Abstract
The lack of effective therapeutics against emerging multi-drug resistant strains of Mycobacterium tuberculosis (Mtb) prompts the identification of novel anti-tuberculosis targets. The essential nature of the peptidoglycan (PG) layer of the mycobacterial cell wall, which features several distinctive modifications, such as the N-glycolylation of muramic acid and the amidation of D-iso-glutamate, makes it a target of particular interest. To understand their role in susceptibility to beta-lactams and in the modulation of host-pathogen interactions, the genes encoding the enzymes responsible for these PG modifications (namH and murT/gatD, respectively) were silenced in the model organism Mycobacterium smegmatis using CRISPR interference (CRISPRi). Although beta-lactams are not included in TB-therapy, their combination with beta-lactamase inhibitors is a prospective strategy to treat MDR-TB. To uncover synergistic effects between the action of beta-lactams and the depletion of these PG modifications, knockdown mutants were also constructed in strains lacking the major beta-lactamase of M. smegmatis BlaS, PM965 (M. smegmatis ΔblaS1) and PM979 (M. smegmatis ΔblaS1 ΔnamH). The phenotyping assays affirmed the essentiality of the amidation of D-iso-glutamate to the survival of mycobacteria, as opposed to the N-glycolylation of muramic acid. The qRT-PCR assays confirmed the successful repression of the target genes, along with few polar effects and differential knockdown level depending on PAM strength and target site. Both PG modifications were found to contribute to beta-lactam resistance. While the amidation of D-iso-glutamate impacted cefotaxime and isoniazid resistance, the N-glycolylation of muramic acid substantially promoted resistance to the tested beta-lactams. Their simultaneous depletion provoked synergistic reductions in beta-lactam MICs. Moreover, the depletion of these PG modifications promoted a significantly faster bacilli killing by J774 macrophages. Whole-genome sequencing revealed that these PG modifications are highly conserved in a set of 172 clinical strains of Mtb, demonstrating their potential as therapeutic targets against TB. Our results support the development of new therapeutic agents targeting these distinctive mycobacterial PG modifications.
Keywords: tuberculosis, antibiotic resistance, peptidoglycan modifications, CRISPR interference, beta-lactams, intracellular survival, host-pathogen interactions, anti-TB targets
1. Introduction
Mycobacterium tuberculosis (Mtb), the etiologic agent of tuberculosis (TB), latently infects one-quarter of the world’s population and causes about 1.5 million deaths annually (WHO, 2021). The emergence and spread of multi-drug (MDR) and extensively-drug resistant (XDR) strains of Mtb has accentuated the demand for novel vaccine candidates and for improved therapeutic regimens (Singh and Chibale, 2021; WHO, 2021). Therefore, the development of innovative anti-TB agents, which depends on the identification of novel drug targets, is imperative (Singh and Chibale, 2021).
The notable success of Mtb as a pathogen relies on its unusual cell wall (CW), comprising a heavily cross-linked and modified network of peptidoglycan (PG), a highly branched arabinogalactan (AG) polysaccharide and typical long-chain mycolic acids (MA) (Jankute et al., 2015; Catalão et al., 2019; Maitra et al., 2019). The mycobacterial PG features unique modifications, of which the N-glycolylation of muramic acid and the amidation of D-iso-glutamate are of particular significance (Catalão et al., 2019; Maitra et al., 2019).
Unlike most bacteria, the PG from Mycobacterium and related genera consists of alternating residues of N-acetylglucosamine and a mixture of muramic acid residues, encompassing N-glycolyl and N-acetyl modifications (Mahapatra et al., 2005; Raymond et al., 2005). The N-glycolylation of muramic acid, catalyzed by the N-acetyl muramic acid hydroxylase (NamH), occurs when the N-acetyl function is converted to the N-glycolyl function, generating N-glycolyl muramic acid (MurNGlyc) (Raymond et al., 2005). Previous studies have probed the role of this PG modification in antibiotic susceptibility (Raymond et al., 2005; Coulombe et al., 2009) and in host-pathogen interactions (Coulombe et al., 2009; Hansen et al., 2014; Wang et al., 2016), inferring that the N-glycolylation of PG promotes resistance to beta-lactams and lysozyme (Raymond et al., 2005) and potentiates the immunogenicity of the mycobacterial CW (Coulombe et al., 2009; Hansen et al., 2014).
Another distinctive mycobacterial PG modification is the amidation of the free carboxyl group of the D-iso-glutamate (D-iso-Glu) residue at the second position of both lipid II and its precursor lipid I (Catalão et al., 2019; Maitra et al., 2019). This PG modification, catalyzed by the MurT/GatD amidotransferase complex, has been characterized in Staphylococcus aureus (Figueiredo et al., 2012; Münch et al., 2012) and in Streptococcus pneumoniae (Morlot et al., 2018). This process begins with the hydrolysis of L-glutamine to generate glutamate and ammonia, catalyzed by GatD (Figueiredo et al., 2012; Münch et al., 2012; Leisico et al., 2018; Morlot et al., 2018). The ammonia is then used to catalyze the amidation of D-iso-Glu into D-iso-glutamine by MurT, a Mur ligase-like protein, in an ATP and Mg2+-dependent manner. The inhibition of D-iso-Glu amidation in S. aureus seems to delay cell division and to increase susceptibility to beta-lactams and lysozyme (Figueiredo et al., 2012; Münch et al., 2012; Figueiredo et al., 2014). Moreover, D-iso-Glu amidated lipid II has been postulated as the preferred substrate for transpeptidation reactions in S. aureus (Figueiredo et al., 2012; Münch et al., 2012), S. pneumoniae (Zapun et al., 2013) and Mtb (Pidgeon et al., 2019). Recently, the enzymatic activity of the MurT/GatD complex has been characterized in Mtb, revealing the co-transcription of the murT/gatD operon and the ATPase activity of MurT (Maitra et al., 2021).
Despite being a successful and relatively safe antibiotic class, beta-lactams have not yet been included in TB-therapeutic regimens because of the inherent resistance of Mtb to this class, fundamentally attributed to a chromosomally encoded beta-lactamase, the impermeable nature of the mycobacterial CW and the predominance of non-classical PG cross-links, only efficiently inhibited by carbapenems (Cordillot et al., 2013; Wivagg et al., 2014; Story-Roller and Lamichhane, 2018). Whereas Mtb encodes a very efficient beta-lactamase BlaC, M. smegmatis produces both a highly active beta-lactamase BlaS and a lowly active cephalosporinase BlaE (Flores et al., 2005). The predominance of non-classical 3→3 cross-links in mycobacterial PG is attributed to the action of L, D - transpeptidases (Ldts) while penicillin binding proteins (PBPs) catalyze the usual 4→3 cross-links (Lavollay et al., 2008; Kumar et al., 2012; Cordillot et al., 2013; Baranowski et al., 2018). Nevertheless, several studies have encouraged the repurposing of carbapenems in combination with beta-lactamase inhibitors (Hugonnet and Blanchard, 2007) to treat MDR-TB infections (Hugonnet et al., 2009; England et al., 2012; De Lorenzo et al., 2013; Cohen et al., 2016; Zhang et al., 2016). Additionally, the World Health Organization (WHO) currently endorses two carbapenems in group C of the drugs used to treat MDR-TB patients on longer therapeutic regimens (WHO, 2020). Recently, our group identified several drivers of resistance/susceptibility to beta-lactams in Mtb (Olivença et al., 2022a; Olivença et al., 2022b), alluding to the potential introduction of beta-lactams in TB therapeutic schemes.
So far, the lack of efficient gene inactivation tools for mycobacteria has hindered the identification of novel vaccine candidates and drug targets. The discovery of CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated proteins) systems has facilitated precise genome editing (Doudna and Charpentier, 2014). CRISPR has recently been repurposed for bacterial transcription regulation through the inactivation of Cas9 by point mutations within its nuclease domains, generating an endonuclease-deficient Cas9 (dCas9) with preserved DNA-binding activity (Bikard et al., 2013; Larson et al., 2013; Qi et al., 2013). In Escherichia coli, CRISPR interference (CRISPRi) employs a dCas9-sgRNA complex to precisely repress any target gene by steric hindrance (Bikard et al., 2013; Qi et al., 2013). The system’s target specificity is determined by the complementarity between the sgRNA and the target sequence and by the presence of a short motif downstream the target sequence, the protospacer adjacent motif (PAM) (Mojica et al., 2009). CRISPRi can prevent transcription initiation whether the promoter region is targeted or block mRNA elongation by RNA polymerase whether the coding sequence is targeted (Bikard et al., 2013; Larson et al., 2013; Qi et al., 2013). To achieve a high knockdown efficiency, the non-template strand of a gene or both strands of its promoter can be targeted (Qi et al., 2013). Several reports have established the use of anhydrotetracycline (ATc)-inducible (Ehrt et al., 2005) CRISPRi systems in mycobacteria, however with associated toxicity and producing a low level of knockdown (Choudhary et al., 2015; Singh et al., 2016). More recently, an ATc-inducible CRISPRi system based on the use of Cas9 from Streptococcus thermophilus (dCas9Sth1), employing several PAM variants, was developed (Rock et al., 2017).
In this work, we implement the dCas9Sth1-based CRISPRi system in M. smegmatis to assert the role of the characteristic modifications of mycobacterial PG, catalyzed by the NamH hydroxylase and the MurT/GatD complex, in susceptibility to beta-lactams and in host-pathogen interactions.
2. Materials and methods
2.1. Bacterial strains, plasmids, cell lines, culture conditions and antibiotics
The bacterial strains, plasmids and cell lines used in this study are listed in Table 1 . To amplify the CRISPRi backbone PLJR962 (Addgene #115162) and for cloning, E. coli was cultured in Luria-Bertani medium (Merck), supplemented with 25 μg/mL of kanamycin (NZYTech) when appropriate. M. smegmatis strains were cultured in Middlebrook 7H9 broth (BD™ Biosciences) supplemented with 0.2% glycerol (ThermoScientific), 0.5% glucose (Sigma-Aldrich) and 0.05% tyloxapol (Sigma-Aldrich) or 7H10 agar (BD™ Biosciences) supplemented with 0.5% glycerol. When necessary, the medium was supplemented with 25 μg/mL of kanamycin. Bacteria were grown at 37°C with (broth) or without (agar) shaking at 180 rpm. To induce sgRNA and dCas9Sth1 expression, 100 ng/mL of ATc were added to mycobacterial cultures every 24 h. The J774.A1 cells were cultured in DMEM media (Gibco) supplemented with 10% fetal bovine serum, 1% sodium pyruvate, 1% L-Glutamine, and 1% HEPES buffer. The growth of J774.A1 cells occurred at 37°C, with 5% CO2. Stocks of amoxicillin (AMX), cefotaxime (CTX), ethambutol (EMB), meropenem (MEM) and isoniazid (INH) (Sigma-Aldrich) were prepared with purified MilliQ water. ATc (Sigma-Aldrich) was prepared at a stock concentration of 10 mg/mL in dimethyl sulfoxide (DMSO) cell culture grade (AppliChem). Potassium clavulanate (CLA) (Sigma-Aldrich) was prepared in phosphate buffer pH 6.0, 0.1 M.
Table 1.
Bacterial strains, plasmids and cell lines used in this study.
| Bacterial strain, plasmid or cell line | Description | Reference or source |
|---|---|---|
| Bacteria | ||
| Escherichia coli | ||
| JM109 | recA1 endA1 gyr96 thi hsdR17 supE44 relA1 Δ(lac-proAB) [F´ traD36 proAB lac q ZΔM15] | Stratagene |
| Mycobacterium smegmatis | ||
| mc2 155 (WT) | High-transformation-efficiency mutant of M. smegmatis ATCC 607 | Snapper et al., 1988 |
| PM965 | M. smegmatis mc2 155 ept-1 rpsL4 ΔblaS1 | Raymond et al., 2005 |
| PM979 | M. smegmatis mc2 155 ept-1 rpsL4 ΔblaS1 ΔnamH | Raymond et al., 2005 |
| Plasmids | ||
| PLJR962 (8881 bp) | Sth1 dcas9 TetR and KanR L5 Int attP for M. smegmatis | Rock et al., 2017 |
| Cell lines | ||
| J774.A1 | Murine-derived monocyte/macrophage cell line | ATCC, TIB-67 |
2.2. Construction of knockdown mutants using CRISPRi
The cloning procedures were executed as previously optimized (Rock et al., 2017; Marques, 2020; Silveiro, 2020; Wong and Rock, 2021). To achieve efficient repression, either the non-template strand or the promoter region of the target genes murT (MSMEG_6276), gatD (MSMEG_6277) and namH (MSMEG_6410) were targeted. Various PAM sequences were identified on the template strand of the target genes, chosen for differential repression regarding PAM strength and target site, and accordingly guided sgRNA design ( Table S1 ). The CRISPRi backbone PLJR962 was amplified in E. coli JM109 and 2 μg of plasmid were digested with BsmBI (ThermoScientific) in the presence of 1 mM DTT (ThermoScientific) for 4 h at 37°C in a PCR block (Applied Biosystems Veriti™). After, the digestion reactions were purified using the QIAquick PCR Purification Kit (Qiagen). To clone the sgRNA targeting sequences into the sgRNA scaffold, two complementary primers were designed ( Table S2 ), synthesized (Eurofins Genomics), annealed, and ligated into PLJR962 using T4 DNA ligase (400 U/µL; NEB) at 22°C for 30 min. To obtain the sgRNA-containing CRISPRi plasmids, E. coli chemically competent cells were transformed with the ligation mixture using a double heat-shock approach, followed by plasmid extraction from the obtained recombinants with the NZYMiniprep kit (NZYTech). The obtainment of the desired sgRNA-containing CRISPRi plasmids was confirmed by Sanger Sequencing. To express the recombinant clones as knockdown mutants in M. smegmatis, competent cells were transformed with 500 ng of recombinant plasmid by electroporation (using 2 mm cuvettes and a Bio-Rad Gene Pulser™ Electroporation System, with pulse settings of 2.5 kV, 1000 Ω resistance and 25 μF capacitance) in the presence of glycerol 10% (Goude and Parish, 2009; Marques, 2020; Silveiro, 2020).
2.3. Phenotyping
To evaluate differences in cell viability after CRISPRi induction, spotting assays were performed as previously described (Marques, 2020; Silveiro, 2020). Briefly, cultures of M. smegmatis were grown to log phase, normalized to a theoretical OD600 of 0.001 and, serially diluted in a 96-well plate. A minimal quantity (5 µL) of each bacterial suspension was plated on solid media with and without ATc and incubated at 37°C for 2 days. At least three biological replicates were performed.
2.4. RNA extraction
Bacterial cultures were grown to log phase, normalized to a theoretical OD600 of 0.001 and, incubated at 37°C with shaking until an OD600 of 0.1 was reached (Marques, 2020; Silveiro, 2020; Wong and Rock, 2021). After an incubation period of 6 h with or without ATc, the cultures were harvested by centrifugation at 3000 g for 5 min, ressuspended in 500 µL of RNAprotect Bacteria Reagent (Qiagen) and, stored at -20°C. When convenient, the cells were thawed and pelleted at 5000 g for 5 min at 4°C. Lysis was promoted by the addition of 350 µL of buffer NR (NZYTech) and 3.5 µL of beta-mercaptoethanol (Merck), followed by intense vortexing. The cells were then disrupted in a BeadBug™ (Benchmark Scientific) by 4 cycles at maximum speed for 1 min, interspersed by resting periods of 1 min on ice. After a spin-down, the lysate was applied into the NZYSpin Homogenization column, and the subsequent steps were performed according to the instructions of the NZYTotal RNA Isolation Kit (NZYTech). The RNA was eluted in 40 µL of DEPC water (Invitrogen) at 60°C. Genomic DNA contamination was avoided by performing a rigorous treatment with TurboDNase (Invitrogen). The quantity and purity of the obtained RNA was determined using Nanodrop™ and the absence of significant DNA contamination was confirmed by performing a PCR on the housekeeping gene sigA using NZYTaq DNA polymerase (NZYTech) followed by 1.5% agarose gel electrophoresis.
2.5. Quantitative reverse-transcription PCR
Purified RNA (100 ng) was used to synthesize cDNA (Singh et al., 2016) by reverse transcription using the NZY First-Strand cDNA Synthesis Kit (NZYTech). Subsequently, the cDNA levels were quantified by qPCR using the NZYSupreme qPCR Green Master Mix (2x), ROX (NZYTech) and a QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems). The qRT-PCR primers were designed using Primer 3 1 to amplify a specific product between 100-250 bps ( Table S3 ). The PCR reaction (40 cycles) proceeded as follows: 95°C for 10 min, 95°C for 15 s, 60°C for 30 s. The amplification efficiency of the primers was verified to be 80-110%. For each run, at least two technical replicates and a negative control were produced, and the mean CT was calculated. Data were analyzed using the Comparative CT method (ΔΔCT method; the fold change is calculated as 2-ΔΔCT) with M. smegmatis WT as the calibrator sample and sigA as a reference gene (Choudhary et al., 2015; Rock et al., 2017; Wong and Rock, 2021). Two biological replicates were utilized for the quantification of the target sequences (Singh et al., 2016).
2.6. Minimum inhibitory concentration determination
The antibiotic susceptibility determination assays were performed for three beta-lactams (amoxicillin, cefotaxime and meropenem), with and without the beta-lactamase inhibitor clavulanate, and for two antimycobacterial agents (isoniazid and ethambutol), as described previously (Marques, 2020; Silveiro, 2020). The antibiotics were prepared at the desired initial concentration (512 μg/mL for beta-lactams and 128 μg/mL for EMB/INH) while clavulanate was used at a fixed concentration of 2.5 μg/mL. Next, the antibiotics were serially diluted 1:2 in supplemented 7H9 media. M. smegmatis cultures were grown to log phase, diluted to a theoretical OD600 of 0.002 and split into two equal volumes with and without 100 ng/mL of ATc. Subsequently, 100 µL of each bacterial suspension was added to the respective wells and, the plates were incubated at 37°C for 3 days. After incubation, the wells were observed for bacterial growth, usually indicated by turbidity or the presence of a bacterial pellet in the bottom of the wells. The dilution at which no bacterial growth was observed corresponds to the annotated MIC value. This assay was performed at least three times for each culture of interest and the median MIC values were calculated. Whenever mentioned, the “breakpoints” refer to the EUCAST non-species related PK/PD breakpoints 2 (version 12) because there are not any currently defined beta-lactam breakpoints for mycobacteria.
2.7. Macrophage infections and intracellular survival assessment
To address the contribution of PG modifications to the intracellular survival of mycobacteria, J774.A1 macrophages were infected with control and mutant strains, as described previously (Anes et al., 2006; Jordão et al., 2008). After several passages and 48 h before infection, the macrophages were seeded in 96-well plates at a density of 12.5 x 103 cells/well. That day, bacterial cultures were diluted to the same OD600 and, incubated at 37°C with shaking until an OD600 of 0.1 was reached. At that point (approximately 24 h before infection), the cultures were incubated with or without 100 ng/mL of ATc. On the day of infection, the cultures were centrifuged at 3000 g for 5 minutes and washed with 5 mL of PBS 1 X (Gibco) (Anes et al., 2006; Jordão et al., 2008). To obtain a single-cell suspension, the cultures were resuspended in DMEM media, subjected to 5 min of ultrasonic bath, and centrifuged at 500 g for 1 min (Pires et al., 2017). The cell media was discarded from the seeding plates and the macrophages were infected with a single-cell suspension of mycobacteria at a multiplicity of infection (MOI; ratio of bacteria/cells) of 5 and, incubated at 37°C with 5% CO2 for different timepoints (1 h; 4 h; 24 h) (Anes et al., 2006; Jordão et al., 2008; Marques, 2020; Silveiro, 2020). Following internalization, the cells were washed twice with PBS 1 X and resuspended in DMEM supplemented with 10 μg/mL gentamicin to kill extracellular bacteria and 100 ng/mL of ATc, when appropriate.
At timepoints 1 h, 4 h and 24 h, the media was discarded, and the macrophages were lysed after 15 min incubation with a 0.05% IGEPAL solution (Pires et al., 2017). Serial dilutions of the resulting lysates were prepared in sterile water and plated onto 7H10 plates, which were incubated for 2 days at 37°C with 5% CO2 (Anes et al., 2006; Jordão et al., 2008). The colony forming units (CFUs) were then counted using an inverted microscope (Nikon TMS Inverted Phase Contrast Microscope). Three biological replicates, each counting three technical replicates, were performed.
2.8. Single nucleotide polymorphism analysis in clinical strains of Mtb
To assess whether the genes encoding the enzymes responsible for the characteristic modifications of mycobacterial PG were conserved in Mtb, data was retrieved from a study recently published by our group that set to uncover beta-lactam susceptibility patterns in a collection of 172 clinical strains of Mtb (Olivença et al., 2022b). Briefly, for each strain, quality improved reads were individually mapped against Mtb H37Rv reference genome (GenBank accession number AL123456.3), and the identification of single nucleotide polymorphisms (SNPs) was performed as previously described (Olivença et al., 2022b). The resulting MIC values were used in this study to uncover susceptibility differences to AMX or MEM, with or without clavulanate, that could be associated with frequent SNPs (with a mutation frequency ≥ 3) in the proposed genes of interest.
2.9. Statistical analysis
The statistical analysis was preformed using GraphPad (version 8.4.3.686) and data are presented as mean ± SEM, except if stated otherwise. Normality testing for qRT-PCR or CFUs data was preformed using Q-Q plots or the Shapiro-Wilk Test, respectively. Multiple comparisons were made using one-way ANOVA, followed by pairwise comparisons of selected groups using the Holm-Sidak test. All the prerequisites of the tests were verified.
3. Results
3.1. Construction of single PG modification knockdown mutants using CRISPR interference
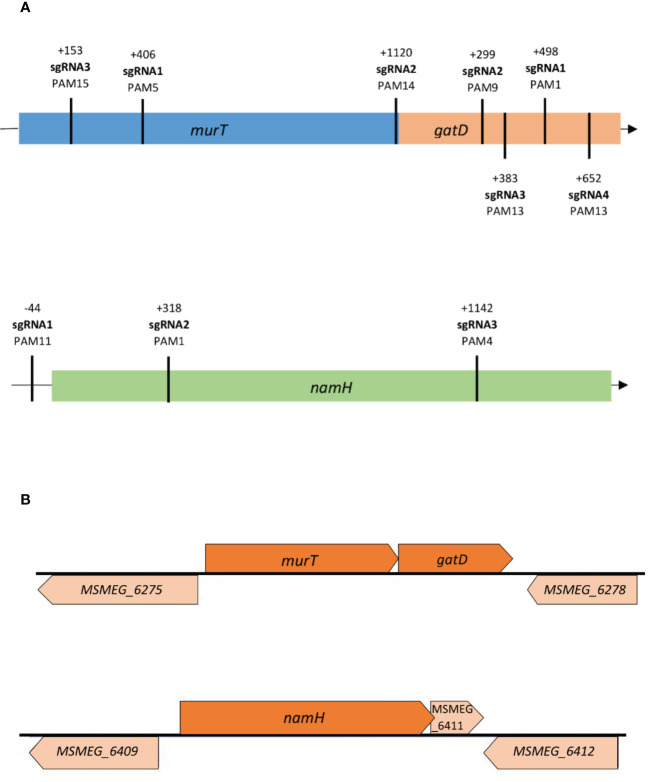
To construct the knockdown mutants, a previously developed ATc-inducible dCas9Sth1-based CRISPRi system, which enables the efficient knockdown of both essential and non-essential genes in mycobacteria, was used (Rock et al., 2017; Wong and Rock, 2021). To achieve efficient repression, either the non-template strand or the promoter region of the target genes murT (MSMEG_6276), gatD (MSMEG_6277) and namH (MSMEG_6410) were targeted. The efficiency of CRISPRi-mediated gene silencing is determined by several factors, namely PAM strength and the PAM position along the targeted gene (Rock et al., 2017; Marques, 2020). Therefore, various PAM sequences were identified on the template strand of the target genes and chosen for differential repression regarding PAM strength and target site ( Figure 1A ). PAM strength is directly proportional to the silencing efficiency, indicated as fold repression, of the possible PAM variants for dCas9Sth1-mediated gene silencing in mycobacteria (Rock et al., 2017; Wong and Rock, 2021). To avoid lethality effects when targeting allegedly essential genes, the essentiality classification of the respective orthologue genes in Mtb was considered. The gene local context was also taken into consideration due to the previously reported probability of downstream polar effects (Choudhary et al., 2015; Singh et al., 2016; Rock et al., 2017) and reverse polarity for operonic genes (Peters et al., 2016), as is the case of our target genes ( Figure 1B ).
Figure 1.
(A) Distribution of sgRNA targeting sequences along the murT/gatD operon and the namH gene in M. smegmatis. Different sites were targeted along the genes to assess the differential repression efficiency of CRISPRi; (B) The gene local context (illustration based on data from the Biocyc database) of the target genes murT, gatD and namH in M. smegmatis needs to be considered due to possible downstream/upstream polar effects. The murT and gatD genes are co-transcribed under regulation of a single promoter. The MSMEG_6275 precedes the murT gene but is transcribed in the opposite direction while the MSMEG_6278 gene occurs after the gatD gene but is also transcribed in the opposite direction. A single promoter guides the conjoint transcription of the namH and MSMEG_6411 genes by RNA polymerase. The MSMEG_6409 precedes the namH gene but is transcribed in the opposite direction while the MSMEG_6412 gene occurs after the MSMEG_6411 gene but is also transcribed in the opposite direction.
3.1.1. Phenotyping of the single mutants through spotting assays
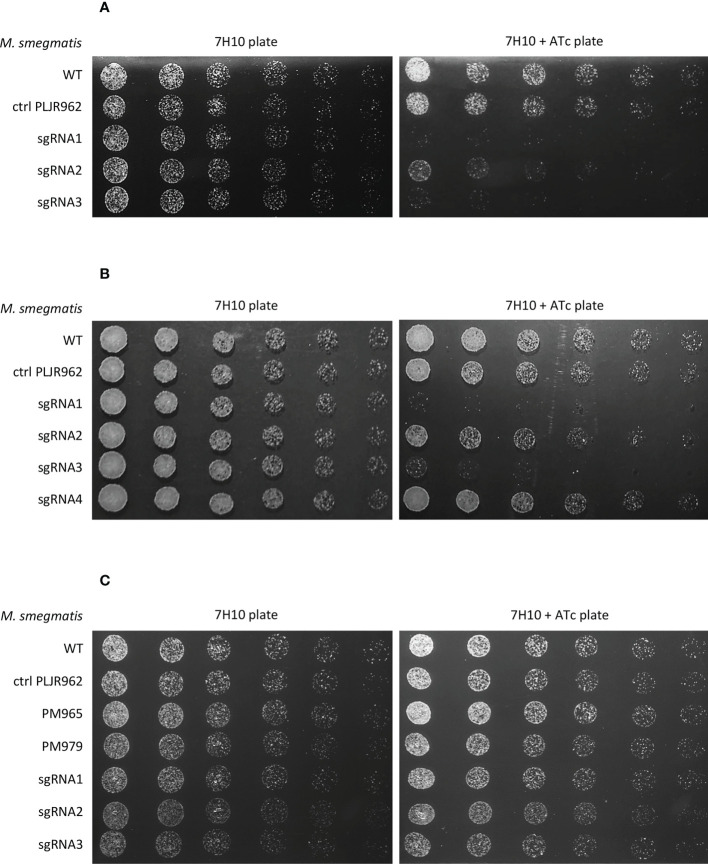
To phenotypically characterize the knockdown mutants, spotting assays were performed. The addition of inducer did not influence the viability of the negative controls M. smegmatis WT and M. smegmatis PLJR962. The results from the spotting assays show that the murT/gatD operon is essential for the viability of M. smegmatis since growth defects were observed for most knockdown mutants in the presence of inducer ( Figure 2A, B ) (Marques, 2020). In the case of murT knockdown (murT -), sgRNA1-murT and sgRNA3-murT were responsible for severe growth defects, while sgRNA2-murT only moderately affected bacterial viability. In the case of gatD knockdown (gatD -), sgRNA1-gatD and sgRNA3-gatD provoked severe growth defects in the spotting assays whereas sgRNA2-gatD caused a slight viability defect and sgRNA4-gatD did not appear to affect viability.
Figure 2.
Spotting assays of all M. smegmatis knockdown mutants (n=3): (A) murT knockdown mutants; (B) gatD knockdown mutants; (C) namH knockdown mutants. Negative controls (M. smegmatis WT, PLJR962 and PM965) and a positive control (M. smegmatis PM979) were included in all experiments. The first spots were normalized to an OD600 of 0.001 and the next were performed by preparing and spotting 2-fold serial dilutions of each culture on agar plates.
MurT comprises two main domains: the first is homologous to the central domain of Mur ligases (Mur_Ligase_M), and the second is a C-terminal domain without known function (DUF1727) (Figueiredo et al., 2012; Münch et al., 2012; Gonçalves et al., 2019; Maitra et al., 2021). The sequence regions that encode the MurT central domain, involved in the binding of the essential cofactors ATP (K59, E108) and Mg2+ (Figueiredo et al., 2012; Maitra et al., 2021) are targeted by sgRNA1-murT and sgRNA3-murT. A sequence alignment of MurT from S. aureus, S. pneumoniae, M. smegmatis and Mtb shows that these sequences are conserved in both MSMEG_6276 and its orthologue in Mtb, Rv3712 ( Figure S1 ) (Marques, 2020). The inhibition of transcription elongation by dCas9Sth1 can generate truncated transcripts, lacking these essential regions, thereby contributing to the low viability phenotype displayed by the respective knockdown mutants (Marques, 2020). The DUF1727 domain has four conserved amino acid regions (I, II, III and IV) that regulate the interaction between MurT and GatD (Gonçalves et al., 2019; Maitra et al., 2021). Although the target site of sgRNA2-murT codes for the DUF1727 domain, the targeted region corresponds to the final nucleotides of murT. Therefore, the RNA polymerase can transcribe almost the entire sequence of the murT gene, possibly leading to the production of a semi-functional protein and, thus, enabling the growth of the respective knockdown mutant (Marques, 2020). In contrast, GatD has a glutaminase domain similar to that of class I-type GATases, possessing conserved residues responsible for its catalytic glutamine amidotransferase activity in GatD (C94, H189) and in MurT (D349). GatD also possesses conserved catalytic residues involved in the sequestration of glutamine (R128) and in the channeling of ammonia to MurT (G59) (Figueiredo et al., 2012; Münch et al., 2012; Leisico et al., 2018; Maitra et al., 2021). A sequence alignment of GatD from S. aureus, S. pneumoniae, M. smegmatis and Mtb shows that these sequences are conserved in both MSMEG_6277 and its orthologue in Mtb, Rv3713 ( Figure S2 ) (Marques, 2020). CRISPRi-mediated interference of gatD with sgRNAs 1 to 3, which target sites near these conserved residues, might result in depleted D-iso-Glu amidation (Marques, 2020). This effect is directly proportional to PAM strength and could explain the growth inhibition phenotypes observed with sgRNA1-gatD and sgRNA3-gatD.
Regarding the target site and PAM strength distribution of the employed sgRNAs, it was observed that the distance from the transcription start site (TSS) directly impacts the phenotype of these mutants, mainly when weak PAMs are employed (Marques, 2020). In the case of murT, both sgRNA2-murT and sgRNA3-murT have weak PAMs, however sgRNA3-murT (PAM15, +153 bp) produced considerably more severe growth defects than sgRNA2-murT (PAM14, +1120 bp), likely due to the target site. In the case of gatD, greater growth defects were observed with sgRNA3-gatD (PAM13, +383 bp) when compared to sgRNA4-gatD (PAM13, +652 bp), although they share the same PAM.
On the other hand, namH is not an essential gene for the viability of M. smegmatis since no growth defects were observed in the presence of inducer ( Figure 2C ) (Silveiro, 2020).
3.1.2. Assessment of CRISPRi-mediated gene knockdown in the single mutants by qRT-PCR
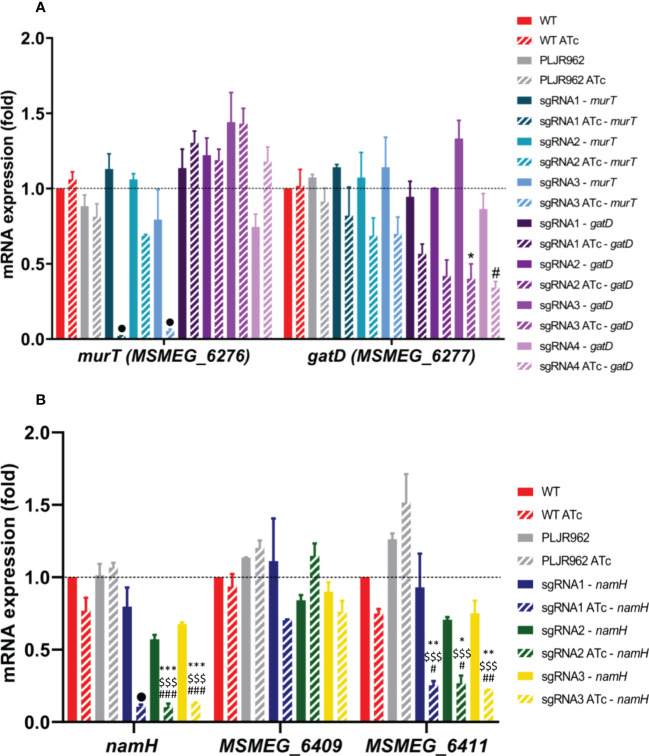
The normalized relative gene expression of the target genes murT, gatD, namH and neighboring genes was determined for selected samples, with and without inducer, at 6 h post-induction ( Figure 3 ).
Figure 3.
Results of qRT-PCR assays for the single mutants, constructed in the M. smegmatis WT strain (n=2): (A) murT (blue shade bars) and gatD (purple shade bars) knockdown mutants; (B) namH knockdown mutants. The graph shows the mean of the relative mRNA expression of genes of interest normalized to sigA, at 6 hours post-induction, with (stripped bars) and without (smooth bars) ATc. The dashed lines show the WT sample as calibrator. Error bars show the standard error of the mean. Multiple comparisons were made using one-way ANOVA, with significance levels: *P < 0.05; **P < 0.01; ***P < 0.001. Significant differences are indicated with symbols: # comparing to the induced WT strain (WT ATc), $ comparing to the induced control M. smegmatis:PLJR962 (PLJR962 ATc), * comparing to the respective uninduced strain. The • symbol indicates a highly significant difference (P < 0.0001) from all controls.
The obtained results show that the murT gene was successfully repressed in most induced murT- mutants ( Figure 3A ). A highly significant knockdown efficiency of murT was found with sgRNA1-murT comparing with WT ATc (53.5-fold; P < 0.0001), PLJR962 ATc (44.6-fold; P < 0.0001) or the respective uninduced mutant (57-fold; P < 0.0001). The same was observed with sgRNA3-murT (ranging from 16.5 to 21.1-fold repression) when compared to all controls (P < 0.0001). In contrast, sgRNA2-murT did not lead to a significant repression of murT. The highest repression of murT was achieved with sgRNA1-murT, being directly justified by PAM strength. Both sgRNA2-murT (PAM14, +1120 bp) and sgRNA3-murT (PAM15, +153 bp) have weak PAMs, however sgRNA3-mediated targeting resulted in a superior knockdown efficiency because sgRNA3-murT targets the beginning of the murT gene. An observable decrease in the mRNA expression of gatD was observed in all induced gatD - mutants ( Figure 3A ). The employed sgRNAs caused a fold repression ranging from 1.7 to 3.3 when compared with the respective uninduced mutants. A significant knockdown efficiency was found with sgRNA3-gatD (P = 0.0103, comparing with the correspondent uninduced mutant) and sgRNA4-gatD (P = 0.0433, comparing with WT ATc). CRISPRi-mediated silencing of the murT and gatD genes did not cause any significant polar effects in neighboring genes ( Figures 3 and S3 ).
Furthermore, the namH gene was also successfully repressed in the induced namH - mutants ( Figure 3B ). A highly significant knockdown efficiency of namH was observed with sgRNA1-namH comparing with WT ATc (7.3-fold; P < 0.0001), PLJR962 ATc (10.1-fold; P < 0.0001) or the respective uninduced mutant (7.5-fold; P < 0.0001). Likewise, highly significant repression differences were also observed with sgRNA2-namH (ranging from 5.6 to 10.5-fold) and sgRNA3-namH (ranging from 5.3 to 8.3 - fold) when compared to all controls (P ≤ 0.0003). The highest repression efficiency was achieved with sgRNA1-namH, which targets the promotor region of the namH gene, therefore inhibiting transcription initiation (Qi et al., 2013). The repression of namH was also more efficient with sgRNA2-namH (PAM1, +318 bp) than with sgRNA3-namH (PAM4, +1142 bp) due to higher PAM strength and to the target site. The knockdown efficiency between these sgRNAs is similar because the fold repression discrepancy between strong PAMs is minimal (Rock et al., 2017). The lack of reverse polarity effects contrasts with the occurrence of significant downstream polar effects in MSMEG_6411 with all sgRNAs ( Figure 3B ).
3.2. Construction of the double and triple mutants
To uncover synergistic effects between the action of beta-lactams and the depletion of the N-glycolylation of muramic acid and/or the D-iso-Glu amidation of PG, knockdown mutants were also constructed in previously modified strains, lacking BlaS, PM965 (M. smegmatis ΔblaS1) and PM979 (M. smegmatis ΔblaS1 ΔnamH) (Raymond et al., 2005).
For the construction of the double mutants, having PM965 as a parental strain, the above knockdown mutants of murT, gatD and namH were considered. M. smegmatis PM965 was transformed with the following sgRNA-containing CRISPRi plasmids targeting the genes of interest: sgRNA1 or sgRNA2 for murT, sgRNA2 or sgRNA4 for gatD and, sgRNA1 or sgRNA3 for namH. To select mutants with medium-high repression vs. medium-low repression, the plasmids were chosen based on theoretical assumptions (differences in PAM strength and target site) (Choudhary et al., 2015; Rock et al., 2017) and on the spotting phenotypes of the single mutants ( Figure 2 ).
To facilitate the identification of synergies between the depletion of NamH and the MurT/GatD enzymatic complex, the triple mutants, using PM979 as a parental strain, were constructed. In this case, only the murT and gatD knockdown mutants were considered since PM979 already lacks the namH gene. The plasmids used for target repression were the same that were used for the double mutants.
3.2.1. Phenotyping of the double and triple mutants through spotting assays
To phenotypically characterize the double and triple mutants, spotting assays were performed.
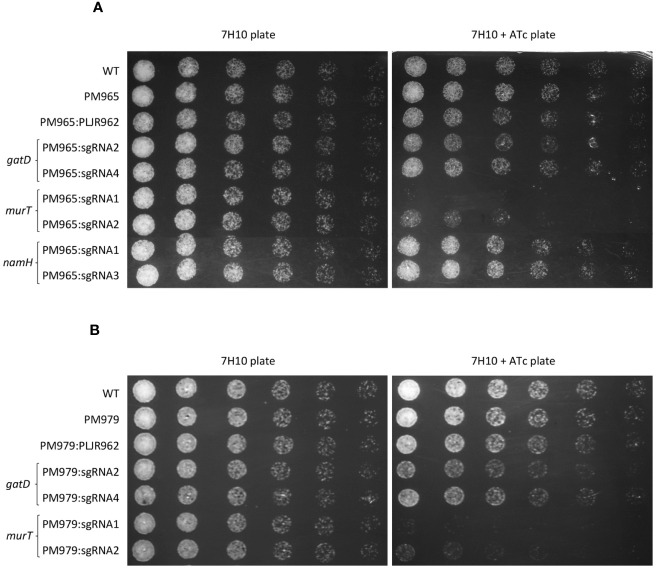
The spotting assays of the double mutants ( Figure 4A ) revealed considerable growth defects for the induced ΔblaS1 murT - mutants, similar to those observed with the induced murT - mutants, with lower viability for sgRNA1-murT (PAM5, +406 bp) compared to sgRNA2-murT (PAM14, +1120bp). The phenotype of the induced ΔblaS1 gatD - mutants was also identical to that observed for the induced gatD- mutants, with slightly more severe growth differences with sgRNA2-gatD (PAM9, +299 bp) than with sgRNA4-gatD (PAM13, +652 bp). Moreover, the phenotype of the induced ΔblaS1 namH - mutants was indistinguishable from that observed for the induced namH - mutants.
Figure 4.
Spotting assays of M. smegmatis double and triple mutants (n=3): (A) double mutants, having PM965 (ΔblaS1) as a parental strain and one target gene repressed; (B) triple mutants, having PM979 (ΔblaS1ΔnamH) as a parental strain and either murT or gatD repressed. Negative and positive controls were included in all experiments. The first spots were normalized to an OD600 of 0.001 and the next were performed by preparing and spotting 2-fold serial dilutions of each culture on agar plates.
The spotting assays of the triple mutants ( Figure 4B ) depicted slight changes in the viability of the induced ΔblaS1 ΔnamH murT - and ΔblaS1 ΔnamH gatD - mutants when compared to the double mutants. These minimal viability changes were mainly observed with the induced PM979:sgRNA2-gatD and ΔblaS1 ΔnamH murT - mutants.
3.2.2. Assessment of CRISPRi-mediated gene knockdown by qRT-PCR
Similarly, to the single mutants, the normalized relative gene expression of genes murT, gatD, namH and neighboring genes was also evaluated for the double and triple mutants in induced and non-induced conditions ( Figure 5 ).
Figure 5.
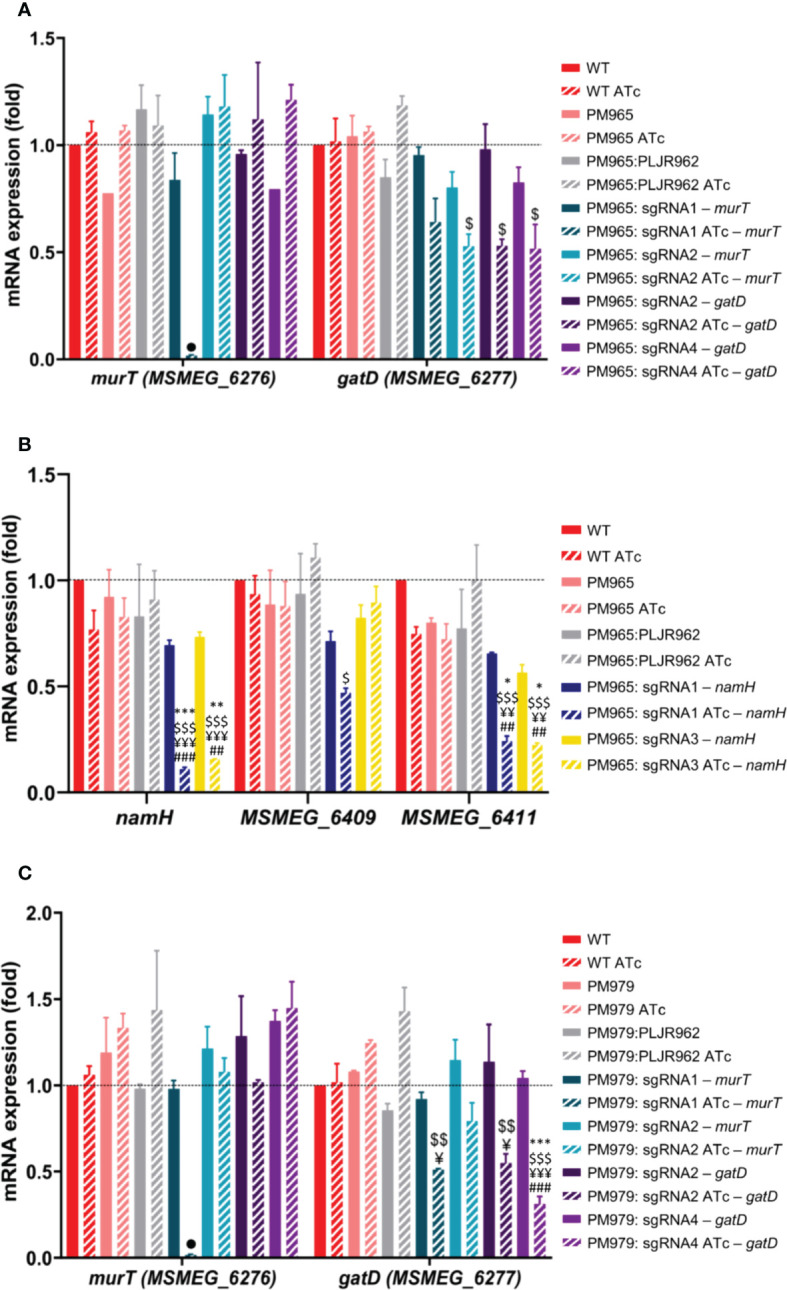
Results of qRT-PCR assays for the double and triple mutants (n=2): (A, B) Double mutants: (A) murT (blue) and gatD (purple) knockdown mutants constructed in PM965; (B) namH knockdown mutants constructed in PM965; (C) Triple mutants: murT (blue) and gatD (purple) knockdown mutants constructed in PM979. The graph shows the mean of the relative mRNA expression of genes of interest normalized to sigA, at 6 hours post-induction, with (stripped bars) and without (smooth bars) ATc. Dashed lines show the WT as calibrator. Error bars show the standard error of the mean. Multiple comparisons were made using one-way ANOVA, *P < 0.05; **P < 0.01; ***P < 0.001. Significant differences are indicated with symbols: # comparing to control M. smegmatis WT ATc, ¥ comparing to controls PM965 ATc or PM979 ATc, $ comparing to PM965:PLJR962 ATc or PM979:PLJR962 ATc, * comparing to the respective uninduced strain. The • symbol indicates a highly significant difference (P < 0.0001) from all controls.
In the case of the induced ΔblaS1 murT - mutants ( Figure 5A ), sgRNA1-murT (PAM5, +406 bp) led to a highly significant repression of murT when compared to WT ATc (64.1-fold; P < 0.0001), PM965 ATc (64.6-fold; P < 0.0001), PM965:PLJR962 ATc (66-fold; P < 0.0001) or the respective uninduced mutant (50.6-fold; P < 0.0001). In contrast, sgRNA2-murT (PAM14, +1120bp) did not lead to a significant repression of murT. Significant polar effects in the mRNA expression of gatD were only observed with sgRNA2-murT when compared to PM965:PLJR962 ATc (2.2-fold; P = 0.0337). In the case of the ΔblaS1 gatD - mutants ( Figure 5A ), a significant repression of gatD was attained with both sgRNA2-gatD (2.2-fold; P = 0.0373) and sgRNA4-gatD (2.3-fold; P = 0.019) when compared to control strain PM965:PLJR962 ATc. Following the repression of either murT/gatD, no upstream or downstream polar effects were found in neighboring genes ( Figures 5 and S4 ).
In the case of the induced ΔblaS1 namH - mutants ( Figure 5B ), a highly significant repression of namH was obtained with both sgRNA1-namH (ranging from 6.3 to 8.2-fold) and sgRNA3-namH (ranging from 4.7 to 5.8-fold repression), when compared to all controls (P ≤ 0.0017). A higher knockdown efficiency was obtained with sgRNA1-namH (PAM11, -44bp) than with sgRNA3-namH (PAM4, +1142 bp) as sgRNA1-namH targets the promoter region and possesses a higher PAM strength. Similarly, to the single mutants, significant downstream polar effects in MSMEG_6411 were observed with both sgRNAs. In terms of reverse polarity, sgRNA1-namH caused a significant decrease in the mRNA expression levels of MSMEG_6409 when compared to PM965:PLJR962 ATc (2.4-fold; P = 0.0322). This might have occurred due to an overlap between the target region and the promoter guiding the transcription of MSMEG_6409, since sgRNA1-namH targets the promoter region of namH and MSMEG_6409 is transcribed in the antisense strand ( Figure 1B ).
Again, a highly significant repression of murT was achieved with sgRNA1-murT (ranging from 56.2 to 82.5–fold repression) when compared to all controls (P < 0.0001), in the case of the induced ΔblaS1 ΔnamH murT - mutants ( Figure 5C ). Contrary to what was observed for the double mutants, significant polar effects in the mRNA levels of gatD were observed with sgRNA1-murT when compared to PM979 ATc (2.4-fold; P = 0.0113) and PM979:PLJR962 ATc (2.8-fold; P = 0.0034). In the case of the induced ΔblaS1 ΔnamH gatD - mutants, gatD was significantly repressed with sgRNA2-gatD when compared to PM979 ATc (2.3-fold; P = 0.0211) and to PM979:PLJR962 ATc (2.6-fold; P = 0.0062) ( Figure 5C ). A highly significant repression of gatD (ranging from 3.3 to 4.6–fold) was also achieved with sgRNA4-gatD, comparing to all controls (P ≤ 0.0009). No significant polar effects were found in neighboring genes following the repression of the murT/gatD operon ( Figure S5 ).
Since the triple mutants have M. smegmatis PM979 as a parental strain (ΔblaS1 ΔnamH), the mRNA levels of namH were not evaluated.
3.3. Minimum inhibitory concentration determination of the constructed mutants
To investigate the role of the D-iso-Glu amidation and of the N-glycolylation of PG in antibiotic susceptibility, the MICs of ethambutol, isoniazid and three beta-lactams, with or without clavulanate, were determined against the constructed mutants and respective control strains ( Figure 6 ). For these experiments, 100 ng/mL of ATc were used since the MIC of ATc against M. smegmatis mc2 155 WT has been defined as 500 ng/mL (Ehrt et al., 2005). Moreover, other studies have also used 100 ng/mL of ATc to induce CRISPRi-mediated silencing in M. smegmatis WT (Singh et al., 2016; Rock et al., 2017), including those employing the dCas9Sth1-based system (Rock et al., 2017).
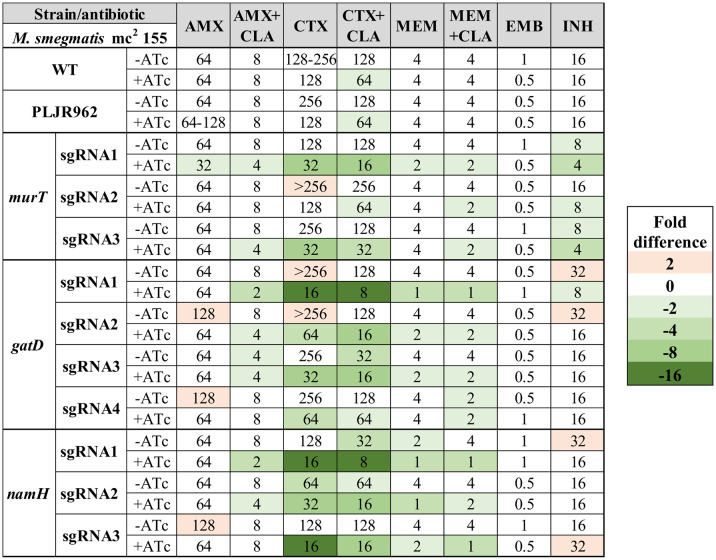
Figure 6.
Heatmap of the fold differences in minimum inhibitory concentrations (MICs) of the knockdown mutants constructed in the M. smegmatis WT strain, with and without ATc. Columns represent the antibiotics and rows represent the strains. The median MIC (in µg/mL) of each antibiotic for each strain is shown in the respective intersection and colours depict an increase (red shades) or decrease (green shades) in the value when compared to the MICs of M. smegmatis WT. AMX, amoxicillin; CLA, clavulanate; CTX, cefotaxime; EMB, ethambutol; INH, isoniazid; MEM, meropenem.
As expected, the observed MICs for the negative controls (M. smegmatis WT, M. smegmatis:PLJR962) were not considerably influenced by the addition of ATc (Marques, 2020; Silveiro, 2020). Notably, the repression of the murT/gatD operon and of the namH gene promoted increased susceptibility to all tested beta-lactams (Marques, 2020; Silveiro, 2020). The MICs of AMX remained mostly constant, as 2-fold differences can occur due to technical variability and are, thus, not considerable. Substantial susceptibility differences (≥ 4-fold) in AMX+CLA were found when gatD was repressed with sgRNA1-gatD or when namH was silenced with sgRNA1-namH. Hence, the correspondent induced knockdown mutants are susceptible to AMX+CLA according to the EUCAST breakpoints ( Table S4 ). In the case of the induced M. smegmatis:PLJR962:sgRNA1-gatD knockdown mutant, the susceptibility gain might be explained by a viability loss, as shown in the phenotyping assays. Unlike CTX or MEM, the addition of CLA to beta-lactams caused marked decreases in the MIC of AMX. The most notable susceptibility differences (ranging from 4 to 16-fold) occurred with CTX (+CLA), being present across all induced knockdown mutants, independently of the target gene. The most extensive increases in susceptibility to CTX (+CLA) occurred when gatD was repressed with sgRNA1-gatD or, when namH was silenced with sgRNA1-namH and sgRNA3-namH. Additionally, the fitness loss of the induced M. smegmatis:PLJR962:sgRNA1-gatD knockdown mutant also provoked a 4-fold decrease in the MIC of MEM (+CLA). In contrast, the repression of namH consistently promoted increased susceptibility to MEM (+CLA), thereby inferring a particular role of NamH in resistance to the carbapenem. Indeed, all knockdown mutants with a meropenem MIC ≤ 2 µg/mL are classified as susceptible to MEM according to the breakpoints ( Table S4 ). The MICs of EMB and INH remained essentially unchanged, with an interesting exception. The repression of murT with sgRNA1-murT and sgRNA3-murT caused a 4-fold decrease in the MIC of INH, which is most likely due to viability loss of the respective mutants.
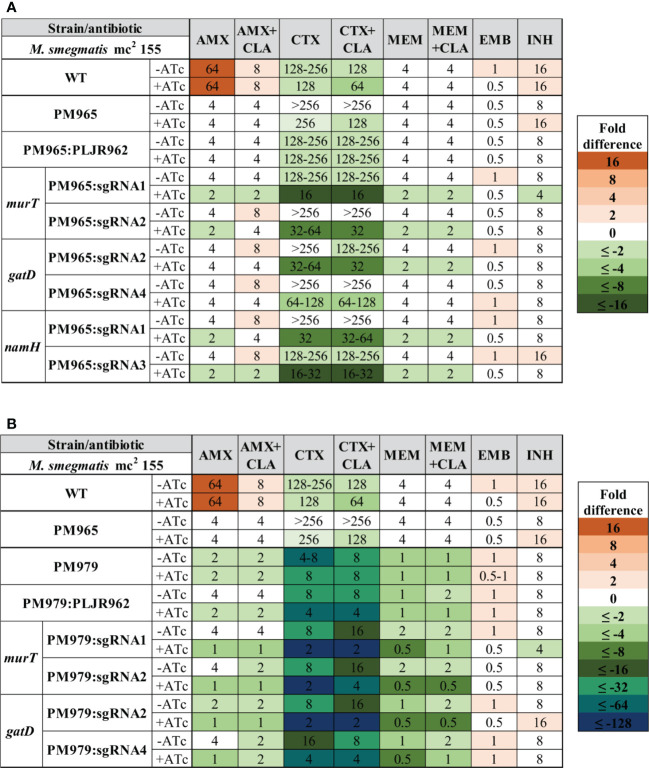
Similarly, the median MICs of the double mutants were determined for the same antibiotics ( Figure 7A ). The MICs of M. smegmatis PM965 and PM965:PLJR962 remained unaltered with the addition of ATc. When compared to M. smegmatis WT, the parental PM965 strain presented substantially lower MICs to all beta-lactams because this strain lacks the major beta-lactamase of M. smegmatis, BlaS (Raymond et al., 2005). The absence of BlaS in conjunction with the repression of murT/gatD and namH promoted increased susceptibility to the tested beta-lactams, although with insubstantial differences in the MICs of AMX (+CLA) and MEM (+CLA). The lack of BlaS in PM965 aided the classification of the induced murT and namH knockdown mutants as susceptible to AMX, according to the breakpoints ( Table S4 ). Both induced M. smegmatis PM965:PLJR962:sgRNA1-murT and PM965:PLJR962:sgRNA1-namH are classified as susceptible to AMX+CLA according to the breakpoints. The most notable susceptibility differences, ranging from 4 to 16-fold in comparison with M. smegmatis WT and PM965, occurred with CTX (+CLA), being present across most induced knockdown mutants. Extensive susceptibility differences (≥16-fold) from the PM965 strain in CTX (+CLA) were found when murT was repressed with sgRNA1-murT or when namH was silenced with sgRNA3-namH. The increased susceptibility of the induced M. smegmatis PM965:PLJR962:sgRNA1-murT knockdown mutant to beta-lactams might be explained by fitness loss, as shown in the phenotyping assays. All induced knockdown mutants constructed in the BlaS-depleted strain except for induced M. smegmatis PM965:PLJR962:sgRNA4-gatD are susceptible to MEM according to the breakpoints ( Table S4 ). The MICs of EMB and INH remained essentially unchanged.
Figure 7.
Heatmap of the fold differences in minimum inhibitory concentrations (MICs) of the double and triple knockdown mutants, with and without ATc. (A) Mutants constructed in M. smegmatis PM965 strain (double mutants); (B) Mutants constructed in the M. smegmatis PM979 strain (triple mutants). Columns represent the antibiotics and rows represent the strains. The median MIC (in µg/mL) of each antibiotic for each strain is shown in the respective intersection and colours depict an increase (red shades) or decrease (green/blue shades) in the value when compared to the MICs of control strain M. smegmatis PM965 (ΔblaS1). AMX, amoxicillin; CLA, clavulanate; CTX, cefotaxime; EMB, ethambutol; INH, isoniazid; MEM, meropenem.
Similarly, the median MICs of the triple mutants, having M. smegmatis PM979 as a parental strain, were determined for the same antibiotics ( Figure 7B ). The MICs of M. smegmatis PM979 were not considerably altered by ATc addition. In fact, the PM979 strain was found to be very susceptible to beta-lactams when compared to the WT and PM965 strains, suggesting a major role for the N-glycolylation of muramic acid in resistance to beta-lactams. The MICs of AMX (+CLA), CTX (+CLA) and MEM+CLA against M. smegmatis PM979:PLJR962 suffered a 2-fold decrease when inducer was added, with the obtained MIC values for AMX (+CLA) and MEM+CLA being equal to those obtained with the PM979 strain in induced conditions. Therefore, the MIC differences observed for the knockdown mutants in CTX (+CLA) cannot be interpreted as a result of their knockdowns alone since these differ in 2-fold from the MIC values obtained with the induced PM979 strain. This reproducible effect is most likely due to dCas9Sth1-associated toxicity as an exogenous protein to the bacterial system. Despite this remark, the additional repression of murT/gatD in the PM979 strain favoured even lower beta-lactam MICs. The cumulative effect of the absence of BlaS and NamH and of the depletion of the D-iso-glutamate amidation of PG provoked increased susceptibility to beta-lactams when compared to the parental strain PM979 (2 or 4-fold differences), and especially when compared to M. smegmatis WT and PM965 (4 to 128-fold differences). Although M. smegmatis PM979 is already very susceptible to beta-lactams when compared to the WT and PM965 strains, the additional repression of murT/gatD favored even lower beta-lactam MICs. When compared to the PM965 strain, all sgRNAs leading to depleted D-iso-Glu amidation caused considerable 4-fold decreases in the MIC of AMX, with comparable results for AMX+CLA. Most sgRNAs caused a 64 to 128-fold reduction in the MIC of CTX (+CLA) compared to PM965 and WT MIC values. Moreover, a noteworthy MEM MIC value of 0.5 µg/mL was obtained with all sgRNAs, differing in 8-fold from the PM965 and WT MIC values. Although the fold differences in the MIC of MEM between the triple mutants and the PM965 control strain seem to be less substantial, the PM965 MIC value for MEM is extremely low in comparison to that of AMX and CTX, making it difficult to attain great fold differences. Furthermore, all induced knockdown mutants are susceptible to AMX (+CLA) and MEM according to the breakpoints ( Table S4 ). Again, the MICs of EMB and INH remained essentially unchanged.
3.4. Macrophage infection and intracellular survival assays of the constructed mutants
To elucidate the contribution of the D-iso-Glu amidation and of the N-glycolylation of PG to mycobacterial intracellular killing, J774 murine macrophages were infected with all strains at a MOI of 5 and, disrupted for the assessment of the intracellular survival of bacteria (in CFUs/mL) at 1 h, 4 h and 24 h post-infection ( Figure 8 ).
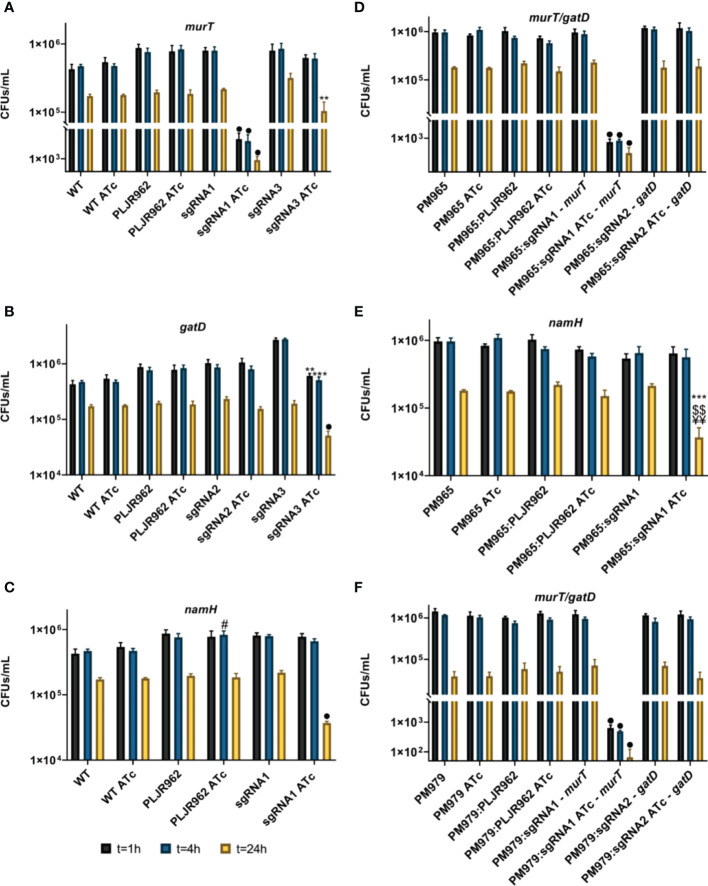
Figure 8.
Logarithmic representation of the mean of bacterial survival, with/without ATc, in CFUs/mL, after infection and disruption of J774.A1 macrophages (n=3). (A–C) Single mutants: (A) murT knockdown mutants; (B) gatD knockdown mutants; (C) namH knockdown mutants; (D, E) Double mutants: (D) murT/gatD knockdown mutants; (C) namH knockdown mutants; (F) Triple mutants, murT/gatD knockdown mutants. Error bars show the standard error of the mean. Multiple comparisons were made using one-way ANOVA: *P < 0.05; **P < 0.01; ***P < 0.001. Significant differences are indicated with symbols: # comparing to control strain WT ATc, ¥ comparing to the controls PM965 ATc or PM979 ATc; $ comparing to M. smegmatis WT/PM965/PM979:PLJR962 ATc, * comparing to the respective uninduced strain. The • symbol indicates a highly significant difference (P < 0.0001) from all controls.
As expected, the intracellular survival of mycobacteria decreased over time (Marques, 2020; Silveiro, 2020). The single mutants’ results show that the mean CFUs/mL of the negative controls M. smegmatis WT and PLJR962 were not significantly influenced by ATc addition ( Figures 8A-C ). The repression of murT with sgRNA1-murT caused a highly significant decrease in the intracellular survival of these bacteria at all timepoints ( Figure 8A ; P < 0.0001, when compared to M. smegmatis WT ATc, PLJR962 ATc or the respective uninduced control). This observation is explained by a strong repression of the murT gene, causing a lethality effect that can be verified by the low viability of the correspondent induced murT - mutant in the phenotyping assays. Besides, the repression of murT with sgRNA3-murT provoked a significant decrease in intracellular bacterial survival at t = 24 h ( Figure 8A ; P = 0.004, comparing with the respective uninduced control) while retaining some viability in the phenotyping assays. A highly significant decrease in the intracellular survival of the induced gatD knockdown mutants at t = 24 h was only attained with sgRNA3-gatD ( Figure 8B ; P < 0.0001, compared to all controls). Furthermore, the knockdown of namH attained with sgRNA1-namH also caused a highly significant decrease in the intracellular survival of bacteria at t = 24 h ( Figure 8C ; P < 0.0001, compared to all controls). These results indicate that both the D-iso-Glu amidation and the N-glycolylation of PG contribute to the intracellular survival of mycobacteria during infection. Moreover, the PM979 strain, lacking both BlaS and NamH, undergoes a significantly faster clearance by J774 macrophages when compared to WT or PM965 ( Figure S6 ).
The mean CFUs/mL of the double ( Figures 8D, E ) and triple mutants ( Figure 8F ) were also determined. The controls of the double mutants (M. smegmatis PM965, M. smegmatis PM965:PLJR962) and of the triple mutants (M. smegmatis PM979, M. smegmatis PM979:PLJR962) did not display any differences in intracellular survival after the addition of inducer. Generally, the obtained results for the knockdown of murT and gatD in the PM965 and PM979 strains were identical to those observed with the single mutants ( Figure 8 ). Unfortunately, the effect of D-iso-Glu amidation on mycobacterial intracellular survival could not be shown using the best candidate sgRNAs targeting murT/gatD leading to significant changes in the CFUs/mL because the respective mutants were not available. Still, the repression of murT in both PM965 and PM979 strains with sgRNA1-murT caused a highly significant decrease in the intracellular survival of these bacteria at all timepoints (P < 0.0001, compared to all controls) ( Figures 8D, F ). Remarkably, the knockdown of namH in the PM965 strain with sgRNA1-namH caused a very significant decrease in the bacterial intracellular survival at t = 24 h when compared to PM965 ATc (P = 0.0018), PM965:PLJR962 ATc (P = 0.0048) or the respective uninduced mutant (P = 0.0008) ( Figure 8E ).
3.5. Single nucleotide variants associated with differential susceptibility to beta-lactams in clinical strains of Mtb
To uncover whether the genes encoding the proteins responsible for the characteristic modifications of mycobacterial PG are of conserved nature, we benefited from the data obtained for 172 Mtb isolates in a previous study (Olivença et al., 2022b), where the susceptibility of the strains to AMX and MEM, with and without clavulanate, was also evaluated.
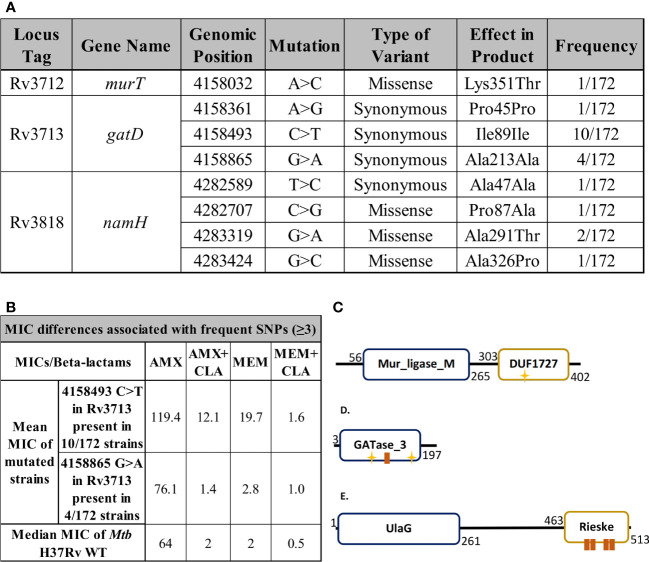
By performing a SNP-based analysis against the H37Rv reference genome, we found the murT, gatD and namH genes to be highly conserved in this specific set of clinical strains of Mtb ( Figure 9A , Tables S5 , S6 ). When a certain SNP was detected in three or more strains, the geometric mean of the MIC of these strains to AMX and MEM was compared to the median MIC of the control strain Mtb H37Rv WT ( Figure 9B ). Among the 172 strains, only one SNP (4158032 snp A>C) was identified for the murT gene, occurring in the region coding for the C-terminal domain of MurT (DUF1727) ( Figure 9C ). Although synonymous mutations do not carry as much importance as missense variants or indels, three synonymous variants were found in gatD. One of these mutations (4158493 snp C>T) was found in 10/172 strains, all of which were classified as lineage 2 strains and had increased beta-lactam MIC values, namely in the case of AMX+CLA and MEM ( Figure 9B ). This mutation occurred in the GATase_3 domain of GatD, near its first catalytic site (C94) ( Figure 9D ). Another one of these mutations (4158865 snp G>A) was found in only 4/172 strains, all of which were classified as lineage 4 strains, however without any considerable MIC differences ( Figure 9B ). Four variants were found for the namH gene, but always at a low frequency so they could not be associated with MIC changes. Two of these mutations (4282589 snp T>C; 4282707 snp C>G) occurred in regions coding for the UlaG domain of NamH whereas the other two (4283319 snp G>A; 4283424 snp G>C) occurred in regions coding for the Rieske domain ( Figure 9E ).
Figure 9.
Single nucleotide polymorphism (SNP) detection and MIC assays. (A) Single nucleotide polymorphisms found in the target genes murT, gatD or namH through the whole genome sequencing of 172 clinical strains of Mtb. Locus tags, gene names, products and positions refer to the H37Rv reference genome (AL123456.3). (B) Geometric mean MIC of clinical strains harbouring frequent single nucleotide polymorphisms (n≥3) compared to the median MIC of Mtb H37Rv WT. (C-E) Protein structures were drawn based on data from the NCBI, Pfam, UniProt and PROSITE databases. The yellow stars represent active sites while the brown boxes represent binding sites. (C) The protein structure of MurT, comprising a central Mur ligase domain and the DUF1727 domain. The D349 residue present in the DUF1727 domain of MurT is important for the catalytic activity of GatD, being part of its catalytic triad. (D) The protein structure of GatD, comprising a GATase_3 domain, responsible for its catalytic glutamine amidotransferase activity (C94, H189). (E) The protein structure of NamH, comprising a beta-lactamase superfamily UlaG domain and a Rieske domain, associated with oxidoreductase reactions.
4. Discussion
The lack of treatment success of drug-resistant TB urges the development of novel antituberculosis agents (Singh and Chibale, 2021). The PG layer of the mycobacterial CW features unique modifications, such as the N-glycolylation of muramic acid and the amidation of D-iso-glutamate (Catalão et al., 2019; Maitra et al., 2019). In this study, these PG modifications were depleted to address their role in beta-lactam susceptibility and in intracellular survival. Given the emergence of MDR-TB and the necessity of novel TB therapeutic regimens, research on the introduction of beta-lactams in TB-therapy is considered important by the WHO (WHO, 2020). Our results, discussed below, support the development of therapeutic agents targeting the mycobacterial PG and suggest that the inhibition of PG polymerization could synergize with beta-lactams to aid the treatment of MDR-TB infections.
The phenotyping assays of the single PG modification knockdown mutants uncovered the essentiality of the murT/gatD operon for the viability of M. smegmatis, as reported by studies with the correspondent Mtb orthologues, Rv3712/Rv3713 (Sassetti et al., 2003; Griffin et al., 2011; DeJesus et al., 2017; Maitra et al., 2021). In contrast, the muramic acid hydroxylase NamH, comprised of the N-terminal UlaG and the C-terminal Rieske domains, was found to be non-essential to the survival of M. smegmatis, also being in line with previous findings in M. smegmatis (Raymond et al., 2005) and in its Mtb orthologue, Rv3818 (Sassetti et al., 2003; Griffin et al., 2011; DeJesus et al., 2017). Overall, the obtained growth defects for sgRNAs with varying PAM strength usually matched the anticipated phenotypes whilst revealing a direct correlation between these and the sgRNA targeting site distance from the TSS.
The qRT-PCR results confirmed the successful repression of all target genes, however with variable efficiency. The repression of gatD had an extremely low efficiency when compared to that of murT, possibly due to differences in target vulnerability (Bosch et al., 2021). The repression efficiency of murT was also found to be higher than that of namH, which could be explained by the fact that sgRNAs targeting essential genes usually lead to a greater knockdown efficiency than sgRNAs targeting non-essential genes (Bosch et al., 2021). In few cases, a reduction in the mRNA levels of the target genes was observed even without inducer, indicating leaky expression. Moreover, an indirect proportionality between the efficiency of repression and the distance between target site and the TSS was uncovered. Although researchers were unable to observe any link between repression efficiency and target site using the same CRISPRi system (Rock et al., 2017; Wong and Rock, 2021), a previous study using a CRISPRi system based on the use of dCas9 from Streptococcus pyogenes (dCas9Spy) is concordant with our results (Choudhary et al., 2015). Despite using the same CRISPRi system, it is noteworthy that the obtained levels of knockdown are not as robust as those previously described for the repression of several target genes in M. smegmatis (11–226 fold) (Rock et al., 2017). An induction time of 6 h was used, which is low in comparison to the 14 h of induction previously reported (Rock et al., 2017). Nevertheless, the CRISPRi system reaches maximal silencing efficiency between one and two cell divisions after ATc addition (Rock et al., 2017), meaning that 6 h of induction time should be sufficient to assess the depletion of target mRNAs. Besides, knockdown efficiency would likely be improved by predepletion in ATc-supplemented medium. Overall, our results show that dCas9Sth1-mediated targeting leads to efficient target inhibition, thus affecting the associated phenotypes.
The double and triple mutants, lacking BlaS, facilitate the discovery of synergistic effects between the action of beta-lactams and the depletion of characteristic PG modifications. The double mutants (ΔblaS1 murT- , ΔblaS1 gatD- , ΔblaS1 namH -) presented a phenotype similar to that of the single knockdown mutants. Likewise, the simultaneous depletion of NamH and MurT/GatD (ΔblaS1 ΔnamH murT- and ΔblaS1 ΔnamH gatD- mutants) caused minimal changes in viability. The obtained knockdown efficiency of murT/gatD and namH was generally comparable between the single, double, and triple mutants, thereby suggesting a successful repression of all target genes independently of the parental strain. Again, an indirect proportionality between repression efficiency and the distance between the target site and the TSS was observed.
The median MICs obtained for the single mutants showed that the depletion of the N-glycolylation and the D-iso-Glu amidation of PG promote increased susceptibility to beta-lactams. Previous studies with S. aureus have also identified the MurT/GatD enzymatic complex as a mechanism of resistance to beta-lactams (Figueiredo et al., 2012; Münch et al., 2012). As expected, the obtained results revealed a direct proportionality between decreases in the MIC and the overall strength of the employed sgRNA. The cumulative absence of BlaS in conjunction with the knockdown of either namH or murT/gatD (double mutants) resulted in increased susceptibility to all beta-lactams. In this case, the most notable susceptibility differences pertaining to WT MICs (16 to 32- fold) occurred with AMX since BlaS is thought to possess substantially superior catalytic affinity for penicillins, when compared to cephalosporins and carbapenems (Solapure et al., 2013). The MIC results of the triple mutants demonstrated that repressing namH is a good strategy to increase susceptibility to beta-lactams. This could be explained by the role that N-glycolylated muropeptides play in strengthening CW stability through hydrogen bonds or by a differential recognition of the N-glycolylated glycan backbone by transpeptidases (Raymond et al., 2005; Silveiro, 2020). Together, the simultaneous depletion of the N-glycolylation of muramic acid and of the amidation of D-iso-Glu seem synergistic in such a way that these mutants present further reduced MIC values. Therefore, the development of a novel anti-TB drug that simultaneously targets the NamH and MurT/GatD proteins would be particularly effective.
Throughout these assays, MEM consistently produced the lowest MIC values when compared to other beta-lactams. While MEM inhibited mycobacterial growth very efficiently, CTX did not. To inhibit PG biosynthesis and kill mycobacteria, all sort of transpeptidases need to be inhibited (Kieser et al., 2015; Story-Roller and Lamichhane, 2018). Both MEM and CTX can inhibit PBPs and Ldts (Dubée et al., 2012) but only MEM can do so efficiently while being an extremely poor substrate to beta-lactamases (Tremblay et al., 2010). This observation supports the recent efforts to repurpose meropenem-clavulanate to treat MDR-TB. Moreover, we found that some reductions in beta-lactam MICs were associated with a loss of viability of the respective induced knockdown mutants, namely when either murT or gatD were targeted. Although not as markedly, this increase in susceptibility to beta-lactams was also seen when weaker sgRNAs were used to target these genes. Furthermore, the most notable susceptibility differences occurred with CTX (+CLA), being present across most induced knockdown mutants. Besides inhibiting PBPs, CTX can also inhibit some Ldts (Dubée et al., 2012), the most active transpeptidases during PG cross-linking in mycobacteria (Lavollay et al., 2008; Kumar et al., 2012; Baranowski et al., 2018). Furthermore, the MurT/GatD enzymatic complex regulates the activity of Ldts in Mtb, thereby determining the levels of PG cross-linking (Pidgeon et al., 2019). Therefore, our observation could be explained by a favorable use of D-iso-Glu amidated PG as a substrate by transpeptidases that are targeted by CTX.
The immune recognition of mycobacterial PG by macrophages occurs through specific immune receptors, the cytosolic Nod-like receptors (NLRs) (Coulombe et al., 2009; Hansen et al., 2014), possibly justifying a more bactericidal response of macrophages. Whereas the recognition of molecules containing D-Glu-m-DAP is achieved by NOD1, PG–derived muramyl dipeptide (MDP) is mostly recognized by NOD2 (Coulombe et al., 2009; Hansen et al., 2014). NOD2-mediated host responses (NMHR) culminate in the production of pro-inflammatory cytokines possibly leading to oxidative burst responses (Coulombe et al., 2009; Hansen et al., 2014). Our intracellular survival assays showed that the depletion of MurT/GatD as well as that of NamH are both associated with an increased clearance rate, possibly due to a higher recognition by host macrophages.
The amidation of D-iso-Glu has been reported to lead to reduced immune recognition by NOD1 (Wang et al., 2016), thereby being an immune evasion strategy that enables mycobacteria to survive inside host macrophages during TB infection (Marques, 2020), being in conformity with our results.
On the other hand, previous studies revealed that N-glycolylated muropeptides are more stimulatory than N-acetylated muropeptides at activating NMHR (Coulombe et al., 2009). Concurring with these observations, other researchers have observed the same while reporting no differences in CFUs/mL between Mtb WT and ΔnamH strains (Hansen et al., 2014). These observations imply that the N-glycolylation of muramic acid is associated with higher immune recognition, which could lead to decreased intracellular survival. However, our results are not concordant with these observations, perhaps because our infection assays were performed with saprophytic M. smegmatis instead of pathogenic Mtb. Nevertheless, our results showed that the N-glycolylation of PG clearly contributes to the increased intracellular survival of M. smegmatis in J774 macrophages. In accordance with our results, a previous publication demonstrated that N-glycolylated muropeptides provoke a reduced NOD2 activation when compared to their N-acetylated counterparts (Wang et al., 2016). Therefore, the role of NamH in the context of immune recognition and host-pathogen interactions remains unclear.
Our results also demonstrated that the triple mutants underwent a faster clearance by J774 macrophages when compared to the single or double mutants. Hence, simultaneously depleting the activity of NamH and of the MurT/GatD protein complex might be a good strategy to facilitate bacilli clearance during TB infection, further underlining the potential of these proteins as novel anti-TB therapeutic targets.
The high prevalence of antibiotic resistance in the TB-therapy setting urges the discovery of conserved drug targets, that is, targets which do not undergo mutation due to the selective pressure associated with the current TB therapeutic schemes. By analyzing the largest dataset of whole genome sequencing data and associated beta-lactam MICs against clinical strains of Mtb to date, our results have found murT, gatD and namH to be highly conserved, implying crucial and unique roles of these genes in the fitness of mycobacteria. Despite being often overlooked, synonymous mutations can lead to changes in the modulation of transcription and translation, thereby resulting in altered protein levels (Bailey et al., 2021). We uncovered one synonymous mutation in gatD (4158493 snp C>T) associated with increased resistance to AMX+CLA and MEM in 10/172 strains. Our results are in conformity with previous reports (Tsolaki et al., 2004; Maitra et al., 2021) and further emphasize the potential of MurT/GatD and NamH as novel anti-TB targets.
Our work is not without limitations. First, CRISPRi is known to cause either reverse or downstream polarity effects in operonic genes (Qi et al., 2013; Choudhary et al., 2015). However, the fact that operonic genes are usually co-transcribed, performing similar molecular functions, somewhat alleviates this problem (Singh et al., 2016; Rock et al., 2017; Bosch et al., 2021). Moreover, the double and triple mutants were constructed in previously modified strains based on theoretical assumptions. Furthermore, the sample of Mtb clinical strains used for the detection of SNPs associated with differences in drug susceptibility involved the overrepresentation of certain phylogenetic lineages in detriment of others (Olivença et al., 2022b). Although we advocate for the development of small molecules able to inhibit these potential antitubercular targets, we are aware that small molecules are not as effective as CRISPRi at inhibiting all enzyme functions (Bosch et al., 2021). At last, substantial differences between M. smegmatis and the pathogen exist and inferred conclusions with the surrogate strain may sometimes not apply to Mtb.
To the best of our knowledge, this is the first study that attempted to simultaneously deplete the activity of MurT/GatD and NamH and phenotypically characterize their conjoint relevance in the modulation of beta-lactam susceptibility and of host-pathogen interactions in mycobacteria. In conclusion, the characteristic modifications of mycobacterial PG were found to promote antibiotic resistance and intracellular survival inside host macrophages, contributing to limited antimicrobial effects and immune evasion. Inhibiting these PG modifications, whether individually or simultaneously, may arise as a strategy to fight TB disease and reduce the duration of TB therapeutic regimens in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Author contributions
CS, MM and MJC conceived and designed the study. CS, MM, DM, and FO performed the experiments. CS, MM, FO, DP, AN, MP, EA and MC analyzed the data. CS wrote the original draft. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors would like to thank everyone who contributed to the writing of this manuscript. We would especially like to thank ADEIM-FFUL (Associação para o Desenvolvimento do Ensino e Investigação em Microbiologia) for financial support. Moreover, we would also like to thank Dr. Martin S. Pavelka, Jr. for supplying the M. smegmatis strains PM965 and PM979 to be used in this study and for providing relevant information about them.
Funding Statement
This work was supported by Fundação para a Ciência e Tecnologia (PTDC/BIA-MIC/31233/2017 to MJC, SFRH/BD/136853/2018 to FO and 2021.05446.BD to CS) and by the European Society of Clinical Microbiology and Infectious Diseases (Research Grant 2018 to MC).
Footnotes
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1089911/full#supplementary-material
References
- Anes E., Peyron P., Staali L., Jordao L., Gutierrez M. G., Kress H., et al. (2006). Dynamic life and death interactions between Mycobacterium smegmatis and J774 macrophages. Cell. Microbiol. 8, 939–960. doi: 10.1111/j.1462-5822.2005.00675.x [DOI] [PubMed] [Google Scholar]
- Bailey S. F., Alonso Morales L. A., Kassen R. (2021). Effects of synonymous mutations beyond codon bias: The evidence for adaptive synonymous substitutions from microbial evolution experiments. Genome Biol. Evol. 13, 1–18. doi: 10.1093/gbe/evab141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowski C., Welsh M. A., Sham L.-T., Eskandarian H. A., Lim H. C., Kieser K. J., et al. (2018). Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. eLife 7, 1–24. doi: 10.7554/eLife.37516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-cas system. Nucleic Acids Res. 41, 7429–7437. doi: 10.1093/nar/gkt520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B., DeJesus M. A., Poulton N. C., Zhang W., Engelhart C. A., Zaveri A., et al. (2021). Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis . Cell 184, 4579–4592.e24. doi: 10.1016/j.cell.2021.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalão M. J., Filipe S. R., Pimentel M. (2019). Revisiting anti-tuberculosis therapeutic strategies that target the peptidoglycan structure and synthesis. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary E., Thakur P., Pareek M., Agarwal N. (2015). Gene silencing by CRISPR interference in mycobacteria. Nat. Commun. 6, 62-67. doi: 10.1038/ncomms7267 [DOI] [PubMed] [Google Scholar]
- Cohen K. A., El-Hay T., Wyres K. L., Weissbrod O., Munsamy V., Yanover C., et al. (2016). Paradoxical hypersusceptibility of drug-resistant Mycobacterium tuberculosis to β-lactam antibiotics. EBioMedicine 9, 170–179. doi: 10.1016/j.ebiom.2016.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordillot M., Dubée V., Triboulet S., Dubost L., Marie A., Hugonnet J.-E., et al. (2013). In vitro cross-linking of Mycobacterium tuberculosis peptidoglycan by l, d-transpeptidases and inactivation of these enzymes by carbapenems. Antimicrobial Agents Chemother. 57, 5940–5945. doi: 10.1128/AAC.01663-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe F., Divangahi M., Veyrier F., De Léséleuc L., Gleason J. L., Yang Y., et al. (2009). Increased NOD2-mediated recognition of n-glycolyl muramyl dipeptide. J. Exp. Med. 206, 1709–1716. doi: 10.1084/jem.20081779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus M. A., Gerrick E. R., Xu W., Park S. W., Long J. E., Boutte C. C., et al. (2017). Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8, 1–17. doi: 10.1128/mBio.02133-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo S., Alffenaar J. W., Sotgiu G., Centis R., D’Ambrosio L., Tiberi S., et al. (2013). Efficacy and safety of meropenem–clavulanate added to linezolid-containing regimens in the treatment of MDR-/XDR-TB. Eur. Respir. J. 41, 1386–1392. doi: 10.1183/09031936.00124312 [DOI] [PubMed] [Google Scholar]
- Doudna J. A., Charpentier E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. doi: 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Dubée V., Triboulet S., Mainardi J. L., Ethève-Quelquejeu M., Gutmann L., Marie A., et al. (2012). Inactivation of Mycobacterium tuberculosis l, d-transpeptidase ldt Mt1 by carbapenems and cephalosporins. Antimicrobial Agents Chemother. 56, 4189–4195. doi: 10.1128/AAC.00665-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S., Guo X. V., Hickey C. M., Ryou M., Monteleone M., Riley L. W., et al. (2005). Controlling gene expression in mycobacteria with anhydrotetracycline and tet repressor. Nucleic Acids Res. 33, e21–e21. doi: 10.1093/nar/gni013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- England K., Boshoff H. I. M., Arora K., Weiner D., Dayao E., Schimel D., et al. (2012). Meropenem-clavulanic acid shows activity against Mycobacterium tuberculosis in vivo . Antimicrobial Agents Chemother. 56, 3384–3387. doi: 10.1128/AAC.05690-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo T. A., Ludovice A. M., Sobral R. G. (2014). Contribution of peptidoglycan amidation to beta-lactam and lysozyme resistance in different genetic lineages of Staphylococcus aureus . Microbial Drug Resistance 20, 238–249. doi: 10.1089/mdr.2014.0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo T. A., Sobral R. G., Ludovice A. M., de Almeida J. M. F., Bui N. K., Vollmer W., et al. (2012). Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus . PloS Pathog. 8, e1002508. doi: 10.1371/journal.ppat.1002508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A. R., Parsons L. M., Pavelka M. S. (2005). Genetic analysis of the β-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to β-lactam antibiotics. Microbiology 151, 521–532. doi: 10.1099/mic.0.27629-0 [DOI] [PubMed] [Google Scholar]
- Gonçalves B. V., Portela R., Lobo R., Figueiredo T. A., Grilo I. R., Ludovice A. M., et al. (2019). Role of MurT c-terminal domain in the amidation of Staphylococcus aureus peptidoglycan. Antimicrobial Agents Chemother. 63 , e00957-19. doi: 10.1128/AAC.00957-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goude R., Parish T. (2009). “Electroporation of mycobacteria,” in Methods in molecular biology, vol. 465. (Totowa, NJ: Humana Press; ), 203–215. [DOI] [PubMed] [Google Scholar]
- Griffin J. E., Gawronski J. D., DeJesus M. A., Ioerger T. R., Akerley B. J., Sassetti C. M. (2011). High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PloS Pathog. 7, e1002251. doi: 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. M., Golchin S. A., Veyrier F. J., Domenech P., Boneca I. G., Azad A. K., et al. (2014). N-glycolylated peptidoglycan contributes to the immunogenicity but not pathogenicity of Mycobacterium tuberculosis . J. Infect. Dis. 209, 1045–1054. doi: 10.1093/infdis/jit622 [DOI] [PubMed] [Google Scholar]
- Hugonnet J.-E., Blanchard J. S. (2007). Irreversible inhibition of the Mycobacterium tuberculosis β-lactamase by clavulanate. Biochemistry 46, 11998–12004. doi: 10.1021/bi701506h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet J.-E., Tremblay L. W., Boshoff H. I., Barry C. E., Blanchard J. S. (2009). Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis . Science 323, 1215–1218. doi: 10.1126/science.1167498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankute M., Cox J. A. G., Harrison J., Besra G. S. (2015). Assembly of the mycobacterial cell wall. Annu. Rev. Microbiol. 69, 405–423. doi: 10.1146/annurev-micro-091014-104121 [DOI] [PubMed] [Google Scholar]
- Jordão L., Bleck C. K. E., Mayorga L., Griffiths G., Anes E. (2008). On the killing of mycobacteria by macrophages 10, 529–548. doi: 10.1111/j.1462-5822.2007.01067.x [DOI] [PubMed] [Google Scholar]
- Kieser K. J., Baranowski C., Chao M. C., Long J. E., Sassetti C. M., Waldor M. K., et al. (2015). Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proc. Natl. Acad. Sci. United States America 112, 13087–13092. doi: 10.1073/pnas.1514135112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Arora K., Lloyd J. R., Lee I. Y., Nair V., Fischer E., et al. (2012). Meropenem inhibits d, d-carboxypeptidase activity in Mycobacterium tuberculosis . Mol. Microbiol. 86, 367–381. doi: 10.1111/j.1365-2958.2012.08199.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M. H., Gilbert L. A., Wang X., Lim W. A., Weissman J. S., Qi L. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8, 2180–2196. doi: 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavollay M., Arthur M., Fourgeaud M., Dubost L., Marie A., Veziris N., et al. (2008). The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l, d -transpeptidation. J. Bacteriol. 190, 4360–4366. doi: 10.1128/JB.00239-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisico F., Vieira V., Figueiredo T. A., Silva M., Cabrita E. J., Sobral R. G., et al. (2018). First insights of peptidoglycan amidation in gram-positive bacteria - the high-resolution crystal structure of Staphylococcus aureus glutamine amidotransferase GatD. Sci. Rep. 8, 5313. doi: 10.1038/s41598-018-22986-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra S., Scherman H., Brennan P. J., Crick D. C. (2005). N glycolylation of the nucleotide precursors of peptidoglycan biosynthesis of Mycobacterium spp. is altered by drug treatment. J. Bacteriol. 187, 2341–2347. doi: 10.1128/JB.187.7.2341-2347.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A., Munshi T., Healy J., Martin L. T., Vollmer W., Keep N. H., et al. (2019). Cell wall peptidoglycan in Mycobacterium tuberculosis: An achilles’ heel for the TB-causing pathogen. FEMS Microbiol. Rev. 43, 548–575. doi: 10.1093/femsre/fuz016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A., Nukala S., Dickman R., Martin L. T., Munshi T., Gupta A., et al. (2021). Characterization of the MurT/GatD complex in Mycobacterium tuberculosis towards validating a novel anti-tubercular drug target. JAC-Antimicrobial Resistance 3, 1–10. doi: 10.1093/jacamr/dlab028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques M. (2020). Significance of mycobacterial peptidoglycan amidation in host immune recognition and antibiotic resistance. master’s dissertation. Portugal (Lisboa) faculty of pharmacy, universidade de lisboa. [Google Scholar]
- Mojica F. J. M., Díez-Villaseñor C., García-Martínez J., Almendros C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. doi: 10.1099/mic.0.023960-0 [DOI] [PubMed] [Google Scholar]
- Morlot C., Straume D., Peters K., Hegnar O. A., Simon N., Villard A.-M., et al. (2018). Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae . Nat. Commun. 9, 3180. doi: 10.1038/s41467-018-05602-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch D., Roemer T., Lee S. H., Engeser M., Sahl H. G., Schneider T. (2012). Identification and in vitro analysis of the GatD/MurT enzyme-complex catalyzing lipid II amidation in Staphylococcus aureus . PloS Pathog. 8, e1002509. doi: 10.1371/journal.ppat.1002509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivença F., Ferreira C., Nunes A., Silveiro C., Pimentel M., Gomes J. P., et al. (2022. a). Identification of drivers of mycobacterial resistance to peptidoglycan synthesis inhibitors. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.985871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivença F., Nunes A., Macedo R., Pires D., Silveiro C., Anes E., et al. (2022. b). Uncovering beta-lactam susceptibility patterns in clinical isolates of Mycobacterium tuberculosis through whole-genome sequencing. Microbiol. Spectr. 10 , e0067422 doi: 10.1128/spectrum.00674-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M., Colavin A., Shi H., Czarny T. L., Larson M. H., Wong S., et al. (2016). A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165, 1493–1506. doi: 10.1016/j.cell.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidgeon S. E., Apostolos A. J., Nelson J. M., Shaku M., Rimal B., Islam M. N., et al. (2019). L, d-transpeptidase specific probe reveals spatial activity of peptidoglycan cross-linking. ACS Chem. Biol. 14, acschembio.9b00427. doi: 10.1021/acschembio.9b00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D., Bernard E. M., Pombo J. P., Carmo N., Fialho C., Gutierrez M. G., et al. (2017). Mycobacterium tuberculosis modulates miR-106b-5p to control cathepsin s expression resulting in higher pathogen survival and poor T-cell activation. Front. Immunol. 8. doi: 10.3389/fimmu.2017.01819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J. B., Mahapatra S., Crick D. C., Pavelka M. S. (2005). Identification of the namH gene, encoding the hydroxylase responsible for the n-glycolylation of the mycobacterial peptidoglycan. J. Biol. Chem. 280, 326–333. doi: 10.1074/jbc.M411006200 [DOI] [PubMed] [Google Scholar]
- Rock J. M., Hopkins F. F., Chavez A., Diallo M., Chase M. R., Gerrick E. R., et al. (2017). Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat. Microbiol. 2, 1–9. doi: 10.1038/nmicrobiol.2016.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C. M., Boyd D. H., Rubin E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84. doi: 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- Silveiro C. (2020). Role of n -glycolylation of the mycobacterial peptidoglycan in host immune recognition and antibiotic resistance. master’s dissertation. Portugal (Lisboa), faculty of pharmacy, universidade de lisboa. [Google Scholar]
- Singh A. K., Carette X., Potluri L.-P., Sharp J. D., Xu R., Prisic S., et al. (2016). Investigating essential gene function in Mycobacterium tuberculosis using an efficient CRISPR interference system. Nucleic Acids Res. 44, e143–e143. doi: 10.1093/nar/gkw625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Chibale K. (2021). Strategies to combat multi-drug resistance in tuberculosis. Accounts Chem. Res. 54, 2361–2376. doi: 10.1021/acs.accounts.0c00878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper S. B., Lugosi L., Jekkel A., Melton R. E., Kieser T., Bloom B. R., et al. (1988). Lysogeny and transformation in mycobacteria: stable expression of foreign genes. Proc. Natl. Acad. Sci. 85, 6987–6991. doi: 10.1073/pnas.85.18.6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solapure S., Dinesh N., Shandil R., Ramachandran V., Sharma S., Bhattacharjee D., et al. (2013). In vitro and In vivo efficacy of β-lactams against replicating and slowly Growing/Nonreplicating Mycobacterium tuberculosis . Antimicrobial Agents Chemother. 57, 2506–2510. doi: 10.1128/AAC.00023-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story-Roller E., Lamichhane G. (2018). Have we realized the full potential of β-lactams for treating drug-resistant TB? IUBMB Life 70, 881–888. doi: 10.1002/iub.1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay L. W., Fan F., Blanchard J. S. (2010). Biochemical and structural characterization of Mycobacterium tuberculosis β-lactamase with the carbapenems ertapenem and doripenem. Biochemistry 49, 3766–3773. doi: 10.1021/bi100232q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolaki A. G., Hirsh A. E., DeRiemer K., Enciso J. A., Wong M. Z., Hannan M., et al. (2004). Functional and evolutionary genomics of Mycobacterium tuberculosis: Insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. United States America 101, 4865–4870. doi: 10.1073/pnas.0305634101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Matsuo Y., Pradipta A. R., Inohara N., Fujimoto Y., Fukase K. (2016). Synthesis of characteristic Mycobacterium peptidoglycan (PGN) fragments utilizing with chemoenzymatic preparation of meso-diaminopimelic acid (DAP), and their modulation of innate immune responses. Organic Biomol. Chem. 14, 1013–1023. doi: 10.1039/C5OB02145F [DOI] [PubMed] [Google Scholar]
- WHO (2020) WHO consolidated guidelines on tuberculosis. module 4: treatment - drug-resistant tuberculosis treatment. Available at: https://www.who.int/publications/i/item/9789240007048 (Accessed September 2, 2022). [PubMed]
- WHO (2021). Global tuberculosis report 2021. Global tuberculosis report 2021. World Health Organization. Available at: https://apps.who.int/iris/handle/10665/346387. [Google Scholar]
- Wivagg C. N., Bhattacharyya R. P., Hung D. T. (2014). Mechanisms of β-lactam killing and resistance in the context of Mycobacterium tuberculosis . J. Antibiotics 67, 645–654. doi: 10.1038/ja.2014.94 [DOI] [PubMed] [Google Scholar]
- Wong A. I., Rock J. M. (2021). CRISPR interference (CRISPRi) for targeted gene silencing in mycobacteria. Methods Mol. Biol. (Clifton N.J.) 2314, 343–364. doi: 10.1007/978-1-0716-1460-0_16 [DOI] [PubMed] [Google Scholar]
- Zapun A., Philippe J., Abrahams K. A., Signor L., Roper D. I., Breukink E., et al. (2013). In vitro reconstitution of peptidoglycan assembly from the gram-positive pathogen Streptococcus pneumoniae . ACS Chem. Biol. 8, 2688–2696. doi: 10.1021/cb400575t [DOI] [PubMed] [Google Scholar]
- Zhang D., Wang Y., Lu J., Pang Y. (2016). In vitro activity of β-lactams in combination with β-lactamase inhibitors against multidrug-resistant Mycobacterium tuberculosis isolates. Antimicrobial Agents Chemother. 60, 393–399. doi: 10.1128/AAC.01035-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .