Abstract
This year marks the 25th anniversary of the coinage of the term metabolome [S. G. Oliver et al., Trends Biotech. 16, 373–378 (1998)]. As the field rapidly advances, it is important to take stock of the progress which has been made to best inform the disciplines future. While a medical-centric perspective on metabolomics has recently been published [M. Giera et al., Cell Metab. 34, 21–34 (2022)], this largely ignores the pioneering contributions made by the plant and microbial science communities. In this perspective, we provide a contemporary overview of all fields in which metabolomics is employed with particular emphasis on both methodological and application breakthroughs made in plant and microbial sciences that have shaped this evolving research discipline from the very early days of its establishment. This will not cover all types of metabolomics assays currently employed but will focus mainly on those utilizing mass spectrometry–based measurements since they are currently by far the most prominent. Having established the historical context of metabolomics, we will address the key challenges currently facing metabolomics and offer potential approaches by which these can be faced. Most salient among these is the fact that the vast majority of mass features are as yet not annotated with high confidence; what we may refer to as definitive identification. We discuss the potential of both standard compound libraries and artificial intelligence technologies to address this challenge and the use of natural variance–based approaches such as genome-wide association studies in attempt to assign specific functions to the myriad of structurally similar and complex specialized metabolites. We conclude by stating our contention that as these challenges are epic and that they will need far greater cooperative efforts from biologists, chemists, and computer scientists with an interest in all kingdoms of life than have been made to date. Ultimately, a better linkage of metabolome and genome data will likely also be needed particularly considering the Earth BioGenome Project.
The term metabolome was coined, by analogy to the transcriptome and proteome, by Steven Oliver and colleagues in a review article on yeast functional genomics published in 1998 (1). A handful of research papers were subsequently published at the turn of the century (2–6), and the primary research article by Fiehn and coworkers in Nature Biotechnology, at the turn of the new millennium, was a seminal paper that demonstrated the utility of gas chromatography-mass spectrometry (GC-MS)-based metabolomics for defining plant phenotypes and for understanding plant mutations. Pertinently, although not mentioned in a recent metabolomics perspective published in Cell Metabolism (7), all of these papers proceeded the earliest mammalian metabolomics studies with which perspective pieces were published only in the early 2000s, and the first research articles arguable did not appear until 2003. While these early plant and microbial studies, similar to their counterparts in mammalian systems, were admittedly descriptive rather than mechanistic in scope, they heralded a widespread adoption of more sophisticated multivariate statistics than had previously been used in earlier studies of metabolism. The plant and microbial sciences adopted these tools in advance of the mammalian metabolomics research field. However, as we will detail below, this, and the fact that large-scale metabolomics was pioneered in plant and microbial sciences, is by no means the unique contribution of these research fields to the discipline of metabolomics. Indeed, key pioneering events in metabolomics research were made in these fields and have played major roles in shaping this rapidly evolving discipline. We will provide a brief overview of these key innovations in the following sections in order to afford a counterpoint to the Giera et al. article (7). We would like to state up front that it is not our objective to criticize this article—which provides fascinating insight and an excellent clinical perspective into many aspects of metabolomics—but rather to highlight the relatively unsung contributions on which many aspects of metabolomics are founded.
Scientific Foundation of Metabolism
Defining our earliest studies of metabolism is a difficult, somewhat nebulous, and subjective task. One of us has previously taken the 13th century philosophical musings of Ibn al-Nafis who stated that “the body and its parts are in a continuous state of dissolution and nourishment, so they are inevitably undergoing permanent change” as a start point (8). While it has also been reported that studies of small molecules can be traced to early traditional Chinese doctors (2000 to 1500 BC) who employed ants to evaluate the glucose content of urine (9), these insects are hardly robust analytical platforms and were limited to a single metabolite. The Giera et al. perspective article, alternatively, rather credits the 18th characterization of the seemingly ubiquitous metabolite urea by Hermann Boerhaave as the cornerstone of metabolic research (7). While we appreciate the value in all of these claims, what also warrants discussion and acknowledgment in this context is the initial isolation of an enzyme from malt extracts by Anselme Payen and Jean-Francois Persoz (10). This was the first enzyme diastase (currently known as amylase) and preceded subsequent, early 19th century, biochemical identification of enzymes from microorganisms (11, 12). Indeed, these works are of particular significance since they provided the mechanistic link between Ibn al-Nafis prescient suggestion/observation of change and Boerhaave’s first identification of a metabolite. While Ibn al-Nafis thesis was generally applicable to all kingdoms of life and Boerhaave arguably identified the first metabolite from human urine, the first isolation of an enzyme and their subsequent biochemical identification were plant and microbial based, respectively. Almost 100 y later, the foundation of metabolomics was at least as diverse with the microbial and plant research communities even taking the lead even though the order of magnitude of the challenge for the latter is considerably higher—as we will outline in the following paragraphs. Following the provision of a broad cross-kingdom context of the development of metabolomics, we will next turn our attention to contemporary developments in metabolomics particularly focusing on how developments emanating from plant and microbial sciences can additionally impact medicinal research. Finally, we will provide a future perspective delineating how we envisage that greater cooperative efforts from biologists, chemists, and computer scientists with an interest in all kingdoms of life, and a better linkage of metabolome and genome data would greatly enhance the prospects of metabolomics to both enhance our fundamental understanding and our ability to improve key biological features across the life sciences.
Ramping Up—Going from Detecting (and Quantifying) a Single or a Handful of Compound(s) to Many—Steps toward Metabolomics
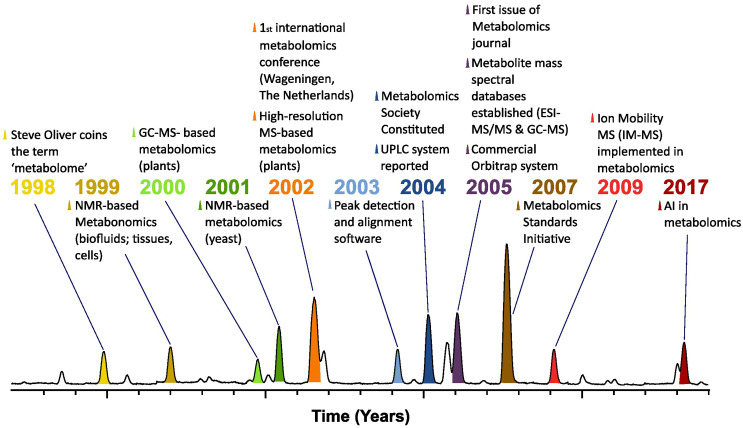
Early metabolite measurements were highly targeted to specific metabolites. Following the development of spectrophotometric approaches to measure enzyme activities came the realization that these assays could be used to determine the levels of the metabolites interconverted by these enzymes. In addition, chromatographic methods were established in order to assay the levels of a range of metabolites of similar structure (13), and radioisotope approaches were used to define the sequence of metabolites in what have become the canonical metabolic pathways across the kingdom of life including the TCA cycle and the Calvin–Benson cycle of photosynthetic tissues (14, 15). At the turn of the century, this changed dramatically with three different mass spectrometry–based techniques being demonstrated to have utility in the (relatively) unbiased detection and quantification of multiple metabolites (Fig. 1). These techniques were all based on MS coupled to different inlet systems with the earliest research reports being on derivatization-based gas chromatography (GC)–mass spectrometry (MS) in Arabidopsis and potato (4, 16, 17) and direct infusion high-resolution mass spectrometry in strawberry and tobacco (5) as well as yeast (18). The former technique allowing the detection of tens to hundreds of metabolites was rapidly adopted in a range of other plant and microbial species as well as eventually by the medicinal sciences. The latter allowing the detection of hundreds to thousands of mass features was subsequently improved via the coupling to liquid chromatography [LC; (19)]—again being rapidly taken up by the plant and microbial sciences and somewhat later by the medicinal sciences. The introduction of ultra-performance liquid chromatography, or UPLC (ultra-performance liquid chromatography) a few years later, was a game changer in metabolomics analysis (20). As compared to high-performance liquid chromatography, the use of smaller particles and higher flow rates in UPLC provides superior resolution, sensitivity, and speed of analysis. The third technique of note capillary electrophoresis (CE)-MS is highly sensitive but not as easily adapted to high-throughput studies–the pioneering study utilizing this technique for metabolomics came from work in the Tomita Laboratory focused on Escherichia coli (21). As an aside, we note that NMR played an important role in early metabolomics studies in plants (22), microbes (23), and medicinal research (24), however, retaining certain advantages is relatively insensitive and slow (when used with complex mixtures) in comparison to mass spectral techniques (Fig. 1). Indeed, given the low-throughput nature of CE-MS and the low sensitivity of NMR approaches, it is unsurprising that chromatography-coupled MS approaches have taken the lead in metabolomics. That this would be the case was probably already clear at the first international metabolomics conference which was held in Wageningen, The Netherlands, in 2002 and centered around plants (25). Indeed, the early prominence of plant and microbial sciences in this field was already documented at the time “metabolomics has been studied in microorganisms and in plants, but little systematic work has so far been performed on the metabolomics of humans or even animals. However, it seems clear that this approach will soon be widely applied” (26). Despite the clarity of the adoption of this technique, it still took several years for articles in the medical field to appear (see citations in ref. 7). To summarize, currently and already for more than a decade high-resolution MS is the leading technology for metabolomics by far (19). The use of this approach for metabolomics, i.e., comprehensive coverage rather than evaluation of predefined targets, is in our opinion what is important. By contrast to the Giera et al. article (7), we do not feel that merely the better targeted detection is a step change rather the possibility to approach biology without preconceived biases. This is not to belittle the contribution of the medical field which has certainly in recent years been instrumental in driving the field forward. Our aim here is rather to illuminate the many contributions of plant and microbial sciences to developing and nurturing the field, a fact that is reflected in the high number of scientists that have had.
Fig. 1.
Timeline and milestones in the metabolomics field. The timeline starts with the definition of the term metabolome by Oliver et al. in 1998 (1). NMR-based metabonomics was shortly reported after this by Nicholson et al. (60). Fiehn et al. (4) provided the first example of GC-MS-based metabolomics (in plants) in 2001, Raamsdonk et al. (6) provided an example of NMR-based metabolomics in yeast, and Aharoni et al. in 2002 demonstrated (in plants) the use of high-resolution MS-based metabolomics (5). At this stage, peak detection and alignment software were employed in metabolomics experiments as, for example, MetAlign (123). Plumb et al. (124) presented a breakthrough in separation technology for metabolomics using UPLC (ultra-performance liquid chromatography). The Golm Metabolome Database (GMD) and Metlin (125, 126) were among the first to establish metabolite mass spectral database for ESI-MS/MS and GC-MS. The introduction of the Orbitrap ion trap mass analyzer was a milestone in metabolomics becoming widely used as of 2005 (127). Later in 2007, the Metabolomics Standards Initiative proposed minimum reporting standards for metabolomics experiments such as for sample preparation, quality control, and metabolite identification (86). Ion mobility MS (IM‐MS) has added another dimension to the separation and identification of molecules and was implemented in metabolomics with first examples published in 2009/2010 (128). Recent years have evidenced a surge in the use of machine learning and other artificial intelligence (AI) technologies to complement both the experimental and analytical aspects in metabolomics (reviewed by refs. 109 and 129–131). This timeline by no means includes all metabolomics-related studies and in the authors opinion reports the most significant advances with this growing research field. We thus apologize for those scientists whose papers were not cited here.
Plants—The Master Chemists
Plant metabolomics has been relatively recently reviewed elsewhere (27) so does not need extensive discussion here. However, several key facets of metabolomics were pioneered in plants, and there are furthermore a number of important features of plants which render them a good model for metabolomics in general (Fig. 1). We have previously estimated that the tree of life contains upward of 1 million different metabolites with the highest proportion of this metabolic diversity being found in plants and fungi with a maximum of 114, 100 being present in a single species (28). Thus far, however, even the most comprehensive methods cannot provide firm upper limits. If we consider that for linear molecules of 700 amu containing just carbon, hydrogen, and oxygen that there will be 1046 possible isomers (29), then the potential complexity in small molecules is huge. Currently, combinations of the best methods available are able to quantify 700 of the 3,700 metabolites predicted to be present in E. coli (30, 31), 500 of the 2,680 metabolites predicted to be present in yeast (32, 33), 8,000 of the 217,920 metabolites predicted to be present in humans (34), and 14,000 of the 2 million metabolites predicted to be metabolites predicted to be present in the plant kingdom (27, 35). Studies of nonmodel species emphasize how little we know with respect to the repertoire of metabolites in plants. In fact, metabolomics of, for example, medicinal plants and exotic species was a major factor driving the technology and pushing it to the limits in terms of metabolite identification (e.g., refs. 36–38). Many of the studies in plants, especially not only in early days of the technology but also to date, exploit metabolomics to obtain insight to uncharted metabolite classes and metabolic pathways (39–41). While the number predicted for humans is very high, it is important to note that this includes metabolites that they ingest (including phytochemicals and drugs and their xenometabolites) and from naturally commensal bacteria and fungi which reside in all mammalian species (42), while plants biosynthesize (at least the vast majority)—of their own metabolites leading them to be labeled—life’s master chemists (43). Another feature of plants is the fact that their sessile nature has led them developing an extensive chemical repertoire providing the base for interactions including defense and resistance strategies against biotic (44) and abiotic (45) stresses, respectively. It would thus appear likely that the immense catalogs of metabolic responses of plant cells to stress could prove highly informative in the study of human disease responses (46). More intimately linked in this respect is the fact that the plant kingdom provides much of our food and medicine with plant metabolomics playing a key role in engineering plants either to be more nutritional or to produce higher levels of bioactive compounds (see, for example, refs. 47 and 48). As we will return to later, expanding such studies to include clinical trials (49, 50) and other disciplines including artificial intelligence (AI) (51) will be instrumental in maximizing their potential. Before looking to the future, however, we would first like to highlight a few key events in the development of metabolomics in which plant and microbial sciences took the lead. Before doing so, we should admit that we have not achieved metabolomics yet in the sense it was initially proposed, i.e., the comprehensive measurement of the metabolite complement of the cell. It can, however, be stated with certainty that we are now in an era of genomics-based metabolism. That said before defining the status quo and defining the challenges that remain, it is important to briefly revisit the early days of metabolomics (see the Infancy of Metabolomics section).
Microbial Metabolomics, Functional Genomics, and Systems Biology
Concomitant with the development and application of metabolomics within the plant sciences, microbiologists were also interested in whether metabolomics could help them understand microbial mechanisms and ascribe function to orphan genes. Indeed, the paper that first used the term “metabolome” (4) was a study investigating the potential of Fourier transform infrared (FT-IR) spectroscopy for discriminating different mutants of the yeast Saccharomyces cerevisiae. In this study, gene deletions were analyzed using metabolomics, and multivariate analysis of these phenotypic fingerprints resulted in coclustering of yeast gene knockouts. The central idea of this functional genomics approach was that when different yeast gene knockouts clustered together, they would do so because the gene deletion had had the same effect on the yeast phenotype (genotype + environment). This “guilt by association” could then be used to ascribe function to unknown genes. Following this, the same authors, in 2001, used NMR spectroscopy with cluster analysis to elucidate the metabolic phenotype of the so-called silent mutations in S. cerevisiae. This pioneering paper showed that even though genetic knockouts in yeast had no effect on the growth rate of the organism (hence were considered silent), that intracellular concentrations of key metabolites in central carbon and nitrogen metabolism changed and that these changes in metabolite flow could be used to reveal the function of genes that do not participate directly in metabolism or its control (6).
One of the challenges with microbial metabolomics is the need to quench metabolism to inactivate any enzymes inside the cells. Popular methods are based on the use of solvents like methanol (52), and these and indeed other quenching methods may cause leakage of metabolites from the bacterial cells. An alternative to measuring the intracellular metabolome (endometabolome) is to use simple filtration (or centrifugation) to remove the cells and thereby measure the metabolic footprint (extracellular metabolome or exometabolome). This was first demonstrated in 2003 for functional genomics in yeast using direct infusion MS for high-throughput analyses (6) and subsequently in the bacterium E. coli with comparison with FT-IR spectroscopy (53). Later in 2017, Uwe Sauer and colleagues would use a similar approach to profile over 3,800 single-gene deletions from the E. coli Keio knockout collection (54), which shows the scale at which modern microbial metabolomics can be conducted. In this massive study, these authors mapped the relative changes in more than 7,000 metabolites and systematically mapped and thereby identified both expected changes following loss of gene function and gene–metabolite interactions that were not represented in metabolic models (54). Using this untargeted metabolomics approach, they predicted additional metabolism-related functions of 72 previously nonannotated genes, which really illustrate the power of metabolomics within bacterial functional genomics. Similarly, studies from the Ralser group in the yeast S. cerevisiae, where targeted quantification of 19 amino acids was undertaken, studied the changes in amino acid metabolism in 4,913 gene deletion strains that were viable in the absence of amino acid supplementation (55). This work revealed that over one third of coding genes contribute to amino acid biosynthesis allowing the correlation of metabolic regulators to their effectors, and these authors identified that chromatin and transport proteins were the largest affected on the amino acid metabolome. In combination, this study like the study of Fuhrer et al., all building on the initial study of Raamsdonk in 2003, showed that metabolomics can be used to quantify “underexplored space in gene function” (55).
Metabolomics as a discipline takes single snapshots of biological systems and therefore misses dynamics and fluxes in pathways. Again, pioneered in the microbial area, researchers have used stable isotope substrates to evaluate intracellular metabolic flux with mass spectrometry and NMR spectroscopy (for a review of last century, see ref. 56). This “chasing” of the 13C label through metabolism can be used to define more accurately metabolic networks with a view to enhancing metabolic engineering in bacteria, for example, fine chemical production (57).
Infancy of Metabolomics
Metabolic profiling—the analysis of scores of metabolites, typically covering metabolites belonging to a similar chemical class (ideally covering multiple metabolic pathways)—proceeded metabolomics and several different births of this discipline with pesticide mode-of-action profiling often being cited as its start point (58), but compelling arguments are made by Giera et al. (7) for studies using the chromatographic techniques established by Archer Martin and Richard Synge (59). NMR-based metabolomics was defined early on by Nicholson, Lindon, and Holmes (1999) as ‘the quantitative measurement of the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification’ (60). While this approach proceeded the development of metabolomics, it was largely constrained to studying response to xenobiotic exposure and in vivo biomarker identification. Nevertheless, metabonomics had a significant conceptual impact on the development of metabolomics, particularly in the early 2000s (Fig. 1). While the advent of metabolic profiling is not so clear that of metabolomics can be more easily pinpointed. As we state above, initial metabolomics studies utilized GC-MS using electron ionization to provide the highly reproducible fragmentation patterns required for large-scale experiments (13). GC-MS has the twin advantages of being relatively sensitive and highly robust meaning that it can routinely measure hundreds of analytes, however, due to its mass range limitations and is restricted to primary metabolism. GC-MS as it is most commonly employed offers little information regarding structure, and as such, the running of standard compounds to identify the analytes is imperative. Its early application was centered on profiling transgenic or environmentally challenged plants (4, 16, 17). We will limit ourselves to two examples. First, the study of Fiehn and coworkers used GC-MS to evaluate 326 analytes in comparing a developmental (stomatal density and distribution) and a metabolic (digalactosyldiacylglycerol synthase) mutant of Arabidopsis with their respective phenotype (4). Unsurprisingly, this study revealed that the metabolic mutant displayed a greater alteration in its metabolome than the developmental one. Second, studies of Roessner et al. (16, 17) that carried out GC-MS profiling of 60 known and 27 unknown metabolites in a range of transgenic potato plants with elevated sucrolytic activity and wild-type tuber material supplied exogenously with varying concentrations of sugars. These transgenic lines had been created in attempt to enhance starch yield but actually resulted in the opposite phenotype. The metabolomics studies revealed that this was due to a massive activation of respiration and amino acid biosynthesis. However, the sugar feedings in conjuncture with principal component analyses (PCA) were further able to demonstrate that this was induced by high levels of hexoses, thereby providing mechanistic insight into the causes behind the failure of this metabolic engineering strategy. Shortly after these studies, the first publication of high-resolution mass spectrometry–based metabolomics was published (5). In this study, direct injection of extracts was followed by either electrospray ionization or atmospheric pressure chemical ionization (in positive and negative modes) to characterize four different developmental stages of strawberry fruit development and ripening and to evaluate the differences in the metabolome evoked by the expression of a transgene in tobacco. Fourier transform ion cyclotron resonance mass spectrometry providing ultra-high mass resolution was employed in these studies. As mentioned above, this technique, when coupled with liquid chromatography, has become the most powerful metabolomics technique of the last decade. Early examples of coupled LC–high-resolution MS included the evaluation of 2,000 different mass signals from roots of wild-type and the chalcone synthase–deficient tt4 mutant Arabidopsis (61) and the use of a broader range of mutants from this pathway as a route toward peak annotation (62–64). In the microbial field, two studies stand out in this time period (6, 21). In the first of these, it was demonstrated how the intracellular concentrations of metabolites can reveal phenotypes for proteins active in metabolic regulation even in the cases of silent genes which do not display a visible phenotype (6). In the more recent of these multiple high-throughput measurements to study the response of E. coli cells to genetic and environmental perturbations revealed E. coli thus seems to use complementary strategies that result in a metabolic network robust against perturbations (21). As covered in detail in their medical perspective of metabolomics, Giera et al. correctly state that key advances in metabolite measurement were clearly made in animal research (7); however, it did not pioneer application of metabolomics with studies of the scale of those highlighted above not appearing until 2003 in the medicinal sciences. Similarly, the integration of metabolomics with other postgenomic technologies occurred in plants (65–67), and as the example above of Ishii et al. (21) demonstrates in E. coli before, it was implemented in medical research. While as we described below the direct mapping of the metabolomics changes to genome regions (68, 69) and latterly genes (for reviews, see refs. 70 and 71) in plants occurred well before analogous experiments in eukaryotic microbes (72) and mammals (73). Another development that we will mention only in passing is the uptake of stable isotope feeding experiments which has brought great insight into plant metabolic engineering strategies (74), although in this instance, many of the plant studies initially were based on methods developed by Katz and coworkers in mammalian studies (75).
Relearning Statistics and the Transition from a Description to Prediction
As for all big data sciences, metabolomics has become intricately linked with biostatistics ranging from simple assessment of statistically significant differences through multivariate analysis to correlation-based network analysis. Such analyses represented a step-change in the way metabolism was evaluated in that they allowed direct comparison of hitherto seemingly unrelated pathways within the self-same extracts, and when the appropriate caution was adopted additionally allowed the comparison of changes in transcript and protein levels with those in metabolite levels. While use of statistical methods such as t tests and ANOVA were commonplace in the study of metabolism, the advent of metabolomics leads to the widespread adoption of techniques such as principal component (PCA) and partial least squares discriminant analyses along with other methods (for details, see ref. 76). As for the chemical analytical techniques themselves, the adoption of these techniques to metabolomics was first seen in the plant and microbial sciences (4–6), although admittedly were rapidly taken up in mammalian studies also (24). Hereafter, the disciplines rapidly diverged in that while the majority of all early metabolomics studies were by nature descriptive, the utility of the results from such studies is arguably more readily apparent from medical studies. For example, it was rapidly realized that a biomarker—even an unknown mass feature—which allows the prediction of the onset of a disease is highly valuable as it may facilitate the early treatment of a disease. Plant and microbial sciences were at this stage more focused on providing comprehensive descriptions of changes to environmental and genetic perturbations and a better understanding of the interconnectivity of metabolism and the link from genotype to phenotype. This information has since proved instrumental in the design of current generation metabolic engineering strategies (74); however, it took a while before the predictive value of metabolomics experiments in plants and microbes was realized. Early studies in plants used metabolic signatures of young Arabidopsis plants to predict their future growth (77), while a more recent example used machine learning bases on the metabolomic profiles of tomato and blueberry populations to predict taste (78, 79). Similarly, in microbes, the predictive power of metabolomics has been used, among other things, to identify mutations in yeast mutant collections (80). Likewise, in biomedical research, the use of biomarkers predictive of the early onset of disease is a massive research field consuming many millions of research dollars and offering great promise in both general and personalized medicine (81, 82).
Directly Coupling the Metabolome to the Genome
We mention above that it is difficult to assess how big the metabolome actually is. This was demonstrated by attempts to do just that in E. coli coming up with a complement of approximately 750 metabolites (83); however, experimental evidence suggested that the metabolome of this species was at least twice this size (21). This case study clearly illustrates that the metabolome is, at least partially, genome independent in that as opposed to proteomics, there is not a linear relationship between the genome and the metabolome. This relationship is complicated by the presence of both horizontal and allosteric control—with the former suggesting that transcripts and metabolites should be correlated but the later governed by effector molecules that may be hard to measure. Having said that utilizing metabolomics to screen large-scale genetic populations has proven instrumental in understanding the genetic architecture of metabolism. Here, the lead was taken by plants with two quantitative trait loci studies being published already in 2006 (68, 69) and many of them later (e.g., refs. 84–86) as well as genome-wide association-based studies establishing prominence since 2010 (70, 71, 87) (Fig. 2). The latter technique was developed in the medicinal sciences, however, its use in combination with metabolomics was not taken up for several years. Indeed, the utilization of metabolomics as part of a multiomics strategy in plants has proven instrumental in both the understanding of how domestication impacted the metabolome (88, 89) and the identification of key genes as future breeding tools (90, 91). Orthogonally, it can be a very powerful tool in the identification of unknown genes since it often is able to distinguish the causal gene underlying accumulation of a metabolite. At least in the case of genes of known function, this can be highly informative. When talking of biological function, it is important to stress that those of the vast majority of the million plus metabolites across the kingdoms of life are unknown or at least only known in very general terms. The study of the metabolic response to stress across broad genetic populations is beginning to address this (45, 91), as are techniques to determine metabolite–protein interactions, which have also been pioneered in plant and microbial systems (92, 93). We will return to such approaches later when we outline what in our opinion are the most important near- and mid-term perspectives for metabolomics. Two further approaches merit discussion here. In the first, machine learning has been used in combination with quantitative proteomics in kinase knockouts in yeast to predict the metabolome of kinase-deficient cells on the basis of enzyme abundance data (94). This study revealed not only that enzyme abundance signatures are highly specific to the various kinases but were able to model the levels of adenosine triphosphate (ATP), adenosine diphosphate (ADP), AMP, adenosine monophosphate, and glycolytic and pentose phosphate pathway intermediates, as well as amino and organic acids, and showed that the (real) measured values correlated significantly with those predicted by the models. These data could subsequently be interrogated in terms of genotype–phenotype interactions intriguingly revealing that over half of the changes in metabolite concentration are attributable to changes in enzyme abundance. A second approach that can aid in identifying nonannotated metabolites (i.e., currently nonidentified metabolite features in MS) is the use of genome-scale metabolic models. While such models have been available for many years (95), they have in general only been confined to primary metabolism for which metabolomics approaches afford good coverage. Given that such modeling approaches for both microbial and plant metabolism have been extensively reviewed (96, 97), we will only briefly describe them here. Essentially, the underlying principle of such approaches is that metabolic networks can be reconstructed on the basis of knowledge of the enzyme encoding complement of the genome—which can be from computational approaches, literature-based findings, and jamborees where consensus from a group of researchers is reached (e.g., as in ref. 98). This naturally requires that the intermediates linking each pair of consecutive enzymes are known. If these are not present in the list of annotated metabolites in a metabolomics experiment, they represent excellent candidates for one of the features that remain nonannotated. This approach suffers somewhat from the fact that genome annotations remain far from perfect and the fact that it excludes nonenzyme-catalyzed reactions but nevertheless can prove effective, especially as most enzyme reactions cause predictable mass differences between metabolites (99). Indeed, since more extensive genome-scale models of secondary metabolism in Streptomyces species were established (100), this method has been utilized to aid in systems metabolic engineering strategies (101). Moreover, the approach has been adopted to study predictive evolution of metabolic pathways in order to identify lineages with enhanced metabolite secretion (102). These studies suggest that, despite the caveats mentioned above, marrying metabolomics with genome-scale modeling of metabolism could prove a major boon in the identification of missing metabolites from cellular metabolic networks.
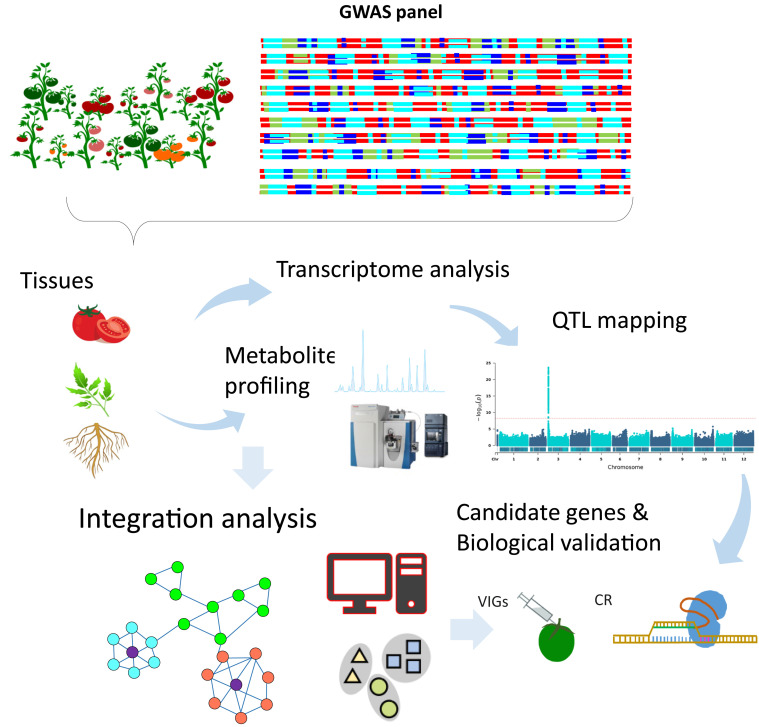
Fig. 2.
An illustrative example of genome-wide association in plants. A population of natural variants harboring sufficient genetic diversity is profiled by metabolomics either in stand-alone experiments or in combination with transcriptomics. These studies not only provide understanding of gene function but also identify key gene/trait associations that can be indicative of protection against biotic or abiotic stresses or of crop yield in plants and of disease susceptibility in mammals.
Expansion of the Coverage of Metabolomics
The extent of metabolome coverage in metabolomics experiments is by no means the most problematic aspect of the technology since its commencement. It has two major weaknesses: The first is the actual metabolites detected in a single extract and analytical run, and the second, the total number of metabolites that could be unambiguously annotated. As an example for the first is that less than a fifth of the approximately 2,700 predicted E. coli metabolites could be detected (27, 28). The situation in the plant kingdom is much worse as less than 1% of the total 2 million predicted metabolites (the latter number is certainly an underestimation) is recovered (27, 35). To overcome this issue and significantly increase the amount of detected metabolites, metabolomics assays should be performed more comprehensibly by using different extraction methods and various types of ionizations, multiple MS, and separation technologies, reducing matrix effects, and improving chromatography. While our capacity to identify metabolites has been improved significantly, a typical LC-MS-based metabolomics experiment will include on average of 1% unambiguously annotated metabolites, the goalposts keep moving with the advent of higher-resolution MS platforms mean that more chemical species can be resolved. Ion mobility (IM) separates ionized compounds based on their mobility through gas in the presence of an electric field. The time it takes for an ionized species to drift (i.e., drift time) can be converted into collision cross-section (CCS) values which are instrument independent and can be employed as an additional criterion determining the confirmation of structures. At the same time, information on isomers, derivatives, and even metabolites from the same chemical class could be of great value in biological studies.
The Metabolomics Standards Initiative for metabolite annotation (103) states that for definitive (Level 1) identification, a standard must be run on the same instrument or instruments and at least two orthogonal characters linked. Thus, the surest way of correctly annotating a metabolite in mass spectrometry–based metabolomics remains matching the properties of an analyte from an extract to those of authenticated purified standards (104, 105). As such, the most facile route to identifying unknown metabolites would be to purchase large standard libraries. However, this presents a huge challenge as many metabolites are not available, and so there are no standards with which to match against. When large standard libraries are available, metabolite identification via spectral matching has proven partially effective with a number of studies with two of particular note in plants being those used for the annotation of lipophilic compounds and specialized metabolites which considerably enhanced coverage of these metabolite classes (104, 105). Complete plant 13C labeling is a different approach for enhancing metabolite identification. In a recent example, Tsugawa et al. (2019) described a metabolome analysis pipeline and metabolites classification for 12 plant species, including model and medicinal plants. They assigned 1,092 structures to 3,604 carbon-determined metabolite ions, 69 of which were found to represent structures not reported earlier. An effective alternative of this approach is the sharing of biological extracts that have been well characterized. For example, it has been shown that there is remarkable conservation in the chromatographic and spectral properties of GC-MS approach that is even independent of the machine type (106). This is not so facile for LC-MS which displays far greater variability in retention times; however, the MassBank database (107) and the sharing of extracts have proven a highly effective manner to (slightly) improve the coverage of various pathways such as, for example, the solenoid glycoalkaloid and polyphenol metabolism of tomato (108). Similar approaches are also being taken in microbial and mammalian metabolomics, although as mentioned above, the problem of coverage is arguably considerably lower in these species.
Ever since the emergence of metabolomics, large efforts have been invested in improving our computational capabilities to identify metabolites in the highest confidence. Such computational tools have been a major driver in metabolite annotation. The long list of metabolomics computational tools developed to date does not allow us to discuss most of them here, and we hence highlight a few recent ones. Especially machine learning–based algorithms generated and implemented in recent years have been a major driver in solving problems of metabolite structure elucidation in metabolomics data analysis, providing major advances in this field (109). Assigning relatedness between metabolites and association with a particular chemical class is the most valuable capability in metabolomics data analysis. It highly complements accurate, single metabolite level annotation particularly when a large set of metabolites is under investigation. A deep learning–based algorithm termed NPClassifier can automatically classify putatively annotated structures into classes and superclasses, particularly natural products (110). MassGenie is another deep learning method that successfully inverts the problem of mass spectrometric molecular identification that is to say, ‘given a mass spectrum can one predict the 2D structure of the molecule from whence it came’ (111). Another program used the Tanimoto similarity index and clustered the metabolites in a form of a tree to subclusters and chemical classes based on their molecular fingerprints (104). The CANOPUS (class assignment and ontology prediction using mass spectrometry) tool predicts close to 2,500 compound classes using fragmentation spectra and employs deep neural networks (DNNs) (112). It explicitly targets compounds lacking spectral or structural reference data reaching very high (i.e., 99.7%) prediction accuracy. The DeepCCS and DarkChem models developed by Plante et al. (113) and Colby et al. (114), respectively, employed machine learning algorithm and models of deep neural network (DNN) to predict CCS obtained in IM-based separation experiments (see above). The CCS value is a valuable addition to the classical spectral matching and increases the confidence in metabolite annotation. Another computational tool for metabolite annotation which is broadly used in the metabolomics community is Global Natural Products Social Molecular Networking that generates the so-called molecular families based on similarity between fragmentation spectra (MS2) of molecules (115). More recently, a complementary tool termed FBMN (standing for feature-based molecular networking) was introduced that uses additional information from MS1 (isotope patterns and retention times) and IM-MS information (115, 116). While the majority of these techniques were pioneered in mammalian research, it is important to note that FBMN has recently greatly benefited from community-based efforts spanning mammalian, microbial, and plant research. Analysis with FBMN advances further metabolite annotation particularly by distinguishing isomers that separate differently in chromatography or IM-MS. An important current challenge is finding a way to maximally utilize NMR in this regard. LC coupled to NMR was long championed as an ideal approach for metabolomics, and in theory, this is still the case. That said, the dearth of published results to support these claims suggests that its application in practice is considerably more difficult than had been anticipated. As an alternative approach to offline approach whereby LC is used to partially purify constituents of complex biological samples prior to their structural determination via NMR would seem likely to be a promising compromise.
Perspective
Despite the relatively low completeness of metabolomics assays as compared to proteomics and certainly transcriptome analysis, the field is expected to make major strides in the coming years toward a more comprehensive examination of metabolomes. Increasing the resolution of metabolomics analysis in terms of resolving metabolites at the spatial level is likely to increase. Particularly in the case of multicellular organisms, metabolomics samples classically include whole organs or tissues. It is apparent that in many cases, metabolites are localized to a particular cell layer and even a few cells with similar identity and function. Thus, sampling specific cells might allow detection while whole organs and tissues results in averaging and significantly reduced metabolite levels consequently leading to lower chances of detection. Accordingly, metabolomics of single cells and moreover the “holy grail” of subcellular compartment analysis could be increasing the amount of yet undetected metabolites.
In recent years, technologies for high-resolution spatial “mapping” of metabolites have been advancing rapidly (117). The spatial resolution of Mass Spectrometry Imaging (MSI) technologies has been improving and currently reaching below 10-mm spatial resolution in some systems. Yet, MSI technologies employed to date suffer from two major drawbacks: i) they cannot reach the required subcellular resolution, and ii) they do not allow efficient production of 3D “imaging” of metabolites from a given sample. The recently developed HybridSIMS (secondary ion mass spectrometry) technology [IonTOF (time-of-flight); (118)] represents the next-generation metabolite “imaging” system as it provides subcellular resolution (nanometer scale) and efficient 3D metabolite mapping. This technology involves time-of-flight secondary ion mass spectrometry (ToF-SIMS) in which a sample is directly irradiated with an accelerated ion beam, and the generated secondary ions are measured using a ToF analyzer and combined with high mass resolution and mass accuracy as well as MS/MS capability mass analyzer (i.e., Orbitrap). The lack of separation in MSI analysis makes high-confidence metabolite identification a major issue, particularly in case of isomers. Coupling IMS to MSI as well as integrating and comparing spectral data from high resolution mass spectrometry (HRMS) of extracts from the same tissue are likely to improve metabolite assignment. Multimodal sampling of the same tissue as, for example, through multiplexed ion beam imaging (MiBI) or nano-desorbtion electrospray ionization (DESI) performing spatial protein mapping, transmission electron microscopy (TEM), fluorescent microscopy, and cytological stains could be assisting in elucidation of unknown structures (119) as could the adoption of orthogonal techniques such as photothermal infrared and Raman which can readily image single bacterial cells (120, 121). Extending metabolomics studies to organisms under multiple conditions (i.e., various biotic and abiotic stresses) and in the course of interactions with other organisms, as well as probing more natural diversity and exotic species, could further increase the number of metabolites detected and comprehensiveness of metabolomics analysis. Precursor feeding experiments could also “expose” pathway intermediates not detected without precursor supplementation, particularly when coupled with stable isotope labeling. One cannot rule out the possibility that some metabolites detected are in fact products of microbial metabolism which modify host metabolites. Thus, metabolomics experiments in the coming years might expose many cases, such as in the human gut, in which some part of the host metabolic repertoire is in fact a result of microbial conversion (42). Such studies might be evolving to the so-called metametabolomics approaches which represent metabolomics of a community rather than interrogation of an individual organism in a given sample. Talking of communities reinforcing the idea of a metabolomics community featuring researchers from all fields of the life sciences will be important for the future. Indeed, while we have come a long way over the last quarter of a century, it is our contention that the remaining challenges are so epic they will need far greater cooperative efforts from biologists, chemists, and computer scientists with an interest in all kingdoms of life than have been made to date. Ultimately, a better linkage of metabolome and genome data will likely also be needed particularly in light of large-scale sequencing projects like the Earth BioGenome Project (122). The use of transcriptomics data to annotate mass spectra has a long history with this approach being used in an early plant metabolomics experiment to define phenylpropanoid metabolism in Arabidopsis (62). The use of both techniques in parallel in characterizing wide natural variance in (plant) species appears to be a highly powerful route to gain insight into the structure of metabolic pathways and their regulation (84). Indeed, despite the lack of an absolute linear connection between the genome and the metabolome, there is much that can be gleaned from the joint study of genomes (and by extension transcriptomes) and metabolomes. One can anticipate that the combination of both platforms will likely provide us with unprecedented datasets that will allow us far greater understanding of both historical and contemporary biology.
Acknowledgments
We gratefully acknowledge Saleh Alseekh and Luis-Alejandro De-Haro for discussions and help with the illustrations. A. A. is the incumbent of the Peter J. Cohn Professorial Chair. We thank the Adelis Foundation, Leona M. and Harry B. Helmsley Charitable Trust, Jeanne and Joseph Nissim Foundation for Life Sciences, Tom and Sondra Rykoff Family Foundation Research, and the Raymond Burton Plant Genome Research Fund for supporting the A.A. laboratory activity.
Author contributions
A.A., R.G., and A.R.F. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
There are no data underlying this work.
References
- 1.Oliver S. G., Winson M. K., Kell D. B., Baganz F., Systematic functional analysis of the yeast genome. Trends Biotechnol. 16, 373–378 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Katona Z. F., Sass P., Molnar P., Simultaneous determination of sugars, sugar alcohols, acids and amino acids in apricots by gas chromatography mass spectrometry. J. Chromatogr. A. 847, 91–102 (1999). [Google Scholar]
- 3.Fraser P. D., Pinto M. E., Holloway D. E., Bramley P. M., Technical advance: Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. Cell Mol. Biol. 24, 551–558 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Fiehn O., et al. , Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18, 1157–1161 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Aharoni A., et al. , Nontargeted metabolome analysis by use of fourier transform ion cyclotron mass spectrometry. OMICS. 6, 217–234 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Raamsdonk L. M., et al. , A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19, 45–50 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Giera M., Yanes O., Siuzdak G., Metabolite discovery: Biochemistry’s scientific driver. Cell Metab. 34, 21–34 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernie A. R., Pichersky E., Focus issue on metabolism: Metabolites, metabolites everywhere. Plant Physiol. 169, 1421–1423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oresic M., Metabolomics, a novel tool for studies of nutrition, metabolism and lipid dysfunction. Nutr. Metab. Cardiovasc. Dis. 19, 816–824 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Hill R., Needham J., The Chemistry of Life: Eight Lectures on the History of Biochemistry (Cambridge University Press, London, 1970). [Google Scholar]
- 11.Kohler R. E., The reception of Eduard Buchner’s discovery of cell-free fermentation. J. History Biol. 5, 327–353 (1972). [DOI] [PubMed] [Google Scholar]
- 12.Harden A., Zymase and alcoholic fermentation. J. Inst. Brew. 11, 2–15 (1905). [Google Scholar]
- 13.Fernie A. R., Trethewey R. N., Krotzky A. J., Willmitzer L., Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5, 763–769 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Kornberg H., Krebs and his trinity of cycles. Nat. Rev. Mol. Cell Biol. 1, 225–228 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Calvin M., “The path of carbon in photosynthesis. Nobel Lecture, 11 December 1961” in Les Prix Nobel in 1961 [Nobel Foundation], Stockholm, G. L., Ed. (Elsevier, Amsterdam, 1961). [Google Scholar]
- 16.Roessner U., et al. , Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13, 11–29 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roessner U., Willmitzer L., Fernie A. R., High-resolution metabolic phenotyping of genetically and environmentally diverse potato tuber systems. Identification of phenocopies. Plant Physiol. 127, 749–764 (2001). [PMC free article] [PubMed] [Google Scholar]
- 18.Allen J., et al. , High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 21, 692–696 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Perez de Souza L., Alseekh S., Scossa F., Fernie A. R., Ultra-high-performance liquid chromatography high-resolution mass spectrometry variants for metabolomics research. Nat. Methods 18, 733–746 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Swartz M. E., UPLC: An introduction and review. J. Liq. Chrom. 28, 1253–1263 (2005). [Google Scholar]
- 21.Ishii N., et al. , Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316, 593–597 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Bailey N. J., Oven M., Holmes E., Nicholson J. K., Zenk M. H., Metabolomic analysis of the consequences of cadmium exposure in Silene cucubalus cell cultures via 1H NMR spectroscopy and chemometrics. Phytochemistry 62, 851–858 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Bundy J. G., Willey T. L., Castell R. S., Ellar D. J., Brindle K. M., Discrimination of pathogenic clinical isolates and laboratory strains of Bacillus cereus by NMR-based metabolomic profiling. FEMS Microbiol. Lett. 242, 127–136 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Griffin J. L., et al. , Metabolic profiling of genetic disorders: A multitissue (1)H nuclear magnetic resonance spectroscopic and pattern recognition study into dystrophic tissue. Analyt. Biochem. 293, 16–21 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Hall R., et al. , Plant metabolomics: The missing link in functional genomics strategies. Plant Cell 14, 1437–1440 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiehn O., Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comparat. Funct. Genom. 2, 155–168 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alseekh S., Fernie A. R., Metabolomics 20 years on: What have we learned and what hurdles remain? Plant J. Cell Mol. Biol. 94, 933–942 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Alseekh S., et al. , Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 18, 747–756 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt-Kopplin P., et al. , Systems chemical analytics: Introduction to the challenges of chemical complexity analysis. Faraday Discussions 218, 9–28 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Guo A. C., et al. , ECMDB: The E. coli metabolome database. Nucleic Acids Res. 41, D625–D630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sajed T., et al. , ECMDB 2.0: A richer resource for understanding the biochemistry of E. coli. Nucleic Acids Res. 44, D495–D501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hautbergue T., Jamin E. L., Debrauwer L., Puel O., Oswald I. P., From genomics to metabolomics, moving toward an integrated strategy for the discovery of fungal secondary metabolites. Nat. Prod. Rep. 35, 147–173 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Ramirez-Gaona M., et al. , YMDB 2.0: A significantly expanded version of the yeast metabolome database. Nucleic Acids Res. 45, D440–D445 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wishart D. S., et al. , HMDB 5.0: The human metabolome database for 2022. Nucleic Acids Res. 50, D622–D631 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito K., Matsuda F., Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 61, 463–489 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Rai A., Saito K., Yamazaki M., Integrated omics analysis of specialized metabolism in medicinal plants. Plant J. Cell Mol. Biol. 90, 764–787 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Jozwiak A., et al. , Plant terpenoid metabolism co-opts a component of the cell wall biosynthesis machinery. Nat. Chem. Biol. 16, 740–748 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Gericke O., et al. , Navigating through chemical space and evolutionary time across the Australian continent in plant genus Eremophila. Plant J. Cell Mol. Biol. 108, 555–578 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Hooft J. J. J., et al. , Linking genomics and metabolomics to chart specialized metabolic diversity. Chem. Soc. Rev. 49, 3297–3314 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Tsugawa H., Rai A., Saito K., Nakabayashi R., Metabolomics and complementary techniques to investigate the plant phytochemical cosmos. Nat. Prod. Rep. 38, 1729–1759 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Tsugawa H., et al. , A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 16, 295–298 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Goodacre R., Metabolomics of a superorganism. J. Nutr. 137, 259s–266s (2007). [DOI] [PubMed] [Google Scholar]
- 43.Eljounaidi K., Lichman B. R., Nature’s chemists: The discovery and engineering of phytochemical biosynthesis. Front. Chem. 8, 596479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenenboim H., Brotman Y., Omic relief for the biotically stressed: Metabolomics of plant biotic interactions. Trends Plant Sci. 21, 781–791 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Obata T., Fernie A. R., The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 69, 3225–3243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones A. M., et al. , The impact of Arabidopsis on human health: Diversifying our portfolio. Cell 133, 939–943 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin C., A role for plant science in underpinning the objective of global nutritional security? Ann. Bot. 122, 541–553 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sreenivasulu N., Fernie A. R., Diversity: Current and prospective secondary metabolites for nutrition and medicine. Curr. Opin. Biotechnol. 74, 164–170 (2021). [DOI] [PubMed] [Google Scholar]
- 49.Liso M., et al. , A bronze-tomato enriched diet affects the intestinal microbiome under homeostatic and inflammatory conditions. Nutrients 10, 1862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butelli E., et al. , Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Barabasi A. L., Menichetti G., Loscalzo J., The unmapped chemical complexity of our diet. Nat. Food 1, 33–37 (2020). [Google Scholar]
- 52.Winder C. L., et al. , Global metabolic profiling of Escherichia coli cultures: An evaluation of methods for quenching and extraction of intracellular metabolites. Analyt. Chem. 80, 2939–2948 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Kaderbhai N. N., Broadhurst D. I., Ellis D. I., Goodacre R., Kell D. B., Functional genomics via metabolic footprinting: Monitoring metabolite secretion by Escherichia coli tryptophan metabolism mutants using FT-IR and direct injection electrospray mass spectrometry. Comparat. Funct. Genom. 4, 376–391 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuhrer T., Zampieri M., Sévin D. C., Sauer U., Zamboni N., Genomewide landscape of gene-metabolome associations in Escherichia coli. Mol. Systems Biol. 13, 907 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mülleder M., et al. , Functional metabolomics describes the yeast biosynthetic regulome. Cell 167, 553–565.e512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittmann C., Heinzle E., Mass spectrometry for metabolic flux analysis. Biotechnol. Bioeng. 62, 739–750 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Fischer E., Sauer U., Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270, 880–891 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Trethewey R. N., Gene discovery via metabolic profiling. Curr. Opin. Biotechnol. 12, 135–138 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Martin A. J., Synge R. L., A new form of chromatogram employing two liquid phases: A theory of chromatography. 2. Application to the micro-determination of the higher monoamino-acids in proteins. Biochem. J. 35, 1358–1368 (1941). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicholson J. K., Lindon J. C., Holmes E., “Metabonomics”: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29, 1181–1189 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Böttcher C., et al. , Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic changes and allows identification of a large number of new compounds in Arabidopsis. Plant Physiol. 147, 2107–2120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tohge T., et al. , Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. Cell Mol. Biol. 42, 218–235 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Tohge T., Fernie A. R., Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protocols 5, 1210–1227 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Achnine L., et al. , Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. Cell Mol. Biol. 41, 875–887 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Urbanczyk-Wochniak E., et al. , Parallel analysis of transcript and metabolic profiles: A new approach in systems biology. EMBO Rep. 4, 989–993 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirai M. Y., et al. , Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 10205–10210 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aharoni A., et al. , Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16, 3110–3131 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schauer N., et al. , Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24, 447–454 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Keurentjes J. J., et al. , The genetics of plant metabolism. Nat. Genet. 38, 842–849 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Alseekh S., Kostova D., Bulut M., Fernie A. R., Genome-wide association studies: Assessing trait characteristics in model and crop plants. Cell. Mol. Life Sci. 78, 5743–5754 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang C., Luo J., Metabolic GWAS-based dissection of genetic bases underlying the diversity of plant metabolism. Plant J. Cell Mol. Biol. 97, 91–100 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Breunig J. S., Hackett S. R., Rabinowitz J. D., Kruglyak L., Genetic basis of metabolome variation in yeast. PLoS Genet. 10, e1004142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nicholson G., et al. , A genome-wide metabolic QTL analysis in europeans implicates two loci shaped by recent positive selection. PLoS Genet. 7, e1002270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sweetlove L. J., Nielsen J., Fernie A. R., Engineering central metabolism–A grand challenge for plant biologists. Plant J. Cell Mol. Biol. 90, 749–763 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Cohen S. M., Rognstad R., Shulman R. G., Katz J., A comparison of 13C nuclear magnetic resonance and 14C tracer studies of hepatic metabolism. J. Biol. Chem. 256, 3428–3432 (1981). [PubMed] [Google Scholar]
- 76.Gromski P. S., et al. , A tutorial review: Metabolomics and partial least squares-discriminant analysis–a marriage of convenience or a shotgun wedding. Analyt. Chimica Acta 879, 10–23 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Meyer R. C., et al. , The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 4759–4764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Colantonio V., et al. , Metabolomic selection for enhanced fruit flavor. Proc. Natl. Acad. Sci. U.S.A. 119, e2115865119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fernie A. R., Alseekh S., Metabolomic selection-based machine learning improves fruit taste prediction. Proc. Natl. Acad. Sci. U.S.A. 119, e2201078119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Allen J., et al. , High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 21, 692–696 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Brindle J. T., et al. , Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using H-1-NMR-based metabonomics. Nat. Med. 8, 1439–1444 (2002). [DOI] [PubMed] [Google Scholar]
- 82.Karczewski K. J., Snyder M. P., Integrative omics for health and disease. Nat. Rev. Genet. 19, 299–310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nobeli I., Ponstingl H., Krissinel E. B., Thornton J. M., A structure-based anatomy of the E.coli metabolome. J. Mol. Biol. 334, 697–719 (2003). [DOI] [PubMed] [Google Scholar]
- 84.Zhu G., et al. , Rewiring of the fruit metabolome in tomato breeding. Cell 172, 249–261.e212 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Garbowicz K., et al. , Quantitative trait loci analysis identifies a prominent gene involved in the production of fatty acid-derived flavor volatiles in tomato. Mol. Plant 11, 1147–1165 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Szymański J., et al. , Analysis of wild tomato introgression lines elucidates the genetic basis of transcriptome and metabolome variation underlying fruit traits and pathogen response. Nat. Genet. 52, 1111–1121 (2020). [DOI] [PubMed] [Google Scholar]
- 87.Chan E. K., Rowe H. C., Hansen B. G., Kliebenstein D. J., The complex genetic architecture of the metabolome. PLoS Genet. 6, e1001198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alseekh S., et al. , Domestication of crop metabolomes: Desired and unintended consequences. Trends Plant Sci. 26, 650–661 (2021). [DOI] [PubMed] [Google Scholar]
- 89.Beleggia R., et al. , Evolutionary metabolomics reveals domestication-associated changes in tetraploid wheat kernels. Mol. Biol. Evol. 33, 1740–1753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tieman D., et al. , A chemical genetic roadmap to improved tomato flavor. Science 355, 391–394 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Zhang F., et al. , Genomic basis underlying the metabolome-mediated drought adaptation of maize. Genome Biol. 22, 260 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sokolowska E. M., Schlossarek D., Luzarowski M., Skirycz A., PROMIS: Global analysis of PROtein-metabolite interactions. Curr. Protoc. Plant Biol. 4, e20101 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Gruber C. H., Diether M., Sauer U., Conservation of metabolic regulation by phosphorylation and non-covalent small-molecule interactions. Cell systems 12, 538–546 (2021). [DOI] [PubMed] [Google Scholar]
- 94.Zelezniak A., et al. , Machine learning predicts the yeast metabolome from the quantitative proteome of kinase knockouts. Cell systems 7, 269–283.e266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Papin J. A., Price N. D., Palsson B., Extreme pathway lengths and reaction participation in genome-scale metabolic networks. Genome. Res. 12, 1889–1900 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heinken A., Basile A., Hertel J., Thinnes C., Thiele I., Genome-scale metabolic modeling of the human microbiome in the era of personalized medicine. Annu. Rev. Microbiol. 75, 199–222 (2021). [DOI] [PubMed] [Google Scholar]
- 97.Clark T. J., Guo L., Morgan J., Schwender J., Modeling plant metabolism: From network reconstruction to mechanistic models. Annu. Rev. Plant Biol. 71, 303–326 (2020). [DOI] [PubMed] [Google Scholar]
- 98.Herrgård M. J., et al. , A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat. Biotechnol. 26, 1155–1160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ohta D., Kanaya S., Suzuki H., Application of Fourier-transform ion cyclotron resonance mass spectrometry to metabolic profiling and metabolite identification. Curr. Opin. Biotechnol. 21, 35–44 (2010). [DOI] [PubMed] [Google Scholar]
- 100.Wang H., et al. , RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput. Biol. 14, e1006541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cho M. K., Lee B. T., Kim H. U., Oh M. K., Systems metabolic engineering of Streptomyces venezuelae for the enhanced production of pikromycin. Biotechnol. Bioengineering 119, 2250–2260 (2022). [DOI] [PubMed] [Google Scholar]
- 102.Jouhten P., et al. , Predictive evolution of metabolic phenotypes using model-designed environments. Mol. Systems Biol. 18, e10980 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sumner L. W., et al. , Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) metabolomics standards initiative (MSI). Metabolomics. 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shahaf N., et al. , The WEIZMASS spectral library for high-confidence metabolite identification. Nat. Commun. 7, 12423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsugawa H., et al. , A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 38, 1159–1163 (2020). [DOI] [PubMed] [Google Scholar]
- 106.Schauer N., et al. , GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 579, 1332–1337 (2005). [DOI] [PubMed] [Google Scholar]
- 107.Horai H., et al. , MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 45, 703–714 (2010). [DOI] [PubMed] [Google Scholar]
- 108.Tohge T., et al. , Exploiting natural variation in tomato to define pathway structure and metabolic regulation of fruit polyphenolics in the Lycopersicum complex. Mol. Plant 13, 1027–1046 (2020). [DOI] [PubMed] [Google Scholar]
- 109.Pomyen Y., et al. , Deep metabolome: Applications of deep learning in metabolomics. Comput. Struct. Biotechnol. J. 18, 2818–2825 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim H. W., et al. , NPClassifier: A deep neural network-based structural classification tool for natural products. J. Nat. Prod. 84, 2795–2807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shrivastava A. D., et al. , MassGenie: A transformer-based deep learning method for identifying small molecules from their mass spectra. Biomolecules 11, 1793 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dührkop K., et al. , Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 39, 462–471 (2021). [DOI] [PubMed] [Google Scholar]
- 113.Plante P. L., et al. , Predicting ion mobility collision cross-sections using a deep neural network: DeepCCS. Anal. Chem. 91, 5191–5199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Colby S. M., Nuñez J. R., Hodas N. O., Corley C. D., Renslow R. R., Deep learning to generate in silico chemical property libraries and candidate molecules for small molecule identification in complex samples. Anal. Chem. 92, 1720–1729 (2020). [DOI] [PubMed] [Google Scholar]
- 115.Nothias L. F., et al. , Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 17, 905–908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aksenov A. A., et al. , Auto-deconvolution and molecular networking of gas chromatography-mass spectrometry data. Nat. Biotechnol. 39, 169–173 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ali A., et al. , Single cell metabolism: Current and future trends. Metabolomics. 18, 77 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Passarelli M. K., et al. , The 3D OrbiSIMS-label-free metabolic imaging with subcellular lateral resolution and high mass-resolving power. Nat. Methods 14, 1175–1183 (2017). [DOI] [PubMed] [Google Scholar]
- 119.Dong Y., Aharoni A., Image to insight: Exploring natural products through mass spectrometry imaging. Nat. Prod. Rep. 39, 1510–1530 (2022). [DOI] [PubMed] [Google Scholar]
- 120.Lima C., et al. , Simultaneous Raman and infrared spectroscopy: A novel combination for studying bacterial infections at the single cell level. Chem. Sci. 13, 8171–8179 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.AlMasoud N., et al. , Discrimination of bacteria using whole organism fingerprinting: The utility of modern physicochemical techniques for bacterial typing. The Analyst 146, 770–788 (2021). [DOI] [PubMed] [Google Scholar]
- 122.Lewin H. A., et al. , The Earth BioGenome project 2020: Starting the clock. Proc. Natl. Acad. Sci. U.S.A. 119, e2115635118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tolstikov V. V., Lommen A., Nakanishi K., Tanaka N., Fiehn O., Monolithic silica-based capillary reversed-phase liquid chromatography/electrospray mass spectrometry for plant metabolomics. Anal. Chem. 75, 6737–6740 (2003). [DOI] [PubMed] [Google Scholar]
- 124.Plumb R. S., et al. , UPLC/MSE; A new approach for generating molecular fragment information for biomarker structure elucidation. Rapid. Commun. Mass Spectrom. 20, 1989–1994 (2006). [DOI] [PubMed] [Google Scholar]
- 125.J. Kopka et al. , GMD@CSB.DB: The Golm metabolome database. Bioinformatics 21, 1635–1638 (2005). [DOI] [PubMed] [Google Scholar]
- 126.Montenegro-Burke J. R., Guijas C., Siuzdak G., METLIN: A tandem mass spectral library of standards. Methods Mol. Biol. 2104, 149–163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hu Q., et al. , The Orbitrap: A new mass spectrometer. J. Mass Spectrom. 40, 430–443 (2005). [DOI] [PubMed] [Google Scholar]
- 128.Kaplan K., et al. , Resistive glass IM-TOFMS. Anal Chem. 82, 9336–9343 (2010). [DOI] [PubMed] [Google Scholar]
- 129.Sen P., et al. , Deep learning meets metabolomics: A methodological perspective. Brief Bioinform. 22, 1531–1542 (2021). [DOI] [PubMed] [Google Scholar]
- 130.Petrick L. M., Shomron N., AI/ML-driven advances in untargeted metabolomics and exposomics for biomedical applications. Cell Rep. Phys. Sci. 3, 100978 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liebal U. W., Phan A. N. T., Sudhakar M., Raman K., Blank L. M., Machine learning applications for mass spectrometry-based metabolomics. Metabolites 10, 243 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no data underlying this work.