Abstract
Tight coordination of growth regulatory signaling is required for intestinal epithelial homeostasis. Protein kinase C α (PKCα) and transforming growth factor β (TGFβ) are negative regulators of proliferation with tumor suppressor properties in the intestine. Here, we identify novel crosstalk between PKCα and TGFβ signaling. RNA-Seq analysis of nontransformed intestinal crypt-like cells and colorectal cancer cells identified TGFβ receptor 1 (TGFβR1) as a target of PKCα signaling. RT-PCR and immunoblot analysis confirmed that PKCα positively regulates TGFβR1 mRNA and protein expression in these cells. Effects on TGFβR1 were dependent on Ras-extracellular signal-regulated kinase 1/2 (ERK) signaling. Nascent RNA and promoter-reporter analysis indicated that PKCα induces TGFβR1 transcription, and Runx2 was identified as an essential mediator of the effect. PKCα promoted ERK-mediated activating phosphorylation of Runx2, which preceded transcriptional activation of the TGFβR1 gene and induction of Runx2 expression. Thus, we have identified a novel PKCα→ERK→Runx2→TGFβR1 signaling axis. In further support of a link between PKCα and TGFβ signaling, PKCα knockdown reduced the ability of TGFβ to induce SMAD2 phosphorylation and cell cycle arrest, and inhibition of TGFβR1 decreased PKCα-induced upregulation of p21Cip1 and p27Kip1 in intestinal cells. The physiological relevance of these findings is also supported by The Cancer Genome Atlas data showing correlation between PKCα, Runx2, and TGFβR1 mRNA expression in human colorectal cancer. PKCα also regulated TGFβR1 in endometrial cancer cells, and PKCα, Runx2, and TGFβR1 expression correlates in uterine tumors, indicating that crosstalk between PKCα and TGFβ signaling may be a common mechanism in diverse epithelial tissues.
Keywords: PKCα, TGFβ, TGFβR1, ERK, Runx2, growth arrest, transcription, intestinal epithelium, colon cancer
The epithelial lining of the intestine and colon is a continuously self-renewing tissue with a turnover time of 2 to 6 days in most adult mammals (1). Tight coordination of cell proliferation, growth arrest, terminal differentiation, and cell death is required for maintenance of intestinal epithelial homeostasis and to prevent disease (2). In the small intestine, cell proliferation is restricted to invaginations in the lamina propria known as crypts of Lieberkühn, while postmitotic functional cells are found on finger-like projections or villi (3, 4). The colon has a similar architecture except that villi are replaced by a flat functional surface epithelium (3, 4). Multipotent stem cells located at the base of intestinal and colonic crypts give rise to transit amplifying cells that continue to divide as they migrate toward the villus/mucosal surface. Near the top of the crypts, cells undergo growth arrest, a prerequisite for differentiation into functional cells. Postmitotic cells are eventually shed at the villus tip/colonic surface. Failure to maintain strict coordination of cell proliferation and growth arrest leads to loss of absorptive function and development of intestinal tumors (2). Although the signals that support stem cell and transit amplifying cell proliferation and fate specification in the crypts are well defined (4), the mechanisms that mediate growth arrest in the upper crypt and the coordination between antiproliferative signaling pathways are not well understood.
Studies from our laboratory and others have identified a role for the serine threonine kinase, protein kinase C α (PKCα), in negative regulation of cell growth in the intestinal epithelium (5, 6). PKCα undergoes hallmarks of activation coincident with growth arrest in both small intestinal and colonic crypts (7, 8, 9, 10). Activation of PKCα in nontransformed intestinal crypt cells in vitro leads to downregulation of D-type cyclins, upregulation of the cyclin-dependent kinase (CDK) inhibitors, p21Cip1 and p27Kip1, and cell cycle withdrawal into G0 (10, 11, 12, 13, 14, 15). The functional consequences of these effects are highlighted by studies in PKCα knockout mice. PKCα deficiency results in increased cell proliferation in the crypt, aberrant expression of proproliferative molecules, and increased intestinal tumorigenesis (7, 16). Furthermore, loss of PKCα is a common characteristic of human colon cancer, and restoration of PKCα expression inhibits the transformed phenotype of colon cancer cells (8, 12, 17, 18, 19).
In addition to PKCα, transforming growth factor β (TGFβ) signaling is an important negative regulator of cell proliferation in epithelial tissues and a critical player in maintenance of intestinal epithelial homeostasis (2, 20). TGFβ ligands (TGFβ1, TGFβ2, and TGFβ3) signal through a transmembrane heteromeric receptor complex consisting of TGFβ receptor 1 (TGFβR1) and TGFβ receptor 2 (TGFβR2). TGFβ initially binds to TGFβR2, leading to TGFβR2-mediated phosphorylation and activation of TGFβR1, which in turn propagates the signal to downstream cytoplasmic proteins (21). In canonical TGFβ signaling, TGFβR1 phosphorylates and activates SMAD2 and SMAD3, which then bind to SMAD4, forming a transcriptional complex that translocates to the nucleus to activate the transcription of TGFβ responsive genes, such as the CDK inhibitors p21Cip1, p27Kip1, and p15Ink4B (21, 22).
Consistent with an antiproliferative role in the intestine, there is an increasing gradient of TGFβ and TGFβ receptor expression from crypt to villus/surface epithelium (23, 24, 25). The importance of TGFβ signaling in maintenance of intestinal homeostasis is also highlighted by the common disruption of the growth inhibitory effects of this pathway in colorectal cancer (CRC) (2). Mutation of TGFβR2 is particularly common in CRC with microsatellite instability and has been linked to CRC progression (20). Germ line mutations in SMAD4 lead to juvenile polyposis syndrome, and SMAD4 mutation is also linked to CRC progression (26, 27). In keeping with its central role in propagation of TGFβ signaling, reduced expression of TGFβR1 is also commonly observed in CRC (28), and TGFβR1 deficiency contributes to the transformed phenotype of CRC cells (29). Notably, analysis of TGFβR1 heterozygous mice indicates that intestinal homeostasis is sensitive to relatively small changes in TGFβR1 expression, with a 40% reduction in TGFβR1 levels markedly increasing tumorigenesis in the APCmin/+ mouse model of intestinal cancer (30). This effect was accompanied by decreased SMAD2 phosphorylation and enhanced proliferation in the normal intestinal epithelium and in tumors (30), indicating that TGFβR1 levels are limiting for growth inhibitory TGFβ signaling in the intestine.
The current study identifies crosstalk between PKCα and TGFβ signaling, with PKCα promoting the expression of TGFβR1 and enhancing TGFβ signaling in intestinal epithelial and CRC cells. Our findings offer new insights into regulation of intestinal homeostasis and the mechanisms underlying PKCα-mediated regulation of cell proliferation and tumorigenesis.
Results
PKCα regulates TGFβR1 expression in epithelial cells
To identify mediators of the antiproliferative and tumor suppressive effects of PKCα signaling, PKCα activity and expression were manipulated in a panel of epithelial cell lines and changes in gene expression were assessed using RNA-Seq. The analysis was performed by activating or silencing PKCα in nontransformed rat intestinal crypt-like cells (IEC-18 cells) and human colon epithelial cells (HCEC cells), as well as PKCα-retaining human colon (HCT-116 cells) and endometrial (SNG-M) cancer cells. IEC-18 cells have relatively low basal PKCα activity; the enzyme was, therefore, activated in these cells using the PKC agonist, phorbol 12-myristate 13-acetate (PMA), and the involvement of PKCα in observed effects was determined using Gö6976, a selective inhibitor of members of the conventional subclass of PKC isozymes (PKCα, βI, βII, γ). Since PKCα is the only conventional PKC expressed in IEC-18 cells, Gö6976 is specific for PKCα in this system ((7, 13), and Fig. S1). The general applicability of identified PKCα targets to intestinal cells was determined by analysis of the effects of PKCα knockdown in HCEC nontransformed colon epithelial cells and HCT-116 colon cancer cells, which have higher levels of basal PKCα activity ((12), and data not shown). SNG-M endometrial cancer cells were also analyzed since PKCα negatively regulates proliferation and tumorigenicity in these cells (31).
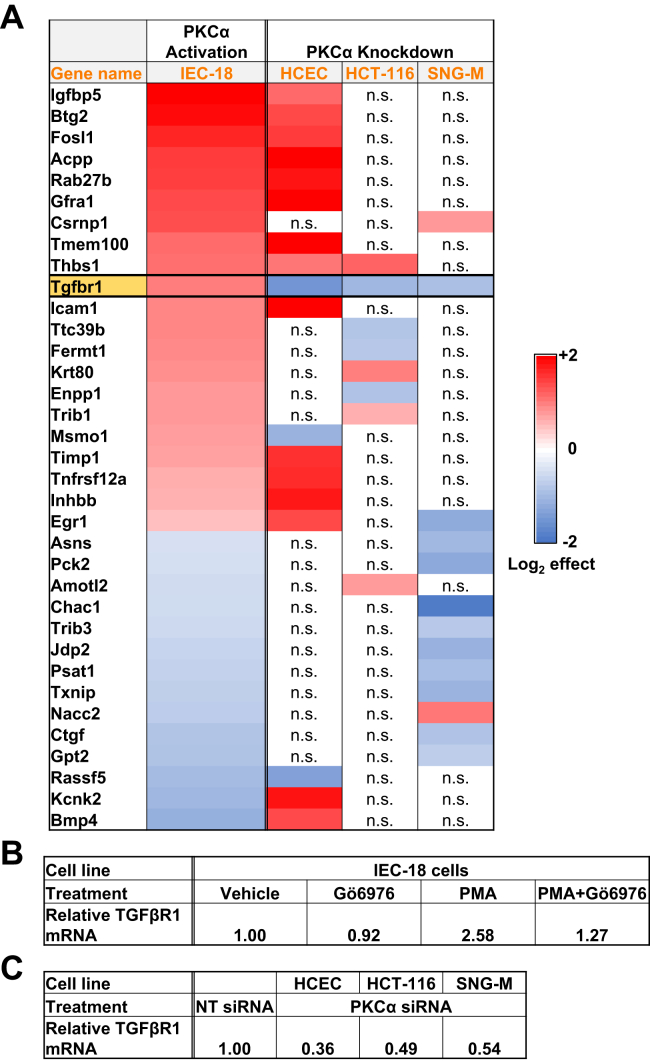
RNA-Seq analysis in IEC-18 cells (Table S1) identified a number of mRNAs encoding growth regulatory genes that showed Gö6976-sensitive regulation by PMA such as bmp4, egr1, fos, hif1a, mtss1, notch3, pak1, tgfbr1, and wnt7b. Although PKCα knockdown in HCEC, HCT-116, or SNG-M cells affected the expression of some of these genes, most genes did not show consistent regulation, presumably reflecting differing signaling contexts in the individual cell lines. Only TGFβR1 mRNA was consistently modulated across all cell lines (Fig. 1 and Tables S1–S4), pointing to a direct relationship between this gene and PKCα signaling. TGFβR1 mRNA was upregulated by PKCα activation in IEC-18 cells (Fig. 1, A and B) and significantly downregulated by PKCα knockdown in HCEC, HCT-116, and SNG-M cells (Fig. 1, A and C), indicating that PKCα acts as a positive regulator of this antiproliferative protein. Based on these findings, the ability of PKCα to regulate TGFβR1 expression was further explored.
Figure 1.
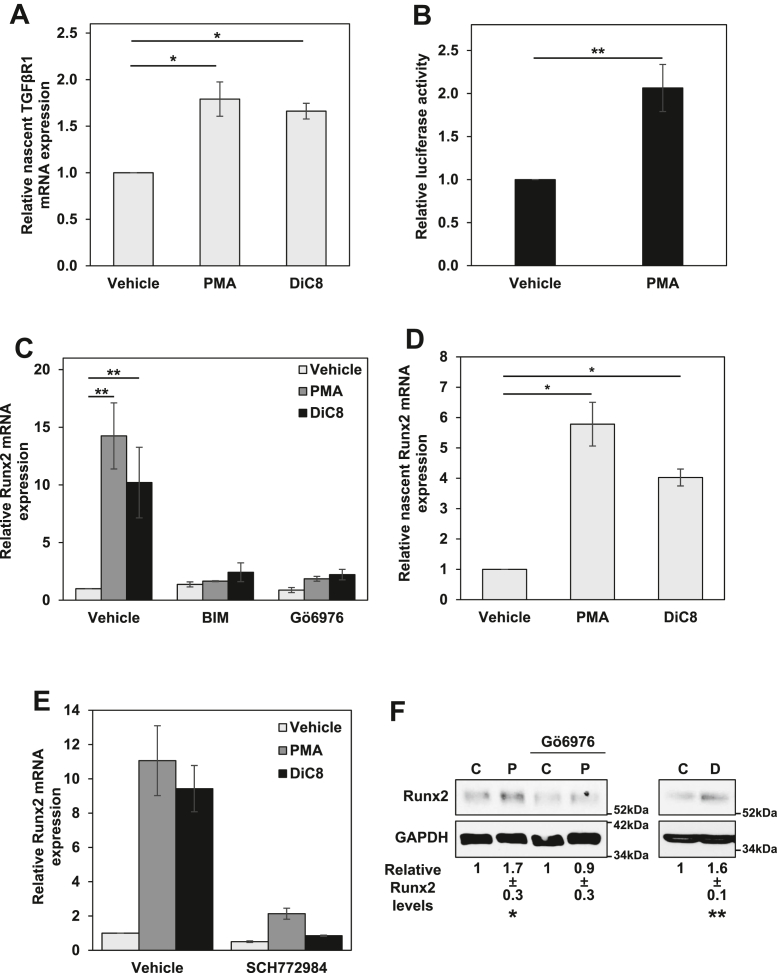
RNA-Seq analysis of the effects of PKCα activation and knockdown in epithelial cells. A, heatmap of the effects of PKCα activation or knockdown in the indicated cell lines. Only mRNAs that showed a significant effect of treatment with the PKCα agonist PMA for 2 h in IEC-18 cells and a significant effect of knockdown in at least one of the other cell lines are listed. Effects of PKCα activation in IEC-18 cells are based on Gö6976 inhibitable PMA activation. See Tables S1–S4 for a full list of mRNAs significantly affected by PMA treatment/PKCα knockdown and for numerical values of the effects. n.s., no significant effect of PKCα knockdown. B, relative expression of TGFβR1 mRNA in IEC-18 cells following the indicated treatments (2 h). C, relative TGFβR1 mRNA expression in cells transfected with the indicated siRNAs. HCEC, human colon epithelial cell; IEC-18, intestinal crypt-like cells; NT, nontargeting; PKCα, protein kinase C α; PMA, phorbol 12-myristate 13-acetate; TGFβR1, transforming growth factor-β receptor 1.
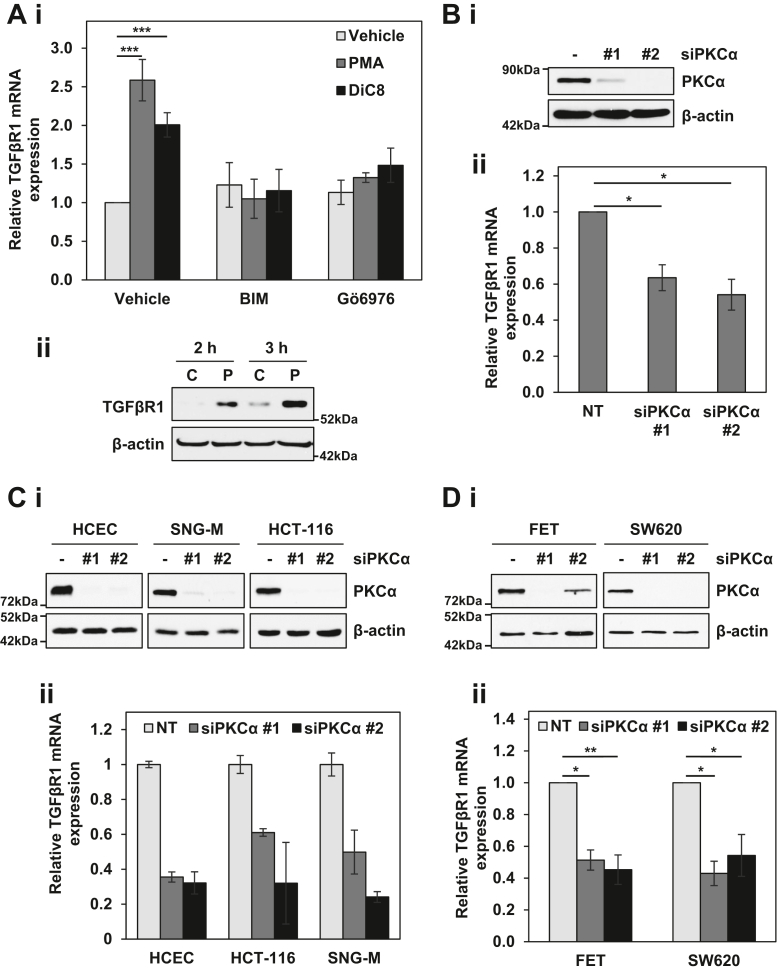
The PKCα-induced upregulation of TGFβR1 in IEC-18 cells seen by RNA-seq was validated by quantitative RT-PCR (RT-qPCR) analysis, which detected a similar 2- to 2.5-fold increase in TGFβR1 mRNA following treatment with PMA (Fig. 2Ai). A similar upregulation was seen when IEC-18 cells were treated with the short chain diacylglycerol, 1,2-dioctanoyl-sn-glycerol (DiC8), which represents a physiological activator of PKCα (Fig. 2Ai). These effects were blocked by two pharmacological inhibitors of the enzyme, the general PKC inhibitor bisindolylmaleimide I (BIM), and Gö6976 (Fig. 2Ai), confirming that they were the result of PKCα activation. Western blot analysis further determined that the increase in TGFβR1 mRNA was accompanied by an increase in TGFβR1 protein (Fig. 2Aii).
Figure 2.
Regulation of TGFβR1 by PKCα signaling. Ai, IEC-18 cells were pretreated with 5 μM BIM or 4 μM Gö6976 prior to addition of vehicle, 100 nM PMA, or 20 μg/ml DiC8. After 2 h, expression of TGFβR1 mRNA was measured by RT-qPCR and normalized to 18S rRNA levels. Aii, IEC-18 cells were treated with 100 nM PMA as indicated, and TGFβR1 protein expression was determined by Western blotting. B–D, the indicated cell lines were transfected with nontargeting (−) or one of two siRNAs targeting PKCα (#1, #2) for 72 h before expression of PKCα protein was determined by Western blotting (i), and relative TGFβR1 mRNA expression was determined by RT-qPCR as in Ai. Data in Ai and Bii are the average ± SEM of three independent experiments, and data in Bi are representative of three independent experiments. Data in C confirm PKCα knockdown and TGFβR1 mRNA downregulation in the samples used for RNA-seq by Western blotting (i) and RT-qPCR (ii), respectively, and data in D are the average ± SEM of 2 (siRNA #1) or 3 (siRNA #2) independent experiments. ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p < 0.001. BIM, bisindolylmaleimide I; DiC8, 1,2-dioctanoyl-sn-glycerol; IEC-18, intestinal crypt-like cells; PKCα, protein kinase C α; PMA, phorbol 12-myristate 13-acetate; TGFβR1, transforming growth factor-β receptor 1.
The ability of PKCα activation to modulate TGFβR1 mRNA levels was further confirmed by PKCα knockdown in multiple cell lines. As shown in Fig. S2, various levels of endogenous TGFβR1 mRNA (Fig. S2A) and protein (Fig. S2B) expression were detected in the intestinal/colon cell lines used in this study. RT-qPCR analysis showed that TGFβR1 mRNA is consistently downregulated by PKCα knockdown in IEC-18 cells (Fig. 2B), and confirmed the downregulation of TGFβR1 mRNA seen by RNA-Seq in HCEC, HCT-116, and SNG-M PKCα knockdown cells (Fig. 2C). A similar ∼2-fold downregulation of TGFβR1 mRNA was observed following PKCα knockdown in two additional PKCα retaining colon cancer cell lines, FET and SW620 (Fig. 2D). Collectively, these data indicate that PKCα positively regulates the expression of TGFβR1 mRNA in intestinal and endometrial cells, pointing to crosstalk between PKCα and TGFβ signaling in epithelial systems.
PKCα upregulates TGFβR1 through the extracellular signal-regulated kinase 1/2 signaling cascade
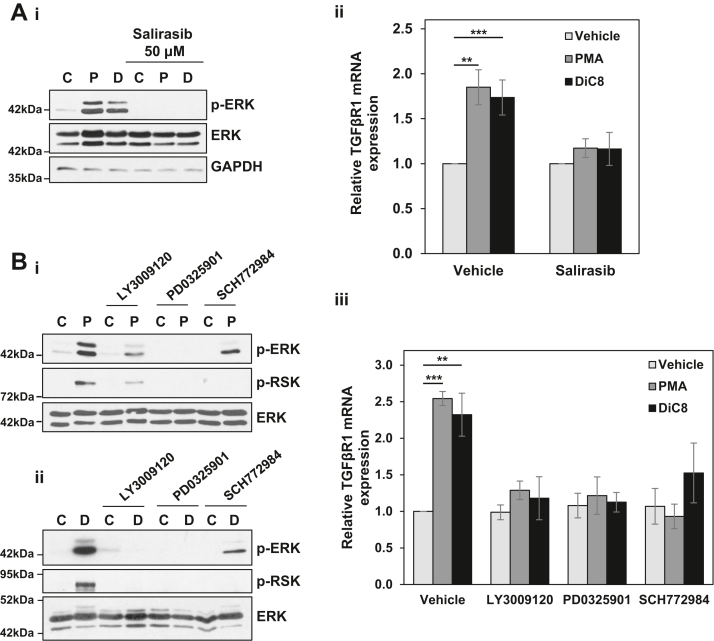
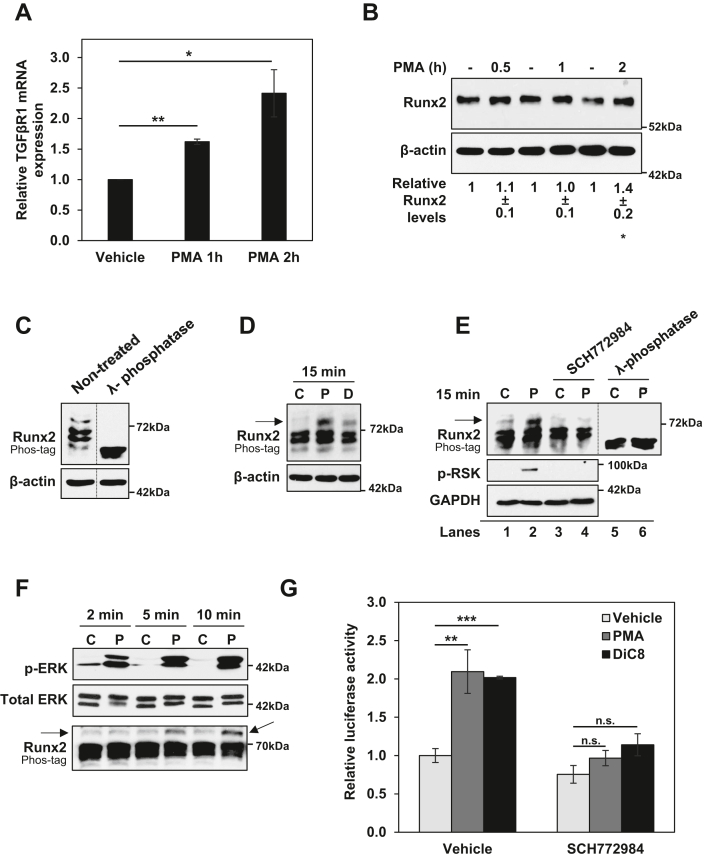
Our previous studies have determined that the growth inhibitory effects of PKCα in intestinal cells are mediated by sustained activation of the RAS-extracellular signal-regulated kinase 1/2 (ERK) pathway (32, 33). To determine if alterations in TGFβ signaling contribute to the ERK-dependent antiproliferative effects of PKCα, we analyzed the effects of inhibiting the ERK pathway on PKCα-induced regulation of TGFβR1 expression in IEC-18 cells. For rigor, we tested the effects of inhibiting multiple steps in this pathway using the ERK inhibitor SCH772984, the mitogen-activated protein kinase kinase 1/2 inhibitor PD0325901, the RAF inhibitor LY3009120, and the Ras inhibitor Salirasib (Fig. 3). Blockade of PKCα-induced activation of ERK by these inhibitors was confirmed by their ability to prevent PMA- or DiC8-induced activating phosphorylation of ERK (pERK) and/or phosphorylation of the ERK substrate RSK (Fig. 3, Ai, and Bi, and ii). The ability of all of these inhibitors to block PMA- and DiC8-induced upregulation of TGFβR1 mRNA (Fig. 3, Aii and Biii) positions TGFβR1 as a downstream component of the antiproliferative PKCα-ERK signaling axis.
Figure 3.
TGFβR1 is regulated by PKCα-ERK signaling. A, serum-starved IEC-18 cells were pretreated with 50 μM Salirasib prior to treatment with vehicle (C), 100 nM PMA (P), or 20 μg/ml DiC8 (D) for 2 h. Expression and phosphorylation of the indicated proteins were assessed by Western blotting (i), and TGFβR1 mRNA levels (normalized to 18S rRNA) were determined by RT-qPCR (ii). B, as in A except that cells in full serum medium were pretreated with 1 μM LY3009120, 10 μM PD0325901, or 1 μM SCH772984. Note that the reduction in ERK phosphorylation in the presence of SCH772984 is an “on-target” effect of this inhibitor that has been attributed to its ability to induce conformational changes in ERK that impair its interaction with MEK (e.g., (80)). Data in Ai, Bi, and Bii are representative of at least three independent experiments. Data in Aii and Biii are the average ± SEM of at least three independent experiments. ∗∗p < 0.01. ∗∗∗p < 0.001. DiC8, 1,2-dioctanoyl-sn-glycerol; ERK, extracellular signal-regulated kinase 1/2; IEC-18, intestinal crypt-like cells; MEK, mitogen-activated protein kinase kinase 1/2; PKCα, protein kinase C α; PMA, phorbol 12-myristate 13-acetate; TGFβR1, transforming growth factor-β receptor 1.
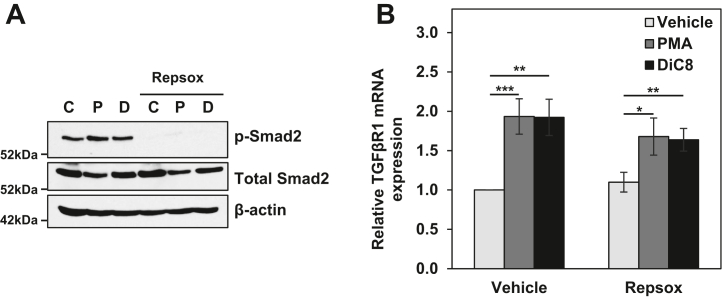
Previous studies have revealed positive feedback of TGFβ signaling on the expression of TGFβR1 mRNA (34, 35). Based on this finding, we explored the requirement for TGFβ signaling in PKCα-induced TGFβR1 expression using the highly specific TGFβR1 inhibitor, Repsox (36, 37). The activity of Repsox was confirmed by its ability to block phosphorylation of the downstream mediator of TGFβ signaling, Smad2 (38), in control as well as PMA- and DiC8-treated cells (Fig. 4A). As shown in Fig. 4B, PKCα agonists were able to induce TGFβR1 expression in the presence of Repsox, indicating that the effect does not require positive feedback from TGFβ signaling. However, there was a consistent, although not statistically significant, reduction in the level of TGFβR1 induction in the presence of Repsox, pointing to the potential for a minor contribution of positive feedback downstream of PKCα-induced upregulation of the receptor. These results were confirmed using the TGFβR1 inhibitor GW788388 (data not shown). Taken together, the data identify PKCα-induced upregulation of TGFβR1 that is independent of TGFβ signaling but, like other antiproliferative effects of PKCα, requires the RAS-ERK signaling cascade.
Figure 4.
Upregulation of TGFβR1 by PKCα does not require TGFβ signaling. IEC-18 cells were pretreated with 50 nM Repsox prior to addition of vehicle (C), 100 nM PMA (P), or 20 μg/ml DiC8 (D) for 2 h. A and B, expression and phosphorylation of the indicated proteins were then assessed by Western blotting (A), and TGFβR1 mRNA levels (normalized to 18S rRNA) were determined by RT-qPCR (B). Data in A are representative of at least three independent experiments, and data in B are the average ± SEM of three independent experiments. ∗p < 0.05. ∗∗p < 0.01. ∗∗∗p < 0.001. DiC8, 1,2-dioctanoyl-sn-glycerol; IEC-18, intestinal crypt-like cells; PKCα, protein kinase C α; PMA, phorbol 12-myristate 13-acetate; TGFβR1, transforming growth factor-β receptor 1.
PKCα transcriptionally activates the TGFβR1 gene and induces the expression and activity of the transcription factor Runx2
To determine if PKCα signaling transcriptionally activates the TGFβR1 gene, we analyzed the effects of PKCα agonists on TGFβR1 mRNA synthesis rates using the Invitrogen 5-ethynyl uridine (EU)-based Click-IT Nascent RNA Capture Kit. As shown in Figure 5A, activation of PKCα with either PMA or DiC8 promoted an ∼1.7-fold increase in TGFβR1 mRNA synthesis in IEC-18 cells by 2 h. The ability of PKCα to upregulate TGFβR1 promoter activity was also tested in reporter assays using a rat TGFβR1 promoter construct driving expression of secreted Gaussia luciferase. This construct encompasses the promoter region from −1529 to −31 relative to the initiation codon of TGFβR1. Analysis of IEC-18 cells using this construct indicated that PMA/PKCα activation increases Gaussia secretion by 2 h (data not shown). However, IEC-18 cells are difficult to transfect, and low levels of Gaussia secretion over the 2 h PMA treatment period led to a signal-to-noise ratio that precluded meaningful quantitative analysis (data not shown). Therefore, further studies were performed using FET CRC cells, since PKCα also regulates TGFβR1 expression in these cells (Fig. 2D). Treatment of transfected FET cells with PMA for 2 h led to an ∼2-fold increase in secretion of Gaussia luciferase (Fig. 5B), confirming that PKCα increases TGFβR1 promoter activity. Together, these data demonstrate that PKCα signaling enhances TGFβR1 expression through enhanced transcription of the TGFβR1 gene.
Figure 5.
PKCα induces ERK-dependent upregulation of TGFβR1 and Runx2 at the transcriptional level.A, IEC-18 cells were treated with vehicle, 100 nM PMA or 20 μg/ml DiC8 for 2 h and incubated with ethynyl uridine (EU) for the final 1 h. Labeled RNA was then isolated using the Click-IT Nascent RNA Capture Kit and quantified by RT-qPCR. Data show levels of nascent TGFβR1 mRNA normalized to nascent 18S rRNA. B, FET colon cancer cells were transfected with a TGFβR1 promoter construct driving expression of Gaussia luciferase. Twenty-four hours after transfection, cells were treated with 100 nM PMA for 2 h. Data show relative luciferase activity from PMA-treated cells normalized to luciferase activity from vehicle-treated cells. C, IEC-18 cells were pretreated with vehicle, 5 μM BIM, or 4 μM Gö6976 prior to addition of PMA (100 nM) or DiC8 (20 μg/ml) for 2 h. Levels of Runx2 mRNA (normalized to 18S RNA) were then assessed by RT-qPCR. D, as in A except that data show relative levels of nascent Runx2 mRNA. E, as in C except that cells were pretreated with vehicle or 1 μM SCH772984 prior to addition of PMA or DiC8. F, IEC-18 cells were pretreated with Gö6976 as indicated prior to addition of vehicle (C), 100 nM PMA (P) or 20 μg/ml DiC8 (D) for 2 h and levels of the indicated proteins were determined by Western blotting. Numbers below the blots represent quantification of Runx2 band intensity relative to loading control (average ± SD from three independent experiments). Data in B, C, and E are averages ± SEM of at least three independent experiments, and data in A and D are averages ± SEM of two independent experiments. ∗p < 0.05. ∗∗p < 0.01. BIM, bisindolylmaleimide I; DiC8, 1,2-dioctanoyl-sn-glycerol; IEC-18, intestinal crypt-like cells; PKCα, protein kinase C α; PMA, phorbol 12-myristate 13-acetate; TGFβR1, transforming growth factor-β receptor 1; ERK, extracellular signal-regulated kinase 1/2; Runx2, runt-related transcription factor 2.
To unveil the mechanism underlying transcriptional induction of TGFβR1 gene expression by PKCα, we leveraged data from RNA-Seq analysis in IEC-18 cells. Notably, this analysis detected Gö6976-sensitive upregulation of runt-related transcription factor 2 (Runx2) (also known as CBFa, PEBP2) by PMA (Table S1). Runx2 was of particular interest since it is a known transcriptional regulator of TGFβR1 in osteoblasts (39, 40), and binding of Runx2 is conserved between rodent and human TGFβR1 promoters (39). Thus, we explored the role of Runx2 as a potential mediator of the effects of PKCα on TGFβR1 transcription. The ability of PKC agonists to upregulate Runx2 was confirmed by RT-qPCR: treatment of IEC-18 cells with PMA or DiC8 led to an ∼10- to 15-fold increase in Runx2 mRNA levels by 2 h (Figs. 5C and S3). As with TGFβR1, the induction of Runx2 was blocked by the general PKC inhibitor BIM and the PKCα-selective inhibitor Gö6976 (Fig. 5C), confirming that the effect was due to PKCα activation. Analysis of nascent RNA using the Click-IT Nascent RNA Capture Kit indicated that upregulation of Runx2 by PMA or DiC8 is at least partially the result of a 4- to 6-fold increase in Runx2 mRNA synthesis (Fig. 5D). PKCα-induced upregulation of Runx2 is also mediated by ERK signaling, as confirmed by the ability of the ERK inhibitor SCH772984 to prevent the increase in Runx2 mRNA promoted by either PMA or DiC8 (Fig. 5E). Using Western blot analysis, we further confirmed that PKCα activation by PMA or DiC8 leads to upregulation of Runx2 protein by 2 h (Fig. 5F). However, this upregulation was modest compared with the increase in Runx2 mRNA. While Runx2 mRNA increased 10- to 15-fold (Fig. 5C), Runx2 protein only increased by ∼1.6-fold in PKC agonist-treated cells, likely reflecting tight regulation of Runx2 at the translational or posttranslational levels (41). Although these data clearly showed that PKCα activation induces the expression of Runx2, time course analysis indicated that the modest upregulation of Runx2 protein occurs after the induction of TGFβR1 gene expression: while upregulation of TGFβR1 mRNA expression was clearly evident by 1 h of PMA treatment (Fig. 6A), upregulation of Runx2 protein was first detected at 2 h (Fig. 6B). These findings, together with the fact that Runx2 was not identified in RNA-Seq analysis of PKCα knockdown cells (Tables S2–S4), indicate that any involvement of Runx2 must require additional levels of regulation.
Figure 6.
PKCα activation increases the transcriptional activity of Runx2 through ERK signaling.A, IEC-18 cells were treated with vehicle or 100 nM PMA for 1 h and 2 h, and TGFβR1 mRNA expression was determined by RT-qPCR. B, IEC-18 cells were treated with vehicle (-) or 100 nM PMA for 0.5 h, 1 h, and 2 h, and Runx2 protein expression was determined by Western blotting. Numbers below the blots indicate relative band intensity (mean ± SD) for Runx2 normalized to corresponding loading control. C, protein extracts from IEC-18 cells were treated with λ-phosphatase as indicated and analyzed by Phos-tag gel electrophoresis (upper panel) or SDS-PAGE (lower panel) and immunoblotted for Runx2 and β-actin. Each panel is from the same blot, and dashed lines indicate where lanes have been rearranged for clarity. D, IEC-18 cells were treated with vehicle (C), 100 nM PMA (P), or 20 μg/ml DiC8 (D) for 15 min and analyzed by Phos-tag gel electrophoresis, SDS-PAGE, and immunoblotting as in C. E, IEC-18 cells were treated with vehicle (C) or 100 nM PMA (P) for 15 min in the presence or absence of 1 μM SCH772984. Extracts were divided and one portion was treated with λ-phosphatase as indicated before analysis by Phos-tag gel electrophoresis (top panels) or SDS-PAGE (middle and lower panels) and immunoblotting for the indicated proteins. Lanes five and six show effects of λ-phosphatase treatment on the extracts shown in lanes 1 and 2, respectively. The dashed line is as in C. F, IEC-18 cells were treated with vehicle (C) or 100 nM PMA (P) for 2 min, 5 min, or 10 min. Phosphorylation/activation of ERK and Runx2 were determined by Western blotting and Phos-tag gel analysis, respectively. G, IEC-18 cells were transfected with p6OSE-luc firefly luciferase reporter along with TK-Renilla luciferase transfection efficiency control. After 24 h, cells were treated with 100 nM PMA or 20 μg/ml DiC8 in the presence or absence of 1 μM SCH772984 for 2 h, and luciferase activity was determined. Data show p6OSE driven firefly luciferase relative to TK-Renilla activity. Data in B–F are representative of at least three independent experiments, and data in A and G are the average ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Arrow in D–F indicates slower mobility species of Runx2 induced by PKC agonist treatment. DiC8, 1,2-dioctanoyl-sn-glycerol; ERK, extracellular signal-regulated kinase 1/2; IEC-18, intestinal crypt-like cells; PKCα, protein kinase C α; PMA, phorbol 12-myristate 13-acetate; Runx2, runt-related transcription factor 2; TGFβR1, transforming growth factor-β receptor 1.
Runx2 activity is regulated by ERK-mediated phosphorylation (42, 43, 44, 45), pointing to the possibility that PKCα-ERK signaling controls Runx2 at the level of its phosphorylation. Changes in Runx2 phosphorylation were, therefore, analyzed using Phos-tag SDS-polyacrylamide gels, which contain phosphate-binding metal chelate complexes that specifically retard migration of phosphorylated proteins during electrophoresis (46). This method of analysis was used because multiple ERK-dependent phosphorylation sites have been associated with Runx2 activation (i.e., Ser240, Ser243, Ser247 Ser301, and Ser319 (42, 43, 44, 45)), and commercial antibodies are not available for these sites. Runx2 from unstimulated cells migrated as multiple bands on Phos-tag gels, which collapsed into a single major band following λ-phosphatase treatment, indicating that the protein is phosphorylated at multiple sites in untreated IEC-18 cells (Fig. 6C). Following PKCα activation by PMA or DiC8, there was an increase in Runx2 phosphorylation, as indicated by the accumulation of a slower migrating species of Runx2 in these gels which is not observed when extracts were treated with λ-phosphatase (Fig. 6, D and E, arrows). Importantly, appearance of this new species was blocked by inhibition of ERK with SCH772984 (Fig. 7E, lanes 1–4), indicating that, as with TGFβR1 induction, Runx2 phosphorylation/activation is downstream of PKCα-ERK signaling. Consistent with the ERK dependence of the effect, increased Runx2 phosphorylation was detected after 5 to 10 min of PKCα activation and thus occurred after ERK activation, which was evident by 2 min of treatment (Fig. 6F). Thus, in contrast to Runx2 upregulation, Runx2 activation precedes induction of TGFβR1 expression, positioning activation of Runx2 rather than its increased expression as a potential mechanism by which PKCα-ERK signaling induces TGFβR1 transcription.
Figure 7.
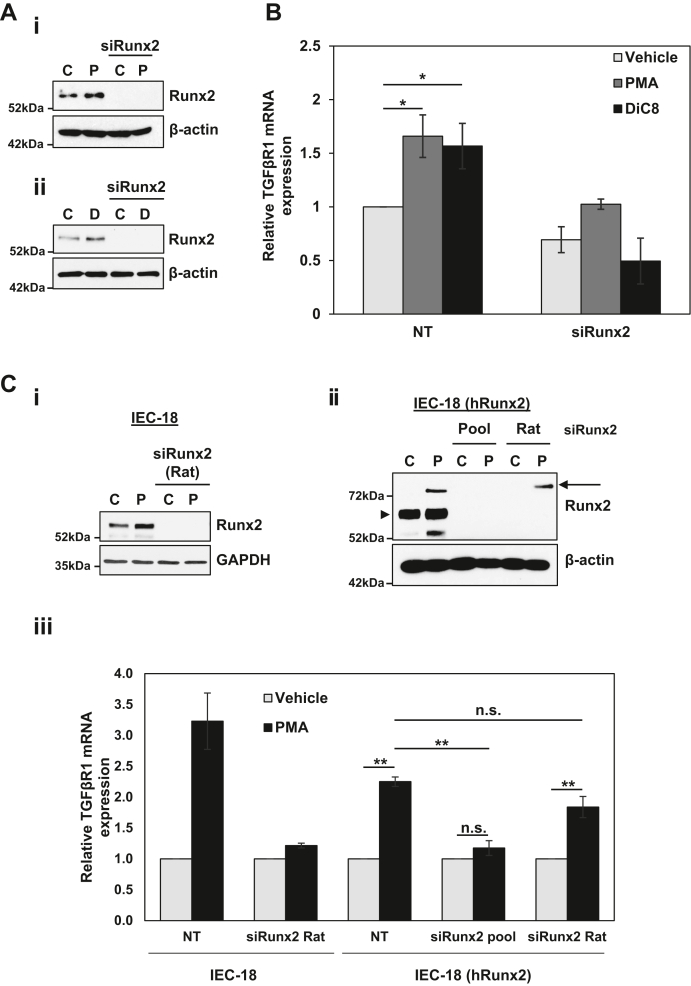
Runx2 is requiredfor PKCα-induced upregulation of TGFβR1. A and B, IEC-18 cells were transfected with nontargeting or Runx2 (rat, mouse, human) targeting ONTARGETplus SMARTPool siRNA for 72 h prior to treatment with 100 nM PMA or 20 μg/ml DiC8. After 2 h, expression of the indicated proteins was determined by Western blotting (Ai,ii) and TGFβR1 mRNA levels (normalized to 18S rRNA) were determined by RT-qPCR (B). C, IEC-18 cells were transfected with nontargeting or rat-specific Runx2-targeting siRNA for 72 h prior to treatment with 100 nM PMA for 2 h, and expression of the indicated proteins was determined by Western blotting (i). IEC-18 cells stably expressing human Runx2 (Origene, RC212936) were transfected with nontargeting siRNA, rat/mouse/human Runx2-targeting ONTARGETplus SMARTPool siRNA, or rat-specific Runx2 siRNA for 72 h prior to treatment with 100 nM PMA for 2 h. Expression of the indicated proteins was determined by Western blotting. Endogenous rat Runx2 (arrowhead) and exogenous human Runx2 (arrow) are indicated (ii). Cells generated as in (i) and (ii) were treated with PMA for 2 h as indicated and TGFβR1 mRNA levels (normalized to 18 S rRNA) were determined by RT-qPCR (iii). Data in A and Ci, and Cii are representative of at least three independent experiments, and data in B and Ciii are the average ± SEM of at least three independent experiments. ∗p < 0.05, ∗∗p < 0.01. DiC8, 1,2-dioctanoyl-sn-glycerol; IEC-18, intestinal crypt-like cells; PKCα, protein kinase C α; PMA, phorbol 12-myristate 13-acetate; Runx2, runt-related transcription factor 2; TGFβR1, transforming growth factor-β receptor 1.
To directly test the ability of PKCα-ERK signaling to induce the transcriptional activity of Runx2, luciferase-reporter assays were performed using p6OSE-luc, a construct which contains six copies of the Runx2 responsive element from the osteocalcin promoter (47) driving expression of firefly luciferase. As shown in Figure 6G, PKCα activation for 2 h led to an ∼2-fold increase in the activity of the reporter. The effect of PMA on p6OSE-luc was blocked by the ERK inhibitor SCH772984 (Fig. 6G); thus, consistent with the effects on Runx2 phosphorylation, these reporter assays show that PKCα-ERK signaling increases the transcriptional activity of Runx2.
Runx2 mediates effects of PKCα on TGFβR1 expression
The role of Runx2 in PKCα-mediated upregulation of TGFβR1 was tested directly using RNAi technology. IEC-18 cells were transfected with Runx2 ON-TARGETplus SMARTPool siRNA, which consists of a pool of four siRNAs that target rat, mouse and human Runx2 (Fig. 7Ai,ii). Runx2 knockdown inhibited PMA- and DiC8-induced upregulation of TGFβR1 mRNA (Fig. 7B), indicating that Runx2 mediates the effects of PKCα on TGFβR1 expression. A requisite role for Runx2 in PKCα-induced TGFβR1 upregulation in IEC-18 cells was confirmed using siRNA that targets rat but not human Runx2 (Fig. 7C, i and iii, left panel). The specificity of the effects of Runx2 knockdown were further confirmed in rescue experiments using IEC-18 cells stably transfected with a vector in which tagged human Runx2 expression is driven by the CMV-promoter (IEC-18 (hRunx2)). Consistent with tight regulation of Runx2 at the protein level (Fig. 5F), expression of the exogenous protein in the stably transfected IEC-18 cells was low to undetectable prior to PMA treatment; however, the tagged human Runx2 protein was clearly detected by anti-Runx2 immunoblotting following PMA treatment for 2 h (Fig. 7Cii, arrow), presumably as a result of the PMA-responsiveness of the CMV promoter in IEC-18 cells (data not shown). The identity of the PMA-induced band as human Runx2 was established by its sensitivity to SmartPool siRNA (Pool) that targets both human and rat Runx2 but not to the rat-specific siRNA (Fig. 7Cii). Importantly, while silencing of both endogenous and exogenous Runx2 with SmartPool siRNA blocked PMA/PKCα-induced upregulation of TGFβR1 mRNA in the stably transfected IEC-18 cells, expression of human Runx2 rescued the ability of PMA/PKCα to induce TGFβR1 expression in the presence of rat-specific siRNA (Fig. 7Ciii).
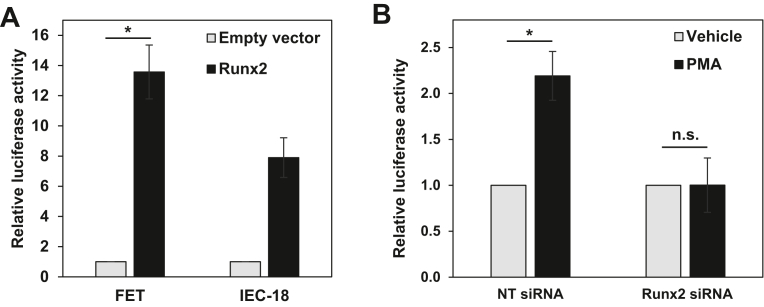
The Runx2 dependence of the effects of PMA/PKCα at the level of TGFβR1 transcription was further supported by promoter-reporter assays using the −1529 to −31 TGFβR1 promoter construct which contains six previously identified Runx2 sites (PS1-6 in ref. (39)). Transient co-transfection with the human Runx2 expression vector enhanced the activity of the TGFβR1 promoter-Guassia luciferase construct, confirming that the TGFβR1 promoter is Runx2-regulated in both IEC-18 and FET cells (Fig. 8A). Furthermore, knockdown of Runx2 blocked the ability of PMA/PKCα to induce the activity of the TGFβR1 promoter construct (Fig. 8B). Collectively, these data confirm that the effects of PMA/PKCα activation on TGFβR1 transcription are mediated by Runx2.
Figure 8.
Runx2 is required for PKCα-induced TGFβR1 promoter activity.A, FET and IEC-18 cells were transfected with empty vector or human Runx2 expression vector, together with TGFβR1 promoter-Gaussia luciferase reporter plasmid, and TGFβR1 promoter activity was measured by Gaussia luciferase assay the following day. B, FET cells were transfected with nontargeting or Runx2 pool siRNA. Forty-eight hours later, cells were transfected with TGFβR1 promoter-reporter plasmid. 24 h after transfection with the reporter plasmid, cells were treated with 100 nM PMA for 2 h, and TGFβR1 promoter activity was measured by Gaussia luciferase assay. Data for FET and IEC-18 cells are averages ± SEM of three or two independent experiments, respectively. ∗p < 0.05. IEC-18, intestinal crypt-like cells; PKCα, protein kinase C α; PMA, phorbol 12-myristate 13-acetate; Runx2, runt-related transcription factor 2; TGFβR1, transforming growth factor-β receptor 1.
PKCα regulates the sensitivity of intestinal cells to TGFβ signaling
Having determined that PKCα regulates TGFβR1 gene expression, the consequences of crosstalk between PKCα and TGFβR1 on TGFβ signaling were explored. Western blot analysis determined that PKCα knockdown leads to an ∼30 to 50% reduction in TGFβR1 protein in CRC cells and IEC-18 crypt-like cells (Fig. 9, A and Bi), a reduction that is comparable with that seen in TGFBR1 mRNA (Fig. 2 and Tables S2–S4). Thus, PKCα loss induces downregulation of TGFβR1 protein, a rate limiting component of TGFβ signaling (30). To examine downstream consequences of PKCα-mediated regulation of TGFβR1 levels, we tested the ability of PKCα knockdown to reduce TGFβ-induced phosphorylation of Smad2, a key transducer of TGFβ-induced signaling that is phosphorylated by activated TGFβR1 (48). FET colon cancer cells were used for this analysis since they represent a TGFβ-sensitive colon cancer cell line (49, 50, 51) that shows low to undetectable basal levels of Smad2 phosphorylation (Fig. 9A) and downregulates TGFβR1 protein following PKCα knockdown (Fig. 9A). Consistent with previous reports (52), TGFβ treatment resulted in robust phosphorylation of Smad2 in FET cells by 30 min (Fig. 9A). However, PKCα knockdown in these cells markedly reduced the ability of TGFβ to induce phosphorylation of Smad2 (Fig. 9A), indicating that PKCα regulates both TGFβR1 expression and TGFβ signaling. Note that our RNA-Seq data did not reveal any consistent effects of PKCα on TGFβR1 ligands that induce phosphorylation of Smad2, including TGFβ1, TGFβ2, TGFβ3, activin, and nodal, further supporting the role of PKCα modulation of TGFβR1 expression in the observed effects.
Figure 9.
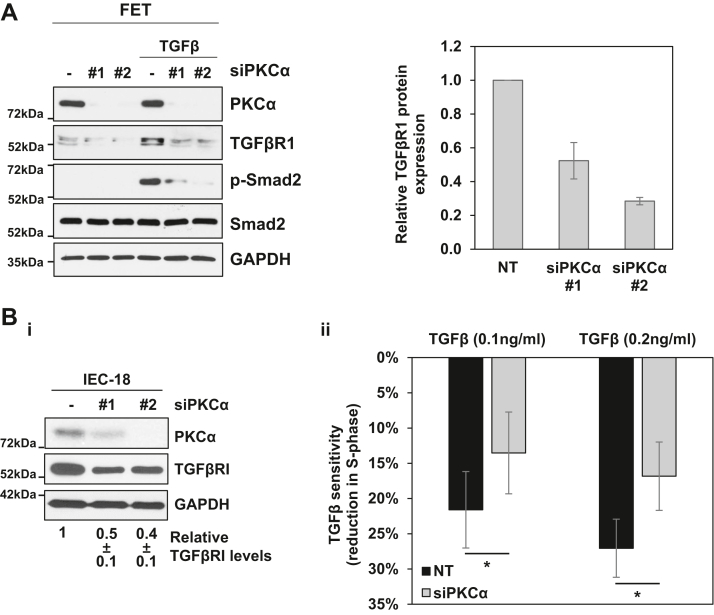
PKCα enhances intestinal cell sensitivity to TGFβ signaling.A, FET cells were transfected with nontargeting siRNA (−) or siRNAs targeting PKCα (#1, #2) for 72 h before addition of 2.5 ng/ml TGFβ1 for 30 min as indicated. Expression and phosphorylation of the indicated proteins was determined by Western blotting. The graph to the right of the blots shows densitometric analysis of relative levels of TGFβR1 normalized to loading control (±s.d., n = 2). Bi, IEC-18 cells were transfected with nontargeting siRNA or siRNA targeting PKCα as in A, and the indicated proteins were detected by Western blotting. Numbers under the blots indicate relative band intensity (mean ± SD) for TGFβR1 normalized to loading control. ii, IEC-18 cells were transfected with siRNA as in A prior to treatment with the indicated concentrations of TGFβ1 for 9 h. Levels of cells in S phase were then determined by flow cytometry and data show the relative reduction in cells in S phase in TGFβ1-treated cells relative to control cells (±SEM). Data are representative (A) or averages (B) of three independent experiments. ∗, p < 0.05. IEC-18, intestinal crypt-like cells; PKCα, protein kinase C α; TGFβ, transforming growth factor β; TGFβR1, transforming growth factor-β receptor 1.
Studies were also performed to determine the impact of PKCα knockdown on TGFβ-induced inhibition of cell cycle progression. FET cells are not suitable for analysis of the cell cycle effects of PKCα because (a) they grow in clusters that cannot be readily disrupted into single cell suspensions for flow cytometric analysis and (b) TGFβ has been shown to induce apoptosis in these cells (53). Therefore, analysis of the effects of PKCα knockdown on TGFβ-induced G1→S phase arrest was performed using IEC-18 cells, which undergo growth inhibition following treatment with 0.1 to 1 ng/ml TGFβ (54, 55). IEC-18 cells were transfected with nontargeting or PKCα-targeting siRNA, and PKCα knockdown and downregulation of TGFβR1 protein were confirmed by Western blot analysis (Fig. 9Bi). The ability of TGFβ treatment (9 h) to reduce the number of cells in S-phase in control and PKCα/TGFβR1-deficient cells was then determined by flow cytometric analysis. Treatment of control IEC-18 cells with 0.1 or 0.2 ng/ml TGFβ led to a 22% and 28% decrease in cells in S phase respectively (Fig. 9Bii), confirming the ability of TGFβ to inhibit G1→S phase progression in these cells. However, the negative effects of TGFβ on the cell cycle were significantly reduced following PKCα knockdown, with only 15% and 18% reduction in S-phase observed following treatment of PKCα siRNA-transfected cells with 0.1 or 0.2 ng/ml TGFβ, respectively (Fig. 9Bii). Thus, consistent with the effects on SMAD2 phosphorylation, PKCα regulates the ability of TGFβ to suppress the growth of intestinal cells.
TGFβR1 signaling contributes to PKCα-induced growth inhibition
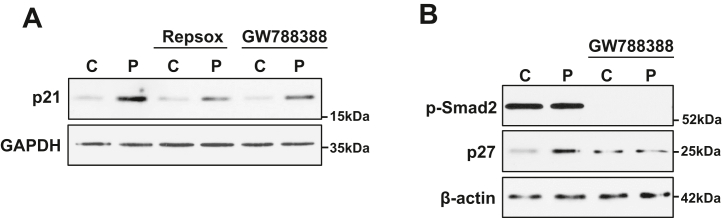
Our previous studies have shown that PKCα-induced growth arrest in intestinal cells involves induction of the CDK inhibitors p21Cip1 and p27Kip1 (10, 13), which are known targets of TGFβ signaling (21, 22). We, therefore, used the TGFβR1 inhibitors Repsox and GW788388 to determine if PKCα-induced effects on p21Cip1 and p27Kip1 involve the TGFβR1 axis identified in this study. As shown in Figure 10, A and B, induction of both p21Cip1 and p27Kip1 was impaired by inhibition of TGFβR1. Although, effects on p21Cip1 were only partial, indicating that additional pathways contribute to the effects of PKCα on this CDK inhibitor, these findings establish TGFβR1 as a downstream component of growth inhibitory PKCα signaling in intestinal epithelial cells.
Figure 10.
TGFβR1 signaling is involved in PKCα-induced p21Cip1and p27Kip1upregulation. IEC-18 cells were pretreated with 50 nM Repsox or 5 μM GW788388 prior to treatment with vehicle (C) or 100 nM PMA (P). A and B, expression of p21Cip1 (A) and p27Kip1 (B), as well as phosphorylation of Smad2 (B), were determined by Western blotting. Data are representative of at least three independent experiments. IEC-18, intestinal crypt-like cells; PKCα, protein kinase C α; PMA, phorbol 12-myristate 13-acetate; TGFβR1, transforming growth factor-β receptor 1.
PKCα expression correlates with levels of Runx2 and TGFβR1 in patient tumors
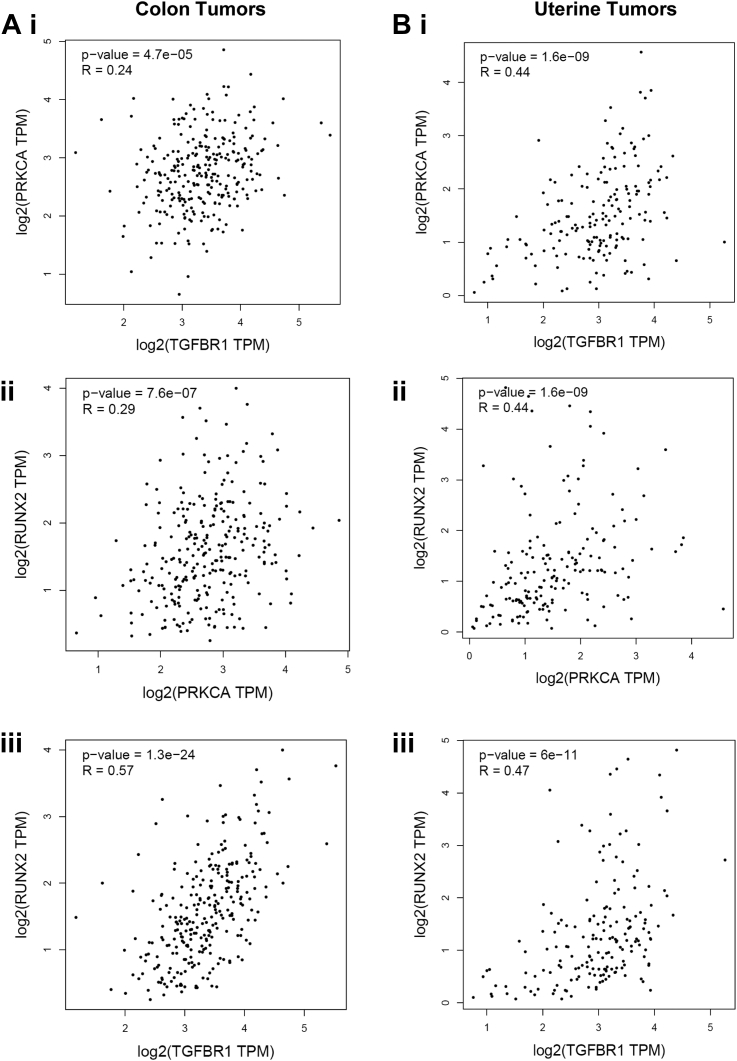
Analysis of TCGA (The Cancer Genome Atlas) gene expression data supports the relevance of the PKCα-Runx2-TGFβR1 axis to human disease. There is a highly significant correlation (p < 0.001) between PKCα and TGFβR1 mRNA expression levels in human colon tumor samples, implying co-occurrence of loss of expression of the two kinases in tumors (Fig. 11Ai). Furthermore, expression of Runx2 correlates with expression of both PKCα and TGFβR1 in these tumors, consistent with a role for this transcription factor in mediating the effects of PKCα on TGFβR1 expression (Fig. 11A, ii and iii). Notably, consistent with the regulation of TGFβR1 by PKCα in SNG-M endometrial cancer cells (Fig. 1, A and C and Table S4), a similar correlation between PKCα, Runx2 and TGFβR1 mRNA was observed in uterine tumors (Fig. 11B). Thus, crosstalk between PKCα and TGFβ signaling may occur in multiple tissues in addition to the intestine.
Figure 11.
Correlation between PKCα, Runx2 and TGFβR1 expression in human tumors.A and B, TCGA data for colon (A) and uterine (B) cancer were analyzed for expression of PKCα and TGFβR1 (i), Runx2 and PKCα (ii), and Runx2 and TGFβR1 (iii) mRNA using the GEPIA web server. The R value is the Spearman’s rank correlation coefficient. PKCα, protein kinase C α; Runx2, runt-related transcription factor 2; TGFβ, transforming growth factor β; TGFβR1, transforming growth factor-β receptor 1; TCGA, The Cancer Genome Atlas.
Discussion
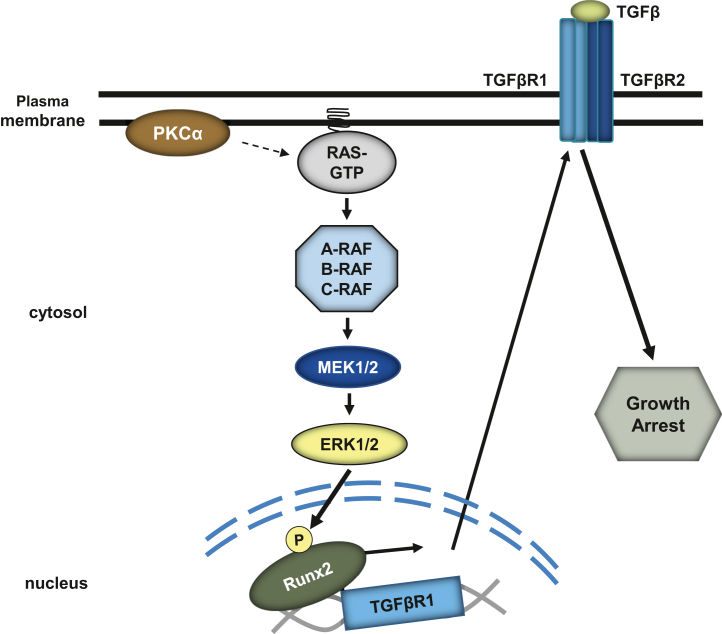
We have determined that PKCα activation positively regulates TGFβR1 expression and signaling in nontransformed intestinal epithelial cells and colon cancer cells, thereby identifying crosstalk between two established antiproliferative signaling pathways in the intestinal epithelium. Our findings are consistent with the ability of PKCα to mediate glucose-induced upregulation of TGFβR1 protein in vascular smooth muscle cells (56), while advancing understanding of TGFβR1 regulation by defining the underlying mechanisms involved. We provide the first evidence that PKCα signaling induces TGFβR1 expression at the level of transcription through activation of Runx2, and we define a novel PKCα→ERK→Runx2→TGFβR1 signaling axis that modulates cellular responses to TGFβ (Fig. 12). The physiological relevance of this signaling axis in the intestinal epithelium is supported by evidence that intestinal cells are highly sensitive to changes in TGFβR1 levels. Haploinsufficiency and decreased allelic expression of TGFβR1 have been linked to increased intestinal tumorigenesis in mice and humans, respectively (28, 30, 57), with a 40% reduction in TGFβR1 expression in intestinal cells of heterozygous mice resulting in a significant decrease in SMAD2 phosphorylation in association with a marked increase in cell proliferation. Notably, similar changes were seen in our study, which demonstrated that a comparable reduction in TGFβR1 expression induced by PKCα knockdown was sufficient to dampen the ability of TGFβ to induce SMAD2 phosphorylation and inhibit cell cycle progression in intestinal cells (Fig. 9). The spatial association between PKCα activation at the crypt–villus junction and increased TGFβR1 expression on the villus further supports the physiological relevance of our findings (8, 9, 23). Thus, we provide the first evidence for crosstalk between PKCα and TGFβ signaling pathways that may be linked to regulation of homeostasis in the intestinal epithelium.
Figure 12.
Model of PKCα regulation of TGFβ signaling. PKCα activation promotes Ras/ERK-dependent phosphorylation/activation of Runx2 to increase TGFβR1 expression and TGFβ-mediated signaling. ERK, extracellular signal-regulated kinase 1/2; PKCα, protein kinase C α; Runx2, runt-related transcription factor 2; TGFβ, transforming growth factor β; TGFβR1, transforming growth factor-β receptor 1.
Consistent with their growth inhibitory role in intestinal epithelial cells, both PKCα and TGFβR1 signaling are tumor suppressive in the intestine (20, 58). TCGA data indicate that mutations in the PKCα (PRKCA) and TGFβR1 (TGFBR1) genes are rare in CRC (≤4% of cases), pointing to the importance of alterations in their expression for the transformed phenotype. Downregulation of PKCα is an early event in CRC progression, seen in aberrant crypt foci and intestinal adenomas, and PKCα loss has been observed in a majority of CRC cases (8, 12, 17, 18). In the case of TGFβR1, a number of polymorphisms, including TGFBR1∗6A and IVS7_24G > A, have been associated with increased risk of CRC in multiple (although not all) studies, supporting the idea that TGFβR1 polymorphisms are a low penetrance risk factor for CRC (59, 60, 61). Interestingly, these polymorphisms do not affect the sequence of the mature receptor; instead, they are associated with reduced levels of the receptor due to decreased allelic expression (12) or miRNA targeting of TGFβR1 (59). Thus, as seen with TGFβR1 haploinsufficiency in mice (59, 60, 61), relatively modest reductions in TGFβR1 levels appear to enhance tumorigenesis in the human intestine. It is notable that somatic acquisition of one of these polymorphisms has been reported in the normal mucosa adjacent to tumors, indicating that, as seen with PKCα, downregulation of TGFβR1 may promote early stages of CRC tumorigenesis (62). A link between PKCα and TGFβR1 in CRC tumorigenesis is seen in the correlation between PKCα and TGFβR1 mRNA levels in CRC patient samples (Fig. 11). Our finding that loss of PKCα leads to a reduction in TGFβR1 expression provides a mechanistic basis for this correlation and supports the idea that regulation of TGFβR1 levels contributes to the tumor suppressive effects of PKCα in the intestine.
Our data also show that PKCα regulates TGFβR1 expression through activation of Runx2 (Figure 6, Figure 7, Figure 8). Consistent with a role of Runx2 as a mediator of PKCα/TGFβR1 crosstalk, Runx2 mRNA levels correlate with both PKCα and TGFβR1 expression in CRC tissues (Fig. 11). Interestingly, while PKCα activation led to a robust increase in Runx2 mRNA levels, the effects on Runx2 protein were modest (Figs. 5 and 6). While the precise mechanism underlying this discrepancy is not known, it has been noted in other systems (e.g., (63)) and likely reflects tight translational and/or posttranslational regulation of the protein (41). PKCα also led to a rapid increase in ERK-mediated activating phosphorylation of Runx2, and analysis of the timing of the effects of PKCα (Fig. 6) indicated that Runx2 activation is the major driver of the initial increase in TGFβR1 transcription, although Runx2 upregulation likely contributes to maintenance of TGFβR1 levels at later times.
A role for Runx2 in PKCα-induced transcriptional activation of TGFβR1 is consistent with the presence of multiple Runx2 consensus binding sites in the TGFβR1 promoter (39) and with data from ChIP-Seq analysis of global Runx2 binding in osteoblasts, which detected Runx2 peaks in the promoter region of TGFBR1 (64, 65). These findings, together with the rapid induction of TGFβR1 mRNA expression (by 1 h), argue for a direct effect of Runx2 on the TGFβR1 promoter, although an indirect mechanism mediated by a Runx2-regulated transcription factor(s) cannot be excluded at this time. Interestingly, the Runx2 promoter also contains multiple Runx2 binding sites, and Runx2 positively regulates its own expression (66, 67), suggesting that the transcriptional upregulation of this gene induced by PKCα may reflect positive feedback downstream of the enhanced ERK-dependent activation of this transcription factor. Ongoing studies are examining the role of Runx2 and other transcription factors known to regulate the Runx2 promoter (e.g., AP1, NF-κB, Sp1, and HIF2A (68, 69, 70, 71)) in the effects of PKCα-ERK signaling.
Although roles for Runx2 in epithelial cells are emerging (64, 65), this study provides the first evidence of a role for this factor in the normal intestinal epithelium. Runx2 is best known as a critical regulator of osteogenesis (72). While PKCα signaling has been linked to osteoblast function (68, 69, 70, 71), its effects in bone have not been attributed to modulation of Runx2. Thus, to our knowledge, this is also the first report to directly link PKCα signaling to enhanced Runx2 activity in any system. Mammalian cells express a family of Runx proteins that includes Runx1 and Runx3 in addition to Runx2 (73). Since Runx proteins recognize the same promoter elements (73), it is possible that Runx1 and/or Runx3 also regulate TGFβR1 expression. Our knockdown experiments in IEC-18 and FET CRC cells confirmed the role of Runx2, while excluding the involvement of Runx1 (Figs. 7 and 8 and Table S1), which is expressed in these cells. However, a role for Runx3 cannot be excluded since Runx3 was not detected in IEC-18 or FET cells (data not shown). Ongoing studies are examining the possible involvement of Runx3 in the antiproliferative and tumor suppressive effects of PKCα in the intestinal epithelium.
Both PKCα and TGFβ signaling have antiproliferative and tumor suppressive activities in a number of epithelial tissues besides the intestine/colon, including the endometrium and skin (74, 75). Our finding that PKCα regulates TGFβR1 expression in endometrial cancer cells (Fig. 1 and Table S4), together with the correlation between PKCα, Runx2, and TGFβR1 mRNA levels in TCGA data for uterine cancer (Fig. 11), indicates that the PKCα→ERK→Runx2→TGFβR1 axis defined in intestinal tissues likely extends to additional epithelial systems. Thus, the current study provides insight into crosstalk between two tumor suppressive signaling pathways that are relevant to regulation of homeostasis and tumorigenesis in multiple tissues.
Experimental procedures
Cell culture and drug treatments
IEC-18 nontransformed rat intestinal epithelial cells (ATCC CRL-1589) were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum (FBS), 10 μg/ml insulin, 4 mM glutamine, and 100 μM sodium pyruvate. HCT-116 (ATCC CCL-247), FET (76), and SW620 (ATCC CCL-227) CRC cells were cultured in RPMI 1640 medium supplemented with 10% FBS and SNG-M endometrial cancer cells (JCRB Cell Bank) were grown in Ham’s:F12 medium supplemented with 10% FBS. For experiments involving Salirasib, cells were serum-starved in medium containing 0.5% FBS for 16 to 18 h prior to addition of the drug. To activate PKCα, cells were treated with 100 nM PMA (Biomol) dissolved in ethanol or 20 μg/ml DiC8 (Cayman Chemical) dissolved in acetonitrile. DiC8 was replaced every hour to compensate for its rapid metabolism in cells (77). TGFβ1 (R&D Systems) was dissolved in 4 mM HCl, 1 mg/ml BSA and added at the indicated concentrations. For inhibitor studies, cells were pretreated with inhibitors (dissolved in DMSO) prior to addition of PKCα agonists or vehicle as follows: 5 μM BIM (Calbiochem), 1 h pretreatment; 4 μM Gö6976 (EMD Millipore), 1 h pretreatment; 50 μM Salirasib (Selleckchem), 2 h pretreatment; 1 μM LY3009120 (MedChemExpress), 1 h pretreatment; 10 μM PD0325901 (Selleckchem), 4 h pretreatment; 1 μM SCH772984 (Cayman Chemical), 1 h pretreatment; 50 nM Repsox (Reprocell), 24 h pretreatment; 5 μM GW788388 (Selleckchem), 1 h pretreatment. Appropriate vehicle was added to all controls: vehicle concentrations were ≤0.2% in all cases.
RNA interference
For PKCα knockdown, cells were transfected with 33 nM (100 pmol) siRNA using RNAiMAX transfection reagent (Invitrogen), and cells were analyzed after 72 h. siRNAs targeting rat/human PKCα mRNA were from ThermoFisher as follows: siRNA #1 - GGAUUGUUCUUUCUUCAUATT; siRNA #2 - GAAGGGUUCUCGUAUGUCATT. For Runx2 knockdown, cells were transfected similarly with 10 nM (33 pmol) ON-TARGETplus Rat/Mouse/Human Runx2 SMARTpool siRNA (Dharmacon, L-082676–02–0005) or Rat Runx2 siRNA (Dharmacon, J-082676–11). Controls were transfected with equivalent levels of ON-TARGETplus nontargeting siRNA (Dharmacon D-001810–01–05). For rescue experiments, IEC-18 cells were transfected with a plasmid in which human Runx2 expression is driven by the CMV promoter (RC212936, Origene), and stable transfectants were selected with 1.5 mg/ml G418.
RNA-seq analysis
Cellular RNA was isolated using the RNAspin mini RNA isolation kit (GE Health). Samples (biological duplicates) included untreated, PMA-treated, 4 μM Gö6976 treated, and PMA + 4 μM Gö6976 treated IEC-18 cells; HCEC cells + nontargeting siRNA, HCEC cells + PKCα siRNA #1, and HCEC cells + PKCα siRNA #2; HCT-116 cells + nontargeting siRNA, HCT116 cells + PKCα siRNA #1, and HCT-116 cells + PKCα siRNA #2; SNGM cells + nontargeting siRNA, SNG-M cells + PKCα siRNA #1, and SNG-M cells + PKCα siRNA #2. cDNA generation and next-generation sequencing was performed by the UNMC Genomics Core. Sequence alignment and quantification were performed by the UNMC Bioinformatics and Systems Biology Core.
Western blot analysis
Cells were rinsed twice with PBS and lysed in SDS lysis buffer (1% SDS, 10 mM Tris-HCl pH 7.4). Equal amounts of protein were subjected to SDS-PAGE, transferred to nitrocellulose membrane, and subjected to Western blotting as we have described (13, 31). Primary antibodies were applied overnight at 4 °C as follows: anti-PKCα (1:10,000; Abcam, ab32376), anti-ERK (1:3000; Cell Signaling, 9102S), anti-phospho-ERK (1:3000; Cell Signaling, 9106S), anti-phospho-Rsk1 (S380) (1:1000; Millipore 04–418); anti-Runx2 (1:500; Santa Cruz Biotechnology, sc-390351), anti-phospho-Smad2 (1:1000; Cell Signaling, 3108T), anti-Smad2 (1:1000; Cell Signaling, 5339T), anti-Smad2/3 (1:1000; Cell Signaling, 8685), anti-TGFβR1 (1:3000; ABclonal Technology, A16983); anti-TGFβR1 (1:1000; Abcam, ab31013), anti-p21Cip1 (1:100; Novus Biologicals, NBP2-29463), anti-p27Kip1 (1:2000; Cell Signaling, 3686T), anti-β-actin (1:10,000; Sigma-Aldrich, A20660), and anti-GAPDH (1:60,000; Cell Signaling, 5174T). Secondary horseradish peroxidase-conjugated goat anti-rabbit antibody (Millipore, AP132P), and goat anti-mouse antibody (Bio-Rad, 170–6516) antibodies were used at 1:1000 and detection used SuperSignal West (Thermo Scientific).
The specificity of the PKC isozyme antibodies used in this study was confirmed through knockdown experiments (e.g., Figs. 2 and 9) and use of pharmacological inhibitors (e.g., Fig. S1). Similar approaches have confirmed the specificity of ERK pathway antibodies (see Fig. 3 and (33)). pSMAD2 antibody specificity was tested using inhibitors of upstream TGFβR1 signaling (e.g., Repsox, GW788388; see Figs. 4 and 10), and the specificity of Runx2 antibodies was confirmed by knockdown experiments and pharmacological inhibition of ERK signaling (e.g., Figs. 6 and 7). Unless otherwise indicated, all Western blot experiments were performed at least three times, with representative blots shown in the figures. Where appropriate, relative signal intensity was quantified from scans of multiple exposures using Image J software (NIH).
Quantitative RT-PCR analysis
Cellular RNA was isolated using Trizol Reagent (ThermoFisher) or RNAspin mini RNA isolation columns (GE Health/Zymo Research). RT-qPCR was performed on 10 ng of RNA using Brilliant II SYBR Green RT-qPCR One-Step Master Mix (Agilent) and a Bio-Rad CFX96 Realtime System. Relative mRNA levels were determined from standard curves using CFX Manager Software (Bio-Rad Laboratories). Primers were as follows: Human TGFβR1: fwd CTATATCTGCCACAACCGCACTGT, rev CGCCACTTTCCTCTCCAAACTTCTC; Rat TGFβR1: fwd GCTTCTCATCGTGTTGGTGG, rev TGAAAAAGGTCCTGTAGTTGGGAG; rat Runx2: fwd GCGCATTCCTCATCCCAGTA, rev GGTGGGGAGGATTGTGTCTG; human and rat/human 18S: fwd CATTGGAGGGCAAGTCTGGTG, rev CTCCCAAGCTCCAACTACGAG. Data are presented normalized to 18S rRNA.
Measurement of nascent mRNA
Labeling and capture of nascent RNA were performed using the Click-iT Nascent RNA Capture Kit (Life Technologies) as instructed by the manufacturer. In brief, cells were labeled with EU for 1 h, and cellular RNA was purified. Newly synthesized, EU-labeled RNA was biotinylated and isolated using Streptavidin magnetic beads. cDNA was generated from bead-bound RNA using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories) and analyzed by quantitative PCR as above using iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories). Normalization to nascent 18 S rRNA and calculation of relative levels used the ΔΔCt method.
Phos-tag gel analysis
Cells were lysed in 1% Ipegal CA-630, 137 mM NaCl, 20 mM Tris-HCl, pH 8.0, with freshly added protease inhibitor cocktail and phosphatase inhibitor cocktails I and II (Sigma) and cleared by centrifugation at 10,000g for 10 min at 4 °C. Protein concentration in the supernatants was determined, and an equal volume of 2x SDS sample buffer was added. For λ-phosphatase treatment, phosphatase inhibitors were omitted, and supernatants were adjusted to 1 mM MnCl2 and treated with 800 units of λ-phosphatase (P0753, New England Biolabs) for 30 min at 30 °C prior to addition of SDS sample buffer. Samples were then subjected to electrophoresis using an 8% SDS-acrylamide resolving gel containing 20 μM Phos-tag reagent (Wako Pure Chemical Industries, AAL-107) and 0.1 mM MnCl2. Following electrophoresis at 60 to 90 V, gels were equilibrated in transfer buffer containing 10 mM EDTA (2 x 10 min) prior to immunoblotting as above.
Promoter-reporter assays
TGFβR1 promoter activity was assessed using a construct containing the TGFβR1 promoter region from −1529 to −31 relative to the initiation codon driving expression of secreted Gaussia luciferase (GeneCopoeia: RPRM57493-LvPG02). IEC-18 or FET cells were transfected with TGFβR1 promoter-Gaussia luciferase plasmid using Lipofectamine 3000 (Thermo Fisher Scientific, L3000001) as described by the manufacturer, except that cells were detached from plates using TrypLE Express (Gibco), and reverse transfection was performed. The transfection mixture was added dropwise to the suspended cells in a 15 ml conical tube to form a single transfection reaction that was then dispensed evenly into 6- or 12-well plates. To examine the effects of Runx2 expression, the TGFβR1 promoter construct was co-transfected with human Runx2 expression vector (RC212936, Origene) or empty vector control, and medium was removed for analysis 16 to 25 h after transfection. For Runx2 knockdown experiments, reverse transfection of cells was performed 24 to 48 h after transfection with Runx2 targeting siRNA as above. For analysis of effects of PMA, medium was removed 16 to 24 h after transfection, and cells were rinsed and then incubated in medium containing vehicle or 100 nM PMA for 2 h before aliquots of the medium were removed for analysis. Gaussia luciferase activity in the medium from triplicate wells was measured using the Pierce Gaussia Luciferase Flash Assay Kit (Thermo Scientific, 16,158) in a Berthold Lumat LB9501 luminometer.
For analysis of Runx2 transcriptional activity, IEC-18 cells in 6-well dishes were transfected with p6OSE-Luc plasmid (a gift from Dr Patricia F. Ducy, Columbia University, NY) together with pRL-TK (Promega) using X-tremeGENE nine DNA Transfection Reagent (Sigma). Firefly and Renilla luciferase activities were measured 24 h after transfection using the Dual Luciferase Reporter Assay System (Promega Corp.) and a Berthold Lumat LB 9501 luminometer. Firefly luciferase activity was normalized to Renilla activity for each sample.
Cell cycle analysis
IEC-18 cells were treated with 0.1 ng/ml or 0.2 ng/ml TGFβ1 for 9 h, trypsinized, fixed in 70% ethanol, stained with Telford Reagent (78) and subjected to flow cytometric analysis in the UNMC Cell Analysis Core Facility. Cell cycle distribution was calculated using FlowJo (FloJo LLC) and Modfit (Verity Software) Software.
Correlation analysis of different genes in tumors
The correlation between PKCα and TGFβR1 mRNA expression, PKCα and Runx2 mRNA expression, and TGFβR1 and Runx2 mRNA expression was determined by Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn/, (79)). Analysis used gene expression profiles from publicly available TCGA and GTEx (Genotype-Tissue Expression) datasets. Nonlog scale was used for calculations, and log-scale axis was used for visualization. Correlations were evaluated using Spearman’s rank correlation analysis.
Statistical analysis and other software
Numerical data are presented as means ± standard error of the mean or standard deviation as appropriate. Student’s t tests were performed using Microsoft Excel software, and statistical significance was determined using an alpha level of 0.05. For presentation, contrast and brightness of scanned images were adjusted using GMU Image Manipulation Program, Adobe Photoshop, or Microsoft PowerPoint Software. All adjustments to contrast and brightness were made equally across the entire blot, and no individual lanes were treated differently than the rest of the blot. Graphs were generated using Microsoft Excel Software, and figures were assembled and annotated in Microsoft PowerPoint and Photoshop Software. Other software used is listed above.
Data availability
All data described in the manuscript are contained within the manuscript or in Supporting Information.
Supporting information
This article contains supporting information. (7, 9)
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Dr Patricia F. Ducy (Columbia University, NY) for providing p6OSE-Luc plasmids; Dr Jixin Dong (University of Nebraska Medical Center) and his laboratory for guidance on Phos-tag gel analysis, and Dr Leah Cook (University of Nebraska Medical Center) and members of the Black laboratory for helpful discussions. Analysis of the correlation of gene expression in human cancer was based on data generated by the TCGA Research Network (https://www.cancer.gov/tcga).
Author contributions
X. L., N. K., and A. R. B. investigation; X. L. and A. R. B. data curation; X. L., M. A., A. R. K., A. R. B., and J. D. B. formal analysis; X. L. A. R. B., and J. D. B. writing-original draft; A. R. K., A. R. B., and J. D. B. conceptualization; A. R. K., A. R. B., and J. D. B. writing–reviewing and editing; A. R. B. and J. D. B. supervision; A. R. B. and J. B. visualization; J. D. B. funding acquisition.
Funding and additional information
This work was supported in part by the National Institutes of Health Grants DK060632, CA054807, CA191894, CA036727, P20GM121316, and a pilot award from P50 CA127297. Support was also received from DOD award W81XWH-20-1-0590. X. L. was supported by a UNMC Dean for Graduate Studies Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Donita Brady
Supporting information
References
- 1.Mayhew T.M., Myklebust R., Whybrow A., Jenkins R. Epithelial integrity, cell death and cell loss in mammalian small intestine. Histol. Histopathol. 1999;14:257–267. doi: 10.14670/HH-14.257. [DOI] [PubMed] [Google Scholar]
- 2.Sancho E., Batlle E., Clevers H. Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- 3.Barker N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 4.Vanuytsel T., Senger S., Fasano A., Shea-Donohue T. Major signaling pathways in intestinal stem cells. Biochim. Biophys. Acta. 2013;1830:2410–2426. doi: 10.1016/j.bbagen.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black A.R., Black J.D. Protein kinase C signaling and cell cycle regulation. Front Immunol. 2012;3:423. doi: 10.3389/fimmu.2012.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black J.D., Affandi T., Black A.R., Reyland M.E. PKCα and PKCδ: friends and rivals. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.102194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao F., Pysz M.A., Curry K.J., Haas K.N., Seedhouse S.J., Black A.R., et al. Protein kinase Calpha signaling regulates inhibitor of DNA binding 1 in the intestinal epithelium. J. Biol. Chem. 2011;286:18104–18117. doi: 10.1074/jbc.M110.208488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verstovsek G., Byrd A., Frey M.R., Petrelli N.J., Black J.D. Colonocyte differentiation is associated with increased expression and altered distribution of protein kinase C isozymes. Gastroenterology. 1998;115:75–85. doi: 10.1016/s0016-5085(98)70367-1. [DOI] [PubMed] [Google Scholar]
- 9.Saxon M.L., Zhao X., Black J.D. Activation of protein kinase C isozymes is associated with post-mitotic events in intestinal epithelial cells in situ. J. Cell Biol. 1994;126:747–763. doi: 10.1083/jcb.126.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey M.R., Clark J.A., Leontieva O., Uronis J.M., Black A.R., Black J.D. Protein kinase C signaling mediates a program of cell cycle withdrawal in the intestinal epithelium. J. Cell Biol. 2000;151:763–778. doi: 10.1083/jcb.151.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pysz M.A., Hao F., Hizli A.A., Lum M.A., Swetzig W.M., Black A.R., et al. Differential regulation of cyclin D1 expression by protein kinase C alpha and signaling in intestinal epithelial cells. J. Biol. Chem. 2014;289:22268–22283. doi: 10.1074/jbc.M114.571554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pysz M.A., Leontieva O.V., Bateman N.W., Uronis J.M., Curry K.J., Threadgill D.W., et al. PKCα tumor suppression in the intestine is associated with transcriptional and translational inhibition of cyclin D1. Exp. Cell Res. 2009;315:1415–1428. doi: 10.1016/j.yexcr.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frey M.R., Saxon M.L., Zhao X., Rollins A., Evans S.S., Black J.D. Protein kinase C isozyme-mediated cell cycle arrest involves induction of p21(waf1/cip1) and p27(kip1) and hypophosphorylation of the retinoblastoma protein in intestinal epithelial cells. J. Biol. Chem. 1997;272:9424–9435. doi: 10.1074/jbc.272.14.9424. [DOI] [PubMed] [Google Scholar]
- 14.Guan L., Song K., Pysz M.A., Curry K.J., Hizli A.A., Danielpour D., et al. Protein kinase C-mediated down-regulation of cyclin D1 involves activation of the translational repressor 4E-BP1 via a phosphoinositide 3-Kinase/Akt-independent, protein phosphatase 2A-dependent mechanism in intestinal epithelial cells. J. Biol. Chem. 2007;282:14213–14225. doi: 10.1074/jbc.M610513200. [DOI] [PubMed] [Google Scholar]
- 15.Hizli A.A., Black A.R., Pysz M.A., Black J.D. Protein kinase C α signaling inhibits cyclin D1 translation in intestinal epithelial cells. J. Biol. Chem. 2006;281:14596–14603. doi: 10.1074/jbc.M601959200. [DOI] [PubMed] [Google Scholar]
- 16.Oster H., Leitges M. Protein kinase Cα but not PKCζ suppresses intestinal tumor formation in ApcMin/+ mice. Cancer Res. 2006;66:6955–6963. doi: 10.1158/0008-5472.CAN-06-0268. [DOI] [PubMed] [Google Scholar]
- 17.Kahl-Rainer P., Karner-Hanusch J., Weiss W., Marian B. Five of six protein kinase C isoenzymes present in normal mucosa show reduced protein levels during tumor development in the human colon. Carcinogenesis. 1994;15:779–782. doi: 10.1093/carcin/15.4.779. [DOI] [PubMed] [Google Scholar]
- 18.Suga K., Sugimoto I., Ito H., Hashimoto E. Down-regulation of protein kinase C-α detected in human colorectal cancer. IUBMB Life. 1998;44:523–528. doi: 10.1080/15216549800201552. [DOI] [PubMed] [Google Scholar]
- 19.Batlle E., Verdu J., Dominguez D., del Mont Llosas M., Diaz V., Loukili N., et al. Protein kinase C-alpha activity inversely modulates invasion and growth of intestinal cells. J. Biol. Chem. 1998;273:15091–15098. doi: 10.1074/jbc.273.24.15091. [DOI] [PubMed] [Google Scholar]
- 20.Jung B., Staudacher J.J., Beauchamp D. Transforming growth factor β; superfamily signaling in development of colorectal cancer. Gastroenterology. 2017;152:36–52. doi: 10.1053/j.gastro.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hata A., Chen Y.-G. TGF-β signaling from receptors to smads. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seoane J. Escaping from the TGFbeta anti-proliferative control. Carcinogenesis. 2006;27:2148–2156. doi: 10.1093/carcin/bgl068. [DOI] [PubMed] [Google Scholar]
- 23.Barnard J.A., Warwick G.J., Gold L.I. Localization of transforming growth factor β; isoforms in the normal murine small intestine and colon. Gastroenterology. 1993;105:67–73. doi: 10.1016/0016-5085(93)90011-z. [DOI] [PubMed] [Google Scholar]
- 24.Avery A., Paraskeva C., Hall P., Flanders K.C., Sporn M., Moorghen M. TGF-Beta expression in the human colon: differential immunostaining along crypt epithelium. Br. J. Cancer. 1993;68:137–139. doi: 10.1038/bjc.1993.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winesett M.P., Ramsey G.W., Barnard J.A. Type II TGFβ receptor expression in intestinal cell lines and in the intestinal tract. Carcinogenesis. 1996;17:989–995. doi: 10.1093/carcin/17.5.989. [DOI] [PubMed] [Google Scholar]
- 26.Zhao M., Mishra L., Deng C.-X. The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018;14:111–123. doi: 10.7150/ijbs.23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow E., Macrae F. A review of juvenile polyposis syndrome. J. Gastroenterol. Hepatol. 2005;20:1634–1640. doi: 10.1111/j.1440-1746.2005.03865.x. [DOI] [PubMed] [Google Scholar]
- 28.Pasche B., Wisinski K.B., Sadim M., Kaklamani V., Pennison M.J., Zeng Q., et al. Constitutively decreased TGFBR1 allelic expression is a common finding in colorectal cancer and is associated with three TGFBR1 SNPs. J. Exp. Clin. Cancer Res. 2010;29:57. doi: 10.1186/1756-9966-29-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Han W., Zborowska E., Liang J., Wang X., Willson J.K.V., et al. Reduced expression of transforming growth factor β type I receptor contributes to the malignancy of human colon carcinoma cells. J. Biol. Chem. 1996;271:17366–17371. doi: 10.1074/jbc.271.29.17366. [DOI] [PubMed] [Google Scholar]
- 30.Zeng Q., Phukan S., Xu Y., Sadim M., Rosman D.S., Pennison M., et al. Tgfbr1 haploinsufficiency is a potent modifier of colorectal cancer development. Cancer Res. 2009;69:678–686. doi: 10.1158/0008-5472.CAN-08-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsu A.H., Lum M.A., Shim K.-S., Frederick P.J., Morrison C.D., Chen B., et al. Crosstalk between PKCα and PI3K/AKT signaling is tumor suppressive in the endometrium. Cell Rep. 2018;24:655–669. doi: 10.1016/j.celrep.2018.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark J.A., Black A.R., Leontieva O.V., Frey M.R., Pysz M.A., Kunneva L., et al. Involvement of the ERK signaling cascade in protein kinase C-mediated cell cycle arrest in intestinal epithelial cells. J. Biol. Chem. 2004;279:9233–9247. doi: 10.1074/jbc.M312268200. [DOI] [PubMed] [Google Scholar]
- 33.Kaur N., Lum M.A., Lewis R.E., Black A.R., Black J.D. A novel antiproliferative PKCα-Ras-ERK signaling axis in intestinal epithelial cells. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.102121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao W., Pan Z., Du X., Zhang J., Li Q. miR-181b-induced SMAD7 downregulation controls granulosa cell apoptosis through TGF-β signaling by interacting with the TGFBR1 promoter. J. Cell Physiol. 2018;233:6807–6821. doi: 10.1002/jcp.26431. [DOI] [PubMed] [Google Scholar]
- 35.Cheng R., Dang R., Zhou Y., Ding M., Hua H. MicroRNA-98 inhibits TGF-β1-induced differentiation and collagen production of cardiac fibroblasts by targeting TGFBR1. Hum. Cell. 2017;30:192–200. doi: 10.1007/s13577-017-0163-0. [DOI] [PubMed] [Google Scholar]
- 36.Gellibert F., Woolven J., Fouchet M.H., Mathews N., Goodland H., Lovegrove V., et al. Identification of 1,5-naphthyridine derivatives as a novel series of potent and selective TGF-beta type I receptor inhibitors. J. Med. Chem. 2004;47:4494–4506. doi: 10.1021/jm0400247. [DOI] [PubMed] [Google Scholar]
- 37.Ichida J.K., Blanchard J., Lam K., Son E.Y., Chung J.E., Egli D., et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji C., Casinghino S., Chang D.J., Chen Y., Javed A., Ito Y., et al. CBFa(AML/PEBP2)-related elements in the TGF-β type I receptor promoter and expression with osteoblast differentiation. J. Cell Biochem. 1998;69:353–363. [PubMed] [Google Scholar]
- 40.Kim K.K., Ji C., Chang W., Wells R.G., Gundberg C.M., McCarthy T.L., et al. Repetitive exposure to TGF-β suppresses TGF-β type I receptor expression by differentiated osteoblasts. Gene. 2006;379:175–184. doi: 10.1016/j.gene.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Jonason J.H., Xiao G., Zhang M., Xing L., Chen D. Post-translational regulation of Runx2 in bone and cartilage. J. Dent Res. 2009;88:693–703. doi: 10.1177/0022034509341629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arumugam B., Vairamani M., Partridge N.C., Selvamurugan N. Characterization of Runx2 phosphorylation sites required for TGF-β1-mediated stimulation of matrix metalloproteinase-13 expression in osteoblastic cells. J. Cell Physiol. 2018;233:1082–1094. doi: 10.1002/jcp.25964. [DOI] [PubMed] [Google Scholar]
- 43.Ge C., Xiao G., Jiang D., Yang Q., Hatch N.E., Roca H., et al. Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. J. Biol. Chem. 2009;284:32533–32543. doi: 10.1074/jbc.M109.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge C., Yang Q., Zhao G., Yu H., Kirkwood K.L., Franceschi R.T. Interactions between extracellular signal-regulated kinase 1/2 and p38 MAP kinase pathways in the control of RUNX2 phosphorylation and transcriptional activity. J. Bone Miner Res. 2012;27:538–551. doi: 10.1002/jbmr.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayes T.K., Neel N.F., Hu C., Gautam P., Chenard M., Long B., et al. Long-term ERK inhibition in KRAS-mutant pancreatic cancer is associated with MYC degradation and senescence-like growth suppression. Cancer Cell. 2016;29:75–89. doi: 10.1016/j.ccell.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kinoshita E., Kinoshita-Kikuta E., Koike T. Phos-tag SDS-PAGE systems for phosphorylation profiling of proteins with a wide range of molecular masses under neutral pH conditions. Proteomics. 2012;12:192–202. doi: 10.1002/pmic.201100524. [DOI] [PubMed] [Google Scholar]
- 47.Ducy P., Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wrana J.L., Attisano L., Wieser R., Ventura F., Massague J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 49.Moses H.L. TGF-beta regulation of epithelial cell proliferation. Mol. Reprod. Dev. 1992;32:179–184. doi: 10.1002/mrd.1080320215. [DOI] [PubMed] [Google Scholar]
- 50.Barnard J.A., Beauchamp R.D., Coffey R.J., Moses H.L. Regulation of intestinal epithelial cell growth by transforming growth factor type beta. Proc. Natl. Acad. Sci. U. S. A. 1989;86:1578–1582. doi: 10.1073/pnas.86.5.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyd F.T., Massague J. Transforming growth factor-beta inhibition of epithelial cell proliferation linked to the expression of a 53-kDa membrane receptor. J. Biol. Chem. 1989;264:2272–2278. [PubMed] [Google Scholar]
- 52.Halder S.K., Beauchamp R.D., Datta P.K. A specific inhibitor of TGF-β receptor kinase, SB-431542, as a potent antitumor agent for human cancers. Neoplasia. 2005;7:509–521. doi: 10.1593/neo.04640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J., Yang L., Yang J., Kuropatwinski K., Wang W., Liu X.-Q., et al. Transforming growth factor β induces apoptosis through repressing the phosphoinositide 3-kinase/AKT/survivin pathway in colon cancer cells. Cancer Res. 2008;68:3152–3160. doi: 10.1158/0008-5472.CAN-07-5348. [DOI] [PubMed] [Google Scholar]
- 54.Zhao J., Buick R.N. Relationship of levels and kinetics of H-ras expression to transformed phenotype and loss of TGF-β1-mediated growth regulation in intestinal epithelial cells. Exp. Cell Res. 1993;204:82–87. doi: 10.1006/excr.1993.1011. [DOI] [PubMed] [Google Scholar]
- 55.Mulder K.M., Segarini P.R., Morris S.L., Ziman J.M., Choi H.G. Role of receptor complexes in resistance or sensitivity to growth inhibition by TGFβ in intestinal epithelial cell clones. J. Cell Physiol. 1993;154:162–174. doi: 10.1002/jcp.1041540120. [DOI] [PubMed] [Google Scholar]
- 56.Lindschau C., Quass P., Menne J., Güler F., Fiebeler A., Leitges M., et al. Glucose-induced TGF-β and TGF-β receptor-1 expression in vascular smooth muscle cells is mediated by protein kinase C-α. Hypertension. 2003;42:335–341. doi: 10.1161/01.HYP.0000087839.72582.DD. [DOI] [PubMed] [Google Scholar]
- 57.Valle L., Serena-Acedo T., Liyanarachchi S., Hampel H., Comeras I., Li Z., et al. Germline allele-specific expression of TGFBR1 confers an increased risk of colorectal cancer. Science. 2008;321:1361–1365. doi: 10.1126/science.1159397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Black A.R., Black J.D. The complexities of PKCα signaling in cancer. Adv. Biol. Reg. 2021;80 doi: 10.1016/j.jbior.2020.100769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xicola R.M., Bontu S., Doyle B.J., Rawson J., Garre P., Lee E., et al. Association of a let-7 miRNA binding region of TGFBR1 with hereditary mismatch repair proficient colorectal cancer (MSS HNPCC) Carcinogenesis. 2016;37:751–758. doi: 10.1093/carcin/bgw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Y., Pasche B. TGF-beta signaling alterations and susceptibility to colorectal cancer. Hum. Mol. Genet. 2007;1:R14–R20. doi: 10.1093/hmg/ddl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pasche B., Pennison M.J., Jimenez H., Wang M. TGFBR1 and cancer susceptibility. Trans. Am. Clin. Climatol Assoc. 2014;125:300–312. [PMC free article] [PubMed] [Google Scholar]
- 62.Bian Y., Knobloch T.J., Sadim M., Kaklamani V., Raji A., Yang G.-Y., et al. Somatic acquisition of TGFBR1∗6A by epithelial and stromal cells during head and neck and colon cancer development. Hum. Mol. Genet. 2007;16:3128–3135. doi: 10.1093/hmg/ddm274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kayed H., Jiang X., Keleg S., Jesnowski R., Giese T., Berger M.R., et al. Regulation and functional role of the Runt-related transcription factor-2 in pancreatic cancer. Br. J. Cancer. 2007;97:1106–1115. doi: 10.1038/sj.bjc.6603984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hojo H., Saito T., He X., Guo Q., Onodera S., Azuma T., et al. Runx2 regulates chromatin accessibility to direct the osteoblast program at neonatal stages. Cell Rep. 2022;40:111315. doi: 10.1016/j.celrep.2022.111315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu H., Whitfield T.W., Gordon J.A., Dobson J.R., Tai P.W., van Wijnen A.J., et al. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol. 2014;15:R52. doi: 10.1186/gb-2014-15-3-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drissi H., Luc Q., Shakoori R., Chuva De Sousa Lopes S., Choi J.Y., Terry A., et al. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell Physiol. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 67.Tou L., Quibria N., Alexander J.M. Transcriptional regulation of the human Runx2/Cbfa1 gene promoter by bone morphogenetic protein-7. Mol. Cell Endocrinol. 2003;205:121–129. doi: 10.1016/s0303-7207(03)00151-5. [DOI] [PubMed] [Google Scholar]
- 68.Zambotti A., Makhluf H., Shen J., Ducy P. Characterization of an osteoblast-specific enhancer element in the CBFA1 gene. J. Biol. Chem. 2002;277:41497–41506. doi: 10.1074/jbc.M204271200. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y., Hassan M.Q., Xie R.-L., Hawse J.R., Spelsberg T.C., Montecino M., et al. Co-Stimulation of the bone-related Runx2 P1 promoter in mesenchymal cells by SP1 and ETS transcription factors at polymorphic purine-rich DNA sequences (Y-repeats) J. Biol. Chem. 2009;284:3125–3135. doi: 10.1074/jbc.M807466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raaz U., Schellinger I.N., Chernogubova E., Warnecke C., Kayama Y., Penov K., et al. Transcription factor Runx2 promotes aortic fibrosis and stiffness in type 2 diabetes mellitus. Circ. Res. 2015;117:513–524. doi: 10.1161/CIRCRESAHA.115.306341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tamiya H., Ikeda T., Jeong J.H., Saito T., Yano F., Jung Y.K., et al. Analysis of the Runx2 promoter in osseous and non-osseous cells and identification of HIF2A as a potent transcription activator. Gene. 2008;416:53–60. doi: 10.1016/j.gene.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 72.Bruderer M., Richards R.G., Alini M., Stoddart M.J. Role and regulation of RUNX2 in osteogenesis. Eur. Cell Mater. 2014;28:269–286. doi: 10.22203/ecm.v028a19. [DOI] [PubMed] [Google Scholar]
- 73.Lund A.H., van Lohuizen M. Runx: a trilogy of cancer genes. Cancer Cell. 2002;1:213–215. doi: 10.1016/s1535-6108(02)00049-1. [DOI] [PubMed] [Google Scholar]
- 74.Miraoui H., Oudina K., Petite H., Tanimoto Y., Moriyama K., Marie P.J. Fibroblast growth factor receptor 2 promotes osteogenic differentiation in mesenchymal cells via ERK1/2 and protein kinase C signaling. J. Biol. Chem. 2009;284:4897–4904. doi: 10.1074/jbc.M805432200. [DOI] [PubMed] [Google Scholar]
- 75.Galea G.L., Meakin L.B., Williams C.M., Hulin-Curtis S.L., Lanyon L.E., Poole A.W., et al. Protein kinase Cα (PKCα) regulates bone architecture and osteoblast activity. J. Biol. Chem. 2014;289:25509–25522. doi: 10.1074/jbc.M114.580365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu S.P., Theodorescu D., Kerbel R.S., Willson J.K., Mulder K.M., Humphrey L.E., et al. TGF-beta 1 is an autocrine-negative growth regulator of human colon carcinoma FET cells in vivo as revealed by transfection of an antisense expression vector. J. Cell Biol. 1992;116:187–196. doi: 10.1083/jcb.116.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lum M.A., Barger C.J., Hsu A.H., Leontieva O.V., Black A.R., Black J.D. Protein kinase cα (PKCα) is resistant to long term desensitization/down-regulation by prolonged diacylglycerol stimulation. J. Biol. Chem. 2016;291:6331–6346. doi: 10.1074/jbc.M115.696211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Telford W.G., King L.E., Fraker P.J. Evaluation of glucocorticoid-induced DNA fragmentation in mouse thymocytes by flow cytometry. Cell Prolif. 1991;24:447–459. doi: 10.1111/j.1365-2184.1991.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 79.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. Gepia: a web server for cancer and normal gene expression profiling and interactive analyses. Nucl. Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chaikuad A., Tacconi E.M., Zimmer J., Liang Y., Gray N.S., Tarsounas M., et al. A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat. Chem. Biol. 2014;10:853–860. doi: 10.1038/nchembio.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in the manuscript are contained within the manuscript or in Supporting Information.