Significance
RNA editing is hypothesized to facilitate adaptive evolution via flexibly diversifying the proteome temporally or spatially. However, direct experimental evidence is lacking. This study unveils the functional importance of conserved missense adenosine-to-inosine (A-to-I) RNA editing (CME) sites in Fusarium graminearum and provides convincing experimental evidence for the adaptive advantages of two CME sites. The first CME site drives the CME5 gene gaining a new important function in ascus and ascospore formation during evolution. Having an editable A at this site is fitter than an uneditable A or a genomically encoded G. The second CME site in the CME11 gene confers a “heterozygote advantage” during ascosporogenesis, meaning that concurrently expressing both edited and unedited versions is more advantageous than either.
Keywords: RNA editing, adaptation, missense editing, sexual reproduction, Fusarium graminearum
Abstract
Adenosine-to-inosine (A-to-I) editing is the most prevalent type of RNA editing in animals, and it occurs in fungi specifically during sexual reproduction. However, it is debatable whether A-to-I RNA editing is adaptive. Deciphering the functional importance of individual editing sites is essential for the mechanistic understanding of the adaptive advantages of RNA editing. Here, by performing gene deletion for 17 genes with conserved missense editing (CME) sites and engineering underedited (ue) and overedited (oe) mutants for 10 CME sites using site-specific mutagenesis at the native locus in Fusarium graminearum, we demonstrated that two CME sites in CME5 and CME11 genes are functionally important for sexual reproduction. Although the overedited mutant was normal in sexual reproduction, the underedited mutant of CME5 had severe defects in ascus and ascospore formation like the deletion mutant, suggesting that the CME site of CME5 is co-opted for sexual development. The preediting residue of Cme5 is evolutionarily conserved across diverse classes of Ascomycota, while the postediting one is rarely hardwired into the genome, implying that editing at this site leads to higher fitness than a genomic A-to-G mutation. More importantly, mutants expressing only the underedited or the overedited allele of CME11 are defective in ascosporogenesis, while those expressing both alleles displayed normal phenotypes, indicating that concurrently expressing edited and unedited versions of Cme11 is more advantageous than either. Our study provides convincing experimental evidence for the long-suspected adaptive advantages of RNA editing in fungi and likely in animals.
RNA editing is an epitranscriptomic modification that alters RNA sequences by base insertions, deletions, or modifications. Adenosine-to-inosine (A-to-I) editing of mRNA, mediated by members of the adenosine deaminase acting on RNA (ADAR) family (1), is the most prevalent type of RNA editing in the animal kingdom. Since the ADAR family is an animal-specific innovation (2), there is no ADAR ortholog in organisms outside the animal kingdom. Nevertheless, A-to-I RNA editing has been found in fungi recently, specifically during sexual reproduction with an unrecognized mechanism (3–8). A-to-I RNA editing seems to be a common feature in Sordariomycetes (9), one of the largest classes of Ascomycota, characterized by perithecial ascomata and inoperculate unitunicate asci.
Because I is recognized as guanosine (G) by translational machinery, A-to-I RNA editing has a similar functional consequence as an A-to-G substitution (1). Editing of protein-coding sequences may lead to nonsynonymous substitutions, possibly affecting the function of proteins. However, unlike genomic mutations, RNA editing is seldom complete and leads to the presence of both edited and unedited transcripts in a cell. The percentage of edited transcripts over total transcripts at a given site (editing level) can be flexibly regulated in a tissue-specific, developmental stage-dependent manner (6, 10). Therefore, nonsynonymous A-to-I RNA editing has long been assumed to have evolutionary advantages over genomic A-to-G substitutions (1, 11), as it increases the intraorganism proteome diversity by creating two protein isoforms per edited site concurrently, which may confer a “heterozygote advantage.” Alternatively, but not mutually exclusively, it can offer an edited version of proteins temporally or spatially that may be advantageous to organisms without affecting the genomically encoded A phenotype in tissues or stages where editing does not occur. However, whether RNA editing is adaptive or not is presently a point of debate. At present, experimental evidence on the adaptive advantages of nonsynonymous RNA editing is inadequate.
In animals, most A-to-I editing sites occur in noncoding regions associated with repetitive elements, and the fraction of nonsynonymous editing sites is low, especially in mammals (3, 12). The vast majority of nonsynonymous editing sites in mammals are functionally unimportant and nonadaptive (13, 14). In Drosophila and coleoid cephalopods, only the evolutionarily conserved nonsynonymous editing sites with higher editing levels were suggested to be adaptive by evolutionary analysis (15–17). Unlike in animals, most of the A-to-I editing sites in fungi are nonsynonymous (3, 4, 6, 7). Evolutionary analysis suggests that the prevalent nonsynonymous editing is generally adaptive in fungi (6, 18). Nevertheless, it is largely unknown which of the nonsynonymous editing sites is functionally important and what adaptive advantage can be gained from these editing events rather than genomic mutations. Experimental studies of the functional and fitness consequences of individual editing sites are challenging and time-extensive but essential for resolving these fundamental questions.
To date, only a small number of strongly edited and conserved A-to-I editing sites have been functionally characterized in animals (1, 19). Although a few cases of editing sites were shown to have a clear effect on protein functions in vitro or in cell lines (20–26), the importance of these editing events for development and normal physiology in organisms has not been determined by genetic targeting to ablate RNA editing. Only three editing sites have been demonstrated to have critical physiological significance using transgenic mice impaired in RNA editing, the Q/R site in GluR-B, the Q2341R site in FLNA, and the I/M site in CaV1.3. The ablation of the Q/R site editing in GluR-B resulted in early death in mice (27), while the mice deficient in the editing of the FLNA Q2341R site led to left ventricular hypertrophy and cardiac remodeling (28). It is worth considering that being functionally important does not necessarily mean that the editing site is adaptive. For example, the Q/R site editing in GluR-B is required to correct for a deleterious G-to-A genomic mutation (29, 30). According to the “harm-permitting model” (16), this type of editing (restorative editing) is not adaptive as having a flexible editable A is no fitter than the original genomically encoded G. Instead of causing defects by disrupting editing, the mice deficient in the editing of the CaV1.3 I/M site had improved spatial learning and memory and neuronal plasticity (31). The selective advantage of this editing site is elusive. To the best of our knowledge, there is no direct experimental evidence for the heterozygote advantages of RNA editing, meaning that concurrently expressing both the edited and unedited isoforms of proteins is more advantageous than either an unedited or an edited one.
In fungi, our previous studies experimentally confirmed that 3 premature stop codon correction (PSC) editing sites are functionally important for sexual reproduction in Fusarium graminearum (7, 32, 33), a predominant causal agent of Fusarium head blight (FHB), one of the most devastating diseases of cereal crops worldwide. Sexual reproduction plays a critical role in the infection cycle of F. graminearum because ascospores discharged from perithecia (sexual fruiting bodies) are the primary inoculum of FHB (34). Although functionally important, these PSC editing sites are thought to be restorative editing and not adaptive. The overwhelming majority of nonsynonymous editing sites in F. graminearum are missense editing that changes one amino acid to a different amino acid (AA). It is yet unknown whether individual missense editing sites are functionally important and adaptive for sexual reproduction. It has been suggested that editing sites conserved among multiple species are likely functionally important and beneficial (35). Previously, we identified 454 conserved A-to-I sites among F. graminearum, Neurospora crassa, and Neurospora tetrasperma (6). Of these, 429 are conserved missense editing (CME) sites. The CME sites with higher editing levels are the best candidates for experimental validation of their biological roles.
In this study, we selected all of the 22 CME sites with editing levels of ≥50% in both F. graminearum and N. crassa for functional characterization in F. graminearum. Functions of the genes with CME sites were first validated by gene deletion. We then investigated the role of individual CME sites in the genes important for sexual reproduction by generating underedited (ue) and overedited (oe) mutants, respectively, using site-specific mutagenesis at the native locus. While only the edited version of proteins encoded by CME5 is fully functional during ascus and ascospore formation, concurrently expressing both edited and unedited versions of Cme11 is important for ascosporogenesis. Conservation of amino acid residues at the CME sites was also revealed across different fungal classes. Taken together, our results provided convincing experimental evidence for the long-suspected adaptive advantages of RNA editing.
Results
Identification and Expression Profiling of Genes Containing CME Sites with Higher Editing Levels.
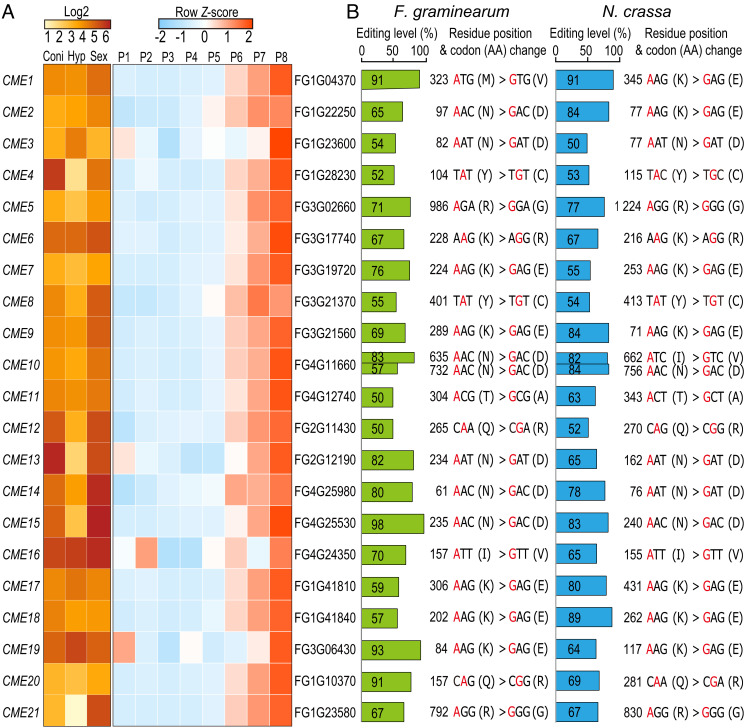
Among the 429 CME sites shared by F. graminearum, N. crassa, and N. tetrasperma (6), 22 of them had editing levels of ≥50% in both F. graminearum and N. crassa (Fig. 1 and SI Appendix, Table S1). These 22 CME sites are in 21 genes (named CME genes) (Fig. 1A), with a single CME site in each CME gene except CME10. Over half of the CME sites have amino acid substitutions with different physicochemical properties, including lysine to glutamate (K-to-E), arginine to glycine (R-to-G), glutamine to arginine (Q-to-R), threonine to alanine (T-to-A), and tyrosine to cysteine (Y-to-C) (Fig. 1B), implying that these CME events potentially influence the function of recoded proteins.
Fig. 1.
Expression patterns of 21 CME genes and editing information of 22 CME sites. (A) Heatmaps showing the log2-normalized or row Z-score scaled TPM values of 21 CME genes from RNA-seq data of conidia (Coni), hyphae (Hyp), and perithecia (Sex) as well as sexual development from 1 to 8 days postfertilization (P1 to P8). See SI Appendix, Table S2 for accession numbers of RNA-seq data used. (B) Editing levels, residue positions, and codon/amino acid (AA) changes of 22 CME sites in F. graminearum and N. crassa. Edited A is marked in red. Editing levels in F. graminearum were calculated from RNA-seq data of the Sex sample (7). Editing levels in N. crassa were the largest editing levels calculated from RNA-seq data of perithecia 3 to 6 days postfertilization (6).
Based on published RNA-seq data (7, 36) (SI Appendix, Table S2), the transcripts of these CME genes were abundant in conidia, vegetative hyphae, and perithecia in F. graminearum, except that a few were expressed at relatively low levels in vegetative hyphae (Fig. 1A). Because A-to-I RNA editing occurs specifically during sexual reproduction, we further examined the expression pattern of these genes with RNA-seq data of sexual development from 1 to 8 days postfertilization (dpf) in F. graminearum. The expression levels of most CME genes were relatively low from 1 to 5 dpf but began to rise at 6 dpf and peaked at 8 dpf (Fig. 1A), suggesting that they may have roles at later stages of sexual development.
Seven CME Genes Are Important for Vegetative Growth.
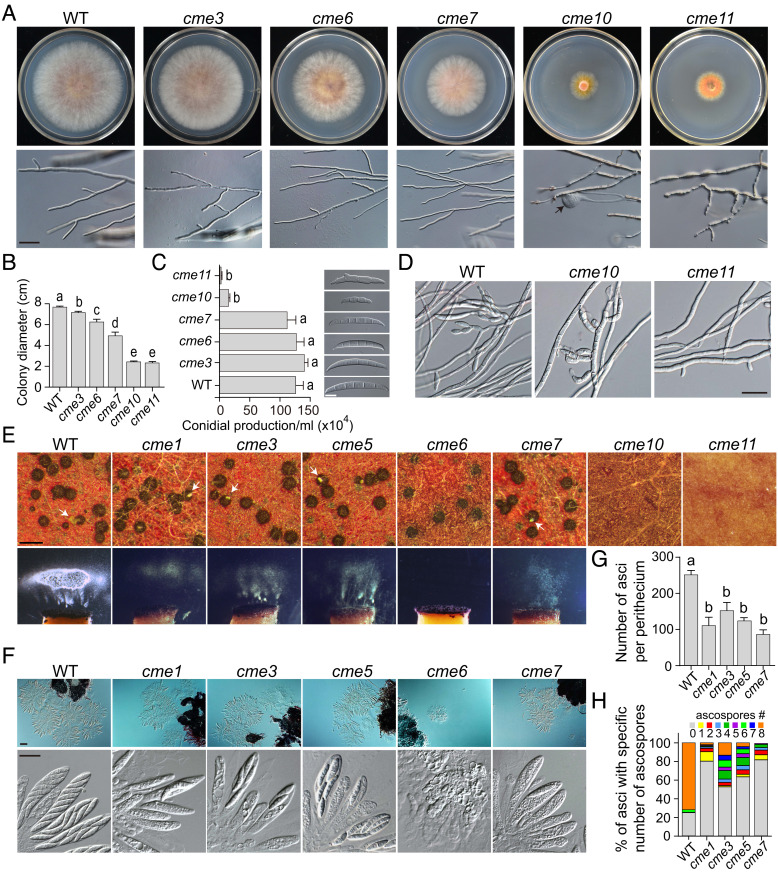
Among the 21 CME genes, deletion mutants of CME4, CME13, CME17, and CME18 have been generated in previous studies (33, 37, 38). For the remaining 17 CME genes, we obtained at least two independent deletion mutants for 15 of them (SI Appendix, Table S3). CME2 and CME20 seem to be essential genes in F. graminearum because we failed to isolate deletion mutants after repeated attempts. The deletion mutants of 10 CME genes were normal in examined phenotypes, while deletion of 7 CME genes resulted in defects in vegetative growth, including CME13 and CME17 reported previously (33, 37) (SI Appendix, Fig. S1 and Table S4). Although the cme3 mutant was only slightly reduced (7%) in the growth rate compared to the wild-type PH-1 strain, the deletion mutants of CME10 and CME11 had severe defects in growth with the growth rate reduced by over 68% (Fig. 2 A and B). The growth rate of cme6 and cme7 mutants was reduced by 19% and 36%, respectively. Although the deletion mutants of CME3, CME6, and CME7 were normal in colony morphology and hyphal branching, the cme10 and cme11 mutants formed colonies with rare aerial hyphae on potato dextrose agar (PDA) plates. In addition, hyphae of the cme10 mutant often had swollen tips. The cme11 mutant produced wavy hyphae with a bigger branching angle. Although the cme3, cme6, and cme7 mutants were normal in conidiation, the cme10 and cme11 mutants produced only a few morphologically abnormal conidia in carboxymethyl cellulose (CMC) cultures (Fig. 2C). Conidia formed by the cme10 mutant were smaller with fewer septa, while conidia of the cme11 mutant were often inappropriately germinated in CMC cultures. Additionally, the cme10 mutant rarely formed clusters of phialides, while the cme11 mutant did not produce phialides (Fig. 2D). Therefore, CME10 and CME11 are crucial for conidiation in F. graminearum. We reintroduced the corresponding full-length genes into these deletion mutants. The resulting complemented strains had similar phenotypes as PH-1 in growth rate, colony morphology, and conidiation (SI Appendix, Fig. S2), indicating that deletion of these genes is responsible for all the phenotypes observed in the mutants.
Fig. 2.
Defects of deletion mutants in vegetative growth, conidiation, and sexual reproduction. (A) Three-day-old PDA cultures and 48-h hyphal tips of PH-1 (WT) and the five marked deletion mutants. The black arrow points to the swollen tip of hyphae. Bar = 20 μm. (B) Colony diameters of 3-d-old PDA cultures of the marked strains. (C) Conidial production of the marked strains was examined by differential interference contrast (DIC) microscopy. Bar = 10 μm. (D) Phialides produced by the marked strains in 72-h carboxymethylcellulose (CMC) cultures were examined by DIC microscopy. Bar = 50 μm. (E) Mating cultures of the marked strains were examined for perithecium formation and ascospore discharge at 8 days postfertilization (dpf). White arrows indicate the ascospore cirrhi. Bar = 0.5 mm. (F) Ascus and ascospore formation of the marked strains were examined by DIC microscopy at 5 dpf (Upper, bar = 50 μm) and 8 dpf (Lower, bar = 20 μm), respectively. (G) The number of asci per perithecium produced by the marked strains at 5 dpf. (H) The percentage of asci containing a specific number of ascospores produced by the marked strains at 8 dpf. For (B), (C), and (G), mean and SD were calculated with data from three independent repeats (n = 3). Different letters indicate significant differences based on one-way ANOVA followed by Turkey’s multiple range test (P < 0.05).
Seven CME Genes Are Important for Sexual Reproduction.
Deletion of seven CME genes resulted in severe defects in sexual reproduction (Fig. 2E). Consistently with their severe growth defects, the cme10 and cme11 mutants failed to form perithecia at 8 dpf on carrot agar. The cme6 mutant generated slightly smaller perithecia without cirrhi formation and spore firing. Elongated asci and matured ascospores were not observed in perithecia at 8 dpf. The cme1, cme3, cme5, and cme7 mutants produced normal perithecia with reduced cirrhi formation and spore firing. Microscopical examination showed that the number of asci per perithecium was reduced in these four mutants (Fig. 2 F and G). Moreover, they often produced fewer than eight ascospores per ascus (Fig. 2 F and H). Particularly, over three-quarters of asci in perithecia of cme1 and cme7 mutants contained no ascospores. The complemented strains exhibited normal phenotypes in sexual reproduction as PH-1 (SI Appendix, Fig. S2). Therefore, CME10 and CME11 are essential for perithecium formation; CME6 is crucial for ascus formation; CME1, CME3, CME5, and CME7 are important for ascus formation and ascosporogenesis in F. graminearum.
Eight CME Sites Are Dispensable for Normal Sexual Development.
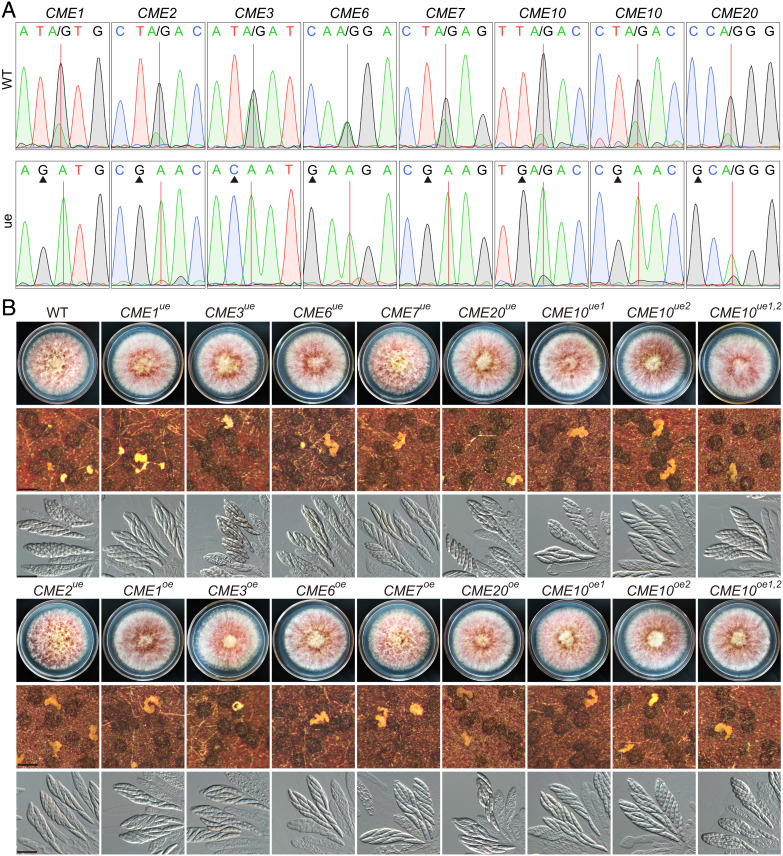
A-to-I RNA editing has strong base preferences, and the preferred bases at −2 to +4 positions of editing sites, especially at the −1 position, are important for editing in F. graminearum (39). Among the ten CME sites in the two essential genes and the seven genes important for sexual development, eight are in the first codon position with preferred thymine (T) at the −1 position of editing sites and the other two were in the second codon position with preferred cytosine (C) at the −2 position of editing sites (SI Appendix, Fig. S3). To characterize the function of these CME sites, we generated underedited mutants by changing the preferred base at the wobble position of the codon immediately preceding the edited codon (the −1 or −2 position of CME sites) into an unpreferred base in situ in PH-1 (SI Appendix, Fig. S3), aiming to abolish or reduce editing without alteration of amino acid sequences. Reverse-transcription (RT)-PCR product sequencing revealed that two peaks (A and G) occurred at the CME sites of CME1, CME2, CME3, CME6, CME7, CME10, and CME20 in Sanger sequencing traces in PH-1 (Fig. 3A). In the underedited mutants, however, only one A peak was observed at the CME sites except for the site in CME20 and the first site in CME10, which also had a minor G peak. These results confirm that editing at the CME sites is abolished or markedly reduced in these underedited mutants. The underedited mutants had no obvious defect in vegetative growth and sexual development (Fig. 3B), suggesting that the edited versions of proteins are not essential for sexual reproduction. We also generated overedited mutants (equivalent to 100% edited) for CME1, CME3, CME6, CME7, CME10, and CME20 by replacing edited A with G in situ (SI Appendix, Fig. S3). All the overedited mutants were normal in vegetative growth and sexual development (Fig. 3B), suggesting that the edited versions of proteins alone are fully functional during sexual reproduction. Because CME10 contains two CME sites, we also simultaneously mutated for both sites. The double underedited (CME10ue1,2) and overedited (CME10oe1,2) mutants had no observed defects in vegetative growth and sexual reproduction (Fig. 3B). Therefore, the eight CME sites in CME1, CME2, CME3, CME6, CME7, CME10, and CME20 are dispensable for normal sexual development.
Fig. 3.
Nonessential functions of 8 CME sites in sexual reproduction. (A) Sequencing traces for flanking sequences of CME sites amplified from cDNA of 7-dpf perithecia of PH-1 (WT) and underedited (ue) mutants. Red lines and black triangles mark CME sites and mutated sites, respectively. (B) Three-day-old PDA cultures and 8-dpf mating cultures of PH-1 (WT) and the marked underedited (ue) and overedited (oe) mutants were examined for colony morphology, perithecium formation, and asci/ascospores morphology. Bar = 0.5 mm (Middle); Bar = 20 μm (Bottom).
The Edited but Not Unedited Version of Cme5 Has Important Functions during Ascus and Ascospore Formation.
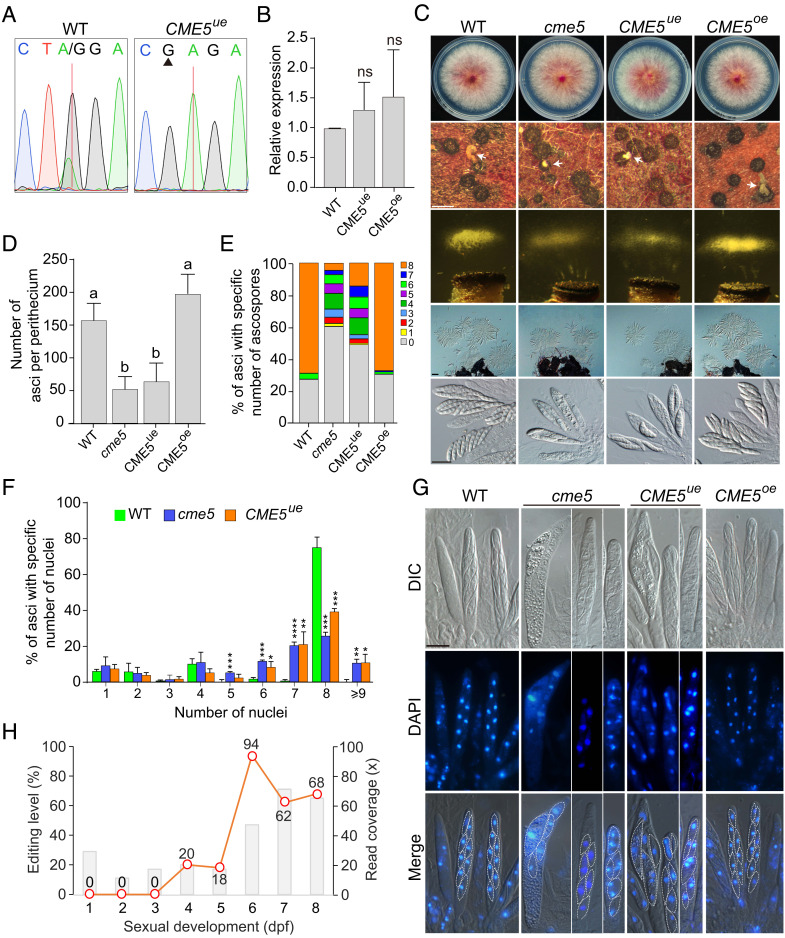
We generated the underedited (CME5ue) and overedited (CME5oe) mutants by site-specific mutagenesis at the native locus of CME5 in PH-1 (SI Appendix, Fig. S3). Editing of CME5 transcripts was abolished in the CME5ue mutant (Fig. 4A). Quantitative real-time RT-PCR analysis showed that the expression level of CME5 in CME5ue and CME5oe mutants was comparable to that in PH-1 (Fig. 4B). The CME5oe mutant was normal in sexual reproduction, but the CME5ue mutant was defective in ascus and ascospore formation (Fig. 4C). Like the cme5 mutant, the CME5ue mutant formed perithecia with smaller and fewer cirrhi. It was also defective in ascospore ejection and produced fewer ascospore masses in ejection assays. The average number of asci per perithecium was reduced by approximately 60% in the CME5ue mutant, and more than 80% of asci had fewer than eight ascospores (Fig. 4 D and E). Therefore, the edited but not unedited version of Cme5 is functional during sexual reproduction, and the CME event plays important roles in ascus and ascospore formation in F. graminearum. Furthermore, DAPI staining revealed that the nuclear division in developing asci at 6 dpf was defective in the CME5ue mutant similar to the cme5 mutant. The fraction of asci with abnormal numbers (5, 6, 7, and >9) of nuclei was increased compared with PH-1 (Fig. 4F). Delimitation of the spore initial in the ascus was observed. Each of the eight nuclei was enclosed by a spore membrane in the CME5oe mutant and PH-1. In the CME5ue mutant and cme5 mutant, however, two or more nuclei enclosed by a spore membrane were often observed (Fig. 4G). Consistently with these observations, the CME site of CME5 had the highest editing level at 6 dpf during sexual development, up to 94% (Fig. 4H). These results indicate that the edited version of Cme5 is important for the nuclear division and ascospore delimitation in the ascus.
Fig. 4.
Defects of the underedited mutant of CME5 during ascus and ascospore formation. (A) Sequencing traces for flanking sequences of the CME site of CME5 amplified from cDNA of 7-dpf perithecia of PH-1 (WT) and the underedited mutant CME5ue. The red line and black triangle mark the CME site and mutated site, respectively. (B) Relative expression of CME5 in perithecia of the marked strains at 7 dpf. Mean and SD were calculated with data from three independent repeats (n = 3). Statistical differences (P < 0.05) relative to PH-1 (WT) are based on the t-test (ns, not significant). (C) Three-day-old PDA cultures and 5-/8-dpf mating cultures of the marked strains were examined for colony morphology, perithecium formation, ascospore discharge, and asci/ascospores morphology. White arrows indicate the ascospore cirrhi. Bar = 0.5 mm (Upper); bar = 50 μm (Middle); bar = 20 μm (Bottom). (D) The number of asci per perithecium produced by the marked strains at 5 dpf. Mean and SD were calculated with data from three independent repeats (n = 3) with at least 30 perithecia examined in each repeat. Different letters indicate significant differences based on one-way ANOVA followed by Turkey’s multiple range test (P < 0.05). (E and F) The percentage of asci containing a specific number of ascospores in the marked strains at 8 dpf (E) or nuclei in the marked strains at 6 dpf (F). Mean and SD were calculated with data from three independent repeats (n = 3) with at least 80 asci examined in each repeat. Significant differences relative to PH-1 (WT) are based on the t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (G) Asci of the marked strains stained with DAPI and examined by epifluorescence microscopy at 6 dpf. The delimitation of ascospores is marked with white dashed lines. Bar = 10 μm. (H) Read coverages (bar chart) and editing levels (line chart) at the CME site of CME5 during sexual development from 1 to 8 dpf.
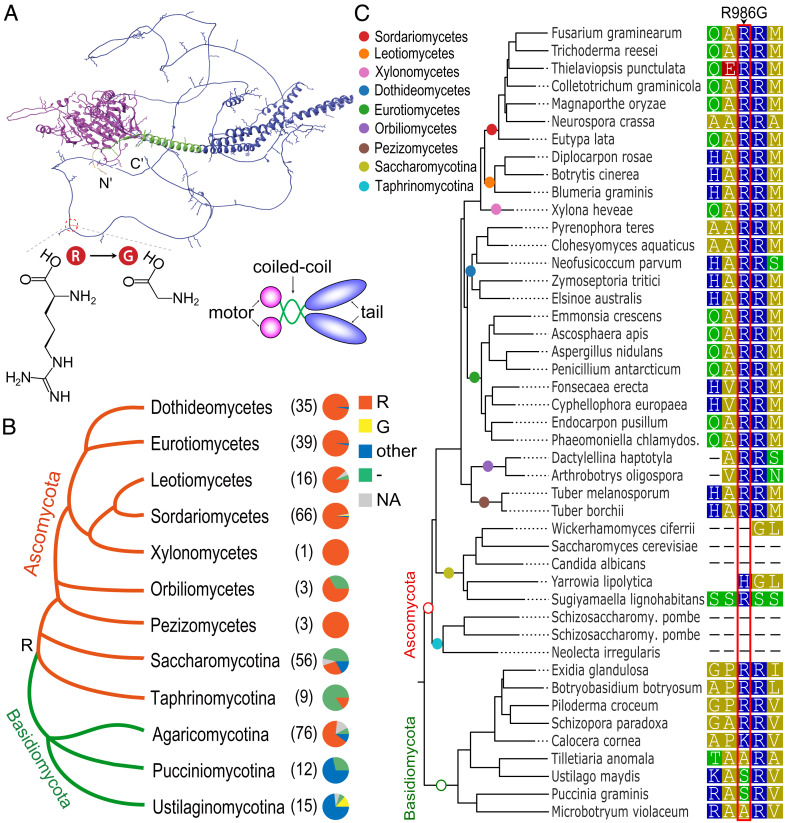
The Preediting Residue of Cme5 Represents the Ancestral State at the CME Site and the Postediting Residue Is Rarely Hardwired into the Genome.
CME5 is orthologous to Saccharomyces cerevisiae KIP3, which encodes a kinesin-8 motor involved in the regulation of microtubule dynamics and spindle organization (40). Like yeast Kip3, Cme5 contains an N-terminal motor domain, followed by a short coiled-coil domain and a C-terminal tail domain (Fig. 5A). Cme5 and its orthologs in filamentous ascomycetes have a relatively long C-terminal tail compared to yeast Kip3 (SI Appendix, Fig. S4). The tail domain contains a high proportion of positively charged lysine (K) and arginine (R) residues (Fig. 5A). The CME site causes a replacement of R986 to glycine (G) in the N-terminal part of the Cme5 tail domain. Orthologs of CME5 are widely distributed in diverse fungal lineages (SI Appendix, Dataset S1). Phylogenetic analysis indicated that the preediting R residue represented the ancestral state at this site in ascomycetes (Fig. 5B). Therefore, according to previous definitions (16), the R986G editing in CME5 is unlikely the restorative editing that converts the amino acid state back to an ancestral state. The R residue at the CME site is conserved during evolution in filamentous ascomycetes. In contrast, the postediting G residue was rarely observed at this site in ascomycetes (Fig. 5B). These results imply that having an editable A at this site leads to higher fitness than a genomically encoded G. Although no significant alteration in the growth rate, conidial production, and virulence was observed for the CME5oe mutant relative to PH-1 and the CME5ue mutant (SI Appendix, Fig. S5), exclusively expressing the edited version of Cme5 may be potentially deleterious in asexual stages where editing does not occur.
Fig. 5.
The 3D structure of Cme5 and conservation of its CME site. (A) Ribbon representation of the AlphaFold structure of Cme5. The N-terminal motor domain, coiled-coil domain, and C-terminal tail domain are shown in magenta, green, and blue, respectively. Positively charged residues are shown as sticks. The N- and C- termini are indicated. The location of the CME site and molecular structures of the preediting residue arginine (R) and postediting residue glycine (G) are shown. A schematic diagram shows the molecular organization of the homodimeric Cme5. (B) Conservation of amino acid residues at the CME site in Cme5 orthologs across different taxa of Ascomycota and Basidiomycota. The dendrogram is drawn based on the MycoCosm tree (46). The number of species in each taxon examined is indicated in the bracket. Each pie chart shows the proportion of fungal species with a preediting residue (R), a postediting residue (G), other types of amino acid residues (other), or missing information (-) at the CME site or without the gene (NA). The inferred ancestral residue R at the CME site in the last common ancestor of ascomycetes is indicated on the dendrogram. (C) Sequence alignment showing amino acid residues at the CME site and neighboring positions in representative species of Ascomycota and Basidiomycota. The phylogenetic tree is drawn based on the gene tree of CME5 orthologs from EnsemblFungi. Colored circles on the branches indicate corresponding taxa. Residues are colored according to their polarity. The CME site is boxed in red.
Concurrently Expressing Both Edited and Unedited Versions of Cme11 Is Required for Ascosporogenesis.
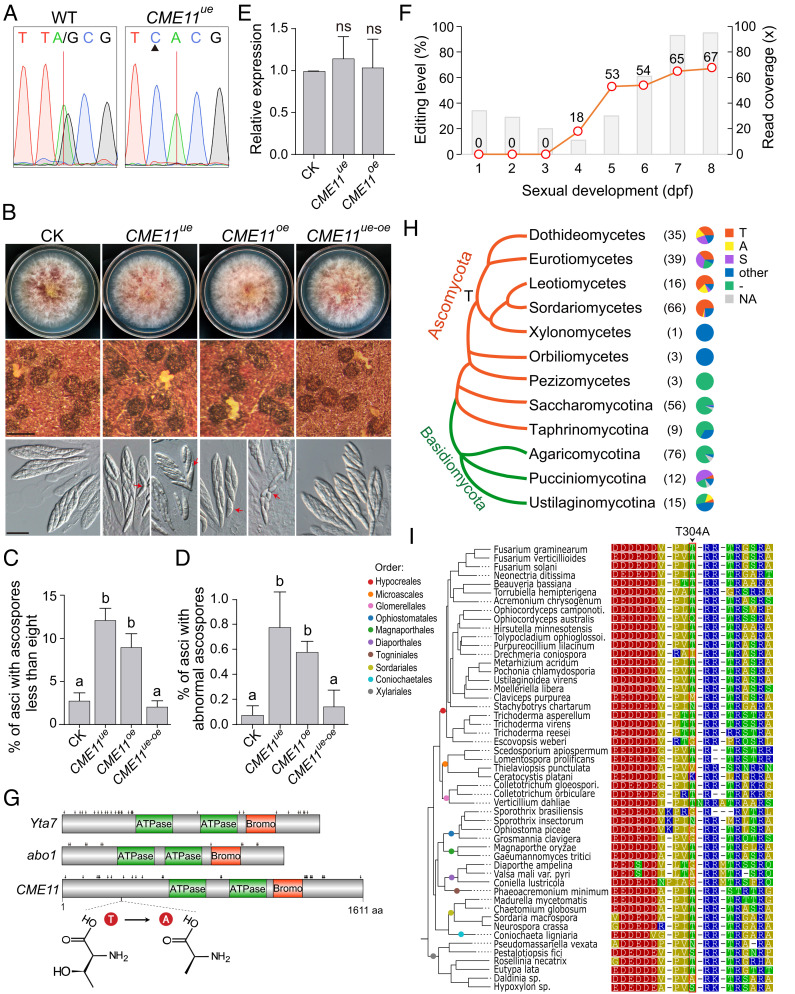
Likewise, we generated both the underedited (CME11ue) and overedited (CME11oe) mutants for CME11 (SI Appendix, Fig. S3). Editing was confirmed to be abolished in the CME11ue mutant (Fig. 6A). Surprisingly, although the growth rate, conidial production, and virulence had no significant alteration, both CME11ue and CME11oe mutants were defective in sexual reproduction (Fig. 6B and SI Appendix, Fig. S6). On average, 12% and 9% of asci in matured perithecia had fewer than eight ascospores, and 0.8% and 0.6% of asci contained ascospores with morphological abnormalities in CME11ue and CME11oe mutants, respectively (Fig. 6 C and D). In contrast, the defects were rarely observed in the control check strain that contains the same selectable marker inserted at the equivalent genomic locus. The expression level of CME11 had no obvious changes in both mutants (Fig. 6E), excluding the possibility that the defect is caused by changes in mRNA stability. We further transformed an overedited allele of CME11 into the CME11ue mutant. The resulting transformant (CME11ue-oe) expressing both underedited and overedited alleles of CME11 showed normal phenotypes in sexual reproduction. Therefore, these results indicate that neither the edited version nor the unedited version of Cme11 is fully functional during sexual reproduction, and ascosporogenesis needs the concurrent expression of both versions of Cme11 in F. graminearum. Compared with CME5, CME11 had a relatively stable, moderate editing level at the CME site during sexual development after 5 dpf (Fig. 6F), which may ensure that both versions of Cme11 are properly expressed during ascosporogenesis.
Fig. 6.
Function and evolution of the CME site in CME11. (A) Sequencing traces for flanking sequences of the CME site of CME11 amplified from cDNA of 7-dpf perithecia of PH-1 (WT) and the underedited mutant CME11ue. The red line and black triangle mark the CME site and mutated site, respectively. (B) Three-day-old PDA cultures and 8-dpf mating cultures of the control check strain (CK) and the underedited (CME11ue) and overedited (CME11oe) mutants were examined for colony morphology, perithecium formation, and ascospore morphology. Red arrows indicate the ascospores with morphological abnormalities. Bar = 0.5 mm (Middle); bar = 20 μm (Bottom). (C and D) The percentage of asci containing less than eight ascospores (C) or containing ascospores with morphological abnormalities (D) for the marked strains at 8 dpf. Mean and SD were calculated with data from three independent repeats (n = 3) with at least 30 perithecia examined in each repeat. Different letters indicate significant differences based on one-way ANOVA followed by Turkey’s multiple range test (P < 0.05). (E) Relative expression of CME11 in perithecia of the marked strains at 7 dpf. Mean and SD were calculated with data from three independent repeats (n = 3). Statistical differences (P < 0.05) relative to CK are based on the t-test (ns, not significant). (F) Read coverages (bar chart) and editing levels (line chart) at the CME site of CME11 during sexual development from 1 to 8 dpf. (G) Conserved domains and phosphorylation sites of S. cerevisiae Yta7, S. pombe abo1, and Cme11. Phosphorylation sites are marked with black arrows. The location of the CME site and molecular structures of the preediting residue threonine (T) and postediting residue alanine (A) are shown. (H) Conservation of amino acid residues at the CME site in Cme11 orthologs across different taxa of Ascomycota and Basidiomycota. The dendrogram is drawn based on the MycoCosm tree (41). The number of species in each taxon examined is indicated in the bracket. Each pie chart shows the proportion of fungal species with a preediting residue (T), a postediting residue (A), a serine residue (S), other types of amino acid residues (other), or missing information (-) at the CME site or without the gene (NA). The inferred ancestral residue T at the CME site in the last common ancestor of leotiomyceta is indicated on the dendrogram. (I) Sequence alignment showing amino acid residues at the CME site and neighboring positions in representative species of Sordariomycetes. The phylogenetic tree is drawn based on the gene tree of CME11 orthologs from EnsemblFungi. Colored circles on the branches indicate corresponding orders in Sordariomycetes. Residues are colored according to their polarity. The CME site is boxed in red.
The Preediting Residue of Cme11 Is Potentially Phosphorylated and Relatively Conserved in Sordariomycetes during Evolution.
CME11 is orthologous to S. cerevisiae YTA7 and Schizosaccharomyces pombe abo1/2, which encode a chromatin-binding ATPase involved in the regulation of chromatin dynamics (41, 42). Cme11 contains two AAA+-type ATPase domains and a TBP7_like bromodomain. The CME site is in the N-terminal region preceding the ATPase domains, resulting in a T304A replacement (Fig. 6G). Notably, the N-terminal regions of Yta7 and Abo1/2 contain many phosphorylated serine (S)/T residues (Fig. 6G). We, therefore, speculated whether the CME site of CME11 was phosphorylated during sexual reproduction. The substitution of T with A by editing could mimic the dephosphorylation of Cme11 at this site. Indeed, the T304 was predicted as a phosphorylated residue in Cme11 by both GPS5.0 (43) and NetPhos-3.1 (44). The T/S residues at this site in its orthologs were also predicted as phosphorylated residues. Therefore, RNA editing fine-tunes the function of CME11 during sexual reproduction possibly via regulating the phosphorylation level at the CME site. To confirm phosphorylation at this site, we generated transformants expressing the CME11-GFP fusion construct. The recombinant Cme11 protein was detected in 16-h hyphae but not in 7-dpf perithecia by western blotting (SI Appendix, Fig. S7), implying that Cme11 is an unstable protein and subjected to regulation specifically during sexual reproduction. We identified 26 phosphorylation sites in Cme11 proteins isolated from hyphae after multiple independent experiments using a triple quadrupole mass spectrometer but did not detect any peptides that cover the T304 (Fig. 6G and SI Appendix, Fig. S7). There may be a special structure blocking enzyme digestion near this site. The orthologs of CME11 are widely distributed in the fungal tree of life, specially conserved in ascomycetes and basidiomycetes (Fig. 6H and SI Appendix, Dataset S1). Phylogenetic analysis suggests that the preediting T residue has arisen at this site at least in the last common ancestor of Dothideomycetes, Eurotiomycetes, Leotiomycetes, and Sordariomycetes (Fig. 6H). The T residue at the CME site was replaced by S in many fungal species in Dothideomycetes and Eurotiomycetes during evolution but generally conserved in Sordariomycetes and Leotiomycetes. Furthermore, T-to-A substitution at this site occurred frequently in Dothideomycetes and Leotiomycetes but rarely in Sordariomycetes (Fig. 6 H and I). The editable A is likely evolutionary maintained in Sordariomycetes to enable the concurrent expression of both versions of Cme11 via RNA editing. The T304 is just located downstream of a stretch of aspartate (D) and glutamate (E) residues and upstream of multiple R residues (Fig. 6I). In fact, the N-terminal tail of Cme11 and its orthologs is rich in both the positively charged R residue and negatively charged D/E residues (SI Appendix, Fig. S8), which are potentially involved in interaction with the chromatin. Phosphorylation of Cme11 at the T304 and other S/T residues in the N-terminal tail possibly regulates dynamic protein-chromatin interactions.
Discussion
Tens of thousands of A-to-I RNA editing sites have been detected specifically during sexual reproduction in fungi. Identifying functional important editing sites from the sea of editing will allow a mechanistic understanding of why RNA editing is advantageous in these cases and how it is co-opted for sexual development in fungi. In this study, we found 2 CME sites in CME5 and CME11 which are both functionally important and adaptive for sexual reproduction in F. graminearum by large-scale gene deletion and site-specific mutagenesis. Note that both CME sites led to amino acid changes with different physicochemical properties.
Deletion of CME5 caused no apparent effect on vegetative growth but significant defects in ascus and ascospore formation in F. graminearum. In S. cerevisiae, however, KIP3 is dispensable for mitotic growth and sexual reproduction (45). The deletion mutant of KipB, the ortholog of KIP3, is also normal in vegetative growth and sexual reproduction in Aspergillus nidulans (47). The sexual stage-specific function of CME5 was most likely to be acquired during evolution. The tail domain of Kip3 promotes the binding of Kip3 to microtubule plus ends and plays a critical role in the destabilizing and stabilizing effects of Kip3 on microtubule dynamics (48). Interestingly, the CME site is just located in the tail domain of Cme5, resulting in an R986G substitution. The edited but not unedited version of Cme5 performed normal function during ascus and ascospore formation, indicating that the R986G editing is crucial for the function of CME5 during sexual reproduction. Because the positively charged residues in the tail domain of the kinesin-8 motors are involved in the binding of the negatively charged tubulin surface (48, 49), the R986G editing potentially regulates the interaction of Cme5 with microtubules. Although also identified in a Pezizomycetes species P. confluens (8), A-to-I RNA editing is common in Sordariomycetes and likely originated independently in the two fungal classes based on our latest evidence. Therefore, the emergence of the R986G editing in Sordariomycetes may promote CME5 to gain a new function in sexual reproduction.
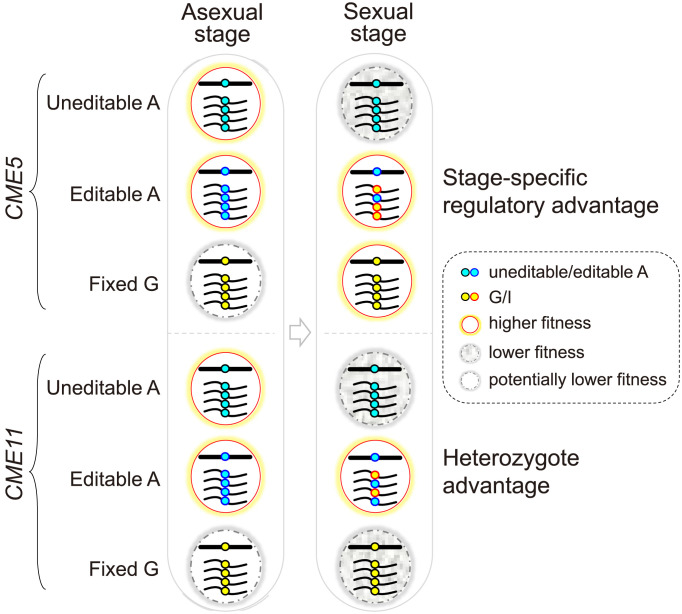
If the edited version of Cme5 is simply fitter than the unedited one during sexual reproduction, it is expected that the postediting G is hardwired into the genome during evolution. However, the preediting residue R986 is evolutionarily conserved, and the postediting G is rarely observed in Sordariomycetes. This raises the question that why CME5 maintains the flexible editable A instead of evolving a genome-encoded G. It is most likely that expressing the edited version of Cme5 in the asexual stage may lead to lower fitness because it is specifically generated and adapted for the sexual stage (Fig. 7). This situation can be perfectly resolved by employing RNA editing, which offers an edited version of proteins specifically during the sexual stage but without affecting the genomically encoded A phenotype in the asexual stage. Although defects of the overedited CME5oe mutant were not observed in vegetative growth, conidiation, and plant infection, our results do not rule out the possibility of more subtle phenotypic consequences under certain conditions. Alternatively, but not mutually exclusively, the unedited version of Cme5 may also have a minor role in sexual reproduction that is not replaced by the edited one. Either way, our results demonstrated the adaptive advantage of the R986G editing site in CME5.
Fig. 7.
Proposed models depicting the adaptive advantages of the CME sites in CME5 and CME11 over genomic mutation. The CME site of CME5 may have a stage-specific regulatory advantage by offering an edited version of Cme5 that has higher fitness in the sexual stage but potentially lower fitness in the asexual stage. The CME site of CME11 confers a “heterozygote advantage,” meaning that concurrently expressing both edited and unedited versions of Cme11 during sexual reproduction is more advantageous than either. Because the edited version of proteins is specifically generated and adapted for the sexual stage, its expression in the asexual stage may be potentially deleterious.
CME11 encodes a conserved AAA ATPase. Its orthologs in yeasts and N. crassa are all important for nucleosome positions as a chromatin remodeler. Deletion of YTA7 caused a progressive shift of nucleosomes toward the 5′-end of genes in S. cerevisiae, while loss of abo1 led to nucleosome disorder and lower nucleosome signals at gene bodies and transposable elements underlying heterochromatin in S. pombe (43, 50). The dim-1 deletion mutant of N. crassa displayed a slow growth rate and atypically spaced nucleosomes throughout the genome (51). In F. graminearum, CME11 may also play a conserved role in nucleosome positioning as the cme11 mutant exhibited severe defects in vegetative growth and both asexual and sexual reproduction. The CME site is located in the N-terminal region of Cme11, resulting in a T304A replacement. Both the underedited and overedited mutants of CME11 were defective in ascosporogenesis, suggesting that normal sexual development in F. graminearum requires the concurrent expression of both versions of Cme11 rather than simply an edited or unedited one. This is experimental evidence of the “heterozygote advantages” of RNA editing (Fig. 7).
Why is expressing both the edited and unedited versions of Cme11 more advantageous than either one? Evolutionary analysis can indicate only whether a fraction of editing sites have heterozygote advantages (proteome diversification) but does not explain how the heterozygote advantages arise. Interestingly, the T304 was predicted as a phosphorylated residue. The T304A editing could therefore reduce the level of phosphorylation at this site. Cme11 at both hypophosphorylated and hyperphosphorylated states may not be normally functional during sexual reproduction. It is, therefore, not hard to understand why having a flexible editable A would be advantageous over either an uneditable A or a fixed G allele. The current study indicates that the heterozygote advantages of RNA editing might be achieved by regulating the levels of protein posttranslational modifications. Considering a high fraction of nonsynonymous editing occurred at the K, R, S, and T sites of proteins in fungi (6, 7), which are potential sites for protein acetylation, methylation, phosphorylation, and ubiquitination, RNA editing may add another layer of regulation to posttranslational modifications of proteins during sexual reproduction.
Besides CME5 and CME11, we found additional seven genes (CME1, CME2, CME3, CME6, CME7, CME10, and CME20) important for F. graminearum. The two potentially essential genes CME2 and CME20 are orthologous to yeast TAD3, an essential gene encoding for the subunit of tRNA-specific adenosine-34 deaminase (52), and yeast KIP1 and CIN8, two functionally redundant genes required for mitotic spindle assembly and chromosome segregation (53), respectively. CME3 is orthologous to yeast DAD2, which encodes a subunit of the DASH complex involved in the positive regulation of microtubule polymerization and attachment of spindle microtubules to the kinetochore (54). DAD2 is essential for cell viability in S. cerevisiae (55). Deletion of DAD2 in S. pome resulted in abnormal chromosome segregation in both mitosis and meiosis (56). Consistently with an important role in meiotic and mitotic chromosome segregation, the cme3 mutant had defects in both growth and ascosporogenesis in F. graminearum. CME6 encodes a regulator of the Skp1-Cul1-Fbx (SCF) complex (57, 58). In A. nidulans, the CME6 ortholog is important for vegetative growth, conidiation, and sexual reproduction (59, 60). Although CME6 is also important for vegetative growth and sexual reproduction, it is dispensable for conidiation in F. graminearum. CME10 is an ortholog of N. crassa ham-2, which encodes a putative transmembrane protein required for hyphal fusion (anastomosis) (61). Since hyphal fusion is essential for growth and development in filamentous fungi, similar to the ham-2 deletion mutant, the cme10 mutant displayed a slow growth rate and failed to produce perithecia and clusters of phialides in F. graminearum. The function of CME1 and CME7 orthologs was not reported previously. We found that CME1 is required specifically during ascus and ascospore formation, while CME7 plays an important role in both vegetative growth and ascospore formation in F. graminearum. The proteins encoded by both genes contain an Smc domain for chromosome segregation ATPase. Cme1 also possesses a centrosome microtubule-binding domain of Cep57 at its C terminus. Therefore, both genes may be involved in cell division. Interestingly, among the 9 CME genes important for F. graminearum, five (CME1, CME3, CME5, CME7, and CME20) are directly associated with cell division. Cell division likely requires dynamic and precise regulation by RNA editing during sexual reproduction.
Except for CME5 and CME11, the CME sites in the other CME genes important for F. graminearum likely play no important role in sexual reproduction. Consistently with these results, all the CME sites without an important role in sexual reproduction led to amino acid changes with similar physicochemical properties, except those of CME7 (K-to-E) and CME20 (Q-to-R). It is important to emphasize that the phenotypes of underedited and overedited mutants have all been assayed under standard laboratory conditions, and only a limited number of parameters have been assessed. It remains to be determined whether the functions of these CME sites during sexual reproduction may only be subtle or under specific conditions.
Materials and Methods
Strain Culture Conditions.
The F. graminearum PH-1 strain and its derived mutants/transformants were routinely cultured on PDA plates at 25 °C. The growth rate and colony morphology were measured after 3 d of growing on PDA plates or 15 d of growing in race tubes. Conidiation was assayed in 5-d-old CMC media. For sexual reproduction, 7-d-old aerial hyphae on carrot agar cultures (250 g/L) were pressed down with sterile 0.1% Tween-20 and cultured under black light at 25 °C. Ten-day-old hyphae on carrot agar cultures were used for cme10 and cme11 mutants due to their severe growth defects. Perithecia, asci, ascospores, and ascospore discharge were examined as described (5, 32). Infection assays with wheat coleoptiles were performed as described (5).
Generation of Gene Deletion and Complemented Strains.
All gene deletion mutants were generated by the split-marker approach (62) as described (63). About 0.8-kb upstream and 0.8-kb downstream fragments of the target gene were amplified and connected to the hygromycin-resistance cassette by overlapping PCR. PCR products were cotransformed into PH-1 protoplasts. Transformants resistant to hygromycin were screened with 300 μg/mL hygromycin-B (H005, MDbio, China) and confirmed by PCR assays. For complementation assays, DNA fragments containing the promoter and coding region of each gene were amplified and cloned into a pFL2 vector by the yeast gap-repair approach (64). The complementation construct was confirmed by DNA sequencing and transformed into protoplasts of corresponding deletion mutants. Transformants harboring complementation constructs were screened with 150 μg/mL geneticin (Sigma-Aldrich, St. Louis, MO) and confirmed by PCR assays. Because defects of deletion mutants of CME1, CME5, CME6, CME7, CME10, and CME11 could not be fully complemented by ectopic expression of the pFL2 complementation constructs, we then adopted a recyclable marker module to delete and complement them as described (39). Deletion mutants generated by the recyclable marker module had indistinguishable phenotypes from mutants generated by previous methods. DNA fragments containing about 1.0-kb upstream and 1.0-kb downstream homologous arms were transformed into the protoplasts of corresponding deletion mutants. Transformants resistant to 25 μg/mL Floxuridine (HY-B0097, MCE, USA) were screened and then assayed by PCR. All gene deletion and complemented strains and primers used were listed in SI Appendix, Tables S3 and Dataset S2, respectively.
Site-Specific Mutagenesis and Allelic Exchange.
Based on codon redundancy and the functional importance of adjacent base preferences for editing (39), underedited alleles of CME genes were generated by mutating the third position of the codon preceding the edited codon by overlapping PCR (SI Appendix, Fig. S3). For editing sites located in the first positions of codons, the preferred T at the −1 position of editing sites was mutated into unpreferred G or C. For editing sites located in the second positions of codons, the preferred C at the −2 position was mutated into unpreferred G. Overedited alleles of CME genes were generated by directly replacing the A with G at editing sites. For allelic exchanges at the native locus, underedited or overedited allelic fragments were fused to the N-terminal region of the hygromycin-resistance gene (hyg) by overlapping PCR. About 1.0-kb downstream fragments of the target gene were amplified and fused to the C-terminal region of hyg by overlapping PCR. Both upstream and downstream homologous arms were cotransformed into PH-1 protoplasts. Transformants resistant to hygromycin were screened by PCR, and desired mutations were confirmed by DNA sequencing. Quantitative PCR was used to confirm the transformants without unintended integration of allelic fragments. The CME11ue-oe transformant was generated by targeted integration of an overedited allele of CME11 into the FG1G36140 locus in CME11ue mutants. DNA fragments containing the overedited allele of CME11, the geneticin-resistance marker, and upstream and downstream homologous arms of FG1G36140 were generated by overlapping PCR and transformed into the protoplasts of CME11ue mutants. Transformants resistant to geneticin were screened with 150 μg/mL geneticin and confirmed by PCR assays. All the strains and primers used were listed in SI Appendix, Tables S3 and Dataset S2, respectively.
RT-PCR.
Total RNA was isolated from 7-dpf perithecia with the TRIzol reagent (Invitrogen). Three biological replicates were prepared for each strain. Complementary DNA (cDNA) synthesis was performed with RevertAid Master Mix (Thermo Scientific) following the manufacturer’s instructions. RT-PCR products were gel-purified and subjected to direct sequencing. Sanger sequencing traces were visualized using SnapGene Viewer 4.3 (https://www.snapgene.com/snapgene-viewer/). Quantitative real-time RT-PCR was performed using the 2−∆∆CT method with the GzUBH gene as an internal control. All the primers used were listed in SI Appendix, Dataset S2.
Identification of Phosphorylation Sites in Cme11.
Phosphorylation sites of Cme11 were identified as described (65). Briefly, the PRP27-CME11-GFP fusion construct was generated by the yeast gap-repair approach and transformed into PH-1 protoplasts. Hygromycin-resistant transformants expressing the fusion constructs were identified by PCR and confirmed by western blotting with the anti-GFP antibody (11814460001, Roche, USA). Total proteins isolated from the transformant were incubated with anti-GFP affinity beads (Smart-Lifesciences, China). Proteins eluted from anti-GFP beads were detected by western blotting and Coomassie blue staining. The Cme11 band was cut out from the SDS-PAGE gel. In-gel digestion was carried out with trypsin (Promega, USA) and/or glutamic acid endopeptidase (GluC) (NEB, USA) enzymes according to the manufacturer’s instructions. Peptides were analyzed by the Thermo Scientific™ TSQ Quantum™ Access MAX triple quadrupole mass spectrometer.
Bioinformatic Analysis.
RNA-seq data of PH-1 (SI Appendix, Table S2) were downloaded from the NCBI SRA database. Low-quality reads and reads containing adapters were removed by Trimmomatic (66) with default settings. The latest genome sequences and gene annotations of PH-1 (36) were obtained from FgBase (http://fgbase.wheatscab.com/). RNA-seq reads were mapped to the PH-1 genome using HISAT2 (67) with the two-step model as described (68). Quality control of RNA-seq alignments was performed with Qualimap 2 (69). The number of reads aligned to each gene (count data) was calculated using featureCounts (70) and normalized by Transcripts Per Million (TPM). Heatmaps of TPM values were plotted by R 4.1.3. Gene orthologs were identified according to the ortholog families in EnsemblFungi and by BLASTp search in the NCBI nr database. Multiple sequence alignments were performed with M-Coffee (71) with default settings. Protein-conserved domains were identified by NCBI CD-Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and visualized by IBS (72). The AlphaFold structure of Cme5 was downloaded from the EMBL-EBI database and visualized by UCSF Chimera 1.16 (73). Phosphorylation sites were predicted by GPS5.0 (44) with the high threshold and NetPhos-3.1 (45) with default settings. Statistical significance tests were performed with GraphPad Prism version 8.0.0 for Windows (San Diego, California USA).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (PDF)
Dataset S02 (PDF)
Acknowledgments
We thank Xueling Huang, Qiong Zhang, and Xiaona Zhou from State Key Laboratory of Crop Stress Biology for Arid Areas for laboratorial assistance. This study was supported by funding from the National Key R&D Program of China (2022YFA1304400) and the National Natural Science Foundation of China (no. 32170200).
Author contributions
K.X. and H.L. designed research; K.X., Y.Z., L.F., Z.Q., and C.F. performed research; Q.W., C.J., J.-R.X., and H.L. contributed new reagents/analytic tools; K.X. and H.L. analyzed data; and K.X., J.-R.X., and H.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Nishikura K., Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grice L. F., Degnan B. M., The origin of the ADAR gene family and animal RNA editing. BMC Evol. Biol. 15, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bian Z., Ni Y., Xu J. R., Liu H., A-to-I mRNA editing in fungi: Occurrence, function, and evolution. Cell. Mol. Life Sci. 76, 329–340 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C., Xu J. R., Liu H., A-to-I RNA editing independent of ADARs in filamentous fungi. RNA Biol. 13, 940–945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun M., et al. , Stage-specific regulation of purine metabolism during infectious growth and sexual reproduction in Fusarium graminearum. New Phytol. 230, 757–773 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Liu H., et al. , A-to-I RNA editing is developmentally regulated and generally adaptive for sexual reproduction in Neurospora crassa. Proc. Natl. Acad. Sci. U.S.A. 114, E7756–E7765 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H., et al. , Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 26, 499–509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teichert I., Dahlmann T. A., Kuck U., Nowrousian M., RNA editing during sexual development occurs in distantly related filamentous ascomycetes. Genome Biol. Evol. 9, 855–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maharachchikumbura S. S. N., et al. , Towards a natural classification and backbone tree for Sordariomycetes. Fungal. Divers 72, 199–301 (2015). [Google Scholar]
- 10.Tan M. H., et al. , Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gommans W. M., Mullen S. P., Maas S., RNA editing: A driving force for adaptive evolution? Bioessays 31, 1137–1145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porath H. T., Knisbacher B. A., Eisenberg E., Levanon E. Y., Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance. Genome Biol. 18, 185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu G., Zhang J., Human coding RNA editing is generally nonadaptive. Proc. Natl. Acad. Sci. U.S.A. 111, 3769–3774 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalk A. M., Taylor S., Heraud-Farlow J. E., Walkley C. R., The majority of A-to-I RNA editing is not required for mammalian homeostasis. Genome Biol. 20, 268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoshan Y., Liscovitch-Brauer N., Rosenthal J. J. C., Eisenberg E., Adaptive proteome diversification by nonsynonymous A-to-I RNA editing in coleoid cephalopods. Mol. Biol. Evol. 38, 3775–3788 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang D., Zhang J., The preponderance of nonsynonymous A-to-I RNA editing in coleoids is nonadaptive. Nat. Commun. 10, 5411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y., et al. , The landscape of A-to-I RNA editome is shaped by both positive and purifying selection. PLoS Genet. 12, e1006191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q., Jiang C., Liu H., Xu J.-R., ADAR-independent A-to-I RNA editing is generally adaptive for sexual reproduction in fungi. bioRxiv [Preprint] (2016). 10.1101/059725 (Accessed 18 June 2016). [DOI]
- 19.Eisenberg E., Levanon E. Y., A-to-I RNA editing–immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Miyake K., et al. , CAPS1 RNA editing promotes dense core vesicle exocytosis. Cell Rep. 17, 2004–2014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L., et al. , Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 19, 209–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeo J., Goodman R. A., Schirle N. T., David S. S., Beal P. A., RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc. Natl. Acad. Sci. U.S.A. 107, 20715–20719 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burns C. M., et al. , Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387, 303–308 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Daniel C., Wahlstedt H., Ohlson J., Bjork P., Ohman M., Adenosine-to-inosine RNA editing affects trafficking of the gamma-aminobutyric acid type A (GABA(A)) receptor. J. Biol. Chem. 286, 2031–2040 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhalla T., Rosenthal J. J., Holmgren M., Reenan R., Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat. Struct. Mol. Biol. 11, 950–956 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Garrett S., Rosenthal J. J., RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science 335, 848–851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higuchi M., et al. , Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Jain M., et al. , RNA editing of Filamin A pre-mRNA regulates vascular contraction and diastolic blood pressure. EMBO J. 37, e94813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian N., Wu X., Zhang Y., Jin Y., A-to-I editing sites are a genomically encoded G: Implications for the evolutionary significance and identification of novel editing sites. RNA 14, 211–216 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kask K., et al. , The AMPA receptor subunit GluR-B in its Q/R site-unedited form is not essential for brain development and function. Proc. Natl. Acad. Sci. U.S.A. 95, 13777–13782 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai J., et al. , Loss of CaV1.3 RNA editing enhances mouse hippocampal plasticity, learning, and memory. Proc. Natl. Acad. Sci. U.S.A. 119, e2203883119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao S., et al. , RNA editing of the AMD1 gene is important for ascus maturation and ascospore discharge in Fusarium graminearum. Sci Rep. 7, 4617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao C., et al. , The meiosis-specific APC activator FgAMA1 is dispensable for meiosis but important for ascosporogenesis in Fusarium graminearum. Mol. Microbiol. 111, 1245–1262 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Trail F., For blighted waves of grain: Fusarium graminearum in the postgenomics era. Plant Physiol. 149, 103–110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu G., Zhang J., In search of beneficial coding RNA editing. Mol. Biol. Evol. 32, 536–541 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu P., et al. , Landscape and regulation of alternative splicing and alternative polyadenylation in a plant pathogenic fungus. New Phytol. 235, 674–689 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Son H., et al. , A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog. 7, e1002310 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang C., et al. , An expanded subfamily of G-protein-coupled receptor genes in Fusarium graminearum required for wheat infection. Nat. Microbiol. 4, 1582–1591 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Feng C., et al. , Uncovering cis-regulatory elements important for A-to-I RNA editing in Fusarium graminearum. mBio 13, e0187222 (2022), 10.1128/mbio.01872-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su X., et al. , Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat. Cell Biol. 15, 948–957 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gradolatto A., et al. , Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics 179, 291–304 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gal C., et al. , Abo1, a conserved bromodomain AAA-ATPase, maintains global nucleosome occupancy and organisation. EMBO Rep. 17, 79–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C., et al. , GPS 5.0: An update on the prediction of kinase-specific phosphorylation sites in proteins. Genom. Proteom. Bioinformat. 18, 72–80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blom N., Sicheritz-Ponten T., Gupta R., Gammeltoft S., Brunak S., Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649 (2004). [DOI] [PubMed] [Google Scholar]
- 45.DeZwaan T. M., Ellingson E., Pellman D., Roof D. M., Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 138, 1023–1040 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grigoriev I. V., et al. , MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–D704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rischitor P. E., Konzack S., Fischer R., The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae. Eukaryot Cell 3, 632–645 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su X., et al. , Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol. Cell 43, 751–763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amos L. A., Schlieper D., Microtubules and maps. Adv. Protein Chem. 71, 257–298 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Lombardi L. M., Ellahi A., Rine J., Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc. Natl. Acad. Sci. U.S.A. 108, E1302–1311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klocko A. D., et al. , Nucleosome positioning by an evolutionarily conserved chromatin remodeler prevents aberrant DNA methylation in Neurospora. Genetics 211, 563–578 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerber A. P., Keller W., An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286, 1146–1149 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Hoyt M. A., He L., Loo K. K., Saunders W. S., Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 118, 109–120 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jenni S., Harrison S. C., Structure of the DASH/Dam1 complex shows its role at the yeast kinetochore-microtubule interface. Science 360, 552–558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y., et al. , The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes. Dev. 16, 183–197 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X., McLeod I., Anderson S., Yates J. R. III, He X., Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24, 2919–2930 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eigentler A., et al. , The impact of Cand1 in prostate cancer. Cancers (Basel) 12, 428 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng S., et al. , Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein Degradation. Plant Cell 16, 1870–1882 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Helmstaedt K., et al. , Recruitment of the inhibitor Cand1 to the cullin substrate adaptor site mediates interaction to the neddylation site. Mol. Biol. Cell 22, 153–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kohler A. M., et al. , Integration of fungus-specific CandA-C1 into a trimeric CandA complex allowed splitting of the gene for the conserved receptor exchange factor of cullinA E3 ubiquitin ligases in Aspergilli. mBio 10, e01094-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiang Q., Rasmussen C., Glass N. L., The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160, 169–180 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Catlett N. L., Lee B.-N., Yoder O., Turgeon B. G., Split-marker recombination for efficient targeted deletion of fungal genes. Fungal. Genet. Newsl. 50, 9–11 (2003). [Google Scholar]
- 63.Wang C., et al. , Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 7, e1002460 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bruno K. S., Tenjo F., Li L., Hamer J. E., Xu J. R., Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3, 1525–1532 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao X., et al. , FgPrp4 kinase is important for spliceosome B-complex activation and splicing efficiency in Fusarium graminearum. PLoS Genet. 12, e1005973 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu H., Xu J. R., Discovering RNA editing events in fungi. Methods Mol. Biol. 2181, 35–50 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Okonechnikov K., Conesa A., Garcia-Alcalde F., Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32, 292–294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Wallace I. M., O’Sullivan O., Higgins D. G., Notredame C., M-Coffee: Combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 34, 1692–1699 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu W., et al. , IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pettersen E. F., et al. , UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (PDF)
Dataset S02 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.