Abstract
Gastric cancer is a leading cause of cancer mortality and health disparities in Latinos. We evaluated gastric intratumoral heterogeneity using multiregional sequencing of >700 cancer genes in 115 tumor biopsies from 32 patients, 29 who were Latinos. Analyses focused on comparisons with The Cancer Genome Atlas (TCGA) and on mutation clonality, druggability, and signatures. We found that only approximately 30% of all mutations were clonal and that only 61% of the known TCGA gastric cancer drivers harbored clonal mutations. Multiple clonal mutations were found in new candidate gastric cancer drivers such as EYS, FAT4, PCDHA1, RAD50, EXO1, RECQL4, and FSIP2. The genomically stable (GS) molecular subtype, which has the worse prognosis, was identified in 48% of our Latino patients, a fraction that was >2.3-fold higher than in TCGA Asian and White patients. Only a third of all tumors harbored clonal pathogenic mutations in druggable genes, with most (93%) GS tumors lacking actionable clonal mutations. Mutation signature analyses revealed that, in microsatellite-stable (MSS) tumors, DNA repair mutations were common for both tumor initiation and progression, while tobacco, POLE, and inflammation signatures likely initiate carcinogenesis. MSS tumor progression was likely driven by aging- and aflatoxin-associated mutations, as these latter changes were usually nonclonal. In microsatellite-unstable tumors, nonclonal tobacco-associated mutations were common. Our study, therefore, contributed to advancing gastric cancer molecular diagnostics and suggests clonal status is important to understanding gastric tumorigenesis. Our findings of a higher frequency of a poor prognosis associated molecular subtype in Latinos and a possible new aflatoxin gastric cancer etiology also advance cancer disparities research.
Significance:
Our study contributes to advancing our knowledge of gastric carcinogenesis, diagnostics, and cancer health disparities.
Introduction
Gastric cancer is the third cause of cancer mortality worldwide (1, 2). The disparity between gastric cancer incidence (∼1 M annual new cases) and mortality (∼760 K annual deaths) remains stubbornly minimal and little progress has been achieved toward treating advanced disease. This minimal disparity is partly explained by difficulties in early detection and the paucity of novel molecularly guided gastric cancer therapies. The Cancer Genome Atlas (TCGA) study showed that most gastric cancers harbor potentially “druggable” mutations (3). Interestingly, several studies have shown that extensive intratumoral heterogeneity (ITH) is present in most malignancies, which evolve by acquiring clonal and/or initiating mutations and subclonal and private progression mutations (4, 5). Understanding the clonal status of key tumor mutations is important to increase our knowledge of tumor evolution and for identifying the most promising and druggable targets. Most ITH studies have relied on bulk sequence analysis of multiple and spatially separated tumor biopsies (4, 5). This multiregional sequencing approach has advantages over single-biopsy-per-sample profiling. Studies have repeatedly shown that one-site/one-time biopsy sampling often misses a significant fraction of mutations (6). Single-biopsy TCGA studies showed that gastric cancers have a high mutation frequency and are likely to show extensive ITH (7). A study of heterogeneity in gastroesophageal adenocarcinoma found extensive mutational differences between primary tumors and metastatic lesions and significant discrepancies, potentially clinically relevant, at different sites within the primary tumor (8). ITH represents a significant challenge for target selection in precision medicine, as it likely explains the failure of most molecularly guided gastric cancer trials. Clonal mutations that drive tumorigenesis are widely considered the optimal drug targets, although nonclonal mutations can be useful targets, especially in combination therapies, if they play a functional role in subclones influencing tumor progression. The primary purpose of our study is to examine gastric cancer ITH patterns and evaluate its implication for tumor evolution and likely response to therapy, with the hypothesis that ITH would be present and help explain the difficulty of finding effective molecularly guided treatments. The study was enriched with patients of Latino ancestry, and we also explored ITH and gastric cancer genetic diversity implications for cancer health disparities in this population.
Materials and Methods
Patient Cohort
Our study cohort included 29 patients with gastric cancer of Latino ancestry (19 patients were recruited by Universidad del Tolima in Colombia, 9 by Instituto Mexicano del Seguro Social, in Mexico, and 1 by University of California, Davis in the United States) and 3 non-Latino Whites (recruited by University of California, Davis in the United States). Patient clinical information is shown in Table 1. Research protocols that were used to recruit human research subjects, who provided written informed consent, adhered to the Common Rule, and were approved by Institutional Review Boards from participating institutions in Colombia, Mexico, and the United States. Tumor biopsies were separated by >3 cm, and normal tissues were obtained from anatomically normal tissue identified during endoscopy or in surgical specimens. Biopsies were snap-frozen, and all patients had their tumors verified by a local surgical pathologist.
TABLE 1.
Patient clinical and assigned molecular subtype information
| Indiv | Type | Hpy | Ctry | Eth | Sex | Age | Hist | Anat | Stage |
|---|---|---|---|---|---|---|---|---|---|
| I_9709 | EBV | N | Col | Lat | M | 78 | NA | B | IV |
| I_26163 | EBV | N | Mex | Lat | M | 72 | I | B | IA |
| Subtotal | 2 | 0 | |||||||
| I_10845 | MSI | Y | Col | Lat | F | 56 | I | A | IV |
| I_8012 | MSI | N | Col | Lat | M | 62 | I | C | IV |
| I_13137 | MSI | N | USA | Wht | M | 68 | D | NA | IB |
| I_20447 | MSI | N | USA | Wht | M | 82 | NA | C | NA |
| Subtotal | 4 | 1 | |||||||
| I_13148 | CIN | N | Col | Lat | F | 37 | D | A | IV |
| I_3755 | CIN | N | Col | Lat | F | 42 | I | A | IIIB |
| I_7245 | CIN | N | Col | Lat | F | 64 | I | A | IIIB |
| I_12808 | CIN | N | Col | Lat | M | NA | NA | NA | IV |
| I_9278 | CIN | N | Col | Lat | M | 60 | I | A | IV |
| I_9297 | CIN | N | Col | Lat | M | 60 | M | C | IIIB |
| I_8164 | CIN | N | Col | Lat | M | 61 | D | A | IIIB |
| I_9296 | CIN | N | Col | Lat | M | 67 | M | A | IIB |
| I_26175 | CIN | N | Mex | Lat | F | 79 | D | A/P | IIIC |
| I_26166 | CIN | N | Mex | Lat | M | 55 | I | A/P | IIIB |
| I_26181 | CIN | N | Mex | Lat | M | 57 | D | B/C | IIIC |
| I_13139 | CIN | N | USA | Wht | F | 74 | D | A/P | NA |
| Subtotal | 12 | 0 | |||||||
| I_9715 | GS | Y | Col | Lat | F | 31 | I | L/A | IIIB |
| I_19344 | GS | Y | Col | Lat | F | 32 | NA | NA | IV |
| I_11981 | GS | Y | Col | Lat | F | 49 | D | L | IB |
| I_19343 | GS | Y | Col | Lat | F | 50 | NA | NA | IV |
| I_6579 | GS | N | Col | Lat | F | 56 | I | A | IV |
| I_9299 | GS | N | Col | Lat | M | 47 | I | B/A | IIB |
| I_9710 | GS | N | Col | Lat | M | 53 | I | A | II |
| I_14885 | GS | N | Col | Lat | M | 78 | I | A | IIB |
| I_26169 | GS | N | Mex | Lat | F | 57 | D | A/P | IIIC |
| I_26167 | GS | N | Mex | Lat | F | 65 | M | B | IV |
| I_26180 | GS | N | Mex | Lat | M | 59 | D | A/P | IIIA |
| I_26171 | GS | N | Mex | Lat | M | 71 | D | B/A | IV |
| I_26172 | GS | N | Mex | Lat | M | 76 | D | A/P | IIIA |
| I_13141 | GS | Y | USA | Lat | M | 55 | NA | B | NA |
| Subtotal | 14 | 5 | |||||||
| Total | 32 | 6 |
Abbreviations: A, antrum; Anat, Anatomic location; B, body; C, Cardia; CIN, Chromosomal instability; Col, Colombia; Ctry, Country; D, diffuse; EBV, Epstein-Barr virus; Eth, Ethnicity; F, Female; GS, Genomically stable; Hist, Histology; Hpy, H. pylori infection; I, intestinal; Indiv = Individual; L, lesser curvature; Lat, Latino; M, Male; M, mixed; Mex, Mexico; MSI, Microsatellite instability; NA, Not available; N, No; P, pylorus; Type, Molecular subtype; Wht, White; Y, Yes.
Somatic Pan-cancer Panel Design
We selected 726 genes based upon known cancer risk (9), recurrence in previous gastric cancer samples, COSMIC Cancer Gene Census, TCGA Gastric cancer studies, and based upon expression list of cancer risk genes were collected from the literature (10–54), Color Genomics test suite (55), and DNA repair pathway genes (Supplementary Table S1) plus TERT promoter, microsatellite instable (MSI) regions, Epstein-Barr virus (EBV) and Helicobacter pylori sequences. Panel probes were designed using Agilent SureSelect XT2 Custom Capture technology (56) with a target capture region of 3.75 Mbp. There were 287 targeted genes that were covered less than 100%, and 24 of these were covered less than 90% (Supplementary Table S2).
DNA Sequencing and Bioinformatics Pipelines
DNA samples were isolated with QIAGEN kits from 168 biopsies of gastric tumors and 37 normal gastric tissue samples adjacent to the tumors of 37 patients. Sequencing was performed on Illumina HiSeq 4000 (57) using paired-end 150 bp sequencing. Scythe (58) version 0.991 and Sickle (59) version 1.33 was utilized for trimming. Based upon DNA-seq quality control (QC), 15 biopsies and 3 patients were removed, gender mismatch QC led to six biopsies and 1 patient removed, and 115 tumor and 32 normal biopsies from 32 patients remained for the final analysis. Reads were aligned to GRCh38 using BWA (60) version 0.7.17. BROAD Institute Best Practices for Variant Calling with the GATK were followed (61). Germline single-nucleotide variants (SNV) were called using the joint variant caller multiSNV (62) version v2.3-15. Somatic variants were called using Mutect2 (63). Variants were annotated using Annovar suite (64). Additional germline and somatic variant filtering was applied as detailed in Supplementary Materials and Methods. A panel of normals was created with variants called in > = 2 samples. MSIsensor (65) was used to predict tumor sample MSI.
A clonality was assigned to each SNV: PRIVATE (only one tumor is a somatic variant), SUBCLONAL (greater than 1 but not all biopsies carry the variant), CLONAL (all tumors have variant), or NONE (applied to aberrant cases). PureCN (66), version 1.16.0 was used to perform copy-number variation (CNV), purity, and ploidy analysis, and biopsy mutation rate estimation (see Supplementary Materials and Methods for more details). Mappability files were created using GEM library (67) version 1.778 beta. Each tumor biopsy's mutation rate was estimated with PureCN. To address possible pseudoheterogeneity due to factors such as allele-specific imbalance or heterogeneous amplification, an in-house heuristic forced variant calling algorithm was used to look for mutant allele reads at levels too low to be called mutant when the same locus was called mutant in a sister biopsy. OncoKB gene list (68, 69) and FDA gastric cancer–targeted therapy genes (70) were combined, leaving 58 druggable genes overlapping our gene targets. Mutation signatures were estimated with deconstructSigs (71). A subset of the V3.2 signatures were used, retaining those deemed relevant to gastric cancer.
Statistical Analysis
The statistical tests used in this study are Student t test for comparing sample means and Fisher exact test for comparing proportions in multiple categories. This study analyzes data from 32 patients and many more biopsies, so the data from patients and biopsies is not independent. The patient count is too low to provide substantial statistical power, so P values are used only when testing individuals independently of biopsies and the study is otherwise more descriptive in nature.
A detailed description of the methods for data processing and the software used for analysis is available in the Supplementary Materials and Methods file.
Data Availability Statement
The data generated in this study were deposited in the European Genome-phenome Archive (Study ID: EGAS00001006650, Dataset ID: EGAD00001009622).
Results
Patient Characteristics
Table 1 shows the main characteristics of the patients analyzed in the study. None of these patients carried germline mutations in Lynch syndrome or hereditary diffuse gastric cancer genes (not shown). Mexican patients had a larger fraction of tumors with diffuse histology (Table 1). Colombian patients, on the other hand, were significantly younger than those from Mexico or United States (55 vs. 66 years, P = 0.020; Student t test). Colombian and Mexican patients had a similar distribution of molecular subtypes. Compared with available TCGA clinical data, our patient population has a similar sex distribution (Table 1; Supplementary Table S3), with 59% of our study patients being male versus 62% in TCGA. Our patients were however younger than those from TCGA (60 vs. 66 years, P = 0.0183; Table 1; ref. 3) and had a higher rate of advanced tumors (76% vs. 45%; Table 1; ref. 3).
Gastric Tumor Genomic Landscape
We evaluated 115 tumor biopsies from 32 patients (Table 1) using our cancer panel (Supplementary Data S1). On average, each tumor biopsy was sequenced at 336X depth and each normal biopsy at 225X (Supplementary Data S2). After filtering out likely false positives, we identified a total of 1,319 different somatic mutations (mean 41/patient), or 1,326 counting recurrences in different patients, or 2,594 counting occurrences in each individual tumor biopsy (mean 23/biopsy), in 473 genes. These included 771 nonsilent coding mutations (635 SNVs and 136 indels; mean 24/patient), or 775 total across patients including recurrences, or 1,543 counting occurrences in each individual tumor biopsy (mean 13/biopsy) occurring in 355 panel-targeted genes (Fig. 1; Supplementary Figs. S1–S8; Supplementary Data S3). For TCGA molecular subtypes, of the 32 patients, 2 were classified as EBV (6.2%), 4 as MSI (12.5%), 12 as chromosomal instability (CIN) (37.5%), and 14 as GS (43.8%). When analyses were restricted to Latinos (n = 29 patients), subtype frequencies were 38% for CIN, 7% for EBV, 48% for GS, and 7% for MSI (Supplementary Table S3), and when our Mexican and Colombian Latino subtypes were compared, there were twice as many Colombian CIN and GS patients (n = 16) as Mexican (n = 8) but the overall difference in subtype proportions was not significant by Fisher exact test (P = 0.8). The subtype frequencies in our Latino patients were significantly different from those in all TCGA patients (P = 0.033; Fisher exact test). Within TCGA's White and Asian patients (n = 238 White, n = 74 Asian), the rate was similar to ours for EBV (8% for Whites, 11% for Asians), higher for MSI (18% for Whites, 20% for Asians) and CIN (61% for Whites, 51% for Asians), and much lower for GS (13% for Whites, 18% for Asians).
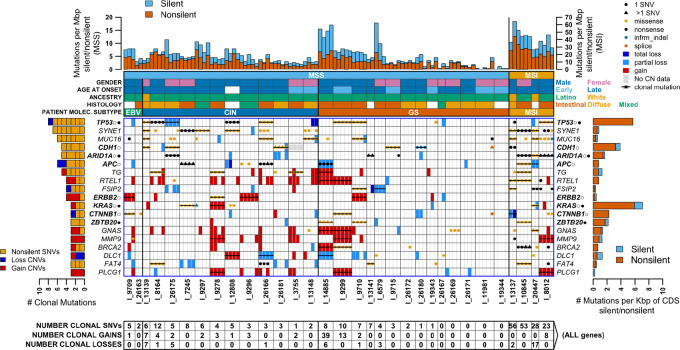
FIGURE 1.
Genes clonally mutated in 3 or more gastric cancer MSS/MSI patients. Gene names shown on both left and right sides are those where at least 3 patients have clonal somatic nonsilent single-nucleotide or short indel (SNV), or copy-number gain/loss (CNV) mutations. Patients (I_#### IDs) are separated by a dark vertical line and patient biopsies within them are separated by a lighter line. SNV and CNV mutations are shown for each gene/biopsy combination. Gains and losses are distinguished by box color, while SNVs are shown as a filled circle or triangle using color to distinguish the mutation type (key at top right; nonsense mutations include stop gain/loss and frameshift). A triangle rather than a circle means a gene has more than one mutation in a patient, and the type shown is the one generally most detrimental. A bar through all biopsies means the mutation is clonal. The left-side bar plot shows the total number of clonal mutations of each type in each gene, with short vertical lines separating individual patients. Numbers in boxes at the bottom give the total number of clonal mutations of each type in each patient in all genes (not just the ones shown). Patient molecular subtypes are indicated by color-coded bars just above the main figure area, and above those are additional color-coded bars indicating patient microsatellite instability status (MSS/MSI), sex, age of onset of gastric cancer (where late onset was defined as occurring at age >50), ancestry (Latino or White), and tumor histology. The top bar plot shows the SNV mutation rate in each biopsy, in the region targeted by the panel, with nonsilent and silent rates distinguished by color. MSS patients have a lower mutation rate and use the y-axis scale on the left, while MSI patients with a higher rate use the right-side scale. The right-side bar plot shows the SNV mutation count in each gene summed over all 115 biopsies, normalized by dividing by the gene CDS length in Kbp, with nonsilent and silent mutations distinguished by color. Gene names are in bold when they were found to be gastric cancer drivers by TCGA gastric cancer study, with open circles denoting driver genes in non-hypermutated (MSS) TCGA samples and filled circles denoting those in hypermutated (MSI) samples.
Somatic Mutation and Phylogenetic Analyses Showed Varying ITH
We next analyzed the distribution of SNVs, indels, and CNVs in the different biopsies of each patient and classified each mutation as clonal (occurring in all individual tumor biopsies from the same patient), subclonal (occurring in more than one but not all individual tumor biopsies), or private (occurring in one individual tumor biopsy only). Supplementary Data S3 shows counts of all somatic SNVs/indels and their clonal status. Of the 1,326 such mutations, 428 were clonal (32%, mean 13/patient), 213 were subclonal (16%, mean 7/patient), and 685 were private (52%, mean 21/patient). The clonal SNV fraction in patients of different molecular subtypes was about the same in GS and CIN (27%; GS mean 5.9/patient; CIN mean 6.9/patient), higher in EBV (34%, mean 8.5/patient), and MSI (36%, mean 61.5/patient). There was at least one clonally mutated gene in 27 of the 32 patients (all 5 patients without clonal changes had the GS subtype). In contrast, nonclonally mutated genes were found in all but 1 patient (Fig. 1; Supplementary Fig. S1). Phylogenetic trees of clonal and nonclonal changes were generated to visualize the evolution of each tumor (Supplementary Fig. S2). These trees graphically illustrate the branched evolution pattern followed by gastric cancers.
Clonally Mutated Genes in Microsatellite-Stable and MSI Patients
In our 28 microsatellite-stable (MSS) patients, we found 15 genes with clonal nonsilent SNVs/indels in multiple patients. Of these, one gene was clonally mutated in 6 patients (TP53), one in 5 patients (CDH1), two in 4 patients (MUC16, SYNE1), and two in 3 patients (APC, TG; Fig. 1; Supplementary Figs. S1 and S3). In addition, even though our targeted approach had limitations to accurately call CNVs, our analyses identified 12 genes that were clonally amplified or deleted in multiple MSS patients, including two with a mixture of amplifications and deletions (DLC1, WRN; Supplementary Fig. S4), one that was clonally amplified in 3 patients (ERBB2), and nine that were clonally amplified in 2 patients each (FGFR2, KLF5, the 8q24 MYC and RECQL4 genes, and the 20q12-13 genes PLCG1, MMP9, GNAS, LAMA5, and RTEL1,Supplementary Figs. S4–S6). Counting both nonsilent SNVs/indels and CNVs, a total of 39 genes were clonally altered in multiple MSS patients. Of those, nine are known TCGA gastric cancer drivers (APC, CDH1, CTNNB1, ERRB2, KRAS, RNF43, ARID1A, and TP53 in MSS tumors, and ZBTB20 in MSI tumors), eight have been identified as gastric cancer drivers in non-TCGA studies (MUC16, DLC1, MMP9, FASN, LAMA5, EGFR, BRCA2, and FGFR2), 14 have been identified as drivers for other cancer types by TCGA (ATAD2, ATR, CSDE1, CSMD3, ELF3, ERBB4, KLF5, TRPA1, TSHZ2, and PLCG1) and non-TCGA studies (GNAS, MYC, SYNE1, and TG), and eight have not been previously identified as gastric cancer drivers (EYS, FAT4, FSIP2, PCDHA1, RAD50, RECQL4, RTEL1, and WRN). These latter clonally mutated genes should be considered candidate gastric cancer driver genes for future studies.
In our 4 MSI patients, we found 17 genes with clonal nonsilent SNV/indels in multiple patients, including 2 in 3 patients (ARID1A, SYNE1) and 15 in 2 patients (ALPK2, ATM, CDC27, CDK12, ESR1, EXO1, FSIP2, KMT2E, LRRK2, MACF1, MUC16, NEB, PIK3CA, RNF111, and RTEL1; Fig. 1; Supplementary Fig. S1 and S5). Five of these genes are known TCGA drivers (ARID1A, ALPK2, PIK3CA, and RNF111 in MSI tumors and MACF1 in MSS tumors), one has been identified as a gastric cancer driver in non-TCGA studies (MUC16), eight are known drivers for other cancer types in TCGA (ATM, CDC27, CDK12, ESR1, KMT2E, and LRRK2) and non-TCGA studies (SYNE1 and NEB), and three have not been previously identified as drivers (EXO1, RTEL1, and FSIP2). These three should also be considered candidate driver genes for future studies.
It is always possible for clonal mutations to be passengers rather than drivers, more so the larger the gene. Using the definition of long gene length as above the 75% of all genes in our panel, we examined the long clonally mutated genes mentioned above for evidence of likely pathogenic changes [defined as a loss-of-function mutation, a known cancer hotspot mutation, annotated as pathogenic in ClinVar (72), amplification of a known oncogene, or complete deletion of the entire gene or its wild-type allele]. Because of the absence of clonal mutations classified as likely pathogenic, nine genes (TG, WRN, PLCG1, LAMA5, RTEL1, FASN, CDK12, LRRK2, and MACF1), were considered unlikely to be candidate gastric cancer drivers. Conversely, in five of these genes, all clonally mutated patients had mutations that were likely pathogenic (CTNNB1, ELF3, ATM, KMT2E, and PIK3CA), lending stronger support to their candidate gastric cancer driver status.
Nonclonally mutated genes are summarized in Supplementary Fig. S7. Genes with a relatively high nonclonal mutation frequency include STK11, GPS2, NDUFB9, and AXIN2. The tumorigenesis role, particularly in cancer progression, of these nonclonally mutated genes should be evaluated in future cancer biology studies.
The Known TCGA Gastric Cancer Drivers are Clonally Heterogeneous
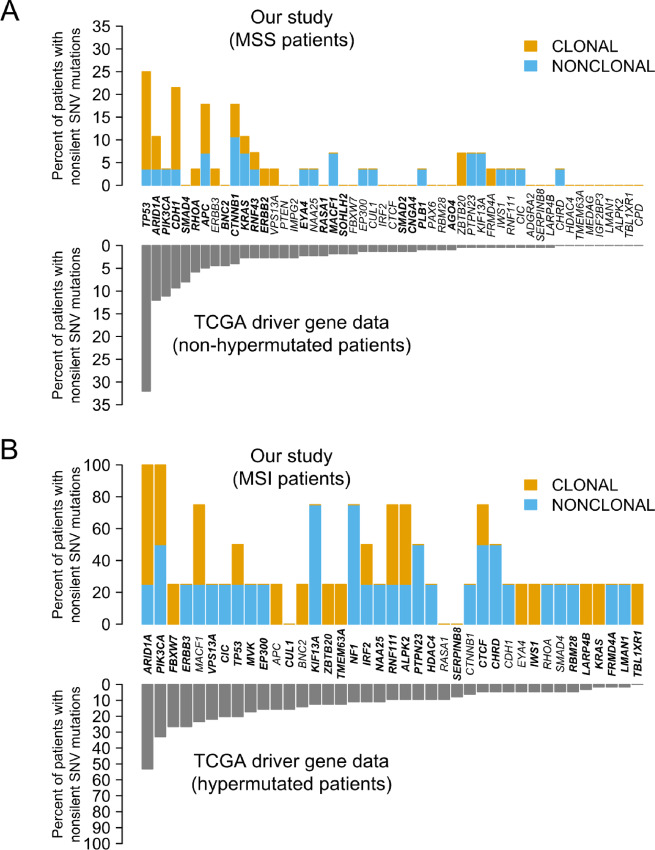
Our panel contained 54 well-covered genes that were identified as gastric cancer drivers by TCGA. We found that 11 of these genes harbored nonsilent clonal SNV/indel mutations in multiple patients (TP53, CDH1, ARID1A, APC, KRAS, CTNNB1, ZBTB20, PIK3CA, RNF111, ALPK2, and MACF1; Fig. 1; Supplementary Fig. S1). In addition, 20 known gastric cancer TCGA drivers had clonal SNV mutations in 1 patient each in our study (Supplementary Fig. S1). Figure 2 contrasts the number of patients having clonal and nonclonal mutations in these known drivers in our study with the number of patients with mutations in TCGA. Most known drivers had both clonal and nonclonal mutations, with half of them clonally mutated in at least 1 patient while the other half was either nonmutated or only nonclonally mutated. Of note, ZBTB20, a driver identified by TCGA in MSI tumors, was clonally mutated only, in multiple patients. In contrast, CIC, NF1, KIF13A, PTPN23, and CHRD (all MSI drivers) were mutated in multiple patients, but always nonclonally. Indeed, after TP53 and ARID1A (clonally mutated in 6 patients each), CDH1 and APC (clonally mutated in 5 patients each), and ERBB2 (clonally mutated/amplified in 4 patients), ZBTB20, KRAS, and BRCA2 were the fourth most common clonally mutated known driver genes in our study. When these analyses were stratified by the patient's country of origin or age of gastric cancer diagnosis (≤50 y vs. >50 y), we found no significant difference in our study. Overall, our clonal analyses of known TCGA drivers support the notion that the driver status of some of these genes is worthy of reexamination in larger ITH studies. Alternatively, as our sample was enriched with tumors from Latino patients, these differences may reflect population differences, as TCGA included mostly Whites in their analyses.
FIGURE 2.
Comparison of TCGA gastric cancer driver mutation frequency with this study. A, Genes significantly mutated in non-hypermutated TCGA samples, compared to MSS patients in our study. B, Similarly, for hypermutated TCGA samples compared with our MSI patients.
ITH in Druggable Genes
Our panel included 58 druggable genes. One-third (n = 11, 34%) of our patients had at least one clonal likely pathogenic change (defined above) in a druggable gene. Clonal likely pathogenic changes in druggable genes were found in all four MSI patients (100%), 5 of the 12 patients with CIN (42%), 1 of the 4 EBV patients (25%), and only 1 of our 14 GS patients (7%). The pathways with the highest number of clonal likely pathogenic changes included tyrosine kinase receptors (ERBB2, EGFR, FGFR2, and FLT4, mutated in 4 patients), homologous recombination repair (BRCA2, ATM, and ATR, mutated in 4 patients), PI3K/AKT/MTOR (PIK3CA and AKT1, mutated in 3 patients), and RAS/RAF/MAPK (KRAS, mutated in 3 patients). Finally, we found six genes with nonclonal pathogenic mutations in 2 or more patients (EGFR, ERBB2, KRAS, NF1, PIK3CA, and STK11;Supplementary Figs. S7 and S8).
Mutation Signatures in MSS Tumors
We were also interested in examining whether different processes could mediate clonal and nonclonal mutations (Supplementary Table S4; Supplementary Figs. S5, S8, and S9). As most of our patients had MSS tumors (28/32), we focused on initial analyses in this group. MSS clonal mutations mainly resulted from signatures SBS1 (deamination), SBS3 (homologous recombination deficiency, HRd), SBS4 (tobacco), SBS5 (age), and SBS10b (POLE mutations, POLEm). Nonclonal mutations, on the other hand, had signatures associated with HRd, tobacco, age, mismatch repair deficiency (MMRd, SBS6, and SBS15), reactive oxygen species damage (ROSd, SBS18), and aflatoxin exposure (SBS24, aflatoxins). As our study was enriched with Latino patients, we compared their signatures with those in TCGA MSS patients (Supplementary Table S4; Supplementary Fig. S9). Deamination and age signatures were found in Latinos and TCGA; however, HRd-, SBS6/MMRd-, ROSd-, and aflatoxins-associated signatures were present only in Latinos.
We also analyzed signatures in CIN and GS tumors separately (Supplementary Table S4; Supplementary Fig. S9). The HRd signature was found in both CIN and GS clonal mutations but not in nonclonal mutations. The POLEm signature was found in clonal mutations of GS but not CIN patients. The SBS15/MMRd was found in both CIN and GS but only in nonclonal mutations. These findings suggest that CIN and GS tumors may result from different mutational processes involved in tumor initiation and progression.
Finally, as GS was our most common subtype, we explored associations between GS tumor signatures and histology in our study and TCGA (Supplementary Table S4). Our study's GS diffuse and intestinal tumor mutations had the age, SBS15/MMRd, and aflatoxins signatures. Age, but not the SBS15/MMRd or aflatoxin signatures, was also detected in TCGA GS diffuse and intestinal tumors. In our study and TCGA, GS diffuse mutations resulted from deamination and POLEm signatures. In GS intestinal tumors, we found SBS6/MMRd in our Latinos and TCGA, while HRd and ROSd were exclusively found in our study. These findings suggest that different mutational processes may affect histologic types in GS tumors.
Mutation Signatures in MSI Tumors
Our study only included 4 MSI patients, so we mainly evaluated mutation signature differences by clonal status. Clonal mutations in MSI tumors were primarily the result of MMRd (SBS15 and SBS26) and deamination. Nonclonal mutations, on the other hand, had signatures associated with deamination, tobacco, and MMRd (SBS6, SBS15, SBS21, and SBS26). These findings suggest, as expected, that MMRd is required for MSI tumor initiation, while tobacco may be important for MSI tumor progression.
Discussion
This study represents a comprehensive investigation of ITH in gastric cancer, a worldwide leading cause of cancer incidence and death (2). Our study showed that gastric cancers are characterized by a complex genetic architecture and suggested the existence of novel driver genes. Our druggable target analysis identified key pathways and genes that often harbor clonal mutations, and that should be prioritized for therapeutic development. Mutational signature analyses suggested that carcinogens likely play a different role in clonal and nonclonal mutations, across molecular and histologic subtypes and that population-specific exposures, such as aflatoxins, may also influence gastric cancer etiology. Therefore, our study reports findings important to understanding gastric cancer etiology, disparities, tumor evolution, and future therapeutic development.
TGCA studies have demonstrated that gastric cancers are among the most genetically diverse tumors (7), with each gastric cancer harboring approximately 500 coding mutations (3). The mutation rate, however, varies greatly between molecular subtypes, with MSI tumors having the highest number of alterations and GS tumors the lowest (3). Consistent with previous work (3), our MSI tumors harbored the highest number of both clonal and nonclonal mutations, followed by CIN, EBV, and GS tumors. These differences in mutation rate are important not only for understanding tumor evolution but also for making the best-informed choice of targeted therapies or immunotherapies. Interestingly, mutation patterns in our study highlighted several important findings about gastric cancer drivers. First, it became evident that the list of gastric cancer driver genes is likely larger than that reported by TGCA. Our results suggested that the clonal mutation status in both known and potentially new candidates is important and should be considered to help validate their “driver” status. In our analyses of known TCGA gastric cancer drivers, for instance, we showed that only approximately 60% had clonal mutations, raising questions about the initiation versus progression “driver” status of genes such as CIC, NF1, KIF13A, PTPN23, and CHRD, which were mutated in multiple patients but always nonclonally. Even though clonal mutations may be the obvious targets for therapies, it is possible that some nonclonal mutations play an important role in cancer progression. Our findings are therefore intriguing and suggest that larger ITH studies should evaluate whether these nonclonally mutated genes are indeed drivers and whether they are involved in tumor progression or only harbor passenger and/or neutral mutations.
An interesting aspect of our analyses is that we identified several recurrent clonally mutated genes. Many of them, such as ATAD2, ATR, BRCA2, CSDE1, CSMD3, DLC1, EGFR, ELF3, ERBB4, FGFR2, KLF5, TRPA1, TSHZ2, GNAS, MYC, and MMP9 for MSS tumors and ATM, CDC27, ESR1, KMT2E, and NEB for MSI tumors, have been previously identified as drivers by TCGA/non-TCGA studies for other cancer types but not for gastric cancer. We also found six genes in MSS tumors (EYS, FAT4, FSIP2, PCDHA1, RAD50, and RECQL4) and two in MSI tumors (EXO1 and FSIP2) that were clonally mutated in multiple patients and have not been previously identified as drivers of gastric cancer or other cancers. Interestingly, many of these new genes are involved in key processes disrupted in gastric tumorigenesis, such as extracellular matrix (EYS) and cell adhesion (FAT4 and PCDHA1, which are protocadherins) or homologous recombination repair (RAD50 and RECQL4). These potentially new gastric cancer driver genes, as well as genes with high nonclonal mutation frequency, such as STK11, GPS2, NDUFB9, and AXIN2, represent good candidates for inclusion in future studies of gastric tumorigenesis.
We and others have shown that genes involved in homologous recombination repair are important in both gastric cancer risk and tumorigenesis (73–75), which is consistent with our observation of multiple patients with clonal nonsilent mutations in ATR, ATM, and BRCA2 and with the mostly clonal nature of the HRd-associated mutation. Our study also found many patients with clonal pathogenic mutations in the RAS/RAF pathway gene KRAS and the PI3K/MTOR/AKT pathway gene PIK3CA, providing further evidence of their importance in gastric cancer biology. While ERBB2 and ERBB3 are both molecular targets of FDA-approved gastric cancer therapies and are known TCGA gastric cancer drivers (3, 76), the high number of patients with mutations in other related tyrosine kinase genes (FGFR2, EGFR/ERBB1, and ERBB4) suggest that these should also be considered important genes in gastric cancer biology. These results suggest clonal status can further identify novel genes that are important in gastric tumor biology.
Gastric cancer targeted therapies have been notorious for their failure in late-stage trials, with only two (targeting ERBB2 and VEGFR2) currently approved by the FDA (76–78). While gastric cancers are characterized by one of the highest mutation rates among all solid malignancies, and TCGA suggested that approximately 70% of them harbor potentially actionable or druggable mutations (3), our ITH analyses indicated that even though a significant fraction of gastric cancers do indeed carry druggable mutations, only about 60% of these tumors have clonal mutations in druggable genes. However, we also found that clonal druggable mutations are closely associated with the molecular subtype, with the good-prognosis MSI subtype having the highest number of druggable targets and the poor-prognosis GS subtype having the lowest (79). Indeed, all 4 MSI patients in our study harbored multiple clonal actionable mutations, while >90% of GS tumors lacked such mutations. This suggests that MSI tumors will be highly amenable to targetable therapy development, which will likely be beneficial for future combinations with immunotherapies (80). GS tumors, on the other hand, remain a significant challenge in drug development, and future efforts should focus on identifying additional targets (such as methylation or synthetically lethal combinations) for preclinical studies and clinical trials. As GS tumors seem more prevalent in Latinos, such studies should involve race and/or ethnic appropriate models and participants (81).
Our mutation signature analyses revealed interesting differences between MSS and MSI tumors and between Latino and TCGA tumors. In MSI tumors, MMRd and deamination likely drive tumor initiation while tumor progression seems to be driven by tobacco-associated mutations. While an association between the MMRd mutation signature and MSI tumorigenesis makes sense, the fact that tobacco may accelerate MSI tumor progression is consistent with previous studies showing a stronger association with gastrointestinal MSI tumors in heavy smokers (82–84) and with the fact that Lynch syndrome patients carrying germline MMR mutations are particularly susceptible to tobacco carcinogens (85). Our signature analyses in MSS tumors, on the other hand, revealed that deamination- and POLEm-associated mutations likely drive tumor initiation; HRd-, tobacco- and age-associated mutations influence both tumor initiation and progression; and MMRd-, ROSd- and aflatoxin-associated mutations play a role in tumor progression. Our separate analyses of CIN and GS tumors revealed that POLEm/SBS10b seems to primarily play a role in GS tumor initiation. Our histologic comparison within GS tumors also revealed consistent associations, in our study and in TCGA, of the importance of deamination in diffuse histology and of MMRd in intestinal histology. Finally, our finding of an aflatoxin mutational signature is also novel and intriguing and suggests that larger and further studies should examine the role of this risk factor (which has been found in Latinos with liver and gallbladder cancers) in gastric tumorigenesis (86–88). This finding, if replicated, also highlights the benefits of racial and/or ethnic diversity in cancer genetics studies.
Gastric tumors are one of the leading causes of cancer health disparities (89, 90). As our study was enriched with Latino patients, our results are important to advance precision health equity in this population. Future studies should evaluate many of the novel findings found in our study and assess whether such patterns are more common in Latinos or are also observed in other populations. Some of our results are particularly puzzling, such as our significant difference in molecular subtypes when compared with TCGA, with our study having a significantly higher prevalence of GS tumors, which are known to have the poorest prognosis in gastric cancer (79) and which are more commonly associated with diffuse histology (3). A high frequency of GS tumors in Latinos, also recently reported in Texas (91), therefore may explain some of the observed disparities in Latino gastric cancer outcomes, and the development of future therapies for this subtype should be a priority in gastric cancer disparity research.
Our observational study however has some limitations. First, it was solely focused on DNA sequence changes in a panel of cancer genes. Genome-wide or exome-wide analyses would likely have led to improved signature analyses. While our study did not have the ability to evaluate methylation or gene expression ITH, our findings showed a striking level of genetic complexity in gastric cancers, which may explain why these tumors are so difficult to treat. We also hope that future functional studies investigate the tumorigenesis role of some of the putative new drivers highlighted in our study. Furthermore, our study was enriched with advanced tumors, which could introduce biases compared with TCGA. Despite these limitations, we believe that our study has several strengths, focused on an understudied population, and reporting several novel findings that will likely contribute to advancing gastric carcinogenesis and disparities.
In sum, we carried out a comprehensive evaluation of gastric tumor genetic diversity. Our study found that Latinos are enriched with a poor prognosis and chemotherapy-resistant subtype that likely account for some of the outcome disparities experienced by Latinos. Our findings showed a striking level of genetic complexity in gastric cancers, explaining why these tumors are so difficult to treat. We hope our results help advance target selection for gastric cancer therapies and aid in understanding gastric tumorigenesis and disparities.
Supplementary Material
Spreadsheet of pan-cancer panel gene composition, with citation codes
Spreadsheet of sample mean sequencing depths
Spreadsheet of counts of patients with SNV/indel mutations in each gene
PDF file of supplementary methods and materials, and supplementary figures and tables
Acknowledgments
We are indebted to all patients who participated in the study. We thank Marcus Riester, developer of PureCN, and Malvina Josephidou, developer of MultiSNV, for considerable support responding in detail to questions about use of their software.
We are grateful for the financial support from Universidad del Tolima, Colombia (Projects 160120516, 470115, 30113, 350113, 160114, 450110, 40218, 250120; contract 398-2017; grant 001-2019, M.E. Bohorquez-Lozano and M. Polanco-Echeverry); MINCIENCIAS, Colombia (grant 850-2019, contract 940-2019, to F. Castro-Valencia); COLCIENCIAS, Colombia (grant 110565843382; contract 204-2015, M.E. Bohorquez-Lozano; Graduate Studentships 647/2014, A.P. Estrada-Florez and, 755/2016, A.A. Guevara-Tique); L'OREAL-UNESCO-ICETEX-COLCIENCIAS, Colombia (project 3900917/2017, A.P. Estrada-Florez); Instituto Mexicano del Seguro Social and Consejo Nacional de Ciencia y Tecnología, México (FIS/IMSS/PROT/PRIO/13/027 and Fronteras de la Ciencia 2015-01-773, J. Torres); AACR (Fellowship 21-40-69-ESTR, A.P. Estrada-Florez); The Auburn Community Endowed Chair in Basic Cancer Research, U.S.(L.G. Carvajal-Carmona); the Heart, BrEast, and BrAin HeaLth Equity Research (HEAL-HER) Program, U.S. (L.G. Carvajal-Carmona) and the U.S. NCI of the NIH (grants R01CA223978, R21CA199631, U54CA233306, and P30CA093373, L.G. Carvajal-Carmona).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
S. Urayama reports other from CellMax, Noah Medical, and Olympus America outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
T.W. Toal: Data curation, software, formal analysis, validation, investigation, visualization, methodology, writing-original draft, writing-review and editing. A.P. Estrada-Florez: Formal analysis, validation, investigation, methodology, writing-review and editing. G.M. Polanco-Echeverry: Investigation, writing-review and editing. R.M. Sahasrabudhe: Validation, investigation, methodology, writing-review and editing. P.C. Lott: Resources, data curation, software, investigation, writing-review and editing. J.J. Suarez-Olaya: Investigation, writing-review and editing. A.A. Guevara-Tique: Investigation, writing-review and editing. S. Rocha: Investigation, writing-review and editing. A. Morales-Arana: Methodology. F. Castro-Valencia: Methodology. S. Urayama: Conceptualization, resources, methodology, writing-review and editing. A. Kirane: Conceptualization, resources, methodology, writing-review and editing. D. Wei: Conceptualization, resources, investigation, writing-review and editing. N. Rios-Sarabia: Conceptualization, resources, investigation, writing-review and editing. R. Medrano: Conceptualization, resources, investigation, writing-review and editing. A. Mantilla: Conceptualization, resources, investigation, writing-review and editing. M. Echeverry de Polanco: Conceptualization, resources, investigation, writing-review and editing. J. Torres: Conceptualization, resources, investigation, writing-review and editing. M.E. Bohorquez-Lozano: Data curation, methodology. L.G. Carvajal-Carmona: Conceptualization, resources, supervision, funding acquisition, writing-review and editing.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Lott PC, Carvajal-Carmona LG. Resolving gastric cancer aetiology: an update in genetic predisposition. Lancet Gastroenterol Hepatol 2018;3:874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81–94. [DOI] [PubMed] [Google Scholar]
- 5. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xue R, Li R, Guo H, Guo L, Su Z, Ni X, et al. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology 2016;150:998–1008. [DOI] [PubMed] [Google Scholar]
- 7. Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science 2015;349:1483–9. [DOI] [PubMed] [Google Scholar]
- 8. Pectasides E, Stachler MD, Derks S, Liu Y, Maron S, Islam M, et al. Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer Discov 2018;8:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rahman N. Realizing the promise of cancer predisposition genes. Nature 2014;505:302–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng S, Cherniack AD, Dewal N, Moffitt RA, Danilova L, Murray BA, et al. Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell 2016;29:723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016;164:550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med 2016;374:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015;161:1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cancer Genome Atlas Research Network. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 2015;372:2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parfenov M, Pedamallu CS, Gehlenborg N, Freeman SS, Danilova L, Bristow CA, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci U S A 2014;111:15544–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014;159:676–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014;26:319–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014;158:929–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang J-Y, Yoshihara K, Tanaka K, Hatae M, Masuzaki H, Itamochi H, et al. Predicting time to ovarian carcinoma recurrence using protein markers. J Clin Invest 2013;123:3740–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cancer Genome Atlas Research Network, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen Y-B, Gonen M, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res 2013;19:3259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Verhaak RGW, Tamayo P, Yang J-Y, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest 2013;123:517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Larman TC, DePalma SR, Hadjipanayis AG, Protopopov A, Zhang J, Gabriel SB, et al. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A 2012;109:14087–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, et al. Landscape of somatic retrotransposition in human cancers. Science 2012;337:967–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Creighton CJ, Hernandez-Herrera A, Jacobsen A, Levine DA, Mankoo P, Schultz N, et al. Integrated analyses of microRNAs demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PLoS One 2012;7:e34546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bolton KL, Chenevix-Trench G, Goh C, Sadetzki S, Ramus SJ, Karlan BY, et al. Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA 2012;307:382–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010;17:510–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, Reimand J, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep 2013;3:2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pritchard CC, Salipante SJ, Koehler K, Smith C, Scroggins S, Wood B, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn 2014;16:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 2013;45:1127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2016;22:105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer 2004;4:177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. COSMIC. Catalogue of Somatic Mutations in Cancer: Cancer Gene census. [cited 2016. Aug 15]. Available from: cancer.sanger.ac.uk
- 51. cBioPortal. cBioPortal for Cancer Genomics. [cited 2016. Sep 5]. Available from: https://www.cbioportal.org
- 52. cBioPortal. Stomach Adenocarcinoma (TCGA, Provisional). [cited 2019. Sep 5]. Available from: https://www.cbioportal.org/study/summary?id=stad_tcga_pan_can_atlas_2018%2Cstad_tcga_pub
- 53. Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The genotype-tissue expression (GTEx) project. Nat Genet 2013;45:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. GTEx. GTEx Portal. [cited 2016. Dec 2]. Available from: https://gtexportal.org/home/
- 55. Color Genomics. Science Powering Color - Color Genomics. [cited 2016. Dec 2]. Available from: https://www.color.com/genomics
- 56. Agilent Technologies. SureSelectXT2 Target Enrichment System for Illumina Paired-End Multiplexed Sequencing. [cited 2016. Dec 2]. Available from: https://www.agilent.com/cs/library/usermanuals/public/G7530-90000.pdf
- 57. Illumina. HiSeq® 3000/HiSeq 4000 Sequencing Systems. [cited 2015. July 12]. Available from: https://www.illumina.com/content/dam/illumina-marketing/documents/products/datasheets/hiseq-3000-4000-specification-sheet-770-2014-057.pdf
- 58. Buffalo V. Scythe. 2014. [cited 2018. April 10]. Available from: https://github.com/vsbuffalo/scythe
- 59. Joshi NA, Fass JN. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33). [cited 2018. April 10]. Available from: https://github.com/vsbuffalo/scythe
- 60. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997. [cited 2018. April 10]. Available from: https://arxiv.org/abs/1303.3997
- 61. BROAD Institute. Best Practices for Variant Calling with the GATK. [cited 2019. Sep 5]. Available from: https://www.broadinstitute.org/partnerships/education/broade/best-practices-variant-calling-gatk-1
- 62. Josephidou M, Lynch AG, Tavare S. multiSNV: a probabilistic approach for improving detection of somatic point mutations from multiple related tumour samples. Nucleic Acids Res 2015;43:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. BROAD Institute. Mutect2. [cited 2019. Sep 5]. Available from: https://gatk.broadinstitute.org/hc/en-us/articles/360037593851-Mutect2
- 64. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014;30:1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Riester M, Singh AP, Brannon AR, Yu K, Campbell CD, Chiang DY, et al. PureCN: copy number calling and SNV classification using targeted short read sequencing. Source Code Biol Med 2016;11:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Derrien T, Estellé J, Sola SM, Knowles DG, Raineri E, Guigó R, et al. Fast computation and applications of genome mappability. PLoS One 2012;7:e30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. OncoKB. OncoKB Precision Oncology Knowledge Base. [cited 2020. Feb 8]. Available from: https://www.oncokb.org/
- 70. Ichikawa H, Nagahashi M, Shimada Y, Hanyu T, Ishikawa T, Kameyama H, et al. Actionable gene-based classification toward precision medicine in gastric cancer. Genome Med 2017;9:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018;46:D1062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sahasrabudhe R, Lott P, Bohorquez M, Toal T, Estrada AP, Suarez JJ, et al. Germline Mutations in PALB2, BRCA1, and RAD51C, which regulate DNA recombination repair, in patients with gastric cancer. Gastroenterology 2017;152:983–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Carvajal-Carmona LG. PALB2 as a familial gastric cancer gene: is the wait over? Lancet Gastroenterol Hepatol 2018;3:451–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Alexandrov LB, Nik-Zainal S, Siu HC, Leung SY, Stratton MR. A mutational signature in gastric cancer suggests therapeutic strategies. Nat Commun 2015;6:8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bang Y-J, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687–97. [DOI] [PubMed] [Google Scholar]
- 77. Wilke H, Muro K, Cutsem EV, Oh S-C, Bodoky G, Shimada Y, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol 2014;15:1224–35. [DOI] [PubMed] [Google Scholar]
- 78. Apicella M, Corso S, Giordano S. Targeted therapies for gastric cancer: failures and hopes from clinical trials. Oncotarget 2017;8:57654–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sohn BH, Hwang J-E, Jang H-J, Lee H-S, Oh SC, Shim J-J, et al. Clinical significance of four molecular subtypes of gastric cancer identified by The Cancer Genome Atlas project. Clin Cancer Res 2017;23:4441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang T, George DJ. Immunotherapy and targeted-therapy combinations mark a new era of kidney cancer treatment. Nat Med 2021;27:586–8. [DOI] [PubMed] [Google Scholar]
- 81. Halmai NB, Carvajal-Carmona LG. Diversifying preclinical research tools: expanding patient-derived models to address cancer health disparities. Trends Cancer 2022;8:291–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang X, Amitay E, Harrison TA, Banbury BL, Berndt SI, Brenner H, et al. Association between smoking and molecular subtypes of colorectal cancer. JNCI Cancer Spectrum 2021;5:pkab056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Slattery ML, Curtin K, Anderson K, Ma KN, Ballard L, Edwards S, et al. Associations between cigarette smoking, lifestyle factors, and microsatellite instability in colon tumors. J Natl Cancer Inst 2000;92:1831–6. [DOI] [PubMed] [Google Scholar]
- 84. Amitay EL, Carr PR, Jansen L, Roth W, Alwers E, Herpel E, et al. Smoking, alcohol consumption and colorectal cancer risk by molecular pathological subtypes and pathways. Br J Cancer 2020;122:1604–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pande M, Lynch PM, Hopper JL, Jenkins MA, Gallinger S, Haile RW, et al. Smoking and colorectal cancer in Lynch syndrome: results from the colon cancer family registry and the University of Texas M. D. Anderson Cancer Center. Clin Cancer Res 2010;16:1331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jiao J, Niu W, Wang Y, Baggerly K, Ye Y, Wu X, et al. Prevalence of aflatoxin-associated TP53R249S mutation in hepatocellular carcinoma in hispanics in South Texas. Cancer Prev Res 2018;11:103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Koshiol J, Gao Y-T, Dean M, Egner P, Nepal C, Jones K, et al. Association of aflatoxin and gallbladder cancer. Gastroenterology 2017;153:488–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ramirez AG, Muñoz E, Parma DL, Michalek JE, Holden AEC, Phillips TD, et al. Lifestyle and clinical correlates of hepatocellular carcinoma in south texas: a matched case-control study. Clin Gastroenterol Hepatol 2017;15:1311–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Klapheke AK, Carvajal-Carmona LG, Cress RD. Racial/ethnic differences in survival among gastric cancer patients in california: gastric cancer survival by race. Cancer Causes Control 2019;30:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Carvajal-Carmona LG. Molecular subtypes and driver mutations in Latinos with gastric cancer: implications for etiological and translational research. In: Ramirez AG, Trapido EJ, editors. Advancing the science of cancer in Latinos. Cham: Springer International Publishing; 2020. p. 89–94. [PubMed] [Google Scholar]
- 91. Wang SC, Yeu Y, Hammer STG, Xiao S, Zhu M, Hong C, et al. Hispanic/Latino patients with gastric adenocarcinoma have distinct molecular profiles including a high rate of germline CDH1 variants. Cancer Res 2020;80:2114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spreadsheet of pan-cancer panel gene composition, with citation codes
Spreadsheet of sample mean sequencing depths
Spreadsheet of counts of patients with SNV/indel mutations in each gene
PDF file of supplementary methods and materials, and supplementary figures and tables
Data Availability Statement
The data generated in this study were deposited in the European Genome-phenome Archive (Study ID: EGAS00001006650, Dataset ID: EGAD00001009622).