Abstract
Flaviviruses are important human pathogens and include dengue (DENV), West Nile (WNV), Yellow fever virus (YFV), Japanese encephalitis (JEV) and Zika virus (ZIKV). DENV, transmitted by mosquitoes, causes diseases ranging in severity from mild dengue fever with non-specific flu-like symptoms to fatal dengue hemorrhagic fever and dengue shock syndrome. DENV infections are caused by four serotypes, DENV1–4, which interact differently with antibodies in blood serum. The incidence of DENV infection has increased dramatically in recent decades and the CDC estimates 400 million dengue infections occur each year, resulting in ~25,000 deaths mostly among children and elderly people. Similarly, ZIKV infections are caused by infected mosquito bites to humans, can be transmitted sexually and through blood transfusions. If a pregnant woman is infected, the virus can cross the placental barrier and can spread to her fetus, causing severe brain malformations in the child including microcephaly and other birth defects. It is noteworthy that the neurological manifestations of ZIKV were also observed in DENV endemic regions, suggesting that pre-existing antibody response to DENV could augment ZIKV infection. WNV, previously unknown in the US (and known to cause only mild disease in Middle East), first arrived in New York city in 1999 (NY99) and spread throughout the US and Canada by Culex mosquitoes and birds. WNV is now endemic in North America. Thus, emerging and re-emerging flaviviruses are significant threat to human health. However, vaccines are available for only a limited number of flaviviruses, and antiviral therapies are not available for any flavivirus. Hence, there is an urgent need to develop therapeutics that interfere with essential enzymatic steps, such as protease in the flavivirus lifecycle as these viruses possess significant threat to future pandemics. In this review, we focus on our E. coli expression of NS2B hydrophilic domain (NS2BH) covalently linked to NS3 protease domain (NS3Pro) in their natural context which is processed by the combined action of both subunits of the NS2B-NS3Pro precursor. Biochemical activities of the viral protease such as solubility and autoproteolysis of NS2BH-NS3Pro linkage depended on the C-terminal portion of NS2BH linked to the NS3Pro domain. Since 2008, we also focus on the use of the recombinant protease in high throughput screens and characterization of small molecular compounds identified in these screens.
1. Introduction
Flaviviruses are important human pathogens and include dengue (DENV), West Nile (WNV), Yellow fever virus (YFV), Japanese encephalitis (JEV), tick-borne encephalitis virus (TBEV), and Zika virus (ZIKV). In the review, we will focus on flaviviruses that are transmitted by mosquitoes. DENV, transmitted by Aedes aegypti and Aedes albopictus mosquitoes, causes diseases ranging in severity from mild dengue fever with non-specific flu-like symptoms to fatal dengue hemorrhagic fever and dengue shock syndrome (Clyde et al., 2006; Gubler, 1998; Halstead, 1988). DENV infections are caused by four serotypes, DENV1–4, which interact differently with antibodies in blood serum. Prior infections of a DENV generate the antibodies that recognize to a DENV of a different serotype but are unable to prevent infection. Instead, these antibodies allow DENV to get into the cells and cause more severe diseases by a process known as antibody-dependent enhancement mechanism (Halstead, 1988). Furthermore, due to a lack of cross-protection by the immune response to one DENV serotype to another serotype, a vaccine requires to be equally protective against all serotypes, which has been difficult to develop. The incidence of DENV infection has increased dramatically in recent decades and the CDC estimates 400 million dengue infections occur each year, resulting in ~25,000 deaths mostly among children and elderly people (Bhatt et al., 2013; Gubler, 2004). Thus, an anti-virus drug treatment effective against all four serotypes as well as pan flavivirus inhibitors are ideal for preventing fatal complications. These broad-spectrum inhibitors would be particularly desirable to prepare for the next flavivirus epidemic, which could emerge from yet unknown or neglected viruses.
Similarly, ZIKV can be transmitted not only by infected mosquitoes, but also from infected people via sexual transmission and through blood transfusion; ZIKV can spread from a pregnant woman to her fetus, causing severe brain malformations in the new-born child including microcephaly and other birth defects (Lazear and Diamond, 2016). It is noteworthy that the neurological manifestations of ZIKV were also observed in DENV endemic regions, suggesting that pre-existing antibody response to DENV could augment ZIKV infection. WNV was previously unknown in the US, and was known to cause only mild disease in the Middle East, first arrived in New York city in 1999 (NY99 strain) and spread throughout the US and Canada by Culex mosquitoes and infected birds. WNV is now endemic in North America (Morrison and Diamond, 2017). Although emerging and re-emerging flaviviruses are significant threat to human health, they have been neglected and did not attract any commercial interest for drug development. This scenario is changing now due to the climatic change and frequent hurricanes and increased prevalence of DENV, WNV, and ZIKV diseases in the US and US territories. Currently, vaccines are available for only a limited number of flaviviruses, and antiviral therapies are not available for any flaviviruses. Hence, there is a grave need to develop therapeutics that interfere with essential steps in the flavivirus lifecycle.
2. Genomic Organization of Flavivirus RNA
Flavivirus RNA genome consists of the 5’ untranslated region (5’ UTR), single open-reading frame (ORF), and 3’ untranslated region (3’ UTR). The 5’ and 3’ UTRs contain RNA stem-loop structures that are essential for viral replicaton (Funk et al., 2010; Goertz and Pijlman, 2015). Flavivirus ORF encodes a single polyprotein which is processed by the host-encoded signalase and the heterodimeric viral NS2B-NS3 protease cotranslationally and post-translationally into mature proteins (reviewed in (Brinton, 2014; Padmanabhan and Strongin, 2010)) (Fig. 1A). Three structural proteins; C, prM and E as well as seven mature non-structural (NS) proteins; NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5 are cleaved by either host signal peptidase or NS3 viral protease. A novel interaction between NS1 and NS4A-2K-NS4B which is required for viral replication but not for formation of the membranous replication organelle, has also been reported by Plaszczyca et al. (Plaszczyca et al., 2019) (see Fig. 1). NS3 and NS5 form a multi-enzyme complex of proteins involved in positive and negative viral RNA syntheses and 5’ capping. Flavivirus NS3 protease has multiple functional domains. The N-terminal domain (aa 1–170) of NS3 codes for the trypsin-like serine protease (Bazan and Fletterick, 1989; Gorbalenya et al., 1989a), followed by a flexible linker of ~20 aa and conserved domains found in RNA helicases (Bartelma and Padmanabhan, 2002; Benarroch et al., 2004; Frick, 2003; Frick et al., 2004; Li et al., 1999; Wu et al., 2005). Similarly, NS5 protein also has multiple functional domains. The N-terminal domain of NS5, has guanine N-7 and ribose 2’-O methylation activities that can methylate GpppA-capped and m(7) GpppA-capped RNAs sequentially, yielding m(7) GpppA and m(7)GpppAm RNA products in the presence of S-adenosyl methionine methyl donor. The conserved motif, K(61)-D(146)-K(182)-E(218) is important for guanine N-7 (Ray et al., 2006) and ribose 2’-O methylation reactions (Egloff et al., 2002; Selisko et al., 2010). N-7 methylation requires only D(146) although other amino acids of the motif facilitate the reaction. The C-terminal domain forms an RNA-dependent RNA polymerase (Gorbalenya et al., 1989b; Issur et al., 2009; Koonin, 1993; Ray et al., 2006; Tay and Vasudevan, 2018; Teramoto et al., 2017; Teramoto et al., 2014; Yon et al., 2005). The viral proteins perform a variety of functions such as viral replication, assembly into mature virions (Apte-Sengupta et al., 2014; Chatel-Chaix and Bartenschlager, 2014; Hodge et al., 2019; Klema et al., 2015; Lescar et al., 2018; Mukhopadhyay et al., 2005; Pierson and Diamond, 2020), evasion of immune response, and disruption cellular homeostatesis in the infected cells (reviewed in (Fischl and Bartenschlager, 2011; Heaton and Randall, 2011; Kirkegaard et al., 2004; Wileman, 2006).
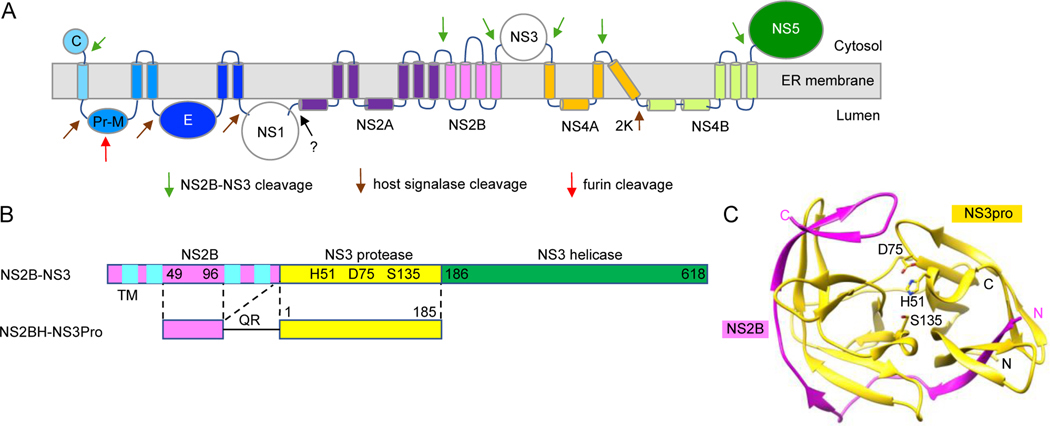
Fig. 1. Flavivirus NS2B-NS3 protease.
A. Flavivirus polyprotein topology and predicted transmembrane domains. Viral protease NS3 cleaves polyprotein from the cytoplasmic side. B. Arrangement of flavivirus DENV2 NS2B-NS3 protease. NS3 protease uses NS2B, a transmembrane (TM) protein with four helices (cyan), as a cofactor. The conserved hydrophilic region of NS2B (NS2BH; 49–96 aa), linked to NS3Pro (yellow), is required for protease activity. The NS2BH-NS3Pro construct used in the HTS is described below. C. Crystal structure of ZIKV protease (PDB code: 5GPI; (Zhang et al., 2016). The protease consists of NS2B peptide (49 aa, pink) and NS3 protease domain (yellow). The catalytic triad, H51, D75 and S135 are shown.
3. Flavivirus protease.
In this regard, the viral protease is an excellent target for drug development. The discovery that NS2B and the N-terminal region of NS3 (NS3Pro) functions as a serine protease involved in the clevage of the polyprotein precursor was made in early studies by sequence homology to the N-terminal region of NS3 (Bazan and Fletterick, 1989; Gorbalenya et al., 1989a), transfection and translation of YFV and DENV2 RNA, and site-directed mutagenesis studies (Chambers et al., 1991; Chambers et al., 1990; Zhang et al., 1992; Zhang and Padmanabhan, 1993). The active viral protease is formed from a hetero-dimeric complex of a 130 amino acid (aa) integral memebrane protein, NS2B, containing four transmembrane (TM) helices linked to the NS3Pro domain (Fig. 1B), which is then self-cleaved at the NS2B-NS3 juction site and dilution-insensitive cleavage of NS2A-NS2B site (Chambers et al., 1990).
In vitro, the active protease complex can be formed from NS2B-NS3Pro precursor with the addition of microsomal membrane in an in vtro coupled transcription/translation system resulting in the cotranslational insertion of NS2B-NS3 precursor into membranes, necessary for the self-cleavage of NS2B-NS3 site. When the central hydrophilic amino acids of NS2B (NS2BH) alone was used as a cofactor for the NS3Pro (Fig. 1B, bottom), addition of membranes was not required for the cis cleavage of NS2BH-NS3Pro site (Clum et al., 1997). Using this information, the NS2BH-NS3Pro precursor containing 49aa of hydrophilic region with the C-terminal 10aa) of NS2B (Fig. 2; shown in red) and the NS3Pro domain, was expressed in E. coli and the recombinant protein was purified by denaturation and refolding. Within the C-terminal 10aa region of NS2B, the hydrophobicity of underlined aa, WYLW, were likely to have contributed to aggregation and localization as insoluble inclusion bodies in E. coli which required denaturation and refolding to enzymatically active protease complex. The in vitro protease assay was established using fluorogenic peptide substrates as well as [35S-Met]-labeled authentic target precursor protein, NS4BFL-NS5(95aa) (Yusof et al., 2000). In subsequent studies, the C-terminal region of NS2BH was shortened to 2aa in DENV2(QR) and ZIKV (KR), and 5aa in WNV (QYTKR) proteases (Abrams et al., 2020; Balasubramanian et al., 2016; Ezgimen et al., 2012; Lai et al., 2013; Mueller et al., 2008; Mueller et al., 2007; Tiew et al., 2012), respectively, for expression, purification, biochemical characterization, and high-throughput screening (HTS) of compound libraries. The recombinant proteins had optimal solubility, yield, and enzymic properties. In another approach, Leung et al described the expression and purification of DENV protease in which NS2BH and NS3Pro were covalently linked by a non-cleavable Gly-rich linker as a single precursor protein, NS2BH-(Gly4-S-Gly4)-NS3Pro (referred here as a “linked protease”) (Leung et al., 2001). The linked protease was highly soluble and enzymatically active in cleaving chromogenic para-nitroanilide (pNA) hexapeptide substrates containing a Lys or Arg at P1 and P2 positions as well as a decapeptide substrate containing four P’ residues which was cleaved between Arg-Ala at the P1 and P1’ positions (Leung et al., 2001). The NS2BH-(Gly4-S-Gly4)-NS3Pro was used in several laboratories for biochemical characterization such as substrate specificity, kinetic parameters of DENV, WNV, and ZIKV as well as for HTS for identification of inhibitors (Abrams et al., 2020; Bera et al., 2007; Chappell et al., 2005; Li et al., 2005; Nall et al., 2004; Niyomrattanakit et al., 2004).
Fig. 2. Alignment of NS2B-NS3pro of DENV1, −2, −3, and −4.
The NS2B is 130aa and NS3Pro domain shown is 180aa after QR (the N-terminal 10 residues of NS3Pro are underlined). The sequences in red represent conserved hydrophilic domain essential for protease activity. The amino acid residues in bold of NS2B upstream of the C-terminus of NS2B are hydrophobic and contribute to insolubility in E, coli expression of DENV2 NS2BH (Yusof et al., 2000).
4. Structure Determinations of “linked” and “unlinked” proteases
The crystal structures of NS2BH-G4SG4-NS3pro were determined in several laboratories (Aleshin et al., 2007; Assenberg et al., 2009; Chandramouli et al., 2010; Chen et al., 2014; Erbel et al., 2006; Noble et al., 2012). The DENV2 NS2BH-(G4-S-G4)-NS3Pro structure was determined in the absence of an inhibitor, whereas the WNV structure contained the substrate peptide-based, Bz-Nle-Lys-Arg-Arg-aldehyde which was covalently linked to catalytic triad Ser135 inhibitor. Comparison of these structures revealed that the NS3Pro domains adopt a chymotrypsin-like fold in both structures with two β-barrels, each formed by six β-strands, with the catalytic triad (His51-Asp75-Ser135) located at the cleft between the two β-barrels (Fig. 1C). The striking difference was observed in the DENV2 structure in the absence of the inhibitor in which the C-terminal aa of NS2BH beyond the residue 76 was disordered and assumed an “open” conformation. On the other hand, in the presence of the inhibitor, the C-terminal region of NS2BH wrapped around the inhibitor to form a closed conformation. Aleshin et al. determined the crystal structure of WNV NS2BH-Gly4-S-Gly4-NS3Pro in the presence and absence of the bovine pancreatic trypsin inhibitor, aprotinin (BPTI) confirming that NS2BH could adopt two distinct conformations (Aleshin et al., 2007). Chandramouli et. al reported the crystal structure of DENV1 NS2BH (aa 49–95 hydrophilic domain) fused to Gly4-S-Gly4-NS3Pro domain which also had the N-terminal deletion of NS3ProΔ11–20 aa (Chandramouli et al., 2010). Assenberg et al. reported the crystal structure of full-length NS3 protein linked to the NS2BH-Gly4-S-Gly4 hydrophilic domain of MVEV (Assenberg et al., 2009). From the biochemical characterization of the protease and the helicase, the authors concluded that the protease domain had little influence on the helicase activity and vice versa. This conclusion from MVEV was not applicable to DENV2, DENV4 and WNV (Chernov et al., 2008; Xu et al., 2005; Yon et al., 2005). The protease domain did seem to influence the helicase activity depending on the native inter-domain region between the protease and the helicase domains. The relative orientations of the protease and the helicase of DENV4 NS3 were also determined by crystallography (Luo et al. 2010; Phoo et al. 2020). For the structure determined by Luo et al. (Luo et al., 2010), only the C-terminal 18 aa of NS2B was linked to the full-length DENV4 NS3 and hence it is inactive as protease. However, the structure of the full-length NS3 protein showed that there were two conformations (I and II) differing with respect to the flexible linker region (169–179aa) between the protease and helicase domains. In conformation II, the helicase domain is more exposed and is active in binding RNA and ATP hydrolysis steps involved in RNA helicase activity (Luo et al., 2010). In the study reported by Phoo et al. (Phoo et al., 2020), the authors compared for the first time the structural features of DENV4 NS2BH and NS3 full-length complexes formed in three different ways: (1) bNS2B47-NS3 (2) cleavable eNS2B47-NS3, and (3) gNS2B47-NS3 containing Gly linker. The authors reported the crystal structures of linked and unlinked NS3 proteases in their free states and in complex with BPTI. Their results showed that NS2BH essentially adopted a closed conformation in the absence of Gly linker which promoted the shift toward an open conformation interfering with the protease activity with little effect on ATPase and helicase activities. In parallel studies, our group used the cleavable NS2BH-NS3Pro domains of DENV and WNV for biochemical and HTS studies in the active ‘closed’ conformation (Balasubramanian et al., 2016; Clum et al., 1997; Mueller et al., 2008; Mueller et al., 2007; Yon et al., 2005; Yusof et al., 2000).
Structure studies performed by NMR in solution have revealed a more complete picture of the dynamics of NS2B-NS3Pro in post-proteolysis state of the enzyme during polyprotein processing. For example, the structure of the last four amino acids of NS2BH cofactor bound at the NS3 active site agrees with the NMR studies and protease activity assays (Phoo et al., 2016). The structure determinations by NMR in solution also revealed that the NS2BH-NS3Pro predominantly exist in closed conformation which offer valuable insight into the conformational changes of the proteases in the absence and presence of substrates and inhibitors which could be useful for the development of potent inhibitors of flavivirus infections (Li and Kang, 2020). NMR spectroscopy has been used to study complexes formed between low molecular weight inhibitors and the WNV NS2B-NS3Pro. These studies revealed that inhibitor binding near the substrate binding site rather than an allosteric site(s) (Su et al., 2009).
5. Identification of direct-acting antivirals against flavivirus proteases.
Direct-acting antivirals would offer specificity, safety and enhanced efficacy for treatment of flavivirus infections compared to non-specific therapeutics such as interferon and ribavirin- based treatments to inhibit flavivirus replication and infection. Viral proteases are an excellent target for anti-viral therapeutics due to their essential function in production of viral proteins (i.e., polyprotein cleavage) and assembly of viral replication complex. Several viral protease inhibitors have been developed into successful therapeutics to treat viral infections. FDA-approved viral inhibitors include for HIV (Lv et. al., 2015) and hepatitis C virus protease inhibitors (telaprevir) (Bukhtiyarova et al., 2001). The flavivirus NS2B-NS3 protease has been a major target for developing inhibitors against flavivirus infections. Peptide-based compounds often exhibit nanomolar affinities for flavivirus proteases but suffer from limited antiviral activities in cellular assays and animal models, which prevents further development into antiviral therapeutics (Abrams et al., 2020; Balasubramanian et al., 2016; Jiang et al., 2022; Lima et al., 2021; Mirza et al., 2022; Nitsche, 2018). Alternatively, HTS of small molecules using in vitro protease activity assays and in silico binding screens have been used to identify inhibitors. We and others have used HTS of large compound libraries and virtual screening, targeting DENV2, WNV, and ZIKV proteases, and identified several compounds with novel chemical scaffolds that not only inhibit the viral protease in vitro but also viral replication in cultured mammalian cells (Mueller et al., 2007). Several of these compounds show a broad-spectrum inhibition for DENV1–4 and WNV NS3 protease (Balasubramanian et al., 2016; Li et al., 2017). Since the activity of NS2B-NS3 protease requires formation of an active protein complex between NS2B and NS3 in the viral replication complex, small molecule inhibitors that target flavivirus NS2B and NS3 interaction is a strategy to treat viral infections.
WNV NS3 protease HTS.
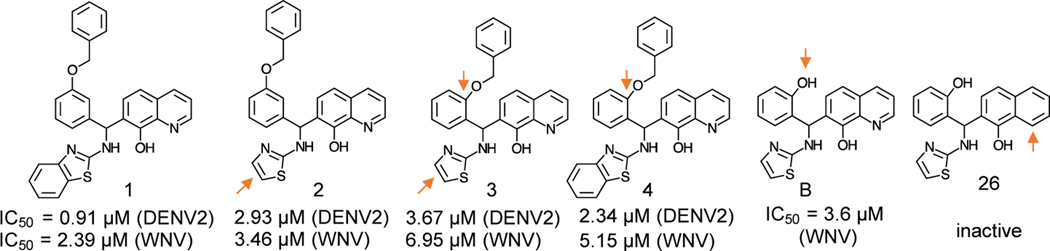
For identification WNV NS3 protease inhibitors, we screened ~32K compounds using WNV NS2BH-NS3Pro in our first HTS (Ezgimen et al., 2012; Mueller et al., 2008). We identified compounds containing the 8-hydroxyquinoline (8-OHQ) scaffold which inhibited both WNV and DENV2 proteases in vitro and Renilla luciferase (Rluc) reporter replicon assays (Mueller et al., 2008) (Fig. 3). The 8-OHQ containing compounds were validated by structure-activity relationships (SAR), and their IC50 values for the 8-OHQ derivatives for the purified DENV2 and WNV protease are shown in Fig. 3. Compound 26 which lacks N1 nitrogen was inactive, confirming that the 8-OHQ ring is required for the inhibitory activity.
Fig 3. WNV protease inhibitors with 8-OHQ scaffold.
IC50 was determined by protease assay using DENV2 and WNV NS2BH-NS3Pro. The positions of substitutions in comparison to compound 1 are indicated by an arrow.
DENV2 NS3 protease HTS.
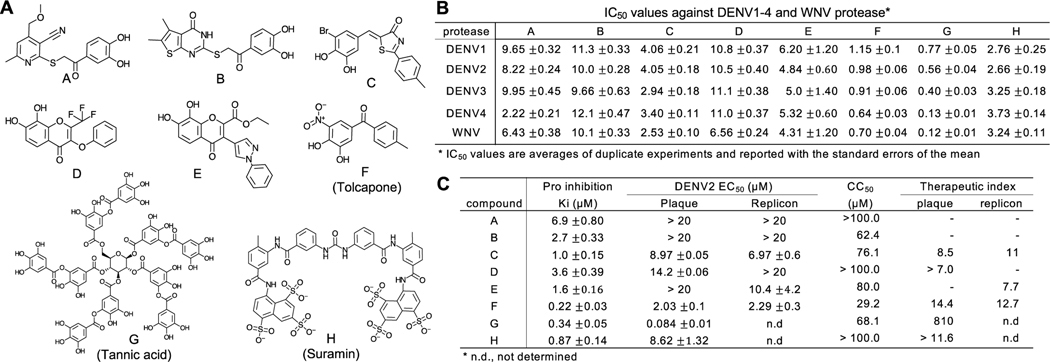
In the second HTS campaign, we screened ~120K compounds in the 384-well HTS format using DENV2 NS2BH-NS3Pro (Balasubramanian et al., 2016). After secondary validation in the 96-well format, we selected 8 ‘hit’ compounds (A-H), 7 of which have the general structure of catechols (1,2-dihydroxybenzne), and analyzed their activity to inhibit the NS3 protease from four DENV serotypes and WNV (Fig 4A–B). The IC50 values range from 120 nM to 10 μM; Five compounds (C, E, F, G, H) had IC50 < 5 μM for all DENV1–4 and WNV protease. We then determined their kinetic constants (Ki) using DENV2 protease, and EC50 by plaque assay and Renilla luciferase (Rluc) reporter replicon assay (Fig. 4C). Compounds C, D, F, G, and H efficiently suppressed virus and replicon replication in cell-based assays (Fig. 4C) (Balasubramanian et al., 2016). All compounds had the cytotoxic concentration (CC50) > 29 μM, and therapeutic index (TI = CC50/EC50) > 7 in DENV2 infection. Although G (tannic acid) and H (Suramin) were identified as protease inhibitors in the HTS and inhibit DENV1–4 and WNV proteases in vitro, they are considered as promiscuous inhibitors and reported to inhibit a variety of viruses (Balasubramanian et al., 2016) and illnesses affecting human health. For example, Suramin is also described as an anti-cancer agent (Marutsuka et al., 1995; Vogelzang et al., 2004), as an anti-HIV reverse transcriptase agent (Mahoney et al., 1990), and that blocked the entry of DENV to target cells; (Chen et al., 1997).
Fig 4. Hit compounds identified by DENV2 NS3 protease HTS.
A. Chemical structures of hit compounds A-H. B. IC50 values of the compounds A-H against DENV1,2,3,4 and WNV NS3 protease. C. Ki, EC50, CC50, and selective index for compound A-H. Ki was measured by in vitro protease assay and EC50 was measured by plaque and replicon assays.
DENV NS3 protease virtual screen.
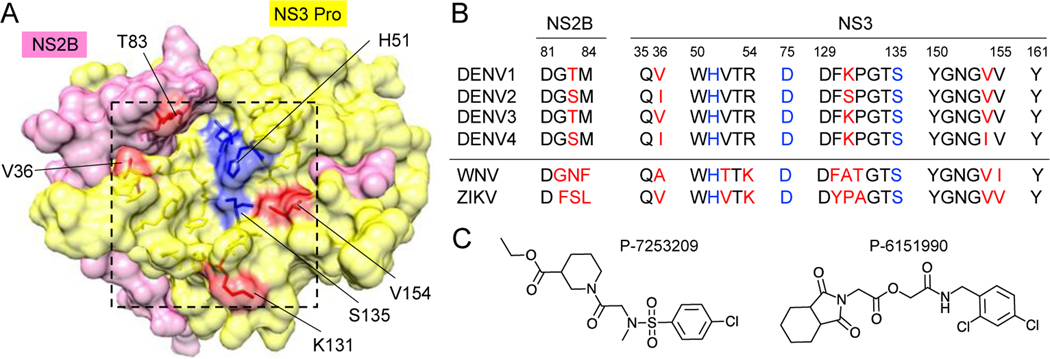
Identification of small molecules that inhibit all four DENV proteases would be desirable for development of pan flavivirus protease inhibitors. Although DENV1–4 NS3 proteases share high sequence identity (67–72 %), the residues near the active site are not identical (Fig 5). Previous DENV2 protease structures were determined using proteins with linked (uncleavable glycine-rich G4-S-G4 linker) NS2B-NS3 protease and did not show electron density for the entire NS2B peptide (open conformation), likely represent inactive conformation. In contrast, the cleaved form of protease (using the construct similar to our group) is in a closed conformation, where entire NS2BH was visible.
Fig 5. Substrate binding site of flavivirus protease.
A. Substrate binding site of DENV NS2B-NS3 protease (boxed) is shown in the same orientation as Fig 1C. The substrate-binding site residues that differ in DENV1–4 protease (B) are colored in red. The active site residues are shown in blue. C. DENV2 protease inhibitors identified by virtual screen.
If the crystal structures of the targets are known, then virtual screens can be employed to identify new compounds that bind to the protein targets by using computational methods. For example, in one study, the search was carried out by using Swiss Similarity tool and molecular docking calculations, molecular dynamics simulations (MD) and free energy calculations with the compounds in the ZINC databases that led to the identification of the compounds with favorable druglike properties (Costa et al., 2022). Rarey et al. described a fast, flexible, and automatic method for docking organic ligands into protein binding sites (Rarey et al., 1996). A new software module, FlexX-Scan, is described which can perform a high throughput, structure-based virtual screening (Schellhammer and Rarey, 2004), which is based on the incremental construction docking tool, FlexX, described by Rarey et al. (Rarey et al., 1996).
Inhibitors targeting ZIKV protease
ZIKV infects and disrupts development in human neural stem cell (NSC) and fetal infection can result in congenital defects and microcephaly. Small molecule inhibitors show different activities in Vero vs. NSC, and thus, compounds need to be screened for their ability to inhibit ZIKV in human NSCs. We have used a combination of HTS and virtual screen to identify ZIKV protease inhibitors (Fig. 6). Selected compounds were tested using in vitro protease assay, Vero cell assay, and NSC assay, and a total of 15 compounds were identified with diverse chemical scaffolds. Thus far, we have characterized three groups of compounds, tetracycline, MK-591, and JNJ404 for their ability to inhibit viral protease and viral infection. All three groups of compounds show activity in Vero cells and NSC.
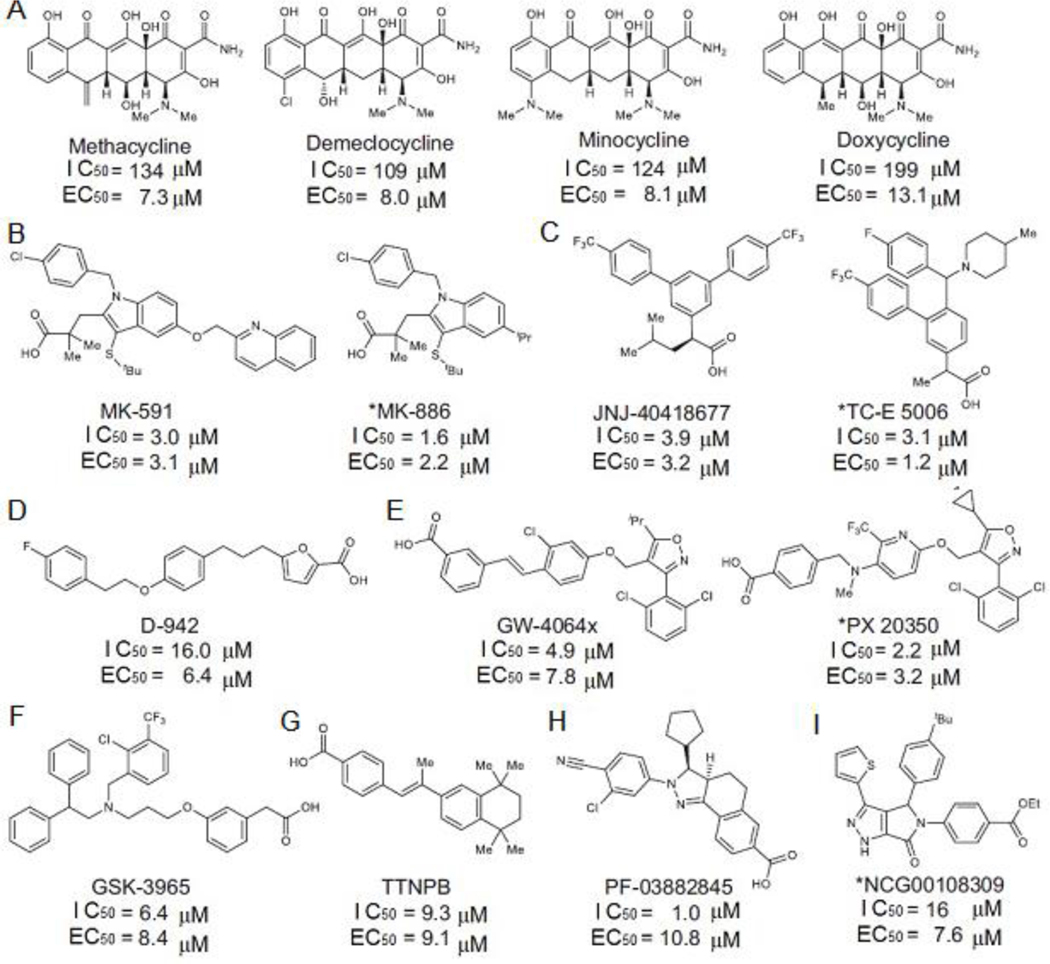
Fig 6. Hits identified from ZIKV protease HTS and viral infection assays in neural stem cells (NSC).
The 15 compounds can be divided into 9 classes of chemicals, A-I. The IC50 values were determined using ZIKV protease and EC50 determined in ZIKV infection assay in NSC.
To identify small molecule inhibitors against ZIKV protease, we used three strategies (Fig 6). First, a pilot HTS was performed using the ZIKV (Brazilian strain) NS2BH-NS3Pro as the target at the National Center for Advancing Translational Sciences (NCATS).(Abrams et al., 2020) In the study, ~2,000 compounds were screened using an AMC-labeled hexapeptide, Ac-VKTGKR-AMC (Ac: acetyl). Three classes of compounds were identified, flavonoids, anthraquinones and tetracyclines. Due to their commercial availability and ability to cross the blood-brain barrier, 11 tetracycline family of drugs were chosen for further study using human NSC assay. Members of the tetracycline family were not effective in inhibiting protease activity in vitro, but four compounds (Fig 6A) suppressed ZIKV replication in NSC with EC50 of 7.3 – 13.1 μM, suggesting that they may have multiple mechanisms of action. Methacycline was most potent (EC50 = 7.3 μM) and reduced the ZIKV in the brain and the ZIKV-induced motor deficits in an immunocompetent mouse model (Abrams et al., 2020) Since the tetracyclines are FDA-approved drugs, it could be quickly brought to clinical practice. Second, in a concurrent quantitative HTS (qHTS) assay in 1054-well plates, a total of 10,807 compounds were screened using in vitro protease assays, from which 272 compounds were selected. Additionally, 83 of known protease inhibitors were tested in the protease assay, and 35 were active. Finally, we used an AI-based QSAR model for virtual screening of 137,083 compounds, and selected 277 predicted inhibitors for protease assay, of which 153 compounds showed inhibition. Thus, a total of 460 hit compounds were identified from the ZIKV protease assay. The hit compounds were next used in consecutive Rluc replicon assay, mCherry infectivity assays, and qRT-PCR in Vero cells. This step eliminated ~95% of the selected compounds, leaving 18 compounds for further analysis in a more physiologically relevant human NSC. Third, the 460 compounds were directly taken to NSC assays. We found that 11 compounds were active in NSC assays. The most active compounds were MK-591 and JNJ-40418677, which have similar IC50 and EC50 values measured in protease activity and NSC-based assays, respectively (3.0 and 3.1 μM for MK-591 and 3.9 and 3.2 μM for JNJ-40418677), suggesting that the compounds inhibit viral replication by blocking viral protease activity (Fig 6B–C) (Abrams et al., 2020). MK-591 was previously identified as a five-lipoxygenase–activating protein (FLAP) inhibitor that would potentially treat a wide range of inflammatory diseases such as asthma (Gurusamy and Abdul, 2019). With its good safety profile, MK-591 is a promising lead compound to treat a ZIKV infection (Abrams et al., 2020)
Within the identified 15 hit compounds from HTS (Fig 6), except four tetracyclines, the 11 compounds show good correlation between the ZIKV protease inhibition (IC50 of 1.0–9.3 μM) and viral replication in NSC (EC50 of 1.2 –10.8 μM), suggesting that they are direct-acting inhibitors of viral replication via the inhibition of viral protease. A new software module, FlexX-Scan has been described in literature to facilitate the structure-based high throughput virtual screening process.
6. Survey of literature between 2018 and 2022 on flavivirus protease functions
Martinez et al. performed a WNV NS2B-NS3Pro enzymatic screen of NIH clinical compound library and identified Zafirlukast, an FDA-approved drug for asthma, and its derivatives, as an inhibitor of WNV NS2B-NS3Pro with an IC50 value of 32 μM. A limited SAR study revealed that replacing the cyclopentenyl with a phenyl moiety improved the inhibition. The mode of inhibition was studied by experimental and computational methods to be by allosteric mechanism by blocking the binding of the NS2B cofactor to NS3pro (Martinez et al., 2018). Yao et al. reported by compound screening and medicinal chemistry, several drug-like broad spectrum inhibitors of flavivirus proteases with IC50 values of ~120 nM, and exhibited antiviral activities in cultured cells with EC68 of ~300–600 nM and in a mouse model for Zika virus. The X-ray structural studies revealed that the inhibitors bind to mostly to an allosteric site, a hydrophobic pocket of DENV NS3 protease and keep it in an open, catalytically inactive conformation (Yao et al., 2019). The placement of the inhibitor in the allosteric pocket is perhaps debatable as discussed by Behnam and Klein (Behnam and Klein, 2020). Bharadwaj et al using virtual screening followed by in vitro assay reported that triterpenoids from the medicinal fungus, Ganodermalucidum, are inhibitors of DENV NS2B-NS3pro. Their analysis was followed by binding affinities and stability calculations using the molecular mechanics / Born surface area method and MD simulations, respectively. Inhibition of viral infection in vitro suggested that Ganodermanontriol is a potent bioactive triterpenoid (Bharadwaj et al., 2019). Kuhl et al. (Kuhl et al., 2021) reported a new class of DENV and WNV inhibitors using chemical synthesis, in vitro SAR studies, DENV2 protease, Renilla remiformis luciferase reporter assays in Hela cells containing NS2BFL-NS3pro.
The DENV protease inhibitor development efforts between 2015–2020 have been reviewed (Murtuja et al., 2021). Kaptein et al. (Kaptein et al., 2021) reported the identification and mode of action of a compound, JNJ-A07, de novo formation of NS3 and NS4B complex which is required for viral replication. The viral protease is indirectly involved in the formation of mature NS3 and NS4B components of the replication complex. The authors show that once the mature NS3/NS4B complex is formed, JNJ-A07 had no effect. JNJ-A07 is a highly potent inhibitor with nanomolar to picomolar potency and high selectivity in various cell lines including the immature dendritic cells (Kaptein et al., 2021). Analogs of JNJ-A07 are being investigated for their antiviral effects. Kuhl et al. reported the discovery of non-basic benzamide derivatives as inhibitors of DENV2pro having sub micromolar efficacy (EC50 = 0.24 μM in plaque reduction assay with a cytotoxicity (CC50) of > 100μM. The compound was stable against liver microsomes and pancreatic enzymes (Kuhl et al., 2021). In a subsequent study, the same group presented synthesis and analysis of benzoxaborole inhibitors of DENV NS2B-NS3pro. The most active compound had an EC50 of 0.54 μM in inhibition of viral replication with no relevant cytotoxicity. The active compound also inhibited SARS-CoV-2 Mpro with a single digit micromolar EC50 (Kuhl et al., 2022). In one study, targeting the NS2B-NS3pro of TBEV, which causes neurological complications like WNV, Akaberi et al., reported peptide inhibitors of the viral protease. The IC50 values of 0.92 μM and 0.25 μM for two tripeptides in the in vitro enzymatic assays. No cell culture experiments like viral infection and cytotoxicity assays were performed (Akaberi et al., 2021). Li et. al. identified niclosamide as an inhibitor of ZIKV NS3pro interaction with NS2B by targeting the interface between the protease subunits. The drug discovery and development efforts for ZIKV and WNV NS2B-NS3pro between 2015–2021 has been recently reviewed (Samrat et al., 2022). Cheng et al. described the discovery of potent DENV NS2B-NS3pro covalent inhibitors containing phenoxymethylphenyl residue (Cheng et al., 2022). They previously reported a covalent inhibitor WSL-01 (IC50=129 nM) and with SAR studies, they improved the potency in two analogs WSL-75 with IC50 of 24.8 nM and WSL-84, IC50 of 32.89 nM. This class of compounds exhibited passive membrane permeability using the precoated tri-layer parallel artificial membrane. The authors presented detailed SAR analysis and MD simulations that reveal binding modes of covalent inhibitors (Cheng et al., 2022). The cytotoxicity of the compounds was measured at 5 μM concentrations although no CC50 values were reported. The same group also identified Montelukast as a competitive inhibitor of DENV and ZIKV replicon replication with EC50 values of 1.03 nM for DENV and 1.14 nM for ZIKV by using an in silico approach (Jiang et al., 2022) involving MD simulations and binding free energy calculations followed by in vitro protease assays and replicon replication assays. Since the potency of this compound is weak in the in vitro protease assays compared to replicon replication assays in DENV and ZIKV-infected cells, Montelukast may not act as a direct-acting antiviral compound as an inhibitors of DENV and ZIKV proteases (Jiang et al., 2022). In another study, using the crystal structure of ZIKV NS2B-NS3pro, a computer-aided structure-based approach was used for screening diverse library of compounds. The top hits were selected based on free energy calculations followed by per-residue decomposition analysis. The selected hits were evaluated for their biological potency using ZIKV protease inhibition assays as well as antiviral activity assays (Mirza et al., 2022). Of the 26 selected compounds, 8 showed inhibition of ZIKV protease >25% at 10μM. Of these selected compounds, only one showed anti-ZIKV activity in cells. The CC50 values were reportedly >100 μM and the EC50 value of 9.79 ± 1.20 μM. Colarusso et al., (2022) described using a SAR study on a series of substrate-like linear tripeptides as non-covalent inhibitors of ZIKV NS2B-NS3Pro. They optimized residues at P1, P2, P3, and N- and C-terminal portions of tripeptides. The reported identification of inhibitors with sub-micromolar potency. Their results indicated phenyl glycine as Arg-mimicking group and benzamide as the C-terminal fragment. Extending the SAR studies, the authors found a series of peptides having a 4-substituted phenylglycine residue at the P1 position gave rise to potent tripeptides showing low nanomolar inhibition of ZIKV protease (IC50=30 nM) with high selectivity against trypsin-like serine proteases with no inhibition of thrombin and low inhibition of trypsin (IC50=203 μM) as well as other flaviviruses such as DENV2 and WNV proteases. In spite of the highly potent in vitro protease activity, the two tri-peptides showed weak activities in inhibiting ZIKV replication in Vero cells with EC50 values of 8.5 μM and 11.2 μM which the authors attributed to low permeability contributed by basic side chains of two Lys residues. In vitro assays against DENV2 and WNV proteases showed good selectivity of the most potent tripeptides as the IC50 values were 0.2 μM and 0.8 μM against DENV2 and WNV proteases.
7. Concluding remarks and future directions.
The race is on to discover potent pan flavivirus protease inhibitors to be ready before a future pandemic strikes the world population. We are seeing a dramatic increase in the number of natural calamities brought by global warming in the US as well as globally. The number of cases and the severity of the disease are likely to increase in future pandemics. It is also heartening to know the research community realizes the dire need in bringing antiviral therapies with the collaboration with big pharmas as seen from the increase in the number of research articles and reviews. We apologize for any omission of other important citations in this review. Research to date focused on identification and development of flavivirus NS3 protease inhibitors using HTS and in silico assays mostly using the isolated NS2BH and NS3 protease domains; thus, other functions of full-length NS2B and NS3 have not been explored as a drug target. In virus-infected cells, the NS3 protein is not cleaved into protease and helicase domains, and thus the full-length NS2B-NS3 is the more authentic target than the NS2BH and NS3 protease domains.
Acknowledgments:
The research in the authors laboratories had been supported by NIH grants, R01 AI32078 (R.P.), U01 AI54776 (Craig E. Cameron and R.P.), R01 AI087856 (K.H.C. and R.P.), R01 AI05985 (K.H.C.), R21 AI154088 (T.T.) and U19 AI171413 (K.H.C).
Abbreviations:
- aa
amino acid
- DENV
Dengue Virus
- DENV1, −2, −3, −4
DENV serotypes 1–4
- HTS
high-throughput screening
- MVEV
Murray Valley Encephalitis Virus
- NS protein
Non-structural protein
- NS2BH
NS2B hydrophilic region
- NS3Pro
NS3 protease domain
- SAR
structure-activity relationships
- TBEV
Tick-borne Encephalitis Virus
- WNV
West Nile Virus
- YFV
Yellow Fever Virus
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams RPM, Yasgar A, Teramoto T, Lee MH, Dorjsuren D, Eastman RT, Malik N, Zakharov AV, Li W, Bachani M, Brimacombe K, Steiner JP, Hall MD, Balasubramanian A, Jadhav A, Padmanabhan R, Simeonov A, Nath A, 2020. Therapeutic candidates for the Zika virus identified by a high-throughput screen for Zika protease inhibitors. Proc Natl Acad Sci U S A 117, 31365–31375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaberi D, Bahlstrom A, Chinthakindi PK, Nyman T, Sandstrom A, Jarhult JD, Palanisamy N, Lundkvist A, Lennerstrand J, 2021. Targeting the NS2B-NS3 protease of tick-borne encephalitis virus with pan-flaviviral protease inhibitors. Antiviral Res 190, 105074. [DOI] [PubMed] [Google Scholar]
- Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC, 2007. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci 16, 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apte-Sengupta S, Sirohi D, Kuhn RJ, 2014. Coupling of replication and assembly in flaviviruses. Curr Opin Virol 9, 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenber R, Mastrangel E, Walte TS, Verm A, Milan M, Owen RJ, Stuar DI, Grime JM, Mancin EJ, 2009. Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J Virol 83, 12895–12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian A, Manzano M, Teramoto T, Pilankatta R, Padmanabhan R, 2016. High-throughput screening for the identification of small-molecule inhibitors of the flaviviral protease. Antiviral Res 134, 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelma G, Padmanabhan R, 2002. Expression, purification, and characterization of the RNA 5’-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology 299, 122–132. [DOI] [PubMed] [Google Scholar]
- Bazan JF, Fletterick RJ, 1989. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology 171, 637–639. [DOI] [PubMed] [Google Scholar]
- Behnam MAM, Klein CDP, 2020. Conformational selection in the flaviviral NS2B-NS3 protease. Biochimie 174, 117–125. [DOI] [PubMed] [Google Scholar]
- Benarroch D, Selisko B, Locatelli GA, Maga G, Romette JL, Canard B, 2004. The RNA helicase, nucleotide 5’-triphosphatase, and RNA 5’-triphosphatase activities of Dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology 328, 208–218. [DOI] [PubMed] [Google Scholar]
- Bera AK, Kuhn RJ, Smith JL, 2007. Functional characterization of cis and trans activity of the Flavivirus NS2B-NS3 protease. J Biol Chem 282, 12883–12892. [DOI] [PubMed] [Google Scholar]
- Bharadwaj S, Lee KE, Dwivedi VD, Yadava U, Panwar A, Lucas SJ, Pandey A, Kang SG, 2019. Discovery of Ganoderma lucidum triterpenoids as potential inhibitors against Dengue virus NS2B-NS3 protease. Sci Rep 9, 19059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI, 2013. The global distribution and burden of dengue. Nature 496, 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton MA, 2014. Replication cycle and molecular biology of the West Nile virus. Viruses 6, 13–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhtiyarova M, Rizzo CJ, Kettner CA, Korant BD, Scarnati HT, King RW, 2001. Inhibition of the bovine viral diarrhoea virus NS3 serine protease by a boron-modified peptidyl mimetic of its natural substrate. Antivir Chem Chemother 12, 367–373. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Grakoui A, Rice CM, 1991. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol 65, 6042–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Weir RC, Grakoui A, McCourt DW, Bazan JF, Fletterick RJ, Rice CM, 1990. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A 87, 8898–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli S, Joseph JS, Daudenarde S, Gatchalian J, Cornillez-Ty C, Kuhn P, 2010. Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J Virol 84, 3059–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell KJ, Nall TA, Stoermer MJ, Fang NX, Tyndall JD, Fairlie DP, Young PR, 2005. Site-directed mutagenesis and kinetic studies of the West Nile Virus NS3 protease identify key enzyme-substrate interactions. J Biol Chem 280, 2896–2903. [DOI] [PubMed] [Google Scholar]
- Chatel-Chaix L, Bartenschlager R, 2014. Dengue virus- and hepatitis C virus-induced replication and assembly compartments: the enemy inside--caught in the web. J Virol 88, 5907–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WN, Loscha KV, Nitsche C, Graham B, Otting G, 2014. The dengue virus NS2B-NS3 protease retains the closed conformation in the complex with BPTI. FEBS Lett 588, 2206–2211. [DOI] [PubMed] [Google Scholar]
- Chen Y, Maguire T, Marks RM, 1997. Dengue Virus Infectivity Depends on Envelope Protein Binding to Target Cell Heparan Sulfate. Nat. Med 3, 8765–8772. [DOI] [PubMed] [Google Scholar]
- Cheng J, Feng S, Zhang Y, Ding T, Jiang H, Zhang Z, Wang J, Wang X, Cheng M, 2022. Discovery of highly potent DENV NS2B-NS3 covalent inhibitors containing a phenoxymethylphenyl residue. Biochem Biophys Res Commun 627, 214–219. [DOI] [PubMed] [Google Scholar]
- Chernov AV, Shiryaev SA, Aleshin AE, Ratnikov BI, Smith JW, Liddington RC, Strongin AY, 2008. The two-component NS2B-NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J Biol Chem 283, 17270–17278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clum S, Ebner KE, Padmanabhan R, 1997. Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J Biol Chem 272, 30715–30723. [DOI] [PubMed] [Google Scholar]
- Clyde K, Kyle JL, Harris E, 2006. Recent advances in deciphering viral and host determinants of dengue virus replication and pathogenesis. J Virol 80, 11418–11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colarusso S, Ferrigno F, Ponzi S, Pavone F, Conte I, Abate L, Beghetto E, Missineo A, Amaudrut J, Bresciani A, Paonessa G, Tomei L, Montalbetti C, Bianchi E, Toniatti C, Ontoria J, 2022. Bioorg Med Chem 57,116631. [DOI] [PubMed] [Google Scholar]
- Costa RAD, Rocha J, Pinheiro AS, Costa A, Rocha E, Silva RC, Goncalves ADS, Santos CBR, Brasil D, 2022. A Computational Approach Applied to the Study of Potential Allosteric Inhibitors Protease NS2B/NS3 from Dengue Virus. Molecules 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B, 2002. An RNA cap (nucleoside-2’-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. Embo J 21, 2757–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U, 2006. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol 13, 372–373. [DOI] [PubMed] [Google Scholar]
- Ezgimen M, Lai H, Mueller NH, Lee K, Cuny G, Ostrov DA, Padmanabhan R, 2012. Characterization of the 8-hydroxyquinoline scaffold for inhibitors of West Nile virus serine protease. Antiviral Res 94, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl W, Bartenschlager R, 2011. Exploitation of cellular pathways by Dengue virus. Curr Opin Microbiol 14, 470–475. [DOI] [PubMed] [Google Scholar]
- Frick DN, 2003. Helicases as antiviral drug targets. Drug News Perspect 16, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN, Rypma RS, Lam AM, Gu B, 2004. The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J Biol Chem 279, 1269–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi PY, Khromykh AA, 2010. RNA structures required for production of subgenomic flavivirus RNA. J Virol 84, 11407–11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goertz GP, Pijlman GP, 2015. Dengue Non-coding RNA: TRIMmed for Transmission. Cell Host Microbe 18, 133–134. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Donchenko AP, Koonin EV, Blinov VM, 1989a. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res 17, 3889–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM, 1989b. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res 17, 4713–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ, 1998. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 11, 480–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler DJ, 2004. The changing epidemiology of yellow fever and dengue, 1900 to 2003: full circle? Comp Immunol Microbiol Infect Dis 27, 319–330. [DOI] [PubMed] [Google Scholar]
- Gurusamy M, Abdul JF, 2019. Lead Optimization Studies Towards Finding NS2B/NS3 Protease Targetspecific Inhibitors as Potential Anti-dengue Drug-like Compounds. Curr Drug Discov Technol 16, 307–314. [DOI] [PubMed] [Google Scholar]
- Halstead SB, 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239, 476–481. [DOI] [PubMed] [Google Scholar]
- Heaton NS, Randall G, 2011. Dengue virus and autophagy. Viruses 3, 1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge K, Kamkaew M, Pisitkun T, Chimnaronk S, 2019. Flavors of Flaviviral RNA Structure: towards an Integrated View of RNA Function from Translation through Encapsidation. Bioessays 41, e1900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, Bisaillon M, 2009. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA 15, 2340–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Zhang Y, Wu Y, Cheng J, Feng S, Wang J, Wang X, Cheng M, 2022. Identification of Montelukast as flavivirus NS2B-NS3 protease inhibitor by inverse virtual screening and experimental validation. Biochem Biophys Res Commun 606, 87–93. [DOI] [PubMed] [Google Scholar]
- Kaptein SJF, Goethals O, Kiemel D, Marchand A, Kesteleyn B, Bonfanti JF, Bardiot D, Stoops B, Jonckers THM, Dallmeier K, Geluykens P, Thys K, Crabbe M, Chatel-Chaix L, Munster M, Querat G, Touret F, de Lamballerie X, Raboisson P, Simmen K, Chaltin P, Bartenschlager R, Van Loock M, Neyts J, 2021. A pan-serotype dengue virus inhibitor targeting the NS3-NS4B interaction. Nature 598, 504–509. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K, Taylor MP, Jackson WT, 2004. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol 2, 301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klema VJ, Padmanabhan R, Choi KH, 2015. Flaviviral Replication Complex: Coordination between RNA Synthesis and 51-RNA Capping. Viruses 7, 4640–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, 1993. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol 74, 733–740. [DOI] [PubMed] [Google Scholar]
- Kuhl N, Lang J, Leuthold MM, Klein CD, 2022. Discovery of potent benzoxaborole inhibitors against SARS-CoV-2 main and dengue virus proteases. Eur J Med Chem 240, 114585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl N, Leuthold MM, Behnam MAM, Klein CD, 2021. Beyond Basicity: Discovery of Nonbasic DENV-2 Protease Inhibitors with Potent Activity in Cell Culture. J Med Chem 64, 4567–4587. [DOI] [PubMed] [Google Scholar]
- Lai H, Sridhar Prasad G, Padmanabhan R, 2013. Characterization of 8-hydroxyquinoline derivatives containing aminobenzothiazole as inhibitors of dengue virus type 2 protease in vitro. Antiviral Res 97, 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Diamond MS, 2016. Zika Virus: New Clinical Syndromes and its Emergence in the Western Hemisphere. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J, Soh S, Lee LT, Vasudevan SG, Kang C, Lim SP, 2018. The Dengue Virus Replication Complex: From RNA Replication to Protein-Protein Interactions to Evasion of Innate Immunity. Adv Exp Med Biol 1062, 115–129. [DOI] [PubMed] [Google Scholar]
- Leung D, Schroder K, White H, Fang NX, Stoermer MJ, Abbenante G, Martin JL, Young PR, Fairlie DP, 2001. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J Biol Chem 276, 45762–45771. [DOI] [PubMed] [Google Scholar]
- Li H, Clum S, You S, Ebner KE, Padmanabhan R, 1999. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol 73, 3108–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lim SP, Beer D, Patel V, Wen D, Tumanut C, Tully DC, Williams JA, Jiricek J, Priestle JP, Harris JL, Vasudevan SG, 2005. Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J Biol Chem 280, 28766–28774. [DOI] [PubMed] [Google Scholar]
- Li Q, Kang C, 2020. Insights into Structures and Dynamics of Flavivirus Proteases from NMR Studies. Int J Mol Sci 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Brecher M, Deng YQ, Zhang J, Sakamuru S, Liu B, Huang R, Koetzner CA, Allen CA, Jones SA, Chen H, Zhang NN, Tian M, Gao F, Lin Q, Banavali N, Zhou J, Boles N, Xia M, Kramer LD, Qin CF, Li H, 2017. Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res 27, 1046–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima CS, Mottin M, de Assis LR, Mesquita N, Sousa BKP, Coimbra LD, Santos KB, Zorn KM, Guido RVC, Ekins S, Marques RE, Proenca-Modena JL, Oliva G, Andrade CH, Regasini LO, 2021. Flavonoids from Pterogyne nitens as Zika virus NS2B-NS3 protease inhibitors. Bioorg Chem 109, 104719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Wei N, Doan DN, Paradkar PN, Chong Y, Davidson AD, Kotaka M, Lescar J, Vasudevan SG, 2010. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J Biol Chem 285, 18817–18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z, Chu Y, Wang Y, 2015. HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV AIDS (Auckl) 7, 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney CW, Azzi A, Huang KP, 1990. Effects of suramin, an anti-human immunodeficiency virus reverse transcriptase agent, on protein kinase C. Differential activation and inhibition of protein kinase C isozymes. J Biol Chem 265, 5424–5428. [PubMed] [Google Scholar]
- Martinez AA, Espinosa BA, Adamek RN, Thomas BA, Chau J, Gonzalez E, Keppetipola N, Salzameda NT, 2018. Breathing new life into West Nile virus therapeutics; discovery and study of zafirlukast as an NS2B-NS3 protease inhibitor. Eur J Med Chem 157, 1202–1213. [DOI] [PubMed] [Google Scholar]
- Marutsuka K, Hasui Y, Asada Y, Naito S, Osada Y, Sumiyoshi A, 1995. Effects of suramin on metastatic ability, proliferation, and production of urokinase-type plasminogen activator and plasminogen activator inhibitor type 2 in human renal cell carcinoma cell line SN12C-PM6. Clin Exp Metastasis 13, 116–122. [DOI] [PubMed] [Google Scholar]
- Mirza MU, Alanko I, Vanmeert M, Muzzarelli KM, Salo-Ahen OMH, Abdullah I, Kovari IA, Claes S, De Jonghe S, Schols D, Schinazi RF, Kovari LC, Trant JF, Ahmad S, Froeyen M, 2022. The discovery of Zika virus NS2B-NS3 inhibitors with antiviral activity via an integrated virtual screening approach. Eur J Pharm Sci 175, 106220. [DOI] [PubMed] [Google Scholar]
- Morrison TE, Diamond MS, 2017. Animal Models of Zika Virus Infection, Pathogenesis, and Immunity. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R, 2008. Identification and biochemical characterization of small-molecule inhibitors of west nile virus serine protease by a high-throughput screen. Antimicrob Agents Chemother 52, 3385–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller NH, Yon C, Ganesh VK, Padmanabhan R, 2007. Characterization of the West Nile virus protease substrate specificity and inhibitors. Int J Biochem Cell Biol 39, 606–614. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kuhn RJ, Rossmann MG, 2005. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 3, 13–22. [DOI] [PubMed] [Google Scholar]
- Murtuja S, Shilkar D, Sarkar B, Sinha BN, Jayaprakash V, 2021. A short survey of dengue protease inhibitor development in the past 6 years (2015–2020) with an emphasis on similarities between DENV and SARS-CoV-2 proteases. Bioorg Med Chem 49, 116415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nall TA, Chappell KJ, Stoermer MJ, Fang NX, Tyndall JD, Young PR, Fairlie DP, 2004. Enzymatic characterization and homology model of a catalytically active recombinant West Nile virus NS3 protease. J Biol Chem 279, 48535–48542. [DOI] [PubMed] [Google Scholar]
- Nitsche C, 2018. Strategies Towards Protease Inhibitors for Emerging Flaviviruses. Adv Exp Med Biol 1062, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyomrattanakit P, Winoyanuwattikun P, Chanprapaph S, Angsuthanasombat C, Panyim S, Katzenmeier G, 2004. Identification of residues in the dengue virus type 2 NS2B cofactor that are critical for NS3 protease activation. J Virol 78, 13708–13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble CG, Seh CC, Chao AT, Shi PY, 2012. Ligand-bound structures of the dengue virus protease reveal the active conformation. J Virol 86, 438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan R, Strongin AY, 2010. Translation and processing of the dengue virus polyprotein, in: Hanley KA, Weaver SC (Eds.), Frontiers in Dengue Virus Research. Caister Academic Press, Norfolk, U.K., pp. 14–33. [Google Scholar]
- Phoo WW, El Sahili A, Zhang Z, Chen MW, Liew CW, Lescar J, Vasudevan SG, Luo D, 2020. Crystal structures of full length DENV4 NS2B-NS3 reveal the dynamic interaction between NS2B and NS3. Antiviral Res 182, 104900. [DOI] [PubMed] [Google Scholar]
- Phoo WW, Li Y, Zhang Z, Lee MY, Loh YR, Tan YB, Ng EY, Lescar J, Kang C,Luo D, 2016. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat Commun 7, 13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Diamond MS, 2020. The continued threat of emerging flaviviruses. Nat Microbiol 5, 796–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaszczyca A, Scaturro P, Neufeldt CJ, Cortese M, Cerikan B, Ferla S, Brancale A, Pichlmair A, Bartenschlager R, 2019. A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLoS Pathog 15, e1007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rarey M, Kramer B, Lengauer T, Klebe G, 1996. A fast flexible docking method using an incremental construction algorithm. J Mol Biol 261, 470–489. [DOI] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY, 2006. West Nile virus 5’-cap structure is formed by sequential guanine N-7 and ribose 2’-O methylations by nonstructural protein 5. J Virol 80, 8362–8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samrat SK, Xu J, Li Z, Zhou J, Li H, 2022. Antiviral Agents against Flavivirus Protease: Prospect and Future Direction. Pathogens 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellhammer I, Rarey M, 2004. FlexX-Scan: fast, structure-based virtual screening. Proteins 57, 504–517. [DOI] [PubMed] [Google Scholar]
- Selisko B, Peyrane FF, Canard B, Alvarez K, Decroly E, 2010. Biochemical characterization of the (nucleoside-2’O)-methyltransferase activity of dengue virus protein NS5 using purified capped RNA oligonucleotides (7Me)GpppAC(n) and GpppAC(n). J Gen Virol 91, 112–121. [DOI] [PubMed] [Google Scholar]
- Su XC, Ozawa K, Qi R, Vasudevan SG, Lim SP, Otting G, 2009. NMR analysis of the dynamic exchange of the NS2B cofactor between open and closed conformations of the West Nile virus NS2B-NS3 protease. PLoS Negl Trop Dis 3, e561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay MYF, Vasudevan SG, 2018. The Transactions of NS3 and NS5 in Flaviviral RNA Replication. Adv Exp Med Biol 1062, 147–163. [DOI] [PubMed] [Google Scholar]
- Teramoto T, Balasubramanian A, Choi KH, Padmanabhan R, 2017. Serotype-specific interactions among functional domains of dengue virus 2 nonstructural proteins (NS) 5 and NS3 are crucial for viral RNA replication. J Biol Chem 292, 9465–9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto T, Boonyasuppayakorn S, Handley M, Choi KH, Padmanabhan R, 2014. Substitution of NS5 N-terminal domain of dengue virus type 2 RNA with type 4 domain caused impaired replication and emergence of adaptive mutants with enhanced fitness. J Biol Chem 289, 22385–22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiew KC, Dou D, Teramoto T, Lai H, Alliston KR, Lushington GH, Padmanabhan R, Groutas WC, 2012. Inhibition of Dengue virus and West Nile virus proteases by click chemistry-derived benz[d]isothiazol-3(2H)-one derivatives. Bioorg Med Chem 20, 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelzang NJ, Karrison T, Stadler WM, Garcia J, Cohn H, Kugler J, Troeger T, Giannone L, Arrieta R, Ratain MJ, Vokes EE, 2004. A Phase II trial of suramin monthly x 3 for hormone-refractory prostate carcinoma. Cancer 100, 65–71. [DOI] [PubMed] [Google Scholar]
- Wileman T, 2006. Aggresomes and autophagy generate sites for virus replication. Science 312, 875–878. [DOI] [PubMed] [Google Scholar]
- Wu J, Bera AK, Kuhn RJ, Smith JL, 2005. Structure of the Flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing. J Virol 79, 10268–10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Sampath A, Chao A, Wen D, Nanao M, Chene P, Vasudevan SG, Lescar J, 2005. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J Virol 79, 10278–10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Huo T, Lin YL, Nie S, Wu F, Hua Y, Wu J, Kneubehl AR, Vogt MB, Rico-Hesse R, Song Y, 2019. Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease. J Am Chem Soc 141, 6832–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yon C, Teramoto T, Mueller N, Phelan J, Ganesh VK, Murthy KH, Padmanabhan R, 2005. Modulation of the nucleoside triphosphatase/RNA helicase and 5’-RNA triphosphatase activities of dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J Biol Chem 280, 27412–27419. [DOI] [PubMed] [Google Scholar]
- Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R, 2000. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem 275, 9963–9969. [DOI] [PubMed] [Google Scholar]
- Zhang L, Mohan PM, Padmanabhan R, 1992. Processing and localization of Dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J Virol 66, 7549–7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Padmanabhan R, 1993. Role of protein conformation in the processing of dengue virus type 2 nonstructural polyprotein precursor. Gene 129, 197–205. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li Y, Loh YR, Phoo WW, Hung AW, Kang C, Luo D, 2016. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 354, 1597–1600. [DOI] [PubMed] [Google Scholar]