Abstract
The extension of life span driven by 40% caloric restriction (CR) in rodents causes trade-offs in growth, reproduction, and immune defense that make it difficult to identify therapeutically relevant CR-mimetic targets. We report that about 14% CR for 2 years in healthy humans improved thymopoiesis and was correlated with mobilization of intrathymic ectopic lipid. CR-induced transcriptional reprogramming in adipose tissue implicated pathways regulating mitochondrial bioenergetics, anti-inflammatory responses, and longevity. Expression of the gene Pla2g7 encoding platelet activating factor acetyl hydrolase (PLA2G7) is inhibited in humans undergoing CR. Deletion of Pla2g7 in mice showed decreased thymic lipoatrophy, protection against age-related inflammation, lowered NLRP3 inflammasome activation, and improved metabolic health. Therefore, the reduction of PLA2G7 may mediate the immunometabolic effects of CR and could potentially be harnessed to lower inflammation and extend the health span.
The beneficial effects of caloric restriction (CR) include enhanced longevity and reduced disease burden (1). However, 40% reduction of calories from a normal ad libitum state in many rodent studies showing life-span extension is associated with increased severity of viral and parasitic infections (2, 3), including mortality from polymicrobial sepsis (4). Impaired immunity may result because energetically expensive functions are dispensed with severe CR as energy resources are diverted toward somatic cell maintenance (5, 6). In addition to host defense, the resident immune system in every organ is required for integration of cellular metabolism, tissue repair, and function. Thus immunological trade-offs may make it difficult to identify clinically relevant beneficial CR mimetics (7). Furthermore, forced extreme CR in nonconsenting animals may elicit stress responses evidenced by increased production of glucocorticoids (5, 8), which can further compromise the immune system by causing thymocyte death and thymic in- volution (9). To address the relevance of CR on human physiology, the Comprehensive Assessment of Long-term Effects of Reducing Intake of Energy (CALERIE) clinical trial was designed to test the long-term effects of 2 years of moderate CR on physiology, aging biomarkers, and predictors of health span and longevity in healthy volunteers (10). The energy intake at baseline (ad libitum state) was evaluated by two consecutive 14-day measures of total daily energy expenditure (TDEE) using doubly labeled water (11). Average %CR over 6-month intervals was retrospectively calculated using the intake-balance method, which involves simultaneous measures of TDEE using doubly labeled water together with changes in body composition (11). This study established that in free-living conditions, humans achieve ~14% sustained CR for 2 years (10-12). This level of voluntary CR in humans in free-living conditions is much lower than the forced 25 to 40% restriction of calories in laboratory animals and may engage unique mechanisms to maintain homeostasis.
Mice on life-long 40% CR maintain thymic lymphopoiesis into late life and have a diverse T cell receptor (TCR) diversity (13), whereas 14% CR in humans reduces the number of circulating lymphocytes and proinflammatory cytokines in the blood (14). Aging of thymus precedes aging of other organs, a process characterized by increased thymic lipid accumulation and loss of T cell production (15). We therefore investigated thymic function at baseline and after 2 years of sustained CR in middle-aged healthy CALERIE participants (Fig. 1A and fig. S1, A to C) using magnetic resonance imaging (MRI) and quantification of signal-joint T cell receptor excision circles (sjTRECs) in blood, which are episomal DNA by-products of TCR recombination and are indicators of recently produced T cells from thymus. Compared with baseline, sustained CR for 2 years significantly (P < 0.05) increased thymic mass (Fig. 1B) as well as total thymic volume in study participants (Fig. 1C). The control participants showed no significant change in thymic volume from baseline to year two (fig. S2A). Compared with baseline, 2 years of CR significantly increased the recent thymic emigrant cells as measured by the presence of sjTRECs in CD4 (Fig. 1D) and CD8 cells (Fig. 1E) in the majority of female (n = 22 and 20, respectively) and male (n = 5 and 5, respectively) study participants. We conducted RNA sequencing of peripheral blood CD4 T cells from study participants to measure the transcriptional changes. However, principal component analysis (PCA) (Fig. 1F) and multidimensional scaling (MDS) analysis (fig. S2C) revealed no effect of CR on gene expression in CD4 T cells. These data may indicate that 14% CR in healthy humans activates a tissue-protective immunometabolic program that can enhance thymic function without altering the transcriptome of CD4 T cells. Reduction in caloric intake induces a decrease in glucose utilization and a switch to fatty acid oxidation and lipolysis (16). Accordingly, the participants in the CALERIE-II study experienced reduction of fat mass (11). Given that adipose tissue also contains a resident immune system that controls inflammation (6), we measured the impact of CR on gene expression in the adipose tissue biopsies. PCA and MDS analysis of whole-transcriptome gene expression in abdominal subcutaneous adipose tissue of study participants revealed that com- pared with baseline, 1 year of CR altered the transcriptome, and this difference was maintained after 2 years of CR (Fig. 1G and fig. S2C). In adipose tissue after CR (Fig. 1H), 233 genes were differentially up-regulated and 131 genes were down-regulated relative to baseline at the year one and year two time points (Fig. 1I). No significant changes in gene expression were found comparing year one and year two after CR (Fig. 1G).
Fig. 1. CR in humans improves thymic function and remodels adipose transcriptome.
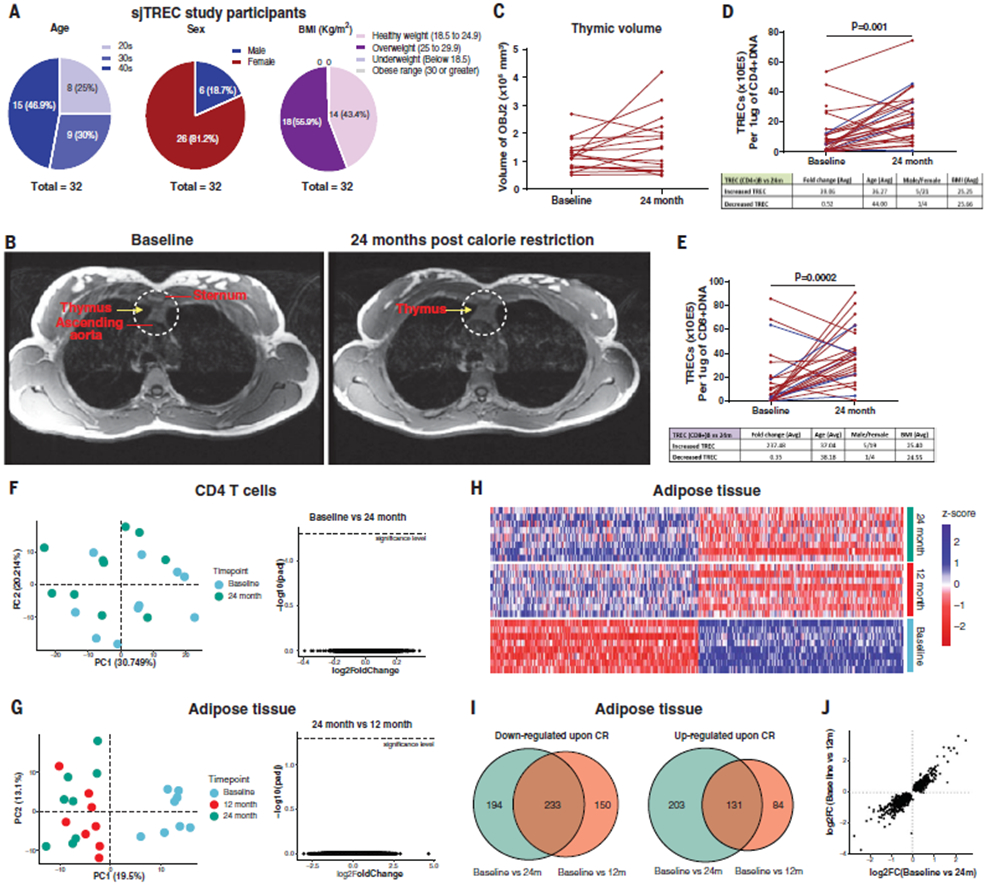
(A) Participant information for the sjTREC study (n = 32). (B) Representative MRI examination image of a female participant performed with the subject head first and supine in a 3 T full-body scanner at the baseline (left) and after 2 years of CR (right). The region of interest depicting the thymus is highlighted by yellow arrows in dotted circle after the fat saturation sequence was applied. (C) Thymic volume of female study participants analyzed by MRI at baseline and after 24 months of CR. (D and E) sjTREC analysis results of CD4 (D) and CD8 (E) T cells at baseline after 2 years of CR. Red line indicates TREC in females; blue line indicates TREC in males. (F), PCA of transcriptional profile of blood CD4 T cells at baseline and after 2 years of CR. Volcano plot depicts the results of differential gene expression analysis of CD4 T cells between baseline and after 2 years of CR. (G) PCA of RNA sequencing of adipose tissue at baseline and after 1 and 2 years of CR (n = 8). Volcano plot depicts the results of differential analysis of subcutaneous adipose tissue between 1 and 2 years of CR (right). Each dot represents a gene. (H) Heatmap of significant differentially expressed genes between baseline and either 1 or 2 years of CR in adipose tissue. (I) Venn diagram illustrating overlaps between genes found to be differential [false discovery rate (FDR), 5%] between baseline and either 1 or 2 years of CR. Overlapping area indicates the genes that are differentially expressed after both 1 and 2 years of CR. (J) Comparison of the log2-scaled fold change between baseline and 1 or 2 years of CR.
Adaptation to CR requires rewiring of immuno- metabolic inputs that control adipose tissue function, which in turn may drive systemic health span effects in humans (6). The top 20 up- and down-regulated genes in human adipose tissue after 14% CR identified transcripts previously not highlighted in rodent studies (Fig. 2A). Transcripts altered at year one of CR (PLA2G7, SPARC, CA3, PLIN5, ACVR1C, CALCRL, PDE3A, DPT, EGFL6, NAMPT, and PPARA) did not change under ad libitum fed conditions for 1 year (fig. S2B). Changes in gene expression in adipose tissue after CR were similar to those observed after bariatric surgery (fig. S3A). Similar gene expression patterns were also observed in a dataset describing twin pairs discordant for physical activity (Fig. 2B), providing an example of another lifestyle intervention that can reprogram the adipose tissue transcriptome.
Fig. 2. CR in humans reveals anti-aging immunometabolic checkpoints.
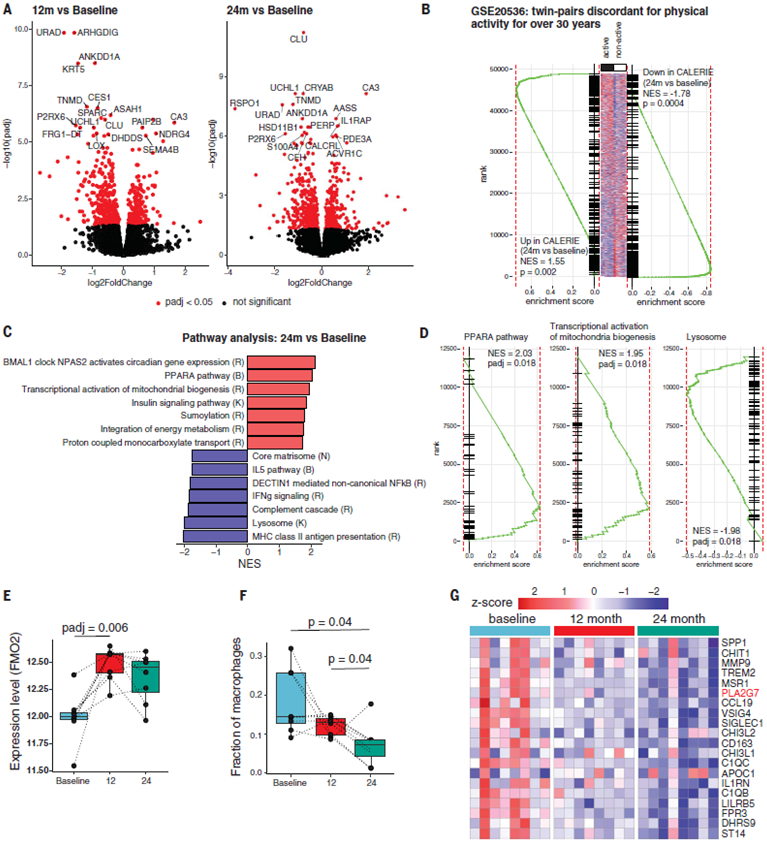
(A) Volcano plots depicting the results of differential gene expression analysis of adipose tissue. Each dot represents a gene. Top 18 significantly up- or down-regulated genes at 1 year (left) or 2 years (right) of CR compared with baseline (n =8). (B) Gene set enrichment analysis (GSEA) of genes up- and down-regulated after 2 years of CR in a signature describing differences between active and nonactive twins (GSE20536). (C) Selected pathways significantly (FDR, 5%) regulated by 2 years of CR based on GSEA. Red and blue bars indicate pathways up- and down-regulated by CR, respectively. Letters correspond to the source of pathway gene set. K, KEGG; R, Reactome; B, BioCarta. (D) Enrichment curves for selected significantly regulated pathways. y-axis denotes rank of the gene in list ordered by log2FC. (E) Changes in the expression level of FMO-2 after CR. Adjusted P values (padj) were calculated in the differential gene expression analysis. (F) Box plots showing estimated macrophage fractions after deconvolution of adipose transcriptome in adipose tissue at baseline and 1 and 2 years after CR. P values were calculated using the paired t test. (G) Expression of genes specific to adipose tissue–resident macrophages shown at baseline and 1 and 2 years after CR. NES corresponds to the normalized enrichment score.
We investigated whether 2 years of CR in humans regulated core pathways previously identified from model organisms that are implicated in nutrient sensing, inflammation, and longevity (1). CR (i) increased mitochondrial biogenesis and the peroxisome proliferator activated receptor (PPAR-α)–driven fatty acid oxidation, including increased expression of components of insulin signaling, suggesting enhanced insulin sensitivity (Fig. 2, C and D); (ii) up-regulated proton-coupled transport of monocarboxylates such as lactate, pyruvate, and ketone bodies (Fig. 2C); (iii) induced the BMAL1 clock pathway in human adipose tissue and sumoylation (Fig. 2C), which are implicated in mediating some of CR’s pro-longevity effects (17, 18); (iv) decreased the expression of components of innate immune activation such as antigen-processing pathways in lysosomes, complement cascade, and noncanonical nuclear factor-kB signaling; (v) decreased the expression of components of extracellular matrix deposition (Fig. 2, C and D) that may reflect lower inflammation; and (vi) increased the gene expression of flavin containing dimethylaniline monoxygenase 2 (FMO-2), an enzyme for which increased activity is associated with enhanced life span in Caenorhabditis elegans (19). FMO proteins (E.C.1.14.13.8 FMO-1 to FMO-5) may have overlapping functions in xenobiotic metabolism, but the relevance of FMO2 to human physiology is unclear (20) (Fig. 2E).In addition, the deconvolution of RNA-sequencing data from whole adipose tissue revealed an anti-inflammatory tissue response with a reduction of macrophage-specific transcripts (Fig. 2F). These data indicate that CR in humans elicits anti-inflammatory pathways that may improve adipose tissue metabolism.
In rodents maintained at the standard sub-thermoneutral housing temperature, CR decreases core body temperature (CBT) to elicit mitochondrial uncoupling, white adipose tissue browning, and thermogenesis to defend the CBT setpoint (1). CALERIE participants in thermo-neutral settings maintained their CBT within the normal physiological range (fig. S3B). CR did not affect the transcriptional signatures of adipose tissue thermogenesis (21) (fig. S3Cand Fig. 2D). The CALERIE participants also did not show any change in uncoupling protein-1 (UCP1) expression in adipose tissue (fig. S3D). Although a general signature characteristic of brown adipose tissue from the Mouse Gene Atlas (21) was up-regulated in human CR samples (fig. S3E, lower panel, and Fig. 2D), this effect was not associated with changes in classical drivers of mitochondrial uncoupling. Thus, in healthy humans, CR does not appear to trigger adaptive thermogenesis and adipose tissue browning.
CR is known to lower inflammation (1, 6, 14). We investigated the impact of CR on gene expression in myeloid cells using bioinformatic deconvolution of adipose tissue RNA-sequencing data. Several proinflammatory-like genes and many transcripts of unknown function in myeloid cells were specifically inhibited after CR (Fig. 2G). Among the top six genes inhibitedatboth1and2yearsofCR.The phospholipase belonging to group VII A platelet activating factor acetylhydrolase (PLA2G7) is secreted by macrophages (22). PLA2G7 can degrade platelet activating factor and generate lysophosphatidylcholine (LysoPC) by degrading OxPAPC (oxidized 1-palmitoyl-2-arachidonyl-sn-glycero-3phosphatidylcholine) (22). Given that CR inhibits PLA2G7 and OxPAPC regulates inflammasome activation, we tested whether depletion of PLA2G7 influences inflammation. In mice, CRISPR-mediated deletion of Pla2g7 (fig. S4A) resulted in its efficient ablation in activated macrophages (Fig. 3A). The control and Pla2g7 knockout (KO) mice were born at normal Mendelian frequencies and displayed similar body weight on a chow diet (Fig. 3B and fig. S4B). PLA2G7-deficient animals were partially protected from high-fat-diet (HFD)–induced weight gain and increased fat mass (Fig. 3B). In addition, compared with their littermate controls, the Pla2g7 KO mice were partially protected from hepatic steatosis, with reduced expression of interleukin (IL)-1β and caspase-1, which are implicated in controlling obesity-induced lipid metabolism and liver dysfunction (23) (Fig. 3, D and E). The reduced weight gain in Pla2g7 KO mice corresponded with increased energy expenditure (Fig. 3, F and G). Analysis of covariance (ANCOVA) further supported that the reduction in PLA2G7 increases TDEE (Fig. 3H). PLA2G7-deficient animals had increased adipose tissue lipolysis as measured by increased concentration of glycerol and free fatty acids (Fig. 3, I and J). Consistent with improved adipose tissue metabolism, the PLA2G7-deficient M1-like macro-phages had lower expression of the proinflammatory cytokines IL-6, IL-12, and tumor necrosis factor a (TNFα) (Fig. 3K). Middle-aged PLA2G7-deficient mice had lower serum concentrations of IL-1b after exposure to lipopolysaccharide (LPS)(Fig. 3L).
Fig. 3. Reduction of PLA2G7 serves as anti-adiposity CR mimic.
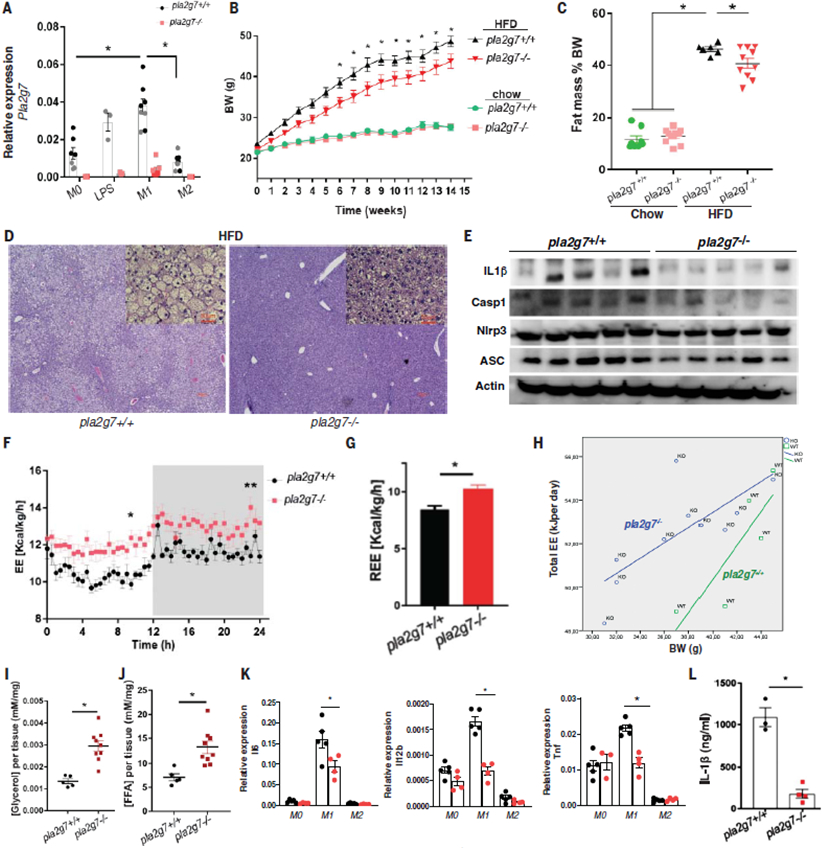
(A) Expression of Pla2g7 in nonpolarized (M0) (n = 7 and 7, respectively), LPS-treated (n = 3 and 3, respectively), M1-polarized (M1) (n = 8 and 7, respectively), and M2-polarized (M2) (n = 8 and 7, respectively) bone marrow–derived macrophages (BMDMs) generated from wild-type (WT) and PLA2G7-deficient mice. (B) Body weight change of WT and Pla2g7 KO mice provided with chow diet (n = 8 and 11, respectively) or HFD (n = 8 and 16, respectively) for 14 weeks. (C) Fat composition of WT and Pla2g7 KO mice (n = 8, 11, 6, and 10, respectively). (D) Representative images of livers from WT and Pla2g7 KO mice with H&E staining. Large images are 10×, small insets are 20×, and each scale bar indicates 50 mm. (E) Immunoblot analysis of inflammasome proteins (IL1B-p17, Caspase 1, NLRP3, and ASC) from livers in WT and Pla2g7-deficient mice fed with HFD. (F and G) Total daily energy expenditure (TDEE) (F) and resting energy expenditure (REE) (G) of pla2g7+/+ and pla2g7−/− mice fed with HFD (n = 6 and 10, respectively). (H) ANCOVA of body weight and EE of WT and Pla2g7 KO mice fed with HFD (n = 6 and 10, respectively). (I and J) Lipolysis assay of visceral adipose tissue from WT and Pla2g7 KO mice fed HFD to detect the release of glycerol (n = 5 and 9, respectively) (I) and free fatty acid (n = 5 and 9, respectively) (J). (K) Quantitative polymerase chain reaction (qPCR) showing the relative expression of the inflammatory genes Il-6, Il-12b, and Tnf to Gapdh in non-polarized (M0) (n = 5 and 4, respectively) and polarized (M1 and M2) BMDMs (n = 5 and 4, respectively) generated from WT and Pla2g7 KO mice. (L) Serum level of IL-1β in LPS-treated WT and Pla2g7 KO mice (12 months). Error bars represent the mean ± SEM. Statistical significance was calculated by a two-tailed unpaired Student’s t test [(G), (I), (J),and(L)] and two-way ANOVA and Holm-Sidak post hoc tests for multiple hypothesis [(A), (B),(C), and (K)]. *P <0.05; **P < 0.005; ***P < 0.001; ****P <0.0001.
Chronic systemic inflammation in aging and Nlrp3 inflammasome activation is associated with age-related functional decline (24). Twenty-four-month-old Pla2g7−/− mice had decreased circulating concentrations of TNF-α and IL-1β without any significant change in IL-6, chemokine C-X-C motif ligand −1 (CXCL1), chemokine C-C motif ligand 2 (CCL2), or C-C motif chemokine 11 (CCL11, also called eotaxin). Figure 4A). Furthermore, aged Pla2g7−/− mice displayed decreased gene expression of IL-1β, increased PPARa mRNA in adipose tissue macrophages, and reduced caspase-1 protein (Fig. 4B), with lower Il-1b and Casp-1 mRNA expression in aged visceral adipose tissue (Fig.4C). Depletion of PLA2G7 in old mice increased the expression of the Ppara and Acvr1c genes (Fig. 4D), which are involved in mitochondrial fatty acid oxidation and were increased in human adipose tissue after CR. Altered abundance of visceral adipose tissue resident immune cells is implicated in inflammaging (6). Loss of PLA2G7 reduced the abun-dance of the B cell, T cell, and macrophage subsets (CD11c+F4/80+CD11b+), which impair adipose metabolism (6), and increased abun-dance of CD206+ F4/80+CD11b+ macrophages and eosinophils (Fig. 4E and fig. S4D), which promote tissue remodeling and responses to metabolic stresses (6).
Fig. 4. Inhibition of PLA2G7 protects against inflammaging and thymic lipoatrophy.
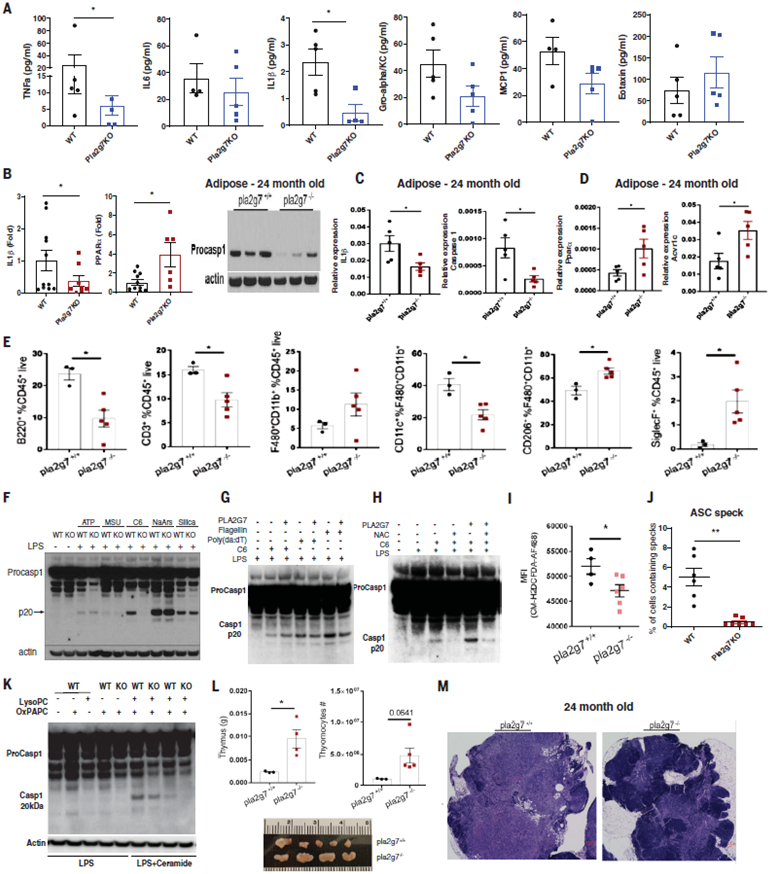
(A) Circulating proinflammatory cytokines (TNF-α, IL-6, IL-1b, Gro-alphaK/C, MCP-1, and Eotaxin) from serum in 24-month-old control and Pla2g7−/− mice (B) qPCR analysis of relative expression of Il-1b and Ppara compared with Gapdh in sorted F4/80+ cells from visceral adipose tissues in 24-month-old control and Pla2g7 KO mice and Caspase 1 protein expression in t he adipose tissue of 24-month-old control and Pla2g7−/− deficient mice (n = 3 and 3, respectively). (C and D) qPCR analysis of the proinflammatory genes Il-1b and Casp1 in the visceral adipose tissue of control and Pla2g7 KO mice. (C and D) The CR-regulated fatty acid oxidation inducers Ppara and Acvr1c in subcutaneous adipose tissue (SAT) of 24-month-old control (C) and Pla2g7 KO mice (n = 5, 5) (D). (E) FACS analysis of proportions of different immune cell populations in the adipose tissue of 24-month-old WT and Pla2g7 KO mice. (F) Representative immunoblot analysis of inflammasome activated by multiple NLRP3 activators in LPS-primed BMDMs from WT and Pla2g7-deficient mice. Inactive caspase-1 (48 kDa), enzymatically active caspase-1 (P20, 20kDa). (G) Western blot analysis of caspase-1 in BMDMs treated with recombinant PLA2G7 (1 mμ/ml) in the presence of flagellin and poly(dA:dT) to activate the NLRC4 and AIM2 inflammasomes. (H) Inflammasome activation measured by caspase-1 Western blot of BMDMs activated with LPS and ceramide and treated in the presence of recombinant PLA2G7 and N-acetyl-cysteine (NAC) (representative of three experiments). (I) Mean fluorescence intensity (MFI) depicting mitochondrial ROS after ceramide-induced NLRP3 inflammasome activation of control and Pla2g7-deficient BMDMs. (n = 4 and 6, respectively). (J) Quantification of ASC speck formation in BMDMs from WT and Pla2G7 mice activated by LPS and ceramide. (K) Caspase 1 expression from BMDM from control and Pla2g7-deficient mice activated by LPS and ceramide in the presence of OxPAPC and LysoPC. (L) Characterization of thymic involution of 24-month-old control WT and Pla2g7 KO mice. Shown are thymus weight (top left)(n = 5), cellularity (top right) (n = 5), and thymic size (bottom) (n =5,4). (M) H&E staining of representative thymi from 24-month-old control mice showing ectopic adipocytes and loss of corticomedullary junction in control mice and preservation of cellularity in Pla2g7 KO mice. Error bars represent the mean ± SEM. Two-tailed unpaired t tests were performed for statistical analysis. *P <0.05.
The PLA2G7-deficient macrophages, when primed with LPS and activated by ceramides, showed reduced activation of caspase-1 (Fig. 4, F to I) and Asc speck formation (Fig. 4J and fig. S4C), which is an indicator of inflammasome assembly (23). Consistent with the role of PLA2G7 in lipid metabolism, the anti-inflammasome effects were specific to ceramides and not to other NLRP3 activators such as ATP, urate crystals, sodium arsenite, and silica particles (Fig. 4F). Neither gain of PLA2G7 (Fig. 4G) nor loss of its function in macrophages (fig. S4E) affected NLRC4 or AIM2 inflammasome activation. PLA2G7-driven ceramide-induced Nlrp3 inflammasome activation was dependent on reactive oxygen species (ROS) (Fig. 4H), and PLA2G7-deficient macrophages showed reduced ROS concentrations when activated by ceramides (Fig. 4I and fig. S4F). When exposed to oxPAPC and LysoPC and primed with LPS and ceramide, the PLA2G7-deficient macro-phages showed decreased inflammasome activation (Fig. 4K). The macrophages exposed to OXPAPC or LysoPC together with recombinant PLA2G7 did not activate the inflammasome (fig. S4G). We suspect that in a tissue microenvironment such as adipose tissue, where macrophages have to process a complex mixture of lipids, PLA2G7 might be an important regulator of the inflammasome. Nlrp3 inflammasome activation by ceramide is also implicated in age-related thymic involution (24). We tested whether loss of PLA2G7 mimicked the beneficial effects of CR on age-related immunological parameters such as thymic lipoatrophy. Twenty-four-month-old PLA2G7-deficient mice (analogous to ~70-year-old humans) had larger thymi and higher thymocyte abundance (Fig. 4L) and were protected from age-related thymic involution (Fig.4M). We propose that reduction of PLA2G7 caused by CR in humans might contribute to better adipose tissue metabolism, lower inflammation, and reduced thymic lipoatrophy.
Although studies in animal models reported divergent effects on immune response, including susceptibility to infections, (2-4), the CALERIE-II analyses indicate that CR in humans over 2-year period may not decrease immunological function. Moreover, 14% sustained CR in humans reduced ectopic lipid and enhanced thymic function in a subset of healthy middle-aged humans. The data from this human study are also relevant regarding controversies emerging from animal studies that questioned CR’s effects on health and inflammation (7). Collectively, our findings demonstrate that sustained CR in humans activates a core transcriptional program that promotes immune function, reduces inflammation, and re-vealsPLA2G7asoneofthepotential mechanisms to mimic the beneficial effects of CR.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the study participants, investigators, and clinical support staff involved in the CALERIE study and D. K. Ingram and T. Horvath for pre submission review of this manuscript.
Funding:
This research was supported in part by the National Institutes of Health (NIH grants AG031797, AG045712, P01AG051459, and AR070811 to V.D.D.); the Glenn Foundation for Medical Research (V.D.D.); Cure Alzheimer’s Fund (V.D.D.); and the Aging Biology Foundation (M.N.A.). The CALERIE study was funded by the National Institute on Aging (grants U01AG022132, U01AG020478, U01AG020487, and U01AG020480).
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: The datasets generated in this study are available from the corresponding author on reasonable request. The RNA-sequencing data have been uploaded at SYNAPSE (www.synapse.org) with Synapse ID syn23667189.
REFERENCES AND NOTES
- 1.Anderson RM, Le Couteur DG, de Cabo R, Gerontol J. A Biol. Sci. Med. Sci 73,1–3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kristan DM, Aging Cell 6, 817–825 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Gardner EM, Gerontol J.ABiol.Sci.Med.Sci 60,688–694 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Sun D, Muthukumar AR, Lawrence RA, Fernandes G, Clin. Diagn. Lab. Immunol 8, 1003–1011 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirkwood TL, Kapahi P, Shanley DP, Anat J. 197, 587–590 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee AH, Dixit VD, Immunity 53, 510–523 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Speakman JR, Mitchell SE, Mol. Aspects Med 32, 159–221 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Patel NV, Finch CE, Neurobiol. Aging 23, 707–717 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Poetschke HL et al. , Carcinogenesis 21, 1959–1964 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Rochon J et al. , J. Gerontol. A Biol. Sci. Med. Sci 66,97–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravussin E et al. , J. Gerontol. A Biol. Sci. Med. Sci 70, 1097–1104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraus WE et al. , Lancet Diabetes Endocrinol. 7, 673–683 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Youm YH, Dixit VD, J. Immunol 183, 3040–3052 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meydani SN et al. , Aging (Albany NY) 8, 1416–1431 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixit VD, Curr. Opin. Immunol 22, 521–528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruss MD, Khambatta CF, Ruby MA, Aggarwal I, Hellerstein MK, Am. J. Physiol. Endocrinol. Metab 298, E108–E116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel SA, Chaudhari A, Gupta R, Velingkaar N, Kondratov RV, FASEB J. 30, 1634–1642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moll L et al. , eLife 7, e38635 (2018).30403374 [Google Scholar]
- 19.Leiser SF et al. , Science 350, 1375–1378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cashman JR, Zhang J, Annu. Rev. Pharmacol. Toxicol 46, 65–100 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Shinoda K et al. , Nat. Med 21, 389–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stafforini DM, Cardiovasc. Drugs Ther 23,73–83 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Swanson KV, Deng M, Ting JP, Nat. Rev. Immunol 19, 477–489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youm YH et al. , Cell Metab. 18, 519–532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


