Summary
Central oscillators are primordial neural circuits that generate and control rhythmic movements 1,2. Mechanistic understanding of these circuits requires genetic identification of the oscillator neurons and their synaptic connections, to target electrophysiological recording and causal manipulation during behavior. Yet, such targeting remains a challenge with mammalian systems. Here, we delimit the oscillator circuit that drives rhythmic whisking, a motor action central to foraging and active sensing in rodents3,4. We found that the whisking oscillator consists of parvalbumin-expressing inhibitory neurons located in the vibrissa intermediate reticular nucleus (vIRtPV) in the brainstem. vIRtPV neurons receive descending excitatory inputs and form recurrent inhibitory connections among themselves. Silencing vIRtPV neurons eliminated rhythmic whisking and resulted in sustained vibrissae protraction. In vivo recording of opto-tagged vIRtPV neurons in awake mice showed that these cells spike tonically when animals are at rest, and transition to rhythmic bursting at the onset of whisking, suggesting that rhythm generation is likely the result of network dynamics, as opposed to intrinsic cellular properties. Importantly, ablating inhibitory synaptic inputs to vIRtPV neurons quenched their rhythmic bursting, impaired the tonic-to-bursting transition and abolished regular whisking. Thus, the whisking oscillator is an all-inhibitory network and recurrent synaptic inhibition plays a key role in its rhythmogenesis.
The vibrissa intermediate reticular nucleus (vIRt), a caudal ventral medulla region situated medial to the compact part of nucleus ambiguus (NA), has previously been shown to be the site of the central oscillator for whisking5. Using kainic acid-induced persistent whisking in anesthetized rats, it was found that vIRt contains neurons that fire at either the protraction or, predominantly, the retraction phase of whisking5,6. Rhythmic whisking is likely driven by an oscillatory signal from inhibitory vIRt neurons that fire during the retraction phase and provide inhibitory inputs onto vibrissa facial motor nucleus (vFMN) neurons, counteracting the tonic excitatory drive (Fig. 1a)7. These studies led to a model in which tonic excitation of vFMN motoneurons leads to protraction, which is stopped by rhythmic inhibition from vIRt oscillator cells that induces retraction, resulting in the back-and-forth whisking of rodents (Fig. 1a)4. Further, rhythmic inhibition from vIRt oscillator cells is thought to be reset by inputs from the breathing center, the preBötzinger complex (preBötC)8, that drives inhalation5,7,9.
Fig. 1. Molecular and functional characterization of PV+ premotor vIRt neurons.
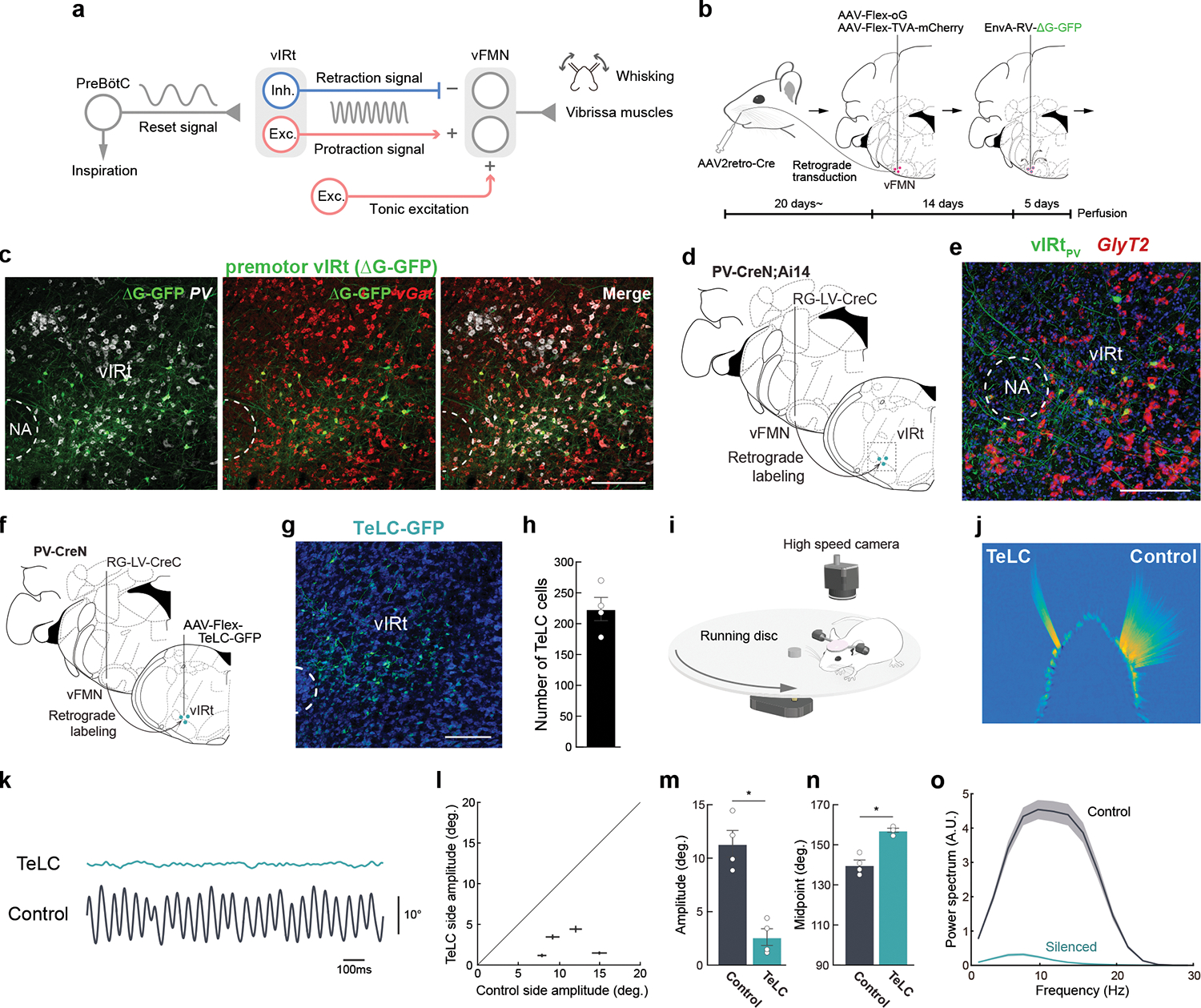
a, Diagram of the proposed whisking generation circuit in the brainstem. B, A three-step monosynaptic rabies virus tracing strategy to label adult vibrissa premotor circuit. C, Representative image of vibrissa premotor neurons in vIRt. PV and vGat mRNA were detected by HCR RNA-FISH. D, Viral-genetic split-Cre strategy for labeling PV+ premotor vIRt neurons. Retrograde lentivirus carrying CreC (RG-LV-CreC) is injected into the vibrissal part of the facial motor nucleus (vFMN) of the PV-CreN;Ai14 animal. Functional Cre is reconstituted only in vibrissal premotor neurons expressing PV. e, Molecular characterization of vIRtPV neurons. vIRtPV neurons expressing tdTomato (shown in green) overlap with GlyT2 (red, 87.9 ± 1.4%, n = 3). f, Strategy for expressing TeLC in vIRtPV neurons using the same split-cre strategy. g, Post hoc histological assessment of TeLC-GFP expression. h, Quantification of TeLC-GFP expressing cells in vIRt (223.8 ± 18.9, n = 4). i, Experimental setup for vibrissa tracking. j, A superimposed image of tracked vibrissae (C2, Left; TeLC-silenced, Right; Control). k, Representative vibrissa angle traces (green; TeLC-silenced, black; Control. Whisking midpoint was subtracted. Protraction is up). l, Plot of whisking amplitude of the TeLC-silenced and control sides. Dots represent individual animals. m, Quantification of whisking amplitude (Control side, 11.4 ± 1.2°; TeLC side, 2.6 ± 0.8°, n=4) n, Quantification of whisking midpoint (Control side, 140.0 ± 2.4°; TeLC side, 157.3 ± 1.0°, n=4.) o, Power spectrum analysis of whisking frequency. Shaded areas are mean ± s.e.m. Data are mean ± s.e.m. * P ≤ 0.05, KS test (m, n). Brain sections were counterstained with Neurotrace Blue (c, g) or DAPI (e). Scale bars, 200 μm.
Oscillator circuits are exceedingly difficult to study in mammalian systems. The lack of molecular identification limits the ability to investigate physically distributed circuits at the neuronal level. Targeting brainstem and spinal cord circuits for in vivo recording in awake behaving animals also adds to the challenge. We therefore set out to find putative molecular markers for vIRt whisking oscillator neurons, with an immediate goal of recording and manipulating identified neurons in awake behaving mice and a subsequent goal of delineating rhythm generation mechanisms.
Subsets of vibrissa premotor neurons in vIRt express parvalbumin (PV)
We used a recently developed three-step monosynaptic rabies virus tracing method that enables transsynaptic tracing of premotor neurons in adult mice (Fig. 1b)10. This method efficiently labeled vibrissa premotor neurons in adult mice and revealed the presence of premotor cells in vIRt (Fig. 1c). Multiplexed Hybridization Chain Reaction (HCR) RNA-Fluorescent in situ hybridization (FISH)11 revealed that 67.0 ± 1.0% (n = 4) of all premotor neurons within vIRt expressed the inhibitory neuronal marker vGat (Fig. 1c), whereas 28.3 ± 2.0% (n = 3) expressed the excitatory neuronal marker vGlut2 (Extended Data Fig. 1a, b). Through candidate marker analysis, we discovered that 69.6 ± 3.4% (n = 4) of these vGat+ premotor vIRt neurons co-expressed Parvalbumin (PV) (Fig. 1c, Extended Data Fig. 1b); henceforth denoted as vIRtPV neurons. To specifically label and manipulate vIRtPV neurons, we employed the split-Cre strategy, taking advantage of the PV-CreN knock-in mouse line in which the N-terminal half of Cre (CreN) is expressed from the PV genomic locus12,13. This involves injecting a retrograde lentivirus (RG-LV) that expresses the C-terminal half Cre (RG-LV-CreC) into the lateral part of the facial nucleus (vFMN) in PV-CreN mice. RG-LV infects neurons from axon terminals14 and thus the injection resulted in CreC expression in premotor neurons that project to vFMN. The full-length Cre can then only be fully reconstituted in PV-positive vibrissa premotor neurons (Fig. 1d). Injection of a viral vector carrying a Cre-dependent construct into the vIRt region, therefore, results in the expression of desired genes specifically in premotor vIRtPV neurons, such as a fluorescent reporter protein (Fig. 1d,e). FISH revealed that most vIRtPV neurons expressed the glycine transporter gene GlyT2 (Fig. 1e; 87.9 ± 1.4 %, n = 3), indicating they are mostly inhibitory cells.
Specific silencing of vIRtPV neurons abolishes rhythmic whisking
To test whether vIRtPV neurons are part of the whisking oscillator, we used the same viral-genetic split-Cre strategy to express tetanus toxin light chain (TeLC)15–17 together with GFP in these neurons (Fig. 1f,g). This resulted in TeLC-GFP being expressed in ~ 200 vIRtPV neurons (Fig. 1h; 224 ± 19 cells, n = 4). Importantly, we observed that TeLC mediated silencing of vIRtPV neurons almost completely abolished the rhythmic movement of vibrissae on the ipsilateral, silenced side (Fig. 1i–l, Supplementary Video 1). Bilateral tracking of C2 vibrissae’s position revealed that on the TeLC-silenced side, the vibrissa remained in a protracted position, indicating that vibrissa retraction was abolished (Fig. 1j), consistent with past unilateral chemical ablation of the vIRt area18. Quantitatively, silencing vIRtPV neurons resulted in minute whisking amplitude (Fig. 1m, 11.4 ± 1.2° in control vs. 2.6 ± 0.8° in TeLC side, n = 4, p = 0.029, KS test); and a protracted midpoint (Fig 1n, 140.0 ± 2.4° in control vs. 157.3 ± 1.0° in TeLC, n = 4, p = 0.029, KS test). The power spectrum analysis showed that the residual small amplitude whisking shifted to lower frequencies (Fig. 1o). The effect of TeLC-silencing was permanent.
We further validated these results by acutely silencing vIRtPV neurons with the expression of an anion-conducting channelrhodopsin (GtACR)19 (Extended Data Fig. 2, n = 6). Optogenetic silencing suppressed rhythmic whisking during the light ON period and resulted in small amplitude whisking, although the effect was weaker than that of TeLC-silencing, likely due to the inefficiency of short duration optogenetic silencing (Extended Data Fig. 2f,g). Collectively, these results identify vIRtPV neurons as the essential constituents for mediating rhythmic retraction.
Excitatory premotor neurons in vIRt (vIRtvGlut2) do not contribute to whisking rhythm generation
Since a subset of premotor vIRt neurons expressed vGlut2 (Extended Data Fig. 1b), we sought to determine whether they also contribute to rhythmic whisking. A viral-genetic intersectional method was used to specifically label and manipulate vGlut2+ premotor neurons in the vIRt, hereafter referred to as vIRtvGlut2. Briefly, in vGlut2-Cre mice, we injected retrograde lentivirus RG-LV-FlpO into the vFMN to infect vibrissa premotor axon terminals, resulting in co-expression of both Cre and FlpO recombinases in vGlut2+ premotor neurons. Subsequently, we injected Cre- and Flp- co-dependent AAV (Con/Fon vectors) into the vIRt to express EYFP or optogenetic silencer halorhodopsin (NpHR3.3) in vIRtvGlut2 neurons (Extended Data Fig. 1lc, e)20,21. We found that vIRtvGlut2 neurons preferentially projected to the dorsolateral and lateral subnuclei of FMN, where motoneurons innervating the vibrissa retractor muscle nasolabialis/maxillolabialis and extrinsic protractor/nares dilator muscle nasolabialis profundus reside, respectively (n = 3, Extended Data Fig. 1d’, d”). Both muscles contract at the onset of inspiration but not during intervening whisks5. In contrast, vIRtvGlut2 neurons showed less innervation of the ventrolateral subnucleus of FMN, which contains motoneurons that drive vibrissa protraction (n = 3, Extended Data Fig. 1d”’). NpHR3.3 mediated optogenetic silencing of vIRtvGlut2 neurons during whisking failed to alter either the amplitude, the midpoint, or the whisking frequency as judged from the peak of the power spectrum (n = 6, Extended Data Fig. 1h–k). Together, these results suggest that vIRtvGlut2 neurons are not involved in whisking rhythm generation. Instead, they may play a role in coordinating vibrissal pad movement with sniffing.
vIRtPV neurons switch from tonic to rhythmic bursting activity pattern at the onset of whisking
In view of the drastic impact of silencing vIRtPV neurons on rhythmic whisking (Fig. 1j–o), we recorded the in vivo activity of vIRtPV neurons in awake whisking mice to assess whether vIRtPV premotor neurons exhibit rhythmic activity during whisking, a required property of central oscillator cells. We first established a preparation in which a head-fixed mouse ran on a wheel while we performed extracellular multi-electrode recordings from the vIRt region, with simultaneous tracking of vibrissa movements. For some recordings, we employed our Split-Cre viral-genetic strategy to express Channelrhodopsin-2 (ChR2) or red-shifted opsin ChRmine22 in vIRtPV neurons (Fig. 2a, Extended data Fig. 3). This allowed us to perform “opto-tagged” recordings of vIRtPV neurons during natural whisking behaviors (Fig. 2b,c). In-vivo identification of vIRtPV neurons was achieved by delivering pulses of 473nm light over the facial nucleus (or 635nm light through the ear canal, for ChRmine experiments), while searching for neurons with reliable short-latency, low jitter, light-induced antidromic spikes (Fig. 2b, e–g). We recorded eight well-isolated and opto-tagged confirmed vIRtPV units from seven mice.
Fig. 2. Response characteristics of vIRtPV premotor neurons during whisking behavior.
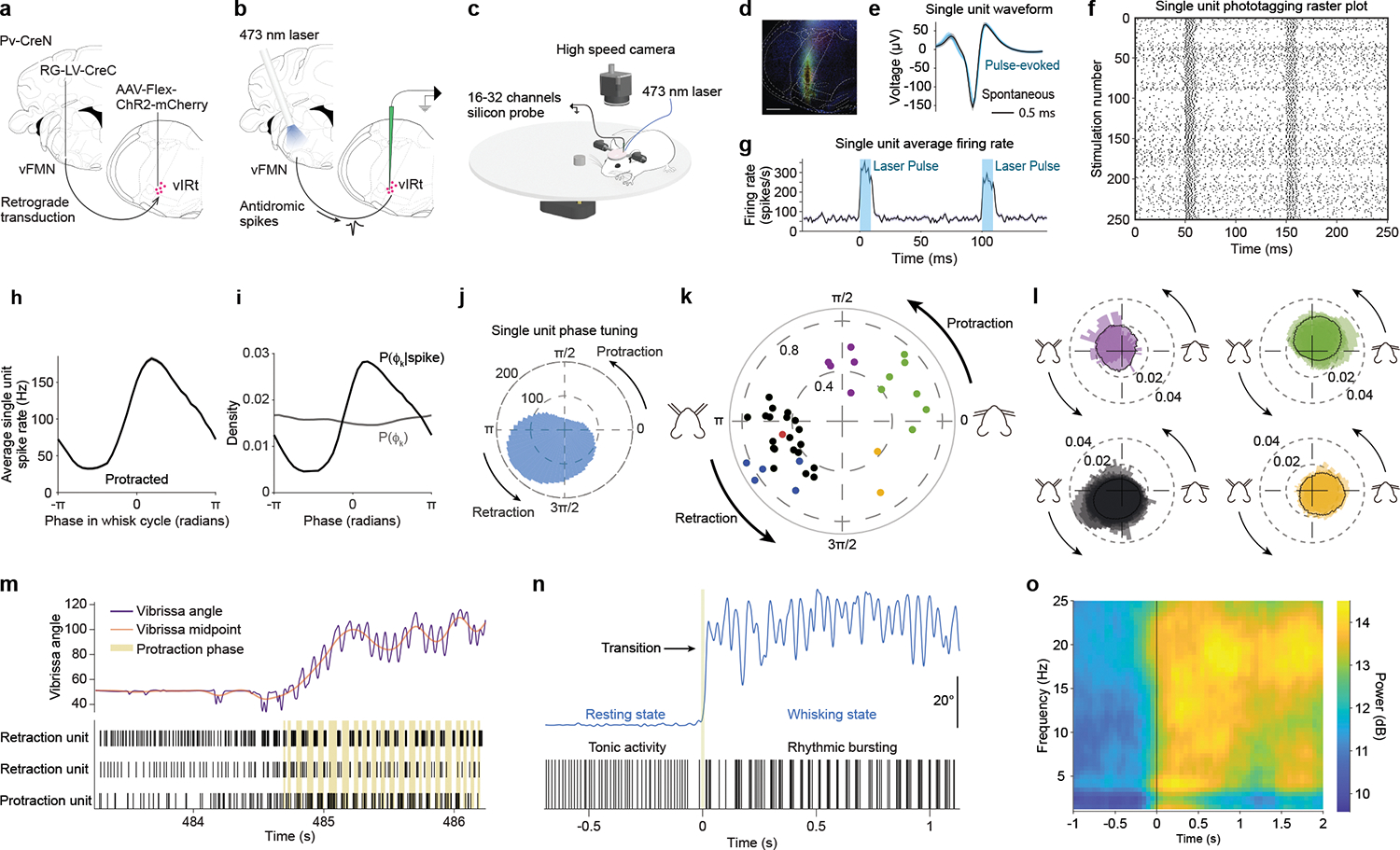
a, Viral-genetic strategy for expressing ChR2 in vIRtPV neurons. b, Opto-tagging strategy. c, Recording and behavioral setup. d, Histological verification of ChR2 expression and recording location. e, f, g, Spike waveform, raster plot, and average laser stimulation response of a opto-tagged single unit. h, i, j, Whisking phase tuning of a opto-tagged single unit. h, Average spike rate across whisking phases. i, Probability of a whisking phase to occur for a given spiking event (black) and probability of each whisking phase (grey). j, Phase tuning for that cell, in polar coordinates. Magnitude in spikes per second. k, Peak magnitude and phase of spike/phase coherence for all tuned cells. Color coding: Black/blue/red, Retraction units; Yellow, Mid-Retraction units; Green, Protraction units; Purple, Mid-Protraction units. Opto-tagged units are shown in blue or red (ChR2 or ChRmine-expressing cells, respectively). l, Normalized average firing rate of Retraction (bottom left), Mid-Retraction (bottom right), Protraction (top right) and Mid-Protraction (top left) groups, expressed as a probability value for each phase bin. Color coding as in k. m, Transition from tonic to rhythmic activity pattern. Top: vibrissa angle trace (purple), overlayed with vibrissa midpoint trace (red). Bottom: raster plot for two retraction and one protraction units. Beige bands mark each protraction phase. n, A high magnification representation of the tonic to rhythmic transition that occurs for a retraction-tuned unit going from resting state to whisking movements. o, Mean time-frequency spiking spectrum for all retraction units, aligned to transition from resting to whisking.
We analyzed both opto-tagged (n=8) and untagged (n=54) whisking-related vIRt neurons as one group; untagged neurons include recordings from mice with no or insufficient opsin expression. Cells were classified according to their tuning with respect to the preferred whisking phase (Fig. 2h–j). Among cells with whisking related responses, a large population (n = 40) was found with a rhythmic bursting pattern time-locked to whisking (Fig. 2k,l). Most cells (26/40 or 65 %) were tuned to the retraction phase with a sharply elevated firing rate that often covered a quadrant of the whisking cycle, starting late in the protraction phase and peaking mid-way through the retraction phase (Fig. 2l, bottom left group). In addition, these cells typically displayed the highest level of spike-phase spectral coherence early in the retraction phase (|C| = 0.55 ± 0.14, mean ± SD, Fig. 2k, black markers, Fig. 2m retraction units).
Remarkably, six of the eight opto-tagged vIRt neurons were tuned to the whisking phase, and all these vIRtPV cells belonged to the “retraction” group (blue or red dots in Fig. 2k, Supplemental video 2), with high spike/phase coherence (|C| = 0.71 ± 0.15). The results suggest that most of vIRtPV neurons are indeed spiking rhythmically and are selectively active during the retraction phase, consistent with these cells playing a central role in generating rhythmic vibrissa retraction7. A smaller group of cells (7/40 or 17.5 % of phase tuned cells) responded during the protraction phase (|C| = 0.60 ± 0.11, Fig. 2k, green markers, Fig. 2m, protraction units) while a minority had responses that occurred at other periods of the whisking cycle (5/40, purple dots, 12.5 % mid-protraction, |C| = 0.45 ± 0.13, 2/40, yellow dots, 5 % mid-late retraction, |C| = 0.50 ± 0.19, Fig. 2k). An unexpected finding was that two out of the eight opto-tagged vIRtPV neurons and ten untagged neurons displayed slower rhythms, with bursting bouts occurring across multiple whisking cycles (Extended Data Fig. 4a,b). These are potentially linked to other orofacial rhythm generators in the brainstem.
Interestingly, the firing patterns of all rhythmic vIRt cells, i.e., both retraction and protraction units and including the six opto-tagged vIRtPV retraction neurons, showed asynchronous tonic firing in non-whisking periods of rest. Rhythmic bursting only appeared at the onset of whisking (Fig. 2m, n, o). This transition is also confirmed by spectral analysis of firing frequencies, which showed that power around 15 to 20 Hz emerged during whisking for units tuned to the whisking phase (Fig. 2o, Extended Data Fig. 4c,e), while “slow” units showed a transition to a slower frequency band, similar to the breathing rhythm (Extended Data Fig. 4d,f). Thus neurons that spike in rhythmic bursts during whisking are seen to transition from tonic firing during non-whisking rest periods to rhythmic bursting at the onset of whisking (Fig. 2m,n). This result strongly suggests that rhythmic firing is not an intrinsic property of vIRtPV neurons, but rather a network property that is enabled, perhaps by a top-down whisking command that shifts the operating point of the network23. This idea was further supported by in vitro recordings of identified vIRtPV neurons in slices. Because vIRtPV cells only express PV in the adult stage, our split-Cre based strategy could only label these cells in adult. Recording from adult brainstem slices is technically highly challenging, we therefore performed cell-attached recording from labeled vIRtPV cells in slices (n=5, Extended Data Fig. 5). Bath application of high potassium (9mM, see Methods), to elevate neuronal excitability, induced vIRPV neurons to burst, but such bursting was not rhythmic, likely due to the severed-network connectivity in slices that was not sufficient to support rhythm generation.
Identifying presynaptic inputs and drivers to vIRtPV oscillator neurons
We next mapped the presynaptic inputs to vIRtPV neurons as a first step to dissect the circuit-level mechanisms that underlie rhythmic bursting generation during the transition from resting to whisking. We made use of the same viral-genetic split-Cre strategy to target vIRtPV neurons with AAV helper viruses, followed by injection of pseudotyped monosynaptic rabies virus (Fig. 3a, n = 3)24–26, to reveal the major sources of presynaptic inputs to vIRtPV neurons (hereafter referred to as pre-vIRtPV cells, Fig. 3c, d, f, and Extended Data Fig. 6). Registration and 3D reconstruction of pre-vIRtPV neurons in Allen Mouse Brain CCF (Allen CCF)27 revealed consistent labeling patterns across samples (Fig. 3d–e, and quantifications shown in 3f). vIRtPV neurons receive bilateral inputs (with ipsilateral dominance) from the IRt, the parvicellular reticular nucleus (PCRt), the gigantocellular reticular formation (Gi), the ventral and dorsal medullary reticular formation (MdV, MdD), and the midbrain reticular nucleus (MRN) located dorsal to the red nucleus (RN) (Fig. 3c, f, Extended Data Fig. 6a). Pre-vIRtPV neurons with a predominantly ipsilateral location were observed in vIRt, preBötC, lateral paragigantocellular nucleus (LPGi), deep cerebellar nuclei (DCN), and a small number in the spinal trigeminal nuclei (SpVO, SpVI, and SpVC) (Fig. 3c, f). vIRtPV neurons also receive inputs from contralateral superior colliculus (SC) and the motor cortex (Fig. 3c, f, Extended Data Fig. 6). Interestingly, MdD, DCN, SC, and MRN were previously identified as premotor areas for vibrissal motoneurons10,28. Co-registration of pre-vIRtPV neurons and vibrissa premotor neurons in the Allen CCF further confirmed that those neurons occupy the same spatial locations within DCN, SC and MRN, suggesting that these areas may act at both premotor and pre-premotor functional levels to coordinate the activity of motoneurons and the whisking oscillator (Fig. 3c, Extended Data Fig. 7)10,28,29. Consistent with this idea, axonal collaterals from labeled pre-vIRtPV neurons were also observed to densely innervate vFMN (Extended Data Fig. 6a). We further characterized neurotransmitter phenotypes of pre-vIRtPV neurons using HCR RNA-FISH (Fig. 3g). We found that SC (vGlut2/ΔG-GFP, 95.4 ± 3.9 % n = 4), MRN (vGlut2/ΔG-GFP 95.4 % ± 3.4%, n = 4) and DCN (vGlut2/ΔG-GFP 100 %, n = 4) pre-vIRtPV neurons were excitatory neurons, while preBötC (vGat/ΔG-GFP, 86.3 ± 1.8 % n = 3), and MdD (vGat/ΔG-GFP, 96.6 ± 1.7 % n = 3) pre-vIRtPV neurons were mostly inhibitory neurons, and regions of dIRt, Gi, and PCRt provide mixed excitatory/inhibitory inputs to vIRtPV neurons (Fig. 3g, see also Extended Data Fig. 6).
Fig. 3. Characterizing presynaptic inputs to vIRtPV neurons (pre-vIRtPV cells).
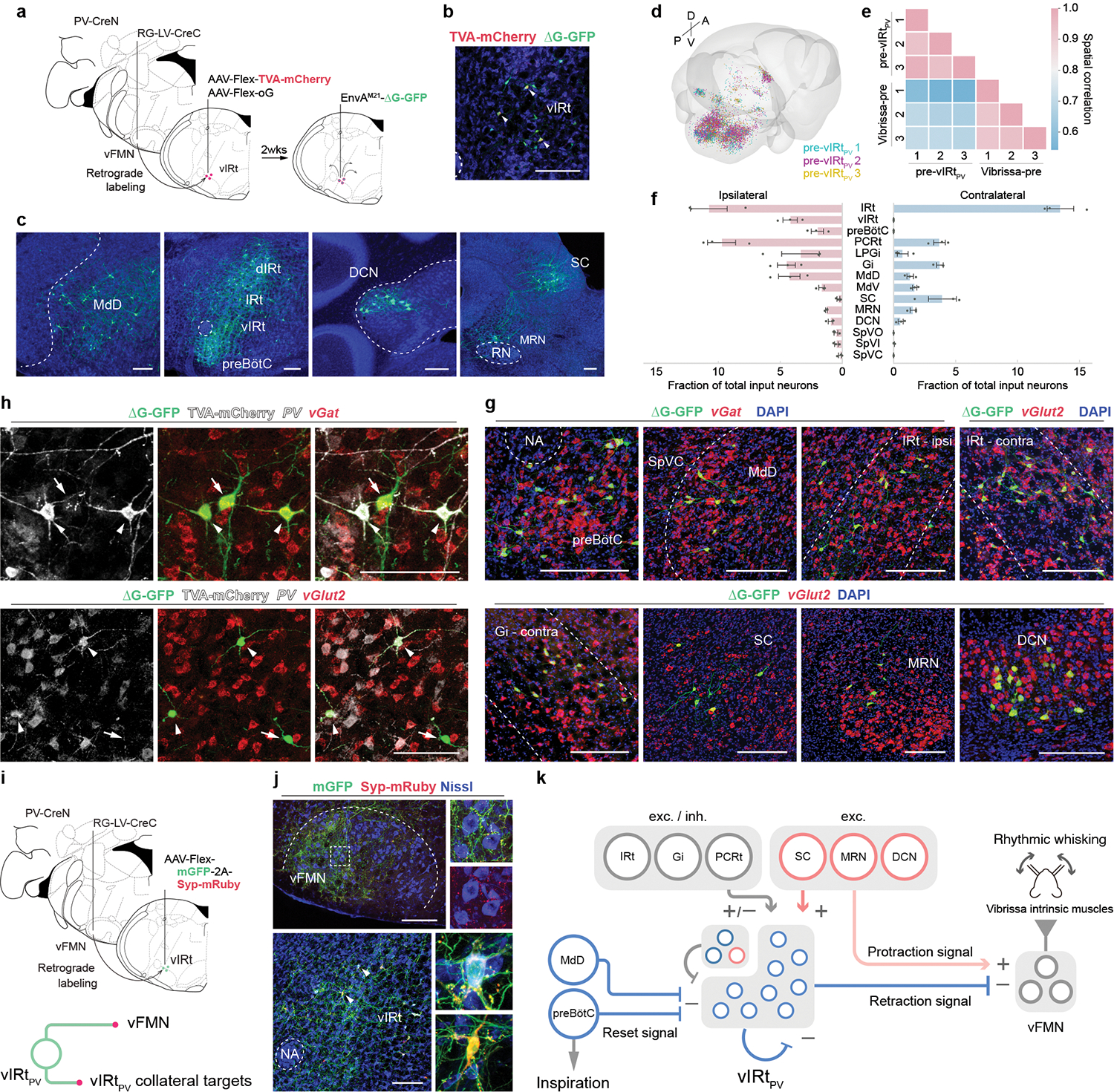
a, Schematic of monosynaptic rabies virus tracing from vIRtPV neurons. b, TVA-mCherry/GFP double-positive source cells (yellow, arrowheads), and GFP single-positive pre-vIRtPV cells in vIRt. c–f, Distribution of pre-vIRtPV neurons. c, Representative images of pre- vIRtPV neurons in the ipsilateral MdD, dIRt, IRt, DCN, preBötC, and contralateral SC, and MRN. d, Reconstructed pre-vIRtPV circuits in Allen CCF (3 mice). e, Cross-correlation analysis of pre-vIRtPV cell positions across animals; pre-vIRtPV neurons (3 mice), and vibrissa premotor neurons (3 mice) as a comparison. f, Quantification of pre-vIRtPV cell number in brain areas. Numbers are normalized by the total number of input neurons. Data are mean ± s.e.m. (n = 3 mice). g, Neurotransmitter characterization of pre- vIRtPV neurons. vGat and vGlut2 mRNA were detected by HCR RNA-FISH. See Extended Data Fig. 6 for quantification. h, Molecular characterization of pre-vIRtPV neurons within vIRt. PV, vGat, vGlut2 mRNA, and TVA-mCherry protein were detected by HCR RNA-FISH and HCR-Immunohistochemistry, respectively. Pre- vIRtPV neurons (arrows) are distinguished from source vIRtPV neurons (arrowheads) by their lack of gray-color labeled axons/dendrites. i, Top, Strategy to identify vIRtPV projections. Bottom, mGFP and Syp-mRuby label somata/axons, and axon terminals, respectively. j, Projections of vIRtPV neurons. Top, axonal projections in vFMN. Bottom, synaptic connections within vIRtPV neurons. Insets, magnified images of the boxed area and the representative vIRtPV neurons receiving mRuby-positive synaptic terminals (arrowheads). Note that strong mGFP-2A-Syp-mRuby expression causes mRuby aggregates in somata. k, Schematic summarizing the presynaptic inputs to vIRtPV. The upper left corner of vIRt indicate PV-/vGat+ (deep blue) and vGlut2+ (red) vIRt neurons. SC, MRN, and DCN provide excitatory inputs to vIRtPV neurons and presumably send simultaneous excitatory protraction signals to vFMN (translucent red). Scale bars, 200 μm (b, c, g, j), 100 μm (h). Sections were counterstained with Neurotrace blue (b, c, j) or DAPI (g, h).
In addition to the above mentioned pre-vIRtPV neurons, we noticed GFP-single-positive cells in the vIRt region near the source vIRtPV neurons; recall that source cells are GFP and TVA-mCherry double-positive (Fig. 3b, c). This suggested that vIRtPV neurons may receive presynaptic inputs from other vIRtPV neurons, i.e., vIRtPV-vIRtPV synapses as a potential source of recurrent connectivity. To gain further insight into this local circuit, we characterized the molecular identity of pre-vIRtPV neurons within vIRt (Fig. 3h). We found that the majority of local presynaptic neurons (GFP+/TVA-mCherry− cells in vIRt) were inhibitory neurons (PV+/vGat+, 49.9 ± 6.4%, n = 5; PV−/vGat+, 22.4 ± 4.6%, n = 5), whereas 15.5 ± 2.7% (n = 4) pre-vIRtPV local inputs were PV−/vGlut2+ excitatory neurons. Note that 67.3 ± 8.3% (n = 5) of labeled cells in vIRt were source cells, suggesting vIRtPV neurons may form dense recurrent inhibitory network within vIRt. To further examine this possibility, we sparsely labeled vIRtPV neurons with mGFP-synaptophysin-mRuby (Fig. 3i)30. In this way, the cell bodies and axons of vIRtPV neurons were labeled by membrane-GFP, while the presynaptic boutons on axon terminals from labeled neurons were visualized by mRuby. Dense mGFP-positive vIRtPV axons were observed in the lateral vFMN as expected, further supporting the labeling specificity (Fig. 3j). Interestingly, numerous synaptophysin-mRuby-positive puncta were observed in vIRt region on both GFP+ (Fig. 3j) and GFP− cells.
Collectively, these results strongly suggest that vIRtPV neurons form inhibitory synapses onto other vIRtPV neurons, likely resulting in a recurrent inhibitory network (Fig. 3j). We do not know the nature of GFP− (vGat+) vIRt cells that receive vIRtPV synapses. Since we and previous recordings in ketamine/xylazine anesthetized mice with kainic-acid induced whisking5,7 found some vIRt rhythmic neurons firing in the protraction phase, one possibility is that GFP− vIRt (vGat+) cells are protraction units, and that vIRtPV retraction neurons form reciprocal inhibitory connections with them. In summary, vIRtPV neurons receive descending excitatory presynaptic inputs from the contralateral SC, MRN, motor cortex, ipsilateral DCN, and inhibitory inputs from the ipsilateral preBötC, MdD and from other vIRtPV and perhaps non-PV vIRt neurons, as well as mixed (excitatory and inhibitory) inputs from IRt and Gi (Fig. 3g, Extended Data Figs. 6 and 7).
Ablating inhibitory post-synapses in vIRtPV neurons abolished high frequency rhythmic whisking
Previous studies have established the prominent role of recurrent inhibition in rhythm generation in several systems1,31–34. Computational modeling further described a key role of inhibition in vIRt rhythm generation (Golomb et al., submitted). To test the causal role of inhibition in vIRtPV neurons rhythmogenesis, we used a strategy to specifically ablate the inhibitory post-synapses in vIRtPV neurons. Using the viral-genetic split-Cre strategy, we expressed a Cre-dependent GFE3-EGFP construct in vIRtPV neurons (Fig. 4a). GFE3 is a Gephyrin-specific ubiquitin ligase that targets the key inhibitory postsynaptic scaffolding protein Gephyrin for degradation, which in turn prevents the clustering of GABAA and glycine receptors35. Neurons expressing GFE3 were previously shown to receive significantly diminished phasic synaptic inhibition35. This manipulation would in principle block all phasic inhibitory inputs to vIRtPV, including those from preBötC and from other vIRtPV neurons (Fig. 4a). Post hoc histological assessment with anti-Gephyrin antibody in vIRtPV-GFE3 animals ascertained the significant reduction of Gephyrin signal in GFP-positive cells compared with GFP-negative neighboring cells (normalized density of gephyrin puncta, in control neighboring neurons is 0.44 ± 0.01, n = 76 cells, n = 4 mice, and in GFE3-GFP 0.14 ± 0.007, n = 78 cells, n = 4 mice, p = 0.0286, KS test, Fig. 4b, c).
Fig. 4. Elimination of inhibitory synaptic inputs onto vIRtPV neurons impairs the generation of intervening whisking and abolishes their rhythmic bursting.
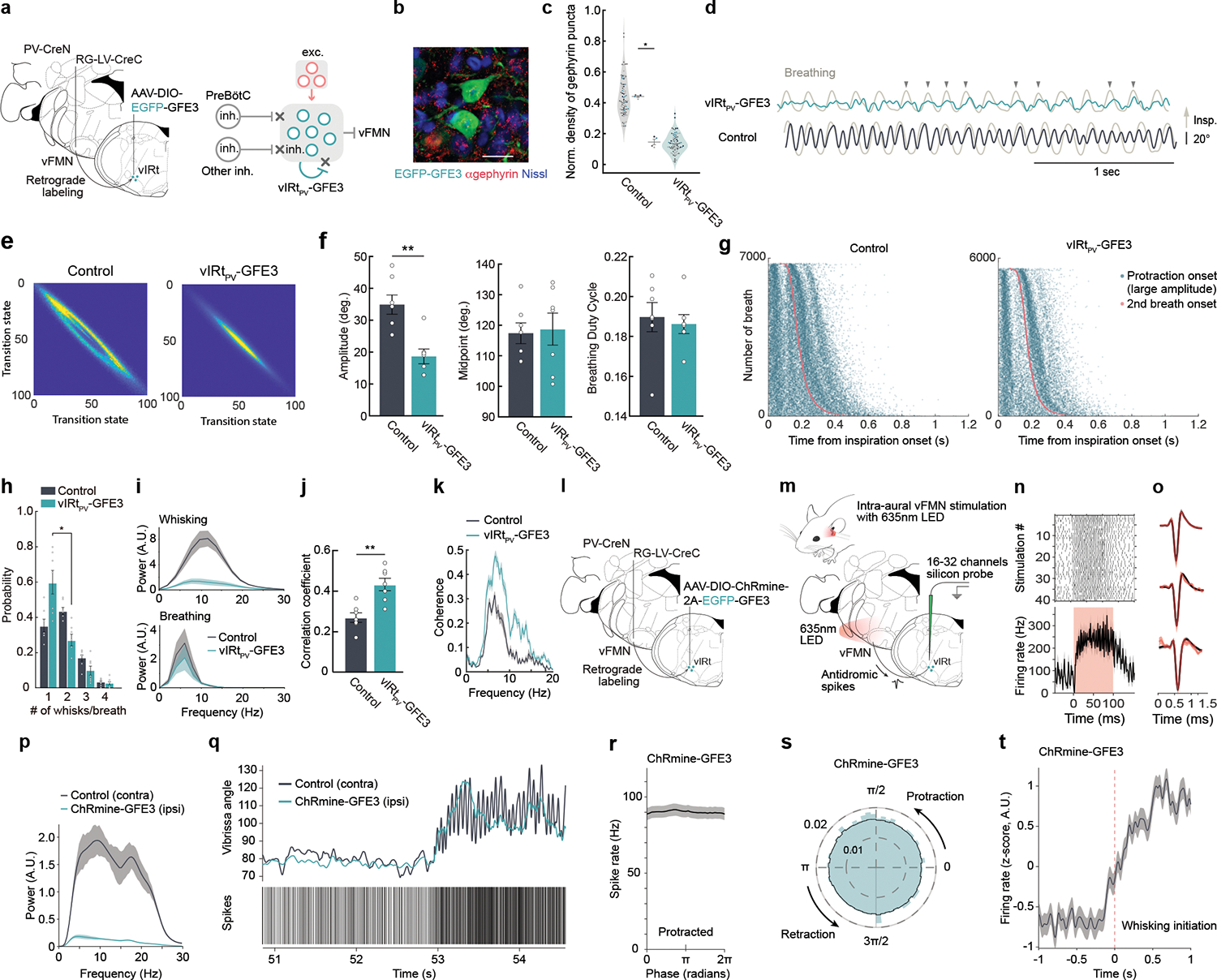
a, Schematic of the GFE3 (green) post-inhibitory synapse ablation experiment. b, Representative image of anti-Gephyrin staining in vIRtPV -GFE3 neurons. Scale bar, 20 μm. c, Quantification of normalized density of Gephyrin puncta (Control/neighboring cells, 0.44 ± 0.006, n = 4 mice, 76 cells; vIRtPV-GFE3, 0.15 ± 0.018, n= 4 mice, 78 cells). Data from individual neurons (dots), color-coded by mice. d, Vibrissa angle and breathing (light-gray) traces from vIRtPV-GFE3 (green) and control (black) mice. Protraction and inspiration are up. Downward arrows, correlated breathing/whisking events. e, Quantification of whisking regularity using state transition analysis. f, Quantification of whisking amplitude (Control, 34.9 ± 3.0°, n=7; vIRtPV-GFE3, 18.7 ± 2.3°, n=7), midpoint (Control, 117.4 ± 3.4°, n =7; vIRtPV-GFE3, 104.1 ± 17. 8°, n =7), and breathing duty cycle (Control, 0.19 ± 0.007, n=7; vIRtPV-GFE3, 0.19 ± 0.004, n=7). g, Raster plots of protraction onset (teal) relative to inspiration onset times (red). Data ordered by breath duration. h, Quantification of the number of whisks per one breathing cycle. i, Power spectrum of whisking and breathing frequency. j, Correlation coefficient of breathing and whisking (Control, 0.27 ± 0.02, n=7; vIRtPV-GFE3, 0.43 ± 0.03, n=7). k, Coherence between whisking and breathing. l,m, Schematics of ChRmine-GFE3 opto-tagging experiments. n, Raster plot and average stimulation response of a opto-tagged vIRtPV-ChRmine-GFE3 single unit. o, Average spike waveforms, before (black) and during (red) light stimulation periods for vIRtPV-ChRmine-GFE3 single units (n=3). p, Power spectrum of ipsilateral (teal) and contralateral (black) whisking frequency for ChRmine-GFE3 mice. q, Unit activity during transition from resting to whisking state. Top: vibrissa angle traces for ipsilateral (teal) and contralateral (black) C2 vibrissae. Bottom: Spike times of a opto-tagged vIRtPV-ChRmine-GFE3 neuron. r, Average spike rate across whisking phases (derived from contralateral vibrissae) for all opto-tagged vIRtPV-ChRmine-GFE3 neurons. s, Same as r, normalized, in polar coordinates. t, z-scored average spike rate of vIRtPV-ChRmine-GFE3 neurons around the time of whisking initiation. Shaded areas are mean ± s.e.m. (i, k, n, o, p, r, t). * P ≤ 0.05. ** P ≤ 0.01, KS test (c, f, i, j), Wilcoxon signed-rank test (g).
After at least three weeks of GFE3 expression, we tracked whisking behavior of subject mice. vIRtPV-GFE3 animals were able to initiate whisking, but their vibrissa movement showed several abnormalities (Fig. 4d–i). First, whisking in air from vIRtPV-GFE3 animals was highly irregular compared to the near sinusoidal pattern observed in control animals. In normal animals, the amplitude of individual whisks does not differ significantly during the whisking epoch (Fig. 4d, bottom trace). In contrast, in vIRtPV-GFE3 animals, larger amplitude whisks were mixed with multiple small-amplitude jitters (Fig. 4d, upper trace, arrowheads). Analysis of whisking regularity with state transition matrix analysis yielded a near elliptical pattern in control mice, indicative of consistent whisking amplitude and frequency from whisk-to-whisk (Fig. 4e, left panel). In contrast, this analysis in vIRtPV-GFE3 mice showed a collapsed eclipse, resulting from the poor relation between current and future vibrissa positions, i.e., irregular whisking (Fig. 4e, right panel). In addition, the overall whisking amplitude was significantly smaller in vIRtPV-GFE3 animals (control, 34.9 ± 3.0°, n = 7; vIRtPV-GFE3, 18.7 ± 2.3°, n = 7, p = 0.0047, KS test, Fig. 4f). All told, perturbing synaptic inhibition onto vIRtPV neurons markedly impaired the generation of regular patterns of synchronous rhythm.
In contrast to TeLC silencing of vIRtPV, that resulted in increased protraction (Fig. 1n), the whisking midpoint was not affected in vIRtPV-GFE3 animals (control, 117.4 ± 3.4°, n = 7; vIRtPV-GFE3, 104.1 ± 17. 8°, n = 7, p = 0.58, KS test, Fig. 4f). One might expect that dis-inhibition of vIRtPV-GFE3 neurons result in stronger net inhibitory output from these cells, and thus larger retraction amplitude, this scenario is only valid if the output of vIRtPV-GFE3 neurons were synchronous. The observed unaffected midpoint but significantly smaller amplitude of whisking (Fig. 4f) suggest that vIRtPV-GFE3 neurons had greater, but desynchronized inhibitory outputs to vFMN neurons. These findings further support the model that lateral, vIRtPV-to-vIRtPV inhibitory synaptic connections play a key role in generating synchronous rhythmic bursting.
A further point is that the frequency of whisking was significantly reduced in vIRtPV-GFE3 animals, and the peak frequency was shifted from around 13 Hz (gray line in Fig. 4i) in control animals to around 6 – 7 Hz in vIRtPV-GFE3 animals (green line in Fig. 4i). The later value is similar to the frequency of breathing (Fig. 4i). Raster plots of protraction and inspiration onset times analysis showed protraction onsets of whisks largely coincided with the inspiration onsets (red line in Fig. 4g) in vIRtPV-GFE3 mice, and there was an obvious lack of intervening whisking, i.e., whisks between breaths, compared to control mice. Indeed, a 1:1 breathing versus whisking ratio was dominant in vIRtPV-GFE3 animals while a 1:2 ratio was dominant in control animals (Fig. 4h). Quantitatively, this led to a correlation of whisking and breathing for vIRtPV-GFE3 animals that is large only in the breathing frequency range (Fig. 4j; control, 0.27 ± 0.02, n = 7; vIRtPV-GFE3, 0.43 ± 0.03, n = 7, p = 0.0082, KS test, Fig. 4k). These data imply that the whisking rhythm in vIRtPV-GFE3 mice was mainly controlled by the slower breathing rhythm, consistent with previous finding that the nasolabialis profundus muscle, which is structurally linked to the mystacial pad, and extrinsic muscles that retract the mystacial pad both receive inputs from breathing oscillators5,7
Synaptic inhibition is required for rhythmic bursting in vIRtPV neurons
Finally, we performed in vivo recordings of opto-tagged vIRtPV-GFE3 neurons to determine whether the whisking phenotypes we observed in vIRtPV-GFE3 mice indeed resulted from loss of rhythmic bursting in these neurons upon blocking synaptic inhibition. To achieve this, we needed to co-express ChR2 and GFE3 in vIRtPV cells. We generated an AAV vector that contains both ChRmine and GFE3: AAV-hSyn-DIO-ChRmine-T2A-EGFP-GFE3. ChRmine made opto-tagged recordings simpler as we could illuminate through the ear canal to activate ChRmine-expressing vIRtPV axons that project to the vFMN (which reside in close proximity to the inner ear) using a 635nm LED22,36 (Fig. 4l–o). Indeed, when we expressed ChRmine alone in vIRtPV neurons (no GFE3 in this case), we recorded a ChRmine-tagged neuron through intra-ear illumination, and this neuron exhibited rhythmic bursting during the retraction phase, similar to other ChR2-tagged neurons (Fig. 2k, Extended Data Fig. 3). When we expressed both ChRmine and GFE3 in vIRtPV neurons and performed in vivo opto-tagged recording, we found, as expected, that the side expressing ChRmine-GFE3 exhibited irregular small amplitude whisking compared to the control side (Fig. 4p,q, Extended Data Fig. 8). Importantly, vIRtPV-ChRmine-GFE3 neurons were no longer capable of firing rhythmic bursts (Fig. 4q–s). Instead, they only tonically increased their firing rates without any phase tuning when mice transitioned from stationary to active whisking (Fig. 4q–t, whisking phase was inferred based on the intact contralateral side). These results provide in vivo evidence that (recurrent) synaptic inhibition is essential for whisking rhythm generation by the vIRtPV oscillator neurons.
DISCUSSION
We identified PV-expressing inhibitory vibrissa premotor neurons in vIRt, vIRtPV, as whisking oscillator neurons and further revealed the putative circuit that underlies synchronous rhythm generation. In vivo recordings in awake mice and in-vitro slice recordings showed that vIRtPV oscillator cells are not intrinsic pace-makers. At rest, vIRtPV neurons have non-synchronized tonic activity (schematic model in Extended Data Fig. 9a; Fig. 2m,n and Extended Data Fig. 5a). At the initiation of whisking, descending tonic excitatory signals activate the vibrissa protractor motoneurons (vFMN), and at the same time excite vIRtPV neurons, which in turn increase recurrent inhibition within the vIRtPV population. Descending excitation may, conceivably, also activate other vIRt neurons, including vIRt-protraction cells. The combination of increased excitation and recurrent inhibition results in an elevation of inhibitory outputs from vIRtPV and other vIRt inhibitory neurons and, importantly, a transition of vIRtPV cells from tonic firing to a synchronous rhythmic bursting mode. Consequently, the periodic inhibitory bursts from vIRtPV suppress the activity of the facial motoneurons. Phasic inhibition from vIRtPV cells results in phasic vibrissa retraction, hence inducing rhythmic whisking movements7 (Extended Data Fig. 9b). Past work showed that during whisking, breathing can reset whisking5. Here we found that inhibitory neurons located in the inspiration oscillator preBötC provide monosynaptic inputs onto vIRtPV neurons. Thus, breathing likely resets whisking through the inhibition of the vIRtPV rhythmic inhibitory signal and the dual inhibition results in the well-documented phenomenon of near-synchronous sniffing and whisking4,5. In addition, breathing oscillators for both inspiration and expiration control nose dilation and mystacial pad retraction motoneurons, which also help to couple the whisking rhythm to breathing5,7 (Extended Data Fig. 9b).
Our results from two loss-of-function experiments further make the case that vIRtPV neurons are required for rhythmic whisking. When we expressed TeLC in vIRtPV neurons, their rhythmic inhibitory outputs were blocked, resulting in persistent vibrissa protraction caused by the tonic excitation of vFMN motoneurons (Extended Data Fig. 9c). In this case, the recruitment of extrinsic retractor muscles by the breathing oscillators cannot overcome the strong tonic activity of intrinsic protractor muscles, resulting in very limited vibrissa movement (Extended Data Fig. 9c). When we blocked synaptic inhibitory inputs onto vIRtPV neurons, through the expression of GFE3, their ability to generate rhythmic bursts in response to descending whisking commands was abolished (Fig. 4). In vIRtPV-GFE3 mice, during whisking periods, the increased descending excitation only resulted on increased tonic firing rate, illustrating the crucial role of external and recurrent inhibitory inputs to the generation of rhythmic bursting. Asynchronous firing of vIRtPV-GFE3 neurons lead to the loss of normal whisking rhythm. Stronger and asynchronous inhibitory outputs onto vFMN protractor motoneurons limited whisking to small amplitude and irregular movements, and less protraction than under TeLC synaptic blocking (Extended Data Fig. 9d). Under the GFE3 condition, as the coupling between expiratory oscillators and mystacial pad retraction motoneurons remained intact, the residual whisking followed the breathing rhythm (Extended Data Fig. 9d). In summary, the whisking oscillator consists of inhibitory neurons that individually, at rest, do not have rhythmogenic property. Rhythm is a network property that emerges with the onset of whisking behavior and critically depends on recurrent synaptic inhibition.
There are many different schemes that nature has used to generate rhythms for the control of movement1,2, even as a result of network phenomena23,37,38. Notably, oscillators for breathing and locomotion in mammals are known to generate rhythmicity based on excitatory interactions, and the blockade of inhibition does not abolish rhythmicity. Thus, inhibition is not obligatory for rhythmogenesis in the known mammalian oscillators1,37,39,40. The crustacean stomatogastric ganglion oscillator contains primarily inhibitory interactions, but the rhythm is produced by pacemakers cells1. By contrast, the whisking vIRt oscillator is arguably the first identified network inhibition-based rhythm generator in mammals. This oscillator produces its own rhythm, but can be rapidly entrained by external, phase-setting inputs, in this case from the preBötC. We further suggest that a network oscillator, as opposed to an oscillator formed from intrinsically rhythmic neurons, may allow the oscillator to rapidly change frequencies and amplitudes in response to sensory feedback or a top-down command41, consistent with the three-fold mode of whisking seen in rodents42. Thus, the vibrissa oscillator can enable highly adaptive motor behaviors beyond whisking per se.
METHODS
Animals
All animal handling and experiment procedures were approved by The Duke University Institutional Animal Care and Use Committee and MIT Committee for Animal Care. Male and female C57B/L6, Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J (Ai14, JAX #007914, Jackson Laboratory), Parvalbumin-CreN knock-in (PV-CreN)12 and vGlut2-Cre knock-in (vGlut2-Cre, JAX #028863) were used in this study.
DNA constructs
pAAV-EF1a-DIO-GtACR2-EYFP and pAAV-hSyn-DIO-ChRmine-T2A-EGFP-GFE3 were constructed by standard molecular cloning procedures using enzymes from New England Biolabs. To generate pAAV-EF1a-DIO-GtACR2-EYFP, NheI and AscI sites were added to the 5’ and 3’ ends of GtACR2-EYFP fragment, respectively, by PCR amplification from pFUGW-hGtACR2-EYFP19 (Addgene #67877). This fragment was inserted into AscI-NheI site of pAAV-Ef1a-DIO-EYFP (Addgene #27056).
To generate pAAV-hSyn-DIO-ChRmine-T2A-EGFP-GFE3, the custom DNA fragment containing a BamHI-double LoxP-flanked inverted ChRmine-T2A-EGFP-GFE3-NcoI was synthesized by Epoch Life Science, Inc. This BamHI-NcoI fragment was inserted into BamHI-NcoI site of pAAV-hSyn-DIO-EGFP (Addgene #50457).
Viruses
AAVs used in this study were AAV2retro-CAG-Cre43 (Boston Children’s Hospital Viral Core), AAV2/8-CAG-Flex-TVA-mCherry25 (Addgene #48332), AAV2/8-CAG-Flex-oG24 (Addgene #74292, Duke Viral Vector Core), AAV2/8-hSyn-FLEX-TeLC-P2A-EYFP-WPRE17 (Addgene #135391), AAV2/8-Ef1a-DIO-hChR2(E123A)-mCherry44 (Addgene #35508), AAV2/8-EF1a-DIO-GtACR2-EYFP19,AAV2/1-hSyn-FLEx-mGFP-2A-Synaptophysin-mRuby30 (Addgene #71760), AAV2/8-Ef1a-DIO_EGFP-GFE335 (Addgene #79871), AAV2/8-CAG-FLEX-EGFP-WPRE (Addgene, #51502), AAV2/8-hSyn-DIO-ChRmine-T2A-EGFP-GFE3 (This paper), AAV2/8-hSyn-Con/Fon-EYFP20 (Addgene, #55650), and AAV2/8-nEF-Con/Fon-NpHR3.3-EYFP21 (Addgene #137152). RG-LV-CreC45, RG-LV-FlpO46, and EnvA(M21)-RV-ΔG-GFP (also called CANE-ΔG-RV) were produced as previously described47.
Surgery
Monosynaptic rabies virus tracing for vFMN premotor neurons in adult mice
A three-step monosynaptic rabies virus tracing was performed as described previously10. Briefly, postnatal day 17 C57B/L6 mice were anesthetized with a mixture of ketamine (100 mg/kg) and xylazine (10 mg/kg). AAV2-retro-Cre was injected into two sites in the mystacial pad (between the B1-B2 and C1-C2 vibrissae, 500nl each) using a volumetric injection system (based on a single-axis oil hydraulic micromanipulator MO-10, Narishige)48 equipped with a beveled glass micropipette (Drummond, 5-000-2005). Twenty days or longer after the first injection, the mice were anesthetized with isoflurane (1~2%), placed in the stereotaxic instrument (Model 963, David Kopf Instruments), and stereotaxically injected with 120 nl of a 1:1 mixture of AAV2/8-CAG-Flex-TVA-mCherry and AAV2/8-CAG-Flex-oG into the lateral part of VII where vibrissa motor neurons reside (vFMN, 5.8 mm posterior, 1.38 mm lateral to the bregma, and 5.2 mm below the brain surface) at the rate of 30nl/min using a beveled glass micropipette equipped with a microsyringe pump (UMP3, WPI) controlled by Micro4 controller (WPI). Fourteen days later, 250 nl of EnvA(M21)-RV-ΔG-GFP (diluted at 1:10 in PBS) was injected into the same injection site. Five days later, the mice were perfused for histological analysis.
Labeling and manipulation of vIRtPV neurons
We initially visualized the whole population of PV-positive vFMN premotor neurons (Fig. 1d, e) by stereotaxically injecting RG-LV-CreC into vFMN (1200 nl, 50 nl/min) of PV-CreN;Ai14 mice. Three weeks later, the mice were perfused for histological analysis of tdTomato-positive cells throughout the brainstem.
To specifically express a desired gene in vIRtPV neurons in a Cre-dependent manner (Fig.1–4), PV-CreN mice were anesthetized, and RG-LV-CreC was stereotaxically injected into vFMN (1200nl, 50 nl/min). Cre-dependent AAV (AAV2/8-hSyn-FLEX-TeLC-P2A-EYFP-WPRE (250 nl), AAV2/8-Ef1a-DIO-hChR2(E123A)-mCherry (200 nl), AAV2/8-EF1a-DIO-GtACR2-EYFP (300 nl), AAV2/1-hSyn-FLEx-mGFP-2A-Synaptophysin-mRuby (200 nl), AAV2/8-Ef1a-DIO_EGFP-GFE3 (200 nl), AAV2/8-hSyn-DIO-ChRmine-T2A-EGFP-GFE3 (300 nl), or AAV2/8-CAG-FLEX-EGFP-WPRE (300nl)) were delivered into vIRt (6.6 mm posterior, 0.9 mm lateral to the bregma, and 4.9 mm below the brain surface) at the rate of 30 nl/min.
Labeling and manipulation of vIRtvGlut2 neurons
vGlut2-Cre mice were anesthetized, and RG-LV-FlpO was stereotaxically injected into vFMN (800 nl, 50 nl/min). AAV2/8-hSyn-Con/Fon-EYFP or AAV2/8 -nEF-Con/Fon-NpHR3.3-EYFP were delivered into vIRt (250 nl, 30nl/min).
Monosynaptic rabies virus tracing from vIRtPV
vIRtPV neurons were transduced with 120 nl of a 1:1 mixture of AAV2/8-CAG-Flex-TVA-mCherry and AAV2/8-CAG-Flex-oG using the procedure described above. Fourteen days later, 250 nl of EnvA(M21)-RV-ΔG-GFP (diluted at 1:10 in PBS) was injected in vIRt. Five days later, the mice were perfused for histological analysis.
For the molecular characterization of pre-vIRtPV neurons within vIRt, a different injection procedure was used to preserve mRNA expression in vIRt. In this procedure, vIRtPV neurons were transduced with 120 nl of a 1:2 mixture of AAV2/8-CAG-Flex-TVA-mCherry (pre-diluted at 1:5 in PBS) and AAV2/8-CAG-Flex-oG. Ten days later, 250 nl of EnvA(M21)-RV-ΔG-GFP (diluted at 1:10 in PBS) was injected in vIRt. Four days later, the mice were perfused for HCR RNA-FISH analysis.
Optic fiber and head-post implantation
For the optogenetic silencing of vIRtPV and vIRtvGlut2 experiments, an optic fiber (200 μm) was implanted above vIRt (6.6 mm posterior, 0.9 mm lateral to the bregma, and 4.1 mm below the brain surface) after injecting AAV2/8-EF1a-DIO-GtACR2-EYFP or AAV2/8 -nEF-Con/Fon-NpHR3.3-EYFP, and affixed to the skull with cyanoacrylate (Super Glue, Loctite) and C&B Metabond (Parkell). For behavior experiments and electrophysiological recordings, a custom-made metal head-post was attached to the skull with cyanoacrylate and C&B Metabond for head-fixation. For opto-tagging experiments using ChR2, an optic fiber was implanted above vFMN after ChR2 expression in vIRtPV neurons employing the viral strategy described above (5.8 mm posterior, 2.75 mm lateral to the bregma with a 20d angle, and 4.5 mm below the brain surface).
Behavior
Mice were head-fixed using plate clamps (Thorlabs) positioned over a running disk made of a transparent plexiglass disk overlaid with a translucent silicone sheet. The disk was fixed to an optical shaft encoder (US Digital, H5-100-NE-S) to measure the rotation of the disk. Before behavior experiments, mice were habituated to the head-fixation and the running disk. One day before behavioral experiments, mice were lightly anesthetized with isoflurane and all vibrissae except for the C row were trimmed. Vibrissae were illuminated by an infrared LED placed below the running disk and vibrissa movement was recorded using a high-speed camera (Basler) at 500 fps. For the GFE3 experiment, breathing was monitored by an air flow sensor (Honeywell, AWM3303V) placed in front of the nose.
Optogenetic silencing experiment
Mice had to run for 1 second to trigger laser onset and then had to continue running for at least 1 second after to be counted as a trial. Continuous 470 nm (GtACR2) or 561 nm (NpHR3.3) laser stimulation (Opto Engine LLC, 7 – 10 mW) was delivered through the implanted optic fiber and which lasted until the end of the run. The optic fiber implantation sites were assessed by post-hoc histological analysis.
In vivo electrophysiology and opto-tagging experiment
Prior to recordings, a small craniotomy (0.7×1 mm) was made over vIRt coordinates (6.6 mm posterior, 0.9 mm lateral to the bregma). The dura was left intact. During acute recordings, the craniotomy was sealed with artificial dura (Dura-gel, Cambridge Neurotech). In between experiments, a silicone elastomer (Kwik-Cast, WPI) was applied to cover and protect the craniotomy. Finally, a small custom-designed 3-printed cap was affixed to the headpost to prevent interference with the fiber optic and the craniotomy area.
In a typical recording session, the mouse was placed on the running wheel, head-fixed and the headpost cap removed. The craniotomy was exposed and covered with sterile saline. For ChR2 opto-tagging experiments, a 473nm laser (Cobolt) was connected to the ferrule. For ChRmine opto-tagging experiments, a 635nm LED (LED635L, Thorlabs) was placed against the ear ipsilateral to the injected side. A 16 or 32 channels silicon probe (Cambridge Neurotech) was then lowered with micromanipulator (Sutter Instruments) about 4.7mm to the IRt region dorsal and medial to preBötC. Recordings were made at multiple steps going further down, while monitoring orofacial activity and responses to trains of laser stimulation. Signals from the silicon probe were filtered (250Hz to 6kHz for spikes, 0.3 to 250 Hz for local field potentials) and recorded at 30kHz on a Cereplex Direct (Blackrock Microsystems). For each recording, video acquisition was piloted and monitored using the open source software Bonsai (bonsai-rx.org49). Within the Bonsai workflow, video frames were triggered through a Teensy 3.2 (PJRC) and video streams encoded with FFmpeg (ffmpeg.org). Laser pulse trains were programmed and triggered with a TTL pulse generator (OTPG-4, Doric Lenses Inc.) controlled by Doric Neuroscience Studio. All TTLs (from the camera, the pulse generator and a master Arduino controlled through Bonsai) were acquired on the Cereplex Direct’s digital inputs at 30kHz. Signals from the rotary encoder and the air flow sensor were recorded on the Cereplex Direct’s analog inputs at 1kHz and 10kHz respectively. At the end of recording sessions, the Dura-gel was covered with Kwik-Cast and the protective cap put back in place.
Slice recordings
For slice recording vIRtPV neurons were labeled in Pv-CreN;Ai14 mice or Pv-CreN mice with AAV2/8-CAG-FLEX-EGFP as described above. Injection and recording were performed at P21–P28 and P49–P56, respectively.
After isofluorane anesthesia and trans-cardial perfusion with NMDG slicing solution, brains were quickly extracted. Coronal brain slices (250 μm) were cut with a vibratome (Leica VT 1200) in ice-cold NMDG slicing solution, kept in NMDG slicing solution (34 °C, 12 min), then moved to ACSF solution (34 °C, 30 min). Then slices were kept in room temperature before the transfer to the recording chamber maintained at 30 °C in ACSF. NMDG slicing solution consisted of (in mM): NMDG 92, HEPES 20, NaHCO3 30, KCl 2.5, NaH2PO4 1.25, thiourea 2, Na-ascorbate 5, Na-pyruvate 3, glucose 25, CaCl2 0.5, MgSO4 10. ACSF consisted of (in mM): NaCl 125, KCl 2.5, NaH2PO4 1.25, glucose 11, NaHCO3 26, MgCl2 1.3, CaCl2 2. Cell-attached recordings were performed in a loose-seal (20~60 MΩ) configuration, using a glass pipette (3–7 MΩ) in voltage clamp mode at holding potential 0 mV. The pipette solution was ACSF with green (A10436, ThermoFisher) or red (A10438, ThermoFisher) fluorescent dye (20 μM). At the end of recording, the membrane was ruptured to fill the recorded cell. Signals were passed through DigiData 1440A and amplified via a Multiclamp 700B amplifier, and recorded with pClamp software (version 11, Molecular Devices). After initial spontaneous firing of 10 min, excitability of the local circuit was increased with bath application of high extracellular K+ concentration (9 mM) and/or a broad inhibitor of voltage-gated potassium, 4-Aminopyridine (100 μM). Detection of action potentials was done in Spike2 (version 7.10c, Cambridge Electronic Design) and further data analysis was performed in Matlab (R2021b, Mathworks).
Histology
Mice were deeply anesthetized with isoflurane overdose and transcardially perfused with 10% sucrose in Milli-Q water, followed by ice-cold 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. The brains were dissected and post fixed in the same fixative overnight at 4°C and freeze-protected in 30% sucrose in phosphate buffer saline (PBS) at 4°C until they sank. The brains were embedded in OCT compound (Sakura Finetek USA, Inc.) and frozen in dry-ice-cooled ethanol.
For monosynaptic rabies virus tracing, eighty μm free-floating coronal sections were made using a cryostat (Leica Biosystems Inc). The sections were briefly washed in PBS and stained with Neurotrace blue fluorescent Nissl stain (1:500, Thermo Fisher Scientific, N21479) in 0.3% Triton-X100/PBS overnight at 4°C. The sections were briefly rinsed with PBS and mounted on slides with Mowiol.
Free-floating fluorescent in situ hybridization was performed as described previously45. Sixty μm free-floating coronal sections containing vIRt area were incubated in 0.3%
Triton-X100/DEPC-treated PBS (DEPC-PBS) at room temperature for 20 min. After washing in PBS, the sections were incubated in proteinase K/DEPC-PBS (5 μg/ml) for 30 min at 37°C. The sections were then acetylated for 10 min. After washing in DEPC-PBS, the sections were incubated in hybridization buffer (5x SSC, 2% Blocking reagent (Sigma, 11096176001), 50% Formamide (Thermo Fisher Scientific, AM9342), 0.1% N-Lauroylsarcosine (NLS, Sigma, 61743), 0.1% SDS (Sigma, 71736)) for 3 hours at 60°C. The sections were then incubated with GlyT2 probe (denatured in hybridization buffer at 80°C for 5 min and snap-cooled on ice for 5 min, 1:1000)28 in the hybridization buffer overnight at 60°C. The sections were then washed in a series of wash buffers (2x SSC, 50% formamide 0.1% NLS at 60°C, 2x SSC, 0.1% NLS at 37°C, and 0.2x SSC, 0.1% NLS at 37°C, twice for 15 min for each wash buffer). The sections were washed in 0.1% Triton-X100/TBS (TBST) for 5 min and blocked in 10% Blocking One (Nacalai USA, Inc, 03953–95)/TBST for 1 hour at room temperature. The sections were incubated with sheep anti-DIG-AP (1:3500, Sigma, 11093274910) and rabbit anti-GFP antibody (1:1000, abcam, ab290) overnight at 4°C. After washing in TBST (15 min × 3), the sections were incubated in 100 mM Tris-HCl (pH8.0) for 5 min and then incubated with Fast Red (SIGMAFAST™, Sigma, F4648) for 90 min at room temperature to develop GlyT2 signal. After rinsing in TBST, the sections were incubated with Donkey anti-rabbit -IgG Alexa Fluor 488 (1:500, Jackson ImmunoResearch, 711-545-152) and DAPI overnight at 4°C. After washing in TBST, the sections were mounted on slides with Mowiol.
Hybridization Chain Reaction (HCR) v3.0 RNA-FISH was performed using reagents from Molecular Instruments. Sixty μm free floating coronal sections were pretreated by the method described previously50 with slight modifications. Briefly, the sections were incubated in 70% Ethanol/PBS overnight at 4°C. After washing in DEPC-PBS (3 min × 2), the sections were treated with 5% SDS/DEPC-PBS for 45 min at room temperature. After rinsing in 2x SSC, the sections were incubated in 2x SSC for 15 min. The sections were then incubated in Probe Hybridization Buffer for 30 min at 37°C for 30 min, followed by incubation with a combination of PV, vGlut2 and vGat probes (Molecular Instruments) overnight at 37°C. After washing in HCR probe wash buffer (15 min × 4 at 37°C), the sections were rinsed in 2x SSC (5 min × 2) and incubated in HCR amplification buffer for 30 min at room temperature. The sections were then incubated two overnight at 25°C with appropriate hairpins conjugated with Alexa fluor 546 or 647 (denatured and snap-cooled according to manufacturer’s instruction) to visualize hybridization signals. After washing in 2x SSC (5 min × 2), the sections were counterstained with DAPI and mounted on slides with Mowiol.
For the molecular characterization of pre-vIRtPV neurons, TVA-mCherry protein was detected by HCR IHC together with PV, vGat, and vGlut2 mRNA. Briefly, after pretreatment with 70% ethanol/PBS and 5% SDS/DEPC-PBS, sections were incubated with rabbit anti-RFP (1:2000, Rockland, 600–401-379) and then incubated with donkey anti-rabbit conjugated with an initiator for B4 amplifier (1:2000, Molecular Instruments). After fixation with 4% PFA/PBS, sections were processed for HCR RNA-FISH as described above. After hybridization, HCR IHC and HCR RNA-FISH signals were visualized with appropriate hairpins conjugated with Alexa fluor 546 or 647. Due to constraints of spectral flexibility of the confocal microscope, PV and TVA-mCherry were visualized with the same fluorophore. PV HCR RNA-FISH only visualizes cell bodies. TVA-mCherry HCR IHC, on the other hand, stains axons and dendrites as well as cell bodies since TVA-mCherry is a membrane protein. Therefore, pre-vIRtPV neurons (GFP+/TVA-mCherry−) were distinguished from source cells (GFP+/PV+/TVA-mCherry+) by their lack of gray color-labeled (pseudo color for PV and TVA-mCrerry) axons/dendrites.
Immunostaining
Anti-gephyrin staining was performed after the GFE3 experiment. Twenty-five μm free-floating coronal sections containing vIRt area were processed with SHIELD post-fixation protocol51 according to the manufacturer’s instruction (LifeCanvas Technologies). Briefly, the sections were incubated in SHIELD-OFF solution for 24 hours at 4°C, followed by incubation in SHIELD-ON buffer for 24 hours at 37°C. The sections were then delipidated in passive clearing solution (LifeCanvas Technologies) for 24 hours at 42°C. After washing in PBS, the sections were immunostained for gephyrin and GFP using a standard immunostaining method.
Antibodies used were rabbit anti-gephyrin (1:500, Synaptic Systems, 147008), chicken anti-GFP (1:1000, Aves labs, GFP-1010), donkey anti-rabbit-Alexa Fluor 555 (1:500, Thermo Fisher Scientific, A32794), and donkey anti-chicken Alexa Fluor 488 (1:500, Jackson ImmunoResearch, 703-545-155). The sections were counterstained with Neurotrace blue and mounted on slides with Mowiol.
Confocal imaging
Histological images were acquired using a confocal microscope LSM700 (Zeiss).
Three-dimensional reconstruction of the pre-vIRtPV circuit
Registration of pre-vIRtPV neurons in the Allen Mouse Brain Common Coordinate Framework (CCFv3)27 was performed as described previously10. The coordinates of registered neurons were converted into Allen CCF coordinates as follows. AP, ML, and DV coordinates were multiplied by 1000, and then 5400 and 5700 were added to AP and ML coordinates, respectively. After the conversion, the neurons were visualized using Brainrender52.
Spatial correlation analysis
The multivariate kernel smoothing density estimation was applied (bandwidth = 1) to the (x, y, z) coordinates of each cell. The resulting kernel density estimation was then vectorized, and the cosine similarity between any two of the mice was calculated to form the correlogram. The coordinates of vibrissa premotor neurons were derived from the published work10.
Analysis of gephyrin-positive synapses
For counting gephyrin-positive synapses, sections were immunostained as described above and imaged by LSM700 using a 40× objective lens with 1.5× zoom. Gephyrin-positive synapses localizing on the cell perimeter of optically sectioned neurons were counted. The number of synapses on each neuron was normalized by dividing by the cell perimeter measured in ImageJ.
Spike data analysis
Spike sorting was performed using Kilosort253 with final curation done with JRClust54. Responsivity to laser pulses (opto-tagging) was quantified using the stimulus-associated spike latency test (SALT55). Cells were classified as tuned to the whisking phase following three statistical measures: first we measured the spike/phase coherence for each whisking epoch using a multi-taper spectral estimation method (Chronux, http://chronux.org/))and computed the confidence level for p=0.01. Then we compared the distribution of spike-occurring whisking phases with the overall distribution of whisking phases (Kuiper two-sample test, p=0.05, CircStat56). From these distributions we calculated the probability of spiking according to the whisking phase. Finally, we performed further circular statistics to assess the phase tuning of each neuron (Rayleigh test for non-uniformity of circular data, p=0.05, CircStat) and obtain descriptive statistics of the periodic activity pattern (CircStat). In addition, we also set a minimum whisking epoch duration of three seconds. For each recording, data for epochs that did not meet all of those criteria were discarded. Phase tuned cells were grouped according to their peak coherence phase (PCP) following these arbitrary limits: Retraction units: PCP >= −30 and <65; Mid-Retraction units: PCP >= 65 and <150; Protraction units: PCP >= 150 and <−115; Mid-Protraction units: PCP >= −115 and <−30. Cells were also classified between fast and slow rhythmic activity, to separate those mainly implicated in whisking from those concerned with slower oscillations such as changes in vibrissa midpoint or breathing. To do so, we computed the power spectral density between 3 and 20Hz (multi-taper spectrum, Chronux) to obtain the median spiking frequency. Cells with a median frequency below 9Hz were classified as slow rhythmic.
Vibrissa tracking and computation of whisking parameters
Vibrissa kinematics were obtained with homemade data processing pipelines based on Janelia Whisker Tracker (Whisk57). The videography processing pipeline provided a set of vibrissa coordinates for each video frame from which whisking parameters (e.g., angle, velocity, midpoint, amplitude) were subsequently obtained. Whisking phase was calculated from the angle trace, band-pass filtering the trace between 8 Hz and 30 Hz (three-pole Butterworth filter run in forward and backward directions), and applying the Hilbert transform 7,58. Whisking parameters were related as: θwhisk (t) = θmidpoint − θamp [1 + cos (2πfwhisk(t) - ϕ0)], with θwhisk the absolute angle, fwhisk the instantaneous whisking frequency, and ϕ0 the phase at the peak spike rate in the whisking cycle. Individual whisks (or, similarly, breathing) cycles were identified by phase resets of the Hilbert transform. A vibrissal motion was identified as protraction or retraction onset if the phase value exceeded ±3.
Power spectrum analysis of whisking traces.
Raw vibrissa angles were bandpass filtered (4th order Butterworth) with low and high frequency cutoffs of 5 and 30 Hz, respectively. A Welch transform was used to calculate the time average spectrogram of the vibrissa signal with the following parameters: nfft=512, window=102, overlap=101, and sampling frequency = 500 Hz. For experiments with trial structure (TeLC, GtACR2, and NpHR3.3 silencing), spectrograms were averaged across animals and trials. GFE3 inhibitory synapse ablation experiments did not have trials, so spectrograms were calculated from the entire experiment duration, and then averaged across animals. The controls for TeLC-silencing were vibrissae on the contralateral side, the controls for GFE3-inhibitory synapse ablation experiments were a separate group of GFP animals, and controls for GtACR2 optogenetic silencing experiments were the same vibrissae whisking in the air during the period before laser illumination. Breathing sensor analysis was mostly the same, except the output of the sensor was first lowpass filtered with a cutoff frequency of 500, and then down-sampled from a sampling rate of 30000 Hz to 500 Hz. Then the bandpass filter before Welch transform used cutoff frequencies of 1 Hz and 15 Hz.
State transition analysis
The recorded raw whisking angles of a single mouse was first corrected by subtracting the midpoint trace from the raw whisking angles. The processed whisking angle trace was then evenly divided and binned into finite states (n = 100) based on the whisking angle ([0.02%, 99.8%] of the whisking angle range). Then for each time point t, we extracted the state i.d. st and the state i.d. st+1 of the next time point t + 1, then the number of the transition from st to st+1 was added one. For all the time points, we calculated how many times the states transitioned from a state si to sj and formed this 2-dimensional histogram into a matrix showing the overall state transitions between each other.
Similarity score
To get the initial similarity between the whisking and breathing patterns in both the control and GFE3 mice, we calculated the similarity score by computing the correlation coefficient between the whisking angle (midpoint corrected) and the breathing signal.
Coherence between whisking and breathing
To investigate the whisking components other than directly coupled to the breathing, we calculated the magnitude-squared coherence between the whisking and breathing signals. The coherence measures the squared correlation coefficient of the cross spectrum density functions of the signals with the frequency range [0.5Hz, 20Hz]. To get the overall oscillation pattern, we calculated the averaged coherence with s.e.m. for all the control and GFE3 mice.
Statistics
Levels of significance are shown as follows: *P 0.05, **P 0.01. The data were tested for normality using quantile plots and/or Shapiro-Wilk normality tests in Prism. Data with non-normal distributions were compared using non-parametric tests. The following statistical tests were used. Fig.1m, Quantification of whisking amplitude (Control vs. vIRtPV-TeLC): p = 0.0286, Kolmogorov–Smirnov (KS) test. Fig. 1n, Quantification of whisking midpoint (Control vs. vIRtPV-TeLC): p = 0.0286, KS test. Fig.4c, Quantification of normalized density of gephyrin puncta on GFE3-positive vIRtPV neurons and neighboring neurons: p = 0.0286, KS test. Fig.4f, Quantification of whisking and breathing parameters (Control vs. vIRtPV-GFE3). Whisking amplitude: p = 0.0047, KS test. Whisking midpoint: p = 0.58, KS test. Breathing duty cycle: p = 0.53, KS test. Fig.4g, Quantification of the number of whisks per one breathing cycle. vIRtPV-GFE3 (1 whisk/breath) vs. vIRtPV-GFE3 (2 whisks/breath): p = 0.0156, Wilcoxon matched-pairs signed-ranks test. Fig.4j, Correlation coefficient of breathing and whisking (Control vs. vIRtPV-GFE3): p = 0.0082, KS test. Extended Data Fig.1i, Quantification of whisking amplitude (vIRtvGlut2-NpHR3.3, Laser-OFF vs. Laser-ON): p = 0.8438, Wilcoxon matched-pairs signed-ranks test. Extended Data Fig.1j, Quantification of whisking midpoint (vIRtvGlut2-NpHR3.3, Laser-OFF vs. Laser-ON): p = 0.1562, Wilcoxon matched-pairs signed-ranks test. Extended Data Fig.2f, Quantification of whisking amplitude (vIRtPV-GtACR2, Laser-OFF vs. Laser-ON): p = 0.0312, Wilcoxon matched-pairs signed-ranks test.
Extended Data
Extended Data Fig. 1. Anatomical and functional characterization of vIRtvGlut2 neurons.
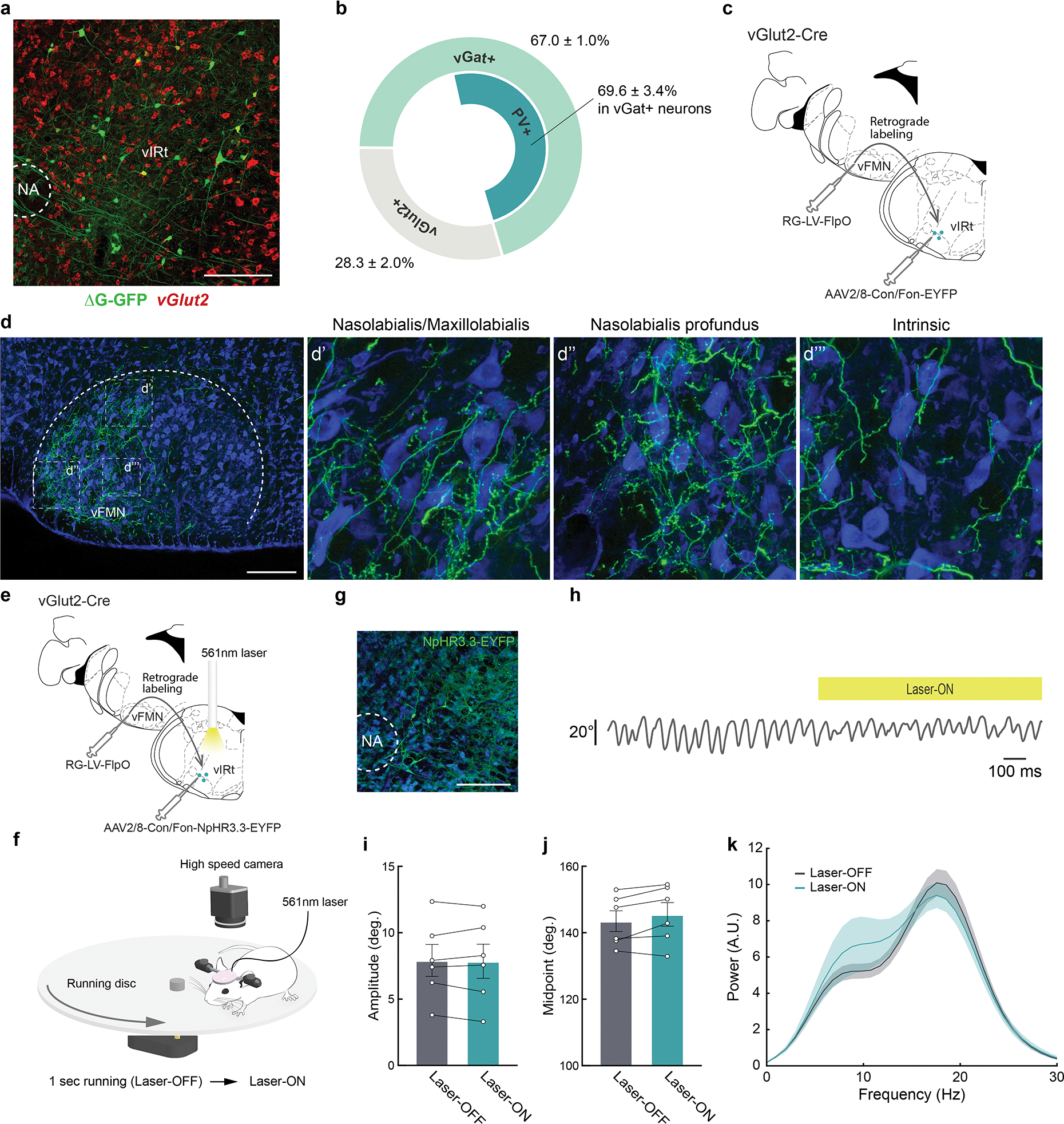
a, Representative image of vibrissa premotor neurons (green) in vIRt traced by a three-step monosynaptic rabies virus tracing method shown in Figure 1. vGlut2 (red) mRNA were visualized by HCR RNA-FISH. b, Molecular identity of premotor vIRt neurons. 67.0 ± 1.0 % (n = 4) and 22.8 ± 1.3 % (n = 3) of premotor vIRt neurons are vGat+ and vGlut2+, respectively. 69.6 ± 3.4% (n = 4) of vGat+ premotor vIRt neurons are PV+. c, Strategy for labeling vGlut2+ premotor vIRt neurons (vIRtvGlut2). Retrograde lentivirus carrying FlpO (RG-LV-FlpO) and AAV carrying Cre- and Flp-dependent-EYFP (AAV2/8-Con/Fon-EYFP) are injected into the vFMN and vIRt of the vGlut2-Cre animal, respectively. c, Projection pattern of vIRtvGlut2 neurons in the vFMN. d’,d”,d”’, High-magnification views of the boxed areas in d. The dorsolateral (d’), lateral (d”), and ventrolateral (d”’) facial motor subnuclei that contain motoneurons innervating the nasolabialis/maxillolabialis, nasolabialis profundus, and intrinsic muscles, respectively. e, A schematic of vIRtvGlut2 silencing experiment. f, Experimental setup for vibrissa tracking. One second running triggers 1 second continuous 561 nm laser stimulation. g, Representative image of NpHR3.3-EYFP expression in vIRt. h, Example vibrissa trace during laser-OFF and -ON period. i, Quantification of whisking amplitude (Laser-OFF, 7.9 ± 1.2° vs. Laser-ON, 7.9 ± 1.3°, n = 6, p = 0.8438, Wilcoxon signed-rank test). j, Quantification of whisking midpoint (Laser-OFF, 53.5 ± 3.1° vs. Laser-ON, 55.5 ± 3.5°, n = 6, p = 0.1562, Wilcoxon signed-rank test). k, Power spectrum analysis of whisking frequency in Laser-OFF and -ON period. Shaded areas are mean ± s.e.m. Data are mean ± s.e.m. Brain sections were counterstained with Neurotrace Blue (d, g).
Extended Data Fig. 2. Optogenetic silencing of vIRtPV neurons also impaired whisking.

a, A schematic of vIRt PV GtACR2 silencing experiment. b, Representative image of GtACR2 expression and the optic fiber track. c, Optic fiber tip placements over vIRt from all AAV-Flex-GtACR2-GFP injected animals. d, Experimental setup for vibrissa tracking. One second running triggers 1 second continuous 473nm laser stimulation. e, Example vibrissa trace during laser-OFF and -ON period. f, Quantification of whisking amplitude (Laser-OFF, 15.6 ± 0.7° vs. Laser-ON, 10.2 ± 1.1°, n = 5, p = 0.0312, Wilcoxon signed-rank test). g, Power spectrum analysis of whisking frequency in Laser-OFF and -ON period. Data are mean ± s.e.m. * P ≤ 0.05. Brain sections were counterstained with Neurotrace Blue (b).
Extended Data Fig. 3. Antidromic opto-tagging of a ChRmine-expressing vIRtPV neuron via light stimulation through the ear.
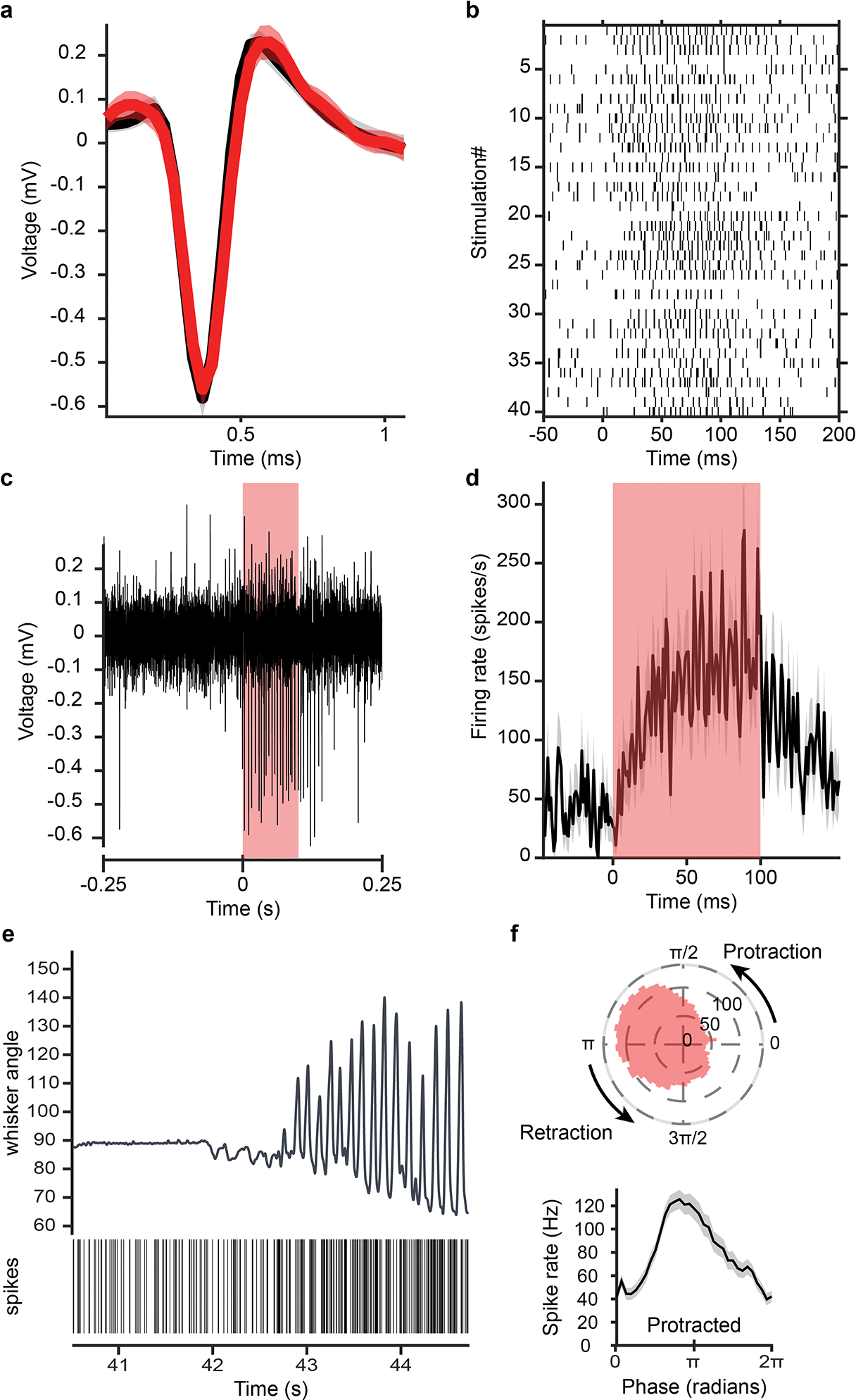
a, Overlapped average waveform, before (black) and during (red) stimulation periods. b, Raster plot of spike times aligned to stimulation onset for 40 light pulses. c, Example single channel recording trace showing antidromic spikes from the opto-tagged unit during a light pulse. d, Firing rate of that unit averaged over 40 light stimulations epochs. e, Unit activity during transition from resting to whisking state. vibrissa angle traces for the ipsilateral C2 vibrissa. Bottom: Spike raster plot for this opto-tagged vIRtPV-ChRmine neuron. f, Phase tuning. Average spike rate across whisking phases for this opto-tagged vIRtPV-ChRmine neuron, in polar (top) and cartesian coordinates (bottom).
Extended Data Fig. 4. Slow oscillation vIRt units and additional analysis of transition from tonic to rhythmic firing of vIRt units.
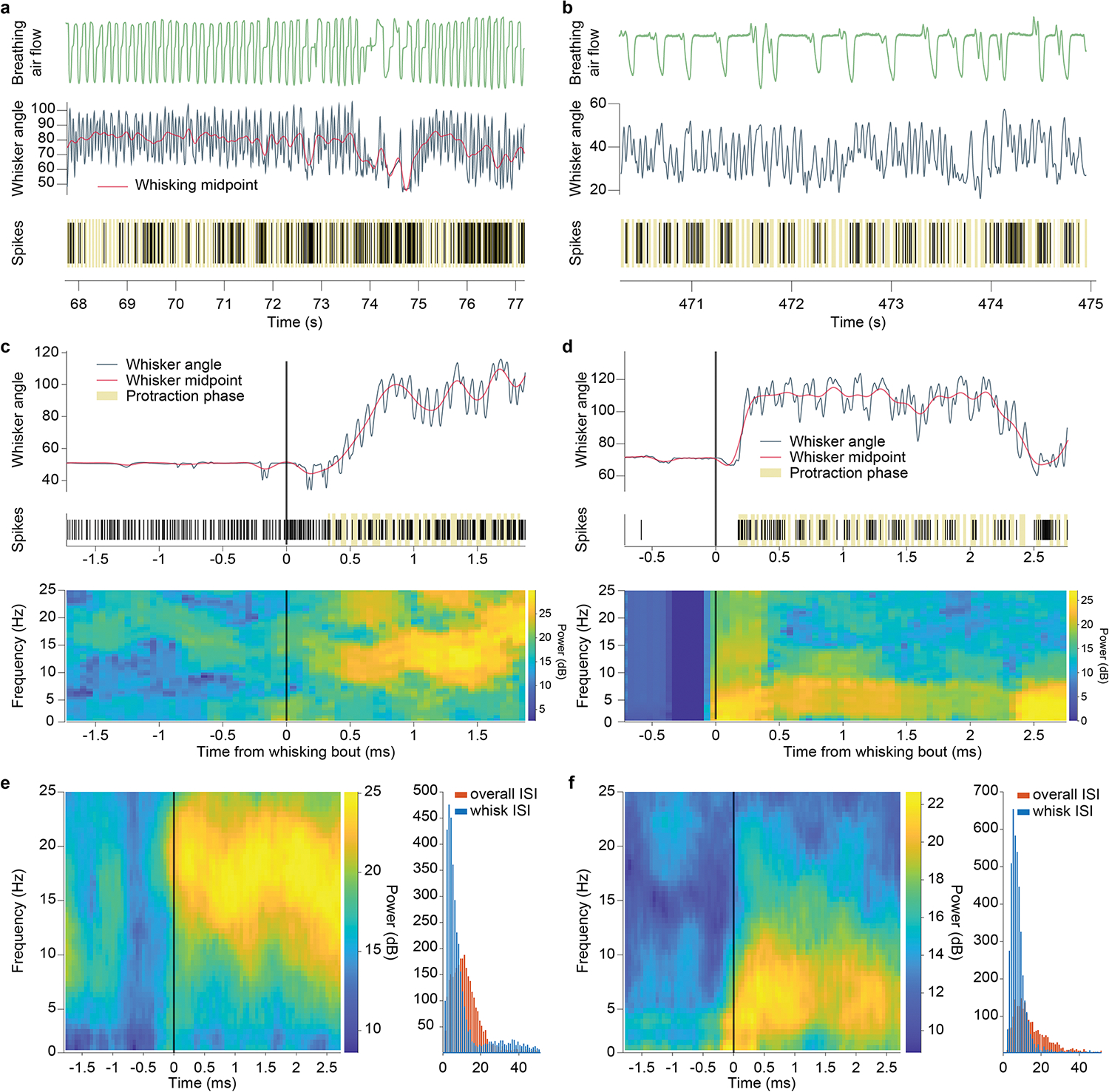
a,b, Two vIRt units with “slow” rhythmic activity patterns. Top: breathing trace. Middle: Vibrissa angle and midpoint traces. Bottom: raster plot of spiking events (protraction phases shown in beige). c, Top: vibrissa trace and raster plot for a retraction unit. Bottom: Time-frequency spectrum of that spiking activity. The transition to rhythmic bursting appears as a high power frequency band, corresponding to whisking frequency. d, Same as c, for a “slow” oscillation unit. Here the transition to whisking shows a low frequency band, similar to vibrissa midpoint variations or breathing rhythm. e, Left: Time-frequency spiking spectrum for a retraction unit, averaged over all whisking bouts. Right: Inter-spike interval histograms for that retraction unit. Red, overall ISI. Blue, ISI during long whisking bouts. The ISI distribution becomes bimodal, with a strong peak at short interval corresponding to bursts. f, same as e, for a “slow” oscillation unit.
Extended Data Fig. 5. Elevated extracellular potassium concentrations induce bursting but not rhythmic activity in vIRtPV neurons.
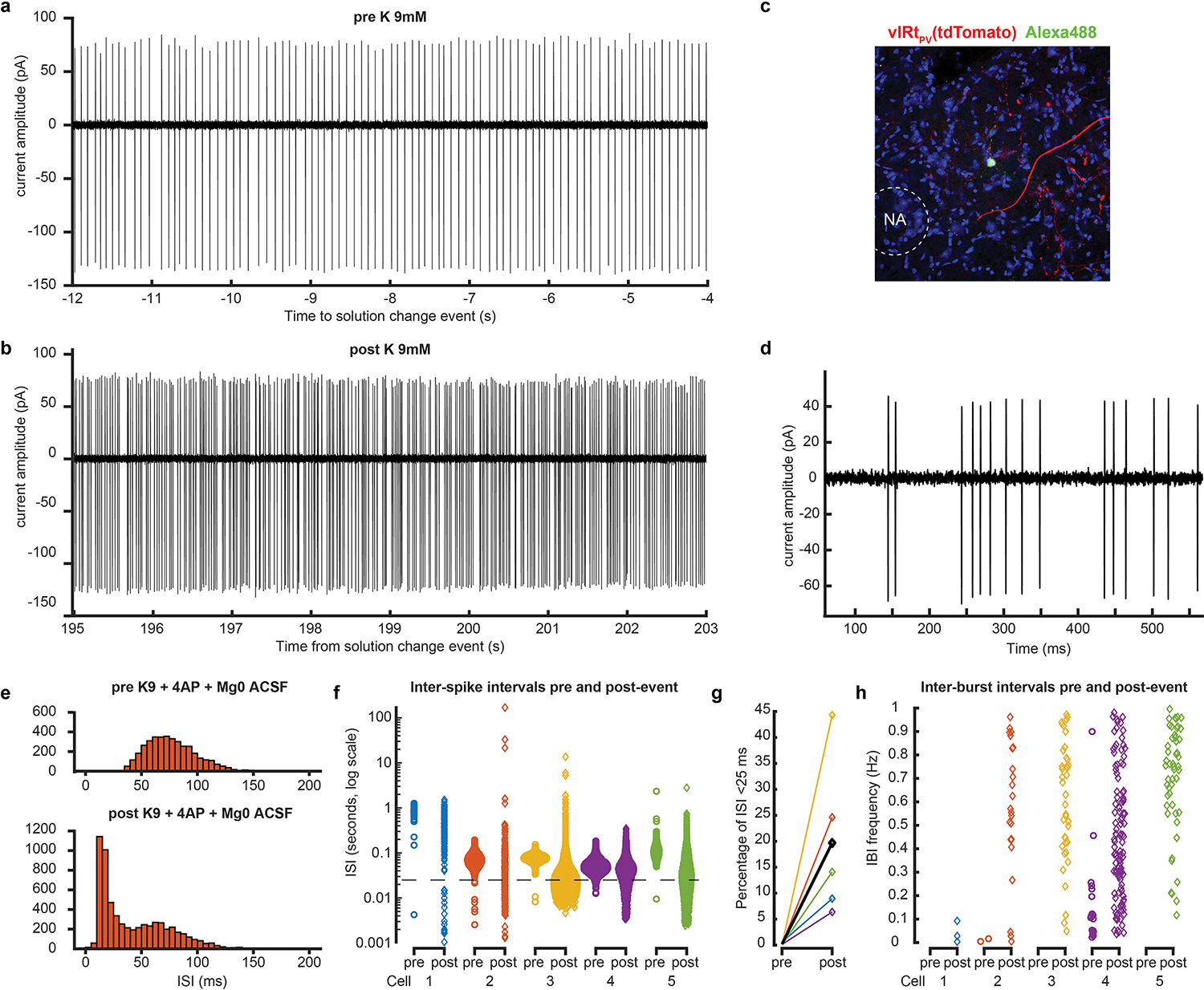
a,b, Cell-attached recording of a vIRtPV unit before (a) and after (b) bath application of K 9mM. c, Representative histological verification of targeted recording. One of the tdTomato-expressing vIRtPV in slice is filled with green Alexa 488 dye from the recording pipette. d, Example of burst, post increase of extracellular potassium concentration. e,f, Inter-spike interval distribution, pre and post increase of extracellular potassium concentration, for one cell (e, pre: top, post: bottom) and all cells (f, ISI shown in log scale. Dash line: 40Hz). g, Percentage of ISIs shorter than 25ms (i.e., above 40Hz), pre and post event, for all cells (average shown in black). h, Inter-burst frequency, for each cell, pre and post. No bursting frequency band is observed for any cell.
Extended Data Fig. 6. . Pre-vIRtPV neurons in the brainstem and motor cortex, and neurotransmitter characterization of pre-vIRtPV neurons.
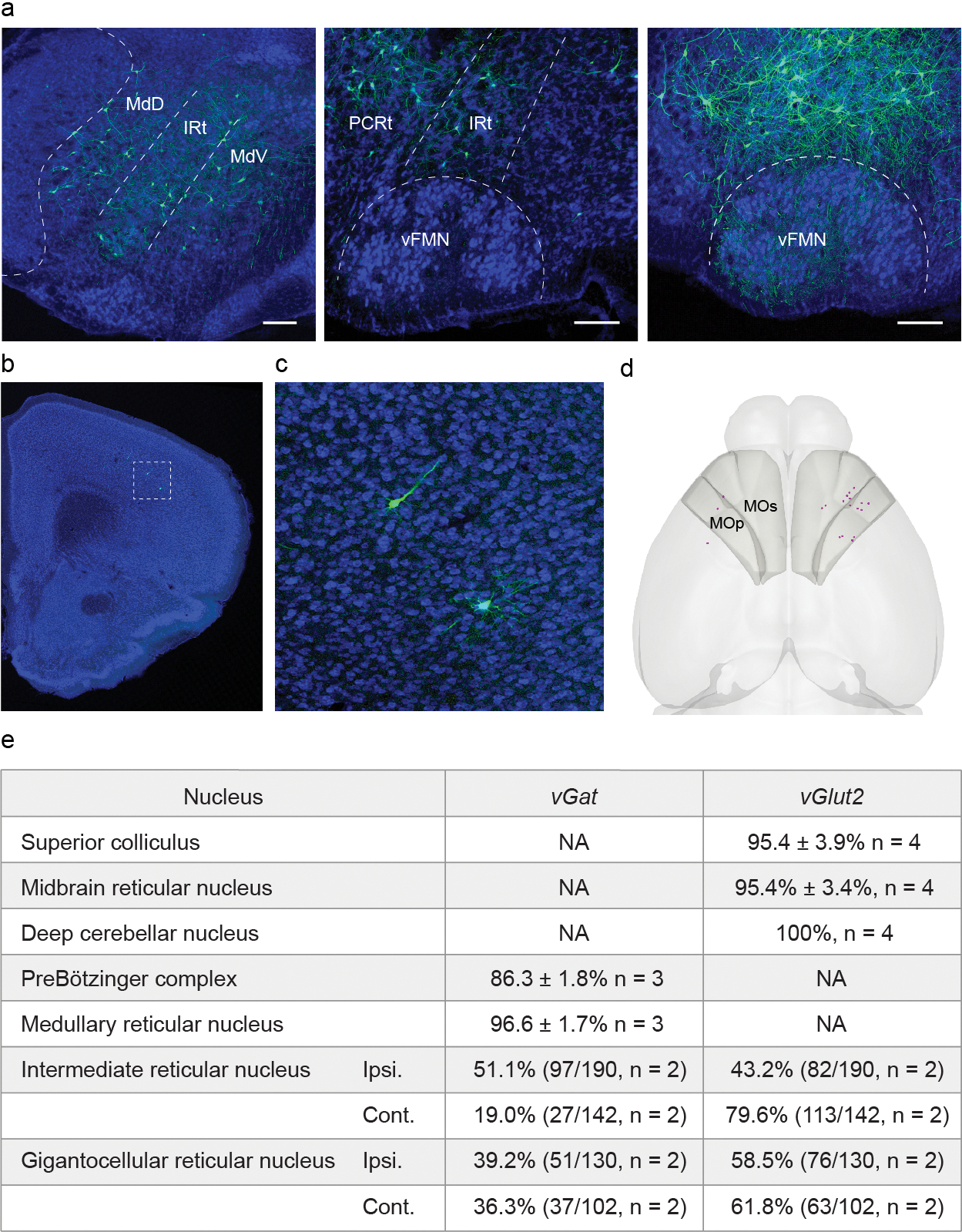
a, Representative image of ΔG-GFP labeled pre-vIRtPVneurons in the brainstem (continued from Fig. 3c). Scale bars, 200 μm. b, Representative image of ΔG-GFP labeled pre-vIRtPV neurons in the cortex. Scale bar, 500 μm. c, Zoomed image of the boxed area in b, d, Representative 3D reconstructed image of labeled pre-vIRtPV neurons in the cortex (magenta). Shaded areas denote the primary motor (MOp) and secondary motor cortices (MOs). e, Neurotransmitter phenotype of pre-vIRtPV neurons determined by fluorescent in situ hybridization or HCR RNA-FISH.
Extended Data Fig. 7. Comparison of the distributions of vIRtPV presynaptic neurons and vibrissal premotor neurons.
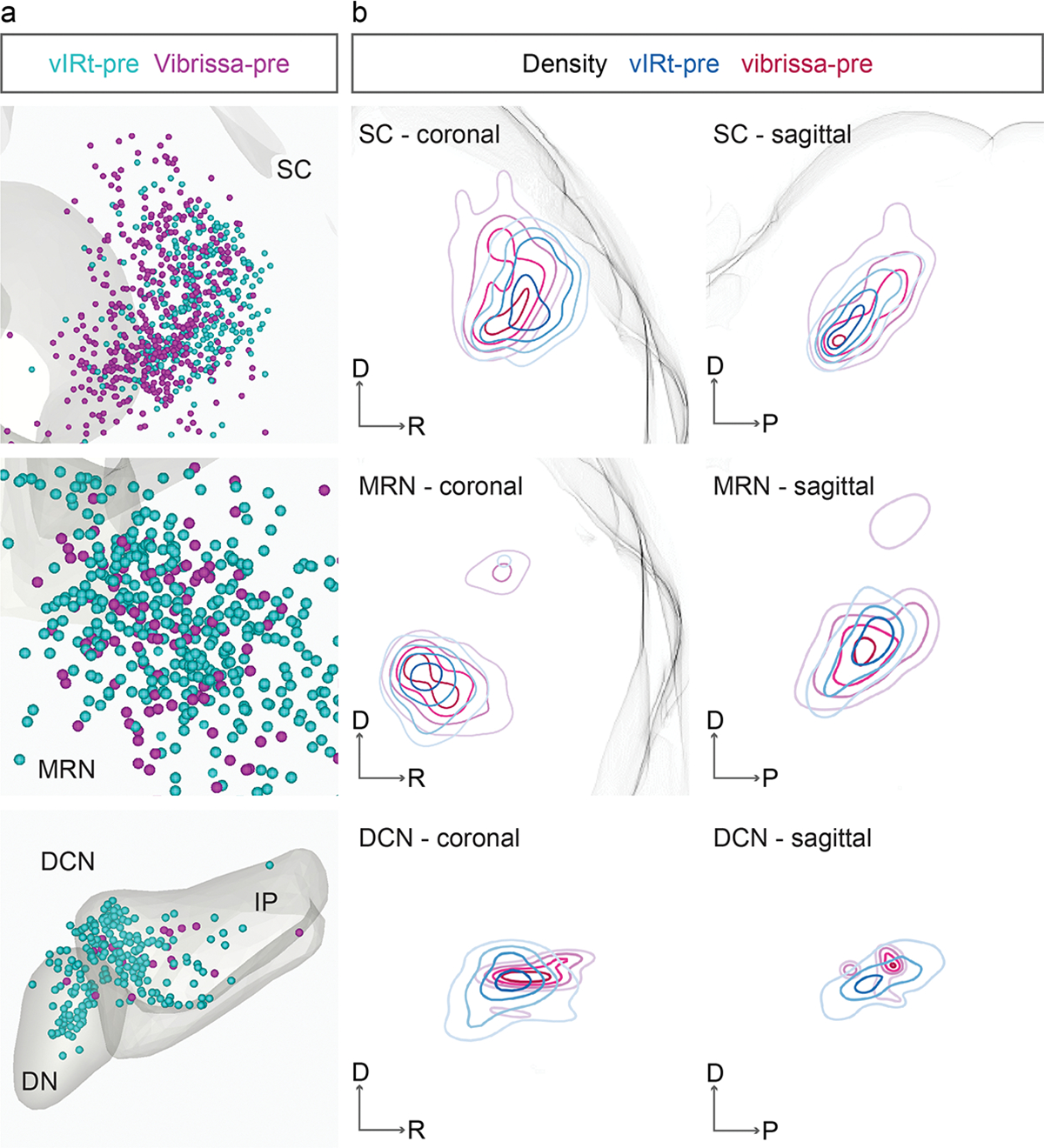
a, Three-dimensionally reconstructed vIRtPV presynaptic neurons (vIRt-pre, cyan, n= 3) and vibrissal premotor neurons (vibrissa-pre, magenta, n = 4) in the Allen Mouse Brain CCF in coronal planes. b, Density analysis of presynaptic neurons (vIRt-pre, cyan, n = 3) and vibrissal premotor neurons (vibrissa-pre, magenta, n = 4) in coronal and sagittal planes. DN, dentate nucleus; IP, interposed nucleus.
Extended Data Fig. 8. Additional results from vIRtPV-ChRmine-GFE3 mice.
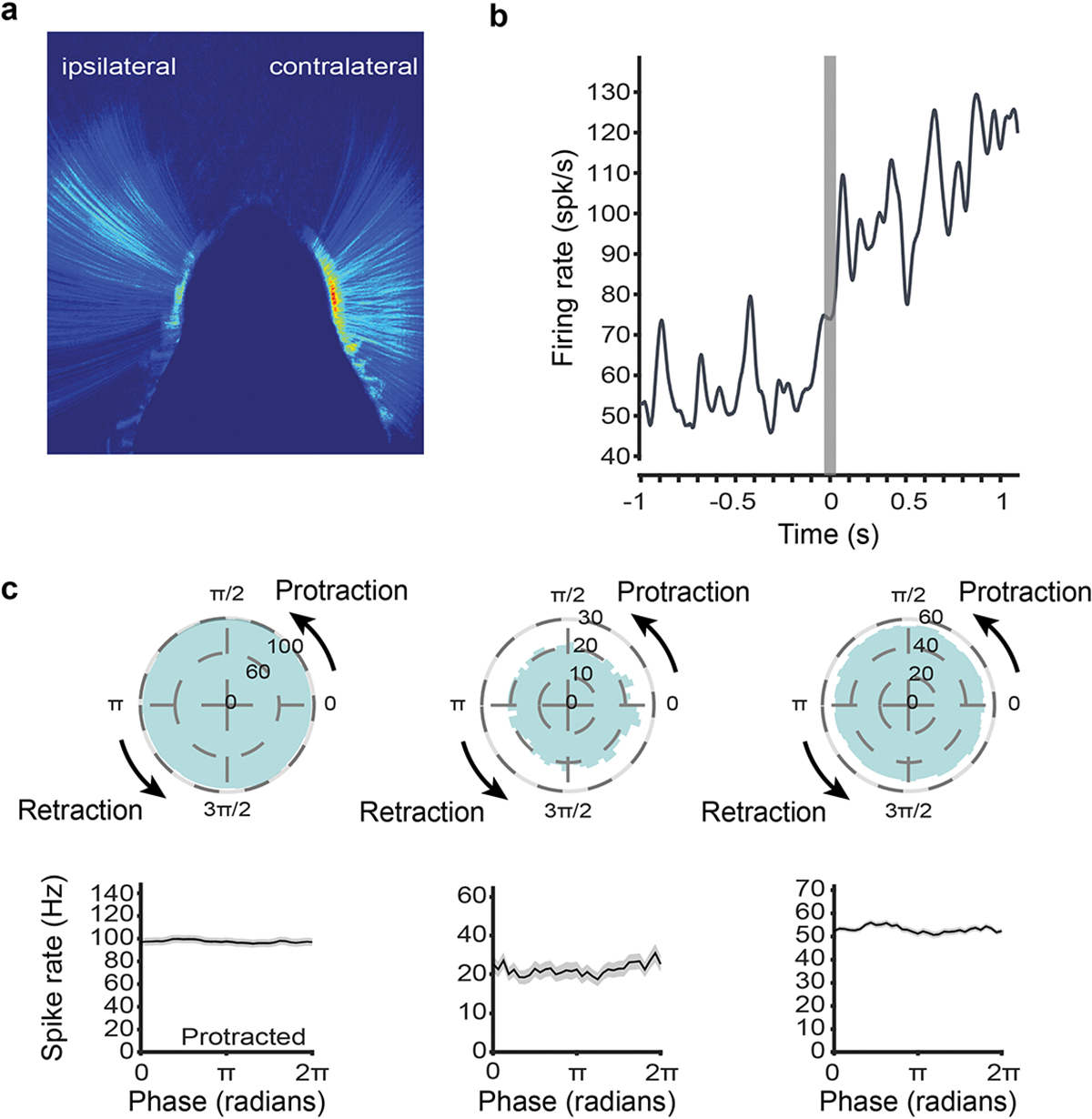
a, Average whisking activity of a vIRtPV-ChRmine-GFE3 mouse, showing the mean of the pixel intensity difference between each consecutive video frame. Blue to red color scale: low to high activity. In contrast to the ipsilateral (GFE3) side, contralateral whisking is highly active across the whole whisking range. b, Increased firing, but no rhythmic activity at whisking initiation for a opto-tagged vIRtPV-ChRmine-GFE3 unit (whisking initiation time determined from contralateral C2 vibrissa). c, Whisking phase tuning of all opto-tagged vIRtPV-ChRmine-GFE3 single units (conventions as in Fig. 2 and 4).
Extended Data Fig. 9. Schematic model of the vIRtPV circuit that generates rhythmic whisking in normal and experimental conditions.
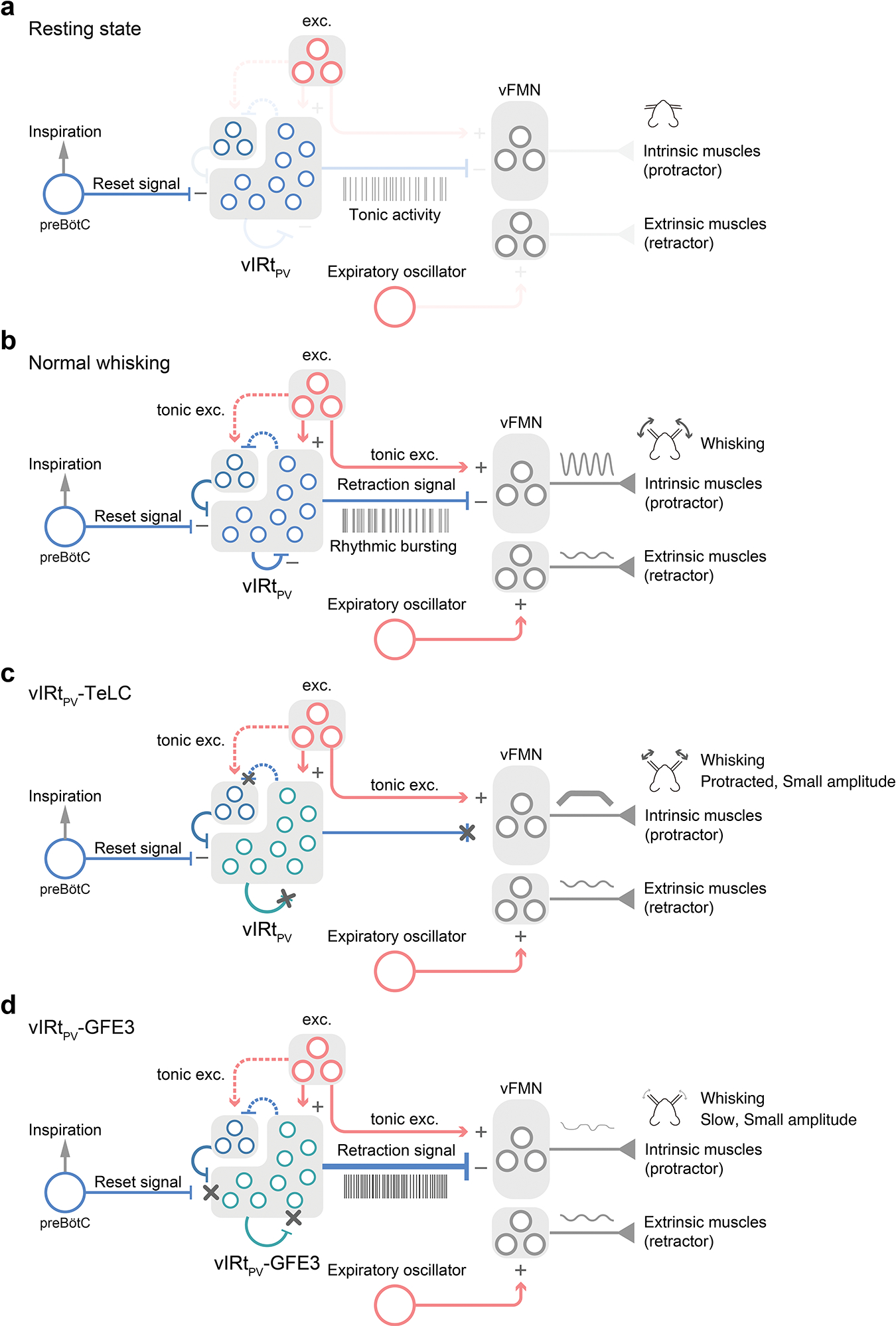
a, In Resting state, vIRtPV neurons show unsynchronized tonic activity. b, In Normal whisking condition, tonic excitatory inputs to vFMN protractor motoneurons protract vibrissae. Concurrently, tonic excitatory inputs to vIRtPV neurons induce recurrent inhibition within vIRtPV and which in turn switches vIRtPV from tonic firing to synchronized rhythmic bursting mode. The rhythmic signal from vIRtPV periodically silences vFMN protractor motoneurons and leads to rhythmic whisking. The rhythmic inhibitory signal from the inspiratory rhythm generator preBötC resets the activity of vIRtPV. The expiratory oscillator activates vFMN retractor motoneurons. c, In vIRtPV-TeLC condition, outputs from vIRtPV are abolished. Lack of inhibition from vIRtPV results in strong continuous activation of vFMN protractor motoneurons and strong protraction of vibrissae. Because of the strong tonic activity of protractor intrinsic muscles, extrinsic retractor muscles play a minor role in vibrissa movement. d, In vIRtPV-GFE3 condition, tonic excitation induces strong unsynchronized tonic inhibitory outputs from vIRtPV to vFMN protractor motoneurons, which results in a less protracted midpoint compared with vIRtPV-TeLC’s. Under this condition, the contribution of expiratory oscillator-extrinsic retractor muscles becomes pronounced. A group of inhibitory neurons in the left top corner of vIRt indicates PV−/vGat+ vIRt neurons. Dotted lines denote putative connections.
Supplementary Material
Supplementary Video 1. TeLC mediated silencing of vIRtPV neurons.
High speed videography showing whisking deficits after tetanus toxin light chain (TeLC) silencing of vIRtPV neurons (left). The control side (right) shows no behavioral deficit. The video is slowed five times from original speed.
Supplementary Video 2. vIRtPV neurons show rhythmic bursting in the whisking retraction phase.
High speed videography showing a mouse whisking while running on a wheel. The whisking phase polar plot is overlaid in the bottom left corner, with the current whisking phase shown as a green bar. Spikes from a simultaneously recorded opto-tagged vIRtPV retraction unit are indicated by an audio cue and red dot on the whisking phase polar plot.
Acknowledgments
We thank members of the Wang laboratory and the aABC U19 BRAIN Circuit Team for helpful discussions and suggestions. We thank Bao-Xia Han and Seonmi Choi for mouse colony maintenance. We thank Boston Children’s Hospital Viral Core for the production of AAV2retro-CAG-Cre. This work is support by NIH grants U19 NS107466, and R01 NS077986.
Footnotes
Competing interests
The authors declare no competing interests.
Code availability
Custom-made scripts used in this Article are available in a GitHub repository at https://github.com/wanglab-neuro/the-whisking-oscillator-circuit.
Data availability
The data are available from the corresponding authors upon reasonable request.
References
- 1.Marder E & Bucher D Central pattern generators and the control of rhythmic movements. Curr Biol 11, R986–996, doi: 10.1016/s0960-9822(01)00581-4 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Marder E & Calabrese RL Principles of rhythmic motor pattern generation. Physiol Rev 76, 687–717, doi: 10.1152/physrev.1996.76.3.687 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Vincent SB The Functions of the Vibrissae in the Behavior of the White Rat. (University of Chicago, 1912). [Google Scholar]
- 4.Welker W Analysis of sniffing of the albino rat 1. Behaviour 22, 223–244 (1964). [Google Scholar]
- 5.Moore JD et al. Hierarchy of orofacial rhythms revealed through whisking and breathing. Nature 497, 205–210, doi: 10.1038/nature12076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore JD, Deschenes M, Kurnikova A & Kleinfeld D Activation and measurement of free whisking in the lightly anesthetized rodent. Nat Protoc 9, 1792–1802, doi: 10.1038/nprot.2014.119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deschenes M et al. Inhibition, Not Excitation, Drives Rhythmic Whisking. Neuron 90, 374–387, doi: 10.1016/j.neuron.2016.03.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith JC, Ellenberger HH, Ballanyi K, Richter DW & Feldman JL Pre-Botzinger complex: a brainstem region that may generate respiratory rhythm in mammals. Science 254, 726–729, doi: 10.1126/science.1683005 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinfeld D, Deschenes M, Wang F & Moore JD More than a rhythm of life: breathing as a binder of orofacial sensation. Nat Neurosci 17, 647–651, doi: 10.1038/nn.3693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takatoh J et al. Constructing an adult orofacial premotor atlas in Allen mouse CCF. Elife 10, doi: 10.7554/eLife.67291 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi HMT et al. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, doi: 10.1242/dev.165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellavance MA et al. Parallel Inhibitory and Excitatory Trigemino-Facial Feedback Circuitry for Reflexive Vibrissa Movement. Neuron 95, 673–682 e674, doi: 10.1016/j.neuron.2017.06.045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P et al. Intersectional Cre driver lines generated using split-intein mediated split-Cre reconstitution. Sci Rep 2, 497, doi: 10.1038/srep00497 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato S et al. A lentiviral strategy for highly efficient retrograde gene transfer by pseudotyping with fusion envelope glycoprotein. Hum Gene Ther 22, 197–206, doi: 10.1089/hum.2009.179 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Link E et al. Tetanus toxin action: inhibition of neurotransmitter release linked to synaptobrevin proteolysis. Biochem Biophys Res Commun 189, 1017–1023, doi: 10.1016/0006-291x(92)92305-h (1992). [DOI] [PubMed] [Google Scholar]
- 16.Schiavo G et al. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature 359, 832–835, doi: 10.1038/359832a0 (1992). [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y et al. Identifying local and descending inputs for primary sensory neurons. J Clin Invest 125, 3782–3794, doi: 10.1172/JCI81156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleinfeld D, Moore JD, Wang F & Deschenes M The Brainstem Oscillator for Whisking and the Case for Breathing as the Master Clock for Orofacial Motor Actions. Cold Spring Harb Symp Quant Biol 79, 29–39, doi: 10.1101/sqb.2014.79.024794 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govorunova EG, Sineshchekov OA, Janz R, Liu X & Spudich JL NEUROSCIENCE. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 349, 647–650, doi: 10.1126/science.aaa7484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenno LE et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods 11, 763–772, doi: 10.1038/nmeth.2996 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenno LE et al. Comprehensive Dual- and Triple-Feature Intersectional Single-Vector Delivery of Diverse Functional Payloads to Cells of Behaving Mammals. Neuron 107, 836–853 e811, doi: 10.1016/j.neuron.2020.06.003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshel JH et al. Cortical layer-specific critical dynamics triggering perception. Science 365, doi: 10.1126/science.aaw5202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinfeld D & Sompolinsky H Associative neural network model for the generation of temporal patterns. Theory and application to central pattern generators. Biophys J 54, 1039–1051, doi: 10.1016/S0006-3495(88)83041-8 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim EJ, Jacobs MW, Ito-Cole T & Callaway EM Improved Monosynaptic Neural Circuit Tracing Using Engineered Rabies Virus Glycoproteins. Cell Rep 15, 692–699, doi: 10.1016/j.celrep.2016.03.067 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamichi K et al. Dissecting local circuits: parvalbumin interneurons underlie broad feedback control of olfactory bulb output. Neuron 80, 1232–1245, doi: 10.1016/j.neuron.2013.08.027 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wickersham IR et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–647, doi: 10.1016/j.neuron.2007.01.033 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q et al. The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell 181, 936–953 e920, doi: 10.1016/j.cell.2020.04.007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takatoh J et al. New modules are added to vibrissal premotor circuitry with the emergence of exploratory whisking. Neuron 77, 346–360, doi: 10.1016/j.neuron.2012.11.010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sreenivasan V, Karmakar K, Rijli FM & Petersen CC Parallel pathways from motor and somatosensory cortex for controlling whisker movements in mice. Eur J Neurosci 41, 354–367, doi: 10.1111/ejn.12800 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beier KT et al. Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell 162, 622–634, doi: 10.1016/j.cell.2015.07.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortin G, Jungbluth S, Lumsden A & Champagnat J Segmental specification of GABAergic inhibition during development of hindbrain neural networks. Nat Neurosci 2, 873–877, doi: 10.1038/13172 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Friesen WO, Poon M & Stent GS An oscillatory neuronal circuit generating a locomotory rhythm. Proc Natl Acad Sci U S A 73, 3734–3738, doi: 10.1073/pnas.73.10.3734 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grillner S Biological pattern generation: the cellular and computational logic of networks in motion. Neuron 52, 751–766, doi: 10.1016/j.neuron.2006.11.008 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Marder E & Bucher D Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol 69, 291–316, doi: 10.1146/annurev.physiol.69.031905.161516 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Gross GG et al. An E3-ligase-based method for ablating inhibitory synapses. Nat Methods 13, 673–678, doi: 10.1038/nmeth.3894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin JY, Knutsen PM, Muller A, Kleinfeld D & Tsien RY ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci 16, 1499–1508, doi: 10.1038/nn.3502 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiehn O Decoding the organization of spinal circuits that control locomotion. Nat Rev Neurosci 17, 224–238, doi: 10.1038/nrn.2016.9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song J et al. Multiple Rhythm-Generating Circuits Act in Tandem with Pacemaker Properties to Control the Start and Speed of Locomotion. Neuron 105, 1048–1061 e1044, doi: 10.1016/j.neuron.2019.12.030 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Janczewski WA, Tashima A, Hsu P, Cui Y & Feldman JL Role of inhibition in respiratory pattern generation. J Neurosci 33, 5454–5465, doi: 10.1523/JNEUROSCI.1595-12.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Del Negro CA, Funk GD & Feldman JL Breathing matters. Nat Rev Neurosci 19, 351–367, doi: 10.1038/s41583-018-0003-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ausborn J, Snyder AC, Shevtsova NA, Rybak IA & Rubin JE State-dependent rhythmogenesis and frequency control in a half-center locomotor CPG. J Neurophysiol 119, 96–117, doi: 10.1152/jn.00550.2017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berg RW & Kleinfeld D Rhythmic whisking by rat: retraction as well as protraction of the vibrissae is under active muscular control. J Neurophysiol 89, 104–117, doi: 10.1152/jn.00600.2002 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Tervo DG et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382, doi: 10.1016/j.neuron.2016.09.021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mattis J et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods 9, 159–172, doi: 10.1038/nmeth.1808 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanek E 4th, Rodriguez E, Zhao S, Han BX & Wang F Supratrigeminal Bilaterally Projecting Neurons Maintain Basal Tone and Enable Bilateral Phasic Activation of Jaw-Closing Muscles. J Neurosci 36, 7663–7675, doi: 10.1523/JNEUROSCI.0839-16.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez E et al. A craniofacial-specific monosynaptic circuit enables heightened affective pain. Nat Neurosci 20, 1734–1743, doi: 10.1038/s41593-017-0012-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakurai K et al. Capturing and Manipulating Activated Neuronal Ensembles with CANE Delineates a Hypothalamic Social-Fear Circuit. Neuron 92, 739–753, doi: 10.1016/j.neuron.2016.10.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petreanu L, Mao T, Sternson SM & Svoboda K The subcellular organization of neocortical excitatory connections. Nature 457, 1142–1145, doi: 10.1038/nature07709 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopes G et al. Bonsai: an event-based framework for processing and controlling data streams. Front Neuroinform 9, 7, doi: 10.3389/fninf.2015.00007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicovich PR et al. Multimodal cell type correspondence by intersectional mFISH in intact tissues. bioRxiv, 525451, doi: 10.1101/525451 (2019). [DOI] [Google Scholar]
- 51.Park YG et al. Protection of tissue physicochemical properties using polyfunctional crosslinkers. Nat Biotechnol, doi: 10.1038/nbt.4281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Claudi F et al. Visualizing anatomically registered data with brainrender. Elife 10, doi: 10.7554/eLife.65751 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pachitariu M, Steinmetz N, Kadir S, Carandini M & Kenneth DH. Kilosort: realtime spike-sorting for extracellular electrophysiology with hundreds of channels. bioRxiv, 061481, doi: 10.1101/061481 (2016). [DOI] [Google Scholar]
- 54.Jun JJ et al. Real-time spike sorting platform for high-density extracellular probes with ground-truth validation and drift correction. bioRxiv, 101030, doi: 10.1101/101030 (2017). [DOI] [Google Scholar]
- 55.Kvitsiani D et al. Distinct behavioural and network correlates of two interneuron types in prefrontal cortex. Nature 498, 363–366, doi: 10.1038/nature12176 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berens P CircStat: A MATLAB Toolbox for Circular Statistics. 2009 31, 21, doi: 10.18637/jss.v031.i10 (2009). [DOI] [Google Scholar]
- 57.Clack NG et al. Automated tracking of whiskers in videos of head fixed rodents. PLoS Comput Biol 8, e1002591, doi: 10.1371/journal.pcbi.1002591 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aljadeff J, Lansdell BJ, Fairhall AL & Kleinfeld D Analysis of Neuronal Spike Trains, Deconstructed. Neuron 91, 221–259, doi: 10.1016/j.neuron.2016.05.039 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1. TeLC mediated silencing of vIRtPV neurons.
High speed videography showing whisking deficits after tetanus toxin light chain (TeLC) silencing of vIRtPV neurons (left). The control side (right) shows no behavioral deficit. The video is slowed five times from original speed.
Supplementary Video 2. vIRtPV neurons show rhythmic bursting in the whisking retraction phase.
High speed videography showing a mouse whisking while running on a wheel. The whisking phase polar plot is overlaid in the bottom left corner, with the current whisking phase shown as a green bar. Spikes from a simultaneously recorded opto-tagged vIRtPV retraction unit are indicated by an audio cue and red dot on the whisking phase polar plot.
Data Availability Statement
The data are available from the corresponding authors upon reasonable request.


