ABSTRACT
Lactococcin A (LcnA), a class IId bacteriocin, induces membrane leakage and cell death by specifically binding to the membrane receptor–mannose phosphotransferase system (man-PTS), as is the case for pediocin-like (class IIa) bacteriocins. The cognate immunity protein of bacteriocins, which protects the producer cell from its own bacteriocin, recognizes and binds to the bacteriocin–man-PTS complex, consequently blocking membrane leakage. We previously deciphered the mode of action and immunity of class IIa bacteriocins. Here, we determined the structure of the ternary complex of LcnA, LciA (i.e., the immunity protein), and its receptor, i.e., the man-PTS of Lactococcus lactis (ll-man-PTS). An external loop on the membrane-located component IIC of ll-man-PTS was found to prevent specific binding of the N-terminal region of LcnA to the site recognized by pediocin-like bacteriocins. Thus, the N-terminal β-sheet region of LcnA recognized an adjacent site on the extracellular side of ll-man-PTS, with the LcnA C-terminal hydrophobic helix penetrating into the membrane. The cytoplasmic cleft formed within the man-PTS Core and Vmotif domains induced by embedded LcnA from the periplasmic side is adopted by the appropriate angle between helices H3 and H4 of the N terminus of LciA. The flexible C terminus of LciA then blocks membrane leakage. To summarize, our findings reveal the molecular mechanisms of action and immunity of LcnA and LciA, laying a foundation for further design of class IId bacteriocins.
IMPORTANCE Class IId (lactococcin-like) bacteriocins and class IIa (pediocin-like) bacteriocins share a few similarities: (i) both induce membrane leakage and cell death by specifically binding the mannose phosphotransferase system (man-PTS) on their target cells, and (ii) cognate immunity proteins recognize and bind to the bacteriocin–man-PTS complex to block membrane leakage. However, class IId bacteriocins lack the “pediocin box” motif, which is typical of class IIa bacteriocins, and basically target only lactococcal cells; in contrast, class IIa bacteriocins target diverse bacterial cells, but not lactococcal cells. We previously solved the structure of class IIa bacteriocin-receptor-immunity ternary complex from Lactobacillus sakei. Here, we determined the structure of the ternary complex of class IId bacteriocin LcnA, its cognate immunity protein LciA, and its receptor, the man-PTS of Lactococcus lactis. By comparing the interactions between man-PTS and class IIa and class IId bacteriocins, this study affords some clues to better understand the specificity of bacteriocins targeting the mannose phosphotransferase system.
KEYWORDS: antibiotic resistance, antimicrobial peptides, mannose phosphotransferase system, man-PTS, bacteriocins, nonpediocin-like/class IId bacteriocins, lactococcin A, LcnA, immunity, LciA, self-protection, class IId bacteriocins
INTRODUCTION
An increasingly wide range of bacteria has developed resistance to antibiotics (1); therefore, it is vital to identify alternative treatments for infections caused by antibiotic-resistant pathogens. Antimicrobial peptides (AMPs) can evidently inhibit the growth of several multidrug-resistant bacteria (2). Bacterial AMPs, or bacteriocins, are peptidic toxins that can inhibit the growth of similar or closely related bacterial strains (3, 4). Organisms are immune to their own bacteriocins, a trait that is mediated by certain immunity proteins (5). Bacteriocins from Gram-positive species can be typically classified into three main classes (6): (i) class I (≤5 kDa), comprising the unusual amino acids lanthionine and methyllanthionine (e.g., nisin); (ii) class II (heat stable, ≤10 kDa), without any posttranslational modifications; and (iii) class III (≥30 kDa), which are high molecular weight bacteriocins. Class II (non-lanthionine-containing) bacteriocins can be further subdivided (7) into (i) IIa, which are pediocin-like linear peptides and often contain one or two disulfide bonds and a conserved “pediocin box” motif (8–10); (ii) IIb of two-peptide bacteriocins, which require the presence of the two components for activity; (iii) IIc, which are cyclic bacteriocins; and (iv) IId, a miscellaneous group of linear peptides that do not fit into any of the other subclasses. Although class IId bacteriocins share no sequence similarity with class IIa bacteriocins, certain members of this group, also known as lactococcin A (LcnA)-like bacteriocins (e.g., LcnA and LcnB [11]; garvicin Q [12]; garvicin A, B, and C [13]; BacSJ [14]; ubericin K [15]; garvicin AG1 and AG2 [16]; and angicin [17]) (Fig. S1), seem to share some common mechanistic features for cell targeting and immunity. Similar to class IIa bacteriocins, LcnA-like bacteriocins use membrane-located components (ManYZ or IIC/IID) of the mannose phosphotransferase system (man-PTS) as a receptor (18). For conferring immunity, immunity proteins form a complex with the man-PTS (receptor) and cognate bacteriocins. However, classes IIa and IId markedly differ from each other in their inhibitory spectra. Class IId bacteriocins exclusively target lactococcal cells, while class IIa bacteriocins, as a group, target cells belonging to diverse bacterial genera, but not lactococcal cells. This difference in target specificity possibly relies on minor differences in membrane-located components of the man-PTS in lactococcal and non-lactococcal cells, i.e., region-α, region-β, and region-γ (19).
Although molecular interactions among class IIa bacteriocins (e.g., pediocin PA-1 and sakacin A) and their cognate man-PTSs have been elucidated (20, 21), it is challenging to deduce the molecular mechanism of LcnA across various subclasses. It is important to elucidate which components of the man-PTS differentiate the recognition of class IIa or LcnA-like bacteriocins, as well as the role of specific immunity proteins. Thus, in this study, we aimed to study the ternary complex between LcnA, the LcnA immunity protein LciA, and the man-PTS of Lactococcus lactis (22).
RESULTS AND DISCUSSION
A ternary complex of LcnA and LciA with the membrane embedded part of the man-PTS of L. lactis.
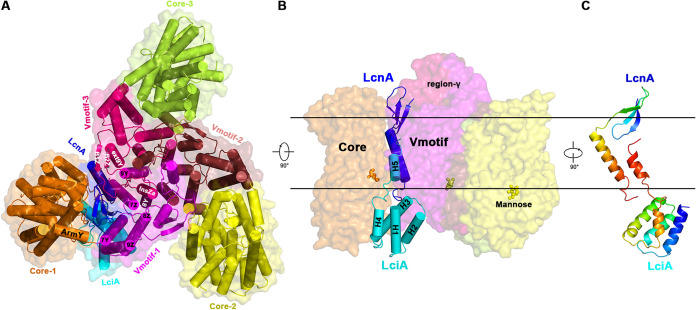
In the LcnA-producing bacteria, LcnA is translated as a precursor peptide that contain 21 residue-long N-terminal leader sequence (Fig. S1). LcnA maturation occurs with the cleavage after the GG motif, and secretion out of the cell by an ABC transporter and peptidase. To obtain the appropriate amount of ternary complex between mature LcnA (residues 22 to 75), immunity protein LciA and man-PTS membrane components of L. lactis (ll-man-PTS) in Escherichia coli for structural study, the signal sequence of MBP (Maltose-Binding-Protein) as well as MBP were fused to the N terminus of mature LcnA to secret the MPB-LcnA chimeric protein into the periplasm. Meanwhile, a flag-tagged LciA and ll-man-PTS were coexpressed in Escherichia coli as well. After anti-MBP affinity resin and size exclusion chromatography purification, peak fractions were concentrated for cryo-electron microscopy (cryo-EM) analysis. A total of 1,529,899 selected particles of approximately 200 kDa yielded a three-dimensional cryo-EM reconstruction at an overall resolution of 2.98 Å (Fig. S2 and Table 1). Similar to the man-PTSs of Listeria monocytogenes (PDB ID: 7VLX, 7VLY) (20) and Lactobacillus sakei (PDB ID: 7XNO) (21), ll-man-PTS can be spatially organized into the Vmotif and Core domains linked through the short helices ArmY and ArmZ (Fig. 1A) (23). Three ll-man-PTSs form a trimer mediated through the Vmotif domains: (i) one within the membrane via the helical bundles TM9Y and TM8Y and (ii) the other outside the membrane via the helical bundle InsZa (or region-γ) in the domain-swapping manner. Considering the different positions of the three substrates away from the inner surface of the membrane (Fig. 1B), three man-PTS protomers have been proposed to exist in three different conformational states (24). Protomers number 2 (yellow) and number 3 (green) are bacteriocin-free and close to the inward-facing states. However, in case of protomer number 1, the binding with LcnA and LciA leads to the opening of the interface between the Core and Vmotif domains (Fig. 1B). The C terminus of LciA (helix H5) penetrates into the same cleft as LcnA but from the opposite side, and the two C-terminal extensions partially overlap in the cleft (Fig. 1C). However, the C-terminal interaction is not strong enough to sustain the complex without the support of the receptor. Therefore, the contact interface between the Core and Vmotif domains is not only the sugar transport pathway of an elevator-type mechanism (23, 24) but also the interaction interface of bacteriocins and immunity proteins.
TABLE 1.
Cryo-electron microscopy data collection, refinement, and validation
| Dataset | LcnA–LciA–man-PTS complex |
|---|---|
| Data collection and processing | |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 50 |
| Defocus range (μm) | −1.5 ~ −1.8 |
| Pixel size (Å) | 1.0742 |
| Symmetry imposed | C1 |
| Final dataset (number of particles) | 1,529,899 |
| Map resolution (Å) FSC01.43 | 2.98 |
| Refinement | |
| Initial model | EMD-32031 |
| Map-sharpening B factor (Å2) | −129.75 |
| Model composition | |
| Nonhydrogen atoms | 16,168 |
| Protein residues | 2,149 |
| Ligands | 3 Mannose |
| R.m.s. deviations | |
| Bond length (Å) | 0.0052 |
| Bond angle (°) | 0.67 |
| Validation | |
| MolProbity score | 2.46 |
| Clash score | 12.12 |
| Poor rotamers (%) | 3.28 |
| Ramachandran plot (%) | |
| Favored | 92.54 |
| Allowed | 7.46 |
| Outliers | 0.0 |
| Model to map fit CC | 0.7797 |
| EMRinger score | 0.872 |
| PDB code | 8HFS |
| EMDB code | EMD-34726 |
FIG 1.
Overall structure of the ternary complex LcnA-LciA-ll-man-PTS. (A) Cartoon and surface representations of the complex viewed from the extracellular side of the membrane. The man-PTS protomer, including ManY/IIC and ManZ/IID is spatially organized into the Vmotif and Core domains (indicated in different colors). Three man-PTSs form a trimer mediated mainly through the Vmotif domains with the interface helices labeled. The class IId bacteriocin LcnA as well as the cognate immunity protein LciA colored in blue and cyan bind only to one of the protomers. (B) View within the plane of the membrane. The spatial position of the complex inside the lipid bilayer is predicted by the PPM server (41). The man-PTSs are represented as surface with the region-γ indicated (19). The substrates, mannose, are shown in ball-and-stick representation. (C) LcnA and LciA shown in rainbow colors, with the N terminus in blue and the C terminus in red.
Role of man-PTS components in the binding orientation of LcnA and pediocin-like bacteriocins.
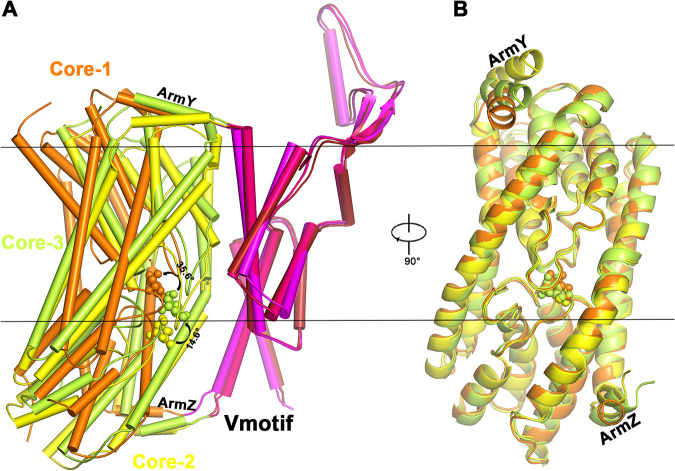
Once bacteriocin-bound (protomer number 1) and bacteriocin-free ll-man-PTSs (protomers number 2 and number 3) are superimposed based solely on their inert Vmotif domains in the membrane, the Core domains are rearranged within the membrane relative to the Vmotif domains (Fig. 2A). In the scope of the elevator mechanism (25), Core-2 is considered to be a minor inward-facing state of Core-3, rotated around 14.6° upwards into the membrane. As the result of LcnA’s insertion from the periplasmic side, Core-1 undergoes a large conformational rotation of 35.6° relative to Core-3, which is bacteriocin-free. Once the Core domains are aligned, they all remain as rigid bodies, except ArmY and ArmZ (Fig. 2B), which are the linkages between the movable Core and inert Vmotif domains (23).
FIG 2.
Structural comparison of protomers number 1, number 2, and number 3 from the man-PTS complex. (A) Structural superposition based on the Vmotif domains. The conformational rotations of the Core domains relative to the Vmotif domains are indicated. Core-2 and Core-3 are bacteriocin-free states. The linkage between Core and Vmotif domains is via ArmY and ArmZ elements. (B) Structural alignment based only on the Core domains. The Core domains rotates as rigid bodies, except the linker helices (ArmY in ManY/IIC and ArmZ in ManZ/IID) between the Core and Vmotif domains.
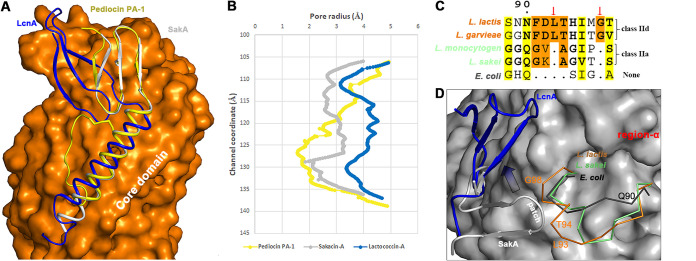
Both class IIa and IId bacteriocins interact using membrane-located components of the man-PTS as a receptor (18). In general, bacteriocins crack the man-PTS like a wedge, resulting in pore formation (Fig. 1B). However, when bacteriocin–Core subcomplexes (LcnA, pediocin PA-1, and SakA) are aligned based only on the Core domains, class IIa bacteriocins (pediocin PA-1 and SakA) and LcnA recognize and bind at slightly different positions on the Core domain (Fig. 3A), which results in distinctly different pore dimensions (Fig. 3B). A study systematically chose several man-PTSs from different genera to report that only a defined phylogenetic group of the man-PTSs confers sensitivity to class IIa bacteriocins (19); moreover, multiple sequence alignments of these man-PTSs revealed at least three regions in IIC and IID proteins: region-α, -β, and -γ. Interestingly, region-α is located in the N-terminal half of IIC of group I and contains the conserved sequence GGQGXXG in the man-PTSs (green bacteria in Fig. 3C). Region-α is spatially close to the putative positively charged pediocin patch (Fig. 3D) (26). L. lactis, which confers sensitivity only to LcnA and not to class IIa bacteriocins, lacks this GGQGXXG sequence; instead, it contains the conserved sequence FDLTHIXGX (orange bacteria in Fig. 3C). From a structural point of view, the insertion of two additional residues (L93 and G98, red arrows in Fig. 3C) directly pushes off a small fragment of helix (T94-G98, orange helix in Fig. 3D) out of the Core surface, inducing clashes with the putative N-terminal part of class IIa bacteriocins. Thus, the N-terminal part of LcnA must seek another recognition site (arrow in Fig. 3D), which is different from the class IIa binding site. The linkage effect within the two functional modules of LcnA (N-terminal region and C-terminal region) transmits into the binding orientation of the C terminus of LcnA, resulting in an overall different position of LcnA on the Core domain compared with that of class IIa bacteriocins (Fig. 3A).
FIG 3.
Effect of the region-α of man-PTS component IIC on the binding orientation of LcnA and pediocin-like bacteriocins. (A) The complex structures between LcnA (blue) and the man-PTS of L. lactis (PDB ID: 8HFS), between pediocin PA-1 (yellow) and the man-PTS of L. monocytogenes (PDB ID: 7VLY), between SakA (white) and the man-PTS of L. sakei (PDB ID: 7XNO) are aligned based on the Core domains, with the Vmotif domains omitted for clarity. (B) Pore radii along the potential transport paths generated by pediocin PA-1 (yellow), sakacin A (white), and LcnA (blue) (42). (C) Alignment of the amino acids of the region-α of man-PTS component IIC, which may define the sensitivity to class IIa or IId bacteriocins. The bacteria susceptible to class IId bacteriocins (L. lactis and L. garvieae) are colored in orange, and that susceptible to class IIa bacteriocins colored in green. Region-α from E. coli is colored in black. The two-residue insertions (L93 and G98) are shown with the red arrows. (D) Structural views of region-α in (C). The region-α’s from E. coli, L. sakei, and L. lactis are represented in ribbon and colored in black, green, orange, respectively. The positively charged “pediocin patch” in the pediocin-like bacteriocins is indicated.
Contact interactions between LcnA and the man-PTS.
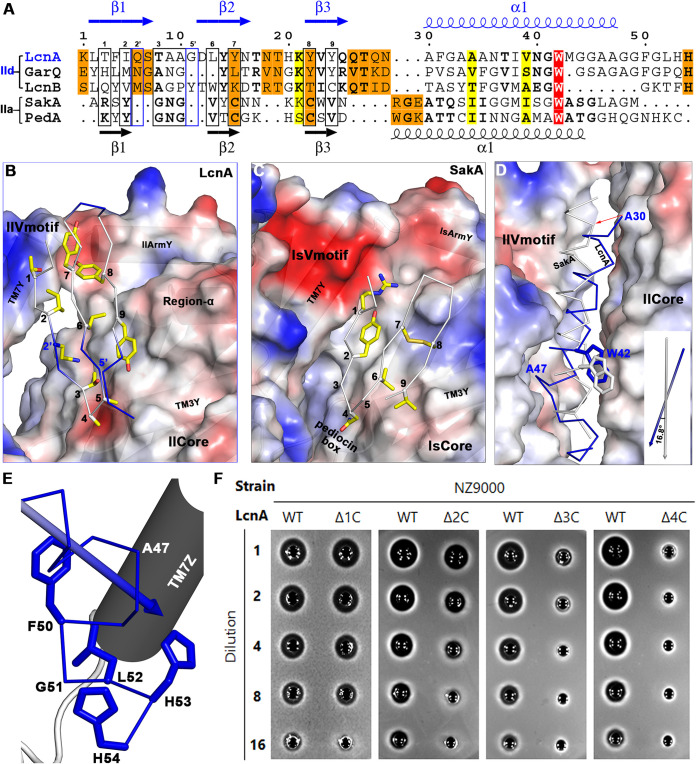
Both class IIa and certain class IId bacteriocins, including LcnA, contain a hydrophilic, cationic, and highly conserved three-stranded antiparallel β-sheet N-terminal region and a less well-conserved hydrophobic/amphiphilic C-terminal α-helical region (Fig. 4A). The N-terminal β-sheet of LcnA is much longer than that of pediocin-like bacteriocins, with two residue insertions in the first (Q6 and S7 at the position 2’) and second (G11 and D12 at the position 5′) strands (Fig. S4). The third strand includes four residues (QTQN) to catch up with the starting point of the α-helical region of LcnA. Surprisingly, the last residue (N29) of the N-terminal region of LcnA skips three residues found in the C-terminal α-helical region of pediocin-like bacteriocins and links directly to Ala30 (Fig. S4).
FIG 4.
Interactions between LcnA and the man-PTS. (A) Structure-based sequence alignment of class IId bacteriocins (LcnA, GarQ, LcnB) and class IIa bacteriocins (SakA, PedA/pediocin PA-1). The secondary structural elements of class IId bacteriocins are indicated above the sequence alignment (blue), and that of class IIa bacteriocins are located below the alignment (black). The hydrophobic residues in the N-terminal region of class IIa bacteriocins with interactions with the receptor are numbered from 1 to 9 and enclosed in the black box. The additional interaction residues contributed from class IId bacteriocins are boxed in blue, with a prime to continue their numbering. (B) Recognition of the N-terminal region of LcnA by the man-PTS of L. lactis. The key interaction secondary structural elements are labeled. The hydrophobic residual numbers are the same as (A). (C) Corresponding binding of the N-terminal region of SakA by the man-PTS of L. sakei. (D) Relative orientation of the C-terminal region of LcnA (blue) and SakA (white). Inset: angle between the helical parts of LcnA and SakA within the membrane. (E) Structural orientation of the C-terminal tail of LcnA. (F) Bactericidal activity of the wild-type (WT) LcnA and C-terminal truncation variants (Δ1C, Δ2C, Δ3C, and Δ4C mean lacking 1, 2, 3, and 4 residues from the C-terminal extremity). Serial dilutions of overnight cultures were spotted (5 μL) on LB plates.
The tip of the first β-hairpin (pediocin box in class IIa bacteriocins) adheres to the carboxyl end of TM3Y (Fig. 4B and C). Because of the steric hindrance of region-α in the ll-man-PTS, the entire N-terminal region of LcnA rotates clockwise toward the Vmotif domain with the pediocin box position as the pivot. Except the nine interaction residues (from No. 1 to No. 9 in Fig. 4C) between the N-terminal region of bacteriocins and class IIa receptor, 2′ and 5′ positions of class IId bacteriocins also contribute to the interactions (Fig. 4B). With changes in the environment of the Core and Vmotif interface positions, the N-terminal residues of LcnA become less hydrophobic to bind with the receptor.
The starting residue Ala30 of LcnA C-terminal helix reclines on the Core domain, and the whole helix transverse 16.8° toward the Vmotif domain ending with Ala47 (Fig. 4D). Trp42 of LcnA delimits the final position of the C-terminal helical penetration in the membrane (blue Trp in Fig. 4D). From G48-G49 onwards, the C-terminal end of LcnA makes a sharp turn deep within the cleft (Fig. 4E). Four truncation variants of LcnA from the C-terminal extremity were constructed (i.e., Δ1C, Δ2C, Δ3C, and Δ4C). The LcnA-susceptible strain NZ9000 was assayed against LcnA truncation variants. Removing one or two residues from the C-terminal end did not affect bactericidal activity, whereas removing three or more residues from the C-terminal end led to the loss of bactericidal activity (Fig. 4F).
Immunity mechanism of LciA.
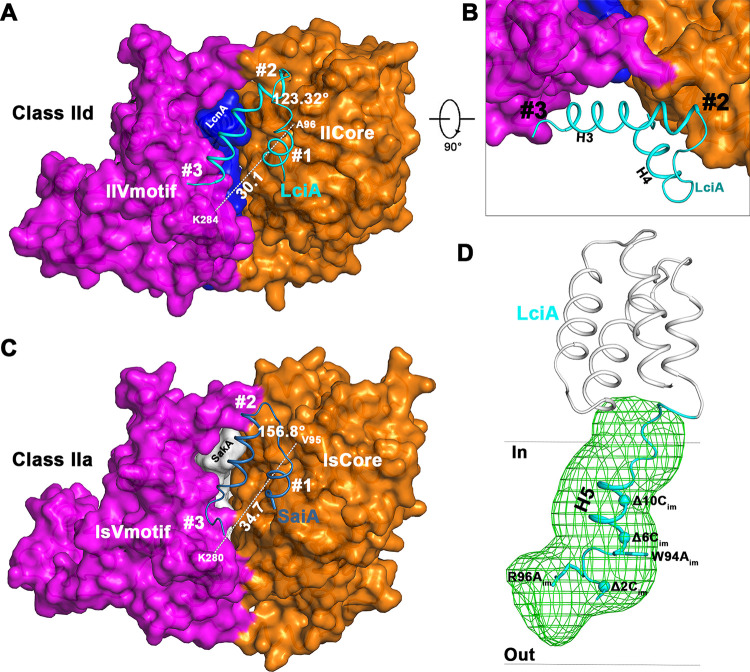
To protect itself against LcnA, the producing strain has to synthesize the immunity protein LciA which blocks the pores (27). The membrane penetration of the C-terminal end of LcnA induces a crevice between the receptor Core and Vmotif domains, which results in a dislocation on the final position of the Core and Vmotif domains against the bacteriocin-free man-PTS (Fig. S5). The last helix hairpin (H3 and H4) in the N-terminal region of LciA, which folds into an antiparallel four helix bundle, recognizes this orientation at three sites: site number 1 with H4, site number 2 with the H3 C terminus, and site number 3 with the H3 N terminus (Fig. 5A and B). The angle between H3 and H4 is 123.32°, which just accommodates the 30.1Å distance between K284 on the Vmotif and A96 on the Core domain. As for class IIa bacteriocin, the size and shape of the C-terminal part of SakA embedded within the receptor are quite different (Fig. 5C), which makes the relative orientation different as well. The distance between K280 on the Vmotif domain and V95 on the Core domain is 34.7Å. To recognize this difference in orientation between the Core and Vmotif domains of ls-man-PTS, the immunity protein of SakA, i.e., SaiA, adjusts the angle between H3 and H4 to 156.8° (Fig. 5C).
FIG 5.
Recognition and immunity mechanism of LciA. (A) Relative orientation of H3 and H4 of LciA matches the inner surface induced by the embedded LcnA. The relationship between the Vmotif and Core domains was measured by the distance between K284 and A96. (B) Detailed sideview of (A). (C) Corresponding binding action between H3 and H4 of SaiA and the man-PTS of L. sakei. (D) Blocking role played by the C-terminal regions of LciA. The truncations or mutations at the C-terminal tail are indicated (28). The transmembrane pore is shown in green mesh as generated using HOLE software (42).
The flexible C-terminal end is vital for LciA functionality. After the N-terminal four-helix bundle of LciA recognizes the internal side of man-PTS induced by LcnA insertion, the flexible C-terminal tail extends into the pore to block bacteriocin-induced membrane leakage (Fig. 5D). Unlike the flexible loop in the C-terminal tail of the pediocin-like immunity protein SaiA, the C-terminal tail of LciA folds into the fifth helix, consequently blocking the pore. P. E. Kristiansen et al. (28) assayed single point mutations (R96Aim and W94Aim) and truncation variants at the C terminus of LciA (Δ2Cim, Δ6Cim, and Δ10Cim) against LcnA to test the degree of protection. They found that single mutations and deletion of the last two residues of the C terminus of LciA resulted in almost the same immunity compared to the wild type LciA. However, the truncation variants of LciA (Δ6Cim or Δ10Cim) almost abolished its immunity activity. From the structural point of view, the skeleton of helix H5 of LciA is the main contributor for blocking the membrane leakage (Fig. 5D). Single mutations or Δ2Cim truncation variant of LciA do not interfere with the integrity of helix H5, therefore, have no effect on the immunity properties. Unfortunately, Δ6Cim or Δ10Cim truncation variants cut off the helix H5 at the half way, inducing the leakage of the membrane and therefore lost of immunity.
Conclusions.
Linear nonpediocin-like one-peptide bacteriocins with no sequence similarity to pediocin-like bacteriocins appear in class IId. LcnA, produced by some L. lactis strains, was among the first representatives of class IId bacteriocins and one of the best characterized. It increases the permeability of target cell membranes in a receptor-mediated manner, thereby dissipating the proton motive force. As with pediocin-like (class IIa) bacteriocins, LcnA binds to mannose phosphotransferase permease in target cell membranes. Compared with the man-PTSs sensitive to class IIa bacteriocins, the two additional residual insertion in an extracellular loop of the man-PTS of L. lactis makes a small fragment of helix extrude away from the Core domain, which prevents the binding of the N-terminal part of LciA at the same position as pediocin-like bacteriocins. As a result, the N-terminal β-sheet of LciA rotates toward the Vmotif domain, with the tip of the first β-hairpin as the pivot. In addition, the N-terminal β-sheet of LcnA is much longer than that of pediocin-like bacteriocins, because of which the starting point of the C-terminal region of LcnA leans more toward the Core domain. The other end of the LcnA helical region must make a sharp turn to prevent penetration into the Vmotif domain. The LcnA immunity protein, which is partly associated with plasma membrane, recognizes and strongly binds to the LcnA–man-PTS complex via H3 and H4 helices. The flexible C-terminal end then extends into the pore to block bacteriocin-induced membrane leakage, thereby presumably preventing LcnA-induced cell killing. In this study, we comprehensively elucidated the structural basis of the mechanisms of action and immunity of the class IId bacteriocin LcnA. Our findings may extend the design scope of bacteriocins beyond class IIa bacteriocins.
MATERIALS AND METHODS
Plasmid construction.
Maltose binding protein (MBP) coded DNA sequence was cloned into the vector pQLinkN (Addgene) with BamHI and NdeI (New England Biolabs) and followed by DNA sequence of mature LcnA from Lactococcus lactis with SalI and NotI (named pQN-MLcnA here). The His-x-6 tag was fused to the C terminus of ll-ManY/IIC, and then inserted into pQLinkN between the BamHI and NotI sites to produce the plasmid pQN-Y. Gene sequence of ll-ManZ/IID was cloned into pQLinkN with BamHI and NotI to obtain the plasmid pQN-Z. The coding sequence of flag tag was fused to the N terminus of LciA. The PCR product was cloned into pQLinkN with BamHI and SalI to generate the plasmid pQN-fLciA. All of these plasmids (pQN-MLcnA, pQN-Y, pQN-Z, and pQN-fLciA) were combined by ligation-independent cloning.
Expression and purification of the ternary complex.
The coexpression of LcnA–LciA–man-PTS was induced in E. coli strain C43 (DE3) by 0.5 mM isopropyl-β-d-thiogalactoside and 1 mM mannose, when the cell density reached OD600 of 1.2. The cells were harvested by centrifugation, and cell pellets were resuspended in lysis buffer (25 mM Tris-HCl [pH 8.0] and 150 mM NaCl), followed by disruption via sonication. The lysate was centrifuged at 12,000 rpm for 10 min, and the supernatant was ultracentrifuged at 41,100 rpm for 1 h. The membrane pellets thus obtained were collected and homogenized in lysis buffer plus 1 mM mannose, and then solubilized with 2% (wt/vol) n-dodecyl-β-d-maltoside (Anatrace) at 4°C for 2 h. The insoluble fraction was precipitated by ultracentrifugation (41,100 rpm) for 30 min at 4°C. The supernatant was collected and loaded onto Ni-NTA affinity resin (Qiagen) three times, and then washed with lysis buffer A plus 20 mM imidazole, 0.2% DDM and 1 mM mannose, followed by elution with lysis buffer A plus 250 mM imidazole, 0.2% DDM and 1 mM mannose. Proteins were purified using amylose resin (NEB) to remove the reductant bacteriocin-free ll-man-PTS. The protein was concentrated, and further applied to the Superose 6 Increase 10/300 GL column (GE Healthcare) re-equilibrated with buffer containing lysis buffer A, 1 mM mannose and 0.07% digitonin. The peak fractions were collected.
Sample preparation for cryo-electron microscopy and data acquisition.
For negative staining, an aliquot of 4 μL sample containing 0.01 mg/mL purified ternary complex was applied on a carbon film grid (Beijing XXBR Technology Co. Ltd.) after plasma glow discharge. The grids were stained with uranyl acetate (2% wt/vol) and stored at room temperature. The negatively stained sample was imaged on a Tecnai Spirit Bio TWIN microscope (Thermo Fisher) operating at 120 kV to verify sample quality.
Aliquots of 4 μL concentrated protein complex (2.8 mg/mL) were loaded onto glow-discharged holey carbon grids (Quantifoil Cu + 2 nm C R1.2/1.3, 300 mesh). The samples were blotted for 3.5 s and plunge-frozen in liquid ethane cooled by liquid nitrogen using Vitrobot Mark IV (Thermo Fisher) at 8°C and 100% humidity. The sample quality of the grids was verified with a Tecnai Arctica 200-kV electron microscope equipped with a K2 camera (Gatan). These grids with optimal ice thickness and particle density were transferred to a Titan Krios electron microscope (Thermo Fisher) operating at 300 kV and equipped with a Gatan Gif Quantum energy filter (slit width 20 eV). Micrographs were recorded using a K3 Summit counting camera (Gatan) in superresolution mode, with nominal magnification of ×810,000. AutoEMation (29) was used for fully automated data collection, and each movie had total accumulate exposure of 50 e−/A2 fractionated in 32 frames of exposure. Micrographs with a calibrated pixel size of 0.5371 Å were 2-fold binned resulting in the final pixel size of 1.0742 Å. Motion correction (MotionCorr2 [30]), CTF estimation (Gctf [31]), and micrograph inspection were automatically executed during data collection. Data collection statistics are summarized in Table 1.
Cryo-electron microscopy image processing.
All particles were automatically picked using Relion-3.0 (32) from 5,823 micrographs. Multiple rounds of 2D classifications (by Relion-3.0) were performed to eventually obtain 45,997,661 good particles. This number were further reduced to 1,529,899 by 3D classifications and high resolution reconstructions at 2.98 Å (EMAutoMask [33]). Reported resolutions were calculated on the basis of the FSC 0.143 criterion (34), and local resolution variations were estimated using ResMap (35).
Model building and structure refinement.
A reconstruction map was used to build a de novo model in EMBuilder (36), which was manually adjusted in COOT (37) and UCSF Chimera (38). Structure refinements were performed using PHENIX in real space with a secondary structure and geometry restraints to prevent structure overfitting (39). Table 1 shows the statistics for 3D reconstruction and model refinement. All structural figures were prepared using PyMol (40).
Bacteriocin assays.
The coding DNA sequence of truncated LcnA was amplified by PCR with pQN-MLcnA as the template. The fragments were cloned into pQlinkN with SalI and NotI. Overexpression of MBP-tagged LcnA and truncation variants was induced with 0.2 mM IPTG in E. coli strain C43 (DE3) at OD600 = 0.8. The cells were harvested after overnight growth at 18°C. Cells were mechanically lysed by sonication with lysis buffer B (50 mM MES pH 6.5, 150 mM NaCl). The pellets were centrifuged at 12,000 rpm for 1h at 4°C. The supernatant was collected and loaded onto Amylose affinity resin three times. The resin was washed with lysis buffer B, before MBP-tagged proteins were eluted using lysis buffer B plus 1% maltose.
Bacteriocin activity was assayed using plate diffusion bioassay. L. lactis NZ9000 (indicator organism) was grown to saturation in liquid media at 30°C. M17 soft agar containing the indicator organism was overlaid on plates; wells were made and filled with bacteriocin solutions at 2-fold dilutions. The plates were then incubated overnight and examined for growth inhibition zones.
Data availability.
The cryo-EM maps and the structures have been deposited to the Electron Microscopy Data Bank (EMD-34726) and the Protein Data Bank (PDB: 8HFS), respectively.
ACKNOWLEDGMENTS
We thank Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for kindly providing the cryo-EM facility support and the computational facility support on the cluster of Bio-Computing Platform. We also thank J. Lei, X. Li, F. Yang, and J. Wen for their technical support.
This work was supported by a grant from the National Natural Science Foundation of China (grant number 32171190).
R.L., J.D., and J.W. conceived the project. R.L., J.D., and Y.Z. designed and performed experiments. J.W. wrote the manuscript. All the authors read and approved the final manuscript.
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
Contributor Information
Jiawei Wang, Email: jwwang@tsinghua.edu.cn.
Charles M. Dozois, INRS Armand-Frappier Sante Biotechnologie Research Centre
REFERENCES
- 1.Urban-Chmiel R, Marek A, Stępień-Pyśniak D, Wieczorek K, Dec M, Nowaczek A, Osek J. 2022. Antibiotic resistance in bacteria—a review. Antibiotics (Basel) 11:1079. 10.3390/antibiotics11081079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haney EF, Mansour SC, Hancock RE. 2017. Antimicrobial peptides: an introduction. Methods Mol Biol 1548:3–22. 10.1007/978-1-4939-6737-7_1. [DOI] [PubMed] [Google Scholar]
- 3.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 4.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 5.Fimland G, Eijsink VGH, Nissen-Meyer J. 2002. Comparative studies of immunity proteins of pediocin-like bacteriocins. Microbiology (Reading) 148:3661–3670. 10.1099/00221287-148-11-3661. [DOI] [PubMed] [Google Scholar]
- 6.Ríos Colombo NS, Chalón MC, Navarro SA, Bellomio A. 2018. Pediocin-like bacteriocins: new perspectives on mechanism of action and immunity. Curr Genet 64:345–351. 10.1007/s00294-017-0757-9. [DOI] [PubMed] [Google Scholar]
- 7.Nissen-Meyer J, Rogne P, Oppegård C, Haugen HS, Kristiansen PE. 2009. Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by gram-positive bacteria. Curr Pharm Biotechnol 10:19–37. 10.2174/138920109787048661. [DOI] [PubMed] [Google Scholar]
- 8.Drider D, Fimland G, Héchard Y, McMullen LM, Prévost H. 2006. The continuing story of class IIa bacteriocins. Microbiol Mol Biol Rev 70:564–582. 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balandin SV, Sheremeteva EV, Ovchinnikova TV. 2019. Pediocin-like antimicrobial peptides of bacteria. Biochemistry (Mosc) 84:464–478. 10.1134/S000629791905002X. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez JM, Martínez MI, Kok J. 2002. Pediocin PA-1, a wide-spectrum bacteriocin from lactic acid bacteria. Crit Rev Food Sci Nutr 42:91–121. 10.1080/10408690290825475. [DOI] [PubMed] [Google Scholar]
- 11.Holo H, Nilssen O, Nes IF. 1991. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J Bacteriol 173:3879–3887. 10.1128/jb.173.12.3879-3887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tymoszewska A, Diep DB, Wirtek P, Aleksandrzak-Piekarczyk T. 2017. The non-lantibiotic bacteriocin garvicin q targets man-pts in a broad spectrum of sensitive bacterial genera. Sci Rep 7:8359. 10.1038/s41598-017-09102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tymoszewska A, Diep DB, Aleksandrzak-Piekarczyk T. 2018. The extracellular loop of Man-PTS subunit IID is responsible for the sensitivity of Lactococcus garvieae to garvicins A, B and C. Sci Rep 8:15790. 10.1038/s41598-018-34087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tymoszewska A, Walczak P, Aleksandrzak-Piekarczyk T. 2020. BacSJ-another bacteriocin with distinct spectrum of activity that targets man-PTS. Int J Mol Sci 21:7860. 10.3390/ijms21217860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heng NC, Burtenshaw GA, Jack RW, Tagg JR. 2007. Ubericin A, a class IIa bacteriocin produced by Streptococcus uberis. Appl Environ Microbiol 73:7763–7766. 10.1128/AEM.01818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maldonado-Barragán A, Alegría-Carrasco E, Blanco MDM, Vela AI, Fernández-Garayzábal JF, Rodríguez JM, Gibello A. 2022. Garvicins AG1 and AG2: two novel class IId bacteriocins of Lactococcus garvieae Lg-Granada. Int J Mol Sci 23:4685. 10.3390/ijms23094685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel V, Olari LR, Jachmann M, Reich SJ, Häring M, Kissmann AK, Rosenau F, Riedel CU, Münch J, Spellerberg B. 2022. The bacteriocin Angicin interferes with bacterial membrane integrity through interaction with the mannose phosphotransferase system. Front Microbiol 13:991145. 10.3389/fmicb.2022.991145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci USA 104:2384–2389. 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kjos M, Nes IF, Diep DB. 2009. Class II one-peptide bacteriocins target a phylogenetically defined subgroup of mannose phosphotransferase systems on sensitive cells. Microbiology (Reading) 155:2949–2961. 10.1099/mic.0.030015-0. [DOI] [PubMed] [Google Scholar]
- 20.Zhu L, Zeng J, Wang C, Wang J. 2022. Structural basis of pore formation in the mannose phosphotransferase system by pediocin PA-1. Appl Environ Microbiol 88:e0199221. 10.1128/AEM.01992-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu L, Zeng J, Wang J. 2022. Structural basis of the immunity mechanisms of pediocin-like bacteriocins. Appl Environ Microbiol 88:e0048122. 10.1128/aem.00481-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erni B. 2013. The bacterial phosphoenolpyruvate: sugar phosphotransferase system (PTS): an interface between energy and signal transduction. J the Iranian Chemical Society 10:593–630. 10.1007/s13738-012-0185-1. [DOI] [Google Scholar]
- 23.Liu X, Zeng J, Huang K, Wang J. 2019. Structure of the mannose transporter of the bacterial phosphotransferase system. Cell Res 29:680–682. 10.1038/s41422-019-0194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeckelmann J-M, Erni B. 2020. The mannose phosphotransferase system (Man-PTS) - Mannose transporter and receptor for bacteriocins and bacteriophages. Biochim Biophys Acta Biomembr 1862:183412. 10.1016/j.bbamem.2020.183412. [DOI] [PubMed] [Google Scholar]
- 25.Drew D, Boudker O. 2016. Shared molecular mechanisms of membrane transporters. Annual Review of Biochemistry 85:543–572. 10.1146/annurev-biochem-060815-014520. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Ludescher RD, Montville TJ. 1997. Electrostatic interactions, but not the YGNGV consensus motif, govern the binding of pediocin PA-1 and its fragments to phospholipid vesicles. Appl Environ Microbiol 63:4770–4777. 10.1128/aem.63.12.4770-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venema K, Haverkort RE, Abee T, Haandrikman AJ, Leenhouts KJ, de Leij L, Venema G, Kok J. 1994. Mode of action of LciA, the lactococcin A immunity protein. Mol Microbiol 14:521–532. 10.1111/j.1365-2958.1994.tb02186.x. [DOI] [PubMed] [Google Scholar]
- 28.Kristiansen PE, Persson C, Fuochi V, Pedersen A, Karlsson GB, Nissen-Meyer J, Oppegård C. 2016. Nuclear magnetic resonance structure and mutational analysis of the lactococcin A immunity protein. Biochemistry 55:6250–6257. 10.1021/acs.biochem.6b00848. [DOI] [PubMed] [Google Scholar]
- 29.Lei J, Frank J. 2005. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J Struct Biol 150:69–80. 10.1016/j.jsb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang K. 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su R, Zhang SY, Wang JW. 2019. Automatically generating mask for mapping out the density map in cryoEM single-particle reconstruction. Progress in Biochemistry and Biophysics 46:1020–1030. [Google Scholar]
- 34.Rosenthal PB, Henderson R. 2003. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333:721–745. 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Kucukelbir A, Sigworth FJ, Tagare HD. 2014. Quantifying the local resolution of cryo-EM density maps. Nat Methods 11:63–65. 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou N, Wang H, Wang J. 2017. EMBuilder: a template matching-based automatic model-building program for high-resolution cryo-electron microscopy maps. Sci Rep 7:2664. 10.1038/s41598-017-02725-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 39.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD. 2018. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74:531–544. 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeLano WL. 2002. The PyMOL molecular graphics system.
- 41.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. 2012. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40:D370–376. 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. 1996. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14:354–360. 376. 10.1016/S0263-7855(97)00009-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.00066-23-s0001.pdf, PDF file, 1.9 MB (1.9MB, pdf)
Data Availability Statement
The cryo-EM maps and the structures have been deposited to the Electron Microscopy Data Bank (EMD-34726) and the Protein Data Bank (PDB: 8HFS), respectively.