Abstract
Most plant-sap feeding insects have obligate relationships with maternally transmitted bacteria. Aphids require their nutritional endosymbiont, Buchnera aphidicola, for the production of essential amino acids. Such endosymbionts are harbored inside of specialized insect cells called bacteriocytes. Here, we use comparative transcriptomics of bacteriocytes between two recently diverged aphid species, Myzus persicae and Acyrthosiphon pisum, to identify key genes that are important for the maintenance of their nutritional mutualism. The majority of genes with conserved expression profiles in M. persicae and A. pisum are for orthologs previously identified in A. pisum to be important for the symbiosis. However, asparaginase which produces aspartate from asparagine was significantly up-regulated only in A. pisum bacteriocytes, potentially because Buchnera of M. persicae encodes its own asparaginase enzyme unlike Buchnera of A. pisum resulting in Buchnera of A. pisum to be dependent on its aphid host for aspartate. One-to-one orthologs that explained the most amount of variation for bacteriocyte specific mRNA expression for both species includes a collaborative gene for methionine biosynthesis, multiple transporters, a horizontally transmitted gene, and secreted proteins. Finally, we highlight species-specific gene clusters which may contribute to host adaptations and/or accommodations in gene regulation to changes in the symbiont or the symbiosis.
Subject terms: Evolution, Genetics
Introduction
The establishment and maintenance of a mutualistic relationship between a eukaryotic host and a microbe involves a series of regulatory changes for both the host and microbe1. These regulatory changes facilitate the integration of both the host’s and the microbe’s physiologies for the successful exchange of symbiotic goods. One of the best studied models of insect-endosymbiont mutualistic interactions is the pea aphid, Acyrthosiphon pisum, and its obligate endosymbiont bacterium, Buchnera aphidicola. This integrated host-symbiont metabolism has been well characterized biochemically and genetically, identifying which enzymes are present and/or expressed by the aphid and its symbiont, Buchnera2–6. Within specialized aphid cells that harbor Buchnera, called bacteriocytes, the aphid’s metabolism is integrated with Buchnera for the production of essential amino acids5,6. Aphid host genes that have been identified to play a role in the regulation and maintenance of this mutualistic relationship involve transporters, a cycle (the GS/GOGAT pathway) that incorporates waste ammonia into amino acids through glutamate, and genes that complement Buchnera’s essential amino acid pathways5–7. Nevertheless, what regulatory mechanisms are conserved and lineage specific between different aphid species is still unclear.
Similar to A. pisum, the green peach aphid, Myzus persicae, belongs to the Macrosiphini tribe within the family Aphididae: Aphidinae8 and is estimated to have diverged from A. pisum ~ 22 million years ago9. While both aphid species are pests and have similar lifecycles their host plant range varies dramatically10. For example, A. pisum is acknowledged as a specialist of Fabaceae host plants, where sympatric aphid populations are divided into a number of different biotypes that specialize on specific Fabaceae species11. In contrast, M. persicae differs from A. pisum in that it is a true generalist herbivore and can feed on 40 different plant families including many economically and agriculturally important crop species12. Furthermore, rapid transcriptional plasticity of multigene families has allowed M. persicae clones to colonize up to 100 species of host plants without genetic specialization10. Given this transcriptional plasticity in response to diverse host plant diets, it is of interest to compare the regulation of bacteriocytes between a closely related aphid specialist and generalist, A. pisum and M. persicae, respectively.
To further understand how aphid gene regulation has evolved to support its symbiotic relationship with Buchnera in bacteriocytes we conducted an interspecies comparative transcriptomics approach between M. persicae and A. pisum. Here we identify conserved and lineage specific mechanisms of aphid host regulation between M. persicae and A. pisum in bacteriocytes when both aphid species develop in a common environment and feed on the same host plant species, Vicia fava, in which both species clones here display high fitness on13–15.
Results
Differential gene expression analysis of symbiosis genes for each aphid species
Total high-quality paired-reads that mapped to the M. persicae genome for bacteriocyte and body samples were an average of ~ 24,022,544 and 37,852,850 reads, respectively (Supplemental Table 1). Total high-quality paired-reads that mapped to the A. pisum genome for bacteriocyte and body samples were an average of ~ 34,163,080 and 34,083,851 reads, respectively (Supplemental Table 1). A total of 5097 genes were significantly differentially expressed (i.e. FDR adjusted p-values were ≤ 0.05 with 1.5 fold change (FC)) in M. persicae bacteriocytes compared to body tissues (Supplemental Tables 2 and 3), where a total of 1986 and 3111 genes were significantly up-regulated and down-regulated, respectively (Supplemental Table 3). For A. pisum, a total of 7325 genes were significantly differentially expressed in A. pisum bacteriocytes compared to body samples (Supplemental Tables 4 and 5), where a total of 3205 and 4120 genes were significantly up-regulated and down-regulated, respectively (Supplemental Table 5).
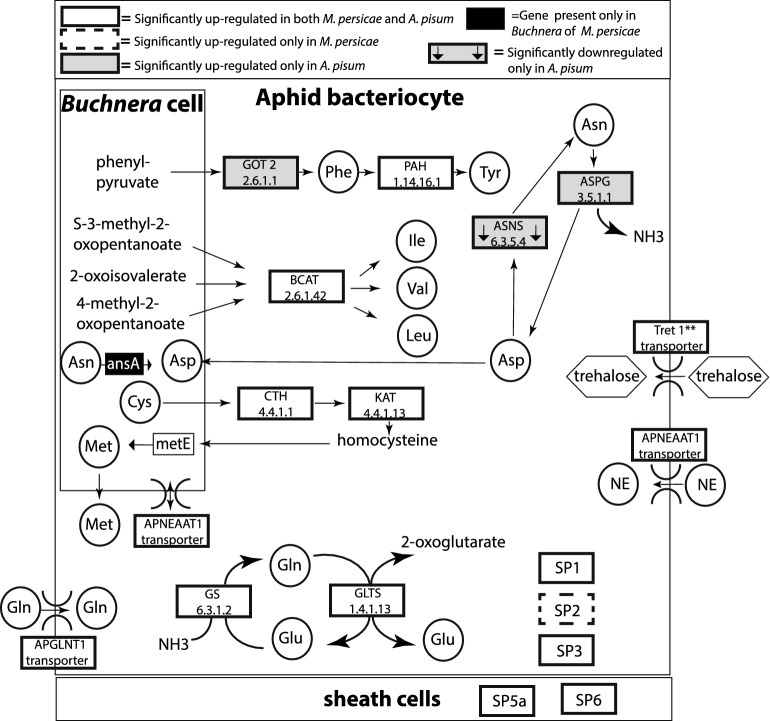
The expression of aphid genes in bacteriocytes that were previously identified in A. pisum to be collaborative in the biosynthesis of Buchnera’s essential amino acids5,6 were examined here for both aphid species (Table 1, Fig. 1). When comparing both M. persicae and A. pisum, six orthologs displayed the same expression pattern where five orthologs were significantly up-regulated in bacteriocytes compared to body tissues (Branched chain amino acid transaminase (BCAT), Phenylalanine 4-monooxygenase (PAH), l-cysteine-S-conjugate thiol-lyase, Cystathionine gamma-lyase (CTH), and Homocysteine S-methyltransferase) and one ortholog was significantly down-regulated in bacteriocytes compared to body tissues for both species (Aspartate transaminase, GOT1) (Table 1, Fig. 1). Species-specific expression patterns for collaborative genes in M. persicae bacteriocytes compared to body samples include the up-regulation of Threonine ammonia-lyase, whereas this ortholog was not significantly up-regulated in A. pisum (Table 1, Fig. 1). For A. pisum species-specific expression patterns for bacteriocytes compared to body samples include the up-regulation of Aspartate transaminase (GOT2), whereas this ortholog was not significantly up-regulated in M. persicae (~ 1.44 FC) (Table 1, Fig. 1).
Table 1.
Expression of collaborative essential amino acid aphid orthologs in bacteriocytes compared to body tissues for two different aphid species. Each row contains a shared ortholog between M. persicae and A. pisum except for N/A which indicates no ortholog was identified between species
| E.C. number | Gene name1 | Pathway | Myzus persicae | Acyrthosiphon pisum | ||||
|---|---|---|---|---|---|---|---|---|
| Gene ID2 | FDR | LogFC | Gene ID2 | FDR | LogFC | |||
| 4.3.1.19 | Threonine ammonia-lyase | Ile | g3539 | 0.004 | 1.86 | 100165866 | 0.881 | − 0.07 |
| 2.6.1.42 | Branched chain amino acid transaminase, BCAT | Ile, Val, Leu | g15579 | 0.000 | 2.20 | 100167587 | 0.000 | 2.96 |
| 2.6.1.1 | Aspartate transaminase, GOT2 | Phe, Tyr | g12035 | 0.072 | 0.53 | 100144899 | 0.000 | 3.21 |
| 2.6.1.1 | Aspartate transaminase, GOT1 | Phe, Tyr | g15167 | 0.001 | − 2.05 | 100163139 | 0.000 | − 3.56 |
| 2.6.1.1 | Aspartate transaminase, GOT1 | Phe, Tyr | N/A | N/A | N/A | 100161812 | nd | nd |
| 2.6.1.1 | Aspartate transaminase, GOT1 | Phe, Tyr | g15165 | nd | nd | N/A | N/A | N/A |
| 2.6.1.1 | Aspartate transaminase, GOT1 | Phe, Tyr | g14986 | 0.282 | 0.39 | 100165255 | 0.399 | 0.31 |
| 1.14.16.1 | Phenylalanine 4-monooxygenase, PAH | Tyr | g26128 | 0.000 | 4.85 | 100166971 | 0.000 | 4.85 |
| 4.4.1.1 | Cystathionine gamma-lyase; CTH | Met | g10610 | 0.000 | 5.57 | 100159197 | 0.000 | 6.48 |
| 4.4.1.1 | Cystathionine gamma-lyase | Met | g10608 | 0.185 | 0.68 | 100159560 | 0.001 | − 1.16 |
| 4.4.1.13 | L-cysteine-S-conjugate thiol-lyase | Met | g6762 | 0.000 | 2.69 | 100164839 | 0.000 | 3.96 |
| 2.1.1.10 | Homocysteine S-methyltransferase | Met | g3958 | 0.000 | 3.04 | 100168557 | 0.000 | 3.26 |
| 2.1.1.10 | Homocysteine S-medfthyltransferase | Met | g9831 | 0.719 | 0.22 | 100159972 | 0.680 | − 0.11 |
1Bolded Gene names indicate orthologs in both aphid species are significantly differentially expressed between bacteriocytes compared to body tissues where both FDR p-value 0.05 and fold change (FC) 1.5 (i.e. LogFC 0.5849)).
2Bolded Gene IDs indicate that the aphid gene is significantly differentially expressed between bacteriocytes compared to body tissues in that aphid species. nd= not detected.
Figure 1.
Gene expression of aphid genes involved in the integrative metabolism with Buchnera. ** indicates four homologs significantly up-regulated in both M. persicae and A. pisum.
Other important aphid genes that were previously identified to be important for A. pisum’s symbiosis with Buchnera include genes that are involved in the recycling of ammonia, biosynthesis of amino donors or intermediates for Buchnera’s essential amino acid pathways, transporters for amino acids or trehalose, and novel secreted proteins5–7,14,16–18 (Table 2). When comparing both M. persicae and A. pisum 13 orthologs were significantly up-regulated in bacteriocytes compared to body tissues for both species and include enzymes involved in the GS/GOGAT cycle (Glutamate synthase (GLTS) and Glutamate-ammonia ligase (GS2)), Ornithine aminotransferase (OAT), the Amino acid transporters (ApGLNT1) and (ApNEAAT1), four Trehalose transporters (TRET1), and four secreted proteins such as SP1, SP3, SP5a, and SP6. Only two orthologs were significantly down-regulated in bacteriocytes compared to body tissues for both species, secreted protein SP4 and Aspartate transaminase (GOT1) (Tables 1 and 2; Fig. 1). Species-specific expression patterns in M. persicae bacteriocytes compared to body tissues include the up-regulation of only one gene, secreted protein SP2, whereas this ortholog was not significantly differentially expressed in A. pisum (Table 2, Fig. 1). Multiple genes in A. pisum displayed species-specific up-regulation in bacteriocytes compared to body tissues for an additional homolog of Glutamate-ammonia ligase (GS2) in A. pisum, Asparaginase, the Mitochondrial 2-oxoglutarate/malate carrier, and an additional homolog of the Trehalose transporter in A. pisum (Table 2, Fig. 1).
Table 2.
Expression of symbiosis-related aphid orthologs in bacteriocytes compared to body tissues in two different aphid species. Each row contains a shared ortholog between M. persicae and A. pisum except for N/A which indicates no ortholog was identified between species.
| Pathway | Myzus persicae | Acyrthosiphon pisum | ||||||
|---|---|---|---|---|---|---|---|---|
| E.C. number | Gene name1 | Gene ID2 | FDR | LogFC | Gene ID2 | FDR | LogFC | |
| Amino acid enzymes | ||||||||
| 1.4.1.14 | Glutamate synthase (GLTS) | Glu | g22515 | 0.000 | 2.04 | 100158883 | 0.000 | 2.07 |
| 6.3.1.2 | Glutamate-ammonia ligase (GS2) | Glu | g15463 | 0.000 | 1.89 | 100160139 | 0.000 | 1.48 |
| 6.3.1.2 | Glutamate-ammonia ligase | Glu | N/A | N/A | N/A | 100165282 | 0.007 | 4.60 |
| 6.3.1.2 | Glutamate-ammonia ligase | Glu | g8271 | 0.401 | 0.81 | N/A | N/A | N/A |
| 2.6.1.13 | Ornithine aminotransferase (OAT) | Glu | g6537 | 0.029 | -1.35 | N/A | N/A | N/A |
| 2.6.1.13 | Ornithine aminotransferase | Glu | g6576 | 0.000 | 3.21 | 100168809 | 0.000 | 4.00 |
| 3.5.1.1 | Asparaginase | Asn | g14033 | 0.131 | 0.48 | 100158730 | 0.000 | -1.52 |
| 3.5.1.1 | Asparaginase | Asn | g8195 | 0.142 | 0.71 | 100164179 | 0.000 | 5.32 |
| 6.3.5.4 | Asparagine synthetase (ASNS) | Asn | g23256 | 0.142 | 1.05 | 100160265 | 0.022 | -0.86 |
| Transporters | ||||||||
| Amino acid transporter (ApGLNT1) | g19243 | 0.000 | 2.38 | 100159667 | 0.000 | 1.40 | ||
| Amino acid transporter (ApNEAAT1) | g25548 | 0.000 | 1.38 | 100168251 | 0.017 | 0.82 | ||
| Mitochondrial 2-oxoglutarate/malate carrier | g11494 | 0.063 | 0.81 | 100159664 | 0.000 | 3.34 | ||
| Trehalose transporter (TRET1) | g27282 | 0.093 | 0.58 | 100159441 | 0.000 | 1.28 | ||
| Trehalose transporter (TRET1) | g22841 | 0.000 | 3.64 | 100165626 | 0.000 | 5.28 | ||
| Trehalose transporter (TRET1) | g11139 | 0.000 | 5.19 | 100169458 | 0.000 | 7.54 | ||
| Trehalose transporter (TRET1) | g16824 | 0.000 | 4.90 | 100161021 | 0.000 | 7.09 | ||
| Trehalose transporter (TRET1) | g12940 | 0.000 | 3.70 | 100169115 | 0.000 | 3.14 | ||
| Secreted proteins | ||||||||
| SP1 | g23997 | 0.000 | 5.78 | 100167607 | 0.000 | 7.94 | ||
| SP2 | g21195 | 0.042 | 1.53 | 100158873 | 0.19 | -0.54 | ||
| SP3 | g18732 | 0.000 | 5.24 | 100164129 | 0.000 | 7.37 | ||
| SP4 | g14959 | 0.009 | -1.41 | 100169357 | 0.000 | -3.07 | ||
| SP5a | g19669 | 0.031 | 1.06 | 100163734 | 0.000 | 2.47 | ||
| SP6 | g11563 | 0.000 | 3.67 | 100160550 | 0.000 | 3.71 | ||
1Bolded Gene names indicate orthologs in both aphid species are significantly differentially expressed between bacteriocytes compared to body tissues where both FDR p-value 0.05 and fold change (FC) (i.e. LogFC 0.5849).
2Bolded Gene IDs indicate that the aphid gene is significantly differentially expressed between bacteriocytes compared to body tissues in that aphid species. nd= not detected.
The differential expression of lineage specific gene clusters was also examined for both M. persicae and A. pisum (Supplemental Table 6). Here we define “A. pisum lineage specific genes” as genes that are not present in M. persicae (non- M. persicae genes) and likewise “M. persicae lineage specific genes” are genes that are not present in A. pisum (non- A. pisum genes). Moreover, we define “conserved genes” here as homologs that are present in both A. pisum and M. persicae. Out of 1053 lineage specific gene clusters in M. persicae (consisting of 4622 genes) a total of 376 genes were significantly differentially expressed between bacteriocytes compared to body tissues. Approximately, 42 percent of these latter genes were significantly up-regulated in bacteriocytes compared to body tissues and include genes primarily associated with gene regulation such as DNA and RNA binding proteins, and uncharacterized genes (Supplemental Table 7). For A. pisum, 580 lineage specific gene clusters were identified (consisting of 1689 genes), and a total of 289 genes from these clusters were significantly differentially expressed between bacteriocytes compared to body tissues (Supplemental Table 8). Approximately, 22% of these latter genes were significantly up-regulated in bacteriocytes compared to body tissues and include genes primarily associated with Balbiani ring genes, death-associated inhibitor of apoptosis 1-like genes, UDP-glucuronosyltransferase genes, lysosomal and kelch associated genes, heat shock genes, ribosomal proteins, and hypothetical genes. (Supplemental Table 8).
KEGG pathway analysis
Gene Set Enrichment Analysis (GSEA) identified seven and two shared pathways in both M. persicae and A. pisum that were significantly positively and negatively enriched, respectively, in bacteriocytes compared to body tissues (Table 3). The positively enriched pathways display hallmarks of metabolically active cells as these pathways are involved in DNA replication, RNA processing, and protein turnover (Table 3). In contrast, negatively enriched pathways that were shared between aphid species were primarily involved in cell signaling and adhesion (Table 3). Species-specific patterns in the enrichment of KEGG pathways were noted where seven and 13 KEGG pathways were significantly positively enriched in only M. persicae or A. pisum bacteriocytes compared to body tissues, respectively, and one and seven KEGG pathways were significantly negatively enriched in only M. persicae or A. pisum bacteriocytes compared to body tissues, respectively (Supplemental Table 9).
Table 3.
GSEA analyses of both aphid species (Mp= M. persicae and Ap= A. pisum) displaying all KEGG pathways that were significantly enriched in bacteriocytes compared to body for both species.
| KEGG pathway significantly enriched in bacteriocyte | Mp NES1 | Ap NES1 |
|---|---|---|
| Positively enriched | ||
| Ribosome | 1.941 | 2.842 |
| Mismatch repair | 1.922 | 1.861 |
| Glycine, serine, and threonine metabolism | 1.755 | 1.607 |
| DNA replication | 1.738 | 2.151 |
| Cysteine and methionine metabolism | 1.688 | 1.348 |
| Proteasome | 1.649 | 2.060 |
| Nucleotide excision repair | 1.556 | 1.759 |
| Negatively enriched | ||
| Neuroactive ligand-receptor interaction | − 1.880 | − 1.593 |
| Cell adhesion molecules | − 1.700 | − 1.523 |
1NES normalized enrichment score; Significance = the normalized p ≤ 0.05 and FDR q ≤ 0.25.
Inter-species comparison of one-to-one orthologs
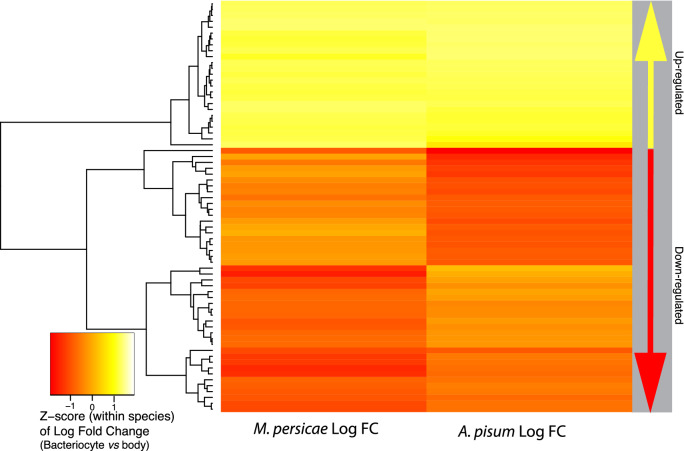
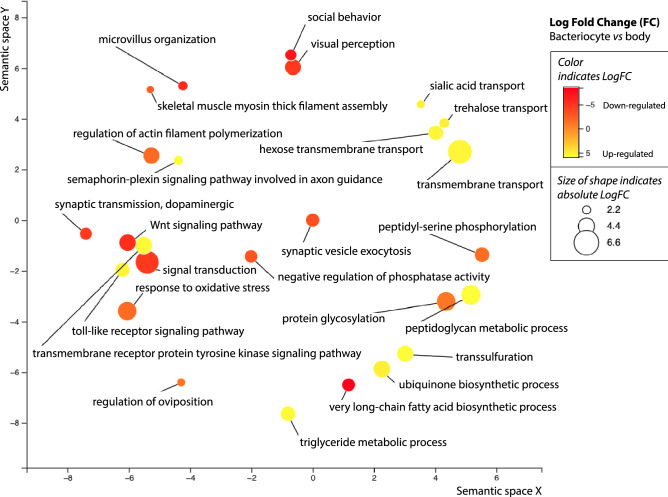
A total of 9465 shared one-to-one orthologs were identified between M. persicae and A. pisum and ~ 23% of these orthologs were significantly expressed in bacteriocytes compared to the body tissue for both aphid species (Fig. 2; Supplemental Table 6). To determine which of these differentially expressed orthologs have the biggest effect in fold-change between bacteriocytes and body for both species similar to Korb et al.19 and Georgiadou et al.20 we conducted PCA where axis 1 and 2 explained ~ 53.3% and 46.7% of the variance in the data, respectively. The top 100 shared orthologs (70 were non-redundant between both axis 1 and 2) with the highest positive and negative correlations with principal components 1 and 2 consist of 25 up-regulated and 45 down-regulated orthologs (Fig. 3, Supplemental Table 10). Symbiosis related genes that were up-regulated in both M. persicae and A. pisum for this subset of orthologs include Cystathionine gamma-lyase (CTH), the secreted proteins SP1 and SP3, the horizontally transferred gene rlpA, and two facilitated Trehalose transporter TRET1 genes (Table 4). Other annotated genes up-regulated in both include multiple acetyl-coenzyme A transporters along with other transporter genes, receptors such as draper and plexins, and the E3 ubiquitin-protein ligase MARCH2 (Table 4). Based on dendrogram clustering of standardized logFC in bacteriocytes compared to body tissues for these 70 paired-orthologs in M. persicae and A. pisum, up-regulated orthologs appear to have very similar logFC profiles (Fig. 3, Supplemental Fig. 1). In contrast, down-regulated orthologs appear to have two distinct logFC profiles with a subset of orthologs having either greater or lower logFC magnitudes in M. persicae or A. pisum (Fig. 3, Supplemental Fig. 1). For example, multiple orthologs that were associated with visual perception were down-regulated at a higher magnitude in M. persicae and orthologs associated with an amino acid transporter, heat shock protein, and peroxidase like protein were down-regulated at a higher magnitude in A. pisum (Supplemental Fig. 1, Table 10). The only GO term significantly enriched for up-regulated orthologs was GO:0055085 for transmembrane transport (p-value = 0.00027) (Fig. 4). Two GO terms were significantly enriched for down-regulated orthologs and include GO:0007601 for visual perception (p-value = 0.00043) and GO:0005524 for ATP binding (p-value = 0.00098) (Fig. 4).
Figure 2.
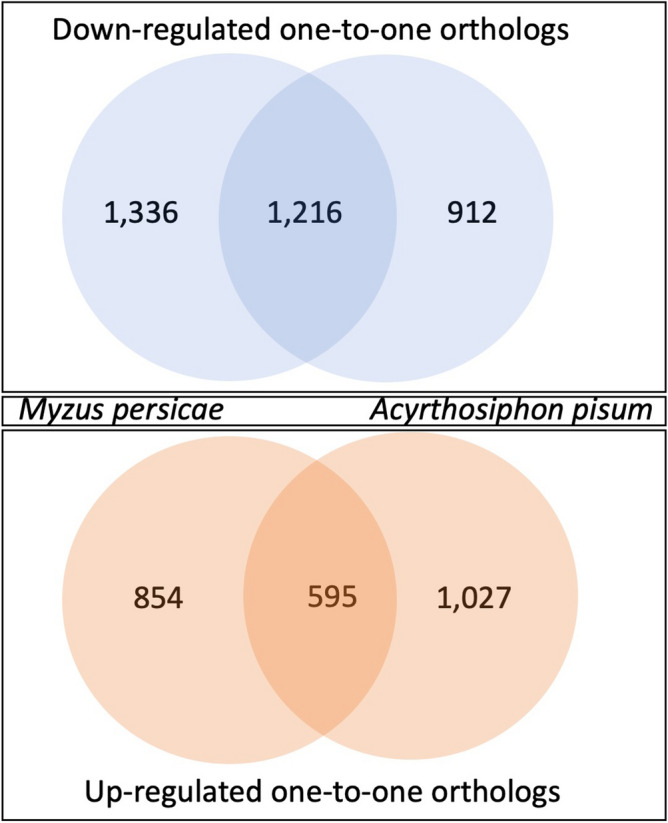
Overview of one-to-one orthologs that have significant differential gene expression in M. persicae and A. pisum for bacteriocytes compared to body tissues, where significance of up- or down-regulation between tissue types is determined for genes with an FDR corrected p-value 0.05 and fold change (FC) 1.5 (i.e. LogFC 0.5849). The shared portion of Venn diagrams indicate that the one-to one ortholog was significantly down-regulated (top panel) or up-regulated (bottom panel) in both aphid species. Un-shared portions of Venn diagrams indicate that the ortholog was only down-regulated (top panel) or up-regulated (bottom panel) in one of the aphid species (left side M. persicae; right side A. pisum).
Figure 3.
Inter-species ortholog comparison of the top 70 one-to-one orthologs with the greatest variance in standardized Log fold-change expression of bacteriocytes compared to body tissues for both M. persicae and A. pisum. “Up-regulated” on the right panel indicates up-regulated in bacteriocytes compared to body tissue and “Down-regulated” indicates down-regulated in bacteriocytes compared to body tissue. Each row represents a shared one-to-one ortholog between M. persicae and A. pisum. See detailed ortholog gene numbers and GO terms in Supplemental Fig. S1.
Table 4.
The 25 aphid ortholog proteins that were identified with PCA as having the greatest magnitude of up-regulation in the bacteriocyte compared to the body for both aphid species.
| Mp gene | Ap gene ID | NCBI protein name | Mp LogFC | Ap LogFC |
|---|---|---|---|---|
| g23997 | 100167607 | SP1 | 5.78 | 7.94 |
| g16265 | 100163669 | RlpA family protein-like precursor | 5.74 | 7.53 |
| g10610 | 100159197 | cystathionine gamma-lyase isoform 1 | 5.57 | 6.48 |
| g11338 | 100160627 | protein draper isoform X1 | 5.55 | 7.06 |
| g25444 | 100568670 | plexin A3 | 5.49 | 7.63 |
| g7735 | 100169049 | uncharacterized protein LOC100169049 isoform X1 | 5.48 | 2.53 |
| g22717 | 100166357 | organic cation transporter protein | 5.40 | 7.55 |
| g7417 | 100575173 | plexin-A3 | 5.40 | 7.60 |
| g18732 | 100164129 | SP3 | 5.25 | 7.37 |
| g20745 | 100168678 | neprilysin-21 isoform X1 | 5.23 | 7.02 |
| g11139 | 100169458 | facilitated trehalose transporter Tret1 | 5.19 | 7.33 |
| g18635 | 100167419 | acetyl-coenzyme A transporter 1 | 5.18 | 7.88 |
| g13201 | 100164813 | major facilitator superfamily domain-containing protein 6 isoform X1 | 5.14 | 6.19 |
| g15906 | 100164131 | acetyl-coenzyme A transporter 1 isoform X1 | 5.12 | 4.95 |
| g11347 | 100161188 | leucine-rich repeat-containing protein 15 | 5.08 | 6.15 |
| g13798 | 100160909 | uncharacterized protein LOC100160909 precursor | 5.01 | 7.05 |
| g24975 | 100162203 | uncharacterized protein LOC100162203 | 4.99 | 6.63 |
| g26344 | 100168152 | sialin | 4.96 | 5.86 |
| g19207 | 100169490 | acetyl-coenzyme A transporter 1 | 4.92 | 7.31 |
| g16824 | 100161021 | facilitated trehalose transporter Tret1 | 4.90 | 7.89 |
| g2999 | 100569384 | E3 ubiquitin-protein ligase MARCH2-like isoform X1 | 4.90 | 6.36 |
| g16263 | 100570509 | uncharacterized protein LOC100570509 | 4.87 | 7.67 |
| g27533 | 100164029 | decaprenyl-diphosphate synthase subunit 2 | 4.83 | 6.80 |
| g16264 | 100570300 | uncharacterized protein LOC100570300 | 4.72 | 8.00 |
| g16266 | 100165005 | uncharacterized protein LOC100165005 precursor | 4.38 | 8.02 |
All proteins in the table were significantly up-regulated in each aphid species (Mp M. persicae and Ap A. pisum) where FDR adjusted p-values were ≤ 0.05 with ≥ 1.5-fold change (FC). Bolded proteins indicate those that are characterized with annotations in NCBI.
Figure 4.
Approximately 54% of the one-to-one orthologs identified in inter-species ortholog analysis have GO term annotations, which are represented as circles in the figure (see Supplemental Table S10 for detail). Multidimensional scaling (MDS) was used in Revigo to reduce the dimensionality of the matrix for these 38 GO terms based on pairwise similarities in biological function. The size of the circle and color indicates the Log fold change of the ortholog (bacteriocyte compared to body) within a unique GO term category.
Discussion
The aphid bacteriome, the organ housing the intracellular symbiont Buchnera and the location of the integrated essential amino acid metabolism, is functionally, structurally, and developmentally conserved among nearly all aphids21–24. Here, we interrogated gene expression profiles of these symbiotic host cells in two aphid species that diverged ~ 22 MYA and detected extensive expression conservation in addition to key species-specific expression changes between A. pisum and M. persicae. The species-specific gene expression changes we detected between both species may contribute to host adaptations and/or accommodations in gene regulation to changes in the symbiont or the symbiosis.
A consequential expression difference we detected between A. pisum and M. persicae bacteriocytes is the significant up-regulation (~ 40X) of Asparaginase (3.5.1.1) only in A. pisum bacteriocytes (Table 2; Fig. 1). This enzyme is critical for the hosts’ integrated metabolism with Buchnera as Asparaginase converts asparagine, which is one of the most abundant non-essential amino acids in the aphid’s sap-diet13, into aspartate and ammonia. Buchnera the nutritional endosymbiont of A. pisum is located inside of bacteriocytes and requires aspartate from the aphid host, because it cannot make it de novo. Further Buchnera needs aspartate for the biosynthesis of the essential amino acids lysine and threonine, which the aphid and Buchnera require for survival5. We predict that this expression difference is because the Buchnera endosymbionts of M. persicae unlike Buchnera of A. pisum encode their own asparaginase gene (ansA); ansA is the only additional essential amino acid biosynthetic gene found in Buchnera M. persicae that is absent in Buchnera A. pisum25 (Fig. 1). In consequence, Buchnera of M. persicae is not entirely dependent on its aphid host for the biosynthesis of aspartate, whereas Buchnera of A. pisum is dependent on its aphid host for access to this important amino acid. It will be of interest of future studies to determine if A. pisum can regulate Buchnera’s production of lysine and threonine by fine-tuning aspartate availability through the aphid encoded Asparaginase and/or the aphid Asparagine synthetase (ASNS) enzyme that converts aspartate back into asparagine. For example, ASNS is significantly down-regulated only in A. pisum indicating that A. pisum bacteriocytes are in high demand for aspartate biosynthesis during this lifestage and environmental condition (Fig. 1).
The overwhelming majority of genes with conserved expression profiles in M. persicae and A. pisum are for orthologs previously identified in A. pisum to be important for the nutritional symbiosis (Tables 1 and 2). For example, in both M. persicae and A. pisum, similar bacteriocyte expression profiles were found for five aphid orthologs BCAT, PAH, CTH, l-cysteine-S-conjugate thiol-lyase, and Homocysteine S-methyltransferase) which were up-regulated in bacteriocytes and complement five of Buchnera’s essential amino acid pathways (isoleucine, valine, leucine, tyrosine, and methionine) (Table 1, Fig. 1). Other notable genes that were previously recognized as important in the A. pisum- Buchnera symbiosis that were up-regulated in both A. pisum and M. persicae bacteriocytes here include ornithine aminotransferase (OAT) providing a potential intermediate for Buchnera’s arginine biosynthesis5, the amino acid transporters ApGLNT1 and ApNEAAT1 that are important in amino acid transport for the symbiosis7,17, Trehalose transporters (TRET1), which may be crucial in providing Buchnera with glucose after trehalose is transported into the bacteriocytes and subsequently converted into glucose14,18,26, and the GS/GOGAT cycle, which is hypothesized to up-grade ammonia into amino donors to help fuel the nitrogen limited nutritional symbiosis5. However, one aphid ortholog (GOT1), which collaborates with Buchnera in the biosynthesis of the amino acid phenylalanine, was only significantly up-regulated in A. pisum bacteriocytes. GOT up-regulation in bacteriocytes convergently occurs in other independently evolved Hemipteran symbioses including the mealybug27, psyllid28, and whitefly29. Therefore, it is possible that in another environmental/host plant diet condition the up-regulation of GOT1 in M. persicae would be less variable in expression and would be significantly up-regulated in bacteriocytes. Alternatively, an uncharacterized enzyme in M. persicae or Buchnera of M. persicae may be responsible for this enzymatic step.
We further identified that bacteriocytes of both A. pisum and M. persicae are metabolically highly active, because mRNA, protein, and DNA replication KEGG pathways were significantly positively enriched in aphid bacteriocytes (Table 3). Our positive enrichment of DNA replication genes in bacteriocytes support recent results from Nozaki and Shigenobu who found a dramatic increase in the number of chromosome sets per bacteriocyte cell (i.e., ploidy) as A. pisum ages after live birth into the reproductive viviparous adult stage30. Ploidy of insect chromosomes is a general feature of bacteriocytes in A. pisum30, M. persicae31, and other diverse insect taxa (reviewed in30). Here we also found that the Glycine, Serine, and Threonine Metabolism and the Cysteine and Methionine Metabolism were positively enriched in bacteriocytes which potentially indicates a high demand for not only the essential amino acids methionine and cysteine but also for methyl group donors for DNA methylation during the DNA replication process. In support of this hypothesis, Pers and Hansen18 found that DNA methylation occurs in bacteriocytes throughout aphid development (1st instar-reproductive adult stage) where A. pisum bacteriocytes are more heterogenous in methylation profiles for the 1st and 2nd instars compared to subsequent lifestages suggesting that there is a high demand for methyl donors via both de novo methylation and maintenance methylation early and in subsequent lifestages, respectively, throughout nymphal and young adult development.
Unique gene expression profile differences between M. persicae and A. pisum bacteriocytes were primarily highlighted by the expression of species-specific gene clusters between M. persicae and A. pisum. These profile differences indicate that there is a species-specific difference in the chromosome metabolism and the apoptotic pathway. For example, NCBI annotated Balbiani ring paralogs, are lineage specific in A. pisum, and consist of 54 paralogous gene copies that are composed of ankyrin repeat domains. Six of these paralogs are significantly enriched in A. pisum bacteriocytes compared to body tissues (Supplemental Table 8). Interestingly, Kwak et al.32 determined that 52 of the copies represented recent, rapid expansions of this gene cluster in A. pisum and homologs of this gene cluster were not found in M. persicae or any of the other Hemipteran species examined in that study, except for Aphis glycines (Supplemental Tables 6 and 8). Based on functional studies in Drosophila, Balbiani ring genes are involved in polytene chromosomes, which are commonly found in fly salivary glands where homologous chromosome copies in the nucleus are aligned and attached to each other, and chromatin of active genes unfold in loops forming puffs33. At this time, it is unclear if polytene chromosomes occur in A. pisum or other aphids. Moreover, it is unclear if these Balbiani ring genes have similar functions in polytene chromosome formation, or alternatively if these A. pisum genes have evolved new functions. This mode of DNA replication, however, can result in advantages for specialized cell types like bacteriocytes, which have a high demand for gene expression. For example, in polytene chromosomes gene amplification can occur for targeted genes because re-replication only occurs for sub-sets of the DNA and under-replication can occur for loci that are targeted for down-regulation34. The trade-off of polytene chromosomes however is an increase in DNA double-strand breaks and errors that can result in the up-regulation of the apoptotic machinery34. Interestingly, a significant positive enrichment for the homologous recombination pathway, a DNA double-strand break repair pathway was only identified in A. pisum bacteriocytes and not M. persicae (Supplemental Table 9).
Regarding apoptotic machinery, the apoptotic pathway was recently annotated and characterized in A. pisum35. Here we found that one of the four effector caspases (Ap-ICE-2) were significantly up-regulated in bacteriocytes compared to body tissues in A. pisum, however none of the other caspases including the initiator caspases or the adaptor protein were significantly differentially expressed at this lifestage and tissue comparison for A. pisum (Supplemental Table 5). Lopes et al.35 predict that the duplicated copies of caspases in A. pisum may have redundant functions, or alternatively may have evolved new functions that are related or unrelated to apoptosis. None of the latter caspase orthologs in M. persicae were differentially expressed, however the adapter protein (g10569) was significantly down-regulated in M. persicae bacteriocytes (Supplemental Tables 3 and 6). Five A. pisum bacteriocyte-specific inhibitors of apoptosis (IAPs) were also characterized in Lopes et al.35 to have bacteriocyte-specific expression in the adult stage of A. pisum and displayed an antiapoptotic role in vivo in a Drosophila model. Here in A. pisum we found one of the five latter genes significantly up-regulated in 4th instar bacteriocytes compared to body tissue (Ap-Deterin-1) (Supplemental Table 5). The one-to-one ortholog of Ap-Deterin-1 in M. persicae (g14656) was also up-regulated significantly here (Supplemental Tables 3 and 6). The up-regulation of this latter IAP in both A. pisum and M. persicae may suggest a conserved role in inhibiting apoptosis in nymphal (4th instar) bacteriocytes and not just the adult stage of aphids. When examining lineage specific apoptosis-related A. pisum genes, which are not encoded in M. persicae, we found that six previously characterized IAPs that belong to paralogy group C (Ap-IAP-C9; Ap-IAP-C14; Ap-IAP-C15; Ap-IAP-C17; Ap-IAP-C18; Ap-IAP-C21) Lopes et al.35 and one “putative inhibitor of apoptosis” gene (115034293), based on BLAST, are significantly up-regulated in bacteriocytes compared to body tissues in A. pisum (Supplemental Table 8). At this time the role of lineage specific IAP genes from group C in A. pisum is unknown.
Novel secreted protein transcripts that are unique to aphids, such as SP1, SP3, SP5a, and SP616, were significantly up-regulated in both aphid species (Table 2). The functional role of these orphan genes with N-terminal signal sequences is not fully understood however they may enter the secretory pathway, and SP1 and SP3 are expressed in bacteriocytes and SP5a and SP6 are expressed in the sheath cells surrounding the bacteriocytes16. Interestingly, our comparative transcriptomics analysis with PCA identified both SP1 and SP3 within the top 70 one-to-one orthologs that explain the greatest amount of variation in fold change for all one-to-orthologs that were significantly differentially expressed in bacteriocytes (N = 2188) for both M. persicae and A. pisum (Table 4). The gene SP1 in M. persicae was found to be regulated in bacteriocytes via miR-92a using a dual luciferase assay36, indicating that miRNA::mRNA interactions may be very important for the regulation of aphid genes that are involved in the aphid-Buchnera mutualism. Other symbiosis genes that fell into this subset of the top 70 one-to-one orthologs included the collaborative gene, CTH, which is involved in methionine biosynthesis with Buchnera (Fig. 1), two trehalose transporter orthologs, and the horizontally transferred gene rlpA (37; Table 4). Nakabachi et al.37 demonstrated that RplA is synthesized as a protein in the maternal bacteriocyte and is transported into Buchnera cells. This horizontally transmitted gene is hypothesized to produce lytic translgycosylase for Buchnera cell wall remodeling and is largely omnipresent in Aphidoidea unlike other horizontally transferred genes that were lost in certain aphid lineages38.
In summary, when controlling for environmental factors, including the same host plant species, both conserved and lineage specific patterns of gene expression were identified in bacteriocytes from M. persicae (clone Fava) and A. pisum (clone LSR1). Since both aphid species display high fitness on this host plant species (V. fava)13,15, and display similar developmental times, at least for the aphid clones used in previous studies and here15,39 we do not predict that bacteriocyte gene expression profiles are due to a stress response. Collaborative genes involved in the nutritional symbiosis were largely conserved between M. persicae and A. pisum however the differential expression of Asparaginase between M. persicae and A. pisum may signify differences in aspartate production in both the aphid bacteriocyte and in Buchnera. Moreover, pathway and lineage specific gene cluster differences between A. pisum and M. persicae in this study may signify species-specific adaptations, including the putative use of polytene chromosomes in A. pisum for the strategic amplification and down-regulation of genes for symbiosis, whereas species-specific genes up-regulated in M. persicae bacteriocytes were primarily composed of proteins containing DNA and RNA regulatory domains. It will be of interest for future studies to determine how plastic bacteriocyte gene expression responses are between aphid species using additional aphid clones in ecology related studies when they feed on different host plant diets, and are exposed to different environmental conditions.
Materials and methods
Insect rearing and RNA sequencing
To control for environmental variation both species, M. persicae and A. pisum, were allowed to develop to 4th instar and fed on the same host plant species, Vicia faba, at 20 °C with a 16:8-h light-dark cycle with ~ 30–40% humidity in Intellus Ultra controller Percival incubators (Percival Scientific, Inc., Perry, IA, USA). Transcriptomic data produced previously for A. pisum using these latter conditions in Kim et al.14 was used for this study. Both studies however were performed around the same time period and collected by the same individual for DK’s Ph.D thesis. For M. persicae, a genetically homogenous strain of M. persicae (Sulzer) from Medina-Ortega and Walker40 was divided into three sub-lines for this study and has been reared on V. faba stably with high vigor for > 5 years in culture (clone Fava). Briefly, similar to A. pisum (clone LSR1) in Kim et al.14 M. persicae was divided into three sub-lines and was reared on V. fava (23 ± 2 days after germination (~ 5 whorls)) for over 20 generations at 20 °C with a 16:8-h light-dark cycle in Intellus Ultra controller Percival incubators (Percival Scientific, Inc., Perry, IA, USA). Approximately 200 aphids that were at 4th instar were dissected from each sub-line to co-collect both bacteriocytes with Buchnera (N = 3 sublines/biological replicates per species) and other body cells without Buchnera (N = 3 sublines/biological replicates per species) using the same methods detailed in Kim et al.14. Developmental times for A. pisum and M. persicae on V. fava at the detailed conditions stated above were similar to Pers and Hansen39 for A. pisum and Hong et al.15 for M. persicae, where both aphid species were at 4th instar on day six. Briefly, pooled total RNA of bacteriocytes and body cells were extracted using the Quick-RNA Microprep kit (Zymo Research, Irvine, CA, USA). Extracted RNA samples were treated with DNase I and purified with the RNA Clean & Concentrator kit (Zymo Research). Strand-specific RNA-seq libraries were generated with poly-A enrichment and sequenced on 1 lane of an Illumina HiSeq 4000 (Illumina, San Diego, CA) with paired-end 150bp reads (see Supplemental Methods for more detail). Reads for all RNA-Seq samples were submitted to the Sequence Read Archive of the National Center for Biotechnology Information (NCBI) under BioProject ID PRJNA866154.
The following RNAseq pipeline was conducted for both A. pisum14 and M. persicae raw RNAseq reads and followed the code detailed in “Dataset_S9_RNAseq_Code” from Pers and Hansen18 (see Supplemental Methods for detail on entire pipeline). First, sequenced RNA reads were quality-checked with FASTQC v.0.11.941 and reads were trimmed using Trimmomatic v.0.3942. The trimmed reads were aligned using HISAT2 v.2.2.143 against the chromosomal assemblies of clone AL4f for A. pisum44 and M. persicae clone O v29. The mapped reads for each gene were quantified as raw read counts using StringTie v.2.2.145, visualized using Principal Components Analyses (PCA) (Supplemental Fig. 2) in R/4.2.046, and differential expression of transcripts between bacteriocytes and body cells was determined in R/4.2.046 using edgeR v.3.28.0 with the exact test47. Similar to Kim et al.14 statistical significance for differentially expressed genes was determined if FDR adjusted p-values were ≤ 0.05 with 1.5-fold change (FC), indicated as “logFC” = log2 fold change between the groups. For KEGG pathway analysis of aphid genes9,44, KO numbers were retrieved using KofamKoala48 and Gene Set Enrichment Analysis (GSEA)49 was used to determine which KEGG pathways were positively or negatively enriched at the normalized p ≤ 0.05 and FDR q ≤ 0.25., as described in Pers and Hansen18.
Identifying orthologous clusters
Orthologous clusters of proteins shared between A. pisum44 and M. persicae9 were identified using default settings in OrthoVenn250 using the longest protein isoforms (see Supplemental Methods for detail). One-to-one orthologs were examined between A. pisum and M. persicae for PCA (see below), and the enrichment of GO terms for orthologs was determined using OrthoVenn2 see Supplemental Methods for detail50. All unshared protein clusters that were unique to A. pisum or M. persicae were examined further for differential gene expression analyses (see above). Proteins were annotated using NCBI annotations for A. pisum clone AL4f and by screening for matches to the NCBI nr database (downloaded on 06/2022) for M. persicae clone O v2 using DIAMOND v.2.0.1351 with an e value cutoff of 10e-10.
Comparative transcriptomics using principal component analysis
To compare patterns of gene expression between species without species-specific variation, specifically focusing on genes associated with symbiosis that are differentially expressed in the bacteriocyte compared to body tissues, we conducted interspecies comparative transcriptomics analysis using PCA following the pipeline of Georgiadou et al.20 (see Supplemental Methods for detailed pipeline on interspecies transcriptomics analyses). Pcord (version 4.25)52 was used for PCA analysis. Similar to Korb et al.19 we obtained the top 50 genes that contributed the most to each principal component axis (axes 1 and 2) by identifying orthologs with the top negative (e.g. 25 genes) and positive (e.g. 25 genes) correlations from the principal components output loading matrix. We then examined the annotations for each ortholog protein (see above) from these top 100 orthologs. To display a heatmap of the logFC for one-to-one orthologs SeqCode’s53 HeatMapper was used and Revigo54 was used to visualize and summarize ortholog GO terms (see Supplemental Methods for more detail).
Supplementary Information
Acknowledgements
We thank Greg Walker for providing the Myzus persicae aphid colony. We also thank DK’s Ph.D thesis committee members (Quinn McFrederick, Kerry Mauck, and Richard Stouthamer) for previous comments on an earlier draft. The sequencing was carried out at the UC Davis Genome Center DNA Technologies and Expression Analysis Core, supported by NIH Shared Instrumentation Grant 1S10OD010786-01. Funding for this research was provided by AH’s startup funds at University of California, Riverside.
Author contributions
J.A. conducted the data and bioinformatic analyses and helped write the manuscript. D.K. helped set up experiments and helped write the manuscript. A.H. helped to design the study, conducted the data and bioinformatic analyses, and helped write the manuscript. All authors contributed to the article and approved the submitted version.
Data availability
Raw RNA sequencing data are available at NCBI under SRA BioProject ID: PRJNA866154 under SAMN30154147, SAMN30154148, SAMN30154149, SAMN30154150, SAMN30154151, SAMN30154152.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-32291-3.
References
- 1.Herrera P, et al. Molecular causes of an evolutionary shift along the parasitism–mutualism continuum in a bacterial symbiont. Proc. Natl. Acad. Sci. USA. 2020;117:21658–21666. doi: 10.1073/pnas.2005536117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 3.Nakabachi A, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium Buchnera. Proc. Natl. Acad. Sci. USA. 2005;102:5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.IAGC Genome Sequence of the Pea Aphid Acyrthosiphon pisum. PLOS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen AK, Moran NA. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. USA. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poliakov A, et al. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol. Cell. Proteomics MCP. 2011;10:M110.007039. doi: 10.1074/mcp.M110.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Price DRG, et al. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc. Natl. Acad. Sci. USA. 2014;111:320–325. doi: 10.1073/pnas.1306068111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nováková E, et al. Reconstructing the phylogeny of aphids (Hemiptera: Aphididae) using DNA of the obligate symbiont Buchnera aphidicola. Mol. Phylogenet. Evol. 2013;68:42–54. doi: 10.1016/j.ympev.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Mathers TC, et al. Chromosome-scale genome assemblies of aphids reveal extensively rearranged autosomes and long-term conservation of the X chromosome. Mol. Biol. Evol. 2021;38:856–875. doi: 10.1093/molbev/msaa246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathers TC, et al. Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome Biol. 2017;18:27. doi: 10.1186/s13059-016-1145-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frantz A, Plantegenest M, Mieuzet L, Simon J-C. Ecological specialization correlates with genotypic differentiation in sympatric host-populations of the pea aphid. J. Evol. Biol. 2006;19:392–401. doi: 10.1111/j.1420-9101.2005.01025.x. [DOI] [PubMed] [Google Scholar]
- 12.Blackman & Eastop. Aphids on the World’s Crops: An Identification and Information Guide, 2nd Edition. Wiley https://www.wiley.com/en-us/Aphids+on+the+World%27s+Crops%3A+An+Identification+and+Information+Guide%2C+2nd+Edition-p-9780471851912.
- 13.Sandström J, Pettersson J. Amino acid composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J. Insect Physiol. 1994;40:947–955. doi: 10.1016/0022-1910(94)90133-3. [DOI] [Google Scholar]
- 14.Kim D, Minhas BF, Li-Byarlay H, Hansen AK. Key transport and ammonia recycling genes involved in aphid symbiosis respond to host-plant specialization. G3 GenesGenomesGenetics. 2018;8:2433–2443. doi: 10.1534/g3.118.200297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong F, et al. Effects of five host plant species on the life history and population growth parameters of Myzus persicae (Hemiptera: Aphididae) J. Insect Sci. 2019;19:15. doi: 10.1093/jisesa/iez094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shigenobu S, Stern D. Aphids evolved novel secreted proteins for symbiosis with bacterial endosymbiont. Proc. R. Soc. B. 2012 doi: 10.1098/rspb.2012.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng H, et al. Trading amino acids at the aphid–Buchnera symbiotic interface. Proc. Natl. Acad. Sci. USA. 2019;116:16003–16011. doi: 10.1073/pnas.1906223116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pers D, Hansen AK. The boom and bust of the aphid’s essential amino acid metabolism across nymphal development. G3 GenesGenomesGenetics. 2021;11:jkab115. doi: 10.1093/g3journal/jkab115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korb J, et al. Comparative transcriptomic analysis of the mechanisms underpinning ageing and fecundity in social insects. Philos. Trans. R. Soc. B. 2021;376:20190728. doi: 10.1098/rstb.2019.0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georgiadou A, et al. Comparative transcriptomic analysis reveals translationally relevant processes in mouse models of malaria. eLife. 2022;11:e70763. doi: 10.7554/eLife.70763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchner P. Endosymbiosis of Animals with Plant Microorganisms. Interscience Publishers; 1965. p. 909. [Google Scholar]
- 22.Braendle C, et al. Developmental origin and evolution of bacteriocytes in the aphid-buchnera symbiosis. PLOS Biol. 2003;1:e21. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran NA, Bennett GM. The tiniest tiny genomes. Annu. Rev. Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 24.Chong RA, Moran NA. Evolutionary loss and replacement of Buchnera, the obligate endosymbiont of aphids. ISME J. 2018;12:898–908. doi: 10.1038/s41396-017-0024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Z, et al. Comparative analysis of genome sequences from four strains of the Buchnera aphidicola Mp endosymbion of the green peach aphid. Myzus persicae. BMC Genomics. 2013;14:917. doi: 10.1186/1471-2164-14-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith TE, Moran NA. Coordination of host and symbiont gene expression reveals a metabolic tug-of-war between aphids and Buchnera. Proc. Natl. Acad. Sci. U. S. A. 2020;117:2113–2121. doi: 10.1073/pnas.1916748117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Husnik F, et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153:1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 28.Sloan DB, et al. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol. Biol. Evol. 2014;31:857–871. doi: 10.1093/molbev/msu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luan J-B, et al. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol. Evol. 2015;7:2635–2647. doi: 10.1093/gbe/evv170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nozaki T, Shigenobu S. Ploidy dynamics in aphid host cells harboring bacterial symbionts. Sci. Rep. 2022;12:9111. doi: 10.1038/s41598-022-12836-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackman, R. L. Reproduction, cytogenetics and development. Aphids Their Biol. Nat. Enemies Control Ed. AK Minks P Harrewijn (1987).
- 32.Kwak Y, Argandona JA, Degnan PH, Hansen AK. Chromosomal-level assembly of Bactericera cockerelli reveals rampant gene family expansions impacting genome structure, function and insect-microbe-plant-interactions. Mol. Ecol. Resour. 2022 doi: 10.1111/1755-0998.13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Björk P, Wieslander L. The Balbiani Ring Story: Synthesis, assembly, processing, and transport of specific messenger RNA–protein complexes. Annu. Rev. Biochem. 2015;84:65–92. doi: 10.1146/annurev-biochem-060614-034150. [DOI] [PubMed] [Google Scholar]
- 34.Stormo BM, Fox DT. Polyteny: Still a giant player in chromosome research. Chromosome Res Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol. 2017;25:201–214. doi: 10.1007/s10577-017-9562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopes MR, et al. Evolutionary novelty in the apoptotic pathway of aphids. Proc. Natl. Acad. Sci. USA. 2020;117:32545–32556. doi: 10.1073/pnas.2013847117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng H, Park JS, Zhai RG, Wilson ACC. microRNA-92a regulates the expression of aphid bacteriocyte-specific secreted protein 1. BMC Res. Notes. 2019;12:638. doi: 10.1186/s13104-019-4665-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikoh N, Nakabachi A. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 2009;7:12. doi: 10.1186/1741-7007-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith TE, Li Y, Perreau J, Moran NA. Elucidation of host and symbiont contributions to peptidoglycan metabolism based on comparative genomics of eight aphid subfamilies and their Buchnera. PLOS Genet. 2022;18:e1010195. doi: 10.1371/journal.pgen.1010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pers D, Hansen AK. The effects of different diets and transgenerational stress on Acyrthosiphon pisum development. Insects. 2019;10:260. doi: 10.3390/insects10090260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medina-Ortega KJ, Walker GP. Faba bean forisomes can function in defense against generalist aphids. Plant Cell Environ. 2015;38:1167–1177. doi: 10.1111/pce.12470. [DOI] [PubMed] [Google Scholar]
- 41.Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- 42.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Park H, Smith TE, Moran NA. Gene family evolution in the pea aphid based on chromosome-level genome assembly. Mol. Biol. Evol. 2019;36:2143–2156. doi: 10.1093/molbev/msz138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R Core Team. R: A Language and Environment for Statistical Computing. (2017).
- 47.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aramaki T, et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu L, et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019;47:W52–W58. doi: 10.1093/nar/gkz333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 52.McCune B, Mefford MJ. PC-ORD: Multivariate Analysis of Ecological Data. MjM Software Design; 1999. [Google Scholar]
- 53.Blanco E, González-Ramírez M, Di Croce L. Productive visualization of high-throughput sequencing data using the SeqCode open portable platform. Sci. Rep. 2021;11:19545. doi: 10.1038/s41598-021-98889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA sequencing data are available at NCBI under SRA BioProject ID: PRJNA866154 under SAMN30154147, SAMN30154148, SAMN30154149, SAMN30154150, SAMN30154151, SAMN30154152.