Background:
Hypertension imposes substantial health and economic burden worldwide. Primary aldosteronism (PA) is one of the most common causes of secondary hypertension, causing cardiovascular events at higher risk compared with essential hypertension. However, the germline genetic contribution to the susceptibility of PA has not been well elucidated.
Method:
We conducted a genome-wide association analysis of PA in the Japanese population and a cross-ancestry meta-analysis combined with UK Biobank and FinnGen cohorts (816 PA cases and 425 239 controls) to identify genetic variants that contribute to PA susceptibility. We also performed a comparative analysis for the risk of 42 previously established blood pressure–associated variants between PA and hypertension with the adjustment of blood pressure.
Results:
In the Japanese genome-wide association study, we identified 10 loci that presented suggestive evidence for the association with the PA risk (P<1.0×10−6). In the meta-analysis, we identified 5 genome-wide significant loci (1p13, 7p15, 11p15, 12q24, and 13q12; P<5.0×10−8), including 3 of the suggested loci in the Japanese genome-wide association study. The strongest association was observed at rs3790604 (1p13), an intronic variant of WNT2B (odds ratio, 1.50 [95% CI, 1.33−1.69]; P=5.2×10−11). We further identified 1 nearly genome-wide significant locus (8q24, CYP11B2), which presented a significant association in the gene-based test (P=7.2×10−7). Of interest, all of these loci were known to be associated with blood pressure in previous studies, presumably because of the prevalence of PA among individuals with hypertension. This assumption was supported by the observation that they had a significantly higher risk effect on PA than on hypertension. We also revealed that 66.7% of the previously established blood pressure–associated variants had a higher risk effect for PA than for hypertension.
Conclusions:
This study demonstrates the genome-wide evidence for a genetic predisposition to PA susceptibility in the cross-ancestry cohorts and its significant contribution to the genetic background of hypertension. The strongest association with the WNT2B variants reinforces the implication of the Wnt/β-catenin pathway in the PA pathogenesis.
Keywords: genome-wide association analysis, hypertension, primary aldosteronism, Wnt/β-catenin pathway
Clinical Perspective.
What Is New?
We revealed the evidence for a genetic predisposition to the risk of primary aldosteronism (PA) through the cross-ancestry genome-wide association study for PA, distinct from other causes of hypertension.
Comparative analysis for the risk effects between PA and hypertension suggested that some of the previously established blood pressure–associated loci might also be derived from their primary effect on the PA risk.
What Are the Clinical Implications?
The elucidation of genetic loci associated with the risk of PA enhances our understanding of the pathogenesis of PA, which will lead to the development of novel treatment approaches and early detection methods.
The strongest association of the WNT2B locus reinforces previous hypotheses for the implication of the Wnt/β-catenin pathway in the pathogenesis of PA.
Future studies scrutinizing genetic risks stratified by the subtypes of PA or somatic mutations in APA will contribute to further delineating the genetic basis behind the pathogenesis of PA.
Editorial, see p 1110
Hypertension affected ~1.28 billion adults ages 30 to 79 years in 2019, imposing a substantial health and economic burden worldwide.1 Primary aldosteronism (PA), characterized by inappropriately elevated aldosterone concentrations despite suppressed renin activity, is one of the major causes of secondary hypertension. Approximately 5% to 10% of all patients with hypertension and 15% to 20% of patients with resistant hypertension are estimated to have PA.2,3 Traditionally, PA has been categorized into 2 main forms: (1) unilateral PA caused by aldosterone-producing adenoma (APA) and (2) bilateral adrenal hyperplasia (BAH), also known as idiopathic hyperaldosteronism. Previous studies have shown the higher cardiovascular risk among patients with PA, either APA or BAH, compared with those with essential hypertension.4 Moreover, elevated aldosterone concentrations impose cardiovascular burden beyond increasing blood pressure (BP).5 Therefore, it is imperative to further delineate the pathogenesis of PA, leading to the development of early detection methods and novel treatment approaches.
During the past decade, the advent of next-generation sequencing has led to the discovery of somatic mutations of several genes in APA, such as KCNJ5, ATP1A1, ATP2B3, CACNA1D, CACNA1H, CLCN2, and CTNNB1.6 In particular, the KCNJ5 mutation is the most frequently detected in Asians and Europeans, which encodes potassium inwardly rectifying channel. Moreover, patients with KCNJ5-mutated APAs are likely to present a clinically pronounced phenotype of PA with increased adverse health outcomes.7,8 On the other hand, the role of germline genetic variants in the pathogenesis of PA remains elusive. Familial forms of PA have been recognized as represented by familial hyperaldosteronism type I, which is caused by an abnormal crossing over CYP11B1 and CYP11B2.9 However, the association of germline genetic variants with the pathogenesis of sporadic PA, which constitutes the majority of PA cases, have not been well elucidated. Previous attempts have mainly focused on candidate gene association analyses.10 A previous genome-wide association study (GWAS) of APA in Swedish individuals identified only 1 suggestive susceptible locus in X-chromosome.11 Furthermore, relationships to the genetics of hypertension have not been well explained.
To elucidate the genetic risk of PA, we performed a GWAS of PA for a Japanese cohort, in which PA was strictly diagnosed in this study. In addition, by using 2 publicly available European biobanks with health registries, UK Biobank (UKB) and FinnGen, we conducted a cross-ancestry meta-analysis (816 PA cases versus 425 239 controls).
Methods
To minimize the possibility of unintentionally sharing information that can be used to reidentify private information, a subset of the data generated for this study is available. The data for controls of the Japanese cohort are deposited at the National Bioscience Database Center Human Database (research ID: hum0311). The UKB data are available at https://www.ukbiobank.ac.uk/. The GWAS summary statistics of FinnGen release 5 is available at https://www.finngen.fi/fi.
Japanese Cohort
Data on patients with PA were collected from Hiroshima University Hospital. PA was defined on the basis of the Japan Endocrine Society guidelines for the diagnosis and treatment of PA. Briefly, after screening of plasma aldosterone concentrations (>12 ng/dL) or the aldosterone to renin ratio (>20), PA was diagnosed when patients were positive for 2 of the 3 confirmatory tests (ie, the captopril challenge test, furosemide upright test, and saline infusion test) according to the clinical guidelines of the Japan Endocrine Society.12 We identified the location of the tumor by computed tomography or magnetic resonance imaging. Adrenal venous sampling was conducted to identify the laterality of aldosterone secretion in all patients with PA. Familial hyperaldosteronism types I to IV and adrenocortical carcinoma were excluded from this study. The presence of CYP11B2 by immunohistochemistry was confirmed in all APA tissues. Genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini QIAcube Kit (Qiagen, Hilden, Germany).
We used individuals enrolled in the BioBank Japan (BBJ) for controls for the PA GWAS and cases and controls for the association test for hypertension. The BBJ is a multi-institutional hospital-based registry composed of DNA, serum, and clinical information of Japanese ancestry with a diagnosis of at least 1 of 47 diseases.13 We included the BBJ individuals who are genotyped and processed using the same manner as individuals with PA.
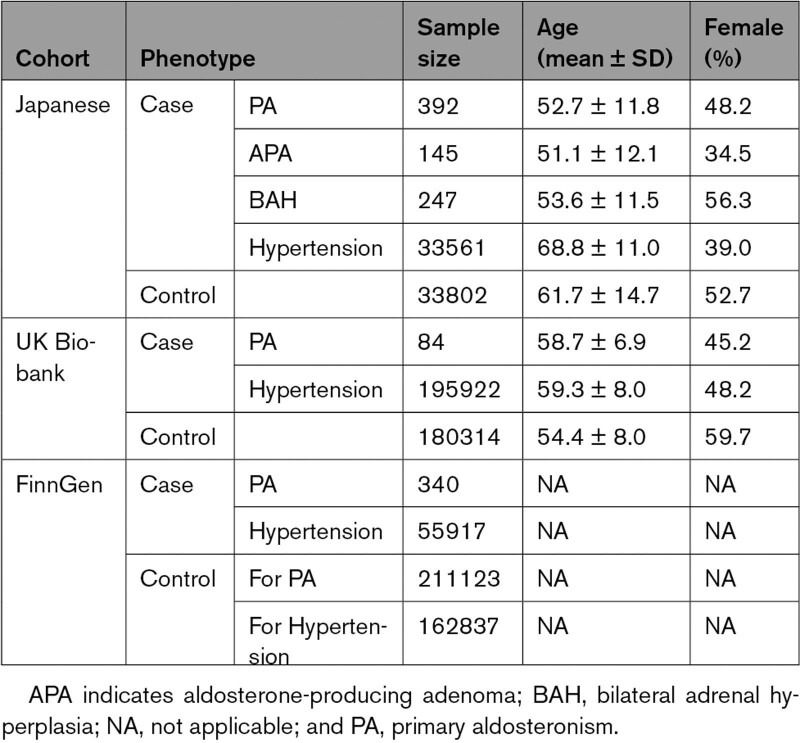
For the PA GWAS, we used individuals with no history of hypertension as the controls to increase the statistical power considering the nonnegligible prevalence of PA among individuals with hypertension.2,3 The history of hypertension was defined as having a diagnosis record of essential hypertension, a history of antihypertensive medication or systolic BP >140 mm Hg or diastolic BP >90 mm Hg. For the association test for hypertension, we used the same controls as those for the PA GWAS. The consistency in association analysis controls between PA and hypertension also helps directly use the analysis results for the estimation of genetic correlation and the comparison of risk effects between PA and hypertension. After the quality control (QC) we subsequently describe, we included 392 PA cases (positive for captopril challenge test, furosemide upright test, and saline infusion test were observed among 308 of 380 [81.1%], 320 of 355 [90.1%], and 278 of 366 [76.0%] patients, respectively), 34 375 hypertension cases, and 33 802 controls in this study.
All Japanese individuals provided written informed consent, as approved by the institutional review board of each institution. This study was approved by the ethical committee of Hiroshima University and Osaka University Graduate School of Medicine.
Genotyping, QC, and Whole-Genome Imputation in the Japanese Cohort
We genotyped the Japanese patients with PA and BBJ cohort using Infinium Asian Screening Array (Illumina, San Diego, CA). We included only the individuals estimated as belonging to the Japan Honshu cluster on the basis of principal components.14
We performed genome-wide genotype imputation to estimate untyped variants computationally. We used the combined reference panel of 1000 Genomes (1KG) Project Phase 3 version 5 genotype (n=2 504) and Japanese whole-genome sequencing data (n=1 037)14,15 as a haplotype reference for genotype imputation. Variants imputed with R-squared >0.7 and a minor allele frequency (MAF) >0.5% were used for subsequent analyses. More detailed procedures for genotyping, QC, and whole genome imputation are provided in Method S1.
UK Biobank
The UKB is composed of health-related information from ~500 000 individuals aged between 40 and 69 years recruited from across the United Kingdom from 2006 to 2010.16 The patient registration process and GWAS data are described elsewhere.16 Briefly, we used the genomic data based on genotyping either by the Applied Biosystems UK BiLEVE Axiom Array or by the Applied Biosystems UK Biobank Axiom Array and imputation using a combination of the Haplotype Reference Consortium, UK10K, and 1KG Phase 3 reference panels.17 Variants imputed with Rsq >0.7 and a MAF >0.5% were used for the analysis. We included only individuals of British ancestry who passed QC (Method S2).
The definition of cases for PA was based on the International Classification of Diseases, Tenth Revision code of E26.0 (primary hyperaldosteronism). The cases for hypertension and controls were defined similarly to those in the Japanese cohort (Method S2). Eventually, we included 84 PA cases, 195 922 hypertension cases, and 180 314 controls in this study.
FinnGen
FinnGen is a large public-private genome research project that collects and analyses genome and health data from Finnish biobanks and digital health record data from Finnish health registries, with its original phenotype definition (mainly using International Classification of Diseases and Anatomical Chemical Therapeutic classification codes).18 We used the GWAS summary statistics of E4_HYPERALDO (based on International Classification of Diseases, Tenth Revision code of E26) for PA (340 PA cases and 211 123 controls) and I9_HYPERTENS (based on International Classification of Diseases, Tenth Revision code of I10-13, 15, I674) for hypertension (55 917 hypertension cases and 162 837 controls) of FinnGen release 5, for which association tests were conducted using SAIGE. Because of the inaccessibility of the individual data, individuals with hypertension were not excluded from the control group in the PA GWAS.
GWAS and Meta-Analysis
We conducted GWAS based on a generalized mixed model of the imputed dosages of each of the variants on case-control status using SAIGE software version 0.44.5.19 SAIGE can control for case-control imbalance that can cause biased estimation of associations. We included age, sex, and the top 10 principal components as covariates in the regression model. For the UKB cohort, we also included the assessment center and genotyping batch as covariates.
We conducted meta-analysis of the GWAS results of the Japanese cohort, UKB, and FinnGen. We only included biallelic variants with MAF >0.005 that were available for all 3 cohorts. The association results for each variant across the studies were combined in a fixed effects inverse variance weighted method using METAL software.20 Heterogeneity in the associations of each variant was assessed using Cochran’s Q statistics and I2 index.21 The genome-wide significance threshold was set at the level of P=5.0×10−8. We assessed the inflation of test statistics based on the genomic control factor λGC.
To test for secondary independent signals in each genome-wide significant region, conditional analysis on its lead variant in the meta-analysis was performed for each cohort, and then the results were meta-analyzed under a fixed effects model. For the FinnGen data, we performed conditional and joint analysis22 using 1KG Phase 3 data as a reference. Variants with P<1.0×10−4 were considered as independent of the lead variant.
In addition, we performed an analysis to evaluate the risk of PA with hypertension as a control (ie, case-case association analysis between PA and hypertension) for the identified PA risk-associated variants using SAIGE. In this analysis, we adjusted for BP to minimize the potential bias caused by reverse causality: ie, patients with elevated BP are more likely to be diagnosed as PA although elevated BP is generally introduced by hyperaldosteronism. We included systolic and diastolic BPs and a history of antihypertensive medication (binarized) in the model as covariates.
Gene-Based Analysis
Gene-based association analysis was performed using Multi-Marker Analysis of GenoMic Annotation23 implemented in FUMA (https://fuma.ctglab.nl/) with its default setting. The analysis was performed for each population separately, and the results were meta-analyzed (Method S3).24 The significance threshold was based on the Bonferroni correction for the number of genes tested (P=0.05/19 364=2.6×10−6). MAGMA gene-set enrichment analysis for tissue expression was also performed using tissue expression profiles based on the GTEx V8 RNA-sequencing (RNA-seq) data.25
Functional Evaluation of Risk-Associated Loci on the Basis of the GTEx Data
We searched for expression and splicing quantitative trait loci (eQTL and sQTL) effects of the lead variant (or a proxy variant when the lead variant was unavailable) of each risk-associated locus using the genotype-tissue expression (GTEx) Portal V8 (https://gtexportal.org/home/).25 When significant effects in the adrenal gland were identified, the colocalization between PA association signals and the molecular quantitative trait loci (QTL) effects was also evaluated using Coloc,26 with a prior probability of colocalization of 10−5 (a default value). Because of the insufficient sample size for non-European GTEx data, colocalization analysis was performed only for the European GWAS meta-analysis.
RNA-Seq and Molecular QTL Analysis in the Tissue of APA
Among the APA individuals included in this study, we reanalyzed RNA-seq data of the APA tissues obtained at adrenalectomy that were used in our previous study (n=19).27 The procedure for RNA extraction was previously described.28
For the alignment and quantification of transcripts, we followed the pipeline provided by the GTEx project (https://github.com/broadinstitute/gtex-pipeline),25 with minimal changes. Remapping of reads based on the variant information was applied to the CYP11B1 and CYP11B2 regions to deal with the sequence homology. More detailed procedures for RNA-seq mapping and quantification of gene expression and splicing events are provided in Method S4.
For cis-eQTL and sQTL analysis, associations were evaluated using fastQTL v2.0,29 based on the additive effect model of the imputed dosage of each variant on the gene expression and splicing event, respectively. We included only age and sex as covariates because of the limited sample size. Variant-gene pairs located within 500 kb of the lead variant of each risk-associated locus were analyzed. Significance thresholds were Bonferroni corrected on the basis of the number of tests (P=0.05/85=5.9×10−4 and 0.05/129=3.9×10−4 for cis-eQTL and sQTL, respectively).
Estimation of Genetic Correlation Between PA and Hypertension
We performed cross-trait linkage disequilibrium score regression to assess the genetic correlation between PA and hypertension.30 We performed QC for the GWAS summary statistics before the analyses and merged with common HapMap3 single nucleotide polymorphisms excluding the major histocompatibility complex region according to general practice for linkage disequilibrium score regression.30 This analysis was not applied to the UKB and FinnGen cohorts because the estimated heritability of PA was converged to negative values or around zero with high SEs even after the meta-analysis of both cohorts, probably because of the insufficient sample sizes.
Evaluation of the Effects of BP-Associated Variants on the Risk of PA and Hypertension
We investigated the genetic risks of variants reported to be associated with BP for PA and hypertension risk. We selected the lead variants associated with BP from the previous independent GWAS results for the Japanese31 to avoid a potentially biased selection of variants caused by the overlaps between the discovery and target samples. We selected variants that were significantly associated with either systolic or diastolic BP (P<5.0×10−8). The variant with the lower P value was selected when the lead variants for the same locus were different between systolic and diastolic BP. We only included variants with MAF >0.005. We obtained 43 loci and lead variants. We excluded only 1 variant that did not even have available proxy variants (r2 of linkage disequilibrium coefficient >0.5) from this analysis. Last, we investigated 42 variants (Excel File S1). The odds ratios (ORs) for hypertension were determined in the same process as the PA GWAS using SAIGE. To estimate the expected proportion of variants having higher ORs for independent traits than for hypertension in the same sample size as the PA GWAS, we performed 1,000 times simulations evaluating the OR of each variant for randomly assigned traits (ie, case-control status was randomly shuffled). In addition, we performed case-case association tests between PA and hypertension including the BP traits as covariates. This analysis was applied only to the Japanese cohort because of the availability of a sufficient number of individual genotype data.
Results
GWAS of PA in the Japanese Cohort
For the Japanese cohort, we performed GWAS for 392 PA cases and 33 802 controls who passed the stringent QC. The demographic characteristics are provided in Table 1. We targeted 8 105 052 autosomal and 223 809 X-chromosome variants that satisfied stringent postimputation QC: MAF >0.5% and R-squared >0.7. The principal components were uniformly distributed across the cases and controls (Figure S1B).
Table 1.
Demographic Characteristics of the Study Participants
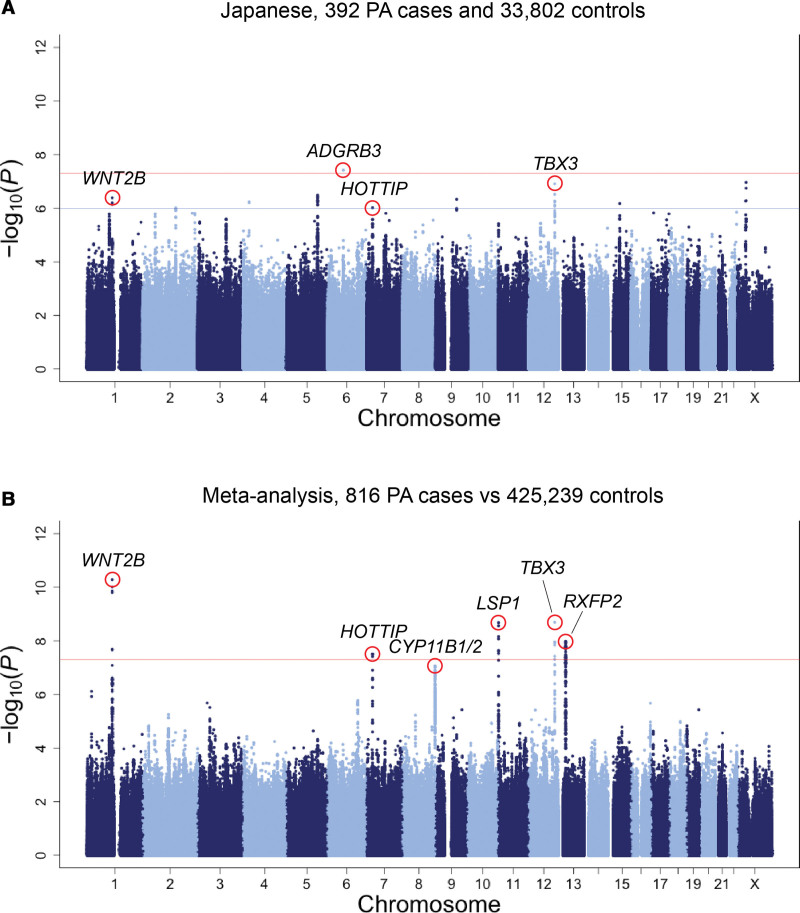
In the GWAS, we identified 1 variant (rs9354826) that was significantly associated with the risk of PA at 6q12, which is in the intronic region of ADGRB3 (OR, 1.53 [95% CI, 1.32−1.79]; P=3.8×10−8; risk allele frequency, 0.55; Figure 1A; Figure S2). In addition, 9 loci presented suggestive evidence for the associations (P<1.0×10−6; Figure 1A; Table S1).
Figure 1.
Manhattan plot for the genome-wide association analysis of primary aldosteronism. Manhattan plots showing −log10(P value) of the genome-wide association study of primary aldosteronism (PA) in the Japanese cohort (A) and meta-analysis (B). The red horizontal line indicates the genome-wide significance threshold (P=5.0×10−8). We also display a suggestive significance threshold (P=1.0×10−6) as the blue horizontal line in the Japanese genome-wide association study.
Cross-Ancestry GWAS Meta-Analysis of PA
We conducted a cross-ancestry meta-analysis combining the Japanese, UKB, and FinnGen GWASs (816 PA cases versus 425 239 controls) for 5 378 110 autosomal and 129 917 X-chromosome variants using the inverse variance weighted method. The quantile-quantile plot of observed versus expected P values for variants showed small inflation of the test statistic (λGC=1.04 in the meta-analysis; Figure S3).
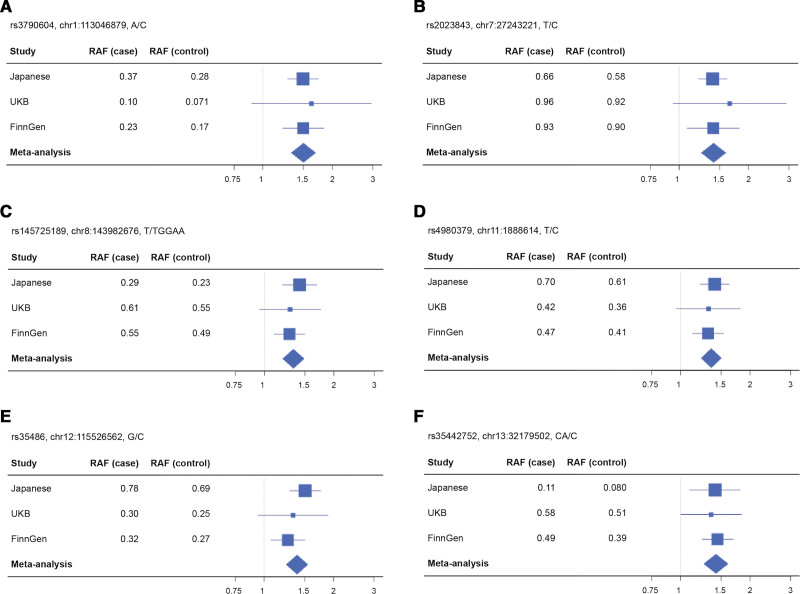
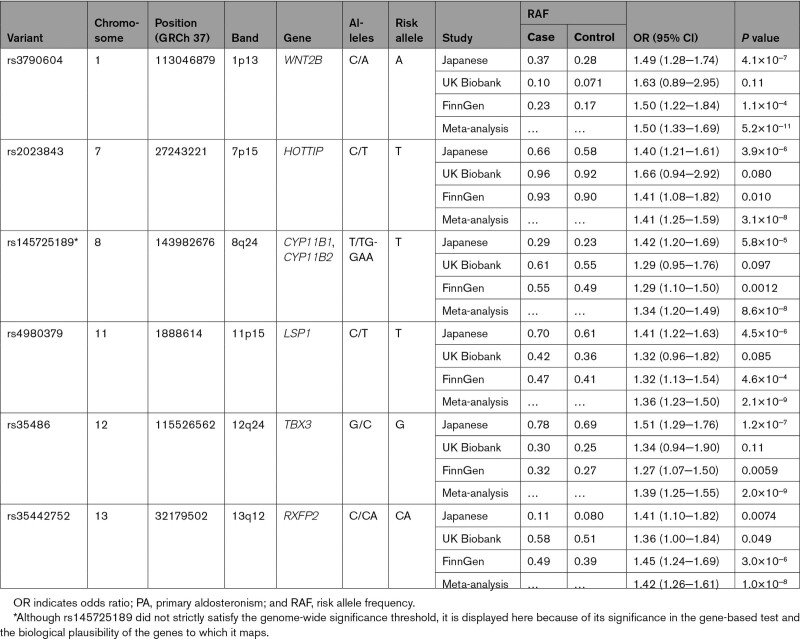
In the cross-ancestry meta-analysis, we identified genome-wide significant association signals at 5 loci (1p13, 7p15, 11p15, 12q24, and 13q12), for which the genes nearest to the lead variants were WNT2B, HOTTIP, LSP1, TBX3, and RXFP2, respectively (P<5.0×10−8; Figures 1B and 2; Table 2; Figure S4). Of the 5 loci, 3 were among those with suggestive associations in the Japanese GWAS (1p13, 7p15, and 12q24). There was no apparent heterogeneity in effects of these loci among studies (Phet≥0.33 and I2≤11). The strongest association was observed at rs3790604 (OR, 1.50 [95% CI, 1.33−1.69]; P=5.2×10−11; risk allele frequency, 0.28, 0.071, and 0.17 in the Japanese, UKB, and FinnGen cohorts, respectively), which maps to the intronic region of WNT2B locus (1p13). In addition, there were nearly genome-wide significant association signals in 8q24 for which the gene nearest to the lead variant was GML, which overlaps with CYP11B1 and CYP11B2 in the genomic position (P=8.8×10−8; Figures 1B and 2; Table 2; Figure S4). We performed a conditional analysis on the lead variant at each locus; however, no additional independent association signals were found (P>1.0×10−4; Figure S5). All the lead variants were located in noncoding regions (Figure S4), and coding variants tagged with them were shown in Table S2. Although there was no apparent intercohort heterogeneity in the effects of the lead variants in the meta-analysis (Phet≥0.33 and I2≤11), the cohort-specific lead variants were different (Figure S4; Table S3). In particular, the FinnGen-specific lead variant for the RFXP2 locus, rs1671966, was genome-wide significant in itself (P=4.6×10−8). The association of rs9354826 (6q12), which was observed in the Japanese GWAS, was not replicated in the UKB and FinnGen cohorts (P>0.05).
Figure 2.
Forest plots showing the odds ratios of lead variants for genome-wide and nearly genome-wide association loci for PA risk in each cohort. Forest plots for the odds ratios (ORs) of rs3790604 (A), rs2023843 (B), rs145725189 (C), rs4980379 (D), rs35486 (E), and rs35442752 (F) on the risk of primary aldosteronism (PA). Each forest plot shows the estimated ORs and 95% CIs from the cohort-specific genome-wide association study results. The variant name, chromosome position, and risk alleles and nonrisk alleles are shown above each plot. The size of the square representing the OR is proportional to the effective sample size of each cohort. RAF indicates risk allele frequency; and UKB, UK Biobank.
Table 2.
Lead Variants for Genome-Wide Significant Loci Associated With PA Risk in the Cross-Ancestry Meta-Analysis
To complement the single marker-based GWAS, we also performed a gene-based association test using MAGMA, which integrated associations of coding and noncoding variants located within each gene.23 Among 19 364 genes, we confirmed significant gene-based associations for WNT2B/ST7L and LSP1, for which single marker-based associations were identified (P<2.6×10−6; Figure S6; Table S4). In addition, we identified significant associations for the genes CYP11B2/GML (P=7.2×10−7 and 5.3×10−7, respectively). In MAGMA gene-set enrichment analysis for tissue expression, the strongest enrichment was identified in the adrenal gland in the Japanese GWAS (P=0.016; Figure S7).
All these risk-associated loci identified in the meta-analysis were reported to be associated with elevated BP in previous large-scale GWASs.31–33 This finding is reasonable, given the potential prevalence of PA among hypertensive individuals.2,3 In general, common BP-associated loci based on different mechanisms from PA (eg, essential hypertension or other secondary hypertension) should basically not be associated with PA status. Nevertheless, these PA risk-associated loci had higher risk effects for PA than for hypertension (Figure S8; Table S5). In addition, to demonstrate the robustness of our findings, we performed a case-case association analysis between PA and hypertension for these variants with the adjustment for BP. We applied this analysis only to the Japanese and UKB cohorts, in which individual genotype and BP information was available, and meta-analyzed their results. All the variants presented significant positive effects on PA against hypertension in the meta-analysis (P<0.01; Table S6). In addition, we evaluated how the exclusion of hypertensive individuals from the control groups, which was performed for the Japanese and UKB cohorts in the main analysis, had an effect on the association of PA risk variants. When the individuals with hypertension were not excluded, the effect sizes weakened, but the effect directions remained in each cohort, and 4 of the 5 variants still showed a genome-wide significant association in the meta-analysis (Table S7).
Stratified Analysis Reveals Distinctive Genetic Features for Each Subtype of PA
We performed stratified GWAS separately for the APA (n=145) and BAH cases (n=247) in the Japanese cohort to investigate whether these subtypes differ in genetic background. In the GWAS of BAH, we identified one locus significantly associated with the risk with a lead variant rs78785501 (2q14), located in the intronic region of DPP10 (OR=3.35, 95% CI=2.21−5.08, P=1.2×10−8, risk allele frequency, 0.069; Figures S9 through S11; Table S8). This variant was not associated with the risk of APA (P=0.35; Figures S9 and S11). We could not investigate its association with PA in the European cohorts because of its rarity (MAF<0.005). No significant subtype differences in the risk effects of the PA risk-associated variants identified in this study were detected in case-case association tests (P>0.05; Figure S11; Table S8). A previous GWAS of APA in Swedish individuals reported a suggestive locus in the X-chromosome with the lead single nucleotide polymorphism rs2224095;11 however, neither this Japanese GWAS stratified for APA nor the GWAS meta-analysis of PA could not replicate its associations (P>0.05; Table S9).
Functional Evaluation of PA Risk-Associated Variants Based on Molecular QTL Analyses
We explored the effects of the PA risk-associated variants by evaluating eQTL and sQTL effects using the GTEx Portal V8 (Table S10). The risk allele at rs35442752 (13q12) was significantly associated with increased expression of RXFP2 in the adrenal gland (P=2.9×10−14). rs145725189 (8q24) had significant sQTL effects for CYP11B1 and CYP11B2 in the adrenal gland (P=5.3×10−41 and 4.0×10−65, respectively). A proxy single nucleotide polymorphism for rs4980379 (rs686722) also had a sQTL effect for LSP1 in the adrenal gland (P=1.8×10−8). The colocalization analysis revealed that the PA association signals strongly colocalized with the RXFP2 eQTL effects (posterior probability, 87.9%; Figure S12).
In addition, we performed cis-eQTL and sQTL analysis for each lead variant using RNA-seq data of the APA tissues obtained at adrenalectomy (n=19).27 The GTEx results suggested that rs145725189 was associated with a splicing event that excises the region between CYP11B1 exon 2 and CYP11B2 exon 1 (ie, trans-splicing; Figure S13). The same intron excision was observed in our analysis. However, more detailed remapping of reads using the variant information revealed that this was most likely a result of mismapping caused by the sequence homology between CYP11B1 and CYP11B2 (Figure S13). On the other hand, the CYP11B1 sQTL effect of rs145725189 that excises CYP11B intron 6 was replicated (P=0.010). The current analysis could not replicate the other effects (P>0.05) or identify additional cis-eQTL or sQTL effects (Padj>0.05; Figure S14).
Significant Genetic Contribution of PA to Hypertension
We evaluated the shared heritability between PA and hypertension in the Japanese cohort using linkage disequilibrium score regression.30 We observed a significant genetic correlation (rg, 0.59, SE, 0.15; P=1.0×10−4).
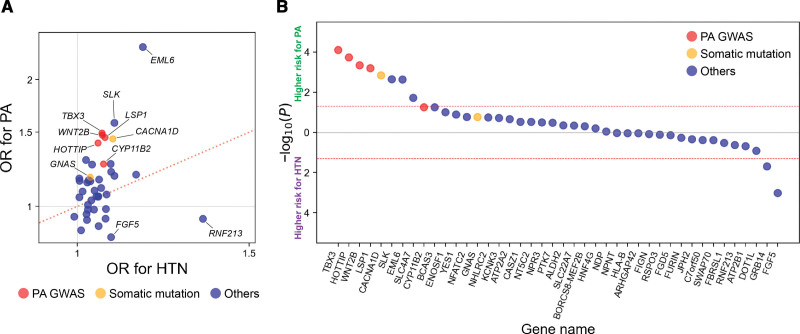
Furthermore, on the basis of the results of the PA GWAS, we hypothesized that some other variants known to be associated with BP might also derive from their primary effects on the risk of PA. Thus, we compared the risk effects of such BP-associated variants between PA and hypertension. We investigated 42 BP-associated variants that have been established in previous independent Japanese GWAS (Excel File S1).31 The directions of its genetic risk effects for hypertension should be consistent with those of elevated BP. Expectedly, all but 1 variant presented positive risk effects on hypertension in our cohort (Figure 3A). On the other hand, the common BP-associated loci based on pathways unrelated to PA should basically not be associated with the PA risk although individuals with PA have elevated BP. However, 66.7% (28 of 42) of the BP-associated variants presented higher ORs for the PA risk than for the hypertension risk (Figure 3A; Excel File S1). The simulation analysis estimated that the expected proportion of BP-associated variants having higher ORs for independent traits than for hypertension was only 25.6% (95% CI, 12.5−38.6). In addition, we performed the case-case association analysis between PA and hypertension with the adjustment of BP to strictly compare the effects of these variants on the PA and hypertension risk. Of all the variants, 61.9% (26 of 42) showed a higher effect on PA than on hypertension, and its proportion increased to 80.0% (8 of 10) when limited to those with nominally significant differences in effects between PA and hypertension (Figure 3B; Excel File S1).
Figure 3.
Comparison of the genetic risk of blood pressure-associated variants for primary aldosteronism and hypertension. A, The plot represents the OR of BP-associated variants for hypertension (horizontal axis) and PA (vertical axis) in the Japanese cohort. B, The plot represents −log10(P value) of the case-case association analysis between PA and hypertension with the adjustment for BP in the Japanese cohort. The vertical axis shows −log10(P value), with the higher risk effects on PA and hypertension shown at the above and below the horizontal axis, respectively. The dots representing individual variants are displayed in the descending order from higher positive association with PA to higher positive association with hypertension. In both panels, the color of dots corresponds to the legend: risk-associated loci for PA identified in this GWAS meta-analysis (red), genes in which somatic mutations are reported to be associated with aldosterone-producing adenoma or aldosterone- and cortisol-cosecreting adrenal tumors (orange), and others (blue). The effect alleles are based on alleles that are reported to increase BP. BP indicates blood pressure; GWAS, genome-wide association study; HTN, hypertension; OR, odds ratio; and PA, primary aldosteronism.
Discussion
Here we presented the evidence for a genetic predisposition to the risk of PA through the initial cross-ancestry meta-analysis of GWASs. We identified 5 genome-wide association loci in the GWAS meta-analysis and 1 nearly genome-wide significant locus, which presented a significant association in the gene-based test. The strongest association was observed at 1p13, in which WNT2B is located. All the loci identified here have been reported to be associated with BP.31–33 If a BP-associated variant increases BP through the pathways unrelated to PA, it should have basically no association with the PA risk, much less a higher risk effect on PA than on hypertension. Nevertheless, all the PA risk-associated loci identified in this study showed significantly positive effects on PA against hypertension in the case-case association test between PA and hypertension even after adjustment for 1-time measurement of BP. The present case-case association analysis would present strict and conservative evaluation because around 15% of individuals with hypertension could have PA.2,3 In addition, because elevated BP in individuals with PA essentially derives from its specific condition of inappropriately elevated aldosterone concentrations, the top risk loci in the GWAS for elevated BP (eg, ALDH2 and ATP2B1) were not preferentially detected in our PA GWAS. Therefore, the overlap of the PA-risk loci with reported BP-associated loci is presumed to be a result of the prevalence of PA among individuals with hypertension, rather increasing the reliability of the results of our GWAS.
WNT2B, a member of the Wnt protein family, is involved in organogenesis and tumorigenesis.34 The Wnt/β-catenin signaling pathway has been shown to be a key factor in the development of normal adrenal cortex, more specifically the zona glomerulosa, and production of aldosterone.35 A previous study showed that mouse model of constitutive β-catenin activation in the adrenal cortex developed adrenal hyperplasia and hyperaldosteronism.36 In addition, the association of aberrant activation of the Wnt/β-catenin pathway with the pathogenesis of APA has been established, and Wnt/β-catenin signaling was actually activated in 70% of APA.37 Our study presented the first evidence that genetic variations in WNT2B are associated with the risk of PA. Given a recent pathological study reporting a larger activation of Wnt/β-catenin signaling among non-KCNJ5 mutant APA compared with KCNJ5 mutant APA,38 future investigations are needed to assess whether WNT2B variants are associated with the risk of APA across somatic mutations including KCNJ5.
Recently, Le Floch et al published a GWAS of PA in a European cohort.39 They reported the robust association of 2 loci mapped to RXFP2 (13q12) and CASZ1 (1p36) and also suggested the association of 2 loci mapped to NDP (Xp11) and LSP1 (11p15) in discovery analysis and meta-analysis, respectively. RXFP2 and LSP1 were overlapped with the loci identified in this study, which enhances the reliability of their association with PA. The lead variants reported in their study were in linkage disequilibrium but different from those identified in our study (r2EUR=0.42 and r2EAS=0.018 between rs1535532 and rs35442752 for RXFP2; r2EUR=0.75 and r2EAS=0.94 between rs2137320 and rs4980389 for LSP1), which might reflect interpopulation differences in genetic structures associated with PA. CASZ1 was one of the previously established BP-associated loci that had a higher effect on PA than on hypertension in our study. RXFP2 encodes a G protein–coupled 7-transmembrane receptor for a relaxin family peptide. Le Floch et al revealed that RXFP2 and CASZ1 expressed in the adrenal gland and their overexpression in adrenocortical cells could modify the basal and stimulated mineralocorticoid output.39 Considering our findings that the PA association signals strongly colocalized with the RXFP2 eQTL effects in the adrenal gland, this would support the hypothesis that its risk variant might have a causative effect on PA through the overexpression of RXFP2. Although they reported the stronger association of the RFXP2 locus with BAH than with APA, there was no apparent heterogeneity between the subtypes in our study.
The roles of the other risk-associated loci might be explained in line with the function or development of the adrenal gland. CYP11B2 is one of the key enzymes for steroidogenesis that is responsible for the catalysis of aldosterone synthesis in the zona glomerulosa of the adrenal cortex.40 The variants in this region might associate with altered splicing. However, more reliable sequencing approaches (eg, long-read sequencing) would be required to determine their functional effects because of the sequence homology between CYP11B1 and CYP11B2. TBX3 is a transcription factor sharing a common DNA-binding domain, T-box, and expresses in the adrenal gland.41 TBX3 acted as a tissue-specific member of β-catenin transcriptional complex.42 HOTTIP is presumed to regulate the expression of HOXA genes, and its dysregulation has been implicated in various tumors.43 HOX genes are expressed in the developing and adult adrenal glands44 and are suggested to promote cell proliferation in adrenocortical tumors.45 DPP10 regulates the expression of voltage-gated potassium channels by binding them, and its expression was reported in the adrenal gland as well as neural tissues.46 Although its functional role might be explained in the context of potassium channels in the adrenal gland, the reason for heterogeneity in the risk effects between APA and BAH remains to be elucidated.
One of the next challenges is to determine how genetic factors are involved in the pathogenesis of PA. The lead variants identified in this study were all located in the noncoding region; however, they might be in linkage disequilibrium with causative coding variants. Given the heterogeneity in linkage disequilibrium structure by ancestry and our results suggesting the intercohort differences in lead variants, multiancestry GWAS from larger cohorts should be warranted. Particularly, this study was limited to common and low-frequency variants, and rare variant analysis using exome or whole genome sequencing would help to perform fine-mapping of each PA risk-associated locus and further delineate the genetic determinants of PA. Furthermore, functional genomics data would also be helpful in elucidating the mechanisms by which risk-associated variants are involved in the pathogenesis of PA through an effect on the expression or function of genes in the adrenal gland. Specifically, investigation of cell type–specific molecular QTL effects using single-cell sequencing will provide further insights beyond our current findings obtained from the GTEx bulk RNA-seq data.
Previous GWASs of BP have suggested the implication of aldosterone-related biology in the genetics of hypertension through the enrichment of the regulatory or functional information of BP-associated loci in the adrenal gland tissue.33,47 Here we revealed that some previously established BP-associated variants had higher risk effects for PA than for hypertension. This result would suggest that part of their associations with BP might derive from their primary effects on PA risk or related biological mechanisms through the renin-angiotensin-aldosterone system. It is interesting that some loci in which somatic mutations are causative of the development of APA, such as CACNA1D48 and GNAS,49 were among this group, indicating their germline genetic contributions to the pathogenesis of PA. These findings motivate us to conduct a GWAS with larger sample sizes as a next step to establish stronger evidence for the role of these genes in PA, distinct from other causes of hypertension. We note the possibility that the 42 BP-associated loci investigated in this study were more likely to be enriched with the PA-related mechanisms because loci with a higher effect on BP would be preferentially detected in the previous BP GWAS. This implies that the current results did not simply indicate that PA explains more than half of the heritability of BP, and its quantitative evaluation should be addressed in future studies.
There are several limitations and future advances in this study. First, the PA risk-associated loci identified in the current study did not satisfy the genome-wide significance threshold in a single cohort, except for the RXFP2 locus. Second, we note the different definitions and guideline of the phenotypes (including confirmatory tests for PA) between cohorts, which might introduce biased estimates in our meta-analysis. In particular, for UKB and FinnGen, the diagnosis was based on health records from registries; thus, not all the individuals were screened for the diagnosis of PA. This might lead to misclassification of PA cases as controls, potentially reducing the statistical power of GWAS. In addition, given the heterogeneity in the sensitivity and specificity for PA confirmatory tests reported in previous studies (captopril challenge test, sensitivity, 70%−100%, specificity, 68%−95%; furosemide upright test, undetermined; saline infusion test, sensitivity, 66%−92%, specificity, 71%−97%),50 we cannot rule out the possibility of misclassification of PA in our cohort. Third, because of the inaccessibility of individual genotype data from FinnGen, we could not exclude hypertensive individuals from the control and perform the PA-hypertension association analysis. The former would lead to the underestimation of the effects and bias in the meta-analysis and comparison of effects between PA and hypertension. Fourth, although APA was confirmed by adrenal tissue, BAH was generally diagnosed from computed tomography imaging and the results of adrenal venous sampling. Thus, some patients labeled as BAH might have bilateral microAPA or aldosterone-producing cell clusters, which might introduce misclassification. Fifth, we had data on PA subtypes only in the Japanese cohort, which did not allow us to conduct a meta-analysis specific to APA and BAH, leading to a lack of statistical power to robustly identify subtype-specific risk-associated variants. Moreover, given that APAs have different histopathological features and regional frequencies according to somatic mutations,51 further insights could be obtained by scrutinizing the genetic risk of APA on the basis of each somatic mutation. Last, the findings obtained in this study were based on the retrospective data and should be validated in prospective cohorts.
In summary, our meta-analysis of GWAS revealed the genome-wide evidence for a genetic predisposition to the susceptibility to PA in the cross-ancestry cohorts. The strongest association with WNT2B variants reinforces the implication of the Wnt/β-catenin pathway in the pathogenesis of PA. Given the pathological heterogeneity among PA phenotypes, stratified analysis with larger sample sizes will provide a more detailed description of the genetic features associated with PA.
Article Information
Acknowledgments
The authors thank all the participants, study coordinators, and investigators involved in this study. The Department of Preventive Medicine for Diabetes and Lifestyle-Related Diseases, Graduate School of Biomedical and Health Sciences, Hiroshima University, is an endowment department, supported with unrestricted grants from OKEIOS Inc and Terumo Corporation.
Sources of Funding
K.O. was supported by Japan Society for the Promotion of Science KAKENHI (21K08557), Mochida Memorial Foundation for Medical and Pharmaceutical Research, Takeda Science Foundation, SENSHIN Medical Research Foundation, and Daiwa Securities Health Foundation. Y. Okada was supported by Japan Society for the Promotion of Science KAKENHI (22H00476), Japan Agency for Medical Research and Development (JP21gm4010006, JP22km0405211, JP22ek0410075, JP22km0405217, JP22ek0109594, JP223fa627002, JP223fa627010, JP233fa627011), Japan Science and Technology Agency Moonshot R&D (JPMJMS2021, JPMJMS2024), Takeda Science Foundation, and Bioinformatics Initiative of Osaka University Graduate School of Medicine. K.K. was supported by Japan Society for the Promotion of Science KAKENHI (21K16058). K.I. was supported by JSPS KAKENHI (21K20900, 22K17392) and the Program for the Development of Next-Generation Leading Scientists With Global Insight (L-INSIGHT), sponsored by the Ministry of Education, Culture, Sports, Science, and Technology.
Disclosures
The Department of Preventive Medicine for Diabetes and Lifestyle-Related Diseases, Graduate School of Biomedical and Health Sciences, Hiroshima University, is an endowment department, supported with unrestricted grants from OKEIOS Inc and Terumo Corporation.
Supplemental Material
Methods S1–S4
Figures S1–S14
Tables S1–S10
Excel File S1
Supplementary Material
Nonstandard Abbreviations and Acronyms
- APA
- aldosterone-producing adenoma
- BAH
- bilateral adrenal hyperplasia
- BBJ
- BioBank Japan
- BP
- blood pressure
- GWAS
- genome-wide association study
- MAF
- minor allele frequency
- OR
- odds ratio
- PA
- primary aldosteronism
- QC
- quality control
- RNA-seq
- RNA-sequencing
- UKB
- UK Biobank
Y. Okada and K. Oki contributed equally.
Supplemental Material, the podcast, and transcript are available with this article at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.122.062349.
For Sources of Funding and Disclosures, see page 1107.
Circulation is available at www.ahajournals.org/journal/circ.
Contributor Information
Tatsuhiko Naito, Email: tnaito@sg.med.osaka-u.ac.jp.
Kosuke Inoue, Email: koinoue@ucla.edu.
Kyuto Sonehara, Email: qsonehara@sg.med.osaka-u.ac.jp.
Ryuta Baba, Email: rtbaba.45@gmail.com.
Takaya Kodama, Email: kktkykk.88@gmail.com.
Yu Otagaki, Email: ytgk1212@gmail.com.
Akira Okada, Email: yokada@sg.med.osaka-u.ac.jp.
Kiyotaka Itcho, Email: kiyo.itcho@gmail.com.
Kazuhiro Kobuke, Email: kazu-kobuke@hiroshima-u.ac.jp.
Shinji Kishimoto, Email: skishimoto@hiroshima-u.ac.jp.
Kenichi Yamamoto, Email: ken.y.2015@ped.med.osaka-u.ac.jp.
Takayuki Morisaki, Email: morisaki@ims.u-tokyo.ac.jp.
Yukihito Higashi, Email: yhigashi@hiroshima-u.ac.jp.
Nobuyuki Hinata, Email: hinata@hiroshima-u.ac.jp.
Koji Arihiro, Email: arihiro@hiroshima-u.ac.jp.
Noboru Hattori, Email: nhattori@hiroshima-u.ac.jp.
References
- 1.Zhou B, Carrillo-Larco RM, Danaei G, Riley LM, Paciorek CJ, Stevens GA, Gregg EW, Bennett JE, Solomon B, Singleton RK, et al. Worldwide trends in hypertension prevalence and progress in treatment and control from 1990 to 2019: a pooled analysis of 1201 population-representative studies with 104 million participants. Lancet. 2021;398:957–980. doi: 10.1016/S0140-6736(21)01330-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown JM, Siddiqui M, Calhoun DA, Carey RM, Hopkins PN, Williams GH, Vaidya A. The unrecognized prevalence of primary aldosteronism. Ann Intern Med. 2020;173:10–20. doi: 10.7326/M20-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funder JW, Carey RM. Primary aldosteronism: where are we now? Where to from here? Hypertension. 2022;79:726–735. doi: 10.1161/HYPERTENSIONAHA.121.18761 [DOI] [PubMed] [Google Scholar]
- 4.Monticone S, D’Ascenzo F, Moretti C, Williams TA, Veglio F, Gaita F, Mulatero P. Cardiovascular events and target organ damage in primary aldosteronism compared with essential hypertension: a systematic review and meta-analysis. Lancet Diabetes Endocrinol. 2018;6:41–50. doi: 10.1016/S2213-8587(17)30319-4 [DOI] [PubMed] [Google Scholar]
- 5.Inoue K, Goldwater D, Allison M, Seeman T, Kestenbaum BR, Watson KE. Serum aldosterone concentration, blood pressure, and coronary artery calcium. Hypertension. 2020;76:113–120. doi: 10.1161/HYPERTENSIONAHA.120.15006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itcho K, Oki K, Ohno H, Yoneda M. Update on genetics of primary aldosteronism. Biomedicines. 2021;9:409. doi: 10.3390/biomedicines9040409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitamoto T, Omura M, Suematsu S, Saito J, Nishikawa T. KCNJ5 mutation as a predictor for resolution of hypertension after surgical treatment of aldosterone-producing adenoma. J Hypertens. 2018;36:619–627. doi: 10.1097/HJH.0000000000001578 [DOI] [PubMed] [Google Scholar]
- 8.Chang Y-Y, Tsai C-H, Peng S-Y, Chen Z-W, Chang C-C, Lee B-C, Liao C-W, Pan C-T, Chen Y-L, Lin L-C, et al. ; TAIPAI Study Group. KCNJ5 somatic mutations in aldosterone-producing adenoma are associated with a worse baseline status and better recovery of left ventricular remodeling and diastolic function. Hypertension. 2021;77:114–125. doi: 10.1161/HYPERTENSIONAHA.120.15679 [DOI] [PubMed] [Google Scholar]
- 9.Lifton RP, Dluhy RG, Powers M, Rich GM, Cook S, Ulick S, Lalouel J-M. A chimaeric llβ-hydroxylase/aldosterone synthase gene causes glucocorticoid-remediable aldosteronism and human hypertension. Nature. 1992;355:262–265. doi: 10.1038/355262a0 [DOI] [PubMed] [Google Scholar]
- 10.Zhang GX, Wang BJ, Ouyang JZ, Deng XY, Ma X, Li HZ, Wu Z, Liu SL, Xu H, Zhang X. Polymorphisms in CYP11B2 and CYP11B1 genes associated with primary hyperaldosteronism. Hypertens Res. 2010;33:478–484. doi: 10.1038/hr.2010.21 [DOI] [PubMed] [Google Scholar]
- 11.Dutta RK, Larsson M, Arnesen T, Heie A, Walz M, Alesina P, Gimm O, Söderkvist P. X-chromosome variants are associated with aldosterone producing adenomas. Sci Rep. 2021;11:10562. doi: 10.1038/s41598-021-89986-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishikawa T, Omura M, Satoh F, Shibata H, Takahashi K, Tamura N, Tanabe A; Task Force Committee on Primary Aldosteronism, The Japan Endocrine Society. Guidelines for the diagnosis and treatment of primary aldosteronism -The Japan Endocrine Society 2009-. Endocr J. 2011;58:711–721. doi: 10.1507/endocrj.ej11-0133 [DOI] [PubMed] [Google Scholar]
- 13.Nagai A, Hirata M, Kamatani Y, Muto K, Matsuda K, Kiyohara Y, Ninomiya T, Tamakoshi A, Yamagata Z, Mushiroda T, et al. Overview of the BioBank Japan Project: study design and profile. J Epidemiol. 2017;27:S2–S8. doi: 10.1016/j.je.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada Y, Momozawa Y, Sakaue S, Kanai M, Ishigaki K, Akiyama M, Kishikawa T, Arai Y, Sasaki T, Kosaki K, et al. Deep whole-genome sequencing reveals recent selection signatures linked to evolution and disease risk of Japanese. Nat Commun. 2018;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akiyama M, Ishigaki K, Sakaue S, Momozawa Y, Horikoshi M, Hirata M, Matsuda K, Ikegawa S, Takahashi A, Kanai M, et al. Characterizing rare and low-frequency height-associated variants in the Japanese population. Nat Commun. 2019;10:4393. doi: 10.1038/s41467-019-12276-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, Reeve MP, Laivuori H, Aavikko M, Kaunisto MA, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508–518. doi: 10.1038/s41586-022-05473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, LeFaive J, VandeHaar P, Gagliano SA, Gifford A, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Ferreira T, Morris AP, Medland SE, Madden PAF, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, et al. ; Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. doi: 10.1038/ng.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bakker PIW, Ferreira MAR, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguet F, Barbeira AN, Bonazzola R, Jo B, Kasela S, Liang Y, Parsana P, Aguet F, Battle A, Brown A, et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itcho K, Oki K, Gomez-Sanchez CE, Gomez-Sanchez EP, Ohno H, Kobuke K, Nagano G, Yoshii Y, Baba R, Hattori N, et al. Endoplasmic reticulum chaperone calmegin is upregulated in aldosterone-producing adenoma and associates with aldosterone production. Hypertension. 2020;75:492–499. doi: 10.1161/HYPERTENSIONAHA.119.14062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishimoto R, Oki K, Yoneda M, Gomez-Sanchez CE, Ohno H, Kobuke K, Itcho K, Kohno N. Gonadotropin-releasing hormone stimulate aldosterone production in a subset of aldosterone-producing adenoma. Medicine (Baltimore). 2016;95:e3659. doi: 10.1097/MD.0000000000003659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JRB, Patterson N, Robinson EB, et al. ; ReproGen Consortium. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakaue S, Kanai M, Tanigawa Y, Karjalainen J, Kurki M, Koshiba S, Narita A, Konuma T, Yamamoto K, Akiyama M, et al. ; FinnGen. A cross-population atlas of genetic associations for 220 human phenotypes. Nat Genet. 2021;53:1415–1424. doi: 10.1038/s41588-021-00931-x [DOI] [PubMed] [Google Scholar]
- 32.Wu Y, Byrne EM, Zheng Z, Kemper KE, Yengo L, Mallett AJ, Yang J, Visscher PM, Wray NR. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat Commun. 2019;10:1891. doi: 10.1038/s41467-019-09572-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surendran P, Feofanova EV, Lahrouchi N, Ntalla I, Karthikeyan S, Cook J, Chen L, Mifsud B, Yao C, Kraja AT, et al. ; LifeLines Cohort Study. Discovery of rare variants associated with blood pressure regulation through meta-analysis of 1.3 million individuals. Nat Genet. 2020;52:1314–1332. doi: 10.1038/s41588-020-00713-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh M, Katoh M. Molecular genetics and targeted therapy of WNT-related human diseases (review). Int J Mol Med. 2017;40:587–606. doi: 10.3892/ijmm.2017.3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drelon C, Berthon A, Sahut-Barnola I, Mathieu M, Dumontet T, Rodriguez S, Batisse-Lignier M, Tabbal H, Tauveron I, Lefrançois-Martinez A-M, et al. PKA inhibits WNT signalling in adrenal cortex zonation and prevents malignant tumour development. Nat Commun. 2016;7:12751. doi: 10.1038/ncomms12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berthon A, Sahut-Barnola I, Lambert-Langlais S, de Joussineau C, Damon-Soubeyrand C, Louiset E, Taketo MM, Tissier F, Bertherat J, Lefrançois-Martinez A-M, et al. Constitutive β-catenin activation induces adrenal hyperplasia and promotes adrenal cancer development. Hum Mol Genet. 2010;19:1561–1576. doi: 10.1093/hmg/ddq029 [DOI] [PubMed] [Google Scholar]
- 37.Berthon A, Drelon C, Ragazzon B, Boulkroun S, Tissier F, Amar L, Samson-Couterie B, Zennaro MC, Plouin PF, Skah S, et al. WNT/β-catenin signalling is activated in aldosterone-producing adenomas and controls aldosterone production. Hum Mol Genet. 2014;23:889–905. doi: 10.1093/hmg/ddt484 [DOI] [PubMed] [Google Scholar]
- 38.De Sousa K, Boulkroun S, Baron S, Nanba K, Wack M, Rainey WE, Rocha A, Giscos-Douriez I, Meatchi T, Amar L, et al. Genetic, cellular, and molecular heterogeneity in adrenals with aldosterone-producing adenoma. Hypertension. 2020;75:1034–1044. doi: 10.1161/HYPERTENSIONAHA.119.14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Floch E, Cosentino T, Larsen CK, Beuschlein F, Reincke M, Amar L, Rossi G, De Sousa K, Baron S, Chantalat S, et al. Identification of risk loci for primary aldosteronism in genome-wide association studies. Nat Commun. 2022;13:5198. doi: 10.1038/s41467-022-32896-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curnow KM, Tusie-Luna M-T, Pascoe L, Natarajan R, Gu J-L, Nadler JL, White PC. The product of the CYP11B2 gene is required for aldosterone biosynthesis in the human adrenal cortex. Mol Endocrinol. 1991;5:1513–1522. doi: 10.1210/mend-5-10-1513 [DOI] [PubMed] [Google Scholar]
- 41.del Valle I, Buonocore F, Duncan AJ, Lin L, Barenco M, Parnaik R, Shah S, Hubank M, Gerrelli D, Achermann JC. A genomic atlas of human adrenal and gonad development. Wellcome Open Res. 2017;2:25. doi: 10.12688/wellcomeopenres.11253.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmerli D, Borrelli C, Jauregi-Miguel A, Söderholm S, Brütsch S, Doumpas N, Reichmuth J, Murphy-Seiler F, Aguet M, Basler K, et al. TBX3 acts as tissue-specific component of the Wnt/β-catenin enhanceosome. Elife. 2020;1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghafouri-Fard S, Dashti S, Taheri M. The HOTTIP (HOXA transcript at the distal tip) lncRNA: review of oncogenic roles in human. Biomed Pharmacother. 2020;127:110158. doi: 10.1016/j.biopha.2020.110158 [DOI] [PubMed] [Google Scholar]
- 44.Neville SE, Baigent SM, Bicknell AB, Lowry PJ, Gladwell RT. HOX gene expression in adult tissues with particular reference to the adrenal gland. Endocr Res. 2002;28:669–673. doi: 10.1081/erc-120016984 [DOI] [PubMed] [Google Scholar]
- 45.Francis JC, Gardiner JR, Renaud Y, Chauhan R, Weinstein Y, Gomez-Sanchez C, Lefrançois-Martinez AM, Bertherat J, Val P, Swain A. HOX genes promote cell proliferation and are potential therapeutic targets in adrenocortical tumours. Br J Cancer. 2021;124:805–816. doi: 10.1038/s41416-020-01166-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takimoto K, Hayashi Y, Ren X, Yoshimura N. Species and tissue differences in the expression of DPPY splicing variants. Biochem Biophys Res Commun. 2006;348:1094–1100. doi: 10.1016/j.bbrc.2006.07.157 [DOI] [PubMed] [Google Scholar]
- 47.Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, et al. ; Million Veteran Program. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50:1412–1425. doi: 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scholl UI, Goh G, Stölting G, De Oliveira RC, Choi M, Overton JD, Fonseca AL, Korah R, Starker LF, Kunstman JW, et al. Somatic and germline CACNA1D calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism. Nat Genet. 2013;45:1050–1054. doi: 10.1038/ng.2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakajima Y, Okamura T, Horiguchi K, Gohko T, Miyamoto T, Satoh T, Ozawa A, Ishii S, Yamada E, Hashimoto K, et al. Gnas mutations in adrenal aldosterone-producing adenomas. Endocr J. 2016;63:199–204. doi: 10.1507/endocrj.EJ15-0642 [DOI] [PubMed] [Google Scholar]
- 50.Naruse M, Katabami T, Shibata H, Sone M, Takahashi K, Tanabe A, Izawa S, Ichijo T, Otsuki M, Omura M, et al. Japan Endocrine Society clinical practice guideline for the diagnosis and management of primary aldosteronism 2021. Endocr J. 2022;69:327–359. doi: 10.1507/endocrj.ej21-0508 [DOI] [PubMed] [Google Scholar]
- 51.Funder JW. Primary aldosteronism. Hypertension. 2019;74:458–466. doi: 10.1161/HYPERTENSIONAHA.119.12935 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.