Abstract
SARS-CoV-2 has been responsible for the major worldwide pandemic of COVID-19. Despite the enormous success of vaccination campaigns, virus infections are still prevalent and effective antiviral therapies are urgently needed. Viroporins are essential for virus replication and release, and are thus promising therapeutic targets. Here, we studied the expression and function of recombinant ORF3a viroporin of SARS-CoV-2 using a combination of cell viability assays and patch-clamp electrophysiology. ORF3a was expressed in HEK293 cells and transport to the plasma membrane verified by a dot blot assay. Incorporation of a membrane-directing signal peptide increased plasma membrane expression. Cell viability tests were carried out to measure cell damage associated with ORF3a activity, and voltage-clamp recordings verified its channel activity. The classical viroporin inhibitors amantadine and rimantadine inhibited ORF3a channels. A series of ten flavonoids and polyphenolics were studied. Kaempferol, quercetin, epigallocatechin gallate, nobiletin, resveratrol and curcumin were ORF3a inhibitors, with IC50 values ranging between 1 and 6 µM, while 6-gingerol, apigenin, naringenin and genistein were inactive. For flavonoids, inhibitory activity could be related to the pattern of OH groups on the chromone ring system. Thus, the ORF3a viroporin of SARS-CoV-2 may indeed be a promising target for antiviral drugs.
Subject terms: Ion channels, Pharmacology, Drug development
Introduction
Coronaviruses (CoVs) belong to the order Nidovirales, family Coronaviridae, and subfamily Coronavirinae1. They are subdivided into four different genera named α-, β-, γ-, and δ-CoVs2. Coronaviruses have been known to infect humans2–4, usually causing mild respiratory infections such as a common cold. However, in the past 20 years, two major outbreaks occurred due to crossover of animal β- coronavirus to humans5. In 2002–03 humans were infected by bat coronavirus resulting in severe acute respiratory syndrome coronavirus (SARS-CoV) and in 2019, a novel coronavirus of bat origin that had spread to humans, had been discovered in Wuhan, China6. This new virus, named SARS-CoV-2, is a member of the β-coronavirus family and is responsible for the ongoing pandemic of COVID-191,7,8.
SARS-CoVs are enveloped, positive sense single-stranded RNA viruses, with a genome of approximately 30 kb arranged into 14 open reading frames (ORF) encoding 31 proteins8–11. Spike (S), envelope (E), membrane (M) and nucleoprotein (N) are the four structural proteins forming the virus capsid. The S protein binds to the host receptor through the receptor-binding domain in the S1 subunit, while S2 subunit is responsible for membrane fusion8. The E protein belongs to the class of viroporins, integral membrane proteins functioning as ion channels and promoting virus release. It was found to be expressed in the ER and the Golgi apparatus forming an ion channel allowing the efflux of cations Na+, K+ and Ca2+, and is required for pathogenesis and stimulation of inflammation8,12–15. The E-protein of SARS-CoV has been characterized thoroughly, showing that its ion channel activity, as well as its interaction with cellular and other virus proteins is essential for virus replication16–23. The M protein is the most abundant structural protein in the viral envelope, the N protein wraps around the RNA forming the nucleocapsid8. Proteins from open reading frame 1, ORF1a and ORF1b, known as the non-structural proteins (NSPs), form the intracellular replicase-transcriptase complex24. In addition to the structural proteins and NSPs, coronaviruses encode several accessory proteins which vary in number, location and size in the different viral subgroups, and are thought to contain additional functions that are often not required for virus replication, but are involved in pathogenicity in the natural host25.
ORF3a (Fig. 1A, B) belongs among the group of viroporins. It is the largest accessory protein (275 amino acids) of SARS-CoV-2 and shares 72.7% identity with the ORF3a protein of SARS-CoV9,12. Despite this high degree of homology, ORF3a was classified as unique to SARS-CoV-224,26, since it was found to be structurally and functionally different from its counterpart in SARS-CoV. The importance of the viroporins of SARS-CoV for virus replication has been studied in detail27, showing that activity of both, E- and ORF3a protein is required for full virus replication and virulence. Simultaneous deletion of both viroporins renders the virus incapable of replication. Deletion of either E- or ORF3a protein resulted in viruses that were viable, but showed reduced replication rate and in vivo toxicity in a mouse model27. While the contribution of the E-protein to virulence of SARS-CoV is stronger, presence of functional ORF3a is nevertheless essential for full activity of the virus. ORF3a is the only viroporin that comprises three transmembrane domains (Fig. 1B), with the N-terminus located at the luminal side of the ER and the C-terminal domain directed to the cytosol28. The ion channel function of ORF3a of SARS-CoV was shown to be unrelated to viral replication in one study27, but associated with pro-apoptotic activity that affected replication in another29. Notably, different animal models were used in these studies. ORF3a activity triggered secretion of apoptotic mediators IL-1beta and NF-κB30,31, and increased production of cellular chemokines32. The cytoplasmic domain of ORF3a was reported to be involved in the induction of pro-apoptotic reactions by activating the mitochondrial p38 MAP kinase death pathway, which could be repressed by a p38 MAPK inhibitor33. Further, ORF3a was shown to induce intracellular membrane rearrangement34. Ion selectivity investigations on ORF3a channels reconstituted in proteoliposomes revealed nonselective cation channel activity with a permeability order of Ca2+ > K+ > Na+ > NMDG+35. ORF3a has been suggested as target for antiviral therapy, and kaempferol and related flavonoids were shown to inhibit the ORF3a channel in vitro36. The anthraquinone derivative emodin is an inhibitor of ORF3a of SARS-CoV, and the related beta-coronavirus HCoV-OC43, and was indeed able to repress virus release from the latter37.
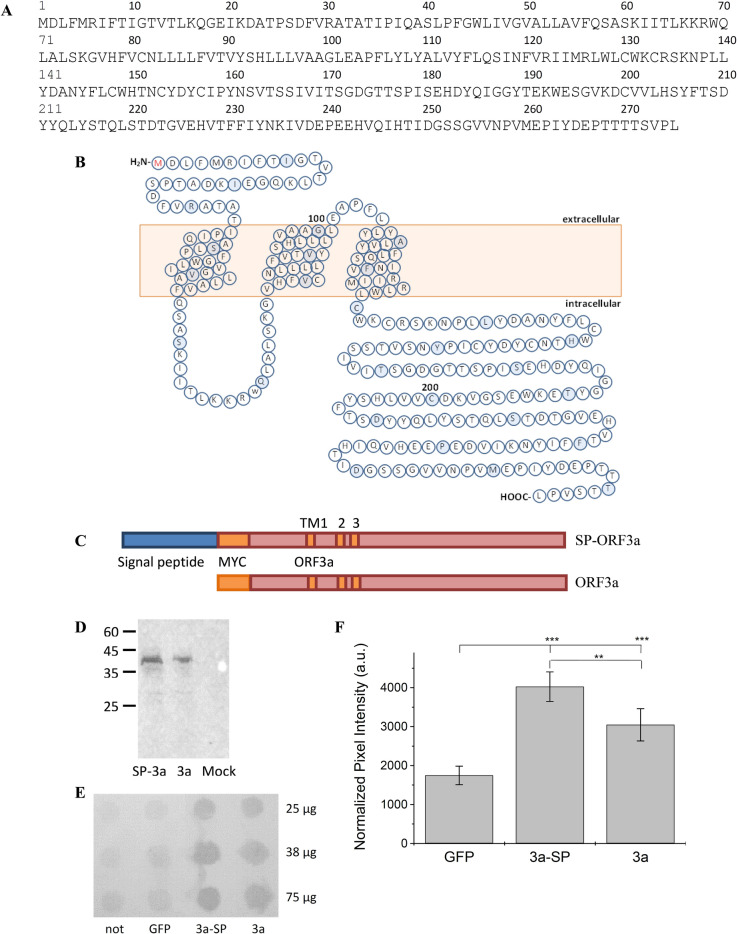
Figure 1.
Sequence, topology and recombinant expression of ORF3a constructs. (A) Sequence of the ORF3a protein (GenBank NC_045512.2). (B) TM topology and amino acid distribution of ORF3a. Positions 100 and 200 are labelled. Every 10th amino acid is coloured. (C) Scheme of SP-MYC-ORF3a and MYC-ORF3a. (D) Western blot confirming the expression of ORF3a SARS-CoV-2 in HEK293 cells. Molecular weights of markers are indicated. Lane 1: HEK293 cells transfected with DNA encoding SP-ORF3a; lane 2: ORF3a without signal peptide; lane 3: mock-transfected HEK293 cells (GFP). Theoretical sizes of SP-ORF3a and ORF3a are 39.8 kDa and 35.6 kDa. Note that both constructs show an immune signal of the same size, indicating efficient cleavage of the signal peptide. One complete blot is shown. (E) Surface Expression Analysis: HEK293 cells were transfected with cDNA encoding ORF3a protein (3a), and ORF3a protein fused to a membrane-directing signal peptide (3a–SP), untransfected (not) and mock-transfected cells were used as controls (GFP); 25, 37.5 and 75 µg of total protein were applied per spot. One complete dot blot from a representative experiment is shown. (F) Quantification of Blot intensities. Two independent experiments were performed and different concentrations of total protein spotted (n = 5). Spot intensities were quantified using ImageJ software, intensities of each spot summated and normalized to the total intensity count of each row. Average ± standard deviation from all determinations is shown, p-values are indicated, ***: p < 0.001, **: p < 0.005 (one-way ANOVA).
ORF3a of SARS-CoV-2 was shown to prevent autophagy, one of the major defence mechanisms of cells in the fight against pathogens38–41. ORF3a prevents fusion of autophagosomes with lysosomes by blocking the interaction of the homotypic fusion and protein sorting (HOPS) complex with the SNARE complex and RAB738. SARS-CoV-2 ORF3a activates the NLRP3 inflammasome42, and has pro-apoptotic activity, reported to be weaker than in case of SARS-CoV43. This reduced apoptotic activity was speculated to account – at least partially – for the lower virulence of SARS-CoV-2 compared to SARS-CoV. Reduced apoptotic reaction may lead to an overall reduced defensive response, allowing the virus to spread more widely into tissue even in case of mild clinical symptoms43. ORF3a of SARS-CoV-2 was shown to downregulate surface expression of MHC-1, thereby preventing detection and destruction of infected cells by CD8(+) T lymphocytes44. ORF3a of SARS-CoV and SARS-CoV-2 interacts with caveolin45,46.
Expression of the ORF3a protein in several in vitro systems describes its localization mainly in the Golgi region46 but also in the ER31, late endosomes38, lysosomes47, and the trans-Golgi network47. In case of SARS-CoV-2, localization of ORF3a in the plasma membrane, as well as in cytosolic compartments has been shown43. Thus, ORF3a is an essential contributor to infection and propagation of SARS-CoV-2, and its structure and function are of ongoing interest26,48–52. Indeed, viroporin inhibitors53 as well as potential inhibitors of ORF3a have been tested in vivo and in vitro36,37, and searched through in-silico approaches54, and potent inhibitors of ORF3a are of interest as antiviral drug candidates.
Given the essential role of viroporins in the virus life cycle, viroporin inhibitors are promising potential antiviral agents12. Classical viroporin inhibitors including amantadine and rimantadine have shown activity against numerous viral channels53,55–61. Another interesting substance class would be phenolics, naturally occurring secondary metabolites found in many plant tissues. They are known for powerful antioxidant, anti-inflammatory, immunomodulatory, and antibacterial activity, as well as for their action against various viruses62. Their antiviral activity involves interference with viral life cycle and pathogenesis63. An additional advantage of many polyphenols is the fact that their beneficial activity in humans is well established, and they can thus easily be repurposed as antiviral therapeutics64,65. Thus, flavonoids and other phenolics may serve as antiviral agents directly, or as lead structures towards the discovery of novel, specific viroporin inhibitors. In this study, we tested seven flavonoids – kaempferol, quercetin, epigallocatechin gallate (ECGC), nobiletin, apigenin, genistein, and naringenin. Given their reported antiviral activity, the phenolics resveratrol66–69, curcumin70–73, as well as 6-gingerol74,75, a component of ginger rhizome76–78 were also included in this study.
We show the inhibition of SARS-CoV-2 ORF3a activity by the classic viroporin inhibitors amantadine and rimantadine and by a series of ten flavonoids and phenolics. Toxicity of recombinantly expressed ORF3a channels was assessed using a cell viability assay, and could be correlated to inhibition of ion channel activity of recombinant ORF3a, measured by patch-clamp electrophysiology. Some of the tested compounds, including kaempferol, quercetin, EGCG, nobiletin, resveratrol and curcumin inhibited ORF3a activity, with IC50 values ranging between 1 and 6 µM, indicating that ORF3a may be a target for antiviral therapy.
Materials and methods
ORF3a DNA constructs
SARS-CoV-2 ORF3a cDNA (gift from Prof. Jun Wang, Rutgers University, NJ, USA), corresponding to the Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1 (GenBank NC_045512.2) (Fig. 1A, B) was cloned into the pRK vector using BamHI and HindIII restriction sites. One construct included the myc-tag (EQKLISEEDL) for verification in Western blotting and dot blot analysis. The second construct included the myc-tag and an additional signal peptide (SP), MWTPRVPPPRPALSFFLLLLLGVTYG, taken from the murine semaphorin-6B precursor (Fig. 1C).
Cell culture and transfection
HEK293 cells (ATCC, LGC Standards GmbH, Wesel, Germany) were cultured in 10 cm tissue culture dishes in Dulbecco's Modified Eagle Medium, low glucose (DMEM, Sigma-Aldrich, Deisenhofen, Germany) supplemented with 10% FBS (Invitrogen, Karlsruhe, Germany) and Penicillin/ Streptomycin (Sigma-Aldrich, Deisenhofen, Germany) at 5% CO2 and 37 °C in a water-saturated atmosphere. For electrophysiological experiments, cells were plated on acetone treated glass coverslips in 24 well plates and transfected one day after passage using 1 µg of target cDNA, 1 µg of green fluorescence protein cDNA and 3 µl polyethyleneimine (PEI) (Sigma-Aldrich, Deisenhofen, Germany) per well. Measurements were performed 2–3 days after transfection.
Membrane preparation
HEK293 cells were grown in 6 cm plates, transfected with 6 µg of cDNA and 20 µg of PEI as described under ‘Cell Culture and Transfection’. HEK293 cells were harvested 3 days after transfection using PBS (1.5 mM KH2PO4, 6.5 mM Na2HPO4, 3.0 mM KCl, 137 mM NaCl) and centrifuged at 2000 g. The pellet was resuspended in hypotonic buffer (20 mM K-Phosphate pH 7.4, 5 mM EDTA, 5 mM EGTA, 1 tablet cOmplete™ Mini Protease Inhibitor Cocktail (Sigma-Aldrich, Deisenhofen, Germany)) and kept on ice for 20 min. Cells were homogenized using a glass cell homogenizer and then centrifuged at 17,000 rpm for 20 min. Pellets were resuspended in hypotonic buffer and total protein concentration was measured prior to SDS PAGE.
Western blot and dot blot analysis
For Western blotting, membrane suspension (30 µg of total protein) in SDS loading dye (2 × loading dye: 0.125 M Tris–HCl pH 6.8, 20% glycerol, 4% SDS, 0.02% bromophenol blue, 0.2 M DTT) were loaded per lane and subjected to SDS-PAGE. We used a combination of a myc-tag polyclonal primary antibody from rabbit and alkaline phosphatase (AP) – conjugated Goat Anti-Rabbit secondary antibody (Proteintech, Martinsried, Germany). The blot was visualized using the reaction of 0.03% nitro blue tetrazolium (NBT) and 0.02% 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) (both Carl Roth, Karlsruhe, Germany) in substrate buffer (100 mM Tris–HCl, pH 9.5; 100 mM NaCl; 5 mM MgCl2).
For the dot blot assay, HEK293 cells were grown in 6 cm plates and transfected as described under “Membrane preparation”. Three days after transfection, culture medium was removed and cells washed with PBS, blocked with fresh full medium for 20 min, then primary antibody was added to the living cells and incubated for 2 h at 37 °C. Secondary antibody was applied for 1 h, cells were harvested, homogenized, protein concentration determined, 25–75 µg of total protein per well were spotted onto a nylon blotting membrane in a dot blot apparatus and immune signal detected. Antibodies and detection reagents were the same as in Western blotting. Pixel intensity of dots was quantified using ImageJ software without further correction of the raw data. For each series of applied protein, pixel intensities for each well were normalized to the total pixel intensity of all peaks of each treatment. The percent of total intensity of each well could then be calculated. Averages were taken from dot blots from two independent expressions using two or three different amounts of total protein (n = 5).
Cell viability assay
120 µl of a HEK293 cell suspension were seeded into a 96-well plate at ~ 60,000 cells per well. Cells were transfected one day after plating. Next, 6.5 µl of inhibitor stock in DMEM were added to achieve final concentrations as required (usually between 1 and 100 µM). Four days after transfection, the MTT assay was performed as described before79. Briefly, culture medium was removed, cells were washed once with PBS, and 100 µl/well of a 0.5% solution of 3-(4,5-dimethyldiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) in PBS was added. Cells were incubated at 37 °C for 2 h. Solution was carefully removed and 100 µl/well of dimethyl sulfoxide (DMSO) were added to dissolve formazan crystals. Absorbance was read in a Victor-3 plate reader (Perkin-Elmer, Berlin, Germany) at a wavelength of 595 nm. Controls included GFP transfected cells to verify efficient transfection and the addition of 200 mM KCl to untransfected cells to induce complete cell death; the 200 mM KCl readings were subtracted from all other data as background correction. Each experimental condition was tested on (i) ORF3a-transfected cells, and (ii) control cells transfected with empty pRK vector. For quantification, viability readings for each condition were averaged. The quotient was calculated and presented in Fig. 3. Each assay was performed in quadruples, a minimum of two independent experiments per condition were carried out (n ≥ 8).
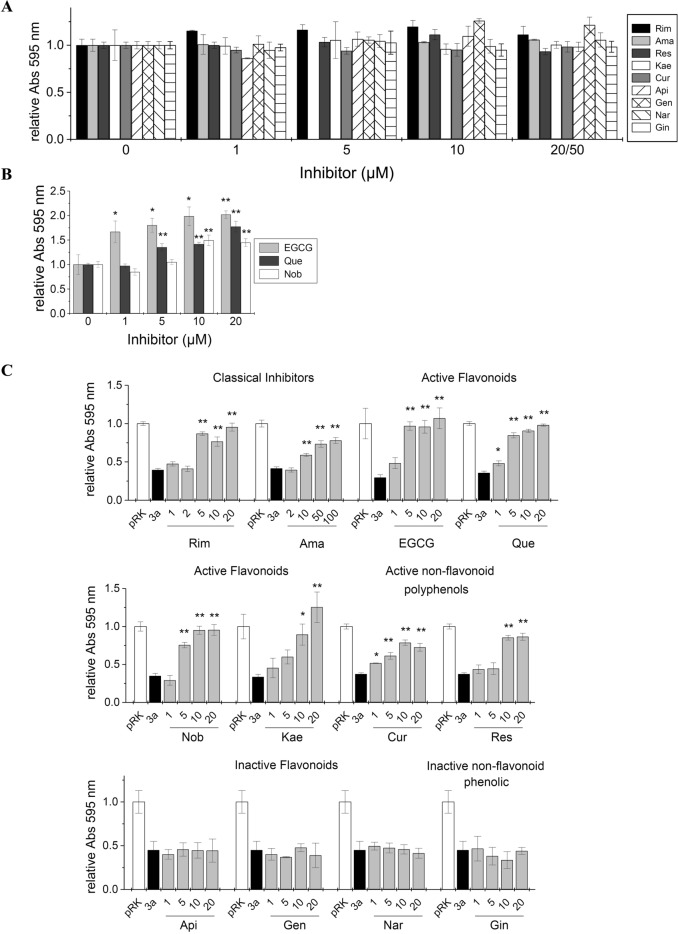
Figure 3.
MTT cell viability assay of ORF3a transfected cells to investigate inhibitory action on ORF3a. (A) HEK293 cells were transfected with empty vector (control) and treated with the indicated inhibitors. Compounds did not affect cell viability. (B) Quercetin, EGCG and nobiletin at concentrations of 5 µM (Que), 10–20 µM (EGCG, Que and Nob) led to increased cell viability, indicating proliferative activity. (C) Viability test of viroporin inhibitors candidates. HEK293 cells were transfected with ORF3a and treated with increasing concentrations of inhibitor. For each data set, the first bar represents the control value (= 1) (white), then relative cell viability after expression ORF3a (black), followed by normalized viability readings at increasing concentrations of inhibitor (grey). (A–C) Significance relative to ORF3a-transfected cells in absence of inhibitor was determined using one-way ANOVA with * = p < 0.05; ** = p < 0.01, no sign = not significant. Abbreviations are as follows: Rim = rimantadine, Ama = amantadine, EGCG = epigallocatechin gallate, Que = quercetin, Nob = nobiletin, Res = resveratrol, Kae = kaempferol, Cur = curcumin, Api = apigenin, Gen = genistein, Nar = naringenin, Gin = 6-gingerol.
Electrophysiological recordings and data analysis
HEK293 cells were plated on glass cover slides; transfected with cDNA constructs of ORF3a or mock and co-transfected with GFP, followed by incubation for 2–3 days to allow protein expression. Cover slides were transferred to a recording chamber, immersed in extracellular buffer and whole-cell currents recorded using a HEKA EPC 10 (NPI electronics, Tamm, Germany) patch-clamp amplifier. Recording pipettes (thin-walled borosilicate glass, 100 × 1.5 mm, 0.86 mm inner diameter, Science Products, Berlin, Germany) were pulled using a Sutter P-1 pipette puller (NPI electronics). For recordings of current–voltage ramps, we used symmetrical external and internal electrophysiology buffers. External buffer consisted of (in mM) 135 NaCl, 5.5 KCl, 2 CaCl2, 1.0 MgCl2 and 10 HEPES (pH adjusted to 7.2 with NaOH); the internal buffer was (in mM) 140 CsCl, 1.0 CaCl2, 2.0 MgCl2, 5.0 EGTA and 10 HEPES (pH adjusted to 7.2 with CsOH). The applied voltage ramp ranged from − 60 mV to + 50 mV in 10 mV steps. For measurements in presence of inhibitor, we added the inhibitor to the extracellular bath before patching cells and recording; a minimum of 6 cells per each concentration of each inhibitor were averaged. Dose–response curves were constructed from the analysis of current differences at − 60 mV, and IC50 values were determined using a non-linear fit to the equation Iobs = Imax / [1 + ([I]/IC50)].
Results
ORF3a constructs and expression of recombinant ORF3a protein
To investigate the ion channel properties of recombinant ORF3a we used a cDNA construct of ORF3a from the SARS-CoV-2 Wuhan wildtype, GenBank accession no NC_045512.2 (gift from Prof. Jun Wang, Rutgers U, NJ, USA) which was cloned into the mammalian expression vector pRK5. Since the main ion channel activity of viroporins is intracellular, related to ER membranes, the late endosome and lysosomes, we cloned ORF3a into the pRK5 vector including a myc-tag for detection in Western blotting (Fig. 1C). No signal peptide for plasma membrane transport was present in this construct which was used to show the effect of active ORF3a expression in cell viability assays on HEK293 cells. Electrophysiological measurements, on the other hand, require transport to the plasma membrane. Here, we introduced a membrane-directing N-terminal signal peptide from the murine semaphorin-6B precursor in addition to the myc-tag to the ORF3a (Fig. 1C). Recombinant expression of ORF3a was confirmed using HEK293 cells that were transfected with cDNA of SP-ORF3a and ORF3a, or a mock construct (GFP), and subjected to SDS PAGE and Western blot analysis three days after transfection (Fig. 1D). Specific signals were observed in Western blots for SP-ORF3a and ORF3a. Both proteins gave an immune signal of the same size, consistent with effective cleavage of the signal peptide during trafficking. We verified surface expression through dot blot analysis on native, living cells. To this end, HEK293 cells were transfected with target cDNA, allowed to express viroporin for three days, followed by application of primary and secondary antibodies directly to the living cells. Under these conditions, only plasma membrane-located ORF3a would be accessible to the antibody (Fig. 1E). When we compared dot blot intensities after subtracting the background signal (GFP), we found a ratio of ~ 1.75 : 1 for SP-ORF3a over ORF3a (Fig. 1F). Apparently, ORF3a – although mainly transported to intracellular membranes – was also found in the plasma membrane, even in absence of a signal peptide. Notably, addition of the semaphorin-6B signal sequence increased the transport to the plasma membrane significantly (Fig. 1F).
Activity of the ORF3a viroporin – cell-based assays
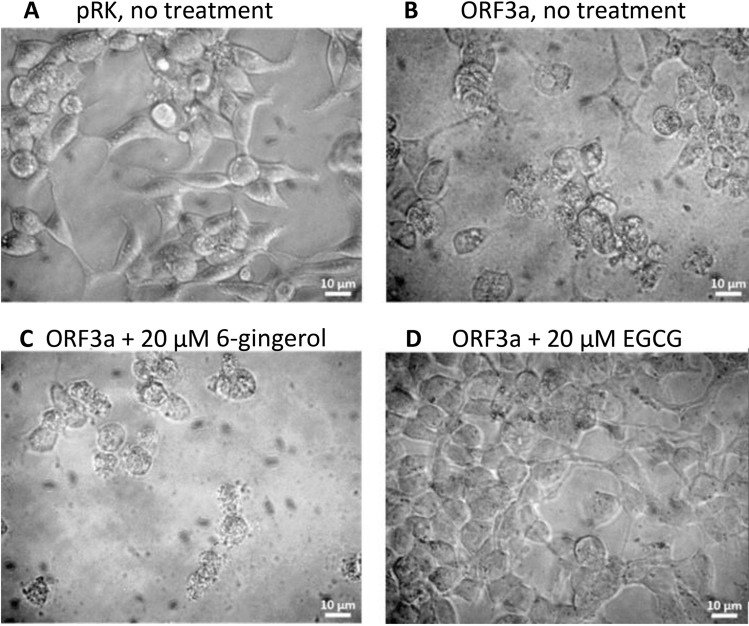
Viroporin activity of ORF3a was analyzed using a cell viability assay system that we had previously established for the viroporin channels of hepatitis (p7), and the E protein of SARS-CoV79,80. Intracellular activity of viroporin channels damages and eventually destroys cells, leading to reduced overall viability of the culture. Treatment of ORF-3a-transfected cells with viroporin inhibitors should restore cell viability79,80. Indeed, morphology and viability/survival of ORF3a transfected cells changed compared to control. Four days after transfection, mock transfected cells looked healthy with normal morphology (Fig. 2A), while ORF3a transfected untreated cells were shrinking and fragmentation into membrane-bound apoptotic bodies could be observed (Fig. 2B). Cells treated with 6-gingerol (inactive against ORF3a) showed the morphology of dying cells (Fig. 2C), while treatment with epigallocatechin gallate (an active inhibitor of ORF3a) resulted in healthy cells with normal morphology (Fig. 2D). Thus, viroporin inhibitors were able to prevent viroporin-induced damage to transfected cells.
Figure 2.
Morphology of HEK293 cells in absence and presence of ORF3a inhibitors. (A) HEK293 cells transfected with empty pRK vector. (B–C): HEK293 cells transfected with ORF3a. (B) untreated; (C) treated with 6-gingerol (inactive against ORF3a); (D) treated with EGCG (active ORF3a inhibitor).
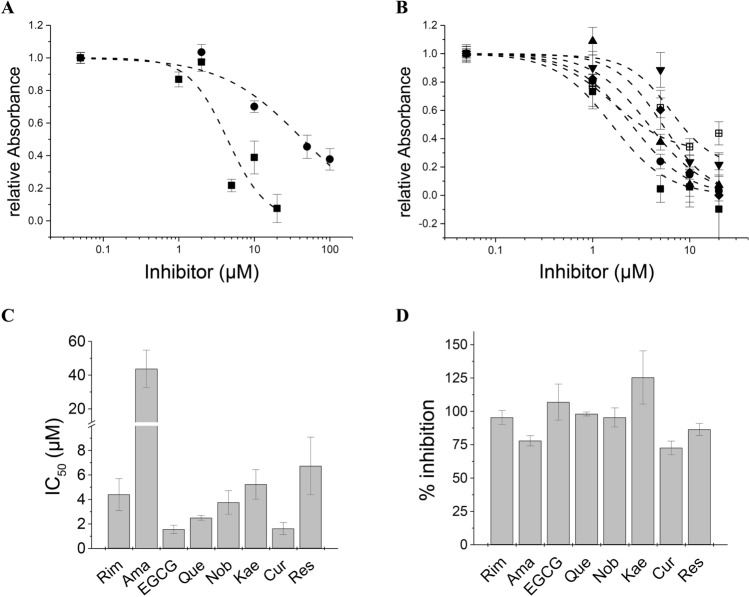
We then used an MTT-based cell viability assay to quantify ORF3a activity and the action of inhibitors and candidate substances on the ORF-3a channel. Inhibitors were applied to ORF-3a-, or mock transfected cells at the same concentrations. In this way, cytotoxic or proliferative effects of the inhibitors could be assessed. Most inhibitors had no effect on cell viability (Fig. 3A). Exceptions were EGCG, quercetin and nobiletin, where concentrations of 5 µM and higher (> 1 µM for ECGC) led to increased cell proliferation as compared to untreated controls (Fig. 3B). We analysed two classical viroporin inhibitors rimantadine (Rim) and amantadine (Ama), a series of seven flavonoids as well as the non-flavonoid phenolics resveratrol (Res), curcumin (Cur), and 6-gingerol (Gin) (Fig. 3C). Rimantadine and amantadine were active inhibitors with IC50 values of 4.4 ± 1.3 µM, and 43.7 ± 11.1 µM, respectively (Fig. 4A, C). EGCG, quercetin (Que), nobiletin (Nob), kaempferol (Kae) and curcumin (Cur) were inhibitors of ORF3a, reversing viroporin-induced cell damage with IC50 values (in µM) of 1.6 ± 0.3 (EGCG), 2.5 ± 0.2 (Que), 3.8 ± 0.9 (Nob), 5.2 ± 1.2 (Kae) and 1.6 ± 0.5 (Cur) (Fig. 4B, C). Resveratrol was an active inhibitor with an IC50 of 6.7 ± 2.3 µM. Most inhibitors showed complete (100%) inhibition of ORF3a activity at 20 µM concentration. Exceptions were amantadine, curcumin and resveratrol with maximal inhibition of 77.9 ± 3.9 µM (Ama), 72.6 ± 5.1 (Cur) and 86.5 ± 4.5 (Res) (Fig. 4D). We found apigenin (Api), genistein (Gen) and naringenin (Nar), as well as 6-gingerol (Gin) to be inactive against ORF3a (Fig. 3C).
Figure 4.
Concentration–response analysis of viroporin inhibition. (A, B) Concentration–response curves. ORF3a activity was calculated from viability data (see Methods), and normalized to viability data in absence of inhibitor. Note that in cases, where cell viability of treated ORF3a expressing HEK293 cells was increased as compared to control, relative absorbances could reach values below 0 in the IC50 graph. (A) Classical viroporin inhibitors; solid squares = rimantadine, solid circles = amantadine (B) Test compounds; solid squares = EGCG, solid circles = quercetin, up triangles = nobiletin, down triangles = resveratrol, diamonds = kaempferol, crossed squares = curcumin. (C) IC50 values of classical inhibitors and flavonoids. Abbreviations are as in Fig. 3. (D) Percentage inhibition was obtained from MTT data at the highest inhibitor concentration. All active inhibitors achieved complete inhibition (~ 100%) except amantadine (78%), curcumin (72%) and resveratrol (86%).
Activity and inhibition of ORF3a – patch-clamp electrophysiology
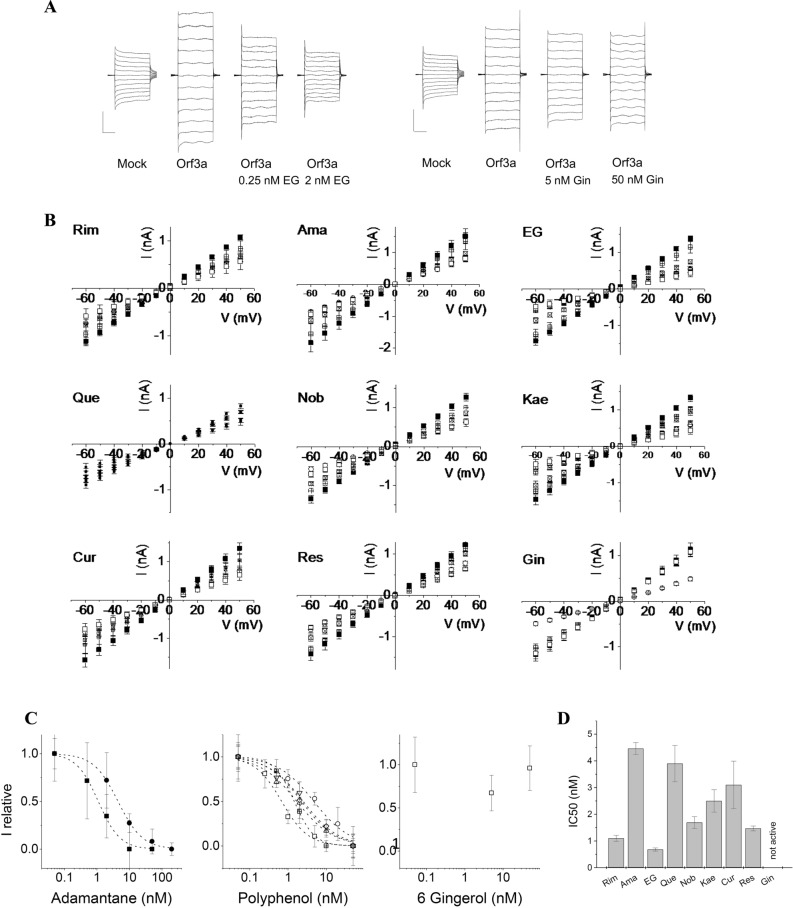
To correlate cell viability data to ion channel activity of ORF3a, we performed patch-clamp electrophysiology for all compounds that showed activity and for one inactive compound, namely 6-gingerol. Mock- or ORF3a-transfected cells were voltage-clamped and current–voltage (IV) ramps in the range from − 60 to + 50 mV were recorded. Presence of active ORF3a resulted in increased currents compared to control cells, due to viroporin channel activity on the cell surface. Currents from 5–6 cells for each condition were averaged. Addition of inhibitor reduced the currents in the IV ramp to control levels in a concentration-dependent manner. Mock transfected cells at − 60 mV gave currents in the range of 0.5 to 0.8 nA, which increased to 1.2–1.5 nA for ORF3a transfected cells. Data sets in presence of inhibitors and corresponding controls were always recorded on the same day and compared (Fig. 5A, B). Results from patch-clamp measurements showed similar trends as those from MTT results: activity of amantadine + (IC50 = 4.5 ± 0.2 nM) was approximately fourfold lower than that of rimantadine (IC50 = 1.1 ± 0.1 nM). IC50 values were 0.7 ± 0.1 nM for EGCG, 3.9 ± 0.7 for quercetin, 1.8 ± 0.3 for nobiletin, 2.4 ± 0.5 for kaempferol, 3.1 ± 0.9 for curcumin, and 1.5 ± 0.1 for resveratrol, while 6-gingerol was inactive (Fig. 5C, D).
Figure 5.
Patch-Clamp Electrophysiology measurements from HEK293 cells expressing SP-ORF3a. (A) IV ramp form − 60 mV to 50 mV was applied to mock-transfected cells and compared to IV ramps from cell transfected with SP-ORF3a in the absence and presence of inhibitors. Currents from ORF3a-expressing cells were increased compared to mock-transfected control cells. Currents were reduced and reached control values upon treatment with classical inhibitors (rimantadine and amantadine) or phenolics. Vertical and horizontal scale bars denote 0.5 nA and 10 ms, respectively. EG = epigallocatechin gallate. (B) Current–voltage relationship of all tested inhibitors. Open circles = mock transfected HEK293 cells; squares = ORF3a expressing cells without inhibitor; crossed squares = small inhibitor concentration (0.5–1 nM), crossed square = intermediate inhibitor concentration (1–5 nM); open squares = highest inhibitor concentration (20–50 nM). (C) IC50 curves from patch-clamp inhibition data: solid squares = rimantadine, solid circles = amantadine, open squares = EGCG, open circles = quercetin, up-triangles = nobiletin, down-triangles = kaempferol, diamonds = curcumin, crossed squares = resveratrol, dotted squares = 6-gingerol. (D) Summary of IC50 values from patch-clamp electrophysiology. Abbreviations are as listed in Fig. 3.
Discussion
Since viroporins are involved in various steps of the virus life cycle including assembly, budding, envelope formation, virus release, and inflammasome activation, they represent a promising target for antiviral drugs 15,59,80. Amantadine and rimantadine are well-characterized viroporin inhibitors, known to be active against a number of viroporins including the influenza M2 ion channel59,60,81, p7 channels of the hepatitis C virus58,82, and E protein of SARS-CoV61,80. To ensure sufficient plasma membrane expression of ORF3a – an essential requirement for patch-clamp electrophysiology – we attached an N-terminal signal peptide derived from murine semaphorin 6B. Plasma membrane expression of ORF3a was verified by Western blot and dot blot analysis. Presence of a membrane-directing signal peptide increased the expression of ORF3a on the plasma membrane.
We determined the activity of known viroporin inhibitors and a series of phenolic compounds against recombinant ORF3a protein using (i) a cell-based assay where viroporin-induced loss of cell viability and its restoration by viroporin inhibitors was quantified, and (ii) patch-clamp electrophysiology where the channel activity of viroporins can be observed directly. Reduced viability of cells expressing the viroporins p7 or E protein have been reported79,80. Both viroporins were shown to activate the NLRP3 inflammasome, in case of the E protein the transport of calcium ions was suggested as reason83,84.
Classical viroporin inhibitors were active against ORF3a. Inhibition was concentration dependent with the potency of rimantadine ~ tenfold and ~ fourfold higher than that of amantadine in cell viability assays and in patch clamp experiments, respectively. Higher potency of rimantadine over amantadine against viroporin channels has been reported before for the inhibition of p7 channels of HCV79,85. Both adamantanes showed inhibitory activity against the E viroporin of SARS-CoV in cell viability assays and patch-clamp measurements. Again, rimantadine was shown to be more potent than amantadine80. Higher antiviral activity of rimantadine in comparison to amantadine has also been reported for SARS-CoV-2 infected Vero E6 cells, where antiviral activity of rimantadine occurred primarily after viral entry rather than prior to viral entry53. Together, these results suggest that adamantane derivatives hold promising therapeutic potential against COVID-19.
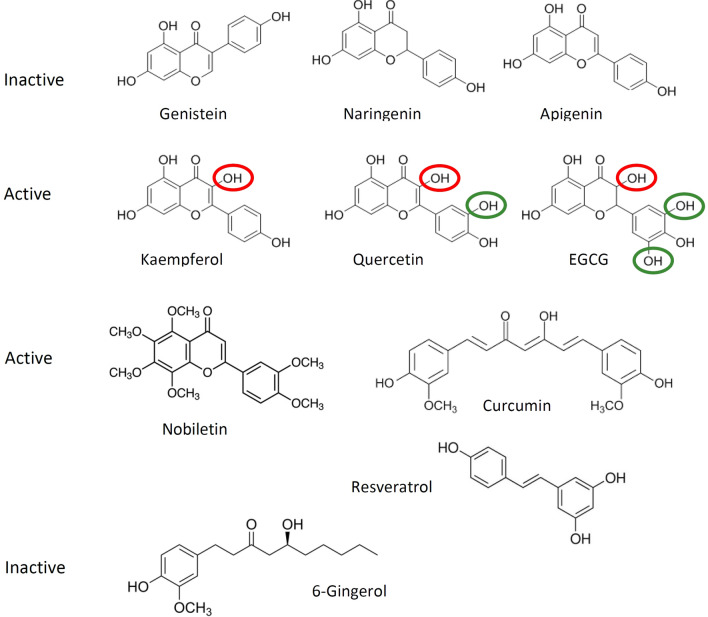
We tested ten phenolics – seven flavonoids, as well as the phenolics resveratrol, curcumin and 6-gingerol – for activity against ORF3a. Six of the tested phenolics (EGCG, quercetin, nobiletin, kaempferol, resveratrol, and curcumin) showed inhibitory activity against ORF3a-induced cytotoxicity, while four (genistein, naringenin, apigenin and 6-gingerol) were inactive. The activity of some natural phenolic compounds and inactivity of others indicates that the observed effects are not due to general antioxidant or metabolic effects of the compounds, but rather that some of these compounds can interact specifically with a distinct binding site on the ORF3a protein. The inactive flavonoids apigenin (flavone), naringenin (flavanone) and genistein (isoflavone) have three OH groups at positions C5 and C7 on ring A and C4’ on ring B (Fig. 6). These hydroxyl functions appear not to be relevant for interaction with ORF3a. In contrast, EGCG, quercetin and kaempferol (all flavonols) have the same basic chromone structure as apigenin, but carry an additional OH group on C3 of ring C (Fig. 6, red highlights). This hydroxy function seemed to be essential for activity against ORF3a, and its absence (as in apigenin) abolishes activity. Quercetin has one additional hydroxyl group on C3′ of ring B, EGCG has two additional hydroxyl groups at C3’ and C5’ of ring B (Fig. 6, highlighted in green). Presence of these hydroxyl groups – in addition to that in ring C – seem to enhance inhibitory potency against ORF3a further. The order of potency against ORF3a of kaempferol (5.23 ± 1.2 µM), quercetin (2.5 ± 0.2 µM) and EGCG (1.56 ± 0.34 µM) could be explained by the additional hydroxyl groups on ring B, where the increased number of available polar interactions or H-bridges appear to correlate with inhibitory potency. In contrast, partial saturation of the chromone skeleton between C2 and C3 (as seen in naringenin and EGCG) had no effect on activity. The role of OH groups on the flavonoid ring system has been studied with respect to antioxidant and protein binding activity65,86,87, but not yet correlated to antiviral effects.
Figure 6.
Structure–activity correlation of tested flavonoids and phenolics. Genistein, naringenin, and apigenin were inactive. Kaempferol, quercetin, EGCG, and nobiletin were active. The OH group encircled in red is essential for activity, those in green enhance activity. Note that nobiletin (no free OH groups), curcumin and resveratrol also inhibited ORF3a activity.
Nobiletin showed potent activity against ORF3a. Although having the same chromone structure as the compounds discussed above, it lacks the essential C3-OH group on ring C, and all OH groups at C5, C6, C7, C8, C3′ and C4′ are methylated. Thus, nobiletin can only accept, but not donate OH bonds, and its polarity is severely reduced compared to the other tested flavonoids. The non-flavonoids, resveratrol (IC50 6.73 ± 2.3 µM) and curcumin (IC50 4.4 ± 1.7 µM) showed activity against ORF3a. In contrast, 6-gingerol was found inactive which would argue against a gingerol-binding site on ORF3a. Taken together, there appears to be a correlation between number and location of OH groups on the chromone ring system, but more structure–activity data are needed before a pharmacophore or a specific site of interaction can be delineated.
Current–voltage recordings confirmed the inhibitory activity for EGCG, quercetin, nobiletin, curcumin and resveratrol, although relative potency of quercetin was lower in the patch-clamp assay. 6-gingerol was also tested and showed to be inactive against ORF3a, as seen in cell viability tests. It was noted that IC50 values of viroporin inhibitors are different from those determined from cellular assays. This effect has been observed for p7 channels of hepatitis C79,82,88, as well as for the E protein of SARS-CoV80. This difference in apparent potency is likely due to availability of the protein to the inhibitor. In cellular assays, the inhibitor (applied extracellularly) has to cross the plasma membrane and may be sequestered or degraded before reaching its intracellular target, while in patch-clamp experiments the target viroporin is directly exposed to the inhibitor in the extracellular buffer.
Activity of plant phenolics against ORF3a has been reported in the literature: kaempferol and its glycosylated derivative juglanin inhibit ORF3a activity of SARS-CoV expressed in Xenopus oocytes, while naringenin and genistein do not36. The same study reported very little inhibition of ORF3a by quercetin36, as also observed here.
Antiviral activity of phenolics has been reported. Consistent with our observations, curcumin effectively inhibited viral replication in SARS-CoV infected Vero E6 cells89. In contrast, 6-gingerol was shown to have only low activity in the same expression system90. Quercetin showed potent anti-inflammatory activity and possible inhibition of the COVID-19-associated cytokine storm91. Clinical studies showed that the administration of nano-encapsulated curcumin to patients infected with COVID-19 resulted in a significant reduction in levels of IL-6 and IL-1β and improvement of patients’ symptoms92. Several in silico analyses suggest that curcumin, and some flavonoids may interact with SARS-CoV-2 proteins24,93,94. Thus, although some of the phenolics studied here (eg 6-gingerol, naringenin, apigenin, and genistein) were not active against ORF3a channels, they may yet possess activity against other proteins of SARS-CoV-2.
The antiviral activity of adamantane derivatives, classical viroporin inhibitors, and ten natural polyphenolics compounds was evaluated in HEK293 transfected with ORF3a of SARS-CoV-2 using cell viability assay and in patch-clamp analysis. Our findings revealed that the adamantane derivatives and six out of ten tested polyphenols have shown high potency against ORF3a and may indeed be promising medicinal agents. A structure–activity pattern for some flavonoids may emerge, which could lead to the identification of a distinct inhibitory site on the ORF3a protein, but further investigation is required. Flavonoids, phenolics and other natural compounds may be a promising source for novel antiviral agents.
Supplementary Information
Acknowledgements
Support by Science and Technology Development Fund (STDF) Egypt, Grant No 45420 is gratefully acknowledged. We are grateful to Prof. Jun Wang, Rutgers University, NJ USA for a gift of ORF3a expression construct. Excellent technical Assistance by Mousa A. Mousa is acknowledged. The authors report no conflict of interest regarding this study.
Author contributions
M.S.F.: design of work, data acquisition and analysis, interpretation of data, revising the manuscript. C.A.S.: data acquisition and analysis, interpretation of data, revising the manuscript. N.O.T.: data acquisition and analysis, interpretation of data, revising the manuscript. H.G.B.: conception, design of work, data acquisition and analysis, interpretation of data, drafting and revising the manuscript. U.B.: conception, design of work, data acquisition and analysis, interpretation of data, drafting, writing and revising the manuscript. All authors have read and approved the submitted manuscript.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-31764-9.
References
- 1.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Grunewald M, Perlman S. coronaviruses: an updated overview of their replication and pathogenesis. In: Maier HJ, Bickerton E, editors. Coronaviruses: Methods and Protocols. Springer US; 2020. pp. 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim YX, Ng YL, Tam JP, Liu DX. Human coronaviruses: a review of virus-host interactions. Diseases. 2016;4:26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren LL, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin. Med. J. (Engl.) 2020;133:1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, et al. A comparative overview of COVID-19, MERS and SARS: review article. Int. J. Surg. 2020;81:1–8. doi: 10.1016/j.ijsu.2020.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naqvi AAT, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redondo N, Zaldívar-López S, Garrido JJ, Montoya M. SARS-CoV-2 accessory proteins in viral pathogenesis: knowns and unknowns. Front. Immunol. 2021;12:708264. doi: 10.3389/fimmu.2021.708264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marra MA, et al. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 11.Vakulenko Y, Deviatkin A, Drexler JF, Lukashev A. Modular evolution of coronavirus genomes. Viruses. 2021;13:1270. doi: 10.3390/v13071270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitinger U, Farag NS, Sticht H, Breitinger HG. Viroporins: Structure, function, and their role in the life cycle of SARS-CoV-2. Int. J. Biochem. Cell Biol. 2022;145:106185. doi: 10.1016/j.biocel.2022.106185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez ME, Carrasco L. Viroporins. FEBS Lett. 2003;552:28–34. doi: 10.1016/S0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 14.Nieva JL, Madan V, Carrasco L. Viroporins: structure and biological functions. Nat. Rev. Microbiol. 2012;10:563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott C, Griffin S. Viroporins: structure, function and potential as antiviral targets. J. Gen. Virol. 2015;96:2000–2027. doi: 10.1099/vir.0.000201. [DOI] [PubMed] [Google Scholar]
- 16.Chen SC, Lo SY, Ma HC, Li HC. Expression and membrane integration of SARS-CoV E protein and its interaction with M protein. Virus Genes. 2009;38:365–371. doi: 10.1007/s11262-009-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimenez-Guardeno JM, et al. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10:e1004320. doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao Y, Tam JP, Liu DX. Viroporin activity of SARS-CoV E protein. Adv. Exp. Med. Biol. 2006;581:199–202. doi: 10.1007/978-0-387-33012-9_34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regla-Nava JA, et al. Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J. Virol. 2015;89:3870–3887. doi: 10.1128/JVI.03566-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruch TR, Machamer CE. The coronavirus E protein: assembly and beyond. Viruses. 2012;4:363–382. doi: 10.3390/v4030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teoh KT, et al. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell. 2010;21:3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verdia-Baguena C, et al. Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology. 2012;432:485–494. doi: 10.1016/j.virol.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson L, Gage P, Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353:294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michel CJ, Mayer C, Poch O, Thompson JD. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020;17:131. doi: 10.1186/s12985-020-01402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, et al. Understanding the role of SARS-CoV-2 ORF3a in viral pathogenesis and COVID-19. Front Microbiol. 2022;13:854567. doi: 10.3389/fmicb.2022.854567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castano-Rodriguez C, et al. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018 doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kern DM, et al. Cryo-EM structure of the SARS-CoV-2 3a ion channel in lipid nanodiscs. bioRxiv. 2021;9:439. [Google Scholar]
- 29.Chan CM, et al. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009;41:2232–2239. doi: 10.1016/j.biocel.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law PTW, et al. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 2005;86:1921–1930. doi: 10.1099/vir.0.80813-0. [DOI] [PubMed] [Google Scholar]
- 32.Kanzawa N, et al. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-kappaB activation. FEBS Lett. 2006;580:6807–6812. doi: 10.1016/j.febslet.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padhan K, Minakshi R, Towheed MAB, Jameel S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J. Gen. Virol. 2008;89:1960–1969. doi: 10.1099/vir.0.83665-0. [DOI] [PubMed] [Google Scholar]
- 34.Freundt EC, et al. The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 2010;84:1097–1109. doi: 10.1128/JVI.01662-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kien, F., Ma, H., Gaisenband, S. D. & Nal, B. in Microbial Pathogenesis: Infection and Immunity (ed Uday Kishore and Annapurna Nayak) Ch. 3, 38–62 (Landes Bioscience and Springer Science+Business Media, 2013).
- 36.Schwarz S, et al. Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Med. 2014;80:177–182. doi: 10.1055/s-0033-1360277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwarz S, Wang K, Yu W, Sun B, Schwarz W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antiviral Res. 2011;90:64–69. doi: 10.1016/j.antiviral.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao G, et al. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell. 2021;56:427–442. doi: 10.1016/j.devcel.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qu Y, et al. ORF3a-mediated incomplete autophagy facilitates severe acute respiratory syndrome coronavirus-2 replication. Front. Cell Dev. Biol. 2021;9:716208. doi: 10.3389/fcell.2021.716208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021;7:31. doi: 10.1038/s41421-021-00268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su WQ, Yu XJ, Zhou CM. SARS-CoV-2 ORF3a induces incomplete autophagy via the unfolded protein response. Viruses. 2021;13:2467. doi: 10.3390/v13122467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu H, et al. SARS-CoV-2 viroporin encoded by ORF3a triggers the NLRP3 inflammatory pathway. Virology. 2022;568:13–22. doi: 10.1016/j.virol.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren Y, et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arshad N, et al. SARS-CoV-2 accessory proteins ORF7a and ORF3a use distinct mechanisms to downregulate MHC-I surface expression. bioRxiv. 2022;204:8. doi: 10.1073/pnas.2208525120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta S, et al. D155Y substitution of SARS-CoV-2 ORF3a weakens binding with Caveolin-1. Comput. Struct. Biotechnol. J. 2022;20:766–778. doi: 10.1016/j.csbj.2022.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Padhan K, et al. Severe acute respiratory syndrome coronavirus ORF3a protein interacts with caveolin. J. Gen. Virol. 2007;88:3067–3077. doi: 10.1099/vir.0.82856-0. [DOI] [PubMed] [Google Scholar]
- 47.Yue Y, et al. SARS-coronavirus open reading frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018;9:904. doi: 10.1038/s41419-018-0917-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azad GK, Khan PK. Variations in ORF3a protein of SARS-CoV-2 alter its structure and function. Biochem. Biophys. Rep. 2021;26:100933. doi: 10.1016/j.bbrep.2021.100933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bianchi M, Borsetti A, Ciccozzi M, Pascarella S. SARS-CoV-2 ORF3a: mutability and function. Int. J. Biol. Macromol. 2021;170:820–826. doi: 10.1016/j.ijbiomac.2020.12.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hassan SS, Attrish D, Ghosh S, Choudhury PP, Roy B. Pathogenic perspective of missense mutations of ORF3a protein of SARS-CoV-2. Virus Res. 2021;300:198441. doi: 10.1016/j.virusres.2021.198441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Issa E, Merhi G, Panossian B, Salloum T, Tokajian S. SARS-CoV-2 and ORF3a: nonsynonymous mutations, functional domains, and viral pathogenesis. Msystems. 2020 doi: 10.1128/mSystems.00266-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Majumdar P, Niyogi S. ORF3a mutation associated with higher mortality rate in SARS-CoV-2 infection. Epidemiol. Infect. 2020;148:e262. doi: 10.1017/S0950268820002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y, et al. Efficacy of ion-channel inhibitors amantadine, memantine and rimantadine for the treatment of SARS-CoV-2 in vitro. Viruses. 2021;13:2082. doi: 10.3390/v13102082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lebedeva NS, et al. Theoretical and experimental study of interaction of macroheterocyclic compounds with ORF3a of SARS-CoV-2. Sci. Rep. 2021;11:19481. doi: 10.1038/s41598-021-99072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dey D, et al. The effect of amantadine on an ion channel protein from Chikungunya virus. PLoS Negl. Trop. Dis. 2019;13:e0007548. doi: 10.1371/journal.pntd.0007548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duff KC, Ashley RH. The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology. 1992;190:485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- 57.Fleming DM. Managing influenza: amantadine, rimantadine and beyond. Int. J. Clin. Pract. 2001;55:189–195. [PubMed] [Google Scholar]
- 58.Griffin SD, et al. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug Amantadine. FEBS Lett. 2003;535:34–38. doi: 10.1016/S0014-5793(02)03851-6. [DOI] [PubMed] [Google Scholar]
- 59.Intharathep P, et al. How amantadine and rimantadine inhibit proton transport in the M2 protein channel. J. Mol. Graph Model. 2008;27:342–348. doi: 10.1016/j.jmgm.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 60.Jing X, et al. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10967–10972. doi: 10.1073/pnas.0804958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Torres J, et al. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007;16:2065–2071. doi: 10.1110/ps.062730007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cione E, et al. Quercetin, epigallocatechin gallate, curcumin, and resveratrol: from dietary sources to human MicroRNA modulation. Molecules. 2019;25:63. doi: 10.3390/molecules25010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panche AN, Diwan AD, Chandra SR. Flavonoids: an overview. J. Nutr. Sci. 2016;5:e47. doi: 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gligorijevic N, et al. Molecular mechanisms of possible action of phenolic compounds in COVID-19 protection and prevention. Int. J. Mol. Sci. 2021;22:12385. doi: 10.3390/ijms222212385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tungmunnithum D, Thongboonyou A, Pholboon A, Yangsabai A. Flavonoids and other phenolic compounds from medicinal plants for pharmaceutical and medical aspects: an overview. Medicines (Basel) 2018;5:93. doi: 10.3390/medicines5030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abba Y, Hassim H, Hamzah H, Noordin MM. Antiviral activity of resveratrol against human and animal viruses. Adv. Virol. 2015;2015:184241. doi: 10.1155/2015/184241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campagna M, Rivas C. Antiviral activity of resveratrol. Biochem. Soc. Trans. 2010;38:50–53. doi: 10.1042/BST0380050. [DOI] [PubMed] [Google Scholar]
- 68.Chen X, et al. Insights into the anti-inflammatory and antiviral mechanisms of resveratrol. Mediat. Inflamm. 2022;2022:7138756. doi: 10.1155/2022/7138756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drago L, Nicola L, Ossola F, De Vecchi E. In vitro antiviral activity of resveratrol against respiratory viruses. J. Chemother. 2008;20:393–394. doi: 10.1179/joc.2008.20.3.393. [DOI] [PubMed] [Google Scholar]
- 70.Jennings MR, Parks RJ. Curcumin as an antiviral agent. Viruses. 2020;12:1242. doi: 10.3390/v12111242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moghadamtousi SZ, et al. A review on antibacterial, antiviral, and antifungal activity of curcumin. Biomed. Res. Int. 2014;2014:186864. doi: 10.1155/2014/186864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pecheur EI. Curcumin against hepatitis C virus infection: Spicing up antiviral therapies with ‘nutraceuticals’? Gut. 2014;63:1035–1037. doi: 10.1136/gutjnl-2013-305646. [DOI] [PubMed] [Google Scholar]
- 73.Thimmulappa RK, et al. Antiviral and immunomodulatory activity of curcumin: a case for prophylactic therapy for COVID-19. Heliyon. 2021;7:e06350. doi: 10.1016/j.heliyon.2021.e06350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hayati RF, et al. [6]-Gingerol inhibits chikungunya virus infection by suppressing viral replication. Biomed. Res. Int. 2021;2021:6623400. doi: 10.1155/2021/6623400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oso BJ, Adeoye AO, Olaoye IF. Pharmacoinformatics and hypothetical studies on allicin, curcumin, and gingerol as potential candidates against COVID-19-associated proteases. J. Biomol. Struct. Dyn. 2022;40:389–400. doi: 10.1080/07391102.2020.1813630. [DOI] [PubMed] [Google Scholar]
- 76.Aboubakr HA, et al. In vitro antiviral activity of clove and ginger aqueous extracts against feline calicivirus, a surrogate for human norovirus. J. Food Prot. 2016;79:1001–1012. doi: 10.4315/0362-028X.JFP-15-593. [DOI] [PubMed] [Google Scholar]
- 77.Chang JS, Wang KC, Yeh CF, Shieh DE, Chiang LC. Fresh ginger (Zingiber officinale) has anti-viral activity against human respiratory syncytial virus in human respiratory tract cell lines. J. Ethnopharmacol. 2013;145:146–151. doi: 10.1016/j.jep.2012.10.043. [DOI] [PubMed] [Google Scholar]
- 78.Kaushik S, Jangra G, Kundu V, Yadav JP, Kaushik S. Anti-viral activity of Zingiber officinale (Ginger) ingredients against the Chikungunya virus. Virusdisease. 2020;31:270–276. doi: 10.1007/s13337-020-00584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Breitinger U, et al. Cell viability assay as a tool to study activity and inhibition of hepatitis C p7 channels. J. Gen. Virol. 2021 doi: 10.1099/jgv.0.001571. [DOI] [PubMed] [Google Scholar]
- 80.Breitinger U, Ali NKM, Sticht H, Breitinger HG. Inhibition of SARS CoV envelope protein by flavonoids and classical viroporin inhibitors. Front. Microbiol. 2021;12:692423. doi: 10.3389/fmicb.2021.692423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salom D, Hill BR, Lear JD, DeGrado WF. pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Breitinger U, Farag NS, Ali NK, Breitinger HG. Patch-clamp study of hepatitis C p7 channels reveals genotype-specific sensitivity to inhibitors. Biophys. J. 2016;110:2419–2429. doi: 10.1016/j.bpj.2016.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nieto-Torres JL, et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Siu KL, et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Griffin SD, et al. Genotype-dependent sensitivity of hepatitis C virus to inhibitors of the p7 ion channel. Hepatology. 2008;48:1779–1790. doi: 10.1002/hep.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kongpichitchoke T, Hsu J-L, Huang T-C. Number of hydroxyl groups on the B-ring of flavonoids affects their antioxidant activity and interaction with phorbol ester binding site of PKCδ C1B domain: in vitro and in silico studies. J. Agric. Food Chem. 2015;63(18):4580–4586. doi: 10.1021/acs.jafc.5b00312. [DOI] [PubMed] [Google Scholar]
- 87.Kumar S, Pandey AK. Chemistry and biological activities of flavonoids: an overview. Sci.WorldJ. 2013;2013:162750. doi: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Farag NS, Breitinger U, El-Azizi M, Breitinger HG. The p7 viroporin of the hepatitis C virus contributes to liver inflammation by stimulating production of Interleukin-1β. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:712–720. doi: 10.1016/j.bbadis.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 89.Rattis BAC, Ramos SG, Celes MRN. Curcumin as a potential treatment for COVID-19. Front. Pharmacol. 2021;12:675287. doi: 10.3389/fphar.2021.675287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kanjanasirirat P, et al. High-content screening of Thai medicinal plants reveals Boesenbergia rotunda extract and its component Panduratin A as anti-SARS-CoV-2 agents. Sci. Rep. 2020;10:19963. doi: 10.1038/s41598-020-77003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Agrawal PK, Agrawal C, Blunden G. Quercetin: antiviral significance and possible COVID-19 integrative considerations. Nat. Prod. Comm. 2020 doi: 10.1177/1934578X20976293. [DOI] [Google Scholar]
- 92.Tahmasebi S, et al. Immunomodulatory effects of nanocurcumin on Th17 cell responses in mild and severe COVID-19 patients. J. Cell Physiol. 2020;236:5325–5338. doi: 10.1002/jcp.30233. [DOI] [PubMed] [Google Scholar]
- 93.Bhowmik D, et al. Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infect. Genet. Evol. 2020;84:104451. doi: 10.1016/j.meegid.2020.104451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mahrosh HS, Mustafa G. An in silico approach to target RNA-dependent RNA polymerase of COVID-19 with naturally occurring phytochemicals. Environ. Dev. Sustain. 2021;23:16674–16687. doi: 10.1007/s10668-021-01373-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.