20). Since the global devastation caused by COVID-19, a great deal of attention and financing have been devoted to COVID-19-related topics, presumably reducing the funding available for fungal infection research. Simultaneously, as the prevalence of COVID-19 increased worldwide, insufficient diagnostic and control programs for fungal infections were performed in clinics, resulting in a double-negative effect on coinfected patients (22). Therefore, how the emerging COVID-19 challenge impacted patients with talaromycosis in countries where it is endemic, especially in poor regions, should be considered, and a series of cohort studies will likely be required after the pandemic.
The talaromycosis disease-causing pathogen, Penicillium marneffei (later renamed Talaromyces marneffei; see below), is the only dimorphic Penicillium species capable of causing systemic mycosis in humans (11). Other members of the Penicillium genus are rarely reported to infect humans or other animals, although a few cases related to special working environments or conditions have been reported recently, including infections with Penicillium chrysogenum (23), Penicillium roqueforti (24), Penicillium cluniae (25), Penicillium digitatum (26), Penicillium notatum (27), and Penicillium stipitatus (28). T. marneffei infection was first discovered over 60 years ago (5, 7, 8); however, only ~920 published articles about T. marneffei were indexed in PubMed as of mid-2022. This low number reflects the relative lack of research on T. marneffei compared to other pathogenic fungi (29): there were ~13,300 articles on Aspergillus fumigatus, ~ 4,430 articles on Histoplasma capsulatum, and ~2,430 articles on Coccidioides immitis/Coccidioides posadasii.
As new methodologies such as molecular genetic technologies and high-throughput profiling methods have been applied to understand the molecular genetics of T. marneffei (30), it is necessary to summarize the recent advances in T. marneffei biology and pathogenicity research and the understanding of talaromycosis. This review discusses the current knowledge on T. marneffei infection in clinical practice, the drugs used in patients, and newly characterized potential antifungal agents. Recent advances in molecular genetics regarding dimorphism regulation, factors affecting T. marneffei infection in response to the extracellular and intracellular milieu, and the interplay between the host immune response and this pathogenic fungus are also discussed. This inclusive review of T. marneffei research will allow us to comprehensively understand the life-threatening severity of talaromycosis, the morphology and pathogenic mechanism regulation of T. marneffei, and the host defense response, revealing a new range of therapeutic targets to combat these life-threatening infections, ultimately alleviating the impact on patients and susceptible populations worldwide.
TALAROMYCOSIS
Severity and Epidemiology of Talaromycosis
The mortality risk associated with talaromycosis can reach 100% if the pathogen is left uncontrolled in HIV-infected patients (31). However, if those individuals are treated promptly with the necessary antifungals, the risk of death is greatly reduced. Multiple retrospective cohort studies and meta-analyses have been performed in recent decades in countries where talaromycosis is endemic to systematically understand the overall severity of talaromycosis. Talaromycosis is one of the most common health problems among HIV-positive persons, and it is estimated that talaromycosis will threaten the lives of 23,117 individuals by the year 2050 (32). In China (including Hong Kong), Vietnam, Thailand, and India, the prevalence of talaromycosis in individuals with HIV ranges from 0.1% to 26.7%, with mortality rates ranging from 8% to 40% (14). Approximately 99% of T. marneffei infections in China were reported in the southern regions: 43% in Guangxi Province and 41% in Guangdong Province. Of these, 88% were found in persons living with HIV (33) (Fig. 2). A total of 16.1% of HIV-positive persons in Guangxi were coinfected with T. marneffei, and these individuals had a substantially higher mortality rate (25.0%) than those not infected with T. marneffei (13.8%) (34). The proportion of persons with HIV infected with T. marneffei in Guangdong was 9.5% to 18.8% (35, 36), and the mortality rate was 14.0% to 25.09% (14, 37). In Vietnam, the proportion of HIV-positive persons infected with T. marneffei was reported to be 4.9% to 8.2%, and the mortality rate was 28.6% to 33% (38–40). The proportion of talaromycosis patients living with HIV in Thailand was as high as 12.26%, and the mortality rate was reported to be 20.7%. These reports delineate that T. marneffei infection is a nonnegligible issue that threatens populations in countries where talaromycosis is endemic and exhibits geographical variations (14) (Fig. 1).
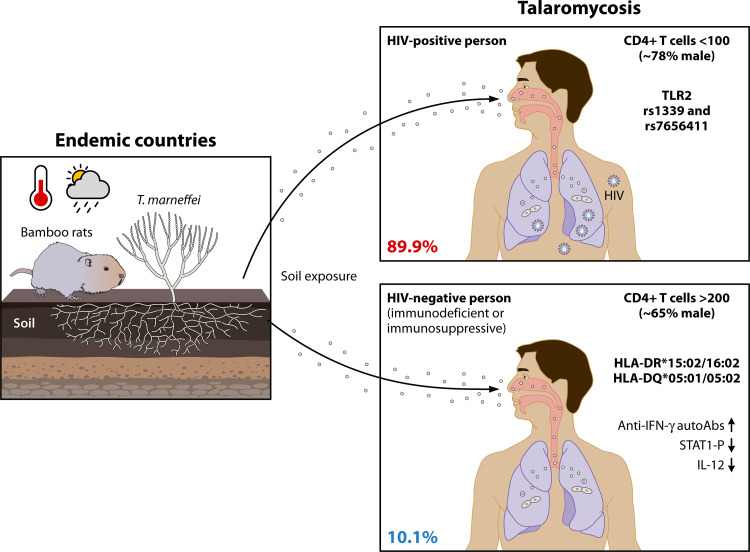
FIG 2.
Talaromycosis in susceptible populations. T. marneffei conidia are recognized as infectious propagules, the transmission of which is reported to be affected by soil exposure and temperature and precipitation parameters (left panel). The proportions of HIV-positive (~89.9%) and HIV-negative (~10.1%) persons infected with the pathogenic fungus T. marneffei are illustrated. Talaromycosis is no longer a disease associated solely with AIDS, although it has a high distribution level in HIV-positive persons (~89.9%). The CD4+ T-cell count in patients with HIV is generally less than 100 cells/μL, whereas the cell count in vulnerable HIV-negative individuals is normal, with values greater than 200 cells/μL. Genotype TLR2 rs1339 and rs7656411 polymorphisms in patients living with HIV are more frequently associated with talaromycosis. Primary immunodeficient HIV-negative populations in Southeast Asia with HLA-DR*15:02/16:02 and HLA-DQ*05:01/05:02 are predisposed to T. marneffei infection, as are individuals with secondary immunosuppressive deficiency.
Areas where talaromycosis is endemic have expanded from southern China, Southeast Asian countries, and northeast India, the traditional primary regions of endemicity, to 34 countries worldwide (6) (Fig. 1). More areas where talaromycosis has the potential to become epidemic, including parts of northern China, eastern India, the Korean Peninsula, Japan, eastern Australia, the central part of the United States, and western Africa (Fig. 1), have been identified from the Maxent ecology model analysis (41). The Maxent model is based on different talaromycosis distribution-driving variables, such as the distribution of Rhizomys (T. marneffei reservoirs; see next section), the HIV/AIDS epidemic, temperature, and precipitation, among which Rhizomys distribution is the key driving factor (41). In addition to those potential risk areas, along with the global travel, other individuals from different countries (i.e., imported epidemic countries shown in Fig. 1) who have visited these areas of endemicity have been diagnosed with T. marneffei infection (6), broadening our understanding of talaromycosis epidemiology and highlighting the need to be aware of the increased risk of talaromycosis worldwide.
In addition to increasing T. marneffei infection rates among individuals with HIV, more HIV-uninfected but immunocompromised patients (10.1%) have been infected since the mid-1990s (6, 10) (Fig. 2). This situation is associated with the expanded utilization of potent immunosuppressive drugs in transplant recipients and autoimmune disease patients. Primary immunodeficient patients with anti-interferon gamma (anti-IFN-γ) autoantibodies (auto-Abs) and secondary immunosuppressed patients (e.g., hematological transplant recipients) who received novel therapies such as anti-CD20 monoclonal antibodies (MAbs) or kinase inhibitors showed a higher incidence of talaromycosis (42, 43). Anti-IFN-γ auto-Ab-induced immunodeficiency was first described in Thailand and the Philippines in 2004 (44, 45), although such immunodeficiency has also been associated with a predisposition to other microorganism infections, such as melioidosis and salmonellosis (46). Patients with higher-titer serum neutralizing anti-IFN-γ auto-Abs that inhibit STAT1 phosphorylation and interleukin 12 (IL-12) production had a severely impaired immune response (47). Moreover, it was later discovered that Asian ethnicity was linked to T. marneffei infection and anti-IFN-γ auto-Ab immunodeficiency (42). The presence of anti-IFN-γ auto-Abs is significantly linked to the HLA-DR*15:02/16:02 and HLA-DQ*05:01/05:02 haplotypes in populations in Southeast Asia (48, 49) (Fig. 2). This association was recently noted to be especially strong in Chinese populations (50), indicating that non-HIV-infected populations with this HLA class haplotype in regions of endemicity may be at higher risk for T. marneffei infection. In addition, improved and novel immunomodulation methods in organ transplantation, particularly renal transplantation (10), and hematological malignancy, anti-IFN-γ auto-Ab and anticancer targeted treatment, such as the application of rituximab (51) and obinutuzumab (52) as anti-CD20 MAbs, were associated with increasing talaromycosis complications (43). Therefore, the dosing regimen and time intervals in immune modulation therapy should be addressed and evaluated in clinical practice, especially in countries where talaromycosis is endemic.
Moreover, with advancements in clinical immunological diagnostics, children with primary immunodeficiency syndromes have also been documented to be vulnerable to T. marneffei infection (10, 53, 54). Approximately 50 pediatric patient cases from Asia have been recorded to date. Immunodeficiency, such as CD40L deficiency, autosomal dominant (AD) hyper-immunoglobulin E (hyper-IgE) syndrome, IL-12/IFN-γ axis deficiency, and other unknown specific immune defects, is increasingly being associated with pediatric talaromycosis in HIV-uninfected patients (54). A substantially higher mortality rate was discovered among HIV-uninfected individuals due to the disease severity and poor outcomes, especially in those presenting with few typical signs and symptoms. China has reported death rates of over 32% among HIV-negative patients. The fatality rate of HIV-uninfected children is much higher, reaching 55% in some cases (53). The mortality rate of extremely advanced HIV-infected patients appears much greater in low-income countries than in industrialized countries (55), consistent with the theory that delayed diagnosis or misdiagnosis may contribute to the elevated mortality rate of HIV-uninfected patients. Tuberculosis is the most common misdiagnosis; 80% of talaromycosis cases are misdiagnosed as tuberculosis (56). Therefore, greater caution should be exercised in diagnosing such infectious diseases; rapid and accurate diagnostic approaches and timely pharmacological interventions are urgently needed.
Reservoirs and Transmission of T. marneffei
Bamboo rats and humans are the most prevalent animal hosts of T. marneffei, although this fungus has also been identified in canines (57–59). However, the route of transmission of pathogenic T. marneffei to humans remains unclear. Human inhalation of airborne conidia from an environmental source, such as soil, followed by distribution to other body sites is a commonly held belief concerning T. marneffei transmission (60). Multiple lines of evidence suggest a quick progression from infection to dissemination in patients. The clinical manifestations of a disseminated infection might arise in just a few weeks (61, 62), although there have been exceptions in patients with protracted latent infections (63, 64). T. marneffei was found in 43 captured adult Rhizomys pruinosus rats in Guangxi, China, with a 100% detection rate, although no obvious pathological alterations were found in any of the rat organs (65). In addition, no isolates were discovered in the fetuses of pregnant rats, suggesting that no vertical transmission occurs, at least among rats (65). Enzootic infection in bamboo rats occurred in the same areas as endemic talaromycosis in humans, and rat species known to be infected include Rhizomys sinensis, R. pruinosus, Rhizomys sumatrensis, and Cannomys badius (66–69), suggesting that humans are infected by rodents or both are infected through their common environment.
The pathogen transmission origin of T. marneffei infection is thought to be the soil in which rodents live, and rodents are natural reservoirs of the fungus (Fig. 2). This assumption is supported by genome characterization studies that revealed that the genotype of T. marneffei from both bamboo rats and humans corresponds to geographical regions (65, 70). However, the lack of success in the endeavor to isolate strains from ambient soil samples, such as bamboo rat burrows and patient residential areas (57), remains puzzling. T. marneffei has been isolated from only two soil samples out of 29 bamboo rat burrows (57, 71) to date. Additionally, a multilocus microsatellite typing (MLMT) analysis of 10 bamboo rat isolates collected from Manipur, India, showed that all 21 microsatellite loci were identical to that of the human T. marneffei isolate CBS 101038, implying that host-to-host transmission is possible (72). Although it is still not able to discriminate among sources of human contamination, advanced multilocus sequence typing (MLST) analysis demonstrated that 43 rodent isolates and 40 human isolates were comparable. It was also revealed that in sensitive populations where humans more frequently come into contact with bamboo rats, no increase in infection rate was detected (67), which is consistent with the finding that bamboo rat exposure or consumption history is not a risk factor for T. marneffei infection (17). However, soil exposure, especially in rainy seasons, is pivotal for T. marneffei infection (73, 74). In addition, one interesting study recently conducted in the Guangdong Province of China suggested that temperature is an environmental factor for talaromycosis in persons with HIV (75) (Fig. 2). Importantly, a high percentage of populations in regions of endemicity are in low-income areas; for example, over half of the populations in Vietnam and Guangxi, China, live in the countryside (17). Farmers are at a much greater risk of developing talaromycosis (i.e., 70 to 90%) than nonfarmers (17, 73), highlighting the severity of talaromycosis among impoverished people.
In addition to spontaneous talaromycosis cases, laboratory-acquired T. marneffei infections have been reported. Thus, talaromycosis has been identified as a cause of laboratory-acquired infection. The first documented human infection case (8), detected before the first natural infection case report in 1973 (9), was a laboratory infection produced by direct inoculation of the fungus into the skin in 1959. Since then, a physician who was unknowingly HIV positive was diagnosed with another laboratory infection in 1994 after visiting a laboratory and occasionally being exposed to a T. marneffei culture (76). These incidents not only support the widely held belief that conidium inhalation is the pathogen’s primary mode of transmission but also emphasize the importance of biosafety in laboratory work with pathogenic T. marneffei.
Clinical Manifestations of Talaromycosis
Talaromycosis demonstrates typical clinical manifestations in patients with HIV, and similar features have been reported in different case series (12, 60, 77) (Table 1). Comparative clinical characteristics of cases in different countries where talaromycosis is endemic over time are analyzed and summarized here (Table 1), including the Thai patients reviewed in the 1990s (78, 79). For example, fever (97%), weight loss (100%), weakness (86%), anemia (86%), and distinctive skin lesions were found among the 36 patients studied between 1998 and 1999 at J. N. Medical Hospital in Manipur, India (81%) (72). In the clinical reports collected from 47 case series from 1994 to 2004 in Hong Kong with a mean patient age of 43 years, fever (96%), anemia (79%), and lymphadenopathy (62%) were the most common clinical manifestations (80). Furthermore, common clinical features with slight differences were revealed in HIV-positive persons with talaromycosis (~78% male) in Vietnam (81), Thailand (77), and China (36, 82, 83) from 2005 until 2018. Typical skin lesions, fever, and weight loss syndromes were observed in the majority of clinical HIV-positive persons. Anemia and diarrhea were also common in approximately half of the infected populations, as were hepatomegaly, lymphadenopathy, splenomegaly, and cough (Table 1). T. marneffei infection has been classified as one of the leading AIDS-defining illnesses in various regions, such as Thailand and Hong Kong, China (11, 80), and talaromycosis is anticipated to become an indicator of AIDS in Southeast Asia.
TABLE 1.
Comparison of the clinical manifestations of talaromycosis in HIV-positive and HIV-negative patients
| Parameter | Data by group and reference no. |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV positive (78% male) |
HIV negative (65% male) |
|||||||||||
| 77 | 60 | 12 | 63 | 72 | 36 | 82 | 83 | 84 | 77 | 10 | 56 | |
| No. of cases | 116 | 160 | 155 | 21 | 36 | 1,079 | 343 | 87 | 15 | 34 | 119 | 162 |
| % males | 68.10 | ~a | ~ | 90.40 | 91.40 | 79.80 | 70.80 | 93.00 | 80.00 | 50.00 | 55.00 | 74.00 |
| % with clinical manifestation | ||||||||||||
| Weight loss | ~ | ~ | 71.60 | 81.00 | 100.00 | 49.80 | 73.80 | ~ | 46.70 | ~ | 28.60 | 29.01 |
| Fever | 87.10 | 95.30 | 98.00 | 95.20 | 97.00 | 85.60 | 87.50 | 93.10 | 66.70 | 64.70 | 74.80 | 82.72 |
| Cutaneous lesions | 40.50 | 60.00 | 69.70 | 76.20 | 81.00 | 44.50 | 46.30 | 35.60 | 46.70 | 41.20 | 44.50 | 53.09 |
| Umbilicated lesions | 38.79 | ~ | ~ | ~ | ~ | ~ | ~ | ~ | 0.00 | 14.70 | ~ | ~ |
| Sweet’s syndrome | 0.00 | ~ | ~ | ~ | ~ | ~ | ~ | ~ | 40.00 | 14.70 | ~ | ~ |
| Cough | 27.60 | 44.50 | 49.70 | 52.40 | ~ | 68.00 | 43.40 | ~ | 80.00 | 23.50 | 42.00 | 85.19 |
| Anemia | 6.90 | 81.00 | 74.80 | 85.70 | 86.00 | 95.60 | ~ | ~ | 0.00 | 0.00 | 39.50 | 44.44 |
| Diarrhea | 12.90 | 22.67 | 23.20 | 19.00 | 22.00 | 44.30 | ~ | ~ | 0.00 | 5.90 | 12.60 | ~ |
| Hepatomegaly | 36.20 | 38.30 | 43.90 | 42.90 | 39.00 | 51.70 | 44.00 | ~ | 0.00 | 17.60 | 19.30 | 37.65 |
| Splenomegaly | 21.60 | 15.50 | 13.50 | 4.76 | ~ | 54.60 | 39.70 | ~ | 0.00 | 5.90 | 16.00 | 37.65 |
| Lymphadenopathy | 31.90 | 51.00 | 52.30 | 42.90 | 33.00 | ~ | 80.20 | 28.70 | 66.70 | 38.20 | 42.00 | 66.05 |
| Arthritis/osteolytic lesions | 0.00 | 0.00 | 7.80 | 0.00 | 0.00 | 46.70 | 0.00 | 0.00 | 46.70 | 14.70 | 34.40 | 24.69 |
~, no corresponding clinical record.
Importantly, depleted CD4+ T lymphocyte number is the most visible clinical characteristic in persons living with HIV. The median CD4+ T-cell counts of HIV-positive persons with talaromycosis are commonly less than 100 cells/μL (36, 77, 78, 80–83) (Fig. 2). In contrast to the depleted numbers in HIV-positive persons, the CD4+ cell count in HIV-negative talaromycosis patients is generally normal, at more than 200 cells/μL (Fig. 2), such as the recorded 640 cells/μL among the 34 patients in Thailand (77). These results are consistent with the premise that the immunological deficit in HIV-negative patients, unlike in those with HIV, is not due to CD4+ lymphocytopenia (77). Intermittent fever, skin lesions, and widespread lymphadenopathy have also been documented in HIV-uninfected patients, among whom 65% are male (Table 1), although the clinical manifestations are not identical. Regarding clinical characteristics, HIV-negative individuals were more likely to develop bone and joint infections, such as arthritis and osteomyelitis (10, 56, 77, 84) (Table 1). However, because comparable lesions have been reported for other systemic infectious diseases, such as cryptococcosis, blastomycosis, African histoplasmosis, and tuberculosis, it is difficult to utilize these syndromes as common criteria for clinical talaromycosis diagnosis. In addition to the nontypical clinical features, the low percentage of T. marneffei infections in HIV-negative patients frequently leads to significant infections being neglected or missed during clinical diagnosis (77), and the lack of attention paid to T. marneffei infection can result in high severity at diagnosis and poor treatment outcomes for T. marneffei-infected HIV-negative patients.
Clinical Pathology of Talaromycosis
Talaromycosis pathology differs among organs and is typically dependent on the host immunological state. AIDS patients with a diffuse infiltrate of lung-laden macrophages and lymphoid depletion frequently exhibit anergic and necrotizing tissue reactions. The liver, lungs, and skin, as well as lymph nodes, bone marrow, and intestines, are often affected (12, 60). Liver biopsy revealed equal proportions of three histological patterns of hepatic lesions, which are diffuse, granulomatous, and mixed types, in 30 HIV-infected patients with disseminated T. marneffei infection in northern Thailand from 1998 to 1999 (85). Abnormal liver function with increased albumin and aspartate aminotransferase (AST) levels and AST/alanine aminotransferase (ALT) ratios are commonly found in talaromycosis patients (86, 87). In the alveolar space of the lungs, aggregate macrophages with localized fibrinous exudate are observed. Typical skin lesions consisting of papules with core necrotic umbilication have historically been considered as a significant clinical diagnostic sign of T. marneffei infection in AIDS patients (88). Both granuloma formation and widespread pathology contribute to the pathogen’s ability to locate and infect HIV-positive persons.
Clinical Diagnosis of Talaromycosis
T. marneffei infection is potentially curable if a clinical diagnosis is made promptly and accurately during the early stages of infection. Most existing procedures for diagnosing T. marneffei infection are pathogen based, such as microscopy and laboratory cultures, serological antigen or antibody detection, and molecular PCR testing. A host-based detection approach based on immune response factors such as antibody testing and IFN-γ release level detection (17) was also developed. The majority of the T. marneffei-infected hosts evaluated by the former detection technique were AIDS patients with characteristic clinical symptoms. Therefore, this pathogen-based diagnostic technique can nearly completely cover clinical talaromycosis diagnosis in HIV-positive individuals, especially in areas of endemicity. However, several documented patients developed talaromycosis years after T. marneffei exposure (2, 89–91), suggesting that T. marneffei can remain latent inside the host and can be reactivated later (64), although there remains a lack of clinical evidence or animal experiments to show the existence of T. marneffei latent infection. The clinically reported latent talaromycosis cases include HIV-positive (2) and HIV-negative individuals, such as organ transplant immunodeficient patients (89, 91) and patients with pulmonary disease (90). All of these patients were found to have positive T. marneffei growth from lung or blood specimens months or years later after their travel to an area where talaromycosis was epidemic, suggesting that T. marneffei can evade the host immune system and persist intracellularly for long periods of time (2, 89–91). Therefore, finding a prediction or quick-detection tool based on the host response is extremely important.
Diagnosis based on pathogen detection.
The gold standard for clinical T. marneffei infection diagnosis is microscopic proof of the presence of the pathogen in tissues, the effective isolation of the fungus from patient specimens, or both. Several microscopy detection methods are available, including laboratory staining of histological structures or the detection of acquired cytology specimens. Grocott methenamine silver or periodic acid-Schiff staining of histopathological sections reveals characteristic round to oval T. marneffei yeast cells that divide by cross-wall formation inside macrophages. Wright’s staining of specimens obtained by fine-needle aspiration, such as lymph node specimens, bone marrow aspiration, touch smears of skin, or lymph node biopsy, shows clear basophilic, spherical, oval, and elliptical yeast cells with distinctively central septation formation characteristics that are diagnostic for T. marneffei (62, 79, 92). Notably, T. marneffei mycological culture is the definitive diagnostic method for pathogen identification in the clinic. This fungus has discriminative culture visualization characteristics due to its distinctive dimorphic growth. The mold-to-yeast form conversion is induced by incubating the growth on brain heart infusion agar at 37°C after growing the mycelia at 25°C on Sabouraud dextrose agar, where it shows a greenish yellow color and a characteristic red diffusible pigment (93). According to the data from different specimens, such as sputum (34%), blood (76%), skin biopsy (90%), bone marrow (100%), and lymph node biopsy (100%), the detection accuracy for laboratory culture can be as high as 100% (11). However, because of the slow growth of T. marneffei, mycological cultivation is classically time-consuming (i.e., at least 6 days for growth) and frequently falls short of the needs of clinical diagnosis, resulting in delayed clinical antifungal therapy. Therefore, more efficient diagnostic methods should be developed to support the auxiliary diagnosis of talaromycosis in the clinic.
A variety of antigen or antibody detection approaches have been used to diagnose T. marneffei infection, such as the immunohistochemical detection of the early-produced monoclonal antibody EB-A1 to galactomannan (GM) predominantly found in Aspergillus spp. and T. marneffei (94, 95). Although the GM assay displayed a cross-reactivity for other fungal species (e.g., Cryptococcus neoformans), a significantly higher GM assay optical density (OD) index was detected for talaromycosis (95), facilitating earlier diagnosis of T. marneffei infection. The β-d-glucan assay is a classically used serodiagnosis method for invasive fungal infections, such as aspergillosis and candidiasis (96). An elevated β-d-glucan serum level was also observed in 82% (9/11) of talaromycosis patients in Japan who had histories of travel to countries where talaromycosis is endemic (97), suggesting that it is a candidate clinical diagnosis method for those settings. Antiglobulins (e.g., rabbit antiglobulins) (98) and murine immunoglobulin M (IgM) monoclonal antibodies (99) were generated through T. marneffei yeast or mycelial cell immunological reactions in various hosts. Antigens from T. marneffei, such as immunogenic yeast proteins (200, 88, 54, and 50 kDa) (100), 38-kDa mycelial protein(s) (101), and cytoplasmic yeast proteins (61, 54, and 50 kDa) (102), can elicit human responses. These are some of the candidates for the development of specific antibodies to identify disseminated T. marneffei infection. When utilizing T. marneffei germinating conidia and yeast for the reaction test, it has been found that the induced immunoglobulin G (IgG) level in infected patients is greater than 1:160, while the level in noninfected patients is less than 1:40. Hence, the proposed approach for identifying IgG antibodies in T. marneffei-infected individuals was evaluated (103). Antibodies with beneficial properties and good prospects for clinical use, monoclonal antibody (MAb) 4D1 (104, 105) and anti-Mp1p (106) MAb, both with high sensitivity (86%) and specificity (100%), were developed in Thailand and Hong Kong, respectively. A rapid lateral-flow immunochromatographic test system (104) and immunochromatographic strip test approach (107) were further developed for talaromycosis diagnosis, with low limits of detection and cross-reactivity and high sensitivity and accuracy in urine specimens, providing a rapid but inexpensive diagnostic method for clinical use. Mp1p antigen testing using matched plasma and urine specimens has been shown to have the highest detection sensitivity, significantly better than blood mycological culture in terms of both sensitivity (88.8% versus 72.8% [P < 0.001, McNemar test]) and diagnosis time (6 h versus 6.6 ± 3.0 days) (106), and the detection results are superior to those of the GM assay, especially for patients with low CD4+ T-cell counts (<50 cells/μL) (108). It is noteworthy that anti-Mp1p MAb is being investigated as a quick diagnostic and screening tool for talaromycosis in a multicenter prospective research project (ClinicalTrials.gov no. NCT04033120) (109), and a commercial antibody version for clinical usage was introduced in China in 2019 (17). As a result, as more translational scientists and firms become involved and more potential diagnostic applications are becoming available, more nonculture-based diagnostics for talaromycosis development can be achieved, thus speeding up the clinical diagnosis process and reducing the damage caused by T. marneffei infection.
Furthermore, molecular approaches based on PCR methodology have been developed for quick diagnosis. The process involves the creation of oligonucleotide primers based on species-specific DNA sequences, such as the commonly used internally transcribed spacer of the 5.8S rRNA gene (ITS1-5.8S-ITS2) (110) and the 18S rRNA (111) and MP1 (112) genes. For pathogen detection, various amplification methods have been developed, including single PCR, nested PCR, one-tube seminested PCR, and rolling-circle amplification (17). However, because large clinical studies are required to determine the efficiency and sensitivity of pathogen detection and prediction, these expensive molecular assays have not yet been widely utilized. Nevertheless, due to the high sensitivity (range from 10% to 100%) and specificity (>95%) of molecular methods in rapid diagnostics, much focus has been placed on them recently (113). Furthermore, next-generation sequencing (NGS) has been developed and increasingly applied in clinical diagnosis, including for fungal infections (114). A total of 11 NGS-diagnosed T. marneffei infection cases have been reported, among which 7 cases were in unexpectedly HIV-negative individuals (114), providing a valuable exploration of the potential application of NGS in the rapid clinical diagnosis of talaromycosis, especially among HIV-uninfected individuals.
Diagnosis or prediction based on the host response.
Talaromycosis patients present with certain syndromes or cytokine secretion disorders, thus providing multiple clinical diagnostic criteria for T. marneffei infection. Stepwise flow cytometric evaluation was used to suggest a pipeline for diagnosing T. marneffei infection in HIV-uninfected pediatric patients based on genetic defect traits or primary immunodeficiency syndromes in the IFN-γ/STAT1 signaling pathway (54). Patterns of high IgM levels but low IgG and immunoglobulin (IgA) levels or very high IgE levels may suggest CD40L deficiency or STAT3 gene mutation. Gain-of-function STAT1 disease is diagnosed by STAT1 hyperphosphorylation in response to IFN-α or IFN-γ and delayed dephosphorylation, whereas IFN-γ receptor deficiency is suggested by STAT1 phosphorylation in response to IFN-γ but a normal response to IFN-α (54). The level of serum anti-IFN-γ auto-Abs is a major detection criterion in adults. Accordingly, the diagnostic pipeline offers a simple and quick diagnostic algorithm for HIV-negative patients and can potentially be employed in the clinic, speeding up the clinical diagnosis of talaromycosis in HIV-uninfected patients.
Recently, a new T. marneffei infection prediction model was developed. In a large-scale cohort study, machine learning was used to produce accurate risk and clinical outcome assessments based on an association analysis between T. marneffei infection and the clinical features presented by HIV-infected patients. Skin lesions, the AST level, the ALT ratio index (AARI), peripheral or abdominal lymphadenopathy (POAL), and CD4+ T-cell number are all good candidate classifiers according to different prediction models. For the early diagnosis of T. marneffei infection, the existing random forest (RF) model combining the patient AST level and the AARI has high classification power (82). The use of prediction models shows that frequently collected data in clinical practice will be usable in the future for accurate and quick diagnostic prediction.
Clinical Treatment of Talaromycosis
Antifungal drugs utilized in clinics.
Amphotericin B (AmB) is the first-line antifungal medicine used for severe talaromycosis (115), followed by weeks to months of azoles such as itraconazole and voriconazole (115) and posaconazole (116). For AIDS patients, international recommendations include intravenous AmB at 0.6 to 0.7 mg/kg of body weight or, where available, 3 to 5 mg/kg of liposomal AmB daily for 2 weeks (115). However, the high cost and difficulties in gaining access to AmB for many Asian patients and the limited supply of the lower-toxicity liposomal AmB formulation have restricted its use in clinical regimens and exacerbated the severity of T. marneffei infection in poor countries where talaromycosis is endemic. Recent clinical investigations revealed that AmB was not superior to itraconazole in terms of mortality or fungicidal activity in the initial clinical treatment (33, 38, 72, 74, 81, 117), although some analyses demonstrated that itraconazole alone is less effective than AmB (118). In addition, there were few differences in medication efficacy between voriconazole and AmB (119), showing that the two are similar as clinical therapy options. Therefore, itraconazole has become a popular choice for treating talaromycosis in countries where talaromycosis is endemic, such as China, Vietnam, and India (120). Since AmB is costly and can cause serious side effects in patients, including renal failure, electrolyte abnormalities, vasculitis, and bone marrow suppression (121), azoles are administered with close clinical monitoring when AmB is unavailable.
Due to the current limitations of fungicides such as azoles and AmB, alternative antifungal targets have been found for potential use in treating T. marneffei infection. These expanded pop genes, for example, encode aspartyl proteases and have been linked to T. marneffei intracellular proliferation after infection (122). The pop genes are orthologs of pep genes, which are associated with the virulence of Candida species (123) and A. fumigatus (124). Notably, clinical antiretroviral medications that target HIV aspartyl protease have an inhibitory effect on Candida albicans (125), suggesting that antiretroviral drugs could potentially be utilized to suppress T. marneffei growth in the host. Furthermore, the growth inhibitory effect of galactose on clinical T. marneffei isolates was discovered by the Yuen group (126). Unlike the usual morphology of elongated hyphae following 1% glucose cultivation at 37°C, the injected conidia cultivated at 1% galactose only barely germinated into ballooning forms within 72 h. Because galactose metabolites are harmful, elucidating the metabolic process could lead to the creation of avirulent T. marneffei mutants and hence to the development of new vaccines.
Many strategies for screening antifungal agents against T. marneffei have been implemented recently (127–131), and such screens have identified the novel fungicidal drug olorofim, which is in the phase IIb clinical trial stage for fungal infection treatment (129), and the traditional Chinese medicine osthole, which exhibits antifungal activity against the yeast form of T. marneffei (130). The screening of antifungal agents against T. marneffei from marine-derived actinomycete extracts revealed one hexane extract, AMA50CH, with effective fungal killing activity in vitro and survival-prolonging activity in a Caenorhabditis elegans infection model (128). In addition, purified proteins derived from the Thai medicinal plants Andrographis paniculata and Rhinacanthus nasutus also exhibited substantial fungal killing capability against T. marneffei (127, 131). These findings not only indicate the potential of these candidates in clinical application but also suggest that additional agents in nature should be identified or tested in talaromycosis therapy in the future.
Clinical drug resistance.
Recurrence or relapse frequently occurs in T. marneffei-infected patients (132) after clinical treatment. Therefore, consolidation treatment and long-term secondary prophylaxis with oral itraconazole are routinely required to prevent the recurrence of this illness after the induction therapy of talaromycosis is completed successfully (133). Since prophylaxis and therapy might continue anywhere from a few weeks to months, the long-term exposure of fungal isolates to azole drugs provides opportunities for the development of drug resistance (134).
Systematic drug susceptibility testing of T. marneffei isolates demonstrated high sensitivity of all isolates to the antifungal drugs AmB, itraconazole, voriconazole, posaconazole, ketoconazole, flucytosine, and terbinafine, while the minimal inhibitory concentration (MIC) values of the drugs fluconazole, anidulafungin, micafungin and caspofungin against T. marneffei were relatively high and exhibited with heterogeneity among isolates (129, 135–137). The drug susceptibility of T. marneffei is consistent with the classical clinically utilized drugs prescribed to talaromycosis patients, and fortunately, no clinical drug-resistant T. marneffei isolates have been discovered yet. In case of future treatment failure, efforts have been made to better understand T. marneffei drug resistance pathways. PmMDR1 and PmMDR3, which code for major facilitator superfamily (MFS) transporters, have been identified as multidrug efflux pumps in T. marneffei and are involved in drug resistance to azoles (e.g., fluconazole and posaconazole), pyrimidine analogs (e.g., flucytosine), and antimalarials (138). Therefore, T. marneffei appears to have the potential to develop drug resistance, similar to other pathogenic fungi (139, 140). More efforts are needed to understand the mechanism of T. marneffei drug resistance and to identify targets to reduce its prevalence.
T. marneffei Coinfection Cases
It is worth noting that coinfections with T. marneffei can be detected in HIV-positive persons at a high frequency, especially coinfection with candidiasis (42%) (75), Pneumocystis pneumonia (19.9%) (75), tuberculosis (15.3%) (75), cytomegalovirus (17.3%) (36), salmonellosis (3.5%) (74), cutaneous herpes (4.6%) (74), and others (74). Tuberculosis, cryptococcosis, and talaromycosis are the top three opportunistic illnesses in persons with HIV, and they all have lethal consequences in these vulnerable populations (11). For example, Mycobacterium intracellulare has been detected in an HIV-positive individual along with T. marneffei (141). Moreover, patients infected with multiple fungal pathogens have been recorded, such as eight HIV-positive patients who were thoroughly investigated and found to have coinfection with T. marneffei and C. neoformans (142). The different pharmacological sensitivities of T. marneffei and C. neoformans to fluconazole and itraconazole have created a serious therapeutic issue in clinical settings, particularly in resource-limited areas (142).
Mixed infections are also commonly reported for HIV-uninfected patients with talaromycosis, such as coinfection with nontuberculous mycobacteria (NTM) (143), Mycobacterium tuberculosis (144), and C. neoformans (142, 145). Both T. marneffei and NTM monoinfections have recently become more common in HIV-negative patients (10, 146, 147). However, these pathogens share common clinical manifestations in patients after infection, such as increased anti-IFN-γ auto-Ab levels in the host (148), making the reliable clinical diagnosis of all infected pathogens challenging. Clinical parameters such as the anti-IFN-γ auto-Ab titer, white blood cell count, neutrophil count, erythrocyte sedimentation rate, and C-reactive protein level were significantly higher in the 22 patients with T. marneffei and NTM coinfection than in patients monoinfected with either T. marneffei or NTM, while the CD4+ T-cell count was lower in a systemic evaluation study conducted in 2021 (143). Therefore, the study provided monitoring criteria and guidelines for detecting coinfection in clinics. Remarkably, 77.3% of these reported coinfected patients were initially diagnosed with only one pathogen infection, highlighting the need for accurate detection of mixed pathogens in susceptible patients (143). Therefore, various types of simultaneous pathogen infections with T. marneffei should be considered during diagnosis in immunocompromised hosts.
MOLECULAR GENETICS OF T. MARNEFFEI
T. marneffei Biology
After the discovery of T. marneffei, a Penicillium-like fungus, by Segretain at the Pasteur Institute in Paris in 1956, its taxonomy was maintained for almost 50 years. T. marneffei was previously classified in the Penicillium subgenus Biverticillium and given the name Penicillium marneffei in honor of the director of the Pasteur Institute (7, 8). However, further phylogenetic research on species relationships revealed that the Penicillium subgenus Biverticillium is distinct from other Penicillium subgenera and that it should be taxonomically separated from Penicillium and grouped with Talaromyces species (104). Therefore, P. marneffei was reclassified into the genus Talaromyces and named T. marneffei in 2011 based on the findings of various DNA-related studies, including mitochondrial genome sequences and other DNA markers, such as the rRNA ITS region and the RNA polymerase (RNAP) II subunit-encoding gene RPB1 (30, 149, 150).
T. marneffei is known for its dimorphic growth mode, mostly caused by thermal switching (Fig. 3). At 37°C, the fungus produces uninucleate yeast cells with ovoid to elongated types that divide by fission rather than budding (93), and diffuse brown (melanized) pigment can be detected (151). T. marneffei demonstrates most of the characteristics of filamentous fungi, including filamentous growth and asexual development. The fungus grows as a multicellular, multinucleated mycelial form at room temperature, with a high growth rate due to the apical expansion of terminal cells and branching. When grown colonies are cultured on agar plates, such as Sabouraud dextrose agar, they are pale gray-green with scattered red pigments. T. marneffei exhibits asexual development throughout the filamentous growth stage under appropriate environmental induction conditions, and infectious propagule conidia with small diameters (3 to 4 μm) are consecutively formed. Conidia are reproductive cells that are frequently pale green in appearance. The pigment, particularly the hyphal dispersed red pigment, is a pivotal diagnostic feature for T. marneffei in mycological culture in the laboratory. The pigment is a secondary metabolite of T. marneffei; 23 polyketide synthase (PKS) genes and two PKS-nonribosomal peptide synthase hybrid genes have been discovered. Phylogenetic analysis shows that these T. marneffei PKSs are distributed evenly across other fungal PKSs, indicating that T. marneffei PKSs did not diverge from lineage-specific gene duplication through a recent expansion and that their functions can be predicted to be homologous with those of other fungal metabolites (152).
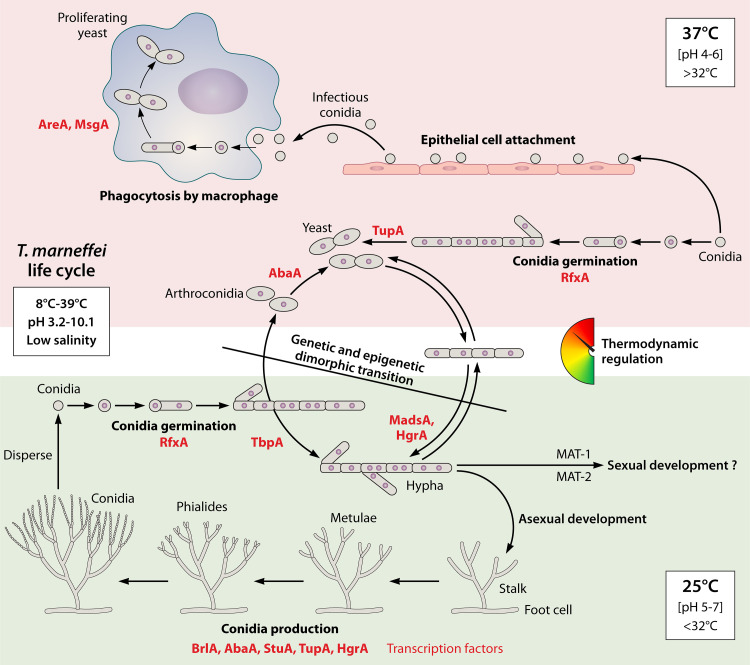
FIG 3.
Life cycle and morphology growth regulation of dimorphic T. marneffei. T. marneffei yeast morphology forms in vitro and in vivo at 37°C (upper panel), while hyphal monogrowth and asexual development occur at 25°C (lower panel). When the temperature is altered, the dimorphic transition between yeast and hypha can occur (middle panel). This transition is thermodynamically regulated with no precise temperature threshold and is under both genetic and epigenetic control (e.g., RNA structural changes) via unidentified mechanisms. T. marneffei can grow under a wide range of culture conditions, including temperatures ranging from 8°C to 39°C, pHs from 3.2 to 10.1, and low salinity, as shown in the illustration. Adequate hyphal development can be achieved when the pH is between 5 and 7 and the temperature is below 32°C. In contrast, pH 4 to 6 and a temperature over 32°C are ideal conditions for yeast development. The conidial germination, hyphal growth, yeast formation, dimorphic transition, and asexual development processes are depicted in the schematic diagram. The identified transcription factors involved in each phase are highlighted with red labels.
Life Cycle of T. marneffei
Despite its thermally dimorphic growth mode, T. marneffei is more closely phylogenetically linked to molds such as Talaromyces, Penicillium, and Aspergillus spp. than to ordinary budding yeasts, such as the budding yeast Saccharomyces cerevisiae, the fission yeast Schizosaccharomyces pombe, and Candida spp. (30, 149). The mitochondrial genome of T. marneffei has been studied and found to be most closely related to that of the model fungus Aspergillus nidulans (153). It is generally known that under 25°C or 37°C culture conditions, the inoculated uninucleate conidia can develop into yeast or hyphal forms (154). T. marneffei produces reproductive conidia in a primitive process of asexual growth comparable to that of Aspergillus spp. Asexual development begins in the presence of abundant air replenishment, such as an air interface. Furthermore, a specific type of hyphal cell known as foot cell is created, frequently followed by multinucleate stalk cells with an enlarged diameter. A secondary stalk cell termed rama is then created constantly, and a septum is formed between these two distinct cell types, which is absent in A. nidulans. Then, budding division occurs, rather than apical development and septation, and a layer of sterigma cells, including metulae and phialides, are produced at the tip of stalk cells. In some circumstances, sterigma cells bud directly from foot cells without stalk generation. However, the quantity of spores is much lower than that produced by A. nidulans, which produces approximately 100 conidia per chain (154, 155) (Fig. 3).
The process from conidial germination to hyphal development, involving isotropic swelling of conidia within the first 6 h and polarized germinated tube creation by 12 h, is unaffected by the culture temperature (Fig. 3). At 48 h after germination, morphological differentiation can be observed, with long (40 μm) vegetative hyphal cells growing at 25°C and highly branched shorter but wider (20 μm) yeast cells growing at 37°C. Furthermore, after 72 h of culture at 37°C, a transitory cell type, uninucleate single arthroconidia, is formed and liberated through cell separation. Finally, these elongated arthroconidia generate small yeast cells (2 to 5 μm) that initiate polarized growth before proceeding to fission division. Notably, a distinct yeast form growth stage progresses inside infected host macrophages, in which conidia germinate directly to yeast forms without the establishment of a hyphal network (156). The in vivo versus in vitro differences show that the morphological transition occurs under different genetic regulatory mechanisms and that yeast and hyphal cells have type-specific cell viabilities or susceptibilities in response to host defense. A typical filamentous development progression begins at 25°C. The germinated conidia tube develops apically, producing lengthy hyphae before forming subapical and branching cells. The vegetative cells are multinucleated, with nuclear and cellular divisions uncoupled during growth, whereas the older subapical cells are often uninucleate. Aside from the production of separate cell types after conidial germination, dimorphic transfer from yeast to hyphae can occur directly under temperature control, and vice versa. Briefly, when yeast cells are transferred from 37 to 25°C, apical development continues, but multinucleate hyphal cells emerge; when hyphal cells are shifted to 37°C, uninucleate arthroconidia and real yeast cells form (154) (Fig. 3). Therefore, the dimorphic switching of T. marneffei is induced by temperature alone, highlighting the importance of the temperature adaptation capabilities of this fungus for its pathogenicity and virulence upon transfer from a low-temperature natural environment to a high-temperature condition inside an infected host. Understanding the environmental conditions that regulate dimorphic switching and the inner molecular genetic control in this pathogenic fungus is important for gaining essential insights into the pathogen and the clinical infection process.
T. marneffei was previously thought to be a strictly asexual fungus (157). However, several lines of evidence have emerged from genome sequence investigations that indicate that this fungus may have a sexual cycle (158–160). (i) The closest mold species, Aspergillus spp., exhibit homothallic or heterothallic sexual cycles. (ii) Crucial genes related to sexual development, such as mating-type genes, genes for meiosis, and genes for pheromone processing enzymes, pheromone receptors, and pheromone response pathways, are also found in the T. marneffei genome. (iii) The SET12 family gene stlA can complement the sexual defect morphology resulting from the steA mutation in A. nidulans, and the gene stuA can compensate for the sexual defect that results from stuA deletion in A. nidulans when the gene is expressed through the A. nidulans promoter rather than its own. These hints point to the existence of a cryptic sexual cycle in T. marneffei, which may require specific developmental conditions that have yet to be identified. Isolates with two different mating types (MAT-1 a box or MAT-2 high-mobility group mating-type genes) (Fig. 3) have been isolated from environments with an unequal distribution. This discovery suggests that T. marneffei can undergo a heterothallic form of sexual development, potentially representing the undiscovered sexual cycle attributable to the unusual correct matching of the two mating types in nature. However, the spatial specificity of the T. marneffei genotype distribution in nature suggests that although a sexual cycle may exist in T. marneffei, it occurs infrequently. Overall, the absence or rarity of sexual reproduction in T. marneffei reveals that the asexual life cycle is the primary mode of reproduction, resulting in stable genome maintenance in T. marneffei.
Environmental Factors Affecting Cell Type Formation
The production of differentiated cell types is triggered by temperature. The optimum growth temperature for T. marneffei is 17 to 28°C, with a wide growth range, 8 to 39°C (Fig. 3). The best growth occurs at 28°C, as evidenced by measuring the growth diameter of the fungus cultured on Sabouraud dextrose agar plates (161). When the temperature exceeds 32°C, the dimorphic transition from hyphae to yeast occurs, with maximum yeast growth at 37 to 39°C. When the temperature exceeds 39°C, fungal development is impeded, but viability can be maintained for up to 1 week at 40°C. Furthermore, because T. marneffei is an intracellular pathogen, host variables (detailed in the section below) significantly impact dimorphic transformation (162, 163). Another crucial component of fungal morphological change is pH. Although T. marneffei can grow at pH 3.2 to 10.1, pH 5 to 7 at 28°C or pH 4 to 6 at 37°C is ideal for yeast or hyphal growth, respectively (Fig. 3). T. marneffei can assimilate a variety of carbon types, including glucose, galactose, maltose, cellobiose, and xyloses; however, the shape of the conidium can be affected (126). On the other hand, high salt concentrations (e.g., >8% NaCl or CaCl2) inhibit T. marneffei (161), in contrast to the case with the well-characterized endemic soil pathogen C. immitis (164). The capacity of C. immitis to flourish at high salinity and high temperatures is compatible with its abundant natural presence in soil, where it competes with other soil microorganisms; therefore, the inability of T. marneffei to tolerate high salinity is consistent with its seemingly restricted survival capability in soil (57), possibly explaining the difficulty of isolating T. marneffei from soil.
Environmental signals affect the development of T. marneffei. Under 0.1% glucose, conidiophores can form within 2 days; however, under carbon-adequate growth conditions, conidiophores are visible only after 5 days (e.g., 1% glucose) (154). Multiple environmental factors that usually stimulate the conidiation process in A. nidulans or other Penicillium spp. have been reported (165), such as light, high osmolarity, nutrient starvation, and calcium replenishment, despite the lack of direct laboratory evidence for factors that contribute to conidiation induction in T. marneffei (165). It is currently unknown whether these elements have a similar effect on T. marneffei conidiation induction. A convincing demonstration of these environmental induction variables and signal sensing systems, which are crucial for T. marneffei virulence, is pivotal for the pathogenic management of those dispersive pathogenic propagules.
Genetic Regulation of Morphological Switching
Morphological transitions are under sophisticated regulation in T. marneffei (Fig. 3; Table 2). The molecular mechanisms governing conidial germination, hyphal and yeast morphogenesis, and dimorphic growth in T. marneffei are currently being investigated, and various signaling pathways have been found (163, 166). At both 25 and 37°C, the conidium germination and polarity establishment signaling pathway, which includes heterotrimeric G protein and Ras signaling, two-component signal sensing, calcium signaling, and downstream mitogen-activated protein kinase (MAPK) signaling cascades, has been extensively studied. In the process of forming diverse cell types, conserved proteins intricately collaborate with distinct downstream effectors. The G protein subunit, as reported for A. nidulans and S. cerevisiae, is also important in T. marneffei conidial germination. The canonical heterotrimeric G protein is composed of three subunits (α, β, and γ), only one of which (the α subunit) is required for T. marneffei conidial germination. In T. marneffei, three subunit-encoding genes, gasA, gasB, and gasC, have been found to encode the G protein α subunit. GasC, but not GasA or GasB, and the Pho GTPase CflA are necessary for conidial germination at either temperature (167), and both are found upstream of the p21-activated kinase (PAK) PkaA. However, PkaA is essential for growth at 37°C but not at 25°C. Specifically, the Ras GTPase RasA is required for CflA or PkaA phosphorylation at 25°C during germination (168) but is not involved in yeast growth at 37°C (169). Disruption of the Ser/Thr protein kinase-encoding gene yakA resulted in early conidial germination with changes in cell wall integrity and chitin deposition through transcriptional regulation of the chitin synthase genes chsB and chsG (170). The path that many downstream effectors take for cell development is under thermodynamic thermoregulation (Fig. 3). Recently, the function of hybrid histidine kinases (HHKs) from two-component systems during germination has been studied. Fungi have expanded sets of HHKs that are classified into 11 classes (i.e., I to XI) and act as alternate signaling sensors to those G proteins (171). The genes hhkJ and slnA (172, 173), which encode class X and class VI HHKs, respectively, have been shown to play roles in conidial germination via interactions with CflA or PkaA. Notably, deleting slnA alters the phosphorylation level of the MAPK component SakA, affecting yeast growth both in vitro and in macrophages (174). Of note, SakA phosphorylation has been shown to affect conidium dormancy status and conidial germination transition in A. nidulans (175). Therefore, the characterization of the function of SakA in T. marneffei conidium maintenance and germination would help elucidate the dormancy of propagule conidia and stress signaling sensing interactions.
TABLE 2.
Identified or described genes participating in T. marneffei growth and morphogenesis
| Growth and morphogenesis path | Gene | Product | Role(s) | Reference(s) |
|---|---|---|---|---|
| Conidium germination at 25°C/37°C | gasC | Gα subunit | Positive regulation of germination | 167 |
| Conidium germination at 25°C/37°C | cflA | Rho GTPase | Gemination, polarized growth of yeast and hypha | 168 |
| Conidium germination at 25°C/37°C | cflB | Rho GTPase | Polarized growth of hypha and conidiophore | 168 |
| Conidium germination at 37°C | pkaA | PAK | Conidium germination | 168 |
| Conidium germination at 25°C | rasA | Ras GTPase | Negative regulation of gemination, polarized growth of yeast and hypha | 168 |
| Conidium germination | hhkJ | HHK (class X) | Conidium germination | 172, 173 |
| Conidium germination | slnA | HHK (class VI) | Conidium germination | 172, 173 |
| Conidium germination | yakA | Ser/Thr protein kinase | Conidium germination | 170 |
| Both yeast and hypha morphogenesis | myoB | Type II myosin | Hyphal, conidiophore, and yeast morphogenesis | 169 |
| Yeast morphogenesis repression at 25°C but not for yeast growth | tupA | Tup1p/GROUCHO-related WD40 transcription factor | Yeast morphogenesis repression | 170 |
| Yeast morphogenesis in macrophages | pkaB | PAK | Yeast morphogenesis during macrophage infection but not in vitro | 162 |
| Yeast morphogenesis | pkaA | PAK | Yeast morphogenesis | 168 |
| Yeast morphogenesis in macrophage | simA | Cytochrome P450 monooxygenase | Essential for yeast cell production during macrophage infection | 250 |
| Yeast morphogenesis | sakA | MAPK | Yeast morphogenesis in vitro and in macrophages | 174 |
| Morphogenesis | rfxA | RFX transcriptional regulator | Essential, required for growth and nuclear division | 180 |
| Hypha morphogenesis | tbpA | TATA-binding transcription factor | Essential for hyphal growth, less significant effect in yeast growth | 181 |
| Morphogenesis | yapA | Basic leucine zipper transcription factor | Delay in both yeast and hypha growth | 178 |
| Hypha morphogenesis | slnA | HHK (class VI) | Hypha morphogenesis | 173 |
| Hypha morphogenesis | drkA | HHK (class III) | Hypha morphogenesis | 173 |
| Dimorphic transition (hypha to yeast) | areA | GATA transcription factor | Facilitates yeast growth under host conditions | 182 |
| Dimorphic transition (hypha to yeast) | abaA | ATTS transcription factor | Hypha-to-yeast form transition | 184 |
| Dimorphic transition (hypha to yeast) | msgA | Dbl homology/BAR domain-containing protein | Maintain yeast growth under host conditions | 183 |
| Dimorphic transition (hyphal growth) | hgrA | C2H2 transcription factor | Hyphal growth inducer | 185 |
| Dimorphic transition (hyphal growth) | madsA | MADS box transcription factor | Hyphal growth | 169 |
| Dimorphic transition (hypha to yeast) | drkA | HHK (class III) | Production of yeast cells (dimorphic switching) | 173 |
| Conidiation | brlA | C2H2 transcription factor BrlA | Conidium formation | 184 |
| Conidiation | abaA | ATTS transcription factor | Conidium formation | 184 |
| Conidiation | stuA | APSES transcription factor | Metula and phialide production | 189 |
| Conidiation | tupA | Tup1p/GROUCHO-related WD40 transcription factor | Conidiation repression | 177 |
| Conidiation | hgrA | C2H2 transcription factor | Conidiation repression | 185 |
| Conidiation | sakA | MAPK | Positive in conidium no. formation | 174 |
| Conidiation | slnA | HHK (class VI) | Promotion of conidiation | 173 |
| Conidiation | drkA | HHK (class III) | Promotion of conidiation | 173 |
| Conidiation | hhkJ | HHK (class X) | Promotion of conidiation | 172, 173 |
| Conidiation | cnaA | Calcineurin homolog | Promotion of conidiation | 192 |
| Conidiation | gasA | Gα subunit | Negative regulation of conidiation | 193 |
| Conidiation | gasC | Gα subunit | Negative regulation of conidiation | 167 |
| Conidiation | rasA | Ras GTPase | Negative regulation of conidiation | 168 |
| Conidiation | yakA | Ser/Thr protein kinase | Conidiation under carbon starvation | 170 |
Several proteins have been identified as regulators of T. marneffei morphology (Fig. 3). Since T. marneffei undergoes cell division to produce hyphal or yeast forms, the myosin-producing gene myoB has been shown to generally affect the morphogenesis of diverse cell types, regardless of whether the cell formation procedure is budding or fission division (176). TupA, a Tup1p-related WD40 repeat transcription factor, is essential for yeast morphological growth repression at 25°C but not for yeast growth (177). The basic leucine zipper (bZip) transcription factor YapA is required for both yeast and hyphal growth (178). RfxA is the only known transcription factor from the regulator factor X (RFX) family that can bind the X box promoter regions in the T. marneffei genome. Because the RFX protein component is substantially conserved from yeast to humans (179), it has been speculated that RfxA is involved in cell cycle and cell division regulation and its effect on T. marneffei morphology has been observed, although the regulatory mechanisms remain unclear (180). The TATA-binding protein TbpA was demonstrated to be essential for hyphal growth, as a tbpA mutant failed to grow at 25°C but showed merely yeast growth reduction at 37°C (181). The demonstrated dimorphic transition factors include AreA, AbaA, MadsA, MsgA, and HgrA. The GATA factor AreA, a nitrogen regulator involved in proteases, and the Dbl homology/BAR domain-containing protein MsgA have been shown to facilitate or maintain yeast growth under host conditions (182, 183). Deletion of the ATTS (AbaA, TEClp, TEF-1 sequence) transcription factor-expressing gene abaA causes a growth defect in the hyphal-to-yeast form transition (184), demonstrating the regulatory role of AbaA in the morphological transition. The madsA gene, which encodes a MADS (MCM1, AG, DEFA and SRF) box transcription factor, has been discovered to be involved in hyphal growth. MadsA overexpression promotes hyphal production even under yeast-growing conditions at 37°C (169). Constitutive overexpression of the hyphal growth inducer C2H2 transcription factor HgrA inhibits conidiation and yeast growth (185), implying that hgrA downregulation is required for appropriate yeast growth inside the host, which is crucial for T. marneffei pathogenicity. Overall, these factors show that the regulation of T. marneffei pathogenesis is inextricably related to morphological growth. Because the morphology regulatory mechanism remains obscure, advanced efforts and approaches should be applied to characterize morphology regulation in T. marneffei.
Aside from factors characterized through molecular validation or genetic analysis, recent high-throughput approaches have revealed additional regulatory pathways, best exemplified by the newly revealed noncoding RNA and RNA structural changes, which are critical in the mycelium-to-yeast transition or temperature-shifting response (169). These findings suggest that epigenetic control may play a role in the dimorphic transition, but the mechanisms are unclear (Fig. 3). MicroRNA (miRNA)-like RNAs, which are dependent on the Dicer enzyme Dcl-2, show differential expression in the yeast and hyphal growth stages (186), as do cell-type-specific proteins such as heat shock protein (HSP) Hsp90 and catalase-peroxidase (187). Although numerous genes relevant to the phase transition have been found, no genes that can specifically produce dimorphism in T. marneffei have been identified to date. Previous experimental results have shown that the thermally dependent dimorphic transition is not a sharp change at a temperature threshold but a programmed or thermodynamic regulation process (169). Therefore, sequential time course monitoring of the process, including transcription and translation regulation, appears to be necessary for uncovering the morphological growth and dimorphic regulation mechanisms.
Genetic Regulation of Asexual Development
The regulation of asexual development has been well demonstrated in A. nidulans (188). Consistent with the discovery in A. nidulans, BrlA and AbaA are key regulators of asexual development in T. marneffei, along with the development modulator StuA (Fig. 3). BrlA is a C2H2 zinc finger protein encoded by the brlA gene. The Andrianopoulos group has shown that BrlA and AbaA have a conserved function in the T. marneffei conidiation process. Loss of brlA or abaA results in a deficiency in conidium production morphology (184). The aforementioned kinase YakA has been shown to be involved in the conidial production process through the transcriptional upregulation of the abaA and brlA genes in cultivation on potato dextrose agar (PDA) medium but not under conditions with an adequate supply of glucose (170). The gene stuA, which encodes a protein in the APSES (Asm1p, Phd1p, Sok2p, Efg1p and StuAp) protein family, is necessary for metula and phialide cell production during asexual development but not for the fission process in yeast form growth. It has also been demonstrated that ectopic expression of T. marneffei stuA can compensate for the conidiation deficiency induced by A. nidulans stuA deletion, highlighting the conserved function of StuA in asexual development (189). Other factors, such as TupA, which functions similarly to Tup1 in S. cerevisiae (190) or RcoA in A. nidulans (191) as a pleiotropic repressor via RNA polymerase II recruitment suppression and chromatin structure modification, are also involved in regulating the development process. Deletion of the tupA gene results in a premature brlA-dependent asexual development phenomenon in solid culture but not in submerged culture (177), demonstrating the repression function of TupA in conidiation (Fig. 3).
In addition to these core regulators, the kinases SakA (174), SlnA (173), and DrkA (173) and the calcineurin CnaA (192) play roles in asexual development. SakA, a stress-activated kinase, is a component of the MAPK stress signaling cascade. The sakA gene mutation not only impairs the T. marneffei stress response but also results in a significantly reduced conidiation formation number (174). SlnA and DrkA are two-component histidine kinases. The deletion of either dlnA or drkA causes delays in asexual development accompanied by reduced brlA expression, and DrkA has a greater influence than SlnA (173). The cnaA mutant had a lower frequency of phialide and conidium formation with an unknown mechanism (192). On the other hand, upstream signaling proteins, such as the Gα subunit of the heterotrimeric G protein GasA (homologous to A. nidulans FadA), are also important regulators of conidiation but not dimorphic switching or growth. It has been shown that GasA negatively affects the expression of the regulators BrlA and AbaA at 25°C (193). Overall, given the importance of kinases in signaling transmission, comprehensive functional characterization of those kinases and phosphatases is crucial for understanding the development and morphology of T. marneffei.
Taken together, these findings show that more investigations should be performed on how the aforementioned factors affect morphology regulation and how a single component can be implicated in distinct morphogenetic programs. Given that transcription factors frequently influence numerous downstream genes, revealing the function of these DNA-binding proteins and the regulatory network of the T. marneffei growth and development cycle is critical. As previously described, putative binding sites for BrlA, AbaA, and StuA have been identified in the promoter region of the rfxA gene, which encodes the regulator RfxA (179), implying a cross-regulatory or hierarchical regulatory network of these fundamental developmental regulators with other pathway factors. Therefore, in addition to the high-throughput techniques used in specialized morphological or developmental programs, such as gene expression (194) or protein expression (182) profiles, a comprehensive characterization of T. marneffei regulators expressed at different growth stages globally is critically needed. Regulator characterization would help us improve the visualization and understanding of the regulatory process by better depicting these regulatory paths, thus improving our currently limited understanding.
INTERPLAY OF T. MARNEFFEI WITH THE HOST IMMUNE SYSTEM
Talaromycosis is an opportunistic mycosis caused by the pathogen T. marneffei that is most prevalent in individuals living with HIV and those with compromised immune systems. Microorganisms have developed a spectrum of strategies in the long-term competition with environmental stresses or host defenses, including the versatile bacterial antiphage systems (195–198). Similarly, T. marneffei has developed sophisticated mechanisms to antagonize immune responses in the interaction of fungal pathogens and human cells (199), and the host immune system has evolved to recognize, activate, and eliminate pathogens. Dimorphic switching and yeast form growth inside host cells are critical pathogenic factors for the dimorphic pathogen T. marneffei. As summarized in the previous section, distinct pathways, such as signal transduction (i.e., drkA, slnA, rasA, cflA, cflB, pkaA, and pkaB), development (i.e., drkA and abaA), cell wall integrity (i.e., drkA and hgrA), and cell division (i.e., myoB and rfxA), are involved. In the case of the host immune response, innate immune systems are the first line of defense against infection by fungal pathogens (199, 200). Macrophages and neutrophils are the dominant phagocytotic alveolar cells involved in T. marneffei infection, facilitating the phagocytosis of the fungus and playing major roles in enabling hematogenous dissemination for invasive infection. A thorough understanding of T. marneffei biology and survival capability within the host and the interaction between host immune system and the fungus would advance our clinical strategies and the search for antifungal targets, allowing effective control of the disease caused by T. marneffei infection.
Host Defense against T. marneffei Infection
The host immune system recognizes pathogen-associated molecular patterns (PAMPs) through specific germ line-encoded pattern recognition receptors (PRRs), including Toll-like receptors (TLRs), C-type lectin receptors (CLRs), NOD-like receptors (NLRs), complement receptors (CRs), and mannose receptors (201, 202). In T. marneffei, identified PAMPs include the Mp1 mannoprotein in conidia and the N-acetyl-β-d-glucosaminyl groups in the yeast cell wall, which are recognized by host unidentified PRR(s) (202). TLRs (i.e., TLR1, TLR2, TLR4, and TLR6), CR3, and dendritic cell (DC)-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin (DC-SIGN) have been characterized to function in T. marneffei recognition by host macrophages or dendritic cells (203–205). It is noteworthy that the single nucleotide polymorphisms (SNPs) of the TLR genes in HIV-positive persons, represented by the TLR2 rs1339 and rs7656411 polymorphisms, have been shown to be associated with susceptibility to T. marneffei infection and severity of the disease (206) (Fig. 2).
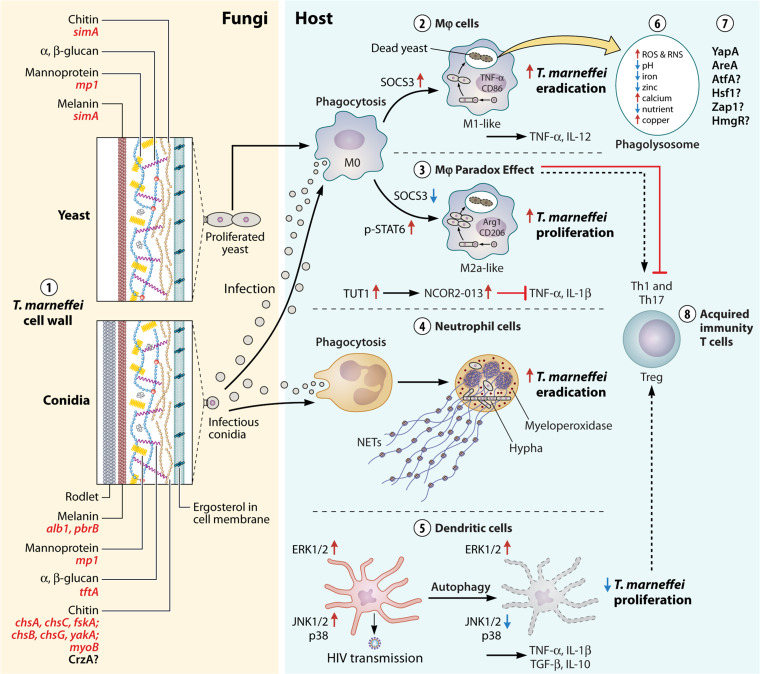
Macrophages can polarize into distinct phenotypes representing different activation states in the immune response and host defense, albeit with instability (207, 208). During pulmonary fungal infections, the most commonly differentiated macrophage types are classical M1-like and alternative M2-like macrophages (207, 208). Inflammatory cytokines, such as IFN-γ, can stimulate the formation of M1-like macrophages in response to fungal infections (209). M1-like macrophages have microbial killing abilities due to the generation of reactive oxygen species (ROS) and reactive nitrogen species (RNS). These fungal cell wall components, such as β-glucan and α-mannan, are essential for the activation of macrophage recognition receptors (e.g., Dectin-1 and Dectin-2) (210, 211), leading to the production of Dectin-1-dependent ROS or cellular cytokines (199). Anti-inflammatory cytokines such as IL-4 and IL-13, IL-1R and TLR, and IL-10 and glucocorticoids promote M2a-like, M2b-like, and M2c-like macrophage polarization, respectively (208). M2a-like cells are one of the M2-like subtypes involved in antifungal responses, but they do not kill invading pathogens as effectively as M1-like cells; instead, M2a-like polarization is primarily involved in promoting the immune response (207) (Fig. 4). Therefore, the polarization state of macrophages has been identified as a possible deciding factor in disease progression.
FIG 4.
Interplay between the pathogenic fungus T. marneffei and host immune cells. On the fungal side (left panel), the cell wall plays a critical role in recognizing and activating the host immune system. Conidia and yeast cell walls are depicted in detail (point 1), with ordered components such as chitin, β-glucan, melanin, and conidium-specific rodlet. The genes associated with those components are underlined in red. Macrophage polarization can be triggered by both infectious conidia and stimulated yeast cells of T. marneffei. Innate immune cells such as macrophages (points 2 and 3), neutrophils (point 4), and dendritic cells (DCs) (point 5) play important roles in the host response. M0 macrophages can be activated to the M1-like and M2a-like polarization subtypes via regulation of the polarization factors SOCS3 and p-STAT6, respectively, after T. marneffei phagocytosis (point 2). TNF-α and CD86 are M1-like activation markers, while Arg-1 and CD206 are M2a-like activation markers (point 3). The main phagocytic cells involved in pathogen eradication are M1-like macrophages and neutrophils, while M2a-like cells appear to provide a protective “hotbed” for fungal proliferation, representing the macrophage paradox effect (points 2, 3, and 4). Along with M2a-like subtype polarization, the alternative splicing of the immune-inflammatory factor NCOR2-013, regulated by TUT1, is also involved, enabling fungal cell proliferation or survival (point 3). Neutrophils have fungicidal activity that partially depends on the enzyme myeloperoxidase (point 4). T. marneffei infection induces the ERK1/2 or JNK1/2 and p38 signaling pathways and autophagy activation in DCs. Activation of ERK1/2 and suppression of JNK1/2 and p38 can activate autophagy and secretion of the cytokines TNF-α, IFN-γ, IL-10, and IL-1β, facilitating the inhibition of pathogen proliferation. Activated DCs can also be hijacked by HIV for its transmission (point 5). The microenvironment of phagolysosomes within macrophages is critical for combating pathogens, as evidenced by enhanced ROS and RNS, acidic pH, decreased iron and zinc ion availability, calcium ion elevation, nutritional deprivation, and heightened copper concentration (point 6). T. marneffei can adjust its gene expression and regulation to compete with the host inside this hostile environment, and this process is potentially governed by numerous transcription factors with unidentified mechanisms (point 7). Furthermore, the activated innate immune response can stimulate or inhibit acquired immunological responses, including Th1-, Th17-, and Treg-related reactions (point 8).
T. marneffei infection has been found to cause macrophages to polarize into the M1-like and M2a-like subtypes (212–215) (Fig. 4). Both the conidium and yeast forms of T. marneffei have been shown to induce the polarization of macrophages into the M1-like phenotype, while with the progression of infection, the M1-like markers tumor necrosis factor alpha (TNF-α) and CD86 gradually decreased, along with the induction of the M2-like markers Arg-1 and CD206 (215). Furthermore, the decrease in CD86 marker levels was demonstrated to result from T. marneffei absorption or uptake of CD86 during the M1-like/M2-like cell type conversion (214). Although the mechanism of cellular CD86 depletion after fungal pathogen infection is unclear, because CD86 level reduction is linked to Th1-cell activity inhibition (216) (Fig. 4), the process is speculated to be significant in aiding fungal pathogen escape from immune cell killing. Notably, not only T. marneffei infection but also cytoplasmic yeast antigen (CYA) stimulation significantly increased the expression of M1-like activation markers, such as inducible nitric oxide synthase (iNOS), NO, IL-12, TNF-α, and M2a-like activation markers (i.e., arginase1 and urea), at an early stage after infection (212) (Fig. 4). Hence, it is proposed that the design and development of an anti-CYA vaccine could potentially be a novel way to prevent T. marneffei infection.
Macrophages are indispensable in the host immune defense after fungal pathogen infection establishment through phagocytosis. However, a macrophage paradox effect (217), in which macrophages provide a niche for fungal pathogens to evade the host’s defense systems, is a common strategy exploited by several pathogens (217–220). In real-time monitoring of the dynamics of the T. marneffei phagocytosis process, Ellett et al. found a nondetectable reduction in fungal cell number after the inoculation of conidia into an established zebrafish model to induce systemic infection (220). These findings suggest that macrophages act as a “hotbed” of protection against conidial parasitism. The strategy of T. marneffei escape from immune systems has been demonstrated through stimulating M2-like macrophage polarization via the degradation of the M1-like polarization factor SOCS3 and hence relieving its inhibitory effect on the M2-like polarization factor p-STAT6 (213). Furthermore, it has been demonstrated that suppressing macrophage inflammatory factors (TNF-α and IL-1β), facilitated by host TuT1 regulation of the alternative splicing process, producing truncated protein NCOR2-013, is vital for T. marneffei survival inside macrophages (221) (Fig. 4). These findings suggest that restricting the access of inhaled T. marneffei propagules to the macrophage niche may improve pathogen elimination during infection establishment. Thus, transient macrophage depletion in patients could be a novel therapeutic strategy for facilitating the eradication of Talaromyces infection or preventing infection in susceptible populations.
NLRP3 activation, which leads to the formation of the NLRP3 inflammasome, can lead to caspase-1-dependent release of proinflammatory cytokines, such as IL-1β and IL-18, and gasdermin D-mediated pyroptotic cell death upon host recognition of T. marneffei (222, 223). It has been shown that in the presence of the caspase-1/-4 inhibitor VX765, both cell death and pyroptosis can be markedly suppressed (86). T. marneffei yeast, but not conidium, stimulation was recently shown to induce NLRP3 inflammasome-triggered production of the cytokines IL-1β and TNF-α by human peripheral blood mononuclear cells (224). The activated inflammasome may also help to boost adaptive Th1 and Th17 immune responses to fungal infection (Fig. 4). Furthermore, one study found that NLRP3- or caspase-1-deficient mice have a higher mortality rate and a higher T. marneffei load than wild-type mice (224), indicating that the NLRP3 inflammasome plays a role in host defenses upon T. marneffei infection.
Neutrophils are another type of host phagocytic cell. Ellett et al. demonstrated that neutrophils have fungicidal activity that partially depends on the enzyme myeloperoxidase (220). Myeloperoxidase deficiency is associated with reduced fungicidal neutrophil extracellular trap (NET) production and elevated fungal infection risk (225, 226) (Fig. 4). Therefore, determining myeloperoxidase levels in susceptible patients may serve as a diagnostic parameter for these patients. On the other hand, the extracellular signal-regulated kinase 1 and 2 (ERK1/2) pathway has been shown to accelerate cell autophagy (227, 228), and T. marneffei phagocytosis can activate the ERK1/2 signaling pathway, inhibiting pathogen survival (229). Furthermore, upon T. marneffei infection, c-Jun N-terminal kinases 1 and 2 (JNK1/2) and the p38 signaling pathway are activated, and autophagy of DCs has been detected, which is linked to the production of the cytokines TNF-α, IFN-γ, IL-10, and IL-1β, as well as pathogen proliferation inhibition (230, 231), implying the importance of DCs in the immune response to T. marneffei infection. Suppressing ERK1/2 or JNK1/2 and p38 results in autophagy inhibition or activation, respectively (230, 231), demonstrating their opposite roles in DC autophagy stimulation (Fig. 4). However, the widespread distribution of DCs in the submucosa and their migration ability can be exploited by HIV for viral dissemination via primary CD4+ T-cell activation, leading to the hypothesis that DC activation is involved in HIV transmission (232, 233). The immune tolerance promotion properties of DCs in infection by T. marneffei yeast cells were further characterized, and the results revealed IL-10 and TGF-β induction and the promotion of regulatory T cell (Treg) expansion (Fig. 4) (234). In addition, T. marneffei infection has been linked to increased HIV-1 trans-infection and viral infection in AIDS patients (Fig. 4) (235).
Additional immune response processes have been studied, such as serum exosome changes (236) and noncoding RNA regulation (237, 238). Three serum exosomal microRNAs (miR-192-5p, miR-194-5p, and miR-1246) involved in immune cell signaling pathways (e.g., transforming growth factor β [TGF-β], cAMP pathway, and others) have been induced in AIDS patients with talaromycosis (236). Hence, these exosome-bound miRNAs have been proposed as potential talaromycosis biomarkers for clinical diagnosis. Bronchial epithelial cells are vital in pathogen invasion and the immune response after the inhalation of T. marneffei, in which the negative effect of long noncoding RNA (lncRNA) lncSSBP1 on the expression of the inflammatory factor IL-6 has been demonstrated (238). The cross-cellular response thus emphasizes the serious impact of fungal pathogen modulation on viral dissemination and infection amplification, both of which affect impairment in HIV-positive individuals. Overall, elucidating host immune response factors, particularly those specific to T. marneffei infection, has pivotal implications for the early detection of fungal infections and the development of antifungal targets.
The Fight between T. marneffei and the Host Immune System
Classically, the asexual reproduction conidia of T. marneffei are recognized as infectious propagules. In HIV-positive individuals and other susceptible populations, infection is established after these conidia enter the lung alveolar cells. T. marneffei, like other pathogenic fungi, has evolved multiple adaptation or counterattack mechanisms to survive in the hostile host environment. These processes include adherence, colonization in the host tissue, proliferation, and escape from or destruction of the host defense system (239). Sialic acid-dependent recognition and glyceraldehyde-3-phosphate dehydrogenase-dependent adhesion of T. marneffei during infection have been reported (240–242). After the establishment of infection, conidia germinate into yeast growth forms, and pathogenic fungi use a variety of strategies to evade the host immune system, including fungal cell wall architecturalization and remodeling, cell signaling transduction, macrophage activation, and cellular environment adaptation, in which the morphology, growth, and primary and secondary metabolism of the fungus are modulated, maximizing its survival (64).
Fungal cell wall recognition and remodeling.
The fungal cell wall is particularly vital for recognizing and breaching the physical barriers of immune cells. Chitin, glucans, mannans, and GMs are the most common PAMPs on cell walls. MyoB is vital for chitin deposition in T. marneffei (176), but how the myosin interacts with the human host is unknown. Among glucans, α- and β-glucan are key components of PAMPs. Gupta discovered that when the major β-1,3/1,4 glucan-encoding gene tftA was mutated, the fungus showed increased binding affinity to cocultured J774 macrophage lines, demonstrating the role of glucan in the adherence of conidia to immune cells (243). Mannoproteins, including the well-identified mannoprotein 1 (Mp1) (244–246) and other unidentified mannoproteins, such as those recognized by the 4D1 antibody, are crucial in the interaction between the fungus and host cells, and corresponding antibodies against these proteins have been developed for detection purposes. Another cell wall component, melanin, is a black or brown pigment located in the cell wall of the majority of fungi (247). The roles of melanin in the virulence of several fungal diseases have been well documented, including the promotion of survival, resistance to host immune systems, and reduced susceptibility to antifungal drugs (248). The genome of T. marneffei contains two common fungal melanin (i.e., DOPA-melanin and DHN-melanin) synthesis genes that require the activity of laccase enzymes (249), implying that this pathogen may produce both types of melanin (250). Deleting the laccase-encoding gene pbrB increases THP1 macrophage phagocytosis and cytokine production (251), indicating that melanin plays a role in immune evasion. Knocking down the alb1 gene, which is related to the biosynthesis of DHN-melanin, results in higher H2O2 susceptibility in conidia and an enhanced survival rate in infected mice (152, 252), demonstrating the importance of this cell wall component in T. marneffei pathogenicity (Fig. 4).
One typical survival strategy for dimorphic fungi is rapid cell wall remodeling during infection to prevent recognition by host phagocytic cells. As characterized in C. albicans, which can switch between yeast and hyphal forms inside the host, β-glucan is concealed by the hyphal cell wall outer layers and is exposed only in the bud scars of yeast cells predisposed to host Dectin-1-mediated recognition and phagocytosis (253). A similar condition has been discovered in T. marneffei. The chitin synthase gene chsE has been detected at induction levels in yeast in comparison to hyphal cells (194). During yeast development, the expression of the cell membrane ergosterol biosynthesis gene ergM is upregulated, suggesting that these cell wall components play a specific role in the interaction of yeast cells with the immunological response (194). The yeast-specific expressed gene simA encodes cytochrome P450 (CYP) (194), an enzyme that functions in cell wall integrity during infection and has been demonstrated to be essential for intracellular yeast growth and survival inside macrophages but had no detectable effect on in vitro growth (254) (Fig. 4).
Signal transduction pathway.
Although the dimorphic transition in T. marneffei is primarily influenced by temperature variations, various T. marneffei morphological types can be observed when the fungus is grown at the same incubation temperature (33°C) but in different intracellular habitats. The filamentous growth of T. marneffei has been observed in neutrophils, whereas yeast forms predominate in macrophages (220). This result hints that one or more unknown temperature-independent host factors inside macrophages contribute to the dimorphic transition of T. marneffei. Mutation of the pakB gene, which encodes a homolog of S. cerevisiae Cla4 p21-activated kinase (PAK), has no effect on yeast cell development in vitro at 37°C but causes abnormal yeast cell formation inside macrophages (Table 2), demonstrating that the pakB mutant is responsive to host signaling rather than temperature. In contrast, the cflA gene, an ortholog of the Cdc42 GTPase-encoding gene cdc42, contributes to the creation of yeast cell types in vitro but is not functional in murine macrophages (162) (Table 2). These findings show that phase transition and morphogenesis in vitro and during host-pathogen interactions in vivo are controlled by different signaling mechanisms. Separating temperature-generated responses from those induced by host signals is critical in understanding the host-pathogen relationship for the dimorphic pathogen T. marneffei. It is critical to determine how host variables influence fungal architecture to gain a better understanding of how fungal pathogens interact with their hosts.
Calcineurin is required for virulence in several fungal pathogens (255–258), participating in pathways related to cation homeostasis, morphological growth, and cell wall integrity. An increase in calcium concentration in macrophages has been detected after T. marneffei infection (259). Calcineurin is a Ca2+-dependent protein phosphatase made up of two subunits: a catalytic subunit (CnaA) and a regulatory subunit (CnaB) (260). Deletion of T. marneffei cnaA abolished pathogenicity in a mouse infection model, likely mediated by increased sensitivity to ROS stress or the destruction of the cell wall structure of the mutant conidia. When the altered conidia are ingested, they are unable to germinate or grow into a yeast form, resulting in a considerably lower fungal burden and increased murine survival capability (192). The phosphorylated calcineurin complex interacts with cell wall proteins directly or indirectly via the transcription factor CrzA (257). The transcription factor CrzA can be further dephosphorylated by the activated calcium complex in the calcium response, allowing the signal to be translocated into the nucleus, where it can activate the transcription of cell wall biosynthesis genes such as β-glucan and chitin-related genes (e.g., chsA, chsC, and fskA) or regulate cell wall homeostasis. However, the interior regulatory mechanism of CrzA remains unclear.
Thermal tolerance establishment.
Inhaled pathogens must first establish thermal homeostasis to survive in host cells due to the high human body temperature. Heat shock proteins (HSPs) are intracellular chaperone proteins that respond to stressful conditions by regulating correct protein folding, transport, and assembly. Many fungal pathogens inside the host have elevated expression of HSP-related genes, such as Hsp30, Hsp70, and Hsp90, which is mediated by the heat shock factor Hsf1 (261, 262). In T. marneffei, elevated expression of Hsp70 (263), Hsp30 (264), and Hsp90 ATPase (182) has been detected in yeast at 37°C or higher heat stress. However, whether these proteins have definite functions in the host intracellular response requires further characterization. Since HSPs play bona fide roles in thermal adaptation and Hsf1 is an evolutionarily conserved important heat shock transcription factor involved in heat response (265), these proteins are hypothesized to have significant functions in the fungus-host interaction, and further investigation in T. marneffei is needed (Fig. 4).
Adaptation of the phagocyte oxidative stress environment.
T. marneffei has been found to stimulate macrophage polarization after infection establishment, as mentioned in the previous section. Upon phagocytosis, oxidative stress is a major native defense mechanism produced by phagocytes to damage intracellular microorganisms. Hence, multiple toxic factors, including reactive oxygen/nitrogen species, hydrolytic enzymes, and toxic metabolites, as well as acidic phagosome conditions, are generated (163, 266). In the 1990s, it was shown that activated macrophages that produce RNS via the l-arginine NO pathway could destroy T. marneffei conidia and impede intracellular fungal reproduction in the J774 murine macrophage model (267). Genes that encode proteins essential for oxidative stress resistance or elimination, such as the superoxide dismutase-encoding gene sodA (268) and the catalase-peroxidase-encoding gene cpeA (269), have demonstrated enhanced expression in T. marneffei yeast cells inside macrophages. In T. marneffei, the stress regulator AtfA (270) has been shown to participate in the oxidative stress response, and when atfA is deleted, the fungus’s intracellular survival capability has a 2-fold reduction. YapA positively regulates the oxidative and nitrosative stress response and the survival capability of the fungus inside macrophages (178). Furthermore, signaling genes such as rttA and sakA are also linked to the oxidative stress response. The aforementioned hyphal growth factor HgrA contributes to resistance against oxidative and cell wall stresses (185), while deletion of the CYP-encoding gene simA results in unexpected H2O2 and NO2 resistance, indicating an as-yet-unidentified role of SimA in the oxidative stress response; however, no effect on macrophage phagolysosomal maturation was observed (254). Hypoxia adaptation in mammalian cells is mostly accomplished through the function of hypoxia-inducible factors (HIFs), which reprogram ATP synthesis from oxidative phosphorylation to glycolysis (271). Although no HIF homolog has been found in lower eukaryotes, analogous metabolic reprogramming control processes have been discovered in fungi, such as the stimulation of glycolysis genes and the suppression of aerobic respiration genes in C. albicans (272–274). Therefore, elucidating glycolysis pathway genes and their expression patterns in the host infection process is also vital for understanding T. marneffei oxidative stress adaptation. Regarding acid pH adaptation, one study revealed that fungal acid phosphatase synthesis inhibits phagocyte respiratory burst and improves T. marneffei survival ability (275). The antimalarial medication 4-aminoquinoline, which has been used to reduce T. marneffei growth, has been demonstrated to be effective in modulating macrophage intracellular pH (276). Hence, identifying the essential mechanisms that cause T. marneffei to respond to or modulate the intracellular acid pH can lead to the discovery of possible antifungal targets for treating talaromycosis (Fig. 4).
T. marneffei extracellular vesicles (EVs) have recently been discovered to play a role in talaromycosis pathogenesis (277). EVs are nanosized lipid bilayer-membrane particles that can transfer chemicals to the extracellular environment. EVs have been identified as pathogenic factors in various fungal infections, such as with C. neoformans (278) and C. albicans (279). T. marneffei-derived EVs contain virulence proteins such as HSPs, MP1, and peroxidase and can induce RAW 264.7 macrophages to release ROS, RNS, and inflammatory factors (e.g., IL-1, IL-6, and IL-10) (277), thus providing new perspectives on fungus-host interaction studies. Therefore, further studies investigating the intracellular communication process to functionally characterize each EV component are needed.
Nutrient starvation adaptation.
Phagocytosed pathogens have been able to reprogram their metabolism pathways to survive the glucose limitation inside the phagosome by the assimilation of other carbon sources, such as amino acids, carboxylic acids, and fatty acids (280). To use the limited resources inside macrophages, the glycolysis metabolism is replaced by the glyoxylate pathway in T. marneffei (281, 282). The downregulation of the glycolytic gene gdpA and the overexpression of the isocitrate lyase gene acuD can be observed as early as 2 h after infection (209). Many pathogenic fungi, such as H. capsulatum (283) and A. fumigatus (284), use amino acids as a source of carbon and nitrogen during their infectious growth. The tyrosine catabolism-dependent yeast form growth of T. marneffei has also been studied (285). The tyrosine catabolism gene hdpA is required for intracellular growth, which is regulated by the GATA-type nitrogen regulator AreA and the within-cluster transcription factor HmgR. Notably, propionyl coenzyme A (propionyl-CoA) is a by-product of branched-chain amino acid breakdown that has the potential to be harmful to both microbes and human cells (286). Lacking the coenzyme B12-dependent methylmalonyl-CoA pathway utilized by mammalian cells, fungi use an alternative metabolic pathway called the methylcitrate cycle (MC cycle) for propionyl-CoA degradation. The importance of the MC cycle gene MCD in T. marneffei pathogenicity has recently been established, with MCD mutation resulting in reduced macrophage elimination (287). Tyrosine, on the other hand, is not only a carbon/nitrogen metabolism substrate but also a precursor for melanin formation, and amino acid metabolism is important for preventing phagolysosome acidification in C. albicans (288), demonstrating the multiple pathway cross-regulation network inside fungal pathogens (Fig. 4).
Iron/zinc acquisition and copper resistance in intracellular growth.
Host cells pose many survival challenges for these phagocytic pathogenic fungi. Another harsh environmental factor that pathogens face is a lack of important trace metals, such as iron and zinc, inside macrophages (289, 290). To thrive in iron-limited circumstances, fungi use high-affinity iron uptake mechanisms such as reductive (RIA) and nonreductive (non-RIA) siderophore-mediated iron assimilation (291). The intracellular growth potential of T. marneffei may be influenced by iron availability via the dysregulation of NO synthesis (292). Both RIA (mediated by FtrA) and non-RIA (controlled by GATA factor SreA) regulation cooperate in iron uptake in distinct cell types of T. marneffei (293), and the glucogenesis factors AcuM and AcuK are also involved in its iron homeostasis regulation (294). The acuK mutant displays higher susceptibility to macrophage killing accompanied by failed growth under iron-limited culture conditions and glucogenic carbon sources (295), demonstrating the function of AcuK in T. marneffei iron assimilation and pathogenesis with heretofore-unidentified regulatory targets. Therefore, the functional elucidation of these transcription factors and others could help determine the iron assimilation regulatory pathway during fungal interplay with the host.
Fungal cells have devised mechanisms to compete with hosts for zinc to obtain these vital micronutrients from the limited nutritional environment. The zinc transporters Zrt1 and Zrt2 and the zinc transport transcriptional regulator Zap1 have been functionally characterized in diverse fungal species (296). Unlike iron transporters, which have received much research attention, zinc transporters have not been studied sufficiently in the pathogenesis of fungal pathogens, although the critical role of zinc assimilation has been identified in A. fumigatus and C. albicans (297). Zinc assimilation and transcription regulation play conserved and essential roles in evolutionarily distant fungal species (297), including A. fumigatus (298). Importantly, the T. marneffei genome contains orthologs with high similarity (>65%) to A. nidulans zapA and zinc transporter-encoding genes, suggesting the significance of Zap1 orthologs and their downstream transporters in T. marneffei for ion assimilation or stress response in pathogenesis, although zinc transporters or regulators have yet to be identified in the T. marneffei genome. It is of great significance to further identify and characterize the zinc acquisition regulation of T. marneffei inside host cells in the future (Fig. 4).
In contrast to the limited availability of iron or zinc for pathogen assimilation, host cells frequently employ copper as an antibacterial weapon, resulting in an excessive copper concentration (299). Different fungal species possess conserved high-affinity copper transporters (i.e., Ctr1, Ctr2, and Ctr3) (300), although some pathogens have lost one or two Ctr protein copies (301). Fungal pathogens have evolved copper detoxification machineries (302) to eliminate the toxicity of copper. Although no copper transporters have been discovered in T. marneffei, the copper/zinc superoxide dismutase SodA is found to be induced in yeast forms inside macrophages (268). Because copper/zinc superoxide dismutase uses intracellular copper to produce powerful antioxidants, copper transport and superoxide dismutase activation must be subjected to synchronized regulation inside the host. The discovery of the expression pattern of such genes in different cell types of T. marneffei and during its interaction with the host will be a significant step forward in understanding virulence mechanisms.
Secondary metabolism products.
In addition to signaling transmission and macrophage environment adaptation, metabolism products have been linked to the pathogenicity of fungal pathogens, such as the secretion of peptide toxins by C. albicans (303) and characterized mycotoxins (e.g., gliotoxin) in A. fumigatus (304, 305). Polyketides are secondary metabolites, including pigments, antibiotics, and mycotoxins, produced by a wide range of microorganisms. T. marneffei has a substantially higher number and variety of PKS genes than other thermally dimorphic pathogenic fungi, such as H. capsulatum (25 versus 1) and C. immitis (25 versus 10) (306), implying that these fungi have diverse virulence mechanisms. Four of the 25 PKS genes that encode products have been described using well-established gene knockout methods. One encodes conidium pigment (252), another encodes the diffusible red pigment produced by hyphae (307), and two encode the yellow mycelial pigment (308). As previously noted, knockout of the DHN-melanin biosynthetic gene alb1 results in the attenuation of virulence in a mouse model (252). PKS12 and PKS11 are required for the synthesis of the T. marneffei yellow pigment components mitorubrinol and mitorubrinic acid (308). Either pks11 or pks12 knockdown or double pks11 and pks12 knockdown led to better survival rates of mice (308) and the reduced survival ability of mutant pathogens inside macrophages. These results demonstrate that these secondary metabolites are vital for T. marneffei virulence (252, 308). Deletion of the oxidative stress regulation factor YapA-encoding gene yapA results in more red pigment production of T. marneffei and its low survival capability in THP1 macrophages (178). Therefore, the functional characterization of more metabolic products and elucidation of the regulation of secondary metabolite synthesis pathways are critical for understanding T. marneffei pathogenesis.
Many more virulence factors are warranted to be further characterized in T. marneffei. Among these, phospholipase B is a well-characterized virulence factor in several pathogenic fungi (309). Elevated expression levels of the phospholipase B-encoding gene plb1 were detected in the yeast growth form of T. marneffei (310), suggesting a possible function in virulence that needs to be clarified in the future. Overall, T. marneffei, like other pathogenic fungi, has evolved several mechanisms in contention with the host, most notably through cell type transition regulation. For example, different cell wall components are expressed in the yeast and hyphal forms of T. marneffei, demonstrating that cell wall components are altered to adapt to host cells and may act as a modified cloak to prevent detection by the host. Additionally, yeast cells are more sensitive than conidia to macrophage-generated NO, and substantially stronger stimulation of macrophage NO production against yeast cells has been detected (311), suggesting that host defense targets the yeast cells rather than the infectious conidia. Therefore, understanding T. marneffei’s pathogenesis requires elucidating the regulatory functions of various cell-type-specific components. Notably, clues from recent transcriptomic, proteomic (182, 240, 312), and phosphoproteomic (313) analyses indicate the expression of cell-type-specific factors, which makes the further characterization of those transcription factors systematically and functionally possible. The established systematic chromatin profiling analytical platform in filamentous fungi (314, 315) provides a good approach for demonstrating the functions of those factors in T. marneffei.
OUTLOOKS
Request for Attention on Talaromycosis
Talaromycosis has been overlooked for a long time; thus, the serious problem of a lack of vigilance regarding talaromycosis and support for talaromycosis research has been raised, and a worldwide plea for support was made recently (17). It should be noted that with the ever-increasing number of travelers and increased population movements around the world, the threat of talaromycosis is no longer limited to specific countries or geographical regions: infections are occurring in more populations worldwide (i.e., 34 countries) (6, 15), including the United States, Australia, France, Japan, Germany, Singapore, and Canada, among others (Fig. 1). Therefore, more research into the biology of the pathogenic fungus T. marneffei and the host-pathogen interplay mechanism is urgently needed to improve the diagnosis and treatment of vulnerable populations.
Several patients with endemic mycoses coinfected with COVID-19 have been systematically reviewed through the end of 2021, including coccidioidomycosis (n = 65), histoplasmosis (n = 11), blastomycosis (n = 1), and paracoccidiomycosis (n = 1), although there are no reported talaromycosis coinfection cases (21). With the current clinical scenario of the effects of the COVID-19 pandemic worldwide since 2020 and the widespread clinical use of dexamethasone for patients with severe COVID-19, the risk of developing mycosis is now uncertain (316, 317). Although pneumonia is a rare symptom among T. marneffei-infected patients, such cases have been reported (318, 319); therefore, caution should be exercised regarding how a chain effect of COVID-19 will manifest in talaromycosis patients. Therefore, it is critical to obtain appropriate research funds and political will to further understand talaromycosis and T. marneffei biology.
Pathogen research and improvements in the clinical diagnosis and therapy of talaromycosis should be prioritized by biological researchers in laboratories and clinical pathologists. It is hoped that through such research, essential pathogen diagnostic reporters and novel potential druggable fungal targets will be discovered, ultimately reducing the burden on patients and vulnerable populations around the world. Of note, one ongoing trial of T. marneffei antigen screening for use in populations at risk for talaromycosis (ClinicalTrials.gov number NCT04033120) may provide opportunities to diminish the disease (109), and a strategy of preemptive prophylactic treatment similar to that used for cryptococcosis (320) can also be considered for T. marneffei infection in high-risk patients or individuals. Co-trimoxazole is an antimicrobial agent that functions in microbial folate biosynthesis and is widely used in the clinic for pathogen inhibition (321, 322). One interesting retrospective study conducted on 3,359 persons with HIV receiving antiretroviral therapy between 2005 and 2016 in Guangxi, China, found that patients with co-trimoxazole prophylaxis had a significantly lower T. marneffei infection rate (i.e., 4.11% versus 7.53%) (323). A further study of the co-trimoxazole inhibition mechanism in T. marneffei revealed that its blocking of dihydropteroic acid synthetase (DHPS), dihydrofolate synthetase (DHFS) and dihydrofolate reductase (DHFR) inhibited fungal growth in an ex vivo THP1 macrophage model (324). These findings support the need for additional epidemiological studies and further research on the antifungal activities of co-trimoxazole in T. marneffei.
More Molecular Genetic Research Tools for T. marneffei Are Needed
For molecular genetic characterization in organisms, the availability of genetic techniques is frequently the limiting step. Although the pathogenic potential of T. marneffei has only recently been recognized, several laboratory molecular genetic research techniques were established around the year 2000. Regarding traditional molecular genetic methods for studying T. marneffei, several gene knockout and other gene-edited mutant strains have been recently developed and have greatly facilitated the understanding of the biology and pathogenicity of T. marneffei. T. marneffei is highly amenable to genetic manipulation, which has sped up the genetic investigation of this fungus. For gene management in T. marneffei, auxotrophic niaD and pyrG selection markers and a dominant selection scheme using the bacterial bleomycin resistance gene have been developed or established, along with a number of reporter genes and inducible promoter systems, such as the lacZ, uidA, and fgp genes, the ethanol-inducible A. nidulans alcA promoter, and the xylose-inducible xylP promoter (189). On this basis, a surge of genetic manipulation tools have been developed, including the RNA interference-mediated gene silencing system (325), homologous integration in the gene mutation system (326), and the CRISPR-Cas9 genome editing system (327). Specifically, the strain constructed by the Andrianopoulos group, which has high homologous integration efficiency (326), improves the efficacy of targeted gene editing, laying the groundwork for more readily available methods to characterize gene functions in T. marneffei.
However, compared to other fungal infections, such as with A. fumigatus, C. albicans, and even the newly discovered pathogen Candida auris, more efforts must be made to develop toolbox applications for T. marneffei. For example, the A. fumigatus FLARE reporter strain allows the real-time imaging of pathogen-host interactions at the single-cell level (328). In a genetic study of the circuitry required for morphogenesis in C. auris, the developed tools for insertional mutant library building were useful (329). The lack of a genetic toolbox for T. marneffei has been a major impediment to understanding T. marneffei morphology and pathogenicity. Therefore, the construction of genetic toolboxes for T. marneffei, such as high-throughput mutant libraries and traceable reporter strains, will be crucial in the development and establishment of analytical tools or platforms. All of these platforms are essential for identifying molecular regulation pathways related to pathogenicity and are the foundations for performing functional genomic investigations.
The Adoption of a Suitable Infection Model Is Critical
The most crucial question to address during the development of an animal model is how close the model is to the natural immunological host in the progression of talaromycosis. To assess this, the pros and cons of existing established models were reviewed in a recent article (330). The in vivo animal models used for studying T. marneffei infection included mouse, zebrafish, Galleria mellonella, and C. elegans, in addition to ex vivo macrophage lines. The main disadvantage of C. elegans model systems is that they are unable to withstand human physiological temperatures (i.e., 37°C). Although G. mellonella can be grown at 37°C and exhibits innate immune responses that are strikingly comparable to those of vertebrates, the lack of an adaptive immune response system (331) makes it a nonideal model for monitoring T. marneffei infection. G. mellonella model systems, however, are still in their infancy. The use of G. mellonella in a detailed study of the host-pathogen interaction was limited (332) due to the lack of genomic information, mature genetic worm mutant establishment procedures, and microarray or RNA interference libraries, among other factors. Due to the physiological and immunological resemblance of zebrafish to humans, pathogen-host infection platforms have been built to study virulence in a wide spectrum of fungal pathogens, including A. fumigatus (333), C. albicans (334), and C. neoformans (335). The genetic tractability and toolbox availability of zebrafish make it possible to meet the need for a deep understanding of diverse immune cell (e.g., macrophages and neutrophils) interactions with pathogens. Researchers have recently become interested in using zebrafish models, within which transgenic strains with distinct immune cells are specifically labeled, to demonstrate the function of innate immune cells during infection establishment in T. marneffei (220, 336).
In comparison to these invertebrate models, mice are the best model for mimicking the natural infection cycle in humans. However, the infection output is determined by many factors, including the pathogen dose implanted, the infection method, and the immune conditions of the mice. The first euthymic and athymic mouse models for pathogenic T. marneffei were developed by the Kudeken group in the early 1990s (337, 338). After high-dose inhalation of T. marneffei in a normal euthymic mouse model through intratracheal microorganism instillation, the mice developed a hyperinflammatory host response driven primarily by CD4+ T cells. Such injection could result in fatal talaromycosis outcomes; however, the exaggerated host-pathogen interaction did not occur with a low-dose instillation of T. marneffei. This model indicated the role of accumulating host CD4+ T cells in the immunological response to pathogenic T. marneffei, which is consistent with the fact that T. marneffei is particularly dangerous to AIDS patients with lymphocytopenia. In addition, the Kudeken group built a congenitally athymic nude mouse model to confirm this finding. However, the optimal inoculation dose to mimic a natural infection is still unclear, as is whether measuring the immune response from a high pathogen dose added directly to the bloodstream can accurately represent a natural infection. To remedy this situation, immunocompromised mouse models, such as athymic nude mice (337), mice with immunosuppression induced through treatment with glucocorticoids (338), and IFN-γ knockout mice (339), have been developed. Injection routes include intravenous inoculation by tail vein injection (339), intranasal instillation, and invasive intratracheal inoculation. The last path appears to be more similar to the natural infection process. However, current bottlenecks for animal models include the impossibility or difficulty of accurately modeling natural infection routes or mimicking the human immune system. Overall, comprehending talaromycosis requires teasing apart the distinct components of host defense systems using a corresponding infection model, and much research is still required to elucidate the host-pathogen interactions. Notably, the recently developed innovative high-throughput profiling spatial transcriptomics method (340, 341) provides a platform for simultaneously analyzing different immune cells in infected patients, enabling the characterization of the host response to T. marneffei infection in situ.
The interaction of coinfected pathogens with T. marneffei inside the host is another key topic that needs thorough study. Multiple concurrent infections have been identified in patients (see “T. marneffei Coinfection Cases” above), although the synergistic and/or antagonistic effects of these pathogens are unclear. Thus, how the host immune system distinguishes among these infecting enemies should be examined. With the development and advancement of cross-species gene expression analysis platforms (342), such as triple transcriptome sequencing (RNA-seq) for investigating coinfection of the pathogenic fungus A. fumigatus and a virus in immune cells (343), both the host immune response and coinfection with different pathogens can be understood spatially and temporally. Critically, this method can also potentially be used to show how the host response to T. marneffei affects the fate of residential HIV in talaromycosis patients living with HIV. Therefore, the interaction between the host and the pathogen will hopefully be understood in the near future through the development of appropriate T. marneffei infection models using sophisticated high-throughput profiling systems.
Genome Sequences of More T. marneffei Clinical Isolates Are Essential
It is clear that the sequences of T. marneffei isolates vary. (i) Divergent biochemical features have been found among different isolates (126, 161), particularly in terms of nutrient assimilation, dimorphic and monomorphic growth differences, and susceptibility to culture temperature and pH variations. (ii) Gene alterations have been observed in numerous environmental and clinical T. marneffei isolates, with acquired or expanded genes such as pop (122) and MadsA (169) accounting for a significant portion of the genome. (iii) Representative assembled genomes for isolates ATCC 18224 and PM1 suggested that these isolates may have distinct chromosomal numbers (i.e., seven versus eight) (169). Specifically, different research groups have used different T. marneffei strains, including the first T. marneffei strain isolate FRR216/ATCC 18224 (7) (which is the derived strain for SPM4139 that has been manipulated to effectively elevate the homologous recombination efficiency), the IFM52703 isolate from China (344), the F4 and CBS 119456 strains from Thailand, and the PM1 isolate from Hong Kong, China (252). Due to a lack of knowledge about the genetic background effects, the use of diverse T. marneffei isolates in different laboratories has prompted the question of whether the results of these studies are comparable, and the issue of how to standardize isolates across different research groups was recently raised. Although the FRR216/ATCC 18224 strain was suggested for use because molecular genetic procedures have been fully developed for this strain, allowing high-quality data to be created, evaluated, and operationally confirmed, data regarding whether the regulation of each isolate is conserved remain inconclusive.
The genetic variation underlying diverse pathogenic fungal isolates has been shown to affect or drive different clinical infection outcomes and drug resistance capacities in multiple fungal pathogens (345, 346). Although genomic and phenotypic differences are widespread across clinical T. marneffei strains, it is unclear whether these differences result in a distinct clinical pathology or variations in host response. Therefore, the characterization of the genomic diversity of T. marneffei and genomic data from multiple studies will help us to gain a full understanding of its pathogenic versatility. We should take advantage of the power of genome-based studies, which are still limited in T. marneffei, to address genomic variation-associated clinical manifestations and identify the core or accessory genetic information of virulence factors for this pathogen, ultimately revealing a new range of potential therapeutic targets to combat these life-threatening infections.
ACKNOWLEDGMENTS
We acknowledge grants from the National Key Research and Development Program of China (2022YFA0912500), the National Natural Science Foundation of China (31925002, 32220103001, and 32200024), and the Shenzhen High-Level Hospital Construction Fund.
Biographies

Fang Wang is a Principal Investigator at the Health Science Center of Shenzhen Institute of Translational Medicine, China, since 2021. She has been working with filamentous fungi since her Ph.D. study in University of Macau, China, from 2016 to 2021. After her graduation, she joined in Shenzhen Institute of Translational Medicine. Now her work focuses on applying genomics and transcriptomics in understanding the virulence and drug resistance regulation of pathogenic fungi (including A. fumigatus and T. marneffei), the interactions between pathogenic bacteria and fungi, and antimicrobial therapy.

RunHua Han is a research associate in the university of Manitoba as of 2022. He has been working in RNA modifications in bacteria and yeast for years at Nankai University, China (Ph.D., 2010–2017) and University of Texas, USA (postdoc, 2017–2021), His general research theme is discovery and characterization of RNA regulators for uncovering complex cellular phenotypes. The critical tools he uses include biochemical approaches, systems biology approaches, genetic studies, large-scale “omics” data analysis, molecular engineering, synthetic biology, and structural analysis. One of the work applications is the manipulation of RNA regulatory systems to engineer more robust cellular phenotypes that can better withstand natural environmental or chemical stresses.

Shi Chen serves as the dean of Shenzhen Institute of Translational Medicine, First Affiliated Hospital of Shenzhen University, China. He was elected as a fellow of the American Academy of Microbiology in 2020. He carried out research at MIT in 2005 and Harvard University in 2007 for postdoctoral work. He joined Wuhan University, China, in 2011 and the First Affiliated Hospital of Shenzhen University, China, in 2021. His research focuses on microbiology, epigenetics, genome engineering, synthetic biology, and innovative drug discovery.
REFERENCES
- 1.Lo Y, Tintelnot K, Lippert U, Hoppe T. 2000. Disseminated Penicillium marneffei infection in an African AIDS patient. Trans R Soc Trop Med Hyg 94:187. 10.1016/S0035-9203(00)90271-2. [DOI] [PubMed] [Google Scholar]
- 2.Waters M, Beliavsky A, Gough K. 2020. Talaromyces marneffei fungemia after travel to China in a Canadian patient with AIDS. CMAJ 192:E92–E95. 10.1503/cmaj.191136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castro-Lainez MT, Sierra-Hoffman M, LLompart-Zeno J, Adams R, Howell A, Hoffman-Roberts H, Fader R, Arroliga AC, Jinadatha C. 2018. Talaromyces marneffei infection in a non-HIV non-endemic population. IDCases 12:21–24. 10.1016/j.idcr.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang P, Chen Y, Xu H, Ding L, Wu Z, Xu Z, Wang K. 2017. Acute disseminated Talaromyces marneffei in an immunocompetent patient. Mycopathologia 182:751–754. 10.1007/s11046-017-0127-7. [DOI] [PubMed] [Google Scholar]
- 5.Cao C, Xi L, Chaturvedi V. 2019. Talaromycosis (penicilliosis) due to Talaromyces (Penicillium) marneffei: insights into the clinical trends of a major fungal disease 60 years after the discovery of the pathogen. Mycopathologia 184:709–720. 10.1007/s11046-019-00410-2. [DOI] [PubMed] [Google Scholar]
- 6.Ning C, et al. 2020. The global distribution, drivers, and burden of talaromycosis 1964–2018, poster 00749. DukeHealth.
- 7.Capponi M, Segretain G, Sureau P. 1956. Pènicillose de Rhizomys sinensis. Bull Soc Pathol Exot 49:418–421. [PubMed] [Google Scholar]
- 8.Segretain G. 1959. Penicillium marneffei n. sp., agent d’une mycose du système réticulo-endothélial. Mycopathol Mycol Appl 11:327–353. 10.1007/BF02089507. [DOI] [PubMed] [Google Scholar]
- 9.DiSalvo AF, Fickling AM, Ajello L. 1973. Infection caused by Penicillium marneffei: description of first natural infection in man. Am J Clin Pathol 60:259–263. 10.1093/ajcp/60.2.259. [DOI] [PubMed] [Google Scholar]
- 10.Chan JF, Lau SK, Yuen KY, Woo PC. 2016. Talaromyces (Penicillium) marneffei infection in non-HIV-infected patients. Emerg Microbes Infect 5:e19. 10.1038/emi.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Supparatpinyo K, Khamwan C, Baosoung V, Nelson KE, Sirisanthana T. 1994. Disseminated Penicillium marneffei infection in southeast Asia. Lancet 344:110–113. 10.1016/s0140-6736(94)91287-4. [DOI] [PubMed] [Google Scholar]
- 12.Duong TA. 1996. Infection due to Penicillium marneffei, an emerging pathogen: review of 155 reported cases. Clin Infect Dis 23:125–130. 10.1093/clinids/23.1.125. [DOI] [PubMed] [Google Scholar]
- 13.Wong KH, Lee SS, Lo YC, Li PC, Ho HF, Sitt WH, Lam TW, Lai KY. 1995. Profile of opportunistic infections among HIV-1 infected people in Hong Kong. Zhonghua Yi Xue Za Zhi (Taipei) 55:127–136. [PubMed] [Google Scholar]
- 14.Qin Y, Huang X, Chen H, Liu X, Li Y, Hou J, Li A, Yan X, Chen Y. 2020. Burden of Talaromyces marneffei infection in people living with HIV/AIDS in Asia during ART era: a systematic review and meta-analysis. BMC Infect Dis 20:551. 10.1186/s12879-020-05260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matos AC, Alves D, Saraiva S, Soares AS, Soriano T, Figueira L, Fraga F, Matos M, Coelho AC. 2019. Isolation of Talaromyces marneffei from the skin of an Egyptian mongoose (Herpestes ichneumon) in Portugal. J Wildl Dis 55:238–241. 10.7589/2017-02-037. [DOI] [PubMed] [Google Scholar]
- 16.Hyde KD, Al-Hatmi AMS, Andersen B, Boekhout T, Buzina W, Dawson TL, Eastwood DC, Jones EBG, de Hoog S, Kang Y, Longcore JE, McKenzie EHC, Meis JF, Pinson-Gadais L, Rathnayaka AR, Richard-Forget F, Stadler M, Theelen B, Thongbai B, Tsui CKM. 2018. The world’s ten most feared fungi. Fungal Divers 93:161–194. 10.1007/s13225-018-0413-9. [DOI] [Google Scholar]
- 17.Narayanasamy S, Dat VQ, Thanh NT, Ly VT, Chan JF-W, Yuen K-Y, Ning C, Liang H, Li L, Chowdhary A, Youngchim S, Supparatpinyo K, Aung NM, Hanson J, Andrianopoulos A, Dougherty J, Govender NP, Denning DW, Chiller T, Thwaites G, van Doorn HR, Perfect J, Le T. 2021. A global call for talaromycosis to be recognised as a neglected tropical disease. Lancet Glob Health 9:e1618–e1622. 10.1016/S2214-109X(21)00350-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roudbary M, Kumar S, Kumar A, Černáková L, Nikoomanesh F, Rodrigues CF. 2021. Overview on the prevalence of fungal infections, immune response, and microbiome role in COVID-19 patients. J Fungi (Basel) 7:720. 10.3390/jof7090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown LB, Spinelli MA, Gandhi M. 2021. The interplay between HIV and COVID-19: summary of the data and responses to date. Curr Opin HIV AIDS 16:63–73. 10.1097/COH.0000000000000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar F, Bignell E, Brown GD, Cook PC, Warris A. 2022. Pathogenesis of respiratory viral and fungal coinfections. Clin Microbiol Rev 35:e00094-21. 10.1128/CMR.00094-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messina FA, Giusiano G, Santiso G. 2022. Endemic mycoses and COVID-19: a review. Curr Fungal Infect Rep 16:98–106. 10.1007/s12281-022-00435-z. [DOI] [Google Scholar]
- 22.Nargesi S, Bongomin F, Hedayati MT. 2021. The impact of COVID-19 pandemic on AIDS-related mycoses and fungal neglected tropical diseases: why should we worry? PLoS Negl Trop Dis 15:e0009092. 10.1371/journal.pntd.0009092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuah CH, Ong YC, Kong BH, Woo YY, Wong PS, Leong KN, Chow TS. 2020. Talaromyces (Penicillium) species infection in the central nervous system. J R Coll Physicians Edinb 50:138–140. 10.4997/JRCPE.2020.211. [DOI] [PubMed] [Google Scholar]
- 24.Radulesco T, Varoquaux A, Ranque S, Dessi P, Michel J, Cassagne C. 2018. A case of fungus ball-type maxillary sinusitis due to Penicillium roqueforti. Mycopathologia 183:439–443. 10.1007/s11046-017-0217-6. [DOI] [PubMed] [Google Scholar]
- 25.Mehta D, Hofacker SA, Villalba JA, Duncan LM, Branda JA, Cañete-Gibas C, Wiederhold N, Moran J, Fathi AT, Chen ST, Cervantes J, Hammond SP. 2021. First reported case of invasive cutaneous Penicillium cluniae infection in a patient with acute myelogenous leukemia: a case report and literature review. Open Forum Infect Dis 8:ofab265. 10.1093/ofid/ofab265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshikata C, Tsurikisawa N, Saito A, Watanabe M, Kamata Y, Tanaka M, Tsuburai T, Mitomi H, Takatori K, Yasueda H, Akiyama K. 2013. Fatal pneumonia caused by Penicillium digitatum: a case report. BMC Pulm Med 13:16. 10.1186/1471-2466-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shokouhi S, Tehrani S, Hemmatian M. 2016. Mixed pulmonary infection with Penicillium notatum and Pneumocystis jiroveci in a patient with acute myeloid leukemia. Tanaffos 15:53–56. [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma B, Nonzom S. 2022. Talaromyces stipitatus, a novel agent causing superficial mycosis in a diabetic patient from North India. Microbes Infect 24:104887. 10.1016/j.micinf.2021.104887. [DOI] [PubMed] [Google Scholar]
- 29.Tsang CC, Lau SKP, Woo PC. 2019. Sixty years from Segretain’s description: what have we learned and should learn about the sasic mycology of Talaromyces marneffei? Mycopathologia 184:721–729. 10.1007/s11046-019-00395-y. [DOI] [PubMed] [Google Scholar]
- 30.Lau SKP, Tsang CC, Woo PCY. 2017. Talaromyces marneffei genomic, transcriptomic, proteomic and metabolomic studies reveal mechanisms for environmental adaptations and virulence. Toxins (Basel) 9:192. 10.3390/toxins9060192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Supparatpinyo K, Nelson KE, Merz WG, Breslin BJ, Cooper CR, Kamwan C, Sirisanthana T. 1993. Response to antifungal therapy by human immunodeficiency virus-infected patients with disseminated Penicillium marneffei infections and in vitro susceptibilities of isolates from clinical specimens. Antimicrob Agents Chemother 37:2407–2411. 10.1128/AAC.37.11.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou L-H, Jiang Y-K, Li R-Y, Huang L-P, Yip C-W, Denning DW, Zhu L-P. 2020. Risk-based estimate of human fungal disease burden, China. Emerg Infect Dis 26:2137–2147. 10.3201/eid2609.200016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Y, Zhang J, Li X, Yang Y, Zhang Y, Ma J, Xi L. 2013. Penicillium marneffei infection: an emerging disease in mainland China. Mycopathologia 175:57–67. 10.1007/s11046-012-9577-0. [DOI] [PubMed] [Google Scholar]
- 34.Jiang J, Meng S, Huang S, Ruan Y, Lu X, Li JZ, Wu N, Huang J, Xie Z, Liang B, Deng J, Zhou B, Chen X, Ning C, Liao Y, Wei W, Lai J, Ye L, Wu F, Liang H. 2019. Effects of Talaromyces marneffei infection on mortality of HIV/AIDS patients in southern China: a retrospective cohort study. Clin Microbiol Infect 25:233–241. 10.1016/j.cmi.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 35.Huang L-F, Tang X-P, Cai W-P, Chen X-J, Lei C-L, Li L-H, Zhang F-C. 2010. An analysis of opportunistic infection in 762 inpatients with human immunodeficiency virus infection in Guangdong areas. Zhonghua Nei Ke Za Zhi 49:653–656. [PubMed] [Google Scholar]
- 36.Ying RS, Le T, Cai WP, Li YR, Luo CB, Cao Y, Wen CY, Wang SG, Ou X, Chen WS, Chen SZ, Guo PL, Chen M, Guo Y, Tang XP, Li LH. 2020. Clinical epidemiology and outcome of HIV-associated talaromycosis in Guangdong, China, during 2011-2017. HIV Med 21:729–738. 10.1111/hiv.13024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang L-F, Tang X-P, Cai W-P, Lei C-L, Zheng F-C, Chen W-L, Ye X-X. 2013. Analysis of death causes of 345 cases with HIV/AIDS in Guangdong area. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 27:57–60. (In Chinese.) [PubMed] [Google Scholar]
- 38.Son VT, Khue PM, Strobel M. 2014. Penicilliosis and AIDS in Haiphong, Vietnam: evolution and predictive factors of death. Med Mal Infect 44:495–501. 10.1016/j.medmal.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen LT, Nguyen MV, Trinh HTX. 2013. P2.025 Etiology of respiratory tract infection in HIV/AIDS patients at the National Hospital of Tropical Diseases (NHTD) Hanoi, Vietnam. Sex Transm Infect 89:A95. 10.1136/sextrans-2013-051184.0290. [DOI] [Google Scholar]
- 40.Louie JK, Chi NH, Thao le TT, Quang VM, Campbell J, Chau NV, Rutherford GW, Farrar JJ, Parry CM. 2004. Opportunistic infections in hospitalized HIV-infected adults in Ho Chi Minh City, Vietnam: a cross-sectional study. Int J STD AIDS 15:758–761. 10.1258/0956462042395159. [DOI] [PubMed] [Google Scholar]
- 41.Wei W, He J, Ning C, Xu B, Wang G, Lai J, Jiang J, Ye L, Liang H. 2021. Maxent modeling for predicting the potential distribution of global talaromycosis. bioRxiv. 10.1101/2021.03.28.437430. [DOI]
- 42.Tang BS-F, Chan JF-W, Chen M, Tsang OT-Y, Mok MY, Lai RW-M, Lee R, Que T-L, Tse H, Li IW-S, To KK-W, Cheng VC-C, Chan EY-T, Zheng B, Yuen K-Y. 2010. Disseminated penicilliosis, recurrent bacteremic nontyphoidal salmonellosis, and burkholderiosis associated with acquired immunodeficiency due to autoantibody against gamma interferon. Clin Vaccine Immunol 17:1132–1138. 10.1128/CVI.00053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan JFW, Chan TSY, Gill H, Lam FYF, Trendell-Smith NJ, Sridhar S, Tse H, Lau SKP, Hung IFN, Yuen K-Y, Woo PCY. 2015. Disseminated infections with Talaromyces marneffei in non-AIDS patients given monoclonal antibodies against CD20 and kinase inhibitors. Emerg Infect Dis 21:1101–1106. 10.3201/eid2107.150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doffinger R, Helbert MR, Barcenas-Morales G, Yang K, Dupuis S, Ceron-Gutierrez L, Espitia-Pinzon C, Barnes N, Bothamley G, Casanova J, Longhurst HJ, Kumararatne DS. 2004. Autoantibodies to interferon-gamma in a patient with selective susceptibility to mycobacterial infection and organ-specific autoimmunity. Clin Infect Dis 38:e10–e14. 10.1086/380453. [DOI] [PubMed] [Google Scholar]
- 45.Höflich C, Sabat R, Rosseau S, Temmesfeld B, Slevogt H, Döcke WD, Grütz G, Meisel C, Halle E, Göbel UB, Volk HD, Suttorp N. 2004. Naturally occurring anti-IFN-gamma autoantibody and severe infections with Mycobacterium cheloneae [sic] and Burkholderia cocovenenans. Blood 103:673–675. 10.1182/blood-2003-04-1065. [DOI] [PubMed] [Google Scholar]
- 46.Kampitak T, Suwanpimolkul G, Browne S, Suankratay C. 2011. Anti-interferon-gamma autoantibody and opportunistic infections: case series and review of the literature. Infection 39:65–71. 10.1007/s15010-010-0067-3. [DOI] [PubMed] [Google Scholar]
- 47.Browne SK, Burbelo PD, Chetchotisakd P, Suputtamongkol Y, Kiertiburanakul S, Shaw PA, Kirk JL, Jutivorakool K, Zaman R, Ding L, Hsu AP, Patel SY, Olivier KN, Lulitanond V, Mootsikapun P, Anunnatsiri S, Angkasekwinai N, Sathapatayavongs B, Hsueh P-R, Shieh C-C, Brown MR, Thongnoppakhun W, Claypool R, Sampaio EP, Thepthai C, Waywa D, Dacombe C, Reizes Y, Zelazny AM, Saleeb P, Rosen LB, Mo A, Iadarola M, Holland SM. 2012. Adult-onset immunodeficiency in Thailand and Taiwan. N Engl J Med 367:725–734. 10.1056/NEJMoa1111160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chi C-Y, Chu C-C, Liu J-P, Lin C-H, Ho M-W, Lo W-J, Lin P-C, Chen H-J, Chou C-H, Feng J-Y, Fung C-P, Sher Y-P, Li C-Y, Wang J-H, Ku C-L. 2013. Anti-IFN-gamma autoantibodies in adults with disseminated nontuberculous mycobacterial infections are associated with HLA-DRB1*16:02 and HLA-DQB1*05:02 and the reactivation of latent varicella-zoster virus infection. Blood 121:1357–1366. 10.1182/blood-2012-08-452482. [DOI] [PubMed] [Google Scholar]
- 49.Ku C-L, Lin C-H, Chang S-W, Chu C-C, Chan JFW, Kong X-F, Lee C-H, Rosen EA, Ding J-Y, Lee W-I, Bustamante J, Witte T, Shih H-P, Kuo C-Y, Chetchotisakd P, Kiertiburanakul S, Suputtamongkol Y, Yuen K-Y, Casanova J-L, Holland SM, Doffinger R, Browne SK, Chi C-Y. 2016. Anti-IFN-gamma autoantibodies are strongly associated with HLA-DR*15:02/16:02 and HLA-DQ*05:01/05:02 across Southeast Asia. J Allergy Clin Immunol 137:945–948.e8. 10.1016/j.jaci.2015.09.018. [DOI] [PubMed] [Google Scholar]
- 50.Guo J, Ning XQ, Ding JY, Zheng YQ, Shi NN, Wu FY, Lin YK, Shih HP, Ting HT, Liang G, Lu XC, Kong JL, Wang K, Lu YB, Fu YJ, Hu R, Li TM, Pan KS, Li XY, Huang CY, Lo YF, Chang IY, Yeh CF, Tu KH, Tsai YH, Ku CL, Cao CW. 2020. Anti-IFN-gamma autoantibodies underlie disseminated Talaromyces marneffei infections. J Exp Med 217:e20190502. 10.1084/jem.20190502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Browne SK, Zaman R, Sampaio EP, Jutivorakool K, Rosen LB, Ding L, Pancholi MJ, Yang LM, Priel DL, Uzel G, Freeman AF, Hayes CE, Baxter R, Cohen SH, Holland SM. 2012. Anti-CD20 (rituximab) therapy for anti-IFN-gamma autoantibody-associated nontuberculous mycobacterial infection. Blood 119:3933–3939. 10.1182/blood-2011-12-395707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Oers MH. 2012. CD20 antibodies: type II to tango? Blood 119:5061–5063. 10.1182/blood-2012-04-420711. [DOI] [PubMed] [Google Scholar]
- 53.Lee PPW, Chan K-W, Lee T-L, Ho MH-K, Chen X-Y, Li C-H, Chu K-M, Zeng H-S, Lau Y-L. 2012. Penicilliosis in children without HIV infection—are they immunodeficient? Clin Infect Dis 54:e8–e19. 10.1093/cid/cir754. [DOI] [PubMed] [Google Scholar]
- 54.Lee PP, Lao-Araya M, Yang J, Chan K-W, Ma H, Pei L-C, Kui L, Mao H, Yang W, Zhao X, Trakultivakorn M, Lau Y-L. 2019. Application of flow cytometry in the diagnostics pipeline of primary immunodeficiencies underlying disseminated Talaromyces marneffei infection in HIV-negative children. Front Immunol 10:2189. 10.3389/fimmu.2019.02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braitstein P, Brinkhof MWG, Dabis F, Schechter M, Boulle A, Miotti P, Wood R, Laurent C, Sprinz E, Seyler C, Bangsberg DR, Balestre E, Sterne JAC, May M, Egger M. Antiretroviral Therapy in Lower Income Countries (ART-LINC) Collaboration, ART Cohort Collaboration (ART-CC) groups. 2006. Mortality of HIV-1-infected patients in the first year of antiretroviral therapy: comparison between low-income and high-income countries. Lancet 367:817–824. [DOI] [PubMed] [Google Scholar]
- 56.He L, Mei X, Lu S, Ma J, Hu Y, Mo D, Chen X, Fan R, Xi L, Xie T. 2021. Talaromyces marneffei infection in non-HIV-infected patients in mainland China. Mycoses 64:1170–1176. 10.1111/myc.13295. [DOI] [PubMed] [Google Scholar]
- 57.Li X, Yang Y, Zhang X, Zhou X, Lu S, Ma L, Lu C, Xi L. 2011. Isolation of Penicillium marneffei from soil and wild rodents in Guangdong, SE China. Mycopathologia 172:447–451. 10.1007/s11046-011-9443-5. [DOI] [PubMed] [Google Scholar]
- 58.Chaiwun B, Vanittanakom N, Jiviriyawat Y, Rojanasthien S, Thorner P. 2011. Investigation of dogs as a reservoir of Penicillium marneffei in northern Thailand. Int J Infect Dis 15:e236–e239. 10.1016/j.ijid.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 59.Gnat S, Łagowski D. 2021. Dogs as reservoir for the endemic pathogen Talaromyces marneffei in the light of new scientific data. Zycie Weterynaryjne 96:825–830. [Google Scholar]
- 60.Wong SY, Wong KF. 2011. Penicillium marneffei infection in AIDS. Patholog Res Int 2011:764293. 10.4061/2011/764293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sirisanthana V, Sirisanthana T. 1995. Disseminated Penicillium marneffei infection in human immunodeficiency virus-infected children. Pediatr Infect Dis J 14:935–940. 10.1097/00006454-199511000-00003. [DOI] [PubMed] [Google Scholar]
- 62.Sirisanthana T. 1997. Infection due to Penicillium marneffei. Ann Acad Med Singap 26:701–704. [PubMed] [Google Scholar]
- 63.Jones PD, See J. 1992. Penicillium marneffei infection in patients infected with human immunodeficiency virus: late presentation in an area of nonendemicity. Clin Infect Dis 15:744. 10.1093/clind/15.4.744. [DOI] [PubMed] [Google Scholar]
- 64.Pruksaphon K, Nosanchuk JD, Ratanabanangkoon K, Youngchim S. 2022. Talaromyces marneffei infection: virulence, intracellular lifestyle and host defense mechanisms. J Fungi (Basel) 8:200. 10.3390/jof8020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cao C, Liang L, Wang W, Luo H, Huang S, Liu D, Xu J, Henk DA, Fisher MC. 2011. Common reservoirs for Penicillium marneffei infection in humans and rodents, China. Emerg Infect Dis 17:209–214. 10.3201/eid1702.100718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ajello L, Padhye AA, Sukroongreung S, Nilakul CH, Tantimavanic S. 1995. Occurrence of Penicillium marneffei infections among wild bamboo rats in Thailand. Mycopathologia 131:1–8. 10.1007/BF01103897. [DOI] [PubMed] [Google Scholar]
- 67.Deng ZL, Yun M, Ajello L. 1986. Human penicilliosis marneffei and its relation to the bamboo rat (Rhizomys pruinosus). J Med Vet Mycol 24:383–389. 10.1080/02681218680000581. [DOI] [PubMed] [Google Scholar]
- 68.Gugnani H, Fisher MC, Paliwal-Johsi A, Vanittanakom N, Singh I, Yadav PS. 2004. Role of Cannomys badius as a natural animal host of Penicillium marneffei in India. J Clin Microbiol 42:5070–5075. 10.1128/JCM.42.11.5070-5075.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li JC, Pan LQ, Wu SX. 1989. Mycologic investigation on Rhizomys pruinous senex in Guangxi as natural carrier with Penicillium marneffei. Chin Med J (Engl) 102:477–485. [PubMed] [Google Scholar]
- 70.Li Y, Luo H, Fan J, Lan X, Liu G, Zhang J, Zhong X, Pang Y, Wang J, He Z. 2019. Genomic analysis provides insights into the transmission and pathogenicity of Talaromyces marneffei. Fungal Genet Biol 130:54–61. 10.1016/j.fgb.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 71.Chariyalertsak S, Vanittanakom P, Nelson KE, Sirisanthana T, Vanittanakom N. 1996. Rhizomys sumatrensis and Cannomys badius, new natural animal hosts of Penicillium marneffei. J Med Vet Mycol 34:105–110. 10.1080/02681219680000161. [DOI] [PubMed] [Google Scholar]
- 72.Ranjana KH, Priyokumar K, Singh TJ, Gupta CC, Sharmila L, Singh PN, Chakrabarti A. 2002. Disseminated Penicillium marneffei infection among HIV-infected patients in Manipur state, India. J Infect 45:268–271. 10.1053/jinf.2002.1062. [DOI] [PubMed] [Google Scholar]
- 73.Chariyalertsak S, Sirisanthana T, Supparatpinyo K, Praparattanapan J, Nelson KE. 1997. Case-control study of risk factors for Penicillium marneffei infection in human immunodeficiency virus-infected patients in northern Thailand. Clin Infect Dis 24:1080–1086. 10.1086/513649. [DOI] [PubMed] [Google Scholar]
- 74.Le T, Wolbers M, Chi NH, Quang VM, Chinh NT, Lan NPH, Lam PS, Kozal MJ, Shikuma CM, Day JN, Farrar J. 2011. Epidemiology, seasonality, and predictors of outcome of AIDS-associated Penicillium marneffei infection in Ho Chi Minh City, Viet Nam. Clin Infect Dis 52:945–952. 10.1093/cid/cir028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y, Deng K. 2021. Environmental risk factors for talaromycosis hospitalizations of HIV-infected patients in Guangzhou, China: case crossover study. Front Med (Lausanne) 8:731188. 10.3389/fmed.2021.731188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hilmarsdottir I, Coutellier A, Elbaz J, Klein JM, Datry A, Guého E, Herson S. 1994. A French case of laboratory-acquired disseminated Penicillium marneffei infection in a patient with AIDS. Clin Infect Dis 19:357–358. 10.1093/clinids/19.2.357. [DOI] [PubMed] [Google Scholar]
- 77.Kawila R, Chaiwarith R, Supparatpinyo K. 2013. Clinical and laboratory characteristics of penicilliosis marneffei among patients with and without HIV infection in Northern Thailand: a retrospective study. BMC Infect Dis 13:464. 10.1186/1471-2334-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vanittanakom N, Sirisanthana T. 1997. Penicillium marneffei infection in patients infected with human immunodeficiency virus. Curr Top Med Mycol 8:35–42. [PubMed] [Google Scholar]
- 79.Vanittanakom N, Cooper CR, Jr, Fisher MC, Sirisanthana T. 2006. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin Microbiol Rev 19:95–110. 10.1128/CMR.19.1.95-110.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu TC, Chan JWM, Ng CK, Tsang DNC, Lee MP, Li PCK. 2008. Clinical presentations and outcomes of Penicillium marneffei infections: a series from 1994 to 2004. Hong Kong Med J 14:103–109. [PubMed] [Google Scholar]
- 81.Vu Hai V, Ngo AT, Ngo VH, Nguyen QH, Massip P, Delmont J, Strobel M, Buisson Y. 2010. Penicilliosis in Vietnam: a series of 94 patients. Rev Med Interne 31:812–818. 10.1016/j.revmed.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 82.Hu J, Li X, Chen T, Liang B, Jiang Z. 2 January 2020. Predictive models for Talaromyces marneffei infection in HIV-infected patients using routinely collected data. Res Sq 10.21203/rs.2.19894/v1. [DOI] [Google Scholar]
- 83.Sun J, Sun W, Tang Y, Zhang R, Liu L, Shen Y, Wang J, Chen J, Qi T, Wang Z, Song W, Lin Y, Xu S, Lu H. 2021. Clinical characteristics and risk factors for poor prognosis among HIV patients with Talaromyces marneffei bloodstream infection. BMC Infect Dis 21:514. 10.1186/s12879-021-06232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li X, Hu W, Wan Q, Lei Q, Sun C, Hou Z, Shrestha N. 2020. Non-HIV talaromycosis: Radiological and clinical analysis. Medicine (Baltimore) 99:e19185. 10.1097/MD.0000000000019185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yousukh A, Jutavijittum P, Pisetpongsa P, Chitapanarux T, Thongsawat S, Senba M, Toriyama K. 2004. Clinicopathologic study of hepatic Penicillium marneffei in Northern Thailand. Arch Pathol Lab Med 128:191–194. 10.5858/2004-128-191-CSOHPM. [DOI] [PubMed] [Google Scholar]
- 86.Wang G, Wei W, Jiang Z, Jiang J, Han J, Zhang H, Hu J, Zhang P, Li X, Chen T, He J, Li Z, Lai J, Liang H, Ning C, Ye L. 2022. Talaromyces marneffei activates the AIM2-caspase-1/-4-GSDMD axis to induce pyroptosis in hepatocytes. Virulence 13:963–979. 10.1080/21505594.2022.2080904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shi M, Lin J, Wei W, Qin Y, Meng S, Chen X, Li Y, Chen R, Yuan Z, Qin Y, Huang J, Liang B, Liao Y, Ye L, Liang H, Xie Z, Jiang J. 2022. Machine learning-based in-hospital mortality prediction of HIV/AIDS patients with Talaromyces marneffei infection in Guangxi, China. PLoS Negl Trop Dis 16:e0010388. 10.1371/journal.pntd.0010388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Supparatpinyo K, Chiewchanvit S, Hirunsri P, Uthammachai C, Nelson KE, Sirisanthana T. 1992. Penicillium marneffei infection in patients infected with human immunodeficiency virus. Clin Infect Dis 14:871–874. 10.1093/clinids/14.4.871. [DOI] [PubMed] [Google Scholar]
- 89.Stathakis A, Lim KP, Boan P, Lavender M, Wrobel J, Musk M, Heath CH. 2015. Penicillium marneffei infection in a lung transplant recipient. Transpl Infect Dis 17:429–434. 10.1111/tid.12377. [DOI] [PubMed] [Google Scholar]
- 90.De Monte A, Risso K, Normand A-C, Boyer G, L’Ollivier C, Marty P, Gari-Toussaint M. 2014. Chronic pulmonary penicilliosis due to Penicillium marneffei: late presentation in a French traveler. J Travel Med 21:292–294. 10.1111/jtm.12125. [DOI] [PubMed] [Google Scholar]
- 91.Hart J, Dyer JR, Clark BM, McLellan DG, Perera S, Ferrari P. 2012. Travel-related disseminated Penicillium marneffei infection in a renal transplant patient. Transpl Infect Dis 14:434–439. 10.1111/j.1399-3062.2011.00700.x. [DOI] [PubMed] [Google Scholar]
- 92.Qin L, Zhao L, Tan C, Chen XU, Yang Z, Mo W. 2015. A novel method of combining periodic acid Schiff staining with Wright-Giemsa staining to identify the pathogens Penicillium marneffei, Histoplasma capsulatum, Mucor and Leishmania donovani in bone marrow smears. Exp Ther Med 9:1950–1954. 10.3892/etm.2015.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Andrianopoulos A. 2020. Laboratory maintenance and growth of Talaromyces marneffei. Curr Protoc Microbiol 56:e97. 10.1002/cpmc.97. [DOI] [PubMed] [Google Scholar]
- 94.Arrese Estrada J, Stynen D, Van Cutsem J, Pierard-Franchimont C, Pierard GE. 1992. Immunohistochemical identification of Penicillium marneffei by monoclonal antibody. Int J Dermatol 31:410–412. 10.1111/j.1365-4362.1992.tb02670.x. [DOI] [PubMed] [Google Scholar]
- 95.Huang Y-T, Hung C-C, Liao C-H, Sun H-Y, Chang S-C, Chen Y-C. 2007. Detection of circulating galactomannan in serum samples for diagnosis of Penicillium marneffei infection and cryptococcosis among patients infected with human immunodeficiency virus. J Clin Microbiol 45:2858–2862. 10.1128/JCM.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karageorgopoulos DE, Vouloumanou EK, Ntziora F, Michalopoulos A, Rafailidis PI, Falagas ME. 2011. β-d-Glucan assay for the diagnosis of invasive fungal infections: a meta-analysis. Clin Infect Dis 52:750–770. 10.1093/cid/ciq206. [DOI] [PubMed] [Google Scholar]
- 97.Yoshimura Y, Sakamoto Y, Lee K, Amano Y, Tachikawa N. 2016. Penicillium marneffei infection with beta-D-glucan elevation: a case report and literature review. Intern Med 55:2503–2506. 10.2169/internalmedicine.55.6173. [DOI] [PubMed] [Google Scholar]
- 98.Kaufman L, Standard PG, Anderson SA, Jalbert M, Swisher BL. 1995. Development of specific fluorescent-antibody test for tissue form of Penicillium marneffei. J Clin Microbiol 33:2136–2138. 10.1128/jcm.33.8.2136-2138.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trewatcharegon S, Chaiyaroj SC, Chongtrakool P, Sirisinha S. 2000. Production and characterization of monoclonal antibodies reactive with the mycelial and yeast phases of Penicillium marneffei. Med Mycol 38:91–96. 10.1080/mmy.38.1.91.96. [DOI] [PubMed] [Google Scholar]
- 100.Vanittanakom N, Mekaprateep M, Sittisombut N, Supparatpinyo K, Kanjanasthiti P, Nelson KE, Sirisanthana T. 1997. Western immunoblot analysis of protein antigens of Penicillium marneffei. J Med Vet Mycol 35:123–131. 10.1080/02681219780001011. [DOI] [PubMed] [Google Scholar]
- 101.Chongtrakool P, Chaiyaroj SC, Vithayasai V, Trawatcharegon S, Teanpaisan R, Kalnawakul S, Sirisinha S. 1997. Immunoreactivity of a 38-kilodalton Penicillium marneffei antigen with human immunodeficiency virus-positive sera. J Clin Microbiol 35:2220–2223. 10.1128/jcm.35.9.2220-2223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jeavons L, Hamilton AJ, Vanittanakom N, Ungpakorn R, Evans EG, Sirisanthana T, Hay RJ. 1998. Identification and purification of specific Penicillium marneffei antigens and their recognition by human immune sera. J Clin Microbiol 36:949–954. 10.1128/JCM.36.4.949-954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yuen KY, Wong SS, Tsang DN, Chau PY. 1994. Serodiagnosis of Penicillium marneffei infection. Lancet 344:444–445. 10.1016/s0140-6736(94)91771-x. [DOI] [PubMed] [Google Scholar]
- 104.Pruksaphon K, Intaramat A, Ratanabanangkoon K, Nosanchuk JD, Vanittanakom N, Youngchim S. 2018. Development and characterization of an immunochromatographic test for the rapid diagnosis of Talaromyces (Penicillium) marneffei. PLoS One 13:e0195596. 10.1371/journal.pone.0195596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pornprasert S, Dettrairat S, Vongchan P, Apichatpiyakul C. 2005. Production of a monoclonal antibody against a yeast secreted antigen of Penicillium marneffei. Southeast Asian J Trop Med Public Health 36:966–969. [PubMed] [Google Scholar]
- 106.Thu NTM, Chan JFW, Ly VT, Ngo HT, Hien HTA, Lan NPH, Chau NVV, Cai J-P, Woo PCY, Day JN, van Doorn R, Thwaites G, Perfect J, Yuen K, Le T. 2021. Superiority of a novel Mp1p antigen detection enzyme immunoassay compared to standard BACTEC blood culture in the diagnosis of talaromycosis. Clin Infect Dis 73:e330–e336. 10.1093/cid/ciaa826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pruksaphon K, Intaramat A, Simsiriwong P, Mongkolsuk S, Ratanabanangkoon K, Nosanchuk JD, Kaltsas A, Youngchim S. 2021. An inexpensive point-of-care immunochromatographic test for Talaromyces marneffei infection based on the yeast phase specific monoclonal antibody 4D1 and Galanthus nivalis agglutinin. PLoS Negl Trop Dis 15:e0009058. 10.1371/journal.pntd.0009058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen X, Ou X, Wang H, Li L, Guo P, Chen X, Cai W, Tang X, Li L. 2022. Talaromyces marneffei Mp1p antigen detection may play an important role in the early diagnosis of talaromycosis in patients with acquired immunodeficiency syndrome. Mycopathologia 187:205–215. 10.1007/s11046-022-00618-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ly VT, Thanh NT, Thu NTM, Chan J, Day JN, Perfect J, Nga CN, Vinh Chau NV, Le T. 2020. Occult Talaromyces marneffei infection unveiled by the novel Mp1p antigen detection assay. Open Forum Infect Dis 7:ofaa502. 10.1093/ofid/ofaa502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun J, Najafzadeh MJ, Zhang J, Vicente VA, Xi L, de Hoog GS. 2011. Molecular identification of Penicillium marneffei using rolling circle amplification. Mycoses 54:e751–e759. 10.1111/j.1439-0507.2011.02017.x. [DOI] [PubMed] [Google Scholar]
- 111.Pongpom M, Sirisanthana T, Vanittanakom N. 2009. Application of nested PCR to detect Penicillium marneffei in serum samples. Med Mycol 47:549–553. 10.1080/13693780802484875. [DOI] [PubMed] [Google Scholar]
- 112.Hien HTA, Thanh TT, Thu NTM, Nguyen A, Thanh NT, Lan NPH, Simmons C, Shikuma C, Chau NVV, Thwaites G, Le T. 2016. Development and evaluation of a real-time polymerase chain reaction assay for the rapid detection of Talaromyces marneffei MP1 gene in human plasma. Mycoses 59:773–780. 10.1111/myc.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ning C, Lai J, Wei W, Zhou B, Huang J, Jiang J, Liang B, Liao Y, Zang N, Cao C, Chen H, Ye L, Liang H. 2018. Accuracy of rapid diagnosis of Talaromyces marneffei: a systematic review and meta-analysis. PLoS One 13:e0195569. 10.1371/journal.pone.0195569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsang CC, Teng JLL, Lau SKP, Woo PCY. 2021. Rapid genomic diagnosis of fungal infections in the age of next-generation sequencing. J Fungi (Basel) 7:636. 10.3390/jof7080636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kaplan JE, Benson C, Holmes KK, Brooks JT, Pau A, Masur H. Centers for Disease Control and Prevention (CDC), National Institutes of Health, HIV Medicine Association of the Infectious Diseases Society of America. 2009. Guidelines for prevention and treatment of opportunistic infections in HIV-infected adults and adolescents: recommendations from CDC, the National Institutes of Health, and the HIV Medicine Association of the Infectious Diseases Society of America. MMWR Recommend Rep 58:1–207; quiz, CE201-204. [PubMed] [Google Scholar]
- 116.Lang Q, Pasheed Chughtai A, Kong WF, Yan HY. 2020. Case report: Successful treatment of pulmonary Talaromyces marneffei infection with posaconazole in a renal transplant recipient. Am J Trop Med Hyg 104:744–747. 10.4269/ajtmh.20-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Larsson M, Nguyen LHT, Wertheim HF, Dao TT, Taylor W, Horby P, Nguyen TV, Nguyen MHT, Le T, Nguyen KV. 2012. Clinical characteristics and outcome of Penicillium marneffei infection among HIV-infected patients in northern Vietnam. AIDS Res Ther 9:24. 10.1186/1742-6405-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Su Q, Ying G, Liang H, Ye L, Jiang J, Liang B, Huang J. 2015. Single use of itraconazole has no effect on treatment for Penicillium marneffei with HIV infection. Arch Iran Med 18:441–445. [PubMed] [Google Scholar]
- 119.Huang W, Li T, Zhou C, Wei F, Cao C, Jiang J. 2021. Voriconazole versus amphotericin B as induction therapy for talaromycosis in HIV/AIDS patients: a retrospective study. Mycopathologia 186:269–276. 10.1007/s11046-021-00533-5. [DOI] [PubMed] [Google Scholar]
- 120.Sheng L, Shen Q, Zhou J. 2021. Efficacy of different antifungal drugs as initial treatment for patients with talaromycosis: a systematic review and meta-analysis. J Mycol Med 31:101108. 10.1016/j.mycmed.2020.101108. [DOI] [PubMed] [Google Scholar]
- 121.Hamill RJ. 2013. Amphotericin B formulations: a comparative review of efficacy and toxicity. Drugs 73:919–934. 10.1007/s40265-013-0069-4. [DOI] [PubMed] [Google Scholar]
- 122.Payne M, Weerasinghe H, Tedja I, Andrianopoulos A. 2019. A unique aspartyl protease gene expansion in Talaromyces marneffei plays a role in growth inside host phagocytes. Virulence 10:277–291. 10.1080/21505594.2019.1593776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Winter MB, Salcedo EC, Lohse MB, Hartooni N, Gulati M, Sanchez H, Takagi J, Hube B, Andes DR, Johnson AD, Craik CS, Nobile CJ. 2016. Global identification of biofilm-specific proteolysis in Candida albicans. mBio 7:e01514-16. 10.1128/mBio.01514-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Monod M, Capoccia S, Léchenne B, Zaugg C, Holdom M, Jousson O. 2002. Secreted proteases from pathogenic fungi. Int J Med Microbiol 292:405–419. 10.1078/1438-4221-00223. [DOI] [PubMed] [Google Scholar]
- 125.De Bernardis F, Tacconelli E, Mondello F, Cataldo A, Arancia S, Cauda R, Cassone A. 2004. Anti-retroviral therapy with protease inhibitors decreases virulence enzyme expression in vivo by Candida albicans without selection of avirulent fungus strains or decreasing their anti-mycotic susceptibility. FEMS Immunol Med Microbiol 41:27–34. 10.1016/j.femsim.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 126.Wong SSY, Ho TYC, Ngan AHY, Woo PCY, Que T-L, Yuen K-Y. 2001. Biotyping of Penicillium marneffei reveals concentration-dependent growth inhibition by galactose. J Clin Microbiol 39:1416–1421. 10.1128/JCM.39.4.1416-1421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jeenkeawpieam J, Yodkeeree S, Andrianopoulos A, Roytrakul S, Pongpom M. 2020. Antifungal activity and molecular mechanisms of partial purified antifungal proteins from Rhinacanthus nasutus against Talaromyces marneffei. J Fungi (Basel) 6:333. 10.3390/jof6040333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sangkanu S, Rukachaisirikul V, Suriyachadkun C, Phongpaichit S. 2021. Antifungal activity of marine-derived actinomycetes against Talaromyces marneffei. J Appl Microbiol 130:1508–1522. 10.1111/jam.14877. [DOI] [PubMed] [Google Scholar]
- 129.Zhang J, Liu H, Xi L, Chang YC, Kwon-Chung KJ, Seyedmousavi S. 2021. Antifungal susceptibility profiles of olorofim (formerly F901318) and currently available systemic antifungals against mold and yeast phases of Talaromyces marneffei. Antimicrob Agents Chemother 65:e00256-21. 10.1128/AAC.00256-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Luo Q, Pan K, Luo H, Cao C. 2019. Evaluation of in vitro antifungal activity of osthole against Talaromyces marneffei in yeast phase. Chin J Dermatol 52:262–265. [Google Scholar]
- 131.Jeenkeawpieam J, Yodkeeree S, Roytrakul S, Pongpom M. 2021. Antifungal properties of protein extracts from Thai medicinal plants to opportunistic fungal pathogens. Walailak J Sci Technol 18:9045. 10.48048/wjst.2021.9045. [DOI] [Google Scholar]
- 132.Supparatpinyo K, Chiewchanvit S, Hirunsri P, Baosoung V, Uthammachai C, Chaimongkol B, Sirisanthana T. 1992. An efficacy study of itraconazole in the treatment of Penicillium marneffei infection. J Med Assoc Thai 75:688–691. [PubMed] [Google Scholar]
- 133.Supparatpinyo K, Perriens J, Nelson KE, Sirisanthana T. 1998. A controlled trial of itraconazole to prevent relapse of Penicillium marneffei infection in patients infected with the human immunodeficiency virus. N Engl J Med 339:1739–1743. 10.1056/NEJM199812103392403. [DOI] [PubMed] [Google Scholar]
- 134.Wiederhold NP. 2017. Antifungal resistance: current trends and future strategies to combat. Infect Drug Resist 10:249–259. 10.2147/IDR.S124918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lau SKP, Lo GCS, Lam CSK, Chow W-N, Ngan AHY, Wu AKL, Tsang DNC, Tse CWS, Que T-L, Tang BSF, Woo PCY. 2017. In vitro activity of posaconazole against Talaromyces marneffei by broth microdilution and etest methods and comparison to itraconazole, voriconazole, and anidulafungin. Antimicrob Agents Chemother 61:e01480-16. 10.1128/AAC.01480-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Imwidthaya P, Thipsuvan K, Chaiprasert A, Danchaivijitra S, Sutthent R, Jearanaisilavong J. 2001. Penicillium marneffei: types and drug susceptibility. Mycopathologia 149:109–115. 10.1023/A:1007245226495. [DOI] [PubMed] [Google Scholar]
- 137.Fang L, Liu M, Huang C, Ma X, Zheng Y, Wu W, Guo J, Huang J, Xu H. 2022. MALDI-TOF MS-based clustering and antifungal susceptibility tests of Talaromyces marneffei isolates from Fujian and Guangxi (China). Infect Drug Resist 15:3449–3457. 10.2147/IDR.S364439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Utami ST, Indriani CI, Bowolaksono A, Yaguchi T, Chen X, Niimi K, Niimi M, Kajiwara S. 2020. Identification and functional characterization of Penicillium marneffei major facilitator superfamily (MFS) transporters. Biosci Biotechnol Biochem 84:1373–1383. 10.1080/09168451.2020.1732185. [DOI] [PubMed] [Google Scholar]
- 139.Cuenca-Estrella M. 2014. Antifungal drug resistance mechanisms in pathogenic fungi: from bench to bedside. Clin Microbiol Infect 20(Suppl 6):54–59. 10.1111/1469-0691.12495. [DOI] [PubMed] [Google Scholar]
- 140.Kontoyiannis DP, Lewis RE. 2002. Antifungal drug resistance of pathogenic fungi. Lancet 359:1135–1144. 10.1016/S0140-6736(02)08162-X. [DOI] [PubMed] [Google Scholar]
- 141.Seok H, Ko J-H, Shin I, Eun YH, Lee S-E, Lee Y-B, Peck KR. 2015. Disseminated Talaromyces marneffei and Mycobacterium intracellulare coinfection in an HIV-infected patient. Int J Infect Dis 38:86–88. 10.1016/j.ijid.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 142.Le T, Hong Chau TT, Kim Cuc NT, Si Lam P, Manh Sieu TP, Shikuma CM, Day JN. 2010. AIDS-associated Cryptococcus neoformans and Penicillium marneffei coinfection: a therapeutic dilemma in resource-limited settings. Clin Infect Dis 51:e65–e68. 10.1086/656685. [DOI] [PubMed] [Google Scholar]
- 143.Qiu Y, Huang J, Li Y, Zeng W, Pan M, Cen J, Zhang H, Sun X, Qu D, Zhang J. 2021. Talaromyces marneffei and nontuberculous mycobacteria co-infection in HIV-negative patients. Sci Rep 11:16177. 10.1038/s41598-021-95686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Qiu Y, Pan M, Yang Z, Zeng W, Zhang H, Li Z, Zhang J. 2022. Talaromyces marneffei and Mycobacterium tuberculosis co-infection in a patient with high titer anti-interferon-gamma autoantibodies: a case report. BMC Infect Dis 22:98. 10.1186/s12879-021-07015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.He S, Lv D, Xu Y, Wu X, Lin L. 2020. Concurrent infection with Talaromyces marneffei and Cryptococcus neoformans in a patient without HIV infection. Exp Ther Med 19:160–164. 10.3892/etm.2019.8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Qiu Y, Feng X, Zeng W, Zhang H, Zhang J. 2021. Immunodeficiency disease spectrum in HIV-negative individuals with talaromycosis. J Clin Immunol 41:221–223. 10.1007/s10875-020-00869-5. [DOI] [PubMed] [Google Scholar]
- 147.Cowman S, van Ingen J, Griffith DE, Loebinger MR. 2019. Non-tuberculous mycobacterial pulmonary disease. Eur Respir J 54:1900250. 10.1183/13993003.00250-2019. [DOI] [PubMed] [Google Scholar]
- 148.Chi C-Y, Lin C-H, Ho M-W, Ding J-Y, Huang W-C, Shih H-P, Yeh C-F, Fung C-P, Sun H-Y, Huang C-T, Wu T-S, Chang C-Y, Liu Y-M, Feng J-Y, Wu W-K, Wang L-S, Tsai C-H, Ho C-M, Lin H-S, Chen H-J, Lin P-C, Liao W-C, Chen W-T, Lo C-C, Wang S-Y, Kuo C-Y, Lee C-H, Ku C-L. 2016. Clinical manifestations, course, and outcome of patients with neutralizing anti-interferon-gamma autoantibodies and disseminated nontuberculous mycobacterial infections. Medicine (Baltimore) 95:e3927. 10.1097/MD.0000000000003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tam EWT, Tsang CC, Lau SK, Woo PCY. 2017. Comparative mitogenomic and phylogenetic characterization on the complete mitogenomes of Talaromyces (Penicillium) marneffei. Mitochondrial DNA B Resour 1:941–942. 10.1080/23802359.2016.1261610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert KA, Peterson SW, Varga J, Frisvad JC. 2011. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol 70:159–183. 10.3114/sim.2011.70.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pruksaphon K, Intaramat A, Ratanabanangkoon K, Nosanchuk JD, Vanittanakom N, Youngchim S. 2020. Diagnostic laboratory immunology for talaromycosis (penicilliosis): review from the bench-top techniques to the point-of-care testing. Diagn Microbiol Infect Dis 96:114959. 10.1016/j.diagmicrobio.2019.114959. [DOI] [PubMed] [Google Scholar]
- 152.Tam EW, Tsang CC, Lau SK, Woo PC. 2015. Polyketides, toxins and pigments in Penicillium marneffei. Toxins (Basel) 7:4421–4436. 10.3390/toxins7114421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Woo PCY, Zhen H, Cai JJ, Yu J, Lau SKP, Wang J, Teng JLL, Wong SSY, Tse RH, Chen R, Yang H, Liu B, Yuen K-y. 2003. The mitochondrial genome of the thermal dimorphic fungus Penicillium marneffei is more closely related to those of molds than yeasts. FEBS Lett 555:469–477. 10.1016/S0014-5793(03)01307-3. [DOI] [PubMed] [Google Scholar]
- 154.Andrianopoulos A. 2002. Control of morphogenesis in the human fungal pathogen Penicillium marneffei. Int J Med Microbiol 292:331–347. 10.1078/1438-4221-00217. [DOI] [PubMed] [Google Scholar]
- 155.Etxebeste O, Garzia A, Espeso EA, Ugalde U. 2010. Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol 18:569–576. 10.1016/j.tim.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 156.Chan YF, Chow TC. 1990. Ultrastructural observations on Penicillium marneffei in natural human infection. Ultrastruct Pathol 14:439–452. 10.3109/01913129009007223. [DOI] [PubMed] [Google Scholar]
- 157.Fisher MC, Hanage WP, de Hoog S, Johnson E, Smith MD, White NJ, Vanittanakom N. 2005. Low effective dispersal of asexual genotypes in heterogeneous landscapes by the endemic pathogen Penicillium marneffei. PLoS Pathog 1:e20. 10.1371/journal.ppat.0010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yuen K-Y, Pascal G, Wong SSY, Glaser P, Woo PCY, Kunst F, Cai JJ, Cheung EYL, Médigue C, Danchin A. 2003. Exploring the Penicillium marneffei genome. Arch Microbiol 179:339–353. 10.1007/s00203-003-0533-8. [DOI] [PubMed] [Google Scholar]
- 159.Borneman AR, Hynes MJ, Andrianopoulos A. 2001. An STE12 homolog from the asexual, dimorphic fungus Penicillium marneffei complements the defect in sexual development of an Aspergillus nidulans steA mutant. Genetics 157:1003–1014. 10.1093/genetics/157.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Woo PCY, Chong KTK, Tse H, Cai JJ, Lau CCY, Zhou AC, Lau SKP, Yuen K-Y. 2006. Genomic and experimental evidence for a potential sexual cycle in the pathogenic thermal dimorphic fungus Penicillium marneffei. FEBS Lett 580:3409–3416. 10.1016/j.febslet.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 161.Cao C, Li R, Wan Z, Liu W, Wang X, Qiao J, Wang D, Bulmer G, Calderone R. 2007. The effects of temperature, pH, and salinity on the growth and dimorphism of Penicillium marneffei. Med Mycol 45:401–407. 10.1080/13693780701358600. [DOI] [PubMed] [Google Scholar]
- 162.Boyce KJ, Schreider L, Andrianopoulos A. 2009. In vivo yeast cell morphogenesis is regulated by a p21-activated kinase in the human pathogen Penicillium marneffei. PLoS Pathog 5:e1000678. 10.1371/journal.ppat.1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Boyce KJ, Andrianopoulos A. 2015. Fungal dimorphism: the switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol Rev 39:797–811. 10.1093/femsre/fuv035. [DOI] [PubMed] [Google Scholar]
- 164.Egeberg RO, Elconin AE, Egeberg MC. 1964. Effect of salinity and temperature on Coccidioides immitis and three antagonistic soil saprophytes. J Bacteriol 88:473–476. 10.1128/jb.88.2.473-476.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Roncal T, Ugalde U. 2003. Conidiation induction in Penicillium. Res Microbiol 154:539–546. 10.1016/S0923-2508(03)00168-2. [DOI] [PubMed] [Google Scholar]
- 166.Boyce KJ, Andrianopoulos A. 2013. Morphogenetic circuitry regulating growth and development in the dimorphic pathogen Penicillium marneffei. Eukaryot Cell 12:154–160. 10.1128/EC.00234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zuber S, Hynes MJ, Andrianopoulos A. 2003. The G-protein alpha-subunit GasC plays a major role in germination in the dimorphic fungus Penicillium marneffei. Genetics 164:487–499. 10.1093/genetics/164.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Boyce KJ, Andrianopoulos A. 2007. A p21-activated kinase is required for conidial germination in Penicillium marneffei. PLoS Pathog 3:e162. 10.1371/journal.ppat.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yang E, Chow W-N, Wang G, Woo PCY, Lau SKP, Yuen K-Y, Lin X, Cai JJ. 2014. Signature gene expression reveals novel clues to the molecular mechanisms of dimorphic transition in Penicillium marneffei. PLoS Genet 10:e1004662. 10.1371/journal.pgen.1004662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Suwunnakorn S, Cooper CR, Kummasook A, Vanittanakom N. 2014. Role of the yakA gene in morphogenesis and stress response in Penicillium marneffei. Microbiology (Reading) 160:1929–1939. 10.1099/mic.0.080689-0. [DOI] [PubMed] [Google Scholar]
- 171.Catlett N.L, Yoder OC, Turgeon BG. 2003. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot Cell 2:1151–1161. 10.1128/EC.2.6.1151-1161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang F, Tao J, Qian Z, You S, Dong H, Shen H, Chen X, Tang S, Ren S. 2009. A histidine kinase PmHHK1 regulates polar growth, sporulation and cell wall composition in the dimorphic fungus Penicillium marneffei. Mycol Res 113:915–923. 10.1016/j.mycres.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 173.Boyce KJ, Schreider L, Kirszenblat L, Andrianopoulos A. 2011. The two-component histidine kinases DrkA and SlnA are required for in vivo growth in the human pathogen Penicillium marneffei. Mol Microbiol 82:1164–1184. 10.1111/j.1365-2958.2011.07878.x. [DOI] [PubMed] [Google Scholar]
- 174.Nimmanee P, Woo PC, Kummasook A, Vanittanakom N. 2015. Characterization of sakA gene from pathogenic dimorphic fungus Penicillium marneffei. Int J Med Microbiol 305:65–74. 10.1016/j.ijmm.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 175.Lara-Rojas F, Sanchez O, Kawasaki L, Aguirre J. 2011. Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol Microbiol 80:436–454. 10.1111/j.1365-2958.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Canovas D, Boyce KJ, Andrianopoulos A. 2011. The fungal type II myosin in Penicillium marneffei, MyoB, is essential for chitin deposition at nascent septation sites but not actin localization. Eukaryot Cell 10:302–312. 10.1128/EC.00201-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Todd RB, Greenhalgh JR, Hynes MJ, Andrianopoulos A. 2003. TupA, the Penicillium marneffei Tup1p homologue, represses both yeast and spore development. Mol Microbiol 48:85–94. 10.1046/j.1365-2958.2003.03426.x. [DOI] [PubMed] [Google Scholar]
- 178.Dankai W, Pongpom M, Youngchim S, Cooper CR, Jr, Vanittanakom N. 2016. The yapA encodes bZIP transcription factor involved in stress tolerance in pathogenic fungus Talaromyces marneffei. PLoS One 11:e0163778. 10.1371/journal.pone.0163778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Emery P, Durand B, Mach B, Reith W. 1996. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res 24:803–807. 10.1093/nar/24.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bugeja HE, Hynes MJ, Andrianopoulos A. 2010. The RFX protein RfxA is an essential regulator of growth and morphogenesis in Penicillium marneffei. Eukaryot Cell 9:578–591. 10.1128/EC.00226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pongsunk S, Andrianopoulos A, Chaiyaroj SC. 2005. Conditional lethal disruption of TATA-binding protein gene in Penicillium marneffei. Fungal Genet Biol 42:893–903. 10.1016/j.fgb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 182.Chandler JM, Treece ER, Trenary HR, Brenneman JL, Flickner TJ, Frommelt JL, Oo ZM, Patterson MM, Rundle WT, Valle OV, Kim TD, Walker GR, Cooper CR. 2008. Protein profiling of the dimorphic, pathogenic fungus, Penicillium marneffei. Proteome Sci 6:17. 10.1186/1477-5956-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Weerasinghe H, Bugeja HE, Andrianopoulos A. 2021. The novel Dbl homology/BAR domain protein, MsgA, of Talaromyces marneffei regulates yeast morphogenesis during growth inside host cells. Sci Rep 11:2334. 10.1038/s41598-020-79593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Borneman AR, Hynes MJ, Andrianopoulos A. 2000. The abaA homologue of Penicillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol Microbiol 38:1034–1047. 10.1046/j.1365-2958.2000.02202.x. [DOI] [PubMed] [Google Scholar]
- 185.Bugeja HE, Hynes MJ, Andrianopoulos A. 2013. HgrA is necessary and sufficient to drive hyphal growth in the dimorphic pathogen Penicillium marneffei. Mol Microbiol 88:998–1014. 10.1111/mmi.12239. [DOI] [PubMed] [Google Scholar]
- 186.Lau SKP, Chow W-N, Wong AYP, Yeung JMY, Bao J, Zhang N, Lok S, Woo PCY, Yuen K-Y. 2013. Identification of microRNA-like RNAs in mycelial and yeast phases of the thermal dimorphic fungus Penicillium marneffei. PLoS Negl Trop Dis 7:e2398. 10.1371/journal.pntd.0002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Xi L, Xu X, Liu W, Li X, Liu Y, Li M, Zhang J, Li M. 2007. Differentially expressed proteins of pathogenic Penicillium marneffei in yeast and mycelial phases. J Med Microbiol 56:298–304. 10.1099/jmm.0.46808-0. [DOI] [PubMed] [Google Scholar]
- 188.Timberlake WE, Marshall MA. 1988. Genetic regulation of development in Aspergillus nidulans. Trends Genet 4:162–169. 10.1016/0168-9525(88)90022-4. [DOI] [PubMed] [Google Scholar]
- 189.Borneman AR, Hynes MJ, Andrianopoulos A. 2002. A basic helix-loop-helix protein with similarity to the fungal morphological regulators, Phd1p, Efg1p and StuA, controls conidiation but not dimorphic growth in Penicillium marneffei. Mol Microbiol 44:621–631. 10.1046/j.1365-2958.2002.02906.x. [DOI] [PubMed] [Google Scholar]
- 190.Edmondson DG, Smith MM, Roth SY. 1996. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev 10:1247–1259. 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 191.Hicks J, Lockington RA, Strauss J, Dieringer D, Kubicek CP, Kelly J, Keller N. 2001. RcoA has pleiotropic effects on Aspergillus nidulans cellular development. Mol Microbiol 39:1482–1493. 10.1046/j.1365-2958.2001.02332.x. [DOI] [PubMed] [Google Scholar]
- 192.Zheng Y-Q, Pan K-S, Latgé J-P, Andrianopoulos A, Luo H, Yan R-F, Wei J-Y, Huang C-Y, Cao C-W. 2019. Calcineurin A is essential in the regulation of asexual development, stress responses and pathogenesis in Talaromyces marneffei. Front Microbiol 10:3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zuber S, Hynes MJ, Andrianopoulos A. 2002. G-protein signaling mediates asexual development at 25 degrees C but has no effect on yeast-like growth at 37 degrees C in the dimorphic fungus Penicillium mameffei. Eukaryot Cell 1:440–447. 10.1128/EC.1.3.440-447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pasricha S, Payne M, Canovas D, Pase L, Ngaosuwankul N, Beard S, Oshlack A, Smyth GK, Chaiyaroj SC, Boyce KJ, Andrianopoulos A. 2013. Cell-type-specific transcriptional profiles of the dimorphic pathogen Penicillium marneffei reflect distinct reproductive, morphological, and environmental demands. G3 (Bethesda) 3:1997–2014. 10.1534/g3.113.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X, You D, Deng Z, Dedon PC. 2007. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol 3:709–710. 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- 196.Xiong X, Wu G, Wei Y, Liu L, Zhang Y, Su R, Jiang X, Li M, Gao H, Tian X, Zhang Y, Hu L, Chen S, Tang Y, Jiang S, Huang R, Li Z, Wang Y, Deng Z, Wang J, Dedon PC, Chen S, Wang L. 2020. SspABCD–SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities. Nat Microbiol 5:917–928. 10.1038/s41564-020-0700-6. [DOI] [PubMed] [Google Scholar]
- 197.Wang L, Jiang S, Deng Z, Dedon PC, Chen S. 2019. DNA phosphorothioate modification—a new multi-functional epigenetic system in bacteria. FEMS Microbiol Rev 43:109–122. 10.1093/femsre/fuy036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Gao L, Altae-Tran H, Böhning F, Makarova KS, Segel M, Schmid-Burgk JL, Koob J, Wolf YI, Koonin EV, Zhang F. 2020. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369:1077–1084. 10.1126/science.aba0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Romani L. 2011. Immunity to fungal infections. Nat Rev Immunol 11:275–288. 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 200.Pathakumari B, Liang G, Liu W. 2020. Immune defence to invasive fungal infections: a comprehensive review. Biomed Pharmacother 130:110550. 10.1016/j.biopha.2020.110550. [DOI] [PubMed] [Google Scholar]
- 201.Erwig LP, Gow NA. 2016. Interactions of fungal pathogens with phagocytes. Nat Rev Microbiol 14:163–176. 10.1038/nrmicro.2015.21. [DOI] [PubMed] [Google Scholar]
- 202.Hoft MA, Duvenage L, Hoving JC. 2022. Key thermally dimorphic fungal pathogens: shaping host immunity. Open Biol 12:210219. 10.1098/rsob.210219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hu Y, Lu S, Xi L. 2020. Murine macrophage requires CD11b to recognize Talaromyces marneffei. Infect Drug Resist 13:911–920. 10.2147/IDR.S237401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ngaosuwankul P, Pongtanalert P, Engering A, Chaiyaroj SC. 2008. Differential gene expression profiles of human monocyte-derived antigen presenting cells in response to Penicillium marneffei: roles of DC-SIGN (CD209) in fungal cell uptake. Asian Pac J Allergy Immunol 26:151–163. [PubMed] [Google Scholar]
- 205.Srinoulprasert Y, Pongtanalert P, Chawengkirttikul R, Chaiyaroj SC. 2009. Engagement of Penicillium marneffei conidia with multiple pattern recognition receptors on human monocytes. Microbiol Immunol 53:162–172. 10.1111/j.1348-0421.2008.00102.x. [DOI] [PubMed] [Google Scholar]
- 206.Wang M, Li L, Xiao S, Chen W, Hu F, Li F, Guo P, Chen X, Cai W, Tang X. 2021. The association of TLR2, TLR3, and TLR9 gene polymorphisms with susceptibility to talaromycosis among Han Chinese AIDS patients in Guangdong. Front Cell Infect Microbiol 11:625461. 10.3389/fcimb.2021.625461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Leopold Wager CM, Wormley FL, Jr.. 2014. Classical versus alternative macrophage activation: the Ying and the Yang in host defense against pulmonary fungal infections. Mucosal Immunol 7:1023–1035. 10.1038/mi.2014.65. [DOI] [PubMed] [Google Scholar]
- 208.Martinez FO, Gordon S. 2014. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 6:13. 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kraaij MD, Vereyken EJF, Leenen PJM, van den Bosch TPP, Rezaee F, Betjes MGH, Baan CC, Rowshani AT. 2014. Human monocytes produce interferon-gamma upon stimulation with LPS. Cytokine 67:7–12. 10.1016/j.cyto.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 210.Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, Brown GD. 2007. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol 8:31–38. 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Vautier S, MacCallum DM, Brown GD. 2012. C-type lectin receptors and cytokines in fungal immunity. Cytokine 58:89–99. 10.1016/j.cyto.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 212.Dai X, Mao C, Lan X, Chen H, Li M, Bai J, Deng J, Liang Q, Zhang J, Zhong X, Liang Y, Fan J, Luo H, He Z. 2017. Acute Penicillium marneffei infection stimulates host M1/M2a macrophages polarization in BALB/C mice. BMC Microbiol 17:177. 10.1186/s12866-017-1086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wei W, Ning C, Huang J, Wang G, Lai J, Han J, He J, Zhang H, Liang B, Liao Y, Le T, Luo Q, Li Z, Jiang J, Ye L, Liang H. 2021. Talaromyces marneffei promotes M2 polarization of human macrophages by downregulating SOCS3 expression and activating the TLR9 pathway. Virulence 12:1997–2012. 10.1080/21505594.2021.1958470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yang D, Shen L-X, Chen R-F, Fu Y, Xu H-Y, Zhang L-N, Liu D-H. 2021. The effect of Talaromyces marneffei infection on CD86 expression in THP-1 cells. Infect Drug Resist 14:651–660. 10.2147/IDR.S297160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lu S, Li D, Xi L, Calderone R. 2019. Interplay of interferon-gamma and macrophage polarization during Talaromyces marneffei infection. Microb Pathog 134:103594. 10.1016/j.micpath.2019.103594. [DOI] [PubMed] [Google Scholar]
- 216.Li J-G, Du Y-M, Yan Z-D, Yan J, Zhuansun Y-X, Chen R, Zhang W, Feng S-L, Ran P-X. 2016. CD80 and CD86 knockdown in dendritic cells regulates Th1/Th2 cytokine production in asthmatic mice. Exp Ther Med 11:878–884. 10.3892/etm.2016.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Price JV, Vance RE. 2014. The macrophage paradox. Immunity 41:685–693. 10.1016/j.immuni.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 218.Wang L, Wu J, Li J, Yang H, Tang T, Liang H, Zuo M, Wang J, Liu H, Liu F, Chen J, Liu Z, Wang Y, Peng C, Wu X, Zheng R, Huang X, Ran Y, Rao Z, Ge B. 2020. Host-mediated ubiquitination of a mycobacterial protein suppresses immunity. Nature 577:682–688. 10.1038/s41586-019-1915-7. [DOI] [PubMed] [Google Scholar]
- 219.Wagener J, MacCallum DM, Brown GD, Gow NA. 2017. Candida albicans chitin increases arginase-1 activity in human macrophages, with an impact on macrophage antimicrobial functions. mBio 8:e01820-16. 10.1128/mBio.01820-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ellett F, Pazhakh V, Pase L, Benard EL, Weerasinghe H, Azabdaftari D, Alasmari S, Andrianopoulos A, Lieschke GJ. 2018. Macrophages protect Talaromyces marneffei conidia from myeloperoxidase-dependent neutrophil fungicidal activity during infection establishment in vivo. PLoS Pathog 14:e1007063. 10.1371/journal.ppat.1007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wei W, Wang G, Zhang H, Bao X, An S, Luo Q, He J, Chen L, Liu Y, Ning C, Lai J, Yuan Z, Chen R, Jiang J, Ye L, Liang H. 13 July 2022. Talaromyces marneffei suppresses human macrophages inflammatory by producing the truncated protein NCOR2-013 via TUT1-regulated alternative splicing. bioRxiv. 10.1101/2022.07.11.499655. [DOI]
- 222.Swanson KV, Deng M, Ting JP. 2019. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol 19:477–489. 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kelley N, Jeltema D, Duan Y, He Y. 2019. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci 20:3328. 10.3390/ijms20133328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ma H, Chan JFW, Tan YP, Kui L, Tsang C-C, Pei SLC, Lau Y-L, Woo PCY, Lee PP. 2021. NLRP3 inflammasome contributes to host defense against Talaromyces marneffei infection. Front Immunol 12:760095. 10.3389/fimmu.2021.760095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lehrer RI, Cline MJ. 1969. Leukocyte myeloperoxidase deficiency and disseminated candidiasis: the role of myeloperoxidase in resistance to Candida infection. J Clin Invest 48:1478–1488. 10.1172/JCI106114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Metzler KD, Fuchs TA, Nauseef WM, Reumaux D, Roesler J, Schulze I, Wahn V, Papayannopoulos V, Zychlinsky A. 2011. Myeloperoxidase is required for neutrophil extracellular trap formation: implications for innate immunity. Blood 117:953–959. 10.1182/blood-2010-06-290171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Pattingre S, Bauvy C, Codogno P. 2003. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J Biol Chem 278:16667–16674. 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- 228.Hu P, Lai D, Lu P, Gao J, He H. 2012. ERK and Akt signaling pathways are involved in advanced glycation end product-induced autophagy in rat vascular smooth muscle cells. Int J Mol Med 29:613–618. 10.3892/ijmm.2012.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Chen R, Li X, Lu S, Ma T, Huang X, Mylonakis E, Liang Y, Xi L. 2014. Role of extracellular signal-regulated kinases 1 and 2 and p38 mitogen-activated protein kinase pathways in regulating replication of Penicillium marneffei in human macrophages. Microbes Infect 16:401–408. 10.1016/j.micinf.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 230.Chen R, Ji G, Xi L, Zhang T, Zheng D, Li X, Ren H. 2018. Role of autophagy in regulating the immune response of dendritic cells to Talaromyces marneffei infection. Microb Pathog 123:120–125. 10.1016/j.micpath.2018.06.044. [DOI] [PubMed] [Google Scholar]
- 231.Chen R, Ji G, Ren H, Liu Z, Feng S, Zhang T, Xi L, Li X. 2020. Activation of autophagy and IL-10 production are regulated by Jun N-terminal kinase 1 and 2 and p38 mitogen activated protein kinase signaling pathways during Talaromyces marneffei infection within dendritic cells. Microb Pathog 139:103891. 10.1016/j.micpath.2019.103891. [DOI] [PubMed] [Google Scholar]
- 232.Lekkerkerker AN, van Kooyk Y, Geijtenbeek TB. 2006. Viral piracy: HIV-1 targets dendritic cells for transmission. Curr HIV Res 4:169–176. 10.2174/157016206776055020. [DOI] [PubMed] [Google Scholar]
- 233.Wilkinson J, Cunningham AL. 2006. Mucosal transmission of HIV-1: first stop dendritic cells. Curr Drug Targets 7:1563–1569. 10.2174/138945006779025482. [DOI] [PubMed] [Google Scholar]
- 234.Tang Y, Zhang H, Xu H, Zeng W, Qiu Y, Tan C, Tang S, Zhang J. 2020. Dendritic cells promote Treg expansion but not Th17 generation in response to Talaromyces marneffei yeast cells. Infect Drug Resist 13:805–813. 10.2147/IDR.S239906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Qin Y, Li Y, Liu W, Tian R, Guo Q, Li S, Li H, Zhang D, Zheng Y, Wu L, Lan K, Wang J. 2011. Penicillium marneffei-stimulated dendritic cells enhance HIV-1 trans-infection and promote viral infection by activating primary CD4+ T cells. PLoS One 6:e27609. 10.1371/journal.pone.0027609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ning Q-Y, Liu N, Wu J-Z, Hu D-F, Wei Q, Zhou J-A, Zou J, Zang N, Li G-J. 2021. Serum exosomal microRNA profiling in AIDS complicated with Talaromyces marneffei infection. Infect Drug Resist 14:4931–4948. 10.2147/IDR.S338321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Li Y, Li M, Luo H, Bai J, Zhang J, Zhong X, Lan X, He Z. 2017. Expression profile of lncRNA in human bronchial epithelial cells response to Talaromyces marneffei infection: a microarray analysis. Microb Pathog 104:155–160. 10.1016/j.micpath.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 238.Li Y, Chen H, Li S, Li Y, Liu G, Bai J, Luo H, Lan X, He Z. 2019. LncSSBP1 functions as a negative regulator of IL-6 through interaction with hnRNPK in bronchial epithelial cells infected with Talaromyces marneffei. Front Immunol 10:2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Faguy DM. 2011. Fungal pathogens: an overview. Radiol Technol 82:321–340. [PubMed] [Google Scholar]
- 240.Lau SKP, Tse H, Chan JSY, Zhou AC, Curreem SOT, Lau CCY, Yuen K-Y, Woo PCY. 2013. Proteome profiling of the dimorphic fungus Penicillium marneffei extracellular proteins and identification of glyceraldehyde-3-phosphate dehydrogenase as an important adhesion factor for conidial attachment. FEBS J 280:6613–6626. 10.1111/febs.12566. [DOI] [PubMed] [Google Scholar]
- 241.Hamilton AJ, Jeavons L, Youngchim S, Vanittanakom N, Hay RJ. 1998. Sialic acid-dependent recognition of laminin by Penicillium marneffei conidia. Infect Immun 66:6024–6026. 10.1128/IAI.66.12.6024-6026.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hamilton AJ, Jeavons L, Youngchim S, Vanittanakom N. 1999. Recognition of fibronectin by Penicillium marneffei conidia via a sialic acid-dependent process and its relationship to the interaction between conidia and laminin. Infect Immun 67:5200–5205. 10.1128/IAI.67.10.5200-5205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gupta A. 2019. Interaction of a pathogen with its host: the Talaromyces marneffei cell wall and its role in pathogenicity. Thesis. The University of Melbourne. [Google Scholar]
- 244.Lam W-H, Sze K-H, Ke Y, Tse M-K, Zhang H, Woo PCY, Lau SKP, Lau CCY, Xu S, Lai P-M, Zhou T, Antonyuk SV, Kao RYT, Yuen K-Y, Hao Q. 2019. Talaromyces marneffei Mp1 protein, a novel virulence factor, carries two arachidonic acid-binding domains to suppress inflammatory responses in hosts. Infect Immun 87:e00679-18. 10.1128/IAI.00679-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Sze K-H, Lam W-H, Zhang H, Ke Y-H, Tse M-K, Woo PCY, Lau SKP, Lau CCY, Cai J-P, Tung ETK, Lo RKC, Xu S, Kao RYT, Hao Q, Yuen K-Y. 2017. Talaromyces marneffei Mp1p is a virulence factor that binds and sequesters a key proinflammatory lipid to dampen host innate immune response. Cell Chem Biol 24:182–194. 10.1016/j.chembiol.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 246.Cao L, Chan CM, Lee C, Wong SS, Yuen KY. 1998. MP1 encodes an abundant and highly antigenic cell wall mannoprotein in the pathogenic fungus Penicillium marneffei. Infect Immun 66:966–973. 10.1128/IAI.66.3.966-973.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Eisenman HC, Casadevall A. 2012. Synthesis and assembly of fungal melanin. Appl Microbiol Biotechnol 93:931–940. 10.1007/s00253-011-3777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Taborda CP, da Silva MB, Nosanchuk JD, Travassos LR. 2008. Melanin as a virulence factor of Paracoccidioides brasiliensis and other dimorphic pathogenic fungi: a minireview. Mycopathologia 165:331–339. 10.1007/s11046-007-9061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Youngchim S, Hay RJ, Hamilton AJ. 2005. Melanization of Penicillium marneffei in vitro and in vivo. Microbiology (Reading) 151:291–299. 10.1099/mic.0.27433-0. [DOI] [PubMed] [Google Scholar]
- 250.Liu D, Wei L, Guo T, Tan W. 2014. Detection of DOPA-melanin in the dimorphic fungal pathogen Penicillium marneffei and its effect on macrophage phagocytosis in vitro. PLoS One 9:e92610. 10.1371/journal.pone.0092610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Sapmak A, Kaewmalakul J, Nosanchuk JD, Vanittanakom N, Andrianopoulos A, Pruksaphon K, Youngchim S. 2016. Talaromyces marneffei laccase modifies THP-1 macrophage responses. Virulence 7:702–717. 10.1080/21505594.2016.1193275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Woo PCY, Tam EWT, Chong KTK, Cai JJ, Tung ETK, Ngan AHY, Lau SKP, Yuen K-Y. 2010. High diversity of polyketide synthase genes and the melanin biosynthesis gene cluster in Penicillium marneffei. FEBS J 277:3750–3758. 10.1111/j.1742-4658.2010.07776.x. [DOI] [PubMed] [Google Scholar]
- 253.Gantner BN, Simmons RM, Underhill DM. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J 24:1277–1286. 10.1038/sj.emboj.7600594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Boyce KJ, De Souza DP, Dayalan S, Pasricha S, Tull D, McConville MJ, Andrianopoulos A. 2018. Talaromyces marneffei simA encodes a fungal cytochrome P450 essential for survival in macrophages. mSphere 3:e00056-18. 10.1128/mSphere.00056-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Fox DS, Cruz MC, Sia RA, Ke H, Cox GM, Cardenas ME, Heitman J. 2001. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol Microbiol 39:835–849. 10.1046/j.1365-2958.2001.02295.x. [DOI] [PubMed] [Google Scholar]
- 256.Chen Y-L, Yu S-J, Huang H-Y, Chang Y-L, Lehman VN, Silao FGS, Bigol UG, Bungay AAC, Averette A, Heitman J. 2014. Calcineurin controls hyphal growth, virulence, and drug tolerance of Candida tropicalis. Eukaryot Cell 13:844–854. 10.1128/EC.00302-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Juvvadi PR, Lamoth F, Steinbach WJ. 2014. Calcineurin-mediated regulation of hyphal growth, septation, and virulence in Aspergillus fumigatus. Mycopathologia 178:341–348. 10.1007/s11046-014-9794-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Fox DS, Heitman J. 2002. Good fungi gone bad: the corruption of calcineurin. Bioessays 24:894–903. 10.1002/bies.10157. [DOI] [PubMed] [Google Scholar]
- 259.Chen R, Ji G, Ma T, Huang X, Ren H, Xi L. 2015. Role of intracellular free calcium in killing Penicillium marneffei within human macrophages. Microb Pathog 83-84:29–34. 10.1016/j.micpath.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 260.Stewart AA, Ingebritsen TS, Manalan A, Klee CB, Cohen P. 1982. Discovery of a Ca2+- and calmodulin-dependent protein phosphatase: probable identity with calcineurin (CaM-BP80). FEBS Lett 137:80–84. 10.1016/0014-5793(82)80319-0. [DOI] [PubMed] [Google Scholar]
- 261.Brown AJ, Leach MD, Nicholls S. 2010. The relevance of heat shock regulation in fungal pathogens of humans. Virulence 1:330–332. 10.4161/viru.1.4.12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Leach MD, Farrer RA, Tan K, Miao Z, Walker LA, Cuomo CA, Wheeler RT, Brown AJP, Wong KH, Cowen LE. 2016. Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans. Nat Commun 7:11704. 10.1038/ncomms11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Kummasook A, Pongpom P, Vanittanakom N. 2007. Cloning, characterization and differential expression of an hsp70 gene from the pathogenic dimorphic fungus, Penicillium marneffei. DNA Seq 18:385–394. 10.1080/10425170701309012. [DOI] [PubMed] [Google Scholar]
- 264.Vanittanakom N, Pongpom M, Praparattanapan J, Cooper CR, Sirisanthana T. 2009. Isolation and expression of heat shock protein 30 gene from Penicillium marneffei. Med Mycol 47:521–526. 10.1080/13693780802566358. [DOI] [PubMed] [Google Scholar]
- 265.Kovács D, Sigmond T, Hotzi B, Bohár B, Fazekas D, Deák V, Vellai T, Barna J. 2019. HSF1Base: a comprehensive database of HSF1 (heat shock factor 1) target genes. Int J Mol Sci 20:5815. 10.3390/ijms20225815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Missall TA, Lodge JK, McEwen JE. 2004. Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts. Eukaryot Cell 3:835–846. 10.1128/EC.3.4.835-846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Cogliati M, Roverselli A, Boelaert JR, Taramelli D, Lombardi L, Viviani MA. 1997. Development of an in vitro macrophage system to assess Penicillium marneffei growth and susceptibility to nitric oxide. Infect Immun 65:279–284. 10.1128/iai.65.1.279-284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Thirach S, Cooper CR, Jr, Vanittanakom P, Vanittanakom N. 2007. The copper, zinc superoxide dismutase gene of Penicillium marneffei: cloning, characterization, and differential expression during phase transition and macrophage infection. Med Mycol 45:409–417. 10.1080/13693780701381271. [DOI] [PubMed] [Google Scholar]
- 269.Pongpom P, Cooper CR, Jr, Vanittanakom N. 2005. Isolation and characterization of a catalase-peroxidase gene from the pathogenic fungus, Penicillium marneffei. Med Mycol 43:403–411. 10.1080/13693780400007144. [DOI] [PubMed] [Google Scholar]
- 270.Nimmanee P, Woo PC, Vanittanakom P, Youngchim S, Vanittanakom N. 2014. Functional analysis of atfA gene to stress response in pathogenic thermal dimorphic fungus Penicillium marneffei. PLoS One 9:e111200. 10.1371/journal.pone.0111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Semenza GL. 2011. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harbor Symp Quant Biol 76:347–353. 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- 272.Askew C, Sellam A, Epp E, Hogues H, Mullick A, Nantel A, Whiteway M. 2009. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog 5:e1000612. 10.1371/journal.ppat.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Setiadi ER, Doedt T, Cottier F, Noffz C, Ernst JF. 2006. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J Mol Biol 361:399–411. 10.1016/j.jmb.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 274.Synnott JM, Guida A, Mulhern-Haughey S, Higgins DG, Butler G. 2010. Regulation of the hypoxic response in Candida albicans. Eukaryot Cell 9:1734–1746. 10.1128/EC.00159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Youngchim S, Vanittanakom N, Hamilton AJ. 1999. Analysis of the enzymatic activity of mycelial and yeast phases of Penicillium marneffei. Med Mycol 37:445–450. 10.1046/j.1365-280x.1999.00235.x. [DOI] [PubMed] [Google Scholar]
- 276.Taramelli D, Tognazioli C, Ravagnani F, Leopardi O, Giannulis G, Boelaert JR. 2001. Inhibition of intramacrophage growth of Penicillium marneffei by 4-aminoquinolines. Antimicrob Agents Chemother 45:1450–1455. 10.1128/AAC.45.5.1450-1455.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Yang B, Wang J, Jiang H, Lin H, Ou Z, Ullah A, Hua Y, Chen J, Lin X, Hu X, Zheng L, Wang Q. 2020. Extracellular vesicles derived from Talaromyces marneffei yeasts mediate inflammatory response in macrophage cells by bioactive protein components. Front Microbiol 11:603183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell 6:48–59. 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Vargas G, Rocha JDB, Oliveira DL, Albuquerque PC, Frases S, Santos SS, Nosanchuk JD, Gomes AMO, Medeiros LCAS, Miranda K, Sobreira TJP, Nakayasu ES, Arigi EA, Casadevall A, Guimaraes AJ, Rodrigues ML, Freire-de-Lima CG, Almeida IC, Nimrichter L. 2015. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell Microbiol 17:389–407. 10.1111/cmi.12374. [DOI] [PubMed] [Google Scholar]
- 280.Haas A. 2007. The phagosome: compartment with a license to kill. Traffic 8:311–330. 10.1111/j.1600-0854.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- 281.Thirach S, Cooper CR, Vanittanakom N. 2008. Molecular analysis of the Penicillium marneffei glyceraldehyde-3-phosphate dehydrogenase-encoding gene (gpdA) and differential expression of gpdA and the isocitrate lyase-encoding gene (acuD) upon internalization by murine macrophages. J Med Microbiol 57:1322–1328. 10.1099/jmm.0.2008/002832-0. [DOI] [PubMed] [Google Scholar]
- 282.Canovas D, Andrianopoulos A. 2006. Developmental regulation of the glyoxylate cycle in the human pathogen Penicillium marneffei. Mol Microbiol 62:1725–1738. 10.1111/j.1365-2958.2006.05477.x. [DOI] [PubMed] [Google Scholar]
- 283.Hwang L, Hocking-Murray D, Bahrami AK, Andersson M, Rine J, Sil A. 2003. Identifying phase-specific genes in the fungal pathogen Histoplasma capsulatum using a genomic shotgun microarray. Mol Biol Cell 14:2314–2326. 10.1091/mbc.e03-01-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Keller S, Macheleidt J, Scherlach K, Schmaler-Ripcke J, Jacobsen ID, Heinekamp T, Brakhage AA. 2011. Pyomelanin formation in Aspergillus fumigatus requires HmgX and the transcriptional activator HmgR but is dispensable for virulence. PLoS One 6:e26604. 10.1371/journal.pone.0026604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Boyce KJ, McLauchlan A, Schreider L, Andrianopoulos A. 2015. Intracellular growth is dependent on tyrosine catabolism in the dimorphic fungal pathogen Penicillium marneffei. PLoS Pathog 11:e1004790. 10.1371/journal.ppat.1004790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Schwab MA, Sauer SW, Okun JG, Nijtmans LGJ, Rodenburg RJT, van den Heuvel LP, Dröse S, Brandt U, Hoffmann GF, Ter Laak H, Kölker S, Smeitink JAM. 2006. Secondary mitochondrial dysfunction in propionic aciduria: a pathogenic role for endogenous mitochondrial toxins. Biochem J 398:107–112. 10.1042/BJ20060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Feng J, He L, Xiao X, Chen Z, Chen C, Chu J, Lu S, Li X, Mylonakis E, Xi L. 2020. Methylcitrate cycle gene MCD is essential for the virulence of Talaromyces marneffei. Med Mycol 58:351–361. 10.1093/mmy/myz063. [DOI] [PubMed] [Google Scholar]
- 288.Vylkova S, Lorenz MC. 2017. Phagosomal neutralization by the fungal pathogen Candida albicans induces macrophage pyroptosis. Infect Immun 85:e00832-16. 10.1128/IAI.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat Rev Microbiol 2:946–953. 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 290.Lonergan ZR, Skaar EP. 2019. Nutrient zinc at the host-pathogen interface. Trends Biochem Sci 44:1041–1056. 10.1016/j.tibs.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Weinberg ED, Miklossy J. 2008. Iron withholding: a defense against disease. J Alzheimers Dis 13:451–463. 10.3233/jad-2008-13409. [DOI] [PubMed] [Google Scholar]
- 292.Taramelli D, Brambilla S, Sala G, Bruccoleri A, Tognazioli C, Riviera-Uzielli L, Boelaert JR. 2000. Effects of iron on extracellular and intracellular growth of Penicillium marneffei. Infect Immun 68:1724–1726. 10.1128/IAI.68.3.1724-1726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Pasricha S, Schafferer L, Lindner H, Joanne Boyce K, Haas H, Andrianopoulos A. 2016. Differentially regulated high-affinity iron assimilation systems support growth of the various cell types in the dimorphic pathogen Talaromyces marneffei. Mol Microbiol 102:715–737. 10.1111/mmi.13489. [DOI] [PubMed] [Google Scholar]
- 294.Pongpom M, Amsri A, Sukantamala P, Suwannaphong P, Jeenkeawpieam J. 2020. Expression of Talaromyces marneffei acuM and acuK genes in gluconeogenic substrates and various iron concentrations. J Fungi (Basel) 6:102. 10.3390/jof6030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Amsri A, Jeenkeawpieam J, Sukantamala P, Pongpom M. 2021. Role of acuK in control of iron acquisition and gluconeogenesis in Talaromyces marneffei. J Fungi (Basel) 7:798. 10.3390/jof7100798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Wilson D. 2015. An evolutionary perspective on zinc uptake by human fungal pathogens. Metallomics 7:979–985. 10.1039/c4mt00331d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Gerwien F, Skrahina V, Kasper L, Hube B, Brunke S. 2018. Metals in fungal virulence. FEMS Microbiol Rev 42:fux050. 10.1093/femsre/fux050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Vicentefranqueira R, Amich J, Marín L, Sánchez C, Leal F, Calera J. 2018. The transcription factor ZafA regulates the homeostatic and adaptive response to zinc starvation in Aspergillus fumigatus. Genes (Basel) 9:318. 10.3390/genes9070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Samanovic MI, Ding C, Thiele D.J, Darwin KH. 2012. Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe 11:106–115. 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Rees EM, Thiele DJ. 2004. From aging to virulence: forging connections through the study of copper homeostasis in eukaryotic microorganisms. Curr Opin Microbiol 7:175–184. 10.1016/j.mib.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 301.Ding C, Yin J, Tovar EMM, Fitzpatrick DA, Higgins DG, Thiele DJ. 2011. The copper regulon of the human fungal pathogen Cryptococcus neoformans H99. Mol Microbiol 81:1560–1576. 10.1111/j.1365-2958.2011.07794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ding C, Festa RA, Chen Y-L, Espart A, Palacios Ò, Espín J, Capdevila M, Atrian S, Heitman J, Thiele DJ. 2013. Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe 13:265–276. 10.1016/j.chom.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, Murciano C, Blagojevic M, Thavaraj S, Förster TM, Hebecker B, Kasper L, Vizcay G, Iancu SI, Kichik N, Häder A, Kurzai O, Luo T, Krüger T, Kniemeyer O, Cota E, Bader O, Wheeler RT, Gutsmann T, Hube B, Naglik JR. 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532:64–68. 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Keller NP. 2019. Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol 17:167–180. 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Sugui JA, Pardo J, Chang YC, Zarember KA, Nardone G, Galvez EM, Müllbacher A, Gallin JI, Simon MM, Kwon-Chung KJ. 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot Cell 6:1562–1569. 10.1128/EC.00141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Collemare J, Billard A, Bohnert HU, Lebrun MH. 2008. Biosynthesis of secondary metabolites in the rice blast fungus Magnaporthe grisea: the role of hybrid PKS-NRPS in pathogenicity. Mycol Res 112:207–215. 10.1016/j.mycres.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 307.Woo PCY, Lam C-W, Tam EWT, Lee K-C, Yung KKY, Leung CKF, Sze K-H, Lau SKP, Yuen K-Y. 2014. The biosynthetic pathway for a thousand-year-old natural food colorant and citrinin in Penicillium marneffei. Sci Rep 4:6728. 10.1038/srep06728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Woo PCY, Lam C-W, Tam EWT, Leung CKF, Wong SSY, Lau SKP, Yuen K-Y. 2012. First discovery of two polyketide synthase genes for mitorubrinic acid and mitorubrinol yellow pigment biosynthesis and implications in virulence of Penicillium marneffei. PLoS Negl Trop Dis 6:e1871. 10.1371/journal.pntd.0001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Ghannoum MA. 2000. Potential role of phospholipases in virulence and fungal pathogenesis. Clin Microbiol Rev 13:122–143. 10.1128/CMR.13.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.He Y, Li L, Hu F, Chen W, Lei H, Chen X, Cai W, Tang X. 2016. Expression and characterization of a Talaromyces marneffei active phospholipase B expressed in a Pichia pastoris expression system. Emerg Microbes Infect 5:e120. 10.1038/emi.2016.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Kudeken N, Kawakami K, Saito A. 1998. Different susceptibilities of yeasts and conidia of Penicillium marneffei to nitric oxide (NO)-mediated fungicidal activity of murine macrophages. Clin Exp Immunol 112:287–293. 10.1046/j.1365-2249.1998.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Stangl K. 2008. Comparative proteomic analysis of phase-switch in the dimorphic fungus, Penicillium marneffei. Thesis. Youngstown State University. [Google Scholar]
- 313.Rowe G. 2011. Analysis of mold and yeast phosphoproteomes in the dimorphic fungus Penicillium marneffei. Thesis. Youngstown State University. [Google Scholar]
- 314.Wang F, Sethiya P, Hu X, Guo S, Chen Y, Li A, Tan K, Wong KH. 2021. Transcription in fungal conidia before dormancy produces phenotypically variable conidia that maximize survival in different environments. Nat Microbiol 6:1066–1081. 10.1038/s41564-021-00922-y. [DOI] [PubMed] [Google Scholar]
- 315.Tan K, Wong KH. 2019. RNA polymerase II ChIP-seq—a powerful and highly affordable method for studying fungal genomics and physiology. Biophys Rev 11:79–82. 10.1007/s12551-018-00497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Delliere S, et al. 2020. Risk factors associated with COVID-19-associated pulmonary aspergillosis in ICU patients: a French multicentric retrospective cohort. Clin Microbiol Infect 27:790.e1–790.e5. 10.1016/j.cmi.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Hoenigl M, Seidel D, Sprute R, Cunha C, Oliverio M, Goldman GH, Ibrahim AS, Carvalho A. 2022. COVID-19-associated fungal infections. Nat Microbiol 7:1127–1140. 10.1038/s41564-022-01172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Bourassa L, Doppalapudi A, Butler-Wu SM. 2019. The brief case: pneumonia caused by Talaromyces marneffei. J Clin Microbiol 57:e01690-18. 10.1128/JCM.01690-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Deesomchok A, Tanprawate S. 2006. A 12-case series of Penicillium marneffei pneumonia. J Med Assoc Thai 89:441–447. [PubMed] [Google Scholar]
- 320.Rajasingham R, Boulware DR. 2012. Reconsidering cryptococcal antigen screening in the U.S. among persons with CD4 <100 cells/mcL. Clin Infect Dis 55:1742–1744. 10.1093/cid/cis725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Smilack JD. 1999. Trimethoprim-sulfamethoxazole. Mayo Clin Proc 74:730–734. 10.4065/74.7.730. [DOI] [PubMed] [Google Scholar]
- 322.Fernández-Villa D, Aguilar MR, Rojo L. 2019. Folic acid antagonists: antimicrobial and immunomodulating mechanisms and applications. Int J Mol Sci 20:4996. 10.3390/ijms20204996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Jiang J, Qin F, Meng S, Nehl EJ, Huang J, Liu Y, Zou J, Dong W, Huang J, Chen H, Zang N, Liang B, Ning C, Liao Y, Luo C, Liu H, Liu X, Wang J, Zhou O, Le T, Ye L, Wu F, Liang H. 2019. Effects of cotrimoxazole prophylaxis on Talaromyces marneffei infection in HIV/AIDS patients receiving antiretroviral therapy: a retrospective cohort study. Emerg Microbes Infect 8:367–376. 10.1080/22221751.2019.1588078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Chen J, et al. 26 April 2022. Role and mechanism of cotrimoxazole resistance to Talaromyces marneffei fungus in vitro. Res Sq 10.21203/rs.3.rs-1546115/v1. [DOI] [Google Scholar]
- 325.Sun J, Li X, Feng P, Zhang J, Xie Z, Song E, Xi L. 2014. RNAi-mediated silencing of fungal acuD gene attenuates the virulence of Penicillium marneffei. Med Mycol 52:167–178. 10.1093/mmy/myt006. [DOI] [PubMed] [Google Scholar]
- 326.Bugeja HE, Boyce KJ, Weerasinghe H, Beard S, Jeziorowski A, Pasricha S, Payne M, Schreider L, Andrianopoulos A. 2012. Tools for high efficiency genetic manipulation of the human pathogen Penicillium marneffei. Fungal Genet Biol 49:772–778. 10.1016/j.fgb.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 327.Zhang X, Hu X, Jan S, Rasheed SM, Zhang Y, Du M, Yang E. 2021. Development of CRISPR-Cas9 genome editing system in Talaromyces marneffei. Microb Pathog 154:104822. 10.1016/j.micpath.2021.104822. [DOI] [PubMed] [Google Scholar]
- 328.Heung LJ, Jhingran A, Hohl TM. 2015. Deploying FLAREs to visualize functional outcomes of host-pathogen encounters. PLoS Pathog 11:e1004912. 10.1371/journal.ppat.1004912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Santana DJ, O’Meara TR. 2021. Forward and reverse genetic dissection of morphogenesis identifies filament-competent Candida auris strains. Nat Commun 12:7197. 10.1038/s41467-021-27545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Weerasinghe H, Payne M, Beard S, Andrianopoulos A. 2016. Organism-wide studies into pathogenicity and morphogenesis in Talaromyces marneffei. Future Microbiol 11:511–526. 10.2217/fmb.16.9. [DOI] [PubMed] [Google Scholar]
- 331.Tsai CJ, Loh JM, Proft T. 2016. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7:214–229. 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Cook SM, McArthur JD. 2013. Developing Galleria mellonella as a model host for human pathogens. Virulence 4:350–353. 10.4161/viru.25240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Knox BP, Deng Q, Rood M, Eickhoff JC, Keller NP, Huttenlocher A. 2014. Distinct innate immune phagocyte responses to Aspergillus fumigatus conidia and hyphae in zebrafish larvae. Eukaryot Cell 13:1266–1277. 10.1128/EC.00080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Kuo Z-Y, Chuang Y-J, Chao C-C, Liu F-C, Lan C-Y, Chen B-S. 2013. Identification of infection- and defense-related genes via a dynamic host-pathogen interaction network using a Candida albicans-zebrafish infection model. J Innate Immun 5:137–152. 10.1159/000347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Tenor JL, Oehlers SH, Yang JL, Tobin DM, Perfect JR. 2015. Live imaging of host-parasite interactions in a zebrafish infection model reveals cryptococcal determinants of virulence and central nervous system invasion. mBio 6:e01425-15. 10.1128/mBio.01425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Pazhakh V, Ellett F, Croker BA, O'Donnell JA, Pase L, Schulze KE, Greulich RS, Gupta A, Reyes-Aldasoro CC, Andrianopoulos A, Lieschke GJ. 2019. β-Glucan-dependent shuttling of conidia from neutrophils to macrophages occurs during fungal infection establishment. PLoS Biol 17:e3000113. 10.1371/journal.pbio.3000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Kudeken N, Kawakami K, Kusano N, Saito A. 1996. Cell-mediated immunity in host resistance against infection caused by Penicillium marneffei. J Med Vet Mycol 34:371–378. 10.1080/02681219680000671. [DOI] [PubMed] [Google Scholar]
- 338.Kudeken N, Kawakami K, Saito A. 1997. CD4+ T cell-mediated fatal hyperinflammatory reactions in mice infected with Penicillium marneffei. Clin Exp Immunol 107:468–473. 10.1046/j.1365-2249.1997.d01-945.x. [DOI] [PubMed] [Google Scholar]
- 339.Sisto F, Miluzio A, Leopardi O, Mirra M, Boelaert JR, Taramelli D. 2003. Differential cytokine pattern in the spleens and livers of BALB/c mice infected with Penicillium marneffei: protective role of gamma interferon. Infect Immun 71:465–473. 10.1128/IAI.71.1.465-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Kleshchevnikov V, Shmatko A, Dann E, Aivazidis A, King HW, Li T, Elmentaite R, Lomakin A, Kedlian V, Gayoso A, Jain MS, Park JS, Ramona L, Tuck E, Arutyunyan A, Vento-Tormo R, Gerstung M, James L, Stegle O, Bayraktar OA. 2022. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat Biotechnol 40:661–671. 10.1038/s41587-021-01139-4. [DOI] [PubMed] [Google Scholar]
- 341.Rao A, Barkley D, França GS, Yanai I. 2021. Exploring tissue architecture using spatial transcriptomics. Nature 596:211–220. 10.1038/s41586-021-03634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Westermann AJ, Vogel J. 2021. Cross-species RNA-seq for deciphering host-microbe interactions. Nat Rev Genet 22:361–378. 10.1038/s41576-021-00326-y. [DOI] [PubMed] [Google Scholar]
- 343.Seelbinder B, Wallstabe J, Marischen L, Weiss E, Wurster S, Page L, Löffler C, Bussemer L, Schmitt A-L, Wolf T, Linde J, Cicin-Sain L, Becker J, Kalinke U, Vogel J, Panagiotou G, Einsele H, Westermann AJ, Schäuble S, Loeffler J. 2020. Triple RNA-Seq reveals synergy in a human virus-fungus co-infection model. Cell Rep 33:108389. 10.1016/j.celrep.2020.108389. [DOI] [PubMed] [Google Scholar]
- 344.Liu DH, Liu XJ, Tan SS. 2007. Analysis of the difference in proteome expression between yeast form and mould form of Penicillium marneffei using SELDI technique. Nan Fang Yi Ke Da Xue Xue Bao 27:59–61. [PubMed] [Google Scholar]
- 345.Barber AE, Sae-Ong T, Kang K, Seelbinder B, Li J, Walther G, Panagiotou G, Kurzai O. 2021. Aspergillus fumigatus pan-genome analysis identifies genetic variants associated with human infection. Nat Microbiol 6:1526–1536. 10.1038/s41564-021-00993-x. [DOI] [PubMed] [Google Scholar]
- 346.Korfanty GA, Dixon M, Jia H, Yoell H, Xu J. 2021. Genetic diversity and dispersal of Aspergillus fumigatus in arctic soils. Genes (Basel) 13:19. 10.3390/genes13010019. [DOI] [PMC free article] [PubMed] [Google Scholar]