Abstract
Recent studies indicate that signaling molecules traditionally associated with central nervous system function play critical roles in cancer. Dopamine receptor signaling is implicated in various cancers including glioblastoma (GBM) and it is a recognized therapeutic target, as evidenced by recent clinical trials with a selective dopamine receptor D2 (DRD2) inhibitor ONC201. Understanding the molecular mechanism(s) of the dopamine receptor signaling will be critical for development of potent therapeutic options. Using the human GBM patient-derived tumors treated with dopamine receptor agonists and antagonists, we identified the proteins that interact with DRD2. DRD2 signaling promotes glioblastoma (GBM) stem-like cells and GBM growth by activating MET. In contrast, pharmacological inhibition of DRD2 induces DRD2-TRAIL receptor interaction and subsequent cell death. Thus, our findings demonstrate a molecular circuitry of oncogenic DRD2 signaling in which MET and TRAIL receptors, critical factors for tumor cell survival and cell death, respectively, govern GBM survival and death. Finally, tumor-derived dopamine and expression of dopamine biosynthesis enzymes in a subset of GBM may guide patient stratification for DRD2 targeting therapy.
Keywords: Dopamine receptor, Glioblastoma, GBM stem cells, MET, TRAIL receptor
Introduction
Glioblastoma (GBM) is the most common and the most lethal brain cancer with no curative therapy available [1]. Challenges inherent in developing effective GBM therapeutics include resistance to standard therapy such as radiation and chemotherapy, limited drug delivery into the tumor due to the Blood-Brain-Barrier (BBB), genetic and molecular heterogeneity, and a subpopulation of stem-like GBM cells (GSCs) [2], [3], [4].
Dopamine signaling in human plays profound physiological roles, including cognition, learning, reward and other behavior and physical functions [5], [6], [7]. In cancer, dopamine receptor signaling has emerged as a potential therapeutic target, as chemical screening studies have demonstrated that dopamine receptor antagonists have anti-proliferative effects against tumor cells [8], [9], [10], [11]. Subsequent studies have shown that inhibition of dopamine receptor signaling exert anti-tumor effects against GBM and a few clinical trials using a selective dopamine receptor D2 (DRD2) inhibitor ONC201 have been conducted [12], [13], [14], [15]. Deeper understanding of the molecular mechanism(s) that governs oncogenic dopamine receptor signaling in cancer is critical for potential therapeutic translation. Toward this goal, we evaluated the effects of dopamine receptor agonists and antagonists on survival, clonogenic growth, and death of GBM cells and elucidated the underlying molecular mechanisms of DRD2-mediated oncogenic signaling and DRD2 inhibition-induced cell death pathway.
Results
DRD2 agonists promote GBM cell proliferation
GBM stem-like cells (GSCs) are critical cell populations that drive GBM propagation and treatment resistance. To investigate DRD2 signaling in GBM, we utilized GSC-enriched primary GBM cells that were functionally validated in assays for self-renewal and tumor propagation, as shown in our previous reports [16], [17], [18], [19].
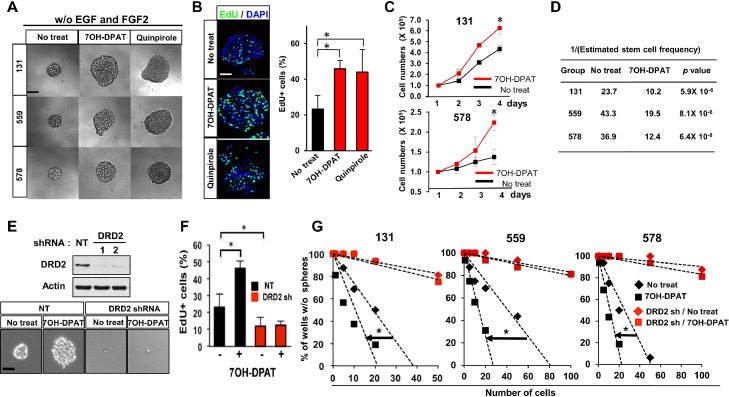
First, to determine whether activation of DRD2 signaling in GBM cells promotes cell survival and proliferation, we treated patient-derived GBM cells with dopamine receptor agonists such as 7-OH-DPAT and quinpirole in the absence of the added growth factors (Fig. 1A and D). Agonist-treated cells formed spheroid-like aggregates, an in vitro indicator of GBM clonogenic growth, more efficiently than vehicle-treated controls (Fig. 1A). Consistent with this, the results from nucleotide analog-incorporation assays, cell counts, and spheroid-formation limiting-dilution assays indicated that short-term proliferation and clonogenic growth of patient-derived GBM cells were significantly increased by dopamine receptor agonists (Fig. 1B–D). Dopamine receptor agonists are known to mainly act through dopamine receptors, which are classified as either dopamine receptor D1-like (DRD1 and DRD5) or DRD2-like (DRD2, DRD3, and DRD4) receptors [20], [21], [22]. Transcriptome analyses showed high levels of DRD2 mRNA in patient-derived GBM cells (n=14) compared to human neural progenitor cells (NPCs) and normal human astrocytes (NHAs) (Fig. S1 and data not shown). The expression levels of DRD 3, 4, and 5 were very low in all samples, whereas DRD1 expression was variable, although much lower than those of DRD2. By immunohistochemical analysis and immunoblot analysis, we found that DRD2 expression was significantly higher in GBM patient specimens compared to non-tumor brain tissue (Fig. S1). To determine whether DRD2-selective agonist 7-OH-DPAT promotes tumor cell proliferation via DRD2, we generated DRD2 knockdown cells derived from three different GBM patients and evaluate the effects of 7-OH-DPAT on cell proliferation and clonogenic growth (Fig. 1E–G). In all cells tested, pro-proliferative effects by 7-OH-DPAT were abrogated by DRD2 knockdown, suggesting that DRD2 is a critical regulator of dopamine receptor agonist-dependent GBM proliferation.
Fig. 1.
Dopamine receptor agonists promote clonogenic GBM growth via DRD2. (A) Representative images of patient-derived GBM cells (131, 559, 578) treated with dopamine receptor agonists (7-OH-DPAT and quinpirole; 2 µM). Single cells derived from primary GBM samples were cultured without EGF and FGF2 in the presence of the above chemicals for 10 days. Scale bar = 50 µm. (B) Tumorspheres derived from 131 patient GBM cells were treated with 7-OH-DPAT or quinpirole for 1 day and processed for 5′-ethynyl-2′-deoxyuridine (EdU) incorporation assays. Nuclei were stained with DAPI (blue). EdU-positive cells (stained green) were counted in three random fields and plotted. Scale bar = 50 µm. Error bars represent SD. * p < 0.01. (C) Cell growth analysis of GBM cells for 4 days of culture treated with 7-OH-DPAT. Error bars represent SD. * p < 0.01. (D) Limiting-dilution assay data to determine clonogenic growth of GBM cells (131, 559, 578) treated with 7-OH-DPAT (2 mM). Stem cell frequency was calculated using extreme limiting-dilution analysis (ELDA) (http://bioinf.wehi.edu.au/software/elda/). P value was calculated using Pairwise tests. (E) Top, Immunoblots of DRD2 in GBM cells transduced with DRD2 shRNA or non-targeting (NT) control shRNA. Actin was used as a loading control. Bottom, representative images of DRD2 knockdown 131 GBM cells treated with or without 7-OH-DPAT. Scale bar = 50 µm. (F) EdU assays were performed to determine the effects of DRD2 knockdown on short-term proliferation and clonogenic growth of GBM cells. Error bars represent SD. * p < 0.01. (G) In vitro tumorsphere-formation limiting-dilution analysis of DRD2 knockdown GBM cells (131, 559, 578) treated with or without 7-OH-DPAT. * p < 0.01.
DRD2 interacts with MET receptor and activates MET signaling in GBM
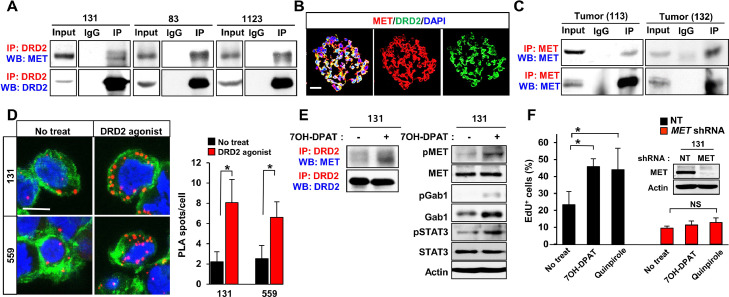
To interrogate the downstream effectors of DRD2 signaling in GBM, we took two independent but complementary approaches: (1) co-immunoprecipitation (IP)-mass spectrometry (MS) to characterize the proteins that interact with DRD2 and (2) phosphorylated protein profiling assays to determine the effector molecules that are modulated by dopamine signaling. By IP-MS and IP immunoblot analyses, we found that DRD2 co-precipitated with MET in protein lysates isolated from patient-derived GBM cells and xenograft tumors (Fig. 2A–C). Consistent with this, immunostaining analysis showed that MET and DRD2 proteins are co-expressed in GBM spheroid cells (Fig. 2B). To confirm a direct molecular interaction between DRD2 and MET in GBM, we performed proximity ligation assays (PLAs) that enable the visualization of protein-protein interactions within a cell. Notably, the interaction between DRD2 and MET proteins in GBM cells was significantly and rapidly increased by treatment with 7-OH-DPAT, as demonstrated by the increased number of PLA spots and by IP immunoblots (Fig. 2D and E). Furthermore, 7-OH-DPAT induced phosphorylation of MET and its immediate downstream effector GAB1, indicating a DRD2-dependent MET activation (Fig. 2E). Conversely, DRD2 agonist-induced signal activation and cell proliferation were abrogated by MET knockdown or by overexpression of a DRD2 mutant that cannot bind to MET (Figs. 2F and S2 A and B), indicating that MET is a key downstream effector of DRD2 signaling in GBM. These data collectively support the notion that DRD2 directly activates MET signaling in GBM.
Fig. 2.
Dopamine receptor agonists promote clonogenic GBM growth via DRD2-MET signaling axis. (A) Co-immunoprecipitation (Co-IP) immunoblots of DRD2 and MET in lysates of patient GBM-derived tumor cells (131, 83, 1123). IgG represents a control antibody used for IPs. For IP-immunoblotting, antibodies used for IP and Western blotting (WB) are labeled in red and blue, respectively. A total of 200 µg of lysates was used for each IP reaction, and total lysates (20 µg) were used as input controls. (B) Representative images of DRD2 and MET in tumorspheres derived from 559 GBM cells. Scale bar = 50 µm. (C) Co-immunoprecipitation (Co-IP) of DRD2 and MET in patient-derived GBM tumors (113 and 132). (D) Representative images of proximity ligation assays (PLAs) using anti-DRD2 and anti-MET antibodies on 131 and 559 GBM cells treated with 7-OH-DPAT for 30 min. Red dots represent the positive signal due to the proximity of the two added antibodies. Alexa Fluor 488-conjugated phalloidin (green) and DAPI (blue) were used to visualize actin cytoskeleton and nuclei, respectively. Red dots were counted in three random fields and quantitated. Scale bar = 20 µm. Error bars represent SD. *p < 0.01. (E) Co-immunoprecipitation (Co-IP) of DRD2 and MET and immunoblot analysis of phosphorylated-MET (pMET), total MET, phosphorylated-GAB1 (pGAB1), and total GAB1, phosphorylated-STAT3 (pSTAT3), total STAT3 in 7-OH-DPAT-treated cells. (F) Effects of MET depletion on DRD2 agonist-mediated GBM proliferation. EdU incorporation assays were performed on the control cells and shRNA-mediated c-MET knockdown cells that were treated with or without DRD2 agonists. Error bars represent SD. * p < 0.01.
GBM-autonomous dopamine-DRD2 signaling
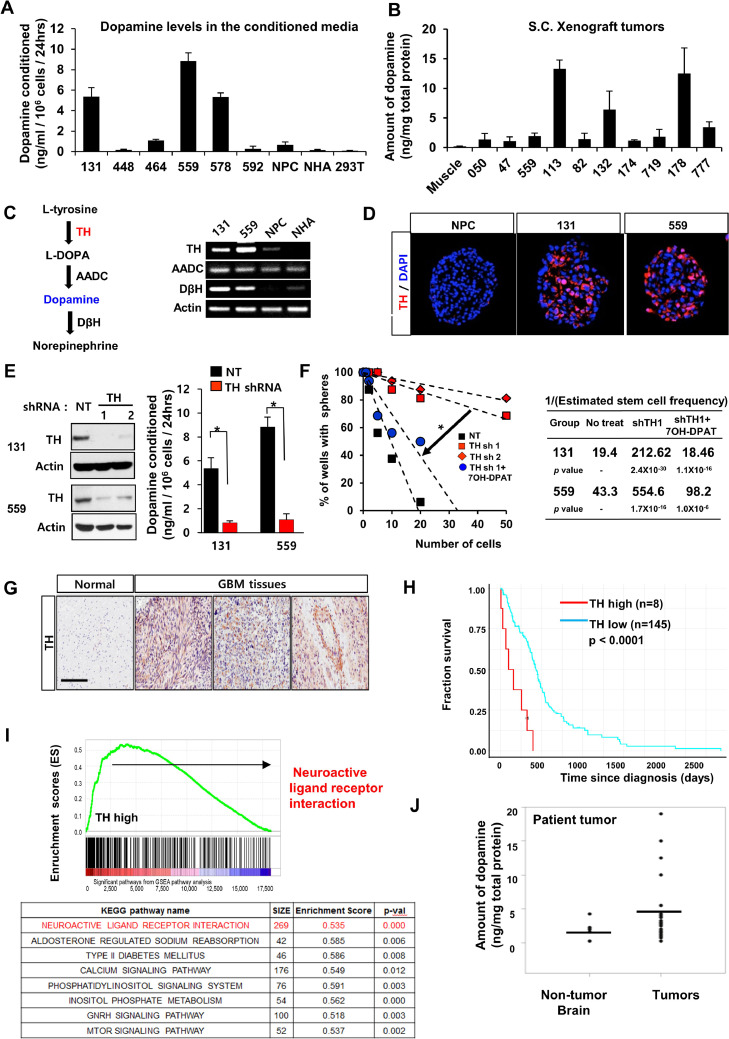
DRD2-dependent proliferation in GBM raises the possibility that tumor cells utilize dopamine, the cognate ligand of DRD2, for GBM growth and survival. GBM grow in a dopamine-rich milieu, i.e., the brain. This leads to a question of how GBM cells in vitro can retain active DRD2 signaling. Because dopamine is secreted by a small subset of rare neuroendocrine tumors in addition to dopaminergic neurons in the normal brain [7,23,24], we hypothesized that GBM tumors may produce dopamine. Dopamine was not detected in fresh media or in conditioned media from normal neural progenitor cells (NPCs) and astrocytes, as determined by dopamine ELISA analysis. In contrast, we found high levels of dopamine in the conditioned media from three primary GBM cells (out of 6 tested; Fig. 3A). Furthermore, GBM-derived subcutaneous tumors contain high levels of dopamine, suggesting a tumor-autonomous dopamine-DRD2 signaling axis (Fig. 3B). To further investigate dopamine secretion by GBM cells, we examined the expression levels of genes that are crucial for dopamine biosynthesis. Tyrosine hydroxylase (TH) catalyzes the conversion from L-tyrosine to L-dopa, which is the rate-limiting step of dopamine synthesis [25] (Fig. 3C). Importantly, 131 and 559 GBM cells express TH mRNA, and their TH expression levels correlated with the levels of dopamine secreted in conditioned media (Fig. 3A–C). TH protein was expressed in GBM-derived spheroids but not in NPCs (Fig. 3 C and D). To determine the role of TH in GBM cells, we expressed shRNA against TH in 131 and 559 GBM cells (Fig. 3E). TH knockdown GBM cells grew similarly compared to the control non-targeting (NT) shRNA-expressing cells when cultured in the presence of EGF and FGF2 (data not shown). However, TH knockdown significantly decreased clonogenic growth of GBM cells without the added mitogens, and treatment with dopamine receptor agonists rescued this phenotype (Fig. 3F), suggesting that the decreased GBM growth by TH knockdown is likely due to the depletion of dopamine. Consistently, alpha-methyl-p-tyrosine (AMPT), a small-molecule TH inhibitor, impaired dopamine secretion and impeded clonogenic growth of 131 GBM cells, similar to TH knockdown (data not shown). Collectively, these data support that a subset of GBMs harbors tumor-autonomous dopamine-DRD2 signaling.
Fig. 3.
A subset of GBMs secrets tumor-derived dopamine. (A) Dopamine levels in the conditioned media cultured with various cells. Six different GBM cells, NPCs, normal human astrocytes (NHAs), and 293T cells were cultured in serum-free conditions for collection of the conditioned media. (B) Quantification of dopamine levels in ten subcutaneous GBM xenograft tumors. Muscle tissues near subcutaneous tumor implantation sites were used as controls. Dopamine content was determined by ELISA analysis. (C) A diagram of dopamine biosynthesis in cells. Expression levels of Tyrosine hydroxylase (TH) mRNA in 131 and 559 GBM cells, NPCs, and NHA determined by RT-PCR analysis. (D) Representative images of TH on 131 and 559 GBM cells compared with NPCs. (E) Immunoblots of TH in GBM cells transduced with TH shRNA or non-targeting (NT) control shRNA. Dopamine secretion of TH knockdown GBM cells (131 and 559). (F) LDA analysis of 131 and 559 cells to determine the effects of TH knockdown on clonogenic growth of GBM cells treated with or without 7OH-DPAT. (G) Representative images of immunohistochemical staining of TH in GBM patient specimens. (H and I) Kaplan-Meier survival curves and Gene set enriched analysis (GSEA) of glioma patients based on the expression levels of TH in the TCGA database. (J) Quantification of dopamine levels in patient specimens and non-tumor brain tissues by ELISA analysis.
To establish the clinical relevance of our findings, we performed immunohistochemical (IHC) analyses on patient GBM tissue microarray (TMA) sections using DRD2 and TH antibodies. DRD2 expression was significantly higher in a majority of patient-derived GBM specimens than in non-tumor brain tissues, further confirmed by immunoblot analyses (Fig. S1). In contrast, only approximately 15% of GBMs (9 out of 60 GBM specimens) have strong immuno-positivity for TH [26] (Fig. 3G). Bioinformatics analysis using the TCGA data set revealed that high levels of TH mRNA correlate with worse prognosis of GBM patients (Fig. 3H). As the TH outlier subgroup (8 out of 153 GBM patients) showed extremely poor survival compared to the remaining group, we interrogated the gene sets that were differentially expressed in this subset of patients and conducted gene set enrichment analysis (GSEA). A top-ranked enriched pathway in the TH outlier group was neuroactive ligand-receptor interaction (GPCR signaling) pathway, raising the possibility of enriched oncogenic neurotransmitter signaling in this GBM subset (Fig. 3I). Finally, we determined dopamine levels in lysates from patient GBM specimens (n=26) and non-tumor brain tissues (n=5). The median values of dopamine content in GBM specimens were slightly higher than those of non-tumor tissues. However, 4 out of 26 GBM specimens harbored more than 5-fold higher levels of dopamine compared to the non-tumor brain tissues (Fig. 3J).
High DRD2 level portends poor survival of the mesenchymal glioma patients
Next, we investigated a potential association between mRNA levels of DRD2 and MET and GBM subtypes in glioma patients using the TCGA and the NIH GBM datasets. DRD2 mRNA expression levels in glioma patients are generally higher than non-tumor bearing brain tissues, however, they have not been directly associated with poor patient survival. We found a moderate but significant positive correlation between mRNA levels of DRD2 and MET mRNA in all glioma patients (Fig. S3A). We then determined whether DRD2 mRNA levels portend the patient survival of specific GBM subtypes. We inferred DRD2-high subset (top 25 % of DRD2 levels in each subtype) and compared overall survival of these patients with the remainder group. Notably, a DRD2-high subset in mesenchymal subtype, but not in proneural subtype, showed significantly poor prognosis (Fig. S3 B to D). These findings suggest that MET-DRD2 signaling is particularly activated in mesenchymal GBM subtype.
DRD2 antagonists inhibit MET activation and induce GBM cell death
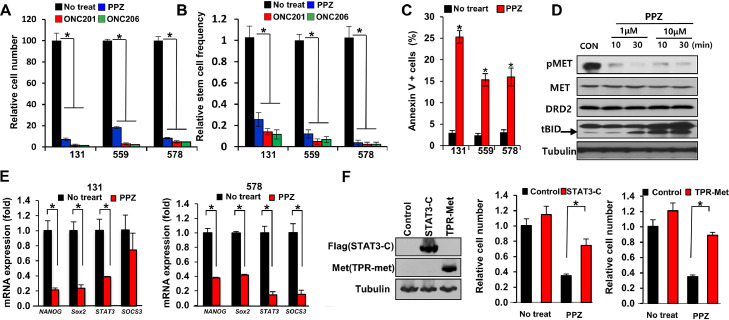
Next, we investigated the molecular mechanisms through which DRD2 inhibition impedes GBM survival and growth. To inhibit DRD2 signaling in GBM, we employed an FDA-approved DRD2 antagonist perphenazine (PPZ) and newly identified selective DRD2 inhibitors (ONC201 and ONC206) [[12], [13], [14], [15],27,28]. Pharmacological DRD2 inhibition significantly impeded the survival and clonogenic growth of GBM cells (Figs. 4 A to C and S4). In addition, DRD2 antagonists potently abolished activation of multiple GBM-promoting effectors such as MET and STAT3, and stem cell regulators including NANOG, SOX2, and SOCS3 (Fig. 4 D and E).
Fig. 4.
DRD2 antagonists induce GBM cell death with MET inactivation. (A) Cell viability of GBM cells (131, 559, 578) cultured with DRD2 antagonists (PPZ (perphenazine; 10 mM), ONC201 (0.5 mM), and ONC206 (0.5 mM)) for 4 days. Error bars represent SD. * p < 0.01. (B) Relative stem cell frequencies of GBM cells (131, 559, 578) treated with DRD2 antagonists. P value was calculated using Pairwise tests. *p <0.01. (C) Levels of annexin V-positive apoptotic GBM cells (131, 559, 578) treated with PPZ. Error bars represent SD. *p <0.01. (D) Western blot analysis of pMET, MET, DRD2 and BID in 131 GBM cells treated with PPZ (1 mM, 10 mM) for 10 and 30 min. (E) RT-PCR analysis of stemness factors (Nanog, Sox2, STAT3, and SOCS3) expression in 131 and 578 cells treated with PPZ. Error bars represent SD. *p <0.01. (F) Effects of constitutively active STAT3-C or TPR-MET mutants on PPZ-treated GBM cells. Expression of STAT3-C and TPR-MET mutants was confirmed by immunoblots (left). Error bars represent SD. *p < 0.01.
By phosphorylated protein arrays, we found that phosphorylated STAT3 in GBM cells was increased by 7-OH-DPAT but decreased by PPZ, compared to untreated control cells (Figs. 2E and S2C). STAT3 and ERK pathways are key downstream effectors of oncogenic signaling pathways including the MET pathway [17,29]. As activities of MET and STAT3 in GBM were inhibited by DRD2 antagonists, we tested whether activation of the above signaling pathways could rescue DRD2 antagonist-induced cell death. We over-expressed constitutively active forms of MET (TPR-MET) or STAT3 (STAT3C) in GBM cells and determined the proliferation kinetics of these cells treated with or without DRD2 antagonists (Fig. 4F). While TPR-MET- or STAT3C-expressing GBM cells grew similarly compared to the controls, these cells exhibited increased survival in the presence of DRD2 antagonists. Together, these data suggest that DRD2 inhibition leads to inactivation of the MET and STAT3 pathways in GBM.
DRD2 antagonist induces TRAIL receptor-mediated GBM cell death
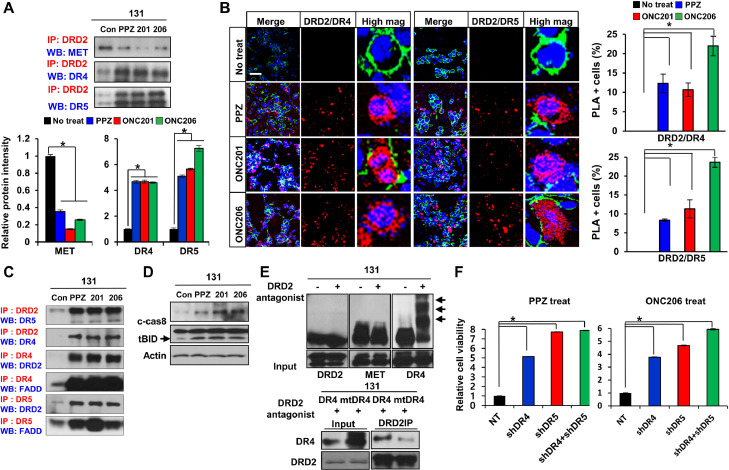
We hypothesized that DRD2 antagonists would alter the downstream signaling cascades of DRD2-interacting proteins. GBM cells were treated with various DRD2 antagonists for 30 min, and IP-immunoblot analyses were performed to determine DRD2-interacting proteins. Notably, DRD2-MET co-IP complexes and MET phosphorylation were greatly decreased in DRD2 antagonist-treated GBM cells. In sharp contrast, TRAIL receptor 1 and 2 (also known as death receptor 4 and 5 (DR4 and DR5) or TNFR10A and TNFR10B, respectively) co-immunoprecipitated with DRD2 (Fig. 5A). These acute and robust interactions between DRD2 and DR4/5 were further confirmed by PLA assays using three different DRD2 antagonists (Fig. 5B). Because TRAIL ligand-TRAIL receptor (DR4 and DR5) signaling is a well-known pathway of extrinsic apoptosis in various cancer types including GBM, we further examined the components of the TRAIL-mediated cell death machinery [30], [31], [32]. By additional IP-immunoblot analyses, we found a significant induction of the DR4/5-FADD immunocomplex, which is a key component of the TRAIL-mediated death-inducing signaling complex (DISC) [33] (Fig. 5C). These data are consistent with molecular cascades of TRAIL-mediated cell death (Fig. 5D); however, TRAIL ligand is not expressed in GBM cells [34,35]. To reconcile this discrepancy, we investigated post-translational modifications of death receptors, some of which are known to stimulate receptor aggregation and trigger TRAIL ligand-independent cell death [36]. Strikingly, the levels of S-palmitoylation of DR4 proteins were significantly increased within 30 min of DRD2 antagonist treatment (Fig. 5E). In contrast, palmitoylation levels of DRD2, MET, and other TNFR superfamily proteins were not affected by DRD2 antagonist treatment (Fig. 5E and data not shown). In addition, ectopic expression of shRNA-mediated DR4/5 depletion rescued GBM cells from DRD2 antagonist-mediated death (Figs. 5F and S5). These data collectively suggest that pharmacological DRD2 inhibition induces GBM cell death via DRD2-DR4/5 interaction and DISC formation in a TRAIL ligand-independent manner.
Fig. 5.
DRD2 antagonists induce TRAILR-mediated cell death via crosstalk between DRD2 and death receptor. (A) Top, IP-immunoblots of DRD2 and MET or DR4/5 using lysates from DRD2 antagonist-treated GBM cells. Bottom, quantitation of IP blots. Error bars represent SD. *p < 0.01. (B) Left, representative images of PLAs using anti-DRD2 with anti-DR4 or anti-DR5 antibodies on 131 GBM cells treated with DRD2 antagonists (PPZ, ONC201, ONC206) for one hour. Red dots represent the positive signal due to the proximity of the two added antibodies. Alexa Fluor 488-conjugated phalloidin (green) and DAPI (blue) were used to visualize the actin cytoskeleton and nuclei, respectively. Right, Quantitation of PLA positive cells. Scale bar = 50 µm. Error bars represent SD. *p < 0.01. (C) IP-immunoblot analysis of DRD2, DR4, DR5, and FADD in 131 GBM cells treated with DRD2 antagonists (PPZ, ONC201, ONC206) for one day. (D) Western blot analysis of cleaved-Caspase8 and BID in 131 cells treated with DRD2 antagonists (PPZ, ONC201, ONC206) for two days. (E) Representative protein palmitoylation assays. Arrows indicate palmitoylated proteins. Palmitoylated signal is only detected for DR4 but not for DRD2 and MET following treatment with DRD2 antagonists for one hour in 131 GBM cells. (F) Cell viability was analyzed in 131 GBM cells transduced with shNT, shDR4, shDR5, and shDR4+shDR5 cultured with the DRD2 antagonists PPZ (1 µM) and ONC206 (0.5 µM) for 2 days. Error bars represent SD. *p < 0.01.
DRD2 targeting extends survival of GBM tumor bearing mice
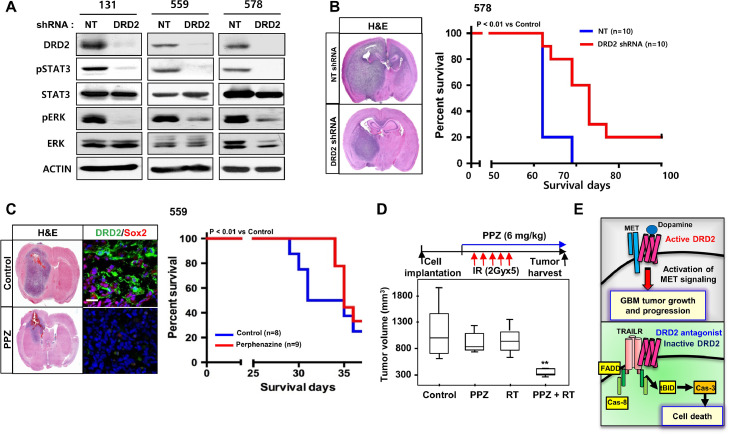
To examine the association between DRD2 and GSC phenotype, we sorted three different GBM patient-derived cells based on DRD2 expression and then evaluated the biology of the sorted populations. DRD2high GBM cells were enriched in stem cell characteristics and possessed enhanced clonogenic capacity compared to matched DRD2 low/- GBM cells, suggesting that DRD2 is a GSC regulator (Fig. S6A). To test whether DRD2 inhibition exhibits in vivo efficacy against GBM, we expressed shRNA against DRD2 in GBM cells (131, 559, 578). DRD2 knockdown GBM cells significantly suppressed phosphorylation of STAT3, ERK, and decreased the levels of stem cell regulators including NANOG, SOX2, and SOCS3 compared to non-targeting (NT) shRNA-expressing cells (Figs. 6A and S6B). Furthermore, DRD2 knockdown significantly extended the survival of tumor-bearing mice (Fig. 6B).
Fig. 6.
DRD2 targeting extends survival of GBM tumor bearing mice. (A) Western blot analysis of DRD2, pSTAT3, STAT3, pERK, and ERK levels in GBM cells (131, 559, 578) after transduction with DRD2 shRNA or with non-targeting (NT) shRNA-expressing lentivirus. β-actin was used as a loading control. (B) Left, representative H&E staining of an orthotopic xenograft tumor. Right, kaplan-Meier survival curves of mice orthotopically implanted with 578 GBM cells transduced with NT shRNA (control, n=10) or DRD2 shRNA (n=10). p < 0.001. (C) Representative images of orthotopic xenograft tumor (559) treated with PPZ. Kaplan-Meier survival curves of mice orthotopically implanted with 559 GBM cells treated with PPZ. p < 0.01. (D) Experimental scheme of irradiation/PPZ combination experiments. GBM83 cell-derived subcutaneous tumors were generated in immunodeficient nude mice. Once tumor size reached 500 mm3, treatments were initiated: mock-control (n=6), PPZ (6 mg/kg body weight, i.p daily, n=6), irradiation (2 Gy daily for 5 days, n=6) or combined PPZ/RT treatment (n=6). Tumor volumes were determined 9 days after treatment and are shown in box-and-whisker plots. *p < 0.01. (E) A schematic model of the DRD2-MET and DRD2-TRAILR signaling axis in GBMs.
We then assessed the therapeutic efficacy of DRD2 antagonist in orthotopic GBM xenograft models. PPZ was chosen because: (1) it is a clinically used anti-psychotic drug for patients with chronic schizophrenia; (2) it crosses the blood-brain barrier (BBB) easily; and 3) it is a DRD2 (and DRD3)-selective antagonist with a binding affinity (Ki) lower than 1 nM for DRD2 [37]. GBM 559 patient-derived tumors were implanted in the brains of nude mice, and PPZ treatment (6 mg/kg body weight, daily injection) was initiated 20 days after tumor inoculation. PPZ-treated mice survived longer than the control tumor-bearing mice, and histological analysis and immunoblot analysis indicated smaller tumor size and decreased proliferation of tumor cells in the treated group (Fig. 6C). Immunofluorescence (IF) analyses revealed that PPZ treatment significantly reduced the numbers of DRD2- and SOX2-expressing GBM cells (Fig. 6C). Finally, we tested whether DRD2 inhibition combined with radiation treatment results in maximal therapeutic benefit. Radiation is the most effective non-surgical therapy currently available for GBM patients; however, its effect is limited. In vivo irradiation or PPZ treatment alone, at the doses tested, modestly delayed tumor growth compared to the control. However, combination index analysis suggested that a combination of radiation and PPZ synergistically suppressed tumor growth in vivo (Fig. 6D).
In summary, our findings illustrate a molecular circuit of DRD2 signaling which controls both survival and death of GBM cells via selective activation of the MET and DR4/5 pathways, respectively (Fig. 6E).
Discussion
Receptor Tyrosine Kinase (RTK) signaling is an essential component of the molecular circuitry of GBM [38,39]. RTKs such as MET can be regulated by the mechanisms independent of their cognate ligand-receptor interactions [40]. Here, we show that DRD2 activates MET-mediated clonogenic growth and stemness of GBM tumors in HGF (a cognate MET ligand)-independent manner. While our study has identified the activation of MET signaling via a DRD2-MET interaction, DRD2 may have a broader role as a “trans-activator” of various RTKs including epidermal growth factor receptor (EGFR) or platelet-derived growth factor receptor (PDGFR) [41], [42], [43], [44].
Levels of TH expression and tumor-derived dopamine appear to be quite variable between tumors. Dopamine levels in the tumor microenvironment, it is possible that basal levels exist in the brain might be sufficient to sustain dopamine-dopamine receptor signaling in GBM. Thus, both autocrine and paracrine-dependent DRD2 signaling in GBM is plausible.
Together with the reports on the NMDA receptor, DRD4, and b-adrenergic receptor [41,[45], [46], [47], [48], [49]], our findings support the notion that a broad repertoire of pro-oncogenic mechanisms utilized by tumor include neurotransmitter signaling. Anti-tumorigenic effects of DRD2 inhibition appear to be quite general in a large number of human GBM cells covering different subtypes and mouse GBM cells with defined genomic alterations, supporting potential therapeutic opportunities. Inhibition of crucial GBM oncogenic signaling pathways by DRD2 antagonists also supports the possibility of the greater efficacy by the combined treatments with currently available tyrosine kinase inhibitors (TKIs). Beyond the traditional role of dopamine receptors primarily in neurons, recent studies have implicated additional roles of DRD2 such as astrocyte-driven DRD2 in neuroinflammation and the stabilization of blood vessels [50], [51], [52]. Extensive investigations to evaluate various roles of the DRD2 signaling are warranted for the potential clinical translation of DRD2 targeting.
Recent studies demonstrated potent anti-cancer effects of ONC201 in orthotopic brain cancer xenograft models and clinical trials [[12], [13], [14], [15],27,28,53,54]. The DRD2-MET and DRD2-TRAIL receptor interactions shown here may guide the development of more specific and effective DRD2 targeting agents, given the recent discovery of high-resolution DRD structures and selective agonists/antagonists [55,56]. In summary, these data indicate an integrated signaling between DRD2 and MET in GBM, with potential clinical implications of targeting this pathway.
Methods
Human GBM specimens and derivatives
Following informed consent, glioblastoma specimens were obtained from patients undergoing surgery in accordance with the Institutional Review Boards. Within hours after surgical removal, tumor specimens were enzymatically dissociated into single cells, following the procedures previously reported [16], [17], [18], [19]. Briefly, tumors classified as GBM were dissociated into single cells and were cultured in Neurobasal medium with N2 and B27 supplements (0.5x each; Invitrogen) and human recombinant bFGF and EGF (25 ng/ml each; R&D systems). All GBM cells used in this study are patient-derived primary GBM cells enriched for stem cell characteristics. Human neural stem/progenitor cells (NPCs) (Lonza and Aruna) and normal human astrocytes (from Lonza) were purchased and cultured as recommended.
Chemicals and antibodies
7-OH-DPAT was obtained from Tocris. Quinpirole, perphenazine, and L-DOPA were purchased from Sigma. ONC201 and ONC206 were obtained from Oncoceutics and purchased from Selleckchem. The following primary antibodies were used: For immunoblotting, anti-DRD1 (324390, 1:2,000 for WB and 1:500 for IF; Calbiochem); anti-DRD2 (SC-5303, 1:1,000 for WB, 1:100 for IF and IP; Santa Cruz), anti-MET (SC-161, 1:1,000; Santa Cruz), anti-pMET (T1234/1235) (3077, 1:1,000 for WB and 1:200 for IF; Cell Signaling), anti-pY705 STAT3 (4113, 1:1,000; Cell Signaling), anti-STAT3 (9139, 1:2,000; Cell Signaling), anti-pERK1/2 (T202/Y204; 4377, 1:2,000; Cell Signaling), anti-ERK1/2 (4695, 1:2000; Cell Signaling), anti-TH (P4010, 1:1,000; Pel-freeze), anti-DR4 (42533, 1:1000 for WB and 1:100 for IP; Cell Signaling), anti-DR5 (8074, 1:1000 for WB and 1:100 for IP; Cell Signaling), anti-FADD (2782, 1:2000 for WB and 1:100 for IP; Cell Signaling), anti-cleaved Caspase8 (9496, 1:1000 for WB; Cell Signaling), anti-BID (2002, 1:1000 for WB; Cell Signaling), anti-V5 (R960-25, 1:4,000; Life Technologies), anti-GAPDH (SC-20327, 1:1,000; Santa Cruz), and anti-β-actin (1:5,000; Sigma-Aldrich). For immunofluorescence, anti-Sox2 (MAB2018, 1:200; R & D Systems) and anti-GFAP (Z0334, 1:500; DAKO) were used.
Immunoblots and immunoprecipitation
Human GBM specimens and tumor cells were lysed in Pierce IP lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol; #87788, Thermo) supplemented with protease inhibitor cocktail (Complete Mini, Roche Diagnostics) and phosphatase inhibitor (#78428, Thermo), incubated on ice for 30 min and cleared by centrifugation at 4°C for 20 min. For immunoprecipitation, protein lysates (200 µg) were incubated with appropriate antibodies overnight at 4°C. Immunocomplexes were washed six times with lysis buffer, eluted by boiling the beads in 2x SDS sample loading buffer for 5 min, and resolved by SDS-polyacrylamide gel electrophoresis. Liquid chromatography-tandem mass spectrometry of the immunoprecipitated samples was performed as previously described [17]. Primary antibodies for immunoblots were incubated with the membrane overnight at 4°C. Protein bands were visualized using ECL Western Blotting Detection Reagents (GE Healthcare) and subjected to densitometry analysis using the ChemiDoc XRS system with Quantity One software (Bio-Rad Laboratories).
Lentivirus production and transduction
Lentiviral plasmids expressing short hairpin RNAs (shRNAs) specific for DRD1, DRD2, TH, MET, DR4, and DR5 were purchased from Sigma-Aldrich (MISSION® shRNA). Lentiviral vectors expressing wild-type DRD2, the DRD2 mutant without the cytoplasmic domain (amino acids 217-370 of DRD2 protein), tpr-MET, and STAT-C and DR4 palmitoylation-defective mutant (DR4 C261-3S) were validated by sequencing and immunoblot analysis. For viral production, 293T cells were co-transfected with a lentiviral expression vector and packaging plasmids (psPAX2 and pCMV-VSV-G) using CalPhos Mammalian Transfection Kit (Clontech). Virus-containing supernatants were collected and concentrated by ultracentrifugation. The titer of each lentivirus was determined by serial dilution.
Tumorsphere-forming limiting-dilution assay
Limiting-dilution assay (LDA) was performed in 96-well plates as previously described [17]. Dissociated cells were seeded at a range of 1 to 100 cells per well without FGF2 and EGF. Spheroid formation was inspected one to two weeks later. Stem cell frequency was calculated using a web-based tool, “ELDA” (extreme limiting-dilution analysis), which is available on the Walter and Eliza Hall Institute of Medical Research web site (http://bioinf.wehi.edu.au/software/elda/).
Cell proliferation assay (Edu staining)
For proliferation assays, 5′-ethynyl-2′-deoxyuridine (Edu) staining was performed using the Click-iT™ EdU imaging kit (Invitrogen) according to the manufacturer's protocol. Edu (10 µM) was added into the culture media for 2 h, and cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS), rinsed with PBS, and then immersed in 30% sucrose in PBS at 4°C for 1 h. Spheres were embedded in Tissue-Tek O.C.T. compound (Sakura Finetek Japan, Tokyo) and sectioned using a CM1950 cryostat (Leica). The sections were washed and incubated with a Click-iT™ reaction cocktail and then visualized with Alexa Fluor 488.
Quantitative real-time RT-PCR
Total RNA was isolated using Trizol Reagent (Invitrogen). Total RNA (2 µg) was used for reverse transcription reactions using the qScript cDNA SuperMix Kit following the manufacturer's instructions (Quanta Biosciences). Real-time RT-PCR was performed on an Applied Biosystems 7900HT cycler using SYBR-Green Mastermix (SA Biosciences) with the following primers:
DRD2 - F: 5′-CGAGCATCCTGAACTTGTGTG-3′
R: 5′-GCFTTATTGAGTCCGAAGAGG-3′
TH - F: 5′-GGGCTGTGTAAGCAGAACG-3′
R: 5′-AAGGCCCGAATCTCAGGCT-3′
NANOG - F: 5′-AATACCTCAGCCTCCAGCAGATG-3′
R: 5′-TGCGTCACACCATTGCTATTCTTC-3′
STAT3 - F: 5′-GGGTGGAGAAGGACATCAGCGGTAA-3′
R: 5′-GCCGACAATACTTTCCGAATGC-3′
SOCS3 - F: 5′-AGACTTCGATTCGGGACCAGCCCC0-3′
R: 5′-GAGCCAGCGTGGATCTGCGC-3′
β-actin - F: 5′-AGAAAATCTGGCACCACACC-3′
R: 5′-AGAGGCGTACAGGGATAGCA-3′
The relative quantification of target gene expression was performed with the standard curve or comparative cycle threshold (CT) method. Results shown were representative of three independent experiments.
Proximity ligation assay (PLA)
Cells were treated with a DRD2 agonist for 30 min or DRD2 antagonists (PPZ, ONC201, and ONC206) for 1 h. Vehicle controls were used as references. Cells were fixed with 4% PFA for 15 min, blocked with 5% donkey serum in PBS containing 0.2% Triton X-100, and incubated overnight with antibodies against anti-MET (SC-8307, 1:250), anti-DRD2 (SC-5303, 1:250), anti-DR4 (42533, 1:200), and/or anti-DR5 (8074, 1:200). The proximity ligation reaction and visualization of signal were performed according to the manufacturer's protocol using the Duolink Detection Kit with PLA PLUS and MINUS probes for mouse and rabbit antibodies (Sigma). DAPI stain was used to detect cell nuclei. Alexa Fluor 488 phalloidin (Life Technologies) was used to visualize the actin cytoskeleton in cells.
Human phospho-kinase array
GBM cells were treated with perphenazine, 7-OH-DPAT, or vehicle control for 24 h. After treatment, cells were rinsed with PBS and lysed. The protein concentration of the lysates was determined using a Biophotometer plus (Eppendorf). The lysates were diluted and incubated with the Human Phospho-Kinase Proteome Profiler Array (R&D Systems) according to the manufacturer's protocol. Briefly, 500 µg of lysates was incubated on each membrane. The intensity of each spot was measured on a ChemiDoc XRS system with Quantity One software (Bio-Rad Laboratories).
Fluorescence-activated cell sorting (FACS)
For isolation of DRD2high and DRD2low/- GBM cells, patient-derived xenograft tumors were dissociated into single-cell suspensions and labeled with anti-DRD2 antibody. These labeled cells were sorted using a FACS Vantage SE flow cytometer (BD). Cell Quest Acquisition and Analysis software (BD) was used to acquire and quantify the fluorescence signal distributions and intensities from individual cells. Sorted DRD2high and DRD2low/- cells were plated into 96 well plates (1 to 100 cells per well) for limiting-dilution assays.
Dopamine ELISA analysis
For quantitative determination of dopamine in patient tumors, xenograft tumors, and conditioned cell culture media, we used a Dopamine Research ELISA kit (Labor Dionostika Nord GmbH & Co) following the manufacturer's instructions. For the quantification of secreted dopamine in cell culture media, the conditioned medium was collected and centrifuged to remove cell debris. The collected supernatant was mixed with sodium metabisulfite (final concentration 4 mM) and quantified. For the detection of dopamine content in patient GBM specimens and their derivative xenograft tumors, tissues were homogenized in lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 4 mM sodium metabisulfite). Lysates were centrifuged at 13,000 rpm at 4°C for 20 min.
GBM xenograft models
All mouse experiments were performed according to the guidelines of the Institutional Animal Care and Use Committees. Six-week-old male BALB/c nude mice were used. GBM cells were dissociated, resuspended in 5 µl of HBSS, and injected intracranially into the striatum of nude mice by using a stereotactic device (Kopf instruments) (coordinates: 2 mm anterior, 2 mm lateral from the bregma, 2 mm depth from the dura) [17,18]. Perphenazine (6 mg/kg body weight, daily) was administered intraperitoneally. When mice developed neurological symptoms (lethargy, ataxia, and seizures) or significant body weight loss, mice were euthanized and processed for histological analysis. For in vivo radiation of tumor-bearing mice, fractionated radiation (2 Gy daily for 5 days) was administered.
Nonradioactive palmitoylation assays
GBM cells were treated with perphenazine or vehicle control for 1 h. For nonradioactive palmitoylation assays, we used a SiteCounter S-Palmitoylated protein kit (Badrilla) following the manufacturer's instructions. After drug treatment, cells were rinsed with PBS, lysed, and thiol blocked with thiol blocking solution at 40°C for 4 h. Following lysis, excess thiol blocking was removed by three sequential ice-cold acetone precipitations, and pellets were resuspended in binding buffer. Samples were incubated with Thioester cleavage reagent for 1 h at room temperature. After flowing through desalting spin columns, site modification of palmitoylated proteins in the samples was performed using Mass Tag reagent for 1 h at room temperature. The proteins were denatured in 2x Laemmli sample buffer at 60°C for 10 min and processed for SDS/PAGE.
Bioinformatics data analysis
The Cancer Genome Atlas (TCGA) database was used to analyze correlations among mRNA expression of DRD2 and MET, TH and dopamine receptors (DRD1-5), with the patient survival, glioma subtypes, and signaling pathways [57], [58], [59], [60]. Pathway and gene ontology analyses were performed using GSEA. Normalized enrichment scores and FDR values were generated by GSEA software. Pearson correlation coefficient was calculated by "cor" function of R and Pearson's Chi-squared Test was conducted using "chisq.test" function of R with default settings.
Statistical analyses
All data were expressed as means ± SD from at least three independent experiments. Quantification of immunopositive cells in immunostaining analyses was carried out using NIH ImageJ software (National Institutes of Health, Bethesda, MD). For the animal survival studies, p values were determined by log-rank test. Student's t-test was used to determine statistical significance. P values less than 0.01 were considered to be significant.
Bioinformatics data analysis
The Cancer Genome Atlas (TCGA) database [57,58] was used to analyze correlations between mRNA expression of TH and Dopamine receptors (DRD1-5), survival, and signaling pathways.
Statistical analyses
All data were expressed as means ± SD from at least three independent experiments. Quantification of immunopositive cells in immunostaining analyses was carried out using NIH imageJ software (National Institutes of Health, Bethesda, MD). For the animal survival studies, p values were determined by log-rank test. Student's t-test was used to determine statistical significance. P values less than 0.05 were considered to be significant.
CRediT authorship contribution statement
Hye-Min Jeon: Conceptualization, Investigation, Visualization, Writing – original draft, Writing – review & editing. Young Taek Oh: Investigation. Yong Jae Shin: Investigation. Nakho Chang: Investigation. Donggeun Kim: Investigation. Donghun Woo: Investigation. Yoon Yeup: Resources. Kyeung Min Joo: Investigation. Heejin Jo: Data curation, Formal analysis. Heekyoung Yang: Investigation. Jin-Ku Lee: Investigation. Wonyoung Kang: Investigation. Jason Sa: Data curation. Won Jun Lee: Data curation. James Hale: Investigation. Justin D. Lathia: Resources. Benjamin Purow: Resources, Writing – review & editing. Myung Jin Park: Validation. Jong Bae Park: Validation. Do-Hyun Nam: Conceptualization, Resources, Supervision. Jeongwu Lee: Conceptualization, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Dr. Josh Allen and Oncoceutics for providing ONC201 and ONC206. This work was supported by NIH R01 CA223370 and Velosano grants (J.L). Additional supports were from the Grant of Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C3418).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2023.100894.
Appendix. Supplementary materials
References
- 1.Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., Curschmann J., Janzer R.C., Ludwin S.K., Gorlia T., Allgeier A., Lacombe D., Cairncross J.G., Eisenhauer E., Mirimanoff R.O., European Organisation for R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Lathia J.D., Mack S.C., Mulkearns-Hubert E.E., Valentim C.L., Rich J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prager B.C., Bhargava S., Mahadev V., Hubert C.G., Rich J.N. Glioblastoma stem cells: driving resilience through chaos. Trends Cancer. 2020;6:223–235. doi: 10.1016/j.trecan.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaulieu J.M., Gainetdinov R.R. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol. Rev. 2011;63:182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- 6.Nutt D.J., Lingford-Hughes A., Erritzoe D., Stokes P.R. The dopamine theory of addiction: 40 years of highs and lows. Nat. Rev. Neurosci. 2015;16:305–312. doi: 10.1038/nrn3939. [DOI] [PubMed] [Google Scholar]
- 7.Snyder S.H. What dopamine does in the brain. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18869–18871. doi: 10.1073/pnas.1114346108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamandis P., Wildenhain J., Clarke I.D., Sacher A.G., Graham J., Bellows D.S., Ling E.K., Ward R.J., Jamieson L.G., Tyers M., Dirks P.B. Chemical genetics reveals a complex functional ground state of neural stem cells. Nat. Chem. Biol. 2007;3:268–273. doi: 10.1038/nchembio873. [DOI] [PubMed] [Google Scholar]
- 9.Dolma S., Selvadurai H.J., Lan X., Lee L., Kushida M., Voisin V., Whetstone H., So M., Aviv T., Park N., Zhu X., Xu C., Head R., Rowland K.J., Bernstein M., Clarke I.D., Bader G., Harrington L., Brumell J.H., Tyers M., et al. Inhibition of dopamine receptor D4 impedes autophagic flux, proliferation, and survival of glioblastoma stem cells. Cancer Cell. 2016;29:859–873. doi: 10.1016/j.ccell.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachlos E., Risueno R.M., Laronde S., Shapovalova Z., Lee J.H., Russell J., Malig M., McNicol J.D., Fiebig-Comyn A., Graham M., Levadoux-Martin M., Lee J.B., Giacomelli A.O., Hassell J.A., Fischer-Russell D., Trus M.R., Foley R., Leber B., Xenocostas A., Brown E.D., et al. Identification of drugs including a dopamine receptor antagonist that selectively target cancer stem cells. Cell. 2012;149:1284–1297. doi: 10.1016/j.cell.2012.03.049. [DOI] [PubMed] [Google Scholar]
- 11.Visnyei K., Onodera H., Damoiseaux R., Saigusa K., Petrosyan S., De Vries D., Ferrari D., Saxe J., Panosyan E.H., Masterman-Smith M., Mottahedeh J., Bradley K.A., Huang J., Sabatti C., Nakano I., Kornblum H.I. A molecular screening approach to identify and characterize inhibitors of glioblastoma stem cells. Mol. Cancer Ther. 2011;10:1818–1828. doi: 10.1158/1535-7163.MCT-11-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arrillaga-Romany I., Chi A.S., Allen J.E., Oster W., Wen P.Y., Batchelor T.T. A phase 2 study of the first imipridone ONC201, a selective DRD2 antagonist for oncology, administered every three weeks in recurrent glioblastoma. Oncotarget. 2017;8:79298–79304. doi: 10.18632/oncotarget.17837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arrillaga-Romany I., Odia Y., Prabhu V.V., Tarapore R.S., Merdinger K., Stogniew M., Oster W., Allen J.E., Mehta M., Batchelor T.T., Wen P.Y. Biological activity of weekly ONC201 in adult recurrent glioblastoma patients. Neuro-Oncol. 2020;22:94–102. doi: 10.1093/neuonc/noz164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall M.D., Odia Y., Allen J.E., Tarapore R., Khatib Z., Niazi T.N., Daghistani D., Schalop L., Chi A.S., Oster W., Mehta M.P. First clinical experience with DRD2/3 antagonist ONC201 in H3 K27M-mutant pediatric diffuse intrinsic pontine glioma: a case report. J. Neurosurg. Pediatr. 2019:1–7. doi: 10.3171/2019.2.PEDS18480. [DOI] [PubMed] [Google Scholar]
- 15.Prabhu V.V., Madhukar N.S., Gilvary C., Kline C.L.B., Oster S., El-Deiry W.S., Elemento O., Doherty F., VanEngelenburg A., Durrant J., Tarapore R.S., Deacon S., Charter N., Jung J., Park D.M., Gilbert M.R., Rusert J., Wechsler-Reya R., Arrillaga-Romany I., Batchelor T.T., et al. Dopamine receptor D5 is a modulator of tumor response to dopamine receptor D2 antagonism. Clin. Cancer Res. 2019;25:2305–2313. doi: 10.1158/1078-0432.CCR-18-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joo K.M., Kim J., Jin J., Kim M., Seol H.J., Muradov J., Yang H., Choi Y.L., Park W.Y., Kong D.S., Lee J.I., Ko Y.H., Woo H.G., Lee J., Kim S., Nam D.H. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 2013;3:260–273. doi: 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Kim E., Kim M., Woo D.H., Shin Y., Shin J., Chang N., Oh Y.T., Kim H., Rheey J., Nakano I., Lee C., Joo K.M., Rich J.N., Nam D.H., Lee J. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W., Park J.K., Fine H.A. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Son M.J., Woolard K., Nam D.H., Lee J., Fine H.A. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gingrich J.A., Caron M.G. Recent advances in the molecular biology of dopamine receptors. Annu. Rev. Neurosci. 1993;16:299–321. doi: 10.1146/annurev.ne.16.030193.001503. [DOI] [PubMed] [Google Scholar]
- 21.Lao C.L., Lu C.S., Chen J.C. Dopamine D3 receptor activation promotes neural stem/progenitor cell proliferation through AKT and ERK1/2 pathways and expands type-B and -C cells in adult subventricular zone. Glia. 2013;61:475–489. doi: 10.1002/glia.22449. [DOI] [PubMed] [Google Scholar]
- 22.Sibley D.R., Monsma F.J., Jr., Shen Y. Molecular neurobiology of dopaminergic receptors. Int. Rev. Neurobiol. 1993;35:391–415. doi: 10.1016/s0074-7742(08)60573-5. [DOI] [PubMed] [Google Scholar]
- 23.Caragher S.P., Shireman J.M., Huang M., Miska J., Atashi F., Baisiwala S., Hong Park C., Saathoff M.R., Warnke L., Xiao T., Lesniak M.S., James C.D., Meltzer H., Tryba A.K., Ahmed A.U. Activation of dopamine receptor 2 prompts transcriptomic and metabolic plasticity in glioblastoma. J. Neurosci. 2019;39:1982–1993. doi: 10.1523/JNEUROSCI.1589-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gimenez-Roqueplo A.P., Dahia P.L., Robledo M. An update on the genetics of paraganglioma, pheochromocytoma, and associated hereditary syndromes. Horm. Metab. Res. 2012;44:328–333. doi: 10.1055/s-0031-1301302. [DOI] [PubMed] [Google Scholar]
- 25.Daubner S.C., Le T., Wang S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011;508:1–12. doi: 10.1016/j.abb.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joo K.M., Kim J., Jin J., Kim M., Seol H.J., Muradov J., Yang H., Choi Y.L., Park W.Y., Kong D.S., Lee J.I., Ko Y.H., Woo H.G., Lee J., Kim S., Nam D.H. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 2013 doi: 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Karpel-Massler G., Ba M., Shu C., Halatsch M.E., Westhoff M.A., Bruce J.N., Canoll P., Siegelin M.D. TIC10/ONC201 synergizes with Bcl-2/Bcl-xL inhibition in glioblastoma by suppression of Mcl-1 and its binding partners in vitro and in vivo. Oncotarget. 2015;6:36456–36471. doi: 10.18632/oncotarget.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ralff M.D., Kline C.L.B., Kucukkase O.C., Wagner J., Lim B., Dicker D.T., Prabhu V.V., Oster W., El-Deiry W.S. ONC201 demonstrates antitumor effects in both triple-negative and non-triple-negative breast cancers through TRAIL-dependent and TRAIL-Independent Mechanisms. Mol. Cancer Ther. 2017;16:1290–1298. doi: 10.1158/1535-7163.MCT-17-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu H., Lee H., Herrmann A., Buettner R., Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 30.Allen J.E., Krigsfeld G., Mayes P.A., Patel L., Dicker D.T., Patel A.S., Dolloff N.G., Messaris E., Scata K.A., Wang W., Zhou J.Y., Wu G.S., El-Deiry W.S. Dual inactivation of Akt and ERK by TIC10 signals Foxo3a nuclear translocation, TRAIL gene induction, and potent antitumor effects. Sci. Transl. Med. 2013;5:171ra17. doi: 10.1126/scitranslmed.3004828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen J.E., Krigsfeld G., Patel L., Mayes P.A., Dicker D.T., Wu G.S., El-Deiry W.S. Identification of TRAIL-inducing compounds highlights small molecule ONC201/TIC10 as a unique anti-cancer agent that activates the TRAIL pathway. Mol. Cancer. 2015;14:99. doi: 10.1186/s12943-015-0346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouyang L., Shi Z., Zhao S., Wang F.T., Zhou T.T., Liu B., Bao J.K. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45:487–498. doi: 10.1111/j.1365-2184.2012.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pennarun B., Meijer A., de Vries E.G., Kleibeuker J.H., Kruyt F., de Jong S. Playing the DISC: turning on TRAIL death receptor-mediated apoptosis in cancer. Biochim. Biophys. Acta. 2010;1805:123–140. doi: 10.1016/j.bbcan.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Frank S., Kohler U., Schackert G., Schackert H.K. Expression of TRAIL and its receptors in human brain tumors. Biochem. Biophys. Res. Commun. 1999;257:454–459. doi: 10.1006/bbrc.1999.0493. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y., Lang F., Xie X., Prabhu S., Xu J., Sampath D., Aldape K., Fuller G., Puduvalli V.K. Efficacy of adenovirally expressed soluble TRAIL in human glioma organotypic slice culture and glioma xenografts. Cell Death. Dis. 2011;2:e121. doi: 10.1038/cddis.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poh T.W., Huang S., Hirpara J.L., Pervaiz S. LY303511 amplifies TRAIL-induced apoptosis in tumor cells by enhancing DR5 oligomerization, DISC assembly, and mitochondrial permeabilization. Cell Death Differ. 2007;14:1813–1825. doi: 10.1038/sj.cdd.4402177. [DOI] [PubMed] [Google Scholar]
- 37.Lieberman J.A., Stroup T.S., McEvoy J.P., Swartz M.S., Rosenheck R.A., Perkins D.O., Keefe R.S., Davis S.M., Davis C.E., Lebowitz B.D., Severe J., Hsiao J.K. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N. Engl. J. Med. 2005;353:1209–1223. doi: 10.1056/NEJMoa051688. [DOI] [PubMed] [Google Scholar]
- 38.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., Beroukhim R., Bernard B., Wu C.J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S.A., Ciriello G., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn G.P., Rinne M.L., Wykosky J., Genovese G., Quayle S.N., Dunn I.F., Agarwalla P.K., Chheda M.G., Campos B., Wang A., Brennan C., Ligon K.L., Furnari F., Cavenee W.K., Depinho R.A., Chin L., Hahn W.C. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Organ S.L., Tsao M.S. An overview of the c-MET signaling pathway. Ther. Adv. Med. Oncol. 2011;3:S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borroto-Escuela D.O., Tarakanov A.O., Guidolin D., Ciruela F., Agnati L.F., Fuxe K. Moonlighting characteristics of G protein-coupled receptors: focus on receptor heteromers and relevance for neurodegeneration. IUBMB Life. 2011;63:463–472. doi: 10.1002/iub.473. [DOI] [PubMed] [Google Scholar]
- 42.Daub H., Weiss F.U., Wallasch C., Ullrich A. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature. 1996;379:557–560. doi: 10.1038/379557a0. [DOI] [PubMed] [Google Scholar]
- 43.Luttrell L.M., Daaka Y., Lefkowitz R.J. Regulation of tyrosine kinase cascades by G-protein-coupled receptors. Curr. Opin. Cell Biol. 1999;11:177–183. doi: 10.1016/s0955-0674(99)80023-4. [DOI] [PubMed] [Google Scholar]
- 44.Prenzel N., Zwick E., Daub H., Leserer M., Abraham R., Wallasch C., Ullrich A. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- 45.Barron T.I., Sharp L., Visvanathan K. Beta-adrenergic blocking drugs in breast cancer: a perspective review. Ther. Adv. Med. Oncol. 2012;4:113–125. doi: 10.1177/1758834012439738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caragher S.P., Hall R.R., Ahsan R., Ahmed A.U. Monoamines in glioblastoma: complex biology with therapeutic potential. Neuro-Oncol. 2018;20:1014–1025. doi: 10.1093/neuonc/nox210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dolma S., Selvadurai H.J., Lan X., Lee L., Kushida M., Voisin V., Whetstone H., So M., Aviv T., Park N., Zhu X., Xu C., Head R., Rowland K.J., Bernstein M., Clarke I.D., Bader G., Harrington L., Brumell J.H., Tyers M., et al. Inhibition of dopamine receptor D4 impedes autophagic flux, proliferation, and survival of glioblastoma stem cells. Cancer Cell. 2016;29:859–873. doi: 10.1016/j.ccell.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., Hanahan D. Hijacking the neuronal NMDAR signaling circuit to promote tumor growth and invasion. Cell. 2013;153:86–100. doi: 10.1016/j.cell.2013.02.051. [DOI] [PubMed] [Google Scholar]
- 49.Magnon C., Hall S.J., Lin J., Xue X., Gerber L., Freedland S.J., Frenette P.S. Autonomic nerve development contributes to prostate cancer progression. Science. 2013;341 doi: 10.1126/science.1236361. [DOI] [PubMed] [Google Scholar]
- 50.Beaulieu J.M., Gainetdinov R.R. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol. Rev. 2011;63:182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- 51.Chakroborty D., Sarkar C., Yu H., Wang J., Liu Z., Dasgupta P.S., Basu S. Dopamine stabilizes tumor blood vessels by up-regulating angiopoietin 1 expression in pericytes and Kruppel-like factor-2 expression in tumor endothelial cells. Proc. Nat. Acad. Sci. U.S.A. 2011;108:20730–20735. doi: 10.1073/pnas.1108696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shao W., Zhang S.Z., Tang M., Zhang X.H., Zhou Z., Yin Y.Q., Zhou Q.B., Huang Y.Y., Liu Y.J., Wawrousek E., Chen T., Li S.B., Xu M., Zhou J.N., Hu G., Zhou J.W. Suppression of neuroinflammation by astrocytic dopamine D2 receptors via alphaB-crystallin. Nature. 2013;494:90–94. doi: 10.1038/nature11748. [DOI] [PubMed] [Google Scholar]
- 53.He Y., Li J., Koga T., Ma J., Dhawan S., Suzuki Y., Furnari F., Prabhu V.V., Allen J.E., Chen C.C. Epidermal Growth Factor Receptor (EGFR) as a molecular determinant of glioblastoma response to dopamine receptor 2 (DRD2) inhibitors. Neuro-Oncol. 2020 doi: 10.1093/neuonc/noaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein M.N., Bertino J.R., Kaufman H.L., Mayer T., Moss R., Silk A., Chan N., Malhotra J., Rodriguez L., Aisner J., Aiken R.D., Haffty B.G., DiPaola R.S., Saunders T., Zloza A., Damare S., Beckett Y., Yu B., Najmi S., Gabel C., et al. First-in-human clinical trial of oral ONC201 in patients with refractory solid tumors. Clin. Cancer Res. 2017;23:4163–4169. doi: 10.1158/1078-0432.CCR-16-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S., Che T., Levit A., Shoichet B.K., Wacker D., Roth B.L. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature. 2018;555:269–273. doi: 10.1038/nature25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S., Wacker D., Levit A., Che T., Betz R.M., McCorvy J.D., Venkatakrishnan A.J., Huang X.P., Dror R.O., Shoichet B.K., Roth B.L. D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science. 2017;358:381–386. doi: 10.1126/science.aan5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowman R.L., Wang Q., Carro A., Verhaak R.G., Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro. Oncol. 2017;19:139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., Beroukhim R., Bernard B., Wu C.J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S.A., Ciriello G., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cancer Genome Atlas Research N., Brat D.J., Verhaak R.G., Aldape K.D., Yung W.K., Salama S.R., Cooper L.A., Rheinbay E., Miller C.R., Vitucci M., Morozova O., Robertson A.G., Noushmehr H., Laird P.W., Cherniack A.D., Akbani R., Huse J.T., Ciriello G., Poisson L.M., Barnholtz-Sloan J.S., et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., Alexe G., Lawrence M., O'Kelly M., Tamayo P., Weir B.A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H.S., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.