Abstract
The human gut microbiome plays a crucial role in human health and has been a focus of increasing research in recent years. Omics-based methods, such as metagenomics, metatranscriptomics, and metabolomics, are commonly used to study the gut microbiome because they provide high-throughput and high-resolution data. The vast amount of data generated by these methods has led to the development of computational methods for data processing and analysis, with machine learning becoming a powerful and widely used tool in this field. Despite the promising results of machine learning-based approaches for analyzing the association between microbiota and disease, there are several unmet challenges. Small sample sizes, disproportionate label distribution, inconsistent experimental protocols, or a lack of access to relevant metadata can all contribute to a lack of reproducibility and translational application into everyday clinical practice. These pitfalls can lead to false models, resulting in misinterpretation biases for microbe–disease correlations. Recent efforts to address these challenges include the construction of human gut microbiota data repositories, improved data transparency guidelines, and more accessible machine learning frameworks; implementation of these efforts has facilitated a shift in the field from observational association studies to experimental causal inference and clinical intervention.
Keywords: gut microbiota, gut microbiome, health, microbiome, eubiosis, dysbiosis, omics, metagenomics, machine learning, supervised learning, unsupervised learning, artificial intelligence
1. The Human Microbiome
In recent years, there has been a significant increase in research on gut microbiota due to the growing understanding of the critical role that gut microbiota plays in human health. The human gastrointestinal tract hosts a diverse community of microorganisms, including bacteria, archaea, fungi, microbial eukaryotes, and viruses, all of which exist in a symbiotic relationship with the human host. This collection of microbes is known as the microbiota, and their genetic material is referred to as the microbiome [1]. In the past, it was believed that the number of cells in the human microbiota was ten times greater than the number of cells in the human body. However, more recent evidence has shown that the ratio is much closer to one-to-one, with a slight advantage for our microbes [2]. The gut microbiome, comprising almost 100 trillion bacteria, has a genome 150 times larger than the human host (3 million vs. approximately 23,000 genes, respectively) [3,4]. In healthy individuals, the host and microbiome maintain a healthy balance referred to as eubiosis, which can be altered to a state of dysbiosis (i.e., an abnormal shift in microbiota compositions) found in several pathological conditions [5,6,7]. The actual association between dysbiosis and the development of disease remains largely unclear, and we expect that defining this connection will be among the greatest medical challenges of the next few decades.
2. Machine Learning and Gut Microbiome
Advances in technology and cost reduction have made it possible to study the previously unexplored landscape of the human gut microbiome at a large scale. Omics-based methods such as metagenomics, metatranscriptomics, and metabolomics are widely used to assess the human gut microbiota [8,9]. These techniques enable high-throughput and high-resolution studies of the overall microbial community [8,9] and approach the microbiome from multiple perspectives. For example, metagenomics techniques (e.g., 16S rRNA gene sequencing or whole-genome shotgun sequencing) provide information about the overall microbial genetic content of the community of interest, and metabolomics measure the concentrations of different compounds produced by that specific community [8,9]. The use of omics-based methods has generated a large amount of data, which has prompted the development of computational methods, such as machine learning, to aid in processing and to analyze this data related to human gut microbiota research [10,11,12]. Machine learning comprises a series of powerful computational tools that have become increasingly important in various fields, including data analysis, computer vision, natural language processing, and predictive modeling [13,14]. Machine learning is a subfield of artificial intelligence that involves the development of algorithms that can learn from data and make predictions or decisions without being explicitly programmed [13,14]. Machine learning algorithms can be divided into two main categories: supervised and unsupervised learning. Supervised learning is the most common type of machine learning, and it involves training an algorithm on a labeled dataset to predict the outcome for new, unseen data. In supervised learning, the algorithm learns to identify patterns or relationships in the data that can be used to make predictions [13,14]. Examples of supervised learning algorithms include linear regression, decision trees, and support vector machines [13,14]. Unsupervised learning is usually employed to discover patterns or structures in unlabeled data [13,14]. Unsupervised learning can help identify patterns or groups in data that may not be obvious [13,14]. Examples of unsupervised learning algorithms include clustering, dimensionality reduction, and anomaly detection [13,14]. Overall, machine learning has become a powerful tool for data analysis and outcome prediction; it can be used to identify patterns and relationships in data that may not be evident, and to make predictions that would be difficult or impossible to define using traditional methods [13,14].
Several studies that analyzed gut microbiota as a potential classifier for diseases showed that microbial features in species, genes, or metabolites could differentiate between cases and healthy subjects or even predict responses to drug treatments, as brilliantly summarized by Marcos-Zambrano et al. [12]. Nevertheless, we will provide a few notable examples in the following text.
Zeller et al. [15] developed a logistic regression model based on gut microbiome composition to discriminate colorectal cancer (CRC) patients from healthy subjects. The authors employed the least absolute shrinkage and selection operator (LASSO) method to withdraw the least informative microbial species from the final method. The authors reported that the AUC-ROC value for the diagnostic model was 0.80, indicating that the model had good performance in distinguishing between patients with CRC and healthy controls. Additionally, the diagnostic model could distinguish between early-stage and advanced-stage CRC with an AUC-ROC value of 0.78. The model demonstrated a performance comparable to that of the fecal occult blood test (FOBT). However, the sensitivity drastically improved when the model was combined with the FOBT (49% increase). Specifically, Fusobacterium nucleatum and Peptostreptococcus stomatis were identified as the most relevant species to the prediction model, as previously found in association studies between CRC and microbiota [16,17].
Derosa et al. [18] tried to determine whether the gut microbiota abundance of several species could discriminate responses to immunotherapy (nivolumab) in a cohort of patients with renal cell cancer. The authors employed partial least square discriminant analysis (PLS-DA), a supervised algorithm that combines feature extraction and discriminant analysis into one algorithm and applies well to high-dimensional data [19]. Their results highlighted that some species (Clostridiales clostridioforme and Clostridiales hathewayi) were associated with drug resistance and with cancer metastasis status. Conversely, other commensal species (Acetobacter senegalensis and Akkermansia muciniphila) were associated with favorable prognosis and increased drug response. Other studies have confirmed that Akkermansia muciniphila could be related to more favorable treatment responses in other cancers, such as non-small-cell lung cancer patients undergoing programmed death-1 (PD-1) immunotherapy [20,21]. These findings prove that computation methods are crucial to discovering possible microbiological signatures to improve disease diagnosis or predict therapeutic responses.
Although classifiers are often employed for predicting categorical variables, such as “healthy” vs. “disease”, regression models are more appropriate for predicting continuous variables, such as metabolite levels. In recent research, regression models have been utilized to predict metabolite levels from microbial features, such as species or genes, and applied in studies examining the association between microbes and metabolites. For instance, Reiman et al. [22] trained a multilayer perceptron (MLP) model to predict metabolite levels based on microbial abundances. The contribution of individual microbes to a given metabolite level was estimated using the weights of the MLP model. An MLP model is an artificial neural network composed of multiple layers of interconnected nodes or perceptrons [23]. These layers are typically arranged in a feedforward structure, where the input is processed through each layer sequentially, and the final layer produces the output [23]. MLP is often used for supervised learning tasks, such as classification and regression, and can be trained using various algorithms, such as backpropagation [23]. The authors found that the MLP model was more accurate at predicting metabolite abundances and identified metabolite levels better than other linear models currently used for individual metabolite predictions. Furthermore, the authors showed that the MLP model could group microbes and metabolites with similar patterns of interaction and functions, which could provide insights into the microbe–metabolite interaction network’s underlying structure and reveal uncharacterized metabolites through “guilt by association”. These findings suggest that using machine learning techniques for integrating and identifying patterns in omics data is crucial to understanding the role of microbes and microbial metabolites in disease progression.
Computational technique design is not confined to data analysis, as illustrated by a recent study that employed machine learning to build a tailored menu for a nutritional intervention trial, recently published in the prestigious New England Journal of Medicine. In their study, Chen et al. [24], performed a randomized controlled trial to define the effect of a microbiota-directed complementary food (MDCF) intervention to treat undernourished children by employing an analysis base on linear mixed-effects models, resulting in a significant superiority in terms of weight gain and restoration of “healthy” microbiota composition.
2.1. Challenges in Current Application
Despite the encouraging results of machine learning techniques for studying the relationship between microbiome and disease, significant challenges still need to be addressed. One of the most critical challenges is the dependence of supervised learning models on the quantity and quality of training data. This dependence can lead to models that lack reproducibility due to small sample sizes, disproportionate label distribution, inconsistent experimental protocols, or a lack of access to relevant metadata, which can all contribute to a lack of reproducibility [25,26]. For example, two meta-analyses found that, while dysbiosis was present in CRC patients, a particular bacterial diversity was peculiar to a given population and not present in other investigations [27,28]. Furthermore, researchers must exercise caution when implementing machine learning, particularly for supervised learning tasks, to prevent pitfalls such as information leaking from the training phase to the test phase [29]. These flaws might result in excessively optimistic models and the misperception of bias as microbe–disease correlations. In the opinion of many experts, recent initiatives aimed at addressing the issues associated with machine learning in microbiome research, including the creation of human gut microbiota data repositories [29,30,31,32,33], improved data disclosure guidelines [34], and more accessible frameworks [35,36], could lead to the development of more accurate and reliable machine learning models, providing valuable insights into the mechanisms underlying microbial dysbiosis and the potential for targeted interventions to improve human health.
2.2. The Importance of Data Repositories and Data Preprocessing
Combining human gut microbiome data repositories with increased transparency in data sharing allows researchers to conduct meta-analyses across various studies, which can lead to the identification of robust biomarkers or indicators of dysbiosis specific to certain diseases [27,28]. In the opinion of many researchers, the availability of preprocessed data in these repositories can minimize technical biases and lower computational costs. Still, they may require more flexibility regarding tool selection or desired output formats for the user. For instance, Pasolli et al. [30] created a curated repository of human gut microbiome data preprocessed using a unified metagenome processing pipeline (e.g., bioBakery [37]). They included whole genome shotgun metagenomic (taxonomic and functional) gene abundance profiles and curated metadata. However, certain methods may require a specific and custom input data format, making the preprocessed data more challenging or incompatible. For example, the Dirichlet multinomial mixture method [38] requires integers data as input, whereas preprocessed relative microbial or relative gene abundances cannot be used as input. Therefore, while preprocessed data repositories (e.g., MGnify [31]) offer more flexibility in downstream analyses, they require more computational resources and expertise in bioinformatics to process.
In the opinion of many researchers, the expansion of public repositories is a crucial step in enabling researchers to address formerly unknown biological topics by providing more human gut microbiome-omics data. This will likely lead to a more significant usage of machine learning techniques on an increasing amount of publicly available omics data. However, we believe that building algorithms from scratch can be time-consuming, prone to error, and not applicable to other clinical settings due to the lack of methodology standardization. Therefore, the use of a machine learning framework, which is a comprehensive collection of tools that supports the analytical process, from data preprocessing to model validation, can be significant in avoiding the most common machine learning errors, making the analysis more efficient and robust. Several machine learning frameworks are available, each written in a different programming language and featuring a variety of modeling techniques; some of these are specifically tailored for microbiome data. From our perspective, the increased accessibility and repeatability of human gut microbiome investigations provided by these frameworks are essential in lowering the danger of overfitting. An excellent example is represented by the framework developed by Topçuoğlu et al. [39], who trained seven models using fecal 16S rRNA sequence data to predict the presence of CRC by creating a reusable, open-source pipeline able to train, validate, and interpret these models. Various machine learning approaches, including logistic regression, support vector machines, decision trees, and random forests, were examined in terms of performance, interpretability, and training time. The logistic regression model was simple, rapid, and interpretable, whereas the random forest model performed best in detecting CRC, but still was difficult to train. Their findings emphasize the need to select a methodological strategy aligned with the study’s aims to balance performance and interpretability. From our point of view, the application of machine learning frameworks has the potential to revolutionize the actual state of the art, but researchers must exercise caution and choose appropriate methodology for their specific research questions and goals.
3. Conclusions
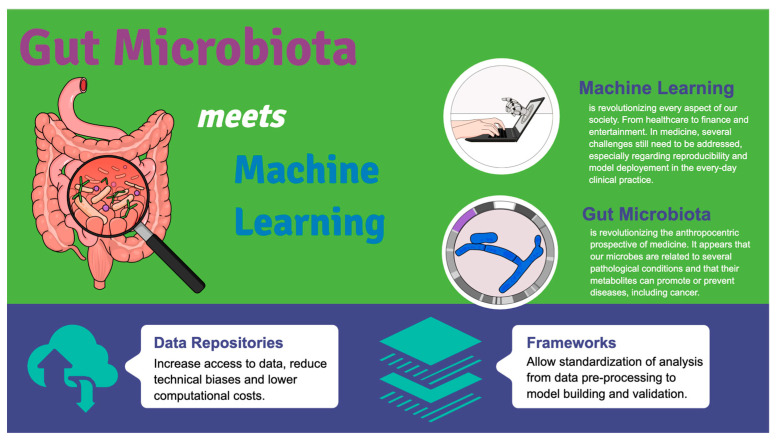
From our perspective, computational techniques, particularly machine learning, have played a crucial role in analyzing the large volume of data produced by multi-omics studies of the human gut microbiota, which has led to the discovery of new associations between microbes and disease [10,11,40] as summarized in Figure 1.
Figure 1.
Gut Microbiota meets machine learning. The increasing data availability due to omics analysis has not been followed by the creation of data repositories, guidelines, and analytical frameworks in the past, which resulted in unsatisfactory reproducibility and reliability. The implementation of such tools has facilitated a shift in the field from observational association studies to experimental causal inference and clinical intervention.
In our opinion, the use of machine learning-based analytical processes relies heavily on data availability and requires expertise in implementation to ensure reproducibility and reliability. Fortunately, recent developments in data repositories [29,30,31,32,33], reporting guidelines [34], and frameworks [35,36] have improved the accessibility and transparency of the data analysis process, making it more efficient and reliable. These advancements have facilitated a shift in the field from observational association studies to experimental causal inference and clinical intervention [41]. We believe that this is an exciting development that holds great promise for the future of microbiome research. We anticipate that computational methods will continue to be essential for the analysis of future experimental data [42,43,44] and will drive the development of microbe-based or microbe-directed clinical interventions [24], primarily when used in conjunction with emerging technologies such as cultivation-free genome sequencing [45] and the manipulation of gut microbial genes [46].
In conclusion, we believe that the continued use of computational methods, particularly machine learning, will be critical in advancing our understanding of the complex relationships between the human gut microbiome and disease. We look forward to further developments in this field and anticipate that these advancements will lead to improved clinical interventions and better health outcomes for patients.
Acknowledgments
This paper is dedicated to the memory of our dear Rita Moretti, who suddenly passed away. All the authors are grateful to Rita Moretti, whose contribution to the current manuscript was substantial and of pivotal importance.
Author Contributions
Conceptualization, M.G., R.M., and C.T.; resources, M.G.; writing—original draft preparation, M.G., R.M., and C.T.; writing—review and editing, M.G., R.M., and C.T.; visualization, M.G., R.M., and C.T.; supervision, C.T. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The Human Microbiome Project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sender R., Fuchs S., Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chassaing B., Etienne-Mesmin L., Gewirtz A.T. Microbiota-liver axis in hepatic disease. Hepatology. 2013;59:328–339. doi: 10.1002/hep.26494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin N., Yang F., Li A., Prifti E., Chen Y., Shao L., Guo J., Le Chatelier E., Yao J., Wu L., et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 5.Giuffrè M., Campigotto M., Campisciano G., Comar M., Crocè L.S. A story of liver and gut microbes: How does the intestinal flora affect liver disease? A review of the literature. Am. J. Physiol. Liver Physiol. 2020;318:G889–G906. doi: 10.1152/ajpgi.00161.2019. [DOI] [PubMed] [Google Scholar]
- 6.Giuffrè M., Moretti R., Campisciano G., Da Silveira A.B.M., Monda V.M., Comar M., Di Bella S., Antonello R.M., Luzzati R., Crocè L.S. You Talking to Me? Says the Enteric Nervous System (ENS) to the Microbe. How Intestinal Microbes Interact with the ENS. J. Clin. Med. 2020;9:3705. doi: 10.3390/jcm9113705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giuffrè M., Gazzin S., Zoratti C., Llido J.P., Lanza G., Tiribelli C., Moretti R. Celiac Disease and Neurological Manifestations: From Gluten to Neuroinflammation. Int. J. Mol. Sci. 2022;23:15564. doi: 10.3390/ijms232415564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguiar-Pulido V., Huang W., Suarez-Ulloa V., Cickovski T., Mathee K., Narasimhan G. Metagenomics, Metatranscriptomics, and Metabolomics Approaches for Microbiome Analysis. Evol. Bioinform. 2016;12:5–6. doi: 10.4137/EBO.S36436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X., Li L., Butcher J., Stintzi A., Figeys D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome. 2019;7:154. doi: 10.1186/s40168-019-0767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghannam R.B., Techtmann S.M. Machine learning applications in microbial ecology, human microbiome studies, and environmental monitoring. Comput. Struct. Biotechnol. J. 2021;19:1092–1107. doi: 10.1016/j.csbj.2021.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodswen S.J., Barratt J.L.N., Kennedy P.J., Kaufer A., Calarco L., Ellis J.T. Machine learning and applications in microbiology. FEMS Microbiol. Rev. 2021;45:fuab015. doi: 10.1093/femsre/fuab015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcos-Zambrano L.J., Karaduzovic-Hadziabdic K., Turukalo T.L., Przymus P., Trajkovik V., Aasmets O., Berland M., Gruca A., Hasic J., Hron K., et al. Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment. Front. Microbiol. 2021;12:634511. doi: 10.3389/fmicb.2021.634511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henn J., Buness A., Schmid M., Kalff J.C., Matthaei H. Machine learning to guide clinical decision-making in abdominal surgery—A systematic literature review. Langenbeck’s Arch. Surg. 2021;407:51–61. doi: 10.1007/s00423-021-02348-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meskó B., Görög M. A short guide for medical professionals in the era of artificial intelligence. NPJ Digit. Med. 2020;3:126. doi: 10.1038/s41746-020-00333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeller G., Tap J., Voigt A.Y., Sunagawa S., Kultima J.R., Costea P.I., Amiot A., Böhm J., Brunetti F., Habermann N., et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castellarin M., Warren R.L., Freeman J.D., Dreolini L., Krzywinski M., Strauss J., Barnes R., Watson P., Allen-Vercoe E., Moore R.A., et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostic A.D., Gevers D., Pedamallu C.S., Michaud M., Duke F., Earl A.M., Ojesina A.I., Jung J., Bass A.J., Tabernero J., et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derosa L., Routy B., Fidelle M., Iebba V., Alla L., Pasolli E., Segata N., Desnoyer A., Pietrantonio F., Ferrere G., et al. Gut Bacteria Composition Drives Primary Resistance to Cancer Immunotherapy in Renal Cell Carcinoma Patients. Eur. Urol. 2020;78:195–206. doi: 10.1016/j.eururo.2020.04.044. [DOI] [PubMed] [Google Scholar]
- 19.Aminu M., Ahmad N.A. Complex Chemical Data Classification and Discrimination Using Locality Preserving Partial Least Squares Discriminant Analysis. ACS Omega. 2020;5:26601–26610. doi: 10.1021/acsomega.0c03362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Routy B., Le Chatelier E., DeRosa L., Duong C.P.M., Alou M.T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 21.Yachida S., Mizutani S., Shiroma H., Shiba S., Nakajima T., Sakamoto T., Watanabe H., Masuda K., Nishimoto Y., Kubo M., et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019;25:968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 22.Reiman D., Layden B.T., Dai Y. MiMeNet: Exploring microbiome-metabolome relationships using neural networks. PLoS Comput. Biol. 2021;17:e1009021. doi: 10.1371/journal.pcbi.1009021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornik K., Stinchcombe M., White H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989;2:359–366. doi: 10.1016/0893-6080(89)90020-8. [DOI] [Google Scholar]
- 24.Chen R.Y., Mostafa I., Hibberd M.C., Das S., Mahfuz M., Naila N.N., Islam M.M., Huq S., Alam M.A., Zaman M.U., et al. A Microbiota-Directed Food Intervention for Undernourished Children. N. Engl. J. Med. 2021;384:1517–1528. doi: 10.1056/NEJMoa2023294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schloss P.D. Identifying and Overcoming Threats to Reproducibility, Replicability, Robustness, and Generalizability in Microbiome Research. Mbio. 2018;9:e00525-18. doi: 10.1128/mBio.00525-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poussin C., Sierro N., Boué S., Battey J., Scotti E., Belcastro V., Peitsch M.C., Ivanov N.V., Hoeng J. Interrogating the microbiome: Experimental and computational considerations in support of study reproducibility. Drug Discov. Today. 2018;23:1644–1657. doi: 10.1016/j.drudis.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Thomas A.M., Manghi P., Asnicar F., Pasolli E., Armanini F., Zolfo M., Beghini F., Manara S., Karcher N., Pozzi C., et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019;25:667–678. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wirbel J., Pyl P.T., Kartal E., Zych K., Kashani A., Milanese A., Fleck J.S., Voigt A.Y., Palleja A., Ponnudurai R., et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019;25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smialowski P., Frishman D., Kramer S. Pitfalls of supervised feature selection. Bioinformatics. 2009;26:440–443. doi: 10.1093/bioinformatics/btp621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasolli E., Schiffer L., Manghi P., Renson A., Obenchain V., Truong D.T., Beghini F., Malik F., Ramos M., Dowd J.B., et al. Accessible, curated metagenomic data through ExperimentHub. Nat. Methods. 2017;14:1023–1024. doi: 10.1038/nmeth.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell A.L., Almeida A., Beracochea M., Boland M., Burgin J., Cochrane G., Crusoe M.R., Kale V., Potter S.C., Richardson L.J., et al. MGnify: The microbiome analysis resource in 2020. Nucleic Acids Res. 2019;48:D570–D578. doi: 10.1093/nar/gkz1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai D., Zhu J., Sun C., Li M., Liu J., Wu S., Ning K., He L.-J., Zhao X.-M., Chen W.-H. GMrepo v2: A curated human gut microbiome database with special focus on disease markers and cross-dataset comparison. Nucleic Acids Res. 2021;50:D777–D784. doi: 10.1093/nar/gkab1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez A., Navas-Molina J.A., Kosciolek T., McDonald D., Vázquez-Baeza Y., Ackermann G., Dereus J., Janssen S., Swafford A.D., Orchanian S.B., et al. Qiita: Rapid, web-enabled microbiome meta-analysis. Nat. Methods. 2018;15:796–798. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirzayi C., Renson A., Furlanello C., Sansone S.-A., Zohra F., Elsafoury S., Geistlinger L., Kasselman L.J., Eckenrode K., van de Wijgert J., et al. Reporting guidelines for human microbiome research: The STORMS checklist. Nat. Med. 2021;27:1885–1892. doi: 10.1038/s41591-021-01552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirbel J., Zych K., Essex M., Karcher N., Kartal E., Salazar G., Bork P., Sunagawa S., Zeller G. Microbiome meta-analysis and cross-disease comparison enabled by the SIAMCAT machine learning toolbox. Genome Biol. 2021;22:93. doi: 10.1186/s13059-021-02306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasolli E., Truong D.T., Malik F., Waldron L., Segata N. Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS Comput. Biol. 2016;12:e1004977. doi: 10.1371/journal.pcbi.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beghini F., McIver L.J., Blanco-Míguez A., Dubois L., Asnicar F., Maharjan S., Mailyan A., Manghi P., Scholz M., Thomas A.M., et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife. 2021;10:e65088. doi: 10.7554/eLife.65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmes I., Harris K., Quince C. Dirichlet Multinomial Mixtures: Generative Models for Microbial Metagenomics. PLoS ONE. 2012;7:e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topçuoğlu B.D., Lesniak N.A., Ruffin M.T., Wiens J., Schloss P.D. A Framework for Effective Application of Machine Learning to Microbiome-Based Classification Problems. Mbio. 2020;11:e00434-20. doi: 10.1128/mBio.00434-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A., Alm E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017;8:1784. doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkinson J.E., Franzosa E.A., Everett C., Li C., Bae S., Berzansky I., Bhosle A., Bjørnevik K., Brennan C.A., Cao Y.G., et al. A framework for microbiome science in public health. Nat. Med. 2021;27:766–774. doi: 10.1038/s41591-021-01258-0. [DOI] [PubMed] [Google Scholar]
- 42.Sanna S., Van Zuydam N.R., Mahajan A., Kurilshikov A., Vila A.V., Võsa U., Mujagic Z., Masclee A.A.M., Jonkers D.M.A.E., Oosting M., et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 2019;51:600–605. doi: 10.1038/s41588-019-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurilshikov A., Medina-Gomez C., Bacigalupe R., Radjabzadeh D., Wang J., Demirkan A., Le Roy C.I., Garay J.A.R., Finnicum C.T., Liu X., et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021;53:156–165. doi: 10.1038/s41588-020-00763-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X., Tong X., Zou Y., Lin X., Zhao H., Tian L., Jie Z., Wang Q., Zhang Z., Lu H., et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat. Genet. 2022;54:52–61. doi: 10.1038/s41588-021-00968-y. [DOI] [PubMed] [Google Scholar]
- 45.Pryszlak A., Wenzel T., Seitz K.W., Hildebrand F., Kartal E., Cosenza M.R., Benes V., Bork P., Merten C.A. Enrichment of gut microbiome strains for cultivation-free genome sequencing using droplet microfluidics. Cell Rep. Methods. 2021;2:100137. doi: 10.1016/j.crmeth.2021.100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin W.-B., Li T.-T., Huo D., Qu S., Li X.V., Arifuzzaman M., Lima S.F., Shi H.-Q., Wang A., Putzel G.G., et al. Genetic manipulation of gut microbes enables single-gene interrogation in a complex microbiome. Cell. 2022;185:547–562.e22. doi: 10.1016/j.cell.2021.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.