ABSTRACT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the viral pathogen responsible for the worldwide coronavirus disease 2019 (COVID-19) pandemic. The novel SARS-CoV-2 ORF8 protein is not highly homologous with known proteins, including accessory proteins of other coronaviruses. ORF8 contains a 15-amino-acid signal peptide in the N terminus that localizes the mature protein to the endoplasmic reticulum. Oligomannose-type glycosylation has been identified at the N78 site. Here, the unbiased molecular functions of ORF8 are also demonstrated. Via an immunoglobulin-like fold in a glycan-independent manner, both exogenous and endogenous ORF8 interacts with human calnexin and HSPA5. The key ORF8-binding sites of Calnexin and HSPA5 are indicated on the globular domain and the core substrate-binding domain, respectively. ORF8 induces species-dependent endoplasmic reticulum stress-like responses in human cells exclusively via the IRE1 branch, including intensive HSPA5 and PDIA4 upregulation, with increases in other stress-responding effectors, including CHOP, EDEM and DERL3. ORF8 overexpression facilitates SARS-CoV-2 replication. Both stress-like responses and viral replication induced by ORF8 have been shown to result from triggering the Calnexin switch. Thus, ORF8 serves as a key unique virulence gene of SARS-CoV-2, potentially contributing to COVID-19-specific and/or human-specific pathogenesis.
IMPORTANCE Although SARS-CoV-2 is basically regarded as a homolog of SARS-CoV, with their genomic structure and the majority of their genes being highly homologous, the ORF8 genes of SARS-CoV and SARS-CoV-2 are distinct. The SARS-CoV-2 ORF8 protein also shows little homology with other viral or host proteins and is thus regarded as a novel special virulence gene of SARS-CoV-2. The molecular function of ORF8 has not been clearly known until now. Our results reveal the unbiased molecular characteristics of the SARS-CoV-2 ORF8 protein and demonstrate that it induces rapidly generated but highly controllable endoplasmic reticulum stress-like responses and facilitates virus replication by triggering Calnexin in human but not mouse cells, providing an explanation for the superficially known in vivo virulence discrepancy of ORF8 between SARS-CoV-2-infected patients and mouse.
KEYWORDS: SARS-CoV-2, ORF8, endoplasmic reticulum stress, Calnexin
INTRODUCTION
Similar to that of other coronaviruses (CoVs), the SARS-CoV-2 genome is composed of 9 open reading frames (ORFs), including ORF1, spike, ORF3, envelope, membrane, ORF6, ORF7, ORF8, and nucleocapsid (1, 2). ORF1 is directly translated by genomic positive RNA to form polyprotein 1a (pp1a) and pp1ab, which are posttranslationally processed into individual nonstructural proteins (NSPs). The latter 8 ORFs encoding 4 structural proteins (spike, envelope, membrane, and nucleocapsid proteins), which are indispensable for viral capsid and nucleic acid packaging, and 4 accessory proteins, which are not commonly regarded to be independently critical for virus replication, are translated from a 3′-nested set of subgenomic mRNA (sgmRNA) species. Most SARS-CoV-2 genes are genetically homologous to those of SARS-CoV. However, the ORF8 protein of SARS-CoV-2 is not highly homologous with ORF8 proteins of other CoVs, including those of two highly pathogenic viruses, SARS-CoV and Middle East respiratory coronavirus (MERS-CoV), or other known proteins. Although SARS-CoV-2 ORF8 has been recently characterized on the basis of its immune antagonistic properties (3–5), with the structure having been identified shortly after the beginning of the SARS-CoV-2 epidemic (6), the mechanistic details of its properties and functions have not been well established.
Similar to glycoproteins in other viruses, the synthesis and proper folding of the CoV spike protein on the host endoplasmic reticulum (ER) depend on various molecular chaperones, such HSPA5 (BiP) and/or Calnexin/Calreticulin, and other factors, including protein disulfide-isomerase (PDI) family members, for proper disulfide bond formation (7–9). Fully mature spike is translocated to the ER and then transitions through the ER-Golgi intermediate compartment, where it interacts with other structural proteins and newly produced genomic RNA, resulting in progeny virus budding (10). In addition to glycoprotein folding and virus budding, the ER is functionally necessary for CoV RNA synthesis: the expression of several NSPs modifies the ER and other host membrane organelles to generate various kinds of viral RNA-synthesizing organelles, which are protective microenvironments for viral genomic RNA replication and sgmRNA transcription (11, 12); thus, robust viral protein and RNA synthesis could induce ER stress in the host cell.
In mammalian cells, three branches of the unfolded protein response (UPR) are activated to alleviate ER stress. Each branch is defined by a class of transmembrane ER resident signaling components, namely, activating transcription factor 6 (ATF6), double-stranded RNA-activated protein kinase–like ER kinase (PERK), and inositol requiring enzyme 1 (IRE1) (13, 14). Specifically, (i) cellular sensing of ER stress results in ATF6 cleavage, and then its N-terminal fragment, ATF6(N), moves into the nucleus to activate UPR target genes; (ii) when activated by ER stress, PERK directly phosphorylates the α-subunit of eukaryotic initiation factor 2 (eIF2α), leading to global mRNA translational attenuation, which alleviates ER stress; (iii) stress-activated IRE1 splices X-box binding protein 1 (XBP1) mRNA and encodes a basic leucine zipper transcription factor named spliced XBP1 (sXBP1), which upregulates UPR target genes, including those functioning in ER-associated protein degradation (ERAD), such as ER degradation enhancing α-mannosidase-like protein (EDEM), those functioning in folding proteins, such as the PDI family, and heat shock protein (HSP) family member HSPA5. Here, we demonstrate that SARS-CoV-2 ORF8 activates ER stress-like responses through the IRE1 branch and promotes virus replication via its interaction with Calnexin.
RESULTS
ORF8 is an oligomannose-type N-glycosylated protein located in the ER.
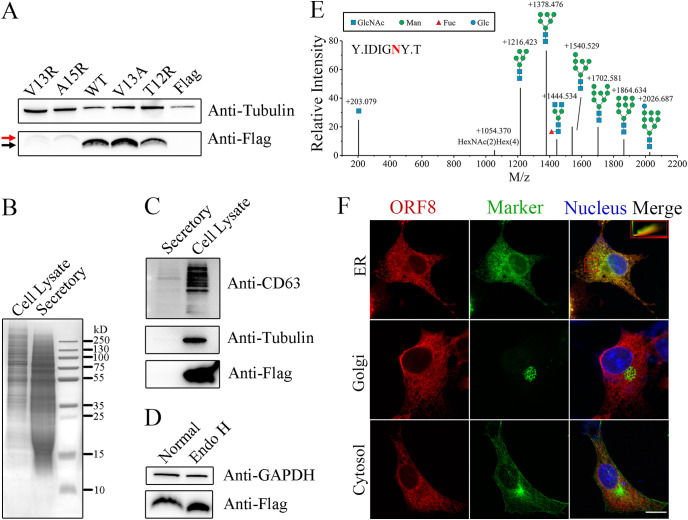
The signal peptide of ORF8 is predicted to include the N-terminal amino acids in positions 1 to 15, using SignalP (Fig. S1A). In general, a signal peptidase tends to recognize the −1 and −3 uncharged indispensable amino acid residues of a cleavage site (15), which have been predicted to correspond to V13 and A15 in ORF8. To experimentally verify the potential signal peptide, we constructed V13R and A15R mutants based on wild type (WT), full-length ORF8 with a C-terminal flag (ORF8-Flag), in which the positive residues commonly block signal peptidase cleavage. A slower mobility shift compared to that of the mature WT protein clearly demonstrates the mutant signals (Fig. 1A); however, mutant expression seems to be largely attenuated, even though we loaded more protein (as demonstrated by the level of the internal reference tubulin), probably due to improper folding, which leads to an unstable structure. In contrast, the mutation in which the indispensable residue is replaced with uncharged amino acids (V13A) and another mutation in which a nonessential residue is replaced with a positive residue (T12R) do not lead to a shift. Thus, the signal peptide of ORF8 is proposed to comprise the N-terminal 1 to 15 residues. Furthermore, ORF8-Flag protein obtained by anti-flag immunoprecipitation was intensively analyzed by mass spectrometry. The identified peptides display almost full coverage, except for sites in the extreme-most N-terminal positions (Fig. S1B). Although residues 9 to 15 are included in few identified peptides, most of the identified sites are located in the 16~121 region of ORF8, as well as the C-terminal Flag tag, and therefore, these sites are regarded as the mature protein. ORF8 purified from E. coli has been thought to possess an interdisulfate bond and form a homodimer (6); however, ORF8 fails to form a stable disulfate bond-dependent dimer in human cells (Fig. S2A).
FIG 1.
Molecular characteristics of the ORF8 protein. (A) The ORF8 protein contains a signal peptide consisting of 15 amino acids at the N terminus. WT ORF8 and mutants with single amino acid mutations, including V13R, A15R, V13A and T12R, were expressed in 293T cells with mock expression (Flag). Uncleaved preproteins containing signal peptides (red arrow) and mature cleaved proteins (black arrow) were identified by western blot (WB) after tricine gel electrophoresis. (B) Secretory proteins are successfully concentrated. ORF8 was expressed in 293F cells, and both concentrated secretory and cell lysate proteins (10 μg) were analyzed by SDS–PAGE with Coomassie blue staining. (C) ORF8 is an intracellular protein. The samples in (B) were analyzed by WB. (D) ORF8 is N-glycosylated. Lysates of ORF8-overexpressing 293T cells were deglycosylated via Endo H treatment, and both treated (Endo H) and untreated (Normal) lysates were analyzed by WB after tricine gel electrophoresis. (E) The glycosylation of ORF8 N78 is mostly of the oligomannose type. ORF8 expressed in 293T cells was immunoprecipitated and analyzed by mass spectrometry. The glycosylation of an N78-containing peptide (“Y.IDIGNY.T” with N78 in red) is identified; the structure and m/z are indicated above each of the relative intensity bars. (F) Subcellular localization of ORF8. COS-7 cells overexpressing ORF8-Flag were analyzed by multiimmunofluorescence using anti-Flag and respective organelle marker antibodies. Left column: ORF8-Flag signals in red; middle column: organelle signals in green; right column: merged signals of the left and the middle with nucleus in blue. The colocalization efficiency of ORF8 and an ER marker (in the chosen area is marked with a red square) is indicated in the upper right rectangle.
Proteins with signal peptides are destined to enter the secretory pathway; alternatively, they can be translocated to certain membrane apparatuses. To determine whether ORF8 is secreted, we expressed ORF8-Flag in 293F cells, collected the cell lysate and concentrated the extracellular secretions in the medium. Although extracellularly secreted proteins are successfully concentrated and include proteins from ~12 kDa to those larger than 250 kDa, only the cell lysate contains a detectable ORF8-Flag signal (Fig. 1B and C), in contrast to CD63 in exosomes (16).
The N78 site of ORF8 shows an N-X-T sequon and therefore is predicted to be N-glycosylated. In addition to the 7 cysteine modifications introduced by the sample treatment procedure of mass spectrometry, another modification is detected at the N78 site (Fig. S1B). Via Endo H glycosidase treatment, deglycosylated ORF8-Flag demonstrate a faster mobility shift (Fig. 1D), revealing that ORF8 is glycosylated with an oligomannose moiety. This result is corroborated by mass spectrometry analysis. The vast majority of the glycosylation species at N78 are of the oligomannose type, including Man9~5GlcNAc2 (Fig. 1E), indicating that the protein is located between the ER and cis-Golgi apparatus, where the glycan is ultimately trimmed by alpha-1,2-mannosidases MAN1A1 and MAN1B1. The subcellular localization of ORF8 is then microscopically demonstrated with an ER marker (Calnexin), a cis-Golgi marker (GM130), and a cytosol marker (β-Tubulin). ORF8 clearly colocalized with the ER marker (Fig. 1F) with a high colocalization coefficient of 0.718. Thus, ORF8 is a nonsecretory ER-retained protein.
ORF8 interacts with Calnexin and HSPA5.
Previous publications have demonstrated the role played by ORF8 as either a type I interferon (IFN) antagonist (3, 5) or as an antagonist of antigen presentation via its inhibition of MHC-I molecule expression (4). Although we perform similar experiments, we fail to obtain consistent results.
Newcastle disease virus expressing GFP (NDV-GFP) assays were performed to identify the effect of ORF8 on type I IFN production and signaling. The replication efficiency of the NDV-GFP virus is an intuitive measure of the amount of IFN produced (and possibly the amount of IFN-stimulated anti-virus molecules) and secreted into culture supernatants, where a high GFP signal indicates low IFN levels and low GFP expression is a read-out for high IFN concentrations (17). The supernatant induced by RIG-I overexpression shows efficient inhibition of NDV-GFP infection, which is caused by the IFN produced in the culture medium (Fig. S2B and S2C). As an established positive control, the supernatant from influenza virus A NS1 protein-overexpressing cells shows a conspicuous antagonistic effect on IFN production in a culture consisting mostly of NDV-GFP-positive cells (Fig. S2B, undiluted and diluted 1:2). In contrast, ORF8 overexpression fails to inhibit IFN production. Moreover, we fail to identify the function of ORF8 in IFN signaling. Specifically, IFNβ incubation leads to robust inhibition of NDV infection in both ORF8-Flag- and Flag-overexpressing Vero cells, demonstrating the failure of ORF8 to suppress IFN signaling (Fig. S2D). Moreover, after IFNβ stimulation, no significant differences in IFN-sensitive response element (ISRE) signaling are observed between ORF8-overexpressing cells and mock (Flag)-overexpressing cells (Fig. S2E). ORF8 overexpression also fails to attenuate the expression and cytoplasmic membrane presentation of MHC I (Fig. S2F and S2G).
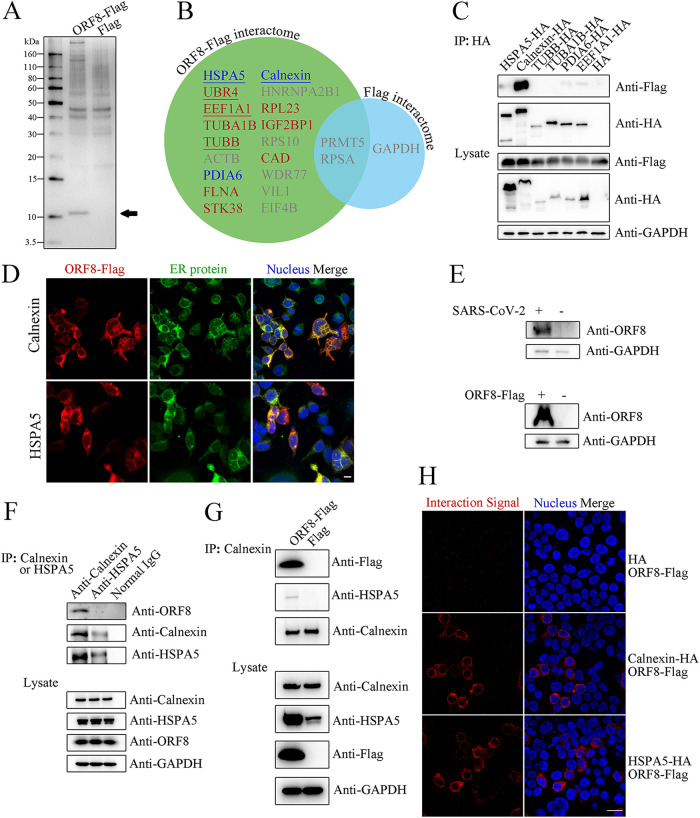
To reveal the unbiased function of SARS-CoV-2 ORF8, the ORF8 interactome was identified. The immunoprecipitation products of ORF8-overexpressing cells show a clear band between 10~15 kDa (Fig. 2A), which corresponds to the molecular weight of ORF8. ORF8-interacting proteins are found to be of molecular weight greater than 60 kDa. Intriguingly, proteins with distinct subcellular localizations are obtained in twice-replicated mass spectrometry results (Fig. 2B), including type A: proteins mainly in ER, including HSPA5 and Calnexin; type B: cytoskeleton proteins, namely, EEF1A1 and TUBB; and type C: proteins with multiple localizations, of which UBR4, an E3 ubiquitin ligase, is identified with a strong signal; although more type A, B, C proteins are identified once with no less than two unique peptides (such as PDIA6, TUBA1B, RPL23, and STK38). We tagged the 7 most frequently identified protein expression constructs (except for UBR4, which is too large) with HA and performed a reverse-immunoprecipitation assay to determine the authenticity of the ORF8 interactome. All the proteins are highly expressed, and among the interactors, Calnexin shows a strong interaction with ORF8 (Fig. 2C). Another ER protein, HSPA5, exhibiting a protein turnover function similar to that of Calnexin, also interacts with ORF8 relatively weakly. Both Calnexin and HSPA5 are highly colocalized with ORF8-Flag in human cells (Fig. 2D). However, in contrast to ORF8-Flag, ORF8-EGFP is dispersed throughout the whole cell and fails to colocalize with Calnexin (Fig. S2H), indicating that EGFP-tag fusion severely affects the subcellular location and, probably, even the protein function. The results are further supported by an endogenous protein–protein interaction test. As several commercial antibodies fail to identify endogenous ORF8 in a WB assay (data not shown), a homemade anti-ORF8 polyclonal antiserum is finally used in the identification of ORF8. Both lysates of SARS-CoV-2-infected cells and ORF8-Flag-overexpressing cells show clear bands at approximately 14 kDa, which correspond with endogenous and exogenous ORF8 proteins, respectively (Fig. 2E). By using the antiserum, the coimmunoprecipitation results support a highly stable endogenous interaction between ORF8 and Calnexin, while only a fairly weak ORF8-HSPA5 interaction is observed (Fig. 2F). Interestingly, the results also indicate that Calnexin and HSPA5 interact in SARS-CoV-2-infected cells, which leads us to wonder whether ORF8 interacts with Calnexin and HSPA5 independently. A further result demonstrates that Calnexin binds HSPA5 exclusively in the presence of ORF8 (Fig. 2G), indicating the actual formation of a Calnexin-ORF8-HSPA5 heterotrimer. Considering the identification of possible ORF8-UBR4 interactions (Fig. 2B) and the fact that various viral proteins target specific E3 ubiquitin ligases to evade IFN-related or MHC-I-related host immune responses (18–20), the basic relationship between ORF8 and ubiquitination is also explored. Compared to treatment with TAK-234 (TAK), a common ubiquitination-inducing agent, overexpression of ORF8 does not affect total ubiquitination levels (Fig. S2I). To provide more solid evidence for the interaction between Calnexin/HSPA5 and ORF8, we performed a proximity ligation assay (PLA), which allows for the in situ visualization of protein–protein interactions per cell. No interaction is viewed between the HA tag and ORF8-Flag, while strong interaction signals are demonstrated for the Calnexin-ORF8 and HSPA5-ORF8 groups (Fig. 2H).
FIG 2.
ORF8 interacts with Calnexin and HSPA5. (A) Products of anti-Flag immunoprecipitation. The immunoprecipitation products from ORF8-Flag- or Flag-overexpressing 293T cell lysates were analyzed by SDS–PAGE with silver staining. The predicted ORF8-Flag band is marked with a black arrow. (B) The ORF8-Flag interactome (in green circle) identified and compared to the Flag interactome (in blue circle). ORF8-interacting proteins predicted to be located in the ER/not in the ER are indicated in blue and red texts, respectively; underscored: ORF8-interacting proteins identified in replicated experiments; gray text: proteins identified with fewer than 2 unique peptides or proteins identified in the control. (C) ORF8 interacts with Calnexin and HSPA5. ORF8-Flag, with each of the interacting protein candidates with an HA tag, was exogenously overexpressed in 293T cells. The products of anti-HA immunoprecipitation were analyzed by WB. (D) Colocalization between ORF8 and endogenous Calnexin/HSPA5. ORF8-Flag-overexpressing 293T cells were analyzed by multiimmunofluorescence using anti-Flag and anti-Calnexin (upper)/anti-HSPA5 (lower) antibodies. Left column: ORF8-Flag signals in red; middle column: ER proteins Calnexin and HSPA5 signals in green; right column: merged signals of left and middle with nucleus in blue. Scale bar: 15 μm. (E) Identification of endogenous (upper) and exogenous (lower) ORF8 by WB using a homemade antiserum. As the a sample for the endogenous WB assay, HeLaACE2+ cells were infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.01 for 48 h. (F) Interaction between endogenous ORF8 and Calnexin/HSPA5. HeLaACE2+ cells were infected with SARS-CoV-2 (at an MOI of 0.1) for 48 h, and the product of either anti-Calnexin (left lane) or anti-HSPA5 (mid lane) immunoprecipitation was analyzed by WB. (G) Identification of a Calnexin-ORF8-HSPA5 heterotrimer. Immunoprecipitation products of ORF8-Flag-overexpressing 293T cells were analyzed by WB. (H) Interaction between ORF8 and Calnexin/HSPA5 by PLA. The HA tag with ORF8-Flag (upper), Calnexin-HA with ORF8-Flag (middle), or HSPA5-HA with ORF8-Flag (lower) was coexpressed in 293T cells, and PLAs were performed to identify the interaction of coexpressed targets. Left column: interaction signals in red; right column: merged signals of interaction and nucleus (in blue) signals. Scale bar: 25 μm.
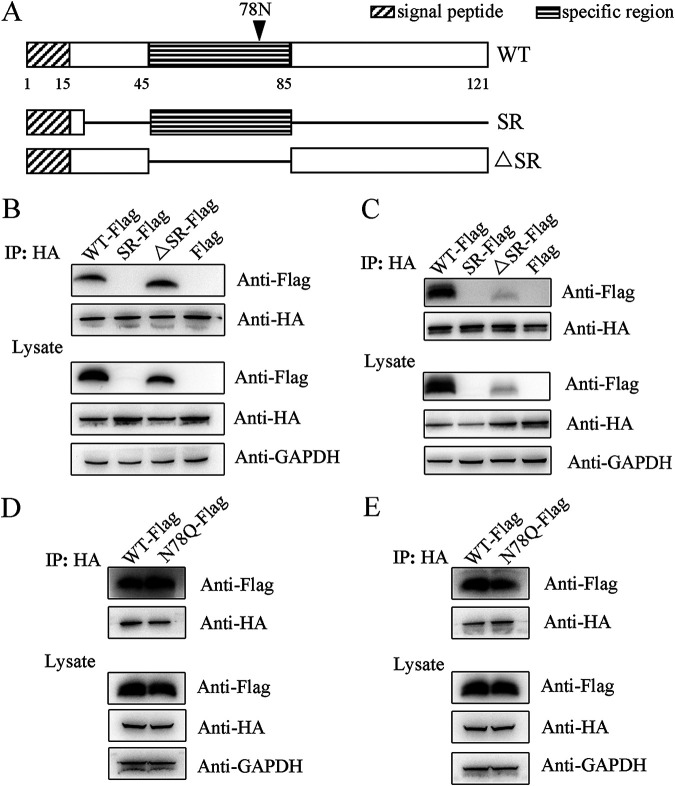
ORF8 interacts with Calnexin and HSPA5 via its immunoglobulin-like fold in a glycan-independent manner.
Except for a specific region in the middle of the polypeptide chain, the published ORF8 structure has revealed an immunoglobulin-like fold (illustrated in Fig. 3A), which is deduced to perform a unique function (6). To elucidate the characteristics of the immunoglobulin-like fold, two ORF8 mutants were generated, of which one contains the specific region (referred to as SR), and the other contains no specific region (ΔSR). Both of them include the N-terminal signal peptide (1 to 20 amino acids) to naturally localize. Both Calnexin and HSPA5 interact with the ΔSR mutant, indicating the importance of the immunoglobulin-like fold, although ΔSR was not as stable as WT ORF8 (Fig. 3B and C). Unfortunately, the expression of the SR mutant is too low to be applied for interaction studies.
FIG 3.
ORF8 interacts with Calnexin and HSPA5 via its immunoglobulin-like fold. (A) Illustration of the domains in WT and mutant ORF8. Rectangles represent the domains as indicated, and solid lines represent the deficient regions. (B-C) ΔSR binds to Calnexin (B) and HSPA5 (C): WT ORF8 (WT-Flag), two mutants (SR-Flag and ΔSR-Flag) or mock (Flag) were coexpressed with Calnexin-HA (B) or HSPA5-HA (C) in 293T cells, and immunoprecipitation products were analyzed by WB. (D–E) The ORF8-Calnexin (D) or ORF8-HSPA5 (E) interactions are independent of N78 glycosylation. WT ORF8 or N78Q (N78Q-Flag) was coexpressed with Calnexin-HA (D) or HSPA5-HA (E) in 293T cells, and immunoprecipitation products were analyzed by WB.
As a lectin dominating glycoprotein turnover, Calnexin interacts with newly synthesized host and viral glycoproteins and facilitates their assembly (8). Although ORF8 is highly glycosylated and is located predominantly in the ER, the interaction between ORF8 and endogenous Calnexin is not based on the N-glycan at the N78 site, as both WT and deglycosylated mutant (N78Q) ORF8 interact with Calnexin with a similar affinity (Fig. 3D). Similarly, deglycosylation exerts a negligible influence on the ORF8-HSPA5 interaction (Fig. 3E).
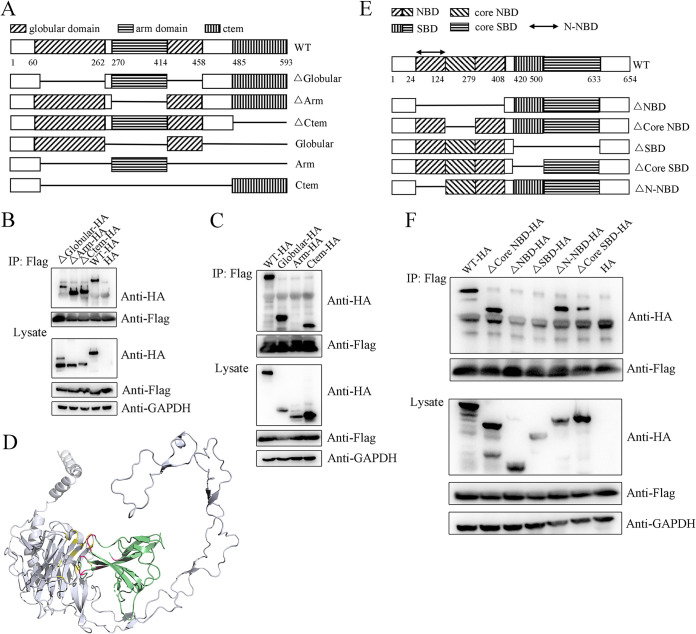
Within the region from the N terminus to the C terminus of the polypeptide chain, mammalian Calnexin is structurally composed of a signal peptide that guides it to the ER membrane, a globular domain with glycan-binding capability that is separated by a long arm domain, and a membrane-integral C terminus (Fig. 4A) (21). To identify the detailed ORF8-interacting domain of Calnexin, three single domain-deficient mutants were generated and referred to as ΔGlobular, ΔArm, and ΔCtem, and their interactions were analyzed by immunoprecipitation. Interestingly, all the mutants interact with ORF8 (Fig. 4B), indicating that calnexin binds ORF8 via multiple domains, although the globular domain deficiency shows the most serious impediment to the interaction. The ORF8 interaction of three other Calnexin mutants containing a single domain, referred to as Globular, Arm, and Ctem, was analyzed further. As expected, both Globular and Ctem interact with ORF8 (Fig. 4C). Notably, the globular mutant shows the highest affinity for ORF8 despite having the lowest expression, indicating that Calnexin binds to ORF8 mainly via the globular domain. Given that the structure of the Calnexin C terminus has not been determined, protein docking between the Calnexin globular domain and the ORF8 immunoglobulin-like fold was performed (Fig. 4D). Full-length ORF8 is properly imbedded into a pocket composed of the Calnexin globular domain and the proximal part of the arm domain with a calculated interaction area of 832.0 Å2 and a Kd = 2.33 × 10−8 M (see Table S1 and Movie S1 for more details). To verify the exact interacting residues, we designed two other ORF8 mutants, referred to as Delta16 and HexA, and used them to explore the interaction between the immunoglobulin-like fold of ORF8 and its target. In the Delta16 mutant, amino acids 37 to 41 and 102 to 112 in ORF8 are deleted, which included C37/C102, and thus, a putative intradisulfate bond and several sites that are predicted to interact with Calnexin are removed. In HexA, probable Calnexin-binding residues H40, F41, F104, D107, E110, and H112 in ORF8 are replaced with A (alanine) residues. Unfortunately, both mutants are expressed at extremely low levels and cannot be used in an interaction study (Fig. S2J), indicating the key effect of the immunoglobulin-like fold in maintaining ORF8 structural homeostasis and its function.
FIG 4.
Specific ORF8-interacting domains of Calnexin and HSPA5. (A) Illustration of domains in WT and mutant Calnexin. (B–C) The ORF8 interaction with Calnexin is not exclusively dependent on a single domain. WT, single-domain-deficient Calnexin mutants (in B: ΔGlobular-HA, ΔArm-HA, ΔCtem-HA), or Calnexin mutants containing a single domain (in C: Globular-HA, Arm-HA, Ctem-HA) were coexpressed in 293T cells with ORF8-Flag, and immunoprecipitation products were analyzed by WB. (D) Protein docking of ORF8-Calnexin. Predicted interacting sites between Calnexin (gray) and ORF8 (green) are shown in yellow and red, respectively. (E) Illustration showing the domains in WT and mutant HSPA5. (F) HSPA5 interacts with ORF8 via both the NBD and SBD. Each of five single-domain-deficient HSPA5 mutants was coexpressed in 293T cells with ORF8-Flag, and immunoprecipitation products were analyzed by WB.
HSPA5 consists of a nucleotide-binding domain (NBD) at the N terminus and a substrate-binding domain (SBD) at the C terminus (22). The NBD contains a functional region referred to herein as the core NBD, which is located in the middle of the NBD and is critical for direct ATP-ADP binding. Similarly, the SBD includes a key region controlling polypeptide binding referred to herein as the core SBD. We generated 5 HSPA5 mutants, including three (partial) NBD-deficient and two (partial) SBD-deficient mutants (Fig. 4E). ΔNBD, ΔCore NBD and ΔN-NBD lack the whole NBD, the core NBD, and the N-terminal part of the NBD, respectively; ΔSBD and ΔCore SBD mutants lack the whole SBD and the core SBD, respectively. All mutants are highly expressed. Interestingly, our results reveal that both the NBD and SBD are indispensable for the HSPA5-ORF8 interaction, but the interaction does not rely exclusively on the core NBD or core SBD (Fig. 4F), although the core SBD is critical for ORF8 binding to a certain degree. Furthermore, the N terminus of the NBD does not affect the interaction. Thus, ORF8 binds HSPA5 via both the C-terminal NBD and N-terminal SBD, which do not seem to be typical HSP-interacting regions.
ORF8 induces ER stress-like responses and facilitates SARS-CoV-2 replication by triggering the Calnexin switch.
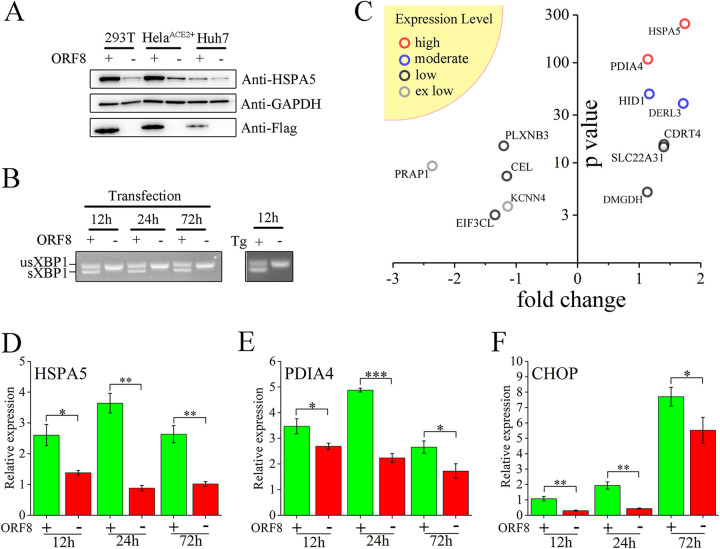
Both Calnexin and HSPA5 are key chaperones critical for the folding of newly synthesized proteins. They are involved in protein turnover, UPR or ER stress-related response regulation, and calcium homeostasis. In 293T cells, ORF8 overexpression induces intensive upregulation of HSPA5, which is a common effector of ER stress (Fig. 5A). In contrast to the fact that all the eIF2α, IREI, and ATF6 branches of ER stress are activated after treatment with thapsigargin (Tg), a commonly used stress-inducing agent, the stress-like outcome induced by ORF8 is generated exclusively via the IREI branch, as demonstrated by the splicing of XBP1 (Fig. 5B, S3A and S3B). The response appears as early as 12 h posttransfection and continues to 72 h posttransfection (Fig. 5B). As revealed by HSPA5 expression, although a positive correlation between ORF8 expression levels and the stress-like response is roughly indicated, a tiny amount of ORF8 is sufficient to induce an intense response (Fig. S3C). The overexpression of an ER-irrelevant EGFP fails to induce HSPA5 upregulation (Fig. S3D). Interestingly, the stress-like response induced by ORF8 is species dependent. In human 293T, HeLaACE2+ and Huh7 cells, ORF8 overexpression leads to a significant response (Fig. 5A). However, in primary mouse embryonic fibroblast (MEF) cells, which are commonly used for ER stress studies (13), ORF8 fails to promote stress via either branch (Fig. S3E and S3F). To identify other cellular effects induced by ORF8 in human cells, a transcriptome comparison between ORF8- and mock-overexpressing cells was performed. Four proteins with high to moderate expression levels are identified, of which HSPA5, derlin-3 (DERL3), and PDIA4 are common ER stress-related molecular effectors (Fig. 5C). Furthermore, the mRNA upregulation of HSPA5 and PDIA4 is demonstrated as early as 12 h posttransfection, which is sustained to 72 h (Fig. 5D and E). ORF8 also induces a consistent increase in the level of CHOP (Fig. 5F), a universal PDIA4 cooperator, and mild upregulation of EDEM expression at 24 h posttransfection (Fig. S3G), both of which are ERAD-associated molecules. In contrast, ORF8 exerts no significant impact on CHOP or EDEM expression in mouse cells (Fig. S3H and S3I).
FIG 5.
ORF8 induces stress-like responses in human cells. (A) ORF8 induces HSPA5 upregulation. ORF8-Flag (ORF8 +) or Flag (ORF8 -) was overexpressed in human cells, as indicated for 60 h, and cell lysates were analyzed by WB. (B) ORF8 induces sustained XBP-1 splicing. RNAs obtained from ORF8-overexpressing or Tg-treated 293T cells at the indicated time posttransfection/treatment were analyzed for XBP-1 splicing detection. (C) Transcriptome abnormalities induced by ORF8. Log2-fold changes (fold change) of gene expression (ORF8 positive versus ORF8 negative) and -log10p value (P value) are shown, including genes with various expression levels in the yellow sector in upper left: high (100 ≤ average FPKM), moderate (10 ≤ average FPKM < 100), low (1 ≤ average FPKM < 1) and extremely (ex) low (average FPKM < 1). (D-F) ORF8 induced upregulation of HSPA5, PDIA4 and CHOP effectors of the stress-like response. RNAs from ORF8-overexpressing 293T cells at the indicated times posttransfection were analyzed by qRT–PCR, and the target genes are shown in the upper left. Significance: *(P ≤ 0.05), **(P ≤ 0.01), and ***(P ≤ 0.001).
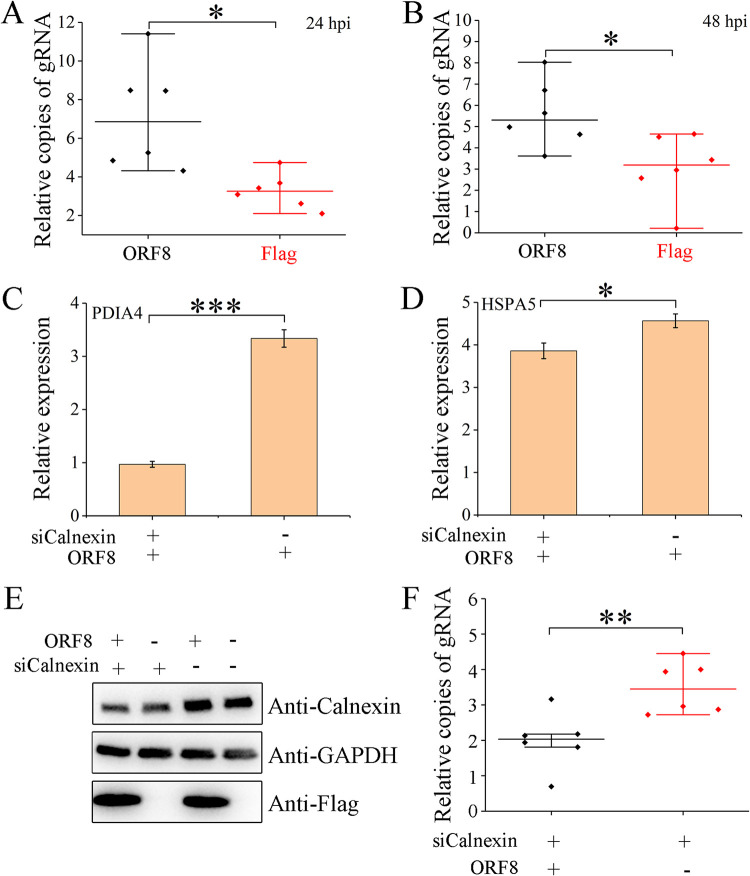
Temporary ER stress usually promotes the expression of chaperone-like molecules to enhance cellular protein folding and ERAD-related responses to eliminate misfolded proteins, which may benefit progeny virus replication; however, sustained ER stress commonly suspends translation via activation of the RNase activity of IRE1, inhibition of the translation initiation factor eIF2a function, or promotion of various types of cell death (23), which is detrimental to virus replication. Thus, we subsequently explore the influence of ORF8 expression with the induced stress-like response on SARS-CoV-2 replication. Overexpression of ORF8 facilitates SARS-CoV-2 replication to a relatively mild but stable degree at both 24 and 48 h postinfection (hpi) (Fig. 6A and B). Based on the sequence of the ORF8 sgmRNA, we designed 6 specific short interfering RNAs (siRNAs) to knockdown ORF8 during SARS-CoV-2 infection (Fig. S4A and Table S3). However, ORF8 is not knocked down successfully, although the siRNA transfection is well-performed (Fig. S4B and S4C). Therefore, the effect of ORF8 knockdown on SARS-CoV-2 replication remains unknown.
FIG 6.
ORF8 induces stress-like responses and facilitates SARS-CoV-2 replication by triggering Calnexin. (A–B) ORF8 overexpression facilitates SARS-CoV-2 replication. ORF8/mock-overexpressing (ORF8/Flag, respectively) HeLaACE2+ cells were infected with SARS-CoV-2 at an MOI of 0.001, and viral genomic RNAs (gRNA) at 24 hpi (A) and 48 hpi (B) in the supernatant were measured by qRT–PCR; each experiment was repeated six times. (C–D) Abnormal expression of Calnexin inhibits the stress-like responses induced by ORF8. Calnexin in ORF8/mock-overexpressing 293T cells is knocked down (siCalnexin+) or mock knocked down (siCalnexin-). The relative expression of PDIA4 (C) or HSPA5 (D) mRNAs was measured by qRT–PCR. A delta-delta CT method was used, and the data were normalized to those of the corresponding Flag-overexpressing groups; each experiment was repeated 3 times. (E) Successful Calnexin knockdown and ORF8 overexpression. (F) ORF8 fails to facilitate SARS-CoV-2 replication under Calnexin-deficient conditions. HeLaACE2+ cells were cotransfected with ORF8 and siCalnexin for 48 h and then infected with SARS-CoV-2 at an MOI of 0.001 for 24 h, followed by gRNA detection as described in (A). Significance of the results shown in (A-D) and (F): *(P ≤ 0.05), **(P ≤ 0.01), and ***(P ≤ 0.001).
As the top interactor of ORF8 is Calnexin, we wonder whether ORF8 induces a stress-like response by interfering with Calnexin function. Relative to the negative control (Flag overexpression), ORF8 leads to an ~3-fold increase in PDIA4 levels (Fig. 6C), while the PDIA4 level is strikingly reduced when Calnexin expression is inhibited. Similarly, a mild decrease in ORF8-induced HSPA5 upregulation is also observed (Fig. 6D). Interestingly, ORF8 fails to promote and inhibit SARS-CoV-2 replication in Calnexin-deficient cells (Fig. 6E and F). Hence, the induction of the stress-like response and the promotion of SARS-CoV-2 replication by ORF8 are highly dependent on normal Calnexin expression.
DISCUSSION
Although SARS-CoV-2 is basically regarded as a homolog of SARS-CoV, with their genomic structure and the majority of their genes being highly homologous, the ORF8 genes of SARS-CoV and SARS-CoV-2 are distinct. With only 121 amino acids, the SARS-CoV-2 ORF8 protein also shows little homology with other viral or host proteins and is regarded as a novel protein, which piqued our interest at the beginning of the coronavirus outbreak. Similar to other nonstructural viral proteins (24, 25), ORF8 has been believed to inhibit type I IFN signaling, probably via the suppression of IRF3 and/or by interacting with HSP90B (3, 5, 26). In addition to acting as an antagonist of innate immunity, ORF8 has been shown to inhibit MHC-Ι and hence plays a role in acquired immune evasion. However, similar to results reported in another publication (27), we are unable to recapitulate the above-mentioned immune antagonist functions of ORF8. We propose that the main difference is that ORF8-GFP was used in the previous studies, while a small flag-tagged ORF8 was used in our study. Through unbiased discovery of the interacting targets of ORF8 by mass spectrometry, further verification via both endogenous and exogenous immunoprecipitation and PLA, we show convincing evidence that ORF8 is located in the ER and interacts with Calnexin to induce various stress-like responses. ORF8 with a relatively large GFP tag, however, is mislocalized and hence probably gains nonexistent functions.
In addition to their functions in virus–host interactions and the resulting virulence enhancement, accessory proteins in CoVs are rarely considered to be indispensable for viral replication in vitro, although some of them can modulate replication by interrupting host pathways (28–30). We discovered that ectopic expression of ORF8 facilitates SARS-CoV-2 replication, which corresponds well with published results indicating the attenuation of viral replication induced by genomic deletion of ORF8 (30, 31). Given that Calnexin universally coordinates the processing of newly synthesized viral glycoproteins by facilitating their folding (7–9) and on the basis of the ORF8-Calnexin interaction revealed in our study, ORF8 is considered to induce stress-like responses and accelerate the proper folding of glycoproteins, probably by stimulating molecular chaperone upregulation (such as HSPA5 and PDIA4 identified here), which ultimately promotes the secretion of mature progeny virus; however, the detailed machinery of spike glycoprotein turnover mediated by the Calnexin-ORF8 interaction needs to be elucidated further. ORF8-induced stress-like responses and virus replication are highly dependent on normal Calnexin expression. Therefore, Calnexin behaves as a molecular switch to control host functions induced by ORF8. Interestingly, the in vivo virulence of the ORF8 protein during SARS-CoV-2 infection seems controversial: previous studies on mouse infection models have indicated that ORF8 is hardly considered to enhance virulence, while clinical data have associated ORF8 with severe symptoms in COVID-19 patients (32, 33). Our study demonstrates that ORF8 functions in human but not mouse cells, which is ascribed to the divergence between human and mouse Calnexin interactors and thus provides a direct explanation for the dispute. Consequently, it is considered that ORF8 is a species-dependent (or even human-exclusive) virulence gene and a putative novel target of antiviral agent design, which probably also contributes to the discrepancy in symptoms between COVID-19 patients and mice infected with an adapted SARS-CoV-2 virus (34, 35).
MATERIALS AND METHODS
Cells, virus, RNA-related, PCR-related, and plasmids.
The generation of the human ACE2-overexpressing cell line, HeLaACE2+, was previously described (36). MEF cell isolation and primary culture followed a published protocol (37). Vero-E6 cells, Vero cells, and 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo). A549 cells were cultured in McCoy's 5A medium (Thermo) supplemented with 50 mg/L tricine (Sigma). All culture medium above was supplemented with 10% FBS (Sigma) and penicillin–streptomycin, and cell growth was performed at 37°C under 5% CO2. 293F cells were cultured in FreeStyle 293 expression medium (Thermo) at 37°C under 8% CO2. SARS-CoV-2 passage was previously described (2). All live SARS-CoV-2-related experiments were approved and performed in the Animal Biosafety Level 3 lab of Changchun Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
RNA extraction, first strain cDNA synthesis, RNA sequencing, and qRT–PCR were previously described (2). RT–PCR was performed using KAPA HiFi polymerase (Roche). For XBP-1 splicing detection, RT–PCR products were separated on a 3% agarose gel. Cells were treated with Tg (Sigma) to induce ER stress or with TAK (MCE) to inhibit ubiquitination as indicated.
A stop codon was generated on the pCAGGS-Flag vector by site-directed mutation of the 3′ flanking the Flag sequence and thus was referred to as pCAGGS-Flag-stop, which only encoded a Flag peptide and was used in all mock-flag transfections. SARS-CoV-2 ORF8 was obtained by RT–PCR using RNA from infected Vero-E6 cells, cloned into the linearized pCAGGS-Flag-stop vector and then fused with a C-terminal Flag tag (referred to as ORF8-Flag) using homologous recombination. ORF8 was then subcloned into the pEGFPN1 (Thermo) plasmid (ORF8-EGFP). For E. coli expression, 16-121 amino acids of ORF8 were codon-optimized, de novo synthesized, and subcloned into the pET28a (Novagen) plasmid and then fused with an N-terminal 6× His tag (His-ORF8). Calnexin, HSPA5, TUBB, TUBA1B, PDIA6, and EEF1A1 were cloned into the pcDNA3 vector with a C-terminal HA tag using RT–PCR and referred to as Calnexin-HA, HSPA5-HA, TUBB-HA, TUBA1B-HA, PDIA6-HA, and EEF1A1-HA, respectively. The N-terminal RIG-I plasmid was previously described (38). NS1 of influenza virus A/PR/8/34 was synthesized de novo and subcloned into the pCAGGS-Flag-stop vector according to publications (24, 39). A canonical enhancer, the IFN-stimulated response element (ISRE, 5′-GAAACTGAAACT-3′) (40), was tandemly repeated 5 times and cloned into a firefly luciferase pGL3p plasmid (Promega) and referred to as pISRE-Luc. A Relina luciferase plasmid, pBind, was purchased from Promega. Mutations of ORF8 (including T12R, V13A, A15R, V13R, ΔSR, SR and N78Q) were generated by one or two rounds of site-directed mutagenesis with the template ORF8-Flag. Mutations of Calnexin (including ΔGlobular, ΔArm, ΔCtem, and Arm) were generated by one or two rounds of site-directed mutagenesis with the template Calnexin-HA. The N terminus and C terminus (with an HA tag) of Calnexin mutations (including Ctem and Globular) were obtained by PCR and ligated into pcDNA3 using homologous recombination. Homologous recombination was performed using the ClonExpress II One Step Cloning kit (Vazyme). Site-directed mutagenesis was performed using the QuikChange II site-directed mutagenesis kit (Agilent) or Q5 site-directed mutagenesis kit (NEB) following the manufacturer’s instructions. ORF8 mutations Delta16 and HexA (with Flag tags) were synthesized de novo. Transfection was performed using Lipofectamine 3000 (Thermo). Detailed information on the primers is provided in Table S1.
NDV-GFP assay and luciferase assay.
Generation of NDV-GFP was described previously (41). To identify the potential role of ORF8 in IFN production, 293T cells in a 6-well plate were transfected with 1.25 μg of N-terminal RIG-I and target plasmids (ORF8-Flag, NS1-Flag, or Flag). Twenty-four hours posttransfection, supernatants were harvested, and serial 2-fold dilutions of the supernatants were used to overlay Vero cells. Twenty-four hours later, Vero cells were infected with 105 TCID50 NDV-GFP per well, and images were taken at 18 h postinfection (hpi) with a fluorescence microscope (Lecia).
To measure the effect on IFN signaling using NDV-GFP, Vero cells in 24-well plates were transfected with 0.5 μg target plasmids (ORF8-Flag or Flag). Twenty-four hours posttransfection, cells were washed with PBS and incubated with or without 1,000 U IFNβ (MCE) for 24 h. Cells in each well were infected with 104 TCID50 NDV-GFP, and images were taken at 18 hpi. For the luciferase assay, Vero cells were cotransfected with 0.20 μg pISRE-Luc, 0.05 μg pBind and 0.25 μg target plasmids. At 24 h posttransfection, cells were washed with PBS and treated with or without 1,000 U IFNβ for another 24 h. Luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Promega).
Expression and purification of ORF8.
The plasmid pET28a-ORF8 was transferred into a BL21(DE3) E.coli strain. Expression was performed by 0.8 mM (final concentration) IPTG induction for 6 h at 37°C. The cells were resuspended in ice-cold PBS and lysed by sonication. The lysate was centrifuged at 18,000 × g for 1 h, and the precipitate was dissolved in lysis buffer (8 M urea in PBS), mixed with 1 mL Ni-NTA resin (Qiagen) by rotation for 2 h at 4°C, washed twice with washing buffer (40 mM imidazole and 8 M urea in PBS), and eluted with elution buffer (300 mM imidazole and 8 M urea in PBS). Protein renaturation was performed by gradient dialysis.
For mammalian expression of ORF8, 293F cells were transfected using ExpiFectamine 293 reagent (Thermo) for 96 h. The medium was successively centrifuged at 300 × g for 5 min, 2,000 × g for 10 min, and 10,000 × g for 30 min, and the supernatant was concentrated by ultrafiltration using Amicon Ultra15 Centrifugal Filter Units (Millipore, nominal molecular weight limit of 3 kDa) to obtain proteins secreted in the medium. Intracellular proteins were obtained as follows.
PAGE, gel staining, and Western blotting.
Cells were lysed with RIPA buffer (50 mM Tris-HCl, pH = 8.0, with 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with 1× cocktail (MCE). Cell lysates were mixed with reducing sample buffers (12% SDS, 6% mercaptoethanol, 30% glycerol, 0.05% Coomassie blue G-250, 150 mM Tris-HCl, pH = 7.0) or nonreducing sample buffers (reducing sample buffers without mercaptoethanol) if mentioned. Deglycosylation was performed by treating 20 μg glycoproteins in lysate with Endo H (NEB). SDS–PAGE was performed with precast Omni-PAGE (Epizyme). Tricin SDS–PAGE was performed following a previous publication (42). Gels were stained with Coomassie blue G-250 or a Fast Silver Stain kit (Beyotime).
Antibodies, immunoprecipitation, immunofluorescence/PLA, and flow cytometry.
To generate homemade anti-ORF8 antiserum, each mouse (female BALB/c, 10 weeks old) was immunized with 100 μg His-ORF8 protein with Freund's complete adjuvant. Booster doses were administered 4, 6, 8, and 10 weeks after the initial immunization using 100 μg His-ORF8 proteins with Freund's incomplete adjuvant. Polyclonal antiserum was obtained from ORF8-immunized mice without further purification. Vendors and usage of commercial antibodies are shown in Table S2.
For immunoprecipitation, cells infected by SARS-CoV-2 at an MOI of 0.1 for 48 h or transfected with plasmids for 60 h were lysed with lysis buffer (20 mM Tris-HCl pH = 8.0, 137 mM NaCl, 1% NP-40, 2 mM EDTA, 0.2 mM Na3VO4, and 1× cocktail). The lysates were preabsorbed by Protein G Sepharose 4 Fast Flow (GE) and then incubated with either antibody-Dynabeads Protein G (Thermo) complex mixture (for anti-HSPA5 or anti-Calnexin immunoprecipitation), anti-Flag M2 Affinity Gel (Sigma, for anti-Flag immunoprecipitation), or monoclonal anti-HA Agarose (Sigma, for anti-HA immunoprecipitation), washed three times with wash buffer (50 mM Tris-HCl pH = 7.4, 150 mM NaCl, 0.1% Triton X-100), and eluted with elution buffer (0.2 M glycine-HCl pH = 2.6).
For immunofluorescence and PLA, cells grown in chamber slides (Thermo) were fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100, and blocked with 2% goat serum. Cells for immunofluorescence were stained with target primary antibodies and goat-anti-rabbit/mouse IgG Alexa Fluor 594 or goat-anti-rabbit/mouse IgG Alexa Fluor 488 (Abcam). For PLA, post blocking cells were incubated with a mixture of anti-HA and anti-Flag antibodies, and other steps were performed following the instructions of the Duolink In Situ Red Starter kit Mouse/Rabbit (Sigma). Images were taken on an LSM 900 confocal microscope (Zeiss).
For flow cytometry, cells were detached with 10 mM EDTA (Sigma) at 48 h posttransfection, transferred into 1% BSA with 0.1% NaN3 in PBS, and stained with FITC anti-human HLA-A, B, C (Biolegend), and 7-AAD viability staining solution (Biolegend). Flow cytometry was performed using CytoFlex (Beckman).
Mass spectrum.
Each sample was mixed with 10 μL 8 M urea, 20 μL deionized water, and 10 mM DTT and incubated at 55°C for 30 min. Then, 40 mM iodoacetamide was added, and the mixture was incubated at room temperature under dark conditions for 30 min. After the pH was adjusted to 8.0 with 500 mM NH4HCO3, trypsin (Promega) and chymotrypsin (for ORF8 identification exclusively) were added at a ratio of sample/enzyme = 30/1 (mass ratio) and incubated at 37°C for 16 h. Then, 0.5 μL of 10% formic acid was added to terminate the enzymatic hydrolysis, and the mixture was lyophilized. The lyophilized polypeptide was added to 30 μL 0.1% trifluoroacetic acid for redissolution. All samples were analyzed by an Orbitrap-Lumos high resolution mass spectrometer (Thermo) coupled with an Easy nLC 1200 (Thermo). The full scan was performed for m/z 350–1550 with a resolution of 120,000 at m/z = 200, and the maximum injection time was 30 ms. The MS/MS scan was performed with higher-energy collision-activated dissociation for m/z 200–2000 with a resolution of 15,000 at m/z = 200, and the maximum injection time was 25 ms. The collision energy was 32%. MS1 tolerance was set to 10 ppm, and MS/MS tolerance was 20 ppm. N-glycan data were analyzed using Byonic (Protein Metrics).
Protein docking.
Structures of human Calnexin was predicted using Alphafold2, of which 60 to 261 and 414 to 457 amino acids were kept and docked with 15 to 45 and 85 to 121 amino acids of ORF8 (Protein Data Bank accession: 7JTL) using ZDOCK. The complex with the highest credibility was displayed, and the protein–protein interaction was analyzed with Discovery Studio. The complex structure, including full-length ORF8 and Calnexin without the C terminus, was presented with PyMOL.
Knockdown.
In total, 6 siRNAs targeting ORF8 sgmRNAs were synthesized and conjugated with FAM, named siORF8-1 to siORF8-6, and cells in 6-well plates were transfected with 60 pmol siORF8 using Lipofectamine 3000 (Thermo). Two siRNAs targeting human calnexin RNA were synthesized and conjugated with FAM, named siCalnexin-1 and siCalnexin-2, and were mixed 1:1 for use. To cotransfect siRNA and plasmids, cells in 12-well plates were transfected with a mixture of 16 pmol siRNAs and 1 μg plasmids using Lipofectamine 3000 (Thermo). For knockdown controls, siNC was used. Detailed information on the siRNAs is provided in Table S3.
Data availability.
The raw sequencing data from this study have been deposited in GenBank under accession number PRJNA875499.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Key Research and Development Project of China (No. 2021YFC2301701).
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
Contributor Information
Hongbin He, Email: yuwei0901@outlook.com.
Yuwei Gao, Email: hongbinhe@sdnu.edu.cn.
Stacey Schultz-Cherry, St. Jude Children's Research Hospital.
REFERENCES
- 1.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Zhao Y, Yan F, Wang T, Sun W, Feng N, Wang W, Wang H, He H, Yang S, Xia X, Gao Y. 2021. Viral and host transcriptomes in SARS-CoV-2-infected human lung cells. J Virol 95:e00600-21. 10.1128/JVI.00600-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J-Y, Liao C-H, Wang Q, Tan Y-J, Luo R, Qiu Y, Ge X-Y. 2020. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res 286:198074. 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Chen Y, Li Y, Huang F, Luo B, Yuan Y, Xia B, Ma X, Yang T, Yu F, Liu J, Liu B, Song Z, Chen J, Yan S, Wu L, Pan T, Zhang X, Li R, Huang W, He X, Xiao F, Zhang J, Zhang H. 2021. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc Natl Acad Sci USA 118:e2024202118. 10.1073/pnas.2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Lu Z, Yang X, Zhou Y, Gao J, Zhang S, Huang S, Cai J, Yu J, Zhao W, Zhang B. 2022. Severe acute respiratory syndrome coronavirus 2 ORF8 protein inhibits type I interferon production by targeting HSP90B1 signaling. Front Cell Infect Microbiol 12:899546. 10.3389/fcimb.2022.899546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flower TG, Buffalo CZ, Hooy RM, Allaire M, Ren X, Hurley JH. 2021. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc Natl Acad Sci USA 118:e2021785118. 10.1073/pnas.2021785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukushi M, Yoshinaka Y, Matsuoka Y, Hatakeyama S, Ishizaka Y, Kirikae T, Sasazuki T, Miyoshi-Akiyama T. 2012. Monitoring of S protein maturation in the endoplasmic reticulum by calnexin is important for the infectivity of severe acute respiratory syndrome coronavirus. J Virol 86:11745–11753. 10.1128/JVI.01250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molinari M, Eriksson KK, Calanca V, Galli C, Cresswell P, Michalak M, Helenius A. 2004. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol Cell 13:125–135. 10.1016/s1097-2765(03)00494-5. [DOI] [PubMed] [Google Scholar]
- 9.Molinari M, Helenius A. 2000. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science 288:331–333. 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- 10.Boson B, Legros V, Zhou B, Siret E, Mathieu C, Cosset F-L, Lavillette D, Denolly S. 2021. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J Biol Chem 296:100111. 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.V'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. 2021. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 19:155–170. 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knoops K, Kikkert M, WOrm S, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-Coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6:e226. 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee A-H, Iwakoshi NN, Glimcher LH. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23:7448–7459. 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oslowski CM, Urano F. 2011. Measuring ER stress and the unfolded protein response using mammalian tissue culture system, p 71–92. In Conn PM (ed), Methods in enzymology, vol 490. Academic Press, New York, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karla A, Lively MO, Paetzel M, Dalbey R. 2005. The identification of residues that control signal peptidase cleavage fidelity and substrate specificity. J Biol Chem 280:6731–6741. 10.1074/jbc.M413019200. [DOI] [PubMed] [Google Scholar]
- 16.Escola J-M, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. 1998. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem 273:20121–20127. 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 17.Park M-S, Shaw ML, Muñoz-Jordan J, Cros JF, Nakaya T, Bouvier N, Palese P, García-Sastre A, Basler CF. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J Virol 77:1501–1511. 10.1128/jvi.77.2.1501-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coscoy L, Ganem D. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci USA 97:8051–8056. 10.1073/pnas.140129797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilton L, Moganeradj K, Zhang G, Chen Y-H, Randall RE, McCauley JW, Goodbourn S. 2006. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J Virol 80:11723. 11732. 10.1128/JVI.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansouri M, Bartee E, Gouveia K, Nerenberg BTH, Barrett J, Thomas L, Thomas G, McFadden G, Früh K. 2003. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J Virol 77:1427–1440. 10.1128/jvi.77.2.1427-1440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrag JD, Bergeron JJM, Li Y, Borisova S, Hahn M, Thomas DY, Cygler M. 2001. The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol Cell 8:633–644. 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Nune M, Zong Y, Zhou L, Liu Q. 2015. Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure 23:2191–2203. 10.1016/j.str.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hetz C. 2012. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13:89–102. 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- 24.Kochs G, García-Sastre A, Martínez-Sobrido L. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 81:7011–7021. 10.1128/JVI.02581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muñoz-Jordán JL, Sánchez-Burgos GG, Laurent-Rolle M, García-Sastre A. 2003. Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci USA 100:14333–14338. 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rashid F, Dzakah EE, Wang H, Tang S. 2021. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res 296:198350. 10.1016/j.virusres.2021.198350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia H, Cao Z, Xie X, Zhang X, Chen JY-C, Wang H, Menachery VD, Rajsbaum R, Shi P-Y. 2020. Evasion of type I interferon by SARS-CoV-2. Cell Rep 33:108234. 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa K, Narayanan K, Wada M, Makino S. 2018. Inhibition of stress granule formation by Middle East respiratory syndrome coronavirus 4a accessory protein facilitates viral translation, leading to efficient virus replication. J Virol 92:e00902-18. 10.1128/JVI.00902-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou P, Wang X, Wang H, Wang T, Yu Z, Xu C, Zhao Y, Wang W, Zhao Y, Chu F, Chang H, Zhu H, Lu J, Zhang F, Liang X, Xingyu L, Wang S, Gao Y, He H. 2023. The ORF7a protein of SARS-CoV-2 initiates autophagy and limits autophagosome-lysosome fusion via degradation of SNAP29 to promote virus replication. Autophagy 19:551–569. 10.1080/15548627.2022.2084686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Zhang X, Liu J, Xia H, Zou J, Muruato AE, Periasamy S, Kurhade C, Plante JA, Bopp NE, Kalveram B, Bukreyev A, Ren P, Wang T, Menachery VD, Plante KS, Xie X, Weaver SC, Shi P-Y. 2022. A live-attenuated SARS-CoV-2 vaccine candidate with accessory protein deletions. Nat Commun 13:4337. 10.1038/s41467-022-31930-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silvas JA, Vasquez DM, Park J-G, Chiem K, Allué-Guardia A, Garcia-Vilanova A, Platt RN, Miorin L, Kehrer T, Cupic A, Gonzalez-Reiche AS, BAkel H, García-Sastre A, Anderson T, Torrelles JB, Ye C, Martinez-Sobrido L. 2021. Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 human ACE2 transgenic mice. J Virol 95:e00402-21. 10.1128/JVI.00402-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young BE, Fong S-W, Chan Y-H, Mak T-M, Ang LW, Anderson DE, Lee CY-P, Amrun SN, Lee B, Goh YS, Su YCF, Wei WE, Kalimuddin S, Chai LYA, Pada S, Tan SY, Sun L, Parthasarathy P, Chen YYC, Barkham T, Lin RTP, Maurer-Stroh S, Leo Y-S, Wang L-F, Renia L, Lee VJ, Smith GJD, Lye DC, Ng LFP. 2020. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet 396:603–611. 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gamage AM, Tan KS, Chan WOY, Liu J, Tan CW, Ong YK, Thong M, Andiappan AK, Anderson DE, Wang DY, Wang L-F. 2020. Infection of human nasal epithelial cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog 16:e1009130. 10.1371/journal.ppat.1009130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu H, Chen Q, Yang G, He L, Fan H, Deng Y-Q, Wang Y, Teng Y, Zhao Z, Cui Y, Li Y, Li X-F, Li J, Zhang N-N, Yang X, Chen S, Guo Y, Zhao G, Wang X, Luo D-Y, Wang H, Yang X, Li Y, Han G, He Y, Zhou X, Geng S, Sheng X, Jiang S, Sun S, Qin C-F, Zhou Y. 2020. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 369:1603–1607. 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. 2020. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 323:1061–1069. 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Hou P, Ma W, Wang X, Wang H, Yu Z, Chang H, Wang T, Jin S, Wang X, Wang W, Zhao Y, Zhao Y, Xu C, Ma X, Gao Y, He H. 2022. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cell Mol Immunol 19:67–78. 10.1038/s41423-021-00807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu L-Q, Lai WS, Stumpo DJ, Blackshear PJ. 2016. mouse embryonic fibroblast cell culture and stimulation. Bio-Protocol 6:e1859. 10.21769/BioProtoc.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, Ma Q, Liu X, Cao C. 2017. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol 91:e02143-16. 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol 79:8004–8013. 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19:623–655. 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Wang C, Feng N, Wang H, Zheng X, Yang S, Gao Y, Xia X, Yin R, Liu X, Hu S, Ding C, Yu S, Cong Y, Ding Z. 2015. Development of a reverse genetics system based on RNA polymerase II for Newcastle disease virus genotype VII. Virus Genes 50:152–155. 10.1007/s11262-014-1137-x. [DOI] [PubMed] [Google Scholar]
- 42.Schägger H. 2006. Tricine–SDS-PAGE. Nat Protoc 1:16–22. 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4 and Tables S1 to S4. Download jvi.00011-23-s0002.pdf, PDF file, 1.7 MB (1.7MB, pdf)
Movie S1. Download jvi.00011-23-s0001.mp4, MP4 file, 8.3 MB (8.1MB, mp4)
Data Availability Statement
The raw sequencing data from this study have been deposited in GenBank under accession number PRJNA875499.