Abstract
Solid tumours display limited response to immunotherapies. By contrast, hematological malignancies exhibit significantly higher response rates to immunotherapies as compared to solid tumors. Among several microenvironmental and biological disparities, the differential expression of unique immune regulatory molecules significantly contributes to the blood cancer’s interaction with immune cells. The self-ligand receptor of the signaling lymphocytic activation molecule family member 7 (SLAMF7), a molecule that is critical in promoting the body’s innate immune cell to detect and engulf cancer cells, is expressed nearly exclusively on the cell surface of hematologic tumours, but not on solid ones. Here, we show that a bispecific nanobioconjugate that decorates SLAMF7 onto the surface of solid tumours induces robust phagocytosis and activates phagocytes cyclic GMP-AMP synthase–stimulator of interferon genes (cGAS-STING) pathway, sensitising the tumours to immune checkpoint blockade. Our findings support an immunological conversion strategy that employs nano-adjuvants to improve the effectiveness of immunotherapies for solid tumours.
Cancer immunotherapy has revolutionised oncologic care and improved the survival of patients diagnosed with terminal malignancies1-4. Despite its clinical benefit, the response rate of cancer immunotherapy , is far from optimal. It is now established that hematologic cancers respond to cancer immunotherapy at a much higher rate than solid tumours5-9. Although the reason for this disparity in cancer immunotherapy response is multifactorial, one possible contributing factor is the distinctive expression of surface proteins on hematologic and solid tumour cells that regulate immune cell functions. One membrane bound protein, signaling lymphocytic activation molecule family member 7 (SLAMF7), was recently reported to be exclusively expressed by hematopoietic cells including cancer cells, but not in cells from solid tumours10. SLAMF7 acts as an “eat me” signal for antigen presenting cells (APCs) and promotes the phagocytosis of cancer cells through the formation of a homodimer complex11. The exclusive expression of SLAMF7 by hematologic cancers was postulated to govern their responsiveness to cancer immunotherapy such as CD47 blockade12-15. Therefore, strategies16,17 that promote the surface presentation of SLAMF7 on solid tumour cells might result in immunologic activity similar to that of hematogic cancers and thereby improve the efficacy of cancer immunotherapy (Figure 1a).
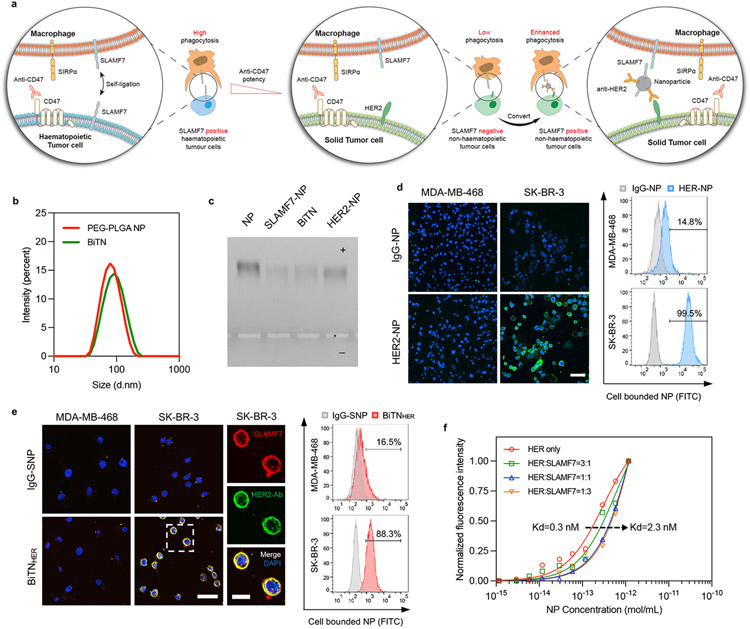
Figure 1. The BiTNHER nanoparticle targets and converts specific non-hematopoietic cancer cells into hematopoietic-like cancer cells.
a, Schematic of the nanoparticle-based conversion strategy via anti-HER2 antibody targeting and SLAMF7 labeling of HER2-expressing cancer cells. b, Size distribution of unconjugated PEG-PLGA nanoparticles (NPs) and BiTNHER measured by dynamic light scattering. The increased size indicated the successful conjugation of antibody/protein onto the NP surface. c, Gel electrophoresis of unconjugated NP and NP conjugated with anti-HER2 antibody (HER-NP), SLAMF7 (S-NP), and both (BiTNHER). Experiment was repeated twice with similar results between repeats. d, HER-NP labeling of the HER2-expressing human breast cancer cell line SK-BR-3. (Left) Confocal images of HER2low MDA-MB-468 cells and HER2high SK-BR-3 cells after incubation with IgG-NP or HER-NP. Green, NPs; blue, DAPI (scale bar, 50 μm). (Right) FACS semiquantitative analysis of binding. e, BiTNHER labeling of HER2-expressing SK-BR-3 cells with SLAMF7. (Left) Confocal images of HER2low MDA-MB-468 cells and HER2high SK-BR-3 cells after incubation with IgG-SLAMF7-NP or HER-SLAMF7-NP. Cells were stained with anti-SLAMF7 antibody and anti-human IgG antibody (scale bar, 50 μm). Higher-magnification image of SK-BR-3 cells in the outlined area (scale bar, 20 μm). (Right) FACS semiquantitative analysis of binding. f, Binding affinity of nanoparticles with different HER:SLAMF7 conjugation ratios to HER2-expressing SK-BR-3 cells. The dissociation constant Kd increased when the conjugation ratio of anti-HER2 antibody decreased.
To test this hypothesis, we developed a universal nanobioconjugate platform, which we termed bispecific tumour-transforming nanoparticles (BiTN). The BiTN is composed of a polymeric core with amine-reactive crosslinkers to conjugate exchangeable tumour-targeting ligands, and SLAMF7. To demonstrate that BiTN can selectively bind to solid tumour cells in a targeted manner, we selected breast cancers that express the human epidermal growth factor receptor 2 (HER2) receptor. We generated BiTN by conjugating polymeric nanoparticles with anti-HER2 antibody and recombinant SLAMF7 (BiTNHER). Attaching anti-HER2 antibody and/or SLAMF7 slightly increased the size and zeta potential of the polymeric nanoparticles (NPs) (Figure 1b and Supplementary Figure S1) but maintained their monodispersity (Figure 1c). The valency of conjugated antibody/SLAMF7 was calculated (Supplemental Figure S1). These BiTN nanoparticles found were to be stable in aqueous solutions with no obvious change of size, polydispersity index (PDI) and protein detachment noted within 72 hours either in PBS or FBS containing media (Supplementary Figure S2). The conjugated anti-HER2 antibody showed selective targeting to HER2-overexpressing human and mouse breast cancer cells (Figure 1d and Supplementary Figure S1), which allowed the successful SLAMF7 labeling of HER2high cells with the bispecific BiTNHER (Figure 1e). Although bispecific conjugation of SLAMF7 to the anti-HER2 antibody conjugated nanoparticles decreased the binding avidity between the NPs and HER2 receptors on the cell surface, as the dissociation constants (Kd) increased with the increased ratio of conjugated SLAMF7:anti-HER2 antibody, the binding efficiency of BiTNHER became much higher than free anti-HER2 antibody owing to the multivalency of NPs18 (Figure 1f).
BiTN sensitises tumour cells to CD47 blockade
To function as an “eat me” signal and induce innate immune responses, SLAMF7 needs to be expressed on target cancer cells as a homotypic receptor and interact with the SLAMF7 on phagocytes for effective recognition and phagocytosis10. For instance, SLAMF7 is exclusively expressed is on hematologic cells, thus making cancer cells of hematopoietic lineage much more sensitive to CD47 blockade–mediated macrophage phagocytosis (Supplementary Figure S3). To adopt SLAMF7-mediated sensitisation of CD47 blockade in solid tumours using BiTN, target cells must first have optimal BiTN binding via receptor-targeting. By varying the ratio of conjugated SLAMF7:anti-HER2 antibody on the NPs, we determined that a 3:1 ratio induced the most robust degree of macrophage phagocytosis of HER2high SK-BR-3 human breast cancer cells compared with unconjugated NPs (Figure 2a). Furthermore, this pro-phagocytic effect required HER2 expression by the target cells, as increased phagocytosis by macrophages was not observed in HER2low MDA-MB-468 human breast cancer cells (Figure 2b). However, HER2 expression was not necessary if SLAMF7 was sufficiently abundant on target cancer cells. Indeed, a high degree of phagocytosis was observed in SLAMF7highHER2low L1210 cells regardless of NP treatment. Inhibition of SLAMF7 ligation between macrophages and cancer cells by anti-SLAMF7 antibody eliminated the pro-phagocytic effect of BiTNHER (Figure 2c). To further substantiate the role of SLAMF7 in promoting macrophage phagocytosis, we generated SLAMF7 overexpressed human breast cancer cell line SK-BR-3SLAMF7+ and MDA-MB-468SLAMF7+, as well as SLAMF7 knock-down in human macrophage THP-1SLAMF7− (Supplementary Figure S4). In contrast to parental cells, both SLAMF7 overexpressing cancer cells exhibited increased phagocytosis by macrophages in response to CD47 blockade. On the other hand, THP-1SLAMF7− cells significantly abrogated BiTN-induced macrophage phagocytosis of HER2high SK-BR-3 cells. Together, these results indicated that the targeted transformation of HER2high breast cancer cells to a SLAMF7high phenotype is essential to initiate phagocytosis by macrophages. Moreover, changing the conjugation of human anti-HER2 antibody to a mouse anti-HER2/neu antibody led to antigen-specific, SLAMF7-mediated phagocytosis in HER2high EO771/E2 and TUBO cells (Figure 2d,e and Supplementary Figure S5-6). The BiTNHER itself could induce moderate macrophage phagocytosis at high concentrations, but it greatly sensitised the HER2high EO771/E2 cells to CD47 blockade as an adjuvant, since the phagocytic activity was further elevated when BiTNHER was combined with anti-CD47 antibody (aCD47) treatment (Supplementary Figure S7). Given that macrophages are often polarised in the tumour microenvironment, which affects their phagocytic ability19, we further tested the pro-phagocytic effect of the BiTNHER and aCD47 combination on M1 macrophages polarised by lipopolysaccharide (LPS) and on M2 macrophages polarised by interleukin-4 (IL-4), the latter phenotypically more resemble that of tumour-associated macrophages. The polarisation of macrophages had little impact on the expression level of SLAMF7 on the surface of the macrophages, or on activation of phagocytosis against EO771/E2 cells mediated by BiTNHER and aCD47 (Supplementary Figure S8).
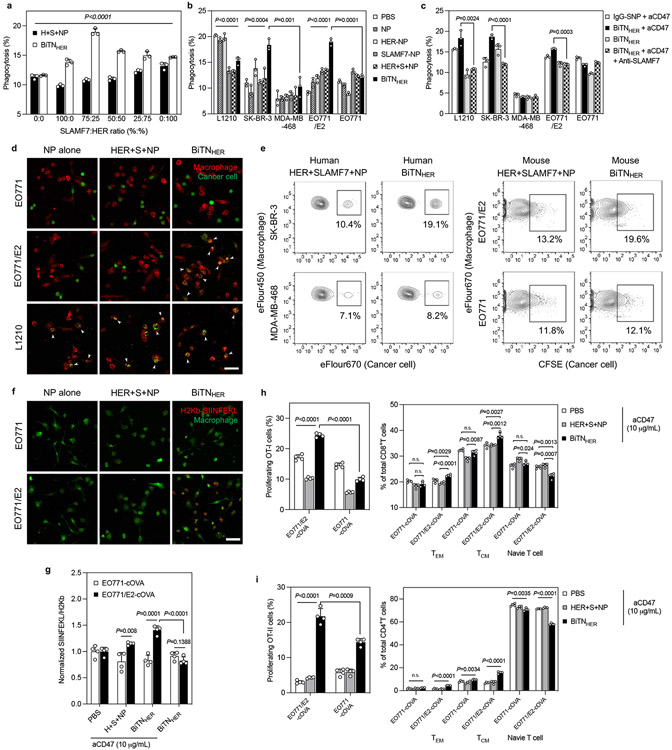
Figure 2. Combination of aCD47 and BiTNHER induces targeted phagocytosis of cancer cells and subsequent macrophage activation.
a, BiTNHER with a 3:1 SLAMF7:HER conjugation ratio had the maximum pro-phagocytosis effect of human THP-1 against HER2-expressing SK-BR-3 cancer cells in the presence of aCD47 (n=3). b, BiTNHER converted HER2/neu-expressing human (SK-BR-3) and mouse (EO771/E2) breast cancer cells into SLAMF7high cells and promoted human THP-1 or mouse (C57BL6 bone marrow) macrophage phagocytosis in the presence of aCD47, comparable with the SLAMF7-expressing mouse leukemia L1210 cells (n=3). c, Anti-SLAMF7 antibody abrogated the pro-phagocytosis effect of BiTNHER and aCD47 on HER2-expressing cancer cells (n=3). d, Phagocytosis of CFSE-labelled HER2low EO771 and HER2high EO771/E2 mouse breast cancer cells and SLAMF7high L1210 mouse leukemia cells by mouse bone marrow macrophages in the presence of aCD47 after treatment with NP alone, NP with unconjugated anti-HER2 antibody and SLAMF7, or BiTNHER. Red, macrophages; green, cancer cells (scale bar, 50 μm). e, BiTNHER with aCD47 promotes macrophage phagocytosis against HER2-expressing breast cancer cells. f, Macrophages had increased antigen presentation of the H2kb-SIINFEKL complex after phagocytosis of BiTNHER -treated HER2-expressing EO771/E2-cOVA cells. Green, macrophages; red, H2kb-SIINFEKL complex (scale bar, 50 μm). g, Combination of BiTNHER and aCD47 increased macrophage antigen presentation of HER2-expressing cancer cells (n=4). h,i, BiTNHER with aCD47 promoted the priming of cOVA antigen-specific T cells (left) and induced a shift in naive T cells towards memory T cells (right) (n=4). For all figures, data are presented as mean±s.e.m.; **P<0.01, ***P<0.001, and ****P<0.0001 by one-way ANOVA with a Bonferroni post hoc correction. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 by unpaired Student’s t-test for the indicated comparisons; n.s., not significant.
Monoclonal antibodies can induce antibody-dependent cellular phagocytosis through engagement of the Fc receptor on macrophages, thus we generated a Fc silenced anti-HER2 antibody to rule out the possibility that the Fc fragments on BiTN were responsible for promoting phagocytosis of tumour cells (Supplementary Figure S9). Fc fragment was digested with pepsin and F(ab’)2 fragment of anti-HER2 antibody was obtained with similar HER2 targeting ability as whole anti-HER2 antibody. We prepared the bispecific nanoconjugate with F(ab’)2 fragment (designated as BiTNHER(aHER2-F(ab’)2)) and with whole IgG antibody (designated as BiTNHER(aHER2-IgG)). BiTNHER(aHER2-F(ab’)2) treated EO771/E2 had slightly lower phagocytosis by macrophage compared with BiTNHER(aHER2-IgG) group, but the Fc silenced NPs still showed potent and selective pro-phagocytosis effects against HER2-positive EO772/E2 cells (Supplementary Figure S9). These results demonstrate that BiTN-mediated macrophage phagocytosis is only partially induced by the Fc engagement from the conjugated antibody, the major pro-phagocytosis effect is mediated through the SLAMF7 labeling of the HER2-positive cancer cell and engaging with SLAMF7 on macrophages.
Macrophages phagocytise cancer cells, presenting tumour-derived antigens and prime T lymphocytes17. To test whether the combination of BiTNHER and aCD47 could bridge these innate and adaptive immune responses, we co-cultured EO771/E2 and EO771 cancer cells expressing cytoplasmic ovalbumin (cOVA) with macrophages and exposed these macrophages with OVA-specific CD4+ (OT-II) and CD8+ (OT-I) T cells. Consistent with the macrophage-phagocytosis findings, enhanced presentation of OVA peptide with major histocompatibility complex I (MHC1, the SIINFEKL-H2kb complex) was observed only on macrophages co-cultured with BiTNHER and aCD47 co-cultured with HER2high EO771/E2-cOVA cells (Figure 2f). The combination treatment induced higher levels of antigen presentation compared with unconjugated NPs, and the selectivity over HER2high cancer cells was abrogated when aCD47 was absent (Figure 2g). We next evaluated the priming of OVA-specific T cells after exposure to APCs. Similarly, the combination treatment led to greater proliferation of both CD4+ and CD8+ T cells from the co-culture system of macrophages and EO771/E2-cOVA cells (Figure 2h,i). Within the activated CD8+ T cells, we found a more significant phenotypic shift from naïve T cells towards effector memory (TEM) and central memory (TCM) T cells in the BiTNHER - and aCD47-treated group compared with aCD47-alone or aCD47- and unconjugated NPs-treated groups (Figure 2h). Although minor shifting was noted in CD4+ T cells, the overall activation level was much less than in CD8+ T cells (Figure 2i).
BiTN enhances antitumour effect of CD47 blockade
Given the strong ability of BiTNHER and aCD47 combination therapy to initiate immune responses in vitro, we next evaluated its antitumour potential in orthotopic mouse breast tumour models by using TUBO breast cancer cells, which express the rodent homolog of the HER2 protein, and 4T1 cells, which lack the receptor. Intratumoural injections of BiTNs resulted in a long retention within the tumour with minimal nanoparticle distribution in peripheral organs (Supplementary Figure S10). A combination of BiTNHER and aCD47 at a predetermined dosage led to significant tumour inhibition compared with monotherapy with BiTNHER or aCD47 in neuhigh TUBO tumours (Figure 3a and Supplementary Figure S11). The combination treatment significantly reduced tumour burden and prolonged the survival of TUBO tumour-bearing mice, with 3 of 6 mice remaining tumour-free (Figure 3c). However, the same antitumour effect was not observed in neulow 4T1 tumour-bearing mice (Figure 3b). To elucidate the tumour-specific inhibitory effect of the combination therapy, we first examined the tumour infiltration of both innate and adaptive immune cells. Treatment with BiTNHER and aCD47 led to increased infiltration of natural killer (NK) cells and DCs but minimal change to the overall number of macrophages in TUBO tumours (Figure 3d and Supplementary Figure S12). However, upon immunostaining ionised calcium binding adaptor molecule 1 (Iba-1), a macrophage-specific calcium-binding protein often used as a marker of macrophage activation, we found the highest levels of activated macrophages in the treated TUBO tumours (Supplementary Figure S13). Treatment with BiTNHER and aCD47 significantly increased the overall number of T cells in TUBO but not in 4T1 tumours (Figure 3e and Supplementary Figure S14). Interestingly, lower percentages of immunosuppressive CD4+ regulatory T cells (Tregs) and higher percentages of interferon γ (IFN-γ)-secreting CD8+ effector T cells were found in TUBO tumours after the combination treatment, relative to untreated TUBO tumours or treated 4T1 tumours (Figure 3f,g). Moreover, depletion of CD8+ T cells completely abolished the antitumour effect of BiTNHER and aCD47 combination therapy (Supplementary Figure S15). This removal of cytotoxic IFN-γ+CD8+ T cells from the tumour microenvironment led to significantly lower infiltration of DCs and more immunosuppressive myeloid-derived suppressor cells (MDSCs) (Supplementary Figure S15). These results suggest that BiTNHER transformed HER2/neuhigh tumours to SLAMF7high tumours, thereby promoting tumour-specific activation of local innate immune responses, as we observed previously in vitro. These results further support the finding that SLAMF7 ligation physically triggers macrophage activation and synergistically promote CD47 blockade to bridge the innate and adaptive immune systems and enhance the elimination of tumour cells14, 20.
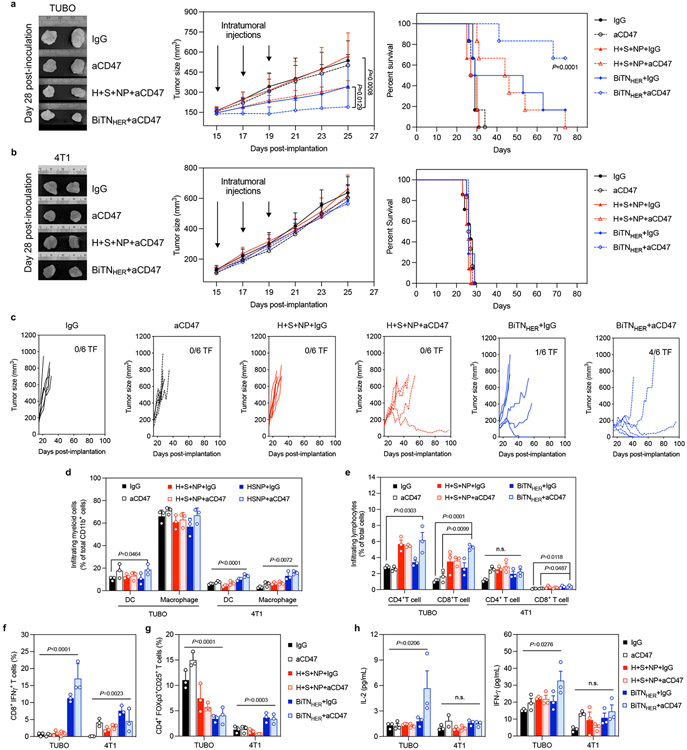
Figure 3. Combined treatment with BiTNHER and aCD47 induces antitumor immune responses in HER2/neu-expressing mouse tumor model.
a,b, Combination of BiTNHER and aCD47 treatment inhibited the growth of HER2/neuhigh TUBO breast cancer cells and prolonged the survival of BALB/c mice, compared with HER2/neulow 4T1 breast cancer cells. ***P<0.001 by log-rank test (n=6). c, Combined treatment with BiTNHER and aCD47 induced the greatest tumor-inhibition effect in HER2/neu-expressing TUBO tumors, with 3 of 6 mice remaining tumor-free (TF). d-g, Immune cell infiltration profiles within HER2/neuhigh TUBO tumors and HER2/neulow 4T1 tumors (n=3). (d) Combined treatment with BiTNHER and aCD47 promoted the infiltration of dendritic cells (DCs) and macrophages within TUBO tumors relative to 4T1 tumors. (e) Combined treatment with BiTNHER and aCD47 increased the infiltration of CD4+ T and CD8+ T cells into TUBO tumors. (f,g) Combined treatment with BiTNHER and aCD47 increased the number of IFNγ-producing CD8+ T cells in TUBO tumors and decreased the number of intratumoral regulatory CD4+ T cells. h, Combined treatment with BiTNHER and aCD47 increased the peripheral-blood cytokine levels in TUBO tumor-bearing mice but not in 4T1 tumor-bearing mice (n=3). For all figures, the data are presented as mean±s.e.m.; *P<0.05, **P<0.01, ****P<0.0001 by one-way ANOVA with a Bonferroni post hoc correction; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 by unpaired Student’s t-test for the indicated comparisons; n.s., not significant.
Locally applied immunotherapies have been reported to induce systemic immune responses due to the so-called cancer-immunity cycle16, 21-23. The cancer-immunity cycle here began with the macrophage phagocytosis mediated by BiTN. The antigen presenting cells then released and presented tumour antigens to naïve T cells, educated them into tumour-killing T cells and activated the adaptive immune response in secondary lymphatic organs like spleen. To assess systemic immune activation, we next tested whether the peripheral blood levels of interleukin-2 (IL-2) and IFN-γ were elevated, as both are required for a proper adaptive immune response24-26. As expected, significant increases in both cytokines were observed in BiTNHER - and aCD47-treated TUBO tumour-bearing mice (Figure 3h). Consistent with this result, the combination treatment led to decreased populations of naïve T cells in the spleens of TUBO-bearing mice; likewise, a significant shift to both the TEM and TCM phenotypes were noted relative to mice bearing TUBO tumours, compared with those bearing 4T1 tumours (Supplementary Figure S16). This selectivity of systemic immunity over the extent of HER2 expression in tumours also reveals a correlation between immunogenicity and the targeted transformation of HER2high tumour to SLAMF7high tumour by BiTNHER treatment.
To ensure the systemic immune activation by BiTNs does not lead to autoimmune toxicity, we performed long-term toxicity study (Supplementary Figure S17). No significant change of blood counts between untreated and treated mice was found. The tumour-bearing mice had slight but not significant increases in the number of lymphocytes after treatment, which is in line with the tumour inhibition effects showing activated systemic immune responses. In addition, no obvious inflammation or lymphocyte infiltrates was found in major organs. These results support the safety profile of BiTN treatment.
The therapeutic effects of the BiTNHER and aCD47 combination were further investigated with HER2high EO771/E2 and HER2low EO771 tumours established in C57BL/6 mice. Unexpectedly, the antitumour effects against the EO771/E2 tumours were not as potent as those against TUBO tumours. Although tumour growth was retarded by the combination treatment compared with untreated or single-component–treated groups, all mice reached the tumour burden end point within 45 days after tumour inoculation (Supplementary Figure S18). In line with these results, we found a much higher expression of programmed death protein 1 (PD1) and programmed death-ligand 1 (PDL1) in EO771/E2 tumours than on treatment-responding TUBO tumours, suggesting a more immunosuppressive tumour microenvironment27-29. Addition of an anti-PD1 antibody (aPD1) greatly enhanced the antitumour effect of BiTNHER with greater tumour growth inhibition (Figure 4a). Similar to the results in TUBO tumours, the triple combination of aPD1, BiTNHER, and aCD47 induced greater local infiltration of tumour-killing immune cells and enhanced systemic immune responses (Supplementary Figures S18-19). Again, these antitumour effects were specific to HER2high EO771/E2 tumours and were not observed in HER2low EO771 tumour-bearing mice. Nevertheless, no significant difference of tumour growth was observed between mice treated with aPD1 alone or aPD1 plus BiTNHER, indicating that aPD1 and BiTNHER lacks the synergetic anti-tumour effect in the absence of aCD47 (Supplementary Figures S20). This result is consistent with what we found in TUBO tumours, where CD47 blockade significantly enhanced the anti-tumour effect of BiTNHER alone (Figure 4). These results further confirm that BiTN works as an adjuvant for CD47 blockade and enhance the solid tumour response to immunotherapy.
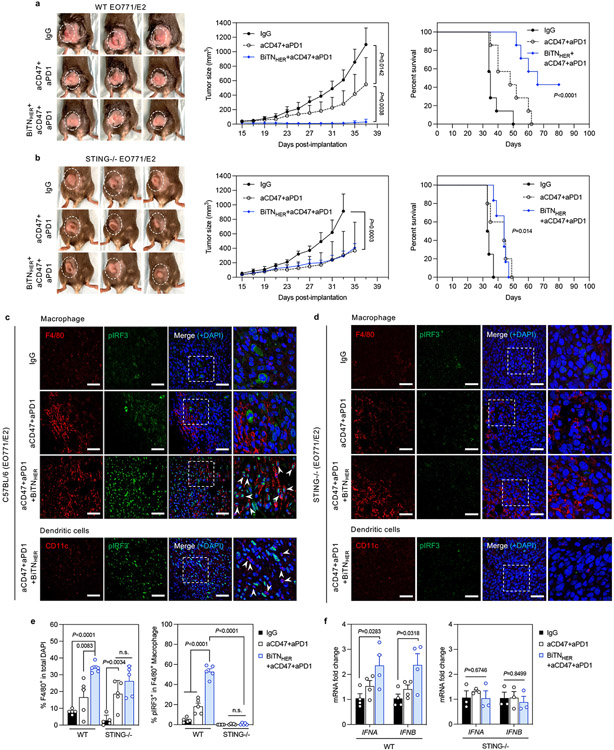
Figure 4. STING activation is involved in the triple-combination-treatment–mediated immune response.
a, Tumor growth was inhibited by the triple-combination treatment and significantly prolonged the survival of EO771/E2 tumor-bearing mice (n=7). b, STING knockout partially abrogated the antitumor effect of the triple-combination treatment in EO771/E2-bearing mice (n=5). c-d, Immunofluorescence staining of EO771/E2 tumors implanted in wild-type (WT) (c) and STING knockout (d) mice (scale bar, 50 μm). Areas within the dashed squares are shown at higher magnification in the images to the right; arrows indicate the nuclear-translocated pIRF3 within F4/80+ macrophage or CD11c+ dendritic cells (scale bar, 20 μm). e, Quantification of infiltrated F4/80+ macrophages and percentage of nuclear pIRF3+ macrophages in the tumors (n=5). f, Expression of type I interferons is elevated in intratumoral F4/80+ macrophages after triple combination treatment (IFNA, interferon α; IFNB, interferon β) (n=4 for WT, n=3 for STING KO). For all figures, data are presented as mean±s.e.m.; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 by two-tailed unpaired Student’s t-test for the indicated comparisons in figure 4a,b, e and f; *P<0.05, ****P<0.0001 by log-rank test; n.s., not significant.
BiTN’s antitumour effects require host cGAS-STING activation
The cyclic GMP-AMP synthase–stimulator of interferon genes (cGAS-STING) pathway is involved in immune activation prompted by combined treatment with CD47 blockade and chemotherapy30, 31. To test whether this innate immune sensor activation is required following combination treatment with BiTN, EO771/E2 tumours were grown in the mammary fat pad of STING knockout (KO) or wild-type (WT) mice and were treated with different regimens. Compared with the WT animals, the antitumour effect of BiTNHER was completely abrogated in STING KO animals, where we found no significant difference in tumour growth after aCD47-plus-aPD1 treatment with or without BiTNHER (Figure 4b). Meanwhile, the triple combination therapy strongly induced the phosphorylation and nuclear translocation of the STING downstream transcription factor IFN regulatory factor 3 (IRF3) in the intratumoural macrophages of WT mice, but not in those of IgG-treated WT mice or STING KO mice (Figure 4c-e). As the major source of type I IFN production32, STING activation led to increased IFNα/β mRNA expression in the intratumoural macrophages isolated from triple combination therapy–treated WT mice, compared with untreated or antibody-only groups (Figure 4f). In contrast, lower IFNα/β mRNA expression was observed in tumour-associated macrophages from the triple combination–treated STING KO mice relative to those from treated WT mice. Moreover, this decreased expression seems to correlate with reduced infiltration of activated macrophages and T cells in treated tumours (Supplementary Figure S21). Since BiTN could promote macrophage phagocytosis of cancer cells, resulting in increased likelihood of activation of STING pathway by released free DNA from digested cancer cells, leading to macrophage activation and upregulation of Iba-1 expression (Figure S13) 33-35. Interestingly, although the total number of tumour-infiltrating DCs was elevated in WT mice, no detectable nuclear translocation of pIRF3 was observed. Expression of type I IFNs from DCs was also not significantly increased after treatment (Supplementary Figure S22), indicating that STING pathway was not activated in DCs at least 24 h after the last treatment. Thus, we speculate that only macrophages were directly engaged by the BiTN to phagocyte cancer cells. They were the first population of cells that responded to the treatment and initiated the subsequent immune responses, followed by DC recruitment and activation 36. Collectively, these results suggest that STING activation in host cells is critical for tumour inhibitory effect of combined BiTN and immunotherapy treatment. Although CD47 and PD1 blockade had moderate antitumour effects, BiTNHER was the necessary component of the combinational therapy that sensitized the tumour to these immune checkpoint inhiibitors37, 38.
Combined immunotherapy with customized BiTN inhibits metastasis
To demonstrate the versatility of BiTN platform for treating solid tumours with distinct molecular profiles, we sought to customised it to target other tumour receptors. We further developed a BiTNFo by exchanging the anti-HER2 antibody with folate to target triple-negative breast cancer (TNBC), where the folate receptor (FR) α is highly expressed39,40, BiTNFo similarly targeted and transformed FRhigh cancer cells into SLAMF7high cells. As expected, incubating 4T1 cancer cells with the optimised BiTNFo and aCD47 led to the greatest phagocytosis by macrophages relative to FRlow TUBO cells (Supplementary Figure S23). The increased optimal ratio of conjugated targeting ligand from 1:3 HER2:SLAMF7 on the BiTNHER to 3:1 folate:SLAMF7 on the BiTNFo may be attributed to the different affinity of ligands for receptors.
We next tested the therapeutic effect of BiTNFo by using a spontaneous 4T1 metastasis mouse model. We treated the primary tumour with intratumourally injected BiTNFo, along with intraperitoneal injections of aCD47 before surgical resection of the primary lesions. Compared with IgG- or aCD47-treated groups, the BiTNFo and aCD47 combination resulted in enhanced inhibition of local disease recurrence (Supplementary Figure S24). Evaluation of tumour infiltrating immune cells revealed increased recruitment of IFNγ+ cytotoxic CD8+ T cells and DCs, with elevated numbers of memory T cells in spleen (Supplementary Figure S24). However, the activated immune response in the BiTNFo-and-aCD47–treated mice did not delay distant metastasis progression or prolonged overall survival (Supplementary Figure S24). When analysing mice that were deemed ‘good responders’ or ‘poor responders’ based on the primary tumour progression after local therapy, we found that the ‘good responders’ had significantly lower expression of PD1 and PDL1 on the tumours, which may have accounted for the increased survival rate of these mice (Supplementary Figure S24). This result may be secondary to high immune resistance through the upregulation of immune checkpoints upon adaptive immune activation41-43, and therefore blockade of the PD1/PDL1 axis may enhance the therapeutic effects of BiTNFo.
As expected, the addition of aPD1 to the treatment promoted the tumour-inhibition effects of both aCD47 alone and aCD47 plus BiTNFo on the primary tumours (Figure 5a,b). In addition, compared with IgG treatment, the triple combination treatment altered the immunosuppressive tumour microenvironment, as demonstrated by increased infiltration of tumour-killing IFNγ+ CD8+ T cells and antigen-presenting DCs with corresponding decreases in tumour-promoting CD4+ Tregs and MDSCs (Figure 5c,d). To further control the formation of metastases after primary tumour removal, we tested continued treatment with BiTNFo, aCD47, and aPD1. In contrast to the previous double combination of aCD47 and BiTNFo, the triple combination treatment led to prolonged inhibition of metastasis burden (size and number). Indeed, 2 of 7 mice had long term survival and remained tumour free (Figure 5e,f). Finally, we did not observe notably acute toxicities from the intravenous administration of BiTNFo (Supplementary Figure S25). These results indicate that BiTN could function as a robust neoadjuvant to generate in situ tumour vaccination via promoting macrophage phagocytosis and inducing systemic inhibition of highly immunosuppressive solid tumours when combined with immune checkpoint blockade. Luciferase has been reported to be weakly immunogenic in Balb/c mice44, which could increase the sensitivity of tumours to checkpoint blockade. However, the immunogenicity of this xeno-antigen alone was insufficient to generate potent immune response as aCD47 and aPD1 only slight prolonged mice survival. In contrast, combination of BiTN with aCD47 and aPD1 strongly boosted the immune system to eliminate residual tumours and prevent metastasis.
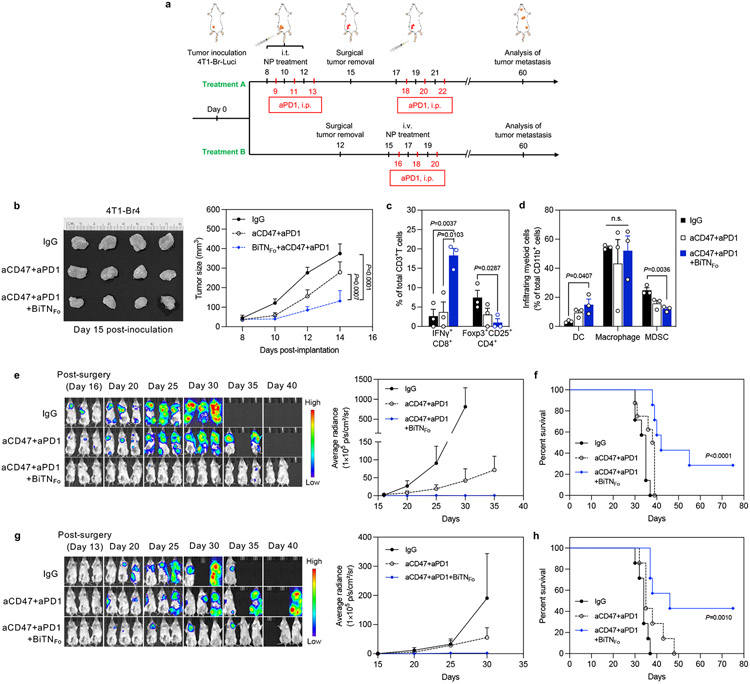
Figure 5. Combination treatment with BiTNFo, aCD47, and aPD1 generated in situ tumor vaccine effects and induced systemic antitumor effects against 4T1 tumor metastasis.
a, Experimental design for establishing 4T1 metastasis model and two treatment strategy. Treatment A, intratumoral pretreatment of primary tumor, followed by additional i.v. injection of BiTNFo after surgery; Treatment B, direct i.v. injection of BiTNFo after surgery without pretreatment. b, The triple combination treatment inhibited the growth of 4T1 primary tumors (n=8). c, The triple combination treatment increased the infiltration of IFNγ+ cytotoxic CD8+ T cells and decreased the infiltration of immunosuppressive CD4+ Treg cells (n=3). d, The triple combination treatment increased the infiltration of dendritic cells (DCs) and decreased the infiltration of myeloid-derived suppressor cells (MDSCs) (n=3). e, IVIS imaging (left) and quantification of whole-body signal (right) showed decreased number and size of tumor metastases after triple combination treatment (n=8). f, Combination treatment induced long-term protection against tumor metastasis, with 2 of 7 surviving mice remaining tumor-free (n=7). g, IVIS imaging (left) and quantification of whole-body signal (right) (n=7). h, Combination treatment induced long-term protection against tumor metastasis (n=7). Data for all figures are presented as mean±s.e.m., with *P<0.05, **P<0.01 by two-tailed unpaired Student t-tests for the indicated comparisons in figure 5b,c,d; *P<0.05, by log-rank test; n.s., not significant.
To further increase the translational relevance of this model, a post-surgery treatment regimen (Treatment B) was performed. In brief, 4T1-Br4-Luci cells were inoculated, and the grown primary tumours were resected on Day 12 without any pretreatment. Starting from Day 15, mice were treated with BiTNFo, naCD47 and aPD1 (Figure 5a). By frequent monitoring of tumour luciferase signal, we found the similar trend of metastasis inhibition and prolonged mice survival induced by the post-surgery treatment of BiTNFo, aCD47 and aPD1 (Figure 5g-h). These results indicated that even without pre-generation of in-situ tumour vaccine by intratumoural treatment, administration of the BiTNFo as an adjuvant therapy was sufficient to eliminate residual tumour cells and reduce systemic diseases when combined with aCD47 and aPD1. These results further strengthened the strategy of BiTN-based immunotherapy and provided solid evidence in its antitumour efficiency in the postoperative setting.
Conclusions
Here, we showed that BiTN platform as a promising nano-adjuvant to enhance the antitumour effect of cancer immunotherapy in solid tumours. Through tumour-receptor-specific customisation and the incorporation of the ‘transforming’ ligand SLAMF7, BiTN targeted and transformed low-immunogenic solid tumour cells into a SLAMF7high phenotype. This transformation led to a markedly enhanced immune response in solid tumours. BiTN effectively promotes tumour cell phagocytosis and antigen presentation by macrophages and primed antigen-specific T cell responses. When combined with CD47 blockade, BiTN greatly enhanced its antitumour effect. Moreover, the addition of PD1 blockade further enhanced systemic T cell immunity against the local recurrence and systemic disease progression of aggressive metastatic tumours. Overall, our study supports a new immune conversion strategy that employs nano-adjuvants to improve the effectiveness of immunotherapies for solid tumours.
Methods
Mice.
Age-matched female C57BL/6, BALB/c, STING knockout (Tmem173gt) (6-8 weeks old) were used for tumour engraftment and tumour growth experiments. OT-I (C57BL/6-Tg(TcraTcrb) 1100Mjb/J) and OT-II (C57BL/6-Tg(TcraTcrb)425Cbn/J) TCR transgenic mice were used for T-cell isolation (female, 6-8 weeks old). All mice were purchased from Jackson Labs and maintained at the institution’s animal facility in a specific pathogen-free environment with an ambient temperature of 22°C and a relative humidity of 50%. All the mice were maintained on a standard diet and water in a 12h:12h light–dark cycle.. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Texas, MD Anderson Cancer Center.
Cell culture.
The human mammary carcinoma cell lines SK-BR-3 (HTB-30) and MDA-MD-468 (HTB-132), the human monocyte cell line THP-1 (TIB-202), the mouse mammary carcinoma cell lines 4T1 (CRL-2539) and EO771 (CRL-3461), and the mouse B-cell lymphocytic leukaemia cell line L1210 (CCL-219) were obtained from the American Type Culture Collection. The mouse mammary TUBO cell line was provided by S. J. Gendler at Mayo Clinic Arizona. The HER2-overexpressing EO771/E2 cell line was a generous gift from Y. Jun at the University of Louisville. SK-BR-3 cells were maintained in McCoy’s 5a medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. EO771/E2 cells were maintained in Dulbecco’s minimum essential medium (DMEM) complete medium supplemented with 0.1 mg mL−1 Geneticin Selective Antibiotic (G418 Sulfate) (ThermoFisher). All other cell lines were cultured in high-glucose DMEM supplemented with 10% FBS and 1% penicillin/streptomycin.
Bone marrow–derived macrophages (BMDMs) were extracted from the bone marrow of C57BL/6 or BALB/c mice femurs according to a previously established protocol45 and maintained in DMEM supplemented with a 30% L929 cell-conditioned medium (Stony Brook Cell Culture/Hybridoma facility), 20% FBS, 1% sodium pyruvate, and 1% penicillin/streptomycin. These macrophages were used for phagocytosis assays without further stimulation unless otherwise specified.
cOVA-expressing EO771 and EO771/E2 tumour cells were developed by pLenti-cOVA transfection according to a previously established protocol46. Cells were maintained in DMEM complete medium supplemented with 0.5 mg mL−1 G418 Sulfate.
The SLAMF7 overexpressing SK-BR-3 and MDA-MB-468 were developed by pLenti-SLAMF7 transfection (OriGene, #RC220985). SK-BR-3 cells were maintained in DMEM complete medium supplemented with 0.5 mg mL−1 G418 Sulfate; MDA-MB-468 cells were maintained in DMEM complete medium supplemented with 0.3 mg mL−1 G418 Sulfate.
The SLAMF7 knock-down THP-1 cells were developed by SLAMF7 silencing siRNA transfection (Thermofisher, #4392421) with DharmaFECT for 48 h after activation overnight.
The 4T1 cell line was transfected with lentivirus encoding the firefly luciferase gene to establish 4T1-Luc cells, which were further developed after four rounds of intracardiac injection/brain cell culture.
Cell cultures were incubated at 37°C in humidified conditions equilibrated with 5% CO2. Cell-line authentication was confirmed by none of the cell lines being listed in the International Cell Line Authentication Committee (ICLAC)’s Database of Contaminated or Misidentified Cell Lines. All cell lines were tested for mycoplasma contamination by using a biochemical method with Hoechst staining and were found to be free of mycoplasma contamination.
Nanoparticle construction.
Poly(D,L-lactic-co-glycolic acid)-b-poly(ethylene glycol) with terminal methoxy groups (PLGA-b-mPEG), terminal N-hydroxysuccinimide esters groups (PLGA-b-PEG-NHS) or terminal folic acid groups (PLGA-b-PEG-Folate) (molecular weight: PEG 5k, PLGA 20k, LA:GA=50:50) (Nanosoft polymers) were used to synthesize the unconjugated nanoparticles (NPs) by nanoprecipitation. Briefly, 1 mg PLGA-b-mPEG polymers were dissolved in dimethyl sulfoxide (DMSO) to 10 mg mL−1 and added dropwise into 1 mL of 10 mM PBS (pH 7.4) under magnetic stirring at 1500 rpm under room temperature for 2 hours. The solution was then centrifuged at 20,000 g for 60 min, and the obtained pellet was washed with PBS twice and resuspended in PBS.
To synthesise conjugated NPs, amine-reactive polymers were directly added into and reacted with PBS solution containing anti-HER2 antibodies (the human monoclonal anti-HER2 antibody trastuzumab from Genentech or the mouse monoclonal anti-HER2/neu antibody clone 7.16.4 from BioXcell), or recombinant SLAMF7 (human recombinant SLAMF7 from MyBioSource #MBS1458102, or mouse recombinant SLAMF7 from Genscript (Lot# U870KEL260-5)). Fc silenced anti-HER2 F(ab’)2 fragment was generated by digesting the whole IgG with pepsin overnight using Pierce F(ab’)2 Preparation Kit per manufacturer’s instruction (Thermofisher, #44988), the F(ab’)2 fragment was further purified by ultracentrifugation with 100 kDa centrifugal filters. Briefly, 0.6 mg PLGA-b-mPEG and 0.4 mg PLGA-b-PEG-NHS polymers were dissolved in DMSO to 10 mg mL−1 and added dropwise into 1 mL of 10 mM PBS (pH 7.4) containing a predetermined ratio of HER/SLAMF7 under magnetic stirring at 1500 rpm under room temperature for 2 hours. The reaction solution was purified as described above to obtain the conjugated NPs.
To synthesise folate-containing SLAMF7-conjugated nanoparticles, amine-reactive polymers were directly added into and reacted with PBS solution containing recombinant SLAMF7 (human recombinant SLAMF7 from MyBioSource, or mouse recombinant SLAMF7 from Genscript). Briefly, 0.6 mg PLGA-b-mPEG, 0.2 mg PLGA-b-PEG-NHS, and 0.2 mg PLGA-b-PEG-Folate polymers were dissolved in DMSO to 10 mg mL−1 and added dropwise into 1 mL of 10 mM PBS (pH 7.4) containing a predetermined amount of SLAMF7 under magnetic stirring at 1500 rpm under room temperature for 2 hours. The reaction solution was purified as described above to obtain the conjugated NPs.
To determine the loading efficiency of antibodies and SLAMF7, the supernatant after the first centrifugation of the reaction solution was collected for analysis. The concentrations of unbound anti-HER2/neu antibodies and SLAMF7 were measured with an immunoglobulin G-1 (IgG1) human or mouse ELISA Kit (ThermoFisher) and SLAMF7 human or mouse ELISA Kit (MyBioSource), respectively. Loading efficiency of anti-HER2/neu antibodies and recombinant SLAMF7, after conjugation, were calculated according to the following equation:
| Equation (1) |
Valency of anti-HER2/neu antibodies and recombinant SLAMF7, after conjugation, were calculated according to the following equations:
| Equation (2) |
| Equation (3) |
Solution-based characterisations of NP size and charge distributions were measured with a Zetasizer Nano instrument (Malvern Panalytical). After conjugation, NPs were subjected to gel electrophoresis with 0.5–1% Agarose gel, run at 100 V for 60 min, and imaged with Gel imager (Bio-Rad).
Fresh prepared BiTN nanoparticles were incubated in PBS or 10% FBS-containing DMEM media at 37°C. Size and PDI were measured at 12, 24, 48 and 72 h; any detachment of loaded antibody/protein was detected by measuring the IgG and SLAMF7 in the supernatant as described above after ultracentrifugation of the NP solution collected at different time points.
HER2+ cell targeting assay.
For microscopy-based assays, tumour cells were seeded in chamber slides (LabTEK) and incubated with anti-HER2 antibody–conjugated nanoparticles (HER-NP) at 37°C for 1 h, with non-targeting IgG1–conjugated nanoparticles (IgG-NP) used as a control. Cells were then washed three times with PBS to remove unbound NPs, and NP-bound cells were detected with AlexaFluor 488-conjugated anti-human (mouse) IgG1 antibody (Invitrogen, #A11013; BioLegend, #405319). Cell nuclei were counterstained with (4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) and the samples were mounted with an antifade mounting medium (ThermoFisher) before imaging. Images were obtained with an Olympus Fluoview FV3000 laser scanning microscope with a ×40 oil emersion objective, numerical aperture (NA) 1.4.
For microplate measurement of binding avidity, fluorescently labeled nanoparticles were prepared by encapsulating coumarin-6 (Cou6) into the NPs during the conjugation. Briefly, 0.6 mg PLGA-b-mPEG and 0.4 mg PLGA-b-PEG-NHS polymers were dissolved in DMSO to 10 mg mL−1 and mixed with 5 μg Cou6. The mixture was added dropwise into 1 mL of 10 mM PBS (pH 7.4) containing a predetermined ratio of HER/SLAMF7, followed by reaction and purification as described above to obtain the fluorescent NP BiTNHER/Cou6. The HER2-overexpressing human mammary carcinoma cell SK-BR-3 was cultured in 96-well cell culture plates and grown to approximately 80% confluence. The cells were then fixed with 1% paraformaldehyde and incubated with various concentrations of BiTNHER/Cou6 containing different HER:SLAMF7 ratios in 5% bovine serum albumin (BSA) -blocking solution for 30 min. Unbound NPs were then removed by three washes with PBS. The fluorescence intensity of Cou6 was measured, and non-fluorescently labeled BiTNHER was used as a control. The dissociation constant KD was obtained by plotting normalised absorbance values versus concentrations of the BiTNHER added to the cell cultures. Nonspecific binding was determined by using unconjugated NPs and IgG-coated NPs.
Cell converting assay.
For the microscopy-based assay, SK-BR-3 and MDA-MB-468 tumour cells were seeded in chamber slides (LabTEK) and grown to approximately 50% confluence. Cells were then incubated with HER/SLAMF7–conjugated NPs (BiTNHER) (HER:SLAMF7 ratio=1:1) at 37°C for 1 h, with non-targeting IgG/SLAMF7-NPs (IgG-SNP) (IgG:SLAMF7 ratio=1:1) used as a control. The cells were then washed three times with PBS to remove unbound NPs and fixed with 1% paraformaldehyde for 15 min. Cells were then blocked with 5% BSA and stained with allophycocyanin-conjugated anti-human SLAMF7 (clone 162.1, BioLegend, #311809, 1:100) and AlexaFluor 488-conjugated anti-human IgG antibody (Invitrogen, #A11013, 1:100) overnight. Cell nuclei were counterstained with DAPI and the samples were mounted with an antifade mounting medium (ThermoFisher) before imaging. Images were obtained with Olympus Fluoview FV3000 laser scanning microscope with a ×40 oil emersion objective, numerical aperture (NA) 1.4.
In vitro phagocytosis assay.
For the flow-cytometry based assay, 5×105 tumour cells labelled with carboxyfluorescein succinimidyl ester (CFSE)-labelled (eBioscience) were seeded overnight in 12-well cell culture plates. The next day, tumour cells were pre-incubated with different treatments for 1 h before the addition of 1.5×106 eFluor 670–labelled (eBioscience) macrophages. Cells were co-cultured for 4 h at 37°C with the different treatments in serum-free or serum-containing medium. The treatment conditions were as follows: unconjugated NPs, NPs conjugated with anti-HER2/neu antibodies or SLAMF7 only, mixture of free antibodies, free SLAMF7 and unconjugated NPs, and BiTNHER of different HER:SLAMF7 ratios (0.4 mg mL−1 NPs, free antibody and SLAMF7 were normalised to the dose of NPs); all treatments included anti-CD47 antibody (10 μg mL−1) unless otherwise specified; for SLAMF7 blocking assay, cells were co-treated with anti-human/mouse SLAMF7 antibody at 50 μg mL−1 (Abcam, ab202840). The cells were then washed three times, collected, and resuspended in PBS containing 1% BSA. Phagocytosis was analysed with a CytoFLEX Flow Cytometer (Beckman Coulter) and calculated as the percentage of CFSE+ cells within eFluor 670+ macrophages. Dead cells were excluded from the analysis by staining with Sytox dead cell staining reagent. For the microscopy-based assay, 1×105 CFSE-labelled tumour cells and 3×105 eFluor 670–labelled macrophage were seeded in chamber slides (LabTEK) and treated as described for the flow-cytometry based assay. Images were obtained with an Olympus Fluoview FV3000 laser scanning microscope with a ×40 oil emersion objective, numerical aperture 1.4.
For pHrodo assay, 1×105 eFluor450-labelled tumour cells were seeded in chamber slides (LabTEK) and treated as described above for 1 h. Then the pHrodo™ Green E. coli BioParticles (Invitrogen, #P35366) were added along with 3×105 macrophage into the treated cancer cells. Cells were co-cultured for 4 h at 37°C, then washed and stained with APC-anti-CD11b (clone M1/70, BioLegend, #101212, 1:100) overnight before imaging with Olympus Fluoview FV3000 laser scanning microscope with a ×60 oil emersion objective, numerical aperture 1.4.
For macrophage activation and phenotype polarisation experiments, human THP-1 or mouse (C57BL/6) BMDMs were activated according to a previously published protocol47. For the classically activated M1 phenotype, macrophages were incubated with BMDMs in complete medium supplemented with 100 ng mL−1 lipopolysaccharide for 48 h. For M2 polarisation, cells were cultured with the BMDMs in complete medium supplemented with 10 ng mL−1 of IL-4 for 48 h.
Antigen presentation.
For the flow-cytometry based assay, cOVA-expressing tumour cells were cocultured with CFSE-labelled macrophage for 3 d with BiTNHER and aCD47. Cells were then washed once with PBS and resuspended in PBS containing 1% BSA, followed by 1 h staining with allophycocyanin-labelled anti-SIINFEK6/H-2Kb (clone 25-D1.16, Biolegend #141606, 1:100). Antigen presentation was assessed with a CytoFLEX Flow Cytometer (Beckman Coulter) and was measured as the percentage of allophycocyanin+ cells within the CFSE-labelled macrophages. For the microscopy-based assay, cells were seeded in chamber slides (LabTEK) and processed as previously described. Images were obtained with Olympus Fluoview FV3000 laser scanning microscope with a ×40 oil emersion objective, numerical aperture 1.4.
T-cell activation.
Spleens from C57BL/6 mice were carefully isolated and washed twice with sterile PBS on ice. Tissues were dissociated through a 70-μm cell strainer by using mechanical force. Cells were centrifuged at 750 g for 4 min and washed twice with cold PBS. Cells were then resuspended in ACK (ammonium-chloride-potassium) lysis buffer (2 mL per spleen) for 2 min to remove erythrocytes, and then washed with 10 mL of PBS to stop lysis. Cells were centrifuged again at 750 g for 4 min, and T cells were isolated by using an EasySep Mouse T cell isolation kit (STEMCELL) according to the manufacturer’s protocol. EO771/E2 and EO71 cells were co-cultured with macrophages under different treatments for 24 h, and then fresh-extracted naïve T cells that had been prestained with CFSE were added and the mixture incubated for another 3 days. Activated T cells were identified with antibodies to CD3, CD4, CD8, CD44, and CD62L and analysed by flow cytometry.
Tumour inhibition.
Orthotopic mammary fat pat tumours were established with mouse breast cancer cells (EO771, EO771/E2 in C57BL/6 or STING KO mice; TUBO, 4T1 in BALB/c mice) by injection of 2×106 cells in 50 μL of PBS. Tumour volumes were measured with calipers and calculated according to an ellipsoid formula (π/6 × length × width2)48. Mice with palpable tumours of similar size were randomly sorted into control and treatment groups on day 14 after implantation. Treatments took place on days 15, 17, and 19 with injections of IgG or aCD47 alone (InVivoMAb anti-mouse CD47 #BE0283 (Clone: MIAP410, BioXcell, USA), 50 μg per intratumoral injection; 100 μg per intraperitoneal injection), anti-HER2 antibody/SLAMF7 mixture with IgG or aCD47; and BiTNHER with IgG or aCD47 (50 μL per intratumoral injection, with an equivalent dose of 4 mg kg−1 BiTNHER). Also, in EO771 or EO771/E2 tumor-bearing mice, treatment with aPD1 (200 μg per intraperitoneal injection) took place on days 16, 18, and 20. If tumor volumes reached over 1000 mm3 or became ulcerated (>5 mm in diameter), the mouse was euthanized by CO2 per institutional policy, thus defining the end point of tumor burden. Statistical comparisons were made with unpaired Student’s t test for nonparametric data. The investigators were not blinded to the group allocation during experiments.
For CD8+ T cell depletion, C57BL/6 or BALB/c mice were injected with 300 μg of InVivoMAb anti-mouse CD8α #BE0117 (Clone: YTS 169.4BioXcell, USA) intraperitoneally every 96 h beginning 2 d before treatment during the study. Depletion was confirmed by flow-cytometry analysis of T cells isolated from murine spleens.
The spontaneous post-surgery tumor metastasis model was established by using luciferase-expressing mouse breast cancer cells 4T1-Br4-Luc in BALB/c mice. Primary tumors were first generated by injection of 1×106 cells in 50 μL of PBS into the mammary fat pad. For Treatment A, on days 8, 10, and 12, the primary tumors were treated with IgG or aCD47 alone (50 μg per intratumoral injection), or BiTNFo with aCD47 (50 μL per intratumoral injection, with 50 μg antibody and an equivalent dose of 10 mg kg−1 BiTNFo). On day 15, mice were anaesthetized, and the primary tumors were resected. On days 16, 18, and 20, mice were treated with IgG or aCD47 alone (100 μg per intraperitoneal injection), or BiTNFo with aCD47 (100 μg antibody per intraperitoneal injection, and 100 μL of an equivalent dose of 40 mg kg−1 BiTNFo). Treatments with aPD1 (200 μg per intraperitoneal injection) took place on day 9, 11, 13, 17, 19, and 21. For Treatment B, tumors were resected on day 12 without any pretreatment. On days 15, 17, and 19, mice were treated with IgG or aCD47 alone (100 μg per intraperitoneal injection), or BiTNFo with aCD47 (100 μg antibody per intraperitoneal injection, and 100 μL of an equivalent dose of 40 mg kg−1 BiTNFo per intravenous injection). Treatments with aPD1 (InVivoMAb anti-mouse PD-1 (CD279) #BE0273 (Clone: 29F.1A12, BioXcell, USA), 200 μg per intraperitoneal injection) took place on day 16, and 20. Every 5 days after surgery, mice were subcutaneously injected with 3 mg D-luciferin (Goldbio) and tumor metastasis was monitored with IVIS imaging (IVIS 200).
Nanoparticle biodistribution after intratumoral injection.
Far-red fluorescently labeled nanoparticles were prepared by encapsulating BODIPY (Lumiprobe, #4E420) into the NPs during the conjugation. Briefly, 0.6 mg PLGA-b-mPEG and 0.4 mg PLGA-b-PEG-NHS polymers were dissolved in DMSO to 10 mg mL−1 and mixed with 5 μg BODIPY. The mixture was added dropwise into 1 mL of 10 mM PBS (pH 7.4) containing anti-HER2 antibody or IgG, followed by reaction and purification as described above to obtain the fluorescent HER-NP/BODIPY or IgG-NP/BODIPY. TUBO-bearing Balb/c mice were used for biodistribution study when tumor size reached 100 mm3. Same amount of nanoparticles (normalised with BODIPY fluorescence intensity) were injected intratumorally and fluorescence signal was monitored and quantified by IVIS imaging at 0.25, 0.5, 1, 2, 4, 8, 12 and 24 h after injection. Mice were sacrificed after the last imaging and major organs were imaged and quantified ex vivo. Tumors were further processed for frozen sections and imaged with Olympus Fluoview FV3000 laser scanning microscope with a ×20 objective, numerical aperture 1.4.
Flow cytometry.
Tumour tissues were carefully resected 3 days after the final treatment (day 22) and dissociated by using a Mouse Tumor Dissociation Kit (Miltenyi). Leukocytes were isolated with CD45 Microbeads on a QuadroMACS magnet (Miltenyi) according to the manufacturer’s instructions. Flow cytometry analysis was done with a CytoFLEX flow cytometer (Beckman Coulter). Fluorescence-labelled antibodies included SLAMF7 (clone 4G2, BioLegend, #152005, 1:100), CD3 (clone 145-2C11, BioLegend, #100308, 1:100), CD4 (clone GK1.5, BioLegend, #100453, 1:100), CD8 (clone 53-6.7, BioLegend, #100741, 1:100), IFN-γ (clone XMG1.2, BioLegend, #505841, 1:50), CD25 (clone 3C7, BioLegend, #101923, 1:50), FoxP3 (clone MF-14, BioLegend, #126405, 1:200), CD44 (clone IM7, BioLegend, #103031, 1:100), CD62L (clone MEL-14, BioLegend, #104435, 1:100), F4/80 (clone BM8, BioLegend, #123109, 1:100), CD11b (clone M1/70, BioLegend, #101239, 1:100), MHC-II (clone M5/114.15.2, BioLegend, #107645, 1:100), CD11c (clone N418, BioLegend, #117333, 1:100), Gr-1 (clone RB6-8C5, BioLegend, #108437, 1:50), NK1.1 (clone PK136, BioLegend, #108736, 1:100), NKp46 (clone 29A1.4, BioLegend, #137612, 1:100); a LIVE/DEAD™ fixable red dead cell stain kit was used to stain dead cells. Specific cell populations were defined as: Teff (CD8+IFN-γ+), Treg (CD4+CD25+FoxP3+), Tem (CD44hiCD62Lneg), Tcm (CD44hiCD62Lhi), DCs (CD11b+CD11chiMHC-IIhi), NK cells (CD3-NKp46+NK1.1+), myeloid-derived suppressor cells (MDSCs) (CD11b+CD11cneg-modMHC-IIneg-modGr-1+), or macrophages (CD11b+CD11cneg-modMHC-IIneg-modF4/80+). Gating strategy is described in Supplementary Figure 26-29.
Cytokine assay.
For peripheral blood collection, mice were killed 3 days after the final treatment and whole-blood samples (~1 mL) were collected via heart puncture. Serum was then obtained from the samples by centrifugation and kept at −20°C for further use. Cytokines were measured with mouse IL-2 or IFN-γ ELISA Kits (BioLegend) according to the manufacturer’s instructions.
Immunohistochemical (IHC) analyses.
Tumour tissues were carefully isolated and fixed in 4% buffered formalin phosphate solution (Fischer Scientific) for 24 h, followed by 70% ethanol solution overnight. The tissues were embedded in paraffin, and 5-μm tissue slices were prepared. For the immunostaining studies, tumour slides were pre-blocked with a blocking solution containing 5% BSA and 10% control normal serum and then stained with primary antibodies including CD4 (clone 4SM95, eBioscience, #14-9766-82, 1:50), CD8 (clone 4SM15, eBioscience, #100702, 1:50), F4/80 (clone CI:A3-1, Abcam, #6640, 1:100), Iba-1 (pAb, Abcam, #ab5076, 1:1000), CD11c (clone N418, eBioscience, #14-0114-82, 1:100), pIRF3 S396 (clone D6O1M, Cell Signaling, #29047, 1:50), PD1 (clone D7D5W, Cell Signaling, #84651, 1:200), and PDL1 (clone D5V3B, Cell Signaling, #64988, 1:200) at 4 °C overnight. After washing, the appropriate fluorescence-labelled secondary antibodies (eBioscience) were added for 2 h at room temperature in the dark. Cell nuclei were counterstained with DAPI and the samples were mounted with an antifade mounting medium (ThermoFisher) before imaging. Fluorescence images were obtained with an Olympus Fluoview FV3000 laser scanning microscope and were further analysed with Image J software.
RNA extraction and quantitative real-time RT-PCR.
Tumour tissues were carefully isolated 24 h after the final treatment (Day 20) and dissociated as described above. Macrophages or DCs were isolated by using F4/80 or CD11c Microbeads, respectively, on a QuadroMACS magnet (Miltenyi) according to the manufacturer’s instructions. Total RNA was extracted from the obtained cell populations by using a PureLink RNA Mini Kit (Applied Biosystems), and cDNA was prepared with High-Capacity cDNA RT Kit (ThermoFisher) according to the manufacturer’s instructions. RT-qPCR was done with the 7500 FAST Real-time PCR System (Applied Biosystems), with TaqMan™ Fast Advanced PCR master Mix (ThermoFisher) and TaqMan FAM dye-labelled probes including GAPDH (Hs02786624_g1, Mm99999915_g1), IFNA1 (Mm03030145_gH), and IFNB1 (Mm00439552_s1) (ThermoFisher). GAPDH was used to normalise signal expression. Fold-change comparisons were done between control and treated samples by using the ΔΔCT method as described elsewhere49.
Autoimmune toxicity study of BiTN.
Female Balb/c mice with same age were randomly divided into 3 groups as healthy mice, healthy mice with subcutaneous injection of NP and aCD47, and TUBO-bearing mice with intratumoral injection of NP and aCD47. The dosage of treatment was same as the dosage for tumor inhibition study and was repeated for three times every other day. On Day 30 after the last treatment, mice were sacrificed, whole-blood samples (~1 mL) were collected via cardiac puncture for complete blood count. Major organs were collected for H&E staining.
Biocompatibility study of BiTNFo.
For in vitro studies of hemolysis, BiTNFo was diluted with PBS to concentrations of 0.01–0.5 mg/mL, mimicking the blood NP concentrations after intravenous injection at a dose of 40 mg/kg. Erythrocytes were isolated from healthy BALB/c mice and incubated with NPs as described previously50. For in vivo studies of toxicity, healthy BALB/c mice were randomly assigned to receive PBS or BiTNFo (n=3). Mice were intravenously injected with 100 μL of PBS or an equivalent dose of 40 mg kg−1 BiTNFo every other day for a total of 3 times. Body weight was recorded every other day from the last injection. At 16 days after the final injection, the mice were killed and whole-blood samples (~1 mL) were collected via heart puncture. Serum was then obtained from the samples by centrifugation and kept at −20°C for further use. Peripheral blood concentrations of urea nitrogen (BUN), aspartate aminotransferase (AST), alanine aminotransferase (ALT), and creatinine were measured by using a mouse BUN colorimetric detection kit (Invitrogen) and mouse AST, ALT, and creatinine with a colorimetric assay kit (Cayman Chemical) according to the manufacturer’s instructions.
Statistical analysis.
Unless otherwise specified, all the experiments were repeated at least twice for 6–8 biological replicates, with similar results between repeats. For in vivo experiments, a sample size of 10 was chosen for each treatment arm unless otherwise specified. This gives at least an 80% power to detect a difference of 50% reduction in tumour volume with a 5% significance level. Data for all bar graphs were plotted by using means and error bars corresponding to standard deviations. For comparisons between three or more independent groups, one-way analysis of variance with Bonferroni post hoc correction was used. For comparisons between two sample groups, unpaired non-parametric Student’s t tests were used. All tests were two-tailed. A P value of <0.05 was considered significant. Statistical analysis was done with GraphPad Prism 9 or Microsoft Excel 2020 software. No animals were excluded from the analysis.
Supplementary Material
Acknowledgements
Supported in part by the Susan G. Komen Foundation Career Catalyst Research Grant CCR19605871 and the National Cancer Institute Grant 1K08 CA241070 to W.J.; Department of Defense Grant W81XWH-19-1-0325 to B.Y.S.K.; and Cancer Center Support (Core) Grant P30 CA016672 from the National Cancer Institute, National Institutes of Health, to The University of Texas MD Anderson Cancer Center (PI: PW Pisters). The authors thank Christine Wogan, MS, ELS, of the Division of Radiation Oncology, MD Anderson Cancer Center, for editorial assistance.
Footnotes
Competing interests
The University of Texas MD Anderson Cancer Center has filed a patent application on the technology and intellectual property reported here for which Y.L., W.J. and B.Y.S.K are inventors. The remaining authors declare no competing interests.
Data availability
The authors declare that data supporting the findings of this study are available within the article and its Supplementary Information. The datasets generated and analysed during the study are publicly available at https://osf.io/jxy9z. Source data are provided with this paper.
References
- 1.Demaria O et al. Harnessing innate immunity in cancer therapy. Nature 574, 45–56 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Vonderheide RH CD47 blockade as another immune checkpoint therapy for cancer. Nat. Med 21, 1122–1123 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Weiskopf K et al. Engineered SIRP alpha Variants as Immunotherapeutic Adjuvants to Anticancer Antibodies. Science 341, 88–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logtenberg MEW, Scheeren FA & Schumacher TN The CD47-SIRPalpha Immune Checkpoint. Immunity 52, 742–752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jalil AR, Andrechak JC & Discher DE Macrophage checkpoint blockade: results from initial clinical trials, binding analyses, and CD47-SIRPalpha structure-function. Antib. Ther 3, 80–94 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W et al. Advances in Anti-Tumor Treatments Targeting the CD47/SIRPalpha Axis. Front. Immunol 11, 18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sikic BI et al. First-in-Human, First-in-Class Phase I Trial of the Anti-CD47 Antibody Hu5F9-G4 in Patients With Advanced Cancers. J. Clin. Oncol 37, 946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ansell SM et al. Phase I Study of the CD47 Blocker TTI-621 in Patients with Relapsed or Refractory Hematologic Malignancies. Clin. Cancer. Res 27, 2190–2199 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Eladl E et al. Role of CD47 in Hematological Malignancies. J. Hematol. Oncol 13, 96 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J et al. SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature 544, 493–497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng M et al. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat. Rev. Cancer 19, 568–586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uger R & Johnson L Blockade of the CD47-SIRPalpha axis: a promising approach for cancer immunotherapy. Expert Opin. Biol. Ther 20, 5–8 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Zhong C et al. Poly(I:C) enhances the efficacy of phagocytosis checkpoint blockade immunotherapy by inducing IL-6 production. J. Leukoc. Biol (2021). [DOI] [PubMed] [Google Scholar]
- 14.Cao X et al. Effect of cabazitaxel on macrophages improves CD47-targeted immunotherapy for triple-negative breast cancer. J. Immunother. Cancer 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang AL et al. Dual targeting of CTLA-4 and CD47 on T-reg cells promotes immunity against solid tumors. Sci. Transl. Med 13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y & Lammers T Combining Nanomedicine and Immunotherapy. Acc. Chem. Res 52, 1543–1554 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan H et al. Multivalent bi-specific nanobioconjugate engager for targeted cancer immunotherapy. Nat. Nanotechnol 12, 763–769 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Weissleder R, Kelly K, Sun EY, Shtatland T & Josephson L Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat. Biotechnol 23, 1418–1423 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Gordon SR et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 545, 495–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pazina T et al. Enhanced SLAMF7 Homotypic Interactions by Elotuzumab Improves NK Cell Killing of Multiple Myeloma. Cancer Immunol. Res 7, 1633–1646 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu K et al. Low-dose X-ray radiotherapy-radiodynamic therapy via nanoscale metal-organic frameworks enhances checkpoint blockade immunotherapy. Nat. Biomed. Eng 2, 600–610 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Shae D et al. Endosomolytic polymersomes increase the activity of cyclic dinucleotide STING agonists to enhance cancer immunotherapy. Nat. Nanotechnol 14, 269–278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X et al. Cancer immunotherapy based on image-guided STING activation by nucleotide nanocomplex-decorated ultrasound microbubbles. Nat. Nanotechnol (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao W, Lin JX & Leonard WJ Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 38, 13–25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morad G, Helmink BA, Sharma P & Wargo JA Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 184, 5309–5337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alizadeh D et al. IFNgamma Is Critical for CAR T Cell-Mediated Myeloid Activation and Induction of Endogenous Immunity. Cancer Discov. 11, 2248–2265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitter MR & Zou W Uncovering the Immunoregulatory Function and Therapeutic Potential of the PD-1/PD-L1 Axis in Cancer. Cancer Research 81, 5141–5143 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer 18, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su S et al. Immune Checkpoint Inhibition Overcomes ADCP-Induced Immunosuppression by Macrophages. Cell 175, 442–457 e423 (2018). [DOI] [PubMed] [Google Scholar]
- 30.von Roemeling CA et al. Therapeutic modulation of phagocytosis in glioblastoma can activate both innate and adaptive antitumour immunity. Nat. Commun 11, 1508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosaka A et al. CD47 blockade enhances the efficacy of intratumoral STING-targeting therapy by activating phagocytes. J. Exp. Med 218 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopfner KP & Hornung V Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol 21, 501–521 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Martin GR, Blomquist CM, Henare KL & Jirik FR Stimulator of interferon genes (STING) activation exacerbates experimental colitis in mice. Sci. Rep 9, 14281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdullah A et al. STING-mediated type-I interferons contribute to the neuroinflammatory process and detrimental effects following traumatic brain injury. J. Neuroinflammation 15, 323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathur V et al. Activation of the STING-Dependent Type I Interferon Response Reduces Microglial Reactivity and Neuroinflammation. Neuron 96, 1290–1302 e1296 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z et al. Immunogenic Cell Death Activates the Tumor Immune Microenvironment to Boost the Immunotherapy Efficiency. Adv. Sci. (Weinh), e2201734 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ & Kroemer G Type I interferons in anticancer immunity. Nat. Rev. Immunol 15, 405–414 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Salvagno C et al. Therapeutic targeting of macrophages enhances chemotherapy efficacy by unleashing type I interferon response. Nat. Cell. Biol 21, 511–521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z et al. Folate receptor alpha associated with triple-negative breast cancer and poor prognosis. Arch. Pathol. Lab. Med 138, 890–895 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Song DG et al. Effective adoptive immunotherapy of triple-negative breast cancer by folate receptor-alpha redirected CAR T cells is influenced by surface antigen expression level. J. Hematol. Oncol 9, 56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aldea M et al. Overcoming Resistance to Tumor-Targeted and Immune-Targeted Therapies. Cancer Discov. 11, 874–899 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Han C et al. Tumor cells suppress radiation-induced immunity by hijacking caspase 9 signaling. Nat. Immunol 21, 546–554 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Tumeh PC et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao JB et al. Preservation of tumor-host immune interactions with luciferase-tagged imaging in a murine model of ovarian cancer. J. Immunother. Cancer 3, 16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
References [for Methods section]
- 45.Qie Y et al. Surface modification of nanoparticles enables selective evasion of phagocytic clearance by distinct macrophage phenotypes. Sci. Rep 6, 26269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan H et al. Multivalent bi-specific nanobioconjugate engager for targeted cancer immunotherapy. Nat. Nanotechnol 12, 763–769 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Mosser DM & Zhang X Activation of murine macrophages. Curr. Protocol. Immunol 83, 14.2.1–14.2.8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiskopf K et al. Engineered SIRPα variants as immunotherapeutic adjuvants to anticancer antibodies. Science 341, 88–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmittgen TD & Livak KJ Analyzing real-time PCR data by the comparative C-T method. Nat. Protoc 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Evans BC et al. Ex vivo red blood cell hemolysis assay for the evaluation of pH-responsive endosomolytic agents for cytosolic delivery of biomacromolecular drugs. J. Vis. Exp 73, 50166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that data supporting the findings of this study are available within the article and its Supplementary Information. The datasets generated and analysed during the study are publicly available at https://osf.io/jxy9z. Source data are provided with this paper.