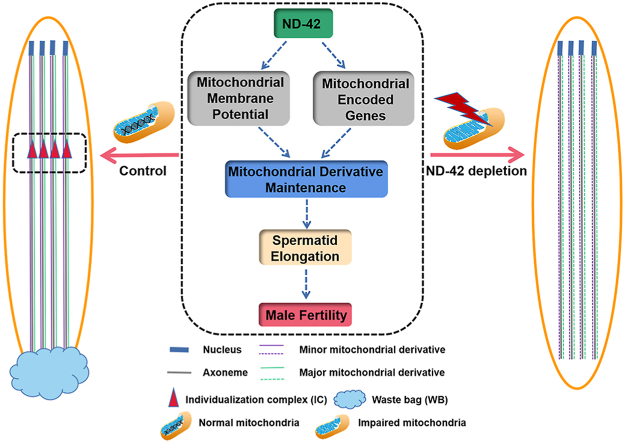
Abstract
During spermatogenesis, mitochondria extend along the whole length of spermatid tail and offer a structural platform for microtubule reorganization and synchronized spermatid individualization, that eventually helps to generate mature sperm in Drosophila. However, the regulatory mechanism of spermatid mitochondria during elongation remains largely unknown. Herein, we demonstrated that NADH dehydrogenase (ubiquinone) 42 kDa subunit (ND-42) was essential for male fertility and spermatid elongation in Drosophila. Moreover, ND-42 depletion led to mitochondrial disorders in Drosophila testes. Based on single-cell RNA-sequencing (scRNA-seq), we identified 15 distinct cell clusters, including several unanticipated transitional subpopulations or differentiative stages for testicular germ cell complexity in Drosophila testes. Enrichments of the transcriptional regulatory network in the late-stage cell populations revealed key roles of ND-42 in mitochondria and its related biological processes during spermatid elongation. Notably, we demonstrated that ND-42 depletion led to maintenance defects of the major mitochondrial derivative and the minor mitochondrial derivative by affecting mitochondrial membrane potential and mitochondrial-encoded genes. Our study proposes a novel regulatory mechanism of ND-42 for spermatid mitochondrial derivative maintenance, contributing to a better understanding of spermatid elongation.
Keywords: ND-42, Single-cell RNA-Sequencing, Spermatid elongation, Mitochondrial derivative, Male fertility
Graphical abstract
1. Introduction
Mature sperm are generated from post-meiotic germ cells through a series of ultrastructural and morphological changes, including the formation of flagellar axonemes, remodeling of mitochondria and nuclei, and polarization of elongating cysts [[1], [2], [3], [4]]. The microtubule-based flagellar axoneme comprises a central pair of microtubules and nine outer doublets, and over 100 proteins have been identified in the axoneme assembly [[4], [5], [6]]. Synchronized spermatid individualization starts after elongation and is essential for maintaining male fertility [7]. Spermatids form an initial individualization complex (IC) that travels caudally down within the cystic bulges (CBs), gathering the bulk of the cytoplasm into waste bags (WBs) at the base of the spermatid cyst [8,9].
In Drosophila testes, mitochondria are essential organelles that undergo dynamic changes in size and shape during spermatogenesis [10]. Mitochondria localize in the cytoplasm of spermatogonia and spermatocytes, and further migrate, aggregate, fuse, and form the Nebenkern near the nucleus of early round spermatids [11]. Spermatids contain two mitochondrial derivatives that elongate along the entire spermatid tails, which are closely related to the axoneme [12]. The major mitochondrial derivative accumulates paracrystalline material and the minor one reduces its size [12,13]. A recent study demonstrates that the sperm leucyl aminopeptidase (S-LAP) family comprises eight closely related proteins that are specifically expressed in testes and are identified as key components of the paracrystalline material in the major mitochondrial derivative [13]. Big bubble 8 (Bb8) is highly expressed in post-meiotic spermatids and is localized to mitochondria [12]. Testes lacking Bb8 lead to megamitochondria, and the paracrystalline material is abnormally distributed in both spermatid mitochondrial derivatives [12]. As described in Sawyer et al., knotted onions (knon) is required for the internal structure of the Nebenkern and elongated spermatid mitochondria [14]. Interestingly, the mitochondria-microtubule linker protein Milton (Milt) is enriched in mitochondria and is required for the sliding movement of microtubules, indicating that mitochondria function to support the microtubule-mediated IC structure [15]. Nevertheless, the exact molecular bases of spermatid mitochondria and individualization remain largely unknown, and further critical factors for mitochondrial derivative maintenance, elongation, and structural composition during spermatid elongation should be identified.
To further understand the regulatory mechanism of spermatid elongation, we identified nicotinamide adenine dinucleotide (NADH) dehydrogenase (ubiquinone) 42 kDa subunit (ND-42) as a key factor related to spermatid mitochondria maintenance. ND-42 is a subunit of the complex I protein in the mitochondrial electron transport chain [16,17]. Herein, we found that ND-42 depletion in spermatogonia resulted in male sterility, mitochondrial derivative maintenance defects, and disordered formation of CBs, WBs, and the IC during spermatid elongation. We performed single-cell RNA sequencing (scRNA-seq) analysis to provide new insights into ND-42-mediated transcriptional regulatory networks in various cell populations in Drosophila testes. We also showed that ND-42 depletion in testes influenced mitochondrial function by affecting mitochondrial membrane potential and mitochondrial-encoded genes. Taken together, our study revealed novel roles and mechanisms for ND-42 in the regulations of spermatid elongation.
2. Results
2.1. ND-42 is required for male fertility and spermatid elongation
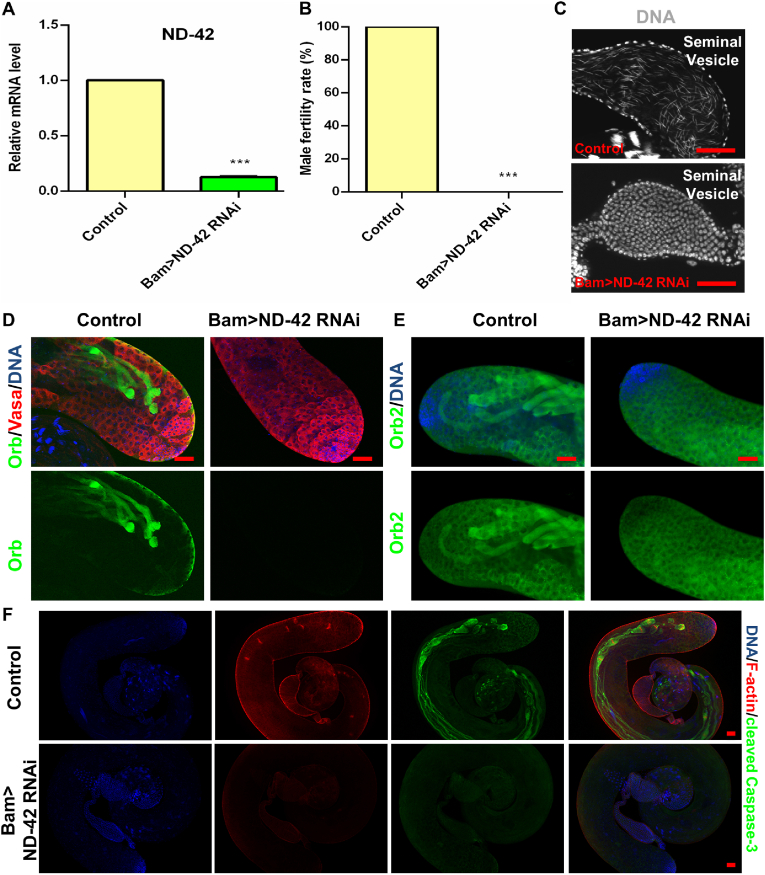
To investigate the mitochondrial function in Drosophila testes, we employed the Gal4/upstream activation sequence (UAS) system, which has been widely used in previous studies [[17], [18], [19], [20], [21]]. Herein, we disrupted the expression levels of several key subunits of mitochondrial complex I (CI) in spermatogonia driven by Bag of marbles (Bam)-Gal4 (Table S1), and their knockdown efficiencies were verified by quantitative real-time reverse transcription PCR (qRT-PCR) in testes (Fig. 1A and Fig. S1). Interestingly, male fertility tests showed that depletions of NADH dehydrogenase (ubiquinone) 23 kDa subunit (ND-23) (male fertility rate: 40%, n = 20), NADH dehydrogenase (ubiquinone) 75 kDa subunit (ND-75) (male fertility rate: 52.9%, n = 17) and mitochondrial NADH-ubiquinone oxidoreductase chain 1 (mt:ND1) (male fertility rate: 94.7%, n = 19) did not cause serious male fertility damage, while ND-42 depletion (male fertility rate: 0%, n = 20) caused male sterility when compared with control group (male fertility rate: 100%, n = 20) (Fig. 1B). Interestingly, ND-42 depletion led to the lack of mature sperm in seminal vesicles, indicating the critical role of ND-42 during testicular spermatogenesis (Fig. 1C). Therefore, we further explore the function of ND-42 during spermatid elongation. Immunofluorescence staining showed that the early-stage germ cells were not affected in Drosophila testes, despite the downregulation of ND-42 in spermatogonia driven by Bam-Gal4 (Fig. S2A). To further explore the function of ND-42, phase-contrast visualization of squashed adult testes demonstrated defects during spermatid elongation and no mature sperm were observed in ND-42 depletion testes when compared with the control group (Fig. S2B), suggesting that ND-42 was specifically required for spermatid elongation. Furthermore, we stained for elongated spermatid markers and found that compared with control testes, both oo18 RNA-binding protein (Orb) and Orb2 proteins were disrupted and not detected at the late stage of spermatogenesis in Bam > ND-42 RNA interference (RNAi) testes (Fig. 1D–E). We next investigated caspase activation along with IC staining, and found normal cleaved-Caspase-3 signals at the onset of spermatid individualization in control testes, while ND-42 depletion led to a lack of CBs, WBs, and the IC in the testes (Fig. 1F). Taken together, these data indicated that ND-42 plays critical roles during elongated spermatid development.
Fig. 1.
Reduction ofND-42in spermatogonia caused male infertility and spermatids elongation defects. (A) Relative ND-42 mRNA level in the control and Bam > ND-42 RNAi testes. Student's t-test was used to evaluate the differences. (B) Male fertility rate (%) in the control and Bam > ND-42 RNAi groups. The ratio was analyzed using the Chi-squared test. (C) DNA staining (Grey) of seminal vesicles. Needle-shaped sperm nuclei were shown in control testes, while no sperm was found in Bam > ND-42 RNAi testes. (D) Vasa (red) and Orb (green) immunostaining at the apex of the control and Bam > ND-42 RNAi testes. (E) Orb2 (green) immunostaining of at the apex of control and Bam > ND-42 RNAi testes. (F) F-actin (red) and cleaved Caspase-3 (green) immunostaining at the apex of the control and Bam > ND-42 RNAi testes. Hoechst was used to stain DNA. ***P < 0.001, scale bar: 50 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.2. ND-42 regulates mitochondrial function in Drosophila testes
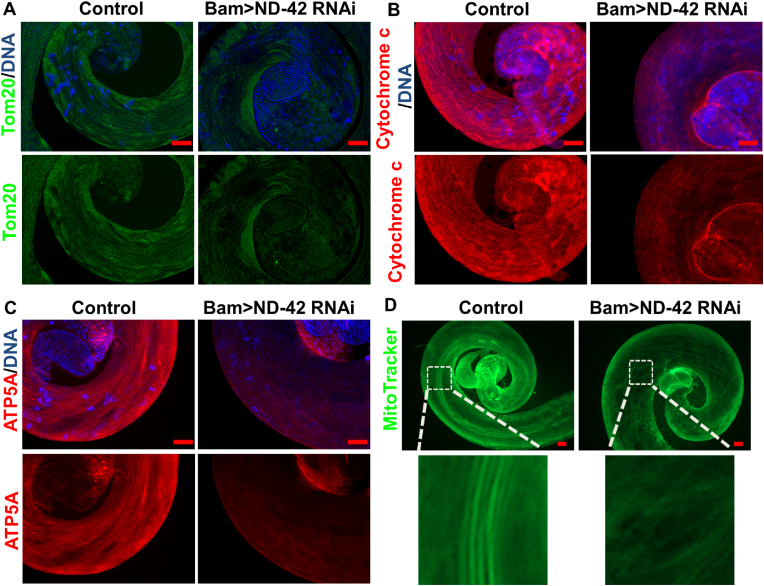
Since ND-42 was a key factor for the mitochondrial electron transport chain, we next validated whether ND-42 was required for mitochondrial function during spermatid elongation. First, we used immunostaining to assess spermatid mitochondria using three independent mitochondria markers: Translocase of outer membrane 20 (Tom20), Cytochrome C, and Bellwether (Blw, also called ‘ATP5A’). We observed the Tom20, Cytochrome C, and ATP5A signals in spermatids mitochondria in control testes and ND-42 depletion testes. In the elongated cysts of the ND-42 group, mitochondrial defects were observed (Fig. 2A–C). MitoTracker staining of testicular cauda showed that the spermatid mitochondria were seriously damaged in ND-42 depletion testes (Fig. 2D), revealing the critical role of ND-42 for the mitochondria functionality.
Fig. 2.
Reduction ofND-42 in spermatogonia affects mitochondrion function in Drosophila testes. (A) Tom20 immunostaining (green) at the tail of the control and Bam > ND-42 RNAi testes. (B) Cytochrome c immunostaining (red) at the tail of the control and Bam > ND-42 RNAi testes. (C) ATP5A immunostaining (red) at the tail of the control and Bam > ND-42 RNAi testes. (D) MitoTracker staining (green) of the tail of the control and Bam > ND-42 RNAi testes. Hoechst staining of DNA appears blue. Scale bar: 50 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.3. scRNA-seq analysis reveals comprehensive cell populations in Drosophila testes
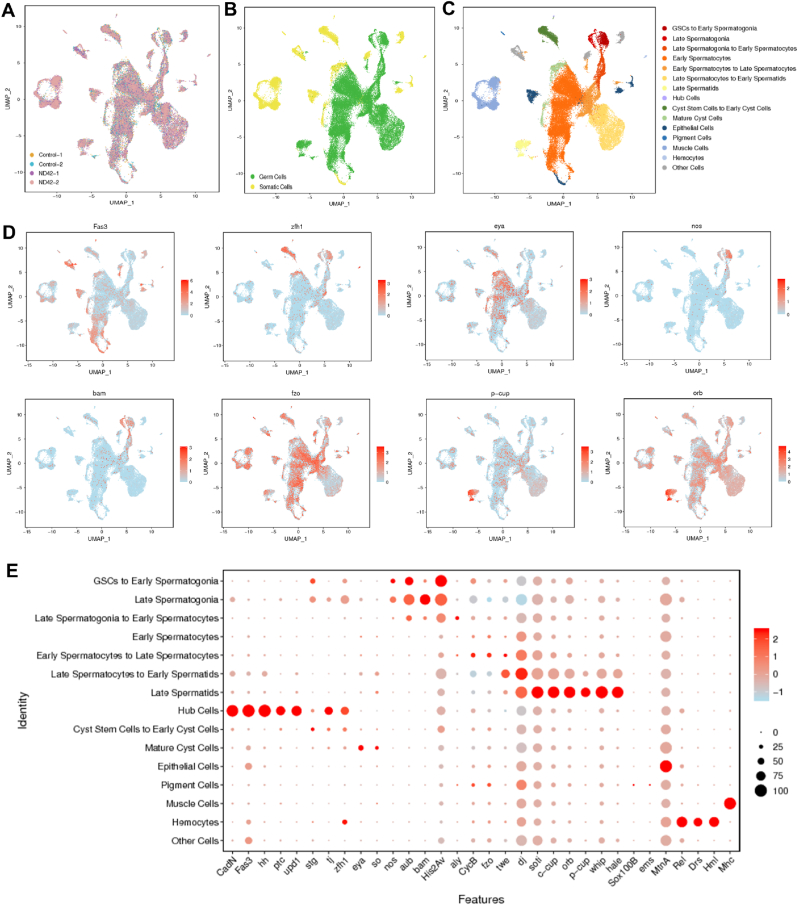
We next profiled cells and performed scRNA-seq by freshly dissecting testes in control and ND-42 groups. An average of 32986 mean reads were mapped per cell and the expression of approximately 15621 genes was detected per cell. After strict filtration, 43162 high-quality cells were used for subsequent bioinformatic analysis. The dimensionality of the gene and/or cell expression matrix was reduced to two primary axes and cells were grouped according to the similarity of their unique gene expression profiles using uniform manifold approximation and projection (UMAP) in Seurat. Moreover, we performed two dissection repeats for each group, and the major cell clusters were independent of batch effects and technical variables, while differences between the two groups were preserved (Fig. 3A and Fig. S3).
Fig. 3.
Clustering and classification of testicular cell types at single-cell resolutions inDrosophila. (A–B) UMAP visualization of testicular cells. (A) Cells are colored for the control and ND-42 groups. (B) Cells are colored for somatic cells and germ cells. (C) UMAP visualization of 15 cell clusters identified in Drosophila testes. (D) UMAP visualization of representative marker genes. (E) Selected marker genes visualized using a dot plot. The diameter of the dot corresponds to the fraction of cells expressing each gene in each cluster (see the scale). Color intensity is representative of the average normalized expression level. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
By grouping similar cells into clusters, UMAP visualized germ cells and somatic cells, and further identified 15 distinct cell populations, covering all expected cell types in Drosophila testes (Fig. 3B–C). The clustering results inferred that germ cells (germline stem cells (GSCs), spermatogonia, spermatocytes, spermatids, and their transition stages), hub cells, cyst stem cells, early cyst cells, mature cyst cells, epithelial cells, pigment cells, muscle cells, and hemocytes were present (Fig. 3C). And the average expression levels of detected genes in each cell cluster were shown in Table S2.
In the UMAP map, clustered cells from each developmental stage were close to each other, which suggested that the progression of cells through spermatogenesis is a continuous process. Recently reported and classical markers were used to identify likely cell types for each cluster (Fig. 3D) [22,23]. Some of these genes have been widely used as markers for specific cell clusters in Drosophila. For instance, Cadherin-N (CadN), Fasciclin 3 (Fas3), hedgehog (hh), and unpaired (upd) are known to have critical roles in hub cells [[24], [25], [26], [27], [28]]. Moreover, the nanos (nos) gene has been used as a GSC marker and is mainly expressed at the early stage of germ cells [29]. Interestingly, the bam gene encodes the key differentiation factor, bag of marbles, and was specifically expressed during spermatogonia transit-amplifying divisions (TA-divisions), which are essential for the switch from proliferation to meiotic differentiation [30]. Cyclin B (CycB) is biased toward mature spermatocytes [31]. Meanwhile, fuzzy onions (fzo) mRNA accumulated in spermatocytes and early spermatids [31,32], while the cup (c-cup and p-cup), dj, and orb genes were highly expressed in spermatids [33,34]. Importantly, several cell populations clustered together (e.g., GSCs to early spermatogonia) probably because of their highly similar transcriptomes, their comparatively low cell numbers in the tissue, and their infrequent expression of cell-specific marker genes [23]. To accurately annotate the cell clusters, the expression patterns of multiple marker genes were confirmed (Fig. 3E and Table S3). Taken together with the above data, we characterized the testicular cell populations and transcriptomic features during spermatogenesis and under the state of ND-42 depletion in Drosophila testes.
2.4. Features of cell subpopulations in Drosophila testes
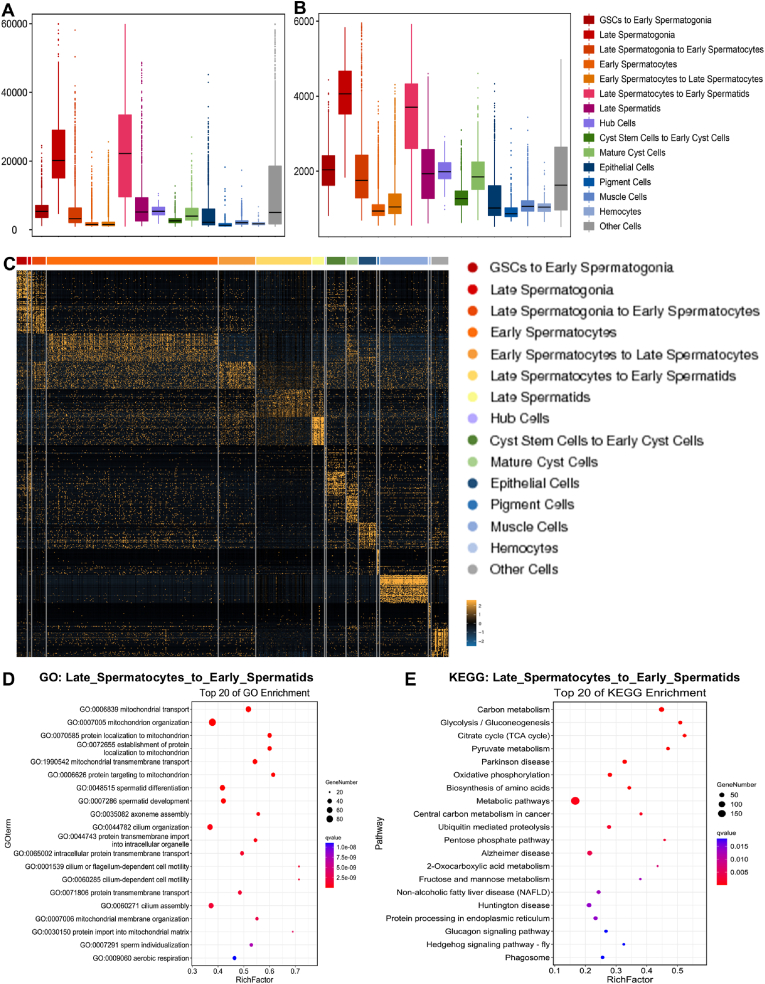
To quantify RNA contents and gene expression in various cell clusters, we counted the number of Unique Molecular Indices (nUMIs) and the number of genes (nGenes), which revealed that clusters Late_spermatogonia and Late_Spermatocytes_to_Early_Spermatids expressed the most UMIs and genes (Fig. 4A–B). We also identified the top 50 most abundantly expressed genes in cell clusters from each cell type, the results of which were consistent with previously identified marker genes (Fig. 4C).
Fig. 4.
Characteristics of gene expression and enrichment in sub-cell clusters ofDrosophilatestes. (A) Quality control of RNA contents for testicular cell clusters. (B) The number of genes identified in testicular cell clusters. (C) Heatmap of the top 50 most abundantly expressed genes in the testicular cell clusters. (D) Top 20 significantly enriched biological processes of GO analysis for the most enriched genes in the Late_Spermatocytes_to_Early_Spermatids cluster. (E) Top 20 of KEGG pathway enrichments for the most enriched genes in the Late_Spermatocytes_to_Early_Spermatids cluster.
To further investigate the transcriptional activation in various cell types, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichments for the most abundantly expressed genes in each specific cell type. Specifically, mitochondrion-mediated biological processes (e.g., mitochondrial transport, oxidative phosphorylation) and spermatogenesis-related processes (e.g., spermatid individualization, spermatid development and differentiation) were highly enriched in the Late_Spermatocytes_to_Early_Spermatids cluster (Fig. 4D–E). These data encompassed the comprehensive genetic module repertoire for the ND-42-mediated mitochondrial function in Drosophila testes.
2.5. Novel insights into ND-42 mediated transcriptional regulation based on scRNA-seq in Drosophila testes
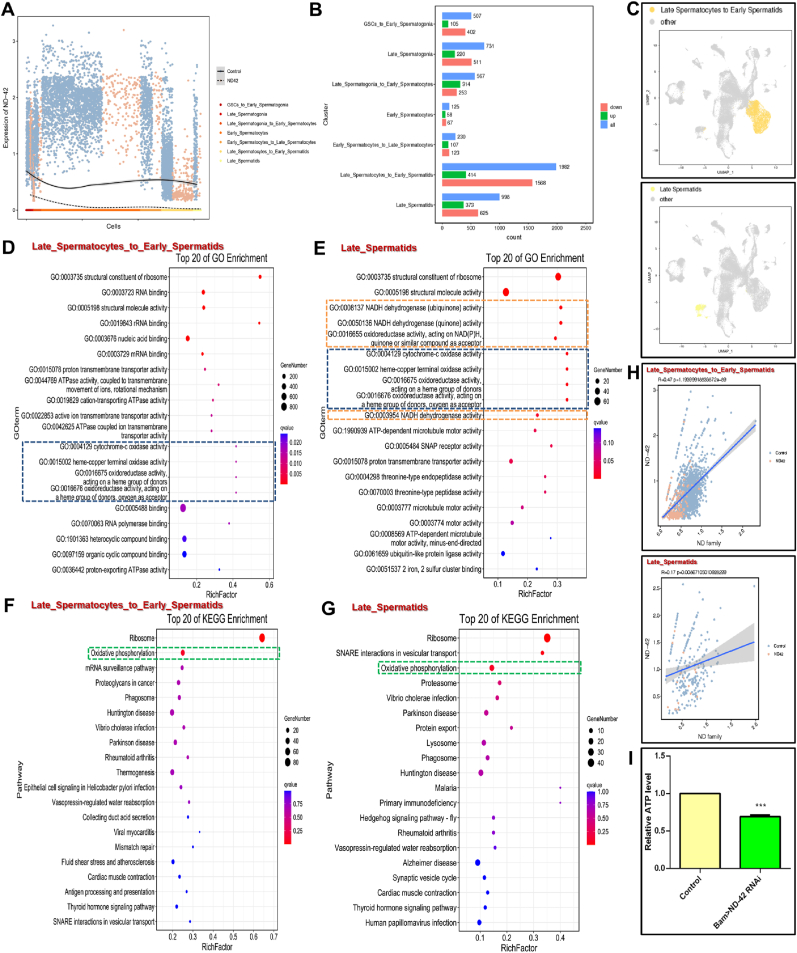
We next investigated the ND-42 expression patterns in germ cell clusters, and found that compared with the control group, ND-42 was dramatically suppressed in multiple stages, especially in the spermatid stage, of spermatogenesis in the ND-42 group (Fig. 5A). To investigate the transcriptional regulation of ND-42 at single-cell resolution, we analyzed differentially expressed genes (DEGs) and their enrichments from each cell cluster, which identified many of DEGs in the Late_Spermatocytes_to_Early_Spermatids cluster (414 upregulated DEGs and 1568 downregulated DEGs) and the Late_Spermatids cluster (373 upregulated DEGs and 625 downregulated DEGs) (Fig. 5B). Moreover, the heterogeneities in these two cell clusters were highlighted by UMAP (Fig. 5C). GO enrichment analysis also showed that mitochondria-related biological events, such as oxidase activity and NADH dehydrogenase activity, were highly enriched at the late stage of spermatogenesis (Fig. 5D–E). Furthermore, KEGG pathway analysis revealed that oxidative phosphorylation played key roles in both the Late_Spermatocytes_to_Early_Spermatids and Late_Spermatids clusters (Fig. 5F–G). We also noticed that several DEGs relevant to the NADH dehydrogenase signature (also named the ‘ND family’) displayed high correlation coefficients with ND-42 in the Late_Spermatocytes_to_Early_Spermatids cluster and the Late_Spermatids cluster, but were dramatically decreased in ND-42 depletion testes (Fig. 5H), suggesting a high potential for maintaining elongated spermatid development via mitochondrial function. We next examined whether ND-42 induced defects in elongated spermatid mitochondria could affect mitochondria-mediated metabolic processes, and found that the relative ATP level was downregulated after disrupting ND-42 in testes (Fig. 5I). Taken together, the scRNA-seq-determined transcriptional regulatory network in each cell cluster identified key roles of ND-42 in mitochondria and its related biological processes during spermatid elongation.
Fig. 5.
Analysis of ND-42 mediated DEGs and their enrichments in cell clusters ofDrosophilatestes. (A) Fitted expression curve of ND-42 in germline cell clusters of the control and ND-42 groups. (B) The DEGs count between the control and ND-42 groups in germline cell clusters during spermatogenesis. (C) UMAP visualization of testicular cells. Cells are colored for the Late_Spermatocytes_to_Early_Spermatids and Late_Spermatids clusters. (D and E) Top 20 GO enrichments for DEGs between the control and ND-42 groups in the Late_Spermatocytes_to_Early_Spermatids (D) and Late_Spermatids (E) clusters. (F and G) Top 20 of KEGG pathway enrichments for the dissimilarity between the control and ND-42 groups in the Late_Spermatocytes_to_Early_Spermatids (F) and Late_Spermatids (G) clusters. (H) The correlations between ND-42 and the ND family in the Late_Spermatocytes_to_Early_Spermatids and Late_Spermatids clusters. (I) The relative ATP level in control and Bam > ND-42 RNAi testes. ***P < 0.001.
2.6. Identification of novel stages in testicular germ cell complexity
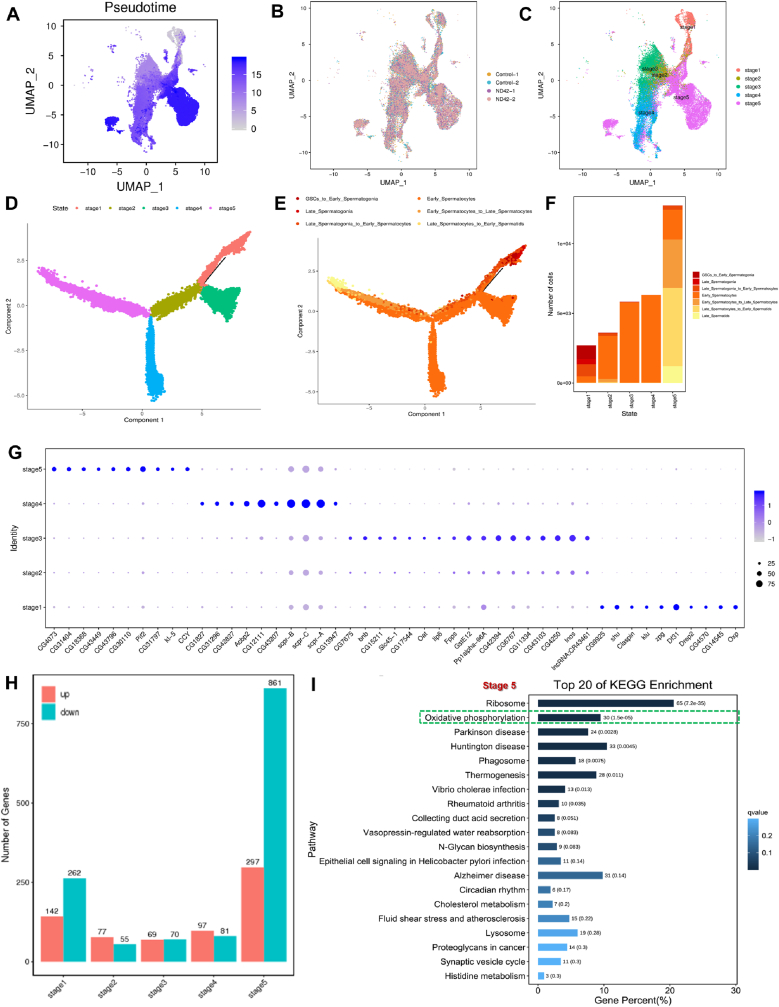
To further investigate the origin and maturation of germ cells, germline subpopulations were re-clustered and pseudotime trajectory analysis was performed based on clustering for the differentiative stages of spermatogenesis (Fig. 6A–B). We identified five novel stages in testicular germ cell development, most of which were consistent with the differentiation trends during spermatogenesis (Fig. 6C–D and Fig. S4A). Cellular component analysis revealed that the early-stage germ cells were mainly clustered in Stage 1 and the late stage of germ cells trended to Stage 5 (Fig. 6E–F and Figs. S4B and S4C). A heatmap of the most highly expressed genes exhibited the overall expression distributions (Fig. S5A), and a DotPlot view highlighted several enriched genes identified in each stage during spermatogenesis (Fig. 6G).
Fig. 6.
Pseudotime trajectory analysis reveals five novel stages during spermatogenesis. (A to C) UMAP visualization of pseudotime trajectory analysis. Cells were colored based on pseudotime (A), sample (B), and stage (C). (D and E) The single-cell trajectory containing five stages. Cells were colored based on their state (D) and cell cluster (E). (F) Cellular component analysis in each stage during spermatogenesis. (G) DotPlot view of selected upregulated enriched genes in each stage during spermatogenesis. (H) The DEGs count between the control and ND-42 groups in each stage during spermatogenesis. (I) Top 20 KEGG pathway enrichments for DEGs between the control and ND-42 groups in Stage 5.
To further explore the molecular regulation among the stage of spermatogenesis, we identified a series of DEGs in different stages, most of which are identified in Stage 5 between the control and ND-42 groups (Fig. 6H). Next, we performed KEGG analysis for Stage 5 germ cells, and identified DEGs in Stage 5 that were highly enriched in oxidative phosphorylation (Fig. 6I). Of special concern, male fertility factor kl5 (Kl-5) has been reported to be involved in flagellated sperm motility and kl-5 mutants failed to assemble the outer dynein arms during spermatid elongation in Drosophila [35,36]. Consistent with our data, kl-5 was highly enriched in Stage 5 and could be used as a key marker for Stage 5 germ cells (Figs. S5B–S5D).
2.7. ND-42 controls mitochondrial derivative maintenance via the regulations of mitochondrial membrane potential and mitochondrial-encoded genes
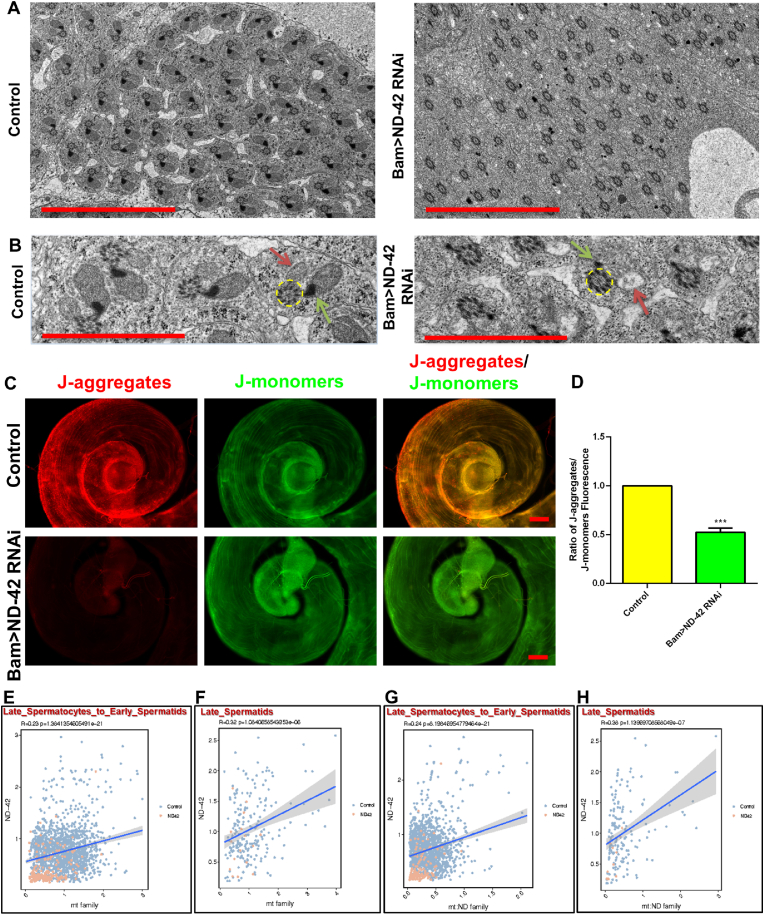
Transmission electron microscopy (TEM) analysis was then performed to further determine how ND-42 depletion affected elongated spermatid subcellular structure. TEM views of ND-42 depletion testes showed that both the major mitochondrial derivative and the minor mitochondrial derivative were destroyed, while the axoneme structure was not affected (Fig. 7A and B). In particular, the major mitochondrial derivative was seriously damaged, with very little condensed paracrystalline material remaining, and the minor mitochondrial derivative was totally lost, leaving only bubble-like structures. These results suggested that ND-42 was essential to maintain the mitochondrial derivatives, but was dispensable for the sperm axoneme assembly in Drosophila testes.
Fig. 7.
Reduction ofND-42 in spermatogonia leads to mitochondrial derivatives defects during elongated spermatids differentiation. (A) Electron microscopy analysis of elongated spermatids in control and Bam > ND-42 RNAi testes. (B) Enlarged views of (A). Representative ultra microstructures are shown as follows: axoneme (yellow circles); major mitochondrial derivatives (green arrows); minor mitochondrial derivatives (red arrows). (C) JC-1 staining of the tail of the control and Bam > ND-42 RNAi testes. (D) Ratio of the fluorescence of J-aggregates to J-monomers. (E and F) Correlations between ND-42 and the mt family in the Late_Spermatocytes_to_Early_Spermatids and Late_Spermatids clusters. (G and H) Correlations between ND-42 and the mt: ND family in the Late_Spermatocytes_to_Early_Spermatids and Late_Spermatids clusters. ***P < 0.001, scale bar: 5 μm for A; 2 μm for B; 50 μm for C. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To further test the functionality of the mitochondria in spermatids, the membrane potential sensitive membrane permeable dye, JC-1, was used to stain mitochondria. JC-1 accumulates in mitochondria in a membrane potential-dependent manner, showing an emission shift from green to red in mitochondria with a membrane potential [12]. There was a marked decrease in the ratio of JC-1 aggregates/JC-1 monomers in the ND-42 depletion testes (Fig. 7C and D).
To further explore the molecular regulations of mitochondrial dysfunctions in elongated spermatids, we further analyzed the relationships between ND-42 and the mitochondrial-encoded genes using scRNA-seq in the late-stage cell populations. We then collected key factors encoded by mitochondria, designated as the mitochondrial genome signature (also named ‘mt: family’) and the mitochondrial NADH-ubiquinone oxidoreductase chain signature (also named ‘mt: ND family’), and used corresponding DEGs in each cell cluster to assess their correlation with ND-42. ND-42 correlated positively with the mt: family and mt: ND family signatures in both Late_Spermatocytes_to_Early_Spermatids and Late_Spermatids clusters (Fig. 7E to H). Collectively, the above results indicated that ND-42 depletion led to defects in mitochondrial derivative maintenance by affecting mitochondrial membrane potential and mitochondrial-encoded genes in elongated spermatids.
3. Discussion
Mitochondria function in energy production, metabolism, cell apoptosis, and differentiation through multiple processes, e.g., oxidative phosphorylation, the tricarboxylic acid cycle, the mitochondrial electron transport chain, and ATP synthase [14,[37], [38], [39]]. It has been shown that mitochondrial defects are associated with an increasingly large proportion of mitochondrial-linked diseases [40]. Previous study showed that the testis-specific gene Cytochrome c oxidase subunit 4-like (COX4L) was a subunit of cytochrome oxidase, functioning in cytochrome c oxidation, oxygen reduction and proton pumping in the testis [41]. COX4L replaced Cytochrome c oxidase subunit 4 (COX4) in the sperm, and loss-of-function of COX4L resulted in male infertility and spermatid mitochondria defects [41,42], providing a plausible explanation for the mitochondrial dysfunction mediated male sterility in Drosophila testes.
Mitochondrial CI is the first and largest electron transport chain complex in mitochondrion, which oxidizes NADH, reduces ubiquinone, and transports protons across the inner membrane [43,44]. In this study, we tested several key subunits of CI in the mitochondrial electron transport chain, and found that depletions of ND-23 and ND-75 led to subfertility injuries and ND-42 depletion caused complete male infertility, indicating key roles of mitochondrial CI in Drosophila testes. ND-23 and ND-75 belong to the nuclear-encoded subunits and ND-42 is found to be a key accessory subunit of mitochondrial CI [45]. High-resolution structures show that many accessory subunits are wrapped around the core subunits and form extended structures containing α-helices, coils and disulfide bonds, which play critical roles for stabilizing the mitochondrial CI [[46], [47], [48]]. Moreover, some accessory subunits exhibit special functions via their protein domains or relevant co-factors [45]. Previous study showed that severe impairment of CI with lengthened youth (Sicily) could interact with cytosolic Heat shock protein 83 (Hsp83, also known as Hsp90) to chaperone the CI subunit ND-42 in the cytoplasm, revealing the roles of Sicily by escorting ND-42 in the cytoplasm prior to its import into the mitochondrion [49]. Garcia CJ et al. also found that NADH dehydrogenase (ubiquinone) 15 kDa subunit (ND-15) was necessary to stabilize or promote incorporation of ND-42 into the mitochondrion CI, indicating that many accessory subunits regulated CI biogenesis [50]. Although testicular co-regulatory factors of ND-42, ND-23 and ND-75 remain largely unknown, we speculate that these proteins may interact with many different co-factors to participate in the regulation of male fertility, and depletions of these genes can lead to male fertility related phenotypes with diverse severities.
As technology develops, scRNA-seq has been widely used in all aspects of research, which enables the construction of single-cell transcriptomic atlases of selected tissues in the reproductive system [[51], [52], [53], [54]]. In Drosophila, Witt et al., produced a single-cell atlas of the transcriptional patterns across cell clusters in adult testes [23]. Based on the single-cell expression patterns, they further revealed that X-chromosome dosage compensation occurred in somatic and pre-meiotic germ cells [55]. Meanwhile, several studies also generated single-cell atlases of differentiating egg chambers and the stem cell compartment, identifying novel markers and transcriptional signatures in adult Drosophila ovaries [[56], [57], [58]]. A recent study provided a common platform (the FlyCell Atlas; https://flycellatlas.org/), which integrates a map of a single-nucleus transcriptome of an entire adult Drosophila, constituting a valuable reference for further gene function study [22].
A logical question is whether specific gene-mediated transcriptional regulation could be investigated at the single-cell resolution in Drosophila testes. In this respect, the present study confirmed the power of single-cell analysis for the assessment of ND-42 mediated specific cell-cluster effects. Among the cell clusters in Drosophila testes, we revealed unanticipated cell type complexity that represents the transitional states of two adjacent differentiated cell types, e.g. cluster Late_Spermatogonia_to_Early_Spermatocytes and cluster Late_Spermatocytes_to_Early_Spermatids. More importantly, recent studies based on scRNA-seq data of human testes found various functions in different states of specific cell types [52,59]. In our study, pseudotime trajectory analysis revealed five novel stages of germ cells, which helped to further reveal as-yet-unknown regulatory signals of spermatogenesis.
Several key events are involved in elongated spermatid differentiation, including the removal of bulk cytoplasm and spermatid individualization [60]. CBs and WBs attached themselves to the IC, which could be detected by phalloidin and bound to actin in elongated spermatids [61]. During the individualization process, nucleoids and cytoplasmic remnants are squeezed to the tail of the elongated spermatids to form WBs for elimination [62]. It has been shown that a caspase-dependent process is activated throughout the whole of the elongated spermatids that exerts nonapoptotic functions for spermatid individualization that result in the formation of individual sperm [4,63]. In this study, we found that ND-42 depletion led to defects in mitochondrial derivative maintenance during elongated spermatid development, ultimately causing male sterility in Drosophila. Surprisingly, we also noticed that cleaved-Caspase-3 was inactivated in the tail of elongated spermatids in ND-42 depletion testes, revealing a novel mechanism for elongated spermatids differentiation via a cleaved-Caspase-3 independent signature.
A previous study suggested that mitochondrial DNA (mtDNA) was eliminated during spermatogenesis in Drosophila, ensuring that mature spermatozoa are devoid of DNA [64]. It has also been well recognized that the elimination of mtDNA was activated by apoptosis-related factors in mitochondria, and mtDNA mutations might result in an unstable mitochondrial genome and male infertility [65]. In this study, we found that disturbing ND-42 also resulted in the downregulation of multiple mitochondrial-encoded genes, e.g., those encoding mitochondrial NADH-ubiquinone oxidoreductase chain subunits, mitochondrial Cytochrome b and Cytochrome c oxidase subunits, which helps to clarify the regulatory mechanisms for spermatid mitochondria-mediated maintenance of male fertility.
By analyzing scRNA-seq data, ND-42 was observed to gradually decrease during spermatogenesis, and reached the lowest level in the Late_Spermatocytes_to_Early_Spermatids and Late_Spermatids clusters after disturbing ND-42 driven by Bam-Gal4 in Drosophila testes. Although we manipulated the levels of ND-42 during spermatogonia TA-divisions, ND-42 specifically caused differentiation defects of elongated spermatids but did not affect the differentiation process of pre-meiotic and meiotic germ cells. A recent study identified that the RNA binding protein, Maca, was specifically expressed in spermatocyte nuclei, while maca mutant males were male-sterile by influencing spermatid individualization formation [66], revealing the wide existence of phenotypic delays in Drosophila testes.
This study provided comprehensive landscapes of transcriptional network through the scRNA-seq approach. We systematically investigated ND-42-mediated mitochondrial function, identifying a novel regulatory mechanism for the differentiation of spermatid elongation in Drosophila, which provided a novel theoretical basis for the clinical diagnosis and treatment of male infertility.
4. Materials and methods
4.1. Fly stocks and crosses
Standard cornmeal molasses agar medium was used to culture all flies, which were incubated at 25 °C with 40–60% relative humidity. The TsingHua Fly Center (THFC, Beijing, China) provided UAS-ND-42 RNAi (#THU3600), UAS-ND-75 RNAi (#TH201500616.S), UAS-ND-23 RNAi (#THU0519), UAS-mt:ND1 RNAi (#TH04748.N) transgenic flies. DH Chen (Institute of zoology, Chinese Academy of Sciences, Beijing, China) kindly provided the Bam-Gal4; △86/ + line.
Male Bam-Gal4; △86/ + flies were crossed with transgenic UAS-RNAi virgins females and incubated at 25 °C until hatching. F1 males with the specific genotype were selected for further analysis. The w1118 line was used as the control. Fertility tests in males were carried out using a single F1 RNAi adult male that was enclosed for 3 days in a cross with three w1118 virgin females, carried out at room temperature.
4.1.1. Phase-contrast microscopy
Phase-contrast microscopy analysis was carried out as described previously [67]. Briefly, testes from the flies were dissected in 1 × phosphate-buffered saline (PBS), followed by several washes. Squashed testes on slides were coverslipped and observed under a phase-contrast microscope.
4.1.2. Confocal imaging and immunostaining
Testes from flies were dissected in 1 × PBS, fixed in 4% paraformaldehyde (PFA) for 30 min, washed three times using 1% PBS-Triton X-100 (PBST), and incubated for 30 min in 5% bovine serum albumin (BSA). The testes were then incubated with primary antibodies (see Table S4 for details) diluted in 5% BSA for 1 h at 25 °C, and washed thrice using 1% PBST. Secondary antibodies conjugated with Cy3 or A647 (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) were diluted with 5% BSA at 1:400, and incubated with the testes samples for 1 h at 25 °C in the dark. Following three washes with 1% PBST, the testes were stained for 5 min using Hoechst 33342 (1:800, C0031, Solarbio, Beijing, China) before finalizing.
The structure of IC in testes was stained using Alexa Fluor™ Plus555 Phalloidin (1:50; A30106, Invitrogen, Waltham, MA, USA). MitoTracker™ Green FM (M7514, Thermo Fisher Scientific, Waltham, MA, USA) was used to label the Mitochondria. JC-1 (C2006, Beyotime, Shanghai, China) was used as a mitochondrial membrane potential marker. Briefly, fly testes were dissected in 1 × PBS and incubated with related working solutions according to the manufacturer's instructions. Testes were then washed three times with 1 × PBS. Images were captured on Zeiss confocal microscope (Carl Zeiss, Oberkochen, Germany) and processed using Adobe Photoshop CS5 software (Adobe, San Jose, CA, USA).
4.1.3. Single-cell suspensions, library preparation and sequencing
Male flies at 2–3 days old were used to profile cells from four independent samples (two control samples vs. two ND-42 samples). Testes (a mixture of 250 testes per sample) were freshly dissected in cold PBS. The cell number and viability of single-cell suspensions were assessed using Trypan Blue staining. A Chromium instrument (10X Genomics, Pleasanton, CA, USA) was used to carry out the single-cell RNA-sequencing. A Chromium Single Cell 3’ Library & Gel Bead Kit v3 (10X Genomics) was used to process the samples. Gene Denovo Biotechnology Co., Ltd (Guangzhou, China) then sequenced the gene expression libraries using a NovaSeq 6000 sequencer (Illumina, San Diego, CA, USA).
4.1.4. scRNA-seq data processing
The raw BCL files were converted to FASTQ files, aligned and the counts quantified using Cell Ranger software (version 3.1.0; 10X Genomics). Reads with low-quality barcodes were discarded, and UMIs were filtered out and mapped to the reference genome. Those reads that mapped uniquely to the transcriptome and intersected an exon by at least 50% were considered for UMI counting. Prior to quantification, the UMI sequences were corrected for sequencing errors, and the EmptyDrops method was used to identify valid barcodes [68]. Cell barcode calling and UMI counting produced the cell by gene matrices. For the downstream analysis of each sample, the cell-by-gene matrices were imported individually into Seurat version 3.1.1 [69]. Those cells having an unusually high number of UMIs (≥60,000) or mitochondrial gene percent (≥20%) were discarded. We also excluded cells with less than 510 or more than 6000 detected genes. Additionally, doublet gel beads in emulsion (GEMs) were also filtered out using DoubletFinder (v2.0.3) [70]. Uniform manifold approximation and projection (UMAP) and t-distributed Stochastic Neighbor Embedding (t-SNE) were generated to visualize the cell clusters. Upregulated enriched genes were determined as significant using the threshold standard of fold change more than 1.28 and a P value lower than 0.01. DEGs were considered significant with a fold change more than 1.50 and the P value lower than 0.05.
4.1.5. Correlation analysis
Correlation analysis was carried out as described previously [53]. The average expression counts belonging to each set of features were used to calculate gene sets, pathways, or signatures via the PercentageFeatureSet function in Seurat. Then, the correlations among these gene sets or signatures were calculated using Pearson correlation, performed directly on the data matrix. Cells with extreme values (expression count = 0) were discarded from the analysis due to their low expression background.
4.1.6. Cell trajectory analysis
Single-cell trajectory was analyzed using a matrix of cells and gene expressions by Monocle (Version2.10.1) [71]. Monocle was used to reduce the space down to one with two dimensions and to order the cells [72]. Using the ordered cells, their trajectory could be visualized in the reduced dimensional space.
4.1.7. Transmission electron microscope (TEM) analysis
Fly testes were dissected and subjected to TEM analysis according to a previously described method [9] using an FEI Tecnai Spirit Biotwin transmission electron microscope (Thermo Fisher Scientific) operated at 100 kV.
4.1.8. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from testes by using the TRIzol Reagent (15596026, Invitrogen, Waltham, MA, USA). First strand cDNA was reverse transcribed from the RNA using a PrimeScript™ II 1st Strand cDNA Synthesis Kit (6210A, Takara). The qPCR step of the qRT-PCR protocol was carried out in a LightCycler® 96 Real-Time PCR System (Roche, Basel, Switzerland) using TB Green Premix Ex TaqII (RR820, Takara). Primer sequences used in this study were shown in Table S5.
4.1.9. ATP measurement
The ATP level was performed using an ATP assay (S0026, Beyotime, Shanghai, China). Briefly, fly testes were dissected in 1 × PBS and 20 testes per sample were added to 200 μL of lysis buffer. After full pyrolysis, the sample was centrifuged at 12,000×g for 5 min at 4 °C, and the supernatant was retained. 100 μL of ATP detection working solution was added to the detection well and incubated for 5 min at room temperature. Then, 20 μL of each sample was added to the detection well and mixed quickly. The relative ATP level was detected using a microplate reader (BioTek Instruments Inc., Friedrichshall, Germany).
4.1.10. Statistical analysis
The mean ± standard error of mean (SEM) was used to present the experimental data. Student's t-test and one-way ANOVA were used to evaluate the differences between the data using GraphPad Prism software Version 6.01 (GraphPad Inc., La Jolla, CA, USA). The ratios were analyzed using the Chi-squared test. *P < 0.05; **P < 0.01; ***P < 0.001.
Funding
This work was supported by The National Key Research and Development Program of China (2021YFC2700200), Natural Science Foundation of Jiangsu Province (BK20221376), National Natural Science Foundation of China (82271633), The Key Research and Development Program of Ningxia Hui Autonomous Region (2021BEG02029), Basic Science Research Program of Nantong City (JC12022006), Research Project of Nantong Health Commission (QA2021016), Gusu Health Talent Program of Suzhou (GSWS2020068), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX20_2794), and the Nantong Key Young Medical Talent Program.
Author contributions
J.Y., B.Z., and F.S. initiated the project, designed the study, coordinated the experiment, and wrote the manuscript. B.Z., Z.L., F-T.S., X.C., Q.H., L.H., H.Y., and Y.F. performed the experiments and provided conceptual inputs for the paper. L.J., X.C., Y.S., J.Y., Z.L., B.Z., X-M.C., and C.S. analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors wish to thank all study participants, research staff, and students who assisted with this work. We would like to thank Chao Tong (Life Sciences Institute, Zhejiang University, Zhejiang, China), Dahua Chen (Institute of zoology, Chinese Academy of Sciences, Beijing, China), and Jianquan Ni (Tsinghua University, Beijing, China) for kindly providing reagents and stocks. We are also grateful to Gene Denovo Biotechnology Co., Ltd (Guangzhou, China) for assisting scRNA-seq and bioinformatics analysis in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102671.
Contributor Information
Jun Yu, Email: yujun9117@126.com.
Bo Zheng, Email: bozheng@njmu.edu.cn.
Fei Sun, Email: sunfei@ntu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.White-Cooper H. Studying how flies make sperm--investigating gene function in Drosophila testes. Mol. Cell. Endocrinol. 2009;306(1–2):66–74. doi: 10.1016/j.mce.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 2.White-Cooper H. Spermatogenesis: analysis of meiosis and morphogenesis. Methods Mol. Biol. 2004;247:45–75. doi: 10.1385/1-59259-665-7:45. [DOI] [PubMed] [Google Scholar]
- 3.Dorogova N.V., Bolobolova E.U., Akhmetova K.A., Fedorova S.A. Drosophila male-sterile mutation emmenthal specifically affects the mitochondrial morphogenesis. Protoplasma. 2013;250(2):515–520. doi: 10.1007/s00709-012-0434-2. [DOI] [PubMed] [Google Scholar]
- 4.Fabian L., Brill J.A. Drosophila spermiogenesis: big things come from little packages. Spermatogenesis. 2012;2(3):197–212. doi: 10.4161/spmg.21798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inaba K. Molecular basis of sperm flagellar axonemes: structural and evolutionary aspects. Ann. N. Y. Acad. Sci. 2007;1101:506–526. doi: 10.1196/annals.1389.017. [DOI] [PubMed] [Google Scholar]
- 6.Vieillard J., et al. Transition zone assembly and its contribution to axoneme formation in Drosophila male germ cells. J. Cell Biol. 2016;214(7):875–889. doi: 10.1083/jcb.201603086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nuwal T., Kropp M., Wegener S., Racic S., Montalban I., Buchner E. The Drosophila homologue of tubulin-specific chaperone E-like protein is required for synchronous sperm individualization and normal male fertility. J. Neurogenet. 2012;26(3–4):374–381. doi: 10.3109/01677063.2012.731119. [DOI] [PubMed] [Google Scholar]
- 8.Cagan R.L. Spermatogenesis: borrowing the apoptotic machinery. Curr. Biol. 2003;13(15):R600–R602. doi: 10.1016/s0960-9822(03)00525-6. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., et al. RSBP15 interacts with and stabilizes dRSPH3 during sperm axoneme assembly in Drosophila. J. Genet Genomics. 2019;46(6):281–290. doi: 10.1016/j.jgg.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Varuzhanyan G., Chan D.C. Mitochondrial dynamics during spermatogenesis. J. Cell Sci. 2020;133(14) doi: 10.1242/jcs.235937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demarco R.S., Eikenes Å.H., Haglund K., Jones D.L. Investigating spermatogenesis in Drosophila melanogaster. Methods. 2014;68(1):218–227. doi: 10.1016/j.ymeth.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vedelek V., Laurinyecz B., Kovács A.L., Juhász G., Sinka R. Testis-specific Bb8 is essential in the development of spermatid mitochondria. PLoS One. 2016;11(8) doi: 10.1371/journal.pone.0161289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurinyecz B., et al. Sperm-Leucylaminopeptidases are required for male fertility as structural components of mitochondrial paracrystalline material in Drosophila melanogaster sperm. PLoS Genet. 2019;15(2) doi: 10.1371/journal.pgen.1007987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawyer E.M., et al. Testis-specific ATP synthase peripheral stalk subunits required for tissue-specific mitochondrial morphogenesis in Drosophila. BMC Cell Biol. 2017;18(1):16. doi: 10.1186/s12860-017-0132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguchi T., Koizumi M., Hayashi S. Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr. Biol. 2011;21(10):805–814. doi: 10.1016/j.cub.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Nandi A., Chowdhuri D.K. Cadmium mediated redox modulation in germline stem cells homeostasis affects reproductive health of Drosophila males. J. Hazard Mater. 2021;402 doi: 10.1016/j.jhazmat.2020.123737. [DOI] [PubMed] [Google Scholar]
- 17.Chen W., et al. CG8005 mediates transit-amplifying spermatogonial divisions via oxidative stress in Drosophila testes. Oxid. Med. Cell. Longev. 2020;2020 doi: 10.1155/2020/2846727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z., et al. Cyst stem cell lineage eIF5 non-autonomously prevents testicular germ cell tumor formation via eIF1A/eIF2γ-mediated pre-initiation complex. Stem Cell Res. Ther. 2022;13(1):351. doi: 10.1186/s13287-022-03025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J., et al. CG6015 controls spermatogonia transit-amplifying divisions by epidermal growth factor receptor signaling in Drosophila testes. Cell Death Dis. 2021;12(5):491. doi: 10.1038/s41419-021-03783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Q., et al. Somatic CG6015 mediates cyst stem cell maintenance and germline stem cell differentiation via EGFR signaling in Drosophila testes. Cell Death Dis. 2021;7(1):68. doi: 10.1038/s41420-021-00452-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M., et al. RpS13 controls the homeostasis of germline stem cell niche through Rho1-mediated signals in the Drosophila testis. Cell Prolif. 2020;53(10) doi: 10.1111/cpr.12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., et al. Fly Cell Atlas: a single-nucleus transcriptomic atlas of the adult fruit fly. Science. 2022;375(6584) doi: 10.1126/science.abk2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witt E., Benjamin S., Svetec N., Zhao L. Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. Elife. 2019;8 doi: 10.7554/eLife.47138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim C., et al. An aminopeptidase in the Drosophila testicular niche acts in germline stem cell maintenance and spermatogonial dedifferentiation. Cell Rep. 2015;13(2):315–325. doi: 10.1016/j.celrep.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitadate Y., Shigenobu S., Arita K., Kobayashi S. Boss/Sev signaling from germline to soma restricts germline-stem-cell-niche formation in the anterior region of Drosophila male gonads. Dev. Cell. 2007;13(1):151–159. doi: 10.1016/j.devcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Amoyel M., Sanny J., Burel M., Bach E.A. Hedgehog is required for CySC self-renewal but does not contribute to the GSC niche in the Drosophila testis. Development. 2013;140(1):56–65. doi: 10.1242/dev.086413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voog J., D'Alterio C., Jones D.L. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature. 2008;454(7208):1132–1136. doi: 10.1038/nature07173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiger A.A., Jones D.L., Schulz C., Rogers M.B., Fuller M.T. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294(5551):2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 29.Li Y., Minor N.T., Park J.K., McKearin D.M., Maines J.Z. Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc. Natl. Acad. Sci. U. S. A. 2009;106(23):9304–9309. doi: 10.1073/pnas.0901452106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Insco M.L., Leon A., Tam C.H., McKearin D.M., Fuller M.T. Accumulation of a differentiation regulator specifies transit amplifying division number in an adult stem cell lineage. Proc. Natl. Acad. Sci. U. S. A. 2009;106(52):22311–22316. doi: 10.1073/pnas.0912454106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker C.C., Fuller M.T. Translational control of meiotic cell cycle progression and spermatid differentiation in male germ cells by a novel eIF4G homolog. Development. 2007;134(15):2863–2869. doi: 10.1242/dev.003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwa J.J., Hiller M.A., Fuller M.T., Santel A. Differential expression of the Drosophila mitofusin genes fuzzy onions (fzo) and dmfn. Mech. Dev. 2002;116(1–2):213–216. doi: 10.1016/s0925-4773(02)00141-7. [DOI] [PubMed] [Google Scholar]
- 33.Barreau C., Benson E., Gudmannsdottir E., Newton F., White-Cooper H. Post-meiotic transcription in Drosophila testes. Development. 2008;135(11):1897–1902. doi: 10.1242/dev.021949. [DOI] [PubMed] [Google Scholar]
- 34.Xu S., Hafer N., Agunwamba B., Schedl P. The CPEB protein Orb2 has multiple functions during spermatogenesis in Drosophila melanogaster. PLoS Genet. 2012;8(11) doi: 10.1371/journal.pgen.1003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zur Lage P., et al. The Drosophila orthologue of the primary ciliary dyskinesia-associated gene, DNAAF3, is required for axonemal dynein assembly. Biol. Open. 2021;10(10) doi: 10.1242/bio.058812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinhauer J., et al. Combover interacts with the axonemal component Rsp3 and is required for Drosophila sperm individualization. Development. 2019;146(17) doi: 10.1242/dev.179275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao S., et al. Mitofusins: from mitochondria to fertility. Cell. Mol. Life Sci. 2022;79(7):370. doi: 10.1007/s00018-022-04386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sousa J.S., D'Imprima E., Vonck J. Mitochondrial respiratory chain complexes. Subcell. Biochem. 2018;87:167–227. doi: 10.1007/978-981-10-7757-9_7. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., Miao J., Wang Y. Mitochondrial regulation in spermatogenesis. Reproduction. 2022;163(4):R55–R69. doi: 10.1530/REP-21-0431. [DOI] [PubMed] [Google Scholar]
- 40.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eslamieh M., Mirsalehi A., Markova D.N., Betrán E. COX4-like, a nuclear-encoded mitochondrial gene duplicate, is essential for male fertility in Drosophila melanogaster. Genes. 2022;13(3):424. doi: 10.3390/genes13030424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wasbrough E.R., et al. The Drosophila melanogaster sperm proteome-II (DmSP-II) J. Proteonomics. 2010;73(11):2171–2185. doi: 10.1016/j.jprot.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Hirst J. Mitochondrial complex I. Annu. Rev. Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 44.Rhooms S., Murari A., Goparaju N.S.V., Vilanueva M., Owusu-Ansah E. Insights from Drosophila on mitochondrial complex I. Cell. Mol. Life Sci. 2020;77(4):607–618. doi: 10.1007/s00018-019-03293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhooms S.K., Murari A., Goparaju N.S.V., Vilanueva M., Owusu-Ansah E. Insights from Drosophila on mitochondrial complex I. Cell. Mol. Life Sci. 2020;77(4):607–618. doi: 10.1007/s00018-019-03293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vinothkumar K.R., Zhu J., Hirst J. Architecture of mammalian respiratory complex I. Nature. 2014;515(7525):80–84. doi: 10.1038/nature13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiedorczuk K., Letts J.A., Degliesposti G., Kaszuba K., Skehel M., Sazanov L.A. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538(7625):406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu J., Vinothkumar K.R., Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536(7616):354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K., et al. The C8ORF38 homologue Sicily is a cytosolic chaperone for a mitochondrial complex I subunit. J. Cell Biol. 2013;200(6):807–820. doi: 10.1083/jcb.201208033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia C.J., Khajeh J., Coulanges E., Chen E.I.J., Owusu-Ansah E. Regulation of mitochondrial complex I biogenesis in Drosophila flight muscles. Cell Rep. 2017;20(1):264–278. doi: 10.1016/j.celrep.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han X., et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172(5):1091–1107. doi: 10.1016/j.cell.2018.02.001. e17. [DOI] [PubMed] [Google Scholar]
- 52.Zhao L., et al. Single-cell analysis of developing and azoospermia human testicles reveals central role of Sertoli cells. Nat. Commun. 2020;11(1):5683. doi: 10.1038/s41467-020-19414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y., et al. High throughput scRNA-seq provides insights into leydig cell senescence induced by experimental autoimmune orchitis: a prominent role of interstitial fibrosis and complement activation. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.771373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi J., et al. Spatio-temporal landscape of mouse epididymal cells and specific mitochondria-rich segments defined by large-scale single-cell RNA-seq. Cell Discov. 2021;7(1):34. doi: 10.1038/s41421-021-00260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Witt E., Shao Z., Hu C., Krause H.M., Zhao L. Single-cell RNA-sequencing reveals pre-meiotic X-chromosome dosage compensation in Drosophila testis. PLoS Genet. 2021;17(8) doi: 10.1371/journal.pgen.1009728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slaidina M., Gupta S., Banisch T.U., Lehmann R. A single-cell atlas reveals unanticipated cell type complexity in Drosophila ovaries. Genome Res. 2021;31(10):1938–1951. doi: 10.1101/gr.274340.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rust K., et al. A single-cell atlas and lineage analysis of the adult Drosophila ovary. Nat. Commun. 2020;11(1):5628. doi: 10.1038/s41467-020-19361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jevitt A., et al. A single-cell atlas of adult Drosophila ovary identifies transcriptional programs and somatic cell lineage regulating oogenesis. PLoS Biol. 2020;18(4) doi: 10.1371/journal.pbio.3000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo J., et al. The dynamic transcriptional cell atlas of testis development during human puberty. Cell Stem Cell. 2020;26(2):262–276. doi: 10.1016/j.stem.2019.12.005. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan X., et al. Drosophila Pif1A is essential for spermatogenesis and is the homolog of human CCDC157, a gene associated with idiopathic NOA. Cell Death Dis. 2019;10(2):125. doi: 10.1038/s41419-019-1398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arama E., Agapite J., Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell. 2003;4(5):687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 62.DeLuca S.Z., O'Farrell P.H. Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell. 2012;22(3):660–668. doi: 10.1016/j.devcel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huh J.R., et al. Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2004;2(1):E15. doi: 10.1371/journal.pbio.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan D.C., Schon E.A. Eliminating mitochondrial DNA from sperm. Dev. Cell. 2012;22(3):469–470. doi: 10.1016/j.devcel.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vertika S., Singh K.K., Rajender S. Mitochondria, spermatogenesis, and male infertility - an update. Mitochondrion. 2020;54:26–40. doi: 10.1016/j.mito.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 66.Zhu L., Fukunaga R. RNA-binding protein Maca is crucial for gigantic male fertility factor gene expression, spermatogenesis, and male fertility, in Drosophila. PLoS Genet. 2021;17(6) doi: 10.1371/journal.pgen.1009655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu J., et al. Identification of seven genes essential for male fertility through a genome-wide association study of non-obstructive azoospermia and RNA interference-mediated large-scale functional screening in Drosophila. Hum. Mol. Genet. 2015;24(5):1493–1503. doi: 10.1093/hmg/ddu557. [DOI] [PubMed] [Google Scholar]
- 68.Lun A., Riesenfeld S., Andrews T., Dao T.P., Gomes T., Marioni J.C. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019;20(1):63. doi: 10.1186/s13059-019-1662-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8(4):329–337. doi: 10.1016/j.cels.2019.03.003. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trapnell C., et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiu X., et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.