Abstract
The current artificial intelligence (AI) models are still insufficient in multi-disease diagnosis for real-world data, which always present a long-tail distribution. To tackle this issue, a long-tail public dataset, “ChestX-ray14,” which involved fourteen (14) disease labels, was randomly divided into the train, validation, and test sets with ratios of 0.7, 0.1, and 0.2. Two pretrained state-of-the-art networks, EfficientNet-b5 and CoAtNet-0-rw, were chosen as the backbones. After the fully-connected layer, a final layer of 14 sigmoid activation units was added to output each disease’s diagnosis. To achieve better adaptive learning, a novel loss (Lours) was designed, which coalesced reweighting and tail sample focus. For comparison, a pretrained ResNet50 network with weighted binary cross-entropy loss (LWBCE) was used as a baseline, which showed the best performance in a previous study. The overall and individual areas under the receiver operating curve (AUROC) for each disease label were evaluated and compared among different models. Group-score-weighted class activation mapping (Group-CAM) is applied for visual interpretations. As a result, the pretrained CoAtNet-0-rw + Lours showed the best overall AUROC of 0.842, significantly higher than ResNet50 + LWBCE (AUROC: 0.811, p = 0.037). Group-CAM presented that the model could pay the proper attention to lesions for most disease labels (e.g., atelectasis, edema, effusion) but wrong attention for the other labels, such as pneumothorax; meanwhile, mislabeling of the dataset was found. Overall, this study presented an advanced AI diagnostic model achieving a significant improvement in the multi-disease diagnosis of chest X-rays, particularly in real-world data with challenging long-tail distributions.
Keywords: Chest X-ray, Lung diseases, Differential diagnoses, Artificial intelligence, Transformer
Introduction
Chest X-ray is still the most commonly used modality for the diagnosis of various thoracic diseases. It is economical and inexpensive, and the equipment is easy to install. Specifically, chest X-ray is an excellent choice to be widely applied in developing or resource-poor areas of the world, where radiology services are highly insufficient. Since the global coronavirus disease-19 (COVID-19) outbreak in 2020, chest X-ray has become a critical imaging application for disease screening worldwide [1]. However, with the surge in chest X-rays during the pandemic, there was a massive increase in imaging data, dramatically overloading frontier radiologists. Driven by this medical demand, many artificial intelligence (AI) diagnostic models, such as convolutional neural networks (CNNs), have been established, which played an essential role in combatting the pandemic [2–4]. They presented good performances in COVID-19 detection, which are even comparable with radiologists [2–4].
However, these AI-based diagnostic models generally have two shortcomings in clinical practice: 1. Lack of independent multi-label classification capabilities. Although most AI models perform well in the diagnosis of a single disease or lesion (e.g., pneumonia or not, with or without lung nodule), real-world imaging diagnosis is usually a multi-label classification task, or so-called “One Check, Many Findings” [5, 6]. Coexisting diseases are more common in real-world scenarios; for example, a typical chest X-ray can reveal more than one disease (e.g., pulmonary infiltration, cardiomegaly). However, multi-disease diagnosis of chest X-rays can be a challenging task for AI models due to the more complex patterns that may be present in the images. Therefore, some solutions can be considered: (1) Combination of multiple pre-trained models for single-disease diagnosis, which is mostly applied by many AI platforms to achieve an apocryphal multi-disease diagnosis, demanding obviously increased computing resources; (2) establishment of an independent multi-label AI diagnostic model by applying state-of-the-art deep learning methods which can effectively reduce computing consumption and accelerate the diagnosis speed. 2. Multi-label long-tail distribution issue. The real-world image samples usually present a long-tailed distribution. Typical negative samples (no findings) constitute the majority of the head category; in contrast, most disease samples fall into the tail categories and can only be collected in a small amount [7–9]. This imbalance makes the model training seriously overfit the negative samples and ignores the disease features in positive samples, leading to useless training. Moreover, because this data imbalance varies among different labels, it further increases the difficulty of accurate classification. That’s why the manual data inclusion or sharing of tailed data to ensure the intra- and inter-class balances is mainly applied in radiological model training [5, 9–11]. However, while a balanced distribution of disease classes may be beneficial for model training, it may not accurately reflect real-world data distribution and compromise the model's generalization to subpopulations [6, 12].
Hence, utilizing a dataset that accurately represents the real-world distribution of diseases, even if it results in class imbalance and multi-disease diagnosis, could offer greater benefits. Suppose effective solutions can be found for the challenges of multi-label classification and long-tailed distribution; in that case, an optimal multi-label diagnostic model for chest X-rays can be established, enabling radiologists to make more accurate diagnoses and improve examination efficiency globally. So far, most studies have focused on AI diagnosis with only 3 to 6 multi-label categories [7]. Although two previous studies explored multi-label classification of 8 and 13 diseases, both presented limited performance of CNNs, with the lowest area under the receiver operating curve (AUROC) of only 0.6 [7, 8]. In this study, we aim to achieve a fourteen-disease classification in a long-tail dataset of chest X-rays, which has rarely been attempted. In order to promote the AI diagnostic performance with increased labels, we adopted three strategies: first, improve algorithms (e.g., self-attention, channel attention) to strengthen learning ability [13, 14]; second, choose or design an appropriate loss (e.g., reweighting, focal loss) to make the learning focus more on the tailed and hard samples [8, 15]; third, using various tricks to promote model convergence and prevent overfitting (e.g., transfer learning, data augmentation) [7, 16, 17].
Methods
Dataset
The enhanced-version ChestX-ray14 public dataset (Link: https://www.kaggle.com/datasets/nih-chest-xrays/data, National Institutes of Health Clinical Center, Bethesda, USA.) as a real-world dataset was used in our study because the ultimate goal of this study is to train a model that generalizes well to new, unseen data in real-world scenarios. This public dataset has undergone privacy-preserving preprocessing and holds a license of CC0 1.0 Universal (CC0 1.0) Public Domain Dedication, which waives copyright interest in scientific work and is dedicated to the worldwide public domain. This dataset contains 112,120 consecutive frontal-view chest X-rays spanning from 1992 to 2015. It includes 14 disease labels identified through using a variety of Natural Language Processing (NLP) techniques mining from related radiological reports. The spectrum of disease labels had the following: “infiltration”, “atelectasis”, “effusion”, “nodule”, “pneumothorax”, “mass”, “consolidation”, “pleural_thickening”, “cardiomegaly”, “emphysema”, “fibrosis”, “edema”, “pneumonia”, and “hernia”. The whole dataset included 60,361 negative chest X-rays (“No findings”); 20,796 images contain two or more disease labels (range: 2–9). The distribution of all disease labels presents apparent long-tail distribution, with disease proportions from 0.2% (label: hernia) to 17.7% (label: infiltration). We randomly divided the entire dataset into train, validation, and test sets with ratios of 0.7, 0.1, and 0.2 for model training, validation, and testing, respectively. The details of the dataset are shown in Table 1.
Table 1.
Summary of the dataset
|
Train set (70%) (count/total, %) |
Validation set (10%) (count/total, %) |
Test set (20%) (count/total, %) |
Total (count/total, %) |
|
|---|---|---|---|---|
| Labels | ||||
| Infiltration |
13,915/78,484 (17.7%) |
1929/11,211 (17.2%) |
4050/22,425 (18.1%) |
19,894/112,120 (17.7%) |
| Atelectasis |
81,06/78,484 (10.3%) |
1133/11,211 (10.1%) |
2320/22,425 (10.3%) |
11,559/112,120 (10.3%) |
| Effusion |
9401/78,484 (12.0%) |
1315/11,211 (11.7%) |
2601/22,425 (11.6%) |
13,317/11,2120 (11.9%) |
| Nodule |
4392/78,484 (5.6%) |
635/11,211 (5.7%) |
1304/22,425 (5.8%) |
6331/11,2120 (5.6%) |
| Pneumothorax |
3730/78,484 (4.8%) |
520/11,211 (4.6%) |
1052/22,425 (4.7%) |
5302/112,120 (4.7%) |
| Mass |
4016/78,484 (5.1%) |
563/11,211 (5.0%) |
1203/22,425 (5.4%) |
5782/112,120 (5.2%) |
| Consolidation |
3244/78,484 (4.1%) |
480/11,211 (4.3%) |
943/22,425 (4.2%) |
4667/112,120 (4.2%) |
| Pleural_Thickening |
2380/78,484 (3.0%) |
339/11,211 (3.0%) |
666/22,425 (3.0%) |
3385/112,120 (3.0%) |
| Cardiomegaly |
1897/78,484 (2.4%) |
277/11,211 (2.5%) |
602/22,425 (2.7%) |
2776/112,120 (2.5%) |
| Emphysema |
1781/78,484 (2.3%) |
266/11,211 (2.4%) |
469/22,425 (2.1%) |
2516/112,120 (2.2%) |
| Fibrosis |
1204/78,484 (1.5%) |
186/11,211 (1.7%) |
296/22,425 (1.3%) |
1686/112,120 (1.5%) |
| Edema |
1623/78,484 (2.1%) |
244/11,211 (2.2%) |
436/22,425 (1.9%) |
2303/112,120 (2.1%) |
| Pneumonia |
997/78,484 (1.3%) |
128/11,211 (1.1%) |
306/22,425 (1.4%) |
1431/112,120 (1.3%) |
| Hernia |
163/78,484 (0.2%) |
28/11,211 (0.2%) |
36/22,425 (0.2%) |
227/112,120 (0.2%) |
| Count of multi-labels in each image | ||||
| 0 label* |
42,197/78,484 (53.8%) |
6055/11,211 (54.0%) |
12,109/22,425 (54.0%) |
60,361/11,2120 (53.8%) |
| 1 label |
21,768/78,484 (27.7%) |
3103/11,211 (27.7%) |
6092/22,425 (27.2%) |
30,963/112,120 (27.6%) |
| 2 labels |
9993/78,484 (12.7%) |
1427/11,211 (12.7%) |
2886/22,425 (12.9%) |
14,306/112,120 (12.8%) |
| 3 labels |
3361/78,484 (4.3%) |
469/11,211 (4.2%) |
1026/22,425 (4.6%) |
4856/112,120 (4.3%) |
| ≥ 4 labels |
1165/78,484 (1.5%) |
157/11,211 (1.4%) |
312/22,425 (1.4%) |
1634/112,120 (1.5%) |
*0 label indicates “no findings”
Networks and Hyperparameters
Baseline
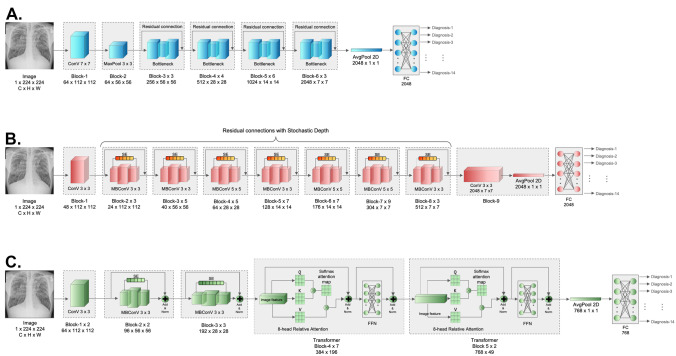
We chose ResNet50 with weighted binary cross-entropy loss () as the baseline, which showed the best performance in the eight-label chest X-ray diagnosis in a previous study [8]. As one of the most used baselines in deep-learning studies, its main contribution is to address the degradation problem. By establishing a “shortcut connection,” or so-called “residual connection,” ResNet allows the original information of the superficial layers to be directly transmitted to the subsequent deeper layers, which eliminates the vanishing gradient issue owing to the excessive depth of the network. In addition, by introducing the Bottleneck structure, ResNet first performs dimensionality reduction through 1 × 1 convolution, followed by a larger kernel convolution, in order to reduce the computational consumption caused by the direct large-kernel convolution process [18]. The ResNet50 adopted in this study comprises 16 Bottleneck residual blocks (Fig. 1A). At the end of the Bottleneck blocks, the output underwent average pooling and flattening; then, the results passed through a fully connected layer (FC) to calculate the diagnosis probability. For the multi-label classifications in our datasets, a final layer of 14 sigmoid activation units was added to output the predicted probability of each disease. To improve the learning in long-tail datasets, was applied following the recommendations from the previous study in the same dataset [8].
Fig. 1.
Schematic diagram of the diagnostic networks in this study. A ResNet50; B EfficientNet-b5; C CoAtNet-0-rw
Study Networks
To enhance the accuracy of multi-disease diagnosis, state-of-the-art (SOTA) deep learning networks can be utilized, incorporating various loss functions and activation functions that address the class imbalance and enhance the interpretability of the models. These networks, such as EffecientNet and CoAtNet, consistently integrate channel attention or Transformer modules to improve their ability to automatically learn the intricate features of image data [19–22]. The channel attention and Transformer modules allow the network to concentrate on the crucial parts of an input image (such as the lungs and mediastinum) and effectively capture complex relationships among different elements, making them particularly beneficial for multi-disease diagnosis in chest X-rays. These SOTA networks have shown exceptional performance in various visual tasks, demonstrating their potential in the field. In this study, we aimed to utilize representative SOTA CNN and CNN + Transformer hybrid architecture networks, including EfficientNet and CoAtNet, which have been widely used for natural image classification tasks in recent years, as our study models [19–22]. These networks have been chosen for their demonstrated efficacy and advanced representation learning abilities, which are crucial in achieving improved multi-disease diagnosis in chest X-rays. Additionally, these networks can be fine-tuned for specific tasks by adjusting the last layers of the network to accommodate the target data, reducing the requirement for extensive reimplementation. To ensure that the parameters between the models are similar, we choose EfficientNet-b5 and CoAtNet-0-rw as our backbones with lightweight designs. The same final layer of 14 sigmoid activation units was added for the multi-label classifications. The architectures of these two networks are elucidated below:
EfficientNet-b5
EfficientNet has been one of the most successful CNNs in recent years [17]. It showed a crushing performance on ImageNet with a spectacular reduction in computing consumption when compared with previous CNNs. Overall, EfficientNet greatly balances the network depth, width, and resolution, leading to an essential breakthrough. In addition to the same residual connection as ResNet, another major contribution of EfficientNet is the joint application of depth-wise separable convolution and the channel attention mechanism named squeeze and excitation (SE) [14, 23]. Without adding too much computation, the depth-wise separable convolution provides a larger number of input and output channels benefiting more feature information extraction [23]. It first increases the number of channels through 1 × 1 convolution; then, the large-kernel convolution is separably performed on each channel, forming the output with the same channel count; finally, the number of channels is reduced to the input size by another 1 × 1 convolution operation, creating an “Inverted Bottleneck” structure. Compared with the conventional convolution operation, the computing consumption is only of the former. As channel attention, SE helps exploit contextual information among different channels [14]. First, the global spatial information is squeezed into a channel descriptor by using the global average pooling; then, after two FC layers with a following sigmoid activation, channel-wise dependencies as a scalar are fully captured; finally, channel-wise multiplication is performed between the scalar and the feature map. Equations (1)–(3) of SE are as follows:
| 1 |
| 2 |
| 3 |
where H, W indicates the height and width of the feature map; is the sigmoid function; suggests the output after two FC layers with the intermediate ReLU activation marked as . The depthwise separable convolution and SE make up a module named MBConV [14, 23]. The EfficientNet-b6 used in this study has a total of 39 MBConV modules. Like ResNet50, the final output passed through a sigmoid activation layer to obtain the multi-label diagnosis probability (Fig. 1B).
CoAtNet-0-rw
Although CNN is still the predominant network in computer vision, Transformer has shown a powerful performance potential since its birth [6]. Compared with CNN, Transformer’s most significant advantage is its larger parameter capacity and global receptive field. On large-scale datasets, Transformer can also achieve the SOTA performance, even better than CNN [21, 22]. However, in datasets with limited sample sizes, such as various medical imaging datasets, CNN still presented a better performance than Transformer owing to its powerful inductive bias capacity [19, 24]. CoAtNet was designed with a CNN + Transformer hybrid architecture that integrates the benefits of local and global receptive fields [19, 21] to combine the advantages of EfficientNet, Transformer, and ResNet. It involves MBConV modules, self-attention, and residual connections. In addition, to better merge CNN and Transformer, the network integrates static convolution kernel parameters in original self-attention equations, also known as relative-attention, achieving three advantages: translation invariance, adaptive input weighting, and global receptive field [19]. The equation of relative-attention (4) is as follows:
| 4 |
where indicates the global spatial space. suggests the position pair. is a trainable scaler that retrieves all static convolutional kernels for all pairs by calculating the pairwise dot product attention. The CoAtNet-0-rw used in this study is a lightweight network with 5 MBConV modules and 9 Transformer modules. After passing through a sigmoid-activation layer, the multi-label diagnosis probabilities are exported (Fig. 1C).
Other hyperparameter settings kept the same (Table 2), including: a batch size of 150 [130 in EfficientNet-b5 owing to the graphics processing unit (GPU) memory limitation), 100 training epochs, an optimizer of Adam, a learning rate (lr) of 5.0e-05. All models were trained on the same cloud GPU platform (gpuhub.com/home). The hardware configuration includes: Nvidia 3090 24G GPU*4, a 60-core Intel(R) Xeon(R) Platinum 8358P central processing unit (CPU), and 360G random access memory (RAM). The training process was carried out using PyTorch distributed parallel computing. All codes have been released on the link: https://github.com/KiwisFraggle/CoAtNet_NIH.
Table 2.
Comparisons of different models in our study
| Models | Params | FLOPs |
Resolution (C*H*W) |
Batch size | Epochs | Optimizer | lr |
|---|---|---|---|---|---|---|---|
| ResNet-50 | 26 M | 4112 M | 1*244*244 | 150 | 100 | Adam | 5.0e − 05 |
| EfficientNet-b5 | 30 M | 2413 M | 1*244*244 | 130* | 100 | Adam | 5.0e − 05 |
| CoAtNet-0-rw | 27 M | 4215 M | 1*244*244 | 150 | 100 | Adam | 5.0e − 05 |
FLOPs floating-point operations, lr learning rate
*Reduced batch size owing to the GPU memory limitation
Loss
This study intended to use two loss strategies (Fig. 2) for the backpropagation:
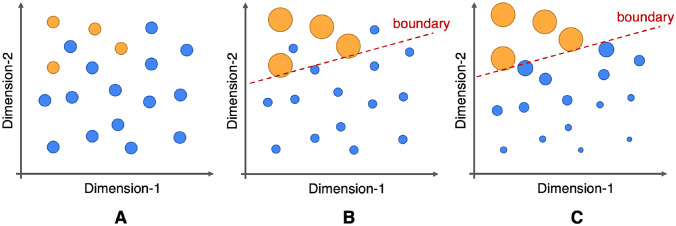
Fig. 2.
The impact of long-tail data distribution on classification and the proposed solution for this study. A For the tail sample (yellow dots), it is difficult for the model to learn the valid classification when using classic binary cross-entropy loss. B increases the weight of tail samples and reduces the weight of head samples (blue dots) through reweighting, which can effectively enhance the learning of tailed categories. C Our design loss () simultaneously increases the weight of tailed data and reduces the contribution of easy head samples, which may help to improve the classification ability of the model when training on a long-tail dataset
Weighted Binary Cross-Entropy Loss
In the multi-label classification task of our datasets, LWBCE was feasible to adjust the long-tail distribution to “rebalance” and to promote the learning of the tailed data [8]. Besides, it holds flexibility when facing different long-tail distributions among various labels. Therefore, the corresponding rebalancing weights should be adjusted for each label to obtain an optimal multi-label diagnosis effect [8]. The specific formula of (7–9) is as follows:
| 5 |
| 6 |
| 7 |
where and indicate the positive label and positive prediction probability calculated from the sigmoid of the output , respectively; was the weight calculated by the positive sample count over the negative sample count; and indicate the label class and sample sequence, respectively.
Our Designed Loss
Considering the lack of hard-sample classification capacity, we designed a novel reweighted loss in the base of , by additionally joining an exponential decay factor of easy negative samples and a nonlinear shifting probability for reducing the contribution of negative samples, which probably makes the training focusing on not only the positive but hard samples [8, 15, 25]. Our loss keeps the capacity to make accurate individual adjustments for each label distribution with different imbalance levels. The specific formula (8–12) is as follows:
| 8 |
| 9 |
| 10 |
| 11 |
| 12 |
Similar to LWBCE, indicates the prediction probability calculated by sigmoid of output , and is the weight calculated by the negative sample count over the positive sample count. is the balanced coefficient to adjust the initial balance at the start of the training, which was set as 0.2 in our study; is an exponential modulating factor on negative samples, which was set as 4.0; a value of 0.05 was set as the nonlinear shifting probability.
Other Training Tricks
In previous attempts, we encountered the issue of non-convergence in model training, which was also met in an earlier study [8]. To address this issue and strengthen the model’s robustness, we applied several additional tricks. First, previous studies suggested that transfer learning with joint data augmentation could improve model accuracy and generalization for multi-label classification tasks [7, 8]. Therefore, we employed the pre-trained weights obtained from ImageNet training as the initialization. Second, we adopted an autoaugment policy, which involves an optimal augmentation strategy established from forced learning [17]. This policy compiles 25 augmentations, including random rotation, shear, sharpness, etc. It can significantly improve the model accuracy and decrease the error rate in various datasets [17]. Moreover, random horizontal flip and patch erasing are additionally applied. Third, we use a learning rate scheduler, OneCycleLR, consisting of 20 epochs spent increasing the lr like a warmup and following 80 epochs with the cosine decay. It can prevent a trap at a local minimum in the training process [26]. Fourth, the parameter weight decay and dropout before the final FC layer was additionally applied to inhibit overfitting in training with a λ value of 0.01 and a dropout probability of 0.5 [27, 28]. At last, the Exponential Moving Average (EMA) was performed with a decay value of 0.9997 to improve the accuracy and robustness of the model [29]. Automatic mixed precision training was applied to reduce memory consumption and accelerate the training process [30].
Performance Evaluations and Statistical Analysis
The statistical analysis was conducted using IBM SPSS Statistics Software (version 26, IBM, New York, USA). Quantitative data were presented as mean ± standard error with a 95% confidence interval. Because the dataset is highly imbalanced, the epoch with the increased accuracy can be achieved simply by identifying samples as the head class (“No findings”), even when the loss persistently elevates. Thus, determining the best model in this study does not rely on maximizing classification accuracy but on minimizing the loss in the validation set. This model selection strategy can also overcome overfitting [31]. To compare the performance of different models, we evaluated the overall and individual AUROC, accuracy, macro precision, macro recall, and macro F1-score, which were compared between different models or labels using repeated measures Analysis Of Variance (ANOVA) tests and AUROC comparison analysis with Bonferroni adjustments [7, 8]. Besides, to explore whether there is any relationship between the classification capacity of the model and positive sample size, the Pearson correlation tests were performed involving AUROC and positive sample ratio. A two-tailed p value less than 0.05 was considered statistically significant.
Lesion Localization and Visual Interpretations
When the model training is completed, we select the model with the highest overall AUROC value; then, we use the group-score-weighted class activation mapping (Group-CAM) to localize the lesions and help visual interpretations. Compared with the commonly used randomized input sampling for explanation (RISE) or gradient-weighted class activation mapping (Grad-CAM), Group-CAM is more convincing and less noisy [32].
Results
Comparisons of AUROCs among different models were summarized in Table 3. After adding multiple tricks as mentioned above, ResNet50 + LWBCE showed a significantly higher AUROC on multi-label classification than the result in a previous study (p = 0.006); in particular, the AUROC of the “Mass” label increased from the reported 0.561 to 0.819 [8]. Second, both SOTA networks, including CoAtNet-0-rw and EfficientNet-b5, presented higher overall AUROCs than ResNet50 (0.826/0.822 vs. 0.811, respectively) when using the same LWBCE, but without significant differences. After applying Lours, both CoAtNet-0-rw and EfficientNet-b5 achieved significantly higher AUROCs than ResNet50 + LWBCE and ResNet50 + Lours (p ≤ 0.037, each), while CoAtNet-0-rw + Lours presented the highest overall AUROC of 0.842. However, different losses rarely affected the performance of ResNet50, unlike the cases of CoAtNet-0-rw and EfficientNet-b5. In addition, the AUROC didn’t show any significant correlations with the positive sample ratio of the label, no matter which model was applied (p > 0.05).
Table 3.
Comparisons of AUROCs among different models
| CoAtNet-0-rw | EfficientNet-b5 | Baseline (ResNet50) | Baseline (ResNet50) | ||||
|---|---|---|---|---|---|---|---|
| Labels | LWBCE | Lours | LWBCE | Lours | LWBCE | Lours | LWBCE – Previous study [8]# |
| AUROC | AUROC | AUROC | AUROC | AUROC | AUROC | AUROC | |
| Atelectasis | 0.795 | 0.833 | 0.795 | 0.812 | 0.776 | 0.791 | 0.707 |
| Cardiomegaly | 0.901 | 0.914 | 0.906 | 0.905 | 0.908 | 0.890 | 0.814 |
| Consolidation | 0.787 | 0.809 | 0.798 | 0.802 | 0.786 | 0.788 | |
| Edema | 0.903 | 0.912 | 0.905 | 0.908 | 0.898 | 0.899 | |
| Effusion | 0.874 | 0.890 | 0.872 | 0.882 | 0.867 | 0.878 | 0.736 |
| Emphysema | 0.927 | 0.940 | 0.906 | 0.912 | 0.872 | 0.871 | |
| Fibrosis | 0.826 | 0.835 | 0.805 | 0.807 | 0.790 | 0.788 | |
| Hernia | 0.865 | 0.791 | 0.826 | 0.837 | 0.847 | 0.768 | |
| Infiltration | 0.686 | 0.715 | 0.699 | 0.712 | 0.688 | 0.705 | 0.613 |
| Mass | 0.827 | 0.856 | 0.827 | 0.832 | 0.819 | 0.823 | 0.561 |
| Nodule | 0.748 | 0.779 | 0.745 | 0.755 | 0.722 | 0.718 | 0.716 |
| Pleural_Thickening | 0.804 | 0.819 | 0.810 | 0.810 | 0.789 | 0.790 | |
| Pneumonia | 0.751 | 0.789 | 0.736 | 0.774 | 0.733 | 0.733 | 0.633 |
| Pneumothorax | 0.870 | 0.902 | 0.878 | 0.886 | 0.857 | 0.859 | 0.789 |
| Overall* | 0.826 ± 0.018 (0.786, 0.866) | 0.842 ± 0.017 (0.805, 0.879)a | 0.822 ± 0.018 (0.784, 0.860) | 0.831 ± 0.016 (0.796, 0.866)b | 0.811 ± 0.018 (0.772, 0.850) | 0.807 ± 0.017 (0.770, 0.844) | 0.696 ± 0.031 (0.623, 0.769)c |
| Pearson correlation with the positive sample ratiod in the test set | −0.517 | −0.385 | −0.421 | −0.432 | −0.442 | −0.269 | −0.260 |
| p value | 0.058 | 0.174 | 0.134 | 0.123 | 0.113 | 0.352 | 0.534 |
aThere are significant differences when compared with ResNet50 + LWBCE (p = 0.037), ResNet50 + Lours (p < 0.001), and results of ResNet50 + LWBCE in previous studies (p = 0.017)
bThere are significant differences when compared with ResNet50 + LWBCE (p = 0.004), ResNet50 + Lours (p = 0.004), and results of ResNet50 + LWBCE in previous studies (p = 0.028)
cThere are significant differences compared to the other six models (p ≤ 0.006)
dThe positive sample ratio was calculated by the count of positive samples over the total sample size. All statistical analyses were performed using repeated measures ANOVA tests with Bonferroni adjustments
#In this previous study, only 8 labels were involved
*Data were presented as mean ± standard error (95% confidence interval)
In addition, CoAtNet-0-rw + Lours shows the highest overall accuracy (0.257), macro precision (0.57), macro recall (0.76), and macro F1-score (0.57) when compared with other models. Similar to the comparison results of AUROCs among different models, the macro F1-score of CoAtNet-0-rw + Lours was significantly higher than ResNet50 + LWBCE and ResNet50 + Lours (p = 0.010 and 0.002, respectively). Besides, the macro F1-scores of EfficientNet-b5 + Lours and CoAtNet-0-rw + LWBCE were also significantly higher than baseline ResNet50 + LWBCE (p = 0.041 and 0.002, respectively). The details are summarized in Table 4.
Table 4.
Other performance evaluations among different models
| CoAtNet-0-rw | EfficientNet-b5 | Baseline (ResNet50) | ||||
|---|---|---|---|---|---|---|
| Labels | LWBCE | Lours | LWBCE | Lours | LWBCE | Lours |
| Accuracy | Accuracy | Accuracy | Accuracy | Accuracy | Accuracy | |
| Atelectasis | 0.689 | 0.772 | 0.731 | 0.714 | 0.650 | 0.740 |
| Cardiomegaly | 0.812 | 0.840 | 0.745 | 0.839 | 0.875 | 0.855 |
| Consolidation | 0.673 | 0.769 | 0.730 | 0.719 | 0.597 | 0.654 |
| Edema | 0.797 | 0.824 | 0.820 | 0.822 | 0.761 | 0.738 |
| Effusion | 0.746 | 0.753 | 0.768 | 0.810 | 0.724 | 0.787 |
| Emphysema | 0.891 | 0.900 | 0.776 | 0.898 | 0.831 | 0.805 |
| Fibrosis | 0.762 | 0.847 | 0.738 | 0.839 | 0.631 | 0.626 |
| Hernia | 0.992 | 0.768 | 0.734 | 0.981 | 0.948 | 0.659 |
| Infiltration | 0.683 | 0.742 | 0.709 | 0.692 | 0.626 | 0.628 |
| Mass | 0.796 | 0.895 | 0.870 | 0.790 | 0.729 | 0.785 |
| Nodule | 0.735 | 0.819 | 0.820 | 0.700 | 0.683 | 0.600 |
| Pleural_Thickening | 0.708 | 0.829 | 0.747 | 0.785 | 0.622 | 0.685 |
| Pneumonia | 0.705 | 0.768 | 0.809 | 0.788 | 0.491 | 0.610 |
| Pneumothorax | 0.803 | 0.894 | 0.797 | 0.858 | 0.819 | 0.788 |
| Overall# | 0.248 | 0.257 | 0.175 | 0.218 | 0.103 | 0.138 |
| Macro precision | Macro precision | Macro precision | Macro precision | Macro precision | Macro precision | |
| Atelectasis | 0.59 | 0.62 | 0.60 | 0.59 | 0.58 | 0.60 |
| Cardiomegaly | 0.55 | 0.56 | 0.54 | 0.56 | 0.57 | 0.56 |
| Consolidation | 0.54 | 0.55 | 0.55 | 0.54 | 0.54 | 0.54 |
| Edema | 0.54 | 0.54 | 0.54 | 0.54 | 0.53 | 0.53 |
| Effusion | 0.64 | 0.64 | 0.65 | 0.66 | 0.63 | 0.65 |
| Emphysema | 0.57 | 0.57 | 0.54 | 0.57 | 0.54 | 0.54 |
| Fibrosis | 0.52 | 0.52 | 0.51 | 0.52 | 0.51 | 0.51 |
| Hernia | 0.55 | 0.50 | 0.50 | 0.52 | 0.51 | 0.50 |
| Infiltration | 0.60 | 0.62 | 0.60 | 0.60 | 0.58 | 0.59 |
| Mass | 0.57 | 0.63 | 0.60 | 0.57 | 0.56 | 0.57 |
| Nodule | 0.55 | 0.57 | 0.57 | 0.54 | 0.54 | 0.53 |
| Pleural_Thickening | 0.53 | 0.55 | 0.53 | 0.54 | 0.53 | 0.53 |
| Pneumonia | 0.51 | 0.52 | 0.52 | 0.51 | 0.51 | 0.51 |
| Pneumothorax | 0.58 | 0.63 | 0.58 | 0.60 | 0.58 | 0.57 |
| Overall* | 0.56 ± 0.01 (0.54, 0.58) | 0.57 ± 0.01 (0.55, 0.60)a | 0.56 ± 0.01 (0.53, 0.58) | 0.56 ± 0.01 (0.54, 0.58) | 0.55 ± 0.01 (0.53, 0.57) | 0.55 ± 0.01 (0.53, 0.58) |
| Macro recall | Macro recall | Macro recall | Macro recall | Macro recall | Macro recall | |
| Atelectasis | 0.72 | 0.76 | 0.74 | 0.72 | 0.71 | 0.72 |
| Cardiomegaly | 0.82 | 0.83 | 0.81 | 0.82 | 0.83 | 0.80 |
| Consolidation | 0.73 | 0.74 | 0.74 | 0.73 | 0.72 | 0.73 |
| Edema | 0.83 | 0.84 | 0.84 | 0.83 | 0.82 | 0.82 |
| Effusion | 0.80 | 0.81 | 0.80 | 0.80 | 0.79 | 0.80 |
| Emphysema | 0.85 | 0.87 | 0.82 | 0.82 | 0.78 | 0.79 |
| Fibrosis | 0.74 | 0.74 | 0.72 | 0.71 | 0.72 | 0.72 |
| Hernia | 0.72 | 0.73 | 0.78 | 0.78 | 0.75 | 0.72 |
| Infiltration | 0.65 | 0.65 | 0.65 | 0.65 | 0.63 | 0.65 |
| Mass | 0.76 | 0.75 | 0.74 | 0.75 | 0.75 | 0.75 |
| Nodule | 0.69 | 0.70 | 0.68 | 0.68 | 0.66 | 0.65 |
| Pleural_Thickening | 0.73 | 0.73 | 0.73 | 0.73 | 0.71 | 0.72 |
| Pneumonia | 0.68 | 0.72 | 0.69 | 0.67 | 0.65 | 0.67 |
| Pneumothorax | 0.80 | 0.81 | 0.80 | 0.80 | 0.78 | 0.78 |
| Overall* | 0.75 ± 0.02 (0.72, 0.79) | 0.76 ± 0.02 (0.73, 0.80)b | 0.75 ± 0.02 (0.72, 0.79) | 0.75 ± 0.02 (0.71, 0.78) | 0.74 ± 0.02 (0.70, 0.77) | 0.74 ± 0.01 (0.70, 0.77) |
| Macro F1-score | Macro F1-score | Macro F1-score | Macro F1-score | Macro F1-score | Macro F1-score | |
| Atelectasis | 0.57 | 0.63 | 0.60 | 0.58 | 0.54 | 0.60 |
| Cardiomegaly | 0.54 | 0.56 | 0.50 | 0.56 | 0.59 | 0.57 |
| Consolidation | 0.48 | 0.53 | 0.51 | 0.51 | 0.44 | 0.47 |
| Edema | 0.51 | 0.53 | 0.53 | 0.53 | 0.49 | 0.48 |
| Effusion | 0.64 | 0.65 | 0.66 | 0.69 | 0.62 | 0.67 |
| Emphysema | 0.59 | 0.60 | 0.51 | 0.59 | 0.53 | 0.52 |
| Fibrosis | 0.47 | 0.51 | 0.46 | 0.50 | 0.41 | 0.41 |
| Hernia | 0.58 | 0.44 | 0.43 | 0.54 | 0.50 | 0.40 |
| Infiltration | 0.59 | 0.62 | 0.61 | 0.60 | 0.56 | 0.56 |
| Mass | 0.58 | 0.66 | 0.63 | 0.57 | 0.53 | 0.57 |
| Nodule | 0.53 | 0.58 | 0.57 | 0.51 | 0.50 | 0.45 |
| Pleural_Thickening | 0.48 | 0.54 | 0.50 | 0.52 | 0.44 | 0.47 |
| Pneumonia | 0.44 | 0.47 | 0.48 | 0.47 | 0.35 | 0.40 |
| Pneumothorax | 0.58 | 0.67 | 0.58 | 0.63 | 0.59 | 0.57 |
| Overall* | 0.54 ± 0.02 (0.51, 0.57)c | 0.57 ± 0.02 (0.53, 0.61)d | 0.54 ± 0.02 (0.50, 0.58) | 0.56 ± 0.02 (0.52, 0.59)e | 0.51 ± 0.02 (0.46, 0.55) | 0.51 ± 0.02 (0.46, 0.56) |
aThere are significant differences when compared with ResNet50 + LWBCE (p = 0.043) and ResNet50 + Lours (p = 0.045).
bThere are differences when compared with ResNet50 + LWBCE (p = 0.051) and ResNet50 + Lours (p = 0.015)
cThere is a significant difference when compared with ResNet50 + LWBCE (p = 0.041)
dThere are significant differences when compared with ResNet50 + LWBCE (p = 0.010) and ResNet50 + Lours (p = 0.002)
eThere are significant differences when compared with ResNet50 + LWBCE (p = 0.002) and ResNet50 + Lours (p = 0.015). All statistical analyses were performed using repeated measures ANOVA tests with Bonferroni adjustments
#It indicates the correct prediction of all 14 labels
*Data were presented as mean ± standard error (95% confidence interval)
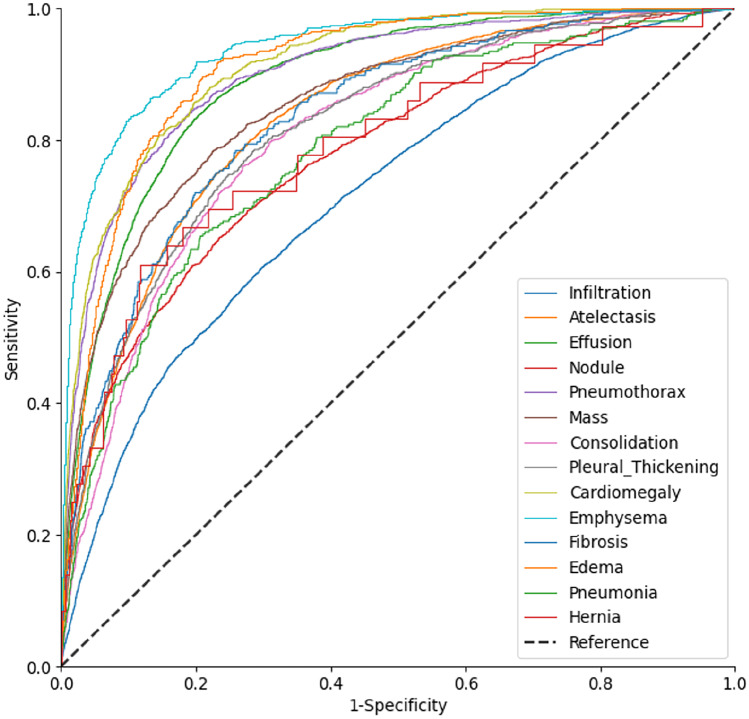
Furthermore, although CoAtNet-0-rw + Lours showed the best overall performance, the AUROC differed significantly among different disease labels, from 0.705 to 0.890 (Fig. 3), with significant differences among part of labels such as emphysema vs. edema (0.939 vs. 0.912, p < 0.001) and cardiomegaly vs. effusion (0.914 vs. 0.889, p < 0.001) (Table 5). Further heatmap visualization of the model showed that, for most disease labels (e.g., atelectasis, edema, effusion), the network could pay close attention to the corresponding areas of the lesions and make an accurate diagnosis (Fig. 4). However, in some cases, such as pneumothorax, the model did not focus on the lesion areas but on the drainage catheter that was used to treat the disease (Fig. 5). Meanwhile, we also noticed that some disease labels in the ChestX-ray14 dataset were inaccurate (Fig. 5).
Fig. 3.
AUROC curves of different label identification by CoAtNet-0-rw + Lours
Table 5.
Comparisons of AUROC among different labels identified by CoAtNet-0-rw + Lours
| Labels | AUROC | Comparisons | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Emphysema | Cardiomegaly | Edema | Pneumothorax | Effusion | Mass | Fibrosis | Atelectasis | Pleural_Thickening | Consolidation | Hernia | Pneumonia | Nodule | Infiltration | ||
| Emphysema | 0.939 | n.s | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | |
| Cardiomegaly | 0.914 | n.s | n.s | n.s | *** | *** | *** | *** | *** | *** | n.s | *** | *** | *** | |
| Edema | 0.912 | *** | n.s | n.s | n.s | *** | *** | *** | *** | *** | n.s | *** | *** | *** | |
| Pneumothorax | 0.902 | *** | n.s | n.s | n.s | *** | *** | *** | *** | *** | n.s | *** | *** | *** | |
| Effusion | 0.889 | *** | *** | n.s | n.s | *** | *** | *** | *** | *** | n.s | *** | *** | *** | |
| Mass | 0.856 | *** | *** | *** | *** | *** | n.s | n.s | *** | *** | n.s | *** | *** | *** | |
| Fibrosis | 0.835 | *** | *** | *** | *** | *** | n.s | n.s | n.s | n.s | n.s | n.s | *** | *** | |
| Atelectasis | 0.833 | *** | *** | *** | *** | *** | n.s | n.s | n.s | n.s | n.s | n.s | *** | *** | |
| Pleural_Thickening | 0.819 | *** | *** | *** | *** | *** | *** | n.s | n.s | n.s | n.s | n.s | *** | *** | |
| Consolidation | 0.809 | *** | *** | *** | *** | *** | *** | n.s | n.s | n.s | n.s | n.s | n.s | *** | |
| Hernia | 0.791 | *** | n.s | n.s | n.s | n.s | n.s | n.s | n.s | n.s | n.s | n.s | n.s | n.s | |
| Pneumonia | 0.789 | *** | *** | *** | *** | *** | *** | n.s | n.s | n.s | n.s | n.s | n.s | *** | |
| Nodule | 0.779 | *** | *** | *** | *** | *** | *** | *** | *** | *** | n.s | n.s | n.s | *** | |
| Infiltration | 0.715 | *** | *** | *** | *** | *** | *** | *** | *** | *** | *** | n.s | *** | *** | |
***A significant difference with a p value less than 0.001; n.s., no significant difference (p > 0.05). Statistical analysis was performed using AUROC comparison analysis with Bonferroni adjustments in SPSS software
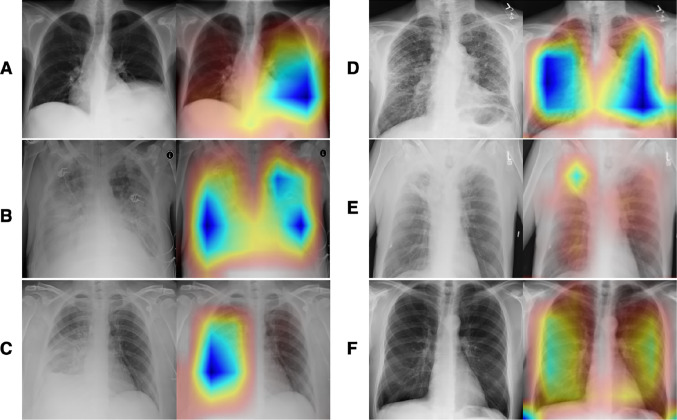
Fig. 4.
An exemplary illustration of accurately predicted cases (left, original images; right, Group-CAM heatmaps). A A case (Image Index: 00000761_010.pgn, label: Atelectasis) showed right attention of atelectasis at the left lower lung. B A case (Image Index: 00012834_049.png, label: Edema|Effusion|Infiltration) showed the right attention to diffused edema, effusion, and infiltration at bilateral lungs. C A case (Image Index: 00012834_049.png, label: Consolidation) showed the right attention of extensive consolidation at the right lower lung. D A case (Image Index: 00014849_011.png, label: Fibrosis) showed the right attention of fibrosis at bilateral lungs. E A case (Image Index: 00009658_002.png, label: Atelectasis|Mass|Pleural_Thickening) showed the right attention of a mass with atelectasis and peripheral pleural thickening at the right upper lung. F A case (Image Index: 00002935_000.png, label: Emphysema) showed the right attention of emphysema at bilateral lungs. Abbreviation: Group-CAM, group-score-weighted class activation mapping
Fig. 5.
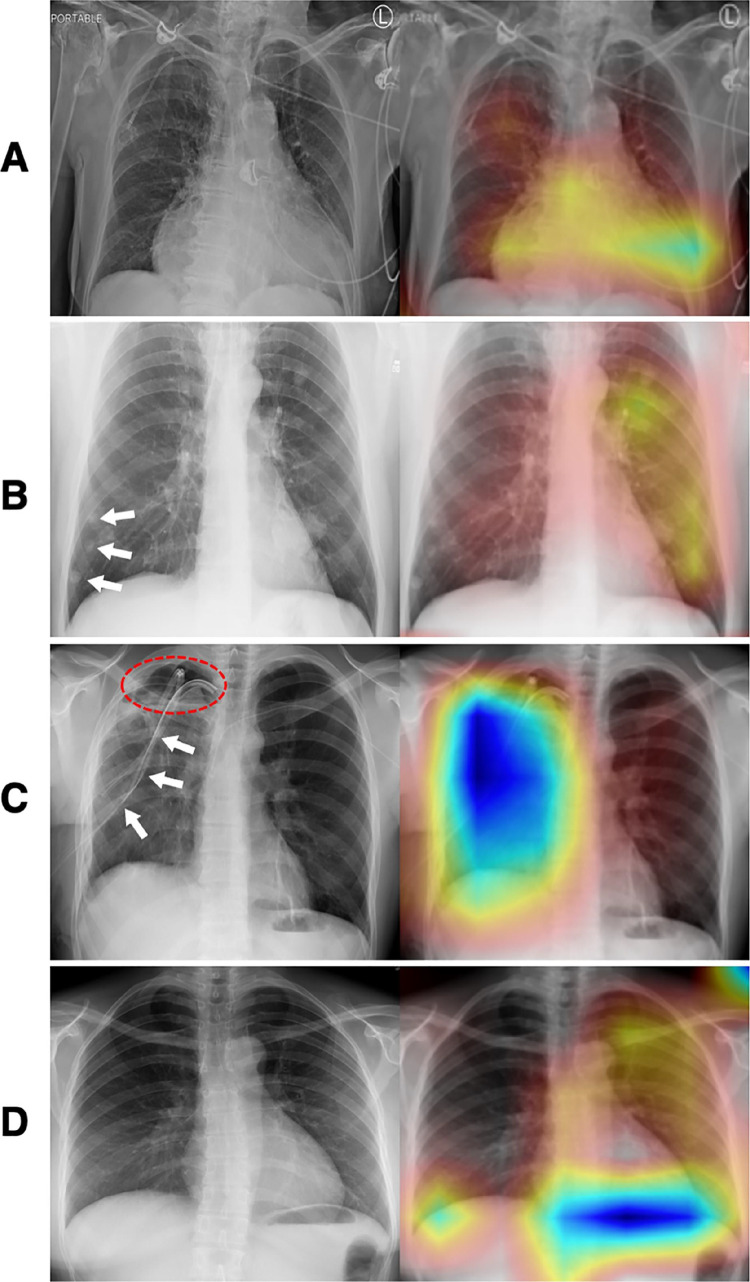
Exemplary illustration with incorrect attention (left, original images; right, Group-CAM heatmaps). A A case (Image Index: 00028948_001.png, label: Cardiomegaly|Hernia|Mass) was predicted only “Cardiomegaly,” which was correctly focused in the heatmap; however, labels “hernia” and “mass” cannot be identified by our professional radiologist from the original image (left). B A case (Image Index: 00003285_001.png, label: Nodule) with correct prediction showed the ignorance of small lung nodules (white arrows) at the right lung. C A case (Image Index: 00000631_004.png, label: Pneumothorax) with correct prediction demonstrated the attention around the thoracic drainage catheter (white arrows) but not the right pneumothorax area (red dash-line circle). D A case (Image Index: 00014234_000.png, label: Pneumonia) presented incorrect attention at the diaphragm, but the original image (left) was identified with no “pneumonia” by our professional radiologist. Abbreviation: Group-CAM, group-score-weighted class activation mapping
Discussion
In this study, we challenged a 14-label AI diagnosis task in a real-world long-tail dataset. To enhance the AI model’s performance, we applied SOTA backbones, a customized loss function (Lours), and several techniques, such as transfer learning and joint data augmentation. As a result, our experiments revealed that CoAtNet-0-rw + Lours achieved the highest overall AUROC and macro F1-score, significantly outperforming the baseline ResNet50 + LWBCE (AUROC: 0.842 vs. 0.811, p = 0.037; macro F1-score: 0.57 vs. 0.51, p = 0.010). In addition, the AUROCs of CoAtNet-0-rw + Lours varied widely across different disease labels (0.705 to 0.890), but no significant correlations were found between the AUROC values and the corresponding positive sample ratios (p ≥ 0.058).
Chest X-rays are still one of the most widely used and cost-effective medical examinations, despite advancements in pulmonary computed tomography (CT) technology. However, AI diagnosis of chest X-rays presents a greater challenge than CT scans due to the fine-grained classification issue [8]. The difficulty in fine-grained classification stems from the need for the model to learn and distinguish very delicate details, such as slight variations in shape, texture, or patterns among different classes. In contrast to CT, these details can be challenging to detect in chest X-rays because they often show subtle changes in grayscale or size and lack apparent morphological and color differences between objects or lesions and lung tissue [33]. As a result, even the trained eyes may struggle to distinguish between different labels, such as nodules and masses or infiltrations and edema. Additionally, the class imbalance problem, which we discussed earlier, exacerbates the challenge of fine-grained classification. Without proper AI techniques, this can result in a biased model that performs well for common diseases but poorly for rare diseases.
In contrast to previous studies, we did not use the conventional hierarchical multi-label method, which relies strongly on human cognition [34]. Instead, we utilized several advanced AI techniques and training tricks, such as depth-wise separable convolution, self-attention, joint data augmentation, class weighting, and tail sample focusing, to address the issue of multi-label imbalance. As a result, we achieved a better performance in the multi-label diagnosis for all 14 diseases than results reported in any previous studies [7, 8]. These AI techniques we used helped yield higher overall and individual AUROCs and macro F1-score compared to the baseline. Regarding the network structures, previous studies have argued that the Transformer structure facilitates higher-level cognition of global receptive fields [21, 22, 35]. Without the aid of large-scale pretraining and datasets, Transformer-based networks were shown to be inferior to CNNs in end-to-end tasks because CNNs have locality learning strategies and thus have a more substantial inductive bias [19, 24]. However, in our experiments, we found that Transformer can catch up or even surpass the performance of the powerful EfficientNet after the fusion with CNNs. CoAtNet, with the addition of Transformer modules connected with prior MBConV modules, has the adaptive learning ability to process long-range image information or lesions with large regions and can obtain better performance [19]. Another advantage of CoAtNet over CNN (e.g., EfficientNet) lies in the transfer learning capability, allowing similar training in further study on 3-dimensional images (e.g., CT) as in the 2-dimensional images (chest X-ray) in this study. Transformer can apply the parameters of its transformer block directly to 3-dimensional data with the same structure due to its global attention. In addition, Transformer has a significantly larger parameter capacity than CNN and is more suitable for more extensive data sizes and complicated data distributions [33, 35]. Therefore, although EfficientNet has the advantage of floating-point operations (FLOPs) with only mild lower AUROC, CoAtNet holds an advantage in future and broader applications.
To further improve the model’s performance, we designed a novel loss (Lours) for training. Theoretically, this loss integrates the advantages of reweighting, hard-sample focus, and a nonlinear shifting probability to reduce the contribution of negative and easy samples [8, 15, 25]. Besides, it can more accurately adjust the long-tail differences between different labels, so AUROCs of various disease labels did not show any significant correlations with the corresponding positive sample ratio. Our results demonstrate that training with Lours improved the performance, and CoAtNet-0-rw + Lours achieved the highest overall AUROC and macro F1-score, both significantly higher than the ResNet50 + LWBCE baseline. While accuracy is not a perfect evaluation metric for imbalanced data, as it can be inflated by over-predicting the majority class (“No findings”), it is worth noting that CoAtNet-0-rw + Lours also achieved the highest accuracy (0.257) among all models. However, this study unexpectedly found little effect of Lours on ResNet50. We speculate that a deep network without an attention mechanism (e.g., channel attention and self-attention) may be insusceptible to our designed loss, which merits further exploration.
Regarding the limitations, this study still has some remaining challenges: (1) Despite efforts to improve classification, the results still show a low overall macro F1-score when diagnosing multi-disease with varying degrees of long-tail label distribution. Data availability remains a significant challenge in chest X-ray research. When facing a large spectrum of more than ten diseases, the sample size of the ChestX-ray14 dataset is still insufficient. (2) The current labeling process, which primarily relies on automated radiology report labelers, leads to potential mislabeling and nonuniformity in the spectrum of diseases in different published chest X-ray datasets (Fig. 5), which negatively affects the performance of the models and the ability to utilize different datasets effectively [36, 37]. (3) In this study we didn’t consider the issues of multiple images from same patient and very small sample sizes in validation and test sets originated from random dataset division. To ensure a comparable experimental setup, we used the same data preprocessing and dataset division as in previous studies [7, 8]. This was done to enable a focus on evaluating and comparing the performance of the proposed deep-learning models for multi-disease diagnosis with these previous networks and setups [7, 8].
In the future, we propose several potential strategies to further increase the accuracy of AI in chest X-ray diagnosis: (1) Federated learning with a standard labeling system using a robust NLP labeling tool for chest X-rays. Federated learning aids in collecting more disease samples from various medical centers and allows model parameters to be shared without original data transfer, addressing the challenges posed by ethical and legal regulations regarding medical privacy when creating a widely accessible public dataset [38]. (2) Implementing multi-modal and cross-modal AI models for comprehensive diagnosis. The routine diagnostic process involves the comprehensive analysis of a patient’s medical history, laboratory investigation results, and chest X-rays before reaching a final diagnosis. This highlights the importance of using multiple sources of information to improve classification accuracy [39]. (3) Utilizing contrastive learning to obtain more accurate representations of the data. This study utilized pre-trained weights from ImageNet, which may somewhat limit the model’s performance on medical datasets. Contrastive learning offers a better self-supervised learning method by using radiology reports as supervision without additional labeling, being able to train the model’s backbone more accurately [33].
Conclusions
This study demonstrated an improved performance in the multi-disease diagnosis of chest X-rays in a long-tailed dataset using a pretrained CNN + Transformer hybrid network named CoAtNet-0-rw. However, the limited sample size of diseases and potential inaccuracies in labeling may have impacted the diagnostic capability of the model. To enhance performance, establishing uniform evaluation criteria for chest X-rays, incorporating multi-modal diagnostic information in training, and adopting contrastive learning techniques have the potential to facilitate federated learning and improve the model’s performance in the future.
Author Contribution
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Yuyang Chen, Yiliang Wan and Feng Pan. The first draft of the manuscript was written by Yuyang Chen and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [Grant Number: 82272083].
Data Availability
The dataset for this study can be found here: https://www.kaggle.com/datasets/nih-chest-xrays/data. The codes are available in the https://github.com/KiwisFraggle/CoAtNet_NIH.
Declarations
Ethics Approval
The dataset for this study holds a license of CC0 1.0 Universal (CC0 1.0) Public Domain Dedication, which waives copyright interest and ethical approval in scientific work and is dedicated to the worldwide public domain.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yuyang Chen, Email: ychen@putnamscience.org.
Yiliang Wan, Email: wan.yl@neusoftmedical.com.
Feng Pan, Email: uh_fengpan@hust.edu.cn.
References
- 1.Jacobi A, Chung M, Bernheim A, Eber C. Portable chest X-ray in coronavirus disease-19 (COVID-19): A pictorial review. Clinical Imaging. 2020;64:35–42. doi: 10.1016/j.clinimag.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rangarajan K, Muku S, Garg AK, Gabra P, Shankar SH, Nischal N, et al. Artificial Intelligence-assisted chest X-ray assessment scheme for COVID-19. European Radiology. 2021;31(8):6039–6048. doi: 10.1007/s00330-020-07628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang G, Liu X, Shen J, Wang C, Li Z, Ye L, et al. A deep-learning pipeline for the diagnosis and discrimination of viral, non-viral and COVID-19 pneumonia from chest X-ray images. Nature Biomedical Engineering. 2021;5(6):509–521. doi: 10.1038/s41551-021-00704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy K, Smits H, Knoops AJ, Korst MB, Samson T, Scholten ET, et al. COVID-19 on chest radiographs: a multireader evaluation of an artificial intelligence system. Radiology. 2020;296(3):E166–E172. doi: 10.1148/radiol.2020201874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan F, Li L, Liu B, Ye T, Li L, Liu D, et al. A novel deep learning-based quantification of serial chest computed tomography in Coronavirus Disease 2019 (COVID-19) Scientific Reports. 2021;11(1):417. doi: 10.1038/s41598-020-80261-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nature Machine Intelligence. 2021;3(3):199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- 7.Albahli S, Rauf HT, Algosaibi A, Balas VE. AI-driven deep CNN approach for multi-label pathology classification using chest X-Rays. PeerJ Computer Science. 2021;7:e495. 10.7717/peerj-cs.495. [DOI] [PMC free article] [PubMed]
- 8.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2017:2097–106. 10.1109/CVPR.2017.369.
- 9.Ferguson AR, Nielson JL, Cragin MH, Bandrowski AE, Martone ME. Big data from small data: data-sharing in the 'long tail' of neuroscience. Nature Neuroscience. 2014;17(11):1442–1447. doi: 10.1038/nn.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ning W, Lei S, Yang J, Cao Y, Jiang P, Yang Q, et al. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nature Biomedical Engineering. 2020;4(12):1197–1207. doi: 10.1038/s41551-020-00633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Deng X, Fu Q, Zhou Q, Feng J, Ma H, et al. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Transactions on Medical Imaging. 2020;39(8):2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 12.Seyyed-Kalantari L, Zhang H, McDermott MB, Chen IY, Ghassemi M. Underdiagnosis bias of artificial intelligence algorithms applied to chest radiographs in under-served patient populations. Nature Medicine. 2021;27(12):2176–2182. doi: 10.1038/s41591-021-01595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Advances in Neural Information Processing Systems. 2017;30. 10.48550/arXiv.1706.03762.
- 14.Hu J, Shen L, Sun G. Squeeze-and-excitation networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2018:7132–41. 10.48550/arXiv.1709.01507.
- 15.Ridnik T, Ben-Baruch E, Zamir N, Noy A, Friedman I, Protter M, et al. Asymmetric loss for multi-label classification. Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV). 2021:82–91. 10.48550/arXiv.2009.14119.
- 16.Weiss K, Khoshgoftaar TM, Wang D. A survey of transfer learning. Journal of Big data. 2016;3(1):1–40. doi: 10.1186/s40537-016-0043-6. [DOI] [Google Scholar]
- 17.Cubuk ED, Zoph B, Mane D, Vasudevan V, Le QV. Autoaugment: Learning augmentation strategies from data. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). 2019:113–23. 10.48550/arXiv.1805.09501.
- 18.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016:770–8. 10.48550/arXiv.1512.03385.
- 19.Dai Z, Liu H, Le QV, Tan M. CoatNet: Marrying convolution and attention for all data sizes. Advances in Neural Information Processing Systems. 2021;34:3965–77. 10.48550/arXiv.2106.04803.
- 20.Tan M, Le Q. EfficientNet: Rethinking model scaling for convolutional neural networks. International Conference on Machine Learning, PMLR. 2019:6105–14. 10.48550/arXiv.1905.11946.
- 21.Tu Z, Talebi H, Zhang H, Yang F, Milanfar P, Bovik A, et al. Maxvit: Multi-axis vision transformer. In: Avidan, S, Brostow, G, Cissé, M, Farinella, GM, Hassner, T (eds) Computer Vision – ECCV 2022 Lecture Notes in Computer Science, vol 13684. 2022. 10.1007/978-3-031-20053-3_27.
- 22.Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint:201011929. 2020. 10.48550/arXiv.2010.11929.
- 23.Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C. MobileNetV2: Inverted residuals and linear bottlenecks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2018:4510–20. 10.48550/arXiv.1801.04381.
- 24.d’Ascoli S, Touvron H, Leavitt ML, Morcos AS, Biroli G, Sagun L. ConViT: Improving vision transformers with soft convolutional inductive biases. Proceedings of the 38th International Conference on Machine Learning, PMLR 139. 2021:2286–96. 10.1088/1742-5468/ac9830.
- 25.Lin T-Y, Goyal P, Girshick R, He K, Dollár P. Focal loss for dense object detection. Proceedings of the IEEE International Conference on Computer Vision (ICCV). 2017:2980–8. 10.48550/arXiv.1708.02002.
- 26.Al-Kababji A, Bensaali F, Dakua SP. Scheduling techniques for liver segmentation: ReduceLRonPlateau vs OneCycleLR. In: Bennour, A, Ensari, T, Kessentini, Y, Eom, S (eds) Intelligent Systems and Pattern Recognition ISPR 2022 Communications in Computer and Information Science, vol 1589. 2022:204–12. 10.1007/978-3-031-08277-1_17.
- 27.Ying X. An overview of overfitting and its solutions. Journal of Physics: Conference Series. 2019;1168(2):022022. 10.1088/1742-6596/1168/2/022022.
- 28.Barrow E, Eastwood M, Jayne C. Selective dropout for deep neural networks. International Conference on Neural Information Processing (ICONIP 2016): Neural Information Processing. 2016:519–28. 10.1007/978-3-319-46675-0_57.
- 29.Wang J, Zhang S. An improved deep learning approach based on exponential moving average algorithm for atrial fibrillation signals identification. Neurocomputing. 2022;513:127–136. doi: 10.1016/j.neucom.2022.09.079. [DOI] [Google Scholar]
- 30.Micikevicius P, Narang S, Alben J, Diamos G, Elsen E, Garcia D, et al. Mixed precision training. arXiv preprint:171003740. 2017. 10.48550/arXiv.1710.03740.
- 31.Li Z, Kamnitsas K, Glocker B. Analyzing overfitting under class imbalance in neural networks for image segmentation. IEEE Transactions on Medical Imaging. 2020;40(3):1065–1077. doi: 10.1109/TMI.2020.3046692. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q, Rao L, Yang Y. Group-CAM: group score-weighted visual explanations for deep convolutional networks. arXiv preprint:210313859. 2021. 10.48550/arXiv.2103.13859.
- 33.Wang Z, Wu Z, Agarwal D, Sun J. MedCLIP: Contrastive learning from unpaired medical images and text. arXiv preprint:221010163. 2022. 10.48550/arXiv.2210.10163. [DOI] [PMC free article] [PubMed]
- 34.Chen H, Miao S, Xu D, Hager GD, Harrison AP. Deep hiearchical multi-label classification applied to chest X-ray abnormality taxonomies. Medical Image Analysis. 2020;66:101811. 10.1016/j.media.2020.101811. [DOI] [PubMed]
- 35.Radford A, Kim JW, Hallacy C, Ramesh A, Goh G, Agarwal S, et al. Learning transferable visual models from natural language supervision. Proceedings of the 38th International Conference on Machine Learning, PMLR 139. 2021:8748–63. 10.48550/arXiv.2103.00020.
- 36.Majkowska A, Mittal S, Steiner DF, Reicher JJ, McKinney SM, Duggan GE, et al. Chest radiograph interpretation with deep learning models: assessment with radiologist-adjudicated reference standards and population-adjusted evaluation. Radiology. 2020;294(2):421–431. doi: 10.1148/radiol.2019191293. [DOI] [PubMed] [Google Scholar]
- 37.Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, et al. CheXpert: A large chest radiograph dataset with uncertainty labels and expert comparison. Proceedings of the AAAI Conference on Artificial Intelligence. 2019;33(01):590–597. doi: 10.1609/aaai.v33i01.3301590. [DOI] [Google Scholar]
- 38.Bai X, Wang H, Ma L, Xu Y, Gan J, Fan Z, et al. Advancing COVID-19 diagnosis with privacy-preserving collaboration in artificial intelligence. Nature Machine Intelligence. 2021;3(12):1081–1089. doi: 10.1038/s42256-021-00421-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Pan F. Multimodal detection of hateful memes by applying a vision-language pre-training model. PLoS One. 2022;17(9):e0274300. 10.1371/journal.pone.0274300. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset for this study can be found here: https://www.kaggle.com/datasets/nih-chest-xrays/data. The codes are available in the https://github.com/KiwisFraggle/CoAtNet_NIH.