Abstract
The use of molecular biomarkers to support disease diagnosis, monitor its progression, and guide drug treatment has gained traction in the last decades. While only a dozen biomarkers have been approved for their exploitation in the clinic by the FDA, many more are evaluated in the context of translational research and clinical trials. Furthermore, the information on which biomarkers are measured, for which purpose, and in relation to which conditions are not readily accessible: biomarkers used in clinical studies available through resources such as ClinicalTrials.gov are described as free text, posing significant challenges in finding, analyzing, and processing them by both humans and machines. We present a text mining strategy to identify proteomic and genomic biomarkers used in clinical trials and classify them according to the methodologies by which they are measured. We find more than 3000 biomarkers used in the context of 2600 diseases. By analyzing this dataset, we uncover patterns of use of biomarkers across therapeutic areas over time, including the biomarker type and their specificity. These data are made available at the Clinical Biomarker App at https://www.disgenet.org/biomarkers/, a new portal that enables the exploration of biomarkers extracted from the clinical studies available at ClinicalTrials.gov and enriched with information from the scientific literature. The App features several metrics that assess the specificity of the biomarkers, facilitating their selection and prioritization. Overall, the Clinical Biomarker App is a valuable and timely resource about clinical biomarkers, to accelerate biomarker discovery, development, and application.
Keywords: Biomarker, Genomic biomarker, Proteomic biomarker, Actionable biomarker, Clinical trial, Text mining
Graphical Abstract
1. Introduction
In the last decades, biomarkers have become pillars in research, healthcare, and drug discovery [1]. Biomarkers provide information about disease prognosis, and progression, and can be used to monitor the efficacy and safety of drug therapy. Although the nature of biomarkers might be very heterogeneous, including proteins, metabolites, cell types, and even physiological variables, like blood pressure, the term is often employed to designate molecular biomarkers [2]. Traditional molecular biomarkers are proteins measured in the blood or other biological fluids, yet more recently, genomic biomarkers1 have emerged as a new type of biomarker. This category includes the detection of alterations in the mRNA levels of genes, or their methylation level, and the presence of single nucleotide polymorphisms [3], [4], [5], [6], [7]. Proteomic and genomic biomarkers are special candidates in the quest to find novel molecular markers due to the role they play in disease development and therefore are also attractive as mechanistic disease biomarkers. Mechanistic biomarkers are preferred over other types because they can be used to monitor more reliably the onset of disease and its progression.
The increasing diversity of technologies to measure molecules, and the decreasing costs of high throughput assays have paved the way for the discovery of new genes and proteins that could serve as biomarkers. Nevertheless, the number of these entities entering the clinic is relatively small, and the rate of approval of new biomarkers is very slow [2], [8]. Resources compiling information about investigational biomarkers are scarce [9], [10] and they do not include data from clinical studies.
The Clinical Trials database (CT.gov from now on) is a public resource providing information on privately and publicly funded clinical studies for a variety of diseases and conditions, carried out across 220 countries. This resource has undergone a process of standardization [11], and a substantial portion of the information about the studies is provided in a structured format, like the conditions (diseases) and the interventions (for example drugs) tested. Different techniques to measure biomarkers are used across clinical studies, and this information is included in the studies provided by CT.gov. Therefore, CT.gov is a rich source of information on the use of biomarkers in clinical studies. Nevertheless, the information on the biomarkers is not structured and is provided across different sections of each study as free text descriptions, making it difficult to find it and its subsequent exploitation. Thus, text mining approaches are required for the identification of biomarkers used in clinical studies from CT.gov and support systematic and large-scale data analysis.
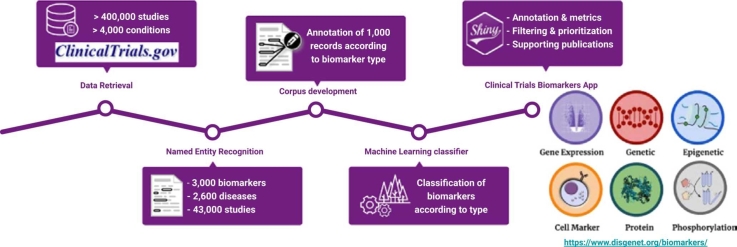
In this contribution, we designed and applied a text mining pipeline to extract information on biomarkers used in clinical trials available on CT.gov and complement it with the structured information contained in this resource, and with information mined from the scientific literature. We employed machine learning approaches to automatically classify the biomarkers in six types according to the methodologies used to measure them by relying on the textual content of sentences in which the biomarker is mentioned. In addition, we implemented different metrics to assess the specificity of biomarkers. Both the biomarker types and the metrics can be used for biomarker selection and prioritization. The data obtained by applying the text mining pipeline to CT.gov was analyzed to uncover patterns of use of biomarkers across therapeutic areas over time, including the biomarker type and their specificity. Finally, we present a new resource, the Clinical Biomarker App (https://www.disgenet.org/biomarkers/), a portal that enables exploring CT.gov-associated biomarkers in novel ways.
2. Methods
2.1. Data
We downloaded the clinical trials data from the Clinical Trials Transformation Initiative (https://aact.ctti-clinicaltrials.org/download, on December 17, 2021).
These data include the name of the conditions (disease, disorder, syndrome, illness, or injury) investigated in the trial. We normalized the condition names provided in the files to Medical Subject Headings (MeSH) identifiers using the Unified Medical Language System (UMLS) Metathesaurus [12].
We queried PubMed using a query to select articles in English on biomarkers in humans ('("Biomarkers"[Mesh]) and hasabstract[text] and english[language] and humans') obtaining 610,000 publications (December 2021). From this list of publications, we kept the ones mentioning the biomarkers identified in the clinical trials dataset (MEDLINE dataset).
2.2. Text mining
Named entity recognition (NER) and relation extraction (RE) were performed using a method based on [13]. Briefly, we performed NER based on a gene dictionary to detect mentions of genes and proteins in the following sections of a clinical trial record from CT.gov: outcomes, outcome measurements, and design outcomes. For MEDLINE publications, we performed NER for genes/proteins and diseases, and RE to identify associations between genes/proteins and diseases.
The classification of the biomarker type according to measure methodology was achieved by machine learning. Two classifiers were developed using 2 manually annotated document corpora. We selected 1500 sentences from CT.gov records with mentions of biomarkers and classified them into 6 different categories: (1) protein biomarkers, which encompasses proteins that are measured in blood, or other biological fluids, or that have been detected using immunohistochemistry, (2) genetic biomarkers, which includes genes that contain polymorphisms, copy number alterations, or chromosomal abnormalities, (3) phosphobiomarkers, that include abnormal protein phosphorylation, (4) epigenetic biomarkers, that undergo changes in DNA methylation patterns, and (5) expression biomarkers, which are genes that are measured as mRNA, and non-coding RNA such as microRNAs, and (6) cell markers, mainly composed of cell surface antigens. We applied a similar annotation procedure to 1500 sentences of the biomarker dataset extracted from MEDLINE abstracts. Thus, we created 2 corpora, one based on CT.gov sentences and the other one from MEDLINE sentences, which were used to develop two different random forest classifiers. The random forest models were used to classify the type of biomarker identified both from CT.gov records and MEDLINE publications. We use the rpart package, via the library tidymodels in R [14]. We performed hyperparameter tuning for the number of tokens (max_tokens), trees (trees), and features (mtry) using a regular grid. We provide more details of the implementation, along with a sample of the developed corpus in this GitHub repository (https://github.com/jpinero/biomarkers).
2.3. Metrics
We use the relative entropy or Kullback–Leibler divergence (DKL) to compare the distribution of disease classes of a particular biomarker to the one observed across all clinical studies (background distribution). The DKL can be used as an indicator of the specificity of the use of biomarkers across therapeutic areas. First, all diseases in clinical trials are classified into therapeutic classes (with a disease potentially belonging to more than one class). Let Q(x) be the fraction of all diseases in class x concerning all disease-class pairs. If a biomarker is tested in T diseases, let P(x) be the fraction of diseases in T within therapeutic class x concerning all disease-class pairs containing any of the T diseases. The Information Content (IC) of a biomarker associated with class x is then the logarithm of the quotient P(x)/Q(x) times P(x). The relative entropy (DKL) of the biomarker is defined as:
We added pseudo counts of 1 to the numerator and denominator to deal with zero-content classes. A relative entropy of 0 means that the 2 distributions are equivalent, while a relative entropy different from 0 means that the distributions differ and therefore the biomarker is more specific for one or several therapeutic areas. For instance, biomarkers measured in a wide range of conditions across different disease classes will have a DKL value close to 0. Contrastingly, proteins more specific to one therapeutic area, like as an example, Neoplasms, will display higher DKL values.
We also computed for each biomarker the Disease Specificity Index (DSI), and the Disease Pleiotropy Index (DPI), as described in [10], [15]. The DSI reflects if a gene is associated with several or fewer diseases. It is computed according to:
Where Nd is the number of diseases associated with the biomarker and NT is the total number of diseases in the dataset.
The rationale for the DPI is similar to the DSI, but we consider if the multiple diseases associated with the biomarker are similar among them (belong to the same MeSH disease class, e.g. Cardiovascular Diseases) or are completely different diseases and belong to different disease classes. The Disease Pleiotropy Index (DPI) is computed according to
Where Ndc is the number of the different MeSH disease classes of the diseases associated with the biomarker and NTC is the total number of MeSH diseases classes in the dataset (22).
2.4. Data analysis
All the analysis was performed with the R software version 4.0. For visualization, we used ggplot2 [16], ComplexUpset [17], and heatmaply [18].
2.5. The Clinical Biomarker App
We created an R Shiny App, that allows searching, filtering, and browsing of the data. In the Clinical Biomarker App, biomarkers are annotated with their gene symbol, name, type of gene, the protein class from the drug target ontology [19], DSI, DPI, pLI (probability of being loss-of-function intolerant from GNOMAD [20]), the number of associated conditions, the number of clinical trials, the number of publications, and the relative entropy, along with the therapeutic area with the highest information content. Conditions are annotated with the semantic type, the number of associated biomarkers, publications, and clinical trials. The Clinical Biomarker App is available at https://www.disgenet.org/biomarkers/. It allows users to explore the data by biomarker, condition, filter by the number of clinical trials, and the number of publications. It also provides annotations for the biomarker and conditions, and their relations, such as the year of the first and last clinical trial, and the year of the first and last publication.
3. Results
The main objective of this contribution was to identify biomarkers measured in clinical trials (CTs), relate them to the conditions in which they are assessed and classify them according to the methodologies used to measure them. For this purpose, we designed and applied a text mining strategy to extract information about biomarkers used in CT.gov and combine it with data from the scientific literature. We also explored the resulting dataset to uncover trends in biomarker discovery. Finally, we made available this information on the Clinical Biomarkers App (https://www.disgenet.org/biomarkers/) to support the analysis of biomarker used in clinical studies.
3.1. The landscape of biomarkers in clinical studies
The text mining methodology here proposed was used to process 398,516 clinical studies available at CT.gov carried out between 1999 and 2021. We found over 43,000 clinical studies measuring 3100 biomarkers for 2600 diseases, establishing 63,000 biomarker-condition pairs. The data obtained were analyzed to gain insight into patterns of biomarker use across clinical studies, therapeutic areas, and the methodologies employed for biomarker measurement. The studies within this dataset are mainly interventional (84%), 58% of them are associated with drugs, while the rest are observational. Regarding the phase of the clinical trial for the interventional studies, we found that biomarkers are used in all the phases of clinical development (19% phase 1, 33% phase 2, 22% phase 3% and 16% phase 4).
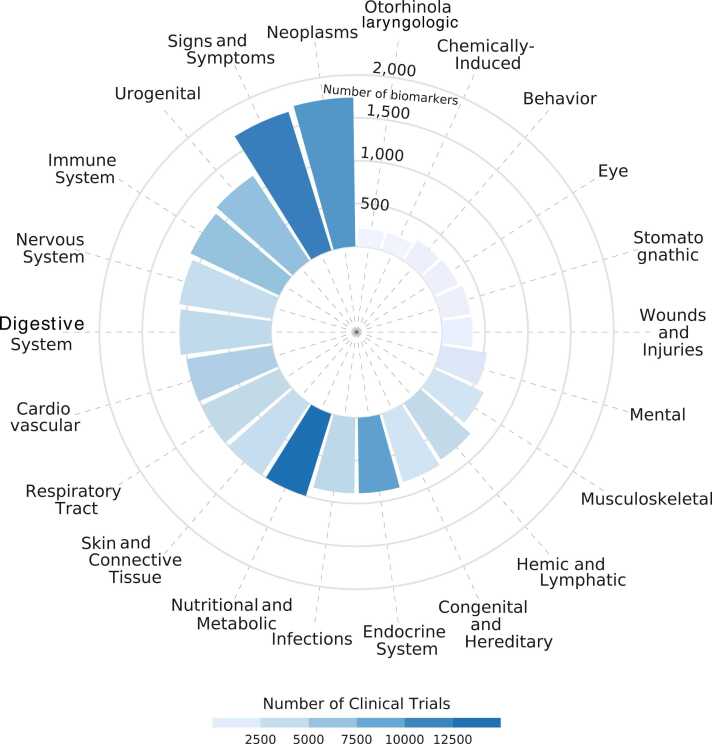
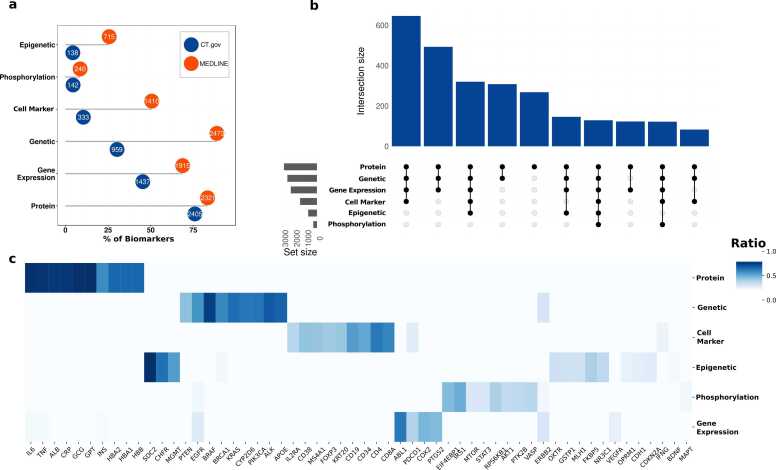
Fig. 1 shows the number of biomarkers found per therapeutic area. Neoplasms is the class with the largest number of biomarkers, followed by Signs and Symptoms. Concerning the total number of diseases in CT.gov, the percentage of clinical trials using biomarkers per therapeutic area is roughly the same: between 10%, except for trials of conditions related to the Endocrine System in which the percentage of studies using biomarkers is 42%, and Nutritional and Metabolic diseases, 34% (Supplementary Table 1).
Fig. 1.
Distribution of biomarkers across therapeutic areas. The color scale is proportional to the number of clinical trials assessing biomarkers in the therapeutic area.
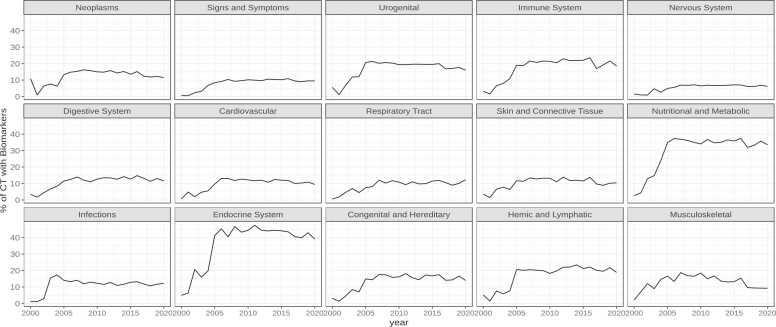
Fig. 2 shows the time evolution of the use of biomarkers across therapeutic areas. In most therapeutic areas there is an increase in the number of studies using biomarkers around 2005, and then it stabilizes at around 10% except for diseases associated with Urogenital and Immune system, which reach 20%, and the Endocrine system and Nutritional and Metabolic conditions, which reach 40%.
Fig. 2.
Use of biomarkers per therapeutic area (as a proportion of clinical trials in the area per year) spanning 20 years of clinical trials.
The conditions with the largest number of CTs are Diabetes Mellitus type 1 and 2, Prostatic and Mammary neoplasms, kidney diseases, and Rheumatoid arthritis, all with over 1000 CTs that use biomarkers. With respect to the number of different biomarkers assessed, different cancer types are at the top of the list, mammary neoplasms being the top with over 1400 different genes and protein biomarkers, followed by colorectal, liver, and prostate. COVID 19 is also at the top of the list, with over 300 different biomarkers, including interleukins, CRP, TNF, and ACE2.
Most biomarkers (84%) are assessed in the context of more than one condition (Supplementary Fig. 1a). Thirty-seven percent of the biomarkers are assessed only in one clinical trial (Supplementary Fig. 1b). The most assessed biomarkers in clinical trials, that are also used to monitor a very large number of conditions are hemoglobin, markers of inflammation, such as CRP, IL6, TNF, IL10, and IFN, enzymes related to liver function such as GPT and AST, and other proteins like insulin, and albumin.
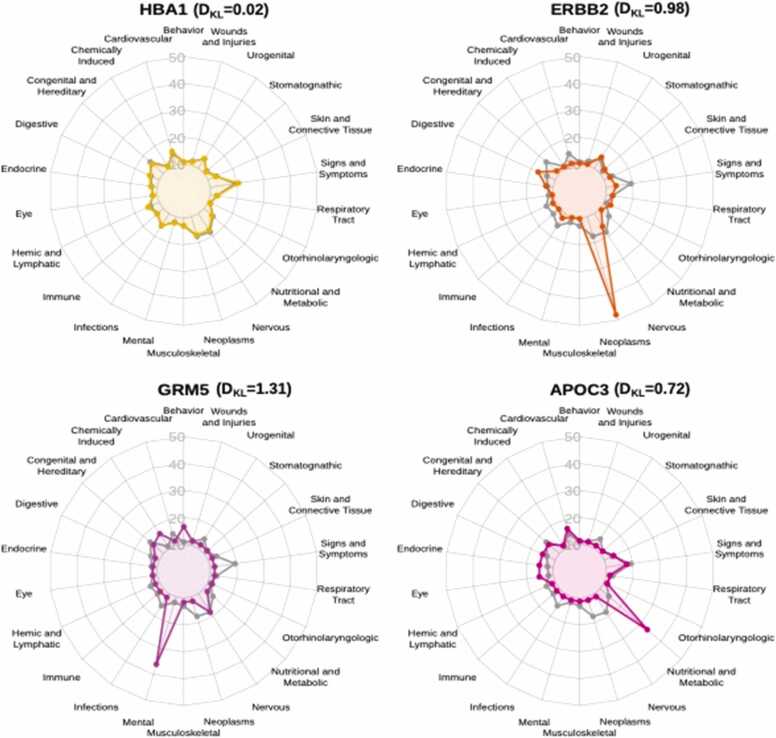
Some biomarkers are usually measured in the context of a specific therapeutic area, while others, like hemoglobin, albumin, and hepatic enzymes are used across practically all therapeutic areas. To quantify the specificity of biomarkers across therapeutic classes, we computed two different metrics: the Disease Pleiotropy Index (DPI), and the relative entropy (DKL). The DPI assesses the fraction of different therapeutic areas in which the biomarker is used. Higher DPI values are obtained for biomarkers used across several therapeutic areas, and vice versa. Nevertheless, the DPI gives the same importance to all therapeutic areas. To assess if a biomarker is associated with several conditions from the same therapeutic area, we employed the concept of relative entropy, which assesses to what extent the distribution of the use of the biomarker across therapeutic classes resembles the background distribution of the whole set of clinical trials. The relative entropy (DKL) is computed by summing up the information content (IC) for each therapeutic class. A relative entropy close to zero indicates that the two distributions in question are very similar, implying that the biomarker is not specific to a therapeutic class. Higher values of relative entropy indicate differences in the two distributions. Fig. 3 shows examples of the distribution of the information content across therapeutic classes for a selection of biomarkers. The relative entropy of Hemoglobin as a biomarker, measured in connection to more than one thousand conditions is very low while ERBB2, GRM5, and APOC3 (all with a higher entropy) exhibit a high IC for neoplasms, infections, and nutritional and metabolic diseases, respectively. An important proportion of the top biomarkers with higher relative entropy values show higher values of IC for neoplasms, and they include cancer drivers such as PTEN, TP53, CCND1, CDK4, and RB1. Other proteins in the high IC for neoplasms group, that also have high IC for conditions belonging to hemic and lymphatic and immune systems include BTK, XPO1, NOTCH1 and BCL6, which are drivers of haematological malignancies (Supplementary Fig. 2). One potential caveat of the comparison using relative entropy is that it is not completely independent of the number of clinical trials involving the biomarkers.
Fig. 3.
Information content per therapeutic class for a selection of four biomarkers. In gray, we show the information content for each therapeutic class of the whole dataset (background distribution Q).
Ninety-eight percent of the biomarkers are protein-coding genes, while less than 2% (60) biomarkers are non-coding RNAs. The microRNA MIR155 is the top ncRNA assessed in 26 conditions belonging to different therapeutic areas: obesity, asthma, arthritis, infertility, and leukemia. Another interesting ncRNA is MIR122, used mainly in the context of liver injuries.
To provide the research context for the biomarkers extracted from CT.gov, we applied text mining approaches to mine MEDLINE publications discussing biomarkers in humans. From the original set of publications annotated with the MeSH tag “biomarker”, we kept only those mentioning the genes and proteins identified in clinical studies by our text mining pipeline, comprising 27,900 articles ranging from 1981 to 2022. We found that 88% of CTs biomarkers are discussed in publications associated with 2200 conditions (Supplementary Fig. 3). Extracting information from scientific publications increased the number of conditions for the biomarkers in 825 additional diseases. Twelve percent of the biomarker-condition pairs were included in this dataset. Several reasons explain this relatively low overlap. First, some pairs of biomarker-condition annotated in CT.gov data use top-level MESH categories, such as cardiovascular diseases, or congenital abnormalities, while diseases retrieved by mining MEDLINE are more specific. Second, some of the pleiotropic biomarkers (hemoglobin, liver enzymes as GOT1, GOT2, and GPT, or markers of inflammation like TNF, CRP and IL6) are associated with the general state of the individual, thus they are not directly associated with the condition evaluated in the clinical trial. Third, restricting the publications to the set annotated with the MeSH tag “biomarker” might cause some articles describing the role of the biomarkers in the disease in the research context to be missed.
To capture different methodologies for measuring the biomarkers we used machine learning approaches. First, we created two corpora, one with the data from CT.gov and one with sentences from the publications discussing biomarkers in MEDLINE. The corpus from CT.gov contained 1300 CTs, including 590 biomarkers and 1490 sentences from the measurement sections. Thirty-one percent of the records were classified as genetic, 26% as protein, 16% as gene expression, 15% as cell marker, 7% as phosphorylation, and 5% as epigenetic. The MEDLINE corpus contained 930 publications, 680 biomarkers, 1100 sentences and 36% of the records were classified as genetic, 31% as protein, 28% as gene expression, 18% as epigenetic,7% as cell marker, and 6% as phosphorylation.
Next, we trained two different random forest models using the matrix containing the term frequency-inverse document frequency of tokens. We tuned the number of tokens for each model, and also the random forest hyperparameters mtry and number of trees. The multiclass accuracy for the model created for CT.gov is 0.83 and for the MEDLINE model was 0.84. The detailed performance of the models is described in Supplementary Table 2. Supplementary Fig. 4 shows the variable importance of the top features in both models. Tokens like phosphorylation, methylation, cell, mRNA, mutations were among the most important words to classify the biomarkers according to methodology.
For the CT.gov dataset, 76% of the biomarkers are measured at the protein level, including the concentration of the protein in fluids, such as blood, but also immunohistochemistry assays. The second category is gene expression (45%) (Fig. 4a). For the MEDLINE dataset, the most populated category is genetic biomarkers (89%) closely followed by proteins (83%). The less populated categories for both datasets are epigenetic and phosphorylation biomarkers, representing 4% for CT.gov (both biomarker types) and 25% and 9% respectively for MEDLINE. These categories are not biomarker-specific: most biomarkers can be measured in several ways: protein, mRNA, or they can be used as genomic biomarkers (Fig. 4b).
Fig. 4.
Different methodologies to measure the biomarkers. a) Distribution of biomarkers in each category. b) Overlaps among biomarker categories. c) Top biomarkers per methodology, measured as the percentage of clinical trials in the category with respect to the total number of clinical trials associated with the biomarker.
The most common biomarkers measured at the protein level are hemoglobin, markers of inflammation, such as CRP, IL6, TNF, enzymes related to liver function such as GPT and GOT2, and other proteins such as insulin, and albumin. Cancer driver genes such as BRAF, ALK, EGFR, PTEN, ERBB2, and EGFR are the top genetic biomarkers together with APOE for nervous and metabolic diseases, and the cytochromes such as CYP2D6, a highly polymorphic gene, involved in the metabolism many drugs commonly used in the clinic [21]. IRS1, VASP, AKT1, STAT3, RPS6KB1, EIF4EBP1, and MTOR are the proteins that are more frequently used as phosphobiomarkers. Over 40 clinical trials assess the methylation status of MGMT. The CD4 membrane glycoprotein of T lymphocytes, used to monitor the status of the immune system, is the most frequent cell marker. Other cell markers that are frequently evaluated belong to the class cluster of differentiation proteins, found on the surface of cells, that allow the identification of cell phenotypes. The most evaluated biomarker at the level transcript is ABL1, for the detection of the BCR-ABL fusion, which is the main driver of some types of leukemias (Fig. 4c).
3.2. The Clinical Biomarker App
To allow researchers the exploration of this new dataset, we have created the Clinical Biomarker App, containing the data mined from CT.gov and complemented with the information extracted from MEDLINE publications, and with metrics and attributes. The app is structured around five different tabs: Biomarkers, Conditions, Summary, Measurements, and Publications.
The Biomarkers tab shows all the biomarkers, along with metrics such as DPI, DSI, pLI, DKL, and the therapeutic area with the highest IC. The Conditions tab displays the number of biomarkers associated with the condition, and the semantic type according to the UMLS. The Summary tab contains the pairs of biomarker-condition found not only in the clinical trial dataset and the MEDLINE dataset. All these tabs also provide information of the number of clinical trials, and publications associated with their reference entities (Biomarker, Condition, or pairs), along with the year of the more recent and latest clinical trial or publication. The user can navigate the specific studies supporting the relationships in each of these tabs by clicking on the number of clinical trials, or publications. The Measurements tab contains the biomarker type, the sentence from which the biomarker has been extracted in the clinical study, with other attributes such as the year, intervention type, and phase. Finally, the Publications tab provides information on the MEDLINE publications mentioning the biomarkers.
3.3. Use case: biomarkers associated with Autistic Disorders
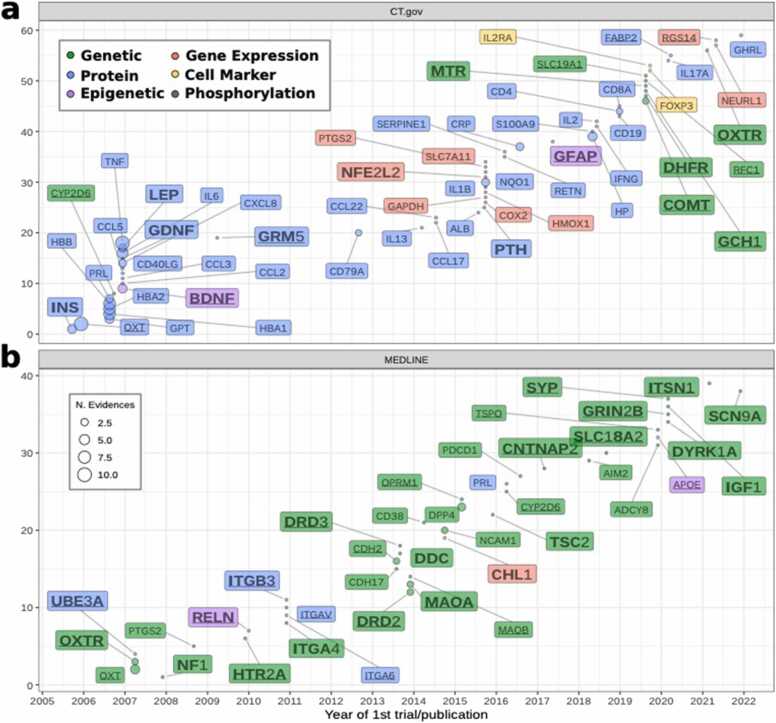
As a use case, we studied the biomarkers associated with Autistic Disorders. There are 93 biomarkers evaluated in the context of Autistic Disorders in the Biomarker App (comprising MeSH:D001321 and MeSH:D000067877). Fifty-nine genes are biomarkers evaluated in CT.gov, and 39 are reported by publications from MEDLINE (Fig. 5). Only 5 biomarkers are shared by the two sets: OXT (oxytocin, 10 CTs and 4 publications), PRL (prolactin, 3CTs and 2 publications), OXTR (oxytocin receptor 1CT and 2 publications), CYP2D6 (cytochrome P450 family 2 subfamily D member 6, 1CT and 1 publication), and PTGS2 (prostaglandin-endoperoxide synthase 2, 1CT and 1 publication). Oxytocin is one of the earliest biomarkers investigated in relation to autism in CT.gov and MEDLINE. This hormone has been tested as a possible treatment for this condition [22]. In fact, 6 out of the 7 studies involving oxytocin (NCT05096676, NCT03640156, NCT03337035, NCT03033784, NCT01256060, and NCT01945957) test the possible effects of the hormone as treatment, while one (NCT01643720) measures the changes in its salivary levels before and after emotionally arousing activities.
Fig. 5.
Autistic Disorders Biomarkers. The figure shows the biomarkers associated with Autistic Disorders. a) biomarkers from CT.gov, and b) biomarkers extracted from MEDLINE. Colors represent the different biomarker classes. Underlined biomarkers are those with therapeutic class “Mental Diseases,” according to their information content. Biomarkers in bold with larger size are those that are currently represented in panels associated with Autism according to the NIH Genetic Testing Registry (https://www.ncbi.nlm.nih.gov/gtr/).
Genetic biomarkers are the main type in the MEDLINE dataset, while proteins dominate the ASD biomarkers from CT. gov. One clinical trial (NCT03152838, The Role of Epigenetic Modifications in Autism Spectrum Disorder) evaluates the methylation levels for BDNF and GFAP, while two publications [23], [24] describe alterations in the level of methylation of RELN and APOE (respectively) in autistic patients. Thirty-three of the 93 biomarkers are included in several panels linked to autism according to the NIH Genetic Testing Registry (https://www.ncbi.nlm.nih.gov/gtr/), and 72 are included in DISGENET plus (https://www.disgenetplus.com/). Twenty-four biomarkers are only found by exploring CT.gov data, (23 only supported by clinical trials information), and while some of them (like HBB, GPT, and cell markers like CD8A) are highly pleiotropic and have low specificity, these results reveal the need of integrating several sources of information to have a complete panorama of the genetic underpinnings of human diseases, and thus, of possible candidates for biomarkers. The five biomarkers with higher relative entropy of the dataset (GRM5, DRD2, DRD3, OXTR, and OXT) have the highest IC for mental disorders, and 4 of them are GPCRs (Fig. 5, underlined biomarkers). See Supplementary Table 3 for the information related to the biomarkers.
4. Conclusions
The rapidly decreasing costs of next-generation sequencing technologies, and the massive generation of omics data have triggered an explosion of information in biomedical research that holds the promise of improving our understanding of human diseases. Central to this promise are molecular biomarkers, that can be used to improve disease diagnosis, stratify patients, guide drug therapy, and monitor the response to interventions. Nevertheless, the rate at which these molecules are discovered and incorporated into the clinical setting has been staggeringly slow. To help fill this gap, in this contribution, we have developed and applied a pipeline to extract fine-grained information about biomarkers measured in clinical studies. Applying this pipeline uncovers over 3000 biomarkers associated with 2600 conditions identified from 43,000 clinical studies from ClinicalTrials.gov. In addition, we created a new classification system for gene and protein biomarkers, useful to identify how biomarkers are measured in the clinical trial (e.g., genetic, epigenetic, phosphorylation, etc.). We have implemented different metrics to help prioritize biomarkers according to their specificity across therapeutic areas. We have made available this information in the Clinical Biomarkers App that enriches the data about biomarkers and their associated conditions with a variety of annotations and metrics that allow filtering and browsing of the information. Text mining of biomarker information from clinical studies available at CT.gov allows for unlocking the data herein provided, paving the way for automatic and comprehensive data analysis and exploration. Finally, although there are around 3000 biomarkers used in clinical studies, a large fraction of the human genome remains to be explored as disease biomarkers in the context of clinical studies. Nevertheless, the advances in genomic and proteomic technologies should facilitate the incorporation of more gene and protein biomarkers into clinical trials. More strikingly, the number of biomarkers assessed in clinical studies is significantly higher than the number of approved biomarkers [2], [8]. In summary, by structuring and organizing data on biomarkers from CT.gov, we aim at supporting the assessment of the evidence of the use of biomarkers in clinical studies.
Authors contributions
J.P., L.I.F., and F.S. conceived the idea. J.P. performed the research and analysis. F.R., P.S.R.F., and P.A. developed the software, R.L.J., J.V.M. analyzed the data. J.P. and LIF wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
CRediT authorship contribution statement
Janet Piñero: Conceptualization, Data curation, Formal analysis, Writing – original draft, Writing – review & editing, Software, Visualization. Pablo S. Rodriguez Fraga: Software. Francesco Ronzano: Software, Methodology. Pablo Accuosto: Software. Jordi Valls-Margarit: Methodology, Formal analysis, Data curation. Ricard Lambea Jane: Software, Visualization. Ferran Sanz: Conceptualization, Supervision, Funding acquisition. Laura I. Furlong: Conceptualization, Writing – original draft, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Janet Piñero, Laura I. Furlong and Ferran Sanz are co-founders and shareholders of MedBioinformatics Solutions SL. Laura I. Furlong is an employee of MedBioinformatics Solutions SL.
Acknowledgements
This work was partially funded by IMI2-JU grants, resources which are composed of financial contributions from the European Union’s Horizon 2020 Research and Innovation Programme and EFPIA [GA: 116030 TransQST and GA: 777365 eTRANSAFE], and the EU H2020 Programme [GA: 871075 Elixir-CONVERGE and GA:964537 RISKHUNT3R, which is part of the ASPIS cluster]; Project 001-P-001647—Valorisation of EGA for Industry and Society funded by the European Regional Development Fund (ERDF) and Generalitat de Catalunya; Agència de Gestió d’Ajuts Universitaris i de Recerca Generalitat de Catalunya [2017SGR00519], and the Institute of Health Carlos III (project IMPaCT-Data, exp. IMP/00019), co-funded by the European Union, European Regional Development Fund (ERDF, “A way to make Europe”). The Research Programme on Biomedical Informatics (GRIB) is a member of the Spanish National Bioinformatics Institute (INB), funded by ISCIII and ERDF (PRB2-ISCIII [PT13/0001/0023, of the PE I + D + i 2013-2016]). The MELIS is a ‘Unidad de Excelencia María de Maeztu’, funded by the MINECO [MDM-2014-0370]. This work reflects only the author’s view and that the IMI2-JU is not responsible for any use that may be made of the information it contains. The European Commission is not responsible for any use that may be made of the information it contains.
Footnotes
https://www.ema.europa.eu/en/documents/scientific-guideline/ich-e-15-establish-definitions-genomic-biomarkers-pharmacogenomics-pharmacogenetics-genomic-data_en.pdf
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.03.014.
Contributor Information
Janet Piñero, Email: janet.pinero@upf.edu.
Laura I. Furlong, Email: laura.furlong@disgenetplus.com.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Gromova M., Vaggelas A., Dallmann G., Seimetz D. Biomarkers: opportunities and challenges for drug development in the current regulatory landscape. Biomark Insights. 2020:15. doi: 10.1177/1177271920974652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rifai N., Gillette M.A., Carr S.A. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. 2006 24:8. [DOI] [PubMed] [Google Scholar]
- 3.Bernard P.S., Parker J.S., Mullins M., Cheung M.C.U., Leung S., Voduc D., et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinnott-Armstrong N., Tanigawa Y., Amar D., Mars N., Benner C., Aguirre M., et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet. 2021;53:185–194. doi: 10.1038/S41588-020-00757-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorokin M., Ignatev K., Poddubskaya E., Vladimirova U., Gaifullin N., Lantsov D., et al. RNA sequencing in comparison to immunohistochemistry for measuring cancer biomarkers in breast cancer and lung cancer specimens. Biomedicines. 2020;Vol 8:114. doi: 10.3390/BIOMEDICINES8050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miura N., Hasegawa J., Shiota G. Serum messenger RNA as a biomarker and its clinical usefulness in malignancies. Clin Med Oncol. 2008;2:511–527. doi: 10.4137/CMO.S379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bock C., Halbritter F., Carmona F.J., Tierling S., Datlinger P., Assenov Y., et al. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol. 2016;34:726–737. doi: 10.1038/nbt.3605. 2016 34:7. [DOI] [PubMed] [Google Scholar]
- 8.Hanash S.M. Why have protein biomarkers not reached the clinic? Genome Med. 2011;3:1–2. doi: 10.1186/GM282/METRICS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wishart D.S., Bartok B., Oler E., Liang K.Y.H., Budinski Z., Berjanskii M., et al. MarkerDB: an online database of molecular biomarkers. Nucleic Acids Res. 2021;49:D1259–D1267. doi: 10.1093/NAR/GKAA1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piñero J., Saüch J., Sanz F., Furlong L.I. The DisGeNET cytoscape app: exploring and visualizing disease genomics data. Comput Struct Biotechnol J. 2021;19:2960–2967. doi: 10.1016/J.CSBJ.2021.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenaerts P., Madre L., Archdeacon P., Califf R.M. The Clinical Trials Transformation Initiative: innovation through collaboration. Nat Rev Drug Discov. 2014;13:797–798. doi: 10.1038/nrd4442. 2014 13:11. [DOI] [PubMed] [Google Scholar]
- 12.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:267D–270D. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bravo Piñero J., Queralt-Rosinach N., Rautschka M., Furlong L.I. Extraction of relations between genes and diseases from text and large-scale data analysis: Implications for translational research. BMC Bioinforma. 2015:16. doi: 10.1186/s12859-015-0472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn M., Wickham H. Tidymodels: a collection of packages for modeling and machine learning using tidyverse principles. 2020.
- 15.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020:48. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wickham H. Springer-Verlag; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 17.Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/BIOINFORMATICS/BTX364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galili T., O’Callaghan A., Sidi J., Sievert C. heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics. 2018;34:1600–1602. doi: 10.1093/BIOINFORMATICS/BTX657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y., Mehta S., Küçük-McGinty H., Turner J.P., Vidovic D., Forlin M., et al. Drug target ontology to classify and integrate drug discovery data. J Biomed Semant. 2017;8:1–16. doi: 10.1186/S13326-017-0161-X/FIGURES/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv. 2019 doi: 10.1101/531210. [DOI] [Google Scholar]
- 21.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharm J. 2005;5:6–13. doi: 10.1038/SJ.TPJ.6500285. [DOI] [PubMed] [Google Scholar]
- 22.Sikich L., Kolevzon A., King B.H., McDougle C.J., Sanders K.B., Kim S.-J., et al. Intranasal oxytocin in children and adolescents with autism spectrum disorder. N Engl J Med. 2021;385:1462–1473. doi: 10.1056/NEJMOA2103583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lintas C., Persico A.M. Neocortical RELN promoter methylation increases significantly after puberty. Neuroreport. 2010;21:114–118. doi: 10.1097/WNR.0B013E328334B343. [DOI] [PubMed] [Google Scholar]
- 24.Liu D., Cao H., Kural K.C., Fang Q., Zhang F. Integrative analysis of shared genetic pathogenesis by autism spectrum disorder and obsessive-compulsive disorder. Biosci Rep. 2019:39. doi: 10.1042/BSR20191942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material