Abstract
The epithelial and interstitial stem cells of the freshwater polyp Hydra are the best-characterized stem cell systems in any cnidarian, providing valuable insight into cell type evolution and the origin of stemness in animals. However, little is known about the transcriptional regulatory mechanisms that determine how these stem cells are maintained and how they give rise to their diverse differentiated progeny. To address such questions, a thorough understanding of transcriptional regulation in Hydra is needed. To this end, we generated extensive new resources for characterizing transcriptional regulation in Hydra, including new genome assemblies for Hydra oligactis and the AEP strain of Hydra vulgaris, an updated whole-animal single-cell RNA-seq atlas, and genome-wide maps of chromatin interactions, chromatin accessibility, sequence conservation, and histone modifications. These data revealed the existence of large kilobase-scale chromatin interaction domains in the Hydra genome that contain transcriptionally coregulated genes. We also uncovered the transcriptomic profiles of two previously molecularly uncharacterized cell types: isorhiza-type nematocytes and somatic gonad ectoderm. Finally, we identified novel candidate regulators of cell type–specific transcription, several of which have likely been conserved at least since the divergence of Hydra and the jellyfish Clytia hemisphaerica more than 400 million years ago.
The advent of highly specialized cell type–specific transcriptional programs played a critical role in the emergence and subsequent diversification of animal life. Decades of research have greatly advanced our understanding of the mechanisms of transcriptional regulation that underlie cell identity in metazoans. However, much of that understanding is based on findings from bilaterian species. Consequently, relatively little is known about transcriptional regulation in nonbilaterian metazoans.
Cnidaria is the sister phylum to Bilateria (Dunn et al. 2014), and despite having diverged more than 500 million years ago, the two clades show extensive homology at the molecular level. These similarities include important aspects of transcriptional regulation: Both cnidarians and bilaterians use combinatorial histone modifications and distal enhancer-like cis-regulatory elements (CREs) (Schwaiger et al. 2014; Reddy et al. 2020; Murad et al. 2021); many transcription factors (TFs) in bilaterians are also present in cnidarians (Technau et al. 2005; Putnam et al. 2007; Babonis and Martindale 2017b); and the target genes of developmentally significant TFs are at least partially conserved across the two clades (Münder et al. 2010; Gufler et al. 2018; Hartl et al. 2019). However, beyond these general similarities, little is known about cnidarian gene regulatory networks and the mechanisms they use to specify and maintain cellular identity. Given Cnidaria's phylogenetic position within Metazoa, research in cnidarians is uniquely positioned to shed light on the evolutionary origins of Bilateria. In addition, many cnidarians possess remarkable abilities of self-repair and self-renewal not found in most bilaterian model systems, with species capable of whole-body regeneration (Trembley 1744; Darling et al. 2005; Bradshaw et al. 2015) and potentially biological immortality (Piraino et al. 1996; Martínez 1998; Schaible et al. 2015). Thus, a thorough characterization of transcriptional regulation in cnidarians can contribute to our understanding of both the origins and fundamental principles of transcriptional regulation of cell type in metazoans and the molecular basis for cnidarian resilience.
Species belonging to the genus Hydra are among the longest-studied and best-characterized cnidarian models, with the first experiments in Hydra dating back to 1744 (Trembley 1744). Hydra has since been used to study patterning (Browne 1909; Gierer and Meinhardt 1972), stem cell biology (David and Murphy 1977; Bode et al. 1987; Bosch and David 1987; David 2012), aging (Martínez 1998; Schaible et al. 2015), regeneration (Trembley 1744), and symbiosis (Fraune and Bosch 2007; Hamada et al. 2018).
One of the strengths of Hydra as a research organism is its simplicity. In contrast to the three life cycle stages—planula, polyp, and medusa—found in their close cnidarian relatives, Hydra species possess only a polyp stage. This polyp is organized along a single oral–aboral axis, with a head made up of a mouth surrounded by a ring of tentacles at the oral pole and an adhesive foot at the aboral pole. Between the head and foot lies the body column, which serves as both the gut and stem cell compartment. The body is made up of two epithelial layers—endoderm and ectoderm—separated by an extracellular matrix. Interspersed throughout both epithelial layers are interstitial cells, which include gland cells, neurons, germ cells, and nematocytes—the specialized stinging cells unique to cnidarians. In adult polyps, ectodermal, endodermal, and interstitial cells constitute three different cell lineages, each supported by their own stem cell population. The simplicity of this system has allowed researchers to identify every cell type in Hydra as well as the developmental trajectories that give rise to them (David 2012; Siebert et al. 2019). However, the gene regulatory networks that coordinate these differentiation events remain poorly understood.
Over the past 15 years, the advent of powerful tools and resources—including a reference genome (Chapman et al. 2010), a single-cell gene expression atlas (Siebert et al. 2019), knockdown techniques (Khalturin et al. 2008; Boehm et al. 2012), and transgenesis (Wittlieb et al. 2006)—has allowed researchers to address topics such as regeneration and patterning at the molecular level. However, complicating the effective use of these tools is the fact that these resources were developed using different genetic backgrounds. Specifically, the currently available and recently improved reference genome (Simakov et al. 2022) was generated using strain 105 of Hydra vulgaris (formerly H. magnipapillata), whereas all transgenic Hydra lines and the single-cell expression atlas were generated using the AEP strain. The AEP and 105 strains belong to two distinct lineages that split ∼16 million years ago, leading to significant sequence divergence that markedly reduces cross-strain mapping efficiencies (Martínez et al. 2010; Schwentner and Bosch 2015; Siebert et al. 2019; Wong et al. 2019; Schenkelaars et al. 2020). This highlights the need for an AEP strain reference genome that would allow researchers to more effectively leverage transgenesis and the single-cell expression atlas.
Another appealing, although currently underused, strength of Hydra is that it is relatively closely related to several other established and emerging laboratory models belonging to the class Hydrozoa, creating opportunities for comparative studies. Recently published genomic and transcriptomic resources, including reference genomes for the green Hydra viridissima (Hamada et al. 2020) and the jellyfish Clytia hemisphaerica (Leclère et al. 2019) as well as a single-cell gene expression atlas of the C. hemisphaerica medusa (Chari et al. 2021), provide valuable reference points for systematic comparative analyses. The Hydra genus is associated with several noteworthy evolutionary gains and losses, including the loss of a medusa stage, the acquisition of stably associated endosymbionts in H. viridissima (Schwentner and Bosch 2015), and the loss of certain types of aboral regeneration in Hydra oligactis (Supplemental Fig. S1; Weimer 1928; Hoffmeister 1991; Grens et al. 1996). Thus, effectively establishing a framework for systematic comparative approaches would greatly enhance our ability to interrogate both the conserved and unique aspects of Hydra biology.
To facilitate comparative genomic research in Hydra, we report two new high-quality genomes, a chromosome-level assembly for the AEP strain of H. vulgaris and a draft assembly for the H. oligactis Innsbruck female12 strain. To leverage these new references to better understand transcriptional regulation in Hydra, we used multiple independent approaches, such as assay for transposase accessible chromatin using sequencing (ATAC-seq), cleavage under targets and tagmentation (CUT&Tag) targeting histone modifications, and phylogenetic footprinting, to annotate CREs in the AEP genome. We also generated Hi-C data that revealed domains of elevated chromatin contact frequency that likely contain transcriptionally coregulated genes. To accompany these new resources, we generated an updated and improved version of the Hydra single-cell atlas using the AEP-strain genome as a reference and subsequently uncovered two previously molecularly uncharacterized cell types: somatic gonad ectoderm and mature isorhiza nematocytes. We then combined our CRE annotations with the AEP single-cell atlas to identify novel candidate regulators of cell type–specific gene coexpression. Finally, we aligned the Hydra single-cell atlas with a Clytia medusa single-cell atlas and identified gene regulatory modules in the interstitial lineage that have likely been conserved over at least 400 million years of evolution (Schwentner and Bosch 2015; Dohrmann and Wörheide 2017). The resources generated in this study, which include a genome browser for the H. oligactis and strain AEP H. vulgaris assemblies, a BLAST server, and an interactive portal for the AEP-mapped Hydra single-cell atlas, are available at the Hydra AEP Genome Project Portal (https://research.nhgri.nih.gov/HydraAEP/).
Results
Generation and annotation of two high-quality Hydra genome assemblies
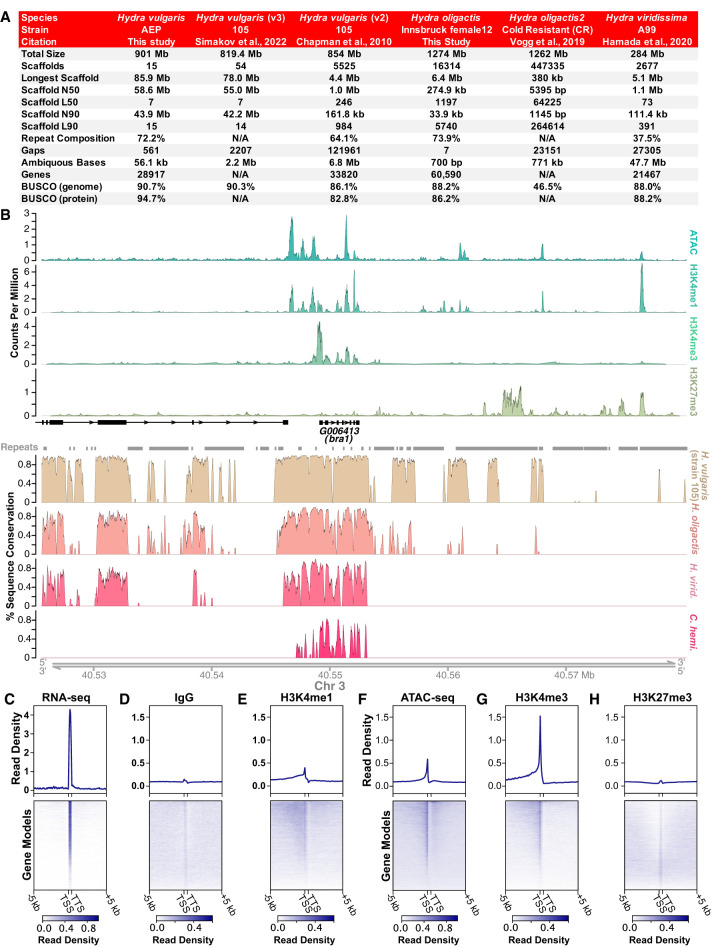
We sequenced, assembled, and annotated a chromosome-level genome assembly for the AEP laboratory strain of H. vulgaris (for details, see Supplemental Material; Supplemental Figs. S1, S2; Supplemental Table S1; Supplemental Data S1). In addition, we generated a high-quality draft genome for the Innsbruck female12 strain of H. oligactis. We were motivated to generate a genome reference for H. oligactis because its phylogenetic position as a sister species to H. vulgaris—along with unique traits such as reduced regenerative capacity (Weimer 1928; Hoffmeister 1991; Grens et al. 1996), a deficient heat shock response (Bosch et al. 1988), and inducible senescence (Yoshida et al. 2006)—makes it valuable for comparative genomic studies of the Hydra genus. The resulting assemblies for H. vulgaris and H. oligactis were of equivalent or greater completeness and contiguity compared with other available hydrozoan genomes (Fig. 1A; Supplemental Table S2).
Figure 1.
New genome assemblies provide improved resources for Hydra molecular biology research. (A) The Hydra vulgaris strain AEP and Hydra oligactis genome assemblies presented in this study are marked improvements on the previously available reference genomes for their respective species. (B) Representative plot of CUT&Tag, ATAC-seq, and genomic conservation tracks centered on the hybra1 gene (Technau and Bode 1999). For the sequencing data, each track represents the signal from pooled biological replicates for the specified library type. A plot of the same locus that includes separate tracks for each CUT&Tag and ATAC-seq biological replicate is presented in Supplemental Figure S4. (C–H) Read distribution for sequencing data centered on AEP assembly gene models. (C) Whole-animal RNA-seq data are strongly enriched in predicted coding sequences. (D) Control IgG CUT&Tag reads show minimal enrichment in or around genes. (E) H3K4me1 is enriched in promoter-proximal regions, but only weakly enriched at transcription start site (TSS). (F) ATAC-seq is enriched at TSS, but also shows some enrichment in more distal regions, likely because ATAC-seq targets both promoters and enhancers. (G) H3K4me3 is strongly enriched at the TSS. (H) H3K27me3 shows minimal or no enrichment near transcribed genes. (TTS) Transcription termination site.
We found that synteny in the strain 105 and AEP genome assemblies was highly conserved, with the notable exception of an ∼5-Mb inversion on Chromosome 8 (Supplemental Fig. S3A,B; Supplemental Table S3). Similarly, the centromeric repeats in the two strains were highly similar, although not identical (Supplemental Fig. S3C; Melters et al. 2013). In addition, this analysis allowed us to place nearly all (36/39) of the unincorporated scaffolds from the strain 105 assembly onto one of the 15 pseudochromosome scaffolds in the AEP assembly (Supplemental Data S2). Similarly, we were able to generate preliminary chromosome assignments for contigs covering 91.3% of the sequence (1.16 out of 1.27 Gb) in the H. oligactis assembly and 32.3% (91.8 out of 284.3 Mb) of a previously published assembly for H. viridissima (Supplemental Fig. S3D,E; Supplemental Data S2; Hamada et al. 2020).
To augment the strain AEP H. vulgaris genome assembly, we also generated genome-wide CRE annotations. To do this, we used the ATAC-seq (Buenrostro et al. 2013; Corces et al. 2017) to map accessible regions of chromatin. We also established a protocol for performing CUT&Tag (Kaya-Okur et al. 2019) in Hydra to globally map multiple histone modifications, including the repressive histone modification H3K27me3 as well as the activating histone modifications H3K4me1 and H3K4me3 (Supplemental Data S3; for details, see Supplemental Material). We validated our results by confirming that they matched the expected distribution patterns of their associated genomic features (Fig. 1B,C; Supplemental Figs. S4–S6).
To supplement our CRE annotations, we performed phylogenetic footprinting (Tagle et al. 1988; Gumucio et al. 1992) by using previously published genomes for the hydrozoans C. hemisphaerica (Leclère et al. 2019), H. viridissima (Hamada et al. 2020), and the 105 strain of H. vulgaris (Chapman et al. 2010)—along with our newly assembled genomes—to generate a cross-species whole-genome alignment that spanned ∼400 million years of hydrozoan evolution (Fig. 1B). Our alignment yielded results that recapitulated the findings from previous manual cross-species alignments of individual Hydra promoter regions (Supplemental Fig. S7; Vogg et al. 2019), supporting the accuracy of our genome-wide approach. We then used our whole-genome alignment to classify genomic features as either conserved or nonconserved (for details, see Supplemental Material). We provide lists of conserved noncoding genomic features in Supplemental Data S3.
Prediction of conserved TF binding sites using phylogenetic footprinting
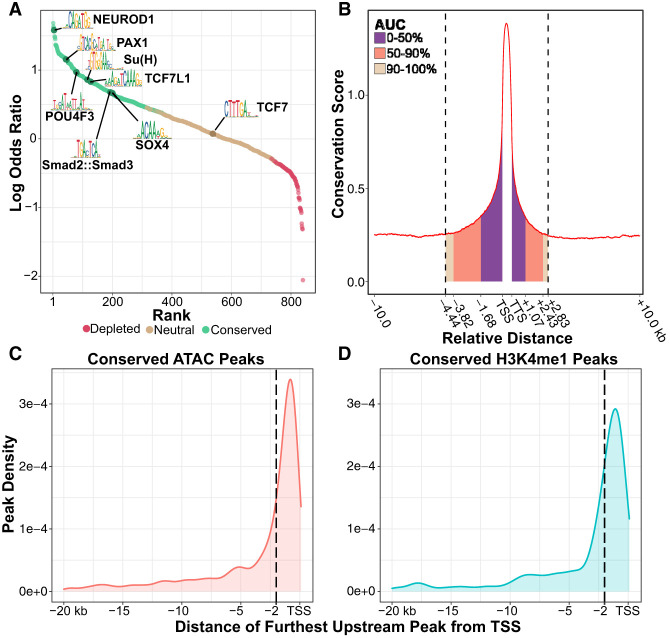
Accurately identifying TF binding sites in CREs is an essential, albeit often challenging, aspect of gene regulatory network characterization. This task is made especially difficult in nonbilaterian metazoans by the lack of specific antibodies needed for conventional TF mapping assays (e.g., ChIP-seq and CUT&RUN). The lack of binding data can even hinder computational approaches for predicting binding sites, as the binding preferences of cnidarian TFs typically must be inferred from data collected from distantly related bilaterians. We therefore sought to evaluate the functional relevance of bilaterian TF binding motifs in Hydra by leveraging phylogenetic footprinting to determine which motifs showed evidence of conservation. Of the 840 motifs considered in our analysis, we found that 384 (45.7%), including those that are bound by numerous conserved and developmentally significant TFs (Fig. 2A), had significantly higher genome-wide conservation rates compared with the shuffled controls (Supplemental Data S4). This suggests that there is extensive conservation of TF binding preferences from cnidarians to bilaterians.
Figure 2.
Phylogenetic footprinting reveals conserved regulatory elements and transcription factor (TF) binding sites across the Hydra genome. (A) Quantification of TF binding motif conservation across four Hydra genomes. A positive log odds value indicates the nonshuffled motif had a higher conservation rate than its shuffled control. Statistical significance was evaluated using a chi-square test with an FDR cutoff of 0.01. (B) The distribution of sequence conservation levels around genes suggests that a sizable minority of promoter-proximal CREs extends farther than 2 kb from the nearest TSS. The conservation score represents the average number of non-AEP hydrozoan genomes that had the same base as the AEP assembly at a given locus. (AUC) Area under the curve; (TSS) transcription start site; (TTS) transcription termination site. Gene bodies were excluded from the AUC calculation. (C,D) Distribution plots summarizing the distance from the furthest upstream CRE for each gene to the predicted target TSS based on either ATAC-seq (C) or H3K4me1 CUT&Tag (D). Dotted vertical lines demarcate 2 kb from the TSS.
Another confounding issue for ab initio TF binding site predictions is that TF binding motifs are typically short and degenerate, leading to high false-positive rates. However, by filtering putative TF binding sites using both our ATAC-seq and phylogenetic footprinting data, we reduced the total number of predicted binding sites genome-wide by >99%, from more than 45 million to 210,122 (Supplemental Data S5). Thus, we simplified the landscape of putative TF binding sites by eliminating loci with a relatively low probability of being bona fide binding sites.
Many Hydra genes are likely regulated by distal regulatory elements
In bilaterians, transcriptional regulation frequently involves long-range interactions between distal CREs and their target promoter, often spanning dozens of kilobases. However, numerous successful reporter lines have been generated in Hydra using only 500–2000 bp of flanking sequence upstream of a gene of interest, motivating some to hypothesize that transcriptional regulation in Hydra is simpler than in bilaterians and primarily regulated by promoter-proximal elements that typically fall within 2 kb of the TSS (Klimovich et al. 2019). However, this hypothesis has not been systematically investigated.
To better understand the distribution of CREs in the Hydra genome, we used our cross-species whole-genome alignment to characterize sequence conservation rates around genes in the AEP assembly (Fig. 2B). We found that flanking noncoding sequences around genes had elevated conservation rates that extended ∼4.4 kb upstream and ∼2.8 kb downstream before falling back to baseline levels. Although we found that most of the elevated sequence conservation fell within 2 kb upstream of the TSS, nearly half of the conservation signal fell outside of that boundary (Fig. 2B). In addition, we found that ∼44% of genes in our analysis had at least one conserved ATAC-seq or H3K4me1 peak further than 2 kb upstream of the TSS (Fig. 2C,D). These results indicate that there are likely many instances in which functionally important CREs lie further than 2 kb from their target gene and highlight the need for functional genomic data to accurately identify promoter regions.
Hydra chromatin is organized into localized contact domains
The three-dimensional organization of DNA molecules in the nucleus is tightly linked to genome regulation (Szabo et al. 2019). Although several cnidarian Hi-C data sets have been published (Supplemental Table S4; Li et al. 2020; Nong et al. 2020; Zimmermann et al. 2020; Simakov et al. 2022), the 3D organization of cnidarian genomes remains largely uncharacterized. We therefore interrogated our Hydra Hi-C data to better understand the 3D architecture of the Hydra genome.
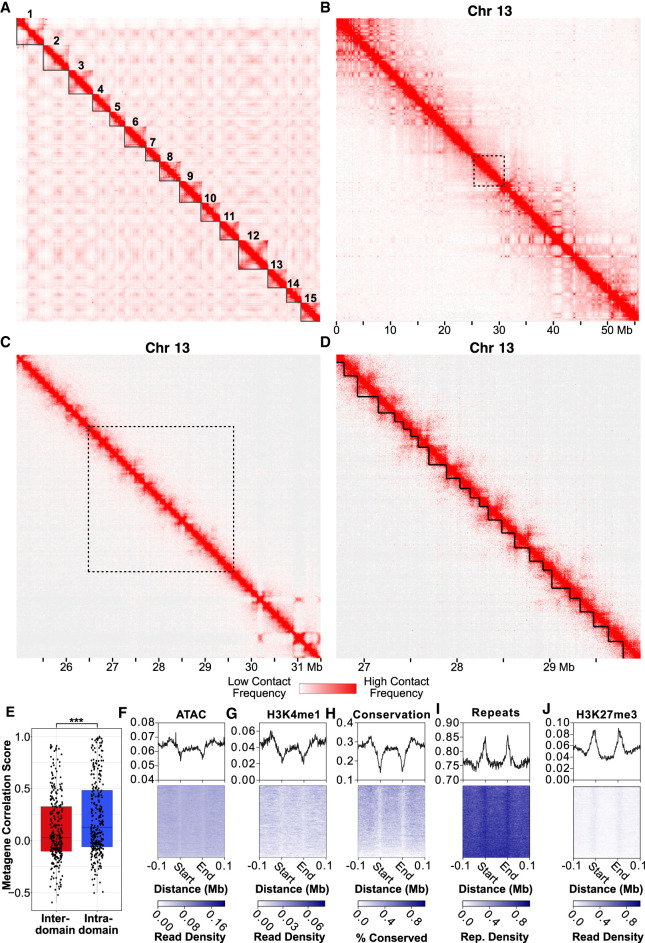
We first examined chromatin interactions at the whole-chromosome scale. We observed signatures of a Rabl-like conformation (Hoencamp et al. 2021), with interactions occurring between centromeres of different chromosomes as well as between centromeres and telomeres within individual chromosomes (Fig. 3A). Compared with previously characterized cnidarian genomes, these interaction patterns appeared unique to Hydra, as we had not observed similar phenomena in other publicly available cnidarian Hi-C data sets. We therefore performed a systematic analysis of inter-chromosomal interactions in cnidarians (for details, see Supplemental Materials) and found that the Hydra genome had significantly elevated levels of inter-centromeric interactions, but not inter-telomeric interactions, relative to other cnidarians (Supplemental Fig. S8). Notably, this change in 3D genome organization appeared to be correlated with the loss of multiple condensin II subunits in hydrozoans (Supplemental Fig. S9; Supplemental Data S6). These lost subunits were shown to inhibit inter-chromosomal interactions in other species (Hoencamp et al. 2021), suggesting that their loss has resulted in the elevated levels of inter-centromeric interactions in Hydra and possibly other hydrozoans. However, the extent to which these interaction patterns are present in other hydrozoan genomes is unknown owing to a lack of Hi-C data from other hydrozoan species.
Figure 3.
Hi-C data reveal hierarchical chromatin architecture in the Hydra genome. (A) Hi-C contact map for the H. vulgaris strain AEP assembly reveals 15 pseudochromosomes with high levels of both inter-chromosomal interactions between presumptive centromeric regions and intra-chromosomal interactions between centromeric and telomeric regions. (B) The chromatin interaction map for Chromosome 13 reveals megabase-scale chromatin compartments. The black dotted lines indicate the region visualized in the subsequent figure panel. (C) Kilobase-scale interaction domains can be found within a single megabase-scale compartment. (D) Representative depiction of predicted kilobase-scale chromatin interaction domains in Hydra (black lines). (E) Boxplot/scatterplot depicting the correlation in expression for adjacent gene pairs show that gene pairs within the same domain (intra-domain pairs) were significantly more similar than pairs that spanned a domain boundary (inter-domain pairs; Welch two-sample t-test P-value = 6.93 × 10−5). (F–J) Predicted domain boundaries fall within regions of heterochromatin. Domain boundaries are associated with reduced chromatin accessibility (F), H3K4me1 (G), and sequence conservation (H) and with elevated repeat element density (I) and H3K27me3 (J).
We next explored intra-chromosomal interactions in our Hi-C data to look for evidence of chromatin domains or loops, which are structures generated by transcriptional regulatory mechanisms in diverse eukaryotic genomes (Szabo et al. 2019; Zheng and Xie 2019). We found that Hydra chromatin is hierarchically organized into megabase-scale domains that contain much smaller kilobase-scale subdomains (Fig. 3B–D). The larger megabase-scale domains showed a checkerboard-like interaction pattern consistent with the A/B compartments observed in other Hi-C data sets (Fig. 3B; Zheng and Xie 2019). Within these A/B compartments were more localized structures that in places resembled the triangle-shaped patterns associated with topologically associating domains (TADs) in other species (Fig. 3C,D). We also occasionally observed contact patterns suggestive of chromatin loops (Supplemental Fig. S10), but such structures were rare.
To determine if the contact domains we observed were associated with transcriptional regulation, we first used a previously established computational pipeline (Ramírez et al. 2018) to predict chromatin domain boundaries (Fig. 3D). Although the resolution of our Hi-C data made it difficult to fully resolve the kilobase scale domains apparent in the Hydra genome, we were nonetheless able to identify 4028 putative contact domains across the AEP assembly with a median size of ∼176 kb using this approach (Supplemental Data S7). We then used the Hydra single-cell atlas (described below) to characterize the expression patterns of genes around the predicted domain boundaries. We found that the cell type–specific expression patterns of adjacent gene pairs that fell within the same contact domain were significantly more correlated than adjacent gene pairs that spanned a domain boundary (Fig. 3E), suggesting that Hydra chromatin contact domains are indeed associated with transcriptional regulation. We also found that chromatin boundaries were depleted of several euchromatin markers—including chromatin accessibility, sequence conservation, and H3K4me1—and enriched in heterochromatin markers such as increased repetitive element density and higher levels of H3K27me3 (Fig. 3F–J). Altogether, these results suggest that, similar to invertebrate bilaterians, Hydra chromatin is organized into large epigenetically regulated domains that contain coregulated genes. In addition, the clear correlation between the predicted location of domain boundaries and other orthogonal data sets such as ATAC-seq and CUT&Tag shows that our domain prediction analysis indeed captured meaningful aspects of chromatin architecture across the Hydra genome.
An updated single-cell RNA-seq atlas for H. vulgaris uncovers the transcriptional profiles of additional cell types
We next used the genomic resources we had generated to interrogate the transcriptional regulation of cell type specification in Hydra, which required access not only to CRE annotations but also to the transcriptomic profiles associated with different Hydra cell types. We previously published a whole-animal single-cell RNA-seq (scRNA-seq) data set for the AEP strain of H. vulgaris that provides an atlas of molecular cell states in adult polyps (Siebert et al. 2019). However, the currently available versions of this data set use either the strain 105 genome or an AEP strain transcriptome as a reference. Both are suboptimal as the transcriptome does not provide information about genomic context, thus hindering any research into transcriptional regulation, and the 105 genome gene models are less complete and have reduced mapping rates when using AEP RNA-seq data (Supplemental Fig. S11; Supplemental Table S2). In addition, there have been substantial improvements in normalization (Hafemeister and Satija 2019), batch-correction (Stuart et al. 2019), and visualization techniques (McInnes et al. 2018) for scRNA-seq data since the Hydra single-cell atlas was initially published. Therefore, we addressed these limitations by reanalyzing the data using the AEP assembly as a reference.
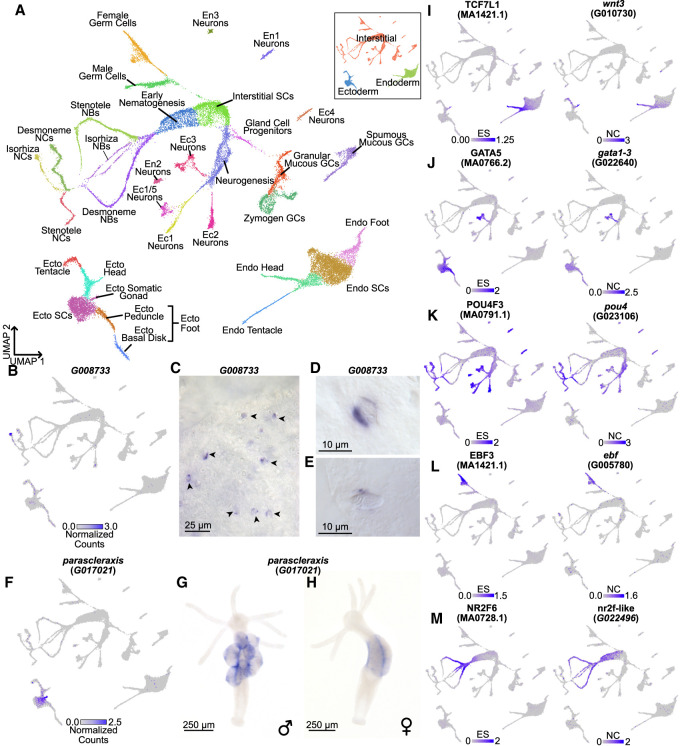
Following mapping and doublet removal (for details, see Supplemental Materials; Supplemental Figs. S12, S13), we recovered 29,339 single-cell transcriptomes that passed our quality control cutoffs, an increase of ∼17.4% compared with the 24,985 transcriptomes presented in the originally published atlas (Siebert et al. 2019). We then used Seurat to perform a Louvain clustering analysis and visualized the results using a uniform manifold approximation and projection (UMAP) dimensional reduction (Fig. 4A; Waltman and Van Eck 2013; McInnes et al. 2018; Hao et al. 2021). We then annotated the resulting clusters using established cell type markers (Supplemental Fig. S14; Siebert et al. 2019). While generating these annotations, we identified two cell types that were not found in previous iterations of the single-cell atlas: isorhiza-type nematocytes and ectodermal male and female somatic gonad cells. We subsequently identified markers of these two populations, which we validated using in situ hybridization (Fig. 4B–H). The isorhiza marker, G008733, has no known functional domains and appears to be specific to brown Hydra. The somatic gonad marker, parascleraxis (G017021), is the ancestral ortholog of two paralogous vertebrate basic helix-loop-helix TFs, paraxis/tcf15 and scleraxis, that regulate muscle differentiation (Freitas et al. 2006; Della Gaspera et al. 2022).
Figure 4.
An updated Hydra single-cell RNA-seq atlas reveals novel regulators of gene coexpression in Hydra. (A) Uniform manifold approximation and projection (UMAP) dimensional reduction of the Hydra single-cell RNA-seq atlas mapped to the AEP reference genome captures virtually all known cell states in adult polyps. Inset shows UMAP colored by the three stem cell lineages in adult Hydra. (NCs) Nematocytes; (NBs) nematoblasts; (SCs) stem cells; (Ecto) ectodermal epithelial cells; (Endo) endodermal epithelial cells; (GCs) gland cells; (Ec) neuron subtypes found in the ectoderm; (En) neuron subtypes found in the endoderm. (B) The gene G008733 is a specific marker for isorhiza nematocytes. (C–E) In situ hybridization targeting G008733 labels isorhiza nematocytes (black arrowheads) in upper body column tissue. (F) scleraxis is a specific marker for ectodermal somatic gonad cells. (G) In situ hybridization targeting scleraxis in male polyps labels ectodermal testes cells. (H) In situ hybridization reveals scleraxis is expressed in egg patches in female polyps. (I–M) Motif enrichment and gene expression patterns reveal candidate regulators of cell state. (I) TCF motif enrichment and wnt3 expression data corroborate the role of TCF/Wnt signaling in epithelial head tissue. (J) GATA motif enrichment and expression data corroborate the role of gata1-3 in aboral epithelial tissue and suggest an additional function in Ec3 neurons. (K) Pou4 motif enrichment and expression data suggest pou4 regulates transcription in differentiating and mature neurons and nematocytes. (L) Ebf motif enrichment and expression data suggest ebf regulates transcription during oogenesis. (M) NR2F motif enrichment and expression data suggest nr2f-like regulates transcription during nematogenesis. Corresponding JASPAR motif IDs are provided in the parenthetical text under the motif names (Fornes et al. 2020). (ES) Enrichment score; (NC) normalized counts.
In summary, we generated an updated scRNA-seq atlas for whole adult Hydra that can now be used in conjunction with the AEP genome assembly. This comprehensively annotated atlas, which incorporates two additional cell types, contains virtually all known cell types in an adult Hydra. We also provide exhaustive lists of marker genes for all clusters (Supplemental Data S8) as well as 56 modules of coexpressed genes (Supplemental Fig. S15; Supplemental Data S9, S10).
Characterizing the evolutionary history of Hydra cell type–specific transcriptomes
The Hydra single-cell atlas captures the transcriptional signatures of virtually all cell states in an adult polyp, which presents a valuable opportunity to gain new insight into the evolutionary history of the transcriptional programs that define cnidarian cell types. The acquisition of novel cellular traits is often accompanied by a concurrent period of genetic innovation (Arendt 2008; Khalturin et al. 2009). This can leave a phylogenetic signature in a cell's transcriptome in the form of an overrepresentation of novel genes that arose during periods of evolutionary change in a cell type's transcriptional program (Domazet-Lošo et al. 2007). Thus, characterizing the age distribution of genes expressed in different cell types can shed light on when those genetic programs arose.
To analyze the relationship between gene age and transcriptional specificity, we first assigned phylostratigraphic ages to Hydra gene families using orthology predictions generated from an OrthoFinder analysis of 44 metazoan proteomes (Supplemental Fig. S16; Supplemental Table S5; Supplemental Data S11; Emms and Kelly 2015, 2019). We then characterized the relative enrichment of genes of a given age across different cell types in our scRNA-seq atlas, revealing clear cell type–specific enrichment patterns (Supplemental Fig. S17A). We also calculated a holistic score, the transcriptome age index (TAI) (Domazet-Lošo and Tautz 2010), for each cell cluster (Supplemental Fig. S17B,C). Consistent with previous reports (Hemmrich et al. 2012), we found that ancient gene families predating Metazoa were most strongly associated with interstitial cells that have a high degree of potency, namely, interstitial stem cells, early neuron and nematocyte progenitors, and germ cells—with interstitial stem cells having the least derived transcriptomic profile overall (Supplemental Fig. S17).
Among differentiated interstitial cell types, both gland cells and neurons were enriched for genes that originated at the base of Metazoa, likely reflecting the ancient origins of their respective transcriptional programs (Supplemental Fig. S17; Smith and Mayorova 2019; Musser et al. 2021). However, neurons also showed enrichment for younger genes, suggesting the existence of cnidarian-specific modifications to neuronal transcription. In contrast, nematocyte transcriptional profiles were generally younger, with nematoblasts (i.e., developing nematocytes) showing stark enrichment for gene families that originated either at the base of Cnidaria or Medusozoa (Supplemental Fig. S17), consistent with the more recent evolutionary origin of nematocytes (Jung et al. 2007; David et al. 2008). The two epithelial lineages were both associated with genes predating Cnidaria, although endodermal cell transcriptomes appeared somewhat older than those in ectodermal cells (Supplemental Fig. S17). Like neurons, both epithelial lineages were also enriched for younger hydrozoan-specific gene families. Overall, our analysis suggests that the transcriptional programs used by interstitial stem cells, germ cells, nematoblasts, and gland cells show relatively little genetic innovation since their initial emergence, whereas epithelial and neuronal transcriptional programs have been more dynamic over the course of cnidarian evolution.
Prediction of Hydra cell fate regulators
We next sought to leverage both the scRNA-seq atlas and the AEP assembly CRE annotations to identify TFs involved in coordinating Hydra cell type–specific transcriptional programs. We had previously explored this question as part of the initial publication of the Hydra atlas using an analysis that combined ATAC-seq from strain 105 polyps with the strain AEP scRNA-seq data (Siebert et al. 2019). Broadly, our approach was first to identify TF binding motifs that were enriched in promoter-proximal CREs associated with a set of coexpressed genes, collectively referred to as a metagene. Then, we predicted candidate regulators by identifying TFs that both had similar expression to the metagene of interest and could plausibly bind one of the enriched motifs. We were motivated to revisit this analysis for two reasons: First, we could use our improved AEP-mapped atlas, and second, our phylogenetic footprinting data would improve our enrichment analysis by eliminating potential TF binding sites that were likely not functionally relevant.
Our motif enrichment analysis identified 336 motifs that were enriched in at least one metagene in the AEP-mapped Hydra single-cell atlas (Supplemental Figs. S15, S18; Supplemental Data S12), and our subsequent coexpression analysis identified 115 TFs as candidate regulators (Fig. 4J–N; Supplemental Fig. S17; Supplemental Data S13). These candidates spanned diverse cell states and included multiple regulators whose function had been previously validated in Hydra, such as TCF/Wnt signaling as a regulator of oral tissue (Fig. 4I; Hobmayer et al. 2000; Broun et al. 2005; Lengfeld et al. 2009; Gee et al. 2010), gata1-3 as a regulator of aboral tissue (Fig. 4J; Ferenc et al. 2021), and zic4 as a regulator of epithelial tentacle tissue (Supplemental Fig. S19J; Vogg et al. 2022). These results validate our analysis as a method for detecting functionally meaningful regulatory relationships underlying cell fate decisions in Hydra. In addition, our analysis identified novel candidate regulators. These included pou4 as a regulator of late stage nematogenesis and neurogenesis (Fig. 4K), ebf as a regulator of oogenesis (Fig. 4L), and nr2f-like as a regulator of early nematogenesis (Fig. 4M).
Systematic comparison of cell type–specific transcription in H. vulgaris and C. hemisphaerica
We next extended our analysis of cell type–specific transcription to another hydrozoan, the jellyfish C. hemisphaerica. Hydra and Clytia, although both hydrozoans, nonetheless show extensive differences at both the genomic and phenotypic level. The most recent common ancestor of Hydra and Clytia lived more than 400 million years ago (Schwentner and Bosch 2015; Dohrmann and Wörheide 2017), and the protein sequence divergence between the two species is roughly equivalent to that of humans and lampreys (Supplemental Fig. S16). Hydra and Clytia also have markedly different life cycles: Hydra have a derived and simplified life cycle that consists only of a polyp stage, whereas Clytia have planula, colonial polyp, and medusa stages, each with distinct morphologies. Because of the extensive divergence between these lineages, identifying molecular commonalities between these two systems provides strong evidence of conservation and, by extension, functional significance.
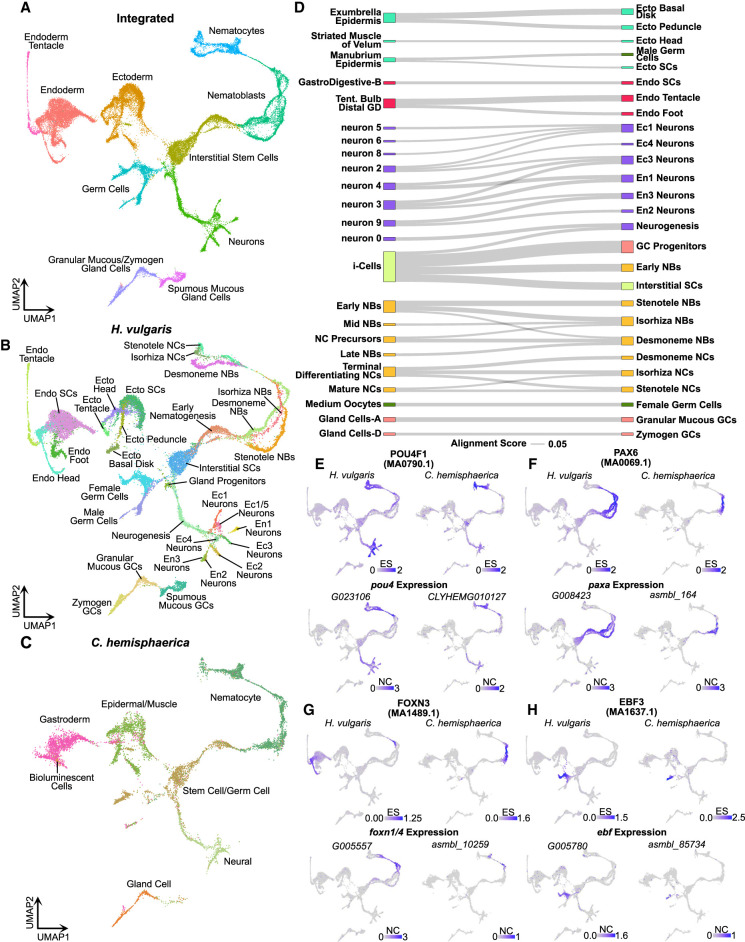
To identify conserved cell type–specific transcriptional patterns in Hydra and Clytia, we used reciprocal principal component analysis to align our Hydra scRNA-seq atlas to a recently published scRNA-seq atlas of the Clytia medusa (Chari et al. 2021). The resulting UMAP representation accurately grouped homologous cell types from the two species (Fig. 5A–C). To assess transcriptional similarities between cell types more quantitatively, we calculated an alignment score (Tarashansky et al. 2021) for all pairwise cross-species cell type comparisons. This revealed extensive similarities between the two species, providing strong evidence of transcriptional conservation across homologous cell types (Fig. 5D). We also calculated a distance metric that quantified the overall degree of transcriptional equivalence between a given cell and similar cells in the other species (Supplemental Fig. S20).
Figure 5.
Aligned Hydra and Clytia single-cell atlases reveal conserved cell type–specific transcriptional regulation. (A–C) UMAP dimensional reduction of aligned Hydra and Clytia medusa single-cell atlases clusters together equivalent cell types from the two species. (D) Sankey plot showing transcriptional similarities between Hydra (right column) and Clytia (left column) cell types highlights extensive similarities among interstitial cell types. The alignment score quantifies the proportion of mutual nearest neighbors for one cell type that are made up of members of another cell type. An alignment score threshold of 0.05 was used to exclude poorly aligned cell types. (NCs) Nematocytes; (NBs) nematoblasts; (SCs) stem cells; (Ecto) ectodermal epithelial cells; (Endo) endodermal epithelial cells; (GCs) gland cells; (Ec) neuron subtypes found in the ectoderm; (En) neuron subtypes found in the endoderm; (Tent.) tentacles; (GD) gastroderm. (E–H) Conserved motif enrichment and gene expression patterns reflect gene regulatory network conservation in hydrozoans. (E) pou4 is a conserved regulator of late stage and mature neurons and nematocytes. (F) paxA is a conserved regulator of nematoblasts. (G) foxn1/4 is a conserved regulator of nematocyte maturation. (H) ebf is a conserved regulator of oogenesis. (ES) Enrichment score; (NC) normalized counts.
Among the three lineages, epithelial cells showed fewer cell type similarities than did interstitial cells (Fig. 5D), consistent with the marked differences in epithelial morphology between polyp and medusa body plans. Nonetheless, we did identify some transcriptional similarities among epithelial cells, suggesting that hydrozoan medusa and polyp body plans are created at least in part through the redeployment of shared transcriptional programs. In addition, Hydra epithelial stem cells had low transcriptional distance scores (Supplemental Fig. S20), potentially indicating the conservation of general epithelial transcriptional signatures despite the lack of direct homologies with individual Clytia epithelial cell types. Interstitial cell types showed more robust conservation, with nearly all Hydra interstitial cell populations showing similarity to at least one Clytia cell type (Fig. 5D). In some cases, there was clear one-to-one homology, such as female germline cells and some gland cell subtypes. In contrast, neuron and nematocyte cell types had either one-to-many or many-to-many patterns of homology.
The Hydra genus has undergone extensive gene loss, likely as a consequence of its simplified life cycle (Chapman et al. 2010; Leclère et al. 2019; Hamada et al. 2020), but the ancestral function of these lost genes has gone largely unexplored. We sought to leverage the Clytia cell atlas to systematically characterize the potential function of genes lost in Hydra. To do this, we calculated a holistic score for each Clytia cell cluster that represented the degree to which that cell type expressed these lost genes (Supplemental Fig. S21). The Clytia cell type with the highest score was the tentacle GFP cell, a bioluminescent cell type located in the medusa tentacle bulb (Fourrage et al. 2014). Among other cell types, gland cell scores were clear outliers, with exceptionally high values across all subtypes. Notably, our cross-species cell type comparison found that the tentacle GFP cell, along with three of the five Clytia gland cell subtypes, did not show strong homology with any Hydra cell types (Fig. 5D). These observations suggest that gene loss in the Hydra genus has been driven, at least in part, by the loss or simplification of cell type–specific transcriptional programs.
Interstitial cell–specific gene regulatory modules are conserved between Hydra and Clytia
Transcriptional similarities between Hydra and Clytia cell types imply the existence of conserved gene regulatory networks. Therefore, we sought to identify the regulators underlying conserved cell type–specific transcription in these two species. To that end, we reapplied the approach we used to identify candidate gene module regulators in Hydra to the Clytia single-cell atlas, albeit with some modifications because of the lack of epigenetic data in Clytia (Supplemental Fig. S22; Supplemental Data S14, S15). We then compared the results from each species to identify commonalities. We found 13 motifs that had similar enrichment patterns in the two species (enrichment correlation score > 0.5) (Supplemental Data S16). Thus, despite the high level of divergence in noncoding sequence between the Clytia and Hydra genomes, we see significant overlap in the motifs associated with conserved gene coexpression modules.
To find candidate regulators of conserved gene coexpression modules, we first sought to identify TFs with similar cell type specificity in Hydra and Clytia. To do this, we identified one-to-one ortholog pairs with correlated expression in the aligned cross-species principal component space (for details, see Supplemental Material). This approach recovered 409 orthologs with highly conserved expression patterns (correlation score > 0.65), including markers for most cell types in the cross-species atlas (Supplemental Figs. S23, S24; Supplemental Data S17). From these 409 orthologs, we identified 30 predicted TFs with conserved cell type–specific expression (Supplemental Fig. S25). Although our analysis did not recover any conserved TFs in epithelial cells (likely because of the relatively poor alignability of the epithelial cell clusters), we did find putative conserved regulators for all interstitial cell types.
To further test if the function of these 30 TFs was conserved from Clytia to Hydra, we manually cross-referenced their expression patterns with our cross-species motif enrichment analysis to identify cases in which both the TF expression pattern and its binding motif enrichment profile were conserved. We identified five TFs that met these stringent conservation criteria (Fig. 5E–H; Supplemental Fig. S26), including regulators of neurogenesis (pou4 and atoh8) (Fig. 5E; Supplemental Fig. S26), nematogenesis (pou4, paxA, and foxn1/4) (Fig. 5E–G), and oogenesis (ebf) (Fig. 5H). Thus, by systematically comparing genomic and transcriptomic data from distantly related hydrozoan species, we were able to identify transcriptional regulators of multiple interstitial cell types that have likely retained their function over at least 400 million years of evolution.
Discussion
Characterizing transcriptional regulation in nonbilaterian metazoans presents significant challenges. In this study, we generated new genomes for H. oligactis and strain AEP H. vulgaris—with the latter being among the most contiguous and best-annotated cnidarian genomes currently available—to facilitate the investigation of hydrozoan transcriptional regulation. By combining our AEP strain assembly with data covering single-cell expression, chromatin accessibility, histone modifications, sequence conservation, and chromatin contact frequency, we were able to perform the most in-depth characterization of transcriptional regulation in a cnidarian to date. These new resources, available at https://research.nhgri.nih.gov/HydraAEP/, provide powerful new tools for future research aimed at unraveling hydrozoan transcriptional regulation.
Evidence of long-range chromatin interactions in Hydra
Consistent with previous characterizations of CREs in cnidarians (Schwaiger et al. 2014; Reddy et al. 2020; Murad et al. 2021), our global maps of histone modifications and chromatin accessibility clearly support the existence of distal enhancer-like regulatory elements in Hydra. This is further supported by our phylogenetic footprinting analysis, which found that many of these putative distal elements were conserved across multiple Hydra species. Nonetheless, we found that Hydra CREs show a strong promoter-proximal bias, with most conserved upstream elements falling within 2 kb of the TSS. This likely explains the relatively high success rate of transgenic Hydra reporter lines generated using just 1–2 kb of upstream sequence (Klimovich et al. 2019). However, there have been some instances, such as with hym-176e and β-catenin, in which short stretches of upstream promoter proximal sequence were not sufficient to fully recapitulate known expression patterns (Hobmayer et al. 2000; Iachetta et al. 2018; Noro et al. 2019). Therefore, the genomic resources generated by this study should facilitate the generation of transgenic reporter lines in the future by allowing researchers to identify likely promoter regions using data collected from the same strain used for transgenesis.
Our characterization of the 3D chromatin architecture of the strain AEP genome provided further evidence that distal chromatin interactions are likely prevalent in Hydra, as we identified thousands of localized chromatin interaction domains that spanned dozens to hundreds of kilobases. The borders of these domains were marked by changes in histone modifications and gene expression patterns, indicating that they were likely related to transcriptional regulation. Thus, Hydra chromatin domains resemble those found in other organisms that lack CTCF-mediated chromatin loops, such as Drosophila and Arabidopsis, where TADs arise passively via the partitioning of heterochromatin and euchromatin into distinct interaction compartments (Rowley et al. 2017; Szabo et al. 2019). However, many of the proteins that localize to TAD boundaries in Drosophila, the system in which non-CTCF-mediated chromatin organization has been best characterized (e.g., BEAF-32, CP190, Chromator, GAF, and M1BP) (Szabo et al. 2019), appear to be absent from the Hydra genome. It therefore remains unclear how domain boundaries are regulated in cnidarians.
A highly conserved feature of chromatin domains in many organisms is that their boundaries often overlap with regions of active chromatin (Szabo et al. 2019). In stark contrast, we found that Hydra domain boundaries generally fell within stretches of heterochromatin. In Drosophila, it was proposed that active regions found at putative domain boundaries are not boundaries at all but rather are small active domains interspersed between larger repressed domains (Rowley et al. 2017). Thus, it may be the case that the heterochromatic signature found at Hydra domain boundaries corresponds to small, repressed regions that we are unable to resolve with our current whole-animal Hi-C data. In the future, the generation of higher resolution Hi-C data from a more homogenous cell population would help clarify the nature and regulation of Hydra domain boundaries.
We also used our Hi-C data to characterize the 3D organization of the Hydra genome at the chromosomal level, which revealed high levels of inter-centromeric interactions. Indeed, we performed a systematic cross-species analysis of available cnidarian Hi-C data sets and found that Hydra had significantly elevated levels of inter-centromeric chromatin interactions relative to other cnidarians, which may have resulted from the loss of a subset of condensin II subunits in the hydrozoan lineage. Alternatively, the increased inter-chromosomal interactions may simply be a byproduct of the increased size of brown Hydra genomes (Wong et al. 2019). Characterizing inter-chromosomal contacts in other hydrozoans, particularly in the green H. viridissima, which has a much smaller genome than H. vulgaris, as well as H. oligactis, which has a larger genome, would help address this question. In addition, the Hi-C data generated from these experiments could be used to generate chromosome-level scaffolds from the available draft genomes for these two species (Hamada et al. 2020), which would greatly facilitate future comparative genomics research within the Hydra lineage.
Deep conservation of hydrozoan cell type–specific transcriptional programs
The stem cell differentiation trajectories in Hydra are the best characterized of any cnidarian, making it well suited for exploring the gene regulatory networks underlying cell fate specification. To that end, we combined the CRE annotations we generated for the AEP assembly with an updated version of the Hydra scRNA-seq atlas to better understand the transcriptional programs directing cell type–specific transcription. In the process of updating the atlas, we recovered ∼17% more single-cell transcriptomes and two additional cell types compared with previous atlas iterations. With the addition of these two previously absent cell types, isorhiza nematocytes and somatic gonad ectoderm, the Hydra single-cell atlas now contains virtually all known cell types in the adult polyp. However, there is likely additional complexity within these two additional cell populations that we are currently unable to resolve. Specifically, we currently cannot differentiate between the two types of mature isorhiza nematocytes, holotrichous and atrichous, nor can we distinguish between male and female somatic gonad. The inability to resolve these subtypes likely results from their relatively low abundance in our data set. The generation of transgenic reporter lines using the markers we provide in this study would greatly facilitate efforts to selectively isolate and transcriptionally profile these cell subtypes.
Our subsequent analysis of the updated atlas provided several insights into the evolution of cell type–specific transcriptional programs in hydrozoans. Consistent with previously published findings (Hemmrich et al. 2012), our phylostratigraphic analysis of the three adult stem cell populations found that the genes transcribed in the epithelial stem cells are substantially younger than those transcribed in interstitial stem cells. Indeed, the transcriptional profiles of the two epithelial stem cell populations and their differentiated progeny were enriched in genes originating at the base of hydrozoa or later. Little is known about the evolution of epithelial cells within hydrozoans, but the topic may merit further study as our analysis suggests these cell types may be a major driver of recent genetic novelty. In contrast, interstitial stem cells had the oldest transcriptional profile of any cell in the Hydra atlas. This finding is consistent with the proposed existence of a deeply conserved genetic program underlying pluripotency in metazoans (Juliano et al. 2010; Sogabe et al. 2019). Thus, although interstitial stem cells are thought to be a derived cell type (Gold and Jacobs 2013), they make use of an evolutionarily ancient transcriptional program.
To better understand the regulation of these cell type–specific transcriptional programs, we integrated our CRE annotations and scRNA-seq data to identify candidate TFs involved in cell fate specification. This analysis recovered previously characterized as well as novel candidate regulators, thus providing an extensive list of candidates for future functional studies across a diverse array of cell types. Notably, in addition to capturing the known functions of previously characterized TFs, our analysis also predicted additional functions that may have been missed by previous studies. Specifically, our work suggests that gata1-3 and zic4 regulate transcription in neurons (Fig. 4J; Supplemental Fig. S19J) in addition to their previously documented roles in epithelial cells (Ferenc et al. 2021; Vogg et al. 2022).
To determine if the composition and regulation of cell type–specific transcription in Hydra are conserved in other hydrozoans, we performed a systematic comparative analysis of the Hydra and Clytia single-cell atlases. This analysis revealed extensive conservation in cell type–specific transcriptional signatures despite the extensive divergence between these two species, which allowed us to identify hundreds of conserved marker genes across all major cell types. However, apart from germ cells and a subset of gland cells, we did not observe clear one-to-one homology among differentiated cell subtypes. This may indicate that although broad cell types (e.g., neuron, nematocyte, gland cell) are well conserved at the transcriptional level, the identities of specific subtypes are not. Indeed, some Clytia cell types, including the tentacle GFP cell and several gland cell subtypes, appear to have been lost in Hydra. One possible hypothesis is that this loss was driven by the simplification of the Hydra life cycle. However, it is currently unclear if such a hypothesis is plausible, as it is not known if the cell types in question are medusa-specific in Clytia. The generation of single-cell atlases for the Clytia planula and polyp stages would help address this question.
Among the cell types that were clearly conserved between Hydra and Clytia, our analysis uncovered robust overlap not only in gene expression but also in predicted transcriptional regulators. Specifically, we identified putative regulators of nematogenesis, neurogenesis, and oogenesis whose gene expression patterns and motif enrichment profiles were conserved from Clytia to Hydra. Given the extensive transcriptional similarities we observed in our aligned cross-species atlas, it is very likely that the relatively small list of conserved regulators we identified in this study is incomplete. The generation of CRE annotations for the Clytia genome would likely increase the sensitivity of this analysis and help reveal additional regulatory conservation.
Among the TFs we identified as having a conserved function in hydrozoans, three of these regulators—namely, pou4, atoh8, and paxA—have been functionally characterized in Nematostella, a cnidarian that diverged from hydrozoans more than 600 million years ago (Schwentner and Bosch 2015; Dohrmann and Wörheide 2017). In all three cases, the reported roles of these TFs in Nematostella are consistent with their predicted functions in hydrozoa based on our analysis (Richards and Rentzsch 2015; Babonis and Martindale 2017a; Tournière et al. 2020). In addition, our predictions regarding pou4 and atoh8 function in hydrozoan neurons are consistent with the well-established roles for these genes in bilaterian nervous systems (Gan et al. 1996; Inoue et al. 2001). Collectively, these findings support the accuracy of our analytical approach and provide insight into the likely ancestral function of these TFs in the last common cnidarian ancestor.
Our analysis also identified novel regulators, including foxn1/4 as a regulator of nematocyte maturation and ebf as a regulator of oogenesis. Although putative functions for these TFs have not, to our knowledge, been previously described, we did find publicly available expression data sets that were consistent with ebf having a conserved role in oogenesis. Specifically, recently published bulk RNA-seq data from the hydrozoan Hydractinia symbiolongicarpus showed that ebf was specifically expressed in polyps undergoing oogenesis (DuBuc et al. 2020). In addition, an scRNA-seq atlas of the zebrafish ovary shows an ebf ortholog, ebf3b, as a marker of female germline stem cells (Liu et al. 2022). Therefore, ebf regulation of oogenesis may predate the split of Bilateria and Cnidaria.
In summary, by taking a comparative approach and leveraging the genomic and transcriptomic data available in Clytia and Hydra, we identified both conserved gene coexpression modules and the TFs that likely regulate them, providing new insight into the transcriptional programs underlying cell identity in hydrozoans.
Methods
Hydra strains and culturing conditions
The following H. vulgaris strains were used in this study: AEP (Martin et al. 1997), 105 (Chapman et al. 2010), inverse watermelon (Glauber et al. 2015), watermelon (Glauber et al. 2015), enGreen1, and operon (Dana et al. 2012). The Innsbruck female12 strain was used for generating the H. oligactis genome assembly. All animals were maintained using standard procedures (Lenhoff and Brown 1970). For details, see Supplemental Material.
H. vulgaris strain AEP genome sequencing, assembly, and annotation
High-molecular-weight (HMW) genomic DNA (gDNA) was extracted from strain AEP H. vulgaris using a Qiagen Gentra Puregene kit. The DNA was then used for the generation of 10x Chromium library using a chromium genome library & gel bead kit v.2, an Oxford Nanopore library using an Oxford Nanopore ligation sequencing kit, and a Pacific Biosciences (PacBio) library using a SMRTbell express template prep kit 2.0. The 10x libraries were sequenced on an Illumina HiSeq X10. The Nanopore libraries were sequenced using an Oxford Nanopore PromethION. The PacBio libraries were sequenced using a PacBio sequel II. Hi-C libraries were prepared using an Arima Hi-C kit and were sequenced on an Illumina NovaSeq 6000. The initial draft assembly was generated with the Nanopore data using Canu (Koren et al. 2017); scaffolding was performed using the 10x and Hi-C data; and error correction and gap-filling was performed using the 10x, PacBio, and Nanopore data. Gene models were generated using BRAKER2 (Brůna et al. 2021) and exonerate (Slater and Birney 2005). For details, see Supplemental Material.
H. oligactis genome sequencing, assembly, and annotation
HMW gDNA was extracted from Innsbruck female12 strain H. oligactis polyps using a Circulomics Nanobind tissue big kit. Sequencing libraries were prepared using an Oxford Nanopore ligation sequencing kit and sequenced using an Oxford Nanopore MinION. The assembly was generated using Flye (Kolmogorov et al. 2019). Gene models were generated using BRAKER2 (Brůna et al. 2021) For details, see Supplemental Material.
ATAC-seq
Whole-animal ATAC-seq was performed on strain AEP H. vulgaris polyps using a previously described protocol (Siebert et al. 2019). The data were analyzed using a previously described pipeline (Siebert et al. 2019). For details, see Supplemental Material.
CUT&Tag
Whole-animal CUT&Tag was performed on strain AEP H. vulgaris polyps using a modified version of the originally published protocol (Kaya-Okur et al. 2019). The data were mapped to the AEP genome assembly using Bowtie 2 (Langmead and Salzberg 2012), and peaks were called using SEACR (Meers et al. 2019). For details, see Supplemental Material.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed on strain AEP H. vulgaris polyps using a previously described protocol (Bode et al. 2009). For details, see Supplemental Material.
Sequence conservation analysis
The whole-genome cross-species alignment was generated using Progressive Cactus (Armstrong et al. 2020). TF binding sites were predicted using FIMO (Grant et al. 2011). For details, see Supplemental Material.
Hydra and Clytia single-cell atlas analysis
Previously published Hydra scRNA-seq data were aligned to the AEP genome assembly using a previously described pipeline (Siebert et al. 2019). The Clytia scRNA-seq data were mapped to gene models for the updated version of the Clytia genome assembly (accessed from the NCBI GenBank database [https://www.ncbi.nlm.nih.gov/genbank/] under accession number CACVBU010000000) using the Cell Ranger pipeline. Normalization, clustering, cross-species alignment, and visualization were performed using Seurat (Hao et al. 2021). Gene coexpression analyses were performed using cNMF (Kotliar et al. 2019). For details, see Supplemental Material.
Data access
We have generated a new Hydra AEP Genome Project Portal (https://research.nhgri.nih.gov/HydraAEP/) that allows users to interact with and download the data generated by this study. The raw sequencing data and assembled genomic sequences data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA816482. Note that the chromosome numbering for the version of the strain AEP H. vulgaris assembly available via GenBank (accession JALDPZ000000000) was changed to be consistent with the numbering used for the strain 105 H. vulgaris assembly (accession JAGKSS000000000) (Simakov et al. 2022). Step-by-step descriptions of all computational analyses conducted as part of this study, including all relevant code, formatted both as markdown and HTML documents, are available at GitHub (https://github.com/cejuliano/brown_hydra_genomes). An archived copy of this repository is also provided as Supplemental Code S1.
Supplementary Material
Acknowledgments
We thank members of the C.E.J., B.H., O.S., and A.D.B. laboratories for valuable research discussions; Ruta Sahasrabudhe, Oanh Nguyen, Diana Burkart-Waco, and Lutz Froenicke from the DNA Technologies and Expression Analysis Core at the UC Davis Genome Center (supported by NIH Shared Instrumentation Grant 1S10OD010786-01) for technical advice and assistance with library preparation and sequencing; Bruce Draper for assistance with imaging; Anh-Dao Nguyen for assistance with generating the genome portal; Vijay Ramani for insight into metazoan chromatin organization; and Birte Mertens for assistance in establishing long-read sequencing in H. oligactis. This work was supported by a National Institutes of Health (NIH) grant R35 GM133689 (to C.E.J.), the National Science Foundation: NSF EDGE 1829158 (to C.E.J.), an Austrian Science Fund (FWF) and Deutsche Forschungsgemeinschaft (DFG) DACH grant I4353 (to O.S.), an Austrian Science Fund (FWF) grant P30347 (to P.L.), and a Fonds National de la Recherche Luxembourg grant 13569708 (to P.B.). This work was also supported in part by the Intramural Research Program of the National Human Genome Research Institute, NIH (ZIA HG000140 to A.D.B.).
Author contributions: J.F.C., S. Siebert, B.H., and C.E.J. designed research; J.F.C., S. Siebert, H.M.L., P.B., A.S.P., M.A., P.L., and O.S. performed research; J.F.C., H.M.L., M.T.F., R.T.M., S. Singh, S.Z., T.G.W., C.E.S., and A.D.B. contributed new analytic tools/resources; J.F.C., S. Siebert, P.B., A.S.P., and O.S. analyzed data; and J.F.C. and C.E.J. wrote the paper with input from O.S., B.H., S. Siebert, and A.D.B.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.277040.122.
Competing interest statement
The authors declare no competing interests.
References
- Arendt D. 2008. The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet 9: 868–882. 10.1038/nrg2416 [DOI] [PubMed] [Google Scholar]
- Armstrong J, Hickey G, Diekhans M, Fiddes IT, Novak AM, Deran A, Fang Q, Xie D, Feng S, Stiller J, et al. 2020. Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature 587: 246–251. 10.1038/s41586-020-2871-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babonis LS, Martindale MQ. 2017a. PaxA, but not PaxC, is required for cnidocyte development in the sea anemone Nematostella vectensis. Evodevo 8: 14. 10.1186/s13227-017-0077-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babonis LS, Martindale MQ. 2017b. Phylogenetic evidence for the modular evolution of metazoan signalling pathways. Philos Trans R Soc Lond B Biol Sci 372: 20150477. 10.1098/rstb.2015.0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode HR, Heimfeld S, Chow MA, Huang LW. 1987. Gland cells arise by differentiation from interstitial cells in Hydra attenuata. Dev Biol 122: 577–585. 10.1016/0012-1606(87)90321-6 [DOI] [PubMed] [Google Scholar]
- Bode H, Lengfeld T, Hobmayer B, Holstein TW. 2009. Detection of expression patterns in hydra pattern formation. In Wnt signaling (ed. Vincan E), pp. 69–84. Humana Press, Totowa, NJ. 10.1007/978-1-60327-469-2_7 [DOI] [PubMed] [Google Scholar]
- Boehm AM, Khalturin K, Anton-Erxleben F, Hemmrich G, Klostermeier UC, Lopez-Quintero JA, Oberg HH, Puchert M, Rosenstiel P, Wittlieb J, et al. 2012. FoxO is a critical regulator of stem cell maintenance in immortal Hydra. Proc Natl Acad Sci 109: 19697–19702. 10.1073/pnas.1209714109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TCG, David CN. 1987. Stem cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. Dev Biol 121: 182–191. 10.1016/0012-1606(87)90151-5 [DOI] [Google Scholar]
- Bosch TC, Krylow SM, Bode HR, Steele RE. 1988. Thermotolerance and synthesis of heat shock proteins: these responses are present in Hydra attenuata but absent in Hydra oligactis. Proc Natl Acad Sci 85: 7927–7931. 10.1073/pnas.85.21.7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw B, Thompson K, Frank U. 2015. Distinct mechanisms underlie oral vs aboral regeneration in the cnidarian Hydractinia echinata. eLife 4: e05506. 10.7554/eLife.05506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun M, Gee L, Reinhardt B, Bode HR. 2005. Formation of the head organizer in hydra involves the canonical Wnt pathway. Development 132: 2907–2916. 10.1242/dev.01848 [DOI] [PubMed] [Google Scholar]
- Browne EN. 1909. The production of new hydranths in Hydra by the insertion of small grafts. J Exp Zool 7: 1–23. 10.1002/jez.1400070102 [DOI] [Google Scholar]
- Brůna T, Hoff KJ, Lomsadze A, Stanke M, Borodovsky M. 2021. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom Bioinform 3: lqaa108. 10.1093/nargab/lqaa108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. 2010. The dynamic genome of Hydra. Nature 464: 592–596. 10.1038/nature08830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari T, Weissbourd B, Gehring J, Ferraioli A, Leclère L, Herl M, Gao F, Chevalier S, Copley RR, Houliston E, et al. 2021. Whole-animal multiplexed single-cell RNA-seq reveals transcriptional shifts across Clytia medusa cell types. Sci Adv 7: eabh1683. 10.1126/sciadv.abh1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14: 959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana CE, Glauber KM, Chan TA, Bridge DM, Steele RE. 2012. Incorporation of a horizontally transferred gene into an operon during cnidarian evolution. PLoS One 7: e31643. 10.1371/journal.pone.0031643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling JA, Reitzel AR, Burton PM, Mazza ME, Ryan JF, Sullivan JC, Finnerty JR. 2005. Rising starlet: the starlet sea anemone, Nematostella vectensis. Bioessays 27: 211–221. 10.1002/bies.20181 [DOI] [PubMed] [Google Scholar]
- David CN. 2012. Interstitial stem cells in Hydra: multipotency and decision-making. Int J Dev Biol 56: 489–497. 10.1387/ijdb.113476cd [DOI] [PubMed] [Google Scholar]
- David CN, Murphy S. 1977. Characterization of interstitial stem cells in hydra by cloning. Dev Biol 58: 372–383. 10.1016/0012-1606(77)90098-7 [DOI] [PubMed] [Google Scholar]
- David CN, Özbek S, Adamczyk P, Meier S, Pauly B, Chapman J, Hwang JS, Gojobori T, Holstein TW. 2008. Evolution of complex structures: minicollagens shape the cnidarian nematocyst. Trends Genet 24: 431–438. 10.1016/j.tig.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Della Gaspera B, Weill L, Chanoine C. 2022. Evolution of somite compartmentalization: a view from Xenopus. Front Cell Dev Biol 9: 790847. 10.3389/fcell.2021.790847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann M, Wörheide G. 2017. Dating early animal evolution using phylogenomic data. Sci Rep 7: 3599. 10.1038/s41598-017-03791-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T, Tautz D. 2010. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468: 815–818. 10.1038/nature09632 [DOI] [PubMed] [Google Scholar]
- Domazet-Lošo T, Brajković J, Tautz D. 2007. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet 23: 533–539. 10.1016/j.tig.2007.08.014 [DOI] [PubMed] [Google Scholar]
- DuBuc TQ, Schnitzler CE, Chrysostomou E, McMahon ET, Febrimarsa, Gahan JM, Buggie T, Gornik SG, Hanley S, Barreira SN, et al. 2020. Transcription factor AP2 controls cnidarian germ cell induction. Science 367: 757–762. 10.1126/science.aay6782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Giribet G, Edgecombe GD, Hejnol A. 2014. Animal phylogeny and its evolutionary implications. Annu Rev Ecol Evol Syst 45: 371–395. 10.1146/annurev-ecolsys-120213-091627 [DOI] [Google Scholar]
- Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16: 157. 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20: 238. 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenc J, Papasaikas P, Ferralli J, Nakamura Y, Smallwood S, Tsiairis CD. 2021. Mechanical oscillations orchestrate axial patterning through Wnt activation in Hydra. Sci Adv 7: eabj6897. 10.1126/sciadv.abj6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, et al. 2020. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 48: D87–D92. 10.1093/nar/gkz1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourrage C, Swann K, Garcia JRG, Campbell AK, Houliston E. 2014. An endogenous green fluorescent protein–photoprotein pair in Clytia hemisphaerica eggs shows co-targeting to mitochondria and efficient bioluminescence energy transfer. Open Biol 4: 130206. 10.1098/rsob.130206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S, Bosch TCG. 2007. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci 104: 13146–13151. 10.1073/pnas.0703375104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas R, Zhang GJ, Cohn MJ. 2006. Evidence that mechanisms of fin development evolved in the midline of early vertebrates. Nature 442: 1033–1037. 10.1038/nature04984 [DOI] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. 1996. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci 93: 3920–3925. 10.1073/pnas.93.9.3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee L, Hartig J, Law L, Wittlieb J, Khalturin K, Bosch TC, Bode HR. 2010. β-Catenin plays a central role in setting up the head organizer in hydra. Dev Biol 340: 116–124. 10.1016/j.ydbio.2009.12.036 [DOI] [PubMed] [Google Scholar]
- Gierer A, Meinhardt H. 1972. A theory of biological pattern formation. Kybernetik 12: 30–39. 10.1007/BF00289234 [DOI] [PubMed] [Google Scholar]
- Glauber KM, Dana CE, Park SS, Colby DA, Noro Y, Fujisawa T, Chamberlin AR, Steele RE, Glauber KM, Dana CE, et al. 2015. A small molecule screen identifies a novel compound that induces a homeotic transformation in Hydra. Development 142: 2081. 10.1242/dev.126235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold DA, Jacobs DK. 2013. Stem cell dynamics in Cnidaria: Are there unifying principles? Dev Genes Evol 223: 53–66. 10.1007/s00427-012-0429-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grens A, Gee L, Fisher DA, Bode HR. 1996. CnNK-2, an NK-2 homeobox gene, has a role in patterning the basal end of the axis in hydra. Dev Biol 180: 473–488. 10.1006/dbio.1996.0321 [DOI] [PubMed] [Google Scholar]
- Gufler S, Artes B, Bielen H, Krainer I, Eder M-K, Falschlunger J, Bollmann A, Ostermann T, Valovka T, Hartl M, et al. 2018. β-Catenin acts in a position-independent regeneration response in the simple eumetazoan Hydra. Dev Biol 433: 310–323. 10.1016/j.ydbio.2017.09.005 [DOI] [PubMed] [Google Scholar]
- Gumucio DL, Heilstedt-Williamson H, Gray TA, Tarlé SA, Shelton DA, Tagle DA, Slightom JL, Goodman M, Collins FS. 1992. Phylogenetic footprinting reveals a nuclear protein which binds to silencer sequences in the human gamma and epsilon globin genes. Mol Cell Biol 12: 4919–4929. 10.1128/mcb.12.11.4919-4929.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, Satija R. 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20: 296. 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M, Schröder K, Bathia J, Kürn U, Fraune S, Khalturina M, Khalturin K, Shinzato C, Satoh N, Bosch TCG. 2018. Metabolic co-dependence drives the evolutionarily ancient Hydra–Chlorella symbiosis. eLife 7: e35122. 10.7554/eLife.35122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M, Satoh N, Khalturin K. 2020. A reference genome from the symbiotic hydrozoan, Hydra viridissima. G3 (Bethesda) 10: 3883–3895. 10.1534/g3.120.401411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. 2021. Integrated analysis of multimodal single-cell data. Cell 184: 3573–3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M, Glasauer S, Gufler S, Raffeiner A, Puglisi K, Breuker K, Bister K, Hobmayer B. 2019. Differential regulation of myc homologs by Wnt/β-Catenin signaling in the early metazoan Hydra. FEBS J 286: 2295–2310. 10.1111/febs.14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmrich G, Khalturin K, Boehm AM, Puchert M, Anton-Erxleben F, Wittlieb J, Klostermeier UC, Rosenstiel P, Oberg HH, Domazet-Lošo T, et al. 2012. Molecular signatures of the three stem cell lineages in Hydra and the emergence of stem cell function at the base of multicellularity. Mol Biol Evol 29: 3267–3280. 10.1093/molbev/mss134 [DOI] [PubMed] [Google Scholar]
- Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, Rothbächer U, Holstein TW. 2000. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature 407: 186–189. 10.1038/35025063 [DOI] [PubMed] [Google Scholar]
- Hoencamp C, Dudchenko O, Elbatsh AMO, Brahmachari S, Raaijmakers JA, van Schaik T, Cacciatore ÁS, Contessoto VG, van Heesbeen RGHP, van den Broek B, et al. 2021. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science 372: 984–989. 10.1126/science.abe2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister S. 1991. Analysis of a foot regeneration deficient strain of Hydra oligactis. Mech Dev 35: 181–192. 10.1016/0925-4773(91)90017-Z [DOI] [PubMed] [Google Scholar]
- Iachetta R, Ambrosone A, Klimovich A, Wittlieb J, Onorato G, Candeo A, D'Andrea C, Intartaglia D, Scotti N, Blasio M, et al. 2018. Real time dynamics of β-catenin expression during Hydra development, regeneration and Wnt signalling activation. Int J Dev Biol 62: 311–318. 10.1387/ijdb.180092ct [DOI] [PubMed] [Google Scholar]
- Inoue C, Bae SK, Takatsuka K, Inoue T, Bessho Y, Kageyama R. 2001. Math6, a bHLH gene expressed in the developing nervous system, regulates neuronal versus glial differentiation. Genes Cells 6: 977–986. 10.1046/j.1365-2443.2001.00476.x [DOI] [PubMed] [Google Scholar]
- Juliano CE, Swartz SZ, Wessel GM. 2010. A conserved germline multipotency program. Development 137: 4113–4126. 10.1242/dev.047969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SH, Ohyanagi H, Hayakawa S, Osato N, Nishimiya-Fujisawa C, Ikeo K, David CN, Fujisawa T, Gojobori T. 2007. The evolutionary emergence of cell type-specific genes inferred from the gene expression analysis of Hydra. Proc Natl Acad Sci 104: 14735–14740. 10.1073/pnas.0703331104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. 2019. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10: 1930. 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Anton-Erxleben F, Sassmann S, Wittlieb J, Hemmrich G, Bosch TCG. 2008. A novel gene family controls species-specific morphological traits in Hydra. PLoS Biol 6: e278. 10.1371/journal.pbio.0060278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. 2009. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet 25: 404–413. 10.1016/j.tig.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Klimovich A, Wittlieb J, Bosch TCG. 2019. Transgenesis in Hydra to characterize gene function and visualize cell behavior. Nat Protoc 14: 2069–2090. 10.1038/s41596-019-0173-3 [DOI] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37: 540–546. 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotliar D, Veres A, Nagy MA, Tabrizi S, Hodis E, Melton DA, Sabeti PC. 2019. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. eLife 8: e43803. 10.7554/eLife.43803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclère L, Horin C, Chevalier S, Lapébie P, Dru P, Peron S, Jager M, Condamine T, Pottin K, Romano S, et al. 2019. The genome of the jellyfish Clytia hemisphaerica and the evolution of the cnidarian life-cycle. Nat Ecol Evol 3: 801–810. 10.1038/s41559-019-0833-2 [DOI] [PubMed] [Google Scholar]
- Lengfeld T, Watanabe H, Simakov O, Lindgens D, Gee L, Law L, Schmidt HA, Özbek S, Bode H, Holstein TW. 2009. Multiple Wnts are involved in Hydra organizer formation and regeneration. Dev Biol 330: 186–199. 10.1016/j.ydbio.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Lenhoff HM, Brown RD. 1970. Mass culture of hydra: an improved method and its application to other aquatic invertebrates. Lab Anim 4: 139–154. 10.1258/002367770781036463 [DOI] [PubMed] [Google Scholar]
- Li Y, Gao L, Pan Y, Tian M, Li Y, He C, Dong Y, Sun Y, Zhou Z. 2020. Chromosome-level reference genome of the jellyfish Rhopilema esculentum. GigaScience 9: giaa036. 10.1093/gigascience/giaa036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Kossack ME, McFaul ME, Christensen LN, Siebert S, Wyatt SR, Kamei CN, Horst S, Arroyo N, Drummond IA, et al. 2022. Single-cell transcriptome reveals insights into the development and function of the zebrafish ovary. eLife 11: e76014. 10.7554/eLife.76014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin VJ, Littlefield CL, Archer WE, Bode HR. 1997. Embryogenesis in Hydra. Biol Bull 192: 345–363. 10.2307/1542745 [DOI] [PubMed] [Google Scholar]
- Martínez DE. 1998. Mortality patterns suggest lack of senescence in hydra. Exp Gerontol 33: 217–225. 10.1016/S0531-5565(97)00113-7 [DOI] [PubMed] [Google Scholar]
- Martínez DE, Iñiguez AR, Percell KM, Willner JB, Signorovitch J, Campbell RD. 2010. Phylogeny and biogeography of Hydra (Cnidaria: Hydridae) using mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol 57: 403–410. 10.1016/j.ympev.2010.06.016 [DOI] [PubMed] [Google Scholar]
- McInnes L, Healy J, Melville J. 2018. UMAP: Uniform Manifold Approximation and Projection for dimension reduction. arXiv:1802.03426. http://arxiv.org/abs/1802.03426
- Meers MP, Tenenbaum D, Henikoff S. 2019. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 12: 42. 10.1186/s13072-019-0287-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Bradnam KR, Young HA, Telis N, May MR, Ruby JG, Sebra R, Peluso P, Eid J, Rank D, et al. 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol 14: R10. 10.1186/gb-2013-14-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münder S, Käsbauer T, Prexl A, Aufschnaiter R, Zhang X, Towb P, Böttger A. 2010. Notch signalling defines critical boundary during budding in Hydra. Dev Biol 344: 331–345. 10.1016/j.ydbio.2010.05.517 [DOI] [PubMed] [Google Scholar]
- Murad R, Macias-Muñoz A, Wong A, Ma X, Mortazavi A. 2021. Coordinated gene expression and chromatin regulation during Hydra head regeneration. Genome Biol Evol 13: evab221. 10.1093/gbe/evab221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Schippers KJ, Nickel M, Mizzon G, Kohn AB, Pape C, Ronchi P, Papadopoulos N, Tarashansky AJ, Hammel JU, et al. 2021. Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. Science 374: 717–723. 10.1126/science.abj2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nong W, Cao J, Li Y, Qu Z, Sun J, Swale T, Yip HY, Qian PY, Qiu JW, Kwan HS, et al. 2020. Jellyfish genomes reveal distinct homeobox gene clusters and conservation of small RNA processing. Nat Commun 11: 3051. 10.1038/s41467-020-16801-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noro Y, Yum S, Nishimiya-Fujisawa C, Busse C, Shimizu H, Mineta K, Zhang X, Holstein TW, David CN, Gojobori T, et al. 2019. Regionalized nervous system in Hydra and the mechanism of its development. Gene Expr Patterns 31: 42–59. 10.1016/j.gep.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Piraino S, Boero F, Aeschbach B, Schmid V. 1996. Reversing the life cycle: medusae transforming into polyps and cell transdifferentiation in Turritopsis nutricula (Cnidaria, Hydrozoa). Biol Bull 190: 302–312. 10.2307/1543022 [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317: 86–94. 10.1126/science.1139158 [DOI] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 9: 189. 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PC, Gungi A, Ubhe S, Galande S. 2020. Epigenomic landscape of enhancer elements during Hydra head organizer formation. Epigenetics Chromatin 13: 43. 10.1186/s13072-020-00364-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GS, Rentzsch F. 2015. Regulation of Nematostella neural progenitors by SoxB, Notch and bHLH genes. Development 142: 3332–3342. 10.1242/dev.123745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, Corces VG. 2017. Evolutionarily conserved principles predict 3D chromatin organization. Mol Cell 67: 837–852.e7. 10.1016/j.molcel.2017.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible R, Scheuerlein A, Dańko MJ, Gampe J, Martínez DE, Vaupel JW. 2015. Constant mortality and fertility over age in Hydra. Proc Natl Acad Sci 112: 15701–15706. 10.1073/pnas.1521002112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkelaars Q, Perez-Cortes D, Perruchoud C, Galliot B. 2020. The polymorphism of Hydra microsatellite sequences provides strain-specific signatures. PLoS One 15: e0230547. 10.1371/journal.pone.0230547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M, Schönauer A, Rendeiro AF, Pribitzer C, Schauer A, Gilles AF, Schinko JB, Renfer E, Fredman D, Technau U. 2014. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res 24: 639–650. 10.1101/gr.162529.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwentner M, Bosch TCG. 2015. Revisiting the age, evolutionary history and species level diversity of the genus Hydra (Cnidaria: Hydrozoa). Mol Phylogenet Evol 91: 41–55. 10.1016/j.ympev.2015.05.013 [DOI] [PubMed] [Google Scholar]
- Siebert S, Farrell JA, Cazet JF, Abeykoon Y, Primack AS, Schnitzler CE, Juliano CE. 2019. Stem cell differentiation trajectories in Hydra resolved at single-cell resolution. Science 365: eaav9314. 10.1126/science.aav9314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simakov O, Bredeson J, Berkoff K, Marletaz F, Mitros T, Schultz DT, O'Connell BL, Dear P, Martinez DE, Steele RE, et al. 2022. Deeply conserved synteny and the evolution of metazoan chromosomes. Sci Adv 8: eabi5884. 10.1126/sciadv.abi5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GSC, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. 10.1186/1471-2105-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Mayorova TD. 2019. Insights into the evolution of digestive systems from studies of Trichoplax adhaerens. Cell Tissue Res 377: 353–367. 10.1007/s00441-019-03057-z [DOI] [PubMed] [Google Scholar]
- Sogabe S, Hatleberg WL, Kocot KM, Say TE, Stoupin D, Roper KE, Fernandez-Valverde SL, Degnan SM, Degnan BM. 2019. Pluripotency and the origin of animal multicellularity. Nature 570: 519–522. 10.1038/s41586-019-1290-4 [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Bantignies F, Cavalli G. 2019. Principles of genome folding into topologically associating domains. Sci Adv 5: eaaw1668. 10.1126/sciadv.aaw1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagle DA, Koop BF, Goodman M, Slightom JL, Hess DL, Jones RT. 1988. Embryonic ε and γ globin genes of a prosimian primate (Galago crassicaudatus): nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J Mol Biol 203: 439–455. 10.1016/0022-2836(88)90011-3 [DOI] [PubMed] [Google Scholar]
- Tarashansky AJ, Musser JM, Khariton M, Li P, Arendt D, Quake SR, Wang B. 2021. Mapping single-cell atlases throughout metazoa unravels cell type evolution. eLife 10: e66747. 10.7554/eLife.66747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U, Bode HR. 1999. HyBra1, a Brachyury homologue, acts during head formation in Hydra. Development 126: 999–1010. 10.1242/dev.126.5.999 [DOI] [PubMed] [Google Scholar]
- Technau U, Rudd S, Maxwell P, Gordon PMK, Saina M, Grasso LC, Hayward DC, Sensen CW, Saint R, Holstein TW, et al. 2005. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet 21: 633–639. 10.1016/j.tig.2005.09.007 [DOI] [PubMed] [Google Scholar]
- Tournière O, Dolan D, Richards GS, Sunagar K, Columbus-Shenkar YY, Moran Y, Rentzsch F. 2020. NvPOU4/Brain3 functions as a terminal selector gene in the nervous system of the cnidarian Nematostella vectensis. Cell Rep 30: 4473–4489.e5. 10.1016/j.celrep.2020.03.031 [DOI] [PubMed] [Google Scholar]
- Trembley A. 1744. Mémoires, pour servir à l'histoire d'un genre de polypes d'eau douce, à bras en forme de cornes. Chez Jean and Herman Verbeek, Geneva, Switzerland. [Google Scholar]
- Vogg MC, Beccari L, Iglesias Ollé L, Rampon C, Vriz S, Perruchoud C, Wenger Y, Galliot B. 2019. An evolutionarily-conserved Wnt3/β-catenin/Sp5 feedback loop restricts head organizer activity in Hydra. Nat Commun 10: 312. 10.1038/s41467-018-08242-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogg MC, Ferenc J, Buzgariu WC, Perruchoud C, Sanchez PGL, Beccari L, Nuninger C, Le Cras Y, Delucinge-Vivier C, Papasaikas P, et al. 2022. The transcription factor Zic4 promotes tentacle formation and prevents epithelial transdifferentiation in Hydra. Sci Adv 8: eabo0694. 10.1126/sciadv.abo0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltman L, Van Eck NJ. 2013. A smart local moving algorithm for large-scale modularity-based community detection. Eur Phys J B 86: 471. 10.1140/epjb/e2013-40829-0 [DOI] [Google Scholar]
- Weimer BR. 1928. The physiological gradients in Hydra I: reconstitution and budding in relation to length of piece and body level in Pelmatohydra oligactis. Physiol Zool 1: 183–230. 10.1086/physzool.1.2.30151045 [DOI] [Google Scholar]
- Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. 2006. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proc Natl Acad Sci 103: 6208–6211. 10.1073/pnas.0510163103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WY, Simakov O, Bridge DM, Cartwright P, Bellantuono AJ, Kuhn A, Holstein TW, David CN, Steele RE, Martínez DE. 2019. Expansion of a single transposable element family is associated with genome-size increase and radiation in the genus Hydra. Proc Natl Acad Sci 116: 22915–22917. 10.1073/pnas.1910106116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Fujisawa T, Hwang JS, Ikeo K, Gojobori T. 2006. Degeneration after sexual differentiation in hydra and its relevance to the evolution of aging. Gene 385: 64–70. 10.1016/j.gene.2006.06.031 [DOI] [PubMed] [Google Scholar]
- Zheng H, Xie W. 2019. The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol 20: 535–550. 10.1038/s41580-019-0132-4 [DOI] [PubMed] [Google Scholar]
- Zimmermann B, Robb SM, Genikhovich G, Fropf WJ, Weilguny L, He S, Chen S, Lovegrove-Walsh J, Hill EM, Ragkousi K, et al. 2020. Sea anemone genomes reveal ancestral metazoan chromosomal macrosynteny. bioRxiv 10.1101/2020.10.30.359448 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.