Significance
For autoimmune type 1 diabetes (T1D) to occur, a primed response for both CD4+ and CD8+ T cells is required. Conventional type 1 dendritic cells (cDC1s) acquire antigens expressed exogenously by other cells and present these antigens as peptides via major histocompatibility complex class 1 molecules (MHC-I), a process termed cross-presentation. Here, we disrupted cDC1 cross-presentation in nonobese diabetic (NOD) mice by deletion of exon 4 of Wdf4. This resulted in the absence of CD8+ T cell priming, the absence of diabetes progression, and severely reduced insulitis; however, CD4+ T cell priming remained intact. These results demonstrate that a primed CD8+ T cell response is required for unrestrained insulitis to occur even when a diabetogenic CD4+ T cell response is preserved.
Keywords: non obese diabetes, type 1 diabetes, cross-presentation, WDFY4, cDC1
Abstract
The events that initiate autoimmune diabetes in nonobese diabetic (NOD) mice remain poorly understood. CD4+ and CD8+ T cells are both required to develop disease, but their relative roles in initiating disease are unclear. To test whether CD4+ T cell infiltration into islets requires damage to β cells induced by autoreactive CD8+ T cells, we inactivated Wdfy4 in nonobese diabetic (NOD) mice (NOD.Wdfy4−/−-) using CRISPR/Cas9 targeting to eliminate cross-presentation by type 1 conventional dendritic cells (cDC1s). Similar to C57BL/6 Wdfy4−/− mice, cDC1 in NOD.Wdfy4−/− mice are unable to cross-present cell-associated antigens to prime CD8+ T cells, while cDC1 from heterozygous NOD.Wdfy4+/− mice cross-present normally. Further, NOD.Wdfy4−/− mice fail to develop diabetes while heterozygous NOD.Wdfy4+/− mice develop diabetes similarly to wild-type NOD mice. NOD.Wdfy4−/− mice remain capable of processing and presenting major histocompatibility complex class II (MHC-II)-restricted autoantigens and can activate β cell-specific CD4+ T cells in lymph nodes. However, disease in these mice does not progress beyond peri-islet inflammation. These results indicate that the priming of autoreactive CD8+ T cells in NOD mice requires cross-presentation by cDC1. Further, autoreactive CD8+ T cells appear to be required not only to develop diabetes, but to recruit autoreactive CD4+ T cells into islets of NOD mice, perhaps in response to progressive β cell damage.
Type 1 diabetes (T1D) is an autoimmune disease targeting insulin-producing pancreatic β cells. Overt diabetes occurs when insulin production becomes insufficient to maintain normal glucose homeostasis. T1D in the nonobese diabetic (NOD) mouse strain shares many molecular, genetic, and cellular features with T1D in humans (1). CD4+ and CD8+ T cells have both been implicated in initiating T1D. In humans, the genetic risk for developing T1D is associated predominantly with major histocompatibility complex class II (MHC-II) (2), suggesting CD4+ T cells may initiate disease. Class II MHC alleles, such as HLA-DQ, with mutations at position 57 of the β chain, impart the major component of susceptibility to diabetes (3) with the highest T1D association for HLA-DQ8. Similarly, in the NOD mouse, the H2-Abg7 class II MHC allele is required for T1D. H2-Abg7 also has a nonaspartic acid residue at position 57 of the β chain, and HLA-DQ8 and H2-Abg7 exhibit similarities in their peptide binding repertoires that frequently include acidic amino acids at the peptide’s p9 residue (4, 5). Other evidence suggests that CD8+ T cells and MHC class I molecules are also involved in T1D. Both CD4+ and CD8+ T cells are required for diabetes to develop in the NOD mouse (6). MHC class I expression is required for T1D initiation and insulitis, suggesting an early role for CD8+ T cells in T1D progression (7). Likewise, in humans, analysis of disease-associated alleles showed that MHC class I alleles HLA-B and HLA-A contribute significantly to T1D susceptibility (8). The recent evidence in humans suggests that autoreactive CD8+ T cells are present in the pancreatic T cell population in healthy individuals (9). Blocking CD8+ T cell activation through Tbx21 perturbation significantly inhibited T1D in several models (10). Finally, CD8+ T cells are the most abundant immune cell found within diabetic human islets of Langerhans and many CD8+ T cell MHC class I antigen epitopes have been confirmed (11, 12).
Using Batf3−/− mice on the NOD background, we previously reported that conventional type 1 dendritic cell (cDC1) are required for initiation of T1D (13). cDC1 are antigen-presenting cells that are specialized for cross-priming cytotoxic CD8+ T cells to exogenously acquired antigen (14). However, we recently showed that cDC1s are also capable of priming CD4+ T cells (15), so that the requirement of cDC1 for developing T1D does not indicate whether T1D in NOD mice is initiated by CD4+ or CD8+ T cells. We recently discovered cross-presentation by cDC1 requires the BEACH domain containing protein WDFY4 (16). Importantly, Wdfy4 deficiency does not impair antigen processing for MHC class II presentation to CD4+ T cells, providing a method to separate a requirement for general cDC1 antigen presentation from a requirement for cross-presentation to CD8+ T cells. In this study, we deleted Wdfy4 directly in NOD mice to evaluate the role of cross-presentation in T1D. Our results show that CD8+ T cell priming by cross-presentation is required for the development of T1D in NOD mice, and that CD4+ T cell priming alone is insufficient to initiate T1D but rather limits islet inflammation to peri-insulitis. These results suggest that the emergence of overt insulitis may require progressive damage of β cells by CD8+ T cells in order to recruit primed autoreactive CD4+ T cells into the islet environment.
Results
NOD.Wdfy4−/− Mice Fail to Cross-present β Cell Antigen to CD8+ T Cells.
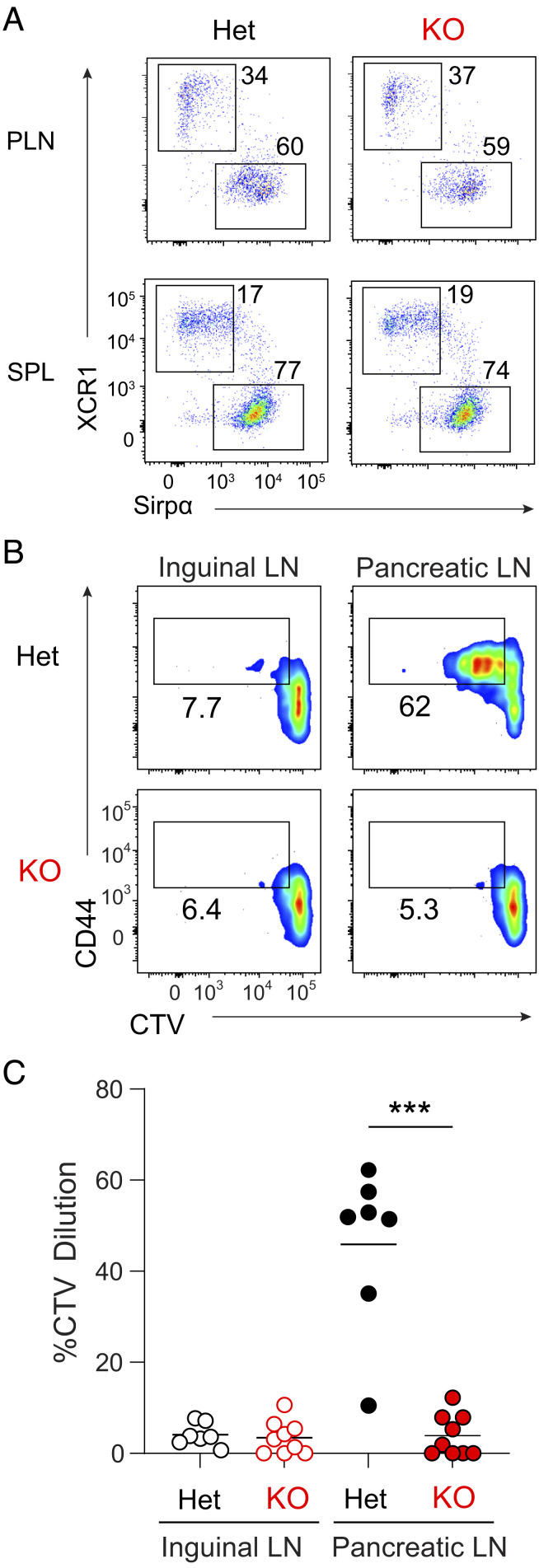
Deletion of Wdfy4 exon 4 causes splicing from exon 3 to exon 5 producing a frame shift that prematurely terminates translation (SI Appendix, Fig. S1 A–C), as previously described (16). NOD.Wdfy4−/− mice develop cDC1 populations and other hematopoietic lineages similar to C57BL/6 Wdfy4−/− mice (Fig. 1A) as previously described (16). We confirmed that NOD.Wdfy4−/− cDC1 do not cross-present cell-associated antigen in vivo using adoptive transfer of 8.3 TCR Tg T cells (Fig. 1 B and C). 8.3 Tg T cells (17, 18) are reactive to peptide residues 206 to 214 of murine islet-specific glucose-6-phosphatase catalytic subunit–related protein (IGRP) presented by H-2Kd (19). In heterozygous NOD.Wdfy4+/− mice, CVT-labeled 8.3 Tg T cells proliferated in pancreatic lymph nodes (PLNs) but not inguinal LNs (ILNs) (Fig. 1 B and C), confirming specific reactivity to IGRP in PLNs, but not ILNs, as expected. By contrast, in NOD.Wdfy4−/− mice, CVT 8.3 Tg T cells failed to proliferate in either PLNs or ILNs (Fig. 1 B and C), indicating lack of proper cross-presentation of IGRP. These results indicate that cDC1 in NOD.Wdfy4−/− mice have the expected inability for cross-presentation.
Fig. 1.
NOD.Wdfy4−/− mice fail to prime β cell reactive 8.3 TCR tg CD8+ T cells. (A) Representative flow plots of PLN (Top) and splenic (Bottom) cDC1 populations from NOD.Wdfy4+/− (Het), NOD.Wdfy4−/−(KO). Gated as B220- TCRβ+CD11c+MHCII+. NOD.Wdfy4+/−, NOD.Wdfy4−/− 6-wk–old female mice were injected intravenously (i.v.) with 106 CTV labeled 8.3 CD45.2 cells. (B) Representative flow plots of proliferating 8.3 CD45.2 T cells 3 d after transfer. (C) Percentages of proliferating 8.3 CD45.2 cells transferred. Data are pooled biologically independent samples from three independent experiments (n = 7 forNOD.Wdfy4+/− (Het) and n = 9 for NOD.Wdfy4−/− (KO). ***P = <0.001 Mann–Whitney test.
NOD.Wdfy4−/− Mice Do Not Develop Diabetes.
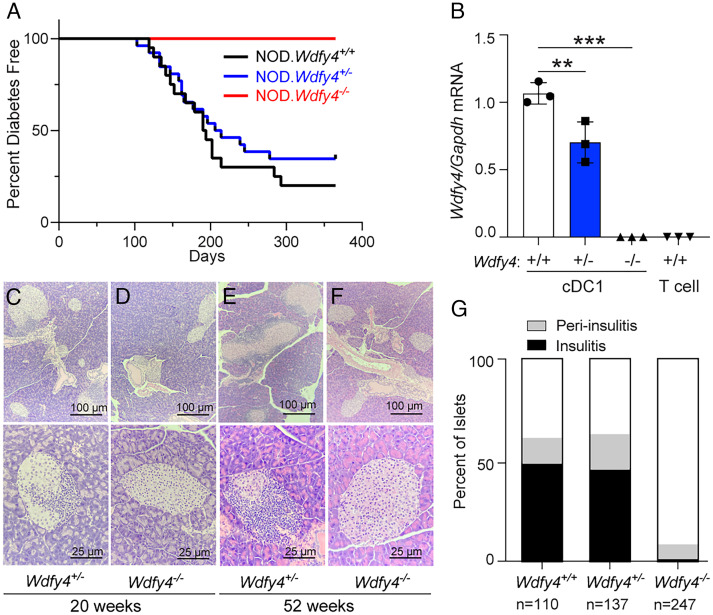
We followed progression to diabetes in NOD.Wdfy4−/−, NOD.Wdfy4−/+, and NOD.Wdfy4+/+ female littermates for 1 y (Fig. 2). The cumulative incidence of diabetes was ∼80% and ~70% in NOD.Wdfy4+/+ and heterozygous NOD.Wdfy4+/− mice respectively (Fig. 2A). By contrast, NOD.Wdfy4−/− females showed no progression to diabetes over the course of a year (Fig. 2A). To confirm that NOD.Wdfy4+/− had decreased levels of Wdfy4, we analyzed transcript by qRT-PCR. NOD.Wdfy4−/− cDC1 and control T cells had no expression of Wdfy4, as expected. However, NOD.Wdfy4+/− had significantly less expression of Wdfy4 transcript as compared to WT controls (Fig. 2B). Therefore, NOD.Wdfy4+/− mice were used as controls for NOD.Wdfy4−/−. The islets in NOD.Wdfy4+/− mice showed typical insulitis and peri-insulitis at both 20 wk and 52 wk (Fig. 2 C and E). By contrast, the islets in NOD.Wdfy4−/− mice showed minimal insulitis at either 20 wk or 52 wk (Fig. 2 D and F). We next analyzed islets from mice that were 52 wk of age (Fig. 2G). Minor peri-insulitis was only occasionally seen and ~1% had insulitis. This contrasted with age-matched NOD.Wdfy4+/+ and NOD.Wdfy4+/− mice where over 60% of all islets had some degree of insulitis (Fig. 2G). Thus, the inactivation of Wdfy4 prevents diabetes and overt insulitis in NOD mice.
Fig. 2.
NOD.Wdfy4−/− mice have reduced insulitis and do not develop diabetes. (A) Diabetes incidence in female NOD.Wdfy4+/+ (n = 20), NOD.Wdfy4+/− (n = 27), and NOD.Wdfy4−/− (n = 17). (B) qRT-PCR for expression of Wdfy4 transcript from sorted cDC1 and control T cells. Dots represent the average of technical replicates from one mouse. Hematoxylin and eosin staining. (C) 20-wk female NOD.Wdfy4+/− islets. (D) 20-wk female NOD.Wdfy4−/− islets. (E) 52-wk female NOD.Wdfy4+/− islets. (F) 52-wk female NOD.Wdfy4−/− islets. (G) Insulitis scoring of 52-wk nondiabetic NOD.Wdfy4+/+ (n = 110), NOD.Wdfy4+/− (n = 137), and NOD.Wdfy4−/− (n = 247) islets. **P = <0.01 and ****P = <0.0001 one-way ANOVA.
NOD.Wdfy4−/− Mice Do Not Develop Lymphocyte Infiltration into Islets.
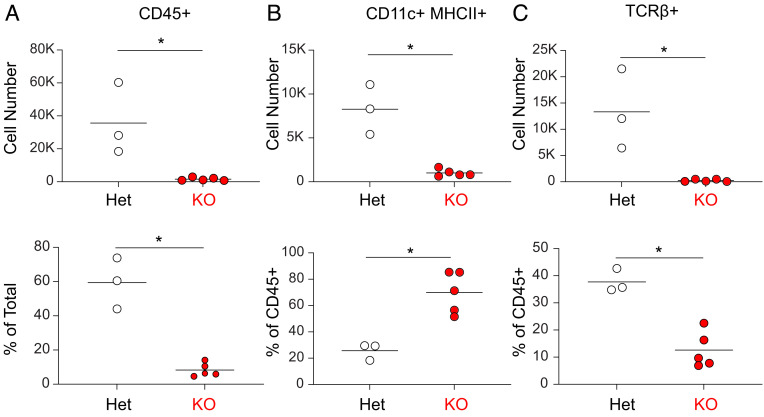
We compared the immune cell infiltrates in islets in NOD.Wdfy4+/− and NOD.Wdfy4−/− mice (Fig. 3 and SI Appendix, Fig. S2). In 12-wk NOD.Wdfy4+/− mice, islets contained high numbers of CD45+ cells, comprising about 60% CD11c+ H2-Abg7+ cells and 40% T cells (Fig. 3 A–C). By contrast, in 12-wk NOD.Wdfy4−/− mice, islets contained only sparse CD45+ cells comprised primarily of CD11c+ H2-Abg7+ cells, but very few T cells (Fig. 3 A–C). Thus, we conclude that NOD mice lacking the capacity for cross-presentation have minimal CD8+ and CD4+ T cell infiltration into islets.
Fig. 3.
NOD.Wdfy4−/− mice have minimal inflammatory cell infiltrate. (A) Graph of absolute cell number (Top) and percentage (Bottom) of CD45+ cells in 12-wk female NOD.Wdfy4−/− (KO) (red circles) and NOD.Wdfy4+/− (Het) (white circles). (B) Graph of absolute cell number (Top) and percentage (Bottom) of CD11c+MHCII+ (gated as in SI Appendix, Fig. S2) cells in 12-wk female NOD.Wdfy4−/− (red, KO) and NOD.Wdfy4+/− (white, Het) islets. (C) Graph of absolute cell number (Top) and percentage (Bottom) of TCRβ+ (gated as in SI Appendix, Fig. S2) cells in 12-wk female NOD.Wdfy4−/− (red, KO) and NOD.Wdfy4+/− (white, Het) islets. Data are pooled biologically independent samples from two independent experiments (n = 3 for NOD.Wdfy4+/− (Het) and n = 5 for NOD.Wdfy4−/− (KO). *P = <0.05 Mann–Whitney test.
β Cell Reactive CD4+ T Cells Are Activated in NOD.Wdfy4−/− Mice.
BDC2.5 Tg T cells (20) recognize a β cell-specific hybrid insulin peptide (21). Previously, we found that BDC2.5 Tg T cells adoptively transferred into NOD.Batf3−/− mice showed severely reduced proliferation in PLNs in vivo compared to WT NOD mice (20). NOD.Batf3−/− mice lack cDC1, and so are unable to prime CD8+ T cells but may also lack an unrecognized requirement for cDC1 in MHC-II restricted antigen presentation to CD4+ T cells. Alternately, the reduced BDC2.5 T cell proliferation could result indirectly from reduced amounts of antigen that may be required if the loss of CD8+ T cell priming led to insufficient amounts of antigen required to drive BDC2.5 Tg T cell proliferation.
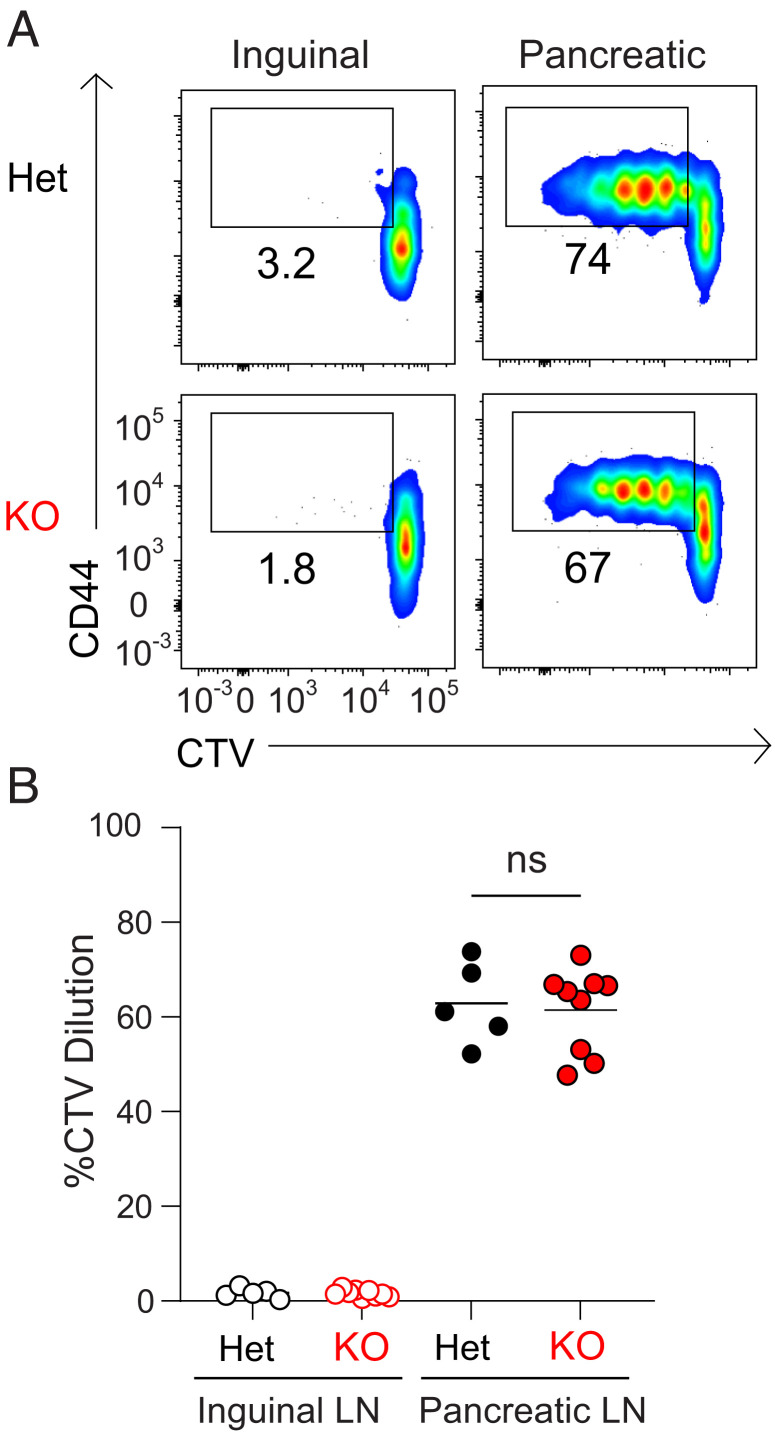
To distinguish these possibilities, we transferred Cell Trace Violet (CTV)-labeled BDC2.5 Tg T cells into mice to determine if autoreactive CD4+ T cells could be primed by cDC1 in the absence of CD8+ T cell cross-priming. We analyzed the proliferation of transferred BDC2.5 Tg T cells in NOD.Wdfy4+/− or NOD.Wdfy4−/− mice. BDC2.5 Tg T cells transferred into NOD.Wdfy4+/− mice proliferated in the PLN, but not in the control ILN (Fig. 4 A and B). BDC2.5 Tg T cells also proliferated after transfer into NOD.Wdfy4−/− mice, and their proliferation was equivalent to heterozygote controls (Fig. 4 A and B). This result demonstrates that CD8+ T cell cross-priming is not required for the priming of autoreactive CD4+ T cells in NOD mice. Together with our results in NOD.Batf3−/− mice, these data suggest that reduced CD4+ T cell priming in NOD.Batf3−/− mice was due to the absence of cDC1. Furthermore, we can conclude that cDC1 are the dominant cell type that primes β cell-specific BDC2.5 autoreactive CD4+ T cells.
Fig. 4.
NOD.Wdfy4−/− mice prime β cell reactive BDC2.5 TCR tg CD4+ T cells. NOD.Wdfy4+/−(Het) and NOD.Wdfy4−/− (KO) 6 wk old female mice were injected intravenously (i.v.) with 106 CTV labeled BDC2.5 CD45.2 cells. (A) Representative flow plots of proliferating BDC2.5 CD45.2 T cells 3 d after transfer. (B) Percentages of proliferating BDC2.5 CD45.2 cells transferred. Data are pooled biologically independent samples from three independent experiments (n = 5 for NOD.Wdfy4+/− (Het) and n = 9 for NOD.Wdfy4−/− (KO). ns = not significant Mann-Whitney test.
Discussion
Our main finding is that selectively inactivating cross-presentation by cDC1 in NOD mice prevents activation of autoreactive CD8+ T cells and averts overt insulitis, but without preventing activation of autoreactive CD4+ T cells and some peri-insulitis. This suggests that autoreactive CD4+ T cells on their own are not sufficient for progression to unrestricted insulitis in NOD mice. cDC1 prime CD8+ T cells through cross-presentation, but are also able to prime CD4+ T cells against cell-associated antigens (15). Thus, the previous finding that NOD.Batf3−/− mice do not develop T1D could have been a result either of the loss of autoreactive CD8+ T cells, or loss of autoreactive CD4+ T cells, or both (13). By contrast, NOD.Wdfy4−/− mice have a defect only in the activation of autoreactive CD8+ T cells, with antigen processing for MHC-II dependent antigens and activation of autoreactive CD4+ T cells being left intact. Since CD8+ T cells are known to be required for T1D in NOD mice (7), the prevention of T1D in NOD.Wdfy4−/− mice is not surprising. However, we were surprised by reduction in insulitis in these mice despite the maintenance of evidence for activation of autoreactive BDC2.5 Tg T cells. These results suggest the infiltration of CD4+ T cells into NOD islets requires additional events beyond their initial activation in PLNs, which are likely to depend on contributions of autoreactive CD8+ T cells.
Autoreactive CD8+ T cells could contribute to the development of insulitis by CD4+ T cells in several ways. For example, damage to β cells by cytolytic CD8+ T cells may be required to recruit CD4+ T cells into the islet. Indeed, recent studies using intravital microscopy have revealed that early lesions in T1D in NOD mice involve infiltration of CD8+ and CD4+ T cells (22), with interactions between T cells and both macrophages and DCs (13, 23). Since we find that BDC2.5 Tg T cells undergo proliferation in PLNs of NOD.Wdfy4−/− mice, it seems that cytolytic damage to islet β cells by CD8+ T cells is not required for the production of islet antigens capable of trafficking to LNs. However, it is conceivable that the proliferation of BDC2.5 in PLNs observed here is not sufficient to fully induce effect CD4+ T cell differentiation that is normally observed in T1D in NOD mice.
The role of cross-presentation in the development of T1D in NOD mice has been unclear. One study suggested that cDC1 are reduced in NOD mice and take on a tolerogenic activity (24). Another study suggested that the activity of cross-presentation by cDC1 in NOD mice is impaired or defective (25). In contrast, our previous results indicated that cDC1 are required for the development of T1D in NOD mice, inconsistent with a tolerogenic function (13). Secondly, our present results indicate that cross-presentation is intact in NOD mice, and in fact is required for the activation and priming of autoreactive CD8+ T cells. In summary, our results suggest that full insulitis leading to T1D in NOD mice involves the coordinated activities of both CD4+ T cells with CD8+ T cells that are activated by cross-presentation by cDC1 in PLNs.
Materials and Methods
Mice.
NOD/ShiLtJ (NOD), NOD.Cg-Tg(TcraBDC2.5,TcrbBDC2.5)1Doi/DoiJ (BDC 2.5), NOD.Cg-Tg(TcraTcrbNY8.3)1Pesa/DvsJ (8.3) mice were obtained from the Jackson Laboratory. NOD.B6-Ptprcb/6908MrkTacJ (NOD.CD45.2) mice were a gift of Dr. Emil Unanue (Washington University in St. Louis). The 8.3 and BDC 2.5 transgenic (Tg) mice were bred to NOD.CD45.2 mice to generate 8.3 CD45.2 and BDC 2.5 CD45.2 mice for T cell transfer. Mice were humanely euthanized by CO2 anesthesia followed by cervical dislocation in accordance with the Guide for the Care and Use of Laboratory Animals of the NIH under approval by the Institutional Animal Care and Use Committee (IACUC) at Washington University School of Medicine (Assurance Number: A3381-01).
Generation NOD.Wdfy4−/− Mice.
NOD.Wdfy4−/− mice were generated essentially as previously described (16) but by directly targeting NOD zygotes in place of C57BL/6 zygotes. These Single guide RNAs (sgRNAs) flanking Wdfy4 exon 4 were identified using CHOPCHOP (http://chopchop.cbu.uib.no/); Wdfy4 gRNA1 (CATGTAGCCTTGAGGTACAT); Wdfy4 gRNA2 (GTCCCCTTTCCTCATAGACT). Single guide RNAs (sgRNAs) were conjugated with Cas9 protein, electroporated into 0.5 d NOD zygotes and transferred into oviducts of pseudopregnant-recipient mice. Offspring were screened for exon 4 deletion using three oligonucleotide primers. A forward oligonucleotide primer located in intron 3 (primer 1) (GTAGGGGTCCAGTTTTGGAGG) and reverse primer located in exon 4 (primer 2) (TCCTGATCCGCGTCACTCTT) were used to confirm the deletion of exon 4 (SI Appendix, Fig. S1). Primer 1 and a reverse oligo located in intron 4 (primer 3) (TGGTTACACACAGCTCGTCC) were used to distinguish the WT and targeted Wdfy4 alleles (SI Appendix, Fig. S1). One founder with complete exon 4 deletion was crossed to wild-type NOD mice and offspring intercrossed to generate experimental NOD.Wdfy4−/− mice and controls. Mice were maintained in a specific pathogen-free facility in accordance with the Guide for the Care and Use of Laboratory Animals of the NIH under approval by the Institutional Animal Care and Use Committee (IACUC) at Washington University School of Medicine (Assurance Number: A3381-01).
Flow Cytometry, Antibodies, and Cell Sorting.
Flow cytometry was performed using a FACSCanto II or FACSAria II (BD Biosciences) essentially as described (13). Data were analyzed using FlowJo software (Tree Star Software).
Pancreatic and inguinal lymph nodes were dispersed using Cell Dissociation Solution Nonenzymatic (Sigma-Aldrich) for 5 min at 37 °C, single cell suspensions treated with 2.4G2 conditioned media (PBS, 1% bovine serum albumin, and 12.5% 2.4G2 in Iscove's modified Dulbecco's medium (IMDM) at 4 °C for 15 min to block Fc receptors. Antibodies included: from BD Biosciences: CD4 (RM4-5), CD8α (53-6.7), CD8β (53-5.8), CD11b (M1/70), B220 (RA3-6B2), CD19 (1D3), CD3 (145-2C11), CD45 (30-F11), Vβ4 (KT4); from Tonbo Biosciences: CD44 (IM7), CD45.1 (A20), CD45.2 (104), CD11c (N418); from Biolegend: anti-rat RT1B (OX-6), XCR1 (ZET), Ter119 (Ter-119), Ly6G (1A8), TCRβ (H57-597), CD3 (145-2C11), CD8 (53-6.7), CD4 (RMA4-5), CD44 (IM7), CD16/32 (93), RT1B (OX-6), Vβ8.1/8.2 (KJ16-133.18); from eBiosciences: CD45.1 (A20), F4/80 (BM8). Cells were stained with fluorescent antibodies and analyzed and/or sorted via a FACSCanto II or FACSAria II (BD Biosciences). Data were analyzed using FlowJo software (Tree Star Software).
RT-qPCR.
Spleens were minced and digested in 5 mL of complete IMDM with 250 μg mL−1 of collagenase B (Roche) and 30 U mL−1 of DNaseI (Sigma) for 30 min at 37 °C with stirring. After digestion, single-cell suspensions were passed through 70-μm strainers and red blood cells were lysed with ammonium chloride-potassium bicarbonate lysis buffer. cDC1 and T cells were sorted and DNase-treated total RNA was prepared with NucleoSpin RNA XS Kit (Macherey-Nagel) and first-strand cDNA synthesis was performed with SuperScript IV Reverse Transcriptase (Invitrogen) using Oligo (dT)25. The relative quantification of gene expression was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) using Luminaris Color HiGreen High ROX qPCR Master Mix (Thermo Scientific). PCR conditions were 2 min at 50 °C, 10 min at 95 °C, followed by 40 three-step cycles consisting of 15 s at 95 °C, 30 s at 60 °C and 30 s at 72 °C. Oligonucleotide primers targeting exon 4 of Wdfy4 are listed as below with Gapdh control primers: Wdfy4F (AAAGGCTGGCAGAAGATGTG); Wdfy4R (ATCCTGATCCGCGTCACTC); mGapdh Qf (ACGGCCGCATCTTCTTGTGCA); mGapdh Qr (ACGGCCAAATCCGTTCACACC).
In Vivo T Cell Proliferation Assay.
The in vivo T cell proliferation assay was performed for BDC2.5 and 8.3 TCR Tg T cells essentially as previously described (13). Briefly, BDC2.5 and 8.3 TCR Tg mouse spleens dispersed into single-cell suspensions, washed, incubated with MagniSortTM SAV negative selection beads (Invitrogen), magnetically separated and sort-purified as B220– CD8– TCRβ+ CD4+ CD45.1+ Vβ4+ (BDC2.5) or B220– CD8+ TCRβ+ CD4– CD45.1+ Vβ8.1/8.2+ (8.3). T cells were stained with 1 μM Cell Trace Violet (CTV) (Invitrogen) for 10 min at 37 °C and quenched with 4 °C IMDM in 10% FCS, 106 labeled T cells injected intravenously into recipient mice. After 3 d, draining pancreatic lymph nodes (PLNs) and inguinal lymph nodes (ILNs) were harvested, dispersed and stained with for CD45.1, CD45.2, Vβ4, 7AAD, CD4, CD44, and TCRβ (BDC2.5 transfer) or CD45.1, CD45.2, Vβ8.1/8.2, 7AAD, CD8, CD44, and TCRβ (8.3 transfer). Cells gated as CD4+ TCRβ+CD45.2+ Vβ4+CD44+ (BDC2.5) or CD8+ TCRβ+CD45.2+ Vβ8.1/8.2+CD44+ (8.3) were analyzed for CTV dilution on a FACs CANTO II.
Diabetes Monitoring.
Blood glucose was monitored daily or weekly by urine glucose readings via Diastix (Ascencia). After two consecutive readings of ≥250 mg/dL mice were considered diabetic.
Islet Isolation and Histology.
Islets were isolated as previously described (13). For histology, pancreata were isolated and placed in neutral buffered formalin for 1 wk, paraffin-embedded, sectioned, and stained with hematoxylin and eosin (H&E).
Insulitis Scoring.
Insulitis was assessed by histological score. Briefly, pancreatic islets from 52-wk–old nondiabetic NOD, NOD.Wdfy4+/−, NOD.Wdfy4−/− (at least 4 mice from each genotype) were examined by H&E to assess the degree of infiltration. Insulitis scoring was performed according to the following criteria: no insulitis, peri-insulitis (peri-insular leukocytic aggregates), and insulitis (leukocytic infiltration into islets).
Statistics.
Statistical analysis was performed using GraphPad Prism software version 8. Unless otherwise noted, a one-way ANOVA or Mann–Whitney test was used to determine significant differences between samples, and all center values correspond to the mean. P ≤ 0.05 was considered statistically significant. Investigators were blinded to the treatments of the mice during sample preparation and data collection.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This publication is solely the responsibility of the authors and does not necessarily represent the official view of the NIH. This work was supported by the NIH (R01AI150297, R01CA248919, R01AI162643, and R21AI164142 to K.M.M., and F30CA247262 to R.W.). S.T.F. is a Cancer Research Institute Irvington Fellows supported by the Cancer Research Institute. S.T.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Author contributions
S.T.F., T.L., J.C., R.A.O., F.O., S.K., T.L.M., and K.M.M. designed research; S.T.F., T.L., J.C., R.A.O., F.O., S.K., and T.L.M. performed research; S.T.F., T.L., J.C., R.A.O., F.O., R.W., S.K., T.L.M., and K.M.M. contributed new reagents/analytic tools; S.T.F., T.L., J.C., R.A.O., F.O., S.K., T.L.M., and K.M.M. analyzed data; and S.T.F., T.L., J.C., R.A.O., F.O., R.W., S.K., T.L.M., and K.M.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article can be found as a preprint on biorxiv.org: doi: 10.1101/2022.09.02.506326.
Reviewers: J.D.K., Cincinnati Children’s Hospital Medical Center; and D.M., Harvard Medical School.
Contributor Information
Stephen T. Ferris, Email: stephen.ferris@health.slu.edu.
Kenneth M. Murphy, Email: kmurphy@wustl.edu.
Data, Materials, and Software Availability
The data generated during and analyzed during the current study are all available in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Makino S., et al. , Breeding of a non-obese, diabetic strain of mice. Jikken Dobutsu 29, 1–13 (1980). [DOI] [PubMed] [Google Scholar]
- 2.Barrett J. C., et al. , Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41, 703–707 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Todd J. A., Bell J. I., McDevitt H. O., HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 329, 599–604 (1987). [DOI] [PubMed] [Google Scholar]
- 4.Suri A., Walters J. J., Gross M. L., Unanue E. R., Natural peptides selected by diabetogenic DQ8 and murine I-A(g7) molecules show common sequence specificity. J. Clin. Invest. 115, 2268–2276 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Lummel M., et al. , Type 1 diabetes-associated HLA-DQ8 transdimer accommodates a unique peptide repertoire. J. Biol. Chem. 287, 9514–9524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bendelac A., Carnaud C., Boitard C., Bach J. F., Syngeneic transfer of autoimmune diabetes from diabetic NOD mice to healthy neonates. Requirement for both L3T4+ and Lyt-2+ T cells. J. Exp. Med. 166, 823–832 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katz J., Benoist C., Mathis D., Major histocompatibility complex class I molecules are required for the development of insulitis in non-obese diabetic mice. Eur. J. Immunol. 23, 3358–3360 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Nejentsev S., et al. , Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 450, 887–892 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bender C., Rajendran S., von Herrath M. G., New insights into the role of autoreactive CD8+ T cells and cytokines in human type 1 diabetes. Front. Endocrinol. (Lausanne) 11, 606434 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esensten J. H., Lee M. R., Glimcher L. H., Bluestone J. A., T-bet-deficient NOD mice are protected from diabetes due to defects in both T cell and innate immune system function. J. Immunol. 183, 75–82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willcox A., Richardson S. J., Bone A. J., Foulis A. K., Morgan N. G., Analysis of islet inflammation in human type 1 diabetes. Clin. Exp. Immunol. 155, 173–181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skowera A., et al. , CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J. Clin. Invest. 118, 3390–3402 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferris S. T., et al. , A minor subset of Batf3-dependent antigen-presenting cells in islets of Langerhans is essential for the development of autoimmune diabetes. Immunity 41, 657–669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildner K., et al. , Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferris S. T., et al. , cDC1 prime and are licensed by CD4(+) T cells to induce anti-tumour immunity. Nature 584, 624–629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theisen D. J., et al. , WDFY4 is required for cross-presentation in response to viral and tumor antigens. Science 362, 694–699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verdaguer J., et al. , Spontaneous autoimmune diabetes in monoclonal T cell nonobese diabetic mice. J. Exp. Med. 186, 1663–1676 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H., et al. , Targeting transcriptional coregulator OCA-B/Pou2af1 blocks activated autoreactive T cells in the pancreas and type 1 diabetes. J. Exp. Med. 218, e20200533 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieberman S. M., et al. , Identification of the beta cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proc. Natl. Acad. Sci. U.S.A. 100, 8384–8388 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz J. D., Wang B., Haskins K., Benoist C., Mathis D., Following a diabetogenic T cell from genesis through pathogenesis. Cell 74, 1089–1100 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Stadinski B. D., et al. , Chromogranin A is an autoantigen in type 1 diabetes. Nat. Immunol. 11, 225–231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohan J. F., et al. , Imaging the emergence and natural progression of spontaneous autoimmune diabetes. Proc. Natl. Acad. Sci. U.S.A. 114, E7776–E7785 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calderon B., Carrero J. A., Unanue E. R., The central role of antigen presentation in islets of Langerhans in autoimmune diabetes. Curr. Opin. Immunol. 26, 32–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welzen-Coppens J. M., Helden-Meeuwsen C. G., Leenen P. J., Drexhage H. A., Versnel M. A., Reduced numbers of dendritic cells with a tolerogenic phenotype in the prediabetic pancreas of NOD mice. J. Leukoc Biol. 92, 1207–1213 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Lee C. N., Lew A. M., Shortman K., Wu L., NOD mice are functionally deficient in the capacity of cross-presentation. Immunol. Cell Biol. 93, 548–557 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The data generated during and analyzed during the current study are all available in the manuscript and/or SI Appendix.