Abstract
The intestine is responsible for nutrient absorption and orchestrates metabolism in different organs during feeding, a process which is partly controlled by intestine-derived hormones. However, it is unclear whether the intestine plays an important role in metabolism during fasting. Here we have identified a novel hormone, famsin, which is secreted from the intestine and promotes metabolic adaptations to fasting. Mechanistically, famsin is shed from a single-pass transmembrane protein, Gm11437, during fasting and then binds OLFR796, an olfactory receptor, to activate intracellular calcium mobilization. This famsin-OLFR796 signaling axis promotes gluconeogenesis and ketogenesis for energy mobilization, and torpor for energy conservation during fasting. In addition, neutralization of famsin by an antibody improves blood glucose profiles in diabetic models, which identifies famsin as a potential therapeutic target for treating diabetes. Therefore, our results demonstrate that communication between the intestine and other organs by a famsin-OLFR796 signaling axis is critical for metabolic adaptations to fasting.
Subject terms: Mechanisms of disease, Cell signalling
Introduction
Mammals have evolved complex metabolic adaptations to survive in response to food deprivation. During prolonged fasting, the liver generates glucose via gluconeogenesis and ketone bodies via ketogenesis as major metabolic fuels for extrahepatic tissues.1,2 Mobilization of these energy sources is critical for metabolic adaptations to fasting and animal survival.1,2 In addition to energy mobilization, many mammals adopt adaptive energy-conserving survival strategies, such as torpor, to decrease core body temperature (Tb) and locomotor activity.3 Hormones derived from different organs, such as ghrelin secreted from the stomach and fibroblast growth factor 21 (FGF21) derived from the liver, orchestrate these energy conservation and mobilization processes to protect animal survival.4–9
The intestine is an important site for nutrient absorption and communication with microbiota and other organs. These processes are at least partially controlled by intestine-derived hormones.10–13 During feeding, the absorption of nutrients is associated with the release of a set of hormones that promote gastric emptying and insulin secretion, such as glucose-dependent insulinotropic polypeptide (GIP) and glucagon-like peptide 1 (GLP1).11–15 However, it is still unclear whether the intestine modulates metabolism in other organs during fasting.
Here, we have identified an intestine-derived hormone, famsin, which has a protective effect during famine by promoting metabolic adaptations to fasting. Famsin is shed from Gm11437, a highly expressed single-pass transmembrane protein (sTMP) in the proximal intestine. Famsin is secreted from the intestine and binds its receptor, OLFR796, to promote glucose and ketone body production for energy mobilization, and torpor for energy conservation to balance the energy budget during fasting. In addition, deficiency of famsin signaling, either by knockout or antibody neutralization, attenuates blood glucose levels, which suggests that famsin signaling is a potential therapeutic target for treating diabetes.
Results
Famsin is a secreted protein
sTMPs on the cell surface are involved in a wide variety of cellular signaling pathways in response to environmental cues and control various biological processes ranging from metabolism, proliferation and apoptosis to immune responses.16,17 They serve as enzymes, adapters, receptors, co-receptors and ligands to fulfill their biological function.16–18 To identify which sTMP-coding genes modulate glucose production in hepatocytes, we created a HepG2 cell line that stably expresses G6pc-Luc, which consists of a luciferase reporter gene linked to the promoter of G6pc, a gene encoding a rate-limiting enzyme in gluconeogenesis.19 The cells were transfected with a siRNA library targeting 941 human sTMP-coding genes and a control vector, RSV-Luc, to monitor transfection efficiency and cell viability (Supplementary information, Fig. S1a). To monitor the screening process, we chose forkhead box protein O1 (FOXO1) and CREB-regulated transcriptional coactivator 2 (CRTC2) as positive regulators of G6pc-Luc activity, and AKT1 and insulin receptor (INSR) as negative regulators19–21 (Supplementary information, Fig. S1b and Table S1). We performed this screen twice with very good reproducibility (Spearman r = 0.96) and identified four genes that either increased G6pc-Luc activity by more than 1.6-fold or decreased it by at least 40% (Supplementary information, Fig. S1b and Table S1).
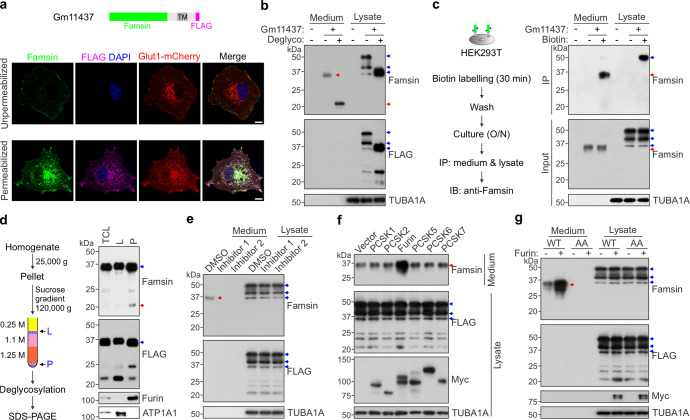
In order to test whether the encoded proteins affect G6pc-Luc activity via secretion, we performed transwell assays by overexpressing the candidate genes in HEK293T cells and measuring G6pc-Luc activity in mouse primary hepatocytes (Supplementary information, Fig. S1a). Overexpression of C17orf78, but not the other three genes, increased G6pc-Luc activity (Supplementary information, Fig. S1c, d), which suggests that C17ORF78 may affect G6pc-Luc activity via secretion. Similar results were obtained by using Gm11437, a mouse ortholog of C17orf78 (Supplementary information, Fig. S1e–g). In addition, immunostaining results showed that the N-terminus of Gm11437 has an extracellular orientation (Fig. 1a).
Fig. 1. Famsin is secreted after cleavage from Gm11437 by furin.
a Immunostaining showing the orientation of FLAG-tagged Gm11437 on the cell membrane in Cos7 cells. The signals from anti-Famsin antibody were observed in both unpermeabilized and permeabilized cells, while the signals from anti-FLAG were only detected in permeabilized cells. These results indicate that the N-terminus of Gm11437 has an extracellular orientation. DAPI, 4,6-diamidino-2-phenylindole; TM, transmembrane. Scale bars, 10 μm. b Immunoblots showing the glycosylation status of famsin in HEK293T cells. The protein extracts from culture medium or whole cell lysate were treated with or without deglycosylation enzymes. The blue arrowheads indicate full-length Gm11437-FLAG and the red arrowheads indicate famsin. Deglyco, deglycosylation. c Left panel, schematic overview showing the biotin labeling of Gm11437 located at the plasma membrane and its subsequent detection by anti-Famsin antibody. O/N, overnight; IP, immunoprecipitation; IB, immunoblotting. Right panel, immunoblots showing cleavage of famsin from plasma membrane-localized Gm11437 in HEK293T cells. The blue arrowheads indicate full-length Gm11437-FLAG and the red arrowheads indicate famsin. d Left panel, schematic diagram showing the subcellular fractionation of HEK293T cells and subsequent detection of famsin by anti-Famsin antibody. Right panel, immunoblots showing cleavage of famsin from cytoplasm-localized Gm11437 in HEK293T cells. The proteins were treated by the deglycosylation mix. The blue arrowheads indicate full-length Gm11437-FLAG and the red arrowheads indicate famsin. TCL, total cell lysate; L, light fraction that includes plasma membranes; P, pellet that includes cellular vesicles. ATP1A1 (Na+/K+-ATPase) is a marker of the plasma membrane fraction. e–g Effect of proprotein convertase inhibitors (5 μM, e) or overexpression of different proprotein convertases (f) on Gm11437 cleavage, and effect of furin on cleavage of WT Gm11437 or Gm11437/AA (g) in HEK293T cells. The blue arrowheads indicate full-length Gm11437-FLAG and the red arrowheads indicate famsin. AA, K190A/R191A.
To characterize the amino acid sequence of secreted Gm11437, we overexpressed Gm11437 in insect cells (Sf9) by a baculovirus expression system and purified the secreted Gm11437 from cell medium (Supplementary information, Fig. S1h–j). The secreted Gm11437, named famsin because of its protective effect during famine conditions, was highly glycosylated (Supplementary information, Fig. S1j). Next, we identified the amino acid sequence of famsin by mass spectrometry (MS) analysis and found that it corresponds to the extracellular 1–191 aa of Gm11437 (Supplementary information, Fig. S1k, l and Table S2). Similarly, highly glycosylated famsin was detected in medium after overexpression of Gm11437 in HEK293T cells (Fig. 1b). To examine whether the cleavage of Gm11437 occurs at the plasma membrane, we labeled the cell surface proteins by biotin and successfully detected biotin-labeled famsin in the medium (Fig. 1c). In addition, the cellular Gm11437 was able to produce famsin (Fig. 1d). These results indicate that famsin can be cleaved from both the plasma membrane-bound and cellular Gm11437.
Secretion of famsin was abrogated by inhibition of proprotein convertases (Fig. 1e). Overexpression of furin, but not other proprotein convertases, dramatically enhanced the level of famsin (Fig. 1f). In fact, furin can cleave wild-type (WT) Gm11437 but not a mutant of Gm11437 (K190A/R191A, AA) (Fig. 1g), which is consistent with the cleavage site of furin.22 Compared to WT Gm11437, the Gm11437/AA mutant lost its stimulating effect on G6pc-Luc activity in the transwell assay, whereas famsin had a stronger effect on G6pc-Luc activity than WT Gm11437 (Supplementary information, Fig. S1f, g). Similar results were also obtained by using C17ORF78 (Supplementary information, Fig. S1c, d). Taken together, these results indicate that famsin is a secreted protein and is released from Gm11437 after cleavage by furin.
Famsin is a fasting-induced hormone
To determine the physiological function of famsin, we performed ELISA analysis and found that the plasma famsin level displays circadian oscillation, with an acute drop coinciding with the onset of feeding23 (Supplementary information, Fig. S2a). Conversely, plasma famsin levels were increased after fasting (Fig. 2a). They reached maximal levels (~700 pM) after 18-h fasting and then dropped following continuous fasting or refeeding, in contrast to glucagon, insulin or ghrelin (Fig. 2a; Supplementary information, Fig. S2b–f). Together, these results indicate that famsin is a fasting-induced hormone.
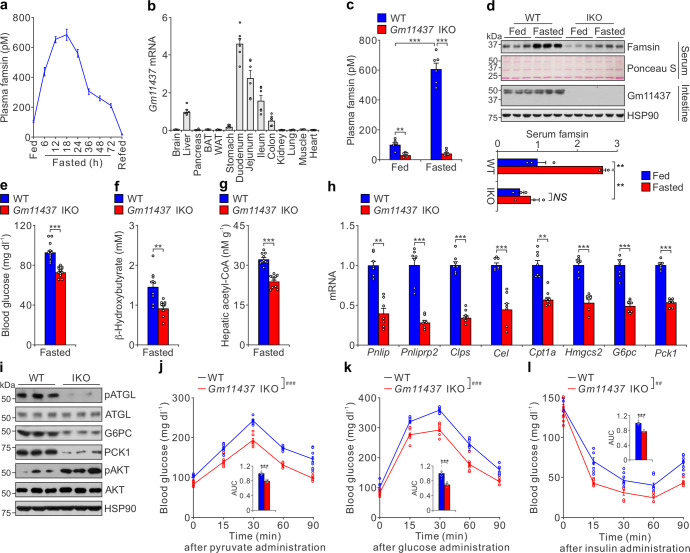
Fig. 2. Famsin is secreted from the intestine and promotes fasting-induced metabolism.
a Effect of ad lib feeding, fasting, and refeeding 1.5 h after fasting on plasma famsin levels. Fasting was started at Zeitgeber time 0 (ZT0). At each indicated time point, plasma famsin was measured by sandwich ELISA assay in six 12-week-old male mice. Data are shown as means ± SEM. b qPCR results showing relative mRNA levels of Gm11437 in different tissues from 8–10-week-old male mice. BAT, brown adipose tissue; WAT, white adipose tissue. Data are shown as means ± SEM. n = 6 mice. c Plasma famsin levels measured by sandwich ELISA assay in WT and Gm11437 IKO 8–10-week-old male mice with ad lib feeding or overnight fasting. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way analysis of variance (ANOVA) followed by Tukey’s test. **P < 0.01, ***P < 0.001. n = 6 mice. d Immunoblots (top panel) and quantification of serum famsin from immunoblots (bottom panel) showing the effect of ad lib feeding or overnight fasting on plasma famsin and intestine Gm11437 levels of 8–10-week-old male mice. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test. **P < 0.01. NS no statistical significance. n = 3. e–g Blood glucose (e), plasma β-hydroxybutyrate (f) and hepatic acetyl-CoA (g) levels from overnight fasted 8-10-week-old WT male mice and Gm11437 IKO male mice. Data are shown as means ± SEM. Comparison of different groups was carried out using unpaired two-tailed Student’s t-test. **P < 0.01, ***P < 0.001. n = 12 mice (e), n = 10 mice (f, g). h qPCR results showing relative mRNA levels of genes involved in lipolysis (Pnlip, Pnliprp2, Clps, Cel), lipid oxidation (Cpt1a), ketogenesis (Hmgcs2) and gluconeogenesis (G6pc, Pck1) from liver extracts of overnight fasted 8–10-week-old WT and IKO male mice. Data are shown as means ± SEM. Comparison of different groups was carried out using unpaired two-tailed Student’s t-test. **P < 0.01, ***P < 0.001. n = 7 mice. i Immunoblots showing the effect of Gm11437 deficiency on hepatic lipolysis and gluconeogenesis from liver extracts of 8–10-week-old male mice. j–l Pyruvate tolerance test (j), glucose tolerance test (k) and insulin tolerance test (l) results from 8–10-week-old WT and Gm11437 IKO male mice. For each test, the relative area under the curve (AUC) is shown in the inset of each panel. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test (curve data, ##P < 0.01, ###P < 0.001) or unpaired two-tailed Student’s t-test (AUC data, ***P < 0.001). n = 8 mice.
Gm11437 is highly expressed in the intestine, especially in the proximal intestine (duodenum) (Fig. 2b). It is slightly expressed in the liver and has no detectable expression in other tissues. Similar to the tissue distribution of Gm11437, C17orf78 is highly expressed in the intestine (Supplementary information, Fig. S2g). To determine which tissue is the major source of circulating famsin, we generated mice with tissue-specific knockout of Gm11437 in the intestine (IKO) or in the liver (LKO) (Supplementary information, Fig. S2h). Plasma famsin levels were increased from ~100 pM to ~600 pM after overnight fasting in both WT mice and LKO mice, while the fasting-induced increase of plasma famsin levels was dramatically decreased in IKO mice (Fig. 2c, d; Supplementary information, Fig. S2i–l). Interestingly, a coincident increase of plasma furin activity during fasting was observed (Supplementary information, Fig. S2m). Strikingly, low glucose levels can increase famsin secretion and furin activity in intestinal organoids (Supplementary information, Fig. S2n–p), which indicates that glucose is a regulator of Gm11437 shedding. Together, these results indicate that circulating famsin is mainly secreted from the intestine during fasting.
To identify which kinds of cells produce famsin, we investigated the distribution of Gm11437 in the intestine by immunostaining. Gm11437 was localized to the basolateral membrane of enterocytes and enteroendocrine cells (EECs) (Supplementary information, Fig. S2q–u). These conclusions were further confirmed by the results from Gm11437-GFP knock-in mice (Supplementary information, Fig. S2t). Gm11437-GFP has a much higher expression in the intestine than in the liver, and no expression in the kidney (Supplementary information, Fig. S2v). These findings are consistent with the Gm11437 tissue distribution evaluated by qPCR (Fig. 2b). In addition, the residual C-terminal fragment of Gm11437 after cleavage was observed in the intestinal extracts by immunoblot (Supplementary information, Fig. S2v), which further confirmed that famsin can be produced by cleavage in mice. To characterize the exact cell types that produce famsin, we used flow cytometry to sort intestinal cells into enterocytes and EECs expressing Gm11437-GFP, and then we used immunoblot analysis to test which of the sorted cell types produce famsin (Supplementary information, Fig. S2w, x). If famsin is produced from the plasma membrane-bound Gm11437, we should observe the residual C-terminal fragment of Gm11437 after cleavage, but not famsin, since famsin will be released out of the cells. Likewise, if famsin is produced within the cells, we should detect both the residual C-terminal fragment of Gm11437 and famsin. We observed robust signals from famsin (detected with anti-famsin antibody) and the residual C-terminal fragment of Gm11437 (detected with anti-GFP antibody) in the lysates of EECs, while a weaker signal from the residual C-terminal fragment of Gm11437 was detected in the lysates of enterocytes (Supplementary information, Fig. S2x). These results indicate that famsin is mainly produced by EECs and can be generated from both the plasma membrane-bound and cellular Gm11437, which is consistent with those results from overexpression of Gm11437 in HEK293T cells (Fig. 1c, d). Taken together, our results indicate that famsin is a fasting-induced hormone mainly derived from the intestine.
Famsin promotes fasting-induced metabolism
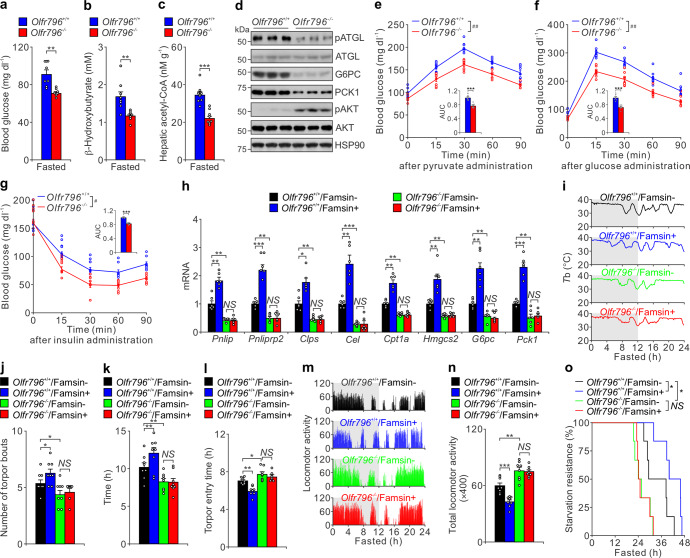
In response to prolonged fasting, the liver produces glucose via gluconeogenesis and ketone bodies as major fuels to fight famine. Mobilization of these energy sources is critical for metabolic adaptations to fasting and animal survival.1,2 Therefore, we investigated glucose and lipid metabolism in the liver to investigate how famsin affects mouse response to fasting. Although similar body weight, food intake, water intake, fat mass, movement, energy expenditure and respiratory exchange ratio (RER) were observed in WT mice and IKO mice during ad libitum (ad lib) feeding, Gm11437 IKO mice showed decreased blood glucose and plasma β-hydroxybutyrate levels after overnight fasting (Fig. 2e, f; Supplementary information, Fig. S3a–d), suggesting that famsin promotes glucose and ketone body production in fasting conditions. In addition, hepatic acetyl-CoA levels in IKO mice were dramatically attenuated (Fig. 2g), which indicates that famsin promotes hepatic lipid oxidation. In contrast to these results, IKO mice had similar blood glucose and hepatic glycogen levels in ad lib feeding conditions (Supplementary information, Fig. S3a). Furthermore, plasma triglyceride levels, hepatic triglyceride content, plasma cholesterol levels, hepatic cholesterol content, plasma free fatty acid levels and plasma glycerol levels were comparable in WT mice and IKO mice (Supplementary information, Fig. S3a). This suggests that IKO affects fasting-induced energy mobilization in the liver but has no obvious effect in adipose tissues.
The decreased expression of gluconeogenic genes (G6pc and Pck1), lipolytic genes (Pnlip, Pnliprp2, Clps, Cel), a ketogenic gene (Hmgcs2) and a gene involved in lipid oxidation (Cpt1a), and the activity of adipose triglyceride lipase (ATGL) measured by phospho-ATGL (pATGL) in the liver further confirmed that IKO resulted in the attenuation of gluconeogenesis, lipolysis, lipid oxidation and ketogenesis (Fig. 2h, i). Decreased glucose production was further confirmed by pyruvate tolerance test (Fig. 2j). This was accompanied by improved insulin sensitivity, measured by glucose tolerance test and insulin tolerance test, in IKO mice (Fig. 2k, l; Supplementary information, Fig. S3a). Together, these results demonstrate that famsin promotes glucose and ketone body production in the liver during fasting.
Famsin promotes torpor and starvation resistance in mice
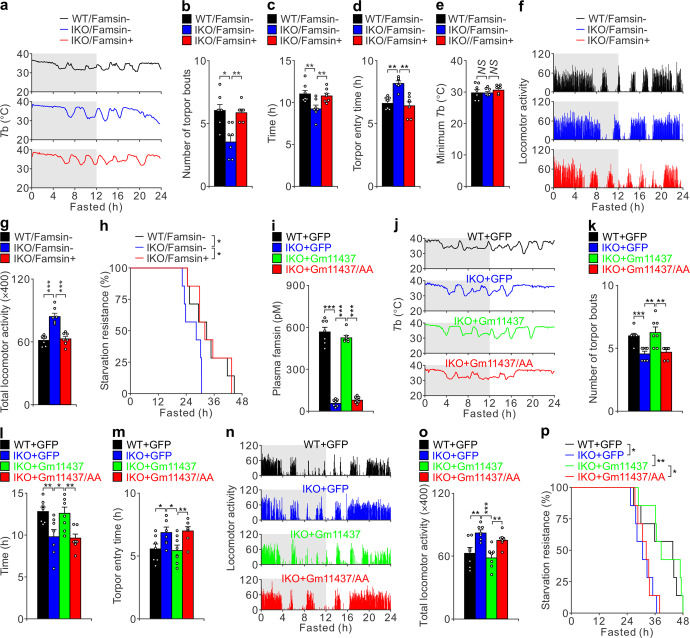
When challenged by food deprivation or harsh environmental conditions, many mammals adopt adaptive energy-conserving survival strategies, such as torpor, to decrease core Tb and locomotor activity for energy conservation.2,3 Since famsin is induced during fasting, we tested its role in fasting-induced torpor, which is characterized by periods of deep hypothermia (Tb of 25–35 °C) for several hours, along with decreases in movement and metabolic rate.3 IKO mice experienced fewer bouts of torpor, shorter duration of Tb < 35 °C and slower entry into torpor (Fig. 3a–d). The minimal Tb was comparable in WT and IKO mice (Fig. 3e). During the periods of hypothermia, the locomotor activity in WT mice was much weaker than that in IKO mice (Fig. 3f, g). Consistent with these results, the expression of lipolytic genes (Pnlip, Pnliprp2, Clps, Cel), which are normally induced by torpor,7,24 was decreased in IKO mice (Fig. 2h). In addition, IKO mice were more susceptible to starvation during fasting than WT mice (Fig. 3h). Together, these results indicate that Gm11437 promotes torpor during fasting.
Fig. 3. Famsin promotes torpor and starvation resistance.
a–h Effect of Gm11437 IKO and famsin on torpor evaluated by Tb (a), torpor frequency (number of torpor bouts, b), time for Tb < 35 °C (c), torpor entry time (d), minimum Tb (e), locomotor activity (f), relative total locomotor activity (g) and starvation resistance (h) of fasted 8-week-old male mice. Failure of starvation resistance was judged as Tb < 28 °C following a quick decrease in Tb below the environmental temperature. 400 μg/kg famsin was intraperitoneally injected after 4 h fasting. The gray and white backgrounds (a, f) indicate 12-h periods of darkness and light, respectively. Data are shown as means ± SEM. Comparison of different groups was carried out using one-way ANOVA followed by Tukey’s test (b–e, g) or log-rank test (h). *P < 0.05, **P < 0.01. NS no statistical significance. n = 7 mice. i–p Effect of adenoviral-expressed WT Gm11437 or Gm11437/AA on torpor evaluated by plasma famsin level (i), Tb (j), torpor frequency (k), time for Tb < 35 °C (l), torpor entry time (m), locomotor activity (n), relative total locomotor activity (o) and starvation resistance (p) of fasted 8-week-old male mice. Failure of starvation resistance was judged as Tb < 28 °C following a quick decrease in Tb below the environmental temperature. The gray and white backgrounds (j, n) indicate 12-h periods of darkness and light, respectively. Data are shown as means ± SEM. Comparison of different groups was carried out using one-way ANOVA followed by Tukey’s test (i, k–m, o) or log-rank test (p). *P < 0.05, **P < 0.01, ***P < 0.001. n = 7 mice.
To test whether famsin affects torpor, we purified famsin from Hi-5 cells and tested its effect on glucose production (Supplementary information, Fig. S3e–k). Famsin enhanced G6pc mRNA levels and elevated fasted blood glucose levels in a dose-dependent manner (Supplementary information, Fig. S3h–k). Compared to ghrelin, which is secreted from the stomach and deepens torpor,4–6 famsin enhances torpor frequency (torpor bouts) without affecting minimal Tb, although both hormones protect mouse resistance to fasting (Supplementary information, Fig. S3l–r). Strikingly, famsin administration by intraperitoneal injection in IKO mice restored the torpor response, starvation resistance, blood glucose levels and the expression of genes involved in gluconeogenesis and ketogenesis (Fig. 3a–h; Supplementary information, Fig. S3s, t). Furthermore, adenoviral-mediated expression of WT Gm11437 but not the Gm11437/AA mutant in the liver restored plasma famsin levels, torpor, starvation resistance, blood glucose and plasma β-hydroxybutyrate levels in IKO mice (Fig. 3i–p; Supplementary information, Fig. S3u–ab). In addition, plasma levels of torpor-inducing hormones (ghrelin, leptin and FGF21)4,6,7,25 or intestine-derived hormones (FGF15 and GLP1)9,12,14 were comparable in WT and IKO mice (Supplementary information, Fig. S3a). These results indicate that circulating famsin, but not the transmembrane protein Gm11437, is critical for fasting-induced torpor and metabolism in mice. Together, our data indicate that famsin promotes mouse adaptations to fasting.
OLFR796 is a receptor of famsin
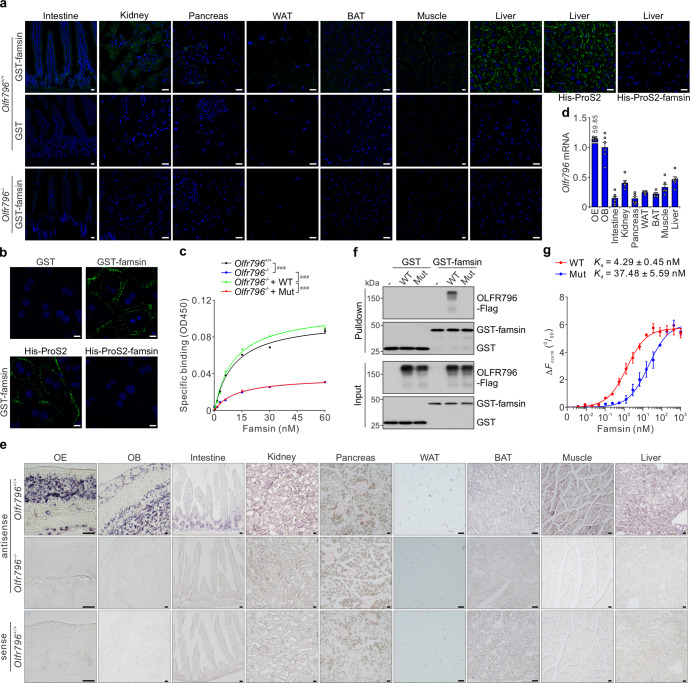
To identify the target tissue of famsin, we performed binding assays on frozen tissue sections by incubating purified GST-famsin with different mouse tissues. GST-famsin gave stronger binding signals in the liver and kidney (Fig. 4a; Supplementary information, Fig. S4a). The binding was specific because competitive preincubation of the sections with His-ProS2-famsin completely abolished the signal in the liver and in mouse primary hepatocytes (Fig. 4a, b; Supplementary information, Fig. S4a). These results were further confirmed by saturation binding studies in mouse primary hepatocytes (Fig. 4c).
Fig. 4. OLFR796 is a receptor of famsin.
a Binding assay on frozen tissue sections from 8–10-week-old Olfr796+/+ and Olfr796–/– male mice. 100 nM GST or GST-famsin was incubated with frozen tissue slices. For competition binding, tissues were pretreated with 5 μM His-ProS2 or His-ProS2-famsin for 30 min. Scale bars, 20 μm. b Binding assay on mouse primary hepatocytes. 100 nM GST or GST-famsin was incubated with hepatocytes. For competition binding, cells were pretreated with 5 μM His-ProS2 or His-ProS2-famsin for 30 min. Scale bars, 10 μm. c Effect of WT OLFR796 or its mutant (Mut, R187D/R195D/E197A) on famsin binding in mouse primary hepatocytes. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test. ###P < 0.001. n = 3. d, e qPCR results (d) and in situ hybridization (e) showing relative mRNA levels of Olfr796 in different tissues from 8–10-week-old male mice. In the pancreas sections, the islets are outlined in red. OE, olfactory epithelium; OB, olfactory bulb. Data are shown as means ± SEM. n = 6 mice (d). f GST pull-down assay showing the interaction between GST-famsin and OLFR796-Flag or its mutant (Mut, R187D/R195D/E197A) in Hi-5 cells. g Quantification of the binding affinity between famsin and WT OLFR796 or its mutant (Mut, R187D/R195D/E197A) by MST. ΔFnorm indicates the change in the normalized fluorescence. Data are shown as means ± SEM. n = 3.
Interestingly, pretreatment of mouse primary hepatocytes with SCH-202676, an inhibitor of G protein-coupled receptors (GPCRs), abolished the effect of famsin on G6pc expression (Supplementary information, Fig. S4b). This indicates that the receptor of famsin is a GPCR. To identify the receptor of famsin, we performed a siRNA screen in HEK293T cells, a human embryonic kidney cell line with strong famsin binding (Supplementary information, Fig. S4c, d). The siRNA library targeting 791 human GPCR-coding genes was used and famsin binding was measured by flow cytometry (Supplementary information, Fig. S4e, f and Table S3). This led us to identify OR10P1 as a candidate receptor of famsin, since knockdown of OR10P1 almost abolished famsin binding in HEK293T cells (Supplementary information, Fig. S4f–h and Table S3).
To investigate whether OLFR796 (mouse ortholog of OR10P1) is a receptor of famsin, we examined the expression of Olfr796 mRNA in different tissues by qPCR and in situ hybridization (ISH). Similar results obtained from qPCR and ISH showed that Olfr796 is highly expressed in olfactory epithelium (OE) and olfactory bulb (OB), and moderately expressed in the liver, kidney and muscle (Fig. 4d, e; Supplementary information, Fig. S4i), which is consistent with the results of ligand binding (Fig. 4a). Interestingly, Olfr796 is also highly expressed in intestinal crypts and islets (Fig. 4e), indicating that OLFR796 may have a special role in those tissues. In support of the conclusion that OLFR796 is a famsin receptor, famsin binding was abolished in Olfr796–/– mice and hepatocytes (Fig. 4a, c, e; Supplementary information, Fig. S4j–m).
A GST pull-down assay showed that GST-famsin purified from E. coli can interact with OLFR796 in Hi-5 cells (Fig. 4f). Further results from microscale thermophoresis (MST) showed that famsin bound to OLFR796 with high affinity (Kd = 4.29 ± 0.45 nM) (Fig. 4g; Supplementary information, Fig. S4n, o). To confirm this interaction, we introduced three mutations (R187D, R195D and E197A) into the extracellular domain of OLFR796 between transmembrane domains 4 and 5, a region previously thought to potentially harbor the ligand-binding activity of olfactory receptors.26,27 Compared to WT OLFR796, the mutant of OLFR796 (Mut) showed a dramatically reduced affinity for famsin (Fig. 4f, g; Supplementary information, Fig. S4n, o), which indicates that these three amino acids are critical for binding of OLFR796 to famsin. Addition of WT OLFR796 but not the famsin-binding-defective mutant of OLFR796 (Mut) restored dose-dependent binding of famsin in Olfr796–/– primary hepatocytes (Fig. 4c; Supplementary information, Fig. S4p, q). Together, all these in vitro and in vivo binding assays identify OLFR796 as a receptor of famsin.
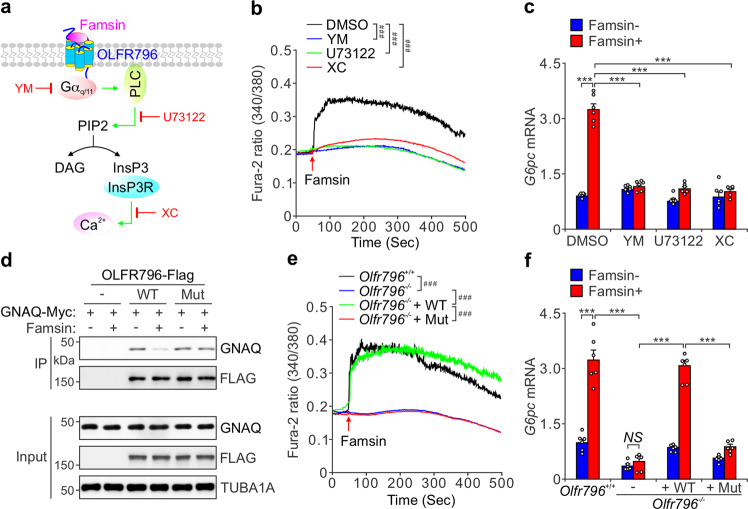
Famsin activates OLFR796-coupled calcium mobilization
Since famsin interacts with OLFR796, we examined the signaling events downstream of OLFR796 by treating hepatocytes with different inhibitors (Fig. 5a). Famsin had no effect on Gαs-coupled cAMP levels; however, it activated Gαq/11-coupled calcium mobilization, since YM-254890, an inhibitor of Gαq/11, abolished its effect (Fig. 5a, b; Supplementary information, Fig. S4r–t). These results were further supported by pretreatment with U73122, an inhibitor of phospholipase C (PLC), and Xestospongin C, an inhibitor of inositol-1,4,5-trisphosphate receptor (InsP3R) (Fig. 5a, b). Since previous results showed that InsP3R-mobilized calcium promotes gluconeogenesis via FoxO1 and CREB-mediated transcription of gluconeogenic genes,28–30 we tested the expression of the gluconeogenic gene G6pc in primary hepatocytes. In consistent with the result of calcium mobilization, famsin enhanced G6pc expression in an InsP3R-dependent manner (Fig. 5c).
Fig. 5. Famsin activates OLFR796-coupled calcium signaling.
a Schematic diagram showing the activation of OLFR796-coupled calcium signaling by famsin, and the targets of different chemicals used in this study. b, c Effect of famsin (30 nM) on calcium mobilization (b) and relative mRNA levels of G6pc (c) in the presence or absence of YM-254890 (YM, 2 μM), U73122 (10 μM) or Xestospongin C (XC, 2 μM) in mouse primary hepatocytes. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test. ###P < 0.001 (b), ***P < 0.001 (c). n = 24–48 cells (b), n = 6 (c). d Co-IP showing the association of GNAQ-Myc with OLFR796-Flag or its mutant (Mut, R187D/R195D/E197A) in HEK293T cells treated with or without famsin (30 nM) for 10 min. e, f Effect of WT OLFR796 or its mutant (Mut, R187D/R195D/E197A) on calcium mobilization (e) and relative mRNA levels of G6pc (f) in mouse primary hepatocytes in the presence or absence of famsin (30 nM). Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test. ###P < 0.001 (e), ***P < 0.001 (f). n = 18–48 cells (e), n = 6 (f).
It was reported that olfactory receptors usually bind to Golf (GNAL), which is specifically expressed in olfactory sensory neurons and other neural tissues, to activate adenylate cyclase and induce cAMP production.31,32 In fact, no detectable GNAL is observed in the liver (Supplementary information, Fig. S4u, v). Since GNAL is not present in the liver, it is possible that OLFR796 binds GNAQ (Gαq) and mobilizes calcium. In support of this notion, OLFR796 can interact with both overexpressed GNAL and GNAQ in HEK293T cells, though GNAL has a stronger association with OLFR796 than GNAQ (Supplementary information, Fig. S4w). Famsin treatment almost abolished the interaction of GNAL and GNAQ with OLFR796 (Supplementary information, Fig. S4w). In addition, famsin decreased the interaction of GNAQ with WT OLFR796 but not the famsin-binding-defective mutant of OLFR796 (Mut) (Fig. 5d). These results were further supported by similar data from famsin-induced calcium mobilization and G6pc expression (Fig. 5e, f; Supplementary information, Fig. S4q). Compared to Olfr796–/– mice, famsin enhanced cAMP levels in the olfactory bulbs but not in the liver of Olfr796+/+ mice (Supplementary information, Fig. S4x, y), which is consistent with the expression profile of GNAL (Supplementary information, Fig. S4u, v). Taken together, all these results demonstrate that famsin activates calcium signaling in hepatocytes in an OLFR796-dependent manner.
OLFR796 mediates famsin-induced metabolic adaptations to fasting
Since OLFR796 is a receptor of famsin, we asked whether Olfr796–/– mice had similar fasting-induced metabolic dysregulation to famsin-deficient mice. Similar to Gm11437 IKO mice, Olfr796–/– mice also showed decreased blood glucose, plasma β-hydroxybutyrate and hepatic acetyl-CoA levels after overnight fasting, and had similar blood glucose and hepatic glycogen levels to Olfr796+/+ mice in ad lib feeding conditions (Fig. 6a–c; Supplementary information, Fig. S5a). Furthermore, Olfr796 knockout had no effect on body weight, food intake, water intake, fat mass, energy expenditure, RER, movement, triglyceride levels, cholesterol levels, plasma free fatty acid levels and plasma glycerol levels (Supplementary information, Fig. S5a–d). The attenuated gluconeogenic capacity, the decreased expression of genes involved in metabolic adaptations to fasting, and the enhanced insulin sensitivity were also manifested in Olfr796–/– mice (Fig. 6d–h; Supplementary information, Fig. S5a). Olfr796 knockout resulted in fewer bouts of torpor, decreased starvation resistance and increased locomotor activity (Fig. 6i–o; Supplementary information, Fig. S5e). In addition, famsin-enhanced effects in WT mice were abolished in Olfr796–/– mice (Fig. 6i–o). These results were supported by the blunted expression of critical genes involved in gluconeogenesis, lipolysis, lipid oxidation and ketogenesis in Olfr796–/– mice (Fig. 6h).
Fig. 6. Olfr796 knockout attenuates metabolic adaptations to fasting.
a–c Blood glucose (a), plasma β-hydroxybutyrate (b) and hepatic acetyl-CoA (c) levels from overnight fasted 8–10-week-old Olfr796+/+ and Olfr796–/– male mice. Data are shown as means ± SEM. Comparison of different groups was carried out using unpaired two-tailed Student’s t-test (a–c). n = 8 mice. d Immunoblots showing the effect of Olfr796 knockout on hepatic lipolysis and gluconeogenesis from liver extracts of 8–10-week-old male mice. e–g Pyruvate tolerance test (e), glucose tolerance test (f) and insulin tolerance test (g) results from 8–10-week-old Olfr796+/+ and Olfr796–/– male mice. The relative AUC is shown in the inset of each panel. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test (curve data, #P < 0.05, ##P < 0.01) or unpaired two-tailed Student’s t-test (AUC data, ***P < 0.001). n = 8 mice. h qPCR results showing relative mRNA levels of genes involved in lipolysis (Pnlip, Pnliprp2, Clps, Cel), lipid oxidation (Cpt1a), ketogenesis (Hmgcs2) and gluconeogenesis (G6pc, Pck1) from liver extracts of overnight fasted 8–10-week-old Olfr796+/+ and Olfr796–/– male mice treated with or without famsin. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test. *P < 0.05, **P < 0.01, ***P < 0.001. NS no statistical significance. n = 6 mice. i–o Effect of Olfr796 knockout on torpor evaluated by Tb (i), torpor frequency (j), time for Tb < 35 °C (k), torpor entry time (l), locomotor activity (m), relative total locomotor activity (n) and starvation resistance (o) of fasted 8-week-old male mice. Failure of starvation resistance was judged as Tb < 28 °C following a quick decrease in Tb below the environmental temperature. 400 μg/kg famsin was intraperitoneally injected after 4 h fasting. The gray and white backgrounds (i, m) indicate 12-h periods of darkness and light, respectively. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test (j–l, n) or log-rank test (o). *P < 0.05, **P < 0.01. NS no statistical significance. n = 6–8 mice.
Torpor is controlled by populations of neurons in the preoptic area (POA), anteroventral periventricular nucleus (AVPe) and suprachiasmatic nucleus (SCN).33–38 Therefore, we tested whether famsin can cross the blood–brain barrier into the cerebrospinal fluid (CSF), and whether OLFR796 is expressed in these neurons. Similar to plasma famsin levels, CSF famsin levels were increased to ~500 pM after fasting, and the increase was even greater after administration of famsin (Supplementary information, Fig. S5f). Olfr796 was expressed in neurons of POA, AVPe and SCN, with the strongest signal in SCN (Supplementary information, Fig. S5g). In addition, famsin enhanced neuronal activation of POA, AVPe and SCN in an OLFR796-dependent manner (Supplementary information, Fig. S5h, i). Taken together, all these results show that Olfr796–/– mice almost phenocopy famsin-deficient mice and OLFR796 is indispensable for famsin-induced metabolic adaptations during fasting.
To evaluate the specific metabolic function of Olfr796 in the liver and avoid any effects from other tissues, we generated mice with tissue-specific knockout of Olfr796 in the liver (LKO) (Supplementary information, Fig. S6a, b). Similar to Olfr796–/– mice, Olfr796 LKO mice also showed decreased blood glucose, plasma β-hydroxybutyrate and hepatic acetyl-CoA levels and enhanced insulin sensitivity after fasting treatment, although other metabolic parameters were similar (Supplementary information, Fig. S6c–j). Olfr796 LKO did not affect torpor, but still slightly decreased mouse starvation resistance (Supplementary information, Fig. S6k–q). These results indicate that modulation of energy mobilization in the liver by a famsin-OLFR796 signaling axis is important for mouse metabolic adaptations to fasting. Furthermore, famsin-enhanced effects in WT mice were abolished in Olfr796 LKO mice, and addition of WT OLFR796 but not the famsin-binding-defective mutant of OLFR796 (Mut) restored the famsin-induced effects (Supplementary information, Fig. S6r–v). Together, these results indicate that both torpor controlled by populations of POA, AVPe and SCN neurons in the hypothalamus and energy mobilization controlled by the liver are essential for metabolic adaptations to fasting.
Neutralization of famsin improves blood glucose homeostasis
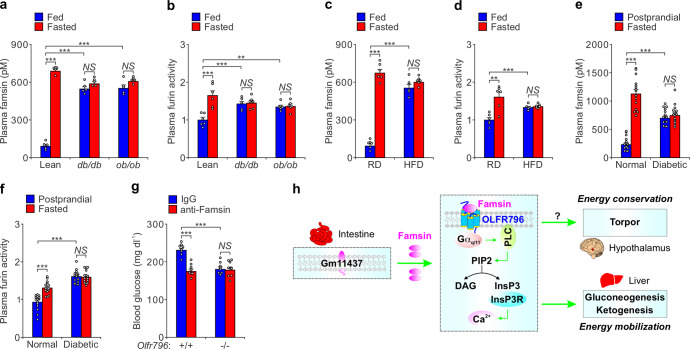
Considering the significant effect of the famsin-OLFR796 signaling axis on fasting-induced glucose metabolism and the role of hepatic gluconeogenesis in the pathogenesis of type 2 diabetes,19 we checked whether this axis contributes to hyperglycemia in obese and/or diabetic models. Although the expression levels of Gm11437 or Olfr796 in lean mice, ob/ob mice, db/db mice and high fat diet (HFD)-induced obese mice are comparable, plasma famsin levels and furin activity were dramatically increased in obese and diabetic ad lib-fed mice (Fig. 7a–d; Supplementary information, Fig. S7a–d). In addition, the fasting-induced increase of plasma famsin levels and furin activity was blunted in obese and diabetic mice (Fig. 7a–d). Similar to these results in mice, diabetic patients had higher plasma famsin levels and furin activity at 2 h after eating and produced less famsin in response to fasting than normal people (Fig. 7e, f; Supplementary information, Fig. S7e–g). Thus, these results suggest that reducing the effect of famsin may improve blood glucose levels in obese and/or diabetic models.
Fig. 7. Neutralization of famsin reduces blood glucose levels in mice.
a, b Plasma famsin levels (a) and plasma furin activity (b) in 8–10-week-old lean, db/db and ob/ob male mice with ad lib feeding or overnight fasting. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test. **P < 0.01, ***P < 0.001. NS no statistical significance. n = 5 mice (a), n = 6 mice (b). c, d Plasma famsin levels (c) and plasma furin activity (d) in regular diet (RD) or HFD-fed 16-week-old male mice with ad lib feeding or overnight fasting. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test. **P < 0.01, ***P < 0.001. NS no statistical significance. n = 6 mice. e, f Plasma famsin levels (e) and plasma furin activity (f) in normal people or patients with type 2 diabetes after overnight fasting or 2 h after eating (postprandial). Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test. ***P < 0.001. NS no statistical significance. n = 15 humans. g Effect of anti-Famsin antibody (200 μg/kg) on blood glucose in Olfr796+/+ and Olfr796–/– male mice fed a HFD for 16 weeks. Data are shown as means ± SEM. Comparison of different groups was carried out using two-way ANOVA followed by Tukey’s test. ***P < 0.001. NS no statistical significance. n = 8 mice. h Famsin, an intestine-derived hormone, binds OLFR796 and promotes energy mobilization via gluconeogenesis and ketogenesis in the liver, and energy conservation via torpor in the hypothalamus during fasting. Famsin activates calcium mobilization in the liver, but the downstream signaling of famsin in the hypothalamus is unclear (indicated with a question mark). Thus, a famsin-OLFR796 signaling axis promotes energy mobilization and conservation, and thereby enhances mouse metabolic adaptations to fasting.
To investigate whether neutralization of famsin by an antibody can decrease blood glucose levels, we tested the ability of the antibody to block famsin activity in primary hepatocytes. In fact, 10 μg/mL antibody abolished G6pc expression induced by 30 nM famsin (Supplementary information, Fig. S7h). We then intraperitoneally injected 200 μg/kg antibody into WT and Olfr796–/– mice fed a HFD for 16 weeks. Neutralization of famsin reduced blood glucose and plasma insulin levels in WT mice but not in Olfr796–/– mice and had no effect on body weight and inflammation evaluated by plasma alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels (Fig. 7g; Supplementary information, Fig. S7i–l). Together, these results indicate that the famsin-OLFR796 axis is a potential target for the treatment of type 2 diabetes.
Discussion
Although it is well known that the intestine, as a major organ of nutrient absorption, modulates metabolism in different organs during feeding, the role of the intestine in fasting-induced metabolic adaptations remains largely unknown. Here, we identified that the fasting-induced hormone famsin is secreted from the intestine and binds its receptor, OLFR796, to promote torpor for energy conservation and fasting-induced enhancement of energy mobilization in mice, thus enhancing mouse metabolic adaptations to fasting (Fig. 7h).
Our results demonstrate the critical roles of the intestine during fasting via famsin-mediated organ–organ communication, although the secretory cells and the cellular site of endogenous famsin production in the intestine remain to be fully characterized. Our results show that famsin can be cleaved from both the cellular-localized and plasma membrane-bound Gm11437 (Fig. 1c, d; Supplementary information, Fig. S2w). The more dispersed cellular distribution of famsin in enterocytes compared to Gm11437 in EECs (Supplementary information, Fig. S2q–u) suggests that some famsin is produced prior to fasting and stored in the intracellular vesicles of EECs. It seems that Gm11437-positive EECs do not belong to the known subtypes of EECs (data not shown). The presence of the cleaved C-terminal fragment of Gm11437 in intestinal extracts indicates that a proportion of famsin is derived from the plasma membrane-bound Gm11437 of EECs and/or enterocytes (Supplementary information, Fig. S2v–x). Although low glucose can promote famsin production in intestinal organoids, it remains unclear which kinds of nutrients or stresses affect famsin release in vivo. It is possible that the identity of the secretory cells and the cellular site of endogenous famsin production may depend on the different environmental cues during the different phases of fasting. Further characterization of famsin production will provide more insight into its physiological regulation and significance. In addition, our study adds to the growing list of olfactory receptors that modulate various metabolic processes.39–47 Future elucidation of the structure of the famsin–OLFR796 complex will provide more insight into how olfactory receptors can recognize both protein and odorant ligands. Considering the wide expression pattern of Olfr796 and the potential contributions of other tissues, such as the kidney and the intestine, to gluconeogenesis during prolonged fasting,48 it will be necessary to generate tissue-specific knockouts of Olfr796 to further investigate the physiological function of this signaling axis.
Previous studies showed that ghrelin in the central nervous system controls torpor.3,4,6 Our results show that the famsin-OLFR796 signaling axis induces neuronal activation and torpor. Notably, famsin and ghrelin affect different aspects of torpor, such as minimal Tb, torpor frequency or torpor entry time (Supplementary information, Fig. S3l–q and Reference4). Although both ghrelin5,8 and famsin are secreted during fasting, the dynamic fluctuation of plasma levels during fasting and the source organs of these hormones are different. This suggests that ghrelin and famsin may have divergent time-dependent roles in energy conservation. Interestingly, both ghrelin and famsin modulate GPCR-coupled calcium signaling.49 A difference is that ghrelin modulates metabolism in peripheral organs indirectly via increasing the release of growth hormone.50 Famsin induces calcium mobilization and glucose production via InsP3R, which is consistent with the reported roles of InsP3R.28–30 It is possible that the famsin-OLFR796 signaling axis may regulate neuronal activation via cAMP signaling but not calcium mobilization because of higher GNAL expression in these neurons.31,32 Of course, it is important to determine how famsin synergizes with other hormones to modulate fasting-induced metabolism.
The hormones in the gut, especially in the duodenum, are critical for weight-independent glucose control by bariatric surgery, though the specific hormones have not yet been identified.51–53 Based on the higher expression of Gm11437 in the proximal intestine and the roles of famsin in energy mobilization, it is possible that famsin is one of the proximal intestine hormones that accounts for improved glucose homeostasis after metabolic surgery. It was previously reported that individuals with higher plasma furin activity have elevated risk of hyperglycemia,54 which is consistent with our observation that plasma famsin levels and furin activity are elevated during fasting. In addition, improvement of blood glucose profiles by blockage of famsin signaling in diabetic models supports the critical role of famsin in modulating glucose homeostasis and identifies famsin as a potential therapeutic target for treating diabetes.
Materials and methods
Human studies
The participants were 15 healthy people and 15 type 2 diabetic patients; each group included 8 females and 7 males aged from 26- to 51-years old. They were recruited at the Second Affiliated Hospital of Zhejiang Chinese Medical University. All participants provided written informed consent. Blood samples from healthy people and type 2 diabetic patients were centrifuged at 4 °C soon after sampling, then separated and stored at –80 °C. The study was approved by the Institution Review Board of Tsinghua University and the Institution Review Board of The Second Affiliated Hospital of Zhejiang Chinese Medical University. Blood glucose was measured using AU5000 (Beckman Coulter). Serum famsin levels were measured using a Sandwich ELISA assay.
Mouse strains and experiments
Mice were housed in a temperature-controlled environment at ~21 °C using a 12-h light/12-h dark cycle (lights on at 7:00 am as ZT0, lights off at 7:00 pm as ZT12) with free access to food and water. Animals had ad lib access to water at all times, and food was only withdrawn if required for an experiment. For high-fat diet feeding experiments, the regular diet (D12450J, Research Diets) was replaced with a diet containing 60 kcal% fat (D12492, Research Diets). For antibody neutralization experiments, an in vitro assay to test the potential of a rat IgG antibody to neutralize famsin was performed. Mice were intraperitoneally injected with anti-famsin antibody (200 μg/kg) or isotype-matched immunoglobulin G (IgG). Animals were maintained with all relevant ethical regulations for animal testing and research. All animal experiments were approved by the Animal Care and Use Committee at Tsinghua University.
B6/JGpt-Lepem1Cd25/Gpt (T001461) and BKS-Leprem2Cd479/Gpt (T002407) mice were purchased from GemPharmatech. Gm11437-EGFP knock-in mice were obtained from Cyagen. Genotypes were confirmed by PCR by using the following primers: forward, 5′-CTCATAAGCACGGTGTTCACATAG-3′; reverse, 5′-GAATGTGCAAAGTGAATTGCAGAG-3′. Ablation of Gm11437 or Olfr796 in zygotes was achieved by injection of a mixture of Cas9 mRNA (100 ng/μL) and two sgRNAs (Gm11437: 5′-ATCAGCCGATAGCTACCCTG-3′ and 5′-GGCTCTGAGGTGACATCTTG-3′; Olfr796: 5′-ACCACCTCGAAGAGATGTGG-3′ and 5′-GGCGTTCCAGAAGAGATGGA-3′; 50 ng/μL each). Cleaved embryos with high quality at the two-cell to blastocyst stage were transferred into the oviduct of matched recipient mice. Heterozygous mice with deleted Gm11437 or Olfr796 were crossed to each other to generate knockout mice. Genotypes were confirmed by PCR by using the following primers: Gm11437: 5′-TCCTCAGGAGGTTGTCTGGA-3′ and 5′-GAGACTAGGGCTAAAAGGGGT-3′; Olfr796: 5′-GTGCTGCACTCCCCCATGTATTT-3′ and 5′-TGCTGGCCGTTCCGAAGAAGAG-3′.
Mice carrying a floxed allele of Gm11437 were obtained from Viewsolid Biotech. For generation of liver-specific or intestine-specific Gm11437 knockout mice, Gm11437fl/fl mice were crossed with mice expressing the Cre-recombinase transgene from the liver-specific albumin promoter (Alb-Cre) or the intestine-specific villin 1 promoter (Vil1-Cre) to delete exon 1. PCR genotyping of Gm11437 knockout mice was performed with primers that detect the following: 3′ LoxP site of the targeted Gm11437 allele: forward, 5′-GTTCTCATTGTTGGCATCAT-3′; reverse, 5′-AGCCTACTGTCACTCTGT-3′. Cre: forward, 5′-GCCTGCATTACCGGTCGATGC-3′; reverse, 5′-CAGGGTGTTATAAGCAATCCC-3′. Internal control for Cre: forward, 5′-CTAGGCCACAGAATTGAAAGATCT-3′; reverse, 5′-GTAGGTGGAAATTCTAGCATCATCC-3′.
Mice carrying a floxed allele of Olfr796 were obtained from GemPharmatech. For generation of liver-specific Olfr796 knockout mice, Olfr796fl/fl mice were crossed with mice expressing the Cre-recombinase transgene from the liver-specific albumin promoter (Alb-Cre) to delete exon 2. PCR genotyping of Olfr796 knockout mice was performed with primers that detect the following: 5′ LoxP site of the targeted Olfr796 allele: forward, 5′-CTTAGCATGTAGCAATCTCAAATTG-3′; reverse, 5′-CAGAAGCTAATTCAACTAAGGCA G-3′. 3′ LoxP site of the targeted Olfr796 allele: forward, 5′-ATCCAAGGTGATATTTCTGGGTC-3′; reverse, 5′-GTTAAAGCAGGAAGTGGGATTG-3′. Cre: forward, 5′-GCCTGCATTACCGGTCGATGC-3′; reverse, 5′-CAGGGTGTTATAAGCAATCCC-3′. All mice were maintained on a C57BL/6J background.
Temperature recording and fasting-induced torpor
Mice were singly housed and implanted abdominally with telemetric temperature and activity probes (G2 E-MITTER, Starr Life Science). After at least five days of recovery, mice were recorded in standard cages placed onto a radiofrequency receiver platform (ER4000, Starr Life Science). Gross movement and core body temperature were measured by using VitalView Telemetry Data Acquisition Software (Starr Life Sciences). Mice were housed in a temperature-controlled environment at ~18 °C with free access to food and water. Following baseline measurements (food and water ad lib), mice were placed in new cages and food was removed at ZT12 to induce torpor. Initial bouts of torpor were observed after ~7 h of fasting. Mouse starvation resistance was defined as a Tb ≥ 28 °C, and failure of starvation resistance was defined as Tb < 28 °C following a quick decrease in Tb below the environmental temperature.55
Indirect calorimetry, physical activity and food intake
Energy expenditure, RER, physical activity, food intake and water intake were simultaneously measured for individually housed mice with a PhenoMaster system (TSE Systems). Mice were allowed to acclimatize in the chambers for at least 24 h. Food and water were provided ad lib in the appropriate devices and measured by the built-in automated instruments. Relative fat mass was measured with an EchoMRI analyzer.
Metabolic studies
Blood glucose values were determined using a LifeScan automatic glucometer. Glucose tolerance tests were performed by intraperitoneal administration of glucose (1 g/kg) after overnight fasting. Insulin tolerance tests were performed by intraperitoneal injection of human regular insulin (1 U/kg) after 5-h fasting. For pyruvate challenge experiments, mice were fasted overnight and injected intraperitoneally with pyruvate (1 g/kg). Triglyceride (TR0100, Sigma) and cholesterol (ab65390, Abcam) levels in the liver and plasma, hepatic glycogen (MAK016, Sigma), hepatic acetyl-CoA (MAK039, Sigma), plasma insulin (10-1247-01, Mercodia), plasma leptin (MOB00B, R&D Systems), plasma glucagon (DGCG0, R&D systems), plasma β-hydroxybutyrate (K632, BioVision), plasma total ghrelin (EZRGRT-91K, EMD Millipore), plasma acyl-ghrelin (EZRGRA-90K, EMD Millipore), plasma FGF21 (MF2100, R&D Systems), plasma FGF15 (HEL154Mu, Cloud-Clone), plasma active GLP1 (EGLP-35K, EMD Millipore), plasma growth hormone (DL-GH-Mu-96T, DLDEVELOP), plasma glycerol (TR0100, Sigma) and plasma free fatty acid levels (294-63601, Wako) were measured according to the manufacturer’s instructions. Plasma ALT and AST levels were measured by the Disposable Veterinary Reagent Dis (AW00869, Seamaty Technology., Ltd.).
Measurement of furin activity
Plasma and intestines were collected from ad lib fed or overnight fasted mice. Isolated mouse intestinal crypts were incubated in the presence of 4 mM or 20 mM glucose for 24 h. The medium was centrifuged at 300× g for 3 min and then concentrated. Furin activity was measured from plasma, intestine lysates, organoid culture medium and organoid lysates using the SensoLyte® Rh110 Furin Activity Assay Kit (AS-72256, AnaSpec Inc.).
Sandwich ELISA
Sandwich ELISA was performed as described previously.43 5 μL plasma (mouse or human) or mouse cerebrospinal fluid (CSF) was used to measure famsin levels. To exclude any potential nonspecific binding of the famsin antibody in ELISA, plasma or CSF from Gm11437 knockout mice was used as an internal blank control. The purified famsin-Flag-His was quantified by BCA protein assay kit and used to generate a standard curve. The calibrated standard curve was then used to quantify plasma or CSF famsin levels in this study. For the mouse famsin sandwich ELISA, a rat monoclonal anti-famsin antibody (1:1000) against mouse Gm11437 amino acids 12–95 was used as the capture antibody, and a rabbit anti-famsin polyclonal antibody (1:2000) against mouse Gm11437 amino acids 14–185 was used as the detection antibody. For the human famsin sandwich ELISA, a mouse polyclonal anti-famsin antibody (1:1000) against human famsin amino acids 15–96 was used as the capture antibody, and a rabbit anti-famsin polyclonal antibody (1:1000) against human famsin amino acids 15–129 was used as the detection antibody. An anti-rabbit secondary antibody linked to HRP (1706515, Bio-Rad) was used to generate a signal. The rat anti-famsin monoclonal antibody and rabbit anti-famsin polyclonal antibody were generated by Beijing Prorevo Biotech Co., Ltd. The mouse anti-famsin polyclonal antibody was generated in the Animal Facility at Tsinghua University.
ISH
Digoxigenin-11-UTP-labeled RNA probes were prepared according to the DIG RNA Labeling Kit instructions (Roche) using SP6 and T7 RNA polymerase. A gene fragment of Olfr796 was amplified using the following primers (forward: CCTAAAACACATGAA, reverse: GTCTTCCCTTTACAGGAA) and cloned into pSPT19.
ISH was performed as previously described.43 Briefly, frozen sections (8 μm) were permeabilized by 20 μg/mL RNase-free Proteinase K and fixed in 4% paraformaldehyde in PBS. Sections were then dehydrated by washing in 25%, 50%, 75% and 100% methanol. Pre-hybridization in hybridization buffer (40% deionized formamide, 4× SSC, 10% dextran sulfate, 1× Denhardt’s solution, 10 mM DTT, 1 mg/mL fragmented salmon testes DNA) for 1 h at 55 °C was followed by overnight hybridization at 55 °C in hybridization buffer containing RNA probes. Unhybridized RNA probes were removed by washing twice with 2× SSC containing 50% formamide at 55 °C for 30 min. The digoxigenin-labeled probe was visualized with DIG Nucleic Acid Detection Kit (Roche).
Intestinal crypt isolation and culture
Intestinal crypts were isolated and cultured as previously described.56 Briefly, the small intestine from 10-week-old male mice was isolated, cut longitudinally and washed twice with cold PBS. Villi were carefully scraped away. The remaining intestine was sliced into small pieces roughly 5 mm in length and incubated in 20 mM EDTA in PBS for 30 min on ice. The pieces were then suspended vigorously in cold PBS. The supernatant was passed through a 70-μm cell strainer (Biologix), and the crypts were enriched by centrifugation at 300× g for 3 min. Then the isolated crypts were embedded in Matrigel (352434, Corning) and seeded into 24-well plates. After polymerization of Matrigel, advanced DMEM/F12 medium (12634028, Thermo Scientific) containing 100 ng/mL penicillin-streptomycin, 1 mM N-acetylcysteine (A9165 Sigma), 1% GlutaMAX-I (35050061, Thermo Scientific), 1% N2 (17502048, Thermo Scientific) and 2% B27 (17504044 Thermo Scientific), 50 ng/mL EGF (CH28, Novoprotein), 100 ng/mL Noggin (C028, Novoprotein) and 500 ng/mL R-spondin 1 (CD83, Novoprotein) was added for crypt culture. The culture medium was replaced by DMEM containing the indicated concentration of glucose on the second day for the famsin secretion assay.
Isolation of intestinal epithelial cells
Cell dissociation and cell sorting were performed as previously described57 with modification. In brief, the small intestine of Gm11437-GFP or Gm11437 IKO mice was isolated and washed in cold PBS. The tissue was sliced into fragments and incubated with 20 mM EDTA in PBS for 30 min on ice. Then, the tissue was mechanically disrupted by pipetting and passed through a 70-μm cell strainer (Biologix). The supernatant was centrifuged at 300× g for 3 min, and dissociated with TrypLE express (Invitrogen) for 1 min at 37 °C. The single-cell suspensions were then passed through a 40-μm filter and resuspended in cold FACS buffer (Advanced DMEM/F12 Invitrogen, 1 mM EDTA, 1% BSA). Cells were stained with anti-ALPI (A6226, Abclonal) antibody and applied to FACSAria™ III (BD Biosciences). Cells were gated on ALPI-high (enterocytes) or ALPI-low, and subsequently sorted into Advanced DMEM/F12.
Cell culture
HEK293T, Cos7 and HepG2 (ATCC) cells were maintained at 37 °C and 5% CO2 in DMEM, containing 10% FBS (HyClone) and 100 mg/mL penicillin-streptomycin. Hi-5 and Sf9 cells were cultured in SIM HF Medium (Sino Biological) and SIM SF Medium (Sino Biological), respectively, at 27 °C. Mouse primary hepatocytes were isolated as previously described43,58 and cultured in M199 medium containing 2% FBS and 0.2% BSA until attached, then continuously cultured in M199 medium without FBS. All cell lines were routinely tested for mycoplasma using a PCR detection kit (Sigma, MP0035).
Biotinylation of cell surface proteins
The assay was performed as previously described.59 HEK293 cells were grown on 6-well plates for 48 h after transient transfection with the appropriate expression plasmid. Prior to biotinylation, cells were washed three times with ice-cold PBS containing 1 mM CaCl2 and 1 mM MgCl2, then biotinylated in the presence of 1 mg/mL sulfo-NHS-LC-Biotin (A39257, Thermo) in the same buffer for 30 min on ice. Biotinylation was stopped by incubating cells for 10 min at 4 °C in the presence of 100 mM glycine in PBS and washing once with PBS. Following biotinylation, cells were cultured at 37 °C in DMEM without FBS for 16 h. Conditioned medium and cell lysates were immunoprecipitated with Streptavidin Agarose (20349, Thermo).
Subcellular fractionation
To obtain the plasma membrane and cytosolic fractions, subcellular fractionation was performed as previously reported60 with modification. Cells were harvested and homogenized in a buffer (20 mM HEPES-KOH, pH 7.2, 400 mM sucrose and 1 mM EDTA) by passing through a 30 G needle until lysis reached ∼90%, as judged by Trypan Blue staining. Homogenates were centrifuged at 25,000× g for 20 min to collect the pellet, which was suspended in 0.75 mL 1.25 M sucrose buffer and overlayed with 0.5 mL 1.1 M and 0.5 mL 0.25 M sucrose buffer. Centrifugation was performed at 120,000× g for 2 h. Two fractions, one at the interface between 0.25 M and 1.1 M sucrose (L fraction) and the pellet on the bottom (P fraction), were collected for the subsequent analysis.
Calcium imaging
Mouse primary hepatocytes were plated on glass coverslips and loaded with 5 μM Fura-2 (F1221, Thermo Fisher) in the presence of 0.025% (w/v) pluronic F127 (P2443, Sigma-Aldrich) in Medium 199 (Gibco) for 30 min. Coverslips were mounted on a laminar flow perfusion chamber (Warner Instruments). Images of Fura-2-loaded cells were collected with a cooled CCD camera while the excitation wavelength was alternated between 340 nm and 380 nm. The ratio of fluorescence intensity at the two excitation wavelengths was calculated after subtracting the background fluorescence. Images were collected and analyzed using the MetaFluor software package. Graphs present average responses from groups of 18–49 individual cells from representative single experiments. All experiments were repeated at least three times with similar results.
Reagents
Glucagon (HY-P0082, 100 nM, MedChemExpress), ghrelin (GC-11583, GlpBio), proprotein convertase inhibitor 1 (537076, 5 μM, Calbiochem), proprotein convertase inhibitor 2 (ALX-260-022, 5 μM, Enzo), SCH-202676 (1400, 10 μM, Tocris), U73122 (S8011, 5 μM, Selleck), YM-254890 (257-00631, 2 μM, Wako), and Xestospongin C (64950, 2 μM, Cayman Chemical) were used in this study.
Plasmids and adenoviruses
Myc-FLAG-tagged CD80 (MR227446), EPHB2 (MR225676), INSRR (MR217196) PCSK2 (MR226625), PCSK6 (MR215393), PCSK7 (MR210558), GNAQ (MR205488) and GNAL (MR207159) were purchased from OriGene Technologies, Inc. V5-tagged PCSK1, Furin and PCSK5 were purchased from GE healthcare. Olfr796 was synthesized by TsingKe Biological Technology, Inc. C17ORF78 and Gm11437 were amplified from a human liver cDNA library and a mouse liver cDNA library, respectively. G6pc-Luc has been described previously.61 Adenoviruses (2 × 108 plaque forming units carrying GFP, WT Gm11437 and Gm11437/AA, WT OLFR796 and mutant) were delivered to 10-week-old male WT and Olfr796 LKO mice by tail vein injection. Mice were injected with adenovirus on day 0 and sacrificed on day 7. All expressed constructs used in this study were confirmed by sequencing.
Luciferase assay and siRNA screening
To test whether overexpressed genes in HEK293T modulate G6pc-Luc in primary hepatocytes via protein secretion, HEK293T cells were plated in the lower chamber and transfected with the candidate genes. Mouse primary hepatocytes were plated in the upper chamber and infected with adenoviral G6pc-Luc and β-gal. 36 h after transfection, hepatocytes in the upper chamber were co-cultured with HEK293T for another 6 h. Luciferase assays were then performed using the Luciferase Reporter Assay System (E1980, Promega) and normalized to the activity of co-infected β-gal.
To identify which sTMPs at the cell membrane affect G6pc-Luc, HepG2 cells with stable expression of G6pc-Luc were transfected with RSV-Luc and siRNAs for human sTMP-coding genes selected from ON-TARGET plus genome-wide siRNA libraries (Dharmacon). 48 h after addition of siRNA, luciferase assays were performed using the Dual Luciferase Reporter Assay System (E1980, Promega) and normalized to co-transfected RSV-Luc activity. To confirm which GPCR is a receptor of famsin, HEK293T cells were transfected with siRNAs for human GPCR-coding genes selected from ON-TARGET plus genome-wide siRNA libraries (Dharmacon). 48 h after addition of siRNA, cells were incubated with GST-famsin (100 nM) for 30 min. The cells were stained with anti-GST (WH117206, Abclonal) antibody and analyzed by FACSCalibur Cytometer (BD Biosciences).
Protein expression and purification
To purify the cleaved products of Gm11437 from medium, Gm11437 was first expressed using the pFastBac baculovirus system (Invitrogen) in Sf9 insect cells. Conditioned medium was collected after 48-h viral infection, diluted with buffer I (20 mM Tris-HCl, pH 6.0) twice and then applied to a cation-exchange column (HiTrap SP FF, Cytiva). The column was washed with 10 column volumes of buffer II (20 mM Tris-HCl, pH 6.0, 50 mM NaCl) and eluted with 20 column volumes of a linear NaCl gradient (from 0.05 M to 0.5 M). The fractions containing secreted Gm11437 fragments were pooled, 6-times diluted with buffer I and reloaded onto the HiTrap SP FF column equilibrated with buffer II. The column was washed with 10 column volumes of buffer III (20 mM Tris-HCl, pH 6.0, 150 mM NaCl) and eluted with 20 column volumes of a linear NaCl gradient (from 0.15 M to 0.3 M). The fractions were pooled, concentrated and further purified by size-exclusion chromatography (Superdex 200 Increase 10/300 GL, Cytiva) in PBS buffer. The peak fractions were pooled and concentrated for subsequent analysis.
To purify famsin expressed and secreted by insect cells, famsin with C-terminal 3× Flag and 6× His tags was expressed using the pFastBac baculovirus system in Hi-5 cells. Conditioned medium was collected after 48-h viral infection and applied to a nickel affinity column (HisTrap excel, Cytiva) equilibrated with equilibration buffer (20 mM NaH2PO4, pH 7.4, 500 mM NaCl). The column was washed with 20 column volumes of wash buffer (20 mM NaH2PO4, pH 7.4, 500 mM NaCl, 10 mM imidazole) and eluted with 15 column volumes of a linear imidazole gradient (from 0.01 M to 0.5 M). The peak fractions were pooled, 10-times diluted with buffer containing 20 mM Tris-HCl, pH 6.0 and then applied to a HiTrap SP FF column. The column was washed with 5 column volumes of buffer containing 20 mM Tris-HCl, pH 6.0, 50 mM NaCl and eluted with 20 column volumes of a linear NaCl gradient (from 0.05 to 0.5 M). The fractions containing famsin were pooled, concentrated and further purified by Superdex 200 Increase 10/300 GL column in PBS buffer. The peak fractions were pooled and concentrated for subsequent experiments. The purified famsin-Flag-His protein was > 90% pure with endotoxin concentration equal to or below 2 EU/mL and was used to perform functional assays in this study.
To purify GST- or His-tagged famsin in bacteria, mouse Famsin was cloned into a pGEX-4T-1 vector or pCold-ProS2 for expression in E. coli. Bacteria expressing GST, GST-famsin, His-ProS2 or His-ProS2-famsin were centrifuged at 7000× g and lysed by homogenization in lysis buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl and 1 mM PMSF. The lysate was centrifuged at 47,850× g for 20 min and the GST or GST-famsin supernatant was incubated with Glutathione Agarose (Pierce Biotechnology) for 2 h at 4 °C. His-ProS2 and His-ProS2-famsin supernatants containing 5 mM imidazole were incubated with cobalt affinity resin for 2 h at 4 °C. GST or GST-famsin was eluted with buffer containing 10 mM reduced glutathione, 150 mM NaCl and 50 mM Tris-HCl, pH 7.4 at 4 °C. His-ProS2 or His-ProS2-famsin was eluted with buffer containing 200 mM imidazole, 150 mM NaCl and 50 mM Tris-HCl, pH 7.4 at 4 °C. Protein solutions were concentrated and further purified by Superdex 200 Increase 10/300 GL column in PBS buffer. The purified proteins were > 90% pure with endotoxin concentration equal to or below 2 EU/mL and were stored for subsequent analysis.
To purify OLFR796 expressed in insect cells, OLFR796 and the famsin-binding-defective OLFR796 mutant (R187D, R195D and E197A) with C-terminal 3× Flag and 6× His tags were expressed using the pFastBac baculovirus system in Hi-5 insect cells. Cells were collected after 72-h viral infection, lysed by homogenization in buffer containing 20 mM Tris-HCl, pH 7.5, 50 mM NaCl and protease inhibitor and centrifuged at 20,216× g for 10 min. The supernatant was then centrifuged at 68,905× g for 60 min and the precipitated membrane fraction was solubilized at 4 °C for 2 h with 2% n-dodecyl-β-d-maltoside and 0.4% sodium cholate. After centrifugation at 68,905× g for 30 min, the supernatant was then applied to cobalt affinity resin (TALON, Takara). The resin was rinsed with wash buffer 1 (W1) containing 20 mM Tris, pH 7.5, 300 mM NaCl, 0.4% n-dodecyl-β-d-maltoside, 0.08% sodium cholate, 10 mM imidazole and protease inhibitors, and rinsed again with wash buffer 2 (W2) containing 20 mM Tris, pH 7.5, 150 mM NaCl, 0.2% n-dodecyl-β-d-maltoside, 0.04% sodium cholate and protease inhibitors. The eluent was collected and applied to anti-Flag M2 affinity resin (Sigma). The resin was rinsed with W2 buffer and the protein was eluted with W2 buffer plus 200 μg/mL FLAG peptide, concentrated and further purified by size-exclusion chromatography (Superdex 200 Increase 5/150 GL, Cytiva) in PBS buffer containing 0.2% n-dodecyl-β-d-maltoside and 0.04% sodium cholate. The peak fractions were pooled and concentrated for subsequent analysis.
GST pull-down assay
Hi-5 cells infected with Flag-OLFR796 for 2 days were collected and lysed in cell lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1% Triton X-100) supplemented with a protease inhibitor cocktail. Post-centrifuged supernatants were mixed with 20 μL glutathione agarose (16101, Thermo) and 200 ng GST or GST-famsin recombinant protein purified from bacteria, and the binding reactions were incubated overnight at 4 °C with rotation. Precipitates were washed extensively with cell lysis buffer. Proteins bound to glutathione beads were eluted, separated on a 12% SDS-PAGE gel and assayed by immunoblotting.
MST assay
The MST assay was performed as previously described.43 The affinity of the purified famsin-Flag-His for OLFR796 or its mutant was measured using the Monolith NT.115 from NanoTemper Technologies. OLFR796 and its mutant were fluorescently labeled according to the manufacturer’s protocol and about 100 nM of labeled protein was used for each assay. A solution of unlabeled famsin-Flag-His was diluted to create an appropriate serial concentration gradient. The samples were loaded into the NanoTemper glass capillaries after incubation at room temperature for 30 min. Measurements were carried out using 20% LED power and 40% MST power. The assays were repeated three times for each affinity measurement. Kd values were calculated using the mass action equation via the NanoTemper Analysis software. For analysis, the change in thermophoresis is expressed as the change in the normalized fluorescence (ΔFnorm), which is defined as Fhot/Fcold.
Saturation binding assay
Famsin-Flag-His, purified from Hi-5 cells, was conjugated with biotin using the EZ-link micro Sulfo-NHS-LC-Biotinylation Kit (21925, Thermo Fisher Scientific). Different doses of famsin-Flag-His labeled with biotin were incubated with wildtype or Olfr796–/– primary hepatocytes with or without overexpression of OLFR796 for 30 min at room temperature. Nonspecific binding was determined using 50-fold excess unconjugated famsin-Flag-His. The cells were washed three times with PBS, followed by addition of streptavidin-HRP (21130, Thermo Fisher Scientific). The absorbance was measured colorimetrically, and the results were normalized to protein content.
cAMP levels
Mouse primary hepatocytes were incubated in the presence or absence of famsin-Flag-His (30 nM) or glucagon (100 nM) for 10 min. cAMP levels were measured from cell lysates or tissue lysates using the Cyclic AMP ELISA Kit (581001, Cayman).
Immunoprecipitation, immunoblotting and immunostaining
Assays were performed as previously described.43,58 For co-immunoprecipitation (co-IP) experiments, the indicated plasmids were transfected into HEK293T cells. After 48 h of transfection, cells were incubated in the presence or absence of 30 nM famsin-Flag-His for 10 min and collected in cell lysis buffer (150 mM NaCl, 50 mM HEPES, pH 7.4, 1% Triton X-100). Flag-tagged OLFR796 and HA-tagged OLFR796 were immunoprecipitated with anti-FLAG® M2 affinity gel (A2220, Sigma) and anti-HA agarose (26181, Thermo Fisher), respectively. All immunoprecipitated samples were washed at least three times with lysis buffer.
For immunoblotting, cells or mouse tissues were homogenized in cell lysis buffer. To analyze the glycosylation status of famsin or Gm11437, protein extracts were treated by Protein Deglycosylation Mix II (P6044S, NEB). To analyze serum famsin levels, plasma proteins were obtained using ProteoMiner Protein Enrichment Kit (1633006, Bio-Rad). Protein concentrations were determined using the BCA Protein Assay Kit (Thermo Fisher, 23227). Samples were loaded on SDS-PAGE gels and then transferred to nitrocellulose membranes. Immunoblotting was done in gelatin buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.05% Tween-20) with the corresponding antibodies. Antibodies were purchased and diluted as follows: anti-pATGL (ab135093, 1:1000), anti-GFP (ab6673, 1:1000) and anti-ATP1A1 (ab7674, 1:1000), Abcam; anti-ATGL (2138S, 1:1000), anti-pAKT (9275S, 1:1000), anti-AKT (4691S, 1:1000), anti-GNAQ (14373, 1:1000) and anti-HSP90 (4874, 1:1000), Cell Signaling Technology; anti-TUBA1A (T6199, 1:10000), anti-Myc (05-419, 1:1000) and anti-FLAG (F1804, 1:5000), Sigma-Aldrich; anti-furin (18413-1-AP, 1:1000), Proteintech; anti-G6PC (A20193, 1:2000), anti-PCK1 (A2036, 1:1000), ant-GNAL (A12804, 1:1000), anti-HA (AE013, 1:1000) and anti-GST (WH117206, 1:2000), Abclonal. The rat monoclonal anti-famsin (1:2500) antibody was generated and purified by Beijing Prorevo Biotech Co., Ltd.
For immunostaining assays, cells or tissue sections (8 μm) were fixed in 4% paraformaldehyde (PFA) solution. Unpermeabilized cells were blocked in PBS with 5% BSA, while permeabilized cells were blocked in PBS with 0.2% Triton X-100 and 5% BSA. Samples were stained overnight with specific primary antibodies in blocking solution at 4 °C, washed three times in PBS and then incubated at room temperature with fluorescent dyes in blocking solution for 1 h. Antibodies were diluted as follows: anti-Famsin (1:1000); anti-FLAG (F1804, 1:5000), Sigma-Aldrich; anti-Fos (ab190289, 1:1000), anti-DCLK1 (ab31704, 1:500) and anti-LYZ (ab108508, 1:500), Abcam; anti-ALPI (A6226, 1:1000) and anti-CLCA1 (A15041, 1:500), Abclonal; anti-CHGB (14968-1-AP, 1:500), Proteintech. A mouse polyclonal anti-famsin antibody against mouse Gm11437 amino acids 1–191 was generated in Tsinghua Animal Facility for immunostaining assay. Samples were incubated in DAPI solution for 10 min to stain the DNA, and then coverslipped. Tiled images were obtained using an epifluorescence microscope (Zeiss) and the exposure time for each channel was kept constant for all slides. Signal intensity was quantified using ImageJ.
To assay famsin binding on cells, 293T cells and primary hepatocytes were plated on glass coverslips. Cells were incubated with 100 nM GST or GST-famsin at 37 °C for 30 min, washed twice with cold PBS, and fixed immediately with buffered 4% PFA. For tissue binding assays, mouse tissues were dissected, fixed with buffered 4% PFA overnight at 4 °C, cryoprotected in 30% sucrose solution overnight, and finally embedded in OCT (Sakura). Frozen tissue slices (8 μm) were incubated with 100 nM GST or GST-famsin for 2 h at room temperature. For competition binding, cells and frozen tissue slices were incubated with 100 nM GST-famsin in the presence of 5 μM His-ProS2 or His-ProS2-famsin for 2 h at room temperature. The fixed cells and frozen tissue slices were stained with anti-GST antibody (WH117206, Abclonal) and Alexa Fluor® 488-conjugated antibodies.
Quantitative PCR
Total RNA from cells or mouse tissues was extracted using a Total RNA Purification kit (Omega). cDNA was obtained using the RevertAid First Strand cDNA Synthesis kit (Thermo). cDNA arrays of human tissues were purchased from Origene (HMRT504). RNA levels were measured with the LightCycler 480 II (Roche) as previously described.43,58 The following primers were used for qPCR: Actb-forward: 5′-GTCCACCCCGGGGAAGGTGA-3′, Actb-reverse: 5′-AGGCCTCAGACCTGGGCCATT-3′; C17orf78-forward: 5′-AACAGCTGCCTGGGATCTTCC-3′, C17orf78-reverse: 5′-TGCTTGACCTTTGGCCTTCTC-3′; Cel-forward: 5′-CTGGCCCAGCACAAAGC-3′, Cel-reverse: 5′-GGGAAAACAGGTAAGAATAGGTCTTG-3′; Clps-forward: 5′-ACCAACACCAACTATGGCATCT-3′, Clps-reverse: 5′-CCAGCTAACTGCGTGATCTCA-3′; Cpt1a-forward: 5′-CAAAGATCAATCGGACCCTAGAC-3′, Cpt1a-reverse: 5′-CGCCACTCACGATGTTCTTC-3′; G6pc-forward: 5′-GTGAATTACCAAGACTCCCAGGACTG-3′, G6pc-reverse: 5′-GATGGAACCAGATGGGAAAGAGGAC-3′; Gm11437-forward: 5′-TTGGCTCTGGAAGGAGAGTGA-3′, Gm11437-reverse: 5′-TAGGAAGATCAGTAATGACTGGCA-3′; Gnal-forward: 5′-CTGAGAACCAGTTCCGGTCA-3′, Gnal-reverse: 5′-CCTTCATCGTCCCACAGCTT-3′; Hmgcs2-forward: 5′-CCGTATGGGCTTCTGTTCAG-3′, Hmgcs2-reverse: 5′-AGCTTTGTGCGTTCCATCAG-3′; Olfr796-forward: 5′-GCCGCAAGGTCTTCTCTACC-3′, Olfr796-reverse: 5′-TATGTGATGCTGGCCGTTCC-3′; Pck1-forward: 5′-ATGTGGCCAGGATCGAAAGCAAGAC-3′, Pck1-reverse: 5′-CTTTCATGCACCCTGGGAACCTGG-3′; Pnlip-forward: 5′-ACAAACAGAAAAACCCGTATCATTAT-3′, Pnlip-reverse: 5′-TGCACATGTCAGATAGCCAGTT-3′; Pnliprp-forward: 5′-CCCCTGTTCCTCCTATGAGAAG-3′, Pnliprp-reverse: 5′-CCATTTTGGGACACCCTTGT-3′; Rpl32-forward: 5′-TCTGGTGAAGCCCAAGATCG-3′, Rpl32-reverse: 5′-CTCTGGGTTTCCGCCAGTT-3′.
MS
To identify the cleavage site of Gm11437, the purified Gm11437 N-terminal products were analyzed by electrospray ionization tandem MS on a Thermo LTQ Orbitrap instrument as previously described.43,58 The MS proteomics data have been deposited in the ProteomeXchange database via the PRIDE partner repository with the dataset identifier PXD024577.
Statistical methods
Age- and weight-matched male mice were randomly assigned for the experiments. No animals were excluded from statistical analyses, and the investigators were not blinded in the studies. All studies were performed on at least three independent occasions. Results are reported as means ± SEM. Comparison of different groups was carried out using two-tailed unpaired Student’s t-test, two-way ANOVA or log-rank test. Differences were considered statistically significant at P < 0.05.
Supplementary information
Acknowledgements
We thank Drs. Haiteng Deng, Zhongchen Xie, Ye-Guang Chen, Tong-Jin Zhao, Shengcai Lin, Liangyou Rui, Y. Eugene Chen, Cheng Zhan, Jay D. Horton, Wei Shen and all lab members for discussion and technical help. We thank Dr. Isabel Hanson for editing. This work was supported by grants from the National Natural Science Foundation of China (91957206 and 82088102), Tsinghua University Initiative Scientific Research Program (2021Z11JCQ016) and the Ministry of Science and Technology of the People’s Republic of China (2021YFA0804801).
Author contributions
A.L., Y.L., X.F., L.J. and Z.L. performed the experiments. J.H., S.W., C.C. and P.H. collected the basic parameters from human blood samples. Y.W. conceived, designed and supervised this study, and wrote the manuscript. All authors reviewed and commented on the manuscript.
Competing interests
Y.W., A.L. and Y.L. have one pending patent application. All other authors declare no competing interests.
Footnotes
These authors contributed equally: Aijun Long, Yang Liu.
Contributor Information
Ping Huang, Email: htyhp_63@163.com.
Yiguo Wang, Email: yiguo@mail.tsinghua.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-023-00782-7.
References
- 1.Cahill GF., Jr Fuel metabolism in starvation. Annu. Rev. Nutr. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 2.Jensen TL, Kiersgaard MK, Sorensen DB, Mikkelsen LF. Fasting of mice: a review. Lab. Anim. 2013;47:225–240. doi: 10.1177/0023677213501659. [DOI] [PubMed] [Google Scholar]
- 3.Geiser F. Metabolic rate and body temperature reduction during hibernation and daily torpor. Annu. Rev. Physiol. 2004;66:239–274. doi: 10.1146/annurev.physiol.66.032102.115105. [DOI] [PubMed] [Google Scholar]
- 4.Gluck EF, Stephens N, Swoap SJ. Peripheral ghrelin deepens torpor bouts in mice through the arcuate nucleus neuropeptide Y signaling pathway. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006;291:R1303–R1309. doi: 10.1152/ajpregu.00232.2006. [DOI] [PubMed] [Google Scholar]
- 5.Kojima M, et al. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402:656–660. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 6.Swoap SJ. The pharmacology and molecular mechanisms underlying temperature regulation and torpor. Biochem. Pharmacol. 2008;76:817–824. doi: 10.1016/j.bcp.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inagaki T, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Mani BK, Zigman JM. Ghrelin as a survival hormone. Trends Endocrinol. Metab. 2017;28:843–854. doi: 10.1016/j.tem.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 2012;26:312–324. doi: 10.1101/gad.184788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021;19:55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 11.Holst JJ, Gribble F, Horowitz M, Rayner CK. Roles of the gut in glucose homeostasis. Diabetes Care. 2016;39:884–892. doi: 10.2337/dc16-0351. [DOI] [PubMed] [Google Scholar]
- 12.Priest C, Tontonoz P. Inter-organ cross-talk in metabolic syndrome. Nat. Metab. 2019;1:1177–1188. doi: 10.1038/s42255-019-0145-5. [DOI] [PubMed] [Google Scholar]
- 13.Small CJ, Bloom SR. Gut hormones and the control of appetite. Trends Endocrinol. Metab. 2004;15:259–263. doi: 10.1016/j.tem.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Drucker DJ. Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metab. 2018;27:740–756. doi: 10.1016/j.cmet.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Müller TD, et al. Glucagon-like peptide 1 (GLP-1) Mol. Metab. 2019;30:72–130. doi: 10.1016/j.molmet.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bugge K, Lindorff-Larsen K, Kragelund BB. Understanding single-pass transmembrane receptor signaling from a structural viewpoint-what are we missing? FEBS J. 2016;283:4424–4451. doi: 10.1111/febs.13793. [DOI] [PubMed] [Google Scholar]
- 17.Pahl, M. C et al. Signalling via single-pass transmembrane proteins. In eLS (John Wiley & Sons Ltd: Chichester, UK, 2013).
- 18.Lichtenthaler SF, Lemberg MK, Fluhrer R. Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments. EMBO J. 2018;37:e99456. doi: 10.15252/embj.201899456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petersen MC, Vatner DF, Shulman GI. Regulation of hepatic glucose metabolism in health and disease. Nat. Rev. Endocrinol. 2017;13:572–587. doi: 10.1038/nrendo.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin HV, Accili D. Hormonal regulation of hepatic glucose production in health and disease. Cell Metab. 2011;14:9–19. doi: 10.1016/j.cmet.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 2012;11:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 23.Challet E. The circadian regulation of food intake. Nat. Rev. Endocrinol. 2019;15:393–405. doi: 10.1038/s41574-019-0210-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Kaasik K, Blackburn MR, Lee CC. Constant darkness is a circadian metabolic signal in mammals. Nature. 2006;439:340–343. doi: 10.1038/nature04368. [DOI] [PubMed] [Google Scholar]
- 25.Gavrilova O, et al. Torpor in mice is induced by both leptin-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA. 1999;96:14623–14628. doi: 10.1073/pnas.96.25.14623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-X. [DOI] [PubMed] [Google Scholar]
- 27.Pilpel Y, Lancet D. The variable and conserved interfaces of modeled olfactory receptor proteins. Protein Sci. 1999;8:969–977. doi: 10.1110/ps.8.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. Inositol-1,4,5-trisphosphate receptor regulates hepatic gluconeogenesis in fasting and diabetes. Nature. 2012;485:128–132. doi: 10.1038/nature10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozcan L, et al. Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metab. 2012;15:739–751. doi: 10.1016/j.cmet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry RJ, et al. Glucagon stimulates gluconeogenesis by INSP3R1-mediated hepatic lipolysis. Nature. 2020;579:279–283. doi: 10.1038/s41586-020-2074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones DT, Reed RR. Golf: an olfactory neuron specific-G protein involved in odorant signal transduction. Science. 1989;244:790–795. doi: 10.1126/science.2499043. [DOI] [PubMed] [Google Scholar]
- 32.Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20:69–81. doi: 10.1016/S0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, et al. Estrogen-sensitive medial preoptic area neurons coordinate torpor in mice. Nat. Commun. 2020;11:6378. doi: 10.1038/s41467-020-20050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hrvatin S, et al. Neurons that regulate mouse torpor. Nature. 2020;583:115–121. doi: 10.1038/s41586-020-2387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi TM, et al. A discrete neuronal circuit induces a hibernation-like state in rodents. Nature. 2020;583:109–114. doi: 10.1038/s41586-020-2163-6. [DOI] [PubMed] [Google Scholar]
- 36.Heller HC, Ruby NF. Sleep and circadian rhythms in mammalian torpor. Annu. Rev. Physiol. 2004;66:275–289. doi: 10.1146/annurev.physiol.66.032102.115313. [DOI] [PubMed] [Google Scholar]
- 37.Ruby NF, Zucker I. Daily torpor in the absence of the suprachiasmatic nucleus in Siberian hamsters. Am. J. Physiol. 1992;263:R353–R362. doi: 10.1152/ajpregu.1992.263.2.R353. [DOI] [PubMed] [Google Scholar]
- 38.van der Vinne V, Bingaman MJ, Weaver DR, Swoap SJ. Clocks and meals keep mice from being cool. J. Exp. Biol. 2018;221:jeb179812. doi: 10.1242/jeb.179812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellono NW, et al. Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell. 2017;170:185–198.e16. doi: 10.1016/j.cell.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang AJ, Ortega FE, Riegler J, Madison DV, Krasnow MA. Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature. 2015;527:240–244. doi: 10.1038/nature15721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffin CA, Kafadar KA, Pavlath GK. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev. Cell. 2009;17:649–661. doi: 10.1016/j.devcel.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shepard BD, et al. A renal olfactory receptor aids in kidney glucose handling. Sci. Rep. 2016;6:35215. doi: 10.1038/srep35215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li E, et al. OLFR734 mediates glucose metabolism as a receptor of asprosin. Cell Metab. 2019;30:319–328. doi: 10.1016/j.cmet.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Orecchioni M, et al. Olfactory receptor 2 in vascular macrophages drives atherosclerosis by NLRP3-dependent IL-1 production. Science. 2022;375:214–221. doi: 10.1126/science.abg3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng J, et al. Autonomous sensing of the insulin peptide by an olfactory G protein-coupled receptor modulates glucose metabolism. Cell Metab. 2022;34:240–255. doi: 10.1016/j.cmet.2021.12.022. [DOI] [PubMed] [Google Scholar]
- 46.Wu C, et al. Olfactory receptor 544 reduces adiposity by steering fuel preference toward fats. J. Clin. Invest. 2017;127:4118–4123. doi: 10.1172/JCI89344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SJ, Depoortere I, Hatt H. Therapeutic potential of ectopic olfactory and taste receptors. Nat. Rev. Drug Discov. 2019;18:116–138. doi: 10.1038/s41573-018-0002-3. [DOI] [PubMed] [Google Scholar]
- 48.Soty M, Gautier-Stein A, Rajas F, Mithieux G. Gut-brain glucose signaling in energy homeostasis. Cell Metab. 2017;25:1231–1242. doi: 10.1016/j.cmet.2017.04.032. [DOI] [PubMed] [Google Scholar]
- 49.Abizaid A, Hougland JL. Ghrelin signaling: GOAT and GHS-R1a take a LEAP in complexity. Trends Endocrinol. Metab. 2020;31:107–117. doi: 10.1016/j.tem.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldstein JL, et al. Surviving starvation: essential role of the ghrelin-growth hormone axis. Cold Spring Harb. Symp. Quant. Biol. 2011;76:121–127. doi: 10.1101/sqb.2011.76.010447. [DOI] [PubMed] [Google Scholar]
- 51.Sinclair P, Brennan DJ, le Roux CW. Gut adaptation after metabolic surgery and its influences on the brain, liver and cancer. Nat. Rev. Gastroenterol. Hepatol. 2018;15:606–624. doi: 10.1038/s41575-018-0057-y. [DOI] [PubMed] [Google Scholar]
- 52.Dimitriadis GK, Randeva MS, Miras AD. Potential hormone mechanisms of bariatric surgery. Curr. Obes. Rep. 2017;6:253–265. doi: 10.1007/s13679-017-0276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubino F, Schauer PR, Kaplan LM, Cummings DE. Metabolic surgery to treat type 2 diabetes: clinical outcomes and mechanisms of action. Annu. Rev. Med. 2010;61:393–411. doi: 10.1146/annurev.med.051308.105148. [DOI] [PubMed] [Google Scholar]
- 54.Fernandez C, et al. Plasma levels of the proprotein convertase furin and incidence of diabetes and mortality. J. Intern. Med. 2018;284:377–387. doi: 10.1111/joim.12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Odegaard JI, et al. Perinatal licensing of thermogenesis by IL-33 and ST2. Cell. 2016;166:841–854. doi: 10.1016/j.cell.2016.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao B, et al. The non-muscle-myosin-II heavy chain Myh9 mediates colitis-induced epithelium injury by restricting Lgr5+ stem cells. Nat. Commun. 2015;6:7166. doi: 10.1038/ncomms8166. [DOI] [PubMed] [Google Scholar]
- 57.Haber AL, et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han J, et al. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature. 2015;524:243–246. doi: 10.1038/nature14557. [DOI] [PubMed] [Google Scholar]
- 59.Le Gall SM, Auger R, Dreux C, Mauduit P. Regulated cell surface pro-EGF ectodomain shedding is a zinc metalloprotease-dependent process. J. Biol. Chem. 2003;278:45255–45268. doi: 10.1074/jbc.M307745200. [DOI] [PubMed] [Google Scholar]
- 60.Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, et al. Targeted disruption of the CREB coactivator Crtc2 increases insulin sensitivity. Proc. Natl. Acad. Sci. USA. 2010;107:3087–3092. doi: 10.1073/pnas.0914897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.