Abstract
Purpose:
Cisplatin (CDDP)-based chemotherapy is a first-line treatment for patients with advanced head and neck squamous cell carcinomas (HNSCC), despite a high rate of treatment failures, acquired resistance, and subsequent aggressive behavior. The purpose of this study was to study the mechanism of CDDP resistance and metastasis in HNSCC. We investigated the role of NRF2 pathway activation as a driven event for tumor progression and metastasis of HNSCC.
Experimental Design:
Human HNSCC cell lines that are highly resistant to CDDP were generated. Clonogenic survival assays and a mouse model of oral cancer were used to examine the impact of NRF2 activation in vitro and in vivo on CDDP sensitivity and development of metastasis. Western blotting, immunostaining, whole-exome sequencing, single-cell transcriptomic and epigenomic profiling platforms were performed to dissect clonal evolution and molecular mechanisms.
Results:
Implantation of CDDP-resistant HNSCC cells into the tongues of nude mice resulted in a very high rate of distant metastases. The CDDP-resistant cells had significantly higher expression of NRF2 pathway genes in the presence of newly acquired KEAP1 mutations, or via epigenomic activation of target genes. Knockdown of NRF2 or restoration of the wild-type KEAP1 genes resensitized resistant cells to CDDP and decreased distant metastasis (DM). Finally, treatment with inhibitor of glutaminase-1, a NRF2 target gene, alleviated CDDP resistance.
Conclusions:
CDDP resistance and development of DM are associated with dysregulated and epigenetically reprogrammed KEAP1–NRF2 signaling pathway. A strategy targeting KEAP1/NRF2 pathway or glutamine metabolism deserves further clinical investigation in patients with CDDP-resistant head and neck tumors.
Translational Relevance.
For more than three decades, cisplatin (CDDP) remains the first-line systemic agent for the treatment of advanced HNSCC and other solid tumors, despite the high prevalence of intrinsic and extrinsic CDDP resistance among patients with these cancer types. Better understanding of how tumor cells evolve into therapy-resistant populations is principal to overcoming treatment failure and maximizing disease control and survival. Here, we demonstrate that hyperactivated KEAP1/NRF2 signaling by CDDP exposure leads not only to acquisition of CDDP resistance, but also an enhanced metastatic phenotype in HNSCC preclinical models which is at least in part supported by either inactivating mutations of KEAP1 or epigenetic reprogramming by NRF2. Our study is an important step towards identifying novel therapeutic strategies to enhance CDDP effectiveness in HNSCC.
Introduction
Head and neck squamous cell carcinoma (HNSCC) not associated with human papillomavirus (HPV) infection remains a leading cause of cancer deaths worldwide with 202,000 cases/year and an estimated 5-year survival of 50% which has not changed for decades (1). Cisplatin (CDDP)-based chemotherapy is used in the treatment of the majority of patients with advanced stage HNSCC despite the introduction of immunotherapy and targeted agents into clinical practice (2). However, CDDP resistance, both intrinsic and acquired, is frequently encountered in clinical practice and has been linked to treatment failure and development of distant metastasis (DM; ref. 3). Therefore, it is critical to understand how tumor cells survive the stress of CDDP or evolve into therapy-resistant populations. We sequenced the TP53 gene from surgical specimens obtained from patients enrolled in clinical trial NRG/RTOG-0234 who were treated surgically for stage II or IV HPV-negative HNSCC with high-risk pathologic features. The patients received radiation plus cetuximab and either CDDP or docetaxel as adjuvant treatment. In this study, patients treated with CDDP demonstrated a higher rate of DM and death from disease in the context of wild-type (WT) TP53 (4). In contrast, this effect was not observed in patients treated with docetaxel indicating that it is a chemotherapy-specific event (4).
These preliminary data led us to hypothesize that head and neck tumors with wtTP53 can develop CDDP resistance and enhanced metastatic potential. To test this hypothesis, we developed CDDP-resistant HNSCC cell lines with WT p53 in vitro (5). Orthotopic implantation of these cells into the tongues of nude mice resulted in a very high rate of DMs. RNA sequencing (RNA-seq) performed in CDDP tumors harvested from the in vivo mouse model showed substantial upregulation of KEAP1/NRF2-dependent genes. Numerous studies have demonstrated that NRF2 (which is generally referred to as NFE2L2) is upregulated by chemotherapeutic drugs including CDDP (6, 7). KEAP1 negatively regulates NRF2, a transcription factor which binds to antioxidant response elements (ARE) on the DNA and initiates the transcription of a number of downstream target genes involved in regulation of redox balance and cellular detoxification (8). Furthermore, whole-exome sequencing (WES) has demonstrated that KEAP1 is mutated at two sites in its BTB (Broad complex, Tramtrack and Bric-a-Brac) domain in the CDDP-resistant cells. The BTB domain mediates binding of KEAP1 to CUL3 and NRF2 (8), suggesting that these mutations are loss-of-function (LOF) mutations which abrogate KEAP1 protein function, thereby activating NRF2 signaling. We also show that short hairpin RNA (shRNA) targeted suppression of NRF2 or overexpression of KEAP1 reversed CDDP resistance in vitro and in vivo in the CDDP-resistant HNSCC cell lines through inhibition of proliferation and induction of ferroptotic cell death. An in vitro single-cell RNA sequencing (scRNA-seq) and single-cell Assay for Transposase-Accessible Chromatin using sequencing (scATAC-seq) analyses provided evidence that most of the KEAP1/NRF2 genes were epigenetically changed in a specific subpopulation of CDDP-resistant cells. Glutamate is the main precursor for glutathione (GSH) synthesis, which is the major cellular antioxidant for maintaining redox homeostasis. Increased NRF2 expression protects tumor cell DNA from intracellular reactive oxygen species (ROS)-mediated damage, in part through NRF2-mediated upregulation of glutamate-cysteine ligase subunits (GCLC and GCLM), the enzymes that catalyze the production of GSH from glutamate (9). Because of this cellular glutamine addiction, glutaminase-1 (GLS1), the rate-limiting enzyme that converts glutamine to glutamate, has emerged as a promising therapeutic target (10). In this study, we demonstrate that the KEAP1/NRF2-mediated CDDP resistance can be overcome in vitro and in vivo with the use of a novel potent, selective, and orally bioavailable GLS1 inhibitor, IACS-6274 (11). This drug is currently under early clinical testing in human solid tumors (NCT03894540; ref. 12).
Materials and Methods
Cell lines and cell culture
The HNSCC parental HN30-P cell line was obtained from an established cell line repository in the laboratory of J.N. Myers (University of Texas MD Anderson Cancer Center) under approved institutional protocols. CDDP-resistant cell line HN30-R8 made from the parental HN30 was obtained from the Laboratory of Dr. Vlad Sandulache (Baylor College of Medicine) in February 2019. The HN30-R8 cell line was established by gradually increasing the CDDP concentration in culture media over 21 weeks following initial CDDP treatment according to a published protocol (5). Cell lines were cultured in DMEM (Gibco), containing 10% FBS, l-glutamine, sodium pyruvate, nonessential amino acids, and vitamin solution. All cell lines were maintained at 37°C in a humidified incubator containing 5% CO2 and periodically tested to ensure a Mycoplasma-free culture environment. All experiments were performed using cells from early passages. The HNSCC cell lines were authenticated using short tandem repeat analysis (13) within 6 months of use for the current study.
Generation of NRF2 knockdown and WT KEAP1 overexpressing stable cell lines
The scrambled control lentiviral shRNA and two different lentiviral NRF2 shRNA particles (sc-37030-V and sc-44332-V), and sc-108060 control shRNA plasmid-A, were all purchased from Santa Cruz Biotech (California). The pLVX-Puro-CMV WT hKEAP1 lentiviral plasmid (Vector ID: VB900000–0026bfu) was obtained from VectorBuilder. The pLVX-M-puro, plasmid #125839 was purchased from Addgene. HN30-R8 cells stably expressing these plasmids were generated as previously described (14), and details of the experiment were described in Supplementary Materials and Methods section.
Clonogenic survival assay
To determine colony formation, HN30-P and HN30-R8 and their derivative cells were seeded in 6-well plates (3 wells/treatment condition) at predetermined densities set for each cell line and exposed to different fixed-ratios of CDDP (0–20 μmol/L) or IACS-6274 (0–5.0 μmol/L). Controls (untreated) received 0.1% DMSO made in culture medium equivalent to the DMSO percentage in other treatment conditions. Cells were cultured for 10 to 14 days and clonogenic cell survival based on the IC50 values was determined as previously described (15).
Analysis of combined drug effects
The combination index (CI) and isobologram analyses evaluating drug synergism were calculated via CalcuSyn Software (Biosoft) using the Chou–Talalay method (16). Details of the experiment were described in the Supplementary Materials and Methods section.
Western blot analysis
According to the IC50 values, minimally toxic and physiologically relevant drug doses of CDDP and IACS-6274 were identified. Briefly, HNSCC cells were treated in 10-cm dishes with CDDP (10 μmol/L) and IACS-6738 (10 μmol/L) either alone or in combination for 12, 24, and 48 hours. Whole cell lysates were prepared, and Western blot analyses were conducted as described previously (15). Antibodies used for Western blotting were described in Supplementary Materials and Methods section.
Incucyte live scratch wound cell migration and invasion assay
Briefly, HN30-P and HN30-R8 cells were grown in a 96-well plate (3×103/well) and treated with various doses of CDDP (0.63–10 μmol/L) for 24 hours. Incucyte Live Scratch Wound and Invasion Assay was performed as described in Supplementary Materials and Methods section.
Orthotopic nude mouse model of oral cancer metastasis and therapy
All animal experimentation was approved by the Institutional Animal Care and use Committee of the University of Texas MD Anderson Cancer Center. Our orthotopic nude mouse tongue model was previously described in the literature (15, 17, 18). To determine the CDDP-resistant HNSCC cell growth kinetics and their response to CDDP in vivo, cells (5×104) were suspended in 30 μL of PBS and injected into the lateral tongues of male athymic nude mice because oral cancer is mostly prevalent in male patients. Mice were then randomized into different groups 8 to 10 days after injection and treatment with vehicle or CDDP (4 mg/kg, i.v., via tail-vein for 4 weeks) was started when tumors reached a range of 3 to 6 mm3 in size. For the in vivo drug combination study, the HN30-R8 CDDP-resistant cells were injected into the mice oral tongues as described above, and treated with either CDDP (4 mg/kg, i.v., once a week) alone, the glutaminase inhibitor, IACS-6274 (100 mg/kg dissolved in methylcellulose, oral gavage, twice a day daily, 5 days/week) alone, or their combination or vehicle (0.5% methylcellulose) for 4 weeks. A total of 10 to 12 mice in each group were used, and 90% of the mice had tumor growth under each condition. Mice were monitored twice a week and their weight and tumor volume were recorded. Tongue tumors were measured with microcalipers, and tumor volume was calculated as A × B2 × (π/6), where A is the longest dimension of the tumor and B is the dimension of the tumor perpendicular to A. Mice were euthanized when they lost more than 20% of their preinjection body weight. During necropsy, cervical lymph nodes and lungs were harvested, formalin-fixed, and subjected to histologic evaluation using hematoxylin and eosin (H&E) staining to identify and count the number and size of metastases. Lymph node and lung metastatic tumor sections from mice injected with HN30-R8 cells were subjected to immunostaining with human leukocyte antigen (HLA) antibody and counterstained with DAPI to confirm the human cell origin of the metastases in these sections.
Mice xenograft bulk RNA-seq
HNSCC cell lines, HN30-P and HN30-R8, were injected in oral tongues of mice and treated with CDDP as described above. When significant CDDP response and tumor growth difference were seen among the treatment groups (at day 42), mice (N = 3) were sacrificed and primary tumors from each group were obtained and frozen in liquid nitrogen. Total RNA was isolated from tumor samples using the RNeasy mini kit reagents (QIAGEN) according to the manufacturer's instructions. For in vivo RNA-seq, total RNA was submitted to the Sequencing and Microarray Facility at MD Anderson Cancer Center for next-generation sequencing (Illumina HiSeq). RNA integrity and confirmation of a lack of genomic DNA contamination was confirmed with Xenome tool classification. Quality control (QC) was then performed using FastQC and FastQ Screen. Sequencing reads were aligned to human reference genome (GRCh38), using TOPHAT2 and its refined classification approach, Xenome. Read counts that aligned to a transcript for each gene region and accurate transcript quantification were calculated using RSEM method. Weakly expressed and noninformative (non-aligned) genes were filtered out before normalization. Data were filtered and then transformed to log2 counts per million and principal component analysis was performed in the R-statistical analysis. Differentially expressed genes (DEG) were identified using linear model likelihood ratio and ANOVA-like tests implemented in the edgeR package with Benjamini–Hochberg multiple testing correction. Significance was set on the basis of the FDR cutoff of 0.05; the significant genes were based on the ANOVA-like test results. Filtering criteria include selection of DEG with an adjusted P value of < 0.05, keeping genes that have at least 50 average reads and 2-fold change in the higher of the two comparison groups (CDDP versus untreated controls), and exclusion of noncoding RNAs. A CDDP resistance gene signature was unbiasedly identified by considering the genes (i.e., up- or downregulated among the groups) based on the above statistical criteria for HN30-R8 cells compared with the parental HN30-P cells. Significant genes regulated by CDDP resistance were analyzed and depicted in the cluster heatmaps.
The Cancer Genome Atlas-HNSCC survival and functional annotation analyses
Transcriptome data on 314 HPV-negative oral cavity squamous cell carcinoma (OCSCC) from The Cancer Genome Atlas (TCGA) was obtained from NCI Genomic Data Commons and analyzed for correlation of the upregulated KEAP1/NRF2/CUL3 (KNC) genes with survival. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of the upregulated KNC genes were performed (19) and details of analyses were described in Supplementary Materials and Methods section.
WES and mutational calling
WES was performed using the Illumina protocol at MD Anderson Cancer Center. Genomic DNA was extracted from HN30-P and HN30-R8 cell lines (1×106 to 2×106) using the QIAamp DNA Kit (QIAGEN) according to the manufacturer's instructions. The WES was carried out as described previously (20–23). The details of the experiment were described in Supplementary Materials and Methods section.
Sanger sequencing
To validate the KEAP1 mutations obtained from the WES, specific flanking intronic primer pairs (KEAP1 forward: 5-ACACTGCAGGTCAAGTACCAG-3; KEAP1 reverse: 5-AGGGATGGACGAGTGCTCATC-3) for the selected KEAP1 mutations were designed using the Primer3 algorithm8 tool. Details of the experiment were described in Supplementary Materials and Methods section.
GSH and ROS levels
Briefly, total GSH reduced was measured after treatment of HN30-P and HN30-R8 cells with IACS-6274 (2.5 μmol/L) for 24 hours as indicated and collected according to the manufacturer's protocol (Promega, V6611). Experimental details were described in Supplementary Materials and Methods section.
Glutamine and glutamate levels in CDDP-resistant HNSCC xenograft model
To determine that the IACS-6274 could inhibit glutaminolysis and engage its targets in vivo, tumors were harvested from mice at 8 or 24 hours following treatment with single-agent IACS-6274 (100 mg/kg/mice) as indicated and snap-frozen until use. Details of the experiments were described in Supplementary Materials and Methods section.
Oil-red staining and ferroptosis experiment
For lipid accumulation, HN30-R8 cells expressing pLenti plasmid with non-targeting control, NRF2 shRNA, and WT hKEAP1 cells were treated with 5 μmol/L CDDP. Oil-red staining experiment was described in detail in Supplementary Materials and Methods section.
In vitro scRNA-seq and scATAC-seq analyses
The HNSCC cells (HN30-P, HN30-R8) were treated with CDDP (2.5 μmol/L) for 48 hours in triplicates and cell suspensions were subjected to scRNA-seq and scATAC-seq analyses as previously described (24–33). Our sequencing library averaged 3,000 cells per sample, 6.5k genes/50k UMIs/115k reads per cell detected. Cell Ranger ATAC tool was used to preprocess (alignment, trimming, and identification of transpose cut sites) the fastq files. We selected only the high-quality cells, in terms of their transcription start site (TSS) scores and number of unique fragments. After the dimensionality reduction (LSI method) and normalization (TF-IDF), we used a graph-based clustering and UMAP which identified 7 unique clusters. Details of the analyses were described in the Supplementary Materials and Methods.
Statistical analysis
The Student t and one-way ANOVA tests were used to analyze in vitro data. For in vivo mouse studies, a two-way ANOVA test was used to compare tumor volumes between control and treatment groups. Survival following drug treatment was analyzed by the Kaplan–Meier method and compared with the log-rank test. All data were expressed as mean ± standard error, and P values of 0.05 or less were considered statistically significant. Sample categorization into “Low” or “High” KNC gene expression was determined by ROC analysis and Youden's Index calculation, considering the 5-year overall survival (OS) as the variable of interest. Associations between categorical variables and survival were determined by the log-rank test, considering a 5-year follow-up interval. For continuous variables, univariate Cox regression analysis was employed and the significance was established by the Wald and likelihood ratio tests. Differences in gene expression between groups were accessed by the Wilcoxon test and were considered significant when P values were lower than 0.05.
Data availability statement
The data generated in this study are available within the article and its Supplementary Data files. Data used in this study that is not included in the paper or supplementary files can be made available upon request from the corresponding author.
Results
Acquired cisplatin resistance enhances tumor formation and DM in vivo in an orthotopic mouse model of oral tongue cancer
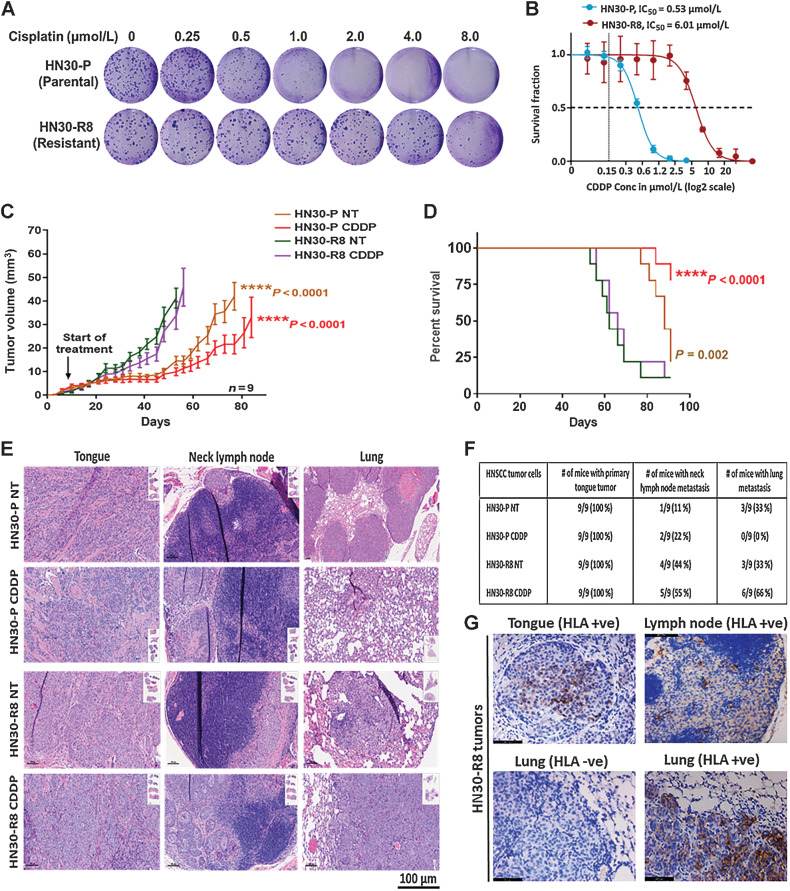
To identify molecules and pathways other than TP53 mediating CDDP resistance and metastases, the HN30-R8 CDDP-resistant HNSCC cell lines were initially generated from an established HNSCC cell line with WT TP53 status (HN30). After chronic growth in escalating CDDP concentrations, the cell line was able to grow in 4 to 10 μmol/L CDDP, at which point a heterogeneous pool of cells (HN30-R8) was created and used for further analyses. These values are approximately 10- to 20-fold greater than the previously described IC50 values for CDDP in a panel of HNSCC cell lines (18). The cells were maintained in growth media containing 8 μmol/L CDDP during that time period, except when CDDP was withdrawn for specific individual experiments. The degree of sensitivity to CDDP treatment in HN30-R8 cells was evaluated using clonogenic survival assay as previously described. HN30-R8 cells demonstrated decreased sensitivity to CDDP and enhanced colony formation (IC50 = 6.0–8.0 μmol/L) compared with their CDDP sensitive HN30-P (parental) cells (IC50 = 0.53 μmol/L; Fig. 1A and B). In addition, a live movie of the Incucyte wound scratch assay showed that CDDP exposure increased the rate of migration in HN30-R8 cells compared with their HN30-P sensitive counterparts (Supplementary Fig. S1). In vivo resistance of HN30-R8 cells was confirmed upon xenotransplantation in nude mice where CDDP treatment had no effect on the growth of HN30-R8 and failed to improve survival of mice. In comparison, CDDP treatment of animals with HN30-P (parental) tumors led to substantial tumor reduction and improved survival following CDDP administration (Fig. 1C and D). Mice harboring HN30-R8 tumors had elevated rates of regional lymph node metastasis and DM to lungs, with the highest DM in CDDP-treated HN30-R8–bearing mice (66%) compared with parental HN30-P treated with CDDP (0%; Fig. 1E and F). Immunostaining with human anti-HLA antibody conformed the human cell origin of the metastases in the lymph nodes and lungs and differentiated them from the possibility of spontaneous mouse lymphomas (Fig. 1G). Taken together, these data suggest that acquired CDDP resistance in HNSCC may be linked to treatment failure and the development of DM in clinical practice.
Figure 1.
Acquired cisplatin resistance enhances tumor formation and DM in vivo in an orthotopic mouse model of oral tongue cancer. HN30-P and HN30-R8 cells were plated on 6-well dishes, treated with various concentrations of CDDP in vitro, and subjected to clonogenic survival assays as indicated. A and B, Representative images of clonogenic survival and curves indicating decreased cisplatin sensitivity in CDDP-resistant HN30-R8 cells. HN30-P and its CDDP-resistant derivative, HN30-R8, were orthotopically injected into the tongues of male athymic nu/nu mice and treated intravenously via tail injection with 4 mg/kg of CDDP for 4 weeks. Tumor growth was routinely monitored with a standard caliper and is reported as tumor volume means ± SEM. C, Tumor growth curves calculated after 4 weeks of injection. Statistical analysis was performed by a one-way ANOVA test. ****, P < 0.0001 CDDP resistant versus sensitive. D, Kaplan–Meier survival curve for OS of mice injected with HN30 parental or CDDP-resistant cells with and without CDDP treatment. E, H&E representative images of the primary tongue, lymph, and lung metastatic tumors. F, Percentage of tumor incidence in tongue, neck lymph nodes, and lungs in mice treated with CDDP. G, Representative images of positive human HLA immunostaining confirming the human cell origin of the metastases in the lymph nodes and lungs. The HLA antibody staining is shown in brown and DAPI counterstain is shown in blue.
Acquisition of cisplatin resistance is accompanied by dysregulation of the KNC pathway in vivo in HNSCC
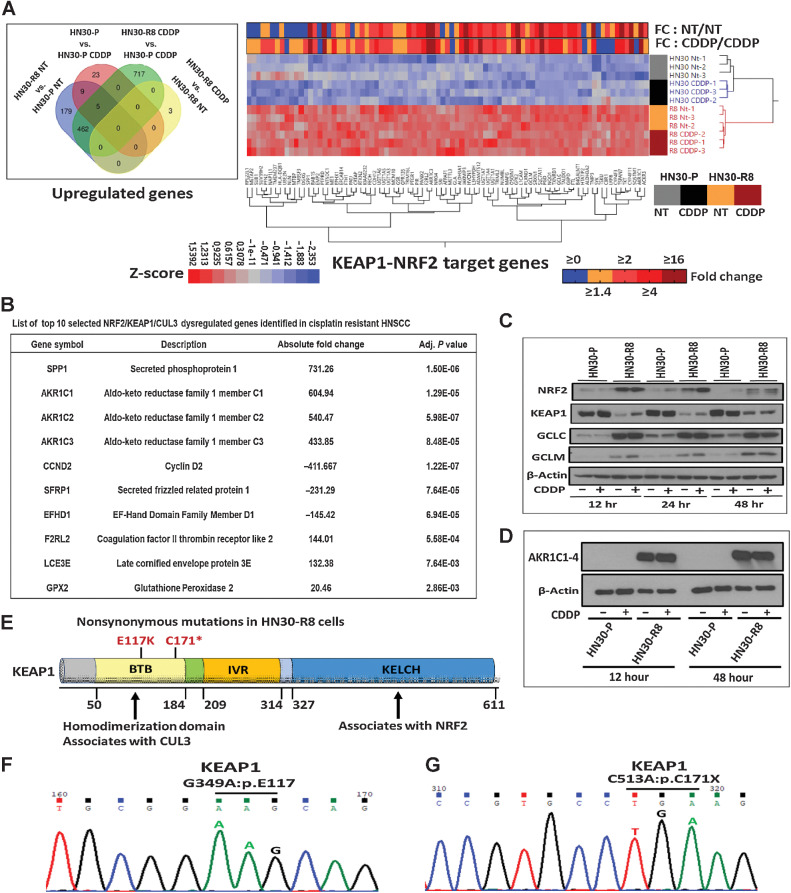
To gain insight into mechanisms of acquired CDDP resistance, we performed unbiased global enrichment genes analysis (i.e., RNA-seq) on primary tumor specimens from mice with HN30-R8 or HN30-P tumors in the presence and absence of CDDP treatment. A CDDP resistance gene signature was unbiasedly identified by taking the genes (i.e., up- or downregulated among the groups) with a Benjamini–Hochberg adjusted P value (q value) less than or equal to 0.05 and a fold change greater than 1.5 for HN30-R8 cells compared with the parental HN30-P cells as described in Materials and Methods. The Venn diagram showed that 462 genes were upregulated basally in untreated (NT) HN30-R8 versus untreated HN30-P that remained upregulated in HN30-R8 CDDP resistant versus HN30-P CDDP treated (Fig. 2A). Approximately, 717 genes were upregulated in HN30-R8 CDDP treated versus HN30-P CDDP treated (Supplementary Table S1). Only 3 genes were more highly expressed in the HN30-R8 after treatment with CDDP (Fig. 2A). No significant increase in gene expression was found in parental HN30-P tumors from mice that received CDDP compared with HN30-P tumors from mice that received only vehicle. However, 85 KNC target genes were differentially expressed and ranked among the top significantly upregulated genes in the list (LogFC >1.5; adj P < 0.05) in the HN30-R8 versus HN30-P tumors, irrespective of CDDP treatment (Fig. 2A). The heatmap shows Z-scores of FPKM expression for KNC genes and upper annotation denotes average fold change of HN30-R8 versus HN30-P untreated (NT) or with CDDP. Because CDDP resistance altered the expression of KEAP1/NRF2 axis genes, we focused on the top 10 regulated genes for further analysis and characterization (Fig. 2B). Western blotting confirmed that total protein levels of NRF2 and its downstream targets GCLC, GCLM, and AKR1C1–3 were highly elevated in HN30-R8 and KEAP1 protein expression severely diminished compared with parental HN30-P (Fig. 2C and D). Expression levels of KEAP1 and NRF2 proteins did not change upon treatment with CDDP in the cells tested (Fig. 2C and D). To examine if the NRF2 target genes are upregulated during metastasis, five formalin-fixed, paraffin-embedded (FFPE) metastatic tumors from the HN30-R8 CDDP-treated group were subjected to RNA-seq according to published method (34, 35) and described in the Supplementary Materials and Methods. Only 8 of the 85 previously NRF2-regulated genes were significantly upregulated in distant metastatic HN30-R8 tumors compared with primary tumors (Supplementary Table S2).
Figure 2.
Acquisition of cisplatin resistance is accompanied by dysregulation of the KNC pathway in vivo in HNSCC. Primary tongue tumor tissues were obtained from mice injected with HN30-P (parental) and HN30-R8 cell lines untreated or treated with CDDP and subjected to RNA-seq analysis as indicated previously. A, Venn diagram showed 462 genes were upregulated basally in untreated (NT) HN30-R8 vs. untreated HN30-P that remained upregulated in resistant cells plus CDDP versus parental cells plus CDDP. A total of 717 genes were upregulated in CDDP-treated HN30-R8 versus HN30-P cells. Only 3 genes were upregulated in HN30-R8 cells treated with CDDP. Approximately 85 KNC target genes were upregulated in the HN30-R8 versus HN30-P cells, irrespective of CDDP treatment. Heatmap shows Z-scores of FPKM expression for 85 KNC genes significantly enriched and upper annotation denotes average fold change of HN30-R8 versus parental HN30-P untreated (NT) or with CDDP-treated cells. B, Table showing the top 10 selected genes including KEAP1/NRF2-regulated genes and their functions in mice tumors. C and D, Expression of KEAP1/NRF2 selected genes validated by Western blot analysis. E, Positions of nonsynonymous mutations on the functional domains of KEAP1 detected in the CDDP-resistant HN30-R8 but not in the parental HN30-P cells using WES analysis as indicated. F and G, Representative Sanger sequencing chromatograms of the KEAP1 mutations shown in E.
Somatic mutations in the KNC pathway occur in roughly 12% of HNSCC, and KEAP1 mutations (36, 37) typically disrupt interaction with NRF2 leading to activation of the latter and upregulation of its downstream target genes (38). Therefore, we performed WES to explore the possible molecular mechanism of NRF2 pathway activation in HN30-P and HN30-R8 cell lines treated with CDDP for 24 hours. We detected about 197 de novo mutations in HN30-R8 CDDP cells that were not seen in the HN30-P parental cell lines (Fig. 2E; Supplementary Table S2). No mutations in traditional tumor suppressors or oncogenes such as TP53, NOTCH1, CDKN2A, or the PIK3CA and RAS oncogenes were detected, nor in cancer drivers previously linked to HNSCC except for KEAP1. Interestingly, two distinct missense (G349A:p.E117K) and truncating stop-gain (C513A:p.C171X) mutations were detected in the BTB binding domain of KEAP1 in HN30-R8 CDDP-resistant cell lines (Fig. 2E; Supplementary Table S3). These mutations were not detected in the HN30-P parental cell lines, suggesting that they were acquired de novo under CDDP pressure or may have been present at a very low frequency in HN30 parental cells and selected for over time with acquisition of CDDP resistance. The presence of KEAP1 mutations was further validated by Sanger sequencing (Fig. 2F and G). GO and KEGG pathway analyses revealed that the top 10 selected KNC genes were significantly enriched in biological processes including daunorubicin metabolic process, doxorubicin metabolic process, steroid hormone and lipid metabolic processes, tissue and epithelium development processes, and metabolism of xenobiotics by cytochrome P450 (Supplementary Fig. S2A). The prognostic value of the top 10 KNC-gene signature was also evaluated in the TCGA-HNSCC patient cohort (N = 314) available in the cBioPortal web. Kaplan–Meier survival plots showed that high expression of this gene signature is significantly correlated with poor survival (P = 0.016 for 5-year survival, and P = 0.023 for disease-free survival) in patients with HPV-negative HNSCC (Supplementary Fig. S2B). These results clearly suggest that genes regulated by the KNC axis are good predictors of poor prognosis in patients with HNSCC, consistent with published data (39). WES has confirmed that HN30-R8 cells remained resistant and did not undergo de novo TP53 mutation during resistance selection. Therefore, we wondered whether the biological impact would be similar in the presence of a mutated TP53, given its prevalence in HNSCC. To explore potential interactions, we turned to the HNSCC TCGA dataset. Both KEAP1 and NRF2 mutations could be found in samples that were either TP53 WT or mutant (Supplementary Fig. S3A), among OCSCC and laryngeal/hypopharyngeal (LHSCC) tumors. Focusing on the OCSCC (Supplementary Fig. S3B), the TP53 status alone was a poor prognosticator of OS. Using our previously published and validated NRF2 gene signature (40), however, we found that OCSCC patients with a high NRF2 activation score had significantly worse OS compared with those with low NRF2 scores (Supplementary Fig. S3C; MS = 35.5 months vs. 65.8 months; P = 0.038). A similar trend of worse survival was found if patients were instead stratified by mutations in the NRF2 pathway, but it didn't reach statistical significance (Supplementary Fig. S3D). When both TP53 status and NRF2 activation score were considered, survival was actually worse for patients with WT TP53 among tumors with high NRF2 activation (Supplementary Fig. S3E). Nevertheless, as expected, patients with high NRF2 activation had worse OS compared with those with low NRF2 activation regardless of TP53 status. Interestingly, tumors with WT TP53 and high NRF2 activation were enriched for NRF2 mutations (Supplementary Fig. S3F and S3G; P = 0.0035), which could explain their worse prognosis. These data suggest that the main driver of poor prognosis appears to be the NRF2 activation state, regardless of TP53 status.
NRF2 targeted suppression or restoration of WT KEAP1 inhibits in vitro and in vivo tumor growth
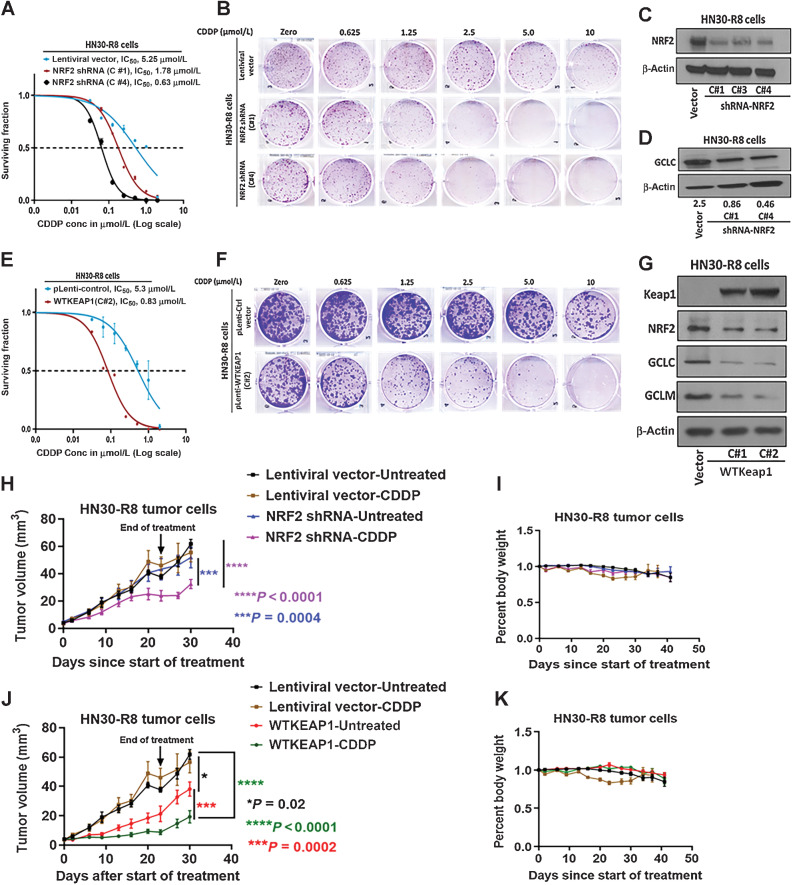
Multiple investigators have linked activation and alterations in the KEAP1/NRF2 pathway to increased tumor aggressiveness (6), reduced response to immunotherapy (41), and CDDP resistance (7). Consistent with these studies, we observed that stable shRNA knockdown (KD) of NRF2 reversed CDDP resistance in HN30-R8 cells, bringing down the IC50 for CDDP from 5.2 μmol/L (control cells infected with empty Lentiviral vector) to as low as 0.6 μmol/L after KD (Fig. 3A–C). Loss of NRF2 protein in several clones (Fig. 3C) was accompanied by a reduction in the protein levels of the downstream target GCLC (Fig. 3D). In an orthogonal approach, we further confirmed the role of altered KEAP1/NRF2 in CDDP resistance by stably restoring a KEAP WT gene to HN30-R8 cells. Infection with cDNA for WT human KEAP1 reduced the CDDP IC50 from 5.3 μmol/L (control cells infected with empty lentiviral vector) to 0.83 μmol/L (Fig. 3E and F), which was accompanied by increased expression of KEAP1 protein and reduction in NRF2, GCLC, and GCLM (Fig. 3G). To evaluate the impact of altered KEAP1/NRF2 on CDDP sensitivity in vivo, we injected the HN30-R8 stably expressing NRF2 shRNA or WT KEAP1 vectors into tongues of NUDE mice followed by CDDP treatment as indicated. Tumor growth kinetics was not significantly different between untreated NRF2 shRNA harboring and vector control cells which were treated or untreated with CDDP (Fig. 3H). However, NRF2 shRNA sensitized HN30-R8 tumors to CDDP treatment and significantly reduced tumor growth when compared with either untreated NRF2 shRNA control group (P < 0.0004) or lentiviral vector treated (P < 0.0001) or untreated control groups (P < 0.0001; Fig. 3H). This result is consistent with published data in cervical cancer cells (42). Body weight was not significantly different between treatment and control groups (Fig. 3I). Similarly, restoration of WT KEAP1 remarkably improved the response to CDDP and resulted in a significant tumor growth inhibition when compared with either untreated WT KEAP1 control group (P = 0.0002) or lentiviral vector treated (P < 0.0001) or untreated control groups (P < 0.0001; Fig. 3J). Interestingly, mice bearing tumors expressing WT KEAP1 alone also showed significant tumor growth reduction (P = 0.02; Fig. 3J) and improved mouse survival (Supplementary Fig. S4). In addition, body weight was not affected and did not differ significantly between treatment and control groups (Fig. 3K). In addition, mice with HN30-R8 tumors expressing NRF2 shRNA or WT KEAP1 showed decreased lung DM upon treatment with CDDP compared with mice with lentiviral vector control with or without CDDP (Supplementary Fig. S5A–S5B).
Figure 3.
NRF2 targeted suppression or restoration of WT KEAP1 inhibits in vitro and in vivo tumor growth. HN30-R8 cells were transduced with NRF2 shRNA and WT KEAP1 constructs and the established stable cell lines were subjected to clonogenic survival assays following CDDP treatment and compared with control transfected cells as described in Methods. A and B, Survival curves and representative clonogenic survival images showing that CDDP resistance is reversed in HN30R8 following NRF2 shRNA KD. C and D, Western blots confirming successful KD of NRF2, associated with decreased GCLC downstream target proteins in clones C#1, C#3, and C#4 respectively. E and F, Survival curves and representative clonogenic survival images showing that CDDP resistance is also reversed in HN30-R8 after restoring WT KEAP1. G, Reduction in downstream targets GCLC and GCLM following the restoration of WT KEAP1 in clones C#1 and C#2, respectively. H–K, Tumor growth curves and percent body weight loss in orthotopic mouse model during the treatment with CDDP. Shown are measurements from oral tongues bearing HN30-R8 cells stably expressing NRF2 shRNA or WT KEAP1 following treatment with CDDP (4 mg/kg) for 4 weeks as indicated. Tumor cells expressing the same lentiviral vectors were used as control and identical for experiments presented in H, I, J, and K. Each treatment group contains 8 to 10 mice. All in vivo data were expressed as ± SEM and P values < 0.05 were considered significant.
Restoration of KEAP1/NRF2 pathways increases cisplatin sensitivity in resistant HNSCC cells through ferroptosis
To determine if KD of NRF2 or forced expression of KEAP1 sensitized HN30-R8 cells to CDDP via apoptosis, the apoptotic markers, PARP and caspase-3 cleavage, were examined after 12, 24, and 48 hours of treatment with CDDP. No cleavage of PARP or caspase-3 was observed in these cells when exposed to CDDP (Supplementary Fig. S6A–S6B). Recent studies have shown that ferroptosis, a newly identified mode of programmed cell death mediated by oxidative stress and lipid peroxidation (43–45) may be triggered by CDDP (46). Therefore, we examined if acquisition of CDDP resistance resulted in decreased ferroptosis in resistant cells upon treatment with CDDP. Compared with cells with lentiviral vector, untreated resistant cells expressing NRF2 shRNA or WT KEAP1 had higher levels of lipid peroxidation and accumulation, and these levels were significantly increased following CDDP exposure (Supplementary Fig. S7A–S7B). Furthermore, Western blot analysis showed that expression of the inhibitory ferroptosis marker GPX4 (glutathione peroxidase 4) was diminished in resistant cells expressing NRF2 shRNA or WT KEAP1 compared with lentiviral control cells (Supplementary Fig. S7C). Ferroptotic cell death in HN30-R8 cells was further confirmed with the use of the ferroptosis inducer RLS3 (inhibitor of GPX4) and inhibitor of ferroptosis, ferrostatin. Consistent with published data (5), clonogenic survival assays showed that ferrostatin did not reverse CDDP toxicity in HN30-R8 cells with lentiviral vector control (Supplementary Fig. S7D and S7G), suggesting that CDDP by itself does not trigger ferroptosis. In addition, treatment with RLS3 did not result in induction of ferroptosis in these cells. However, CDDP and RLS3 were able to trigger significant ferroptosis in the cells expressing NRF2 shRNA or WT KEAP1 vectors as demonstrated by the ability of ferrostatin to reverse CDDP and RLS toxicities in these cells (Supplementary Fig. S7E–S7G). These results are consistent with a recent publication which showed that NRF2 KD or KEAP1 overexpression increased ferroptotic cell death in F98 glioma cells (47). Taken together, our data clearly indicate that upregulation of the KEAP1/NRF2 pathway is associated with resistance to ferroptosis during acquisition of CDDP resistance in HNSCC cells.
scRNA-seq revealed enrichment of a KNC gene signature in cisplatin-resistant HNSCC cells
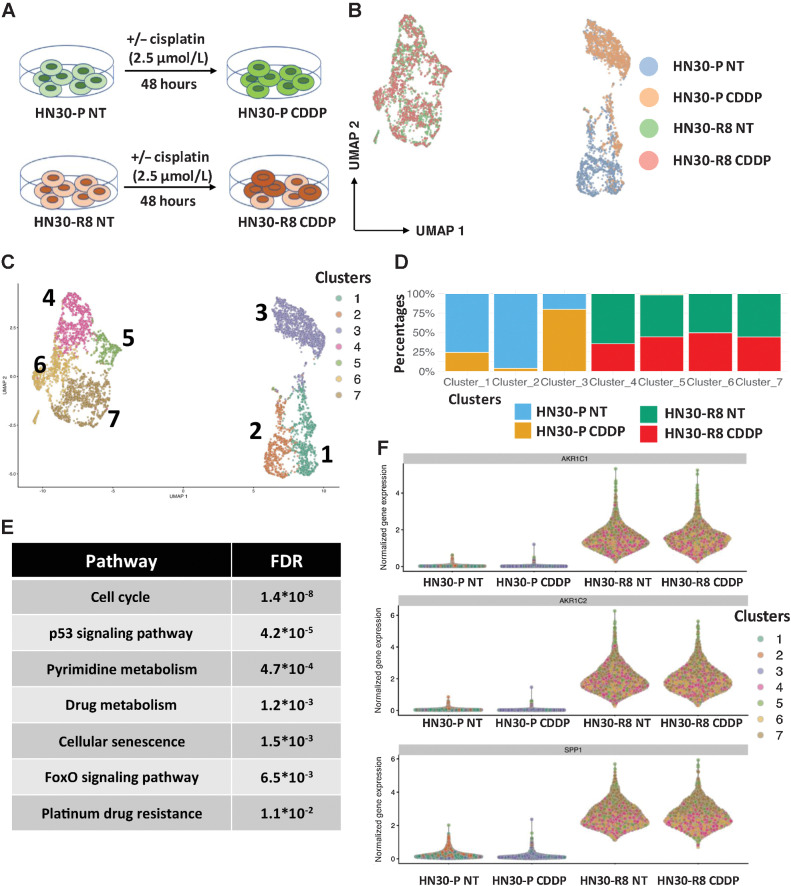
To deeply understand the evolution of the CDDP-resistant cells, we performed scRNA-seq and scATAC-seq analysis on HN30 CDDP-sensitive cells (HN30-P) along with the CDDP-resistant line (HN30-R8) with or without treatment with CDDP in vitro (Fig. 4A). For the scRNA-seq, our sequencing library averaged 3,000 cells per sample which is capable of detecting 6.5k genes/50k UMIs/115k reads per cell. Graph-based clustering (UMAP) showed 7 different clusters identifiable from the scRNA-seq data from these 4 cell populations (Fig. 4B and C). Interestingly, CDDP treatment induced a transcriptomically distinct phenotype as obvious from the differentiation of CDDP-treated parental HN30 cells (Cluster 3 vs. 1 and 2) from parental cell lines in the UMAP space (Fig. 4B–D). Similarly, we noted distinct clusters for HN30-R8 cells in comparison to HN30-P cells. Pathway analysis revealed activation of the cell cycle, p53 signaling, senescence, platinum resistance, and FoxO signaling pathway in the CDDP-resistant Cluster 3 (Fig. 4E). We found multiple genes of the KNC pathways to be upregulated upon CDDP treatment in parental HN30-P cells and in CDDP-resistant HN30-R8 cells (versus parental; Fig. 4F). These data suggest transcriptionally distinct clusters of cells arise upon CDDP treatment as well as during acquired resistance over time.
Figure 4.
scRNA-seq revealed enrichment of a KNC gene signature in cisplatin-resistant HNSCC cells. A, Experimental model in which HN30-P and HN30-R8 cells were collected after 48 hours of treatment with an optimal dose of 2.5 μmol/L CDDP which causes minimal apoptotic cell death and then processed for single-cell gene expression profiling as described in Methods. B and C, UMAP clustering analysis of all 4 cell lines tested, where B shows the cell labels and C shows the clusters. D, Relative representation of each cell sample in cell clusters. E, Pathway analysis of genes activated upon CDDP treatment in HN30-P parental cell lines. F, Violin plot showing upregulated expression of selected KNC primary target genes in HN30-R8 cells with and without CDDP treatment.
scATAC-seq analysis identified epigenomic features of cisplatin resistance
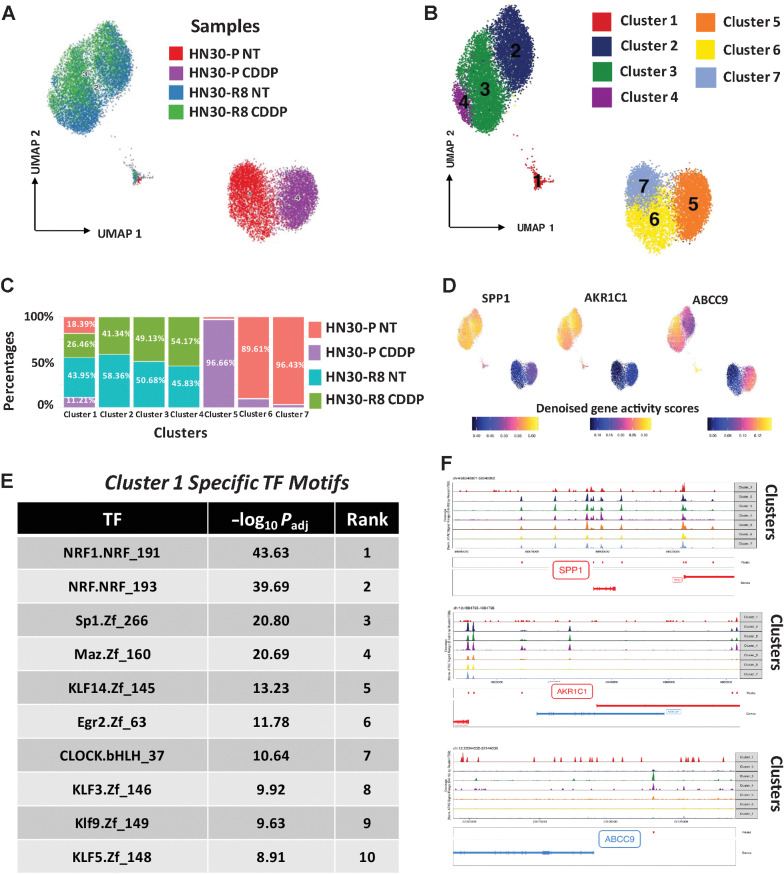
NRF2, a master regulator of the anti-oxidative stress response (48) is a transcription factor which is likely to regulate the epigenome. To better understand the chromatin accessibility differences at enhancers and promoters, we performed scATAC-seq studies in the same samples as above with an average of 3,000 cells per sample. Cell Ranger ATAC tools (alignment, trimming, and identification of transpose cut sites) were used to preprocess the fastq files. We selected only the high-quality cells, in terms of their TSS scores and number of unique fragments. After the dimensionality reduction (LSI method) and normalization (TF-IDF), UMAP-based clustering identified 7 unique populations (Fig. 5A and B). Clusters 6 and 7 were enriched in HN30 parental untreated line (HN30-P NT); Cluster 5 in parental CDDP-treated line (HN30-P CDDP); and Clusters 2, 3, and 4 intermingled in the CDDP-resistant HN30-R8 untreated (HN30-R8 NT) and CDDP-treated (HN30-R8 CDDP) cell lines (Fig. 5A–C). Importantly, we noted a unique subpopulation “Cluster 1” which consisted of cells from all 4 conditions (albeit a lower number of cells). The cells in this cluster displayed stem cell–like features including enrichment of epithelial–mesenchymal transition (EMT), hypoxia, p53, and xenobiotic metabolism pathways (HALLMARK pathway analysis, FDR < 0.01). It also harbored high expression of KNC genes AKR1C1, AKR1C2, AKR1C3, and SPP1 (Fig. 5D; Supplementary Table S4A). This cluster also expressed cell surface markers such as ABCC9, LPAR4, LILRA4, KIR3DL3, SSTR1, and CLEC14a (Supplementary Table S4B). In this Cluster 1, the topmost two motifs present in the enriched peaks belonged to NRF transcription factors (TF; Fig. 5E). Also, this cluster contains motifs for various Krüppel-like factor family members which are known to be key regulators of stem cell function (49). We also observed ATAC-seq peaks for KNC genes and ABCC9 among others in the resistant clusters (Fig. 5F). This raises the possibility that an NRF2-driven epigenetically primed subpopulation with stem-like features may associate with, and potentially contribute to, the CDDP resistance. Taken together, these data suggest that activation of the KNC pathway enhances cellular oxidative stress response to CDDP and drives CDDP resistance and metastasis through a novel epigenetic mechanism.
Figure 5.
scATAC-seq analysis identified epigenomic features of cisplatin resistance. Parental HN30-P and CDDP-resistant HN30-R8 cells were treated with CDDP (2.5 μmol/L) and collected after 48 hours of treatment with an optimal dose of 2.5 μmol/L CDDP which causes minimal apoptotic cell death and then processed for scATAC gene expression profiling as indicated. A, UMAP clustering analysis and scATAC cell labels of all four cell lines tested. B, Identification of 7 unique scATAC clusters in all 4 cells. C, Relative representation of each cell sample in cell clusters. D, Gene scores of KNC pathway targets and ABCC9 in specific clusters in CDDP-resistant HN30-R8 cells. E, Specific transcriptional (TF) motifs overrepresented in the peaks enriched in Cluster 1. F, Tracks showing aggregate ATAC-seq peaks for annotated genes in observed 7 clusters.
Glutaminase synthase inhibition disrupts redox balance and overcomes acquired cisplatin resistance in vitro and in vivo
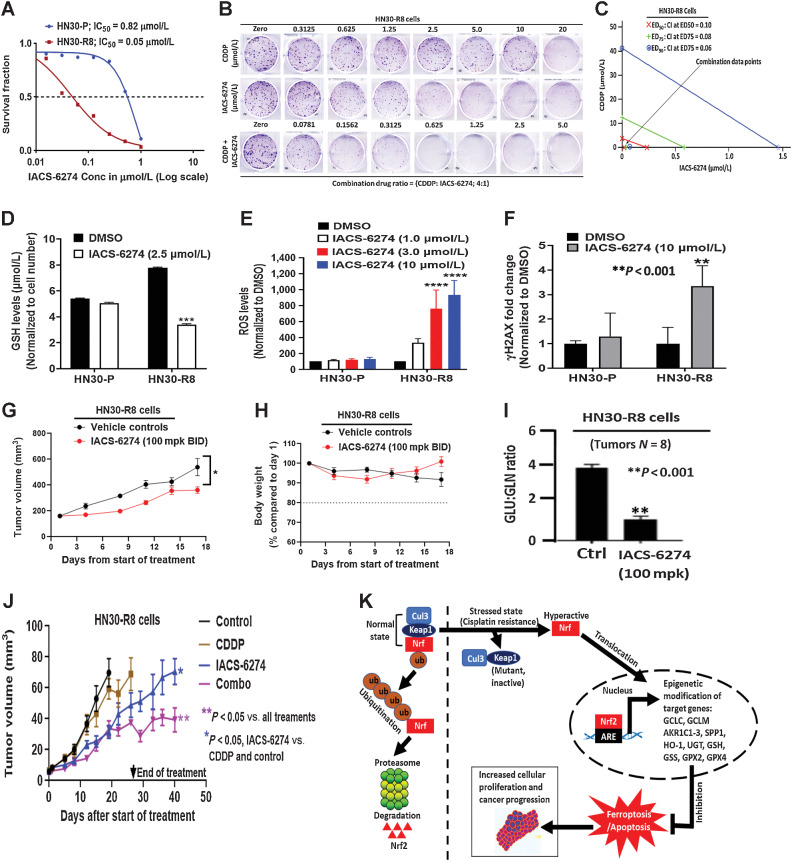
Our data support a hypothesis that NRF2 hyperactivation is required to develop CDDP resistance in HNSCC. One way to reverse acquired CDDP resistance would be to pharmacologically target one of the metabolic steps required for effective GSH synthesis or utilization that drives NRF2-dependent CDDP neutralization. The enzyme GLS1 is needed for glutamine utilization and conversion to glutamate, an important precursor amino acid for GSH synthesis (50). Clonogenic survival assays demonstrated that treatment with the GLS1 inhibitor, IACS-6274, resulted in higher growth inhibition in HN30-R8 cells (IC50 = 0.05 μmol/L) compared with parental HN30-P (IC50 = 0.82 μmol/L; Fig. 6A). We next examined whether IACS-6274 synergized with CDDP treatment in HN30-R8 cells, using the CI method of Chou and Talalay (16). An isobologram plot of effective doses (ED50; 50% inhibition), ED75 (75% inhibition), and ED90 (90% inhibition) obtained from Fig. 6B, showed that the CI at each inhibitory concentration is less than 1.0, indicating strong synergism following combination treatment in these cells (Fig. 6C). We hypothesized that acquired CDDP resistance is likely a function of oxidative stress generation, and to test this, we measured in vitro cellular changes in GSH, ROS, and γH2AX in HN30-P and HN30-R8 cells following IACS-6274 treatment as described in Methods. IACS-6274 significantly decreased the GSH levels and increased global ROS levels in HN30-R8 but not in HN30-P cells in a dose-dependent manner (Fig. 6D and E). Consistent with these results, IACS-6274 greatly increased γH2AX at the higher dose tested in HN30-R8 compared with parental HN30-P cells (Fig. 6F). These results suggest that IACS-6274 antitumor effects are linked to the generation of oxidative stress response. To demonstrate that IACS-6274 could robustly inhibit its targets and reduce glutaminolysis in vivo, a group of mice (N = 8) was first injected subcutaneously with HN30-R8 cells and tumors were randomized across groups by size. Animals were treated with IACS-6274 at a dose of 100 mg/kg orally, twice a day as indicated. Single-agent IACS-6274 activity was observed with this dosing regimen and was well tolerated (Fig. 6G and H). Tumors were also harvested and snap-frozen after 8 hours and analyzed to determine the ratio of glutamine and glutaminase metabolites. Treatment with the drug for the time indicated significantly decreased glutamate/glutamine (GLU/GLN) ratio, indicating target engagement with the dose used (Fig. 6I). We next tested whether the IACS-6274 could sensitize CDDP-resistant HNSCC cells to CDDP treatment in vivo. Therefore, the effect of IACS-6274 either alone or in combination with CDDP, was assessed in an orthotopic nude mouse model injected with HN30-R8 cells in the tongue. Unlike subcutaneous injection, orthotopic oral injection model described above, IACS-6274 (100 mg/kg, orally, twice daily, 5 times a week) alone resulted in significant tumor growth reduction compared with untreated control or CDDP-treated mice (P < 0.05; Fig. 6J), suggesting the positive impact of the tumor microenvironment on the drug response. The combination of these drugs significantly inhibited oral tumor growth, when compared with all treatment groups (P < 0.05; Fig. 6J). Furthermore, IACS-6274 alone and in combination with CDDP improved animal survival compared with animals in the untreated control and CDDP treatment groups (P < 0.05; Supplementary Fig. S8A). None of the mice in the single or combination treatment arms showed more than 10% body weight loss or any signs of drug toxicity (Supplementary Fig. S8B), suggesting that the combination of drugs was well tolerated during the study.
Figure 6.
Glutaminase synthase inhibition disrupts redox balance and overcomes acquired cisplatin resistance in vitro and in vivo. A, IC50 values obtained from clonogenic survival curves showing that HN30-R8 are more sensitive to the glutaminase synthase inhibitor (IACS-6274) in vitro compared with their parental HN30-P counterparts. B, Representative images of clonogenic survival assays in HN30-R8 cell lines treated with combination of CDDP and IACS-6274. C, Isobolograms assessed by Chou and Talalay CI indicate strong synergism with combination of CDDP and IACS-6274 in HN30-R8 cells. The CI < 1.0 indicates synergism. Addition of IACS-6274 to CDDP resulted in CI = 0.10 at the effective dose (ED50) that killed 50% of the cells following treatment with the two drugs. D, Reduced levels of GSH (***, P < 0.001) in HN30-R8 cells following treatment with IACS-6274. E, ROS levels are significantly elevated (****, P = 0.0001) after treatment with various doses of IACS-6274 in HN30-R8 compared with HN30-P cells in vitro. F, Treatment with IACS-6274 increases expression of the DNA damage response marker, H2AX, in HN30-R8 compared with HN30-P cells. Mice injected with HN30-R8 cells were treated with IACS-6274 as described in Methods. G, Single-agent IACS-6274 activity was observed with 100 mg/kg twice a day dosing regimen (*, P = 0.03 two-way ANOVA) and was well tolerated (H). Treatment with IACS-6274 for 8 hours decreases GLU/GLN ratio indicating target engagement with the dose used (I). Nude male mice were injected orally with the HN30-R8 CDDP-resistant cells and treated with vehicle control, CDDP, IACS-6274 alone and in combination with CDDP as described in Methods. J, IACS-6274 showed single agent activity, enhanced CDDP sensitivity, and decreased tumor growth. K, Proposed signaling model of acquired CDDP resistance in HNSCC. Under non-stressed conditions, KEAP1 directs ubiquitin-mediated degradation of NRF2 via proteasomes. Under oxidative stress generated by CDDP resistance, KEAP1 is mutated, NRF2 is stabilized and promotes transcription of ARE-containing genes associated with epigenetic modification of its downstream target genes. The hyperactivated NRF2 can protect the HNSCC cells from oxidative stress via suppression of ferroptotic and/or apoptotic cell death leading to enhanced cellular proliferation and accelerated tumor progression.
Discussion
Cisplatin was introduced into clinical practice in the 1970s, and it remains an essential treatment regimen for advanced stage HNSCC to decrease the rates of locoregional recurrence and DM (51). Regardless of its importance and extensive research into this therapeutic agent, the mechanisms of CDDP resistance remain unclear. Although many putative genomic drivers and proteomic events have been linked to intrinsic or acquired CDDP resistance, there is no cohesive principle that explains the molecular mechanism(s) of resistance or identifying strategies to overcome it in resistant tumors. Using human HNSCC tumor lines with WT TP53, we generated clones that are highly proliferative and resistant to CDDP in the laboratory. We observed an increased incidence of DM in a mouse orthotopic tongue tumor using these CDDP-resistant cell lines, in agreement with human clinical data that patients with TP53 WT HNSCC failing CDDP treatment have a higher rate of DM (4). We also provide strong evidence that acquired CDDP resistance in HNSCC is mechanistically linked to an upregulated gene signature that constitutes the KNC pathway consistent with other recent publications (39, 52). Remarkably, this appears to be driven at least in part through selection and enrichment of cell populations with two acquired and distinct KEAP1 mutations resulting in hyperactivated NRF2 signaling. We think that these mutations are likely LOF mutations, occurring within the BTB binding domain of KEAP1 that is required for homodimerization and binding with the CUL3. As a result, KEAP1 adopts an altered conformation and becomes incapable of binding and recruiting CUL3 to degrade NRF2. Therefore, NRF2 becomes stabilized and localized in the nucleus to enhance ARE target gene expression. Consequently, mutant KEAP1 drives HNSCC CDDP resistance and progression via KEAP1/NRF2 pathway hyperactivation. In this study, the expression levels of the KEAP1, NRF2, and their downstream targets remain unchanged in vitro upon treatment with CDDP, particularly in the parental cell line. This is probably due to a slowed and more organized adaptation to higher levels of oxidative stress to avoid swift metabolic disturbance in these cells that are readily sensitive to CDDP. KD of NRF2 with shRNA or restoration of WT KEAP1 in vitro and in vivo significantly resensitized resistant cells to CDDP and decreased the incidence of DM suggesting to us that activation of NRF2 pathway is a permissive step to acquired CDDP resistance which may be necessary but not sufficient to confer this resistance. It is not clear why CDDP-resistant tumors expressing NRF2 shRNA or WT KEAP1 constructs continued to grow slowly in most experimental conditions following cessation of CDDP treatment in vivo. These tumors were established from heterogenous pooled cells and probably some populations had incomplete NRF2 KD and/or low WT KEAP1 expression levels that resulted in slow growth characteristics.
We have also shown that increased sensitivity to CDDP in NRF2 KD or overexpressing WT KEAP1 resistant cells is likely driven by activation of ferroptosis, a programmed cell death mechanism dependent on lipid accumulation and peroxidation via utilization of iron (46). These results further suggest that the KEAP1/NRF2-related anti-oxidative stress in CDDP-resistant HNSCC cells is strongly associated with ferroptosis suppression. Currently, it is not clear how acquired CDDP resistance increases the incidence of DM. SPP1 (secreted phosphoprotein 1, also called osteopontin, OPN) is a primary NRF2 target gene overexpressed and correlated with poor survival in many cancers including HNSCC (53). SPP1 functions as an extracellular matrix oncoprotein involved in cellular migration and tumor metastasis (54). Whole transcriptome RNA-seq performed on FFPE sections from mice tissues revealed that the SPP1 was significantly upregulated in DM site compared with the primary site in the HN30-R8 CDDP–treated group (Supplementary Table S2). Future experiments are needed to determine if activation of SPP1 is a major driver event in CDDP resistance and metastasis.
Somatic mutations in the KNC pathway occur in small portion of HNSCC (36, 37). Therefore, alternative mechanisms must exist to regulate KEAP1 and NRF2 expression. Recent studies have suggested that the KEAP1/NRF2 signaling can be regulated by epigenetic mechanisms in cancers (55, 56). Consistent with these studies, we showed that multiple KEAP1/NRF2 target genes were epigenetically regulated at the transcriptional level in the CDDP-resistant HNSCC cells. More importantly, our scATAC-seq revealed that Cluster 1 displayed enriched peaks with transcriptional motifs containing stem cell–like features, EMT surface markers, and high expression of KNC genes such as AKR1C1, AKR1C2, AKR1C3, and SPP1. These findings are supported by a recent study which demonstrated that EMT and cancer stem cells lead to acquired CDDP resistance and tumor recurrence through epigenetic changes in HNSCC (57). Although our data suggest that a unique epigenomic reprogramming contributes to evolution of CDDP-resistant clones in HNSCC, it remains unclear how this occurs and therefore further in vitro and in vivo molecular characterization is necessary to understand this mechanistic link. We speculate that it is largely driven by NRF2 which may act as a core/pioneer transcription factor in this scenario. Moreover, because Cluster 1 cells are epigenetically different, it would be crucial to isolate cells from this cluster and determine if a clonal population with a specific enhancer or repressor modulates the stem cell–like features and KNC genes and drives the resistance to CDDP. It would also be important to determine the epigenome maps and chromatin state of Cluster 1 cells through profiling 6 histone modification marks that have been previously used to characterize essential epigenetic elements and combinatorial chromatin states in normal tissues (30, 31). Okazaki and colleagues have shown that NRF2 KD decreases the H3K27Ac deposition, suggesting that NRF2 contributes to enhancer formation (58). Our bulk-level ATAC-seq analysis suggests that there is an immediate chromatin opening following CDDP treatment in HN30-P cells, and a long-term chromatin opening in the CDDP-resistant HN30-R8 cells in parallel to the acquisition of resistance (Supplementary Table S5A-B). Chromatin immunoprecipitation sequencing analyses will be needed to identify the histone marks and TF DNA binding motifs that drive acquisition of CDDP resistance in our HNSCC cell line models.
It is entirely possible that multiple mechanisms exist, which enable tumor cells to become resistant to CDDP. This includes increased localization of CDDP to the lysosomal compartment and cellular efflux as suggested by others (59–61). Using fluorescent probes, we previously observed increased localization of CDDP to the lysosome in HN30-R8 cells accompanied by a reduction in total cellular CDDP compared with CDDP-sensitive parental HN30-P cells (59). An emerging body of evidence has linked expression of the copper (Cu) transporter ATP7B to lysosomal sequestering and efflux of CDDP in exosomal bodies (60). Interestingly, we observed a significant increase in the in vivo expression of ATP7B in CDDP-resistant HN30-R8 cells, consistent with multiple resistance mechanisms (Supplementary Fig. S9). Given the large number of DEGs between HN30-R8 and parental cells, efforts are now underway to comprehensively identify all the different pathways and genes that may contribute to the CDDP-resistant phenotype, using pooled LOF CRISPRi library screens. It has been reported that KEAP1 mutations in cancer cells can cause increased cellular antioxidant capacity and dependency on exogenous glutamine and as such become sensitive to glutamine deprivation (62). Consistent with this study, we demonstrated that CDDP resistance in HNSCC could be robustly reversed with the novel GLS1 inhibitor, IACS-6274, with concomitant increase in glutamine levels relative to glutamate. Therefore, our data implicate that the KEAP1/NRF2 pathway inactivation provides a specific vulnerability to blockade of glutamine metabolism under CDDP-resistant conditions in HNSCC. One limitation of this study is the use of one CDDP-resistant cell line model and validation in more CDDP-resistant cell lines is necessary.
In conclusion, our data provide evidence that chronic CDDP treatment selects for resistant tumors with an expression profile dominated by KEAP1/NRF2 dysregulated genes and epigenetic reprograming. This may contribute to induction of acquired CDDP resistance and development of DM in HNSCC. Our data also suggest that inhibition of KEAP1/NRF2 pathway or GLS1 are potential approaches to sensitize CDDP-resistant HNSCC tumors.
Supplementary Material
Supplementary Materials and Methods
Cisplatin resistant HNSCC cells migrate faster than their Cisplatin sensitive counterparts.
GO and KEGG pathway analysis of top KEAP1/NRF2/CUL3 (axis regulated genes and their association with clinical outcome in HNSCC.
Relationship between TP53 status and the NRF2/KEAP1/CUL3 pathway in OCSCC TCGA samples.
Restoration of wildtype KEAP1 improves survival in mice.
NRF2 targeted suppression or restoration of wild type KEAP1 decreases distant metastasis in cisplatin resistant HNSCC cells.
Cisplatin sensitivity following knockdown of NRF2 or restoration of wildtype KEAP1 in cisplatin resistant HNSCC cells is not mediated through apoptosis.
Restoration of KEAP1/NRF2 pathways increases cisplatin sensitivity in resistant HNSCC cells through ferroptosis.
IACS-6274 improves survival and is well tolerated in combination with cisplatin in mice.
In vivo expression of cisplatin transporters in CDDP resistant HNSCC tumors.
In vivo RNA sequencing and pair-wise comparison between 4 distinct groups in HN30-P and HN30-R8 HNSCC tumors.
NRF2 regulated genes significantly upregulated in distant metastasis compared to paired primary tumors in HN30-R8 CDDP group.
Whole exome sequencing showing mutational status in the parental HN30-P versus HN30-R8 cisplatin resistant cell lines.
Supplementary Table S4A. The KNC genes enriched in cluster 1 in HNSCC cells; Supplementary Table S4B. Gene activity score of cell surface markers in cluster 1 in HNSCC cells.
Supplementary Table S5A. Bulk-level single cell ATAC sequencing analysis showing immediate (short-term) chromatin openings (peaks) in HN30-P cells following CDDP treatment; Supplementary Table S5B. Bulk-level single cell ATAC sequencing analysis showing long-term chromatin openings (peaks) gained in cisplatin-resistant HN30-R8 cells following CDDP treatment.
Acknowledgments
This work was supported by the NIH/NIDCR R01DE024601 (to A.A. Osman and J.N. Myers) and NCI U54CA274321 (to A.A. Osman and J.N. Myers) and Cellular Imaging Core, Bioinformatics Shared Resource, and the Reverse Phase Protein Array (RPPA) Core, which are supported by the NIH through MD Anderson's Cancer Center Support Grant (P30CA016672).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
J. Kovacs reports a patent for IACS-6274 issued and is an employee of and shareholder in Aktis Oncology. T. Heffernan reports personal fees from Cullgen Inc. and Roivant Discovery, as well as other support from Boehringer Ingelheim, Taiho Pharmaceuticals, Blueprint Medicines, and Schrodinger outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
A.A. Osman: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. E. Arslan: Conceptualization, resources, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. M. Bartels: Conceptualization, data curation, formal analysis, validation, investigation, methodology. C. Michikawa: Conceptualization, formal analysis, investigation, visualization, methodology, writing–review and editing. A. Lindemann: Conceptualization, formal analysis, investigation, methodology. K. Tomczak: Conceptualization, software, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. W. Yu: Resources, investigation, methodology. V. Sandulache: Conceptualization, resources, formal analysis, investigation, methodology, writing–review and editing. W. Ma: Data curation, software, formal analysis, investigation, methodology. L. Shen: Data curation, software, formal analysis, investigation, methodology. J. Wang: Data curation, software, formal analysis, investigation, methodology. A.K. Singh: Data curation, investigation, methodology. M.J. Frederick: Data curation, software, formal analysis, investigation, visualization, methodology. N.D. Spencer: Formal analysis, validation, investigation, methodology. J. Kovacs: Data curation, formal analysis, validation, investigation, methodology. T. Heffernan: Data curation, formal analysis, validation, investigation, methodology. W.F. Symmans: Formal analysis, methodology. K. Rai: Conceptualization, resources, data curation, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–review and editing. J.N. Myers: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–review and editing.
References
- 1. Sandulache VC, Michikawa C, Kataria P, Gleber-Netto FO, Bell D, Trivedi S, et al. High-risk TP53 mutations are associated with extranodal extension in oral cavity squamous cell carcinoma. Clin Cancer Res 2018;24:1727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferris RL, Blumenschein G, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous cell carcinoma of the head and neck. N Engl J Med 2016;375:1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo TW, Saiyed F, Yao CMKL, Kiong KL, Martinez J, Sacks R, et al. Outcomes of patients with oropharyngeal squamous cell carcinoma treated with induction chemotherapy followed by concurrent chemoradiation compared with those treated with concurrent chemoradiation. Cancer 2021;127:2916–25. [DOI] [PubMed] [Google Scholar]
- 4. Michikawa C, Torres-Saavedra PA, Silver NL, Harari PM, Kies MS, Rosenthal DI, et al. Evolutionary action score of TP53 analysis in pathologically high-risk HPV-negative head and neck cancer from a phase II clinical trial: NRG oncology RTOG 0234. Adv Radiat Oncol 2022;7:100989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu W, Chen Y, Putluri N, Coarfa C, Robertson MJ, Putluri V, et al. Acquisition of cisplatin resistance shifts head and neck squamous cell carcinoma metabolism toward neutralization of oxidative stress. Cancers 2020;12:1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goeman F, De Nicola F, Scalera S, Sperati F, Gallo E, Ciuffreda L, et al. Mutations in the KEAP1–NFE2L2 pathway define a molecular subset of rapidly progressing lung adenocarcinoma. J Thorac Oncol 2019;14:1924–34. [DOI] [PubMed] [Google Scholar]
- 7. Jaramillo MC, Zhang DD. The emerging role of the Nrf2–Keap1 signaling pathway in cancer. Genes Dev 2013;27:2179–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Panieri E, Saso L. Inhibition of the NRF2/KEAP1 axis: a promising therapeutic strategy to alter redox balance of cancer cells. Antioxid Redox Signal 2021;34:1428–83. [DOI] [PubMed] [Google Scholar]
- 9. Wu WL, Papagiannakopoulos T. The pleiotropic role of the KEAP1/NRF2 pathway in cancer. Annu Rev Cancer Biol 2020;:413–35. [Google Scholar]
- 10. Boysen G, Jamshidi-Parsian A, Davis MA, Siegel ER, Simecka CM, Kore RA, et al. Glutaminase inhibitor CB-839 increases radiation sensitivity of lung tumor cells and human lung tumor xenografts in mice. Int J Radiat Biol 2019;95:436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soth MJ, Le K, Di Francesco ME, Hamilton MM, Liu G, Burke JP, et al. Discovery of IPN60090, a clinical stage selective glutaminase-1 (GLS1) inhibitor with excellent pharmacokinetic and physicochemical properties. J Med Chem 2020;63:12957–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yap TA, Dumbrava EE, Rodon Ahnert J, Hong DS, Pant S, Karp DD, et al. First-in-human biomarker-driven phase I trial of the potent and selective glutaminase-1 (GLS1) inhibitor IACS-6274 (IPN60090) in patients (Pts) with molecularly selected advanced solid tumors. J Clin Oncol 2021;39:3001. [Google Scholar]
- 13. Zhao M, Sano D, Pickering CR, Jasser SA, Henderson YC, Clayman GL, et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin Cancer Res 2011;17:7248–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lindemann A, Patel AA, Tang L, Tanaka N, Gleber-Netto FO, Bartels MD, et al. Combined inhibition of Rad51 and Wee1 enhances cell killing in HNSCC through induction of apoptosis associated with excessive DNA damage and replication stress. Mol Cancer Ther 2021;20:1257–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Osman AA, Monroe MM, Ortega Alves MV, Patel AA, Katsonis P, Fitzgerald AL, et al. Wee-1 kinase inhibition overcomes cisplatin resistance associated with high-risk TP53 mutations in head and neck cancer through mitotic arrest followed by senescence. Mol Cancer Ther 2015;14:608–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 2006;58:621–81. [DOI] [PubMed] [Google Scholar]
- 17. Tanaka N, Patel AA, Wang J, Frederick MJ, Kalu NN, Zhao M, et al. Wee-1 kinase inhibition sensitizes high-risk HPV+ HNSCC to apoptosis accompanied by downregulation of MCl-1 and XIAP antiapoptotic proteins. Clin Cancer Res 2015;21:4831–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Osman AA, Neskey DM, Katsonis P, Patel AA, Ward AM, Hsu TK, et al. Evolutionary action score of TP53 coding variants is predictive of platinum response in head and neck cancer patients. Cancer Res 2015;75:1205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 20. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. 1000 genome project data processing subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. KDD-96 Proceedings 1996.
- 26. Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 2012;22:1813–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, et al. Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell 2015;162:1051–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 2010;28:817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li J, Fu C, Speed TP, Wang W, Symmans WF. Accurate RNA sequencing from formalin-fixed cancer tissue to represent high-quality transcriptome from frozen tissue. JCO Precis Oncol 2018;2:PO.17.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marczyk M, Fu C, Lau R, Du L, Trevarton AJ, Sinn BV, et al. The impact of RNA extraction method on accurate RNA sequencing from formalin-fixed, paraffin-embedded tissues. BMC Cancer 2019;19:1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sheth S, Farquhar DR, Schrank TP, Stepp W, Mazul A, Hayward M, et al. Correlation of alterations in the KEAP1/CUL3/NFE2L2 pathway with radiation failure in larynx squamous cell carcinoma. Laryngoscope Invest Otolaryngol 2021;6:699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. The Cancer Genome Atlas Network (348 collaborators). Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hast BE, Cloer EW, Goldfarb D, Li H, Siesser PF, Yan F, et al. Cancer-derived mutations in KEAP1 impair NRF2 degradation but not ubiquitination. Cancer Res 2014;74:808–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Namani A, Rahaman MM, Chen M, Tang X. Gene-expression signature regulated by the KEAP1/NRF2/CUL3 axis is associated with a poor prognosis in head and neck squamous cell cancer. BMC Cancer 2018;18:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahmed KM, Veeramachaneni R, Deng D, Putluri N, Putluri V, Cardenas MF, et al. Glutathione peroxidase 2 is a metabolic driver of the tumor immune microenvironment and immune checkpoint inhibitor response. J Immunother Cancer 2022;10:e004752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhu H, Xie D, Yu Y, Yao L, Xu B, Huang L, et al. KEAP1/NFE2L2 mutations of liquid biopsy as prognostic biomarkers in patients with advanced non–small cell lung cancer: results from two multicenter, randomized clinical trials. Front Oncol 2021;11:659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ma X, Zhang J, Liu S, Huang Y, Chen B, Wang D. Nrf2 knockdown by shRNA inhibits tumor growth and increases efficacy of chemotherapy in cervical cancer. Cancer Chemother Pharmacol 2012;69:485–94. [DOI] [PubMed] [Google Scholar]
- 43. Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan V, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156:317–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hirschhorn T, Stockwell BR. The development of the concept of ferroptosis. Free Radic Biol Med 2019;133:130–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo J, Xu B, Han Q, Zhou H, Xia Y, Gong C, et al. Ferroptosis: a novel antitumor action for cisplatin. Cancer Res Treat 2018;50:445–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fan Z, Wirth A-K, Chen D, Wruck CJ, Rauh M, Buchfelder M, et al. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis 2017;6:e371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vomund S, Schäfer A, Parnham MJ, Brüne B, von Knethen A. Nrf2, the master regulator of anti-oxidative responses. Int J Mol Sci 2017;18:2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McConnell BB, Yang VW. Mammalian Krüppel-like factors in health and diseases. Physiol Rev 2010;90:1337–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Galan-Cobo A, Sitthideatphaiboon P, Qu X, Kovacs JJ, Poteete A, Tong P, et al. LKB1 and KEAP1/NRF2 pathways cooperatively promote metabolic reprogramming with enhanced glutamine dependence in KRAS-mutant lung adenocarcinoma. Cancer Res 2019;79:3251–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bauml JM, Vinnakota R, Anna Park Y-H, Bates SE, Fojo T, Aggarwal C, et al. Cisplatin every 3 weeks versus weekly with definitive concurrent radiotherapy for squamous cell carcinoma of the head and neck. J Natl Cancer Inst 2019;111:490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Islam SS, Qassem K, Islam S, Parag RR, Rahman MZ, Farhat WA, et al. Genetic alterations of Keap1 confers chemotherapeutic resistance through functional activation of Nrf2 and Notch pathway in head and neck squamous cell carcinoma. Cell Death Dis 2022;13:696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bie T, Zhang X. Higher expression of SPP1 predicts poorer survival outcomes in head and neck cancer. J Immunol Res 2021;2021:8569575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang KX, Denhardt DT. Osteopontin: role in immune regulation and stress responses. Cytokine Growth Factor Rev 2008;19:333–45. [DOI] [PubMed] [Google Scholar]
- 55. Guo Y, Yu S, Zhang C, Tony Kong AN. Epigenetic regulation of Keap1-- Nrf2 signaling. Free Rad Biol and Med 2015;88:337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS One 2010;5:e8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de Oliveira JL, Milan TM, Bighetti-Trevisan RL, Fernandes RR, Leopoldino AM, de Almeida LO. Epithelial–mesenchymal transition and cancer stem cells: a route to acquired cisplatin resistance through epigenetics in HNSCC. Oral Dis 2022;1–15. [DOI] [PubMed] [Google Scholar]
- 58. Okazaki K, Anzawa H, Liu Z, Ota N, Kitamura H, Onodera Y, et al. Enhancer remodeling promotes tumor-initiating activity in NRF2-activated non–small cell lung cancers. Nat Commun 2020;11:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pham D, Deter CJ, Reinard MC, Gibson GA, Kiselyov K, Yu W, et al. Using ligand-accelerated catalysis to repurpose fluorogenic reactions for platinum or copper. ACS Cent Sci 2020;6:1772–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Safaei R, Larson BJ, Cheng TC, Gibson MA, Otani S, Naerdemann W, et al. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther 2005;4:1595–604. [DOI] [PubMed] [Google Scholar]
- 61. Ciarimboli G. Membrane transporters as mediators of cisplatin effects and side effects. Scientifica 2012;2012:473829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sayin VI, LeBoeuf SE, Singh SX, Davidson SM, Biancur D, Guzelhan BSet al. Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. eLife 2017;6:e28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Cisplatin resistant HNSCC cells migrate faster than their Cisplatin sensitive counterparts.
GO and KEGG pathway analysis of top KEAP1/NRF2/CUL3 (axis regulated genes and their association with clinical outcome in HNSCC.
Relationship between TP53 status and the NRF2/KEAP1/CUL3 pathway in OCSCC TCGA samples.
Restoration of wildtype KEAP1 improves survival in mice.
NRF2 targeted suppression or restoration of wild type KEAP1 decreases distant metastasis in cisplatin resistant HNSCC cells.
Cisplatin sensitivity following knockdown of NRF2 or restoration of wildtype KEAP1 in cisplatin resistant HNSCC cells is not mediated through apoptosis.
Restoration of KEAP1/NRF2 pathways increases cisplatin sensitivity in resistant HNSCC cells through ferroptosis.
IACS-6274 improves survival and is well tolerated in combination with cisplatin in mice.
In vivo expression of cisplatin transporters in CDDP resistant HNSCC tumors.
In vivo RNA sequencing and pair-wise comparison between 4 distinct groups in HN30-P and HN30-R8 HNSCC tumors.
NRF2 regulated genes significantly upregulated in distant metastasis compared to paired primary tumors in HN30-R8 CDDP group.
Whole exome sequencing showing mutational status in the parental HN30-P versus HN30-R8 cisplatin resistant cell lines.
Supplementary Table S4A. The KNC genes enriched in cluster 1 in HNSCC cells; Supplementary Table S4B. Gene activity score of cell surface markers in cluster 1 in HNSCC cells.
Supplementary Table S5A. Bulk-level single cell ATAC sequencing analysis showing immediate (short-term) chromatin openings (peaks) in HN30-P cells following CDDP treatment; Supplementary Table S5B. Bulk-level single cell ATAC sequencing analysis showing long-term chromatin openings (peaks) gained in cisplatin-resistant HN30-R8 cells following CDDP treatment.
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data files. Data used in this study that is not included in the paper or supplementary files can be made available upon request from the corresponding author.